Abstract
Dynamic functional connectivity (DFC) aims to maximize resolvable information from functional brain scans by considering temporal changes in network structure. Recent work has demonstrated that static, i.e. time-invariant resting-state and task-based FC predicts individual differences in behavior, including attention. Here, we show that DFC predicts attention performance across individuals. Sliding-window FC matrices were generated from fMRI data collected during rest and attention task performance by calculating Pearson’s r between every pair of nodes of a whole-brain atlas within overlapping 10–60s time segments. Next, variance in r values across windows was taken to quantify temporal variability in the strength of each connection, resulting in a DFC connectome for each individual. In a leave-one-subject-out-cross-validation approach, partial-least-square-regression (PLSR) models were then trained to predict attention task performance from DFC matrices. Predicted and observed attention scores were significantly correlated, indicating successful out-of-sample predictions across rest and task conditions. Combining DFC and static FC features numerically improves predictions over either model alone, but the improvement was not statistically significant. Moreover, dynamic and combined models generalized to two independent data sets (participants performing the Attention Network Task and the stop-signal task). Edges with significant PLSR coefficients concentrated in visual, motor, and executive-control brain networks; moreover, most of these coefficients were negative. Thus, better attention may rely on more stable, i.e. less variable, information flow between brain regions.
Keywords: dynamic functional connectivity, predictive modeling, sustained attention, individual differences, partial least squares regression
1. Introduction
Functional connectivity (FC) probes the network organization of the human brain by quantifying temporal dependencies between signals from voxels, regions, or networks (Fox and Raichle, 2007). FC studies have traditionally focused on revealing group-wise differences in disordered and healthy populations (Fox and Raichle, 2007) and decoding the functional magnetic resonance imaging (fMRI) signal correlates of cognitive states (Shirer et al., 2012), for which inter-individual variability was mostly considered as a source of noise. However, recent work has demonstrated that FC patterns are unique to individuals and relatively stable across cognitive states including task performance and rest, akin to neural “fingerprints” (Finn et al., 2015; Cole et al., 2014; Noble et al., 2017; Gratton et al., 2018). Furthermore, FC patterns predict individual traits, aptitudes, clinical diagnoses, and treatment outcomes (Arbabshirani et al., 2017; Rosenberg et al., 2016a, b), leading to the emergence of a new personalized neuroscience geared toward behavioral prediction (Rosenberg et al., 2016; Shen et al., 2017; Kessler et al, 2016; Poole et al., 2016; O’Halloran et al., 2018; Galeano-Weber et al., 2017; Reinen et al., 2018; Woo et al., 2017; Gabrieli et al., 2015).
In tandem, a promising subfield known as dynamic FC (DFC) has gained momentum. Traditionally, FC is considered in a “static” sense in that all time points in a fMRI scan are taken to produce a single measure of average FC. Despite its success in characterizing behaviorally relevant individual variation, static FC (SFC) loses information at a temporal scale, since functional brain networks exhibit spontaneous dynamic fluctuations over scan time (Chang and Glover, 2010; Hutchison, 2013). Recently, Matsui and colleagues (2016) have corroborated a neurophysiological underpinning of observed FC nonstationarity by aligning hemodynamic- and calcium-based neuronal signals in mice and ascribing temporal FC fluctuations to transient co-activation patterns. A growing body of studies now considers fluctuations in FC across time statistically valid and physiologically informative.
The most well-adopted method to measure DFC is to slide a temporal window across time points in the scan, and compute a correlation matrix within each resultant window (Hutchison et al., 2013; Allen et al., 2013; Rashid et al., 2016; see Preti et al., 2016 for a review of DFC methods). This outputs a three-dimensional stack of windowed FC matrices, which is then reduced into summary statistics using one of two approaches. The state-based approach tracks recurring spatial FC configurations over time. To this end, windowed FC series are first concatenated across subjects, and then clustered with an algorithm such as k-means to identify canonical centroid “states”. This allows higher-order summary statistics to be taken with respect to the states as fundamental units, such as dwell time in the identified states (Jones et al., 2012; Li et al., 2017), or a metastate vector representing the various states’ contributions to each windowed segment (Rashid et al., 2016). The edge-based approach, in contrast, takes temporal features for each functional connection or “edge” as the fundamental unit. An operation such as standard deviation (Kucyi et al., 2013; Laufs et al., 2014) or coefficient of variation (Gonzalez-Castillo et al., 2014) is directly applied along the windowed FC stack to estimate the dynamic properties of each edge, yielding a DFC matrix of the same dimension as an SFC matrix.
DFC measured with sliding window correlations and other approaches (Chang and Glover, 2010; Yaesoubi et al., 2015a; Xu and Lindquist, 2015; Lindquist et al., 2014) has been promising in discovering variation in brain function across groups and individuals (see Preti et al., 2016 for a review of DFC applications). FC variability and state occupancies have been used as a demographic marker, showing significant differences across age (Hutchison and Morton, 2015) and gender (Yaesoubi et al., 2015a). Studies of consciousness have related FC variability between specific regions to frequency of daydreaming (Kucyi and Davis, 2014), mental flexibility (Yang et al., 2014) and attentional task performance (Madhyastha et al., 2015). Furthermore, cross-validated clinical neuromarkers have been derived from DFC: most prominently, individuals with schizophrenia were found to spend more time at rest in less lower-connectivity states (Du et al., 2016; Damaraju et al., 2014); more generally, Rashid et al., 2016 demonstrated improved SVM classification of control, schizophrenic, and bipolar disorder individuals by using DFC than SFC features. DFC has further yielded clinical applications in autism (Price et al., 2014), mild cognitive impairment (Wee et al., 2016a, 2013), and post-traumatic stress disorder (Li et al., 2014).
Despite these results, the robustness of the sliding-window correlations method has been called into question, especially in resting-state studies. Hindriks and colleagues (2016) showed that sliding-window correlations cannot capture statistically significant DFC in the resting-state. Laumann and colleagues (2016) further demonstrated that most of the measured resting-state FC variability is attributable to sampling variability, being indistinguishable from simulated data with stationary FC by design, and to sources of non-stationary physiological noise including head motion. Thus, there is a call for more rigorous statistical testing for DFC, more comprehensive correction of head motion, richer behavioural validation of DFC, and corroboration of resting-state DFC discoveries through task-based fMRI.
The shift in FC literature toward individual predictive modeling, in conjunction with the development of DFC knowledge, offers a unique opportunity to explore the utility of DFC, particularly standard sliding window-based approaches, in predicting individual behaviour. We applied a recently developed cross-validated machine-learning pipeline, connectome-based predictive modeling (“CPM”), to predict individual behavior scores from brain activity (Finn et al., 2015; Rosenberg et al., 2016; Shen et al., 2017). In CPM, a brain-behavior regression model is trained on a database of individuals scanned under task-based or resting-state fMRI, and fitted to brain data of novel individuals to predict a behavior score. Using whole-brain SFC as model input, CPM accurately predicted individual differences in sustained attention (Rosenberg et al., 2016), neuroticism and extraversion (Hsu et al., 2018), and cognitive impairment related to Alzheimer’s Disease (Lin et al., 2018), reliably generalizing across independent datasets. Here, we demonstrate that DFC during task performance and rest successfully predicts individual differences in sustained attention. Using sliding-window correlations, we quantify the dynamics of functional connections, or edges. These DFC statistics, obtained under either task performance or resting state, are used as CPM features to successfully predict individual differences in performance across three attention tasks. Moreover, incorporating DFC features consistently improves predictions upon SFC alone, and this result generalizes across independent datasets.
To maintain attention toward a specific task requires individuals to resist mind-wandering and external distraction, which could be mediated by FC fluctuations (Hutchison 2013b). In light of this, one might hypothesize that sustained attention benefits from reduced temporal variability in functional networks underlying executive processing, working memory, perception, internal cognition and other task demands. Indeed, analyzing the spatial distribution of the significantly predictive networks, we find that improved sustained attention is predicted by reduced temporal variability of edge strength in the majority of edges, with notable exceptions in the frontoparietal network. Overall, better attention may rely on more stable (less variable) information flow between regions processing ongoing tasks, and DFC may complement SFC in characterizing individual differences in attention.
2. Materials and Methods
2.1. Data acquisition
1. Internal validation dataset: gradual-onset continuous performance task (gradCPT)
Participants:
The internal validation sample described here was originally described in previous work (Rosenberg et al., 2016). In brief, thirty-one adults from Yale University and the New Haven community were recruited. Six were excluded due to excessive head motion, and data from only the remaining twenty-five were used for subsequent analyses.
Task paradigm and stimuli:
The gradual-onset continuous performance task (gradCPT; Rosenberg et al, 2013; Esterman et al., 2013) for sustained attention was conducted. Stimuli were grayscale pictures of city or mountain scenes. Each picture gradually transitioned to the next over an 800-ms duration. Participants were instructed to press a button to city scenes (90% of the stimuli) and withhold response to mountain scenes.
Task-based and resting-state scan procedure:
Following an MPRAGE anatomical run, two 6-min resting-state runs and three 13.44-min task runs were conducted. Each task run consisted of an 8-s fixation period followed by four 3-min gradCPT task blocks (thus 225 stimuli per block), interleaved with 32-s break periods. Fixation and break periods are excluded in subsequent analyses. Block-wise task performance was quantified with sensitivity (d’), i.e. the inverse of the standard normal cumulative distribution function of the false alarm rate subtracted from that of the hit rate. An individual’s overall sustained attention performance was quantified as the mean d’ over all usable task blocks.
Image parameters and preprocessing:
fMRI data were acquired at the Yale Magnetic Resonance Research Center (3T Siemens Trio TIM system equipped with a 32 channel head coil) using multiband echo-planar imaging (EPI) with the following parameters: repetition time (TR) = 1,000 ms, echo time (TE) = 30 ms, flip angle = 62°, acquisition matrix = 84 × 84, in-plane resolution = 2.5 mm2, 51 axial-oblique slices parallel to the anterior commissure–posterior commissure (AC–PC) line, slice thickness = 2.5 mm, multiband 3, acceleration factor = 2. MPRAGE parameters were as follows: TR = 2530 ms, TE = 2.77 ms, flip angle = 7°, acquisition matrix = 256 × 256, in-plane resolution = 1.0 mm2, slice thickness = 1.0 mm, 176 sagittal slices. Slice timing correction was not applied due to the use of multiband. The 2D T1-weighted structural image was also acquired with the same slice prescription as the EPI images. fMRI data were preprocessed using BioImage Suite and custom MATLAB (Mathworks) scripts, and motion-corrected with SPM8 (http://www.fil.ion.ucl.ac.uk/spm/software/spm8/). The data processing steps steps were: (1) conversion of dicom to nifti files; (2) motion correction of functional images with SPM; (3) skull-stripping of high-resolution anatomical and coplanar images; (4) registration of the functional atlas from MNI space into single-subject space via a concatenation of a linear and nonlinear registration between the functional images, high-resolution anatomical scan, coplanar scan, and MNI brain; (5) regression of nuisance parameters including linear and quadratic drift, cerebrospinal fluid signal, white matter signal, global signal, and a 24-parameter motion model (6 motion parameters, 6 temporal derivatives, and their squares); and (6) temporal smoothing with a zero mean unit variance Gaussian filter (cutoff frequency = .12Hz).
Functional runs with excessive head translation (> 2 mm) translation or rotation (> 3°) were excluded from analysis; these included a resting run from two participants and a task run from 5 participants.
2. External validation dataset 1: Attention Network Task (ANT)
Participants:
Fifty-six adults from Yale University and the New Haven community were recruited, of which eight were excluded for excessive head motion, and a further four were excluded due to missing task runs, leaving forty-four subjects with usable data. This sample is described in detail in previous work (Rosenberg et al., 2018).
Task paradigm and stimuli:
ANT administration replicated Fan et al. (2005) except for increased inter-trial intervals (ITIs) (Fan et al., 2005). ANT trials began with a 200-ms cue period. Each trial had equal probabilities of being center-cue, spatial-cue, or no-cue. On central-cue trials, an asterisk appeared in the center of the screen to alert subjects to an upcoming target; on spatial-cue trials, the asterisk appeared above or below the center fixation cross to indicate where on the screen the upcoming target would appear; no cue appeared on no-cue trials. After a variable period of time (0.3–11.8 seconds; mean = 2.8 seconds), five horizontal arrows appeared above or below the central fixation cross. Participants were instructed to press a button indicating whether the center arrow pointed left or right. On congruent trials (50%), the central target arrow pointed in the same direction as the flanking distractor arrows; on incongruent trials (50%), the target pointed in the opposite direction. After a button press (or after two seconds if no response was detected), the arrows disappeared and a variable ITI (5–17 seconds; mean = 8 seconds) began. Each of the six ANT runs consisted of two buffer trials followed by 36 task trials. Each run consisted of six examples of each of the six trial types (combinations of central-cue/spatial-cue/no-cue and congruent/incongruent). Trial order was counterbalanced within and across runs. ANT performance was measured with the variability of correct- trial RTs (Rosenberg et al., 2018), multiplied by −1, so that a higher score indicated better performance.
Image parameters and preprocessing:
MRI data were collected at the Yale Magnetic Resonance Research Center on the same system as the gradCPT dataset. Experimental sessions began with a high-resolution anatomical scan, followed by two 6-min resting scans and six 7:05-minute ANT runs. Participants fixated on a center cross during rest runs, and were asked to respond quickly and accurately on task runs. Rest runs included 360 whole-brain volumes and task runs included 425 volumes. MRI scan parameters and preprocessing procedures were the same as those in the gradCPT dataset. Runs with excessive head translation (>2mm), rotation (>3°), or mean framewise displacement “FD” (>0.15mm) were excluded from analysis. As a result, one task run was excluded from three participants, and two task runs were excluded from one participant. All participants had two rest runs with acceptable levels of motion.
Note that in the gradCPT sample, FD was not used as an explicit exclusion criterion because FD in all included runs was <.11 mm. Framewise displacement was added as an exclusion criteria for the Attention Network Task (ANT) and stop-signal task (SST; see below) datasets as an additional conservative control.
3. External validation dataset 2: stop-signal task (SST)
Participants:
We used a dataset of healthy adults originally collected by Farr and colleagues (Farr et al., 2014a, b) and analyzed in our previous work (Rosenberg et al., 2016b). These data included 24 healthy adults given a single 45 mg dose of methylphenidate approximately 40 min before an fMRI scan session and 92 healthy adults scanned as part of a control cohort. Detailed participant information can be found in previous studies (Farr et al., 2014a, b). Data exclusion was performed as described in Rosenberg et al., 2016b.
Task paradigm and stimuli:
Subjects performed a stop-signal task in the scanner (Farr et al., 2014a). In each trial, a dot first appeared in the center of the screen, and after a random interval of one to five seconds, the dot changed into a circle, representing a “go” signal, upon which participants were instructed to press a button as quickly as possible. Once the button was pressed, or after one second if no response was detected, the circle disappeared. A 2-second interval preceded the next trial. ~25% of trials were stop trials, in which an “X”, the “stop” signal, replaced the go signal after it appeared. Participants were instructed to withhold response whenever they saw a stop signal. The stop-signal delay (time between the go and stop-signal onsets) was stair-cased within-subject so that each participant was able to withhold response on approximately half of stop trials. As described previously (Rosenberg et al., 2016b), task performance was assessed with go rate, the percent of trials a subject correctly responded to a go signal.
Image parameters and preprocessing:
MRI data were collected at the Yale Magnetic Resonance Research Center as in the above datasets. Images were acquired using an EPI sequence with the following parameters: TR = 2,000 ms, TE = 25 ms, flip angle = 85°, field of view (FOV) = 220 × 220 mm, acquisition matrix = 64 × 64, 32 axial slices parallel to the AC–PC line; slice thickness = 4 mm (no gap). A high-resolution T1-weighted structural gradient echo scan was acquired with the following parameters: TR = 2530 ms, TE = 3.66 ms, flip angle = 7°, FOV = 256 × 256 mm, acquisition matrix = 256 × 256, 176 slices parallel to the AC–PC line, slice thickness = 1 mm.
fMRI data were preprocessed as described in the gradCPT section above. fMRI runs with excessive head motion, defined a priori as > 2 mm translation, > 3° rotation, or > 0.15 mm mean FD were excluded from analysis. Next, following visual inspection of pitch, roll, and yaw head motion time courses courses, four resting-state runs with obvious movements near the start or end were cropped to remove 49–105 volumes associated with excessive motion. Finally, to eliminate differences in mean frame-to-frame displacement between the methylphenidate and control groups, participants in the methylphenidate group with the highest mean frame-to-frame displacement were excluded one by one until groups were matched on all motion measures. The final rest cohort included 16 individuals administered methylphenidate and 56 controls, and the final task cohort included 19 individuals administered methylphenidate and 64 controls (Rosenberg et al., 2016b).
2.2. Static and dynamic FC connectome construction (“Step 1”, Figure 1)
Figure 1: Method for predicting individual behavior using dynamic functional connectivity.
Step 1: Take the variance of sliding-window FC matrices to build individual DFC (and SFC) matrices. Step 2: Using static/dynamic/combined FC matrices as input, train PLSR model and test on novel samples. Internal validation: Train on n-1 gradCPT subjects, test on left-out gradCPT subject. External validation: Train on all gradCPT subjects, test on SST and ANT subjects.
First, functional network nodes were defined using the Shen 268-node functional brain atlas, which covers the cortex, subcortex and cerebellum (Shen et al., 2013). This atlas was transformed from MNI space to individual space with transformations estimated using intensity-based registration algorithms in BioImage Suite. Each subject’s preprocessed voxel-wise time series were then averaged to produce 268 nodal time series, normalized to zero mean and unit variance.
Next, for each individual in each data set, we constructed static functional connectivity matrices by computing the Pearson’s correlation coefficient (r) between every possible pair of nodal time series, concantenated across rest runs or task blocks. The resulting values were Fisher z-transformed, leading to one resting-state and one task-based 268-by-268 SFC matrix per subject (Rosenberg et al., 2016).
After generating SFC matrices, we generated DFC matrices for each individual with the use of a tapered sliding window. Compared to a rectangular window, tapering reduces the weight of boundary time points in calculating windowed-FC matrices, increasing the sensitivity to outliers in the detection of DFC, leading to a smoothening of the DFC timecourse. This ensures that the inclusion or exclusion of instantaneous noisy observations do not appear as a sudden change in the DFC timecourse (Preti et al., 2016; Allen et al., 2014). Here, six Tukey-shaped windows with r = 0.5 (i.e. the first and last 1/4 of the entire window length consist of segments of a phase-shifted cosine with period 2r = 1), and respective widths of w=10, 20, 30, 40, 50, 60 TRs were created and slid down each rest run or task block in steps of 1TR. Within each windowed segment, pairwise Fisher’s z-transformed r values were again taken to generate a FC matrix. This led to a DFC timecourse for each edge, or a 268-by-268-by-#{windows} array, within each rest run or task block. The DFC timecourses were concatenated across the 2 rest runs or 12 task blocks. Finally, variance (σ2) was calculated along the concatenated DFC timecourses to quantify the temporal variability of edge strength, leading to a rest-based and a task-based 268-by-268 DFC matrix per subject corresponding to each of the six window size values.
2.3. Predictive model: training, optimization, and cross-validation (“Step 2”, Figure 1)
Partial least squares regression (PLSR) highlights relationships between variables by representing them in latent components. Our previous work (Yoo et al., 2018) justified PLSR as the method of choice in building models to predict attention scores from static FC patterns. In this work, we expanded upon Yoo and colleagues by allowing the number of PLS components to be a tunable parameter in our predictive algorithm. To that end, we specifically used the kernel PLSR algorithm, which was developed to construct nonlinear regression models in high-dimensional feature spaces (Rosipal and Trejo, 2001). Kernel PLSR is preferred over its counterpart, the SIMPLS algorithm, because it optimally represents significant variance in the feature space with a lower number of components, while retaining the same results for single-component predictions. DFC was the new feature space in our models, and results were compared to previous SFC benchmarks. Combined models were then created to determine whether DFC supplements information from SFC to yield improved predictions. Predictive models were trained and tested on both resting-state and task-based fMRI data in order to show maximal generalizability across cognitive states.
Static and dynamic models were first constructed within the gradCPT “internal validation” sample, under a leave-one-out-cross-validation (LOOCV) framework. In each iteration of LOOCV, the 25 subjects were split into a training set (24) and a testing set (1); then, a PLSR model was trained to predict sensitivity (d’) from SFC and DFC matrices in the training set, and applied on the left-out subject’s corresponding matrix to predict their d’. Spearman’s rank correlation coefficient (ρ) was taken between predicted and observed d’ scores across iterations to quantify model performance.
Two parameters were allowed to vary across the LOOCV iterations: in the dynamic model, w (window size) could take on values from {10,20,30,40,50,60TR}; in all models, n (number of PLS components to retain) could range from 1 to the number of subjects. To optimize each out-of-sample prediction, the values of these parameters were tuned in a nested LOOCV procedure: an inner LOOCV loop was run to determine the value of w and n that maximized ρ in that particular training set.
Combined models were then constructed by concatenating each individual’s SFC matrix with their DFC matrix corresponding to the optimal w value and re-applying PLSR; once again, n was optimized in a nested LOOCV loop, and ρ was taken between predicted and observed d’.
Finally, static, dynamic, and combined predictive models trained on the internal datasets were applied to the ANT and SST “external validation” datasets. Since each round of LOOCV results in a different PLSR model, median values of w and n among the 25 iterations were used to define an overall PLSR model, which was applied, completely unchanged, to predict d’ in ANT and SST subjects. ρ was then taken between predicted d’ scores and observed attention scores (- RT variability and go rate, respectively) to quantify model performance.
2.4. Similarity of individual static and dynamic matrices
If a FC measure shows no replicability within an individual across scans, cognitive states, and time, it cannot provide robust information inherent to the individual. Thus, an FC measure must first exhibit reliability within individuals before it can have utility for behavioral prediction. To characterize the intra-subject reliability of static and dynamic matrices, similarity 1) between each individual’s static/dynamic task-based/resting-state matrices across the conducted scan runs, 2) between each individual’s SFC matrix and DFC matrix (from 20TR windows, which was determined as the optimal window length for prediction, see Results) under task-based and resting-state scans, and 3) between each individual’s task-based matrix and resting-state matrix for both static and dynamic FC, were quantified with Pearson’s r.
2.5. Feature-level statistical head motion correction
Head motion is a nonstationary source of non-physiological noise. This can be especially detrimental to sliding-window fMRI analyses, since windows across time may be affected differently by noise artifacts, contaminating the observed DFC properties (Laumann, 2016). To demonstrate that model performance was not fully attributable to motion confounds, we included a further feature-level statistical head motion correction, in addition to the head motion correction carried out in preprocessing steps. We linearly regressed static functional connectivity (SFC) and dynamic functional connectivity (DFC) matrix values on three motion parameters: maximum displacement, mean frame-to-frame displacement, and rotation, across subjects, observed during corresponding rest or task scans. Then, we ran predictive modelling using the residuals of SFC and DFC matrices in the internal validation sample, now controlled for motion.
2.6. Significance testing
To obtain the null distribution for each model’s ρ, a 103-iteration permutation test was performed by shuffling the d’ labels and re-applying the predictive modeling procedure described above. The one-tailed p-value associated with each ρ value (denote ρ*) was the proportion of permutations yielding a ρperm ≥ ρ*.
2.7. Neuroanatomical analysis of predictive edges
We first grouped the 268 nodes into eight intrinsic connectivity networks (e.g., default mode, frontoparietal), defined previously using the Shen atlas (Shen et al., 2013). This localizes each edge within an intersection of two networks (including a network’s intersection with itself). We next conducted two analyses to investigate the neuroanatomical distribution of features driving PLSR model predictions.
Firstly, we counted the number of significantly predictive edges within each network pair. A 105-iteration permutation test (separate from those described in the previous section) was conducted on 20TR-window task-based DFC connectomes in the internal validation sample, to identify the edges whose temporal variability had significant PLSR coefficients (denote ß*). The null distribution for each edge’s ß was obtained by shuffling the d’ labels in 105 permutations, and fitting a PLSR model to predict these shuffled d’ values from all 25 DFC connectomes in the dataset. For edges with positive ß*, p was calculated as the proportion of permutations with ßperm ≥ ß*; for edges with negative ß*, p was calculated as the proportion of permutations with ßperm ≤ ß* (hence, one-tailed p-values are reported). Edges with p < 0.05 were considered significant dynamic features. We then simply counted the number of significant edges within each network pair.
Secondly, edge coefficients in the PLSR model were summed within pairs of intrinsic connectivity networks, regardless of their significance values. This considers the effect of raw magnitudes of edge coefficients on model predictions.
3. Results
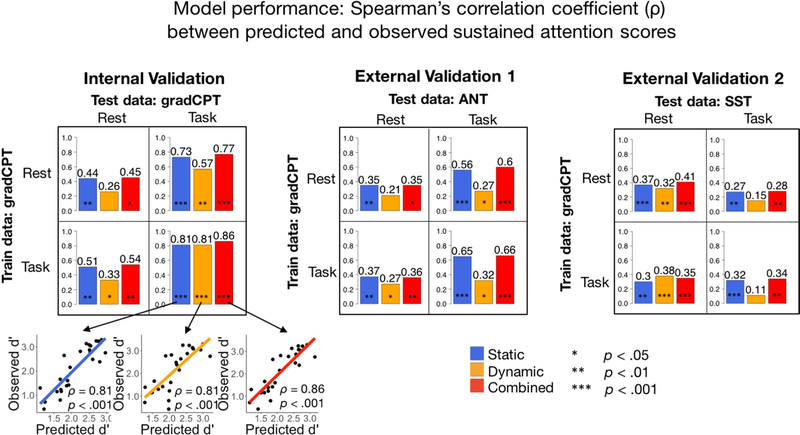
Performance of predictive models was assessed with Spearman’s correlation (ρ) between predicted d’ and observed d’ (internal validation set), or between predicted d’ and observed RT variability or Go-rate (external validation sets) (Figure 2). There was a total of 3 functional connectivity feature sets (static/dynamic/combined) × 3 independent datasets × 2 training conditions (rest/task) × 2 testing conditions (rest/task) = 36 PLSR models.
Figure 2: Prediction performance of static, dynamic and combined FC models across task across datasets.
Bar levels and values indicate Spearman’s ρ between predicted d’ and observed behavior score (d’ in gradCPT, response time variability in ANT, and go-rate in SST). Statistical significance is assessed with a 1000-iteration permutation test detailed in the Methods section. See Table 1 for more detailed performance and significance metrics.
3.1. Dynamic models predict attention across datasets
In the internal validation sample, PLSR models trained on resting-state DFC matrices significantly predicted left-out individuals’ d’ scores from task-based DFC matrices, whereas those trained on task-based DFC matrices significantly predicted left-out individuals’ d’ scores from both resting-state and task-based DFC matrices (p < .05; Figure 2, Table 1). Models trained on task matrices numerically outperformed models trained on rest matrices (tested on rest: ρ = .46 vs. 34; tested on task: ρ = .81 vs. .57), mirroring previous findings in SFC-based CPM (Rosenberg et al., 2016; Yoo et al., 2018).
Table 1: Amount of test-set variance (R2) explained by static, dynamic and combined FC PLSR models across datasets.
For internal validation, we took where y represents observed d’, predicted d’, and the mean of observed d’ across subjects. For external validation, R2 was quantified as the square of the Pearson correlation between predicted d’ and observed attention scores (inverted for ANT).
| Internal Validation: Gradual-onset Continuous Performance Task (gradCPT) | ||||
| Training data | Rest | Task | ||
| Testing data | Rest | Task | Rest | Task |
| Static model | 0.14 | 0.33 | 0.12 | 0.68 |
| Dynamic model | 0.20 | 0.29 | 0.19 | 0.57 |
| Combined model | 0.11 | 0.38 | 0.13 | 0.69 |
| External Validation 1: Attention Network Task (ANT) | ||||
| Training data | Rest | Task | ||
| Testing data | Rest | Task | Rest | Task |
| Static model | 0.11 | 0.35 | 0.12 | 0.41 |
| Dynamic model | 0.07 | 0.07 | 0.09 | 0.12 |
| Combined model | 0.12 | 0.37 | 0.13 | 0.39 |
| External Validation 2: Stop-signal Task (SST) | ||||
| Training data | Rest | Task | ||
| Testing data | Rest | Task | Rest | Task |
| Static model | 0.16 | 0.25 | 0.09 | 0.31 |
| Dynamic model | 0.12 | 0.00 | 0.17 | 0.00 |
| Combined model | 0.19 | 0.14 | 0.13 | 0.14 |
In the external validation samples, dynamic models were trained on either task or rest matrices using the full gradCPT sample, and applied, completely unchanged, to the independent SST and ANT samples. The gradCPT model trained on task-based dynamics generalized to predict SST performance from resting-state test data and ANT performance from both resting-state and task-based test data; the gradCPT model trained on resting-state dynamics generalized to predict SST performance from resting-state test data and ANT performance from task-based test data. Thus, task-based and resting-state models each showed generalizability to at least one unseen task-based and resting-state dataset. Static models generally outperformed dynamic models, with the exceptions of the model trained and tested on task-based gradCPT data (r = 0.81 for both static and dynamic), and the model trained on resting-state gradCPT and tested on task-based SST data (r = 0.30 for static; r = 0.38 dynamic). (Figure 2). Trends in the amount of test-set variance (R2) explained by static, dynamic and combined FC PLSR models across datasets closely mirrored those in Spearman’s ρ (Table 1).
We applied separate Bonferroni corrections for the 4 p-values (p < 0.0125) in the internal validation sample and for the 8 p-values (p < .006025) in the external validation samples corresponding to the dynamic models. Of the 12 tests, all except 2 (gradCPT rest-rest; ANT rest-rest) were originally significant (p < 0.05). Of the 10 significant tests, 6 survived Bonferroni correction (gradCPT rest-task, task-task; ANT task-rest; SST rest-task, task-rest, task-task). We note that Bonferroni is the most conservative way to correct for multiple comparisons; moreover, the tests are not independent, due to the overlap between the training data across tests. Hence, while this procedure is very stringent against our hypothesis, we still observe significant prediction power after applying it.
Related DFC work by Leonardi and colleagues (2014) have recommended applying high-pass filtering on fMRI timecourses to prevent spurious fluctuations slower than the window size from affecting estimated within-window correlations. Supplementary Table S1 shows the results of internal validation tests after applying a high-pass filter with cut-off frequency (eg. 0.05Hz for 20TR windows) to both rest and task fMRI timecourses before generating DFC and combined FC matrices. The models remained significantly predictive, suggesting that observed DFC is attributable to meaningful, non-spurious correlations.
Additionally, GSR is a controversial preprocessing step (Ciric et al., 2017). For a comprehensive control, we replicated the internal validation tests without GSR, with results enclosed in Supplementary Table S2. 2 of the 3 originally significant dynamic models remained significant, and combined models continued to outperform static models.
3.2. Combined models numerically, but not significantly, outperformed static functional connectivity models across datasets
In all internal and external validation analyses, combined models, which included both SFC and DFC features, significantly predicted d’ from task-based and resting-state data (Figure 2). Combined models performed numerically equivalent to or better than static models in all cases except when trained on task gradCPT and tested on rest ANT data. The difference between Spearman’s ρ from static and combined FC models did not reach significance (p < 0.05) under Steiger’s Z-test for testing the statistical significance of the difference between dependent correlations (Revelle, 2018).
We next carried out a control analysis, substituting DFC with BOLD variability features (for each of the 268 nodes) and then combining them with SFC models. We observe that combined SFC-BOLD variability models underperformed baseline SFC models in all cases, even where BOLD variability models were statistically significant (see Supplementary Table S3). Thus, while simple brain features may predict behavior on their own, they can become a source of noise and hurt model performance when added to pre-existing models with better features. The small but consistent numerical improvement from combining SFC and DFC models strongly suggests this is not the case with DFC. Furthermore, combined SFC-DFC models outperformed combined SFC-BOLD variability models, even where BOLD variability outperforms DFC (trained on task, tested on rest). This result was significant (p < 0.05) under Steiger’s Z-test for models trained on rest or task and tested on task, suggesting that DFC adds significantly more value than noise.
3.3. Optimal parameter choices for prediction performance
Window size values were allowed to vary between {10,20,30,40,50,60TR} across the gradCPT internal validation iterations. Within each iteration, a nested LOOCV procedure was run to select the window size that optimized Spearman’s ρ between predicted and observed d’. 20TR emerged as the optimal window size in 93% of the iterations. Thus, 20TR was overwhelmingly favored over other values in predicting attention. We furthermore conducted a post-hoc analysis on the effect of variable window sizes on prediction performance (Supplementary Figure S1) and on the distribution of values in the resultant DFC matrices (Supplementary Figure S2). Despite being suboptimal, results from longer windows (or at least 30TR windows) were comparable to those from 20TR windows.
In previous DFC work, 30–60s windows were reported to capture transient FC not found in larger window sizes (Hutchison et al., 2013). It should thus be emphasized that 20TR windows may fall into potentially spurious dynamics. Our study motivates future DFC work to assess the statistical rigor of 20TR windows in capturing transient neuronal, non-spurious dynamics, since they exhibit maximal utility in predicting behavior here.
The optimal number of PLSR components ranged from 1 to 6 across iterations (Supplementary Table S4). Generally, PLSR models trained on rest data were optimally represented in a lower number of components than those trained on task data.
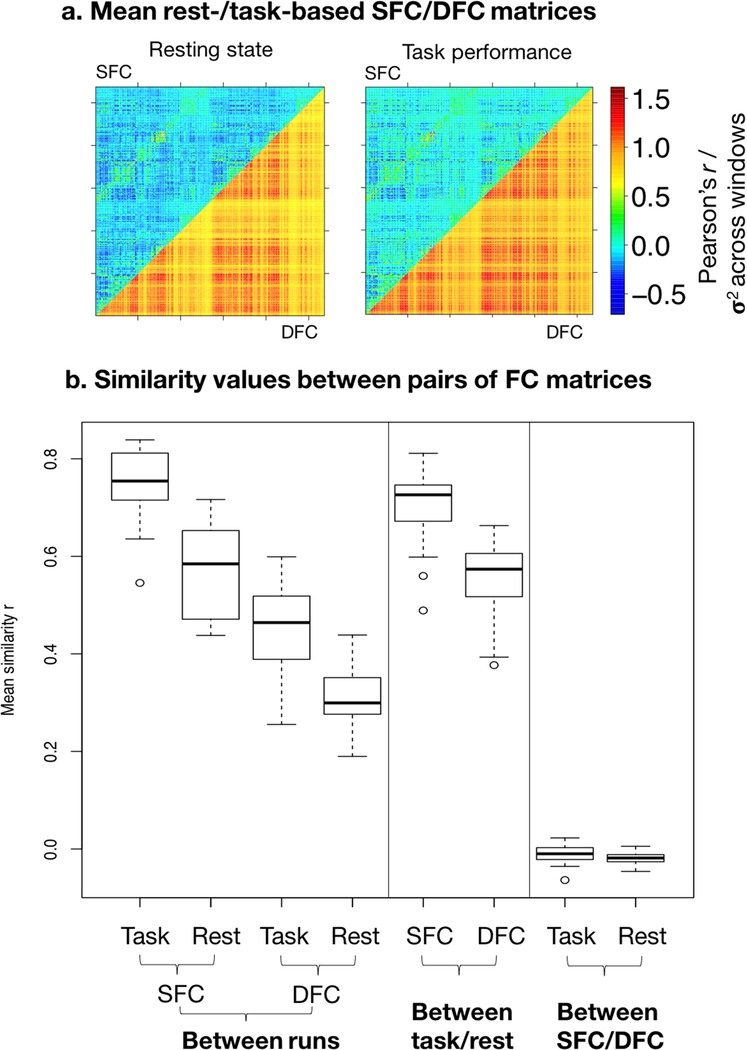
3.4. Similarity between FC measures within individuals
To characterize the intra-subject reliability of static and dynamic measures, we quantified the similarity 1) between each individual’s static/dynamic task-based/resting-state matrices across scan runs, 2) between each individual’s static matrix and dynamic matrix under task-based and resting-state scans, and 3) between each individual’s task-based matrix and resting-state matrix for both static and dynamic functional connectivity, with Pearson’s r (Figure 3). For this analysis, we used DFC matrices from 20TR windows, the optimal window length for prediction (Supplementary Figure S1). As previously found with static matrices (task SFC across runs: mean=0.751, range={0.546,0.839}, sd=0.0719; rest SFC across runs: mean=0.569, range={0.438,0.716}, sd=0.092), dynamic matrices were positively correlated across task-based (mean=0.459, range={0.255,0.599}, sd=0.089) and across resting-state (mean=0.309, range={0.190,0.439}, sd=0.059) fMRI runs, suggesting that edge variability is consistent within individuals across scans. Moreover, robust similarity across task performance and resting state is observed in both static (mean=0.708, range={0.489, 0.811}, sd=0.075) and dynamic (mean=0.555, range={0.377, 0.663}, sd=0.077) matrices, suggesting that individual functional connectivity dynamics are common to resting-state and task performance, and could constitute functional connectivity profiles inherent to individuals. However, it is possible that this similarity in part reflects non-stationary noise common across rest and task. Conversely, static and dynamic matrices were highly dissimilar under both rest (mean=−0.010, range={−0.064,0.023}, sd=0.018) and task (mean=−0.020, range={−0.046,0.005}, sd=0.012), suggesting minimal information overlap between the two measures (Figure 3).
Figure 3: a. Mean static and dynamic functional connectivity matrices across subjects.
In both figures, the upper triangle contains entries from the mean SFC matrix across subjects, which are Fisher-z transformed Pearson’s r values; the lower triangle contains entries from the mean DFC matrix, which are variability (σ2) values of Fisher-z transformed Pearson’s r values across sliding windows, upscaled by 3× for better visual display. Left: resting-state matrix; right: task-based matrix. The diagonal is excluded as it spans the connections of nodes to themselves. By definition, static matrices include both positive and negative values, while dynamic matrices include only positive values. b. Mean similarity between FC measures within subjects. Similarity is quantified by Pearson’s r, and boxplot shows distribution of similarity across individuals. From left to right – 1. between task-based SFC matrices across scan runs, 2. between resting-state SFC matrices across runs, 3. between task-based DFC matrices across runs, 4. between resting-state DFC matrices across runs, 5. between task-based and resting-state SFC matrices, 6. between task-based and resting-state DFC matrices, 7. between resting-state SFC and DFC matrices, 8. between task-based SFC and DFC matrices.
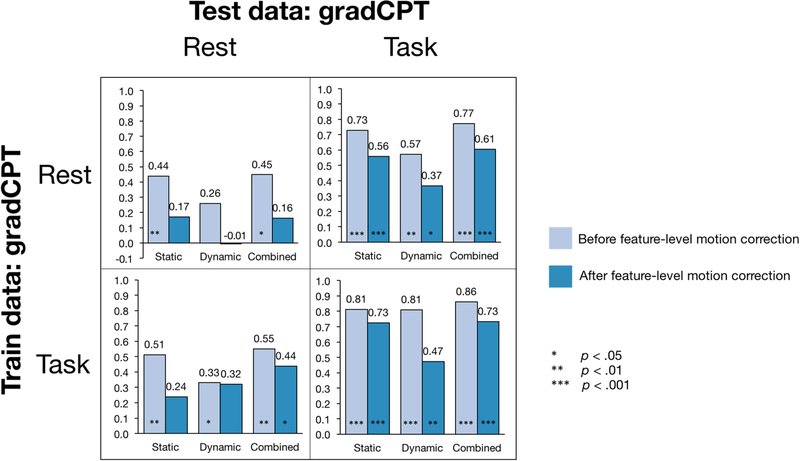
3.5. Statistical correction for head motion
As described in the methods section, we conducted a highly stringent feature-level statistical head motion correction, in addition to the preprocessing steps employed, by regressing SFC and DFC matrices on three head motion parameters: maximum displacement, mean frame-to-frame displacement, and maximum rotation, and re-running CPM with the residuals. Figure 4 compares model performance before and after the addition of this conservative feature-level correction step. Among all models tested on rest data, only the combined model trained on task data retained significant predictive power. Since the reduction in performance was common to static and dynamic models, it is more likely a statistical outcome of feature exclusion than a unique effect of head motion nonstationarity. On the other hand, models tested on task data retained statistical significance despite a reduction in numerical performance.
Figure 4: The effect of statistical head-motion correction on prediction performance.
Correction is conducted on gradCPT dataset, across task and rest conditions. The gridplot compares prediction performance before and after additional feature-level head motion correction (based on data corrected for motion during preprocessing) for all internal validations: static, dynamic, and combined, trained and tested on task or on rest. Bar levels and values indicate Spearman’s ρ between predicted d’ and observed behavior score.
3.6. Functional anatomy of predictive networks in the DFC attention model
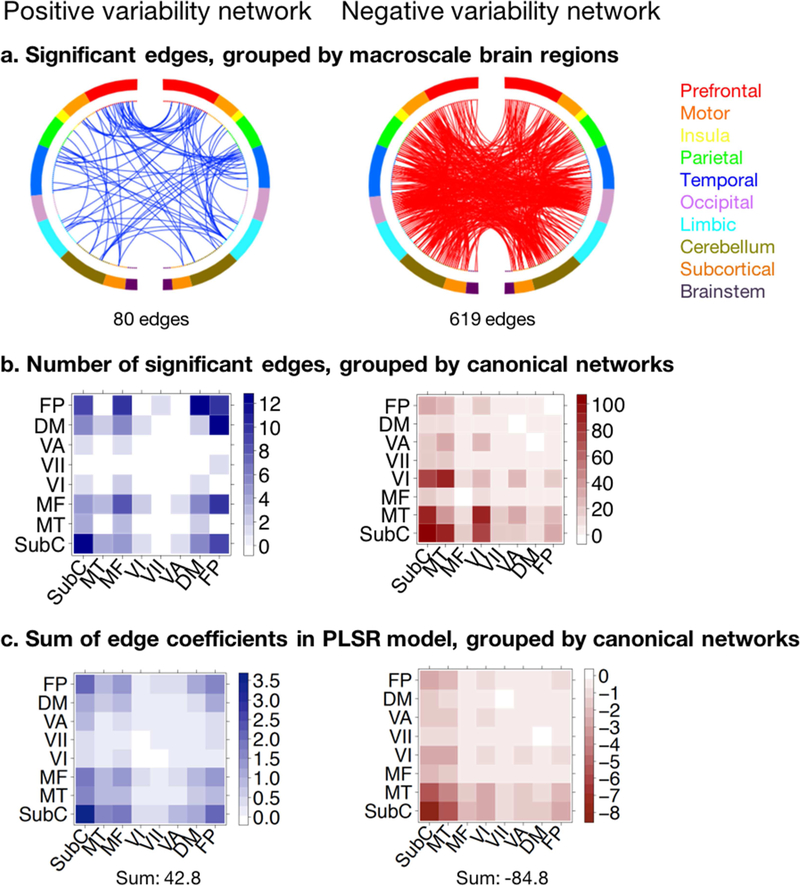
Connections whose temporal variability significantly contributed to the dynamic model were identified with a 1000-iteration permutation test as detailed in the Methods section (Figure 5). Out of a total of 35,778 edges, temporal variability in strength of 80 edges had significant positive coefficients (“positive variability network”), whereas that of 619 edges had significant negative coefficients (“negative variability network”) in the task-based PLSR model predicting d’ scores, suggesting that sustained attention is generally afforded by more stable connections across brain regions. To summarize the anatomical properties of these predictive edges, we grouped them into pairs of intrinsic connectivity networks defined previously using the 268-node Shen atlas (Shen et al., 2013) (Figure 5b).
Figure 5: Functional anatomy of predictive networks in the DFC attention model.
Positive variability (blue) and negative variability (red) networks span the edges whose temporal variability had respectively significant positive and significant negative PLSR coefficients in the DFC model predicting d’ from gradCPT task-based data. a. Distribution across macroscale brain regions. Macroscale regions include prefrontal cortex, motor cortex, insula, parietal, temporal, occipital, limbic (including the cingulate cortex, amygdala and hippocampus), cerebellum, subcortical (thalamus and striatum) and brainstem. Online visualization tool for making circle brain plots accessible via https://bioimagesuiteweb.github.io/webapp/connviewer.html. b. Distribution within and across canonical networks. Canonical networks include subcortical-cerebellum (SubC), motor (MT), medial frontal (MF), visual I (VI), visual II (VII), visual association (VA), default mode (DM), frontoparietal (FP) (Finn et al., 2015). c. Sum of all positive and negative edge coefficients in PLSR dynamic model, regardless of significance, grouped into canonical networks.
The positive network was mostly dominated by connections within the subcortical network, within the frontoparietal network, and between frontoparietal and other networks, but was otherwise relatively sparse. This finding supplements previous DFC work by Douw and colleagues, which found that higher across-window variability of frontoparietal and default mode networks was associated with greater out-of-scanner cognitive flexibility (Douw et al., 2016), which has been shown to predict inter-individual sustained attention (Figueroa et al., 2014). The negative network was more evenly distributed across brain networks but connections within and between visual, motor, and subcortical networks were the most prominent constituents. These networks are likely heavily engaged in information processing during the demanding gradCPT attention task, mediating visual processing, executive control, attentional resource allocation, and motor response.
Furthermore, to account for the raw magnitude of each edge’s coefficient in the PLSR models, including that of edges that were not individually significantly predictive, we summed the positive or negative coefficients across the edges constituting each network intersection (Figure 5c). We observed remarkably similar patterns to those observed in the original network distribution of significant edges: the sum of positive edge weights was high across frontoparietal and subcortical networks, and the sum of negative edge weights was high across visual, motor and subcortical networks in the negative network; moreover, the total sum of negative coefficients significantly exceeds that of the positive coefficients. Spearman’s rank correlation between number of significant edges and sum of edge coefficient magnitudes across network pairs was 0.81 for the positive and −0.89 for the negative networks respectively.
In previous work (Rosenberg et al., 2016), we isolated a network of edges whose overall strength predicted sustained attention (“strength network”); in this work, we examine a network in which temporal variability of edge strength predicted attention (“variability network”). To probe possible interactions between the networks, we looked for their intersections. Only 26 edges had strength and variability values that both predicted attention (all 26 were in the negative variability network; 20 were in the positive and 6 in the negative strength network), suggesting minimal overlap between the two networks, and likely diverging mechanisms underlying the effects of edge strength and variability on attention.
4. Discussion
DFC significantly predicts individual attention
The study of functional connectivity using fMRI recently harbored two novel developments: 1) an extension from group-level comparison studies to individual predictive modeling of behavior, and 2) the emergence of a subfield concerning the dynamic, time-varying properties of FC. While several studies have already trodden the intersection of the two to relate DFC to individual differences in clinical symptoms and behavior (Qin et al., 2015; Jin et al. 2017; Rashid et al. 2016; Falahpour et al., 2016; Laufs et al., 2014; see Preti et al., 2016 for a review of DFC applications), this study is the first to date to build a fully cross-validated DFC-based model predicting behavior in novel subjects and independently acquired datasets completely, across resting-state and task-based data. Here, individual whole-brain DFC significantly predicted attention task performance, with task-based and resting-state models each generalizing to at least one unseen task-based and resting-state dataset, demonstrating external validity. We furthermore demonstrated that as with SFC, DFC connectomes are reliable within individuals, showing replicability across scan sessions and cognitive states. This complements work by Liu and colleagues (2018), who used DFC “chronnectomes” to reliably identify individuals. Thus, future “fingerprinting” studies can investigate whether incorporating the dynamics of FC markers confers statistically significant utility in predictive modeling of behavior.
Combining static and dynamic features numerically, but not significantly, improves prediction performance
Including multiple feature classes in predictive models may help maximize predictive power and holistically uncover different neural mechanisms underlying behavior (Rosenberg et al., 2018). This study provides an instance of integrating feature subtypes, showing that combining static and dynamic FC models leads to numerically improved prediction performance over either model alone. Suggesting that DFC offers additional predictive value for attention, SFC and DFC matrices are highly dissimilar within each subject, and, there is negligible overlap between the significantly predictive features (edges) in static and in dynamic models. Further statistical testing to delineate the unique contributions of static and dynamic networks to predictive models poses an interesting avenue for future work.
Lower whole-brain FC temporal variability predicts better sustained attention across individuals
In addition to improving the predictive power of connectome-based models of behavior, the incorporation of novel fMRI features can help better characterize the functional architecture of attention and cognition. Previously, we found that mean strength in specific sets of functional brain connections predicts individual attention ability across datasets (Rosenberg et al., 2016). Here, we discover a separate “variability network” of brain connections whose temporal variability in strength, captured across sliding windows, predicts individual differences in attention. In the vast majority of edges, higher variability predicts worse attention. This suggests that sustained attention is generally afforded by increased stability of functional connections, regardless of the role of their overall strengths. As evidenced by the high dissimilarity between SFC and DFC connectomes, the mean and temporal variability of connection strength are minimally related features. Considering that fluctuations in FC networks on the order of 0.3 ~ 90 Hz index visual attention, working memory, and sensorimotor integration (Hutchison et al., 2013b), as well as transitions between on-task and off-task attention (Mittner et al., 2014), one would expect stable FC patterns to accompany more consistent attention allocation. The frontoparietal network emerged as a significant exception to this pattern of results: Predictive models included more intra- and inter-frontoparietal edges whose variability predicted better attention than edges whose variability predicted worse attention. In light of previous findings that higher temporal variability of frontoparietal and default mode network connections was associated with greater out-of-scanner cognitive flexibility (Douw et al., 2016), which has been shown to predict sustained attention (Figuero et al., 2014), the frontoparietal edges may play a role in an executive control system whose connectivity is continuously optimized to process ongoing tasks.
We previously found that the same SFC “strength networks” predicting individual differences in attention also differentiated healthy adults given methylphenidate (Ritalin) and unmediated controls (Rosenberg et al., 2016b), suggesting that these networks are malleable and potentially modulate attentional ability within individuals. Similarly, future work can extend DFC-based biomarkers, including FC temporal variability, to predict within-subject attentional variation. This would allow us to investigate whether FC variability in the identified networks relate to behavior at both across-subject and within-subject levels.
Methodological implications of an edge-based approach to sliding windows correlations
As one of the first to construct DFC-based predictive models of individual behaviour, this study highlights several methodological choices that bear implications for future DFC work. The sliding-window correlations method usually outputs a stack of windowed correlation matrices, which can be processed in either a state-based or an edge-based approach (See Introduction). Briefly, while the former considers recurring spatial functional connectivity configurations over time, the latter considers temporal properties of spatially localized edges. Here, we took the edge-based approach, summarizing DFC properties with edge strength variability across windows. We observed several advantages with this approach. First, the summary statistics take the same dimensionality as static FC connectomes, facilitating comparison between static and dynamic brain features. Second, taking the variance of windowed FC matrices is a computationally straightforward operation, allowing efficient construction of DFC connectomes. Third, the specific effects of edge-wise DFC on attention are easily interpreted, yielding novel insights about its cognitive architecture. Nevertheless, future DFC work can provide complementary insights by leveraging the state-based approach, which is in a sense more “dynamic” in that the summary statistics themselves may vary over time. The state-based approach also retains the same order of information as SFC: although allowed to change over time, the brain feature being tracked remains FC strength, a more traditional measure. Thus, in studies predicting a time-varying behaviour construct or cognitive state shifts within an individual, rather than a static score between individuals, the state-based approach confers unique advantages. This work motivates integrating these two approaches, and investigating how changes in attentional performance over time relate to transient FC configurations.
Head motion corrections
We imposed highly stringent statistical corrections for head motion. Our former approach was to exclude any edges that significantly correlated with head motion parameters across individuals (e.g., Hsu et al., 2017). This study explored an even more stringent method of regressing out head motion variance across individuals and used the residuals to perform CPM analyses. Expectedly, predictions were less effective, especially when applied to rest data, while predictions applied to task data remained highly significant. This provides a promising basis on which to corroborate resting-state DFC findings with task-based fMRI, since resting-state DFC studies have been the primary recipient of criticism for head motion confounds. However, although regression-based corrections are useful to demonstrate significant predictions, we believe that they are overly conservative. Because the goal of CPM is to predict individual differences in behavior, which cannot be explained by head motion in the current samples, excessively stringent corrections reduce meaningful signal and underestimate predictive performance.
Future directions
In almost all internal and external validations, DFC models alone numerically underperformed SFC models. While SFC measured with Pearson’s correlations is a well-established predictor of individual behavior, DFC represented as temporal variability is a relatively recent concept. By introducing a neuromarker that successfully predicts behavior, this study motivates the discovery of potentially more complicated DFC measures that have higher prediction utility. Moreover, this study frames DFC as a higher-order statistic derived upon SFC, in that we are still building upon the same measure of FC strength despite applying it to shortened time segments. As a result, it is difficult to expect it to outperform the original, lower-order feature. Nevertheless, it is possible that mean FC strength simply bears stronger predictive utility than FC strength variability in predicting attention, which also motivates the search for other DFC markers.
As with other CPM studies, the present study follows a data-driven approach to predictive modeling. Rather than postulating specifically localized biomarkers for individual differences a priori, we use machine learning to assess the data and search for predictive features, in doing so reducing experimental bias while escaping the limitations of inherent knowledge in generating hypotheses (van Helden, 2013). A downside of the data-driven framework is that it precludes a thorough study into the dynamical phenomena underlying the success of models: without introducing hypothesis-driven priors, we could not go much beyond isolating the networks whose strength variability predicted attention. Beyond predicting individual attention scores, follow-up DFC work could aim to explain the neural correlates of attentional experience, including shifts in attention or changes in performance, across individuals.
This study offers a proof of concept in using multiple feature classes to improve predictions. This is especially pertinent to connectome-based predictive modeling studies, where the value of abundant functional and structural brain data is mainly limited by the complexity of modeling algorithms. In this case, we elected to re-run the PLSR algorithm on individual concatenated static and dynamic FC matrices, allowing the contributions of significant predictors to be straightforwardly analyzed. However, penalized regression and dimensionality reduction strategies offer viable alternatives to assign feature weights and eliminate redundant predictors (Rosenberg et al., 2018). Since feature engineering is a field in its own right in machine learning, future work in connectome-based predictive modeling may benefit from emerging algorithms that integrate multiple brain features without losing biological interpretability. At the same time, this study demonstrated that combined models numerically outperformed models with a single feature class. Further testing is required to evaluate whether dynamic functional connectivity features measured during rest, task performance, or other conditions such as naturalistic viewing confer statistically significant improvements in behavioral prediction across a range of cognitive domains.
Supplementary Material
Acknowledgements
This project was supported by the National Institute of Health (NIH) grant MH108591, R01AA021449, and R01DA023248 and by the National Science Foundation grant BCS 1558497 and BCS1309260. S.Z was supported by the NIH grant K25DA040032.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Ethics statement
All participants of this study gave written informed consent in accordance with the Yale University Human Subjects Committee.
Data and code availability statement
The code supporting the findings of this study are available from the corresponding author upon request.
References
- Allen EA , Damaraju E, Plis SM, Erhardt EB, Eichele T, Calhoun VD, 2014. Tracking whole-brain connectivity dynamics in the resting state. Cereb. Cortex 24 (March (3)), 663–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbabshirani MR, Plis S, Sui J, Calhoun VD, 2017. Single subject prediction of brain disorders in neuroimaging: Promises and pitfalls. NeuroImage 145, 137–165. doi: 10.1016/j.neuroimage.2016.02.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Glover GH, 2010. Time-frequency dynamics of resting-state brain connectivity measured with fMRI. NeuroImage 50 (March (1)), 81–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciric R, Wolf DH, Power JD, Roalf DR, Baum GL, Ruparel K, … Satterthwaite TD, 2017. Benchmarking of participant-level confound regression strategies for the control of motion artifact in studies of functional connectivity. NeuroImage, 154, 174–187. doi: 10.1016/j.neuroimage.2017.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole MW, Bassett DS, Power JD, Braver TS, & Petersen SE, 2014. Intrinsic and task-evoked network architectures of the human brain. Neuron, 83(1), 238–251. doi: 10.1016/j.neuron.2014.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damaraju E, Allen E, Belger A, Ford J, McEwen S, Mathalon D, Mueller B, Pearlson G, Potkin S, Preda A, Turner J, Vaidya J, van Erp T, Calhoun V, 2014. Dynamic functional connectivity analysis reveals transient states of dysconnectivity in schizophrenia. NeuroImage: Clin 5 (July), 298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douw L, Wakeman DG, Tanaka N, Liu H, Stufflebeam SM, 2016. State-dependent variability of dynamic functional connectivity between frontoparietal and default networks relates to cognitive flexibility. J. Neurosci (December (17)), 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Pearlson GD, Yu Q, He H, Lin D, Sui J, Wu L, Calhoun VD, 2016. Interaction among subsystems within default mode network diminished in schizophrenia patients: a dynamic connectivity approach. Schizophr. Res 170 (1), 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esterman M, Noonan SK, Rosenberg M, Degutis J, 2013. In the zone or zoning out? Tracking behavioral and neural fluctuations during sustained attention. Cereb. Cortex 23, 2712–2723. [DOI] [PubMed] [Google Scholar]
- Falahpour M, Thompson WK, Abbott AE, Jahedi A, Mulvey ME, Datko M, Liu TT, Müller RA, 2016. Underconnected, but not broken? Dynamic functional connectivity MRI shows underconnectivity in autism is linked to increased intraindividual variability across time. Brain Connect 6 (June (5)), 403–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, McCandliss BD, Fossella J, Flombaum JI, Posner MI, 2005. The activation of attentional networks. NeuroImage 26, 471–479. doi: 10.1016/j.neuroimage.2005.02.004 [DOI] [PubMed] [Google Scholar]
- Farr OM, Hu S, Matuskey D, Zhang S, Abdelghany O, Li CR, 2014a. The effects of methylphenidate on cerebral activations to salient stimuli in healthy adults. Exp. Clin. Psychopharmacol 22, 154–165. doi: 10.1037/a0034465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr OM, Zhang S, Hu S, Matuskey D, Abdelghany O, Malison RT, Li C-SR, 2014b. The effects of methylphenidate on resting-state striatal, thalamic and global functional connectivity in healthy adults. Int. J. Neuropsychopharmacol 17, 1177–1191. doi: 10.1017/S1461145714000674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa IJ, Youmans RJ, Shaw TH, 2014. Cognitive flexibility and sustained attention: See something, say something (even when it’s not there). Proceedings of the Human Factors and Ergonomics Society Annual Meeting, 58, 954–958. [Google Scholar]
- Finn ES, Shen X, Scheinost D, Rosenberg MD, Huang J, Chun MM, Papademetris X, Constable RT, 2015. Functional connectome fingerprinting: identifying individuals using patterns of brain connectivity. Nat. Neurosci 18, 1664–1671. doi: 10.1038/nn.4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Raichle ME, 2007. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat. Rev. Neurosci 8, 700–711. doi: 10.1038/nrn2201 [DOI] [PubMed] [Google Scholar]
- Gabrieli JDE, Ghosh SS, Whitfield-Gabrieli S, 2015. Prediction as a Humanitarian and Pragmatic Contribution from Human Cognitive Neuroscience. Neuron 85, 11–26. doi: 10.1016/j.neuron.2014.10.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber EMG, Hahn T, Hilger K, Fiebach CJ, 2017. Distributed patterns of occipitoparietal functional connectivity predict the precision of visual working memory. NeuroImage 2017;146:404–418. doi: 10.1016/j.neuroimage.2016.10.006. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Castillo J, Handwerker DA, Robinson ME, Hoy CW, Buchanan LC, Saad ZS, Bandettini PA, 2014. The spatial structure of resting state connectivity stability on the scale of minutes. Front. Neurosci 8 (June (8)), 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratton C, Laumann TO, Nielsen AN, Greene DJ, Gordon EM, Gilmore AW, Petersen SE, 2018. Functional Brain Networks Are Dominated by Stable Group and Individual Factors, Not Cognitive or Daily Variation. Neuron, 98(2), 439–452.e5. doi: 10.1016/j.neuron.2018.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindriks R, Adhikari M, Murayama Y, Ganzetti M, Mantini D, Logothetis N, Deco G, 2016. Can sliding-window correlations reveal dynamic functional connectivity in resting-state fMRI? NeuroImage 127 (February), 242–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu W-T, Rosenberg MD, Scheinost D, Constable RT, & Chun MM, 2018. Resting-state functional connectivity predicts neuroticism and extraversion in novel individuals. Social Cognitive and Affective Neuroscience, 13(2), 224–232. doi: 10.1093/scan/nsy002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison RM, Morton JB, 2015. Tracking the Brain’s functional coupling dynamics over development. J. Neurosci 35 (April (17)), 6849–6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison RM, Womelsdorf T, Allen EA, Bandettini PA, Calhoun VD, Corbetta M, Della Penna S, Duyn JH, Glover GH, Gonzalez-Castillo J, Handwerker DA, Keilholz S, Kiviniemi V, Leopold DA, de Pasquale F, Sporns O, Walter M, Chang C, 2013. Dynamic functional connectivity: Promise, issues, and interpretations. NeuroImage 80, 360–378. doi: 10.1016/j.neuroimage.2013.05.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Jia H, Lanka P, Rangaprakash D, Li L, Liu T, Hu X, Deshpande G, 2017. Dynamic brain connectivity is a better predictor of PTSD than static connectivity. Hum. Brain Mapp, 38: 4479–4496. doi: 10.1002/hbm.23676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Vemuri P, Murphy MC, Gunter JL, Senjem ML, Machulda MM, Przybelski SA, Gregg BE, Kantarci K, Knopman DS, Boeve BF, Petersen RC, Jack CR, 2012. Non-stationarity in the resting brain’s modular architecture. PLoS One 7 (June (6)), e39731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler RC, van Loo HM, Wardenaar KJ, Bossarte RM, Brenner LA, Cai T, Zaslavsky AM, 2016. Testing a machine-learning algorithm to predict the persistence and severity of major depressive disorder from baseline self-reports. Molecular Psychiatry, 21(10), 1366–1371. doi: 10.1038/mp.2015.198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucyi A, Davis KD, 2014. Dynamic functional connectivity of the default mode network tracks daydreaming. NeuroImage 100 (October), 471–480. [DOI] [PubMed] [Google Scholar]
- Kucyi A, Salomons TV, Davis KD, 2013. Mind wandering away from pain dynamically engages antinociceptive and default mode brain networks. Proc. Natl. Acad. Sci 110 (November (46)), 18692–18697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufs H, Rodionov R, Thornton R, Duncan JS, Lemieux L, Tagliazucchi E, 2014. Altered fMRI connectivity dynamics in temporal lobe epilepsy might explain seizure semiology. Front. Neurol 5 (September), 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laumann TO, et al. , Snyder AZ, Mitra A, Gordon EM, Gratton C, Adeyemo B, Gilmore AW, Nelson SM, Berg JJ, Greene DJ, 2016. On the stability of bold fmri correlations. Cereb. Cortex [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhu D, Jiang X, Jin C, Zhang X, Guo L, Zhang J, Hu X, Li L, Liu T, 2014a. Dynamic functional connectomics signatures for characterization and differentiation of PTSD patients. Hum. Brain Mapp 35 (April (4)), 1761–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zhang D, Liang A, Liang B, Wang Z, Cai Y, … Liu M, 2017. High transition frequencies of dynamic functional connectivity states in the creative brain. Scientific Reports, 7, 46072. doi: 10.1038/srep46072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Rosenberg MD, Yoo K, Hsu TW, O’Connell TP, & Chun MM, 2018. Resting-State Functional Connectivity Predicts Cognitive Impairment Related to Alzheimer’s Disease. Frontiers in Aging Neuroscience, 10, 94. doi: 10.3389/fnagi.2018.00094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist MA, Xu Y, Nebel MB, Caffo BS, 2014. Evaluating dynamic bivariate correlations in resting-state fMRI: a comparison study and a new approach. NeuroImage 101 (November), 531–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Liao X, Xia M, He Y, 2018. Chronnectome fingerprinting: Identifying individuals and predicting higher cognitive functions using dynamic brain connectivity patterns. Hum Brain Mapp, 39: 902–915. doi: 10.1002/hbm.23890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhyastha TM, Askren MK, Boord P, Grabowski TJ, 2015. Dynamic connectivity at rest predicts attention task performance. Brain Connect 5 (February (1)), 45–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Murakami T, Ohki K, 2017. Neuronal origin of the temporal dynamics of spontaneous BOLD activity correlation. bioRxiv, 10.1101/169698 [DOI] [PubMed] [Google Scholar]
- Mittner M, Boekel W, Tucker AM, Turner BM, Heathcote A, Forstmann BU, 2014. When the brain takes a break: a modelbased analysis of mind wandering. J Neurosci 34:16286–16295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble S, Scheinost D, Finn ES, Shen X, Papademetris X, McEwen SC, Bearden CE, Addington J, Goodyear B, Cadenhead KS, Mirzakhanian H, Cornblatt BA, Olvet DM, Mathalon DH, McGlashan TH, Perkins DO, Belger A, Seidman LJ, Thermenos H, Tsuang MT, van Erp TGM, Walker EF, Hamann S, Woods SW, Cannon TD, Constable RT, 2017. Multisite reliability of MR-based functional connectivity. NeuroImage 146, 959–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Halloran L, Cao Z, Ruddy K, Jollans L, Albaugh MD, Aleni A, Whelan R, 2018. Neural circuitry underlying sustained attention in healthy adolescents and in ADHD symptomatology. NeuroImage, 169, 395–406. doi: 10.1016/j.neuroimage.2017.12.030 [DOI] [PubMed] [Google Scholar]
- Poole VN, Robinson ME, Singleton O, DeGutis J, Milberg WP, McGlinchey RE, Esterman M, 2016. Intrinsic functional connectivity predicts individual differences in distractibility. Neuropsychologia, 86, 176–182. doi: 10.1016/j.neuropsychologia.2016.04.023 [DOI] [PubMed] [Google Scholar]
- Preti MG, Bolton TAW, Van De Ville D, 2016. The dynamic functional connectome: state-of-the-art and perspectives. NeuroImage, 1053–8119 (2016). doi: 10.1016/j.neuroimage.2016.12.061 [DOI] [PubMed] [Google Scholar]
- Price T, Wee C-Y, Gao W, Shen D, 2014. Multiple-network classification of childhood autism using functional connectivity dynamics. In: Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, pp. 177–184. [DOI] [PubMed] [Google Scholar]
- Qin J, Chen S-G, Hu D, Zeng L-L, Fan Y-M, Chen X-P, Shen H, 2015. Predicting individual brain maturity using dynamic functional connectivity. Front. Hum. Neurosci 9 (July), (Article 418). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revelle W, 2018. psych: Procedures for Personality and Psychological Research, Northwestern University, Evanston, Illinois, USA, https://CRAN.R-project.org/package=psychVersion=1.8.4. [Google Scholar]
- Rashid B, Arbabshirani MR, Damaraju E, Cetin MS, Miller R, Pearlson GD, Calhoun VD, 2016. Classification of schizophrenia and bipolar patients using static and dynamic resting-state fmri brain connectivity. NeuroImage 134, 645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinen JM, Chén OY, Hutchison RM, Yeo BTT, Anderson KM, Sabuncu MR, Holmes AJ, 2018. The human cortex possesses a reconfigurable dynamic network architecture that is disrupted in psychosis. Nature Communications, 9(1), 1157. doi: 10.1038/s41467-018-03462-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M, Noonan S, DeGutis J, Esterman M, 2013. Sustaining visual attention in the face of distraction: a novel gradual-onset continuous performance task. Atten. Percept. Psychophys 75, 426–439. [DOI] [PubMed] [Google Scholar]
- Rosenberg MD, Finn ES, Scheinost D, Constable RT, Chun MM, 2017. Characterizing Attention with Predictive Network Models. Trends Cogn. Sci 21, 290–302. doi: 10.1016/j.tics.2017.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg MD, Finn ES, Scheinost D, Papademetris X, Shen X, Constable RT, Chun MM, 2016a. A neuromarker of sustained attention from whole-brain functional connectivity. Nat. Neurosci 19, 165–171. doi: 10.1038/nn.4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg MD, Zhang S, Hsu W-T, Scheinost D, Finn ES, Shen X, Constable RT, Li C-SR, Chun MM, 2016b. Methylphenidate Modulates Functional Network Connectivity to Enhance Attention. J. Neurosci 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg MD, Hsu W-T, Scheinost D, Todd Constable R, Chun MM, 2017. Connectome-based Models Predict Separable Components of Attention in Novel Individuals. Journal of Cognitive Neuroscience, 30(2), 160–173. 10.1162/jocn_a_01197 [DOI] [PubMed] [Google Scholar]
- Shen X, Finn ES, Scheinost D, Rosenberg MD, Chun MM, Papademetris X, Constable RT, 2017. Using connectome-based predictive modeling to predict individual behavior from brain connectivity. Nat. Protoc 12, 506–518. doi: 10.1038/nprot.2016.178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Tokoglu F, Papademetris X, Constable RT, 2013. Groupwise whole-brain parcellation from resting-state fMRI data for network node identification. Neuroimage 82, 403–415. doi: 10.1016/j.neuroimage.2013.05.081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirer WR, Ryali S, Rykhlevskaia E, Menon V, Greicius MD, 2012. Decoding Subject-Driven Cognitive States with Whole-Brain Connectivity Patterns. Cerebral Cortex (New York, NY) 2012;22(1):158–165. doi: 10.1093/cercor/bhr099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Helden P, 2013. Data-driven hypotheses. EMBO Reports, 14(2), 104. doi: 10.1038/embor.2012.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee C-Y, Yang S, Yap P-T, Shen D, 2013. Temporally dynamic resting-state functional connectivity networks for early mci identification. In: Proceedings of the International Workshop on Machine Learning in Medical Imaging. Springer, pp. 139–146. [Google Scholar]
- Wee C-Y, Yang S, Yap P-T, Shen D, 2016a. Sparse temporally dynamic resting-state functional connectivity networks for early MCI identification. Brain Imaging Behav 10 (June (2)), 342–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo CW, Chang LJ, Lindquist MA, Wager TD, 2017. Building better biomarkers: brain models in translational neuroimaging. Nat. Neurosci 20, 365–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Lindquist MA, 2015. Dynamic connectivity detection: an algorithm for determining functional connectivity change points in fMRI data. Front. Neurosci 9 (September). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaesoubi M, Miller RL, Calhoun VD, 2015b. Mutually temporally independent connectivity patterns: a new framework to study the dynamics of brain connectivity at rest with application to explain group difference based on gender. NeuroImage 107 (February), 85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Craddock RC, Margulies DS, Yan C. g., Milham MP, 2014. Common intrinsic connectivity states among posteromedial cortex subdivisions: insights from analysis of temporal dynamics. NeuroImage 93 (June), 124–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo K, Rosenberg MD, Hsu WT, Zhang S, Li CSR, Scheinost D, Chun MM, 2018. Connectome-based predictive modeling of attention: Comparing different functional connectivity features and prediction methods across datasets. NeuroImage, 167, 11–22. doi: 10.1016/j.neuroimage.2017.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.