Abstract
Reduced lung function predicts mortality and is key to the diagnosis of chronic obstructive pulmonary disease (COPD). In a genome-wide association study in 400,102 individuals of European ancestry, we define 279 lung function signals, 139 of which are new. In combination, these variants strongly predict COPD in independent patient populations. Furthermore, the combined effect of these variants showed generalizability across smokers and never-smokers, and across ancestral groups. We highlight biological pathways, known and potential drug targets for COPD and, in phenome-wide association studies, autoimmune-related and other pleiotropic effects of lung function associated variants. This new genetic evidence has potential to improve future preventive and therapeutic strategies for COPD.
Editorial summary:
A genome-wide association study in over 400,000 individuals identifies 139 new signals for lung function. These variants can predict chronic obstructive pulmonary disease in independent, trans-ethnic cohorts.
Introduction
Impaired lung function is predictive of mortality1 and is the key diagnostic criterion for chronic obstructive pulmonary disease (COPD). Globally, COPD accounted for 2.9 million deaths in 20162, being one of the key causes of both Years of Life Lost and Years Lived with Disability worldwide3. Determinants of maximally attained lung function and of lung function decline can influence the risk of developing COPD. Tobacco smoking is the single largest risk factor for COPD, although other environmental exposures and genetic makeup are important4,5. Genetic variants associated with lung function and COPD susceptibility can provide etiological insights, assisting with risk prediction, as well as drug target identification and validation6. Whilst there has been considerable progress in identifying genetic markers associated with lung function and risk of COPD4,7–19 seeking a high yield of associated genetic variants is key to progressing knowledge because: (i) implication of multiple molecules in each pathway will be needed to build an accurate picture of the pathways underpinning development of COPD; (ii) not all proteins identified will be druggable and; (iii) combining information across multiple variants can improve prediction of disease susceptibility.
Through new detailed quality control and analyses of spirometric measures of lung function in UK Biobank and expansion of the SpiroMeta Consortium, we undertook a large genome-wide association study of lung function. Our study entailed a near seven-fold increase in sample size over previous studies of similar ancestry to address the following aims: (i) to generate a high yield of genetic markers associated with lung function; (ii) to confirm and fine-map previously reported lung function signals; (iii) to investigate the putative causal genes and biological pathways through which lung function associated variants act, and their wider pleiotropic effects on other traits; and (iv) to generate a weighted genetic risk score for lung function and test its association with COPD susceptibility in individuals of European and other ancestries.
Results
139 new signals for lung function
We increased the sample size available for the study of quantitative measures of lung function in UK Biobank by refining the quality control of spirometry based on recommendations of the UK Biobank Outcomes Adjudication Working Group (Supplementary Note). Genome-wide association analyses of forced expired volume in 1 second (FEV1), forced vital capacity (FVC) and FEV1/FVC were undertaken in 321,047 individuals in UK Biobank (Supplementary Table 1) and in 79,055 individuals from the SpiroMeta Consortium (Supplementary Tables 2 and 3). A linear mixed model implemented in BOLT-LMM20 was used for UK Biobank to account for relatedness and fine-scale population structure (Online Methods). A total of 19,819,130 autosomal variants imputed in both UK Biobank and SpiroMeta were analyzed. Peak expiratory flow (PEF) was also analyzed genome-wide in UK Biobank and up to 24,218 samples from SpiroMeta. GWAS results in UK Biobank were adjusted for the intercept of LD score regression21, but SpiroMeta and the meta-analysis were not, as intercepts were close to 1.00 (Online Methods). All individuals included in the genome-wide analyses were of European ancestry (Supplementary Figure 1 and Supplementary Note).
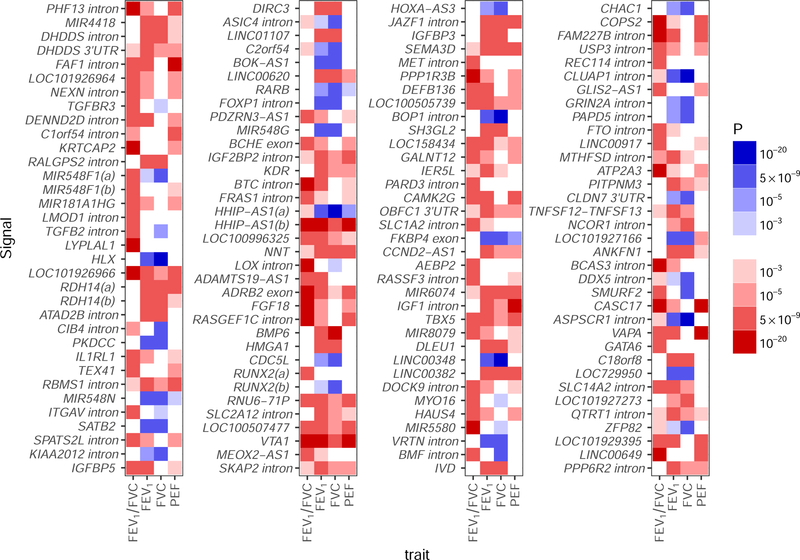
To maximize statistical power for discovery of new signals, whilst maintaining stringent significance thresholds to minimize reporting of false positives, we adopted a study design incorporating both two-stage and one-stage approaches (Figure 1). In the two-stage analysis, 99 new distinct signals, defined using conditional analyses22, were associated with one or more traits at P<5×10−9 (23) in UK Biobank and showed association (P<10−3) with a consistent direction of effect in SpiroMeta (“Tier 1” signals, Supplementary Figure 2; Supplementary Table 4). In the one-stage analysis, we meta-analyzed UK Biobank and SpiroMeta (up to 400,102 individuals) and 40 additional new distinct signals associated with one or more lung function traits reaching P<5×10−9 were identified (Supplementary Figure 2, Supplementary Table 4) that were also associated with P<10−3 separately in UK Biobank and in SpiroMeta, with consistent direction of effect (“Tier 2” signals). An additional 323 autosomal signals were significantly associated with one or more lung function traits in the meta-analysis of UK Biobank and SpiroMeta (P<5×10−9) and reached P<10−3 for association in only one of UK Biobank or SpiroMeta (“Tier 3” signals, Supplementary Table 5). Analysis of chromosome X variants in 359,226 individuals (321,027 UK Biobank and 38,199 SpiroMeta15) gave an additional five Tier 3 signals. Only the 139 signals meeting Tier 1 and Tier 2 criteria were followed up further. The strength and direction of association of the sentinel variant (the variant in each signal with the lowest P value) for these 139 new signals across all 4 lung function traits are shown in Figure 2. Of the 139 signals, 131 were associated with at least two lung function traits at P<10−3, eight signals were unique to FEV1/FVC and no signals were unique to FEV1, FVC or PEF at this threshold.
Figure 1: Study design.
Tier 1 signals had P<5×10−9 in UK Biobank and P<10−3 in SpiroMeta with consistent direction of effect.
Tier 2 signals had P<5×10−9 in the meta-analysis of UK Biobank and SpiroMeta with P<10−3 in UK Biobank and P<10−3 in SpiroMeta with consistent directions of effect. Signals with P<5×10−9 in the meta-analysis of UK Biobank and SpiroMeta, and that had consistent directions of effect but did not meet P<10−3 in both cohorts were reported as Tier 3.
Figure 2: Strength and direction of association across four lung function traits for 139 novel signals:
Signals are in chromosome and genomic position order from top to bottom then left to right. Red indicates a decrease in the lung function trait; blue indicates an increase. All effects are aligned to the allele associated with decreased FEV1/FVC, hence the FEV1/FVC column is only red or white. P-values are from the meta-analysis of UK Biobank and SpiroMeta (n=400,102). The scale points are thresholds used for (i) confirmation in 2-stage analysis and 1-stage analysis (P<10−3); (ii) confirmation of association of previous signals (P<10−5); (iii) signal selection in 2-stage and 1-stage analysis (P<5×10−9); capped at (P<10−20). FEV1, forced expired volume in 1 second; FVC, forced vital capacity; PEF, peak expiratory flow
We assessed whether any of these 139 signals associated with lung function could be driven via an underlying association with smoking behavior (Online Methods). Only rs193686 (Supplementary Table 6) was associated with smoking behavior. Whilst rs193686 was associated with smoking initiation (P=9.18×10−6), the allele associated with smoking initiation was associated with increased lung function in never smokers (FEV1/FVC P=5.28×10−10, Supplementary Table 7). Therefore, this signal was retained for further analysis.
A total of 279 signals of association for lung function
Of 157 previously published autosomal signals of association with lung function and COPD3,6–18, 142 were associated at P<10−5 in UK Biobank (Online Methods, Supplementary Figure 3, Supplementary Table 8). Two sentinel variants (rs1689510 and rs11134789) were associated with smoking initiation (Supplementary Table 6), but were also associated with lung function in never smokers (Supplementary Table 7). SNP rs17486278 at CHRNA5 and rs11667314 near CYP2A6 were each associated with cigarettes per day (Supplementary Table 6); neither were significantly associated with lung function among never smokers and so were excluded from further analysis. This brings the total number of distinct signals of association with lung function to 279 (Supplementary Table 9). None of these variants showed interaction with ever-smoking status (P>1.8×10−4, Online Methods, Supplementary Table 7). Using the effect estimates, allele frequencies and assuming a total heritability of 40%24,25 (Online Methods), we calculated that the 140 previously reported lung function signals showing association in this study (UK Biobank P<10−5) explained 5.0%, 3.4%, 9.2% and 4.5% of the estimated heritability of FEV1, FVC, FEV1/FVC and PEF, respectively. The 139 new signals reported here, explain an additional 4.3%, 3.3%, 3.9% and 3.3% of the estimated heritability, respectively.
Identification of putative causal genes
Bayesian refinement was undertaken for each signal, using the meta-analysis of UK Biobank and SpiroMeta, to identify the set of variants that were 99% likely to contain the underlying causal variant (assuming the causal variant has been analyzed, Online Methods, Supplementary Table 10, Supplementary Data 1 and Supplementary Data 2).
To identify putative causal genes for each signal, we identified deleterious variants and variants associated with gene expression (expression quantitative trait loci (eQTLs)) or protein levels (protein quantitative trait loci (pQTLs)) within each 99% credible set for all new and previously reported signals outside the HLA region (Online Methods).
There were 25 SNPs, located in 22 unique genes, which were annotated as potentially deleterious (Online Methods, Supplementary Table 11). Amongst our new signals, there were 10 variants annotated as deleterious in 9 different genes: DOCK9 (rs117633128), CEP72 (rs12522955), BCHE (rs1799807), DST (rs11756977), KIAA0753 (rs2304977, rs9889363), LRRC45 (rs72861736), BTC (rs11938093), C2orf54 (rs6709469) and IER5L (rs184457). Of these, the missense variant in BCHE (rs1799807) had the highest posterior probability (0.996) in its respective credible set, was low frequency (minor allele frequency (MAF)=1.95%) and results in an amino acid change from aspartic acid (D) to glycine (G), known to affect the function of the encoded butyrylcholinesterase enzyme by altering substrate binding26. The two common missense variants in KIAA0753 were within the credible set of new signal rs4796334. KIAA0753, CEP72 and LRRC45 all encode proteins with a role in ciliogenesis or cilia maintenance27–31, and all are highly expressed in the airway epithelium32.
Variants in the 99% credible sets were queried in three eQTL resources to identify associations with gene expression in lung33–35 (n=1,111; Supplementary Table 12), blood36 (n=4,896) and a subset of Genotype-tissue Expression (GTEx)37 tissues (max n=388, Online Methods). The tissues included from GTEx were lung and blood, plus nine tissues containing smooth muscle (Online Methods). The latter were chosen based on previous reports of enrichment of lung function GWAS signals in smooth muscle-containing tissues18,38. We identified 88 genes, implicated by 58 of the 279 signals, for which the most significant SNP associated with expression of that gene in the respective eQTL resource was within one of the 99% credible sets (Supplementary Table 13).
We checked credible set variants for association with protein levels in a pQTL study39 comprising SNP associations for 3,600 plasma proteins (Online Methods). We found five proteins with a sentinel pQTL contained within our lung function credible set: ECM1, THBS4, NPNT, C1QTNF5 and SCARF2 (Supplementary Table 14).
In total, 107 putative causal genes were identified (Table 1), amongst which, we highlight 75 for the first time as putative causal genes for lung function (43 implicated by a new signal and 32 newly implicated by a previous signal18).
Table 1:
Genes implicated using gene expression data, protein level data and functional annotation
| Gene | Phenotype | Other traits | Novel Tier/ Previous | Sentinel SNP | Position (b37) | COPD risk/alt | Functionally implicated genes |
|---|---|---|---|---|---|---|---|
| DHDDS (intron) | FVC | FEV1 | Tier 2 | rs9438626 | 1:26,775,367 | G/C | DHDDS† |
| DHDDS (3’-UTR) | FEV1 | Tier 1 | rs12096239 | 1:26,796,922 | C/G | HMGN2†, DHDDS† | |
| NEXN (intron) | FEV1/FVC | Tier 1 | rs9661687 | 1:78,387,270 | T/C | NEXN† | |
| DENND2D (intron) | FEV1/FVC | FEV1 | Tier 1 | rs9970286 | 1:111,737,398 | G/A | CEPT1†, CHI3L2†, DRAM2† |
| C1orf54 (intron) | PEF | Tier 1 | rs11205354 | 1:150,249,101 | C/A | MRPS21†, RPRD2†, ECM1‡ | |
| KRTCAP2 | FEV1/FVC | Tier 1 | rs141942982 | 1: 155153537 | T/C | THBS4‡ | |
| RALGPS2 (intron) | FEV1 | FVC | Tier 1 | rs4651005 | 1:178,719,306 | C/T | ANGPTL1† |
| LMOD1 (intron) | FEV1/FVC | Tier 2 | rs4309038 | 1:201,884,647 | G/C | SHISA4† | |
| ATAD2B (intron) | FVC | FEV1 | Tier 2 | rs13009582 | 2:24,018,480 | G/A | UBXN2A† |
| PKDCC | FVC | FEV1 | Tier 1 | rs4952564 | 2:42,243,850 | A/G | PKDCC† |
| ITGAV (intron) | FEV1/FVC | Tier 1 | rs2084448 | 2:187,530,520 | C/T | ITGAV† | |
| SPATS2L (intron) | FEV1/FVC | Tier 2 | rs985256 | 2:201,208,692 | C/A | SPATS2L† | |
| C2orf54 | FVC | Tier 1 | rs6437219 | 2:241,844,033 | C/T | C2orf54†* | |
| MIR548G | FVC | FEV1 | Tier 1 | rs1610265 | 3:99,420,192 | T/C | FILIP1L† |
| BCHE (exon) | FEV1/FVC | Tier 1 | rs1799807 | 3:165,548,529 | C/T | BCHE* | |
| BTC (intron) | FEV1/FVC | FEV1 | Tier 1 | rs62316310 | 4:75,676,529 | G/A | BTC* |
| LOC100996325 | FEV1 | FEV1/FVC | Tier 1 | rs11739847 | 5:609,661 | A/G | CEP72* |
| RNU6–71P | FEV1 | FEV1/FVC, PEF | Tier 1 | rs2894837 | 6:56,336,406 | G/A | DST* |
| JAZF1 (intron) | FEV1 | FVC, PEF | Tier 1 | rs1513272 | 7:28,200,097 | C/T | JAZF1† |
| MET (intron) | FEV1/FVC | Tier 2 | rs193686 | 7:116,431,427 | T/C | MET† | |
| IER5L | FEV1 | Tier 2 | rs967497 | 9:131,943,843 | G/A | CRAT†, PPP2R4†, IER5L* | |
| DOCK9 | FEV1/FVC | Tier 1 | rs11620380 | 13:99,665,512 | A/C | DOCK9* | |
| CHAC1 | FVC | Tier 1 | rs4924525 | 15:41,255,396 | A/C | INO80†, CHP1†, RAD51† | |
| ATP2A3 | FEV1/FVC | Tier 1 | rs8082036 | 17:3,882,613 | G/C | ATP2A3† | |
| PITPNM3 | FEV1 | Tier 2 | rs4796334 | 17:6,469,793 | A/G | KIAA0753†*, TXNDC17†, PITPNM3† | |
| TNFSF12-TNFSF13 | FEV1 | Tier 2 | rs4968200 | 17:7,448,457 | C/G | TNFSF13†, SENP3† | |
| NCOR1 (intron) | FVC | Tier 2 | rs34351630 | 17:16,030,520 | C/T | ADORA2B†, TTC19† | |
| ASPSCR1 (intron) | FVC | FEV1 | Tier 1 | rs59606152 | 17:79,952,944 | C/T | LRRC45* |
| C18orf8 | FVC | FEV1 | Tier 1 | rs303752 | 18:21,074,255 | A/G | C18orf8† |
| ZFP82 | FVC | Tier 2 | rs2967516 | 19:36,881,643 | A/G | ZFP14†, ZFP82† | |
| MFAP2 | FEV1/FVC | FVC, PEF | Previous | rs9435733 | 1:17,308,254 | C/T | MFAP2† |
| LOC101929516 | FEV1/FVC | FEV1, PEF | Previous | rs755249 | 1:39,995,074 | T/C | PABPC4† |
| TGFB2 | PEF | Previous | rs6604614 | 1:218,631,452 | C/G | TGFB2† | |
| TRAF3IP1 | FEV1 | FEV1/FVC | Previous | rs6710301 | 2:239,441,308 | C/A | ASB1* |
| SLMAP (intron) | FEV1 | FVC, FEV1/FVC, PEF | Previous | rs6445932 | 3:57,879,611 | T/G | SLMAP† |
| RSRC1 (intron) | FVC | FEV1 | Previous | rs12634907 | 3:158,226,886 | G/A | RSRC1† |
| GSTCD (intron) | FEV1 | FVC, FEV1/FVC | Previous | rs11722225 | 4:106,766,430 | T/C | INTS12† |
| NPNT (intron) | FEV1/FVC | FEV1, FVC, PEF | Previous | rs34712979 | 4:106,819,053 | A/G | NPNT†‡ |
| AP3B1 (intron) | FVC | Previous | rs425102 | 5:77,396,400 | G/T | AP3B1† | |
| SPATA9 | FEV1/FVC | Previous | rs987068 | 5:95,025,146 | C/G | RHOBTB3† | |
| P4HA2-AS1 | FVC | Previous | rs3843503 | 5:131,466,629 | A/T | SLC22A5†, P4HA2†, C1QTNF5‡ | |
| CYFIP2 (intron) | FEV1/FVC | FEV1, PEF | Previous | rs11134766 | 5:156,908,317 | T/C | ADAM19† |
| ADAM19 (intron) | FEV1/FVC | FEV1, PEF | Previous | rs11134789 | 5:156,944,199 | A/C | ADAM19†* |
| DSP (intron) | FEV1/FVC | Previous | rs2076295 | 6:7,563,232 | T/G | DSP† | |
| MIR588 | FVC | FEV1 | Previous | rs6918725 | 6:126,990,392 | T/G | CENPW† |
| GPR126 (exon) | FEV1/FVC | FVC, PEF | Previous | rs17280293 | 6:142,688,969 | A/G | GPR126* |
| C1GALT1 (intron) | FEV1/FVC | Previous | rs4318980 | 7:7,256,490 | A/G | C1GALT1† | |
| QSOX2 (3’-UTR) | FVC | FEV1 | Previous | rs7024579 | 9:139,100,413 | T/C | QSOX2† |
| DNLZ (intron) | FVC | Previous | rs4073153 | 9:139,259,349 | G/A | SNAPC4†, CARD9†, INPP5E† | |
| CDC123 (intron) | FEV1/FVC | FEV1, FVC, PEF | Previous | rs7090277 | 10:12,278,021 | T/A | NUDT5† |
| MYPN (intron) | FVC | FEV1 | Previous | rs10998018 | 10:69,962,954 | A/G | MYPN* |
| EML3 (intron) | FEV1 | FVC | Previous | rs71490394 | 11:62,370,155 | G/A | EEF1G†, ROM1†*, EML3†* |
| ARHGEF17 (intron) | FEV1/FVC | FEV1 | Previous | rs2027761 | 11:73,036,179 | C/T | FAM168A†, ARHGEF17†* |
| RAB5B (intron) | FEV1 | Previous | rs1689510 | 12:56,396,768 | C/G | CDK2† | |
| LRP1 (intron) | FEV1/FVC | PEF | Previous | rs11172113 | 12:57,527,283 | T/C | LRP1† |
| FGD6 (intron) | FEV1/FVC | Previous | rs113745635 | 12:95,554,771 | T/C | FGD6† | |
| RPAP1 | FEV1/FVC | Previous | rs2012453 | 15:41,840,238 | G/A | ITPKA†, LTK†, TYRO3†, RPAP1† | |
| AAGAB | FVC | Previous | rs12917612 | 15:67,491,274 | A/C | AAGAB†, SMAD3†, IQCH† | |
| THSD4 (intron) | FEV1/FVC | FEV1, PEF | Previous | rs1441358 | 15:71,612,514 | G/T | THSD4† |
| IL27 | FEV1 | Previous | rs12446589 | 16:28,870,962 | A/G | SBK1†, TUFM†, CCDC101†, SULT1A1†, SULT1A2†*, SH2B1†, NPIPL1†, CLN3†, ATXN2L†, EIF3C† | |
| MMP15 (intron) | FEV1/FVC | Previous | rs11648508 | 16:58,063,513 | G/T | MMP15† | |
| SSH2 (intron) | FEV1/FVC | PEF | Previous | rs2244592 | 17:28,072,327 | A/G | EFCAB5† |
| FBXL20 (intron) | FVC | FEV1 | Previous | rs8069451 | 17:37,504,933 | C/T | CRKRS†, FBXL20† |
| MAPT-AS1 | FEV1 | FVC, PEF | Previous | rs79412431 | 17:43,940,021 | A/G | LRRC37A4†, MAPT* |
| TSEN54 (intron) | FEV1 | Previous | rs9892893 | 17:73,525,670 | G/T | CASKIN2†, TSEN54* | |
| LTBP4 (exon) | FEV1/FVC | PEF | Previous | rs34093919 | 19:41,117,300 | G/A | LTBP4* |
| ABHD12 (intron) | FEV1 | Previous | rs2236180 | 20:25,282,608 | C/T | PYGB†* | |
| UQCC1 (5’-UTR) | FVC | FEV1, PEF | Previous | rs143384 | 20:34,025,756 | G/A | UQCC1†, GDF5† |
| SLC2A4RG (intron) | FVC | FEV1 | Previous | rs4809221 | 20:62,372,706 | A/G | LIME1† |
| SCARF2 (intron) | FEV1 | FEV1/FVC | Previous | rs9610955 | 22:20,790,723 | C/G | SCARF2*‡ |
Genes implicated by eQTL signals: Lung eQTL (n=1,111) and Blood eQTL (n=4,896) datasets and eleven GTEx (V7) tissues were screened: Artery Aorta (n=267), Artery Coronary (n=152), Artery Tibial (n=388), Colon Sigmoid (n=203), Colon Transverse (n=246), Esophagus Gastroesophageal Junction (n=213), Esophagus Muscularis (n=335), Lung (n=383), Small Intestine Terminal Ileum (n=122), Stomach (n=237), and Whole Blood (n=369); see Supplementary Table 13 for direction of gene expression for the COPD risk (FEV1/FVC reducing) allele.
Genes implicated by pQTL signals: pQLT look up in 3,600 plasma proteins (n up to 3,300).
Genes implicated because they contain a deleterious variant (Supplementary Table 11).
“Other traits” column lists the other lung function traits for which the sentinel was associated at P<5×10−9 in the meta-analysis of UK Biobank and SpiroMeta.
In total, 107 putative causal genes were identified: 8 by both a deleterious variant and an eQTL signal (including KIAA0753 implicated by two deleterious variants), 1 (NPNT) by both an eQTL and a pQTL signal, 1 (SCARF2) by both a deleterious variant and a pQTL signal, 13 by a deleterious variant only, 81 by an eQTL signal only and 3 by a pQTL signal only
Pathway analysis
We tested whether these 107 putative causal genes were enriched in gene sets and biological pathways (Online Methods), finding an enrichment of genes in elastic fiber and extracellular matrix organization pathways, and a number of gene ontologies including gene sets relating to the cytoskeleton and processes involved in ciliogenesis (Supplementary Table 15). Whilst the enrichment in elastic fiber-related pathways is consistent with our previous study18, enrichment in these pathways was further supported in this analysis by two new genes, ITGAV (at a new signal) and GDF5 (a newly implicated gene for a previously reported signal), and by strengthened eQTL evidence for TGFB2 and MFAP2 at two previously reported signals. The presence of TGFB2, GDF5 and SMAD3 in our list of 107 genes resulted in enrichment of a TGF-β superfamily signalling pathway (TGF-Core) and related gene ontology terms (Supplementary Table 15).
Functional enrichment analyses
Using stratified LD-score regression40, we showed that FEV1/FVC and FVC heritability is significantly enriched at variants overlapping histone marks that are specific to lung, fetal lung, and smooth muscle-containing cell lines. SNPs that overlap with H3K4me1 marks that are specific to fetal lung correspond to 6.99% of the input SNPs yet explain 57.09% (P=2.85×10−25) and 35.84% (P=4.19×10−21) of the SNP-chip heritability for FEV1/FVC and FVC, respectively (Supplementary Table 16).
We also tested enrichment of (i) FEV1/FVC and (ii) FVC SNPs at DNase I hypersensitive site (DHS) hotspots using GARFIELD41 (Online Methods). For FEV1/FVC results, we see significant enrichment across most cell lines with increased fold-enrichment in fetal and adult lung, fetal muscle and fibroblasts (Supplementary Figure 4a). For FVC, we see similar broad significant enrichment without evidence of increased enrichment in a subset of tissues (Supplementary Figure 4b) suggesting that SNPs influencing FVC may act via more complex and broader developmental pathways.
We used DeepSEA42 to identify whether our signals were predicted to have a chromatin effect in lung-related cell lines. We identified 10 signals (including 5 new signals) for which the SNP with the largest posterior probability of being causal also had a significant predicted effect on a DHS in lung-related cells (Supplementary Table 17). This included a new signal near SMURF2 (rs11653958).
Drug targets
All 107 putative causal genes were investigated for known gene-drug interactions43 (Supplementary Table 18). We highlight two examples of new genetic signals implicating targets for drugs in development for indications other than COPD. One of our new signals is an eQTL for ITGAV. ITGAV encodes a component of the αvβ6 integrin heterodimer, which is inhibited by a monoclonal antibody in development for pulmonary fibrosis (NCT01371305) and for which the small molecule GSK3008348 (NCT03069989) is an antagonist44. Integrins have an emerging role as local activators of TGFβ and specifically the avb6 integrin heterodimer can activate latent-TGFβ45. In our study, the allele associated with reduced expression of ITGAV (Supplementary Table 13) was associated with increased lung function (Supplementary Table 9) suggesting that inhibitors of αvβ6 integrin might also have a beneficial effect in COPD. Another new signal is associated with expression of TNFSF13 (synonym APRIL), which encodes a cytokine of the TNF ligand family. Atacicept blocks B cell stimulation by TNFSF13 (as well as by BLyS) and reduced systemic lupus erythematosus disease activity in a recent Phase IIb trial46. In our study, the allele associated with decreased expression of TNFSF13 was associated with reduced FEV1, indicating that vigilance for pulmonary consequences of atacicept may be warranted.
Association with FEV1/FVC and COPD in multiple ancestries
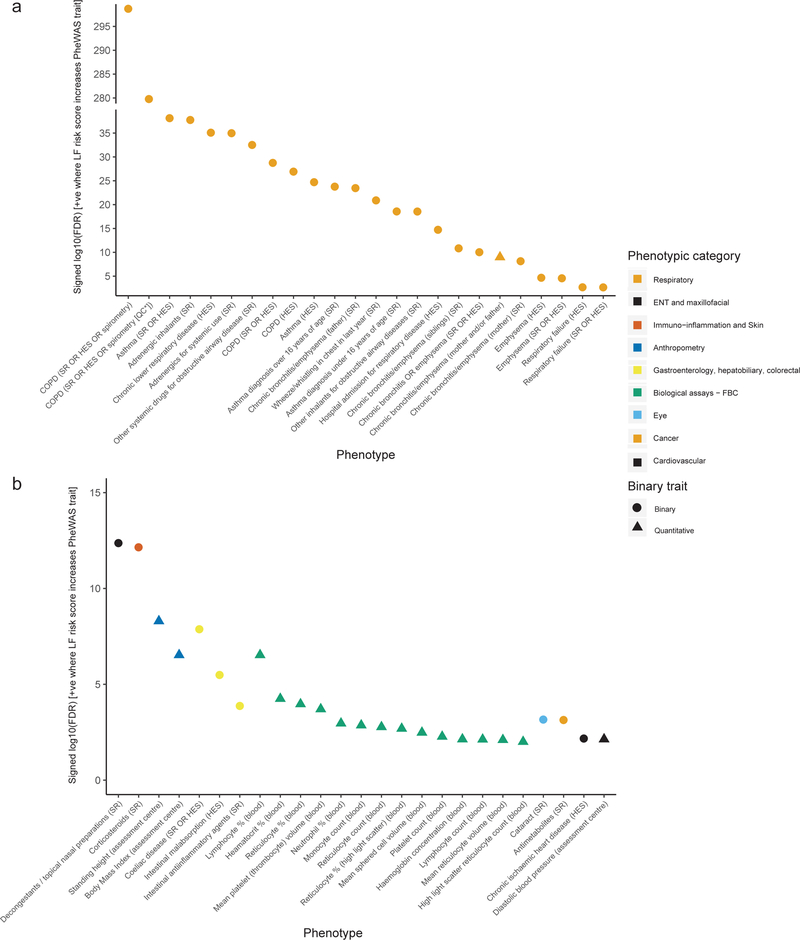
We constructed a genetic risk score (GRS) weighted by FEV1/FVC effect sizes comprising all 279 sentinel variants, and tested for association with FEV1/FVC and GOLD Stage 2–4 COPD (FEV1/FVC<0.7 and FEV1<80% predicted) in different ancestry groups in UK Biobank, and China Kadoorie Biobank (Online Methods, Supplementary Table 19). UK Biobank participants of non-European ancestry were not included in the discovery analyses. The GRS was associated with a significant decrease in lung function, and corresponding significant increase in COPD risk in each of the independent ancestry groups (Figure 3a).
Figure 3: Association of weighted genetic risk score (wGRS) with COPD and FEV1/FVC.
a. Association of the wGRS with FEV1/FVC and COPD in UK Biobank (UKB) and China Kadoorie Biobank (CKB) (Supplementary Table 19). Left-hand axis: standard deviation (SD) change in FEV1/FVC per SD increase in wGRS (light grey bars, N=total sample size). Right-hand axis: the translation of this effect to COPD (GOLD stage 2–4) odds ratio (OR) per SD increase in wGRS in the same individuals for UKB ancestries with >100 COPD cases (dark grey bars, N=number of cases + number of controls. Whiskers represent 95% confidence intervals. Some variants in the wGRS were discovered in UKB Europeans, therefore UKB Europeans are shown for reference only (far left, ‘Discovery sample’). All other ancestral groups are independent to UKB Europeans.
b. OR for COPD per SD increase in wGRS in six study groups. COPD was defined using GOLD 2–4 criteria (Supplementary Table 21: means and SDs of risk scores). The vertical black line indicates the null effect (OR=1). The point estimate of each study is represented by a box proportional to study weight; whiskers represent 95% confidence intervals. The diamond represents a fixed effect meta-analysis of the five European-ancestry groups, the width of which represents the 95% confidence interval (I2 statistic=0).
c. OR for COPD according to deciles of the wGRS, with decile 1 (the 10% of individuals with the lowest GRS) as the reference group. Each point represents a meta-analysis of results for a given comparison (e.g. decile 2 vs reference, decile 3 vs reference, etc.) in five external European-ancestry study groups (COPDGene, ECLIPSE, GenKOLS, SPIROMICS, NETT-NAS). Deciles were calculated and models were run in each group separately. Error bars show 95% confidence intervals (Supplementary Table 22).
We tested for a GRS interaction with smoking in European ancestry individuals in UK Biobank47. No statistical interaction was seen for FEV1/FVC (interaction term −0.002 per SD change in GRS, 95% CI: [0.009, 0.005], P=0.532), whilst the findings for COPD were consistent with a slightly smaller effect of the GRS in ever-smokers (odds ratio (OR) for ever-smoking-GRS interaction term per SD change in GRS 0.96, 95% CI: [0.92, 0.99], P=0.015).
The association of the GRS with COPD susceptibility was additionally tested in five independent COPD case-control studies (Supplementary Table 20, Online Methods). Similar effect size estimates were seen across each of the 5 European ancestry studies (Figure 3b); in the meta-analysis of these studies (n=6,979 cases and 3,915 controls), the odds ratio for COPD per standard deviation of the weighted GRS was 1.55 (95% CI: [1.48, 1.62]), P=2.87×10−75 (Supplementary Table 21). The GRS was also associated with COPD in individuals of African-American ancestry in COPDGene (P=8.36×10−7), albeit with a smaller effect size estimate, odds ratio=1.26 (95% CI: [1.15, 1.37]).
To aid clinical interpretation, we divided individuals in each of the five European ancestry COPD case-control studies into deciles, according to their value of the weighted GRS. The odds ratio for COPD in members of the highest GRS decile compared to the lowest GRS decile was 4.73 (95% CI: [3.79, 5.90]), P=3.00×10−43 (Figure 3c, Supplementary Table 22). We calculated the population attributable risk fraction (Supplementary Note) and estimated that the proportion of COPD cases attributable to risk scores above the first GRS decile was 54.6% (95% CI: [50.6%, 58.4%]).
Incorporation of the GRS into a risk model already comprising available clinical information (age, sex, height and pack-years of smoking in COPDGene non-Hispanic Whites) led to a statistically significant (P=3.33×10−10), yet modest, increase in the area under the curve, from 0.751 to 0.771 (Supplementary Note). Based on our estimated GRS relative risk and absolute risk estimates of COPD48, one would expect the highest GRS risk decile group of smokers to have an absolute risk of developing COPD by approximately 70 years of age of 82.4%, versus 17.4% for the lowest GRS decile (Supplementary Note).
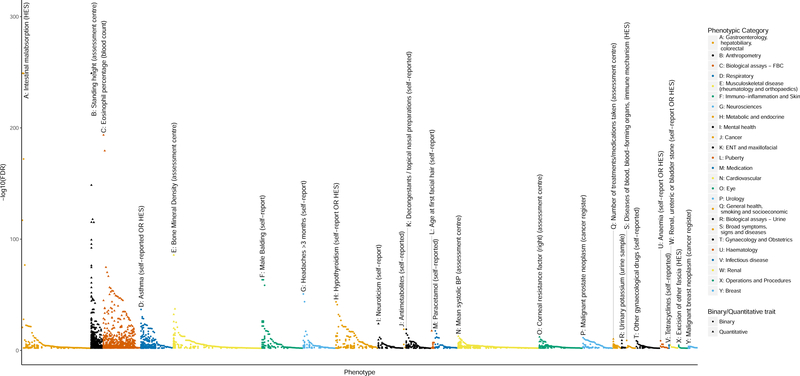
Pleiotropy and phenome-wide association studies
As phenome-wide association studies (PheWAS) can provide evidence mimicking pharmacological interventions of drug targets in humans and informing drug development49, we undertook a PheWAS of 2,411 phenotypes in UK Biobank (Online Methods, Figure 4, Supplementary Table 23); 226 of the 279 sentinel variants were associated (false discovery rate (FDR)<1%) with one or more traits and diseases (excluding quantitative lung function traits). Eighty-five of the lung function signals were associated with standing height. In order to investigate whether the genetic association signals for lung function were driven by incomplete adjustment for height, we tested for correlation of effects on lung function in UK Biobank and height in a meta-analysis of UK Biobank and the GIANT consortium for 246 of the 279 signals that had a proxy variant in GIANT50; there was no significant correlation (Supplementary Figure 5). Additionally, the PheWAS identified associations with body composition measures such as fat free mass (54 SNPs) and hip circumference (40 SNPs), as well as muscle strength (32 SNPs, grip strength). One hundred and fourteen of the 279 SNPs were associated with several quantitative measures of blood count, including eosinophil counts and percentages (25 SNPs). Twenty-five of our SNPs were also associated with asthma including 12 SNPs associated both with asthma and eosinophil measures (Supplementary Table 24). Eight of these SNPs were in linkage disequilibrium (LD, r2>0.1) with a SNP reported for association with asthma in previously published genome-wide association studies. We compared our observed effect sizes with those estimated after exclusion of all self-reported asthma cases and observed similar estimates (Supplementary Figure 6) suggesting that the lung function associations we report are not driven by asthma.
Figure 4: Individual PheWAS with 279 variants (traits passing FDR 1% threshold).
Separate association of 279 variants with 2,411 traits (FDR<1%) in UK Biobank (n up to 379,337). In each category, the trait with the strongest association, i.e. highest –log10(FDR), is shown first, followed by other traits in that category in descending order of –log10(FDR). Categories are colour-coded, and outcomes are denoted with a circular or triangular point, according to whether they were coded as binary or quantitative. The top association per-category is labelled with its rsID number, and a plain English label describing the trait. The letter at the beginning of each label allows easy cross-reference with the categories labelled in the legend. Zoomed in versions of each category with visible trait names and directionality are available in Supplementary Figure 10. These plots have signed log10(FDR) values, where a positive values indicates that a positive SNP-trait association is concordant with the risk allele for reduced lung function (as measured by lower FEV1/FVC). Tabulated results of all SNP-trait PheWAS associations associated at an FDR of<1% are available in Supplementary Table 23.
We examined the specificity of genetic associations, given the potential for this to predict specificity of drug target modification, and found that 53 of the 279 signals were associated only with lung function and COPD-related traits. In contrast, three of our 279 signals were associated with over 100 traits across multiple categories – among these rs3844313, a known intergenic signal near HLA-DQB1 was associated with 163 traits, and also had the strongest signal in the PheWAS, which was for association with intestinal malabsorption and celiac disease.
In our 279-variant weighted GRS PheWAS analysis (Supplementary Table 25), we found association with respiratory traits including COPD, chronic bronchitis, emphysema, respiratory failure, corticosteroid use and both pediatric and adult-onset asthma (Figure 5a). The GRS was also associated with non-respiratory traits including celiac disease, an intestinal autoimmune disorder (Figure 5b). These pleiotropic effects on risk of autoimmune diseases was further confirmed by analysis of previously reported GWAS (Online Methods, Supplementary Table 26) which showed overlapping single variant associations with Crohn’s disease, ulcerative colitis, psoriasis, systemic lupus erythematosus, IgA nephropathy, pediatric autoimmune disease and type 1 diabetes.
Figure 5: PheWAS with genetic risk score (traits passing FDR 1% threshold).
Association of 279 variant weighted genetic risk score with 2,453 traits (FDR<1%) in UK Biobank (n up to 379,337). In each panel, the category with the strongest association, i.e. highest –log10(FDR), is shown first, followed by all other associations in that category, ordered by descending order of –log10(FDR). Sample sizes varied across traits and are available in Supplementary Table 25, along with the full summary statistics for each association, plus details of categorisation and plain English labels for each trait. Trait categories are colour coded, and outcomes are denoted with a circular or triangular point, according to whether they were coded as binary or quantitative. The sign of the log10(FDR) value is positive where an increase in the risk score (i.e. greater risk of COPD, reduced lung function) is associated with a positive effect estimate for that trait. *QC refers to spirometry passing ERS/ATS criteria. SR=self-report; HES=Hospital Episode Statistics.
a. Associations with respiratory traits.
b.Associations with all other traits. ENT=Ear, Nose and Throat; FBC=Full Blood Count.
Discussion
The large sample size of our study, achieved by our refinement of the spirometry in UK Biobank and inclusion of the substantially expanded SpiroMeta consortium data set, has doubled the yield of lung function signals to 279. Fine-mapping of all new and previously reported signals, together with gene and protein expression analyses with improved tissue specificity and stringency, has implicated new genes and pathways, highlighting the importance of cilia development, TGF-β signalling via SMAD3, and elastic fibers in the etiology of airflow obstruction. Many of the genes and pathways reported here contain druggable targets; we highlight examples where the genetic variants mimicking therapeutic modulation of targets may have opposing effects on lung function. We have developed and applied the first weighted GRS for lung function and tested it in independent COPD case-control studies. Our GRS shows stronger association and larger effect size estimates than a previous GRS in European ancestry populations18, as well as generalizability to other ancestry groups. We undertook the first comprehensive PheWAS for lung function signals, and report genetic variants with apparent specificity of effects and others with pleiotropic effects that might indicate shared biological pathways between different diseases.
For the first time in a GWAS of lung function, we report an enrichment of genes involved in ciliogenesis (including KIAA0753, CDK2 and CEP72). Defects in primary cilia as a result of highly deleterious mutations in essential genes result in ciliopathies known to affect multiple organ systems. We found an enrichment of genes with a role in centriolar replication and duplication, core processes in primary and motile cilia formation. Mutations in KIAA0753 cause the ciliopathies Joubert Syndrome and Orofaciodigital Syndrome28. Reduced airway motile cilia function impacting mucus clearance is a feature of COPD, but it has not been clear whether this is causal or the consequence of damage by external factors such as smoking or infection. Our findings suggest that impaired ciliary function might be a driver of the disease process. We have previously shown enrichment of rare variants in cilia-related genes in heavy smokers without airflow obstruction51.
New signals, implicating ITGAV and GDF5, as well as stronger support for TGFB2 and MFAP2 as likely causal genes, provide new genetic support for the importance of elastic fiber pathways in lung function and COPD18. The elastic fibers of the extracellular matrix are known to be disrupted in COPD52. As the breakdown of elastic fibers by neutrophil elastase leads to emphysema in individuals with alpha1-antitrypsin deficiency, we also assessed the association with the SERPINA1 Z allele, which was not associated with FEV1/FVC in our study (rs28929474, P=0.109 in UK Biobank).
Smoking and genetic risk both have important effects on lung function and COPD. For lung function, we found no interaction between smoking and individual variants, and for FEV1/FVC no interaction between smoking status and the weighted GRS. However, for COPD a weak smoking-GRS interaction was observed. Whilst the weighted GRS showed a strong association with COPD susceptibility, and a high attributable risk, we do not claim that this would represent an appropriate method of screening for COPD risk. Importantly, our findings demonstrate the high absolute risk among genetically susceptible smokers (82.4% by approximately 70 years of age).
We used two complementary study designs to maximize sample size for discovery and ensure robustness of findings by requiring independent support for association. Furthermore, through additional analysis of the spirometry data in UK Biobank and substantial expansion of the SpiroMeta consortium, we have markedly increased samples sizes to almost seven times those included in previous studies. As no lower MAF threshold was applied in our analyses, an overall threshold of P<5×10−9, as recommended for re-sequencing analyses of European ancestry individuals23, was applied. We identified the largest number of new signals in our more stringent two-stage design (“Tier 1”, 99 new signals). Amongst the signals that we report as “Tier 3” (and did not include in further analyses), all reached P<10−3 in UK Biobank and 183 met a less stringent threshold of P<0.05 in SpiroMeta.
Our study is the first to investigate genome-wide associations with PEF. PEF is determined by various physiological factors including lung volume, large airway caliber, elasticity of the lung and expiratory muscle strength, is used for monitoring asthma, and was incorporated in a recently evaluated clinical score for diagnosing COPD and predicting acute exacerbations of COPD53. Overall, 133 of the 279 signals were also associated with PEF (P<10−5) and for 15 signals (including 4 new signals), PEF was the most significantly associated trait. Of note, a signal near SLC26A9, a known cystic fibrosis modifier gene54, was highly significantly associated with PEF in UK Biobank (P=3.97×10−66) and nominally significant in SpiroMeta (P=6.93×10−3), with consistent direction of effect, but did not meet the Tier 2 criteria. This could reflect the limited power for PEF in SpiroMeta (up to 24,218 for PEF compared to 79,055 for the other traits).
Examining associations of a given genetic variant with a wide range of human phenotypes is a valuable tool in therapeutic target validation. As in our PheWAS, it can highlight variants which show associations with one or more respiratory traits that might be expected to demonstrate greater target specificity than variants associated with many traits. Additionally, in some instances, association with multiple traits may indicate the relevance of drug repurposing. Association of a given SNP with multiple traits does not necessarily imply shared etiology, and further investigation is warranted. Our GRS PheWAS assesses broader genetic overlap between lung function and other traits and supports the evidence for some shared genetic determinants with autoimmune diseases.
In summary, our study has doubled the number of signals for lung function and provides new understanding and resources of utility for the development of therapeutics. The 279-variant GRS we constructed was associated with a 4.73-fold increased relative risk of moderate-severe COPD between highest and lowest deciles, such that one would expect over 80% of smokers in the highest genetic risk decile to develop COPD. The GRS was also predictive of COPD across multiple ancestral groups. Our PheWAS highlights both expected and unexpected associations relevant to respiratory and other systemic diseases. Investigating the nature of the pleiotropic effects of some of these variants will be of benefit for drug target identification and validation.
Online Methods
Study Design Overview and rationale
For the two-stage approach, we first selected distinct signals of association (defined using conditional analyses) with one or more traits achieving P<5×10−9 in UK Biobank only (maximum n=321,047). A threshold of P<5×10−9 was selected to maximize stringency and for consistency with currently recommended genome-wide significance thresholds for re-sequencing analyses of European ancestry individuals23. We reported as new those signals which additionally met P<10−3 in SpiroMeta (N effective>70% of n up to 79,055; see Supplementary Note and Supplementary Figure 7 for power calculations), with consistent directions of effect. We term these “Tier 1” signals, as they meet our highest level of stringency. Methods for conditional analyses and determining novelty are described below.
For the one-stage approach, we selected distinct signals of association (defined using conditional analyses) with one or more traits reaching P<5×10−9 in the meta-analysis of UK Biobank and SpiroMeta (maximum n=400,102), reporting as new those with a consistent direction of effect that additionally met P<10−3 in both UK Biobank and SpiroMeta. We term these signals “Tier 2”, as they meet our second-highest level of stringency.
All signals meeting either set of criteria described above, and that had not been previously published, were reported as new association signals for lung function. Signals that reached P<5×10−9 in the meta-analysis of UK Biobank and SpiroMeta, had a consistent direction of effect in UK Biobank and SpiroMeta, but that did not reach P<10−3 in both UK Biobank and SpiroMeta are presented as “Tier 3”, and were not included in further analyses.
Data for chromosome X were available for 321,027 European individuals in UK Biobank and 38,199 individuals from SpiroMeta (1000 Genomes Project Phase 1 imputation).55
Please see the ‘Life Sciences Reporting Summary’.
UK Biobank
The UK Biobank resource is described elsewhere (see URLs). Individuals were selected for inclusion in this study if they: (i) had complete data for age, sex, height and smoking status; (ii) had spirometry meeting quality control requirements (based on analyses of acceptability, reproducibility and blow curve metrics; Supplementary Note); (iii) had genome-wide imputed data and; (iv) were of European ancestry based on genetic data (Supplementary Note; Supplementary Figure 1). Genotyping was undertaken using the Affymetrix Axiom® UK BiLEVE and UK Biobank arrays13. Genotypes were imputed to the Haplotype Reference Consortium panel56 (Supplementary Note), and retained if minor allele count≥3 and imputation quality (info)>0.5. In total, 321,047 individuals were included in our analyses (Supplementary Table 1).
Residuals from linear regression of each trait (FEV1, FVC, FEV1/FVC and PEF) against age, age2, sex, height, smoking status (ever/never) and genotyping array were ranked and inverse-normal transformed, giving normally distributed Z-scores. These Z-scores were used for genome-wide association testing under an additive genetic model using BOLT-LMM v2.320. Principal components were not included as BOLT-LMM uses a linear mixed model to account for relatedness and fine-scale population structure.
Linkage disequilibrium (LD) score regression implemented in LDSC21 was used to estimate test statistic inflation due to confounding. Genomic control was applied, adjusting test statistics by LD score regression intercepts: 1.12 for FEV1, 1.14 for FVC, 1.19 for FEV1/FVC and 1.13 for PEF (Supplementary Figure 8; Supplementary Table 27), acknowledging that this might be over-conservative for UK Biobank.
SpiroMeta consortium
The SpiroMeta consortium meta-analysis comprised a total of 79,055 individuals from 22 studies. Thirteen studies (n=21,436) were imputed to the 1000 Genomes Project Phase 1 panel55 (B58C, BHS1&2, three Croatian studies [CROATIA-Korcula, CROATIA-Split and CROATIA-Vis], Health 2000, KORA F4, KORA S3, LBC1936, NSPHS, ORCADES, SAPALDIA and YFS) and 9 studies (n=61,682) were imputed to the Haplotype Reference Consortium (HRC) panel57 (EPIC [obese cases and population-based studies], GS:SFHS, NFBC1966, NFBC1986, PIVUS, SHIP, SHIP-TREND, UKHLS and VIKING). See Supplementary Tables 2 and 3 for abbreviation definitions, study characteristics, and details of genotyping platforms, imputation panels and methods). Measurements of spirometry for each study are described in the Supplementary Note.
In each study, linear regression models were fitted for each trait (FEV1, FEV1/FVC, FVC and where available, PEF), with adjustment for age, age2, sex and height. For studies with unrelated individuals, models were fitted separately in ever and never smokers, with additional adjustment for ancestral principal components. Studies with related individuals fitted mixed models in all individuals to account for relatedness, with ever smoking status as a covariate.
In all studies, residuals were rank-based inverse normal transformed and used as the phenotype for association testing, under an additive genetic model (Supplementary Table 3).
In the study-level results, variants were excluded if they had a low minor allele count (MAC) (Supplementary Table 3) or imputation quality (info)<0.3. In studies of unrelated individuals, ever and never smokers’ results were combined using inverse-variance weighted meta-analysis. Genomic control was applied to all study-level results, before combining results across all studies using inverse-variance weighted meta-analysis. LD score regression intercepts for the meta-analysis were close to 1.00 (Supplementary Figure 8; Supplementary Table 27), therefore genomic control was not applied.
Meta-analyses
A total of 19,819,130 variants (imputed or genotyped) in both UK Biobank and SpiroMeta were meta-analyzed, using inverse-variance weighted fixed effect meta-analysis. No further genomic control was applied as LD score regression intercepts were close to 1.00 (Supplementary Table 27).
Selection of new signals using conditional analyses
All SNPs ±1 Mb were extracted around each sentinel variant. We performed stepwise conditional analysis to select independently associated SNPs within each 2-Mb region, using GCTA58. LD was estimated for UK Biobank from the same individuals used in discovery, and for SpiroMeta, from an unrelated subset of 48,943 UK Biobank individuals18. Secondary signals identified within each 2-Mb region were required to meet Tier 1 or Tier 2 criteria (described above) after conditioning on the primary sentinel variant. A combined list of distinct lung function signals was then made across the four phenotypes, FEV1, FVC, FEV1/FVC and PEF, as follows: where sentinel variants for 2 signals for different phenotypes were in high LD (r2>0.5), we retained the most significant variant; where 2 signals were in moderate LD (0.1>r2>0.5), we retained variants if, after conditional analysis, they still met the Tier 1 or Tier 2 threshold; for signals in low LD (r2<0.1) we retained both variants. We then used the same criteria to identify a subset of new signals which were distinct from previously published independent signals (see below).
Assessment of previously reported lung function signals
We identified 184 autosomal signals from previous GWAS of lung function and COPD1,4–14. After LD pruning (only keeping signals with LD of r2<0.1), we removed 24 non-independent SNPs, leaving 160 previously reported independent signals. Of 6 previously reported signals in the HLA region, we included only the 3 independent lung function HLA signals reported from conditional analysis using all imputed HLA genotypes18: AGER (rs2070600), HLA-DQB1 (rs114544105) and near ZNF184 (rs34864796), leaving 157 autosomal signals.
We confirmed association of previously reported signals in our data if they met any of three criteria: (i) the previously reported sentinel was associated (P<10−5) with any lung function trait in UK Biobank; (ii) a proxy for the previously reported sentinel with r2>0.5 was associated (P<10−5) with any lung function trait in UK Biobank; (iii) a proxy for the previously reported sentinel with r2>0.1 was associated with any lung function trait meeting tier 1 or tier 2 criteria (Supplementary Figure 3).
Effect on COPD susceptibility – genetic risk score in multiple ancestries
To test association of all lung function signals with COPD susceptibility, we constructed a 279-variant weighted GRS comprising the 139 novel and 140 previously reported signals; we used the previously reported sentinel SNP for published signals. Weights were derived using the FEV1/FVC decreasing (generally COPD risk increasing) alleles. For previously reported signals (n=140), effect sizes from UK Biobank were used as weights for the 94 signals that were not discovered using UK Biobank data. Weights were taken from SpiroMeta for 46 signals where UK Biobank was included in the discovery of those signals. For novel signals, weights were taken from SpiroMeta for two-stage (tier 1) signals (n=99), and the smallest absolute effect size from either UK Biobank or SpiroMeta was used for one-stage (tier 2) signals (n=40) (Supplementary Table 28). This approach was taken in order to derive conservative weights, thus reducing the likelihood of bias by winner’s curse. For the weighted GRS the number of risk alleles at each variant was multiplied by its weight.
The GRS was first calculated in unrelated individuals (KING kinship coefficient of<0.0884) within 6 ancestral groups of UK Biobank: Europeans, South Asians, Africans, Chinese, Mixed African and Europeans, and Mixed Other (total sample of unrelated individuals across six ancestries: 323,001) using PLINK. Weights and alleles were as described above. COPD was defined as FEV1/FVC<0.7 and FEV1<80% predicted, i.e. GOLD stage 2–4 categorization. Associations with the GRS were then tested using COPD (in ancestral groups with at least 100 COPD cases) and FEV1/FVC as the outcomes.
We also calculated the GRS in individuals from the China Kadoorie Biobank (CKB). Four of the 279 SNPs were unavailable in CKB (rs1800888, rs56196860, rs72724130 and rs77672322), and for 12 SNPs, proxies were used (minimum r2=0.3). Analyses were undertaken in all COPD GOLD stage 2–4 cases (FEV1/FVC<0.7 and FEV1<0.8 of the predicted value: 6,013 cases and 69,567 controls), against an unbiased set of population controls. The GRS was also tested for association with FEV1/FVC in CKB (n=72,796).
Logistic regression of COPD case-control status with the GRS in UK Biobank and China Kadoorie Biobank assumed an additive genetic effect and was adjusted for age, age2, sex, height, and smoking (Supplementary Table 19). Ten principal components were included in UK Biobank analyses. In China Kadoorie Biobank, analyses were stratified by geographical regions, then meta-analyzed using an inverse-variance fixed effect model. Linear models assessing the association with FEV1/FVC were fitted using the transformed outcome used in the main GWAS analysis.
We then tested association in 5 European-ancestry COPD case-control studies: COPDGene (Non-Hispanic White Population) (3,068 cases, 2,110 controls), ECLIPSE (1,713 cases, 147 controls), GenKOLS (836 cases, 692 controls), NETT-NAS (374 cases, 429 controls) and SPIROMICS (988 cases, 537 controls) (Supplementary Table 20). We also tested this GRS in the COPDGene African American population study (910 cases, 1,556 controls). Logistic regression models using COPD as outcome and the GRS as exposure were adjusted for age, age2, sex, height, and principal components (Supplementary Table 21, Supplementary Figure 9). Single variant associations of the 279 SNPs with COPD are in Supplementary Table 29.
Next, we divided individuals in the external COPD case-control studies into deciles, according to their values of the weighted GRS (undertaken separately by study group). For each decile, logistic models were fitted, comparing the risk of COPD for members of the decile compared to those in the lowest decile (i.e. those with lowest values of the weighted GRS). Covariates were as for COPD analyses. Results were combined across European-ancestry study groups by fixed effect meta-analysis (Supplementary Table 22).
Effects on smoking behavior
As our discovery GWAS in UK Biobank was adjusted for ever smoking status, and not for pack years of smoking (this information was missing for 32% of smokers), we evaluated whether any lung function association signals might be driven by an association with smoking behavior, by testing for association with smoking initiation (123,890 ever smokers vs. 151,706 never smokers) and cigarettes per day (n=80,015) in UK Biobank (see Supplementary Note). We also tested for association with lung function in never smokers only (n=173,658). We excluded signals associated with smoking behavior (Supplementary Table 6) but not with lung function in never smokers.
Smoking interaction
For associated variants (new and previously reported), we repeated association testing for lung function separately in UK Biobank and SpiroMeta (up to 176,701 ever smokers and 197,999 never smokers), and tested for an interaction effect with smoking using the Welch test (Supplementary Note). A threshold of P<1.79×10−4 (Bonferroni corrected for 279 tests) indicated significance.
We also tested for interaction between the weighted GRS and smoking, within 303,619 unrelated individuals of European ancestry in UK Biobank, using COPD and FEV1/FVC as outcomes (FEV1/FVC was pre-adjusted for age, age2, sex, and height, and the residuals transformed as per the main GWAS analysis). For COPD (defined as FEV1/FVC<0.7, and FEV1<80% predicted) a logistic model was fitted:
COPD ~ genotyping array + 10 principal components + age + age2 + sex + height + smoking status + weighted risk score + (smoking status × weighted risk score).
For FEV1/FVC, a linear model was fitted:
FEV1/FVC ~ genotyping array + 10 principal components + smoking status + weighted risk score + (smoking status x weighted risk score).
Proportion of variance explained
We calculated the proportion of variance explained by the previously reported (n=140) and new variants (n=139) associated with lung function using the formula:
where n is the number of variants, fi and βi are the frequency and effect estimate of the i’th variant, and V is the phenotypic variance (always 1 as our phenotypes were inverse-normal transformed). We used the same conservative effect estimates (β) used as GRS weights for the 279 GRS variants, derived from either UK Biobank or SpiroMeta effect estimates (described above). Our previously published estimate of proportion of variance explained18 used UK Biobank effect estimates. We assumed a heritability of 40%24,25 to estimate the proportion of additive polygenic variance.
Fine-mapping
A Bayesian method59 was used to fine-map lung-function-associated signals to the set of variants that were 99% likely to contain the underlying causal variant (assuming that the causal variant was analyzed). This was undertaken for new signals and for previously reported signals reaching P<10−5 in UK Biobank. For previously reported signals, the sentinel variant from the current UK Biobank analysis was used, instead of the previously reported variant. We used a value of 0.04 for the prior W in the approximate Bayes factor formula60. Effect sizes and standard errors for fine-mapping were obtained from inverse-variance weighted meta-analysis of UK Biobank and SpiroMeta (maximum n=400,102). Signals in the HLA region were not included.
Implication of potentially causal genes
Annotation of deleterious variants
Variants in the 99% credible sets were checked for predicted functional effect if they were annotated as “exonic”, “splicing”, “ncRNA_exonic”, “5’-UTR” or “3’-UTR” (untranslated region) by ANNOVAR61. We then used SIFT, PolyPhen-2 (implemented using the Ensembl GRCh37 Variant Effect Predictor, see URLs) and FATHMM62 to annotate missense variants, and CADD (also implemented using VEP) to annotate non-coding variation. Variants were annotated as deleterious if they were labelled ‘deleterious’ by SIFT, ‘probably damaging’ or ‘possibly damaging’ by PolyPhen-2, ‘damaging’ by FATHMM (specifying the ‘Inherited Disease’ option of the ‘Coding Variants’ method, and using the ‘Unweighted’ prediction algorithm) or had a CADD scaled score ≥2018. The union of the four methods was taken to establish the number of potentially deleterious variants and their unique genes.
Gene expression and protein levels
At 276 of 279 (3 HLA signals excluded) signals, the sentinel variant and 99% credible set59 were used to query three eQTL resources: lung eQTL (n=1,111)13, blood eQTL (n=4,896)63 and GTEx (V7; with maximum n=388, depending on tissue: ‘Artery Aorta’ (n=267), ‘Artery Coronary’ (n=152), ‘Artery Tibial’ (n=388), ‘Colon Sigmoid’ (n=203), ‘Colon Transverse’ (n=246), ‘Esophagus Gastroesophageal Junction’ (n=213), ‘Esophagus Muscularis’ (n=335), ‘Lung’ (n=383), ‘Small Intestine Terminal Ileum’ (n=122), ‘Stomach’ (n=237), and ‘Whole Blood’ (n=369))64, and one blood pQTL resource (n=3,301)39.
A gene was classified as a ‘putative causal gene’ if the sentinel SNP or any SNP in the respective 99% credible set was associated with expression of this gene or its protein levels (FDR<5% for eQTL, P<5.03×10−8 for pQTL [276 tests at 3,600 proteins]) and if the GWAS sentinel SNP or any SNP in the respective 99% credible set was the variant most strongly associated with expression of the respective gene or level of the respective protein (i.e. the sentinel eQTL/pQTL SNP) in one or more of the eQTL and pQTL data sets.
Pathway analysis
We tested for enrichment of genes identified via functional annotation, gene expression or protein level analyses in pathway and gene set ontology databases using ConsensusPathDB.65 Pathways or gene sets represented entirely by genes implicated by the same association signal were excluded. Gene sets and pathways with FDR<5% are reported.
Functional enrichment analyses
We carried out stratified LD score regression to identify significant enrichment of heritability at variants overlapping histone marks (e.g. H3K4me1, H3K4me3) specific to lung, foetal lung, and smooth muscle-containing (e.g. colon, stomach) cell lines, using methods specified by Finucane et al.40
We separately selected FEV1/FVC and FVC associated SNPs passing two thresholds (P<5×10−5 and P<5×10−9 in the meta-analysis) as input to GARFIELD41 to test for enrichment of our signals for 424 DHS hotspot annotations derived from 55 different tissues in the RoadMap Epigenomics and ENCODE projects.
Using DeepSEA42, we analyzed all SNPs in the 99% credible set for predicted chromatin effects. We reported effects for any chromatin effect and lung-related cell line with an E-value<0.05 (i.e. the expected proportion of SNPs with a larger predicted effect based on empirical distributions of predicted effects for 1000 Genomes SNPs) and an absolute difference in probability of>0.1 (threshold for “high confidence”) between the reference and alternative allele.
Drug targets
Genes identified as potentially causal using eQTL, pQTL or variant annotation were interrogated against the gene-drug interactions table of the Drug-Gene Interactions Database (DGIDB) (see URLs). Drugs were mapped to CHEMBL IDs (see URLs), and indications (MeSH headings) were added.
Phenome-wide association studies
To identify whether the 279 signals were associated with other traits and diseases, the weighted GRS was calculated in up to 379,337 UK Biobank samples, and a phenome-wide association study (PheWAS) was performed, with the GRS as the exposure. Traits included UK Biobank baseline measures (from questionnaires and physical measures), self-reported medication usage, and operative procedures, as well as those captured in Office of Population Censuses and Surveys codes from the electronic health record. We also included self-reported disease variables and those from hospital episode statistics (ICD-10 codes truncated to three-character codes and combined in block and chapter groups), combining these where possible to maximize power. The GRS analysis included 2,453 traits, and the single-variant analysis contained 2,411 traits (traits with>200 cases were included for the single-variant PheWAS, whereas traits with>50 cases were included in the GRS PheWAS). Analyses were conducted in unrelated European-ancestry individuals (KING kinship coefficient <0.0442), and were adjusted for age, sex, genotyping array, and ten principal components. Logistic and linear models were fitted for binary and quantitative outcomes, respectively. False discovery rates were calculated according to the number of traits in the GRS and single-variant PheWAS (2,453 or 2,411, respectively).
In addition, the sentinel variants 99% credible set variants were queried against the GWAS catalog66 (see URLs) and GRASP67 (see URLs) for associations at P<5×10−8. Associations relating to methylation, expression, metabolite or protein levels, as well as lung function and COPD, were not included.
Supplementary Material
Acknowledgments
This research has been conducted using the UK Biobank Resource under applications 648, 4892 and 26041. L. Wain holds a GSK/British Lung Foundation Chair in Respiratory Research. M. Tobin is supported by a Wellcome Trust Investigator Award (WT202849/Z/16/Z). M. Tobin and L. Wain have been supported by the MRC (MR/N011317/1). The research was partially supported by the NIHR Leicester Biomedical Research Centre; the views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health. I. Hall: The research was partially supported by the NIHR Nottingham Biomedical Research Centre; the views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health. This research used the ALICE and SPECTRE High Performance Computing Facilities at the University of Leicester. Additional acknowledgments and funding details for other co-authors and contributing studies (including the SpiroMeta consortium) can be found in the Supplementary Note.
Footnotes
Competing Interests Statement
The following authors report potential conflicts of interest:
K. Song: Kijoung Song is an employee of GlaxoSmithKline and may own company stock.
Z. Chen: reports grants from GSK and Merck.
J. Danesh: John Danesh reports personal fees and non-financial support from Merck Sharp & Dohme (MSD) and Novartis, and grants from British Heart Foundation, European Research Council, MSD, NIHR, NHS Blood and Transplant, Novartis, Pfizer, UK MRC, Wellcome Trust, and AstraZeneca.
J. Hoffman: Joshua D. Hoffman is an employee of GlaxoSmithKline and may own company stock.
N. Locantore: Nicholas Locantore is an employee and shareholder of GSK.
J. Maranville: Joseph C. Maranville was a Merck employee during this study, and is now a Celgene employee.
D. Nickle: David C Nickle has been a Merck & Co. employee during this study and is now an employee at Biogen Inc.
H. Runz: Heiko Runz has been a Merck & Co. employee during this study and is now an employee at Biogen Inc.
I. Sayers: Ian Sayers has received support from GSK and BI.
R. Tal-Singer: Ruth Tal-Singer is an employee and shareholder of GlaxoSmithKline.
M. van den Berge: Maarten van den Berge reports grants paid to the University from Astra Zeneca, TEVA, GSK, Chiesi, outside the submitted work.
J. Whittaker: John C. Whittaker is an employee of GlaxoSmithKline and may own company stock.
L. Yerges-Armstrong: Laura M. Yerges-Armstrong is an employee of GlaxoSmithKline and may own company stock.
H. Schulz: Helmholtz Center Munich funded by the German Federal Ministry of Education and Research (BMBF) and by the State of Bavaria, Competence Network Asthma and COPD (ASCONET), network COSYCONET (subproject 2, BMBF FKZ 01GI0882) funded by the German Federal Ministry of Education and Research (BMBF)
E. Silverman: In the past three years, Edwin K. Silverman received honoraria from Novartis for Continuing Medical Education Seminars and grant and travel support from GlaxoSmithKline.
A. Butterworth: Adam S. Butterworth reports grants from Merck, Pfizer, Novartis, Biogen and AstraZeneca and personal fees from Novartis.
R. Scott: Robert A Scott is an employee and shareholder in GlaxoSmithKline.
R. Walters: Robin G. Walters reports that the China Kadoorie Biobank study has received grant support from GSK.
M. Cho: Michael H. Cho has received grant support from GSK.
I. Hall: Ian P. Hall has funded research collaborations with GSK, Boehringer Ingelheim and Orion.
M. Tobin: Martin D. Tobin receives funding from GSK for a collaborative research project, outside of the submitted work.
L. Wain: Louise V. Wain receives funding from GSK for a collaborative research project, outside of the submitted work.
Data availability statement
SpiroMeta GWAS summary statistics and UK Biobank GWAS summary statistics are available online via LD-Hub (http://ldsc.broadinstitute.org/ldhub/). Single-variant PheWAS results are available by request to the corresponding authors. The newly derived spirometry variables are available from UK Biobank (http://www.ukbiobank.ac.uk/).
References
- 1.Young RP, Hopkins R & Eaton TE Forced expiratory volume in one second: not just a lung function test but a marker of premature death from all causes. Eur Respir J 30, 616–22 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Global, regional, and national age-sex specific mortality for 264 causes of death, 1980–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet 390, 1151–1210 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet 390, 1211–1259 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hobbs BD et al. Genetic loci associated with chronic obstructive pulmonary disease overlap with loci for lung function and pulmonary fibrosis. Nat Genet 49, 426–432 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salvi SS & Barnes PJ Chronic obstructive pulmonary disease in non-smokers. Lancet 374, 733–43 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Nelson MR et al. The support of human genetic evidence for approved drug indications. Nat Genet 47, 856–60 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Wilk JB et al. A genome-wide association study of pulmonary function measures in the Framingham Heart Study. PLoS Genet 5, e1000429 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Repapi E et al. Genome-wide association study identifies five loci associated with lung function. Nat Genet 42, 36–44 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hancock DB et al. Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat Genet 42, 45–52 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soler Artigas M. et al. Genome-wide association and large-scale follow up identifies 16 new loci influencing lung function. Nat Genet 43, 1082–90 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho MH et al. A genome-wide association study of COPD identifies a susceptibility locus on chromosome 19q13. Hum Mol Genet 21, 947–57 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loth DW et al. Genome-wide association analysis identifies six new loci associated with forced vital capacity 46, 669–77 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wain LV et al. Novel insights into the genetics of smoking behaviour, lung function, and chronic obstructive pulmonary disease (UK BiLEVE): a genetic association study in UK Biobank. Lancet Respir Med 3, 769–81 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lutz SM et al. A genome-wide association study identifies risk loci for spirometric measures among smokers of European and African ancestry. BMC Genet 16, 138 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soler Artigas M. et al. Sixteen new lung function signals identified through 1000 Genomes Project reference panel imputation. Nat Commun 6, 8658 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hobbs BD et al. Exome Array Analysis Identifies a Common Variant in IL27 Associated with Chronic Obstructive Pulmonary Disease 194, 48–57 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson V et al. Meta-analysis of exome array data identifies six novel genetic loci for lung function [version 3; referees: 2 approved]. Wellcome Open Research 3(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wain LV et al. Genome-wide association analyses for lung function and chronic obstructive pulmonary disease identify new loci and potential druggable targets. Nat Genet 49, 416–425 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wyss AB et al. Multiethnic Meta-analysis Identifies New Loci for Pulmonary Function. bioRxiv (2017).
- 20.Loh PR et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat Genet 47, 284–90 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bulik-Sullivan BK et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nature Genetics 47, 291 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang J et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet 44, 369–75, S1–3 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pulit SL, de With SA & de Bakker PI Resetting the bar: Statistical significance in whole-genome sequencing-based association studies of global populations. Genet Epidemiol 41, 145–151 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Palmer LJ et al. Familial aggregation and heritability of adult lung function: results from the Busselton Health Study. Eur Respir J 17, 696–702 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Wilk JB et al. Evidence for major genes influencing pulmonary function in the NHLBI family heart study. Genet Epidemiol 19, 81–94 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Benyamin B et al. GWAS of butyrylcholinesterase activity identifies four novel loci, independent effects within BCHE and secondary associations with metabolic risk factors. Hum Mol Genet 20, 4504–14 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hammarsjo A, Wang Z, Vaz R & Taylan F Novel KIAA0753 mutations extend the phenotype of skeletal ciliopathies 7, 15585 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stephen J et al. Mutations in KIAA0753 cause Joubert syndrome associated with growth hormone deficiency. Hum Genet 136, 399–408 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loukil A, Tormanen K & Sütterlin C The daughter centriole controls ciliogenesis by regulating Neurl-4 localization at the centrosome. The Journal of Cell Biology 216, 1287–1300 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He R et al. LRRC45 is a centrosome linker component required for centrosome cohesion. Cell Rep 4, 1100–7 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Conkar D et al. The centriolar satellite protein CCDC66 interacts with CEP290 and functions in cilium formation and trafficking 130, 1450–1462 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uhlén M et al. Tissue-based map of the human proteome. Science 347(2015). [DOI] [PubMed] [Google Scholar]
- 33.Hao K et al. Lung eQTLs to help reveal the molecular underpinnings of asthma. PLoS Genet 8, e1003029 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lamontagne M et al. Refining susceptibility loci of chronic obstructive pulmonary disease with lung eqtls. PLoS One 8, e70220 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Obeidat M et al. GSTCD and INTS12 regulation and expression in the human lung. PLoS One 8, e74630 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Westra HJ et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet 45, 1238–1243 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–60 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kundaje A et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–30 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun BB et al. Genomic atlas of the human plasma proteome. Nature 558, 73–79 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Finucane HK et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet 47, 1228–35 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iotchkova V et al. Discovery and refinement of genetic loci associated with cardiometabolic risk using dense imputation maps. Nat Genet 48, 1303–1312 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou J & Troyanskaya OG Predicting effects of noncoding variants with deep learning-based sequence model. Nat Methods 12, 931–4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cotto KC et al. DGIdb 3.0: a redesign and expansion of the drug–gene interaction database. Nucleic Acids Research 46, D1068–D1073 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Slack R et al. P112 Discovery of a Novel, High Affinity, Small Molecule αvβ6 Inhibitor for the Treatment of Idiopathic Pulmonary Fibrosis. QJM: An International Journal of Medicine 109, S60–S60 (2016). [Google Scholar]
- 45.Raab-Westphal S, Marshall JF & Goodman SL Integrins as Therapeutic Targets: Successes and Cancers. Cancers (Basel) 9, e110 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Merrill JT et al. Efficacy and Safety of Atacicept in Patients With Systemic Lupus Erythematosus: Results of a Twenty-Four-Week, Multicenter, Randomized, Double-Blind, Placebo-Controlled, Parallel-Arm, Phase IIb Study. Arthritis Rheumatol 70, 266–276 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aschard H et al. Evidence for large-scale gene-by-smoking interaction effects on pulmonary function. Int J Epidemiol 46, 894–904 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lokke A, Lange P, Scharling H, Fabricius P & Vestbo J Developing COPD: a 25 year follow up study of the general population. Thorax 61, 935–9 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pulley JM et al. Accelerating Precision Drug Development and Drug Repurposing by Leveraging Human Genetics. ASSAY and Drug Development Technologies 15, 113–119 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yengo L et al. Meta-analysis of genome-wide association studies for height and body mass index in ~700,000 individuals of European ancestry. bioRxiv (2018). [DOI] [PMC free article] [PubMed]
- 51.Wain LV et al. Whole exome re-sequencing implicates CCDC38 and cilia structure and function in resistance to smoking related airflow obstruction. PLoS Genet 10, e1004314 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Black PN et al. Changes in elastic fibres in the small airways and alveoli in COPD. Eur Respir J 31, 998–1004 (2008). [DOI] [PubMed] [Google Scholar]
- 53.Martinez FJ et al. A New Approach for Identifying Patients with Undiagnosed Chronic Obstructive Pulmonary Disease. Am J Respir Crit Care Med 195, 748–756 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Strug LJ et al. Cystic fibrosis gene modifier SLC26A9 modulates airway response to CFTR-directed therapeutics. Hum Mol Genet 25, 4590–4600 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.The 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature 467, 1061 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bycroft C et al. Genome-wide genetic data on ~500,000 UK Biobank participants. bioRxiv (2017).
- 57.McCarthy S et al. A reference panel of 64,976 haplotypes for genotype imputation. Nature genetics 48, 1279–1283 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang J, Lee SH, Goddard ME & Visscher PM GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 88, 76–82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wakefield J Reporting and interpretation in genome-wide association studies. Int J Epidemiol 37, 641–53 (2008). [DOI] [PubMed] [Google Scholar]
- 60.van de Bunt M, Cortes A, Brown MA, Morris AP & McCarthy MI Evaluating the Performance of Fine-Mapping Strategies at Common Variant GWAS Loci. PLoS Genet 11, e1005535 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang K, Li M & Hakonarson H ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38, e164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shihab HA et al. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum Mutat 34, 57–65 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jansen R et al. Conditional eQTL analysis reveals allelic heterogeneity of gene expression. Hum Mol Genet 26, 1444–1451 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Battle A, Brown CD, Engelhardt BE & Montgomery SB Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kamburov A, Stelzl U, Lehrach H & Herwig R The ConsensusPathDB interaction database: 2013 update. Nucleic Acids Res 41, D793–800 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.MacArthur J et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Research 45, D896–D901 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leslie R, O’Donnell CJ & Johnson AD GRASP: analysis of genotype-phenotype results from 1390 genome-wide association studies and corresponding open access database. Bioinformatics 30, i185–94 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.