Delaney et al. show that the unstructured domain cofactor LIN-65 is essential for the stable formation of heterochromatic foci. It stably interacts with the H3K9 methyltransferase MET-2/SETDB1 and modulates MET-2 nuclear localization, activity, and H3K9me-mediated silencing in C. elegans.
Abstract
The segregation of the genome into accessible euchromatin and histone H3K9-methylated heterochromatin helps silence repetitive elements and tissue-specific genes. In Caenorhabditis elegans, MET-2, the homologue of mammalian SETDB1, catalyzes H3K9me1 and me2, yet like SETDB1, its regulation is enigmatic. Contrary to the cytosolic enrichment of overexpressed MET-2, we show that endogenous MET-2 is nuclear throughout development, forming perinuclear foci in a cell cycle–dependent manner. Mass spectrometry identified two cofactors that bind MET-2: LIN-65, a highly unstructured protein, and ARLE-14, a conserved GTPase effector. All three factors colocalize in heterochromatic foci. Ablation of lin-65, but not arle-14, mislocalizes and destabilizes MET-2, resulting in decreased H3K9 dimethylation, dispersion of heterochromatic foci, and derepression of MET-2 targets. Mutation of met-2 or lin-65 also disrupts the perinuclear anchoring of genomic heterochromatin. Loss of LIN-65, like that of MET-2, compromises temperature stress resistance and germline integrity, which are both linked to promiscuous repeat transcription and gene expression.
Introduction
Organismal development, somatic tissue differentiation, and germline immortality require both genome stability and the proper spatiotemporal regulation of transcription. One of the dominant features of higher eukaryotic genomes is the prevalence of repetitive sequences, which must be transcriptionally silenced to preserve genome stability (Zeller et al., 2016). Essential for this repression is the methylation of nucleosomes on histone H3 lysine 9 (H3K9me), which allows such domains to be segregated away from active euchromatin (Martens et al., 2005; Towbin et al., 2012; Padeken and Heun, 2014; Lemaître and Bickmore, 2015). In addition to repetitive or constitutive heterochromatin, terminally differentiated cells restrict their capacity to assume alternative cell fates through the segregation of silenced tissue-specific genes into heterochromatic domains, which are also marked by H3K9me3 (Meister et al., 2010b; Solovei et al., 2013; Zeller et al., 2016; Nicetto et al., 2019). The spatial segregation of heterochromatin requires H3K9me2/me3 (Towbin et al., 2012) and components of the inner nuclear membrane (INM), including nuclear lamins and associated factors (Solovei et al., 2013; Gonzalez-Sandoval et al., 2015; Hübner et al., 2015).
H3K9 methylation in Caenorhabditis elegans is established by two conserved histone methyltransferases (HMTs). MET-2 (Andersen and Horvitz, 2007), a homologue of mammalian SETDB1/2, mediates mono- and dimethylation, while SET-25 shares homology with Suv39H1/2 and G9a and mediates H3K9me3 along with me1 and me2 (Towbin et al., 2012). Histones bearing H3K9me2 or me3 in C. elegans are bound by heterochromatin protein 1 (HP1) homologues HPL-1 and HPL-2, as well as by the MBT domain protein LIN-61 (Koester-Eiserfunke and Fischle, 2011; Ahringer and Gasser, 2018). All three readers contribute to transcriptional repression. A fourth H3K9me reader in worms is the INM protein CEC-4, which has an HP1-like chromodomain that mediates the perinuclear tethering heterochromatin in embryos, without conferring repression (Gonzalez-Sandoval et al., 2015).
Unlike the situation in mammals, C. elegans embryos lacking detectable H3K9 methylation, i.e., met-2 set-25 double mutants, are viable and differentiate (Towbin et al., 2012; Zeller et al., 2016). However, they show temperature-dependent defects in development and a loss of fertility when cultured at ≥20°C (Garrigues et al., 2015; Zeller et al., 2016). The absence of H3K9me leads to the transcription of all classes of repetitive elements, RNA:DNA hybrid accumulation, and increased mutation rates, particularly over repeats, which is the likely driver of the DNA damage–induced germline apoptosis that renders these animals sterile (Zeller et al., 2016). Similar sterility phenotypes have been observed in worms lacking key “readers” of H3K9 methylation, such as the HP1-like factors HPL-1 and HPL-2 (McMurchy et al., 2017).
Recent observations have suggested that heterochromatin may possess liquid droplet–like properties dependent on the concentration of the H3K9me reader HP1, which is thought to contribute to membrane-free separation of chromatin domains (Larson et al., 2017; Strom et al., 2017). Droplets form in part through weak interactions between intrinsically disordered domains found in heterochromatin-associated proteins (Frege and Uversky, 2015; Mittag and Parker, 2018). HP1 accumulation in heterochromatin has been proposed to depend in part on this property of phase separation.
Whereas HP1 proteins recognize H3K9 methylation, it is unclear how MET-2 itself is targeted to repetitive regions that need to be transcriptionally repressed. Previous results suggested that an overexpressed mCherry::MET-2 fusion protein localized to the cytoplasm of early C. elegans embryos (Towbin et al., 2012). In contrast, we show here, by tagging the endogenous met-2 locus to generate a fully functional fusion, that MET-2 is enriched in the nucleus at the nuclear periphery in irregular-sized foci, throughout C. elegans development. These foci appear and disappear with the cell cycle and seem to be generated by weak intermolecular interactions. Surprisingly, MET-2 focus formation is not dependent on known heterochromatic H3K9me readers, but rather on the physical association of MET-2 with the highly disordered protein LIN-65. Loss of LIN-65 recapitulates phenotypes associated with met-2 deletion, including extensive reduction in H3K9me2 levels, aberrant transcription of repetitive DNA and tissue-specific genes, loss of heterochromatin anchoring at the INM, and temperature-dependent sterility (Ahringer and Gasser, 2018; Padeken et al., 2019). Interestingly, MET-2 and LIN-65 foci are dispersed by temperature stress, and both proteins are required for recovery from heat shock. Taken together, these findings suggest that MET-2 concentrates into foci through interaction with LIN-65 to ensure the appropriate methylation of histones in heterochromatin and its subsequent sequestration at the INM. This, in turn, ensures the transcriptional repression that is necessary to maintain germline integrity.
Results
The H3K9 methyltransferase MET-2 accumulates in nuclear foci
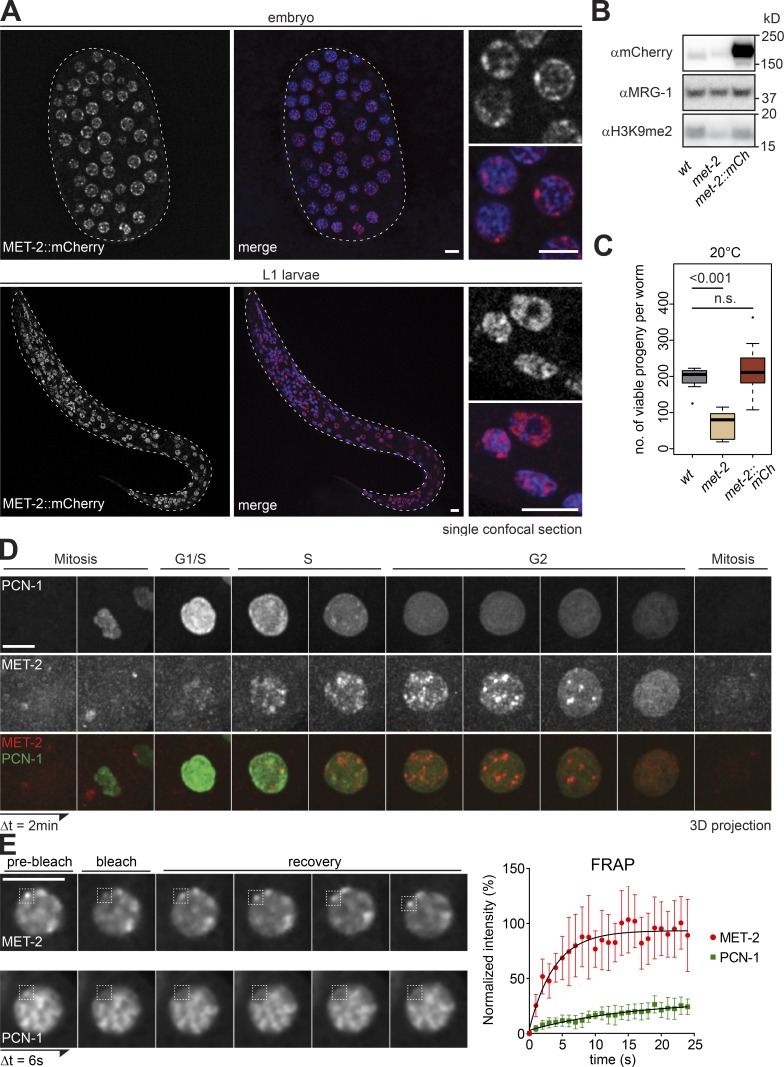
To study MET-2 regulation under physiological conditions, we tagged the endogenous met-2 locus with a C-terminal flag::mcherry cassette. In contrast to earlier results obtained with an overexpressed mCherry-tagged MET-2 (Towbin et al., 2012), MET-2::FLAG::mCherry expressed from its endogenous promoter was enriched in the nucleus throughout embryonic and larval development (Fig. 1 A). RNAi knockdown of met-2 in the MET-2 fusion expressing strain confirmed that the mCherry signal arises from MET-2 (Fig. S1 A). Moreover, the total levels of H3K9me2, repeat repression, and reproductive brood size in the MET-2::FLAG:mCherry strain (Fig. 1, B and C; and Fig. S1, B and C) were similar to those in wild-type worms (Zeller et al., 2016), confirming that the tagged protein is functional.
Figure 1.
A functional MET-2::mCh fusion forms nuclear foci through C. elegans development. (A) Confocal sections of MET-2::FLAG::mCherry (red) and Hoechst (blue) in a pregastrulation C. elegans embryo and L1 stage larva. Bar = 5 µm. (B) Western blot of whole-cell lysates of wild-type (N2), met-2(n4256), and met-2::flag::mcherry(gw1419) embryos blotted for H3K9me2, mCherry, and MRG-1 as a control (quantified in Fig. S1 B). (C) Number of viable progeny of strains used in B cultured at 20°C (N = 2, n = 50; P values calculated using two-sided Wilcoxon signed-rank test). (D) Representative Z-projected images from time-lapse series (2-min intervals) of early embryos, showing MET-2::FLAG::mCherry foci in S-G2 phase, identified by GFP::PCN-1. For movies, see Videos 1 and 2. Bar = 5 µm. (E) Left: Representative images of MET-2::FLAG::mCherry or GFP::PCN-1 FRAP over time. Interval between images, 6 s. Bar = 5 µm. Right: One representative experiment of three biological replicas is quantified. Error bars indicate 95% confidence interval. n = 9 and 12 nuclei, respectively.
In embryos, MET-2::FLAG::mCherry was concentrated in a highly variable number of nuclear foci (Fig. 1 A). Given that early embryonic cells are undergoing extremely rapid cellular division, one possible source of this variability was the cell cycle. To test this, we crossed the MET-2::FLAG::mCherry tag into worms expressing the PCNA homologue PCN-1 fused to GFP (gfp::pcn-1), to monitor MET-2 localization with respect to DNA replication. We found that MET-2 was excluded from the chromatin during mitosis and G1 phase but accumulated in bright nuclear foci during S phase, peaking in G2 (Fig. 1 D). This is consistent with a report showing that in mammalian cells, the MET-2 homologue SETDB1 is associated with histone chaperone CAF1, which deposits nucleosomes on newly replicated DNA (Loyola et al., 2009; Cheloufi et al., 2015; Yang et al., 2015).
The cell cycle–dependent formation of foci led us to examine the dynamics of MET-2 association with the foci. Using FRAP, we selectively ablated mCherry or GFP signal within independent MET-2 or PCN-1 foci and measured recovery of signal over time. We found that the MET-2 signal recovers completely and rapidly (half-time of recovery, 2.64 s), in contrast to PCN-1, which marks replication foci (half-time of recovery, 10.87 s; Fig. 1 E). This suggests that MET-2 has a high exchange rate between focal and nucleoplasmic populations, a characteristic attributed as well to the droplet-forming heterochromatin factor HP1 (Larson et al., 2017; Strom et al., 2017).
Consistent with the notion that heterochromatin becomes more stable as the replication cycle slows and cell type–specific transcription increases, we found that the MET-2 signal became more intense at later stages of embryonic development (Fig. 2 A). To test whether MET-2 foci form on heterochromatin, we made use of a well-characterized GFP-tagged heterochromatic reporter, gwIs4, which consists of an integrated array (n ≅ 280 copies) of a cassette containing the ubiquitously expressed baf-1 promoter and GFP-LacI fusion gene (Meister et al., 2010b; Towbin et al., 2010). The array is silenced by repressive histone modifications including H3K9me2/me3 and H3K27me3, and it lacks active marks such as H3K4me3. The MET-2::FLAG::mCherry foci coincided with the large heterochromatin domain, which is anchored at the nuclear periphery in an H3K9me-dependent manner (Fig. 2 B; Towbin et al., 2012; Gonzalez-Sandoval et al., 2015). By immunostaining, we find that MET-2 also colocalizes with H3K9me2, which marks endogenous heterochromatin in embryos (Fig. S1 D; Pearson correlation coefficient r = 0.7).
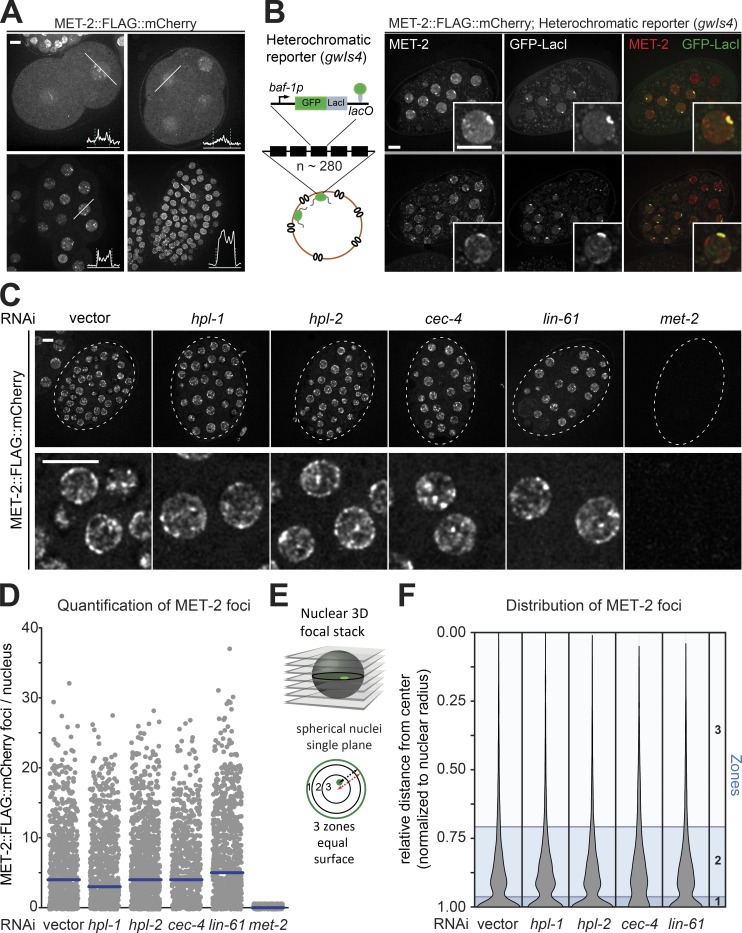
Figure 2.
Integrity and prevalence of MET-2 foci are independent of canonical H3K9me readers. (A) Live-cell images of 2-, 4-, 20-, and ∼100-cell embryos expressing MET-2::FLAG::mCherry, with line profiles averaged over a 10-pixel-width zone for mCherry intensity. Dashed lines indicate peaks of LEM-2::GFP signal. Bar = 5 µm. (B) Live-cell images of embryos expressing met-2::flag::mcherry and gfp::lacI from the integrated heterochromatic array (∼280 copies) of gwIs4 [baf-1p::GFP-lacI::let-858 3′UTR; myo-3p::RFP]. Assay visualized through GFP-lacI binding to lacO site (Meister et al., 2010b). (C) Example images of MET-2::FLAG::mCherry in 50-cell-stage embryos treated with RNAi against hpl-1, hpl-2, cec-4, lin-61, met-2, or empty vector (vector). The boxed region is enlarged in the lower row. Bar = 5 µm. (D) Quantitation of MET-2::FLAG::mCherry foci in the embryos shown in C using automated image analysis. N = 3, nuclei scored: n (vector) = 873, n (hpl-1) = 948, n (hpl-2) = 1,043, n (cec-4) = 675, n (lin-61) = 881, and n (met-2) = 589. (E) Scheme of nuclear zoning assay (Meister et al., 2010a). See Materials and methods for explanation of zones; zone 1 is the most peripheral. (F) Violin plot showing the cumulative spread of distances of MET-2::FLAG::mCherry foci to the nuclear periphery. Each measurement is normalized to the radius. Zones calculated as in E are shown in blue. N = 3, nuclei scored: n (vector) = 4,822, n (hpl-1) = 4,852, n (hpl-2) = 6,108, n (cec-4) = 3,991, and n (lin-61) = 6,126.
The dynamic nature of the MET-2 foci is reminiscent of recent findings suggesting that the H3K9me3 ligand HP1 can drive liquid–liquid phase separation of heterochromatin in both human and Drosophila melanogaster cells (Larson et al., 2012; Strom et al., 2017). Similar to Drosophila HP1a, we found that the MET-2 foci were dispersed by treatment with 1,6-hexanediol (Fig. S1, E and F), a compound known to disrupt liquid-like condensates by blocking weak hydrophobic interactions (Kroschwald et al., 2017; Strom et al., 2017). In contrast, PCN-1 replication foci were unaffected by 1,6-hexanediol treatment (Fig. S1, E and F). We conclude that endogenous MET-2 is concentrated in heterochromatic foci that are governed by weak interactions and whose abundance fluctuates through the cell cycle. The foci also show spatial dynamics within the nucleoplasm, as illustrated by time-lapse imaging (Videos 1 and 2).
MET-2 nuclear foci form independently of canonical H3K9me readers
MET-2 contains no predicted methyl-lysine recognition motifs that might lead to being bound to H3K9me within heterochromatin. We therefore asked whether any of the known H3K9me readers is essential for the targeting of MET-2 into foci and/or to chromatin (Elgin and Reuter, 2013; Ahringer and Gasser, 2018). Following RNAi against genes encoding the C. elegans HP1 family members HPL-1 and HPL-2, the chromodomain protein CEC-4 or the MBT domain protein LIN-61 (Figs. S1 A and S2 A), we imaged embryos expressing MET-2::FLAG::mCherry. In the same embryos, we also monitored GFP::PCN-1 to generate an independent nuclear mask. Quantitation of nuclear MET-2 foci by automated image analysis showed no loss of MET-2 foci by the knockdown of any of the tested H3K9me readers (Fig. 2, C and D). We assessed the subnuclear distribution of MET-2 foci following RNAi using the three-zone scoring method (Fig. 2 E; Meister et al., 2010a), which entails measuring the distance from the MET-2 focus to the nuclear rim in a single plane of focus and dividing this by the nuclear diameter in that plane. The resulting ratios are binned into three zones of equal surface (Fig. 2, E and F). This analysis showed that MET-2 foci are enriched in the peripheral-most zone, in both control cells and cells treated with RNAi against the HP1 homologues or LIN-61 (Fig. 2 F). Only loss of the perinuclear anchor protein, CEC-4, reduced the enrichment of MET-2 foci at the nuclear rim (Fig. 2, D and F), although loss of CEC-4 did not affect the formation of MET-2 foci or their average number (Fig. 2, D and F). This is consistent with previous work showing that CEC-4 is required for the perinuclear anchoring of H3K9me-marked chromatin (Gonzalez-Sandoval et al., 2015). We conclude that MET-2 foci form independently of canonical heterochromatin readers and depend on CEC-4 for perinuclear anchoring.
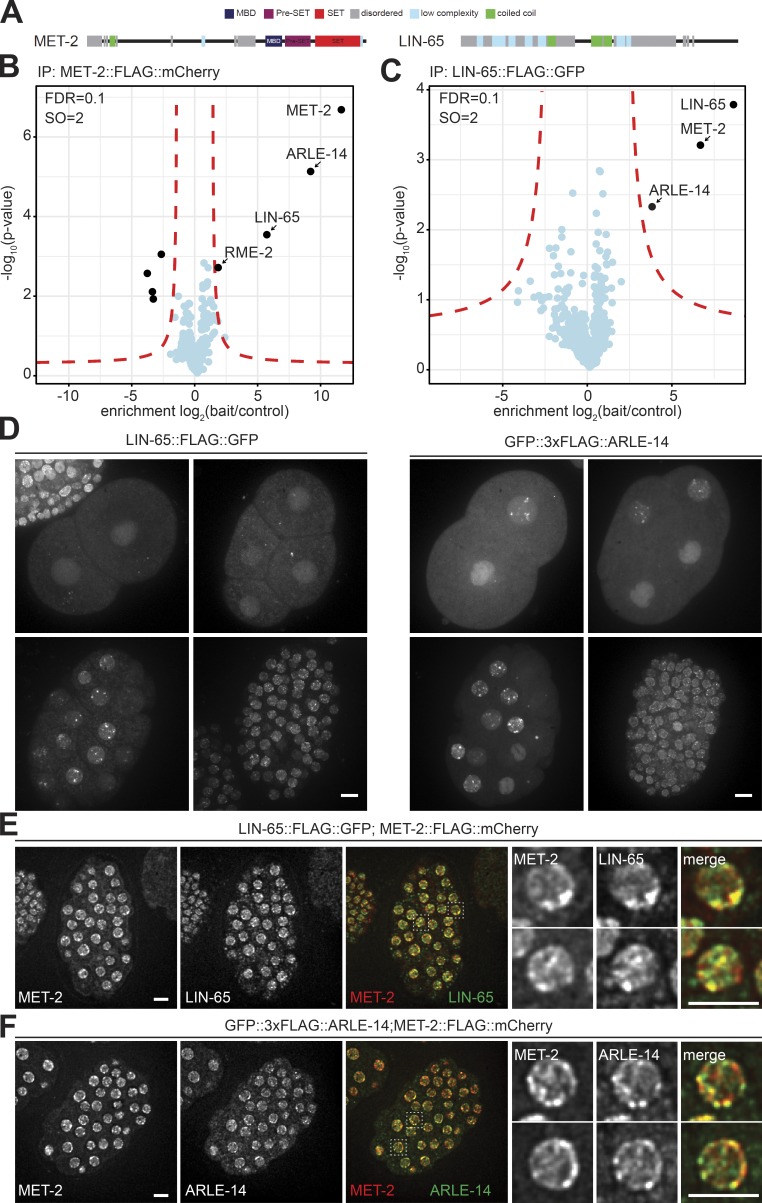
LIN-65 and ARLE-14 interact with MET-2 at nuclear heterochromatic foci
Since depletion of the conserved H3K9me readers did not affect MET-2 localization, we took an unbiased approach and performed MET-2 immunoprecipitation coupled to mass spectrometry (MS) to identify novel ligands that might help target MET-2. Using the FLAG tag to pull down MET-2::FLAG::mCherry and any tightly bound proteins, we found a clear enrichment for only two proteins: LIN-65 and ARLE-14 (Fig. 3, A and B) over an untagged control strain. Structurally, LIN-65 was reported to be related to ATF7IP/MCAF1 (Mutlu et al., 2018). Up to 70% of the protein is intrinsically disordered, being composed of interspersed low-complexity domains embedded in two long unstructured stretches. These two blocks are separated by a short coiled-coil motif and are flanked at the protein’s C terminus by a folded domain (Fig. 3 A; Mutlu et al., 2018). Initially lin-65 was described as a synMuvB gene, a classification identifying loci involved in the repression of the RAS pathway in C. elegans (Ceol et al., 2006). Interestingly, met-2 falls into the same synMuvB category (Poulin et al., 2005; Tischler et al., 2006). In addition, lin-65 was implicated in the mitochondrial stress response, where it appears to regulate the repression of stress response genes by promoting H3K9me2 (Tian et al., 2016). ARLE-14, on the other hand, is the worm homologue of a conserved but uncharacterized mammalian protein called adenosine 5′-diphosphate-ribosylation factor–like GTPase 14 effector protein (ARL14EP; Fig. S2 B).
Figure 3.
MET-2 binds and colocalizes in nuclear foci with LIN-65 and ARLE-14. (A) Scheme of structured and disordered domains of MET-2 and LIN-65 proteins. (B) FLAG-LC-MS/MS of MET-2 endogenously tagged with FLAG::mCherry. N = 3 biological replicates. Statistical analysis was done with Perseus (red lines indicate an FDR ≤ 0.1 and curve bend [SO] = 2). (C) FLAG-LC-MS/MS of LIN-65 endogenously tagged with FLAG::GFP. N = 3 biological replicates. Statistical analysis was done as in A. (D) Live-cell images of 2-, 4-, 20-, and ∼100-cell-stage embryos expressing LIN-65::FLAG::GFP or GFP::3xFLAG::ARLE-14. Bar = 5 µm. (E) Localization of MET-2 and LIN-65 in embryos with endogenous met-2 tagged with flag::mcherry (gw1419; red) and endogenous lin-65 tagged with flag::GFP (gw1578; green). Bar = 5 µm. (F) Localization of MET-2 and ARLE-14 in embryos with endogenous met-2 tagged with flag::mcherry(gw1419; red) and endogenous arle-14 tagged with 3xflag::GFP (gw1623; green). Bar = 5 µm.
To confirm the pulldown experiments, we tagged the full-length endogenous lin-65 gene with C-terminal flag::gfp (Ceol et al., 2006) and performed reciprocal FLAG pulldowns from embryos. Strikingly, the only proteins that were significantly enriched with LIN-65 were again MET-2 and ARLE-14 (Fig. 3 C). Further, the pulldown of an endogenously N-terminally tagged GFP::3xFLAG::ARLE-14 fusion identified only MET-2 and LIN-65 as interacting partners (Fig. S2 C). These reciprocal interaction studies suggest that ARLE-14 and LIN-65 simultaneously bind MET-2. We note that ARLE-14 and MET-2 were consistently recovered in stoichiometric amounts, while LIN-65 was substoichiometric (Fig. S2 D). This suggests either that LIN-65 is more labile, or else not all MET-2 and ARLE-14 molecules are bound by LIN-65.
Both LIN-65 and ARLE-14 showed accumulation in nuclear foci throughout embryogenesis, similar to MET-2 (Fig. 3 D, compare to Fig. 2 A). To further characterize LIN-65 and ARLE-14 localization with respect to MET-2, we introduced either GFP-tagged arle-14 or lin-65 alleles into the met-2::mcherry background. Both LIN-65 and ARLE-14 showed a high degree of colocalization with MET-2 (Fig. 3, E and F; Pearson correlation coefficient of r = 0.65 and r = 0.74, respectively). While not all MET-2 foci colocalized with a LIN-65 signal, and not all ARLE-14 foci colocalized with MET-2, the strong correlation nonetheless confirms that MET-2 tends to be associated with LIN-65 and ARLE-14 in vivo.
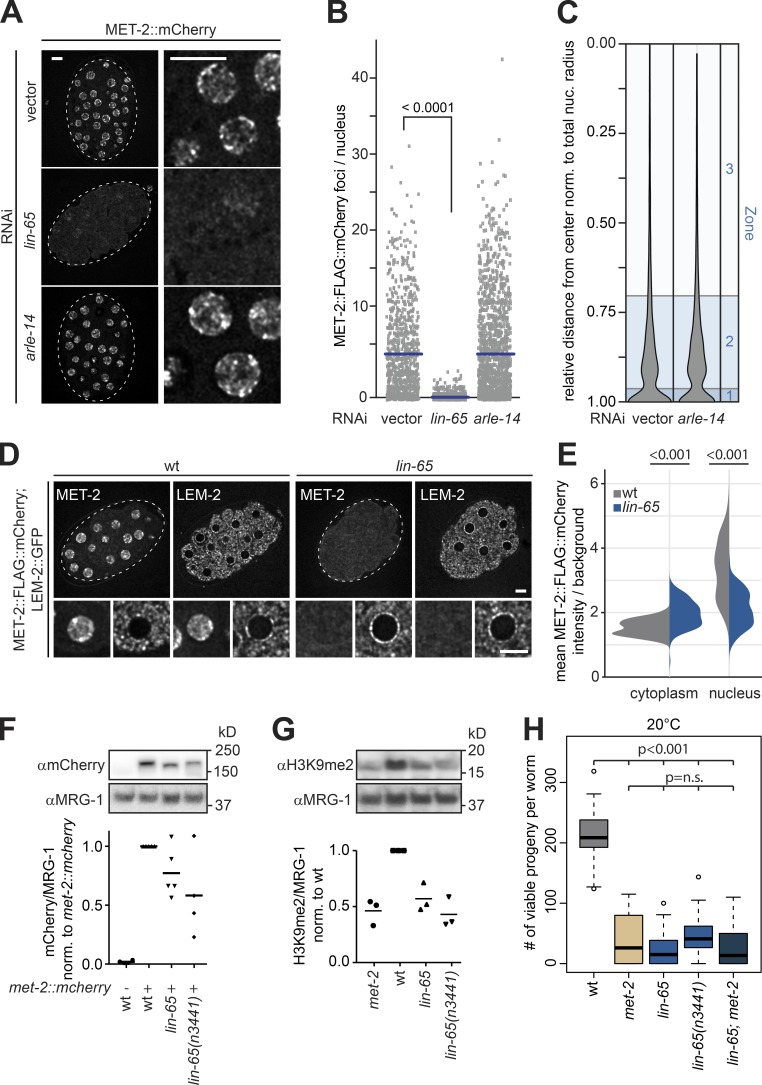
MET-2 accumulation in nuclear foci is dependent on LIN-65
The interaction and colocalization results for MET-2, ARLE-14, and LIN-65 suggested that ARLE-14 and/or LIN-65 might regulate MET-2 function. In mammals, a LIN-65–related factor, ATF7IP, was suggested to stabilize the MET-2 homologue, SETDB1 (Timms et al., 2016), and to stimulate HMT activity in vitro (Wang et al., 2003). Consistently, RNAi knockdown of lin-65 in C. elegans embryos led to an almost complete loss of MET-2 foci (Fig. 4, A and B), while the knockdown of arle-14 had no measurable impact on the number or the perinuclear localization of MET-2 foci (Fig. 4, A–C).
Figure 4.
LIN-65, but not ARLE-14, is required for MET-2 localization, focus formation, and stability. (A) Confocal images of MET-2::FLAG::mCherry-expressing embryos after RNAi for lin-65, arle-14, or empty vector (vector). Bar = 5 µm. (B) Quantitation of MET-2::FLAG::mCherry foci in embryos as shown in A using automated image analysis. N = 3, n (vector) = 873, n (lin-65) = 1,294, and n (arle-14) = 1,209; P values are indicated above scatter plots and were calculated using a Kruskal–Wallis test with Dunn’s correction for multiple testing. (C) Quantitation of MET-2::FLAG::mCherry foci distance to nuclear rim as shown in A using the three-zone method (see Fig. 2 and Meister et al. [2010a]). N = 3, n (vector) = 4,822, and n (arle-14) = 7,397. (D) Images of MET-2::FLAG::mCherry in wild-type and lin-65(gw1465) mutant embryos with nuclear rim visualized by LEM-2::GFP. Bar = 5 µm. (E) Quantitation of MET-2::FLAG::mCherry fluorescence intensity in the cytoplasm and the nucleus in wild-type and lin-65(gw1465) embryos (n = 118, P values were calculated using two-sided Wilcoxon signed-rank test and indicated above violin plots). (F) Example Western blot (top) and quantification (bottom) of MET-2::FLAG::mCherry protein levels in wild-type, lin-65(gw1465), and lin-65(n3441) embryos from Western blots (N = 4) probed as indicated and normalized to endogenous MRG-1 abundance in wild-type embryos. P values in met-2::FLAG::mCherry(gw1419) versus wild-type: lin-65(gw1465) = 0.042, lin-65(n3441) = 0.036; bars indicate mean signal. (G) As in F, except that H3K9me2 levels are quantified, in wild-type, met-2(n4256), lin-65(gw1465), lin-65;met-2, and lin-65(n3441) embryos. N = 3, P values versus wild-type: met-2 = 0.015, lin-65(gw1465) = 0.028, lin-65;met-2 = 0.018, lin-65(n3441) = 0.007; bars indicate mean signal. (H) Number of viable progeny of strains indicated in G cultured at 20°C (N = 2, n = 50; P values were calculated using two-sided ANOVA and indicated above boxplots).
To understand the interdependence of LIN-65, ARLE-14, and MET-2, we monitored each protein in embryos by microscopy, after knockdown of either of the other two partners. RNAi efficiency was confirmed by both fluorescence microscopy and Western blot (Fig. S3, A and B). Intriguingly, when met-2 is depleted by RNAi, the fluorescent signals for LIN-65 and ARLE-14 proteins drop to roughly the same point as if the worms had been treated with RNAi against their own transcripts (Fig. S3 A). This argues that the stability of LIN-65 and ARLE-14 is strongly dependent on MET-2. This could be due to either protein–protein interaction or methylation of the ligands by MET-2. Indeed, G9a has been shown to methylate ATF7IP (Tsusaka et al., 2018). Intriguingly, knockdown of lin-65 eliminated the LIN-65 signal and resulted in the loss of both MET-2 and ARLE-14 foci (Fig. S3, A and B), although the cells retained weak fluorescence of MET-2 and ARLE-14 dispersed throughout the nucleus and cytoplasm (Figs. 4 D and S3 A). This argues that LIN-65 is crucial for focus formation, but that the other two proteins persist in the absence of LIN-65. Interestingly, knockdown of arle-14 affected neither MET-2 nor LIN-65 foci (Fig. S3 A). This argues that LIN-65 and MET-2 are interdependent and act upstream of ARLE-14.
To examine the loss of MET-2 foci in the absence of LIN-65 quantitatively, we generated a novel null allele, lin-65(gw1465) which eliminates most of the open reading frame. We will henceforth refer to this allele as the lin-65 mutant (Fig. S4 A). Both the newly generated deletion and a previously characterized nonsense allele, lin-65(n3441), which truncates the 728–amino acid protein at position 534 (W534X; Fig. S4 A; Ceol et al., 2006), caused loss of nuclear MET-2 foci (Figs. 4 D and S4 B).
We next tested whether loss of LIN-65 altered met-2 gene expression, the correct localization of MET-2 to the nucleus, or its stability in the nucleus. We found no changes in the transcript levels of met-2 or lin-65 in lin-65 or met-2 mutant embryos, respectively (Fig. S4 C). To determine whether loss of LIN-65 disrupts the nuclear targeting of MET-2, we measured the mean fluorescence intensity of MET-2::FLAG::mCherry in defined nuclear and cytoplasmic zones in wild-type and lin-65 mutant embryos. Embryos mutant for lin-65 showed a reduction of nuclear MET-2 signal (Fig. 4 E; mean 4.5 ± 0.8-fold/background enrichment in wild-type vs. 2.5 ± 0.4-fold/background in lin-65), but an increased cytoplasmic MET-2 signal (Fig. 4 E; 1.6 ± 0.2-fold/background in wild-type vs. 2.2 ± 0.3-fold/background in lin-65). We monitored the same increase in cytoplasmic MET-2 after lin-65 RNAi (Fig. S4 D). The mislocalization of MET-2 was also accompanied by an overall 40–50% drop in MET-2 protein level in both lin-65 mutant backgrounds, as determined by immunoblot (Fig. 4 F). This is consistent with data suggesting that the loss of mammalian ATF7IP leads to a partial degradation of nuclear SETDB1 (Timms et al., 2016).
LIN-65 is predominantly composed of disordered or low-complexity domains, which are a common feature of proteins that drive formation of phase-separated condensates (Frege and Uversky, 2015; Mittag and Parker, 2018). Not surprisingly, as shown above for MET-2 foci (Fig. S1, E and F), LIN-65 foci were also sensitive to 1,6-hexanediol treatment (Fig. S3 C). This supports the argument that liquid–liquid phase separation may contribute to the formation of these foci. In conclusion, the unstructured protein LIN-65 is necessary for the formation of heterochromatic MET-2 foci in nuclei, and its loss led to a loss of both MET-2 stability and its enrichment in the nucleus. ARLE-14 localization is similarly dependent on LIN-65, although it does not contribute to MET-2 targeting, as far as we can tell. While both LIN-65 and MET-2 foci are sensitive to 1,6-hexanediol, further in vitro studies are needed to determine if the unstructured domains of LIN-65 can drive focus formation by liquid–liquid phase separation.
Loss of lin-65 up-regulates genes repressed by MET-2
To test whether LIN-65 is required for MET-2’s enzymatic function, we measured H3K9me2 levels in wild-type embryos and embryos of the appropriate mutant genotypes. Intriguingly, embryos deficient for lin-65 showed a reduction of H3K9me2 to a similar level as met-2(n4256) embryos (Fig. 4 G; hereafter called the met-2 mutant). We note that the loss of met-2 alone causes a strong temperature-dependent reduction in germline viability (Garrigues et al., 2015; Padeken et al., 2019), similar to that observed in the met-2 set-25 double mutant (Zeller et al., 2016). Although loss of LIN-65 had only a partial effect on MET-2 protein levels, the lin-65 deletion and lin-65(n3441) point mutant reduced fertility at 20°C to the same degree as the met-2 mutant (Fig. 4 H), showing complete sterility and larval arrest at 26°C (Fig. S4 E). Given the similarity of progeny numbers from the single mutants and the lin-65;met-2 double mutant, it is likely that met-2 and lin-65 are epistatic for germline stability; in other words, robust MET-2 activity requires LIN-65–mediated nuclear accumulation and focus formation.
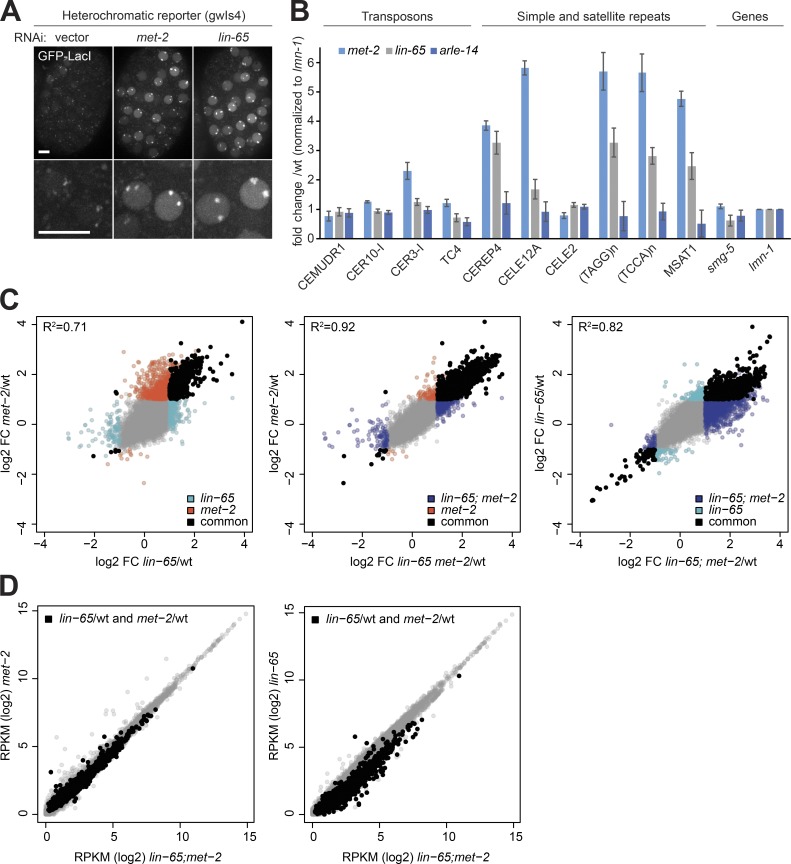
We monitored the effect of lin-65 mutation on transcriptional silencing by RNA-seq and RNA-qPCR and the heterochromatic array. First, lin-65 RNAi resulted in a loss of repression of the heterochromatic reporter gwIs4, yielding a level of GFP-LacI fluorescence that was roughly similar to the level after met-2 RNAi (Fig. 5 A). Furthermore, reverse transcription–quantitative PCR (RT-qPCR) performed on total RNA isolated from lin-65 mutants showed a loss of silencing of met-2–dependent microsatellite and simple repeats (Fig. 5 B; CeREP4, CELE12A, (TAGG)n, (TCCA)n, and MSAT1). Transposons that do not depend on MET-2 for repression (Fig. 5 B; CEMUDR1, CER10-I, CER3-I, and TC4; Padeken et al., 2019), as well as two single-copy genes, also showed independence of LIN-65 (smg-5 and lmn-1; Fig. 5 B). No transcriptional changes were detected in an arle-14 deletion mutant (arle-14(tm6748); Fig. 5 B). Elsewhere, we have shown that the aberrant expression of microsatellite and simple repeats correlates with the loss of germline integrity in the met-2 mutant (Padeken et al., 2019), which explains the parallel infertility of lin-65 and met-2 mutants. Interestingly, however, the level of satellite derepression in the lin-65 mutant was on average ∼55% of that observed in the met-2 mutant (Fig. 5 B), suggesting that MET-2 has residual activity in the absence of LIN-65.
Figure 5.
Transcripts up-regulated in absence of lin-65 correlate with and are largely epistatic to met-2-induced changes. (A) Representative image of embryos bearing reporter gwIs4, which expresses GFP::LacI when derepressed, following RNAi against met-2, lin-65, or empty vector. Bar = 5 µm. (B) RT-qPCR on total RNA isolated from wild-type, lin-65(gw1465), met-2(n4256), and arle-14(tm6748) mutant embryos for selected repetitive elements showing the fold change in expression over wild-type (N = 4; error bars indicate the standard deviation). Classes of repetitive elements are indicated above the graph. (C and D) Changes of genes in met-2(n4256), lin-65(gw1465), and met-2;lin-65 embryos determined by total RNA-seq (N = 3). (C) Correlation between the fold change (FC, log2) over wild-type for gene expression between met-2 and lin-65 single and double mutants. Genes significantly changed compared with wild-type (FDR <0.05; fold change >2 or <−2) are colored according to the genotype (lin-65 = cyan; met-2 = red; met-2 = purple), and genes significantly changed in two compared genotypes are black. Pearson correlations are indicated as R2 in the graphs. (D) Expression level of genes in reads per kilobase per million (RPKM in log2) in met-2 and lin-65 single mutants (y axis) compared with the lin-65;met-2 double mutant (x axis). Black mark genes significantly changed in both met-2 and lin-65 single mutants.
To examine the degree to which met-2 and lin-65 transcriptional control correlates genome-wide, we performed RNA-seq from total RNA isolated from early embryos of wild-type, lin-65, met-2, and lin-65;met-2 mutants cultured at 20°C. Data generated from three independent biological replicates demonstrated that genes aberrantly expressed in lin-65 and met-2 single mutants correlate significantly (Pearson coefficient = 0.71; Fig. 5 C; see Fig. S5 for replicate correlation). Loss of met-2, however, resulted in the derepression of genes that were not significantly up-regulated in the lin-65 mutant (1,941 genes derepressed at least twofold in the met-2 vs. 973 in the lin-65 mutants; false discovery rate [FDR] < 0.01; Fig. 5 C), although 86% of the genes up-regulated in the lin-65;met-2 double mutant were also up-regulated in the met-2 single mutant (Pearson coefficient = 0.92; Fig. 5 C). Comparing the lin-65 single mutant to the lin-65;met-2 double mutant confirms that a subset of the met-2–sensitive genes are not significantly up-regulated upon loss of LIN-65 (Fig. 5 C). Among genes significantly up-regulated in both single mutants, we observed a higher expression level in the lin-65;met-2 double mutant compared with lin-65 alone, but there was no additional increase over the derepression scored in met-2 alone (Fig. 5 D). Taken together, these results argue that LIN-65 functions in gene repression almost exclusively through interaction with MET-2, and that the dispersion of MET-2 foci observed in the lin-65 mutant is sufficient to derepress a large subset of MET-2–dependent loci. Loss of lin-65 does not mimic a complete loss of MET-2 function, as some coding regions that are repressed by MET-2 do not seem to require LIN-65. We propose that LIN-65 enhances the heterochromatin association of MET-2 and therefore its activity at these sites.
MET-2 and LIN-65 are reversibly dispersed by heat shock but are essential for recovery
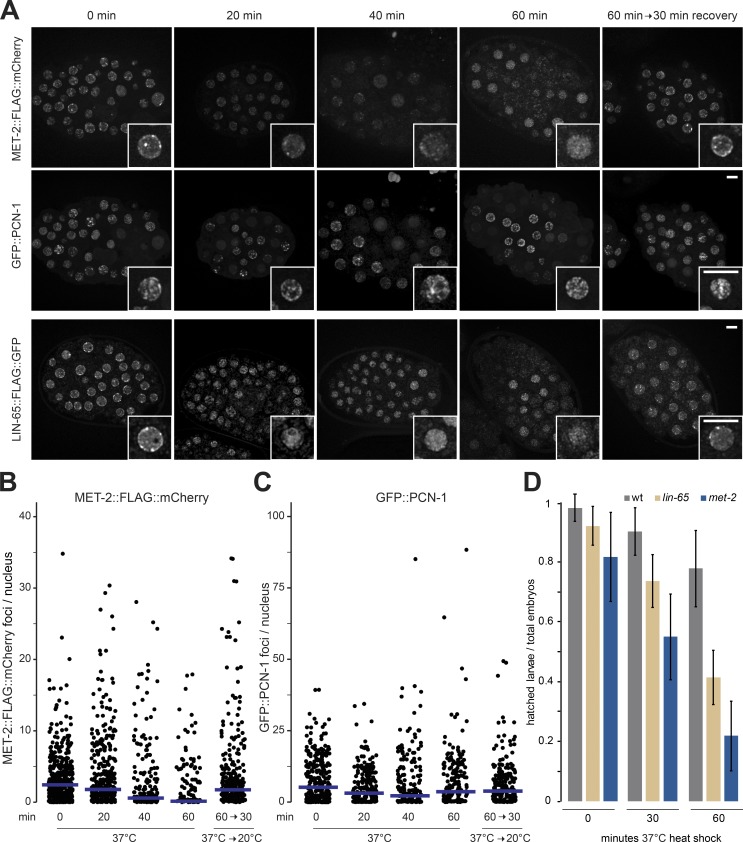
LIN-65 has no obvious enzymatic function on its own (Fig. 3 A), yet the analysis of MET-2 foci and RNA-seq in the lin-65 mutant suggests that it is able to stabilize and localize MET-2. LIN-65 has also been shown to be essential for the stress response in C. elegans (Tian et al., 2016). Intriguingly, H3K9me-deficient mutants show temperature sensitivity for both germline viability and developmental delay (Zeller et al., 2016). We therefore examined the behavior of MET-2 foci in embryos under heat-shock conditions, to see if the role of LIN-65 in stress survival is linked to MET-2 focus dynamics. We observed the gradual dispersal of MET-2::mCherry foci during a 1-h exposure of embryos to 37°C (Fig. 6, A and B). This does not reflect a general disruption of nuclear organization, as GFP::PCN-1 localization did not change upon heat-shock in the same cells (Fig. 6, A and C). Importantly MET-2 foci reappeared when embryos were shifted back to 20°C for 30 min after the 1-h exposure to 37°C (Fig. 6, A and B). If this loss of MET-2 foci is due to its interaction with LIN-65, we expected to observe a similar loss of LIN-65 foci upon heat shock. This was the case: LIN-65::GFP foci exhibited the same dispersal and recovery during the described heat shock treatment as MET-2::mCherry (Fig. 6 A). To see whether the dispersal and recovery of MET-2 and LIN-65 foci are physiologically relevant, we quantified the hatching rate 1 d after a 30- or 60-min heat shock exposure. While 80 ± 14% of the wild-type embryos hatched even after 60 min at 37°C, only 31 ± 10% of the met-2 and 48 ± 5% of the lin-65 mutant embryos hatched (Fig. 6 D). We conclude that MET-2 and LIN-65 are dynamically regulated during stress, and that they both contribute to organismal recovery from heat shock exposure.
Figure 6.
MET-2 and LIN-65 are essential to recover from temperature stress. (A) Representative images of MET-2::FLAG::mCherry, GFP::PCN-1, and LIN-65::FLAG::GFP embryos subjected to 37°C heat shock for the indicated length of time (N = 3, n = 60 embryos). (B and C) Quantitation of MET-2::FLAG::mCherry (B) and GFP::PCN-1 (C) foci/nucleus from A; blue bars indicate mean foci number. (D) Mean survival of animals 24 h after embryonic heat shock. N = 2, n (wt 0 min) = 520, n (wt 30 min) = 463, n (wt 60 min) = 436, n (met-2 0 min) = 213, n (lin-65 0 min) = 238, n (lin-65 30 min) = 239, n (lin-65 60 min) = 243, n (met-2 30 min) = 229, and n (met-2 60 min) = 210; error bars indicate standard deviation.
Loss of met-2 or lin-65 compromises heterochromatin anchoring to the nuclear periphery
In addition to forming foci, heterochromatin has a propensity to be sequestered at the INM, which in C. elegans embryos is mediated by H3K9 methylation and the chromodomain protein, CEC-4 (Gonzalez-Sandoval et al., 2015). Previous work has shown that integrated heterochromatic arrays and the arms of C. elegans autosomes are associated with the INM, and that this anchoring is lost when all H3K9 methylation is lost, i.e., upon ablation of both H3K9 HMTs, MET-2 and SET-25 (Ikegami et al., 2010; Meister et al., 2010b; Towbin et al., 2010, 2012; Gonzalez-Sandoval et al., 2015). On the other hand, the presence of either SET-25 or MET-2 was shown to be sufficient to allow gwIs4 array tethering to CEC-4 at the INM (Fig. 6 A; Towbin et al., 2012). Given the dominant effect of met-2 ablation alone on the expression of repeats and genes (Padeken et al., 2019), and the enrichment of MET-2 at the INM (Fig. 2 F), we next examined the localization of endogenous sequences in the single mutants, set-25 and met-2. Moreover, if it were entirely MET-2 dependent, then it would be reasonable to expect that it is also sensitive to the loss of LIN-65.
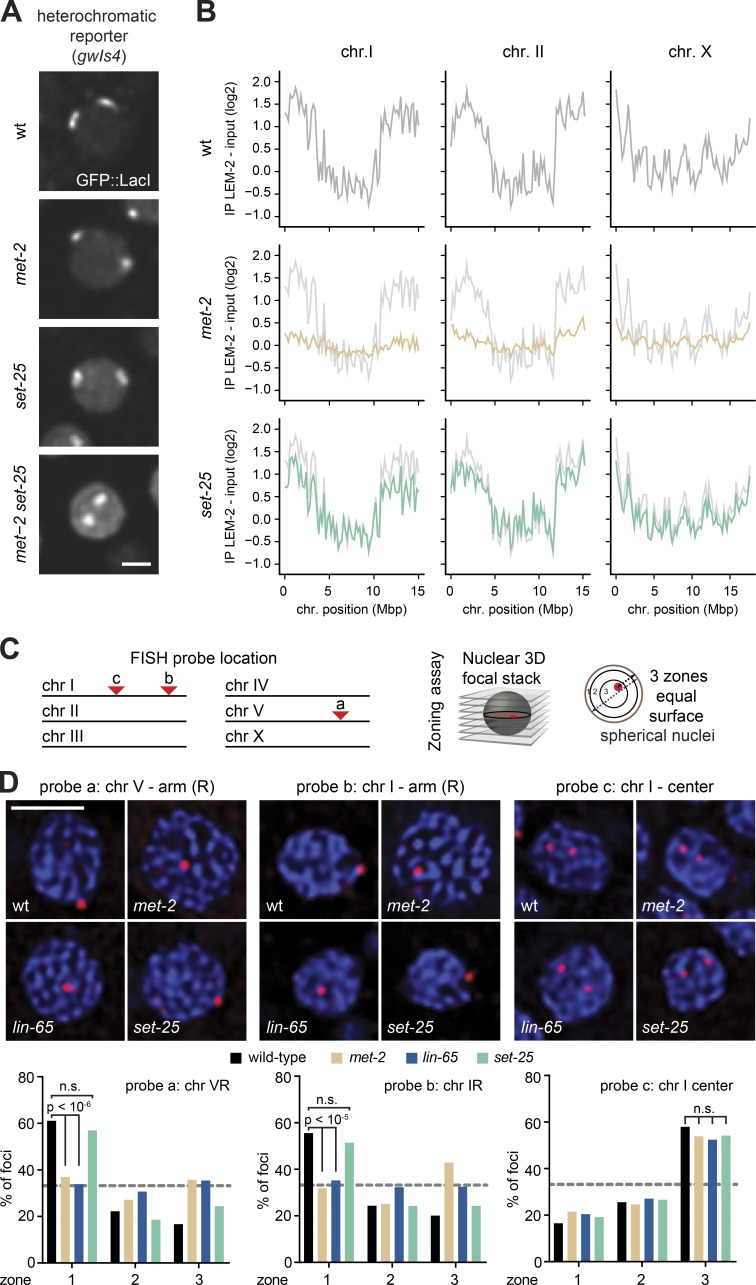
To test this, we performed chromatin immunoprecipitation sequencing (ChIP-seq) experiments with the INM-associated protein LEM-2, a well-established proxy for nuclear lamin-associated chromatin (Gerstein et al., 2010; Ikegami et al., 2010; Towbin et al., 2012; Gonzalez-Sandoval et al., 2015; Morales-Martínez et al., 2015). In wild-type embryos, both arms of the autosomes and the left arm of the X chromosome are enriched for interactions with the periphery, while the chromosome centers are depleted (Ikegami et al., 2010; Figs. 7 B and S5 B). Intriguingly, mutants that have lost histone H3K9me3, owing to ablation of set-25 (Towbin et al., 2012), largely retained enrichment of the chromosome arms at the periphery (Figs. 7 B and S5 B). In contrast, the ablation of met-2 alone completely abolished the preferential association of autosomal arms with LEM-2, indicating a shift from the INM, and similarly compromised the enrichment of mid-chromosome domains in the nuclear core (Figs. 7 B and S5 B). This argues that H3K9 trimethylation is not essential for peripheral anchoring, while MET-2–mediated H3K9me2 is crucial. This is consistent with our finding that MET-2–catalyzed H3K9me2 often replaces H3K9me3 on repetitive sequences in the set-25 mutant, whereas the opposite is not true (Padeken et al., 2019). We suggest that the peripheral enrichment of MET-2 foci observed in Fig. 2 reflects and may contribute to the propagation of spatially separated, perinuclear heterochromatin domains.
Figure 7.
MET-2 and LIN-65, but not SET-25, drive the perinuclear association of heterochromatin. (A) Confocal images of GFP::LacI tagged gwIs4 heterochromatic array in wild-type, met-2(n4256), or set-25(n5021) embryonic nuclei. Neither met-2 nor set-25 deletion delocalize the array, whereas met-2 set-25 double mutants do (Towbin et al., 2012). Bar = 2 µm. (B) LEM-2 ChIP in wild-type (gray), met-2(n4256) (orange), or set-25(n5021) (green), on mixed embryos from the indicated mutant lines grown at 20°C (averaged from three biological replicates) for chromosomes I, II, and X. See Fig. S5 for other chromosomes. Mean signals averaged over 100-kb bins. (C) Left: Scheme of C. elegans chromosomes with FISH probe position. Right: Scheme of three-zone assay used to score FISH probe location; see Fig. 2 and Meister et al. (2010a). (D) Top: Confocal images of a single focal plane of wild-type, met-2(n4256), lin-65(gw1465), or set-25(n5021) nuclei from mid-stage embryos, probed by FISH for the indicated loci (red) and DAPI (blue). Bottom: Quantitation of FISH signals by three-zone scoring; gray line = random distribution or 33%. The χ2 test compares changes in zonal enrichments. Foci were scored from multiple nuclei and embryos in >2 replicates (for probe 1: wt, n = 563; nmet-2, n = 394; lin-65, n = 593; set-25, n = 676; for probe 2: wt, n = 423; met-2, n = 282; lin-65, n = 502; set-25, n = 276; for probe 3: wt, n = 504; met-2, n = 256; lin-65, n = 391; set-25, n = 349. Bar = 2 µm.
We confirmed the loss of heterochromatin anchoring in the met-2 mutant embryos by FISH, using probes for two regions in the arms of chromosomes V and I which showed a clear enrichment of LEM-2 in the ChIP-seq (probes a and b) and one probe in mid-chromosome I that was depleted in the LEM-2 ChIP-seq (probe c; Fig. 7 C). The three-zone assay was used to quantify the subnuclear position of these loci in mid-stage embryos (50–100 cells). In this assay, all measurements were performed in the plane of focus. The distance between the signal and the nuclear periphery was divided by the nuclear diameter, as described above, such that a random distribution would yield 33% in each zone (Fig. 7 C). Quantitation of the FISH signal positions confirmed that the distal arm regions (probes a and b) were enriched in the perinuclear zone 1 in wild-type embryos, and that they lost this enrichment in the met-2, but not the set-25, mutant (Fig. 7 D). The central probe c showed internal enrichment in wild-type, met-2, and set-25 mutant embryos (Fig. 7 D). Given that LIN-65 is essential for the formation of MET-2 foci and for full repression of repetitive sequences (Fig. 5 B), we also checked whether the loss of LIN-65 would compromise the perinuclear positioning of the heterochromatic probes. Indeed, we found an identical loss of anchoring upon loss of LIN-65 as with ablation of MET-2 (Fig. 7 D). Thus, the interaction between MET-2 and LIN-65 is critical for focus formation, peripheral anchoring, and gene repression mediated by H3K9 methylation despite the presence of the second H3K9 HMT, SET-25.
Discussion
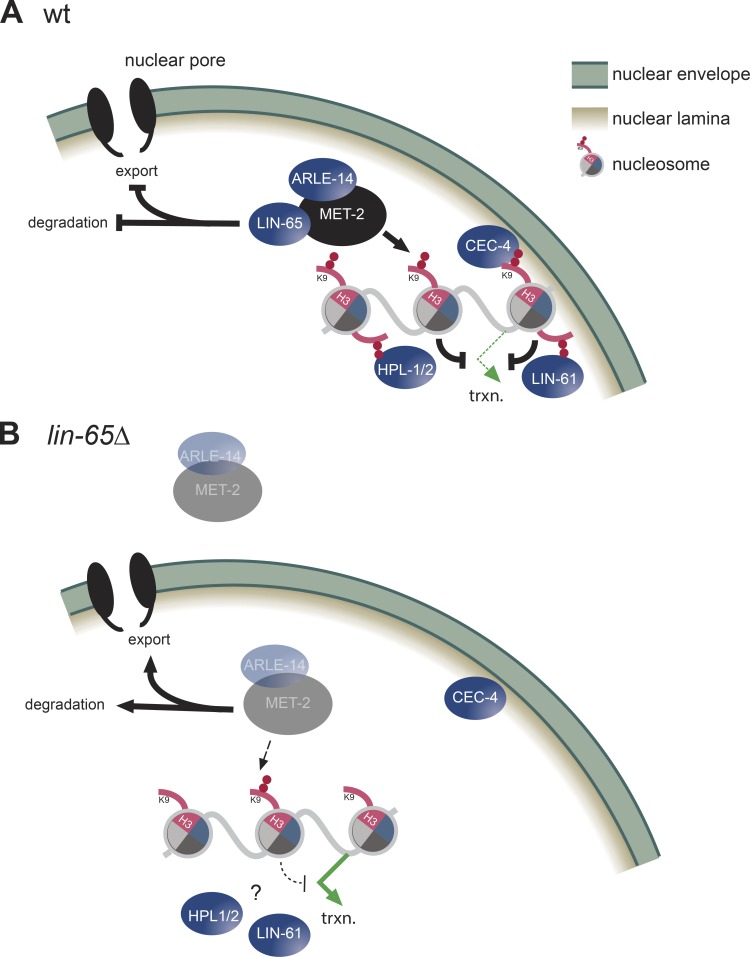
Heterochromatin formation through the methylation of histone H3K9 is an essential process that ensures genome stability and germline integrity by silencing repetitive elements and tissue-specific genes (Zeller et al., 2016; Ahringer and Gasser, 2018; Nicetto et al., 2019). Both repetitive elements and tissue-specific genes that are repressed by H3K9me2/me3 are known to coalesce into discrete subnuclear domains that are sequestered away from active domains and are often enriched at the INM or around the nucleolus (Padeken and Heun, 2014; Lemaître and Bickmore, 2015). We show here that the H3K9 methyltransferase MET-2/SETDB1 is also concentrated in perinuclear foci, together with two cofactors LIN-65 and ARLE-14. These MET-2/LIN-65 foci appear to be critical for MET-2’s ability to silence both repetitive elements and genes, and correlate with the anchoring of heterochromatin at the nuclear periphery. The MET-2 focus formation and perinuclear anchoring, as well as H3K9me2-mediated repression of repeats and genes, depend on LIN-65 (Fig. 8).
Figure 8.
LIN-65 targets a trimeric ARLE-14/LIN-65/MET-2 complex to heterochromatin to form foci. (A) The essential role of the unstructured protein LIN-65 for MET-2 focus formation on heterochromatic loci, and its role in import and stabilization, are shown. MET-2–mediated H3K9me2 is necessary for repeat and tissue specific gene repression. (B) The repercussions of LIN-65 loss are shown.
LIN-65 and ARLE-14 were identified by protein pulldown and MS as the two dominant interacting partners of MET-2 in C. elegans embryos (Figs. 3 and S2; Mutlu et al., 2018). In our hands, ARLE-14 ablation produces none of the phenotypes that we have linked to met-2 or lin-65 mutation (Figs. 5 B and S3). Although several high-throughput MS studies have identified ARL14EP, the mammalian homologue of ARLE-14, as a putative interactor of SETDB1 or SETDB2, to date there remains no clear function ascribed to it (Guruharsha et al., 2011; Rolland et al., 2014; Hübner et al., 2015). In contrast to ARLE-14, the loss of LIN-65 resulted in the mislocalization of MET-2, the dispersion of MET-2 foci, and a compensatory increase in cytosolic enzyme coupled with an overall drop in MET-2 protein level (Fig. 8).
With respect to H3K9 methylation and gene silencing, the impact of a lin-65 mutation was similar to that of met-2 deletion: H3K9me2 was reduced and transcripts from MET-2–targeted loci were up-regulated (Fig. 5). However, the loss of MET-2 clearly derepressed more genes than the loss of LIN-65. By comparing the up-regulated transcripts in the lin-65 and met-2 single mutants with those misregulated in the double met-2;lin-65 mutant, we conclude that residual MET-2 activity persists in the lin-65 mutant, even though MET-2 focus formation and perinuclear attachment at the INM is lost. Indeed, the genes misregulated in the lin-65 mutant have a strong positive correlation, but incomplete overlap, with those up-regulated in the met-2 mutant (Pearson coefficient = 0.7; Fig. 5). Whether LIN-65 helps target MET-2 to specific sequences, or simply enhances its activity by creating zones that are enriched in the targets of H3K9 methylation, requires further study. Importantly, even though MET-2 retains residual activity in the lin-65 mutant, LIN-65, like MET-2, is essential for germline integrity and resistance to temperature stress.
It has been proposed that LIN-65 is a functional homologue of the mammalian activating transcription factor 7 interacting protein (ATF7IP), although direct sequence identity is low (Mutlu et al., 2018). Like LIN-65, ATF7IP (also known as MCAF1 or AM) is a highly disordered protein that interacts with the H3K9 HMT, SETDB1 (Fujita et al., 2003; Wang et al., 2003), and may regulate its stability (Timms et al., 2016). Similarly, in Drosophila, the ATF7IP homologue WDE binds to SETDB1 and is required for its proper function (Koch et al., 2009; Yu et al., 2015). In mammals, it is proposed that ATF7IP both promotes H3K9 methylation and binds transcriptional activators (Timms et al., 2016; Fukuda et al., 2018). Similarly, LIN-65 may have functions beyond the spatial regulation of MET-2. In Tian et al. (2016), evidence is presented showing that LIN-65 is necessary for the nuclear accumulation of a mitochondrial unfolded protein response (UPRmt) transcription factor. This is an intriguing suggestion, as we found ∼30 genes that are down-regulated in the lin-65 mutant and not in met-2–deficient embryos, suggesting that LIN-65 may serve not only to facilitate repression, but also in gene induction (Fig. 5). Consistently, not all LIN-65 foci coincide with MET-2 foci (Fig. 3 E). Thus, despite the dependence of MET-2 activity on LIN-65 in heterochromatin, the proteins may have distinct roles in gene induction at other sites.
Cell cycle and early embryonic dynamics of MET-2 and heterochromatic foci
We note that MET-2 foci are dynamic during the cell cycle, and that the turnover of MET-2 in these foci is very rapid (Fig. 1 and Videos 1 and 2). MET-2 foci form during S-G2 phase and persist through G2, consistent with evidence in mammalian systems which argues that H3K9 methylation is deposited by a CAF1/SETDB1 complex to restore heterochromatic states in postreplicative nuclei (Loyola et al., 2009; Yang et al., 2015). As the cell cycle time increases in later stages of C. elegans development, the stability of heterochromatic foci increases, and these foci become persistent in the nondividing, differentiated cells of the L1 larva. As noted in a recent study of MET-2 in C. elegans (Mutlu et al., 2018), the nuclear enrichment of this HMT is less pronounced in 2–4-cell embryos than at later stages. This may reflect the much shorter cell cycle (∼5 min per S phase) in 2-cell embryos (Bao et al., 2008) or else may reflect the presence of developmental regulators, such as those proposed to control early mammalian development. In mammals, the establishment of H3K9 methylation is a crucial event that helps silence retrotransposons as well as imprinted genes in very early stages of embryogenesis (Svoboda et al., 2004; Macfarlan et al., 2012; Fadloun et al., 2013).
The importance of heterochromatic focus formation is not restricted to embryogenesis, and MET-2–bound heterochromatic foci persist in differentiated somatic cell nuclei (Fig. 1 A; Towbin et al., 2012; Garrigues et al., 2015; Zeller et al., 2016; Ahringer and Gasser, 2018). Recent work proposes that heterochromatin foci form by a phase separation mechanism that relies on HP1 (Larson et al., 2017; Strom et al., 2017), and which depends on other intrinsically disordered domain factors in the nucleus (Frege and Uversky, 2015; Cho et al., 2018; Sabari et al., 2018). Similar liquid phase demixing has been demonstrated for DNA damage foci (Altmeyer et al., 2015) and enhancer–promoter interactions (Boija et al., 2018; Sabari et al., 2018). The characteristics of dynamic subnuclear foci are rapid assembly and dissolution and a membrane-free accumulation of specific factors, all which are features demonstrated for the MET-2 foci we characterize in this article. While there is no evidence that MET-2 would directly carry out liquid–liquid phase separation, LIN-65 has structural characteristics that would predict involvement in such events. Moreover, MET-2 residency in heterochromatin foci is highly dynamic (Fig. 1 E), and MET-2/LIN-65 foci are governed by weak interactions that are sensitive to 1,6-hexanediol (Figs. S1 E and S3 C) and heat stress (Fig. 6). Thus, liquid–liquid phase separation triggered by LIN-65 could explain the concentration of MET-2 in heterochromatic foci. Nonetheless, a better understanding of the principles that control such interactions and characterization of the LIN-65 and MET-2 proteins in vitro is required to substantiate whether such a mechanism underlies MET-2 focus formation. We note that none of the other well-known C. elegans readers of H3K9me, including the worm HP1 homologues, HPL-1 and HPL-2, LIN-61, and CEC-4, affects MET-2 enrichment in the nuclear foci, although CEC-4 mediates the perinuclear sequestration of the foci in embryos (Fig. 2 F; Gonzalez-Sandoval et al., 2015).
LIN-65 and MET-2 interaction in foci may be linked to stress survival
LIN-65 and MET-2 are interdependent for their proper localization and stability, and both lin-65 and met-2 mutants show temperature-dependent loss of fertility and sensitivity to temperature stress (37°C; Figs. 4, 6, and S3). The dispersal of MET-2 and LIN-65 foci during temperature stress and their subsequent recovery after a shift back to 20°C indicates that this mutual dependence may function as a switch that either turns off genes as an essential survival mechanism or is required for de novo H3K9me targeting to restore silencing efficiently after stress-induced derepression. The absence of met-2 or lin-65 leads to temperature-stress induced lethality. We speculate that this principle may be broadly applicable to conditions of stress or aging, given that H3K9me levels decrease both upon aging (Larson et al., 2012; Tsurumi and Li, 2012; Brunet and Rando, 2017) and upon certain types of nutritional stress (Sidler et al., 2017). Intriguingly, LIN-65 and MET-2 have been implicated in the genome regulation that mediates the unfolded protein response provoked upon mitochondrial dysfunction (UPRmt; Tian et al., 2016). LIN-65 and MET-2 promote the accumulation of DVE-1, a transcription factor that promotes stress response survival, into nuclear foci (Tian et al., 2016). Thus, the LIN-65 interaction with MET-2 may be key not only to ensure H3K9me2 deposition, but also to regulate H3K9me2 loss and selected gene activation in response to stress. It will be important to identify LIN-65 protein interactors under conditions of stress, namely heat shock and UPRmt, and/or during recovery from environmental insults. This may provide insight into the mechanisms through which essential epigenetic marks like H3K9me are modulated by environmental cues.
Materials and methods
Strains and transgenics, RNAi, and hexanediol treatment
Strains used in this experiment are listed in Table S1. Unless otherwise indicated, experiments were performed using early embryos isolated from animals cultured at 20°C.
GW1562 was made using single guide RNAs (sgRNAs) targeting the 5′ and 3′ end of the lin-65 reading frame which were cloned into pIK198 (Katic et al., 2015). For GW1419 and GW1618, we tagged the endogenous gene at the C termini of met-2 with FLAG::TEV::mCherry and of lin-65 with FLAG::TEV::GFP, respectively, by using the SapTrap system essentially as described by Schwartz and Jorgensen (2016). The sgRNA was cloned into pIK198 (Katic et al., 2015) and the repair construct into pMLS257 (Schwartz and Jorgensen, 2016). For GW1623, HDR template was made from PCR fragments containing GFP, TEV, and 3xFLAG from pDD282 (Dickinson et al., 2015) that were fused using overlap PCR using primers that contained >30-bp homology arms to the arle-14 N terminus. This HDR template was injected with synthetic crRNA and tracrRNA (Dharmacon) preloaded in recombinant Cas-9(IDT) as previously described (Paix et al., 2015). sgRNA and crRNA sequences are provided in Table S2.
RNAi feeding experiments were performed described previously (Kamath et al., 2001; Timmons et al., 2001). Briefly, synchronized L1s were transferred to nematode growth media plates containing 1 mM IPTG and 100 µg/ml carbenicillin, seeded with the indicated RNAi, and grown to adulthood. Embryos from these animals were used in subsequent analyses. For 1,6-hexanediol treatment, MET-2::FLAG::mCh, GFP::PCN-1, and LIN-65::GFP embryos from synchronous cultures were incubated with 10% 1,6-hexanediol diluted in M9 buffer containing 0.1% Triton X-100 for 2 min before imaging. Images were acquired and analyzed as described in Live microscopy.
Western immunoblotting
Embryos were lysed with a Fast Prep 24-5G Benchtop Homogenizer (MP Biomedicals) using 0.5 mm zirconia/silicon beads (BioSpec) in RIPA buffer in the presence of cOmplete EDTA-free protease inhibitors (Roche) and 1 mM DTT at 4°C. Lysates were treated with 5 µl Benzonase (Sigma) for 1 h at 4°C with end-over-end rotation. 10 µg total protein was separated on Bolt 4–12% Bis-Tris Plus gels (Thermo Fisher Scientific) and transferred to PVDF (Bio-Rad). Blots were blocked in PBS plus 0.5% Tween-20 with 5% powdered milk. Antibodies used included 1:500 mouse anti-H3K9me2 MABI0317 (MBL; Kimura et al., 2008), 1:1,000 rabbit anti-RFP antibody Pre-adsorbed (Rockland), and 1:2,000 rabbit anti-MRG-1 (49130002; Novus Biologicals). Blots were treated with antibodies diluted in blocking buffer overnight at 4°C. After three washes, blots were reblocked and exposed to secondary antibodies coupled to HRP. After three subsequent washes, ECL (Millipore) was applied, and signal was detected using an Imager 600 (GE).
Brood size
Total progeny from animals were scored as previously described (Zeller et al., 2016). Briefly, young adults were plated singly at 20°C or 26°C, where indicated to test temperature sensitivity. They were transferred to fresh plates every 24 h for 3 d, after which total progeny was counted.
MS
Samples were prepared at 4°C. Embryos were lysed as above in TAP buffer (150 mM NaCl, 20 mM Tris-HCl, pH 7.5, 0.5% NP-40, 1 mM EDTA, 10% glycerol, 2× cOmplete-EDTA-free protease inhibitors [Roche], and 1 mM DTT). After bead beating, lysates were sonicated using a Bioruptor Plus (Diagenode) for 15 cycles, 15 s on, 30 s off. Lysates were cleared by centrifugation at 21,000 g for 10 min. 3 mg cleared lysates were rotated overnight at 4°C with 10 µl anti-FLAG M2 beads (Sigma) prewashed in TAP buffer. Beads were washed four times with TAP buffer, then three times in 20 mM Tris, pH 7.5, and 150 mM NaCl. Bound proteins were digested on beads with 0.2 µg trypsin overnight at 37°C plus 5 h with fresh trypsin, and then analyzed as described previously (Ostapcuk et al., 2018). Briefly, peptides were acidified to 0.8% with TFA, then liquid chromatography/tandem MS (LC-MS/MS) was performed on an Easy-nLC 1000 two-column setup (Thermo Fisher Scientific). Peptides were then loaded with 2% acetonitrile and 0.1% formic acid on a 75 µm × 2 cm Acclaim PepMap Trap, C18, 2 µm, 100 Å column. We used a 50 µm × 15 cm ES801 C18, 2 µm, 100 Å column (Thermo Fisher Scientific), with a DPV ion source (New Objective) with the following linear gradient: 0–3 min, 2–6% B in A; 3–43 min, 6–22%, 43–52 min, 22–28%; 52–60 min, 28–36%; 60–61 min, 36–80%; and 61–75 min, 80%. A consisted of 0.1% formic acid in H2O, and B, 0.1% formic acid in MeCN, at 45°C with a 150 µl/min flow rate. Acquisitions were made at 120,000 resolution in the Orbitrap and top T (3 s) method with HCD fragmentation for precursors and fragments according to the manufacturer’s instructions (Thermo Fisher Scientific). Results were analyzed with MaxQuant version 1.5.3.8 (Cox and Mann, 2008). Perseus version 1.5.2.6 was used for statistical analysis using the Andromeda search engine (Cox et al., 2011). To generate a volcano plot, significance lines for FDR = 0.1 with a curve bend of 2.0 were determined (Tusher et al., 2001; Ostapcuk et al., 2018). Analysis was then exported and plotted using R.
ChIP
ChIP experiments were performed as previously described (Zeller et al., 2016). In brief, embryos were harvested from synchronized animals in three replicates. 40 µg of chromatin was incubated overnight with 3 µg of anti-LEM-2 antibody (Novus Biologicals) coupled to Dynabeads sheep anti-rabbit IgG (Invitrogen), in FA buffer (50 mM Hepes/KOH, pH 7.5, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, and 150 mM NaCl) containing 1% SDS. Antibody-bound chromatin was washed for 3 × 5 min with FA buffer; 5 min with FA buffer with 1 M NaCl; 10 min with FA buffer with 500 mM NaCl; 5 min with TEL buffer (0.25 M LiCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA, and 10 mM Tris-HCl, pH 8.0), and 2 × 5 min with TE. Complexes were eluted in 1% SDS in TE with 250 mM NaCl at 65°C for 15 min. Samples and inputs were treated with 20 µg of RNase A for 30 min at 37°C and 20 µg of proteinase K for 1 h at 55°C. Cross-links were reversed by overnight incubation at 65°C. DNA was subsequently purified using Zymo DNA purification columns (Zymo Research).
Libraries were prepared as previously described (Zeller et al., 2016) using the NEBNext ultra DNA library prep kit for Illumina (7370; NEB) and the NEBNext Multiplex Oligos for Illumina (E7335; NEB), according to the manufacturer’s recommendations without size selection. Libraries were indexed and amplified using 12 PCR cycles, following manufacturer’s recommendations. Libraries were further purified with Agencourt AmPure XP beads (A63881; Beckman). Library size range and concentration were determined using a BioAnalyzer 2100 (Agilent Technologies) and Qubit (Invitrogen) instrument, respectively. Equimolar amounts of indexed libraries were pooled and sequenced on a HiSeq 2500 (Illumina) in rapid mode (Paired-End 50). Reads were aligned to the C. elegans genome (ce10) with the R package QuasR using Bowtie.
Read density was calculated by tiling the genome into 200-bp nonoverlapping windows and using the qCount function of the QuasR package to quantify the number of reads in each window. Differences in read depths between samples were normalized by dividing each sample by total reads and multiplying by average library size. Log2 expression levels were determined after addition of a pseudocount of 1 [y = log2(x + 1)]. Results are displayed as the mean enrichment of immunoprecipitation − input (log2).
RNA expression analysis
RNA was isolated from embryos as described previously (Zeller et al., 2016). Briefly, embryos resuspended in Trizol (Invitrogen) were freeze cracked four times, and then extracted with phenol-chloroform followed by isopropanol precipitation. Ribosomal RNA was depleted using Ribo-Zero Gold kit (Epicentre). Libraries were produced using Total RNA Sequencing ScriptSeq kit (Illumina). Single-end 50-bp reads were generated using an Illumina HiSeq 2500. Reads were analyzed as described previously (Towbin et al., 2012) using Wormbase (WS190). cDNA for qPCR was generated using Superscript IV reverse transcription (Invitrogen) followed by amplification using PowerUp SYBR green Master Mix (Thermo Fisher Scientific). Primer sequences used in qPCR experiments are previously reported (Zeller et al., 2016).
Live microscopy and Immunofluorescence (IF)
Animal morphology images were taken at 20°C with a Leica M205 FA microscope. Fluorescent images of live animals or embryos were captured in M9 buffer on 2% agarose pads at 20°C for all experiments except heat-shock, which were imaged at 37°C. Unless otherwise stated, IF and live images were captured using Visiview software (Visitron) on a confocal spinning-disk system: AxioImager M1 (Zeiss) with a Yokogawa CSU-X1 scanhead (Yokogawa), a Rolera Thunder camera (Photometrics), and an α plan-NEOFLUAR 100×/1.45 oil objective (Zeiss). For IF, bleached embryos were fixed for 5 min in 1% formaldehyde before spotting onto poly-l-lysine–coated coverslips, freezing on dry ice, and storing at −80°C. When needed, slides were freeze cracked and immediately fixed in a −20°C 100% ethanol (EtOH) bath for 2 min and then dried. After washing 3 × 5 min with PBS + 0.25% Triton X-100 (PBS-T) and blocking for 1 h with PBS-T and 2% milk, slides were incubated overnight at 4°C in a humid chamber with primary antibodies in PBS-T and 2% milk: 1:500 mouse anti-H3K9me2 MABI0317 (MBL) and 1:500 polyclonal rabbit anti-RFP (Rockland). After washing 3 × 5 min with PBS-T, slides were incubated for 1 h at RT in a humid chamber with secondary antibodies in PBS-T and 2% milk: 1:1,000 goat anti-mouse Alexa Fluor 488 (A11001; Invitrogen) and 1:1,000 donkey anti-rabbit Alexa Fluor 555 (A31572; Invitrogen). DNA was counterstained with DAPI (1:2,000) in PBS-T for 10 min, washed 3 × 5 min with PBS-T, and mounted with ProLong Gold Antifade (Thermo Fisher Scientific). All images were deconvolved with Huygens Professional version 18.04 (Scientific Volume Imaging) and the Huygens remote manager (http://www.huygens-rm.org/wp/). Colocalization was quantified using the integrated colocalization tool in the Huygens remote manager with automatic background and threshold detection.
We quantified nuclear foci using the KNIME Analytics Platform (Dietz and Berthold, 2016). In summary, nuclei were detected using a seeded watershed segmentation on the PCN-1 (GFP) channel. For foci detection, we used a Laplacian-of-Gaussian detector from TrackMate (Tinevez et al., 2017; fmi-ij2-plugins-0.2.5, https://doi.org/10.5281/zenodo.1173536) on the MET-2 (mCherry) channel. The distance of foci to the nuclear periphery was measured by computing a Euclidean distance map on the 3D nucleus mask and measuring its intensity for the coordinates of each spot. Foci outside of a nucleus were ignored in the analysis. Subsequently, we normalized the distance value of each spot to the size of the containing nucleus, i.e., the maximum of the distance map, and inverted the distance value to be in line with previous analyses, resulting in a “normalized distance from center” that was plotted per condition using the R package ggplot2 (https://ggplot2.tidyverse.org/).
FISH
For FISH, formaldehyde fixation and freeze cracking were done as for IF, immediately followed by fixing in −20°C EtOH baths: 70% for 2 min, 80% for 2 min, 95% for 2 min, and 100% for 2 min. After washing 3 × 5 min with PBS-T, slides were treated for 30 min at 37°C with 1 mg/ml RNaseA. Slides were again washed with PBS-T and refixed in EtOH baths. After drying, the FISH probe was added, denatured for 3 min at 95°C, and then incubated overnight at 37°C. Slides were then washed 3 × 5 min in 2× SSC/50% formamide (prewarmed to 42°C), 3 × 5 min in 2× SSC (prewarmed to 42°C), and 1 × 10 min in 1× SSC (prewarmed to 42°C). DNA was counterstained with DAPI (1:2,000) in 4× SSC for 10 min, followed by 3 × 5 min washes in 2× SSC and mounting with ProLong Gold Antifade Mountant (Thermo Fisher Scientific). Images were captured at 20°C using Visiview software (Visitron) on a confocal spinning-disk system: Ti2-E Eclipse (Nikon) with a Yokogawa CSU-W1 scanhead (Yokogawa), an iXon Ultra 888 camera (Andor), and a CFI Plan Apochromat Lambda 100×/1.45 oil objective (Nikon).
FISH probes were generated using fosmids (probe 1, WRM0637cA03; probe 2, WRM0635aE09; and probe 3, WRM0626aC03) as a template for nick translation using the FISH Tag DNA kit with the Alexa Fluor 555 dye (F32948; Thermo Fisher Scientific). Probe was prepared according to manufacturer’s instructions until the final precipitation. Approximately 1 µg of prepared probe was precipitated with 0.4 mg of salmon sperm DNA, 15 µl KAc (3 M, pH 5.2), and 350 µl EtOH for 20 min at −20°C. After centrifugation, the DNA pellet was washed with 70% EtOH and resuspended in 160 µl of deionized formamide. The probe was then denatured for 10 min at 72°C, chilled on ice for 5 min, and then mixed with 160 µl of 2× hybridization buffer (4× SSC, 4 mg/ml BSA, and 20% dextran sulfate). Prepared probes were stored at −20°C until needed.
Assessment of peripheral localization was performed using a well-established three-zone method that is described in detail in Gonzalez-Sandoval et al. (2015) and Meister et al. (2010a). The zoning assay exploits two facts: first, that confocal image resolution is better in x–y than in z, and second, that a tagged locus can usually be assigned to specific plane of a through-focus stack of images. The quantitation of localization is performed as follows. A stack of 50–70 focal planes is taken through a spherical nucleus for which the nuclear envelope is marked by mCherry and the chromatin locus of interest by GFP. In the plane of focus where the GFP spot is brightest, we measure the diameter of the nucleus (through the spot) and the distance from the spot to the nearest point on the nuclear envelope. That ratio can be compared from sample to sample no matter whether the plane is near the equator or near either pole of the sphere. The disc of the sphere in which the locus is found is divided into three zones of equal surface, each containing 33% of the area. Cavalieri’s principle proves that each zone of a disc represents one third of the volume of the sphere. A randomly distributed focus scored in a population of nuclei will yield 33% in each of the three zones.
Due to the so-called z-stretch, which limits resolution in z, we do not score spots that are in the uppermost and lowermost planes, and we generally eliminate the top and bottom 20% of the sphere with a procedure called “decapping.” As long as the focal stacks capture nuclei in all orientations, this does not bias results; indeed, decapping improves the accuracy of the zone measurements (Meister et al., 2010a).
FRAP
FRAP experiments were done using the FRAP settings in Visiview Software (Visitron). Live embryos in M9 buffer were imaged in a single plane every 1 s for 30 frames, with five prebleach images followed by bleaching (500 ms at 8% laser power), and 25 post-bleach images. Imaging for FRAP was done at 20°C on a confocal spinning-disk system: Axio Observer (Zeiss) equipped with a Plan-Apochromat 100×/1.4 oil objective (Zeiss), a CSU-W1 scan-head (Yokogawa), a Prime 95B sCMOS camera (Photometrics), a 488- or 561-nm Obis laser (Coherent) for illumination, an homogenizer for shading compensation (Visitron), and a FRAP scan-head (Visitron). FRAP was performed with the 405 nm iBeam smart laser (Toptica) attached to the FRAP module. Fluorescence intensity was measured using Fiji (Schindelin et al., 2012). Background signal was subtracted, and all values were normalized relative to the first prebleach intensity (100%) and immediately post-bleach intensity (0%). Focal signal was also corrected for loss due to photobleaching (using signal from an adjacent nucleus) and for internal loss (using signal from adjacent region of the same nucleus). A nonlinear regression curve for FRAP recovery was generated in Prism (GraphPad Software) using an exponential one-phase association with least squares fit. Half-time to recovery was determined from the regression curves.
Accession codes
All datasets from this study have been uploaded to the Gene Expression Omnibus under accession no. GSE122341.
Online supplemental material
Fig. S1 shows MET-2::FLAG::mCherry functionality, by RNAi, H3K9me2 Western blot, RT-PCR of repeat expression, and H3K9me2 IF, as well as MET-2::FLAG::mCherry sensitivity to 1,6-hexanediol. Fig. S2 shows RNAi efficiency by RT-qPCR, ARLE-14 interactome, and iBAQ values of MET-2, LIN-65, and ARLE-14 interactions. Fig. S3 summarizes the interdependencies between MET-2, LIN-65, and ARLE-14; RNAi efficiency against lin-65 and arle-14; and the sensitivity of LIN-65::GFP foci to 1,6 hexanediol. Fig. S4 supports the mislocalization of MET-2 upon loss of LIN-65 shown in Fig. 4. Fig. S5 shows the correlation between RNA-seq replicas from Fig. 5 and additional LEM-2 ChIP-seq tracks for chromosomes III–V. Videos referred to in Fig. 1 are available as Videos 1 and 2. Table S1 lists all strains, and Table S2 lists the gRNAs used in this study.
Supplementary Material
Acknowledgments
We thank H. Kimura for the H3K9me2 antibody, the Gasser laboratory for critical reading of the manuscript, and Jan Eglinger and Laurent Gelman of the FMI imaging and microscopy facility for excellent help with image analysis. We thank the FMI Functional Genomics, Protein Structure and Protein Analysis facilities, and European Molecular Biology Laboratory GeneCore for advice and support.
We thank the Caenorhabditis Genetics Center of the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440) for strains. J. Padeken and S.P. Methot are both supported by European Molecular Biology Organization Long Term Fellowships. The research is funded by European Research Council and Swiss National Science Foundation grants.
The authors declare no competing financial interests.
Author contributions: C.E. Delaney, S.P. Methot, M. Guidi, and J. Padeken planned experiments, performed them, carried out analysis where necessary, and helped make figures and write the text. I. Katic provided extensive helpful advice and contributed to strain generation. S.M. Gasser evaluated data, provided advice, secured funding, and helped write the paper.
References
- Ahringer J., and Gasser S.M.. 2018. Repressive Chromatin in Caenorhabditis elegans: Establishment, Composition, and Function. Genetics. 208:491–511. 10.1534/genetics.117.300386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmeyer M., Neelsen K.J., Teloni F., Pozdnyakova I., Pellegrino S., Grøfte M., Rask M.-B.D., Streicher W., Jungmichel S., Nielsen M.L., and Lukas J.. 2015. Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat. Commun. 6:8088 10.1038/ncomms9088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen E.C., and Horvitz H.R.. 2007. Two C. elegans histone methyltransferases repress lin-3 EGF transcription to inhibit vulval development. Development. 134:2991–2999. 10.1242/dev.009373 [DOI] [PubMed] [Google Scholar]
- Bao Z., Zhao Z., Boyle T.J., Murray J.I., and Waterston R.H.. 2008. Control of cell cycle timing during C. elegans embryogenesis. Dev. Biol. 318:65–72. 10.1016/j.ydbio.2008.02.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boija A., Klein I.A., Sabari B.R., Dall’Agnese A., Coffey E.L., Zamudio A.V., Li C.H., Shrinivas K., Manteiga J.C., Hannett N.M., et al. 2018. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell. 175:1842–1855.e16. 10.1016/j.cell.2018.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A., and Rando T.A.. 2017. Interaction between epigenetic and metabolism in aging stem cells. Curr. Opin. Cell Biol. 45:1–7. 10.1016/j.ceb.2016.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceol C.J., Stegmeier F., Harrison M.M., and Horvitz H.R.. 2006. Identification and classification of genes that act antagonistically to let-60 Ras signaling in Caenorhabditis elegans vulval development. Genetics. 173:709–726. 10.1534/genetics.106.056465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloufi S., Elling U., Hopfgartner B., Jung Y.L., Murn J., Ninova M., Hubmann M., Badeaux A.I., Euong Ang C., Tenen D., et al. 2015. The histone chaperone CAF-1 safeguards somatic cell identity. Nature. 528:218–224. 10.1038/nature15749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho W.K., Spille J.H., Hecht M., Lee C., Li C., Grube V., and Cisse I.I.. 2018. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science. 361:412–415. 10.1126/science.aar4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., and Mann M.. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26:1367–1372. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., and Mann M.. 2011. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10:1794–1805. 10.1021/pr101065j [DOI] [PubMed] [Google Scholar]
- Dickinson D.J., Pani A.M., Heppert J.K., Higgins C.D., and Goldstein B.. 2015. Streamlined Genome Engineering with a Self-Excising Drug Selection Cassette. Genetics. 200:1035–1049. 10.1534/genetics.115.178335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz C., and Berthold M.R.. 2016. KNIME for Open-Source Bioimage Analysis: A Tutorial. Adv. Anat. Embryol. Cell Biol. 219:179–197. 10.1007/978-3-319-28549-8_7 [DOI] [PubMed] [Google Scholar]
- Elgin S.C., and Reuter G.. 2013. Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harb. Perspect. Biol. 5:a017780 10.1101/cshperspect.a017780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadloun A., Le Gras S., Jost B., Ziegler-Birling C., Takahashi H., Gorab E., Carninci P., and Torres-Padilla M.E.. 2013. Chromatin signatures and retrotransposon profiling in mouse embryos reveal regulation of LINE-1 by RNA. Nat. Struct. Mol. Biol. 20:332–338. 10.1038/nsmb.2495 [DOI] [PubMed] [Google Scholar]
- Frege T., and Uversky V.N.. 2015. Intrinsically disordered proteins in the nucleus of human cells. Biochem. Biophys. Rep. 1:33–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N., Watanabe S., Ichimura T., Ohkuma Y., Chiba T., Saya H., and Nakao M.. 2003. MCAF mediates MBD1-dependent transcriptional repression. Mol. Cell. Biol. 23:2834–2843. 10.1128/MCB.23.8.2834-2843.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda K., Okuda A., Yusa K., and Shinkai Y.. 2018. A CRISPR knockout screen identifies SETDB1-target retroelement silencing factors in embryonic stem cells. Genome Res. 28:846–858. 10.1101/gr.227280.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigues J.M., Sidoli S., Garcia B.A., and Strome S.. 2015. Defining heterochromatin in C. elegans through genome-wide analysis of the heterochromatin protein 1 homolog HPL-2. Genome Res. 25:76–88. 10.1101/gr.180489.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein M.B., Lu Z.J., Van Nostrand E.L., Cheng C., Arshinoff B.I., Liu T., Yip K.Y., Robilotto R., Rechtsteiner A., Ikegami K., et al. modENCODE Consortium . 2010. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 330:1775–1787. 10.1126/science.1196914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Sandoval A., Towbin B.D., Kalck V., Cabianca D.S., Gaidatzis D., Hauer M.H., Geng L., Wang L., Yang T., Wang X., et al. 2015. Perinuclear Anchoring of H3K9-Methylated Chromatin Stabilizes Induced Cell Fate in C. elegans Embryos. Cell. 163:1333–1347. 10.1016/j.cell.2015.10.066 [DOI] [PubMed] [Google Scholar]
- Guruharsha K.G., Rual J.F., Zhai B., Mintseris J., Vaidya P., Vaidya N., Beekman C., Wong C., Rhee D.Y., Cenaj O., et al. 2011. A protein complex network of Drosophila melanogaster. Cell. 147:690–703. 10.1016/j.cell.2011.08.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hübner B., Lomiento M., Mammoli F., Illner D., Markaki Y., Ferrari S., Cremer M., and Cremer T.. 2015. Remodeling of nuclear landscapes during human myelopoietic cell differentiation maintains co-aligned active and inactive nuclear compartments. Epigenetics Chromatin. 8:47 10.1186/s13072-015-0038-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami K., Egelhofer T.A., Strome S., and Lieb J.D.. 2010. Caenorhabditis elegans chromosome arms are anchored to the nuclear membrane via discontinuous association with LEM-2. Genome Biol. 11:R120 10.1186/gb-2010-11-12-r120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R.S., Martinez-Campos M., Zipperlen P., Fraser A.G., and Ahringer J.. 2001. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2:RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katic I., Xu L., and Ciosk R.. 2015. CRISPR/Cas9 Genome Editing in Caenorhabditis elegans: Evaluation of Templates for Homology-Mediated Repair and Knock-Ins by Homology-Independent DNA Repair. G3 (Bethesda). 5:1649–1656. 10.1534/g3.115.019273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H., Hayashi-Takanaka Y., Goto Y., Takizawa N., and Nozaki N.. 2008. The organization of histone H3 modifications as revealed by a panel of specific monoclonal antibodies. Cell Struct. Funct. 33:61–73. 10.1247/csf.07035 [DOI] [PubMed] [Google Scholar]
- Koch C.M., Honemann-Capito M., Egger-Adam D., and Wodarz A.. 2009. Windei, the Drosophila homolog of mAM/MCAF1, is an essential cofactor of the H3K9 methyl transferase dSETDB1/Eggless in germ line development. PLoS Genet. 5:e1000644 10.1371/journal.pgen.1000644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koester-Eiserfunke N., and Fischle W.. 2011. H3K9me2/3 binding of the MBT domain protein LIN-61 is essential for Caenorhabditis elegans vulva development. PLoS Genet. 7:e1002017 10.1371/journal.pgen.1002017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroschwald S., Maharana S., and Simon A. Hexanediol: a chemical probe to investigate the material properties of membrane-less compartments. Matters. 2017. [DOI] [Google Scholar]
- Larson A.G., Elnatan D., Keenen M.M., Trnka M.J., Johnston J.B., Burlingame A.L., Agard D.A., Redding S., and Narlikar G.J.. 2017. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature. 547:236–240. 10.1038/nature22822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson K., Yan S.J., Tsurumi A., Liu J., Zhou J., Gaur K., Guo D., Eickbush T.H., and Li W.X.. 2012. Heterochromatin formation promotes longevity and represses ribosomal RNA synthesis. PLoS Genet. 8:e1002473 10.1371/journal.pgen.1002473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaître C., and Bickmore W.A.. 2015. Chromatin at the nuclear periphery and the regulation of genome functions. Histochem. Cell Biol. 144:111–122. 10.1007/s00418-015-1346-y [DOI] [PubMed] [Google Scholar]
- Loyola A., Tagami H., Bonaldi T., Roche D., Quivy J.P., Imhof A., Nakatani Y., Dent S.Y., and Almouzni G.. 2009. The HP1alpha-CAF1-SetDB1-containing complex provides H3K9me1 for Suv39-mediated K9me3 in pericentric heterochromatin. EMBO Rep. 10:769–775. 10.1038/embor.2009.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan T.S., Gifford W.D., Driscoll S., Lettieri K., Rowe H.M., Bonanomi D., Firth A., Singer O., Trono D., and Pfaff S.L.. 2012. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 487:57–63. 10.1038/nature11244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens J.H., O’Sullivan R.J., Braunschweig U., Opravil S., Radolf M., Steinlein P., and Jenuwein T.. 2005. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 24:800–812. 10.1038/sj.emboj.7600545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurchy A.N., Stempor P., Gaarenstroom T., Wysolmerski B., Dong Y., Aussianikava D., Appert A., Huang N., Kolasinska-Zwierz P., Sapetschnig A., et al. 2017. A team of heterochromatin factors collaborates with small RNA pathways to combat repetitive elements and germline stress. eLife. 6:e21666 10.7554/eLife.21666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister P., Gehlen L.R., Varela E., Kalck V., and Gasser S.M.. 2010a Visualizing yeast chromosomes and nuclear architecture. Methods Enzymol. 470:535–567. 10.1016/S0076-6879(10)70021-5 [DOI] [PubMed] [Google Scholar]
- Meister P., Towbin B.D., Pike B.L., Ponti A., and Gasser S.M.. 2010b The spatial dynamics of tissue-specific promoters during C. elegans development. Genes Dev. 24:766–782. 10.1101/gad.559610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittag T., and Parker R.. 2018. Multiple Modes of Protein-Protein Interactions Promote RNP Granule Assembly. J. Mol. Biol. 430:4636–4649. 10.1016/j.jmb.2018.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Martínez A., Dobrzynska A., and Askjaer P.. 2015. Inner nuclear membrane protein LEM-2 is required for correct nuclear separation and morphology in C. elegans. J. Cell Sci. 128:1090–1096. 10.1242/jcs.164202 [DOI] [PubMed] [Google Scholar]
- Mutlu B., Chen H.M., Moresco J.J., Orelo B.D., Yang B., Gaspar J.M., Keppler-Ross S., Yates J.R., Hall D.H., Maine E.M., and Mango S.E.. 2018. Regulated nuclear accumulation of a histone methyltransferase times the onset of heterochromatin formation in C. elegans embryos. Sci. Adv. 4:eaat6224 10.1126/sciadv.aat6224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicetto D., Donahue G., Jain T., Peng T., Sidoli S., Sheng L., Montavon T., Becker J.S., Grindheim J.M., Blahnik K., et al. 2019. H3K9me3-heterochromatin loss at protein-coding genes enables developmental lineage specification. Science. 363:294–297. 10.1126/science.aau0583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostapcuk V., Mohn F., Carl S.H., Basters A., Hess D., Iesmantavicius V., Lampersberger L., Flemr M., Pandey A., Thomä N.H., et al. 2018. Activity-dependent neuroprotective protein recruits HP1 and CHD4 to control lineage-specifying genes. Nature. 557:739–743. 10.1038/s41586-018-0153-8 [DOI] [PubMed] [Google Scholar]
- Padeken J., and Heun P.. 2014. Nucleolus and nuclear periphery: velcro for heterochromatin. Curr. Opin. Cell Biol. 28:54–60. 10.1016/j.ceb.2014.03.001 [DOI] [PubMed] [Google Scholar]
- Padeken J., Zeller P., Towbin B., Katic I., Kalck V., Methot S., and Gasser S.M.. 2019. Synergistic lethality between BRCA1 and H3K9me2 loss reflects satellite derepression. Genes Dev. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A., Folkmann A., Rasoloson D., and Seydoux G.. 2015. High Efficiency, Homology-Directed Genome Editing in Caenorhabditis elegans Using CRISPR-Cas9 Ribonucleoprotein Complexes. Genetics. 201:47–54. 10.1534/genetics.115.179382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin G., Dong Y., Fraser A.G., Hopper N.A., and Ahringer J.. 2005. Chromatin regulation and sumoylation in the inhibition of Ras-induced vulval development in Caenorhabditis elegans. EMBO J. 24:2613–2623. 10.1038/sj.emboj.7600726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland T., Taşan M., Charloteaux B., Pevzner S.J., Zhong Q., Sahni N., Yi S., Lemmens I., Fontanillo C., Mosca R., et al. 2014. A proteome-scale map of the human interactome network. Cell. 159:1212–1226. 10.1016/j.cell.2014.10.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabari B.R., Dall’Agnese A., Boija A., Klein I.A., Coffey E.L., Shrinivas K., Abraham B.J., Hannett N.M., Zamudio A.V., Manteiga J.C., et al. 2018. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 361:eaar3958 10.1126/science.aar3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M.L., and Jorgensen E.M.. 2016. SapTrap, a Toolkit for High-Throughput CRISPR/Cas9 Gene Modification in Caenorhabditis elegans. Genetics. 202:1277–1288. 10.1534/genetics.115.184275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidler C., Kovalchuk O., and Kovalchuk I.. 2017. Epigenetic Regulation of Cellular Senescence and Aging. Front. Genet. 8:138 10.3389/fgene.2017.00138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovei I., Wang A.S., Thanisch K., Schmidt C.S., Krebs S., Zwerger M., Cohen T.V., Devys D., Foisner R., Peichl L., et al. 2013. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell. 152:584–598. 10.1016/j.cell.2013.01.009 [DOI] [PubMed] [Google Scholar]
- Strom A.R., Emelyanov A.V., Mir M., Fyodorov D.V., Darzacq X., and Karpen G.H.. 2017. Phase separation drives heterochromatin domain formation. Nature. 547:241–245. 10.1038/nature22989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda P., Stein P., Anger M., Bernstein E., Hannon G.J., and Schultz R.M.. 2004. RNAi and expression of retrotransposons MuERV-L and IAP in preimplantation mouse embryos. Dev. Biol. 269:276–285. 10.1016/j.ydbio.2004.01.028 [DOI] [PubMed] [Google Scholar]
- Tian Y., Garcia G., Bian Q., Steffen K.K., Joe L., Wolff S., Meyer B.J., and Dillin A.. 2016. Mitochondrial Stress Induces Chromatin Reorganization to Promote Longevity and UPR(mt). Cell. 165:1197–1208. 10.1016/j.cell.2016.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L., Court D.L., and Fire A.. 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 263:103–112. 10.1016/S0378-1119(00)00579-5 [DOI] [PubMed] [Google Scholar]
- Timms R.T., Tchasovnikarova I.A., Antrobus R., Dougan G., and Lehner P.J.. 2016. ATF7IP-Mediated Stabilization of the Histone Methyltransferase SETDB1 Is Essential for Heterochromatin Formation by the HUSH Complex. Cell Reports. 17:653–659. 10.1016/j.celrep.2016.09.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinevez J.Y., Perry N., Schindelin J., Hoopes G.M., Reynolds G.D., Laplantine E., Bednarek S.Y., Shorte S.L., and Eliceiri K.W.. 2017. TrackMate: An open and extensible platform for single-particle tracking. Methods. 115:80–90. 10.1016/j.ymeth.2016.09.016 [DOI] [PubMed] [Google Scholar]
- Tischler J., Lehner B., Chen N., and Fraser A.G.. 2006. Combinatorial RNA interference in Caenorhabditis elegans reveals that redundancy between gene duplicates can be maintained for more than 80 million years of evolution. Genome Biol. 7:R69 10.1186/gb-2006-7-8-r69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin B.D., Meister P., Pike B.L., and Gasser S.M.. 2010. Repetitive transgenes in C. elegans accumulate heterochromatic marks and are sequestered at the nuclear envelope in a copy-number- and lamin-dependent manner. Cold Spring Harb. Symp. Quant. Biol. 75:555–565. 10.1101/sqb.2010.75.041 [DOI] [PubMed] [Google Scholar]
- Towbin B.D., González-Aguilera C., Sack R., Gaidatzis D., Kalck V., Meister P., Askjaer P., and Gasser S.M.. 2012. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell. 150:934–947. 10.1016/j.cell.2012.06.051 [DOI] [PubMed] [Google Scholar]
- Tsurumi A., and Li W.X.. 2012. Global heterochromatin loss: a unifying theory of aging? Epigenetics. 7:680–688. 10.4161/epi.20540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsusaka T., Kikuchi M., Shimazu T., Suzuki T., Sohtome Y., Akakabe M., Sodeoka M., Dohmae N., Umehara T., and Shinkai Y.. 2018. Tri-methylation of ATF7IP by G9a/GLP recruits the chromodomain protein MPP8. Epigenetics Chromatin. 11:56 10.1186/s13072-018-0231-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher V.G., Tibshirani R., and Chu G.. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA. 98:5116–5121. 10.1073/pnas.091062498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., An W., Cao R., Xia L., Erdjument-Bromage H., Chatton B., Tempst P., Roeder R.G., and Zhang Y.. 2003. mAM facilitates conversion by ESET of dimethyl to trimethyl lysine 9 of histone H3 to cause transcriptional repression. Mol. Cell. 12:475–487. 10.1016/j.molcel.2003.08.007 [DOI] [PubMed] [Google Scholar]
- Yang B.X., El Farran C.A., Guo H.C., Yu T., Fang H.T., Wang H.F., Schlesinger S., Seah Y.F., Goh G.Y., Neo S.P., et al. 2015. Systematic identification of factors for provirus silencing in embryonic stem cells. Cell. 163:230–245. 10.1016/j.cell.2015.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Gu J., Jin Y., Luo Y., Preall J.B., Ma J., Czech B., and Hannon G.J.. 2015. Panoramix enforces piRNA-dependent cotranscriptional silencing. Science. 350:339–342. 10.1126/science.aab0700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller P., Padeken J., van Schendel R., Kalck V., Tijsterman M., and Gasser S.M.. 2016. Histone H3K9 methylation is dispensable for Caenorhabditis elegans development but suppresses RNA:DNA hybrid-associated repeat instability. Nat. Genet. 48:1385–1395. 10.1038/ng.3672 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.