Abstract
We attempted to generate de novo resistance to a newly described biocidal complex, ITC (iodo-thiocyanate complex), and to levofloxacin (LVX) in Escherichia coli ATCC 25922, by means of selective chemostat culture. We measured resistance by determining the minimum inhibitory concentrations (MICs) for these agents. E. coli underwent 20-day parallel adaptive evolution routes under no antimicrobial selection, and gradually increasing ITC and LVX selection pressure. Long-term exposure of E. coli to ITC did not induce resistance to ITC, or cross-resistance to LVX. No distinct mutational pattern was evidenced from whole-genome sequence (WGS)-based comparisons of ITC-challenged and unchallenged bacterial populations. Moreover, the exposed E. coli population could not survive a 2 × MIC challenge of ITC. By contrast, resistance to LVX was rapidly induced (on day 1 the MIC had increased 16-fold), selected for (by day 14 the MIC had increased 64-fold) and enriched with a highly characteristic genome mutational pattern. WGS of this evolving population revealed that the majority of mutations appeared in the genes of LVX target proteins (GyrA, ParC, ParE) and drug influx (OmpF). This study suggests that the usage of ITC may not trigger the emergence of facile resistance or cross-resistance, in contrast to common antibiotics.
Introduction
Antibiotic resistance is a major global health problem. Preventing the emergence of resistance requires a thorough understanding of the underlying causes. Mechanisms of bacterial resistance to antibiotics, or other antimicrobial agents, have been well documented. Some bacteria possess intrinsic resistance towards antibiotics as natural structural and functional properties, while many others acquire resistance, mediated by several biochemical mechanisms (e.g. to stop its entrance, to pump it out, to break it down and to inactivate its target), by gene sharing between bacteria, or de novo as a result of specific gene mutations (spontaneous or induced by an agent)1. Though development of antibiotic resistance is a natural phenomenon and inevitable, the emergence of widespread, multi-agent resistance in many bacteria, including important pathogens, has been accelerated by the use, misuse and abuse of antibiotics and other antimicrobial agents, including triclosan, triclocarbon and others2. Preservation of antibiotics, underpinned by the availability of novel antimicrobial biocides that avoid the emergence of antibiotic cross-resistance, is thus an important societal goal.
We recently described a new biocidal complex, which we named the iodo-thiocyanate complex (ITC), and reported on its antibacterial properties3. In parallel, we assessed the in vitro cytotoxic, hemolytic and genotoxic potential of ITC by comparison with common antiseptics, which suggested the feasibility for use of ITC as a biocide/antiseptic4. Generally, a biocide is described as a chemical agent, usually broad-spectrum, that inactivates microorganisms5. The term biocide is mainly defined by its usage6 and includes disinfectants, antiseptics and preservatives7,8. Biocides generally have multiple targets in the bacterial cell and the cell death is caused by combined detrimental effects9. Moreover, the target sites can differ according to the biocide concentration applied and interactions resulting with cell death are not always clearly defined10. Our study showed that ITC is an oxidative mixture containing reactive oxygen (ROS) and iodine species that possess biocidal properties3. These reactive species appeared to interact with bacterial DNA and ribosomes, however, like other biocidal agents, the exact cause of cell death induced by ITC was difficult to pinpoint. Taking into account that such reactive species provide direct and indirect mechanisms for mutagenesis11, the potential for the development of bacterial resistance toward ITC, and other antimicrobials, is of concern in the context of its potential application. Indeed, there is increasing concern that extensive biocide usage can be responsible for the selection and maintenance of antibiotic-resistant bacteria in various environments12. Accumulated experimental evidence suggests that bacteria that have become less sensitive to a biocide (because of exposure to the biocide) also show decreased susceptibilities to antibiotics. Household pine oil cleaner (POC) resistant mutants of Staphylococcus aureus isolated on media containing POC also demonstrated reduced susceptibility to the cell wall-active antibiotics vancomycin and oxacillin13. The sequential growth of Salmonella typhimurium in gradually increasing concentrations of triclosan in the growth medium resulted in a population that was highly resistant (2000-fold) to triclosan and 8-fold resistant to ampicillin14. Population of Pseudomonas aeruginosa subjected to increasing levels of benzalkonium chloride (BKC) selection pressure in long-term continuous culture carried a mutant variant with >12-fold decreased sensitivity to BKC and 256-fold increased resistance to fluoroquinolone antibiotic ciprofloxacin15.
In recent years, substantial research efforts have been directed towards understanding bacterial adaptation and resistance to different types of biocides16. As the modes of action of biocides are generally difficult to distinguish, the detailed mechanisms underpinning acquired bacterial resistance to biocides also remain uncertain12. From the genetic point of view, biocide resistance can arise via mutation or by the acquisition of genetic elements bearing resistance genes. From the mechanistic point of view, because of the multiplicity of target sites, resistance to biocides is less common acquired via target inactivation mechanisms. More commonly, biocide resistance can be developed due to the inactivation of the biocide and the reduction of its intracellular concentration to a level that is not harmful to the bacterial cell, that is to say, reduced influx, or enhanced efflux of a biocide8,17,18. Considering the common shared mechanisms for resistance towards antibiotics and biocides, there is a likelihood of cross-resistance between biocides and antibiotics.
Since resistance is a response adaptation of bacteria to strong natural selection imposed by the application of antibiotics and biocides, understanding the adaptive processes in pathogen populations and, in particular, characterizing the genetic changes that turn drug-sensitive bacteria into resistant variants, is essential to address the problem of resistance. Laboratory evolution experiments have been successfully used to study pathogen adaptation to antibiotics during the evolutionary process, in real time and under precisely controlled conditions19. Chemostat-based continuous flow systems are one of the generally applied experimental evolution techniques20. In a chemostat vessel, the bacterial population can be maintained in exponential growth phase continuously by the regular inflow of fresh media and simultaneous outflow of waste culture21. Bacterial populations in these continuous cultures become highly heterogeneous in remarkably short time periods existing as a temporal, spatial, physiological and genetic heterogeneous complexity22. Although mutations in chemostat populations will occur naturally in the presence, or absence, of a mutagenic agent, their evolution may be “directed” by the addition of a selector (such as an antimicrobial drug) and the population may be enriched with a desired phenotype (such as antimicrobial-resistant)23. In such settings, the evolving pathogen population may be continuously challenged by a temporal gradient of drug concentrations to promote the sequential emergence and fixation of multiple resistance mutations leading to increasingly higher resistance levels24,25. Chemostat continuous culture systems may thus simulate bacterial evolution in the host environment, in response to an antimicrobial treatment. As would be expected at an infection site, the pathogens in continuous cultures are grown at high densities, contain physiologically refractory planktonic (persisters) and biofilm subpopulations and antimicrobials are supplied in a continuous manner26.
Microbial experimental evolution studies have been revolutionized by recent advances in nucleic acid sequencing technologies. It is now technically and financially feasible to associate the phenotypic response to the underlying genetic changes during an experimental evolution study. Combining experimental evolution with whole-genome sequencing of evolved populations may uncover the genetic basis of laboratory adaptation to antimicrobials and the emergence of antimicrobial resistance27. This approach may reveal how many mutations underlie antimicrobial resistance, how are they distributed across the genome (coding versus noncoding, synonymous versus nonsynonymous mutations) and through time (mutations rise and fix at early or later stages of adaptation) and which target genes contribute to resistance28.
In this study, we used enrichment chemostats in attempts to generate de novo resistance to the ITC biocide and to assess its relationship to cross-resistance to a bactericidal antibiotic in a model organism. E. coli ATCC 25922 was used as the model organism, as it is well-characterized at a molecular and physiological level and a complete annotated genome sequence is available. Levofloxacin was selected as the antibiotic of choice as increased resistance to LVX may be acquired in E. coli under chemostat settings23 and the mechanisms of resistance toward LVX are well documented. Starting with an isogenic drug-sensitive E. coli ATCC 25922, we exposed the evolving population to increasing levels of ITC or LVX. To prevent culture washout, the concentrations of the antimicrobials were increased in a stepwise manner, with increases applied when the density of the culture had almost completely returned to the pre-antimicrobial treatment level15. We monitored the development of ITC and LVX-resistant phenotypes by changes in minimum inhibitory concentration profiles. In parallel, we followed ITC and antibiotic resistance emergence in unexposed (control) populations. Next, we performed WGS analysis of planktonic polymorphic populations sampled over time and compared starter, intermediate and final populations to reveal possible mutational determinants of ITC and LVX resistance.
Results
Growth and phenotypic resistance profiles of chemostat-derived cultures
The E. coli ATCC 25922 was gradually adapted to increasing concentrations of ITC and LVX over 20-days of selective chemostat culture, with the aim of enriching the population with antimicrobial-resistant bacteria. The emergence of drug resistance in these exposed cultures was then compared with unexposed culture.
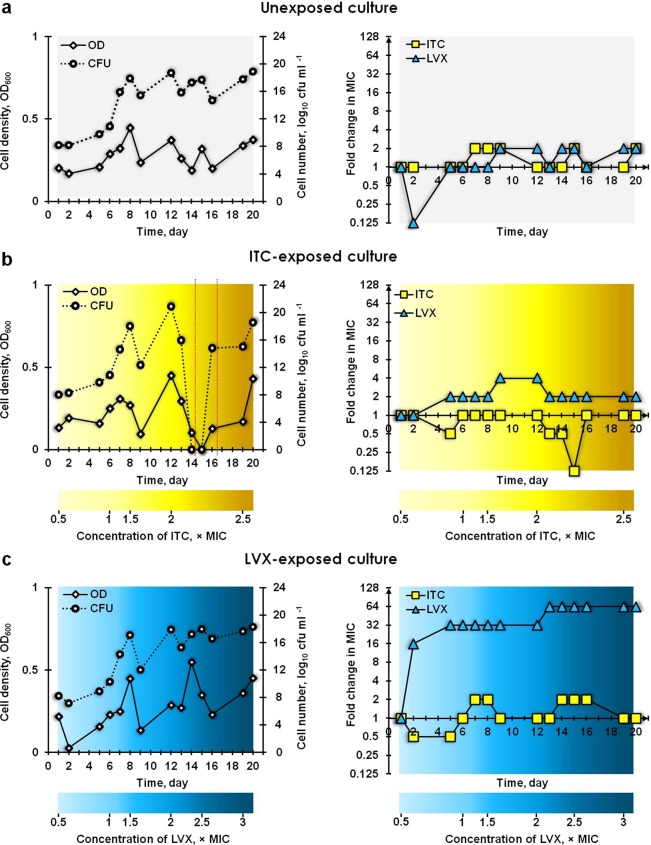
The growth and MIC profiles of three cultures are summarized in Fig. 1. The cell density and the number of cells fluctuated in the chemostat without added selection pressure (Fig. 1a). Such oscillations can be due to stochastic experimental measures or can be attributed to population-wide die-offs and recoveries29. The MIC values of ITC and LVX towards this unexposed continuous culture did not alter substantially between the first and last day (day 20) of cultivation (Fig. 1a; Supplementary Table S1). This indicated that the culture, which was not exposed to any antimicrobial agent, did not develop resistance toward the ITC and LVX during the 20 days of evolution.
Figure 1.
Growth and MIC profiles of continuous cultures of E. coli ATCC 25922: (a) control unexposed, exposed to (b) ITC and exposed to (c) LVX. The optical density (OD; 600 nm), viable cell counts (colony forming units; cfu ml−1) and minimum inhibitory concentrations (MICs; µg ml−1) of ITC and LVX were measured daily for parallel populations evolving under no antimicrobial selection, selection with ITC and LVX. On the last day, the concentrations of ITC and LVX added to the medium reservoirs corresponded to 2.5× and 3× their original MICs, respectively. The fold change in MIC was the ratio of the MIC obtained on the particular sampling day to the MIC recorded on day 1. The area delineated with red dotted lines represent the batch state of ITC-exposed culture.
The cell density and cell count measurements of the ITC-exposed culture (Fig. 1b) indicated that, despite the administration of a media containing 0.5 and 1 × MIC of ITC into the chemostat vessel, the E. coli ATCC 25922 grew exponentially up to day 8. The E. coli population declined in response to a 1.5 × MIC supplementation of ITC on day 8, but resumed growth during the following days. Subsequently, a 2 × MIC of ITC was applied into the vessel on day 12, which caused growth to abruptly cease – the culture density decreased progressively reaching 0 on day 15, while no viable cells were detectable on either day 14 or 15. On day 15, the feeding of the ITC-exposed culture was paused overnight. Following this, the culture resumed growth, possibly as a result of i) physiologically antimicrobial-refractory wall-adhering survivors reseeding the planktonic population and re-establishing a population which is as sensitive to antimicrobials as a naïve planktonic population26; and/or ii) physiologically antimicrobial-refractory non-dividing or slowly-dividing persisters re-growing a new population as sensitive to antimicrobials as naïve bacteria30. During the batch state (the sector delineated with red dotted lines; Fig. 1b), the ITC-exposed culture further revived to the density of the antimicrobial-free culture due to the cell-associated decline in the effective concentration of ITC in the culture26. As the 2 × MIC concentration of ITC waned, the bacterial population recovered after the preceding clearance. However, the recovered population was not as sensitive towards replenished 2 × MIC dose of ITC compared to the population preceding the clearance, as it did not encounter gradually increasing ITC stress over 13 days. In consequence, the culture outlived the 2 × MIC of ITC. Regarding the MIC profile, there was no increase in the ITC MIC in samples tested throughout the culture experiment, despite prolonged exposure to ITC (Fig. 1b; Supplementary Table S1). However, a decrease in MIC was noted on days 13, 14 and 15 compared to the samples tested on days 9 and 12, which may have been an artefact created as a result of culture decline and low starting inoculum density for MIC determinations. No increase was recorded either in the MIC of LVX in the ITC-exposed culture, although 4-fold fluctuations were noted on days 9 and 12. These observations indicated that exposure to ITC did not induce resistance to it, nor did it induce cross-resistance to LVX. In addition, ITC at a concentration of 2 × MIC was able to eliminate the planktonic bacterial population despite 13 days of prior exposure to ITC (days 14, 15; Fig. 1b).
On day 1, feeding the E. coli ATCC 25922 culture with medium containing 0.5 × MIC of LVX, inhibited the growth of the culture and had also triggered a 16-fold increase in LVX MIC by day 2 (Fig. 1c; Supplementary Table S1), indicating a rapid adaptation of the culture to LVX. Subsequently, the culture continued to grow exponentially and the MIC against LVX increased further (32-fold that of the inoculum by day 5), indicating that the culture was being enriched with more resistant variants. The subsequently applied stepwise challenge with LVX caused the culture density and cell number to fluctuate in a saw-tooth pattern – the introduction of a 2 × MIC LVX selected for higher resistance, leading to a 64-fold increase in LVX MIC by day 13 of culture. The MIC of ITC towards the LVX-exposed culture did not alter substantially during the experiment (Fig. 1c; Supplementary Table S1). These results indicated that a stepwise challenge of an E. coli ATCC 25922 culture, with increasing concentrations of LVX, rapidly selected for LVX-resistant phenotypes, failed to eliminate the planktonic population and did not induce cross-resistance against ITC.
Genotypic profile of chemostat-derived cultures
We performed whole-genome sequencing on samples of the planktonic polymorphic populations isolated over the course of the chemostat selection experiments and determined the allelic frequencies of all mutations that reached at least 5% frequencies within an individual population. We sequenced, using the Illumina platform, the genomes of mixed populations from unexposed chemostat culture from day 1 (start), day 14 (intermediate) and day 20 (end), ITC-exposed culture and LVX-exposed culture on days 14 and 20, and then screened for mutations using the BRESEQ pipeline. Our principal focus was on data obtained from thawed cultures (denoted TC), but we also sequenced revived cultures (denoted RC) for quality assurance. Mutations identified as being shared among thawed and revived cultures are available as Venn diagrams in Supplementary Fig. S1.
The total number and the type of genetic alterations identified in the chemostat-derived thawed cultures are shown in Fig. 2, mutations identified in thawed cultures at final day of evolution is presented in Fig. 3, while the detailed information on predicted mutations is provided in Supplementary Table S2. BRESEQ detected several large insertions and amplifications in the samples, but as all occurred in repeat regions, they were not considered in this analysis to avoid any concern about inaccurate short-read alignment. The number of total mutations in all treatment groups increased over time, reaching the highest totals in the LVX and ITC-exposed day 20 populations (Fig. 2). The counts of nonsynonymous substitutions also increased over time in all the test populations. This is important in that it shows the accumulation of random mutations with chemostat experiment.
Figure 2.
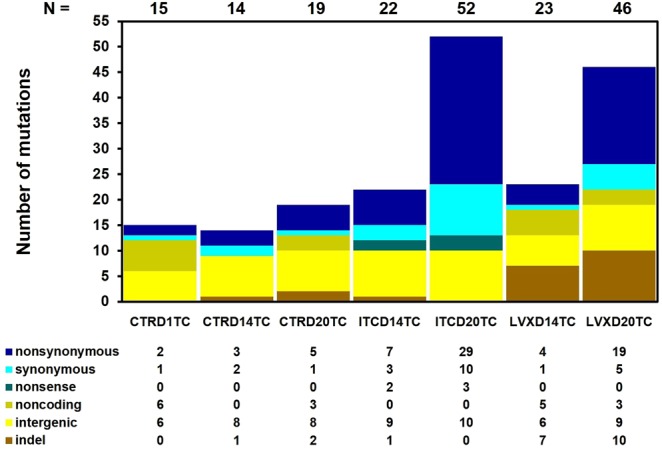
Mutational spectrum of chemostat-derived E. coli ATCC 25922 polymorphic population samples. “Metagenomes” of selection-free (CTR day 1, 14, 20), ITC-exposed (day 14 and 20) and LVX-exposed (day 14 and 20) thawed cultures were sequenced using the Illumina platform and polymorphisms were identified by BRESEQ. The total number of mutations (N) in each culture is shown above each column. The types of mutations present in each culture are presented below the columns. Base substitutions are presented as nonsynonymous (a point mutation altering the amino acid sequence of a protein), synonymous (a point mutation not altering the amino acid), nonsense (a point mutation resulting in a premature termination codon), noncoding (a mutation in noncoding RNA genes) and intergenic mutations; indels are deletions and insertions of ≤20 nt size.
Figure 3.
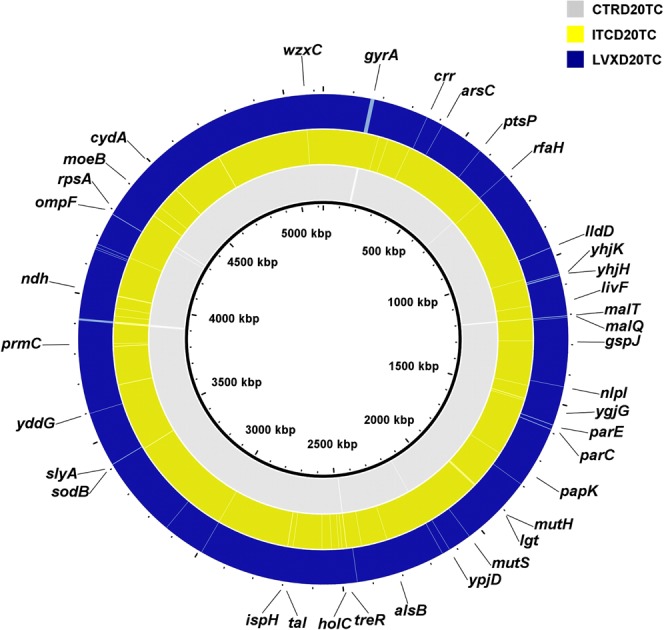
Mutations identified in chemostat-derived E. coli ATCC 25922 polymorphic populations. Genome visualization and comparison of final day unexposed, ITC-exposed and LVX-exposed populations to E. coli reference genome (CP009072) was performed using BRIG. From inner to outer rings: reference genome, unexposed thawed culture at day 20 (CTRD20TC), ITC-exposed thawed culture at day 20 (ITCD20TC) and LVX-exposed thawed culture at day 20 (LVXD20TC). The gaps shown indicate where the sequence data from chemostat populations differ from the reference sequence.
The inoculum population, which was started from a single isogenic colony and was grown in a batch mode for 24 h (CTRD1TC) already bore a collection of mutations by comparison with the reference E. coli ATCC 25922 isolate (Fig. 2; Supplementary Table S2). The majority of these mutations occurred in noncoding (6/14), or intergenic regions (6/14). Most interestingly, two nonsynonymous single nucleotide polymorphisms (SNPs) were also observed. Notably, the CTRD1TC population was already polymorphic containing subpopulations with a common LVX conferring mutation at position 87 for the GyrA subunit of DNA gyrase. The same codon in gyrA was a subject to two mutations: D87G at 64.3% and D87H at 31.8% frequencies (Supplementary Table S2). These two mutations were also present in the unexposed population on days 14 and 20, however, their frequencies fluctuate, and we cannot predict whether the mutations would have ultimately been lost or retained. Similarly, the population revived from the unexposed day 1 culture (CTRD1RC) contained subpopulations bearing repeated gyrA D87G and D87H mutations at relatively similar frequencies (Supplementary Table S2). In any case, according to the MIC profile, the unexposed polymorphic population did not display or develop resistance towards LVX throughout the culture history (Fig. 1a; Supplementary Table S1). In addition, these two nonsynonymous gyrA D87 mutations were not present in the ITC and LVX-exposed populations on days 14 and 20. The unexposed culture on days 14 and 20 (CTRD14TC and CTRD20TC) had a slightly different set of mutations by comparison with day 1. Of interest, perhaps, are the nonsynonymous A221P substitution in malT gene (MalT, the transcriptional activator of the maltose regulon; Fig. 3; Supplementary Table S2), 6 nt deletion in treR coding (99–104/948 nt) region (TreR, the repressor of operons involved in trehalose transport and degradation under osmotic stress; Fig. 3; Supplementary Table S2) and an A → G substitution in the intergenic region between the dicarboxylate symporter family protein/inner membrane protein Alx (Supplementary Table S2). These changes were present only in the CTRD14 and CTRD20 samples (in both thawed and revived cultures), at >15% frequencies.
Although the ITC-exposed culture did not show decreased susceptibility towards ITC or LVX, we sought genomic alterations that could potentially contribute to the development of ITC resistance, or antibiotic cross-resistance. We examined the mutation list (Supplementary Table S2) of ITC-exposed populations to identify genetic determinants, which may represent possible mutational targets in this evolved population. In most cases, however, we do not know if a particular mutation inactivates, activates, or otherwise acts on the activity of the gene product. The E. coli ATCC 25922 population, which was under selection pressure through exposure to ITC, accumulated a higher number of nonsynonymous and synonymous SNPs over time compared to the other cultures (Fig. 2). The frequencies at which these mutations occur in the ITC-exposed populations are, however, generally low (Supplementary Table S2). Distinctively, the ITC-exposed populations contained subpopulations, which had nonsense mutations in genes, such as the PTS system mannose/fructose/sorbose IID component family protein gene (W264* change in all ITC-exposed populations at 7 to 12% frequencies; Fig. 3; Supplementary Table S2), or the conserved hypothetical protein under DR76_2342 locus tag gene (W105* change in ITCD20TC and ITCD20RC populations at 7.7% and 5.8% frequencies, respectively; Fig. 3; Supplementary Table S2), or in the DNA mismatch repair endonuclease MutH gene (E148* change in ITCD14TC at 9.5%, in ITCD144C at 8.3% and ITCD20TC at 33.2% frequencies, respectively; Fig. 3; Supplementary Table S2). The ITCD20TC and ITCD20RC populations harboured a small proportion of subpopulations with an additional W108R mutH substitution (at 7.9% and 6.2% frequencies, respectively; Supplementary Table S2). MutH is the endonuclease of the DNA methyl-directed mismatch-repair (MMR) pathway, which is a pathway critical for the avoidance of mutations and maintenance of replicative fidelity. MMR defective bacteria have reduced ability to repair DNA damage, and are thus more prone to develop and accumulate mutations, including those conferring antibiotic resistance31. These mutations only existed in a small fraction of the population, however, and it is unknown whether finally they would become fixed in a population.
The key mechanisms of LVX resistance are chromosomal mutations that alter the A and B subunits of the target proteins, DNA gyrase (encoded by gyrA and gyrB) and topoisomerase IV (encoded by parC and parE); decreased intracellular drug accumulation due to increased drug efflux (mutations in acrRAB, marRAB and soxSR operons)32; or changes in outer membrane proteins (such as OmpF, OmpC, OmpD and OmpA)33. In our study, the population that was evolved under LVX selection carried a point mutation in the gyrA gene at a frequency of 100% on days 14 and 20 (Fig. 3; Supplementary Table S2). The nonsynonymous G to A substitution resulted in a S83L amino acid change, which is the most frequent mutation observed in quinolone-resistant E. coli34. This was a unique mutation, which occurred only in LVX-exposed whole populations (including the revived cultures). In addition, all the LVX-exposed populations uniquely possessed a parC G78D nonsynonymous substitution (LVXD14TC at 12.7%, LVXD14RC at 21.4%, LVXD20TC at 18.2% and LVXD20RC at 13.3% frequencies; Supplementary Table S2). The G78D change was an amino acid position (amino acid positions numbered according to E. coli) also known to be associated with LVX resistance in both Gram-positive and Gram-negative bacteria, although reports on this change are less frequent than changes in S80 or E84 of parC35. LVXD20TC also had other mutations occurring in the genes specific to quinolone resistance, such as nonsynonymous A426V, R432S substitutions and a 3 nt insertion in coding (1373/1893 nt) region of parE, as well as a 4 nt insertion in the coding (34/1089 nt) region of ompF gene (Fig. 3; Supplementary Table S2). The role that reduced expression, or the complete absence, of the OmpF porin plays in conferring fluoroquinolone resistance has been reported previously36,37. Our results indicated that decreased susceptibility and resistance to LVX in the LVX-exposed polymorphic culture were associated with multiple mutations in the gyrA, parC, parE and ompF genes. It is worth noting that LVXD20TC had a F36C nonsynonymous change in mutS gene (9.2%; Fig. 3; Supplementary Table S2). MutS is another protein of the MMR system, which recognizes and binds to mispaired nucleotides and enables the further action of the MMR machinery. It was shown that, as a result of mutS F36A substitution, MutS failed to recognize the mismatch site thus failing to initiate the repair38. In addition, LVXD20TC had elevated numbers of mutations (Fig. 2), perhaps, as a result of this mutS F36C modification. The LVX-exposed populations also had an increased number of insertions and deletions (indels). Interestingly, LVX-subjected populations also had several nucleotide insertions in the yhjK (LVXD20TC and LVXD20RC) and yhjH (LVXD14TC, LVXD20TC and LVXD20RC) genes (Fig. 3; Supplementary Table S2). YhjK is a predicted c-di-GMP phosphodiesterase (PDE), while YhjH is a c-di-GMP PDE involved in regulation of the switch from flagellar motility to sessile behaviour. Deletion of the yhjH gene and an insertion in yhjH gene was shown to impair the swimming motility of E. coli39 and contribute to the transition from planktonic to biofilm lifestyle. The 6 nt deletion in coding region (727–732/2706 nt) of malT gene, is also noteworthy (Fig. 3; Supplementary Table S2), perhaps as a modification to facilitate the adaptation of E. coli to nutrient limiting environment in chemostat40.
Discussion
The potential for the development of resistance to an antimicrobial is an important factor in determining its suitability for therapeutic or environmental applications. We have previously shown that the repeated exposure of E. coli, P. aeruginosa, S. aureus and methicillin-resistant S. aureus to sub-inhibitory levels of ITC during serial passage of batch cultures did not generate ITC resistance3. In the present study, we used selection/enrichment chemostat culture to generate mutants with decreased susceptibility to ITC, coupled with WGS to identify those mutations underpinning resistance. Antimicrobial resistance may develop and increase via the gradual accumulation of multiple mutations41. To study this, we gradually challenged the evolving population in the chemostat with antimicrobials. Three separate enrichment continuous cultures of drug-susceptible E. coli ATCC 25922 were operated for 20 days to select for ITC and LVX-resistant mutants, which were then compared with selection-free culture. We measured the resistance using the MIC metric – resistance occurs when the population is able to grow in the environment of higher concentration of an antimicrobial25.
Planktonic chemostat populations grown without added selection pressure and with ITC selection pressure yielded populations with no altered susceptibility towards either ITC or LVX (Fig. 1a, b; Supplementary Table S1). These results indicate that ITC did not cause the emergence of resistance to itself, or the emergence of cross-resistance to the antibiotic LVX. However, attention should be given to the fact that there can be a possibility of population-level variability in terms of resistance, as only a minute fraction of the population was sampled. Further, ITC at 2 × MIC dose was able to eradicate a 1020 cfu ml−1 load of bacteria (Fig. 1b), a concentration much higher, for example, than the bacterial concentration in wound fluid allowing the progression of wound healing (<106 cfu ml−1)42. This signifies that ITC may be suitable to disinfect heavily contaminated sites without risk of the facile emergence of resistance, even at very high cell densities. Nevertheless, we should note here that while ITC could eliminate a “weakened” population which was under the gradually increasing ITC stress over a long time period, it could not eliminate the resurrected ITC stress-“rested” population at the finish of this evolution experiment (Fig. 1b). On the other hand, the derivative chemostat culture, grown under LVX selection pressure, rapidly gained resistance towards LVX (MIC of LVX increased 64-fold by day 20), but no cross-resistance was observed towards ITC (Fig. 1c; Supplementary Table S1). LVX also failed to eradicate the bioburden at a 3 × MIC dose. Complete genome sequencing of these populations provided information about the mechanisms underlying the acquired phenotypes.
ITC is a biocidal mixture of reactive oxygen and iodine species. These reactive species can impair diversity of cellular macromolecules in bacteria and thus trigger protective oxidative stress responses towards these stressors. Moreover, these species alone can induce resistance-promoting responses in bacteria. The oxidative agents also promote the expression of multidrug efflux systems, likely to mitigate the impact of these stressors. Likewise, antioxidant mechanisms can be involved in response to exposure of ROS-based antimicrobials. Hence, oxidative stress responses may contribute to antimicrobial resistance in several ways43. Poole has reviewed, in detail, the evidence supporting links between the oxidative stress response and antimicrobial resistance in bacteria, and summarized the mechanisms of oxidative stress-induced resistance43. These include, amongst others, the redox-responsive regulator SoxSR of multidrug efflux system AcrAB-TolC in E. coli; e.g. reactive agents can cause direct mutations leading to constitutive soxS expression and elevated acrAB expression and, thus, antimicrobial resistance. We examined the genome sequences of ITC-exposed populations (Supplementary Table S2) for significant (nonsynonymous, nonsense) nucleotide variations, including those involving amino acid change or amino acid sequence termination, and compared with stress-inducible antimicrobial resistance mechanisms (Table 1 from reference)43. We could not identify any determinants, which could have contributed to oxidative (or stress in general) response-mediated resistance of the ITC-exposed culture to ITC and/or LVX.
On the other hand, cross-resistance between differing antimicrobial agents may arise when they act on the same target, access to their specific targets by a common way, or initiate the cell death by a common pathway. As a result, resistance to one antibacterial agent co-occurs with resistance to another agent15,44. LVX is a member of the fluoroquinolone class of antibiotics with broad antimicrobial activity against clinically relevant bacteria causing respiratory, skin and genitourinary tract infections. Following regulatory approval in the mid-1990s, LVX became one of the most widely prescribed antibiotics in the world45. The heavy prescription of LVX has led to increased resistance. LVX resistance is thought to be a result of gradually acquired changes, and several mutations are required to produce a high level of resistance46. In Gram-negative bacteria, resistance mutations firstly occur in gyrA, with “hot spots” for mutation at amino acid positions 83 and 87. Once this initial mutation has reduced the susceptibility of resistant bacteria to LVX, additional mutations in target genes parC, parE, or else, mutations causing reduced drug accumulation, can further augment the resistance47. Indeed, in our study, the day 14 LVX-exposed population was entirely composed of gyrA S83L mutants (occurrence 100%), while a proportion of the population also carried the parC G78D mutation (Supplementary Table S2). By day 20, the LVX-exposed population was further enriched with parE and ompF mutants (Fig. 3; Supplementary Table S2). These results indicate that, the acquisition of high level LVX resistance (e.g. a 64-fold increase in MIC) was related to mutations in genes encoding the three protein targets and mutations in uptake protein. In general, combining chemostat and WGS was a productive approach to generate, follow and understand the evolution of LVX resistance in E. coli ATCC 25922. ITC-exposed populations, by contrast, did not possess any common mutations conferring LVX resistance (Supplementary Table S2). In populations with the size described in these experiments, spontaneous mutations are virtually certain to occur at any position in the genome, including in the target genes of LVX. The fact that these mutations were not selected under ITC-selection pressure means that they were not (sufficiently) beneficial under ITC selection.
The MMR pathway safeguards the genome by correcting the base mismatches arising as a result of replication errors48. Recent studies suggest that MMR may also be important in the response to oxidative DNA damage49. Defects in MMR result in increased rates of mutations in organisms ranging from bacteria to humans. MMR-defective bacteria are considered to be “hypermutable” and to express a “mutator” phenotype (“mutator” strains). Mutator phenotypes, in general, result from alterations in genes coding for DNA repair enzymes (mutS, mutL, mutH and mutU). The genes affected in the studied mutator strains are mutS, mutL, mutH and mutU in decreasing order50. The E. coli ATCC 25922 population, which was grown under ITC selection, harboured a nonsense mutation in mutH gene on day 14 (detected in thawed culture (ITCD14TC) at a frequency of 9.5% and in 4 °C culture (ITCD144C) at a frequency of 8.3%) and on day 20 [detected at a frequency of 33.2% in thawed culture (ITCD20TC), but not in a revived culture (ITCD20RC)]. The LVXD20TC population also harboured the F36C nonsynonymous change in mutS gene at a frequency of 9.2% (Fig. 3; Supplementary Table S2). The ITCD20TC and LVXD20TC cultures, indeed, had an elevated number of mutations by comparison to the other samples (Fig. 2); however, we cannot definitively ascribe this to mutH or mutS mutations. These mutations only represented a small fraction of the total; but to be able to produce a mutator phenotype they must be genetically dominant. At present, there are mixed views about the link between hypermutation and emergence of antimicrobial resistance50. Schaaff and co-workers (2002) found that vancomycin resistance could be reached much more quickly in a mutator background (mutS knockout mutant) versus nonmutator (5 versus 19 passages, respectively)51. However, in our study during the timeline of the selection experiments, the ITC-exposed population did not develop elevated MIC values for ITC and LVX (Fig. 1b; Supplementary Table S1), while the LVX-exposed population showed elevated MIC values towards LVX (which was due to other primary mutations rather than mutS), but not toward ITC. These mutations may have appeared too late in the selection to become common in the populations. On the contrary, O’Neill and co-authors (2002) have concluded that stable hypermutation or transient increases in mutation frequency were probably not involved in the emergence of antibiotic resistance in S. aureus, despite the fact that a mutS-inactive S. aureus could have a 78-fold higher mutation frequency compared to the wild type strain52.
The overall conclusions of this study are that we were unable to generate de novo resistance to ITC in a drug-sensitive E. coli ATCC 25922, despite 20 days of selective evolution, and, thus, we could not identify mutations that would have yielded ITC resistance. Mutations in the gene of DNA mismatch repair endonuclease MutH may be implicated in elevated mutation numbers and result in mutator phenotype, thus contributing to antimicrobial resistance; however, these mutations did not become prominent in the evolved ITC-exposed population. By contrast, resistance to LVX quickly built up in the E. coli ATCC 25922 culture exposed to increasing concentrations of LVX, apparently driven by mutations in the genes coding for the target proteins (GyrA, ParC, ParE) and reduced drug accumulation (OmpF). Judging from the example of LVX, combining experimental evolution and whole-genome sequencing can provide a deeper understanding of the principles of how resistance emerges under different selective pressures.
The lack of emergence of facile resistance to ITC in E. coli ATCC 25922, and the lack of cross-resistance to LVX are encouraging with respect to the potential for applications of ITC as a biocidal agent in a variety of settings. An understanding of the possible mechanisms, if any, of ITC resistance, however, requires further longer-term investigations. The dynamics and determinants of potential ITC resistance should also be tested in numerous parallel cultures of a range of other bacteria, and the potential for cross-resistance studied on a range of important antibiotics.
Materials and Methods
Bacteria, growth media and antimicrobials
The parental strain used in this study was the drug-susceptible E. coli ATCC 25922. All the other cultures were derivatives generated by means of chemostat culturing. The lyophilized strain was resuscitated on fresh Lennox agar (Sigma-Aldrich) and the growth from a single colony was transferred into lysogeny broth (LB; Sigma-Aldrich). This single colony suspension was used to inoculate three parallel chemostat vessels in order to ensure that the parental strain utilized throughout the continuous culturing was genetically homogeneous at the start. The continuous cultures were grown in quarter-strength LB medium to ensure nutrient limitation. The developed cultures were stored at −20 °C in 20% glycerol.
LVX (Fluka) stock solution was prepared from powder in sterile deionized water (dH2O) and was filter-sterilized; ITC stock solution was prepared as previously described3. Briefly, to prepare 1% stock solution of ITC, hydrogen peroxide, potassium iodide and potassium thiocyanate aqueous solutions were mixed at 1:1:1 (v/v/v) ratio. Stock solutions of ITC and LVX were kept at 4 °C and reused for the duration of the experiments.
Chemostat configuration and operation
A schematic diagram of the chemostat setup is illustrated in Supplementary Fig. S2 and was based on a design published previously23. Continuous cultures were performed in glass culture vessels (200 × 45 mm) with 100 ± 2 ml working volume equipped with the medium break, by which the media and the air were supplied. The oxygen was delivered into the vessel by pumping filter-sterilized air at a rate of ca. 100 ml min−1. The cultures were mixed by bubbling with sterile air. Medium flow from the reservoir to the culture vessel was controlled by a peristaltic pump (Watson-Marlow 505U). The working volume of the chemostat was fixed by the height of the central overflow tube. Fresh feed was introduced at constant flow rate and culture waste was removed at the same rate according to hydraulic principles. Chemostats were operated at a dilution rate of 0.4 hr−1. This rate of dilution will result in a generation time of 1.73 h per generation53. Samples were taken using a sterile syringe attached to the sampling port. Chemostat vessels were submerged in a temperature-controlled water bath and operated at 37 °C.
Three individual vessels (designated as selection-free, ITC and LVX-selection chemostats) were brought to their working volumes and inoculated from a single colony suspension of E. coli to obtain a final cell concentration of 106 cfu ml−1. Cultivations were started in batch mode for 24 h to reach culture saturation, and then switched to chemostat mode by initiating the feeding. Meantime, the MICs of ITC and LVX toward this isogenic pre-culture were determined, and these values were used to infer the initial antimicrobial concentrations to be added in the chemostat feeds. Subsequently, two media feeds were supplemented separately with ITC and LVX at their 0.5 × MIC (7.81 and 0.024 µg ml−1, respectively). Antimicrobial concentrations in the media feed were increased stepwise by 0.5 × MIC increments when OD600 of ITC and LVX-exposed cultures reached to the value recorded prior the addition of the antimicrobial. Each chemostat culture was continuously operated for 20 days (282 generations). Increases in the feed concentrations of ITC and LVX to 2.5 × MIC and 3 × MIC, respectively, were made in the respective feeds during the chemostat course. Daily 2 ml samples were taken from chemostat cultures and used to test for a variety of parameters, such as OD600 readings, viable cell counts, broth MIC determinations, culture preservation and WGS of mixed planktonic population samples.
Culture density, cell number and minimum inhibitory concentrations
The cell density of 1 ml chemostat-derived culture was measured using a spectrophotometer (Spectronic 20 Genesys) at a wavelength of 600 nm. Viable cell number was determined by colony counts on antibiotic-free agar plates after appropriate dilutions in phosphate-buffered saline. MICs for LVX and ITC in quarter-strength LB were measured by following growth in 96-well plates, as described previously3. In short, 3 inocula from a daily chemostat sample were prepared by taking aliquots from a planktonic population sample and diluting to cell density of 106 colony forming units per ml (cfu ml−1) in three separate tubes. These 3 inocula at 5 × 105 cfu ml−1 final density were challenged with ITC and LVX at serial 2-fold dilutions in 96-well plates. Microplates were incubated in culture chamber at 37 °C over 24 h. The MIC was defined as the lowest concentration of antimicrobial agent preventing the appearance of turbidity. The median of the triplicate measurements were reported as an MIC. For each of the chemostat cultures, the MIC change was calculated as the ratio of the MIC at sampling day to the MIC of starting day. Chemostat-derived E. coli cultures were considered to be resistant to ITC and/or LVX when a sustained >4-fold MIC increase from initial MIC was noted54. The actual MIC values from triplicate measurements of isogenic pre-culture at day 0, chemostat introduced cultures at day 1 and unexposed, ITC-exposed and LVX-exposed cultures throughout 20 days are presented in Supplementary Table S1. The observed differences between the MICs of isogenic pre-culture at day 0 and chemostat introduced cultures at day 1 might exist due to environmental conditions.
Whole-genome sequencing of chemostat-derived polymorphic populations and mutation calling
The focal populations for WGS were unexposed chemostat culture from day 1, 14 and 20, ITC-exposed culture from day 14 and 20, and LVX-exposed culture from day 14 and 20. Aliquots (1 ml) of frozen glycerol stocks of planktonic polymorphic population samples (and not single colonies) collected from three chemostat cultures were thawed and immediately pelleted for genomic DNA isolation [further referred as thawed culture (TC)]. The samples represented approximately 1% of the total population in the chemostat. Likewise, a small aliquot was used to revive the frozen samples via overnight growth in 1 ml LB at 37 °C and pelleted for gDNA extraction [further referred as revived culture (RC)]. We could not revive day 14 ITC-exposed culture as a result of the total clearance of bacteria by ITC (Fig. 1b), and the leftover of the ITC-exposed day 14 culture earlier used for OD, CFU, MIC measurements and later kept at 4 °C was used for gDNA extraction and downstream bioinformatics analysis instead [further indicated as 4 °C culture (4 C)]. gDNA was extracted using QIAamp DNA Mini Kit (Qiagen), DNA concentration was quantified by Qubit Fluorometer (Life Technologies) and DNA integration was assessed by 0.75% agarose gel electrophoresis.
The genomes were sequenced using the Illumina-based platform of the MicrobesNG service (http://www.microbesng.uk) to generate 2 × 250 bp paired-end reads. For fourteen sequenced samples (6 control, 4 ITC and 4 LVX), the average coverage (over the 5130767 bp of the reference genome) ranged from 53–200×. Reads were adapter trimmed using Trimmomatic and the quality assessed using in-house scripts combined with SAMtools, BEDtools, and BWA-MEM via MicrobesNG. These reads were then aligned onto the E. coli ATCC 25922 reference genome (accession no. CP009072) and putative mutations were identified with the BRESEQ re-sequencing pipeline. This pipeline can detect SNPs, small indels (≤20 nt), as well as structural variations, such as large indels, duplications, mobile element insertions or other rearrangements, in polymorphic population samples55–58. As the samples were “metagenomic”, that is to say, a mixture of DNA from different individuals, BRESEQ was run at polymorphism mode with default parameters. BRESEQ uses a Kolmogorov-Smirnov test for bias in the qualities of bases supporting the new variant, and Fisher’s exact test for bias in the strands of reads supporting the new variant, in both cases rejecting the mutation if it was biased at a p-value = 0.05 significance level. The variation in reads at a given position was classified as a true population polymorphism (as opposed to sequencing error) when ≥5% of the reads differed from the others for a given allele.
The initial lists of predicted mutations by BRESEQ were manually edited. Mutations in repeat regions may not be fully predicted due to limitations of short-read DNA sequencing data. For that reason, we identified the positions of repeat regions in the reference genome using NUCmer (alignment of multiple closely related nucleotide sequences in MUMmer software) and BLASTN (alignment of somewhat similar sequences in National Center for Biotechnology Information website) to perform self-comparison. SNPs and small indel mutations, which had a BLASTN match in several locations, as well as large indels within repetitive regions or genes with multiple copy numbers, were ignored to avoid any concern about inaccurate read alignment57,59. We also discarded the mutations that were present to 100% in all samples (which were likely to be the difference between the reference E. coli ATCC 25922 and the starting isolate), considering only the mutations that accumulated throughout the selection experiment. A circular genomic map for the genomes of unexposed, ITC-exposed and LVX-exposed final day (day 20) thawed cultures were plotted using the BLAST Ring Image Generator (BRIG) software60.
Supplementary information
Acknowledgements
This work was funded by Irish Research Council Government of Ireland Postgraduate Scholarship Scheme (GOIPG/2013/584). We would like to acknowledge MicrobesNG (http://www.microbesng.uk) team for excellent genome sequencing service. We also wish to sincerely thank Dr. Leighton Pritchard (The James Hutton Institute, Dundee, Scotland) and Dr. Fiona Brennan (Teagasc, Johnstown Castle, Co. Wexford, Ireland) for advice on genome sequencing and for valuable scientific discussion. Huge thanks to Nick Waters (NUI, Galway) for his assistance in bioinformatics.
Author Contributions
L.T. designed and completed the experiments, analysed the data, performed the bioinformatics analyses, wrote and reviewed the manuscript. L.T. and G.T.A.F. built the chemostat system. G.T.A.F. and V.OF. contributed to the conception and design of the experimental work and the interpretation of the results. L.T., V.OF., R.F. and G.T.A.F. critically reviewed and edited the manuscript.
Data Availability
Raw sequencing reads from thawed and revived samples have been deposited in the European Nucleotide Archive’s Sequence Read Archive (Accession No. PRJEB27402 and PRJEB27390, respectively). The authors declare that all other relevant data generated or analysed during this study are included in this published article and its Supplementary Information files.
Competing Interests
Dr. Friel is an employee of Westway Health and owns shares in the company. Prof. O’Flaherty also owns shares in the company. Dr. Tonoyan and Dr. Fleming declare no potential conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Lilit Tonoyan, Email: lilit.tonoyan@gmail.com.
Vincent O’Flaherty, Email: vincent.oflaherty@nuigalway.ie.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-019-38925-9.
References
- 1.Walsh C. Molecular mechanisms that confer antibacterial drug resistance. Nature. 2000;406:775–781. doi: 10.1038/35021219. [DOI] [PubMed] [Google Scholar]
- 2.FDA Safety and effectiveness of consumer antiseptics; topical antimicrobial drug products for over-the-counter human use; proposed amendment of the tentative final monograph; reopening of administrative record. Federal Register. 2013;78:76444–76478. [Google Scholar]
- 3.Tonoyan, L., Fleming, G. T. A., Mc Cay, P. H., Friel, R. & O’Flaherty, V. Antibacterial potential of an antimicrobial agent inspired by peroxidase-catalyzed systems. Front Microbiol8, 10.3389/fmicb.2017.00680 (2017). [DOI] [PMC free article] [PubMed]
- 4.Tonoyan L, et al. In vitro comparative cytotoxicity study of a novel biocidal iodo-thiocyanate complex. Toxicol in Vitro. 2018;50:264–273. doi: 10.1016/j.tiv.2018.03.014. [DOI] [PubMed] [Google Scholar]
- 5.McDonnell G, Russell AD. Antiseptics and disinfectants: Activity, action, and resistance. Clin Microbiol Rev. 1999;12:147–179. doi: 10.1128/CMR.12.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rossmoore HW. Handbook of Biocide and Preservative Use. Netherlands: Springer; 2012. [Google Scholar]
- 7.Fraise AP. Biocide abuse and antimicrobial resistance – a cause for concern? J Antimicrob Chemother. 2002;49:11–12. doi: 10.1093/jac/49.1.11. [DOI] [PubMed] [Google Scholar]
- 8.Chapman JS. Biocide resistance mechanisms. Int Biodeterior Biodegradation. 2003;51:133–138. doi: 10.1016/S0964-8305(02)00097-5. [DOI] [Google Scholar]
- 9.Maillard JY. Bacterial target sites for biocide action. J Appl Microbiol. 2002;92:16S–27S. doi: 10.1046/j.1365-2672.92.5s1.3.x. [DOI] [PubMed] [Google Scholar]
- 10.Fraise, A. P., Lambert, P. A. & Maillard, J. Y. Russell, Hugo & Ayliffe’s Principles and Practice of Disinfection, Preservation & Sterilization. (Wiley, 2008).
- 11.Dwyer DJ, Kohanski MA, Collins JJ. Role of reactive oxygen species in antibiotic action and resistance. Curr Opin Microbiol. 2009;12:482–489. doi: 10.1016/j.mib.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Russell AD. Antibiotic and biocide resistance in bacteria: Introduction. J Appl Microbiol. 2002;92:1S–3S. doi: 10.1046/j.1365-2672.92.5s1.14.x. [DOI] [PubMed] [Google Scholar]
- 13.Price CTD, Singh VK, Jayaswal RK, Wilkinson BJ, Gustafson JE. Pine oil cleaner-resistant Staphylococcus aureus: Reduced susceptibility to vancomycin and oxacillin and involvement of sigb. Appl Environ Microbiol. 2002;68:5417–5421. doi: 10.1128/aem.68.11.5417-5421.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karatzas KAG, et al. Prolonged treatment of Salmonella enterica serovar Typhimurium with commercial disinfectants selects for multiple antibiotic resistance, increased efflux and reduced invasiveness. J Antimicrob Chemother. 2007;60:947–955. doi: 10.1093/jac/dkm314. [DOI] [PubMed] [Google Scholar]
- 15.Mc Cay PH, Ocampo-Sosa AA, Fleming GT. Effect of subinhibitory concentrations of benzalkonium chloride on the competitiveness of Pseudomonas aeruginosa grown in continuous culture. Microbiology. 2010;156:30–38. doi: 10.1099/mic.0.029751-0. [DOI] [PubMed] [Google Scholar]
- 16.Russell AD. Bacterial adaptation and resistance to antiseptics, disinfectants and preservatives is not a new phenomenon. J Hosp Infect. 2004;57:97–104. doi: 10.1016/j.jhin.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 17.Poole K. Mechanisms of bacterial biocide and antibiotic resistance. J Appl Microbiol. 2002;92:55S–64S. doi: 10.1046/j.1365-2672.92.5s1.8.x. [DOI] [PubMed] [Google Scholar]
- 18.Maillard JY. Antimicrobial biocides in the healthcare environment: Efficacy, usage, policies, and perceived problems. Ther Clin Risk Manag. 2005;1:307–320. [PMC free article] [PubMed] [Google Scholar]
- 19.Jansen G, Barbosa C, Schulenburg H. Experimental evolution as an efficient tool to dissect adaptive paths to antibiotic resistance. Drug Resist Updat. 2013;16:96–107. doi: 10.1016/j.drup.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 20.Fleming G, Dawson MT, Patching JW. The isolation of strains of Bacillus subtilis showing improved plasmid stability characteristics by means of selective chemostat culture. J Gen Microbiol. 1988;134:2095–2101. doi: 10.1099/00221287-134-8-2095. [DOI] [PubMed] [Google Scholar]
- 21.Ziv, N., Brandt, N. J. & Gresham, D. The use of chemostats in microbial systems biology. J Vis Exp, 10.3791/50168 (2013). [DOI] [PMC free article] [PubMed]
- 22.Ferenci T. Bacterial physiology, regulation and mutational adaptation in a chemostat environment. Adv microb physiol. 2008;53:169–229. doi: 10.1016/s0065-2911(07)53003-1. [DOI] [PubMed] [Google Scholar]
- 23.Fleming GT, McCarthy DM, Colombet N, Patching JW. The effect of levofloxacin concentration on the development and maintenance of antibiotic-resistant clones of Escherichia coli in chemostat culture. J Ind Microbiol Biotechnol. 2002;29:155–162. doi: 10.1038/sj.jim.7000295. [DOI] [PubMed] [Google Scholar]
- 24.Palmer AC, Kishony R. Understanding, predicting and manipulating the genotypic evolution of antibiotic resistance. Nat Rev Genet. 2013;14:243–248. doi: 10.1038/nrg3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lukačišinová M, Bollenbach T. Toward a quantitative understanding of antibiotic resistance evolution. Curr Opin Biotech. 2017;46:90–97. doi: 10.1016/j.copbio.2017.02.013. [DOI] [PubMed] [Google Scholar]
- 26.Udekwu KI, Levin BR. Staphylococcus aureus in continuous culture: A tool for the rational design of antibiotic treatment protocols. PLoS One. 2012;7:e38866. doi: 10.1371/journal.pone.0038866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Köser CU, Ellington MJ, Peacock SJ. Whole-genome sequencing to control antimicrobial resistance. Trends Genet. 2014;30:401–407. doi: 10.1016/j.tig.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dettman JR, et al. Evolutionary insight from whole-genome sequencing of experimentally evolved microbes. Mol Ecol. 2012;21:2058–2077. doi: 10.1111/j.1365-294X.2012.05484.x. [DOI] [PubMed] [Google Scholar]
- 29.Cornejo OE, Rozen DE, May RM, Levin BR. Oscillations in continuous culture populations of Streptococcus pneumoniae: Population dynamics and the evolution of clonal suicide. Proc R Soc B. 2009;276:999–1008. doi: 10.1098/rspb.2008.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 31.Woodford N, Ellington MJ. The emergence of antibiotic resistance by mutation. Clin Microbiol Infect. 2007;13:5–18. doi: 10.1111/j.1469-0691.2006.01492.x. [DOI] [PubMed] [Google Scholar]
- 32.Zayed AA-F, Essam TM, Hashem A-GM, El-Tayeb OM. ‘Supermutators’ found amongst highly levofloxacin-resistant E. coli isolates: A rapid protocol for the detection of mutation sites. Emerg Microbes Infect. 2015;4:e4. doi: 10.1038/emi.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Correia S, Poeta P, Hebraud M, Capelo JL, Igrejas G. Mechanisms of quinolone action and resistance: Where do we stand? J Med Microbiol. 2017;66:551–559. doi: 10.1099/jmm.0.000475. [DOI] [PubMed] [Google Scholar]
- 34.Ruiz J. Mechanisms of resistance to quinolones: Target alterations, decreased accumulation and DNA gyrase protection. J Antimicrob Chemother. 2003;51:1109–1117. doi: 10.1093/jac/dkg222. [DOI] [PubMed] [Google Scholar]
- 35.Weigel LM, Anderson GJ, Tenover FC. DNA gyrase and topoisomerase IV mutations associated with fluoroquinolone resistance in Proteus mirabilis. Antimicrob Agents Chemother. 2002;46:2582–2587. doi: 10.1128/AAC.46.8.2582-2587.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fàbrega A, Martin RG, Rosner JL, Tavio MM, Vila J. Constitutive SoxS expression in a fluoroquinolone-resistant strain with a truncated SoxR protein and identification of a new member of the marA-soxS-rob regulon, mdtG. Antimicrob Agents Chemother. 2010;54:1218–1225. doi: 10.1128/aac.00944-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tavío MDM, et al. Mechanisms involved in the development of resistance to fluoroquinolones in Escherichia coli isolates. J Antimicrob Chemother. 1999;44:735–742. doi: 10.1093/jac/44.6.735. [DOI] [PubMed] [Google Scholar]
- 38.Junop MS, Yang W, Funchain P, Clendenin W, Miller JH. In vitro and in vivo studies of MutS, MutL and MutH mutants: Correlation of mismatch repair and DNA recombination. DNA Repair. 2003;2:387–405. doi: 10.1016/S1568-7864(02)00245-8. [DOI] [PubMed] [Google Scholar]
- 39.Girgis HS, Liu Y, Ryu WS, Tavazoie S. A comprehensive genetic characterization of bacterial motility. PLoS Genet. 2007;3:1644–1660. doi: 10.1371/journal.pgen.0030154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Notley L, Ferenci T. Differential expression of mal genes under cAMP and endogenous inducer control in nutrient-stressed Escherichia coli. Mol Microbiol. 1995;16:121–129. doi: 10.1111/j.1365-2958.1995.tb02397.x. [DOI] [PubMed] [Google Scholar]
- 41.Toprak E, et al. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat Genet. 2012;44:101–105. doi: 10.1038/ng.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bowler PG, Duerden BI, Armstrong DG. Wound microbiology and associated approaches to wound management. Clin Microbiol Rev. 2001;14:244–269. doi: 10.1128/CMR.14.2.244-269.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poole K. Bacterial stress responses as determinants of antimicrobial resistance. J Antimicrob Chemother. 2012;67:2069–2089. doi: 10.1093/jac/dks196. [DOI] [PubMed] [Google Scholar]
- 44.Chapman JS. Disinfectant resistance mechanisms, cross-resistance, and co-resistance. Int Biodeterior Biodegradation. 2003;51:271–276. doi: 10.1016/S0964-8305(03)00044-1. [DOI] [Google Scholar]
- 45.Noel GJ. A review of levofloxacin for the treatment of bacterial infections. Clin Med Insights Ther. 2009;1:433–458. doi: 10.4137/CMT.S28. [DOI] [Google Scholar]
- 46.Fàbrega A, Madurga S, Giralt E, Vila J. Mechanism of action of and resistance to quinolones. Microb Biotechnol. 2009;2:40–61. doi: 10.1111/j.1751-7915.2008.00063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jacoby GA. Mechanisms of resistance to quinolones. Clin Infect Dis. 2005;41(Suppl 2):S120–126. doi: 10.1086/428052. [DOI] [PubMed] [Google Scholar]
- 48.Hsieh P, Yamane K. DNA mismatch repair: Molecular mechanism, cancer, and ageing. Mech Ageing Dev. 2008;129:391–407. doi: 10.1016/j.mad.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bridge G, Rashid S, Martin SA. DNA mismatch repair and oxidative DNA damage: Implications for cancer biology and treatment. Cancers. 2014;6:1597–1614. doi: 10.3390/cancers6031597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eliopoulos GM, Blázquez J. Hypermutation as a factor contributing to the acquisition of antimicrobial resistance. Clin Infect Dis. 2003;37:1201–1209. doi: 10.1086/378810. [DOI] [PubMed] [Google Scholar]
- 51.Schaaff F, Reipert A, Bierbaum G. An elevated mutation frequency favors development of vancomycin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46:3540–3548. doi: 10.1128/aac.46.11.3540-3548.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O’Neill AJ, Chopra I. Insertional inactivation of mutS in Staphylococcus aureus reveals potential for elevated mutation frequencies, although the prevalence of mutators in clinical isolates is low. J Antimicrob Chemother. 2002;50:161–169. doi: 10.1093/jac/dkf118. [DOI] [PubMed] [Google Scholar]
- 53.Dykhuizen DE, Hartl DL. Selection in chemostats. Microbiol Rev. 1983;47:150–168. doi: 10.1128/mr.47.2.150-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.D’Lima L, et al. No decrease in susceptibility to NVC-422 in multiple-passage studies with methicillin-resistant Staphylococcus aureus, S. aureus, Pseudomonas aeruginosa, and Escherichia coli. Antimicrob Agents Chemother. 2012;56:2753–2755. doi: 10.1128/aac.05985-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barrick JE, et al. Identifying structural variation in haploid microbial genomes from short-read resequencing data using breseq. BMC Genomics. 2014;15:1039. doi: 10.1186/1471-2164-15-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barrick JE, Lenski RE. Genome-wide mutational diversity in an evolving population of Escherichia coli. Cold Spring Harb Symp Quant Biol. 2009;74:119–129. doi: 10.1101/sqb.2009.74.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deatherage DE, Barrick JE. Identification of mutations in laboratory evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol. 2014;1151:165–188. doi: 10.1007/978-1-4939-0554-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deatherage, D. E., Traverse, C. C., Wolf, L. N. & Barrick, J. E. Detecting rare structural variation in evolving microbial populations from new sequence junctions using breseq. Front Genet5, 10.3389/fgene.2014.00468 (2014). [DOI] [PMC free article] [PubMed]
- 59.Tenaillon O, et al. Tempo and mode of genome evolution in a 50,000-generation experiment. Nature. 2016;536:165–170. doi: 10.1038/nature18959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alikhan N-F, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing reads from thawed and revived samples have been deposited in the European Nucleotide Archive’s Sequence Read Archive (Accession No. PRJEB27402 and PRJEB27390, respectively). The authors declare that all other relevant data generated or analysed during this study are included in this published article and its Supplementary Information files.