The molecular chaperone “trigger factor” of plant chloroplasts functions in biogenesis of soluble proteins, and its absence leads to substantial reorganization of the chloroplast energy household.
Abstract
Biochemical processes in chloroplasts are important for virtually all life forms. Tight regulation of protein homeostasis and the coordinated assembly of protein complexes, composed of both imported and locally synthesized subunits, are vital to plastid functionality. Protein biogenesis requires the action of cotranslationally acting molecular chaperones. One such chaperone is trigger factor (TF), which is known to cotranslationally bind most newly synthesized proteins in bacteria, thereby assisting their correct folding and maturation. However, how these processes are regulated in chloroplasts remains poorly understood. We report here functional investigation of chloroplast-localized TF (TIG1) in the green alga (Chlamydomonas reinhardtii) and the vascular land plant Arabidopsis (Arabidopsis thaliana). We show that chloroplastic TIG1 evolved as a specialized chaperone. Unlike other plastidic chaperones that are functionally interchangeable with their prokaryotic counterpart, TIG1 was not able to complement the broadly acting ortholog in Escherichia coli. Whereas general chaperone properties such as the prevention of aggregates or substrate recognition seems to be conserved between bacterial and plastidic TFs, plant TIG1s differed by associating with only a relatively small population of translating ribosomes. Furthermore, a reduction of plastidic TIG1 levels leads to deregulated protein biogenesis at the expense of increased translation, thereby disrupting the chloroplast energy household. This suggests a central role of TIG1 in protein biogenesis in the chloroplast.
Maintaining and balancing proteome homeostasis is indispensable for all cells and subcellular compartments. This includes the correct integration of newly synthesized polypeptides into an existing (sub)proteome, refolding of stress-denatured proteins, and guiding aberrant proteins into proteolytic clearance (Tyedmers et al., 2010; Balchin et al., 2016). To achieve these tasks, all species have acquired a diverse set of evolutionary conserved molecular chaperones that provide assistance during virtually the entire lifespan of a protein (Hartl et al., 2011). Newly synthesized polypeptides emerge vectorially during translation, and folding can only be completed once the nascent polypeptide is fully released from the ribosome (Gloge et al., 2014; Balchin et al., 2016). Several molecular chaperones have been found to directly associate with translating ribosomes in order to provide a protective environment that prevents premature folding or unwanted interactions in a crowded intercellular molecular environment (Frydman, 2001; Preissler and Deuerling, 2012). In bacteria, the predominant ribosome-associated chaperone is trigger factor (TF), which fluctuates on and off ribosomes in an ATP-independent manner (Hoffmann et al., 2010). In Escherichia coli, TF (EcTF) is found in a 2- to 3-fold molar excess relative to the abundance of ribosomes, thus providing sufficient molecules to assist in the biogenesis of all synthesized proteins (Lill et al., 1988; Teter et al., 1999). The interaction between TF and the ribosome involves the conserved “signature motif” (43GlyPheArgxGlyxxPro50) and two surrounding alpha-helices embedded in the TF N-terminus. TF binds the ribosomal proteins uL23 and uL29 (Ban et al., 2014) that position the chaperone in close vicinity to the ribosomal exit tunnel in order to embrace emerging nascent polypeptides (Kristensen and Gajhede, 2003; Ferbitz et al., 2004; Schlünzen et al., 2005; Merz et al., 2008; Kramer et al., 2009; Deeng et al., 2016). Nascent chains are bound by TF via the C-terminal domain, which shapes the backbone of the molecule with an open cavity and two protruding arms. In this way, substrates are accommodated in a protective environment in the interior of TF to prevent misfolding and aggregation (Hoffmann et al., 2010; Deeng et al., 2016). The C-terminus is the largest and major chaperone module of TF (Kramer et al., 2004), and substrate binding appears to involve multiple sites across the entire cavity for both hydrophobic and hydrophilic substrate interactions (Merz et al., 2006; Saio et al., 2014). The third domain of TF is the peptidyl-prolyl cis-trans isomerase (PPIase) or so-called “head domain,” which is in the middle of the amino acid sequence but opposite of the N-terminal “tail domain” in the three-dimensional structure (Ferbitz et al., 2004). To date, the contribution of the PPIase domain is not fully understood.
Despite its abundance and evident importance, deletion of EcTF causes no obvious mutant phenotype under physiological growth conditions. However, under cold conditions, TF mutants propagate slower compared to the growth of wild-type cells (Kandror and Goldberg, 1997), and simultaneous deletion of TF and the major heat-shock protein 70, DnaK (ΔtigΔdnaK) is synthetic lethal at growth temperatures > 30°C, and widespread protein aggregation has been observed in these mutant cells (Deuerling et al., 1999; Teter et al., 1999; Genevaux et al., 2004; Martinez-Hackert and Hendrickson, 2009; Calloni et al., 2012). Global characterization of the EcTF substrate pool showed that the chaperone binds nascent polypeptides longer than 100 amino acids belonging to proteins targeted to the cytoplasm and to the outer membrane, whereas nascent polypeptides with inner membrane localization are not bound (Oh et al., 2011). The chaperone was shown to recognize and bind substrates at small stretches of aromatic and hydrophobic amino acids to prevent premature cotranslational folding (Patzelt et al., 2001; Kaiser et al., 2006; Hoffmann et al., 2012; Mashaghi et al., 2013; Saio et al., 2014; Nilsson et al., 2016). Thus, binding of TF to nascent polypeptides is thought to reduce the rate of cotranslational folding, which subsequently improves the efficiency and fidelity of de novo folding (Agashe et al., 2004). Further downstream protein folding is mediated by a cascade of molecular chaperones, including the heat shock proteins (Hsps) Hsp90s, Hsp70s, and chaperonins (Hoffmann et al., 2010; Preissler and Deuerling, 2012; Pechmann et al., 2013; Balchin et al., 2016).
Besides the existence of TF in bacteria, orthologous forms are also found in chloroplasts of eukaryotic cells. Chloroplasts contain a small genome that encodes only a minor fraction of approximately 60 to 100 proteins of the entire ∼3,000 chloroplast-localized proteins. However, these chloroplast-encoded proteins are major subunits of central protein complexes involved in gene expression and photosynthesis (Allen et al., 2011). Gene expression and biogenesis of these proteins is achieved by many components that are still most closely related to their cyanobacterial ancestors, including RNA polymerase, the 70S ribosome, and diverse chaperones (Gray, 1993; Trösch et al., 2015; Zoschke and Bock, 2018). Whereas bacterial TF has been extensively studied, only limited information is available concerning its role and properties within the chloroplast. We have recently reported biophysical and structural characterization of chloroplast-localized TF (TIG1) from the green alga Chlamydomonas reinhardtii and the vascular land plant Arabidopsis thaliana (Chlamydomonas and Arabidopsis hereafter, respectively). We could show by in-solution small-angle X-ray scattering and subsequent ab initio modeling that plastidic TIG1s have a dragon-like shape, similar to the conformation of bacterial TF, albeit with slightly altered domain arrangement and flexibility (Ries et al., 2017). This structural conservation, despite low amino acid sequence homology of less than 18% amino acid identity relative to that of EcTF, illustrates a remarkable evolutionary robustness of chaperone conformations across various kingdoms of life (Ries et al., 2017). Based on this information, we focused our research on investigating the function of chloroplast TIG1s in Chlamydomonas and Arabidopsis.
Here, TIG1s of both plant species were functionally compared with bacterial TF by complementation of the E. coli ΔtigΔdnaK mutant, ribosome-binding assays in bacteria, and comparison of in vitro chaperone activity. In addition, substrate specificity of the three chaperones was compared by hybridizing the proteins to peptide arrays derived from putative chloroplast TIG1 substrates. Our data suggest that chloroplast TIG1s exhibit similar overall chaperone activities like bacterial TF; however, unlike other chloroplast chaperones, plastidic TIG1s are not able to fully replace the function of bacterial TF. This indicates a more specialized function of these proteins in plants. The phenotypic analysis of plastidic tig1 mutants showed that chloroplasts with reduced levels of TIG1 display altered protein synthesis and a perturbed energy household, which suggests a specific role of these chaperones in protein biogenesis.
RESULTS
Chloroplast TIG1 Is Functionally Different from Bacterial TF
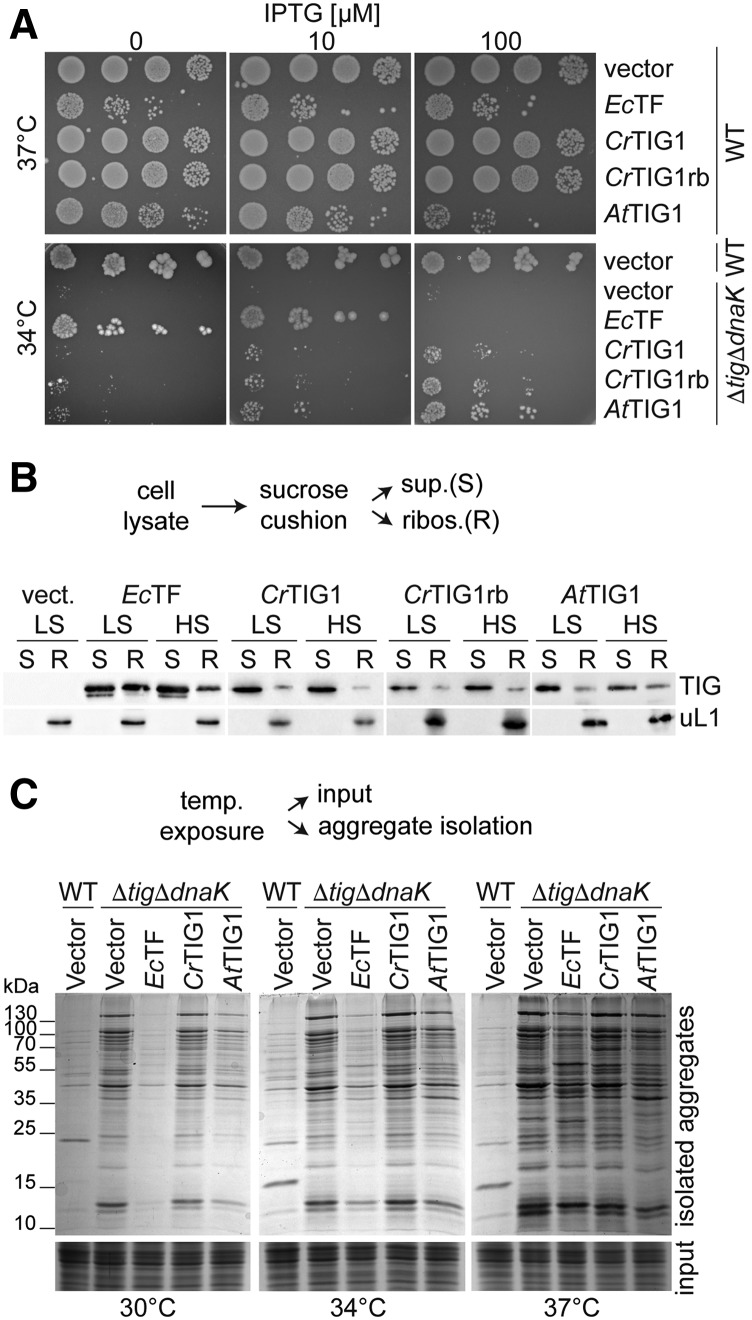
We have previously shown that chloroplast TIG1s have an overall similar protein conformation as their orthologous bacterial counterpart (Ries et al., 2017). However, it is not clear whether TIG1 has similar functions as those of bacterial TF. To test this, we constructed plasmids for the expression of Chlamydomonas (CrTIG1) and Arabidopsis (AtTIG1) TF in E. coli ΔtigΔdnaK cells and investigated if the respective plastidic chaperones rescued the reported growth defects at temperatures above 30°C. In addition, a hybrid of CrTIG1 was expressed that harbors the ribosome-binding signature motif of EcTF (discussed below). Expression of EcTF served as a control. Expression of the different constructs in wild-type and ΔtigΔdnaK strains was induced in log-phase, and viability was determined by growth on plates containing different isopropyl β-d-1-thiogalactopyranoside (IPTG) concentrations. IPTG is a functional analog of lactose, which triggers transcription through the lac operon promoter. Since IPTG is nonhydrolysable by E. coli cells, increasing IPTG concentrations is suited to enhancing lac promoter-dependent expression. Wild-type-containing plates were grown at 37°C. Consistent with previous reports (Kramer et al., 2004), high levels of expressed EcTF were toxic in E. coli cells, suggesting that overexpression of EcTF causes cell division defects (Patzelt et al., 2001; Schaffitzel et al., 2001). This effect was visible even in the absence of IPTG (Fig. 1A, top), presumably due to the leakiness of the IPTG-regulated promoter of pTrc-plasmids but was fully abolished when cells were grown on 2% Glc, which is known to tightly repress the promoter (Supplemental Fig. S1A). Indeed, leaky expression of EcTF is substantially higher compared with endogenous EcTF amounts (Supplemental Fig, S1B). AtTIG1 expression at higher IPTG concentrations also had mild toxic effects in wild-type E. coli cells (Fig. 1A, top). This effect was not observed for the CrTIG1 variants. Plates with ΔtigΔdnaK strains were incubated at 34°C, which is lethal for cells without TF (Fig. 1A). Interestingly, accumulation of chloroplast TIG1 in ΔtigΔdnaK strains could only partially rescue the growth defect of the mutants at 34°C (Fig. 1A, bottom). Despite sufficient protein levels of the chloroplast TIG1s (Supplemental Fig. S1B), mutant cells expressing either AtTIG1 or CrTIG1 still showed a severe growth defect upon growth at 34°C (compare EcTF, AtTIG1, and CrTIG1 at 10 µm; Fig. 1A, bottom).
Figure 1.
Chloroplast trigger factor function differs from that of its bacterial isoform. A, Complementation analysis of chloroplast trigger factor (TF) in E. coli. Wild-type (WT) MC4100 (top) and ΔtigΔdnaK cells (bottom) were transformed with the empty vector or constructs carrying EcTF, mature wild-type CrTIG1, CrTIG1 with the EcTF ribosome-binding signature motif (CrTIG1rb), and mature wild-type AtTIG1. Cells were spotted in serial dilutions onto Luria broth (LB) plates with different IPTG concentrations and grown overnight at 37°C and 34°C, respectively. Control plates containing Glc for full suppression of leaky expression are shown in Supplemental Figure S1A. B, E. coli ΔtigΔdnaK cells carrying the vectors described in A were grown to logarithmic phase, and TF production was induced by the addition of 20 µm IPTG for EcTF and 250 µm IPTG for the plastidic TIG1s. Cells were harvested in the presence of 100 µg/mL chloramphenicol and cleared of cell debris, and ribosome-nascent chains were isolated by centrifugation through a Suc cushion. Low-salt conditions (LS) contained 100 mm KCl and high-salt conditions (HS) 750 mm KCl. Supernatant fractions (S) and a 5× concentrate of the ribosomal pellet (R) were separated by SDS-PAGE and immunodetected with the indicated antibodies (uL1c, chloroplast ribosomal protein L1). C, E. coli transformed with the same constructs as in A, exclusive of CrTIG1rb, were grown for 4 h at the indicated temperatures, and expression was induced with IPTG as described for B. Insoluble protein aggregates were isolated from equal concentrations of cell lysates (input) and visualized by 10% SDS-PAGE and Coomassie staining. For expression control of the individual TF constructs, see Supplemental Figure S2. All immunoblots are representative of at least three biological replicates.
We next investigated if chloroplast TIG1 proteins associate with translating E. coli ribosomes in vivo. ΔtigΔdnaK mutant strains harboring plasmids for the expression of EcTF, CrTIG1, CrTIG1rb, or AtTIG1 were grown to midlogarithmic phase, and their expression was induced by IPTG. Immunoblot analysis of CrTIG1- and AtTIG1-expressing cells and Coomassie staining of EcTF-expressing cells confirmed that all proteins were expressed at the temperatures tested (Supplemental Fig. S2, A–C). Cosedimentation of the recombinant TF species with bacterial ribosomal particles was determined by centrifugation of cell lysate through a Suc cushion. Comparable to a previous study (Hesterkamp et al., 1997), we detected clear cosedimentation of EcTF in ribosome-containing pellet fractions, which was diminished when the ionic strength was increased from 100 to 750 mm KCl during lysis and centrifugation through the Suc cushion (Fig. 1B). By contrast, lower amounts of CrTIG1, CrTIG1rb, and AtTIG1 cosedimented with E. coli ribosomes (Fig. 1B; Supplemental Fig. S2D).
Simultaneous deletion of both dnaK and tig in E. coli not only leads to a severe growth defect at elevated temperatures but also causes profound protein aggregation, which progressively intensifies with increasing growth temperatures (Fig. 1C; Genevaux et al., 2004; Kramer et al., 2004; Merz et al., 2006). Thus, we systematically tested if chloroplast TFs can suppress protein aggregation in the mutant strain. Whereas viability at higher temperatures is restored upon mild EcTF expression, aggregate formation is only fully prevented at 30°C, and aggregate formation intensifies with increasing temperatures (Fig. 1C; Martinez-Hackert and Hendrickson, 2009), which might be caused by the lack of DnaK. Interestingly, AtTIG1 appeared to prevent aggregate accumulation to some extent, which was most evident at 30°C but also detectable at 34°C and 37°C (Fig. 1C). By contrast, aggregate formation was not influenced by CrTIG1 (Fig. 1C).
In Vitro Chaperone Activity of Plastidic TF
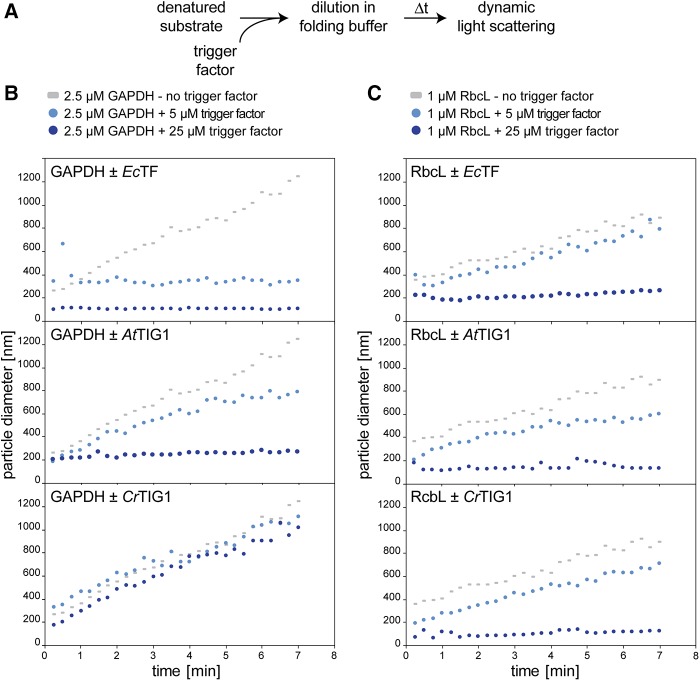
To further investigate and specify the capacity of TIG1 to prevent aggregate formation, in vitro chaperone assays were performed with denatured substrate proteins. Previously, it has been shown that bacterial TF efficiently prevents aggregation of glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Huang et al., 2000; Kramer et al., 2004). Accordingly, rabbit GAPDH was denatured, diluted to a final concentration of 2.5 µm in folding buffer, and the kinetics of aggregate formation were followed by dynamic light scattering (DLS), which monitors the hydrodynamic particle size in solution (Fig. 2A; Hassan et al., 2015). In the absence of chaperones, the size of GAPDH particles linearly increased over the measured time span, indicating protein aggregation. Addition of a 2-fold molar excess of EcTF reduced aggregate formation, whereas a 10-fold excess essentially suppressed aggregate formation (Fig. 2B, top). Also, AtTIG1 showed aggregate-preventing activity, albeit less than that of EcTF. Here, a 2-fold excess of AtTIG1 reduced the formation of large GAPDH particles only to some extent (Fig. 2B, middle). Addition of CrTIG1 to denatured GAPDH did not prevent aggregate formation (Fig. 2B, bottom).
Figure 2.
Trigger factor prevents protein aggregate formation. Prevention of GAPDH and RbcL aggregation by the addition of purified trigger factor (TF). A, Flow chart of experimental method. Chemically denatured, purified rabbit muscle GAPDH or Chlamydomonas RbcL were diluted to final concentrations of 2.5 µm and 1 µm, respectively, in folding buffer in the absence or presence of the indicated concentrations of EcTF, AtTIG1, or CrTIG1. B and C, Changes of hydrodynamic particle size (given as distribution widths of z-average diameters), as derived by DLS over a 7-min time course at 25°C. To aid comparison between plots, the same datasets for GAPDH and RbcL without TF are included in each plot. For control experiments with bovine serum albumin instead of TF, see Supplemental Figure S3. All graphs are representative of at least three biological replicates.
We next examined if plastidic TFs have a more specific chaperone activity tailored to proteins synthesized in the chloroplast. Thus, similar aggregation assays were performed with the purified and denatured large subunit (RbcL) of the Chlamydomonas ribulose-1,5-bisphosphate-carboxylase/oxygenase complex. Comparable to GAPDH, 1 µm of renatured RbcL massively aggregated in the absence of chaperones, which was prevented upon the addition of 25 µm EcTF (Fig. 2C, top). Interestingly, for RbcL, both AtTIG1 and CrTIG1 showed a comparable, if not better, chaperone function comparable to that of EcTF (Fig. 2C, middle and bottom). Hence, chloroplast TFs, and particularly CrTIG1, seem not suited to chaperone the broad substrate spectrum of EcTF but rather exhibit specificity for chloroplast-encoded substrates.
The Binding Specificity of Chloroplast TIG1 to Polypeptide Sequences
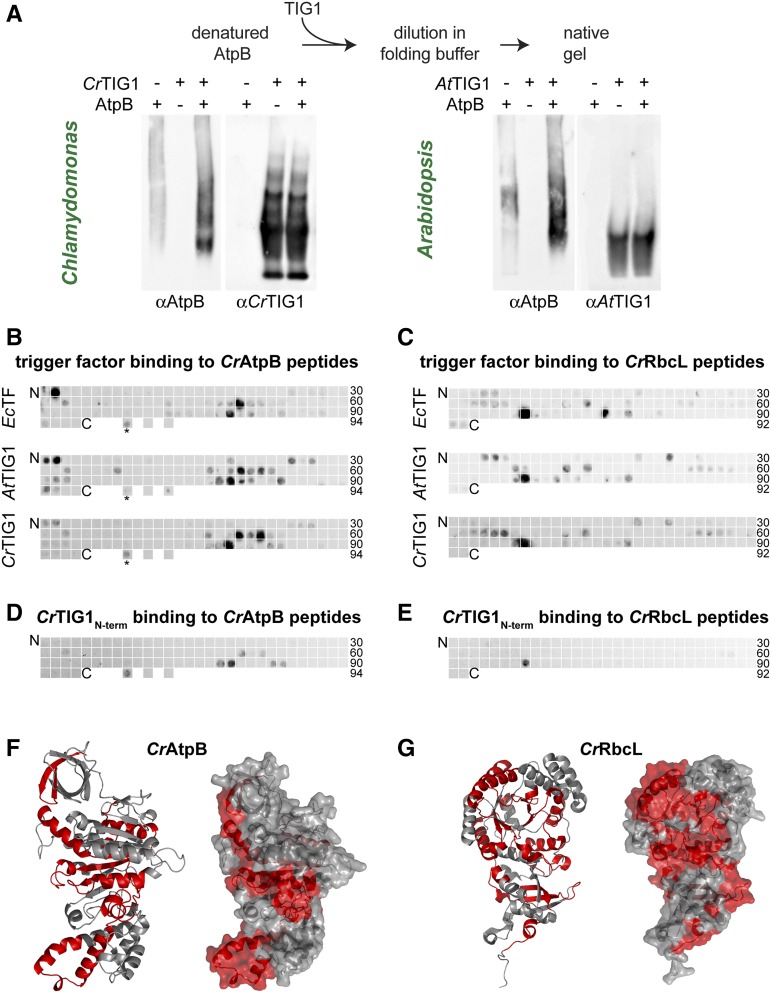
Since we could show that TIG1 of both plant species prevent aggregation of denatured RbcL, we assessed whether TIG1 processes other proteins that are synthesized in the stroma. To this end, Chlamydomonas AtpB, a subunit of the ATP synthase CF1 head domain, was heterologously expressed and purified. AtpB was denatured and subsequently renatured in the presence or absence of either CrTIG1 or AtTIG1 and subsequently analyzed by native polyacrylamide gel electrophoresis (PAGE) and immunoblotting. Without a chaperone present, AtpB showed a faint smear, indicating that AtpB formed amorphous aggregate species that only partially entered the gel (Fig. 3A, left lanes). In the presence of both TIG1 species, larger quantities of AtpB were detectable. CrTIG1 and AtTIG1 showed different migration patterns in native gels. AtTIG1 migrated as one prominent band, whereas CrTIG1 migrated as three rather well-defined bands, which is consistent with our previous study, where we showed that AtTIG1 is mainly monodisperse and CrTIG1 can form oligomers at higher Mr (Ries et al., 2017). Since both TIG1 species were added in 5-fold molar excess, it is difficult to detect a band comigrating with AtpB (Fig. 3A, right lanes).
Figure 3.
Bacterial and eukaryotic trigger factor bind peptides of similar properties. Bacterial and plastidic trigger factor (TF) binding to specific amino acid sequence stretches within putative TIG1 substrates. A, TF binds and processes unfolded AtpB in vitro. Purified AtpB protein was denatured and diluted to 0.4 µm in folding buffer in the presence and absence of 2 µm CrTIG1 or AtTIG1 (5-fold excess of the chaperone, as in Fig. 2C). Samples were separated by 4% native polyacrylamide gels and immunoblotted with the indicated antibodies. Immunoblot is representative of three biological replicates. B and C, Binding of EcTF, CrTIG1, and AtTIG1 to 20-meric peptide libraries of Chlamydomonas chloroplast-encoded AtpB (beta subunit of the plastidic ATP synthase; B) and RbcL (large subunit of Rubisco; C). Peptide frames are shifted by five amino acids; the numbers indicate the last peptide spot of a row. As a control, the TufA peptide bound by EcTF (Patzelt et al., 2001; indicated by an asterisk) and the adjacent downstream sequence (not binding EcTF) was spotted at three separated locations of the last row of the AtpB blot. D and E, Binding of the CrTIG1 protein N-terminal domain to the CrAtpB (D) and CrRbcL (E) peptide libraries described above. For specificity of the applied antibodies, see Supplemental Figure S5. Hybridization was done for each protein species in two independent replicates. F and G, Ribbon (left) and surface (right) presentation of AtpB (F) and RbcL (G). Peptides bound by all three TF species (B and C) are labeled in red. AtpB was modeled according to the spinach crystal structure (Groth, 2002). The RbcL structure was derived from Taylor et al. (2001).
We further investigated whether TIG1 recognizes different binding sites within substrates than that of EcTF, which was shown to exhibit specificity for short sequence motifs enriched in basic and aromatic amino acids (Patzelt et al., 2001). We applied a similar peptide-scanning approach comprising the full sequences of the two chloroplast-encoded Chlamydomonas proteins, RbcL and AtpB. The Arabidopsis and Chlamydomonas proteins display high amino acid sequence conservation (89% and 81% sequence identity, respectively; Trösch et al., 2018), which allowed using CrTIG1 and AtTIG1 on the same peptide arrays. The arrays consisted of synthesized and cellulose-bound 20-meric peptides that were shifted by frames of five amino acids along the full polypeptide sequence of RbcL and AtpB, hence presenting all putative binding sites in the linear protein sequence. In addition, a sequence stretch of E. coli elongation factor Tu (TufA) was bound to three spots, one of which presented a previously shown binding site of EcTF (Patzelt et al., 2001). The arrays were incubated with all three TF species independently (EcTF, CrTIG1, AtTIG1), extensively washed, and immunodetected with the respective antibodies. Overall, we observed that all three TF species bound to similar regions on RbcL and AtpB (Fig. 3, B and C). Importantly, some peptides appeared to be specifically bound by one or two of the three TF species. This finding potentially indicates variations in substrate binding specificity, which is expected for specific functions within the individual protein homeostasis networks. Peptides bound by the chaperones were not bound by the unrelated mCherry protein, indicating that these peptides were specifically bound by the TFs (Supplemental Fig. S4). For EcTF it has been shown that the N-terminus does not contribute to substrate binding outside of the ribosomal context (Patzelt et al., 2001; Saio et al., 2014). Consistently, we observed that the purified N-terminus of CrTIG1 showed substantially less binding to the tested peptides, especially to RbcL peptides (Fig. 3, D and E). However, some peptides, including the E. coli TufA peptide, were bound by the fragment, indicating that at least in the CrTIG1 chaperone, the N-terminus contributes to the binding to some peptides. Since the antibody showed weaker affinity due to fewer epitopes, we applied a 10-fold excess of the N-terminal fragment compared to the amount of full-length CrTIG1 (Supplemental Fig. S5). Despite the specialized functional adaptation of chloroplast TF and the low sequence conservation of the three chaperone species (Ries et al., 2017), substrate recognition and the overall binding affinity appear surprisingly conserved between the prokaryotic and the eukaryotic orthologs. Interestingly, these TF-bound peptides are not only found in the hydrophobic core of the substrate conformation, as observed for E. coli TF substrates (Patzelt et al., 2001), but also partially at the surface of chloroplast-encoded substrate proteins (i.e. Rubisco and the ATP synthase; Fig. 3, F and G).
Chloroplast TIG1 Binding to Ribosome-Nascent-Chain Complexes
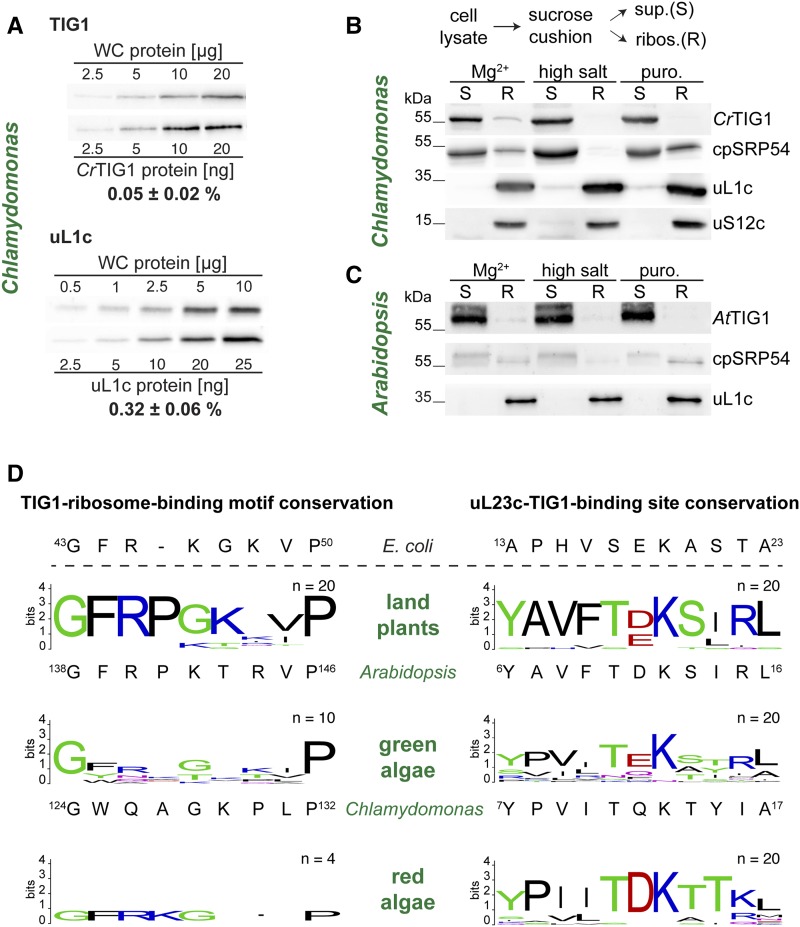
In order to provide sufficient TF protein for the pool of translating ribosomes in E. coli cells, EcTF is expressed at an abundance exceeding even that of the cellular ribosomal population (50 µm EcTF versus 20 µm ribosomes; Lill et al., 1988). We therefore investigated whether TIG1 is also in molar excess relative to the abundance of chloroplast ribosomes. Quantitative immunoblotting with Chlamydomonas cell lysates and a standard of purified uL1c and CrTIG1 proteins indicated that the chloroplast ribosomal protein uL1c represented about 0.32% of total cellular protein, whereas the concentration of CrTIG1 was only 0.05% (Fig. 4A). With these numbers, we estimated that uL1c and CrTIG1 are present at concentrations of ∼16 and ∼1.4 µm in Chlamydomonas chloroplasts, respectively. Because of the almost undetectable levels of uL1c in the non-ribosome-containing supernatant (Fig. 4B), it can be assumed that concentrations of plastidic ribosomes can be approximated from uL1c. Hence, the molar ratio of TIG1 to ribosomes in the Chlamydomonas chloroplast is only 1:11 and thus much lower than that in bacteria.
Figure 4.
Ribosome association of trigger factor in chloroplasts. A, Quantitative immunoblotting of CrTIG1 and chloroplast ribosomal protein uL1c. Recombinant Chlamydomonas TIG1 and uL1c were produced in E. coli, purified, and loaded in dilution series. Chlamydomonas whole-cell proteins were loaded at the indicated amounts. All samples were separated on 12% SDS-polyacrylamide gels and analyzed by immunoblotting. Relative protein abundances were determined from n = 3 independent biological replicates and given as means with standard deviations. B and C, Ribosome cosedimentation assays of Chlamydomonas (B) and Arabidopsis (C) lysates. Ribosomes were isolated under three different conditions: ribosome-stabilizing condition (“Mg2+” buffer, 50 mm HEPES, pH 8.0; 25 mm KCl; 10 mm MgCl2; 1 mm DTT; 0.25× Protease-Inhibitor) supplemented with 100 µg/mL CAP, 100 µg/mL cycloheximide, and 200 µg/mL Heparin; “high salt”-mediated release of ribosome-associated factors (high ionic strength buffer, 50 mm HEPES, pH 8.0; 800 mm KCl; 10 mm MgCl2; 1 mm DTT; 0.25× Protease-Inhibitor); “Puro.” release of nascent chains by addition of 1 mm Puromycin (buffer, 50 mm HEPES, pH 8.0; 25 mm KCl; 10 mm MgCl2; 1 mm DTT). Precleared cell lysates were separated into non-ribosome-containing supernatant (S) and ribosome pellet (R) by centrifugation through a 25% (w/v) Suc cushion, and fractions were separated on a 12% SDS-polyacrylamide gel. Note that R was concentrated 5× compared to S. Enrichment of trigger factor (TIG1) in the ribosomal fraction upon chemical cross linking is given in Supplemental Figure S6. Immunoblots are representative of at least three biological replicates. D, WebLogo representation of the TF “ribosome-binding signature motif” and the putative TF-binding site of the ribosomal protein uL23c. Comparison between land plants, green algae, and red algae. Sequence conservation is indicated by the height of stacks (bits); height of amino acid letters represents their relative frequency at the indicated position. Low letter height of red algal symbols is due to the low number of available sequences. All analyzed sequence identifiers are given in Supplemental Table S2. The respective amino acid sequence of E. coli, Chlamydomonas, and Arabidopsis is given below the diagrams. Note that for the plastidic TIG1 sequences, amino acids were counted from the translational start site of the chloroplast precursor sequence (including the transit peptide). For a phylogenetic comparison of uL23 between different species, see Supplemental Figure S7.
To test whether TIG1 is ribosome associated, ribosome-nascent chain complexes (RNCs) were isolated by sedimentation of Chlamydomonas and Arabidopsis cell lysates through a Suc cushion. In parallel, ribosome-associated proteins were washed off by increasing the ionic strength from 25 to 800 mm KCl or by releasing the nascent chains into the supernatant fraction by pretreatment with puromycin. As done for the E. coli assays, Chlamydomonas cultures were briefly preincubated with 100 µg/mL of the translation elongation inhibitor chloramphenicol (CAP) to prevent the dissociation of RNCs during harvest. Leaves from young Arabidopsis plants were flash frozen after excision. Consistent with the assays in E. coli (Fig. 1B), only minor fractions of < 2% of the TIG1 chaperones were detectable in the ribosomal fraction in both species (Fig. 4, B and C). Immunoblotting with the chloroplast signal recognition particle 54 (cpSRP54) served as a control, since cpSPR54 was previously shown to associate with RNCs in land plants (Ziehe et al., 2017). Here, cpSRP54 was found in the ribosomal fractions with a slightly higher ratio of pellet to supernatant signal than that detected for TF (Fig. 4, B and C). High salt treatment fully abolished RNC binding of both TIG1 and cpSRP54, whereas the dissociation of nascent chains by puromycin only released TIG1 and not cpSRP54. This suggested that TF might not directly interact with ribosomes as observed in bacteria (Kramer et al., 2009), whereas cpSRP54 might be able to contact ribosomes (Fig. 4, B and C). Since the interactions between TIG1 and the ribosomes might have been lost during sample preparations, ribosome-TIG1 cosedimentation was assayed in cross linked cell lysates. To this end, 2 mm dithio-bis(succinimidyl propionate) was added to both Chlamydomonas and Arabidopsis samples directly after lysis. In addition, Chlamydomonas complexes were cross linked in vivo by the addition of 0.37% formaldehyde prior to harvesting. Importantly, cross linking strongly increased the signal of RNC-associated TF both in Chlamydomonas and Arabidopsis samples (Supplemental Fig. S6A). In vivo cross linking improved the signal even further, indicating that chloroplast RNCs rapidly dissociate during or shortly after lysis (Supplemental Fig. S6A). However, quantification of the immunoblot signals of RNC-associated versus non-ribosome-bound chloroplast TF indicated that even upon in vivo cross linking, less than 6% of all chloroplast TIG1 cosedimented with ribosomes (Supplemental Fig. S6B). This is remarkably lower than the ∼30% EcTF binding to translating ribosomes (Hoffmann et al., 2010).
Several studies have reported that the signature motif GFRxGxxP is essential for EcTF-ribosome binding (Kramer et al., 2002; Kristensen and Gajhede, 2003; Ferbitz et al., 2004). With the weak ribosome association of chloroplast TIG1, we questioned whether this signature motif is strictly conserved in plastidic TIG1. To elucidate this, the consensus motif was analyzed by sequence alignment and WebLogo generation (Crooks et al., 2004) for 20 land-plant, 10 green-algae, and four red-algae species (Fig. 4D). Land plants show a striking conservation of this GFRPxxxP motif that is highly similar to the bacterial motif (e.g. 75% amino acid identity between the motifs of EcTF and AtTIG1) but with a Pro at the ninth instead of the eighth position (Fig. 4D, top plot). The motif of green algal TIG1s has a similar length as that of land plants; however, it is far more divergent, showing high conservation only for the first Gly and the ninth Pro (e.g. 25% amino acid identity between the motifs of EcTF and AtTIG1; Fig. 4D, middle plot). Interestingly, the four sequenced red algal genomes encode for TFs with a signature motif of the same length as the bacterial counterpart and a consensus sequence of GFRKGxxP (Fig. 4D, bottom plot), confirming our earlier finding that red algal chloroplast TIG1s are more closely related to bacterial TFs than TIG1s from the green lineage (Ries et al., 2017). Thus, the TF contact site on uL23c might have evolved complementarily in green algae. Our phylogenetic analysis of bacteria, algae, and land plants revealed that uL23c sequences from the green lineage fall into a separate clade than that of prokaryotic uL23 sequences. Land plant uL23c sequences displayed relatively high conservation, whereas uL23c sequences of green algae are more diverse (Supplemental Fig. S7). Thus, both uL23 sequences and the respective TIG1 ribosome binding motif seem to be evolutionary less constrained in green algae compared with that in land plants and bacteria. The interaction between uL23 and EcTF is mediated by a hydrogen bond between Glu-18 in uL23 and Arg-45 in the EcTF molecule (Ferbitz et al., 2004). Whereas the respective Arg in TIG1 is conserved, Glu seems frequently replaced by Asp in uL23c sequences of land plants (Fig. 4D). Again, green algae show no strict conservation of this amino acid. In the 20 green algal uL23c sequences analyzed, Glu was most frequently found at this position; however, Chlamydomonas uL23c contains a Gln at that position, which is unlikely to form a stable interaction with the respective Gln in the CrTIG1 ribosome binding site (Fig. 4D). Thus, it is possible that a putative contact between plastidic TIG1s and the ribosome is structurally different from that in bacteria.
Since chloroplast TIG1s seem to have a low affinity for translating ribosomes, we asked whether TIG1 might serve additional tasks in plastids and thus interact with other proteins outside of the context of cotranslational nascent polypeptide binding. Using the Chlamydomonas ΔCr-tig1 mutants (described below), we exploited a previously established approach termed “quantitative immunoprecipitation combined with knockdown” (Selbach and Mann, 2006; Heide et al., 2009) to obtain insights into the putative TIG1 interactome. With quantitative immunoprecipitation combined with knockdown, proteins isolated by coimmunoprecipitation (co-IP) with a protein of interest are quantitatively compared by mass spectrometry between a metabolically labeled wild-type strain and an unlabeled knockdown/mutant strain, in which the levels of the protein of interest are reduced. The identification of interactors is based on the assumption that reduced target proteins in the mutant also result in reduced coprecipitating interacting proteins. Unspecific protein interactions result in precipitating contaminants that are measured at equal abundances in both the wild-type and mutant sample. Chlamydomonas wild-type cells were grown in medium containing 15NH4Cl as sole nitrogen source to completely label the proteome (Mühlhaus et al., 2011). The ΔCr-tig1 mutant was grown in 14NH4Cl. Eluates of mutant and wild-type co-IPs with affinity-purified TIG1 antibody were pooled and measured by mass spectrometry, and 14N/15N ratios were estimated for all identified proteins. (Supplemental Fig. S8). In two different experiments with three respective independent co-IPs, we were able to precipitate the respective TIG1 target protein in similar ratios as observed in immunoblotting of cell lysates (Fig. 5, A and B). However, despite cross linking, none of the identified coprecipitating proteins showed similar Δtig1 mutant/wild-type ratios as the target protein but rather scattered around ratios of 1 (Supplemental Fig. S8). Whereas this method is not suited to catch low-abundant and transient interactions such as nascent polypeptides, these data suggest that TIG1 does not engage in other functions such as binding of mature proteins or complex assembly as postulated for the bacterial ortholog (Hoffmann et al., 2010).
Figure 5.
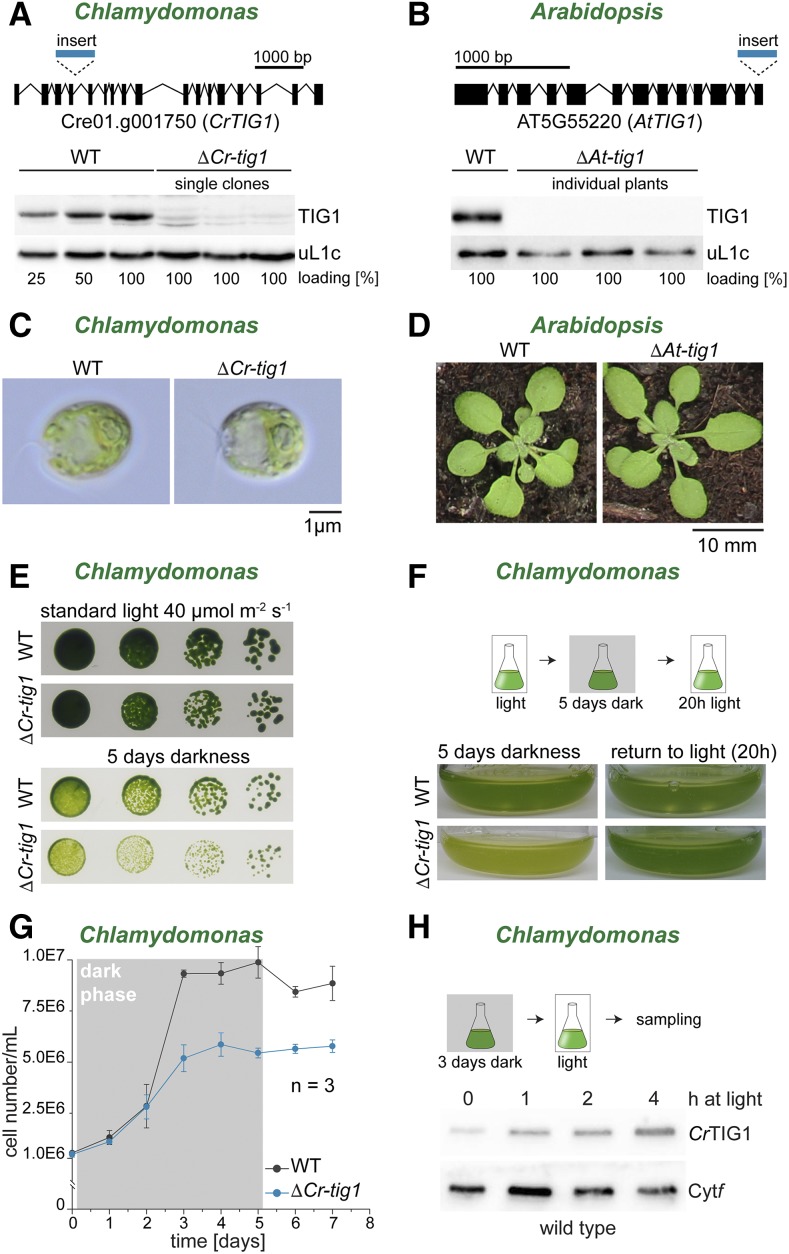
Chloroplast trigger factor mutants show reduced growth upon prolonged dark exposure. Characterization of Chlamydomonas and Arabidopsis TIG1 mutants. A, The Chlamydomonas tig1 mutant LMJ.RY0402.205185 (ΔCr-tig1) carries a paromomycin cassette insertion within the fourth intron of the trigger factor gene (top, lines indicate introns, black boxes mark exons, scale bar indicates distance of 1,000 bp), which results in a strong reduction of CrTig1 protein accumulation (bottom). B, In the Arabidopsis tig1 mutant SALK_037730 (ΔAt-tig1), the last three exons and two introns are replaced by the T-DNA insertion (top, lines indicate introns, black boxes mark exons, scale bar indicates distance of 1,000 bp), which abolishes AtTIG1 expression to protein levels below the detection limit by immunoblotting (bottom). C, Images of Chlamydomonas wild-type CC4533 (Li et al., 2016) and ΔCr-tig1 grown on a rotatory shaker at 25°C and 50 µmol of photons m−2 s−1. Scale bar indicates 1 µm. D, Photographs of 3-week-old Arabidopsis wild-type Col-0 and ΔAt-tig1 mutants grown in soil. E, Reduced growth of ΔCr-tig1 upon prolonged dark exposure. Twenty-microliter volumes of serial dilutions from 106 to 103 wild-type and mutant cells was spotted onto TAP agar plates and grown for 7 d, incubated under light or darkness. F, Mixotrophically grown cultures of wild type and ΔCr-tig1 were kept for 5 d in the dark and were subsequently exposed for 20 h under 30 µmol of photons m−2 s−1 light. G, Growth curve of Chlamydomonas wild type and ΔCr-tig1 during prolonged darkness. Cultures were diluted to equal cell numbers, adapted for 3 h under illumination, grown for 5 d during complete darkness, and transferred back to 30 µmol of photons m−2 s−1 light. Cell numbers are given in nEm nonsuperscript format for n·10m. Data are means and standard deviations of n = 3 independent biological replicates. H, CrTIG1 protein accumulation after 3 d of complete darkness (time point 0) and at the indicated time points upon exposure to 80 µmol of photons m−2 s−1 light. WT, Wild type; Cytf, cytochrome f. All figures are representative of at least three biological replicates.
Deletion of Chloroplast TIG1 Results in a Perturbed Energy Balance in Chlamydomonas Cells
We next examined whether the deletion or down-regulation of chloroplast TIG1 protein resulted in specific phenotypes. The Chlamydomonas mutant library (Li et al., 2016) contains a clone (LMJ.RY0402.205185; termed ΔCr-tig1 herein) that carries a paromomycin resistance cassette inserted into the fourth intron of the TIG1 gene (Fig. 5A). The correct insertion was confirmed by PCR and sequencing of the regions flanking the insertion cassette (Supplemental Fig. S9). Immunoblotting showed a reduction to 15% of wild-type levels of the TIG1 protein in all investigated single clones (Fig. 5A; Supplemental Fig. S9C). For an independent knockdown of CrTIG1, a constitutive artificial microRNA (amiRNA) approach was used, targeting a sequence spanning parts of the 11th and 12th exon (Supplemental Fig. S10A). Screening of more than 200 clones yielded only transformants with a mild reduction of TIG1 levels (∼60% of wild-type levels; Supplemental Fig. S10B). For Arabidopsis, a homoplastic knockout line with a T-DNA (transfer DNA) insertion (Alonso et al., 2003) replacing the last exons was ordered and confirmed by PCR and immunoblotting (SALK_037730; Figure 5B). In these mutants (ΔAt-tig1 herein), TIG1 protein was reduced below the detection limit (Fig. 5B). Under standard growth conditions, no phenotype was detectable for Chlamydomonas and Arabidopsis mutants (Fig. 5, C and D), which agrees with previous findings in E. coli (Deuerling et al., 1999; Teter et al., 1999). We further tested ΔCr-tig1 mutants under various other conditions (mixotrophic versus photoautotrophic growth, high light at 800 µmol of photons m−2 s−1, heat stress at 38°C and cold stress at 4°C); however, no obvious growth difference was detected between wild-type and mutant strains (Supplemental Fig. S11). Strikingly, mixotrophically grown ΔCr-tig1 mutants were paler than the wild type when plates and liquid cultures were exposed to prolonged darkness (i.e. 5 d; Fig. 5, E and F). This phenotype in the dark was fully reverted by 20 h light exposure, indicating that the ΔCr-tig1 mutant was not irreversibly affected during darkness (Fig. 5F). We next determined cell division rates during dark exposure. By supplementing growth medium with acetate as an external carbon source, Chlamydomonas strains can propagate during darkness, albeit at lower growth rates (Yang et al., 2015; Fig. 5G). During the first 2 d of dark exposure, both wild-type and ΔCr-tig1 cells divided at similar rates. Whereas wild-type cultures reached the stationary phase at a density of 1 × 107 cells/mL, ΔCr-tig1 cultures arrested growth at a density of only ∼5 × 106 cells/mL (Fig. 5G). Importantly, our tig1 amiRNA mutant with only a mild reduction of CrTIG1 protein levels showed a similar trend, but reduced cell division was detectable already after 24 h in darkness (Supplemental Fig. S12A). During the dark phase, chlorophyll per cell values behaved similar between wild type and ΔCr-tig1 mutants with peaks after 2 d in the dark and a decline during subsequent days (Supplemental Fig. S12B). Thus, it could be envisioned that CrTIG1 has a specific function during heterotrophic growth in the dark. However, CrTIG1 protein levels are rather low during prolonged growth in darkness, and expression is induced after illumination of cultures (Fig. 5H). In addition, in datasets of diurnally grown and synchronized cells (Zones et al., 2015), CrTIG1 transcript levels are upregulated during the light phase with a peak 2 h after the onset of day and shows lowest levels during darkness. This expression pattern is comparable with the transcript changes of nuclear-encoded ribosomal proteins of chloroplast 70S particles, pointing to a higher requirement of TIG1 in de novo protein biogenesis, at least in the light (Supplemental Fig. S13).
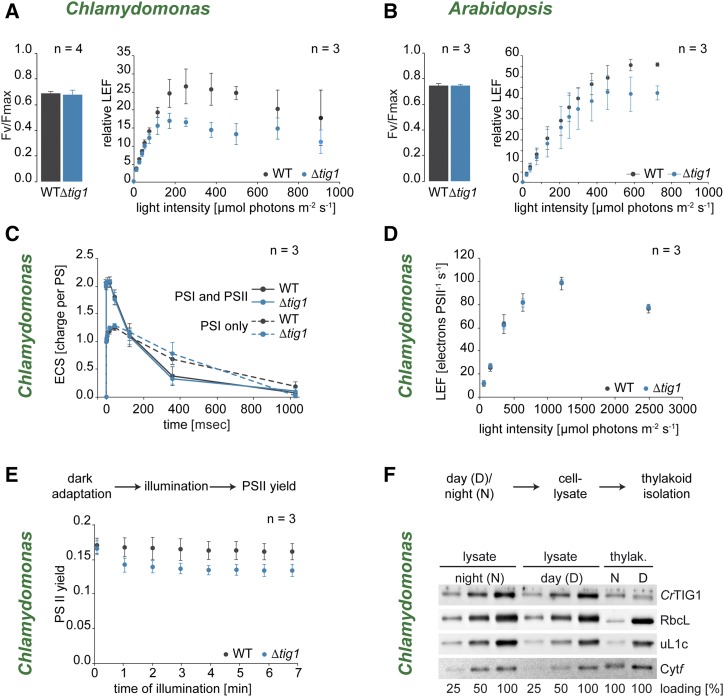
If cells were grown under moderate light (i.e. 30 µmol of photons m2 s−1) PSII maximum quantum yield was comparable between wild type and mutants of both Chlamydomonas and Arabidopsis (Fig. 6, A and B), suggesting that PSII function is not directly impacted by TIG1 deletion. In contrast, estimate of steady-state linear electron flow (LEF) shows a severe decrease under saturating light levels in Chlamydomonas strains with reduced TIG1 protein (Fig. 6A; Supplemental Fig. S14). A similar trend was observed in Arabidopsis mutants (Fig. 6B). To determine whether the decreased LEF associated with TIG1 mutation might be caused by defects in thylakoid electron and proton transport, we measured absorbance changes (electrochromic shift [ECS]) produced by a short illumination generating a single charge separation in PSI and PSII. ECSs were identical for wild-type and ΔCr-tig1, meaning the amplitude was twice as large in the PSII + PSI samples (untreated) than in PSI alone (PSII inhibited by 3-(3,4-dichlorophenyl)-1,1-dimethylurea [DCMU]), and kinetics of ECS accumulation and decay were similar. This suggests that the complexes involved in the photosynthetic light reactions (i.e. PSII, Cytb6f, PSI, ATP synthase) are normally accumulated and functional upon deletion of the chaperone (Fig. 6C). In addition, when measured upon short illumination, we observed LEF was similar in wild type and ΔCr-tig1 mutant both in light-limited and light-saturated regimes (Fig. 6D), indicating an apparent contradiction. In fact, diminished LEF in the measurements was only evident during prolonged high-light periods (Fig. 6E) when thylakoid reactions become tightly wired with the carbon assimilation cycle (Radmer and Kok, 1976; Chaux et al., 2017), suggesting a shift in light utilization in the mutants. This notion was supported by fluorescence measurements on cells grown under low, moderate, or high light (Supplemental Fig. S15), which indicated that PSII maximum quantum yield is also affected in the mutant cells under saturating light levels, most likely by downstream limitations or metabolic compensation due to the lack of TIG1. Furthermore, we examined if CrTIG1 colocalized to membranes with plastidic ribosomes during the day-night alterations. During the light phase, ribosomal protein uL1c showed a stronger signal in thylakoid membrane fraction compared with that in darkness (Fig. 6F), presumably due to the spatial synthesis of integral subunits of the thylakoid membrane (Chua et al., 1976). This pattern was not observed for CrTIG1, which is mainly in the nonmembrane fraction (Fig. 6F). Taken together, these data suggest that the photosystems are fully functional in ΔCr-tig1 mutants but that LEF is affected by metabolic processes, pointing to a misregulation of the chloroplast energy household in mutant cells.
Figure 6.
Photosynthetic electron flow is altered in chloroplast trigger factor mutants. Characterization of photosynthetic activity in Chlamydomonas and Arabidopsis TIG1 mutants. A, Left: photosynthetic activity of PSII determined by maximum quantum yield of fluorescence (Fv/Fmax). Right: photosynthetic linear electron flow (LEF) in relation to downstream metabolic processes. PSII fluorescence yield was measured after 2 min illumination at each light intensity in mixotrophically grown Chlamydomonas wild-type and ΔCr-tig1 (Δtig1) cells. B, Left: photosynthetic activity of PSII determined by maximum quantum yield (Fv/Fmax). Right: photosynthetic LEF in relation to downstream metabolic processes. PSII yield was measured after 2 min preillumination at each light intensity of Arabidopsis wild type and ΔAt-tig1 (Δtig1) mutants grown under long-day conditions (14-h light, 10-h dark). C, Electrochromic shift of carotenoids (ECS) was triggered by flash excitations of both PSII and PSI (solid lines) or PSI only (in the presence of PSII inhibitors, dashed lines), which generate a transmembrane electrochemical gradient. Data were normalized to one charge separation per photosystem. Electrochemical gradient is then dissipated through ATP synthase. Prior to the experiment, mixotrophically grown Chlamydomonas wild-type and ΔCr-tig1 (Δtig1) cells were dark-adapted for about 2 h. D, LEF in transitory regime independent of the carbon assimilation cycle. PSII yield was measured after 5 s illumination at each light intensity, interspaced with 30-s dark periods in mixotrophically grown Chlamydomonas wild-type and ΔCr-tig1 (Δtig1) cells. E, Time course of PSII yield during a 7-min illumination at 800 µmol of photons m−2 s−1, measured by chlorophyll fluorescence in mixotrophically grown Chlamydomonas wild-type and ΔCr-tig1 (Δtig1). F, Cells were grown photoautotrophically for 72 h in diurnal cycles of 12-h dark and 12-h light. Samples were collected 3 h after onset of night (N) or day (D), and thylakoids (Thylak.) were isolated. Thylakoid and lysate samples were adjusted to equal protein concentrations and visualized by separation on a 12% SDS-polyacrylamide gel and immunoblotted with the indicated antibodies. All values are given as mean values of the indicated independent biological replicates. Error bars denote standard deviations. Immunoblot in F is representative of two independent biological replicates.
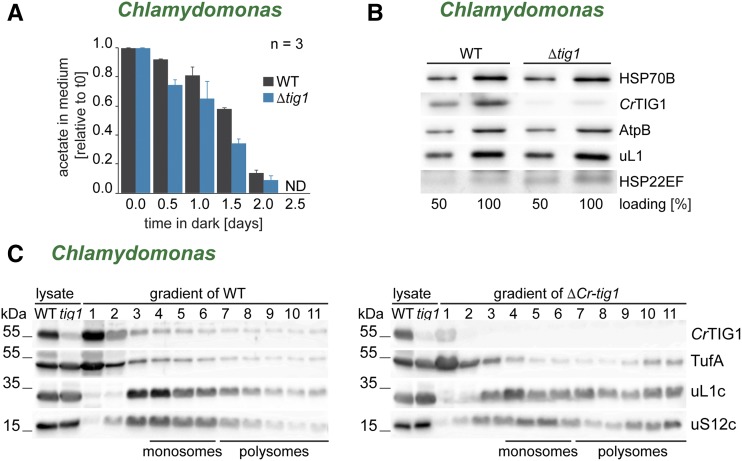
The modulated energy homeostasis of TIG1 mutants might also explain the observation that cell division is arrested earlier in ΔCr-tig1 mutants in the dark. Upon prolonged dark exposure, levels of acetate, the sole carbon source for heterotrophically growing cultures, declined rapidly in the medium (Fig. 7A). The time by which acetate was consumed below the detection limit of high performance liquid chromatography (HPLC) quantification correlated well with when ΔCr-tig1 cultures reached stationary phase. Interestingly, acetate concentrations declined faster in the medium of ΔCr-tig1 cultures than in the medium of the wild type (Fig. 7A). Starch accumulation and degradation in the dark behaved similar between wild-type and mutant cells, indicating that starch metabolism is not impaired in the mutant (Supplemental Fig. S16A). However, when cells were grown photoautotrophically under diurnal cycles, ΔCr-tig1 accumulated more starch during the light phase, which was degraded to similarly low levels until the end of the night phase (Supplemental Fig. S16B). Thus, deletion of TF seemed to cause an increased energy demand, which leads to a mutant phenotype only once external energy sources are limiting. One explanation for the higher energy consumption in the mutant lines might be an increased energy demand for the plastidic protein homeostasis network due to errors in protein biogenesis and folding. Hence, expression levels of stress-inducible plastidic chaperones were tested. Surprisingly, none of the major chaperones (i.e. HSP70B; Fig. 7B) were significantly upregulated in mutant cells, as observed, for example, during severe proteotoxic stress (Schroda et al., 2015). However, we repeatedly noticed a very mild up-regulation of the small Hsp HSP22E/F, which has previously been shown to be a highly sensitive marker for protein unfolding in chloroplasts (Rütgers et al., 2017b). We further tested mutant lines for increased protein misfolding and aggregation with a focus on chloroplast-encoded proteins. To this end, protein aggregates were isolated from Chlamydomonas cells and investigated by Coomassie staining and immunoblotting. Whereas heat exposure at 42°C led to widespread accumulation of proteins in insoluble aggregates, none of these showed increased aggregation in the mutant (Supplemental Fig. S17). We further tested whether reduced levels of TIG1 impact chloroplast protein synthesis rates. To address this, the distribution between plastidic monosomes and polysomes in cross linked Chlamydomonas wild-type and tig1 mutant cell lysates was examined by Suc density centrifugation and subsequent immunoblotting. Indeed, we consistently found a slight shift toward more polysomes in the ΔCr-tig1 strain as judged by the distribution of the ribosomal protein uL1c of the large subunit, uS12c of the small subunit, and the TufA (Fig. 7C; Supplemental Fig. S18), suggesting that the up-regulation of chloroplast protein biogenesis might be one factor for the increased energy demand of Δtig1 mutant cells.
Figure 7.
Chlamydomonas trigger factor mutants suffer from a disturbed energy balance in the chloroplast. Cells lacking chloroplast trigger factor show a higher energy demand. A, Acetate consumption of wild-type and ΔCr-tig1 (Δtig1) strains during heterotrophic growth in darkness. Remaining acetate in medium was quantified by HPLC and plotted relative to that at time point 0, which indicates freshly diluted cells. ND, not detectable. Data are means and standard deviations of n = 3 independent biological replicates. B, Immunoblot comparison of chloroplast molecular chaperone expression between Chlamydomonas wild-type and ΔCr-tig1 cells (Δtig1). C, Polysome analysis of Chlamydomonas wild-type and ΔCr-tig1 cells (Δtig1). Immunoblot of ribosomes (uL1c and uS12c), distribution of CrTIG1, and the TufA in fractions. Expected positions of monosomes and polysomes in the gradient are marked below the blots. All immunoblots are representative of at least three biological replicates (one additional replicate is shown in Supplemental Fig. S18).
DISCUSSION
In this study, we have investigated the function of chloroplast TIG1s from an alga and a vascular land plant and functionally compared their properties to that of their bacterial isoform. We show by in vitro chaperone assays and peptide arrays that overall molecular chaperone properties are conserved between bacterial and plastidic TF species. However, EcTF seems to exhibit an affinity to a broader substrate spectrum compared with that of AtTIG1 and especially that of CrTIG1 (Fig. 2). This is supported by our in vivo assays with the E. coli ΔtigΔdnaK mutant, in which both plastidic isoforms did not sufficiently suppress aggregate formation at higher growth temperatures (Fig. 1C). The biggest physiological difference between the bacterial and plastidic chaperones was observed in the nature of ribosome binding. EcTF was previously shown to exist in a 2-fold excess over bacterial ribosomes, and one-third of the EcTF population is engaged in ribosome association at a ratio of one chaperone per ribosome (Lill et al., 1988). Plastidic TIG1 exists in a 1:11 ratio with ribosomes (Fig. 4A), and our ribosome-binding assays showed that the affinity is far lower in chloroplasts, where only ≤ 6% of the TIG1 population cosedimented with ribosomes. This association appears more transient than in E. coli and is only evident upon chemical cross linking (Fig. 4B; Supplemental Fig. S6). Consistently, heterologous expression of AtTIG1 and CrTIG1 in E. coli cells also showed weak association with bacterial ribosomes (Fig. 1B), and both chaperones were not able to fully revert the strong growth defect of the ΔtigΔdnaK mutant (Fig. 1A).
The Cotranslational Role of Chloroplast TF
In bacteria, the most prominent function of TF is its cotranslational role of preventing premature nascent chain folding (Hoffmann et al., 2010; Preissler and Deuerling, 2012; Balchin et al., 2016). This interaction is mediated through the signature binding motif of TF and the ribosomal protein uL23 (Kramer et al., 2009; Hoffmann et al., 2012). Consistently, TIG1s of land plants show a highly conserved ribosome-binding signature motif, which may promote contact with ribosomes. Red algae show an even closer homology of this sequence to bacteria. In green algae, however, this motif is considerably less conserved (Fig. 4D), arguing against a requirement of this motif for TIG1 function. Our ribosome cosedimentation assays further support a rather minor contribution of the ribosome-binding motif, since ribosome affinity of TIG1 was equally low in Chlamydomonas and Arabidopsis (Fig. 4, B and C; Supplemental Fig. S6) despite the obvious differences of this motif between these two species (Fig. 4D). Moreover, despite the high conservation of the AtTIG1 ribosome-binding signature motif, the chaperone only poorly associated with translating ribosomes in E. coli cells (Fig. 1B). Also, the introduction of the ribosome binding motif of EcTF into CrTIG1 did not increase the ribosome affinity in E. coli (Fig. 1B). It is possible that the binding interfaces of TIG1 and uL23c have coevolved similarly. In E. coli uL23, Glu 18 is essential for TF binding (Kramer et al., 2002; Ferbitz et al., 2004). In land plant chloroplasts, both Glu and Asp residues are found at this position, whereas Glu is more conserved in green algae. The amino acid residues surrounding Glu or Asp differ strongly from the sequence stretch in E. coli (Fig. 4D). Thus, the interaction between plastidic TIG1 and the ribosome has drastically changed during evolution, as suggested also by the slightly different conformation and orientation of the N-terminal domains of plastidic TIG1s (Ries et al., 2017). Of note, chloroplast-encoded prokaryotic-type uL23 seems not uniformly present in chloroplast ribosomes of all species. On one hand, it was shown that this uL23c is essential in Nicotiana tabacum (Fleischmann et al., 2011), and we described its synthesis in the chloroplast of Chlamydomonas and Arabidopsis through our recent ribosome-profiling analysis (Trösch et al., 2018). On the other hand, reading frames of chloroplast-encoded uL23c are disrupted in some species of the Caryophyllidae and Rosidae families; here, a nuclear-encoded uL23c variant is used instead (Moore et al., 2010; Weng et al., 2016). The acquisition of this eukaryotic-type uL23 leads to severe changes within the architecture of the ribosome exit tunnel of spinach chloroplast ribosomes, and the lack of a structural hairpin within this uL23 may circumvent at least cpSRP54 binding to the ribosomal exit tunnel in spinach (Bieri et al., 2017; Graf et al., 2017; Perez Boerema et al., 2018).
The relatively small proportion of TIG1 that cosediments with plastidic ribosomes might be explained by the distribution of chloroplast-encoded proteins within the organelle. Of the 66 and 72 annotated protein-coding genes of the Chlamydomonas and Arabidopsis plastome, respectively, more than 30 highly expressed genes (Trösch et al., 2018) encode for constituents of integral thylakoid membrane complexes involved in photosynthesis-related processes. A cotranslational role of chloroplast TF for these integral thylakoid proteins seems rather unlikely since (1) CrTIG1 appears not to follow the cycling onto and off thylakoid membranes during diurnal growth as described for ribosomes (Chua et al., 1973, 1976; Fig. 6F), (2) photosynthesis seems not impaired in ΔCr-tig1 mutants (Fig. 6, C and D), (3) proteins of the inner membrane are rather depleted in EcTF substrate lists (Oh et al., 2011), and (4), the routes of protein targeting to the inner membrane do not involve the bacterial TF (Castanié-Cornet et al., 2014). Thus, only a relatively small subpopulation of mainly soluble stromal RNCs may require assistance by this chaperone, which is in line with our finding that chloroplast TIG1 levels are in substoichiometric ratios compared with the abundance of chloroplast ribosomes (Fig. 4A), unlike in bacteria (Lill et al., 1988). This would also explain why eukaryotic TIG1 was not preserved in mitochondria, since the small genome of this organelle mainly encodes for hydrophobic integral membrane proteins (D’Souza and Minczuk, 2018). However, the low amount of ribosome-associated TIG1 in chloroplasts in our experiments might underestimate the actual kinetics of TIG1 cycling on and off ribosomes, since interactions might be disrupted during sample preparation.
Increased Energy Demand of Chloroplast TF Mutants
Under standard growth conditions, the down-regulation of TIG1 results in very subtle phenotypes with only a very minor effect on growth and development. This might not be surprising since the deletion of the orthologous chaperone in E. coli also shows no phenotype at ambient temperatures (summarized in Hoffmann et al., 2010). Since deletion of EcTF leads to decreased cell viability under cold conditions (Kandror and Goldberg, 1997), we tested various environmental changes such as high light, heat, and cold. At least for ΔCr-tig1 mutants, none of these conditions influenced viability and growth (Supplemental Fig. S11). However, ΔCr-tig1 arrested cell division earlier than wild type in prolonged darkness (Fig. 5G; Supplemental Fig. S12A). In the presence of acetate as an external carbon source and oxic conditions, Chlamydomonas cells are not affected by prolonged darkness and even maintain photosynthetically competent thylakoid membranes through heterotrophic metabolism (Yang et al., 2015). Thus, the most obvious explanation of the ΔCr-tig1 dark phenotype would be a defect of specific processes during darkness, such as light-independent chlorophyll biogenesis (Cahoon and Timko, 2000), or a defect in mitochondrial processes required for acetate metabolism (Yang et al., 2015). However, mutants are able to metabolize acetate in the dark, which thus excludes defects of mitochondria in these strains (Fig. 7A). Our data rather suggest the predominant function of TIG1 during light, albeit specific defects of chloroplast metabolism during darkness cannot be ruled out at this point. We show that TIG1 is mainly expressed in the light (Fig. 5H), and transcript levels resemble the dynamics of plastidic ribosomes (Supplemental Fig. S13). Other chloroplast molecular chaperones, which are also involved in the coping with heat stress (i.e. HSP22E/F, HSP70B, HSP90C, and CLPB3; Supplemental Fig. S13; Schroda et al., 2015), instead peaked sharply with the onset of light, suggesting an important contribution of these chaperones toward protein refolding upon the transition into the light, whereas TIG1 appears instead to be involved in protein biogenesis. It is interesting that wild-type cells seemed to take longer to resume growth and chlorophyll biogenesis upon reillumination after prolonged dark exposure (Fig. 5, F and G; Supplemental Fig. S12, A and B), which may either point to affected chlorophyll biogenesis or altered state transitions in the mutant.
The increased assimilation of acetate during prolonged darkness does not result in altered starch turnover (Supplemental Fig. S16A). However, reduced growth of ΔCr-tig1 mutants is observed once acetate levels are depleted from the medium (Fig. 7A) and cellular starch levels are low (Supplemental Fig. S16A), suggesting that the earlier growth arrest of ΔCr-tig1 mutants (Fig. 5G) is caused by higher energy demand when energy supply is strictly limited (Yang et al., 2015). In the light, energy is not limiting, and consequently no growth defect is seen upon TF depletion (Fig. 5, C–E). Our measurements of photosynthesis activity exclude that the misregulated energy balance stems from direct defects in one of the components involved in the photosynthetic light reaction. Maximum quantum efficiency of PSII is indifferent between wild type and Δtig1 mutants (Fig. 6, A and B; Supplemental Fig. S14), the kinetics of all photosynthetic complexes are indifferent (Fig. 6C), and the electron transport rate is unchanged under low light intensities (when the thylakoid components are rate-limiting) or if LEF is quantified with short light exposures and dark regeneration times between the measured points (Fig. 6D). Strikingly, in both ΔCr-tig1 cells and Arabidopsis mutants, LEF was reduced when electron transport measurements were conducted with longer light exposure times (Fig. 6, A and B; Supplemental Fig. S14). The reduction was most prominent at higher light intensities, at which metabolic processes such as the Calvin-Benson cycle are rate-limiting steps. This might point to specific defects in downstream metabolic pathways. Hindered consumption of ATP and nicotinamide adenine dinucleotide phosphate, the products of the light reaction, directly affects LEF (Rochaix, 2011). However, the lack of evident defects in both Chlamydomonas and Arabidopsis Δtig1 mutants during photoautotrophic growth in the light argues against strong defects of the Calvin-Benson cycle. A plausible explanation for the reduced LEF would be a shift toward cyclic electron flow (CEF). Under normal conditions, CEF serves as compensatory electron flux around PSI in order to maintain the correct stoichiometry of nicotinamide adenine dinucleotide phosphate and ATP, which is required for carbon fixation by the Calvin-Benson cycle (Kramer and Evans, 2011). CEF has been shown to be increased during ATP-demanding situations such as photorespiration (Rochaix, 2011), high light (Takahashi et al., 2009), or drought (Kohzuma et al., 2009). Thus, the observed decrease of LEF might be the cause of upregulated CEF to compensate for the increased energy (i.e. ATP) demand in the tig1 mutants. Interestingly, Strand et al. (2017) isolated an Arabidopsis mutant with constitutively elevated CEF levels, which turned out to lack a plastidic tRNA-editing enzyme required for efficient codon recognition (tRNA adenosine deaminase Arg). Inefficient translation in these mutants led to altered stoichiometries of plastidic complexes, and the authors proposed that miscoordination of plastidic protein levels results in the up-regulation of CEF. Accordingly, lack of chloroplast TIG1 could likewise affect the energy balance in the chloroplast and lead to increased CEF. Here, nascent polypeptide misfolding, as expected to occur upon deletion of a protein biogenesis factor, might cause a compensatory up-regulation of chloroplast translation (Fig. 7C), which in turn is an energetically costly event (Buttgereit and Brand, 1995; Russell and Cook, 1995). Alternatively, the observed shift toward more polysomes in the tig1 mutants might be a consequence of reduced elongation rates, which may point to a direct cross talk between chloroplast polypeptide synthesis and subsequent protein maturation as postulated for other cotranslationally acting factors (Pechmann et al., 2013). Future radioactive pulse labeling or ribosome profiling experiments might help to better detangle the specific defects caused by deletion of this chaperone.
CONCLUSION
Altogether, these data shed light on the function of chloroplast TIG1 and suggest that plastidic TIG1 functions as a central modulator for the correct maturation of nascent chloroplast-encoded proteins. Some chaperone properties, such as the binding specificity to polypeptide stretches enriched in hydrophobic and basic amino acids and its aggregate preventing function, seem conserved between prokaryotic and eukaryotic TF. However, plastidic TIG1 seemed to have functionally diverged from its bacterial ancestor. In contrast to other chloroplast chaperones, such as HSP70B (Willmund et al., 2007, 2008) and the CPN60 chaperonins (Bai et al., 2015), TIG1 is not capable of complementing its respective counterpart in E. coli. It remains to be investigated in future studies why only a minor fraction of the cellular TIG1 population is engaged in transient nascent polypeptide binding at the ribosome and if the major function of TIG1 may rather be associated with nascent chain release from the ribosome. Alternatively, TIG1 may also contribute to protein biogenesis by associating with imported nascent polypeptides that derive from the cytosol. The phenotypic consequences resulting from TIG1 down-regulation or deletion are only subtle under saturating light conditions but lead to severe growth arrest once energy is limited.
MATERIALS AND METHODS
Cells and Culture Conditions
If not stated elsewhere, experiments were conducted with the Chlamydomonas reinhardtii strain cw15 CF185 (Schroda et al., 1999). ΔCr-tig1 mutant strain LMJ.RY0402.205185 and the respective wild type (CC4533) were received from the CLiP library (https://www.chlamylibrary.org; Li et al., 2016). Unless specified otherwise, cultures were grown photomixotrophically in Tris-acetate phosphate (TAP) medium (Harris et al., 1974) on a rotary shaker at 25°C and under an illumination of 50 µmol of photons m−2s−1. For photoautotrophic growth, cells were grown in minimal medium (Mettler et al., 2014). Cell densities were determined using a Z2 Coulter Counter (Beckman Coulter). For prolonged growth in darkness, logarithmically grown wild-type and mutant cells were diluted in TAP medium to a density of 3 to 5 × 105 cells/mL, acclimated for 3 h under illumination and transferred into dark. Induction of CrTIG1 after dark to light shift was done as described in Willmund and Schroda (2005). For thylakoid association of CrTIG1 during diurnal cycles, cells were grown photoautotrophically in a 12-h day–12-h night cycle.
Arabidopsis (Arabidopsis thaliana) seeds were ordered from the Nottingham Arabidopsis Stock Centre (ΔAt-tig1 T-DNA insertion line SALK_ 037730; Alonso et al., 2003). Arabidopsis plants were grown under long-day conditions (22°C, 14 h light at 120 µmol of photons m−2s−1, 10 h dark).
Arabidopsis and Chlamydomonas Δtig1 mutants were characterized by PCR (Supplemental Table S1) and immunoblotting. ΔCr-tig1 mutants were separated into single clones prior to analysis.
Plasmid Construction and Protein Purification
Cloning and purification of mature proteins of EcTF, CrTIG1, and AtTIG1 was described in Ries et al. (2017). For TF mutant complementation, plasmids were constructed as described earlier (Kramer et al., 2004). Resulting plasmids were termed pFW149 for EcTF, pFW150 for CrTIG1, and pFW151 for AtTIG1. For the expression of plant TIG1s in E. coli, sequences were not codon optimized. In addition, the putative ribosome-binding motif of CrTIG1 124Gly-Trp-Glu-Ala-Gly-Lys-Pro-Leu-132Pro was mutated by site-directed mutagenesis to 124Gly-Phe-Arg-Ala-Gly-Lys-Pro-Leu-132Pro in order to mimic the ribosome binding signature motif of E. coli (Ferbitz et al., 2004) giving pFW152. For expression of the CrTIG1 N-terminal domain (68Ala–247Gly) pFW13 (Ries et al., 2017) was modified by PCR. For heterologous expression of RbcL, the full Chlamydomonas coding sequence was amplified from genomic DNA, digested with NcoI and EcoRI, and cloned into NcoI/EcoRI-cut petDuet-1 (Novagen) yielding pFW75. pFW75 was expressed in ER2566 E. coli cells (NEB), and the protein was purified from inclusion bodies. In brief, cell pellets were resuspended in 40 mm Tris-HCl, pH 8.0, 0.25 m Suc, 1% (v/v) Triton X-100 supplemented with complete protease inhibitor cocktail (Roche), 0.5 mg/mL lysozyme, and incubated on ice for 30 min. After addition of 0.5 m EDTA, cells were disrupted by sonification, and inclusion bodies were harvested by multiple rounds of centrifugation (60 min, 22,000g) and washing with 40 mm Tris-HCl, pH 8.0, 0.25 m Suc, 10 mm EDTA, 1% (v/v) Triton X-100, 2 m urea. The final protein pellet was resuspended in 40 mm Tris-HCl, pH 8.0, 6 m GdnHCl, 1 mm EDTA, 5 mm dithiothreitol (DTT). Mature cpSRP54 (Cre11.g479750, lacking the predicted chloroplast transit peptide) was cloned from Chlamydomonas cDNA, cut with NdeI and EcoRI, and cloned into pTyb21 (NEB) resulting pFW55. pFW55 was transformed into ER2566 cells, cpSRP54 expression was induced, and the fusion-protein purified via chitin affinity resins according to the manufacturer’s instructions (NEB). Full-length atpB was amplified by PCR, digested with BamHI and HindIII, and cloned into BamHI/HindIII-cut petDuet-1 (Novagen) yielding pFW117. Expression and purification were done according to Willmund and Schroda (2005). All primers are listed in Supplemental Table S1.
E. coli Complementation Assays
E. coli strain MC4100 and ΔtigΔdnaK derivatives were transformed with the empty vector TRC99B or vectors pFW149, pFW150, pFW151, pFW152 containing the different TF species, respectively. Cells were grown in Luria broth medium supplemented with 100 µg/mL (w/v) ampicillin at 37°C (MC4100) or 28°C (ΔtigΔdnaK). Samples with 5 µL of cells were spotted in dilution series from 104 to 101 cells/mL on Luria broth plates containing 100 µg/mL (w/v) ampicillin and indicated amounts of IPTG, and growth was monitored at 34°C and 37°C, respectively. Glc plates contained 2% (w/v) Glc and no IPTG.
Aggregate Isolation from E. coli Cells
E. coli from the complementation were grown to logarithmic phase for MC4100 at 37°C and ΔtigΔdnaK at 28°C, TF expression was induced by addition of 20 µm IPTG for the EcTF construct and 250 µm for the other constructs and transferred to a water bath at the respective temperatures. After 4 h, cells were harvested and quickly chilled on ice. Aggregation preparations were done as described by Tomoyasu et al. (2001). The final protein aggregate pellet was dissolved in 6 m urea and supplemented with 1 volume of sample buffer (125 mm Tris-HCl pH 6.8, 20% (v/v) glycerol, 100 mm DTT, 4% (w/v) sodium dodecyl sulfate (SDS) and 0.005% (w/v) bromophenol blue before separation by SDS-PAGE and Coomassie staining.
DLS
For aggregate formation assays, 125 µM rabbit-muscle GAPDH (Sigma) or 50 µm RbcL was denatured at 4°C for at least 16 h in unfolding buffer (3 m GdnHCl, 100 mm potassium phosphate buffer, pH 7.5, 1 mm EDTA, 5 mm DTT). For renaturation, unfolded GAPDH or RbcL was diluted to final concentrations of 2.5 µm and 1 µm, respectively, in folding buffer (100 mm potassium phosphate buffer, pH 7.5, 1 mm EDTA, 5 mm DTT) in the presence or absence of TF variants. TF concentrations examined were 0, 5, 25 µm. DLS measurements were carried out in a 60-µL quartz glass cuvette with a 3-mm × 3-mm cross section (Hellma Analytics, Müllheim, Germany) at 25°C using a Zetasizer Nano S90 (Malvern Instruments, Malvern, UK) equipped with a 633 nm He-Ne laser and a photodetector mounted at an angle of 90°. Each sample was measured twice, once with the attenuator position automatically optimized for determination of size distributions and a second time with maximum open attenuator position to ensure comparability of total scattering intensities. The effects of buffer components on the viscosity and refraction index of the solvent were accounted for during data analysis. Nonnegatively constrained least-square functions were applied to fit autocorrelation functions and to yield intensity-weighted particle size distributions (Hassan et al., 2015). Cumulant analysis yielded z-average particle diameters and associated polydispersity indices (PDIs). Graphs show distribution widths of z-average diameters, σ, calculated as σ = √zPDIz.
Isolation of Ribosome-TF Complexes Ex Vivo
For Chlamydomonas, cells were lysed in MgCl2 buffer (50 mm HEPES, pH 8.0; 25 mm KCl; 10 mm MgCl2; 100 µg/mL CAP; 100 µg/mL cycloheximide; 1 mm DTT; 200 µg/µL Heparin [Sigma]; Protease-Inhibitor; Ribolock [1:200; Thermo]) or 1 mm Puromycin (without Heparin, Ribolock, and elongation inhibitors; 20 min at 24°C) or in high-salt buffer with 800 mm KCl. Cells were lysed in the presence of 1% (v/v) Triton X-100 with a Potter-Elvehjem. The cleared lysate was layered on 25% (w/v) Suc cushion prepared in appropriate buffer. Ribosomes were pelleted by centrifugation in a Sorvall Discovery M150 SE (Hitachi) at 204,000g (72,000 rpm) and 4°C for 20 min with a S150AT rotor. Afterward, the supernatant was collected, and the ribosome pellet was dissolved in 20% of the loaded sample volume. From Arabidopsis, ribosomes were isolated as described for Chlamydomonas. Plants were grown for 3 to 4 weeks (short day) on soil and directly harvested in liquid nitrogen. The tissue was ground thoroughly in liquid nitrogen and 250 mg plant material was dissolved in appropriate buffer containing 1% (w/v) n-dodecyl-D-maltoside.
E. coli ΔtigΔdnaK strains carrying an empty vector or plasmids for the expression of different TF variants were grown to logarithmic phase at 28°C. TF expression was induced by addition of 20 µm IPTG to the EcTF strain and 250 µm IPTG to the other strains and incubated for 3 h at 28°C. Prior to harvesting, translation was arrested by addition of 100 µg/mL CAP for 3 min. Cells were quickly chilled on ice, centrifuged at 4°C, and resuspended in buffer A (50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 10 mm MgCl2, 10% [w/v] Suc, 100 µg/mL CAP). Cells were lysed by several rounds of freezing and thawing including 1 mg/mL lysozyme. Lysates were precleared by centrifugation at 15,000g for 15 min at 4°C, and equal protein amounts of supernatants were layered on Suc cushions. Low-salt cushions contained 20% (w/v) Suc, 20 mm Tris-HCl, pH 7.5, 100 mm KCl, 10 mm MgCl2, 2 mm DTT, and 100 µg/mL CAP; high-salt Suc cushions contained 20% (w/v) Suc, 20 mm Tris-HCl, pH 7.5, 750 mm KCl, 10 mm MgCl2, 2 mm DTT, and 100 µg/mL CAP. Ribosomes were pelleted as described above. Supernatants were collected and ribosome pellets were resuspended in 1/5 volume of the loaded lysate.
Polysome Analysis
Logarithmically grown wild-type and ΔCr-tig1 cells were harvested, lysed, and cross linked directly with 2 mm dithio-bis(succinimidyl propionate) for 30 min at 4°C. Cross linking was quenched with 100 mm Tris pH 8.0 for 15 min at 4°C. Precleared lysates were layered on a 7%–47% Suc gradient and centrifuged for 2.5 h at 35,000 rpm at 4°C in a SW41 rotor (Beckman coulter). Gradients were split into 1 mL fractions, precipitated with chloroform/methanol, and analyzed by SDS-PAGE and immunoblotting.
Screening for TF Binding Sites by Peptide Arrays
20-meric peptides for the Chlamydomonas proteins RbcL and AtpB were synthesized on a cellulose membrane as described previously (Weckbecker et al., 2012). The amino acid frame was shifted by five amino acids from one spot to the next, covering the full sequence of the respective proteins. As positive controls, three peptides covering the sequence stretch RHTPFFKGYRPQFYFRTTDVTGTIELPEGVEMVMPGDNIKMVVTLIHPIA of E. coli TufA, which has previously been shown to contain a EcTF binding site (Patzelt et al., 2001), were spotted accordingly. The membrane was soaked with methanol for 2 min at room temperature, washed 3× for 2 min with H2O, and equilibrated in binding buffer (31 mm Tris-HCl, pH 7.5, 170 mm NaCl, 6.4 mm KCl, 0.05% [v/v] Tween 20, 5% [w/v] Suc) for 20 min. The membrane was incubated for 3 h with 0.5 μm of recombinantly purified EcTF, CrTIG1, CrTIG1-N terminus, and AtTIG1 protein, respectively, in binding buffer. The membrane was washed twice for 10 min with binding buffer and twice with Tris-buffered saline (31 mm Tris-HCl, pH 7.5, 170 mm NaCl, 6.4 mm KCl) before transfer to nitrocellulose membrane and immunoblotting with the respective TF antibodies. The sequence of the spots that were bound to all TF species was visualized with PyMOL in the crystal structure of RbcL (Taylor et al., 2001) and a structural model of AtpB (Groth, 2002).
Measurements of Photosynthetic Activity
For Chlamydomonas, maximum fluorescence quantum yield of PSII (Fv/Fm) was determined with a pulse amplitude-modulated Mini-PAM-II fluorometer (Heinz Walz, Effeltrich, Germany). On a Suspension Cuvette KS-2500, 3 to 3.5 × 106 cell/mL were adapted to dark for 5 min and measured with 1 s saturating pulse (6,000 μmol photons m−2 s−1, gain value = 4). LEF was determined from PSII yield (YII) at increasing light intensity (PAR) by LEF = (YII) × PAR × 0.5 × 0.84. JTS-10 (BeamBio, France) was used for ECS measurements and thorough chlorophyll fluorescence measurements (Fig. 6, C–E) as described in Bailleul et al. (2010). Cells were sampled during exponential phase from three independent cultures. For Figure 6D, increasing light intensities were applied for 5 s, interspaced with 30 s dark, and stirring. At each intensity, a saturating flash was given at t = 5 s, allowing for calculation of PSII yield. Cells were then poisoned with PSII inhibitor DCMU (10 µm) and exposed to the same light treatment. Photochemical rates were calculated as the inverse of fluorescence rise half-lives in DCMU. LEF was hence calculated as the product of PSII yields and photochemical rates at each intensity. For Figure 6E, cells were dark-adapted for 5 min and subsequently exposed to a 7-min illumination at 800 µmol of photons m−2 s−1 superimposed with saturating flashes at 5 s and then every minute, allowing the calculation of PSII yield over time. For Figure 6C, cells were harvested by centrifugation and resuspended in Ficoll to avoid sedimentation. ECS was recorded after single-turnover saturating laser flash. Each measurement was averaged over 15 repeats interspaced by 10 s dark (not taken into account in the error bars, which account for biological triplicates). ECS signals were recorded at 520 and 546 nm; shown are the difference between 520 and 546 nm to eliminate contribution of cytochromes redox signals.
Acetate Quantification in Cultures
For acetate quantification in Chlamydomonas wild-type and ΔCr-tig1 cultures, aliquots were removed, cells were pelleted by centrifugation, and the supernatant was transferred to new tubes. Equal volumes (10 µL) of the respective supernatant were resolved by HPLC with a 1090M Series II (Hewlett-Packard, Waldbronn) on a LiChrospher 100 RP 18 column (125 × 4 mm, 5 µm particle size) in 0.1% (v/v) H3PO4 and 1% (v/v) acetonitrile. Peaks were quantified relative to the peaks of acetate in fresh TAP medium.
Miscellaneous
Thylakoids from Chlamydomonas cells were isolated as described before (Zerges and Rochaix, 1998). Protein extractions, SDS-PAGE, immunoblotting, and antibody affinity purification was published earlier (Willmund and Schroda, 2005). Immunodetection was done using enhanced chemiluminescence and the FUSION-FX7 Advance imaging system (PEQLAB). Immunoblots were quantified with ImageJ according to (https://imagej.nih.gov/ij/). For SDS-PAGE loading, protein samples were adjusted based on protein concentrations determined by Bradford (Bio-Rad) or BCA (Pierce) according to the manufacturer’s manual. Relative cellular concentrations of CrTIG1 and uL1c were determined by loading dilution series of respective purified protein and CC1690 Chlamydomonas whole-cell protein samples. Signal intensities of the whole-cell samples were quantified and compared relative to the signal obtained for the purified proteins. Molar concentrations were calculated assuming reported chloroplast volumes of 129 µm3 (http://bionumbers.hms.harvard.edu) and measured protein concentrations of 20 pg per Chlamydomonas cell. Antiserum against cpSRP54 was raised against full-length, mature cpSRP54 in rabbits. Antibodies described earlier were against CrTIG1, AtTIG1, uL1c (Ries et al., 2017), EcTF (Hesterkamp et al., 1996), uS12c and TufA (Ramundo et al., 2013), HSP70B (Schroda et al., 1999), CF1β(AtpB; Lemaire and Wollman, 1989), cytochrome F (Pierre and Popot, 1993), and RbcL (Rütgers et al., 2017a).
For TF-substrate-renaturing assays and native PAGE, purified AtpB was denatured for 16 h in 3 m urea and 20 mm Tris, pH 8.0 at 4°C. The protein was then diluted to 0.4 µm in refolding buffer (100 mm KPO4, pH 7.5, 1 mm EDTA, 5 mm DTT, 20% [v/v] glycerol, 0.005% [w/v] Ponceau) in the presence or absence of 2 µm purified TIG1 protein, incubated for 5 min at 25°C, spun for 10 min at 1,900g, and separated on a 4% native gel (Douglas et al., 2011).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries as listed in Supplemental Table S2.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Supporting data for Figure 1.
Supplemental Figure S2. Expression control of bacterial and chloroplast TFs in E. coli cells.
Supplemental Figure S3. Aggregate formation of GAPDH and RbcL is not prevented by addition of BSA.
Supplemental Figure S4. Control for Figure 3: peptide arrays do not bind proteins unspecifically.
Supplemental Figure S5. Specificity of antibody directed against the different TF species.
Supplemental Figure S6. Ribosome-association of chloroplast TF is stabilized by chemical cross linking.
Supplemental Figure S7. Phylogenetic analysis of bacterial-type ribosomal protein uL23.
Supplemental Figure S8. Chloroplast TF is not involved in stable protein-protein interactions.
Supplemental Figure S9. Locus validation of the Chlamydomonas tig1 mutant.
Supplemental Figure S10. amiRNA-induced knockdown of Chlamydomonas TF.
Supplemental Figure S11. Temperature shifts and high light do not affect Chlamydomonas tig1 mutant growth.
Supplemental Figure S12. Reduced growth of Chlamydomonas tig1 ami-mutant during prolonged dark exposure and chlorophyll of ΔCr-tig1 mutants.
Supplemental Figure S13. Transcript accumulation of selected Chlamydomonas chloroplast genes during diurnal growth.
Supplemental Figure S14. Linear photosynthetic electron flow is also affected in Chlamydomonas tig1 ami-mutants.
Supplemental Figure S15. Chlorophyll fluorescence of Chlamydomonas wild-type and tig1 cells grown at different light intensities.
Supplemental Figure S16. Starch content of Chlamydomonas wild type and ΔCr-tig1 mutants.
Supplemental Figure S17. Chlamydomonas TF reduction does not cause severe accumulation of unfolded protein aggregates.
Supplemental Figure S18. Chlamydomonas TF mutants show increased plastidic polysomes.
Supplemental Table S1. Primers used for cloning.
Supplemental Table S2. Sequence information about analyzed TF and ribosomal protein L23.
Acknowledgments
The authors thank Bernd Bukau and Günter Kramer for antiserum against EcTF, E. coli strains MC4100 and ΔtigΔdnaK, and advice for complementation assays and Jean-David Rochaix and Silvia Ramundo for antibodies against uS12c and TufA. They thank Francis-André Wollman for discussion of the data, Anja Meffert for technical assistance with the HPLC analyses, and Lisa Ohler for help with the Arabidopsis mutant characterization. For critical reading of this manuscript, the authors thank Karolin Willmund. The authors declare that they have no conflicts of financial interest concerning the contents of this article.
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft (grant CRC 894) to M.J., the Carl-Zeiss fellowship to F.R., and the Deutsche Forschungsgemeinschaft (grants TRR175-A05 and WI3477/2-1) to F.W.
Articles can be viewed without a subscription.
References
- Agashe VR, Guha S, Chang H-C, Genevaux P, Hayer-Hartl M, Stemp M, Georgopoulos C, Hartl FU, Barral JM (2004) Function of trigger factor and DnaK in multidomain protein folding: increase in yield at the expense of folding speed. Cell 117: 199–209 [DOI] [PubMed] [Google Scholar]
- Allen JF, de Paula WB, Puthiyaveetil S, Nield J (2011) A structural phylogenetic map for chloroplast photosynthesis. Trends Plant Sci 16: 645–655 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Bai C, Guo P, Zhao Q, Lv Z, Zhang S, Gao F, Gao L, Wang Y, Tian Z, Wang J, et al. (2015) Protomer roles in chloroplast chaperonin assembly and function. Mol Plant 8: 1478–1492 [DOI] [PubMed] [Google Scholar]
- Bailleul B, Cardol P, Breyton C, Finazzi G (2010) Electrochromism: A useful probe to study algal photosynthesis. Photosynth Res 106: 179–189 [DOI] [PubMed] [Google Scholar]
- Balchin D, Hayer-Hartl M, Hartl FU (2016) In vivo aspects of protein folding and quality control. Science 353: aac4354. [DOI] [PubMed] [Google Scholar]
- Ban N, Beckmann R, Cate JH, Dinman JD, Dragon F, Ellis SR, Lafontaine DL, Lindahl L, Liljas A, Lipton JM, et al. (2014) A new system for naming ribosomal proteins. Curr Opin Struct Biol 24: 165–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieri P, Leibundgut M, Saurer M, Boehringer D, Ban N (2017) The complete structure of the chloroplast 70S ribosome in complex with translation factor pY. EMBO J 36: 475–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttgereit F, Brand MD (1995) A hierarchy of ATP-consuming processes in mammalian cells. Biochem J 312: 163–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon AB, Timko MP (2000) yellow-in-the-dark mutants of Chlamydomonas lack the CHLL subunit of light-independent protochlorophyllide reductase. Plant Cell 12: 559–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calloni G, Chen T, Schermann SM, Chang HC, Genevaux P, Agostini F, Tartaglia GG, Hayer-Hartl M, Hartl FU (2012) DnaK functions as a central hub in the E. coli chaperone network. Cell Reports 1: 251–264 [DOI] [PubMed] [Google Scholar]
- Castanié-Cornet MP, Bruel N, Genevaux P (2014) Chaperone networking facilitates protein targeting to the bacterial cytoplasmic membrane. Biochim Biophys Acta 1843: 1442–1456 [DOI] [PubMed] [Google Scholar]
- Chaux F, Burlacot A, Mekhalfi M, Auroy P, Blangy S, Richaud P, Peltier G (2017) Flavodiiron proteins promote fast and transient O2 photoreduction in Chlamydomonas. Plant Physiol 174: 1825–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua NH, Blobel G, Siekevitz P, Palade GE (1973) Attachment of chloroplast polysomes to thylakoid membranes in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 70: 1554–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua NH, Blobel G, Siekevitz P, Palade GE (1976) Periodic variations in the ratio of free to thylakoid-bound chloroplast ribosomes during the cell cycle of Chlamydomonas reinhardtii. J Cell Biol 71: 497–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: A sequence logo generator. Genome Res 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza AR, Minczuk M (2018) Mitochondrial transcription and translation: Overview. Essays Biochem 62: 309–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeng J, Chan KY, van der Sluis EO, Berninghausen O, Han W, Gumbart J, Schulten K, Beatrix B, Beckmann R (2016) Dynamic behavior of trigger factor on the ribosome. J Mol Biol 428: 3588–3602 [DOI] [PubMed] [Google Scholar]
- Deuerling E, Schulze-Specking A, Tomoyasu T, Mogk A, Bukau B (1999) Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature 400: 693–696 [DOI] [PubMed] [Google Scholar]
- Douglas NR, Reissmann S, Zhang J, Chen B, Jakana J, Kumar R, Chiu W, Frydman J (2011) Dual action of ATP hydrolysis couples lid closure to substrate release into the group II chaperonin chamber. Cell 144: 240–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferbitz L, Maier T, Patzelt H, Bukau B, Deuerling E, Ban N (2004) Trigger factor in complex with the ribosome forms a molecular cradle for nascent proteins. Nature 431: 590–596 [DOI] [PubMed] [Google Scholar]
- Fleischmann TT, Scharff LB, Alkatib S, Hasdorf S, Schöttler MA, Bock R (2011) Nonessential plastid-encoded ribosomal proteins in tobacco: A developmental role for plastid translation and implications for reductive genome evolution. Plant Cell 23: 3137–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman J. (2001) Folding of newly translated proteins in vivo: The role of molecular chaperones. Annu Rev Biochem 70: 603–647 [DOI] [PubMed] [Google Scholar]
- Genevaux P, Keppel F, Schwager F, Langendijk-Genevaux PS, Hartl FU, Georgopoulos C (2004) In vivo analysis of the overlapping functions of DnaK and trigger factor. EMBO Rep 5: 195–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloge F, Becker AH, Kramer G, Bukau B (2014) Co-translational mechanisms of protein maturation. Curr Opin Struct Biol 24: 24–33 [DOI] [PubMed] [Google Scholar]
- Graf M, Arenz S, Huter P, Dönhöfer A, Novácek J, Wilson DN (2017) Cryo-EM structure of the spinach chloroplast ribosome reveals the location of plastid-specific ribosomal proteins and extensions. Nucleic Acids Res 45: 2887–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW. (1993) Origin and evolution of organelle genomes. Curr Opin Genet Dev 3: 884–890 [DOI] [PubMed] [Google Scholar]
- Groth G. (2002) Structure of spinach chloroplast F1-ATPase complexed with the phytopathogenic inhibitor tentoxin. Proc Natl Acad Sci USA 99: 3464–3468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EH, Boynton JE, Gillham NW (1974) Chloroplast ribosome biogenesis in Chlamydomonas. Selection and characterization of mutants blocked in ribosome formation. J Cell Biol 63: 160–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M (2011) Molecular chaperones in protein folding and proteostasis. Nature 475: 324–332 [DOI] [PubMed] [Google Scholar]
- Hassan PA, Rana S, Verma G (2015) Making sense of Brownian motion: Colloid characterization by dynamic light scattering. Langmuir 31: 3–12 [DOI] [PubMed] [Google Scholar]
- Heide H, Nordhues A, Drepper F, Nick S, Schulz-Raffelt M, Haehnel W, Schroda M (2009) Application of quantitative immunoprecipitation combined with knockdown and cross-linking to Chlamydomonas reveals the presence of vesicle-inducing protein in plastids 1 in a common complex with chloroplast HSP90C. Proteomics 9: 3079–3089 [DOI] [PubMed] [Google Scholar]
- Hesterkamp T, Hauser S, Lütcke H, Bukau B (1996) Escherichia coli trigger factor is a prolyl isomerase that associates with nascent polypeptide chains. Proc Natl Acad Sci USA 93: 4437–4441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesterkamp T, Deuerling E, Bukau B (1997) The amino-terminal 118 amino acids of Escherichia coli trigger factor constitute a domain that is necessary and sufficient for binding to ribosomes. J Biol Chem 272: 21865–21871 [DOI] [PubMed] [Google Scholar]
- Hoffmann A, Bukau B, Kramer G (2010) Structure and function of the molecular chaperone Trigger Factor. Biochim Biophys Acta 1803: 650–661 [DOI] [PubMed] [Google Scholar]
- Hoffmann A, Becker AH, Zachmann-Brand B, Deuerling E, Bukau B, Kramer G (2012) Concerted action of the ribosome and the associated chaperone trigger factor confines nascent polypeptide folding. Mol Cell 48: 63–74 [DOI] [PubMed] [Google Scholar]
- Huang GC, Li ZY, Zhou JM, Fischer G (2000) Assisted folding of D-glyceraldehyde-3-phosphate dehydrogenase by trigger factor. Protein Sci 9: 1254–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser CM, Chang HC, Agashe VR, Lakshmipathy SK, Etchells SA, Hayer-Hartl M, Hartl FU, Barral JM (2006) Real-time observation of trigger factor function on translating ribosomes. Nature 444: 455–460 [DOI] [PubMed] [Google Scholar]
- Kandror O, Goldberg AL (1997) Trigger factor is induced upon cold shock and enhances viability of Escherichia coli at low temperatures. Proc Natl Acad Sci USA 94: 4978–4981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohzuma K, Cruz JA, Akashi K, Hoshiyasu S, Munekage YN, Yokota A, Kramer DM (2009) The long-term responses of the photosynthetic proton circuit to drought. Plant Cell Environ 32: 209–219 [DOI] [PubMed] [Google Scholar]
- Kramer DM, Evans JR (2011) The importance of energy balance in improving photosynthetic productivity. Plant Physiol 155: 70–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer G, Rauch T, Rist W, Vorderwülbecke S, Patzelt H, Schulze-Specking A, Ban N, Deuerling E, Bukau B (2002) L23 protein functions as a chaperone docking site on the ribosome. Nature 419: 171–174 [DOI] [PubMed] [Google Scholar]
- Kramer G, Rutkowska A, Wegrzyn RD, Patzelt H, Kurz TA, Merz F, Rauch T, Vorderwülbecke S, Deuerling E, Bukau B (2004) Functional dissection of Escherichia coli trigger factor: Unraveling the function of individual domains. J Bacteriol 186: 3777–3784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer G, Boehringer D, Ban N, Bukau B (2009) The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat Struct Mol Biol 16: 589–597 [DOI] [PubMed] [Google Scholar]
- Kristensen O, Gajhede M (2003) Chaperone binding at the ribosomal exit tunnel. Structure 11: 1547–1556 [DOI] [PubMed] [Google Scholar]
- Lemaire C, Wollman FA (1989) The chloroplast ATP synthase in Chlamydomonas reinhardtii. II. Biochemical studies on its biogenesis using mutants defective in photophosphorylation. J Biol Chem 264: 10235–10242 [PubMed] [Google Scholar]
- Li X, Zhang R, Patena W, Gang SS, Blum SR, Ivanova N, Yue R, Robertson JM, Lefebvre PA, Fitz-Gibbon ST, et al. (2016) An indexed, mapped mutant library enables reverse genetics studies of biological processes in Chlamydomonas reinhardtii. Plant Cell 28: 367–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill R, Crooke E, Guthrie B, Wickner W (1988) The “trigger factor cycle” includes ribosomes, presecretory proteins, and the plasma membrane. Cell 54: 1013–1018 [DOI] [PubMed] [Google Scholar]
- Martinez-Hackert E, Hendrickson WA (2009) Promiscuous substrate recognition in folding and assembly activities of the trigger factor chaperone. Cell 138: 923–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashaghi A, Kramer G, Bechtluft P, Zachmann-Brand B, Driessen AJ, Bukau B, Tans SJ (2013) Reshaping of the conformational search of a protein by the chaperone trigger factor. Nature 500: 98–101 [DOI] [PubMed] [Google Scholar]
- Merz F, Hoffmann A, Rutkowska A, Zachmann-Brand B, Bukau B, Deuerling E (2006) The C-terminal domain of Escherichia coli trigger factor represents the central module of its chaperone activity. J Biol Chem 281: 31963–31971 [DOI] [PubMed] [Google Scholar]
- Merz F, Boehringer D, Schaffitzel C, Preissler S, Hoffmann A, Maier T, Rutkowska A, Lozza J, Ban N, Bukau B, et al. (2008) Molecular mechanism and structure of Trigger Factor bound to the translating ribosome. EMBO J 27: 1622–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettler T, Mühlhaus T, Hemme D, Schöttler MA, Rupprecht J, Idoine A, Veyel D, Pal SK, Yaneva-Roder L, Winck FV, et al. (2014) Systems analysis of the response of photosynthesis, metabolism, and growth to an increase in irradiance in the photosynthetic model organism Chlamydomonas reinhardtii. Plant Cell 26: 2310–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Soltis PS, Bell CD, Burleigh JG, Soltis DE (2010) Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc Natl Acad Sci USA 107: 4623–4628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlhaus T, Weiss J, Hemme D, Sommer F, Schroda M (2011) Quantitative shotgun proteomics using a uniform 15N-labeled standard to monitor proteome dynamics in time course experiments reveals new insights into the heat stress response of Chlamydomonas reinhardtii. Mol Cell Proteomics 10: M110.004739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson OB, Müller-Lucks A, Kramer G, Bukau B, von Heijne G (2016) Trigger Factor reduces the force exerted on the nascent chain by a cotranslationally folding protein. J Mol Biol 428: 1356–1364 [DOI] [PubMed] [Google Scholar]
- Oh E, Becker AH, Sandikci A, Huber D, Chaba R, Gloge F, Nichols RJ, Typas A, Gross CA, Kramer G, et al. (2011) Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell 147: 1295–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzelt H, Rüdiger S, Brehmer D, Kramer G, Vorderwülbecke S, Schaffitzel E, Waitz A, Hesterkamp T, Dong L, Schneider-Mergener J, et al. (2001) Binding specificity of Escherichia coli trigger factor. Proc Natl Acad Sci USA 98: 14244–14249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechmann S, Willmund F, Frydman J (2013) The ribosome as a hub for protein quality control. Mol Cell 49: 411–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez Boerema A, Aibara S, Paul B, Tobiasson V, Kimanius D, Forsberg BO, Wallden K, Lindahl E, Amunts A (2018) Structure of the chloroplast ribosome with chl-RRF and hibernation-promoting factor. Nat Plants 4: 212–217 [DOI] [PubMed] [Google Scholar]
- Pierre Y, Popot JL (1993) Identification of two 4-kDa miniproteins in the cytochrome b6f complex from Chlamydomonas reinhardtii. C R Acad Sci III 316: 1404–1409 [PubMed] [Google Scholar]
- Preissler S, Deuerling E (2012) Ribosome-associated chaperones as key players in proteostasis. Trends Biochem Sci 37: 274–283 [DOI] [PubMed] [Google Scholar]
- Radmer RJ, Kok B (1976) Photoreduction of O2 primes and replaces CO2 assimilation. Plant Physiol 58: 336–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramundo S, Rahire M, Schaad O, Rochaix JD (2013) Repression of essential chloroplast genes reveals new signaling pathways and regulatory feedback loops in chlamydomonas. Plant Cell 25: 167–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries F, Carius Y, Rohr M, Gries K, Keller S, Lancaster CRD, Willmund F (2017) Structural and molecular comparison of bacterial and eukaryotic trigger factors. Sci Rep 7: 10680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix JD. (2011) Regulation of photosynthetic electron transport. Biochim Biophys Acta 1807: 375–383 [DOI] [PubMed] [Google Scholar]
- Russell JB, Cook GM (1995) Energetics of bacterial growth: Balance of anabolic and catabolic reactions. Microbiol Rev 59: 48–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rütgers M, Muranaka LS, Mühlhaus T, Sommer F, Thoms S, Schurig J, Willmund F, Schulz-Raffelt M, Schroda M (2017a) Substrates of the chloroplast small heat shock proteins 22E/F point to thermolability as a regulative switch for heat acclimation in Chlamydomonas reinhardtii. Plant Mol Biol 95: 579–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rütgers M, Muranaka LS, Schulz-Raffelt M, Thoms S, Schurig J, Willmund F, Schroda M (2017b) Not changes in membrane fluidity but proteotoxic stress triggers heat shock protein expression in Chlamydomonas reinhardtii. Plant Cell Environ 40: 2987–3001 [DOI] [PubMed] [Google Scholar]
- Saio T, Guan X, Rossi P, Economou A, Kalodimos CG (2014) Structural basis for protein antiaggregation activity of the trigger factor chaperone. Science 344: 1250494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffitzel E, Rüdiger S, Bukau B, Deuerling E (2001) Functional dissection of trigger factor and DnaK: Interactions with nascent polypeptides and thermally denatured proteins. Biol Chem 382: 1235–1243 [DOI] [PubMed] [Google Scholar]
- Schlünzen F, Wilson DN, Tian P, Harms JM, McInnes SJ, Hansen HA, Albrecht R, Buerger J, Wilbanks SM, Fucini P (2005) The binding mode of the trigger factor on the ribosome: Implications for protein folding and SRP interaction. Structure 13: 1685–1694 [DOI] [PubMed] [Google Scholar]
- Schroda M, Vallon O, Wollman FA, Beck CF (1999) A chloroplast-targeted heat shock protein 70 (HSP70) contributes to the photoprotection and repair of photosystem II during and after photoinhibition. Plant Cell 11: 1165–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroda M, Hemme D, Mühlhaus T (2015) The Chlamydomonas heat stress response. Plant J 82: 466–480 [DOI] [PubMed] [Google Scholar]
- Selbach M, Mann M (2006) Protein interaction screening by quantitative immunoprecipitation combined with knockdown (QUICK). Nat Methods 3: 981–983 [DOI] [PubMed] [Google Scholar]
- Strand DD, Livingston AK, Satoh-Cruz M, Koepke T, Enlow HM, Fisher N, Froehlich JE, Cruz JA, Minhas D, Hixson KK, et al. (2017) Defects in the expression of chloroplast proteins leads to H2O2 accumulation and activation of cyclic electron flow around photosystem I. Front Plant Sci 7: 2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Milward SE, Fan DY, Chow WS, Badger MR (2009) How does cyclic electron flow alleviate photoinhibition in Arabidopsis? Plant Physiol 149: 1560–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor TC, Backlund A, Bjorhall K, Spreitzer RJ, Andersson I (2001) First crystal structure of Rubisco from a green alga, Chlamydomonas reinhardtii. J Biol Chem 276: 48159–48164 [DOI] [PubMed] [Google Scholar]
- Teter SA, Houry WA, Ang D, Tradler T, Rockabrand D, Fischer G, Blum P, Georgopoulos C, Hartl FU (1999) Polypeptide flux through bacterial Hsp70: DnaK cooperates with trigger factor in chaperoning nascent chains. Cell 97: 755–765 [DOI] [PubMed] [Google Scholar]
- Tomoyasu T, Mogk A, Langen H, Goloubinoff P, Bukau B (2001) Genetic dissection of the roles of chaperones and proteases in protein folding and degradation in the Escherichia coli cytosol. Mol Microbiol 40: 397–413 [DOI] [PubMed] [Google Scholar]
- Trösch R, Mühlhaus T, Schroda M, Willmund F (2015) ATP-dependent molecular chaperones in plastids—More complex than expected. Biochim Biophys Acta 1847: 872–888 [DOI] [PubMed] [Google Scholar]
- Trösch R, Barahimipour R, Gao Y, Badillo-Corona JA, Gotsmann VL, Zimmer D, Mühlhaus T, Zoschke R, Willmund F (2018) Commonalities and differences of chloroplast translation in a green alga and land plants. Nat Plants 4: 564–575 [DOI] [PubMed] [Google Scholar]
- Tyedmers J, Mogk A, Bukau B (2010) Cellular strategies for controlling protein aggregation. Nat Rev Mol Cell Biol 11: 777–788 [DOI] [PubMed] [Google Scholar]
- Weckbecker D, Longen S, Riemer J, Herrmann JM (2012) Atp23 biogenesis reveals a chaperone-like folding activity of Mia40 in the IMS of mitochondria. EMBO J 31: 4348–4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng ML, Ruhlman TA, Jansen RK (2016) Plastid-nuclear interaction and accelerated coevolution in plastid ribosomal genes in Geraniaceae. Genome Biol Evol 8: 1824–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmund F, Schroda M (2005) HEAT SHOCK PROTEIN 90C is a bona fide Hsp90 that interacts with plastidic HSP70B in Chlamydomonas reinhardtii. Plant Physiol 138: 2310–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmund F, Mühlhaus T, Wojciechowska M, Schroda M (2007) The NH2-terminal domain of the chloroplast GrpE homolog CGE1 is required for dimerization and cochaperone function in vivo. J Biol Chem 282: 11317–11328 [DOI] [PubMed] [Google Scholar]
- Willmund F, Dorn KV, Schulz-Raffelt M, Schroda M (2008) The chloroplast DnaJ homolog CDJ1 of Chlamydomonas reinhardtii is part of a multichaperone complex containing HSP70B, CGE1, and HSP90C. Plant Physiol 148: 2070–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Catalanotti C, Wittkopp TM, Posewitz MC, Grossman AR (2015) Algae after dark: mechanisms to cope with anoxic/hypoxic conditions. Plant J 82: 481–503 [DOI] [PubMed] [Google Scholar]
- Zerges W, Rochaix JD (1998) Low density membranes are associated with RNA-binding proteins and thylakoids in the chloroplast of Chlamydomonas reinhardtii. J Cell Biol 140: 101–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziehe D, Dünschede B, Schünemann D (2017) From bacteria to chloroplasts: Evolution of the chloroplast SRP system. Biol Chem 398: 653–661 [DOI] [PubMed] [Google Scholar]
- Zones JM, Blaby IK, Merchant SS, Umen JG (2015) High-resolution profiling of a synchronized diurnal transcriptome from Chlamydomonas reinhardtii reveals continuous cell and metabolic differentiation. Plant Cell 27: 2743–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoschke R, Bock R (2018) Chloroplast translation: structural and functional organization, operational control, and regulation. Plant Cell 30: 745–770 [DOI] [PMC free article] [PubMed] [Google Scholar]