Abstract
In the last 15 years, several classes of small regulatory RNAs have been identified, uncovering the widespread impact of non-coding elements in the human genome on cell homeostasis and human diseases. MicroRNAs (miRNAs) are a family of small, non-coding RNAs, which exert silencing of mRNA targets in a sequence-dependent fashion. Many miRNAs are specifically expressed in the central nervous system, where they display roles in differentiation, neuronal survival, neuronal plasticity and learning. On the other hand, deregulated miRNA/mRNA expression networks are deeply involved in neurodegeneration.
Recent findings suggest a role for miRNAs in the pathogenesis of motor neuron diseases. In particular, cell-specific changes in miRNA profile are involved in the motor neuron disease phenotype and might be implicated in their selective vulnerability. Exploitation of noncoding RNAs, in particular miRNAs, for therapeutic strategies is being assessed for implementing current therapies. In this regard, the neuroprotective potential of certain miRNAs could represent a promising potential tool to improve therapies for motor-neuron diseases. This review focuses on emerging roles of miRNAs in motor neuron diseases and on their impact on neuron life-span and integrity.
Keywords: Motor neuron diseases, microRNAs, RNA therapeutics
1. Introduction
MicroRNAs (miRNAs) are a class of small non-coding RNAs first described in Caenorhabditis elegans that are involved in gene silencing [1,2]. They are ∼18–25 nt long and act as regulators of gene expression by orchestrating a wide range of biological processes, such as development, cell proliferation and differentiation, apoptosis and cell metabolism [1,2]. Remarkably, miRNAs are also involved in the pathogenesis of several human diseases, such as cancer, metabolic disorders and neurodegenerative diseases.
The process of miRNA biogenesis (reviewed by Ha and Kim, 2014) [3] begins with transcription by RNA polymerase II, giving rise to primary transcripts (pri-miRNAs), capped and polyadenylated [4]. These transcripts are processed in the nucleus by the RNase III family enzyme Drosha in complex with DGCR8 protein [5], thus generating a ∼70-nt-long precursor miRNA (pre-miRNA) that is transported to the cytoplasm by exportin 5 [6]. Dicer, another RNase III enzyme, cleaves the pre-miRNA in the cytoplasm to generate a ∼20 bp miRNA duplex. Depending on the specific miRNA sequence, one of the strands of this duplex is loaded into the miRNA-induced silencing complex (RISC), whereas the other strand (the passenger strand) is usually released and degraded [3].
The active miRNA-containing RISC (miRISC) is then recruited to target mRNAs harboring complementary seed sequences within their 3′UTRs. In this way, miRNAs suppress the translation and/or promote the degradation of target genes [7].
MiRNAs display tissue-specific functions based on their cell- and organ-specific expression patterns. In the nervous system miRNAs regulate in time and space gene expression circuits essential for neural differentiation, development and activity. Notably, depending on the specific context, miRNAs can display either protective or disease-promoting effects. The manipulation of miRNAs might therefore offer novel therapeutic opportunities.
In this review, we provide an overview of the most relevant miRNAs involved in motor neuron (MN) development, focusing on how miRNA-mediated control of gene expression fail in the pathogenesis of neuromuscular disorders, such as spinal muscular atrophy (SMA) and amyotrophic lateral sclerosis (ALS).
2. miRNA dynamics in neurogenesis and motor neuron determination
The transition from progenitor cells to mature neurons is fine-tuned by changes in gene expression programs achieved both transcriptionally and post-transcriptionally. The prominent role played by miRNAs in orchestrating this developmental process was first highlighted by Dicer mutations in zebrafish, leading to severe defects in terminal differentiation of neuronal subtypes [8]. In mice, depletion of miRNAs in the developing neocortex (E10) results in impaired differentiation of newborn neurons, whereas neuronal progenitor proliferation is unaffected [9]. Thus, miRNAs are dispensable in the early neural fate decision, but critically involved in the proper expression of a differentiated neuronal phenotype.
At the onset of neurogenesis, the cerebral cortex is composed by proliferating neuroepithelial cells that self-renew by symmetric divisions [10]. At embryonic day 11.5 (E11.5) in mouse, these cells start expressing the transcription factor Pax6 and become apical progenitors, also named radial glia cells [11], which divide asymmetrically to self-renew and originate neurons, either directly or through generation of Tbr2-positive intermediate progenitors [12]. These intermediate progenitors occupy the subventricular zone and divide symmetrically to generate post-mitotic neurons that migrate to the cortical plate [13]. One of the most studied miRNAs in this context is miR-9. In zebrafish, miR-9 promotes neurogenesis and diminishes the pool of neural progenitor cells pool by targeting different components of the fibroblast growth factor (FGF) signaling pathway and anti-neurogenic basic helix-loop-helix transcription factors [14]. In mice, miR-9 overexpression induces neuronal differentiation via direct inhibition of the nuclear TLX receptor, also known as NR2E1 [15]. TLX acts as an upstream positive regulator of the Wnt signaling pathway [16], essential for neuronal progenitor self-renewal. The pro-neurogenic function of miR-9, and similarly miR-124, involves also decreased expression of BAF53a [17], a component of the BAF complex implicated in the recruitment of the Polycomb repressive complex to chromatin and in the establishment of H3K27me3 repressive marks [18,19]. Specific BAF complexes are assembled during neurogenesis: the subunits of the npBAF complex (including BAF53a) are essential for neural-progenitor proliferation, and mice with reduced expression of these genes have defects in neural-tube closure [20]; in contrast, BAF53b and the nBAF complex are essential for post-mitotic neural development and dendritic morphogenesis [21].
Let-7b is a key regulator of neural stem cell proliferation and differentiation. It is significantly up-regulated during neural differentiation and suppresses the expression of TLX and cyclin D1 by binding to the 3′ untranslated region (UTR) of their mRNAs [22]. Notably, increased expression of let-7b leads to reduced neural stem cell proliferation and accelerates neural differentiation, whereas antisense-knockdown of let-7b results in increased neural stem cell proliferation [22]. Interestingly, TLX activity is also inhibited by miR-137, which targets and is targeted by the TLX co-repressor histone lysine-specific demethylase 1 (LSD1) [23]. Thus, miR-137 forms a feedback regulatory loop with TLX and LSD1 to orchestrate the dynamic switch between neural stem cell proliferation and differentiation.
The exit of neural cells from the cell cycle is achieved by miR-20a/20b and miR-23, which negatively regulate cyclin D1 levels via direct binding to its 3′UTR [24]. Cyclin D1 expression is indirectly targeted also by miR-15b through targeting of Tet methylcytosine dioxygenase 3 (TET3), a regulator of the methylation status of the cyclin D1 promoter [25].
During late neurogenesis and in the early postnatal development different types of glial cells are generated from neuronal precursor cells [26]. Recent studies support a role for miRNAs in the development of these glial lineages. For example, it has been shown that miR-219, miR-338 and miR-138 are specifically enriched in oligodendrocyte precursors and play a functional role in oligodendrocyte differentiation [22,27]. Thus, miRNAs not only settle major cell fate decisions (e.g. neuronal versus glial differentiation), but also determine in time and space the expression of specific glial and neural cell types, for example whether a neuron becomes a motor neuron (MN) versus an interneuron.
In the developing spinal cord, specification of neuronal cell fate is established by morphogen gradients that guide the patterning of progenitor domains, giving rise to specific neuronal types [28]. Spinal MNs are a group of cholinergic neurons located in the ventral horn of the spinal cord that control locomotion. The ventral spinal cord can be divided into five progenitor domains, giving rise to different interneurons and to MNs. The borders of the progenitor domains are established through the repressive interactions between transcription factors expressed in the neighboring domains [29]. The combinatorial action of these transcription factors allows the specification of each cell type. In particular, the progenitors of spinal MNs and interneurons are specified by the repressive transcription factor Olig2, engaged in a cross-interaction with the transcription factor of the Iroquois homeobox gene family Irx3 [[30], [31], [32]]. Notably, miR17∼92 cluster inhibits the expression of Olig2, which is enriched in the progenitors of spinal MNs, to promote the specification of interneurons [33]. Mice lacking miRNAs, or just the miR-17–92 cluster, display defects in the specification of interneurons [33]. Conversely, miR-218, which is enriched in MNs, suppresses interneuron development by selectively targeting interneuron-specific genes [34]. As progenitor cells exit the cell cycle, transcription factors that promote the differentiation of distinct interneurons and MNs are up-regulated [35]. Two LIM-homeodomain factors, LIM homeobox 3 (Lhx3) and Islet-1 (Isl1), are co-expressed in differentiating MNs [35]. MiR-218 is up-regulated by Isl1-Lhx3 at the onset of MN differentiation and contribute to MN identity by targeting genes that promote non-MN functions [34].
Spinal MNs are organized in anatomical columns in a segmental manner along the rostro-caudal axis, which is controlled by dosage of homeobox (HOX) proteins and co-factors [36]. The HOX accessory factor FoxP1 coordinates MN subtype identity and connectivity [37]. Notably, miR-9 is transiently expressed during the differentiation of spinal cord MNs and regulate the expression levels of FoxP1 by targeting its 3′-UTR [38]. Moreover, in spinal MNs miR-9 targets Onecut1 (OC1), critically involved in the specification of MN in the developing spinal cord [39].
By using an in vitro model of human spinal MN development, it has been shown that miR-375 is strongly activated during spinal motor neurogenesis and its expression is specific to MNs [40]. Knockdown of miR-375 significantly impairs MN differentiation, highlighting its essential role in MN development. Interestingly, miR-375 expression is lower in highly proliferative neural precursors, such as neuroepithelial cells and MN progenitors, but higher in post-mitotic MNs, which display also reduced expression of miR-375 targets relevant for the exit from a proliferative state, such as PAX6, REST and CCND2 [40].
Collectively, the examples discussed above highlight the role played by specific miRNAs in neuronal development and in the determination of different neuronal sub-types. Given the critical function of miRNAs in these developmental processes, we can gather how subtle change or defects in miRNA profiles may result in inappropriate neuronal development, including premature neuronal cell death, that is a hallmark of neurodegenerative disorders.
3. Dysregulation of miRNAs in motor neuron diseases
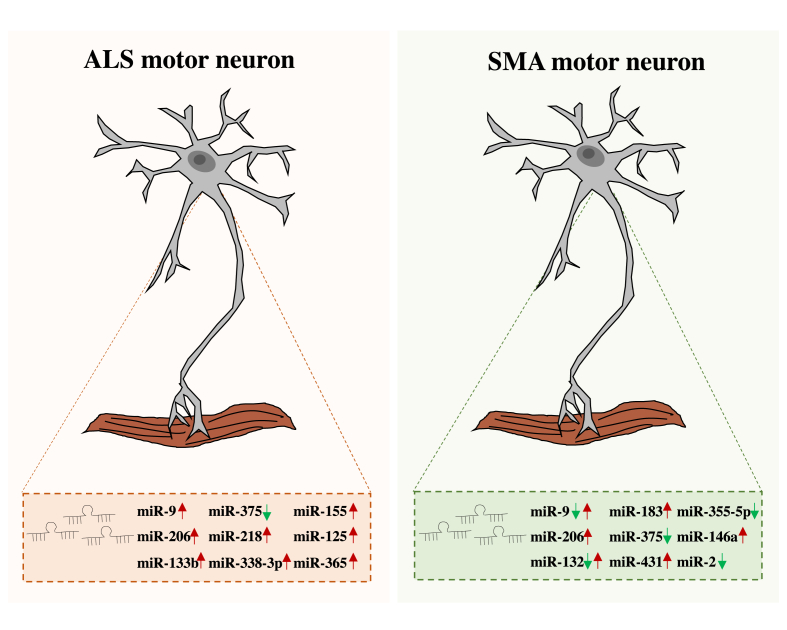
Motor neuron diseases (MNDs) result from the progressive degeneration and death of MNs. The two most studied MNDs are the childhood genetic disease spinal muscular atrophy (SMA) and the adult-onset neurodegenerative disease amyotrophic lateral sclerosis (ALS) [41,42]. Both diseases involve neuromuscular dysfunction progressively leading to fatal paralysis. miRNAs play a crucial role in the development and in the function of MNs. Alterations in these functions contribute to MND and neurodegeneration [43] (see Fig. 1).
Fig. 1.
Schematic representation of miRNAs involved in MNDs. In the left panel is illustrated the expression profile of miRNAs dysregulated in ALS MNs while in the right panel are listed the miRNAs affected in SMA MNs. The green arrows represent miRNAs down-regulated in the disease. Red arrows indicate miRNAs up-regulated in the diseases. In SMA disease, miR-9 and miR-132 have been found differentially dysregulated in the spinal cord and skeletal muscle of affected motor units. In particular, their expression is decreased in the spinal cord and increased in the skeletal muscle (double arrows).
3.1. MiRNAs in spinal muscular atrophy
SMA is a neurodegenerative disorder that affects the lower MNs and leads to progressive muscular weakness, atrophy and paralysis. It is one of the most common autosomal recessive diseases of childhood with an incidence of 1 in 11000 live births [44,45]. The disease is classified in three types based on the severity of symptoms and the age of onset [46]. The type 1, called Werdnig-Hoffmann disease, is the most severe form that occurs in 60% of patient. Type II is the intermediate form, occurring in 27% of SMA patients. The Kugelberg-Wellander is the type III and is the mildest form [47,48].
SMA is caused by mutations in the survival motor neuron 1 gene (SMN1), located on chromosome 5q13 [49]. The SMN1 homologous pseudogene, SMN2, is located on the same chromosome and differs from SMN1 only for 5 nucleotides, with a C to T transition leading to an exonic splicing silencer (ESS), that causes the skipping of exon 7 and reduced expression of the functional protein [50,51].
95% of SMA cases are characterized by the homozygous absence of SMN1 [44]. Only 10% of the SMN2 transcripts encodes a functional protein that can balance the absence of SMN1 [52]. It has been observed a relationship between the abundance of SMN2 copies and the severity of the pathology [53]. The more severe forms typically carry one to two copies of SMN2, while the milder forms of SMA hold three to four copies of the gene [53,54].
SMN protein plays a key role in multiple fundamental aspects of RNA processing, including the assembly of the spliceosome and the biogenesis of ribonucleoprotein. SMN is also involved in miRNA biogenesis, by interacting with the fragile X mental retardation protein (FMRP) [55], the KH-type splicing regulatory protein (KSRP) [56] and the fused in sarcoma/translocated in liposarcoma (FUS/TLS) protein [57].
Here, we report some miRNAs involved in the pathogenesis of SMA.
As mentioned above, miR-9 is one of the most abundant miRNAs in the central nervous system (CNS), involved in several aspects relevant for proper neuronal development [38,39]. Genetic ablation of Dicer1 leads to denervation and muscular atrophy, with loss of spinal MNs and alteration in the expression levels of neurofilament subunits in mice [43]. This phenotype is mainly due to the dysregulation of miR-9, which is an upstream regulator of the neurofilament mRNAs [43]. Down-regulation of SMN protein expression up-regulates miR-9 expression. Conversely, forced SMN protein expression down-regulates miR-9 [58]. Importantly, miR-9 expression negatively correlates with SMN protein level in SMA patients [58]. In particular, miR-9 is down-regulated in the spinal cord of SMA I mice, while its expression is increased in the skeletal muscle. Treatment with antisense oligonucleotides (ASOs) modulating SMN2 splicing is sufficient to normalize miR-9 levels in skeletal muscle but not in the spinal cord. High levels of miR-9 were also detected in the serum of SMA mice and were down-regulated upon ASO treatment, suggesting that alterations in SMN protein expression affects miR-9 levels [59].
miR-183 is up-regulated in fibroblasts, MNs and cortical neurons in human and mouse models of SMA [60]. In particular, miR-183 is up-regulated in the neurites of SMN-deficient neurons, where it regulates the translation of mTOR by binding to its 3′UTR. Inhibition of miR-183 expression in the spinal cord of SMA mouse models prolongs the survival of MNs and improves motor function. These interesting results provide a direct link between miRNA metabolism and the mTOR signaling pathway in the pathology of SMA [60].
As mentioned above, miR-375 plays an important role in MN development [40]. Interestingly, SMA MNs display down-regulation in the expression of miR-375 and up-regulation of p53 protein levels. As a consequence, SMA MNs are more susceptible to DNA damage, suggesting that miR-375 could play a protective role in MNs by targeting p53 [40].
miR-431 is expressed in the brain and spinal cord during embryonic development [61] and it is highly dysregulated in SMN knockdown models [62]. In particular, Wertz and colleagues [62] showed increased expression of miR-431 in MNs upon SMN deficiency, due to the down-regulation of mRNA targets involved in the regulation of neurite length [62].
miR-206 is predominantly expressed in the skeletal muscle [63], but it is also expressed in spinal MNs, where it is involved in the regeneration of neuromuscular junction (NMJ) after injury [64]. Up-regulation of miR-206 in SMNΔ7 mice, an intermediate model of SMA, rescues the innervation of NMJ [65]. In SMA mice, the levels of miR-132, previously shown to be involved in dendritic maturation [66], synaptic function [67] and neovascularization [68], are reduced in the spinal cord and increased in the serum [59]. Importantly, ASO treatment is sufficient to stabilize miR-132 levels [59].
miR-335-5p, which is involved in self-renewal and differentiation of mouse embryonic stem cells (mESCs) [69], is down-regulated in SMNΔ7 neural progenitor cells, probably for the increased proliferation activity observed in SMA cells [70]. Moreover, the down-regulation of miR-335-5p in SMA cells was also observed during MN differentiation. In fact, before differentiation human induced pluripotent stem cells (iPSCs) do not show any difference in miR-335-5p expression [71].
miR-146a is up-regulated in SMA iPSC-derived astrocytes and SMNΔ7 mouse spinal cord. The increase of miR-146a was sufficient to induce MN loss, while the inhibition of miR-146a prevented this loss, thus suggesting that alteration in the astrocyte production induced by miR-146a could be relevant for SMA pathogenesis [72].
Reduced SMN activity in C. elegans affects miR-2 expression, which in turn induces MNs to produce more M2 muscarinic receptor (M2R) ortholog GAR-2. Increased M2R activity is deleterious and consistent with a subset of the NMJ defects observed in SMA models across species. Importantly, M2R levels are increased also in SMN-deficient mouse, and M2R inhibition ameliorates axon outgrowth defects in MNs [73].
Taken together these studies link functionally and mechanistically SMN to the miRNA pathway. Decreased SMN levels lead to down-regulation of specific miRNAs and, consequently, increased expression of protein with deleterious effects on the survival of MNs.
3.2. MiRNAs in amyotrophic lateral sclerosis
In contrast to SMA, ALS affects both cortical and spinal MNs [74]. ALS is a late-onset MND that leads to the degeneration of MNs in the brain and spinal cord, thus causing muscle atrophy and leading to paralysis and death [74]. 90% of ALS cases are classified as sporadic (sALS), showing no family history neither genetic component, whereas 10% of cases are defined as familial (fALS), because the patients affected show gene mutations that lead to the neurodegeneration. The most relevant mutations causing ALS are found in the chromosome 9 open reading frame 72 (C9orf72), in the Cu2+/Zn2+ superoxide dismutase 1 (SOD1), TAR DNA-binding protein 43 (TARDBP), and fused in sarcoma/translocated in liposarcoma (FUS/TLS) [75,76]. The mechanisms driving ALS are composite and include oxidative stress, neuroinflammation, altered protein and RNA metabolism, impaired structure and signaling at the NMJ [74].
Increasing evidences suggest that deregulation of miRNA biogenesis and changes in their expression could contribute to MN damage and progression of the disease [77]. Recently, it has been shown a reduction in the global levels of miRNAs in both familial and sporadic cases of ALS, resulting from accelerated RNA turnover or cytoplasmic aggregation of RNA-binding proteins (RBP) [78]. Indeed, the RBPs TDP-43 and FUS contribute to different steps of miRNA biogenesis [79]. TDP-43 stimulates miRNA processing by interacting with Drosha and Dicer complexes and by promoting the cleavage of pri- and pre-miRNAs. Delocalization of TDP-43 in cytoplasmic aggregates impairs the processing of TDP-43–regulated miRNAs by Drosha and Dicer [80]. Moreover, ALS-associated mutations in TDP-43 gene affect the interaction between Drosha and FUS, leading to a decrease in Drosha stability and causing neuronal cytotoxicity [81]. Furthermore, the expression of TDP-43 target microRNAs was altered in the cerebrospinal fluid (CSF) and serum of sporadic ALS patients [82], confirming the relevance of this RBP in the impairment of miRNA network in ALS disease. FUS, as well as other members of the FET (FUS, EWS, TAF15) family, interacts with the microprocessor complex during miRNA biogenesis [5] and binds specific pri-miRNAs, thus enhancing the recruitment of Drosha [83]. ALS‐associated mutations in FUS gene lead to decreased nuclear localization of the protein and its cytoplasmic sequestration, thus possibly altering different steps of miRNA biogenesis [83].
An ALS-associated mutation within the 3′UTR of FUS transcript has been shown to alter miR-141/200a binding, thus disrupting a feedback loop between FUS and these miRNAs and resulting in the aberrant accumulation of FUS protein [84]. Notably, the miR-141/200 family fine tunes FET protein expression in differentiating neuronal cells [85], and transfection of antagomiR-141 abolished the onset of neuronal differentiation induced by retinoic acid (RA), together with suppressing the down-regulation of FUS and EWS [85]. These results suggest that miR-141 controls a general neuron differentiation program that includes concomitant down-regulation of FUS and EWS expression [85]. It has been shown that FUS mutations cause a downstream impairment of miRNAs production that could alter RNA metabolism and increase MN vulnerability. Indeed, it has been observed a reduction of miR-375 levels and an increase of its target mRNAs, such as p53 and other pro-apoptotic genes, in both FUS mutant MNs and in the spinal MNs of ALS patients [86]. These data are consistent with MNs-overexpressing miR-375 protected from DNA-damage induced apoptosis, as described above [40].
Several studies highlighted the mechanism of dying back in ALS, in which MN degeneration begins by distal nerve terminals from the NMJ and continues retrogradely along the motor axon [87]. Indeed, evidences in both SOD1-G93A ALS mouse model and in samples of ALS patients suggest that the NMJ and nerve terminals are degraded before the MN death [88]. To this regard, some miRNAs specifically expressed in the skeletal muscle (indicated as myomiRs) are involved in ALS progression. In particular, miR-206 has been shown to act as a key regulator of the signaling between MNs and NMJ [64]. In SOD1-G93A mouse model, miR-206 delays the onset of the disease, while its genetic ablation in mice induces faster disease progression, delayed reinnervation and regeneration of NMJ, followed by accelerated atrophy of skeletal muscle [64,89]. Collectively these studies suggest that miR-206 is important for regeneration of neuromuscular synapses, possibly by suppressing histone deacetylase 4 (HDAC4), and inducing the fibroblast growth factor signaling pathway [64]. At the onset of the disease, up-regulation of miR-206 in SOD1-G93A mouse is consistent with an increased expression of miR-133b, which plays a pivotal role in the regeneration of NMJ after nerve injury [64,89]. miR-133b belongs to miR-133 family, that includes also miR-133a, recently shown to be involved in ALS [90]. These data suggest that these myomiRs could play a protective role by promoting the formation of new synapses, thus delaying the progression of ALS.
Numerous evidences indicate that loss of MNs can be due to alterations in the neighboring non-neuronal cells, the glial cells. A pathological mechanism that affects glial cells is the neuroinflammation, in which recent studies have highlighted the emerging role of specific miRNAs [91]. These miRNAs, called immune-miRs, act by modulating the expression of inflammatory genes linked to the switch of microglia from an anti-inflammatory phenotype (M2), characteristic of the early state of disease, towards a pro-inflammatory phenotype (M1), typical of the late state of the disease [92]. In SOD1-G93A mouse model, miR-125b and miR-365 are up-regulated and reduce the expression levels of the signal transducer and activator of transcription 3 (STAT3) and of interleukin-6 (IL-6). Down-regulation of these two pathways promotes pro-inflammatory signals and increases the expression levels of the tumor necrosis factor alpha (TNF-α) [93]. Interestingly, also miR-155 is associated with this neuroinflammatory response; in fact, it is up-regulated in SOD1-G93A microglia as well as in mouse and human CSF [94]. In particular, miR-155 has been shown to promote the recruitment of macrophagic cells, to increase the pro-inflammatory cytokine secretion, and to reduce the transforming growth factor beta 1 (TFG- β1) production [94]. Indeed, genetic ablation of miR-155 decreases the recruitment of monocytes in the spinal cord of SOD1-G93A mice and promotes a longer survival in ALS animal model [95].
Among the miRNAs associated with neuronal differentiation, some are implicated in the progression of MND. For example miR-9, a regulator of cell fate during neurogenesis, is up-regulated in the spinal cord of SOD1-G93A mouse, especially in the ventral horn, that is the main area involved in neurodegeneration [96], suggesting a possible contribution to ALS pathology.
Recently, a high throughput approach to identify MN-enriched miRNAs in physiological and pathological conditions, revealed higher levels of miR-218 (a MN-specific miRNA) at the end-stage of ALS than in pre-symptomatic rat CSF [97]. MiR-218 is released extracellularly from MNs in ALS rats and incorporated by the astrocytes, where it directly targets and down-regulates the glutamate transporter EAAT2 (excitatory amino acid transporter 2) [98]. Inhibition of miR-218 with ASOs in ALS mice mitigates the loss of EAAT2, thus reducing excitotoxicity in MNs [98]. These studies support a model in which glial dysfunction substantially contributes to ongoing neuronal loss and disease progression.
In ALS spinal MNs, expression of miRNAs that control stoichiometry of neurofilament subunits has been found altered. This dysregulation contributes to axonal degeneration, neuro-pathological cytoplasmic inclusion and eventually MN death [99].
4. MiRNAs as novel biomarkers and therapeutic perspectives
The identification of novel biomarkers for early diagnosis of MNDs and for monitoring their progression is essential for treatment. MiRNAs are now representing novel clinical biomarkers for prognosis of several diseases. They can be clinically detected in many body fluids, such as CSF, serum/plasma, saliva, and urine [100], by using noninvasive methods. These characteristics, coupled with the rapidly evolving improvements in technologies that facilitate the detection of RNA species from small amounts of biological material, have contributed to the strong interest towards the study of miRNAs as potential biomarkers also for MNDs.
To this regard, Catapano and colleagues [59] investigated the differential expression of miR-9, miR-206, and miR-132 in spinal cord, skeletal muscle and serum from SMA mice and patients. MiRNA changes in the serum of SMA mice precede their changes in spinal cord and skeletal muscle tissues, and also the clinical symptoms, thus suggesting the efficacy of this method for SMA prognosis [59]. However, no correlation was reported between the level of the three mentioned miRNAs in the serum and the motor functional ability of the patients, highlighting the variability among patients as a limit in the use of these miRNAs as biomarkers for SMA. Larger group of patients are needed to increase the power of biomarker studies [52,59].
Expression profile of miRNAs in skeletal muscle of ALS patients identified 11 miRNAs differentially expressed. Of these, only miR-424 and miR-206 were overexpressed both in the skeletal muscle and in the plasma [101]. Importantly, their expression correlated with clinical deterioration, highlighting their potential as prognostic markers for ALS [101]. Notably, riluzole treatment did not affect miRNA expression profile in ALS patients [102].
In a recent study performed in the SOD1-G93A mouse model, miR-206 was flagged as a potential circulating biomarker candidate of the disease as it exhibited strong up-regulation in the serum of mice and in ALS patients [64,103]. MiR-206 was significantly up-regulated in the pre-symptomatic stages of SOD1-G93A mice, making it an interesting biomarker candidate for early diagnosis of ALS. The downside was the fact that a similar increase in miR-206 expression was also observed in other pathologies, including the Duchenne Muscular Dystrophy [104], Alzheimer Disease, cerebral ischemia [105,106], schizophrenia [107], and upon cell exposure to environmental toxins [108], further reinforcing the importance of identifying specific circulating biomarkers to properly discriminate CNS conditions.
Expression profiles of 911 human miRNAs using microarray technology identified miR338-3p as specifically and significantly up-regulated in leukocytes from sALS patients [109]. Remarkably, miR-338-3p was previously shown to target the glutamate transporter EAAT2, that is responsible of the glutamate clearance described in CSF of ALS patients [110]. Furthermore, 30 miRNAs have been identified significantly downregulated in the serum of fALS patients, the majority of them already dysregulated in pre-symptomatic subjects carrying mutations causative of the disease [111]. More recently, a high-throughput sequencing approach, followed by an integrated bioinformatics/biostatistics analysis, identified 38 miRNAs significantly downregulated in sALS [112]. Remarkably, some of the identified miRNAs were conserved across studies, thus pointing at them as circulating biomarkers critical for early prognosis of clinical deterioration and relevant for future therapeutic efforts.
To this regard, miRNAs have been proposed as therapeutic targets for MN diseases. Possible strategies include the overexpression of miRNAs as mimics or their down-regulation using antisense oligonucleotides (ASOs), antagomiRs or Locked Nucleic Acid antimiRs [52,113]. Advances in technologies to deliver RNA molecules in vivo have made miRNA-based therapeutics feasible. These constructs display several modifications in the RNA backbone to provide higher stability and protection from nucleases [114]. In particular, the ability of selected miRNAs to target multiple mRNAs altered in the disease makes these molecules interesting candidates as therapeutics (in the form of miRNA mimics) or as targets of therapeutics (in the form of antagomiRs). To this regard, up-regulation of miR‐375 and miR-183 could be exploited to confer MN protection from increased susceptibility to DNA damage‐induced apoptosis [40] and to improve MN function and survival [60]. In addition, therapeutic up-regulation of miR‐206 could ameliorate and preserve NMJ function [64], thus representing a potential additional therapy for SMA. Koval and collaborators [95] developed ASOs against miR-155, a miRNA up-regulated in both human and mouse ALS CSF. Notably, inhibition of miR-155 increased mice survival by 10 days and disease duration by 15 days [95], highlighting miRNAs as potential therapeutic targets also for ALS pathology.
Recently, it was developed the first vertebrate system able to manipulate SMN function by stable miR-mediated knockdown technology [115]. The induced expression of an engineered anti-smn1 miRNA in zebrafish MNs was sufficient to reproduce the phenotype observed in the SMA model, including abnormal MN development, poor motor function and premature death [115]. The same approach with engineered miRNAs could be therapeutically exploited to target aberrant transcripts up-regulated in the disease.
Two years ago, the first-ever approved treatment for SMA became effective. Several decades of intensive research efforts have culminated in the global approval of nusinersen, marketed as Spinraza (Biogen), the first disease-modifying therapy for SMA, approved by the FDA in the USA the 23rd of December 2016 and by the EMA in EU the 30th of May 2017. Nusinersen is an ASO targeting SMN2 exon 7 and increasing its inclusion, thus rescuing SMN protein expression [116]. Interestingly treatment with ASO targeting SMN2 splicing is sufficient to restore miR-9, miR-132 and miR-206 levels in SMA mice [59]. MiRNA therapeutics could be used to improve and ameliorate the effect of Spinraza on SMA patients.
To conclude, miRNAs could represent a potential therapeutic approach for both SMA and ALS treatment and could be used as a novel strategy to delay disease progression.
5. Concluding remarks
In vitro and in vivo experiments have clearly demonstrated the neuroprotective potential of certain miRNAs, as well as the value of specific miRNAs in ameliorating disease symptoms and progression in animal models [64,95]. Hence, modified miRNAs, ASOs, miRNA mimics and antagomiRs represent a promising therapeutic strategy in association with more targeted therapies. Nevertheless, the therapeutic potential of miRNAs still remains largely unexplored.
The application of high throughput RNA sequencing technologies, single-cell qRT-PCR and proteomic approaches has allowed comprehensive comparison of MNs generated via different strategies, providing additional insights into the biology of human spinal MNs. Although the recent progress in producing and understanding MNs has been remarkable, substantial challenges still remain open. The intricated miRNA networks contributing to MND pathogenesis are still far to be completely unraveled, to fully exploit their potential as novel therapeutic tools.
Funding
This work was supported by grants from the Associazione Italiana Ricerca sul Cancro (AIRC) [IG17278 to M.P.P.]; from Ministry of Health “Ricerca Corrente” and “5x1000 Anno 2017” to Fondazione Santa Lucia.
Conflicts of interest
None declared.
References
- 1.Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 2.Wightman B., Ha I., Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75(5):855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 3.Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15(8):509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 4.Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Rådmark O., Kim S., Kim V.N. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 5.Gregory R.I., Yan K.P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N., Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432(7014):235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 6.Bohnsack M.T., Czaplinski K., Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10(2):185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krol J., Loedige I., Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11(9):597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 8.Giraldez A.J., Cinalli R.M., Glasner M.E., Enright A.J., Thomson J.M., Baskerville S., Hammond S.M., Bartel D.P., Schier A.F. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308(5723):833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 9.Rajman M., Schratt G. MicroRNAs in neural development: from master regulators to fine-tuners. Development. 2017;144(13):2310–2322. doi: 10.1242/dev.144337. [DOI] [PubMed] [Google Scholar]
- 10.Götz M., Huttner W.B. The cell biology of neurogenesis. Nat. Rev. Mol. Cell Biol. 2005;6(10):777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- 11.Götz M., Stoykova A., Gruss P. Pax6 controls radial glia differentiation in the cerebral cortex. Neuron. 1998;21(5):1031–1044. doi: 10.1016/S0896-6273(00)80621-2. [DOI] [PubMed] [Google Scholar]
- 12.Englund C., Fink A., Lau C., Pham D., Daza R.A., Bulfone A., Kowalczyk T., Hevner R.F. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J. Neurosci. 2005;25(1):247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noctor S.C., Martínez-Cerdeño V., Ivic L., Kriegstein A.R. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat. Neurosci. 2004;7(2):136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- 14.Leucht C., Stigloher C., Wizenmann A., Klafke R., Folchert A., Bally-Cuif L. MicroRNA-9 directs late organizer activity of the midbrain-hindbrain boundary. Nat. Neurosci. 2008;11(6):641–648. doi: 10.1038/nn.2115. [DOI] [PubMed] [Google Scholar]
- 15.Zhao C., Sun G., Li S., Shi Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat. Struct. Mol. Biol. 2009;16(4):365–371. doi: 10.1038/nsmb.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qu Q., Sun G., Li W., Yang S., Ye P., Zhao C., Yu R.T., Gage F.H., Evans R.M., Shi Y. Orphan nuclear receptor TLX activates Wnt/beta-catenin signalling to stimulate neural stem cell proliferation and self-renewal. Nat. Cell Biol. 2010;12(1):31–40. doi: 10.1038/ncb2001. sup pp. 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoo A.S., Staahl B.T., Chen L., Crabtree G.R. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460(7255):642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho L., Ronan J.L., Wu J., Staahl B.T., Chen L., Kuo A., Lessard J., Nesvizhskii A.I., Ranish J., Crabtree G.R. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc. Natl. Acad. Sci. U. S. A. 2009;106(13):5181–5186. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho L., Miller E.L., Ronan J.L., Ho W.Q., Jothi R., Crabtree G.R. esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat. Cell Biol. 2011;13(8):903–913. doi: 10.1038/ncb2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lessard J., Wu J.I., Ranish J.A., Wan M., Winslow M.M., Staahl B.T., Wu H., Aebersold R., Graef I.A., Crabtree G.R. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55(2):201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu J.I., Lessard J., Olave I.A., Qiu Z., Ghosh A., Graef I.A., Crabtree G.R. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron. 2007;56(1):94–108. doi: 10.1016/j.neuron.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 22.Zhao C., Sun G., Li S., Lang M.F., Yang S., Li W., Shi Y. MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc. Natl. Acad. Sci. U. S. A. 2010;107(5):1876–1881. doi: 10.1073/pnas.0908750107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun G., Ye P., Murai K., Lang M.F., Li S., Zhang H., Li W., Fu C., Yin J., Wang A., Ma X., Shi Y. miR-137 forms a regulatory loop with nuclear receptor TLX and LSD1 in neural stem cells. Nat. Commun. 2011;2:529. doi: 10.1038/ncomms1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghosh T., Aprea J., Nardelli J., Engel H., Selinger C., Mombereau C., Lemonnier T., Moutkine I., Schwendimann L., Dori M., Irinopoulou T., Henrion-Caude A., Benecke A.G., Arnold S.J., Gressens P., Calegari F., Groszer M. MicroRNAs establish robustness and adaptability of a critical gene network to regulate progenitor fate decisions during cortical neurogenesis. Cell Rep. 2014;7(6):1779–1788. doi: 10.1016/j.celrep.2014.05.029. [DOI] [PubMed] [Google Scholar]
- 25.Lv X., Jiang H., Liu Y., Lei X., Jiao J. MicroRNA-15b promotes neurogenesis and inhibits neural progenitor proliferation by directly repressing TET3 during early neocortical development. EMBO Rep. 2014;15(12):1305–1314. doi: 10.15252/embr.201438923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kriegstein A., Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu. Rev. Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dugas J.C., Cuellar T.L., Scholze A., Ason B., Ibrahim A., Emery B., Zamanian J.L., Foo L.C., McManus M.T., Barres B.A. Dicer1 and miR-219 Are required for normal oligodendrocyte differentiation and myelination. Neuron. 2010;65(5):597–611. doi: 10.1016/j.neuron.2010.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jessell T.M. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat. Rev. Genet. 2000;1(1):20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- 29.Lee S.K., Pfaff S.L. Transcriptional networks regulating neuronal identity in the developing spinal cord. Nat. Neurosci. 2001;4(Suppl):1183–1191. doi: 10.1038/nn750. [DOI] [PubMed] [Google Scholar]
- 30.Cai J., Chen Y., Cai W.H., Hurlock E.C., Wu H., Kernie S.G., Parada L.F., Lu Q.R. A crucial role for Olig2 in white matter astrocyte development. Development. 2007;134(10):1887–1899. doi: 10.1242/dev.02847. [DOI] [PubMed] [Google Scholar]
- 31.Novitch B.G., Chen A.I., Jessell T.M. Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron. 2001;31(5):773–789. doi: 10.1016/S0896-6273(01)00407-X. [DOI] [PubMed] [Google Scholar]
- 32.Zhou Q., Anderson D.J. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109(1):61–73. doi: 10.1016/S0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- 33.Chen J.A., Huang Y.P., Mazzoni E.O., Tan G.C., Zavadil J., Wichterle H. Mir-17-3p controls spinal neural progenitor patterning by regulating Olig2/Irx3 cross-repressive loop. Neuron. 2011;69(4):721–735. doi: 10.1016/j.neuron.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thiebes K.P., Nam H., Cambronne X.A., Shen R., Glasgow S.M., Cho H.H., Kwon J.S., Goodman R.H., Lee J.W., Lee S., Lee S.K. Corrigendum: miR-218 is essential to establish motor neuron fate as a downstream effector of Isl1-Lhx3. Nat. Commun. 2015;6:8227. doi: 10.1038/ncomms9227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Helms A.W., Johnson J.E. Specification of dorsal spinal cord interneurons. Curr. Opin. Neurobiol. 2003;13(1):42–49. doi: 10.1016/S0959-4388(03)00010-2. [DOI] [PubMed] [Google Scholar]
- 36.Dasen J.S., Liu J.P., Jessell T.M. Motor neuron columnar fate imposed by sequential phases of Hox-c activity. Nature. 2003;425(6961):926–933. doi: 10.1038/nature02051. [DOI] [PubMed] [Google Scholar]
- 37.Dasen J.S., De Camilli A., Wang B., Tucker P.W., Jessell T.M. Hox repertoires for motor neuron diversity and connectivity gated by a single accessory factor, FoxP1. Cell. 2008;134(2):304–316. doi: 10.1016/j.cell.2008.06.019. [DOI] [PubMed] [Google Scholar]
- 38.Otaegi G., Pollock A., Hong J., Sun T. MicroRNA miR-9 modifies motor neuron columns by a tuning regulation of FoxP1 levels in developing spinal cords. J. Neurosci. 2011;31(3):809–818. doi: 10.1523/JNEUROSCI.4330-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luxenhofer G., Helmbrecht M.S., Langhoff J., Giusti S.A., Refojo D., Huber A.B. MicroRNA-9 promotes the switch from early-born to late-born motor neuron populations by regulating Onecut transcription factor expression. Dev. Biol. 2014;386(2):358–370. doi: 10.1016/j.ydbio.2013.12.023. [DOI] [PubMed] [Google Scholar]
- 40.Bhinge A., Namboori S.C., Bithell A., Soldati C., Buckley N.J., Stanton L.W. MiR-375 is essential for human spinal motor neuron development and may Be involved in motor neuron degeneration. Stem Cells. 2016;34(1):124–134. doi: 10.1002/stem.2233. [DOI] [PubMed] [Google Scholar]
- 41.Burghes A.H., Beattie C.E. Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick? Nat. Rev. Neurosci. 2009;10(8):597–609. doi: 10.1038/nrn2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ling S.C., Polymenidou M., Cleveland D.W. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79(3):416–438. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haramati S., Chapnik E., Sztainberg Y., Eilam R., Zwang R., Gershoni N., McGlinn E., Heiser P.W., Wills A.M., Wirguin I., Rubin L.L., Misawa H., Tabin C.J., Brown R., Chen A., Hornstein E. miRNA malfunction causes spinal motor neuron disease. Proc. Natl. Acad. Sci. U. S. A. 2010;107(29):13111–13116. doi: 10.1073/pnas.1006151107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sugarman E.A., Nagan N., Zhu H., Akmaev V.R., Zhou Z., Rohlfs E.M., Flynn K., Hendrickson B.C., Scholl T., Sirko-Osadsa D.A., Allitto B.A. Pan-ethnic Carrier screening and prenatal diagnosis for spinal muscular atrophy: clinical laboratory analysis of >72,400 specimens. Eur. J. Hum. Genet. 2012;20(1):27–32. doi: 10.1038/ejhg.2011.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Faravelli I., Nizzardo M., Comi G.P., Corti S. Spinal muscular atrophy--recent therapeutic advances for an old challenge. Nat. Rev. Neurol. 2015;11(6):351–359. doi: 10.1038/nrneurol.2015.77. [DOI] [PubMed] [Google Scholar]
- 46.Ogino S., Leonard D.G., Rennert H., Wilson R.B. Spinal muscular atrophy genetic testing experience at an academic medical center. J. Mol. Diagn. 2002;4(1):53–58. doi: 10.1016/S1525-1578(10)60680-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Melki J., Lefebvre S., Burglen L., Burlet P., Clermont O., Millasseau P., Reboullet S., Bénichou B., Zeviani M., Le Paslier D. De novo and inherited deletions of the 5q13 region in spinal muscular atrophies. Science. 1994;264(5164):1474–1477. doi: 10.1126/science.7910982. [DOI] [PubMed] [Google Scholar]
- 48.Ogino S., Wilson R.B., Gold B. New insights on the evolution of the SMN1 and SMN2 region: simulation and meta-analysis for allele and haplotype frequency calculations. Eur. J. Hum. Genet. 2004;12(12):1015–1023. doi: 10.1038/sj.ejhg.5201288. [DOI] [PubMed] [Google Scholar]
- 49.Lefebvre S., Bürglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80(1):155–165. doi: 10.1016/S0378-1119(97)00510-6. [DOI] [PubMed] [Google Scholar]
- 50.Bürglen L., Lefebvre S., Clermont O., Burlet P., Viollet L., Cruaud C., Munnich A., Melki J. Structure and organization of the human survival motor neurone (SMN) gene. Genomics. 1996;32(3):479–482. doi: 10.1006/geno.1996.0147. [DOI] [PubMed] [Google Scholar]
- 51.Monani U.R., Lorson C.L., Parsons D.W., Prior T.W., Androphy E.J., Burghes A.H., McPherson J.D. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum. Mol. Genet. 1999;8(7):1177–1183. doi: 10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- 52.Magri F., Vanoli F., Corti S. miRNA in spinal muscular atrophy pathogenesis and therapy. J. Cell Mol. Med. 2018;22(2):755–767. doi: 10.1111/jcmm.13450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feldkötter M., Schwarzer V., Wirth R., Wienker T.F., Wirth B. Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am. J. Hum. Genet. 2002;70(2):358–368. doi: 10.1086/338627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shababi M., Lorson C.L., Rudnik-Schöneborn S.S. Spinal muscular atrophy: a motor neuron disorder or a multi-organ disease? J. Anat. 2014;224(1):15–28. doi: 10.1111/joa.12083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Piazzon N., Rage F., Schlotter F., Moine H., Branlant C., Massenet S. In vitro and in cellulo evidences for association of the survival of motor neuron complex with the fragile X mental retardation protein. J. Biol. Chem. 2008;283(9):5598–5610. doi: 10.1074/jbc.M707304200. [DOI] [PubMed] [Google Scholar]
- 56.Tadesse H., Deschênes-Furry J., Boisvenue S., Côté J. KH-type splicing regulatory protein interacts with survival motor neuron protein and is misregulated in spinal muscular atrophy. Hum. Mol. Genet. 2008;17(4):506–524. doi: 10.1093/hmg/ddm327. [DOI] [PubMed] [Google Scholar]
- 57.Yamazaki T., Chen S., Yu Y., Yan B., Haertlein T.C., Carrasco M.A., Tapia J.C., Zhai B., Das R., Lalancette-Hebert M., Sharma A., Chandran S., Sullivan G., Nishimura A.L., Shaw C.E., Gygi S.P., Shneider N.A., Maniatis T., Reed R. FUS-SMN protein interactions link the motor neuron diseases ALS and SMA. Cell Rep. 2012;2(4):799–806. doi: 10.1016/j.celrep.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang L.T., Chiou S.S., Liao Y.M., Jong Y.J., Hsu S.H. Survival of motor neuron protein downregulates miR-9 expression in patients with spinal muscular atrophy. Kaohsiung J. Med. Sci. 2014;30(5):229–234. doi: 10.1016/j.kjms.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 59.Catapano F., Zaharieva I., Scoto M., Marrosu E., Morgan J., Muntoni F., Zhou H. Altered levels of MicroRNA-9, -206, and -132 in spinal muscular atrophy and their response to antisense oligonucleotide therapy. Mol. Ther. Nucleic Acids. 2016;5(7):e331. doi: 10.1038/mtna.2016.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kye M.J., Niederst E.D., Wertz M.H., Gonçalves I.o.C., Akten B., Dover K.Z., Peters M., Riessland M., Neveu P., Wirth B., Kosik K.S., Sardi S.P., Monani U.R., Passini M.A., Sahin M. SMN regulates axonal local translation via miR-183/mTOR pathway. Hum. Mol. Genet. 2014;23(23):6318–6331. doi: 10.1093/hmg/ddu350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wheeler G., Ntounia-Fousara S., Granda B., Rathjen T., Dalmay T. Identification of new central nervous system specific mouse microRNAs. FEBS Lett. 2006;580(9):2195–2200. doi: 10.1016/j.febslet.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 62.Wertz M.H., Winden K., Neveu P., Ng S.Y., Ercan E., Sahin M. Cell-type-specific miR-431 dysregulation in a motor neuron model of spinal muscular atrophy. Hum. Mol. Genet. 2016;25(11):2168–2181. doi: 10.1093/hmg/ddw084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rao P.K., Kumar R.M., Farkhondeh M., Baskerville S., Lodish H.F. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc. Natl. Acad. Sci. U. S. A. 2006;103(23):8721–8726. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Williams A.H., Valdez G., Moresi V., Qi X., McAnally J., Elliott J.L., Bassel-Duby R., Sanes J.R., Olson E.N. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science. 2009;326(5959):1549–1554. doi: 10.1126/science.1181046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Valsecchi V., Boido M., De Amicis E., Piras A., Vercelli A. Expression of muscle-specific MiRNA 206 in the progression of disease in a Murine SMA model. PLoS One. 2015;10(6) doi: 10.1371/journal.pone.0128560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Magill S.T., Cambronne X.A., Luikart B.W., Lioy D.T., Leighton B.H., Westbrook G.L., Mandel G., Goodman R.H. microRNA-132 regulates dendritic growth and arborization of newborn neurons in the adult hippocampus. Proc. Natl. Acad. Sci. U. S. A. 2010;107(47):20382–20387. doi: 10.1073/pnas.1015691107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kawashima H., Numakawa T., Kumamaru E., Adachi N., Mizuno H., Ninomiya M., Kunugi H., Hashido K. Glucocorticoid attenuates brain-derived neurotrophic factor-dependent upregulation of glutamate receptors via the suppression of microRNA-132 expression. Neuroscience. 2010;165(4):1301–1311. doi: 10.1016/j.neuroscience.2009.11.057. [DOI] [PubMed] [Google Scholar]
- 68.Anand S., Majeti B.K., Acevedo L.M., Murphy E.A., Mukthavaram R., Scheppke L., Huang M., Shields D.J., Lindquist J.N., Lapinski P.E., King P.D., Weis S.M., Cheresh D.A. MicroRNA-132-mediated loss of p120RasGAP activates the endothelium to facilitate pathological angiogenesis. Nat. Med. 2010;16(8):909–914. doi: 10.1038/nm.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schoeftner S., Scarola M., Comisso E., Schneider C., Benetti R. An Oct4-pRb axis, controlled by MiR-335, integrates stem cell self-renewal and cell cycle control. Stem Cells. 2013;31(4):717–728. doi: 10.1002/stem.1315. [DOI] [PubMed] [Google Scholar]
- 70.Luchetti A., Ciafrè S.A., Murdocca M., Malgieri A., Masotti A., Sanchez M., Farace M.G., Novelli G., Sangiuolo F. A perturbed MicroRNA expression pattern characterizes embryonic neural stem cells derived from a severe mouse model of spinal muscular atrophy (SMA) Int. J. Mol. Sci. 2015;16(8):18312–18327. doi: 10.3390/ijms160818312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Murdocca M., Ciafrè S.A., Spitalieri P., Talarico R.V., Sanchez M., Novelli G., Sangiuolo F. SMA human iPSC-derived motor neurons show perturbed differentiation and reduced miR-335-5p expression. Int. J. Mol. Sci. 2016;17(8) doi: 10.3390/ijms17081231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sison S.L., Patitucci T.N., Seminary E.R., Villalon E., Lorson C.L., Ebert A.D. Astrocyte-produced miR-146a as a mediator of motor neuron loss in spinal muscular atrophy. Hum. Mol. Genet. 2017;26(17):3409–3420. doi: 10.1093/hmg/ddx230. [DOI] [PubMed] [Google Scholar]
- 73.O'Hern P.J., do Carmo G Gonçalves I., Brecht J., López Soto E.J., Simon J., Chapkis N., Lipscombe D., Kye M.J., Hart A.C. Decreased microRNA levels lead to deleterious increases in neuronal M2 muscarinic receptors in Spinal Muscular Atrophy models. Elife. 2017;6 doi: 10.7554/eLife.20752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Taylor J.P., Brown R.H., Cleveland D.W. Decoding ALS: from genes to mechanism. Nature. 2016;539(7628):197–206. doi: 10.1038/nature20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hardiman O., Al-Chalabi A., Chio A., Corr E.M., Logroscino G., Robberecht W., Shaw P.J., Simmons Z., van den Berg L.H. Amyotrophic lateral sclerosis. Nat. Rev. Dis. Primers. 2017;3:17071. doi: 10.1038/nrdp.2017.85. [DOI] [PubMed] [Google Scholar]
- 76.Svetoni F., Frisone P., Paronetto M.P. Role of FET proteins in neurodegenerative disorders. RNA Biol. 2016;13(11):1089–1102. doi: 10.1080/15476286.2016.1211225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rinchetti P., Rizzuti M., Faravelli I., Corti S. MicroRNA metabolism and dysregulation in amyotrophic lateral sclerosis. Mol. Neurobiol. 2018;55(3):2617–2630. doi: 10.1007/s12035-017-0537-z. [DOI] [PubMed] [Google Scholar]
- 78.Emde A., Eitan C., Liou L.L., Libby R.T., Rivkin N., Magen I., Reichenstein I., Oppenheim H., Eilam R., Silvestroni A., Alajajian B., Ben-Dov I.Z., Aebischer J., Savidor A., Levin Y., Sons R., Hammond S.M., Ravits J.M., Möller T., Hornstein E. Dysregulated miRNA biogenesis downstream of cellular stress and ALS-causing mutations: a new mechanism for ALS. EMBO J. 2015;34(21):2633–2651. doi: 10.15252/embj.201490493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bicker S., Schratt G. MicroRNAs in ALS: small pieces to the puzzle. EMBO J. 2015;34(21):2601–2603. doi: 10.15252/embj.201592805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kawahara Y., Mieda-Sato A. TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proc. Natl. Acad. Sci. U. S. A. 2012;109(9):3347–3352. doi: 10.1073/pnas.1112427109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim K.Y., Lee H.W., Shim Y.M., Mook-Jung I., Jeon G.S., Sung J.J. A phosphomimetic mutant TDP-43 (S409/410E) induces Drosha instability and cytotoxicity in Neuro 2A cells. Biochem. Biophys. Res. Commun. 2015;464(1):236–243. doi: 10.1016/j.bbrc.2015.06.125. [DOI] [PubMed] [Google Scholar]
- 82.Freischmidt A., Müller K., Ludolph A.C., Weishaupt J.H. Systemic dysregulation of TDP-43 binding microRNAs in amyotrophic lateral sclerosis. Acta Neuropathol. Commun. 2013;1:42. doi: 10.1186/2051-5960-1-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Morlando M., Dini Modigliani S., Torrelli G., Rosa A., Di Carlo V., Caffarelli E., Bozzoni I. FUS stimulates microRNA biogenesis by facilitating co-transcriptional Drosha recruitment. EMBO J. 2012;31(24):4502–4510. doi: 10.1038/emboj.2012.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dini Modigliani S., Morlando M., Errichelli L., Sabatelli M., Bozzoni I. An ALS-associated mutation in the FUS 3'-UTR disrupts a microRNA-FUS regulatory circuitry. Nat. Commun. 2014;5:4335. doi: 10.1038/ncomms5335. [DOI] [PubMed] [Google Scholar]
- 85.Svetoni F., De Paola E., La Rosa P., Mercatelli N., Caporossi D., Sette C., Paronetto M.P. Post-transcriptional regulation of FUS and EWS protein expression by miR-141 during neural differentiation. Hum. Mol. Genet. 2017;26(14):2732–2746. doi: 10.1093/hmg/ddx160. [DOI] [PubMed] [Google Scholar]
- 86.De Santis R., Santini L., Colantoni A., Peruzzi G., de Turris V., Alfano V., Bozzoni I., Rosa A. FUS mutant human Motoneurons display altered transcriptome and microRNA pathways with implications for ALS pathogenesis. Stem Cell Rep. 2017;9(5):1450–1462. doi: 10.1016/j.stemcr.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dadon-Nachum M., Melamed E., Offen D. The "dying-back" phenomenon of motor neurons in ALS. J. Mol. Neurosci. 2011;43(3):470–477. doi: 10.1007/s12031-010-9467-1. [DOI] [PubMed] [Google Scholar]
- 88.Fischer L.R., Culver D.G., Tennant P., Davis A.A., Wang M., Castellano-Sanchez A., Khan J., Polak M.A., Glass J.D. Amyotrophic lateral sclerosis is a distal axonopathy: evidence in mice and man. Exp. Neurol. 2004;185(2):232–240. doi: 10.1016/j.expneurol.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 89.Kovanda A., Leonardis L., Zidar J., Koritnik B., Dolenc-Groselj L., Ristic Kovacic S., Curk T., Rogelj B. Differential expression of microRNAs and other small RNAs in muscle tissue of patients with ALS and healthy age-matched controls. Sci. Rep. 2018;8(1):5609. doi: 10.1038/s41598-018-23139-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sobuś A., Baumert B., Litwińska Z., Gołąb-Janowska M., Stępniewski J., Kotowski M., Pius-Sadowska E., Kawa M.P., Gródecka-Szwajkiewicz D., Peregud-Pogorzelski J., Dulak J., Nowacki P., Machaliński B. Safety and feasibility of lin- cells administration to ALS patients: a novel view on humoral factors and miRNA profiles. Int. J. Mol. Sci. 2018;19(5) doi: 10.3390/ijms19051312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gaudet A.D., Fonken L.K., Watkins L.R., Nelson R.J., Popovich P.G. MicroRNAs: roles in regulating neuroinflammation. Neuroscientist. 2018;24(3):221–245. doi: 10.1177/1073858417721150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cunha C., Gomes C., Vaz A.R., Brites D. Exploring new inflammatory biomarkers and pathways during LPS-induced M1 polarization. Mediat. Inflamm. 2016;2016:6986175. doi: 10.1155/2016/6986175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Parisi C., Arisi I., D'Ambrosi N., Storti A.E., Brandi R., D'Onofrio M., Volonté C. Dysregulated microRNAs in amyotrophic lateral sclerosis microglia modulate genes linked to neuroinflammation. Cell Death Dis. 2013;4:e959. doi: 10.1038/cddis.2013.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Louafi F., Martinez-Nunez R.T., Sanchez-Elsner T. MicroRNA-155 targets SMAD2 and modulates the response of macrophages to transforming growth factor-{beta} J. Biol. Chem. 2010;285(53):41328–41336. doi: 10.1074/jbc.M110.146852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Koval E.D., Shaner C., Zhang P., du Maine X., Fischer K., Tay J., Chau B.N., Wu G.F., Miller T.M. Method for widespread microRNA-155 inhibition prolongs survival in ALS-model mice. Hum. Mol. Genet. 2013;22(20):4127–4135. doi: 10.1093/hmg/ddt261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhou F., Guan Y., Chen Y., Zhang C., Yu L., Gao H., Du H., Liu B., Wang X. miRNA-9 expression is upregulated in the spinal cord of G93A-SOD1 transgenic mice. Int. J. Clin. Exp. Pathol. 2013;6(9):1826–1838. eCollection 2013. [PMC free article] [PubMed] [Google Scholar]
- 97.Hoye M.L., Koval E.D., Wegener A.J., Hyman T.S., Yang C., O'Brien D.R., Miller R.L., Cole T., Schoch K.M., Shen T., Kunikata T., Richard J.P., Gutmann D.H., Maragakis N.J., Kordasiewicz H.B., Dougherty J.D., Miller T.M. MicroRNA profiling Reveals marker of motor neuron disease in ALS models. J. Neurosci. 2017;37(22):5574–5586. doi: 10.1523/JNEUROSCI.3582-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hoye M.L., Regan M.R., Jensen L.A., Lake A.M., Reddy L.V., Vidensky S., Richard J.P., Maragakis N.J., Rothstein J.D., Dougherty J.D., Miller T.M. Motor neuron-derived microRNAs cause astrocyte dysfunction in amyotrophic lateral sclerosis. Brain. 2018;141(9):2561–2575. doi: 10.1093/brain/awy182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Campos-Melo D., Hawley Z.C.E., Strong M.J. Dysregulation of human NEFM and NEFH mRNA stability by ALS-linked miRNAs. Mol. Brain. 2018;11(1):43. doi: 10.1186/s13041-018-0386-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gilad S., Meiri E., Yogev Y., Benjamin S., Lebanony D., Yerushalmi N., Benjamin H., Kushnir M., Cholakh H., Melamed N., Bentwich Z., Hod M., Goren Y., Chajut A. Serum microRNAs are promising novel biomarkers. PLoS One. 2008;3(9):e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.de Andrade H.M., de Albuquerque M., Avansini S.H., de S Rocha C., Dogini D.B., Nucci A., Carvalho B., Lopes-Cendes I., França M.C. MicroRNAs-424 and 206 are potential prognostic markers in spinal onset amyotrophic lateral sclerosis. J. Neurol. Sci. 2016;368:19–24. doi: 10.1016/j.jns.2016.06.046. [DOI] [PubMed] [Google Scholar]
- 102.Waller R., Goodall E.F., Milo M., Cooper-Knock J., Da Costa M., Hobson E., Kazoka M., Wollff H., Heath P.R., Shaw P.J., Kirby J. Serum miRNAs miR-206, 143-3p and 374b-5p as potential biomarkers for amyotrophic lateral sclerosis (ALS) Neurobiol. Aging. 2017;55:123–131. doi: 10.1016/j.neurobiolaging.2017.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Toivonen J.M., Manzano R., Oliván S., Zaragoza P., García-Redondo A., Osta R. MicroRNA-206: a potential circulating biomarker candidate for amyotrophic lateral sclerosis. PLoS One. 2014;9(2) doi: 10.1371/journal.pone.0089065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Roberts T.C., Godfrey C., McClorey G., Vader P., Briggs D., Gardiner C., Aoki Y., Sargent I., Morgan J.E., Wood M.J. Extracellular microRNAs are dynamic non-vesicular biomarkers of muscle turnover. Nucleic Acids Res. 2013;41(20):9500–9513. doi: 10.1093/nar/gkt724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jeyaseelan K., Lim K.Y., Armugam A. MicroRNA expression in the blood and brain of rats subjected to transient focal ischemia by middle cerebral artery occlusion. Stroke. 2008;39(3):959–966. doi: 10.1161/STROKEAHA.107.500736. [DOI] [PubMed] [Google Scholar]
- 106.Shioya M., Obayashi S., Tabunoki H., Arima K., Saito Y., Ishida T., Satoh J. Aberrant microRNA expression in the brains of neurodegenerative diseases: miR-29a decreased in Alzheimer disease brains targets neurone navigator 3. Neuropathol. Appl. Neurobiol. 2010;36(4):320–330. doi: 10.1111/j.1365-2990.2010.01076.x. [DOI] [PubMed] [Google Scholar]
- 107.Hansen T., Olsen L., Lindow M., Jakobsen K.D., Ullum H., Jonsson E., Andreassen O.A., Djurovic S., Melle I., Agartz I., Hall H., Timm S., Wang A.G., Werge T. Brain expressed microRNAs implicated in schizophrenia etiology. PLoS One. 2007;2(9):e873. doi: 10.1371/journal.pone.0000873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang B., Pan X. RDX induces aberrant expression of microRNAs in mouse brain and liver. Environ. Health Perspect. 2009;117(2):231–240. doi: 10.1289/ehp.11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.De Felice B., Guida M., Coppola C., De Mieri G., Cotrufo R. A miRNA signature in leukocytes from sporadic amyotrophic lateral sclerosis. Gene. 2012;508(1):35–40. doi: 10.1016/j.gene.2012.07.058. [DOI] [PubMed] [Google Scholar]
- 110.Rothstein J.D. Therapeutic horizons for amyotrophic lateral sclerosis. Curr. Opin. Neurobiol. 1996;6(5):679–687. doi: 10.1016/S0959-4388(96)80103-6. [DOI] [PubMed] [Google Scholar]
- 111.Freischmidt A., Müller K., Zondler L., Weydt P., Volk A.E., Božič A.L., Walter M., Bonin M., Mayer B., von Arnim C.A., Otto M., Dieterich C., Holzmann K., Andersen P.M., Ludolph A.C., Danzer K.M., Weishaupt J.H. Serum microRNAs in patients with genetic amyotrophic lateral sclerosis and pre-manifest mutation carriers. Brain. 2014;137(Pt 11):2938–2950. doi: 10.1093/brain/awu249. [DOI] [PubMed] [Google Scholar]
- 112.Liguori M., Nuzziello N., Introna A., Consiglio A., Licciulli F., D'Errico E., Scarafino A., Distaso E., Simone I.L. Dysregulation of MicroRNAs and target genes networks in peripheral blood of patients with sporadic amyotrophic lateral sclerosis. Front. Mol. Neurosci. 2018;11:288. doi: 10.3389/fnmol.2018.00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Basak I., Patil K.S., Alves G., Larsen J.P., Møller S.G. microRNAs as neuroregulators, biomarkers and therapeutic agents in neurodegenerative diseases. Cell. Mol. Life Sci. 2016;73(4):811–827. doi: 10.1007/s00018-015-2093-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rupaimoole R., Slack F.J. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017;16(3):203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 115.Laird A.S., Mackovski N., Rinkwitz S., Becker T.S., Giacomotto J. Tissue-specific models of spinal muscular atrophy confirm a critical role of SMN in motor neurons from embryonic to adult stages. Hum. Mol. Genet. 2016;25(9):1728–1738. doi: 10.1093/hmg/ddw044. [DOI] [PubMed] [Google Scholar]
- 116.Hua Y., Sahashi K., Rigo F., Hung G., Horev G., Bennett C.F., Krainer A.R. Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature. 2011;478(7367):123–126. doi: 10.1038/nature10485. [DOI] [PMC free article] [PubMed] [Google Scholar]