Abstract
Eukaryotic mRNAs contain a 5′ leader sequence preceding the main open reading frame (mORF) and, depending on the species, 20%–50% of eukaryotic mRNAs harbor an upstream ORF (uORF) in the 5′ leader. An unknown fraction of these uORFs encode sequence conserved peptides (conserved peptide uORFs, CPuORFs). Experimentally validated CPuORFs demonstrated to regulate the translation of downstream mORFs often do so in a metabolite concentration-dependent manner. Previous research has shown that most CPuORFs possess a start codon context suboptimal for translation initiation, which turns out to be favorable for translational regulation. The suboptimal initiation context may even include non-AUG start codons, which makes CPuORFs hard to predict. For this reason, we developed a novel pipeline to identify CPuORFs unbiased of start codon using well-annotated sequence data from 31 eudicot plant species and rice. Our new pipeline was able to identify 29 novel Arabidopsis thaliana (Arabidopsis) CPuORFs, conserved across a wide variety of eudicot species of which 15 do not initiate with an AUG start codon. In addition to CPuORFs, the pipeline was able to find 14 conserved coding regions directly upstream and in frame with the mORF, which likely initiate translation on a non-AUG start codon. Altogether, our pipeline identified highly conserved coding regions in the 5′ leaders of Arabidopsis transcripts, including in genes with proven functional importance such as LHY, a key regulator of the circadian clock, and the RAPTOR1 subunit of the target of rapamycin (TOR) kinase.
Keywords: 5′-UTR, translation, translational initiation, translational stalling, uORF
INTRODUCTION
Gene expression is regulated particularly at the level of transcription, translation, and protein stability. Translational regulation is of increasing interest as transcript levels do not necessarily correlate with protein levels (Conrads et al. 2005; Gibon et al. 2006; Bianchini et al. 2008; Merchante et al. 2017). Translation can be regulated both globally and in a transcript-specific manner. Transcript-specific translation regulation usually involves specific features of the mRNA, often present in the 5′ leader sequence of the mRNA (also referred to as the 5′ untranslated region [5′-UTR]) (Merchante et al. 2017). Most, if not all, eukaryotic mRNAs contain a 5′ leader, and 20%–50% of eukaryotic mRNAs contain upstream open reading frames (uORFs) (Kochetov 2008). Termination of translation of these uORFs usually leads to dissociation of the ribosome from the mRNA, thereby inhibiting translation of the downstream main open reading frame (mORF). In some cases, the small ribosomal subunit remains attached to the mRNA following translation termination, allowing it to continue scanning and re-initiate on a downstream start codon (Von Arnim et al. 2014; Browning and Bailey-Serres 2015; Merchante et al. 2017). In case the start codon region of the uORF is unfavorable, scanning ribosomes are prone to skipping such start codons, resulting in increased mORF translation. Recognition of a start codon depends on the start codon and its surrounding nucleotides, also known as the start codon context. An optimal start codon context contains an adenine at the −3 position and guanine at the +4 position relative to the A+1UG codon (Browning and Bailey-Serres 2015; Diaz de Arce et al. 2018). Start codons deviating from this optimal context are less likely to be recognized by the preinitiation complex (PIC). The PIC therefore continues to scan to the next start site a phenomenon called leaky scanning. Most mORFs preceded by a uORF are translated due to leaky scanning of the uORF start codons.
uORFs generally repress mORF translation in a sequence-independent manner, due to reducing the number of ribosomes reaching the mORF. However, in silico analyses of the Arabidopsis thaliana (Arabidopsis) genome have identified around 80 uORFs that encode sequence conserved peptides (conserved peptide uORFs, CPuORFs), indicating biological relevance (Hayden and Jorgensen 2007; Jorgensen and Dorantes-Acosta 2012; Takahashi et al. 2012; Vaughn et al. 2012). Indeed, several such CPuORFs were shown to regulate the translation of the downstream mORF in a metabolite-dependent manner (Imai et al. 2008; Rahmani et al. 2009; Alatorre-Cobos et al. 2012; Laing et al. 2015; Merchante et al. 2017). This metabolite-regulated translation (MRT) results in a regulatory circuit where the concentration of a specific metabolite together with a CPuORF determines the level of translation of the mORF and therefore the protein level.
A well-studied example of MRT involving a CPuORF is the translation of S1 group of bZIP transcription factors. These bZIPs control amino acid- and sugar metabolism, and resource allocation (Hanson et al. 2008; Ma et al. 2011; Thalor et al. 2012; Dröge-Laser and Weiste 2018). Translation of S1-group bZIPs is regulated by their upstream CPuORFs. Increasing sucrose levels promote CPuORF-dependent ribosome stalling (Wiese et al. 2004; Rahmani et al. 2009; Weltmeier et al. 2009; Hou et al. 2016; Yamashita et al. 2017) and, as a consequence due to steric hindrance on the mRNA, stalled ribosomes prevent translation of the downstream mORF. Similar mechanisms have also been proposed for other metabolites. Examples in Arabidopsis include (phospho)choline (Tabuchi et al. 2006; Alatorre-Cobos et al. 2012), polyamines (Imai et al. 2008; Uchiyama-Kadokura et al. 2014; Guerrero-González et al. 2016), and ascorbate (Laing et al. 2015).
Interestingly, in most plant CPuORFs the start codon context is suboptimal for translation initiation (Rahmani et al. 2009; Alatorre-Cobos et al. 2012; Guerrero-González et al. 2016). Moreover, the CPuORF of the GDP-l-galactose phosphorylase (GGP) gene, involved in ascorbate-mediated MRT, does not initiate translation with an AUG codon but likely with an ACG start codon. Substituting the ACG start codon of the GPP CPuORF with an AUG drastically reduced the amount of GGP protein produced, already during noninhibitory (low ascorbate) conditions (Laing et al. 2015). These results suggest that a suboptimal start codon context is a requirement for CPuORF-regulated translation of mORFs through a leaky scanning mechanism. This mechanism allows for sufficient translation of the mORF in noninhibitory conditions and at the same time inhibition of mORF translation during inhibitory conditions by stalled ribosomes. Recently it was shown that non-AUG codons can initiate translation with equal efficiency compared to AUG codons in a suboptimal context (Diaz de Arce et al. 2018). Non-AUG translational initiation is not limited to uORF translation. Main ORFs can also be translated from non-AUG start codons although with lower efficiency (Gordon et al. 1992; Riechmann et al. 1999; Depeiges et al. 2006; Wamboldt et al. 2009; Simpson et al. 2010). An interesting example is the translation initiation of the organellar DNA polymerase POLγ2, where the initiation on a canonical AUG start codon results in protein localization to the chloroplast; however, when translation is initiated on an upstream CUG codon a N-terminal localization signal is added, resulting in localization to the mitochondrion (Christensen et al. 2005). Another study revealed that translation of FCA, a key regulator of flowering time, consistently uses CUG as a translation initiation site; however, the function for non-AUG translation initiation remains unknown (Simpson et al. 2010).
Given the regulatory importance of CPuORFs, attempts have been made to identify more genes regulated by CPuORFs using the available full-genome and transcriptome sequence data. Previous in silico studies searching for plant CPuORFs, all focused on uORFs containing an AUG start codon (Hayden and Jorgensen 2007; Tran et al. 2008; Jorgensen and Dorantes-Acosta 2012; Takahashi et al. 2012). However, recent findings have shown that the use of alternative initiation codons by CPuORFs can be important for translational regulation, which is so far not addressed at the genome level. In addition, no studies have systematically searched for all noncanonical mORF translational initiation events in plants using conservation in sequence data (Simpson et al. 2010).
To determine the extent of non-AUG initiation in CPuORFs and mORFs in Arabidopsis, we have explored all 5′ leader sequences of Arabidopsis mRNAs for upstream conserved coding regions (uCCRs) regardless of initiation codon. We performed an extensive BLASTp search using all regions in between and around stop codons in the 5′ leaders of Arabidopsis and those of 31 other plant species. Using this method combined with filters to remove false positives, we discovered with high confidence 25 novel CPuORFs, five CPuORFs with a stop codon in the mORF, and 14 uCCRs directly upstream and in frame with the mORF, which potentially extend the N terminus of the mORF.
RESULTS
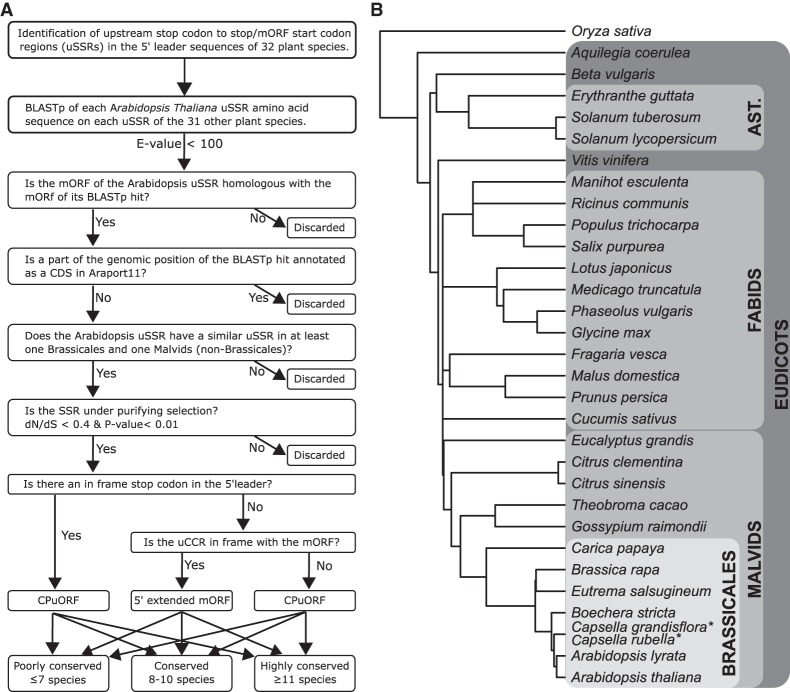
In this study, we developed a pipeline to search for uCCRs in the 5′ leader of mRNAs, such as CPuORFs and 5′ extensions of mORFs with non-AUG start codons (Fig. 1A). Previous pipelines searching for CPuORFs searched for uORFs initiating translation with AUG start codons, while recent evidence has shown that translation of CPuORFs and mORFs does not necessarily initiate with an AUG start codon (Simpson et al. 2010; Laing et al. 2015; Hsu et al. 2016; Ivanov et al. 2018). Stop codons, on the other hand, are bound to three codon types: UGA, UAA, and UAG. Consequently, the maximum length of a uCCR in the 5′ leader can be defined as from the transcriptional start site to a stop codon, from a stop codon to another stop codon or from a stop codon to the mORF start codon. Therefore, as a first step to search for uCCRs in the 5′ leader, the regions surrounding and in frame with stop codons were extracted from Arabidopsis mRNA (Araport11) (stop to stop region or stop to mORF start region, hereafter referred to as uSSR). In total, 330,566 uSSRs were retrieved from the Arabidopsis genome from 39,650 transcripts (including splice variants) of 22,118 genes (Supplemental Table S1).
FIGURE 1.
(A) Schematic overview of the pipeline used to identify upstream conserved coding regions. (B) Species tree indicating species whose genomic data were used in this study. Branch lengths were retrieved from TimeTree.org (Hedges et al. 2015). (*) No data were present for Capsella grandisflora (Hedges et al. 2015).
Most of these uSSRs were likely established between randomly formed stop codons and thus lack biological relevance. Conservation is a good indication for biological relevance and therefore uSSRs were evaluated for similarity with uSSRs from other species. For this, additional uSSRs were extracted from seven Brassicales species, five Malvids species (other than Brassicales), 12 Fabids species, Vitis vinifera, three species of the asterid clade, Beta vulgaris, Aquilegia coerulea, and Oryza sativa (Fig. 1B; Supplemental Table S2). These species are similar enough to identify recently evolved uCCRs and most have well-annotated transcriptomes, although it should be noted that the quality of the annotation varies. This resulted in a total of 8,272,590 uSSRs. Next, all uSSR sequences were translated into amino acid sequences and the Arabidopsis uSSRs were aligned with the uSSRs of the 31 other species using BLASTp. The BLASTp E-value threshold was set to 100 to include identification of shorter uCCRs. This resulted in 263,522 Arabidopsis uSSRs with at least one BLASTp hit with an uSSR from another species (Supplemental Table S1).
In general, the function of a uCCR in the 5′ leader of a mRNA is directly linked with the function of its downstream mORF. CPuORFs generally regulate the translation of their own mORF, and uCCRs directly upstream and in frame with the mORF are likely part of the full-length protein. Following this reasoning, biologically conserved uSSRs must share a conserved mORF if their biologic function is conserved. For that reason, uSSR hits without homologous mORFs were discarded in our pipeline. Protein sequences of mORFs with a BLASTp E-value <1 × 10−5 were considered homologs. 112,295 Arabidopsis uSSRs from 27,768 transcripts and 15,345 genes did have homologous mORFs (Supplemental Table S1).
uCCRs in 5′ leaders are not restricted to CPuORFs and 5′ extended mORFs. For instance, during splicing, fragments of the mORF can end up in the 5′ leader, or a mORF of a different gene can overlap with the 5′ leader of another. These fragments are likely conserved, since they are part of the mORF of another transcript; however, they do not have biological relevance in the 5′ leader. To eliminate these uSSRs, the Arabidopsis genomic region of each Arabidopsis uSSR BLASTp hit was evaluated for annotation of a CDS in Araport11. If the Araport11 CDS overlaps with the uSSR BLASTp hit, the hit was removed. This way, 849 genes, from the Arabidopsis genome, with potential uCCRs were discarded leaving 100,843 uSSRs from 14,964 genes (Supplemental Table S1).
Conservation across different clades is a good indication of biological function. We therefore compared the Arabidopsis uSSRs with those of species in different clades. The vast majority (99,907) of the 100,843 uSSRs had a hit with an uSSR from one of the seven Brassicales species, while only 3303 uSSRs have a hit with one of the other 25 species. Arabidopsis is closely related to the other species of the Brassicales order. The nucleotide sequence between these species has not diverged much, which in turn leads to similarities on the amino acid level even if there is no evolutionary constraint and are therefore false positives. To overcome this problem, all Arabidopsis uSSRs that do not have at least one similar uSSR in a species of the Malvids clade, outside of the Brassicales species, were considered less conserved and therefore removed from the list (Fig. 1A). The species in the Malvids clade, outside of Brassicales, are divergent enough for the nonconserved uSSRs to differ with Arabidopsis uSSRs on the amino acid level and in the meantime not too divergent if a uCCR has evolved recently. About 99% of the 100,843 Arabidopsis uSSRs did not fulfill this requirement, leaving 1307 conserved Arabidopsis uSSRs for further analysis.
The 1307 uSSRs are derived from 883 Arabidopsis transcripts (including splice variants), meaning that on average each of these transcripts has 1.48 uSSRs. For example, the 5′ leader of bZIP11 mRNA still has four candidate uSSRs. After inspection, one of these uSSRs is indeed the CPuORF, while the others were (partially) overlapping the CPuORF but in a different frame. Since conservation at the amino acid level indirectly leads to conservation at the nucleotide level, translated nucleotide sequences in a different frame might also be indirectly conserved. Moreover, conserved nucleotide motifs (e.g., RNA binding protein recognition sites) presents a similar situation, where the amino acids sequence corresponding to the conserved nucleotide sequence seems conserved based on underlying nucleotide sequence conservation. To have an indication of the coding potential of the nucleotide sequence, all remaining full-length uSSRs were aligned and checked for purifying selection using codeML of the PAML program (see Materials and Methods for detailed information). To determine a threshold for the dN/dS (ω) and P-value, previously identified CPuORFs were used as a positive control. When the threshold of the ω-value was set lower than 0.4, the amount of previously identified CPuORFs also discovered by our pipeline started to decrease drastically, and the same accounts for a P-value lower than 0.01 (Supplemental Fig. S1). Therefore, the threshold was set to an ω-value of 0.4 and P-value of 0.01, all uSSRs which had a value above either one of these thresholds were removed. In total, 416 of the 1307 uSSRs have coding potential from 373 transcripts of 213 genes (Supplemental Fig. S1B; Supplemental Table S1).
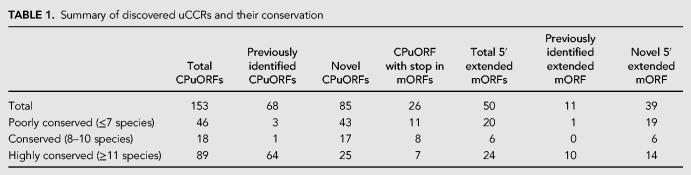
Of these 213 genes with uCCRs, 153 contain a CPuORFs and 75 uCCRs do not have a stop codon before the annotated start codon of the mORF. Of the latter, 50 were in frame with the annotated start codon, and likely initiate the mORF on a non-AUG start codon (hereafter referred to as 5′ extended mORF), while 26 uCCRs were not in frame with the mORF but contained a stop codon in the mORF and are thus considered CPuORFs (Table 1). Some of the uCCRs were only conserved in very few species, which reduces the likelihood of biological relevance of these uCCRs. Therefore, the uCCRs were divided into three different classes based on how well spread the uCRR was among the species tested: poorly conserved (present in seven or less species), conserved (present in 8–10 species), and highly conserved (present in 11 or more species) (Table 1). The majority (58%) of the discovered CPuORFs fall in the highly conserved category (Table 1), which is also the case for the 5′ extended mORFs (48%), but not for the CPuORFs with the stop codon in the mORF where only seven are highly conserved out of the 26 in total. Of the latter, two are “regular” CPuORFs whose stop codon was removed by splicing. To reduce the risk of including uCCRs lacking biological relevance, only the highly conserved uCCRs are considered “true” uCCRs and will be further discussed in this article (Table 2; Supplemental Data 1). We realize that these are very strict criteria and likely biologically relevant uCCRs are missed and therefore, the full list is presented in Supplemental Data 1.
TABLE 1.
Summary of discovered uCCRs and their conservation
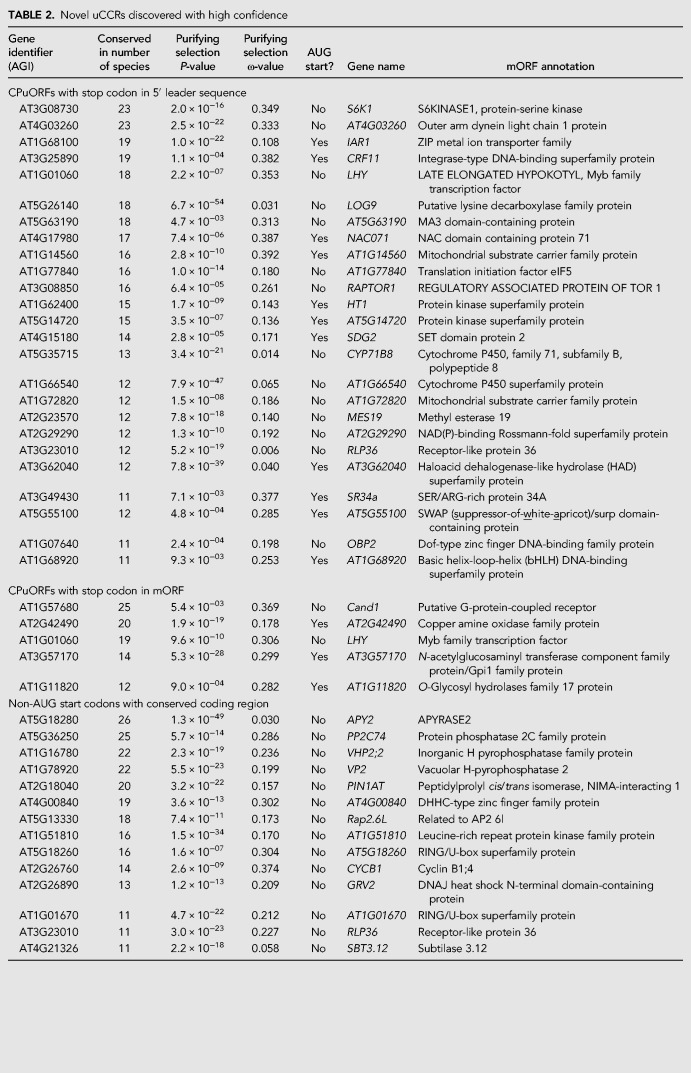
TABLE 2.
Novel uCCRs discovered with high confidence
Genome annotation between the species varies, and lack of 5′ leader annotation could lead to classification as poorly conserved. To get an idea if genome annotation played a role in classification of the discovered CPuORFs, we determined in each species whether the most conserved mORF homolog of each CPuORF (lowest E-value) possesses a 5′ leader. The homologs of poorly conserved CPuORFs has annotated 5′ leader in on average 22.4 species in our data set, while this average was 21.9 for the conserved CPuORFs and 22.1 for the highly conserved CPuORFs (Supplemental Fig. S2). Thus, the classification of the CPuORFs depending on conservation is not biased based on variable annotation quality.
Of the 89 CPuORFs, 62 were previously identified using bioinformatics studies (Hayden and Jorgensen 2007; Jorgensen and Dorantes-Acosta 2012; Takahashi et al. 2012; Vaughn et al. 2012). In addition, the pipeline was able to identify both GGP paralogues which do not initiate with a canonical AUG codon (Liang et al. 2016). This means that our pipeline was able to find about 80% of the total 78 genes containing CPuORF previously identified. Unsuccessful identification of the 14 remaining CPuORFs could be due to novel annotation of coding regions (three CPuORFs), not fulfilling the requirements for purifying selection (five CPuORFs), or poor conservation (six CPuORFs) (Supplemental Table S3).
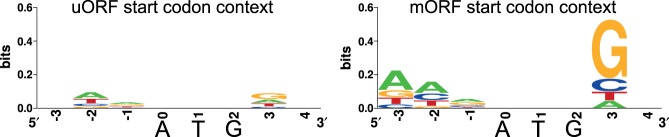
Translational regulation via CPuORFs requires a suboptimal start codon context as leaky scanning ensures sufficient mORF translation during noninhibitory conditions (Hummel et al. 2009; Laing et al. 2015). Our pipeline detected 56 CPuORF homology groups that likely initiate with an AUG and 15 with a non-AUG start codon. Moreover, analysis of the start codon context of the AUG initiating CPuORFs shows that most have a suboptimal start codon context compared to their mORFs (Fig. 2). The high number of non-AUG start codons and nonoptimal AUG start codon context are in line with the hypothesis that CPuORFs require leaky scanning to allow for sufficient translation of the mORF in noninhibitory conditions.
FIGURE 2.
Start codon context of all highly conserved CPuORFs with an AUG start codon (left) and their downstream mORFs discovered in this study (right). Logos were created using weblogo.berkeley.edu.
Simpson et al. (2010) discovered that translation of FCA (AT4G16280) initiates at a CUG codon upstream of the annotated mORF start codon (Simpson et al. 2010). The same study searched for other genes whose translation could initiate at a CUG followed by an additional guanine and looked at conservation between the CUG start codon and the mORF start. Ten genes with uCCRs between CUG and mORF start codons were discovered by Simpson et al. (2010) and later an additional three were discovered by Vaughn et al. (2012). Of these 14 uCCRs, 10 were found in our study (AT1G21000 did not pass the purifying selection requirements and AT1G32700 and AT2G20680 were only conserved in Brassicales). Interestingly, the FCA noncanonical initiation site was judged as poorly conserved in this study, indicating that our threshold is very strict (Table 1; Supplemental Data 1).
Next, we evaluated whether the uCCRs are indeed translated using ribosome footprint data (Merchante et al. 2015). Ribosome footprints are established by incubating cycloheximide-treated cell lysates with RNAses. The RNA protected by the ribosomes is sequenced and represents the ribosome “footprints.” AT5G36250 contains a highly conserved coding region between a non-AUG and mORF start codon and was discovered in this study (Table 2; Supplemental Data 1; Supplemental Fig. S3). This uCCR likely initiates with a CUG (codon context AUCCUGGC) in Brassicales while the other species have an ACG conserved (codon context c/gUgACGGC). Many ribosome footprint reads are present in between the CUG and AUG start codon in Arabidopsis, confirming translation of this region (Fig. 3A).
FIGURE 3.
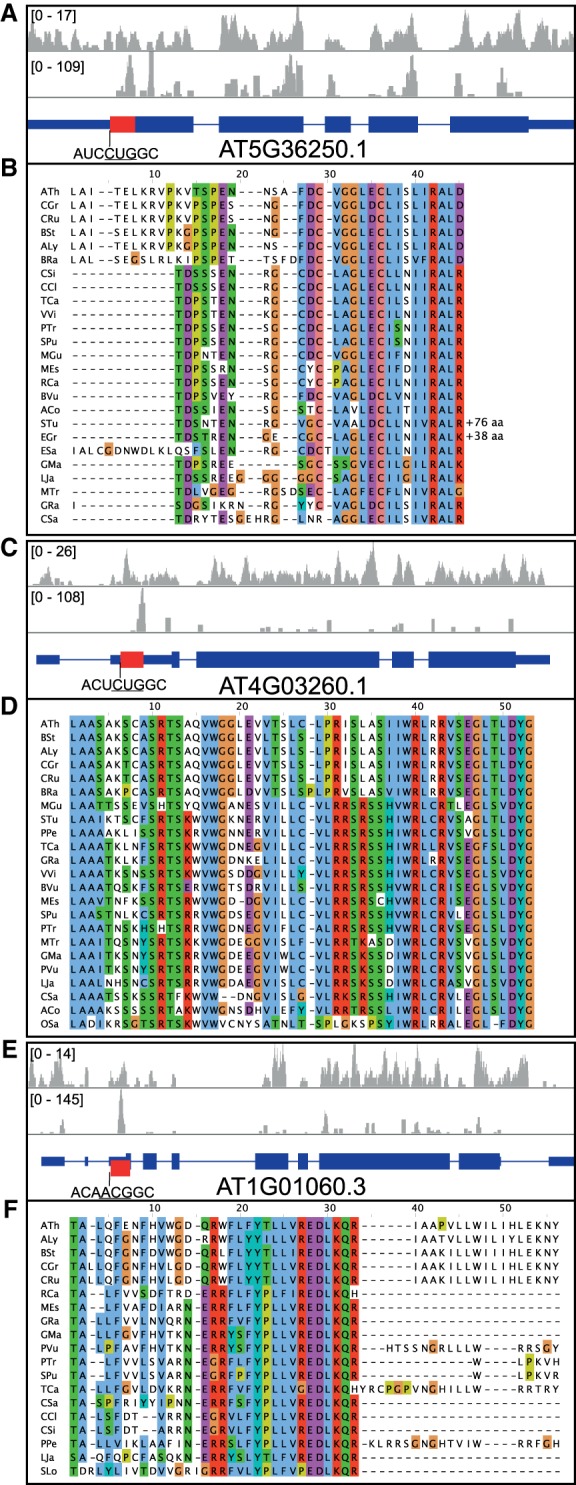
Ribosome footprint data confirms translation of uCCRs. (A,C,E) RNA-sequencing results from Merchante et al. (2015), from total RNA (top) or ribosome footprints (middle), and the mRNA architecture (bottom), where the thicker bars represent the ORFs with the discovered uCCR in red, the thinner bars indicate other regions on the transcript, and the lines represent introns from three different genes: AT5G36250 (A,B), AT4G03260 (C,D), and AT1G01060 (LHY) (E,F). The red bar in panel E indicates a uCCR that is out of frame with the mORF. (B,D,F) Sequence alignments of the discovered uCCRs, aligned using MAFFT v. 7.307 (FFT-NS-2) and displayed using Jalview v. 2.10. See Supplemental Table S2 for species abbreviations.
The non-AUG start codons on uCCRs that are in frame with the mORF are prone to leaky scanning, meaning that translation can initiate on both the non-AUG start codon and the regular AUG start codon. This can result in the production of two protein isoforms, with one having an additional amino acid sequence at the N terminus. Signal peptides are often present at the N terminus of a protein, therefore proteins with and without localization signal could be produced (Christensen et al. 2005; Wamboldt et al. 2009). To evaluate the possibility of dual localization on the genes of the newly discovered 5′ extended mORFs, mORFs with non-AUG and with AUG start codon were tested on subcellular localization using three different programs. This analysis revealed that AT4G00840 might localize to the mitochondrion when translated by an upstream GUG start codon and to the endoplasmic reticulum when translated by the canonical AUG start codon. AT5G36250 is predicted to localize to chloroplast if translated by the canonical AUG start codon and cytoplasm when translated by the non-AUG start codon (Supplemental Table S4). Additionally, AT2G25110, previously discovered by Vaughn et al. (2012) but not evaluated on localization, is predicted to be localized to the ER if translated canonically, while it is predicted to localize to the chloroplast or mitochondrion when translated from the noncanonical start codon (Supplemental Table S4).
Translational repression via CPuORFs usually involves stalling of the ribosome on the uORF (Rahmani et al. 2009; Merchante et al. 2017). Often, stalling occurs near the stop codon of the CPuORF; however, this is not a prerequisite (Uchiyama-Kadokura et al. 2014; Hou et al. 2016; Peviani et al. 2016; Yamashita et al. 2017). AT4G03260 encoding for outer arm dynein light chain 1 protein, contains a CPuORF that could initiate using a CUG start codon (context ACTCUGGC) and has a highly conserved stop codon (Fig. 3; Supplemental Fig. S4). Interestingly, ribosome footprint data show a large peak on the stop codon of this CPuORFs, much higher than peaks in the mORF, indicating ribosome stalling and translational repression of the mORF (Fig. 3). One of the Arabidopsis genes that we predict to contain a CPuORF without a stop codon before the mORF is AT1G01060, which encodes the clock associated protein LHY (late elongated hypocotyl). Interestingly, in about half of the investigated species LHY does not have a stop codon before the mORF, while the other half does, with a conserved position. The amino acid sequence of the uORF is conserved exactly until this conserved stop codon (Fig. 3; Supplemental Figs. S5, S6). Arabidopsis ribosome footprint data reveal a large peak precisely at the position where the conservation stops and where a stop codon is present in many species (Fig. 3). It seems that ribosome stalling can occur independent of the presence of a stop codon in Arabidopsis. In summary, ribosome footprinting information suggests that many of the predicted CPuORFs present biological functionality under the plant growth conditions used in the ribosomal experiments.
DISCUSSION
In this study, a pipeline was developed to search for biological relevant uORFs and other upstream coding regions. Most previous studies identifying upstream coding regions either use conservation between different species or ribosome footprinting (ribo-seq) data (Cvijović et al. 2007; Hayden and Jorgensen 2007; Hayden and Bosco 2008; Tran et al. 2008; Selpi et al. 2009; Takahashi et al. 2012; Skarshewski et al. 2014; Hu et al. 2016; Hsu et al. 2016; Spealman et al. 2018). The latter approach has the advantage of revealing experimentally proven translated regions but often lack proof for biological relevance. Biologically irrelevant sequences are expected to diverge faster than sequences under selective constraint. Therefore, using conservation is an excellent tool for discovering novel biologically relevant uCCRs. Compared to previous studies identifying conserved plant uCCRs, the current pipeline benefits from three main features: (i) it only takes stop codons into account for its search for uCCRs, enabling it to find uCCRs with non-AUG start codons, (ii) the pipeline is able to find both CPuORFs and conserved 5′ extended mORFs, and (iii) it uses 32 species, mostly annotated with RNA-sequencing data, to confirm the evolutionary conservation and thereby biological relevance of the identified uCCRs (Fig. 4).
FIGURE 4.
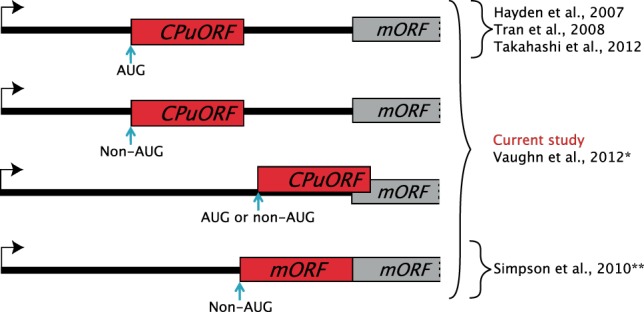
Overview of the types of upstream conserved coding regions discovered by our pipeline compared to previous studies. (*) Vaughn et al. (2012) searched for nucleotide conservation and indirectly discovered some uCCRs. (**) Simpson et al. (2010) only searched for 5′ extended mORFs initiating on a CUG start codon with a guanine at +4 position.
Using our pipeline, we discovered 29 novel CPuORFs with high confidence (Table 2). The majority of the novel discovered CPuORFs likely initiate with a non-AUG start codon. Previous studies searching for CPuORFs were restricted to AUG initiating uORFs and thereby overlooked these CPuORFs. With this study we show that there are at least 15 CPuORFs that do not initiate with an AUG start codon. Moreover, previous CPuORF searches required a stop codon to be present in the 5′ leaders; however, we show that at least five CPuORFs have very conserved amino acids sequences in the 5′ leader of Arabidopsis mRNAs directly upstream of the mORF but in a different frame. Finally, 14 CPuORFs do contain an AUG codon in the sequence. Likely, the high number of species and the use of well-annotated 5′ leaders resulted in the discovery of these novel CPuORFs (Fig. 4).
In addition to the 29 novel CPuORFs, we discovered 64 of the 78 previously identified CPuORFs with high confidence (discovered in over 10 species) (Jorgensen and Dorantes-Acosta 2012; Takahashi et al. 2012; Vaughn et al. 2012; Laing et al. 2015). In total, 22% of the CPuORF homology groups does not initiate with an AUG start codon, meaning they are prone to leaky scanning. In addition, we show that the CPuORFs that do initiate with an AUG start codon have a poor start codon context (Fig. 2). These results are in line with the hypothesis that CPuORFs require leaky scanning to allow for sufficient translation of the mORF in noninhibitory conditions.
Our pipeline did not identify 14 of the 78 previously identified CPuORFs. This could partly be due to altered annotation of mORFs in the Araport11 annotation compared to previous versions. This is exemplified by two CPuORFs that overlap with the mORF of another splice variant while one CPuORF overlaps with the mORF of another gene (Supplemental Table S3). Each of these genes have an Arabidopsis paralogue with a similar CPuORF but these, however, do not have a mORF annotated at the position of the CPuORF. It is likely that either the CPuORF of the paralogs or the overlapping mORF is miss-annotated. As new splice variants are continuously being discovered, we cannot exclude that some of the newly discovered CPuORFs are part of a to be discovered splice variant. Five other, previously identified, CPuORFs were not detected because the sequences did not fulfill the requirements for purifying selection, which is a measure for the coding potential of a nucleotide sequence. A challenge in this test was presented by not knowing the start codon of the uCCRs. The full-length amino acid sequences of the uSSRs were aligned and used to check for purifying selection. However, the N-terminal region of this sequence might not be translated into a protein and could skew the result to a more neutral selection. On the other hand, the nontranslated part of the uSSR is usually not conserved and therefore introduces many gaps (Supplemental Figs. S3–S5). CodeML, the program used to calculate the dN/dS and likelihood ratio, cannot use amino acid positions where a gap is present in the alignment. Therefore, most of the nontranslated parts are not used for the analysis. Unsuccessful identification of the CPuORFs could also be due to too strict thresholds (ω < 0.4 and χ2 P-value < 0.01). In fact, results of the dN/dS analyses performed by Hayden and Jorgensen (2007) of four of the five CPuORFs would not have passed our threshold (Hayden and Jorgensen 2007). However, when the threshold was more relaxed more false positives were identified, like uSSRs that overlapped a CPuORF in a different frame (data not shown).
Finally, six previously identified CPuORFs were not detected by our pipeline because they did not pass the conservation criteria. Two of these were not conserved in Malvids, while the other four passed purifying selection criteria but were not conserved in over 10 species (Supplemental Table S3). Except for O. sativa all species used in this analysis are part of the eudicot clade and the majority is rosid (Fig. 1B). Most of the species in this clade are divergent enough from Arabidopsis to differ at nucleotide level, while similar enough to identify recently evolved uCCRs. We decided to call an uSSR a CPuORF if the Arabidopsis uSSR had at least one BLASTp hit with a uSSR from 10 other species. This threshold is based on the number of remaining, previously identified CPuORFs and manual inspection of the alignments (Supplemental Data 1; Supplemental Figs. S3–S5). We realize that this threshold is strict, however we prefer for the pipeline to result in false negatives over false positives. In addition, it should be noted that the annotation quality of the species differ, meaning that if an uCCR was not discovered in a certain species it does not necessarily mean that it is absent. Therefore, discovered CPuORFs conserved in less than 11 species may still be biological relevant (Supplemental Data 1). Altogether, most of the previously identified CPuORFs were also identified in this study including the two previously identified and biologically confirmed non-AUG initiating CPuORFs (Laing et al. 2015). All of the previously experimentally confirmed CPuORFs were identified in our study. Due to our strict criteria, few CPuORFs were not identified in our study which would have been identified with more relaxed criteria. However, the strict criteria make us confident of the biological relevance of the identified CPuORFs including the 29 CPuORFs not previously identified. Fourteen out of 78 previously identified CPuORFs are not identified using our approach. At least three of these have newly annotated splicing variants and the CPuORFs may be parts of the main ORFs.
Translation of multiple non-AUG CPuORFs was confirmed by ribosome footprint data from Merchante et al. (2015) (Fig. 3). For example, AT4G03260 has a CPuORF likely initiating with an ACG start codon. The ribosome footprint data shows a large peak exactly at the position of the stop codon of the CPuORF. Ribosome toeprinting, in vitro translation assays and ribosome footprinting analyses have shown that ribosome stalling on uORFs regulating mORF translation often occurs near stop codons (Uchiyama-Kadokura et al. 2014; Yamashita et al. 2014, 2017; Peviani et al. 2016; Tanaka et al. 2016). Therefore, it is likely that ribosomes stall near the stop codon of the AT4G03260 CPuORF in the conditions used by Merchante et al. (2015).
Moreover, increasing evidence shows that metabolites which induce ribosome stalling are sensed inside the ribosomal exit tunnel (Imai et al. 2008; Bischoff et al. 2014; Kakehi et al. 2015; Arenz et al. 2016). The ribosomal exit tunnel is a 100 Å cavity which fits about 40 amino acids (Kowarik et al. 2002). Interestingly, the length of the majority of the CPuORFs is less than 40 amino acids, when looked at the first conserved amino acid up to the stop codon (Supplemental Fig. S7). This is in line with the hypothesis that CPuORFs fulfill their function inside the ribosomal exit tunnel.
A well-known example of ribosomes stalling induced by a metabolite are the CPuORFs of the S1 group of bZIP protein genes, where ribosomes stall in the presence of sucrose (Hummel et al. 2009; Rahmani et al. 2009; Juntawong et al. 2014; Yamashita et al. 2017). CPuORFs of S1 bZIPs only allow mORF translation when sucrose levels are low. It has previously been described that two distinct CPuORFs are present in genes of the core sucrose response network in the mRNAs encoding T6P phosphatases (TPP) and SnRK1 activating kinases (SnAK) (Jorgensen and Dorantes-Acosta 2012). This study uncovered that the mRNA of RAPTOR1 (regulatory-associated protein of TOR), that controls the master regulator target of rapamycin (TOR), also contains a CPuORF and likely initiates with a CUG. TOR enhances mORF reinitiation after uORF translation via phosphorylation of S6 KINASE 1 (S6K1). A previous search for CPuORFs using genomic data from five cereal species discovered a CPuORF in the S6K mRNA; however, this search did not discover the S6K1 CPuORF in Arabidopsis (Tran et al. 2008). Our pipeline discovered the S6K1 uCCR in both eudicots and monocots. However, the methionine start codon was lost in all Brassicaceae species, with no clear near-cognate start codon in the uCCR sequence (Supplemental Fig. S8). It is therefore possible that the CPuORF in S6K1 mRNA has lost its functionality in Brassicaceae. Nevertheless, these results suggest that each of these CPuORFs is involved in sensing a specific molecule, allowing for a system to respond quickly to changes in metabolite levels and energy availability.
The mRNA encoding LHY also possesses a highly conserved CPuORF. The position of the stop codon is very conserved in about half of the 19 species where the uORF is detected, which could not be allocated to a specific clade (Fig. 3). Moreover, the extended part of the uORF in the other species is not conserved outside the Brassicales. Arabidopsis ribosome footprint data from Merchante et al. (2015) shows a large peak exactly on the position where the conservation stops and where a stop codon is present in half of the species, indicating ribosome stalling (Fig. 3). LHY is a key player in regulation of the circadian clock. Previous work on Arabidopsis lhy-1, a mutant constitutively expressing LHY mRNA, revealed that protein accumulation was increased within 30 min following exposure to light (Kim et al. 2003). As light did not alter mRNA concentrations or protein turnover in this mutant, it was concluded that light must regulate LHY translation. The plants used by Merchante et al. (2015) for their ribosome footprinting experiments were grown in darkness, where CPuORF-mediated ribosome stalling would be expected to inhibit LHY protein production (Merchante et al. 2015). The observed ribosome stalling at the LHY CPuORF in the dark supports the hypothesis that light abolishes stalling, resulting in LHY protein production. Further research on the regulatory function of this CPuORF could uncover the photosensory mechanism involved.
The CPuORF of LHY is one of the five CPuORFs discovered in this study that has its stop codon in the mORF, but in a different frame than the mORF (Table 2). The other four CPuORFs could also induce stalling as exemplified by the CPuORF present on the LHY mRNA. Possibly, these uCCRs are prone to ribosomal frameshifting, a process where the ribosome shifts its frame during the elongation phase of translation of an ORF (Kurian et al. 2011; Yordanova et al. 2015; Gao and Simon 2016; Meydan et al. 2017). To our knowledge, ribosomal frameshifting in plants has only been shown on viral RNA (Gao and Simon 2016). In other kingdoms, however, ribosomal frameshifting on endogenous RNA can be used for production of two proteins of one gene or for translation regulation through metabolites (Kurian et al. 2011; Yordanova et al. 2015; Gao and Simon 2016). The CPuORF on the mRNA of AT3G57170 is a possible candidate for ribosomal frameshifting in plants. The CPuORF encodes for a 145 amino acid long peptide, which is nearly fully conserved (Supplemental Fig. S3). Peptides inducing ribosome stalling are usually much shorter, therefore a stalling mechanism is unlikely. Moreover, the context of the annotated start codon of the mORF of AT3G57170 is not optimal (UGCAUGAT), reducing the likelihood of this being the true start codon of this gene. Further investigation of this mRNA might reveal the first example of ribosomal frameshifting on an endogenous RNA in plants.
Noncanonical start codon initiation for mORF translational can be used for dual localization of a protein (Christensen et al. 2005; Wamboldt et al. 2009). We evaluated this possibility on the newly discovered uCCRs. mORFs with and without noncanonical start codon were tested on subcellular localization using three programs. The results revealed that AT5G36250 and AT4G00840 might localize to a different subcellular location when translated by the alternative start codon (Supplemental Table S4). Additionally, AT2G25110, encoding for stromal-derived factor 2 (SDF2), is predicted to localize to the ER when translated from the annotated AUG, but when translated from a GUG upstream, it is predicted to localize to the chloroplast or mitochondrion (Supplemental Table S4). SDF2 is involved in correct protein folding after ER stress and the AUG mORF protein has been shown to translocate to the ER (Schott et al. 2010). However, SDF2 localization experiments including the alternative uCCR are lacking. Similar to DNA polymerase POLγ2, SDF2 might have a biological function in two different subcellular compartments.
In conclusion, this study demonstrated that our pipeline is a powerful tool to identify AUG and non-AUG initiating CPuORFs and 5′ extended mORFs with high confidence. Our pipeline is robust enough to identify very small peptides such as the CPuORF in AT3G08850 (15 aa) and AT4G15180 (11 aa). Therefore, our methodology could be adapted to detect small peptides, such as peptide hormones or conserved peptides on long noncoding RNAs (Hsu et al. 2016; Bazin et al. 2017). Moreover, bioinformatics analyses on non-AUG CPuORFs in other kingdoms are lacking and our methodology could be used to fill this gap (Cvijović et al. 2007; Hayden and Bosco 2008; Selpi et al. 2009; Skarshewski et al. 2014). Most importantly, the new findings will serve as a basis for further research in translational regulation in plants, a field where progress is rapid due to novel technologies such as precise ribosome footprinting.
MATERIALS AND METHODS
Retrieving upstream stop to stop regions (uSSR)
In a first step, cDNA sequences were retrieved for 32 plant species from public repositories (Fig. 1; Supplemental Table S2). Then, mORFs (main ORF) were defined as the longest ORF (ATG to an in frame stop codon) using getorf (EMBOSS package version 6.5.7.0). Everything upstream of the mORF was considered the 5′ leader and sequences around and in frame with the stop codons in the 5′ leaders were retrieved using getorf. These regions contain three types of sequences: (i) sequences from the transcriptional start site to the next in frame stop codon, (ii) sequences from a stop codon to the next in frame stop codon, or (iii) sequences from a stop codon to the mORF start codon. These sequences will for now all be designated as uSSR (for upstream stop to stop region or stop mORF start region). Arabidopsis uSSRs were aligned with uSSRs retrieved from each of the 31 other species using BLASTp (blast version 2.6.0) after translating the uSSRs to amino acid sequences, alignments with an E-value <100 were considered a hit (Fig. 1A).
Second, all the Arabidopsis mORF protein sequences were compared with each mORF sequence of the 31 other species using BLASTp sequence similarity searches. mORFs with an E-value below 1 × 10−5 were considered homologous. If an uSSR from one of the 31 species has a BLASTp hit with one of the Arabidopsis uSSR but the downstream mORFs are not homologs, the uSSR hit was removed.
Third, the Arabidopsis genomic position of each uSSR BLASTp hit was compared with annotations (Araport11 release 201606) on the same position. If a coding DNA sequence (CDS) was annotated on that position, the BLASTp hit was removed. This way, hits caused by alternative splicing events or overlapping genes were removed.
Conservation across multiple species was checked in the fourth step. If an Arabidopsis uSSR was not similar to at least one uSSR in the Brassicales and one uSSR in the Malvids (outside of Brassicales) clade, the uSSR was discarded (Fig. 1B). The remaining uSSRs were categorized in three groups depending on the degree of conservation: Poorly conserved (similar uSSRs in seven or less species [including Arabidopsis]), conserved (similar uSSRs in 8–10 species [including Arabidopsis]), or highly conserved (similar uSSRs in 11 or more species [including Arabidopsis]).
Finally, the uCCRs were divided into three categories: (i) CPuORFs (includes a stop codon in the 5′ leader), (ii) CPuORFs without a stop codon before the mORF and out of frame with the mORF, and (iii) uCCRs in frame with the mORF that does not contain a stop codon in the 5′ leader and initiates on a non-AUG start codon (hereafter referred to as 5′ extended mORFs).
Purifying selection
The amino acid sequence of each uSSR set (all uSSRs of BLASTp hits of a certain Arabidopsis uSSR) were aligned using MAFFT (version 7.307, FFT-NS-2 method). Then the conserved region was assigned using the program “cons” from the EMBOSS package (version 6.6.6.0) with “-setcase” as 0.75 multiplied by the number of sequences in the alignment. If a sequence in the alignment did not have >50% of the consensus amino acids (capitalized letters from cons output), the sequence was removed. The codons corresponding to the amino acids of the remaining uSSRs were then retrieved which served as input for PAML.
Tree topologies were calculated per uSSR set using the corresponding mORFs of each uSSR. Amino acid sequences of the mORFs were first aligned using MAFFT (version 7.307, FFT-NS-2 method) and then trimmed using trimAl (version 1.4.rev15) with the gap score threshold set at 0.25. Tree topologies were retrieved from these alignments using RAxML (version 8.2.10) using 100 bootstraps with the model set to “PROTGAMMAAUTO.”
Assessment for purifying selection was performed by calculating the dN/dS ratios using PAML (version 4.9d) according to the protocol in Nekrutenko et al. (2002). In brief, the ratios between nonsynonymous and synonymous substitutions (dN/dS) were calculated twice, first with dN/dS fixed at one and then as a free parameter using the codeml program from the PAML package (version 4.9d). The maximum likelihood ratios were used to calculate the likelihood ratio which in turn was used to calculate the P-value from the χ2 distribution with one degree of freedom. All uSSR sets with a P-value higher than 0.01 or a dN/dS higher than 0.4 were removed.
Subcellular localization predictions
First, noncanonical start codons of the 5′ extended mORFs were annotated by manual inspection, taking conservation and similarity to the plant “Kozak's sequence” (AxxAUGGc) into account. mORFs with and without alternative start codon were translated into amino acids, where the alternative start codon was translated into a methionine (see Supplemental Data 2 for FASTA file). These sequences were loaded into predotar v1.04 (https://urgi.versailles.inra.fr/predotar/), TargetP v1.1 (http://www.cbs.dtu.dk/services/TargetP/), and iPSORT (http://ipsort.hgc.jp/) to predict the subcellular localization.
Ribosome footprinting data analysis
Indexed BAM files with ribosome footprinting data from Merchante et al. (2015) were kindly provided by Julia Bailey-Serres and Maureen Hummel (University of California). The reads were viewed using the Integrative Genome Viewer (IGV, version 2.4) and screenshots are shown in Figure 3.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
The authors are grateful to Julia Bailey-Serres and Maureen Hummel for the critical comments on the manuscript and for the sharing of ribosome footprinting data. We thank Bio4Energy, a Strategic Research Environment appointed by the Swedish government and ALW-NOW (grant no. ALWOP.2015.115) for supporting this work.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.067983.118.
Freely available online through the RNA Open Access option.
REFERENCES
- Alatorre-Cobos F, Cruz-Ramírez A, Hayden CA, Pérez-Torres C-A, Chauvin A-L, Ibarra-Laclette E, Alva-Cortés E, Jorgensen RA, Herrera-Estrella L. 2012. Translational regulation of Arabidopsis XIPOTL1 is modulated by phosphocholine levels via the phylogenetically conserved upstream open reading frame 30. J Exp Bot 63: 5203–5221. 10.1093/jxb/ers180 [DOI] [PubMed] [Google Scholar]
- Arenz S, Bock L V, Graf M, Innis CA, Beckmann R, Grubmüller H, Vaiana AC, Wilson DN. 2016. A combined cryo-EM and molecular dynamics approach reveals the mechanism of ErmBL-mediated translation arrest. Nat Commun 7: 12026 10.1038/ncomms12026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazin J, Baerenfaller K, Gosai SJ, Gregory BD, Crespi M, Bailey-Serres J. 2017. Global analysis of ribosome-associated noncoding RNAs unveils new modes of translational regulation. Proc Natl Acad Sci 114: E10018–E10027. 10.1073/pnas.1708433114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchini C, Pastore A, Pelucchi S, Torreggiani E, Lambertini E, Marchesi E, Magri E, Frasson C, Querzoli P, Piva R. 2008. Sex hormone receptor levels in laryngeal carcinoma: a comparison between protein and RNA evaluations. Eur Arch Otorhinolaryngol 265: 1089–1094. 10.1007/s00405-008-0589-9 [DOI] [PubMed] [Google Scholar]
- Bischoff L, Berninghausen O, Beckmann R. 2014. Molecular basis for the ribosome functioning as an L-tryptophan sensor. Cell Rep 9: 469–475. 10.1016/j.celrep.2014.09.011 [DOI] [PubMed] [Google Scholar]
- Browning KS, Bailey-Serres J. 2015. Mechanism of cytoplasmic mRNA translation. Arabidopsis Book 13: e0176 10.1199/tab.0176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen AC, Lyznik A, Mohammed S, Elowsky CG, Elo A, Yule R, Mackenzie SA. 2005. Dual-domain, dual-targeting organellar protein presequences in Arabidopsis can use non-AUG start codons. Plant Cell 17: 2805–2816. 10.1105/tpc.105.035287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrads KA, Yi M, Simpson KA, Lucas DA, Camalier CE, Yu LR, Veenstra TD, Stephens RM, Conrads TP, Beck GR Jr. 2005. A combined proteome and microarray investigation of inorganic phosphate-induced pre-osteoblast cells. Mol Cell Proteomics 4: 1284–1296. 10.1074/mcp.M500082-MCP200 [DOI] [PubMed] [Google Scholar]
- Cvijović M, Dalevi D, Bilsland E, Kemp GJ, Sunnerhagen P. 2007. Identification of putative regulatory upstream ORFs in the yeast genome using heuristics and evolutionary conservation. BMC Bioinformatics 8: 295 10.1186/1471-2105-8-295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depeiges A, Degroote F, Espagnol MC, Picard G. 2006. Translation initiation by non-AUG codons in Arabidopsis thaliana transgenic plants. Plant Cell Rep 25: 55–61. 10.1007/s00299-005-0034-0 [DOI] [PubMed] [Google Scholar]
- Diaz de Arce AJ, Noderer WL, Wang CL. 2018. Complete motif analysis of sequence requirements for translation initiation at non-AUG start codons. Nucleic Acids Res 46: 985–994. 10.1093/nar/gkx1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dröge-Laser W, Weiste C. 2018. The C/S1 bZIP network: a regulatory hub orchestrating plant energy homeostasis. Trends Plant Sci 23: 422–433. 10.1016/j.tplants.2018.02.003 [DOI] [PubMed] [Google Scholar]
- Gao F, Simon AE. 2016. Multiple cis-acting elements modulate programmed -1 ribosomal frameshifting in Pea enation mosaic virus. Nucleic Acids Res 44: 878–895. 10.1093/nar/gkv1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y, Usadel B, Blaesing OE, Kamlage B, Hoehne M, Trethewey R, Stitt M. 2006. Integration of metabolite with transcript and enzyme activity profiling during diurnal cycles in Arabidopsis rosettes. Genome Biol 7: R76 10.1186/gb-2006-7-8-r76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon K, Fütterer J, Hohn T. 1992. Efficient initiation of translation at non-AUG triplets in plant cells. Plant J 2: 809–813. [PubMed] [Google Scholar]
- Guerrero-González ML, Ortega-Amaro MA, Juárez-Montiel M, Jiménez-Bremont JF. 2016. Arabidopsis polyamine oxidase-2 uORF is required for downstream translational regulation. Plant Physiol Biochem 108: 381–390. 10.1016/j.plaphy.2016.08.006 [DOI] [PubMed] [Google Scholar]
- Hanson J, Hanssen M, Wiese A, Hendriks MM, Smeekens S. 2008. The sucrose regulated transcription factor bZIP11 affects amino acid metabolism by regulating the expression of ASPARAGINE SYNTHETASE1 and PROLINE DEHYDROGENASE2. Plant J 53: 935–949. 10.1111/j.1365-313X.2007.03385.x [DOI] [PubMed] [Google Scholar]
- Hayden CA, Jorgensen RA. 2007. Identification of novel conserved peptide uORF homology groups in Arabidopsis and rice reveals ancient eukaryotic origin of select groups and preferential association with transcription factor-encoding genes. BMC Biol 5: 32 10.1186/1741-7007-5-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden CA, Bosco G. 2008. Comparative genomic analysis of novel conserved peptide upstream open reading frames in Drosophila melanogaster and other dipteran species. BMC Genomics 9: 61 10.1186/1471-2164-9-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges SB, Marin J, Suleski M, Paymer M, Kumar S. 2015. Tree of life reveals clock-like speciation and diversification. Mol Biol Evol 32: 835–845. 10.1093/molbev/msv037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou C, Lee W, Chou H, Chen A, Chou S, Chen H. 2016. Global analysis of truncated RNA ends reveals new insights into ribosome stalling in plants. Plant Cell 28: 2398–2416. 10.1105/tpc.16.00295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PY, Calviello L, Wu HL, Li FW, Rothfels CJ, Ohler U, Benfey PN. 2016. Super-resolution ribosome profiling reveals unannotated translation events in Arabidopsis. Proc Natl Acad Sci 113: E7126–E7135. 10.1073/pnas.1614788113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Merchante C, Stepanova A, Alonso J, Heber S. 2016. Genome-wide search for translated upstream open reading frames in Arabidopsis thaliana. IEEE Trans Nanobioscience 15: 148–157. 10.1109/TNB.2016.2516950 [DOI] [PubMed] [Google Scholar]
- Hummel M, Rahmani F, Smeekens S, Hanson J. 2009. Sucrose-mediated translational control. Ann Bot 104: 1–7. 10.1093/aob/mcp086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai A, Komura M, Kawano E, Kuwashiro Y, Takahashi T. 2008. A semi-dominant mutation in the ribosomal protein L10 gene suppresses the dwarf phenotype of the acl5 mutant in Arabidopsis thaliana. Plant J 56: 881–890. 10.1111/j.1365-313X.2008.03647.x [DOI] [PubMed] [Google Scholar]
- Ivanov IP, Shin BS, Loughran G, Tzani I, Young-Baird SK, Cao C, Atkins JF, Dever TE. 2018. Polyamine control of translation elongation regulates start site selection on antizyme inhibitor mRNA via ribosome queuing. Mol Cell 254–264.e6. 10.1016/j.molcel.2018.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen RA, Dorantes-Acosta AE. 2012. Conserved peptide upstream open reading frames are associated with regulatory genes in angiosperms. Front Plant Sci 3: 191 10.3389/fpls.2012.00191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juntawong P, Girke T, Bazin J, Bailey-Serres J. 2014. Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc Natl Acad Sci 111: E203–E212. 10.1073/pnas.1317811111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakehi JI, Kawano E, Yoshimoto K, Cai Q, Imai A, Takahashi T. 2015. Mutations in ribosomal proteins, RPL4 and RACK1, suppress the phenotype of a thermospermine-deficient mutant of Arabidopsis thaliana. PLoS One 10: e0117309 10.1371/journal.pone.0117309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Song HR, Taylor BL, Carré IA. 2003. Light-regulated translation mediates gated induction of the Arabidopsis clock protein LHY. EMBO J 22: 935–944. 10.1093/emboj/cdg075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochetov AV. 2008. Alternative translation start sites and hidden coding potential of eukaryotic mRNAs. Bioessays 30: 683–691. 10.1002/bies.20771 [DOI] [PubMed] [Google Scholar]
- Kowarik M, Küng S, Martoglio B, Helenius A. 2002. Protein folding during cotranslational translocation in the endoplasmic reticulum. Mol Cell 10: 769–778. 10.1016/S1097-2765(02)00685-8 [DOI] [PubMed] [Google Scholar]
- Kurian L, Palanimurugan R, Gödderz D, Dohmen RJ. 2011. Polyamine sensing by nascent ornithine decarboxylase antizyme stimulates decoding of its mRNA. Nature 477: 490–494. 10.1038/nature10393 [DOI] [PubMed] [Google Scholar]
- Laing WA, Martínez-Sánchez M, Wright MA, Bulley SM, Brewster D, Dare AP, Rassam M, Wang D, Storey R, Macknight RC, et al. 2015. An upstream open reading frame is essential for feedback regulation of ascorbate biosynthesis in Arabidopsis. Plant Cell 27: 772–786. 10.1105/tpc.114.133777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Shen W, Sun H, Migawa MT, Vickers TA, Crooke ST. 2016. Translation efficiency of mRNAs is increased by antisense oligonucleotides targeting upstream open reading frames. Nat Biotechnol 34: 875–880. 10.1038/nbt.3589 [DOI] [PubMed] [Google Scholar]
- Ma J, Hanssen M, Lundgren K, Hernández L, Delatte T, Ehlert A, Liu CM, Schluepmann H, Dröge-Laser W, Moritz T, et al. 2011. The sucrose-regulated Arabidopsis transcription factor bZIP11 reprograms metabolism and regulates trehalose metabolism. New Phytol 191: 733–745. 10.1111/j.1469-8137.2011.03735.x [DOI] [PubMed] [Google Scholar]
- Merchante C, Brumos J, Yun J, Hu Q, Spencer KR, Enríquez P, Binder BM, Heber S, Stepanova AN, Alonso JM. 2015. Gene-specific translation regulation mediated by the hormone-signaling molecule EIN2. Cell 163: 684–697. 10.1016/j.cell.2015.09.036 [DOI] [PubMed] [Google Scholar]
- Merchante C, Stepanova AN, Alonso JM. 2017. Translation regulation in plants: an interesting past, an exciting present and a promising future. Plant J 90: 628–653. 10.1111/tpj.13520 [DOI] [PubMed] [Google Scholar]
- Meydan S, Klepacki D, Karthikeyan S, Margus T, Thomas P, Jones JE, Khan Y, Briggs J, Dinman JD, Vázquez-Laslop N, et al. 2017. Programmed ribosomal frameshifting generates a copper transporter and a copper chaperone from the same gene. Mol Cell 65: 207–219. 10.1016/j.molcel.2016.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrutenko A, Makova KD, Li WH. 2002. The KA/KS ratio test for assessing the protein-coding potential of genomic regions: an empirical and simulation study. Genome Res 12: 198–202. 10.1101/gr.200901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peviani A, Lastdrager J, Hanson J, Snel B. 2016. The phylogeny of C/S1 bZIP transcription factors reveals a shared algal ancestry and the pre-angiosperm translational regulation of S1 transcripts. Sci Rep 6: 30444 10.1038/srep30444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmani F, Hummel M, Schuurmans J, Wiese-Klinkenberg A, Smeekens S, Hanson J. 2009. Sucrose control of translation mediated by an upstream open reading frame-encoded peptide. Plant Physiol 150: 1356–1367. 10.1104/pp.109.136036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Ito T, Meyerowitz EM. 1999. Non-AUG initiation of AGAMOUS mRNA translation in Arabidopsis thaliana. Mol Cell Biol 19: 8505–8512. 10.1128/MCB.19.12.8505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott A, Ravaud S, Keller S, Radzimanowski J, Viotti C, Hillmer S, Sinning I, Strahl S. 2010. Arabidopsis stromal-derived factor 2 (SDF2) is a crucial target of the unfolded protein response in the endoplasmic reticulum. J Biol Chem 285: 18113–18121. 10.1074/jbc.M110.117176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selpi S, Bryant CH, Kemp GJ, Sarv J, Kristiansson E, Sunnerhagen P. 2009. Predicting functional upstream open reading frames in Saccharomyces cerevisiae. BMC Bioinformatics 10: 451 10.1186/1471-2105-10-451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson GG, Laurie RE, Dijkwel PP, Quesada V, Stockwell PA, Dean C, Macknight RC. 2010. Noncanonical translation initiation of the Arabidopsis flowering time and alternative polyadenylation regulator FCA. Plant Cell 22: 3764–3777. 10.1105/tpc.110.077990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarshewski A, Stanton-Cook M, Huber T, Al Mansoori S, Smith R, Beatson SA, Rothnagel JA. 2014. uPEPperoni: an online tool for upstream open reading frame location and analysis of transcript conservation. BMC Bioinformatics 15: 36 10.1186/1471-2105-15-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spealman P, Naik AW, May GE, Kuersten S, Freeberg L, Murphy RF, McManus J. 2018. Conserved non-AUG uORFs revealed by a novel regression analysis of ribosome profiling data. Genome Res 28: 214–222. 10.1101/gr.221507.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi T, Okada T, Azuma T, Nanmori T, Yasuda T. 2006. Posttranscriptional regulation by the upstream open reading frame of the phosphoethanolamine N-methyltransferase gene. Biosci Biotechnol Biochem 70: 2330–2334. 10.1271/bbb.60309 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Takahashi A, Naito S, Onouchi H. 2012. BAIUCAS: a novel BLAST-based algorithm for the identification of upstream open reading frames with conserved amino acid sequences and its application to the Arabidopsis thaliana genome. Bioinformatics 28: 2231–2241. 10.1093/bioinformatics/bts303 [DOI] [PubMed] [Google Scholar]
- Tanaka M, Sotta N, Yamazumi Y, Yamashita Y, Miwa K, Murota K, Chiba Y, Hirai MY, Akiyama T, Onouchi H, et al. 2016. The minimum open reading frame, AUG-stop, induces boron-dependent ribosome stalling and mRNA degradation. Plant Cell 28: 2830–2849. 10.1105/tpc.16.00481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalor SK, Berberich T, Lee SS, Yang SH, Zhu X, Imai R, Takahashi Y, Kusano T. 2012. Deregulation of sucrose-controlled translation of a bZIP-type transcription factor results in sucrose accumulation in leaves. PLoS One 7: e33111 10.1371/journal.pone.0033111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran MK, Schultz CJ, Baumann U. 2008. Conserved upstream open reading frames in higher plants. BMC Genomics 9: 361 10.1186/1471-2164-9-361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama-Kadokura N, Murakami K, Takemoto M, Koyanagi N, Murota K, Naito S, Onouchi H. 2014. Polyamine-responsive ribosomal arrest at the stop codon of an upstream open reading frame of the AdoMetDC1 gene triggers nonsense-mediated mRNA decay in Arabidopsis thaliana. Plant Cell Physiol 55: 1556–1567. 10.1093/pcp/pcu086 [DOI] [PubMed] [Google Scholar]
- Vaughn JN, Ellingson SR, Mignone F, Arnim Av. 2012. Known and novel post-transcriptional regulatory sequences are conserved across plant families. RNA 18: 368–384. 10.1261/rna.031179.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Arnim AG, Jia Q, Vaughn JN. 2014. Regulation of plant translation by upstream open reading frames. Plant Sci 214: 1–12. 10.1016/j.plantsci.2013.09.006 [DOI] [PubMed] [Google Scholar]
- Wamboldt Y, Mohammed S, Elowsky C, Wittgren C, de Paula WB, Mackenzie SA. 2009. Participation of leaky ribosome scanning in protein dual targeting by alternative translation initiation in higher plants. Plant Cell 21: 157–167. 10.1105/tpc.108.063644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weltmeier F, Rahmani F, Ehlert A, Dietrich K, Schütze K, Wang X, Chaban C, Hanson J, Teige M, Harter K, et al. 2009. Expression patterns within the Arabidopsis C/S1 bZIP transcription factor network: availability of heterodimerization partners controls gene expression during stress response and development. Plant Mol Biol 69: 107–119. 10.1007/s11103-008-9410-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese A, Elzinga N, Wobbes B, Smeekens S. 2004. A conserved upstream open reading frame mediates sucrose-induced repression of translation. Plant Cell 16: 1717–1729. 10.1105/tpc.019349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Y, Kadokura Y, Sotta N, Fujiwara T, Takigawa I, Satake A, Onouchi H, Naito S. 2014. Ribosomes in a stacked array: elucidation of the step in translation elongation at which they are stalled during S-adenosyl-L-methionine-induced translation arrest of CGS1 mRNA. J Biol Chem 289: 12693–12704. 10.1074/jbc.M113.526616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Y, Takamatsu S, Glasbrenner M, Becker T, Naito S, Beckmann R. 2017. Sucrose sensing through nascent peptide-meditated ribosome stalling at the stop codon of Arabidopsis bZIP11 uORF2. FEBS Lett 591: 1266–1277. 10.1002/1873-3468.12634 [DOI] [PubMed] [Google Scholar]
- Yordanova MM, Wu C, Andreev DE, Sachs MS, Atkins JF. 2015. A nascent peptide signal responsive to endogenous levels of polyamines acts to stimulate regulatory frameshifting on antizyme mRNA. J Biol Chem 290: 17863–17878. 10.1074/jbc.M115.647065 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.