Abstract
An unbiased sample preparation free of interferents (i.e., competing analytes, detergents, plastics) is critical to any lipid MS workflow. Here we present a novel three-phase lipid extraction (3PLE) technique using a single-step liquid-liquid extraction (LLE) that allows both extraction and fractionation of lipids by polarity. 3PLE is composed of one aqueous and two organic phases. The upper organic phase is enriched in neutral lipids (triacylglycerols and cholesteryl esters), while the middle organic phase contains the major glycerophospholipids. Thin-layer chromatography, radioactive labeling, and MS were used to confirm lipid partitioning. 3PLE efficiency was demonstrated for bovine liver, human pooled plasma, mouse liver, mouse brain, and mouse white adipose tissue. Compared with the gold-standard Bligh/Dyer LLE, 3PLE showed significant advantages. For direct-infusion workflows, there was a decrease in ion suppression with a corresponding increased number of lipid species identified. For LC/MS workflows, increased signal intensities were observed for lower-abundance lipid species such as phosphatidic acid and phosphatidylserine. 3PLE also proved to be a valuable tool for fatty acid profiling by GC/MS, allowing for the separate identification of neutral and polar fatty acids.
Keywords: mass spectrometry, lipids, chemistry, fatty acids, phospholipids, triglyceride, liquid chromatography, gas chromatography, direct infusion
Lipids are biomolecules that are essential to life and play various roles in diverse cellular functions such as signaling, energy storage, and membrane permeability and integrity (1–4). They are classified into different categories, classes, and subclasses, with a wide spectrum of chemical and structural variants (5). For example, phospholipids have an amphipathic nature compared with cholesteryl esters (CEs) and triacylglycerols (TAGs), which are among the most hydrophobic lipid species. Ceramides and diacylglycerols (DAGs) have polarities that fall in between the previous examples. This range of hydrophobicity adds complexity to the lipid analysis. For this reason, an effective method that decreases bias while isolating a representative sample of lipids from biological matrices is critical for a lipidomic analysis.
Lipidomic profiling by MS has become a widely used tool in lipid biochemistry. Recent advances in MS have led to better sensitivity, improved mass accuracy, and novel methods for broad lipid profiling. ESI is commonly used in MS but is particularly sensitive to ion suppression and matrix effects. Therefore, coextraction of interferents, such as nonlipid biomolecules, leads to increased complexity of MS spectra, decreased sensitivity, and reduced lipid identification (2, 6, 7). The importance of sample preparation prior to lipidomic analysis is critical, as it can negatively influence the analytical measurements. Ensuring the quality of lipid extracts is an important step because improper sample preparation will yield questionable or misleading results. Liquid-liquid extractions (LLEs) and solid-phase extractions (SPEs) are the most common sample preparation techniques used in lipidomics and are implemented depending on analytical strategy; LLE is favored for global lipidomic analysis (8–10), while SPE is predominately used for targeted methods when the fractionation and/or isolation of a specific lipid class or classes is necessary. However, SPE can be variable, time-consuming, and challenging to automate. The efficiency of LLE depends on the partitioning of lipids into an organic phase. The most commonly used extraction techniques are the Folch (11) and Bligh/Dyer (BD) (12) methods, which are based on a solvent system containing chloroform, methanol, and water at different ratios. Lipids are extracted into the chloroform phase, while most of the proteins, sugars, salts, and some hydrophilic lipids remain in the water/methanol phase. These two-phase partitioning systems have proven to be very efficient in extracting a broad range of lipid classes from different biological matrices and are often referred to as the gold standard for bulk lipid analysis. In the past few years, a modified BD (mBD) method has been used for lipid extractions. Chloroform is replaced by dichloromethane, a less toxic alternative with a similar lipid extraction efficiency (2, 8, 13, 14). A drawback of these extraction techniques is that chloroform or dichloromethane has a higher density than the water/methanol layer and consequently is the bottom layer. Therefore, to recover the lipid extract, a pipette must cross the water/methanol layer and the interface, which is rich in nonlipid species (salts, proteins, sugars), resulting in an inevitable contamination of the lipid extract. To overcome this difficulty, alternative extraction methods have been developed using solvents such as methyl-tert-butyl ether (7) and butanol/methanol (15), in which the density of the organic layer is less then water, making it the upper phase (UP). These methods have been validated in a variety of sample matrices and with some lipid classes but may be limited with untested conditions for lipid species (6, 16). If new matrices are used, methods must be revalidated regarding lipid class diversity, total lipid extraction, and reproducibility, and the results must be compared with the extraction efficiency of well-established methods such as the mBD and Folch methods.
In this work, we present a three-phase liquid extraction (3PLE) method that extracts and separates lipids by polarity in a single LLE. The solvent system is a mixture of hexanes, methyl acetate, acetonitrile, and water in a 4:4:3:4 ratio, respectively. This allows the formation of three phases, one aqueous phase (bottom) and two organic phases [middle phase (MID) and UP]. Most neutral lipids partition into the UP, while the most polar lipids are present in the MID. This solvent system was first described by Shibusawa et al. (17) and was used to separate multiple organic compounds with a wide range of hydrophobicity using counter-current chromatography. However, to our knowledge, this is the first time that this method has been applied to the extraction and separation of lipids from biological matrices. The extraction efficiency of 3PLE was tested with several different matrices, including tissues and biological fluids. The results are alike or better than those obtained by mBD extraction. Moreover, 3PLE shows several advantages over mBD for direct-infusion, LC/MS, and GC/MS workflows. By partitioning the neutral and polar lipid content between two organic phases, ion suppression decreases, resulting in an increase in sensitivity and subsequently increased the number of lipid species identified by direct-infusion and LC/MS techniques. 3PLE also provides another dimension to classic total fatty acid (TFA) profiling studies performed by GC/MS (18, 19) by distinguishing the fatty acyl composition of neutral versus polar lipids. Like other LLE techniques, 3PLE can be readily automated by liquid-handling robots, allowing for an efficient extraction method for high-throughput lipidomic analysis.
MATERIALS AND METHODS
Chemicals and materials
All solvents were either HPLC or LC/MS grade and purchased from Sigma-Aldrich (St. Louis, MO). Bovine liver extract and SPLASH™ LipidoMix™ standards, a mixture of deuterated lipids from major lipid classes found in biological samples (supplemental Table S1), were purchased from Avanti Polar Lipids (Alabaster, AL). FA standards [FA(16:0{2H31}), FA(18:1ω9{2H5}), and FA(20:4ω6{2H8})] were purchased from Cayman Chemical (Ann Arbor, MI). A mixture of 26 FAs (GLC-490) were purchased from Nu-Chek Prep (Elysian, MN) and used as the reference standard for the GC/MS technique.
All lipid extractions were performed in 16 × 100 mm glass tubes with polytetrafluoroethylene-lined caps (Fisher Scientific, Pittsburgh, PA). Glass Pasteur and serological pipettes were used to minimize leaching of polymers and plasticizers. Micropipettes equipped with solvent-resistant plasticware tips (Mettler-Toledo, Columbus, OH) were used for pipetting volumes smaller than 1 ml. For the addition of lipid standards, an eVol® precision pipette equipped with a glass syringe (Trajan Scientific, Austin, TX) was used. Animal tissues were collected from animals that were euthanized under protocols approved by the University of Texas Southwestern Medical Center Institutional Animal Care and Use Committee.
Sample preparation
Approximately 100 mg of tissue [liver, brain, and white adipose tissue (WAT)] was weighed and transferred to a 2.0 ml prefilled Bead Ruptor tube (2.8 mm ceramic beads; Omni International, Kennesaw, GA), in which 1 ml methanol-dichloromethane (1:2; v/v) was added using a serological pipette. Tissue was homogenized with a Bead Ruptor (Omni International) for 50 s (5.5 mps, 3 cycles, 10 s/cycle, 5 s dwell time). The homogenates were transferred to glass tubes and diluted to a final concentration of 20 mg/ml using methanol-dichloromethane (1:2; v/v). Aliquots of 25 µl (equivalent to 0.5 mg homogenized tissue) were transferred to fresh glass tubes for immediate LLE.
Plasma (10 μl) was transferred to a glass tube. Prior to LLE, proteins were precipitated by adding 2 ml methanol-dichloromethane (1/1; v/v). Samples were vortexed and then centrifuged for 5 min at 1,635 g. The supernatant was transferred to a new tube with a Pasteur pipette, dried under N2, and immediately resuspended in the LLE conditions. The protein precipitation in plasma is an optional step; the native sample can directly undergo LLE. For LLE methods such as mBD, in which the organic phase is at the bottom, precipitating the proteins prior to LLE extraction can be advantageous because it greatly reduces the presence of precipitate between the aqueous and organic phase.
Liquid-liquid extractions
All LLEs were performed at room temperature (including centrifugation) to maintain consistent solubility and phase separation. 3PLEs began by adding 1 ml hexane, 1 ml methyl acetate, 0.75 ml acetonitrile, and 1 ml water using serological pipettes to a glass tube containing the sample. The mixture was vortexed for 5 s and then centrifuged at 2,671 g for 5 min, resulting in the separation of three distinct liquid phases. The UP and MID organic layers were collected into separate glass tubes with Pasteur pipettes and dried under N2. The 3PLE MID clean fraction was obtained by reextracting 3PLE MID. Briefly, 1 ml hexane was added to the 3PLE MID extract. The mixture was vortexed for 5 s and then centrifuged at 2,671 g for 5 min, resulting in the separation of two distinct liquid phases. The bottom phase (3PLE MID clean) was collected in a fresh glass tube with a Pasteur pipette and dried under N2.
For the mBD (12) extraction, 1 ml each of methanol, dichloromethane, and water was added to a glass tube containing the sample. The mixture was vortexed for 5 s and centrifuged at 2,671 g for 5 min, resulting in two distinct liquid phases. The organic phase (bottom phase) was collected in a fresh glass tube with a Pasteur pipette and dried under N2.
For direct-infusion analysis, dried extracts were reconstituted in 600 µl dichloromethane-methanol-isopropanol (2:1:1; v/v/v) containing 8 mM NH4F and 33 µl/ml 3:50 diluted SPLASH LipidoMix internal standard. For LC/MS analysis, dried extracts were reconstituted in 400 µl of starting eluent conditions containing 33 µl/ml 3:50 diluted SPLASH LipidoMix internal standard. Proper dilutions were performed when necessary. The internal standard solution was added using an eVol, and the samples were reconstituted in the abovementioned solvents using a micropipette equipped with solvent-resistant plastic tips.
Lipid profiling by direct-infusion MS/MSall workflow
Lipid extracts were infused into a quadrupole TOF TripleTOF™ 6600+ mass spectrometer (SCIEX, Framingham, MA) via a customized InfusePAL HTS-xt autosampler (LEAP Technologies, Morrisville, NC). ESI source parameters were as follows: ion source gas 1 (GS1) and gas 2 (GS2) set to 25 and 55 psi, respectively; curtain (Cur) gas set to 20 psi; source temperature of 300°C; and ion-spray voltage of 5,500 and −4,500 V in the positive and negative ionization mode, respectively. GS1 and GS2 were zero-grade air, while Cur and collisional-activated dissociation (CAD) gas were nitrogen. Optimal declustering potential and collision energy settings were 120 V and 40 eV for the positive ionization mode and −90 V and −50 eV for the negative ionization mode. Samples were infused for 3 min at a flow rate of 10 µl/min. MS/MSall analysis was performed by collecting product-ion spectra at each unit mass from 200 to 1,200 Da (sequentially adjusted for the mass defect of hydrogen) with an accumulation time of 0.3 s per mass. Analyst® TF version 1.7.1 software (SCIEX) was used for TOF MS and MS/MSall data acquisition. Chronos XT® software (Axel Semrau, Srockhovel, Germany) was used to control the InfusePAL system. Data were analyzed by the MarkerView (SCIEX) peak-picking algorithm, which converts the MS/MSall spectra (mass tolerance window of 0.3 Da and an intensity threshold of 30) data into a numeric list containing precursor/fragment pair intensities, followed by species identification using LipPy, an in-house script. This script provides instrument quality control information, isotopic peak corrections, lipid species identification, data normalization, and basic statistics. A detailed description of LipPy can be found in the supplemental data.
Phospholipid profiling by LC/MS/MS
Phospholipids were analyzed by LC/MS/MS using a SCIEX API 5000 triple quadrupole mass spectrometer equipped with a Shimadzu LC-20AD XR HPLC system (Columbia, MD) and a Supelco Ascentis 150 × 2.1 mm, 5 µm silica column (Bellefonte, PA). The phospholipid profiling method was previously described by Martin et al. (20). Data were acquired by switching between the positive and negative mode in a Q3 scan profile. Data in the positive mode were obtained within a mass range of 450 to 900 Da in 0.1 Da steps with a total scan time of 3.7 s. ESI source parameters were as follows: ion GS1 set at 40 psi, Cur gas set at 20 psi, temperature of 300°C, declustering potential of 100, and entrance potential of 10. Data in the negative mode were collected within a mass range of 450 to 950 Da in 0.1 Da steps with a total scan time of 3.2 s. ESI source parameters were as follows: ion GS1 set at 40 psi, Cur gas set at 20 psi, temperature of 300°C, declustering potential of −100, and entrance potential of −5. GS1 and GS2 were zero-grade air, while Cur and CAD gases were nitrogen. For the LC conditions, the mobile phases used were hexane-isopropanol (30:40; v/v) (A) and hexane-isopropanol-water (30:40:7; v/v/v) with 5 mM ammonium acetate (B). The initial mobile phase was 25% B at a flow rate of 0.2 ml/min. This was held for 5 min and then increased to 60% over 10 min and then 95% over 5 min. The mobile phase was held at 95% B for 20 min and then was reequilibrated to 25% B and held for 14 min. The column oven temperature was set to 25°C. The LC/MS data were analyzed using MultiQuant software (SCIEX). Identified lipids were normalized to their corresponding internal standard (i.e., each lipid of a specific class was normalized to its corresponding lipid class internal standard).
Neutral lipid profiling by LC/MS/MS
Neutral lipids were analyzed by LC/MS/MS using a SCIEX API 5000 triple quadrupole mass spectrometer equipped with a Shimadzu LC-20AD XR HPLC system and a 150 × 2.1 mm, 5 µm Supelco Ascentis silica column. Neutral lipids were analyzed based on methods previously described by Hutchins, Barkley, and Murphy (21). Data were acquired in positive mode under a Q3 scan. The profile was obtained within a mass range of 550 to 1,000 Da in a 0.1 Da step with a total scan time of 4 s. ESI source parameters were as follows: ion GS1 set at 40 psi, Cur gas set at 10 psi, temperature of 150°C, and declustering potential of 60. GS1 and GS2 were zero-grade air, while Cur and CAD gas were nitrogen. The mobile phases were isooctane (A) and methyl-tert-butyl/isooctane (50:50; v/v) (B). A solution of 10 mM ammonium acetate in water-acetonitrile (5:95; v/v) was delivered postcolumn at a flow rate of 0.03 ml/min. The initial mobile phase was 5% B at a flow rate of 0.2 ml/min. These conditions were held for 8 min and then increased to 30% B over 17 min and 90% B over 4 min. The mobile phase was held for 6 min at 90% B and then was reequilibrated to 5% B and held for 10 min. The column oven temperature was set to 25°C.The LC/MS data were analyzed using MultiQuant software (SCIEX). Identified lipids were normalized to their corresponding internal standard (i.e., each lipid of a specific class was normalized to its corresponding lipid class internal standard).
FA profiling by GC/MS
FA profiles were generated by a modified GC/MS method previously described by Quehenberger, Armando, and Dennis (19). Briefly, the lipid extract was spiked with 100 μl of 0.5 µg/ml FA standards [FA(16:0{2H31}), FA(20:4ω6{2H8}), and FA(22:6ω3{2H5})] in methanol using an eVol pipette and then hydrolyzed in 1 ml of 0.5 M potassium hydroxide solution prepared in methanol at 80°C for 1 h. Hydrolyzed FAs were extracted by adding 1 ml each of dichloromethane and water to the sample in the hydrolysis solution. The mixture was vortexed and centrifuged at 2,671 g for 5 min, and the organic phase (bottom phase) was collected in a fresh glass tube using a Pasteur pipette and dried under N2. Dried lipid extract was resuspended in 50 µl 1% triethylamine in acetone and derivatized with 50 µl 1% pentafluorobenzyl bromide in acetone at room temperature for 25 min in capped glass tubes. Solvents were dried under N2, and samples were resuspended in 500 µl isooctane using a micropipette. Samples were analyzed using an Agilent 7890/5975C (Santa Clara, CA) by electron capture negative ionization equipped with a DB-5MS column (40 m × 0.180 mm; 0.18 µm film thickness) from Agilent. The hydrogen (carrier gas) flow rate was 1.6 ml/min, and the injection port temperature was set at 300°C. The sample injection volume was 1 µl. The initial oven temperature was set at 150°C and then increased to 200°C at 25°C/min. This was followed by an increase of 8°C/min until a temperature of 300°C was reached and held for 2.2 min for a total run time of 16.7 min. FAs were analyzed in selected ion-monitoring mode. Supplemental Table S2 provides a list of the different FAs monitored. The FA data were normalized to the internal standards. FAs with carbon lengths of ≤18, 20, and 22 were normalized to FA(16:0{2H31}), FA(20:4ω6{2H8}), and FA(22:6ω3{2H5}), respectively. Data were processed using MassHunter software (Agilent).
TLC and radioactive labeling
TLC for phospholipid and neutral lipid separation was performed on channeled hard-layer silica gel plates (Analtech, Newark, DE). Lipid content from 50 mg mouse liver was extracted by both mBD and 3PLE. Lipid extracts were dried under N2, resuspended in 50 µl chloroform, and spotted on the silica plate along with standards using a micropipette. Phospholipids were resolved with an eluent system composed of chloroform-methanol-ammonium hydroxide (60:35:8; v/v/v), while neutral lipids were resolved using a solvent system of hexane-ethanol-acetic acid (80:20:1; v/v/v). Spots were visualized with iodine and identified by comparing their retention factors with those obtained with standards.
For the radioactive labeling, mouse livers were homogenized in 1× PBS solution at a concentration of 50 mg/ml. Aliquots of 50 mg liver homogenates were transferred to separate tubes in which 0.5 µCi phosphocholine (PC) l-a-dioleoyl (specific activity: 55 mCi/mmol; American Radiolabeled Chemicals, Inc., Saint Louis, MO), 0.25 µCi diolein (specific activity: 55 mCi/mmol; American Radiolabeled Chemicals, Inc.), or 0.2 µCi [carboxy-14C]triolein (specific activity: 108.8 mCi/mmol; PerkinElmer, Waltham, MA) was added to the homogenates. Samples were then extracted by mBD or 3PLE as previously described. Each fraction was dried, resuspended in scintillation cocktail (3a70B Complete Counting Cocktail; Research Products International, Mount Prospect, IL), and counted on an LS6500 liquid scintillation counter (Beckman Coulter, Brea, CA). Samples extracted by mBD were set to 100%, and the 3PLE UP and MID were compared with the signal obtained by mBD. All samples were compared back to a nonextracted sample to confirm extraction efficiency.
Statistical analysis
The means (n = 4) and standard errors were calculated to assess intrareplicate variability. Comparisons between groups of interest were performed using Student’s t-test. P < 0.05 and P < 0.01 were considered significant.
RESULTS
Lipid partitioning in 3PLE
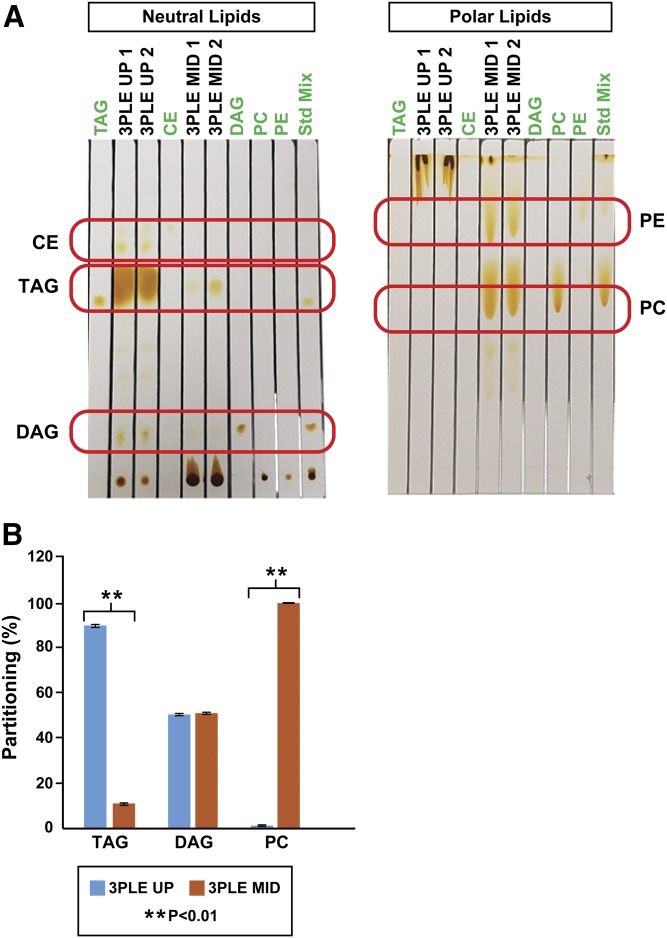
Previous observations have shown that 3PLE allows lipid extraction and partitioning based on polarity. To study the lipids partitioning between both organic phases, non-MS (TLC and radioactive labeling) and MS-based techniques (direct-infusion MS/MSall) were used. The TLC results showed that CE species are present in the 3PLE UP but absent from the MID (Fig. 1A). Most TAGs and DAGs are present in the UP; however, low-intensity spots with the same retention factor were observed in the MID. The radioactive labeling experiments (Fig. 1B) showed that >90% of TAGs partitioned to the UP, while the DAGs partitioned equally between the UP and MID. The data support nearly complete partitioning of CE and TAGs to the UP. Samples with a large amount of TAGs (e.g., adipose, VLDL, lipid droplets) will have a greater amount of TAGs partitioning into the MID. In these sample types, the MID can be reextracted using hexane to approach 100% recovery. Radioactive labeling experiments showed that PCs partition to the MID, and ≤2% will partition to the UP. TLC analysis showed that there was no evidence of PC and PE in the UP (Fig. 1A). These data support nearly complete partitioning of phospholipids into the 3PLE MID, with the phospholipids being the major component of the MID.
Fig. 1.
Mouse liver lipids extracted by BD and 3PLE. A: Neutral and polar lipid content analyzed by TLC. B: Radiolabeled standards with scintillation counting. Values are expressed as means and standard errors (n = 4; **P < 0.01).
3PLE technique validation
This work focused on the analytical advantages of 3PLE relative to a biphasic LLE for lipidomic workflows. For this reason, only a qualitative validation of the technique was performed.
3PLE was validated by using multiple analytical measurements. First, a commercially available matrix-free bovine liver lipid extract was reextracted using 3PLE and mBD, and the lipid profiles were compared. This approach allows the evaluation of lipid extraction efficiency and assesses whether lipid yields were influenced by using a different solvent system. After establishing that both extraction techniques delivered similar results, several biological samples (tissues and fluids) were extracted using 3PLE and mBD. This allowed us to assess whether 3PLE efficiency was matrix-dependent. Last, 3PLE homogenates were analyzed using different MS techniques, including direct infusion, LC/MS, and GC/MS. This approach allowed us to assess 3PLE versatility along different lipidomic workflows.
Lipid extraction efficiency
Direct-infusion MS/MSall workflow.
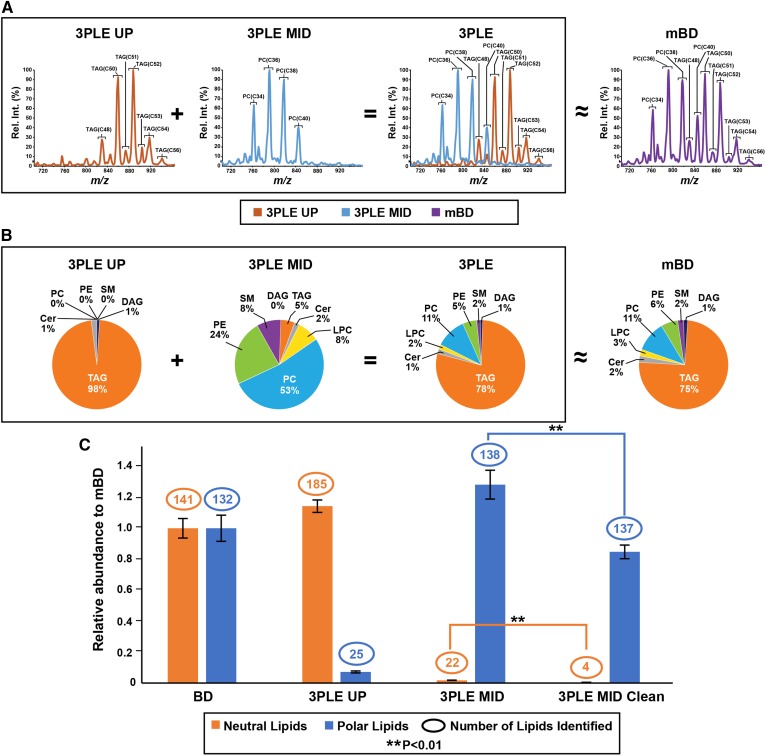
A commercially available matrix-free bovine liver lipid extract standard was used to compare lipid extraction efficiency between mBD and 3PLE. Bovine liver extracts were reextracted using both extraction techniques and analyzed by direct-infusion MS/MSall (Fig. 2) and LC/MS (Fig. 3). Direct-infusion MS/MSall is a data-independent acquisition method in which a predefined mass range of precursors are sequentially fragmented in a unit-based resolution stepwise fashion, and all the respective product ions are collected in both the positive and negative ionization modes (22). As with other direct-infusion techniques, this is a rapid and high-throughput method used to characterize global lipidomic profiles from biological samples. For the direct-infusion technique, bovine liver mBD, 3PLE UP, and 3PLE MID lipid extracts were infused into a quadrupole TOF and analyzed by MS/MSall. Figure 2A contains an MS/MSall total ion chromatogram scan that was restricted to 700–960 Da, which represents the majority of TAG and PC species present in the bovine liver. TAG species were only detected in the 3PLE UP, while PC species were only detected in the 3PLE MID. When the 3PLE UP and MID profiles are overlaid they mirror the spectrum obtained by mBD. These data were also confirmed with other neutral and polar lipids (Fig. 2B). To determine whether 3PLE affects the quantity of identified lipids, the number of neutral and polar lipids found in 3PLE and mBD were plotted in Fig. 2C. The data represent the sum of the signal intensities normalized to the mBD results. 3PLE resulted in i) a greater number of neutral and polar lipids identified in the extracts and ii) slightly higher but not significantly different signal intensities compared with mBD. Figure 2C shows that a small number of polar and neutral lipids are extracted in the 3PLE UP and MID, respectively. The partitioning of some polar lipids to the UP and neutral lipids to the MID was observed for the most abundant PCs and TAGs and represented 2% to 3% of the total signal for those lipid species. Data on the five most abundant PC and TAG lipid species found in the sample and their respective partitioning in the 3PLE phases are presented in supplemental Fig. S1A and B. To obtain a more complete separation of neutral lipids into the UP and polar lipids into the MID, a second extraction of each phase can be performed. For example, the 3PLE MID can be isolated and reextracted with hexanes for the nearly complete removal of the neutral lipid content (Fig. 2C).
Fig. 2.
Bovine liver extracts analyzed by direct-infusion MS/MSall in the positive ionization mode. Data were obtained by the BD and 3PLE techniques. A: MS/MSall total ion chromatogram obtained from BD, 3PLE UP, and 3PLE MID lipid extracts. Data shown are restricted to a mass window of 710–960 Da and are the most representative PC and TAG species found in bovine liver extracts. B: Lipid composition from different bovine liver lipid extracts obtained by 3PLE UP, 3PLE MID, and BD. The 3PLE lipid profile was obtained by the sum of 3PLE UP and 3PLE MID contributions. Identified lipid classes comprising less than 1% of total lipid composition are labeled as 0%. C: Neutral and polar lipid content of BD, 3PLE UP, 3PLE MID, and 3PLE MID clean lipid extracts. The 3PLE MID clean is a reextraction of the 3PLE MID using hexane to reduce or remove the neutral lipid content from the MID faction. The y-axis represents relative abundance to BD signal intensities obtained by direct-infusion MS/MSall. Values are expressed as means and standard errors (n = 4; **P < 0.01). Cer, ceramide; LPC, lysophosphatidylcholine.
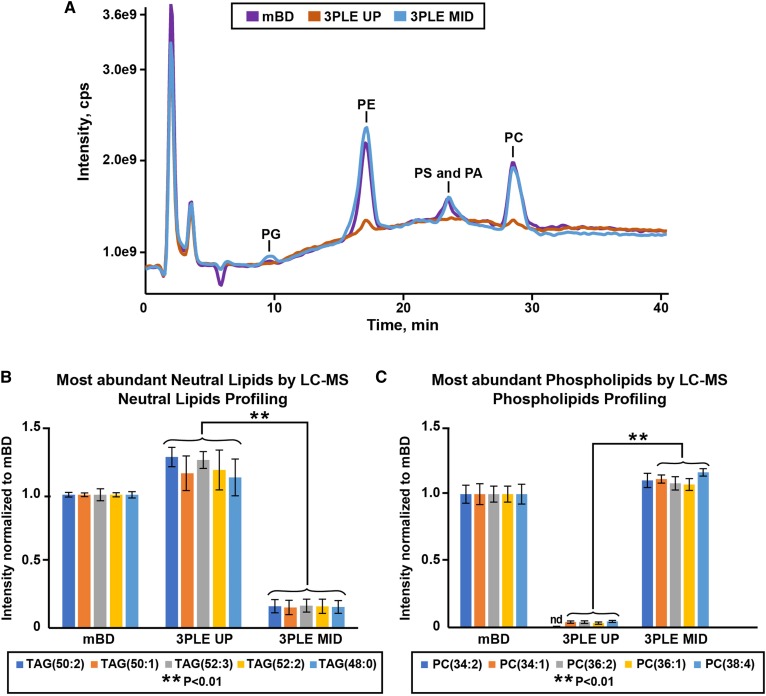
Fig. 3.
Bovine liver extracts analyzed by LC/MS. Data were obtained by the BD and 3PLE techniques. A: LC/MS analysis of phospholipids using the Q1 scan in the negative ionization mode. B: Most abundant neutral lipid species found in bovine liver BD, 3PLE UP, and 3PLE MID lipid extracts analyzed by LC/MS neutral lipid profiling in the positive ionization mode. Values are expressed as means and standard errors (n = 4; **P < 0.01). C: Most abundant polar lipid species found in bovine liver BD, 3PLE UP, and 3PLE MID lipid extracts analyzed by LC/MS phospholipid profiling in the negative ionization mode. Values are expressed as means and standard errors (n = 4; **P < 0.01). nd, not detected; PA, phosphatidic acid; PG, phosphatidylglycerol;PS, phosphatidylinositol.
LC/MS/MS workflow.
Figure 3A and the reconstructed ion chromatogram of PC(38:4) and PE(36:3) shown in supplemental Fig. S2 were obtained using the phospholipid profiling method. These data support the presence of phospholipids in both the mBD and 3PLE MID and confirmed their absence from the 3PLE UP. Moreover, peak intensities between mBD and MID extracts are similar, thus indicating identical lipid recovery. The five most abundant phospholipids and neutral lipids found in bovine liver extract using the phospholipid and neutral lipid LC/MS profiling methods are presented in Fig. 3B and C. As observed by direct-infusion MS/MSall, TAGs and PCs are the most abundant neutral and polar lipid species detected in bovine liver. Residual neutral lipids and polar lipids were observed in the MID and UP, respectively. This can be resolved by an additional reextraction step, as previously shown with the direct-infusion data.
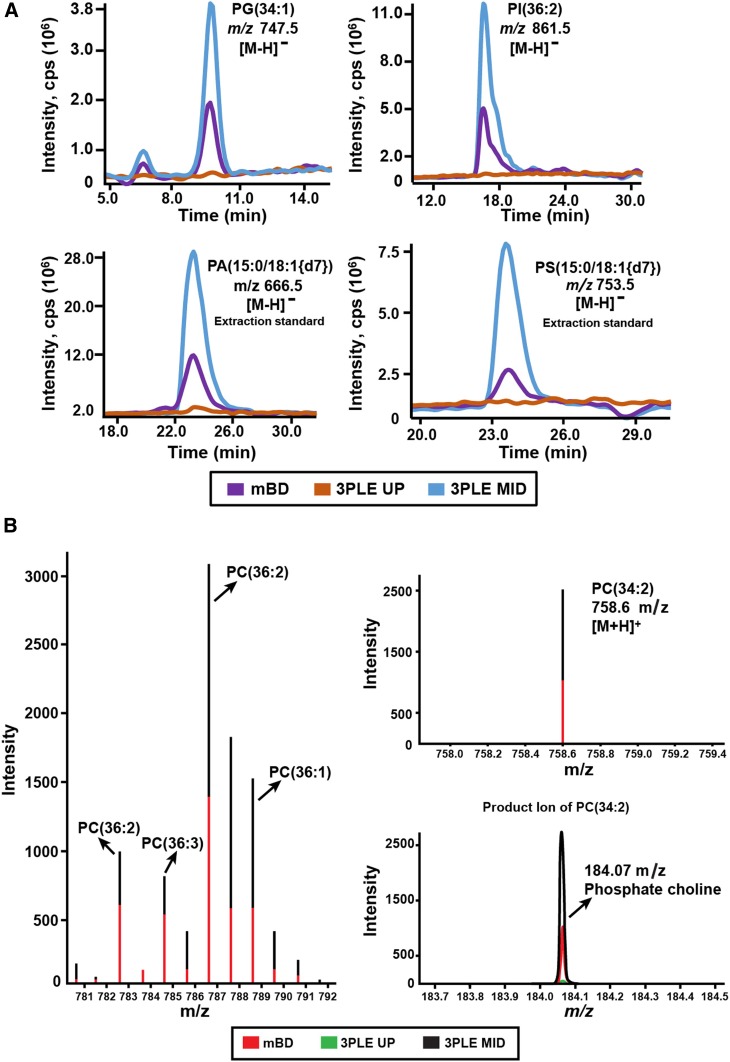
LC/MS revealed increased signal intensities for some lipid species in 3PLE extracts compared with mBD, most notably in phospholipid profiling in the negative ionization mode (Fig. 4A). A 2- to 3-fold signal increase was observed not only in bovine liver endogenous species [phosphatidylglycerol (34:1) and phosphatidylinositol (36:2)] but also in phospholipid standards [phosphatidic acid (15:0/18:1{2H7}) and phosphatidylserine (15:0/18:1{2H7})] spiked into the matrix prior to LLE.
Fig. 4.
A: Extracted ion chromatograms obtained by LC/MS phospholipid profiling of bovine liver samples. Lipids were extracted by BD and 3PLE. PG(34:1) and PI(36:2) are two endogenous phospholipid species identified in the matrix. PA(15:0/18:1{2H7}) and PS(15:0/18:1{2H7}) are two phospholipid standards spiked in the samples prior to the lipid extraction. The represented data were acquired in the negative ionization mode. B: Total ion chromatogram and product ion mass spectra of PC(34:2) obtained by direct-infusion MS/MSall in the positive ionization mode. The bovine liver lipids were extracted by BD and 3PLE. PA, phosphatidic acid;PG, phosphatidylglycerol; PI, phosphatidylinositol; PS, phosphatidylinositol.
Lipid extraction efficiency in complex biological matrices
We assessed the lipid extraction efficiency of 3PLE in homogenates of mouse WAT, liver, and brain and in human pooled plasma (HPP). These matrices have distinct lipid profiles that lead to different analytical challenges. The results obtained were compared with those from the mBD extraction technique.
Mouse white adipose tissue.
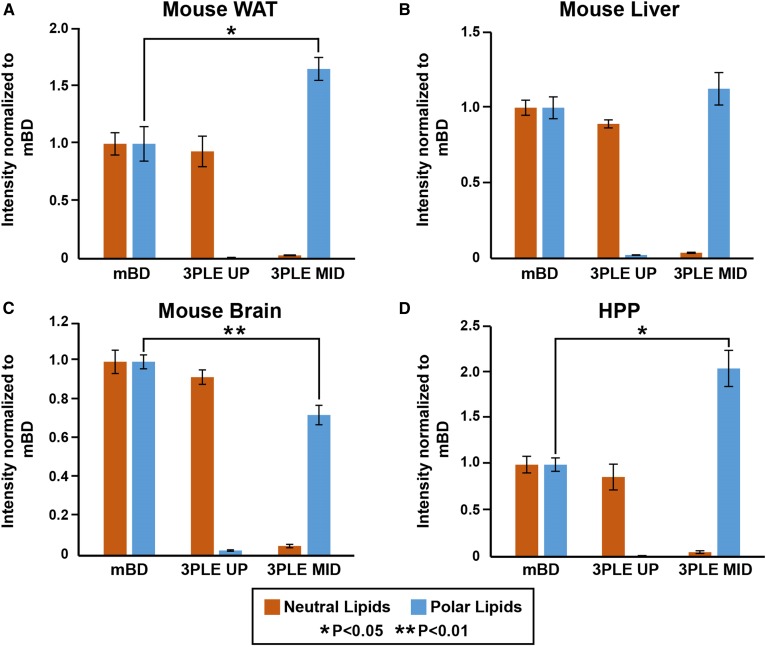
Figure 5A shows the direct-infusion MS/MSall data from WAT. There were no significant differences between the 3PLE UP and mBD neutral lipid content. However, a significant increase (P < 0.05) of approximately 1.8-fold in the total signal intensity of all polar lipids identified in the MID was observed relative to mBD. Moreover, when comparing the five most abundant phospholipid species found in WAT (Fig. 6), a significant increase in signal intensity (P < 0.01) was observed for all five. Some PC species showed a 2- to 3-fold signal intensity increase compared with mBD extraction results. This increase in signal intensity was related to a decrease in ion suppression. The LC/MS results (Fig. 7) showed a more modest increase in some of the PC species’ signal intensities, with no significant differences compared with mBD. There was no evidence of phospholipids in the 3PLE UP fraction. There were also no significant differences between the 3PLE UP and mBD relative to the neutral lipid content (Fig. 7).
Fig. 5.
Lipid profiles of mouse (A) WAT, (B) liver, (C) brain, and (D) HPP obtained by direct-infusion MS/MSall. Lipid content was extracted by BD and 3PLE. Values are expressed as means and standard errors (n = 4; *P < 0.05 and **P < 0.01). nd, not detected.
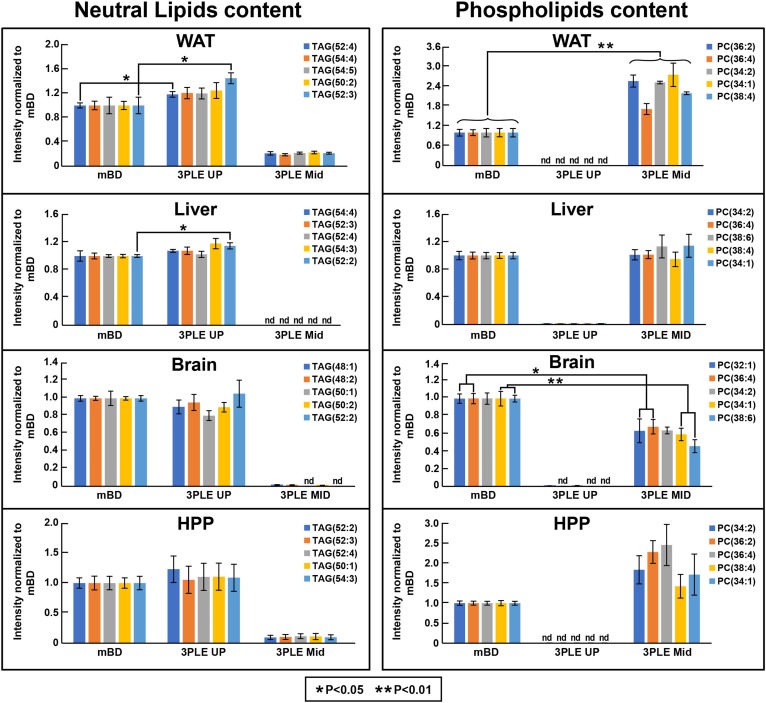
Fig. 6.
Top five most abundant neutral and polar lipids found in mouse WAT, liver, brain, and HPP. Data were obtained by direct-infusion MS/MSall. Lipid content was extracted by BD and 3PLE. Values are expressed as means and standard errors (n = 4; *P < 0.05 and **P < 0.01). nd, not detected.
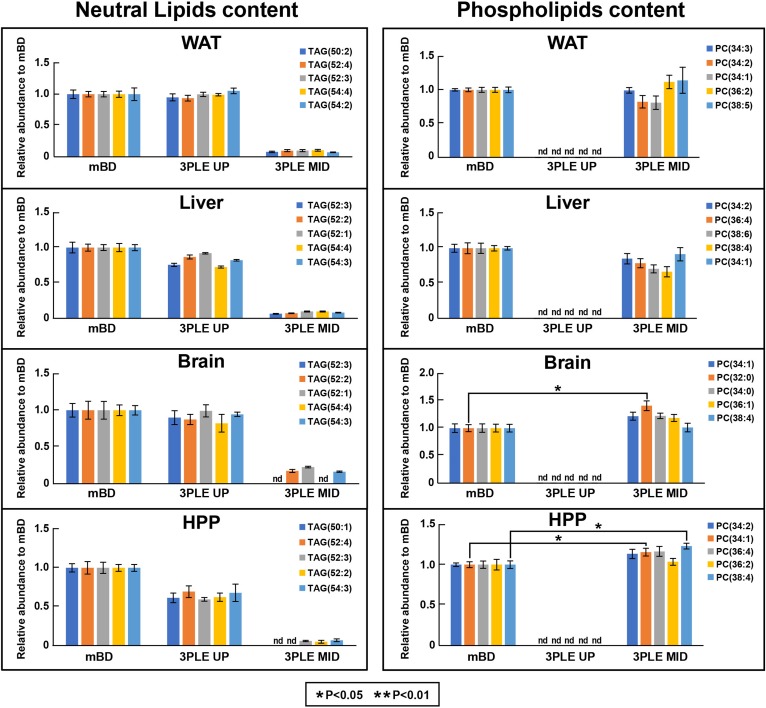
Fig. 7.
Top five most abundant neutral and polar lipids found in mouse WAT, liver, brain, and HPP. Data were obtained by LC/MS. The lipid content was extracted by BD and 3PLE. Values are expressed as means and standard errors (n = 4; *P < 0.05). nd, not detected.
Mouse liver.
Figure 5B represents the direct-infusion MS/MSall data obtained from mouse liver. We observed a slight increase, even though not significant, in polar lipid signal intensity for the 3PLE MID relative to BD. For the most abundant phospholipids, no significant differences between the 3PLE MID and mBD on both direct-infusion and LC/MS results (Figs. 6, 7) were observed. Likewise, there were no significant differences observed between 3PLE UP and mBD neutral lipid content (Fig. 5B). Relative to the most abundant neutral lipids, results also showed no significant differences between 3PLE and mBD results, with the exception of the TAG(52:2) species in the direct-infusion data (Fig. 6).
Mouse brain.
Direct-infusion data (Fig. 5C) showed no significant differences between the neutral lipid content of the 3PLE UP and mBD. However, significant changes (P < 0.01) were found between the polar lipid content of the mBD and 3PLE MID. Although the number of phospholipid species identified in the 3PLE MID (103 phospholipid species) was higher than those identified in the BD extracts (73 phospholipid species), the total signal intensity for the phospholipid fraction was significantly lower (P < 0.01) in the 3PLE MID extracts. However, a decrease in polar lipid signal intensity was not observed in the LC/MS data (Fig. 6). The data showed no significant differences between mBD and 3PLE MID content, with the exception of PC(32:0) species, which showed a significant increase (P < 0.05) relative to mBD data.
Human pooled plasma.
Similar to the other matrices studied, there were no significant differences found in 3PLE UP and mBD neutral lipid content. However, a significant increase (P < 0.05) in the total signal intensity for the polar lipids was identified in the 3PLE MID relative to mBD. When comparing the five most abundant phospholipid signal intensities (Fig. 6), no significant changes between both methods were observed. The same was true for the most abundant neutral lipids. LC/MS data (Fig. 7) also showed no significant differences relative to the most abundant neutral lipids between the 3PLE UP and mBD. However, a significant increase in signal intensity (P < 0.05) for PC(34:1) and PC(38:4) was observed for species in the 3PLE MID relative to the mBD homogenates.
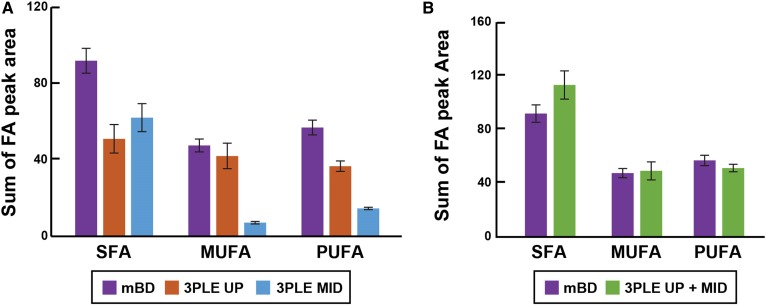
Figure 8A shows that the FAs identified by GC/MS in HPP are organized based on their saturation level and the LLE technique used. The mBD data are limited to the TFA content, while the 3PLE data revealed FA distributions in both the polar and neutral lipids (mostly TAG). The 3PLE UP and MID FA contributions can be summed to obtain the TFA composition derived by mBD (Fig. 8B), further illustrating that the lipid extraction efficiencies of mBD and 3PLE are identical, and no significant differences were found between these two methods.
Fig. 8.
FA profile of HPP measured by GC/MS. A: FA content of lipid extracts measured by BD and 3PLE. B: Comparison between the FA profiles obtained by BD and by the sum of both 3PLE organic phases (UP and MID). Values are expressed as means and standard errors (n = 4). SFA, saturated fatty acid.
DISCUSSION
3PLE allows lipid fractionation based on polarity. The UP is enriched in neutral lipids, while the MID contains glycerophospholipids and other polar lipid species (Figs. 1–4). Although 3PLE was not developed as an alternative method to SPE, both techniques were compared in our laboratory and showed similar lipid separation efficiency by polarity (data not shown). The mBD extraction was used as a reference method to validate 3PLE. The mBD has proven to be an effective method for bulk lipid extraction in different matrices and is commonly used for broad lipidomic profiling analysis. 3PLE efficiency was first evaluated with a commercially available bovine liver extract. Lipids were reextracted from the sample using mBD and 3PLE techniques and analyzed by direct-infusion MS/MSall (Fig. 2) and LC/MS (Fig. 3). The direct-infusion results confirmed what was previously shown with the TLC and radiolabeling experiments (Fig. 1): that most neutral lipids were present in the 3PLE UP and the polar lipid content was mainly present in the 3PLE MID (Fig. 2). Moreover, the lipid profile obtained after summing 3PLE UP and MID contributions was identical to that obtained using mBD. 3PLE not only mirrored the lipid composition obtained by mBD but also added the benefit of separating two major lipid classes. An increase in the number of neutral and polar lipids found in the 3PLE UP and MID, respectively, compared with the mBD (Fig. 2C) was observed. Nevertheless, even though the signal intensities of neutral and polar lipids are slightly higher in 3PLE extracts, they are not statistically different from the signals obtained with mBD. The data did not show an improved lipid recovery by 3PLE; therefore, 3PLE reduced ion suppression due to a decreased presence of the extraction matrix. In addition to the sample matrix, the lipid composition of the sample can affect the ionization of other lipid classes. For example, PC ionizes more efficiently and causes partial ion suppression of TAG. By removing the polar lipids from the more neutral TAG content, TAG suppression significantly decreased, leading to a 30% increase in the number of neutral lipid species identified (Fig. 2C). The number of polar lipids identified in the 3PLE MID also increased, though to a lesser degree. Matrix effects are known to potentially affect ionization. In separating the lipids by polarity, it is expected that the nonlipid ubiquitous anthropogenic contaminants (plasticizers, polymers, etc.) are likely also divided between the two organic phases, leading to a decrease in the observed background signals.
The neutral and polar lipid partitioning between both the UP and MID is not perfectly resolved. Results showed that a small percentage of neutral and polar lipids will also be present in the MID and UP, respectively (Figs. 2C, 6, 7). This contribution can be almost completely removed after a second extraction of each phase. For example, Fig. 2C and supplemental Fig. S1 show that the reextraction of the MID significantly decreases (P < 0.01) the presence of neutral lipids in this extract. However, the introduction of an extra analytical step can also bring disadvantages. Polar lipid signal intensities in the reextracted MID phase (3PLE MID clean) are significantly lower (P < 0.01) compared with the 3PLE MID (Fig. 2C, supplemental Fig. 1B). This can be explained in part by losses of analyte during the reextraction process. However, the number of lipid species identified in both the 3PLE MID and 3PLE MID clean were very similar. One should evaluate the necessity of a second reextraction, taking into consideration the advantages and disadvantages discussed here.
The LC/MS data (Figs. 3, 4, supplemental Fig. S2) confirmed the direct-infusion findings: most neutral and polar lipids were present in the 3PLE UP and MID, respectively. Moreover, 3PLE lipid signal intensities were identical or higher than those obtained by mBD. For some lipid species an increase in signal intensities was observed compared with mBD (Fig. 4A). This increased sensitivity in the negative ionization mode might be related to a decrease in the background signal. For example, as previously shown by the direct-infusion data, 3PLE extracts delivered a simpler matrix than mBD, thus increasing the quality of lipid analysis.
Overall, 3PLE was very effective in extracting lipids from bovine liver extracts. In a relatively simple matrix, 3PLE showed extraction efficiency similar to or better than that of mBD. 3PLE produced a simpler matrix, decreasing ion suppression and the presence of interferents and thus improving the data obtained in both direct-infusion and LC/MS workflows.
Regardless of the analytical approach (i.e., direct infusion, LC/MS, or GC/MS), the success of a lipidomic analysis relies on the efficiency of the lipid extraction from different biological matrices. The lipid extraction efficiency of the 3PLE described here was tested in mouse WAT, liver, brain, and HPP samples. These matrices contain different lipid profiles and therefore pose different analytical challenges. For example, the WAT is composed of 60% to 85% lipid by mass, of which 90% to 99% are TAGs (23). This massive neutral lipid content complicates polar lipid content analysis in WAT, especially by direct-infusion MS techniques, due to differences in class abundance and ion suppression. The 3PLE results showed that by separating neutral lipids from polar lipids, polar lipid signal intensities increased, thereby increasing the method sensitivity and data quality of direct-infusion MS/MSall analysis (Fig. 4B). We also observed i) a significant increase (P < 0.05) in the total signal intensity of all polar lipids found in the 3PLE MID relative to mBD (Fig. 5A) and ii) a significant increase (P < 0.01) in the signal intensity of the five most abundant phospholipids found in WAT (Fig. 6). This increase in signal intensity is related to a decrease in ion suppression. In this example, the primary cause for ion suppression was the large neutral lipid content present in WAT. On the other hand, LC/MS techniques, which are less prone to ion suppression, showed similar phospholipid signal intensities between the mBD and 3PLE (Fig. 7). The polar lipid content of WAT completely partitioned to the 3PLE MID, with no evidence of polar lipids found in the 3PLE UP. This partitioning is related to the low polar lipid content in WAT.
The liver plays an important role in lipid metabolism and is the center for FA and cholesterol biosynthesis and lipoprotein secretion. Its lipid composition can vary with diet and between animal models, but in general it is composed of approximately 75% TAGs, 20% phospholipids, and 5% other lipid species (CEs, free FAs) (24). The lipid composition of liver is more diverse than WAT, which makes it a good model to evaluate 3PLE. Data obtained by direct infusion and LC/MS show that 3PLE lipid extraction efficiency was similar to mBD, with the advantage of having both neutral and lipid content separated (Figs. 5B, 6, 7).
A thorough understanding of brain lipid metabolism is limited. It is one of the most lipid-rich organs in the body and has a very unique lipid composition (25). The brain is particularly rich in sphingolipids, but much of its lipid composition is phospholipids (26, 27). Similar to the other matrices, there was no difference in the neutral lipid content of the 3PLE UP and mBD. However, a significant decrease (P < 0.01) in total polar lipid signal intensities in the 3PLE MID compared with mBD (Fig. 5C) was observed. Remarkably, there were more phospholipid species found in the 3PLE MID than in the mBD results. Moreover, this trend was not observed in the LC/MS data, which showed that the polar lipid content of the 3PLE MID and mBD was identical (Fig. 7). These results lead to the assumption that differences observed in direct-infusion data are likely related to matrix effects, which are known to be more challenging in direct-infusion techniques.
In 2010, the LIPID MAPS consortium characterized the lipid composition of human plasma using the National Institute of Standards and Technology Standard Reference Material 1950 (Metabolites in Frozen Human Plasma). As part of the LIPIDS MAPS eicosanoid core, Quehenberger et al. (19) developed a GC/MS-based analytical method for free FA and TFA profiling in biological samples. For the TFA analysis of HPP, lipids are extracted from plasma using the mBD, hydrolyzed in methanolic potassium hydroxide solution, derivatized with pentafluorobenzyl bromide, and analyzed by GC/MS (electron capture negative ionization). Results give TFA content of HPP lipids with no further information about their lipid source. Using 3PLE, neutral and polar lipids are extracted and separated by polarity prior to hydrolysis. This allows profiling of neutral and polar lipids independently, providing greater insights about FA distribution and abundance within different lipid components (Fig. 8A). Furthermore, there are no significant differences between the FA profile obtained by mBD and 3PLE (with both organic phases summed together; Fig. 8B), resulting in no loss of information during the analysis. The advantages of 3PLE in the GC/MS FA profiling technique are remarkable, adding a new level of detail that can be explored and furthering the study and understanding of lipid metabolism and metabolic diseases. Relative to the direct-infusion data, a significant increase was observed (P < 0.05) in the 3PLE MID polar lipid signal intensities compared with mBD (Fig. 5D). As previously discussed for other matrices, these differences are related to a decrease in ion suppression.
3PLE is a versatile technique that can extract lipids from a variety of biological matrices having different degrees of complexity. Furthermore, this technique can be used as a sample preparation step for most MS-based lipidomic workflows. With a lipid extraction and recovery efficiency similar to (and in some examples better than) mBD, 3PLE demonstrates several advantages over other lipid LLE biphasic methods. By separating lipids into two distinct organic phases (neutral and polar lipid fractions), 3PLE results in less complex lipid extract matrices. Separating these two classes is advantageous for i) direct-infusion techniques (by decreasing ion suppression and thereby increasing data quality and the number of lipid species identified) and ii) LC/MS techniques (by decreasing the background in MS spectra, resulting in increased sensitivity). 3PLE can also be successfully applied to the FA profiling of HPP by GC/MS, allowing the determination of polar and neutral lipid FA content. This provides a clear advantage compared with the classic FA profiling method, which only delivers total FA content of lipid extracts. 3PLE is capable of extracting and separating lipids by polarity in a single LLE with the same efficiency as the most commonly used lipid extraction techniques. However, these advantages also lead to a drawback because the number of samples to be analyzed doubles. This only becomes a concern when both a neutral and polar lipid profile are needed. One should decide whether the advantages in 3PLE data quality compensate for the increase in analysis time. The 3PLE method stands as an alternative to other LLEs used today and is especially useful for direct-infusion platforms.
Supplementary Material
Acknowledgments
The authors thank Nancy Heard and Chelsea Burroughs for assistance in preparing figures and the Center for Human Nutrition and Molecular Genetics at the University of Texas Southwestern for valuable discussions and suggestions.
Footnotes
Abbreviations:
- BD
- Bligh/Dyer
- CAD
- collisional-activated dissociation
- CE
- cholesteryl ester
- Cur
- curtain
- DAG
- diacylglycerol
- GS1
- gas source 1
- GS2
- gas source 2
- HPP
- human pooled plasma
- LLE
- lipid-lipid extraction
- mBD
- modified Bligh/Dyer
- MID
- middle phase
- PC
- phosphocholine
- PE
- phosphatidylethanolamine
- SPE
- solid-phase extraction
- TAG
- triacylglycerol
- TFA
- total fatty acid
- UP
- upper phase
- WAT
- white adipose tissue
- 3PLE
- three-phase liquid extraction
J.G.M. is supported in part by National Institutes of Health Grant HL20948.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Yang K., and Han X.. 2016. Lipidomics: techniques, applications, and outcomes related to biomedical sciences. Trends Biochem. Sci. 41: 954–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Han X. 2016. Lipidomics: Comprehensive Mass Spectrometry of Liquids. John Wiley and Sons, Hoboken, NJ. [Google Scholar]
- 3.Murphy R. C. 2014. Tandem Mass Spectrometry of Lipids: Molecular Analysis of Complex Lipids. Royal Society of Chemistry, London. [Google Scholar]
- 4.Ridgway N., and McLeod R.. 2015. Biochemistry of Lipids, Lipoproteins and Membranes. 6th ed. Elsevier, Amsterdam, the Netherlands. [Google Scholar]
- 5.Fahy E., Subramaniam S., Brown H. A., Glass C. K., Merrill A. H. Jr., Murphy R. C., Raetz C. R., Russell D. W., Seyama Y., Shaw W., et al. 2005. A comprehensive classification system for lipids. J. Lipid Res. 46: 839–861. [DOI] [PubMed] [Google Scholar]
- 6.Liebisch G., Ekroos K., Hermansson M., and Ejsing C. S.. 2017. Reporting of lipidomics data should be standardized. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 1862: 747–751. [DOI] [PubMed] [Google Scholar]
- 7.Matyash V., Liebisch G., Kurzchalia T. V., Shevchenko A., and Schwudke D.. 2008. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 49: 1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cajka T., and Fiehn O.. 2016. Toward merging untargeted and targeted methods in mass spectrometry-based metabolomics and lipidomics. Anal. Chem. 88: 524–545. [DOI] [PubMed] [Google Scholar]
- 9.Rustam Y. H., and Reid G. E.. 2018. Analytical challenges and recent advances in mass spectrometry based lipidomics. Anal. Chem. 90: 374–397. [DOI] [PubMed] [Google Scholar]
- 10.Li M., Yang L., Bai Y., and Liu H.. 2014. Analytical methods in lipidomics and their applications. Anal. Chem. 86: 161–175. [DOI] [PubMed] [Google Scholar]
- 11.Folch J., Lees M., and Stanley G. H. S.. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226: 497–509. [PubMed] [Google Scholar]
- 12.Bligh E. G., and Dyer W. J.. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 13.Cequier-Sánchez E., Rodriguez C., Ravelo A. G., and Zarate R.. 2008. Dichloromethane as a solvent for lipid extraction and assessment of lipid classes and fatty acids from samples of different natures. J. Agric. Food Chem. 56: 4297–4303. [DOI] [PubMed] [Google Scholar]
- 14.Carlson L. A. 1985. Extraction of lipids from human whole serum and lipoproteins and from rat liver tissue with methylene chloride-methanol: a comparison with extraction with chloroform-methanol. Clin. Chim. Acta. 149: 89–93. [DOI] [PubMed] [Google Scholar]
- 15.Löfgren L., Stahlman M., Forsberg G. B., Saarinen S., Nilsson R., and Hansson G. I.. 2012. The BUME method: a novel automated chloroform-free 96-well total lipid extraction method for blood plasma. J. Lipid Res. 53: 1690–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reis A., Rudnitskaya A., Blackburn G. J., Mohd Fauzi N., Pitt A. R., and Spickett C. M.. 2013. A comparison of five lipid extraction solvent systems for lipidomic studies of human LDL. J. Lipid Res. 54: 1812–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shibusawa Y., Yamakawa Y., Noji R., Yanagida A., Shindo H., and Ito Y.. 2006. Three-phase solvent systems for comprehensive separation of a wide variety of compounds by high-speed counter-current chromatography. J. Chromatogr. A. 1133: 119–125. [DOI] [PubMed] [Google Scholar]
- 18.Quehenberger O., Armando A. M., Brown A. H., Milne S. B., Myers D. S., Merrill A. H., Bandyopadhyay S., Jones K. N., Kelly S., Shaner R. L., et al. 2010. Lipidomics reveals a remarkable diversity of lipids in human plasma. J. Lipid Res. 51: 3299–3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quehenberger O., Armando A. M., and Dennis E. A.. 2011. High sensitivity quantitative lipidomics analysis of fatty acids in biological samples by gas chromatography-mass spectrometry. Biochim. Biophys. Acta. 1811: 648–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin S. A., Gijon M. A., Voelker D. R., and Murphy R. C.. 2014. Measurement of lysophospholipid acyltransferase activities using substrate competition. J. Lipid Res. 55: 782–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hutchins P. M., Barkley R. M., and Murphy R. C.. 2008. Separation of cellular nonpolar neutral lipids by normal-phase chromatography and analysis by electrospray ionization mass spectrometry. J. Lipid Res. 49: 804–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simons B., Kauhanen D., Sylvanne T., Tarasov K., Duchoslav E., and Ekroos K.. 2012. Shotgun lipidomics by sequential precursor ion fragmentation on a hybrid quadrupole time-of-flight mass spectrometer. Metabolites. 2: 195–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Body D. R. 1988. The lipid composition of adipose tissue. Prog. Lipid Res. 27: 39–60. [DOI] [PubMed] [Google Scholar]
- 24.Kotronen A., Seppanen-Laakso T., Westerbacka J., Kiviluoto T., Arola J., Ruskeepaa A. L., Yki-Jarvinen H., and Oresic M.. 2010. Comparison of lipid and fatty acid composition of the liver, subcutaneous and intra-abdominal adipose tissue, and serum. Obesity (Silver Spring). 18: 937–944. [DOI] [PubMed] [Google Scholar]
- 25.Betsholtz C. 2015. Lipid transport and human brain development. Nat. Genet. 47: 699–701. [DOI] [PubMed] [Google Scholar]
- 26.Bourre J. M. 2010. Diet, brain lipids, and brain functions: polyunsaturated fatty acids, mainly omega-3 fatty acids. In Handbook of Neurochemistry and Molecular Neurobiology: Neural Lipids. Lajtha A., Tettamanti G., and Goracci G., editors. Springer US, Boston: 409–441. [Google Scholar]
- 27.Hirabayashi Y. 2012. A world of sphingolipids and glycolipids in the brain—novel functions of simple lipids modified with glucose. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 88: 129–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.