Indole is a degradation product of tryptophan that functions as a signaling molecule in many bacteria. This includes Vibrio cholerae, where indole was shown to regulate biofilm and type VI secretion in nontoxigenic environmental isolates.
KEYWORDS: cholera, indole, biofilms, virulence regulation
ABSTRACT
Indole is a degradation product of tryptophan that functions as a signaling molecule in many bacteria. This includes Vibrio cholerae, where indole was shown to regulate biofilm and type VI secretion in nontoxigenic environmental isolates. Indole is also produced by toxigenic V. cholerae strains in the human intestine, but its significance in the host is unknown. We investigated the effects of indole on toxigenic V. cholerae O1 El Tor during growth under virulence inducing conditions. The indole transcriptome was defined by RNA sequencing and showed widespread changes in the expression of genes involved in metabolism, biofilm production, and virulence factor production. In contrast, genes involved in type VI secretion were not affected by indole. We subsequently found that indole repressed genes involved in V. cholerae pathogenesis, including the ToxR virulence regulon. Consistent with this, indole inhibited cholera toxin and toxin-coregulated pilus production in a dose-dependent manner. The effects of indole on virulence factor production and biofilm were linked to ToxR and the ToxR-dependent regulator LeuO. The expression of leuO was increased by exogenous indole and linked to repression of the ToxR virulence regulon. This process was dependent on the ToxR periplasmic domain, suggesting that indole was a ToxR agonist. This conclusion was further supported by results showing that the ToxR periplasmic domain contributed to indole-mediated increased biofilm production. Collectively, our results suggest that indole may be a niche-specific cue that can function as a ToxR agonist to modulate virulence gene expression and biofilm production in V. cholerae.
INTRODUCTION
Vibrio cholerae is a Gram-negative bacterial pathogen and the causative agent of the acute diarrheal disease cholera. V. cholerae is an inhabitant of aquatic ecosystems and is acquired by ingestion of V. cholerae-contaminated food or water. After ingestion, V. cholerae colonizes the small intestine where it produces virulence factors, including toxin-coregulated pilus (TCP) and cholera toxin (CT), that result in the production of a severe secretory diarrhea. The expression of these virulence genes is under the control of ToxR, a membrane-bound regulatory protein that functions as the masthead of the ToxR virulence regulon.
The ToxR regulon is a hierarchal regulatory system that activates virulence gene expression following host entry. The in vivo stimuli that activate the ToxR regulon are unclear, but in vitro it has been shown to be modulated by a wide variety of environmental cues, including temperature, pH, osmolarity, oxygen, and bile salts (1–4). The induction of the regulon begins with two cytoplasmic proteins, AphA and AphB, which activate expression of the tcpPH operon (5). TcpP then binds with ToxR at the toxT promoter to initiate toxT expression (6). Thereafter, ToxT directly activates expression of the ctxAB and tcpABQCRDSTEF operons, which encode CT and TCP, respectively. Once V. cholerae is established in the small intestine, it replicates to high cell density before disseminating in the secretory diarrhea. Although much has been discerned about the transition of V. cholerae from the environment into the gastrointestinal tract, little is known about V. cholerae exiting the host. Several studies have documented a phenotypic shift that occurs late during infection that is characterized by a downregulation of virulence genes and upregulation of genes that contribute to both a hyperinfectious phenotype and enhanced fitness in aquatic environments (7, 8). These late infection phenotypes are likely critical for the life cycle of toxigenic V. cholerae, contributing both to epidemic spread and to survival and persistence in aquatic ecosystems.
Recent studies suggest that ToxR responds to extracellular cues to regulate the expression of genes that are important for adaptation to growth at high cell density. Given that V. cholerae replicates to high cell density during human infection, these results suggest that ToxR could contribute to the expression of genes during late infection. This conclusion was supported by the observation that the cyclic dipeptide metabolite cyclo(Phe-Pro) (cFP) accumulates to high concentrations at the stationary phase in the culture supernatants of some V. cholerae strains (9). cFP has been shown to signal through the periplasmic domain of ToxR to activate the expression of the LysR-family transcription factor leuO (10). LeuO then contributes to diverse phenotypes that are associated with late infection, including regulating genes involved in dissemination and transmission (e.g., biofilm, acid tolerance, antimicrobial resistance, and CT/TCP production) (10–14). The fact that leuO is positively regulated by ToxR (12) and maximally expressed at stationary phase (10) is consistent with this hypothesis. In addition, the requirement for ToxR in biofilm production (15), a phenotype that is associated with human shed V. cholerae (16), further bolsters this idea.
ToxR is a membrane-associated protein that contains a periplasmic sensing domain that is linked to a cytoplasmic DNA binding domain by a single transmembrane-spanning domain (17). The activity of ToxR is enhanced by interaction with another membrane-associated protein, ToxS (18). ToxR is thought to function as an environmental sensor that transduces environmental cues into transcriptional responses (3). There is accumulating evidence suggesting that low-molecular-weight compounds, both endogenously produced and exogenous, influence ToxR activity via interaction with its periplasmic domain (PPD) (10, 12, 19). Genetic studies from our laboratory showed that ToxR-dependent induction of leuO and ompU in response to cyclic dipeptides and bile salts was dependent on the presence of the PPD (10, 12), but the mechanism by which they activated ToxR was unclear. However, recent biochemical studies put forth a new model that can explain our genetic results (19). These studies showed that ToxR agonists bind to and destabilize the ToxR PPD. The destabilization of the PPD facilitates the formation of heterodimers between the ToxR PPD and the periplasmic domain of ToxS, which results in ToxR activation. Altogether, these findings support the conclusion that ToxR is an environmental sensor that responds to environmental cues via its PPD to effect the expression of adaptive responses.
Resistance-nodulation-division (RND) efflux systems are ubiquitous transporters in Gram-negative bacteria. The RND systems are critical for multiple antibiotic resistance due to their broad substrate specificity, which provides cross-resistance to multiple antimicrobial compounds (20). However, the RND systems also affect diverse phenotypes in bacteria, which suggests that they contribute to unknown physiological functions. This is true in V. cholerae, where RND-mediated efflux is required for virulence gene expression and cell homeostasis (21, 22). Recently, we documented that the V. cholerae RND efflux systems were linked to the ToxR-dependent regulation of virulence factor production (23). We found that in the absence of RND-mediated efflux, leuO transcription increased and led to repression of the ToxR regulon. This suggested a model whereby ToxR downregulated virulence genes in response to the accumulation of unknown endogenous metabolites that are normally exported from the cell by RND transporters.
The link between RND efflux and CT and TCP production suggested that products of V. cholerae metabolism may function in a feedback circuit to regulate virulence. Although the native substrates of the RND efflux systems are largely unknown, previous studies suggested that indole was a substrate for the V. cholerae VexAB RND efflux system (24). Indole is a bioactive metabolite produced by many bacteria, including V. cholerae. Indole has been shown to affect diverse bacterial phenotypes (25) and has been proposed to function as an interkingdom signaling molecule. Indole is produced in the human gut by the resident flora and is thought to enhance the barrier functions of the intestinal epithelium (26–28). V. cholerae also produces large amounts of indole during human infection (29). In V. cholerae indole is generated during stationary phase from the breakdown of tryptophan by the enzyme tryptophanase (tnaA). In environmental strains of V. cholerae, exogenous indole was shown to promote biofilm production in addition to affecting the expression of other genes (30). The observations that indole is produced during V. cholerae infection of humans and that biofilm contributes to hyperinfectivity and environmental survival (16, 31) suggest the possibility that indole may function as a signal in vivo that influences gene expression during infection.
We examined here the effect of indole on toxigenic V. cholerae. We found that indole was produced by a tnaA-dependent process and accumulated to a maximum concentration of ∼0.6 mM in the extracellular supernatant at early stationary phase. RNA sequencing of V. cholerae exposed to indole revealed that indole affected diverse physiological processes, including the activation of genes involved in biofilm production and the repression of genes involved in virulence. Subsequent analysis showed that indole-dependent activation of ToxR resulted in increased leuO expression. The upregulation of leuO contributed to indole-mediated repression of the ToxR regulon and increased biofilm formation. Altogether, our findings suggest that indole may function as an environmental cue during the life cycle of toxigenic V. cholerae.
RESULTS
Indole production.
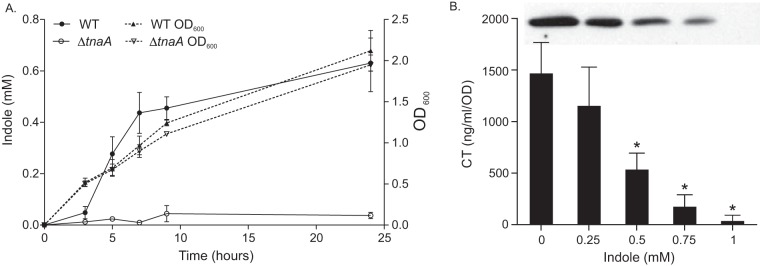
Indole production during V. cholerae human infection was first reported in the late 1800s and has been used as a diagnostic (29). To confirm that the O1 El Tor strain N16961 produced indole, we cultured strain JB58 and an isogenic tnaA deletion mutant in Luria-Bertani (LB) broth and quantified indole production over time. The results showed that indole production began at mid-log phase and reached the maximal concentration of ∼0.6 mM in stationary phase (Fig. 1A). These results are similar to findings reported for both environmental V. cholerae (30) and other O1 strains (32). Similar levels of indole production were also reported among the Enterobacteriaceae (33, 34). Based on these results, we selected 0.5 mM indole as a physiologically relevant concentration to assess the effects of indole on V. cholerae. We noted that indole has been reported to be toxic at high concentrations (35), but 0.5 mM indole does not produce deleterious effects on V. cholerae (30).
FIG 1.
Indole is produced at stationary phase and inhibits virulence factor production. (A) WT strain JB58 and an isogenic ΔtnaA mutant were cultured in LB medium at 37°C with shaking. Culture aliquots were collected at the indicated times; cell growth was assessed (right axis, triangle symbols), and indole production was quantified (left axis, circle symbols). The presented data show the means ± the standard deviations (SD) for at least three independent experiments. (B) V. cholerae strain JB58 was grown under AKI conditions with the indicated amounts of indole before CT and TcpA production (inset) were assayed. The CT data are the means ± the SD of three independent experiments. Statistical significance was determined using one-way analysis of variance (ANOVA) comparing each mean to that of the methanol control (*, P < 0.05).
Indole inhibits CT and TcpA production.
The observation that V. cholerae produced indole at a high cell density suggested that indole could be an environmental cue that contributed to virulence repression during infection. To test this, we quantified the effects of indole on CT and TCP production by culturing wild-type (WT) strain JB58 under AKI conditions in the presence of increasing concentrations of indole (Fig. 1B). The results showed an indole dose-dependent decrease in CT and TCP production beginning at 0.25 mM indole. CT production was reduced by ∼65% at 0.5 mM and ∼80% at 0.75 mM. CT and TcpA production was abolished at 1 mM indole. From these results, we concluded that indole was a virulence inhibitor.
Determination of the indole transcriptome.
Since the results presented above suggested that indole was a virulence inhibitor, we sought to determine the mechanism by which indole inhibited CT and TCP production. We therefore performed RNA sequencing (RNA-seq) on V. cholerae during growth under virulence gene inducing conditions in the presence or absence 0.5 mM indole. This identified 143 differentially expressed genes: 74 upregulated genes and 69 downregulated genes (Table S2). The largest category of differentially regulated genes was those of unknown function (45 genes), followed by metabolism (23 genes), pathogenesis (23 genes), and transport and binding (20 genes) (Table 1).
TABLE 1.
Functional categories of indole-responsive genes
| Functional group | No. of genes |
||
|---|---|---|---|
| Upregulated | Downregulated | Total | |
| Amino acid biosynthesis | 4 | 0 | 4 |
| Biosynthesis of cofactors, prosthetic groups, and carriers | 0 | 1 | 1 |
| Cell envelope | 6 | 2 | 8 |
| Cellular processes | 2 | 1 | 3 |
| Conserved, hypothetical, and unknown | 23 | 22 | 45 |
| DNA replication, recombination, and repair | 0 | 1 | 1 |
| Metabolism | 21 | 2 | 23 |
| Pathogenesis | 2 | 21 | 23 |
| Purines, pyrimidines, nucleosides, and nucleotides | 1 | 0 | 1 |
| Protein fate | 0 | 5 | 5 |
| Regulatory functions | 3 | 6 | 9 |
| Transport and binding proteins | 12 | 8 | 20 |
| Total | 74 | 69 | 143 |
Consistent with these results, the genes involved in CT and TCP production were largely downregulated by indole (i.e., ctxAB and tcpABQCRDSTEF operons; Table S2). Among the regulatory genes in the ToxR virulence regulon, toxT was identified as being repressed, suggesting that indole affected toxT transcription. We suspect that we did not identify genes that are upstream of toxT as being differentially expressed because the ToxR regulon had not been fully induced due to the early time point used for RNA collection. Interestingly, the genes involved in the production of multifunctional-auto processing repeats-in-toxin (MARTX) were also repressed by indole. The mechanisms regulating MARTX are unknown but MARTX was previously shown to be regulated according to growth (36, 37), suggesting that indole could play a role in MARTX expression. Collectively, these results confirmed the CT/TCP bioassays and suggested that indole affects the expression of the ToxR regulon.
Previous studies using nontoxigenic environmental V. cholerae isolates showed that indole upregulated the expression of genes involved in biofilm production and production of the type VI secretion system (T6SS) (30). Consistent with these studies, we also observed that indole increased the expression of genes involved in biofilm production (Table S2). Vibrio polysaccharide (VPS) is the major carbohydrate component of V. cholerae biofilm. The genes involved in VPS production, the vps genes, are clustered in two loci, vpsABCDEFGHIJK (VCO917-VC0927) and vpsLMNOPQ (VC0934-VC0939) (38), and several genes within each of these two clusters (vpsU, vpsA-vpsB, and vpsL-vpsN) were upregulated in the presence of indole. In addition, the expression of the genes encoding the biofilm matrix proteins Bap1, RbmA and RbmC were increased by indole (39). We did not observe changes in the expression of genes involved in type VI secretion. This is in contrast to what was observed in environmental V. cholerae isolates (30). The reasons for this are unknown, but it may be due to strain variation since toxigenic V. cholerae strains, such as N16961 used in this study, do not typically express the type VI secretion systems in vitro (40).
Indole represses toxT expression.
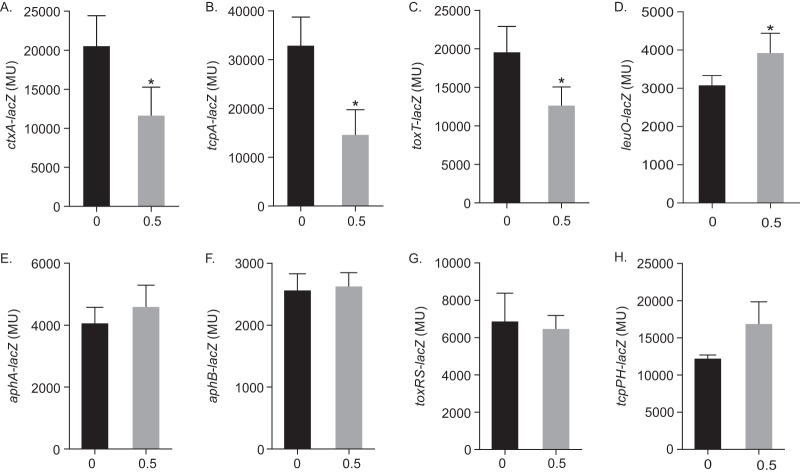
The RNA-seq data suggested that indole inhibited CT and TCP production by repressing the ToxR regulon. Since the ToxR regulon is a hierarchical regulatory system, we sought to determine at which point indole was acting in the ToxR regulon. To test this, we cultured WT strain JB58 carrying lacZ transcriptional reporters for each of the regulatory genes in the ToxR regulon under AKI conditions in the presence or absence of 0.5 mM indole and quantified reporter expression as β-galactosidase activity. We first examined the effect of indole on ctxAB-lacZ and tcpA-lacZ expression (Fig. 2A and B). We observed that the addition of indole resulted in 1.7- and 2-fold reductions in ctxA and tcpA promoter activities, respectively. This finding was consistent with the CT and TcpA results presented in Fig. 1. Since ctxA and tcpA promoters are directly activated by ToxT, we tested whether indole affected toxT expression. Consistent with the RNA-seq results, the presence of indole reduced toxT expression by ∼1.5-fold (Fig. 2C). We next tested the genes that are upstream of toxT in the ToxR regulon. The results showed that the expression of aphA, aphB, tcpP, and toxR was unchanged in the presence of 0.5 mM indole (Fig. 2E to H). The expression of tcpP appeared to slightly increase in the presence of indole, but the increase was not significant. Overall, these results indicate that indole inhibited CT and TCP production by repressing toxT transcription.
FIG 2.
Indole represses toxT expression. (A to H) V. cholerae JB58 strains carrying the indicated transcriptional reporters were cultured under AKI conditions for 5 h in the presence or absence of 0.5 mM indole, and the gene expression was quantified. The presented results are means ± the SD for at least three independent experiments. Statistical significance was determined using a t test comparing each mean to the no-indole control (*, P < 0.05). MU, Miller units.
LeuO contributes to virulence repression by indole.
The results presented above suggested that indole repressed toxT transcription, but not aphA, aphB, tcpPH, or toxRS transcription. This suggested that indole may be functioning through another regulator. The RNA-seq results identified nine regulatory genes as being differentially expressed in indole (Table 1), including the LysR-family transcription factor leuO, which was upregulated 1.5-fold in the presence of indole (Table S2). LeuO is a global regulator that has been linked to multiple V. cholerae phenotypes, including ToxR regulon repression (10, 12, 13). We therefore investigated whether LeuO contributed to the indole-dependent virulence repression. We first confirmed that indole increased leuO transcription by comparing leuO-lacZ expression in WT strain JB58 during growth under AKI conditions in the presence or absence of 0.5 mM indole. The results showed that leuO expression was significantly increased with the addition of indole (Fig. 2D), confirming the RNA-seq results.
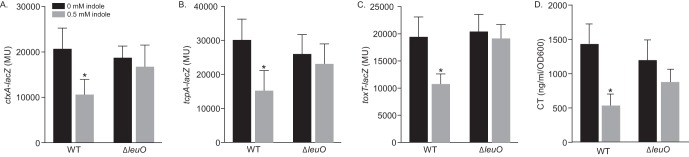
Expression of leuO has been correlated with downregulation of the ToxR regulon (10, 23). Therefore, we examined if LeuO contributed to the indole-dependent repression of the ToxR regulon. To test this, we compared toxT-lacZ, ctxA-lacZ, and tcpA-lacZ expression in the WT and in an isogenic ΔleuO mutant during growth under AKI conditions in the presence or absence of indole. The results showed that indole caused an ∼2-fold decrease in the expression of toxT, ctxA, and tcpA in the WT, whereas the negative effects of indole on the expression of all three genes were abated in the leuO-negative strain (Fig. 3A to C). These results suggested that LeuO contributed to the indole-dependent inhibition of virulence gene expression.
FIG 3.
LeuO contributes to indole-dependent virulence repression. V. cholerae strain JB58 and an isogenic leuO deletion strain carrying (A) ctxA-lacZ, (B) tcpA-lacZ, or (C) toxT-lacZ reporters were cultured under AKI conditions for 5 h in the presence or absence of 0.5 mM indole, and gene expression was quantified by determining the β-galactosidase activity. (D) WT and an isogenic ΔleuO mutant were cultured overnight in AKI medium supplemented with 0 or 0.5 mM indole before CT production was quantified by GM1 ELISA. The data are presented as means ± the SD for three independent experiments. Statistical significance was determined using two-way ANOVA, comparing each mean to the methanol control (*, P ≤ 0.05).
We hypothesized that if indole inhibited CT and TCP production via LeuO, then deletion of leuO would restore virulence factor production in cells grown in the presence of indole. We tested this by quantifying CT production in WT and ΔleuO strains during growth under AKI conditions in the presence of 0.5 mM indole. The results showed that the presence of indole resulted in ∼60% reduction in CT production in the WT. In contrast, the negative effects of indole on CT production were partially suppressed in the ΔleuO mutant (Fig. 3D). The fact that leuO deletion did not completely restore CT production in the presence of indole suggests that indole has additional effects on virulence factor production independent of LeuO.
Indole is a ToxR agonist.
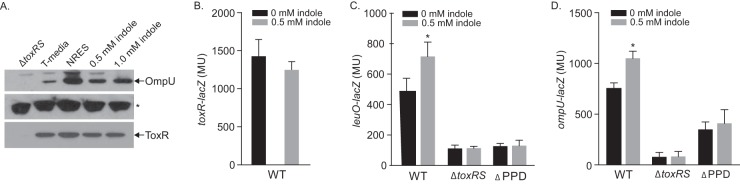
The finding that indole activated leuO transcription led us to hypothesize that indole was a ToxR agonist (41–43). To test this, we examined the effect of indole on OmpU production in WT strain JB58 during growth in minimal T medium. The amino acid cocktail NRES was included as a negative control (NRES promotes OmpU production by activating toxR transcription [44], not by altering ToxR activity). The results showed that indole increased OmpU production in a dose-dependent manner (Fig. 4A). To discriminate if the increase in OmpU was due to activation of preformed ToxR or due to increased ToxR production, we performed ToxR Western blotting (Fig. 4A). The results showed that indole did not affect ToxR abundance, whereas NRES supplementation increased ToxR and OmpU production as previously reported (44). Consistent with this, exogenous indole also did not affect toxR transcription (Fig. 4B). Taken together, these results suggested that indole was a ToxR agonist that increased OmpU production by enhancing the activity of preformed ToxR.
FIG 4.
Indole is an agonist of ToxR activity. (A) ToxR and OmpU Western blots. WT strain JB58 was cultured in T medium supplemented with amino acids (NRES) or increasing concentrations of indole for 4 h, and culture aliquots were collected, normalized by determining the OD600, and used for Western blotting with an anti-ToxR (bottom) or an anti-OmpU (top and middle) polyclonal antibody. A ΔtoxRS mutant grown in T medium was included as a marker for ToxR and OmpU. The asterisk indicates a nonspecific band that was used as a loading control. (B) Indole does not affect toxR expression. WT strain JB58 carrying a toxRS-lacZ reporter was cultured in T medium with the indicated indole concentrations for 4 h when β-galactosidase was quantified. The data are means ± the SD for three independent experiments. (C and D) Indole increases the expression of ToxR-dependent genes. The indicated V. cholerae strains harboring leuO-lacZ or ompU-lacZ reporters were cultured under the conditions described above, and gene expression was quantified by determining the β-galactosidase activity. The data are means ± the SD for at least three independent experiments. Statistical significance was determined by two-way ANOVA (*, P ≤ 0.05 relative to the methanol control).
The ToxR PPD is required for agonist-dependent activation of leuO transcription (10, 12, 23). Therefore, we tested whether the ToxR PPD was required for indole-dependent leuO induction. To do this, we cultured the WT strain JB58 and the isogenic ΔtoxRS (DT733) and toxRΔPPD (SS4) strains in minimal T medium containing 0.5 mM indole for 4 h. We then quantified leuO-lacZ and ompU-lacZ expression. The results showed that indole supplementation increased leuO and ompU expression ∼1.5-fold (Fig. 4C and D) and that the expression of both genes was abrogated in the toxRΔPPD mutant. From this, we concluded that indole activity was dependent on the periplasmic domain of ToxR. Since the addition of indole to T medium did not affect toxR expression (Fig. 4B), these results suggested that exogenous indole activated ToxR by a process that was dependent on its periplasmic sensing domain.
ToxR and LeuO mediate indole-dependent biofilm production.
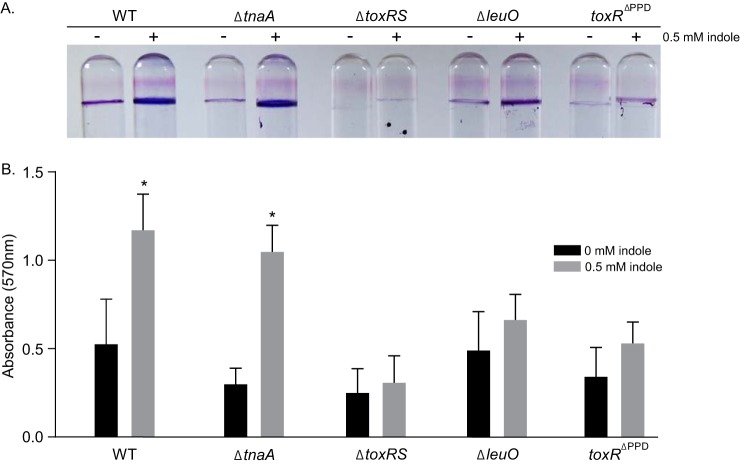
Indole was previously shown to activate biofilm formation in environmental V. cholerae isolates (30). This phenotype appears to be conserved in toxigenic V. cholerae, as evidenced by the indole-dependent increase in transcription of vps and biofilm matrix genes in the RNA-seq results (Table S2). Since LeuO and ToxR were shown to contribute to biofilm formation (14, 45), we examined whether they contributed to indole-dependent biofilm activation. To test this, we compared the effects of indole on biofilm production in WT and ΔleuO, ΔtoxRS, and ΔtnaA mutant strains (Fig. 5). The results showed that indole increased biofilm formation in WT relative to the no indole control. Biofilm production in an indole-negative ΔtnaA mutant was reduced relative to the WT strain, suggesting that endogenous indole production functions in a feedback loop to enhance biofilm production. The biofilm defect associated with the loss of tnaA was complemented by adding indole back to the growth medium, confirming previous results (30).
FIG 5.
Effects of indole on biofilm formation. Biofilm assays were performed with WT strain JB58 and the indicated isogenic mutant strains in the presence or absence of indole. Crystal violet-stained borosilicate tubes (A) and quantifications of the acetic acid-solubilized dye (B) are shown for the indicated strains after 48 h of static growth in LB medium supplemented with 0 or 0.5 mM indole at room temperature. The stained test tubes are representative of at least three independent experiments, with each assay being performed in triplicate. The absorbance data are presented as means ± the SD for three experiments. Statistical significance was determined using two-way ANOVA (*, P ≤ 0.05 relative to the methanol control).
In contrast, the ΔtoxRS mutant displayed a biofilm defect regardless of culture conditions, confirming the requirement for ToxR in biofilm production (19). Biofilm production in the ΔleuO mutant was reduced compared to the WT, and indole addition partially restored biofilm production (relative to the WT plus indole). Since the ToxR PPD was required for indole-dependent leuO induction (Fig. 4C), we tested whether the PPD was also required for biofilm production. Therefore, we assayed biofilm production in a toxRΔPPD mutant. In the absence of indole the toxRΔPPD mutant, in which leuO expression is impaired (23), produced similar amounts of biofilm as the ΔleuO mutant. However, in contrast to the leuO mutant, the addition of exogenous indole did not restore biofilm production in the toxRΔPPD mutant (Fig. 5). From these results, we concluded that LeuO and the periplasmic signal-sensing domain of ToxR contribute to indole-dependent regulation of biofilm formation.
DISCUSSION
Indole is a bioactive molecule that is widespread in nature and thought to be an intraspecies, interspecies and interkingdom signaling molecule. Mammals do not produce indole, but indole is present at high concentrations in their gut, being produced by the resident flora. V. cholerae also produces indole during human infection (29), and as in most organisms, the function of indole in V. cholerae biology is unclear. Here, we document that indole has broad physiological effects on V. cholerae and suggest that indole may function as a niche-specific signaling molecule in V. cholerae.
Indole is produced through the degradation of tryptophan by tryptophanase (TnaA). The tnaA gene is regulated by catabolite repression in the Enterobacteriaceae (46), making it predominantly expressed at the stationary phase. Consistent with this, we observed maximal indole production in V. cholerae during the transition to stationary phase, where the maximal indole concentration reached ∼0.6 mM (Fig. 1). These results mirror previous studies with environmental V. cholerae isolates (30). We did not observe indole production from V. cholerae cultured under virulence-inducing conditions in AKI medium. Since indole production is dependent on exogenous tryptophan (47), we speculate that this was due to low levels of tryptophan in AKI medium. This conclusion was supported by the observation that tryptophan addition to AKI resulted in indole production (data not shown). The production of indole appears to be relevant in vivo as indole is present in cholera patients (29). Consistent with in vivo indole production, tnaA expression was also upregulated in infant rabbit and infant mouse colonization models (48). The latter findings suggest that the results reported in this study may be relevant during V. cholerae growth in vivo.
V. cholerae shed from animal and human hosts is primed for survival in aquatic ecosystems and for the infection of new hosts (8, 49). However, the factors that contribute to the development of these phenotypes are not known. Transcriptional profiling of cholera stool samples revealed a physiological state characterized by increased expression of genes involved in amino acid biosynthesis, nutrient transport, and metabolism (7, 50). Here, we observed that indole regulated similar functional groups. Indole activated the expression of genes involved in tryptophan biosynthesis and acquisition of carbon sources such as sialic acid (VC1779, or siaP) and glycerol (VCA0137, or glpT). In aquatic environments, V. cholerae faces nutrient limitation; thus, indole-dependent induction of these genes may facilitate storage of nutrients to survive under nutrient-limiting conditions. Notable genes activated by indole included glycerol kinase glpK (VCA0774), the cadaverine/lysine transporter cadB (VC0280), and chitin binding protein gbpA (VCA0811). Expression of glpK is increased during late infection and is important for V. cholerae persistence in pond water (8). CadB functions in V. cholerae acid tolerance response, an adaptive phenotype that was reported to increase V. cholerae infectivity (51). GbpA is a colonization factor that was reported to mediate adherence to human intestinal cells and chitinous organisms in aquatic environments (52, 53). Overall, the transcriptional results are consistent with indole impacting physiological processes that have been observed during late infection.
Transcriptional profiling of cholera stool samples also revealed the repression of the ToxR virulence regulon (7, 54). Here, we propose that indole repressed virulence gene expression by two independent mechanisms that converged on toxT. The first mechanism was dependent on ToxR activation of leuO transcription. ToxR positively regulates leuO expression in response to environmental stimuli (e.g., bile salts and cyclic dipeptides). Multiple studies suggest that these compounds affect the activation state of ToxR by interacting with its PPD (10, 12, 19, 23). This is evidenced by recent studies showing that bile salts directly bind to the PPD (19). This binding destabilizes the PPD and promotes heterodimer formation with ToxS to activate ToxR. We suspect that indole may function by a similar mechanism to activate ToxR. Indole has been suggested to be a protein folding inhibitor (55, 56). This was documented for AsqR, a LuxR-type regulator, where indole blocked AsqR folding to inhibit its interaction with quorum-sensing molecules (56). In the case of ToxR, if indole blocked PPD folding, it would presumably promote interaction of the PPD with ToxS and lead to the formation of an activated ToxR. The facts that ToxR-mediated expression of leuO in the presence of extracellular indole was dependent on the PPD (Fig. 4) and that the PPD was dispensable for OmpU and CT/TCP production (23, 57) are consistent with this hypothesis, but additional work is required to confirm this. Based on these results, we propose that indole is a ToxR agonist.
Deletion of leuO only partially restored CT production during growth in the presence of indole (Fig. 3). This suggested that indole also inhibited virulence factor production by a second LeuO-independent mechanism. Although we were unable to determine this mechanism, we note that virulence regulation is linked to central metabolism via ToxT. Diminished respiration and tricarboxylic acid (TCA) cycle activity were reported to increase toxT expression (58). ToxT was also reported to repress a number of genes involved in carbon metabolism (15), suggesting a feedback loop between central metabolism and ToxT. Since metabolism genes represented one of the largest categories of indole-regulated genes (Table 1), it is possible that indole-dependent effects on metabolism may have contributed to virulence repression. Among the indole-regulated metabolism genes were genes involved in the TCA cycle (sdhCD) and glycolysis (gapA and lld) that were previously reported to be regulated by ToxT (15). This suggests that indole may regulate V. cholerae virulence by altering cell metabolism. We previously reported that ToxR can function as a metabolic sensor (23). This conclusion was supported by the fact that the TCA cycle intermediate malate impacts ToxR activation. Malate production was also previously linked to toxT expression (58). Thus, we speculate that indole-mediated effects on metabolism may lead to the generation and accumulation of metabolites that inhibit virulence by directly impacting ToxT or through metabolite-dependent effects on ToxR activation.
The ability to form biofilms is critical to the V. cholerae life cycle. Biofilm-associated V. cholerae is present in rice-water stool (16), and biofilm formation has been reported to be required for the hyperinfectious phenotype observed in human- and animal-shed V. cholerae (31). Biofilm production genes were the most highly upregulated genes in cells grown in the presence of indole (Table S2). This finding corroborates a previous study showing that indole promoted biofilm formation in environmental V. cholerae isolates (30). Here, we extended these studies by documenting that indole-mediated biofilm production was dependent upon ToxR and LeuO (Fig. 5). ToxR was shown to directly regulate the transcription of vps genes (19), whereas leuO deletion was reported to diminish biofilm production (14). Our results are consistent with these findings. Interestingly, we observed that deletion of the ToxR PPD diminished biofilm production. Given that ToxR PPD was previously shown to be dispensable for ompU expression and for induction of the ToxR virulence regulon (23, 57), this finding indicates that there are differential requirements for the PPD at specific ToxR target promoters. The observation that the ToxR PPD was required for indole-promoted biofilm production provides additional support for the conclusion that ToxR senses indole via its PPD.
In addition to promoting biofilm production, indole was reported to activate the expression of the type VI secretion system in environmental V. cholerae isolates (30). This is an intriguing result in light of the high concentrations of indole that are present in the human gut and recent studies showing the contribution of the T6SS to V. cholerae pathogenesis (59). Here, we were not able to confirm indole-dependent T6SS activation in the O1 El Tor strain N16961. We suspect that this may be due to strain-specific differences since most toxigenic strains do not exhibit T6SS activity in vitro (40). Thus, it is still possible that indole affects T6SS production in vivo. Additional work will be required to address this.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strains, plasmids and oligonucleotides used in this study are listed in Table S1. V. cholerae O1 El Tor ΔlacZ strain N16961 (JB58) was used as the WT (60) in all experiments unless otherwise noted. Escherichia coli strains EC100λpir and SM10λpir were hosts for cloning experiments and conjugation, respectively. All bacterial strains were maintained at –80°C in 25% glycerol and grown in lysogeny broth or agar (61). V. cholerae was grown in AKI broth for induction of the ToxR regulon, as previously described (62). Minimal T medium was prepared as previously described (44). Amino acids (Fisher Scientific) were dissolved in water and used at final concentration of 12.5 mM total amino acids. Antibiotics were used in the following concentrations: streptomycin (Sm), 100 μg/ml; carbenicillin (Cb), 100 μg/ml; and kanamycin (Km), 25 μg/ml. A fresh indole (Acros Organics) stock solution was prepared in methanol on the day of each experiment; control cultures received an equivalent volume of methanol.
Mutant construction.
The allelic-exchange vector pWM91::ΔtnaA was created by crossover PCR, as previously described (63). Briefly, VCA0161-F1/-R2 and VCA0161-F2/-R1 PCR primer pairs were used in separate reactions with N16961 genomic DNA. The resulting ∼1-kb PCR amplicons were pooled and then used as the template for a second PCR using flanking VCA0161-F1/-R1 PCR primers. The resulting ∼2-kb amplicon was restricted with SacI and BamHI endonucleases before being ligated with similarly digested pWM91 to generate pWM91::ΔtnaA. Deletion of tnaA was accomplished by conjugating pWM91::ΔtnaA into V. cholerae strain JB58 and selecting cointegrants for Cb/Sm resistance. Sm- and Cb-resistant cointegrants were then plated on no-salt LB agar plates containing 5% sucrose, and sucrose-resistant and Cb-sensitive colonies were then screened by PCR using the VCA0161-F1/-R1 primers to confirm tnaA deletion.
RNA sequencing.
V. cholerae strain JB58 was grown under AKI conditions with the addition of 0.5 mM indole or an equivalent amount of methanol for 3.5 h when the total RNA was isolated using TRIzol according to the manufacturer’s directions (Invitrogen) and further purified using a RNeasy kit with in column DNase treatment (Qiagen). The resulting total RNA was then processed and sequenced by the University of Pittsburgh Health Sciences Sequencing Core at Children’s Hospital of Pittsburgh. Briefly, the RNA samples were quantified using a Qubit 2.0 fluorimeter (Thermo Scientific), and the RNA integrity was assessed using an Agilent TapeStation 2220 system. The Ribo-Zero rRNA removal kit (Illumina) was used to deplete rRNA, and sequencing libraries were prepared and barcoded using a TruSeq Stranded Total RNA Library Prep kit (Illumina). The resulting libraries were pooled, and single-end sequencing was performed on an Illumina NextSeq 500 using a High-Output 75 cycle kit. The resulting FASTQ files from three independent experiments were mapped to the N16961 reference genome (61) using CLC Genomics Workbench (v10.1; Qiagen) and the default mapping parameters (i.e., mismatch cost, 2; insertion cost, 3; deletion cost, 3; length fraction, 0.8; similarity fraction, 0.8; strand specificity, both; maximum hits for a read, 10; expression value, total counts). Sample normalization (via the TMM method [64]) and identification of differentially expressed genes were accomplished using the Differential Expression for RNA-Seq tool in CLC Genomics Workbench. Genes showing a ≥1.5-fold difference in expression and a false discovery rate P value of ≤0.05 were defined as differentially expressed genes.
Quantification of indole production in V. cholerae.
WT strain JB58 and an isogenic ΔtnaA mutant were inoculated 1:100 into LB broth, followed by incubation with shaking at 37°C. Culture aliquots were then collected at the indicated times and subjected to centrifugation to separate the cells from the culture supernatant. Aliquots of the supernatants were then used to quantify indole production using a hydroxylamine-based indole assay, as previously described (65). Briefly, freshly prepared indole standards ranging from 0 to 1 mM were prepared in methanol. In a microtiter plate, 100-μl portions of the indole standards and V. cholerae culture supernatants were added in triplicate, followed by incubation for 15 min at room temperature with 5.3 M NaOH and 0.3 M hydroxylamine hydrochloride (NH2OH-HCl). After incubation, 125 μl of 2.7 M sulfuric acid (H2SO4) was added to each well, and the plates were incubated at room temperature for up to 30 min before indole production was measured spectrophotometrically in a BioTek XL808 plate reader at 530 nm. The concentration of indole in V. cholerae culture supernatants was then determined relative to the standard curve.
Gene reporter assays.
β-Galactosidase assays were performed as follows. Overnight cultures of V. cholerae strain JB58 carrying the indicated lacZ promoter reporters were diluted 1:5,000 into AKI medium containing 0.5 mM indole or an equivalent amount of methanol and incubated at 37°C under AKI growth conditions. Culture aliquots were then collected in triplicate at 5 h to quantify β-galactosidase production, as previously described (66). Gene expression assays done in minimal T medium were performed as follows: 1-ml portions of overnight cultures of the indicated V. cholerae strains carrying toxRS-lacZ, ompU-lacZ, or leuO-lacZ reporters were pelleted and washed twice in T medium before being subcultured at a 1:100 dilution into fresh T medium containing 0.5 mM indole or an equivalent amount of methanol at 37°C with shaking. Culture aliquots were collected in triplicate at 4 h (optical density at 600 nm [OD600] ∼0.3) to quantify the β-galactosidase activity.
Indole agonist activity.
An OmpU/OmpT porin-switching assay was used to assess whether indole was a ToxR agonist (44). ToxR directly regulates ompU and ompT expression, activating ompU and repressing ompT (42, 43). In rich medium (and in vivo) OmpU is predominantly produced. In contrast, OmpT predominates in minimal medium. However, the addition of specific additives to minimal media causes a porin switch that results in increased OmpU production. This porin switch is mediated by two independent mechanisms: (i) increased toxR expression or (ii) ToxR protein activation (44). The addition of an amino acid cocktail of asparagine, arginine, glutamate, and serine (NRES) to minimal medium results in porin switching due to increased toxR transcription. The concomitant increase in ToxR protein is enough to activate ompU transcription and repress ompT transcription. The second mechanism is agonist dependent, whereby adding ToxR agonists to minimal medium activates preformed ToxR proteins (i.e., it does not affect toxR expression or ToxR abundance). This has been demonstrated with bile salts and cFP, which are thought to interact with the ToxR PPD to facilitate the formation of an activated conformation (10, 12, 19).
ToxR and OmpU immunoblotting.
V. cholerae strain JB58 and an isogenic ΔtoxRS mutant were grown overnight in minimal T medium. The overnight cultures were then diluted 1:100 into fresh T medium containing the amino acid cocktail NRES (12.5 mM total amino acids), 0.5 or 1 mM indole, or an equivalent amount of methanol. The resulting cultures were then grown for 4 h (OD600 of ∼0.5) before aliquots were collected and normalized by cell density. Equal volumes from each sample were then suspended in Laemmli solubilization buffer, followed by heating at 100°C for 10 min before being resolved on an SDS–10% PAGE gel. The production of ToxR and OmpU was determined by Western immunoblotting with antisera against ToxR or OmpU, respectively. Proteins on Western blots were visualized using the SuperSignal West Pico chemiluminescent detection kit (Pierce Biotechnology).
CT and TCP quantification.
V. cholerae strain JB58 and an isogenic ΔleuO mutant were grown with or without the indicated concentrations of indole for 16 h under AKI conditions, after which culture aliquots were collected for quantification of CT and TCP. CT production was quantified by a GM1 enzyme-linked immunosorbent assay (ELISA) as previously described using purified CT (Sigma) as a standard (21). TCP production was determined by Western immunoblotting as previously described using polyclonal antisera against TcpA, the pilin subunit of TCP (21). Proteins on Western blots were visualized using a SuperSignal West Pico chemiluminescent detection kit (Pierce Biotechnology).
Biofilm assays.
Biofilm formation was assessed by inoculating the indicated V. cholerae strains at a 1:1,000 ratio into 1 ml of LB medium containing 0 or 0.5 mM indole in 13-by-100-mm borosilicate glass tubes. Triplicate cultures for each test strain and test condition were then incubated at room temperature without shaking for 48 h. Biofilm formation was then visualized by decanting the culture media and staining the glass-adherent biofilm with 1% crystal violent solution. The test tubes were then washed three times with distilled water before biofilm production was quantified by destaining the crystal violet-stained biofilms with 5% acetic acid and measuring the absorbance of the resulting solution at 570 nm.
RNA sequencing data.
The RNA-seq data have been deposited with links to BioProject accession number PRJNA374569 in the NCBI BioProject database.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the National Institutes of Health (NIH) under award AI132460. M.F.H. was supported by NIH predoctoral award AI129381 and training grant AI049820.
The content is solely the responsibility of the authors.
We acknowledge Greg Buchan for constructing the tnaA mutant.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00776-18.
REFERENCES
- 1.Hung DT, Mekalanos JJ. 2005. Bile acids induce cholera toxin expression in Vibrio cholerae in a ToxT-independent manner. Proc Natl Acad Sci U S A 102:3028–3033. doi: 10.1073/pnas.0409559102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller VL, Mekalanos JJ. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol 170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skorupski K, Taylor RK. 1997. Control of the ToxR virulence regulon in Vibrio cholerae by environmental stimuli. Mol Microbiol 25:1003–1009. doi: 10.1046/j.1365-2958.1997.5481909.x. [DOI] [PubMed] [Google Scholar]
- 4.Fan F, Liu Z, Jabeen N, Birdwell LD, Zhu J, Kan B. 2014. Enhanced interaction of Vibrio cholerae virulence regulators TcpP and ToxR under oxygen-limiting conditions. Infect Immun 82:1676–1682. doi: 10.1128/IAI.01377-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kovacikova G, Skorupski K. 1999. A Vibrio cholerae LysR homolog, AphB, cooperates with AphA at the tcpPH promoter to activate expression of the ToxR virulence cascade. J Bacteriol 181:4250–4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hase CC, Mekalanos JJ. 1998. TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci U S A 95:730–734. doi: 10.1073/pnas.95.2.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Merrell DS, Butler SM, Qadri F, Dolganov NA, Alam A, Cohen MB, Calderwood SB, Schoolnik GK, Camilli A. 2002. Host-induced epidemic spread of the cholera bacterium. Nature 417:642–645. doi: 10.1038/nature00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schild S, Tamayo R, Nelson EJ, Qadri F, Calderwood SB, Camilli A. 2007. Genes induced late in infection increase fitness of Vibrio cholerae after release into the environment. Cell Host Microbe 2:264–277. doi: 10.1016/j.chom.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park DK, Lee KE, Baek CH, Kim IH, Kwon JH, Lee WK, Lee KH, Kim BS, Choi SH, Kim KS. 2006. Cyclo(Phe-Pro) modulates the expression of ompU in Vibrio spp. J Bacteriol 188:2214–2221. doi: 10.1128/JB.188.6.2214-2221.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bina XR, Taylor DL, Vikram A, Ante VM, Bina JE. 2013. Vibrio cholerae ToxR downregulates virulence factor production in response to cyclo(Phe-Pro). mBio 4:e00366-13. doi: 10.1128/mBio.00366-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ante VM, Bina XR, Bina JE. 2015. The LysR-type regulator LeuO regulates the acid tolerance response in Vibrio cholerae. Microbiology 161:2434–2443. doi: 10.1099/mic.0.000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ante VM, Bina XR, Howard MF, Sayeed S, Taylor DL, Bina JE. 2015. Vibrio cholerae leuO transcription is positively regulated by ToxR and contributes to bile resistance. J Bacteriol 197:3499–3510. doi: 10.1128/JB.00419-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bina XR, Howard MF, Ante VM, Bina JE. 2016. Vibrio cholerae LeuO links the ToxR regulon to expression of lipid A remodeling genes. Infect Immun 84:3161–3171. doi: 10.1128/IAI.00445-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moorthy S, Watnick PI. 2005. Identification of novel stage-specific genetic requirements through whole genome transcription profiling of Vibrio cholerae biofilm development. Mol Microbiol 57:1623–1635. doi: 10.1111/j.1365-2958.2005.04797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies BW, Bogard RW, Young TS, Mekalanos JJ. 2012. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for Vibrio cholerae virulence. Cell 149:358–370. doi: 10.1016/j.cell.2012.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faruque SM, Biswas K, Udden SM, Ahmad QS, Sack DA, Nair GB, Mekalanos JJ. 2006. Transmissibility of cholera: in vivo-formed biofilms and their relationship to infectivity and persistence in the environment. Proc Natl Acad Sci U S A 103:6350–6355. doi: 10.1073/pnas.0601277103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller VL, Taylor RK, Mekalanos JJ. 1987. Cholera toxin transcriptional activator toxR is a transmembrane DNA binding protein. Cell 48:271–279. doi: 10.1016/0092-8674(87)90430-2. [DOI] [PubMed] [Google Scholar]
- 18.DiRita VJ, Mekalanos JJ. 1991. Periplasmic interaction between two membrane regulatory proteins, ToxR and ToxS, results in signal transduction and transcriptional activation. Cell 64:29–37. doi: 10.1016/0092-8674(91)90206-E. [DOI] [PubMed] [Google Scholar]
- 19.Midgett CR, Almagro-Moreno S, Pellegrini M, Taylor RK, Skorupski K, Kull FJ. 2017. Bile salts and alkaline pH reciprocally modulate the interaction between the periplasmic domains of Vibrio cholerae ToxR and ToxS. Mol Microbiol 105:258–272. doi: 10.1111/mmi.13699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Bambeke F, Glupczynski Y, Plesiat P, Pechere JC, Tulkens PM. 2003. Antibiotic efflux pumps in prokaryotic cells: occurrence, impact on resistance and strategies for the future of antimicrobial therapy. J Antimicrob Chemother 51:1055–1065. doi: 10.1093/jac/dkg224. [DOI] [PubMed] [Google Scholar]
- 21.Bina XR, Provenzano D, Nguyen N, Bina JE. 2008. Vibrio cholerae RND family efflux systems are required for antimicrobial resistance, optimal virulence factor production, and colonization of the infant mouse small intestine. Infect Immun 76:3595–3605. doi: 10.1128/IAI.01620-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kunkle DE, Bina XR, Bina JE. 2017. The Vibrio cholerae VexGH RND efflux system maintains cellular homeostasis by effluxing vibriobactin. mBio 8:e00126-17. doi: 10.1128/mBio.00126-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bina XR, Howard MF, Taylor-Mulneix DL, Ante VM, Kunkle DE, Bina JE. 2018. The Vibrio cholerae RND efflux systems impact virulence factor production and adaptive responses via periplasmic sensor proteins. PLoS Pathog 14:e1006804. doi: 10.1371/journal.ppat.1006804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor DL, Ante VM, Bina XR, Howard MF, Bina JE. 2015. Substrate-dependent activation of the Vibrio cholerae vexAB RND efflux system requires vexR. PLoS One 10:e0117890. doi: 10.1371/journal.pone.0117890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee JH, Lee J. 2010. Indole as an intercellular signal in microbial communities. FEMS Microbiol Rev 34:426–444. doi: 10.1111/j.1574-6976.2009.00204.x. [DOI] [PubMed] [Google Scholar]
- 26.Bansal T, Alaniz RC, Wood TK, Jayaraman A. 2010. The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc Natl Acad Sci U S A 107:228–233. doi: 10.1073/pnas.0906112107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chimerel C, Emery E, Summers DK, Keyser U, Gribble FM, Reimann F. 2014. Bacterial metabolite indole modulates incretin secretion from intestinal enteroendocrine L cells. Cell Rep 9:1202–1208. doi: 10.1016/j.celrep.2014.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimada Y, Kinoshita M, Harada K, Mizutani M, Masahata K, Kayama H, Takeda K. 2013. Commensal bacterium-dependent indole production enhances epithelial barrier function in the colon. PLoS One 8:e80604. doi: 10.1371/journal.pone.0080604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pollitzer R. 1955. Cholera studies. 3. Bacteriology. Bull World Health Organ 12:777–875. [PMC free article] [PubMed] [Google Scholar]
- 30.Mueller RS, Beyhan S, Saini SG, Yildiz FH, Bartlett DH. 2009. Indole acts as an extracellular cue regulating gene expression in Vibrio cholerae. J Bacteriol 191:3504–3516. doi: 10.1128/JB.01240-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamayo R, Patimalla B, Camilli A. 2010. Growth in a biofilm induces a hyperinfectious phenotype in Vibrio cholerae. Infect Immun 78:3560–3569. doi: 10.1128/IAI.00048-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nuidate T, Tansila N, Saengkerdsub S, Kongreung J, Bakkiyaraj D, Vuddhakul V. 2016. Role of indole production on virulence of Vibrio cholerae using Galleria mellonella larvae model. Indian J Microbiol 56:368–374. doi: 10.1007/s12088-016-0592-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gaimster H, Cama J, Hernandez-Ainsa S, Keyser UF, Summers DK. 2014. The indole pulse: a new perspective on indole signaling in Escherichia coli. PLoS One 9:e93168. doi: 10.1371/journal.pone.0093168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li G, Young KD. 2013. Indole production by the tryptophanase TnaA in Escherichia coli is determined by the amount of exogenous tryptophan. Microbiology 159:402–410. doi: 10.1099/mic.0.064139-0. [DOI] [PubMed] [Google Scholar]
- 35.Kim J, Park W. 2015. Indole: a signaling molecule or a mere metabolic byproduct that alters bacterial physiology at a high concentration? J Microbiol 53:421–428. doi: 10.1007/s12275-015-5273-3. [DOI] [PubMed] [Google Scholar]
- 36.Olivier V, Haines GK III, Tan Y, Satchell KJ. 2007. Hemolysin and the multifunctional autoprocessing RTX toxin are virulence factors during intestinal infection of mice with Vibrio cholerae El Tor O1 strains. Infect Immun 75:5035–5042. doi: 10.1128/IAI.00506-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olivier V, Salzman NH, Satchell KJ. 2007. Prolonged colonization of mice by Vibrio cholerae El Tor O1 depends on accessory toxins. Infect Immun 75:5043–5051. doi: 10.1128/IAI.00508-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teschler JK, Zamorano-Sanchez D, Utada AS, Warner CJ, Wong GC, Linington RG, Yildiz FH. 2015. Living in the matrix: assembly and control of Vibrio cholerae biofilms. Nat Rev Microbiol 13:255–268. doi: 10.1038/nrmicro3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fong JC, Yildiz FH. 2007. The rbmBCDEF gene cluster modulates development of rugose colony morphology and biofilm formation in Vibrio cholerae. J Bacteriol 189:2319–2330. doi: 10.1128/JB.01569-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miyata ST, Kitaoka M, Wieteska L, Frech C, Chen N, Pukatzki S. 2010. The Vibrio cholerae type VI secretion system: evaluating its role in the human disease cholera. Front Microbiol 1:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Champion GA, Neely MN, Brennan MA, DiRita VJ. 1997. A branch in the ToxR regulatory cascade of Vibrio cholerae revealed by characterization of toxT mutant strains. Mol Microbiol 23:323–331. doi: 10.1046/j.1365-2958.1997.2191585.x. [DOI] [PubMed] [Google Scholar]
- 42.Crawford JA, Kaper JB, DiRita VJ. 1998. Analysis of ToxR-dependent transcription activation of ompU, the gene encoding a major envelope protein in Vibrio cholerae. Mol Microbiol 29:235–246. doi: 10.1046/j.1365-2958.1998.00925.x. [DOI] [PubMed] [Google Scholar]
- 43.Li CC, Crawford JA, DiRita VJ, Kaper JB. 2000. Molecular cloning and transcriptional regulation of ompT, a ToxR-repressed gene in Vibrio cholerae. Mol Microbiol 35:189–203. doi: 10.1046/j.1365-2958.2000.01699.x. [DOI] [PubMed] [Google Scholar]
- 44.Mey AR, Craig SA, Payne SM. 2012. Effects of amino acid supplementation on porin expression and ToxR levels in Vibrio cholerae. Infect Immun 80:518–528. doi: 10.1128/IAI.05851-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kazi MI, Conrado AR, Mey AR, Payne SM, Davies BW. 2016. ToxR antagonizes H-NS regulation of horizontally acquired genes to drive host colonization. PLoS Pathog 12:e1005570. doi: 10.1371/journal.ppat.1005570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yanofsky C, Horn V, Gollnick P. 1991. Physiological studies of tryptophan transport and tryptophanase operon induction in Escherichia coli. J Bacteriol 173:6009–6017. doi: 10.1128/jb.173.19.6009-6017.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li G, Young KD. 2015. A new suite of tnaA mutants suggests that Escherichia coli tryptophanase is regulated by intracellular sequestration and by occlusion of its active site. BMC Microbiol 15:14. doi: 10.1186/s12866-015-0346-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mandlik A, Livny J, Robins WP, Ritchie JM, Mekalanos JJ, Waldor MK. 2011. RNA-seq-based monitoring of infection-linked changes in Vibrio cholerae gene expression. Cell Host Microbe 10:165–174. doi: 10.1016/j.chom.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alam A, Larocque RC, Harris JB, Vanderspurt C, Ryan ET, Qadri F, Calderwood SB. 2005. Hyperinfectivity of human-passaged Vibrio cholerae can be modeled by growth in the infant mouse. Infect Immun 73:6674–6679. doi: 10.1128/IAI.73.10.6674-6679.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Larocque RC, Harris JB, Dziejman M, Li X, Khan AI, Faruque AS, Faruque SM, Nair GB, Ryan ET, Qadri F, Mekalanos JJ, Calderwood SB. 2005. Transcriptional profiling of Vibrio cholerae recovered directly from patient specimens during early and late stages of human infection. Infect Immun 73:4488–4493. doi: 10.1128/IAI.73.8.4488-4493.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Angelichio MJ, Merrell DS, Camilli A. 2004. Spatiotemporal analysis of acid adaptation-mediated Vibrio cholerae hyperinfectivity. Infect Immun 72:2405–2407. doi: 10.1128/IAI.72.4.2405-2407.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhowmick R, Ghosal A, Das B, Koley H, Saha DR, Ganguly S, Nandy RK, Bhadra RK, Chatterjee NS. 2008. Intestinal adherence of Vibrio cholerae involves a coordinated interaction between colonization factor GbpA and mucin. Infect Immun 76:4968–4977. doi: 10.1128/IAI.01615-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stauder M, Huq A, Pezzati E, Grim CJ, Ramoino P, Pane L, Colwell RR, Pruzzo C, Vezzulli L. 2012. Role of GbpA protein, an important virulence-related colonization factor, for Vibrio cholerae’s survival in the aquatic environment. Environ Microbiol Rep 4:439–445. doi: 10.1111/j.1758-2229.2012.00356.x. [DOI] [PubMed] [Google Scholar]
- 54.Bina J, Zhu J, Dziejman M, Faruque S, Calderwood S, Mekalanos J. 2003. ToxR regulon of Vibrio cholerae and its expression in vibrios shed by cholera patients. Proc Natl Acad Sci U S A 100:2801–2806. doi: 10.1073/pnas.2628026100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim J, Hong H, Heo A, Park W. 2013. Indole toxicity involves the inhibition of adenosine triphosphate production and protein folding in Pseudomonas putida. FEMS Microbiol Lett 343:89–99. doi: 10.1111/1574-6968.12135. [DOI] [PubMed] [Google Scholar]
- 56.Kim J, Park W. 2013. Indole inhibits bacterial quorum sensing signal transmission by interfering with quorum sensing regulator folding. Microbiology 159:2616–2625. doi: 10.1099/mic.0.070615-0. [DOI] [PubMed] [Google Scholar]
- 57.Crawford JA, Krukonis ES, DiRita VJ. 2003. Membrane localization of the ToxR winged-helix domain is required for TcpP-mediated virulence gene activation in Vibrio cholerae. Mol Microbiol 47:1459–1473. doi: 10.1046/j.1365-2958.2003.03398.x. [DOI] [PubMed] [Google Scholar]
- 58.Minato Y, Fassio SR, Wolfe AJ, Hase CC. 2013. Central metabolism controls transcription of a virulence gene regulator in Vibrio cholerae. Microbiology 159:792–802. doi: 10.1099/mic.0.064865-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao W, Caro F, Robins W, Mekalanos JJ. 2018. Antagonism toward the intestinal microbiota and its effect on Vibrio cholerae virulence. Science 359:210–213. doi: 10.1126/science.aap8775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Newton WA, Morino Y, Snell EE. 1965. Properties of crystalline tryptophanase. J Biol Chem 240:1211–1218. [PubMed] [Google Scholar]
- 61.Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Umayam L, Gill SR, Nelson KE, Read TD, Tettelin H, Richardson D, Ermolaeva MD, Vamathevan J, Bass S, Qin H, Dragoi I, Sellers P, McDonald L, Utterback T, Fleishmann RD, Nierman WC, White O, Salzberg SL, Smith HO, Colwell RR, Mekalanos JJ, Venter JC, Fraser CM. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Iwanaga M, Yamamoto K, Higa N, Ichinose Y, Nakasone N, Tanabe M. 1986. Culture conditions for stimulating cholera toxin production by Vibrio cholerae O1 El Tor. Microbiol Immunol 30:1075–1083. doi: 10.1111/j.1348-0421.1986.tb03037.x. [DOI] [PubMed] [Google Scholar]
- 63.Bina JE, Mekalanos JJ. 2001. Vibrio cholerae tolC is required for bile resistance and colonization. Infect Immun 69:4681–4685. doi: 10.1128/IAI.69.7.4681-4685.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Robinson MD, Oshlack A. 2010. A scaling normalization method for differential expression analysis of RNA-Seq data. Genome Biol 11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Darkoh C, Chappell C, Gonzales C, Okhuysen P. 2015. A rapid and specific method for the detection of indole in complex biological samples. Appl Environ Microbiol 81:8093–8097. doi: 10.1128/AEM.02787-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.