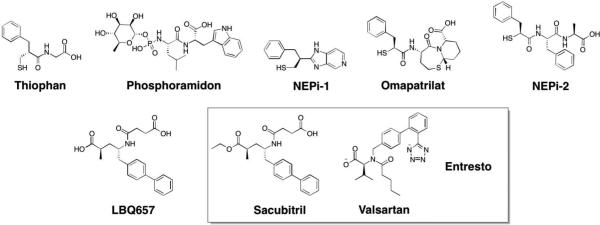
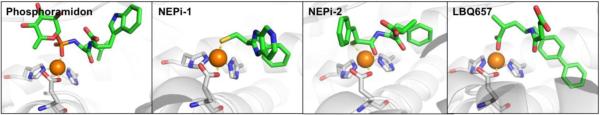
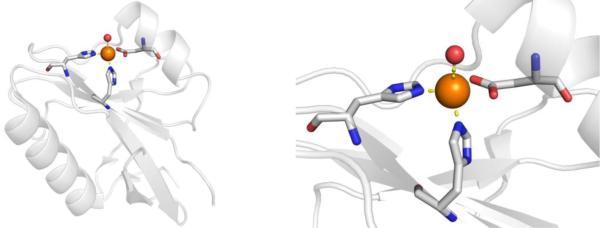
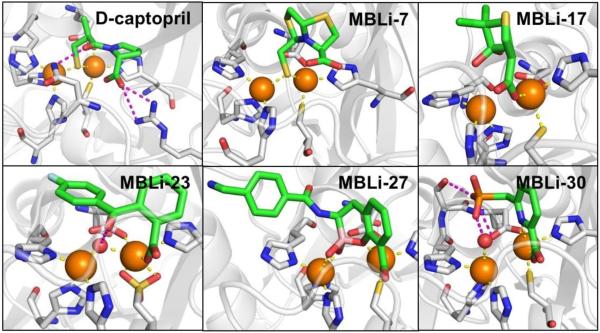
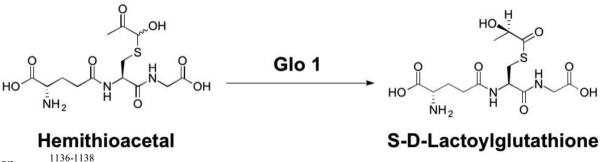
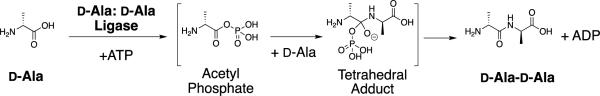
Abstract
Metalloenzymes are central to a wide scope of essential biological activities, including nucleic acid modification, protein degradation, and many others. The role of metalloenzymes in these processes also makes them central for the progression of many diseases, and as such, makes metalloenzymes attractive targets for therapeutic intervention. Increasing awareness of the role metalloenzymes play in disease and their importance as a class of targets has amplified interest in the development of new strategies to develop inhibitors and ultimately useful drugs. In this review, we provide a broad overview of several drug discovery efforts focused on metalloenzymes and attempt to map out the current landscape of high-value metalloenzyme targets.
1. Introduction
1.1. Metalloenzymes as Medicinal Targets
Many proteins require metal ions to properly function and as such are referred to as metalloproteins. Generally, the role of metal ions in metalloproteins fall into two broad categories: structural and functional. Structural metal ions are required for proper folding of a protein, with a canonical example being the zinc finger proteins. In zinc finger proteins, the Zn2+ ion serves to transform an unstructured polypeptide into a properly folded protein domain capable of nucleic acid-protein or protein-protein binding.1 Structural metal ions, via their influence on protein assembly, can also serve in a regulatory capacity. Functional metal ions are found at the active site of metalloenzymes and carry out a diverse range of processes, such as electron transfer, substrate recognition/binding, and catalysis that together serve a wide variety of biological functions. For example, the role of metal ions as conduits for electron transfer is represented by metalloproteins that utilize well studied Cu centers, Fe-S clusters, or Fe-heme (i.e., cytochrome) co-factors.2 In some cases, these redox centers can also serve a dual role as catalytic sites.
When the functional metal ion serves to promote catalysis, the metalloprotein can be categorized as a metalloenzyme. The ubiquitous roles of metalloenzymes in biology also results in metalloenzymes playing central roles in the propagation of many diseases. This can be due to the overexpression, enhanced activation, or misregulation of an endogenous metalloenzyme. In other cases, such as metallo-beta-lactamases or viral endonucleases, the normal, primary function of the metalloenzyme serves to proliferate a pathogenic infection. The metalloenzymes involved in the proliferation of human disease are the subject of this review. More specifically, those metalloenzymes that are validated targets, or where the biological role of the metalloenzyme supports the case for therapeutic intervention, are of greatest interest for the development of metalloenzyme inhibitors. An excellent 2016 review by Liao and co-workers3 highlighted a number metalloenzyme targets of interest and the state of inhibitor development for these targets. The collection presented here is structured similarly, but covers a broader range of potential targets. After a brief discussion of recent drug approvals and online resources, the subsequent sections will discuss different metalloenzymes (or class of metalloenzymes) as therapeutic targets. Metalloenzyme targets are organized by enzyme commission (EC) numbers and for each potential target, the role of the metalloenzyme in biology and disease, protein and active site structure, state of inhibitor development, and future prospects are discussed. Two metalloenzymes, carbonic anhydrases (Section 1.4) and matrix metalloproteinases (Section 1.5), are discussed in concise sections prior to the remaining metalloenzyme sections. These two metalloenzymes represent the earliest and most comprehensive efforts to develop metalloenzyme inhibitors and are placed at the beginning of the review to provide context for the remaining sections. Given the vast literature on both targets, the sections on carbonic anhydrases and matrix metalloproteinases are rather short, with many excellent reviews are available elsewhere. Given the large number of potential targets, this review is not intended to be comprehensive, but does attempt to show the breadth, current state, and value of the field. This review is largely focused on the primary published literature, with fewer examples taken from the patent literature. Metalloproteins where metal ions serve a structural or other non-catalytic role will not be discussed in this review; however, these metalloproteins may also be viable therapeutic targets and the reader is referred to other publications on this subject.4,5
1.2. Scope of Metalloenzyme Targets
An early review by Solomon in 1996 stated 52% of all proteins in the Protein Data Bank (Section 1.3) included a metal ion.6 A 2008 study using the Metal MACiE database (Section 1.3), suggested ~40% of enzymes with known structures were metal-dependent.7 Another review by Robinson in 2009 states that nearly half of all enzymes require a metal ion for proper function.8 Collectively, the literature suggests that number of enzymes that can be categorized as metalloenzymes is between ~40–50%.
The majority of metalloenzyme inhibitors are small molecules (i.e., not biologics), and hence only small molecule inhibitors will be discussed in this review. The vast majority of FDA-approved drugs that target metalloenzymes are reported to act via coordination of the inhibitor to the catalytic active site metal ion. This is true for inhibitors that have been reported from both academic and pharmaceutical laboratories, including those that have not entered clinical trials. The term metal-binding pharmacophore (MBP, also often referred to as a metal-binding group, MBG, in the literature) will be used to refer to the functional group in a metalloenzyme inhibitor responsible for binding the active site metal ion. Apart from some deliberate efforts, particularly in the realm of matrix metalloproteinases (Section 1.5), to develop non-coordinating inhibitors, metalloenzyme drug development has been focused on compounds that form dative (i.e., coordinate) bonds to the catalytic metal center. As such, the development of MBP-containing inhibitors will be the emphasis of this review.
An examination of FDA-approved drugs over the last five years (2013–2017, Table 1) shows that 181 drugs received FDA approval, of which 137 are new molecular entities (NMEs) and 44 are biologics license applications (BLAs). Interestingly, of the 137 NMEs, 13 target metalloenzymes, representing ~7% of the FDA-approved drugs in the last five years (Figure 1). An additional 17 NMEs of the last five years are kinase inhibitors, which contain Mg2+ ions that act as a co-substrate with ATP, but these inhibitors, like other reported kinase inhibitors, do not engage in Mg2+ binding.9,10 With kinase inhibitors representing >9% of the 181 drugs approved by the FDA in the last five years, perhaps there should be more efforts to develop metal binding inhibitors against these high-value targets.
Table 1.
FDA-Approved Drugs by Year.
| Year | Number of FDA-Approved Drugs | NMEs for Metalloenzyme Targets | Ref. | ||||
|---|---|---|---|---|---|---|---|
| NME | BLA | Total | Not Metal-Binding | Metal-Binding | Total | ||
| 2013 | 25 | 2 | 27 | 1 | 2 | 3 | 11,12 |
| 2014 | 30 | 11 | 41 | 2 | 3 | 5 | 13,14 |
| 2015 | 33 | 12 | 45 | 0 | 3 | 3 | 15,16 |
| 2016 | 15 | 7 | 22 | 0 | 1 | 1 | 17,18 |
| 2017 | 34 | 12 | 46 | 1 | 0 | 1 | 19,20 |
| 2013–2017 | 137 | 44 | 181 | 4 | 9 | 13 | This work |
| All | ~1565 | N/A | N/A | N/A | ~64 | N/A | 3 |
Figure 1.
FDA-approved inhibitors of metalloenzyme between 2013 and 2017. Metal-binding pharmacophores (MBPs) are shown in red.
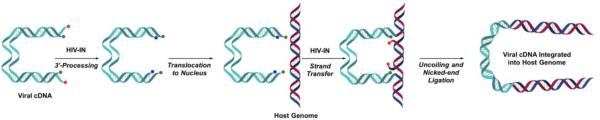
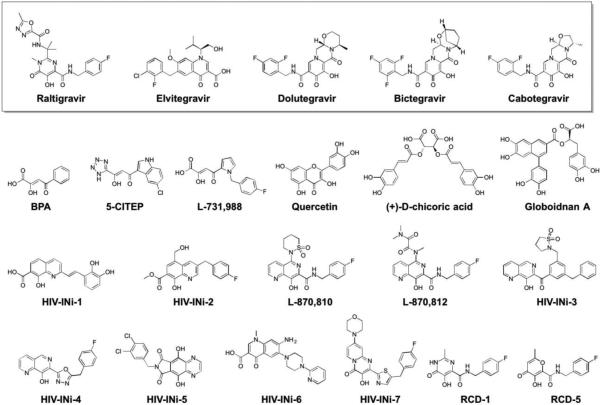
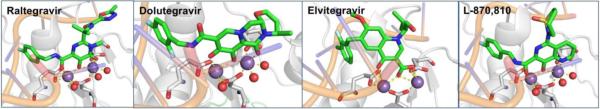
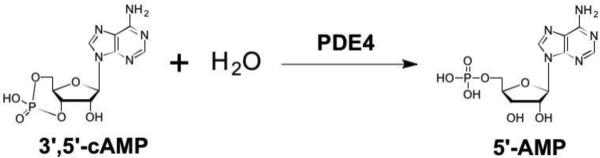
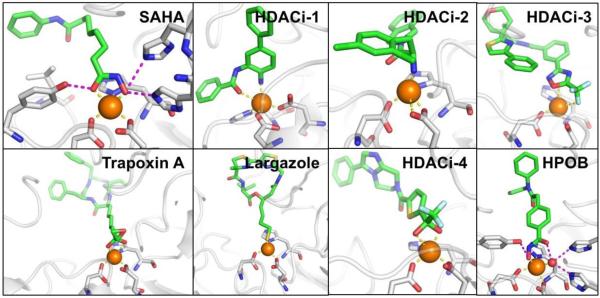
The 13 NMEs (Figure 1) target six different metalloenzyme classes, including HIV-1 integrase (Section 5.1), histone deacetylase (Section 7.3), phosphodiesterase 4 (Section 5.4), tryptophan hydroxylase,21 and heme proteins (Section 10). Two NMEs were approved as histone deacetylase inhibitors and another two as HIV-1 integrase inhibitors, all four of which possess prominent MBPs for binding the active site metal ions in these targets. The two NMEs targeting histone deacetylases, belinostat and panobinostat,14,16 feature the ubiquitous, bidentate hydroxamic acid MBP. In contrast, the HIV-1 integrase inhibitors dolutegravir and elvitegravir have more complex and diverse MBPs; however, both use a triad of donor atoms to bind the dinuclear active site of HIV-1 integrase (Section 5.1). One of the HIV-1 integrase inhibitors, elvitegravir, achieved earlier FDA-approval in 2012 as a component of the drug Stribild, a combination therapy consisting of elvitegravir, cobicistat (cytochrome P450 inhibitor), and tenofovir disoproxil (reverse transcriptase inhibitors), and emtricitabine (reverse transcriptase inhibitors).22 In 2015, elvitegravir was again approved by the FDA as an active ingredient in the combination drug Genvoya. Two phosphodiesterase 4 inhibitors gained FDA-approval, apremilast and crisaborole, the latter of which has been confirmed as metal-binding (Section 5.4). Crisaborole, which features a boronic acid MBP, is the first and only metal-binding inhibitor of phosphodiesterase 4 to receive FDA approval to date.
Telotristat ethyl is a new FDA-approved tryptophan hydroxylase (TPH) inhibitor for the treatment of carcinoid syndrome diarrhea. TPH is a non-heme Fe metalloenzyme that hydroxylates tryptophan to 5-hydroxytryptophan. Both in vivo and in vitro, telotristat ethyl is hydrolyzed from an ethyl ester to a carboxylic acid, called telotristat.23 There is no crystal structure of telotristat bound to TPH, but this molecule is not anticipated to utilize metal binding as it lacks any obvious MBP.24 Nearly all inhibitors developed for TPH are not metal binding, with a few exceptional catechol compounds that chelate to the Fe ion;25 however, to the best of our knowledge, no metal binding TPH inhibitors have progressed to clinical trials.
In the last five years, six NMEs targeting heme proteins have emerged. Of these, four contain an imidazole, triazole, or thiazole ring where a nitrogen atom likely coordinates to the active site heme center. Three of these (Efinaconazole, Isavuconzale, Luliconazole) are antifungals that target lanosterol 14α-demethylase.3 The fourth (Ritonavir) is a component of a drug whose other active ingredients inhibit hepatitis C virus proteins. Ritonavir targets cytochrome P4503A4 and is used to increase exposure of the hepatitis C virus inhibitors. A crystal structure of Ritonavir bound to cytochrome P4503A4 shows the nitrogen atom of the thiazole ring in Ritonavir coordinating to the heme group.26 Riociguat also targets a heme enzyme, soluble guanylate cyclase (sGC), to treat pulmonary hypertension, but does not utilize metal binding. Riociguat is a stimulator (not an inhibitor) of sGC and is expected to synergize with the substrate NO.27 The mechanism of action of miltefosine (a cytochrome c oxidase inhibitor and anti-leishmanial agent) is unknown, but it has no obvious MBPs and structures with this miltefosine and its target are not available.14
In the last five years, nine NMEs targeting one or more of three essential proteins involved in RNA replication of hepatitis C virus (HCV) have gained FDA approval. These nine NMEs include inhibitors of NS5B, NS5A, and/or NS3/4A, all of which contain structural metal centers. NS5B is an RNA dependent RNA polymerase that contains an Mg2+-nucleotide binding site. NS5A is a dimeric protein in which each dimer contains a structural Zn2+ ion bound by four cysteine residues.28 NS3/4A is a serine protease that contains a structural Zn2+ ion coordinated by three cysteine residues as well as a histidine residue through a water bridge.29
Beyond the last five years of FDA drug approvals (Table 1), two recent studies provide useful information on the number of metal binding metalloenzyme inhibitors for all FDA-approved drugs. One of these studies identified ~1,453 NMEs as of December 31, 2013, with this number including drugs no longer marketed.30 The second study by Liao and coworkers identified 60 metal binding metalloenzyme inhibitors up through its date of publication in 2016.3 Taking data from these two studies and combining them with the analysis of FDA-approved drugs in the last five years, as well as the targets in this review, it can be estimated that as of December 31, 2017 that ~1,565 NMEs have received FDA approval. Of the ~1,565 NMEs, ~62 (~4%) target metalloenzymes and utilize metal binding (Table 2). This analysis is based on the 2016 review by Liao, which lists 60 FDA-approved drugs are metalloenzyme inhibitors.3 This figure was updated to 62 FDA-approved metalloenzyme inhibitors based on recent FDA approvals since the publication of the review by Liao. These ~62 drugs do not target 62 distinct metalloenzymes; rather, these inhibitors only target a small sampling of metalloenzyme classes (Table 2). Considering there are >1500 FDA-approved drugs (both NMEs and BLAs), with only ~62 drugs targeted to metalloenzymes, this indicates that <4% of drugs engage metalloenzymes and that the metalloenzyme target space is vastly underexploited. Despite their ubiquity, the number of metalloenzymes that are the validated targets of FDA-approved therapeutics is relatively small, representing a vast and underexploited target space.31,32
Table 2.
FDA-Approved Metal Binding Metalloenzyme Inhibitors. Adapted and updated from Liao et al.3
| EC number | Metalloenzyme Target | FDA-Approved, Metal-Binding Metalloenzyme Inhibitors (~64 Total) | Number of Inhibitors (%) |
|---|---|---|---|
| 1.1 | Lanosterol 14α-demethylase | Albaconazole, Bifonazole, Butoconazole, Clotrimazole, Econazole, Efinaconazole, Fenticonazole, Fluconazole, Isavuconazole, Isoconazole, Itraconazole, Ketoconazole, Luliconazole, Miconazole, Omoconazole, Oxiconazole, Posaconazole, Ravuconazole, Sertaconazole, Sulconazole, Terconazole, Tioconazole, Voriconazole | 23 (~37%) |
| 1.14 | Cytochrome P4503A4 | Ritonavir | 1 (~1.5%) |
| Tyrosinase | Hydroquinone | 1 (~1.5%) | |
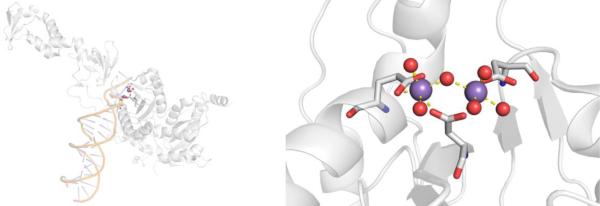
| 3.1 | HIV-1 integrase | Elvitegravir, Dolutegravir, Raltegravir | 3 (~5%) |
| Phosphodiesterase 4 | Crisaborole | 1 (~1.5%) | |
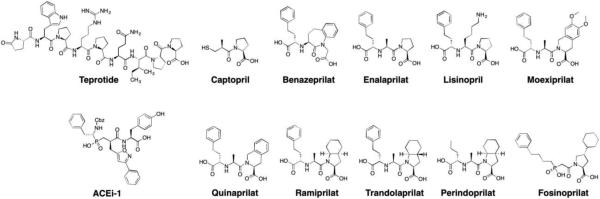
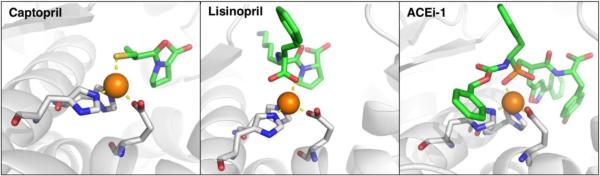
| 3.4 | Angiotensin converting enzymea | Benazepril, Captopril, Enalapril, Fosinopril, Lisinopril, Moexipril, Perindopril, Quinapril, Ramipril, Trandolapril | 10 (~16%) |
| Matrix metalloproteinase | Periostat | 1 (~1.5%) | |
| 3.5 | Histone deacetylase | Belinostat, Panobinostat, Romidepsin, Vorinost | 4 (~6%) |
| Lipoxygenase | Zileuton | 1 (~1.5%) | |
| 4.2 | Carbonic anhydrase | Acetazolamide, Bendroflumethiazide, Benzthiazide, Brinzolamide, Chlorothiazide, Cyclothiazide, Diazoxide, Diclofenamide, Dorzolamide, Ethoxzolamide, Furosemide, Hydrochlorothiazide, Hydroflumethiazide, Methazolamide, Methyclothiazide, Quinethazone, Topiramate, Trichlormethiazide, Zonisamide | 19 (~30%) |
In the review by Liao, Imidapril and Cilazapril are also listed as ACE inhibitors. We removed them from our table because they are not FDA-approved; however, they are approved in other countries.
Despite the overwhelming majority of metalloenzyme inhibitors utilizing coordinate bonds between the inhibitor and the active site metal center as a key interaction, the scope of functional groups used in inhibitors to form these important interactions is surprisingly small. A review of matrix metalloproteinase (MMP) inhibitors from 1999 by Whittaker provided a comprehensive report on the scope of compounds that had been explored up to that time.33 Although substantial chemical diversity could be found among the MMP inhibitors reported, nearly all of the compounds relied on a hydroxamic acid as the functional group (i.e., MBP) to bind to the Zn2+ ion in the MMP active site.33 The disproportionate utilization of the hydroxamic acid MBP for use in MMP inhibitors likely originates from earlier studies that utilized hydroxamic acid functionalized resins to purify MMP proteins from cells.33 Despite this innocuous origin, the hydroxamic acid evolved into the ‘go-to’ MBP for many future (and present) drug discovery efforts against metalloenzymes. Inhibitors that utilize hydroxamic acid MBPs are pervasive in metalloenzyme drug discovery programs, including MMPs, histone deacetylases, and many others. The nearly universal use of the hydroxamic acid MBP in metalloenzyme drug development will be highlighted in the enzyme sections below. Beyond hydroxamic acids, the scope of MBPs explored for metalloenzyme inhibition is rather limited, but will be highlighted throughout this review.
1.3. Database Resources
There are several online resources and databases for fragment- and structure-based drug discovery. Among them, the PDB, BRENDA, MetLigDB, CCDC, and others have information that is useful for identifying and examining metalloenzyme inhibitors. The Protein Data Bank (PDB, https://www.rcsb.org) is a comprehensive repository of macromolecules, currently containing >137,000 biomolecule structures, and it is particularly useful for querying structures for specific proteins and ligands.34 The PDB is a tremendously valuable resource; however, a shortcoming of the PDB in the context of metalloenzyme inhibitors is its limited search parameters specific to metalloproteins and metal-bound ligands. To overcome this limitation, the European Bioinformatics Institute (EMBL-EBI) developed PDBeMotif (http://www.ebi.ac.uk/pdbe-site/pdbemotif/), which is a database derived from the PDB that can be queried by ligand, binding sites, and small 3D structural and sequence motifs, including the ability to search by coordination geometry of metal ions in the protein structure.35 PDBeMotif has not been updated since October 2015; however, users can still upload their own protein structures to PDBeMotif for structural analysis and annotation.
The BRaunschweig ENzyme DAtabase (BRENDA, https://www.brenda-enzymes.org) is an online resource that contains comprehensive enzyme information on ~83,000 enzymes, including ~206,000 enzyme ligands.36 The database is regularly updated via manually extracting information from the primary literature and using text mining procedures as well as integration of external data and prediction algorithms.36 This results in a database of comprehensive enzyme information, including (but not limited to) substrates and products, inhibitors, cofactors, metal ions, Ki and IC50 values of inhibitors, and enzyme structures. Users can search the database using text- and structure-based queries, as well explore the database by enzyme classification, metabolic pathways, protein folding, and more. While BRENDA offers invaluable information on enzymes, it suffers the same limitations as the PDB in the context of metalloenzyme inhibitors: both have limited ability to query databases with parameters exclusive to metalloenzymes.
To this end, other databases have been created that exclusively catalogue metalloproteins. These include the Metalloprotein Ligand Interaction Database (MetLigDB, http://silver.sejong.ac.kr/MetLigDB/home.html), the Metal Mechanism, Annotation and Classification in Enzymes (Metal MACiE), the Prosthetic Centers and Metal Ions in Protein Active Sites (PROMISE) database, and the Metalloprotein Database (MDB).37–40 The Metal MACiE, PROMISE, and MDB have apparently been discontinued and their websites are no longer accessible; however, MetLigDB, created in 2011 at Sejong University, is still accessible. MetLigDB was constructed with the goal of aiding in the design of new metalloenzyme inhibitors. It contains over 400 different PDB structures for ~200 different small molecule ligands and 250 different metalloenzymes. Users may view the entire list of structures and can organize them according to MBP, by metal ion, by function, or by EC number. After selecting a specific entry, a results page provides basic information on the protein, as well as interactive structures of the metal-ligand coordination and overall protein. The information on each entry in MetLigDB is limited and the literature references are not complete, but more information can be obtained by searching the PDB using the PDB code provided for each entry. A current examination found that MetLigDB does not include structures of some recent FDA-approved drugs, suggeting the database has not been updated for several years. Despite these limitations, MetLigDB is a useful resource for its ability to organize search results according to MBP and metal ion, which lends itself to assisting in the development of new metalloenzyme inhibitors as the creators intended.
In the development of new MBPs, it can be useful to examine their metal coordination modes in small molecule crystal structures prior to using these fragments in screening campaigns with proteins. The Cambridge Structural Database (CSD) from the Cambridge Crystallographic Data Centre (CCDC, https://www.ccdc.cam.ac.uk) is a comprehensive repository of small molecule structures. This database may be useful for querying MBP motifs to identify their coordination modes and geometry in small molecule structures. This database does not include information on protein structures.
Overall, the PDB and BRENDA are comprehensive and up-to-date databases of biomolecule structures and enzyme information. As such, they serve as useful starting points for surveying structures of and information on metalloenzymes and ligands. More specific information on metal-ligand coordination environments and properties may be gained from MetLigDB; however, this resource is not up-to-date. Finally, the CCDC may be useful for examining the metal coordination mode of MBPs prior to screening potential against metalloenzyme targets.
1.4. Carbonic Anhydrase (CA, EC 4.2.1.1)
Function in Biology and Disease.
Carbonic anhydrases (CAs) were the first Zn2+-dependent metalloenzymes characterized. CAs catalyze the reversible hydration of carbon dioxide to form the bicarbonate anion. This reaction is important for a number of biological processes, including regulation of blood pH and CO2 content. Being the oldest known Zn2+-dependent metalloenzyme, inhibitors of CAs have long been studied for uses as diverse as diuretics and anticancer agents. The most widespread use of CA inhibitors is in the treatment of glaucoma, with ~20 FDA-approved CA inhibitors reported to date. A 2009 book, co-edited and co-authored by C.T. Supuran (one of the leading investigators of CA inhibitors), extensively reviews CAs and the development of CA inhibitors, as well as several other Zn2+-dependent metalloenzymes. The reader is referred to this excellent resource for additional details.41
Active Site Structure.
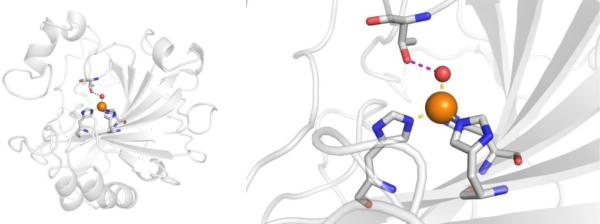
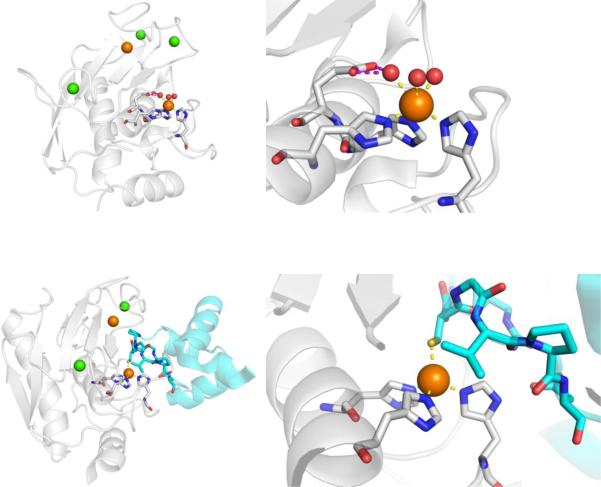
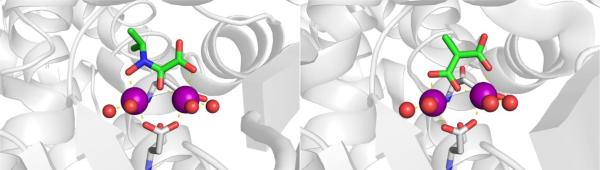
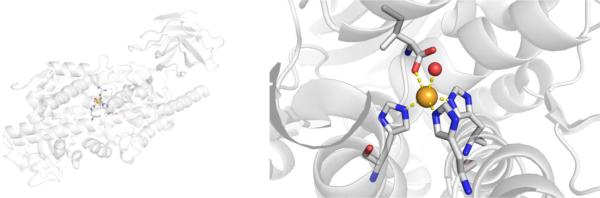
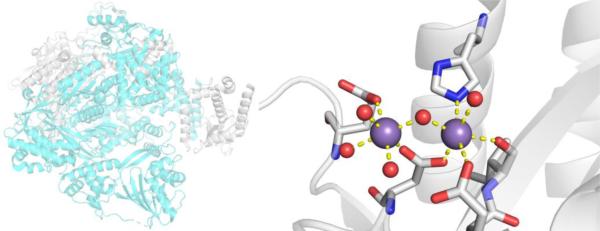
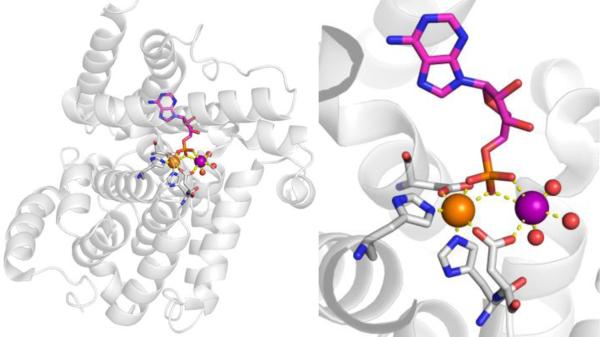
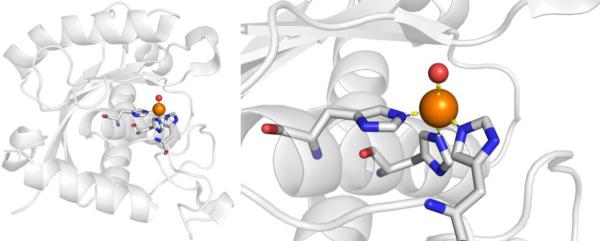
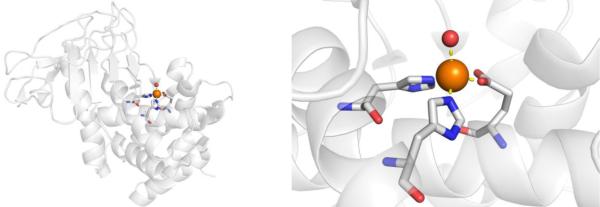
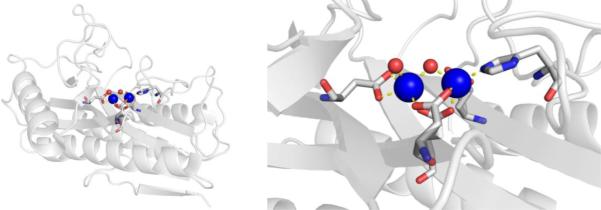
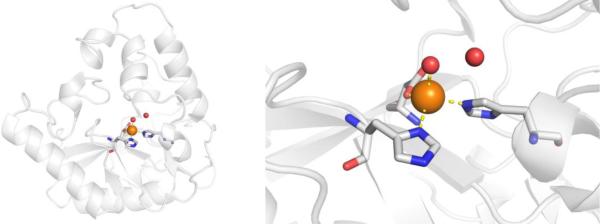
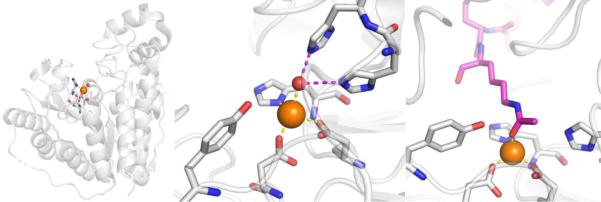
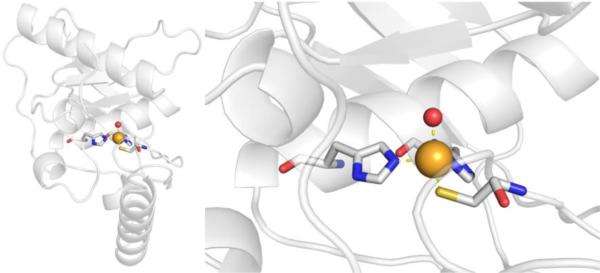
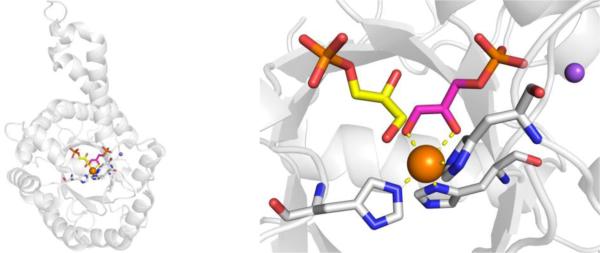
CAs are found in a broad range of organisms, with those found in mammals (~16 in total) being of the α-CA form.41 As a canonical example, human carbonic anhydrase II (hCAII) is a monomeric metalloenzyme ~29 kDa in size. The core of the protein is a twisted β-sheet dividing the protein approximately in half (Figure 2). The active site of the metalloenzyme is comprised of two halves, with a hydrophobic wall on one side and an ordered water network on the other. The active site of hCAII contains a catalytic Zn2+ ion at the bottom of an ~15 Å deep cleft, where the metal ion is bound by three His residues and a water molecule in a tetrahedral geometry (Figure 2).42 The metal-bound water is also stabilized by a hydrogen bond to a neighboring residue (Thr199), which is important for the catalytic activity of the enzyme.
Figure 2.
Stucture of hCAII (PDB 1CA2). The protein backbone is shown in a ribbon style, with coordinating residues and the hydrogen-bonding Thr199 shown in sticks and colored by element (carbons in gray). The Zn2+ ion is shown as an orange sphere and the coordinated water molecule is shown as a red sphere. Metal-ligand coordination bonds are shown as dashed yellow lines and hydrogen bonds as dashed magenta lines. The stylistic features of this figure are used throughout this review. All protein images were generated using PyMOL (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC).
Inhibitor Development.
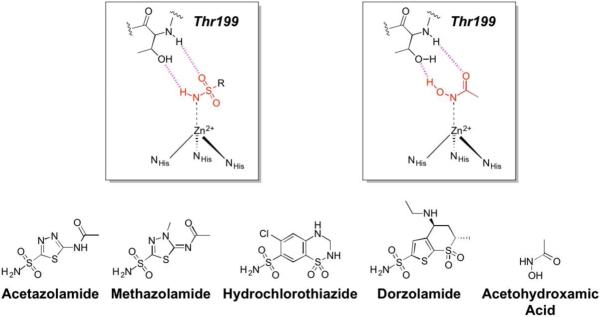
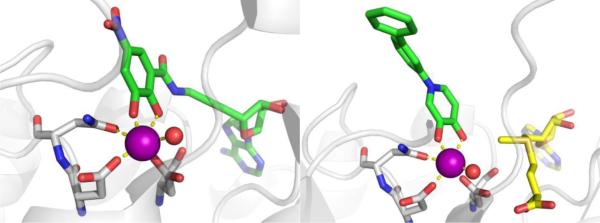
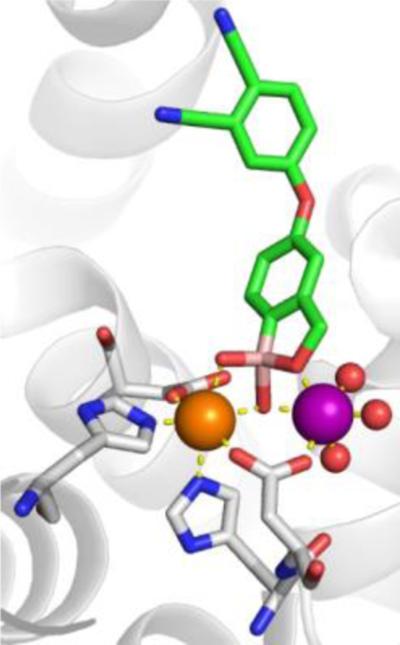
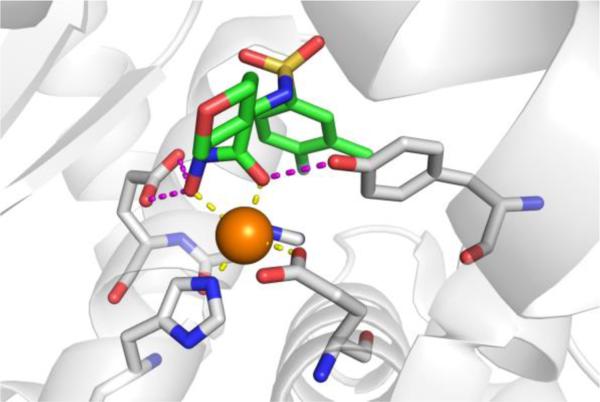
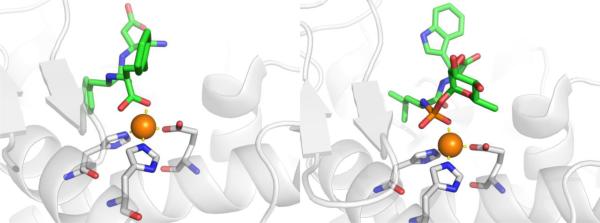
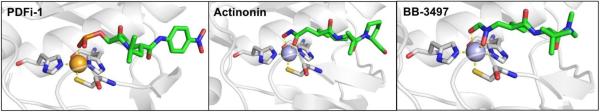
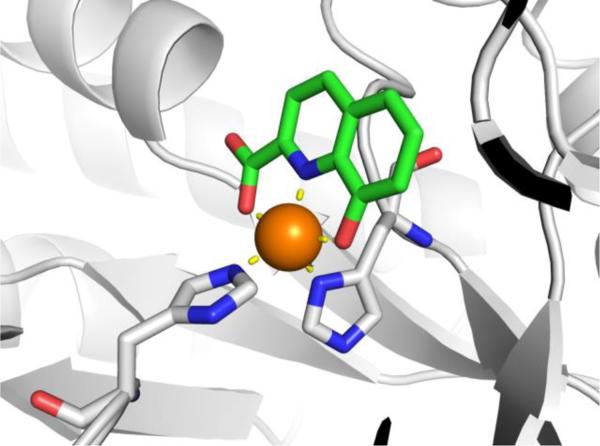
An incredibly diverse range of compounds have been identified as CA inhibitors (Figure 3). That stated, the most active, and clinically successful CA inhibitors have relied on a highly privileged sulfonamide MBP scaffold. Sulfonamide-based inhibitors, and their isosteres (e.g., sulfamates, sulfamides), have a very high affinity for CA binding. The mode of binding of sulfonamide MBP inhibitors is exemplified by FDA-approved acetazolamide (Figure 3) bound to human carbonic anhydrase 2 (hCAII, Figure 4).43 Acetazolamide displaces the metal-bound water molecule and binds to the Zn2+ ion via the deprotonated nitrogen atom of the sulfonamide MBP. This mode of coordination maintains the active site metal ion in a 4-coordinate, tetrahedral geometry. Importantly, the nitrogen and oxygen atoms of the sulfonamide engage in strong, bipartite hydrogen bonding with the nearby Thr199 residue (Figure 4). The combination of strong ligation by the anionic nitrogen donor atom and stabilization by an array of hydrogen bonding interactions has made sulfonamides the dominant, privileged MBP scaffold for CA inhibitors. Interesting, unlike hydroxamic acids, which are the dominant MBP used in inhibitors of MMPs (Section 1.5) and many other metalloenzymes, sulfonamides are generally not active against metalloenzymes other than CAs and have not been widely employed as MBPs beyond CAs. Considering that sulfonamide inhibitors of CAs predate hydroxamic acid inhibitors of MMPs, it is the unique selectivity of the sulfonamide-CA interaction that likely prevented this MBP from becoming more widespread, as became the case with hydroxamic acid compounds.
Figure 3.
Inhibitors of CA. Structures in the boxes show the mode of binding to the catalytic Zn2+ ion for a sulfonamide (left) and acetohydroxamic acid (right) with MBPs highlighted in red and hydrogen bonds in magenta.
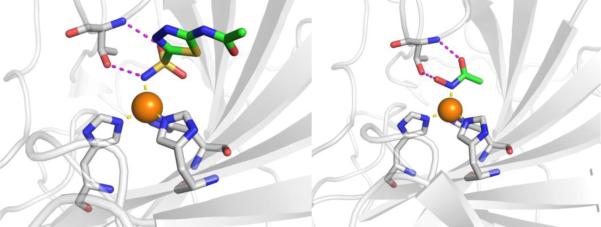
Figure 4.
Left: Stucture of acetazolamide bound to hCAII (PDB 3HS4). Right: Stucture of acetohydroxamic acid bound to hCAII (PDB 1AM6). Inhibitors are shown as sticks and colored by element (carbons in green). The protein backbones are shown in a ribbon style, with coordinating residues shown as sticks and colored by element (carbons in gray). The Zn2+ ions are shown as orange spheres. Metal-ligand coordination bonds are shown as dashed yellow lines and hydrogen bonds as dashed magenta lines. The stylistic features of this figure are used throughout this review.
The unique and highly complementary interaction of sulfonamides and CA can be further illustrated by examining the mode of binding of acetohydroxamic acid, a simple hydroxamic acid MBP fragment (Figure 3). Hydroxamic acids, as shown throughout this review (Section 1.5), typically bind to metalloenzyme active sites in a bidentate, chelating mode using the two oxygen atoms of the hydroxamic acid MBP. However, when bound to CA, acetohydroxamic acid adopts a highly unusual monodentate mode of binding, whereby coordination to the Zn2+ ion occurs via the deprotonated nitrogen atom of the hydroxamate MBP (Figure 4).44 This odd mode of hydroxamic acid binding becomes more readily explained by the extensive hydrogen bonding interactions acetohydroxamic acid engages in with Thr199. The hydrogen bonding within the tight CA active site drives the unusual coordination mode of acetohydroxamic acid, such that this fragment binds in a manner that reflects that of the preferred sulfonamide MBPs.
Current and Future Prospects.
With more than a dozen CA inhibitors in clinical use today, it is clear that CA is a viable and valuable target. Despite studies into many different uses of CA inhibitors, their most common clinical use is for glaucoma. Most CA inhibitors, being dependent on the sulfonamide MBP, are not particularly isozyme selective, but rather broad spectrum CA inhibitors. The discovery of new CA inhibitors is generally focused on improved isoform selectivity that might prove valuable for a wider range of indications.45 Some CA isoforms, specifically hCAIX and hCAXII, have become more prominent targets due to their role in cancer and tumor survival. To this end, Phase I clinical trials (NCT02215850) have been successful with a sulfonamide compound (SLC-0111) in patients with solid tumors. Phase II clinical trials (NCT03450018) are slated to begin in 2018 for subjects who test positive for hCAIX in metastatic pancreatic ductal cancer. Other scaffolds have been examined as isoform selective CA inhibitors, although nothing has progressed into the clinic as far as those that possess the sulfonamide MBP. Interestingly, the unique fit of the sulfonamide MBP for the CA active site prevented its wide dissemination and adoption among metalloenzyme drug discovery efforts. Rather, the use of hydroxamic acids in MMP inhibitors became the ‘go-to’ MBP for many metalloenzyme programs, becoming a key factor in the progression (and sometimes lack thereof) of the field of metalloenzyme inhibition.
1.5. Matrix Metalloproteinase (MMP, EC 3.4.24)
Function in Biology and Disease.
Matrix metalloproteinases (MMPs) are a class of hydrolytic Zn2+-dependent metalloenzymes that serve, in large part, to breakdown and restructure the extracellular matrix (e.g., collagen) during processes such as embryogenesis and wound healing. Greater than 20 MMPs are known in humans and extensive reviews of the biological role of MMPs have been published elsewhere.46 Their role in tissue restructuring processes inspired the development of MMP inhibitors that could serve as agents for treating diseases where restructuring has gone awry, such as arthritis. MMP inhibitors were also extensively explored as anticancer agents, because of the role of MMPs in tumor metastasis and angiogenesis. Extensive reviews on the development of MMP inhibitors are available.33,47 Indeed, the search for MMP inhibitors represents the among the most exhaustive and extensive efforts to develop metalloenzyme inhibitors, and ‘set the stage’ for many metalloenzyme drug discovery efforts that followed.
Active Site Structure.
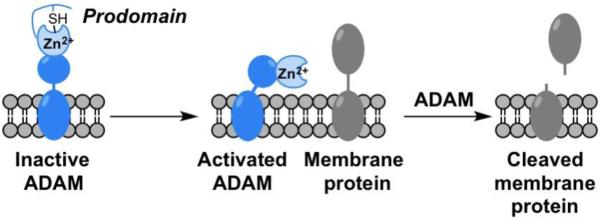
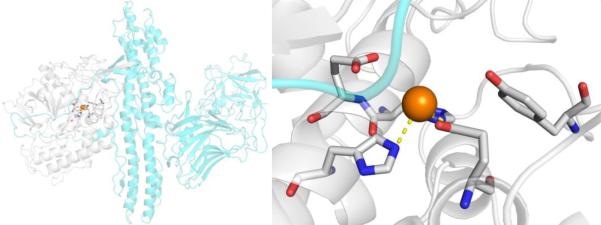
MMPs have a somewhat complex and multidomain protein structure, the details of which can be found elsewhere.33 The catalytic domain of MMPs consists of ~165 residues with a β-sheet wrapped around several α-helices to give an overall spherical shape to the domain (Figure 5). The protein contains two Zn2+ ions (one structural and one catalytic) and several structural Ca2+ ions. The catalytic Zn2+ ion is identifiable by the binding motif HEXXHXXGXXH, which contains three His residues that bind the catalytic Zn2+ ion. The Zn2+ ion is bound in a 6-coordinate, octahedral geometry by the three His ligands and three water molecules (Figure 5). The water molecules are activated by coordination to the Lewis acidic Zn2+ ion and serves as a nucleophile for amide bond cleavage in the mechanism of MMP peptide hydrolysis. One of the Zn2+-bound water molecules is hydrogen-bonded to a conserved Glu residue that serves to stabilize the nucleophile and serve as a general acid/base. MMPs are expressed as zymogens, self-inactivated enzymes, where a prodomain blocks catalytic activity. MMPs are activated by cleavage of this prodomain to produce the active MMP enzyme. Importantly, the MMP prodomain blocks the catalytic site, in part, by displacement of the Zn2+-bound water molecules and direct coordination of a Cys thiolate residue (Figure 5). This highlights the importance the use of metal ion coordination, even by Nature, for the inhibition of this metalloenzyme.
Figure 5.
Top: Structure of MMP-12 (PDB 2OXU). Bottom: Structure of proMMP-9 with the prodomain shown in cyan (PDB 5UE3). Ca2+ and Zn2+ are shown as green and orange spheres, respectively.
Inhibitor Development.
The development of MMP inhibitors is the canonical example of a metalloenzyme drug discovery program. During the 1990s, several major pharmaceutical companies had a substantial program on MMP inhibitors.33 The influence these programs would have on the chemistry and general attitudes toward metalloenzyme drug discovery has persisted until the present day. There are exhaustive reviews on MMP inhibitors,47 so only some representative highlights will be discussed here.
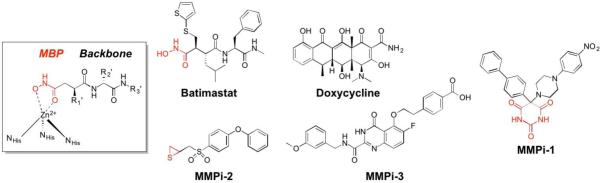
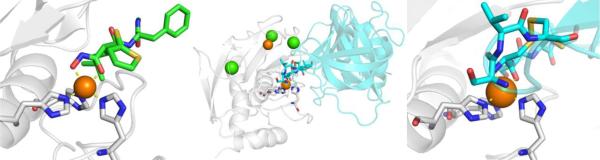
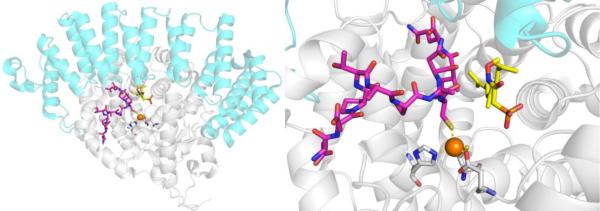
MMP inhibitors, like many other metalloenzyme inhibitors can be coarsely divided into two components: the MBP and the so-called ‘backbone’ (Figure 6). Among the reported MMP inhibitors, a rich chemical diversity of backbone substituents can be found, as efforts were made to obtain ever more potent and selective compounds.33,47 As mentioned earlier, the development of MMP inhibitors was largely, although not entirely, dominated by hydroxamic acid MBPs. Simple hydroxamic acids have a rather low affinity for the MMP Zn2+ ion;48 nonetheless, thousands of hydroxamic acid MMP inhibitors were reported and a number entered clinical trials, the first of which was batimastat (Figure 6), which began Phase I trials in 1994, proceeded to Phase III trials, but never achieved clinical approval.49 As shown in Figure 7, structural studies revealed that batimastat, like other hydroxamic acid MMP inhibitors, binds to the active site Zn2+ ion in a chelating, bidentate fashion using the two oxygen donor atoms from the MBP. This changes the coordination number (from 6 to 5) and geometry of the Zn2+ ion, when compared to the free form of the enzyme (Figure 5). Binding of the inhibitor to the Zn2+ ion displaces the metal-bound waters, occludes the active site, and thereby stops catalytic activity. Interestingly, the binding of batimastat and other hydroxamic acid MMP inhibitors shows a notable resemblance of MMP inhibition by tissue inhibitors of matrix metalloproteinases (TIMPs).50 The four TIMPs (TIMP1-4, ~25kDa) are endogenous inhibitors of MMPs that regulate their activity by binding to the Zn2+ active site using the terminal amino acid (Cys1) as a bidentate O,N-donor ligand (the Cys thiol is part of a disulfide in TIMPs, Figure 7).51 The determination that TIMPs also utilize direct metal coordination as a key interaction for inhibition of MMPs further highlights the importance of metal binding in the development of MMP inhibitors and metalloenzyme inhibitors more broadly. Beyond hydroxamic acids, a relatively limited number of other MBPs were explored in the development of MMP inhibitors (e.g., pyrimidine triones, MMPi-1, Figure 6). Unconventional approaches were also pursued in the development of MMP inhibitors, including ‘reactive’ thirane MBPs (MMPi-2)52 and inhibitors that lacked a MBP moiety altogether (MMPi-3).53 Ultimately, despite the reliance on the low affinity hydroxamic acid MBP, MMP inhibitors achieved a very high level of activity (with IC50 values commonly <10 nM) and selectivity (even with respect to different MMP isoforms). In addition, many important drug discovery methods, including fragment-based drug discovery (FBDD) and structure-activity relationships by nuclear magnetic resonance (SAR-by-NMR),54,55 were applied early on to MMP inhibitor discovery. Despite these efforts, only one MMP inhibitor (doxycycline, Figure 6) has been approved by the FDA for use in periodontal disease; however, this inhibitor is not a novel compound, but rather a repurposed antibiotic.
Figure 6.
Inhibitors of MMPs. The generic structure of a hydroxamic acid inhibitor (in box) is shown bound to a schematic of the MMP active site Zn2+ ion. MBPs are highlighted in red.
Figure 7.
Left: Structure of batimastat bound to MMP-12 (PDB 1JK3). Center and Right: Structure of MMP-3 complexed with TIMP-1 (cyan) (PDB 1GXD). In addition to the active site Zn2+ ion, a structural Zn2+ ion (orange) and structural Ca2+ ions (green) are shown.
Current and Future Prospects.
After several intense decades of MMP drug discovery, today few, if any, efforts to discover and develop clinically useful MMP inhibitors are underway.47 The cessation of these programs can be directly attributed the many potent and selective MMP inhibitors that ultimately failed in clinical trials. Many clinical trials reported a lack of efficacy or dose-limiting side effects that often manifest as muscloskeletal pain (i.e., tendinitis, which is consistent with the biological role of MMPs).46 Originally thought to be primarily involved in tissue restructuring, subsequent studies have revealed that the biological role of MMPs is complex and multifaceted. A recent article by Murphy nicely summarizes what she refers to as the MMP ‘roller coaster’.46 As MMP inhibitors began to fail in clinical trials, it became apparent that the activity of MMPs went far beyond tissue restructuring, and included effects on cytokines and chemokines that had diverse roles including immunity, inflammation, and other processes. The lack of appreciation for these other MMP roles, combined with potential shortcomings of the available clinical trial protocols, ultimately resulted in a relatively abrupt abandonment of this target class. Ultimately, the legacy of MMP inhibitor development are the chemical design principles behind MMP inhibitors. On the positive side, these vast efforts established the ability to design highly selective and active inhibitors against a metalloenzyme target, rich structure-activity relationships (SARs), and one of the most comprehensive resources (both intellectually and chemically) on the inhibition of a metalloenzyme. Vast libraries of hydroxamic acid based MMP inhibitors were developed that were later applied to other metalloenzyme targets of interest. On the negative side, the over-reliance on the hydroxamic acid MBP and clinical failing of MMP inhibitors left lingering questions about the feasibility of metalloenzyme inhibition for many decades.
2. Reductoisomerases
2.1. Human Alcohol Dehydrogenase (ADH, EC 1.1.1.1)
Function in Biology and Disease.
Human alcohol dehydrogenase (ADH) is primarily found in the liver, and plays a critical role as the first step of ethanol metabolism.56 ADH is not to be confused with aldehyde dehydrogenase, which uses the same abbreviation and is also involved in ethanol metabolism. ADH is a Zn2+-dependent metalloenzyme that oxidizes ethanol to acetaldehyde,57 which is the causative agent of hangover symptoms after the consumption of alcohol.58 ADH is able to act on other alcohol substrates, leading to toxic and potentially deadly metabolites; for example, ADH oxidizes methanol to formaldehyde, and initates the oxidation of ethylene glycol to glycolic and oxalic acids.59 Efforts to develop ADH inhibitors to treat methanol and ethylene glycol poisoning have led to the development of a FDA-approved metalloenzyme inhibitor that acts by directly binding to catalytic Zn2+ ion.60
The mechanism of ADH oxidation is proposed to start with binding of coenzyme NAD+ followed by binding of the alcohol substrate to the catalytic Zn2+ ion through the oxygen atom of the alchohol, which displaces a native Zn2+-bound water. Alcohol deprotonation then occurs, which triggers an electron cascade resulting in generation of the aldehyde, as well as a hydride transfer to NAD+, forming NADH. Then both the product aldehyde and NADH dissociate from the active site, and the cycle starts anew.61
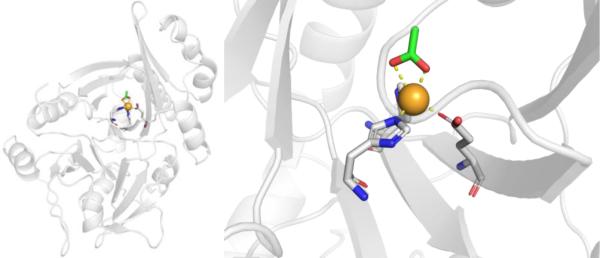
Protein and Active Site Structure.
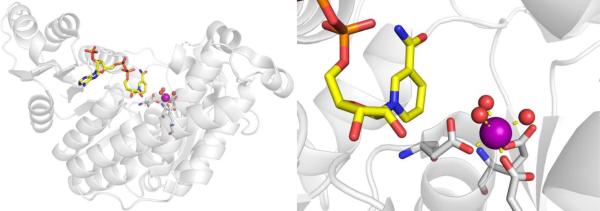
ADH is a NAD+-dependent, Zn2+-dependent homodimeric metalloenzyme with each subunit containing both a structural and a catalytic Zn2+ site. The majority of the active site is occupied by the NAD+ cofactor, so that the available area for substrates to bind is rather small at ~7–10 Å wide and 15 Å deep.62 The catalytic Zn2+ ion is located at the bottom of this active site pocket, and is coordinated by Cys46, His67, Cys174, and a water/hydroxyl ligand in a tetrahedral coordination geometry (Figure 8).63 The structural Zn2+ site is coordinated by four Cys residues in a tetrahedral geometry.56
Figure 8.
Structure of human ADH with NAD+ cofactor bound (yellow), highlighting both structural and catalytic Zn2+ sites (ß1 isoform, PDB 1HDX). The water bound to the catalytic Zn2+ ion was not observed crystallographically.
Inhibitor Development.
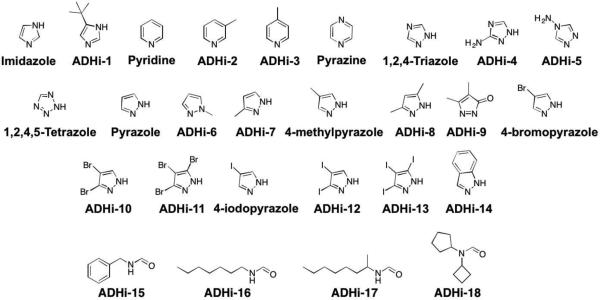
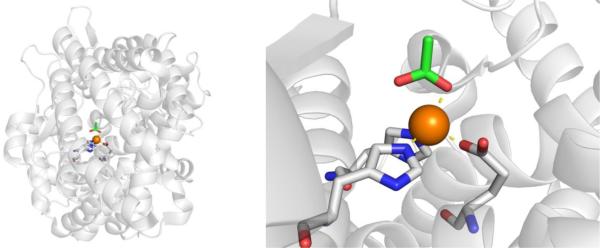
Efforts to develop inhibitors against ADH have been limited because unlike inhibition of aldehyde dehydrogenase, inhibition of alcohol dehydrogenase is not useful in managing alcoholism. ADH inhibition prolongs the half-life of ethanol and can lead to elevated levels of ethanol after alcoholic drink consumption.64 However, in terms of preventing methanol and ethylene glycol poisoning, the development of ADH inhibitors has been successful, and has led to the FDA-approved therapeutic 4-methylpyrazole, which is commercially known as Fomepizole (Figure 9).65
Figure 9.
Inhibitors of ADH.
Early inhibitor development began with a kinetic study of horse liver ADH, where imidazole was identified as a weak inhibitor due to its ability to coordinate Zn2+.66 Follow-up work on imidazole derivatives capable of monodentate coordination lead to the discovery of the pyrazole scaffold as a potent inhibitor of horse liver ADH with an IC50 value of 0.2 μM (Figure 9). Other imidazole isosteres, including pyridine, pyrazine, triazole, tetrazole, and pyrazolone were explored, but none of these scaffolds had comparable activity to pyrazole (Table 3).
Table 3.
Activity (Ki values) of ADH inhibitors. All compounds tested against horse liver ADH, unless otherwise indicated.67,68
| Compound | Inhibition (μM) |
|---|---|
| ADHi-1 | 280 |
| ADHi-2 | 290 |
| ADHi-3 | 250 |
| ADHi-4 | 14000 |
| ADHi-5 | 69000 |
| ADHi-6 | 17000 |
| ADHi-7 | 125 |
| ADHi-8 | 7100 |
| ADHi-9 | N.A. |
| ADHi-10 | 20 |
| ADHi-11 | 20 |
| ADHi-12 | 30 |
| ADHi-13 | 10 |
| ADHi-14 | 14 |
| 4-Bromopyrazole | 0.020, 0.290a |
| Imidazole | 7600 |
| 4-Iodopyrazole | 0.020, 0.120a |
| 4-Methylpyrazole (Fomepizole) | 0.080, 0.210a |
| Pyrazole | 0.2, 2.6a |
| Pyridine | 2100 |
| 1,2,4,5-Tetrazole | 3100 |
| 1,2,4-Triazole | 410 |
Ki value against human ADH
Further derivatization of the pyrazole scaffold showed that methyl substitution (ADHi-6, ADHi-7, Figure 9) is not tolerated and sharply curtails inhibitory activity (Table 3). However, 4-methylpyrazole, which is the only FDA-approved ADH inhibitor, gives a substantial improvement in binding over pyrazole, with an IC50 value of 80 nM. A preference for substitution at the 4-position holds true for halogen derivatives such as 4-bromopyrazole and 4-iodopyrazole as well, which both have an IC50 value of ~20 nM. These 4-derivatized pyrazole inhibitors have a slightly lower activity against human ADH when compared to horse liver ADH, but still show excellent inhibition with Ki values of 210 nM, 290 nM, and 120 nM for 4-methylpyrazole, 4-bromopyrazole, and 4-iodopyrazole, respectively.67 The halogen compounds 4-bromopyrazole and 4-iodopyrazole were found to produce toxicity in animals, but 4-methylpyrazole (Fomepizole) attained FDA approval in 1997 as a treatment for methanol and ethylene glycol poisioning.65
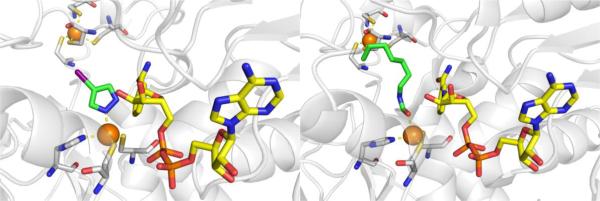
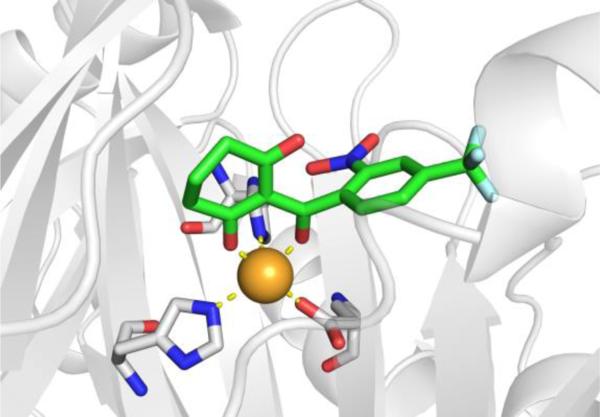
There is no crystal structure reported of 4-methylpyrazole bound to the active site of human ADH. However, 4-iodopyrazole has been crystallized with various isoforms of human ADH (Figure 10)69 and binds in the expected manner with a single nitrogen atom coordinating to the catalytic Zn2+ ion.70 It has been inferred that 4-methylpyrazol binds in an identical fashion to 4-iodopyazole.
Figure 10.
Left: Structure of 4-iodopyrazole bound to human ααADH (PDB 1DEH). Right: Structure of ADHi-16 bound to human ß2ß2ADH (PDB 1U3V).
There are different isoforms of human ADH with slight variations within the active site.71,72 4-Methylpyrazole has been shown to inhibit all of these isoforms, with Ki values typically ranging from 0.1 – 2.1 μM.71 For the purpose of developing inhibitors capable of isoform-specific inhibition of ADH, a series of formamide-based transition state mimetics of ADH have been developed (Figure 9, Table 4).73 These compounds were not intended to replace 4-methylpyrazpole as a treatment for methanol poisoning, but to act as a tool for studying the metabolic effects of selectively inhibiting the different ADH isoenzymes. By maintaining the formamide warhead to bind the Zn2+ ion and varying the capping group, inhibitors were identified that could distinguish between different human ADH isoenzymes (Table 4). Crystallographic studies confirmed that these formamide-based inhibitors bind by coordinating to the catalytic Zn2+ ion through the aldehyde warhead (Figure 10).74
Table 4.
Activity (IC50 values in μM) of isoenzyme-selective ADH inhibitors.73
| Compound | α α | β 2 β 2 | γ 2 γ 2 | π π | σ σ |
|---|---|---|---|---|---|
| ADHi-15 | 31 | 0.33 | 4.9 | 110 | 11 |
| ADHi-16 | 3.6 | 0.33 | 12 | 11 | 0.74 |
| ADHi-17 | 7.0 | 1.7 | 0.41 | 40 | 100 |
| ADHi-18 | 0.36 | 10000 | 47 | 360 | 1100 |
Current and Future Prospects.
At present, there is little need for the development of additional ADH inhibitors, as ADH is not known to be relevant to any disease state outside of incidental methanol and ethylene glycol poisonings and 4-methylpyrazole is FDA approved for these incidents. However, the history of the development of 4-methylpyrazole as an ADH inhibitor is a useful example of the utility of metalloenzyme inhibitors. The development of 4-methylpyrazole itself is an example of even a simple MBP serving as a therapeutic, identified from imidazole, then utilizing small substituents to modulate and improve affinity, and finally simple derivatization to obtain the needed efficacy and low toxicity.65,75 Overall, ADH inhibition by 4-methylpyrazole, which is essentially a standalone MBP core scaffold, highlights the validity and utility of inhibiting metalloenzymes using metal-binding compounds.
2.2. 1-Deoxy-D-xylulose 5-Phosphate Reductoisomerase (DXR, DOXP Reductoisomerase, EC 1.1.1.267)
Function in Biology and Disease.
Malaria is an infection caused by the P. falciparum parasite. The parasite resides and reproduces in the red blood cells, and the produced chemical waste builds up in the blood stream, leading to symptoms such as diarrhea, fever, vomiting, and death.76 In 2016, the World Health Organization reported a staggering 216 million cases of malaria worldwide (with almost half a million related deaths), mainly effecting people residing in Africa and Southeast Asia.77 P. falciparum has become highly resistant to many conventional antimalarial drugs, including amodiaquine, sulphadoxine-pyrimethamine, and chloroquine, creating a need for new treatments for malaria.78
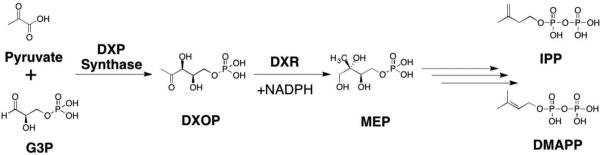
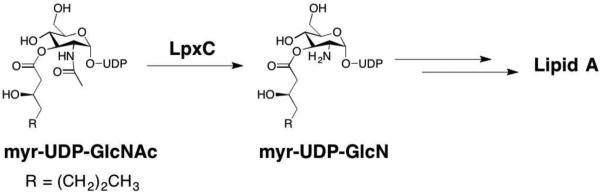
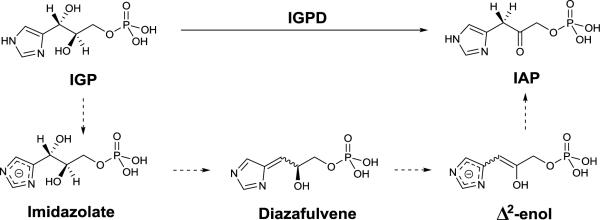
A target of interest for treating malaria includes inhibiting the non-mevalonate pathway (NMP) for isoprenoid biosynthesis (Figure 11). This essential pathway is used by the vast majority of bacteria and apicomplexan protozoa for the conversion of pyruvate and glyceraldehyde-3-phosphate (G3P) into isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP). IPP and DMAPP are two precursors for the biosynthesis of >35,000 isoprenoids, which are important for protein degradation, cell membrane maintenance, glycoprotein biosynthesis, hormone-based signaling, protein anchoring, N-glycosylation, and other processes.79–81 Importantly, the NMP pathway is not present in humans,78 making it an attractive antimalarial (P. falciparum) and antibacterial (M. tuberculosis, E. coli) target. Knockout studies show the enzyme required for the second step of the NEP pathway, 1-deoxyxylulose-5-phosphate reductoisomerase (DXR or DOXP Reductoisomerase), is essential for isoprenoid biosynthesis making it a viable target.82–85
Figure 11.
The non-mevalonate pathway (NMP) for isoprenoid biosynthesis.
Protein and Active Site Structure.
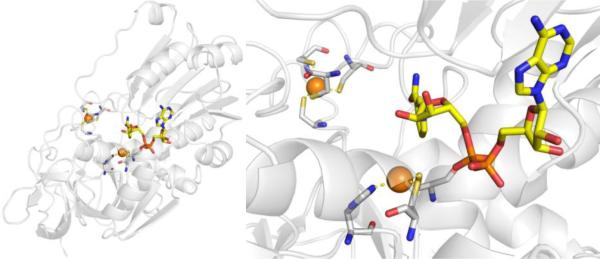
DXR catalyzes the isomerization of 1-deoxy-D-xylulose 5-phosphate (DOXP) to 2-C-methyl-D-erythritol 4-phosphate (MEP) utilizing NADPH and a divalent cation, either Mg2+or Mn2+, as cofactors.78,79,85,86 It is not clear which cation is found in native DXR, as studies have shown that Mn2+ results in higher enzymatic activity and increased enzyme stability, but Mg2+ may be the relevant cation due to its greater abundance in vivo.78,87 Due to interest in DXR as both an antibacterial and antimalarial target, inhibitor development and crystallographic structural studies have been performed on DXR derived from various sources (P. falciparum DXR, PfDXR; M. tuberculosis DXR, MtDXR; E. coli DXR, EcDXR); however, the overall protein structure and catalytic mechanism of DXR are similar across different organisms78,79,88–93 and PfDXR will be used as a representative case study here.
The structure of PfDXR with Mn2+ and NADPH bound reveals two subunits of an asymmetric unit forming a homodimer (each monomer containing 488 residues, MW ~47 kDa).78 The monomer has two large domains separated by a cleft containing a deep pocket, a linker region, and a C-terminal domain (Figure 12).78 The larger domain (residues 77–230) is responsible for NADPH binding, while the smaller domain (residues 231–369) and linker region is the site of catalytic activity. The divalent ion is ligated by the side chains of Asp231, Glu233, and Glu315, and three water molecules in an octahedral coordination geometry (Figure 12).78 Residues Glu233 and Glu315 are further stabilized by the side chain of Lys312 with the mobile hydrophobic loop in an open loop confirmation. Comparison of inhibitor-free with inhibitor-bound DXR reveals a loop close confirmation in the ladder, suggesting the mobile loops function as a gate keeper and undergoes an induced fit upon substrate/inhibitor binding.78,92 In depth mechanistic and protein structure information is detailed elsewhere.79
Figure 12.
Structure of PfDXR with NADPH cofactor (yellow) (PDB 3AU8).
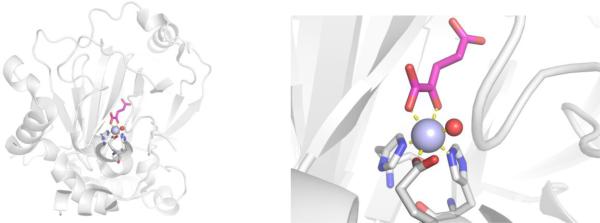
Inhibitor Development.
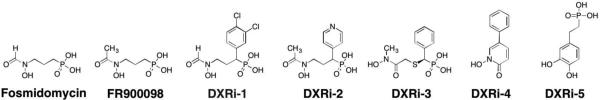
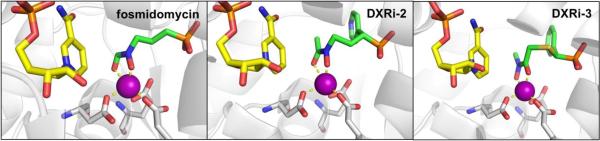
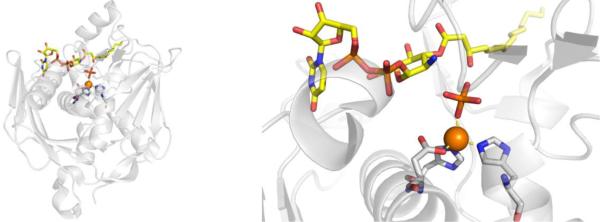
In 1980, it was discovered that fosmidomycin, a natural product found in Streptomyces lavendulae culture containing a hydroxamic acid MBP, and its formyl derivative (FR900098) inhibited EcDXR with an IC50 value between 8.2 nM – 30 nM (Figure 13).94,95 Follow-up studies found that both inhibitors were also potent against PfDXR (IC50 = 90 – 370 nM), suppressed the growth of multidrug-resistant P. falciparum bacteria in vitro, and rescued infected mice in vivo.96 The structure of PfDXR complexed with fosmidomycin and NADPH reveals the hydroxamic acid MBP displacing the bound water molecules and coordinating to the divalent ion, which is further ligated by residues Asp231, Glu233, and Glu315 in a distorted trigonal bipyramid coordination geometry (Figure 14).78 The terminal phosphonate group resides in a hydrophilic pocket and forms an intricate network of H-bonds with the side chains of Ser270, Ser306, Asn311, Lys312, His293, and two water molecules.78,79 Comparison of the residue numbering, coordination geometry, and key binding interactions between EcDXR, MtDXR, and PfDXR complexed with fosmidomycin is available elsewhere.79
Figure 13.
Inhibitors of DXR.
Figure 14.
Structure of fosmidomycin (PDB 3AU9), DXRi-2 (PDB 4GAE), and DXRi-3 (PDB 4KP7) bound to PfDXR with NADPH cofactor (yellow).
While fosmidomycin showed remarkable antimalarial activity in preclinical studies,97–100 its short half-life (~1.5 h) in plasma and poor bioavailability (due to being highly ionized at pH 7.4) precluded its development in the clinical setting.101 In addition, organisms that lack the glycerol-3-phosphate transporter (GlpT, such as M. tuberculosis) are not susceptible to fosmidomycin due to their inability to actively transport the drug across the cell membrane.102,103 To address these shortcomings, DXR inhibitor development has largely focused on achieving better PK properties by making more lipophilic compounds. DXR inhibitors can be grouped into three classes including modification on the phosphate group,101,104–108 the linker,95,104,109–114 and the hydroxamic acid MBP,101,107,108,110–112,115–119 and have been evaluated for both antibacterial and antimalarial activity.
Maintaining the hydroxamic acid MBP and replacing the phosphonate group of fosmidomycin or FR900098 to increase lipophilicity has been an important focus in inhibitor development. The phosphonate group has been replaced by isosteres such as bisphosphonate, phosphate, carboxylate, sulfamate, sulphonate, or has been deleted entirely from the inhibitor.101,104–108 Unfortunately, most of these compounds showed a decrease in activity when compared to fosmidomycin and FR900098, and no further bacterial studies were pursued. Another large class of fosmidomycin analogues modified the linker between the hydroxamic acid and phosphonate, either by increasing/decreasing the linker length (with three methylene groups considered the optimum linker length) or by adding bulky, lipophilic substituents,104,110–114 of which DXRi-195 and DXRi-2109 are examples of this strategy (Figure 13). An early study examining α-substituted fosmidomycin derivatives yielded DXRi-1, with improved antimalarial activity against P. falciparum in vitro (IC50 = 28 – 90 nM) compared to that of fosmidomycin (IC50 = 0.36 – 1.1 μM).95 Enhanced in vitro activity was attributed to better cellular uptake due to incorporation of the lipophilic ring. Further exploration of varying the linker by the installation of a pyridine ring yielded DXRi-2 (Figure 13) as potent inhibitor of PfDXR (Ki = 13 nM) and EcDXR (Ki = 82 nM).109 Additionally, DXRi-2 was non-cytotoxic to human cells and displayed antimalarial activity against the growth of multi-drug resistant P. falciparum (EC50 = 460 – 630 nM). The structure of PfDXR complexed with DXRi-2 and NADPH reveals the hydroxamic acid MBP coordinated to the central Mn2+ ion in a bidentate manner resulting in a distorted trigonal bipyramid coordination geometry (Figure 14). The phosphonate group is situated in the hydrophilic pocket forming hydrogen bonds and electrostatic interactions. The pyridine ring of DXRi-2 is situated in the closed hydrophobic loop pocket defined by residues His293, Trp296, Met298, Cys338, and Pro358, and the pyridine nitrogen further interacts with the side chain of Cys33 and forms π-π stacking interaction with the indole ring of Trp296.109 Similar interactions are observed in other fosmidomycin derivatives and are considered important for inhibitor activity.
While fosmidomycin derivatives with modified linker and phosphonate motifs have been well explored, little attention has been placed on improving the hydroxamate MBP. Some derivatives have eliminated the MBP altogether, utilized a modified hydroxamic acid MBP (DXRi-3, DXRi-4), or replaced the hydroxamic acid with a catechol MBP (DXRi-5).101,107,108,110–112,115–119 The majority of modified hydroxamic acid MBPs include the utilization of a N-hydroxy-N-methylacetamide motif instead of the N-hydroxy-N-methylformamide presented in fosmidomycin (DXRi-3).112 DXRi-3 (as an enantiomeric mixture) was found to be a potent inhibitor of EcDXR and PfDXR (IC50 = 8.2 and 24 nM, respectively), a moderate inhibitor against MtDXR (IC50 = 280 nM), and effective at inhibiting growth of Plasmodium in vitro. Separation and screening of the two enantiomers revealed the S-(+) enantiomer of DXRi-3 as the active compound (IC50 = 5.1 – 120 nM against EcDXR, MtDXR and PfDXR). The structure of DXRi-3 bound to PfDXR reveals the MBP binding in a bidentate manner to the Mn2+ creating a trigonal bipyramid coordination geometry (Figure 14).112 Metal coordination, as well as other active site interactions are similar to that of fosmidomycin and DXRi-2. Another modification of the hydroxamic acid MBP includes the use of 1-hydroxypyridin-2-one MBP (DXRi-4, Figure 13).107 It was suggested that a cyclized hydroxamic acid derivative would be non-hydrolyzable, making it a better MBP for drug development compared to its hydroxamate counterparts. While DXRi-4 showed only modest in vitro enzymatic activity against EcDXR (IC50 = 1.4 μM), the inhibitor showed antibacterial activity against both Gram-negative and Gram-positive bacteria (MIC = 20 – 100 μM against E. coli, P. aeruginosa, B. anthracis, and M. luteus) and was non-cytotoxic to human cells.107 No structure of DXRi-4 bound to DXR has been reported; however, the MBP is expected to bind in a bidentate manner similar to the hydroxamic acids in other inhibitors. Lastly, attempts have been made to replace the hydroxamic acid MBP with a catechol scaffold (DXRi-5).107,108,116 Specifically, while DXRi-5 showed modest inhibition of EcDXR (IC50 = 4.5 μM), it did not exhibit antibacterial activity (MIC >1000 μM against E. coli, P. aeruginosa, B. anthracis, M. luteus).107 No crystallographic analysis or further inhibitor development have been reported on these catechol-based DXR inhibitors.
Current and Future Prospects.
Inhibitor development for DXR can be divided into three parts: modification of the hydrophilic terminal phosphate motif, the linker, and the hydroxamic acid MBP. Inhibitor development has focused not only on improving activity, but also enhancing lipophilicity for better cellular uptake. Unfortunately, replacing the phosphonate group with other isosteres did not yield more potent inhibitors, and identifying suitable isosteres for the terminal phosphonate group remains challenging. Most inhibitor optimization has focused on modifying the fosmidomycin linker, with the addition of aromatic substituents being the most successful. Finally, few attempts have been made to replace the hydroxamic acid with other MBPs. The hydroxamic acid, and its N-hydroxy-N-methylacetamide derivative, are routinely used; however, these MBPs possess a poor pharmacokinetic profile, readily undergoing hydrolysis and glucuronidation. Catechol MBP inhibitors have also been published, but little information on their mode of binding is available. While there have been steady reports on the development of new DXR inhibitors, most inhibitors are structurally similar to fosmidomycin and bear a hydroxamic acid MBP. Future efforts in the development of inhibitors of DXR might benefit from a focus on introducing new MBPs and more structurally diverse compounds that can reduce the overall polarity of the inhibitors.
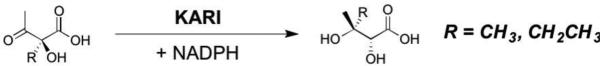
2.3. Ketol-acid Reductoisomerase (KARI, EC 1.1.1.86)
Function in Biology and Disease.
The biosynthetic pathways of branched chain amino acids (valine, leucine, and isoleucine) are essential for organisms that must biosynthesize them. Plants, bacteria, and fungi extensively utilize these pathways, while animals do not (which obtain these amino acids from food), thus the enzymes that comprise these pathways represent attractive targets for herbicides, antibiotics, and antifungals.120,121 The second central enzyme in the biosynthetic pathways of these amino acids is the metalloenzyme ketol-acid reductoisomerase (KARI), which catalyzes the reduction and isomerization of 2-acetolactate and 2-aceto-2-hydroxybutyrate to 2,3-dihydroxyisovalerate and 2,3-dihydroxy-3-methylvalerate, respectively. KARI uses NADPH as a co-factor (Figure 15) and has been studied as a herbicidal, antifungal, and antibacterial target.121 Although it has gathered less attention in the biosynthetic pathway of branched amino acids compared to other enzymes,122 it still holds potential as a viable target. Based on biological studies in E. coli and S. pneumoniae, the gene cluster (ilvC) which encodes for KARI was determined to be important for optimal growth and virulence and is suspected to be important for M. tuberculosis growth and virulence.123–125 KARI has also been found to be essential for optimal virulence of F. graminearum, a fungus responsible for billions of dollars in agricultural losses yearly.126 Given the rapid onset of resistance to current antibiotics and herbicides, KARI has potential as a target for inhibitor development due to its widespread role in many infectious organisms and pests, while not being present in humans.
Figure 15.
Reactions catalyzed by KARI, a central enzyme in the branched chain amino acid biosynthetic pathways. 2,3-Dihydroxyisovalerate (R = CH3) is utilized in the biosynthesis of valine and leucine, while 2,3-dihydroxy-3-mathylvalerate (R = CH2CH3) is utilized as a precursor for isoleucine.
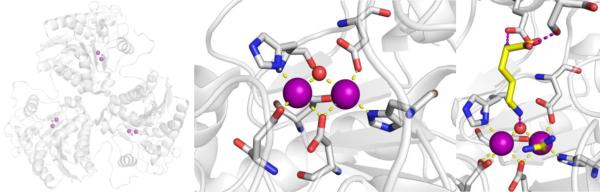
Protein and Active Site Structure.
The metalloenzyme KARI is a dinuclear Mg2+-dependent enzyme which requires NAPDH for catalysis.121,127 KARIs are divided into two classes, I and II, with class I being ~150 residues shorter than class II (Figure 16).128 All KARIs possess two domains: a N-terminal Rossmann fold domain (for binding NADPH) and a C-terminal α-helical domain with either a partial or complete figure-of-eight knot fold, from which the distinction between classes I and II is derived.128 Sequence homology between classes I and II is significantly lower than within each class, although the residues involved in NADPH and metal binding are highly conserved among all KARIs.128,129 Fungal, and the majority of bacterial KARIs belong to class I, while class II includes all plant and remaining bacterial KARIs.130 Based on structural data it is thought that for the active sites of class I KARIs to be formed, two monomers dimerize to form a complete figure-of-eight knot fold, resulting in two active sites with both monomers contributing to each active site. For class II KARIs each monomer forms its own active site; however, class II KARIs have also been shown to form a homodimer resulting in two active sites.130
Figure 16.
Structure of M. tuberculosis class I KARI (PDB 4YPO).
The active site for KARIs is found at the interface between the C-terminal and N-terminal domains.121 In the dinuclear active site of KARIs both Mg2+ ions are coordinated by acidic Glu and Asp residues with the remaining coordination sites occupied by water. For example, in the M. tuberculosis KARI (class I), the active site metals are coordinated by Asp188 and Glu192 from one monomer and Glu224 and Glu228 from the other monomer, as well as free water molecules resulting in an octahedral coordination geometry at each metal center (Figure 16).131 The structure of the active site is such that NADPH binds near the active site metal ions and the substrate (Figure 16) so it can provide a hydride as per the proposed mechanism.130 Based on structural comparison of apo- and holo-class I and class II KARIs, it is suggested there may be conformational changes upon Mg2+ and NADPH binding, as well as upon inhibitor binding.128,132
Inhibitor Development.
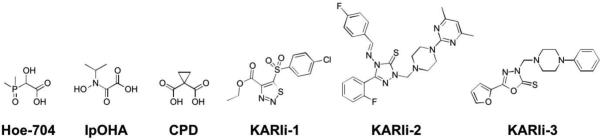
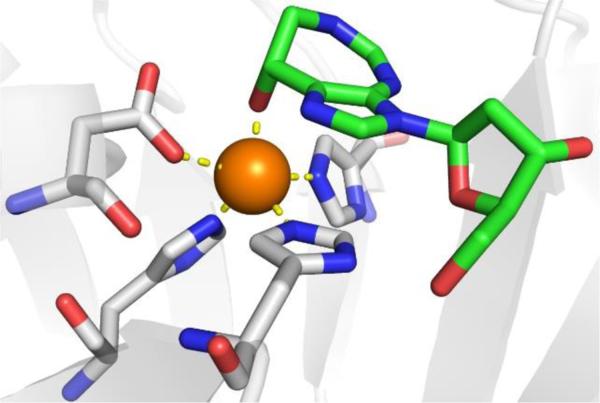
Inhibitor discovery for KARIs began in 1988 with the discovery of the experimental herbicide, 2-(dimethylphosphinoyl)-2-hydroxyacetic acid (Hoe-704, Figure 17).133 Hoe-704 was not originally designed as an inhibitor of KARIs, but KARI inhibition was implicated in its herbicidal and antibacterial properties, with a Ki value of 800 nM against E. coli KARI. After the discovery of Hoe-704, a series of compounds were designed to mimic the reaction intermediate, the most potent of which was N-hydroxy-N-isopropyloxamate (IpOHA, Figure 17).134 IpOHA utilizes a hydroxamic acid MBP and was found to be a very slow, but tight binder with a KD value of 22–38 pM against E. coli KARI. IpOHA was also found to possess in vivo activity against M. tuberculosis.134 Another transition state mimic, cyclopropane-1,1-dicarboxylate (CPD) uses a dicarboxylate MBP and was suggested to mimic a reaction intermediate of methyl transfer. CPD has an IC50 value of 4.7 μM against E. coli KARI,135 but attempts to make CPD derivatives have generally resulted in compounds with poorer activity than CPD.136,137 From a HTS campaign, a family of compounds containing thiadiazoles (KARIi-1) were found to be potent covalent inhibitors (Kapp = 750 nM) of S. oleracea KARI, via a covalent modification at Cys498 based on protease digestion mass spectrometry analysis.138 A mechanism for the covalent modification via a Michael addition of the Cys498 side chain to the C-C double bond of the heterocycle in KARIi-1 has been proposed. Additionally, a series of compounds using a piperazine scaffold derivatized with various heterocycles (KARIi-2, KARIi-3) were tested as O. sativa KARI inhibitors, with the most active inhibitors exhibiting generally modest activity;139,140 it is unclear if these compounds interact with the active site metal ions.
Figure 17.
Inhibitors of KARI.
The first crystal structure of KARI with an inhibitor bound to the active site was reported in 1997, with IpOHA bound to the S. oleracea KARI.130 Later structures of both IpOHA and CPD bound to the active site of S. aureus KARI were reported (Figure 18).141 IpOHA bound to S. aureus KARI showed that IpOHA coordinates to both active site Mg2+ ions in a bidentate manner forming a 5-membered chelate ring to each active site metal. IpOHA utilizes a hydroxamate/carboxylate combined MBP with all Zn-O bond distances being between 2.1 and 2.3 Å. This mode of coordination was essentially identical to that of IpOHA bound to S. oleracea KARI.130 For CPD bound to S. aureus KARI, the inhibitor coordinates in a bidentate fashion to one active site Mg2+ utilizing only one carboxylate group forming a 4-membered chelate ring. Coordination to the other active site Mg2+ is achieved in a bidentate fashion utilizing both carboxylates, resulting in a 6-membered chelate ring, with all CPD Zn-O bond distances in the structure being between 2.0 and 2.1 Å. The cyclopropyl group of CPD and the isopropyl moiety of IpOHA both make contacts with a hydrophobic region of the active site.
Figure 18.
Left: Structure of IpOHA bound to S. aureus KARI (PDB 6AQJ). Right: Structure of CPD bound to S. aureus KARI (PDB 5W3K).
Current and Future Prospects.
The most active and earliest KARI inhibitors have focused on transition state mimics, which have produced hydroxamic and carboxylic acid based inhibitors. Some of these molecules demonstrate excellent enzyme inhibition, show herbicidal activity, and hinder bacterial growth. Both IpOHA and CPD can be further derivatized, most obviously around the isopropyl and cyclopropyl groups, but it is unclear if such efforts would yield more useful compounds. Based on the encouraging target validation data and the lack of chemical diversity utilized for inhibitors, KARIs arguably remain an underexplored area of metalloenzyme target space for the development of potential antibiotics and antifungals.
3. Oxidoreductases (EC 1.13, EC 1.14)
3.1. 4-Hydroxyphenylpyruvate dioxygenase (HPPD, EC 1.13.11.27)
Function in Biology and Disease.
4-Hydroxyphenylpyruvate dioxygenase (HPPD) is found in nearly all aerobic organisms, with the exception of a few Gram-negative bacteria, and is responsible for catalyzing the second step of L-tyrosine catabolism.142 HPPD is an oxygen- and non-heme Fe2+-dependent metalloenzyme that converts 4-hydroxyphenylpyruvate to 2,5-dihydroxyphenylacetate (also known as homogentisate).143 In animals, this conversion is used to modulate blood tyrosine concentrations, whereas in plants, this conversion is used to ultimately produce plastoquinone and tocopherol, which are necessary cofactors for electron transport in photosynthesis.144 Several studies to elucidate the mechanism of HPPD have culminated in a proposed mechanistic scheme that involves substrate coordination, oxygen-mediated decarboxylation, followed by an alkyl ring group migration, and finally ring hydroxylation to produce the homogentisate product.145 The importance of HPPD in producing cofactors necessary for plant photosynthesis has led to agricultural interest in developing herbicides that act via HPPD inhibition, which ultimately results in plant bleaching and death due to the inability to sustain chlorophyll production.146,147
There are several hereditary human diseases, namely type I tyrosinemia and alkaptonuria, which occur due to misregulation of later steps in tyrosine catabolism. Type I tyrosinemia is a rare and usually fatal orphan disease that primarily affects infants and children;148 whereas the onset of alkaptonuria does not usually occur until adulthood.149 Interestingly, it has been found that one of the early discovered herbicidal HPPD inhibitors, 2-[2-nitro-4-(trifluoromethyl)benzoyl]-1,3-cyclohexanedione (NTBC), is also able to inhibit human HPPD, which prevents formation and subsequent build-up of the toxic metabolites associated with these diseases. In an unusual and bold study, NTBC (Figure 20) bypassed all standard clinical trials and was tested with overwhelming success in an infant patient severely afflicted with type I tyrosinemia.150 After additional testing against other infant patients, NTBC attained FDA approval in 2002 as a HPPD inhibitor under the drug name Nitisinone.151
Figure 20.
Inhibitors of HPPD.
Protein and Active Site Structure.
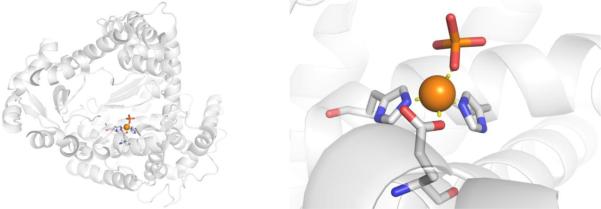
HPPD is a homodimeric, molecular, non-heme Fe2+ metalloenzyme with a molecular weight ranging from ~40–50 kDa depending on the species of origin. Across different species, the active site of HPPD is highly conserved, and consists of a catalytic Fe2+ ion coordinated by a His, His, Glu motif along with three labile water molecules in an octahedral geometry (Figure 19).143,152 The 4-hydroxyphenylpyruvate substrate of HPPD has never been crystallized in the active site, but is believed to initially bind to the catalytic Fe2+ through displacement of two of the coordinating water molecules.143,144
Figure 19.
Structure of HPPD from Psuedomonas fluorescens (PDB 1CJX). A molecule of acetate (green, from the crystallization buffer) is coordinated to the active site Fe2+ ion.
Inhibitor Development.
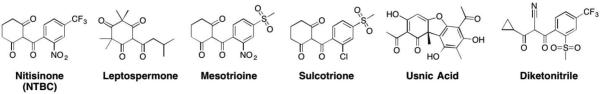
There has only been one class of inhibitors explored against HPPD, namely the triketones. These inhibitors were discovered serendipitously in 1977,153 as an extract from bottlebrush plants, resulting in the isolation of leptospermone as an herbicidal agent (Figure 20). Efforts to derivatize and optimize the leptospermone scaffold led to the development of mesotrioine, sulcotrione, and diketonitrile, which are all used as commercial herbicides (Figure 20).147,154,155 Usnic acid is a natural product secreted by lichen that has also been found to inhibit HPPD. IC50 values for these herbicidal inhibitors vary, as they were developed against different plants with different HPPD enzymes, making comparisons difficult.143
A crystal structure of NTBC bound to the active site of S. avermitilis HPPD confirms that the triketones inhibit by directly coordinating to the catalytic Fe2+ ion, displacing two of the three labile water molecules (Figure 21).1441H NMR studies have shown that the exocyclic enol form of NTBC is the preferred tautomer over the keto form.143,144 After NTBC is bound to HPPD molecular oxygen is no longer able to bind.144 It is presumed that other herbicidal inhibitors of HPPD (Figure 20) bind to the HPPD active site in a similar fashion.
Figure 21.
Structure of NTBC (Nitisinone) bound to S. avermitilis HPPD (PDB 1T47).
NTBC was discovered to have therapeutic effects in treating type I tyrosinemia.156 Again, in an act of serendipity, during a study to determine the target of herbicidal triketones, NTBC was tested on rats rather than plants, which resulted in heighted levels of blood plasma tyrosine as well as higher levels of 4-hydroxyphenylpyruvate (the substrate of HPPD) in rat urine samples. These observations lead researchers to conclude that HPPD was the target of NTBC, which was later confirmed with various in vitro assays.156 Later trials on human liver HPPD found NTBC to have an excellent IC50 value of 5 nM,150 and NTBC was immediately and successfully tested as an inhibitor of HPPD to treat infants with hereditary type I tyrosenemia.148
Current and Future Prospects.
To date, only the triketone class of inhibitors has been extensively studied against this HPPD. Triketone HPPD inhibitors have been extremely successful through herbicidal applications, and in particular, the triketone NTBC has been FDA approved for treating type I tyrosinemia in infant patients. Furthermore, NTBC mediated inhibition of HPPD to treat alkaptonuria has entered Phase III clinical trials.157 The discovery of NTBC as an inhibitor of HPPD is unique and unorthodox, as NTBC went from use as an herbicide, to testing in rats, to testing against human liver HPPD in vitro, and then rapidly transitioned to useful human clinical applications. Considering that only one MBP scaffold has been explored against HPPD, there is certainly room for further inhibitor development. That stated, there is likely to be little need for additional human HPPD inhibitors, although other classes of HPPD inhibitors could find applications in the agricultural field, or could potentially be explored as new classes of antibiotics and antimicrobials. In that regard, HPPD could remain an interesting target that merits further exploration.
3.2. Arachidonate 5-Lipoxygenase (5-LO, 5-LOX, ALOX5, EC 1.13.11.34)
Function in Biology and Disease.
Leukotrienes (LTs) are fatty acid mediators of inflammatory innate immune responses. During LT biosynthesis, arachidonic acid (AA) is released from membrane phospholipids and is metabolized by 5-lipoxygenase (5-LO). 5-LO catalyzes the conversion of arachidonic acid into 5(S)-hydroperoxyeicosatetraenoic acid (5-HPETE) and leukotriene A4 with the help of the 5-lipoxygenase-activating protein (FLAP, Figure 22).158,159 Leukotriene A4 is subsequently converted into either leukotriene B4 or leukotriene C4. These mature leukotrienes, when secreted from the cell, modulate the innate immune response. Stimulation of leukotriene B4 receptor (BLT) initiates a chemotactic response in neutrophils (i.e., neutrophil recruitment) towards the site of inflammation, whereas stimulation of cysteinyl leukotriene receptor 1 (CysLT1) by leukotriene C4 metabolites induces smooth muscle contraction, increased vascular permeability and vasodilation, and eosinophil recruitment.160
Figure 22.
The 5-lipoxygenase pathway. Beginning from AA, 5-LO catalyzes the sequential synthesis of 5-HPETE and leukotriene A4. Leukotriene A4 is then further elaborated to generate leukotrienes B4 and Cys-leukotrienes, which are signaling molecules in the innate immune response.
5-LO is a non-heme iron-dependent metalloenzyme which employs a single Fe3+ ion in catalysis.159,161 5-LO is inactive in the ferrous state, and must be activated (generally by lipid hydroperoxides) to yield the active, ferric state. This enzyme is classified as an Fe-dioxygenase and catalyzes the incorporation of molecular oxygen into the cis C5–C6 alkene of arachidonic acid. Over a two-step total reaction pathway, 5-LO oxidizes arachidonic acid to 5-HPETE, and subsequently dehydrates the peroxide intermediate to form the epoxide leukotriene A4 (Figure 22). 5-LO is also capable of reducing the peroxide intermediate to the corresponding alcohol under certain conditions.161
5-LO derived arachidonic acid metabolites have been shown to be potent mediators of inflammatory response with implications in disease. Over the last three decades, investigations have demonstrated that the 5-LO pathway plays a role in the development of diseases including allergic disorders such as asthma,162 inflammatory disorders such as inflammatory bowel disease, rheumatoid arthritis, psoriasis,163,164 and some cardiovascular disorders.165 Moreover, there is evidence that the 5-LO pathway may be involved in tumorigenesis in prostate cancer and certain forms of leukemia.166 5-LO is expressed in nearly all blood leukocytes which is the ultimate biological cause of its key role in cell-mediated inflammatory response.167 Thus, as one of the rate-limiting enzymes in the biosynthesis of LTs, 5-LO represents an attractive target for therapeutic intervention against many inflammation-related disease pathways.
Protein Active Site and Structure.
5-LO contains two primary domains: a C-terminal catalytic domain and an N-terminal C2-like β-barrel domain.168 The catalytic domain contains the active site wherein the Fe3+ cofactor is coordinated by His367, His372, and His550, as well as the carboxylate group of the C-terminal Ile673, and one water molecule (Figure 23). Asn554 has also been reported to coordinate to the metal center;161,169,170 however, these interactions have not been observed crystallographically in human 5-LO. In all available crystal structures, the Fe-O (Asn554) bond length is found to be between 3.1–3.5 Å, which is too long to be considered a coordination bond, although long range electrostatic or solvent mediated contacts are possible at this distance. The N-terminal domain is a regulatory domain which controls catalytic activity in response to various lipids, as well as to Ca2+ and Mg2+ cations. 5-LO activity is strongly stimulated by Ca2+, which binds to the N-terminal C2 domain and increases affinity of 5-LO towards both AA and activating lipid hydroperoxide species.167 Glyceride lipids likewise stimulate the regulatory domain by impeding 5-LO regulation by other protein mediators.
Figure 23.
Structure of 5-LO (PDB 3V99).
Inhibitor Development.
5-LO inhibitors are generally classified into three main groups: redox-active inhibitors, Fe-coordinating inhibitors, and non-redox-non-coordinating inhibitors, which primarily make interactions with the periphery of the active site or the FLAP binding domain.171 However, the dearth of structural data on 5-LO can makes classification of these inhibitors difficult or ambiguous. Most reported 5-LO inhibitors bind within the catalytic domain and are competitive inhibitors, although some redox-active inhibitors display poor binding affinities, as their mechanism of inhibition is not competitive, but rather redox inactivation of the iron center. Redox-active 5-LO inhibitors include various lipophilic reducing agents, many of which are derived from natural product isolates. These include polyphenols, flavonoids, coumarins, and catechol natural products. It is possible that several inhibitors classified as redox-active inhibitors may also be Fe-coordinating (e.g., catechol natural products). Redox-active inhibitors act on 5-LO by reducing and maintaining the active site iron in the ferrous state, which inhibits enzyme activity. Many of these are highly efficient inhibitors in vitro, but lack suitable PK properties and/or selectivity for 5-LO. Coupled with off-target effects on other biological redox systems, or even a tendency to generate reactive oxygen species (ROS), this class of inhibitor has been largely relegated for use in biological model systems.171,172
Non-redox-non-coordinating inhibitors of 5-LO bind competitively with the AA or lipid hydroperoxide binding sites, and in some cases, interfere with FLAP binding. These inhibitors do not interact with the active site metal ion and encompass a structurally diverse set of molecules.173,174 Some of these inhibitors have also been found to bind to allosteric sites of 5-LO. A wide variety of nonsteroidal anti-inflammatory drugs (NSAIDs) are members of this class. The similarity in substrate binding pockets between 5-LO and COX enzymes results in many NSAID inhibitors showing non-specific inhibition against 5-LO.
Fe-coordinating inhibitors directly coordinate to the active site metal cation. While several classes of 5-LO inhibitors are presumed to be metal coordinating, based on the SAR associated with the MBP moieties present in the inhibitor, there are no published crystal structures of 5-LO with an inhibitor bound. Hence, inhibitors that contain a MBP will be discussed here as proposed Fe-coordinating inhibitors. Interestingly, there are some studies where non-specific 5-LO/COX inhibitors or COX-specific inhibitors have been re-purposed with the addition of a MBP or redox-active moieties to more effectively inhibit 5-LO.
To date, Zileuton (Figure 24) is the only FDA approved 5-LO inhibitor. Zileuton is a member of the most successful class of metal binding inhibitors of 5-LO, namely the hydroxyurea and hydroxamic acid based inhibitors. Compounds such as BWB70C (Figure 24) show good cellular- and protein-based efficacy with reported EC50 values ranging between 2–5 μM.175 Further structural elaboration by Abbot laboratories ultimately resulted in the use of ‘reverse’ hydroxamate (i.e., acyl N-hydroxylamides) MBPs, generating compounds A-63162 and A-64077 (Zileuton), which show IC50 values against 5-LO of 0.24–0.51 μM and 0.56–2.3 μM, respectively.163,176 While these inhibitors showed poorer activity against 5-LO when compared to BWB70C, they were more stable and less toxic in cell-based studies, with Zileuton eventually receiving FDA approval for the prophylaxis and chronic treatment of asthma.177 However, Zileuton still suffers from many PK liabilities including significant hepatic toxicity and a short biological half-life, primarily due to its reverse hydroxamate MBP.164 Tenidap (Figure 24) is a 5-LO inhibitor that is also active against COX-2 (IC50 = 36.7 μM and 8.4 μM, respectively) that was developed by Pfizer as a treatment for rheumatoid arthritis. While Tenidap does not contain a hydroxamate MBP, the putative MBP employed bears structural resemblance to, and was developed from, hydroxamate-containing inhibitors. Employing this MBP in lieu of hydroxamates or hydroxyureas was found to diminish the characteristic hepatotoxicity associated with hydroxamate groups. However, in 1996 the FDA rejected Tenidap, citing renal toxicity, which was ultimately attributed to metabolites of the thiophene ring and not the MBP.178
Figure 24.
Inhibitors of 5-LO.
In the past 10 years, several 5-LO inhibitors that may be iron coordinating have entered clinical investigation.160 Atreleuton (VIA-2291) is an N-hydroxyurea derivative that also uses a reverse hydroxamate MBP. Atreleuton completed Phase II clinical trials for the treatment of cardiovascular disease and vascular inflammation. However, a Phase III trial for the treatment of asthma was recently terminated citing toxicity concerns.160 Setileuton (MK-0633) is a functionalized coumarin-based 5-LO inhibitor that may coordinate to the active site iron center via an oxa-3,4-diazole ring and/or α-trifluoroethanol group (Figure 24). Setileuton recently concluded Phase II clinical trials for use in treating atherosclerosis, asthma, and COPD. However, all of these studies have been discontinued as risk-benefit analysis did not support clinical utility of MK-0633 due to potential hepatotoxicity.179,180 Flavocoxid, a mixture of two naturally derived polyphenolic flavonoids (catechin and baicalin), is currently marketed as a ‘medical food’ or nutritional supplement and has recently completed several Phase I and Phase II trials as a treatment for osteoarthritis. This drug mixture shows efficacy against 5-LO, but has also been shown to inhibit COX-2. In spite of positive outcomes in completed trials, there have been some reports of acute hepatotoxicity in elderly women, prompting the FDA to request a voluntary recall of flavocoxid supplements until NME approval.181
As with many metalloenzymes, a large number of natural products have been identified as inhibitors of 5-LO. Many of these inhibitors are polyphenolic and employ a catechol moiety as a likely MBP (Figure 24). These are relatively promiscuous and weak binders, but some polyphenol 5-LO inhibitors show very good activity (e.g., both chemical components of flavocoxid). Baicalen, a naturally occurring flavone isolated from the roots of members of the genus Scutellaria, has been shown to inhibit lipoxygenases with an IC50 value of 0.6–3.8 μM.182 The related compounds aesculetin and caffeic acid have both been found to inhibit 5-LO in screens of natural product isolates. Aesculetin, a derivative of caffeic acid isolated from various natural sources such as chicory, has been found to inhibit 5-LO with an IC50 value of 0.1–1.0 μM.183 Similarly, caffeic acid, which is found in relatively high abundance in coffee and the bark of several eucalyptus species, displays similar IC50 values against 5-LO as aesculetin, but also shows very good activity against human colorectal cancer cells (EC50 = 0.8 μM) and some human breast cancer cell lines (EC50 = 11.4 μM) in inhibiting 5-LO mediated cell proliferation.184
Based on the efficacy of caffeic acid at inhibiting lipoxygenases in vitro and in vivo, extensive synthetic effort was expended to optimize the activity and stability of caffeic acid derivatives.185 It was found that the catechol moiety was essential for activity, indicating that these groups were probable MBPs, although they may be acting through a redox-inhibition mechanism. Further structural elaborations resulted in the discovery of cinnamyl-3,4-dihydroxy-α-cyanocinnamate (CDC, Figure 24), which was very active against 5-LO in protein based assays (IC50 = 9–25 nM) and showed efficacy at inhibiting lipoxygenase activity in human leukocytes and monocytes (EC50 = 0.45–0.8 μM).186
Although most 5-LO inhibitors studied contain hydroxamate, catechol, or carboxylic acid MBPs, over the past ~30 years, some novel metal-binding groups have been explored in the literature. In 2011, a new class of inhibitors contacting triazole or oxadiazole rings as potential metal-coordinating moieties were identified by Amira Pharmaceuticals.187 Further work on these scaffolds resulted in the development of compounds 5-LOi-1 and 5-LOi-2 (Figure 24). Interestingly, the addition of the α-trifluoroethanol group to the azole ring both increased activity of these compounds (EC50 <0.3 μM against human leukocytes).187 More recently, 5-LOi-3 was identified in a click chemistry-based library screen. Noting that triazoles can inhibit 5-LO, ~15 triazole compounds were screened, ultimately identifying 5-LOi-3 as a lead inhibitor with an IC50 value of 33 μM.188
The dual inhibition of both the COX and 5-LO pathways has been another therapeutic goal over the past ~20 years. Single inhibition of the 5-LO pathway does not totally inhibit the production of proinflammatory prostaglandins in most cells because of the orthogonal COX pathway. Because these two enzyme classes both act on arachidonic acid, there is a high degree of active site similarity, although COX enzymes employ a heme iron center rather than the non-heme iron in 5-LO. Recently, the strategy of covalently linking a MBP to nonspecific NSAIDs or to COX inhibitors has led to the development of many dual inhibitors. Using this approach, a derivative of the COX-2 selective inhibitor, celecoxib, was synthesized using a 1,2-N-hydroxypyridinone MBP in place of a toluyl moiety (5-LOi-4).189,190 5-LOi-4 uses a hydropyridinone MBP and displays an IC50 value of 0.35 μM against 5-LO and ~10 μM against COX-1 and COX-2, with excellent oral anti-inflammatory activity comparable to celecoxib or ibuprofen in mouse models. Further work derivatizing celecoxib-like inhibitor cores with MBPs resulted in compounds such as 5-LOi-5, licofelone, and Tepoxalin.178 The incorporation of a hydroxamate MBP with a flexible linker onto a modified celecoxib core resulted in compound 5-LOi-5 (IC50 = 0.45–1.07 μM),187 which was more active than celecoxib in protein based assays and in rat neutrophils. Similarly, Tepoxalin, which is approved for veterinary use as a dual 5-LO and COX inhibitor, employs a N-methyl hydoxamate MBP on a flexible linker (5-LO IC50 = 1.7 μM; COX-2 IC50 = 4.2 μM).178
Current and Future Prospects.
Despite ongoing efforts to develop novel, specific 5-LO inhibitors, Zileuton remains the only FDA approved drug for 5-LO despite its toxicity and unfavorable PK profile.179,180 Hepatotoxicity has been a recurring issue for 5-LO inhibitors explored in clinical trials, particularly among inhibitors sharing hydroxamic acid or hydroxyurea MBP groups. In spite of many clinical candidates in Phase I and II trials in the past decade, overall progress in the field has stalled after the failures of Zileuton ‘me-too’ inhibitors, with few novel inhibitor classes being disclosed. New efforts towards the development of pan-inhibitors of 5-LO and COX enzymes has received attention, although few inhibitors have shown efficacy or safety profiles better than currently approved NSAIDs. Interest remains in exploring novel inhibitors for 5-LO inhibitors, as evidenced by several inhibitors that have been explored in clinical trials over the past 10 years. The development of lipoxygenase inhibitor based on reported COX inhibitor scaffolds is also intriguing; however, the field could benefit from obtaining structures of inhibitors complexed with 5-LO (i.e., enabling structure-based drug design, SBDD) and perhaps the use of alternative MBPs that have fewer ADME and toxicity liabilities.
3.3. Jumonji-domain Lysine Demethylase (JmjC KDMs, EC 1.14.11.27)
Function in Biology and Disease.
Post-translational modification of the amino-terminal domain of histones influence chromatin structure and dynamics, the lifetimes of chromatin components, and can mediate chromatin binding interactions.191,192 Histone modifications, including acetylation, methylation, phosphorylation, ubiquitination, and others, play a complex and vital role in eukaryotic transcriptional regulation and the epigenetic process of the cell.192–194 The misregulation of modification patterns have been linked to major human diseases, including mental disorders and cancers.194–198 Drugs, such as azacitidine and decitabine, which inhibit DNA methyltransferase, are currently used for the treatment of myelodysplastic syndrome, a disorder which develops prior to acute myeloid leukaemia.199,200
Prior to the discovery of histone N-methylated lysine residue demethylases (KDMs), the N-methylation modification of DNA and histones were thought to be an irreversible process. Since the discovery of the first KDM (KDM1A) in 2004,201 >20 human KDMs have been identified. Irregular or deregulated KDM activity leads to abnormal gene transcription and is linked to both cancer and neurological disorders.202–205 Depending on the mechanism of catalytic activity, KDMs are classified into two families: KDM1, also known as lysine-specific demethylases (LSDs), and KDM2-7, known as jumonji domain-containing KDMs (JmjC KDMs). The KDM1 family utilizes a flavin adenine dinucleotide (FAD) cofactor for the catalysis of mono- or di-methylated lysine residues, while JmjC KDMs are Fe2+-dependent metalloenzymes that share a conserved catalytic domain (i.e., the jumonji domain) and utilizes α-ketoglutaric acid (2OG) and molecular oxygen as cofactors to catalyze the demethylation of mono-, di-, and tri-methylated lysine residues (Figure 25). Detailed mechanism of action and classification of KDM families can be found elsewhere.202,204,206 The overexpression of certain JmjC KDMs have been correlated with cancer (including breast and testicular cancer),207 and the misregulation of JmjC KDMs are associated with the development of neurodevelopmental disease, including autisms spectrum disorder, X-linked mental retardation, and Kabuki syndrome,208 making JmjC KDMs a new, attractive therapeutic target. While a first generation KDM1 inhibitor, tranylcypromine, is approved by the FDA for the treatment of mood and anxiety disorders, there are currently no approved drugs which target JmjC KDMs.
Figure 25.
JmjC KDMs are Fe2+-dependent metalloenzymes that utilizes α-ketoglutaric acid (2OG) and oxygen as cofactors to catalyze the demethylation of mono-, di-, and tri-methylated lysine residues of histones.
Protein and Active Site Structure.
Members of the JmjC KDMs belong to the superfamily of Fe2+/2OG-dependent oxygenases (>60 human proteins).205 Other members of the 2OG dependent oxygenase family that have jumonji catalytic domains are of medicinal value. For example, the JmjC hydroxylases, which include factor inhibiting hypoxia inducible factor FIH and JmjC oxygenases (5JMJD6, JMJD5, JMJD7, JMJD8) have been explored as therapeutic targets. Comparison of the domain structural features and catalytic reactions of these enzymes are summarized elsewhere.205,209 Notably, other members of the 2OG oxygenases family, specifically those involved in hypoxic response pathway for the regulation of human genes such as JmjC hydroxylases FIH and hypoxia-inducible factor (HIF) prolyl 4-hydroxylases (PHDs) are commonly used as counter screens to determine the selectivity of JmjC KDM inhibitors. PHDs are related to the collagen prolyl hydroxylases subfamily of 2OG oxygenases and are also the target of current inhibitor development.210
The JmjC KDMs comprises of >20 human enzymes, and can further be classified into five subfamilies, including KDM2/7, KDM3, KDM4, KDM5, and KDM6 depending on JmjC domain sequence homology and overall architecture.202,205,209 The JmjC domain of JmjC KDMs is defined as the region of catalytic activity of JmjC KDMs (where the Fe2+ and 2OG bind) and has been the focus of inhibitor development; however, it is important to note that the specificity of various KDMs for their substrates is not only dictated by the JmjC domain, but also the histone-binding pocket, additional non-catalytic “helper domains” in the tertiary structure, as well as the other components in the multimeric complex in which the KDM resides.209,211
In general, the JmjC domain adopts a distorted double stranded b-helix (DSBH) fold core comprised of eight anti-parallel β-strands, with the Fe2+ binding residues at the more open end of the DSBH barrel (Figure 26).211–213 The DSBH barrel provides the metal coordinating and 2OG binding ligands. The Fe2+ in the JmjC domain is ligated by conserved residues His, Asp/Glu, and His (HxD/ExH) from the HSBH barrel, the 2-oxo and 1-carboxylate oxygen atoms of 2OG, and a water molecule in an overall octahedral coordination geometry. The terminal carboxylic acid (5-carboxylate) of 2OG is further stabilized by a basic residue (typically a Lys or Arg residue) and forms additional H-bonding interactions with side-chain donor residues, including Tyr in KDM4 and Thr in KDM6A.205,211,212 The structure of 2OG bound to KDM4D shows the 340 residue protein (~40 kDa) with the JmjC domain containing a Ni2+ ion (substituted for Fe2+), ligated by residues His192, Gly194, His280, the bidentate 2OG, and a water molecule (Figure 26).214
Figure 26.
Structure of KDM4D (Ni2+ in active site) with 2OG bound (magenta, PDB 4HON).
Inhibitor Development.
Rapid advancements in purification and crystallization, understanding the catalytic mechanism and selectivity of JmjC KDMs, and assays to evaluate inhibitor activity has led to a rise in JmjC KDM inhibitors, with several excellent reviews available.215,216 The first JmjC KDM inhibitors can be classified as 2OG substrate analogues with a carboxylic acid MBP, of which N-oxalyclicine (NOG) is a representative example (Figure 27). NOG is an N-oxalyl amide analogue of 2OG and a competitive inhibitor, displacing 2OG from the active site with an IC50 value of between 24 and 78 μM against KDM4E, depending on assay conditions.217 Unfortunately, due to the structural similarity of NOG to 2OG, NOG not only inhibits all tested JmjC KDMs, but is also a broad-spectrum inhibitor against PHD1-3 and FIH.217–222 In an attempt to achieve more selective inhibitors of JmjC KDM, the histone-binding hydrophobic pocket near the JmjC-containing domain has been exploited.223–225 A series of NOG derivatives bearing hydrophobic backbones were synthesized and found to be good inhibitors of JmjC KDM4E with IC50 values as low as 5.4 μM, with no inhibition of FIH or PHD2.224 The structure of KDM4A complexed with the representative inhibitor KDMi-1 (IC50 = 37 μM) reveals the 2-oxo and 1-carboxylate MBP groups of KDMi-1 bind in a bidentate manner to the active site Ni2+ ion (a surrogate for the native Fe2+), which is further ligated by the active site His188, Gly190, His276, and a water molecule in a octahedral coordination geometry (Figure 28). Metal coordination by KDMi-1 is identical to that found for NOG214 and 2OG.226 The aromatic backbone of KDMi-1 occupies the large hydrophobic pocket, interfering with the histone substrate and is proposed to aid in JmjC KDM specificity; however, no other JmjC KDMs were screened to verify selectivity. Related inhibitors bearing a carboxylic acid MBP have also been described.223,227
Figure 27.
Inhibitors of JmjC KDMs.
Figure 28.
Structure of: KDMi-1 bound to KDM4A (Ni2+ in active site, PDB 2WWJ); 2,6-PDCA bound to KDM4A (Ni2+ in active site, PDB 2VD7); KDMi-4 bound to KDM6B (Ni2+ in active site, PDB 2XXZ); and GSK-J1 bound to KDM6B (Co2+ in active site, PDB 4ASK).
A second collection of JmjC KDM inhibitors employ a hydroxamic acid MBP, including repurposed histone deacetylase (HDAC) inhibitors (Section 7.3) such as TSA and SAHA (Figure 97). Although TSA and SAHA were shown to inhibit KDM4E (IC50 values of 28 and 14 μM, respectively),217 they inhibited HDACs more effectively,228 raising questions of selectivity.215,216 Nonetheless, some hydroxamic acids, such as KDMi-2, have been developed as potent and selective inhibitors. KDMi-2 shows good activity against KDM4A and KDM4C (IC50 values of 3 μM and 1 μM, respectively), and are >100-fold more selective for KDM4A and KDM4C compared to PHD1 and PHD2.229 Although no structures of this class of inhibitors bound to JmjC KDM4 are available, molecular modeling suggest the hydroxamic acid is coordinated to the Fe2+ in a bidentate manner. The selectivity of KDMi-2 is attributed an optimal 8-linker chain, followed by the terminal dimethylamino group, which can fill the hydrophobic substrate channel and interact with residues Asp137 and Tyr179.229 Interestingly, the replacement of the terminal dimethylamino group by a cyclopropane yields KDM2/7 selective inhibitors.230 The cyclopropane derivative exerted antiproliferative activity against cancer cells, whilst KDMi-2 did not. Other hydroxamic acid MBP inhibitors have also been reported.229–232
Figure 97.
Inhibitors of HDACs.
A third class of MBPs which have been investigated for inhibitor development utilize a pyridine motif, including pyridine dicarboxylates, 8-hydroxyquinoline, and bipyridine MBPs. Through structure-guided drug design, pyridine-2,4-dicarboxylic acid (2,6-PDCA, Figure 27), a broad spectrum 2OG oxygenase inhibitor,217,233 was optimized to yield KDMi-3, an KDM4E selective inhibitor.234 Installation of a bulky substituent at the 3-position of the pyridine ring allowed for the selective inhibition for larger binding pocket of KDM4E (IC50 = 2.5 μM) over the smaller binding pocket found in that of PHD2 (IC50 >400 μM).234 The structure of KDM4A bound to 2,6-PDCA reveals the pyridine carboxylate MBP occupying the 2OG site, binding in a bidentate manner to the active site Ni2+ (again, as a surrogate for Fe2+). The Ni2+ is further ligated by residues His188, Glu190, His276, and a water molecule in an octahedral coordination geometry (Figure 28).217 The pyridine ring forms hydrophobic interactions with the active site pocket Tyr177, Phe185, Trp208, while the 4-carboxylate motif makes similar H-bonding interactions to that of the 5-carboxylate of 2OG, binding to Lys206 and Tyr132. Bearing some structural similarity to 2,6-PDCA, 8-hydroxyquinolines (KDMi-4) have also been investigated. Through HTS, KDMi-4 was found to have comparable activity against KDM4E (IC50 = 200 nM) as 2,6-PDCA (IC50 = 180 nM) and KDMi-4 inhibited cellular histone demethylase activity in vitro with an EC50 value of 87 μM.233 The broad spectrum of KDMi-4 against a wide panel of KDM subfamilies had hindered its development into a clinically relevant drug.219 The structure of KDMi-4 bound to KDM4233 and KDM6B have been solved and show the same metal binding mode (Figure 28), with the quinolone nitrogen atom and deprotonated 8-hydroxy oxygen atom of the 8-hydroxyquinoline MBP bound in a bidentate manner to the Ni2+ ion, which is also bound by residues Glu1389 and His1387 (Glu190 and His188 of KDM4A) and two water molecules in an octahedral coordination geometry. Notably, in both structures the hydroxyl motif of KDMi-4 causes displacement of the ligating His1467 (His276 in KDM4A), resulting in the loss of a Ni2+-His bond. Additionally, similar to 2,6-PDCA and 2OG, the non-metal binding carboxylic acid makes H-bonding interactions with conserved Lys and Tyr residues. Additional studies utilizing the 8-hydroxyquinoline MBP for inhibitor development and photoaffinity probes have been reported.235,236
Inhibitors bearing a bipyridine MBP have also been of interest, such as GSK-J1 (Figure 27). Through HTS and structure-guided drug design, GSK-J1 was discovered as a highly selective inhibitor for KDM6B (IC50 = 60 nM), while inactive against a panel of other JmjC KDMs, protein kinases, and other unrelated protiens.237 The ester pro-drug of GSK-J1, GSK-J4, which is hydrolyzed by macrophage esterases to yield the active inhibitor, was shown to reduce lipopolysaccharide-induced proinflammatory cytokine production by human primary macrophages (a process that depends on both KDM6A and KDM6B).237 Follow up studies which screened GSK-J1 against a panel of twelve JmjC KDMs revealed GSK-J1 also inhibited KDM5B and KDM5C, albeit with 5- to 10-fold weaker activity.238 The structure of GSK-J1 bound to human KDM6B reveals the bipyridine MBP bound in a bidentate manner to the Co2+ (used as a surrogate for Fe2+), which is further ligated by His1390, Glu1492, and two water molecules in an octahedral coordination geometry (Figure 28).237 Notably, the metal ion is shifted and results in loss of the native His1470-metal coordination, with His1470 instead hydrogen bonded to a metal-bound water molecule. Further key interactions include the propionic acid of GSK-J1 binding to Lys1381, Thr1387, and Asn1480 (similar to that of 2OG) and the aromatic ring of GSK-J1 situated in the histone peptide pocket. Additional studies have shown GSK-J4 has anticancer activity against acute lymphoblastic leukemia and pediatric brainstem glioma,239,240 and inhibited the survival of ovarian cancer stem cells.241
Current and Future Prospects.
The post-translational modifications of histones affect various aspects of chromatin, and play a vital role in eukaryotic transcriptional regulation and the epigenetic process of the cell. Modifications include acetylation, methylation, phosphorylation, and ubiquitination, with histone demethylases being recently recognized as playing an important role in the development of cancer and mental disodrders.215 The Fe2+-dependent JmjC KDMs, which utilize 2-oxoglutarate (2OG) and oxygen for the demethylation of histone targets has emerged as a promising target.215 The development of subfamily specific inhibitors will help determine the relevance of JmjC KDMs in the regulation of cellular process, as well as their potential as therapeutics. Inhibitors bearing various MBPs, including carboxylic acids, hydroxamic acids, and bipyridine have been identified; however, these small molecule inhibitors bind primarily to the highly conserved JmjC-domain catalytic site, which has yet to be demonstrated to give satisfactory JmjC KDM subfamily specificity. In fact, these inhibitors are commonly broad spectrum 2OG oxygenase inhibitors, being able to inhibit FIH and PHDs. However, given the selectivity of JmjC KDMs for demethylation of their histone targets, the development of JmjC KDM specific inhibitors should be viable. This would require inhibitors to not only bind to the Fe2+ and occupy the 2OG binding pocket, but to also extend into the corresponding histone-binding sites. A successful case of this includes GSK-J1, which uses a bipyrdine MBP to coordinate to the metal ion, but also a hydrophobic substituent that branches into the histone-binding pocket. GSK-J1 was found to be selective for KDM5 and KDM6 subfamilies (due to the high structural homology of the catalytic domain between the two subfamilies),238 while showing no significant inhibition against other JmjC KDMs. Future JmjC KDMs inhibitors would not only require optimal MBPs for metal binding and inhibitor occupancy of the 2OG site, but also exploitation of the JmjC KDM specific histone-binding pocket.
3.4. Tyrosinase (TYR, EC 1.14.18.1)
Function in Biology and Disease.
The biopolymer melanin is the pigment responsible for the coloration of human eyes, hair, and skin. Under normal conditions melanin is extremely beneficial and aids in the photo-protection of human skin.242 However in melanoma, the most dangerous form of skin cancer, melanin production or ‘melanogenesis’ can be deregulated,243 which can cause an attenuation of the effectiveness of radiotherapy, chemotherapy, phototherapy, and immunotherapy.244 Melanogenesis and its attenuation of common therapies has been observed in the clinic, with melanin production being correlated with decreased survival rates of patients with stage III and IV melanoma.245 Evidence suggests that by decreasing melanin production in melanoma, the therapeutic efficacy of ionizing radiation and chemotherapy is improved.246,247 Additionally, overproduction of melanin is implicated in a range of dermatological conditions including post inflammatory melanoderma and age spots.248,249 Given the implications in melanoma and breadth of dermatological conditions in which melanin is involved, there is considerable interest in therapeutic strategies to modulate the production of melanin.
Melanins are biosynthesized in specialized pigment producing cells called melanocytes, via the melanogenesis pathway. The first enzyme in the melanogenesis pathway is the dinuclear Cu-dependent metalloenzyme tyrosinase (TYR), which catalyzes the rate limiting step in melanin biosynthesis.250 TYR catalyzes the conversion of L-tyrosine to L-DOPA using molecular oxygen, followed by the oxidation of L-DOPA to L-DOPAquinone (Figure 29). L-DOPAquinone is then further transformed, either by a spontaneous reaction or via the aid of the remaining enzymes in the melanogenesis pathway, into two main classes of melanins: eumelanin and pheomelanin.251,252 Additionally, TYR can also oxidize diphenolic catechols to their corresponding quinones.251 Therefore, TYR is regarded as a promising target for the pharmaceutical regulation of melanin production.
Figure 29.
TYR converts L-tyrosine to L-DOPA using oxygen, which it then oxidizes to L-DOPAquinone.
Protein and Active Site Structure.
The metalloenzyme TYR is classified as a copper protein with a type-3 copper center and is generally found throughout the phylogenetic scale from bacteria to humans.6 As a type-3 copper center, the active site contains two copper ions, each coordinated by three His residues (Figure 30), with the coordinating motif being highly conserved among TYR enzymes from different organisms. The oxidation state of the active site ions cycles between Cu+ and Cu2+ when deoxygenated and oxygenated, respectively (Figure 29).2 Although the active site metal ions and coordinating residues are conserved, there is low sequence identity between human TYR (hTYR) and those from other organisms. For example, hTYR shares a ~10% sequence homology with the commonly studied TYR from the mushroom A. bisporus (AbTYR).243,253 When analyzing the second coordination sphere of the active site (within ~6 Å of the Cu ions) from multiple TYRs, there is ~58% sequence identity, suggesting some structural differences at the active site between TYRs from different sources.254 To date, there is no available crystal structure of hTYR, although it is known to be a monomeric glycosylated type-I transmembrane protein ~67 kDa in size.255,256 Until very recently recombinant expression of hTYR has been difficult,257 which has resulted in AbTYR serving as a surrogate in inhibition and structural studies (largely due ease of purification and commercial availability). As with all TYRs, the active site in AbTYR is a dinuclear copper site with each Cu ion coordinated by three His residues. It was discovered that His85, one of the His residues coordinating an active site Cu, possess a covalent modification from a thioether side chain of Cys83 (Figure 30).258 Whether this modification is present in the active site of mammalian TYRs, including hTYR, is presently unknown.259
Figure 30.
Structure of AbTYR (PDB 2Y9W), with lectin-like fold subunit not shown.
Inhibitor Development.
Inhibition of TYR has been a significant area of interest and many inhibitors of AbTYR have been identified from a variety of natural and synthetic sources, with several comprehensive reviews available.260–262 Some inhibitors, such as flavonoids, are thought to coordinate to the dinuclear active site based on spectroscopic studies,263 while the mode of binding for other inhibitors is unknown. One of the more well studied TYR inhibitors, which is used as a skin whitening agent, is the natural product kojic acid (Figure 31).264 Kojic acid exhibits mixed inhibition265 with moderate activity against TYR enzymes. Specifically, kojic acid activity against Bacillus megaterium TYR (BmTYR, Ki = 3.5 μM) and Agaricus bisporus TYR (AbTYR, Ki = 4.3 μM) have been reported.253,265 An X-ray structure of kojic acid bound to BmTYR showed the inhibitor interacting with active site residues, but surprisingly not bound to the active site metal ions.266 In a related study, kojic acid was observed engaging in a hydrogen bonding interaction with a Cu-bridging water molecule, but again not directly coordinated to the active site metal ions of BmTYR (Figure 32).265 The observed modes of kojic acid binding is in contradiction with previously proposed inhibition mechanisms that involve metal coordination, which were based on spectroscopic studies of model complex.267 It is suggested that both modes may explain the mixed inhibition kojic acids exhibits. In spite of its use as a skin whitening agent, kojic acid exhibits significantly lower activity against hTYR (Ki = 350 μM).253
Figure 31.
Inhibitors of TYR.
Figure 32.
Structure of kojic acid bound to BmTYR (PDB 5I38). Kojic acid is hydrogen bonded (dashed magenta line) to a bridging water molecule and is not coordinated to the active site metal ions.
Another inhibitor of TYR is hydroquinone, which is a FDA approved skin whitening agent (Figure 31).264 An X-ray structure of hydroquinone bound to BmTYR (reconstituted with Zn2+ instead of Cu2+ to prevent hydroquinone oxidation)265 revealed a hydroxyl group of hydroquinone is coordinated with one of the active site Zn2+ ions with a Zn-O bond distance of ~2.2 Ǻ. Hydroquinone also engages in a hydrogen bonding interaction with Asn205. Interestingly, when hydroquinone was analyzed against hTYR it was much less active (IC50 >4 mM)268 than against BmTYRs (IC50 = 32 μM).265 Testing using in vivo skin models showed hydroquinone caused a decrease in melanogenesis at relatively low concentrations (IC50 <40 μM) when compared to its in vitro activity against hTYR. It was proposed that hydroquinone may inhibit melanogenesis via a different pathway than TYR inhibition. In spite of their effectiveness and use as skin whitening agents, both hydroquinone and kojic acid generally lacked satisfactory inhibitory activity against hTYR.253,268 In addition, these agents have been reported to have undesirable side effects in clinical applications.264,269 One clear problem in evaluating these and other TYR inhibitors is that the majority of studies are performed on non-human forms of TYR,262 which share low sequence identity with the human enzyme.254,262 Given this low sequence identity, it makes it difficult to correlate observed inhibitory activity between non-human and human TYR.
In an effort to develop novel, specific, and potent inhibitors against hTYR, one study examined the natural product family, thujaplicins, as potential inhibitors.270 A preliminary SAR was observed based on the activity of the α, β, and γ-thujaplicin (TYRi-1, TYRi-2, TYRi-3, respectively, Figure 31). TYRi-1 demonstrated the poorest activity (IC50 >1 mM), while TYRi-2 yielded moderate activity (IC50 = 9 μM), and TYRi-3 exhibited the best inhibitory activity (IC50 = ~1 μM).270 The binding mode of TYRi-3 was modeled in a homology model of the hTYR active site, which indicated the hydroxyl and carbonyl coordinated to the active site metal ions and the isopropyl moiety making hydrophobic interactions with other parts of the active site. The poorer activity of TYRi-1 and TYRi-2 was explained by their isopropyl moieties causing steric clashes with the active site. The natural product tropolone, which is the cyclic core of thujaplicin (but lacks the isopropyl moiety), has also been studied as a TYR inhibitor. A crystal structure was obtained with tropolone bound to the active site of AbTYR, although it does not coordinate to the dinuclear active site Cu ions.258
The natural product family of aurones are also hTYR inhibitors.271 Aurones are a sub-class of flavonoids, which as mentioned above are a class of TYR inhibitors.260 One of the most active aurones analyzed was TYRi-4 (IC50 = 38.4 μM, Figure 31). The SAR around this scaffold suggested that the aurones are not engaged in binding the active site Cu ions. A subsequent study focused on introducing MBPs to the aurone scaffold.272 The N-hydroxypyridinone MBP (TYRi-5, Figure 31) alone has some inhibitory activity (Ki = 128 μM) against hTYR, with subsequent attachment to an aurone scaffold yielding TYRi-6, which showed increased activity (Ki = 350 nM) against hTYR. TYRi-6 was evaluated against human MNT-1 melanoma cells and showed in vivo activity by decreasing melanin biosynthesis (IC50 = 85.3 μM) while exhibiting low overall cytotoxicity. These studies represent a potential path to the development of novel hTYR inhibitors for clinical anticancer therapeutics.
Current and Future Prospects.
Development of TYR inhibitors has largely been performed using non-human TYR, which is a potential roadblock for inhibitors to translate into the clinic. Despite this, hydroquinone is approved for topical use only even though it has undesirable side effects and its safety has been questioned by regulatory agencies.273 Another roadblock for developing inhibitors of hTYR is that the structure of hTYR is unknown and homology modeling is used in lieu of crystallographic studies. Recent studies claim large scale expression of hTYR, along with some preliminary X-ray crystal structure data at low resolution (~3.5 Å), which if improved would be a significant boon to the field.253,257 Despite the lack of a hTYR structure, potent inhibitors such as TYRi-3 and TYRi-6 have been identified. Encouragingly, TYRi-6 has shown in vivo activity for reducing melanogenesis in human melanoma cells while displaying low cytotoxicity. These compounds may represent new scaffolds for improved activity and PK properties that may reinvigorate clinical investigations.
4. Transferases (EC 2.X)
4.1. Catechol-O-methyltransferase (COMT, EC 2.1.1.6)
Function in Biology and Disease.
Parkinson’s disease (PD) is the second most common neurodegenerative disorder of the central nervous system (CNS), affecting ~1% of individuals over the age of 60 and ~3% for those over the age of 80.274 While the specific cause for PD is unknown, longitudinal studies have found an increased risk for PD associated with exposure to pesticides, history of melanoma, usage of methamphetamines, and traumatic brain injury.275 Symptoms of PD (as well as many other CNS disorders such as schizophrenia and depression) are associated with reduced dopamine levels in the brain.276–279 Intake of levodopa (L-DOPA), a direct metabolic precursor of dopamine, is an effective way to modulate L-DOPA levels for alleviating the symptoms of PD. However, the efficacy of L-DOPA is challenged by catechol-O-methyltransferase (COMT). COMT is responsible for the elimination of biologically active or toxic catechols, and regulates the amount of available dopamine.280 COMT, a Mg2+-dependent metalloenzyme, transfers the methyl group of cofactor S-adenosyl-L-methionine (SAM) to the 3-O-hydroxyl of dopamine (Figure 33).281 Co-administration of COMT inhibitors with L-DOPA has proven an effective method for maintaining the efficacy of L-DOPA and increasing dopamine levels.278 COMT inhibitors (such as tolcapone and entacapone) are currently co-administered clinically with L-DOPA for the treatment of PD; however, these inhibitors have poor PK properties and more effective inhibitors are still desired.
Figure 33.
Reaction catalyzed by COMT. The O-methylation of dopamine by COMT with co-substrate SAM leads to 3-methoxytyramine, which inactivates dopamine as a treatment for PD.
Protein and Active Site Structure.
Two isoforms of COMT exists, a soluble (S-COMT) and membrane-bound (MB-COMT) form. S-COMT is the predominate form in most tissue with the highest levels found in the liver and kidneys. S-COMT is located in the cytosol and it inactivates endogenous and xenobiotic catechols. MB-COMT is anchored to the rough endoplasmic reticulum, found predominantly in the brain, and inactivates catecholaminergic neurotransmitters such as dopamine and norepinephrine.282,283 The isoforms of COMT vary by 50 residues, specifically an extension found in the MB-COMT form, which is the signal sequence for membrane anchoring.280
Human COMT (264 – 271 residues long, ~30 kDa) is a single domain α/β-folded structure composed of a seven-stranded β-sheet core, flanked by two sets of α-helices (Figure 34).280 A crystal structure of cofactor SAM and DNC (3,5-dinitrocatecol, a dopamine mimic and early inhibitor of COMT, Ki = 8 – 24 nM)284,285 bound to COMT reveals a SAM binding domain, as well as the catalytic site. In the SAM binding domain, the SAM methionine sulfur atom is oriented toward the catalytic site in proximity to the 3-O-hydroxyl group to be methylated. The active site Mg2+ is ligated by the side chains of Asp141, Asp169, Asn170, the two catechol hydroxyls of DNC, and a water molecule in an octahedral coordination geometry (Figure 34).286,287 Additionally, a hydrophobic wall consisting of residues Trp38, Trp143, and Pro174 adjacent to the catalytic site determines the regioselectivity for 3-O-methylation over 4-O-methylation of the catechol hydroxyl groups. Analysis of the regioselectivity of the COMT enzyme have been described in detail.280,288,289
Figure 34.
Structure of DNC (green) and SAM (yellow) bound to COMT (PDB 3BWM).
Inhibitor Development.
Unsurprisingly, inhibitors bearing a catechol MBPs constitute the largest, most clinically advanced class of COMT inhibitors.282,288,290–309 In the 1960s, the first generation of COMT inhibitors appeared that contained a catechol MBP.295–302 U-0521 (Figure 35) is the most archetypal and potent inhibitor of this class (Ki = 7.8 μM); however, U-0521 displayed low selectivity (also inhibiting tyrosine hydroxylase), poor activity, and high toxicity. In addition, first generation inhibitors were often substrates for COMT themselves and underwent O-methylation. This resulted in the formation of toxic O-methylated metabolites, and precluded these inhibitors from clinical development.310 Second generation COMT inhibitors bearing a nitro-catechol MBP were introduced as more potent inhibitors, as well as poorer substrates of COMT. The addition of a nitro-group at the ortho-position to one of the hydroxyl groups lowers the pKa of the adjacent hydroxyl group, thereby preventing O-methylation.293 Clinically advanced second generation inhibitors include, but are not limited to, nitecapone, entacapone, and tolcapone (Figure 35). Nitecapone, entacapone, and tolcapone exhibited good activity against COMT (IC50 = 18 nM, 10 nM, and 36 nM, respectively), low acute toxicity, and high selectivity for COMT versus other catecholamine metabolizing enzymes (such as tyrosine hydroxylase, DβH, MAO-A, and MAO-B).280,282,293,294,311,312 Although, entacapone and tolcapone (trade names Comtan and Tasmar) are approved drugs for the treatment of PD, they lack bioavailability and are toxic, limiting their overall clinical usefulness (see discussion below). Extensive reviews on the history, recent developments, and clinical data of these inhibitors can be found elsewhere.282,287
Figure 35.
Inhibitors of COMT.
Another class of COMT inhibitors bearing the catechol MBP are bisubstrate inhibitors (COMTi-1, Figure 35). Bisubstrate inhibitors are rationally designed to link a SAM analogue with a dopamine analogue, and mimic their binding activity during catalysis.288 Various derivatives of bisubstrate inhibitors have been synthesized, with modifications to the linker length, nitro-group position, ribose motif, and adenine motif,303–309 with COMTi-1 being the most potent example reported date (IC50 = 9 nM).309 In vivo data for bisubstrate inhibitors is lacking, so their clinical potential remains unclear. Crystallographic data of COMTi-1 bound to COMT (Figure 36) reveals COMTi-1 occupying both SAM and dopamine binding pockets.303 The dihydroxyl group of the catechol MBP binds in a bidentate manner to the Mg2+ ion, which is ligated by the sides chains of Asp141, Asp169, Asn170, and a water molecule forming an octahedral coordination geometry (similar to that of DNC and other first- and second-generation inhibitors).
Figure 36.
Left: Structure of COMTi-1 bound to COMT (PDB 1JR4). Right: Structure of COMTi-3 bound to COMT with SAM (yellow) (PDB 4XUC).
In addition to the catechol MBP, tropolone (COMTi-2, Figure 35),313 and hydroxypyridone MBPs282,314–316 (COMTi-3, COMTi-4, COMTi-5, Figure 35) have also been explored. These inhibitors are able to bind in a bidentate manner to Mg2+ ion analogous to catechol MBPs, but have the advantage that they do not undergo O-methylation. Inhibitors bearing a tropolone MBP exhibited modest activity against COMT (COMTi-2, Ki = 8.9 μM), and have not been pursued as clinical candidates.282,313,317 In contrast, inhibitors bearing a hydroxypyridone MBP (COMTi-3, COMTi-4, COMTi-5) have achieved IC50 values of 8 nM.316 X-ray crystallographic analysis of COMTi-3 bound to COMT reveals the hydroxypyridone MBP coordinates in a bidentate fashion to the Mg2+ ion resulting in an octahedral coordination geometry identical to that of the catechol-based inhibitors (Figure 36).315 Further optimization and characterization of hydroxypyridone MBP compounds yielded COMTi-5 (IC50 = 8 nM), disclosed by Merck in a patent for the symptomatic treatment of neurological and psychiatric disorders where COMT is involved;316 however, its clinical efficacy has not yet been validated.
Current and Future Prospects.
While structurally diverse inhibitors of COMT have been developed, most rely on a catechol or hydroxypyridone MBP motifs for metal binding. The first generation COMT inhibitors exhibited weak in vitro activity, poor selectivity, and toxicity. The need to increase activity and eliminate O-methylation by COMT led to the development of second generation inhibitors, including nitecapone, entacapone, and tolcapone. These inhibitors are quite potent and some are used clinically. While nitecapone completed Phase I clinical trials, it was replaced by entacapone, which was judged to be a more efficacious clinical candidate.282 Since the late 1990s, entacapone has been approved for the adjunctive treatment of PD; however, its low bioavailability and short duration of action has limited its overall clinical effectiveness.282,318 Tolcapone, another approved drug used for the adjunctive treatment of PD, exhibits liver toxicity issues and its usage has been limited to a last resort drug when patients are unresponsive to other treatment methods.319 The design and synthesis of bisubstituted inhibitors yielded good in vitro activity, but their clinical efficacy has yet to be determined. Inhibitors bearing a hydroxypyridone MBP have been extensively studied, but their clinical efficacy has also yet to be validated.
The desire to develop a potent COMT inhibitor for treating PD has led to major advancements in understanding the COMT enzyme and the mechanism of action of various inhibitors; however, further development with new molecular entities, including new MBPs (beyond catechols, tropolones, and hydroxypyridone) is merited based on the drawbacks of current compounds such as poor inhibitory activity and low oral bioavailability (due to glucuronidation and sulfation).320 Inhibitors with alternative MBPs should be structurally distinct from the diphenolic motif, but also capable of bidentate metal coordination. Further research into new MBPs which incorporate the SAM substrate group may yield the breakthrough needed for more active and selective COMT inhibitors.
4.2. Farnesyl transferase (FTase, EC 2.5.1.58)
Function in Biology and Disease.
Farnesyl transferase (FTase) belongs to the prenyltransferase class of enzymes, and functions to transfer a farsenyl signaling moiety onto a variety of substrate proteins (Figure 37). FTase has garnered significant attention over the past ~30 years due to its potential as a target for anticancer therapeutics.321 FTase is a signaling protein responsible for activating several enzyme classes, including, but not limited to, members of the RAS family and the RAS superfamily, laminin, phosphorylase kinase, and transducing rhodopsin kinase.322 The ability of FTase to activate the RAS proteins is critical, as this FTase action is necessary for RAS proteins to function.323 The RAS proteins, especially KRAS, are a heavily studied class of metalloenzyme for cancer therapy, as mutations in these proteins are extremely prevalent in the majority of cancer serotypes,324 to the point that many cancers are dependent on KRAS for survival.321 Correspondingly, there has been a tremendous interest in developing FTase inhibitors to preemptively block RAS function to inhibit tumor growth and potentially treat cancer.
Figure 37.
Reaction catalyzed by FTase.
Early studies on rodents demonstrated the potential of FTase inhibitors, as nude mice bearing the tumorigenic HRAS cancer model were successfully treated and cured with FTase inhibitors.325 However, to great disappointment, FTase inhibitors failed to provide benefit in late stage Phase III clinical trials as cancer therapeutics.326–328 Similar to MMPs (Section 1.5), FTase inhibitors were quite potent, but failed because the biology regarding KRAS was not fully understood.321 It was found that although inhibition of FTase prevents FTase from farsenylating and activating KRAS, other enzymes within the prenyltransferase family (geranylgeranyltransferase, GGTase) are able to fill the void and activate KRAS, providing an alternative activation pathway to FTase.329–331 This secondary activation was not observed in rodent models because these models used HRAS (the dominant tumor-driver in rodent) instead of KRAS (the dominant tumor-driver in humans), and HRAS cannot undergo the same secondary activation mechanisms.332 It is worth noting that GGTase is also a Zn2+-dependent metalloenzyme that is structurally similar to FTase; however, inhibition of GGTase has been shown to be lethal in rodents, making dual inhibition of FTase and GGTase an unviable route in cancer therapy.322,333
Protein and Active Site Structure.
FTase is a heterodimeric Zn2+-dependent metalloenzyme responsible for transferring a phosphorylated isopreneoid substrate to the C-terminus of protein substrates bearing a CAAX terminal motif, where C is Cys, A is either Ala or Phe, and X is any amino acid. As shown in Figure 38, both subunits are mainly comprised of α-helices, with the α-subunit enveloping the β-subunit. The β-subunit contains the catalytic Zn2+ ion, which is coordinated by His362, Asp297, Cys299, and a water molecule in a tetrahedral geometry.334 Interestingly, there is also a loosely coordinated Mg2+ ion within the active site that is believed to aid in enzyme catalysis.335 The mechanism of FTase farsenylation begins with coordination of the farsenyl diphosphate substrate, which is necessary for binding of the second substrate, namely the CAAX peptide. The CAAX peptide is coordinated to the Zn2+ though the Cys sulfur atom, and then ligation of the farnesyl and peptide occur. Finally, a second farnesyl diphosphate molecule enters the active site to trigger dissociation of the newly formed farnesylated peptide.336
Figure 38.
Structure of FTase with farsenyl diphosphate (yellow) and peptide substrate magenta bound (PDB 1TN6). The α-subunit is shown in cyan and the β-subunit in gray.
Inhibitor Development.
Having been studied for ~30 years, there exists a substantial body of literature on FTase inhibitors.337,338 Among the most prominent inhibitors within the field summarized in Figure 39. α-Hydroxyfarnesyl phosphonic acid and L-744,832 are significant as they represent the earliest FTase inhibitor classes, whereas lonafarnib and tipifarnib were evaluated in Phase III clinical trials. While α-hydroxyfarnesyl phosphonic acid and lonafarnib contain functional groups capable of metal coordination, crystal structures of these inhibitors show that they do not coordinate to the catalytic Zn2+ but bind elsewhere within the FTase active site.339 In contrast, L-744,832 and tipifarnib have been crystallography demonstrated to inhibit FTase by coordination to the active site Zn2+ ion.
Figure 39.
Inhibitors of FTase. Only L-744,832 and tipifarnib have been confirmed to inhibit via metal coordination.
L-744,832 is based on the amino acid sequence CVFM, and was designed as a mimic of the native KRAS tetrapeptide CAAX motif that FTase acts on.340 Surprisingly, L-744,832 is not farsenylated by FTase, a property which has been attributed to the Phe residue.341 L-744,832 has an IC50 value around 0.240 μM, depending on the carboxylate pro-drug used.322 The structure of L-744,832 bound to FTase shows the compound acts as a competitive inhibitor and coordinates to the active site Zn2+ through the Cys sulfur atom (Figure 40).342 This thiol tetrapeptide inhibitor also showed cellular activity with the ability to inhibit the growth of >70% of cancer lines when administered at concentrations of 2–20 μM.343 Unfortunately, L-744,832 failed Phase I clinical trials as this it displayed adverse neutropenia, thrombocytopenia, fatigue, and vomiting.344,345
Figure 40.
Left: Crystal structure of L-744,832 (green) and farsenyl diphosphate (yellow) bound to FTase (PDB 1JCQ). Right: Crystal structure of tipifarnib (green) and farsenyl diphosphate (yellow) bound to FTase (PDB 1SA4).
Tipifarnib (previously known as R115777) was discovered through HTS by the Janssen Research Foundation through a combination of library screening and computer aided drug design.346 A crystal structure of tipifarnib bound to FTase shows the compound coordinates to the catalytic Zn2+ ion using the nitrogen atom of an imidazole group (Figure 40). Tipifarnib has an IC50 value of 7.9 nM, and was found to inhibit cell proliferation in 75% of human tumor cell lines bearing RAS mutations when administered at concentrations ≤100 nM.347 Unfortunately, tipifarnib failed to improve patient outcomes in late stage Phase III.326–328
Current and Future Prospects.
FTase functions as a signaling enzyme that activates protein substrates by attaching a farnesyl group though a Zn2+ mediated-mechanism. In particular, FTase has garnered significant interest due to its ability to activate the notorious cancer target KRAS; and it was widely speculated that FTase inhibitors would serve to upstream inhibit KRAS function. Unfortunately, FTase inhibitors have failed in Phase III clinical trials due to insufficient ability to improve patient outcomes with advanced cancer, regardless of whether these drugs were administered alone or in combination.321 This lack of efficacy was due to KRAS activation by alternate prenylation mechanisms, which was not detected in rodent models. While the interest in developing inhibitors of human FTase has understandably diminished, there is still potential for FTase inhibitors to be used in other disease states affected by FTase signaling, such as cardiovascular disease, Parkinson’s disease, Alzheimer’s disease, and schizophrenia.348 Furthermore, as FTase is found in many pathogens, there has recently been a burgeoning interest in developing FTase inhibitors as potential antibiotics and antimicrobials to target parasitic diseases including malaria, African sleeping sickness, and Chagas disease.349,350
4.3. Mobilized Colistin Resistance-11 (MCR-1, EC N.A.)
Function in Biology and Disease.
Colistin (a.k.a., Polymyxin E) is considered to be one of the drugs of last resort against antibiotic resistant bacteria, and operates by destroying the bacterial outer membrane.351 Due to the structure of the bacterial outer membrane, the outer membrane surface has an overall negative charge from the phosphate backbone of the lipopolysaccharide coating. Colistin contains five terminal amine residues that are protonated at physiological pH, leaving colistin with an overall positive charge (Figure 41). This creates an electrostatic attraction allowing colistin to bind the outer membrane surface, so that once the amine enriched portion of the molecule is bound, the hydrophobic tail is free to incorporate into the bacterial lipid layer, creating micropores that weaken the cell membrane, eventually leading to cell death.352
Figure 41.
Left: Structure of colistin with the amines necessary for electrostatic binding circled in cyan, and the hydrophobic tail for lipid intercalation colored in red. Colistin disrupts the structure of bacterial cell membranes. Right: Reaction catalyzed by MCR-1 to generate resistance against colistin. The transferred phosphoethanol amine is highlighted in red. R1 and R2 are fatty acids.
Historically, the development of resistance to colistin was relatively rare, as bacteria had to individually develop resistance against this antibiotic through random chromosomal mutations. However, changing usage, including the production of ~12,000 tones of colistin in 2015 primarily for agricultural use, have led to the development of Mediated Colistin Resistance-1 (MCR-1).353 MCR-1 is plasmid-born and is easily communicable among multiple bacteria populations. Since its initial detection in China in 2015, MCR-1 has rapidly spread worldwide. As of early 2017, there have been reports of MCR-1 detected in animals in 28 countries, and reports of human MCR-1 in human isolates in 29 countries.354 Furthermore, MCR-1 has been detected with other antibiotic resistance mechanisms, most notably NDM-1 (Section 7.5),355 leading to the fear that MCR-1 will produce pan-resistant bacteria. Therefore, there is a strong interest in preemptively developing inhibitors against MCR-1 to alleviate the burden of resistant bacteria.
MCR-1 is a Zn2+-dependent metalloenzyme that operates as a phosphoethanolamine transferase to confer resistance to colistin (Figure 41). Specifically, MCR-1 places a terminal amine group on the phosphate backbone of the outer membrane, which at physiological pH, changes the overall charge of the bacterial outer membrane from negative to positive. The resulting positively charged outer membrane serves to electrostatically repel colistin so that the antibiotic can no longer interact with the outer membrane to cause cell death.
Protein and Active Site Structure.
MCR-1 is a large enzyme that is anchored to the inner membrane by an α-helix bundle domain that is tethered to the catalytic domain (Figure 42).356 While MCR-1 has been confirmed to be a Zn2+-dependent metalloenzyme via mass spectrometry experiments,357 as well as incubation with EDTA, which abolished activity in cellular assays,356 the literature is currently unresolved whether MCR-1 possesses a mononuclear or dinuclear Zn2+ active site. The first published crystal structures of the catalytic domain of MCR-1 showed a dimeric structure with a dinuclear Zn2+ active site, where the second Zn2+ serves as a bridge connecting the two enzyme units.357,358 Additional crystallography studies have captured both mononuclear and dinuclear Zn2+ forms of MCR-1 (Figure 42), with the mononuclear Zn2+ form as a single enzyme unit, and the dinuclear Zn2+ form again as a dimeric enzyme.356 It is also unknown whether MCR-1 functions as a dimer, or if the enzyme is actually functional as a monomer. In the case of the mononuclear Zn2+ form, the active site consists of a large flat plane, with the catalytic Zn2+ ion coordinated by His466, Asp465, Glu246, and phosphorylated Thr285 (Figure 42). Thr phosphorylation is reported to be an intermediate in phosphoethanolamine transfer.356 In the dinuclear Zn2+ form, the active site is more constrained with ZnA remains coordinated by His466, D465, and E246, and ZnB coordinated by His478, His395, and the Glu300 from a second MCR-1 unit, with both Zn2+ ions coordinated in a tetrahedral geometry. Mutation of Glu300 to Ala found that the catalytic activity of MCR-1 remained unchanged,359 suggesting that the ZnB site is not required for catalysis, and lends credence to the argument that MCR-1 preferentially functions as a mononuclear metalloenzyme, with ZnA as the site of catalytic activity.
Figure 42.
Top: Structure of the mononuclear form of MCR-1 (PDB 5LRN). Bottom: Structure of the dinuclear form of MCR-1 (PDB 5LRM). Glu300 that completes the coordination of the second Zn2+ is highlighted in magenta.
Inhibitor Development.
To the best of our knowledge, there have been no efforts to develop inhibitors against MCR-1, with the closest example being the use of EDTA to demonstrate the Zn2+-dependence of MCR-1.356 The lack of inhibitors against MCR-1 can largely be attributed to the recent discovery of this metalloenzyme in 2015,353 and even more recent release of crystallographic data in the last few years.356–359 Furthermore, there also have not been any reports of assay development for the in vitro screening of potential inhibitors against MCR-1. Other phosphoethanolamine transferases, such as lipooligosaccharide phosphoethanolamine transferase A (LptA), bear 40% sequence identity to MCR-1.358 In vitro assays for LptA use a substrate comprised of a p-nitrophenol chromophore conjugated to phosphoethanolamine,360 and it is possible that this same substrate can be utilized to monitor MCR-1 activity for the purposes of inhibitor development.
Current and Future Prospects.
MCR-1 is a Zn2+-dependent metalloenzyme that confers antibiotic resistance to colistin. Since the recent discovery of MCR-1 in China in 2015, this resistance mechanism has spread worldwide and has the potential to lead to superbug type infections. While MCR-1 has been confirmed to be a metalloenzyme, debates remain over whether the active site contains one or two Zn2+ ions, although present evidence suggests a mononuclear site. As MCR-1 has only recently been discovered, there has been no progress in inhibitor development against this target. Further work to elucidate the structure and mechanism of MCR-1, as well as vitro assay development, are needed to help speed inhibitor discovery against this rising target of interest.
5. Phosphodiesterases (EC 3.1)
5.1. HIV-1 Integrase (HIV IN, EC 3.1.21)
Function in Biology and Disease.
Human immunodeficiency virus integrase (HIV IN) facilitates the importation and integration of viral cDNA into the human host-cell genome, which is the final step in HIV viral infection.361 After virus-cell fusion and entry of the viral core into the host cytoplasm, the double-stranded RNA viral genome is converted into double-stranded cDNA by the HIV reverse transcriptase. This viral cDNA is the substrate for HIV IN, which catalyzes the incorporation of the viral genome via two distinct reactions: 3’-processing (3’P)362 and strand transfer (ST).363 During 3’P, HIV IN binds to and selectively cleaves the last two nucleotides (GT) from both 3’-terminal ends of the cDNA viral genome to generate two CA-3’-hydroxyl ends (Figure 43). Following 3’P, the IN-cDNA complex is translocated to the cell nucleus. ST occurs when the activated IN-cDNA complex binds to a transcriptionally active region of the host genome in a site non-specific fashion.361 Once bound, the activated 3’-hydroxyl DNA ends are inserted into the host-genome via IN-catalyzed phosphodiester transesterification reactions. After integration, the remaining nicked ends are ligated by host-cell DNA-repair machinery.361
Figure 43.
Schematic representation of the 3′-processing and strand transfer reactions catalyzed by HIV IN. After 3′-processing the activated IN-cDNA (cyan ribbons) complex is translocated to the nucleus where the 3′-hydroxyl moieties (blue dots) insert into the host genome (red and blue ribbons) through the process known as strand transfer. The nicked reaction complex is subsequently ligated by the host cell in the last step.
Millions of new HIV infections and acquired immunodeficiency syndrome (AIDS) related-deaths occur annually, in spite of the recent advances in HIV treatment and prophylaxis. Highly-active antiretroviral therapy (HAART) has increased the quality of life and duration of life for many individuals who suffer from HIV infection, but problems of toxicity related patient non-adherence and multi-drug resistance continue to necessitate the development of new and better HIV antiretrovirals. To address these problems HIV IN was identified as an attractive drug target, as there is no mammalian HIV IN counterpart. Since the mid-1990s HIV IN has been highly studied361,364–366 and several HIV IN inhibitors have undergone clinical trials, with three HIV IN inhibitors currently approved for the treatment of AIDS.
Protein and Active Site Structure.
HIV IN proteins are highly conserved among retroviruses, and all retroviral HIV INs can be structurally divided into three canonical domains: an N-terminal Zn2+-binding domain, a catalytic core domain, and a C-terminal DNA-binding domain.367 HIV IN is a 32 kDa protein that in solution exists in a dynamic equilibrium between monomers, dimers, tetramers, and higher order oligomers. Monomers are reportedly inactive in vitro, whereas dimers are able to catalyze 3’P and ST (Figure 44).368 The N-terminal domain is comprised of residues 1–51 and contains a conserved, structural HHCC zinc-finger like domain.369 The catalytic core domain, which is comprised of residues 51–212, contains the metal-dependent endonuclease-transferase active site. The C-terminal domain is comprised of residues 213–288 and is involved in non-specific DNA binding during the integration ST process.361,366 The HIV IN active site contains three essential and highly-conserved acidic residues: Asp64, Asp116, and Glu152, which comprise the DDE catalytic motif. The DDE motif is the site of metal binding, and can bind one or two Mn2+ or Mg2+ cations. Both Mn2+ and Mg2+ are catalytically active, with two divalent metal cations required for catalysis, but Mg2+ is considered the biologically relevant metalloform.367 Removal or substitution of any member of the DDE domain results in greatly diminished catalytic activity.370
Figure 44.
Structure of PFV (PDB 2L3R), a structural homologue of HIV IN. Viral cDNA bound is shown as beige and purple tubes.
To date, no full-length crystal structure of HIV IN has been reported, although there are several reported crystal structures of truncated HIV IN domains, including the catalytic domain. While these truncated structures give some insight into ligand binding, their utility is limited as truncated constructs do not properly bind to the cDNA which makes up an important portion of the holoenzyme active site.366 It has become common in the field to use the more readily crystallized prototype foamy virus (PFV) as a stand-in for structural analysis of integrase inhibitors.366 The overall structure of PFV is similar to HIV IN, and the ligand interaction motifs observed between HIV IN inhibitors and PFV are expected to be similar to those that occur with HIV IN. Hence, co-crystal structures with PFV provide valuable information for the design of HIV IN inhibitors.371,372 As shown in Figure 44, MgA is coordinated by both carboxylic acid oxygens of Glu220, Asp128, two water molecules, and a bridging water/hydroxide in an overall octahedral geometry. MgB is also coordinated by a bridging water/hydroxide, as well as Asp128, Asp185, and three water molecules in an overall octahedral geometry.
Inhibitor Development.
HIV IN stands out among therapeutic metalloenzyme targets as one of the most successful class of FDA approved metalloenzyme inhibitors. Notably, HIV IN inhibitors also use a more diverse array of MBPs when compared to most other metalloenzyme drug discovery programs. Currently, three HIV IN inhibitors are approved for use in HIV infection (raltigravir,373 elvitegravir,374 dolutegravir375) and two other inhibitors are in late stage Phase III clinical trials (bictegravir,376 cabotegravir377). As seen in Figure 45, the chemical structures of these five inhibitors share many similarities, particularly for dolutegravir, bictegravir, and cabotegravir. The success of these inhibitors has led to the recent development of many ‘me-too’ inhibitors.374 While hundreds of HIV IN inhibitors have been reported in the past three decades, we will briefly review representative chemotypes of metal-binding HIV IN inhibitors and general trends focused on more recent reports. More comprehensive reviews on HIV IN inhibitors can be found elsewhere.364,366
Figure 45.
Inhibitors of HIV IN. Compounds shown boxed are FDA-approved inhibitors or in Phase III clinical trials.
The first promising lead inhibitors against HIV IN were compounds containing a β-diketo acid group as a MBP, adjacent to an aryl group that was found to stack favorably with the terminal nucleotide(s) of the IN-cDNA complex.378 Beginning from the fragment lead benzoylpyruvic acid (BPA, Figure 45), 5-CITEP was identified as a lead inhibitor of HIV IN, with an IC50 value of 2.2 μM.379 5-CITEP became the first inhibitor to be co-crystalized with a truncated catalytic domain of HIV IN. However, the active site of HIV IN in the co-crystal structure was only partially metallated and in this structure the inhibitor did not bind to the active site metal ions. This was likely an artifact of the crystallization conditions, as well as due to use of the truncated construct without accompanying cDNA (as evidenced by later structural data). Continued development of diketo acid HIV IN inhibitors resulted in the identification of L-731,988, which displayed an IC50 value 0.17 μM and an EC95 value of 9.6 μM. Further work on this scaffold resulted in several lead compounds with low IC50 values.380 The development of diketo acid HIV IN inhibitors has continued; however, absorption, permeability, and toxicity problems associated with the diketo acid functional group has limited the efficacy of this class of HIV IN inhibitor. Subsequent efforts turned to identifying isosteres of the diketo acid group with improved PK properties.
Distinct from the diketo acids are polyphenols and similar hydroxylated aromatic compounds comprise a large class of HIV IN inhibitors. Many of these compounds were identified through natural product HTS campaigns. In one such example (Figure 45), methanol extracts of Eucalyptus globoidea buds was found to have activity against HIV IN. Isolation of the bioactive compound identified Globoidnan A as a modest inhibitor of HIV IN with an IC50 value of 0.64 μM.381 The majority of this class of molecules share a common dicatechol motif, such as the hydroxycinnamic acid derivative (+)-D-chicoric acid (IC50 = 0.5 μM),382 and the flavonoid quercetin (IC50 = 11 μM).383 Although the exact mode of inhibition is not well understood, and it remains unclear whether the catechol moieties engage in metal coordination, SAR analysis on this class of compounds has shown that activity is directly related to the catechol moiety and that blocking or modulating this functional group severely affects inhibitory activity.384,385
8-Hydroxyquinoline is a well known ligand for divalent metal cations and this scaffold has been explored for HIV IN inhibitors. Early work with 8-hydroxyquinoline inhibitors identified derivatives of 8-hydroxyquiniline-7-carboxylic acid (8HQ7CA) as fragment-like inhibitors of HIV IN (Figure 45). While this scaffold appears structurally distinct from diketo acid-based inhibitors, the 8HQ7CA core is in fact a bioisostere of the diketo acid moiety that mimics the acid, enol, and keto groups in a rigid, coplanar structure. Much of the SAR identified in the diketo acid inhibitor work was directly applied to the 8-hydroxyquiniline scaffold with great success.
Merging 8HQ7CA with the catechol moiety common among natural product inhibitors of HIV IN identified several styrylquinoline inhibitors with good activity against IN, including HIV-INi-1 (IC50 = 0.03 μM, Figure 45).386,387 A similar 8HQ7CA scaffold reported by Shionogi, which introduced what would become the canonical 4-fluorobenzyl moiety, displayed good HIV IN inhibition (HIV-INi-2, IC50 = 0.2 μM) and overcame physiochemical liabilities of the catechol group and improved cell permeability over related compounds.365 Lead optimization of 8HQ7CA by Merck resulted in naptheridine-compounds such as L-870,810 (IC50 = 8–15 nM, Figure 45), which was the second HIV IN inhibitor to enter clinical trials.388 L-870,810 was found to coordinate to the active site metal centers in a bridging fashion, with the naphthyridine hydroxyl group bridging the two metal ions and the vicinal nitrogen and amide carbonyl moieties coordinating to either metal center. MgA is coordinated in a 6-membered chelate ring by the amid carbonyl and the naphthyridine hydroxyl group, whereas MgB is coordinated as a five-membered chelate ring by the bridging hydroxyl group and the pyridine-like nitrogen of the naphthyridine ring (Figure 46). Further structural optimization, resulted in compound L-870,812 (IC50 = 40 nM). While this lead exhibited decreased activity against HIV IN, L-870,812 displayed greatly improved PK properties in humans.389 While clinical trials of L-870,812 were ultimately terminated due to long-term toxicity related issues in higher mammals, the SAR gained from this campaign, particularly with regards to the oxadiazole-carboxamide moiety, was directly applied in the development of raltegravir, which was the first HIV IN inhibitor to gain FDA approval.
Figure 46.
Structure of Raltegravir (PDB 3OYA), Dolutegravir (PDB 3S3N), Elvitegravir (PDB 3L2U), and L-870,810 (PDB 3OYF) bound to human PFV as a surrogate HIV IN. Bound nucleic acids are shown as faded beige and purple tubes.
Other work on the 8-hydroxyquinoline scaffolds focused on removal of the 7-carboxylic acid moiety present in early inhibitors, as this moiety can be a liability for in vivo activity, particularly the associated low cellular permeability. While some groups found that conversion of the carboxylic acid to an amide was generally tolerated, other groups sought to replace this group with isosteres. Based on the keto group present in diketo acid scaffolds, aryl ketone-based 8-hydroxynaptheridine inhibitors displayed very good activity against HIV IN. One compound, HIV-INi-3 (Figure 45), displayed an IC50 value of 0.01 μM in protein based assays, and excellent antiviral activity (EC95 = 0.39 μM) with minimal cytotoxicity. This compound also identified the cyclic sulfonamide as a key moiety.390 Other work on isostere replacement on the 8HQ7CA core found that the carboxylic acid moiety can be successfully replaced by certain oxadiazole and triazole rings. HIV-INi-4, which replaced the 8HQ7CA carboxylate with a 4-fluorobenzyl-functionalized oxadiazole ring, showed both good inhibition values (IC50 = 0.042 μM, EC50 = 0.13 μM) and improved permeability relative to the parent compound.391 Tricyclic, N-functionalized hydroxyphthalimide-based mimics of 8HQ7CA were also explored. Structural optimization of every ring position of the two aromatic rings identified HIV-INi-5 as an optimized inhibitor, which displayed submicromolar activity in both protein and tissue culture-based assays (IC50 = 0.11 μM, EC50 = 0.85 μM).392
Quinolone carboxylic acid MBPs have also been successfully implemented in the development of HIV IN inhibitors. The most successful of this group is elvitegravir (IC50 = 7.2 nM, EC50 = 0.9 nM, Figure 45), which employs a modified hydroxypyrimidinone carboxylate MBP.393 Elvitegravir employs a metal binding carboxylate group that coordinates to one metal center as a bidentate ligand, which is distinct from the more typical coordination motif seen in other classes of inhibitors (Figure 46). Elvitegravir binds to MgA as a 6-membered chelate ring through the pyridinone carbonyl and one carboxylate oxygen, with the same carboxylate oxygen bridging to MgB. MgB is coordinated in a bidentate fashion by the carboxylate in a 4-membered ring, which while not unusual for carboxylate metal ligands in general, but is unusual among other integrase inhibitors. Related work identified compound HIV-INi-6, which displayed good inhibition in protein based assays and an EC50 value of 0.1 μM in human lymphoblastoid cell lines.394
Hydroxypyrimidinones and hydroxypyridinones comprise the most successful class of HIV IN inhibitors, with two FDA approved drugs and several Phase III candidates all bearing analogous MBPs. During the development of raltegravir, the majority of optimization efforts focused on improving inhibitor interactions with the protein backbone, as opposed to the metal center or bound cDNA. Extensive work ultimately revealed cyclic sulfonamides and oxadiazole-carboxamide moieties to have ideal inhibition values as well as acceptable PK properties; however, a plethora of similar, highly-active inhibitors with poorer PK properties have also been disclosed.395–397 Following the success of raltegravir, synthetic efforts shifted away from the N-methylpyrimidinone scaffold employed in raltegravir towards the development of bicyclic pyrimidinones and eventually pyridinones (e.g., elvitegravir and dolutegravir; Figure 45). The inclusion of a second, -5, -6, or 7-membered saturated ring to the pyrimidinone structure was exhaustively explored. This effort revealed bicyclic pyrimidinone inhibitors with moderated activity.398 Building on this report, a series of aromatic, substituted biaryl-pyrimidinones were explored which displayed good activity (IC50 = 1–10 nM) and efficacy in cell studies.399 In an attempt to understand and optimize metal binding interactions, various azole rings were employed as amide isosteres, as seen in compound HIV-INi-7 (Figure 45). Ultimately, thiazole and oxazole rings showed improved activity as compared to the parent amide (IC50 = 20 nM vs. 59 nM, respectively) while maintaining excellent activity in cell-based assays.400 The continuation of these efforts resulted in the development of dolutegravir as an FDA-approved, second generation HIV IN inhibitor. The structure of dolutegravir bound to PFV have been solved and shows dolutegravir binds to the active site metal ions of PFV in a bridging fashion (Figure 46). MgA is coordinated in a five-membered chelate ring through the pyridinone carbonyl and the bridging pyridinone phenol, resulting in a distorted octahedral overall coordination geometry. MgB is coordinated by the bridging phenolic oxygen atom as well as the proximal cyclic amide carbonyl oxygen in a six-membered chelate ring. This results in an overall octahedral coordination geometry for MgB. Dolutegravir requires only single daily dosing, as do the structurally related candidates bictegravir and cabotegravir, which are currently in late-stage clinical trials as extended release formulations and for use against raltegravir resistant mutants.
Emerging from the development of many, diverse HIV IN inhibitors, a singular, canonical pharmacophore has emerged that is shared by most active inhibitors. The pharmacophore consists of inhibitors with three key components that make key interactions with: the metal centers, the nucleic acids, and the protein active site. One study attempted to isolate and probe the effect of changes to the MBP on inhibition. A series of structurally analogous inhibitors were designed that only varied in the MBP.401 Termed raltegravir-chelator derivatives (RCDs), 21 compounds were prepared all with identical backbones, but different MBPs. From these 21 compounds, two (RCD-1, RCD-5; Figure 45) were found to inhibit HIV IN with IC50 <1 μM. More importantly, the series of RCD inhibitors displayed a wide range of inhibitory activities (IC50 = 0.5–300 μM) that could be interpreted based on subtle changes solely at the MBP.
Current and Future Prospects.
While there exists no published co-crystal structure of HIV IN, the use of PFV integrase as a stand-in has given insight to the binding of HIV IN inhibitors that is consistent with the wealth of established inhibitor SAR and the representative pharmacophore model. The majority of the new development in the field have focused on ‘me-too’ inhibitors and 3rd-generation inhibitors designed to overcome specific-mutation resistance.366 This is due, in part, to the wealth of established literature and the tremendous success of second and third generations HIV IN inhibitors, such as dolutegravir and elvitegravir.
While the field of HIV IN inhibitor development is now quite mature and will likely not see continued development to the extent it has previously, work on this target has greatly enriched the field of metalloenzyme inhibitor development as a whole. The variety and diversity of inhibitors and MBP chemotypes is beyond that generally explored for other metalloenzyme targets. Indeed, the success seen in this field is likely due in large part due to the thorough exploration of different MBPs, which should be more widely applied to the other metalloenzyme targets discussed in this review.
5.2. HIV-1 Reverse Transcriptase-associated Ribonuclease H (HIV RNaseH, EC 3.1.26.13)
Function in Biology and Disease.
Reverse transcription is an essential process in the human immunodeficiency virus (HIV-1) lifecycle. Upon viral infection, the HIV-1 reverse transcriptase (HIV RT) converts the single-stranded viral RNA genome to double-stranded cDNA, which is then translocated to the host-cell nucleus and subsequently incorporated into the host genome by the HIV IN (Section 5.1).403 HIV RT catalyzes two distinct reactions: RNA- and DNA-dependent DNA polymerization of viral cDNA, and RNase H activity which selectively degrades the RNA strand of RNA:DNA hybrid reverse transcription intermediates.404 Both of these reactions are critical for viral replication.
Reverse transcription of the viral genome begins from the positive-sense RNA strand and is initiated by a host-cell tRNALys3 (the tRNA corresponding to the lysine-3 codon) hybridizing to the primer binding site (PBS) near the 5’-end of the HIV RNA genome. The tRNA serves as a primer for reverse transcription to the 5’-end of the RNA strand, followed by subsequent degradation of the RNA portion of the generated RNA:DNA hybrid intermediate by the RT RNase H domain. This negative-sense, short single-stranded DNA (-sssDNA) is then transferred by the RT to the 3’-end of either template RNA strand of the original viral RNA genome. After strand transfer, the copied -sssDNA is elongated to generate a complete negative-sense cDNA chain. During reverse transcription, the RNase H domain simultaneously degrades the original positive-sense RNA template, leaving only two purine-rich regions, known as polypurine tracks (PPTs), which serve as primers for positive-sense cDNA synthesis. After elongation of these PPT primers, a second strand transfer event occurs in which the complementary PBS domain of the nascent positive-sense cDNA is translocated to the 3’-PBS domain of the negative-sense cDNA chain. Bidirectional elongation of this complex continues, coupled with intermediate degradation of the remaining RNA PPT primer by the RNase H domain, until the proviral cDNA genome is complete. The reader is referred to existing references for a complete description of RNase H mechanism and function.405–407
HIV derived AIDS remains a global health concern, with millions of new HIV infections and AIDS related deaths occurring annually. Currently, HAART remains the only effective treatment for AIDS; however, emerging resistance to even the newest drugs demands the continuing development of new HIV antiretrovirals. The pivotal role of RT in the viral lifecycle has made this enzyme a key target in HAART therapies, and several FDA approved inhibitors of RT are currently employed in AIDS therapy. Although HIV RT is a multifunctional enzyme with two distinct active sites, all currently approved inhibitors only target RT polymerase activity. Hence, the RNase H domain of HIV RT remains a promising target for novel RT inhibitors.
Protein Active Site and Structure.
The HIV RT functions as a heterodimer comprised of the p66 and p51 subunits. Both subunits are expressed from the same viral pol gene, and p51 is derived directly from p66;408,409 proteolytic cleavage by the HIV protease of a single p66 protein produces the p51 and p15 polypeptides. In spite of their perfect sequence homology, p66 and p51 adopt significantly different secondary structure when in the active dimer complex. The p66 subunit contains the active site for both polymerase and RNase H function, while p51 plays a more structural role with the analogous ‘active site’ residues buried within the subunit.
The p66 subunit is structurally similar to other polymerases, and contains the three canonical polymerase subdomains:410 a ‘finger’ domain (residues 1–85, 118–155), a ‘palm’ domain (residues 86–117, 156–237), and a ‘thumb’ domain (residues 238–318). The connection domain (residues 319–426) and the RNase H domain (residues 427–560) form the remainder of the subunit.410,411 X-ray crystal structures of the HIV RT (Figure 47) have shown the RNase H domain to be comprised of a central five-stranded mixed β-sheet flanked by four asymmetric α-helices. The polymerase and RNase H domains are located ~18 base-pairs from each other and are catalytically interdependent.403
Figure 47.
Structure of HIV RT p66 (gray) and p51 (cyan) heterodimer bound to DNA (shown as beige and purple tubes, PDB 3KJV).
RT RNase H has been shown to degrade RNA both during DNA polymerization and in the absence of DNA synthesis.412 Based on how the RT interacts with RNA:DNA hybrids throughout viral cDNA synthesis, three distinct phosphodiester cleavage modes have been identified: DNA 3’-directed, internal/endonucleic, and RNA 5’-directed cleavage.413 DNA 3’-directed cleavage occurs when a DNA 3’-end of a RNA:DNA hybrid is recognized by the RT, and the RNase H activity occurs ~15–20 bp away from the 3’-end. This type of cleavage generally occurs in concert with DNA reverse transcription, but can also be polymerization-independent. RNA 5’-directed cleavage occurs when RNA 5’-end of a RNA:DNA hybrid is recognized by the RT, and the RNase H activity occurs in a window of ~13–19 bp downstream of the 5’-end. This form of RNase H activity is completely independent of polymerization. Internal or endonucleic cleavage is independent of polymerization activity and is not positioned by a DNA 3’- or an RNA 5’-end. Rather, this cleavage activity is typical of other RNase Hs where RNA sequence is the major directing factor for cleavage site.413,414
The catalytic activity of nucleotide cleavage in the RT RNase H generally follows an SN2 mechanism in which an activated hydroxide anion attacks the scissile phosphate group in the RNA backbone. To carry out this reaction the RT RNase H requires either Mn2+ or Mg2+ cations to activate a nucleophilic hydroxide anion and to stabilize the reaction intermediates, which classifies the enzyme as a metal-dependent transposase.415 Although crystal structures have shown both one and two metal cations present in the active site,416 several studies have demonstrated that this RNase H achieves catalysis via a two-metal ion-dependent mechanism.417–420 This is in contrast with other RNase Hs such as E. coli RNase H which requires only a single divalent metal cation.421 The active site of the RNase H domain is characterized by the highly conserved DEDD metal coordinating motif, comprised of Asp549, Glu478, Asp443, and Asp498. MnA (or MgA) is coordinated by Asp498, Glu478, and Asp443, while MnB (or MgB) is coordinated by Asp443 and Asp549, with water molecules occupying the remaining coordination sites with both metal centers in a distorted octahedral geometry (Figure 47). In the native structure, a single hydroxide anion bridges the two metal centers. The conserved DEDD fold is essential for catalytic function, and mutation of any of the coordinating residues completely abolishes activity.422–424
Inhibitor Development.
Natural products were among the first class of molecules found to inhibit RNase H function in protein based assays.425 Anionic compounds such as heparin, xylan polysulfate, dextran sulfate, and other similar microbial extracts were found to inhibit RNase H activity with IC50 values ranging from 0.1–8 nM.425 However, these compounds were shown to interfere with virus entry and not with RNase H activity when assayed against live virus.403 Small molecule natural products were also found to be active inhibitors of HIV RNase H. 2-Hydroxyquinone metabolites from sea sponges (RNaseHi-1, -2, and -3, Figure 48) were found to have IC50 values of 1–50 μM against RNase H, which was shown to be dependent on metal coordination and not secondary redox cycling activity (which can occur with quinones in biochemical assays). Synthetic derivatives of RNaseHi-1 in which the hydroxyl group ortho to the carbonyl group was modified such that it could no longer chelate a metal cation showed complete loss of efficacy against RNase H in enzyme based assays.426 Many other polyhydroxylated natural products have been identified as modest inhibitors of HIV RNase H, usually with IC50 values between 1–50 μM. These have been reviewed in depth,403 and while interesting compounds, they did not produce notable activity against live virus.
Figure 48.
Inhibitors of HIV RNase H.
One interesting class of natural product-based inhibitors of HIV RNase H are hydroxytropolones. HTS identified β-thujaplicinol and manicol as potent and selective inhibitors of RT RNase H activity (Figure 48). These compounds were show to inhibit RNase H with IC50 values of 0.2 μM and 1.5 μM, respectively, while failing to inhibit polymerase activity.427 These compounds have also been shown to be effective at inhibiting other dinuclear viral metalloenzymes, such as HIV IN (Section 5.1).428 This is likely related to the ability of this pharmacophore to simultaneously coordinate to both active-site metal cations (Figure 49).429 As shown in Figure 49, β-thujaplicinol coordinates to the active-site metal centers in a symmetric fashion, with the central carbonyl oxygen acting as a bridging atom, with both hydroxyl groups coordinating to their adjacent metal center as a five-membered chelate ring. The charge and shape of this class of inhibitors is thought to mimic transition state intermediates, and is likely the underlying cause of their high activity.429
Figure 49.
Structure of β-Thujaplicinol (PDB 3K2P), RNaseHi-5 (PDB 3HYF), RNaseHi-7 (PDB 5UV5), RNaseHi-15 (PDB 3LP0), and RNaseHi-20 (PDB 4QAG) bound to RNase H.
Nucleotide-analogue polymerase inhibitors are among currently approved treatments for AIDS, and several have been approved as RT polymerase inhibitors. Some of these have been found to also inhibit the RNase H function of RT. In particular, AZT-monophosphate was shown to inhibit RNase H more effectively than other nucleotide monophosphates. Interestingly, the inhibitory activity of AZT was dependent on the metalloform of RNase H employed in the assay (i.e., Mn2+ or Mg2+). The IC50 for the Mn2+ metalloform was found to be 5,000 μM while the IC50 for the Mg2+ metalloform was found to be 50 μM.430 Related nucleotides, 2’,3’-dideoxyadenosine 5’-monophosphate (ddAMP) and 2’,3’-dideoxyguanosine 5’-monophosphate (ddGMP), were also found to inhibit RNase H activity, but only against the Mn2+ metalloform (IC50 = 220 μM and 450 μM, respectively).431
In the late 1990s, hydrazone derivatives were identified as dual inhibitors of both RT polymerase and RNase H activity (Figure 48). RNAseHi-4 (BBNH) was reported to inhibit both functions of RT, with an IC50 value of 3.5 μM.432 Interestingly this compound was shown to have an EC50 value of 1.5 μM against live virus in cord-blood mononuclear cells. The good antiviral activity of this compound was proposed to be due, in part, to the ability of this compound to inhibit both functions of RT. Further studies with this and structurally related compounds suggested that these compounds bound in various pockets of the RT but that RNase H inhibition was likely due in part to active site inhibition through metal binding.433
Diketo acid inhibitors were first identified as inhibitors of HIV IN (Figure 48).434 Given the structural similarities of the HIV IN and RNase H active sites, many diketo acid inhibitors have been repurposed for the development of RNase H inhibitors.435 The diketo acid based inhibitor BTDBA was reported to selectively inhibit RNase H function in RT while sparing polymerization activity, with an IC50 value of 3.2 μM. ITC experiments were employed to show that BTDBA binding to RT is independent of nucleic acid but strictly dependent on the presence of Mn2+.436 While BTDBA inhibited RT RNase H in protein based assays, it failed to inhibit viral replication in cell culture. This is largely due to the poor PK and permeability profile of diketo acids; the inhibitor RDS 1643, which employs an ethyl ester pro-moiety of the diketo acid, was found to inhibit RNase H activity with an IC50 value of 13 μM and was effective against various strains of HIV-1, with an EC50 value of 7.3–18.9 μM.
Based on the three-oxygen metal coordinating diketo acid pharmacophore identified in RNase H inhibitors and inhibitors of similar dinuclear DNA processing enzymes (e.g., HIV IN), hydroxypyrimidinone carboxylic acid inhibitors of RNase H were explored (Figure 48).437 This MBP is the same employed in the FDA approved HIV IN inhibitor raltegravir (Figure 45). In contrast to HIV IN, where amides are observed to maintain equivalent activity as analogous carboxylic acids, the free carboxylic acid of RNaseHi-5 and similar compounds were found to be far more active against RNase H than ester or amide derivatives. These compounds were found to coordinate to both Mn2+ ions as a multidentate bridged complex, with the hydroxypyrimidinone carbonyl oxygen acting as a bridging atom (Figure 49), and to be very selective for HIV RNase H over human RNase H.437 Compound RNaseHi-5 was found to inhibit RNase H with an IC50 value of 1.2 μM, but was inactive against the virus in whole cell assays, likely due to poor permeability originating from the carboxylate group. The structurally related hydroxypyridinone carboxylic acid MBP was identified for HIV and is employed in inhibitors such as dolutegravir (Figure 45). Using the hydroxypyridinone scaffold as a starting point, a study identified RNaseHi-6 as an inhibitor of RNase H with an IC50 value of 2.7 μM and an EC50 value of 10 μM.438
N-Hydroximides, which were originally identified as inhibitors of the influenza PAN endonuclease (Section 5.3), were also found to be active against HIV RNase H. Compound RNaseHi-7 was found to inhibit RNase H activity with an IC50 value of 0.65 μM, and several similar fragment molecules employing the same MBP were also found to inhibit RNase H activity with IC50 values of 0.4–1 μM.439–441 SAR around the N-hydroximide MBP (Figure 48) indicated that the N-hydroxy group of RNaseHi-8 is essential for activity. Structural studies confirmed the N-hydroxy group acts as the bridging ligand between the two Mn2+ or Mg2+ metal ions with each proximal carbonyl group coordinating one metal ion (Figure 49). This coordination motif is similar to the coordination motif observed with hydroxytropolone inhibitors; however, the N-hydroximide MBP itself is far less active than the hydroxytropolone MBP. The differences in pKa and charge of the overall complex potentially explain this disparity despite the very similar coordination geometry. Compounds RNaseHi-9 and RNaseHi-10 were completely inactive against the RNase H as they were unable to bind the metal centers in a similar manner. Interestingly, further SAR of the MBP indicated that the enol-tautomer was the active species and that blocking this tautomer (e.g., RNaseHi-11, RNaseHi-12) rendered the inhibitor inactive. Further synthetic development of this class of compounds resulted in RNaseHi-13 which selectively inhibits RNase H activity (IC50 = 0.061 μM) and can inhibit HIV replication in cell studies.421 Related work employing an N-hydroxypyrimidione MBP produced RNaseHi-14 that showed good RNase H inhibitory activity (IC50 = 0.15 μM), but this compound did not show efficacy against HIV replication in cells.442,443
In 2010, N-hydroxy napthyridinones were identified as a class of metal coordinating inhibitors of RNase H (Figure 48). The compound RNaseHi-14 was identified as a lead, with an IC50 value of 0.11 μM. The structure of RNaseHi-15 with RNase H showed the inhibitor bound to both Mn2+ ions with the N-hydroxy group reported as a bridging ligand; however, closer inspection of deposited structures seems to indicate that each Mn2+ is only coordinated by one donor atom.444 As shown in Figure 49, the pyridine-like nitrogen of RNaseHi-14 coordinates to MnA in a four-coordinate geometry (coordinating water molecules were not reported in the X-ray structure). The N-hydroxyl group was found to be too far removed from MnA to engage in coordination bonding (bond length >2.9 Å). MnB is coordinated by the N-hydroxy group, with all other ligand donor atoms too far removed from the metal center to engage in coordinate bonds.444 Further SAR identified RNaseHi-15 which displayed an IC50 value of 45 nM. In separate work GSK5750, which shares the same MBP, was found to inhibit RNase H with an IC50 value of 0.33 μM.409 Other structurally related pyridopyrimidione inhibitors (RNaseHi-17), which were originally identified as HIV IN inhibitors, were identified in a cross-screen of HIV IN inhibitors and was found to have a modest IC50 value of 10 μM against RNase H.445 Deconstructive SAR showed that the MBP core alone had a similar IC50 value (RNaseHi-18, IC50 = 11 μM). Merging this MBP core with SAR generated from N-hydroxy napthyridinone inhibitors produced RNaseHi-19, which exhibits good RNase H inhibitory activity (IC50 = 0.05 μM) and was active in antiviral assays (EC50 = 0.25 μM).
Finally, in other recent reports, HTS screening identified a dihydroxycoumarin fragment as a novel RNase H inhibitor chemotype,446 culminating in compound RNaseHi-21 (Figure 48), which showed good RNase H inhibition (IC50 = 0.2 μM). Structural studies of the MBP fragment from RNaseHi-21 revealed coordination to MnB via the catechol moiety as a bidentate bridged complex, with the pyridinone endocyclic oxygen potentially making electrostatic interactions with MnA (RNaseHi-20, Figure 49).
Current and Future Prospects.
HIV-1 RNase H remains an attractive target for future drug development, as the increasing emergence of multi-drug resistance and toxicity related treatment non-compliance in patients necessitate the development of novel and more well-tolerated therapies against HIV infections. Many chemotypes have been identified in the past decade which have been shown to inhibit the RNase H biochemically (i.e., in enzyme based assays), but few of these classes of compounds have shown any significant activity against live virus in cell-based assays. HTS and FBDD have both proven effective tool in identifying novel inhibitor chemotypes, which when coupled with recent advances in X-ray crystallography for this target have made lead design more accessible and have likely instigated the recent diversification of reported inhibitor chemotypes. A co-targeting approach against both RNase H and HIV IN has also immerged as many inhibitors show efficacy against both targets due to the similarity in metal active sites. Further study and repurposing of HIV IN inhibitors to target RNase H may lead to effective inhibitors.
There is a need for a more systematic approach to the development of RNase H inhibitors. Particularly, more early-stage analysis of PK properties in lead design will be necessary to overcome the lack of cellular efficacy present in the literature. This is particularly true for inhibitors that employ very polar MBPs. In fact, despite of the large number of identified MBP scaffolds that show good inhibition against RNase H, there have been very few studies identifying MBPs that have been optimized for more ideal PK properties or to overcome poor cell efficacy and permeance. Nevertheless, prospects for this field of study remain good for the development of new AIDS therapy targeting RNase H.
5.3. Influenza RNA-dependent RNA polymerase PA N-terminal endonuclease domain (PAN Endonuclease, EC 3.1.21)
Function in Biology and Disease.
The influenza virus is a respiratory pathogen that is responsible for annual seasonal illness. These epidemics cause substantial morbidity and mortality, particularly for the elderly and other high-risk individuals, and are responsible for a significant financial burden worldwide. While influenza infections themselves are generally not life-threatening, secondary opportunistic infection can easily occur in infected individuals, and these infections are the root cause of most influenza-related deaths. Vaccinations can provide significant protection against the influenza virus, but must be annually re-administered and often lack efficacy due to rapid viral mutation or emergence of pandemic strians.448,449 Rapid mutation and antigenic variation also contribute to the advent of resistance to currently available antivirals.450–452 Several small molecule influenza antivirals have been approved by the FDA, namely, M2 ion-channel blockers and neuraminidase inhibitors. In addition, two influenza RNA polymerase inhibitors, favipiravir and baloxavir marboxi, have recently been approved for use in Japan.453,454 However, some of these drugs are limited by viral resistance and a time-dependent effectiveness (i.e., duration of time between infection and being treated with a therapeutic).453,455–458 Considering this, there is still a need for the development of new drugs to combat influenza infection.
The influenza virus belongs to the orthomyxoviridae family that all contain a segmented single-stranded negative sense RNA genome.459 Within the viral capsid, each RNA segment binds to a single heterotrimeric RNA-dependent RNA polymerase complex, which is comprised of three district subunits (PA, PB1, PB2).460,461 The polymerase complex is responsible for viral genome replication, but cannot generate the 5′-mRNA cap necessary for eukaryotic translation.462,463 In order to transcribe viral RNA (vRNA) into mRNA recognizable by the host cell, the polymerase complex sequesters host mRNA which contains a 5′-mRNA cap and then cleaves the capped mRNA 8–14 nucleotides downstream. This capped mRNA fragment then serves as a primer for viral transcription, and this mechanism is commonly referred to as ‘cap-snatching’ (Figure 50).463–465 The influenza virus RNA-dependent RNA polymerase, particularly the metal-dependent PAN endonuclease domain of this enzyme, is an attractive target for the development of new influenza antivirals, as the PAN endonuclease domain is both indispensable for the viral lifecycle and highly conserved among various circulating and historic influenza strains.
Figure 50.
Cap-snatching mechanism employed by the influenza RNA polymerase. The PB1 subunit (green) binds the viral RNA genome (green ribbon), while the PB2 subunit (blue) binds to the 5′-cap of host mRNA (blue ribbon). The host mRNA is cleaved by the PAN endonuclease subunit (gray) to generate a primer for vRNA translation.
Protein Active Site and Structure.
The PA subunit contains two primary sub-domains, with the N-terminal domain (first 196 residues) containing the PAN endonuclease active site and the C-terminal domain playing a structural role. Upon 5′-cap binding of a host mRNA to the PB2 subunit, the nucleotide chain is cleaved 8–14 nucleotides downstream of the cap by the viral PAN endonuclease domain.463–465 The sequestered, capped RNA segment then serves as a primer for viral RNA synthesis by the PB1 domain of the polymerase complex. Structural studies have shown the PAN endonuclease contains a dinuclear metal active site which binds to two Mg2+ or Mn2+ cations (Figure 51).463 The metal cations are bound in a pocket and coordinated by residues His41, Glu80, Asp108, Glu119, and Ile120.463,466 MnA coordinates to His41, the backbone carbonyl of Ile120, and the carboxylates of Asp108 and Glu119, as well as two water molecules in an octahedral geometry. MnB is likewise coordinated by the carboxylate of Asp108 as well as the carboxylate of Glu80 with four bound water molecules in an octahedral geometry (Figure 51). A bound water molecule or hydroxide anion is the catalytic nucleophile employed in endonucleic hydrolysis of the phosphodiester backbone of RNA or DNA, and both metal centers stabilize the transition-state intermediate via coordination. Both metal ions are essential for catalysis and most reported inhibitors PAN endonuclease function by coordinating the metal ions.467–471
Figure 51.
Left: Structure of the influenza A RNA-dependent RNA polymerase heterotrimer complex. The PA subunit is colored gray, the PB1 and PB2 subunits are colored cyan (PDB 4WSB). Right: Structure of the active site of the PA subunit (PDB 5DES).
Inhibitor Development.
There are currently no FDA approved PAN endonuclease inhibitors; however, in the past ~10 years, several classes of small molecules that inhibit PAN endonuclease activity have been reported, with one inhibitor, baloxavir marboxil, currently in Phase III clinical trials (NCT02954354, NCT02949011). One common structural characteristic of nearly all reported PAN endonuclease inhibitors is a two oxygen atom metal coordination motif. Mn2+ is a hard Lewis acid and is likely to make more favorable coordination interactions with harder Lewis base oxygen donor atoms as opposed to softer Lewis bases such as nitrogen or sulfur. This trait is shared with similar viral Mn2+- and Mg2+-dependent metalloenzymes such as HIV IN (Section 5.1) and HIV RNaseH (Section 5.2). Beyond the similarity shared by the MBPs of reported PAN endonuclease inhibitors, a wide variety of chemical space has been explored, with many inhibitors well characterized by both biochemical and crystallographic studies.
As with many metalloenzyme targets, natural product polyphenols and catechols were among the first reported inhibitors of PAN endonuclease. Among these natural product inhibitors, epigallocatechin gallate (EGCG) has been extensively studied (Figure 52). EGCG is a major polyphenol component of green tea that has been found to inhibit PAN endonuclease activity with IC50 values between 1–10 μM and EC50 values between 20–50 μM (Table 7).472,473 Deconstructive SAR indicated that the galloyl group is crucial for inhibitory activity; derivatives that blocked bidentate coordination by the phenolic moieties lost substantial activity. This provided evidence that the galloyl group was responsible for metal binding, which was validated by subsequent X-ray structures (Figure 53).467 Through a combination of pharmacophore docking, virtual screening, and HTS, several other catechol-based inhibitors of PAN endonuclease have been identified. Thalidomide, a hypnotic/sedative drug, was also identified as a weak inhibitor of PAN endonuclease.474 Addition of phenylethylcatechol groups to thalidomide to produce compounds such as PANi-1 (Figure 52) resulted in a large increase in activity, likely through the addition of the catechol MBP. These inhibitors were assayed directly against H1N1 virus in cell-saving assays and PANi-1 displayed an EC50 value of 24 μM.474 Several N-acylhydrazone inhibitors containing catechol or galloyl MBPs (PANi-2, PANi-3) have also displayed good activity against PAN endonuclease. Interestingly, these molecules contained two potential MBPs (catechol/galloyl and acylhydrazone moieties); however, structural studies have since confirmed that the catechol group is the preferred MBP. Compound PANi-2 and its catechol analogue PANi-3 were found to be active inhibitors of PAN endonuclease with IC50 values of 14 μM and 0.7 μM, respectively. While PANi-3 was more active, it was also highly cytotoxic. However, PANi-2 was found to have minimal cytotoxicity (CC50 >100 μM) and displayed an EC50 value of 18 μM.475 Subsequent work on this scaffold has recently resulted in compound PANi-4 with an IC50 value of 8.7 μM and improved cellular efficacy with an EC90 value of 3.5 μM in MDCK cells.476
Figure 52.
Inhibitors of PAN endonuclease.
Table 7.
Activity (IC50 values) of PAN endonuclease inhibitors.
| Compound | Inhibition (μM) | Ref. |
|---|---|---|
| PANi-1 | 26a | 474 |
| PANi-2 | 14 | 475 |
| PANi-3 | 0.7 | 475 |
| PANi-4 | 8.7 | 476 |
| PANi-5 | ~1 | 479,480 |
| PANi-6 | 12.7 | 485 |
| PANi-7 | 15 | 434 |
| PANi-8 | 0.5 | 481 |
| PANi-9 | 0.15 | 470 |
| PANi-10 | >50 | 468 |
| PANi-11 | 0.011 | 482 |
| PANi-12 | 0.940 | 483 |
| PANi-13 | 0.036 | 483 |
| PANi-14 | 0.014 | 483 |
| Baloxavir Marboxil | 0.0005–0.0065a | 486 |
| BPA | 21.3 | 473,477 |
| EGCG | 22–28a | 472 |
| Flutamide | 5.1 | 479,480 |
| L-731,988 | 0.8 | 477 |
| L-742,001 | 0.43 | 478 |
| RO-7 | 0.005–0.015a | 484 |
| Thalidomide | >100a | 474 |
EC50 value.
Figure 53.
Structure of EGCG (PDB 4AWM), L-742,001 (PDB 5CGV), PANi-10 (PDB 4E5I), and PANi-11 (PDB 4M5U) bound to PAN.
Diketo acids have been previously identified as potent inhibitors of dinuclear metalloenzymes, such as HIV-1 integrase (Section 5.1).477,478 Based on these reports, both benzoylpyruvic acid (BPA) and compound L-731,988 (Figure 52), which were initially identified as HIV IN inhibitors, were found to be inhibitors of PAN endonuclease.477,478 Further development of this scaffold ultimately produced L-742,001 as one of the most active inhibitors of PAN endonuclease (IC50 = 0.43 μM, EC50 = 0.35 μM). Interestingly, inhibitory activity was not appreciably altered when the N-benzyl moiety of L-742,001 was replaced with a cyclohelxylmethyl, chlorobenzyl, or benzenesylfonamide moiety.479 Crystal structures of this class of inhibitor bound to PAN endonuclease reveals that the diketo acid moiety coordinates to both active site metal centers simultaneously, and that aryl and alkyl moieties interact favorably with Ala20 and Tyr24 (Figure 53). Unfortunately, these inhibitors suffer from a poor PK and toxicity profiles, which has limited their clinical development.
Flutamide (Figure 52) is a fungal metabolite originally isolated from Delitschia confertaspora, that was found to have moderate activity against influenza virus. Biochemical analysis indicated that flutamide inhibited the RNA polymerase PAN endonuclease domain (Table 7), with structural studies confirming a metal binding mode of inhibition.479,480 Modifications to the N-hydroxyl group or the olefin resulted in dramatic decreases in activity, further illustrating the metal coordination mechanism of inhibition. Structural elaboration in which one isopropyl moiety is replaced by p-substituted benzyl groups resulted in compound PANi-5 that displayed an IC50 value of ~1 μM. However, PANi-5 and related compounds were found to be cytotoxic. Scaffold-hoping produced compounds PANi-6 and PANi-7 (IC50 = 12.7 μM and 15 μM, respectively) that employ a similar MBP, but show markedly lower cytotoxicity.
Pyridinone-based inhibitors of influenza PAN endonuclease have become a very successful class of inhibitors, with the only inhibitor to enter clinical trials being a member of this class. The first reports of hydroxypyridinone inhibitors of PAN endonuclease were largely scaffold hopping exercises employing different core MBPs. These molecules exhibited analogous metal binding and were identified through a variety of methods, but all capitalized on very similar SAR trends to generate highly active inhibitors. A series of 2,3-hydroxyquinolone inhibitors were identified based on the high activity of the parent MBP (IC50 = 24 μM), with 6- or 7-position p-fluorobenzyl substituted derivatives (PANi-8) being the most active derivatives with an IC50 value of 0.5 μM.481 Structurally related 5,6-hydroxypyrimidinone inhibitors were found to likewise be active against PAN endonuclease, such as PANi-9 (IC50 = 0.15 μM). PANi-9 employs a phenyl tetrazole moiety (Figure 52) that makes favorable interactions with basic residues near the active site pocket.470 The tetrazole chemical functionality has since become a characteristic motif shared by many active inhibitors of this class. Structurally related 4-carboxy-5,6-hydroxypyrimidinones (PANi-10, IC50 = 5 μM) have also been explored. However, these compounds were found to have similar or reduced activity against PAN endonuclease as compared to non-carboxylate hydroxypyrimidinone inhibitors (e.g., PANi-9).468
Crystallographic fragment screening, in which protein crystals are soaked with high concentrations of fragment molecules as a means of inhibitor discovery, was employed to identify a 5-bromo-2,3-hydroxypyridinone MBP scaffold as a potential inhibitor of PAN endonuclease with an IC50 value of 16 μM. Hit-to-lead modification ultimately resulted in PANi-11, which was found to inhibit PAN endonuclease activity with an IC50 value of 11 nM.471,482 In another study, screening of a designer MBP library consisting of ~300 fragments against PAN endonuclease identified pyromeconic acid derivatives as active inhibitors.483 Guided by modeling, derivatives of pyromeconic acid (PANi-12) and the structurally related 3,4-hydroxypyridinone (PANi-13) were prepared and found to inhibit PAN endonuclease with IC50 values of 940 nM and 36 nM, respectively. Using a fragment merging strategy, compound PANi-14 was obtained with superior activity against PAN endonuclease (IC50 = 14 nM) and an EC50 value of 2.1 μM against live virus in MDCK cells.
Very recently, novel inhibitors of PAN endonuclease have been disclosed by Shionogi. These fused ring inhibitors are based on a 2,3-hydroxypyridinone core MBP. Compound RO-7 (Figure 52) was first disclosed as a tool compound and was found to have very high efficacy against live virus in cell saving assays (EC50 = 5–15 nM).484 The clinical candidate, Baloxavir Marboxil, inhibits PAN endonuclease and influenza virus with similar activity as RO-7. This drug candidate is currently in Phase III studies (NCT02954354, NCT02949011) and has shown promising results as a combination therapy with oseltamivir (a.k.a., Tamiflu).
Current and Future Prospects.
In the past decade, many new structural classes of PAN endonuclease inhibitors have been reported, with a large number of these inhibitors showing good activity in protein based assays. The availability of co-crystal structures has facilitated the development of highly active inhibitors, and will play an increasingly crucial role as the field moves towards inhibitors that are rationally designed to maintain high enzymatic activity, but with improved biologically active. Although a variety of MBPs have been explored, increasing numbers of studies have found hydroxypyridinone and similar scaffolds to be highly active scaffolds for inhibitor development, with the first PAN endonuclease inhibitors to enter clinical trials using these MBPs. Much like the early days of HIV IN inhibitor development (Section 5.1), the field of influenza PAN endonuclease inhibitor development is primed to rapidly grow in the next decade in the effort to identify clinically useful compounds.
5.4. Phosphodiesterase-4 (PDE4, EC 3.1.4.53)
Function in Biology and Disease.
Phosphodiesterases (PDE) hydrolyze cyclic adenosine monophosphate (cAMP) and/or cyclic guanosine monophosphate (cGMP) (Figure 54) and are thereby responsible for regulating intracellular levels of these second messengers.487,488 cAMP and cGMP bind to and activate protein kinases A and G (PKA and PKG), which regulate important cellular functions.488 Decreased concentrations of cyclic nucleotides due to degradation by PDE is implicated in several human diseases, including heart failure, depression, asthma, inflammation and erectile dysfunction; therefore, inhibitors of PDEs are considered of therapeutic importance.488 The PDE superfamily includes 11 isoenzyme families of which PDE4, -7, and -8 are cAMP-specific enzymes; PDE5, -6, and -9 are cGMP-specific enzymes, and PDE1, -2, -3, -10, and -11 hydrolyze both cyclic nucleotides.489 Most drug discovery efforts target PDE3, -4, or -5.487,488
Figure 54.
Reaction catalyzed by PDE4.
This chapter focuses on PDE4 because a metal-binding PDE4 inhibitor has recently obtained FDA-approval (Figure 1). PDE4 is present in the brain, testes, leukocytes, skeletal, visceral and vascular muscles and is involved in inflammation, vascular and visceral muscle tone, depression, and reproduction.490 Four different genes encode PDE4, including PDE4A, PDE4B, PDE4C, PDE4D, and there are 20 different isoforms.487 Although the catalytic domain of PDE4 isoenzymes is highly conserved, each gene fulfills a nonredundant role in controlling cell function.491 For example, PDE4B inhibitors have shown to be particularly useful in treating inflammatory diseases.487,488,491,492
Protein and Active Site Structure.
PDEs consist of an N-terminal regulatory domain (~151 residues), a conserved catalytic domain (~337 residues), and a C-terminal domain (~78 residues).493 The catalytic domain (Figure 55) consists of 17 α-helices divided into three subdomains, including an N-terminal cyclin-fold region, a linker region, and a C-terminal helical bundle.488 The active site lies at the interface of these three subdomains, and it contains four subsites called the metal-binding site, core pocket, hydrophobic pocket, and lid region.488 There are two metal ions in the metal-binding site and their identity is unclear.488,494 Metal soaking experiments combined with anomalous X-ray diffraction experiments indicate that one metal is a Zn2+ ion, and it is coordinated by residues Asp392, His238, His274, and Asp275.493 The second metal is likely Mn2+ but could also be Mg2+, Ca2+, or a mixture of these ions in vivo.493 The Mn2+ ion is coordinated by Asp275 and water molecules.493 In the AMP-bound structure, both metals are also coordinated to oxygen atoms of the AMP phosphate.493 In the absence of ligand, the metals are bridged by a water molecule.495
Figure 55.
Structure of the catalytic domain of PDE4B with AMP (magenta) bound (PDB 1ROR).
A catalytic mechanism was postulated with the assumption that the in vivo active site structure consists of Zn2+ and Mg2+ (or Ca2+) bridged by an activated hydroxide.493,494 This activated hydroxide may perform a nucleophilic attack on the phosphorous of cAMP, triggering an associative, dissociative, or hybrid mechanism of cAMP hydrolysis.493 These mechanisms require protonation of the O3’ leaving group, which is likely satisfied by His234, a strictly conserved residue among PDEs.493,494
Inhibitor Development.
The development of PDE4 inhibitors to treat respiratory inflammatory diseases is an established field.496 The vast majority of these inhibitors are not metal-binding; however, a metal-binding drug, crisaborole (IC50 = 110 nM), was granted FDA approval in 2016 (Figure 1). The most well-characterized PDE4 inhibitor is rolipram (Figure 56).488 Although it is still used in research, rolipram failed clinical trials for causing significant gastrointestinal side effects.497 Rolipram binds to two different conformers of PDE4 with Ki values of ~500 nM and ~2 nM;498 therefore, the conformers were named the low-affinity PDE4 (LPDE4) and high-affinity PDE4 (HPDE4), respectively.499,500 Different proportions of LPDE4 and HPDE4 exist in tissues and cell types.500,501 Most immune and inflammatory cells contain the LPDE4 conformation, and some of the gastrointestinal side effects of rolipram are due to HPDE4 inhibition, not LPDE4 inhibition.500,501 This evidence has led to developing inhibitors with selectivity for LPDE4 over HPDE4,488,499–502 which has resulted in FDA-approved drugs that inhibit PDE4 with improved therapeutic indices (Figure 56), such as roflumilast (IC50 = 0.8 nM),503 cilomilast (IC50 = 13 nM),498 and apremilast (IC50 = 74 nM).504 These PDE4 inhibitors, as well as rolipram, feature an alkylated catechol moiety that is commonly present in PDE4 inhibitors. The alkylated catechol moiety occupies the same pocket as the adenine ring in AMP and forms hydrogen bonds with Gln443.495 There are several other non-metal-binding PDE4 inhibitors that are in clinical development or FDA-approved.487,488,492,502 Incorporating a hydroxamic acid into the rolipram scaffold increases activity, as exemplified by PDE4i-1, which has an IC50 value of ~2 nM compared to IC50 ~10–36 nM of rolipram.505 These values were determined by measuring inhibition against PDE4A, -B, and D; the conformer of PDE4 (LPDE4 or HPDE4) is not reported.505 Additional experiments indicated that, these hydroxamic acid compounds were implicated in off-target activity and were not further developed.495,506
Figure 56.
Inhibitors of PDE4.
A subsequent study on a library of boron-containing small molecules507 produced a series of benzoxaboroles with inhibitory activity against cytokine release and PDE4.508 The 5-phenoxybenzoborole compounds were particularly effective against PDE4.508 Crisaborole (IC50 = 110 nM) was further tested for selectivity over PDE isozymes, including PDE1-11.508 AN2728 showed some inhibitory activity against PDE1A3, PDE3Cat, PDE7A1 but no inhibition of other isozymes at 10 μM.508 The conformer of PDE4 (LPDE4 or HPDE4) used in this study is not reported, but the measured IC50 value of rolipram is 860 nM, which shows the boron-containing inhibitors are more potent than rolipram.508 A crystal structure of one of these compounds (PDE4i-2) bound to PDE4 reveals the mode of binding (Figure 57).495 The boron center is observed to be in a hydrated, tetrahedral configuration, with a hydroxyl group on the boron atom bridging the Zn2+ and Mg2+ ions. Superimposing the structures of PDE4 with the boron-based inhibitor and cAMP in the active site shows that the tetrahedral boron overlaps with the phosphate of cAMP; therefore, the hydrated boron mimics the transition state of cAMP hydrolysis.495 The presence of a tetrahedral boron atom is essential for the activity of these inhibitors as evidenced by significant loss of inhibitory activity when the oxaborole ring is substituted with a lactol ring.495 Boron-based inhibitors employing a similar binding motif to a bimetal center have been developed for arginase (Section 7.2).509 Crisaborole was granted FDA approval in 2016 to treat atopic dermatitis (a.k.a., eczema) and is the first successful metal-binding PDE4 inhibitor to receive FDA approval. It is also under development to treat psoriasis.490
Figure 57.
Structure of PDE4i-2 bound to PDE4 (PDB 3O0J). The FDA-approved inhibitor crisaborole is proposed to bind in an identical fashion to the metal centers. Zn2+ and Mn2+ are shown as orange and purple spheres, respectively.
Current and Future Prospects.
There are many PDE4 inhibitors in clinical development or that have gained FDA approval. The most recent to receive FDA approval are apremilast and crisaborole in 2014 and 2016, respectively. Initial attempts to create metal-binding PDE4 inhibitors used the ubiquitous hydroxamic acid MBP, but did not achieve clinical advancement. Crisaborole, which contains a cyclic boronate that binds to the activated water molecule bridging the Zn2+ and Mn2+ active site metals, is the first successful metal-binding PDE4 inhibitor. The tetrahedral boron in crisaborole mimics the transition state of cAMP hydrolysis similar to boron-based inhibitors developed for arginase (Section 7.2). Time will tell how the market impact of crisaborole compares to other PDE4 drugs; however, the success of the boron-based PDE4 inhibitors opens doors for new MBP-containing inhibitors of PDE4. Also, the use of the cyclic boronates in the development of inhibitors for metalloenzymes containing a dinuclear metal centers, such as PDE4 and arginase, highlights the potential of this interesting MBP. Examination of patent literature on inhibitors developed against PDE isoenzymes suggests that this is a fruitful field.496 While the greatest number of patents have and continue to be filed for PDE4 inhibitors, there is also demonstrated interest in developing inhibitors against PDE5 and PDE10 as well as PDE1 and PDE2 more recently.496 Currently, metal-binding inhibitors only exist for PDE4; however, similarity of the PDE active site suggests that the same boron-based MBP could be used to develop selective inhibitors for other PDE isoenzymes.
6. Peptidases (EC 3.4)
6.1. Angiotensin-Converting Enzyme (ACE, EC 3.4.15.1)
Function in Biology and Disease.
Hypertension, as defined by a systolic blood pressure ≥130 mmHg or a diastolic blood pressure ≥80 mmHg, affects a total of ~1.4 billion people worldwide, with the number of people impacted doubling in the past 40 years.510 Hypertension leads to various cardiovascular and renal diseases, including congestive heart failure, left ventricular systolic dysfunction, myocardial infarction, diabetic nephropathy, and cardiovascular death.511–514 The first-line therapy for the treatment of hypertension and related diseases is the administration of angiotensin-converting enzyme (ACE) inhibitors. ACE is a Zn2+-dependent dipeptidyl carboxyl metallopeptidase that plays a pivotal role in simultaneously regulating the renin‐angiotensin system (RAS) and kallikrein-kinin system (Figure 58). In RAS, ACE cleaves decapeptide angiotensin I to octapeptide angiotensin II, a bioactive peptide with vasoconstrictive properties;515–517 in the kallikrein-kinin system, ACE degrades bradykinin, an inflammatory mediator with smooth muscle contraction, vascular permeability, and vascodilation properties.518–520 Inhibition of ACE for the suppression of angiotensin II formation and bradykinin degradation is regarded as an effective method for the treatment of hypertension and related diseases.
Figure 58.
Reaction catalyzed by ACE in the regulation of the Renin-angiotensin system (RAS) and kallikrein-kinin system.
There are currently ten FDA approved ACE drugs, each varying in their PK and ADME profiles.521,522 Side effects from the prolonged usage of ACE drugs include dry cough and angioedema. These side effects are associated with elevated levels of bradykinin, which stem from a mixed inhibition of both the C- and N-domains of ACE.523–526 To tackle these shortcomings and improve the efficacy and safety of administered drugs, there is a need for domain-specific inhibitors. Additionally, vasopeptidase inhibitors, dual inhibitors of ACE and neutral endopeptidase (NEP, Section 6.7) are also of interest.527–529
Protein and Active Site Structure.
Human ACE exists in two isoforms (encoded by a single gene): as a glycoprotein in somatic tissue (somatic ACE) or as a glycoform in testis cells (tACE).530,531 Somatic ACE is a type-I membrane-anchored dipeptidyl carboxypeptidase and bears two homologous extracellular catalytic binding sites, the N- and C-domain. While both domains bear high sequence homology (>60%), similar overall structures, and a catalytic Zn2+ active site (with the same ligating residues),532 the domains differ in substrate and inhibitor specificity, physiological function, and chloride-dependent activation profiles.533,534 In vitro, both domains are capable of hydrolyzing angiotensin I and bradykinin with similar efficacy; however, in vivo, angiotensin I is primarily hydrolyzed via the C-domain while bradykinin is degraded by both domains.535–537 Furthermore, domain specific inhibition and expression in transgenic mice revealed the C-domain as dominate for controlling blood pressure and cardiovascular functions.532,537,538 Thus, the C-domain is the dominate and primary target for treatment of hypertension. Selectively targeting the C-domain, while sparing the N-domain, has been shown to prevent the buildup of bradykinin and reduce side effects.532 tACE, an isozyme identical to the C-domain of somatic ACE (from residue 37 to the C-terminus),539 has been used for crystallographic and domain specific-inhibition studies.
X-ray crystal structures of human tACE reveals a 588-residue protein of ~70 kDa. tACE is composed of twenty seven α-helices and six β-strands, two of which are located near the active site, and adopts an overall ellipsoid shape.535 The active site is buried ~10 Å from the surface, and is located within a central groove consisting of four a-helices (α13, α14, α15, α17) and one β-strand (β4). The Zn2+ ion is coordinated to residues His383, His387, and Glu411 and an acetate ion (from the crystallization medium) in a tetrahedral coordination geometry (Figure 59). Additional structural details of tACE, including the role of chloride ions, can be found elsewhere.535
Figure 59.
Structure of native tACE with the active site buried in the central groove (PDB 1O8A). An acetate ion (green) is bound to the Zn2+ ion.
Inhibitor Development.
Since ACE inhibitors were first introduced in 1981 for the treatment of refractory hypertension, numerous inhibitors of ACE have since been accepted in the clinic for the treatment of cardiovascular and renal diseases.514 Indeed, ACE inhibitors represent one of the greatest success stories for metalloenzyme inhibitors. Current FDA approved inhibitors can be categorized into three classes based on the MBP: sulfhydryl-, carboxyl-, and phosphoryl-based inhibitors.
During the time period of 1965–1970, teprotide (Figure 60) was isolated from the snake venom of Bothrops jararaca and identified as the first potent inhibitor of ACE.540–542 Although teprotide possessed antihypertensive properties (IC50 = 250 nM),543 poor oral bioavailability precluded further clinical development.544 Utilizing teprotide and a surrogate Zn2+-dependent metalloenzyme model (carboxypeptidase A), a series of dipeptide proline analogues bearing carboxylic acid and sulfhydryl MBPs were designed, synthesized, and screened against ACE. This led to the discovery of the first orally active ACE inhibitor, captopril (Ki = 1.7 nM, IC50 = 6.9 nM, Figure 60).545–548 Captopril is among the first FDA approved metalloenzyme inhibitors, the only inhibitor of ACE bearing a sulfhydryl MBP, and is still used today as a benchmark compound for metalloenzyme inhibition. The X-ray crystal structure of captopril bound to tACE reveals the sulfhydryl MBP of captopril binding in a monodentate fashion to the Zn2+ ion, which is further ligated by the residues His283, His387, and Glu411 in a tetrahedral coordination geometry (Figure 61).549 Other key active site interactions include H-bonding of the central carbonyl motif (between the sulfhydryl MBP and proline) with residues His513 and His353, and the terminal carboxylic acid group of proline with Tyr520, Gln281, and Lys511.
Figure 60.
Inhibitors of ACE. All but teprotide and ACEi-1 are FDA approved for the treatment of cardiovascular and renal diseases.
Figure 61.
Structures of tACE with: captopril (PDB 1UZF), lisinopril (PDB 1O86), and ACEi-1 (PDB 2XY9).
The largest class of ACE inhibitors include captopril analogues bearing a carboxylic acid MBP. These inhibitors include benazeprilat, enalaprilat, lisinopril, moexiprilat, quinaprilat, ramiprilat, trandolaprilat and perindoprilat (Figure 60).522,547,550–553 With the exception of lisinopril, all carboxylic acid MBP inhibitors are administered as an ester-prodrug that are hydrolyzed in the liver into the active carboxylic acid metabolite (Figure 60). The Ki values for these inhibitors range from 0.4 – 11.0 nM, depending on the assay conditions used;522,547,550–555 however, these inhibitors are usually tested against somatic ACE protein (bearing both C- and N-domains), so the specificity of the inhibitors towards the two domains are not well characterized. The structure of lisinopril bound to tACE reveals the carboxylic acid MBP of lisinopril bound in a monodentate manner to the Zn2+ ion, which is further ligated by residues His383, His387, and Glu411 in a tetrahedral coordination geometry (Figure 61).535 Other key interactions include: the unbound oxygen atom of the carboxylic acid MBP forms a H-bond with Glu384, the phenylpropyl motif makes a van der Waals interactions with Val518, the lysyl amine forms a weak binding interactions with the nearby Glu162, and the proline carboxylate forms a H-bond with Lys511 and Tyr520.
Another clinically approved class of ACE inhibitors utilize a phosphoryl MBP, and were initially developed as means to selectivity inhibit the C- and N-domains. The phosphoryl MBP is generally a weaker ligand for Zn2+ compared to that of the sulfhydryl and carboxylic acid MBPs, potentially allowing for greater tuning of the inhibitor backbone to target specific residues in the active site of the C- and N-domains.537,555,556 ACEi-1 (Figure 60) was found to be a potent inhibitor for ACE C-domain (Ki = 0.65 nM) and exhibited >230-fold selectivity for the C-domain over the N-domain.555 ACEi-1 lowered the arterial blood pressure of hypertensive rats and led to an increase in bradykinin levels. Other derivatives bearing a phospsphoryl MBP inhibitors have been investigated and are described elsewhere.555,557,558 The structure of ACEi-1 complexed with tACE reveals the phosphoryl MBP binding in a monodentate fashion to the Zn2+ ion, which is further ligated by side chains of His383, His387, and Glu411 (Figure 61).558 Further key interactions of ACEi-1 include the nitrogen atom of the isoxazole ring bound to Asp415 through a water molecule, the oxygen atom of the isoxazole ring interacting with the coordinating residue His383, and the terminal isoxazole ring and tyrosine form favorable interactions with a hydrophobic pocket consisting of residues Val380, Phe527, and Phe457.558 While there are no available crystal structures of fosinoprilat (the only FDA approved drug utilizing a phosphoryl MBP) bound to ACE, fosinoprilat is expected to have a similar metal coordination geometry to that of ACEi-1. In contrast to the C-domain selectivity of ACEi-1, fosinoprilat is 6-fold more selective for the N-domain.559
Current and Future Prospects.
ACE inhibitors are a tremendous success in the field of metalloenzyme inhibition, being vital for the treatment of hypertension and various cardiovascular and renal diseases. There are currently ten FDA approved inhibitors of ACE, with varying PK and ADME properties. These drugs operate by coordinating to the active site Zn2+ ion using sulfhydryl-, carboxyl-, or phosphoryl MBPs. While these drugs are effective for the inhibition of ACE, they are generally not domain specific, leading to adverse side effects such as coughing and angioedema that are associated with the undesired build-up of bradykinin. One potential solution to prevent bradykinin buildup is to only inhibit the C-domain of ACE via domain-specific inhibitors. These compounds inhibit the C-domain with minimal activity towards the N-domain, allowing the N-domain to continue in the degradation of bradykinin, thereby preventing harmful substrate accumulation.532,537,560 Notably, while the therapeutic potential for N-domain specific inhibitors have not been clinically validated, the N-domain has a high specificity for the hydrolysis (~50 times faster compared to that of C-domain) for AcSDKP (N-acetyl-seryl-aspartyl-lysyl-proline), a negative regulator of hematopoietic stem cell differentiation and proliferation.536,561,562 Another mode of therapy through ACE inhibition would be through the use of vasopeptidase inhibitors. Vasopeptidase inhibitors, such as omapatrilat (Section 6.7), inhibits both ACE and NEP; however, similar to current FDA approved inhibitors, omapatrilat is not domain selective and has resulted in higher instances of angioedema in clinical trials.528 This suggests some motivation to continue to develop domain-specific inhibitors of ACE.
6.2. A Disintegrin and Metalloproteinase (ADAM, EC 3.4.24.46)
Function in Biology and Disease.
The metzincin superfamily of metalloproteinases includes three subfamilies: astacins, matrix metalloproteinases (MMPs), and adamalysins. The latter is comprised of A Disintegrin And Metalloproteinases (ADAMs), snake venom metalloproteinases (SVMPs), and ADAMs containing thrombospondin motifs (ADAMTS).563 All members of the adamlysin family have the following domains: prodomain, metalloproteinase, disintegrin, and cysteine-rich domain.563 Additionally, ADAMs, which were previously referred to as the metalloproteinase/disintegrin/cysteine-rich (MDC) family, contain a transmembrane and cytoplasmic domain.
ADAMs are transmembrane metalloendopeptidases involved in many biological processes such as cell adhesion, migration, proteolysis, and signaling.564 The ADAM metalloproteinase domain is activated when the prodomain is cleaved.565 Subsequently, activated ADAMs cleave the extracellular domain of membrane proteins through a process called ectodomain shedding. Proteolysis occurs at the catalytic Zn2+ site in the metalloproteinase domain (Figure 62). The water molecule coordinated to the Zn2+ ion is deprotonated by a conserved glutamic acid (Glu346). The oxygen atom of the resulting hydroxide anion performs nucleophilic attack on a carbonyl in the backbone of the substrate, leading to proteolysis.566
Figure 62.
Reaction catalyzed by ADAMs. ADAMs are activated by cleavage of the prodomain. The activated ADAM then behaves as a sheddase by cleaving the extracellular domain of transmembrane proteins. The resulting cleaved ectodomains are involved in processes such as signaling.
Substrates of ADAMs include growth factors, receptors and their ligands, cytokines, cell adhesion molecules, and more.563 Ectodomain shedding of these substrates is important for regulating function of many cell surface proteins; however, dysregulation of shedding can contribute to autoimmune disease, cardiovascular diseases, neurodegenerative diseases, infection, inflammation, and cancer.567 The most widely studied ADAMs for potential as therapeutic targets are ADAM-10 and ADAM-17. ADAM-17, which is the best characterized, is more commonly referred to as Tumor Necrosis Factor-α Convertase (TACE).568 Elevated tumor necrosis factor-α (TNF-α) levels are implicated in many diseases, including inflammatory disorders, cardiovascular diseases, skin disorders, and cancer.567
Protein and Active Site Structure.
ADAMs across multiple species have been catalogued, 38 in total, including 25 expressed in humans.564 About half the ADAMs encoded in the human genome do not contain a Zn2+-binding signature in the metalloproteinase domain. The biological function of these ADAMs likely depends on protein folding or protein-protein interactions instead of catalysis.564 The other ADAMs, which are the focus of this section, contain the catalytic Zn2+ binding signature HEXXHXXGXX(H/D) in their metalloproteinase domain.569
ADAMs contain ~750 amino acids divided among the several domains mentioned above.564 The metalloproteinase domain (Figure 63) has roughly 200 amino acids and contains the active site Zn2+ ion. Except for a few loop regions, the secondary and tertiary structures of the metalloproteinase domain of different ADAMs is conserved: there is a five-stranded β-sheet (the N-terminus) and four α-helices (the C-terminus).569 The Zn2+ ion in the activated ADAM metalloproteinase domain is coordinated by a water molecule and the imidazole side chain of His345, His349, and His355(within the HEXXHXXGXX(H/D) motif.569 The conserved Glu346 serves to deprotonate the catalytic water molecule during proteolysis. The average Zn2+-NHis bond distance is ~2.1 Å.569 There is also a conserved methionine turn (a.k.a., ‘Met-turn’) at one end of the Zn2+ binding site.569,570 The Met-turn is a tight 1,4-turn that is structurally and spatially conserved in all members of the Metzincin family.570
Figure 63.
Structure of the ADAM metalloproteinase domain from ADAM-33 (PDB 1R55).
Inhibitor Development.
Of all the ADAMs implicated in human diseases, the majority of drug discovery efforts have targeted ADAM-17 (TACE) and, to a lesser extent, ADAM-10, both of which will be the focus of this section. Although many ADAM inhibitors have reached clinical trials, there are no FDA-approved ADAM inhibitors due to selectivity or efficacy shortcomings.568 The most widely investigated approach to drugging ADAMs is the synthesis of low molecular weight inhibitors that bind to the Zn2+ ion in the active site.571 A critical consideration for such efforts is inhibitor selectivity over MMPs as well as over other ADAMs, which is achieved by synthesizing inhibitors that occupy the appropriate subsites within the ADAM active site.568,571 Figure 64 and Table 8 show the structures and inhibition data for inhibitors. There is an extensive literature on the inhibitors of ADAMs and TACE with complete reviews available elsewhere.568,571,572
Figure 64.
Inhibitors of TACE and ADAM-10.
Table 8.
Activity of TACE and ADAM-10 inhibitors.
| Compound | Target | Inhibition (nM) | Ref. |
|---|---|---|---|
| ADAMi-1 | TACE | Ki = 2.8 | 573 |
| ADAMi-2 | TACE | Ki = 1.4 | 574 |
| ADAMi-3 | TACE | IC50 = 9 | 575 |
| ADAMi-4 | TACE | IC50 = 0.5 | 576 |
| ADAMi-5 | TACE | IC50 = 6.0 | 577 |
| ADAMi-6 | TACE | Ki = 10 | 578 |
| ADAMi-7 | TACE | IC50 = 80 | 579 |
| ADAMi-8 | TACE | Ki = 143 | 580 |
| ADAMi-9 | TACE | Ki = 0.86 | 581 |
| ADAMi-10 | TACE | Ki = 5 | 582 |
| ADAMi-11 | TACE | Ki = 8 | 583 |
| ADAMi-12 | ADAM-10, TACE | Ki = 12 (TACE) | 583 |
| ADAMi-13 | ADAM-10 | IC50 = 97 | 584 |
| ADAMi-14 | ADAM-10 | IC50 = 26 | 585 |
| DPC-333 | TACE | IC50 = 133 | 586 |
| GI254023X | ADAM-10 | IC50 = 5.3 | 587,588 |
| GW280264X | ADAM-10, TACE | IC50 = 11.5 (ADAM-10), 8.0 (TACE) | 587,588 |
| IK682 | TACE | Ki = 0.56 | 589 |
| INCB3619 | ADAM-10, TACE | IC50 = 22 (ADAM-10), 14 (TACE) | 584 |
| INCB7839 | ADAM-10, TACE | IC50 = 320 | 590 |
| TMI-005 (apratastat) | TACE | EC50 = 87 | 591 |
| TMI-2 | TACE | IC50 = 2 | 592,593 |
Some of the earliest work on TACE inhibitors is based on marimastat (Figure 64), a broad spectrum MMP inhibitor (Section 1.5) that features a bidentate hydroxamate MBP.568 A derivative of marimastat containing a macrocycle and a biphenyl moiety, ADAMi-1, was reported with an IC50 value of 2.8 nM.573 The biphenyl moiety fits the S1’ pocket, thereby achieving >100-fold selectivity over a broad panel of MMPs.573 Shortly thereafter, a γ-lactam hydroxamic acid inhibitor partly developed from marimastat, called IK682, was reported.589 IK682 has a hydrogen bond donor oxygen on a 2-pyrrolidinone that properly orients the inhibitor toward the S1’ subsite and a (2-methyl-quinolin-4-yl)methoxy group to fill the S1’ subsite (Figure 65).589 IK682 is a highly active TACE inhibitor with a Ki value of 0.56 nM against TACE.589 IK682 shows strong selectivity over a broad panel of MMPs (Ki >100 nM) as well as ADAM-33 (Ki = 15 nM).589,594 Further optimization of IK682 led to DPC-333, which has an IC50 value of 113 nM and entered Phase II clinical trials for rheumatoid arthritis.572,586 A series of β-aminohydroxamic acid inhibitors, such as ADAMi-2 with an IC50 value of 1.4 nM, show selectivity over MMPs as well as ADAM-10 and several ADAMTSs. ADAMi-2 proceeded to preclinical studies for rheumatoid arthritis.572,574 ADAMi-2 fits into the active site of TACE, including the S1’ pocket, similar to IK682 and DPC-333 (Figure 65).568,574
Figure 65.
Structures of TACE with: IK682 (PDB 2FV5), ADAMi-4 (PDB 3LE9), ADAMi-6 (PDB 3B92), ADAMi-8 (PDB 3EWJ), ADAMi-9 (PDB 3LGP), and ADAMi-11 (PDB 3E8R).
Other scaffolds with a hydroxamate MBP include a butynyloxy group, as exemplified by TMI-2, where the phenyl ring occupies the S1’ pocket and the butynyl group points toward the S3’ pocket.592,593 The butynyloxy group cannot be properly accommodated by shorter or longer MMP channels; thus, these scaffolds have high selectivity over MMPs and ADAM-10.592,593 TMI-2 has an IC50 value of 2 nM and proceeded to preclinical studies for rheumatoid arthritis.592,593 A related structure, TMI-5 (apratastat), has an EC50 value of 87 nM against TACE.591 Apratatstat entered Phase II clinical trials to treat rheumatoid arthritis, but it failed due to lack of efficacy.591
Non-hydroxamate TACE inhibitors have been explored to avoid the toxicity associated with hydroxamate metabolic byproducts. For example, ADAMi-3 with an IC50 value of 9 nM, features a succinimide in which the carbonyl oxygen coordinates to the Zn2+ ion in a monodentate fashion, as predicted by molecular modeling.575,595 A related set of inhibitors have a hydantoin MBP in which a nitrogen atom in the hydantoin ring coordinates to the Zn2+ ion in a monodentate fashion.576,596 The best of this series is ADAMi-4, which has an IC50 value of 0.5 nM and high selectivity over a broad panel of MMPs, as well as ADAM-10 (Figure 65).576
Other efforts to avoid the hydroxamate MBP include using a reverse hydroxamate MBP, as in ADAMi-5 (IC50 = 6.0 nM), which demonstrates selectivity over a broad panel of MMPs.577 Additionally, a thiol-based inhibitor with a butynyloxy group, ADAMi-6 (Ki = 10 nM) was found to be surprisingly selective given its small size.578 In an X-ray crystal structure of ADAMi-6 bound to TACE, the thiol binds to the Zn2+ ion, the phenyl ring occupies the S1’ pocket, and the butynyloxy group points toward the S3’ pocket (Figure 65).568 Carboxylate MBPs have also been explored, in compounds such as ADAMi-7 and ADAMi-8.579,580 ADAMi-7 has an IC50 value of 80 nM and ADAMi-8 has a Ki value of 143 nM.579,580 An X-ray crystal structure of ADAMi-8 bound in the active site of TACE shows that the carboxylate binds to the Zn2+ ion in a pseudo-bidentate fashion with a Zn-O bond of 1.9 Å and a weak Zn-O contact of 2.5 Å (Figure 65).580
Another class of inhibitors contain tartrate MBPs is exemplified by ADAMi-9. The tartrate group binds in an unusual tridentate fashion to the Zn2+ ion (Figure 65).597 ADAMi-9 has a Ki value of 0.86 nM and is selective over MMPs and ADAM-10.581 To increase the bioavailability of this scaffold, a 2-heteroaryl substituted pyrrolidines were added.582 One such derivative is ADAMi-10, which maintains selectivity over ADAM-10, has an inhibition constant of 5 nM, and has improved oral bioavailability compared to earlier tartrate inhibitors.582
The subsites that an inhibitor occupies in the ADAM active site is critical for achieving selectivity among ADAM homologues, as demonstrated by pairs of inhibitors across several classes. For example, a pair of hydroxamic acid inhibitors, ADAMi-11 and ADAMi-12, differ in the stereochemistry of a cyclopropyl ring. This stereochemistry causes ADAMi-11 to project the quinoline group through S1’ toward the S3’ subsite and ADAMi-12 to project its quinoline group in the opposite direction through S1 toward S3 (Figure 65).583 Both inhibitors are selective over MMPs; however, only ADAMi-11 is selective for TACE, while ADAMi-12 inhibits TACE and ADAM-10.583 This suggests that targeting the ‘primed’ subsites may be useful in achieving selectivity against various ADAMs, while targeting the unprimed sites is useful for creating selectivity over MMPs.583 This is further demonstrated by compounds ADAMi-13, INCB3619, ADAMi-14, INCB7839, as well as the reverse hydroxamate inhibitors GI254023X and GW280264X.584,585,587 INCB7839, a dual ADAM-10/TACE inhibitor, has advanced to clinical trials.590
Current and Future Prospects.
The two ADAM inhibitors evaluated in clinical trials used a hydroxamic acid MBP and have since been discontinued due to hepatotoxicity and/or lack of efficacy.568 Another hydroxamate inhibitor, INCB7839, is entering Phase I and II clinical trials. It remains to be seen if INCB7839 will suffer from the same issues as previous hydroxamate inhibitors. To overcome the limitations of hydroxamate inhibitors, much of the recent work on ADAM inhibitors has explored other MBPs that include carboxylates, hydantoins, reverse hydroxamates, and thiols, as well as tartrates that display a highly unusual tridentate binding mode. Many of these inhibitors with alternative MBPs are still being optimized for adequate activity and PK properties. The most challenging aspect of developing ADAM inhibitors is achieving selectivity among ADAM homologues and over MMPs. Several examples of altering stereochemistry such that the inhibitor occupies specific subsites within the active site has yielded selective ADAM inhibitors. Given the development of non-hydroxamate MBPs and selective inhibitors for TACE, opportunities remain to develop selective compounds that target TACE to treat a large variety of diseases, such as cancer, diabetes, rheumatoid arthritis, and Alzheimer’s disease.598 Perhaps understanding of how to produce potent, selective, and non-toxic TACE inhibitors can be applied to other ADAMs, of which many are implicated in human diseases.
6.3. Anthrax Lethal Factor (LF, EC 3.4.24.83)
Function in Biology and Disease.
The disease anthrax is caused by Bacillus anthracis, an endemic zoonotic soil bacteria, and has long been relevant to both modern medicine and human history. Virgil clearly described cases of this disease during the Roman era,599 and anthrax has even been attributed to the ancient plagues of Egypt.599,600 Historically, anthrax has been an animal disease primarily affecting livestock, only infecting people in contact with contaminated animals. In 1876, Robert Koch used B. anthracis to prove the germ theory of disease by demonstrating that this bacteria was able to cause human disease.601 Louis Pasteur developed the first vaccine from live attenuated bacteria for human use by treating the microbes with “oxygen”, or what is now known as potassium dichromate.
Anthrax remains highly relevant in modern times, where it has been used in biological warfare programs. There have been reports of weaponized anthrax releases, both accidental and intentional. Japan first used anthrax against Manchuria in 1940,600 and later the Sverdlovsk incident of 1979 in the former Soviet Union occurred when a military facility accidentally released aerosolized anthrax resulting in the death of 64 people.602 Anthrax experiments by the United Kingdom in 1942 left the Scottish Gruinard Island uninhabitable until 1990. Most recently in 2001, a series of bioterrorism attacks in the United States involved the mailing of anthrax spores, resulting in five deaths.603
Anthrax has been a bacterium of choice for state bioweapon programs because of its ease of dissemination through aerosolization, high rates of mortality when inhaled, and the intrinsic durability of the bacteria that can survive in exceedingly harsh environments. B. anthracis is able to form an endospore that is extremely hardy, and is resistant to UV and gamma radiation, freezing, boiling, as well as most common chemical disinfectants, including bleach and ethanol. Inhalational anthrax presents itself as a respiratory infection, resulting in death ~48 days from symptom onset.604 Left untreated, inhalational anthrax has mortality rates of 100%, and even when treated with antibiotics the mortality rate is 70%.603
Anthrax infections can be treated with antibiotics, such as penicillins and doxycycline,600 but antibiotics alone are insufficient as the bacteria produce two extremely lethal, bipartite protein toxins: lethal toxin (LeTx) and edema toxin (EdTx).605 These toxins have been shown to persist in the body after the bacterial source has been eliminated through antibiotic treatment. Indeed, injection of either of these toxins into rodents, even in the absence of the anthrax bacteria, is lethal. One study demonstrated that LeTx targets cardiomycytes and smooth muscle cells (except for hepatocytes), whereas EdTx specifically targets hepatocytes, so that B. anthracis induces lethality via the targeting of two distinct and vital systems.606 In particular, LF is currently known to selectively target and hydrolyze members of the mitogen-activated protein kinase kinase (MAPKK) family.607–609 While the biology of the MAPKKs is not fully understood, it is known these MAPKK proteins play a role in directing protein-protein interactions in signaling complexes, and the action of hydrolysis by LF eventully results in cell lysis.607,610
The LeTx complex is comprised of the proteins lethal factor (LF) and protective antigen (PA), whereas the EdTx is comprised of the proteins edema factor (EF) and PA. The common protein between LeTx and EdTx, PA (whose name derives from its use in the development of vaccines against anthrax) is a pore-forming subunit used for cellular entry by both EF and LF. PA is an ~83 kDa monomeric protein that binds to human cell surfaces and is subsequently hydrolyzed by host furin family proteases, resulting in the removal of a small portion of the PA N-terminus. The remaining ~63 kDa C-terminus of the PA spontaneously self-assembles with six other monomers to form a heptameric pore that allows for cellular entry by LF and EF. Therefore, in the following text, LeTx will refer to the combination of LF and PA, whereas LF will refer to studies performed on the lethal factor protein alone (in the absence of PA). In particular, in vitro studies are usually performed with LF, but in vivo studies use the LeTx combination.
During B. anthracis infections, LF is expressed at fivefold higher concentrations than EF, with LF concentrations reaching 10–35 μg/mL within ~48 hours in a rabbit infection model.611–613 This suggests that LF is more relevant than EF in contributing to the lethality of the anthrax disease state. Indeed, mice genetically modified for LeTx resistance are more likely to survive anthrax infections,606 indicating that the anthrax lethal factor represents a strong target for inhibition to treat otherwise deadly anthrax infections.
Protein and Active Site Structure.
Anthrax LF is a ~90 kDa mononuclear Zn2+ metalloprotease that catalyzes the hydrolysis of the MAPKK family (Figure 66). LF is comprised of four distinct protein domains: domain I is largely made of α-helices and is vital for PA binding, whereas domains II, III, and IV are conjoined to form the substrate binding groove, with domain IV as the site of catalytic. LF was long suspected to be a Zn2+-dependent metalloenzyme as studies demonstrating incubation with EDTA and o-phenanthroline blocked enzymatic activity. This was confirmed in 2001 when the first crystal structure of anthrax LF was released.610
Figure 66.
Structure of LF. Left: Composite image depicting the full structure of LF with the peptide substrate in magenta (composite of PDB 1JKY and 1J7N). Right: Active site of LF, with hydrogen bonding (dashed magenta lines) from neighboring residues shown (PDB 1J7N).
While LF does contain the characteristic metalloprotease motif of His686–Glu687–X–X–His690, the Glu687 residue does not participate in metal binding, as it is 4.07 Å610 away from the catalytic Zn2+. Instead, Glu735, which is located further in the sequence, participates in Zn2+ coordination.610,614 Thus, the catalytic Zn2+ is bound by His690, His686, and Glu735, with a hydroxyl completing the tetrahedral geometry.615 Glu687, along with Tyr728, are believed to play a large role in substrate recognition and intermediate stabilization, and are necessary for LF activity.614 Importantly, a crystal structure of LF complexed with the uncleaved natural MAPKK peptide substrate has also been obtained (Figure 66). In this complex, Pro10 of the substrate is the closest scissile bond to the Zn2+ ion at a distance of 6 Å, and is postulated to make a 90° swing to fit into the S1 pocket for hydrolysis to occur.
Inhibitor Development.
The release of the anthrax LF crystal structure in 2001610 spurred inhibitor development against this target.604 By far the most studied class of inhibitor against LF utilizes the hydroxamic acid MBP (Figure 67, Table 9). This class of inhibitor coordinates to the Zn2+ ion through the hydroxamate MBP in the typical bidentate manner, which effectively blocks the active site. The first inhibitor of this kind was reported in 2002, which developed a colorimetric assay for LF based on the native substrate. Hydroxamic acids appended onto various substrate analogues generated LFi-1 with an excellent Ki value of 1 nM.616 It is postulated that this long, Arg enriched chain fits in the substrate binding groove in addition to aiding in cellular uptake. Efforts to shorten the peptide chain lead to the discovery of LFi-2, a known MMP inhibitor (Section 1.5) with a Ki value of 2.1 μM against LF.617 Work done by Merck to improve LFi-2 led to the discovery of LFi-3 (Ki = 24 nM), which has been shown to be effective at increasing survival rates of rabbits infected with different strains of anthrax, especially when co-administered with antibiotics.618 Crystal structure analysis of LFi-3 shows this inhibitor binds to the catalytic Zn2+ through the hydroxamic acid, and then proceeds to make a myriad of interactions within the active site (Figure 68). LFi-3 was seminal in the field of LF inhibitors, and has been the structural starting point for many additional campaigns to discover other hydroxamic based LF inhibitors.619–622 For example, a three-part study to further elaborate LFi-3 culminated in the discovery of LFi-4 and LFi-5 (Ki = 0.31 nM and Ki = 0.27 nM, respectively).620,623,624 Both LFi-4 and LFi-5 were found to be effective at rescuing animals from exposure to LeTx, with LFi-4 being the most effective in a rat model.620 In a follow-up study, LFi-5 was used in a combination with ciprofloxacin in B. anthracis infected rodents and was shown to provide protection when LFi-5 was administered twice a day.625 Computational modeling has been used to develop LF inhibitors based on LFi-3,622 and incorporation of a proline backbone has been shown to improve inhibitor PK properties.619 Recent trends have begun to shift away from the hydroxamic acid MBP due to PK liabilities. In one example, hydroxamic acid compound LFi-10 (Ki = 1 nM) had poor PK properties in mice, which was shown to be due to LF induced hydrolysis of the hydroxamic acid MBP in LFi-10 upon prolonged incubation.58 Double methylation of the hydroxamic acid lead to compound LFi-11, which has presumably weakened or no metal-binding ability, but drastically improved PK and maintained activity with a Ki = 6.4 nM. It is surprising that LFi-11 with poor metal binding ability displays such strong activity, but it may be that the affinity of LFi-11 derives largely from backbone interactions with LF. There is a body of literature focused on non-metal binding small inhibitors of LF, which instead of fitting in the metal-binding pocket, instead bind in the long substrate groove. Several compounds of this sort have been reported to have IC50 values in the low nanomolar range, which may explain the good activity of LFi-11.604,605
Figure 67.
Inhibitors of LF. The binding orientation of benzothiazole inhibitors (LFi-24, LFi-25, LFi-26) are shown as originally published and as proposed based on a more likely coordination mode.
Table 9.
Activity of LF inhibitors.
| Compound | Inhibition (μM) | Ref. |
|---|---|---|
| LFi-1 | Ki = 0.001 | 616 |
| LFi-2 | Ki = 2.1 | 617 |
| LFi-3 | Ki = 0.024 | 618 |
| LFi-4 | Ki = 0.00031 | 620 |
| LFi-5 | Ki = 0.00027 | 620 |
| LFi-6 | IC50 = 1.6 | 622 |
| LFi-7 | IC50 = 5.6 | 622 |
| LFi-8 | IC50 = 4.4 | 622 |
| LFi-9 | IC50 = 1.4 | 619 |
| LFi-10 | Ki = 0.0014 | 626 |
| LFi-11 | Ki = 0.0064 | 626 |
| LFi-12 | Ki = 3 | 627 |
| LFi-13 | Ki > 100 | 627 |
| LFi-14 | Ki > 100 | 627 |
| LFi-15 | Ki = 1.7 | 627 |
| LFi-16 | IC50 = 70 | 627 |
| LFi-17 | Ki = 6.0 | 627 |
| LFi-18 | IC50 ~2 | 628 |
| LFi-19 | IC50 = 50 | 628 |
| LFi-20 | IC50 = 260 | 629 |
| LFi-21 | IC50 >6000 | 629 |
| LFi-22 | IC50 = 13.9 | 629 |
| LFi-23 | IC50 = 5.0 | 630 |
| LFi-24 | IC50 = 15 | 621 |
| LFi-25 | IC50 = 21 | 621 |
| LFi-26 | IC50 > 100 | 621 |
| EGCG | IC50 = 0.097 | 631 |
Figure 68.
Structure of LFi-3 bound to LF (PDB 1YQY).
Exploration of alternate MBPs against LF was catalyzed by the 2004 discovery that epigallocatechin gallate (EGCG) was able to inhibit LF with an IC50 value of 97 nM (Figure 67).631 While EGCG has not been shown to be a metal-binding inhibitor of LF, EGCG has good activity in rescuing animal models from LeTx injection631 and has prompted other groups to probe polyphenol based MBPs for LF inhibitors. In 2005, a series of N-oleoyldopamine compounds bearing catechol-based warheads were explored against LF.627 Of these, the catechol (LFi-12) and gallate (LFi-15) warheads were found to have the best activity with Ki values of 2.2 μM and 1.7 μM, respectively. Although no structures have been published, these N-oleoyldopamine compounds are presumed to act by binding to the catalytic Zn2+ ion though the catechol group, which is consistent with a loss of activity for derivatives LFi-13 and LFi-14, which lack metal binding groups. Other catechol containing inhibitors were discovered by a matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry based screen,628 which identified the hydrazine catechol LFi-18 (IC50 = 2 μM).
Another metal-binding inhibitor campaign used a FBDD approach to identify small MBPs,629 which found the thiohydroxypyridinone MBP to be active against LF (LFi-20, IC50 = 260 μM, Figure 67). In particular, thiohydroxypyridinones (LFi-20) were shown to have stronger binding to LF than hydroxypyridinones (LFi-21), which showed no inhibition against LF. The high activity of thiohydroxypyridinones compared to hydroxypyridinones readily follows the trends of Lewis hard-soft acid base theory, with the relatively soft Zn2+ preferring the soft sulfur donor over the hard oxygen donor. In this study, derivatized thiohydroxypyridinone compound LFi-22 was found to have an IC50 value of 13.9 μM. As a follow-up, thiohydroxypyridinones bearing thioamide linkers opposed to amide linkers were found to be further improved, with LFi-23 having an IC50 value of 5.0 μM.630
Finally, in another study to develop metal-binding inhibitors against LF, a series of benzothiazole thiol compounds were investigated.621 After discovering an initial lead (LFi-24, Figure 67) through a HTS campaign, this study used molecular docking to suggest their lead was a mimic of the Merck hydroxamic acid lead LFi-3. This model proposes the thiophene moiety as a metal binding mimic of the hydroxamic acid MBP, and that the thiophene group is crucial for metal-binding. This hypothesis was supported by SAR, finding the thiol was vital for inhibition in that neither the analogs such as LFi-26 had any inhibitory activity. However, metal binding of a thiophene through a carbon donor atom to Zn2+ is highly unlikely; a more reasonable metal binding mode for LFi-24 is proposed in Figure 67. This proposed orientation coordinates through the nitrogen heteroatom on the benzothiazole and the deprotonated nitrogen of the sulfonamide. This mode should be preferred for metal-binding as it results in a monoanionic, bidentate ligand architecture, and forms a stable, six-member coordination ring. Ultimately, a macromolecular structure is needed to verify the metal-binding orientation of compound LFi-24 with LF.
Current and Future Prospects.
LF remains a valuable target for combating anthrax as a biological warfare and bioterrorism agent. Currently there are no inhibitors against LF in human clinical trials, despite several compounds showing good activity in animal models. This can partially be attributed to the presence of vaccines available against anthrax, which helps mitigate the threat and burden of this disease. While a substantial body of work has examined hydroxamic acid based inhibitors against LF, there has been a comparatively small amount of work on alternative MBPs. It is likely that exploring alternate MBPs will lead to new inhibitor scaffolds against LF. Any such new MBP could be combined with the backbones already optimized from hydroxamic acid compounds, and be used to develop new, potent inhibitors against LF.
6.4. Botulinum Neurotoxin (BoNT, E.C. 3.4.24.69)
Function in Biology and Disease.
Botulinum neurotoxin (BoNT), the source of Botulism disease, is secreted by the Gram-positive Clostridium botulinum bacteria, and is the deadliest substance known to humankind, with a single gram of the toxin being able to kill over 1 million people.632 As such, many nations have developed bioweapon technology involving Botulism, making BoNT a highly relevant target for anti-biological warfare programs. Despite its inherent lethality, there are relatively few cases of Botulism disease each year, with virtually all reports involving improper food preparation resulting in C. botulinum contamination.633 The earliest documented cases of Botulism date to the 1800s,634 with examples of bioterrorism involving Botulism including the Japanese invasion of Manchuria in the 1930s, the Iraqi production and stockpiling of BoNT, and the failed Aum Shinrikyo attacks in Japan from 1990–1995.632
BoNT acts by disrupting acetylcholine neurotransmission resulting in muscle flaccidity and paralysis. More specifically, the soluble N-ethylmaleimide sensitive factor attachment protein receptor (SNARE) proteins are responsible for allowing membrane fusion to release the acetylcholine enclosed in synaptic vesicles. BoNT cleaves these SNARE proteins, which prevents assembly of the synaptic vesicle complex and blocks acetylcholine release, making muscles unable to contract.635,636 Eventually, the resulting paralysis spreads to the muscles of the upper body system causing death. Treatment for Botulism includes equine-derived antibodies in anticipation of exposure, and post-exposure dosage with antibiotics to eliminate the C. botulinum infection. Currently there is no approved inhibitor for BoNT itself, with recovery time being the only way to restore damaged nerves.637 Despite its toxicity, BoNT has a substantial role in medicine, where it is widely used as the active ingredient in Botox for eliminate facial wrinkles. In Botox, BoNT is administered at drastically diluted concentrations of 0.005% of a lethal oral dose to avoid toxicity.632 Other medical uses include BoNT as a treatment for dysphonia, various types of spasm, hypersalivation, as well as hyperhydrosis.638 However, as the looming threat of BoNT based biological attacks remain, it is vital to develop therapeutics against this toxin.
Protein and Active Site Structure.
There are several serotypes of BoNT, termed A-F, with serotypes A, B, E, and F being the most relevant for human disease.639 In particular, BoNTA is the most toxic and therefore the most important in relation to human health. All of the BoNT serotypes act by cleaving various components of the SNARE proteins, but different serotypes act by hydrolyzing different SNARE proteins. All BoNT isoforms are ~150 kDa in size and are comprised of a single peptide chain. This chain is made of two domains, a heavy chain used for cellular entry (residues Ala448-Leu1295), and a light chain that contains the catalytic domain (residues Pro1-Lys437).640 This catalytic light chain is a mononuclear Zn2+-dependent metalloprotease that selectively hydrolyzes the SNARE proteins.641 The first crystal structure of BoNTA was published in 1998 (Figure 69), and shows the BoNTA active site is buried 20–24 Å deep, accessible via a 12×15×62 Å channel.642 The catalytic Zn2+ ion is coordinated by the His222, His226, Glu261, and a water molecule to complete the tetrahedral geometry at the metal ion. Residues Glu223 and Tyr365 are located in close proximity to the catalytic Zn2+ and are believed to play important roles in substrate binding and hydrolysis.
Figure 69.
Crystal structure of BoNTA depicting the light chain in gray and the heavy chain in cyan (PDB 3BTA).
Inhibitor Development.
To date, there appears to be no comprehensive literature review of BoNT inhibitor development. This Section will focus on the most pertinent metal-binding inhibitors of BoNT. Unless otherwise stated, inhibitors described in this Section are against BoNTA, as this serotype is considered the most relevant to human health and has historically been the focus of most drug development studies.
Compounds that act through a metal-striping mechanism comprised the earliest inhibitors of BoNT. EDTA, DTPA, and TPEN were all used to pre-treat tissues for protection against BoNT (Figure 70), and TPEN and DTPA were found to have similar apparent EC50 values of 20–40 μM, whereas EDTA was slightly less active with an apparent EC50 value a >160 μM.660 Later, the benzimidazole containing BoNTi-1 (IC50 = 5–10 μM) was reported, and is thought to act by a metal-stripping mechanism. The structure of BoNTi-1 bound to BoNTB show the inhibitor coordinated to an additional, second Zn2+ ion within the active site, while the catalytic Zn2+ remains in place but is perturbed by the displacement of the coordinating Glu residue (Figure 71).643 This unusual mode of metal-aided inhibition has been observed in other metalloenzyme systems (Section 6.6); however, optimization of activity and selectivity of these inhibitors is underdeveloped.
Figure 70.
Inhibitors of BoNT.
Figure 71.
Structure of: BoNTi-1 bound to BoNTB (note, the coordinating Glu is displaced from the catalytic Zn2+) (PDB 1G9D), BoNTi-9 bound to BoNTA (PDB 2IMA), and BoNTi-21 bound to BoNTA (PDB 5V8P).
One of the earliest discovered metal-binding BoNTA inhibitor classes were based on phosphonic acid MBPs (Figure 70). These compounds were discovered by seeking to repurpose known MMP inhibitors against BoNT.645 Naturally occurring phosphoramidon (IC50 = 14.7 mM against BoNTB) was a poor inhibitor, but when administered to mouse nerve tissue at 200 μM, was found to protect against lethal doses of both BoNTA and BoNTB. Phosphoramidon was originally designed to act as a metal-binding MMP inhibitor (Section 1.5) and NEP inhibitor (Section 6.7), and is presumed to similarly inhibit BoNT by binding to the catalytic Zn2+ ion through the phosphonic acid group. Subsequent work to replace the synthetically liable rhamnose moiety resulted in the discovery of compounds BoNTi-2, BoNTi-3, and BoNTi-4, of which the phenyl substituted BoNTi-4 was found to be the most effective with a poor IC50 value of 8.1 mM against BoNTB.644 It is worth noting that phosphoramidon was found to be more effective at protecting against BoNT in the tissue model645 than what would be expected from the poor activity reported in vitro enzymatic assay.644 This discrepancy between the in vitro enzymatic assay and cell culture and tissue models has been noted elsewhere, and suggests that there are relevant differences between the truncated form of BoNT used in in vitro assays and the intact form of BoNT used in in vivo assays that are not yet fully understood with respect to drug development.657
The most prolifically explored class of inhibitor against BoNT uses the hydroxamic acid MBP (Figure 70). However, the earliest tested hydroxamic acid, BoNTi-5, showed no activity at protecting mouse nerve tissue from BoNTA/B exposure when administered at 75 μM.645 Later, BoNTi-6, which was designed based on molecular dynamic simulations, was the first reported hydroxamic acid based BoNTA inhibitor to have protective activity with a Ki value of 12 μM.646
The subsequent development of compound BoNTi-9 marked an important milestone in inhibitor development and this compound has become a standard for evaluating new BoNTA inhibitors.647 Initial use of an in situ based screening approach lead to the discovery of BoNTi-7 and BoNTi-8, with IC50 values of 15 and 3 μM, respectively. The structural features of BoNTi-7 and BoNTi-8 were merged to give a synergistic improvement in activity, resulting in compound BoNTi-9. A crystal structure of BoNTi-9 bound to BoNTA shows that BoNTi-9 binds with the hydroxamic acid coordinating the catalytic Zn2+ ion, and the inhibitor backbone engaged in multiple interactions with residues around the active site (Figure 71).662 In particular, the aromatic of BoNTi-9 ring makes stacking interactions with several Phe residues, and the o-chlorine of BoNTi-9 makes a strong halogen-cation interaction with a neighboring Arg. This halogen-cation interaction is not present in BoNTi-7, and likely explains why BoNTi-9 has much improved activity. In vivo mouse studies found BoNTi-9 to have limited efficacy at protecting 16% of animals from lethal doses of BoNTA when administered at 1 mM doses, with the survivors showing no signs of Botulism disease.663
Additional investigations led to the discovery of an arginine-hydroxamic acid conjugate BoNTi-10 that displays 75% inhibition of BoNTA at 50 μM.648,664 Use of LC-MS based screening identified adamantane hydroxamic acid BoNTi-11, which was found to have a Ki of 460 nM.649 Follow-up studies to improve the activity of BoNTi-11 resulted in the discovery of BoNTi-12 with an improved Ki of 27 nM; however, this compound was determined to have poor PK and was unable to cross cell membranes to provide protection against BoNT.650 Still other approaches have focused on occupying large amounts of three-dimensional space in the protein active site, and use the same hydroxamic acid MBP.652,653
Other compounds have used sulfur MBPs to exploit the mixed hard-soft Lewis acidic character of the Zn2+ active site ion in BoNT (Figure 70). It is worth noting that the FDA approved ACE inhibitor L-captopril (Section 6.1), which is often used as a probe inhibitor for Zn2+-dependent metalloenzymes, was not found to be effective at protecting mouse tissue from BoNT.645 Instead, the first reported sulfur-based inhibitor to show inhibitory activity against BoNTA in vitro, BoNTi-18, was a truncated peptide substrate with a D-cysteine added for metal-binding interactions.654 BoNTi-18 had a Ki value of 1.8 μM, and many other inhibitors follow a similar tactic of using thiol MBPs with a substrate mimetic peptide, as illustrated by BoNTi-19. Breaking away from peptides, other small molecule sulfur-based BoNT inhibitors, such as BoNTi-20 had modest activity.656 A co-crystal structure of BoNTi-21 confirms that the inhibitor acts through a metal-binding mechanism with the sulfur coordinating the catalytic Zn2+ in a tetrahedral geometry, with the rest of the compound making similar interactions as BoNTi-9 (Figure 71). Work to improve the PK of BoNTi-20 led to BoNTi-21, which is less active (IC50 = 13.7 μM) when compared to BoNTi-20, but was reported to have much better activity in vivo. Mice treated with 0.1 mL of a 10 mg/mL solution of BoNTi-21 were able to prolong the onset of Botulism symptoms by ~18 hours.657 This study again highlights the discrepancy between in vitro enzymatic activity and in vivo results for BoNT inhibitors.
The latest metal-binding motif to be used in BoNT inhibitors has been the 8-hydroxyquinoline (8HQ) scaffold (Figure 70). While 8HQ itself was found to have essentially no activity against BoNTA in vitro, derivatives containing the 8HQ scaffold have displayed good activity against BoNT in vivo.661 The first 8HQ BoNT inhibitor was discovered through a virtual screen of an NCI library followed by a HPLC-based protease assay screen, which initially identified BoNTi-22 as having 52% inhibition against BoNTA at 20 μM.658 A brief SAR study around BoNTi-22 led to the discovery of several other BoNT inhibitors.665–667 Most notable of these was BoNTi-23.659 This study found that the 2-position only accommodates small substitutients, and that the 5-position represents another site to further elaborate inhibitors. Furthermore, BoNTi-23 was also found to be effective in cell models as well as mouse muscle models at protecting from BoNTA exposure. Lastly, the natural product lomofungin, which contains the 8HQ scaffold, was also found to inhibit BoNTA with a Ki value of 6.7 μM, but this compound is not believed to act through a metal-binding mechanism as kinetic studies found lomofungin to have a noncompetitive mode of binding.661
Current and Future Prospects.
BoNT, especially BoNTA, has been shown to persist in the neural cytosol for months after the initial exposure, where the neurotoxin evades normal degradation pathways and continues to hydrolyze SNARE peptides, prolonging the paralytic effects.668 Therefore, it is of great interest to develop inhibitors against this highly lethal enzyme. While many drug discovery efforts have been made against BoNT, no BoNT inhibitors have been tested in clinical trials or reached FDA approval. BoNT has historically been a very difficult enzyme to target,646 in part, because the active site is highly flexible making predictions for rational SAR difficult.653,657,662,667,669,670 Furthermore, in vitro assays typically use the truncated light chain form of BoNT, with the heavy chain removed for safety; however, this single chain is less active than the native, two chain form.641 The truncated enzyme used for in vitro assays is often not representative on the full-length metalloenzyme activity, and inhibitors generally display different activity in cellular and in vivo screening.651,657,658,671,672 Greater efforts into developing accurate in vitro assays and screening protocols would greatly benefit BoNT inhibitor advancement. Additionally, the SAR around inhibitor development has been rather flat in that most small molecule inhibitors have activity >1 μM, indicating that there is room for inhibitor improvement. It is worth noting that there is limited availability equine-derived antibodies to treat BoNT exposure; however these must be administered with a narrow therapeutic window of approximately 24 hours post exposure before the toxin is taken up within the nerve synapse.633,657,671,673 In the event of an unexpected bioterrorism attack involving BoNT, prophylactic antibiotic treatment is not realistic, and would have catastrophic impact on civilian areas. Therefore, it is of great importance to improve assay screening protocls for BoNT in order to develop new inhibitors aginst this enzyme.
6.5. Elastase B (LasB, pseudolysin, EC 3.4.24.26)
Function in Biology and Disease.
The emergence of antibiotic resistance mechanisms (such as β-lactamases) is an immediate threat to public health, and the discovery of new antibacterial drugs are of urgent need. Pseudomonas aeruginosa is a Gram-negative bacterium isolated from nosocomial settings, causing acute infections in immunocompromised patients and individuals suffering from cystic fibrosis.674,675 The survival of the bacteria, as well as the ability of the bacteria to infect the host, relies heavily on virulence factors, such as bacterial adhesion molecules, toxins, and proteases.676 LasB (also known as elastase or pseudolysin) is a key virulence factor excreted by P. aeruginosa. The production of LasB plays a two-fold disruptive role in pseudomonal infections: first, LasB damages the tissue of host cells by enzymatically hydrolyzing various components of the extracellular matrix and degrading biomolecules involved in innate immunity including immunoglobulins, complement factors, and cytokines.677,678 The destruction of host tissue liberates nutrients for bacterial growth and accelerates the proliferation of P. aeruginosa. Additionally, the production of LasB is required for swarming, a migration of highly differentiate bacterial cells,679 and biofilm production.680 Biofilms consists of exopolysaccharide matrices and act as a as a protective layer for the bacteria. Biofilms block the access of antibiotics and immune response agents, and leads to an increased bacterial resistance to antimicrobials (resistance within biofilms can be enhanced as much as 1000-fold).681–684 The defense and proliferation virulence mechanisms of LasB make it an attractive antimicrobial target for the treatment of P. aeruginosa. Inhibitors of LasB are effective in delaying the progression of corneal tissue destruction,685–688 and reducing the severity of lung infections.689 There are currently no approved drugs for targeting LasB for the treatment of P. aeruginosa infections.
Protein and Active Site Structure.
LasB is a Zn2+-dependent metalloenzyme belonging to the bacterial neutral metalloprotease family. LasB is a protein of ~33 kDa with an overall tertiary structure similar to that of thermolysin, albiet only sharing 28% sequence homology with thermolysin.690,691 The secondary structure of LasB is constructed primarily of antiparallel β-sheets, with the C-terminus bearing an α-helix (Figure 72). Two helices and the loop which connects them (from residues 135 to 178) span the active site and provide the ligands for Zn2+ coordination. The Zn2+ ion is ligated by His140, His144, Glu164, and a water molecule in a tetrahedral coordination geometry (Figure 72). Similar to thermolysin, the water molecule is polarized by Zn2+ and stabilized by hydrogen bonding by a conserved Glu141 in the second coordination sphere.690 Two hydrophobic binding pockets (S1’ and S2’) in the active site have been the focus of inhibitor development. The deeper S1’ pocket is the principle determinate of specificity while the S2’ pocket is displays lower substrate selectivity.692 Inhibitors with large aromatic residues that target the S1’ and S2’ pockets are most active against LasB.
Figure 72.
Structure of LasB (PDB 1EZM).
Inhibitor Development.
Peptide derivatives bearing a 2-mercaptoacetyl MBP constitute a major family of LasB inhibitors (LasBi-1 and LasBi-2, Figure 73).685,686,692–694 In 1982, a series of peptide (Leu and Phe) derivatives bearing various MBPs (hydroxamic acid, phosphoryl, and thiol) were synthesized and screened against LasB.685 Inhibitors bearing a 2-mercaptoacetyl MBP were identified as active inhibitors of LasB (Ki = 0.2 – 34 μM), with LasBi-1 achieving a Ki value of 0.2 μM. Comparison of LasB inhibition by LasBi-1 and the dipeptide Phe-Leu (a non-MBP bearing derivative) revealed a 2000-fold increase in activity for LasBi-1 (Ki = 400 μM), validating the importance of metal binding. Further in vitro studies showed LasBi-1 effectivity prevented the degradation of cartilage proteoglycan.685 Injection of LasBi-1 and the antibiotic gentamicin in rabbit eyes affected with Pseudomonas keratitis, significantly reducing the infection compared to those treated with gentamicin alone.687,688 There are no structures of LasBi-1 bound to LasB available, but one of two binding modes is anticipated. One possibility is the 2-mercaptoacetyl motif can bind in a bidentate fashion to the Zn2+ ion, forming a trigonal bipyramidal coordination geometry (similar to that of 2-mercaptoacetyl MBP inhibitors bound to NEP, Section 6.7). A second possible binding mode includes the thiol group of the 2-mercaptoacetyl MBP binding in a monodentate fashion to the Zn2+ ion, resulting in a tetrahedral coordination geometry. An extensive 2-mercaptoacetyl MBP dipeptide library containing 160 inhibitors with various basic, aromatic, large aliphatic, and acidic side chains has been synthesized.693 This library revealed inhibitors possessing large hydrophobic side chains in the P1’ and P2’ positions (corresponding to the S1’ and S2’ subsites) are most active for LasB inhibition, with futher studies producing LasBi-2 with a Ki value of 41 nM (Figure 73). However, no further studies were performed with these 2-mercaptoacetyl dipeptide inhibitors. Although the backbone of LasB inhibitors has been explored and optimized, the usage of a 2-mercaptoacetyl MBP has remained largely unchanged.
Figure 73.
Inhibitors of LasB.
In order to identify a non-peptide small molecule inhibitor of LasB, a library of MBPs was screened against LasB. This screen revealed 3-hydroxy-1,2-dimethylpyridine-4(1H)-thione (LasBi-3) and tropolone (LasBi-5) as hits with moderate IC50 values of 81 μM and 387 μM, respectively.695,696 LasBi-3 is expected to coordinate to the Zn2+ through the O,S donor pair in a bidentate fashion, resulting in a five-coordinate trigonal bipyramid coordination geometry. Elaboration of the LasBi-3 MBP hit by the addition of a biphenyl substituent at the endocyclic nitrogen position resulted in an inhibitor with 30-fold increase in activity (LasBi-4, IC50 = 2.7 μM). LasBi-4 displayed anti-swarming properties against P. aeruginosa in a bacterial phenotypic assay, indicative of on-target activity against LasB.695 However, the indiscriminate inhibition of LasBi-3 and LasBi-4 of other Zn2+-dependent metalloenzymes (MMPs, anthrax LF) precluded further development of LasBi-4.697 In order to develop more potent and selective inhibitors of LasB, the tropolone MBP (LasBi-5) was investigated. Based on known metal coordination of LasBi-5 against Zn2+-dependent thermolysin and SAR from LasBi-5 analogs, the O,O donor pair is expected to bind bidentate to the Zn2+ in a trigonal bipyramid coordination geometry.696 LasBi-5 was further elaborated to yield LasBi-6 (IC50 = 1.2 μM), a very hydrophobic, but active non-peptide inhibitor of LasB. LasBi-6 was selective for LasB when compared to a small panel of metalloenzymes (MMP-2, MMP-9, hCAII, tyrosinase), and displayed antiswarming properties against P. aeruginosa.
Unfortunately, no structures of the aforementioned inhibitors complexed with LasB are available; however, two structures each bearing a novel MBP, have been reported. This includes LasBi-7, bearing a carboxylic acid MBP and LasBi-8, bearing a phosphonate MBP (Figure 73). In both structures, the MBP is bound monodentate fashion to the Zn2+ ion, which is further ligated by residues His140, His144, and Glu164 in a tetrahedral coordination geometry (Figure 74).
Figure 74.
Left: Structure of LasBi-7 bound to LasB (PDB 1U4G). Right: Structure of LasBi-8 bound to LasB (PDB 3DBK).
Current and Future Prospects.
Drugs that target bacterial virulence factors have been coined as a “second generation” antibiotic approach. While bactericidal antibiotics have traditional been effective, their clinical usefulness is rapidly diminishing due to the evolution of bacterial antibiotic resistance mechanisms and a slow progress in the development of new antibiotics. Antibiotics that target bacterial virulence factors should be bacteriostatic, and provide many potential advantages over current antibiotics, including preservation of the host endogenous microbiome and exerting weaker selective pressure. However, due to the specificity of the drug against the target virulence factor(s), clinicians must precisely identify the bacterial infection, resulting in a narrow window of opportunity for treatment.676 Furthermore, the economic incentive for pharmaceutical companies to develop narrow-spectrum antibiotics may be even less than that of broad-spectrum inhibitors. As such, the potential of this second generation approach is to be determined, but there is growing interest in these types of antibiotics.676
LasB is a key virulence factor excreted by P. aeruginosa, causing tissue damage to the host cells and required for bacterial swarming and biofilm production. Inhibition of LasB has proven to be an effective method for treatment of Pseudomonas keratitis and reduce the severity of lung infections. To date, extensive libraries of 2-mercaptoacetyl MBP dipeptides have been synthesized to yield LasBi-2 as the most potent inhibitor reported to date (Ki = 0.04 μM). Despite efforts to optimize the inhibitor backbone, the 2-mercaptoacetyl MBP has remained dominant for this target. Additionally, the lack of structural data (only three crystal structures are available for LasB) also creates a roadblock to inhibitor development. Future work in LasB inhibitor development would benefit from additional crystallographic data, investigating alternative MBPs, and utilizing other non-peptide based backbones. Interestingly, in 2017, the CARB-X consortium awarded the biotech company Antabio $8.9 million in funding to pursue a program on LasB inhibitors. This program seeks to develop a first-in-class small molecule inhibitor of LasB which could be co-administered antibiotics for the treatment of P. aeruginosa infections in patients suffering from cystic fibrosis. While no results have been disclosed, this effort may help validate the potential of inhibiting virulence factors, specifically LasB, as a new antibacterial target.
6.6. Methionine Aminopeptidase (MetAP, EC 3.4.11.18)
Function in Biology and Disease.
Angiogenesis, or the development new blood vessels from existing vasculature, is an attractive cancer target for complementary treatments to cytotoxic therapies.698,699 For cancerous tissues to grow beyond a size of 2–3 mm3, as well as to metastasize, direct access to nutrients and the vascular system is required and supplied by new blood vessels.698 One metalloenzyme that has been linked to angiogenesis is human type-2 methionine aminopeptidase (hMetAP2).700 Methionine aminopeptidases (MetAPs) are responsible for cleaving the N-terminal methionyl residue from all translated proteins when the second amino acid is Ala, Cys, Gly, Pro, Ser, Thr, or Val (with certain exceptions) using a coordinated water molecule (Figure 75).701 This modification is estimated to occur for at least 50% of all proteins, and prevention of this modification causes a blockade of further N-terminal modifications which is suspected to be the mode of hMetAP2 inhibition that leads to blockage of angiogenesis. Additionally, hMetAP2 inhibition is implicated as a treatment for obesity based on patients in preliminary clinical trials exhibiting significant weight loss, as dynamic angiogenesis is involved in adipose tissue maintenance.702 Due to MetAPs catalyzing an important translational modification, it is unsurprising they are ubiquitous in most life forms. As such, they have been found to also be valid targets for antibiotics, based on knockdown studies of MetAP that result in decreased proliferation and/or cell death.703,704 Based on the broad range of clinical implication there has been a significant interest in targeting human and bacterial MetAPs.
Figure 75.
The reaction catalyzed by MetAPs is the N-terminal Met excision of translated proteins.
Protein and Active Site Structure.
MetAPs are ubiquitous throughout the phylogenetic scale and were found to possess a “pita-bread” fold (Figure 76).705,706 This protein class was initially divided into two classes, MetAP1 and MetAP2, based on sequence alignments.707 The distinction between the two classes is MetAP2 enzymes possess an additional ~60 residues in an α-helical subdomain in the catalytic domain. MetAPs are further differentiated by the absence (subclass a) or presence (subclass b) of an N-terminal domain.708 For example E. coli MetAP (EcMetAP) is a type 1 subclass a enzyme, while hMetAP2 is a type 2 subclass b enzyme (although the N-terminal domain is frequently truncated for structural studies of hMetAP2). The metal coordinating residues are highly conserved across eukaryotic and prokaryotic organisms.707 In MetAPs, two Asp, two Glu, and one His residues are involved in metal coordination. The coordination geometry at the dinuclear active site of E. coli MetAP is different for each metal ion. From the structure of Co2+ constituted EcMetAP (Figure 76),709 CoA is coordinated in a monodentate fashion by Asp108, His171, Glu204, Glu235, and a bridging water producing distorted bipyramidal geometry. The coordination geometry at CoB is distorted octahedral with Asp108, Glu235, a bridging water, and a terminal water coordinating in a monodentate fashion and Asp97 coordinating in a bidentate fashion. This dinuclear active site coordination motif is also observed in hMetAP2 as the coordinating residues are highly conserved among MetAPs.
Figure 76.
Structure of Co2+ reconstituted EcMetAP (PDB 2MAT).
The overall sequence of MetAPs at the active site are very similar with the structures generally being the same,706 although comparisons between EcMetAP and hMetAP2 show some structural differences, largely due to the additional residues in the α-helical subdomain of hMetAP2.710 MetAPs are generally characterized as dinuclear metalloenzymes, but have been shown to be catalytically competent with one equivalent of an activating metal ion in the case of EcMetAP.711 As a class, MetAPs are catalytically competent with a wide range of first row transition metals, including Mn2+, Fe2+, Co2+, Ni2+, and Zn2+, although MetAPs from different organismal sources can be activated by some of these metals and not others.712 Co2+ has frequently been utilized to reconstitute the enzyme, particularly in earlier inhibition and structural studies (Figure 76).713 However, biochemical studies suggest that Mn2+ is the native cofactor of hMetAP2,714 while Fe2+ is the suggested native cofactor of EcMetAP.715
Inhibitor Development.
The significant interest in hMetAP2 as an anticancer target began with the serendipitous discovery of the natural product fumagillin (Figure 77) being a potent angiogenesis inhibitor.716 It was quickly determined that type II MetAPs were the target of fumagillin with the compound acting as a covalent inhibitor of the hMetAP2. Synthetic analogs, such as TNP-470, resulted in very active inhibitors (IC50 = 1.3 nM).700,717 TNP-470 was tested in cancer clinical trials but ultimately failed due to neurotoxicity.718 Beloranib is another fumagillin analog that was developed for weight loss in patients with Prader-Willi syndrome (Figure 77). Unfortunately, beloranib was terminated in Phase III clinical trials due to significant side effects and patient deaths.719 Although fumagillin based covalent inhibitors have failed in clinical trials, hMetAP2 has remained a target of significant interest, especially with the development of reversible inhibitors that bind the active site metal ions.
Figure 77.
Inhibitors of MetAP.
Another natural product class, bengamides collected from sea sponges, were found to have potent anti-tumor activities.720 In an effort to identify their mode of action, synthetic derivatives of bengamides were synthesized with improved solubility and proteomics studies were performed, identifying hMetAP2 as the target for this class of natural products.721 Two bengamides derivatives, LAF153 and LAF389 (Figure 77), showed good activity (LAF389, IC50 = 800 nM) against Co2+ reconstituted hMetAP2 (Co-hMetAP2). A crystal structure of LAF153 was obtained in complex with Co-hMetAP2 (Figure 78) that revealed LAF153 coordinates both active site metals via three hydroxyl groups, resulting in bidentate coordination of both active site metal ions. Additionally, the t-butyl moiety makes significant hydrophobic contacts with the active site while the caprolactam ring is largely solvent exposed. Unfortunately, in patients with advanced cancer, LAF389 was unsuccessful due to cardiovascular toxicity.722
Figure 78.
Structures of hMetAP2 with: LAF153 (PDB 1QZY), A311263 (PDB 1R5G), MetAPi-2 (PDB 2ADU), MetAPi-4 (PDB 2OAZ), MetAPi-8 (PDB 5JI6), and MetAPi-10 (PDB 5LYX). Crystal structures of EcMetAP with: MetAPi-16 (PDB 1XNZ), MetAPi-18 (PDB 4A6W), MetAPi-20 (PDB 3D27). Co2+ ions shown as blue spheres and Mn2+ ions shown as magenta spheres.
Being an aminopeptidase, efforts were made to utilize previous aminopeptidase inhibitor scaffolds for MetAP inhibitor development.723,724 Substituted 3-amino-hydroxamides and acylhydrazines were developed and found to be potent inhibitors of MetAP. Three of the more potent inhibitors against Mn2+-reconstituted hMetAP2 (Mn-hMetAP2) were A357300 (IC50 = 110 nM), A311263 (IC50 = 30 nM), and A320282 (IC50 =180 nM) (Figure 77). These three inhibitors were also crystalized with Mn-hMetAP2 and all showed similar bidentate coordination motifs to both active site ions, as exemplified by A311263 (Figure 78). Further testing of in vivo studies with A357300 showed significant anti-angiogenesis and anticancer activity.725 Attempts to modify the 3-amino-2-hydroxyamide scaffold by substituting in a hydroxamic acid MBP, resulted in compounds that were less active than the parent scaffold but exhibited higher activity against EcMetAP.726
HTS has also been employed to identify lead compounds for MetAP inhibitor development. One such a campaign lead to the identification of 4-aryl-1,2,3-triazoles (Figure 77),727 which exhibited good activity with simple derivatives such as 4-methyl-phenyl-1,2,3-triazole (MetAPi-1, Ki = 15 nM) against the Co-hMetAP2. However, the activity of MetAPi-1 was much poorer when tested against Mn-hMetAP2 (Ki = 4.4 μM) and similarly in vivo activity for this compound was poor. A crystal structure was obtained showing a similar derivative 3-methyl-phenyl-1,2,3-triazole (MetAPi-2, Ki = 18 nM against Co-hMetAP2) binding to the active site of Co-hMetAP2 (Figure 78). From the structure, it is observed that two nitrogen atoms of the triazole MBP bind to and bridge the active site metal ions. Additionally, a third nitrogen atom appears to engage in hydrogen bonding interactions with a nearby His residue. The SAR for the aryl substituents was fairly consistent with small hydrophobic (methyl or halogen) being preferred over small hydrophilic ones (amine or hydroxyl). Another HTS campaign yielded a similarly related 1,2,4-triazole scaffold, 3-anilino-5-benzylthio-1,2,4-triazole (MetAPi-3, Figure 77), as an excellent Co-hMetAP2 inhibitor (Ki = 500 pM).728 SAR of the scaffold was fairly successful with modulations in the electronic nature of the aniline substituents causing moderate changes in activity, although deviation from a benzyl substituent at the thiobenzyl position exhibited either similar or poorer activity. The activity of these 1,2,4-traizole derivatives was consistent with previous 1,2,3-triazoles, in that they exhibited a 2–3 order of magnitude loss in activity against Mn-hMetAP2. A crystal structure of Co-hMetAP2 with MetAPi-4 (Ki = 2 nM) bound to the active site was obtained (Figure 78), which showed the 1,2,4-triazole MBP in a similar coordination motif as observed with the 1,2,3-triazole based inhibitors. A separate HTS campaign yielded a 1,2,4-triazole containing drug candidate, which unfortunately did not exhibit in vivo activity.745 Structurally similar to the 1,2,4-triazole based inhibitors, oxadiazoles have also been explored as MetAP2 inhibitors.746
Another class of hMetAP2 inhibitors that were discovered and developed from screening campaigns are anthranilic acid sulfonamides, which were further developed through the aid of structural studies.729 The lead inhibitor from the screen was 5-bromo-2-((4-chlorophenyl)sulfonamido)benzoic acid (MetAPi-5, Figure 77) and was evaluated against Mn-hMetAP2 (IC50 = 9.1 μM). A crystal structure with MetAPi-5 bound to Mn-hMetAP2 was obtained showing that the carboxylic acid is bound in a monodentate fashion to MnA. Significant gains in activity were gained by modifying the anthranilic portion of the MetAPi-5 and the replacement of the chloro-phenyl substituent for a fluoro-phenyl substituent. Unfortunately, addition of human serum albumin caused significant decreases in inhibitory activity (~2–3 order of magnitude decrease). To address this issue, further modification produced MetAPi-6 (IC50 = 19 nM),730 which exhibited good in vivo activity (EC50 = 36 nM), as well as significantly lower binding to human serum albumin.
A FBDD approach focusing on indazole scaffolds was used to develop hMetAP2 inhibitors for treatment of obesity.731 A lead compound of interest was a 6-bromo-1H-indazole (MetAPi-7, Figure 77), which exhibited activity against Mn-hMetAP2 (IC50 = 1.1 μM). No crystal structure was reported with the lead scaffold, but modeling predicted a nitrogen from the indazole MBP engaging in monodentate binding with an active site Mn. Utilizing the proposed mode of binding, addition of a range of substituents at the 4- and 6-positions of the indazole ring generated a strong SAR that was validated by a crystal structure obtained with 4-(3-methylpyridin-4-yl)-6-(trifluoromethyl)-1H-indazole (MetAPi-8, IC50 = 11 nM), with the indazole MBP coordinated to an active site Mn2+ in a monodentate fashion (Figure 78). The structure also showed the second indazole nitrogen was engaged in a hydrogen bonding interaction with a metal bound water as well. Further modification of the 4-position yielded bioactive compounds that exhibited weight loss activity when tested in mouse models. The work on indazoles lead to a pyrazolo[4,3-b]indole as a potential scaffold to develop into a hMetAP2 inhibitor for targeting obesity.747 This scaffold was shown to have the same interactions with the active site metals, in addition to efficacy in vivo.
Another HTS campaign was reported where elaborated purines were identified as Mn-hMetAP2 inhibitors.732 The lead purine compound achieved a modest gain (<10-fold) in activity, with attachment of a 5-isoquinoline being the most active (MetAPi-9, IC50 = 230 nM, Figure 77). Elaboration of the purine with even a methyl group resulted in decreased activity. Additional efforts were put into modifying the core purine scaffold, which was suspected to be the metal binding motif. These modification modulated the electronics of metal binding and resulted in a more potent scaffold [1,2,4]triazolo[1,5-a]pyrimidine (MetAPi-10, IC50 = 38 nM). Structural analysis of purine derivatives MetAPi-11 (IC50 = 700 nM) and MetAPi-10 bound to Mn-hMetAP2 (Figure 78) showed the purine and pyrimidine MBPs were coordinating in a monodentate fashion using a nitrogen atom to each active site Mn2+, with other substituents effectively sitting in the same positions within the active site.
Additional campaigns have yielded pyridine pyrimidine based inhibitors of Co-hMetAP2.733 This family of inhibitors are structurally related to MetAPi-12 (IC50 = 0.8 μM, Figure 77) and exhibited IC50 values in the range of ~1 to ~10 μM. No structure of these scaffolds bound to hMetAP2 was provided; however, a crystal structure of Co2+ reconstituted hMetAP1 with a similar pyridine pyrimidine inhibitor shows the compound coordinating a third Co2+ ion in the active site. A further screening campaign discovered that the approved antibiotic nitroxoline (Figure 77) was a very active inhibitor of Mn-hMetAP2 (IC50 = 55 nM), which showed in vivo activity against cancer mouse models.744 Based on the high bioavailability of this compound in the urine, it has potential as a treatment against certain bladder cancers.
Apart from targeting hMetAP2, bacterial MetAP inhibitors have also been widely studied and categorized into several inhibitor classes including: 1,2,4-triazoles, biaryl chelators, aryl carboxylic acids, quinolines, peptides, thiazoles, and bengamides (Figure 77).748 Some of these inhibitor classes have been structurally analyzed against hMetAPs and exhibit similar modes of binding against bacterial MetAPs. This is typified by the 1,2,4-triazole based inhibitors, similar to MetAPi-3 and MetAPi-4, which exhibit inhibitory activity against different metalloisoforms of MetAPs from M. tuberculosis,749 S. aureus,750 A. baumannii,751 and B. pseudomallei.752 Structural characterization of a 1,2,4-triazole species bound to the active site of MetAP1c from M. tuberculosis (MtMetAPlc) showed the triazole coordinating both active site Ni2+ atoms in a bridging fashion, in a similar manner as MetAPi-4 when coordinating the active site metals in Co-hMetAP2 (Figure 78).753
The biaryl chelator class of inhibitors has been structurally analyzed against both human and bacterial MetAPs. Included in this class are metal binding scaffolds such as pyridinylpyrimidine (MetAPi-12),733,754 2-(pyridin-2-yl)-1H-imidazo[4,5-b]pyridine (MetAPi-13),735 and 4-(1H-imidazo[4,5-b]pyridin-2-yl)thiazole (MetAPi-14, Figure 77).735 Generally, this class of inhibitors has been analyzed against Co-EcMetAP and typically displays IC50 values in the ~0.1–5 μM range, with MetAPi-14 being one of the more active scaffolds (IC50 = 78 nM).735 A crystal structure of a similar biaryl chelator to MetAPi-14 bound to Co-EcMetAP showed the inhibitor coordinating a third, exogenous Co2+ ion in the active site.755 This recruitment of a third metal ion in MetAP has been observed in other structures and is suspected to be an artifact of crystallization. MetAPi-13 exhibited ~250-fold selectivity for EcMetAPs (IC50 = 110 nM) over human hMetAPs.734
The quinoline class of bacterial MetAP inhibitors has focused on the 8-hydroxy and 8-amino sulfonamide scaffolds (Figure 77). Crystallization of MetAPi-15 with MetAP showed the compound bound in a bidentate manner to a third auxiliary Mn2+ atom in the active site.736 Again, as with MetAPi-14, this extra metal ion was likely due to crystallization conditions. MetAPi-15 as well as certain derivatives, exhibited some metalloisoform selectivity towards Ni2+ and Co2+ reconstituted EcMetAP (IC50 = 184 nM and 137 nM, respectively). The 8-hydroxyquinoline scaffold was tested for inhibitory activity against Co2+ reconstituted B. pseudomallei MetAP (Co-BpMetAP) using nitroxoline (IC50 = 60 nM).743 Substitution of the nitro group with alternative functional groups (such as a sulfonate) resulted in decreased activity. The 8-hydroxyquinoline scaffold, specifically nitroxoline, is generally not selective for bacterial over human MetAPs.
The bengamide class of bacterial MetAP inhibitors has also been analyzed against MtMetAP.756 Derivatives of this class lacking the caprolactam ring were active against the different metalloisoforms of MtMetAps, with some selectivity for Mn-MtMetAP (IC50 = ~0.2 - ~1.0 μM); however, these inhibitors only showed modest in vivo activity. Crystallographic analysis shows that this class of compounds coordinate the Mn-MtMetAP1c active site metals in the same manner as in the LAF153 bound hMetAP2 structure (Figure 78). Similar binding was observed with a peptide based inhibitor utilizing a 3-amino-2-hydroxy amino acid motif (similar to A311263, Figure 77).757 In terms of inhibitory activity, these peptide-derived inhibitors did not exhibit metalloisoform selectivity, with modest activity against EcMetAP (IC50 = 10–20 μM) reconstituted with Co2+, Mn2+, Zn2+, or Ni2+.758
Efforts to develop metalloisoform selective inhibitors for EcMetAP were undertaken beginning with a HTS campaign to find fragments with metalloisoform selectivity.737 A compound library was screened against Co-EcMetAP and Mn-EcMetAP, with hits being subsequently tested against the Mn2+, Fe2+, Co2+, and Ni2+ metalloisoforms of EcMetAP. From these studies, the aryl carboxylic acids (MetAPi-16) were found to be selective for Mn-EcMetAP with good activity (IC50 = 240 nM). Crystallographic studies with Mn-EcMetAP and MetAPi-16 show that the aryl carboxylate MBP coordinates one of the active site Mn2+ in a bidentate manner (Figure 78).
To develop inhibitors with alternative metalloisoform selectivity against EcMetAP, hydroxamic acid MBPs were utilized.738 Direct replacement of the carboxylate on MetAPi-16 for a hydroxamic acid resulted in MetAPi-17 (Figure 77) with better inhibitory activity against all tested metalloisoforms (e.g., Fe-EcMetAP, IC50 = 116 μM vs. 0.79 μM). Efforts to increase the metalloisoform selectivity for hydroxamic acid aryl inhibitors for Fe-EcMetAP were successful by varying the heterocyclic 5-membered ring.739 Structural studies with Mn-EcMetAP were performed with MetAPi-18 (Figure 77) showing the hydroxamic acid MBP coordinated in a bidentate manner to an active site Mn2+, with the N-hydroxyl group bridging both active site ions (Figure 78). Further attempts to develop Fe-EcMetAP inhibitors included a HTS campaign against Fe-EcMetAP.740 The screen yielded compounds with catechol MBPs, with MetAPi-19 (Figure 77) being the lead hit against Fe-EcMetAP (IC50 = 13 μM) and exhibited no significant inhibitory activity against Co- or Mn-EcMetAP. Crystallography studies with a catechol containing inhibitor MetAPi-20 (Figure 77), which exhibited similar activity for Fe-EcMetAP (IC50 =13 μM), but lower metalloisoform selectivity than MetAPi-19, showed the catechol motif coordinated to an active site Mn2+ in a bidentate fashion, with one phenolic oxygen bridging both active site ions. Catechol inhibitors exhibited activity against E. coli strains, verifying that iron is likely the relevant metal ion for EcMetAP in vivo.
Finally, thiazole containing MetAP inhibitors are largely comprised of three main scaffolds: N-(2-thiazolyl)picolinamide (MetAPi-21), N-(2-thiazolyl)thiazole-4-carboxamide (MetAPi-22), and N-(2-thiazolyl)oxamide (MetAPi-23) (Figure 77). This class of bacterial MetAP inhibitors has largely been characterized against Co-EcMetAP. The core scaffold in MetAPi-21 was found to have modest activity against Co-MetAP (IC50 = 5 μM), with further derivatization increasing in vitro activity but decreasing in vivo activity.741 Replacement of the pyridine moiety of MetAPi-21 with a thiazole resulted in MetAPi-22 and improved activity (IC50 = 110 nM), although further derivatization generally yielded similar to modest (~10-fold) improvements in activity and the compounds did not possess as potent in vivo activity.742,759 The thiazole oxamide scaffold present in MetAPi-23 was tested against Co-EcMetAP and found to be active with an IC50 value of 67 nM.737 Unfortunately, the only crystal structure of a thiazole containing inhibitor, with MetAPi-23 bound to Co-EcMetAP, shows coordination of a third, auxiliary Co2+.760
Current and Future Prospects.
hMetAP2 as a clinical target remains attractive for anticancer and weight loss interventions. However, further development of hMetAP2 inhibitors will require overcoming side effects that have been observed clinically. In the case of Beloranib, a pharmacologically improved fumagillin derivative,761 patient deaths in an obesity study group were the cause of its Phase III trial termination, even though the trials showed extremely promising results for obesity treatments.719 Novel fumagillin derivatives may remain viable prospects as there are several Phase I clinical trials examining new derivatives. In addition, a Phase I clinical trial from Merck is examining an undisclosed hMetAP2 inhibitor for treatment of advanced solid tumors. Finally, bacterial MetAPs are potential antibacterial targets, where the development of species selective inhibitors remains important. However, the promiscuous nature of bacterial MetAPs in terms of the range of metals that activate them, may prove to be an impediment to effectively developing potent antibiotics against this target.
6.7. Neutral Endopeptidase (Neprilysin, NEP, EC 3.4.24.11)
Function in Biology and Disease.
Atrial natriuretic peptide (ANP) is a 28 residue C-terminal peptide hormone with diuretic, natriuretic, and vasodilatory properties.762 Administration of ANP as a method of treatment for patients with systematic hypertension, chronic renal failure, as well as chronic heart failure (CHF) has been successful; however, poor oral absorption and rapid clearance of the polypeptide from circulation have hindered this treatment as an effective mode of therapy.762–766 The physiological degradation of ANP is caused by a Zn2+-dependent mammalian type II integral membrane endopeptidase, known as neutral endopeptidase (NEP). NEP, originally extracted and purified from the brush border membranes of rabbit kidneys, is a member of the M13 family of metalloproteases and involved in the cleavage of biologically active peptides, such as ANP, opioid peptides, amyloid β-protein, brandykinin, and gastrin.767,768 NEP cleaves ANP at the N-terminus of hydrophobic or aromatic residues (such as between Cys and Phe) via the mechanism of carboyxpeptidase A and thermolysin-like protease.767–770 The inhibition of NEP has shown to extend the half-life of ANP, prolonging its diuretic and vasodilatory properties, and is a useful form of treatment for hypertension and CHF.767 Inhibitors of NEP have been investigated in combination therapy with angiotensin-converting enzyme (ACE) inhibitors (Section 6.1) as vasopeptidase inhibitor. The vasopeptidase inhibitor Entresto is currently used to help reduce the risk of cardiovascular death in heart failure patients.771 The inhibition of NEP provides an anti-hypertensive and anti-fibrotic effects, while inhibition of ACE lowers the blood pressure of patients.
Protein and Active Site Structure.
The structure of the extracellular domain of human NEP (~81 kDa) bears two largely α-helical subdomains.771,772 Both subdomains, along with inter-domain linker fragments, form a spherical cavity ~20 Å in diameter in which the active site is buried. The N subdomain is 321 residues long and contains the Zn2+ ion active site, while the C-domain is 286 residues long and proposed to act as a gatekeeper by restricting substrate active site access.772 In the active site of soluble rabbit NEP (the only structure available of NEP not bound to an inhibitor), the catalytic Zn2+ is ligated by residues His584, His588, and Glu647, and a phosphate ion a tetrahedral coordination geometry (Figure 79). This active site coordination geometry is same to that of human NEP. In addition to the Zn2+ ion cofactor, there are three main hydrophobic substituent binding pockets: S1, S1’, and S2’.773,774 Targeting these pockets with various bulky aromatic substituents been the major focus in inhibitor development.
Figure 79.
Structure of rabbit NEP with phosphate ion bound (PDB 4XBH).
Inhibitor Development.
In 1980, investigation of the inhibitory activity of various di-, tri-, tetra-, and penta-peptides led to thiorphan, the first synthetic inhibitor of NEP (Ki = 4.7 nM, Figure 80).775 Thiorphan was a known inhibitor of thermolysin, which is unsurprising given the similarities between the active site of these two enzymes (although NEP and thermolysin do not share sequence homology).768 A description of an unpublished crystal structure (no entry in the PDB is available) of thiorphan bound to NEP states that the inhibitor is bound in the active site pocket of NEP;776 however, no description of metal binding is provided. Another early inhibitor was phosphoramidon (Ki = 2 nM, Figure 80).777 Co-crystallization of human NEP and phosphoramidon reveals coordination in a monodentate fashion via a single oxygen atom of the N-phosphoryl moiety to the Zn2+ ion in an overall tetrahedral coordination geometry at the metal center (Figure 81).772 The three subsites, S1, S1’, and S2’ are fully occupied by the phosphoramidon.
Figure 80.
Inhibitors of NEP.
Figure 81.
Structure of NEP with: phosphoramidon (PDB 1DMT), NEPi-1 (PDB 1Y8J), NEPi-2 (PDB 1R1J), and LBQ657 (PDB 5JMY).
The majority of NEP inhibitors have a strong resemblance to thiorphan, utilizing either a thiol MBP as monodentate ligand or a mercaptoacetyl MBP as a bidentate ligand.773,776,778–781 An example of such includes NEPi-1, a bicyclic inhibitor bearing a thiol MBP discovered via structure-based drug design.776 Utilizing the unpublished crystal structure of NEP complexed with thiorphan, a small library of five bicyclic inhibitors bearing a thiol MBP were designed and synthesized. From this library, NEPi-1 was discovered one of the two most potent inhibitors, with an IC50 value of 40 nM. A derivative where the thiol MBP was protected by a benzyl group only showed 30% inhibition at a concentration of 100 μM, validating the need for a thiol motif for metal binding. A structure of NEP complexed with NEPi-1 shows the thiol MBP coordinated in a monodentate manner to the Zn2+ ion, which retains the overall tetrahedral coordination geometry of the metal center (Figure 81).776 Key bonding interactions from the central bicyclic scaffold include: H-bonding with the side chains of Asn542 and Arg717, π-π stacking with the side chain of His711, edge-to-face type interaction with the side chain of Trp693, and H-bonding with the side chain of Arg102.776 Additionally, only the S1’ pocket is occupied by the benzyl group of NEPi-1.
Various inhibitors bearing a mercaptoacetyl MBP have also been of interest due to their bidentate metal binding capabilities. Inhibitors such as omapatrilat and pseudo-peptides (NEPi-2, Figure 80), have achieved excellent activity, with IC50 or Ki values of <10 nM.778–781 The structure of NEPi-2 (Ki = 2.3 nM) bound to human NEP reveals the mercaptoacetyl MBP binding Zn2+ in a bidentate manner through the sulfur and oxygen atoms of the mercaptoacetyl MBP, resulting in a distorted trigonal bipyramid geometry (Figure 81).778 The two phenyl rings of NEPi-2 occupy the hydrophobic S1 and S1’ subsites, while the terminal L-alanine residue of NEPi-2 occupies the S2’ subsite, forming H-bonds between the inhibitor backbone and the side chains of Ala543, Arg717, and Asn542.778 Of the inhibitors bearing a mercaptoacetyl MBP, omapatrilat is the most studied and reached Phase III clinical trials;782 however, clinical trials were discontinued due to the adverse side effect of angioedema.
Other inhibitors utilizing a carboxylic acid, hydroxamic acid, and phosphinic acid MBPs to achieve bidentate metal binding have also been prepared, with the dicarboxylic acid compound LBQ657 being the most successful in drug development.762,771,772,783–786 LBQ657 was identified from a series of dicarboxylic acids that were synthesized and screened against NEP.784 LBQ657 exhibited excellent inhibitory activity in vitro (IC50 = 5 nM) and in vivo. The ethyl-ester prodrug of LBQ657 (sacubitril) showed promising PK profiles in three animal species (rats, dogs and monkeys), and resulted in elevated levels of plasma ANP in rats and dogs after oral administration.784 The structure of human NEP complexed with LBQ657 reveals the catalytic Zn2+ ion ligated by an oxygen atom from the carboxylate group of LBQ657 (Figure 81).771 Additionally, all three hydrophobic subsites are occupied: in the S1 pocket, the methyl group of LBQ657 makes hydrophobic interactions with the side chains of Phe544; the biphenyl group of LBQ657 forms hydrophobic interactions with the Trp693 and Phe106 residues residing in the S1’ subsite; and, lastly, the terminal carboxylic acid of LBQ657 forms H-bonding interactions with the Arg110 and Arg102 in the S2’ subsite.771
Current and Future Prospects.
ANP is a peptide hormone used for the treatment of systematic hypertension, chronic renal failure, and CF due to its diuretic, natriuretic, and vasodilatory properties; however, the efficacy of ANP is challenged by NEP, a Zn2+-dependent metalloenzyme responsible for the hydrolysis of ANP and rendering the drug ineffective. A method to extend the half-life of ANP, allowing for prolonged diuretic and vasodilatory properties, includes the development of NEP inhibitors. Early reported inhibitors of NEP include thiorphan, which utilizes a thiol MBP, and phosphoramidon, which utilizes a phosphonate MBP. Subsequent developed inhibitors of NEP bear a thiol or mercaptoacetyl MBP, of which Omapatrilat is the most clinically advanced. Omapatrilat was found to inhibit NEP and ACE with similar activity and has been suggested as a promising treatment for antihypertensive agent and CHF;762,787–791 however, after a series of clinical trials, including OCTAVE (Omapatrilat Cardiovascular Treatment Assessment Versus Enalapril),792,793 OPERA (Omapatrilat in Persons with Enhanced Risk of Atherosclerotic Events)794, OVERTURE (Omapatrilat Versus Enalapril Randomized Trial of Utility in Reducing Events),795 and IMPRESS (Inhibition of Metallo Protease by BMS-186716 in a Randomized Exercise and Symptoms Study in Subjects with Heart Failure),796 it was concluded that a risk for the side effect angioedema outweighed the benefits of omaparilat as a therapeutic, and omaparilat was no longer pursued as a clinical drug. A detailed review of omaparilat and other vasopeptidase inhibitors can be found elsewhere.782,797
Recently, the synergistic combination of sacubitril (prodrug of LBQ657) and Valsartan (known together as Entresto, Figure 80) was approved by the FDA. Entresto was reviewed under the priority review program of the FDA, received fast track designation, and was approved in 2015 for the treatment of patients with heart failure.798 The recent development of LBQ657 as an inhibitor of NEP and vasopeptidase inhibitor of clinical relevance validates NEP as a therapeutic target. Current inhibitors of NEP mostly bear a thiol, mercaptoacetyl, or carboxylic acid MBP; however, more MBPs should be investigated to yield inhibitors with reduced side effects, such as hypotension observed with Entresto.799 Additionally, improving the MBP of vasopeptidase inhibitors which have failed clinical trials (e.g., such as switching the MBP of omaparilat) may be a useful strategy for improving the efficacy and PK properties of these drugs.
6.8. 19S Proteasome Regulatory Subunit Rpn11 (EC 3.4.19.12)
Function in Biology and Disease.
Multiple myeloma (MM) is the second most common haematological malignancy in the U.S. and Europe, accounting for ~1% of all malignancies and ~10% of the hematologic malignancies.800,801 MM is caused by the uncontrolled proliferation of malignant plasma cells, resulting in end-organ damage (hypercalcemia, renal insufficiency, anemia, or skeletal lesions).800,802 Currently, there is no cure for MM, but treatment of MM by proteasome inhibitors has proven to extend patient survival rates.803,804
The proteasome plays a vital role in maintaining protein homeostasis and quality control by degrading unwanted or damaged proteins within eukaryotic cells.805,806 Inhibition of the proteasome within the ubiquitin-proteasine system (UPS) prevents degradation of proapoptotic proteins, which triggers the programmed cell death of malignant cells. The degradation of proteins by the UPS involves a complex pathway in which the protein destined for degradation undergoes several rounds of ubiquitination, resulting in the addition of a polyubiquitin chain. The polyubiquitinated chain of the tagged protein serves for substrate recognition by the 19S regulatory particle (RP), followed by translocation and degradation of the tagged protein by the 20S core particle (CP).805,807,808 Inhibition of the β5 proteolytic active site of the 20S CP has proven to be an effective method for the treatment for MM, with several FDA approved drugs (bortezomib, carfilzomib, and ixazomib).809–811 Success in 20S CP inhibition has led to the development of other inhibitors of the 20S CP subunit.812–815 Although the aforementioned drugs have improved the survival rate of MM patients, drug resistance is commonly observed resulting in relapse801 and creating a need for additional MM therapeutic targets.
The JAMM (JAB1/MPN/Mov34 metalloenzymes) domain of regular particle number 11 (Rpn11) is found within the 19S RP. Rpn11 is a metalloprotease in the greater deubiquitinating enzyme (DUB) family.816 Rpn11 is an isopeptidase responsible for the removal of the polyubiquitinated chain from tagged substrates prior to entry into the 20S CP for degradation (Figure 82). The polyubiquitin chain is further cleaved into monomeric ubiquitin by other DUBs and released into the cytosol to be recycled.817 Inhibition of the JAMM domain of Rpn11 prevents polyubiquitin cleavage, which blocks translocation of the tagged protein into the 20S CP, and stops further downstream protein degradation mechanisms.818–821 Isopeptidase activity by the JAMM domain is found in other human proteins, including Csn5 subunit of the COP9 signalosome, AMSH, AMSH-LP, the BRCC36 subunit of BRISC, MPND and MYSM1.822–828 Inhibition of Rpn11 has been demonstrated to be a viable target for the treatment of MM, leading to cell apoptosis of MM cells and overcoming proteasome inhibitor resistance.829–832
Figure 82.
Reaction performed by Rpn11. Rpn11 removes the polyubiquitinated chain from tagged substrates prior to entry into the 20S CP for degradation.
Protein and Active Site Structure.
The structure of the JAMM domain from several proteins have been reported.828 A crystal structure of the JAMM domain from A. fulgidus AF2198 (AfJAMM, Figure 83) reveals eight-stranded β sheets (β1–β8) flanked by two α helices (α1 and α2). The longer α1 resides between the β1 and β2, while the shorter α2 resides between β4 and β5. The Zn2+ active site is located on the convex surface formed by β2-β4 and α2, and is adjacent to a loop that connects the end of β4 to the beginning of α2 (Figure 83).825 The JAMM domain contains a conserved EXnHS/THX7SXXD sequence (also known as a JAMM motif), in which the Zn2+ ion is coordinated to His67, His69, Asp80, and one water molecule in a tetrahedral geometry. The water molecule, stabilized by hydrogen bonding with a conserved Glu in the second coordination sphere, acts as a Lewis base in the hydrolysis mechanism.816,825,833 Although bearing little sequence homology, the HEXXH metalloprotease motif and core fold of AfJAMM are similar to those found in thermolysin, S. caepitosus Zn2+ endoprotease (ScNP), and neurolysin.816,825
Figure 83.
Structure of AfJAMM (PDB 1R5X).
Inhibitor Development.
Inhibitors of various 19S RP subunits (such as ubiquitin receptors) have been limited,834–838 with inhibitors that specifically target Rpn11 scarce.807,839 A first-in-class Rpn11 inhibitor was discovered utilizing a FBDD method. Screening of a MBP library against Rpn11 revealed a single active MBP hit, 8-thioquinoline (Rpn11i-1, IC50 = 2.5 μM, Figure 84).839 The efficiency and effectiveness of utilizing a small library screen for MBP identification was validated when a HTS effort using >300,000 compounds revealed an 8-thioquinoline derivative as the only hit.807 Heterocyclic analogues of Rpn11i-1 where the metal coordinating nitrogen and sulfur atoms were moved, removed, or modified, such that they obstructed metal binding, resulted in inactive compounds, supporting a proposed bidentate model of metal coordination by the nitrogen and sulfur atoms of Rpn11i-1. Elaboration of Rpn11i-1 yielded the inhibitor capzimin with an IC50 value of 390 nM (Figure 84).807,839 Capzimin showed >5-fold selectivity for the JAMM domain of Rpn11 over other JAMM proteases and >100-fold selectivity for Rpn11 over a small panel of Zn2+-dependent metalloenzymes. Treatment of cancer cell lines with capzimin resulted in the accumulation of polyubiquitinated substrates and blocked cell proliferation in culture, including cell lines shown to be bortezomib resistant.807,839
Figure 84.
Inhibitors of Rpn11.
In addition to capzimin, thiolutin, an antibiotic and anti-angiogenic compound produced by Streptomyces, was also identified as an inhibitor of Rpn11 (Figure 84).840 Inhibition studies performed with and without reducing agent dithiothreitol (DTT) revealed only the DTT containing reaction resulted in Rpn11 inhibition by thiolutin, suggesting reduced thiolutin (Rpn11i-2) as the inhibitor of Rpn11. Rpn11i-2 inhibits Rnp11 with an IC50 value of 530 nM and successfully blocks the removal of ubiquitin from the tagged enzymatic substrate. Although the mode of metal binding by thiolutin has not been established, the compound likely uses a thiol donor to bind the active site Zn2+ ion in Rpn11. Rpn11i-2 inhibited other JAMM metalloproteases (Csn5, AMSH, and BrCC36), and thermolysin.840
Current and Future Prospects.
Current FDA approved drugs for the treatment of MM inhibit the UPS pathway by binding to the 20S CP; however, the efficacy of these drugs is challenged by the emergence of drug resistance. The Zn2+-dependent JAMM domain of Rpn11 subunit in the 19S RP represents an attractive new target for drug development. Capzimin represents a first-in-class inhibitor of Rpn11. Discovery of a second inhibitor, thiolutin, further supports Rpn11 inhibitor development as a viable route to MM treatment. Inhibitor development for Rpn11 represents a new method for proteasome inhibition and is an untapped opportunity with few inhibitors currently available. Capzimin and thiolutin both display only modest inhibitory activity and both utilize thiol groups that may represent a PK liability. Further development of more potent compounds that use alternative MBPs for inhibitors of Rpn11 are clearly justified.
6.9. VanX (EC 3.4.13.22)
Function in Biology and Disease.
Cell wall synthesis is a critical process for bacterial survival, and therefore represents the target of many classes of current antibiotics. One of the unique aspects of cell wall synthesis is the action of the transpeptidase that hydrolyzes the substrate D-Ala-D-Ala as part of the formation of the peptidoglycan cross layer. D-Ala-D-Ala is a non-native peptide sequence in the human biome, making targeting of this single step a highly successful strategy for antibiotics. ß-lactam antibiotics inhibit the action of the transpeptidase (Section 7.5), whereas vancomycin binds to the D-Ala-D-Ala peptide substrate. The action of vancomycin binding to the D-Ala-D-Ala pentapeptide (Figure 85) physically blocks the substrate and prevents further peptidoglycan crosslinking, stopping cell wall growth, and ultimately results in cellular death. As such, vancomycin is a powerful glycopeptide antibiotic active against most Gram-positive bacteria, and is often considered a drug of last resort. Vancomycin was first discovered by Dr. Edmund Kornfield at Eli Lilly in 1952,841,842 and was approved by the FDA in 1958.
Figure 85.
Mechanism of resistance to vancomycin. Left: Schematic of vancomycin binding to D-Ala-D-Ala. The hydrogen bond eliminated in resistant bacteria is colored red. Right: Schematic of vancomycin binding to D-Ala-D-Lactate. The site of the mutation is highlighted in cyan.
Vancomycin binds to the D-Ala-D-Ala substrate through five hydrogen bonds, sterically blocking the transpeptidase from any further action on the substrate (Figure 85). However, in vancomycin-resistant bacterial strains, the D-Ala-D-Ala is replaced with a D-Ala-D-Lactate (Figure 85), where a single atom replacement from nitrogen to oxygen results in a 1000-fold loss of binding activity that renders vancomycin completely ineffective as an antibiotic. Loss of vancomycin activity from this single atom mutation is due to a combination of the loss of favorable H-bonding interactions, as well as the creation of new repulsive lone-pair interactions between the vancomycin carbonyl and substrate ester.843 SAR studies indicated that the loss of the H-bonding to the D-Ala-D-Lactate resistant substrate had a smaller impact on vancomycin binding than lone-pair repulsion. Attempts have been made to synthesize vancomycin derivatives that address this resistance mechanism,844 but none so far have been evaluated in clinical trials.
Five proteins have been found to be necessary to confer bacterial resistance to vancomycin.845 These proteins are encoded in a transposon that can be converted into a plasmid for horizontal gene transfer, which allows for rapid transmission of vancomycin resistance. Two of these proteins, VanS and VanR are sensor proteins that in the presence of vancomycin, turn on transcription of VanH, VanA, and VanX, which then serve to shunt the peptidoglycan precursors from the native D-Ala-D-Ala to the resistant D-Ala-D-Lactate pathway. In this process, VanH generates the D-lactate, and VanA ligates D-Lactate to D-Ala, generating the vancomycin resistant peptidoglycan precursor. Because the cellular machinery is still producing the native D-Ala-D-Ala substrate, VanX serves to selectively hydrolysis the native D-Ala-D-Ala sequence to prevent the native terminus from being used in the peptidoglycan layer.846 This prevents incorporation of D-Ala-D-Ala and results in the vancomycin resistant D-Ala-D-Lactate phenotype. Knockout studies of any one of these Van components have been shown to restore vancomycin activity, and in particular knockout of VanX results in bacteria that express both D-Ala-D-Lactate as well as D-Ala-D-Ala.846,847 These VanX knockout bacteria are susceptible to inhibition by vancomycin, making VanX an intriguing target for drug discovery campaigns.847
Protein and Active Site Structure.
VanX is a mononuclear Zn2+-dependent metalloenzyme with an active site designed to specifically accommodate and hydrolyze D-Ala-D-Ala (Figure 86).847 Currently there is only one crystal structure of VanX published in the PDB.848 The VanX active site is fairly small at ~150 Å3, and is very hydrophilic with many polar residues designed to give high specificity for the unique D-Ala-D-Ala peptide. The catalytic Zn2+ ion is coordinated by two histidine residues and an aspartic acid, with either one or two additional water molecules completing the coordination sphere. The crystal structure clearly indicates only one water molecule coordinated to the Zn2+ center; however, EXAFs data support an argument for a 5-coordinate Zn2+ ion with two bound water molecules.849 Regardless of how many water molecules are coordinated to the Zn2+ ion in the VanX resting state, the D-Ala-D-Ala hydrolysis mechanism is proposed to involve a substrate polarization via binding to the Lewis acidic Zn2+ ion, which then results in a nucleophilic attack to insert one of the water molecules into the amide bond, resulting in hydrolysis of the dipeptide.
Figure 86.
Structure of VanX (PDB 1R44). An additional unbound water molecule (red sphere) is shown near the metal center.
Inhibitor Development.
The first inhibitors of VanX were reported in 1995, and used a phosphinate group to both mimic the peptide substrate intermediate and to bind the active site Zn2+ ion by displacing Zn2+-coordinated water (Figure 87, Table 12).847 Interestingly, it was found that the phosphinate based inhibitor VanXi-2 (Ki = 0.47 μM) was far more active than the phosphonate-based inhibitor VanXi-1 (Ki = 300 μM). X-ray crystallography showed that while both compounds have the same coordination to the active site Zn2+ through the oxygen atoms of the phosphorous in a trigonal bipyramidal geometry, the phosphonate based VanXi-1 adopts an energetically less favorable position that results in the loss of a H-bond interaction (note that these structures were published in 1998, but there is no entry in the PDB). Attempts to further improve phosphinate based inhibitors resulted in the incorporation of an ethyl tail that improved the binding affinity in VanXi-4 (Ki = 0.35 μM).850 Later efforts to generate new H-bond interactions explored isosteres of the phosphinate scaffold that replaced the methylene carbon with either a nitrogen or sulfur atom. These compounds did not show any improvement, with the amine linker having worse activity (VanXi-6, Ki = 36 μM), and the thioether (VanXi-7) having no activity whatsoever.851
Figure 87.
Inhibitors of VanX.
Table 12.
Activity (Ki values) of VanX inhibitors.
| Compound | Inhibition (μM) | Ref. |
|---|---|---|
| VanXi-1 | 300 | 847 |
| VanXi-2 | 0.47 | 847 |
| VanXi-3 | 0.35a | 850 |
| VanXi-4 | 1.7a | 850 |
| VanXi-5 | 0.81a | 850 |
| VanXi-6 | 36 | 851 |
| VanXi-7 | N.A. | 851 |
| VanXi-8 | 30b | 852 |
| VanXi-9 | 10.7b | 853,854 |
| VanXi-10 | 113b | 853,854 |
| VanXi-11 | N.A.b | 853,854 |
| VanXi-12 | 79 | 855 |
| VanXi-13 | 13 | 855 |
| VanXi-14 | 430 | 855 |
| VanXi-15 | 1.8 | 855 |
| VanXi-16 | 0.32 | 855 |
| VanXi-17 | 0.19 | 855 |
| VanXi-18 | 17 | 855 |
| VanXi-19 | 0.73 | 855 |
| VanXi-20 | 122 | 855 |
| VanXi-21 | 6.84 | 856 |
| VanXi-22 | 2.74 | 856 |
Compounds isolated and tested as racemic mixtures.
Kirr value.
Covalent inhibitors of VanX have also been investigated (VanXi-8 to VanXi-11, Figure 87). These compounds are substrate mimics that upon hydrolysis by VanX release a highly reactive electrophile that covalently binds to the enzyme. While the exact nucleophilic residue is presently unknown, it is understood that this covalent binding effectively blocks the active site and prevent further action.852 Moving the difluoromethyl group from the para position in compound VanXi-8 to the ortho position in VanXi-9 was found to improve the stability of the leaving electrophile and improved the Kirr from 22 μM to 10.7 μM.853,854 Unfortunately, additional attempts to induce selectivity for the unknown nucleophilic residue(s) were unsuccessful, and this approach of covalent inhibitors against VanX has not progressed further.
In 1996, a series of simple thiol containing molecules were investigated as VanX inhibitors to take advantage of the high affinity of sulfur for the Zn2+ ion (VanXi-12 to VanXi-20, Figure 87). There is no structural data on the binding of these compounds, but it may be assumed that these thiols coordinate via one or more sulfur atoms. Of these inhibitors, VanXi-17 was found to be the most active with a Ki value of 0.19 μM, but no follow-up effort has been reported on these compounds.855 Two hydroxypyridinethione compounds, VanXi-21 and VanXi-22 have been reported to have modest activity (Ki = 6.84 and Ki = 2.74 μM, respectively). Again, there is no crystallographic data on these inhibitors, but they are expected to bind in a bidentate fashion to the catalytic Zn2+ ion. Both VanXi-21 and VanXi-22 were demonstrated to be active against vancomycin resistant Enterococcus faecium.856 However, there is some concern around selectivity, as the same compounds (VanXi-21 and VanXi-22) have been reported as inhibitors of HDACs (Section 7.3) and MBLs (Section 7.5).
Current and Future Prospects.
VanX plays a key role in providing resistance to the antibiotic vancomycin. While VanX is a promising target to restore activity of vancomycin against resistant bacteria, inhibitor development has been limited, with only phosphinate based, covalent inhibitors, and sulfur containing compounds being widely explored. Few of these compounds show activity with IC50 values of <1 mM, and none have entered clinical trials. Perhaps one impediment in the development of VanX inhibitors has been the difficulty in expressing, purifying, and handling VanX. Indeed, literature reports describing VanX assays have been withdrawn due to protein contamination from a difficult purification that gave false positive assay readouts.857,858 Furthermore, VanX has been reported to have tendencies to form aggregates, which compounds the difficulty in enzyme purification and screening;849 this aggregate nature is evidenced by the hexameric structure of VanX found in X-ray crystallography.848 Overall, there is a need for better enzyme purification techniques in order for further inhibitor development of VanX inhibitors.
7. Non-Peptide C-N Hydrolases (EC 3.5)
7.1. Adenosine Deaminase (ADA, EC 3.5.4.4)
Function in Biology and Disease.
Adenosine deaminase (ADA, also known as adenosine aminohydrolase) is involved in purine metabolism.859 Specifically, it irreversibly deaminates adenosine to inosine and 2′-deoxyadenosine to 2’-deoxyinosine by chemically changing the amino group to a keto group (Figure 88).859 The inosine products of ADA activity are competitive inhibitors with the substrates.859 ADA is typically found in the cytosol and on the cellular surface of nearly all human tissues, but the highest levels exist in the lymphoid tissue, including B and T lymphocytes.859 The primary function of ADA is development and maintenance of the immune system. ADA deficiency causes severe combined immunodeficiency disease (ADA-SCID) due to impairment of B-cell and T-cell development.860 Conversely, inhibition of ADA has been demonstrated to be a useful therapy in some viral infections and lymphoproliferative disorders, such as lymphoma and leukemia, as well as in modulating immune response in B and T cell malignancies.861 As such, ideal ADA inhibitors should be reversible and have a limited lifetime to not impact healthy ADA activity.862 Although several ADA inhibitors have gone to clinical trials to treat various cancers, the only FDA approved inhibitor is Nipent (a.k.a., pentostatin) for the treatment of hairy cell leukemia.
Figure 88.
Deaminase reaction catalyzed by ADA.
Protein and Active Site Structure.
The amino acid sequence of ADAs in humans and bacteria is highly conserved particularly at the active site.859 ADA exists as two isozymes, ADA1 and ADA2, which differ in primary sequence but catalyze the same reaction.859 ADA1 is found in all cells (with highest expression in lymphocytes and monocytes), and ADA2 is found only in monocytes.863 ADA1 exists in two isoforms: a monomer of ~33 kDa (small form) and a dimer of ~280 kDa (large form).863 The extra mass of the large form is from a combining protein, which connects two ADA1 monomers.863 The small form of ADA1, which is the target of most drug discovery efforts, is composed of eight central β-barrels and eight peripheral α-helices that create a (β/α)8 barrel. The active site is buried deep in the enzyme and contains a Zn2+ ion coordinated by His15, His17, His214, Asp295, and a water/hydroxide molecule in a tetrahedral geometry (Figure 89). During turnover, the substrate is stabilized in the active site by hydrogen bonding to Glu217, Gly184, Asp19, and Asp296, with deamination occurring by a stereospecific addition-elimination.859,864 The Zn2+ ion serves as a strong electrophile and drives the bound water/hydroxide to perform nucleophilic attack on the substrate creating a tetrahedral intermediate (Figure 88). Ultimately, ammonia is eliminated from the substrate upon formation of the ketone product.
Figure 89.
Structure of ADA (PDB 1VFL).
Inhibitor Development.
Various classes of ADA inhibitors have been extensively explored. The major classes are described herein, including representatives from each class. Important for discussing inhibitor binding are the two conformations of ADA, the open and closed form,865 both of which are accessed during substrate turnover. Structural investigations show that inhibitors can bind to either conformation, depending on whether a specific water-trigger site is occupied.865 In the closed conformation, the active site forms a ‘boot shape’ (Figure 90), in which the trigger-water site is unoccupied by the inhibitor, and the inhibitor directly interacts with Glu217.865 This is exemplified by the inhibitor 1-deazaadenosine (1-DAA, Ki = 660 nM, Figure 91). In the open conformation, the trigger-water site is occupied by the inhibitor, and two water molecules form a hydrogen bonding bridge between Glu217 and the inhibitor, as illustrated with the EHNA (Ki ~5 nM, Figure 90).865 Another key residue in the determination of the conformation is Phe65, which points into or away from the active site in the closed and open forms, respectively. Movement of Phe65 is implicated in the downstream structural rearrangements that ultimately close or open the active site.865
Figure 90.
Left: Structure of 1-deazaadenosine (1-DAA) bound to the closed conformation of ADA (PDB 1ADD). In closed conformation, the inhibitor directly interacts with Glu217 and the water-trigger site is unoccupied by the inhibitor. Right: Structure of EHNA bound to the open conformation of ADA (PDB 2Z7G). In open conformation, the inhibitor and Glu217 interact via hydrogen bonds with bridging water molecules, and the water-trigger site is occupied.
Figure 91.
Inhibitors of ADA.
Transition state analogue inhibitors of ADA have a chiral tetrahedral carbon positioned in the active site toward the Zn2+ ion resembling the intermediate in the deaminase reaction mechanism. A hydroxyl group on the tetrahedral carbon coordinates to the Zn2+ center (Figure 92), with the stereochemistry of these inhibitors being crucial: only the R-diastereomer fits in the active site and inhibits ADA.859 Inhibitors of this class include coformycin (Ki = 0.01 nM) and pentostatin (Ki = 0.0025 nM). These natural products were first reported in the 1960s and 1970s as ADA inhibitors.866,867 Adechlorin (Ki = 0.53 nM) and adecypenol (Ki = 4.7 nM) are also natural product inhibitors of ADA; they have the same metal binding mode as coformycin and pentostatin, but their sugar groups contains a chlorine or cyclopentene ring, respectively,868,869 and they are less active than coformycin and pentostatin. Coformycin and pentostatin are still two of the most active inhibitors of ADA known, and pentostatin is the only ADA inhibitor with FDA approval.
Figure 92.
Structure of pentostatin bound to ADA (PDB 2PGR).
The first total synthesis of pentostatin was reported in 1979, which opened doors for exploring modifications to the coformycin and pentostatin scaffolds.870 Modifications to the structure while maintaining the tetrahedral carbon atom with a hydroxyl group resulted in compounds such as ADAi-1 (Ki = 61 μM), which had poorer activity.871 More recently, a series of analogs containing imidazo[4,5-e][1,2,4]triazepine were prepared to explore the interactions responsible for the tight binding of pentostatin and coformycin.872 Eight derivatives were synthesized with several changes, including: a lack of sugar moiety, which weakens the binding interaction by removing stabilizing hydrogen bonds; inclusion of alkyl groups at position 1; an additional nitrogen atom at position 7 to reduce nucleophilicity of the hydroxyl at position 8; and, addition of methyl group at position 6 and 7 to avoid rearrangement of the ring system.872 Compound ADAi-2 (Ki = 12 μM) in this series was the most active (Figure 91). Overall, these inhibitors have inhibition constants many orders of magnitude worse than pentostatin.872 Their poorer activity is attributed to the lack of a sugar moiety, which forms several hydrogen bonds to protein residues. No structural data has been reported for these inhibitors, so it is unclear how these inhibitors bind to ADA and if the active site adopts closed or open conformation.
Another class of ADA inhibitors are substrate analogues that bind in the active site like adenosine. These inhibitors do not bind to the Zn2+ ion and they feature a planar ring system. The inhibitors differ from substrates in that they lack a protonatable nitrogen required for the deaminase reaction, and hence, cannot be deaminated. Among the inhibitors in this class, changes to the sugar moiety and the amine were explored (e.g., 1-DAA, Figure 91).873,874 Substitution of an amine hydrogen with a methyl, hydroxyl, and cylopropyl group were also tested, such as ADAi-3 (Ki = 250 nM),874 but these inhibitors demonstrate similarly modest activity. Although no structural data is available for these inhibitors, it can be inferred with reasonable confidence that the inhibitors occupy the active site in a similar orientation as adenosine. Many other substrate analogs have been synthesized as ADA inhibitors, including non-nucleoside compounds, but none of these utilize metal binding, nor show particularly exceptional activity.875–881 Indeed, none of these ADA inhibitors have been advanced to clinical trials.
Current and Future Prospects.
Substantial effort has been made to develop therapeutically useful, small molecule inhibitors of ADA. ADA inhibitors include transition state analogues, substrate analogues, and non-nucleosides, and many of these investigations are based on natural products that inhibit ADA. The transition state analogs, such as pentostatin, are the most potent and utilize metal-binding. Pentostatin is the only FDA-approved drug that is an ADA inhibitor. Pentostatin is marketed as Nipent and is used to treat hairy cell leukemia; however, it no longer appears to be on patent.882 Despite the clinical use of pentostatin, other ADA inhibitors that have made it to clinical trials have failed due to off- and on-target toxicity, and to the best of our knowledge, no new substrate analogues or non-nucleoside inhibitors have entered clinical trials. The failure of ADA inhibitors in clinical trials combined with the the fact that Nipent has gone off patent and may now be sold in a cheaper, generic form suggests that ADA is no longer a promising target for the development of new therapeutics.
7.2. Arginase 1 (Arg1, EC 3.5.3.1)
Function in Biology and Disease.
Arginase was initially described in 1904 for its ability to hydrolyze the substrate L-arginine into L-ornithine and urea,883,884 and is the oldest known Mn2+-dependent metalloenzyme. Due to its importance in the urea cycle, arginase is a ubiquitous protein conserved across all branches of life with some variant found in all organisms.885 For the sake of simplicity, as well as its relevance towards human health, this Section will focus on human arginase unless otherwise stated. There are two types of human arginase: arginase-1 (Arg1) which dwells in the cytosol, and arginase-2 (Arg2), which is located in the mitochondria. Arg1 is primarily localized in the liver, where it plays a critical role as the last step of the urea cycle. However, Arg1 is also distributed throughout other body tissues, albeit at lower concentrations than found in the liver, where it plays various homeostatic roles.
In addition to its role in the urea cycle, Arg1 is used to regulate the immune system through the hydrolysis of L-arginine. L-Arginine is considered a semi-essential amino acid, in that some cells, including T-cells, are unable to produce this amino acid and require an extracellular source. In the absence of L-arginine, T-cells lose their T-cell receptor and become unable to proliferate.886 It is currently unknown why the absence of L-arginine produces such drastic effects on T-cells. After an immune response occurs, there is a heightened level of T-cells, so Arg1 is produced by immature myeloid cells (IMCs) to allow the body to enter a healing phase.887 Cancer tumors take advantage of this cycle and recruit IMCs and activate them into myeloid derived suppressor cells for their anti-proliferative effect against T-cells.888 The resulting overexpression of Arg1 creates a tumor microenvironment devoid of L-arginine and inhibits T-cell proliferation, allowing the cancer tumor to evade the immune system. Arg1 overexpression is linked to many other disease states, including but not limited to hypertension, diabetic vascular disease, atherosclerosis, myocardial ischemia injury, ischemic stroke, Alzheimer’s disease, multiple sclerosis, diabetic retinopathy, and erectile dysfunction,889 suggesting that Arg1 inhibitors could be of great benefit to a range of conditions.
Protein and Active Site Structure.
Arg1 is a homotrimeric metalloenzyme, with each unit bearing a dinuclear Mn2+ active site designed to cleave its substrate, L-arginine into L-ornithine and urea.890 The Arg1 active site consists of a deep, narrow pocket ~15 Å in depth. The entrance of the pocket contains an amino acid binding region that provides substrate specificity, whereas the Mn2+ ions, which are responsible for substrate hydrolysis, are located in the bottom of the pocket, ~3.3 Å apart from each other.891 Each Mn2+ ion is coordinated to one His and two Asn residues, with an additional two Asn residues bridging between the Mn2+ ions.509 MnA is coordinated in a 5-coordinate square pyramidal geometry and MnB is coordinated in distorted octahedral geometry.892 The coordination sphere of each Mn2+ ion is completed by a bridging hydroxyl that is inserted in the guanidine moiety during L-arginine hydrolysis (Figure 93).
Figure 93.
Structure of human Arg1 (PDB 1HQG). Images of the active site are shown with (center) and without (right) bound L-ornithine and urea (yellow).
Typical of most metal ion mediated substrate hydrolysis mechanisms, the Mn2+ ions mainly serve to raise the pKa of the bridging hydroxyl to aid nucleophilic attack. Indeed, human Arg1 has an optimum pH of 9.4.893 Mutagenesis within the active site to disrupt and displace metal coordination of MnA only slow the rate of substrate hydrolysis, and do not affect the rate of substrate binding, indicating that the function of the metal is to activate the hydroxyl for nucleophilic attack, and not act as a site of substrate coordination.890 Therefore, the mechanism of Arg1 L-arginine hydrolysis is believed to involve the bridging hydroxyl ligand being inserted into the guanidine group. This generates a tetrahedral intermediate, which is followed by a series of proton transfer steps resulting in protonation of the amine leaving group on the substrate, and finally triggering an electron cascade that cleaves the L-arginine into urea and L-ornithine.509
Inhibitor Development.
Both iNOS and Arg1 share L-arginine as a common substrate, but instead of the hydrolytic cleavage of Arg1, iNOS catalyzes the oxidation of L-arginine to N-hydroxyarginase (NOHA).894 Interestingly, NOHA serves as a native Arg1 inhibitor with a Kd value of 3.6 μM (Figure 94, Table 13), so that the production of NOHA by iNOS is natively used to regulate Arg1. NOHA binds the Arg1 active site as a transition state mimic, with the hydroxyl group binding in a bridging fashion between the Mn2+ ions and displacing the metal-bound hydroxyl ion (Figure 95).895 However, the crystal structure of NOHA bound to Arg1 shows that the amino acid residues at the entrance of the active site dictate the preferred binding interactions, so that in order to maintain these highly stable interactions at the entrance to the active site, the Mn2+ centers are actually perturbed into a less favorable position to accommodate the bulk of the NOHA within the active site cavity (Figure 95). Most notably, the hydroxyl group on NOHA is pushed an additional 1.7 Å deeper into the active site than the native bridging hydroxyl, which results in the Mn2+ ions being pushed further apart to ~3.9 Å in the NOHA bound structure. Furthermore, Asp232, which normally coordinates both Mn2+ ions, is shifted to a distance of ~3.1 Å away from MnA where it is no longer able to effectively coordinate MnA, resulting in a shift from a square pyramidal metal coordinating geometry to distorted tetrahedral geometry (Figure 95).
Figure 94.
Inhibitors of Arg1.
Table 13.
Activity of Arg1 inhibitors. All values are against human Arg1 unless otherwise noted.
| Compound | Inhibition (μM) | Ref. |
|---|---|---|
| Arg1i-1 | IC50 = 1000 | 895 |
| Arg1i-2 | IC50 = 300 | 895 |
| Arg1i-3 | IC50 = 5000 | 895 |
| Arg1i-4 | IC50 (pH 7.4) = 40, IC50 (pH 9.0) = 400a | 899 |
| Arg1i-5 | IC50 = 5a | 897 |
| Arg1i-6 | IC50 = 2a | 897 |
| Arg1i-7 | IC50 = 230b | 898 |
| Arg1i-8 | IC50 = 11b | 898 |
| Arg1i-9 | Ki = 90b | 900 |
| Arg1i-10 | Kd = 52b | 900 |
| Arg1i-11 | Ki = 4 | 901 |
| Arg1i-12 | Kd = 60 | 902 |
| Arg1i-13 | Ki = 6a | 903 |
| Arg1i-14 | N.A. | 904 |
| Arg1i-15 | Kd = 0.88 | 905 |
| Arg1i-16 | Kd = 34 | 905 |
| Arg1i-17 | IC50 = 0.223 | 906 |
| Arg1i-18 | IC50 = 0.017 | 907 |
| ABH | Kd = 0.005, IC50 = 1.45 | 906,908 |
| BEC | Kd = 0.270 | 908 |
| NOHA | Kd (pH 8.5) = 3.6, IC50 = 20 | 895,909 |
| Nor-NOHA | Kd (pH 8.5) = 0.517, IC50 = 1 | 895,909 |
Rat liver Argl;
Bovine liver Argl.
Figure 95.
Structures of Arg1 with: NOHA (PDB 1HQF), nor-NOHA (PDB 1HQH), ABH (PDB 2AEB), Arg1i-9 (PDB 1R1O), Argli-12 (PDB 3F80), and Argli-13 (PDB 1TF5). Hydrogen bonding (dashed magenta lines) with amino carboxylate groups is shown for select inhibitors.
In an effort to alleviate the strain on the metal environment, the effect of chain length was examined with the derivative N-hydroxy-nor-arginine (nor-NOHA), which has one less carbon than NOHA. nor-NOHA is a better Arg1 inhibitor (Kd = 0.52 μM) than NOHA (Kd = 3.6 μM). An X-ray crystal structure (Figure 95) shows that nor-NOHA maintains the same favorable amino acid interactions at the active site entrance as NOHA, but does not create perturbations at the dinuclear metal center.895 nor-NOHA still binds the metal site with a hydroxyl displacing the native bridging hydroxyl group, but the nor-NOHA hydroxyl is only 0.8 Å deeper into the active site than the native hydroxyl, keeping the nor-NOHA hydroxyl in a ‘sweet spot’ not too far from where the native hydroxyl would reside. Furthermore, in the nor-NOHA Arg1 structure, the Mn2+ ions are only displaced from 3.3 Å to 3.5 Å, and Asp232 remains within coordination distance of MnA in the nor-NOHA structure. While nor-NOHA is considered one of the best Arg1 inhibitors, the PK of nor-NOHA severely limits the potential for clinical development. nor-NOHA has extremely rapid elimination in rats with a half-life of 15–30 minutes after intravenous or intraperitoneal injection.896 While many other derivatives of the NOHA scaffold have been explored, none of these compounds are nearly as active as NOHA and nor-NOHA (Figure 94, Table 13).895,897,898
Since 1945, boric acid has been known to inhibit Arg1, and was considered one of the best Arg1 inhibitor for decades. In aqueous solution, boric acid exists in both the borate and boric acid forms, with both forms acting as noncompetitive, irreversible Arg1 inhibitors. Inhibition studies of borate with L-ornithine lead to the discovery of 2(S)-amino-6-boronohexanoic acid (ABH, Figure 94) as powerful and novel new class of Arg1 inhbitors.910 ABH is a slow binding irreversible inhibitor that operates by mimicking the Arg1 transition state. Once ABH enters the active site and the boronic acid moiety is positioned across the Mn2+ ions, the metal-bound hydroxyl group inserts into the boronic acid.908 This creates an uncleavable tetrahedral transition state mimic that remains trapped in the active site, rendering Arg1 unable to hydrolyze its L-arginine substrate (Figure 95). It should be noted that at pH values >9.0 the boronic acid warhead of ABH takes on the borate form with a different mechanism of action. The borate is instead a competitive inhibitor that binds Arg1 by displacing the metal coordinated hydroxyl. At pH values <9.0, derivatized boronic acid inhibitors maintain the hydroxyl insertion mechanism. Other attempts have been made to mimic this transition state analog by varying the terminal functional group into other ‘suicide’ substrates, but no studies have yielded active compounds.903,909
Recent trends in developing new Arg1 inhibitors has shifted towards further exploration and elaboration of the core ABH scaffold, with efforts on exploration of alternate warheads, different linkers, and further elaboration of the amino acid terminus. Early efforts focused on attempts to replace the boronic acid warhead of ABH with bioisosteres capable of mimicking the tetrahedral transition state while maintaining metal coordination. As such, ABH analogues with isostere warheads including a terminal sulfonamide (Arg1i-9, Kd = 52 μM; Arg1i-10, Kd = 52 μM),900 2-aminoimidazole (Arg1i-11, Ki = 4.0 μM),901 and a nitro group (Arg1i-12, Kd = 60 μM),902 were tested, but all compounds showed no improvement compared to ABH (Table 13). Other warheads designed to mimic the electrophilic nature of ABH include aldehyde (Arg1i-13, Kd = 60 μM),903 and epoxide moieties (Arg1i-14).904 Once again, while these warheads were shown to bind to the active site Mn2+, none of them were more active than the initial ABH lead (Table 13). Modifications to the chain length of ABH resulted in the discovery of S-(2-boronoethyl)-L-cysteine (BEC, Figure 94) that has poorer activity (Kd = 270 nM), likely because the shorter chain places the boronic acid warhead further away from the Mn2+ cluster.908 Finally, a large body of work on derivatization of the amino acid terminus of ABH has been reported. Building from the amino acid terminus increases binding affinity, this was first accomplished in Arg1i-15 and Arg1i-16 (IC50 = 0.38 – 34 μM).905 Later work focused on building further off the amino acid terminus, and resulted in Argli-17 with an IC50 value of 223 nM.906 This good binding is attributed to a decrease in entropy, from the inclusion of more hydrophobic functional groups. Shifting from the piperidine in Argli-17 to the tropane in Argli-18 further improved the activity (IC50 = 17 nM), making it among the most potent Arg1 inhibitors reported to date.907
In addition to these reported compounds, Calithera Biosciences and Incyte Corporation currently have an ABH-based boronic acid inhibitor in Phase I and II clinical trials. This ongoing study is focusing on co-administrating lead compound INCB001158911 with known cancer drugs oxaliplatin, leucovorin, 5-fluorouracil, gemcitabine, cisplatin, and pacilitaxel, as well as the antibody pembrolizumab to look for a synergistic effect in treating tumors. Finally, other studies have described a host of miscellaneous inhibitors of non-human Arg1, including flavonoids, purines, retrovirus inhibitors (e.g., raltegravir, dolutegravir, Figure 45), but none of these have had the same level of success against Arg1 as the boronic acid based inhibitors.896,912
Current and Future Prospects.
Human Arg1 completes the last step of the urea cycle in the liver, and plays a myriad of roles in maintaining homeostasis throughout the rest of the body. Misregulation of Arg1 is linked to numerous diseases, including but not limited to cancer, where the overexpression of Arg1 allows the cancer tumor to evade the immune system. Inhibition of Arg1 is predicted to restore native immune activity against cancer tumors and has implications for combination therapies. Indeed, there is currently one compound (INCB001158) in Phase I and II clinical trials being co-administered with a variety of known anticancer drugs. While the boronic acid warhead has seen great success against Arg1, little work has been done on exploring other MBPs. The use of novel MBPs remains an untapped area and would likely lead to other novel inhibitors against Arg1.
7.3. Histone Deacetylase (HDAC, EC 3.5.1.98)
Function in Biology and Disease.
Histone deacetylases (HDACs, a.k.a., lysine deacetylases, KDACs) are a class of regulatory enzymes that function to deacetylate histone Lys residues as a part of DNA transcriptional regulation.913 This deacetylation action results in a positive charge on the terminal amine of the Lys at physiological pH, so that the histone binds more tightly to the negatively charged DNA phosphate backbone through electrostatic interactions.914 In turn, the more tightly bound histone represses DNA transcription, allowing for regulation of gene expression.
This mechanism of gene silencing through HDAC activity is utilized by cancer tumors to downregulate factors that suppress tumor growth. Thus, inhibition of HDACs has been determined a viable route to reverse the abnormal epigenetic state within the cancer tumor environment.913 Currently the biology behind the effect of HDAC inhibition on cancer is not fully understood.915 However, it is known that HDAC inhibition typically induces morphological changes resulting in the selective apoptosis of cancer tumors over healthy cells.916 HDAC inhibition is a validated method of cancer treatment, as there are currently four HDAC inhibitors with FDA approval for the treatment of cancer, with several additional compounds in clinical trials.
Protein and Active Site Structure.
There are currently 18 known human HDAC isoforms, broken into four distinct classes. Class I HDACs 1, 2, 3, and 8; class IV contains only HDAC 11; and class II is broken into two subgroups, where class IIa contains HDACs 4, 5, and 7, and class IIb contains HDACs 6 and 10.917 These different classes were developed based on structural homology to other known classes of yeast HDAC. Finally, class III HDACs are the sirtuins 1–7, which depend on NAD+ as a cofactor, and unlike the other classes are not metal dependent proteins.918
Overall, the HDAC family bears a characteristic α/β fold that is shared with the arginase family of enzymes (Section 7.2).919 HDACs are typically monomeric, with the exception of HDAC6, which is heterodimeric with two catalytic domains, CD1 and CD2. The active site is partially conserved across all HDACs with a substrate binding channel ~10 Å deep, but there is variation in the entrance to the active site across the isoforms. This variation is the source of the different isoform selectivity, and many inhibitors attempt to exploit these differences for selective inhibitors. At the bottom of the substrate binding channel is the catalytic region containing the Zn2+ ion necessary for substrate lysis. This Zn2+ binding region is conserved across the HDAC isoforms, with the catalytic Zn2+ ion coordinated by Glu705, Glu612, His614, and a nucleophilic water in a distorted tetrahedral geometry (Figure 96). The mechanism of HDACs involves the acetylated lysine entering the active site, coordination to the Zn2+ ion through the carbonyl of the acetyl group, then attack of the nucleophilic water Zn2+-bound water on the carbonyl, creating a gem-diol intermediate. A series of proton transfers, largely mediated by two adjacent His and a Tyr, ultimately result in the deprotection of the lysine substrate, and release of the acetyl group as acetic acid.920
Figure 96.
Left and Middle: Structure of HDAC6 (PDB 5EEM). Right: Acetylated substrate (magenta, prior to lysis) bound to HDAC8 (PDB 3EWF).
Inhibitor Development.
As HDACs have been a highly sought after cancer target for ~30 years, there has been a substantial amount of inhibitor development against these enzymes. As described above, because the catalytic Zn2+ ion is essential for HDAC activity, virtually all inhibition campaigns have incorporated some form of MBP as the basis for inhibitor activity through coordination to the catalytic Zn2+ within the active site. This inhibitor design approach has meet great success, with four HDAC inhibitors approved for treatments of various cancers, and many more in clinical trials. (Figure 97, Table 14).
Table 14.
Activity of HDAC inhibitors.
| Compound | Inhibition (nM) | Ref. |
|---|---|---|
| HDACi-1 | IC50 = 900, HDAC2 | 921 |
| HDACi-2 | IC50 = 90, HDAC8 | 922 |
| HDACi-3 | IC50 = 36, HDAC7 | 923 |
| HDACi-4 | IC50 = 367, HDAC4 | 924 |
| HDACi-5 | IC50 = 60, HDAC6 | 925 |
| HDACi-6 | IC50 = 680, HDAC6 | 926 |
| HDACi-7 | IC50 = 306, HDAC6 | 926 |
| HDACi-8 | IC50 = 356, HDAC6 | 927 |
| HDACi-9 | IC50 = 150, HDAC6 | 928 |
| HDACi-10 | IC50 = 4,200, HDAC6 | 928 |
| HDACi-11 | IC50 = 8,800, HDAC6 | 928 |
| Belinostat | IC50 = 40–125, HDAC 1, 2, 3, 6 | 929 |
| Entinostat | IC50 = 250, HDAC2 | 930 |
| HPOB | IC50 = 56, HDAC6 | 931 |
| Panobinostat | IC50 = 3–60, HDAC 1, 2, 3, 6 | 929 |
| Romidepsin | EC50 = 0.02–0.2 | 932 |
| SAHA | IC50 = 10–50, HDAC 1, 2, 3, 6 | 933,934 |
| Trapoxin A | IC50 = 0.82, HDAC1 | 935,936 |
| Trichostatin A | IC50 = 1, HDAC6 | 937 |
Suberoylanilide hydroxamic acid (Vorinostat, SAHA, Figure 97) has enjoyed a place of prominence in the field of HDAC inhibitors. SAHA was among the earliest reported HDAC inhibitors, and its general structure has remained the basis of inhibitor development for countless drug development campaigns. The path to SAHA was unusual, especially considering that SAHA was developed through phenotypic screening, before HDACs were known to be the in vivo target of the compound.228 The discovery of SAHA began with the simple observation that dimethyl sulfoxide (DMSO) turned erythroleukemia cells red, which suggested the presence of hemoglobin,938 and eventually led to the realization that polar small molecules could induce cellular growth arrest and differentiation in already transformed cells.939 Follow-up work led to diamide alkyl compounds with optimization of a six-carbon linker between amide groups.228 Subsequent observations implicated metal coordination, which prompted coversion of the diamides into bis(hydroxamic acid) compounds.228 Finally, replacement of one of the hydroxamic acids with a phenyl group produced SAHA.940 The target of SAHA was identified to be HDACs two years later in 1998,941 and SAHA became the first-in-class FDA approved HDAC inhibitor in 2007.942 Interestingly, the natural product trichostatin A (Figure 97) has a striking structural resemblance to SAHA and was independently identified as an inhibitor of HDACs.943
The majority of inhibitor development against HDACs has since been based on the SAHA scaffold, and has resulted in the development of Belinostat, and Pabinostat. SAHA and these other hydroxamic acid inhibitors have been crystallographically confirmed to bind to the HDAC active site with the hydroxamic acid coordinating to the catalytic Zn2+ as a monoanionic ligand through the carbonyl and hydroxyl donor atoms. The Zn2+-bound water is displaced by the inhibitor leaving the metal center in a trigonal bipyramidal geometry, with the phenyl ring of SAHA acting as a capping group to block off the entrance of the HDAC active site (Figure 98).920 Overall, the majority of hydroxamate HDAC inhibitors consist of the same structure of the hydroxamate MBP, a flexible linker, and a capping group designed to impart isoform selectivity. Extensive efforts have gone into developing HDAC isoform selective inhibitors through modification of the capping group.944–947
Figure 98.
Structures of: SAHA bound to HDAC6 (PDB 5EEI); HDACi-1 bound to HDAC2 (PDB 3MAX); HDACi-2 bound to HDAC8 (PDB 3SFH); HDACi-3 bound to HDAC7 (PDB 3ZNR); Trapoxin A bound to HDAC1 (PDB 5VI6); Largazole bound to HDAC8 (PDB 3RQD); HDACi-4 bound to HDAC4 (PDB 2VQJ); HPOB bound to HDAC6 (PDB 5EF7). Hydrogen bonding (dashed magenta lines) are shown for select inhibitors.
The macrocyclic depsipeptide romidepsin (Figure 97) was first reported as an HDAC inhibitor in 1998.948 The disulfide bond undergoes reduction within the cellular environment to free the thiol MBP for metal binding.949 Indeed, the disulfide can be viewed a sort of natural protecting group or prodrug. While romidepsin has not been crystallized with an HDAC, the structural analogue largazole, has been crystallized with HDAC8.950 Largazole binds HDAC8 through the terminal thiol coordinating to the catalytic Zn2+ in tetrahedral geometry,950 and it can be assumed through structural similarity that romidepsin binds in a similar fashion (Figure 98).
Apart from FDA approved compounds, there are currently eight HDAC inhibitors in clinical trials. As shown in Figure 97, the majority of these HDAC inhibitors are hydroxamic acid based, with large amounts of variation within the “capping group”, which is designed to impart selectivity between the HDAC isoforms. The majority of these compounds are hydroxamate based, so they likely inhibit and bind HDACs in a similar manner to SAHA. That stated, at least one hydroxamate inhibitor is reported to have a mode of metal binding differing from the other hydroxamic acids, namely N-hydroxy-4-(2-[(2-hydroxyethyl)(phenyl)amino]-2-oxoethyl)benzamide (HPOB, Figure 97). HPOB has been reported to be selective for HDAC6 with an IC50 value of 56 nM.931 Crystallographically, HPOB has been found to bind in an unusual manner that does not result in displacing the native Zn2+-bound water. HPOB coordinates to the Zn2+ ion solely through the hydroxyl donor atom, with the hydroxamic carbonyl instead making H-bond contacts with the Zn2+-bound water (Figure 98).951 It has been hypothesized that the shorter linker length of HPOB combined with this unusual metal binding mode may be the source of HDAC selectivity for HPOB.
In contrast to the plethora of hydroxamic acid based HDAC inhibitors, there are several HDAC inhibitors that have been developed with other MBPs. The structures of several of these inhibitors bound to HDACs have been determined (Figure 98). The inhibitor entinostat (IC50 = 250 nM against HDAC2) has never been co-crystallized, but its analogue, HDACi-1 has been with HDAC2.921 HDACi-1 binds to the catalytic Zn2+ through an anilinic nitrogen atom and the carbonyl oxygen atom generating a trigonal bipyramidal geometry. Within this structure, the phenyl ring of the amide extends into a seldom-explored region of the HDAC active site called the ‘foot pocket’. It is assumed that entinostat binds in a similar fashion with trigonal bipyramidal geometry. Interesting another nitrogen donor atom based inhibitor, HDACi-2 (IC50 = 90 nM), instead binds through a singular sp3 amine donor atom placing the Zn2+ ion in a tetrahedral geometry.922
Among the other unusual MBPs reported for the HDAC scaffold is the trifluoromethyloxadiazolyl group of HDACi-3.923 This compound does not coordinate to the Zn2+ through traditional donor atoms, but rather is reported to display a cation-π interaction with the Zn2+ ion, which produces a compound has an IC50 value of 36 nM against HDAC7. The ‘coordinating’ ring is 3.2 Å away from the Zn2+, which is a typical distance seen in aromatic ring and cation-π interactions, and the fluorine atom is 3.0 Å away from the Zn2+ (Figure 98).952 The structure shows that the related HDACi-4 has weak, if any, interactions with the Zn2+ ion, suggesting that this unorthodox mode of active site interaction is not representative of true metal coordination.
Other inhibitors bearing carbonyl moieties, such as Trapoxin A (IC50 = 0.82 nM against HDAC2),953 have been shown to undergo nucleophilic attack by the Zn2+-bound catalytic water. This generates a gem-diolate MBP that coordinates to the Zn2+, which acts as a mimic of the tetrahedral reaction intermediate. In particular, because Trapoxin A is such a superb inhibitor, it was initially believed to be a covalent HDAC inhibitor that utilized the epoxide moiety to bind to the HDAC active site.935,936 Crystallography with HDAC8 revealed that Trapoxin A does not covalently modify the HDAC active site, and that the epoxide remains intact (Figure 98).953 Additional studies using mass spectrometry as well as isothermal titration calorimetry further confirmed the crystallographic evidence that Trapoxin A does not covalently modify HDAC.953
Finally, there are several compounds that contain known MBPs that are highly likely to inhibit through coordination to the active site Zn2+, but have not been crystallographically confirmed to have a metal-binding mechanism of action. The sulfonamide HDACi-5 (Figure 97) was used as a mimic of the general structure of SAHA, but replaces the hydroxamic acid with a terminal sulfonamide group as the MBP.925 Other reports utilized the hydroxypyridinethione scaffold,926 specifically a 2-hydroxy-pyridiene-3-thione was selected over other hydroxy pyridienthiones scaffolds based on computational docking, which predicted the heterocyclic nitrogen to be favorably positioned for further derivatization. The 2-hydroxy-pyridiene-3-thione MBP (HDACi-6, Figure 97) has an IC50 value of 680 nM, and further elaboration resulted in HDACi-7 with a slightly improved IC50 value of 306 nM.927 Alternatively, a 1-hydroxy-pyridine-2-thione MBP (HDACi-9, Figure 97) showed an IC50 value of 150 nM.928 Unfortunately, the carboxylic acid in HDACi-9 was likely very important for interactions within the active site, as amide derivatization off the carboxylic acid resulted in a drastic loss of activity in HDACi-10 and HDACi-11 with IC50 values of 4,200 nM and IC50 = 8,800 nM, respectively.
Current and Future Prospects.
HDACs are a class of epigenetic regulatory enzymes that function to deacetylate lysine residues. This act of deacetylation results in electrostatic interactions causing histones to coil more tightly around DNA, effectively silencing certain regions. Cancer tumors take advantage of this HDAC action to down regulate factors that signal apoptosis in malignant cells. Thus, HDACs have been a target of interest in anti-cancer programs to restore these apoptosis pathways. This strategy of HDAC inhibition has been meet with substantial success, with four FDA approved HDAC inhibitors used in cancer treatment. In particular, SAHA is noteworthy story as this first-in-class therapeutic was not developed through traditional rational SAR, but by phenotypic analysis. Additionally, all of these FDA approved HDAC inhibitors contain MBPs that take advantage of the binding to the catalytic Zn2+ ion, which further affirms the strategy of metalloenzyme inhibition through metal-coordination.
Outside of the relation to anticancer therapeutics, HDACs have been shown to capable of deacetyling substrates outside of histones, indicating that HDAC inhibition can be of benefit in diseases beyond cancer. Indeed, studies have shown that HDACs are involved in regulating inflammation, and can potentially be explored as treatment for autoimmune disorders.954 Additionally, HDACs play a role in neurodegenerative conditions and HDAC inhibition may provide a route to treat these disorders.955 Interestingly, the ability of HDAC inhibitors to modulate DNA activation is also being explored against HIV in order to re-activate and clear latent pools of the retrovirus.956,957 Due to the multifaceted roles of HDACs, they remain an important target class in metalloenzyme inhibition. Recent trends in the field of HDAC inhibition have focused on improving isoform selectivity, for example by variation of the capping group, which is believed to be important for future advancement and greater clinical adoption of this class of therapeutics.
7.4. UDP-3-O-Acyl-N-acetylglucosamine Deacetylase (LpxC, EC 3.5.1.108)
Function in Biology and Disease.
The “ESKAPE” pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) are the leading cause of nosocomial infections worldwide and have developed various drug-resistant mechanisms, including drug inactivation, modification of drug binding sites, and changes in cell permeability.958 The Gram-negative bacteria cell envelope comprises of an outer membrane (OM) in addition to the cell membrane and a peptidoglycan layer. The OM is composed of lipopolysaccharides (LPSs), proteins, and phospholipids, with the LPS acting as a selective barrier to prevent the entrance of antibiotics and other exogenous material.959 Lipid A (also known as endotoxin) is a glucosamine-based saccharolipid and the hydrophobic membrane anchor motif of LPS.960,961 Lipid A is vital to the growth and survival of the vast majority of Gram-negative pathogens, and disruption of lipid A biosynthesis has shown to increase antibiotic susceptibility.960–962 The mammalian immune system has picomolar sensitivity for the detection of lipid A that can lead to fatal inflammatory responses associated with septic shock (such as hyper-production of inflammation mediators and tissue factor production).963–967 Hence, the development of lipid A biosynthesis inhibitors is an attractive and promising target for tackling antibiotic resistant Gram-negative pathogens, as well as reducing the complications of septic shock.
Lipid A biosynthesis, also known as the Raetz pathway, consists of a series of nine enzymes for the biosynthesis of lipid A and is conserved in most Gram-negative pathogens.960,968 UDP-3-O-acyl-N-acetylglucosamine deacetylase (LpxC) catalyzes the first committed step in the Raetz pathway, and is a Zn2+-dependent metalloenzyme responsible for the deacetylation of UDP-3-O-[(3R)-3-hydroxymyristoyl]-N-acetyl-alpha-D-glucosamine (myr-UDP-GlcNAc) to produce UDP-3-O-[(3R)-3-hydroxymyristoyl]-alpha-D-glucosamine (myr-UDP-GlcN, Figure 99).960,969,970 LpxC is one of the two most conserved enzymes of the Raetz pathway and bears no human homology,968 making it an attractive target for the development of new antibiotics.971 Efforts both in academia and the pharmaceutical industry have been undertaken to develop potent and clinically useful inhibitors of LpxC; to date, none have successfully entered the drug market.
Figure 99.
LpxC is responsible for the deacetylation of myr-UDP-GlcNAc to produce myr-UDP-GlcN.
Protein and Active Site Structure.
E. coli LpxC is the most divergent orthologue amongst the known LpxCs, sharing 34% sequence homology with A. aeolicus LpxC, 57% sequence homology with P. aeruginosa LpxC, and >92% sequence homology with S. typhimurium and Y. pestis LpxC.972 Alignment of the backbone α-carbons of LpxC from various species yield a RMS deviation of ~0.8 to 1.4 Å,973 revealing a conserved tertiary structure. Despite the conserved tertiary structures amongst the LpxC from various species, the development of broad-spectrum inhibitors of LpxC has proved challenging.
The X-ray crystal structure of E. coli LpxC bound to reaction product myr-UDP-GlcN and a phosphate ion (from the crystallization medium) revels a 305-residue protein (~34 kDa) in two tightly packed “β-α-α-β sandwich” domains (Figure 100).973 Domain I contains Insert I (a small three-stranded β-sheet) while Domain II contains Insert II (β–α–β structure). The catalytic site is situated in a cleft between the interface of the two inserts and contains two pockets: a hydrophobic tunnel (defined by Insert II and the focus for inhibitor development) and a UDP-binding pocket. The active site of LpxC contains a Zn2+ ion coordinated to the residues His79, His238, Asp242, and a phosphate ion (from the crystallization buffer) in a tetrahedral coordination geometry. In addition to the phosphate anion being stabilized by the side chains of Thr191, His265, and Glu78 in the active site, it further interacts with the 2-amino and myristoyl 3-hydroxyl groups of myr-UDP-GlcN residing in the UDP-binding pocket.973
Figure 100.
Structure of myr-UDP-GlcN product (yellow) and phosphate ion bound to E. coli LpxC (PDB 4MDT).
Inhibitor Development.
Several decades of research have led to the discovery of various LpxC inhibitors. Inhibitor development has primarily focused on identifying compounds that can effectively target a wide range of Gram-negative pathogens with good cell permeability. While a variety of inhibitor backbones have been investigated to probe the hydrophobic tunnel, very few MBPs have been explored. Many LpxC inhibitors bearing a hydroxamic acid MBP are reported in literature972,974–985 and disclosed in patents,986–989 and representative inhibitors are discussed below. Often inhibitors are screened against a variety of pathogens, making it difficult for a direct comparison of their efficacy.
A library of over 200 phenyloxazoline inhibitors bearing a hydroxamic acid MBP were synthesized and screened against E. coli LpxC.990,991 L-161,240 (Figure 101), was identified as a potent inhibitor of E. coli LpxC (Ki = 50 nM), showed antibacterial activity against E. coli (MIC = 1 – 3 μg/mL), and rescued mice from fatal E. coli infections (EC50 = 15 mg/kg).990 However, L-161,240 showed no activity against P. aeruginosa (MIC >100 μg/mL) and other clinically relevant pathogens.971,990 The resistance from P. aeruginosa stems from variance in the intrinsic enzyme active site pocket rather than differences between membrane structure or efflux pumps, and encouraged further development of a broad-spectrum LpxC inhibitor. TU-514 (Figure 101), a substrate analogue, was later reported as a modest inhibitor of E. coli and A. aeolicus LpxC (IC50 = 7.2 and 7.0 μM).992 A. aeolicus LpxC shares the least sequence homology with E. coli LpxC (~34%) and was resistant to the inhibition by L-161,204, hence TU-514 validated the concept of developing broad-spectrum LpxC inhibitors. However, the limited cell permeability of TU-514 prohibited antibacterial activity and usefulness for clinical development.
Figure 101.
Inhibitors of LpxC.
Further development of inhibitors of LpxC utilize a variety of backbones to expand the efficiency of the antibiotic to reach a broader spectrum of Gram-negative pathogens and increase cell permeability while maintaining the hydroxamic acid MBP. Representative inhibitors include sulfonamide inhibitor BB-78485 and the threonine-based inhibitors CHIR-090 and LPC-009 (Figure 101)972,993–997 BB-78485 was a potent inhibitor of E. coli LpxC (IC50 = 160 nM, MIC = 1 μg/mL) and inhibited a broader spectrum of Gram-negative pathogens compared to that of L-161,240. BB-78485 was effective against pathogens Burkholderia cepacia, Serratia marcescens (MIC = 1 μg/mL), and others.993 Unfortunately, no activity was observed for P. aeruginosa (MIC >32 μg/mL), and BB-78485 was not effective enough to be of clinical relevance. CHIR-090, a slow, tight-binding inhibitor of LpxC from various Gram-negative species (Ki = 1–4 nM, for E. coli, P. aeruginosa, and A. aeolicus), exhibited antibacterial activity against both E. coli and P. aeruginosa that was as effective as the antibiotic ciprofloxacin and tobramycin.986,994–996 The X-ray crystal structure of CHIR-090 bound to P. aeruginosa shows the hydroxamic acid MBP binding in a bidentate manner to the Zn2+ ion, which is further ligated by the residues His78, His237, and Asp241 (P. aeruginosa residue numbering) in a trigonal bipyramid coordination geometry (Figure 102).998 A comparison of CHIR-090 bound to P. aeruginosa with CHIR-090 bound to A. aeolicus995 and Y. enterocolitica999 LpxC reveal identical MBP binding. Unfortunately, CHIR-090 was not as effective in inhibiting Rhizobium leguminosarum LpxC (Ki = 340 nM) compared to E. coli LpxC (Ki = 4 nM) as a result of a Gly-to-Ser point mutation present in the Insert II of R. leguminosarum.995,996 This mutation resulted in a narrower hydrophobic tunnel, causing van der Waal clashes of the tunnel with the distal phenyl ring of CHIR-090, and is the source of CHIR-090 resistance by R. leguminosarum (Figure 102).995 The resistance of R. leguminosarum LpxC highlights concerns of possible resistance against LpxC inhibitors by a single point mutation.
Figure 102.
Left: Structure of CHIR-090 bound to P. aeruginosa LpxC (PDB 5U39). Right: Structure of LPC-009 bound to P. aeruginosa LpxC (PDB 3P3E).
To design inhibitors that could fit into the hydrophobic tunnel and expand their spectrum of antibiotic activity, linear diacetylene motifs were investigated.972,997 LPC-009 was found to be a potent inhibitor of R. leguminosarum LpxC knock in-strain of E. coli (W3110RL), reducing the MIC difference between W3110RL and wild-type E. coli (W3110) to 126-fold (compared to the 600-fold difference exhibited by CHIR-090) and successfully overcame the resistance mechanism of R. leguminosarum LpxC. Additionally, LPC-009 does not display time-dependent inhibition (Ki = 0.18 nM against E. coli LpxC), shows 4-fold enhanced antibacterial activity against E. coli (MIC = 0.05 μg/mL) when compared to CHIR-090, and shows a 2- to 64-fold enhancement in antibacterial activity against the growth of eight additional Gram-negative pathogens compared to that of CHIR-090.972 X-ray crystallographic analysis of LPC-009 bound to P. aeruginosa LpxC revealed the hydroxamic acid of LPC-009 binding in a similar fashion of that CHIR-090 with the hydroxamic acid MBP binding bidentate fashion to the Zn2+ ion in a trigonal bipyramid coordination geometry (Figure 102). The linear diphenyl-acetylene group of LPC-009 is able to penetrate through the hydrophobic passage of R. leguminosarum and reduce the steric clash induced by the Gly-to-Ser point mutation. Unfortunately, LPC-009 did not display inhibitor activity against A. baumannii, a pathogen associated with nosocomial infections, and was replaced by other broader-spectrum diacetylene derivatives.1000 Lastly, hydroxamic acids bearing a pyridone methylsulfone motif have also been reported by Pfizer, such as PF 05081090 (Figure 101).976 PF 05081090 was a potent inhibitor of P. aeruginosa LpxC (IC50 = 1.1 nM) and displayed an MIC value of 0.008 μg/mL against P. aeruginosa.978 Additionally, PF 05081090 displayed an MIC90 values of 0.25 – 1.0 μg/mL against P. aeruginosa, E. coli, K. pneumoniae, and others. Unfortunately, PF 05081090 was not active against A. baumannii (MIC90 >64 μg/mL), hindering development into a clinically useful drug.989
The first and only inhibitor of LpxC to enter clinical trials was ACHN-975 (Figure 101). ACHN-975 was reported by Achaogen Inc. as potent inhibitor of P. aeruginosa LpxC (IC50 = 0.02 nM).1001 ACHN-975 successfully inhibited the growth of 98.6% of 998 P. aeruginosa isolates (including isolates that were antibiotic resistant, MIC ≤1 μg/mL), showed antibacterial activity against 98.3% of an additional 120 Gram-negative isolates, including LpxC from that of E. coli, K. pneumoniae and Y. enterocolitica (MIC ≤ 4 μg/mL), and yielded synergistic effects when administered with vancomycin in infected mice.989,1001 Unfortunately, despite promising pre-clinical results, Phase I clinical trials of ACHN-975 was terminated after one year due to inflammation at the infusion site of some subjects. This side effect is not believed to be target related, and details regarding the pre-clinical and Phase I clinical studies of ACHN-975 is described elsewhere.989
Non-hydroxamate LpxC inhibitors include hydantoin,1000 phosphinic acid, and mercapto MBPs.1002 While these inhibitors generally exhibit moderate activity against E. coli LpxC (IC50 ≥4 μM), they lacked detailed SAR analysis and their potential for as a broad-spectrum antibiotic is not determined. Recently, an inhibitor of LpxC that uses a hydroxypyrone MBP, LpxCi-1 (Figure 101),1003,1004 displayed an IC50 <0.10 μM against E. coli LpxC. Unfortunately, LpxCi-1 showed modest antibacterial activity when screened against E. coli, S. aureus, and P. aeruginosa (MIC >32 μg/mL).1003 Other disclosed hydroxypyrone inhibitors of LpxC showed inhibition against K. pneumonia (IC50 ≥10 nM) in vitro, but were ineffective when evaluated against E. coli LpxC and P. aeruginosa LpxC (MIC >32 μg/mL). Although no structural data is presently available, the hydroxypyrone MBP is expected to bind in a bidentate manner (through the phenolic and carbonyl oxygens) to the Zn2+ ion, similar to that of the previously presented hydroxamate inhibitors.
Current and Future Prospects.
Due to the rapid emergence of multidrug resistant Gram-negative bacteria, development of antibiotics with novel mechanisms of action is of urgent need. LpxC inhibitors have been developed in both academic and pharmaceutical labs; however, pre-clinical advancement of most inhibitors has stalled due to a lack of broad spectrum activity. The only inhibitor to successfully enter clinical trials was ACHN-975, but the program was terminated due to inflammation at the infusion site of the patents, and there are currently no inhibitors of LpxC in clinical development. The desire to design a broad-spectrum inhibitor has led to extensive investigation of various backbones that can target the hydrophobic tunnel of LpxC. In contrast, optimization of the hydroxamic acid MBP has received far less attention. The majority of inhibitors reported to date utilize a hydroxamic acid MBP, therefore the development of inhibitors with new MBPs is warranted. Further research should not only investigate in new MBPs but also inhibitors that can bind to the UPD-binding pocket, as current inhibitors leave the UPD-binding pocket unoccupied. Development of non-hydroxamic acid MBP inhibitors that can bind both sites (the UDP-binding pocket and the hydrophobic passage) may provide added binding interactions needed for more potent and broad spectrum inhibition.
7.5. Metallo-β-Lactamase (MBL, E.C. 3.5.2.6)
Function in Biology and Disease.
Since their accidental discovery by Fleming in 1928, β-lactam drugs have become the most successful and commonly prescribed class of antibiotics. Their powerful anti-bacterial activity stems from their ability to block peptidoglycan synthesis through inhibition of the unique bacterial transpeptidase enzyme, which disrupts and weakens the cell wall, eventually leading to cellular lysis and death (Figure 103).1005
Figure 103.
The mechanism of β-lactam inhibition of transpeptidase enzyme. The right portion displays the normal action of transpeptidase crosslinking, whereas the left pathway displays the mechanism of covalent inhibition by a generic β-lactam.
Unfortunately, the overuse of these antibiotics has created a natural selection pressure for bacteria to adapt and develop resistance mechanisms to ß-lactam drugs.1006 Some of these resistance mechanisms include efflux pumps, mutations of target proteins, and most problematic, the emergence of ß-lactamase enzymes.1007 ß-lactamases confer bacterial resistance through hydrolysis of the ß-lactam bond that is crucial for the activity of these drugs.1007 There are several classes of ß-lactamases, where classes A, C, and D rely on an active site serine residue to accomplish their hydrolytic activity.1006 Class A is selective for penicillins, C for cephalosporins, and D for oxacillins.1006 Class B is unique among the ß-lactamases in that these consist of metallo-ß-lactamases (MBLs) that rely on one or more active site Zn2+ ions to achieve ß-lactam cleavage. Of the MBLs, New Delhi Metallo-ß-Lactamase-1 (NDM-1), Verona Integrase Metallo-ß-Lactamase-2 (VIM-2), and Imipenemase-1 (IMP-1), and have been the most prevalent in terms of posing a threat to human health.1008 Of these, NDM-1 is generally regarded as the MBL of greatest concern as it is the most easily communicable and has been shown to be capable of hydrolyzing all clinically relevant ß-lactams,1009 including the carbapenems, which are used as a last resort against resistant infections. Therefore, this Section will focus on NDM-1 as a representative of the MBL family (unless otherwise noted).
NDM-1 is thought to have originated within the Xanthomonas/Psuedoxanthomonas species of rice plant pathogenic bacteria,1010 but it has since been documented in multiple members of the Enterobacteriaceae family, the Acinetobacter family, and as well as the Pseudomonas family.1011 NDM-1 was first reported in 2008 in a Swedish patient visiting New Delhi,1012 and has since been documented worldwide, with most NDM-1 infections predominately located in India.1013 NDM-1 is easily communicable between different bacteria populations as it is carried on blaNDM-1,1012 a plasmid capable of horizontal gene transfer. In addition to conferring ß-lactam resistance, the blaNDM-1 plasmid is also known to encode resistance to many other common antibiotics, including aminoglycosides, fluoroquinolones, macrolides, and sulfonamides, so that NDM-1 bearing infections are often difficult to treat as they display pan-antibiotic resistance.1014,1015 In light of rapid transmission and widespread resistance to conventional antibiotic treatments, NDM-1 harboring pathogens have been labeled as “superbugs” and are widely considered to be of grave concern towards human health.1016
Protein and Active Site Structure.
Typical of other MBLs, NDM-1 relies on two active site Zn2+ ions to achieve ß-lactam hydrolysis (Figure 104). Within the active site, ZnA2+ is bound by His122, His120, and His189 in a tetrahedral geometry, while ZnB2+ is bound by a Cys208, His250, and Asp124 in a distorted tetrahedral geometry. The two metal ions are bridged by a hydroxyl anion.1017 ZnA2+ is tightly bound, but ZnB2+ is weakly coordinated with a Kd value of 2 μM.1018 The active site of NDM-1 surrounding the dinuclear center consists of a long, shallow, solvent-exposed groove1019 with a large β-hairpin loop that is closed during substrate binding and catalysis; this loop is largest in NDM-1 compared to other MBLs, such as VIM-2 and IMP-1.1020 In addition, the active site of NDM-1 structure is more flexible than other MBLs, which allows NDM-1 to accommodate such a broad range of substrates, including the carbapenems.1021 The mechanism of ß-lactam cleavage involves coordination of the ß-lactam substrate to the Zn2+ ions, which polarizes the ß-lactam, making it more electrophilic and susceptible to nucleophilic attack. The intrinsic Lewis acidity of the active site Zn2+ ions lowers the pKa of the bridging hydroxyl,1022 making it more nucleophilic and effectively catalyzing insertion of the hydroxyl into the carbonyl, thus cleaving the ß-lactam bond.
Figure 104.
Structure of NDM-1 (PDB 3SPU).
Inhibitor Development.
Currently there are no FDA approved drugs that target NDM-1 or any other MBL. NDM-1 bearing infections have been historically been treated through the administration of nitrofurantoin1023 or by antibiotics of last resort (e.g., colistin) in the hopes of exploiting any antibiotic resistance not covered by the powerful blaNDM-1 cassette.1024 These treatments are not expected to be viable long-term solutions as growing resistance these therapeutics has already been reported; in particular with the recent development of MCR-1 (Section 4.3).353
Unfortunately, NDM-1 has been a difficult target for inhibitor development; mainly due to the aforementioned flexibility of the active site.1008 The vast majority of the inhibitor development against NDM-1 has focused on the incorporation of some form of MBP in an effort to directly target the Zn2+ ions that are responsible for the catalytic activity of this metalloenzyme (Figure 105, Table 15).1008
Figure 105.
Inhibitors of MBLs (NDM-1).
Table 15.
Activity of MBL inhibitors. Values are against NDM-1 unless otherwise noted.
| Compound | Inhibition (μM) | Ref. |
|---|---|---|
| MBLi-1 | IC50 = 0.09a | 1025 |
| MBLi-2 | Ki = 0.22b | 1026 |
| MBLi-3 | Ki = 0.4c | 1027 |
| MBLi-4 | IC50 = 290d | 1028 |
| MBLi-5 | IC50 = 9.44 | 1029 |
| MBLi-6 | IC50 = 1.05 | 1029 |
| MBLi-7 | IC50 = 23 | 1030 |
| MBLi-8 | IC50 = 232 | 1031 |
| MBLi-9 | IC50 = 12 | 1032 |
| MBLi-10 | IC50 = 7.1 | 1033 |
| MBLi-11 | IC50 = 12 | 1033 |
| MBLi-12 | IC50 = 0.27a | 1034 |
| MBLi-13 | 18% inhibition at 50 μMa | 1034 |
| MBLi-14 | IC50 = 68a | 1034 |
| MBLi-15 | IC50 = 0.42 | 1035 |
| MBLi-16 | IC50 = 3.57 | 1036 |
| MBLi-17 | IC50 = 122a | 1037 |
| MBLi-18 | IC50 ~ 100 | 1038 |
| MBLi-19 | Ki = 24 | 1039 |
| MBLi-20 | IC50 = 87.9 | 1040 |
| MBLi-21 | IC50 = 280 | 1041 |
| MBLi-22 | IC50 = 0.27a | 1042 |
| MBLi-23 | IC50 = 14b | 1031 |
| MBLi-24 | IC50 = 20b | 1043 |
| MBLi-25 | IC50 ~ 2 | 1044 |
| MBLi-26 | IC50 = 0.004 | 1045 |
| MBLi-27 | IC50 = 0.029 | 1045 |
| MBLi-28 | IC50 = 32.4 | 1046 |
| MBLi-29 | IC50 = 6.6 | 1047 |
| MBLI-30 | IC50 = 0.374 | 1048 |
| AMA | IC50 = 4 | 1049 |
| D-captopril | IC50 = 20.1 | 1050 |
| L-captopril | IC50 = 157.4 | 1050 |
| epi-D-captopril | IC50 = 64.6 | 1050 |
| epi-L-captopril | IC50 > 500 | 1050 |
| 2,3-dimercaprol | IC50 = 1.3 | 1051 |
| DPA | IC50 = 0.41 | 1052 |
| DL-thiorphan | IC50 = 1.8 | 1051 |
Inhibition activity against IMP-1.
Inhibition activity against VIM-2.
Inhibition activity against the L1 MBL.
Inhibition activity against the B1a2 MBL.
Thiol MBP inhibitors are most widely explored class of compounds against NDM-1. Many of these thiol-based inhibitors include a carboxylic acid (or carboxylate bioisostere) to mimic the transition state of the native hydrolyzed substrate, such as MBLi-1, MBLi-2, and MBLi-3, which displayed IC50 values ranging from 90–400 nM against various MBLs.1025–1027 Compound MBLi-4 was tested against the MBL Bla2 with a reported IC50 value of 290 μM.1028 The rhodanine containing MBLi-5 was discovered through X-ray crystallography to have an interesting mechanism of inhibition through decomposition into MBLi-6 as the active inhibitor.1029 MBLi-6 binds with the thiol bridging between the Zn2+ ions, and the carboxylic acid also coordinated to ZnB2+, so that ZnA2+ is in a tetrahedral geometry, and ZnB2+ is in a trigonal bipyramidal geometry (Figure 106).1029 Similarly, the bisthizolidine MBLi-7 was found to primarily bind by bridging through the terminal thiol.1030 Although it contains a thiol ligand, MBLi-8 (IC50 = 232 μM) coordinates both Zn2+ ions, but with a nitrogen of the triazole ring coordinated to ZnA+ and the sulfur atom coordinated to ZnB2+ in a non-bridging fashion (Figure 106).1031 A derivative of MBLi-8, MBLi-9, was reported to have an IC50 value of 12 μM against NDM-1.1032
Figure 106.
Structures of: D-captopril bound to VIM-2 (PDB 4C1E), MBLi-7 bound to NDM-1 (PDB 4U4L), MBLi-17 bound to SPM (PDB 5NDB), MBLi-23 bound to VIM-2 (PDB 5ACX), MBLi-27 bound to VIM-2 (PDB 5FQC), and MBLi-30 bound to IMP-1 (PDB 5HH4).
Other studies have focused on the repurposing of approved drugs as MBL inhibitors. L-Captopril, which is approved as an ACE inhibitor (Section 6.1) has been one of the most commonly studied MBL inhibitors, with an IC50 value of 157 μM against NDM-1.1050 Interestingly, the enantiomer D-captopril has been found to be much more active against NDM-1 with an IC50 value of 20 μM against NDM-1.1050 While D-captopril has never been crystallized in NDM-1, it has been crystallized in VIM-2, and this structure reveals that the inhibitor binds with the terminal thiol bridging between the two Zn2+ ions (Figure 106).1050 The remainder of the inhibitor is oriented to point towards ZnB2+ so that the carboxylic acid on D-captopril is optimally placed to make interactions with a nearby Arg residue.1050 For the structure of L-captopril bound to NDM-1, Zn2+ binding is maintained, but the carboxylic acid is positioned away from a cationic residue, thus resulting in the lower activity of L-captopril compared to D-captopril.1053 Additionally, epi-L-captopril and epi-D-captopril were found to follow the same trend with epi-D-captopril being more active compared to epi-L-captopril.1050
Another study examining FDA-approved drugs containing thiol moieties (Figure 105),1051 included thiomandelic acid, D,L-captopril, D,L-thiorphan, N-acetylcysteine, meso-2,3-dimercaptosuccinic acid, 2,3-dimercaprol, D-(-)-penicillamine, glutathione, L-cysteine, zofenoprilat, and tiopronin, showed that D,L-thiorphan and 2,3-dimercaprol were the most active with IC50 values of 1.8 μM and 1.3 μM against NDM-1, respectively. While neither of these inhibitors were crystallized within the NDM-1 active site, tiopronin has been. Tiopronin has a unique binding mode for a terminal thiol against NDM-1 in that while most of the other thiol based inhibitors (MBLi-6, MBLi-7, MBLi-15, MBLi-16, L-captopril, and D-captopril) adopt a bridging coordination mode, tiopronin only coordinates to ZnB2+ through the thiol and amide nitrogen atom generating a trigonal bipyramidal geometry (Figure 106).1051
MBLi-10 and MBLi-11 were prepared as carboxylic acid bioisosteres of the earlier reported MBLi-1 (Figure 105), but both were far less active than MBLi-1.1033 While neither of these compounds were structurally characterized in the active site of NDM-1, a thio-acetylated version of MBLi-10 was crystallized in the active site of VIM-2, and it was found that the sulfur still preferentially adopts a bridged coordination mode between the Zn2+ ions.1033 Other thiol based inhibitors of MBLs include compounds published as inhibitors of VanX (Section 6.9) and HDACs (Section 7.3), namely MBLi-12, MBLi,-13 and MBLi-14.1034 None of these compounds showed particularly notable activity as MBL inhibitors (Table 15).
Other MBL inhibitors include MBLi-15, which was discovered through in silico screening and has an IC50 value of 0.42 μM against NDM-1.1035 Crystal structure analysis of an analogue of MBLi-15 in VIM-2 shows that the thiol-based inhibitor also adopts a bridged mode of coordination with the sulfur binding mu between the two Zn2+ ions.1035 Finally, the most recently reported thiol-based inhibitor, MBLi-16 was designed based on L-captopril, and has an IC50 = 3.57 against NDM-1.1036 Crystallographic analysis of MBLi-16 in VIM-2 shows that MBLi-16 also shares the typical binding mode of bridging coordination through the thiol between the Zn2+ ions.1036
Beyond the thiol MBP, the vast majority of MBL inhibitors have utilized a carboxylic acid (or carboxylic acid bioisosteres) as the MBP, so that the carboxylate serves to both coordinate the metal and function as a transition state mimic of the native ß-lactam substrate. One of the earliest of these transition state mimics, MBLi-17, contains the core of a carbapenem antibiotic, but lacks the nitrogen of the ß-lactam, rendering the inhibitor unhydrolyzable.1037 MBLi-17 is a relatively weak inhibitor (IC50 = 122 μM against IMP-1),1037 and has been shown to bind the São Paulo Metallo-ß-lactamase (SPM) through ZnB2+.1054 A related approach to generating ß-lactam mimics resulted in MBLi-18, which replaces the ß-lactam carbonyl with a phosphate group, but activity against NDM-1 was modest (IC50 ~100 μM).1038 Another simple carboxylic acid inhibitor described was MBLi-19 with a Ki value of 24 μM.1039 A complex polyketide inhibitor, MBLi-20, is a natural product isolated from Penicillium sp. also had relatively modest (IC50 = 88 μM against NDM-1).1040
Other work examined a series of carboxylic acids and tetrazoles appended to a thiophene core, and found compounds with a wide range of activities (e.g., MBLi-21, IC50 = 280 μM against NDM-1; MBLi-22, IC50 = 270 nM against IMP-1; MBLi-23, IC50 = 14 μM against VIM-2).1041,1042 Both a derivative of MBLi-22 and MBLi-23 have been structurally characterized bound to MBLs, which show both carboxylate groups engaged in Zn2+ coordination.1031 Other inhibitors, such as MBLi-24 (IC50 = 20 μM against VIM-2) have been shown to adopt a binding pose similar to MBLi-8, but with the triazole binding between the Zn2+ ions, and the carboxylic acid coordinated to Zn22+.1043 Later work replaced the carboxylic acid that is important for ZnB2+ coordination with an aldehyde, resulting in the covalent inhibitor MBLi-25, with an IC50 ~2 μM against NDM-1.1044 While there is no crystal structure of MBLi-25, this inhibitor is thought to act by undergoing nucleophilic attack at the aldehyde by a nearby Lys residue to form an imine that locks the compound in place.
Recently, there has been an interest in the use of boronic acid based inhibitors against MBLs. In particular, the cyclic boronate MBLi-26 was originally developed and patented by Novartis,1055 and was later reported to have an impressive IC50 value of 4 nM against NDM-1,1045 making MBLi-26 the most active reported NDM-1 inhibitor to date. Crystal structure analysis of MBLi-27, which is an analog of MBLi-26, in VIM-2 shows that the cyclic boronate binds in a pose reminiscent of the native substrate, with the carboxylic acid coordinated to ZnB2+ in trigonal bipyramidal geometry, and the boronate moiety coordinated to ZnA2+ also in trigonal bipyramidal geometry.1045 The terminal amine group then flexes to make favorable contacts with a Glu residue in the back wall of the MBL active site.
Outside of a metal-binding mode of action, a few metal-stripping MBL inhibitors have been explored. In particular, the fungal natural product Aspergillomarasmine A (AMA) was reported to have an IC50 value of 4 μM against NDM-1, and was demonstrated to reverse carbapenem resistance in mice.1049 However, AMA is structurally similar to EDTA and has been shown inhibit NDM-1 via metal stripping rather than formation of a stable complex with NDM-1.1049,1056 This is not an ideal mechanism of inhibition as it is difficult to develop selective or targeted metal stripping agents, resulting in an inhibition mechanism that is prone to off-target effects. Other reported metal-stripping compounds include MBLi-28, which has an IC50 value of 6.6 μM against NDM-1.1047
Finally, in a FBDD program, it was demonstrated that a metal-stripping agent could be transformed into a selective inhibitor that forms a ternary complex with NDM-1.1052 This work took the known metal-stripping compound dipicolinic acid (DPA, IC50 = 400 nM against NDM-1), and appended an aniline group to develop MBLi-29 with an IC50 value of 80 nM against NDM-1.1052 Evaluation of MBLi-29 using1H NMR, EPR, equilibrium dialysis, intrinsic tryptophan fluorescence, and UV-vis spectroscopy, confirmed this MBLi-29 forms a ternary complex with NDM-1.1052 No structure of the DPA-derived MBLi-29 with a MBL is presently available, but the structure and activity of a DPA derivative was recently reported (MBLi-30, Figure 106).1048
Current and Future Prospects.
The efficacy of the current generation of antibiotics is rapidly waning due to the rise of antibiotic resistant bacteria; and it is predicted that these resistant bacteria will cause more human deaths than cancer by the year 2050. To that end, it is vital to develop either new antibiotic drugs, or develop methodologies to extend the usefulness of currently existing antibiotic compounds. One such example of the latter is clavulanic acid, a ß-lactam based covalent inhibitor that acts by binding to the active site of serine-ß-lactamases, thereby preventing the premature hydrolysis of ß-lactams. Clavulanic acid (under the name Augmentin) has been FDA approved since 1984, demonstrating that serine ß-lactamase inhibition has long been a viable route for maintaining efficacy of ß-lactam antibiotics.1057 This well-established methodology of serine ß-lactamase inhbition provides highly compeling evidence that MBL inhibitors will have similar effects, and suggests that the development of MBL inhibtors will be of great importance in combating the rise of antibiotic resistant bacteria.
Despite the significant body of work examining MBL inhibitors, no compounds have succeeded in attaining FDA approval to combat MBL resistance. The most widely explored class of MBL inhibitor use a terminal thiol MBP, but these compounds have a relatively flat SAR. Increased exploration in the diversity of MBPs could be a potential source of new, more active inhibitors, as evidenced by recent work with cyclic boronate compounds. It is likely that exploration of other classes of MBPs, especially those capable of bidentate (or even tridentate) metal coordination, will lead to more active MBL inhibitors. It is worth noting that the most active reported class of MBL inhibitors, the cyclic boronates, has been shown to be capable of inhibiting both serine- and metallo-ß-lactamases, with MBLi-27 as a particularly promising compound.1058 Historically, broad-spectrum MBL inhibition has been a challenging feat, so that this recent development represents a welcome turn in the tide of the battle against antibiotic resistant bacteria.
7.6. Peptide Deformylase (PDF, EC 3.5.1.88)
Function in Disease and Biology.
Emerging resistance to currently approved antibiotics is an area of growing global concern. Therefore, an emphasis on developing novel classes of antibiotics that target new mechanisms of action has become an important strategy to counter the growing antibiotic resistance of approved therapeutics. A biochemical process that has potential to be targeted with a novel class of antibiotics is bacterial protein post-translational modification.1059 The process of protein translation in bacterial and mammalian cells is highly conserved, with a key difference being the general utilization of N-formylmethionine by bacterial cells as the initiating residue.1060 Mammalian cells generally utilize a non-formylated methionyl residue as the initiating residue. The removal of this formyl group in the bacterial protein maturation process is necessary for N-terminal methionyl excision,1061 which affects protein folding and further N-terminal modification.701 Targeting the metalloenzyme responsible for this transformation (Figure 107), peptide deformylase (PDF), could lead to a novel class of antibiotics.1059 This is validated by the deletion of genes responsible for PDF expression resulting in static bacterial growth or cell death.1061,1062 More recently, PDF has been found in the mitochondria of humans and been proposed to be a potential target in cancer.1063 However the majority of focus on PDF has been as an antibacterial target.
Figure 107.
PDF catalyzes the removal of the N-formyl group from translated bacterial proteins.
Protein and Active Site Structure.
PDF is highly conserved across bacteria, generally being found as a monomeric protein (Figure 108) with a relative size of ~200 AA. Sequence homology between isolated PDFs from various organisms varies greatly, although the active site sequence and structure is largely conserved.1064 Three conserved active site motifs are found in PDFs: motif 1 (Gly-Hyd-Gly-Hyd-Ala-Ala-XXX-Gln), motif 2 (Glu-Gly-Cys-Lys-Ser), and motif 3 (His-Glu-Hyd-Asp-His), where Hyd is any hydrophobic residue and XXX is any residue. In the active site, the catalytic metal ion possesses a tetrahedral geometry being coordinated two His residues from motif 3, the Cys residue from motif 2, and a water molecule (Figure 108).1065 PDFs were initially thought to be Zn2+-dependent enzymes,1066 but upon isolating Fe2+ reconstituted PDF and determining that the iron metalloform had significantly higher activity than zinc form,1067 PDF is now generally regarded as an Fe2+-dependent metalloenzyme.1059 Other first row transition metal metalloforms of PDF have been shown to be catalytically active, including those with Co2+ and Ni2+.1068–1070 The structures of Fe-PDF (Figure 108), Ni-PDF, and Zn-PDF are all nearly identical.1065,1071 From crystal structures complexed with N-terminal methionine tripeptides, a hydrophobic pocket for substrate recognition (Met side chain) was observed.1065 The active site around the metal ion is quite small, preventing any substrate other than a formylated, terminal residue from binding.1072 A significant number of structural and inhibition studies utilize Ni-PDF as a surrogate for Fe-PDF, due to its similar activity and structure, but lack of oxidative sensitivity found with Fe-PDF.1068
Figure 108.
Structure of E. coli PDF reconstituted with Fe2+ (PDB 1BSZ).
Inhibitor Development.
Due to the function of the PDFs, peptide based inhibitors have been a logical place for PDF inhibitor development to begin. An early PDF inhibitor was designed from a peptidyl high affinity substrate.1073 The inhibitor utilized a tetrahedral phosphonate transition state analog (PDFi-1, Figure 109) and possessed modest activity (Ki = 37 μM). Crystallographic studies of Co2+ and Zn2+ reconstituted E. coli PDF (Co-EcPDF and Zn-EcPDF) with PDFi-1 both showed the phosphonate MBP binding to the active site metal in a monodentate manner via a H-phosphonate oxygen atom (Figure 110).1074 The binding mode of PDFi-1 inspired a variety of other potential MBPs to be tested. Increased activity was found with a thiol MBP,1075 with certain thiol-peptidyl derivatives possessing very good activity in vitro against Fe2+ reconstituted E. coli PDF (Fe-EcPDF) (PDFi-2, Ki = 19 nM).1076 Other MBPs on a peptidyl scaffold included an aldehyde (PDFi-3) which exhibited modest activity against Co-EcPDF (IC50 = 10 μM).1077 Hydroxamic acid MBPs attached to peptidyl scaffolds were determined to possess very good activity, based on the identification of actinonin (Figure 109) as a potent inhibitor of different metalloforms of EcPDF (Zn-, Ni-, and Fe-EcPDF, IC50 = 90, 3, and 0.8 nM, respectively).1078–1080 The structure of actinonin bound to Ni-EcPDF was determined, which showed the hydroxamic acid MBP coordinated the active site Ni2+ in bidentate manner (Figure 110).1081 Since the discovery of the hydroxamic acid motif being a potent scaffold for PDFs, the landscape for PDF inhibitors has shifted greatly toward the use of this MBP.
Figure 109.
Inhibitors of PDF.
Figure 110.
Structure of: PDFi-1 bound to Zn-EcPDF (PDB 1BSK), Actinonin bound to Ni-EcPDF (PDB 1G2A), and BB-3497 bound to Ni-EcPDF (PDB 1G27).
An area of significant interest has been attachment of the hydroxamic acid MBP to peptidyl derivatives similar to actinonin.1082,1083 Other inhibitor development efforts have been focused on non-peptidic hydroxamic acid (and ‘reverse’ hydroxamic acid) derivatives, which have resulted in inhibitors with good activity (PDFi-4, Fe-EcPDF IC50 = 16 nM, Figure 109).1084 A reverse hydroxamic acid inhibitor, BB-3497 (Ni-EcPDF IC50 = 7 nM, Figure 109),1081 possessed better PK properties than actinonin and displayed potent antimicrobial in vivo activity. Structural characterization of BB-3497 bound to Ni-EcPDF showed the reverse hydroximic acid MBP coordinating in a similar bidentate motif as to hydroxamic acid inhibitors (Figure 110). A related MBP, which uses a hydrazide in place of hydroxylamine in the hydroxamate group (PDFi-5), resulted in poor activity against Co-EcPDF (Ki = 200 μM), but still coordinates the active site metal ion based on spectroscopic evidence.1085 Finally, a N-hydroxy urea inhibitor (PDFi-6) was found to exhibit modest inhibitory activity against S. aureus PDF (IC50 = 2.2 μM).1086
Non-hydroxamate MBPs, such as carboxylic acids, have been used in PDF inhibitors, albeit to a more limited extent. A novel scaffold containing a carboxylic acid (PDFi-7, Figure 109) was identified by screening Ni-EcPDF showing good activity (IC50 = 0.94 μM) but no in vivo activity.1087 Attempts to modify the carboxylate with an amide or ester resulted in at least a 100-fold loss in activity, indicating the carboxylate was important for activity likely via metal coordination. Biaryl scaffolds containing acidic moieties such as, carboxylic acids (PDFi-8), tetrazoles (PDFi-9), and acyl sulfonamides (PDFi-10) (Figure 109) were analyzed against Zn-EcPDF and possessed some inhibitory activity (IC50 = 22.8, 3.9, and 10 μM respectively).1088 It is unknown if the acidic moieties of these inhibitors interact with the active site metal ion or if they inhibit PDF by binding elsewhere; no in vivo activity was reported for these inhibitors. Another potential metal binding inhibitor identified was 5,5’-disulfanediylbis(1,3,4-thiadiazol-2-amine) (PDFi-11, Figure 109), which possessed moderate inhibitory activity against Ni-EcPDF (Ki = 3.7 μM).1089 The inhibitor was predicted to coordinate to the active site metal via an exocyclic amine and an endocyclic nitrogen based on modeling studies.
Current and Future Prospects.
PDF inhibitors represent a novel class of potential antibiotics with validated cellular biology. An earlier review provides promising in vivo and PK analysis of hydroxamate based PDF inhibitors, as well as inhibitors that entered Phase I clinical trials.1082 Recently, a hydroxamate based inhibitor, GSK 1322322, similar to BB-3497 has completed Phase I clinical trials.1090 The compound then went on to demonstrate efficacy in Phase II trials for bacterial skin infections.1091 Unfortunately, there are no reports of PDF inhibitors moving beyond Phase II clinical trials; this may be due, in part, to concerns over the development of bacterial resistance to PDF inhibitors.1092 Also, the identification of a human version of PDF in the mitochondria may complicate the development of antibiotic PDF inhibitors. Despite some shortcomings, PDF remains a promising target for the development of antibiotics with promising clinical data and validated cellular biology.
8. Lyases (EC 4.X)
8.1. Class II Fructose-1,6-bisphosphate aldolase (FBP-aldolase) (EC 4.1.2.13)
Function in Biology and Disease.
Tuberculosis (TB) is ranked the ninth leading cause of death worldwide.1093 In 2016, the World Health Organization (WHO) estimated that 10.4 million people were infected with tuberculosis (TB) and that there were 1.3 million TB related deaths.1093 Additionally, it is estimated that one-third of the world’s population carries latent M. tuberculosis.1094 There are currently ten FDA approved drugs for the treatment of TB, and the first-line treatment of drugs include isoniazid, rifampin, ethambutol, and pyrazinamide; however, the overall effectiveness of the current treatment of TB, Directly Observed Therapy, Short Course (DOTS), has declined from 95 to 85% due to the emergence of multidrug resistant TB (MDR-TB) and extensively drug-resistant TB (XDR-TB).1093 Hence, a novel drug for treating TB is of urgent need. One attractive target for the treatment of M. tuberculosis includes the inhibition of fructose 1,6-bisphosphate aldolase (FBA). FBA catalyzes the reversible aldol condensation of dihydroxyacetone phosphate (DHAP) with glyceraldehyde 3-phosphate (G3P) to produce fructose 1,6-bisphosphate (FBP) in glycolysis, gluconeogenesis, the Calvin cycle, and is crucial for cellular metabolism (Figure 111).1095,1096 There are two distinct enzyme classes of FBAs, depending on their mechanism of catalysis and prevalence among species. The class I FBAs utilize an active site lysine residue to generate a covalent Schiff base with DHAP during catalysis, and is found in higher order organisms such as humans. Class II FBAs are metalloenzymes which utilize a Zn2+ ion to polarize and stabilize the keto carbonyl group of DHAP for catalysis and is found in pathogenic microbes (i.e., protozoa, bacteria, and fungi). The detailed mechanism of class II FBA is described elsewhere.1097 It is important to note that while M. tuberculosis, E. coli, S. galbus, and other autotrophic prokaryotes can express both classes of FBAs, knockout studies of the class II FBA gene has resulted in nonviable bacteria.1098,1099 Class I and class II FBAs share no significant sequence homology (~15%),1096,1100 making inhibition of class II FBA an attractive target for the treatment of M. tuberculosis and other protozoan infections.
Figure 111.
FBA catalyzes the reversible aldol condensation of dihyroxyacetone phosphate (DHAP) with glyceraldehyde 3-phosphate (G3P) to produce fructose 1,6-bisphosphate (FBP).
Protein and Active Site Structure.
Class II FBAs can be further classified into two families (a and b), according to their sequence homology and their oligomeric state. There is a ~40% sequence similarity between members of each family, and ~25% – 30% sequence similarity across all class II FBAs.1097,1101 Class IIa FBAs include those extracted from M. tuberculosis and E. coli, while class IIb FBAs include B. anthracis. Class IIa FBAs generally function as homodimers, with a total molecular weight of ~78 kDa.1096 The protein has a (α/β)8 TIM barrel fold with an inner lining of eight β-sheets surrounded by eight α-helixes, and an active site at the C-terminal end of the barrels.1102 The X-ray crystal structure of DHAP and G3P bound to M. tuberculosis FBA (MtFBA) reveals a narrow and hydrophilic binding pocket, bearing a catalytic Zn2+ ion and a Na+ ion (Figure 112). The oxygen atoms of the β-hydroxyl ketone of DHAP bind in a bidentate manner to the Zn2+ ion, which is further ligated by His110, His226, and His264, resulting in a trigonal bipyramid geometry.1097 Additionally, the terminal phosphate group of DHAP branches into the hydrophilic binding pocket, forming H-bond interactions with the Na+.
Figure 112.
Structure of DHAP (substrate, magenta) and G3P (yellow) bound to MtFBA (PDB 3EKZ). A Na+ ion is shown in purple.
Inhibitor Development.
In 1973, the first-in-class inhibitor of class II FBAs, phosphoglycolohydroxymate (PHG, Figure 113) was reported.1103,1104 PGH is a DHAP substrate analogue and a potent competitive inhibitor of class II FBAs with a Ki value of 10 nM.1103,1105,1106 An X-ray crystal structure reveals that the hydroxamic acid MBP of PGH binds the FBA active site Zn2+ ion in a bidentate binding mode resulting in a trigonal bipyramidal geometry similar to DHAP,1102,1107 while the terminal phosphate group of PGH also branches into the active site hydrophilic pocket. Although PGH is a potent inhibitor of FBA, its instability in water, which is not uncommon among hydroxamic acids that can hydrolyze to release toxic hydroxylamines, hindered further development.1107,1108 Subsequent development of class II FBA inhibitors include substrate (DHAP) analogues or transition state mimics, but have largely maintained the use of a hydroxamic acid MBP with a phosphoglycolate skeleton.1105,1107–1113 Inhibitors in this class include N-sulfonyl hydroxamate, hydrazine, hexitol bis-phosphate, and hydroxamate based derivatives (e.g., FBAi-1, FBAi-2, Figure 113), with the most potent inhibitors achieving Ki value as low as 4 nM against FBA from various sources.1105,1107,1108,1110–1112 In addition to the disadvantages of a hydroxamic acid MBP, the use of a polar phosphate group has also hindered the development of these inhibitors. Due to the limited ability of polar phosphate groups to cross biological membranes, these inhibitors showed no inhibition against the growth of cultivated pathogens (at concentrations as high as 1 mM).1098,1107,1112 Lastly, the structural similarity of these compounds to DHAP have led to a lack of specificity for class II FBA over class I FBA.1097
Figure 113.
Inhibitors of FBA.
Through structure-based drug design and virtual screening, inhibitors which do not contain the hydroxamic acid MBP or phosphoglycolate skeleton have been discovered. This includes the use of MBPs such as dipicolinic acid,1109 8-hydroxyquinoline,1098 and hydrazine (FBAi-3, FBAi-4, FBAi-5, FBAi-6, Figure 113).1114 2-Carboxy-8-hydroxyquinoline is a non-completive inhibitor (FBAi-4, IC50 = 10 μM) against MtFBA and showed effectively no inhibition against class I FBA.1098 X-ray crystallography, biochemical, and biophysical studies show FBAi-4 binds in a tridentate manner to the active site Zn2+ ion and displaces ligating residue His212, resulting in a trigonal bipyramid coordination geometry (Figure 114). FBAi-4 is not positioned to block the substrate pocket, but instead, displaces residues His212 and Val164 to yield a protein conformational change which blocks substrate access.1098 Beyond these initial findings, no further development of the 8-hydroxyquinoline MBP in inhibitors of MtFBA has been investigated. Another set of recently published inhibitors include phenylhydrazone compounds (FBAi-5, FBAi-6, Figure 113).1114 Utilizing virtual HTS, a hydrazine lead (FBAi-5, IC50 =17 μM) was found to have antifungal effects against C. albicans (CaFBA), an isoform bearing an active site highly similar to that of MtFBA. Various phenylhydrazone derivatives were developed from this hit, with the most potent inhibitor FBAi-6 achieving an IC50 value of 2.7 μM against CaFBA. Through computational docking, the hydrazone nitrogen atom is predicted to coordinate to the Zn2+ ion in monodentate fashion, resulting in a tetrahedral coordination geometry;1114 however, there is no experimental evidence to verify this mode of binding. Phenylhydrazone inhibitors exhibited moderate antifungal activities against C. glabrata and synergistic antifungal activities in combination antifungal drug fluconazole.
Figure 114.
Structure of FBAi-4 bound to MtFBA (PDB 4LV4). The loop that includes the displaced His212 residue is not resolved in the structure.
Current and Future Prospects.
Previous research regarding FBA had primarily focused on understanding the FBA catalytic mechanism of action, including the various intermediates and ternary complexes. This has resulted in a large class of DHAP analogues and transition state mimics as inhibitors of FBA. Of these, inhibitors bearing hydroxamic acid MBPs and phosphoglycolate motifs claim majority. While these inhibitors achieved good activity, they are limited by a lack of selectivity for class II over class I FBAs and limited diffusivity across biological membranes.1098,1113 Many of these inhibitors are unable to inhibit the growth of cultivated pathogens and efforts on these class II FBA inhibitors have stalled. Recent work has diverged away from transition state/DHAP inhibitor mimics, and focused on developing compounds with new MBPs.1098,1109,1114 While these inhibitors displayed poorer inhibition against class II FBAs, these compounds may not have the same limitations as the transition state/DHAP inhibitor mimics. Additionally, inhibitors such as FBAi-4 present a novel, non-covalent mechanism of inhibition, presenting a new therapeutic potential. Incorporation of polar side chains in the derivatization of the FBAi-4 MBP may be useful for targeting the neighboring hydrophilic pocket, such as making H-bonds with the Na2+ ion, and achieve greater inhibitory activity. There is room for new MBPs to be explored as inhibitors of class II FBA to achieve compounds that are selective for class II FBA, and overcome the other shortcomings of existing inhibitors. Given the value and novelty of targeting FBA for the treatment of TB, a well understood catalytic mechanism, and a vast repertoire of structural data, more effort toward the development of FBA inhibitors is merited.
8.2. Imidazoleglycerol-phosphate dehydratase (IGPD, 4.2.1.19)
Function in Biology and Disease.
Imidazoleglycerol-phosphate dehydratase (IGPD) is a dinuclear Mn2+-dependent hydro-lyase that plays an essential role in histidine biosynthesis and has been identified as a herbicidal, antifungal, and antibiotic target. It catalyzes the dehydration of (2R,3S)-2,3-dihydroxy-3-(1H-imidazol-5-yl)propyl dihydrogen phosphate (2R3S IGP) to 3-(1H-imidazol-4-yl)-2-oxopropyl dihydrogen phosphate (IAP). This process is the sixth step in histidine biosynthesis in both prokaryotes and eukaryotes, with IGPD generally conserved across species and isoforms, particularly at the active site.1115,1116 This makes IGPD an attractive target for the development of cytostatic agents that may have efficacy against both bacterial and fungal microbes, while sparing mammalian cells, as mammals do not synthesize histidine, but acquire histidine through their diet. A majority of IGPD inhibitor studies have focused on the development of broad-spectrum herbicides, particularly because early transition state analogue inhibitors were found to be highly effective at inhibiting plant germination and development;1117–1120 however, recent research has indicated that IGPD may be a viable bacteriostatic agent.1116
The generation of IAP from IGP proceeds through four discrete steps (Figure 115), each of which is catalyzed by IGPD. The first step in catalysis is the deprotonation of the imidazole moiety of IGP.1121 The highly basic imidazolate intermediate is stabilized by the active site Mn2+ cations, which allows for the subsequent dehydration of the α-hydroxyl group and formation of the diazafulvene intermediate. Proton abstraction at the β-carbon generates the Δ2-enol intermediate, which can then tautomerize to form IAP.
Figure 115.
IGPD catalyzes the dehydration of IGP to IAP as part of histidine biosynthesis. Upon binding of IGP and deprotonation of the imidazole ring, the active site Mn2+ cations stabilize the negatively charged imidazolate intermediate and facilitate the dehydration of IGP to diazafulvene. Proton-abstraction at the β-carbon of diazafulvene generates the Δ2-enol which can tautomerize to form IAP.
Protein and Active Site Structure.
IGPD exists as a 24-mer, with two Mn2+ cations in each active site. The active site is found at the interface of three different monomers, referred to as A, A′, and C. Both Mn2+ ions are bound in an octahedral geometry in the absence of substrate, with each ion coordinated by three His residues, an Asp residue, and two water molecules. The metal active site possesses an inter-subunit 2-fold rotational pseudosymmetry in which both the A and A′ chains provide two histidine residues and an aspartate to one Mn2+ ion, and one histidine to the other Mn2+ ion (Figure 116).1122 Each monomer also possess an intra-subunit 2-fold rotational pseudosymmetry axis, which allows for the formation of the unusual bioactive 24-mer complex.
Figure 116.
Structure of IGPD 24-mer (PDB 4QNK). In the 24-mer, each individual subunit is colored differently for clarity. In the active site, the A, A′, and C chains are depicted in grey, cyan, and yellow, respectively.
The phosphate binding site is surrounded by a cluster of highly conserved basic residues that hydrogen bond to the phosphate moiety of the substrate, and originate from the A and C chains (Figure 116). The residues involved in phosphate binding are dynamic, and multiple conformational changes are observed in this pocket as the substrate moves towards the metal ions during catalysis. Unlike some other Mn2+-dependent metalloenzymes, where the divalent cation(s) may be exchanged with metals such as Zn2+ and Mg2+ and still maintain activity, IGPD has a mechanistic requirement for Mn2+;7,1115,1123 however, Co2+ and Cd2+ metalloforms have been reported to support some reduced enzymatic activity.1124
Inhibitor Development.
There are two main pockets that comprise a majority of the IGPD active site: the metal binding pocket and the phosphate binding pocket. The phosphate binding pocket is flanked by dynamic basic residues, which makes drugging this pocket with large or non-acidic moieties difficult. Indeed, nearly every reported inhibitor of IGPD employs a phosphate, phosphonate, or carboxylate to interact with this pocket (Figure 117). The metal binding pocket is designed to accommodate a bridging imidazole/imidazolate moiety, and most reported inhibitors coordinate both active site Mn2+ anions with a diazole or triazole ring. Many inhibitors include a Lewis-basic moiety (e.g., hydroxyl, amine, or amide group) proximal to the azole ring which may also coordinate to the Mn2+; however, little exploration of different MBPs has been reported.
Figure 117.
Inhibitors of IGPD.
The majority of reported inhibitors are structural mimics of diazafulvene or proposed diazafulvene transition state intermediates. These inhibitors typically contain three essential functional groups: an azole ring, a Lewis-basic hydrogen bond donor/acceptor, and a phosphate or phosphonate group (Figure 117).1117,1118,1120,1125–1129 The most active reported inhibitor, Zeneca 1, inhibits IGPD with an IC50 value of 0.6 nM. A structure of Zeneca 1 with IGPD shows the compound uses two triazole nitrogen atoms and a β-hydroxyl group to form coordinate bonds with the active site metal centers, and the phosphonate group making key hydrogen bonding interactions with Arg99 (Figure 118).1117 There are ample reports of substitutions of these three primary functional groups; however, major modifications or deletions of one or more of these groups greatly reduces activity (Table 16). Replacing the imidazole moiety with a pyridine or pyrimidine ring1117 or replacing the phosphate group with a malonate moiety1125 both result in a substantial loss in activity as seen with IGPDi-4. Expansion or contraction of the linker length between the central hydrogen bond donor/acceptor moiety and the phosphate causes a >100-fold loss of activity as illustrated by compound IGPDi-2 (IC50 >1 μM).1125 Likewise, removal of the hydrogen bond donor/acceptor moiety is not tolerated (IGPDi-3). However, the absolute position of this moiety may be varied between the α- and β-carbons without substantial changes in activity.1117,1118,1120 Simple modifications of the central hydrogen bond donor/acceptor moiety to amines, amides, or esters is also tolerated (IGPDi-5, IGPDi-6), but associated with a loss of 10- to 50-fold activity relative to Zeneca 1.1117,1128 Nevertheless, modification of this moiety allows for further synthetic exploration of new active site pockets and provides a handle for the modification of physiochemical properties for this class of inhibitor.1128 It remains unclear if compounds containing an amine or amide group are able to coordinate to the Mn2+ ion in the same manner as diazafulvene, as structural characterization of theses inhibitors has not been reported. However, it is likely that hydroxyl, amine, amide, and ester groups at this position are able to engage in hydrogen bonding interactions with the proximal A’ Glu21 residue. This may explain why modifications of this kind are far more tolerated than modifications at either the azole or phosphate functional groups.
Figure 118.
Structure of (R)-Zeneca 1 bound to IGPD (PDB 5NDX). The A, A′, and C chains are depicted in grey, cyan, and yellow, respectively.
Table 16.
Activity for IGPD inhibitors.
There are few reports of IGPD inhibitors that are not transition state mimics, and none of these inhibitors have been structurally characterized with IGPD. The majority of these studies identified new inhibitors via computational approaches. In one report,1129 a representative pharmacophore model was generated from a structural ensemble of previously published inhibitors. This model, which was unfortunately biased by the overwhelming number of transition state mimics present in the literature, was used as a query structure to search several commercial 3-D chemical databases. Compounds sharing this pharmacophore were purchased and screened against IGPD, which revealed a novel pyrrole aldehyde chemotype (IGPDi-9, Figure 117) with moderate inhibitory activity. A more recent and extensive computational screen identified several novel inhibitors of IGPD through a virtual fragment screening of the ZINC database, employing previously published crystal structures of bacterial IGPD.1116 Docking and subsequent MD simulations identified compounds that likely bound to a long, somewhat hydrophobic channel adjacent to the metal-binding pocket of the active site. Several of these compounds were predicted to coordinate to at least one metal ion via a carboxylate or similar moiety, although docking and MD simulations involving metal coordination were acknowledged to be difficult and often not predictive.1116 Several identified inhibitors were screened against both wild-type and multi-resistant strains of E. coli and S. aureus grown on minimal media. Both IGPDi-11 and IGPDi-12 were found to inhibit bacterial growth, and were particularly effective against S. aureus.
Current and Future Prospects.
IGPD has been well established as a viable broad-spectrum herbicidal target, and more recent studies have demonstrated its potential as a novel bacteriostatic target. A majority of reported inhibitors mimic transition state intermediates with great success; however, there is ample room for expansion to more drug-like, non-transition state mimic inhibitors. The poor physiochemical properties associated with current transition state mimics has limited the application of IGPD inhibitors beyond topical herbicidal indications, although recent reports have sought to remedy this. Repurposing existing inhibitors from related targets with dinuclear Mn2+ or Mg2+ active sites (Sections 5.1, 5.2) may be beneficial in the field of IGPD inhibitor development. Despite the somewhat unique coordination environment of the IGPD metal centers, the lack of chemical diversity among reported inhibitors may be remedied by this adoptive approach and may allow for exciting new developments in the field. This might also allow for the exploration of the novel active site binding pockets that have been identified in recent computational screening. Excellent work in IGPD structural biology has recently been published that will serve as a boon for further inhibitor development,1115 as until recently no crystal structures were available and nearly all inhibitor development was driven solely by chemical SAR. As both herbicide resistance and antimicrobial resistance continue to increase worldwide, IGPD inhibitor development will likely prove to be a valuable field of study.
8.3. Human Glyoxalase I (Glo1, EC 4.4.1.5)
Function in Biology and Disease.
The metalloenzyme human glyoxalase I (Glo1) has been linked to tumor viability, growth, and drug resistance in various forms of cancer.1130,1131 Glo1 is the first of two enzymes in the glyoxalase system, along with the coenzyme glutathione. This system is primarily responsible for the detoxification of the endogenous reactive metabolite class 2-oxoaldehydes, the most prevalent being methylglyoxal (MG), which derives from degradation of triosephosphates (glycolysis), ketone body metabolism, threonine degradation, and glycated protein fragmentation.1132 The detoxification begins with MG reacting with glutathione creating the hemithioacetal, which is isomerized by Glo1 to the S-D-lactoylglutathione, and is finally hydrolyzed by glyoxalase II, releasing D-lactate and glutathione (Figure 119).1133 The rapid rate of growth and glycolysis in many tumor lines, leads to increased MG production; therefore, increased Glo1 activity is associated with increased tumor survival and growth rates. Additionally, multi-drug resistant leukemias are known to exhibit increased Glo1 expression,1134 with multi-drug resistance being reversible upon cotreatment with a Glo1 inhibitor. In addition to its potential value as an anticancer target, Glo1 has potential as a target for Plasmodium falciparum (the parasite responsible for malaria),1135 as well as a treatment for neurological conditions such as anxiety.1136–1138
Figure 119.
Catalytic mechanism of Glo1, which isomerizes the hemithioacetal, formed by glutathione reacting with methylglyoxal and other 2-oxoaldehydes.
Protein and Active Site Structure.
The functional form of the metalloenzyme Glo1 is a dimer with two identical subunits A and B (Figure 120).1139 Glo1 possesses 2-fold internal symmetry and subsequently two active sites. Based on metal analysis of Glo1 purified from human erythrocytes, it has been determined that Glo1 is a Zn2+-dependent metalloenzyme, with a Zn2+ ion at both active sites.1140,1141 Each active site Zn2+ ion is coordinated by Gln33 and Glu99 from one monomer and His126 and Glu172 from the other monomer (Figure 120) placing the Zn2+ ion in an octahedral coordination geometry, with two open coordination sites occupied by exchangeable water molecules, which are displaced upon substrate binding.1142–1145 Upon substrate binding, the glutathione portion of the substrate makes significant contacts with a glutathione binding site (Figure 120).1139 The γ-glutamyl residue of glutathione engages in polar interactions with protein residues, specifically a salt bridge with Arg37 and hydrogen bonding with Asn103 and Arg122. The active site of Glo1 also possesses a hydrophobic pocket proximal to the active site metal with a volume of ~70 Å3, where the hydrophobic moiety of glutathione conjugates are situated (Figure 120).1139 Based on structural studies with transition state intermediate mimetics bound to the active site metal, it is thought that Glu172 is displaced form the coordination sphere of the Zn2+ ion upon substrate binding, although this displacement mechanism is not supported by computational studies.1146,1147 The yeast isoform of Glo1 was used as a surrogate for the human isoform in many earlier inhibition studies,1148,1149 but human Glo1 is now utilized in most current inhibition and structural studies.
Figure 120.
Structure of human Glo1 with a glutathione derivative (yellow) bound (PDB 1FRO).
Inhibitor Development.
Some of the earliest inhibitors of Glo1 were focused on a glutathione scaffold, with the addition of undecorated alkyl substituents on the thiol group (Figure 121, Table 17).1149,1150 It was found that as the alkyl substituent was increased in length, a significant gain in activity was observed with the most active being S-octyl-glutathione (Glo1i-1) with an IC50 value of 20 μM against yeast Glo1. Quickly after the importance of hydrophobic substituents in glutathione derivatives was discovered, a series of Aryl-glutathione derivatives were reported with the most potent being S-(p-bromobenzyl)glutathione (Glo1i-2) with an IC50 value of 9 μM against yeast Glo1.1151 Unfortunately, these compounds were not active in vivo, but upon esterification of the carboxylic acids groups in vivo activity was observed.1152,1153 It took >25 years from the development of glutathione derivatives as Glo1 inhibitors before the first crystal structure of human Glo1 with a glutathione derivative to be reported, which confirmed the importance of the hydrophobic interactions with the hydrophobic pocket near the Zn2+ active site.1139 Additionally, the glutathione portion of the inhibitor was shown to make considerable polar interactions with the enzyme. Subsequent studies of glutathione derivatives included attempts to modify the glutathione moiety to decrease metabolic susceptibility and further modification of the aryl substituent, with both attempts yielding inhibitors with poorer activity.1154,1155 Additional attempts to create product mimetic inhibitors, based on α-hydroxythiol esters, were unsuccessful.1156
Figure 121.
Inhibitors of Glo1.
Table 17.
Activity of Glol inhibitors. All values shown are against human Glol, unless otherwise noted.
| Compound | Inhibition (μM) | Ref. |
|---|---|---|
| Glo1i-1 | IC50 = 20a | 1150 |
| Glo1i-2 | IC50 = 9a | 1151 |
| Glo1i-3 | Ki = ~400a | 1157 |
| Glo1i-4 | IC50 = 120 | 1158 |
| Glo1i-5 | IC50 = 280 | 1158 |
| Glo1i-6 | IC50 = 530 | 1158 |
| Glo1i-7 | IC50 = 210 | 1158 |
| Glo1i-8 | IC50 = 3.5 | 1159 |
| Glo1i-9 | IC50 = 34a | 1160 |
| Glo1i-10 | IC50= 154a | 1160 |
| Glo1i-11 | Ki = l.7 | 1161 |
| Glo1i-12 | Ki = 0.01 | 1146 |
| Glo1i-13 | Ki = 0.001 | 1162 |
| Glo1i-14 | IC50 = 1.19 | 1163 |
| Glo1i-15 | IC50 = 0.011 | 1163 |
| Baicalein | Ki = 0.183 | 1164 |
| Indomethacin | Ki = 18.1b | 1165 |
| Methyl-gerfelin | Ki = 0.23b | 1166 |
| Myricetin | IC50 = 5.0 | 1167 |
| Zopolrestat | Ki = 1.2 | 1168 |
Value reported against yeast Glo1.
Value reported against mouse Glo1.
Following the success of substrate mimetics as inhibitors of Glo1, transition state mimicking inhibitors were investigated (Figure 121, Table 17). Inspired partly by a study of natural product inhibitors,1169,1170 and by the observation that Glo1 utilized an enediol intermediate, some proposed transition state mimics were tested against yeast Glo1. More active inhibitors included maltol (Glo1i-3, yeast Glo1 Ki = ~400 μM), squaric acid (Glo1i-4, human Glo1 IC50 = 120 μM), and 2,3-dihydroxy-benzoic acid (Glo1i-5, IC50 = 280 μM).1157,1158 An expansion of potential transition state mimetics, including 2,3-dihydroxypyridine (Glo1i-6, human Glo1 IC50 = 530 μM) and polyphenolic benzohydroxamic acids such as 2,3,4-trihydroxybenzohydroxamic acid (Glo1i-7, human Glo1 IC50 = 210 μM), were reported.1158 Other transition state mimetic inhibitors include coumarins and flavones, with the most active being 7,8-dihydroxy-4-phenylcoumarin (Glo1i-8, human Glo1 IC50 = 3.5 μM) and myricetin (human Glo1 IC50 = 5 μM).1159,1167 Further exploration of transition state mimetics was performed analyzing methylated and nonmethylated enediol transition state mimics, which in combination with spectral data indicated that the enediol mimicking moiety of many transition state based inhibitors is involved in metal coordination.1171 A computational analysis of previously published transition state mimics, demonstrated that the most important motif for activity was a α-hydroxy-α,β-unsaturated carbonyl group attached to a fused ring carbon, such as that present in flavones like myricetin.1172 A later study reexamined previous inhibitors of yeast Glo1 and reported that some of the inhibitors were significantly less active than previously thought against human Glo1, specifically pyrogallol (not shown) and Glo1i-4.1160 This study was also the first report of 1,2-naphthalenediol (Glo1i-9, yeast Glo1 IC50 = 34 μM), tropolone (Glo1i-10, yeast Glo1 IC50 = 154 μM), and select tropolone derivatives as inhibitors of yeast Glo1. After a period of publications focusing on transition state mimetics, newer reports shifted focus to applying transition state moieties to a glutathione backbone.
The enediol transition state mimetic, hydroxamic acid, when combined with glutathione, resulted in S-(N-hydroxy-N-methylcarbamoy1)glutathione (Glo1i-11, Figure 121), which exhibited a Ki value of 1.7 μM against human Glo1.1161,1173 A significant gain in inhibitory activity was achieved when hydrophobic derivatives of Glo1i-11 were analyzed against human Glo1, the most active being S-[N-Hydroxy-N-(4-iodophenyl)carbamoyl]glutathione (Glo1i-12) with a Ki value of 10 nM.1146,1161 The mode of binding of the hydroxamic acid MBP in these glutathione derivatives was characterized shortly afterwards, with the hydroxamic acid coordinating the active site Zn2+ ion in a bidentate manner, the hydrophobic substituent situated in the hydrophobic pocket, and the glutathione making significant polar contacts with the protein (Figure 122).1146 Interestingly upon the reaction state intermediate mimic binding the active site metal, the coordinating residue Glu172 no longer coordinates the active site metal, which provided some insight into the reaction mechanism. In vivo testing of esterified derivatives of these inhibitors exhibited activity using in vivo mouse tumor models.1174 Further work on these derivatives included successful modification of the peptide portion of the glutathione moiety to decrease its metabolic susceptibility to γ-glutamyltransferase, while maintaining similar inhibitory activity against yeast Glo1.1175 To improve the activity of this class of inhibitors, a dimer of Glo1i-12 connected via various polymeric linkers at the γ-glutamyl-NH3, provided increased inhibitory activity.1176,1177 A fairly recent development to this class of inhibitor was the replacement of the para-halogenated phenyl on Glo1i-12 for a meta-ethynyl phenyl (Glo1i-13) as well as the replacement of the thioether sulfur for an alkyl carbon increased the activity yielding a Ki value of 1.0 nM against human Glo1.1162 Attempts have been made to use alternative MBPs with non-sulfur containing glutathione analogs, such as β-ketoesters and reverse hydroxamic acids, but these compounds possess lower activity than the analogous hydroxamic acid containing inhibitors.1178,1179
Figure 122.
Structure of: Glo1i-12 bound to human Glo1 (PDB 1QIN); methyl-gerfelin bound to mouse Glo1 (PDB 2ZA0); baicalein bound to mouse Glo1 (PDB 4X2A); Glo1i-15 bound to human Glo1 (PDB 3VW9); indomethacin bound to mouse Glo1 (PDB 4KYK); zopolrestat bound to mouse Glo1 (PDB 4KYH).
Natural products have been a longstanding source of inhibitors for Glo1 including, but not limited to, curcumins,1180 anthocyanidins,1181 flavonoids,1182 and phenolic trans-stillbenes.1183 Computational analysis of natural products, such as myricetin, has also led to the determination of novel scaffolds with lower in vitro activity that show inhibition of cancer cell proliferation.1184,1185 One of the more potent natural product inhibitors of Glo1, is methyl gerfelin, with a Ki of ~230 nM against mouse Glo1 (Figure 121).1166 A crystal structure of mouse Glo1 with methyl gerfelin is shown in Figure 122. The structure shows that the inhibitor coordinates the active site Zn2+ ion via a catechol MBP in a bidentate manner. Another crystal structure of mouse Glo1 has also been reported with the flavonoid baicalein (human Glo1 Ki = 183 nM, Figure 121) coordinated to the active site Zn2+ ion via a related pyrogallol MBP (Figure 122).1164
HTS was applied to the development of human Glo1 inhibitors, which yielded the molecule 4,6-diphenyl-N-hydroxypyridinone (Glo1i-14), which utilizes the 1,2-hydroxypyridinone MBP, with an IC50 value of 1.19 μM (Figure 121).1163 Substitution of the 4-position phenyl resulted in poorer activity, while replacement of the 6-position phenyl yielded improved activity especially with an azaindole, and upon addition of a N-propyl-methoxy resulted in the most active inhibitor Glo1i-15 with an IC50 value of 11 nM. A crystal structure of Glo1i-15 bound to Glo1 shows the 4-phenyl substituent situated in the hydrophobic pocket, the N-hydroxypyridinone MBP bound in a bidentate manner to the Zn2+ ion, and the 6-position azaindole interacting with the glutathione binding residues and solvent (Figure 122).
Additional efforts to inhibit human Glo1 have reported the repurposing of already approved pharmaceuticals (Figure 121). Such studies have included examining indomethacin (mouse Glo1 Ki = 18 uM) and zopolrestat (human Glo1 Ki = 1.2 uM).1165,1168 Crystal structures of both molecules bound to the active site of mouse Glo1 have been obtained showing that neither inhibitor binds to the active site Zn2+ ion (Figure 122).1168 In the structure with zopolrestat bound, the inhibitor binds at the opening of the active site and interacts with the glutathione binding pocket. The structure with indomethacin bound shows the inhibitor binding deeper in the active site relative to zopolrestat, but does not bind in the hydrophobic pocket.
Current and Future Prospects.
Significant efforts have been put into the development of Glo1 inhibitors, but despite >50 years of investigation no compounds have entered the clinic. Esterified glutathione derivatives including those containing hydroxamic acid MBPs have shown efficacy in vivo. However, they also have significant metabolic liabilities as glutathione derivatives and use of the hydroxamic acid MBP, with some attempts to mitigate these liabilities.1155,1179 The development of potent inhibitors using a hydroxypyridinone MBP suggests significant progress, but no follow up in vivo studies have been reported. The validity of Glo1 as a cancer target has been recently discussed along with some of the challenges for an inhibitor to enter the clinic.1131 The main challenges presented included the limited activity and fast clearance of esterified glutathione derivatives, potential metabolic resistance to increased levels of MG, and the lack of tumor sensitivity markers for Glo1 inhibitors. Nonetheless, Glo1 remains a target for cancer with validated cell biology and pharmacology,1131,1186 although a deviation from previously utilized scaffolds such as glutathione may prove necessary to have an inhibitor reach the clinic.
9. C-N Ligases (EC 6.3)
9.1. D-Alanine:D-Alanine Ligase (Ddl, EC 6.3.2.4)
Function in Biology and Disease.
The integrity of the peptidoglycan membrane is vital to the survival of both Gram-positive and Gram-negative bacteria, and consists of alternating N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) units. Interruption of the peptidoglycan biosynthesis leads to compromised cell integrity, an inability to resist high internal osmotic pressure, and cell lysis.1187 Antibiotics that target various stages of the peptidoglycan synthesis mechanisms (such as β-lactams and vancomycin) have been extensively studied. While β-lactams bind to the penicillin binding protein and inhibit transpeptidase, vancomycin binds to the D-ala-D-ala termini of peptidoglycan chains and blocks transpeptidase. Both antibiotics are used clinically for treatment of bacterial infections; however, their efficacy is challenged by β-lactamase (Section 7.5) and vancomycin resistance (Section 6.9). Another viable target in peptidoglycan biosynthesis includes the ATP-dependent dinuclear Mg2+-dependent metalloenzyme D-ala:D-ala ligase (Ddl). Ddl is responsible for the assembly of the D-ala-D-ala precursor required for the biosynthesis of the MurNAc peptide chain. Ddl has no human counterpart, making it an attractive candidate for antibiotics; in fact, Ddl is currently targeted by D-cycloserine (DCS) for the treatment of tuberculosis.1188 The catalytic mechanism of Ddl includes ATP cofactor binding to the free enzyme, followed by the first D-ala substrate. Phosphorylation of the D-ala carboxylate by the γ-phosphate of the ATP generates an acyl phosphate intermediate and an ADP byproduct. The acetyl phosphate is then attacked by a second D-ala substrate to yield the dipeptide D-ala-D-ala product (Figure 123).1189–1192 The intermediate steps of acyl phosphate and tetrahedral adduct are the basis of which most Ddl inhibitors are designed.
Figure 123.
Reaction catalyzed by D-Ala-D-ala ligase (Ddl). Ddl generates a D-ala-D-Ala dipeptide precursor from D-alanine and ATP.
In addition to Ddl itself being an antibiotic target, the Ddl variants D-ala-D-lac ligase (VanA, VanB) and D-ala-D-ala dipepeptidase (VanX, Section 6.9), are also attractive targets for broad-spectrum inhibition.1193–1197 In D-Ala-D-lac ligase, a D-lactate is substituted for the D-alanine as the second precursor and produces D-ala-D-lac instead of D-ala-D-ala. This reduces the binding affinity of vancomycin to the dipeptide by ~1000-fold and renders resistance to vancomycin.1198 In D-ala-D-ala dipepeptidase, vanmyocin resistance is achieved by hydrolysis of D-ala-D-ala, thereby reducing the pool of dipeptide precursor available for peptidoglycan synthesis.1197 Hence, the development of broad spectrum inhibitors for Ddl and Van are an attractive method for targeting antibiotic-resistant bacterial infections. Currently, D-cycloserine (DCS) is used as second-line therapeutic treatment for multidrug-resistant (MDR) and extensively drug-resistant (XDR) strains of M. tuberculosis.1188
Protein and Active Site Structure.
Ddl from both Gram-negative and -positive bacteria have been characterized, with EcDdlB (one of the two Ddl isoforms from E. Coli) being the most extensively studied.1199 EcDdlB consists of 306-residues (~34 kDa) in three α+β domains, with the ATP binding site located between the β-sheets of the central and C-terminal domains, and the acylphosphate binding site between the central and N-terminal domains (Figure 124).1192 A crystal structure of EcDdlB complexed with products ADP, D-ala-D-ala, and a putative carbonate ion reveals two Mg2+ ions (MgA and MgB) in the active site.1196 The Mg2+ ions are 3.6 Å apart and coordinate with the oxygen atoms of the α- and β-phosphates of ADP. MgA is ligated by the side chains of Glu270 and Asn272, the carbonate ion, a water molecule, and the α-phosphate of ADP in an octahedral coordination geometry. MgB is ligated by the side chains of Asp257 and Glu270 (bridging MgA and MgB), the carbonate ion, a water molecule, and the α- and β-phosphates of ADP in an octahedral coordination geometry.
Figure 124.
Structure of EcDdl bound to D-ala-D-ala product (cyan), ADP (magenta), and putative carbonate ion (yellow) (PDB 4C5B).
Inhibitor Development.
Based on structural characteristics, inhibitors of Ddl can be classified into three main categories, including: D-ala substrate analogues, transition state analogues, and others. D-Cycloserine (DCS, Ki = 22 μM, IC50 = 300 μM, Figure 125) is a D-ala substrate analogue reported in 1962 as the first potent inhibitor of Ddl and is the most clinically advanced to date.1200–1203 DCS is currently used as second-line drug for patients with MDR and XDR strains of M. tuberculosis.1188 Although DCS exhibits broad-spectrum antibiotic activity (also targeting alanine racemase, another enzyme utilized for synthesis of MurNAc),1202,1204,1205 its use in the clinical setting has been almost eliminated due to a short shelf life (~24 months), high dosages required, and undesired side effects such as drug-induced psychosis.1188,1206,1207 The description of an unpublished structure revealed DCS binding in the D-ala substrate pocket as a noncompetitive inhibitor of ATP;1204 however, recent studies have shown that phosphorylation of DCS by Ddl to Ddli-1 (a transition state analogue, Figure 125) to be the actual active inhibitor.1196 Other D-ala substrate analogues have been investigated and are described in detail elsewhere.1202
Figure 125.
Inhibitors of Ddl.
DCS is phosphorylated by Ddl in situ to yield active inhibitor Ddli-1.1196 Ddli-1 resembles acetyl phosphate intermediate of the native mechanism (Figure 124) and inhibits via a metal binding interaction. The structure of Ddli-1 and ADP bound to EcDdlB reveals Ddli-1 situated in the D-ala binding pocket, with two oxygens of the phosphate MBP coordinating to the two Mg2+ ions (Figure 126). MgA is ligated by the side chains of residues of Glu270 and Asn272, a putative water molecule (not resolved in the crystal structure), the phosphate MBP of Ddli-1, and the α-phosphate of ADP in an octahedral coordination geometry. MgB is ligated by the side chains of Asp257 and Glu270, a water molecule, the phosphate MBP of Ddli-1, and both α- and β-phosphates of ADP also in an octahedral coordination geometry.
Figure 126.
Left: Structure of Ddli-1 (green) and ADP (magenta) bound to EcDdl (PDB 4C5A). Right: Structure of Ddli-5 (green) and ADP (magenta) bound to EcDdlB (PDB 1IOW).
Similar to the mechanism in which DCS undergoes phosphorylation to yield inhibitor Ddli-1, another class of Ddl compounds bearing a phosphate or phosphinate motif that also undergo phosphorylation to yield transition state analogue inhibitors are known.1203,1208–1213 These inhibitors (Ddli-2 and Ddli-4) are ATP-dependent and phosphorylated by Ddl in situ to give the acyl phosphate or tetrahedral adduct intermediate analogue (Ddli-3 and Ddli-5, Figure 125), where the resulting phosphate MBP can tightly bind to the Mg2+ ions in the active site.1211,1214 Through the design and synthesis of phosphinic acid dipeptide analogous, precursors Ddli-2 (IC50 = 4 μM) and Ddli-4 (IC50 = 35 μM, Ki = 3 nM) were reported as ATP-dependent irreversible inhibitors of Ddl.1203,1208 Although these inhibitors were more potent than Ddli-1 against EcDdl, they showed modest antibacterial activity in vitro (MIC = 4–128 μg/mL) against a panel of Gram-positive and Gram-negative bacteria. The lack of antibacterial activity was attributed the limited cell permeability of the polar phosphate and phosphinate motifs and has preclude their development in the clinical setting. Interestingly, co-administration of Ddli-4 with fluoro-D-alanine (a D-ala racemase inhibitor that is inactive when used alone) against P. aeruginosa led to a potentiation of the fluoro-D-alanine drug.1203 The structure of Ddli-5 and ADP bound to EcDdlB (Figure 126) reveals the phosphate MBP of Ddli-5 and the α- and β-phosphate groups of ADP ligating the active site Mg2+ ions. MgA is ligated by the side chains of Glu270 and Asn272, a water molecule, the phosphate MBP of Ddli-5, and the α-phosphate of ADP in a distorted octahedral coordination geometry. MgB is ligated by the side chains of Asp257 and Glu270, a water molecule, the phosphate MBP of Ddli-5, and both α- and β-phosphates of ADP in an octahedral coordination geometry.1192 This binding motif is the same as that of Ddli-1 and other transition state analogue inhibitors.
Other inhibitors that do not fit in the two aforementioned categories have been discovered via virtual screening or HTS methods. Among these other inhibitors, those that do not include a MBP, with an ambiguous mechanism of action,1215–1220 or which allosterically inhibit Ddl1221 will not be discussed. Utilizing computer aided drug discovery, a cycloproply-based amino acid inhibitor Ddli-6 (Ki of 12.5 μM) was identified (Figure 125).1222 The carboxylic acid MBP of Ddli-6 is proposed to bind bidentate the Mg1 ion, while forming strong H-bonding interactions with side chains of Glu15, Arg255, and Gly276; however, no structural data is presently available to validate the mode of inhibition.
A small library of semicarbazides, aminocarbonyldiazenecarboxylates, hydrazinedicarboxamides, and diazenedicarboxamides have been designed, synthesized, and screened for inhibitor activity against EcDdlB.1223 Diazenedicarboxamides, Ddli-7 and Ddli-8 (IC50 = 15 and 111 μM, respectively, Figure 125) were found to be potent inhibitors of Ddl with moderate antimicrobial activity against E. coli and S. aureus (MIC = 64–256 μg/mL). Computational docking revealed the carbonyl groups of Ddli-8 interacting with Mg2+ ions; however, no structural data is available to verify this. Due to poor SAR between the observed IC50 values and MICs, diazenedicarboxamides were not further pursued as inhibitors of Ddl.
Current and Future Prospects.
Since the discovery of penicillin, targeting Gram-negative and Gram-positive bacterial infections by disruption the peptidoglycan synthesis pathway has been a validated mode of action for many antibiotics. Antibiotics which target the extracellular stages of peptidoglycan synthesis (β-lactams, vancomycin) have been extensively studied; however, overuse of such antibiotics has led to the production of β-lactamase and vancomycin resistance. Antibiotics which target the intracellular stages of peptidoglycan synthesis have been less well studied. Ddl is responsible for the assembly of D-ala-D-ala dipeptide precursor for MurNAc synthesis and represents another possible target for inhibiting this pathway. The Ddl inhibitor DCS is a current antibiotic for the treatment of tuberculosis; however, its limitations have almost eliminated its use in the clinical setting, and an effective alternatives are still needed. The majority of Ddl inhibitors date back more than 30 years and heavily focused on transition state analogue and D-ala analogue inhibitors. These inhibitors undergo phosphorylation in situ to yield a phosphate MBP, which coordinates to the active site Mg2+ ions. However, due to poor cellular uptake, these inhibitors lack antibacterial activity in vitro and were not pursued as a clinical drug. Recently, novel Ddl inhibitors have been discovered via virtual or HTS screening. The mechanism of action of these inhibitors is often not validated, and commonly neglect to address the presence and importance of the Mg2+ ions in the active site. The development of inhibitors for Ddl, specifically identifying other MBPs capable of binding to a dinuclear Mg2+ active site, is an untapped area. Design and synthesis of inhibitors bearing new MBPs could produce more active inhibitors of Ddl, overcome issues with poor cell permeability and uptake, and yield a broad-spectrum inhibitor for Ddl.
10. Heme-dependent Metalloenzymes
Heme-dependent proteins encompass an enormous number of important enzymes, many of which are potential therapeutic targets. There are many possible targets, with many of the most prominent heme targets involving the development of small molecule inhibitors as antifungals. These antifungals are in wide clinical use and exhaustive reviews can be found elsewhere.1224,1225 Herein, the full scope of potential heme targets will not be covered; however, two related enzymes indoleamine 2,3-dioxygenase (IDO) and tryptophan 2,3-dioxygenase (TDO) will be described as a topical case study. Both of these enzymes are currently of high interest in the field of immunomodulation therapy and can serve as representative examples of the importance of heme targets in metalloenzyme inhibition.
10.1. Indoleamine 2,3-dioxygenase (IDO, EC 1.13.11.52) and tryptophan 2,3-dioxygenase (TDO, EC 1.13.11.11)
Function in Biology and Disease.
Indoleamine 2,3-dioxygenase (IDO) and tryptophan 2,3-dioxygenase (TDO) are heme dioxygenases that catalyze the initial and rate-determining step in the catabolism of tryptophan into N-formylkynurenine and other metabolites, including kynurenine, kynurenic acid, 3-hydroxy-kynurenine, and 3-hydroxy anthralinic acid (Figure 127).1226 Both enzymes are cytosolic and share a similar active site structure. The mechanism of these enzymes has been thoroughly investigated, whereby the enzymes utilize dioxygen (IDO can also utilize the superoxide anion) to oxidatively cleave the pyrrole ring of tryptophan between the C2 and C3 carbon atoms. Despite active site and mechanistic similarities, IDO has a wider substrate scope (including serotonin, tryptamine, melatonin) than TDO. In addition, IDO is widely expressed in many tissues types, while TDO is largely confined to the liver.1226 Most important to disease relevance, IDO and TDO are immunosuppressive with respect to T-lymphocytes, both by depriving these cells of tryptophan, but also via the aforementioned metabolites of tryptophan catabolism that are also known to suppress T-cell function.1227 Hence, in recent years, IDO (both isoforms IDO1 and IDO2), and to a lesser extent TDO, have become increasingly attractive targets for immunotherapy intervention primarily for cancer.
Figure 127.
Reaction catalyzed by IDO and TDO.
Protein and Active Site Structure.
Despite their common mechanism and the reaction they catalyze, the structure of IDO and TDO vary substantially. IDO is a monomeric glycoprotein of ~45kDa, while TDO is a ~191kDa tetramer and the two enzymes share only ~10% sequence homology. IDO is largely α-helical in structure with a C-terminal domain that contains the active site and a smaller N-terminal domain. As shown in Figure 128,1228 the Trp substrate binds in the distal heme pocket, with the indole ring roughly perpendicular to the heme face and surrounded by several, neighboring hydrophobic residues, including F226, L234, F163, V130, and Y126. The proximal face of the heme center is occupied by an axial His346 ligand that is coordinated to the iron center. The ferrous form is the active form of the enzyme1226 and proximal axial coordination site is open for dioxygen binding (in Figure 128 this is occupied by a cyanide ligand).
Figure 128.
Structure of human IDO1 with tryptophan substrate (yellow) and cyanide ligand bound (PDB 5WMU).
Inhibitor Development.
Since the role of IDO and TDO in cancer immune evasion has become apparent, inhibitor development has gained increasing attention. Indeed, there are several IDO inhibitors in Phase I and II clinical trials, and at least one other in Phase III trials, often in combination with other therapeutics, including biologics.1229 For brevity, only a discussion of a few of the most clinically advanced inhibitors will be provided here.
Three of the most advanced inhibitors are shown in Figure 129: Epacadostat (Phase I, II, III), Indoximod (Phase I, II), and NLG919 (a.k.a., GDC-0919, Phase I combination therapy).1230 Docking studies suggest that Indoximod does not utilize heme binding to inhibit IDO, but rather occupies the Trp binding site (Figure 128); however, no crystallographic data for Indoximod bound to IDO is present available. In contrast, both Epacadostat and NLG919 bind to the heme center using different MBPs. For Epacadostat, the X-ray structure with human IDO reveals that oxygen atom of the hydroxyamidine group binds to the axial coordination site on the distal side of the heme (Figure 130) and is stabilized by several interamolecular hydrogen bonds and a hydrogen bond with Ala264.1228 The halogenated benzene ring of Epacadostat occupies the same binding pocket as the indole ring of the native Trp substrate, while the furazan and sulfonamide groups occupy the site where the aminocarboxylate portion of Trp reside (Figure 130). Like Epacadostat, NLG919 binds to the distal axial site of the heme in IDO1, but uses the nitrogen atom from the imidazoleisoindole fused ring system (Figure 130).1231 Note that NLG919 has two chiral centers, and all four possible diastereomers (collective IC50 = 38 nM) were used for co-crystallization, but only two diastereomers (the R,S- and S,S-stereoisomers) could be modeled with the electron density obtained (bound in similar conformations).1231 The imidazoleisoindole ring and 1-cyclohexylethanol group occupy the indole ring and aminocarboxylate groups of the native substrate Trp substrate, respectively. Although both Epacadostat and NLG919 bind to the distal axial coordination site of IDO, structural evidence has been found for other inhibitors that bind to alternative sites on the proximal face of the heme.1228 Detailed reviews on these and other inhibitors, as well as their clinical progress, can be found elsewhere.1232
Figure 129.
Inhibitors of IDO.
Figure 130.
Left: Structure of Epacadostat bound to human IDO1 (PDB 5WN8). Right: Structure of NLG919 bound to human IDO1 (PDB 5EK3).
Current and Future Prospects.
Interest in IDO and TDO inhibitors is likely to increase in the immediate future, with the growing enthusiasm over immunotherapeutic approaches to cancer treatment. Previous inhibitor development has focused on mimicking the tryptophan substrate or targeting the heme-cofactor. Many investigations are examining IDO and TDO inhibitors in combination with other chemotherapeutic agents. Unfortunately, the recent setback of nine phase 3 IDO trails (which combine epacadostate with other agents, such as Opdivo by Bristol-Myer Squibb and Keytruda by Merck) has led to substantial questioning of the robustness of this target space. Recently, an alternative class of IDO inhibitors with a novel mode of inhibition has been proposed.1233 These inhibitors displace the heme-cofactor and bind to the apo-form of IDO and provide a new direction of IDO inhibitor development. Inhibitors that use different MBPs to bind the heme center, as well as inhibitors that do not bind to the heme center at all (in the proximal site), are both likely to be the focus of future studies. Vigorous activity in the biotechnology sector, with substantial acquisitions of IDO inhibitor assets suggest a continuing robust interest in this target space.
Conclusions and Outlook
The opportunity for having a significant impact on human health via the development of small molecule metalloenzyme inhibitors remains a fertile landscape. Herein, we have described over three dozen different metalloenzyme targets, highlighting the biological and pathogenic role of each metalloenzyme, drug discovery efforts, structural characterization of inhibitor-metalloenzyme complexes, and the current developmental/clinical status of each target. Even having covered a wide range of targets involved in every disease arena, including antibiotics, antivirals, anticancer drugs, and others, the scope of our efforts here are not complete. There are dozens of other targets that were not included in this review, including prolyl 4-hydroxylases (PHDs), collagen prolyl 4-hydroxylase (CP4H), RNA polymerase, DNA gyrase, antifungal heme targets, and numerous others, which bodes well for the many opportunities that remain untapped for this field. There also may be many opportunities to interfere with metal ion trafficking to metalloenzymes, or to target the metal ion centers in non-catalytic metalloproteins. By applying the methods of modern medicinal chemistry, including structure-, fragment-, and computation-based drug discovery and design, HTS,1234 and others, more of these targets should become increasingly tractable and druggable. Greater augmentation of these methods with tools from the field of bioinorganic chemistry (e.g., spectroscopic methods such as electron paramagnetic resonance spectroscopy, X-ray absorption spectroscopy, etc.) is merited, as use of these tools can greatly aid in understanding the binding, mechanism of inhibition, and design of inhibitors. Challenges remain in the field, which if addressed, would further accelerate these efforts. For example, computational methods (including docking methods) still struggle with accurately predicting metal-inhibitor interactions in a robust and time-effective manner, but ongoing efforts are rapidly advancing this field as well.1235 Also, despite the tremendous clinical success of some metalloenzyme inhibitors, further studies on the effect of these compounds on metal ion trafficking and distributions (e.g., the so-called ‘metallome’) are needed to ease concerns around the safety and selectivity of these therapeutics. Indeed, consideration of other approaches, such as allosteric inhibitors that do not bind the active site metal ion offer additional options for metalloenzyme drug development. Finally, by applying the knowledge and knowhow of bioinorganic chemistry to the development of metalloenzyme inhibitors, as our laboratory has done for >15 years,32 the productivity of this field can be further accelerated and discoveries to improve the human condition can be uncovered.
Table 5.
Activity (IC50 values) of HIV IN inhibitors.
| Compound | Inhibition (μM) | Ref. |
|---|---|---|
| HIV-INi-1 | 0.03 | 386,387 |
| HIV-INi-2 | 0.2 | 365 |
| HIV-INi-3 | 0.01 | 390 |
| HIV-INi-4 | 0.042 | 391 |
| HIV-INi-5 | 0.11 | 392 |
| HIV-INi-6 | 0.1a | 394 |
| HIV-INi-7 | 0.02 | 400 |
| Chicoric Acid | 0.5 | 382 |
| 5-CITEP | 2.2 | 379 |
| Elvitegravir | 0.007 | 393 |
| Globoidnan A | 0.64 | 381 |
| L-731,988 | 0.17 | 402 |
| L-870,810 | <0.015 | 388 |
| Quercetin | 11 | 383 |
| RCD-1 | 1 | 401 |
| RCD-5 | 0.55 | 401 |
EC50 value.
Table 6.
Activity (IC50 values) of RNase-H inhibitors.
| Compound | Inhibition (μM) | Ref. |
|---|---|---|
| RNaseHi-1 | 15 | 426 |
| RNaseHi-2 | 1 | 403 |
| RNaseHi-3 | 50 | 403 |
| RNaseHi-4 | 3.5 | 432 |
| RNaseHi-5 | 1.2 | 437 |
| RNaseHi-6 | 2.7 | 438 |
| RNaseHi-7 | 0.65 | 441 |
| RNaseHi-8 | 0.43 | 439,440 |
| RNaseHi-9 | >50 | 439,440 |
| RNaseHi-10 | >50 | 439,440 |
| RNaseHi-11 | >50 | 439,440 |
| RNaseHi-12 | >50 | 439 |
| RNaseHi-13 | 0.061 | 421 |
| RNaseHi-14 | 0.15 | 444 |
| RNaseHi-15 | 0.11 | 444 |
| RNaseHi-16 | 0.045 | 444 |
| RNaseHi-17 | 10 | 445 |
| RNaseHi-18 | 11 | 445 |
| RNaseHi-19 | 0.05 | 446 |
| RNaseHi-20 | >10 | 446 |
| RNaseHi-21 | 0.2 | 446 |
| AZT-MP | 50 | 431 |
| BTDBA | 3.2 | 436 |
| GSK5750 | 0.33 | 445 |
| Manicol | 1.5 | 427 |
| RDS 1643 | 13 | 447 |
| β-Thujaplicinol | 0.2 | 427 |
Table 10.
Activity of BoNTA inhibitors. Unless otherwise noted, all values are against BoNTA.
| Compound | Inhibition (μM) | Ref. |
|---|---|---|
| BoNTi-1 | IC50 = 5–10 | 643 |
| BoNTi-2 | IC50 > 12000a | 644 |
| BoNTi-3 | IC50 > 12000a | 644 |
| BoNTi-4 | IC50 = 8100a | 644 |
| BoNTi-5 | N.A. | 645 |
| BoNTi-6 | Ki = 12 | 646 |
| BoNTi-7 | IC50 = 15 | 647 |
| BoNTi-8 | IC50 = 3.1 | 647 |
| BoNTi-9 | IC50 = 0.400, Ki = 0.300 | 647 |
| BoNTi-10 | 75% inhibition at 50 μM | 648 |
| BoNTi-11 | IC50 = 1, Ki = 0.460 | 649 |
| BoNTi-12 | IC50 = 0.030, Ki = 0.027 | 650 |
| BoNTi-13 | Ki = 0.760 | 651 |
| BoNTi-14 | Ki = 0.160 | 652 |
| BoNTi-15 | Ki = 4.6 | 653 |
| BoNTi-16 | Ki = 6.1 | 653 |
| BoNTi-17 | Ki = 6.3 | 653 |
| BoNTi-18 | Ki = 1.8 | 654 |
| BoNTi-19 | Ki = 0.330 | 655 |
| BoNTi-20 | IC50 = 4.8 | 656 |
| BoNTi-21 | IC50 = 13.7 | 657 |
| BoNTi-22 | 52% inhibition at 20 μM | 658 |
| BoNTi-23 | IC50 = 3.3 | 659 |
| EDTA | EC50 >160 | 660 |
| DTPA | EC50 = 20–40 | 660 |
| 8HQ | IC50 >500 | 661 |
| L-captopril | N.A. | 645 |
| Lomofungin | Ki = 6.7 | 661 |
| Phosphoramidon | IC50 = 14700a | 644,645 |
| TPEN | EC50 = 20–40 | 660 |
Inhibition against BoNTB.
Inhibition against BoNTF.
Table 11.
Activity of MetAP inhibitors.
| Compound | Inhibition (nM) | Ref. |
|---|---|---|
| MetAPi-1 | Ki = 15, Co-hMetAP2; Ki = 4400, Mn-hMetAP2 | 727 |
| MetAPi-2 | Ki = 18, Co-hMetAP2 | 727 |
| MetAPi-3 | Ki = 0.5, Co-hMetAP2 | 728 |
| MetAPi-4 | Ki = 2, Co-hMetAP2 | 728 |
| MetAPi-5 | IC50 = 9100, Mn-hMetAP2 | 729 |
| MetAPi-6 | IC50 = 19, Mn-hMetAP2 | 730 |
| MetAPi-7 | IC50 = 1100, Mn-hMetAP2 | 731 |
| MetAPi-8 | IC50 = 11, Mn-hMetAP2 | 731 |
| MetAPi-9 | IC50 = 230, Mn-hMetAP2 | 732 |
| MetAPi-10 | IC50 = 38, Mn-hMetAP2 | 732 |
| MetAPi-11 | IC50 = 700, Mn-hMetAP2 | 732 |
| MetAPi-12 | IC50 = 800, Co-hMetAP2 | 733 |
| MetAPi-13 | IC50 = 110, Co-EcMetAP | 734 |
| MetAPi-14 | IC50 = 78, Co-EcMetAP | 735 |
| MetAPi-15 | IC50 = 184, Ni-EcMetAP; IC50 = 137, Co-EcMetAP | 736 |
| MetAPi-16 | IC50 = 240, Mn-EcMetAP; IC50 = 116000, Fe-EcMetAP | 737,738 |
| MetAPi-17 | IC50 = 790, Fe-EcMetAP | 738 |
| MetAPi-18 | IC50 = 950, Fe-EcMetAP | 739 |
| MetAPi-19 | IC50 = 13000, Fe-EcMetAP | 740 |
| MetAPi-20 | IC50 = 12900, Fe-EcMetAP | 740 |
| MetAPi-21 | IC50 = 5000, Co-EcMetAP | 741 |
| MetAPi-22 | IC50 = 110, Co-EcMetAP | 742 |
| MetAPi-23 | IC50 = 67, Co-EcMetAP | 737 |
| A311263 | IC50 = 30, Mn-hMetAP2 | 724 |
| A320282 | IC50 =180, Mn-hMetAP2 | 724 |
| A357300 | IC50 = 110, Mn-hMetAP2 | 724 |
| LAF389 | IC50 = 800, Co-hMetAP2 | 721 |
| Nitroxoline | IC50 = 55, Mn-hMetAP2; IC50 = 60, Co-BpMetAP | 743,744 |
| TNP-470 | IC50 =1.3, Co-hMetAP2 | 717 |
ACKNOWLEDGMENT
S.M.C. would like to thank the many coworkers, colleagues, and collaborators – past, present, and future – that have contributed to advancing and improving our studies on metalloenzyme inhibitors. The authors thank Stephanie M. Duggan, Christian Perez, Ryjul W. Stokes for assisting in the editing, reviewing, and proofreading of this manuscript. This work was supported by the National Institutes of General Medical Sciences (R01-GM098435). The development of MBL inhibitors in the laboratory of S.M.C. was supported by the National Institutes of General Medical Sciences (R01-GM111926). The development of Rpn11 inhibitors in the laboratory of S.M.C. was supported by the National Cancer Institute (R01-CA164803). S.M.C. is a co-founder, has an equity interest, and receives income as member of the Scientific Advisory Board for Forge Therapeutics and is a co-founder, has an equity interest, and a member of the Scientific Advisory Board for Cleave Biosciences. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies.
Biography
Allie Y. Chen grew up in Temple City, California. She received her B.S. in Chemistry from the University California, Irvine in 2014, and performed research in nanoparticles antitoxins with Prof. Kenneth J. Shea. She then joined the graduate program at U.C. San Diego in 2014, where she is currently a doctoral candidate in the laboratory of Prof. Seth M. Cohen.. Her current research includes developing metalloprotein inhibitors in the field of bioinorganic and medicinal chemistry.
Rebecca N. Adamek grew up near Seattle, Washington. She received her B.S. in Chemistry from California State University, Fullerton in 2014, where she graduated magna cum laude with University Honors, and performed Undergraduate research with Prof. Nicholas T. Salzameda. She then joined the graduate program at the University of California, San Diego, where she was a fellow of the Chemistry-Biology Interface Graduate Training Program, and is currently a doctoral candidate in the laboratory of Prof. Seth M. Cohen. Her research interests include bioinorganic, organic, and medicinal chemistry.
Benjamin L. Dick grew up near Rochester, New York. He received his B.S. and M.S. in Chemistry from the University of Rochester in 2013, and performed research with Prof. Kara L. Bren. He then joined the graduate program at U.C. San Diego in 2014, where he is currently a doctoral candidate in the laboratory of Prof. Seth M. Cohen. During his time in Prof. Cohen’s laboratory he has been a trainee in the Molecular Biophysics Training Program at U.C. San Diego. His research interests include bioinorganic, biophysical, inorganic, and medicinal chemistry.
Cy V. Credille grew up near Provo, Utah. He received his B.S. in Biochemistry from Brigham Young University in 2012, where he did research on antimicrobial peptide mimetics, surface coatings, and natural product synthesis in the laboratory of Professor Paul B. Savage. He then joined the research group of Prof. Seth M. Cohen at U.C. San Diego in 2012 where he is currently a doctoral candidate researching fragment-based methods to inhibit metalloenzymes, with an emphasis on developing inhibitors of the influenza RNA-dependent polymerase complex.
Christine N. Morrison grew up near Detroit, Michigan. She received her B.S. in Chemistry from the University of Michigan in 2010, performing research with Prof. Adam J. Matzger. The following year, she was a Fulbright scholar at the Karlsruhe Institute of Technology in Karlsruhe, Germany in the laboratory of Prof. Dr. Annie K. Powell. She then earned her Ph.D. in Chemistry in 2017 from Caltech with a National Science Foundation Graduate Research Fellowship under the direction of Prof. Douglas C. Rees. She is currently a U.C. President’s and Ruth L. Kirschstein postdoctoral fellow in the laboratory of Prof. Seth M. Cohen at U.C. San Diego. Her research interests include bioinorganic, biophysical, and medicinal chemistry.
Seth M. Cohen is a native of the San Fernando Valley of Los Angeles, California. He received a B.S. in Chemistry and B.A. in Political Science from Stanford University in 1994. He completed his Ph.D. in Chemistry at U.C. Berkeley under the direction of Prof. Kenneth N. Raymond and performed postdoctoral studies with Prof. Stephen J. Lippard at M.I.T. He started his independent career at U.C. San Diego in 2001, served as Chair of the Department of Chemistry and Biochemistry at U.C. San Diego from 2012–2015, and is presently the Leslie Orgel Scholar of Inorganic Chemistry at U.C. San Diego. His research interests are in the areas of inorganic, bioinorganic, medicinal, materials, and supramolecular chemistry.
References
- (1).Laity JH; Lee BM; Wright PE Zinc Finger Proteins: New Insights into Structural and Functional Diversity. Curr. Opin. Struct. Biol 2001, 11, 39–46. [DOI] [PubMed] [Google Scholar]
- (2).Solomon EI; Heppner DE; Johnston EM; Ginsbach JW; Cirera J; Qayyum M; Kieber-Emmons MT; Kjaergaard CH; Hadt RG; Tian L Copper Active Sites in Biology. Chem. Rev 2014, 114, 3659–3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Yang Y; Hu X-Q; Li Q-S; Zhang X-X; Ruan B-F; Xu J; Liao C Metalloprotein Inhibitors for the Treatment of Human Diseases. Curr. Top. Med. Chem 2016, 16, 384–396. [DOI] [PubMed] [Google Scholar]
- (4).Heffern MC; Kurutz JW; Meade TJ Spectroscopic Elucidation of the Inhibitory Mechanism of Cys2His2 Zinc Finger Transcription Factors by Cobalt(III) Schiff Base Complexes. Chem. Eur. J 2013, 19, 17043–17053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Louie AY; Meade TJ A Cobalt Complex that Selectively Disrupts the Structure and Function of Zinc Fingers. Proc. Natl. Acad. Sci. U. S. A 1998, 95, 6663–6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Solomon EI; Sundaram UM; Machonkin TE Multicopper Oxidases and Oxygenases. Chem. Rev 1996, 96, 2563–2605. [DOI] [PubMed] [Google Scholar]
- (7).Andreini C; Bertini I; Cavallaro G; Holliday GL; Thornton JM Metal Ions in Biological Catalysis: From Enzyme Databases to General Principles. JBIC, J. Biol. Inorg. Chem 2008, 13, 1205–1218. [DOI] [PubMed] [Google Scholar]
- (8).Waldron KJ; Rutherford JC; Ford D; Robinson NJ Metalloproteins and Metal Sensing. Nature 2009, 460, 823–830. [DOI] [PubMed] [Google Scholar]
- (9).Knape MJ; Ballez M; Burghardt NC; Zimmermann B; Bertinetti D; Kornev AP; Herberg FW Divalent Metal Ions Control Activity and Inhibition of Protein Kinases. Metallomics 2017, 9, 1576–1584. [DOI] [PubMed] [Google Scholar]
- (10).Ferguson FM; Gray NS Kinase Inhibitors: The Road Ahead. Nature Rev. Drug Discover 2018, 8, 96–120. [DOI] [PubMed] [Google Scholar]
- (11).Mullard A 2013 FDA Drug Approvals. Nat. Rev. Drug Discovery 2014, 13, 85–89. [DOI] [PubMed] [Google Scholar]
- (12).Administration, U. S. F. D. In Drug Innovation 2013.
- (13).Mullard A 2014 FDA Drug Approvals. Nat. Rev. Drug Discovery 2015, 14, 77–81. [DOI] [PubMed] [Google Scholar]
- (14).Administration, U. S. F. D. In Drug Innovation 2014.
- (15).Mullard A 2015 FDA Drug Approvals. Nat. Rev. Drug Discovery 2016, 15, 73–76. [DOI] [PubMed] [Google Scholar]
- (16).Administration, U. S. F. D. In Drug Innovation 2015.
- (17).Mullard A 2016 FDA Drug Approvals. Nat. Rev. Drug Discovery 2017, 16, 73–76. [DOI] [PubMed] [Google Scholar]
- (18).Administration, U. S. F. D. In Drug Innovation 2016.
- (19).Mullard A 2017 FDA Drug Approvals. Nat. Rev. Drug Discovery 2018, 1–5. [DOI] [PubMed] [Google Scholar]
- (20).Administration, U. S. F. D. In Drug Innovation 2017.
- (21).Waloen K; Kleppe R; Martinez A; Haavik J Tyrosine and Tryptophan Hydroxylases as Therapeutic Targets in Human Disease. Exp. Opin. Ther. Targ 2017, 21, 167–180. [DOI] [PubMed] [Google Scholar]
- (22).Parczewski M Stribild, as a Novel Option of Integrase Inhibitor Based Single Tablet Regimen for the Treatment of HIV Infection. HIV & AIDS Rev 2014, 13, 101–105. [Google Scholar]
- (23).Lapuerta P; Zambrowicz B; Fleming D; Wheeler D; Sands A Telotristat Etiprate, a Novel Inhibitor of Serotonin Synthesis for the Treatment of Carcinoid Syndrome. Clin. Invest 2015, 5, 447–456. [Google Scholar]
- (24).Windahl MS; Petersen CR; Christensen HEM; Harris P Crystal Structure of Tryptophan Hydroxylase with Bound Amino Acid Substrate. Biochemistry 2008, 47, 12087–12094. [DOI] [PubMed] [Google Scholar]
- (25).Erlandsen H; Flatmark T; Stevens RC; Hough E Crystallographic Analysis of the Human Phenylalanine Hydroxylase Catalytic Domain with Bound Catechol Inhibitors at 2.0 AÅ Resolution. Biochemistry 1998, 37, 15638–15646. [DOI] [PubMed] [Google Scholar]
- (26).Sevrioukova IF High-Level Production and Properties of the Cysteine-Depleted Cytochrome P450 3A4. Biochemistry 2017, 56, 3058–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Lian TY; Jiang X; Jing ZC Riociguat: A Soluble Guanylate Cyclase Stimulator for the Treatment of Pulmonary Hypertension. Drug. Des. Devel. Ther 2017, 11, 1195–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Tellinghuisen TL; Marcotrigiano J; Gorbalenya AE; Rice CM The NS5A Protein of Hepatitis C Virus Is a Zinc Metalloprotein. J. Biol. Chem 2004, 279, 48576–48587. [DOI] [PubMed] [Google Scholar]
- (29).Sharma A; Gupta SP; Siddiqui AA; Sharma N HCV NS3/4A Protease and its Emerging Inhibitors. J. Anal. Pharm. Res 2017, 4, 00108. [Google Scholar]
- (30).Kinch MS; Haynesworth A; Kinch SL; Hoyer D An Overview of FDA-Approved New Molecular Entities: 1827–2013. Drug Discovery Today 2014, 19, 1033–1039. [DOI] [PubMed] [Google Scholar]
- (31).Rouffet M; Cohen SM Emerging Trends in Metalloprotein Inhibition. Dalton Trans 2011, 40, 3445–3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Cohen SM A Bioinorganic Approach to Fragment-Based Drug Discovery Targeting Metalloenzymes. Acc. Chem. Res 2017, 50, 2007–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Whittaker M; Floyd CD; Brown P; Gearing AJH Design and Therapeutic Application of Matrix Metalloproteinase Inhibitors. Chem. Rev 1999, 99, 2735–2776. [DOI] [PubMed] [Google Scholar]
- (34).Berman HM; Westbrook J; Feng Z; Gilliland G; Bhat TN; Weissig H; Shindyalov IN; Bourne PE The Protein Data Bank. Nucleic Acids Res 2000, 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Golovin A; Henrick K MSDmotif: Exploring Protein Sites and Motifs. BMC Bioinf. 2008, 9, 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Placzek S; Schomburg I; Chang A; Jeske L; Ulbrich M; Tillack J; Schomburg D BRENDA in 2017: New Perspectives and New Tools in BRENDA. Nucleic Acids Res. 2017, 45, D380–D388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Degtyarenko K; C. T. North A; B. C. Findlay J PROMISE: A Database of Bioinorganic Motifs. Nucleic Acids Res 1999, 27, 233–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Castagnetto JM; Hennessy SW; Roberts VA; Getzoff ED; Tainer JA; Pique ME MDB: The Metalloprotein Database and Browser at The Scripps Research Institute. Nucleic Acids Res 2002, 30, 379–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Andreini C; Bertini I; Cavallaro G; Holliday GL; Thornton JM Metal-MACiE: A Database of Metals Involved in Biological Catalysis. Bioinformatics 2009, 25, 2088–2089. [DOI] [PubMed] [Google Scholar]
- (40).Choi H; Kang H; Park H MetLigDB: A Web-Based Database for the Identification of Chemical Groups to Design Metalloprotein Inhibitors. J. Appl. Crystallogr 2011, 44, 878–881. [Google Scholar]
- (41).Supuran CT; Winum J-Y Drug Design of Zinc-Enzyme Inhibitors; John Wiley & Sons, Inc: Hoboken, New Jersey, 2009. [Google Scholar]
- (42).Eriksson AE; Jones TA; Liljas A Refined Structure of Human Carbonic Anhydrase II at 2.0 A Resolution. Proteins 1988, 4, 274–282. [DOI] [PubMed] [Google Scholar]
- (43).Sippel KH; Robbins AH; Domsic J; Genis C; Agbandje-McKenna M; McKenna R High-resolution Structure of Human Carbonic Anhydrase II Complexed with Acetazolamide Reveals Insights into Inhibitor Drug Design. Acta Crystallogr., Sect.F 2009, 65, 992–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Scolnick LR; Clements AM; Liao J; Crenshaw L; Hellberg M; May J; Dean TR; Christianson DW Novel Binding Mode of Hydroxamate Inhibitors to Human Carbonic Anhydrase II. J. Am. Chem. Soc 1997, 119, 850–851. [Google Scholar]
- (45).Supuran CT Advances in Structure-based Drug Discovery of Carbonic Anhydrase Inhibitors. Expert Opin. Drug Discovery 2017, 12, 61–88. [DOI] [PubMed] [Google Scholar]
- (46).Murphy G Riding the Metalloproteinase Roller Coaster. J. Biol. Chem 2017, 292, 7708–7718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Vandenbroucke RE; Libert C Is There New Hope for Therapeutic Matrix Metalloproteinase Inhibition? Nat. Rev. Drug Dis 2014, 13, 904–927. [DOI] [PubMed] [Google Scholar]
- (48).Puerta DT; Lewis JA; Cohen SM New Beginnings for Matrix Metalloproteinase Inhibitors: Identification of High-Affinity Zinc-Binding Groups. J. Am. Chem. Soc 2004, 126, 8388–8389. [DOI] [PubMed] [Google Scholar]
- (49).Rothenberg ML; Nelson AR; Hande KR New Drugs on the Horizon: Matrix Metalloproteinase Inhibitors. Stem Cells 1999, 17, 237–240. [DOI] [PubMed] [Google Scholar]
- (50).Jackson HW; Defamie V; Waterhouse P; Khokha R TIMPs: Versatile Extracellular Regulators in Cancewr. Nat. Rev. Cancer 2017, 17, 38–53. [DOI] [PubMed] [Google Scholar]
- (51).Morgunova E; Tuuttila A; Bergmann U; Tryggvason K Structural Insight Into the Complex Formation of Latent Matrix Metalloproteinase 2 with Tissue Inhibitor of Metalloproteinase 2. Proc. Natl. Acad. Sci. U. S. A 2002, 99, 7417–7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Ikejiri M; Bernardo MM; Bonfil RD; Toth M; Chang M; Fridman R; Mobashery S Potent Mechanism-based Inhibitors for Matrix Metalloproteinases. J. Biol. Chem 2005, 280, 33992–34002. [DOI] [PubMed] [Google Scholar]
- (53).Nara H; Sato K; Naito T; Mototani H; Oki H; Yamamoto Y; Kuno H; Santou T; Kanzaki N; Terauchi J; Uchikawa O; Kori M Discovery of Novel, Highly Potent, and Selective Quinazoline-2-carboxamide-Based Matrix Metalloproteinase (MMP)-13 Inhibitors without a Zinc Binding Group Using a Structure-Based Design Approach. J. Med. Chem 2014, 57, 8886–8902. [DOI] [PubMed] [Google Scholar]
- (54).Hajduk PJ; Sheppard G; Nettesheim DG; Olejniczak ET; Shuker SB; Meadows RP; Steinman DH; Carrera GM Jr.; Marcotte PA; Severin J; Walter K; Smith H; Gubbins E; Simmer R; Holzman TF; Morgan DW; Davidsen SK; Summers JB; Fesik SW Discovery of Potent Nonpeptide Inhibitors of Stromelysin Using SAR by NMR. J. Am. Chem. Soc 1997, 119, 5818–5827. [Google Scholar]
- (55).Hajduk PJ; Shuker SB; Nettesheim DG; Craig R; Augeri DJ; Betebenner D; Albert DH; Guo Y; Meadows RP; Xu L; Michaelides M; Davidsen SK; Fesik SW NMR-Based Modification of Matrix Metalloproteinase Inhibitors with Improved Bioavailability. J. Med. Chem 2002, 45, 5628–5639. [DOI] [PubMed] [Google Scholar]
- (56).Parkin G Synthetic Analogues Relevant to the Structure and Function of Zinc Enzymes. Chem. Rev 2004, 104, 699–767. [DOI] [PubMed] [Google Scholar]
- (57).Lieber CS Alcohol and the Liver - 1994 Update. Gastroenterology 1994, 106, 1085–1105. [DOI] [PubMed] [Google Scholar]
- (58).Koppaka V; Thompson DC; Chen Y; Ellermann M; Nicolaou KC; Juvonen RO; Petersen D; Deitrich RA; Hurley TD; Vasiliou V Aldehyde Dehydrogenase Inhibitors: a Comprehensive Review of the Pharmacology, Mechanism of Action, Substrate Specificity, and Clinical Application. Pharmacol. Rev 2012, 64, 520–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Jacobsen D; McMartin KE Antidotes for Methanol and Ethylene Glycol Poisoning. J. Toxicol., Clin. Toxicol 1997, 35, 127–143. [DOI] [PubMed] [Google Scholar]
- (60).Baud FJ; Galliot M; Astier A; Bien DV; Garnier R; Likforman J; Bismuth C Treatment of Ethylene-Glycol Poisoning with Intravenous 4-Methylpyrazole. N. Engl. J. Med 1988, 319, 97–100. [DOI] [PubMed] [Google Scholar]
- (61).Hammes-Schiffer S; Benkovic SJ Relating Protein Motion to Catalysis. Annu. Rev. Biochem 2006, 75, 519–541. [DOI] [PubMed] [Google Scholar]
- (62).Eklund H; Horjales E; Vallee BL; Jornvall H Computer-Graphics Interpretations of Residue Exchanges between the Alpha-Subunit, Beta-Subunit and Gamma-Subunit of Human-Liver Alcohol-Dehydrogenase Class-I Isozymes. Eur. J. Biochem 1987, 167, 185–193. [DOI] [PubMed] [Google Scholar]
- (63).Ryde U The Coordination of the Catalytic Zinc Ion in Alcohol Dehydrogenase Studied by Combined Quantum-Chemical and Molecular Mechanics Calculations. J. Comput. -Aided Mol. Des 1996, 10, 153–164. [DOI] [PubMed] [Google Scholar]
- (64).Salaspuro MP; Lindros KO; Pikkarainen PH Effect of 4-Methylpyrazole on Ethanol Elimination Rate and Hepatic Redox Changes in Alcoholics with Adequate or Inadequate Nutrition and in Non-Alcoholic Controls. Metab., Clin. Exp 1978, 27, 631–639. [DOI] [PubMed] [Google Scholar]
- (65).Barceloux DG; Krenzelok EP; Olson K; Watson W American Academy of Clinical Toxicology Practice Guidelines on the Treatment of Ethylene Glycol Poisoning. J. Toxicol., Clin. Toxicol 1999, 37, 537–560. [DOI] [PubMed] [Google Scholar]
- (66).Theorell H; Mckinleymckee JS Liver Alcohol Dehydrogenase .2. Equilibrium Constants of Binary and Ternary Complexes of Enzyme, Coenzyme, and Caprate, Isobutyramide and and Imidazole. Acta Chem. Scand 1961, 15, 1811–1833. [Google Scholar]
- (67).Li TK; Theorell H Human Liver Alcohol Dehydrogenase - Inhibition by Pyrazole and Pyrazole Analogs. Acta Chem. Scand 1969, 23, 892–902. [DOI] [PubMed] [Google Scholar]
- (68).Theorell H; Yonetani T On the Effects of Some Heterocyclic Compounds on the Enzymic Activity of Liver Alcohol Dehydrogenase. Acta Chem. Scand 1969, 23, 255–260. [DOI] [PubMed] [Google Scholar]
- (69).Davis GJ; Bosron WF; Stone CL; OwusuDekyi K; Hurley TD X-ray Structure of Human Beta(3)Beta(3) Alcohol Dehydrogenase - The Contribution of Ionic Interactions to Coenzyme Binding. J. Biol. Chem 1996, 271, 17057–17061. [DOI] [PubMed] [Google Scholar]
- (70).Xie PGT; Hurley TD Methionine-141 Directly Influences the Binding of 4-Methylpyrazole in Human Sigma Sigma Alcohol Dehydrogenase. Protein Sci 1999, 8, 2639–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Bosron WF; Li TK Catalytic Properties of Human Liver Alcohol Dehydrogenase Isoenzymes. Enzyme 1987, 37, 19–28. [DOI] [PubMed] [Google Scholar]
- (72).Niederhut MS; Gibbons BJ; Perez-Miller S; Hurley TD Three-Dimensional Structures of the Three Human Class I Alcohol Dehydrogenases. Protein Sci 2001, 10, 697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Schindler JF; Berst KB; Plapp BV Inhibition of Human Alcohol Dehydrogenases by Formamides. J. Med. Chem 1998, 41, 1696–1701. [DOI] [PubMed] [Google Scholar]
- (74).Gibbons BJ; Hurley TD Structure of Three Class I Human Alcohol Dehydrogenases Complexed with Isoenzyme Specific Formamide Inhibitors. Biochemistry 2004, 43, 12555–12562. [DOI] [PubMed] [Google Scholar]
- (75).Makar AB; Tephly TR Inhibition of Monkey Liver Alcohol Dehydrogenase by 4-Methylpyrazole. Biochem. Med 1975, 13, 334–342. [DOI] [PubMed] [Google Scholar]
- (76).White NJ; Pukrittayakamee S; Hien TT; Faiz MA; Mokuolu OA; Dondorp AM Malaria. Lancet 2014, 383, 723–735. [DOI] [PubMed] [Google Scholar]
- (77).WHO World Malaria Report 2017, 2017.
- (78).Umeda T; Tanaka N; Kusakabe Y; Nakanishi M; Kitade Y; Nakamura KT Molecular Basis of Fosmidomycin’s Action on the Human Malaria Parasite Plasmodium falciparum. Sci. Rep 2011, 1, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Murkin AS; Manning KA; Kholodar SA Mechanism and Inhibition of 1-Deoxy-D-xylulose-5-Phosphate Reductoisomerase. Bioorg. Chem 2014, 57, 171–185. [DOI] [PubMed] [Google Scholar]
- (80).Hunter WN The Non-mevalonate Pathway of Isoprenoid Precursor Biosynthesis. J. Biol. Chem 2007, 282, 21573–21577. [DOI] [PubMed] [Google Scholar]
- (81).Rohdich F; Bacher A; Eisenreich W Perspectives in Anti-infective Drug Design. The Late Steps in the Biosynthesis of the Universal Terpenoid Precursors, Isopentenyl Diphosphate and Dimethylallyl Diphosphate. Bioorg. Chem 2004, 32, 292–308. [DOI] [PubMed] [Google Scholar]
- (82).Rodriguez-Concepcion M; Campos N; Maria Lois L; Maldonado C; Hoeffler JF; Grosdemange-Billiard C; Rohmer M; Boronat A Genetic Evidence of Branching in the Isoprenoid Pathway for the Production of Isopentenyl Diphosphate and Dimethylallyl Diphosphate in Escherichia coli. FEBS Lett 2000, 473, 328–332. [DOI] [PubMed] [Google Scholar]
- (83).Schwender J; Muller C; Zeidler J; Lichtenthaler HK Cloning and Heterologous Expression of a cDNA Encoding 1-Deoxy-D-xylulose-5-phosphate Reductoisomerase of Arabidopsis thaliana. FEBS Lett 1999, 455, 140–144. [DOI] [PubMed] [Google Scholar]
- (84).Kobayashi K; Ehrlich SD; Albertini A; Amati G; Andersen KK; Arnaud M; Asai K; Ashikaga S; Aymerich S; Bessieres P; Boland F; Brignell SC; Bron S; Bunai K; Chapuis J; Christiansen LC; Danchin A; Debarbouille M; Dervyn E; Deuerling E; Devine K; Devine SK; Dreesen O; Errington J; Fillinger S; Foster SJ; Fujita Y; Galizzi A; Gardan R; Eschevins C; Fukushima T; Haga K; Harwood CR; Hecker M; Hosoya D; Hullo MF; Kakeshita H; Karamata D; Kasahara Y; Kawamura F; Koga K; Koski P; Kuwana R; Imamura D; Ishimaru M; Ishikawa S; Ishio I; Le Coq D; Masson A; Mauel C; Meima R; Mellado RP; Moir A; Moriya S; Nagakawa E; Nanamiya H; Nakai S; Nygaard P; Ogura M; Ohanan T; O’Reilly M; O’Rourke M; Pragai Z; Pooley HM; Rapoport G; Rawlins JP; Rivas LA; Rivolta C; Sadaie A; Sadaie Y; Sarvas M; Sato T; Saxild HH; Scanlan E; Schumann W; Seegers JF; Sekiguchi J; Sekowska A; Seror SJ; Simon M; Stragier P; Studer R; Takamatsu H; Tanaka T; Takeuchi M; Thomaides HB; Vagner V; van Dijl JM; Watabe K; Wipat A; Yamamoto H; Yamamoto M; Yamamoto Y; Yamane K; Yata K; Yoshida K; Yoshikawa H; Zuber U; Ogasawara N Essential Bacillus subtilis Genes. Proc. Natl. Acad. Sci. U. S. A 2003, 100, 4678–4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Takahashi S; Kuzuyama T; Watanabe H; Seto H A 1-Deoxy-D-xylulose 5-phosphate Reductoisomerase Catalyzing the Formation of 2-C-methyl-D-erythritol 4-phosphate in an Alternative Nonmevalonate Pathway for Terpenoid Biosynthesis. Proc. Natl. Acad. Sci. U. S. A 1998, 95, 9879–9884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Proteau PJ 1-Deoxy-D-xylulose 5-phosphate Reductoisomerase: an Overview. Bioorg. Chem 2004, 32, 483–493. [DOI] [PubMed] [Google Scholar]
- (87).Cai G; Deng L; Fryszczyn BG; Brown NG; Liu Z; Jiang H; Palzkill T; Song Y Thermodynamic Investigation of Inhibitor Binding to 1-Deoxy-D-Xylulose-5-Phosphate Reductoisomerase. ACS Med. Chem. Lett 2012, 3, 496–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Reuter K; Sanderbrand S; Jomaa H; Wiesner J; Steinbrecher I; Beck E; Hintz M; Klebe G; Stubbs MT Crystal Structure of 1-Deoxy-D-xylulose-5-phosphate Reductoisomerase, a Crucial Enzyme in the Non-mevalonate Pathway of Isoprenoid Biosynthesis. J. Biol. Chem 2002, 277, 5378–5384. [DOI] [PubMed] [Google Scholar]
- (89).Yajima S; Nonaka T; Kuzuyama T; Seto H; Ohsawa K Crystal Structure of 1-Deoxy-D-xylulose 5-phosphate Reductoisomerase Complexed with Cofactors: Implications of a Flexible Loop Movement Upon Substrate Binding. J. Biochem 2002, 131, 313–317. [DOI] [PubMed] [Google Scholar]
- (90).Steinbacher S; Kaiser J; Eisenreich W; Huber R; Bacher A; Rohdich F Structural Basis of Fosmidomycin Action Revealed by the Complex with 2-C-methyl-D-erythritol 4-phosphate Synthase (IspC). Implications for the Catalytic Mechanism and Anti-malaria Drug Development. J. Biol. Chem 2003, 278, 18401–18407. [DOI] [PubMed] [Google Scholar]
- (91).Mac Sweeney A; Lange R; Fernandes RP; Schulz H; Dale GE; Douangamath A; Proteau PJ; Oefner C The Crystal Structure of E.coli 1-Deoxy-D-xylulose-5-phosphate Reductoisomerase in a Ternary Complex with the Antimalarial Compound Fosmidomycin and NADPH Reveals a Tight-binding Closed Enzyme Conformation. J. Mol. Biol 2005, 345, 115–127. [DOI] [PubMed] [Google Scholar]
- (92).Yajima S; Hara K; Iino D; Sasaki Y; Kuzuyama T; Ohsawa K; Seto H Structure of 1-Deoxy-D-xylulose 5-phosphate Reductoisomerase in a Quaternary Complex with a Magnesium ion, NADPH and the Antimalarial Drug Fosmidomycin. Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun 2007, 63, 466–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (93).Henriksson LM; Unge T; Carlsson J; Åqvist J; Mowbray SL; Jones TA Structures of Mycobacterium tuberculosis 1-Deoxy-D-xylulose-5-phosphate Reductoisomerase Provide New Insights into Catalysis. J. Biol. Chem 2007, 282, 19905–19916. [DOI] [PubMed] [Google Scholar]
- (94).Kuzuyama TS, T.; Takahashi S; Seto H Fosmidomycin, a Specific Inhibitor of 1-Deoxy-d-xylulose 5-phosphate Reductoisomerase in the Nonmevalonate Pathway for Terpenoid Biosynthesis. Tetrahedron Lett 1998, 39, 7913–7916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).Haemers T; Wiesner J; Van Poecke S; Goeman J; Henschker D; Beck E; Jomaa H; Van Calenbergh S Synthesis of Alpha-substituted Fosmidomycin Analogues as Highly Potent Plasmodium falciparum Growth Inhibitors. Bioorg. Med. Chem. Lett 2006, 16, 1888–1891. [DOI] [PubMed] [Google Scholar]
- (96).Jomaa H; Wiesner J; Sanderbrand S; Altincicek B; Weidemeyer C; Hintz M; Turbachova I; Eberl M; Zeidler J; Lichtenthaler HK; Soldati D; Beck E Inhibitors of the Nonmevalonate Pathway of Isoprenoid Biosynthesis as Antimalarial Drugs. Science 1999, 285, 1573–1576. [DOI] [PubMed] [Google Scholar]
- (97).Borrmann S; Adegnika AA; Matsiegui PB; Issifou S; Schindler A; Mawili-Mboumba DP; Baranek T; Wiesner J; Jomaa H; Kremsner PG Fosmidomycin-clindamycin for Plasmodium falciparum Infections in African children. J. Infect. Dis 2004, 189, 901–908. [DOI] [PubMed] [Google Scholar]
- (98).Borrmann S; Lundgren I; Oyakhirome S; Impouma B; Matsiegui PB; Adegnika AA; Issifou S; Kun JF; Hutchinson D; Wiesner J; Jomaa H; Kremsner PG Fosmidomycin Plus Clindamycin for Treatment of Pediatric Patients Aged 1 to 14 Years with Plasmodium falciparum Malaria. Antimicrob. Agents Chemother 2006, 50, 2713–2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99).Borrmann S; Adegnika AA; Moussavou F; Oyakhirome S; Esser G; Matsiegui PB; Ramharter M; Lundgren I; Kombila M; Issifou S; Hutchinson D; Wiesner J; Jomaa H; Kremsner PG Short-course Regimens of Artesunate-Fosmidomycin in Treatment of Uncomplicated Plasmodium falciparum Malaria. Antimicrob. Agents. Chemother 2005, 49, 3749–3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (100).Oyakhirome S; Issifou S; Pongratz P; Barondi F; Ramharter M; Kun JF; Missinou MA; Lell B; Kremsner PG Randomized Controlled Trial of Fosmidomycin-clindamycin Versus Sulfadoxine-pyrimethamine in the Treatment of Plasmodium falciparum Malaria. Antimicrob. Agents Chemother 2007, 51, 1869–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (101).Deng L; Endo K; Kato M; Cheng G; Yajima S; Song Y Structures of 1-Deoxy-D-Xylulose-5-Phosphate Reductoisomerase/Lipophilic Phosphonate Complexes. ACS Med. Chem. Lett 2011, 2, 165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (102).Dhiman RK; Schaeffer ML; Bailey AM; Testa CA; Scherman H; Crick DC 1-Deoxy-D-xylulose 5-phosphate Reductoisomerase (IspC) from Mycobacterium tuberculosis: Towards Understanding Mycobacterial Resistance to Fosmidomycin. J. Bacteriol 2005, 187, 8395–8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (103).Brown AC; Parish T Dxr is Essential in Mycobacterium tuberculosis and Fosmidomycin Resistance is Due to a Lack of Uptake. BMC Microbiol. 2008, 8, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (104).Zingle C; Kuntz L; Tritsch D; Grosdemange-Billiard C; Rohmer M Isoprenoid Biosynthesis Via the Methylerythritol Phosphate Pathway: Structural Variations Around Phosphonate Anchor and Spacer of Fosmidomycin, a Potent Inhibitor of Deoxyxylulose Phosphate Reductoisomerase. J. Org. Chem 2010, 75, 3203–3207. [DOI] [PubMed] [Google Scholar]
- (105).Woo YH; Fernandes RPM; Proteau PJ Evaluation of Fosmidomycin Analogs as Inhibitors of the Synechocystis Sp. PCC6803 1-Deoxy-d-xylulose 5-phosphate Reductoisomerase. Bioorg. Med. Chem 2006, 14, 2375–2385. [DOI] [PubMed] [Google Scholar]
- (106).Yajima S; Hara K; Sanders JM; Yin F; Ohsawa K; Wiesner J; Jomaa H; Oldfield E Crystallographic Structures of Two Bisphosphonate:1-Deoxyxylulose-5-phosphate Reductoisomerase Complexes. J. Am. Chem. Soc 2004, 126, 10824–10825. [DOI] [PubMed] [Google Scholar]
- (107).Deng L; Sundriyal S; Rubio V; Shi ZZ; Song Y Coordination Chemistry Based Approach to Lipophilic Inhibitors of 1-deoxy-D-xylulose-5-phosphate Reductoisomerase. J. Med. Chem 2009, 52, 6539–6542. [DOI] [PubMed] [Google Scholar]
- (108).Andaloussi M; Lindh M; Bjorkelid C; Suresh S; Wieckowska A; Iyer H; Karlen A; Larhed M Substitution of the Phosphonic Acid and Hydroxamic Acid Functionalities of the DXR Inhibitor FR90098: an Attempt to Improve the Activity Against Mycobacterium tuberculosis. Bioorg. Med. Chem. Lett 2011, 21, 5403–5407. [DOI] [PubMed] [Google Scholar]
- (109).Xue J; Diao J; Cai G; Deng L; Zheng B; Yao Y; Song Y Antimalarial and Structural Studies of Pyridine-containing Inhibitors of 1-Deoxyxylulose-5-phosphate Reductoisomerase. ACS Med. Chem. Lett 2013, 4, 278–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (110).Behrendt CT; Kunfermann A; Illarionova V; Matheeussen A; Pein MK; Grawert T; Kaiser J; Bacher A; Eisenreich W; Illarionov B; Fischer M; Maes L; Groll M; Kurz T Reverse Fosmidomycin Derivatives Against the Antimalarial Drug Target IspC (Dxr). J. Med. Chem 2011, 54, 6796–6802. [DOI] [PubMed] [Google Scholar]
- (111).Brucher K; Illarionov B; Held J; Tschan S; Kunfermann A; Pein MK; Bacher A; Grawert T; Maes L; Mordmuller B; Fischer M; Kurz T Alpha-Substituted Beta-oxa Isosteres of Fosmidomycin: Synthesis and Biological evaluation. J. Med. Chem 2012, 55, 6566–6575. [DOI] [PubMed] [Google Scholar]
- (112).Kunfermann A; Lienau C; Illarionov B; Held J; Gräwert T; Behrendt CT; Werner P; Hähn S; Eisenreich W; Riederer U; Mordmüller B; Bacher A; Fischer M; Groll M; Kurz T IspC as Target for Antiinfective Drug Discovery: Synthesis, Enantiomeric Separation, and Structural Biology of Fosmidomycin Thia Isosters. J. Med. Chem 2013, 56, 8151–8162. [DOI] [PubMed] [Google Scholar]
- (113).Sooriyaarachchi S; Chofor R; Risseeuw MD; Bergfors T; Pouyez J; Dowd CS; Maes L; Wouters J; Jones TA; Van Calenbergh S; Mowbray SL Targeting an Aromatic Hotspot in Plasmodium falciparum 1-Deoxy-d-xylulose-5-phosphate Reductoisomerase with beta-Arylpropyl Analogues of Fosmidomycin. ChemMedChem 2016, 11, 2024–2036. [DOI] [PubMed] [Google Scholar]
- (114).Chofor R; Sooriyaarachchi S; Risseeuw MD; Bergfors T; Pouyez J; Johny C; Haymond A; Everaert A; Dowd CS; Maes L; Coenye T; Alex A; Couch RD; Jones TA; Wouters J; Mowbray SL; Van Calenbergh S Synthesis and Bioactivity of Beta-Substituted Fosmidomycin Analogues Targeting 1-Deoxy-D-xylulose-5-phosphate Reductoisomerase. J. Med. Chem 2015, 58, 2988–3001. [DOI] [PubMed] [Google Scholar]
- (115).Deng L; Diao J; Chen P; Pujari V; Yao Y; Cheng G; Crick DC; Prasad BVV; Song Y Inhibition of 1-Deoxy-d-Xylulose-5-Phosphate Reductoisomerase by Lipophilic Phosphonates: SAR, QSAR, and Crystallographic Studies. J. Med. Chem 2011, 54, 4721–4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (116).Zingle C; Tritsch D; Grosdemange-Billiard C; Rohmer M Catechol-rhodanine Derivatives: Specific and Promiscuous Inhibitors of Escherichia coli Deoxyxylulose Phosphate Reductoisomerase (DXR). Bioorg. Med. Chem 2014, 22, 3713–3719. [DOI] [PubMed] [Google Scholar]
- (117).San Jose G; Jackson ER; Haymond A; Johny C; Edwards RL; Wang X; Brothers RC; Edelstein EK; Odom AR; Boshoff HI; Couch RD; Dowd CS Structure-Activity Relationships of the MEPicides: N-Acyl and O-Linked Analogs of FR900098 as Inhibitors of Dxr from Mycobacterium tuberculosis and Yersinia pestis. ACS Infect. Dis 2016, 2, 923–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (118).Munier M; Tritsch D; Krebs F; Esque J; Hemmerlin A; Rohmer M; Stote RH; Grosdemange-Billiard C Synthesis and Biological Evaluation of Phosphate Isosters of Fosmidomycin and Analogs as Inhibitors of Escherichia coli and Mycobacterium smegmatis 1-Deoxyxylulose 5-phosphate Reductoisomerases. Bioorg. Med. Chem 2017, 25, 684–689. [DOI] [PubMed] [Google Scholar]
- (119).Chofor R; Risseeuw MD; Pouyez J; Johny C; Wouters J; Dowd CS; Couch RD; Van Calenbergh S Synthetic Fosmidomycin Analogues with Altered Chelating Moieties Do Not Inhibit 1-Deoxy-d-xylulose 5-phosphate Reductoisomerase or Plasmodium falciparum Growth in Vitro. Molecules 2014, 19, 2571–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (120).Singh BK; Shaner DL Biosynthesis of Branched Chain Amino Acids: From Test Tube to Field. Plant Cell 1995, 7, 935–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (121).Dumas R; Biou V; Halgand F; Douce R; Duggleby RG Enzymology, Structure, and Dynamics of Acetohydroxy Acid Isomeroreductase. Acc. Chem. Res 2001, 34, 399–408. [DOI] [PubMed] [Google Scholar]
- (122).McCourt JA; Duggleby RG Acetohydroxyacid Synthase and its Role in the Biosynthetic Pathway for Branched-Chain Amino Acids. Amino Acids 2006, 31, 173–210. [DOI] [PubMed] [Google Scholar]
- (123).Vejborg RM; de Evgrafov MR; Phan MD; Totsika M; Schembri MA; Hancock V Identification of Genes Important for Growth of Asymptomatic Bacteriuria Escherichia Coli in Urine. Infect. Immun 2012, 80, 3179–3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (124).Kim GL; Lee S; Luong TT; Nguyen CT; Park SS; Pyo S; Rhee DK Effect of Decreased BCAA Synthesis Through Disruption of ilvC Gene on the Virulence of Streptococcus Pneumoniae. Arch. Pharmacal Res 2017, 40, 921–932. [DOI] [PubMed] [Google Scholar]
- (125).Griffin JE; Gawronski JD; Dejesus MA; Ioerger TR; Akerley BJ; Sassetti CM High-Resolution Phenotypic Profiling Defines Genes Essential for Mycobacterial Growth and Cholesterol Catabolism. PLoS Pathog 2011, 7, e1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (126).Liu X; Wang J; Xu J; Shi J FgIlv5 is Required for Branched-Chain Amino Acid Biosynthesis and Full Virulence in Fusarium Graminearum. Microbiology 2014, 160, 692–702. [DOI] [PubMed] [Google Scholar]
- (127).Tadrowski S; Pedroso MM; Sieber V; Larrabee JA; Guddat LW; Schenk G Metal Ions Play an Essential Catalytic Role in the Mechanism of Ketol-Acid Reductoisomerase. Chem. - Eur. J 2016, 22, 7427–7436. [DOI] [PubMed] [Google Scholar]
- (128).Ahn HJ; Eom SJ; Yoon HJ; Lee BI; Cho H; Suh SW Crystal Structure of Class I Acetohydroxy Acid Isomeroreductase from Pseudomonas Aeruginosa. J. Mol. Biol 2003, 328, 505–515. [DOI] [PubMed] [Google Scholar]
- (129).Dumas R; Butikofer MC; Job D; Douce R Evidence for two Catalytically Different Magnesium-Binding Sites in Acetohydroxy Acid Isomeroreductase by Site-Directed Mutagenesis. Biochemistry 1995, 34, 6026–6036. [DOI] [PubMed] [Google Scholar]
- (130).Biou V; Dumas R; Cohen-Addad C; Douce R; Job D; Pebay-Peyroula E The Crystal Structure of Plant Acetohydroxy Acid Isomeroreductase Complexed with NADPH, Two Magnesium Ions and a Herbicidal Transition State Analog Determined at 1.65 A Resolution. EMBO J 1997, 16, 3405–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (131).Lv Y; Kandale A; Wun SJ; McGeary RP; Williams SJ; Kobe B; Sieber V; Schembri MA; Schenk G; Guddat LW Crystal Structure of Mycobacterium tuberculosis Ketol-acid Reductoisomerase at 1.0 A Resolution - a Potential Target for Anti-tuberculosis Drug Discovery. FEBS J 2016, 283, 1184–1196. [DOI] [PubMed] [Google Scholar]
- (132).Leung EW; Guddat LW Conformational Changes in a Plant Ketol-Acid Reductoisomerase upon Mg(2+) and NADPH Binding as Revealed by Two Crystal Structures. J. Mol. Biol 2009, 389, 167–182. [DOI] [PubMed] [Google Scholar]
- (133).Schulz A; Sponemann P; Kocher H; Wengenmayer F The Herbicidally Active Experimental Compound Hoe 704 is a Potent Inhibitor of the Enzyme Acetolactate Reductoisomerase. FEBS Lett 1988, 238, 375–378. [DOI] [PubMed] [Google Scholar]
- (134).Aulabaugh A; Schloss JV Oxalyl Hydroxamates as Reaction-Intermediate Analogues for Ketol-Acid Reductoisomerase. Biochemistry 1990, 29, 2824–2830. [DOI] [PubMed] [Google Scholar]
- (135).Gerwick BC; Mireles LC; Eilers RJ Rapid Diagnosis of ALS/AHAS-Resistant Weeds. Weed Technol 1993, 7, 519–524. [Google Scholar]
- (136).Liu XH; Chen PQ; Wang BL; Li YH; Wang SH; Li ZM Synthesis, Bioactivity, Theoretical and Molecular Docking Study of 1-Cyano-N-Substituted-Cyclopropanecarboxamide as Ketol-Acid Reductoisomerase Inhibitor. Bioorg. Med. Chem. Lett 2007, 17, 3784–3788. [DOI] [PubMed] [Google Scholar]
- (137).Liu XH; Pan L; Tan CX; Weng JQ; Wang BL; Li ZM Synthesis, Crystal Structure, Bioactivity and DFT Calculation of New Oxime Ester Derivatives Containing Cyclopropane Moiety. Pestic. Biochem. Physiol 2011, 101, 143–147. [Google Scholar]
- (138).Halgand F; Vives F; Dumas R; Biou V; Andersen J; Andrieu JP; Cantegril R; Gagnon J; Douce R; Forest E; Job D Kinetic and Mass Spectrometric Analyses of the Interactions Between Plant Acetohydroxy Acid Isomeroreductase and Thiadiazole Derivatives. Biochemistry 1998, 37, 4773–4781. [DOI] [PubMed] [Google Scholar]
- (139).Wang BL; Zhang LY; Liu XH; Ma Y; Zhang Y; Li ZM; Zhang X Synthesis, Biological Activities and SAR Studies of New 3-Substitutedphenyl-4-Substitutedbenzylideneamino-1,2,4-Triazole Mannich Bases and bis-Mannich Bases as Ketol-Acid Reductoisomerase Inhibitors. Bioorg. Med. Chem. Lett 2017, 27, 5457–5462. [DOI] [PubMed] [Google Scholar]
- (140).Zhang Y; Liu XH; Zhan YZ; Zhang LY; Li ZM; Li YH; Zhang X; Wang BL Synthesis and Biological Activities of Novel 5-Substituted-1,3,4-Oxadiazole Mannich Bases and bis-Mannich Bases as Ketol-Acid Reductoisomerase Inhibitors. Bioorg. Med. Chem. Lett 2016, 26, 4661–4665. [DOI] [PubMed] [Google Scholar]
- (141).Patel KM; Teran D; Zheng S; Kandale A; Garcia M; Lv Y; Schembri MA; McGeary RP; Schenk G; Guddat LW Crystal Structures of Staphylococcus Aureus Ketol-Acid Reductoisomerase in Complex with Two Transition State Analogues that Have Biocidal Activity. Chem. - Eur. J 2017. [DOI] [PubMed] [Google Scholar]
- (142).Gunsior M; Ravel J; Challis GL; Townsend CA Engineering p-Hydroxyphenylpyruvate Dioxygenase to a p-Hydroxymandelate Synthase and Evidence for the Proposed Benzene Oxide Intermediate in Homogentisate Formation. Biochemistry 2004, 43, 663–674. [DOI] [PubMed] [Google Scholar]
- (143).Moran GR 4-Hydroxyphenylpyruvate Dioxygenase. Arch. Biochem. Biophys 2005, 433, 117–128. [DOI] [PubMed] [Google Scholar]
- (144).Brownlee JM; Johnson-Winters K; Harrison DHT; Moran GR Structure of the Ferrous Form of (4-Hydroxyphenyl)pyruvate Dioxygenase from Streptomyces avermitilis in Complex with the Therapeutic Herbicide, NTBC. Biochemistry 2004, 43, 6370–6377. [DOI] [PubMed] [Google Scholar]
- (145).Crouch NP; Adlington RM; Baldwin JE; Lee MH; MacKinnon CH A Mechanistic Rationalisation for the Substrate Specificity of Recombinant Mammalian 4-Hydroxyphenylpyruvate Dioxygenase (4-HPPD). Tetrahedron 1997, 53, 6993–7010. [Google Scholar]
- (146).Munne-Bosch S; Alegre L The Function of Tocopherols and Tocotrienols in Plants. Crit. Rev. Plant Sci 2002, 21, 31–57. [Google Scholar]
- (147).Pallett KE; Cramp SM; Little JP; Veerasekaran P; Crudace AJ; Slater AE Isoxaflutole: The Background to its Discovery and the Basis of its Herbicidal Properties. Pest Manage. Sci 2001, 57, 133–142. [DOI] [PubMed] [Google Scholar]
- (148).Lindstedt S; Holme E; Lock EA; Hjalmarson O; Strandvik B Treatment of Hereditary Tyrosinaemia Type I by Inhibition of 4-Hydroxyphenylpyruvate Dioxygenase. Lancet 1992, 340, 813–817. [DOI] [PubMed] [Google Scholar]
- (149).Phornphutkul C; Introne WJ; Perry MB; Bernardini I; Murphey MD; Fitzpatrick DL; Anderson PD; Huizing M; Anikster Y; Gerber LH; Gahl WA Natural History of Alkaptonuria. N. Engl. J. Med 2002, 347, 2111–2121. [DOI] [PubMed] [Google Scholar]
- (150).Lock EA; Ellis MK; Gaskin P; Robinson M; Auton TR; Provan WM; Smith LL; Prisbylla MP; Mutter LC; Lee DL From Toxicological Problem to Therapeutic Use: The Discovery of the Mode of Action of 2-(2-nitro-4-trifluoromethylbenzoyl)-1,3-cyclohexanedione (NTBC), its Toxicology and Development as a Drug. J. Inherited Metab. Dis 1998, 21, 498–506. [DOI] [PubMed] [Google Scholar]
- (151).McKiernan PJ Nitisinone in the Treatment of Hereditary Tyrosinaemia Type 1. Drugs 2006, 66, 743–750. [DOI] [PubMed] [Google Scholar]
- (152).Serre L; Sailland A; Sy D; Boudec P; Rolland A; Pebay-Peyroula E; Cohen-Addad C Crystal Structure of Pseudomonas fluorescens 4-Hydroxyphenylpyruvate Dioxygenase: An Enzyme Involved in the Tyrosine Degradation Pathway. Structure 1999, 7, 977–988. [DOI] [PubMed] [Google Scholar]
- (153).Beaudegnies R; Edmunds AJF; Fraser TEM; Hall RG; Hawkes TR; Mitchell G; Schaetzer J; Wendeborn S; Wibley J Herbicidal 4-Hydroxyphenylpyruvate Dioxygenase Inhibitors-A Review of the Triketone Chemistry Story From a Syngenta Perspective. Bioorg. Med. Chem 2009, 17, 4134–4152. [DOI] [PubMed] [Google Scholar]
- (154).Mitchell G; Bartlett DW; Fraser TEM; Hawkes TR; Holt DC; Townson JK; Wichert RA Mesotrione: A New Selective Herbicide for Use in Maize. Pest Manage. Sci 2001, 57, 120–128. [DOI] [PubMed] [Google Scholar]
- (155).Romagni JG; Meazza G; Nanayakkara NPD; Dayan FE The Phytotoxic Lichen Metabolite, Usnic Acid, is a Potent Inhibitor of Plant p-Hydroxyphenylpyruvate Dioxygenase. FEBS Lett 2000, 480, 301–305. [DOI] [PubMed] [Google Scholar]
- (156).Lee DL; Prisbylla MP; Cromartie TH; Dagarin DP; Howard SW; Provan WM; Ellis MK; Fraser T; Mutter LC The Discovery and Structural Requirements of Inhibitors of p-Hydroxyphenylpyruvate Dioxygenase. Weed Sci 1997, 45, 601–609. [Google Scholar]
- (157). https://clinicaltrials.gov/ct2/show/NCT01390077?term=Nitisinone%2C+NTBC&rank=1.
- (158).Steinhilber D; Hofmann B Recent Advances in the Search for Novel 5-Lipoxygenase Inhibitors. Basic Clin. Pharmacol. Toxicol 2014, 114, 70–77. [DOI] [PubMed] [Google Scholar]
- (159).Samuelsson B; Rouzer CA; Matsumoto T Human Leukocyte 5-Lipoxygenase: An Enzyme Possessing Dual Enzymatic Activities and a Multicomponent Regulatory System. Adv. Prostaglandin, Thromboxane, Leukotriene Res. 1987, 17A, 1–11. [PubMed] [Google Scholar]
- (160).Bruno F; Spaziano G; Liparulo A; Roviezzo F; Nabavi SM; Sureda A; Filosa R; D’Agostino B Recent Advances in the Search For Novel 5-Lipoxygenase Inhibitors For the Treatment of Asthma. Eur. J. Med. Chem 2017. [DOI] [PubMed] [Google Scholar]
- (161).Radmark O Arachidonate 5-Lipoxygenase. J. Lipid Mediators Cell Signal 1995, 12, 171–184. [DOI] [PubMed] [Google Scholar]
- (162).Montuschi P Leukotrienes, Antileukotrienes and Asthma. Mini-Rev. Med. Chem 2008, 8, 647–656. [DOI] [PubMed] [Google Scholar]
- (163).Werz O; Steinhilber D Therapeutic Options for 5-Lipoxygenase Inhibitors. Pharmacol. Ther 2006, 112, 701–718. [DOI] [PubMed] [Google Scholar]
- (164).Zaman K; Hanigan MH; Smith A; Vaughan J; Macdonald T; Jones DR; Hunt JF; Gaston B Endogenous S-Nitrosoglutathione Modifies 5-Lipoxygenase Expression in Airway Epithelial Cells. Am. J. Respir. Cell Mol. Biol 2006, 34, 387–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (165).Funk CD Prostaglandins and Leukotrienes: Advances in Eicosanoid Biology. Science 2001, 294, 1871–1875. [DOI] [PubMed] [Google Scholar]
- (166).Chen Y; Hu Y; Zhang H; Peng C; Li S Loss of the Alox5 Gene Impairs Leukemia Stem Cells and Prevents Chronic Myeloid Leukemia. Nat. Genet 2009, 41, 783–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (167).Radmark O; Werz O; Steinhilber D; Samuelsson B 5-Lipoxygenase: Regulation of Expression and Enzyme Activity. Trends Biochem. Sci 2007, 32, 332–341. [DOI] [PubMed] [Google Scholar]
- (168).Gilbert NC; Bartlett SG; Waight MT; Neau DB; Boeglin WE; Brash AR; Newcomer ME The Structure of Human 5-Lipoxygenase. Science 2011, 331, 217–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (169).Radmark O Arachidonate 5-lipoxygenase. Prostaglandins Other Lipid Mediators 2002, 68–69, 211–234. [DOI] [PubMed] [Google Scholar]
- (170).Pergola C; Werz O 5-Lipoxygenase Inhibitors: A Review of Recent Developments and Patents. Expert. Opin. Ther. Pat 2010, 20, 355–375. [DOI] [PubMed] [Google Scholar]
- (171).Ford-Hutchinson AW; Gresser M; Young RN 5-Lipoxygenase. Annu. Rev. Biochem 1994, 63, 383–417. [DOI] [PubMed] [Google Scholar]
- (172).McMillan RM; Walker ER Designing Therapeutically Effective 5-Lipoxygenase Inhibitors. Trends Pharmacol. Sci 1992, 13, 323–330. [DOI] [PubMed] [Google Scholar]
- (173).Werz O 5-Lipoxygenase: Cellular Biology and Molecular Pharmacology. Curr. Drug Targets: Inflammation Allergy 2002, 1, 23–44. [DOI] [PubMed] [Google Scholar]
- (174).Werz O; Steinhilber D Development of 5-Lipoxygenase Inhibitors-Lessons From Cellular Enzyme Regulation. Biochem. Pharmacol 2005, 70, 327–333. [DOI] [PubMed] [Google Scholar]
- (175).Hussey HJ; Tisdale MJ Inhibition of Tumour Growth by Lipoxygenase Inhibitors. Br. J. Cancer 1996, 74, 683–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (176).Summers JB; Gunn BP; Martin JG; Martin MB; Mazdiyasni H; Stewart AO; Young PR; Bouska JB; Goetze AM; Dyer RD; Brooks DW; Carter GW Structure-Activity Analysis of a Class of Orally Active Hydroxamic Acid Inhibitors of Leukotriene Biosynthesis. J. Med. Chem 1988, 31, 1960–1964. [DOI] [PubMed] [Google Scholar]
- (177).Wenzel SE; Kamada AK Zileuton: The First 5-Lipoxygenase Inhibitor for the Treatment of Asthma. Ann. Pharmacother 1996, 30, 858–864. [DOI] [PubMed] [Google Scholar]
- (178).Hwang SH; Wecksler AT; Wagner K; Hammock BD Rationally Designed Multitarget Agents Against Inflammation and Pain. Curr. Med. Chem 2013, 20, 1783–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (179).Wasfi YS; Villaran C; de Tilleghem Cle B; Smugar SS; Hanley WD; Reiss TF; Knorr BA The Efficacy and Tolerability of MK-0633, a 5-Lipoxygenase Inhibitor, in Chronic Asthma. Respir. Med 2012, 106, 34–46. [DOI] [PubMed] [Google Scholar]
- (180).Bernstein JA; Liu N; Knorr BA; Smugar SS; Hanley WD; Reiss TF; Greenberg S MK-0633, A Potent 5-Lipoxygenase Inhibitor in Chronic Obstructive Pulmonary Disease. Respir. Med 2011, 105, 392–401. [DOI] [PubMed] [Google Scholar]
- (181).Chalasani N; Vuppalanchi R; Navarro V; Fontana R; Bonkovsky H; Barnhart H; Kleiner DE; Hoofnagle JH; Network D-ILI Acute Liver Injury due to Flavocoxid (Limbrel), a Medical Food for Osteoarthritis. Ann. Intern. Med 2012, 156, 857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (182).Deschamps JD; Kenyon VA; Holman TR Baicalein is a Potent In Vitro Inhibitor Against Both Reticulocyte 15-Human and Platelet 12-Human Lipoxygenases. Bioorg. Med. Chem. Lett 2006, 14, 4295–4301. [DOI] [PubMed] [Google Scholar]
- (183).Sekiya K; Okuda H; Arichi S Selective Inhibition of Platelet Lipoxygenase by Esculetin. Biochim. Biophys. Acta 1982, 713, 68–72. [PubMed] [Google Scholar]
- (184).Bishayee K; Khuda-Bukhsh AR 5-Lipoxygenase Antagonist Therapy: A New Approach Towards Targeted Cancer Chemotherapy. Acta Biochim. Biophys. Sin 2013, 45, 709–719. [DOI] [PubMed] [Google Scholar]
- (185).Cho H; Ueda M; Tamaoka M; Hamaguchi M; Aisaka K; Kiso Y; Inoue T; Ogino R; Tatsuoka T; Ishihara T; et al. Novel Caffeic Acid Derivatives: Extremely Potent Inhibitors of 12-Lipoxygenase. J. Med. Chem 1991, 34, 1503–1505. [DOI] [PubMed] [Google Scholar]
- (186).Pergola C; Jazzar B; Rossi A; Buehring U; Luderer S; Dehm F; Northoff H; Sautebin L; Werz O Cinnamyl-3,4-Dihydroxy-alpha-Cyanocinnamate Is a Potent Inhibitor of 5-Lipoxygenase. J. Pharmacol. Exp. Ther 2011, 338, 205–213. [DOI] [PubMed] [Google Scholar]
- (187).Hofmann B; Steinhilber D 5-Lipoxygenase Inhibitors: A Review of Recent Patents (2010–2012). Expert Opin. Ther. Pat 2013, 23, 895–909. [DOI] [PubMed] [Google Scholar]
- (188).Farjadmand F; Arshadi H; Moghimi S; Nadri H; Moradi A; Eghtedari M; Jafarpour F; Mahdavi M; Shafiee A; Foroumadi A Synthesis and Evaluation of Novel Quinazolinone-1,2,3-Triazoles as Inhibitors of Lipoxygenase. J. Chem. Res 2016, 188–191. [Google Scholar]
- (189).Chowdhury MA; Abdellatif KRA; Dong Y; Das D; Suresh MR; Knaus EE Synthesis of Celecoxib Analogues Possessing a N-Difluoromethyl-1,2-dihydropyrid-2-one 5-Lipoxygenase Pharmacophore: Biological Evaluation as Dual Inhibitors of Cyclooxygenases and 5-Lipoxygenase with Anti-Inflammatory Activity. J. Med. Chem 2009, 52, 1525–1529. [DOI] [PubMed] [Google Scholar]
- (190).Chowdhury MA; Abdellatif KRA; Dong Y; Das D; Suresh MR; Knaus EE Synthesis of Celecoxib Analogs that Possess a N-hydroxypyrid-2(1H)one 5-Lipoxygenase Pharmacophore: Biological Evaluation as Dual Inhibitors of Cyclooxygenases and 5-Lipoxygenase with Anti-Inflammatory Activity. Bioorg. Med. Chem. Lett 2008, 18, 6138–6141. [DOI] [PubMed] [Google Scholar]
- (191).Suganuma T; Workman JL Signals and Combinatorial Functions of Histone Modifications. Annu. Rev. Biochem 2011, 80, 473–499. [DOI] [PubMed] [Google Scholar]
- (192).Zentner GE; Henikoff S Regulation of Nucleosome Dynamics by Histone Modifications. Nat. Struct. Mol. Biol 2013, 20, 259–266. [DOI] [PubMed] [Google Scholar]
- (193).Duan GY; Walther D The Roles of Post-translational Modifications in the Context of Protein Interaction Networks. PLoS Comput. Biol 2015, 11, e1004049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (194).Berdasco M; Esteller M Aberrant Epigenetic Landscape in Cancer: How Cellular Identity Goes Awry. Dev. Cell 2010, 19, 698–711. [DOI] [PubMed] [Google Scholar]
- (195).Greer EL; Shi Y Histone Methylation: a Dynamic Mark in Health, Disease and Inheritance. Nat. Rev. Genet 2012, 13, 343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (196).Iwase S; Shi Y Histone and DNA Modifications in Mental Retardation. Prog. Drug Res 2011, 67, 147–173. [DOI] [PubMed] [Google Scholar]
- (197).Chi P; Allis CD; Wang GG Covalent Histone Modifications - Miswritten, Misinterpreted and Mis-erased in Human Cancers. Nat. Rev. Cancer 2010, 10, 457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (198).Albert M; Helin K Histone Methyltransferases in Cancer. Semin. Cell Dev. Biol 2010, 21, 209–220. [DOI] [PubMed] [Google Scholar]
- (199).Momparler RL; Cote S; Momparler LF; Idaghdour Y Inhibition of DNA and Histone Methylation by 5-Aza-2’-Deoxycytidine (Decitabine) and 3-Deazaneplanocin-A on Antineoplastic Action and Gene Expression in Myeloid Leukemic Cells. Front. Oncol 2017, 7, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (200).Momparler RL; Cote S; Momparler LF; Idaghdour Y Epigenetic Therapy of Acute Myeloid Leukemia Using 5-Aza-2’-deoxycytidine (Decitabine) in Combination with Inhibitors of Histone Methylation and Deacetylation. Clin. Epigenet 2014, 6, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (201).Shi Y; Lan F; Matson C; Mulligan P; Whetstine JR; Cole PA; Casero RA; Shi Y Histone Demethylation Mediated by the Nuclear Amine Oxidase Homolog LSD1. Cell 2004, 119, 941–953. [DOI] [PubMed] [Google Scholar]
- (202).Pedersen MT; Helin K Histone Demethylases in Development and Disease. Trends Cell Biol 2010, 20, 662–671. [DOI] [PubMed] [Google Scholar]
- (203).Cloos PAC; Christensen J; Agger K; Helin K Erasing the Methyl Mark: Histone Demethylases at the Center of Cellular Differentiation and Disease. Genes Dev 2008, 22, 1115–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (204).Rotili D; Mai A Targeting Histone Demethylases: A New Avenue for the Fight against Cancer. Genes Cancer 2011, 2, 663–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (205).Johansson C; Tumber A; Che K; Cain P; Nowak R; Gileadi C; Oppermann U The Roles of Jumonji-type Oxygenases in Human Disease. Epigenomics 2014, 6, 89–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (206).Walport LJ; Hopkinson RJ; Schofield CJ Mechanisms of Human Histone and Nucleic Acid Demethylases. Curr. Opin. Chem. Biol 2012, 16, 525–534. [DOI] [PubMed] [Google Scholar]
- (207).Shi Y Histone Lysine Demethylases: Emerging Roles in Development, Physiology and Disease. Nat. Rev. Genet 2007, 8, 829–833. [DOI] [PubMed] [Google Scholar]
- (208).Fueyo R; Garcia MA; Martinez-Balbas MA Jumonji Family Histone Demethylases in Neural Development. Cell Tissue Res 2015, 359, 87–98. [DOI] [PubMed] [Google Scholar]
- (209).Markolovic S; Leissing TM; Chowdhury R; Wilkins SE; Lu X; Schofield CJ Structure-function Relationships of Human JmjC Oxygenases-demethylases Versus Hydroxylases. Curr. Opin. Struct. Biol 2016, 41, 62–72. [DOI] [PubMed] [Google Scholar]
- (210).Yeh TL; Leissing TM; Abboud MI; Thinnes CC; Atasoylu O; Holt-Martyn JP; Zhang D; Tumber A; Lippl K; Lohans CT; Leung IKH; Morcrette H; Clifton IJ; Claridge TDW; Kawamura A; Flashman E; Lu X; Ratcliffe PJ; Chowdhury R; Pugh CW; Schofield CJ Molecular and Cellular Mechanisms of HIF prolyl Hydroxylase Inhibitors in Clinical Trials. Chem. Sci 2017, 8, 7651–7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (211).Pilka ES; James T; Lisztwan JH Structural Definitions of Jumonji Family Demethylase Selectivity. Drug Discovery Today 2015, 20, 743–749. [DOI] [PubMed] [Google Scholar]
- (212).Aik W; McDonough MA; Thalhammer A; Chowdhury R; Schofield CJ Role of the jelly-roll Fold in Substrate Binding by 2-Oxoglutarate Oxygenases. Curr. Opin. Struct. Biol 2012, 22, 691–700. [DOI] [PubMed] [Google Scholar]
- (213).Clifton IJ; McDonough MA; Ehrismann D; Kershaw NJ; Granatino N; Schofield CJ Structural Studies on 2-Oxoglutarate Oxygenases and Related Double-stranded beta-helix Fold Proteins. J. Inorg. Biochem 2006, 100, 644–669. [DOI] [PubMed] [Google Scholar]
- (214).Krishnan S; Trievel RC Structural and Functional Analysis of JMJD2D Reveals Molecular Basis for Site-specific Demethylation Among JMJD2 Demethylases. Structure 2013, 21, 98–108. [DOI] [PubMed] [Google Scholar]
- (215).Hoffmann I; Roatsch M; Schmitt ML; Carlino L; Pippel M; Sippl W; Jung M The Role of Histone Demethylases in Cancer Therapy. Mol. Oncol 2012, 6, 683–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (216).Thinnes CC; England KS; Kawamura A; Chowdhury R; Schofield CJ; Hopkinson RJ Targeting Histone Lysine Demethylases — Progress, Challenges, and the Future. Biochim. Biophys. Acta 2014, 1839, 1416–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (217).Rose NR; Ng SS; Mecinovic J; Lienard BM; Bello SH; Sun Z; McDonough MA; Oppermann U; Schofield CJ Inhibitor Scaffolds for 2-Oxoglutarate-dependent Histone Lysine Demethylases. J. Med. Chem 2008, 51, 7053–7056. [DOI] [PubMed] [Google Scholar]
- (218).Cunliffe CJ; Franklin TJ; Hales NJ; Hill GB Novel Inhibitors of Prolyl 4-Hydroxylase 3. Inhibition by the Substrate-Analog N-Oxaloglycine and Its Derivatives. J. Med. Chem 1992, 35, 2652–2658. [DOI] [PubMed] [Google Scholar]
- (219).Hopkinson RJ; Tumber A; Yapp C; Chowdhury R; Aik W; Che KH; Li XS; Kristensen JBL; King ONF; Chan MC; Yeoh KK; Choi H; Walport LJ; Thinnes CC; Bush JT; Lejeune C; Rydzik AM; Rose NR; Bagg EA; McDonough MA; Krojer TJ; Yue WW; Ng SS; Olsen L; Brennan PE; Oppermann U; Muller S; Klose RJ; Ratcliffe PJ; Schofield CJ; Kawamura A 5-Carboxy-8-hydroxyquinoline is a Broad Spectrum 2-oxoglutarate Oxygenase Inhibitor which Causes Iron Translocation. Chem. Sci 2013, 4, 3110–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (220).Tian YM; Yeoh KK; Lee MK; Eriksson T; Kessler BM; Kramer HB; Edelmann MJ; Willam C; Pugh CW; Schofield CJ; Ratcliffe PJ Differential Sensitivity of Hypoxia Inducible Factor Hydroxylation Sites to Hypoxia and Hydroxylase Inhibitors. J. Biol. Chem 2011, 286, 13041–13051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (221).Ivan M; Haberberger T; Gervasi DC; Michelson KS; Gunzler V; Kondo K; Yang H; Sorokina I; Conaway RC; Conaway JW; Kaelin WG Jr. Biochemical Purification and Pharmacological Inhibition of a Mammalian Prolyl Hydroxylase Acting on Hypoxia-inducible Factor. Proc. Natl. Acad. Sci. U. S. A 2002, 99, 13459–13464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (222).McDonough MA; McNeill LA; Tilliet M; Papamicael CA; Chen QY; Banerji B; Hewitson KS; Schofield CJ Selective Inhibition of Factor Inhibiting Hypoxia-inducible Factor. J. Am. Chem. Soc 2005, 127, 7680–7681. [DOI] [PubMed] [Google Scholar]
- (223).Hamada S; Kim TD; Suzuki T; Itoh Y; Tsumoto H; Nakagawa H; Janknecht R; Miyata N Synthesis and Activity of N-Oxalylglycine and its Derivatives as Jumonji C-Domain-containing Histone Lysine Demethylase Inhibitors. Bioorg. Med. Chem. Lett 2009, 19, 2852–2855. [DOI] [PubMed] [Google Scholar]
- (224).Rose NR; Woon EC; Kingham GL; King ON; Mecinovic J; Clifton IJ; Ng SS; Talib-Hardy J; Oppermann U; McDonough MA; Schofield CJ Selective Inhibitors of the JMJD2 Histone Demethylases: Combined Nondenaturing Mass Spectrometric Screening and Crystallographic Approaches. J. Med. Chem 2010, 53, 1810–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (225).Rose NR; McDonough MA; King ON; Kawamura A; Schofield CJ Inhibition of 2-Oxoglutarate Dependent Oxygenases. Chem. Soc. Rev 2011, 40, 4364–4397. [DOI] [PubMed] [Google Scholar]
- (226).Ng SS; Kavanagh KL; McDonough MA; Butler D; Pilka ES; Lienard BM; Bray JE; Savitsky P; Gileadi O; von Delft F; Rose NR; Offer J; Scheinost JC; Borowski T; Sundstrom M; Schofield CJ; Oppermann U Crystal Structures of Histone Demethylase JMJD2A Reveal Basis for Substrate Specificity. Nature 2007, 448, 87–91. [DOI] [PubMed] [Google Scholar]
- (227).Tarhonskaya H; Nowak RP; Johansson C; Szykowska A; Tumber A; Hancock RL; Lang P; Flashman E; Oppermann U; Schofield CJ; Kawamura A Studies on the Interaction of the Histone Demethylase KDM5B with Tricarboxylic Acid Cycle Intermediates. J. Mol. Biol 2017, 429, 2895–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (228).Marks PA; Breslow R Dimethyl Sulfoxide to Vorinostat: Development of this Histone Deacetylase Inhibitor as an Anticancer Drug. Nat. Biotechnol 2007, 25, 84–90. [DOI] [PubMed] [Google Scholar]
- (229).Hamada S; Suzuki T; Mino K; Koseki K; Oehme F; Flamme I; Ozasa H; Itoh Y; Ogasawara D; Komaarashi H; Kato A; Tsumoto H; Nakagawa H; Hasegawa M; Sasaki R; Mizukami T; Miyata N Design, Synthesis, Enzyme-inhibitory Activity, and Effect on Human Cancer Cells of a Novel Series of Jumonji Domain-containing Protein 2 Histone Demethylase Inhibitors. J. Med. Chem 2010, 53, 5629–5638. [DOI] [PubMed] [Google Scholar]
- (230).Suzuki T; Ozasa H; Itoh Y; Zhan P; Sawada H; Mino K; Walport L; Ohkubo R; Kawamura A; Yonezawa M; Tsukada Y; Tumber A; Nakagawa H; Hasegawa M; Sasaki R; Mizukami T; Schofield CJ; Miyata N Identification of the KDM2/7 Histone Lysine Demethylase Subfamily Inhibitor and Its Antiproliferative Activity. J. Med. Chem 2013, 56, 7222–7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (231).Rose NR; Woon EC; Tumber A; Walport LJ; Chowdhury R; Li XS; King ON; Lejeune C; Ng SS; Krojer T; Chan MC; Rydzik AM; Hopkinson RJ; Che KH; Daniel M; Strain-Damerell C; Gileadi C; Kochan G; Leung IK; Dunford J; Yeoh KK; Ratcliffe PJ; Burgess-Brown N; von Delft F; Muller S; Marsden B; Brennan PE; McDonough MA; Oppermann U; Klose RJ; Schofield CJ; Kawamura A Plant Growth Regulator Daminozide is a Selective Inhibitor of Human KDM2/7 Histone Demethylases. J. Med. Chem 2012, 55, 6639–6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (232).Luo X; Liu Y; Kubicek S; Myllyharju J; Tumber A; Ng S; Che KH; Podoll J; Heightman TD; Oppermann U; Schreiber SL; Wang X A Selective Inhibitor and Probe of the Cellular Functions of Jumonji C Domain-containing Histone Demethylases. J. Am. Chem. Soc 2011, 133, 9451–9456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (233).King ON; Li XS; Sakurai M; Kawamura A; Rose NR; Ng SS; Quinn AM; Rai G; Mott BT; Beswick P; Klose RJ; Oppermann U; Jadhav A; Heightman TD; Maloney DJ; Schofield CJ; Simeonov A Quantitative High-throughput Screening Identifies 8-Hydroxyquinolines as Cell-active Histone Demethylase Inhibitors. PLoS One 2010, 5, e15535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (234).Thalhammer A; Mecinovic J; Loenarz C; Tumber A; Rose NR; Heightman TD; Schofield CJ Inhibition of the Histone Demethylase JMJD2E by 3-Substituted Pyridine 2,4-Dicarboxylates. Org. Biomol. Chem 2011, 9, 127–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (235).Liang Y; Vogel JL; Arbuckle JH; Rai G; Jadhav A; Simeonov A; Maloney DJ; Kristie TM Targeting the JMJD2 Histone Demethylases to Epigenetically Control Herpesvirus Infection and Reactivation from Latency. Sci. Transl. Med 2013, 5, 167ra165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (236).Rotili D; Altun M; Kawamura A; Wolf A; Fischer R; Leung IKH; Mackeen MM; Tian YM; Ratcliffe PJ; Mai A; Kessler BM; Schofield CJ A Photoreactive Small-Molecule Probe for 2-Oxoglutarate Oxygenases. Chem. Biol 2011, 18, 642–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (237).Kruidenier L; Chung CW; Cheng ZJ; Liddle J; Che KH; Joberty G; Bantscheff M; Bountra C; Bridges A; Diallo H; Eberhard D; Hutchinson S; Jones E; Katso R; Leveridge M; Mander PK; Mosley J; Ramirez-Molina C; Rowland P; Schofield CJ; Sheppard RJ; Smith JE; Swales C; Tanner R; Thomas P; Tumber A; Drewes G; Oppermann U; Patel DJ; Lee K; Wilson DM A Selective Jumonji H3K27 Demethylase Inhibitor Modulates the Proinflammatory Macrophage Response. Nature 2012, 488, 404–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (238).Heinemann B; Nielsen JM; Hudlebusch HR; Lees MJ; Larsen DV; Boesen T; Labelle M; Gerlach LO; Birk P; Helin K Inhibition of Demethylases by GSK-J1/J4. Nature 2014, 514, E1–2. [DOI] [PubMed] [Google Scholar]
- (239).Hashizume R; Andor N; Ihara Y; Lerner R; Gan HY; Chen XY; Fang D; Huang X; Tom MW; Ngo V; Solomon D; Mueller S; Paris PL; Zhang ZG; Petritsch C; Gupta N; Waldman TA; James CD Pharmacologic Inhibition of Histone Demethylation as a Therapy for Pediatric Brainstem Glioma. Nat. Med. (N. Y., NY, U. S.) 2014, 20, 1394–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (240).Burchfield JS; Li QT; Wang HY; Wang RF JMJD3 as an Epigenetic Regulator in Development and Disease. Int. J. Biochem. Cell Biol 2015, 67, 148–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (241).Sakaki H; Okada M; Kuramoto K; Takeda H; Watarai H; Suzuki S; Seino S; Seino M; Ohta T; Nagase S; Kurachi H; Kitanaka C GSKJ4, A Selective Jumonji H3K27 Demethylase Inhibitor, Effectively Targets Ovarian Cancer Stem Cells. Anticancer Res 2015, 35, 6607–6614. [PubMed] [Google Scholar]
- (242).Brenner M; Hearing VJ The Protective Role of Melanin Against UV Damage in Human Skin. Photochem. Photobiol 2008, 84, 539–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (243).Buitrago E; Hardre R; Haudecoeur R; Jamet H; Belle C; Boumendjel A; Bubacco L; Reglier M Are Human Tyrosinase and Related Proteins Suitable Targets for Melanoma Therapy? Curr. Top. Med. Chem 2016, 16, 3033–3047. [DOI] [PubMed] [Google Scholar]
- (244).Slominski AT; Carlson JA Melanoma Resistance: A Bright Future for Academicians and a Challenge for Patient Advocates. Mayo Clin. Proc 2014, 89, 429–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (245).Brozyna AA; Jozwicki W; Carlson JA; Slominski AT Melanogenesis Affects Overall and Disease-Free Survival in Patients with Stage III and IV Melanoma. Hum. Pathol 2013, 44, 2071–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (246).Brozyna AA; VanMiddlesworth L; Slominski AT Inhibition of Melanogenesis as a Radiation Sensitizer for Melanoma Therapy. Int. J. Cancer 2008, 123, 1448–1456. [DOI] [PubMed] [Google Scholar]
- (247).Slominski A; Zbytek B; Slominski R Inhibitors of Melanogenesis Increase Toxicity of Cyclophosphamide and Lymphocytes Against Melanoma Cells. Int. J. Cancer 2009, 124, 1470–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (248).Bahadoran P; Levine N; Burk C; Protein M; Nordlund JJ; Groysman V; Bangert S; Fulk CS; Rubenzik R; Urabe K; Nakayama J; Hori Y; Lamerson CL; Breneman DL In The Pigmentary System; Blackwell Publishing Ltd: 2007, p 907–978. [Google Scholar]
- (249).Unver N; Freyschmidt-Paul P; Horster S; Wenck H; Stab F; Blatt T; Elsasser HP Alterations in the Epidermal-Dermal Melanin Axis and Factor XIIIa Melanophages in Senile Lentigo and Ageing Skin. Br. J. Dermatol 2006, 155, 119–128. [DOI] [PubMed] [Google Scholar]
- (250).Hearing VJ; Jimenez M Mammalian Tyrosinase-the Critical Regulatory Control Point in Melanocyte Pigmentation. Int. J. Biochem 1987, 19, 1141–1147. [DOI] [PubMed] [Google Scholar]
- (251).Sanchez-Ferrer A; Rodriguez-Lopez JN; Garcia-Canovas F; Garcia-Carmona F Tyrosinase: a Comprehensive Review of its Mechanism. Biochim. Biophys. Acta 1995, 1247, 1–11. [DOI] [PubMed] [Google Scholar]
- (252).Halaban R; Patton RS; Cheng E; Svedine S; Trombetta ES; Wahl ML; Ariyan S; Hebert DN Abnormal Acidification of Melanoma Cells Induces Tyrosinase Retention in the Early Secretory Pathway. J. Biol. Chem 2002, 277, 14821–14828. [DOI] [PubMed] [Google Scholar]
- (253).Fogal S; Carotti M; Giaretta L; Lanciai F; Nogara L; Bubacco L; Bergantino E Human Tyrosinase Produced in Insect Cells: a Landmark for the Screening of New Drugs Addressing its Activity. Mol. Biotechnol 2015, 57, 45–57. [DOI] [PubMed] [Google Scholar]
- (254).Haudecoeur R; Gouron A; Dubois C; Jamet H; Lightbody M; Hardre R; Milet A; Bergantino E; Bubacco L; Belle C; Reglier M; Boumendjel A Investigation of Binding-Site Homology Between Mushroom and Bacterial Tyrosinases by Using Aurones as Effectors. ChemBioChem 2014, 15, 1325–1333. [DOI] [PubMed] [Google Scholar]
- (255).Nishioka K Particulate Tyrosinase of Human Malignant Melanoma. Solubilization, Purification Following Trypsin Treatment, and Characterization. Eur. J. Biochem 1978, 85, 137–146. [DOI] [PubMed] [Google Scholar]
- (256).Kwon BS; Haq AK; Pomerantz SH; Halaban R Isolation and Sequence of a cDNA Clone for Human Tyrosinase that Maps at the Mouse c-Albino Locus. Proc. Natl. Acad. Sci. U. S. A 1987, 84, 7473–7477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (257).Lai X; Soler-Lopez M; Wichers HJ; Dijkstra BW Large-Scale Recombinant Expression and Purification of Human Tyrosinase Suitable for Structural Studies. PLoS One 2016, 11, e0161697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (258).Ismaya WT; Rozeboom HJ; Weijn A; Mes JJ; Fusetti F; Wichers HJ; Dijkstra BW Crystal Structure of Agaricus Bisporus Mushroom Tyrosinase: Identity of the Tetramer Subunits and Interaction with Tropolone. Biochemistry 2011, 50, 5477–5486. [DOI] [PubMed] [Google Scholar]
- (259).VanGelder CWG; Flurkey WH; Wichers HJ Sequence and Structural Features of Plant and Fungal Tyrosinases. Phytochemistry 1997, 45, 1309–1323. [DOI] [PubMed] [Google Scholar]
- (260).Kim YJ; Uyama H Tyrosinase Inhibitors from Natural and Synthetic Sources: Structure, Inhibition Mechanism and Perspective for the Future. Cell. Mol. Life Sci 2005, 62, 1707–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (261).Lee SY; Baek N; Nam TG Natural, Semisynthetic and Synthetic Tyrosinase Inhibitors. J. Enzyme Inhib. Med. Chem 2016, 31, 1–13. [DOI] [PubMed] [Google Scholar]
- (262).Pillaiyar T; Manickam M; Namasivayam V Skin Whitening Agents: Medicinal Chemistry Perspective of Tyrosinase Inhibitors. J. Enzyme Inhib. Med. Chem 2017, 32, 403–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (263).Kubo I; Kinst-Hori I; Chaudhuri SK; Kubo Y; Sanchez Y; Ogura T Flavonols from Heterotheca Inuloides: Tyrosinase Inhibitory Activity and Structural Criteria. Bioorg. Med. Chem 2000, 8, 1749–1755. [DOI] [PubMed] [Google Scholar]
- (264).Fisk WA; Agbai O; Lev-Tov HA; Sivamani RK The Use of Botanically Derived Agents for Hyperpigmentation: a Systematic Review. J. Am. Acad. Dermatol 2014, 70, 352–365. [DOI] [PubMed] [Google Scholar]
- (265).Deri B; Kanteev M; Goldfeder M; Lecina D; Guallar V; Adir N; Fishman A The Unravelling of the Complex Pattern of Tyrosinase Inhibition. Sci. Rep 2016, 6, 34993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (266).Sendovski M; Kanteev M; Ben-Yosef VS; Adir N; Fishman A First Structures of an Active Bacterial Tyrosinase Reveal Copper Plasticity. J. Mol. Biol 2011, 405, 227–237. [DOI] [PubMed] [Google Scholar]
- (267).Battaini G; Monzani E; Casella L; Santagostini L; Pagliarin R Inhibition of the Catecholase Activity of Biomimetic Dinuclear Copper Complexes by Kojic Acid. JBIC, J. Biol. Inorg. Chem 2000, 5, 262–268. [DOI] [PubMed] [Google Scholar]
- (268).Kolbe L; Mann T; Gerwat W; Batzer J; Ahlheit S; Scherner C; Wenck H; Stab F 4-n-Butylresorcinol, a Highly Effective Tyrosinase Inhibitor for the Topical Treatment of Hyperpigmentation. J. Eur. Acad. Dermatol. Venereol 2013, 27 Suppl 1, 19–23. [DOI] [PubMed] [Google Scholar]
- (269).Engasser PG Ochronosis Caused by Bleaching Creams. J. Am. Acad. Dermatol 1984, 10, 1072–1073. [DOI] [PubMed] [Google Scholar]
- (270).Yoshimori A; Oyama T; Takahashi S; Abe H; Kamiya T; Abe T; Tanuma S Structure-Activity Relationships of the Thujaplicins for Inhibition of Human Tyrosinase. Bioorg. Med. Chem 2014, 22, 6193–6200. [DOI] [PubMed] [Google Scholar]
- (271).Okombi S; Rival D; Bonnet S; Mariotte AM; Perrier E; Boumendjel A Discovery of Benzylidenebenzofuran-3(2H)-one (Aurones) as Inhibitors of Tyrosinase Derived from Human Melanocytes. J. Med. Chem 2006, 49, 329–333. [DOI] [PubMed] [Google Scholar]
- (272).Haudecoeur R; Carotti M; Gouron A; Maresca M; Buitrago E; Hardre R; Bergantino E; Jamet H; Belle C; Reglier M; Bubacco L; Boumendjel A 2-Hydroxypyridine-N-oxide-Embedded Aurones as Potent Human Tyrosinase Inhibitors. ACS Med. Chem. Lett 2017, 8, 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (273).Draelos ZD Skin Lightening Preparations and the Hydroquinone Controversy. Dermatol. Ther 2007, 20, 308–313. [DOI] [PubMed] [Google Scholar]
- (274).Lee A; Gilbert RM Epidemiology of Parkinson Disease. Neurol. Clin 2016, 34, 955–965. [DOI] [PubMed] [Google Scholar]
- (275).Ascherio A; Schwarzschild MA The Epidemiology of Parkinson’s Disease: Risk Factors and Prevention. Lancet Neurol 2016, 15, 1257–1272. [DOI] [PubMed] [Google Scholar]
- (276).Apud JA; Weinberger DR Treatment of Cognitive Deficits Associated with Schizophrenia: Potential Role of Catechol-O-methyltransferase Inhibitors. CNS Drugs 2007, 21, 535–557. [DOI] [PubMed] [Google Scholar]
- (277).Riederer P; Gerlach M; Muller T; Reichmann H Relating Mode of Action to Clinical Practice: Dopaminergic Agents in Parkinson’s Disease. Parkinsonism Relat. Disord 2007, 13, 466–479. [DOI] [PubMed] [Google Scholar]
- (278).Calne DB Treatment of Parkinson’s Disease. N. Engl. J. Med 1993, 329, 1021–1027. [DOI] [PubMed] [Google Scholar]
- (279).Fava M; Rosenbaum JF; Kolsky AR; Alpert JE; Nierenberg AA; Spillmann M; Moore C; Renshaw P; Bottiglieri T; Moroz G; Magni G Open Study of the Catechol-O-methyltransferase Inhibitor Tolcapone in Major Depressive Disorder. J. Clin. Psychopharmacol 1999, 19, 329–335. [DOI] [PubMed] [Google Scholar]
- (280).Mannisto PT; Kaakkola S Catechol-O-methyltransferase (COMT): Biochemistry, Molecular Biology, Pharmacology, and Clinical Efficacy of the New Selective COMT Inhibitors. Pharmacol. Rev 1999, 51, 593–628. [PubMed] [Google Scholar]
- (281).Muller T Catechol-O-methyltransferase Inhibitors in Parkinson’s Disease. Drugs 2015, 75, 157–174. [DOI] [PubMed] [Google Scholar]
- (282).Kiss LE; Soares-da-Silva P Medicinal Chemistry of Catechol O-methyltransferase (COMT) Inhibitors and Their Therapeutic Utility. J. Med. Chem 2014, 57, 8692–8717. [DOI] [PubMed] [Google Scholar]
- (283).Szegedi A; Rujescu D; Tadic A; Müller MJ; Kohnen R; Stassen HH; Dahmen N The Catechol-O-methyltransferase Val108/158Met Polymorphism Affects Short-term treatment Response to Mirtazapine, But Not to Paroxetine in Major Depression. Pharmacogenomics J. 2004, 5, 49. [DOI] [PubMed] [Google Scholar]
- (284).Byrne JM; Tipton KF Nitrocatechol Derivatives as Inhibitors of Catechol-O-Methyltransferase. Biochem. Soc. Trans 1996, 24, 64S. [DOI] [PubMed] [Google Scholar]
- (285).Perez RA; Fernandez-Alvarez E; Nieto O; Piedrafita FJ Inhibition of Catechol-O-methyltransferase by 1-Vinyl Derivatives of Nitrocatechols and Nitroguaiacols. Kinetics of the Irreversible Inhibition by 3-(3-Hydroxy-4-methoxy-5-nitro benzylidene)-2,4-pentanedione. Biochem. Pharmacol 1993, 45, 1973–1981. [DOI] [PubMed] [Google Scholar]
- (286).Rutherford K; Le Trong I; Stenkamp RE; Parson WW Crystal Structures of Human 108V and 108M Catechol O-Methyltransferase. J. Mol. Biol 2008, 380, 120–130. [DOI] [PubMed] [Google Scholar]
- (287).Bonifacio MJ; Palma PN; Almeida L; Soares-da-Silva P Catechol-O-methyltransferase and Its Inhibitors in Parkinson’s Disease. CNS Drug Rev 2007, 13, 352–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (288).Ma Z; Liu H; Wu B Structure-based Drug Design of Catechol-O-methyltransferase Inhibitors for CNS Disorders. Br. J. Clin. Pharmacol 2014, 77, 410–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (289).Axelrod J Methylation Reactions in the Formation and Metabolism of Catecholamines and Other Biogenic Amines. Pharmacol. Rev 1966, 18, 95–113. [PubMed] [Google Scholar]
- (290).Giles RE; Miller JW The Catechol-O-methyl Transferase Activity and Endogenous Catecholamine Content of Various Tissues in the Rat and the Effect of Administration of U-0521 (3’,4’-Dihydroxy-2-methyl Propiophenone). J. Pharmacol. Exp. Ther 1967, 158, 189–194. [PubMed] [Google Scholar]
- (291).Giles RE; Miller JW Studies on the Potentiation of the Inotropic Actions of Certain Catecholamines by U-0521 (3’,4’-Dihydroxy-alpha-methyl Propiophenone). J. Pharmacol. Exp. Ther 1967, 157, 55–61. [PubMed] [Google Scholar]
- (292).Rice PJ; Abraham ST; Huang NY; Doman RJ Extraneuronal Uptake Inhibitor U-0521 Decreases Contractile Responses in Rat Vas Deferens. Gen. Pharmacol 1997, 29, 437–439. [DOI] [PubMed] [Google Scholar]
- (293).Backstrom R; Honkanen E; Pippuri A; Kairisalo P; Pystynen J; Heinola K; Nissinen E; Linden IB; Mannisto PT Synthesis of Some Novel Potent and Selective Catechol O-methyltransferase Inhibitors. J. Med. Chem 1989, 32, 841–846. [DOI] [PubMed] [Google Scholar]
- (294).Zurcher G; Keller HH; Kettler R; Borgulya J; Bonetti EP; Eigenmann R; Da Prada M Ro 40–7592, a Novel, Very Potent, and Orally Active Inhibitor of Catechol-O-methyltransferase: a Pharmacological Study in Rats. Adv. Neurol 1990, 53, 497–503. [PubMed] [Google Scholar]
- (295).Porter CC; Watson LS; Titus DC; Totaro JA; Byer SS Inhibition of DOPA Decarboxylase by the Hydrazino Analog of Alpha-methylDOPA. Biochem. Pharmacol 1962, 11, 1067–1077. [DOI] [PubMed] [Google Scholar]
- (296).Burkard WP; Gey KF; Pletscher A A New Inhibitor of Decarboxylase of Aromatic Amino Acids. Experientia 1962, 18, 411–412. [DOI] [PubMed] [Google Scholar]
- (297).Muguet D; Broussolle E; Chazot G Apomorphine in Patients with Parkinson’s Disease. Biomed. Pharmacother 1995, 49, 197–209. [DOI] [PubMed] [Google Scholar]
- (298).Sonnenblick EH; Frishman WH; LeJemtel TH Dobutamine: a New Synthetic Cardioactive Sympathetic Amine. N. Engl. J. Med 1979, 300, 17–22. [DOI] [PubMed] [Google Scholar]
- (299).Caruana MP; Heber M; Brigden G; Raftery EB Effects of Fenoldopam, a Specific Dopamine Receptor Agonist, on Blood Pressure and Left Ventricular Function in Systemic Hypertension. Br. J. Clin. Pharmacol 1987, 24, 721–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (300).Frohlich ED Methyldopa. Mechanisms and Treatment 25 Years Later. Arch. Intern. Med 1980, 140, 954–959. [DOI] [PubMed] [Google Scholar]
- (301).Popa VT Clinical Pharmacology of Adrenergic Drugs. J. Asthma 1984, 21, 183–207. [DOI] [PubMed] [Google Scholar]
- (302).Bowman WC; Rodger IW Actions of the Sympathomimetic Bronchodilator, Rimiterol (R798), on the Cardiovascular, Respiratory and Skeletal Muscle Systems of the Anaesthetized Cat. Br. J. Pharmacol 1972, 45, 574–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (303).Lerner C; Ruf A; Gramlich V; Masjost B; Zurcher G; Jakob-Roetne R; Borroni E; Diederich F X-ray Crystal Structure of a Bisubstrate Inhibitor Bound to the Enzyme Catechol-O-methyltransferase: A Dramatic Effect of Inhibitor Preorganization on Binding Affinity. Angew. Chem., Int. Ed. Engl 2001, 40, 4040–4042. [DOI] [PubMed] [Google Scholar]
- (304).Paulini R; Lerner C; Jakob-Roetne R; Zurcher G; Borroni E; Diederich F Bisubstrate Inhibitors of the Enzyme Catechol O-Methyltransferase (COMT): Efficient Inhibition Despite the Lack of a Nitro Group. ChemBioChem 2004, 5, 1270–1274. [DOI] [PubMed] [Google Scholar]
- (305).Ellermann M; Jakob-Roetne R; Lerner C; Borroni E; Schlatter D; Roth D; Ehler A; Rudolph MG; Diederich F Molecular Recognition at the Active Site of Catechol-O-Methyltransferase: Energetically Favorable Replacement of a Water Molecule Imported by a Bisubstrate Inhibitor. Angew. Chem., Int. Ed 2009, 48, 9092–9096. [DOI] [PubMed] [Google Scholar]
- (306).Paulini R; Trindler C; Lerner C; Brändli L; Schweizer WB; Jakob-Roetne R; Zürcher G; Borroni E; Diederich F Bisubstrate Inhibitors of Catechol O-Methyltransferase (COMT): the Crucial Role of the Ribose Structural Unit for Inhibitor Binding Affinity. ChemMedChem 2006, 1, 340–357. [DOI] [PubMed] [Google Scholar]
- (307).Ellermann M; Paulini R; Jakob-Roetne R; Lerner C; Borroni E; Roth D; Ehler A; Schweizer WB; Schlatter D; Rudolph MG; Diederich F Molecular Recognition at the Active Site of Catechol-O-methyltransferase (COMT): Adenine Replacements in Bisubstrate Inhibitors. Chem. - Eur. J 2011, 17, 6369–6381. [DOI] [PubMed] [Google Scholar]
- (308).Ellermann M; Lerner C; Burgy G; Ehler A; Bissantz C; Jakob-Roetne R; Paulini R; Allemann O; Tissot H; Grunstein D; Stihle M; Diederich F; Rudolph MG Catechol-O-methyltransferase in Complex with Substituted 3’-Deoxyribose Bisubstrate Inhibitors. Acta Crystallogr., Sect. D: Biol. Crystallogr 2012, 68, 253–260. [DOI] [PubMed] [Google Scholar]
- (309).Lerner C; Masjost B; Ruf A; Gramlich V; Jakob-Roetne R; Zurcher G; Borroni E; Diederich F Bisubstrate Inhibitors for the Enzyme Catechol-O-methyltransferase (COMT): Influence of Inhibitor Preorganisation and Linker Length Between the Two Substrate Moieties on Binding Affinity. Org. Biomol. Chem 2003, 1, 42–49. [DOI] [PubMed] [Google Scholar]
- (310).Masri MS; Booth AN; DeEds F O-Methylation in Vitro of Dihydroxy- and Trihydroxy-phenolic Compounds by Liver Slices. Biochim. Biophys. Acta 1962, 65, 495–500. [Google Scholar]
- (311).Merello M; Lees AJ; Webster R; Bovingdon M; Gordin A Effect of Entacapone, a Peripherally Acting Catechol-O-Methyltransferase Inhibitor, on the Motor Response to Acute Treatment with Levodopa in Patients with Parkinson’s Disease. J. Neurol., Neurosurg. Psychiatry 1994, 57, 186–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (312).Lotta T; Vidgren J; Tilgmann C; Ulmanen I; Melen K; Julkunen I; Taskinen J Kinetics of Human Soluble and Membrane-Bound Catechol O-Methyltransferase: A Revised Mechanism and Description of the Thermolabile Variant of the Enzyme. Biochemistry 1995, 34, 4202–4210. [DOI] [PubMed] [Google Scholar]
- (313).Belleau B; Burba J Tropolones: a Unique Class of Potent Non-competitive Inhibitors of S-Sdenosylmethionine-cathechol Methyltransferase. Biochim. Biophys. Acta 1961, 54, 195–196. [DOI] [PubMed] [Google Scholar]
- (314).Zhao Z; Harrison ST; Schubert JW; Sanders JM; Polsky-Fisher S; Zhang NR; McLoughlin D; Gibson CR; Robinson RG; Sachs NA; Kandebo M; Yao L; Smith SM; Hutson PH; Wolkenberg SE; Barrow JC Synthesis and Optimization of N-Heterocyclic Pyridinones as Catechol-O-methyltransferase (COMT) Inhibitors. Bioorg. Med. Chem. Lett 2016, 26, 2952–2956. [DOI] [PubMed] [Google Scholar]
- (315).Harrison ST; Poslusney MS; Mulhearn JJ; Zhao Z; Kett NR; Schubert JW; Melamed JY; Allison TJ; Patel SB; Sanders JM; Sharma S; Smith RF; Hall DL; Robinson RG; Sachs NA; Hutson PH; Wolkenberg SE; Barrow JC Synthesis and Evaluation of Heterocyclic Catechol Mimics as Inhibitors of Catechol-O-methyltransferase (COMT). ACS Med. Chem. Lett 2015, 6, 318–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (316).Wolkenberg S; Barrow JC; Harrison ST; Trotter BW; Nanda KK; Manley PJ; Zhao Z 2011.
- (317).Broch OJ The in Vivo Effect of Tropolone on Noradrenaline Metabolism and Catechol-O-Methyl Transferase Activity in Tissues of the Rat. Acta Pharmacol. Toxicol 1973, 33, 417–428. [DOI] [PubMed] [Google Scholar]
- (318).Keranen T; Gordin A; Karlsson M; Korpela K; Pentikainen PJ; Rita H; Schultz E; Seppala L; Wikberg T Inhibition of Soluble Catechol-O-methyltransferase and Single-dose Pharmacokinetics After Oral and Intravenous Administration of Entacapone. Eur. J. Clin. Pharmacol 1994, 46, 151–157. [DOI] [PubMed] [Google Scholar]
- (319).Borges N Tolcapone in Parkinson’s Disease: Liver Toxicity and Clinical Efficacy. Expert Opin. Drug Saf 2005, 4, 69–73. [DOI] [PubMed] [Google Scholar]
- (320).Wu B; Kulkarni K; Basu S; Zhang S; Hu M First-pass Metabolism via UDP-glucuronosyltransferase: a Barrier to Oral Bioavailability of Phenolics. J. Pharm. Sci 2011, 100, 3655–3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (321).Berndt N; Hamilton AD; Sebti SM Targeting Protein Prenylation for Cancer Therapy. Nat. Rev. Cancer 2011, 11, 775–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (322).Shen MY; Pan PC; Li YY; Li D; Yu HD; Hou TJ Farnesyltransferase and Geranylgeranyltransferase I: Structures, Mechanism, Inhibitors and Molecular Modeling. Drug Discovery Today 2015, 20, 267–276. [DOI] [PubMed] [Google Scholar]
- (323).Jackson JH; Cochrane CG; Bourne JR; Solski PA; Buss JE; Der CJ Farnesol Modification of Kirsten-Ras Exon 4b-Protein Is Essential for Transformation. Proc. Natl. Acad. Sci. U. S. A 1990, 87, 3042–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (324).Bos JL Ras Oncogenes in Human Cancer - a Review. Cancer Res 1989, 49, 4682–4689. [PubMed] [Google Scholar]
- (325).Kohl NE; Omer CA; Conner MW; Anthony NJ; Davide JP; Desolms SJ; Giuliani EA; Gomez RP; Graham SL; Hamilton K; Handt LK; Hartman GD; Koblan KS; Kral AM; Miller PJ; Mosser SD; Oneill TJ; Rands E; Schaber MD; Gibbs JB; Oliff A Inhibition of Farnesyltransferase Induces Regression of Mammary and Salivary Carcinomas in Ras Transgenic Mice. Nat. Med 1995, 1, 792–797. [DOI] [PubMed] [Google Scholar]
- (326).Van Cutsem E; de Velde HV; Karasek P; Oettle H; Vervenne WL; Szawlowski A; Schoffski P; Post S; Verslype C; Neumann H; Safran H; Humblet Y; Ruixo JP; Ma Y; Von Hoff D Phase III Trial of Gemcitabine Plus Tipifarnib Compared with Gemcitabine Plus Placebo in Advanced Pancreatic Cancer. J. Clin. Oncol 2004, 22, 1430–1438. [DOI] [PubMed] [Google Scholar]
- (327).Rao S; Cunningham D; de Gramont A; Scheithauer W; Smakal M; Humblet Y; Kourteva G; Iveson T; Andre T; Dostalova J; Illes A; Belly R; Perez-Ruixo JJ; Park YC; Palmer PA Phase III Double-Blind Placebo Controlled Study of Farnesyl Transferase Inhibitor R115777 in Patients with Refractory Advanced Colorectal Cancer. J. Clin. Oncol 2004, 22, 3950–3957. [DOI] [PubMed] [Google Scholar]
- (328).Harousseau JL; Martinelli G; Jedrzejczak WW; Brandwein JM; Bordessoule D; Masszi T; Ossenkoppele GJ; Alexeeva JA; Beutel G; Maertens J; Vidriales MB; Dombret H; Thomas X; Burnett AK; Robak T; Khuageva NK; Golenkov AK; Tothova E; Mollgard L; Park YC; Bessems A; De Porre P; Howes AJ; Investigators F-A- A Randomized Phase 3 Study of Tipifarnib Compared with Best Supportive Care, Including Hydroxyurea, in the Treatment of Newly Diagnosed Acute Myeloid Leukemia in Patients 70 Years or Older. Blood 2009, 114, 1166–1173. [DOI] [PubMed] [Google Scholar]
- (329).Whyte DB; Kirschmeier P; Hockenberry TN; NunezOliva I; James L; Catino JJ; Bishop WR; Pai JK K- and N-Ras are Geranylgeranylated in Cells Treated with Farnesyl Protein Transferase Inhibitors. J. Biol. Chem 1997, 272, 14459–14464. [DOI] [PubMed] [Google Scholar]
- (330).Lerner EC; Zhang TT; Knowles DB; Qian YM; Hamilton AD; Sebti SM Inhibition of the Prenylation of K-Ras, but not H- or N-Ras, is Highly Resistant to CAAX Peptidomimetics and Requires both a Farnesyltransferase and a Geranylgeranyltransferase I Inhibitor in Human Tumor Cell Lines. Oncogene 1997, 15, 1283–1288. [DOI] [PubMed] [Google Scholar]
- (331).Sun JZ; Qian YM; Hamilton AD; Sebti SM Both Farnesyltransferase and Geranylgeranyltransferase I Inhibitors are Required for Inhibition of Oncogenic K-Ras Prenylation but Each Alone is Sufficient to Suppress Human Tumor Growth in Nude Mouse Xenografts. Oncogene 1998, 16, 1467–1473. [DOI] [PubMed] [Google Scholar]
- (332).Cox AD; Der CJ; Philips MR Targeting RAS Membrane Association: Back to the Future for Anti-RAS Drug Discovery? Clin. Cancer Res 2015, 21, 1819–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (333).Lobell RB; Liu D; Buser CA; Davide JP; DePuy E; Hamilton K; Koblan KS; Lee Y; Mosser S; Motzel SL; Abbruzzese JL; Fuchs CS; Rowinsky EK; Rubin EH; Sharma S; Deutsch PJ; Mazina KE; Morrison BW; Wildonger L; Yao SL; Kohl NE Preclinical and Clinical Pharmacodynamic Assessment of L-778,123, a Dual Inhibitor of Farnesyl:Protein Transferase and Geranylgeranyl:Protein Transferase Type-I. Mol. Cancer Ther 2002, 1, 747–758. [PubMed] [Google Scholar]
- (334).Park HW; Boduluri SR; Moomaw JF; Casey PJ; Beese LS Crystal Structure of Protein Farnesyltransferase at 2.25 Angstrom Resolution. Science 1997, 275, 1800–1804. [DOI] [PubMed] [Google Scholar]
- (335).Zhang FL; Casey PJ Influence of Metal Ions on Substrate Binding and Catalytic Activity of Mammalian Protein Geranylgeranyltransferase Type-I. Biochem. J 1996, 320, 925–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (336).Long SB; Casey PJ; Beese LS Reaction Path of Protein Farnesyltransferase at Atomic Resolution. Nature 2002, 419, 645–650. [DOI] [PubMed] [Google Scholar]
- (337).Moorthy NSHN; Sousa SF; Ramos MJ; Fernandes PA Farnesyltransferase Inhibitors: A Comprehensive Review Based on Quantitative Structural Analysis. Curr. Med. Chem 2013, 20, 4888–4923. [DOI] [PubMed] [Google Scholar]
- (338).Shen Y; Qiang SS; Ma ST The Recent Development of Farnesyltransferase Inhibitors as Anticancer and Antimalarial Agents. Mini-Rev. Med. Chem 2015, 15, 837–857. [DOI] [PubMed] [Google Scholar]
- (339).Strickland CL; Weber PC; Windsor WT; Wu Z; Le HV; Albanese MM; Alvarez CS; Cesarz D; del Rosario J; Deskus J; Mallams AK; Njoroge FG; Piwinski JJ; Remiszewski S; Rossman RR; Taveras AG; Vibulbhan B; Doll RJ; Girijavallabhan VM; Ganguly AK Tricyclic Farnesyl Protein Transferase Inhibitors: Crystallographic and Calorimetric Studies of Structure-Activity Relationships. J. Med. Chem 1999, 42, 2125–2135. [DOI] [PubMed] [Google Scholar]
- (340).Reiss Y; Goldstein JL; Seabra MC; Casey PJ; Brown MS Inhibition of Purified P21ras Farnesyl - Protein Transferase by Cys-Aax Tetrapeptides. Cell 1990, 62, 81–88. [DOI] [PubMed] [Google Scholar]
- (341).Brown MS; Goldstein JL; Paris KJ; Burnier JP; Marsters JC Tetrapeptide Inhibitors of Protein Farnesyltransferase - Amino-Terminal Substitution in Phenylalanine-Containing Tetrapeptides Restores Farnesylation. Proc. Natl. Acad. Sci. U. S. A 1992, 89, 8313–8316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (342).Long SB; Hancock PJ; Kral AM; Hellinga HW; Beese LS The Crystal Structure of Human Protein Farnesyltransferase Reveals the Basis for Inhibition by CaaX Tetrapeptides and their Mimetics. Proc. Natl. Acad. Sci. U. S. A 2001, 98, 12948–12953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (343).Sepp-Lorenzino L; Ma Z; Rands E; Kohl NE; Gibbs JB; Oliff A; Rosen N A Peptidomimetic Inhibitor of Farnesyl:Protein Transferase Blocks the Anchorage-Dependent and -Independent Growth of Human Tumor Cell Lines. Cancer Res 1995, 55, 5302–5309. [PubMed] [Google Scholar]
- (344).Britten CD; Rowinsky EK; Soignet S; Patnaik A; Yao SL; Deutsch P; Lee Y; Lobell RB; Mazina KE; McCreery H; Pezzuli S; Spriggs D A Phase I and Pharmacological Study of the Farnesyl Protein Transferase Inhibitor L-778,123 in Patients with Solid Malignancies. Clin. Cancer Res 2001, 7, 3894–3903. [PubMed] [Google Scholar]
- (345).Sebti SM; Hamilton AD Farnesyltransferase and Geranylgeranyltransferase I Inhibitors and Cancer Therapy: Lessons from Mechanism and Bench-to-Bedside Translational Studies. Oncogene 2000, 19, 6584–6593. [DOI] [PubMed] [Google Scholar]
- (346).Venet M R115777 as Farnesyl Protein Transferase Inhibitor: Synthesis and SAR. Abstr. Pap. - Am. Chem. Soc 2001, 222, U639–U640. [Google Scholar]
- (347).End DW; Smets G; Todd AV; Applegate TL; Fuery CJ; Angibaud P; Venet M; Sanz G; Poignet H; Skrzat S; Devine A; Wouters W; Bowden C Characterization of the Antitumor Effects of the Selective Farnesyl Protein Transferase Inhibitor R115777 In Vivo and In Vitro. Cancer Res 2001, 61, 131–137. [PubMed] [Google Scholar]
- (348).Wang JY; Yao X; Huang J New Tricks for Human Farnesyltransferase Inhibitor: Cancer and Beyond. MedChemComm 2017, 8, 841–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (349).Eastman RT; Buckner FS; Yokoyama K; Gelb MH; Van Voorhis WC Fighting Parasitic Disease by Blocking Protein Farnesylation. J. Lipid Res 2006, 47, 233–240. [DOI] [PubMed] [Google Scholar]
- (350).Sharma K A Review on Plasmodium falciparum-Protein Farnesyltransferase Inhibitors as Antimalarial Drug Targets. Curr. Drug Targets 2017, 18, 1676–1686. [DOI] [PubMed] [Google Scholar]
- (351).Falagas ME; Kasiakou SK Colistin: The Revival of Polymyxins for the Management of Multidrug-Resistant Gram-Negative Bacterial Infections. Clin. Infect. Dis 2005, 40, 1333–1341. [DOI] [PubMed] [Google Scholar]
- (352).Velkov T; Thompson PE; Nation RL; Li J Structure-Activity Relationships of Polymyxin Antibiotics. J. Med. Chem 2010, 53, 1898–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (353).Liu YY; Wang Y; Walsh TR; Yi LX; Zhang R; Spencer J; Doi Y; Tian GB; Dong BL; Huang XH; Yu LF; Gu DX; Ren HW; Chen XJ; Lv LC; He DD; Zhou HW; Liang ZS; Liu JH; Shen JZ Emergence of Plasmid-Mediated Colistin Resistance Mechanism MCR-1 in Animals and Human Beings in China: A Microbiological and Molecular Biological Study. Lancet Infect. Dis 2016, 16, 161–168. [DOI] [PubMed] [Google Scholar]
- (354).Al-Tawfiq JA; Laxminarayan R; Mendelson M How Should We Respond to the Emergence of Plasmid-Mediated Colistin Resistance in Humans and Animals? Int. J. Infect. Dis 2017, 54, 77–84. [DOI] [PubMed] [Google Scholar]
- (355).Karaiskos I; Souli M; Galani I; Giamarellou H Colistin: Still a Lifesaver for the 21st Century? Expert Opin. Drug Metab. Toxicol 2017, 13, 59–71. [DOI] [PubMed] [Google Scholar]
- (356).Hinchliffe P; Yang QE; Portal E; Young T; Li H; Tooke CL; Carvalho MJ; Paterson NG; Brem J; Niumsup PR; Tansawai U; Lei L; Li M; Shen ZQ; Wang Y; Schofield CJ; Mulholland AJ; Shen JZ; Fey N; Walsh TR; Spencer J Insights into the Mechanistic Basis of Plasmid-Mediated Colistin Resistance from Crystal Structures of the Catalytic Domain of MCR-1. Sci. Rep 2017, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (357).Ma GX; Zhu YF; Yu ZC; Ahmad A; Zhang HM High Resolution Crystal Structure of the Catalytic Domain of MCR-1. Sci. Rep 2016, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (358).Stojanoski V; Sankaran B; Prasad BVV; Poirel L; Nordmann P; Palzkill T Structure of the Catalytic Domain of the Colistin Resistance Enzyme MCR-1. BMC Biol 2016, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (359).Hu ML; Guo JB; Cheng QP; Yang ZQ; Chan EWC; Chen S; Hao Q Crystal Structure of Escherichia coli Originated MCR-1, a Phosphoethanolamine Transferase for Colistin Resistance. Sci. Rep 2016, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (360).Wanty C; Anandan A; Piek S; Walshe J; Ganguly J; Carlson RW; Stubbs KA; Kahler CM; Vrielink A The Structure of the Neisserial Lipooligosaccharide Phosphoethanolamine Transferase A (LptA) Required for Resistance to Polymyxin. J. Mol. Biol 2013, 425, 3389–3402. [DOI] [PubMed] [Google Scholar]
- (361).Pommier Y; Johnson AA; Marchand C Integrase Inhibitors to Treat HIV/AIDS. Nat. Rev. Drug Discovery 2005, 4, 236–248. [DOI] [PubMed] [Google Scholar]
- (362).Benard C; Zouhiri F; Normand-Bayle M; Danet M; Desmaele D; Leh H; Mouscadet JF; Mbemba G; Thomas CM; Bonnenfant S; Le Bret M; d’Angelo J Linker-Modified Quinoline Derivatives Targeting HIV-1 Integrase: Synthesis and Biological Activity. Bioorg. Med. Chem. Lett 2004, 14, 2473–2476. [DOI] [PubMed] [Google Scholar]
- (363).Chi G; Neamati N; Nair V Inhibition of the Strand Transfer Step of HIV-1 Integrase by Non-Natural Dinucleotides. Bioorg. Med. Chem. Lett 2004, 14, 4815–4817. [DOI] [PubMed] [Google Scholar]
- (364).Rogolino D; Carcelli M; Sechi M; Neamati N Viral Enzymes Containing Magnesium: Metal Binding as a Successful Strategy in Drug Design. Coord. Chem. Rev 2012, 256, 3063–3086. [Google Scholar]
- (365).Ingale KB; Bhatia MS HIV-1 Integrase Inhibitors: A Review of Their Chemical Development. Antiviral Chem. Chemother 2011, 22, 95–105. [DOI] [PubMed] [Google Scholar]
- (366).Li Y; Xuan S; Feng Y; Yan A Targeting HIV-1 Integrase With Strand Transfer Inhibitors. Drug Discovery Today 2015, 20, 435–449. [DOI] [PubMed] [Google Scholar]
- (367).Pommier Y; Marchand C; Neamati N Retroviral Integrase Inhibitors Year 2000: Update and Perspectives. Antiviral Res 2000, 47, 139–148. [DOI] [PubMed] [Google Scholar]
- (368).Alian A; Griner SL; Chiang V; Tsiang M; Jones G; Birkus G; Geleziunas R; Leavitt AD; Stroud RM Catalytically-Active Complex of HIV-1 Integrase With a Viral DNA Substrate Binds Anti-Integrase Drugs. Proc. Natl. Acad. Sci. U. S. A 2009, 106, 8192–8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (369).Zheng RL; Jenkins TM; Craigie R Zinc Folds the N-terminal Domain of HIV-1 Integrase, Promotes Multimerization, and Enhances Catalytic Activity. Proc. Natl. Acad. Sci. U. S. A 1996, 93, 13659–13664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (370).Vangent DC; Groeneger AAMO; Plasterk RHA Mutational Analysis of the Integrase Protein of Human-Immunodeficiency-Virus Type-2. Proc. Natl. Acad. Sci. U. S. A 1992, 89, 9598–9602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (371).Metifiot M; Maddali K; Johnson BC; Hare S; Smith SJ; Zhao XZ; Marchand C; Burke TR Jr.; Hughes SH; Cherepanov P; Pommier Y Activities, Crystal Structures, and Molecular Dynamics of Dihydro-1H-isoindole Derivatives, Inhibitors of HIV-1 Integrase. ACS Chem. Biol 2013, 8, 209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (372).Desimmie BA; Demeulemeester J; Suchaud V; Taltynov O; Billamboz M; Lion C; Bailly F; Strelkov SV; Debyser Z; Cotelle P; Christ F 2-Hydroxyisoquinoline-1,3(2H,4H)-diones (HIDs), Novel Inhibitors of HIV Integrase With a High Barrier to Resistance. ACS Chem. Biol 2013, 8, 1187–1194. [DOI] [PubMed] [Google Scholar]
- (373).Summa V; Petrocchi A; Bonelli F; Crescenzi B; Donghi M; Ferrara M; Fiore F; Gardelli C; Gonzalez Paz O; Hazuda DJ; Jones P; Kinzel O; Laufer R; Monteagudo E; Muraglia E; Nizi E; Orvieto F; Pace P; Pescatore G; Scarpelli R; Stillmock K; Witmer MV; Rowley M Discovery of Raltegravir, a Potent, Selective Orally Bioavailable HIV-Integrase Inhibitor for the Treatment of HIV-AIDS Infection. J. Med. Chem 2008, 51, 5843–5855. [DOI] [PubMed] [Google Scholar]
- (374).Serrao E; Odde S; Ramkumar K; Neamati N Raltegravir, Elvitegravir, and Metoogravir: The Birth of “me-too” HIV-1 Integrase Inhibitors. Retrovirology 2009, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (375).Hare S; Smith SJ; Metifiot M; Jaxa-Chamiec A; Pommier Y; Hughes SH; Cherepanov P Structural and Functional Analyses of the Second-Generation Integrase Strand Transfer Inhibitor Dolutegravir (S/GSK1349572). Mol. Pharmacol 2011, 80, 565–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (376).Tsiang M; Jones GS; Goldsmith J; Mulato A; Hansen D; Kan E; Tsai L; Bam RA; Stepan G; Stray KM; Niedziela-Majka A; Yant SR; Yu H; Kukolj G; Cihlar T; Lazerwith SE; White KL; Jin HL Antiviral Activity of Bictegravir (GS-9883), a Novel Potent HIV-1 Integrase Strand Transfer Inhibitor with an Improved Resistance Profile. Antimicrob. Agents Chemother 2016, 60, 7086–7097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (377).Hassounah SA; Alikhani A; Oliveira M; Bharaj S; Ibanescu RI; Osman N; Xu HT; Brenner BG; Mesplede T; Wainberg MA Antiviral Activity of Bictegravir and Cabotegravir Against Integrase Inhibitor-Resistant SIVmac239 and HIV-1. Antimicrob. Agents Chemother 2017, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (378).Pais GCG; Burke TR Novel Aryl Diketo-Containing Inhibitors of HIV-1 Integrase. Drugs Future 2002, 27, 1101–1111. [Google Scholar]
- (379).Goldgur Y; Craigie R; Cohen GH; Fujiwara T; Yoshinaga T; Fujishita T; Sugimoto H; Endo T; Murai H; Davies DR Structure of the HIV-1 Integrase Catalytic Domain Complexed With an Inhibitor: A Platform for Antiviral Drug Design. Proc. Natl. Acad. Sci. U. S. A 1999, 96, 13040–13043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (380).Wai JS; Egbertson MS; Payne LS; Fisher TE; Embrey MW; Tran LO; Melamed JY; Langford HM; Guare JP; Zhuang LG; Grey VE; Vacca JP; Holloway MK; Naylor-Olsen AM; Hazuda DJ; Felock PJ; Wolfe AL; Stillmock KA; Schleif WA; Gabryelski LJ; Young SD 4-Aryl-2,4-dioxobutanoic Acid Inhibitors of HIV-1 Integrase and Viral Replication in Cells. J. Med. Chem 2000, 43, 4923–4926. [DOI] [PubMed] [Google Scholar]
- (381).Ovenden SPB; Yu J; Wan SS; Sberna G; Tait RM; Rhodes D; Cox S; Coates J; Walsh NG; Meurer-Grimes BM Globoidnan A: A Lignan From Eucalyptus globoidea Inhibits HIV Integrase. Phytochemistry 2004, 65, 3255–3259. [DOI] [PubMed] [Google Scholar]
- (382).Lin ZW; Neamati N; Zhao H; Kiryu Y; Turpin JA; Aberham C; Strebel K; Kohn K; Witvrouw M; Pannecouque C; Debyser Z; De Clercq E; Rice WG; Pommier Y; Burke TR Chicoric Acid Analogues as HIV-1 Integrase Inhibitors. J. Med. Chem 1999, 42, 1401–1414. [DOI] [PubMed] [Google Scholar]
- (383).Fesen MR; Kohn KW; Leteurtre F; Pommier Y Inhibitors of Human-Immunodeficiency-Virus Integrase. Proc. Natl. Acad. Sci. U. S. A 1993, 90, 2399–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (384).Burke TR; Fesen MR; Mazumder A; Wang J; Carothers AM; Grunberger D; Driscoll J; Kohn K; Pommier Y Hydroxylated Aromatic Inhibitors of Hiv-1 Integrase. J. Med. Chem 1995, 38, 4171–4178. [DOI] [PubMed] [Google Scholar]
- (385).Zhao H; Neamati N; Mazumder A; Sunder S; Pommier Y; Burke TR Arylamide Inhibitors of HIV-1 Integrase. J. Med. Chem 1997, 40, 1186–1194. [DOI] [PubMed] [Google Scholar]
- (386).Mouscadet JF; Desmaele D Chemistry and Structure-Activity Relationship of the Styrylquinoline-Type HIV Integrase Inhibitors. Molecules 2010, 15, 3048–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (387).Mekouar K; Mouscadet JF; Desmaele D; Subra F; Leh H; Savoure D; Auclair C; d’Angelo J Styrylquinoline Derivatives: A New Class of Potent HIV-1 Integrase Inhibitors that Block HIV-1 Replication in CEM Cells. J. Med. Chem 1998, 41, 2846–2857. [DOI] [PubMed] [Google Scholar]
- (388).Hazuda DJ; Anthony NJ; Gomez RP; Jolly SM; Wai JS; Zhuang LH; Fisher TE; Embrey M; Guare JP; Egbertson MS; Vacca JP; Huff JR; Felock PJ; Witmer MV; Stillmock KA; Danovich R; Grobler J; Miller MD; Espeseth AS; Jin LX; Chen IW; Lin JH; Kassahun K; Ellis JD; Wong BK; Xu W; Pearson PG; Schleif WA; Cortese R; Emini E; Summa V; Holloway MK; Young SD A Naphthyridine Carboxamide Provides Evidence for Discordant Resistance Between Mechanistically Identical Inhibitors of HIV-1 Integrase. Proc. Natl. Acad. Sci. U. S. A 2004, 101, 11233–11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (389).Guare JP; Wai JS; Gomez RP; Anthony NJ; Jolly SM; Cortes AR; Vacca JP; Felock PJ; Stillmock KA; Schleif WA; Moyer G; Gabryelski LJ; Jin LX; Chen IW; Hazuda DJ; Young SD A Series of 5-Aminosubstituted 4-Fluorobenzyl-8-hydroxy-[1,6]naphthyridine-7-carboxamide HIV-1 Integrase Inhibitors. Bioorg. Med. Chem. Lett 2006, 16, 2900–2904. [DOI] [PubMed] [Google Scholar]
- (390).Zhuang LC; Wai JS; Embrey MW; Fisher TE; Egbertson MS; Payne LS; Guare JP; Vacca JP; Hazuda DJ; Felock PJ; Wolfe AL; Stillmock KA; Witmer MV; Moyer G; Schleif WA; Gabryelski LJ; Leonard YM; Lynch JJ; Michelson SR; Young SD Design and Synthesis of 8-Hydroxy-[1,6]naphthyridines as Novel Inhibitors of HIV-1 Integrase in Vitro and in Infected Cells. J. Med. Chem 2003, 46, 453–456. [DOI] [PubMed] [Google Scholar]
- (391).Johns BA; Weatherhead JG; Allen SH; Thompson JB; Garvey EP; Foster SA; Jeffrey JL; Miller WH The Use of Oxadiazole and Triazole Substituted Naphthyridines as HIV-1 Integrase Inhibitors. Part 1: Establishing the Pharmacophore. Bioorg. Med. Chem. Lett 2009, 19, 1802–1806. [DOI] [PubMed] [Google Scholar]
- (392).Verschueren WG; Dierynck I; Amssoms KIE; Hu LL; Boonants PMJG; Pille GME; Daeyaert FFD; Hertogs K; Surleraux DLNG; Wigerinck PBTP Design and Optimization of Tricyclic Phtalimide Analogues as Novel Inhibitors of HIV-1 Integrase. J. Med. Chem 2005, 48, 1930–1940. [DOI] [PubMed] [Google Scholar]
- (393).Sato M; Motomura T; Aramaki H; Matsuda T; Yamashita M; Ito Y; Kawakami H; Matsuzaki Y; Watanabe W; Yamataka K; Ikeda S; Kodama E; Matsuoka M; Shinkai H Novel HIV-1 Integrase Inhibitors Derived From Qinolone Antibiotics. J. Med. Chem 2006, 49, 1506–1508. [DOI] [PubMed] [Google Scholar]
- (394).Cecchetti V; Parolin C; Moro S; Pecere T; Filipponi E; Calistri A; Tabarrini O; Gatto B; Palumbo M; Fravolini A; Palu G 6-Aminoquinolones as New Potential Anti-HIV Agents. J. Med. Chem 2000, 43, 3799–3802. [DOI] [PubMed] [Google Scholar]
- (395).Summa V; Petrocchi A; Matassa VG; Gardelli C; Muraglia E; Rowley M; Paz OG; Laufer R; Monteagudo E; Pace P 4,5-Dihydroxypyrimidine Carboxamides and N-alkyl-5-hydroxypyrimidinone Carboxamides are Potent, Selective HIV Integrase Inhibitors With Good Pharmacokinetic Profiles in Preclinical Species. J. Med. Chem 2006, 49, 6646–6649. [DOI] [PubMed] [Google Scholar]
- (396).Gardelli C; Nizi E; Muraglia E; Crescenzi B; Ferrara M; Orvieto F; Pace P; Pescatore G; Poma M; Ferreira MDRR; Scarpelli R; Homnick CF; Ikemoto N; Alfieri A; Verdirame M; Bonelli F; Paz OG; Taliani M; Monteagudo E; Pesci S; Laufer R; Felock P; Stillmock KA; Hazuda D; Rowley M; Summa V Discovery and Synthesis of HIV Integrase Inhibitors: Development of Potent and Orally Bioavailable N-methyl Pyrimidones. J. Med. Chem 2007, 50, 4953–4975. [DOI] [PubMed] [Google Scholar]
- (397).Ferrara M; Fiore F; Summa V; Gardelli C Development of 2-pyrrolidinyl-N-methyl Pyrimidones as Potent and Orally Bioavailable HIV Integrase Inhibitors. Bioorg. Med. Chem. Lett 2010, 20, 5031–5034. [DOI] [PubMed] [Google Scholar]
- (398).Muraglia E; Kinzel O; Gardelli C; Crescenzi B; Donghi M; Ferrara M; Nizi E; Orvieto F; Pescatore G; Laufer R; Gonzalez-Paz O; Di Marco A; Fiore F; Monteagudo E; Fonsi M; Felock PJ; Rowley M; Summa V Design and Synthesis of Bicyclic Pyrimidinones as Potent and Orally Bioavailable HIV-1 Integrase Inhibitors. J. Med. Chem 2008, 51, 861–874. [DOI] [PubMed] [Google Scholar]
- (399).Jones ED; Vandegraaff N; Le GA; Choi N; Issa W; Macfarlane K; Thienthong N; Winfield LJ; Coates JAV; Lu L; Li XM; Feng XA; Yu CJ; Rhodes DI; Deadman JJ Design of a Series of Bicyclic HIV-1 Integrase Inhibitors. Part 1: Selection of the Scaffold. Bioorg. Med. Chem. Lett 2010, 20, 5913–5917. [DOI] [PubMed] [Google Scholar]
- (400).Le GA; Vandegraaff N; Rhodes DI; Jones ED; Coates JAV; Thienthong N; Winfield LJ; Lu L; Li XM; Yu CJ; Feng XA; Deadman JJ Design of a Series of Bicyclic HIV-1 Integrase Inhibitors. Part 2: Azoles: Effective Metal Chelators. Bioorg. Med. Chem. Lett 2010, 20, 5909–5912. [DOI] [PubMed] [Google Scholar]
- (401).Agrawal A; DeSoto J; Fullagar JL; Maddali K; Rostami S; Richman DD; Pommier Y; Cohen SM Probing Chelation Motifs in HIV Integrase Inhibitors. Proc. Natl. Acad. Sci. U. S. A 2012, 109, 2251–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (402).Hazuda DJ; Felock P; Witmer M; Wolfe A; Stillmock K; Grobler JA; Espeseth A; Gabryelski L; Schleif W; Blau C; Miller MD Inhibitors of Strand Transfer That Prevent Integration and Inhibit HIV-1 Replication in Cells. Science 2000, 287, 646–650. [DOI] [PubMed] [Google Scholar]
- (403).Tramontano E; Di Santo R HIV-1 RT-associated RNase H Function Inhibitors: Recent Advances in Drug Development. Curr. Med. Chem 2010, 17, 2837–2853. [DOI] [PubMed] [Google Scholar]
- (404).Sarafianos SG; Marchand B; Das K; Himmel DM; Parniak MA; Hughes SH; Arnold E Structure and Function of HIV-1 Reverse Transcriptase: Molecular Mechanisms of Polymerization and Inhibition. J. Mol. Biol 2009, 385, 693–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (405).Arts EJ; Le Grice SF Interaction of Retroviral Reverse Transcriptase With Template-Primer Duplexes During Replication. Prog. Nucleic Acid Res. Mol. Biol 1998, 58, 339–393. [DOI] [PubMed] [Google Scholar]
- (406).Rausch JW; Le Grice SF ’Binding, Bending and Bonding’: Polypurine Tract-Primed Initiation of Plus-Strand DNA Synthesis in Human Immunodeficiency Virus. Int. J. Biochem. Cell. Biol 2004, 36, 1752–1766. [DOI] [PubMed] [Google Scholar]
- (407).Hughes SH Reverse Transcription of Retroviruses and LTR Retrotransposons. Microbiol. Spectrum 2015, 3, MDNA3-0027-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (408).Cao L; Song W; De Clercq E; Zhan P; Liu X Recent Progress in the Research of Small Molecule HIV-1 RNase H Inhibitors. Curr. Med. Chem 2014, 21, 1956–1967. [DOI] [PubMed] [Google Scholar]
- (409).Wang X; Gao P; Menendez-Arias L; Liu X; Zhan P Update on Recent Developments in Small Molecular HIV-1 RNase H Inhibitors (2013–2016): Opportunities and Challenges. Curr. Med. Chem 2017. [DOI] [PubMed] [Google Scholar]
- (410).Yu F; Liu X; Zhan P; De Clercq E Recent Advances in the Research of HIV-1 RNase H Inhibitors. Mini-Rev. Med. Chem 2008, 8, 1243–1251. [DOI] [PubMed] [Google Scholar]
- (411).Kohlstaedt LA; Wang J; Friedman JM; Rice PA; Steitz TA Crystal Structure at 3.5 A Resolution of HIV-1 Reverse Transcriptase Complexed with an Inhibitor. Science 1992, 256, 1783–1790. [DOI] [PubMed] [Google Scholar]
- (412).Schultz SJ; Zhang M; Champoux JJ Specific Cleavages by RNase H Facilitate Initiation of Plus-Strand RNA Synthesis by Moloney Murine Leukemia Virus. J. Virol 2003, 77, 5275–5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (413).Champoux JJ; Schultz SJ Ribonuclease H: Properties, Substrate Specificity and Roles in Retroviral Reverse Transcription. FEBS J 2009, 276, 1506–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (414).Schultz SJ; Champoux JJ RNase H Activity: Structure, Specificity, and Function in Reverse Transcription. Virus Res 2008, 134, 86–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (415).Keck JL; Goedken ER; Marqusee S Activation/attenuation Model for RNase H. A One-Metal Mechanism with Second-Metal Inhibition. J. Biol. Chem 1998, 273, 34128–34133. [DOI] [PubMed] [Google Scholar]
- (416).Huang H; Chopra R; Verdine GL; Harrison SC Structure of a Covalently Trapped Catalytic Complex of HIV-1 Reverse Transcriptase: Implications for Drug Resistance. Science 1998, 282, 1669–1675. [DOI] [PubMed] [Google Scholar]
- (417).Nowotny M; Gaidamakov SA; Crouch RJ; Yang W Crystal Structures of RNase H Bound to an RNA/DNA Hybrid: Substrate Specificity and Metal-dependent Catalysis. Cell 2005, 121, 1005–1016. [DOI] [PubMed] [Google Scholar]
- (418).Nowotny M; Gaidamakov SA; Ghirlando R; Cerritelli SM; Crouch RJ; Yang W Structure of Human RNase h1 Complexed with an RNA/DNA Hybrid: Insight into HIV Reverse Transcription. Mol. Cell 2007, 28, 264–276. [DOI] [PubMed] [Google Scholar]
- (419).Steitz TA; Steitz JA A General Two-Metal-ion Mechanism for Catalytic RNA. Proc. Natl. Acad. Sci. U. S. A 1993, 90, 6498–6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (420).Steitz TA Structural biology - A Mechanism for all Polymerases. Nature 1998, 391, 231–232. [DOI] [PubMed] [Google Scholar]
- (421).Billamboz M; Bailly F; Lion C; Touati N; Vezin H; Calmels C; Andreola ML; Christ F; Debyser Z; Cotelle P Magnesium Chelating 2-Hydroxyisoquinoline-1,3(2H,4H)-diones, as Inhibitors of HIV-1 Integrase and/or the HIV-1 Reverse Transcriptase Ribonuclease H Domain: Discovery of a Novel Selective Inhibitor of the Ribonuclease H Function. J. Med. Chem 2011, 54, 1812–1824. [DOI] [PubMed] [Google Scholar]
- (422).Mizrahi V; Usdin MT; Harington A; Dudding LR Site-Directed Mutagenesis of the Conserved Asp-443 and Asp-498 Carboxy-Terminal Residues of Hiv-1 Reverse-Transcriptase. Nucleic Acids Res 1990, 18, 5359–5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (423).Mizrahi V; Brooksbank RL; Nkabinde NC Mutagenesis of the Conserved Aspartic-Acid-443, Glutamic-Acid-478, Asparagine-494, and Aspartic-Acid-498 Residues in the Ribonuclease H-Domain of P66/P51 Human-Immunodeficiency-Virus Type-I Reverse-Transcriptase - Expression and Biochemical-Analysis. J. Biol. Chem 1994, 269, 19245–19249. [PubMed] [Google Scholar]
- (424).Klumpp K; Mirzadegan T Recent Progress in the Design of Small Molecule Inhibitors of HIV RNase H. Curr. Pharm. Des 2006, 12, 1909–1922. [DOI] [PubMed] [Google Scholar]
- (425).Moelling K; Schulze T; Diringer H Inhibition of Human Immunodeficiency Virus Type-1 Rnase-H by Sulfated Polyanions. J. Virol 1989, 63, 5489–5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (426).Loya S; Tal R; Kashman Y; Hizi A Illimaquinone, a Selective Inhibitor of the RNase H Activity of Human Immunodeficiency Virus Type 1 Reverse Transcriptase. Antimicrob. Agents Chemother 1990, 34, 2009–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (427).Budihas SR; Gorshkova I; Gaidamakov S; Wamiru A; Bona MK; Parniak MA; Crouch RJ; McMahon JB; Beutler JA; Le Grice SFJ Selective Inhibition of HIV-1 Reverse Transcriptase-Associated Ribonuclease H Activity by Hydroxylated Tropolones. Nucleic Acids Res 2005, 33, 1249–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (428).Semenova EA; Johnson AA; Marchand C; Davis DA; Yarchoan R; Pommier Y Preferential Inhibition of the Magnesium-Dependent Strand Transfer Reaction of HIV-1 Integrase by Alpha-Hydroxytropolones. Mol. Pharmacol 2006, 69, 1454–1460. [DOI] [PubMed] [Google Scholar]
- (429).Himmel DM; Maegley KA; Pauly TA; Bauman JD; Das K; Dharia C; Clark AD; Ryan K; Hickey MJ; Love RA; Hughes SH; Bergqvist S; Arnold E Structure of HIV-1 Reverse Transcriptase with the Inhibitor beta-Thujaplicinol Bound at the RNase H Active Site. Structure 2009, 17, 1625–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (430).Tan CK; Civil R; Mian AM; So AG; Downey KM Inhibition of the Rnase H Activity of Hiv Reverse-Transcriptase by Azidothymidylate. Biochemistry 1991, 30, 4831–4835. [DOI] [PubMed] [Google Scholar]
- (431).Zhan XY; Tan CK; Scott WA; Mian AM; Downey KM; So AG Catalytically Distinct Conformations of the Ribonuclease-H of Hiv-1 Reverse-Transcriptase by Substrate Cleavage Patterns and Inhibition by Azidothymidylate and N-Ethylmaleimide. Biochemistry 1994, 33, 1366–1372. [DOI] [PubMed] [Google Scholar]
- (432).Borkow G; Fletcher RS; Barnard J; Arion D; Motakis D; Dmitrienko GI; Parniak MA Inhibition of the ribonuclease H and DNA polymerase activities of HIV-1 reverse transcriptase by N-(4-tert-butylbenzoyl)-2-hydroxy-1-naphthaldehyde hydrazone. Biochemistry 1997, 36, 3179–3185. [DOI] [PubMed] [Google Scholar]
- (433).Sluis-Cremer N; Arion D; Parniak MA Destabilization of the HIV-1 Reverse Transcriptase Dimer Upon Interaction with N-acyl Hydrazone Inhibitors. Mol. Pharmacol 2002, 62, 398–405. [DOI] [PubMed] [Google Scholar]
- (434).Parkes KEB; Ermert P; Fassler J; Ives J; Martin JA; Merrett JH; Obrecht D; Williams G; Klumpp K Use of a Pharmacophore Model to Discover a New Class of Influenza Endonuclease Inhibitors. J. Med. Chem 2003, 46, 1153–1164. [DOI] [PubMed] [Google Scholar]
- (435).Tarrago-Litvak L; Andreola ML; Fournier M; Nevinsky GA; Parissi V; de Soultrait VR; Litvak S Inhibitors of HIV-1 Reverse Transcriptase and Integrase: Classical and Emerging Therapeutical Approaches. Curr. Pharm. Des 2002, 8, 595–614. [DOI] [PubMed] [Google Scholar]
- (436).Shaw-Reid CA; Munshi V; Graham P; Wolfe A; Witmer M; Danzeisen R; Olsen DB; Carroll SS; Embrey M; Wai JS; Miller MD; Cole JL; Hazuda DJ Inhibition of HIV-1 Ribonuclease H by a Novel Diketo acid, 4-[5-(benzoylamino)thien-2-yl]-2,4-dioxobutanoic Acid. J. Biol. Chem 2003, 278, 2777–2780. [DOI] [PubMed] [Google Scholar]
- (437).Kirschberg TA; Balakrishnan M; Squires NH; Barnes T; Brendza KM; Chen XW; Eisenberg EJ; Jin WL; Kutty N; Leavitt S; Liclican A; Liu Q; Liu XH; Mak J; Perry JK; Wang M; Watkins WJ; Lansdon EB RNase H Active Site Inhibitors of Human Immunodeficiency Virus Type 1 Reverse Transcriptase: Design, Biochemical Activity, and Structural Information. J. Med. Chem 2009, 52, 5781–5784. [DOI] [PubMed] [Google Scholar]
- (438).Kankanala J; Kirby KA; Liu F; Miller L; Nagy E; Wilson DJ; Parniak MA; Sarafianos SG; Wang ZQ Design, Synthesis, and Biological Evaluations of Hydroxypyridonecarboxylic Acids as Inhibitors of HIV Reverse Transcriptase Associated RNase H. J. Med. Chem 2016, 59, 5051–5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (439).Klumpp K; Hang JQ; Rajendran S; Yang YL; Derosier A; In PWK; Overton H; Parkes KEB; Cammack N; Martin JA Two-Metal Ion Mechanism of RNA Cleavage by HIV RNase H and Mechanism-Based Design of Selective HIV RNase H Inhibitors. Nucleic Acids Res 2003, 31, 6852–6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (440).Hang JQ; Rajendran S; Yang YL; Li Y; In PWK; Overton H; Parkes KEB; Cammack N; Martin JA; Klumpp K Activity of the Isolated HIV RNase H Domain and Specific Inhibition by N-hydroxyimides. Biochem. Biophys. Res. Commun 2004, 317, 321–329. [DOI] [PubMed] [Google Scholar]
- (441).Kirby KA; Myshakina NA; Christen MT; Chen Y-L; Schmidt HA; Huber AD; Xi Z; Kim S; Rao RK; Kramer ST; Yang Q; Singh K; Parniak MA; Wang Z; Ishima R; Sarafianos SG A 2-Hydroxyisoquinoline-1,3-dione Active Site RNase H Inhibitor Binds in Multiple Modes to HIV-1 Reverse Transcriptase. Antimicrob. Agents Chemother 2017, 61, e01351–01317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (442).Tang J; Liu F; Nagy E; Miller L; Kirby KA; Wilson DJ; Wu BL; Sarafianos SG; Parniak MA; Wang Z 3-Hydroxypyrimidine-2,4-diones as Selective Active Site Inhibitors of HIV Reverse Transcriptase-Associated RNase H: Design, Synthesis, and Biochemical Evaluations. J. Med. Chem 2016, 59, 2648–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (443).Tang J; Maddali K; Dreis CD; Sham YY; Vince R; Pommier Y; Wang ZQ N-3 Hydroxylation of Pyrimidine-2,4-diones Yields Dual Inhibitors of HIV Reverse Transcriptase and Integrase. ACS Med. Chem. Lett 2011, 2, 63–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (444).Williams PD; Staas DD; Venkatraman S; Loughran HM; Ruzek RD; Booth TM; Lyle TA; Wai JS; Vacca JP; Feuston BP; Ecto LT; Flynn JA; DiStefano DJ; Hazuda DJ; Bahnck CM; Himmelberger AL; Dornadula G; Hrin RC; Stillmock KA; Witmer MV; Miller MD; Grobler JA Potent and Selective HIV-1 Ribonuclease H Inhibitors Based on a 1-hydroxy-1,8-naphthyridin-2(1H)-one Scaffold. Bioorg. Med. Chem. Lett 2010, 20, 6754–6757. [DOI] [PubMed] [Google Scholar]
- (445).Velthuisen EJ; Johns BA; Gerondelis P; Chen Y; Li M; Mou K; Zhang WW; Seal JW; Hightower KE; Miranda SR; Brown K; Leesnitzer L Pyridopyrimidinone Inhibitors of HIV-1 RNase H. Eur. J. Med. Chem 2014, 83, 609–616. [DOI] [PubMed] [Google Scholar]
- (446).Himmel DM; Myshakina NS; Ilina T; Van Ry A; Ho WC; Parniak MA; Arnold E Structure of a Dihydroxycoumarin Active-Site Inhibitor in Complex with the RNase H Domain of HIV-1 Reverse Transcriptase and Structure-Activity Analysis of Inhibitor Analogs. J. Mol. Biol 2014, 426, 2617–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (447).Tramontanoa E; Espositoa F; Badasa R; DiSanto R; Costi R; LaColla P 6-[1-(4-Fluorophenyl)methyl-1H-pyrrol-2-yl)]-2, 4-dioxo-5-hexenoic acid ethyl ester a novel diketo acid derivative which selectively inhibits the HIV-1 viral replication in cell culture and the ribonuclease H activity in vitro. Antiviral Res 2005, 65, 117–124. [DOI] [PubMed] [Google Scholar]
- (448).Nichol KL; Treanor JJ Vaccines for Seasonal and Pandemic Influenza. J. Infect. Dis 2006, 194 (Suppl. 2), S111–118. [DOI] [PubMed] [Google Scholar]
- (449).Subbarao K; Joseph T Scientific Barriers to Developing Vaccines Against Avian Influenza Viruses. Nat. Rev. Immunol 2007, 7, 267–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (450).Monod A; Swale C; Tarus B; Tissot A; Delmas B; Ruigrok RW; Crepin T; Slama-Schwok A Learning from Structure-Based Drug Design and New Antivirals Targeting the Ribonucleoprotein Complex for the Treatment of Influenza. Expert Opin. Drug Discovery 2015, 10, 345–371. [DOI] [PubMed] [Google Scholar]
- (451).Drake JW Rates of Spontaneous Mutation among Rna Viruses. Proc. Natl. Acad. Sci. U. S. A 1993, 90, 4171–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (452).Watanabe T; Kawaoka Y Influenza Virus-Host Interactomes as a Basis for Antiviral Drug Development. Curr. Opin. Virol 2015, 14, 71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (453).Li TCM; Chan MCW; Lee N Clinical Implications of Antiviral Resistance in Influenza. Viruses 2015, 7, 4929–4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (454).Furuta Y; Gowen BB; Takahashi K; Shiraki K; Smee DF; Barnard DL Favipiravir (T-705), a Novel Viral RNA Polymerase Inhibitor. Antiviral Res 2013, 100, 446–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (455).Bright RA; Shay DK; Shu B; Cox NJ; Klimov AI Adamantane Resistance Among Influenza A Viruses Isolated Early During the 2005–2006 Influenza Season in the United States. JAMA, J. Am. Med. Assoc 2006, 295, 891–894. [DOI] [PubMed] [Google Scholar]
- (456).Amarelle L; Lecuona E; Sznajder JI Anti-Influenza Treatment: Drugs Currently Used and Under Development. Arch. Bronconeumol 2017, 53, 19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (457).Collins PJ; Haire LF; Lin YP; Liu JF; Russell RJ; Walker PA; Skehel JJ; Martin SR; Hay AJ; Gamblin SJ Crystal Structures of Oseltamivir-resistant Influenza Virus Neuraminidase Mutants. Nature 2008, 453, 1258–U1261. [DOI] [PubMed] [Google Scholar]
- (458).Moscona A Global Transmission of Oseltamivir-Resistant Influenza. N. Engl. J. Med 2009, 360, 953–956. [DOI] [PubMed] [Google Scholar]
- (459).Huang TS; Palese P; Krystal M Determination of Influenza-Virus Proteins Required for Genome Replication. J. Virol 1990, 64, 5669–5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (460).Fodor E The RNA Polymerase of Influenza A Virus: Mechanisms of Viral Transcription and Replication. Acta Virol. (Engl. Ed.) 2013, 57, 113–122. [DOI] [PubMed] [Google Scholar]
- (461).Resa-Infante P; Jorba N; Coloma R; Ortin J The Influenza Virus RNA Synthesis Machine: Advances in its Structure and Function. RNA Biol 2011, 8, 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (462).Plotch SJ; Bouloy M; Ulmanen I; Krug RM A Unique Cap(m7GpppXm)-dependent Influenza Virion Endonuclease Cleaves Capped RNAs to Generate the Primers that Initiate Viral RNA Transcription. Cell 1981, 23, 847–858. [DOI] [PubMed] [Google Scholar]
- (463).Dias A; Bouvier D; Crepin T; McCarthy AA; Hart DJ; Baudin F; Cusack S; Ruigrok RW The Cap-snatching Endonuclease of Influenza Virus Polymerase Resides in the PA Subunit. Nature 2009, 458, 914–918. [DOI] [PubMed] [Google Scholar]
- (464).Yuan PW; Bartlam M; Lou ZY; Chen SD; Zhou J; He XJ; Lv ZY; Ge RW; Li XM; Deng T; Fodor E; Rao ZH; Liu YF Crystal Structure of an Avian Influenza Polymerase PA(N) Reveals an Endonuclease Active Site. Nature 2009, 458, 909–912. [DOI] [PubMed] [Google Scholar]
- (465).Guilligay D; Tarendeau F; Resa-Infante P; Coloma R; Crepin T; Sehr P; Lewis J; Ruigrok RWH; Ortin J; Hart DJ; Cusack S The Structural Basis for Cap Binding by Influenza Virus Polymerase Subunit PB2. Nat. Struct. Mol. Biol 2008, 15, 500–506. [DOI] [PubMed] [Google Scholar]
- (466).Rao P; Yuan WM; Krug RM Crucial Role of CA cleavage Sites in the Cap-snatching Mechanism for Initiating Viral mRNA Synthesis. EMBO J 2003, 22, 1188–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (467).Kowalinski E; Zubieta C; Wolkerstorfer A; Szolar OHJ; Ruigrok RWH; Cusack S Structural Analysis of Specific Metal Chelating Inhibitor Binding to the Endonuclease Domain of Influenza pH1N1 (2009) Polymerase. PLoS Pathog. 2012, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (468).DuBois RM; Slavish PJ; Baughman BM; Yun MK; Bao J; Webby RJ; Webb TR; White SW Structural and Biochemical Basis for Development of Influenza Virus Inhibitors Targeting the PA Endonuclease. PLoS Pathog. 2012, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (469).Nakazawa M; Kadowaki SE; Watanabe I; Kadowaki Y; Takei M; Fukuda H PA Subunit of RNA Polymerase as a Promising Target for Anti-Influenza Virus Agents. Antiviral Res 2008, 78, 194–201. [DOI] [PubMed] [Google Scholar]
- (470).Sagong HY; Bauman JD; Patel D; Das K; Arnold E; LaVoie EJ Phenyl Substituted 4-Hydroxypyridazin-3(2H)-ones and 5-Hydroxypyrimidin-4(3H)-ones: Inhibitors of Influenza A Endonuclease. J. Med. Chem 2014, 57, 8086–8098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (471).Bauman JD; Patel D; Baker SF; Vijayan RSK; Xiang A; Parhi AK; Martinez-Sobrido L; LaVoie EJ; Das K; Arnold E Crystallographic Fragment Screening and Structure-Based Optimization Yields a New Class of Influenza Endonuclease Inhibitors. ACS Chem. Biol 2013, 8, 2501–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (472).Song JM; Lee KH; Seong BL Antiviral Effect of Catechins in Green Tea on Influenza Virus. Antiviral Res 2005, 68, 66–74. [DOI] [PubMed] [Google Scholar]
- (473).Kuzuhara T; Iwai Y; Takahashi H; Hatakeyama D; Echigo N Green Tea Catechins Inhibit the Endonuclease Activity of Influenza A Virus RNA Polymerase. PLoS Curr. 2009, 1, RRN1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (474).Iwai Y; Takahashi H; Hatakeyama D; Motoshima K; Ishikawa M; Sugita K; Hashimoto Y; Harada Y; Itamura S; Odagiri T; Tashiro M; Sei Y; Yamaguchi K; Kuzuhara T Anti-influenza Activity of Phenethylphenylphthalimide Analogs Derived From Thalidomide. Bioorg. Med. Chem 2010, 18, 5379–5390. [DOI] [PubMed] [Google Scholar]
- (475).Chen E; Swift RV; Alderson N; Feher VA; Feng GS; Amaro RE Computation-Guided Discovery of Influenza Endonuclease Inhibitors. ACS Med. Chem. Lett 2014, 5, 61–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (476).Carcelli M; Rogolino D; Gatti A; De Luca L; Sechi M; Kumar G; White SW; Stevaert A; Naesens L N-acylhydrazone Inhibitors of Influenza Virus PA Endonuclease with Versatile Metal Binding Modes. Sci. Rep 2016, 6, 31500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (477).Stevaert A; Nurra S; Pala N; Carcelli M; Rogolino D; Shepard C; Domaoal RA; Kim B; Alfonso-Prieto M; Marras SA; Sechi M; Naesens L An Integrated Biological Approach to Guide the Development of Metal-chelating Inhibitors of Influenza Virus PA Endonuclease. Mol. Pharmacol 2015, 87, 323–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (478).Tomassini J; Selnick H; Davies ME; Armstrong ME; Baldwin J; Bourgeois M; Hastings J; Hazuda D; Lewis J; McClements W; et al. Inhibition of Cap (m7GpppXm)-Dependent Endonuclease of Influenza Virus by 4-substituted 2,4-dioxobutanoic Acid Compounds. Antimicrob. Agents Chemother 1994, 38, 2827–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (479).Tomassini JE; Davies ME; Hastings JC; Lingham R; Mojena M; Raghoobar SL; Singh SB; Tkacz JS; Goetz MA A Novel Antiviral Agent which Inhibits the Endonuclease of Influenza Viruses. Antimicrob. Agents Chemother 1996, 40, 1189–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (480).Singh SB; Tomassini JE Synthesis of Natural Flutimide and Analogous Fully Substituted Pyrazine-2,6-diones, Endonuclease Inhibitors of Influenza Virus. J. Org. Chem 2001, 66, 5504–5516. [DOI] [PubMed] [Google Scholar]
- (481).Sagong HY; Parhi A; Bauman JD; Patel D; Vijayan RSK; Das K; Arnod E; LaVoie EJ 3-Hydroxyquinolin-2(1H)-ones As Inhibitors of Influenza A Endonuclease. ACS Med. Chem. Lett 2013, 4, 547–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (482).Parhi AK; Xiang A; Bauman JD; Patel D; Vijayan RSK; Das K; Arnold E; LaVoie EJ Phenyl Substituted 3-hydroxypyridin-2(1H)-ones: Inhibitors of Influenza A Endonuclease. Bioorg. Med. Chem 2013, 21, 6435–6446. [DOI] [PubMed] [Google Scholar]
- (483).Credille CV; Chen Y; Cohen SM Fragment-Based Identification of Influenza Endonuclease Inhibitors. J. Med. Chem 2016, 59, 6444–6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (484).Jones JC; Marathe BM; Lerner C; Kreis L; Gasser R; Pascua PNQ; Najera I; Govorkova EA A Novel Endonuclease Inhibitor Exhibits Broad-Spectrum Anti-Influenza Virus Activity In Vitro. Antimicrob. Agents Chemother 2016, 60, 5504–5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (485).Zoidis G; Giannakopoulou E; Stevaert A; Frakolaki E; Myrianthopoulos V; Fytas G; Mavromara P; Mikros E; Bartenschlager R; Vassilaki N; Naesens L Novel indole–flutimide Heterocycles with Activity Against Influenza PA Endonuclease and Hepatitis C Virus. MedChemComm 2016, 7, 447–456. [Google Scholar]
- (486).Heo YA Baloxavir: First Global Approval. Drugs 2018, 78, 693–697. [DOI] [PubMed] [Google Scholar]
- (487).Boswell-Smith V; Spina D; Page CP Phosphodiesterase Inhibitors. Br. J. Pharmacol 2006, 147, S252–S257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (488).Jeon YH; Heo Y-S; Kim CM; Hyun Y-L; Lee TG; Ro S; Cho JM Phosphodiesterase: Overview of Protein Structures, Potential Therapeutic Applications and Recent Progress in Drug Development. Cell. Mol. Life Sci 2005, 62, 1198–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (489).Mehats C; Andersen CB; Filopanti M; Jin SL; Conti M Cyclic Nucleotide Phosphodiesterases and their Role in Endocrine Cell Signaling. Trends Endocrinol. Metab 2002, 13, 29–35. [DOI] [PubMed] [Google Scholar]
- (490).Moustafa F; Feldman SR A Review of Phosphodiesterase-Inhibition and the Potential Role for Phosphodiesterase 4 Inhibitors in Clinical Dermatology. Dermatol. Online J 2014, 20, 22608. [PubMed] [Google Scholar]
- (491).Jin SL; Ding SL; Lin SC Phosphodiesterase 4 and its Inhibitors in Inflammatory Diseases. Chang Gung Med. J 2012, 35, 197–210. [DOI] [PubMed] [Google Scholar]
- (492).Wittmann M; Helliwell PS Phosphodiesterase 4 Inhibition in the Treatment of Psoriasis, Psoriatic Arthritis and Other Chronic Inflammatory Diseases. Dermatol. Ther 2013, 3, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (493).Xu RX; Rocque WJ; Lambert MH; Vanderwall DE; Luther MA; Nolte RT Crystal Structures of the Catalytic Domain of Phosphodiesterase 4B Complexed with AMP, 8-Br-AMP, and Rolipram. J. Mol. Biol 2004, 337, 355–365. [DOI] [PubMed] [Google Scholar]
- (494).Xu RX; Hassell AM; Vanderwall D; Lambert MH; Holmes WD; Luther MA; Rocque WJ; Milburn MV; Zhao Y; Ke H; Nolte RT Atomic Structure of PDE4: Insights into Phosphodiesterase Mechanism and Specificity. Science 2000, 288, 1822–1825. [DOI] [PubMed] [Google Scholar]
- (495).Freund YR; Akama T; Alley MRK; Antunes J; Dong C; Jarnagin K; Kimura R; Nieman JA; Maples KR; Plattner JJ; Rock F; Sharma R; Singh R; Sanders V; Zhou Y Boron-Based Phosphodiesterase Inhibitors show Novel Binding of Boron to PDE4 Bimetal Center. FEBS Lett 2012, 586, 3410–3414. [DOI] [PubMed] [Google Scholar]
- (496).Svensson F; Bender A; Bailey D Fragment-Based Drug Discovery of Phosphodiesterase Inhibitors. J. Med. Chem 2018, 61, 1415–1424. [DOI] [PubMed] [Google Scholar]
- (497).Zhu J; Mix E; Winblad B The Antidepressant and Antiinflammatory effects of Rolipram in the Central Nervous System. CNS Drug Rev 2001, 7, 387–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (498).Torphy TJ; Barnette MS; Underwood DC; Griswold DE; Christensen SB; Murdoch RD; Nieman RB; Compton CH ArifloTM(SB 207499), a Second Generation Phosphodiesterase 4 Inhibitor for the Treatment of Asthma and COPD: From Concept to Clinic. Pulm. Pharmacol. Ther 1999, 12, 131–135. [DOI] [PubMed] [Google Scholar]
- (499).Souness JE; Rao S Proposal for Pharmacologically Distinct Conformers of PDE4 Cyclic AMP Phosphodiesterases. Cell Signal 1997, 9, 227–236. [DOI] [PubMed] [Google Scholar]
- (500).Atta-Ur-Rahman; Reitz AB Frontiers in Medicinal Chemistry; Bentham Science Publishers Ltd, 2004. [Google Scholar]
- (501).Szmuszkovicz J; Gupta SP; Kundu B; Khare SK; Rastogi SK; Spence P; Barnette M S. Progress in Drug Research; Birkhäuser Verlag: Basel, 1999; Vol. 53. [Google Scholar]
- (502).Carzaniga L; Amari G; Rizzi A; Capaldi C; De Fanti R; Ghidini E; Villetti G; Carnini C; Moretto N; Facchinetti F; Caruso P; Marchini G; Battipaglia L; Patacchini R; Cenacchi V; Volta R; Amadei F; Pappani A; Capacchi S; Bagnacani V; Delcanale M; Puccini P; Catinella S; Civelli M; Armani E Discovery and Optimization of Thiazolidinyl and Pyrrolidinyl Derivatives as Inhaled PDE4 Inhibitors for Respiratory Diseases. J. Med. Chem 2017, 60, 10026–10046. [DOI] [PubMed] [Google Scholar]
- (503).Hatzelmann A; Schudt C Anti-Inflammatory and Immunomodulatory Potential of the Novel PDE4 Inhibitor Roflumilast in Vitro. J. Pharmacol. Exp. Ther 2001, 297, 267–279. [PubMed] [Google Scholar]
- (504).Schett G; Sloan VS; Stevens RM; Schafer P Apremilast: A Novel PDE4 Inhibitor in the Treatment of Autoimmune and Inflammatory Diseases. Ther. Adv. Musculoskelet. Dis 2010, 2, 271–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (505).Kleinman EF; Campbell E; Giordano LA; Cohan VL; Jenkinson TH; Cheng JB; Shirley JT; Pettipher ER; Salter ED; Hibbs TA; DiCapua FM; Bordner J Striking Effect of Hydroxamic Acid Substitution on the Phosphodiesterase Type 4 (PDE4) and TNFα Inhibitory Activity of Two Series of Rolipram Analogues: Implications for a New Active Site Model of PDE4. J. Med. Chem 1998, 41, 266–270. [DOI] [PubMed] [Google Scholar]
- (506).Wang H; Liu Y; Chen Y; Robinson H; Ke H Multiple Elements Jointly Determine Inhibitor Selectivity of Cyclic Nucleotide Phosphodiesterases 4 and 7. J. Biol. Chem 2005, 280, 30949–30955. [DOI] [PubMed] [Google Scholar]
- (507).Baker SJ; Zhang Y-K; Akama T; Lau A; Zhou H; Hernandez V; Mao W; Alley MRK; Sanders V; Plattner JJ Discovery of a New Boron-Containing Antifungal Agent, 5-Fluoro-1,3-dihydro-1-hydroxy-2,1-benzoxaborole (AN2690), for the Potential Treatment of Onychomycosis. J. Med. Chem 2006, 49, 4447–4450. [DOI] [PubMed] [Google Scholar]
- (508).Akama T; Baker SJ; Zhang YK; Hernandez V; Zhou H; Sanders V; Freund Y; Kimura R; Maples KR; Plattner JJ Discovery and Structure-Activity Study of a Novel Benzoxaborole Anti-Inflammatory Agent (AN2728) for the Potential Topical Treatment of Psoriasis and Atopic Dermatitis. Bioorg. Med. Chem. Lett 2009, 19, 2129–2132. [DOI] [PubMed] [Google Scholar]
- (509).Christianson DW Arginase: Structure, Mechanism, and Physiological Role in Male and Female Sexual Arousal. Acc. Chem. Res 2005, 38, 191–201. [DOI] [PubMed] [Google Scholar]
- (510).Bloch MJ Worldwide Prevalence of Hypertension Exceeds 1.3 Billion. J. Am. Soc. Hypertens 2016, 10, 753–754. [DOI] [PubMed] [Google Scholar]
- (511).Levy D; Larson MG; Vasan RS; Kannel WB; Ho KK The Progression from Hypertension to Congestive Heart Failure. JAMA, J. Am. Med. Assoc 1996, 275, 1557–1562. [PubMed] [Google Scholar]
- (512).Moser M; Hebert PR Prevention of Disease Progression, Left Ventricular Hypertrophy and Congestive Heart Failure in Hypertension Treatment Trials. J. Am. Coll. Cardiol 1996, 27, 1214–1218. [DOI] [PubMed] [Google Scholar]
- (513).Palano F; Paneni F; Sciarretta S; Tocci G; Volpe M The Progression from Hypertension to Congestive Heart Failure. Recenti Prog. Med 2011, 102, 461–467. [DOI] [PubMed] [Google Scholar]
- (514).Bicket DP Using ACE Inhibitors Appropriately. Am. Fam. Physician 2002, 66, 461–468. [PubMed] [Google Scholar]
- (515).Folkow B; Johansson B; Mellander S The Comparative Effects of Angiotensin and Noradrenaline on Consecutive Vascular Sections. Acta Physiol. Scand 1961, 53, 99–104. [DOI] [PubMed] [Google Scholar]
- (516).Saxena PR Interaction Between the Renin-angiotensin-aldosterone and Sympathetic Nervous Systems. J. Cardiovasc. Pharmacol 1992, 19, S80–88. [DOI] [PubMed] [Google Scholar]
- (517).Padfield PL; Morton JJ Effects of Angiotensin II on Arginine-vasopressin in Physiological and Pathological Situations in Man. J. Endocrinol 1977, 74, 251–259. [DOI] [PubMed] [Google Scholar]
- (518).Proud D; Kaplan AP Kinin Formation: Mechanisms and Role in Inflammatory Disorders. Annu. Rev. Immunol 1988, 6, 49–83. [DOI] [PubMed] [Google Scholar]
- (519).Vanhoutte PM Endothelium and Control of Vascular Function. State of the Art Lecture. Hypertension 1989, 13, 658–667. [DOI] [PubMed] [Google Scholar]
- (520).Stein JH; Congbalay RC; Karsh DL; Osgood RW; Ferris TF The Effect of Bradykinin on Proximal Tubular Sodium Reabsorption in the Dog: Evidence for Functional Nephron Heterogeneity. J. Clin. Invest 1972, 51, 1709–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (521).Nguyen Q; Dominguez J; Nguyen L; Gullapalli N Hypertension Management: an Update. Am. Health Drug Benefits 2010, 3, 47–56. [PMC free article] [PubMed] [Google Scholar]
- (522).Chrysant SG; Chrysant GS Pharmacological and Clinical Profile of Moexipril: a Concise Review. J. Clin. Pharmacol 2004, 44, 827–836. [DOI] [PubMed] [Google Scholar]
- (523).Antonios TF; MacGregor GA Angiotensin Converting Enzyme Inhibitors in Hypertension: Potential Problems. J. Hypertens 1995, 13, S11–16. [DOI] [PubMed] [Google Scholar]
- (524).Israili ZH; Hall WD Cough and Angioneurotic Edema Associated with Angiotensin-converting Enzyme Inhibitor Therapy. A Review of the Literature and Pathophysiology. Ann. Intern. Med 1992, 117, 234–242. [DOI] [PubMed] [Google Scholar]
- (525).Sica DA Angiotensin-Converting Enzyme Inhibitors Side Effects-Physiologic and Non-Physiologic Considerations. J. Clin. Hypertens 2004, 6, 410–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (526).Hanif K; Bid HK; Konwar R Reinventing the ACE Inhibitors: Some Old and New Implications of ACE Inhibition. Hypertens. Res 2010, 33, 11–21. [DOI] [PubMed] [Google Scholar]
- (527).Weber MA Vasopeptidase Inhibitors. Lancet 2001, 358, 1525–1532. [DOI] [PubMed] [Google Scholar]
- (528).Campbell DJ Vasopeptidase Inhibition: a Double-edged Sword? Hypertension 2003, 41, 383–389. [DOI] [PubMed] [Google Scholar]
- (529).Zaman MA; Oparil S; Calhoun DA Drugs Targeting the Renin-angiotensin-aldosterone System. Nat. Rev. Drug Discovery 2002, 1, 621–636. [DOI] [PubMed] [Google Scholar]
- (530).Esther CR Jr.; Marino EM; Bernstein KE The Role of Angiotensin-converting Enzyme in Blood Pressure Control, Renal Function, and Male Fertility. Trends Endocrinol. Metab 1997, 8, 181–186. [DOI] [PubMed] [Google Scholar]
- (531).Hagaman JR; Moyer JS; Bachman ES; Sibony M; Magyar PL; Welch JE; Smithies O; Krege JH; O’Brien DA Angiotensin-converting Enzyme and Male Fertility. Proc. Natl. Acad. Sci. U. S. A 1998, 95, 2552–2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (532).Acharya KR; Sturrock ED; Riordan JF; Ehlers MR Ace Revisited: a New Target for Structure-based Drug Design. Nat. Rev. Drug Discovery 2003, 2, 891–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (533).Wei L; Clauser E; Alhenc-Gelas F; Corvol P The Two Homologous Domains of Human Angiotensin I-Converting Enzyme Interact Differently with Competitive Inhibitors. J. Biol. Chem 1992, 267, 13398–13405. [PubMed] [Google Scholar]
- (534).Yates CJ; Masuyer G; Schwager SL; Akif M; Sturrock ED; Acharya KR Molecular and Thermodynamic Mechanisms of the Chloride-Dependent Human Angiotensin-I-converting Enzyme (ACE). J. Biol. Chem 2014, 289, 1798–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (535).Natesh R; Schwager SL; Sturrock ED; Acharya KR Crystal Structure of the Human Angiotensin-converting Enzyme-lisinopril Complex. Nature 2003, 421, 551–554. [DOI] [PubMed] [Google Scholar]
- (536).Junot C; Gonzales MF; Ezan E; Cotton J; Vazeux G; Michaud A; Azizi M; Vassiliou S; Yiotakis A; Corvol P; Dive V RXP 407, a Selective Inhibitor of the N-domain of Angiotensin I-converting Enzyme, Blocks in vivo the Degradation of Hemoregulatory Peptide Acetyl-Ser-Asp-Lys-Pro with No Effect on Angiotensin I Hydrolysis. J. Pharmacol. Exp. Ther 2001, 297, 606–611. [PubMed] [Google Scholar]
- (537).Georgiadis D; Beau F; Czarny B; Cotton J; Yiotakis A; Dive V Roles of the Two Active Sites of Somatic Angiotensin-converting Enzyme in the Cleavage of Angiotensin I and Bradykinin: Insights from Selective Inhibitors. Circ. Res 2003, 93, 148–154. [DOI] [PubMed] [Google Scholar]
- (538).Fuchs S; Xiao HD; Cole JM; Adams JW; Frenzel K; Michaud A; Zhao H; Keshelava G; Capecchi MR; Corvol P; Bernstein KE Role of the N-terminal Catalytic Domain of Angiotensin-converting Enzyme Investigated by Targeted Inactivation in Mice. J. Biol. Chem 2004, 279, 15946–15953. [DOI] [PubMed] [Google Scholar]
- (539).Ehlers MR; Fox EA; Strydom DJ; Riordan JF Molecular Cloning of Human Testicular Angiotensin-converting Enzyme: the Testis Isozyme is Identical to the C-Terminal Half of Rndothelial Angiotensin-Converting Enzyme. Proc. Natl. Acad. Sci. U. S. A 1989, 86, 7741–7745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (540).Ferreira SH A Bradykinin-Potentiating Factor (Bpf) Present in the Venom of Bothrops Jararca. Br. J. Pharmacol. Chemother 1965, 24, 163–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (541).Ferreira SH; Bartelt DC; Greene LJ Isolation of Bradykinin-Potentiating Peptides from Bothrops Jararaca Venom. Biochemistry 1970, 9, 2583–2593. [DOI] [PubMed] [Google Scholar]
- (542).Ondetti MA; Williams NJ; Sabo E; Pluscec J; Weaver ER; Kocy O Angiotensin-converting Enzyme Inhibitors from the Venom of Bothrops jararaca. Isolation, Elucidation of Structure, and Synthesis. Biochemistry 1971, 10, 4033–4039. [DOI] [PubMed] [Google Scholar]
- (543).Schaefer B Natural Products in the Chemical Industry; WILEY-VCH Verlag, 2014. [Google Scholar]
- (544).Newman DJ; Cragg GM; Snader KM The Influence of Natural Products Upon Drug Discovery. Nat. Prod. Rep 2000, 17, 215–234. [DOI] [PubMed] [Google Scholar]
- (545).Cushman DW; Cheung HS; Sabo EF; Ondetti MA Design of Potent Competitive Inhibitors of Angiotensin-converting Enzyme. Carboxyalkanoyl and Mercaptoalkanoyl Amino Acids. Biochemistry 1977, 16, 5484–5491. [DOI] [PubMed] [Google Scholar]
- (546).Ondetti MA; Rubin B; Cushman DW Design of Specific Inhibitors of Angiotensin-converting Enzyme: New Class of Orally Active Antihypertensive Agents. Science 1977, 196, 441–444. [DOI] [PubMed] [Google Scholar]
- (547).Waterfall JF A Review of the Preclinical Cardiovascular Pharmacology of Cilazapril, a New Angiotensin Converting Enzyme Inhibitor. Br. J. Clin. Pharmacol 1989, 27, S139–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (548).Smith CG; Vane JR The Discovery of Captopril. FASEB J 2003, 17, 788–789. [DOI] [PubMed] [Google Scholar]
- (549).Natesh R; Schwager SL; Evans HR; Sturrock ED; Acharya KR Structural Details on the Binding of Antihypertensive Drugs Captopril and Enalaprilat to Human Testicular Angiotensin I-converting Enzyme. Biochemistry 2004, 43, 8718–8724. [DOI] [PubMed] [Google Scholar]
- (550).Chevillard C; Jouquey S; Bree F; Mathieu MN; Stepniewski JP; Tillement JP; Hamon G; Corvol P Compared Properties of Trandolapril, Enalapril, and Their Diacid Metabolites. J. Cardiovasc. Pharmacol 1994, 23, S11–15. [PubMed] [Google Scholar]
- (551).Kaplan HR; Taylor DG; Olson SC Quinapril: Overview of Preclinical Data. Clin. Cardiol 1990, 13, 6–12. [DOI] [PubMed] [Google Scholar]
- (552).Edling O; Bao G; Feelisch M; Unger T; Gohlke P Moexipril, a New Angiotensin-converting Enzyme (ACE) Inhibitor: Pharmacological Characterization and Comparison with Enalapril. J. Pharmacol. Exp. Ther 1995, 275, 854–863. [PubMed] [Google Scholar]
- (553).Ceconi C; Francolini G; Olivares A; Comini L; Bachetti T; Ferrari R Angiotensin-converting Enzyme (ACE) Inhibitors Have Different Selectivity for Bradykinin Binding Sites of Human Somatic ACE. Eur. J. Pharmacol 2007, 577, 1–6. [DOI] [PubMed] [Google Scholar]
- (554).DeForrest JM; Waldron TL; Harvey C; Scalese B; Rubin B; Powell JR; Petrillo EW; Cushman DW Fosinopril, a Phosphinic Acid Inhibitor of Angiotensin I Converting Enzyme: in Vitro and Preclinical in Vivo Pharmacology. J. Cardiovasc. Pharmacol 1989, 14, 730–736. [DOI] [PubMed] [Google Scholar]
- (555).Jullien N; Makritis A; Georgiadis D; Beau F; Yiotakis A; Dive V Phosphinic Tripeptides as Dual Angiotensin-converting Enzyme C-domain and Endothelin-converting Enzyme-1 Inhibitors. J. Med. Chem 2010, 53, 208–220. [DOI] [PubMed] [Google Scholar]
- (556).Dive V; Cotton J; Yiotakis A; Michaud A; Vassiliou S; Jiracek J; Vazeux G; Chauvet MT; Cuniasse P; Corvol P RXP 407, a Phosphinic Peptide, is a Potent Inhibitor of Angiotensin I Converting Enzyme Able to Differentiate Between Its Two Active Sites. Proc. Natl. Acad. Sci. U. S. A 1999, 96, 4330–4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (557).Masuyer G; Akif M; Czarny B; Beau F; Schwager SL; Sturrock ED; Isaac RE; Dive V; Acharya KR Crystal Structures of Highly Specific Phosphinic Tripeptide Enantiomers in Complex with the Angiotensin-I Converting Enzyme. FEBS J 2014, 281, 943–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (558).Akif M; Schwager Sylva L.; Anthony Colin S.; Czarny B; Beau F; Dive V; Sturrock Edward D.; Acharya KR Novel Mechanism of Inhibition of Human Angiotensin-I-converting Enzyme (ACE) by a Highly Specific Phosphinic Tripeptide. Biochem. J 2011, 436, 53–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (559).Azizi M; Massien C; Michaud A; Corvol P In Vitro and in Vivo Inhibition of the 2 Active Sites of ACE by Omapatrilat, a Vasopeptidase Inhibitor. Hypertension 2000, 35, 1226–1231. [DOI] [PubMed] [Google Scholar]
- (560).Messerli FH; Nussberger J Vasopeptidase Inhibition and Angio-oedema. Lancet 2000, 356, 608–609. [DOI] [PubMed] [Google Scholar]
- (561).Rasoul S; Carretero OA; Peng H; Cavasin MA; Zhuo J; Sanchez-Mendoza A; Brigstock DR; Rhaleb NE Antifibrotic Effect of Ac-SDKP and Angiotensin-converting Enzyme Inhibition in Hypertension. J. Hypertens 2004, 22, 593–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (562).Rousseau A; Michaud A; Chauvet MT; Lenfant M; Corvol P The Hemoregulatory Peptide N-Acetyl-Ser-Asp-Lys-Pro is a Natural and Specific Substrate of the N-Terminal Active Site of Human Angiotensin-converting Enzyme. J. Biol. Chem 1995, 270, 3656–3661. [DOI] [PubMed] [Google Scholar]
- (563).Reiss K; Saftig P The “A Disintegrin And Metalloprotease” (ADAM) Family of Sheddases: Physiological and Cellular Functions. Semin. Cell. Dev. Biol 2009, 20, 126–137. [DOI] [PubMed] [Google Scholar]
- (564).Edwards DR; Handsley MM; Pennington CJ The ADAM Metalloproteinases. Mol. Aspects Med 2008, 29, 258–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (565).Howard L; Maciewicz RA; Blobel CP Cloning and Characterization of ADAM28: Evidence for Autocatalytic Pro-domain Removal and for Cell Surface Localization of Mature ADAM28. Biochem. J 2000, 348 Pt 1, 21–27. [PMC free article] [PubMed] [Google Scholar]
- (566).Groot AJ; Vooijs MA The Role of Adams in Notch Signaling. Adv. Exp. Med. Biol 2012, 727, 15–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (567).Saftig P; Reiss K The “A Disintegrin And Metalloproteases” ADAM10 and ADAM17: Novel Drug Targets with Therapeutic Potential? Eur. J. Cell Biol 2011, 90, 527–535. [DOI] [PubMed] [Google Scholar]
- (568).Georgiadis D; Yiotakis A Specific Targeting of Metzincin Family Members with Small-Molecule Inhibitors: Progress toward a Multifarious Challenge. Bioorg. Med. Chem 2008, 16, 8781–8794. [DOI] [PubMed] [Google Scholar]
- (569).Orth P; Reichert P; Wang W; Prosise WW; Yarosh-Tomaine T; Hammond G; Ingram RN; Xiao L; Mirza UA; Zou J; Strickland C; Taremi SS; Le HV; Madison V Crystal Structure of the Catalytic Domain of Human ADAM33. J. Mol. Biol 2004, 335, 129–137. [DOI] [PubMed] [Google Scholar]
- (570).Tallant C; García-Castellanos R; Baumann U; Gomis-Rüth FX On the Relevance of the Met-turn Methionine in Metzincins. J. Med. Chem 2010, 285, 13951–13957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (571).Duffy MJ; Mullooly M; O’Donovan N; Sukor S; Crown J; Pierce A; McGowan PM The ADAMs Family of Proteases: New Biomarkers and Therapeutic Targets for Cancer? Clin. Proteomics 2011, 8, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (572).Moss ML; Sklair-Tavron L; Nudelman R Drug Insight: Tumor Necrosis Factor-Converting Enzyme as a Pharmaceutical Target for Rheumatoid Arthritis. Nat. Clin. Pract. Rheumatol 2008, 4, 300–309. [DOI] [PubMed] [Google Scholar]
- (573).Xue C-B; Voss ME; Nelson DJ; Duan JJW; Cherney RJ; Jacobson IC; He X; Roderick J; Chen L; Corbett RL; Wang L; Meyer DT; Kennedy K; DeGrado WF; Hardman KD; Teleha CA; Jaffee BD; Liu R-Q; Copeland RA; Covington MB; Christ DD; Trzaskos JM; Newton RC; Magolda RL; Wexler RR; Decicco CP Design, Synthesis, and Structure−Activity Relationships of Macrocyclic Hydroxamic Acids That Inhibit Tumor Necrosis Factor α Release in Vitro and in Vivo. J. Med. Chem 2001, 44, 2636–2660. [DOI] [PubMed] [Google Scholar]
- (574).Ott GR; Asakawa N; Lu Z; Anand R; Liu RQ; Covington MB; Vaddi K; Qian M; Newton RC; Christ DD; Trzaskos JM; Duan JJ Potent, Exceptionally Selective, Orally Bioavailable Inhibitors of TNF-Alpha Converting Enzyme (TACE): Novel 2-Substituted-1H-benzo[d]imidazol-1-yl)methyl)benzamide P1’ Substituents. Bioorg. Med. Chem. Lett 2008, 18, 1577–1582. [DOI] [PubMed] [Google Scholar]
- (575).Sheppeck JE 2nd; Gilmore JL; Yang A; Chen XT; Xue CB; Roderick J; Liu RQ; Covington MB; Decicco CP; Duan JJ Discovery of Novel Hydantoins as Selective Non-Hydroxamate Inhibitors of Tumor Necrosis Factor-Alpha Converting Enzyme (TACE). Bioorg. Med. Chem. Lett 2007, 17, 1413–1417. [DOI] [PubMed] [Google Scholar]
- (576).Yu W; Tong L; Kim SH; Wong MK; Chen L; Yang DY; Shankar BB; Lavey BJ; Zhou G; Kosinski A; Rizvi R; Li D; Feltz RJ; Piwinski JJ; Rosner KE; Shih NY; Siddiqui MA; Guo Z; Orth P; Shah H; Sun J; Umland S; Lundell DJ; Niu X; Kozlowski JA Biaryl Substituted Hydantoin Compounds as TACE Inhibitors. Bioorg. Med. Chem. Lett 2010, 20, 5286–5289. [DOI] [PubMed] [Google Scholar]
- (577).Kamei N; Tanaka T; Kawai K; Miyawaki K; Okuyama A; Murakami Y; Arakawa Y; Haino M; Harada T; Shimano M Reverse Hydroxamate-Based Selective TACE Inhibitors. Bioorg. Med. Chem. Lett 2004, 14, 2897–2900. [DOI] [PubMed] [Google Scholar]
- (578).Bandarage UK; Wang T; Come JH; Perola E; Wei Y; Rao BG Novel Thiol-Based TACE Inhibitors, Part 2: Rational Design, Synthesis, and SAR of Thiol-Containing Aryl Sulfones. Bioorg. Med. Chem. Lett 2008, 18, 44–48. [DOI] [PubMed] [Google Scholar]
- (579).Park K; Gopalsamy A; Aplasca A; Ellingboe JW; Xu W; Zhang Y; Levin JI Synthesis and Activity of Tryptophan Sulfonamide Derivatives as Novel Non-Hydroxamate TNF-Alpha Converting Enzyme (TACE) Inhibitors. Bioorg. Med. Chem 2009, 17, 3857–3865. [DOI] [PubMed] [Google Scholar]
- (580).Guo Z; Orth P; Wong SC; Lavey BJ; Shih NY; Niu X; Lundell DJ; Madison V; Kozlowski JA Discovery of Novel Spirocyclopropyl Hydroxamate and Carboxylate Compounds as TACE Inhibitors. Bioorg. Med. Chem. Lett 2009, 19, 54–57. [DOI] [PubMed] [Google Scholar]
- (581).Li D; Popovici-Muller J; Belanger DB; Caldwell J; Dai C; David M; Girijavallabhan VM; Lavey BJ; Lee JF; Liu Z; Mazzola R; Rizvi R; Rosner KE; Shankar B; Spitler J; Ting PC; Vaccaro H; Yu W; Zhou G; Zhu Z; Niu X; Sun J; Guo Z; Orth P; Chen S; Kozlowski JA; Lundell DJ; Madison V; McKittrick B; Piwinski JJ; Shih NY; Shipps GW Jr.; Siddiqui MA; Strickland CO Structure and Activity Relationships of Tartrate-Based TACE Inhibitors. Bioorg. Med. Chem. Lett 2010, 20, 4812–4815. [DOI] [PubMed] [Google Scholar]
- (582).Dai C; Li D; Popovici-Muller J; Zhao L; Girijavallabhan VM; Rosner KE; Lavey BJ; Rizvi R; Shankar BB; Wong MK; Guo Z; Orth P; Strickland CO; Sun J; Niu X; Chen S; Kozlowski JA; Lundell DJ; Piwinski JJ; Shih NY; Siddiqui MA 2-(2-Aminothiazol-4-yl)Pyrrolidine-Based Tartrate Diamides as Potent, Selective and Orally Bioavailable TACE Inhibitors. Bioorg. Med. Chem. Lett 2011, 21, 3172–3176. [DOI] [PubMed] [Google Scholar]
- (583).Zhu Z; Mazzola R; Sinning L; McKittrick B; Niu X; Lundell D; Sun J; Orth P; Guo Z; Madison V; Ingram R; Beyer BM Discovery of Novel Hydroxamates as Highly Potent Tumor Necrosis Factor-Alpha Converting Enzyme Inhibitors, Part I: Discovery of Two Binding Modes. J. Med. Chem 2008, 51, 725–736. [DOI] [PubMed] [Google Scholar]
- (584).Zhou B-BS; Peyton M; He B; Liu C; Girard L; Caudler E; Lo Y; Baribaud F; Mikami I; Reguart N; Yang G; Li Y; Yao W; Vaddi K; Gazdar AF; Friedman SM; Jablons DM; Newton RC; Fridman JS; Minna JD; Scherle PA Targeting ADAM-Mediated Ligand Cleavage to Inhibit HER3 and EGFR Pathways in Non-Small Cell Lung Cancer. Cancer Cell 2006, 10, 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (585).Burns DM; He C; Li Y; Scherle P; Liu X; Marando CA; Covington MB; Yang G; Pan M; Turner S; Fridman JS; Hollis G; Vaddi K; Yeleswaram S; Newton R; Friedman S; Metcalf B; Yao W Conversion of an MMP-Potent Scaffold to an MMP-Selective HER-2 Sheddase Inhibitor via Scaffold Hybridization and Subtle P1’ Permutations. Bioorg. Med. Chem. Lett 2008, 18, 560–564. [DOI] [PubMed] [Google Scholar]
- (586).Qian M; Bai SA; Brogdon B; Wu JT; Liu RQ; Covington MB; Vaddi K; Newton RC; Fossler MJ; Garner CE; Deng Y; Maduskuie T; Trzaskos J; Duan JJ; Decicco CP; Christ DD Pharmacokinetics and Pharmacodynamics of DPC 333 ((2R)-2-((3R)-3-amino-3{4-[2-methyl-4-quinolinyl)methoxy]phenyl}-2-oxopyrrolidinyl)-N-hydroxy-4-methylpentanamide)), a Potent and Selective Inhibitor of Tumor Necrosis Factor Alpha-Converting Enzyme in Rodents, Dogs, Chimpanzees, and Humans. Drug Metab. Dispos 2007, 35, 1916–1925. [DOI] [PubMed] [Google Scholar]
- (587).Andreas L; Christian H; Millard HL; Neil B; Robert CA; Bickett DM; Leesnitzer MA; Becherer JD Metalloproteinase Inhibitors for the Disintegrin-Like Metalloproteinases ADAM10 and ADAM17 that Differentially Block Constitutive and Phorbol Ester-Inducible Shedding of Cell Surface Molecules. Comb. Chem. High Throughput Screening 2005, 8, 161–171. [DOI] [PubMed] [Google Scholar]
- (588).Herzog C; Haun RS; Ludwig A; Shah SV; Kaushal GP ADAM10 Is the Major Sheddase Responsible for the Release of Membrane-associated Meprin A. J. Biol. Chem 2014, 289, 13308–13322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (589).Niu X; Umland S; Ingram R; Beyer BM; Liu YH; Sun J; Lundell D; Orth P IK682, a Tight Binding Inhibitor of TACE. Arch. Biochem. Biophys 2006, 451, 43–50. [DOI] [PubMed] [Google Scholar]
- (590).Friedman S; Levy R; Garrett W; Doval D; Bondarde S; Sahoo T; Lokanatha D; Julka P; Shenoy K; Nagarkar R; Bhattacharyya G; Kumar K; Nag S; Mohan P; Narang N; Raghunadharao D; Walia M; Yao W; Li J; Emm T; Yeleswaram S; Scherle P; Newton R Clinical Benefit of INCB7839, a Potent and Selective Inhibitor of ADAM10 and ADAM17, in Combination with Trastuzumab in Metastatic HER2 Positive Breast Cancer Patients. Cancer Res 2009, 69, 5056. [Google Scholar]
- (591).Thabet MM; Huizinga TW Drug Evaluation: Apratastat, a Novel TACE/MMP Inhibitor for Rheumatoid Arthritis. Curr. Opin. Investig. Drugs 2006, 7, 1014–1019. [PubMed] [Google Scholar]
- (592).Zhang Y; Hegen M; Xu J; Keith JC; Jin G; Du X; Cummons T; Sheppard BJ; Sun L; Zhu Y; Rao VR; Wang Q; Xu W; Cowling R; Nickerson-Nutter CL; Gibbons J; Skotnicki J; Lin L-L; Levin J Characterization of (2R, 3S)-2-({[4-(2-butynyloxy)phenyl]sulfonyl}amino)-N,3-dihydroxybutanamide, a Potent and Selective Inhibitor of TNF-α Converting Enzyme. Int. J. Immunopharmacol 2004, 4, 1845–1857. [DOI] [PubMed] [Google Scholar]
- (593).Levin JI; Chen JM; Cheung K; Cole D; Crago C; Santos ED; Du X; Khafizova G; MacEwan G; Niu C; Salaski EJ; Zask A; Cummons T; Sung A; Xu J; Zhang Y; Xu W; Ayral-Kaloustian S; Jin G; Cowling R; Barone D; Mohler KM; Black RA; Skotnicki JS Acetylenic TACE Inhibitors, Part 1: SAR of the Acyclic Sulfonamide Hydroxamates. Bioorg. Med. Chem. Lett 2003, 13, 2799–2803. [DOI] [PubMed] [Google Scholar]
- (594).Zou J; Zhang R; Zhu F; Liu J; Madison V; Umland SP ADAM33 Enzyme Properties and Substrate Specificity. Biochemistry 2005, 44, 4247–4256. [DOI] [PubMed] [Google Scholar]
- (595).Sheppeck JE 2nd; Tebben A; Gilmore JL; Yang A; Wasserman ZR; Decicco CP; Duan JJ A Molecular Modeling Analysis of Novel Non-Hydroxamate Inhibitors of TACE. Bioorg. Med. Chem. Lett 2007, 17, 1408–1412. [DOI] [PubMed] [Google Scholar]
- (596).Yu W; Guo Z; Orth P; Madison V; Chen L; Dai C; Feltz RJ; Girijavallabhan VM; Kim SH; Kozlowski JA; Lavey BJ; Li D; Lundell D; Niu X; Piwinski JJ; Popovici-Muller J; Rizvi R; Rosner KE; Shankar BB; Shih NY; Siddiqui MA; Sun J; Tong L; Umland S; Wong MK; Yang DY; Zhou G Discovery and SAR of Hydantoin TACE Inhibitors. Bioorg. Med. Chem. Lett 2010, 20, 1877–1180. [DOI] [PubMed] [Google Scholar]
- (597).Rosner KE; Guo Z; Orth P; Shipps GW Jr.; Belanger DB; Chan TY; Curran PJ; Dai C; Deng Y; Girijavallabhan VM; Hong L; Lavey BJ; Lee JF; Li D; Liu Z; Popovici-Muller J; Ting PC; Vaccaro H; Wang L; Wang T; Yu W; Zhou G; Niu X; Sun J; Kozlowski JA; Lundell DJ; Madison V; McKittrick B; Piwinski JJ; Shih NY; Arshad Siddiqui M; Strickland CO The Discovery of Novel Tartrate-Based TNF-Alpha Converting Enzyme (TACE) Inhibitors. Bioorg. Med. Chem. Lett 2010, 20, 1189–1193. [DOI] [PubMed] [Google Scholar]
- (598).Gooz M ADAM-17: The Enzyme That Does It All. Crit. Rev. Biochem. Mol. Biol 2010, 45, 146–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (599).Dirckx JH Virgil on Anthrax. Am. J. Dermatopathol 1981, 3, 191–195. [DOI] [PubMed] [Google Scholar]
- (600).Dixon TC; Meselson M; Guillemin J; Hanna PC Anthrax. N. Engl. J. Med 1999, 341, 815–826. [DOI] [PubMed] [Google Scholar]
- (601).Koch R The Etiology of Anthrax, Based on the Life History of Bacilhs anthacis. Beitr. Biol. Pflanz 1876, 2, 277–310. [Google Scholar]
- (602).Meselson M; Guillemin J; Hughjones M; Langmuir A; Popova I; Shelokov A; Yampolskaya O The Sverdlovsk Anthrax Outbreak of 1979. Science 1994, 266, 1202–1208. [DOI] [PubMed] [Google Scholar]
- (603).Holty JEC; Bravata DM; Liu H; Olshen RA; McDonald KM; Owens DK Systematic Review: A Century of Inhalational Anthrax Cases from 1900 to 2005. Ann. Intern. Med 2006, 144, 270–280. [DOI] [PubMed] [Google Scholar]
- (604).Kumar BVSS; Malik S; Grandhi P; Dayam R; Sarma JARP Anthrax Lethal Factor Inhibitors as Potential Countermeasure of the Infection. Curr. Top. Med. Chem 2014, 14, 1977–1989. [DOI] [PubMed] [Google Scholar]
- (605).Goldberg AB; Turk BE Inhibitors of the Metalloproteinase Anthrax Lethal Factor. Curr. Top. Med. Chem 2016, 16, 2350–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (606).Liu SH; Zhang Y; Moayeri M; Liu J; Crown D; Fattah RJ; Wein AN; Yu ZX; Finkel T; Leppla SH Key Tissue Targets Responsible for Anthrax-Toxin-Induced Lethality. Nature 2013, 501, 63–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (607).Vitale G; Bernardi L; Napolitani G; Mock M; Montecucco C Susceptibility of Mitogen-Activated Protein Kinase Kinase Family Members to Proteolysis by Anthrax Lethal Factor. Biochem. J 2000, 352, 739–745. [PMC free article] [PubMed] [Google Scholar]
- (608).Vitale G; Pellizzari R; Recchi C; Napolitani G; Mock M; Montecucco C Anthrax Lethal Factor Cleaves the N-Terminus of MAPKKs and Induces Tyrosine/Threonine Phosphorylation of MAPKs in Cultured Macrophages. Biochem. Biophys. Res. Commun 1998, 248, 706–711. [DOI] [PubMed] [Google Scholar]
- (609).Duesbery NS; Webb CP; Leppla SH; Gordon VM; Klimpel KR; Copeland TD; Ahn NG; Oskarsson MK; Fukasawa K; Paull KD; Vande Woude GF Proteolytic Inactivation of MAP-Kinase-Kinase by Anthrax Lethal Factor. Science 1998, 280, 734–737. [DOI] [PubMed] [Google Scholar]
- (610).Pannifer AD; Wong TY; Schwarzenbacher R; Renatus M; Petosa C; Bienkowska J; Lacy DB; Collier RJ; Park S; Leppla SH; Hanna P; Liddington RC Crystal Structure of the Anthrax Lethal Factor. Nature 2001, 414, 229–233. [DOI] [PubMed] [Google Scholar]
- (611).Dal Molin F; Fasanella A; Simonato M; Garofolo G; Montecucco C; Tonello F Ratio of Lethal and Edema Factors in Rabbit Systemic Anthrax. Toxicon 2008, 52, 824–828. [DOI] [PubMed] [Google Scholar]
- (612).Mabry R; Brasky K; Geiger R; Carrion R; Hubbard GB; Leppla S; Patterson JL; Georgiou G; Iverson BL Detection of Anthrax Toxin in the Serum of Animals Infected with Bacillus anthracis by Using Engineered Immunoassays. Clin. Vaccine Immunol 2006, 13, 671–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (613).Sirard JC; Mock M; Fouet A The Three Bacillus anthracis Toxin Genes are Coordinately Regulated by Bicarbonate and Temperature. J. Bacteriol 1994, 176, 5188–5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (614).Tonello F; Naletto L; Romanello V; Dal Molin F; Montecucco C Tyrosine-728 and Glutamic Acid-735 are Essential for the Metalloproteolytic Activity of the Lethal Factor of Bacillus anthracis. Biochem. Biophys. Res. Commun 2004, 313, 496–502. [DOI] [PubMed] [Google Scholar]
- (615).Dalkas GA; Chasapis CT; Gkazonis PV; Bentrop D; Spyroulias GA Conformational Dynamics of the Anthrax Lethal Factor Catalytic Center. Biochemistry 2010, 49, 10767–10769. [DOI] [PubMed] [Google Scholar]
- (616).Tonello F; Seveso M; Marin O; Mock M; Montecucco C Pharmacology - Screening Inhibitors of Anthrax Lethal Factor. Nature 2002, 418, 386–386. [DOI] [PubMed] [Google Scholar]
- (617).Turk BE; Wong TY; Schwarzenbacher R; Jarrell ET; Leppla SH; Collier RJ; Liddington RC; Cantley LC The Structural Basis for Substrate and Inhibitor Selectivity of the Anthrax Lethal Factor. Nat. Struct. Mol. Biol 2004, 11, 60–66. [DOI] [PubMed] [Google Scholar]
- (618).Shoop WL; Xiong Y; Wiltsie J; Woods A; Guo J; Pivnichny JV; Felcetto T; Michael BF; Bansal A; Cummings RT; Cunningham BR; Friedlander AM; Douglas CM; Patel SB; Wisniewski D; Scapin G; Salowe SP; Zaller DM; Chapman KT; Scolnick EM; Schmatz DM; Bartizal K; MacCoss M; Hermes JD Anthrax Lethal Factor Inhibition. Proc. Natl. Acad. Sci. U. S. A 2005, 102, 7958–7963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (619).Calugi C; Trabocchi A; Lalli C; Guarna A D-Proline-Based Peptidomimetic Inhibitors of Anthrax Lethal Factor. Eur. J. Med. Chem 2012, 56, 96–107. [DOI] [PubMed] [Google Scholar]
- (620).Jiao GS; Kim S; Moayeri M; Crown D; Thai A; Cregar-Hernandez L; McKasson L; Sankaran B; Lehrer A; Wong T; Johns L; Margosiak SA; Leppla SH; Johnson AT Antidotes to Anthrax Lethal Factor Intoxication. Part 3: Evaluation of Core Structures and Further Modifications to the C2-Side Chain. Bioorg. Med. Chem. Lett 2012, 22, 2242–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (621).Johnson SL; Chen LH; Barile E; Emdadi A; Sabet M; Yuan HB; Wei J; Guiney D; Pellecchia M Structure-Activity Relationship Studies of a Novel Series of Anthrax Lethal Factor Inhibitors. Bioorg. Med. Chem 2009, 17, 3352–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (622).Kurbanov EK; Chiu TL; Solberg J; Francis S; Maize KM; Fernandez J; Johnson RL; Hawkinson JE; Walters MA; Finzel BC; Amin EA Probing the S2’ Subsite of the Anthrax Toxin Lethal Factor Using Novel N-Alkylated Hydroxamates. J. Med. Chem 2015, 58, 8723–8733. [DOI] [PubMed] [Google Scholar]
- (623).Jiao GS; Kim S; Moayeri M; Cregar-Hernandez L; McKasson L; Margosiak SA; Leppla SH; Johnson AT Antidotes to Anthrax Lethal Factor Intoxication. Part 1: Discovery of Potent Lethal Factor Inhibitors with In Vivo Efficacy. Bioorg. Med. Chem. Lett 2010, 20, 6850–6853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (624).Kim S; Jiao GS; Moayeri M; Crown D; Cregar-Hernandez L; McKasson L; Margosiak SA; Leppla SH; Johnson AT Antidotes to Anthrax Lethal Factor Intoxication. Part 2: Structural Modifications Leading to Improved in Vivo Efficacy. Bioorg. Med. Chem. Lett 2011, 21, 2030–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (625).Moayeri M; Crown D; Jiao GS; Kim S; Johnson A; Leysath C; Leppla SH Small-Molecule Inhibitors of Lethal Factor Protease Activity Protect against Anthrax Infection. Antimicrob. Agents Chemother 2013, 57, 4139–4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (626).Li F; Chvyrkova I; Terzyan S; Wakeham N; Turner R; Ghosh AK; Zhang XJC; Tang J Inhibition of Anthrax Lethal Factor: Lability of Hydroxamate as a Chelating Group. Appl. Microbiol. Biotechnol 2012, 94, 1041–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (627).Gaddis BD; Avramova LV; Chmielewski J Inhibitors of Anthrax Lethal Factor. Bioorg. Med. Chem. Lett 2007, 17, 4575–4578. [DOI] [PubMed] [Google Scholar]
- (628).Min DH; Tang WJ; Mrksich M Chemical Screening by Mass Spectrometry to Identify Inhibitors of Anthrax Lethal Factor. Nat. Biotechnol 2004, 22, 717–723. [DOI] [PubMed] [Google Scholar]
- (629).Lewis JA; Mongan J; McCammon JA; Cohen SM Evaluation and Binding-Mode Prediction of Thiopyrone-Based Inhibitors of Anthrax Lethal Factor. ChemMedChem 2006, 1, 694–697. [DOI] [PubMed] [Google Scholar]
- (630).Agrawal A; de Oliveira CAF; Cheng YH; Jacobsen JA; McCammon JA; Cohen SM Thioamide Hydroxypyrothiones Supersede Amide Hydroxypyrothiones in Potency against Anthrax Lethal Factor. J. Med. Chem 2009, 52, 1063–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (631).Dell’Aica I; Dona M; Tonello F; Piris A; Mock M; Montecucco C; Garbisa S Potent Inhibitors of Anthrax Lethal Factor from Green Tea. EMBO Rep 2004, 5, 418–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (632).Arnon SS; Schechter R; Inglesby TV; Henderson DA; Bartlett JG; Ascher MS; Eitzen E; Fine AD; Hauer J; Layton M; Lillibridge S; Osterholm MT; O’Toole T; Parker G; Perl TM; Russell PK; Swerdlow DL; Tonat K; Biodefense WGC Botulinum Toxin as a Biological Weapon - Medical and Public Health Management. JAMA, J. Am. Med. Assoc 2001, 285, 1059–1070. [DOI] [PubMed] [Google Scholar]
- (633).Kostrzewa RM; Kostrzewa RA; Kostrzewa JP Botulinum Neurotoxin: Progress in Negating its Neurotoxicity; and in Extending its Therapeutic Utility via Molecular Engineering. Mini Review. Peptides 2015, 72, 80–87. [DOI] [PubMed] [Google Scholar]
- (634).Erbguth FJ; Naumann M Historical Aspects of Botulinum Toxin - Justinus Kerner 1786–1862 and the “Sausage Poison.”. Neurology 1999, 53, 1850–1853. [DOI] [PubMed] [Google Scholar]
- (635).Blasi J; Chapman ER; Link E; Binz T; Yamasaki S; Decamilli P; Sudhof TC; Niemann H; Jahn R Botulinum Neurotoxin-a Selectively Cleaves the Synaptic Protein Snap-25. Nature 1993, 365, 160–163. [DOI] [PubMed] [Google Scholar]
- (636).Schiavo G; Benfenati F; Poulain B; Rossetto O; Delaureto PP; Dasgupta BR; Montecucco C Tetanus and Botulinum-B Neurotoxins Block Neurotransmitter Release by Proteolytic Cleavage of Synaptobrevin. Nature 1992, 359, 832–835. [DOI] [PubMed] [Google Scholar]
- (637).Mann JM; Martin S; Hoffman R; Marrazzo S Patient Recovery from Type-a Botulism - Morbidity Assessment Following a Large Outbreak. Am. J. Public Health 1981, 71, 266–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (638).Montecucco C; Molgo J Botulinal Neurotoxins: Revival of an Old Killer. Curr. Opin. Pharmacol 2005, 5, 274–279. [DOI] [PubMed] [Google Scholar]
- (639).Montal M Botulinum Neurotoxin: A Marvel of Protein Design. Annu. Rev. Biochem 2010, 79, 591–617. [DOI] [PubMed] [Google Scholar]
- (640).Krieglstein KG; Dasgupta BR; Henschen AH Covalent Structure of Botulinum Neurotoxin Type-a - Location of Sulfhydryl-Groups, and Disulfide Bridges and Identification of C-Termini of Light and Heavy-Chains. J. Protein Chem 1994, 13, 49–57. [DOI] [PubMed] [Google Scholar]
- (641).Schiavo G; Rossetto O; Santucci A; Dasgupta BR; Montecucco C Botulinum Neurotoxins are Zinc Proteins. J. Biol. Chem 1992, 267, 23479–23483. [PubMed] [Google Scholar]
- (642).Lacy DB; Tepp W; Cohen AC; DasGupta BR; Stevens RC Crystal Structure of Botulinum Neurotoxin Type A and Implications for Toxicity. Nat. Struct. Biol 1998, 5, 898–902. [DOI] [PubMed] [Google Scholar]
- (643).Eswaramoorthy S; Kumaran D; Swaminathan S A Novel Mechanism for Clostridium botulinum Neurotoxin Inhibition. Biochemistry 2002, 41, 9795–9802. [DOI] [PubMed] [Google Scholar]
- (644).Adler M; Nicholson JD; Starks DF; Kane CT; Cornille F; Hackley BE Evaluation of Phosphoramidon and Three Synthetic Phosphonates for Inhibition of Botulinum Neurotoxin B Catalytic Activity. J. Appl. Toxicol 1999, 19, S5–S11. [DOI] [PubMed] [Google Scholar]
- (645).Deshpande SS; Sheridan RE; Adler M A Study of Zinc-Dependent Metalloendopeptidase Inhibitors as Pharmacological Antagonists in Botulinum Neurotoxin Poisoning. Toxicon 1995, 33, 551–557. [DOI] [PubMed] [Google Scholar]
- (646).Park JG; Sill PC; Makiyi EF; Garcia-Sosa AT; Millard CB; Schmidt JJ; Pang YP Serotype-Selective, Small-Molecule Inhibitors of the Zinc Endopeptidase of Botulinum Neurotoxin Serotype A. Bioorg. Med. Chem 2006, 14, 395–408. [DOI] [PubMed] [Google Scholar]
- (647).Boldt GE; Kennedy JP; Janda KD Identification of a Potent Botulinum Neurotoxin A Protease Inhibitor Using In Situ Lead Identification Chemistry. Org. Lett 2006, 8, 1729–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (648).Boldt GE; Kennedy JP; Hixon MS; McAllister LA; Barbieri JT; Tzipori S; Janda KD Synthesis, Characterization and Development of a High-Throughput Methodology for the Discovery of Botulinum Neurotoxin A Inhibitors. J. Comb. Chem 2006, 8, 513–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (649).Capkova K; Hixon MS; McAllister LA; Janda KD Toward the Discovery of Potent Inhibitors of Botulinum Neurotoxin A: Development of a Robust LC MS Based Assay Operational from Low to Subnanomolar Enzyme Concentrations. Chem. Commun 2008, 3525–3527. [DOI] [PubMed] [Google Scholar]
- (650).Silhar P; Silvaggi NR; Pellett S; Capkova K; Johnson EA; Allen KN; Janda KD Evaluation of Adamantane Hydroxamates as Botulinum Neurotoxin Inhibitors: Synthesis, Crystallography, Modeling, Kinetic and Cellular Based Studies. Bioorg. Med. Chem 2013, 21, 1344–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (651).Pang YP; Vummenthala A; Mishra RK; Park JG; Wang SH; Davis J; Millard CB; Schmidt JJ Potent New Small-Molecule Inhibitor of Botulinum Neurotoxin Serotype A Endopeptidase Developed by Synthesis-Based Computer-Aided Molecular Design. PloS One 2009, 4, e7730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (652).Stowe GN; Silhar P; Hixon MS; Silvaggi NR; Allen KN; Moe ST; Jacobson AR; Barbieri JT; Janda KD Chirality Holds the Key for Potent Inhibition of the Botulinum Neurotoxin Serotype A Protease. Org. Lett 2010, 12, 756–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (653).Thompson AA; Jiao GS; Kim S; Thai A; Cregar-Hernandez L; Margosiak SA; Johnson AT; Han GW; O’Malley S; Stevens RC Structural Characterization of Three Novel Hydroxamate-Based Zinc Chelating Inhibitors of the Clostridium botulinum Serotype A Neurotoxin Light Chain Metalloprotease Reveals a Compact Binding Site Resulting from 60/70 Loop Flexibility. Biochemistry 2011, 50, 4019–4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (654).Schmidt JJ; Stafford RG; Bostian KA Type A Botulinum Neurotoxin Proteolytic Activity: Development of Competitive Inhibitors and Implications for Substrate Specificity at the S1’ Binding Subsite. FEBS Lett 1998, 435, 61–64. [DOI] [PubMed] [Google Scholar]
- (655).Schmidt JJ; Stafford RG A High-Affinity Competitive Inhibitor of Type A Botulinum Neurotoxin Protease Activity. FEBS Lett 2002, 532, 423–426. [DOI] [PubMed] [Google Scholar]
- (656).Moe ST; Thompson AB; Smith GM; Fredenburg RA; Stein RL; Jacobson AR Botulinum Neurotoxin Serotype A Inhibitors: Small-Molecule Mercaptoacetamide Analogs. Bioorg. Med. Chem 2009, 17, 3072–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (657).Jacobson AR; Adler M; Silvaggi NR; Allen KN; Smith GM; Fredenburg RA; Stein RL; Park JB; Feng XC; Shoemaker CB; Deshpande SS; Goodnough MC; Malizio CJ; Johnson EA; Pellett S; Tepp WH; Tzipori S Small Molecule Metalloprotease Inhibitor with In Vitro, Ex Vivo and In Vivo Efficacy Against Botulinum Neurotoxin Serotype A. Toxicon 2017, 137, 36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (658).Roxas-Duncan V; Enyedy I; Montgomery VA; Eccard VS; Carrington MA; Lai HG; Gul N; Yang DCH; Smith LA Identification and Biochemical Characterization of Small-Molecule Inhibitors of Clostridium botulinum Neurotoxin Serotype A. Antimicrob. Agents Chemother 2009, 53, 3478–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (659).Bremer PT; Adler M; Phung CH; Singh AK; Janda KD Newly Designed Quinolinol Inhibitors Mitigate the Effects of Botulinum Neurotoxin A in Enzymatic, Cell-Based, and ex Vivo Assays. J. Med. Chem 2017, 60, 338–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (660).Simpson LL; Coffield JA; Bakry N Chelation of Zinc Antagonizes the Neuromuscular Blocking Properties of the 7 Serotypes of Botulinum Neurotoxin as Well as Tetanus Toxin. J. Pharmacol. Exp. Ther 1993, 267, 720–727. [PubMed] [Google Scholar]
- (661).Eubanks LM; Silhar P; Salzameda NT; Zakhari JS; Feng XC; Barbieri JT; Shoemaker CB; Hixon MS; Janda KD Identification of a Natural Product Antagonist against the Botulinum Neurotoxin Light Chain Protease. ACS Med. Chem. Lett 2010, 1, 268–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (662).Silvaggi NR; Boldt GE; Hixon MS; Kennedy JP; Tzipori S; Janda KD; Allen KN Structures of Clostridium Botulinum Neurotoxin Serotype A Light Chain Complexed with Small-Molecule Inhibitors Highlight Active-Site Flexibility. Chem. Biol 2007, 14, 533–542. [DOI] [PubMed] [Google Scholar]
- (663).Eubanks LM; Hixon MS; Jin W; Hong S; Clancy CM; Tepp WH; Baldwin MR; Malizio CJ; Goodnough MC; Barbieri JT; Johnson EA; Boger DL; Dickerson TJ; Janda KD An In Vitro and In Vivo Disconnect Uncovered through High-Throughput Identification of Botulinum Neurotoxin A Antagonists. Proc. Natl. Acad. Sci. U. S. A 2007, 104, 2602–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (664).Fu ZJ; Chen S; Baldwin MR; Boldt GE; Crawford A; Janda KD; Barbieri JT; Kim JJP Light Chain of Botulinum Neurotoxin Serotype A: Structural Resolution of a Catalytic Intermediate. Biochemistry 2006, 45, 8903–8911. [DOI] [PubMed] [Google Scholar]
- (665).Lai HG; Feng MH; Roxas-Duncan V; Dakshanamurthy S; Smith LA; Yang DCH Quinolinol and Peptide Inhibitors of Zinc Protease in Botulinum Neurotoxin A: Effects of Zinc Ion and Peptides on Inhibition. Arch. Biochem. Biophys 2009, 491, 75–84. [DOI] [PubMed] [Google Scholar]
- (666).Caglic D; Krutein MC; Bompiani KM; Barlow DJ; Benoni G; Pelletier JC; Reitz AB; Lairson LL; Houseknecht KL; Smith GR; Dickerson TJ Identification of Clinically Viable Quinolinol Inhibitors of Botulinum Neurotoxin A Light Chain. J. Med. Chem 2014, 57, 669–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (667).Harrell WA; Vieira RC; Ensel SM; Montgomery V; Guernieri R; Eccard VS; Campbell Y; Roxas-Duncan V; Cardellina JH; Webb RP; Smith LA A Matrix-Focused Structure-Activity and Binding Site Flexibility Study of Quinolinol Inhibitors of Botulinum Neurotoxin Serotype A. Bioorg. Med. Chem. Lett 2017, 27, 675–678. [DOI] [PubMed] [Google Scholar]
- (668).Whitemarsh RCM; Tepp WH; Johnson EA; Pellett S Persistence of Botulinum Neurotoxin A Subtypes 1–5 in Primary Rat Spinal Cord Cells. PLoS One 2014, 9, e90252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (669).Burnett JC; Ruthel G; Stegmann CM; Panchal RG; Nguyen TL; Hermone AR; Stafford RG; Lane DJ; Kenny TA; McGrath CF; Wipf P; Stahl AM; Schmidt JJ; Gussio R; Brunger AT; Bavari S Inhibition of Metalloprotease Botulinum Serotype A from a Pseudo-Peptide Binding Mode to a Small Molecule that is Active in Primary Neurons. J. Biol. Chem 2007, 282, 5004–5014. [DOI] [PubMed] [Google Scholar]
- (670).Silhar P; Lardy MA; Hixon MS; Shoemaker CB; Barbieri JT; Struss AK; Lively JM; Javor S; Janda KD C-Terminus of Botulinum A Protease Has Profound and Unanticipated Kinetic Consequences upon the Catalytic Cleft. ACS Med. Chem. Lett 2013, 4, 283–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (671).Kumaran D; Adler M; Levit M; Krebs M; Sweeney R; Swaminathan S Interactions of a Potent Cyclic Peptide Inhibitor with the Light Chain of Botulinum Neurotoxin A: Insights from X-ray Crystallography. Bioorg. Med. Chem 2015, 23, 7264–7273. [DOI] [PubMed] [Google Scholar]
- (672).Zuniga JE; Hammill JT; Drory O; Nuss JE; Burnett JC; Gussio R; Wipf P; Bavari S; Brunger AT Iterative Structure-Based Peptide-Like Inhibitor Design against the Botulinum Neurotoxin Serotype A. PloS One 2010, 5, e11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (673).Cai S; Singh BR Strategies to Design Inhibitors of Clostridium botulinum Neurotoxins. Infect. Disord.: Drug Targets 2007, 7, 47–57. [DOI] [PubMed] [Google Scholar]
- (674).Mesaros N; Nordmann P; Plesiat P; Roussel-Delvallez M; Van Eldere J; Glupczynski Y; Van Laethem Y; Jacobs F; Lebecque P; Malfroot A; Tulkens PM; Van Bambeke F Pseudomonas Aeruginosa: Resistance and Therapeutic Options at the Turn of the New Millennium. Clin. Microbiol. Infect 2007, 13, 560–578. [DOI] [PubMed] [Google Scholar]
- (675).Cohen TS; Prince A Cystic Fibrosis: a Mucosal Immunodeficiency Cyndrome. Nat. Med 2012, 18, 509–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (676).Clatworthy AE; Pierson E; Hung DT Targeting Virulence: a New Paradigm for Antimicrobial Therapy. Nat. Chem. Biol 2007, 3, 541–548. [DOI] [PubMed] [Google Scholar]
- (677).Johnson DA; Carter-Hamm B; Dralle WM Inactivation of Human Bronchial Mucosal Proteinase Inhibitor by Pseudomonas aeruginosa Elastase. Am. Rev. Respir. Dis 1982, 126, 1070–1073. [DOI] [PubMed] [Google Scholar]
- (678).Leidal KG; Munson KL; Johnson MC; Denning GM Metalloproteases from Pseudomonas aeruginosa Degrade Human RANTES, MCP-1, and ENA-78. J. Interferon Cytokine Res 2003, 23, 307–318. [DOI] [PubMed] [Google Scholar]
- (679).Lai S; Tremblay J; Deziel E Swarming Motility: a Multicellular Behaviour Conferring Antimicrobial Resistance. Environ. Microbiol 2009, 11, 126–136. [DOI] [PubMed] [Google Scholar]
- (680).Kamath S; Kapatral V; Chakrabarty AM Cellular Function of Elastase in Pseudomonas aeruginosa: Role in the Cleavage of Nucleoside Diphosphate Kinase and in Alginate Synthesis. Mol. Microbiol 1998, 30, 933–941. [DOI] [PubMed] [Google Scholar]
- (681).Prosser BL; Taylor D; Dix BA; Cleeland R Method of Evaluating Effects of Antibiotics on Bacterial Biofilm. Antimicrob. Agents Chemother 1987, 31, 1502–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (682).Xu KD; McFeters GA; Stewart PS Biofilm Resistance to Antimicrobial Agents. Microbiology 2000, 146, 547–549. [DOI] [PubMed] [Google Scholar]
- (683).Mah TF; O’Toole GA Mechanisms of Biofilm Resistance to Antimicrobial Agents. Trends Microbiol 2001, 9, 34–39. [DOI] [PubMed] [Google Scholar]
- (684).Costerton JW; Lewandowski Z; Caldwell DE; Korber DR; Lappin-Scott HM Microbial Biofilms. Annu. Rev. Microbiol 1995, 49, 711–745. [DOI] [PubMed] [Google Scholar]
- (685).Kessler E; Israel M; Landshman N; Chechick A; Blumberg S In Vitro Inhibition of Pseudomonas aeruginosa Elastase by Metal-chelating Peptide Derivatives. Infect. Immun 1982, 38, 716–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (686).Burns FR; Paterson CA; Gray RD; Wells JT Inhibition of Pseudomonas aeruginosa Elastase and Pseudomonas Keratitis Using a Thiol-based Peptide. Antimicrob. Agents Chemother 1990, 34, 2065–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (687).Spierer A; Kessler E The Effect of 2-Mercaptoacetyl-L-phenylalanyl-L-leucine, a Specific Inhibitor of Pseudomonas aeruginosa Elastase, on Experimental Pseudomonas keratitis in Rabbit Eyes. Curr. Eye Res 1984, 3, 645–650. [DOI] [PubMed] [Google Scholar]
- (688).Kessler E; Spierer A; Blumberg S Specific Inhibition of Pseudomonas aeruginosa Elastase Injected Intracorneally in Rabbit Eyes. Invest. Ophthalmol. Visual Sci 1983, 24, 1093–1097. [PubMed] [Google Scholar]
- (689).Sokol PA; Kooi C; Hodges RS; Cachia P; Woods DE Immunization with a Pseudomonas aeruginosa Elastase Peptide Reduces Severity of Experimental Lung Infections due to P. aeruginosa or Burkholderia cepacia. J. Infect. Dis 2000, 181, 1682–1692. [DOI] [PubMed] [Google Scholar]
- (690).Thayer MM; Flaherty KM; McKay DB Three-dimensional Structure of the Elastase of Pseudomonas aeruginosa at 1.5-A resolution. J. Biol. Chem 1991, 266, 2864–2871. [DOI] [PubMed] [Google Scholar]
- (691).Matthews BW; Colman PM; Jansonius JN; Titani K; Walsh KA; Neurath H Structure of Thermolysin. Nature (London), New Biol 1972, 238, 41–43. [DOI] [PubMed] [Google Scholar]
- (692).Cathcart GR; Quinn D; Greer B; Harriott P; Lynas JF; Gilmore BF; Walker B Novel Inhibitors of the Pseudomonas aeruginosa Virulence Factor LasB: a Potential Therapeutic Approach for the Attenuation of Virulence Mechanisms in Pseudomonal Infection. Antimicrob. Agents Chemother 2011, 55, 2670–2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (693).Cathcart GR; Gilmore BF; Greer B; Harriott P; Walker B Inhibitor Profiling of the Pseudomonas aeruginosa Virulence Factor LasB using N-Alpha Mercaptoamide Template-based Inhibitors. Bioorg. Med. Chem. Lett 2009, 19, 6230–6232. [DOI] [PubMed] [Google Scholar]
- (694).Zhu J; Cai X; Harris TL; Gooyit M; Wood M; Lardy M; Janda KD Disarming Pseudomonas aeruginosa Virulence Factor LasB by Leveraging a Caenorhabditis Elegans Infection Model. Chem. Biol 2015, 22, 483–491. [DOI] [PubMed] [Google Scholar]
- (695).Garner AL; Struss AK; Fullagar JL; Agrawal A; Moreno AY; Cohen SM; Janda KD 3-Hydroxy-1-alkyl-2-methylpyridine-4(1H)-thiones: Inhibition of the Pseudomonas aeruginosa Virulence Factor LasB. ACS Med. Chem. Lett 2012, 3, 668–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (696).Fullagar JL; Garner AL; Struss AK; Day JA; Martin DP; Yu J; Cai X; Janda KD; Cohen SM Antagonism of a Zinc Metalloprotease Using a Unique Metal-chelating Scaffold: Tropolones as Inhibitors of P. aeruginosa Elastase. Chem. Commun 2013, 49, 3197–3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (697).Agrawal A; Johnson SL; Jacobsen JA; Miller MT; Chen LH; Pellecchia M; Cohen SM Chelator Fragment Libraries for Targeting Metalloproteinases. ChemMedChem 2010, 5, 195–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (698).Folkman J Seminars in Medicine of the Beth Israel Hospital, Boston. Clinical Applications of Research on Angiogenesis. N. Engl. J. Med 1995, 333, 1757–1763. [DOI] [PubMed] [Google Scholar]
- (699).Teicher BA; Holden SA; Ara G; Sotomayor EA; Huang ZD; Chen YN; Brem H Potentiation of Cytotoxic Cancer Therapies by Tnp-470 Alone and with Other Anti-Angiogenic Agents. Int. J. Cancer 1994, 57, 920–925. [DOI] [PubMed] [Google Scholar]
- (700).Sin N; Meng LH; Wang MQW; Wen JJ; Bornmann WG; Crews CM The Anti-Angiogenic Agent Fumagillin Covalently Binds and Inhibits the Methionine Aminopeptidase, MetAP-2. Proc. Natl. Acad. Sci. U. S. A 1997, 94, 6099–6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (701).Giglione C; Boularot A; Meinnel T Protein N-terminal Methionine Excision. Cell. Mol. Life Sci 2004, 61, 1455–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (702).Cao Y Angiogenesis Modulates Adipogenesis and Obesity. J. Clin. Invest 2007, 117, 2362–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (703).Chang SY; McGary EC; Chang S Methionine Aminopeptidase Gene of Escherichia coli is Essential for Cell Growth. J. Bacteriol 1989, 171, 4071–4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (704).Miller CG; Kukral AM; Miller JL; Movva NR PepM is an Essential Gene in Salmonella typhimurium. J. Bacteriol 1989, 171, 5215–5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (705).Bazan JF; Weaver LH; Roderick SL; Huber R; Matthews BW Sequence and Structure Comparison Suggest That Methionine Aminopeptidase, Prolidase, Aminopeptidase-P, and Creatinase Share a Common Fold. Proc. Natl. Acad. Sci. U. S. A 1994, 91, 2473–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (706).Liu S; Widom J; Kemp CW; Crews CM; Clardy J Structure of Human Methionine Aminopeptidase-2 Complexed with Fumagillin. Science 1998, 282, 1324–1327. [DOI] [PubMed] [Google Scholar]
- (707).Arfin SM; Kendall RL; Hall L; Weaver LH; Stewart AE; Matthews BW; Bradshaw RA Eukaryotic Methionyl Aminopeptidases - 2 Classes of Cobalt-Dependent Enzymes. Proc. Natl. Acad. Sci. U. S. A 1995, 92, 7714–7718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (708).Bradshaw RA; Brickey WW; Walker KW N-terminal Processing: The Methionine Aminopeptidase and N Alpha-Acetyl Transferase Families. Trends Biochem. Sci 1998, 23, 263–267. [DOI] [PubMed] [Google Scholar]
- (709).Lowther WT; Orville AM; Madden DT; Lim S; Rich DH; Matthews DW Escherichia coli Methionine Aminopeptidase: Implications of Crystallographic Analyses of the Native, Mutant, and Inhibited Enzymes for the Mechanism of Catalysis. Biochemistry 1999, 38, 7678–7688. [DOI] [PubMed] [Google Scholar]
- (710).Ma ZQ; Xie SX; Huang QQ; Nan FJ; Hurley TD; Ye QZ Structural Analysis of Inhibition of E-coli Methionine Aminopeptidase: Implication of Loop Adaptability in Selective Inhibition of Bacterial Enzymes. BMC Struct. Biol 2007, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (711).Ye QZ; Xie SX; Ma ZQ; Huang M; Hanzlik RP Structural Basis of Catalysis by Monometalated Methionine Aminopeptidase. Proc. Natl. Acad. Sci. U. S. A 2006, 103, 9470–9475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (712).Lowther WT; Matthews BW Metalloaminopeptidases: Common Functional Themes in Disparate Structural Surroundings. Chem. Rev 2002, 102, 4581–4607. [DOI] [PubMed] [Google Scholar]
- (713).Benbassat A; Bauer K; Chang SY; Myambo K; Boosman A; Chang S Processing of the Initiation Methionine from Proteins - Properties of the Escherichia-Coli Methionine Aminopeptidase and Its Gene Structure. J. Bacteriol 1987, 169, 751–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (714).Wang JY; Sheppard GS; Lou PP; Kawai M; Park C; Egan DA; Schneider A; Bouska J; Lesniewski R; Henkin J Physiologically Relevant Metal Cofactor for Methionine Aminopeptidase-2 is Manganese. Biochemistry 2003, 42, 5035–5042. [DOI] [PubMed] [Google Scholar]
- (715).D’souza VM; Holz RC The Methionyl Aminopeptidase from Escherichia coli can Function as an Iron(II) Enzyme. Biochemistry 1999, 38, 11079–11085. [DOI] [PubMed] [Google Scholar]
- (716).Ingber D; Fujita T; Kishimoto S; Sudo K; Kanamaru T; Brem H; Folkman J Synthetic Analogues of Fumagillin that Inhibit Angiogenesis and Suppress Tumour Growth. Nature 1990, 348, 555–557. [DOI] [PubMed] [Google Scholar]
- (717).Griffith EC; Su Z; Niwayama S; Ramsay CA; Chang YH; Liu JO Molecular Recognition of Angiogenesis Inhibitors Fumagillin and Ovalicin by Methionine Aminopeptidase 2. Proc. Natl. Acad. Sci. U. S. A 1998, 95, 15183–15188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (718).Kruger EA; Figg WD TNP-470: An Angiogenesis Inhibitor in Clinical Development for Cancer. Expert Opin. Invest. Drugs 2000, 9, 1383–1396. [DOI] [PubMed] [Google Scholar]
- (719).McCandless SE; Yanovski JA; Miller J; Fu C; Bird LM; Salehi P; Chan CL; Stafford D; Abuzzahab MJ; Viskochil D; Barlow SE; Angulo M; Myers SE; Whitman BY; Styne D; Roof E; Dykens EM; Scheimann AO; Malloy J; Zhuang DL; Taylor K; Hughes TE; Kim DD; Butler MG Effects of MetAP2 Inhibition on Hyperphagia and Body Weight in Prader-Willi Syndrome: A Randomized, Double-blind, Placebo-controlled Trial. Diabetes, Obes. Metab 2017, 19, 1751–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (720).Thale Z; Kinder FR; Bair KW; Bontempo J; Czuchta AM; Versace RW; Phillips PE; Sanders ML; Wattanasin S; Crews P Bengamides Revisited: New Structures and Antitumor Studies. J. Org. Chem 2001, 66, 1733–1741. [DOI] [PubMed] [Google Scholar]
- (721).Towbin H; Bair KW; DeCaprio JA; Eck MJ; Kim S; Kinder FR; Morollo A; Mueller DR; Schindler P; Song HK; van Oostrum J; Versace RW; Voshol H; Wood J; Zabludoff S; Phillips PE Proteomics-Based Target Identification: Bengamides as a New Class of Methionine Aminopeptidase Inhibitors. J. Biol. Chem 2003, 278, 52964–52971. [DOI] [PubMed] [Google Scholar]
- (722).Dumez H; Gall H; Capdeville R; Dutreix C; van Oosterom AT; Giaccone G A Phase I and Pharmacokinetic Study of LAF389 Administered to Patients with Advanced Cancer. Anticancer Drugs 2007, 18, 219–225. [DOI] [PubMed] [Google Scholar]
- (723).Umezawa H; Aoyagi T; Suda H; Hamada M; Takeuchi T Bestatin, an Inhibitor of Aminopeptidase-B, Produced by Actinomycetes. J. Antibiot 1976, 29, 97–99. [DOI] [PubMed] [Google Scholar]
- (724).Sheppard GS; Wang JY; Kawai M; BaMaung NY; Craig RA; Erickson SA; Lynch L; Patel J; Yang F; Searle XB; Lou PP; Park C; Kim KH; Henkin J; Lesniewski R 3-Amino-2-hydroxyamides and Related Compounds as Inhibitors of Methionine Aminopeptidase-2. Bioorg. Med. Chem. Lett 2004, 14, 865–868. [DOI] [PubMed] [Google Scholar]
- (725).Wang J; Sheppard GS; Lou P; Kawai M; BaMaung N; Erickson SA; Tucker-Garcia L; Park C; Bouska J; Wang YC; Frost D; Tapang P; Albert DH; Morgan SJ; Morowitz M; Shusterman S; Maris JM; Lesniewski R; Henkin J Tumor Suppression by a Rationally Designed Reversible Inhibitor of Methionine Aminopeptidase-2. Cancer Res 2003, 63, 7861–7869. [PubMed] [Google Scholar]
- (726).Hu XB; Zhu JG; Srivathsan S; Pei DH Peptidyl Hydroxamic Acids as Methionine Aminopeptidase Inhibitors. Bioorg. Med. Chem. Lett 2004, 14, 77–79. [DOI] [PubMed] [Google Scholar]
- (727).Kallander LS; Lu Q; Chen W; Tomaszek T; Yang G; Tew D; Meek TD; Hofmann GA; Schulz-Pritchard CK; Smith WW; Janson CA; Ryan MD; Zhang GF; Johanson KO; Kirkpatrick RB; Ho TF; Fisher PW; Mattern MR; Johnson RK; Hansbury MJ; Winkler JD; Ward KW; Veber DF; Thompson SK 4-Aryl-1,2,3-triazole: A Novel Template for a Reversible Methionine Aminopeptidase 2 Inhibitor, Optimized to Inhibit Angiogenesis In Vivo. J. Med. Chem 2005, 48, 5644–5647. [DOI] [PubMed] [Google Scholar]
- (728).Marino JP Jr.; Fisher PW; Hofmann GA; Kirkpatrick RB; Janson CA; Johnson RK; Ma C; Mattern M; Meek TD; Ryan MD; Schulz C; Smith WW; Tew DG; Tomazek TA Jr.; Veber DF; Xiong WC; Yamamoto Y; Yamashita K; Yang G; Thompson SK Highly Potent Inhibitors of Methionine Aminopeptidase-2 Based on a 1,2,4-triazole Pharmacophore. J. Med. Chem 2007, 50, 3777–3785. [DOI] [PubMed] [Google Scholar]
- (729).Kawai M; BaMaung NY; Fidanze SD; Erickson SA; Tedrow JS; Sanders WJ; Vasudevan A; Park C; Hutchins C; Comess KM; Kalvin D; Wang J; Zhang Q; Lou P; Tucker-Garcia L; Bouska J; Bell RL; Lesniewski R; Henkin J; Sheppard GS Development of Sulfonamide Compounds as Potent Methionine Aminopeptidase Type II Inhibitors with Antiproliferative Properties. Bioorg. Med. Chem. Lett 2006, 16, 3574–3577. [DOI] [PubMed] [Google Scholar]
- (730).Sheppard GS; Wang JY; Kawai M; Fidanze SD; BaMaung NY; Erickson SA; Barnes DM; Tedrow JS; Kolaczkowski L; Vasudevan A; Park DC; Wang GT; Sanders WJ; Mantei RA; Palazzo F; Tucker-Garcia L; Lou PP; Zhang Q; Park CH; Kim KH; Petros A; Olejniczak E; Nettesheim D; Hajduk P; Henkin J; Lesniewski R; Davidsen SK; Bell RL Discovery and Optimization of Anthranilic Acid Sulfonamides as Inhibitors of Methionine Aminopeptidase-2: A Structural Basis for the Reduction of Albumin Binding. J. Med. Chem 2006, 49, 3832–3849. [DOI] [PubMed] [Google Scholar]
- (731).Cheruvallath Z; Tang M; McBride C; Komandla M; Miura J; Ton-Nu T; Erikson P; Feng J; Farrell P; Lawson JD; Vanderpool D; Wu Y; Dougan DR; Plonowski A; Holub C; Larson C Discovery of Potent, Reversible MetAP2 Inhibitors Via Fragment Based Drug Discovery and Structure Based Drug Design-Part 1. Bioorg. Med. Chem. Lett 2016, 26, 2774–2778. [DOI] [PubMed] [Google Scholar]
- (732).Heinrich T; Buchstaller HP; Cezanne B; Rohdich F; Bomke J; Friese-Hamim M; Krier M; Knochel T; Musil D; Leuthner B; Zenke F Novel Reversible Methionine Aminopeptidase-2 (MetAP-2) Inhibitors Based on Purine and Related Bicyclic Templates. Bioorg. Med. Chem. Lett 2017, 27, 551–556. [DOI] [PubMed] [Google Scholar]
- (733).Hu XY; Addlagatta A; Matthews BW; Liu JO Identification of Pyridinylpyrimidines as Inhibitors of Human Methionine Aminopeptidases. Angew. Chem., Int. Ed 2006, 45, 3772–3775. [DOI] [PubMed] [Google Scholar]
- (734).Altmeyer MA; Marschner A; Schiffmann R; Klein CD Subtype-selectivity of Metal-Dependent Methionine Aminopeptidase Inhibitors. Bioorg. Med. Chem. Lett 2010, 20, 4038–4044. [DOI] [PubMed] [Google Scholar]
- (735).Schiffmann R; Neugebauer A; Klein CD Metal-mediated Inhibition of Escherichia coli Methionine Aminopeptidase: Structure-Activity Relationships and Development of a Novel Scoring Function for Metal-ligand Interactions. J. Med. Chem 2006, 49, 511–522. [DOI] [PubMed] [Google Scholar]
- (736).Huang M; Xie SX; Ma ZQ; Hanzlik RP; Ye QZ Metal Mediated Inhibition of Methionine Aminopeptidase by Quinolinyl Sulfonamides. Biochem. Biophys. Res. Commun 2006, 339, 506–513. [DOI] [PubMed] [Google Scholar]
- (737).Ye QZ; Xie SX; Huang M; Huang WJ; Lu JP; Ma ZQ Metalloform-Selective Inhibitors of Echerichia coli Methionine Aminopeptidase and X-ray Structure of a Mn(II)-form Enzyme Complexed with an Inhibitor. J. Am. Chem. Soc 2004, 126, 13940–13941. [DOI] [PubMed] [Google Scholar]
- (738).Huang QQ; Huang M; Nan FJ; Ye QZ Metalloform-selective Inhibition: Synthesis and Structure-activity Analysis of Mn(II)-Form-Selective Inhibitors of Escherichia coli Methionine Aminopeptidase. Bioorg. Med. Chem. Lett 2005, 15, 5386–5391. [DOI] [PubMed] [Google Scholar]
- (739).Huguet F; Melet A; de Sousa RA; Lieutaud A; Chevalier J; Maigre L; Deschamps P; Tomas A; Leulliot N; Pages JM; Artaud I Hydroxamic Acids as Potent Inhibitors of FeII and MnII E. coli Methionine Aminopeptidase: Biological Activities and X-ray Structures of Oxazole Hydroxamate-EcMetAP-Mn Complexes. ChemMedChem 2012, 7, 1020–1030. [DOI] [PubMed] [Google Scholar]
- (740).Wang WL; Chai SC; Huang M; He HZ; Hurley TD; Ye QZ Discovery of Inhibitors of Escherichia coli Methionine Aminopeptidase with the Fe(II)-form Selectivity and Antibacterial Activity. J. Med. Chem 2008, 51, 6110–6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (741).Luo QL; Li JY; Liu ZY; Chen LL; Li J; Qian Z; Shen Q; Li Y; Lushington GH; Ye QZ; Nan FJ Discovery and Structural Modification of Inhibitors of Methionine Aminopeptidases from Escherichia coli and Saccharomyces cerevisiae. J. Med. Chem 2003, 46, 2631–2640. [DOI] [PubMed] [Google Scholar]
- (742).Cui YM; Huang QQ; Xu J; Chen LL; Li JY; Ye QZ; Li J; Nan FJ Identification of Potent Type I MetAP Inhibitors by Simple Bioisosteric Replacement. Part 1: Synthesis and Preliminary SAR Studies of Thiazole-4-carboxylic Acid Thiazol-2-ylamide Derivatives. Bioorg. Med. Chem. Lett 2005, 15, 3732–3736. [DOI] [PubMed] [Google Scholar]
- (743).Wangtrakuldee P; Byrd MS; Campos CG; Henderson MW; Zhang Z; Clare M; Masoudi A; Myler PJ; Horn JR; Cotter PA; Hagen TJ Discovery of Inhibitors of Burkholderia pseudomallei Methionine Aminopeptidase with Antibacterial Activity. ACS Med. Chem. Lett 2013, 4, 699–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (744).Shim JS; Matsui Y; Bhat S; Nacev BA; Xu J; Bhang HE; Dhara S; Han KC; Chong CR; Pomper MG; So A; Liu JO Effect of Nitroxoline on Angiogenesis and Growth of Human Bladder Cancer. J. Natl. Cancer. Inst 2010, 102, 1855–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (745).Garrabrant T; Tuman RW; Ludovici D; Tominovich R; Simoneaux RL; Galemmo RA Jr.; Johnson DL Small Molecule Inhibitors of Methionine Aminopeptidase Type 2 (MetAP-2). Angiogenesis 2004, 7, 91–96. [DOI] [PubMed] [Google Scholar]
- (746).Sun J; Li MH; Qian SS; Guo FJ; Dang XF; Wang XM; Xue YR; Zhu HL Synthesis and Antitumor Activity of 1,3,4-oxadiazole Possessing 1,4-benzodioxan Moiety as a Novel Class of Potent Methionine Aminopeptidase Type II Inhibitors. Bioorg. Med. Chem. Lett 2013, 23, 2876–2879. [DOI] [PubMed] [Google Scholar]
- (747).McBride C; Cheruvallath Z; Komandla M; Tang M; Farrell P; Lawson JD; Vanderpool D; Wu Y; Dougan DR; Plonowski A; Holub C; Larson C Discovery of Potent, Reversible MetAP2 Inhibitors Via Fragment Based Drug Discovery and Structure Based Drug design-Part 2. Bioorg. Med. Chem. Lett 2016, 26, 2779–2783. [DOI] [PubMed] [Google Scholar]
- (748).Helgren TR; Wangtrakuldee P; Staker BL; Hagen TJ Advances in Bacterial Methionine Aminopeptidase Inhibition. Curr. Top. Med. Chem 2016, 16, 397–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (749).Lu JP; Ye QZ Expression and Characterization of Mycobacterium tuberculosis Methionine Aminopeptidase Type 1a. Bioorg. Med. Chem. Lett 2010, 20, 2776–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (750).Oefner C; Douangamath A; D’Arcy A; Hafeli S; Mareque D; Mac Sweeney A; Padilla J; Pierau S; Schulz H; Thormann M; Wadman S; Dale GE The 1.15 Angstrom Crystal Structure of the Staphylococcus aureus Methionyl-Aminopeptidase and Complexes with Triazole Based Inhibitors. J. Mol. Biol 2003, 332, 13–21. [DOI] [PubMed] [Google Scholar]
- (751).Yuan H; Chai SC; Lam CK; Howard Xu H; Ye QZ Two Methionine Aminopeptidases from Acinetobacter baumannii are Functional Enzymes. Bioorg. Med. Chem. Lett 2011, 21, 3395–3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (752).Wangtrakuldee P; Byrd MS; Campos CG; Henderson MW; Zhang Z; Clare M; Masoudi A; Myler PJ; Horn JR; Cotter PA; Hagen TJ Discovery of Inhibitors of Burkholderia pseudomallei Methionine Aminopeptidase with Antibacterial Activity. ACS Med Chem Lett 2013, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (753).Lu JP; Chai SC; Ye QZ Catalysis and Inhibition of Mycobacterium tuberculosis Methionine Aminopeptidase. J. Med. Chem 2010, 53, 1329–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (754).Kishor C; Arya T; Reddi R; Chen XC; Saddanapu V; Marapaka AK; Gumpena R; Ma DW; Liu JO; Addlagatta A Identification, Biochemical and Structural Evaluation of Species-Specific Inhibitors against Type I Methionine Aminopeptidases. J. Med. Chem 2013, 56, 5295–5305. [DOI] [PubMed] [Google Scholar]
- (755).Schiffmann R; Heine A; Klebe G; Klein CDP Metal Ions as Cofactors for the Binding of Inhibitors to Methionine Aminopeptidase: A Critical View of the Relevance of in Vitro Metalloenzyme Assays. Angew. Chem., Int. Ed 2005, 44, 3620–3623. [DOI] [PubMed] [Google Scholar]
- (756).Lu JP; Yuan XH; Yuan H; Wang WL; Wan B; Franzblau SG; Ye QZ Inhibition of Mycobacterium tuberculosis Methionine Aminopeptidases by Bengamide Derivatives. ChemMedChem 2011, 6, 1041–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (757).Evdokimov AG; Pokross M; Walter RL; Mekel M; Barnett BL; Amburgey J; Seibel WL; Soper SJ; Djung JF; Fairweather N; Diven C; Rastogi V; Grinius L; Klanke C; Siehnel R; Twinem T; Andrews R; Curnow A Serendipitous Discovery of Novel Bacterial Methionine Aminopeptidase Inhibitors. Proteins 2007, 66, 538–546. [DOI] [PubMed] [Google Scholar]
- (758).Li JY; Chen LL; Cui YM; Luo QL; Li J; Nan FJ; Ye QZ Specificity for Inhibitors of Metal-Substituted Methionine Aminopeptidase. Biochem. Biophys. Res. Commun 2003, 307, 172–179. [DOI] [PubMed] [Google Scholar]
- (759).Cui YM; Huang QQ; Xu J; Chen LL; Li JY; Ye QZ; Li J; Nan FJ Identification of Potent Type I MetAPs Inhibitors by Simple Bioisosteric Replacement. Part 2: SAR Studies of 5-heteroalkyl Substituted TCAT Derivatives. Bioorg. Med. Chem. Lett 2005, 15, 4130–4135. [DOI] [PubMed] [Google Scholar]
- (760).Xie SX; Huang WJ; Ma ZQ; Huang M; Hanzlik RP; Ye QZ Structural Analysis of Metalloform-Selective Inhibition of Methionine Aminopeptidase. Acta Crystallogr., Sect. D: Struct. Biol 2006, 62, 425–432. [DOI] [PubMed] [Google Scholar]
- (761).Kim EJ; Shin WH General Pharmacology of CKD-732, a New Anticancer Agent: Effects on Central Nervous, Cardiovascular, and Respiratory System. Biol. Pharm. Bull 2005, 28, 217–223. [DOI] [PubMed] [Google Scholar]
- (762).Nawarskas J; Rajan V; Frishman WH Vasopeptidase Inhibitors, Neutral Endopeptidase Inhibitors, and Dual Inhibitors of Angiotensin-converting Enzyme and Neutral Endopeptidase. Heart Dis 2001, 3, 378–385. [DOI] [PubMed] [Google Scholar]
- (763).Cody RJ; Atlas SA; Laragh JH; Kubo SH; Covit AB; Ryman KS; Shaknovich A; Pondolfino K; Clark M; Camargo MJ; et al. Atrial Natriuretic Factor in Normal Subjects and Heart Failure Patients. Plasma Levels and Renal, Hormonal, and Hemodynamic Responses to Peptide Infusion. J. Clin. Invest 1986, 78, 1362–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (764).Weidmann P; Gnadinger MP; Ziswiler HR; Shaw S; Bachmann C; Rascher W; Uehlinger DE; Hasler L; Reubi FC Cardiovascular, Endocrine and Renal Effects of Atrial Natriuretic Peptide in Essential Hypertension. J. Hypertens. 1986, 4, S71–83. [PubMed] [Google Scholar]
- (765).Janssen WM; de Zeeuw D; van der Hem GK; de Jong PE Antihypertensive Effect of a 5-day Infusion of Atrial Natriuretic Factor in Humans. Hypertension 1989, 13, 640–646. [DOI] [PubMed] [Google Scholar]
- (766).Windus DW; Stokes TJ; Morgan JR; Klahr S The Effects of Atrial Peptide in Humans with Chronic Renal Failure. Am. J. Kidney Dis 1989, 13, 477–484. [DOI] [PubMed] [Google Scholar]
- (767).Mangiafico S; Costello-Boerrigter LC; Andersen IA; Cataliotti A; Burnett JC Jr. Neutral Endopeptidase Inhibition and the Natriuretic Peptide System: an Evolving Strategy in Cardiovascular Therapeutics. Eur. Heart J 2013, 34, 886–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (768).Barrett AJ, Rawlings ND, Woessner JF Handbook of Proteolytic Enzymes; Elsevier Science, 2012. [Google Scholar]
- (769).Kerr MA; Kenny AJ The Purification and Specificity of a Neutral eEndopeptidase from Rabbit Kidney Brush Border. Biochem. J 1974, 137, 477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (770).Kerr MA; Kenny AJ The Molecular Weight and Properties of a Neutral Metallo-Endopeptidase from Rabbit Kidney Brush Border. Biochem. J 1974, 137, 489–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (771).Schiering N; D’Arcy A; Villard F; Ramage P; Logel C; Cumin F; Ksander GM; Wiesmann C; Karki RG; Mogi M Structure of Neprilysin in Complex with the Active Metabolite of Sacubitril. Sci. Rep 2016, 6, 27909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (772).Oefner C; D’Arcy A; Hennig M; Winkler FK; Dale GE Structure of Human Neutral Endopeptidase (Neprilysin) Complexed with Phosphoramidon. J. Mol. Biol 2000, 296, 341–349. [DOI] [PubMed] [Google Scholar]
- (773).Sahli S; Stump B; Welti T; Schweizer WB; Diederich F; Blum-Kaelin D; Aebi JD; Böhm H-J A New Class of Inhibitors for the Metalloprotease Neprilysin Based on a Central Imidazole Scaffold. Helv. Chim. Acta 2005, 88, 707–730. [Google Scholar]
- (774).Voisin S; Rognan D; Gros C; Ouimet T A Three-dimensional Model of the Neprilysin 2 Active Site Based on the X-ray Structure of Neprilysin. Identification of Residues Involved in Substrate Hydrolysis and Inhibitor Binding of Neprilysin 2. J. Biol. Chem 2004, 279, 46172–46181. [DOI] [PubMed] [Google Scholar]
- (775).Roques BP; Fournie-Zaluski MC; Soroca E; Lecomte JM; Malfroy B; Llorens C; Schwartz JC The Enkephalinase Inhibitor Thiorphan Shows Antinociceptive Activity in Mice. Nature 1980, 288, 286–288. [DOI] [PubMed] [Google Scholar]
- (776).Sahli S; Frank B; Schweizer WB; Diederich F; Blum-Kaelin D; Aebi JD; Böhm H-J; Oefner C; Dale GE Second-Generation Inhibitors for the Metalloprotease Neprilysin Based on Bicyclic Heteroaromatic Scaffolds: Synthesis, Biological Activity, and X-Ray Crystal-Structure Analysis. Helv. Chim. Acta 2005, 88, 731–750. [Google Scholar]
- (777).Fulcher IS; Matsas R; Turner AJ; Kenny AJ Kidney Neutral Endopeptidase and the Hydrolysis of Enkephalin by Synaptic Membranes Show Similar Sensitivity to Inhibitors. Biochem. J 1982, 203, 519–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (778).Oefner C; Roques BP; Fournie-Zaluski MC; Dale GE Structural Analysis of Neprilysin with Various Specific and Potent Inhibitors. Acta Crystallogr., Sect. D: Biol. Crystallogr 2004, 60, 392–396. [DOI] [PubMed] [Google Scholar]
- (779).Poras H; Patouret R; Leiris S; Ouimet T; Fournie-Zaluski MC; Roques BP Substituted Alpha-mercaptoketones, New Types of Specific Neprilysin Inhibitors. Bioorg. Med. Chem. Lett 2017, 27, 3883–3890. [DOI] [PubMed] [Google Scholar]
- (780).Robl JA; Cimarusti MP; Simpkins LM; Brown B; Ryono DE; Bird JE; Asaad MM; Schaeffer TR; Trippodo NC Dual MetalloproteaseInhibitors. 6. Incorporation of Bicyclic and Substituted Monocyclic Azepinones as Dipeptide Surrogates in Angiotensin-converting Enzyme/Neutral Endopeptidase Inhibitors. J. Med. Chem 1996, 39, 494–502. [DOI] [PubMed] [Google Scholar]
- (781).Robl JA; Sulsky R; Sieber-McMaster E; Ryono DE; Cimarusti MP; Simpkins LM; Karanewsky DS; Chao S; Asaad MM; Seymour AA; Fox M; Smith PL; Trippodo NC Vasopeptidase Inhibitors: Incorporation of Geminal and Spirocyclic Substituted Azepinones in Mercaptoacyl Dipeptides. J. Med. Chem 1999, 42, 305–311. [DOI] [PubMed] [Google Scholar]
- (782).Venugopal J Pharmacological Modulation of the Natriuretic Peptide System. Expert Opin. Ther. Pat 2003, 13, 1389–1409. [Google Scholar]
- (783).Trapani AJ; De Lombaert S; Beil ME; Bruseo CW; Savage P; Chou M; Jeng AY CGS 34043: a Non-peptidic, Potent and Long-acting Dual Inhibitor of Endothelin Converting Enzyme-1 and Neutral Endopeptidase 24.11. Life Sci 2000, 67, 1025–1033. [DOI] [PubMed] [Google Scholar]
- (784).Ksander GM; Ghai RD; deJesus R; Diefenbacher CG; Yuan A; Berry C; Sakane Y; Trapani A Dicarboxylic Acid Dipeptide Neutral Endopeptidase Inhibitors. J. Med. Chem 1995, 38, 1689–1700. [DOI] [PubMed] [Google Scholar]
- (785).De Lombaert S; Erion MD; Tan J; Blanchard L; el-Chehabi L; Ghai RD; Sakane Y; Berry C; Trapani AJ N-Phosphonomethyl Dipeptides and Their Phosphonate Prodrugs, a New Generation of Neutral Endopeptidase (NEP, EC 3.4.24.11) Inhibitors. J. Med. Chem 1994, 37, 498–511. [DOI] [PubMed] [Google Scholar]
- (786).Ksander GM; Diefenbacher CG; Yuan AM; Clark F; Sakane Y; Ghai RD Enkephalinase Inhibitors. 1. 2,4-Dibenzylglutaric Acid Derivatives. J. Med. Chem 1989, 32, 2519–2526. [DOI] [PubMed] [Google Scholar]
- (787).Robl JA; Sun C-Q; Stevenson J; Ryono DE; Simpkins LM; Cimarusti MP; Dejneka T; Slusarchyk WA; Chao S; Stratton L; Misra RN; Bednarz MS; Asaad MM; Cheung HS; Abboa-Offei BE; Smith PL; Mathers PD; Fox M; Schaeffer TR; Seymour AA; Trippodo NC Dual Metalloprotease Inhibitors: Mercaptoacetyl-Based Fused Heterocyclic Dipeptide Mimetics as Inhibitors of Angiotensin-Converting Enzyme and Neutral Endopeptidase. J. Med. Chem 1997, 40, 1570–1577. [DOI] [PubMed] [Google Scholar]
- (788).Trippodo NC; Robl JA; Asaad MM; Bird JE; Panchal BC; Schaeffer TR; Fox M; Giancarli MR; Cheung HS Cardiovascular Effects of the Novel Dual Inhibitor of Neutral Endopeptidase and Angiotensin-converting Enzyme BMS-182657 in Experimental Hypertension and Heart Failure. J. Pharmacol. Exp. Ther 1995, 275, 745–752. [PubMed] [Google Scholar]
- (789).Gonzalez Vera W; Fournie-Zaluski MC; Pham I; Laboulandine I; Roques BP; Michel JB Hypotensive and Natriuretic Effects of RB 105, a New Dual Inhibitor of Angiotensin Converting Enzyme and Neutral Endopeptidase in Hypertensive Rats. J. Pharmacol. Exp. Ther 1995, 272, 343–351. [PubMed] [Google Scholar]
- (790).Trippodo NC; Robl JA; Asaad MM; Fox M; Panchal BC; Schaeffer TR Effects of Omapatrilat in Low, Normal, and High Renin Experimental Hypertension. Am. J. Hypertens 1998, 11, 363–372. [DOI] [PubMed] [Google Scholar]
- (791).Nawarskas JJ; Anderson JR Omapatrilat: a Unique New Agent for the Treatment of Cardiovascular Disease. Heart Dis 2000, 2, 266–274. [PubMed] [Google Scholar]
- (792).Coats AJS Omapatrilat- the Story of Overture and Octave. Int. J. Cardiol 2002, 86, 1–4. [DOI] [PubMed] [Google Scholar]
- (793).Pickering TG Effects of Stress and Behavioral Interventions in Hypertension: the Rise and Fall of Omapatrilat. J. Clin. Hypertens 2002, 4, 371–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (794).Kostis JB; Cobbe S; Johnston C; Ford I; Murphy M; Weber MA; Black HR; Plouin PF; Levy D; Mancia G; Larochelle P; Kolloch RE; Alderman M; Ruilope LM; Dahlof B; Flack JM; Wolf R Design of the Omapatrilat in Persons with Enhanced Risk of Atherosclerotic Events (OPERA) Trial. Am. J. Hypertens 2002, 15, 193–198. [DOI] [PubMed] [Google Scholar]
- (795).Packer M; Califf RM; Konstam MA; Krum H; McMurray JJ; Rouleau JL; Swedberg K Comparison of Omapatrilat and Enalapril in Patients with Chronic Heart Failure: the Omapatrilat Versus Enalapril Randomized Trial of Utility in Reducing Events (OVERTURE). Circulation 2002, 106, 920–926. [DOI] [PubMed] [Google Scholar]
- (796).Rouleau JL; Pfeffer MA; Stewart DJ; Isaac D; Sestier F; Kerut EK; Porter CB; Proulx G; Qian C; Block AJ Comparison of Vasopeptidase Inhibitor, Omapatrilat, and Lisinopril on Exercise Tolerance and Morbidity in Patients with Heart Failure: IMPRESS Randomised Trial. Lancet 2000, 356, 615–620. [DOI] [PubMed] [Google Scholar]
- (797).Zanchi A; Maillard M; Burnier M Recent Clinical Trials with Omapatrilat: New Developments. Curr. Hypertens. Rep 2003, 5, 346–352. [DOI] [PubMed] [Google Scholar]
- (798).Fala L Entresto (Sacubitril/Valsartan): First-in-Class Angiotensin Receptor Neprilysin Inhibitor FDA Approved for Patients with Heart Failure. Am. Health Drug Benefits 2015, 8, 330–334. [PMC free article] [PubMed] [Google Scholar]
- (799).Sible AM; Nawarskas JJ; Alajajian D; Anderson JR Sacubitril/Valsartan: A Novel Cardiovascular Combination Agent. Cardiol. Rev 2016, 24, 41–47. [DOI] [PubMed] [Google Scholar]
- (800).Kyle RA; Rajkumar SV Treatment of Multiple Myeloma: a Comprehensive Review. Clin. Lymphoma Myeloma 2009, 9, 278–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (801).Dimopoulos MA; Richardson PG; Moreau P; Anderson KC Current Treatment Landscape for Relapsed and/or Refractory Multiple Myeloma. Nat. Rev. Clin. Oncol 2015, 12, 42–54. [DOI] [PubMed] [Google Scholar]
- (802).Palumbo A; Anderson K Multiple Myeloma. N. Engl. J. Med 2011, 364, 1046–1060. [DOI] [PubMed] [Google Scholar]
- (803).Kisselev AF; van der Linden WA; Overkleeft HS Proteasome Inhibitors: an Expanding Army Attacking a Unique Target. Chem. Biol 2012, 19, 99–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (804).Moreau P; Richardson PG; Cavo M; Orlowski RZ; San Miguel JF; Palumbo A; Harousseau JL Proteasome Inhibitors in Multiple Myeloma: 10 years Later. Blood 2012, 120, 947–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (805).Glickman MH; Ciechanover A The Ubiquitin-Proteasome Proteolytic Pathway: Destruction for the Sake of Construction. Physiol. Rev 2002, 82, 373–428. [DOI] [PubMed] [Google Scholar]
- (806).Finley D Recognition and Processing of Ubiquitin-Protein Conjugates by the Proteasome. Annu. Rev. Biochem 2009, 78, 477–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (807).Li J; Yakushi T; Parlati F; Mackinnon AL; Perez C; Ma Y; Carter KP; Colayco S; Magnuson G; Brown B; Nguyen K; Vasile S; Suyama E; Smith LH; Sergienko E; Pinkerton AB; Chung TDY; Palmer AE; Pass I; Hess S; Cohen SM; Deshaies RJ Capzimin is a Potent and Specific Inhibitor of Proteasome Isopeptidase Rpn11. Nat. Chem. Biol 2017, 13, 486–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (808).Goldberg AL Functions of the Proteasome: from Protein Degradation and Immune Surveillance to Cancer Therapy. Biochem. Soc. Trans 2007, 35, 12–17. [DOI] [PubMed] [Google Scholar]
- (809).Muz B; Ghazarian RN; Ou M; Luderer MJ; Kusdono HD; Azab AK Spotlight on Ixazomib: Potential in the Treatment of Multiple Myeloma. Drug Des. Devel. Ther 2016, 10, 217–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (810).Field-Smith A; Morgan GJ; Davies FE Bortezomib (Velcade) in the Treatment of Multiple Myeloma. Ther. Clin. Risk. Manag 2006, 2, 271–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (811).Stewart AK; Rajkumar SV; Dimopoulos MA; Masszi T; Spicka I; Oriol A; Hajek R; Rosinol L; Siegel DS; Mihaylov GG; Goranova-Marinova V; Rajnics P; Suvorov A; Niesvizky R; Jakubowiak AJ; San-Miguel JF; Ludwig H; Wang M; Maisnar V; Minarik J; Bensinger WI; Mateos MV; Ben-Yehuda D; Kukreti V; Zojwalla N; Tonda ME; Yang X; Xing B; Moreau P; Palumbo A; Investigators A Carfilzomib, Lenalidomide, and Dexamethasone for Relapsed Multiple Myeloma. N. Engl. J. Med 2015, 372, 142–152. [DOI] [PubMed] [Google Scholar]
- (812).van der Linden WA; Willems LI; Shabaneh TB; Li N; Ruben M; Florea BI; van der Marel GA; Kaiser M; Kisselev AF; Overkleeft HS Discovery of a Potent and Highly beta1 Specific Proteasome Inhibitor from a Focused Library of Urea-containing Peptide Vinyl Sulfones and Peptide Epoxyketones. Org. Biomol. Chem 2012, 10, 181–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (813).Desvergne A; Genin E; Marechal X; Gallastegui N; Dufau L; Richy N; Groll M; Vidal J; Reboud-Ravaux M Dimerized Linear Mimics of a Aatural Cyclopeptide (TMC-95A) are Potent Noncovalent Inhibitors of the Eukaryotic 20S Proteasome. J. Med. Chem 2013, 56, 3367–3378. [DOI] [PubMed] [Google Scholar]
- (814).Geurink PP; van der Linden WA; Mirabella AC; Gallastegui N; de Bruin G; Blom AE; Voges MJ; Mock ED; Florea BI; van der Marel GA; Driessen C; van der Stelt M; Groll M; Overkleeft HS; Kisselev AF Incorporation of Non-Natural Amino Acids Improves Cell Permeability and Potency of Specific Inhibitors of Proteasome Trypsin-like Sites. J. Med. Chem 2013, 56, 1262–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (815).Kawamura S; Unno Y; List A; Mizuno A; Tanaka M; Sasaki T; Arisawa M; Asai A; Groll M; Shuto S Potent Proteasome Inhibitors Derived from the Unnatural Cis-cyclopropane Isomer of Belactosin A: Synthesis, Biological Activity, and Mode of Action. J. Med. Chem 2013, 56, 3689–3700. [DOI] [PubMed] [Google Scholar]
- (816).Shrestha RK; Ronau JA; Davies CW; Guenette RG; Strieter ER; Paul LN; Das C Insights into the Mechanism of Deubiquitination by JAMM Deubiquitinases from Cocrystal Structures of the Enzyme with the Substrate and Product. Biochemistry 2014, 53, 3199–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (817).Tanaka K The Proteasome: Overview of Structure and Functions. Proc. Jpn. Acad., Ser. B 2009, 85, 12–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (818).Verma R; Aravind L; Oania R; McDonald WH; Yates JR 3rd; Koonin EV; Deshaies RJ Role of Rpn11 Metalloprotease in Deubiquitination and Degradation by the 26S Proteasome. Science 2002, 298, 611–615. [DOI] [PubMed] [Google Scholar]
- (819).Reyes-Turcu FE; Wilkinson KD Polyubiquitin Binding and Disassembly by Deubiquitinating Enzymes. Chem. Rev 2009, 109, 1495–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (820).Koulich E; Li X; DeMartino GN Relative Structural and Functional Roles of Multiple Deubiquitylating Proteins Associated with Mammalian 26S Proteasome. Mol. Biol. Cell 2008, 19, 1072–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (821).Guterman A; Glickman MH Complementary Roles for Rpn11 and Ubp6 in Deubiquitination and Proteolysis by the Proteasome. J. Biol. Chem 2004, 279, 1729–1738. [DOI] [PubMed] [Google Scholar]
- (822).McCullough J; Clague MJ; Urbe S AMSH is an Endosome-associated Ubiquitin Isopeptidase. J. Cell. Biol 2004, 166, 487–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (823).Nijman SM; Luna-Vargas MP; Velds A; Brummelkamp TR; Dirac AM; Sixma TK; Bernards R A Genomic and Functional Inventory of Deubiquitinating Enzymes. Cell 2005, 123, 773–786. [DOI] [PubMed] [Google Scholar]
- (824).Zhu P; Zhou W; Wang J; Puc J; Ohgi KA; Erdjument-Bromage H; Tempst P; Glass CK; Rosenfeld MG A Histone H2A Deubiquitinase Complex Coordinating Histone Acetylation and H1 Dissociation in Transcriptional Regulation. Mol. Cell 2007, 27, 609–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (825).Ambroggio XI; Rees DC; Deshaies RJ JAMM: a Metalloprotease-like Zinc Site in the Proteasome and Signalosome. PLoS Biol 2004, 2, 113–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (826).Sobhian B; Shao G; Lilli DR; Culhane AC; Moreau LA; Xia B; Livingston DM; Greenberg RA RAP80 targets BRCA1 to Specific Ubiquitin Structures at DNA Damage Sites. Science 2007, 1198–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (827).Cope GA; Suh GS; Aravind L; Schwarz SE; Zipursky SL; Koonin EV; Deshaies RJ Role of Predicted Metalloprotease Motif of Jab1/Csn5 in Cleavage of Nedd8 from Cul1. Science 2002, 298, 608–611. [DOI] [PubMed] [Google Scholar]
- (828).Sato Y; Yoshikawa A; Yamagata A; Mimura H; Yamashita M; Ookata K; Nureki O; Iwai K; Komada M; Fukai S Structural Basis for Specific Ceavage of Lys 63-linked Polyubiquitin Chains. Nature 2008, 455, 358–362. [DOI] [PubMed] [Google Scholar]
- (829).Song Y; Li S; Ray A; Das DS; Qi J; Samur MK; Tai YT; Munshi N; Carrasco RD; Chauhan D; Anderson KC Blockade of Deubiquitylating Enzyme Rpn11 Triggers Apoptosis in Multiple Myeloma Cells and Overcomes Bortezomib Resistance. Oncogene 2017, 36, 5631–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (830).Cenci S; Oliva L; Cerruti F; Milan E; Bianchi G; Raule M; Mezghrani A; Pasqualetto E; Sitia R; Cascio P Pivotal Advance: Protein Synthesis Modulates Responsiveness of Differentiating and Malignant Plasma Cells to Proteasome Inhibitors. J. Leukoc. Biol 2012, 92, 921–931. [DOI] [PubMed] [Google Scholar]
- (831).Deshaies RJ Proteotoxic Crisis, the Ubiquitin-proteasome System, and Cancer Therapy. BMC Biol 2014, 12, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (832).Buckley SM; Aranda-Orgilles B; Strikoudis A; Apostolou E; Loizou E; Moran-Crusio K; Farnsworth CL; Koller AA; Dasgupta R; Silva JC; Stadtfeld M; Hochedlinger K; Chen EI; Aifantis I Regulation of Pluripotency and Cellular Reprogramming by the Ubiquitin-proteasome System. Cell Stem Cell 2012, 11, 783–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (833).Yao T; Cohen RE A Cryptic Protease Couples Deubiquitination and Degradation by the Proteasome. Nature 2002, 419, 403–407. [DOI] [PubMed] [Google Scholar]
- (834).Lim HS; Archer CT; Kodadek T Identification of a Peptoid Inhibitor of the Proteasome 19S Regulatory Particle. J. Am. Chem. Soc 2007, 129, 7750–7751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (835).Lim HS; Cai D; Archer CT; Kodadek T Periodate-triggered Cross-linking Reveals Sug2/Rpt4 as the Molecular Target of a Peptoid Inhibitor of the 19S Proteasome Regulatory Particle. J. Am. Chem. Soc 2007, 129, 12936–12937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (836).Trader DJ; Simanski S; Kodadek T A Reversible and Highly Selective Inhibitor of the Proteasomal Ubiquitin Receptor Rpn13 is Toxic to Multiple Myeloma Cells. J. Am. Chem. Soc 2015, 137, 6312–6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (837).Pisani L; Muncipinto G; Miscioscia TF; Nicolotti O; Leonetti F; Catto M; Caccia C; Salvati P; Soto-Otero R; Mendez-Alvarez E; Passeleu C; Carotti A Discovery of a Novel Class of Potent Coumarin Monoamine Oxidase B Inhibitors: Development and Biopharmacological Profiling of 7-[(3-Chlorobenzyl)oxy]-4-[(methylamino)methyl]-2H-chromen-2-one methanesulfonate (NW-1772) as a Highly Potent, Selective, Reversible, and Orally Active Monoamine Oxidase B Inhibitor. J. Med. Chem 2009, 52, 6685–6706. [DOI] [PubMed] [Google Scholar]
- (838).Anchoori RK; Karanam B; Peng S; Wang JW; Jiang R; Tanno T; Orlowski RZ; Matsui W; Zhao M; Rudek MA; Hung CF; Chen X; Walters KJ; Roden RB A Bis-benzylidine Piperidone Targeting Proteasome Ubiquitin Receptor RPN13/ADRM1 as a Therapy for Cancer. Cancer Cell 2013, 24, 791–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (839).Perez C; Li J; Parlati F; Rouffet M; Ma Y; Mackinnon AL; Chou TF; Deshaies RJ; Cohen SM Discovery of an Inhibitor of the Proteasome Subunit Rpn11. J. Med. Chem 2017, 60, 1343–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (840).Lauinger L; Li J; Shostak A; Cemel IA; Ha N; Zhang Y; Merkl PE; Obermeyer S; Stankovic-Valentin N; Schafmeier T; Wever WJ; Bowers AA; Carter KP; Palmer AE; Tschochner H; Melchior F; Deshaies RJ; Brunner M; Diernfellner A Thiolutin is a Zinc Chelator that Inhibits the Rpn11 and other JAMM Metalloproteases. Nat. Chem. Biol 2017, 13, 709–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (841).Levine DP Vancomycin: A History. Clin. Infect. Dis 2006, 42 Suppl 1, S5–12. [DOI] [PubMed] [Google Scholar]
- (842).Moellering RC Jr. Vancomycin: A 50-Year Reassessment. Clin. Infect. Dis 2006, 42 Suppl 1, S3–4. [DOI] [PubMed] [Google Scholar]
- (843).McComas CC; Crowley BM; Boger DL Partitioning the Loss in Vancomycin Binding Affinity for D-Ala-D-Lac into lost H-Bond and Repulsive Lone Pair Contributions. J. Am. Chem. Soc 2003, 125, 9314–9315. [DOI] [PubMed] [Google Scholar]
- (844).Crowley BM; Boger DL Total Synthesis and Evaluation of [Psi[CH2NH]Tpg4]Vancomycin Aglycon: Reengineering Vancomycin for Dual D-Ala-D-Ala and D-Ala-D-Lac Binding. J. Am. Chem. Soc 2006, 128, 2885–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (845).Walsh CT; Fisher SL; Park IS; Prahalad M; Wu Z Bacterial Resistance to Vancomycin: Five Genes and One Missing Hydrogen Bond Tell the Story. Chem. Biol 1996, 3, 21–28. [DOI] [PubMed] [Google Scholar]
- (846).Reynolds PE; Depardieu F; Dutka-Malen S; Arthur M; Courvalin P Glycopeptide Resistance Mediated by Enterococcal Transposon Tn1546 Requires Production of VanX for Hydrolysis of D-alanyl-D-alanine. Mol. Microbiol 1994, 13, 1065–1070. [DOI] [PubMed] [Google Scholar]
- (847).Wu Z; Wright GD; Walsh CT Overexpression, Purification, and Characterization of VanX, a D-, D-Dipeptidase which is Essential for Vancomycin Resistance in Enterococcus faecium BM4147. Biochemistry 1995, 34, 2455–2463. [DOI] [PubMed] [Google Scholar]
- (848).Bussiere DE; Pratt SD; Katz L; Severin JM; Holzman T; Park CH The Structure of VanX Reveals a Novel Amino-Dipeptidase Involved in Mediating Transposon-Based Vancomycin Rsesistance. Mol. Cell 1998, 2, 75–84. [DOI] [PubMed] [Google Scholar]
- (849).Breece RM; Costello A; Bennett B; Sigdel TK; Matthews ML; Tierney DL; Crowder MW A Five-Coordinate Metal Center in Co(II)-Substituted VanX. J. Biol. Chem 2005, 280, 11074–11081. [DOI] [PubMed] [Google Scholar]
- (850).Wu Z; Walsh CT Phosphinate Analogs of D-, D-Dipeptides: Slow-Binding Inhibition and Proteolysis Protection of VanX, a D-, D-Dipeptidase Required for Vancomycin Resistance in Enterococcus faecium. Proc. Natl. Acad. Sci. U. S. A 1995, 92, 11603–11607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (851).Crowder MW Combating Vancomycin Resistance in Bacteria: Targeting the D-ala-D-ala Dipeptidase VanX. Infect. Disord.: Drug Targets 2006, 6, 147–158. [DOI] [PubMed] [Google Scholar]
- (852).Araoz R; Anhalt E; Rene L; Badet-Denisot MA; Courvalin P; Badet B Mechanism-Based Inactivation of VanX, a D-alanyl-D-alanine Dipeptidase Necessary for Vancomycin Resistance. Biochemistry 2000, 39, 15971–15979. [DOI] [PubMed] [Google Scholar]
- (853).Yaouancq L; Rene L; Tran Huu Dau ME; Badet B Toward Synthesis of Alpha-Alkyl Amino Glycines (A3G), New Amino Acid Surrogates. J. Org. Chem 2002, 67, 5408–5411. [DOI] [PubMed] [Google Scholar]
- (854).Yaouancq L; Anissimova M; Badet-Denisot MA; Badet B Design and Evaluation of Mechanism-Based Inhibitors of D-Alanyl-D-Alanine Dipeptidase VanX. Eur. J. Org. Chem 2002, 3573–3579. [Google Scholar]
- (855).Wu Z; Walsh C Dithiol Compounds: Potent, Time-Dependent Inhibitors of VanX, a Zinc-Dependent D,D-dipeptidase Required for Vancomycin Resistance in Enterococcus faecium. J. Am. Chem. Soc 1996, 118, 1785–1786. [Google Scholar]
- (856).Muthyala R; Rastogi N; Shin WS; Peterson ML; Sham YY Cell Permeable VanX Inhibitors as Vancomycin Re-sensitizing Agents. Bioorg. Med. Chem. Lett 2014, 24, 2535–2538. [DOI] [PubMed] [Google Scholar]
- (857).Golich FC; Sigdel T; Breece RM; Detar L; Herron LR; Crowder MW L-Alanine-p-Nitro Anilide is not a Substrate for VanX. Anal. Biochem 2004, 331, 398–400. [DOI] [PubMed] [Google Scholar]
- (858).Brandt JJ; Chatwood LL; Yang KW; Crowder MW Continuous Assay for VanX, the D-Alanyl-D-Alanine Dipeptidase Required for High-Level Vancomycin Resistance. Anal. Biochem 1999, 272, 94–99. [DOI] [PubMed] [Google Scholar]
- (859).Cristalli G; Costanzi S; Lambertucci C; Lupidi G; Vittori S; Volpini R; Camaioni E Adenosine Deaminase: Functional Implications and Different Classes of Inhibitors. Med. Res. Rev 2001, 21, 105–128. [DOI] [PubMed] [Google Scholar]
- (860).Aldrich MB; Blackburn MR; Kellems RE The Importance of Adenosine Deaminase for Lymphocyte Development and Function. Biochem. Biophys. Res. Commun 2000, 272, 311–315. [DOI] [PubMed] [Google Scholar]
- (861).Glazer RI Adenosine Deaminase Inhibitors: Their Role in Chemotherapy and Immunosuppression. Cancer Chemother. Pharmacol 1980, 4, 227–235. [DOI] [PubMed] [Google Scholar]
- (862).Smyth JF; Prentice HG; Proctor S; Hoffbrand AV Deoxycoformycin in the Treatment of Leukemias and Lymphomasa. Ann. N. Y. Acad. Sci 1985, 451, 123–128. [DOI] [PubMed] [Google Scholar]
- (863).Ungerer JPJ; Oosthuizen HM; Retief JH; Bissbort SH Significance of Adenosine Deaminase Activity and Its Isoenzymes in Tuberculous Effusions. CHEST 1994, 106, 33–37. [DOI] [PubMed] [Google Scholar]
- (864).Gleeson MP; Burton NA; Hillier IH The Mechanism of Adenosine Deaminase Catalysis Studied by QM/MM Calculations: The Role of Histidine 238 and the Activity of the Alanine 238 Mutant. Phys. Chem. Chem. Phys 2003, 5, 4272–4278. [Google Scholar]
- (865).Kinoshita T; Tada T; Nakanishi I Conformational Change of Adenosine Deaminase during Ligand-Exchange in a Crystal. Biochem. Biophys. Res. Commun 2008, 373, 53–57. [DOI] [PubMed] [Google Scholar]
- (866).Sawa T; Fukagawa Y; Homma I; Takeuchi T; Umezawa H Mode of Inhibition of Coformycin on Adenosine Deaminase. J. Antibiot 1967, 20, 227–231. [PubMed] [Google Scholar]
- (867).Agarwal RP; Spector T; Parks RE Tight-Binding Inhibitors: IV. Inhibition of Adenosine Deaminases by Various Inhibitors. Biochem. Pharmacol 1977, 26, 359–367. [DOI] [PubMed] [Google Scholar]
- (868).Omura S; Imamura N; Kuga H; Ishikawa H; Yamazaki Y; Okano K; Kimura K; Takahashi Y; Tanaka H Adechlorin, a new Adenosine Deaminase Inhibitor Containing Chlorine: Production, Isolation and Properties. J. Antibiot 1985, 38, 1008–1015. [DOI] [PubMed] [Google Scholar]
- (869).Omura S; Ishikawa H; Kuga H; Imamura N; Taga S; Takahashi Y; Tanaka H Adecypenol, a Unique Adenosine Deaminase Inhibitor Containing Homopurine and Cyclopentene Rings: Taxonomy, Production and Enzyme Inhibition. J. Antibiot 1986, 39, 1219–1224. [DOI] [PubMed] [Google Scholar]
- (870).Putt SR; Baker DC Total Synthesis of Pentostatin, the Potent Inhibitor of Adenosine Deaminase. J. Am. Chem. Soc 1979, 101, 6127–6128. [Google Scholar]
- (871).Montgomery JA; Thomas HJ; Zell AL; Einsphar HM; Bugg CE Study on the Inhibition of Adenosine Deaminase. J. Med. Chem 1985, 28, 1751–1753. [DOI] [PubMed] [Google Scholar]
- (872).Reayi A; Hosmane RS Inhibition of Adenosine Deaminase by Novel 5:7 Fused Heterocycles Containing the Imidazo[4,5-e][1,2,4]triazepine Ring System: A Structure−Activity Relationship Study. J. Med. Chem 2004, 47, 1044–1050. [DOI] [PubMed] [Google Scholar]
- (873).Cristalli G; Eleuteri A; Franchetti P; Grifantini M; Vittori S; Lupidi G Adenosine Deaminase Inhibitors: Synthesis and Structure Activity Relationships of Imidazole Analogs of Erythro-9-(2-hydroxy-3-nonyl)adenine. J. Med. Chem 1991, 34, 1187–1192. [DOI] [PubMed] [Google Scholar]
- (874).Cristalli G; Vittori S; Eleuteri A; Volpini R; Camaioni E; Lupidi G; Mahmood N; Bevilacqua F; Palu G Synthesis and Biological Evaluation of N6-Cycloalkyl Derivatives of 1-Deazaadenine Nucleosides: A New Class of Anti-Human Immunodeficiency Virus Agents. J. Med. Chem 1995, 38, 4019–4025. [DOI] [PubMed] [Google Scholar]
- (875).Ujjinamatada RK; Phatak P; Burger AM; Hosmane RS Inhibition of Adenosine Deaminase by Analogues of Adenosine and Inosine, Incorporating a Common Heterocyclic Base, 4(7)-Amino-6(5)H-imidazo[4,5-d]pyridazin-7(4)one. J. Med. Chem 2008, 51, 694–698. [DOI] [PubMed] [Google Scholar]
- (876).Schaeffer HJ; Schwender CF Enzyme Inhibitors: Bridging Hydrophobic and Hydrophilic Regions on Adenosine Deaminase with some 9-(2-hydroxy-3-alkyl)adenines. J. Med. Chem 1974, 17, 6–8. [DOI] [PubMed] [Google Scholar]
- (877).Barankiewicz J; Danks AM; Abushanab E; Makings L; Wiemann T; Wallis RA; Pragnacharyulu PVP; Fox A; Marangos PJ Regulation of Adenosine Concentration and Cytoprotective Effects of Novel Reversible Adenosine Deaminase Inhibitors. J. Pharmacol. Exp. Ther 1997, 283, 1230–1238. [PubMed] [Google Scholar]
- (878).Antonini I; Cristalli G; Franchetti P; Grifantini M; Martelli S; Lupidi G; Riva F Adenosine Deaminase Inhibitors: Synthesis of Deaza Analogues of Erythro-9-(2-hydroxy-3-nonyl)adenine. J. Med. Chem 1984, 27, 274–278. [DOI] [PubMed] [Google Scholar]
- (879).Cristalli G; Franchetti P; Grifantini M; Vittori S; Lupidi G; Riva F; Bordoni T; Geroni C; Verini MA Adenosine Deaminase Inhibitors: Synthesis and Biological Activity of Deaza Analogues of Erythro-9-(2-hydroxy-3-nonyl)adenine. J. Med. Chem 1988, 31, 390–393. [DOI] [PubMed] [Google Scholar]
- (880).Cristalli G; Eleuteri A; Volpini R; Vittori S; Camaioni E; Lupidi G Adenosine Deaminase Inhibitors: Synthesis and Structure-Activity Relationships of 2-Hydroxy-3-nonyl Derivatives of Azoles. J. Med. Chem 1994, 37, 201–205. [DOI] [PubMed] [Google Scholar]
- (881).Terasaka T; Okumura H; Tsuji K; Kato T; Nakanishi I; Kinoshita T; Kato Y; Kuno M; Seki N; Naoe Y; Inoue T; Tanaka K; Nakamura K Structure-Based Design and Synthesis of Non-Nucleoside, Potent, and Orally Bioavailable Adenosine Deaminase Inhibitors. J. Med. Chem 2004, 47, 2728–2731. [DOI] [PubMed] [Google Scholar]
- (882).Dillman RO Pentostatin (Nipent) in the Treatment of Chronic Lymphocyte Leukemia and Hairy Cell Leukemia. Expert Rev. Anticancer. Ther 2004, 4, 27–36. [DOI] [PubMed] [Google Scholar]
- (883).Kossel A; Dakin HD Further Examinations of Fermentative Urea Formation. Hoppe-Seyler’s Z. Physiol. Chem 1904, 42, 181–188. [Google Scholar]
- (884).Kossel A; Dakin HD Arginase. Hoppe-Seyler’s Z. Physiol. Chem 1904, 41, 321–331. [Google Scholar]
- (885).Jenkinson CP; Grody WW; Cederbaum SD Comparative Properties of Arginases. Comp. Biochem. Physiol., Part B: Biochem. Mol. Biol 1996, 114, 107–132. [DOI] [PubMed] [Google Scholar]
- (886).Baniyash M TCR Zeta-Chain Downregulation: Curtailing an Excessive Inflammatory Immune Response. Nat. Rev. Immunol 2004, 4, 675–687. [DOI] [PubMed] [Google Scholar]
- (887).Bronstein-Sitton N; Cohen-Daniel L; Vaknin I; Ezernitchi AV; Leshem B; Halabi A; Houri-Hadad Y; Greenbaum E; Zakay-Rones Z; Shapira L; Baniyash M Sustained Exposure to Bacterial Antigen Induces Interferon-Gamma-Dependent T Cell Receptor Zeta Down-Regulation and Impaired T Cell Function. Nat. Immunol 2003, 4, 957–964. [DOI] [PubMed] [Google Scholar]
- (888).Serafini P; De Santo C; Marigo I; Cingarlini S; Dolcetti L; Gallina G; Zanovello P; Bronte V Derangement of Immune Responses by Myeloid Suppressor Cells. Cancer Immunol. Immunother 2004, 53, 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (889).Caldwell RB; Toque HA; Narayanan SP; Caldwell RW Arginase: An Old Enzyme with New Tricks. Trends Pharmacol. Sci 2015, 36, 395–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (890).Shishova EY; Di Costanzo L; Emig FA; Ash DE; Christianson DW Probing the Specificity Determinants of Amino Acid Recognition by Arginase. Biochemistry 2009, 48, 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (891).Kanyo ZF; Scolnick LR; Ash DE; Christianson DW Structure of a Unique Binuclear Manganese Cluster in Arginase. Nature 1996, 383, 554–557. [DOI] [PubMed] [Google Scholar]
- (892).Christianson DW; Cox JD Catalysis by Metal-Activated Hydroxide in Zinc and Manganese Metalloenzymes. Annu. Rev. Biochem 1999, 68, 33–57. [DOI] [PubMed] [Google Scholar]
- (893).Kuhn NJ; Ward S; Piponski M; Young TW Purification of Human Hepatic Arginase and Its Manganese (Ii)-Dependent and Ph-Dependent Interconversion between Active and Inactive Forms - a Possible Ph-Sensing Function of the Enzyme on the Ornithine Cycle. Arch. Biochem. Biophys 1995, 320, 24–34. [DOI] [PubMed] [Google Scholar]
- (894).Bronte V; Serafini P; Mazzoni A; Segal DM; Zanovello P L-Arginine Metabolism in Myeloid Cells Controls T-Lymphocyte Functions. Trends Immunol 2003, 24, 302–306. [DOI] [PubMed] [Google Scholar]
- (895).Cox JD; Cama E; Colleluori DM; Pethe S; Boucher JL; Mansuy D; Ash DE; Christianson DW Mechanistic and Metabolic Inferences from the Binding of Substrate Analogues and Products to Arginase. Biochemistry 2001, 40, 2689–2701. [DOI] [PubMed] [Google Scholar]
- (896).Pudlo M; Demougeot C; Girard-Thernier C Arginase Inhibitors: A Rational Approach Over One Century. Med. Res. Rev 2017, 37, 475–513. [DOI] [PubMed] [Google Scholar]
- (897).Custot J; Boucher JL; Vadon S; Guedes C; Dijols S; Delaforge M; Mansuy D N Omega-Hydroxyamino-Alpha-Amino Acids as a New Class of Very Strong Inhibitors of Arginases. JBIC, J. Biol. Inorg. Chem 1996, 1, 73–82. [Google Scholar]
- (898).Moali C; Brollo M; Custot J; Sari MA; Boucher JL; Stuehr DJ; Mansuy D Recognition of Alpha-Amino Acids Bearing Various C=NOH Functions by Nitric Oxide Synthase and Arginase Involves Very Different Structural Determinants. Biochemistry 2000, 39, 8208–8218. [DOI] [PubMed] [Google Scholar]
- (899).Cama E; Pethe S; Boucher JL; Han SF; Emig FA; Ash DE; Viola RE; Mansuy D; Christianson DW Inhibitor Coordination Interactions in the Binuclear Manganese Cluster of Arginase. Biochemistry 2004, 43, 8987–8999. [DOI] [PubMed] [Google Scholar]
- (900).Cama E; Shin H; Christianson DW Design of Amino Acid Sulfonamides as Transition-State Analogue Inhibitors of Arginase. J. Am. Chem. Soc 2003, 125, 13052–13057. [DOI] [PubMed] [Google Scholar]
- (901).Ilies M; Di Costanzo L; North ML; Scott JA; Christianson DW 2-Aminoimidazole Amino Acids as Inhibitors of the Binuclear Manganese Metalloenzyme Human Arginase I. J. Med. Chem 2010, 53, 4266–4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (902).Zakharian TY; Di Costanzo L; Christianson DW (S)-2-Amino-6-Nitrohexanoic Acid Binds to Human Arginase I through Multiple Nitro-Metal Coordination Interactions in the Binuclear Manganese Cluster. J. Am. Chem. Soc 2008, 130, 17254–17255. [DOI] [PubMed] [Google Scholar]
- (903).Shin H; Cama E; Christianson DW Design of Amino Acid Aldehydes as Transition-State Analogue Inhibitors of Arginase. J. Am. Chem. Soc 2004, 126, 10278–10284. [DOI] [PubMed] [Google Scholar]
- (904).Zakharian TY; Di Costanzo L; Christianson DW Synthesis of (2S)-2-Amino-7,8-Epoxyoctanoic Acid and Structure of its Metal-Bridging Complex with Human Arginase I. Org. Biomol. Chem 2008, 6, 3240–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (905).Ilies M; Di Costanzo L; Dowling DP; Thorn KJ; Christianson DW Binding of Alpha,Alpha-Disubstituted Amino Acids to Arginase Suggests New Avenues for Inhibitor Design. J. Med. Chem 2011, 54, 5432–5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (906).Van Zandt MC; Whitehouse DL; Golebiowski A; Ji MK; Zhang M; Beckett RP; Jagdmann GE; Ryder TR; Sheeler R; Andreoli M; Conway B; Mahboubi K; D’Angelo G; Mitschler A; Cousido-Siah A; Ruiz FX; Howard EI; Podjarny AD; Schroeter H Discovery of (R)-2-amino-6-borono-2-(2-(piperidin-1-yl)ethyl)hexanoic acid and Congeners as Highly Potent Inhibitors of Human Arginases I and II for Treatment of Myocardial Reperfusion Injury. J. Med. Chem 2013, 56, 2568–2580. [DOI] [PubMed] [Google Scholar]
- (907).Golebiowski A; Whitehouse D; Beckett RP; Van Zandt M; Ji MK; Ryder TR; Jagdmann E; Andreoli M; Lee Y; Sheeler R; Conway B; Olczak J; Mazur M; Czestkowski W; Piotrowska W; Cousido-Siah A; Ruiz FX; Mitschler A; Podjarny A; Schroeter H Synthesis of Quaternary Alpha-Amino Acid-Based Arginase Inhibitors via the Ugi Reaction. Bioorg. Med. Chem. Lett 2013, 23, 4837–4841. [DOI] [PubMed] [Google Scholar]
- (908).Di Costanzo L; Sabio G; Mora A; Rodriguez PC; Ochoa AC; Centeno F; Christianson DW Crystal Structure of Human Arginase I at 1.29-A Resolution and Exploration of Inhibition in the Immune Response. Proc. Natl. Acad. Sci. U. S. A 2005, 102, 13058–13063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (909).Di Costanzo L; Ilies M; Thorn KJ; Christianson DW Inhibition of Human Arginase I by Substrate and Product Analogues. Arch. Biochem. Biophys 2010, 496, 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (910).Baggio R; Elbaum D; Kanyo ZF; Carroll PJ; Cavalli RC; Ash DE; Christianson DW Inhibition of Mn-2(2+)-Arginase by Borate Leads to the Design of a Transition State Analogue Inhibitor, 2(S)-Amino-6-Boronohexanoic Acid. J. Am. Chem. Soc 1997, 119, 8107–8108. [Google Scholar]
- (911).Gross MI; Steggerda SM; Li W; Calithera Biosciences, Inc: U.S.A., 2016. [Google Scholar]
- (912).Lisi L; Pizzoferrato M; Miscioscia FT; Topai A; Navarra P Interactions Between Integrase Inhibitors and Human Arginase 1. J. Neurochem 2017, 142, 153–159. [DOI] [PubMed] [Google Scholar]
- (913).Mai A; Massa S; Rotili D; Cerbara I; Valente S; Pezzi R; Simeoni S; Ragno R Histone Deacetylation in Epigenetics: An Attractive Target for Anticancer Therapy. Med. Res. Rev 2005, 25, 261–309. [DOI] [PubMed] [Google Scholar]
- (914).Marmorstein R Protein Modules that Manipulate Histone Tails for Chromatin Regulation. Nat. Rev. Mol. Cell. Bio 2001, 2, 422–432. [DOI] [PubMed] [Google Scholar]
- (915).Minucci S; Pelicci PG Histone Deacetylase Inhibitors and the Promise of Epigenetic (and more) Treatments for Cancer. Nat. Rev. Cancer 2006, 6, 38–51. [DOI] [PubMed] [Google Scholar]
- (916).Bolden JE; Peart MJ; Johnstone RW Anticancer Activities of Histone Deacetylase Inhibitors. Nat. Rev. Drug Discovery 2006, 5, 769–784. [DOI] [PubMed] [Google Scholar]
- (917).Lombardi PM; Cole KE; Dowling DP; Christianson DW Structure, Mechanism, and Inhibition of Histone Deacetylases and Related Metalloenzymes. Curr. Opin. Struct. Biol 2011, 21, 735–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (918).Xu WS; Parmigiani RB; Marks PA Histone Deacetylase Inhibitors: Molecular Mechanisms of Action. Oncogene 2007, 26, 5541–5552. [DOI] [PubMed] [Google Scholar]
- (919).Dowling DP; Di Costanzo L; Gennadios HA; Christianson DW Evolution of the Arginase Fold and Functional Diversity. Cell. Mol. Life Sci 2008, 65, 2039–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (920).Hai Y; Christianson DW Histone Deacetylase 6 Structure and Molecular Basis of Catalysis and Inhibition. Nat. Chem. Biol 2016, 12, 741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (921).Bressi JC; Jennings AJ; Skene R; Wu YQ; Melkus R; De Jong R; O’Connell S; Grimshaw CE; Navre M; Gangloff AR Exploration of the HDAC2 Foot Pocket: Synthesis and SAR of Substituted N-(2-aminophenyl)benzamides. Bioorg. Med. Chem. Lett 2010, 20, 3142–3145. [DOI] [PubMed] [Google Scholar]
- (922).Whitehead L; Dobler MR; Radetich B; Zhu YY; Atadja PW; Claiborne T; Grob JE; McRiner A; Pancost MR; Patnaik A; Shao WL; Shultz M; Tichkule R; Tommasi RA; Vash B; Wang P; Stams T Human HDAC Isoform Selectivity Achieved via Exploitation of the Acetate Release Channel with Structurally Unique Small Molecule Inhibitors. Bioorg. Med. Chem 2011, 19, 4626–4634. [DOI] [PubMed] [Google Scholar]
- (923).Lobera M; Madauss KP; Pohlhaus DT; Wright QG; Trocha M; Schmidt DR; Baloglu E; Trump RP; Head MS; Hofmann GA; Murray-Thompson M; Schwartz B; Chakravorty S; Wu ZN; Mander PK; Kruidenier L; Reid RA; Burkhart W; Turunen BJ; Rong JX; Wagner C; Moyer MB; Wells C; Hong X; Moore JT; Williams JD; Soler D; Ghosh S; Nolan MA Selective Class IIa Histone Deacetylase Inhibition via a Nonchelating Zinc-Binding Group. Nat. Chem. Biol 2013, 9, 319–325. [DOI] [PubMed] [Google Scholar]
- (924).Bottomley MJ; Lo Surdo P; Di Giovine P; Cirillo A; Scarpelli R; Ferrigno F; Jones P; Neddermann P; De Francesco R; Steinkuhler C; Gallinari P; Carfi A Structural and Functional Analysis of the Human HDAC4 Catalytic Domain Reveals a Regulatory Structural Zinc-Binding Domain. J. Biol. Chem 2008, 283, 26694–26704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (925).Wahhab A; Smil D; Ajamian A; Allan M; Chantigny Y; Therrien E; Nguyen N; Manku S; Leit S; Rahil J; Petschner AJ; Lu AH; Nicolescu A; Lefebvre S; Montcalm S; Fournel M; Yan TP; Li ZM; Besterman JM; Deziel R Sulfamides as Novel Histone Deacetylase Inhibitors. Bioorg. Med. Chem. Lett 2009, 19, 336–340. [DOI] [PubMed] [Google Scholar]
- (926).Patil V; Sodji QH; Kornacki JR; Mrksich M; Oyelere AK 3-Hydroxypyridin-2-thione as Novel Zinc Binding Group for Selective Histone Deacetylase Inhibition. J. Med. Chem 2013, 56, 3492–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (927).Sodji QH; Patil V; Kornacki JR; Mrksich M; Oyelere AK Synthesis and Structure-Activity Relationship of 3-Hydroxypyridine-2-thione-Based Histone Deacetylase Inhibitors. J. Med. Chem 2013, 56, 9969–9981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (928).Muthyala R; Shin WS; Xie JS; Sham YY Discovery of 1-Hydroxypyridine-2-thiones as Selective Histone Deacetylase Inhibitors and their Potential Application for Treating Leukemia. Bioorg. Med. Chem. Lett 2015, 25, 4320–4324. [DOI] [PubMed] [Google Scholar]
- (929).Khan N; Jeffers M; Kumar S; Hackett C; Boldog F; Khramtsov N; Qian XZ; Mills E; Berghs SC; Carey N; Finn PW; Collins LS; Tumber A; Ritchie JW; Jensen PB; Lichenstein HS; Sehested M Determination of the Class and Isoform Selectivity of Small-Molecule Histone Deacetylase Inhibitors. Biochem. J 2008, 409, 581–589. [DOI] [PubMed] [Google Scholar]
- (930).Jones P; Altamura S; De Francesco R; Paz OG; Kinzel O; Mesiti G; Monteagudo E; Pescatore G; Rowley M; Verdirame M; Steinkuhler C A Novel Series of Potent and Selective Ketone Histone Deacetylase Inhibitors with Antitumor Activity in Vivo. J. Med. Chem 2008, 51, 2350–2353. [DOI] [PubMed] [Google Scholar]
- (931).Lee JH; Mahendran A; Yao YS; Ngo L; Venta-Perez G; Choy ML; Kim N; Ham WS; Breslow R; Marks PA Development of a Histone Deacetylase 6 Inhibitor and its Biological Effects. Proc. Natl. Acad. Sci. U. S. A 2013, 110, 15704–15709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (932).Ito T; Ouchida M; Morimoto Y; Yoshida A; Jitsumori Y; Ozaki T; Sonobe H; Inoue H; Shimizu K Significant Growth Suppression of Synovial Sarcomas by the Histone Deacetylase Inhibitor FK228 in Vitro and in Vivo. Cancer Lett 2005, 224, 311–319. [DOI] [PubMed] [Google Scholar]
- (933).Daniel KB; Sullivan ED; Chen Y; Chan JC; Jennings PA; Fierke CA; Cohen SM Dual-Mode HDAC Prodrug for Covalent Modification and Subsequent Inhibitor Release. J. Med. Chem 2015, 58, 4812–4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (934).Day JA; Cohen SM Investigating the Selectivity of Metalloenzyme Inhibitors. J. Med. Chem 2013, 56, 7997–8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (935).Furumai R; Komatsu Y; Nishino N; Khochbin S; Yoshida M; Horinouchi S Potent Histone Deacetylase Inhibitors Built from Trichostatin A and Cyclic Tetrapeptide Antibiotics Including Trapoxin. Proc. Natl. Acad. Sci. U. S. A 2001, 98, 87–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (936).Kijima M; Yoshida M; Sugita K; Horinouchi S; Beppu T Trapoxin, an Antitumor Cyclic Tetrapeptide, Is an Irreversible Inhibitor of Mammalian Histone Deacetylase. J. Biol. Chem 1993, 268, 22429–22435. [PubMed] [Google Scholar]
- (937).Bradner JE; West N; Grachan ML; Greenberg EF; Harggarty SJ; Warnow T; Mazitschek R Chemical Phylogenetics of Histone Deacetylases. Nat. Chem. Biol 2010, 7, 238–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (938).Friend C; Scher W; Holland JG; Sato T Hemoglobin Synthesis in Murine Virus-Induced Leukemic Cells in-Vitro - Stimulation of Erythroid Differentiation by Dimethyl Sulfoxide. Proc. Natl. Acad. Sci. U. S. A 1971, 68, 378–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (939).Tanaka M; Levy J; Terada M; Breslow R; Rifkind RA; Marks PA Induction of Erythroid Differentiation in Murine Virus-Infected Erythroleukemia Cells by Highly Polar Compounds. Proc. Natl. Acad. Sci. U. S. A 1975, 72, 1003–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (940).Richon VM; Webb Y; Merger R; Sheppard T; Jursic B; Ngo L; Civoli F; Breslow R; Rifkind RA; Marks PA Second Generation Hybrid Polar Compounds are Potent Inducers of Transformed Cell Differentiation. Proc. Natl. Acad. Sci. U. S. A 1996, 93, 5705–5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (941).Richon VM; Emiliani S; Verdin E; Webb Y; Breslow R; Rifkind RA; Marks PA A Class of Hybrid Polar Inducers of Transformed Cell Differentiation Inhibits Histone Deacetylases. Proc. Natl. Acad. Sci. U. S. A 1998, 95, 3003–3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (942).Mann BS; Johnson JR; Cohen MH; Justice R; Pazdur R FDA Approval Summary: Vorinostat for Treatment of Advanced Primary Cutaneous T-cell Lymphoma. Oncologist 2007, 12, 1247–1252. [DOI] [PubMed] [Google Scholar]
- (943).Yoshida M; Kijima M; Akita M; Beppu T Potent and Specific Inhibition of Mammalian Histone Deacetylase Both in Vivo and in Vitro by Trichostatin A. J. Biol. Chem 1990, 265, 17174–17179. [PubMed] [Google Scholar]
- (944).Paris M; Porcelloni M; Binaschi M; Fattori D Histone Deacetylase Inhibitors: From Bench to Clinic. J. Med. Chem 2008, 51, 1505–1529. [DOI] [PubMed] [Google Scholar]
- (945).Moradei O; Maroun CR; Paquin I; Vaisburg A Histone Deacetylase Inhibitors: Latest Developments, Trends and Prospects. Curr. Med. Chem.: Anti-Cancer Agents 2005, 5, 529–560. [DOI] [PubMed] [Google Scholar]
- (946).Manal M; Chandrasekar MJN; Priya JG; Nanjan MJ Inhibitors of Histone Deacetylase as Antitumor Agents: A Critical Review. Bioorg. Chem 2016, 67, 18–42. [DOI] [PubMed] [Google Scholar]
- (947).Wang XX; Wan RZ; Liu ZP Recent Advances in the Discovery of Potent and Selective HDAC6 Inhibitors. Eur. J. Med. Chem 2018, 143, 1406–1418. [DOI] [PubMed] [Google Scholar]
- (948).Nakajima H; Kim YB; Terano H; Yoshida M; Horinouchi S FR901228, a Potent Antitumor Antibiotic, is a Novel Histone Deacetylase Inhibitor. Exp. Cell Res 1998, 241, 126–133. [DOI] [PubMed] [Google Scholar]
- (949).Furumai R; Matsuyama A; Kobashi N; Lee KH; Nishiyama N; Nakajima I; Tanaka A; Komatsu Y; Nishino N; Yoshida M; Horinouchi S FK228 (Depsipeptide) as a Natural Prodrug that Inhibits Class I Histone Deacetylases. Cancer Res 2002, 62, 4916–4921. [PubMed] [Google Scholar]
- (950).Cole KE; Dowling DP; Boone MA; Phillips AJ; Christianson DW Structural Basis of the Antiproliferative Activity of Largazole, a Depsipeptide Inhibitor of the Histone Deacetylases. J. Am. Chem. Soc 2011, 133, 12474–12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (951).Hai Y; Christianson DW Histone Deacetylase 6 Structure and Molecular Basis of Catalysis and Inhibition. Nat. Chem. Biol 2016, 12, 741-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (952).Bissantz C; Kuhn B; Stahl M A Medicinal Chemist’s Guide to Molecular Interactions. J. Med. Chem 2010, 53, 5061–5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (953).Porter NJ; Christianson DW Binding of the Microbial Cyclic Tetrapeptide Trapoxin A to the Class I Histone Deacetylase HDAC8. ACS Chem. Biol 2017, 12, 2281–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (954).Shakespear MR; Halili MA; Irvine KM; Fairlie DP; Sweet MJ Histone Deacetylases as Regulators of Inflammation and Immunity. Trends Immunol 2011, 32, 335–343. [DOI] [PubMed] [Google Scholar]
- (955).Chuang DM; Leng Y; Marinova Z; Kim HJ; Chiu CT Multiple Roles of HDAC Inhibition in Neurodegenerative Conditions. Trends Neurosci 2009, 32, 591–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (956).Shirakawa K; Chavez L; Hakre S; Calvanese V; Verdin E Reactivation of Latent HIV by Histone Deacetylase Inhibitors. Trends Microbiol 2013, 21, 277–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (957).Archin NM; Espeseth A; Parker D; Cheema M; Hazuda D; Margolis DM Expression of Latent HIV Induced by the Potent HDAC Inhibitor Suberoylanilide Hydroxamic Acid. AIDS Res. Hum. Retroviruses 2009, 25, 207–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (958).Santajit S; Indrawattana N Mechanisms of Antimicrobial Resistance in ESKAPE Pathogens. Biomed Res Int 2016, 2016, 2475067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (959).Silhavy TJ; Kahne D; Walker S The Bacterial Cell Envelope. Cold Spring Harbor Perspect. Biol 2010, 2, a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (960).Raetz CR; Reynolds CM; Trent MS; Bishop RE Lipid A Modification Systems in Gram-Negative Bacteria. Annu. Rev. Biochem 2007, 76, 295–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (961).Delcour AH Outer Membrane Permeability and Antibiotic Resistance. Biochim. Biophys. Acta 2009, 1794, 808–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (962).Vuorio R; Vaara M The Lipid A Biosynthesis Mutation LpxA2 of Escherichia coli Results in Drastic Antibiotic Supersusceptibility. Antimicrob. Agents Chemother 1992, 36, 826–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (963).Beutler B; Cerami A Tumor Necrosis, Cachexia, Shock, and Inflammation: a Aommon Mediator. Annu. Rev. Biochem 1988, 57, 505–518. [DOI] [PubMed] [Google Scholar]
- (964).Drake TA; Cheng J; Chang A; Taylor FB Jr. Expression of Tissue Factor, Thrombomodulin, and E-selectin in Baboons with Lethal Escherichia coli Sepsis. Am. J. Pathol 1993, 142, 1458–1470. [PMC free article] [PubMed] [Google Scholar]
- (965).Li A; Chang AC; Peer GT; Hinshaw LB; Taylor FB Jr. Comparison of the Capacity of rhTNF-alpha and Escherichia coli to Induce Procoagulant Activity by Baboon Mononuclear Cells in Vivo and in Vitro. Shock 1996, 5, 274–279. [DOI] [PubMed] [Google Scholar]
- (966).Dinarello CA Interleukin-1, Interleukin-1 Receptors and Interleukin-1 Receptor Antagonist. Int. Rev. Immunol 1998, 16, 457–499. [DOI] [PubMed] [Google Scholar]
- (967).Parrillo JE Mechanisms of Disease- Pathogenic Mechanisms of Septic Shock. N. Engl. J. Med 1993, 328, 1471–1477. [DOI] [PubMed] [Google Scholar]
- (968).Raetz CR; Whitfield C Lipopolysaccharide Endotoxins. Annu. Rev. Biochem 2002, 71, 635–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (969).Young K; Silver LL; Bramhill D; Cameron P; Eveland SS; Raetz CR; Hyland SA; Anderson MS The envA Permeability/cell Division Gene of Escherichia coli Encodes the Second Enzyme of Lipid A Biosynthesis. UDP-3-O-(R-3-Hydroxymyristoyl)-N-acetylglucosamine Deacetylase. J. Biol. Chem 1995, 270, 30384–30391. [DOI] [PubMed] [Google Scholar]
- (970).Anderson MS; Robertson AD; Macher I; Raetz CR Biosynthesis of lipid A in Escherichia coli: Identification of UDP-3-O-[(R)-3-hydroxymyristoyl]-alpha-D-glucosamine as a Precursor of UDP-N2,O3-bis[(R)-3-hydroxymyristoyl]-alpha-D-glucosamine. Biochemistry 1988, 27, 1908–1917. [DOI] [PubMed] [Google Scholar]
- (971).Mdluli KE; Witte PR; Kline T; Barb AW; Erwin AL; Mansfield BE; McClerren AL; Pirrung MC; Tumey LN; Warrener P; Raetz CR; Stover CK Molecular Validation of LpxC as an Antibacterial Drug Target in Pseudomonas aeruginosa. Antimicrob. Agents Chemother 2006, 50, 2178–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (972).Lee CJ; Liang X; Chen X; Zeng D; Joo SH; Chung HS; Barb AW; Swanson SM; Nicholas RA; Li Y; Toone EJ; Raetz CR; Zhou P Species-specific and Inhibitor-dependent Conformations of LpxC: Implications for Antibiotic Design. Chem. Biol 2011, 18, 38–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (973).Clayton GM; Klein DJ; Rickert KW; Patel SB; Kornienko M; Zugay-Murphy J; Reid JC; Tummala S; Sharma S; Singh SB; Miesel L; Lumb KJ; Soisson SM Structure of the Bacterial Deacetylase LpxC Bound to the Nucleotide Reaction Product Reveals Mechanisms of Oxyanion Stabilization and Proton Transfer. J. Biol. Chem 2013, 288, 34073–34080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (974).Kline T; Andersen NH; Harwood EA; Bowman J; Malanda A; Endsley S; Erwin AL; Doyle M; Fong S; Harris AL; Mendelsohn B; Mdluli K; Raetz CR; Stover CK; Witte PR; Yabannavar A; Zhu S Potent, Novel in Vitro Inhibitors of the Pseudomonas aeruginosa Deacetylase LpxC. J. Med. Chem 2002, 45, 3112–3129. [DOI] [PubMed] [Google Scholar]
- (975).Pirrung MC; Tumey LN; McClerren AL; Raetz CR High-throughput Catch-and-release Synthesis of Oxazoline Hydroxamates. Structure-activity Relationships in Novel Inhibitors of Escherichia coli LpxC: in Vitro Enzyme Inhibition and Antibacterial Properties. J. Am. Chem. Soc 2003, 125, 1575–1586. [DOI] [PubMed] [Google Scholar]
- (976).Montgomery JI; Brown MF; Reilly U; Price LM; Abramite JA; Arcari J; Barham R; Che Y; Chen JM; Chung SW; Collantes EM; Desbonnet C; Doroski M; Doty J; Engtrakul JJ; Harris TM; Huband M; Knafels JD; Leach KL; Liu S; Marfat A; McAllister L; McElroy E; Menard CA; Mitton-Fry M; Mullins L; Noe MC; O’Donnell J; Oliver R; Penzien J; Plummer M; Shanmugasundaram V; Thoma C; Tomaras AP; Uccello DP; Vaz A; Wishka DG Pyridone Methylsulfone Hydroxamate LpxC Inhibitors for the Treatment of Serious Gram-negative Infections. J. Med. Chem 2012, 55, 1662–1670. [DOI] [PubMed] [Google Scholar]
- (977).Brown MF; Reilly U; Abramite JA; Arcari JT; Oliver R; Barham RA; Che Y; Chen JM; Collantes EM; Chung SW; Desbonnet C; Doty J; Doroski M; Engtrakul JJ; Harris TM; Huband M; Knafels JD; Leach KL; Liu S; Marfat A; Marra A; McElroy E; Melnick M; Menard CA; Montgomery JI; Mullins L; Noe MC; O’Donnell J; Penzien J; Plummer MS; Price LM; Shanmugasundaram V; Thoma C; Uccello DP; Warmus JS; Wishka DG Potent Inhibitors of LpxC for the Treatment of Gram-negative Infections. J. Med. Chem 2012, 55, 914–923. [DOI] [PubMed] [Google Scholar]
- (978).McAllister LA; Montgomery JI; Abramite JA; Reilly U; Brown MF; Chen JM; Barham RA; Che Y; Chung SW; Menard CA; Mitton-Fry M; Mullins LM; Noe MC; O’Donnell JP; Oliver RM 3rd; Penzien JB; Plummer M; Price LM; Shanmugasundaram V; Tomaras AP; Uccello DP Heterocyclic Methylsulfone Hydroxamic Acid LpxC Inhibitors as Gram-negative Antibacterial Agents. Bioorg. Med. Chem. Lett 2012, 22, 6832–6838. [DOI] [PubMed] [Google Scholar]
- (979).Tomaras AP; McPherson CJ; Kuhn M; Carifa A; Mullins L; George D; Desbonnet C; Eidem TM; Montgomery JI; Brown MF; Reilly U; Miller AA; O’Donnell JP LpxC Inhibitors as New Antibacterial Agents and Tools for Studying Regulation of Lipid A Biosynthesis in Gram-negative Pathogens. mBio 2014, 5, e01551–e01614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (980).Kurasaki H; Tsuda K; Shinoyama M; Takaya N; Yamaguchi Y; Kishii R; Iwase K; Ando N; Nomura M; Kohno Y LpxC Inhibitors: Design, Synthesis, and Biological Evaluation of Oxazolidinones as Gram-negative Antibacterial Agents. ACS Med. Chem. Lett 2016, 7, 623–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (981).Dmitrii VK; Ralph H Insights into the Zinc-dependent Deacetylase LpxC: Biochemical Properties and Inhibitor Design. Curr. Top. Med. Chem 2016, 16, 2379–2430. [DOI] [PubMed] [Google Scholar]
- (982).Zhang J; Chan A; Lippa B; Cross JB; Liu C; Yin N; Romero JA; Lawrence J; Heney R; Herradura P; Goss J; Clark C; Abel C; Zhang Y; Poutsiaka KM; Epie F; Conrad M; Mahamoon A; Nguyen K; Chavan A; Clark E; Li TC; Cheng RK; Wood M; Andersen OA; Brooks M; Kwong J; Barker J; Parr IB; Gu Y; Ryan MD; Coleman S; Metcalf CA 3rd Structure-based Discovery of LpxC Inhibitors. Bioorg. Med. Chem. Lett 2017, 27, 1670–1680. [DOI] [PubMed] [Google Scholar]
- (983).Zhang J; Zhang L; Li X; Xu W UDP-3-O-(R-3-hydroxymyristoyl)-N-Acetylglucosamine Deacetylase (LpxC) Inhibitors: A New Class of Antibacterial Agents. Curr. Med. Chem 2012, 19, 2038–2050. [DOI] [PubMed] [Google Scholar]
- (984).Mansoor UF; Vitharana D; Reddy PA; Daubaras DL; McNicholas P; Orth P; Black T; Siddiqui MA Design and Synthesis of Potent Gram-negative Specific LpxC Inhibitors. Bioorg. Med. Chem. Lett 2011, 21, 1155–1161. [DOI] [PubMed] [Google Scholar]
- (985).Li XC; McClerren AL; Raetz CRH; Hindsgaul O Mapping the Active Site of the Bacterial Enzyme LpxC Using Novel Carbohydrate-based Hydroxamic Acid Inhibitors. J. Carbohydr. Chem 2005, 24, 583–609. [Google Scholar]
- (986).Anderson NH; Bowman J; Erwin A; Harwood E; Kline T; Mdluli K; Ng S; Pfister KB; Shawar R; Wagman A; Emeryville CC, Ed. 2004.
- (987).Raju BG; O’Dowd H; Gao H; Patel DV; Trias J; Inc, N. U. P., Ed. 2008. [Google Scholar]
- (988).Dobler MR; Lenoir F; Parker DT; Peng Y; Piizzi G; Wattanasin S; AG N, Ed. 2010.
- (989).Kalinin DV; Holl R LpxC Inhibitors: a Patent Review (2010–2016). Expert Opin. Ther. Pat 2017, 27, 1227–1250. [DOI] [PubMed] [Google Scholar]
- (990).Onishi HR; Pelak BA; Gerckens LS; Silver LL; Kahan FM; Chen MH; Patchett AA; Galloway SM; Hyland SA; Anderson MS; Raetz CR Antibacterial Agents That Inhibit Lipid A Biosynthesis. Science 1996, 274, 980–982. [DOI] [PubMed] [Google Scholar]
- (991).Chen M-H; Steiner MG; de Laszlo SE; Patchett AA; Anderson MS; Hyland SA; Russell Onishi H; Silyer LL; Raetz CRH Carbohydroxamido-oxazolidines: Antibacterial Agents That Target Lipid A Biosynthesis. Bioorg. Med. Chem. Lett 1999, 9, 313–318. [DOI] [PubMed] [Google Scholar]
- (992).Jackman JE; Fierke CA; Tumey LN; Pirrung M; Uchiyama T; Tahir SH; Hindsgaul O; Raetz CR Antibacterial Agents that Target Lipid A Biosynthesis in Gram-negative Bacteria. Inhibition of Diverse UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine deacetylases by Substrate Analogs Containing Zinc Binding Motifs. J. Biol. Chem 2000, 275, 11002–11009. [DOI] [PubMed] [Google Scholar]
- (993).Clements JM; Coignard F; Johnson I; Chandler S; Palan S; Waller A; Wijkmans J; Hunter MG Antibacterial Activities and Characterization of Novel Inhibitors of LpxC. Antimicrob. Agents Chemother 2002, 46, 1793–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (994).McClerren AL; Endsley S; Bowman JL; Andersen NH; Guan Z; Rudolph J; Raetz CR A Slow, Tight-binding Inhibitor of the Zinc-dependent Deacetylase LpxC of Lipid A Biosynthesis with Antibiotic Activity Comparable to Ciprofloxacin. Biochemistry 2005, 44, 16574–16583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (995).Barb AW; Jiang L; Raetz CR; Zhou P Structure of the Deacetylase LpxC Bound to the Antibiotic CHIR-090: Time-dependent Inhibition and Specificity in Ligand Binding. Proc. Natl. Acad. Sci. U. S. A 2007, 104, 18433–18438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (996).Barb AW; McClerren AL; Snehelatha K; Reynolds CM; Zhou P; Raetz CR Inhibition of Lipid A Biosynthesis as the Primary Mechanism of CHIR-090 Antibiotic Activity in Escherichia coli. Biochemistry 2007, 46, 3793–3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (997).Liang X; Lee CJ; Chen X; Chung HS; Zeng D; Raetz CR; Li Y; Zhou P; Toone EJ Syntheses, Structures and Antibiotic Activities of LpxC Inhibitors Based on the Diacetylene Scaffold. Bioorg. Med. Chem 2011, 19, 852–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (998).Piizzi G; Parker DT; Peng Y; Dobler M; Patnaik A; Wattanasin S; Liu E; Lenoir F; Nunez J; Kerrigan J; McKenney D; Osborne C; Yu D; Lanieri L; Bojkovic J; Dzink-Fox J; Lilly MD; Sprague ER; Lu Y; Wang H; Ranjitkar S; Xie L; Wang B; Glick M; Hamann LG; Tommasi R; Yang X; Dean CR Design, Synthesis, and Properties of a Potent Inhibitor of Pseudomonas aeruginosa Deacetylase LpxC. J. Med. Chem 2017, 60, 5002–5014. [DOI] [PubMed] [Google Scholar]
- (999).Cole KE; Gattis SG; Angell HD; Fierke CA; Christianson DW Structure of the Metal-dependent Deacetylase LpxC from Yersinia enterocolitica Complexed with the Potent Inhibitor CHIR-090. Biochemistry 2011, 50, 258–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1000).Titecat M; Liang X; Lee C-J; Charlet A; Hocquet D; Lambert T; Pagès J-M; Courcol R; Sebbane F; Toone EJ; Zhou P; Lemaitre N High Susceptibility of MDR and XDR Gram-negative Pathogens to Biphenyl-diacetylene-based Difluoromethyl-allo-threonyl-hydroxamate LpxC Inhibitors. J. Antimicrob. Chemother 2016, 71, 2874–2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1001).Kasar R; Linsell MS; Aggen JB; LU Q; Wang D; CHURCH T; Moser HE; Patten PA 2012.
- (1002).Pirrung MC; Tumey LN; Raetz CR; Jackman JE; Snehalatha K; McClerren AL; Fierke CA; Gantt SL; Rusche KM Inhibition of the Antibacterial Target UDP-(3-O-Acyl)-N-acetylglucosamine Deacetylase (LpxC): Isoxazoline Zinc Amidase Inhibitors Bearing Diverse Metal Binding Groups. J. Med. Chem 2002, 45, 4359–4370. [DOI] [PubMed] [Google Scholar]
- (1003).Cohen SM; Puerta DT; Perez C 2015.
- (1004).Nammalwar B; Teng M; Taganov K; Puerta DT 2017.
- (1005).Lee W; McDonough MA; Kotra L; Li ZH; Silvaggi NR; Takeda Y; Kelly JA; Mobashery S A 1.2-A Snapshot of the Final Step of Bacterial Cell Wall Biosynthesis. Proc. Natl. Acad. Sci. U. S. A 2001, 98, 1427–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1006).Drawz SM; Bonomo RA Three Decades of Beta-Lactamase Inhibitors. Clin. Microbiol. Rev 2010, 23, 160–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1007).Fisher JF; Meroueh SO; Mobashery S Bacterial Resistance to Beta-Lactam Antibiotics: Compelling Opportunism, Compelling Opportunity. Chem. Rev 2005, 105, 395–424. [DOI] [PubMed] [Google Scholar]
- (1008).Rotondo CM; Wright GD Inhibitors of Metallo-Beta-Lactamases. Curr. Opin. Microbiol 2017, 39, 96–105. [DOI] [PubMed] [Google Scholar]
- (1009).Nordmann P; Poirel L; Walsh TR; Livermore DM The Emerging NDM Carbapenemases. Trends Microbiol 2011, 19, 588–595. [DOI] [PubMed] [Google Scholar]
- (1010).Sekizuka T; Matsui M; Yamane K; Takeuchi F; Ohnishi M; Hishinuma A; Arakawa Y; Kuroda M Complete Sequencing of the bla(NDM-1)-positive IncA/C Plasmid from Escherichia coli ST38 Isolate Suggests a Possible Origin from Plant Pathogens. PLoS One 2011, 6, e25334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1011).Wailan AM; Paterson DL The Spread and Acquisition of NDM-1: A Multifactorial Problem. Expert Rev. Anti-Infect. Ther 2014, 12, 91–115. [DOI] [PubMed] [Google Scholar]
- (1012).Yong D; Toleman MA; Giske CG; Cho HS; Sundman K; Lee K; Walsh TR Characterization of a New Metallo-Beta-Lactamase Gene, bla(NDM-1), and a Novel Erythromycin Esterase Gene Carried on a Unique Genetic Structure in Klebsiella pneumoniae Sequence Type 14 from India. Antimicrob. Agents Chemother 2009, 53, 5046–5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1013).Berrazeg M; Diene S; Medjahed L; Parola P; Drissi M; Raoult D; Rolain J New Delhi Metallo-Beta-Lactamase Around the World: an eReview using Google Maps. Euro. Surveill 2014, 19. [DOI] [PubMed] [Google Scholar]
- (1014).Woodford N; Turton JF; Livermore DM Multiresistant Gram-Negative Bacteria: The Role of High-Risk Clones in the Dissemination of Antibiotic Resistance. FEMS Microbiol. Rev 2011, 35, 736–755. [DOI] [PubMed] [Google Scholar]
- (1015).Walsh TR Emerging Carbapenemases: A Global Perspective. Int. J. Antimicrob. Agents 2010, 36 Suppl 3, S8–14. [DOI] [PubMed] [Google Scholar]
- (1016).Moellering RC Jr. NDM-1-A Cause for Worldwide Concern. N. Engl. J. Med 2010, 363, 2377–2379. [DOI] [PubMed] [Google Scholar]
- (1017).Kim Y; Cunningham MA; Mire J; Tesar C; Sacchettini J; Joachimiak A NDM-1, the Ultimate Promiscuous Enzyme: Substrate Recognition and Catalytic Mechanism. FASEB J 2013, 27, 1917–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1018).Thomas PW; Zheng M; Wu SS; Guo H; Liu DL; Xu DG; Fast W Characterization of Purified New Delhi Metallo-beta-lactamase-1. Biochemistry 2011, 50, 10102–10113. [DOI] [PubMed] [Google Scholar]
- (1019).Zhang H; Hao Q Crystal Structure of NDM-1 Reveals a Common Beta-Lactam Hydrolysis Mechanism. FASEB J 2011, 25, 2574–2582. [DOI] [PubMed] [Google Scholar]
- (1020).Aitha M; Moller AJ; Sahu ID; Horitani M; Tierney DL; Crowder MW Investigating the Position of the Hairpin Loop in New Delhi Metallo-Beta-Lactamase, NDM-1, during Catalysis and Inhibitor Binding. J. Inorg. Biochem 2016, 156, 35–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1021).Palzkill T Metallo-Beta-Lactamase Structure and Function. Ann. N. Y. Acad. Sci 2013, 1277, 91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1022).Yang H; Aitha M; Marts AR; Hetrick A; Bennett B; Crowder MW; Tierney DL Spectroscopic and Mechanistic Studies of Heterodimetallic Forms of Metallo-Beta-Lactamase NDM-1. J. Am. Chem. Soc 2014, 136, 7273–7285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1023).Rogers BA; Sidjabat HE; Silvey A; Anderson TL; Perera S; Li J; Paterson DL Treatment Options for New Delhi Metallo-Beta-Lactamase-Harboring Enterobacteriaceae. Microb. Drug Resist 2013, 19, 100–103. [DOI] [PubMed] [Google Scholar]
- (1024).Garonzik SM; Li J; Thamlikitkul V; Paterson DL; Shoham S; Jacob J; Silveira FP; Forrest A; Nation RL Population Pharmacokinetics of Colistin Methanesulfonate and formed Colistin in ritically Ill Patients from a Multicenter Study Provide Dosing Suggestions for Various Categories of Patients. Antimicrob. Agents Chemother 2011, 55, 3284–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1025).Concha NO; Janson CA; Rowling P; Pearson S; Cheever CA; Clarke BP; Lewis C; Galleni M; Frere JM; Payne DJ; Bateson JH; Abdel-Meguid SS Crystal Structure of the IMP-1 Metallo Beta-Lactamase from Pseudomonas Aeruginosa and its Complex with a Mercaptocarboxylate Inhibitor: Binding Determinants of a Potent, Broad-Spectrum Inhibitor. Biochemistry 2000, 39, 4288–4298. [DOI] [PubMed] [Google Scholar]
- (1026).Yamaguchi Y; Jin W; Matsunaga K; Ikemizu S; Yamagata Y; Wachino JI; Shibata N; Arakawa Y; Kurosaki H Crystallographic Investigation of the Inhibition Mode of a VIM-2 Metallo-Beta-Lactamase from Pseudomonas aeruginosa by a Mercaptocarboxylate Inhibitor. J. Med. Chem 2007, 50, 6647–6653. [DOI] [PubMed] [Google Scholar]
- (1027).Lassaux P; Hamel M; Gulea M; Delbruck H; Mercuri PS; Horsfall L; Dehareng D; Kupper M; Frere JM; Hoffmann K; Galleni M; Bebrone C Mercaptophosphonate Compounds as Broad-Spectrum Inhibitors of the Metallo-Beta-Lactamases. J. Med. Chem 2010, 53, 4862–4876. [DOI] [PubMed] [Google Scholar]
- (1028).Schlesinger SR; Bruner B; Farmer PJ; Kim SK Kinetic Characterization of a Slow-Binding Inhibitor of Bla2: Thiomaltol. J. Enzyme Inhib. Med. Chem 2013, 28, 137–142. [DOI] [PubMed] [Google Scholar]
- (1029).Brem J; van Berkel SS; Aik W; Rydzik AM; Avison MB; Pettinati I; Umland KD; Kawamura A; Spencer J; Claridge TDW; McDonough MA; Schofield CJ Rhodanine Hydrolysis Leads to Potent Thioenolate Mediated Metallo-Beta-Lactamase Inhibition. Nat. Chem 2014, 6, 1084–1090. [DOI] [PubMed] [Google Scholar]
- (1030).Gonzalez MM; Kosmopoulou M; Mojica MF; Castillo V; Hinchliffe P; Pettinati I; Brem J; Schofield CJ; Mahler G; Bonomo RA; Llarrull LI; Spencer J; Vila AJ Bisthiazolidines: A Substrate-Mimicking Scaffold as an Inhibitor of the NDM-1 Carbapenemase. ACS Infect. Dis 2015, 1, 544–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1031).Christopeit T; Carlsen TJO; Helland R; Leiros HKS Discovery of Novel Inhibitor Scaffolds against the Metallo-beta-lactamase VIM-2 by Surface Plasmon Resonance (SPR) Based Fragment Screening. J. Med. Chem 2015, 58, 8671–8682. [DOI] [PubMed] [Google Scholar]
- (1032).Sevaille L; Gavara L; Bebrone C; De Luca F; Nauton L; Achard M; Mercuri P; Tanfoni S; Borgianni L; Guyon C; Lonjon P; Turan-Zitouni G; Dzieciolowski J; Becker K; Benard L; Condon C; Maillard L; Martinez J; Frere JM; Dideberg O; Galleni M; Docquier JD; Hernandez JF 1,2,4-Triazole-3-thione Compounds as Inhibitors of Dizinc Metallo-beta-lactamases. ChemMedChem 2017, 12, 972–985. [DOI] [PubMed] [Google Scholar]
- (1033).Skagseth S; Akhter S; Paulsen MH; Muhammad Z; Lauksund S; Samuelsen O; Leiros HKS; Bayer A Metallo-Beta-Lactamase Inhibitors by Bioisosteric Replacement: Preparation, Activity and Binding. Eur. J. Med. Chem 2017, 135, 159–173. [DOI] [PubMed] [Google Scholar]
- (1034).Shin WS; Bergstrom A; Bonomo RA; Crowder MW; Muthyala R; Sham YY Discovery of 1-Hydroxypyridine-2(1H)-thione-6-carboxylic Acid as a First-in-Class Low-Cytotoxic Nanomolar Metallo -Lactamase Inhibitor. ChemMedChem 2017, 12, 845–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1035).Cain R; Brem J; Zollman D; McDonough MA; Johnson RM; Spencer J; Makena A; Abboud MI; Cahill S; Lee SY; McHugh PJ; Schofield CJ; Fishwick CWG In Silico Fragment-Based Design Identifies Subfamily B1 Metallo-beta-lactamase Inhibitors. J. Med. Chem 2018, 61, 1255–1260. [DOI] [PubMed] [Google Scholar]
- (1036).Liu S; Jing L; Yu ZJ; Wu C; Zheng Y; Zhang E; Chen Q; Yu Y; Guo L; Wu Y; Li GB ((S)-3-Mercapto-2-Methylpropanamido)Acetic Acid Derivatives as Metallo-Beta-Lactamase Inhibitors: Synthesis, Kinetic and Crystallographic Studies. Eur. J. Med. Chem 2018, 145, 649–660. [DOI] [PubMed] [Google Scholar]
- (1037).Johnson JW; Gretes M; Goodfellow VJ; Marrone L; Heynen ML; Strynadka NCJ; Dmitrienko GI Cyclobutanone Analogues of beta-Lactams Revisited: Insights into Conformational Requirements for Inhibition of Serine- and Metallo-beta-Lactamases. J. Am. Chem. Soc 2010, 132, 2558–2560. [DOI] [PubMed] [Google Scholar]
- (1038).Yang KW; Feng L; Yang SK; Aitha M; LaCuran AE; Oelschlaeger P; Crowder MW New Beta-Phospholactam as a Carbapenem Transition State Analog: Synthesis of a Broad-Spectrum Inhibitor of Metallo-Beta-Lactamases. Bioorg. Med. Chem. Lett 2013, 23, 5855–5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1039).Livermore DM; Mushtaq S; Morinaka A; Ida T; Maebashi K; Hope R Activity of Carbapenems with ME1071 (Disodium 2,3-Diethylmaleate) Against Enterobacteriaceae and Acinetobacter spp. with Carbapenemases, Including NDM Enzymes. J. Antimicrob. Chemother 2013, 68, 153–158. [DOI] [PubMed] [Google Scholar]
- (1040).Gan ML; Liu YF; Bai YL; Guan Y; Li L; Gao RM; He WY; You XF; Li YH; Yu LY; Xiao CL Polyketides with New Delhi Metallo-beta-lactamase 1 Inhibitory Activity from Penicillium sp. J. Nat. Prod 2013, 76, 1535–1540. [DOI] [PubMed] [Google Scholar]
- (1041).Shen BZ; Yu Y; Chen H; Cao X; Lao XZ; Fang YL; Shi Y; Chen J; Zheng H Inhibitor Discovery of Full-Length New Delhi Metallo-beta-Lactamase-1 (NDM-1). PLoS One 2013, 8, e62955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1042).Hiraiwa Y; Saito J; Watanabe T; Yamada M; Morinaka A; Fukushima T; Kudo T X-Ray Crystallographic Analysis of IMP-1 Metallo-Beta-Lactamase Complexed with a 3-Aminophthalic Acid Derivative, Structure-Based Drug Design, and Synthesis of 3,6-Disubstituted Phthalic Acid Derivative Inhibitors. Bioorg. Med. Chem. Lett 2014, 24, 4891–4894. [DOI] [PubMed] [Google Scholar]
- (1043).Christopeit T; Yang KW; Yang SK; Leiros HKS The Structure of the Metallo-Beta-Lactamase VIM-2 in Complex with a Triazolylthioacetamide Inhibitor. Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun 2016, 72, 813–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1044).Christopeit T; Albert A; Leiros HKS Discovery of a Novel Covalent Non-Beta-Lactam Inhibitor of the Metallo-Beta-Lactamase NDM-1. Bioorg. Med. Chem 2016, 24, 2947–2953. [DOI] [PubMed] [Google Scholar]
- (1045).Brem J; Cain R; Cahill S; McDonough MA; Clifton IJ; Jimenez-Castellanos JC; Avison MB; Spencer J; Fishwick CWG; Schofield CJ Structural Basis of Metallo-Beta-Lactamase, Serine-Beta-Lactamase and Penicillin-Binding Protein Inhibition by Cyclic Boronates. Nat. Commun 2016, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1046).Santucci M; Spyrakis F; Cross S; Quotadamo A; Farina D; Tondi D; De Luca F; Docquier JD; Prieto AI; Ibacache C; Blazquez J; Venturelli A; Cruciani G; Costi MP Computational and Biological Profile of Boronic Acids for the Detection of Bacterial Serine- and Metallo-Beta-Lactamases. Sci. Rep 2017, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1047).Falconer SB; Reid-Yu SA; King AM; Gehrke SS; Wang WL; Britten JF; Coombes BK; Wright GD; Brown ED Zinc Chelation by a Small-Molecule Adjuvant Potentiates Meropenem Activity in Vivo against NDM-1-Producing Klebsiella pneumoniae. ACS Infect. Dis 2015, 1, 533–543. [DOI] [PubMed] [Google Scholar]
- (1048).Hinchliffe P; Tanner CA; Krismanich AP; Labbe G; Goodfellow VJ; Marrone L; Desoky AY; Calvopiña K; Whittle EE; Zeng F; Avison MB; Bols NC; Siemann S; Spencer J; Dmitrienko GI Structural and Kinetic Studies of the Potent Inhibition of Metallo-β-lactamases by 6-Phosphonomethylpyridine-2-carboxylates. Biochemistry 2018, 57, 1880–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1049).King AM; Reid-Yu SA; Wang W; King DT; De Pascale G; Strynadka NC; Walsh TR; Coombes BK; Wright GD Aspergillomarasmine A Overcomes Metallo-Beta-Lactamase Antibiotic Resistance. Nature 2014, 510, 503–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1050).Brem J; van Berkel SS; Zollman D; Lee SY; Gileadi O; McHugh PJ; Walsh TR; McDonough MA; Schofield CJ Structural Basis of Metallo-beta-Lactamase Inhibition by Captopril Stereoisomers. Antimicrob. Agents Chemother 2016, 60, 142–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1051).Klingler FM; Wichelhaus TA; Frank D; Cuesta-Bernal J; El-Delik J; Muller HF; Sjuts H; Gottig S; Koenigs A; Pos KM; Pogoryelov D; Proschak E Approved Drugs Containing Thiols as Inhibitors of Metallo-Beta-Lactamases: Strategy To Combat Multidrug-Resistant Bacteria. J. Med. Chem 2015, 58, 3626–3630. [DOI] [PubMed] [Google Scholar]
- (1052).Chen ALY; Thomas PW; Stewart AC; Bergstrom A; Cheng ZS; Miller C; Bethel CR; Marshal SH; Credille CV; Riley CL; Page RC; Bonomo RA; Crowder MW; Tierney DL; Fast W; Cohen SM Dipicolinic Acid Derivatives as Inhibitors of New Delhi Metallo-beta-lactamase-1. J. Med. Chem 2017, 60, 7267–7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1053).King DT; Worrall LJ; Gruninger R; Strynadka NCJ New Delhi Metallo-beta-Lactamase: Structural Insights into beta-Lactam Recognition and Inhibition. J. Am. Chem. Soc 2012, 134, 11362–11365. [DOI] [PubMed] [Google Scholar]
- (1054).Abboud MI; Kosmopoulou M; Krismanich AP; Johnson JW; Hinchliffe P; Brem J; Claridge TDW; Spencer J; Schofield CJ; Dmitrienko GI Cyclobutanone Mimics of Intermediates in Metallo-beta-Lactamase Catalysis. Chemistry 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1055).Burns CJ; Goswami R; Jackson RW; Lessen T; Li W; Pevear D; Tirunahari PK; Xu H; Novartis International Pharmaceutical Ltd: 2010. [Google Scholar]
- (1056).Yoshizumi A; Ishii Y; Livermore DM; Woodford N; Kimura S; Saga T; Harada S; Yamaguchi K; Tateda K Efficacies of Calcium-EDTA in Combination with Imipenem in a Murine Model of Sepsis caused by Escherichia coli with NDM-1 Beta-Lactamase. J. Infect. Chemother 2013, 19, 992–995. [DOI] [PubMed] [Google Scholar]
- (1057).Shlaes DM New beta-lactam-beta-lactamase Inhibitor Combinations in Clinical Development. Ann. N.Y. Acad. Sci 2013, 1277, 105–114. [DOI] [PubMed] [Google Scholar]
- (1058).Cahill ST; Cain R; Wang DY; Lohans CT; Wareham DW; Oswin HP; Mohammed J; Spencer J; Fishwick CWG; McDonough MA; Schofield CJ; Brema J Cyclic Boronates Inhibit All Classes of -Lactamases. Antimicrob. Agents Chemother. 2017, 61, e02260–02216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1059).Pei D Peptide Deformylase: a Target for Novel Antibiotics? Expert Opin. Ther. Targets 2001, 5, 23–40. [DOI] [PubMed] [Google Scholar]
- (1060).Kozak M Comparison of Initiation of Protein Synthesis in Procaryotes, Eucaryotes, and Organelles. Microbiol. Rev 1983, 47, 1–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1061).Solbiati J; Chapman-Smith A; Miller JL; Miller CG; Cronan JE Jr. Processing of the N Termini of Nascent Polypeptide Chains Requires Deformylation Prior to Methionine Removal. J. Mol. Biol 1999, 290, 607–614. [DOI] [PubMed] [Google Scholar]
- (1062).Mazel D; Pochet S; Marliere P Genetic Characterization of Polypeptide Deformylase, a Distinctive Enzyme of Eubacterial Translation. EMBO J 1994, 13, 914–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1063).Escobar-Alvarez S; Goldgur Y; Yang G; Ouerfelli O; Li Y; Scheinberg DA Structure and Activity of Human Mitochondrial Peptide Deformylase, a Novel Cancer Target. J. Mol. Biol 2009, 387, 1211–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1064).Meinnel T; Lazennec C; Villoing S; Blanquet S Structure-Function Relationships Within the Peptide Deformylase Family. Evidence for a Conserved Architecture of the Active Site Involving Three Conserved Motifs and a Metal Ion. J. Mol. Biol 1997, 267, 749–761. [DOI] [PubMed] [Google Scholar]
- (1065).Becker A; Schlichting I; Kabsch W; Groche D; Schultz S; Wagner AF Iron Center, Substrate Recognition and Mechanism of Peptide Deformylase. Nat. Struct. Biol 1998, 5, 1053–1058. [DOI] [PubMed] [Google Scholar]
- (1066).Meinnel T; Blanquet S Evidence that Peptide Deformylase and Methionyl-Trna(F)(Met) Formyltransferase are Encoded Within the Same Operon in Escherichia-Coli. J. Bacteriol 1993, 175, 7737–7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1067).Rajagopalan PTR; Yu XC; Pei DH Peptide Deformylase: A New Type of Mononuclear Iron Protein. J. Am. Chem. Soc 1997, 119, 12418–12419. [Google Scholar]
- (1068).Groche D; Becker A; Schlichting I; Kabsch W; Schultz S; Wagner AFV Isolation and Crystallization of Functionally Competent Escherichia coli Peptide Deformylase Forms Containing Either Iron or Nickel in the Active Site. Biochem. Biophys. Res. Commun 1998, 246, 342–346. [DOI] [PubMed] [Google Scholar]
- (1069).Ragusa S; Blanquet S; Meinnel T Control of Peptide Deformylase Activity by Metal Cations. J. Mol. Biol 1998, 280, 515–523. [DOI] [PubMed] [Google Scholar]
- (1070).Rajagopalan PTR; Grimme S; Pei DH Characterization of Cobalt(II)-Substituted Peptide Deformylase: Function of the Metal Ion and the Catalytic Residue Glu-133. Biochemistry 2000, 39, 779–790. [DOI] [PubMed] [Google Scholar]
- (1071).Becker A; Schlichting I; Kabsch W; Schultz S; Wagner AF Structure of Peptide Deformylase and Identification of the Substrate Binding Site. J. Biol. Chem 1998, 273, 11413–11416. [DOI] [PubMed] [Google Scholar]
- (1072).Meinnel T; Blanquet S Enzymatic Properties of Escherichia coli Peptide Deformylase. J. Bacteriol 1995, 177, 1883–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1073).Hu YJ; Rajagopalan PTR; Pei DH H-phosphonate Derivatives as Novel Peptide Deformylase Inhibitors. Bioorg. Med. Chem. Lett 1998, 8, 2479–2482. [DOI] [PubMed] [Google Scholar]
- (1074).Hao B; Gong WM; Rajagopalan PTR; Zhou Y; Pei DH; Chan MK Structural Basis for the Design of Antibiotics Targeting Peptide Deformylase. Biochemistry 1999, 38, 4712–4719. [DOI] [PubMed] [Google Scholar]
- (1075).Meinnel T; Patiny L; Ragusa S; Blanquet S Design and Synthesis of Substrate Analogue Inhibitors of Peptide Deformylase. Biochemistry 1999, 38, 4287–4295. [DOI] [PubMed] [Google Scholar]
- (1076).Huntington KM; Yi T; Wei Y; Pei D Synthesis and Antibacterial Activity of Peptide Deformylase Inhibitors. Biochemistry 2000, 39, 4543–4551. [DOI] [PubMed] [Google Scholar]
- (1077).Durand DJ; Gordon Green B; O’Connell JF; Grant SK Peptide Aldehyde Inhibitors of Bacterial Peptide Deformylases. Arch. Biochem. Biophys 1999, 367, 297–302. [DOI] [PubMed] [Google Scholar]
- (1078).Chen DZ; Patel DV; Hackbarth CJ; Wang W; Dreyer G; Young DC; Margolis PS; Wu C; Ni ZJ; Trias J; White RJ; Yuan Z Actinonin, a Naturally Occurring Antibacterial Agent, is a Potent Deformylase Inhibitor. Biochemistry 2000, 39, 1256–1262. [DOI] [PubMed] [Google Scholar]
- (1079).Gordon JJ; Kelly BK; Miller GA Actinonin: An Antibiotic Substance Produced by an Actinomycete. Nature 1962, 195, 701–702. [DOI] [PubMed] [Google Scholar]
- (1080).East SP In Antimicrobials: New and Old Molecules in the Fight Against Multi-resistant Bacteria; Marinelli F, Genilloud O, Eds.; Springer: Berlin, 2014, p 287–305. [Google Scholar]
- (1081).Clements JM; Beckett RP; Brown A; Catlin G; Lobell M; Palan S; Thomas W; Whittaker M; Wood S; Salama S; Baker PJ; Rodgers HF; Barynin V; Rice DW; Hunter MG Antibiotic Activity and Characterization of BB-3497, a Novel Peptide Deformylase Inhibitor. Antimicrob. Agents Chemother 2001, 45, 563–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1082).Jain R; Chen D; White RJ; Patel DV; Yuan Z Bacterial Peptide Deformylase Inhibitors: a New Class of Antibacterial Agents. Curr. Med. Chem 2005, 12, 1607–1621. [DOI] [PubMed] [Google Scholar]
- (1083).Sangshetti JN; Khan FA; Shinde DB Peptide Deformylase: a New Target in Antibacterial, Antimalarial and Anticancer Drug Discovery. Curr. Med. Chem 2015, 22, 214–236. [DOI] [PubMed] [Google Scholar]
- (1084).Apfel C; Banner DW; Bur D; Dietz M; Hirata T; Hubschwerlen C; Locher H; Page MG; Pirson W; Rosse G; Specklin JL Hydroxamic Acid Derivatives as Potent Peptide Deformylase Inhibitors and Antibacterial Agents. J. Med. Chem 2000, 43, 2324–2331. [DOI] [PubMed] [Google Scholar]
- (1085).Grant SK; Green BG; Kozarich JW Inhibition and Structure-Activity Studies of Methionine Hydroxamic Acid Derivatives with Bacterial Peptide Deformylase. Bioorg. Chem 2001, 29, 211–222. [DOI] [PubMed] [Google Scholar]
- (1086).Thorarensen A; Deibel MR Jr.; Rohrer DC; Vosters AF; Yem AW; Marshall VD; Lynn JC; Bohanon MJ; Tomich PK; Zurenko GE; Sweeney MT; Jensen RM; Nielsen JW; Seest EP; Dolak LA Identification of Novel Potent Hydroxamic Acid Inhibitors of Peptidyl Deformylase and the Importance of the Hydroxamic Acid Functionality on Inhibition. Bioorg. Med. Chem. Lett 2001, 11, 1355–1358. [DOI] [PubMed] [Google Scholar]
- (1087).Jayasekera MM; Kendall A; Shammas R; Dermyer M; Tomala M; Shapiro MA; Holler TP Novel Nonpeptidic Inhibitors of Peptide Deformylase. Arch. Biochem. Biophys 2000, 381, 313–316. [DOI] [PubMed] [Google Scholar]
- (1088).Green BG; Toney JH; Kozarich JW; Grant SK Inhibition of Bacterial Peptide Deformylase by Biaryl Acid Analogs. Arch. Biochem. Biophys 2000, 375, 355–358. [DOI] [PubMed] [Google Scholar]
- (1089).Berg AK; Yu QF; Qian SY; Haldar MK; Srivastava DK Solvent-Assisted Slow Conversion of a Dithiazole Derivative Produces a Competitive Inhibitor of Peptide Deformylase. Biochim. Biophys. Acta, Proteins Proteomics 2010, 1804, 704–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1090).Naderer OJ; Dumont E; Zhu J; Kurtinecz M; Jones LS Single-Dose Safety, Tolerability, and Pharmacokinetics of the Antibiotic GSK1322322, a Novel Peptide Deformylase Inhibitor. Antimicrob. Agents Chemother 2013, 57, 2005–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1091).Corey R; Naderer OJ; O’Riordan WD; Dumont E; Jones LS; Kurtinecz M; Zhu JZ Safety, Tolerability, and Efficacy of GSK1322322 in the Treatment of Acute Bacterial Skin and Skin Structure Infections. Antimicrob. Agents Chemother 2014, 58, 6518–6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1092).Silver LL Challenges of Antibacterial Discovery. Clin. Microbiol. Rev 2011, 24, 71–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1093).World Health Organization, Global Tuberculosis Report 2017. Global Tuberculosis Report 2017 2017, 1–262.
- (1094).Houben RMGJ; Dodd PJ The Global Burden of Latent Tuberculosis Infection: A Re-estimation Using Mathematical Modelling. PLoS Medicine 2016, 13, e1002152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1095).Rutter WJ Evolution of Aldolase. Fed. Proc 1964, 23, 1248–1257. [PubMed] [Google Scholar]
- (1096).Qamar S; Marsh K; Berry A Identification of Arginine 331 as an Important Active Site Residue in the Class II Fructose-1,6-bisphosphate Aldolase of Escherichia coli. Protein Sci 1996, 5, 154–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1097).Pegan SD; Rukseree K; Franzblau SG; Mesecar AD Structural Basis for Catalysis of a Tetrameric Class IIa Fructose 1,6-Bisphosphate Aldolase from Mycobacterium tuberculosis. J. Mol. Biol 2009, 386, 1038–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1098).Capodagli GC; Sedhom WG; Jackson M; Ahrendt KA; Pegan SD A Noncompetitive Inhibitor for Mycobacterium tuberculosis’s Class IIa Fructose 1,6-Bisphosphate Aldolase. Biochem 2014, 53, 202–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1099).Pegan SD; Rukseree K; Capodagli GC; Baker EA; Krasnykh O; Franzblau SG; Mesecar AD Active Site Loop Dynamics of a Class IIa Fructose 1,6-Bisphosphate Aldolase from Mycobacterium tuberculosis. Biochemistry 2013, 52, 912–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1100).Naismith JH; Ferrara JD; Bailey S; Marshall K; Dauter Z; Wilson KS; Habash J; Harrop SJ; Berry AJ; Hunter WN Initiating a Crystallographic Study of a Class II Fructose-1,6-bisphosphate Aldolase. J. Mol. Biol 1992, 225, 1137–1141. [DOI] [PubMed] [Google Scholar]
- (1101).Plaumann M; Pelzer-Reith B; Martin WF; Schnarrenberger C Multiple Recruitment of Class-I Aldolase to Chloroplasts and Eubacterial Origin of Eukaryotic Class-II Aldolases Revealed by cDNAs from Euglena gracilis. Curr. Genet 1997, 31, 430–438. [DOI] [PubMed] [Google Scholar]
- (1102).Hall DR; Leonard GA; Reed CD; Watt CI; Berry A; Hunter WN The Crystal Structure of Escherichia coli Class II Fructose-1, 6-bisphosphate Aldolase in Complex with Phosphoglycolohydroxamate Reveals Details of Mechanism and Specificity. J. Mol. Biol 1999, 287, 383–394. [DOI] [PubMed] [Google Scholar]
- (1103).Collins KD An Activated Intermediate Analogue. The Use of Phosphoglycolohydroxamate as a Stable Analogue of a Transiently Occurring Dihydroxyacetone Phosphate-derived Enolate in Enzymatic Catalysis. J. Biol. Chem 1974, 249, 136–142. [PubMed] [Google Scholar]
- (1104).Lewis DJ; Lowe G Phosphoglycollohydroxamic Acid: an Inhibitor of Class I and II Aldolases and Triosephosphate Isomerase. A Potential Antibacterial and Antifungal Agent. Chem. Commun 1973, 0, 713–715. [Google Scholar]
- (1105).Fonvielle M; Weber P; Dabkowska K; Therisod M New Highly Selective Inhibitors of Class II Fructose-1,6-Bisphosphate Aldolases. Bioorg. Med. Chem. Lett 2004, 14, 2923–2926. [DOI] [PubMed] [Google Scholar]
- (1106).Galkin A; Li Z; Li L; Kulakova L; Pal LR; Dunaway-Mariano D; Herzberg O Structural Insights Into the Substrate Binding and Stereoselectivity of Giardia Fructose-1,6-bisphosphate Aldolase. Biochemistry 2009, 48, 3186–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1107).Fonvielle M; Coincon M; Daher R; Desbenoit N; Kosieradzka K; Barilone N; Gicquel B; Sygusch J; Jackson M; Therisod M Synthesis and Biochemical Evaluation of Selective Inhibitors of Class II Fructose Bisphosphate Aldolases: Towards New Synthetic Antibiotics. Chem. - Eur. J 2008, 14, 8521–8529. [DOI] [PubMed] [Google Scholar]
- (1108).Daher R; Coincon M; Fonvielle M; Gest PM; Guerin ME; Jackson M; Sygusch J; Therisod M Rational Design, Synthesis, and Evaluation of New Selective Inhibitors of Microbial Class II (Zinc Dependent) Fructose Bis-phosphate Aldolases. J. Med. Chem 2010, 53, 7836–7842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1109).Labbe G; Krismanich AP; de Groot S; Rasmusson T; Shang M; Brown MD; Dmitrienko GI; Guillemette JG Development of Metal-chelating Inhibitors for the Class II Fructose 1,6-Bisphosphate (FBP) Aldolase. J. Inorg. Biochem 2012, 112, 49–58. [DOI] [PubMed] [Google Scholar]
- (1110).Gavalda S; Braga R; Dax C; Vigroux A; Blonski C N-Sulfonyl Hydroxamate Derivatives as Inhibitors of Class II Fructose-1,6-diphosphate Aldolase. Bioorg. Med. Chem. Lett 2005, 15, 5375–5377. [DOI] [PubMed] [Google Scholar]
- (1111).Mabiala-Bassiloua CG; Zwolinska M; Therisod H; Sygusch J; Therisod M Separate Synthesis and Evaluation of Glucitol Bis-phosphate and Mannitol Bis-phosphate, as Competitive Inhibitors of Fructose Bis-phosphate Aldolases. Bioorg. Med. Chem. Lett 2008, 18, 1735–1737. [DOI] [PubMed] [Google Scholar]
- (1112).Mabiala-Bassiloua CG; Arthus-Cartier G; Hannaert V; Therisod H; Sygusch J; Therisod M Mannitol Bis-phosphate Based Inhibitors of Fructose 1,6-Bisphosphate Aldolases. ACS Med. Chem. Lett 2011, 2, 804–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1113).Desvergnes S; Courtiol-Legourd S; Daher R; Dabrowski M; Salmon L; Therisod M Synthesis and Evaluation of Malonate-based Inhibitors of Phosphosugar-metabolizing Enzymes: Class II Fructose-1,6-bis-phosphate Aldolases, Type I Phosphomannose Isomerase, and Phosphoglucose Isomerase. Bioorg. Med. Chem 2012, 20, 1511–1520. [DOI] [PubMed] [Google Scholar]
- (1114).Han X; Zhu X; Hong Z; Wei L; Ren Y; Wan F; Zhu S; Peng H; Guo L; Rao L; Feng L; Wan J Structure-Based Rational Design of Novel Inhibitors Against Fructose-1,6-Bisphosphate Aldolase from Candida albicans. J. Chem. Inf. Model 2017, 57, 1426–1438. [DOI] [PubMed] [Google Scholar]
- (1115).Bisson C; Britton KL; Sedelnikova SE; Rodgers HF; Eadsforth TC; Viner RC; Hawkes TR; Baker PJ; Rice DW Crystal Structures Reveal that the Reaction Mechanism of Imidazoleglycerol-Phosphate Dehydratase is Controlled by Switching Mn(II) Coordination. Structure 2015, 23, 1236–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1116).Henriksen ST; Liu J; Estiu G; Oltvai ZN; Wiest O Identification of Novel Bacterial Histidine Biosynthesis Inhibitors Using Docking, Ensemble Rescoring, and Whole-Cell Assays. Bioorg. Med. Chem 2010, 18, 5148–5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1117).Schweitzer BA; Loida PJ; Thompson-Mize RL; CaJacob CA; Hegde SG Design and Synthesis of Beta-Carboxamido Phosphonates as Potent Inhibitors of Imidazole Glycerol Phosphate Dehydratase. Bioorg. Med. Chem. Lett 1999, 9, 2053–2058. [DOI] [PubMed] [Google Scholar]
- (1118).Mori I; Fonnepfister R; Matsunaga S; Tada S; Kimura Y; Iwasaki G; Mano J; Hatano M; Nakano T; Koizumi S; Scheidegger A; Hayakawa K; Ohta D A Novel Class of Herbicides - Specific Inhibitors of Imidazoleglycerol Phosphate Dehydratase. Plant Physiol 1995, 107, 719–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1119).Mori I; Iwasaki G; Hayakawa K Rational Design of a New Herbicide by Inhibition of Histidine Biosynthesis - Design and Synthesis of Inhibitors of Imidazole Glycerol Phosphate Dehydratase (IGPD). J. Synth. Org. Chem. Jpn 1996, 54, 514–524. [Google Scholar]
- (1120).Mori I; Iwasaki G; Kimura Y; Matsunaga SI; Ogawa A; Nakano T; Buser HP; Hatano M; Tada S; Hayakawa K Synthesis of Inhibitors of Imidazole Glycerol Phosphate Dehydratase. J. Am. Chem. Soc 1995, 117, 4411–4412. [Google Scholar]
- (1121).Walba H; Isensee RW Acidity Constants of Some Arylimidazoles and Their Cations. J. Org. Chem 1961, 26, 2789–2791. [Google Scholar]
- (1122).Sinha SC; Chaudhuri BN; Burgner JW; Yakovleva G; Davisson VJ; Smith JL Crystal Structure of Imidazole Glycerol-Phosphate Dehydratase - Duplication of an Unusual Fold. J. Biol. Chem 2004, 279, 15491–15498. [DOI] [PubMed] [Google Scholar]
- (1123).Foster AW; Osman D; Robinson NJ Metal Preferences and Metallation. J. Biol. Chem 2014, 289, 28095–28103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1124).Petersen J; Hawkes TR; Lowe DJ The Metal-Binding Site of Imidazole Glycerol Phosphate Dehydratase; EPR and ENDOR Studies of the Oxo-Vanadyl Enzyme. JBIC, J. Biol. Inorg. Chem 1997, 2, 308–319. [Google Scholar]
- (1125).Lindell SD; Earnshaw CG; Wright BJ; Carver DS; OMahony MJ; SavilleStones EA Synthesis of Inhibitors of Imidazole Glycerol Phosphate Dehydratase. Bioorg. Med. Chem. Lett 1996, 6, 547–552. [Google Scholar]
- (1126).Norcross RD; vonMatt P; Kolb HC; Bellus D Synthesis of Novel Cyclobutylphosphonic Acids as Inhibitors of Imidazole Glycerol Phosphate Dehydratase. Tetrahedron 1997, 53, 10289–10312. [Google Scholar]
- (1127).Cox JM; Hawkes TR; Bellini P; Ellis RM; Barrett R; Swanborough JJ; Russell SE; Walker PA; Barnes NJ; Knee AJ; Lewis T; Davies PR The Design and Synthesis of Inhibitors of Imidazoleglycerol Phosphate Dehydratase as Potential Herbicides. Pestic. Sci 1997, 50, 297–311. [Google Scholar]
- (1128).Jin Y; Zhao HQ; Lu HZ; Kuemmel CM; Zhang JJ; Wang DQ Synthesis and Herbicidal Activity of Novel 1-(Diethoxyphosphoryl)-3-(4-one-1H-1,2,3-triazol-1-yl)-propan-2-yl Carboxylic Esters. Molecules 2015, 20, 1088–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1129).Schweitzer BA; Loida PJ; CaJacob CA; Chott RC; Collantes EM; Hegde SG; Mosier PD; Profeta S Discovery of Imidazole Glycerol Phosphate Dehydratase Inhibitors Through 3-D Database Searching. Bioorg. Med. Chem. Lett 2002, 12, 1743–1746. [DOI] [PubMed] [Google Scholar]
- (1130).Hutschenreuther A; Bigl M; Hemdan NY; Debebe T; Gaunitz F; Birkenmeier G Modulation of GLO1 Expression Affects Malignant Properties of Cells. Int. J. Mol. Sci 2016, 17, 2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1131).Rabbani N; Xue M; Weickert MO; Thornalley PJ Multiple Roles of Glyoxalase 1-Mediated Suppression of Methylglyoxal Glycation in Cancer Biology-Involvement in Tumour Suppression, Tumour Growth, Multidrug Resistance and Target for Chemotherapy. Semin. Cancer. Biol 2017. [DOI] [PubMed] [Google Scholar]
- (1132).Rabbani N; Xue M; Thornalley PJ Methylglyoxal-Induced Dicarbonyl Stress in Aging and Disease: First Steps Towards Glyoxalase 1-Based Treatments. Clin. Sci 2016, 130, 1677–1696. [DOI] [PubMed] [Google Scholar]
- (1133).Thornalley PJ The Glyoxalase System in Health and Disease. Mol. Aspects Med 1993, 14, 287–371. [DOI] [PubMed] [Google Scholar]
- (1134).Sakamoto H; Mashima T; Kizaki A; Dan S; Hashimoto Y; Naito M; Tsuruo T Glyoxalase I is Involved in Resistance of Human Leukemia Cells to Antitumor Agent-Induced Apoptosis. Blood 2000, 95, 3214–3218. [PubMed] [Google Scholar]
- (1135).Vander Jagt DL; Hunsaker LA; Campos NM; Baack BR D-Lactate Production in Erythrocytes Infected with Plasmodium Falciparum. Mol. Biochem. Parasitol 1990, 42, 277–284. [DOI] [PubMed] [Google Scholar]
- (1136).Williams R. t.; Lim JE; Harr B; Wing C; Walters R; Distler MG; Teschke M; Wu C; Wiltshire T; Su AI; Sokoloff G; Tarantino LM; Borevitz JO; Palmer AA A Common and Unstable Copy Number Variant is Associated with Differences in Glo1 Expression and Anxiety-Like Behavior. PLoS One 2009, 4, e4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1137).Distler MG; Plant LD; Sokoloff G; Hawk AJ; Aneas I; Wuenschell GE; Termini J; Meredith SC; Nobrega MA; Palmer AA Glyoxalase 1 Increases Anxiety by Reducing GABAA Receptor Agonist Methylglyoxal. J. Clin. Invest 2012, 122, 2306–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1138).Hovatta I; Tennant RS; Helton R; Marr RA; Singer O; Redwine JM; Ellison JA; Schadt EE; Verma IM; Lockhart DJ; Barlow C Glyoxalase 1 and Glutathione Reductase 1 Regulate Anxiety in Mice. Nature 2005, 438, 662–666. [DOI] [PubMed] [Google Scholar]
- (1139).Cameron AD; Olin B; Ridderstrom M; Mannervik B; Jones TA Crystal Structure of Human Glyoxalase I-Evidence for Gene Duplication and 3D Domain Swapping. EMBO J 1997, 16, 3386–3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1140).Ridderstrom M; Mannervik B Optimized Heterologous Expression of the Human Zinc Enzyme Glyoxalase I. Biochem. J 1996, 314 (Pt 2), 463–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1141).Aronsson AC; Marmstal E; Mannervik B Glyoxalase I, a Zinc Metalloenzyme of Mammals and Yeast. Biochem. Biophys. Res. Commun 1978, 81, 1235–1240. [DOI] [PubMed] [Google Scholar]
- (1142).Sellin S; Eriksson LE; Mannervik B Fluorescence and Nuclear Relaxation Enhancement Studies of the Binding of Glutathione Derivatives to Manganese-Reconstituted Glyoxalase I from Human Erythrocytes. A Model for the Catalytic Mechanism of the Enzyme Involving a Hydrated Metal Ion. Biochemistry 1982, 21, 4850–4857. [DOI] [PubMed] [Google Scholar]
- (1143).Sellin S; Rosevear PR; Mannervik B; Mildvan AS Nuclear Relaxation Studies of the Role of the Essential Metal in Glyoxalase I. J. Biol. Chem 1982, 257, 10023–10029. [PubMed] [Google Scholar]
- (1144).Sellin S; Eriksson LE; Aronsson AC; Mannervik B Octahedral Metal Coordination in the Active Site of Glyoxalase I as Evidenced by the Properties of Co(II)-Glyoxalase I. J. Biol. Chem 1983, 258, 2091–2093. [PubMed] [Google Scholar]
- (1145).Sellin S; Eriksson LE; Mannervik B Electron Paramagnetic Resonance Study of the Active Site of Copper-Substituted Human Glyoxalase I. Biochemistry 1987, 26, 6779–6784. [DOI] [PubMed] [Google Scholar]
- (1146).Cameron AD; Ridderstrom M; Olin B; Kavarana MJ; Creighton DJ; Mannervik B Reaction Mechanism of Glyoxalase I Explored by an X-ray Crystallographic Analysis of the Human Enzyme in Complex with a Transition State Analogue. Biochemistry 1999, 38, 13480–13490. [DOI] [PubMed] [Google Scholar]
- (1147).Himo F; Siegbahn PE Catalytic Mechanism of Glyoxalase I: A Theoretical Study. J. Am. Chem. Soc 2001, 123, 10280–10289. [DOI] [PubMed] [Google Scholar]
- (1148).Frickel EM; Jemth P; Widersten M; Mannervik B Yeast Glyoxalase I is a Monomeric Enzyme with Two Active Sites. J. Biol. Chem 2001, 276, 1845–1849. [DOI] [PubMed] [Google Scholar]
- (1149).Kermack WO; Matheson NA The Effects of Some Analogues of Glutathione on the Glyoxalase System. Biochem. J 1957, 65, 48–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1150).Vince R; Wadd WB Glyoxalase Inhibitors as Potential Anticancer Agents. Biochem. Biophys. Res. Commun 1969, 35, 593–598. [DOI] [PubMed] [Google Scholar]
- (1151).Vince R; Daluge S; Wadd WB Studies on the Inhibition of Glyoxalase I by S-Substituted Glutathiones. J. Med. Chem 1971, 14, 402–404. [DOI] [PubMed] [Google Scholar]
- (1152).Lo TW; Thornalley PJ Inhibition of Proliferation of Human Leukaemia 60 Cells by Diethyl Esters of Glyoxalase Inhibitors in Vitro. Biochem. Pharmacol 1992, 44, 2357–2363. [DOI] [PubMed] [Google Scholar]
- (1153).Conroy PJ Carcinostatic Activity of Methylglyoxal and Related Substances in Tumour-Bearing Mice. Ciba. Found. Symp 1978, 271–300. [DOI] [PubMed] [Google Scholar]
- (1154).Kester MV; Reese JA; Norton SJ Mouse Liver Glyoxalase I Inhibition by S-Substituted Glutathiones. J. Med. Chem 1974, 17, 413–416. [DOI] [PubMed] [Google Scholar]
- (1155).Vince R; Wolf M; Sanford C Glutaryl-S-(p-Bromobenzyl)-L-Cysteinylglycine. A Metabolically Stable Inhibitor of Glyoxalase I. J. Med. Chem 1973, 16, 951–953. [DOI] [PubMed] [Google Scholar]
- (1156).Hall SS; Doweyko LM; Doweyko AM; Zilenovski JS Synthesis and Evaluation of Alpha-Hydroxythiol Esters as Antitumor Agents and Glyoxalase I Inhibitors. J. Med. Chem 1977, 20, 1239–1242. [DOI] [PubMed] [Google Scholar]
- (1157).Douglas KT; Nadvi IN Inhibition of Glyoxalase I: a Possible Transition-State Analogue Inhibitor Approach to Potential Antineoplastic Agents? FEBS Lett 1979, 106, 393–396. [DOI] [PubMed] [Google Scholar]
- (1158).Brandt RB; Brandt ME; April ME; Thomson C Inhibitors of Glyoxalase I in Vitro. Biochem. Med 1983, 29, 385–391. [DOI] [PubMed] [Google Scholar]
- (1159).Brandt RB; Laux JE; Yates SW; Boots MR; Thomson C; Edge C Inhibition of Glyoxalase I in Vitro by Coumarin and Coumarin Derivatives. Int. J. Quantum Chem 1986, 30, 155–165. [Google Scholar]
- (1160).Barnard JF; Honek JF Investigation on Glyoxalase I Inhibitors. Biochem. Biophys. Res. Commun 1989, 165, 118–124. [DOI] [PubMed] [Google Scholar]
- (1161).Murthy NS; Bakeris T; Kavarana MJ; Hamilton DS; Lan Y; Creighton DJ S-(N-Aryl-N-Hydroxycarbamoyl)glutathione Derivatives are Tight-Binding Inhibitors of Glyoxalase I and Slow Substrates for Glyoxalase II. J. Med. Chem 1994, 37, 2161–2166. [DOI] [PubMed] [Google Scholar]
- (1162).Jin T; Zhai J; Liu X; Yue Y; Huang M; Li Z; Ni C; Deng Q; Sang Y; Yao Z; Zhang H; Hu X; Zheng ZB Design, Synthesis and Biological Evaluation of Potent Human Glyoxalase I Inhibitors. Chem. Pharm. Bull 2017, 65, 455–460. [DOI] [PubMed] [Google Scholar]
- (1163).Chiba T; Ohwada J; Sakamoto H; Kobayashi T; Fukami TA; Irie M; Miura T; Ohara K; Koyano H Design and Evaluation of Azaindole-Substituted N-Hydroxypyridones as Glyoxalase I Inhibitors. Bioorg. Med. Chem. Lett 2012, 22, 7486–7489. [DOI] [PubMed] [Google Scholar]
- (1164).Zhang H; Zhai J; Zhang L; Li C; Zhao Y; Chen Y; Li Q; Hu XP In Vitro Inhibition of Glyoxalase I by Flavonoids: New Insights from Crystallographic Analysis. Curr. Top. Med. Chem 2016, 16, 460–466. [DOI] [PubMed] [Google Scholar]
- (1165).Sato S; Kwon Y; Kamisuki S; Srivastava N; Mao Q; Kawazoe Y; Uesugi M Polyproline-Rod Approach to Isolating Protein Targets of Bioactive Small Molecules: Isolation of a New Target of Indomethacin. J. Am. Chem. Soc 2007, 129, 873–880. [DOI] [PubMed] [Google Scholar]
- (1166).Kawatani M; Okumura H; Honda K; Kanoh N; Muroi M; Dohmae N; Takami M; Kitagawa M; Futamura Y; Imoto M; Osada H The Identification of an Osteoclastogenesis Inhibitor Through the Inhibition of Glyoxalase I. Proc. Natl. Acad. Sci. U. S. A 2008, 105, 11691–11696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1167).Brandt RB; Laux J; Thomson C; Johnson MA; Gross M Inhibition of Glyoxalase I in Vitro by Flavones. Int. J. Quantum Chem 1984, 26, 195–200. [Google Scholar]
- (1168).Zhai J; Zhang H; Zhang L; Zhao Y; Chen S; Chen Y; Peng X; Li Q; Yuan M; Hu X Zopolrestat as a Human Glyoxalase I Inhibitor and its Structural Basis. ChemMedChem 2013, 8, 1462–1464. [DOI] [PubMed] [Google Scholar]
- (1169).Chimura H; Nakamura H; Takita T; Takeuchi T; Umezawa H The Structure of a Glyoxalase I Inhibitor and its Chemical Reactivity with SH-Compounds. J. Antibiot 1975, 28, 743–748. [DOI] [PubMed] [Google Scholar]
- (1170).Takeuchi T; Chimura H; Hamada M; Umezawa H; Yoshioka O A Glyoxalase I Inhibitor of a New Structural Type Produced by Streptomyces. J. Antibiot 1975, 28, 737–742. [DOI] [PubMed] [Google Scholar]
- (1171).Douglas KT; Gohel DI; Nadvi IN; Quilter AJ; Seddon AP Partial Transition-State Inhibitors of Glyoxalase I from Human Erythrocytes, Yeast and Rat Liver. Biochim. Biophys. Acta 1985, 829, 109–118. [DOI] [PubMed] [Google Scholar]
- (1172).Klopman G; Dimayuga ML Computer-Automated Structure Evaluation of Flavonoids and Other Structurally Related Compounds as Glyoxalase I Enzyme Inhibitors. Mol. Pharmacol 1988, 34, 218–122. [PubMed] [Google Scholar]
- (1173).Hamilton DS; Creighton DJ Inhibition of Glyoxalase I by the Enediol Mimic S-(N-Hydroxy-N-Methylcarbamoyl)glutathione. The Possible Basis of a Tumor-Selective Anticancer Strategy. J. Biol. Chem 1992, 267, 24933–24936. [PubMed] [Google Scholar]
- (1174).Sharkey EM; O’Neill HB; Kavarana MJ; Wang H; Creighton DJ; Sentz DL; Eiseman JL Pharmacokinetics and Antitumor Properties in Tumor-Bearing Mice of an Enediol Analogue Inhibitor of Glyoxalase I. Cancer Chemother. Pharmacol 2000, 46, 156–166. [DOI] [PubMed] [Google Scholar]
- (1175).More SS; Vince R A Metabolically Stable Tight-Binding Transition-State Inhibitor of Glyoxalase-I. Bioorg. Med. Chem. Lett 2006, 16, 6039–6042. [DOI] [PubMed] [Google Scholar]
- (1176).Sang Y; Shi Q; Mo M; Ni C; Li Z; Liu B; Deng Q; Creighton DJ; Zheng ZB Novel Bivalent Inhibitors with Sub-Nanomolar Affinities Towards Human Glyoxalase I. Bioorg. Med. Chem. Lett 2015, 25, 4724–4727. [DOI] [PubMed] [Google Scholar]
- (1177).Zheng ZB; Creighton DJ Bivalent Transition-State Analogue Inhibitors of Human Glyoxalase I. Org. Lett 2003, 5, 4855–4858. [DOI] [PubMed] [Google Scholar]
- (1178).More SS; Vince R Design, Synthesis, and Binding Studies of Bidentate Zn-Chelating Peptidic Inhibitors of Glyoxalase-I. Bioorg. Med. Chem. Lett 2007, 17, 3793–3797. [DOI] [PubMed] [Google Scholar]
- (1179).More SS; Vince R Inhibition of Glyoxalase I: the First Low-Nanomolar Tight-Binding Inhibitors. J. Med. Chem 2009, 52, 4650–4656. [DOI] [PubMed] [Google Scholar]
- (1180).Santel T; Pflug G; Hemdan NY; Schafer A; Hollenbach M; Buchold M; Hintersdorf A; Lindner I; Otto A; Bigl M; Oerlecke I; Hutschenreuther A; Sack U; Huse K; Groth M; Birkemeyer C; Schellenberger W; Gebhardt R; Platzer M; Weiss T; Vijayalakshmi MA; Kruger M; Birkenmeier G Curcumin Inhibits Glyoxalase 1: a Possible Link to its Anti-Inflammatory and Anti-Tumor Activity. PLoS One 2008, 3, e3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1181).Takasawa R; Saeki K; Tao A; Yoshimori A; Uchiro H; Fujiwara M; Tanuma S Delphinidin, a Dietary Anthocyanidin in Berry Fruits, Inhibits Human Glyoxalase I. Bioorg. Med. Chem 2010, 18, 7029–7033. [DOI] [PubMed] [Google Scholar]
- (1182).Takasawa R; Takahashi S; Saeki K; Sunaga S; Yoshimori A; Tanuma S Structure-Activity Relationship of Human GLO I Inhibitory Natural Flavonoids and Their Growth Inhibitory Effects. Bioorg. Med. Chem 2008, 16, 3969–3975. [DOI] [PubMed] [Google Scholar]
- (1183).Takasawa R; Akahane H; Tanaka H; Shimada N; Yamamoto T; Uchida-Maruki H; Sai M; Yoshimori A; Tanuma SI Piceatannol, a Natural Trans-Stilbene Compound, Inhibits Human Glyoxalase I. Bioorg. Med. Chem. Lett 2017, 27, 1169–1174. [DOI] [PubMed] [Google Scholar]
- (1184).Takasawa R; Tao A; Saeki K; Shionozaki N; Tanaka R; Uchiro H; Takahashi S; Yoshimori A; Tanuma S Discovery of a New Type Inhibitor of Human Glyoxalase I by Myricetin-Based 4-Point Pharmacophore. Bioorg. Med. Chem. Lett 2011, 21, 4337–4342. [DOI] [PubMed] [Google Scholar]
- (1185).Takasawa R; Shimada N; Uchiro H; Takahashi S; Yoshimori A; Tanuma S TLSC702, a Novel Inhibitor of Human Glyoxalase I, Induces Apoptosis in Tumor Cells. Biol. Pharm. Bull 2016, 39, 869–873. [DOI] [PubMed] [Google Scholar]
- (1186).Santarius T; Bignell GR; Greenman CD; Widaa S; Chen L; Mahoney CL; Butler A; Edkins S; Waris S; Thornalley PJ; Futreal PA; Stratton MR GLO1-A Novel Amplified Gene in Human Cancer. Genes, Chromosomes Cancer 2010, 49, 711–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1187).Vollmer W; Blanot D; de Pedro MA Peptidoglycan Structure and Architecture. FEMS Microbiol. Rev 2008, 32, 149–167. [DOI] [PubMed] [Google Scholar]
- (1188).Caminero JA; Sotgiu G; Zumla A; Migliori GB Best Drug Treatment for Multidrug-resistant and Extensively Drug-resistant Tuberculosis. Lancet Infect. Dis 2010, 10, 621–629. [DOI] [PubMed] [Google Scholar]
- (1189).Bugg TD; Braddick D; Dowson CG; Roper DI Bacterial Cell Wall Assembly: Still an Attractive Antibacterial Target. Trends Biotechnol 2011, 29, 167–173. [DOI] [PubMed] [Google Scholar]
- (1190).Mullins LS; Zawadzke LE; Walsh CT; Raushel FM Kinetic Evidence for the Formation of D-alanyl Phosphate in the Mechanism of D-Alanyl-D-alanine Ligase. J. Biol. Chem 1990, 265, 8993–8998. [PubMed] [Google Scholar]
- (1191).Fan C; Moews PC; Walsh CT; Knox JR Vancomycin Resistance: Structure of D-Alanine:D-alanine Ligase at 2.3 A Resolution. Science 1994, 266, 439–443. [DOI] [PubMed] [Google Scholar]
- (1192).Fan C; Park IS; Walsh CT; Knox JR D-Alanine:D-alanine Ligase: Phosphonate and Phosphinate Intermediates with Wild Type and the Y216F Mutant. Biochemistry 1997, 36, 2531–2538. [DOI] [PubMed] [Google Scholar]
- (1193).Handwerger S; Pucci MJ; Volk KJ; Liu J; Lee MS Vancomycin-resistant Leuconostoc Mesenteroides and Lactobacillus Casei Synthesize Cytoplasmic Peptidoglycan Precursors that Terminate in Lactate. J. Bacteriol 1994, 176, 260–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1194).Marshall CG; Broadhead G; Leskiw BK; Wright GD D-Ala-D-ala Ligases from Glycopeptide Antibiotic-producing Organisms are Highly Homologous to the Enterococcal Vancomycin-resistance Ligases VanA and VanB. Proc. Natl. Acad. Sci. U. S. A 1997, 94, 6480–6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1195).Park IS; Walsh CT D-Alanyl-D-lactate and D-Alanyl-D-alanine Synthesis by D-Alanyl-D-alanine Ligase from Vancomycin-resistant Leuconostoc Mesenteroides. Effects of a Phenylalanine 261 to Tyrosine Mutation. J. Biol. Chem 1997, 272, 9210–9214. [DOI] [PubMed] [Google Scholar]
- (1196).Batson S; de Chiara C; Majce V; Lloyd AJ; Gobec S; Rea D; Fülöp V; Thoroughgood CW; Simmons KJ; Dowson CG; Fishwick CWG; de Carvalho LPS; Roper DI Inhibition of D-Ala:D-Ala ligase Through a Phosphorylated form of the Antibiotic D-Cycloserine. Nat. Commun 2017, 8, 1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1197).Gold HS Vancomycin-resistant Enterococci: Mechanisms and Clinical Observations. Clin. Infect. Dis 2001, 33, 210–219. [DOI] [PubMed] [Google Scholar]
- (1198).Bugg TDH; Wright GD; Dutkamalen S; Arthur M; Courvalin P; Walsh CT Molecular-Basis for Vancomycin Resistance in Enterococcus-Faecium Bm4147 - Biosynthesis of a Depsipeptide Peptidoglycan Precursor by Vancomycin Resistance Proteins VanH and VanA. Biochemistry 1991, 30, 10408–10415. [DOI] [PubMed] [Google Scholar]
- (1199).Zawadzke LE; Bugg TD; Walsh CT Existence of Two D-Alanine:D-alanine Ligases in Escherichia coli: Cloning and Sequencing of the ddlA Gene and Purification and Characterization of the DdlA and DdlB Enzymes. Biochemistry 1991, 30, 1673–1682. [DOI] [PubMed] [Google Scholar]
- (1200).Neuhaus FC; Lynch JL Studies on the Inhibition of D-Alanyl-D-alanine Synthetase by the Antibiotic D-Cycloserine. Biochem. Biophys. Res. Commun 1962, 8, 377–382. [DOI] [PubMed] [Google Scholar]
- (1201).Neuhaus FC; Lynch JL The Enzymatic Synthesis of D-Alanyl-D-alanine. III. On the Inhibition of D-Alanyl-D-alanine Synthetase by the Antibiotic D-Cycloserine. Biochemistry 1964, 3, 471–480. [DOI] [PubMed] [Google Scholar]
- (1202).Neuhaus FC; Hammes WP Inhibition of Cell Wall Biosynthesis by Analogues and Alanine. Pharmacol. Ther 1981, 14, 265–319. [DOI] [PubMed] [Google Scholar]
- (1203).Parsons WH; Patchett AA; Bull HG; Schoen WR; Taub D; Davidson J; Combs PL; Springer JP; Gadebusch H; Weissberger B; et al. Phosphinic Acid Inhibitors of D-Alanyl-D-alanine Ligase. J. Med. Chem 1988, 31, 1772–1778. [DOI] [PubMed] [Google Scholar]
- (1204).Prosser GA; de Carvalho LP Kinetic Mechanism and Inhibition of Mycobacterium tuberculosis D-Alanine:D-alanine Ligase by the Antibiotic D-Cycloserine. FEBS J 2013, 280, 1150–1166. [DOI] [PubMed] [Google Scholar]
- (1205).Lambert MP; Neuhaus FC Mechanism of D-cycloserine Action: Alanine Racemase from Escherichia coli W. J. Bacteriol 1972, 110, 978–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1206).Lu PL; Peng CF; Hwang JJ; Chen YH Activity of Twelve Second-line Antimicrobial Agents Against Mycobacterium tuberculosis in Taiwan. J. Chemother 2008, 20, 202–207. [DOI] [PubMed] [Google Scholar]
- (1207).Behera C; Krishna K; Singh HR Antitubercular Drug-induced Violent Suicide of a Hospitalised Patient. BMJ Case Rep. 2014, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1208).Ellsworth BA; Tom NJ; Bartlett PA Synthesis and Evaluation of Inhibitors of Bacterial D-Alanine:D-alanine Ligases. Chem. Biol 1996, 3, 37–44. [DOI] [PubMed] [Google Scholar]
- (1209).Lacostel AC-G, A.; Vo-Quang L; Vo-Quang Y; Le Goffic F Time-dependent Inhibition of Streptococcus faecalis D-Alanine:D-alanine Ligase by α-Aminophosphonamidic Acids Eur. J. Med. Chem 1991, 26, 255–260. [Google Scholar]
- (1210).Chakravarty PK; Greenlee WJ; Parsons WH; Patchett AA; Combs P; Roth A; Busch RD; Mellin TN (3-Amino-2-oxoalkyl)phosphonic Acids and Their Analogues as Novel Inhibitors of D-Alanine:D-Alanine Ligase. J. Med. Chem 1989, 32, 1886–1890. [DOI] [PubMed] [Google Scholar]
- (1211).Duncan K; Walsh CT ATP-Dependent Inactivation and Slow Binding Inhibition of Salmonella typhimurium D-Alanine:D-alanine Ligase (ADP) by (Aminoalkyl)phosphinate and Aminophosphonate Analogues of D-Alanine. Biochemistry 1988, 27, 3709–3714. [DOI] [PubMed] [Google Scholar]
- (1212).Vo-Quang Y; Carniato D; Vo-Quang L; Lacoste AM; Neuzil E; Le Goffic F (1-Amino-2-propenyl) Phosphonic Acid, an Inhibitor of Alanine Racemase and D-Alanine:D-Alanine Ligase. J. Med. Chem 1986, 29, 579–581. [DOI] [PubMed] [Google Scholar]
- (1213).Vo-Quang Y; Gravey AM; Simonneau R; Vo-Quang L; Lacoste AM; Le Goffic F Towards New Inhibitors of D-Alanine:D-alanine Ligase: The Synthesis of 3-Amino Butenylphosphonic and Aminophosphonamidic Acids. Tetrahedron Lett 1987, 28, 6167–6170. [Google Scholar]
- (1214).McDermott AE; Creuzet F; Griffin RG; Zawadzke LE; Ye QZ; Walsh CT Rotational Resonance Determination of the Structure of an Enzyme-inhibitor Complex: Phosphorylation of an (Aminoalkyl)phosphinate Inhibitor of D-Alanyl-D-alanine Ligase by ATP. Biochemistry 1990, 29, 5767–5775. [DOI] [PubMed] [Google Scholar]
- (1215).Sova M; Cadez G; Turk S; Majce V; Polanc S; Batson S; Lloyd AJ; Roper DI; Fishwick CW; Gobec S Design and Synthesis of New Hydroxyethylamines as Inhibitors of D-Alanyl-D-lactate Ligase (VanA) and D-Alanyl-D-alanine ligase (DdlB). Bioorg. Med. Chem. Lett 2009, 19, 1376–1379. [DOI] [PubMed] [Google Scholar]
- (1216).Wu D; Kong Y; Han C; Chen J; Hu L; Jiang H; Shen X D-Alanine:D-alanine Ligase as a New Target for the Flavonoids Quercetin and Apigenin. Int. J. Antimicrob. Agents 2008, 32, 421–426. [DOI] [PubMed] [Google Scholar]
- (1217).Triola G; Wetzel S; Ellinger B; Koch MA; Hübel K; Rauh D; Waldmann H ATP Competitive Inhibitors of D-Alanine–D-Alanine Ligase Based on Protein Kinase Inhibitor Scaffolds. Bioorg. Med. Chem 2009, 17, 1079–1087. [DOI] [PubMed] [Google Scholar]
- (1218).Vehar B; Hrast M; Kovač A; Konc J; Mariner K; Chopra I; O’Neill A; Janežič D; Gobec S Ellipticines and 9-Acridinylamines as Inhibitors of D-Alanine:D-alanine Ligase. Bioorg. Med. Chem 2011, 19, 5137–5146. [DOI] [PubMed] [Google Scholar]
- (1219).Kovac A; Konc J; Vehar B; Bostock JM; Chopra I; Janezic D; Gobec S Discovery of New Inhibitors of D-Alanine:D-alanine Ligase by Structure-based Virtual Screening. J. Med. Chem 2008, 51, 7442–7448. [DOI] [PubMed] [Google Scholar]
- (1220).Putty S; Rai A; Jamindar D; Pagano P; Quinn CL; Mima T; Schweizer HP; Gutheil WG Characterization of D-boroAla as a Novel Broad-spectrum Antibacterial Agent Targeting D-Ala-D-ala Ligase. Chem. Biol. Drug Des 2011, 78, 757–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1221).Liu S; Chang JS; Herberg JT; Horng MM; Tomich PK; Lin AH; Marotti KR Allosteric Inhibition of Staphylococcus aureus D-Alanine:D-Alanine Ligase Revealed by Crystallographic Studies. Proc. Natl. Acad. Sci. U. S. A 2006, 103, 15178–15183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1222).Besong GE; Bostock JM; Stubbings W; Chopra I; Roper DI; Lloyd AJ; Fishwick CW; Johnson AP A de Novo Designed Inhibitor of D-Ala-D-ala Ligase from E. coli. Angew. Chem., Int. Ed. Engl 2005, 44, 6403–6646. [DOI] [PubMed] [Google Scholar]
- (1223).Kovač A; Majce V; Lenaršič R; Bombek S; Bostock JM; Chopra I; Polanc S; Gobec S Diazenedicarboxamides as Inhibitors of D-Alanine-D-alanine Ligase (Ddl). Bioorg. Med. Chem. Lett 2007, 17, 2047–2054. [DOI] [PubMed] [Google Scholar]
- (1224).Allen D; Wilson D; Drew R; Perfect J Azole Anitfungals: 35 Years of Invasive Fungal Infection Management. Expert Rev. Anti Infect. Ther 2015, 13, 787–798. [DOI] [PubMed] [Google Scholar]
- (1225).Ngo HX; Garneau-Tsodikova S; Green KD A Complex Game of Hide and Seek: The Search for New Antifungals. Med. Chem. Commun 2016, 7, 1285–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1226).Sono M; Roach MP; Coulter ED; Dawson JH Heme-Containing Oxygenases. Chem. Rev 1996, 96, 2841–2887. [DOI] [PubMed] [Google Scholar]
- (1227).Moon YW; Hajjar J; Hwu P; Naing A Targeting the Indoleamine 2,3-Dioxygenase Pathway in Cancer. J. ImmunoTher. Cancer 2015, 3, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1228).Lewis-Ballester A; Pham KN; Batabyal D; Karkashon S; Bonanno JB; Poulos TL; Yeh, S.-R. Structural Insights into Substrate and Inhibitor Binding Sites in Human Indoleamine 2,3-dioxygenase 1. Nat. Commun 2017, 8, 1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1229).Sheridan C IDO Inhibitors Move Center Stage in Immuno-oncology. Nat. Biotechnol 2015, 33, 321–322. [DOI] [PubMed] [Google Scholar]
- (1230).Burris HA; Gordon MS; Hellmann MD; LoRusso P; Emens LA; Hodi FSA Phase Ib Dose Escalation Study of Combined Inhibition of IDO1 (GDC-0919) and PD-L1 (atezolizumab) in Patients (pts) with Locally Advanced or Metastatic Solid Tumors. J. Clin. Oncol 2017, 35, 105. [Google Scholar]
- (1231).Peng Y-H; Ueng S-H; Tseng C-T; Hung M-S; Song J-S; Wu J-S; Liao F-Y; Fan Y-S; Wu M-H; Hsiao W-C; Hsueh C-C; Lin S-Y; Cheng C-Y; Tu C-H; Lee L-C; Cheng M-F; Shia K-S; Shih C; Wu S-Y Important Hydrogen Bond Networks in Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitor Design Revealed by Crystal Structures of Imidazoleisoindole Derivatives with IDO1. J. Med. Chem 2016, 59, 282–293. [DOI] [PubMed] [Google Scholar]
- (1232).Brochez L; Chevolet I; Kruse V The Rationale of Indoleamine 2,3-Dioxygenase Inhibition for Cancer Therapy. Eur. J. Cancer 2017, 76, 167–182. [DOI] [PubMed] [Google Scholar]
- (1233).Nelp MT; Kates PA; Hunt JT; Newitt JA; Balog A; Maley D; Zhu X; Abell L; Allentoff A; Borzilleri R; Lewis HA; Lin Z; Seitz SP; Yan C; Groves JT Immune-modulating Enzyme Indoleamine 2,3-Dioxygenase is Effectively Inhibited by Targeting its apo-form. Proc. Natl. Acad. Sci. U.S.A 2018, 115, 3249–3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1234).Baggio C; Cerofolini L; Fragai M; Luchinat C; Pellecchia M HTS by NMR for the Identification of Potent and Selective Inhibitors of Metalloenzymes. ACS Med. Chem. Lett 2018, 9, 137–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1235).Riccardi L; Genna V; DeVivo M Metal-ligand Interactions in Drug Design. Nat. Rev. Chem 2018, 2, 100–112. [Google Scholar]