Abstract
Stochasticity is a key characteristic of intracellular processes such as gene regulation and chemical signalling. Therefore, characterizing stochastic effects in biochemical systems is essential to understand the complex dynamics of living things. Mathematical idealizations of biochemically reacting systems must be able to capture stochastic phenomena. While robust theory exists to describe such stochastic models, the computational challenges in exploring these models can be a significant burden in practice since realistic models are analytically intractable. Determining the expected behaviour and variability of a stochastic biochemical reaction network requires many probabilistic simulations of its evolution. Using a biochemical reaction network model to assist in the interpretation of time-course data from a biological experiment is an even greater challenge due to the intractability of the likelihood function for determining observation probabilities. These computational challenges have been subjects of active research for over four decades. In this review, we present an accessible discussion of the major historical developments and state-of-the-art computational techniques relevant to simulation and inference problems for stochastic biochemical reaction network models. Detailed algorithms for particularly important methods are described and complemented with Matlab® implementations. As a result, this review provides a practical and accessible introduction to computational methods for stochastic models within the life sciences community.
Keywords: biochemical reaction networks, stochastic simulation, Monte Carlo, Bayesian inference, approximate Bayesian computation
1. Introduction
Many biochemical processes within living cells, such as regulation of gene expression, are stochastic [1–5]; that is, randomness or noise is an essential component of living things. Internal and external factors are responsible for this randomness [6–9], particularly within systems where low copy numbers of certain chemical species greatly affect the system dynamics [10]. Intracellular stochastic effects are key components of normal cellular function [11] and have a direct influence on the heterogeneity of multicellular organisms [12]. Furthermore, stochasticity of biochemical processes can play a role in the onset of disease [13,14] and immune responses [15]. Stochastic phenomena, such as resonance [16], focusing [17] and bistability [18–20], are not captured by traditional deterministic chemical rate equation models. These stochastic effects must be captured by appropriate theoretical models. A standard approach is to consider a biochemical reaction network as a well-mixed population of molecules that diffuse, collide and react probabilistically. The stochastic law of mass action is invoked to determine the probabilities of reaction events over time [21,22]. The resulting time-series of biochemical populations may be analysed to determine both the average behaviour and variability [23]. This powerful approach to modelling biochemical kinetics can be extended to deal with more biologically realistic settings that include spatial heterogeneity with molecular populations being well-mixed only locally [24–27].
In practice, stochastic biochemical reaction network models are analytically intractable meaning that most approaches are entirely computational. Two distinct, yet related, computational problems are of particular importance: (i) the forwards problem that deals with the simulation of the evolution of a biochemical reaction network forwards in time and (ii) the inverse problem that deals with the inference of unknown model parameters given time-course observations. Over the last four decades, significant attention has been given to these problems. Gillespie et al. [28] describe the key algorithmic advances in the history of the forwards problem and Higham [29] provides an accessible introduction connecting stochastic approaches with deterministic counterparts. Recently, Schnoerr et al. [23] provide a detailed review of the forwards problem with a focus on analytical methods. Golightly & Wilkinson [30], Toni et al. [31] and Sunnåker et al. [32] review techniques relevant to the inverse problem.
Given the relevance of stochastic computational methods to the life sciences, the aim of this review is to present an accessible summary of computational aspects relating to efficient simulation for both the forwards and inverse problems. Practical examples and algorithmic descriptions are presented and aimed at applied mathematicians and applied statisticians with interests in the life sciences. However, we expect the techniques presented here will also be of interest to the wider life sciences community. Electronic supplementary material provides clearly documented code examples (available from GitHub https://github.com/ProfMJSimpson/Warne2018) using the Matlab® programming language.
2. Biochemical reaction networks
We provide an algorithmic introduction to stochastic biochemical reaction network models. In the literature, rigorous theory exists for these stochastic modelling approaches [33]. However, we focus on an informal definition useful for understanding computational methods in practice. Relevant theory on the chemical master equation, Markov processes and stochastic differential equations is not discussed in any detail (see [22,34,35] for accessible introductions to these topics).
Consider a domain, for example, a cell nucleus, that contains a number of chemical species. The population count for a chemical species is a non-negative integer called its copy number. A biochemical reaction network model is specified by a set of chemical formulae that determine how the chemical species interact. For example, X + 2Y → Z + W states ‘one X molecule and two Y molecules react to produce one Z molecule and one W molecule’. If a chemical species is involved in a reaction, then the number of molecules required as reactants or produced as products are called stoichiometric coefficients. In the example, Y has a reactant stoichiometric coefficient of two, and Z has a product stoichiometric coefficient of one.
2.1. A computational definition
Consider a set of M reactions involving N chemical species with copy numbers X1(t), …, XN(t) at time t. The state vector is an N × 1 vector of copy numbers, X(t) = [X1(t), …, XN(t)]T. This represents the state of the population of chemical species at time t. When a reaction occurs, the copy numbers of the reactants and products are altered according to their respective stoichiometric coefficients. The net state change caused by a reaction event is called its stoichiometric vector. If reaction j occurs, then a new state is obtained by adding its stoichiometric vector, νj, that is,
| 2.1 |
where t− denotes the time immediately preceding the reaction event. The vectors ν1, …, νM are obtained through , where and are, respectively, vectors of the reactant and product stoichiometric coefficients of the chemical formula of reaction j. Equation (2.1) describes how reaction j affects the system.
Gillespie [21,33] presents the fundamental theoretical framework that provides a probabilistic rule for the occurrence of reaction events. We shall not focus on the details here, but the essential concept is based on the stochastic law of mass action. Informally,
| 2.2 |
The tacit assumption is that the system is well mixed with molecules equally likely to be found anywhere in the domain. The right-hand side of equation (2.2) is typically expressed as aj(X(t))dt, where aj(X(t)) is the propensity function of reaction j. That is,
| 2.3 |
where the positive constant is known as the kinetic rate parameter.1 Equations (2.1)–(2.3) are the main concepts needed to consider computational methods for the forwards problem. Importantly, equations (2.1) and (2.2) indicate that the possible model states are discrete, but state changes occur in continuous time.
2.2. Two examples
We now provide some representative examples of biochemical reaction networks that will be used throughout this review.
2.2.1. Mono-molecular chain
Consider two chemical species, A and B, and three reactions that form a mono-molecular chain,
| 2.4 |
with kinetic rate parameters k1, k2 and k3. We adopt the convention that indicates the reactions are part of an open system involving external chemical processes that are not explicitly represented in equation (2.4). Given the state vector, X(t) = [A(t), B(t)]T, the respective propensity functions are
| 2.5 |
and the stoichiometric vectors are
| 2.6 |
This mono-molecular chain is interesting since it is a part of a general class of biochemical reaction networks that are analytically tractable, though they are only applicable to relatively simple biochemical processes [36].
2.2.2. Enzyme kinetics
A biologically applicable biochemical reaction network describes the catalytic conversion of a substrate, S, into a product, P, via an enzymatic reaction involving enzyme, E. This is described by Michaelis–Menten enzyme kinetics [37,38],
| 2.7 |
with kinetic rate parameters, k1, k2 and k3. This particular enzyme kinetic model is a closed system. Here, we have the state vector X(t) = [E(t), S(t), C(t), P(t)]T, propensity functions
| 2.8 |
and stoichiometric vectors
| 2.9 |
Since the first chemical formula involves two reactant molecules, there is significantly less progress that can be made without computational methods.
See MonoMolecularChain.m and MichaelisMenten.m for example code to generate useful data structures for these biochemical reaction networks. These two biochemical reaction networks have been selected to demonstrate two stereotypical problems. In the first instance, the mono-molecular chain model, the network structure enables progress to be made analytically. The enzyme kinetic model, however, represents a more realistic case in which computational methods are required. The focus on these two representative models is done so that the exposition is clear.
3. The forwards problem
Given a biochemical reaction network with known kinetic rate parameters and some initial state vector, X(0) = x0, we consider the forwards problem. That is, we wish to predict the future evolution. Since we are dealing with stochastic models, all our methods for dealing with future predictions will involve probabilities, random numbers and uncertainty. We rely on standard code libraries2 for generating samples from uniform (i.e. all outcomes equally likely), Gaussian (i.e. the bell curve), exponential (i.e. time between events) and Poisson distributions (i.e. number of events over a time interval) and do not discuss algorithms for generating samples from these distributions. This enables the focus of this review to be on algorithms specific to biochemical reaction networks.
There are two key aspects to the forwards problem: (i) the simulation of a biochemical reaction network that replicates the random reaction events over time and (ii) the calculation of average behaviour and variability among all possible sequences of reaction events. Relevant algorithms for dealing with both these aspects are reviewed and demonstrated.
3.1. Generation of sample paths
Here, we consider algorithms that deal with the simulation of biochemical reaction network evolution. These algorithms are probabilistic, that is, the output of no two simulations, called sample paths, of the same biochemical reaction network will be identical. The most fundamental stochastic simulation algorithms (SSAs) for sample path generation are based on the work of Gillespie [21,39,40], Gibson & Bruck [41] and Anderson [42].
3.1.1. Exact stochastic simulation algorithms
Exact SSAs generate sample paths, over some interval t ∈ [0, T], that identically follow the probability laws of the fundamental theory of stochastic chemical kinetics [33]. Take a sufficiently small time interval, [t, t + dt), such that the probability of multiple reactions occurring in this interval is zero. In such a case, the reactions are mutually exclusive events. Hence, based on equations (2.2) and (2.3), the probability of any reaction event occurring in [t, t + dt) is the sum of the individual event probabilities,
where a0(X(t)) = a1(X(t)) + · · · + aM(X(t)) is the total reaction propensity function. Therefore, if we know that the next reaction occurs at time s ∈ [t, t + dt), then we can: (i) randomly select a reaction with probabilities a1(X(s))/a0(X(s)), …, aM(X(s))/a0(X(s)) and (ii) update the state vector according to the respective stoichiometric vector, ν1, …, νM.
All that remains for an exact simulation method is to determine the time of the next reaction event. Gillespie [21] demonstrates that the time interval between reactions, Δt, may be treated as a random variable that is exponentially distributed with rate a0(X(t)), that is, Δt ∼ Exp(a0(X(t))). Therefore, we arrive at the most fundamental exact SSA, the Gillespie direct method:
-
(1)
initialize, time t = 0 and state vector X = x0;
-
(2)
calculate propensities, a1(X), …, aM(X), and a0(X) = a1(X) + · · · + aM(X);
-
(3)
generate random sample of the time to next reaction, Δt ∼ Exp(a0(X));
-
(4)
if t + Δt > T, then terminate the simulation, otherwise go to step 5;
-
(5)
randomly select integer j from the set {1, …, M} with , ;
-
(6)
update state, X = X + νj, and time t = t + Δt, then go to step 2.
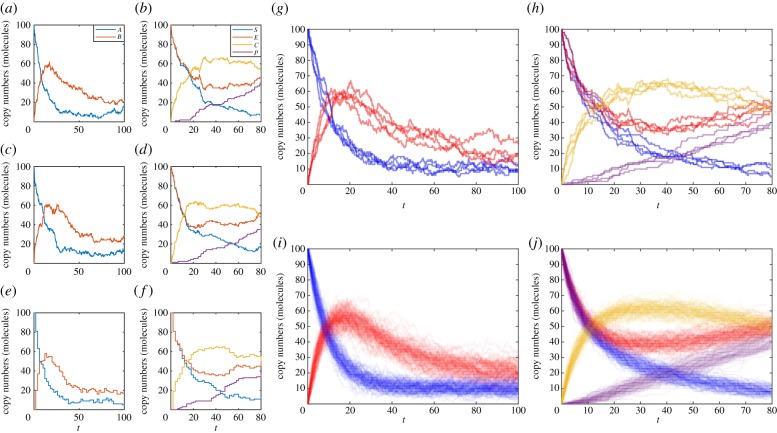
An example implementation, GillespieDirectMethod.m, and example usage, DemoGillespie.m, are provided. Figure 1 demonstrates sample paths generated by the Gillespie direct method for the mono-molecular chain model (figure 1a) and the enzyme kinetic model (figure 1b).
Figure 1.
Examples of exact sample paths of the mono-molecular chain model using the (a) Gillespie direct method and (c) modified next reaction method; similarly exact sample paths of the enzyme kinetics model using the (b) Gillespie direct method and (d) modified next reaction method. Approximate sample paths may be computed with less computational burden using the tau-leaping method with τ = 2, at the expense of accuracy: (e) the mono-molecular chain model and (f) the enzyme kinetics model. Every sample path will be different; as demonstrated by four distinct simulations of (g) the mono-molecular chain model and (h) the enzyme kinetics model. However, trends are revealed when 100 simulations are overlaid to reveal states of higher probability density using (i) the mono-molecular chain model and (j) the enzyme kinetics model. The mono-molecular chain model simulations are configured with parameters k1 = 1.0, k2 = 0.1, k3 = 0.05, and initial state A(0) = 100, B(0) = 0. The enzyme kinetics model simulations are configured with parameters k1 = 0.001, k2 = 0.005, k3 = 0.01, and initial state E(0) = 100, S(0) = 100, C(0) = 0, P(0) = 0. (Online version in colour.)
A mathematically equivalent, but more computationally efficient, exact SSA formulation is derived by Gibson & Bruck [41]. Their method independently tracks the next reaction time of each reaction separately. The next reaction to occur is the one with the smallest next reaction time, therefore no random selection of reaction events is required. It should, however, be noted that the Gillespie direct method may also be improved to yield the optimized direct method [43] with similar performance benefits. Anderson [42] further refines the method of Gibson and Bruck by scaling the times of each reaction so that the scaled times between reactions follow unit-rate exponential random variables. This scaling allows the method to be applied to more complex biochemical reaction networks with time-dependent propensity functions; however, the recently proposed Extrande method [44] is computationally superior. In Anderson’s approach, scaled times are tracked for each reaction independently with tj being the current time at the natural scale of reaction j. This results in the modified next reaction method:
-
(1)
initialize, global time t = 0, state vector X = x0 and scaled times t1 = t2 = · · · = tM = 0;
-
(2)
generate M first reaction times, s1, …, sM ∼ Exp(1);
-
(3)
calculate propensities, a1(X), …, aM(X);
-
(4)
rescale time to next reaction, Δtj = (sj − tj)/aj(X) for j = 1, 2, …, M;
-
(5)
choose reaction k, such that Δtk = min{Δt1, …, ΔtM};
-
(6)
if t + Δtk > T terminate simulation, otherwise go to step 7;
-
(7)
update rescaled times, tj = tj + aj(X)Δtk for j = 1, …, M, state X = X + νk and global time t = t + Δtk;
-
(8)
generate scaled next reaction time for reaction k, Δsk ∼ Exp(1);
-
(9)
update next scaled reaction time sk = sk + Δsk, and go to step 3.
An example implementation, ModifiedNextReactionMethod.m, and example usage, DemoMNRM.m, are provided. Figure 1 demonstrates sample paths generated by the modified next reaction method for the mono-molecular chain model (figure 1c) and the enzyme kinetic model (figure 1d). Note that the sample paths are different from those generated using the Gillespie direct method, despite the random number generators being initialized the same way. However, both represent exact sample paths, that is, sample paths that exactly follow the dynamics of the biochemical reaction network.
While the Gillespie direct method and the more efficient modified next reaction method and optimized direct method represent the most fundamental examples of exact SSAs, other advanced methods are also available to further improve the computational performance for large and complex biochemical reaction networks. Particular techniques include partial-propensity factorization [45], rejection-based methods [46,47] and composition-rejection [48] methods. We do not discuss these approaches, but we highlight them to indicate that efficient exact SSA method development is still an active area of research.
3.1.2. Approximate stochastic simulation algorithms
Despite some computational improvements provided by the modified next reaction method [41,42], all exact SSAs are computationally intractable for large biochemical populations and with many reactions, since every reaction event is simulated. Several approximate SSAs have been introduced in an attempt to reduce the computational burden while sacrificing accuracy.
The main approximate SSA we consider is also developed by Gillespie [39] almost 25 years after the development of the Gillespie direct method. The key idea is to evolve the system in discrete time steps of length τ, hold the propensity functions constant over the time interval [t, t + τ) and count the number of reaction events that occur. The state vector is then updated based on the net effect of all the reaction events. The number of reaction events within the interval can be shown to be a random variable distributed according to a Poisson distribution with mean aj(X(t))τ. If Yj denotes the number of reaction j events in [t, t + τ) then Yj ∼ Po(aj(X(t))τ). The result is the tau-leaping method:
-
(1)
initialize, time t = 0 and state Z = x0;
-
(2)
if t + τ > T then terminate simulation, otherwise continue;
-
(3)
calculate propensities, a1(Z), …, aM(Z);
-
(4)
generate reaction event counts, Yj ∼ Po(aj(Z)τ) for j = 1, …, M;
-
(5)
update state, Z = Z + Y1ν1 + · · ·YMνM, and time t = t + τ;
-
(6)
go to step 2.
Note that we use the notation Z(t) to denote an approximation of the true state X(t). An example implementation, TauLeapingMethod.m, and example usage, DemoTauLeap.m, are provided. Figure 1 demonstrates sample paths generated by the tau-leaping method for the mono-molecular chain model (figure 1e) and the enzyme kinetic model (figure 1f). Note that there is a visually obvious difference in the noise patterns of the tau-leaping method sample paths and the exact SSA sample paths (figure 1a–d).
The tau-leaping method is the only approximate SSA that we will explicitly discuss as it captures the essence of what approximations try to achieve; trading accuracy for improved performance. Several variations of the tau-leaping method have been proposed to improve accuracy, such as adaptive τ selection [49,50], implicit schemes [51] and the replacement of Poisson random variables with binomial random variables [52]. Hybrid methods that combine exact SSAs and approximations that split reactions into different time scales are also particularly effective for large scale networks with reactions occurring on very different time scales [53–55]. Other approximate simulation approaches are, for example, based on a continuous state chemical Langevin equation approximation [40,56,57] and employ numerical schemes for stochastic differential equations [35,58].
3.2. Computation of summary statistics
Owing to the stochastic nature of biochemical reaction networks, one cannot predict with certainty the future state of the system. Averaging over n sample paths can, however, provide insights into likely future states. Figure 1g,h shows that there is considerable variation in four independent sample paths, n = 4, of the mono-molecular chain and enzyme kinetic models. However, there is still a qualitative similarity between them. This becomes more evident for n = 100 sample paths, as in figure 1i,j. The natural extension is to consider average behaviour as n → ∞.
3.2.1. Using the chemical master equation
From a probability theory perspective, a biochemical reaction network model is a discrete-state, continuous-time Markov process. One key result for discrete-state, continuous-time Markov processes is, given an initial state, X(t) = x0, one can describe how the probability distribution of states evolves. This is given by the chemical master equation [33],3
| 3.1 |
where P(x, t|x0) is the probability that X(t) = x given X(0) = x0. Solving the chemical master equation provides an explicit means of computing the probability of being in any state at any time. Unfortunately, solutions to the chemical master equation are only known for special cases [36,59].
However, the mean and variance of the biochemical reaction network molecule copy numbers can sometimes be derived without solving the chemical master equation. For example, for the mono-molecular chain model equation (2.4), one may use equation (3.1) to derive the following system of ordinary differential equations (see the electronic supplementary material):
| 3.2 |
| 3.3 |
| 3.4 |
| 3.5 |
| 3.6 |
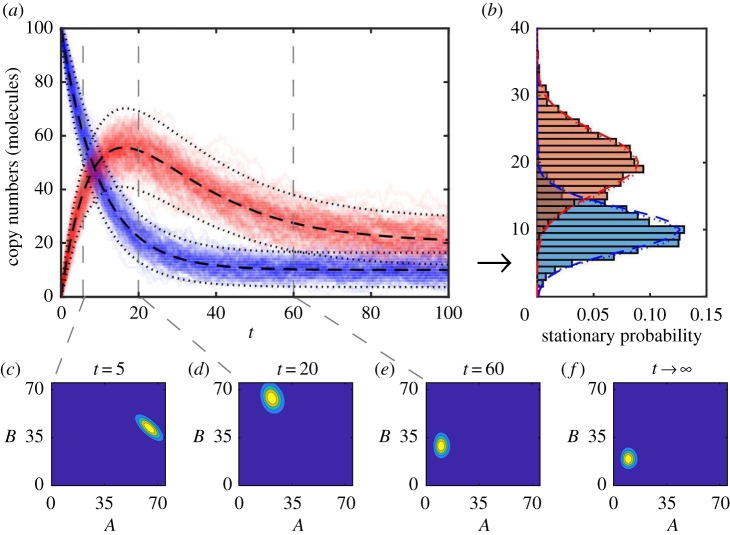
where Ma(t) and Va(t) (Mb(t) and Vb(t)) are the mean and variance of the copy number A(t) (B(t)) over all possible sample paths. Ca,b(t) is the covariance between A(t) and B(t). Equations (3.2)–(3.6) are linear ordinary differential equations that can be solved analytically and the solution is plotted in figure 2a superimposed with a population of sample paths.
Figure 2.
(a) The chemical master equation mean (black dashed) ± two standard deviations (black dots) of copy numbers of A (blue) and B (red) chemical species are displayed over simulated sample paths to demonstrate agreement. (b) The stationary distributions of A and B computed using: long running, T = 1000, simulated sample paths (blue/red histograms); Gaussian approximation (blue/red dashed) using long time limits of chemical master equation mean and variances; and the long time limit of the full chemical master equation solution (blue/red dots). Transient chemical master equation solution at times (c) t = 5, (d) t = 20 and (e) t = 60. (f) Chemical master equation solution stationary distribution. The parameters are k1 = 1.0, k2 = 0.1, k3 = 0.05, and initial state is A(0) = 100, B(0) = 0. (Online version in colour.)
The long time limit behaviour of a biochemical reaction network can also be determined. For the mono-molecular chain, as t → ∞ we have: Ma(t) → k1/k2; Va(t) → k1/k2; Mb(t) → k1/k3; Vb(t) → k1/k3; and Ca,b(t) → 0. We can approximate the long time steady-state behaviour, called the stationary distribution, of the mono-molecular chain model as two independent Gaussian random variables. That is, as t → ∞, A(t) → A∞ with . Similarly, B(t) → B∞ with . This approximation is shown in figure 2b against histograms of sample paths generated using large values of T (see example code, DemoStatDist.m).
In the case of the mono-molecular chain model, the chemical master equation is analytically tractable [36,59]. However, the solution is algebraically complicated and non-trivial to evaluate (see the electronic supplementary material). The full chemical master equation solution in figure 2c–e and the true stationary distribution of two independent Poisson distributions is shown in figure 2f. The true stationary distribution is also compared with the Gaussian approximation in figure 2b; the approximation is reasonably accurate. However, we could have also reasonably surmised the true stationary distribution by noting that, for a Poisson distribution, the mean is equal to the variance.
3.2.2. Monte Carlo methods
The chemical master equation can yield insight for special cases; however, for practical problems, such as the enzyme kinetic model, the chemical master equation is intractable and numerical methods are required. Here, we consider numerical estimation of the mean state vector at a fixed time, T.
In probability theory, the mean of a distribution is defined via an expectation,
| 3.7 |
where Ω is the set of all possible states. It is important to note that the methods we describe here are equally valid for a more general expectations of the form where f is some function that satisfies certain conditions [60].
We usually cannot compute equation (3.7) directly since Ω is typically infinite and the chemical master equation is intractable. However, exact SSAs provide methods for sampling the chemical master equation distribution, X(T) ∼ P(x, T|x0). This leads to the Monte Carlo estimator
| 3.8 |
where X(T)(1), …, X(T)(n) are n independent sample paths of the biochemical reaction network of interest (see example implementation MonteCarlo.m).
Unlike equation (3.7), the Monte Carlo estimates, such as equation (3.8), are random variables for finite n. This incurs a probabilistic error. A common measure of the accuracy of a Monte Carlo estimator, , of some expectation, , is the mean-square error that evaluates the average error behaviour and may be decomposed as follows:
| 3.9 |
Equation (3.9) highlights that there are two sources of error in a Monte Carlo estimator, the estimator variance and bias, and much of the discussion that follows deals with how to balance both these error sources in a computationally efficient manner.
Through analysis of the mean-square error of an estimator, the rate at which the mean-square error decays as n increases can be determined. Hence, we can determine how large n needs to be to satisfy the condition
| 3.10 |
where c is a positive constant and h is called the error tolerance.
Since , the bias term in equation (3.9) is zero and we call an unbiased estimator of . For an unbiased estimator, the mean-square error is equal to the estimator variance. Furthermore, , so the estimator variance decreases linearly with n, for sufficiently large n. Therefore, . That is, to halve h, one must increase n by a factor of four. This may be prohibitive with exact SSAs, especially for biochemical reaction networks with large variance. In the context of the Monte Carlo estimator using an exact SSA, equation (3.8), for n sufficiently large, the central limit theorem (CLT) states that (see Wilkinson [22] for a good discussion on the CLT).
Computational improvements can be achieved by using an approximate SSA, such as the tau-leaping method,
| 3.11 |
where Z(T)(1), …, Z(T)(n) are n independent approximate sample paths of the biochemical reaction network of interest (see the example implementation MonteCarloTauLeap.m).
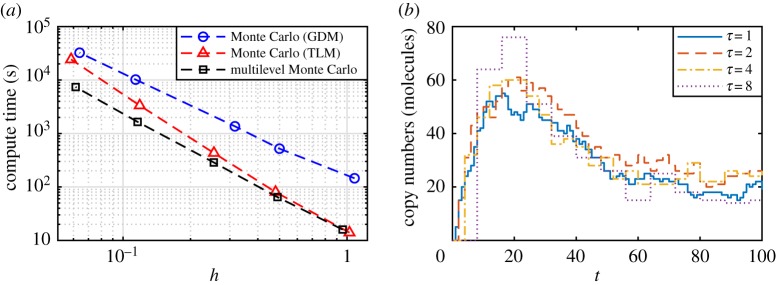
Note that . Since in general, we call a biased estimator. Even in the limit of n → ∞, the bias term in equation (3.9) may not be zero, which incurs a lower bound on the best achievable error tolerance, h, for fixed τ. However, it has been shown that the bias of the tau-leaping method decays linearly with τ [61,62]. Therefore, to satisfy the error tolerance condition (equation (3.10)) we not only require n ∝ 1/h2 but also τ ∝ h. That is, as h decreases, the performance improvement of Monte Carlo with the tau-leaping method reduces by a factor of τ, because the computational cost of the tau-leaping method is proportional to 1/τ. In figure 3a, the decay of the computational advantage in using the tau-leaping method for Monte Carlo over the Gillespie direct method is evident, and eventually the tau-leaping method will be more computationally burdensome than the Gillespie direct method. By the CLT, for large n, we have .
Figure 3.
(a) Improved performance from MLMC when estimating at T = 100 using the mono-molecular chain model with parameters k1 = 10, k2 = 0.1, k3 = 0.5, and initial condition A(0) = 1000, B(0) = 0; the computational advantage of the tau-leaping method (TLM; red triangles dashed) over the Gillespie direct method (GDM; blue circles dashed) for Monte Carlo diminishes as the required error tolerance decreases. The MLMC method (black squares dashed) exploits the correlated tau-leaping method to obtain sustained computational efficiency. (b) Demonstration of correlated tau-leaping method simulations for nested τ steps; sample paths using a step of τ = 2 (red dashed), τ = 4 (yellow dashed-dotted), and τ = 8 (purple dots) are all correlated with the same τ = 1 trajectory (blue solid). Computations are performed using an Intel® CoreTM i7-5600U CPU (2.6 GHz). (Online version in colour.)
The utility of the tau-leaping method for accurate (or exact) Monte Carlo estimation is identified by Anderson & Higham [63] through extending the idea of multilevel Monte Carlo (MLMC) originally proposed by Giles for stochastic differential equations [60,64]. Consider a sequence of L + 1 tau-leaping method time steps τ0, τ1, …, τL, with τℓ < τℓ−1 for ℓ = 1, …, L. Let Zℓ(T) denote the state vector of a tau-leaping method approximation using time step τℓ. Assume τL is small enough that is a good approximation of . Note that for large τℓ (small ℓ), sample paths are cheap to generate, but inaccurate; conversely, small τℓ (large ℓ) results in computationally expensive, but accurate sample paths.
We can write
| 3.12 |
Importantly, the final estimator on the right of equation (3.12), called the multilevel telescoping summation, is equivalent in bias to . At first glance, equation (3.12) looks to have complicated the computational problem and inevitably decreased performance of the Monte Carlo estimator. The insight of Giles [64], in the context of stochastic differential equation models for financial applications, is that the bias correction terms may be computed using Monte Carlo approaches that involve generating highly correlated sample paths in estimation of each of the correction terms, thus reducing the variance in the bias corrections. If the correlation is strong enough, then the variance decays such that few of the most accurate tau-leaping method sample paths are required; this can result in significant computational savings.
A contribution of Anderson & Higham [63] is an efficient method of generating correlated tau-leaping method sample path pairs (Zℓ(T), Zℓ−1(T)) in the case when τℓ = τℓ−1/δ for some positive integer scale factor δ. The algorithm is based on the property that the sum of two Poisson random variables is also a Poisson random variable. This enables the sample path with τℓ−1 to be constructed as an approximation to the sample path with τℓ directly. Figure 3b demonstrates tau-leaping method sample paths of B(t) in the mono-molecular chain model with τ = 2, 4, 8 generated directly from a tau-leaping method sample path with τ = 1. The algorithm can be thought of as generating multiple approximations of the same exact biochemical reaction network sample path. The algorithm is the correlated tau-leaping method:
-
(1)
initialize time t = 0, and states Zℓ, Zℓ−1 corresponding to sample paths with τ = τℓ and τ = τℓ−1, respectively;
-
(2)
if t + τℓ > T, then terminate simulation, otherwise continue;
-
(3)
calculate propensities for path Zℓ, a1(Zℓ), …, aM(Zℓ);
-
(4)
if t/τℓ is not an integer multiple of δ, then go to step 6, otherwise continue;
-
(5)
calculate propensities for path Zℓ−1, a1(Zℓ−1), …, aM(Zℓ−1), initialize intermediate state ;
-
(6)for each reaction j = 1, …, M;
-
6.1calculate virtual propensities, bj,1 = min{aj(Zℓ), aj(Zℓ−1)}, bj,2 = aj(Zℓ) − bj,1 and bj,3 = aj(Zℓ−1) − bj,1;
-
6.2generate virtual reaction event counts, Yj,1 ∼ Po(bj,1τℓ), Yj,2 ∼ Po(bj,2τℓ) and Yj,3 ∼ Po(bj,3τℓ);
-
6.1
-
(1)
set, Zℓ = Zℓ + (Y1,1 + Y1,2)ν1 + · · · + (YM,1 + YM,2)νM;
-
(1)
set, ;
-
(1)
if (t + τℓ)/τℓ is an integer multiple of δ, then set ;
-
(1)
update time t = t + τℓ, and go to step 2.
See example implementation, CorTauLeapingMethod.m, and example usage, DemoCorTauLeap.m.
Given the correlated tau-leaping method, Monte Carlo estimation can be applied to each term in equation (3.12) to give
| 3.13 |
where are n0 independent tau-leaping method sample paths with τ = τ0, and (Zℓ(T)(1), are nℓ paired correlated tau-leaping method sample paths with time steps τ = τℓ, τ = τℓ−1 and τℓ−1 = δτℓ for each bias correction ℓ = 1, 2, …, L. Given an error tolerance, h, it is possible to calculate an optimal sequence of sample path numbers n0, n1, …, nL such that the total computation time is optimized [63–65]. The results are shown in figure 3a for a more computationally challenging parameter set of the mono-molecular chain model. See the example implementation, MultilevelMonteCarlo.m and DemoMonteCarlo.m, for the full performance comparison.
As formulated here, MLMC results in a biased estimator, though it is significantly more efficient to reduce the bias of this estimator than by direct use of the tau-leaping method. If an unbiased estimator is required, then this can be achieved by correlating exact SSA sample paths with approximate SSA sample paths. Anderson & Higham [63] demonstrate a method for correlating tau-leaping method sample paths and modified next reaction method sample paths, and Lester et al. [65] demonstrate correlating tau-leaping method sample paths and Gillespie direct method sample paths. Further refinements such as adaptive and non-nested τℓ steps are also considered by Lester et al. [66], a multilevel hybrid scheme is developed by Moraes et al. [67] and Wilson & Baker [68] use MLMC and maximum entropy methods to generate approximations to the chemical master equation.
3.3. Summary of the forwards problem
Significant progress has been made in the study of computational methods for the solution to the forwards problem. As a result, forwards problem is relatively well understood, particularly for well-mixed systems, such as the biochemical reaction network models we consider in this review.
An exact solution to simulation is achieved though the development of exact SSAs. However, if Monte Carlo methods are required to determine expected behaviours, then exact SSAs can be computationally burdensome. While approximate SSAs provide improvements is some cases, highly accurate estimates will still often become burdensome since very small time steps will be required to keep the bias at the same order as the estimator standard deviation. In this respect, MLMC methods provide impressive computational improvements without any loss in accuracy. Such methods have become popular in financial applications [60,69]; however, there have been fewer examples in a biological setting.
Beyond the Gillespie direct method, the efficiency of sample path generation has been dramatically improved through the advancements in both exact SSAs and approximate SSAs. While approximate SSAs like the tau-leaping method provide computational advantages, they also introduce approximations. Some have noted that the error in these approximations is likely to be significantly lower than the modelling error compared with the real biological systems [57]. However, there is no general theory or guidelines as to when approximate SSAs are safe to use for applications.
We have only dealt with stochastic models that are well-mixed, that is, spatially homogeneous. The development of robust theory and algorithms accounting for spatial heterogeneity is still an active area of research [23,28,70]. The model of biochemical reaction networks, based on the chemical master equation, can be extended to include spatial dynamics through the reaction–diffusion master equation [26]. However, care must be taken in its application because the kinetics of the reaction–diffusion master equation depend on the spatial discretization and it is not always guaranteed to converge in the continuum limit [24–26,70,71]. We refer the reader to Gillespie et al. [28] and Isaacson [26] for useful discussions on this topic.
State-of-the-art Monte Carlo schemes, such as MLMC methods, have the potential to significantly accelerate the computation of summary statistics for the exploration of the range of biochemical reaction network behaviours. However, these approaches are also known to perform poorly for some biochemical reaction network models [63]. An open question is related to the characterization of biochemical reaction network models that will benefit from an MLMC scheme for summary statistic estimation. Furthermore, to our knowledge, there has been no application of the MLMC approach to the spatially heterogeneous case. The potential performance gains make MLMC a promising space for future development.
4. The inverse problem
When applying stochastic biochemical reaction network models to real applications, one often wants to perform statistical inference to estimate the model parameters. That is, given experimental data, and a biochemical reaction network model, the inverse problem seeks to infer the kinetic rate parameters and quantify the uncertainty in those estimates. Just as with the forwards problem, an enormous volume of the literature has been dedicated to the problem of inference in stochastic biochemical reaction network models. Therefore, we cannot cover all computational methods in detail. Rather we focus on a computational Bayesian perspective. For further reading, the monograph by Wilkinson [22] contains very accessible discussions on inference techniques in a systems biology setting, also the monographs by Gelman et al. [72] and Sisson et al. [73] contain a wealth of information on Bayesian methods more generally.
4.1. Experimental techniques
Typically, time-course data are derived from time-lapse microscopy images and fluorescent reporters [57,74,75]. Advances in microscopy and fluorescent technologies are enabling intracellular processes to be inspected at unprecedented resolutions [76–79]. Despite these advances, the resulting data never provide complete observations since: (i) the number of chemical species that may be observed concurrently is relatively low [57]; (ii) two chemical species might be indistinguishable from each other [30]; and (iii) the relationships between fluorescence levels and actual chemical species copy numbers may not be direct, in particular, the degradation of a protein may be more rapid than that of the fluorescent reporter [75,80]. That is, inferential methods must be able to deal with uncertainty in observations.
For the purposes of this review, we consider time-course data. Specifically, we suppose the data consist of nt observations of the biochemical reaction network state vector at discrete points in time, . That is, , where Y(t) represents an observation of the state vector sample path X(t). To model observational uncertainty, it is common to treat observations as subject to additive noise [23,30,31,74], so that
| 4.1 |
where A is a K × N matrix and ξ is a K × 1 vector of independent Gaussian random variables. The observation vectors, Y(t), are K × 1 vectors, with K ≤ N, reflecting the fact that only a sub-set of chemical species of X(t) are generally observed, or possibly only a linear combination of chemical species [30]. The example code, GenerateObservations.m, simulates this observation process (equation (4.1)) given a fully specified biochemical reaction network model.
4.1.1. Example data
The computation examples given in this review are based on two synthetically generated data sets, corresponding to the biochemical reaction network models given in §2. This enables the comparison between inference methods and the accuracy of inference.
The data for inference on the mono-molecular chain model (equation (2.4)) are taken as perfect observations, that is, K = N, A = I and . A single sample path is generated for the mono-molecular chain model with true parameters, θtrue = [1.0, 0.1, 0.05], and initial condition A(0) = 100 and B(0) = 0 using the Gillespie direct method. Observations are made using equation (4.1) applied at nt = 4 discrete times, t1 = 25, t2 = 50, t3 = 75 and t4 = 100. The resulting data are given in the electronic supplementary material.
The data for inference on the enzyme kinetic model equation (2.7) assumes incomplete and noisy observations. Specifically, only the product is observed, so K = 1, A = [0, 0, 0, 1]. Furthermore, we assume that there is some measurement error, ; that is, the error standard deviation is two product molecules. The data are generated using the Gillespie direct method with true parameters θtrue = [0.001, 0.005, 0.01] and initial condition E(0) = 100, S(0) = 100, C(0) = 0 and P(0) = 0. Equation (4.1) is evaluated at nt = 5 discrete times, t1 = 0, t2 = 20, t3 = 40, t4 = 60 and t5 = 80 (including an observation of the initial state), yielding the data in the electronic supplementary material.
4.2. Bayesian inference
Bayesian methods have been demonstrated to provide a powerful framework for the design of experiments, model selection and parameter estimation, especially in the life sciences [81–88]. Given observations, Yobs, and a biochemical reaction network model parametrized by the M × 1 real-valued vector of kinetic parameters, θ = [k1, k2, …, kM]T, the task is to quantify knowledge of the true parameter values in light of the data and prior knowledge. This is expressed mathematically through Bayes’ theorem,
| 4.2 |
The terms in equation (4.2) are interpreted as follows: p(θ|Yobs) is the posterior distribution, that is, the probability4 of parameter combinations, θ, given the data, Yobs; p(θ) is the prior distribution, that is, the probability of parameter combinations before taking the data into account; p(Yobs|θ) is the likelihood, that is, the probability of observing the data given a parameter combination; and p(Yobs) is the evidence, that is, the probability of observing the data over all possible parameter combinations. Assumptions about the parameters and the biochemical reaction network model are encoded through the prior and the likelihood, respectively. The evidence acts as a normalization constant, and ensures the posterior is a true probability distribution.5
First, consider the special case Y(t) = X(t), that is, the biochemical reaction network state can be perfectly observed at time t. In this case, the likelihood is
| 4.3 |
where the function P is the solution to the chemical master equation (3.1) and t0 = 0 [89,90]. It should be noted that, due to the stochastic nature of X(t), this perfect observation case is unlikely to recover the true parameters. Regardless of this issue, since the likelihood depends on the solution to the chemical master equation, the exact Bayesian posterior will not be analytically tractable in any practical case. In fact, even for the mono-molecular chain model, there are problems since the evidence term is not analytically tractable. The example code, DemoDirectBayesCME.m, provides an attempt at such a computation, though this code is not practical for an accurate evaluation. Just as with the forwards problem, we must defer to sampling methods and Monte Carlo.
4.3. Sampling methods
For this review, we focus on the task of estimating the posterior mean,
| 4.4 |
where Θ is the space of possible parameter combinations. However, the methods presented here are applicable to other quantities of interest. For example, the posterior covariance matrix,
| 4.5 |
is of interest as it provides an indicator of uncertainty associated with the inferred parameters. Marginal distributions are extremely useful for visualization: the marginal posterior distribution of the jth kinetic parameter is
| 4.6 |
where Θj ⊂ Θ is the parameter space excluding the jth dimension.
Just as with Monte Carlo for the forwards problem, we can estimate posterior expectations (shown here for equation (4.4), but the method may be similarly applied to equations (4.5) and (4.6)) using Monte Carlo,
| 4.7 |
where θ(1), …, θ(m) are independent samples from the posterior distribution. In the remainder of this section, we focus on computational schemes for generating such samples. We assume throughout that it is possible to generate samples from the prior distribution.
It is important to note that the sampling algorithms we present are not directly relevant to statistical estimators that are not based on expectations, such as maximum-likelihood estimators or the maximum a posteriori. However, these samplers can be modified through the use of data cloning [91,92] to approximate these effectively. Estimator variance and confidence intervals may also be estimated using bootstrap methods [93,94].
4.3.1. Exact Bayesian sampling
Assuming the likelihood can be evaluated, that is, the chemical master equation is tractable, a naive method of generating m samples from the posterior is the exact rejection sampler:
-
(1)
initialize index i = 0;
-
(2)
generate a prior sample θ* ∼ p(θ);
-
(3)
calculate acceptance probability α = p(Yobs|θ*);
-
(4)
with probability α, accept θ(i+1) = θ* and i = i + 1;
-
(5)
if i = m, terminate sampling, otherwise go to step 2.
Unsurprisingly, this approach is almost never viable as the likelihood probabilities are often extremely small. In the code example, DemoExactBayesRejection.m, the acceptance probability is never more than 9 × 10−15.
The most common solution is to construct a type of stochastic process (a Markov chain) in the parameter space to generate mn steps, . An essential property of the Markov chain used is that its stationary distribution is the target posterior distribution. This approach is called Markov chain Monte Carlo (MCMC), and a common method is the Metropolis–Hastings method [95,96]:
-
(1)
initialize n = 0 and select starting point θ(0);
-
(2)
generate a proposal sample, θ* ∼ q(θ|θ(n));
-
(3)calculate acceptance probability
-
(4)
with probability α, set θ(n+1) = θ*, and with probability 1 − α, set θ(n+1) = θ(n);
-
(5)
update time, n = n + 1;
-
(6)
if n > mn, terminate simulation, otherwise go to step 2.
The acceptance probability is based on the relative likelihood between two parameter configurations, the current configuration θ(n) and a proposed new configuration θ*, as generated by the user-defined proposal kernel q(θ|θ(n)). It is essential to understand that since the samples, , are produced by a Markov chain we cannot treat the mn steps as mn independent posterior samples. Rather, we need to take mn to be large enough that the mn steps are effectively equivalent to m independent samples.
One challenge in MCMC sampling is the selection of a proposal kernel such that the size of mn required for the Markov chain to reach the stationary distribution, called the mixing time, is small [97]. If the variance of the proposal is too low, then the acceptance rate is high, but the mixing is poor since only small steps are ever taken. On the other hand, a proposal variance that is too high will almost always make proposals that are rejected, resulting in many wasted likelihood evaluations before a successful move event. Selecting good proposal kernels is a non-trivial exercise and we refer the reader to Cotter et al. [98], Green et al. [99] and Roberts & Rosenthal [100] for detailed discussions on the wide variety of MCMC samplers including proposal techniques. Other techniques used to reduce correlations in MCMC samples include discarding the first mb steps, called burn-in iterations, or sub-sampling the chain by using only every mhth step, also called thinning. However, in general, the use of thinning decreases the statistical efficiency of the MCMC estimator [101,102].
Alternative approaches for exact Bayesian sampling can also be based on importance sampling [22,103]. Consider a random variable that cannot be simulated, X ∼ p(x), but suppose that it is possible to simulate another random variable, Y ∼ q(y). If X, Y ∈ Ω and p(x) = 0 whenever q(y) = 0, then
| 4.8 |
where Y(1), …, Y(m) are independent samples of q(Y). Using equation (4.8), one can show that if the distributions of the target, p(x), and the proposal, q(y), are similar, then a collection of samples, Y(1), …, Y(m), can be used to generate m approximate samples from p(x). This is called importance resampling:
-
(1)
generate samples Y(1), …, Y(m) ∼ q(y);
-
(2)
compute weights w(1) = p(Y(1))/q(Y(1)), …, w(m) = p(Y(m))/q(Y(m)); for i = 1, 2, …, m;
-
(3)
generate samples {X(1), …, X(m)} by drawing from {Y(1), …, Y(m)} with replacement using probabilities .
In Bayesian applications, the prior is often very different from the posterior. In such a case, importance resampling may be applied using a sequence of intermediate distributions. This is called sequential importance resampling and is one approach from the family of sequential Monte Carlo (SMC) [104] samplers. However, like the Metropolis–Hastings method, these methods also require explicit calculations of the likelihood function in order to compute the weights, thus all of these approaches are infeasible for practical biochemical reaction networks. Therefore, we only present more practical forms of these methods later.
More recently, it has been shown that the MLMC telescoping summation can accelerate the computation of posterior expectations when using MCMC [105,106] or SMC [107]. The key challenges in these applications is the development of appropriate coupling strategies. We do not cover these technical details in this review.
4.3.2. Likelihood-free methods
Since exact Bayesian sampling is rarely tractable, due to the intractability of the chemical master equation, alternative, likelihood-free sampling methods are required. Two main classes of likelihood-free methods exist: (i) so-called pseudo-marginal MCMC and (ii) approximate Bayesian computation (ABC). The focus of this review is ABC methods, though we first briefly discuss the pseudo-marginal approach.
The basis of the pseudo-marginal approach is to use a MCMC sampler (e.g. Metropolis–Hastings method), but replace the explicit likelihood evaluation with a likelihood estimate obtained through Monte Carlo simulation of the forwards problem [108]. For example, a direct unbiased Monte Carlo estimator is
where for j = 1, 2, …, n are independent sample paths of the biochemical reaction network of interest observed discretely at times . The most successful class of pseudo-marginal techniques, particle MCMC [109], apply a SMC sampler to approximate the likelihood and inform the MCMC proposal mechanism. The particle marginal Metropolis–Hastings method is a popular variant [30,109]. However, the recent model based proposals variant also holds promise for biochemical reaction networks specifically [110].
A particularly nice feature of pseudo-marginal methods is that they are unbiased,6 that is, the Markov chain will still converge to the exact target posterior distribution. This property is sometimes called an ‘exact approximation’ [22,30]. Unfortunately, the Markov chains in these methods typically converge more slowly than their exact counterparts. However, computational improvements have been obtained through application of MLMC [111].
Another popular likelihood-free approach is ABC [73,112–114]. ABC methods have enabled very complex models to be investigated, that are otherwise intractable [32]. Furthermore, ABC methods are very intuitive, leading to wide adoption within the scientific community [32]. Applications are particularly prevalent in the life sciences, especially in evolutionary biology [112,113,115–117], cell biology [118–120], epidemiology [121], ecology [122,123] and systems biology [31,90].
The basis of ABC is a discrepancy metric ρ(Yobs, Sobs) that provides a measure of closeness between the data, Yobs, and simulated data Sobs generated through stochastic simulation of the biochemical reaction network with simulated measurement error. Thus, acceptance probabilities are replaced with a deterministic acceptance condition, ρ(Yobs, Sobs) ≤ ε, where ε is the discrepancy threshold. This yields an approximation to Bayes’ theorem,
| 4.9 |
The key insight here is that the ability to produce sample paths of a biochemical reaction network model, that is, the forwards problem, enables an approximate algorithm for inference, that is, the inverse problem, regardless of the intractability or complexity of the likelihood. In fact, a formula for the likelihood need not even be known.
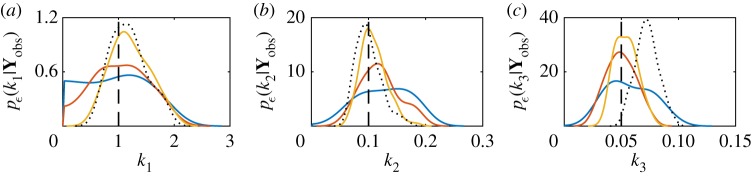
The discrepancy threshold determines the level of approximation; however, under the assumption of model and observation error, equation (4.9) can be treated as exact [124]. As ε → 0 then p(θ|ρ(Yobs, Sobs) ≤ ε) → p(θ|Yobs) [125,126]. Using data for the mono-molecular chain model, we can demonstrate this convergence, as shown in figure 4. The marginal posterior distributions are shown for each parameter, for various values of ε and compared with the exact marginal posteriors. The discrepancy metric used is
| 4.10 |
where S(t) is simulated data generated using the Gillespie direct method and equation (4.1). The example code, DemoABCConvergence.m, is used to generate these marginal distributions.
Figure 4.
Convergence of ABC posterior to the true posterior as ε → 0 for the mono-molecular chain inference problem. Marginal posteriors are plotted for ε = 50 (blue solid), ε = 25 (red solid), ε = 12.5 (yellow solid) and ε = 0 (black dotted). Here, the ε = 0 case corresponds to the exact likelihood using the chemical master equation solution. (a) Marginal posteriors for k1, (b) marginal posteriors for k2 and (c) marginal posteriors for k3. The true parameter values (black dashed) are k1 = 1.0, k2 = 0.1 and k3 = 0.05. Note that the exact Bayesian posterior does not recover the true parameter for k3. The priors used are , and . (Online version in colour.)
For any ε > 0, ABC methods are biased, just as the tau-leaping method is biased for the forwards problem. Therefore, a Monte Carlo estimate of a posterior summary statistic, such as equation (4.7), needs to take this bias into account. Just like the tau-leaping method, the rate of convergence in mean-square of ABC-based Monte Carlo is degraded because of this bias [125,126]. Furthermore, as the dimensionality of the data increases, small values of ɛ are not computationally feasible. In such cases, the data dimensionality may be reduced by computing lower dimensional summary statistics [32]; however, these must be sufficient statistics in order to ensure the ABC posterior still converges to the correct posterior as ε → 0. We refer the reader to Fearnhead & Prangle [126] for more detail on this topic.
4.3.3. Samplers for approximate Bayesian computation
We now focus on computational methods for generating m samples, , from the ABC posterior equation (4.9) with ε > 0 and discrepancy metric as given in equation (4.10). Throughout, we denote s(Sobs; θ) as the process for generating simulated data given a parameter vector; this process is identical to the processes used to generate our synthetic example data.
In general, the ABC samplers are only computationally viable if the data simulation process is not computationally expensive, in the sense that it is feasible to generate millions of sample paths. However, this is not always realistic, and many extensions exist to standard ABC in an attempt to deal with these more challenging cases. The lazy ABC method [127] applies an additional probability rule that terminates simulations early if it is likely that ρ(Yobs, Sobs) > ε. The approximate ABC method [128,129] uses a small set of data simulations to construct an approximation to the data simulation process.
The most notable early applications of ABC samplers are Beaumont et al. [115], Pritchard et al. [112] and Taveré et al. [113]. The essential development of this early work is that of the ABC rejection sampler:
-
(1)
initialize index i = 0;
-
(2)
generate a prior sample θ* ∼ p(θ);
-
(3)
generate simulated data, ;
-
(4)
if , accept and set i = i + 1, otherwise continue;
-
(5)
if i = m, terminate sampling, otherwise go to step 2.
There is a clear connection with the exact rejection sampler. Note that every accepted sample of the ABC posterior corresponds to at least one simulation of the forwards problem as shown in figure 5. As a result, the computational burden of the inverse problem is significantly higher than the forwards problem, especially for small ε. The example code, ABCRejectionSampler.m, provides an implementation of the ABC rejection sampler.
Figure 5.
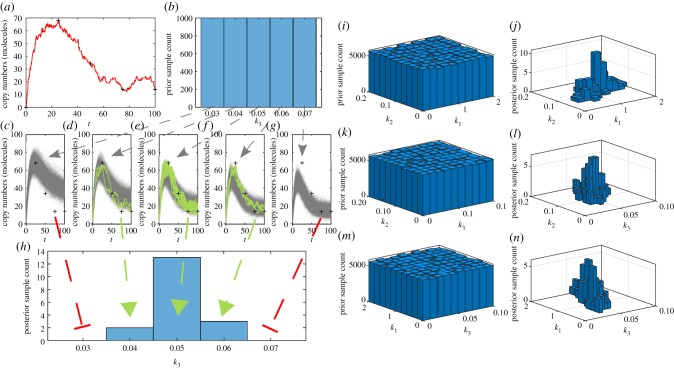
Demonstration of the ABC rejection sampler method using the mono-molecular chain model dataset. (a) Experimental observations (black crosses) obtained from a true sample path of B(t) in the mono-molecular chain model (red line) with k1 = 1.0, k2 = 0.1 and k3 = 0.05, and initial conditions, A(0) = 100 and B(0) = 0. (b) Prior for inference on k3. (c–g) Stochastic simulation of many choices of k3 drawn from the prior, showing accepted (solid green) and rejected (solid grey) sample paths with ε = 15 (molecules). (h) ABC posterior for k3 generated from accepted samples. (i–n) Bivariate marginal distributions of the full ABC inference problem on θ = {k1, k2, k3}. (Online version in colour.)
Unfortunately, for small ε, the computational burden of the ABC rejection sampler may be prohibitive as the acceptance rate is very low (this is especially an issue for biochemical reaction networks with highly variable dynamics). For example in figure 4, m = 100 posterior samples takes approximately one minute for ε = 25, but nearly ten hours for ε = 12.5. However, for ε = 12.5 the marginal ABC posterior for k3 has not yet converged.
Marjoram et al. [130] provide a solution via an ABC modification to the Metropolis–Hastings method. This results in a Markov chain with the ABC posterior as the stationary distribution. This method is called ABC Markov chain Monte Carlo (ABCMCMC):
-
(1)
initialize n = 0 and select starting point ;
-
(2)
generate a proposal sample, ;
-
(3)
generate simulated data, ;
-
(4)
if , then set and go to step 7, otherwise continue;
-
(5)calculate acceptance probability
-
(6)
with probability α, set , and with probability 1 − α, set ;
-
(7)
update time, n = n + 1;
-
(8)
if n > mn, terminate simulation, otherwise go to step 2.
An example implementation, ABCMCMCSampler.m, is provided.
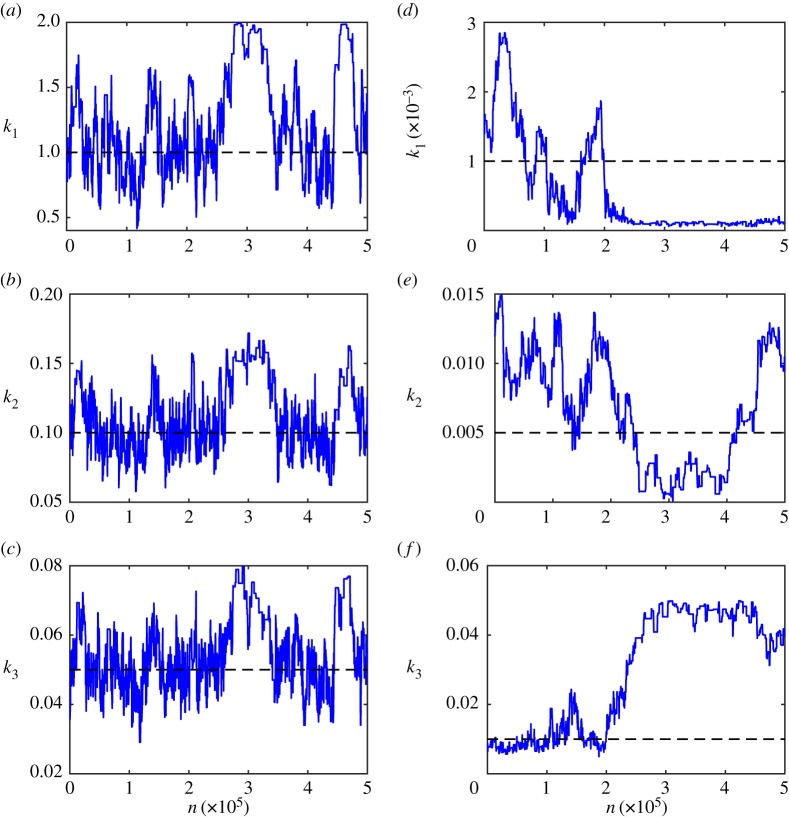
Just as with the Metropolis–Hastings method, the efficacy of the ABCMCMC rests upon the non-trivial choice of the proposal kernel, . The challenge of constructing effective proposal kernels is equally non-trivial for ABCMCMC as for the Metropolis–Hastings method. Figure 6 highlights different Markov chain behaviours for heuristically chosen proposal kernels based on Gaussian random walks with variances that we alter until the Markov chain seems to be mixing well.
Figure 6.
mn = 500 000 steps of ABCMCMC for: (a–c) the mono-molecular chain model; and (d–f ) the enzyme kinetics model. True parameter values (dashed black) are shown.
For the mono-molecular chain model (figure 6a–c), ABCMCMC seems to be performing well with the Markov chain state moving largely in the regions of high posterior probability. However, for the enzyme kinetics model (figure 6d–f), the Markov chain has taken a long excursion into a region of low posterior probability (figure 6f) where it is a long way from the true parameter value of k3 = 0.01 for many steps. Such an extended path into the low probability region results in significant bias in this region and many more steps of the algorithm are required to compensate for this [131,132]. Just as with exact MCMC, burn-in samples and thinning may be used to reduce correlations but may not improve the final Monte Carlo estimate.
To avoid the difficulties in ensuring efficient ABCMCMC convergence, Sisson et al. [132] developed an ABC variant of SMC. Alternative versions of the method are also designed by Beaumont et al. [131] and Toni et al. [31]. The fundamental idea is to use sequential importance resampling to propagate mp samples, called particles, through a sequence of R + 1 ABC posterior distributions defined through a sequence of discrepancy thresholds, ε0, ε1, …, εR with εr > εr+1 for r = 0, 1, …, R − 1 and ε0 = ∞ (that is, is the prior). This results in ABCSMC:
-
(1)
initialize r = 0 and weights for i = 1, …, mp;
-
(2)
generate mp particles from the prior, , for i = 1, 2, …, mp;
-
(3)
set index i = 0;
-
(4)
randomly select integer, j, from the set {1, …, mp} with ;
-
(5)
generate proposal, ;
-
(6)
generate simulated data, ;
-
(7)
if , go to step 4, otherwise continue;
-
(8)
set , and i = i + 1;
-
(9)
if i < mp, go to step 4, otherwise continue;
-
(10)
set index i = 0;
-
(11)
randomly select integer, j, from the set {1, …, mp} with ;
-
(12)
set and i = i + 1;
-
(13)
if i < mp, go to step 11, otherwise continue;
-
(14)
set and r = r + 1;
-
(15)
if r < R, go to step 3, otherwise terminate simulation.
An implementation of the ABCSMC method is provided in ABCSMCSampler.m.
The ABCSMC method avoids some issues inherent in ABCMCMC such as strong sample correlation and long excursions into low probability regions. However, it is still the case that the number of particles, mp, often needs to be larger than the desired number of independent samples, m, from the ABC posterior with εR. This is especially true when there is a larger discrepancy between two successive ABC posteriors in the sequence. Although techniques exist to adaptively generate the sequence of acceptance thresholds [133]. Also note that ABCSMC still requires a proposal kernel to mutate the particles at each step and the efficiency of the method is affected by the choice of proposal kernel. Just as with ABCMCMC, the task of selecting an optimal proposal kernel is non-trivial [134].
The formulation of ABCSMC using a sequence of discrepancy thresholds hints that MLMC ideas could also be applicable. Recently, a variety of MLMC methods for ABC have been proposed [135–137]. All of these approaches are similar in their application of the multilevel telescoping summation to compute expectations with respect to ABC posterior distributions,
| 4.11 |
where ε0, ε1, …, εL is a sequence of discrepancy thresholds with εℓ > εℓ+1 for all ℓ = 0, 1, …, L − 1.
Unlike the forwards problem, no exact solution has been found for the generation of correlated ABC posterior pairs, , with ℓ = 1, …, L. Several approaches to this problem have been proposed: Guha & Tan [135] use a sequence of correlated ABCMCMC samplers in a similar manner to Dodwell et al. [105] and Efendiev et al. [106]; Jasra et al. [136] apply the MLSMC estimator of Beskos et al. [107] directly to equation (4.11); and Warne et al. [137] develop a MLMC variant of ABC rejection sampler through the introduction of an approximation based on the empirical marginal posterior distributions.
The coupling scheme of Warne et al. [137] is the simplest to present succinctly. The scheme is based on the inverse transform method for sampling univariate distributions. Given random variable X ∼ p(x), the cumulative distribution function is defined as
Note that 0 ≤ F(z) ≤ 1. If F(z) has an inverse function, F−1(u), and U(1), U(2), …, U(m) are independent samples of , then independent samples from p(x) can be generated using X(1) = F−1(U(1)), X(2) = F−1(U(2)), …, X(m) = F−1(U(m)).
Given samples, , an estimate for the posterior marginal cumulative distribution functions can be obtained using
where is the jth component of for all j = 1, …, M, and if x ≤ z and otherwise. Assuming we already have an estimate of the posterior marginal cumulative distribution functions for acceptance threshold εℓ−1, then we apply the transform . This results in a coupled marginal pair since it is equivalent to using the inverse transform method to generate and using the same uniform sample . These approximately correlated samples can be obtained using:
-
(1)
generate sample from level ℓ ABC posterior using the ABC rejection sampler;
-
(2)
set for j = 1, 2, …, M;
-
(3)
set .
Thus, the ABC multilevel Monte Carlo (ABCMLMC) [137] method proceeds though computing the Monte Carlo estimate to equation (4.11),
| 4.12 |
where are m0 independent samples using ABC rejection sampler and , are mℓ independent sample pairs using the approximately correlated ABC rejection sampler for ℓ = 1, 2, …, L. It is essential that each bias correction term in equation (4.12) be estimated in ascending order to ensure the marginal CDF estimates for εℓ−1 are available for use in the coupling scheme for the ℓth bias correction term. The complete algorithm is provided in the electronic supplementary material and the example code is provided in ABCMLMC.m. We refer the reader to Warne et al. [137] for further details.
Provided successive ABC posteriors are similar in correlation structure, then the ABCMLMC method accelerates ABC rejection sampler through reduction of the number of mL samples required for a given error tolerance. However, additional bias is introduced by the approximately correlated ABC rejection sampler since the telescoping summation is not strictly satisfied. In practice, this affects the choice of the acceptance threshold sequence [137].
We provide a comparison of the ABC rejection sampler, ABCMCMC, ABCSMC and ABCMLMC methods as presented here. Posterior means are computed for both the mono-molecular model and enzyme kinetics model inference problems along with the CIs for each estimate7 (see electronic supplementary material). Computation times and parameter estimates are provided in tables 1 and 2. Example code, DemoABCMethodsMonoMol.m and DemoABCMethodsMichMent.m, is provided. Algorithm configurations are supplied in the electronic supplementary material.
Table 1.
Comparison of ABC methods for the mono-molecular inference problem. Estimates of the posterior mean are given along with the CIs. Signed relative errors are shown for the confidence interval limits with respect to the true parameter values k1 = 1, k2 = 0.1, and k3 = 0.05. Computations are performed using an Intel® CoreTM i7-5600U CPU (2.6 GHz).
| method | posterior mean estimate | relative error | compute time |
|---|---|---|---|
| ABC rejection | 7268 (s) | ||
| sampler | |||
| ABCMCMC | 8059 (s) | ||
| ABCSMC | 580 (s) | ||
| ABCMLMC | 1327 (s) | ||
Table 2.
Comparison of ABC methods for the enzyme kinetics model inference problem. Estimates of the posterior mean are given along with the CIs. Signed relative errors are shown for the confidence interval limits with respect to the true parameter values k1 = 0.001, k2 = 0.005 and k3 = 0.01. Computations are performed using an Intel® CoreTM i7-5600U CPU (2.6 GHz).
| method | posterior mean estimate | relative error | compute time |
|---|---|---|---|
| ABC rejection | 2037 (s) | ||
| sampler | |||
| ABCMCMC | 2028 (s) | ||
| ABCSMC | 342 (s) | ||
| ABCMLMC | 795 (s) | ||
Note that no extensive tuning of the ABCMCMC, ABCSMC or ABCMLMC algorithms is performed, thus these outcomes do not reflect a detailed and optimized implementation of the methods. Such a benchmark would be significantly more computationally intensive than the comparison of Monte Carlo methods for the forwards problem (figure 3). The computational statistics literature demonstrates that ABCMCMC [130], ABCSMC [131,132] and ABCMLMC [135–137] can be tuned to provide very competitive results on a given inference problem. However, a large number of trial computations are often required to achieve this tuning, or more complex adaptive schemes need to be exploited [100,133]. Instead, our comparisons represent a more practical guide in which computations are kept short and almost no tuning is performed, thus giving a fair comparison for a fixed, short computational budget.
For both inference problems, ABCSMC and ABCMLMC produce more accurate parameter estimates in less computation time than ABC rejection sampler and ABCMCMC. ABCSMC performs better on the mono-molecular chain model and ABCMLMC performs better on the enzyme kinetic model. This is not to say that ABCMCMC cannot be tuned to perform more efficiently, but it does indicate that ABCMCMC is harder to tune without more extensive testing. An advantage of ABCMLMC is that it has fewer components that require tuning, since the sample numbers may be optimally chosen, and the user need only provide a sequence of acceptance thresholds [137]. However, the ability to tune the proposal kernel in ABCSMC can be a significant advantage, despite the challenge of determining a good choice for such a proposal [133,134].
4.4. Summary of the inverse problem
Bayesian approaches for uncertainty quantification and parameter inference have proven to be powerful techniques in the life sciences. However, in the study of intracellular biochemical processes, the intractability of the likelihood causes is a significant computational challenge.
A crucial advance towards obtaining numerical solutions in this Bayesian inverse problem setting has been the advent of likelihood-free methods that replace likelihood evaluations with stochastic simulation. Through such methods, especially those based on ABC, a direct connection between the forwards and inverse problems is made explicit. Not only is efficient forwards simulation key, but information obtained through each new sample path must be effectively used. While ABC rejection sampler ignores the latter, advanced methods like ABCMCMC, ABCSMC and ABCMLMC all provide different mechanisms for incorporating this new information. For a specific inverse problem, however, it will often not be clear which method will perform optimally. In general, significant trial sampling must be performed to get the most out of any of these algorithms.
The application of MLMC methods to ABC-based samplers is a very new area of research. There are many open questions that relate to the appropriateness of various approximations and coupling schemes for a given inference problem. The method of Warne et al. [137] is conceptually straightforward; however, it is known that additional bias is incurred through the coupling scheme. Alternative coupling approximations that combine MLMC and MCMC [135] or SMC [136] may be improvements, or a hybrid scheme could be derived through a combination of these methods. Currently, it is not clear which method is the most widely applicable.
5. Summary and outlook
Stochastic models of biochemical reaction networks are routinely used to study various intracellular processes, such as gene regulation. By studying the forwards problem, stochastic models can be explored in silico to gain insight into potential hypotheses related to observed phenomena and inform potential experimentation. Models may be calibrated and key kinetic information may be extracted from time-course data through the inverse problem. Both the forward and inverse problems are, in practice, reliant upon computation.
Throughout this review, we deliberately avoid detailed theoretical derivations in favour of practical computational or algorithmic examples. Since all of our code examples are specifically developed within the user friendly Matlab® programming environment, our codes do not represent optimal implementations. Certainly, there are software packages available that implement some of these techniques and others that we have not discussed. For example, stochastic simulation is available in COPASI [138], StochKit [139,140], StochPy [141] and STEPS [142], and ABC-based inference is available in ABC-SysBio [143], abctools [144] and ABCtoolbox [145].
Throughout this review, we have focused on two biochemical reaction network models, the mono-molecular chain model for which the chemical master equation can be solved analytically, and the enzyme kinetics model with an intractable chemical master equation. However, the majority of our Matlab® implementations are completely general and apply to an arbitrary biochemical reaction network model. The biochemical reaction network construction functions, MonoMolecularChain.m and MichaelisMenten.m, demonstrate how to construct a biochemical reaction network data structure that most of our scripts and functions may directly use, with the only exceptions being those that relate to the analytical solution to the chemical master equation of the mono-molecular chain model. Therefore, our code may be viewed as a useful collection of prototype implementations that may be easily applied in new contexts.
The techniques we present here are powerful tools for the analysis of real biological data and design of experiments. In particular, these methods are highly relevant for the reconstruction of genetic regulatory networks from gene expression microarray data [146–148]. Cell signalling pathways can also be analysed using mass spectroscopy proteomic data [149,150]. Furthermore, Bayesian optimal experimental design using stochastic biochemical reaction networks is key to reliable characterization of light-induced gene expression [151], a promising technique for external genetic regulation in vivo [152,153].
This review provides researchers in the life sciences with the fundamental concepts and computational tools required to apply stochastic biochemical reaction network models in practice. Through practical demonstration, the current state-of-the-art in simulation and Monte Carlo methods will be more widely and readily applied by the broader life sciences community.
Supplementary Material
Acknowledgements
Computational resources where provided by the eResearch Office, Queensland University of Technology. We thank the two referees for helpful and insightful comments.
Endnotes
These are not ‘rates’ but scale factors on reaction event probabilities. A ‘slow’ reaction with low kinetic rate may still occur rapidly, but the probability of this event is low.
We use the Statistics and Machine Learning Toolbox within the Matlab® environment for generating random samples from any of the standard probability distributions.
In the theory of Markov processes, this equation is known as the Kolmogorov forward equation.
Since θ and Yobs are real-valued, we are not really dealing with probability distribution functions, but rather probability density functions. We will not continue to make this distinction. However, the main technicality is that it no longer makes sense to say ‘the probability of θ = θ0’ but rather only ‘the probability that θ is close to θ0’. The probability density function must be integrated over the region ‘close to θ0’ to yield this probability.
That is, the probability of an event that encompasses all possible outcomes is one.
Provided the method of estimating the likelihood is unbiased, or has bias that is independent of θ.
We report the 95% CI as an accuracy measure for the Monte Carlo estimate of the true posterior mean. This does not quantify parameter uncertainty that would be achieved through posterior covariances and credible intervals.
Data accessibility
The Matlab® code examples and demonstration scripts are available from GitHub https://github.com/ProfMJSimpson/Warne2018.
Authors' contributions
D.J.W., R.E.B. and M.J.S. designed the review. D.J.W. performed the review. D.J.W., R.E.B. and M.J.S. contributed analytical tools. D.J.W. developed algorithm implementations. D.J.W., R.E.B. and M.J.S. wrote the article.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by the Australian Research Council (DP170100474). R.E.B. is a Royal Society Wolfson Research Merit Award holder, would like to thank the Leverhulme Trust for a Research Fellowship and also acknowledges the BBSRC for funding via grant no. BB/R000816/1.
References
- 1.Abkowitz JL, Catlin SN, Guttorp P. 1996. Evidence that hematopoiesis may be a stochastic process in vivo. Nat. Med. 2, 190–197. ( 10.1038/nm0296-190) [DOI] [PubMed] [Google Scholar]
- 2.Arkin A, Ross J, McAdams HH. 1998. Stochastic kinetic analysis of developmental pathway bifurcation in phage l-infected Escherichia coli cells. Genetics 149, 1633–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaern M, Elston TC, Blake WJ, Collins JJ. 2005. Stochasticity in gene expression: from theories to phenotypes. Nat. Rev. Genet. 9, 451–464. ( 10.1038/nrg1615) [DOI] [PubMed] [Google Scholar]
- 4.McAdams HH, Arkin A. 1997. Stochastic mechanisms in gene expression. Proc. Natl Acad. Sci. USA 94, 814–819. ( 10.1073/pnas.94.3.814) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raj A, van-Oudenaarden A. 2008. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 135, 216–226. ( 10.1016/j.cell.2008.09.050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elowitz MB, Levine AJ, Siggia ED, Swain PS. 2002. Stochastic gene expression in a single cell. Science 297, 1183–1186. ( 10.1126/science.1070919) [DOI] [PubMed] [Google Scholar]
- 7.Keren L, van-Dijk D, Weingarten-Gabbay S, Davidi D, Jona G, Weinberger A, Milo R, Segal E. 2015. Noise in gene expression is coupled to growth rate. Genome Res. 25, 1893–1902. ( 10.1101/gr.191635.115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soltani M, Vargas-Garcia CA, Antunes D, Singh A. 2016. Intercellular variability in protein levels from stochastic expression and noisy cell cycle processes. PLoS Comput. Biol. 12, e1004972 ( 10.1371/journal.pcbi.1004972) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taniguchi Y, Choi PJ, Li G-W, Chen H, Babu M, Hearn J, Emili A, Xie XS. 2010. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 329, 533–538. ( 10.1126/science.1188308) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fedoroff N, Fontana W. 2002. Small numbers of big molecules. Science 297, 1129–1131. ( 10.1126/science.1075988) [DOI] [PubMed] [Google Scholar]
- 11.Eldar A, Elowitz MB. 2010. Functional roles for noise in genetic circuits. Nature 467, 167–173. ( 10.1038/nature09326) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith S, Grima R. 2018. Single-cell variability in multicellular organisms. Nat. Commun. 9, 345 ( 10.1038/s41467-017-02710-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feinberg AP. 2014. Epigenetic stochasticity, nuclear structure and cancer: the implications for medicine. J. Intern. Med. 276, 5–11. ( 10.1111/joim.12224) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta P, Fillmore CM, Jiang G, Shapira SD, Tao K, Kuperwasser C, Lander ES. 2011. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell 146, 633–644. ( 10.1016/j.cell.2011.07.026) [DOI] [PubMed] [Google Scholar]
- 15.Satija R, Shalek AK. 2014. Heterogeneity in immune responses: from populations to single cells. Trends Immunol. 35, 219–229. ( 10.1016/j.it.2014.03.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moss F, Pei X. 1995. Neurons in parallel. Nature 376, 211–212. ( 10.1038/376211a0) [DOI] [PubMed] [Google Scholar]
- 17.Paulsson J, Berg OG, Ehrenberg M. 2000. Stochastic focusing: fluctuation-enhanced sensitivity of intracellular regulation. Proc. Natl Acad. Sci. USA 97, 7148–7153. ( 10.1073/pnas.110057697) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bressloff PC. 2017. Stochastic switching in biology: from genotype to phenotype. J. Phys. A: Math. Theor. 50, 133001 ( 10.1088/1751-8121/aa5db4) [DOI] [Google Scholar]
- 19.Thattai M, van-Oudenaarden A. 2001. Intrinsic noise in gene regulatory networks. Proc. Natl Acad. Sci. USA 98, 8614–8619. ( 10.1073/pnas.151588598) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tian T, Burrage K. 2006. Stochastic models for regulatory networks of the genetic toggle switch. Proc. Natl Acad. Sci. USA 103, 8372–8377. ( 10.1073/pnas.0507818103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gillespie DT. 1977. Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem. 81, 2340–2361. ( 10.1021/j100540a008) [DOI] [Google Scholar]
- 22.Wilkinson DJ. 2012. Stochastic modelling for systems biology, 2nd edn Boca Raton, FL: CRC Press. [Google Scholar]
- 23.Schnoerr D, Sanguinetti G, Grima R. 2017. Approximation and inference methods for stochastic biochemical kinetics—a tutorial review. J. Phys. A: Math. Theor. 50, 093001 ( 10.1088/1751-8121/aa54d9) [DOI] [Google Scholar]
- 24.Erban R, Chapman J. 2009. Stochastic modelling of reaction-diffusion processes: algorithms for bimolecular reactions. Phys. Biol. 6, 046001 ( 10.1088/1478-3975/6/4/046001) [DOI] [PubMed] [Google Scholar]
- 25.Fange D, Berg OG, Sjöberg P, Elf J. 2010. Stochastic reaction-diffusion kinetics in the microscopic limit. Proc. Natl Acad. Sci. USA 107, 19 820–19 825. ( 10.1073/pnas.1006565107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Isaacson SA. 2008. Relationship between the reaction-diffusion master equation and particle tracking models. J. Phys. A: Math. Theor. 41, 065003 ( 10.1088/1751-8113/41/6/065003) [DOI] [Google Scholar]
- 27.Turner TE, Schnell S, Burrage K. 2004. Stochastic approaches for modelling in vivo reactions. Comput. Biol. Chem. 28, 165–178. ( 10.1016/j.compbiolchem.2004.05.001) [DOI] [PubMed] [Google Scholar]
- 28.Gillespie DT, Hellander A, Petzold LR. 2013. Perspective: stochastic algorithms for chemical kinetics. J. Chem. Phys. 138, 170901 ( 10.1063/1.4801941) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Higham DJ. 2008. Modeling and simulating chemical reactions. SIAM Rev. 50, 347–368. ( 10.1137/060666457) [DOI] [Google Scholar]
- 30.Golightly A, Wilkinson DJ. 2011. Bayesian parameter inference for stochastic biochemical network models using particle Markov chain Monte Carlo. Interface Focus 1, 807–820. ( 10.1098/rsfs.2011.0047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toni T, Welch D, Strelkowa N, Ipsen A, Stumpf MPH. 2009. Approximate Bayesian computation scheme for parameter inference and model selection in dynamical systems. J. R. Soc. Interface 6, 187–202. ( 10.1098/rsif.2008.0172) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sunnåker M, Busetto AG, Numminen E, Corander J, Foll M, Dessimoz C. 2013. Approximate Bayesian computation. PLoS Comput. Biol. 9, e1002803 ( 10.1371/journal.pcbi.1002803) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gillespie DT. 1992. A rigorous derivation of the chemical master equation. Phys. A 188, 404–425. ( 10.1016/0378-4371(92)90283-V) [DOI] [Google Scholar]
- 34.Erban R, Chapman SJ, Maini PK. 2007 A practical guide to stochastic simulation of reaction-diffusion processes. (http://arxiv.org/abs/0704.1908. )
- 35.Higham DJ. 2001. An algorithmic introduction to numerical simulation of stochastic differential equations. SIAM Rev. 43, 525–546. ( 10.1137/S0036144500378302) [DOI] [Google Scholar]
- 36.Jahnke T, Huisinga W. 2007. Solving the chemical master equation for monomolecular reaction systems analytically. J. Math. Biol. 54, 1–26. ( 10.1007/s00285-006-0034-x) [DOI] [PubMed] [Google Scholar]
- 37.Michaelis L, Menten ML. 1913. Die kinetik der invertinwirkung. Biochem. Z. 49, 333–369. [Google Scholar]
- 38.Rao CV, Arkin AP. 2003. Stochastic chemical kinetics and the quasi-steady-state assumption: application to the Gillespie algorithm. J. Chem. Phys. 118, 4999–5010. ( 10.1063/1.1545446) [DOI] [Google Scholar]
- 39.Gillespie DT. 2001. Approximate accelerated simulation of chemically reacting systems. J. Chem. Phys. 115, 1716–1733. ( 10.1063/1.1378322) [DOI] [Google Scholar]
- 40.Gillespie DT. 2000. The chemical Langevin equation. J. Chem. Phys. 113, 297–306. ( 10.1063/1.481811) [DOI] [Google Scholar]
- 41.Gibson MA, Bruck J. 2000. Efficient exact stochastic simulation of chemical systems with many species and many channels. J. Phys. Chem. 104, 1876–1889. ( 10.1021/jp993732q) [DOI] [Google Scholar]
- 42.Anderson DF. 2007. A modified next reaction method for simulating chemical systems with time dependent propensities and delays. J. Chem. Phys. 127, 214107 ( 10.1063/1.2799998) [DOI] [PubMed] [Google Scholar]
- 43.Cao Y, Li H, Petzold L. 2004. Efficient formulation of the stochastic simulation algorithm for chemically reacting systems. J. Chem. Phys. 121, 4059–4067. ( 10.1063/1.1778376) [DOI] [PubMed] [Google Scholar]
- 44.Voliotis M, Thomas P, Grima R, Bowsher R. 2016. Stochastic simulation of biomolecular networks in dynamic environments. PLoS Comput. Biol. 12, e1004923 ( 10.1371/journal.pcbi.1004923) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Indurkhya S, Beal J. 2010. Reaction factoring and bipartite update graphs accelerate the Gillespie algorithm for large-scale biochemical systems. PLoS ONE 5, e8125 ( 10.1371/journal.pone.0008125) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thanh VH, Priami C, Zunino R. 2014. Efficient rejection-based simulation of biochemical reactions with stochastic noise and delays. J. Chem. Phys. 141, 134116 ( 10.1063/1.4896985) [DOI] [PubMed] [Google Scholar]
- 47.Thanh VH. 2017. Stochastic simulation of biochemical reactions with partial-propensity and rejection-based approaches. Math. Biosci. 292, 67–75. ( 10.1016/j.mbs.2017.08.001) [DOI] [PubMed] [Google Scholar]
- 48.Slepoy A, Thompson AP, Plimpton SJ. 2008. A constant-time kinetic Monte Carlo algorithm for simulation of large biochemical reaction networks. J. Chem. Phys. 128, 205101 ( 10.1063/1.2919546) [DOI] [PubMed] [Google Scholar]
- 49.Cao Y, Gillespie DT, Petzold LR. 2005. Avoiding negative populations in explicit Poisson tau-leaping. J. Chem. Phys. 123, 054104 ( 10.1063/1.1992473) [DOI] [PubMed] [Google Scholar]
- 50.Cao Y, Gillespie DT, Petzold LR. 2006. Efficient step size selection for the tau-leaping simulation method. J. Chem. Phys. 124, 044109 ( 10.1063/1.2159468) [DOI] [PubMed] [Google Scholar]
- 51.Rathinam M, Petzold LR, Cao Y, Gillespie DT. 2003. Stiffness in stochastic chemically reacting systems: the implicit tau-leaping method. J. Chem. Phys. 119, 12 784–12 794. ( 10.1063/1.1627296) [DOI] [Google Scholar]
- 52.Tian T, Burrage K. 2004. Binomial leap methods for simulating stochastic chemical kinetics. J. Chem. Phys. 121, 10 356–10 364. ( 10.1063/1.1810475) [DOI] [PubMed] [Google Scholar]
- 53.Cao Y, Gillespie DT, Petzold LR. 2005. The slow-scale stochastic simulation algorithm. J. Chem. Phys. 122, 014116 ( 10.1063/1.1824902) [DOI] [PubMed] [Google Scholar]
- 54.E W, Liu D, Vanden-Eijnden E. 2005. Nested stochastic simulation algorithm for chemical kinetic systems with disparate rates. J. Chem. Phys. 123, 194107 ( 10.1063/1.2109987) [DOI] [PubMed] [Google Scholar]
- 55.Marchetti L, Priami C, Thanh VH. 2016. HRSSA: efficient hybrid stochastic simulation for spatially homogeneous biochemical reaction network. J. Comput. Phys. 317, 301–317. ( 10.1016/j.jcp.2016.04.056) [DOI] [Google Scholar]
- 56.Cotter SL, Erban R. 2016. Error analysis of diffusion approximation methods for multiscale systems in reaction kinetics. SIAM J. Sci. Comput. 38, B144–B163. ( 10.1137/14100052X) [DOI] [Google Scholar]
- 57.Wilkinson DJ. 2009. Stochastic modelling for quantitative description of heterogeneous biological systems. Nat. Rev. Genet. 10, 122–133. ( 10.1038/nrg2509) [DOI] [PubMed] [Google Scholar]
- 58.Burrage K, Burrage PM, Tian T. 2004. Numerical methods for strong solutions of stochastic differential equations: an overview. Proc. R. Soc. Lond. A 460, 373–402. ( 10.1098/rspa.2003.1247) [DOI] [Google Scholar]
- 59.Gadgil C, Lee CH, Othmer HG. 2005. A stochastic analysis of first-order reaction networks. Bull. Math. Biol. 67, 901–946. ( 10.1016/j.bulm.2004.09.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Giles MB. 2015. Multilevel Monte Carlo methods. Acta Numer. 24, 259–328. ( 10.1017/S096249291500001X) [DOI] [Google Scholar]
- 61.Anderson DF, Ganguly A, Kurtz TG. 2011. Error analysis of tau-leap simulation methods. Ann. Appl. Probab. 21, 2226–2262. ( 10.1214/10-AAP756) [DOI] [Google Scholar]
- 62.Li T. 2007. Analysis of explicit tau-leaping schemes for simulating chemically reacting systems. Multiscale Model. Simul. 6, 417–436. ( 10.1137/06066792X) [DOI] [Google Scholar]
- 63.Anderson DF, Higham DJ. 2012. Multilevel Monte Carlo for continuous time Markov chains, with applications in biochemical kinetics. Multiscale Model. Simul. 10, 146–179. ( 10.1137/110840546) [DOI] [Google Scholar]
- 64.Giles MB. 2008. Multilevel Monte Carlo path simulation. Oper. Res. 56, 607–617. ( 10.1287/opre.1070.0496) [DOI] [Google Scholar]
- 65.Lester C, Baker RE, Giles MB, Yates CA. 2016. Extending the multi-level method for the simulation of stochastic biological systems. Bull. Math. Biol. 78, 1640–1677. ( 10.1007/s11538-016-0178-9) [DOI] [PubMed] [Google Scholar]
- 66.Lester C, Yates CA, Giles MB, Baker RE. 2015. An adaptive multi-level simulation algorithm for stochastic biological systems. J. Chem. Phys. 142, 024113 ( 10.1063/1.4904980) [DOI] [PubMed] [Google Scholar]
- 67.Moraes A, Tempone R, Vilanova P. 2016. A multilevel adaptive reaction-splitting simulation method for stochastic reaction networks. SIAM J. Sci. Comput. 38, A2091–A2117. ( 10.1137/140972081) [DOI] [Google Scholar]
- 68.Wilson D, Baker RE. 2016. Multi-level methods and approximating distribution functions. AIP Adv. 6, 075020 ( 10.1063/1.4960118) [DOI] [Google Scholar]
- 69.Higham DJ. 2015. An introduction to multilevel Monte Carlo for option valuation. Int. J. Comput. Math. 82, 2347–2360. ( 10.1080/00207160.2015.1077236) [DOI] [Google Scholar]
- 70.Smith S, Grima R. 2016. Breakdown of the reaction-diffusion master equation with nonelementary rates. Phys. Rev. E 93, 052135 ( 10.1103/PhysRevE.93.052135) [DOI] [PubMed] [Google Scholar]
- 71.Gillespie DT, Petzold LR, Seitaridou E. 2014. Validity conditions for stochastic chemical kinetics in diffusion-limited system. J. Chem. Phys. 140, 054111 ( 10.1063/1.4863990) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gelman A, Carlin JB, Stern HS, Dunston DB, Vehtari A, Rubin DB. 2014. Bayesian data analysis, 3rd edn London, UK: Chapman & Hall/CRC. [Google Scholar]
- 73.Sisson SA, Fan Y, Beaumont M. 2018. Handbook of approximate Bayesian computation, 1st edn London, UK: Chapman & Hall/CRC. [Google Scholar]
- 74.Finkenstädt B. et al. 2008. Reconstruction of transcriptional dynamics from gene reporter data using differential equations. Bioinformatics 24, 2901–2907. ( 10.1093/bioinformatics/btn562) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Iafolla MAJ, Mazumder M, Sardana V, Velauthapillai T, Pannu K, McMillen DR. 2008. Dark proteins: effect of inclusion body formation on quantification of protein expression. Proteins: Struct. Funct. Bioinf. 72, 1233–1242. ( 10.1002/prot.22024) [DOI] [PubMed] [Google Scholar]
- 76.Bajar BT, Wang ES, Zhang S, Lin MZ, Chu J. 2016. A guide to fluorescent protein FRET pairs. Sensors 16, 1488 ( 10.3390/s16091488) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen B-C. et al. 2014. Lattice light sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science 346, 1257998 ( 10.1126/science.1257998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Leung BO, Chou KC. 2011. Review of super-resolution fluorescent microscopy for biology. Appl. Spectrosc. 65, 967–980. ( 10.1366/11-06398) [DOI] [PubMed] [Google Scholar]
- 79.Sahl SJ, Hell SW, Jakobs S. 2017. Fluorescence nanoscopy in cell biology. Nat. Rev. Mol. Cell Biol. 18, 685–701. ( 10.1038/nrm.2017.71) [DOI] [PubMed] [Google Scholar]
- 80.Vittadello ST, McCue SW, Gunasingh G, Haass NK, Simpson MJ. 2018. Mathematical models for cell migration with real-time cell cycle dynamics. Biophys. J. 114, 1241–1253. ( 10.1016/j.bpj.2017.12.041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Browning AP, McCue SW, Simpson MJ. 2017. A Bayesian computational approach to explore the optimal duration of a cell proliferation assay. Bull. Math. Biol. 79, 1888–1906. ( 10.1007/s11538-017-0311-4) [DOI] [PubMed] [Google Scholar]
- 82.Ellison AM. 2004. Bayesian inference in ecology. Ecol. Lett. 7, 509–520. ( 10.1111/j.1461-0248.2004.00603.x) [DOI] [Google Scholar]
- 83.Liepe J, Filippi S, Komorowski M, Stumpf MPH. 2013. Maximizing the information content of experiments in systems biology. PLoS Comput. Biol. 9, e1002888 ( 10.1371/journal.pcbi.1002888) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Maclaren OJ, Byrne HM, Fletcher AG, Maini PK. 2015. Models, measurement and inference in epithelial tissue dynamics. Math. Biosci. Eng. 12, 1321–1340. ( 10.3934/mbe.2015.12.1321) [DOI] [PubMed] [Google Scholar]
- 85.Maclaren OJ, Parker A, Pin C, Carding SR, Watson AJ, Fletcher AG, Byrne HM, Maini PK. 2017. A hierarchical Bayesian model for understanding the spatiotemporal dynamics of the intestinal epithelium. PLoS Comput. Biol. 13, e1005688 ( 10.1371/journal.pcbi.1005688) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vanlier J, Tiemann CA, Hilbers PAJ, van-Riel NAW. 2014. Optimal experiment design for model selection in biochemical networks. BMC Syst. Biol. 8, 20 ( 10.1186/1752-0509-8-20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Warne DJ, Baker RE, Simpson MJ. 2018. Using experimental data and information criteria to guide model selection for reaction–diffusion problems in mathematical biology. bioRxiv. ( 10.1101/444679) [DOI] [PubMed] [Google Scholar]
- 88.Warne DJ, Baker RE, Simpson MJ. 2017. Optimal quantification of contact inhibition in cell populations. Biophys. J. 113, 1920–1924. ( 10.1016/j.bpj.2017.09.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Browning AP, Haridas P, Simpson MJ. 2019. A Bayesian sequential learning framework to parameterise continuum models of melanoma invasion into human skin. Bull. Math. Biol. 81, 676–698. ( 10.1007/s11538-018-0532-1) [DOI] [PubMed] [Google Scholar]
- 90.Wilkinson DJ. 2011. Parameter inference for stochastic kinetic models of bacterial gene regulation: a Bayesian approach to systems biology. In Bayesian statistics 9: Proc. 9th Valencia International Meeting, pp. 679–706. Oxford, UK: Oxford University Press.
- 91.Lele SR, Dennis B, Lutscher F. 2007. Data cloning: easy maximum likelihood estimation for complex ecological models using Bayesian Markov chain Monte Carlo methods. Ecol. Lett. 10, 551–563. ( 10.1111/j.1461-0248.2007.01047.x) [DOI] [PubMed] [Google Scholar]
- 92.Picchini U, Anderson R. 2017. Approximate maximum likelihood estimation using data-cloning ABC. Comput. Stat. Data Anal. 105, 166–183. ( 10.1016/j.csda.2016.08.006) [DOI] [Google Scholar]
- 93.Efron B. 1979. Bootstrap methods: another look at the jackknife. Ann. Stat. 7, 1–26. ( 10.1214/aos/1176344552) [DOI] [Google Scholar]
- 94.Rubin DB. 1981. The Bayesian bootstrap. Ann. Stat. 9, 130–134. ( 10.1214/aos/1176345338) [DOI] [Google Scholar]
- 95.Hastings WK. 1970. Monte Carlo sampling methods using Markov chains and their applications. Biometrika 57, 97–109. ( 10.2307/2334940) [DOI] [Google Scholar]
- 96.Metropolis N, Rosenbluth AW, Rosenbluth MN, Teller AH, Teller E. 1953. Equation of state calculations by fast computing machines. J. Chem. Phys. 21, 1087–1092. ( 10.1063/1.1699114) [DOI] [Google Scholar]
- 97.Geyer CJ. 1992. Practical Markov chain Monte Carlo. Stat. Sci. 7, 473–483. ( 10.1214/ss/1177011137) [DOI] [Google Scholar]
- 98.Cotter SL, Roberts GO, Stuart AM, White D. 2013. MCMC methods for functions: modifying old algorithms to make them faster. Stat. Sci. 28, 424–446. ( 10.1214/13-STS421) [DOI] [Google Scholar]
- 99.Green PJ, Łatuszyński K, Pereyra M, Robert CP. 2015. Bayesian computation: a summary of the current state, and samples backwards and forwards. Stat. Comput. 25, 835–862. ( 10.1007/s11222-015-9574-5) [DOI] [Google Scholar]
- 100.Roberts GO, Rosenthal JS. 2009. Examples of adaptive MCMC. J. Comput. Graph. Stat. 18, 349–367. ( 10.1198/jcgs.2009.06134) [DOI] [Google Scholar]
- 101.Link WA, Eaton MJ. 2011. On thinning of chains in MCMC. Methods Ecol. Evol. 3, 112–115. ( 10.1111/j.2041-210X.2011.00131.x) [DOI] [Google Scholar]
- 102.MacEachern SN, Berliner LM. 1994. Subsampling the Gibbs sampler. Am. Stat. 48, 188–190. ( 10.1080/00031305.1994.10476054) [DOI] [Google Scholar]
- 103.Glynn PW, Iglehart DL. 1989. Importance sampling for stochastic simulations. Manage. Sci. 35, 1367–1392. ( 10.1287/mnsc.35.11.1367) [DOI] [Google Scholar]
- 104.Del Moral P, Doucet A, Jasra A. 2006. Sequential Monte Carlo samplers. J. R. Stat. Soc. Ser. B 68, 411–436. ( 10.1111/j.1467-9868.2006.00553.x) [DOI] [Google Scholar]
- 105.Dodwell TJ, Ketelsen C, Scheichl R, Teckentrup AL. 2015. A hierarchical multilevel Markov chain Monte Carlo algorithm with applications to uncertainty quantification in subsurface flow. SIAM/ASA J. Uncertainty Quantif. 3, 1075–1108. ( 10.1137/130915005) [DOI] [Google Scholar]
- 106.Efendiev Y, Jin B, Michael P, Tan X. 2015. Multilevel Markov chain Monte Carlo method for high-contrast single-phase flow problems. Commun. Comput. Phys. 17, 259–286. ( 10.4208/cicp.021013.260614a) [DOI] [Google Scholar]
- 107.Beskos A, Jasra A, Law K, Tempone R, Zhou Y. 2017. Multilevel sequential Monte Carlo samplers. Stoch. Process. Appl. 127, 1417–1440. ( 10.1016/j.spa.2016.08.004) [DOI] [Google Scholar]
- 108.Andrieu C, Roberts GO. 2009. The pseudo-marginal approach for efficient Monte Carlo computations. Ann. Stat. 37, 697–725. ( 10.1214/07-AOS574) [DOI] [Google Scholar]
- 109.Andrieu C, Doucet A, Holenstein R. 2010. Particle Markov chain Monte Carlo methods. J. R. Stat. Soc. B 72, 269–342. ( 10.1111/j.1467-9868.2009.00736.x) [DOI] [Google Scholar]
- 110.Pooley CM, Bishop SC, Marion G. 2015. Using model-based proposals for fast parameter inference on discrete state space, continuous-time Markov processes. J. R. Soc. Interface 12, 20150225 ( 10.1098/rsif.2015.0225) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jasra A, Kamatani K, Law K, Zhou Y. 2018. Bayesian static parameter estimation for partially observed diffusions via multilevel Monte Carlo. SIAM J. Sci. Comput. 40, A887–A902. ( 10.1137/17M1112595) [DOI] [Google Scholar]
- 112.Pritchard JK, Seielstad MT, Perez-Lezaun A, Feldman MW. 1999. Population growth of human y chromosomes: a study of y chromosome microsatellites. Mol. Biol. Evol. 16, 1791–1798. ( 10.1093/oxfordjournals.molbev.a026091) [DOI] [PubMed] [Google Scholar]
- 113.Tavaré S, Balding DJ, Griffiths RC, Donnelly P. 1997. Inferring coalescence times from DNA sequence data. Genetics 145, 505–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Blum MGB. 2010. Approximate Bayesian computation: a nonparametric perspective. J. Am. Stat. Assoc. 105, 1178–1187. ( 10.1198/jasa.2010.tm09448) [DOI] [Google Scholar]
- 115.Beaumont MA, Zhang W, Balding DJ. 2002. Approximate Bayesian computation in population genetics. Genetics 162, 2025–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ratmann O, Donker G, Meijer A, Fraser C, Koelle K. 2012. Phylodynamic inference and model assessment with approximate Bayesian computation: influenza as a case study. PLoS Comput. Biol. 8, e1002835 ( 10.1371/journal.pcbi.1002835) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wilkinson RD, Steiper ME, Soligo C, Martin RD, Yang Z, Tavaré S. 2011. Dating primate divergences through an integrated analysis of palaeontological and molecular data. Syst. Biol. 60, 16–31. ( 10.1093/sysbio/syq054) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Johnston ST, Simpson MJ, McElwain DLS, Binder BJ, Ross JV. 2014. Interpreting scratch assays using pair density dynamics and approximate Bayesian computation. Open Biol. 4, 140097 ( 10.1098/rsob.140097) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ross RJH, Baker RE, Parker A, Ford MJ, Mort RL, Yates CA. 2017. Using approximate Bayesian computation to quantify cell–cell adhesion parameters in a cell migratory process. npj Syst. Biol. Appl. 3, 9 ( 10.1038/s41540-017-0010-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Vo BN, Drovandi CC, Pettit AN, Simpson MJ. 2015. Quantifying uncertainty in parameter estimates for stochastic models of collective cell spreading using approximate Bayesian computation. Math. Biosci. 263, 133–142. ( 10.1016/j.mbs.2015.02.010) [DOI] [PubMed] [Google Scholar]
- 121.Tanaka MM, Francis AR, Luciani F, Sisson SA. 2006. Using approximate Bayesian computation to estimate tuberculosis transmission parameter from genotype data. Genetics 173, 1511–1520. ( 10.1534/genetics.106.055574) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Csillery K, Blum MGB, Gaggiotti OE, François O. 2010. Approximate Bayesian computation (ABC) in practice. Trends Ecol. Evol. 25, 410–418. ( 10.1016/j.tree.2010.04.001) [DOI] [PubMed] [Google Scholar]
- 123.Stumpf MPH. 2014. Approximate Bayesian inference for complex ecosystems. F1000Prime Rep. 6, 60 ( 10.12703/P6-60) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wilkinson RD. 2013. Approximate Bayesian computation (ABC) gives exact results under the assumption of model error. Stat. Appl. Genet. Mol. Biol. 12, 129–142. ( 10.1515/sagmb-2013-0010) [DOI] [PubMed] [Google Scholar]
- 125.Barber S, Voss J, Webster M. 2015. The rate of convergence for approximate Bayesian computation. Electron. J. Stat. 9, 80–195. ( 10.1214/15-EJS988) [DOI] [Google Scholar]
- 126.Fearnhead P, Prangle D. 2012. Constructing summary statistics for approximate Bayesian computation: semi-automatic approximate Bayesian computation. J. R. Stat. Soc. B 74, 419–474. ( 10.1111/j.1467-9868.2011.01010.x) [DOI] [Google Scholar]
- 127.Prangle D. 2016. Lazy ABC. Stat. Comput. 26, 171–185. ( 10.1007/s11222-014-9544-3) [DOI] [Google Scholar]
- 128.Buzbas EO, Rosenberg NA. 2015. AABC: approximate approximate Bayesian computation for inference in population-genetic models. Theor. Popul. Biol. 99, 31–42. ( 10.1016/j.tpb.2014.09.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lambert B, MacLean AL, Fletcher AG, Combes AN, Little MH, Byrne HM. 2018. Bayesian inference of agent-based models: a tool for studying kidney branching morphogenesis. J. Math. Biol. 76, 1673–1697. ( 10.1007/s00285-018-1208-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Marjoram P, Molitor J, Plagnol V, Tavaré S. 2003. Markov chain Monte Carlo without likelihoods. Proc. Natl Acad. Sci. USA 100, 15 324–15 328. ( 10.1073/pnas.0306899100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Beaumont MA, Cornuet J-M, Marin J-M, Robert CP. 2009. Adaptive approximate Bayesian computation. Biometrika 96, 983–990. ( 10.1093/biomet/asp052) [DOI] [Google Scholar]
- 132.Sisson SA, Fan Y, Tanaka MM. 2007. Sequential Monte Carlo without likelihoods. Proc. Natl Acad. Sci. USA 104, 1760–1765. ( 10.1073/pnas.0607208104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Drovandi CC, Pettitt AN. 2011. Estimation of parameters for macroparasite population evolution using approximate Bayesian computation. Biometrics 67, 225–233. ( 10.1111/j.1541-0420.2010.01410.x) [DOI] [PubMed] [Google Scholar]
- 134.Filippi S, Barnes CP, Cornebise J, Stumpf MPH. 2013. On optimality of kernels for approximate Bayesian computation using sequential Monte Carlo. Stat. Appl. Genet. Mol. Biol. 12, 87–107. ( 10.1515/sagmb-2012-0069) [DOI] [PubMed] [Google Scholar]
- 135.Guha N, Tan X. 2017. Multilevel approximate Bayesian approaches for flows in highly heterogeneous porous media and their applications. J. Comput. Appl. Math. 317, 700–717. ( 10.1016/j.cam.2016.10.008) [DOI] [Google Scholar]
- 136.Jasra A, Jo S, Nott D, Shoemaker C, Tempone R. 2017 Multilevel Monte Carlo in approximate Bayesian computation. (http://arxiv.org/abs/1702.03628. )
- 137.Warne DJ, Baker RE, Simpson MJ. 2018. Multilevel rejection sampling for approximate Bayesian computation. Comput. Stat. Data Anal. 124, 71–86. ( 10.1016/j.csda.2018.02.009) [DOI] [Google Scholar]
- 138.Hoops S. et al. 2006. COPASI—a complex pathway simulator. Bioinformatics 22, 3067–3074. ( 10.1093/bioinformatics/btl485) [DOI] [PubMed] [Google Scholar]
- 139.Li H, Cao Y, Petzold LR, Gillespie DT. 2008. Algorithms and software for stochastic simulation of biochemical reacting systems. Biotechnol. Prog. 24, 56–61. ( 10.1021/bp070255h) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sanft KR, Wu S, Roh M, Fu J, Lim RK, Petzold LR. 2011. StochKit2: software for discrete stochastic simulation of biochemical systems with events. Bioinformatics 27, 2457–2458. ( 10.1093/bioinformatics/btr401) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Maarleveld TR, Olivier BG, Bruggeman FJ. 2013. StochPy: a comprehensive, user-friendly tool for simulating stochastic biological processes. PLoS ONE 8, e79345 ( 10.1371/journal.pone.0079345) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hepburn I, Chen W, Wils S, De Schutter E. 2012. STEPS: efficient simulation of stochastic reaction–diffusion models in realistic morphologies. BMC Syst. Biol. 6, 36 ( 10.1186/1752-0509-6-36) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Liepe J, Kirk P, Filippi S, Toni T, Barnes CP, Stumpf MPH. 2014. A framework for parameter estimation and model selection from experimental data in systems biology using approximate Bayesian computation. Nat. Protoc. 9, 439–456. ( 10.1038/nprot.2014.025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Nunes MA, Prangle D. 2015. abctools: an R package for tuning approximate Bayesian computation analysis. R J. 7, 189–205. [Google Scholar]
- 145.Wegmann D, Leuenberger C, Neuenschwander S, Excoffier L. 2010. ABCtoolbox: a versatile toolkit for approximate Bayesian computations. BMC Bioinf. 11, 116 ( 10.1186/1471-2105-11-116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Cohen M, Kicheva A, Ribeiro A, Blassberg R, Page KM, Barnes CP, Briscoe J. 2015. Ptch1 and Gli regulate Shh signalling dynamics via multiple mechanisms. Nat. Commun. 6, 6709 ( 10.1038/ncomms7709) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Gao NP, Ud-Dean SMM, Gandrillon O, Gunawan R. 2018. SINCERITIES: inferring gene regulatory networks from time-stamped single cell transcriptional expression profiles. Bioinformatics 34, 258–266. ( 10.1093/bioinformatics/btx575) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ocone A, Haghverdi L, Mueller NS, Theis FJ. 2015. Reconstructing gene regulatory dynamics from high-dimensional single-cell snapshot data. Bioinformatics 31, i89–i96. ( 10.1093/bioinformatics/btv257) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Tian T. 2010. Stochastic models for inferring genetic regulation from microarray gene expression data. Biosystems 99, 192–200. ( 10.1016/j.biosystems.2009.11.002) [DOI] [PubMed] [Google Scholar]
- 150.Wang J, Wu Q, Hu XT, Tian T. 2016. An integrated approach to infer dynamic protein-gene interactions: a case study of the human P53 protein. Methods 110, 3–13. ( 10.1016/j.ymeth.2016.08.001) [DOI] [PubMed] [Google Scholar]
- 151.Ruess J, Parise F, Milias-Argeitis A, Khammash M, Lygeros J. 2015. Iterative experiment design guides the characterization of a light-inducible gene expression circuit. Proc. Natl Acad. Sci. USA 112, 8148–8153. ( 10.1073/pnas.1423947112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Jayaraman P, Devarajan K, Chua TK, Zhang H, Gunawan E, Poh CL. 2016. Blue light-mediated transcriptional activation and repression of gene expression in bacteria. Nucleic Acids Res. 44, 6994–7005. ( 10.1093/nar/gkw548) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yamada M, Suzuki Y, Nagasaki SC, Okuno H, Imayoshi I. 2018. Light control of the Tet gene expression system in mammalian cells. Cell Rep. 25, 487–500. ( 10.1016/j.celrep.2018.09.026) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Matlab® code examples and demonstration scripts are available from GitHub https://github.com/ProfMJSimpson/Warne2018.