Significance
Actin filaments assemble into ordered networks able to exert forces and shape cells. In response, filaments are exposed to mechanical stress which can potentially modulate their interactions with regulatory proteins. We developed in vitro tools to manipulate single filaments and study the impact of mechanics on the activity of actin depolymerizing factor (ADF)/cofilin, the central player in actin disassembly. While tension has almost no effect, curvature enhances severing by ADF/cofilin. We also discovered a mechanism that boosts the severing of anchored filaments: When binding to these filaments, ADF/cofilin locally increases their natural helicity, generating a torque that accelerates filament fragmentation up to 100-fold. As a consequence, interconnected filament networks are severed far more efficiently than independent filaments.
Keywords: cytoskeleton, mechanotransduction, actin dynamics, microfluidics
Abstract
Proteins of the actin depolymerizing factor (ADF)/cofilin family are the central regulators of actin filament disassembly. A key function of ADF/cofilin is to sever actin filaments. However, how it does so in a physiological context, where filaments are interconnected and under mechanical stress, remains unclear. Here, we monitor and quantify the action of ADF/cofilin in different mechanical situations by using single-molecule, single-filament, and filament network techniques, coupled to microfluidics. We find that local curvature favors severing, while tension surprisingly has no effect on cofilin binding and weakly enhances severing. Remarkably, we observe that filament segments that are held between two anchoring points, thereby constraining their twist, experience a mechanical torque upon cofilin binding. We find that this ADF/cofilin-induced torque does not hinder ADF/cofilin binding, but dramatically enhances severing. A simple model, which faithfully recapitulates our experimental observations, indicates that the ADF/cofilin-induced torque increases the severing rate constant 100-fold. A consequence of this mechanism, which we verify experimentally, is that cross-linked filament networks are severed by cofilin far more efficiently than nonconnected filaments. We propose that this mechanochemical mechanism is critical to boost ADF/cofilin’s ability to sever highly connected filament networks in cells.
A number of essential cellular processes rely on the regulated assembly and disassembly of actin filament networks (1, 2). The main proteins responsible for actin filament (F-actin) disassembly are the members of the actin depolymerizing factor (ADF)/cofilin protein family (3–5). ADF/cofilin binds to ADP–F-actin in a cooperative manner, leading to the formation of ADF/cofilin domains (6–10). These domains make filaments locally more flexible, for both bending and twisting (11–14), and shorten their right-handed helical pitch (without changing their length) (15–17). Filaments consequently sever at, or near, domain boundaries (8–10, 18–25). ADF/cofilin-saturated filament fragments do not sever, since they contain no domain boundaries, but they depolymerize from both ends. In particular, ADF/cofilin-decorated filaments have barbed ends that can hardly elongate or get capped, and thus depolymerize extensively, even in the presence of monomeric actin or capping proteins (10, 26).
We have recently measured the rate constants of these different binding, severing, and depolymerizing reactions (10). These results were obtained, as for many in vitro characterizations, by monitoring filaments that were barely constrained mechanically. In contrast, most filaments in cells are part of interconnected, or cross-linked, networks, and are exposed to various mechanical stresses. The specific activity of ADF/cofilin in this context is unclear.
Mechanical stress has long been proposed to potentially enhance severing by cofilin (27–31). Filaments immobilized on coverslips were reported to sever preferentially in bent regions when exposed to actophorin, a member of the ADF/cofilin family found in amoeba (27). Tension has been reported to protect filaments from ADF/cofilin binding and severing (32), and so has, very recently, formin-induced filament torsion (33). A recent theoretical study proposes that buckled filaments are easier to sever, while twisting a filament would mostly favor the dissociation of cofilin (31).
In addition to the external application of mechanical stress, seemingly passive mechanical constraints such as filament anchoring may also play a role. For instance, it has been demonstrated that the number of severing events induced by cofilin increased with the density of anchoring points to the coverslip surface (34). The authors interpreted their observation by proposing that severing was enhanced because anchors made it more difficult for filaments to relax structural strain induced by cofilin binding. In cells as well as in vitro, filaments cross-linked into bundles by fascin have been reported to sever faster than individual filaments when exposed to ADF/cofilin, and several explanations have been proposed, including a contribution of mechanical constraints (35).
A primary aspect is that, according to structural data, ADF/cofilin domains locally change the helical pitch of actin filaments (15–17). When filaments are anchored or cross-linked, their overall twist is constrained and this feature thus appears to be in conflict with ADF/cofilin binding. Existing data thus indicate that twist constraints and torque are likely to be key parameters affecting cofilin activity. Whether they contribute to favor or hinder cofilin binding, and/or severing, and to what extent, are all open questions.
Here, we investigate how ADF/cofilin binding and severing are affected by mechanical tension, by bending, and by constraints applied on the filament’s twist. We show that cofilin generates a torsional stress when binding to twist-constrained filaments, leading to a drastic enhancement of their severing.
Results and Discussion
Cofilin Binding Appears to Induce a Torsional Stress on Actin Filaments Which Cannot Freely Rotate Around Their Main Axis.
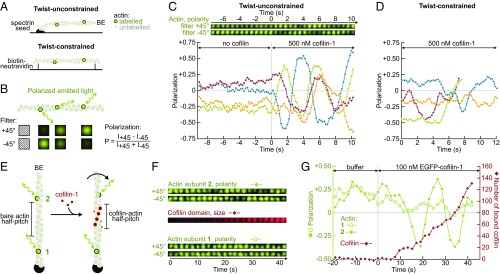
To directly assess the effect of ADF/cofilin binding on filament torsion, we monitored the polarization of the light emitted by single labeled actin subunits (36, 37), incorporated within filaments that were anchored by either one or two ends in a microfluidics chamber (Fig. 1). In the absence of cofilin, the polarization index of labeled subunits fluctuated mildly around a constant value, indicating that these subunits remained pointing in a fixed direction. When exposed to cofilin, the polarization index began to vary, reflecting the rotation of the subunits’ orientation around the filament’s main axis (for 13 out of 15, and 12 out of 25 observed subunits on filaments anchored by one or two ends, respectively. Fig. 1 C and D shows subsets of four representative measurements for each condition). Variations of the polarization index were more pronounced and more regular when only one filament end was anchored (Fig. 1 and SI Appendix, Fig. S1E). These observations are in agreement with numerical simulations that we performed (SI Appendix, Fig. S1) assuming that cofilin-decorated regions have a 25% shorter right-handed helical pitch (15) and are 18-fold more compliant to twist than bare regions (11).
Fig. 1.
Direct visualization of the torsional stress induced by cofilin binding to anchored actin filaments. (A) In a microfluidics chamber, filaments with a low fraction of labeled subunits are either polymerized from surface-anchored spectrin-actin seeds (single anchor, twist is unconstrained), or anchored by their sides via biotin–neutravidin bonds (between two anchoring sites, the twist is constrained). (B) The polarization of the emitted light indicates the orientation of a single actin subunit. The polarization P = (I+45 − I−45)/(I+45 + I−45) is determined by measuring the emitted intensity through two different polarization filters (I+45 and I−45). (C, Top) For a twist-unconstrained filament, time-lapse of the fluorescent intensities measured for a single actin subunit through the two polarization filters. (C, Bottom) Variation of the polarization P over time, measured for a single labeled subunit on four different twist-unconstrained filaments. The green curve corresponds to the same subunit as the time-lapse shown above. From time t = 0 onward, 500 nM cofilin was injected. Here only, filaments were polymerized for 15–20 min but not aged further. F-actin was partially in a ADP state to slow down cofilin binding (see SI Appendix, Fig. S2 for full ADP-F-actin). (D) Variation of the polarization P over time, measured for a single labeled subunit on four different twist-constrained filaments, exposed to 500 nM cofilin from time t = 0 onward. (E) Sketched from above: ADF/cofilin binding shortens the right-handed helical pitch and thus rotates the filament segment located between the ADF/cofilin domain and the free barbed end (BE), including subunit 2, while subunit 1’s orientation does not change. (F) Time-lapses of the fluorescent intensities measured in the configuration sketched in E: the two labeled actin subunits, each seen through the two polarization filters, as well as the total intensity of the growing EGFP–cofilin-1 domain positioned between these two labeled subunits. (G) Polarization of the same two labeled actin subunits, compared with the estimated number of cofilin monomers bound between these two subunits. From time t = 0 onward, 250 nM EGFP–cofilin-1 was injected. On this specific example, subunit 2 (filled green symbols) made one full rotation at t ∼ 30 s, when ∼60 EGFP–cofilin-1 molecules had bound the filament.
Filament rotation induced by cofilin binding could be most clearly characterized by monitoring the appearance and growth of a labeled cofilin-1 domain, between the anchored filament end and a labeled actin subunit (Fig. 1 E–G). Within the resolution of our experiment, this subunit began to rotate when the fluorescent signal from the cofilin-1 domain was first detected between the anchoring point and the subunit. We have calibrated the fluorescence intensity of EGFP–cofilin-1, and could thus estimate that one full turn was achieved when 91 ± 32 (std, n = 10 filaments) cofilin molecules were bound between the labeled actin subunit and the anchoring point. This number is close to what one would deduce from the reported reduction in the right-handed helical pitch for cofilin-decorated filaments, which leads to an estimated 80 cofilins to cause one full turn. Consistently, we found the rotation velocity of the filaments to be correlated with cofilin concentration, which modulates domain nucleation and growth rate (SI Appendix, Fig. S2).
These results show that filaments with a free end rotate around their main axis upon cofilin binding, thereby preventing the application of torsional stress. In contrast, cofilin domains decorating a filament segment between two anchoring points will impose a mechanical torque on this segment: Both bare and decorated regions will be undertwisted relative to their natural helicity (SI Appendix, Fig. S1B).
Twist-Constrained Filaments Are Severed Faster by ADF/Cofilin.
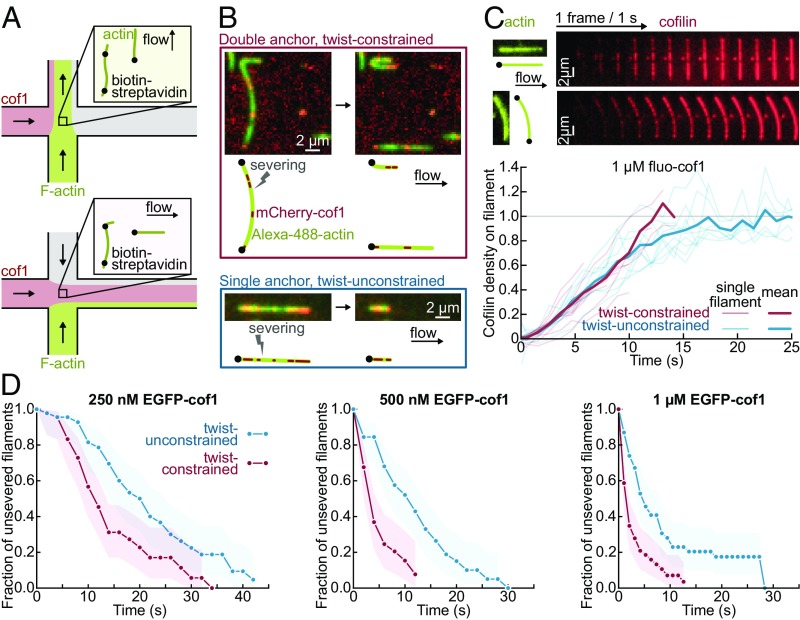
We next sought to examine the consequences of this mechanical torque. To do so, we compared the action of cofilin on filaments held between two anchoring points (i.e., twist-constrained, thus experiencing a torque as cofilin binds) with its action on filaments held by a single anchoring point (i.e., free to rotate, and thus not subjected to torque). These two configurations were achieved simultaneously in the same microfluidics chamber, by anchoring sparsely biotinylated filaments with one flow direction and then exposing them to labeled cofilin-1 with an orthogonal flow direction (Fig. 2A). We monitored the increase in the fluorescence signal of EGFP–cofilin-1 on each population and found that cofilin binds equally fast to twist-unconstrained or twist-constrained filaments (Fig. 2C).
Fig. 2.
Constraining the twist of actin filaments leads to a faster severing by cofilin. (A) Experimental setup, seen from above. Sparsely biotinylated F-actin is injected across the chamber and binds neutravidin–biotin-PLL-PEG. A perpendicular flow (containing ADF or cofilin) reveals the position of anchored points along the filament. (B) Examples of severing events for actin segments anchored at one (blue, Bottom) or both ends (red, Top). Severing occurs at cofilin-1 domain borders. (C) Cofilin-1 binding is unaffected by twist constraint. (Top) Raw data for two filaments, with unconstrained and constrained twist. (Bottom) Mean cofilin density averaged over 10 filaments for each condition. Condition: 1 µM EGFP–cofilin-1 from time t = 0 onward. Images acquired using total internal reflection fluorescence microscopy (TIRFM). (D) Actin segments with constrained twist fragment faster. Fraction of unsevered actin segments for twist-constrained and unconstrained, exposed to different cofilin concentrations from time t = 0 onward. The survival fraction is calculated over 49 filaments for each condition, blindly selected with the same length distributions (SI Appendix, Supplementary Methods and Information) and <L> = 6.6 ± 1.6 µm (std). Shadows represent 95% confidence intervals.
We measured the survival fraction of unsevered filaments in each population (excluding events observed near the anchoring points) and found that twist-constrained filaments were severed significantly faster (Fig. 2D). Severing occurred near domain boundaries, both on unconstrained (83% of severing events, n = 24) and constrained filaments (93% of severing events, n = 28) (Fig. 2B). In the absence of cofilin, no significant severing was observed in either population (SI Appendix, Fig. S3). We also verified that, after a severing event on a twist-constrained filament, the two resulting single-anchored filament fragments exhibited the same, lower severing rate as filaments in the twist-unconstrained population (SI Appendix, Fig. S4). The enhanced severing was also observed with ADF or at pH 7.0 (SI Appendix, Fig. S5).
It thus appears that cofilin severing is enhanced by the torsional stress induced by cofilin binding to filament segments between two anchoring points. Here, local filament curvature (which we specifically address in the next section) does not appear to contribute to this enhancement of severing, because the curvature of double-anchored filaments is very weak (with a typical curvature radius of more than 10 µm; see Fig. 2B and SI Appendix) and because these filaments sever at the same rate when no curvature is imposed, in the absence of flow (SI Appendix, Fig. S7B). Sharp bends can occur near the anchoring points (Fig. 2B), but these regions were excluded from our analysis.
Filament Bending Enhances Severing by Cofilin, While Tension Has Almost No Effect.
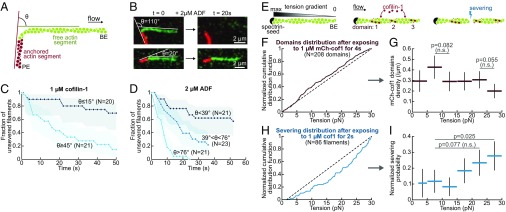
We next examined if an externally applied stress could alter the severing rate. Due to the helical nature of the actin filament, twist and bending are coupled (38) and we thus expected sharp filament bends to also enhance severing by cofilin (19, 29, 31). To test this idea, we anchored short phalloidin-stabilized filaments to the bottom of the flow chamber and, thanks to the flow, we imposed a different direction to the unanchored filaments that elongated from them (Fig. 3A). We found that larger angular differences between the anchored and free segments, which correspond to higher local curvatures near the anchored segment, led to faster severing by ADF/cofilin in that region (Fig. 3 A–D). We also compared these results to severing on straight filament portions (SI Appendix, Fig. S6). The same effect was observed in experiments where we did not use phalloidin to stabilize the anchored filaments segments (SI Appendix, Fig. S6).
Fig. 3.
ADF/cofilin-induced severing is faster on bent filaments but barely affected by tension. (A) To assess the effect of curvature, a segment of biotin–F-actin is stabilized with rhodamine–phalloidin and left to bind the neutravidin–biotin-poly-L-lysine-PEG surface with a random orientation (red). A second segment, that does not bind the surface, is then polymerized from this seed with Alexa-488–actin (green). Only severing events taking place in the curved green region were considered. (B) Example of two filaments. The angle θ is defined as the angle between the flow direction and direction of the anchored segment. (C and D) ADF/cofilin-induced severing rate is significantly larger on curved filaments. The filament population was split into two or three subsets of equal size and different angle θ. The free filament segment (green) was on average 5.5 µm long (std 1 µm). The flow gradient was kept low (300/s), resulting in a tension of about 0.2 pN in the curved region. Shadows represent 95% confidence intervals (SI Appendix, Supplementary Methods and Information). We performed simple exponential fits to compare the severing rate between populations. (C) A 1 µM cofilin-1 solution was injected continuously from time t = 0. n = 20, 21 from low to high angles. We calculated a fivefold difference in the severing rate between the two populations. The two curves differ significantly (P = 0.0005, log-rank test). (D) A 2 µM ADF solution was injected continuously from time t = 0. n = 21, 23, 22 from low to high angles. We calculated a 10-fold difference in the severing rate between the two extreme populations, whose curves differ significantly (P < 0.0001, log-rank test). (E) Sketch of the experiment. The viscous drag of the fluid on a filament anchored by only one end generates a gradient of tension. (F and G) Distribution of cofilin domains along actin filaments exposed to 1 µM mCherry-cofilin-1 for 4 s. n = 208 domains over 30 filaments. <L> = 31 ± 5 µm. Flow gradient: 16,200/s. (F) Cumulative distribution of domains over the tension gradient. The distribution has been normalized to take into account the nonlinearity of the tension gradient and the differences in filament length (SI Appendix). The dashed line indicates a homogeneous distribution. (G) Density of cofilin domains on the different tension ranges. Red horizontal bars indicate the tension range. Black vertical bars indicate the 95% confidence interval (binomial confidence interval, SI Appendix). Statistical significance: Fisher’s exact test (SI Appendix). (H and I) Distribution of severing events along actin filaments exposed to 1 µM unlabeled cofilin-1 for 2 s. n = 86 filaments. <L> = 37 ± 7 µm. As it is difficult to distinguish severing events near the free BE from depolymerization, we excluded all events occurring in the first 5 pixels (<2 pN). (H) Cumulative distribution of severing events over the tension gradient. The distribution has been normalized to take into account the nonlinearity of the tension gradient (SI Appendix). Dashed line indicates a homogeneous distribution. (I) Effective probability for a severing event to occur in a tension range. Blue horizontal bars indicate the tension range. Black vertical bars indicate the 95% confidence interval (binomial confidence interval, SI Appendix). Statistical significance: one-sample binomial test (SI Appendix).
The flowing solutions also put filaments under tension. When filaments are anchored by a single point, they are exposed to a tension gradient (39), which we have modulated up to a maximum tension of 30 pN by varying the flow rate (SI Appendix and SI Appendix, Fig. S8). We found that the local tension had no effect on the binding of cofilin (Fig. 3 F and G), while severing was slightly favored on regions exposed to the highest tensions (above 25 pN, Fig. 3 H and I). Filaments anchored between two points, perpendicular to the flow, are exposed to a nearly uniform tension (SI Appendix). On these twist-constrained filaments, we observed no effect of tension (up to 13 pN) on the cofilin severing rate and severing events were homogeneously distributed (SI Appendix, Fig. S7B).
Globally, our results on tension differ from those previously published by Hayakawa et al. (32), reporting that both cofilin binding and severing were hindered by filament tension, as low as 3.4 pN. To further test our results, we have repeated our experiments probing lower force ranges, using different isoforms (ADF, cofilin-2, cytoplasmic actin), and anchoring filament barbed ends with gelsolin (SI Appendix, Fig. S7). These experiments all confirmed that, in our assays, filament tension had no significant effect on ADF/cofilin activity. Perhaps our experiments fail to show an effect of tension because they lack some of the conditions used by Hayakawa et al. (32), such as the anchoring of filaments to surfaces with inactivated myosins, which may induce specific conformational changes upon the application of force, or the use of rhodamine-labeled actin (from Cytoskeleton, Inc.) with a possibly high labeling fraction (not specified), which may give rise to a specific mechanical response of the filament.
In cells, there is evidence of a mechanosensitive disassembly of filaments (40). In this context, filament tension may affect other factors modulating the activity of cofilin, such as tropomyosins (41).
A Simple Model Accounts for the Torque-Induced Enhancement of Severing.
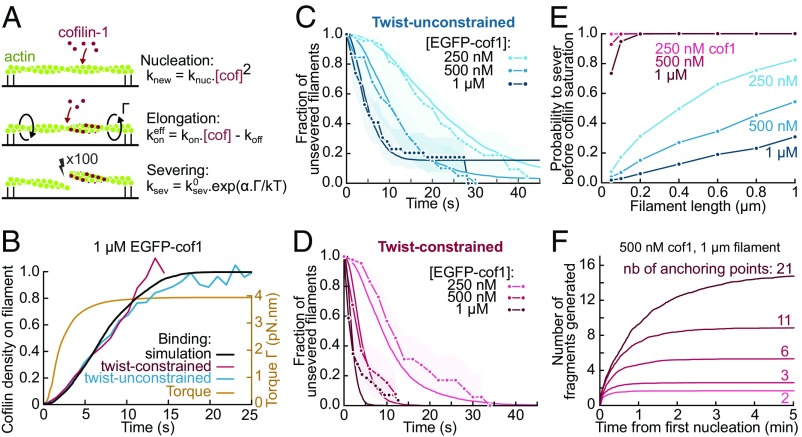
To further describe and quantify the enhanced severing of twist-constrained filaments by ADF/cofilin, we have recapitulated our results in the following model (summarized in Fig. 4A, and detailed in SI Appendix), which we compared with our experimental data thanks to numerical simulations. To account for ADF/cofilin cooperative binding, we assume domain nucleation to follow a quadratic dependence on cofilin concentration, and grow with the rate constants that we have previously measured (10). When twist is constrained, ADF/cofilin domains nucleate and grow with the same rates as on twist-unconstrained filaments, as indicated by our observations (Fig. 2C). A simple energy balance also supports this hypothesis: We can estimate that the energy benefit of cofilin binding is much larger than its torque-induced energy cost (SI Appendix).
Fig. 4.
Model for the torque-induced enhancement of severing by cofilin. (A) Sketch and summary of the model used for simulations. (B) Computed cofilin density and resulting torque on twist-constrained filaments. The experimental curves for cofilin density (in red and blue) come from Fig. 2C. The maximum torque applied on twist-constrained filaments corresponds to an undertwist of ∼5 rotations per micrometer. (C and D) Fit of the fraction of unsevered filaments in free (C) and constrained twist (D) conditions. k0sev = 2.4 × 10−2 s−1 was determined by fitting the data at 1 µM cofilin with unconstrained twist, and α = 5 was then determined by fitting the data at 1 µM cofilin with constrained twist. All other curves were simulated using these same parameters, with no further adjustment. The experimental curves are from Fig. 2D. Simulations were performed over 50-fold larger samples of filaments with the same lengths. Shadows represent 95% confidence intervals (SI Appendix, Supplementary Methods and Information). (E) Probability for a filament to sever before being saturated by cofilin-1 depending on its length and cofilin concentration, when twist is free (blue curves) or constrained (red curves). Each probability was computed by simulating 1,000 filaments. (F) Simulated number of actin fragments generated by cofilin, on filaments anchored by at least their two ends plus additional points, randomly positioned along their length. Each curve shows the mean over 100 simulations. The curves reach a plateau when the filament is saturated by cofilin.
The growth of a cofilin domain applies a mechanical torque Γ on the double-anchored filament. Using published values of torsional stiffness for (stiffer) bare and (softer) cofilin-decorated F-actin (11), we can compute Γ, which is uniform throughout the filaments, as a function of the cofilin coverage ratio (SI Appendix). We find that, due to the greater flexibility of cofilin-decorated regions, this torque rapidly reaches its maximum value (Fig. 4B).
Severing occurs at domain boundaries. Fitting the survival fraction for twist-unconstrained filaments (Fig. 4C) allowed us to determine the zero-torque severing rate constant k0sev. Since actin is partially labeled here, this severing rate constant is larger than the one we have measured in earlier work on unlabeled F-actin (10). We assumed that torque increased the severing rate exponentially, following a modified Bell model:
where α, quantifying the torque sensitivity of severing, was the only unknown parameter and was determined by fitting the experimental survival fractions for twist-constrained filaments (Fig. 4D). Our model appears to be in very good agreement with our experimental data.
We can estimate the maximum cofilin-induced torque to be ∼3.9 pN nm (Fig. 4B), based on published values of torsional stiffness, which are on the order of 10−27 N m2 (11). Recent computations (31) indicate that these numbers correspond to intersubunit torsional rigidities and that the filament torsional rigidity would be ∼10-fold larger (42, 43). Our observation, using polarization microscopy, that individual subunits located micrometers away from the anchored end of the filament have a well-defined orientation (Fig. 1), appears consistent with these larger values of torsional rigidity. These values would lead to a larger estimate of the maximum torque and thus to a lower value of α, but our conclusion would remain: As cofilin domains nucleate and grow on twist-constrained filaments, they rapidly generate a torque, thereby enhancing the severing rate per cofilin domain.
Based on our computations, the severing rate per domain is increased over 20-fold when 10% of the filament is decorated by cofilin, 50-fold when 20% is decorated, and up to 100-fold when the filament is nearly saturated by cofilin. However, as in the absence of torque, a fully decorated filament will not sever because it lacks domain boundaries.
Constraining a Filament’s Twist Allows It to Sever Before Being Saturated by Cofilin.
Since severing occurs at the boundaries between cofilin domains and bare filament regions, cofilin-saturated filaments do not sever (8, 10). Thus, a factor that will determine the number of severing events is their ability to occur before the filament is fully decorated by cofilin (44). Enhancing severing with torsional stress not only allows it to occur faster, it may also allow it to happen on segments that would otherwise not sever at all. We expected this effect to be more pronounced on short filaments, which are more prone to become saturated without severing. This situation is certainly common in cells, where filament segments between cross-links can be a few hundred nanometers long. However, individual severing events are difficult to resolve at such short length scales in our experiments.
Therefore, to investigate and quantify this point further, we have performed numerical simulations using our knowledge of the different reaction rates. We found that the torque-induced amplification of severing indeed allows cofilin to sever filaments in conditions where they would otherwise reach saturation without being severed (Fig. 4E). The difference is particularly strong for short filaments, which will be faster to saturate with cofilin. Note that, in this race against saturation, the enhancement of severing is made particularly effective by the fact that a significant torsional stress is already imposed by low densities of cofilin (Fig. 4B). Similarly, multiplying anchoring points allows cofilin to break filaments into more fragments (Fig. 4F).
This result explains why severing is more efficient when filaments are immobilized on a coverslip densely coated with myosins (34). As speculated by the authors of this work, cofilin binding generates a torsional stress which cannot relax when filaments are immobilized on a surface, leading to an enhanced severing rate. We show here that, within our range of concentrations, every filament segment between two anchoring points is likely to be severed before being saturated by cofilin, thanks to this torsional stress.
Cross-Linked Networks Sever Much Faster than Unconnected Filaments.
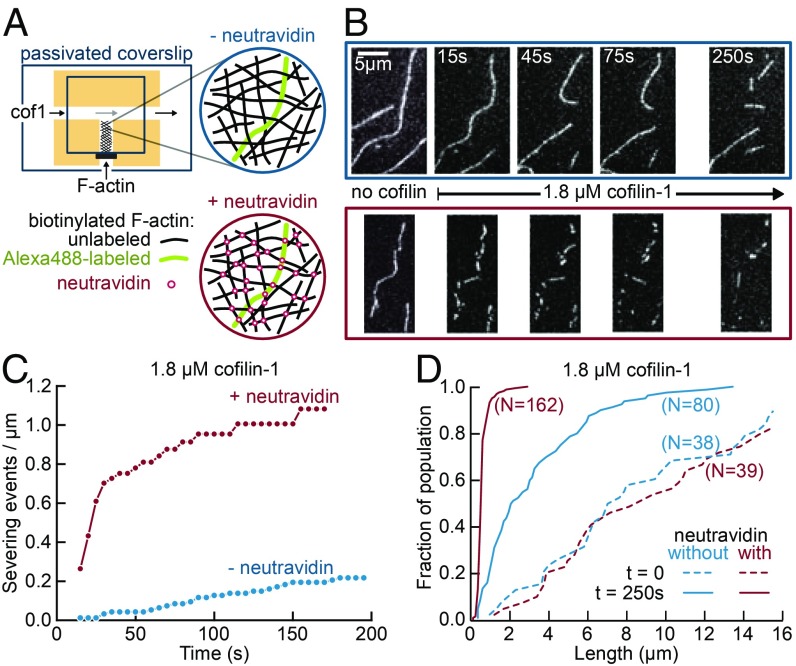
Our observations and calculations suggest that the rate and extent of severing by cofilin will be greater in networks of cross-linked filaments than equivalent populations of free filaments. To test this prediction, we have performed experiments on filament networks in T-shaped flow chambers (Fig. 5A). Preformed biotinylated actin filaments, with a 20:1 unlabeled-to-labeled filament ratio, were injected in the short end of the T-shaped chamber, which was then sealed, thereby creating a dead end which contained the filaments. We made similar observations where all of the filaments were fluorescently labeled, but having only a fraction of labeled filaments allows one to monitor and quantify single events (45).
Fig. 5.
Enhanced severing of interconnected actin filament networks. (A) Experimental setup, using T-shaped chambers, seen from above. A solution of biotinylated F-actin, containing one fluorescently labeled filament for 20 unlabeled filaments, was first injected in the short channel. The filaments could either be left nonconnected (Top, blue) or be cross-linked by injecting neutravidin through the main channel (Bottom, red). Cofilin-1 (unlabeled) was then injected in the main channel and F-actin severing was observed in the same region 500 µm away from the channel junction. All solutions were supplemented with 0.15% methylcellulose to maintain filaments close to the surface, and 0.25% BSA to maintain a good surface passivation. (B) Time-lapse of individual Alexa-488–labeled filaments, within a meshwork of unlabeled filaments, before and after filling the main channel with 1.8 µM cofilin (at time t = 0). Filaments are either nonconnected (Top, blue) or cross-linked by neutravidin (Bottom, red). (C). Quantification of severing events, cumulated over time, for nonconnected (blue, 10 filaments with a total initial length of 102 µm) and interconnected (red, five filaments with a total initial length of 60 µm) filaments, following the introduction of 1.8 µM cofilin in the main channel. (D) Cumulative length distributions (i.e., showing the fraction of a filament population having a length smaller than a given value) before exposure to cofilin (dashed lines) and 250 s after flowing 1.8 µM cofilin in the main channel (solid lines), for nonconnected (blue, initial population of 38 filaments, final population of 80 observable filament fragments) and interconnected filaments (red, initial population of 39 filaments, final population of 162 observable filament fragments).
Methylcellulose was present in the buffer, to maintain the filaments close to the passivated surface at the bottom of the chamber, thus forming a dense, quasi-bidimensional filament network. Different solutions could then be flowed in the main channel of the chamber, and their components could diffuse into the chamber dead end, without mechanically perturbing the filament population. We first introduced either a neutravidin solution or buffer in the main channel, to either cross-link filaments or not. In this experiment, the cross-links are thus artificially mediated by biotin and neutravidin, and are certainly stronger than what would be typically encountered in cells. We then flowed a solution of cofilin in the main channel, and observed its impact on the filaments. In each experiment, we monitored filaments in the same region of the chamber, 500 µm away from the channel junction.
Upon exposure to cofilin, the fates of the two filament populations were dramatically different, with the interconnected filaments experiencing far more severing (Fig. 5 B–D). We have quantified the severing events in each population (Fig. 5C) and we can estimate that, shortly after flowing in cofilin, interconnected filaments severed more than 30-fold faster than nonconnected filaments (with initial severing rates of ∼0.001 and 0.035 events per micrometer per second, for nonconnected and interconnected filaments, respectively). On longer timescales, when the filaments were saturated by cofilin, many filaments of a few micrometers in length could be observed in the nonconnected network, while only submicrometer fragments remained of the interconnected network (Fig. 5D). By creating new filament ends, severing also promotes the depolymerization of the filaments in the network: 250 s after flowing in cofilin, only 22% of the cross-linked F-actin remained visible (the rest being either fully depolymerized or in fragments too small to be detected) while 73% of the nonconnected F-actin was still visible.
Compared with our single-filament observations, severing appears to take place slower in our network experiment, possibly due to the diffusion and consumption of the finite cofilin pool by the dense actin filament network in the closed, T-shaped microchamber [such a depletion was recently reported in branched actin networks (46)], and to the presence of methylcellulose. Nonetheless, our experimental observations are in good quantitative agreement with the results of our simulations, which were based on our measured rates and our model (Fig. 4). From the observed filament density, and taking into account that there were 20 unlabeled filaments for every labeled filament, we could estimate that the cross-link density in our experiment was on the order of 1 µm−1. According to Fig. 4E we could thus expect that interconnected filaments would typically experience one severing event per micrometer, while most of the equivalent segments in nonconnected filaments will saturate and not sever. This is indeed what we observed: After 200 s, interconnected filaments cumulated a bit more than one severing event per micrometer, while nonconnected filaments were still several micrometers long on average (Fig. 5 C and D).
Implications for Actin Disassembly in Cells.
We show here that torsional stress and bending enhance filament severing by cofilin. These observations are consistent with early reports showing that, in the absence of cofilin, imposing a torque (42) or sharp bends (47) to actin filaments makes it easier to break them by applying tension. In our study, however, the torque is applied by cofilin itself as it binds to twist-constrained filaments, and the resulting torque is enough to dramatically increase the severing rate at the boundaries of cofilin domains. In cells, where filaments are typically interconnected and are not free to rotate, this mechanism is likely to play an important role. In particular, since its consequences are more drastic on densely connected filaments, it may modulate the disassembly of filament networks based on their cross-link density.
In cells, additional effects may come from the application of a torque to actin filaments by other factors. A recent theoretical study predicts that undertwisting an actin filament, beyond the maximum of ∼5 rotations per micrometer that cofilin can induce on its own, would lead to an enhancement of cofilin dissociation, and that overtwisting would have a stronger effect (31). Torque may be applied to filaments as they are elongated by formins which are unable to rotate, and this formin-induced torque was recently reported to protect filaments from cofilin (33). These results suggest that formin-induced torque can reach higher values than the cofilin-induced torque we report here, and future studies will be needed to further explore the specificities of the different means to apply a torque to actin filaments.
The enhancement of severing by a cofilin-induced torque is a very general mechanism since all it requires is for filaments to be constrained in twist. This situation arises whenever filaments are anchored, or cross-linked, regardless of the molecular nature of the cross-links, even though the strength of the cross-linking bonds is likely to modulate the global outcome. Our results are sufficient to explain why severing by cofilin is enhanced when filaments are bundled by fascin (35). Consistently, when filaments are bundled by a crowding agent, without cross-links that would constrain their twist, no enhancement of severing is observed (48). Other factors, specific to different cross-linkers, may also modulate severing, in addition to the generation of a torque. For instance, severing may be further enhanced by cofilin discontinuities due to its competition with cross-linkers, or by local changes in stiffness due to the presence of cross-links (21). Bulky cross-linkers (49) or very tight filament packing may also alter cofilin’s access to the sides of the filaments.
The torque generated by the binding of cofilin onto twist-constrained filaments may also affect the binding of other regulatory proteins, such as tropomyosins (41, 50) or Aip1 (9, 23, 51), and thereby modulate the competition or the cooperative binding of these proteins. Cofilin-induced torque on interconnected filaments is thus likely to have consequences beyond the enhanced severing we report here, and may play an essential regulatory role in cells.
Materials and Methods
Detailed experimental procedures and data analysis used in this study are described in SI Appendix, Supplementary Methods and Information.
Proteins and Buffer.
Actin was purified from rabbit muscle and labeled on surface lysines with Alexa-488- or Alexa-568-succinimidyl ester. Recombinant human cofilin-1 and ADF were expressed in Escherichia coli and purified. All experiments were performed in F-buffer with 50 mM KCl [5 mM Tris⋅HCl pH 7.8, 50 mM KCl, 1 mM MgCl2, 0.2 mM EGTA, 0.2 mM ATP, 10 mM DTT, and 1 mM 1,4-Diazabicyclo[2.2.2]octane (DABCO)].
Microscopy Experiments.
Actin filaments were aged for at least 15 min after polymerizing to have 99.9% ADP-actin (52), except for the experiments in Fig. 1. Microfluidics experiment were performed following the lines of our initial microfluidics experiments (52) where filaments were anchored by one end only to the coverslip surface, at the bottom of a flow chamber otherwise made of poly dimethyl siloxane.
Data Analysis.
A Kaplan–Meier algorithm was applied to determine survival functions from the observation of individual events.
Simulations.
Numerical simulations followed a Gillespie algorithm, and programs were written in Python.
Supplementary Material
Acknowledgments
We thank Martin Lenz for fruitful discussions, as well as Arnaud Echard, Nicolas Minc, Alexis Gautreau, and Christophe Le Clainche for their critical reading of an early draft of this manuscript. We thank all members of the G.R.-L./A.J. laboratory for their help, and especially Emiko Suzuki for initiating measurements on polarized fluorophore emission. We acknowledge funding from the Human Frontier Science Program (Grant RGY0066 to G.R.-L.), the French ANR (Grant Muscactin to G.R.-L.), the European Research Council (Grant StG-679116 to A.J.), and Fondation ARC pour la Recherche sur le Cancer (Postdoctoral Fellowship to H.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. E.M.D.L.C. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1812053116/-/DCSupplemental.
References
- 1.Blanchoin L, Boujemaa-Paterski R, Sykes C, Plastino J. Actin dynamics, architecture, and mechanics in cell motility. Physiol Rev. 2014;94:235–263. doi: 10.1152/physrev.00018.2013. [DOI] [PubMed] [Google Scholar]
- 2.Pollard TD. Actin and actin-binding proteins. Cold Spring Harb Perspect Biol. 2016;8:a018226. doi: 10.1101/cshperspect.a018226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bamburg JR, Harris HE, Weeds AG. Partial purification and characterization of an actin depolymerizing factor from brain. FEBS Lett. 1980;121:178–182. doi: 10.1016/0014-5793(80)81292-0. [DOI] [PubMed] [Google Scholar]
- 4.Andrianantoandro E, Pollard TD. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol Cell. 2006;24:13–23. doi: 10.1016/j.molcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein BW, Bamburg JR. ADF/cofilin: A functional node in cell biology. Trends Cell Biol. 2010;20:187–195. doi: 10.1016/j.tcb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De La Cruz EM. Cofilin binding to muscle and non-muscle actin filaments: Isoform-dependent cooperative interactions. J Mol Biol. 2005;346:557–564. doi: 10.1016/j.jmb.2004.11.065. [DOI] [PubMed] [Google Scholar]
- 7.Cao W, Goodarzi JP, De La Cruz EM. Energetics and kinetics of cooperative cofilin-actin filament interactions. J Mol Biol. 2006;361:257–267. doi: 10.1016/j.jmb.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 8.Suarez C, et al. Cofilin tunes the nucleotide state of actin filaments and severs at bare and decorated segment boundaries. Curr Biol. 2011;21:862–868. doi: 10.1016/j.cub.2011.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gressin L, Guillotin A, Guérin C, Blanchoin L, Michelot A. Architecture dependence of actin filament network disassembly. Curr Biol. 2015;25:1437–1447. doi: 10.1016/j.cub.2015.04.011. [DOI] [PubMed] [Google Scholar]
- 10.Wioland H, et al. ADF/cofilin accelerates actin dynamics by severing filaments and promoting their depolymerization at both ends. Curr Biol. 2017;27:1956–1967.e7. doi: 10.1016/j.cub.2017.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prochniewicz E, Janson N, Thomas DD, De la Cruz EM. Cofilin increases the torsional flexibility and dynamics of actin filaments. J Mol Biol. 2005;353:990–1000. doi: 10.1016/j.jmb.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 12.McCullough BR, Blanchoin L, Martiel J-L, De la Cruz EM. Cofilin increases the bending flexibility of actin filaments: Implications for severing and cell mechanics. J Mol Biol. 2008;381:550–558. doi: 10.1016/j.jmb.2008.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfaendtner J, De La Cruz EM, Voth GA. Actin filament remodeling by actin depolymerization factor/cofilin. Proc Natl Acad Sci USA. 2010;107:7299–7304. doi: 10.1073/pnas.0911675107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan J, et al. Molecular origins of cofilin-linked changes in actin filament mechanics. J Mol Biol. 2013;425:1225–1240. doi: 10.1016/j.jmb.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGough A, Pope B, Chiu W, Weeds A. Cofilin changes the twist of F-actin: Implications for actin filament dynamics and cellular function. J Cell Biol. 1997;138:771–781. doi: 10.1083/jcb.138.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galkin VE, et al. Remodeling of actin filaments by ADF/cofilin proteins. Proc Natl Acad Sci USA. 2011;108:20568–20572. doi: 10.1073/pnas.1110109108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huehn A, et al. The actin filament twist changes abruptly at boundaries between bare and cofilin-decorated segments. J Biol Chem. 2018;293:5377–5383. doi: 10.1074/jbc.AC118.001843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De La Cruz EM. How cofilin severs an actin filament. Biophys Rev. 2009;1:51–59. doi: 10.1007/s12551-009-0008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCullough BR, et al. Cofilin-linked changes in actin filament flexibility promote severing. Biophys J. 2011;101:151–159. doi: 10.1016/j.bpj.2011.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elam WA, Kang H, De La Cruz EM. Competitive displacement of cofilin can promote actin filament severing. Biochem Biophys Res Commun. 2013;438:728–731. doi: 10.1016/j.bbrc.2013.07.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elam WA, Kang H, De la Cruz EM. Biophysics of actin filament severing by cofilin. FEBS Lett. 2013;587:1215–1219. doi: 10.1016/j.febslet.2013.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang H, et al. Site-specific cation release drives actin filament severing by vertebrate cofilin. Proc Natl Acad Sci USA. 2014;111:17821–17826. doi: 10.1073/pnas.1413397111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jansen S, et al. Single-molecule imaging of a three-component ordered actin disassembly mechanism. Nat Commun. 2015;6:7202. doi: 10.1038/ncomms8202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ngo KX, Kodera N, Katayama E, Ando T, Uyeda TQP. Cofilin-induced unidirectional cooperative conformational changes in actin filaments revealed by high-speed atomic force microscopy. eLife. 2015;4:e04806. doi: 10.7554/eLife.04806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elam WA, et al. Phosphomimetic S3D cofilin binds but only weakly severs actin filaments. J Biol Chem. 2017;292:19565–19579. doi: 10.1074/jbc.M117.808378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wioland H, Jegou A, Romet-Lemonne G. Quantitative variations with pH of Actin Depolymerizing Factor/cofilin’s multiple actions on actin filaments. Biochemistry. 2019;58:40–57. doi: 10.1021/acs.biochem.8b01001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maciver SK, Zot HG, Pollard TD. Characterization of actin filament severing by actophorin from Acanthamoeba castellanii. J Cell Biol. 1991;115:1611–1620. doi: 10.1083/jcb.115.6.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ressad F, et al. Kinetic analysis of the interaction of actin-depolymerizing factor (ADF)/cofilin with G- and F-actins. Comparison of plant and human ADFs and effect of phosphorylation. J Biol Chem. 1998;273:20894–20902. doi: 10.1074/jbc.273.33.20894. [DOI] [PubMed] [Google Scholar]
- 29.De La Cruz EM, Martiel J-L, Blanchoin L. Mechanical heterogeneity favors fragmentation of strained actin filaments. Biophys J. 2015;108:2270–2281. doi: 10.1016/j.bpj.2015.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De La Cruz EM, Gardel ML. Actin mechanics and fragmentation. J Biol Chem. 2015;290:17137–17144. doi: 10.1074/jbc.R115.636472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schramm AC, et al. Actin filament strain promotes severing and cofilin dissociation. Biophys J. 2017;112:2624–2633. doi: 10.1016/j.bpj.2017.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayakawa K, Tatsumi H, Sokabe M. Actin filaments function as a tension sensor by tension-dependent binding of cofilin to the filament. J Cell Biol. 2011;195:721–727. doi: 10.1083/jcb.201102039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mizuno H, Tanaka K, Yamashiro S, Narita A, Watanabe N. Helical rotation of the diaphanous-related formin mDia1 generates actin filaments resistant to cofilin. Proc Natl Acad Sci USA. 2018;115:E5000–E5007. doi: 10.1073/pnas.1803415115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pavlov D, Muhlrad A, Cooper J, Wear M, Reisler E. Actin filament severing by cofilin. J Mol Biol. 2007;365:1350–1358. doi: 10.1016/j.jmb.2006.10.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Breitsprecher D, et al. Cofilin cooperates with fascin to disassemble filopodial actin filaments. J Cell Sci. 2011;124:3305–3318. doi: 10.1242/jcs.086934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sase I, Miyata H, Ishiwata S, Kinosita K., Jr Axial rotation of sliding actin filaments revealed by single-fluorophore imaging. Proc Natl Acad Sci USA. 1997;94:5646–5650. doi: 10.1073/pnas.94.11.5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mizuno H, et al. Rotational movement of the formin mDia1 along the double helical strand of an actin filament. Science. 2011;331:80–83. doi: 10.1126/science.1197692. [DOI] [PubMed] [Google Scholar]
- 38.De La Cruz EM, Roland J, McCullough BR, Blanchoin L, Martiel J-L. Origin of twist-bend coupling in actin filaments. Biophys J. 2010;99:1852–1860. doi: 10.1016/j.bpj.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jégou A, Carlier M-F, Romet-Lemonne G. Formin mDia1 senses and generates mechanical forces on actin filaments. Nat Commun. 2013;4:1883. doi: 10.1038/ncomms2888. [DOI] [PubMed] [Google Scholar]
- 40.Tojkander S, Gateva G, Husain A, Krishnan R, Lappalainen P. Generation of contractile actomyosin bundles depends on mechanosensitive actin filament assembly and disassembly. eLife. 2015;4:e06126. doi: 10.7554/eLife.06126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gateva G, et al. Tropomyosin isoforms specify functionally distinct actin filament populations in vitro. Curr Biol. 2017;27:705–713. doi: 10.1016/j.cub.2017.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsuda Y, Yasutake H, Ishijima A, Yanagida T. Torsional rigidity of single actin filaments and actin-actin bond breaking force under torsion measured directly by in vitro micromanipulation. Proc Natl Acad Sci USA. 1996;93:12937–12942. doi: 10.1073/pnas.93.23.12937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yasuda R, Miyata H, Kinosita K., Jr Direct measurement of the torsional rigidity of single actin filaments. J Mol Biol. 1996;263:227–236. doi: 10.1006/jmbi.1996.0571. [DOI] [PubMed] [Google Scholar]
- 44.Chan C, Beltzner CC, Pollard TD. Cofilin dissociates Arp2/3 complex and branches from actin filaments. Curr Biol. 2009;19:537–545. doi: 10.1016/j.cub.2009.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murrell MP, Gardel ML. F-actin buckling coordinates contractility and severing in a biomimetic actomyosin cortex. Proc Natl Acad Sci USA. 2012;109:20820–20825. doi: 10.1073/pnas.1214753109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mogilner A, et al. 2018. Reconstitution of the equilibrium state of dynamic actin networks. bioRxiv:10.1101/437806. Preprint, posted October 8, 2018.
- 47.Arai Y, et al. Tying a molecular knot with optical tweezers. Nature. 1999;399:446–448. doi: 10.1038/20894. [DOI] [PubMed] [Google Scholar]
- 48.Michelot A, et al. Actin-filament stochastic dynamics mediated by ADF/cofilin. Curr Biol. 2007;17:825–833. doi: 10.1016/j.cub.2007.04.037. [DOI] [PubMed] [Google Scholar]
- 49.Huang S, et al. Arabidopsis VILLIN1 generates actin filament cables that are resistant to depolymerization. Plant Cell. 2005;17:486–501. doi: 10.1105/tpc.104.028555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Christensen JR, et al. Competition between Tropomyosin, Fimbrin, and ADF/cofilin drives their sorting to distinct actin filament networks. eLife. 2017;6:e23152. doi: 10.7554/eLife.23152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nadkarni AV, Brieher WM. Aip1 destabilizes cofilin-saturated actin filaments by severing and accelerating monomer dissociation from ends. Curr Biol. 2014;24:2749–2757. doi: 10.1016/j.cub.2014.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jégou A, et al. Individual actin filaments in a microfluidic flow reveal the mechanism of ATP hydrolysis and give insight into the properties of profilin. PLoS Biol. 2011;9:e1001161. doi: 10.1371/journal.pbio.1001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.