ABSTRACT
Microtubules (MTs) are important for cellular structure, transport of cargoes and segregation of chromosomes and organelles during mitosis. The stochastic growth and shrinkage of MTs, known as dynamic instability, is necessary for these functions. Previous studies to determine how individual MT-associated proteins (MAPs) affect MT dynamics have been performed either through in vivo studies, which provide limited opportunity for observation of individual MTs or manipulation of conditions, or in vitro studies, which focus either on purified proteins, and therefore lack cellular complexity, or on cell extracts made from genetically intractable organisms. In order to investigate the ensemble activities of all MAPs on MT dynamics using lysates made from a genetically tractable organism, we developed a cell-free assay for budding yeast lysates using total internal reflection fluorescence (TIRF) microscopy. Lysates were prepared from yeast strains expressing GFP-tubulin. MT polymerization from pre-assembled MT seeds adhered to a coverslip was observed in real time. Through use of cell division cycle (cdc) and MT depolymerase mutants, we found that MT polymerization and dynamic instability are dependent on the cell cycle state and the activities of specific MAPs.
KEY WORDS: Microtubule, Reconstitution, Dynamic instability, Kinesin
Summary: A new in vitro assay for measuring the growth and dynamics of single microtubules within the complexity of total budding yeast soluble protein.
INTRODUCTION
Microtubules (MTs) are polar cytoskeletal tracks that have many crucial functions in cells, including trafficking of cargoes, maintenance of cell shape and partitioning of genetic material to daughter cells during mitosis and meiosis (Hirokawa and Tanaka, 2015). These diverse functions are achieved through the plasticity of MT structural, biochemical and dynamic properties that can vary between cell types, between cell cycle stages or even between MTs at a given time within a single cell. Mechanisms that control MT dynamics include the stochastic growing and shrinking of ends intrinsic to the polymer (known as dynamic instability), the combined forces of motor proteins acting on the microtubules and the activities of microtubule-associated proteins (MAPs) that act as polymerases, depolymerases, stabilizers and destabilizers (Bowne-Anderson et al., 2015).
Information about MT dynamics and the effects of their MAPs have historically come from in vivo or in vitro studies. In vivo studies on dynamics are limited to either bulk analysis through such approaches as fluorescence recovery after photobleaching (FRAP) (Salmon et al., 1984) and speckle analysis (Grego et al., 2001; Waterman-Storer et al., 1998) or analysis of single MTs that can be resolved by light microscopy, usually at the cell periphery (Shaw et al., 1997). Dynamics at MT plus-ends at the cell periphery in interphase cells or astral MTs in dividing cells can be measured, but this excludes the plus-ends of most interphase and all kinetochore and interpolar MTs. Dynamics studies using reconstituted in vitro systems have produced stunning insights into such processes as spindle assembly (Sawin and Mitchison, 1991) and plus-end regulation (Li et al., 2012; Moriwaki and Goshima, 2016). Another productive approach for study of MT dynamics is genetics, which has been particularly valuable for discovery of key microtubule dynamics regulators (Pasqualone and Huffaker, 1994; Wang and Huffaker, 1997), which tend to be of low abundance and therefore difficult to identify by biochemical means. Although in vivo, in vitro and genetic approaches have been extremely productive, the combination of two or more of these, such as genetics and in vitro reconstitution, promises to achieve a level of analysis greater than could be achieved with any single approach alone. Development of a budding yeast lysate system for MT dynamics studies would allow genetics to be combined with the full complexity of total cellular protein in an open system reflecting the activities of the full panoply of yeast proteins expressed in yeast by comparing activities of extracts prepared from mutants of MAP and cell cycle control genes.
We developed an in vitro assay that reconstitutes MT dynamics within the high complexity of the total soluble protein content of a cell. This approach overcomes some of the inherent limitations of previous studies and can elucidate and examine the emergent properties of multiple MAPs acting on MTs. Our assay utilizes cleared lysate prepared from yeast strains and observes the polymerization and plus-end dynamics of MTs in vitro. Budding yeast is an ideal organism for in vitro reconstitution of MT dynamics for several reasons: its MT network is relatively simple but well studied; there is a fairly complete components list; and mutants of every key structural and regulatory protein are readily available (Moore et al., 2009; Winey and Bloom, 2012). Budding yeast includes homologs of many metazoan proteins and results are relevant to these organisms. Our system utilizes pre-formed MT seeds, allowing specific analysis of plus-end MT dynamics for readily resolved single MTs in the absence of rate-limiting nucleation reactions.
RESULTS
Creation of lysates and imaging chambers
Our goal was to create a protein system that enabled us to visualize single MT dynamics within the complexity of the cellular milieu. To that end, we combined aspects of classical in vivo genetics with the expediency, control and accessibility of an in vitro assay. We achieved this by preparing cleared lysates from budding yeast strains that natively express GFP-tubulin (Straight et al., 1997). We wanted to examine MT dynamics in lysates from actively growing cells, so strains were grown to late log phase before being harvested by centrifugation. To create a concentrated product, cells were washed with water during harvest to remove remaining medium. Any remaining liquid on top of the cell pellet was aspirated off. The concentrated cell pellet was apportioned and flash-frozen with liquid nitrogen and then crushed using a cryogenic impact mill, avoiding addition of any buffer or other liquid. The resulting lysate was highly concentrated (protein concentration of 81.5±8.5 mg ml−1).
Preparation of microscope slides, cover glasses and cellular lysates are discussed in detail in Materials and Methods. Briefly, a passivated coverslip was affixed to a microscope slide with strips of double-sided tape to create a flow chamber (Bieling et al., 2010). A biotin–streptavidin system was used to adhere rhodamine-labeled, GMPCPP-stabilized porcine MT seeds to the coverslip. Whole-cell lysates from strains natively expressing GFP-tubulin were buffered with 10× PEM solution, cleared of insoluble material by ultracentrifugation and supplemented with ATP and GTP. This mixture was then allowed to flow into the chamber. The microtubules were imaged by total internal reflection fluorescence (TIRF) microscopy at 28°C in an environmental chamber. The samples were imaged using 561 nm and 488 nm laser illumination every 5 s for 10 min. In lysates capable of polymerizing MTs, we found that by the time we could find an appropriate area to image, the GFP-tubulin had already begun to assemble off the rhodamine-labeled seeds. The activity of the lysates remained stable for up to 45 min of observation.
Initial experiments using lysate of wild-type strains growing asynchronously did not regularly exhibit any growing MTs on the rhodamine-labeled seeds. This was a puzzling result, as MTs are present and dynamic in all cells, but the observation is not without precedent. Previous work with actin showed that in vitro assembly of filaments was highly dependent on the cell cycle stage of the harvested cells (Miao et al., 2013). Microtubule dynamics across phyla are also known to fluctuate throughout the cell cycle to accommodate the variety of distinct functions they serve at each stage of the cell cycle (Rusan et al., 2001). To test this possibility, we sought to arrest the cultures at different points of the cell cycle before harvesting. Strains with temperature-sensitive alleles of cell-division cycle genes were used to arrest cells at different cell cycle stages. When utilizing these strains, incubation at the restrictive temperature for 3 h yielded >95% of cells arrested at either G1 (cdc28-4), S phase (cdc7-1), metaphase (cdc23-1) or in late anaphase (cdc15-2) (Goranov et al., 2009). We conducted our assay with these lysates and observed MTs growing from the seeds for all cases, except when the cells had been arrested in G1 (Fig. 1A). From here, we analyzed MT dynamics by generating kymographs and measuring the rates of growth and shrinking and the overall dynamicity (dimers s−1) for individual MTs.
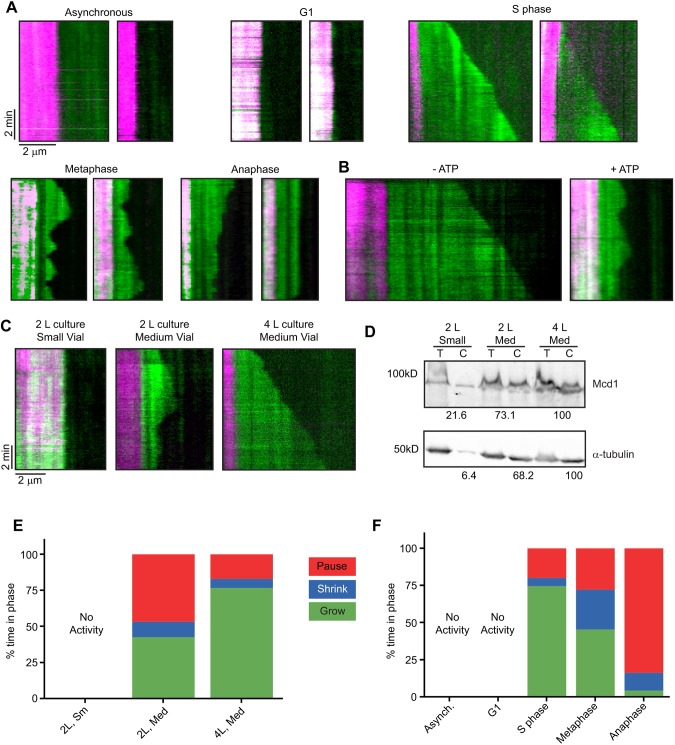
Fig. 1.
Conditions for assembly and dynamics of MTs in lysate. (A) Two examples of kymographs showing GFP-Tub1 (green) assembly from rhodamine-labeled MT seeds (magenta) and in lysates from each stage in the cell cycle. Minus ends are on the left, plus ends on the right. Time on the y-axis, 5 s pixel−1, totaling 10 min, starting from the top. (B) Kymographs of S phase lysate without additional ATP (left) and with 0.9 mM ATP. (C) Effect of culturing and grinding conditions on MT behavior in S phase-arrested lysate. (D) Western blot probing for levels of tubulin and Mcd1, a nuclear protein, in S phase lysates. Total lysate (T) and lysate cleared by ultracentrifugation (C) were analyzed. The first row of numbers indicates the percentage of Mcd1 present in the cleared lysate compared with the total lysate, normalized to the 4 l/medium vial lane. The second row of numbers indicates the percent of GFP-Tub1 present in the cleared lysate compared with the total lysate, normalized to the 4 l/medium vial lane. (E) MT growth profiles in S phase lysates based on culture volume and milling vial size. (F) MT growth profiles for lysates by cell cycle. The percentage of time spent in phases for the entire population of MTs is compared across stages of the cell cycle. MT measurements were pooled from three lysates generated from independent harvests of cells. MT counts for each condition are listed in Table 1.
Lysate optimization for reproducibility
To obtain interpretable results using this assay it was essential to have a high degree of reproducibility, especially when comparing dynamics properties across a large number of genetic backgrounds. It quickly became apparent that a number of factors could affect the dynamics of samples made independently from the same strain. These factors included added nucleotide, protein concentration and lysing conditions during milling.
Initially, only excess GTP was added to the lysate to maintain the GTPase activity necessary for MT polymerization (Carlier and Pantaloni, 1981). However, because many protein kinases, MAPs and the MT motors dynein and kinesin (in particular kinesin-8 and kinesin-14 family members) have direct effects on MT dynamics and require ATP for activity (Gupta et al., 2006; Maddox et al., 2003; Sproul et al., 2005; Varga et al., 2006), we investigated the need for exogenous ATP. We observed the MT dynamics of three independent preparations of lysates from S phase-arrested cells in the presence or absence of additional 0.9 mM ATP (Fig. 1B). In one of three lysates, without ATP added, MTs underwent constant growth without catastrophe whereas the other two had cycles of catastrophe and rescue. After exogenous ATP was added, all three preparations showed cycles of polymerization and depolymerization. Variable levels of endogenous ATP in the lysate clearly change the dynamics of MTs. Therefore, excess ATP and GTP were added for all experiments.
We hypothesized that the efficiency of milling could affect the protein concentration in lysates, which in turn might affect the dynamics of MTs assembled in the assay. Variability in milling can arise at several steps. The SPEX 6875 Cryogenic Impact Mill used for these studies allows for three different sizes of vial and corresponding impactors and has several settings for the duration and intensity of milling. Even though we had standardized the duration and intensity of our milling protocol (see Materials and Methods), different sizes of impactors had been used for different sizes of cell harvests, based on the manufacturer's instructions. To test the effect of these variables on reproducibility, we created several independent lysates from the same cdc7-1 parent strain using different culture volumes. These harvests were then milled in the recommended vial and impactor set for the corresponding harvested mass, according to the SPEX manual. The variability in MT activity between these samples was surprising. Samples ranged from having no detectable activity to assembling MTs that only grew and paused; other samples had MTs that underwent growth, pausing and shrinking (Fig. 1C). By quantifying protein levels using the Bradford assay and western blotting (Fig. 1D), we found that milling cells from a 2 l culture with the small milling set was less efficient in recovering α-tubulin than using the medium milling set on a 4 l culture. A correlation was discovered between protein concentration and these distinct phenotypes (Fig. 1E).
We wanted to determine whether nuclear protein levels might also be sensitive to changes in culture and milling volumes and therefore contribute to variations in the protein concentration of lysates. In budding yeast, the nuclear membrane stays intact throughout the cell cycle, creating separate compartments for microtubules with distinct subpopulations of MAPs in the cytoplasm and in the nucleus. To assess levels of nuclear proteins present in the cleared lysate, we compared protein extracts from cdc7-1 samples created using two different sizes of vials and impactors and two different culture volumes (2 l and 4 l). We then assayed the lysates for the presence of the cohesin complex subunit Mcd1 (Guacci et al., 1997), which localizes inside the nucleus, and α-tubulin, present in both compartments (Fig. 1D). We compared the amount of Mcd1 in the cleared lysate with the amount in total lysate. We found that when using the small impactor and vial set, the cleared lysate had only 23.1% of the Mcd1 found in the total lysate, whereas cleared lysate prepared using the medium impactor and vial set had 78.3% of the Mcd1 found in total lysate. When we increased the volume of cell culture from 2 l to 4 l and used the medium impactor and vial, essentially all of the Mcd1 was found in the cleared lysate. We then examined MT growth in these lysates (Fig. 1E). We did not observe any MTs growing from seeds in the 2 l sample prepared in the small vial. The increase in culture volume and resulting inclusion of more nuclear proteins and tubulin in the lysate significantly changed the behavior of MTs in our assay. MTs in lysate from a 2 l culture milled with a medium impactor exhibited growth only for 42.6% of observed time, whereas MTs in lysate from a 4 l culture grew 76.6% of the time. These results indicated that using a larger and heavier impactor and milling a greater mass of cells were necessary to recover nuclear proteins efficiently in the cleared lysates. This result can be attributed to increased efficiency in lysing cells and/or organelles.
To ensure consistent protein composition and concentration levels across all preparations, we standardized our harvest at several steps. First, 4 l cultures were grown to a standard density. Strains harvested without arrest were grown to late log-phase (OD600≈0.7). If the strains were to be arrested by temperature shift, they were first cultured to an OD600 of about 0.35 and then shifted to the restrictive temperature for 3 h. Second, exactly 4 g of cells was used when grinding cells using the cryogenic impact mill for lysate preparation, as this was the minimum mass of material harvested from 4 l of culture under our conditions. Third, milling was only carried out using the medium vial and impactor set. This procedure controlled for any effects on lysis efficiency as a result of spatial constraints of the impactor and sample within the sample vials.
Cell-cycle dependence of MT activity
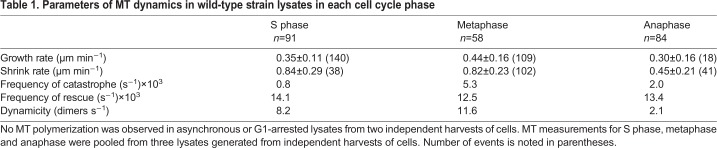
As mentioned above, MT growth and dynamics were only observed when otherwise wild-type cultures were arrested at different stages of the cell cycle (Fig. 1A, Movies 1–3). Asynchronous and G1-arrested lysates did not consistently show any appreciable GFP-tubulin signal growing off the rhodamine-labeled seeds. Table 1 lists the rates of growth and shrinkage, the frequency of catastrophe and rescue, and the overall dynamicity of the S phase-, metaphase- and anaphase-arrested lysates. In addition to the above measurements, we pooled the times of growth, shrinking and pausing for each population of MTs to create growth profiles for the respective cell cycle stages (Fig. 1F). These data support previous findings that all MT dynamics parameters are dependent upon the cell cycle.
Table 1.
Parameters of MT dynamics in wild-type strain lysates in each cell cycle phase
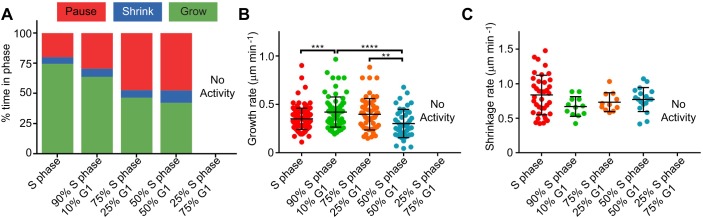
The lack of assembled MTs in asynchronous and G1-arrested lysates led us to hypothesize that an inhibitor of polymerization might be present in these lysates but not in lysates from later arrest points in the cell cycle. We further postulated that a mixture of G1 and S phase lysates might have characteristics intermediate between the original lysates if key factors are titratable, or might exhibit the characteristics of G1 lysate if the inhibitor acts in a dominant manner. We tested this by preparing two lysates arrested in different cell cycle stages, mixing them at different ratios and incubating for 5 min before adding nucleotides and flowing them onto slides. S phase and G1 lysates were mixed at 9:1, 3:1, 1:1 and 1:3 ratios, respectively, run through our assay and analyzed by kymographs. The 9:1 lysate mixture showed less growing time compared with S phase lysate (from 74.5% down to 63.7%) and an increase in shrinking time (5.6% to 6.7%) and pausing time (19.9% to 29.6%) (Fig. 2A). Interestingly, the growth rate increased from 0.35±0.11 to 0.42±0.16 μm min−1 (Fig. 2B). When the amount of S phase lysate was decreased to a 3:1 ratio, the growth rate was only slightly faster than in S phase (0.40±0.16 μm min−1) and the time spent paused was doubled to 47.4%. An equal mixture of lysate slowed the growth rate slightly to 0.30±0.15 μm min−1 and increased the amount of time the MTs were shrinking (5.6% to 10.3%) or pausing (19.9% to 47.6%). Importantly, none of the shrinkage rates in these experiments were statistically different from each other (Fig. 2C). When the ratio was reversed to 1 part S phase and 3 parts G1 lysate, there were no measurable MTs grown from the seeds, similar to G1 lysate. These data show that G1-arrested lysate has a dominant effect on MT dynamics when constituting 75% of a mixture with S phase-arrested lysate, but that the effect becomes titratable when constituting 50% or less of the mixture.
Fig. 2.
MT activity in mixed lysates is titratable. (A) MT growth profiles for mixed lysates. The 1:3 S:G1 lysate lacks any MT activity. (B) Growth rates of MTs in mixed lysates compared with wild-type lysates arrested in S phase. (C) MT shrinkage rates in S phase-arrested and mixed lysates. None are statistically different from the others, with the exception of the 1:3 S:G1 lysate. MT counts for each condition were 9:1=30, 3:1=25, 1:1=30. **P<0.01, ***P<0.001 and ****P<0.0001.
Association of MAPs with dynamic MTs
We next determined whether our assay is appropriate for studies on the association of MAPs with MTs. We used an RFP-tagged clone of the yeast homolog of the EB1 tip-tracking protein, BIM1, to follow its localization with MTs as they assembled in S phase lysate (Fig. 3A, Movie 4). Bim1-TagRFP-T was found along the entire length of the MTs, but concentrated at both growing and shrinking plus-ends, as described for in vivo (Wolyniak et al., 2006) and in vitro experiments (Zimniak et al., 2009). We also investigated the association and translocation of kinesin motors along these MTs by utilizing a strain that expressed KIP3-TagRFP-T and GFP-TUB1. Observation of lysates from cells arrested in metaphase showed that Kip3-TagRFP-T bound to a MT and moved towards the plus end (Fig. S1 and Movie 5). Kymograph analysis of Kip3-TagRFP-T molecules showed that these motors moved to the end of the MT and accumulated there as previously reported (Gupta et al., 2006). We calculated the rate of Kip3-TagRFP-T movement to be 2.8 μm min−1, similar to reported rates (Varga et al., 2009). These observations indicate that endogenous MAPs localize in this assay in a similar way to that reported in live cells.
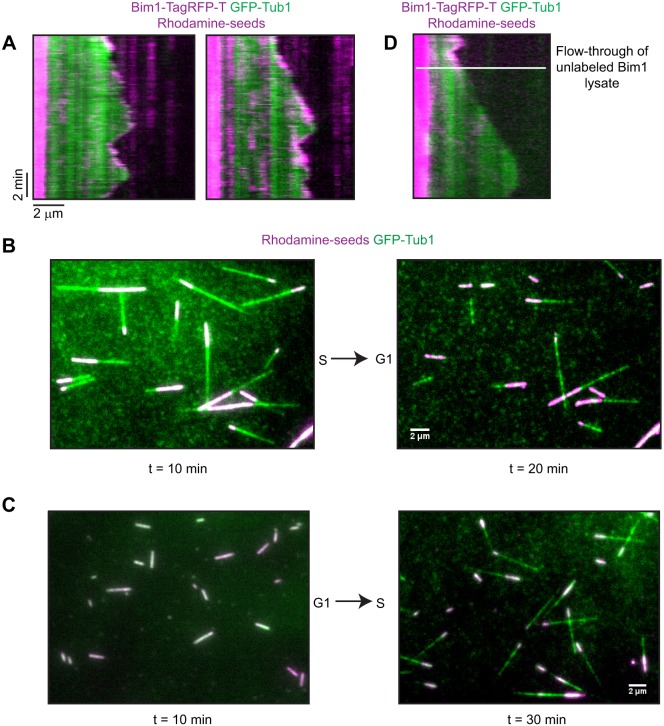
Fig. 3.
MAPs dynamically associate with MTs in lysates. (A) Kymograph of GFP-Tub1 (green) with Bim1-TagRFP-T and rhodamine-labeled seeds (magenta). Bim1-TagRFP-T is along MTs and decorates the plus end during growth and can be present on shrinking ends. (B) Fields of MTs in flow-through experiments. Dynamic MTs in S phase lysate were arrested and some began to shrink back to the seeds after flow through of G1 lysate. (C) In the reciprocal experiment, there were no MTs growing from seeds after 10 min in G1 lysate. At 20 min after flow-through of S phase lysate, MTs had grown and were dynamic. (D) Kymograph of S phase-arrested lysate expressing Bim1-TagRFP-T being replaced with S phase-arrested lysate with untagged Bim1 (white line).
Next, we determined whether our assay allows analysis of dynamic MAP exchange when different lysates are flowed sequentially over assembled MTs. For these experiments, two different lysates were thawed and cleared simultaneously before the addition of nucleotide. The first lysate was loaded into the chamber and the reaction was observed for 10 min so the MTs could assemble and become dynamic. Next, a new field of MTs was found and imaged for 2 min. Then, nucleotide was added to the second lysate and this mixture was flowed into the chamber, replacing the first lysate. The slide was mounted on the microscope during the whole procedure. Imaging of the same field resumed immediately, allowing us to follow the effects on one population of MTs as the lysate was changed. In control experiments of S phase lysate followed by fresh S phase lysate, we observed MTs that continued to have dynamic MT activity (Fig. S2). However, MTs assembled in an S phase lysate showed arrested growth and began to shrink back to the seeds when G1 lysate was flowed into the chamber (Fig. 3B). The opposite was seen for the reciprocal shift experiment in which MT seeds in G1 lysate began to assemble MTs after the addition of S phase lysate (Fig. 3C).
These effects could be explained if the exchange of lysates were sufficient to dissociate and/or exchange MAPs on the MTs. We tested this possibility by starting our assay with a BIM1-TagRFP-T lysate from S phase-arrested cells and then flowing onto the slide a similarly arrested lysate from cells that express unlabeled Bim1. In this sequence of events, the Bim1-TagRFP-T signal was lost from the MTs after flow-through of the second lysate (Fig. 3D).
Role of motor depolymerases
Having determined that MT polymerization and dynamics are dependent on the cell cycle stage of the cells from which lysates are prepared, we next investigated the possible cause of the absence of MT assembly in our assay in asynchronous and G1-arrested lysates. We tested the possibility that MT depolymerases prevent MT polymerization in the lysates. Two MT depolymerases have been identified in budding yeast, Kip3 (kinesin-8) (Gupta et al., 2006; Varga et al., 2009) and Kar3 (kinesin-14) (Endow et al., 1994). We examined MT dynamics in the absence of these proteins in asynchronous and cell cycle-arrested lysates.
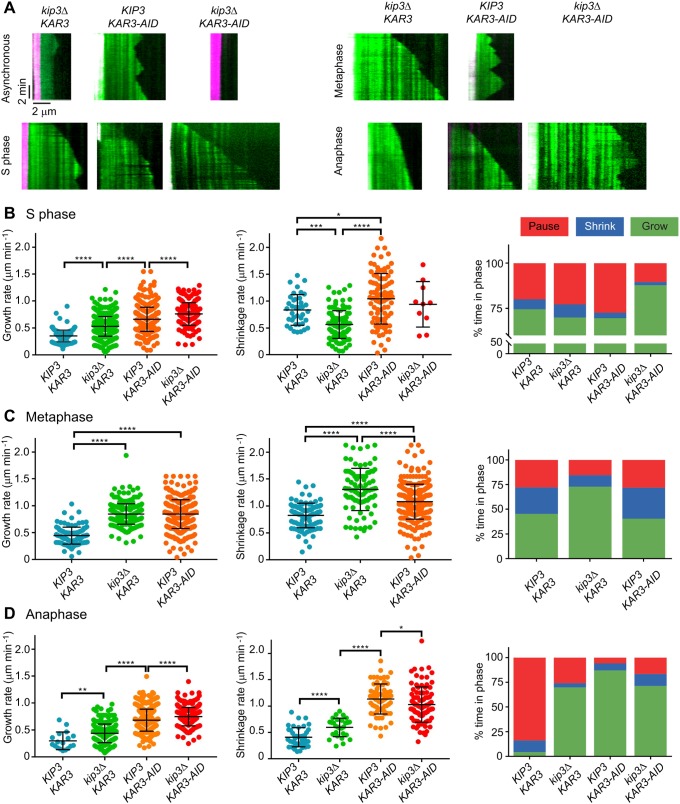
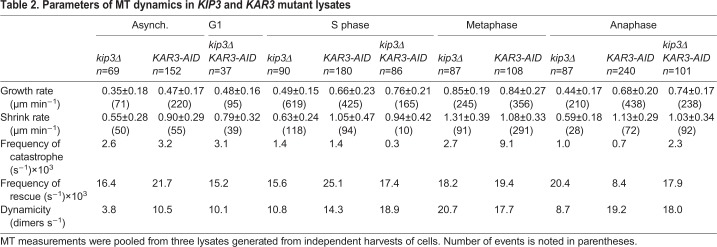
Kip3 is a highly processive plus-end-directed kinesin that destabilizes the plus end of the MT (Gupta et al., 2006). Absence of Kip3 in cells leads to longer cytoplasmic MTs (cMTs) as well as longer spindles that do not properly breakdown until after telophase (DeZwaan et al., 1997; Huyett et al., 1998; Woodruff et al., 2010). We deleted KIP3 in our strains to determine whether it is responsible for preventing MT assembly in lysates prepared from asynchronous and G1-arrested cells, and to determine how it affects MT dynamics in lysates arrested in other cell cycle stages. Surprisingly, the asynchronous kip3Δ lysates showed little MT assembly (Fig. 4A). The MTs that were observed had muted dynamics (0.35±0.18 μm min−1 growth rate and 0.55±0.28 μm min−1 shrinkage rate; Table 2). In keeping with our previous results, the G1-arrested kip3Δ lysates lacked any MT activity. However, in S phase-arrested kip3Δ lysates, the shrinkage rate decreased 25% (from 0.84±0.29 to 0.63±0.24 μm min−1) without any major change in the assembly profile. This result is in contrast to the increase in shrinkage rates for both the metaphase-arrested and anaphase-arrested kip3Δ lysates (from 0.82±0.23 to 1.31±0.39 μm min−1 and 0.45±0.21 to 0.59±0.18 μm min−1, respectively). Another interesting finding was that the growth rate increased in kip3Δ compared with the wild type for lysates arrested at S phase (from 0.35±0.11 to 0.49±0.15 μm min−1), metaphase (from 0.44±0.16 to 0.85±0.19 μm min−1) and anaphase (from 0.30±0.16 to 0.44±0.17 μm min−1). Moreover, in S phase kip3Δ lysates, MTs spent less time growing (70.0%) and more time shrinking (7.2%) or pausing (22.8%) than in wild-type lysates (Fig. 4B). This result was in contrast to those for metaphase- and anaphase-arrested kip3Δ lysates, wherein MTs spent most of their time growing (Fig. 4C,D). The catastrophe frequencies mirrored this change in growth profile for all three phases (Table 2). Based on these results, we conclude that, in our assay, Kip3 contributes to MT destabilization, but it alone does not prevent MT growth in lysates from G1-arrested cells. Additionally, the effects of Kip3 on MT dynamics are dependent on the cell cycle stage.
Fig. 4.
MT dynamics depend upon the cell cycle and motor activity in lysates. (A) Kymographs of kip3Δ, KAR3-AID and kip3Δ KAR3-AID lysates made from cells that were treated with auxin (KAR3-AID genotype cells) and were either asynchronous or arrested in S phase, metaphase or anaphase. Rhodamine-labeled seeds in magenta and GFP-Tub1 in green. (B,C,D) Growth rates, shrinkage rates and growth profiles for lysates made from cells with these genotypes in S phase (B), metaphase (C) and anaphase (D). MT measurements were pooled from three lysates generated from independent harvests of cells. MT counts for each condition are listed in Table 2. *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001.
Table 2.
Parameters of MT dynamics in KIP3 and KAR3 mutant lysates
The other budding yeast protein suggested to have MT depolymerase activity is Kar3. This kinesin-14 moves predominantly towards MT minus-ends (Molodtsov et al., 2016) and is involved in kinetochore capture (Tanaka et al., 2005) and spindle formation and stability (Hoyt et al., 1993; Saunders et al., 1997). Early work with Kar3 suggested that it has MT depolymerase activity (Chu et al., 2005; Endow et al., 1994; Sproul et al., 2005), but recent work from Mieck and colleagues has questioned this possibility (Mieck et al., 2015). Though KAR3 is not necessary for cell viability, kar3Δ strains exhibit mitotic delays (Meluh and Rose, 1990). To avoid any complications from cell cycle defects in KAR3 mutants, we used the auxin-induced degron (AID) system to enable tight control over the depletion of Kar3 activity from yeast (Fallis et al., 2009; Morawska and Ulrich, 2013). Our western blots showed that most Kar3-9myc-AID (referred to as Kar3-AID) protein is depleted upon treatment with indole acetic acid (IAA) for 15 min (Fig. S3). We paired our KAR3-AID allele with cdc mutants and added IAA during the last 30 min of a 3 h temperature shift. Interestingly, asynchronous KAR3-AID lysates displayed robust MT growth and dynamics (0.47±0.17 μm min−1 growth rate, 0.90±0.29 μm min−1 shrinkage rate; Table 2). However, G1-arrested lysates depleted of Kar3 did not assemble MTs. In S phase-arrested Kar3-depleted lysates, the growth (0.66±0.23 μm min−1) and shrinkage (1.05±0.47 μm min−1) rates increased 188.5% and 125%, respectively, compared with wild-type lysates, and the time spent pausing increased (27.4%) at the expense of shrink time (3.0%) (Fig. 4B). For lysates of metaphase-arrested cells, we found that depletion of Kar3 led to an increase in growth rate similar to the rate of kip3Δ lysates (0.84±0.27 μm min−1) and a slight increase in shrinkage rate compared with the wild type (1.08±0.33 μm min−1). Though the growth profile of this lysate appeared very similar to that of the wild type at the same arrested stage, the overall dynamics were elevated (Table 2). Dramatic differences were seen in anaphase-arrested lysates depleted of Kar3. Both the growth rate and shrinkage rate increase significantly over wild-type and kip3Δ rates (0.68±0.20 and 1.13±0.29 μm min−1, respectively) (Fig. 4D). In lysate of the Kar3-depleted anaphase-arrested cells, MTs spent most of their time (87.0%) growing and only seemed to change this behavior once they intersected or overlapped with another MT. This phenomenon became more prevalent over time. These data suggest that Kar3 has different roles throughout the cell cycle, with the most pronounced contribution to MT dynamics observed in anaphase.
We next analyzed MT dynamics in lysates made from kip3Δ KAR3-AID strains. The kip3Δ kar3Δ phenotype has not been reported previously because of a synthetic lethal genetic interaction (Cottingham and Hoyt, 1997), although a kip3ts kar3Δ analysis did not show any morphological defects in MTs (DeZwaan et al., 1997). Surprisingly, lysates from the asynchronous culture did not support MT assembly (Fig. 4A). However, lysates prepared from G1-arrested kip3Δ KAR3-AID lysate supported MT assembly from the seeds with moderate growth and shrinkage rates (0.48±0.16 and 0.79±0.32 μm min−1, respectively), and the MTs only grew about 55% of the time. In lysates prepared from the double mutant arrested in S phase, the MTs grew at a rate over twice that of the wild-type lysate (0.76±0.21 μm min−1). S phase lysate lacking Kar3 and Kip3 spent <2% of the time shrinking (Fig. 4B). The dynamicity of MTs in the S phase-arrested double mutants was the highest of any S phase lysate observed (18.9 dimers s−1). For anaphase lysates lacking these two motors, the growth rate doubled and the shrinkage rate increased 2.5-fold (0.74±0.17 and 1.03±0.34 μm min−1, respectively) compared with anaphase-arrested wild-type lysate, but the frequencies of catastrophe and rescue remained similar. Despite this, MTs in the double mutant lysate spent 71.3% of the time growing, 12.1% shrinking and 16.6% pausing, which was quite different from wild-type lysates in anaphase, but not that different from the single kip3Δ mutant (Fig. 4D). Based on our observations, we conclude that these two kinesin-family proteins contribute to MT destabilization, but in different capacities. When both are missing, MTs grow more often compared with wild-type lysates and, for single mutant lysates, and at faster rates. Unexpectedly, in the absence of these motors, MTs also depolymerize at a faster rate in anaphase lysates. Thus, their MT-associated activities are more complex than previously appreciated.
Two types of Kar3 heterodimers behave distinctly during the cell cycle
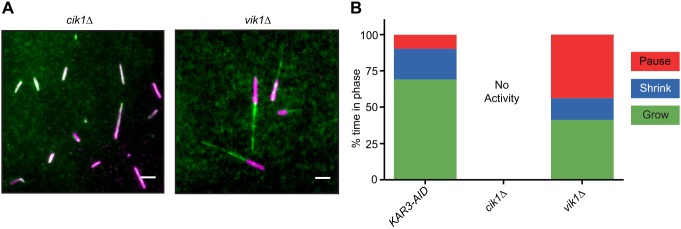
The Kar3 kinesin family protein is unique in that it has two distinct binding partners that create two different heterodimers with distinct cellular localizations and functions (Manning et al., 1999; Page et al., 1994; Shanks et al., 2001). Cik1 has a nuclear localization sequence that sequesters the Kar3-Cik1 heterodimer in the nucleus during vegetative growth, where it acts on nuclear MTs (nMTs) (Manning et al., 1999). Conversely, in mating cells, Cik1 is transcribed from an alternate start site, which omits the NLS, sending the heterodimer to the cytoplasmic face of the spindle pole body (SPB) (Benanti et al., 2009) where it forms the cMT array necessary for karyogamy (Hepperla et al., 2014). Neither of these functions requires depolymerase activity. Kar3's other binding partner is Vik1. Although both binding partners resemble kinesin-like proteins, Vik1 lacks the NLS found on Cik1 and its motor homology domain is truncated. Function of the Kar3-Vik1 heterodimer is not well understood, but its localization seems to be limited to the cytoplasmic face of the SPB and along cMTs (Manning et al., 1999). Using our assay, we attempted to dissect the different activities of these two heterodimers by creating single deletion mutants of CIK1 or VIK1 in our strains and prepared lysates from each strain. In lysates from asynchronously grown cik1Δ cells, there was no MT growth, similar to other asynchronous lysates (Table 3). However, in lysates prepared from asynchronously grown vik1Δ cells, MTs grew as well as they did in lysates from asynchronously grown KAR3-AID cells (Fig. 5A). These data indicate that in the context of a cellular lysate, Kar3-Vik1 (but not Kar3-Cik1) possesses depolymerase activity. Moreover, comparing the MT growth profiles for lysates prepared from asynchronously grown KAR3-AID and vik1Δ cells, MTs were found to spend less time growing (69.0% and 41.2%, respectively) and more time pausing (21.2% and 43.7%, respectively) and shrinking (9.8% and 15.0%, respectively) when Vik1 was absent (Fig. 5B). Thus, both Kar3-Cik1 and Kar3-Vik1 heterodimers contribute to MT dynamics regulation.
Table 3.
Parameters of MT dynamics in cik1Δ and vik1Δ asynchronous lysates
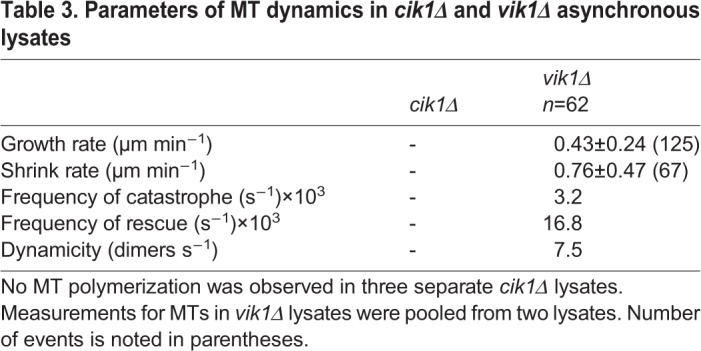
Fig. 5.
Kar3 binding partners alter MT polymerization activity. (A) Fields of rhodamine-labeled seeds (magenta) and GFP-Tub1 (green) using asynchronous cik1Δ and vik1Δ lysates. Scale bars: 2 μm. (B) MT growth profiles for KAR3-AID, cik1Δ and vik1Δ lysates from asynchronous cultures. KAR3-9myc-AID data from asynchronous lysate is presented again for comparison. No MT polymerization was observed in three separate cik1Δ lysates. Measurements for MTs in vik1Δ lysates were pooled from two lysates. A total of 62 MTs were counted.
DISCUSSION
MT dynamics assay using budding yeast cleared lysates
We created an assay that allows the dynamics of individual MTs to be analyzed in the context of a budding yeast cleared lysate made from total cellular proteins. Studies of MT dynamics often involve in vivo imaging of MTs or in vitro analysis of activities of purified proteins. Studies using Xenopus extracts have also made major contributions (Belmont et al., 1990). Each approach has advantages and disadvantages. In vivo imaging reveals the biological behavior of MTs, but often suffers from the inability to distinguish individual MTs. Studies of purified proteins reveal the functional capacity of individual proteins or small collections of proteins, but typically do not account for the full complexity of proteins and regulators. Use of Xenopus extracts allows the impact of the full complexity of the cytoplasm to be explored, but it can be challenging to identify the roles of individual proteins because of incomplete knockdown of activity using RNAi or antibodies. An extract system from budding yeast has great promise because activities can be explored in the total cellular protein complexity, and the functions of individual proteins can be tested by making extracts from mutants. Moreover, mutants can be exploited to explore several different cell cycle stages, and proteins can be fluorescently tagged and expressed at endogenous levels for visualization in the extract system.
Our lysate preparation procedure excludes any non-native material and minimizes the amount of added liquid. Frozen cells are pelleted and then crushed using a cryogenic impact mill, creating a lysate that is expected to contain the full complement of soluble cellular proteins. Minimizing protein dilution by adding only concentrated buffer and protease inhibitors can better mimic the native conditions within a cell. The specific components added were intended to maintain conditions suitable for MT dynamics despite the potential disruption of vacuoles, mitochondria, peroxisomes and other organelles that might potentially release proteases, hydrolyze nucleotides or lower the pH of the lysate. The importance of maintaining a high protein concentration for reproducibility might be due to near-threshold levels of activity at lower protein concentrations, minor differences therefore resulting in substantial variation in activity, or might be the result of other differences related to lysis efficiency.
Our assay was specifically developed to enable analysis of plus-end dynamics and not nucleation. Ultracentrifugation removes membranes and large cellular debris from the lysates so SPBs, the yeast MT organizing centers, are not likely to be present. The assay bypasses the nucleation steps of MT assembly because stable MT seeds are affixed onto a coverslip. Cleared lysate was supplemented with excess ATP to keep ATPases including motor proteins and protein kinases active; GTP was added for tubulin assembly. When the lysates were added to the seeds, microtubules could grow and exhibit dynamics without the complication of a nucleation step. In budding yeast, the minus ends of MTs are anchored into the SPB and are probably not dynamic (Bergman et al., 2012; Byers et al., 1978; Maddox et al., 2000). In our assay, we mainly observed MT growth only on one end of the seeds. Only rarely did tubulin assemble off both ends of a seed. The dynamics of the two ends were easily distinguishable by the lower dynamicity of the minus end. Only the plus end was measured in these cases.
To preserve physiological conditions, we monitored MT dynamics using a GFP-tagged copy of TUB1, one of the two α-tubulin genes in yeast. Use of endogenously tagged tubulin has the advantage of preserving the physiologically relevant mix of tubulin isotypes and any relevant post-translational modifications. Recent work supports the ‘tubulin-code’ hypothesis: that the differences in MT and MAP behavior between cell types can be a direct result of differences in the levels of tubulin isotypes incorporated into MTs and the actions of acetylases, methylases and detyrosinases (Garnham and Roll-Mecak, 2012; Sirajuddin et al., 2014; Vemu et al., 2017). In S. cerevisiae, levels of the α-tubulins Tub1 and Tub3 affect the dynamics of the MTs containing them (Bode et al., 2003). Another advantage of using a homogeneous extract system is that recent work has shown that MAPs from one species can interact differently with tubulins from different species (Howes et al., 2018; Kollman et al., 2015; Podolski et al., 2014). For these reasons, our system is likely to have advantages regarding preservation of physiological MT dynamic behavior and regulation.
Cell cycle dependence of MT dynamics
A few key conclusions can be drawn from these data. First and foremost is that MT dynamics in our reconstituted system approach known values (within one- to twofold) collected from in vivo data (Kosco et al., 2001). Table 1 reports the observed rates of growth and shrinkage, the frequencies of rescue and catastrophe and the overall dynamicity of MTs during different phases of the cell cycle. Combined with the growth profiles shown in Fig. 1F, these values approximate those reported for live cells. However, MT dynamics in our assay are significantly less than those reported using purified protein reconstitution systems that can recapitulate all phases of polymerization dynamics (Moriwaki and Goshima, 2016). Additionally, comparison of catastrophe events in our study versus those observed in previous in vitro systems demonstrates that kymographs of MTs from our assay show a slower rate of shrinking during catastrophe and that MTs typically undergo a rescue before reaching the GMPCPP-stabilized seed (Figs 1, 3 and 4). A catastrophe that almost instantaneously results in disassembly completely back to the seed has commonly been reported. Our system more closely resembles what is observed in vivo and in fully reconstituted systems than previously reported in vitro systems.
Our analysis is admittedly complicated in two ways by the fact that we have created a system without nucleo-cytoplasmic boundaries and spatial cues. First, we assay nuclear and cytoplasmic activities together. Because budding yeast undergo closed mitosis, the composition of MTs in the nucleus can be different from those found in the cytoplasm. Our method of lysis pools all available tubulin and MAPs, possibly creating unnatural mixtures of proteins and protein modifications. The pools of MAPs acting on nMTs and cMTs normally remain separate throughout the cell cycle. Second, the activity of some MAPs and the overall behavior of MTs has been shown to be spatially regulated in the cytoplasm to facilitate specific cellular functions (Estrem et al., 2017; Fukuda et al., 2014). Our assay lacks spatial cues other than proximity to other MTs. We can, nevertheless, speculate on the possible biological underpinnings of our results with the caveat that, in the future, it might be important to develop an approach that separates the cytoplasmic and nuclear pools of proteins. During S phase, cMTs grow rapidly so they can probe the cytoplasmic space in order to be captured by Myo2 and transported towards the bud neck (Hwang et al., 2003; Yeh et al., 2000; Yin et al., 2000). This could be why we mainly observed growing MTs. Meanwhile, kinetochore microtubules (kMTs) growing from the SPB probe the nuclear space and become captured by kinetochores (Huang and Huffaker, 2006; Tanaka et al., 2005). This process occurs earlier in the cell cycle than in cells with open mitosis, which might also place a premium on MT growth. In metaphase cells, the kMTs must fluctuate between growing and shrinking states in order to align the sister chromatids at the metaphase plate (Kosco et al., 2001; Sprague et al., 2003); this could explain the increased dynamics observed in our system. During late anaphase, kMTs are short and stable after they have been disassembled during chromosome separation. This reflects the prolonged pause time observed in lysates representing this stage. As for the interpolar microtubules (ipMTs), they are mostly stable in arrested cdc15-2 mutants because they have slid across each other to push the poles to the daughter cells, although some muted plus-end dynamics continue through anaphase (Fridman et al., 2009; Higuchi and Uhlmann, 2005; Rizk et al., 2014). Accordingly, the growth profiles for anaphase-arrested lysates were paused for >80% of the observed time, possibly reflecting a blended state of the dynamics observed in vivo for kMTs and ipMTs.
Despite the similarities between MTs observed in live cells and those in our assay during later parts of the cell cycle, it was curious that MTs did not assemble in lysates made from asynchronous and G1-arrested cells (with the exception of kip3Δ KAR3-AID). Clearly, MTs exist and are dynamic in the cytoplasm during G1. There is evidence that in budding yeast the nuclear MTs during this stage of the cell cycle are firmly attached to kinetochores and are not dynamic (Dorn et al., 2005; Jin et al., 2000; O'Toole et al., 1999). This MT behavior could be modulated through MAPs that are only present in either the cytoplasm or the nucleus. Recent work suggests that the nuclear-localized fraction of Stu2, the yeast XMAP215 homolog, is in part responsible for maintaining the short length and muted dynamics of nMTs during mating (van der Vaart et al., 2017). Results from our lysate mixing experiments (Fig. 2) suggest that there is a titratable factor that can inhibit the growth and formation of MTs. The sequential flow-through experiments also point to a factor (or factors) in G1 lysates that can stop the growth of existing MTs that will then depolymerize all the way back to the seeds. In the future, it will be important to use genetics and our extract system to identify this factor or factors.
In summary, these studies reveal pronounced changes in MT dynamics with different cell cycle stages, the molecular and mechanistic basis for which can now be determined using our budding yeast lysate system.
Emergent properties of two kinesin depolymerases
One approach for investigating the basis for the absence of MT assembly in lysates from asynchronous and G1-arrested cells is to remove the two known MT depolymerases genetically. Budding yeast has a kinesin-8 (Kip3) and a kinesin-14 (Kar3), which have both been shown to destabilize MTs (Endow et al., 1994; Gupta et al., 2006). In our assay, deleting the KIP3 locus resulted in only modest MT dynamics in asynchronous lysates and no detectable MT growth in G1 lysates (Fig. 4). However, in asynchronous lysates, knocking down Kar3 protein with a KAR3-AID allele showed robust MT assembly and dynamics. Interestingly, lysate from asynchronous kip3Δ KAR3-AID cells lacks the ability to assemble MTs, unlike both single-mutant asynchronous lysates. The intriguing interplay of Kip3 and Kar3 activities became even more apparent when the cells were arrested in S phase. When Kar3 is the only depolymerase present, MTs shrunk for a greater portion of time. Without Kar3, MTs paused longer and shrunk faster. Lysate generated from S phase-arrested cells lacking both depolymerases spent more time growing and had an increased disassembly rate. These observations are consistent with the possibility that Kip3 increases the disassembly rate in S phase-arrested lysates. However, there are further layers of complexity that need to be investigated because the microtubule disassembly rates of kip3Δ lysates in other cell cycle phases increase (Table 2), as previously reported in cells (Fukuda et al., 2014; Gupta et al., 2006; Su et al., 2013). Examination of the MT growth rates in these lysates showed that they increase in either single mutant, and even more so in the double mutant. An increase in growth rate in the absence of depolymerases suggests that Kar3 and Kip3 not only promote MT disassembly, but also modulate assembly rates.
This new technique begins to address questions that have been difficult to answer using in vivo approaches alone. Kar3 has long been thought to exhibit MT depolymerase activity. Direct evidence of this activity has thus far eluded the field, perhaps due to the presence of two different Kar3 heterodimers (Kar3-Cik1 and Kar3-Vik1) in separate cellular localizations and due to cell-cycle-dependent contexts. In our assay, the presence of MT assembly in vik1Δ but not cik1Δ asynchronous cell lysates indicates that the Kar3-Vik1 heterodimer has MT depolymerase activity at some point in the cell cycle, probably during G1. It is now important to investigate the contributions of the activity of these two heterodimers at different points in the cell cycle in order to determine the nature of this phenotype.
Our assay has successfully reconstituted MT dynamics in cell lysate from a genetically tractable organism. We have shown that MT behavior is greatly influenced by the cell cycle stage, with roles for depolymerases. Although the mixing of nuclear and cytoplasmic contents is a complication, this system can be further exploited to dissect nuclear and cytoplasmic regulatory mechanisms, both of which are probably reflected in our observations. This is particularly valuable for nuclear MT regulation because this MT population is very difficult to visualize in live cells. Another consideration is that, in budding yeast, each SPB emanates three to five cytoplasmic MTs in G1 (Byers and Goetsch, 1975) or two during mitosis (O'Toole et al., 1999), and about 20 nuclear MTs, and this major MT population cannot be readily observed in vivo. Because we are observing single MTs in the complexity of soluble cellular lysates, the emergent properties of protein populations on MTs can be studied. We have shown that MAPs can be visualized and tracked on MTs in this system. During our work, we also observed MT bundling in parallel and anti-parallel orientations, and MTs zippering together after meeting at their plus ends (results not shown), suggesting that there is even more potential for elucidating biologically important MT-based activities using the complex protein environment of yeast lysate. Every yeast gene has been disrupted, so it is possible to assay, in an open extract system, the MT-related activities of essentially all yeast proteins and to adapt the assay for the study of additional cellular processes.
MATERIALS AND METHODS
Yeast strains, culturing and harvesting
Yeast strains used in this study can be found in Table S1. Fluorescent and degradation tags were integrated by homologous recombination as previously described. Strains were grown in standard rich medium (YPD) at 30°C, or at 25°C if they contained a temperature-sensitive allele. Strains containing the KAR3-9myc-AID allele were treated with 250 μM 3-indole acetic acid (Sigma-Aldrich) in DMSO and buffered with 50 mM potassium phosphate buffer at pH 6.2 for the 30 min just prior to harvesting.
For lysate generation, strains were grown overnight in starter cultures and then diluted into two parallel 2 l cultures of YPD and grown until OD600≈0.7, or OD600≈0.35 if cultures were to be shifted for arrest. To arrest cells, cultures were shifted to 37°C for 3 h before being harvested. Strains that contained an auxin-inducible degron had a final concentration of 50 mM potassium phosphate buffer pH 6.2 and 250 μM indole acetic acid in DMSO added for the last 30 min of culturing. Cells were then harvested by serial centrifugation at 6000 rpm in a Sorvall RC5B with a SLA-3000 rotor for 10 min at 4°C. Cells were then resuspended in ddH2O, transferred to a 50 ml conical tube and pelleted in a Thermo Fisher Scientific CR3i table-top centrifuge for 3 min at 3000 rpm. This wash and pelleting process was repeated. After the second wash, all standing moisture was removed from the cell pellet by aspiration. Cells were then flash-frozen in liquid nitrogen and stored at −80°C.
Generation of whole cell lysates
Frozen cells (4 g) were weighed and placed into a medium-sized SPEX vial that had been pre-chilled in liquid nitrogen. Milling consisted of a 3 min pre-chill followed by 10 cycles comprising 3 min of grinding at 30 impacts per second (15 cps) and 1 min of rest. The sample vial remained submerged in liquid nitrogen throughout the process. Powdered lysate was collected in a 50 ml conical tube and stored at −80°C. Lysate preparations were stable for >1 year.
Generating rhodamine-labeled tubulin seed mix
Previously isolated porcine tubulin was cycled to ensure the absence of nonfunctional tubulin. This was mixed with both biotin-conjugated and rhodamine-labeled porcine tubulin (Cytoskeleton Inc.) resuspended in PEM (80 mM PIPES pH 6.9, 1 mM EGTA, 1 mM MgCl2). Final concentrations were 5 mg ml−1 unlabeled tubulin, 1 mg ml−1 biotin-labeled tubulin and 1 mg ml−1 rhodamine-labeled tubulin. GMPCPP (Jena Biosciences) was added to a final concentration of 1 mM. Aliquots were flash-frozen in liquid nitrogen and stored at −80°C.
Cleaning of glass slides and passivation of coverslips
Prior to use, microscope slides (Corning Inc.) were washed in acetone for 15 min and then in 100% ethanol for 15 min. Slides were left to air dry before being stored in an airtight container. Cover glass (1.5 thickness; Corning Inc.) was first cleaned by submerging in isopropanol and then subjected to 20 min of sonication. Coverslips were then washed twice with ddH2O for 1 min and then in 70% ethanol for 1 min. The coverslips were then blow-dried with nitrogen gas and placed into a ceramic coverslip holder. This step was followed by 10 min of illumination in a plasma cleaner chamber (Harrick Plasma PDC-32G). A 0.1 mg ml−1 solution of PLL-g-PEG:PEG(3.4)-biotin (50%:50%) (SuSoS AG) in 10 mM HEPES was prepared and 50 μl drops were placed onto Parafilm. Coverslips were placed on top of the drops and covered in a humidity chamber for 1 h. The passivated cover glass was then washed for 2 min in PBS and rinsed in ddH2O for 1 min. The coverslips were again air-dried with nitrogen and stored in an airtight container at 4°C. Passivated coverslips could be used up to 2 weeks later.
Assembly of flow chamber
Double-sided tape was cut lengthwise into thirds and the strips were placed in parallel on the center of a cleaned microscope slide to create two equally sized channels. A passivated coverslip was placed on top of the tape and a cotton-tipped applicator was used to press gently on the coverslip at the lines of contact with the tape to ensure a continuous bond and prevent leaks. A single channel was first washed with 30 μl PEM and wicked through with Whatman filter paper. Neutravidin (Invitrogen) was diluted 1:400 in PEM and 30 μl was pulled through the channel. The slide was then kept in a humidified chamber for 10 min. The channel was then washed twice with 30 μl of Pluronic™ F-127 (Invitrogen) diluted to 0.1% in PEM. This assembly was again incubated under humidity for 10 min. During this incubation, porcine tubulin was assembled into seeds by incubating Tubulin Seed Mix (above) at 37°C for 10 min. A 10× oxygen scavenging (OS) solution was freshly prepared by mixing 10 μl of PEM with 5 μl of 40× glucose oxidase+catalase (8 mg ml−1 glucose oxidase, 1.4 mg ml−1 catalase in PEM) and 5 μl of 40×gGlucose+2–mercaptoethanol (180 mg ml−1 glucose, 20% 2-mercaptoethanol in PEM). MT seed mix was made by addition of 0.5 μl of assembled seeds to 36.5 μl PEM, 5 μl 5 mg ml−1 casein, 5 μl 10× OS and 2.5 μl 2% methylcellulose. MT seed mix (20 μl) was then flowed through the channel. Excess MT seed mix was placed on the ends of the channel to prevent drying of the channel. The slide was incubated at 37°C for 5 min. The channel was then washed with 50 μl of warm PEM (1× OS solution diluted with PEM at 37°C), leaving it ready for addition of lysate.
Preparation of whole cell lysates for assay
To prepare lysate for use in the assay, 0.22 g of powdered lysate was weighed out into a 1.5 ml tube pre-chilled in liquid nitrogen. Then, 25 μl of cold 10× PEM (800 mM PIPES pH 6.9, 10 mM MgCl2, 10 mM EGTA) and 0.5 μl of Protease Inhibitor Cocktail IV (Calbiochem) were added to the lysate and spun down briefly. Lysate was thawed on ice for 10 min before loading into the pre-chilled polycarbonate ultracentrifuge tube. Lysate was then cleared of insoluble material by spinning at 346,000 g for 25 min at 4°C. After the spin, 20 μl of cleared lysate was aliquoted into a fresh 1.5 ml tube pre-chilled on ice. Cleared lysates were stable for >1 h. Before use, 1 μl each of 20 mM ATP and 20 mM GTP (both in PEM) were added to cleared lysate before flowing through the prepared chamber.
Immunoblotting
To compare relative protein concentrations in lysates, equal amounts of thawed and diluted lysate powder were run on a 10% polyacrylamide gel. Preparation of cleared lysate is described above. Total lysate was prepared by adding 220 µl of cold 2× PEM and 0.5 µl of Protease Inhibitor Cocktail IV to 0.22 g of lysate powder. The mixture was solubilized with an equal volume of 2× SDS sample buffer, boiled for 1 min and briefly centrifuged before the supernatant was loaded onto the gel. Immunoblotting was performed with 1:10,000 rabbit anti-Mcd1 (gift from Dr Vincent Guacci, University of California, Berkeley) and 1:1000 rat anti-α-tubulin (Santa Cruz Biotechnology, 5C-53030). Band intensities were measured with Image Studio Lite (LI-COR).
For tracking the amount of Kar3-9myc-AID protein in cells, cultures were grown to mid-log phase in rich media and treated with 250 µM indole acetic acid in DMSO and buffered with 50 mM potassium phosphate buffer at pH 6.2. Cells were then harvested over a time course by centrifugation in a Thermo Fisher CR3i table-top centrifuge for 3 min at 3000 rpm. Cell pellets were resuspended in 1 ml 20% trichloroacetic acid (TCA, Sigma-Aldrich) and transferred to a 2 ml screw-top tube. Cells were again pelleted in a microfuge at top speed for 2 min. The supernatant was removed and the cell pellet was resuspended in 200 µl 20% TCA. Approximately 200 µl of 425–600 µm acid-washed glass beads (Sigma-Aldrich) was added to the tube, which was then agitated on a vortexer for 10 min at 4°C. To collect lysate, a hole in the bottom of the tube was made with a 25 G needle and the entire tube was placed in a 5 ml round-bottom tube. This apparatus was then centrifuged at 2500 rpm for 3 min in the table-top centrifuge. Beads were washed with 200 µl 5% TCA and collected in the same 5 ml tube, twice. The pellet in the 5 ml tube was resuspended in the standing liquid and transferred to a 1.5 ml tube. This was spun at 5000 rpm for 10 min in a microcentrifuge. The supernatant was discarded and the pellet was resuspended in 2× SDS sample buffer. Tris base (1 M) was used to neutralize any remaining TCA. Equivalent OD amounts were then separated electrophoretically on a 10% polyacrylamide gel and transferred to nitrocellulose. 9E10 mouse anti-myc primary antibody (prepared from hybridoma supernatant) was used at 1:750 dilution for detecting KAR3-9myc-AID protein.
TIRF microscopy
Once lysate flowed through the prepared chamber, the slide was loaded onto an Olympus IX81 inverted microscope with an environmental chamber pre-warmed to 28°C. Images were acquired with a 100× PlanApo objective (NA 1.45) and an Orca CCD camera (Hamamatsu) using Metamorph software (Molecular Devices). TIRF was used to illuminate a single plane of the field with 488 nm and 561 nm light every 5 s for 10 min.
Sequential lysate flow assays
For sequential flow experiments, double-stick tape was used to make a perpendicular chamber across the microscope slide. A passivated rectangular coverslip was then placed on top of the tape to form a chamber that ran the entire width of the slide, with overhangs on both sides. The whole assembly was flipped with the coverslip-side down in order to flow solutions through chamber as described above. All flow solution volumes were increased by 10 μl to account for the larger volume of the chamber. To observe initial behavior, 30 μl of the first lysate was flowed through the chamber and imaged for 10 min. This was followed by 40 μl of the second lysate while the sample remained mounted on the microscope.
Image and data analysis
Image files were analyzed using Fiji (NIH). Kymographs were constructed from all MTs whose entire length was trackable for the entire movie after registration (StackReg; Thévenaz et al., 1998). Dynamics parameters were calculated as described by Moriwaki and Goshima (2016). To calculate values, data from independent technical and biological repeats from one genotype were pooled unless otherwise indicated. Growth and shrinkage rates are reported as mean±standard deviation. Statistical significance was determined using an unpaired t-test. Values are reported as *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001.
Supplementary Material
Acknowledgements
The authors would like to thank Itziar Ibarlucea Benitez for discussion and development of methods. We also thank members of the Drubin lab for discussion on experiments and analysis. Strains with temperature-sensitive alleles of CDC genes were a gift from Wei Guo (University of Pennsylvania).
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: Z.J.B., J.W., D.G.D., G.B.; Methodology: Z.J.B., J.W.; Validation: Z.J.B., J.W.; Formal analysis: Z.J.B., J.W.; Investigation: Z.J.B., J.W.; Writing - original draft: Z.J.B., D.G.D., G.B.; Writing - review & editing: Z.J.B., J.W., D.G.D., G.B.; Visualization: Z.J.B., J.W.; Funding acquisition: G.B.
Funding
This work was supported by the National Institutes of Health (Grant R01 GM 47842) to G.B. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.219386.supplemental
References
- Belmont L. D., Hyman A. A., Sawin K. E. and Mitchison T. J. (1990). Real-time visualization of cell cycle-dependent changes in microtubule dynamics in cytoplasmic extracts. Cell 62, 579-589. 10.1016/0092-8674(90)90022-7 [DOI] [PubMed] [Google Scholar]
- Benanti J. A., Matyskiela M. E., Morgan D. O. and Toczyski D. P. (2009). Functionally distinct isoforms of Cik1 are differentially regulated by APC/C-mediated proteolysis. Mol. Cell 33, 581-590. 10.1016/j.molcel.2009.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman Z. J., Xia X., Amaro I. A. A. and Huffaker T. C. (2012). Constitutive dynein activity in she1 mutants reveals differences in microtubule attachment at the yeast spindle pole body. Mol. Biol. Cell 23, 2319-2326. 10.1091/mbc.e12-03-0223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling P., Telley I. A., Hentrich C., Piehler J. and Surrey T. (2010). Fluorescence Microscopy Assays on Chemically Functionalized Surfaces for Quantitative Imaging of Microtubule, Motor, and +Tip Dynamics. Methods Cell Biol. 95, 555-580. 10.1016/S0091-679X(10)95028-0 [DOI] [PubMed] [Google Scholar]
- Bode C. J., Gupta M. L., Suprenant K. A. and Himes R. H. (2003). The two α-tubulin isotypes in budding yeast have opposing effects on microtubule dynamics in vitro. EMBO Rep. 4, 94-99. 10.1038/sj.embor.embor716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowne-Anderson H., Hibbel A. and Howard J. (2015). Regulation of microtubule growth and catastrophe: unifying theory and experiment. Trends Cell Biol. 25, 769-779. 10.1016/j.tcb.2015.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B. and Goetsch L. (1975). Behavior of spindles and spindles plaques in the cell cyle and conjugation of Saccharomyces cerevisiae. J. Bacteriol. 124, 511-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B., Shriver K. and Goetsch L. (1978). The role of spindle pole bodies and modified microtubule ends in the initiation of microtubule assembly in Saccharomyces cerevisiae. J. Cell Sci. 30, 331-352. [DOI] [PubMed] [Google Scholar]
- Carlier M. F. and Pantaloni D. (1981). Kinetic analysis of guanosine 5′-triphosphate hydrolysis associated with tubulin polymerization. Biochemistry 20, 1918-1924. 10.1021/bi00510a030 [DOI] [PubMed] [Google Scholar]
- Chu H. M. A., Yun M., Anderson D. E., Sage H., Park H.-W. and Endow S. A. (2005). Kar3 interaction with Cik1 alters motor structure and function. EMBO J. 24, 3214-3223. 10.1038/sj.emboj.7600790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottingham F. R. and Hoyt M. A. (1997). Mitotic spindle positioning in Saccharomyces cerevisiae is accomplished by antagonistically acting microtubule motor proteins. Cell 138, 1041-1053. 10.1083/jcb.138.5.1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeZwaan T. M., Ellingson E., Pellman D. and Roof D. M. (1997). Kinesin-related KIP3 of Saccharomyces cerevisiae is required for a distinct step in nuclear migration. J. Cell Biol. 138, 1023-1040. 10.1083/jcb.138.5.1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn J. F., Jaqaman K., Rines D. R., Jelson G. S., Sorger P. K. and Danuser G. (2005). Yeast kinetochore microtubule dynamics analyzed by high-resolution three-dimensional microscopy. Biophys. J. 89, 2835-2854. 10.1529/biophysj.104.058461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow S. A., Kang S. J., Satterwhite L. L., Rose M. D., Skeen V. P. and Salmon E. D. (1994). Yeast Kar3 is a minus-end microtubule motor protein that destabilizes microtubules preferentially at the minus ends. EMBO J. 13, 2708-2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrem C., Fees C. P. and Moore J. K. (2017). Dynein is regulated by the stability of its microtubule track. J. Cell Biol. 216, 2047-2058. 10.1083/jcb.201611105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallis A., Nishimura K., Fukagawa T., Takisawa H., Kakimoto T. and Kanemaki M. (2009). An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods 53, 1689-1699. 10.1038/nmeth.1401 [DOI] [PubMed] [Google Scholar]
- Fridman V., Gerson-Gurwitz A., Movshovich N., Kupiec M. and Gheber L. (2009). Midzone organization restricts interpolar microtubule plus-end dynamics during spindle elongation. EMBO Rep. 10, 387-393. 10.1038/embor.2009.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda Y., Luchniak A., Murphy E. R. and Gupta M. L. (2014). Spatial control of microtubule length and lifetime by opposing stabilizing and destabilizing functions of kinesin-8. Curr. Biol. 24, 1826-1835. 10.1016/j.cub.2014.06.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnham C. P. and Roll-Mecak A. (2012). The chemical complexity of cellular microtubules: tubulin post-translational modification enzymes and their roles in tuning microtubule functions. Cytoskeleton 69, 442-463. 10.1002/cm.21027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goranov A. I., Cook M., Ricicova M., Ben-Ari G., Gonzalez C., Hansen C., Tyers M. and Amon A. (2009). The rate of cell growth is governed by cell cycle stage. Genes Dev. 23, 1408-1422. 10.1101/gad.1777309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grego S., Cantillana V. and Salmon E. D. (2001). Microtubule treadmilling in vitro investigated by fluorescence speckle and confocal microscopy. Biophys. J. 81, 66-78. 10.1016/S0006-3495(01)75680-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V., Koshland D. and Strunnikov A. (1997). A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell 91, 47-57. 10.1016/S0092-8674(01)80008-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M. L., Carvalho P., Roof D. M. and Pellman D. (2006). Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat. Cell Biol. 8, 913-923. 10.1038/ncb1457 [DOI] [PubMed] [Google Scholar]
- Hepperla A. J., Willey P. T., Coombes C. E., Schuster B. M., Gerami-Nejad M., McClellan M., Mukherjee S., Fox J., Winey M., Odde D. J. et al. (2014). Minus-end-directed kinesin-14 motors align antiparallel microtubules to control metaphase spindle length. Dev. Cell 31, 61-72. 10.1016/j.devcel.2014.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi T. and Uhlmann F. (2005). Stabilization of microtubule dynamics at anaphase onset promotes chromosome segregation. Nature 433, 171-176. 10.1038/nature03240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N. and Tanaka Y. (2015). Kinesin superfamily proteins (KIFs): various functions and their relevance for important phenomena in life and diseases. Exp. Cell Res. 334, 16-25. 10.1016/j.yexcr.2015.02.016 [DOI] [PubMed] [Google Scholar]
- Howes S. C., Geyer E. A., LaFrance B., Zhang R., Kellogg E. H., Westermann S., Rice L. M. and Nogales E. (2018). Structural and functional differences between porcine brain and budding yeast microtubules. Cell Cycle 17, 278-287. 10.1080/15384101.2017.1415680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt M. A., He L., Totis L. and Saunders W. S. (1993). Loss of function of Saccharomyces cerevisiae kinesin-related CIN8 and KIP1 is suppressed by KAR3 motor domain mutations. Genetics 135, 35-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B. and Huffaker T. C. (2006). Dynamic microtubules are essential for efficient chromosome capture and biorientation in S. cerevisiae. J. Cell Biol. 175, 17-23. 10.1083/jcb.200606021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyett A., Kahana J., Silver P., Zeng X. M. and Saunders W. S. (1998). The Kar3p and Kip2p motors function antagonistically at the spindle poles to influence cytoplasmic microtubule numbers. J. Cell Sci. 111, 295-301. [DOI] [PubMed] [Google Scholar]
- Hwang E., Kusch J., Barral Y. and Huffaker T. C. (2003). Spindle orientation in Saccharomyces cerevisiae depends on the transport of microtubule ends along polarized actin cables. J. Cell Biol. 161, 483-488. 10.1083/jcb.200302030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q. W., Fuchs J. and Loidl J. (2000). Centromere clustering is a major determinant of yeast interphase nuclear organization. J. Cell Sci. 113, 1903-1912. [DOI] [PubMed] [Google Scholar]
- Kollman J. M., Greenberg C. H., Li S., Moritz M., Zelter A., Fong K. K., Fernandez J.-J., Sali A., Kilmartin J., Davis T. N. et al. (2015). Ring closure activates yeast γTuRC for species-specific microtubule nucleation. Nat. Struct. Mol. Biol. 22, 132-137. 10.1038/nsmb.2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosco K. A., Pearson C. G., Maddox P. S., Wang P. J., Adams I. R., Salmon E. D., Bloom K. and Huffaker T. C. (2001). Control of microtubule dynamics by Stu2p is essential for spindle orientation and metaphase chromosome alignment in yeast. Mol. Biol. Cell 12, 2870-2880. 10.1091/mbc.12.9.2870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moriwaki T., Tani T., Watanabe T., Kaibuchi K. and Goshima G. (2012). Reconstitution of dynamic microtubules with drosophila XMAP215, EB1, and sentin. J. Cell Biol. 199, 849-862. 10.1083/jcb.201206101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox P. S., Bloom K. S. and Salmon E. D. (2000). The polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae. Nat. Cell Biol. 2, 36-41. 10.1038/71357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox P. S., Stemple J. K., Satterwhite L., Salmon E. D. and Bloom K. (2003). The minus end-directed motor Kar3 is required for coupling dynamic microtubule plus ends to the cortical shmoo tip in budding yeast. Curr. Biol. 13, 1423-1428. 10.1016/S0960-9822(03)00547-5 [DOI] [PubMed] [Google Scholar]
- Manning B. D., Barrett J. G., Wallace J. A., Granok H. and Snyder M. (1999). Differential regulation of the Kar3p kinesin-related protein by two associated proteins, Cik1p and Vik1p. J. Cell Biol. 144, 1219-1233. 10.1083/jcb.144.6.1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh P. B. and Rose M. D. (1990). KAR3, a kinesin-related gene required for yeast nuclear fusion [published erratum appears in Cell 1990 May 4;61(3):548]. Cell 60, 1029-1041. 10.1016/0092-8674(90)90351-E [DOI] [PubMed] [Google Scholar]
- Miao Y., Wong C. C. L., Mennella V., Michelot A., Agard D. A., Holt L. J., Yates J. R. and Drubin D. G. (2013). Cell-cycle regulation of formin-mediated actin cable assembly. Proc. Natl. Acad. Sci. USA 110, E4446-E4455. 10.1073/pnas.1314000110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieck C., Molodtsov M. I., Drzewicka K., van der Vaart B., Litos G., Schmauss G., Vaziri A. and Westermann S. (2015). Non-catalytic motor domains enable processive movement and functional diversification of the kinesin-14 kar3. eLife 2015, 1-23. 10.7554/eLife.04489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molodtsov M. I., Mieck C., Dobbelaere J., Dammermann A., Westermann S. and Vaziri A. (2016). A force-induced directional switch of a molecular motor enables parallel microtubule bundle formation. Cell 167, 539-552.e14. 10.1016/j.cell.2016.09.029 [DOI] [PubMed] [Google Scholar]
- Moore J. K., Magidson V., Khodjakov A. and Cooper J. A. (2009). The spindle position checkpoint requires positional feedback from cytoplasmic microtubules. Curr. Biol. 19, 2026-2030. 10.1016/j.cub.2009.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morawska M. and Ulrich H. D. (2013). An expanded tool kit for the auxin-inducible degron system in budding yeast. Yeast 30, 341-351. 10.1002/yea.2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriwaki T. and Goshima G. (2016). Reconstitution of three-phase microtubule polymerisation dynamics. J. Cell Biol. 215, 357-368. 10.1083/jcb.201604118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole E. T., Winey M. and McIntosh J. R. (1999). High-voltage electron tomography of spindle pole bodies and early mitotic spindles in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 10, 2017-2031. 10.1091/mbc.10.6.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page B. D., Satterwhite L. L., Rose M. D. and Snyder M. (1994). Localization of the Kar3 kinesin heavy chain-related protein requires the Cik1 interacting protein. J. Cell Biol. 124, 507-519. 10.1083/jcb.124.4.507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualone D. and Huffaker T. C. (1994). STU1, a suppressor of a β-tubulin mutation, encodes a novel and essential component of the yeast mitotic spindle. J. Cell Biol. 127, 1973-1984. 10.1083/jcb.127.6.1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podolski M., Mahamdeh M. and Howard J. (2014). Stu2, the budding yeast XMAP215/Dis1 homolog, promotes assembly of yeast microtubules by increasing growth rate and decreasing catastrophe frequency. J. Biol. Chem. 289, 28087-28093. 10.1074/jbc.M114.584300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizk R. S., DiScipio K. A., Proudfoot K. G. and Gupta M. L. (2014). The kinesin-8 Kip3 scales anaphase spindle length by suppression of midzone microtubule polymerization. J. Cell Biol. 204, 965-975. 10.1083/jcb.201312039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusan N. M., Fagerstrom C. J., Yvon A. M. and Wadsworth P. (2001). Cell cycle-dependent changes in microtubule dynamics in living cells expressing green fluorescent protein-α tubulin. Mol. Biol. Cell 12, 971-980. 10.1091/mbc.12.4.971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon E. D., Leslie R. J., Saxton W. M., Karow M. L. and McIntosh J. R. (1984). Spindle microtubule dynamics in sea urchin embryos: analysis using a fluorescein-labeled tubulin and measurements of fluorescence redistribution after laser photobleaching. J. Cell Biol. 99, 2165-2174. 10.1083/jcb.99.6.2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders W., Lengyel V. and Hoyt M. A. (1997). Mitotic spindle function in Saccharomyces cerevisiae requires a balance between different types of kinesin-related motors. Mol. Biol. Cell 8, 1025-1033. 10.1091/mbc.8.6.1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin K. E. and Mitchison T. J. (1991). Mitotic spindle assembly by two different pathways in vitro. J. Cell Biol. 112, 925-940. 10.1083/jcb.112.5.925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks R. M. Q., Kamieniecki R. J. and Dawson D. S. (2001). The Kar3-interacting protein Cik1p plays a critical role in passage through meiosis I in Saccharomyces cerevisiae. Genetics 159, 939-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw S. L., Yeh E., Maddox P., Salmon E. D. and Bloom K. (1997). Astral microtubule dynamics in yeast: a microtubule-based searching mechanism for spindle orientation and nuclear migration into the bud. J. Cell Biol. 139, 985-994. 10.1083/jcb.139.4.985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirajuddin M., Rice L. M. and Vale R. D. (2014). Regulation of microtubule motors by tubulin isotypes and post-translational modifications. Nat. Cell Biol. 16, 335-344. 10.1038/ncb2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague B. L., Pearson C. G., Maddox P. S., Bloom K. S., Salmon E. D. and Odde D. J. (2003). Mechanisms of microtubule-based kinetochore positioning in the yeast metaphase spindle. Biophys. J. 84, 3529-3546. 10.1016/S0006-3495(03)75087-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sproul L. R., Anderson D. J., Mackey A. T., Saunders W. S. and Gilbert S. P. (2005). Cik1 targets the minus-end Kinesin depolymerase Kar3 to microtubule plus ends. Curr. Biol. 15, 1420-1427. 10.1016/j.cub.2005.06.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight A. F., Marshall W. F., Sedat J. W. and Murray A. W. (1997). Mitosis in living budding yeast: anaphase A but no metaphase plate. Science 277, 574-578. 10.1126/science.277.5325.574 [DOI] [PubMed] [Google Scholar]
- Su X., Arellano-Santoyo H., Portran D., Gaillard J., Vantard M., Thery M. and Pellman D. (2013). Microtubule-sliding activity of a kinesin-8 promotes spindle assembly and spindle-length control. Nat. Cell Biol. 15, 948-957. 10.1038/ncb2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Mukae N., Dewar H., van Breugel M., James E. K., Prescott A. R., Antony C. and Tanaka T. U. (2005). Molecular mechanisms of kinetochore capture by spindle microtubules. Nature 434, 987-994. 10.1038/nature03483 [DOI] [PubMed] [Google Scholar]
- Thévenaz P., Ruttimann U. E. and Unser M. (1998). A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process. 7, 27-41. 10.1109/83.650848 [DOI] [PubMed] [Google Scholar]
- van der Vaart B., Fischböck J., Mieck C., Pichler P., Mechtler K., Medema R. H. and Westermann S. (2017). TORC1 signaling exerts spatial control over microtubule dynamics by promoting nuclear export of Stu2. J. Cell Biol. 216, 3471-3484. 10.1083/jcb.201606080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga V., Helenius J., Tanaka K., Hyman A. A., Tanaka T. U. and Howard J. (2006). Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat. Cell Biol. 8, 957-962. 10.1038/ncb1462 [DOI] [PubMed] [Google Scholar]
- Varga V., Leduc C., Bormuth V., Diez S. and Howard J. (2009). Kinesin-8 motors act cooperatively to mediate length-dependent microtubule depolymerization. Cell 138, 1174-1183. 10.1016/j.cell.2009.07.032 [DOI] [PubMed] [Google Scholar]
- Vemu A., Atherton J., Spector J. O., Moores C. A. and Roll-Mecak A. (2017). Tubulin isoform composition tunes microtubule dynamics. Mol. Biol. Cell. 28, 3564-3572. 10.1091/mbc.e17-02-0124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P. J. and Huffaker T. C. (1997). Stu2p: a microtubule-binding protein that is an essential component of the yeast spindle pole body. J. Cell Biol. 139, 1271-1280. 10.1083/jcb.139.5.1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman-Storer C. M., Desai A., Bulinski J. C. and Salmon E. D. (1998). Fluorescent speckle microscopy, a method to visualize the dynamics of protein assemblies in living cells. Curr. Biol. 8, 1227-1230. 10.1016/S0960-9822(07)00515-5 [DOI] [PubMed] [Google Scholar]
- Winey M. and Bloom K. (2012). Mitotic spindle form and function. Genetics 190, 1197-1224. 10.1534/genetics.111.128710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolyniak M. J., Blake-Hodek K., Kosco K., Hwang E., You L. and Huffaker T. C. (2006). The regulation of microtubule dynamics in Saccharomyces cerevisiae by three interacting plus-end tracking proteins. Mol. Biol. Cell 17, 2789-2798. 10.1091/mbc.e05-09-0892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff J. B., Drubin D. G. and Barnes G. (2010). Mitotic spindle disassembly occurs via distinct subprocesses driven by the anaphase-promoting complex, Aurora B kinase, and kinesin-8. J. Cell Biol. 191, 795-808. 10.1083/jcb.201006028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E., Yang C., Chin E., Maddox P., Salmon E. D., Lew D. J. and Bloom K. (2000). Dynamic positioning of mitotic spindles in yeast: role of microtubule motors and cortical determinants. Mol. Biol. Cell 11, 3949-3961. 10.1091/mbc.11.11.3949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Pruyne D., Huffaker T. C. and Bretscher A. (2000). Myosin V orientates the mitotic spindle in yeast. Nature 406, 1013-1015. 10.1038/35023024 [DOI] [PubMed] [Google Scholar]
- Zimniak T., Stengl K., Mechtler K. and Westermann S. (2009). Phosphoregulation of the budding yeast EB1 homologue Bim1p by Aurora/Ipl1p. J. Cell Biol. 186, 379-391. 10.1083/jcb.200901036 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.