Abstract
Different types of effector and memory T lymphocytes are induced and maintained in protective or pathological immune responses. Here we characterized two human CD4+ TH17 helper cell subsets that, in the recently activated state, could be distinguished on the basis of their expression of the anti-inflammatory cytokine IL-10. IL-10+ TH17 cells upregulated a variety of genes encoding immunoregulatory molecules, as well as genes characteristic of tissue-resident T cells. In contrast, IL-10- TH17 cells maintained a pro-inflammatory gene expression profile and upregulated the expression of homing receptors that guide recirculation from tissues to blood. Expression of the transcription factor c-MAF was selectively upregulated in IL-10+ TH17 cells, and it was bound to a large set of enhancer-like regions and modulated the immunoregulatory and tissue-residency program. Our results identify c-MAF as a relevant factor that drives two highly divergent post-activation fates of human TH17 cells and provide a framework with which to investigate the role of these cells in physiology and immunopathology.
Introduction
Upon antigen recognition on stimulatory dendritic cells, naive CD4+ and CD8+ T cells proliferate and differentiate into effector cells capable of migrating to peripheral tissues and of performing protective functions. Once antigen has been eliminated, part of the primed T cells persist as circulating central and effector memory T cells that can provide enhanced responses upon re-exposure to their cognate antigen in secondary lymphoid organs or peripheral tissues, respectively1. It is now well established that some of the T cells entering tissues, in particular of the CD8+ effector T cells entering epithelial and mucosal barriers, remain in the tissue and form a pool of resident memory T cells that can promptly respond and provide protective immunity independently of T cells recruited from blood2,3.
T cell effector function is largely mediated through the release of pro-inflammatory cytokines. T helper cells that produce IL-17 (TH17 cells) can induce recruitment of neutrophils and trigger production of pro-inflammatory cytokines and chemokines by a broad range of cellular targets. Although these effector functions confer TH17 cells the ability to protect against certain extracellular bacteria and fungi, a deregulated TH17 response can induce severe tissue damage and chronic inflammation. Several mechanisms have evolved to limit the immune response to pathogens: for instance, interleukin-10 (IL-10) is a potent anti-inflammatory cytokine with a non-redundant role in restraining inflammatory responses in vivo, thereby preventing damage to the host4. In addition to IL-10, activated effector T cells can upregulate the expression of a number of inhibitory receptors that limit costimulatory signals to dampen the immune response5–7. For example, CTLA-4 can inhibit T cell activation intrinsically by outcompeting CD28 for binding to CD80 and CD86, while PD-1 engagement by PD-L1 or PD-L2 triggers an inhibitory signal.
We previously reported that IL-10 production is a characteristic of human TH17 cells that have been primed by Staphylococcus aureus, but not of TH17 cells that have been primed by Candida albicans, which instead co-express IL-17A and interferon-γ (IFN-γ)8. Interestingly, IL-17A and IL-10 production by S. aureus-specific TH17 cells was temporally regulated, with IL-17A being produced within hours of antigenic stimulation and IL-10 being produced only by 3–5 days after T cell antigen receptor (TCR) triggering. These results led us to hypothesize that antigenic stimulation of memory T cells may set in motion a post-activation gene expression program that can reveal further T cell heterogeneity relevant for in vivo regulation of the immune response.
Results
IL-10 production is a property of a human TH17 cell subset
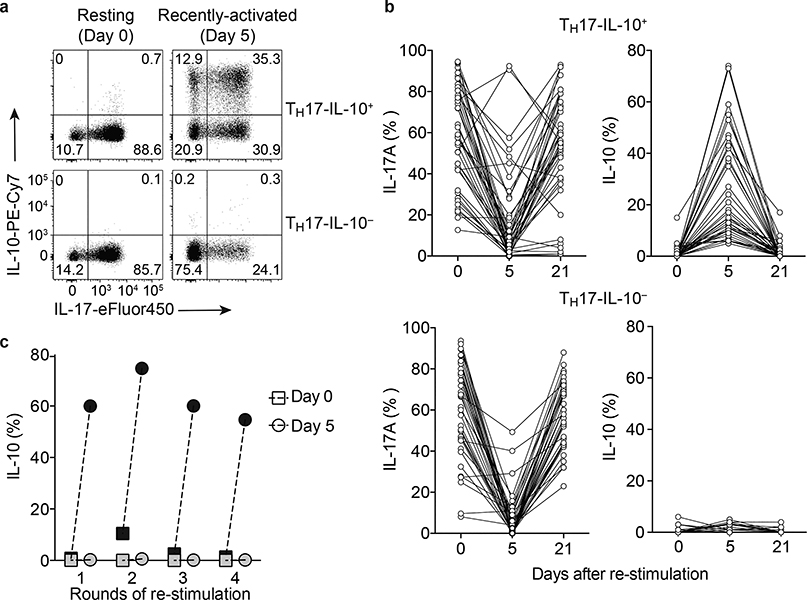
A large number of human TH17 clones were isolated from CCR6+CCR4+CXCR3- memory T cells or from IL-17A-producing CCR6+CXCR3- T cells (Supplementary Fig. 1a). Cytokine production was measured in T cell clones in the resting state (Day 0) and in the recently activated state (Day 5 following re-stimulation with CD3 and CD28 antibodies). On Day 0, all TH17 clones produced IL-17A but no IL-10 (Fig. 1a,b). However, on Day 5 following re-stimulation, the TH17 clones showed a heterogeneous pattern of cytokine production. About 25% of the clones acquired the capacity to produce IL-10, concomitant with downregulation of IL-17A (referred to as TH17-IL-10+), while the remaining clones downregulated IL-17A but did not acquire the capacity to produce IL-10 (referred to as TH17-IL-10-) (Fig. 1a,b). When reverted to a resting state (Day 21 following re-stimulation), the clones re-acquired the ability to produce IL-17A and, in the case of TH17-IL-10+ clones, lost the capacity to produce IL-10 (Fig. 1b). Importantly, production of IL-10 was observed over repeated rounds of stimulation (Fig. 1c), indicating that TH17-IL-10+ cells maintain memory of IL-10 expression. On Day 0 and Day 5, the TH17-IL-10- clones produced significantly more IFN-γ, IL-22 and GM-CSF than TH17-IL-10+ clones (Supplementary Fig. 1b).
Figure 1. Transient production of IL-10 is a stable feature of a subset of human memory TH17 cells.
a,b. Production of IL-17 and IL-10 in TH17 clones analyzed in the resting state (Day 0 and Day 21) and in the recently activated state (Day 5) as measured by intracellular cytokine staining. The clones were divided according to their ability to produce IL-10 on Day 5. Representative staining of a TH17-IL-10+ clone (upper panel) and a TH17-IL-10- clone is shown in (a) and data from several TH17-IL-10+ and TH17-IL-10- clones representative of more than 15 experiments performed are shown in (b). The percentage of TH17-IL-10+ clones isolated from CCR6+CCR4+CXCR3– T cells in 13 experiments performed with different donors was 24.67 ± 3.22 (mean ± s.e.m). A similar frequency of TH17-IL-10+ clones was obtained from 4 experiments performed with different donors in which clones were isolated from IL-17-producing CCR6+CXCR3– T cells (25.6 ± 7.87%, mean ± s.e.m.). c. The capacity of TH17-IL-10+ clones (black symbols) and TH17-IL-10- clones (grey symbols) to produce IL-10 was evaluated after repeated rounds of re-stimulation with anti-CD3 and anti-CD28 antibodies, both on Day 0 (square) and on Day 5 (circle); at least 21 days separated two following re-stimulations. Data are representative of 4 independent experiments. In panel c and in all subsequent experiments, the TH17 cells analyzed were pools of TH17 clones that comprised ≥ 50% IL-17+ cells in the resting state.
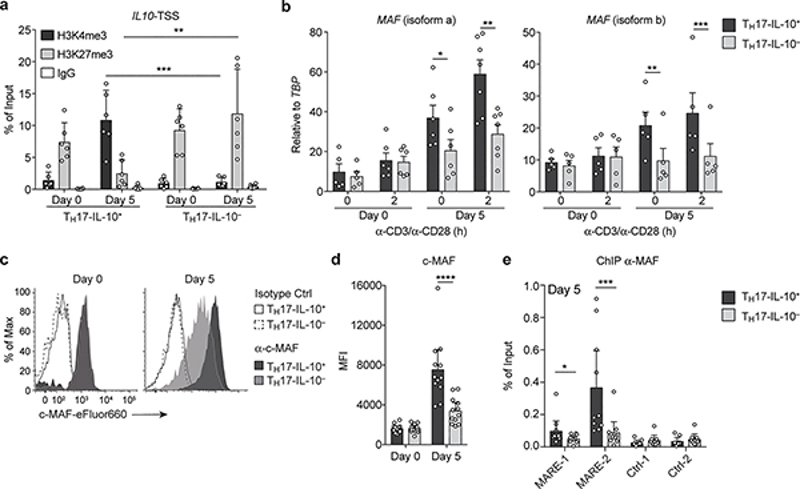
To investigate the mechanisms that underlie the delayed kinetics of IL-10 production in Day 5-activated TH17 cells, we evaluated histone modifications at the IL10 transcriptional start site (TSS) by chromatin immunoprecipitation (ChIP) and measured the expression of c-MAF, a transcription factor that regulates IL10 transcription in several immune cells9–12. In Day 0-resting TH17-IL-10+ and TH17-IL-10- cells the IL10 TSS was associated with high content of H3K27me3 (Fig. 2a), indicative of a repressed chromatin. In contrast, in Day 5-activated TH17-IL-10+ cells, but not in TH17-IL-10- cells, the IL10 TSS was associated with abundant H3K4me3, indicative of a transcriptionally active chromatin (Fig. 2a). Day 0 TH17 cells exhibited low expression of c-MAF mRNA (both isoform a and isoform b) and protein, but the amount of c-MAF selectively increased in Day 5 TH17-IL-10+ cells (Fig. 2b-d). Furthermore, binding of c-MAF to a previously characterized c-MAF binding site (MAF Responsive Element, MARE-2) on the IL10 promoter11,13 was significantly higher in Day 5 IL-10+ TH17 cells compared to IL-10- TH17 cells (Fig. 2e). Collectively, these data indicate that consequent to a first antigenic stimulation, a subset of human memory TH17 cells undergoes changes in chromatin conformation that are accompanied by the upregulation of c-MAF (and possibly other transcription factors) and the capacity to produce the anti-inflammatory cytokine IL-10.
Figure 2. Production of IL-10 by Day 5-activated TH17 cells requires chromatin remodeling and c-MAF up-regulation.
a. Abundance of histone modifications associated with gene transcription (H3K4me3) or repression (H3K27me3) at the IL10 TSS analyzed by ChIP in Day 0-resting and Day 5-activated TH17-IL-10+ and TH17-IL-10- cells. Data are represented as mean + 95% c.i. (n = 6). b. Expression of MAF isoforms a and b was measured in Day 0 and Day 5 TH17-IL-10+ and TH17-IL-10- clones by qPCR and normalized to the TATA-box binding protein gene (TBP). Shown are individual values and mean + s.e.m. (isoform a: Day 0, n = 5; Day 0 + 2h and Day 5, n = 6; Day 5 + 2h, n = 7; isoform b, n = 5). c. Expression of c-MAF as detected by intracellular staining in representative Day 0 and Day 5 TH17-IL-10+ and TH17-IL-10- cells. d. Mean fluorescence Intensity (MFI) of c-MAF staining in Day 0 and Day 5 IL-10+ and IL-10- TH17 clones. Data are represented as mean + 95% c.i., with each dot indicating a T cell clone pool (n = 12). e. c-MAF binding to two regions located in the IL10 promoter as determined by ChIP. A developmentally repressed locus (Ctrl_1) and a pericentromeric DNA repeat region (Ctrl_2) were used as negative controls. Data are represented as mean + 95% c.i., with each dot indicating a T cell clone pool (n = 10). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, as determined by ratio paired t test. Data are representative of at least 3 independent experiments.
M1 and M2 polarization by TH17-IL-10- and TH17-IL-10+ cells
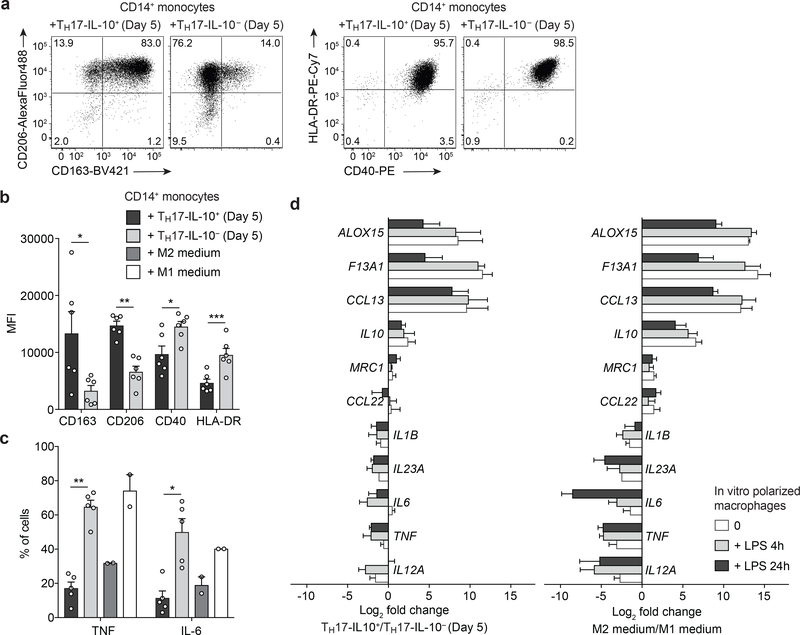
Since TH17-IL-10+ and TH17-IL-10- cells produce a different array of cytokines we evaluated their capacity to polarize monocytes toward M1 macrophages, which produce pro-inflammatory cytokines and nitrogen radicals for increased anti-microbial activity, or M2 macrophages, which inhibit production of inflammatory mediators and promote tissue homeostasis and repair14,15. Monocytes co-cultured with Day 5 TH17-IL-10+ cells rapidly upregulated the M2-associated markers CD163 and CD206, while the same cells co-cultured with Day 5 TH17-IL-10- cells upregulated the M1-associated markers CD40 and HLA-DR and produced high amounts of TNF and IL-6 (Fig. 3a-c). In addition, monocytes cultured in the presence of conditioned supernatants from Day 5 TH17-IL-10- or TH17-IL-10+ cells differentiated into macrophages with constitutive and lipopolysaccharide (LPS)-induced gene expression comparable to that of macrophages differentiated using M1- or M2-inducing cytokines, respectively (Fig. 3d). M2-like macrophages induced by TH17-IL-10+ cells expressed genes for biosynthesis of pro-resolving mediators (e.g. ALOX15 encoding arachidonate 15-lipoxigenase) and promoting tissue repair (e.g. F13A1 encoding Coagulation Factor XIII A chain, promoting wound healing and angiogenesis), while M1-like macrophages induced by TH17-IL-10- cells expressed genes encoding for pro-inflammatory cytokines, such as IL-12, IL-23, IL-1β, IL-6 and TNF16,17. Thus, recently activated TH17-IL- 10+ and TH17-IL-10- cells can promote M2 and M1 differentiation, respectively, indicating they may differentially modulate the immune microenvironment.
Figure 3. TH17-IL-10− and TH17-IL-10+ cells differentially polarize monocytes to M1 and M2 macrophages.
CD14+ monocytes were sorted from PBMCs and co-cultured with allogeneic Day 5-activated TH17-IL-10+ or TH17-IL-10- cells or in medium containing M1- or M2-inducing cytokines. a,b. Expression of M1- and M2-associated surface markers was determined after 24 h culture by flow cytometry (a, representative plots; b, mean + s.e.m.; n = 6). c. Production of TNF and IL-6 by CD14+CD4- monocytes assessed by intracellular staining after 48 h culture (mean + s.e.m.; TH17, n = 5; M1/M2 medium, n = 2). d. CD14+ monocytes were cultured in the presence of supernatants from Day 5 TH17-IL-10+or TH17- IL-10- cells or in M1/M2 medium for 7 days and then stimulated with LPS for the indicated time points. RNA was extracted and the expression of M1- and M2-associated genes was measured by qPCR (mean + s.e.m.; TH17 supernatants, n = 4; M1/M2 medium, n = 3). Genes are ranked according to their differential expression in M1/M2 medium-macrophages stimulated with LPS for 4 hours. *P < 0.05; **P < 0.01; ***P < 0.001, as determined by ratio paired t test. Data are from 3 (a-c) and 2 (d) independent experiments.
Regulatory or pro-inflammatory program in TH17 cell subsets
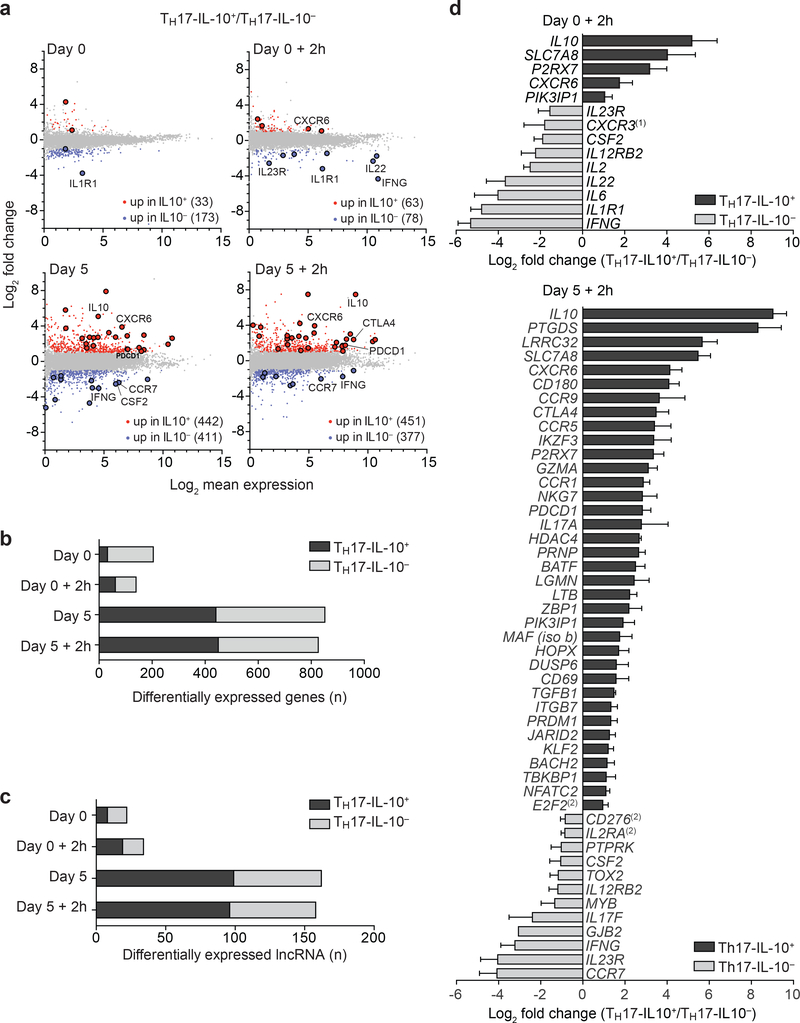
To investigate the differences between the two TH17 subsets on a genome-wide scale, we performed RNA-seq on Day 0 and Day 5, before and after stimulation for 2 h with CD3 and CD28 antibodies. Using the heuristic expression cut-off of 2 FPKM, 295 genes were found differentially expressed (fold change ≥ 2) between Day 0 TH17-IL-10+ and TH17-IL-10- clones, while this number increased to 1,185 between Day 5 TH17-IL-10+ and TH17-IL-10- clones (Fig. 4a,b). Genes expressed more abundantly in TH17-IL-10+ cells were significantly enriched in biological processes involving regulation of immune responses, whereas genes expressed more abundantly in TH17-IL-10- cells were enriched in processes linked to stress, external cues and chemotactic signals (P < 1 × 10–5, hypergeometric distribution) (Supplementary Fig. 2a).
Figure 4. Distinct transcriptional programs in Day 5-activated TH17-IL-10+ and TH17- IL-10− cells.
a. MA plots of RNA-seq analysis of Day 0-resting and Day 5-activated TH17- IL-10+ and TH17-IL-10− cells, before or after 2 h stimulation with CD3/CD28 antibodies. Differentially expressed genes are marked in red (fold change ≥ 2) or blue (fold change ≤ 0.5). The genes selected for validation are highlighted by larger dots. b,c. Number of differentially expressed protein-coding genes (b) and lncRNAs (c) in the indicated samples. Data represent the average of two independent experiments. d. qPCR validation of a set of differentially expressed genes in Day 0 (upper panel) and Day 5 (lower panel) TH17-IL-10+ and TH17-IL-10− cells (mean + s.e.m; n ≥ 3, P-value ≤ 0.05 as determined by paired t test) ((1) P-value = 0.06; (2) fold change ≥ 1.8). In b, c and d, black bars indicate genes more expressed in TH17-IL-10+ cells and grey bars genes more expressed in TH17-IL-10− cells.
A further hint for different gene regulation in TH17-IL-10+ and TH17-IL-10- clones came from the analysis of lncRNAs, non-protein coding regulatory transcripts that are differentially expressed in functionally distinct T cell subsets18,19. The number of lncRNAs differentially expressed between Day 0 TH17-IL-10+ and TH17-IL-10- clones was limited to 40 but increased to > 200 in Day 5 clones (Fig. 4c). Genes proximal to lncRNAs found in TH17-IL-10- clones are involved in T cell differentiation and activation, while genes proximal to lncRNAs found in TH17-IL-10+ clones are mainly involved in positive and negative modulation of the immune response and cell death (Supplementary Fig. 2b).
The differential expression of selected regulatory and pro-inflammatory genes was validated by qPCR (Fig. 4d). In addition to IL-10 and c-MAF, TH17-IL-10+ clones preferentially expressed regulatory genes such as TGFB1, CTLA4, and PDCD1, as well as IKZF3 encoding the transcription factor AIOLOS, which controls IL-10 expression20, and genes involved in regulatory T (Treg) cell function and regulation of TH17/Treg cell balance, such as BACH221, LRRC32 (GARP)22,23, LGMN (Legumain)24 and P2RX725. In addition, TH17-IL-10+ cells expressed genes encoding for effector molecules, such as GZMA, LTB (lymphotoxins), NKG7 and PTGDS (prostaglandin D synthase). Conversely, activated TH17- IL-10- clones preferentially expressed pro-inflammatory genes, such as IFNG, CSF2, IL22, IL23R, IL1R1, IL12RB2, IL2 and IL2RA (Supplementary Table 1). Genes controlling tissue homing and residency were also differentially expressed in the two TH17 subsets. TH17-IL- 10+ cells abundantly expressed CD69, CXCR6, CCR9 and ITGB7 (integrin β7), as well as DUSP6 and PRDM1 (BLIMP-1), that have been associated with tissue-resident T cells3,26,27. Conversely, TH17-IL-10- expressed high amounts of CCR7, which drives T cells from tissues to lymph nodes via afferent lymphatics28. The differential expression of CTLA-4, PD-1, CD25, CD69, CXCR6 and CCR7 in Day 5 TH17-IL-10+ and TH17-IL-10- cells was confirmed at the protein level (Supplementary Fig. 3a,b). Although several genes highly expressed in TH17-IL-10+ have been associated with Treg cell function, we could not detect any difference in FOXP3 expression between TH17-IL-10+ and TH17-IL-10- cells (Supplementary Fig. 3b). Of note, compared to TH17-IL-10- cells, TH17-IL-10+ cells expanded less when stimulated with CD3 and CD28 antibodies (Supplementary Fig. 3c). Taken together, the above findings indicate a new level of substantial heterogeneity in TH17 cells that becomes evident when the cells are in a recently activated state, with one subset acquiring anti-inflammatory, regulatory and tissue-residency properties and a second subset acquiring pro-inflammatory and recirculating properties.
TH17-associated genes are modulated by IL-27 and IL-1β
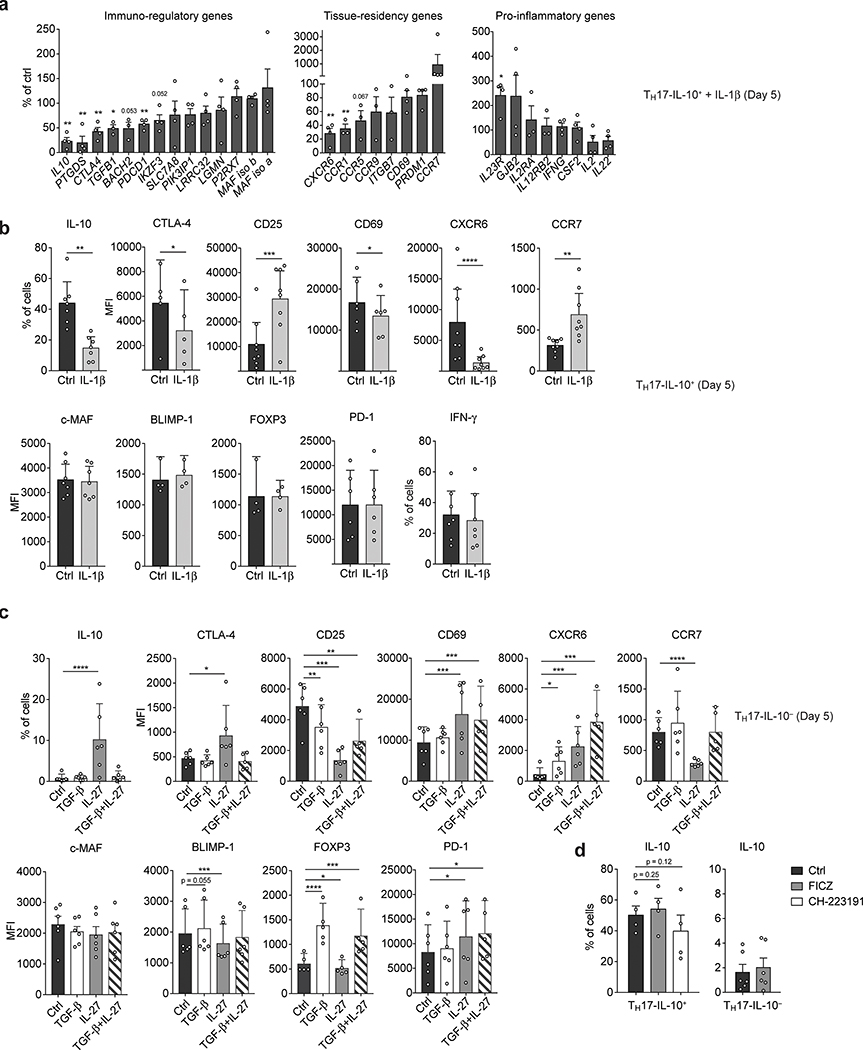
IL-1β induces IFN-γ and inhibits IL-10 production in TH17 cells while TGF-β and IL-27 induce IL-10 expression and cooperate with the transcription factor aryl hydrocarbon receptor (AHR) to promote the differentiation of regulatory Tr1 lymphocytes8,11. Activation in the presence of IL-1β induced the downregulation of IL10 and of the immunoregulatory and tissue residency genes CTLA4, PTGDS, TGFB1 and CXCR6 in TH17-IL-10+ cells, while expression of IL23R, CD25 and CCR7 was upregulated, both at the mRNA and protein level (Fig. 5a,b). Activation of TH17-IL-10- cells in the presence of IL-27, but not TGF-β, induced the expression of immunoregulatory and tissue-residency genes and at the same time repressed the surface expression of CCR7 (Fig. 5c). Conversely, neither the induction of AHR in TH17-IL-10- cells nor its inhibition in TH17-IL-10+ cells altered IL-10 production (Fig. 5d). Together these data indicate that the expression of TH17-IL-10+ and TH17-IL-10- associated genes can be, at least in part, differentially regulated by IL-1β and IL-27.
Figure 5. IL-27 and IL-lβ antagonistically promote the expression of TH17-IL-10+ and TH17-IL-10- associated genes.
a,b. TH17-IL-10+ cells were polyclonally activated with or without IL-1β (10 ng/ml) and the expression of representative immunoregulatory, tissue- residency and pro-inflammatory genes was measured on Day 5 at mRNA level by qPCR, after 2 h re-stimulation with CD3/CD28 antibodies (a, mean + s.e.m; n > 3), and at protein level by flow cytometry (b, mean + 95% c.i.; CXCR6, CD25 and CCR7, n = 8; IL-10, IFN-γ and c-MAF, n = 7; CD69 and PD-1, n = 6; CTLA-4, n = 5; BLIMP-1 and FOXP3, n = 4), after 5 h stimulation with PMA+I. c. TH17-IL-10- cells were polyclonally activated in presence of IL-27 (25 ng/ml), TGF-β (2 ng/ml) or a combination of the two cytokines and the expression of the indicated proteins was assessed on Day 5 by flow cytometry, after 5 h stimulation with PMA+I (mean + 95% c.i.; n = 6; surface markers TGF-β+IL-27, n = 5). d. TH17-IL-10+ (n = 4) and TH17-IL-10- (n = 6) cells were polyclonally activated in the presence of AHR antagonist (CH-223191, 3 μΜ) or agonist (FICZ, 100 nM) and IL-10 production was measured on Day 5 by flow cytometry, following 5 h PMA+I stimulation (mean + s.e.m.). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, as determined by ratio paired t test (a) or ratio paired t test (b-d).
TH17 transcriptional profiles and disease states
To validate in vivo the gene signatures identified for the two types of TH17 cells, we compared their transcriptional profiles with publicly available transcriptomic datasets from two TH17-mediated autoimmune disease patient cohorts. Gene set enrichment analysis (GSEA) of transcriptomic datasets from ileal biopsies of Crohn’s disease (CD) patients (GSE5794529) and healthy controls showed a significant association with the TH17-IL-10- signature, which increased with severity of inflammation (assessed on the basis of histology) (P < 0.05, familywise error rate) (Supplementary Fig. 4a). Interestingly, MAF expression in ileal biopsies from CD patients negatively correlated with the degree of intestinal inflammation and ulceration (Supplementary Fig. 4c). GSEA of transcriptomic datasets of PBMCs obtained from juvenile rheumatoid arthritis (RA) patients (GSE1402)30 and healthy controls revealed a dominant gene expression signature associated with TH17-IL-10+ cells in the latter, whereas gene signatures associated with TH17-IL-10- cells were identified in PBMCs and, in particular, synovial fluid mononuclear cells from the RA patients (Supplementary Fig. 4b). These data suggest that TH17-IL-10- cells, but not TH17-IL-10+ cells, may contribute to the progression of chronic inflammatory diseases.
c-MAF binds to bona fide gene enhancers in TH17-IL-10+ cells
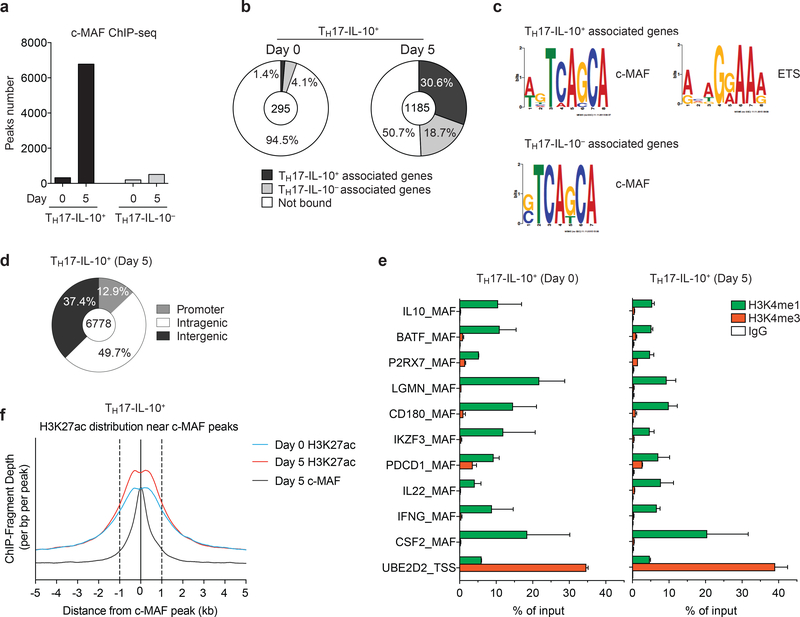
In search for a mechanism that may control the immunoregulatory and tissue-residency transcriptional program in Day 5-activated TH17-IL-10+ cells, we investigated the role of c- MAF that is selectively upregulated in these cells. We immunoprecipitated c-MAF in Day 0 and Day 5 TH17-IL-10+ and TH17-IL-10- cells and performed deep sequencing (ChIP-seq). DNA binding of c-MAF was very limited in Day 0 TH17-IL-10- cells and TH17-IL-10+ and Day 5 TH17-IL-10- cells (192, 323 and 509 peaks, respectively) (Fig. 6a). In contrast, c-MAF bound extensively to DNA in Day 5 TH17-IL-10+ cells (6,778 peaks, MACS P-value < 10-10). DNA binding motif analysis on the immunoprecipitated sequences revealed MAF as top scored TF binding site (Supplementary Fig. 5a), supporting the quality of ChIP-seq. In addition, the analysis recognized binding motifs of known (AP-1, ETS, NFAT) and novel (RUNX, TCF) interactors, which potentially bind with c-MAF to chromatin in a cooperative manner.
Figure 6. c-MAF extensively binds bona fide enhancers associated to immune-related genes in TH17-IL-10+ cells.
a. Number of c-MAF peaks found in Day 0-resting and Day 5- activated TH17-IL-10+ and TH17-IL-10- cells, as assessed by ChIP-seq (MACS P-value ≤ 10 × 10−10). b. Pie chart showing the portion of differentially expressed c-MAF-bound genes in Day 0 and Day 5 TH17-IL-10+ cells. The portion of genes associated with TH17-IL-10+ or TH17-IL-10- is shown in black and grey, respectively; c-MAF not bound genes are shown in white. c. Position weight matrix (PWM) of DNA motives in TH17-IL-10+ and TH17-IL-10- associated genes. d. Genomic distribution of c-MAF peaks; promoters were defined as a −2.5 kb to +1 kb window around gene TSSs. e. ChIP-qPCR analysis of H3K4me1 and H3K4me3 levels on a selected number of c-MAF bound regions in Day 0 and Day 5 TH17-IL-10+ cells (mean + s.e.m.; n = 2). Each region is named according to the associated gene. f. H3K27ac distribution in a 10 kb window around c-MAF peaks, in Day 0 or Day 5 TH17-IL-10+ cells. Data represent the average of two independent experiments.
We then assigned each c-MAF peak to the genes located within 185 kb, a median size of genomic interaction domains31. One or two genes were assigned to 2,833 and 3,275 c- MAF peaks, respectively, identifying a total of 5,078 genes, representing about 21% of all transcripts identified in Day 5 TH17-IL-10+ cells. Gene ontology analysis on the associated genes revealed that c-MAF preferentially binds in proximity to genes involved in regulation of lymphocyte activation and in immune response (Supplementary Fig. 5b). Most of the validated immunoregulatory and tissue residency-associated genes (e.g. IL10, CTLA4, PDCD1, IKZF3, PRDM1, CD69, and CXCR6) had one or more c-MAF peaks within the analyzed genomic window (Supplementary Table 2 and Supplementary Fig. 6). By matching the genomic c-MAF binding pattern with the transcriptional profile, we found that about 50% of the genes differentially expressed between Day 5 TH17-IL-10+ and TH17-IL- 10- cells (584 out of 1,185) were associated to a c-MAF peak, while only 5.4% of the genes differentially expressed in Day 0 cells (16 out of 295) were associated to a c-MAF peak (Fig. 6b). Among the 584 c-MAF-associated genes, 362 were more expressed in Day 5 TH17-IL- 10+ cells, where c-MAF protein is more abundant and extensively binds chromatin. However, the remaining 222 genes were more transcribed in Day 5 TH17-IL-10- cells (Fig. 6b). The analysis of the immunoprecipitated sequences revealed an enrichment of ETS binding sites flanking (± 250 bp window) c-MAF peaks in proximity of genes more expressed in TH17-IL- 10+ but not in proximity of genes more expressed in TH17-IL-10- cells (Fig. 6c).
Most (87.1%) of c-MAF binding sites in Day 5 TH17-IL-10+ cells were localized outside gene promoters (Fig. 6d). To test the possibility that these sites may represent enhancers, we performed ChIP experiments targeting histone modifications associated with enhancer elements and analyzed a subset of c-MAF bound regions. All the regions tested by ChIP-qPCR were associated with a high content of H3K4me1 and low H3K4me3 (Fig. 6e), which are characteristics of enhancer regions32,33. In this context, c-MAF exploits a preexisting chromatin landscape since these bona fide enhancers were present in both Day 0 and Day 5 TH17-IL-10+ cells. Moreover, analysis of H3K27ac ChIP-seq in Day 0 and Day 5 TH17-IL-10+ cells demonstrated that c-MAF binding occurred at active chromatin regions and that H3K27ac abundance in these regions was maximal in Day 5 cells (Fig. 6f). With this analysis, we also identified a novel putative enhancer about 12 kb upstream the IL10 TSS, that is conserved between human and mouse34 (Supplementary Fig. 5c, d). Furthermore, the activation status of this enhancer, monitored by H3K27ac and H3K27me3 modification35, reflected IL10 expression in TH17-IL-10+ cells (Supplementary Fig. 5e). Collectively these data indicate that c-MAF, by binding to putative enhancers, modulates the immunoregulatory and tissue-residency transcriptional program in recently activated TH17-IL-10+ cells, behaving mostly as a transcriptional co-activator, but acting also as a transcriptional co-repressor in a context-dependent manner.
c-MAF promotes the transcriptional program in TH17-IL-10+ cells
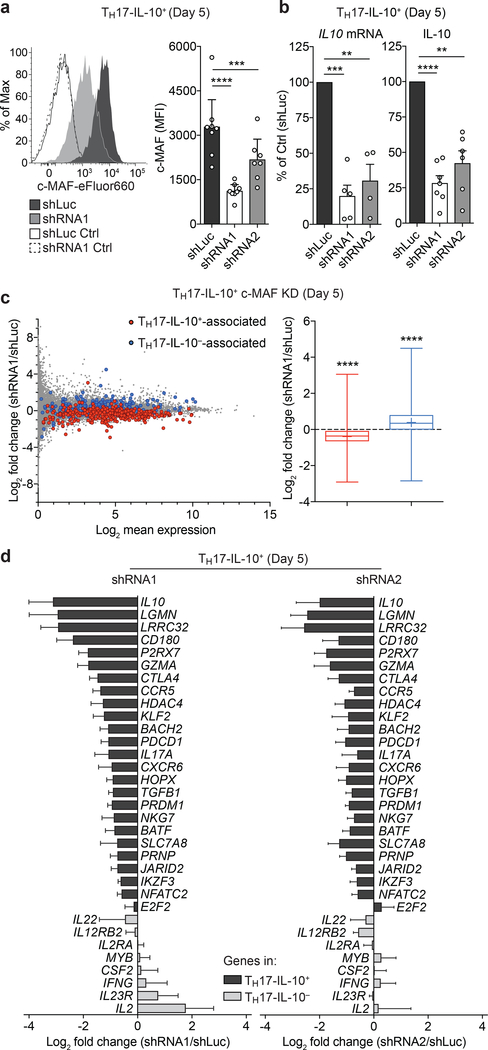
To directly assess the role of c-MAF in the control of the immunoregulatory and tissue-residency transcriptional program in TH17-IL-10+ cells, we tested the effect of c-MAF depletion in Day 5-activated TH17-IL-10+ cells. As expected, depletion of c-MAF by shRNA resulted in significant reduction of IL-10 expression at mRNA and protein levels (Fig. 7a,b). A global analysis on c-MAF depleted cells by RNA-seq showed that TH17-IL-10+-associated and TH17-IL-10--associated genes segregated in two groups (Fig. 7c, left) that were down-regulated and up-regulated, respectively (Fig. 7c, right). These data show that c-MAF is essential for the transcription of some TH17-IL-10+-associated genes (27 genes, 24 down-regulated and 3 upregulated, with fold change ≥ 2), including relevant immunoregulatory and tissue-residency genes, such as IL10, LGMN, LRRC32 and CCR9, and modulates the expression of a consistent proportion of the transcriptional program in Day 5 TH17-IL-10+ (110 genes, 84 down-regulated and 26 upregulated, with fold change ≥ 1.33). In addition, we validated the altered expression of selected TH17-IL-10+ and TH17-IL-10−-associated genes in c-MAF-depleted TH17-IL-10+ cells by qPCR in two separate biological replicates and through the use of an independent short hairpin RNA (shRNA). The expression of most of the TH17-IL-10+-associated genes tested was impaired upon c-MAF knock-down, including genes that did not fulfil the selection criteria applied to the RNA-seq analysis, while transcription of some TH17-IL-10--associated genes was up-regulated (Fig. 7d).
Figure 7. c-MAF promotes the immunoregulatory and tissue-residency transcriptional program in Day 5-activated TH17-IL-10+ cells.
a. c-MAF depletion in Day 5-activated TH17-IL-10+ clones by shRNAs. Left panel depicts a representative c-MAF staining in TH17- IL-10+ cells transduced with control shRNA (shLuc) or c-MAF-specific shRNAs (shRNA1). Right panel shows the shRNA-mediated downregulation of c-MAF levels in TH17-IL-10+ clones, as detected by intracellular staining. Two independent c-MAF-targeting shRNAs (shRNAl and shRNA2) were used. Data are represented as mean + 95% c.i., with each dot indicating a T cell clone pool (shLuc and shRNA1 n = 8, shRNA2 n = 7). b. IL-10 expression as determined by qPCR (left panel, mean + s.e.m.) and intracellular cytokine staining (ICCS, right panel, mean + 95% c.i.) in TH17-IL-10+ clones transduced with control shRNA (shLuc) or c-MAF-targeting shRNA1 and shRNA2. Each dot indicates a T cell clone pool (qPCR: shRNA1 n = 5, shRNA2 n = 4, ICCS: shRNA1 n = 7, shRNA2 n = 6). c. MA plots of RNA-seq analysis of Day 5 c-MAF-depleted TH17-IL-10+, after 2 h stimulation with CD3/CD28 antibodies (left panel). Genes associated with TH17-IL-10+ or TH17-IL-10- are highlighted in red and blue, respectively. Changes in expression of genes associated with TH17-IL-10+ and TH17-IL-10- upon c-MAF depletion are summarized in a box (interquartiles, with a line indicating the median value) and whiskers (min to max values) plot (right panel, mean indicated as a “+”). Data represent the average of two independent experiments. d. Expression of c-MAF-dependent genes in Day 5 c-MAF-depleted TH17-IL10+ cells, as measured by qPCR. Black and grey bars indicate genes associated with TH17-IL10+ or TH17-IL10- cells, respectively. Shown is the average log2 fold change of the two indicated shRNAs compared to the shLuc control (mean + s.e.m.; n = 4). **P < 0.01; ***P < 0.001; ****P < 0.0001, as determined by ratio paired t test (a), paired t test (b) and Wilcoxon matched-pairs signed rank test (c).
We next asked whether the selected set of genes associated with TH17-IL-10+ or TH17-IL-10- cells would be affected by ectopic expression of c-MAF in TH17-IL-10- cells. Over-expression of the isoform c-MAF-b, but not the isoform c-MAF-a, led to a modest up- regulation of IL-10 production (Supplementary Fig. 7a,b). The different outcome of c- MAF-a and c-MAF-b may result from differences in protein levels or stability or in interaction with dimerization partners. Addition of IL-27 increased IL-10 production in c- MAF-b overexpressing TH17-IL-10- cells (Supplementary Fig. 7c), suggesting it can synergize in the induction of IL-10 expression. Other TH17-IL-10+-associated genes, such as LGMN, SLC7A8 and CXCR6, were also up-regulated upon c-MAF-a or c-MAF-b overexpression, while other genes associated with TH17-IL-10- cells, such as IFNG, IL22, CSF2, and IL12RB2, were downregulated, both at mRNA and protein level (Supplementary Fig. 7e,f).
Overall, the above analyses indicate that c-MAF is largely necessary to set an immunoregulatory and tissue residency program in TH17-IL-10+ cells but it is not sufficient to induce the same transcriptional program in TH17-IL-10- cells. Day 5-activated TH17-IL- 10- lymphocytes failed to up-regulate but expressed c-MAF; however, its expression is not accompanied by binding of the transcription factor to chromatin. This finding may be in part due to reduced chromatin accessibility of c-MAF binding sites in TH17-IL-10- cells compared to TH17-IL-10+ cells (Supplementary Fig. 8a). Furthermore, our data indicating that c-MAF binds to a set of pre-existing enhancers in Day 5 TH17-IL-10+ cells, suggest that it lacks pioneering factor activity. Therefore, c-MAF forced expression cannot be sufficient per se to overcome the steric hindrance imposed by chromatin in TH17-IL-10− cells.
DNA motif enrichment analysis of H3K27ac subset-specific regions (Supplementary Fig. 8b) highlights the possibility that the antagonistic expression of several transcription factor pairs in Day 5-activated TH17-IL-10+ and TH17-IL-10- cells may regulate the transcriptional programs of immunoregulatory and pro-inflammatory TH17 cells (Supplementary Fig. 8c-e). Indeed, the lack of c-MAF binding may also result from the high expression of FOS proteins (FOS and FOSB) in Day 5 TH17-IL-10- cells (Supplementary Fig. 8e). Both c-MAF and FOS transcription factors can form heterodimers with JUN36,37 and have partially overlapping DNA binding consensus motifs (Supplementary Fig. 8d), suggesting they may compete for dimerization partners and binding to the same genomic loci. Finally, c-MAF may need to cooperate with other transcription factors for optimal gene expression; the reduced expression of IKZF3 and BATF in Day 5 TH17-IL-10- cells (Supplementary Fig. 8e) may explain why c-MAF is not sufficient to fully convert them into TH17-IL-10+ cells.
Discussion
Circulating memory T cells are recruited to inflamed tissues where, upon antigen recognition, they rapidly produce inflammatory cytokines, such as IFN-γ and IL-17. Previous in vivo studies in mice have shown that this initial stimulation leads to dynamic changes that modulate the capacity of the T cell to respond to further antigenic stimulation38. In this study, we used an in vitro system that mimics the temporally distinct phases of T cell activation to reveal a new level of heterogeneity in human TH17 cells that becomes evident when recently activated cells respond to a second antigenic stimulation. A TH17 subset maintains pro-inflammatory activity and acquires recirculating properties, while another subset acquires anti-inflammatory, immunoregulatory and tissue-residency properties. The latter transcriptional program is substantially modulated by the upregulation of c-MAF that binds to a large number of non-promoter regions with enhancer-like features, acting as transcriptional co-activator or co-repressor in a context dependent manner.
The distinctive feature of immunoregulatory TH17 cells is their ability to produce IL-10, a potent anti-inflammatory cytokine that restrains inflammatory responses thus preventing damage to the host4. Indeed, IL10-deficient mice develop spontaneous intestinal inflammation, and loss-of-function mutations in the IL-10 receptor lead to infantile onset inflammatory bowel disease in humans39,40. It is interesting to compare and contrast the immediate inflammatory response, which is characterized by the production of IL-17 that occurs a few hours after antigenic stimulation, with the late anti-inflammatory response characterized by the production of IL-10. Previous studies have shown that in human IL-10- producing TH1 cells, the IL10 locus is highly methylated and that these cells lack memory for IL-10 expression41. Our study extends these findings by showing that in resting TH17 cells the IL10 TSS is associated with high H3K27me3 levels, indicating a repressed chromatin. However, we show that immunoregulatory TH17 cells maintain memory of IL-10 expression which correlates with a selective upregulation of c-MAF expression. Conversely, the lack of IL-10 production by pro-inflammatory TH17 cells is associated with the inability of these cells to upregulate c-MAF upon stimulation. Recently activated TH17-IL-10+ cells differ from the TH17-derived Tr1-like cells (exTH17) that have been recently described42, as they set in motion a temporally defined immunoregulatory program that is independent from AHR activation and at the end of which the cells recover the original ability to produce IL-17A.
An intriguing finding of our study is that recently activated TH17-IL-10+ cells acquire markers characteristic of tissue-resident memory T (TRM) cells, a lymphocyte memory subset that resides in non-lymphoid tissues2. TRM cells have been best defined in the CD8+ compartment2, but recent evidence indicates that, upon local antigen recognition, also CD4+ T cells can establish transient or permanent residency in non-lymphoid tissues3,43. However, TH17-IL-10+ cells also upregulate KLF2 and its target gene S1PR1 whose downregulation in TRM cells is important to promote T cell retention in tissues2. It is possible that activated TH17-IL-10+ cells may represent TRM-committed precursors that require, in order to fully differentiate to TRM, additional cues from the tissues that are absent in our in vitro system. Although more work will be required to define the function of TH17-IL-10+ cells in vivo, it is tempting to speculate that, after the acute inflammatory response, c-MAF expressing TH17 cells may differentiate to TRM-like cells and, by extending their residency in tissues, contribute to the resolution of inflammation through production of IL-10, induction of lipid mediators and tissue repair, either directly or through the release of cytokines that induce monocyte differentiation to M2 macrophages.
Our study implicates c-MAF in the modulation of a broad immunoregulatory and tissue-residency program in TH17-IL-10+ cells and is coherent with its recently described role in mediating iTreg-dependent immunological tolerance in the gut44. This conclusion is supported by the finding that c-MAF binds in proximity of a relevant proportion of genes that are differentially expressed in recently activated TH17 cell subsets and by the finding that the TH17-IL-10+ transcriptional signature was impaired upon c-MAF depletion. Notably, c-MAF does not behave as a pioneering transcription factor, but it exploits a pre-existing enhancer landscape, suggesting that cooperation with other transcription factors is required. Cooperative binding to regulatory genomic regions may also account for the different transcriptional outcome of c-MAF binding in proximity to immunoregulatory or proinflammatory genes where it acts as a co-activator or co-repressor, respectively. In this context, c-MAF shares several features with FOXP3 which has been recently shown to bind pre-existing active enhancer elements45 in proximity to genes that are over-or under-expressed in Treg cells compared to memory T cells and whose transcriptional activity depends on the interacting partners46. The identification of the transcriptional regulators cooperating with c-MAF would allow a deeper understanding of the molecular mechanisms underlying the heterogeneity of TH17 cells.
Our findings in human memory TH17 cells are in line with the established role of c- MAF in controlling IL-10 production in various TH subsets9,10,11. A recent study by O’Garra and colleagues47 showed that, in mouse TH1 and TH2 disease models, c-Maf deletion leading to reduced IL-10 production is accompanied by enhanced inflammation. In contrast, in the TH17 EAE model, c-Maf deletion resulted in decreased disease severity due to the ability of c-MAF to negatively regulate Il2, resulting in increased TH17 and decreased Treg differentiation. We also found that c-MAF negatively regulates IL2 expression in already differentiated human memory TH17-IL-10+ cells, an effect which is however transient and dependent on the T cell activation state. Thus, c-MAF may regulate T cell function at priming as well as at the effector phase. Together, these data are consistent with positive, negative and context-specific effects of c-MAF on T cell differentiation and function.
While the main focus of our study was on the characterization of immunoregulatory TH17-IL-10+ cells, our findings also define novel properties of pro-inflammatory TH17 cells. Upon TCR stimulation, pro-inflammatory TH17 cells express high levels of CCR7, which may drive these cells out of inflamed tissues into lymphatics or to tertiary lymphoid tissues28,48. In addition, these cells induce polarization of monocytes to M1 macrophages that, by producing IL-1β and TNF, can suppress IL-10 production by T cells8,20, potentially enabling a self-sustaining paracrine circuit that could establish and maintain an inflammatory environment.
While we show that the pro-inflammatory and immunoregulatory phenotypes are a stable feature of two circulating subsets of memory TH17 cells, we and others have shown that these cells are not fixed and maintain a considerable level of plasticity, at least in vitro. IL-10-producing TH17 cells can be converted to pro-inflammatory TH17 cells when exposed to IL-1β and TNF that irreversibly switch off IL-10 production, while increasing production of IFN-γ and GM-CSF8,49. IL-1β exposure impairs also the transcription of other genes associated with TH17-IL-10+ cells, while IL-27 can promote, in a reciprocal fashion, their expression in TH17-IL-10- cells. Considered the strong enrichment of the TH17-IL-10-- associated transcriptional signature in autoimmune diseases, a deeper understanding of the stimuli and signaling pathways that may induce the immunoregulatory program in pro-inflammatory TH17 cells could provide a new strategy for therapeutic interventions in autoimmune pathologies.
Several of the genes which we found highly expressed in TH17-IL-10+ cells have been associated with the “non-pathogenic” phenotype of mouse TH17 cells, while those highly expressed in TH17-IL-10- cells have been associated with “pathogenic” mouse TH17 cells50-52. TH17 cells are known to play an important role in host defense against fungi and extracellular bacteria and in the maintenance of gut homeostasis53–56, but they are also involved in tissue inflammation and autoimmune diseases, such as multiple sclerosis, inflammatory bowel disease (IBD), psoriasis and rheumatoid arthritis57–62. The pathogenic role of TH17 and other IL-17-producing cells remains to be fully elucidated to help explaining why anti-IL-17-based therapies are efficacious in some autoimmune diseases but ineffective or even detrimental in others63. The identification of two TH17 subsets with programmed opposing post-activation fates provides a new paradigm for investigating the role of these cells in physiology and immunopathology.
Online Methods
Blood samples and cell sorting.
Blood from healthy donors was obtained from the Swiss Blood Donation Centres of Basel and Lugano, and used in compliance with the Federal Office of Public Health (authorization n. A000197/2 to F.S.). PBMCs were isolated from buffy-coat by Ficoll-Paque Plus (GE Healthcare Life Sciences) gradient and CD4+ T cells were enriched by positive selection with magnetic microbeads (Miltenyi Biotec) to an average of 97.5 % purity. Memory T helper cells, including CD45RA-CCR7+ central memory T cells and CD45RA-CCR7- and CD45RA+CCR7- effector memory T cells, were then FACS sorted after exclusion of CD45RA+CCR7+ naive T cells and CD8+ CD14+ CD19+ CD56+ CD25+ cells. The following antibodies were used for FACS-based sorting: anti- CD45RA-Qdot655 (Life Technologies; clone: MEM-56); anti-CCR7-BV421 (BioLegend; clone: G043H7); anti-CCR6-PE (BD Biosciences; clone: 11A9) or anti-CCR6-BV605 (BioLegend; clone: G034E3); anti-CCR4-PE-Cy7 (BD Biosciences; clone: 1G1); anti- CXCR3-PE-Cy5 or anti-CXCR3-APC (BD Biosciences; clone: 1C6); anti-CD8-FITC or anti- CD8-PE-Cy5 (Beckman Coulter; clone: B9.11); anti-CD25-FITC or anti-CD25-PE-Cy5 (Beckman Coulter; clone: B1.49.9); anti-CD14-FITC or anti-CD14-PE-Cy5 (Beckman Coulter; clone: RMO52); anti-CD19-FITC (BD Biosciences, clone: HIB19) or anti-CD19- PE-Cy5 (Beckman Coulter; clone: J3–119); CD56-PE-Cy5 (Beckman Coulter; clone: N901). Viable IL-17-producing T cells were FACS sorted using the cytokine secretion assay (Miltenyi Biotec) following 3 h of phorbol 12-myristate 13-acetate (0.2 μΜ) and Ionomycin (1 μg/ml) (both from Sigma-Aldrich) stimulation, according to the manufacturer’s instructions. Cell sorting was performed using a FACSAria III device (BD Biosciences).
T cell culture.
T cells were cultured in RPMI-1640 medium supplemented with 1% (v/v) glutaMAX™-I, 1% (v/v) non-essential amino acids, 1 mM sodium pyruvate, 50 μM β-mercaptoethanol, penicillin (50 U/ml), streptomycin (50 μg/ml), 1% (v/v) kanamycin (all from Gibco, Life Technologies) and 5% (v/v) human serum (Swiss Blood Center Basel) (hereafter referred as complete medium). 500 U/ml IL-2 (produced in house from IL-2T6 culture supernatants) was added to the culture medium for long term cultures. Single T cell clones were isolated by limiting dilution in 384-well plates (0.65 cells/well) and expanded with allogeneic irradiated (45 Gy) feeders (2.5 χ 104/well) and phytohemagglutinin (PHA, 1 μg/ml; Remel) in IL-2- (500 U/ml) containing complete medium. Where indicated, T cells were stimulated with plate-bound antibodies to CD3 (156 ng/cm2, clone: TR66) and CD28 (156 ng/cm2 clone: CD28.2, BD Biosciences). T cell clones were analyzed in the resting state, defined as Day 0 (at least 21 days after cloning or re-stimulation) and in the recently activated state after stimulation in CD3 and CD28 antibody-coated plates for 48 h, and additional 3 days culture in uncoated plates, defined as Day 5. Day 0-resting and Day 5- activated T cell clones were stimulated for 5 h with PMA and Ionomycin (for protein analysis) or for 2 h with plate-bound CD3 and CD28 antibodies (for RNA analysis). Where indicated, T cells were stimulated in presence of IL-1β (BioLegend, cat. num. 579402), IL-27 (R&D Systems, cat. num. 2526-IL-010), TGF-β (R&D Systems, cat. num. 240-B-002), FICZ (Enzo Life Sciences, cat. num. BML-GR206–0100) or CH-223191 (Sigma-Aldrich, cat. num. C8124).
Monocyte differentiation.
CD14+ monocytes were magnetic-sorted from PBMCs. 105 monocytes were co-cultured with 5 X 104 allogeneic Day 5-activated TH17-IL-10+ or TH17- IL-10- cells in 100 μl complete medium + 100 μl Day 5-activated TH17 cell supernatant in flat-bottom 96-well plate for 24 h or 48 h. Alternatively, 106 CD14+ monocytes were cultured in the presence of supernatants from Day 5-activated TH17-IL-10+ or TH17-IL-10- cells (500 μl complete medium + 500 μl supernatant) in 24 well plate for 7 days. After 48 h 500 μl of TH17 cell-conditioned medium was added to the culture and 700 μl supernatant was replaced every 2 days with fresh supernatant. On day 7, monocytes were washed twice to remove nonadherent cells and stimulated with 10 ng/ml LPS (TLRgrade™, Alexis Biochemicals) for 4 h and 24 h. In parallel, CD14+ monocytes were in vitro polarized to M1 and M2 macrophages by recombinant cytokines. M1, GM-CSF 5 ng/ml + IFN-γ 50 ng/ml; M2, M-CSF 20 ng/ml + IL-4 20 ng/ml + IL-10 20 ng/ml + TGF-β 4 ng/ml.
Flow cytometry.
The expression of surface markers was analyzed by staining T cells for 15 min at 20 °C plus 5 min on ice in PBS supplemented with 0.5% (v/v) human serum. Monocytes were stained in PBS supplemented with 3% (v/v) heat-inactivated human serum and 2 mM EDTA. For intracellular cytokine staining (ICCS), cells were stimulated for 5 h with PMA (0.2 μM) and Ionomycin (1 μg/ml) in the presence of brefeldin A (BFA, 10 μg/ml) (all from Sigma-Aldrich) for the last 2.5 h. To exclude dead cells from the analysis, cells were stained prior to fixation with the LIVE/DEAD® fixable aqua or near-IR fluorescent dyes (Life Technologies). Cells were then fixed and permeabilized with Cytofix/Cytoperm (BD Biosciences) according to manufacturer’s instructions. The following conjugated antibodies were used for analysis: anti-CD4-AlexaFluor700 (BioLegend, clone: OKT4); anti- CD14-FITC or anti-CD14-PE-Cy5 (Beckman Coulter, clone: RMO52); anti-CTLA-4-APC (BioLegend, clone: L3D10); anti-PD-1-BV785 (BioLegend, clone: EH12.2H7); anti-CD40- PE (BioLegend, clone: 5C3); anti-CD163-BV421 (BioLegend, clone: GHI/61); anti-CD206- AlexaFluor488 (BioLegend, clone: 15–2); anti-HLA-DR-PE-Cy7 (BD Bioscience, clone: G46–6); anti-CD186 (CXCR6)-PECy7 (BioLegend, clone: K041E5); anti-CD69-PE (Beckman Coulter, clone TP1.55.3); anti-CD25-FITC (Beckman Coulter, clone: B1.49.9) or anti-CD25-BV785 (BioLegend, clone BC96); anti-CCR7-BV421 (BioLegend; clone: G043H7); anti-IL-17A-eF660 or anti-IL-17A-eF450 (eBioscience, clone: eBio64DEC17); anti-IFN-γ-FITC or anti-IFN^-PE or anti-IFN-γ-APC (BD Biosciences, clone: B27); anti-IL- 4-PE (BD Biosciences, clone: 8D4–8) or anti-IL-4-APC (BD Biosciences, clone: MP4– 25D2); anti-IL-22-PerCP-eF710 (eBioscience, clone: 22URTI); anti-IL-10-PE (BD Biosciences, clone: JES3–19F1) or anti-IL-10-PE-Cy7 (BioLegend, clone: JES3–9D7); anti- GM-CSF-PE (R&D Systems, clone: 6804); anti-IL-6-AlexaFluor700 (eBioscience, clone: MQ2–13A5); anti-TNF-PE (BD Biosciences, clone: MAb11). For c-MAF, FOXP3 and BLIMP-1 staining, cells were fixed (30 min) and permeabilized (30 min) at 20°C using the transcription factor staining buffer set (eBioscience), according to manufacturer’s instructions. Transcription factors staining was performed in permeabilization buffer (30 min at 20 °C) using the following antibodies: anti-c-MAF-eFluor660 (eBioscience, clone: sym0F1), anti-FOXP3-FITC (eBioscience, clone PCH101); anti-BLIMP-1-PE (SantaCruz Biotechnology, clone C-21). Stained cells were analyzed using a BD FACSCanto™ I or a BD LSRFortessa™ (BD Biosciences) and flow cytometry data were analyzed with FlowJo software (Tree Star).
Gene expression analysis.
Total RNA was extracted from at least 106 cells using the E.Z.N.A.® Total RNA Kit I (Omega Bio-tek), according to manufacturer’s instructions. The eluted RNA was incubated at 65 °C for 10 min to resolve secondary structures prior to the cDNA synthesis and quantified by NanoDrop™ 2000c (Thermo Scientific). cDNA was synthesized using the qScript™ cDNA SuperMix (Quanta Biosciences) according to manufacturer’s instructions. Relative gene expression was measured by qPCR with the KAPA SYBR® FAST qPCR master mix (Kapa Biosystems) on an ABI PRISM 7900HT Fast System (Applied Biosystems) and normalized to TATA-binding protein (TBP). Primer specificity was validated by single-peak dissociation curve profile.
RNA-sequencing.
RNA sequencing on TH17-IL-10+ and TH17-IL-10- clone pools was conducted by the Genomics Platform of the Broad Institute (Cambridge, USA) in compliance with the institutional review board of Brigham and Women’s Hospital (IRB protocol 2014P000124/BWH to H.L.W.). Briefly, 250 ng for each sample of total RNA isolated from T cell clone pools with the E.Z.N.A.® Total RNA kit I (Omega Bio-tek) was submitted to the Genomics Platform for high coverage (50 million paired-end reads), Tru-Seq standard, strand-specific RNA-sequencing. RNA-seq reads generated by the Genomics Platform were quality checked and processed with Tuxedo RNA-Seq pipeline64 to align reads to human genome assembly GRCh37 and quantify transcripts. Two different gene data sets were extracted from the RNA-seq data that referred to 63,809 genes: a protein-coding data set and a lncRNA data set. GENCODE v19 annotation was used to filter the data (gencode.v19.chr_patch_hapl_scaff.annotation.gtf available on http://www.gencodegenes.org/). For the protein coding set, we excluded from the list, those genes annotated as mitochondrial genes, genes encoding TCRs, pseudogenes and non-protein coding transcripts. This resulted in a protein coding set of 23,744 genes. Comparison of gene expression was performed by analysis of fragments per kilobase of exon per million fragments mapped (FPKM) values. Differentially expressed genes were defined using the ratio of detected fold changes (IL-10+/IL-10-) for two donors. Genes with a mean ratio ≥ 2 or ≤0.5 were considered (ratio ≥ 1.8 or ≤ 0.555 in the two independent experiments). From this filtered list, only the genes with a mean FPKM value for IL-10+ or IL-10- ≥ 2 were considered. To select specifically expressed lncRNA genes, we first select the transcripts annotated as “lincRNA”, “sense_intronic”, “antisense” and “nonsense_ mediated_decay”. In total 14,035 transcripts were identified as lncRNA elements. As previously described, only the differentially expressed transcripts were selected. All measurements were log2 transformed and a visual representation of differential gene expression was generated with MA-plots. Genes with a log2 mean ratio between −10 and 10 and mean expression above 1 are shown (10,657 “Day 0”, 10,626 “Day 0 + 2h”, 11,081 “Day 5” and 10,899 “Day 5 + 2h”. Each lncRNA was associated to the proximal protein-coding gene, regardless of its transcriptional sense, to perform a Gene Ontology (GO) enrichment analysis for biological process terms. Metacore™ (Thomson Reuters) software was used to perform the GO analyses. The top 20 (or top 10) ranked GO biological process terms were selected according to their p-value. GO terms were then clustered based on their semantic similarity analysis by G-SESAME (gene semantic similarity analysis and measurement) tool18. This analysis results in a symmetric matrix in which each value represents a score for similarity between GO term pairs. To group all GO terms with the common GO parent, the G-SESAME semantic similarity matrices were subjected to a hierarchical clustering using the heatmap.2 function from ‘gplots’ package in R (Euclidean metric and complete aggregation method). RNA sequencing on c-MAF-depleted TH17-IL-10+ cells was conducted by IGA Technology Services (Udine, Italy), following the same protocols and analysis pipeline.
Gene set enrichment analysis (GSEA).
Gene expression datasets were downloaded from the Gene Expression Omnibus website (http://www.ncbi.nlm.nih.gov/geo/). To analyze and compare the expression of single genes, data were median normalized and log2 transformed. When gene expression was covered by multiple microarray probes, data were collapsed to the maximum probe. For the generation of consistent gene identifiers across datasets the DAVID65 Gene ID Conversion Tool was used (https://david.ncifcrf.gov/conversion.jsp). The GSEA 3.0 analysis tool66 provided by the Broad Institute (http://software.broadinstitute.org/gsea/) was used for the analysis of enrichments of TH17 cell-associated gene signatures and leading edge genes. The following parameters were set to run the enrichment test: 1000 phenotype permutations, weighted enrichment statistic, signal to noise ranked genes, collapsing of probe sets to the maximum probe. GSEA enrichment results were reported as normalized enrichment score and familywise enrichment (FWER) P-value. FWER P-values smaller than 0.05 were considered significant. For processing and visualization of gene expression data GENE-E (https://software broadinstitute.org/GENE-E/) and Morpheus (https://software.broadinstitute.org/morpheus/) analysis tools were used. The following publicly available datasets were analyzed in this study: GSE57945 (Core Ileal Transcriptome in Paediatric Crohn Disease); GSE1402 (Juvenile rheumatoid arthritis expression profiles in mononuclear cells).
Chromatin immunoprecipitation (ChIP) and ChIP-seq.
ChIP was performed as previously described67. Briefly, 2–4 X 106 cells for histone modifications and 20–50 X 106 cells for TF ChIPs were fixed in 1% formaldehyde (Sigma-Aldrich) for exactly 10 min at 20 °C under rotation. Crosslinking reaction was quenched by the addition of 125 mM Tris-HCl pH 7.6 for 5 min at 20 °C. Cells were washed 3 times with ice-cold PBS. The cell pellet was then resuspended in 300 μl (histone modifications) or 1500 μl (TFs) RIPA buffer (without Triton X-100) supplemented with protease inhibitors (Halt™ Protease Inhibitor Cocktail, Thermo Scientific) and frozen at −80 °C. Chromatin was sheared with a Bioruptor® Plus bath sonicator (Diagenode) applying the following settings: 5 X 10 cycles 30 sec ON and 60 sec OFF, high intensity, at 4 °C. Every 10 cycles samples were briefly vortexed and spun down. The shearing of DNA into 200- to 300-bp fragments was evaluated by electrophoresis on a 1.5% agarose gel. Prior to IP, 1% Triton X-100 was added to the sample and an aliquot was stored at −20 °C as input control. Antibody-bound Protein G dynabeads™ (Life Technologies) were added to each sample and immunoprecipitation was performed overnight at 4 °C on a rotating wheel. The day after, dynabeads were thoroughly washed and the eluted DNA was subjected to RNAse A and proteinase K treatment before overnight de-crosslinking at 65 °C. Finally, de-crosslinked DNA was purified with SPRI magnetic beads and eluted in 10 mM Tris-Cl pH 8.0. ChIP DNA was quantified by dsDNA HS assay kit (Life Technologies, cat. num. Q32854) on a Qubit 2.0 device. ChIP-seq libraries were prepared as previously described67. Illumina-compatible universal and indexed adapters and amplification primers were synthesized by Microsynth AG. Sequencing was performed by IGA Technology Services (Udine, Italy) on an Illumina HiSeq platform. About 20 and 40 million unique single-end 50 bp reads were generated for ChIP and Input samples, respectively. ChIP-seq data were analyzed with the Fish the ChIP’s automated genomic annotation tool68 and visualized through the Integrative Genomics Viewer v2.3 (https://www.broadinstitute.org/igv/). Enriched DNA motives in c-MAF ChIP-seq datasets were identified by MEME-ChIP69. c-MAF peaks were assigned by proximity to the genes located within 185 kb using the GREAT web-based software70. H3K27ac distribution in proximity to c-MAF peaks was computed with the annotatePeaks.pl command from HOMER software (v.4.9.1), using the genomic coordinates of c-MAF peaks from the human hg19 genome assembly as reference and applying the “-hist 10” and “-size 10000” options. ChIP- qPCR for ChIP-seq validation or histone modifications analyses was performed using the Fast SYBR® Green master mix (Applied Biosystems) on an ABI PRISM 7900HT Fast System (Applied Biosystems). Data were normalized to ChIP Input (% of Input). The following ChIP grade antibodies were used for immunoprecipitation: anti-c-MAF (Santa Cruz Biotechnology, cat. num. sc-7866 X; M-153); anti-H3K4me1 (Abcam, cat. num. ab8895); anti-H3K4me3 (Active Motif, cat. num. 39159); anti-H3K27me3 (Millipore, cat. num. ABE44); anti-H3K27ac (Abcam, cat. num. ab4729).
Identification of nucleosome-free regions.
Nucleosome-free regions (NFR) have been called from H3K27ac ChIP-seq datasets using the findPeaks command from Homer (Hypergeometric Optimization of Motif EnRichment) (version 4.9.1) applying the “-nfr” and “-style histone” options71. NFRs that overlapped or resided within 7.5 kb from H3K27ac peaks were identified with Bedtools (version 2.25.0) and selected for further analyses.
Lentiviral transduction.
The pCDH-EF1-MCS-T2A-copGFP vector (CD526A-1) for ectopic gene expression was purchased from SBI Systems Biosciences; the pALPS-GFP- miR30 vector for shRNA-mediated knock-down was kindly provided by S. Monticelli (IRB, Bellinzona). Lentiviral particles were produced by transiently co-transfecting HEK293FT cells with the shRNA- or gene-transfer vector together with the psPAX2 (Addgene plasmid 12260) and pMD2.G (Addgene plasmid 12259) packaging vectors, as previously described72. Briefly, HEK293FT cells were co-transfected with a cocktail of transfer vector, psPAX, and pMD2.G at a 4:3:1 ratio in Opti-MEM, using polyethylenimine (PEI). Viral particles were harvested 72 h post-transfection and concentrated by centrifugation on a sucrose gradient. Concentrated lentivirus was added to T clone pools concomitantly to polyclonal stimulation.
Statistical analysis.
GraphPad Prism (version 7) and Microsoft Excel (for Mac 2011) were used to perform experimental statistical analysis. The statistical significance of differences between data groups was determined by two-tailed paired t test or ratio paired t test at the 95% confidence level, unless differently indicated. Exact n values and error bars used are provided in the figure legends.
Reporting Summary.
Additional technical details and further information on experimental design is available in the Life Science Reporting Summary.
Data availability.
The data that support the findings of this study are available from the corresponding authors upon request.
Supplementary Material
Acknowledgments
We thank S. Monticelli for providing reagents and for critical reading of the manuscript and I. Barozzi for discussion on ChIP-seq analyses. This work was supported by the European Research Council (grant 323183 PREDICT to F.S.), the Swiss National Science Foundation (grants 149475 and 170213 to F.S.) and the National Institutes of Health (P01 grant NS076410 to H.L.W.). F.S. and the Institute for Research in Biomedicine are supported by the Helmut Horten Foundation.
Footnotes
Competing Financial Interests Statement
The authors declare no competing interests.
Accession Codes
NGS data that support the findings of this study have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE101389 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE101389).
References
- 1.Sallusto F, Lenig D, Forster R, Lipp M & Lanzavecchia A Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 401, 708–712 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Mueller SN & Mackay LK Tissue-resident memory T cells: local specialists in immune defence. Nat Rev Immunol 16, 79–89 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Park CO & Kupper TS The emerging role of resident memory T cells in protective immunity and inflammatory disease. Nat Med 21, 688–697 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ouyang W, Rutz S, Crellin NK, Valdez PA & Hymowitz SG Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu Rev Immunol 29, 71–109 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Chen L & Flies DB Molecular mechanisms of T cell co-stimulation and coinhibition. Nat Rev Immunol 13, 227–242 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Attanasio J & Wherry EJ Costimulatory and Coinhibitory Receptor Pathways in Infectious Disease. Immunity 44, 1052–1068 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baumeister SH, Freeman GJ, Dranoff G & Sharpe AH Coinhibitory Pathways in Immunotherapy for Cancer. Annu Rev Immunol 34, 539–573 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Zielinski CE et al. Pathogen-induced human TH17 cells produce IFN-gamma or IL- 10 and are regulated by IL-1beta. Nature 484, 514–518 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Saraiva M & O’Garra A The regulation of IL-10 production by immune cells. Nature Rev Immunol 10, 170–181 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Xu J et al. c-Maf regulates IL-10 expression during TH17 polarization. J Immunol 182, 6226–6236 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Apetoh L et al. The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nat Immunol 11, 854861 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pot C et al. Cutting edge: IL-27 induces the transcription factor c-Maf, cytokine IL- 21, and the costimulatory receptor ICOS that coordinately act together to promote differentiation of IL-10-producing Tr1 cells. J Immunol 183, 797–801 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao S, Liu J, Song L & Ma X The protooncogene c-Maf is an essential transcription factor for IL-10 gene expression in macrophages. J Immunol 174, 3484–3492 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mosser DM & Edwards JP Exploring the full spectrum of macrophage activation. Nature Rev Immunol 8, 958–969 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez FO & Gordon S The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000 Prime Rep 6, 13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinez FO, Gordon S, Locati M & Mantovani A Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol 177, 7303–7311 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Beyer M et al. High-resolution transcriptome of human macrophages. PloS One 7, e45466 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ranzani V et al. The long intergenic noncoding RNA landscape of human lymphocytes highlights the regulation of T cell differentiation by linc-MAF-4. Nat Immunol 16, 318–325 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang W et al. DDX5 and its associated lncRNA Rmrp modulate TH17 cell effector functions. Nature 528, 517–522 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Evans HG et al. TNF-alpha blockade induces IL-10 expression in human CD4+ T cells. Nat Commun 5, 3199 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roychoudhuri R et al. BACH2 represses effector programs to stabilize T(reg)- mediated immune homeostasis. Nature 498, 506–510 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang R et al. Expression of GARP selectively identifies activated human FOXP3+ regulatory T cells. Proc Natl Acad Sci USA 106, 13439–13444 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hahn SA et al. Soluble GARP has potent antiinflammatory and immunomodulatory impact on human CD4(+) T cells. Blood 122, 1182–1191 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Probst-Kepper M et al. GARP: a key receptor controlling FOXP3 in human regulatory T cells. J Cell Mol Med 13, 3343–3357 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schenk U et al. ATP inhibits the generation and function of regulatory T cells through the activation of purinergic P2X receptors. Sci Signal 4, ra12 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Mackay LK et al. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science 352, 459–463 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Mackay LK et al. The developmental pathway for CD103(+)CD8+ tissue-resident memory T cells of skin. Nat Immunol 14, 1294–1301 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Bromley SK, Thomas SY & Luster AD Chemokine receptor CCR7 guides T cell exit from peripheral tissues and entry into afferent lymphatics. Nat Immunol 6, 895–901 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Haberman Y et al. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J Clin Invest 124, 3617–3633 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bames MG et al. Gene expression in juvenile arthritis and spondyloarthropathy: pro-angiogenic ELR+ chemokine genes relate to course of arthritis. Rheumatology (Oxford) 43, 973–979 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Rao SS et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heintzman ND et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39, 311–318 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Heintzman ND et al. Histone modifications at human enhancers reflect global cell- type-specific gene expression. Nature 459, 108–112 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ciofani M et al. A validated regulatory network for TH17 cell specification. Cell 151, 289–303 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heinz S, Romanoski CE, Benner C & Glass CK The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol 16, 144–154 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kerppola TK & Curran T Maf and Nrl can bind to AP-1 sites and form heterodimers with Fos and Jun. Oncogene 9, 675–684 (1994). [PubMed] [Google Scholar]
- 37.Kataoka K, Noda M & Nishizawa M Maf nuclear oncoprotein recognizes sequences related to an AP-1 site and forms heterodimers with both Fos and Jun. Mol Cell Biol 14, 700–712 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Honda T et al. Tuning of antigen sensitivity by T cell receptor-dependent negative feedback controls T cell effector function in inflamed tissues. Immunity 40, 235–247 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuhn R, Lohler J, Rennick D, Rajewsky K & Muller W Interleukin-10- deficient mice develop chronic enterocolitis. Cell 75, 263–274 (1993). [DOI] [PubMed] [Google Scholar]
- 40.Glocker EO et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. New Engl J Med 361, 2033–2045 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong J et al. IL-10 is excluded from the functional cytokine memory of human CD4+ memory T lymphocytes. J Immunol 179, 2389–2396 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Gagliani N et al. TH17 cells transdifferentiate into regulatory T cells during resolution of inflammation. Nature 523, 221–225 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turner DL & Farber DL Mucosal resident memory CD4 T cells in protection and immunopathology. Front Immunol 5, 331 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu M et al. c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature 554, 373–377 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samstein RM et al. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell 151, 153–166 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kwon HK, Chen HM, Mathis D & Benoist C Different molecular complexes that mediate transcriptional induction and repression by FoxP3. Nat Immunol 18, 1238–1248 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gabrysova L et al. c-Maf controls immune responses by regulating disease-specific gene networks and repressing IL-2 in CD4(+) T cells. Nat Immunol 19, 497–507 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Forster R et al. CCR7 coordinates the primary immune response by establishing functional microenvironments in secondary lymphoid organs. Cell 99, 23–33 (1999). [DOI] [PubMed] [Google Scholar]
- 49.El-Behi M et al. The encephalitogenicity of T(H)17 cells is dependent on IL-1- and IL-23-induced production of the cytokine GM-CSF. Nat Immunol 12, 568–575 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gaublomme JT et al. Single-Cell Genomics Unveils Critical Regulators of TH17 Cell Pathogenicity. Cell 163, 1400–1412 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee Y et al. Induction and molecular signature of pathogenic TH17 cells. Nat Immunol 13, 991–999 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ghoreschi K et al. Generation of pathogenic T(H)17 cells in the absence of TGF- beta signalling. Nature 467, 967–971 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Milner JD et al. Impaired T(H)17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature 452, 773–776 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Puel A et al. Chronic mucocutaneous candidiasis in humans with inborn errors of interleukin-17 immunity. Science 332, 65–68 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raffatellu M et al. Simian immunodeficiency virus-induced mucosal interleukin-17 deficiency promotes Salmonella dissemination from the gut. Nat Med 14, 421–428 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee JS et al. Interleukin-23-Independent IL-17 Production Regulates Intestinal Epithelial Permeability. Immunity 43, 727–738 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bettelli E, Korn T, Oukka M & Kuchroo VK Induction and effector functions of T(H)17 cells. Nature 453, 1051–1057 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kebir H et al. Human TH17 lymphocytes promote blood-brain barrier disruption and central nervous system inflammation. Nat Med 13, 1173–1175 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fujino S et al. Increased expression of interleukin 17 in inflammatory bowel disease. Gut 52, 65–70 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kobayashi T et al. IL23 differentially regulates the Th1/TH17 balance in ulcerative colitis and Crohn’s disease. Gut 57, 1682–1689 (2008). [DOI] [PubMed] [Google Scholar]
- 61.Wilson NJ et al. Development, cytokine profile and function of human interleukin 17-producing helper T cells. Nat Immunol 8, 950–957 (2007). [DOI] [PubMed] [Google Scholar]
- 62.Nakae S, Nambu A, Sudo K & Iwakura Y Suppression of immune induction of collagen-induced arthritis in IL-17-deficient mice. J Immunol 171, 6173–6177 (2003). [DOI] [PubMed] [Google Scholar]
- 63.Patel DD & Kuchroo VK TH17 Cell Pathway in Human Immunity: Lessons from Genetics and Therapeutic Interventions. Immunity 43, 1040–1051 (2015). [DOI] [PubMed] [Google Scholar]
- 64.Trapnell C et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7, 562–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang da W, Sherman BT & Lempicki RA Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 66.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Blecher-Gonen R et al. High-throughput chromatin immunoprecipitation for genome-wide mapping of in vivo protein-DNA interactions and epigenomic states. Nat Protoc 8, 539–554 (2013). [DOI] [PubMed] [Google Scholar]
- 68.Barozzi I, Termanini A, Minucci S & Natoli G Fish the ChIPs: a pipeline for automated genomic annotation of ChIP-Seq data. Biol Direct 6, 51 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Machanick P & Bailey TL MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27, 1696–1697 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McLean CY et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28, 495–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Heinz S et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mayoral RJ & Monticelli S Stable overexpression of miRNAs in bone marrow- derived murine mast cells using lentiviral expression vectors. Methods Mol Biol 667, 205–214 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon request.