Significance
African populations have been underrepresented in human genomics research yet are important for understanding modern human origins and the genetic basis of adaptive traits. Here we analyze a genome-wide dataset in 840 ethnically and geographically diverse Africans. We find that geographically distant hunter-gatherer populations from East Africa share unique common ancestry and we see strong signatures of local adaptation near genes that play a role in immune response, as well as lipid and glucose metabolism.
Keywords: African hunter-gatherers, African diversity, population genetics, natural selection, human evolution
Abstract
Anatomically modern humans arose in Africa ∼300,000 years ago, but the demographic and adaptive histories of African populations are not well-characterized. Here, we have generated a genome-wide dataset from 840 Africans, residing in western, eastern, southern, and northern Africa, belonging to 50 ethnicities, and speaking languages belonging to four language families. In addition to agriculturalists and pastoralists, our study includes 16 populations that practice, or until recently have practiced, a hunting-gathering (HG) lifestyle. We observe that genetic structure in Africa is broadly correlated not only with geography, but to a lesser extent, with linguistic affiliation and subsistence strategy. Four East African HG (EHG) populations that are geographically distant from each other show evidence of common ancestry: the Hadza and Sandawe in Tanzania, who speak languages with clicks classified as Khoisan; the Dahalo in Kenya, whose language has remnant clicks; and the Sabue in Ethiopia, who speak an unclassified language. Additionally, we observed common ancestry between central African rainforest HGs and southern African San, the latter of whom speak languages with clicks classified as Khoisan. With the exception of the EHG, central African rainforest HGs, and San, other HG groups in Africa appear genetically similar to neighboring agriculturalist or pastoralist populations. We additionally demonstrate that infectious disease, immune response, and diet have played important roles in the adaptive landscape of African history. However, while the broad biological processes involved in recent human adaptation in Africa are often consistent across populations, the specific loci affected by selective pressures more often vary across populations.
Genetic, archaeological, and linguistic evidence reflect a complex demographic history for populations in Africa. Anatomically modern humans emerged in Africa ∼300 kya (1–3) and lived in Africa for tens of thousands of years before a subset migrated out of Africa 80–40 kya (4). Many studies have focused on when, where, and how modern humans colonized the rest of the globe, but relatively few have characterized prehistoric demography within Africa during the Late Pleistocene 70–10 kya (4). This is likely because the archaeological and paleo-biological record is incomplete during that time period (5), linguistic reconstruction does not extend much beyond 10 kya (6), and recent demographic events, such as historical migrations, complicate genomic signatures of older population movements and interactions.
Relatively more is known about population histories in Africa during the recent past due to linguistic reconstructions. One of the most striking recent demographic events in Africa was the expansion of Bantu peoples (speakers of Bantu languages) from West Africa accompanying agricultural innovation in the Neolithic ∼5 kya (7). This expansion, commonly referred to as the “Bantu expansion,” significantly impacted the landscape of genetic and cultural diversity in Africa (8, 9). While Bantu languages, which belong to the Niger-Congo (NC) language family, are widely spoken across Africa, languages belonging to two additional language families, Nilo-Saharan (NS) and Afro-Asiatic (AA), are spoken by populations primarily located in central, eastern, and northern Africa who practice pastoralism and agriculture (10). A fourth language family, Khoisan, which contains click phonemes, includes several languages spoken by hunter-gatherer populations in southern Africa, as well as two languages spoken by hunter-gatherer populations in eastern Africa, the Hadza and Sandawe (11). While the Sandawe language has been identified as linguistically more similar to the Khoisan languages spoken in southern Africa than it is to the Hadza language, the inclusion of the latter two languages within the Khoisan language family is generally contentious, arguably because the relationships between the eastern and southern African Khoisan languages are older than 10 kya (12).
Several languages spoken throughout Africa remain unclassified and are considered “language isolates.” One such example is the Shabo language, also referred to as Mikeyir, spoken by the people of Ethiopia who self-identify as Sabue (also known as Sabu). While proto-Shabo is thought to be an early branch of the NS languages, the classification of Shabo into any linguistic family is unresolved (13, 14). The language spoken by the Dahalo, also referred to as Sanye, of Kenya is another such example. Some linguists classify Dahalo as AA, but it also shares a dental click phoneme with Khoisan languages (12). The shared presence of clicks has led linguists to hypothesize that the Dahalo share recent common ancestry with the Hadza and Sandawe or that ancestors of the Dahalo, speaking a proto-Dahalo language, came into contact with and subsequently borrowed linguistic features from individuals speaking a proto-Khoisan language in East Africa (15). The archaeological evidence for a common ancestry of East African hunting-gathering (HG) populations and the Khoisan-speaking populations of southern Africa is debated: while there has been some evidence of a genetic connection, the archaeological data are not conclusive (16–19). However, there have been no prior genetic studies of the Sabue or Dahalo. Here, we examine the genetic relationships of the Sabue, Dahalo, and the Khoisan-speaking populations of eastern Africa to shed light on the history of East African HG populations.
In addition to the linguistic diversity found in Africa—over 2,000 languages are spoken in the continent—African populations practice diverse subsistence strategies (11). As previously noted, agricultural technologies spread throughout sub-Saharan Africa with the Bantu expansion 5–3 kya. Before that, pastoralism spread from northeastern Africa southward into central and eastern Africa 6–3 kya (10). Populations speaking Khoisan languages, including the Hadza and Sandawe, engage, or until recently engaged, in an HG subsistence strategy. The Dahalo, Boni, El Molo, Yaaku, Sengwer, and Ogiek populations living in Kenya, and the Wata from Ethiopia, also practice an HG lifestyle, as do the Sabue of Ethiopia. Anthropologists have debated whether these East African HG populations represent distinct groups or whether they represent descendants of communities that were displaced due to past political, economic, and social phenomena (20). Other African populations traditionally practicing HG include the western (e.g., Biaka, Baka, Bakola, Bedzan) and eastern (e.g., Mbuti) rain forest hunters and gatherers (WRHG and ERHG, respectively), commonly referred to as “Pygmies,” who have adopted the languages of neighboring populations, and the Khoisan-speaking San from southern Africa. Here, we analyze the genetic diversity of 16 ethnic groups in Africa that practice a foraging life-style to better understand their relationships with each other and with neighboring populations.
Taken together, linguistic, archaeological, and genetic data have led to a proposed wide range for Khoisan-speaking HG populations throughout southern and eastern Africa, extending from Ethiopia to southern Africa (6, 15, 21, 22). However, this hypothesis remains contentious, and the origins of East African HG populations remain unknown largely because of the limits of linguistic reconstruction, archaeological data, and sparse sampling of genomic diversity in East Africa (12, 16). To explore this question further, we have genotyped 724 individuals from 46 diverse ethno-linguistic populations living in central and eastern Africa with the Illumina 1M-Duo SNP array (Fig. 1A), including all of the eastern African and WRHG populations described above. We merged these data with publicly available data from population samples including Mbuti ERHG living in the Democratic Republic of Congo, San living in Namibia and South Africa, Mandenka living in Senegal, and Mozabite living in Algeria (23, 24). In total, the merged dataset is comprised of 840 individuals sampled from 50 populations living throughout sub-Saharan Africa and genotyped for a set of ∼621,000 markers present on all platforms (SI Appendix, Table S1).
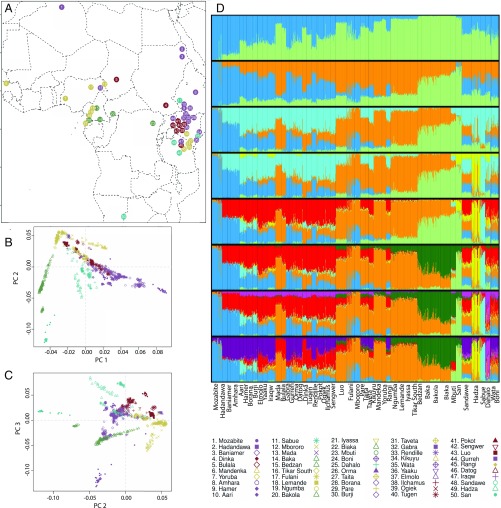
Fig. 1.
Geographic distribution of populations studied and summaries of population structure. (A) The geographic distribution of populations included in the study presented on a map of Africa. The legend indicates the colors assigned to each language family and the number and unique combination of color and symbol for each ethno-linguistic population. (B) PCA was performed using individuals’ genotypes; PC1, which explains 2.11% of the genotypic variance and shows a North–South cline, was plotted against PC2, which explains 0.91% of the genotypic variance and separates individuals with NC ancestry. (C) Hadza and Sabue individuals cluster at one extreme end of PC3, which explains 0.73% of variance in individuals’ genotypes; NS-speaking individuals are also found clustering near the Hadza and Sabue. (D) Population structure was inferred using the STRUCTURE software using 20,000 unlinked loci; results are shown from K = 2 to K = 9, the latter of which was identified as having the best, most stable fit to the data. The STRUCTURE analysis revealed K = 9 AAC. Supporting the PCA, two AAC’s corresponded to NC ancestry (orange); that is, correlated with the Bantu expansion, and North African ancestry (blue). In addition, the other AACs identify structure between HG populations: San (light green), WRHG (dark green), Hadza (yellow), Dahalo (light purple), and Sabue (light blue). Results from K = 2 to K = 8 are discussed in SI Appendix.
Results
Genome-Wide Patterns of Diversity.
To characterize genome-wide patterns of diversity in Africa, we employed principal components analysis (PCA) of individuals at 621,000 biallelic SNPs (25, 26) (Fig. 1 B and C and SI Appendix, Fig. S1). The first principal component (PC1), which explains 2.11% of the genotypic variance, is well predicted by a linear model with latitude, longitude, and linguistic affiliation variables (R2 = 0.86; P < 1.0 × 10−16 ) (SI Appendix, Fig. S2A). On one extreme end of the PC1 axis are North African Mozabite (Algeria) individuals, and on the other end of the axis are Mbuti ERHG (Democratic Republic of Congo) and San (southern Africa) individuals (Fig. 1B). We also observed a good fit between PC2, which explains 0.91% of the genotypic variance, and a linear model with latitude, longitude, and linguistic affiliation variables (adjusted R2 = 0.58, P < 1.0 × 10−16), albeit less strongly than PC1 (SI Appendix, Fig. S2B). Individuals speaking NC languages are represented at one end of the PC2 axis and San individuals at the other end. Thus, geography and language are significantly correlated with patterns of genetic variation in Africa.
To explore our hypothesis of a possible common ancestry of the Hadza, Sandawe, Sabue, and Dahalo, heretofore referred to as the eastern HG (EHG), we tested whether they cluster more closely to each other in the PCA compared with other populations. We observed that they cluster significantly closer to each other than to any other populations on PC1 and PC2 based on a comparison of Euclidean distances among EHG and among EHG and non-EHG individuals (Wilcoxon rank-sum test: W = 52,704,648; P < 1.0 × 10−16) (SI Appendix, Fig. S3A). In addition, the Sabue, Hadza, and Dinka individuals significantly cluster together at one extreme of PC3 (Wilcoxon rank-sum test, W = 30,247.5; P < 1.0 × 10−16) (Fig. 1C and SI Appendix, Fig. S3B). These observations are consistent with possible shared ancestry between the Hadza and Sabue, and some evidence for shared ancestry of these populations with the Dinka (NS language) (27), as well as linguistic evidence supporting a relationship between the Shabo language and proto-NS (13). PCs explaining a smaller proportion of the genetic variance in the data are presented in SI Appendix, Fig. S1.
We explored patterns of population structure in African population samples using STRUCTURE analysis (28) (Fig. 1D) with a set of 20,000 SNPs, pruned to reduce linkage disequilibrium (LD). We also used haplotype clusters inferred by BEAGLE (29) as a k-allele system at the same 20,000 loci (SI Appendix, Fig. S4). We found that K = 9 was the number of ancestral allele clusters (AAC) that consistently produced the highest data likelihoods across runs for both genotypes (SI Appendix, Fig. S5A) and haplotypes (SI Appendix, Fig. S5B) without producing multiple modes (i.e., inferring different ancestral allele clusters across runs). Additionally, we observed lower variance in likelihood scores at K = 9 compared with higher values of K. At K = 9 (Fig. 1D), we find that individuals from populations that speak languages belonging to the same language family have significantly similar AAC proportions (Mantel test: M = 0.473; P = 0.001) (SI Appendix). However, several population samples are distinguished by unique AACs at K = 9. These include the North African Mozabite (Fig. 1D, dark blue) who have the greatest proportion of Saharan ancestry compared with other populations (Wilcoxon rank-sum test: W = 191; P < 1.0 × 10−16). The other AACs at K = 9 distinguish HG populations: San (Fig. 1D, light green), WRHG and ERHG (Fig. 1D, dark green), Hadza (Fig. 1D, yellow), Dahalo (Fig. 1D, pink), and Sabue (Fig. 1D, light blue) populations, respectively. Distinct AACs corresponding to HG populations may be explained by genetic drift caused by isolation of these populations or persistently small effective population sizes (Ne) (30). Unlike other EHG populations, the Sandawe are not enriched for a particular AAC at K = 9; rather, they have considerable AA (Fig. 1D, dark purple) and NC (Fig. 1D, orange) ancestry: 37.1% and 26.4% on average, respectively. In contrast, the Elmolo, Yaaku, Boni, Wata, Ogiek, and Sengwer from East Africa share ancestry with neighboring agriculturalist or pastoralist populations. Patterns of clustering at lower AACs, which support ancient common ancestry between the rain forest HG and San and between the Hadza and Sabue, as well as the genetic relationship between AACs based on the inferred ancestral allele frequencies, are described in the SI Appendix.
Historical changes in population size contributes to contemporary genetic variation; therefore, we used patterns of LD decay to estimate Ne in population samples with at least 10 individuals (SI Appendix, Figs. S6 and S7A) (31). Several of the EHG, the Hadza, Dahalo, and Sabue, have relatively low estimates of Ne (∼9,000–11,000), consistent with their relatively smaller census population sizes (∼1,000–3,000) (32). In contrast, the Sandawe and WRHG have maintained relatively higher Ne (on the order of 17,000 and 19,000, respectively), consistent with their larger census sizes (∼30K) (33, 34). The estimates of Ne from LD in the Hadza, WRHG, and Sandawe are consistent with estimates of Ne based on levels of genetic diversity from whole-genome sequence data in the same populations (35). The largest Ne estimates are for agriculturalist and pastoralist populations, which is also consistent with prior studies (35, 36).
To examine the influence of demographic history on patterns of haplotype sharing within populations, we examined sharing of identity-by-descent (IBD) regions, which are stretches of DNA between individuals inherited from a common ancestor, and runs of homozygosity (ROH), which are stretches of DNA that are identical between the two haploid chromosomes of an individual. For each population, we calculated the average of the total IBD between all pairs of individuals, which we refer to as cumulative IBD (cIBD) and cumulative ROH (cROH) within individuals. Comparing cROH with cIBD for each population (SI Appendix, Fig. S7B), we find that the Hadandawa-Beja have the greatest cROH (195 cM) but are only the 15th highest for cIBD (45 cM); the high cROH is consistent with the documented practice of consanguineous marriages in this population (37). In contrast, the Hadza have the greatest cIBD (398 cM) as well as the second greatest cROH (158 cM). The presence of elevated cIBD and cROH in the Hadza is consistent with a small census size (∼1,000), a low Ne and long-term endogamy (24, 35).
Historical Relationships Among African Populations.
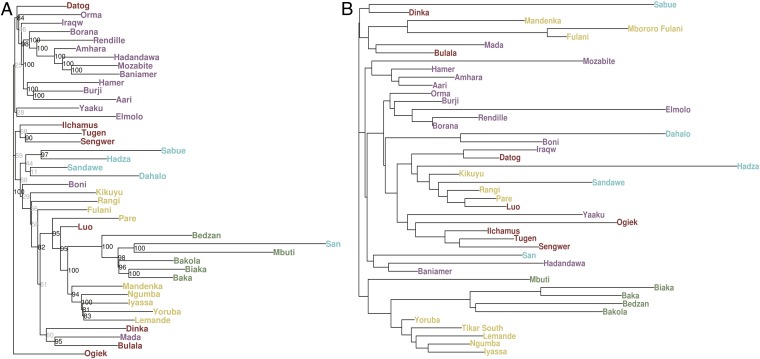
We reconstructed a population tree using pairwise estimates of genetic distance based on the FST statistic (Fig. 2A). We assessed statistical support for internal nodes in the neighbor-joining (NJ) population tree by bootstrapping loci with 1,000 replicates (SI Appendix). Broadly, the tree reflects geographic residence and linguistic affiliation as observed in the previous results. In addition, four geographically dispersed EHG populations—the Hadza, Sabue, Sandawe, and Dahalo—form a clade. The Hadza form a subclade with the Sabue with 97% bootstrap support. Support for inclusion of the Dahalo and Sandawe in the EHG clade is lower; however, examination of the bootstraps shows that this is because >80% of the replicates show the Dahalo cluster with the Boni, a neighboring population with whom they share recent contact (see IBD results below). As noted above, the linguistic relationships among these populations are unclear and contentious. While evidence for recent common ancestry between the neighboring Hadza and Sandawe has previously been shown (17, 24, 27), our results represent genetic evidence for a uniquely shared common ancestry of these populations with the Dahalo and Sabue from Kenya and Ethiopia, respectively. It is also noteworthy that other HG populations from central and southern Africa cluster with high bootstrap support: the San and Mbuti form a clade despite being geographically isolated from each other, and both form a clade with the WRHG, supporting results from PCA (Fig. 1 B and C) and STRUCTURE analyses (Fig. 1D) (18, 36, 38, 39). In contrast, other HG populations from East Africa (i.e., Ogiek, Dorobo, and so forth) cluster together with neighboring agriculturalist or pastoralist populations.
Fig. 2.
Population trees. (A) An NJ population tree was inferred using estimates of pairwise genetic distances between populations based on FST values scaled by Ne. Populations largely cluster by geography or language affiliation, with the notable exceptions of the clade consisting of the Hadza, Sabue, Sandawe, and Dahalo and the clade consisting of the WRHG, ERHG, and San, whose populations cluster together despite being geographically distant. (B) An NJ population tree based on pairwise distances based on the ratio of within-population to between-population haplotype sharing (i.e., IBD); this statistic is more sensitive to recent demographic events, such as gene flow than FST. The EHG cluster most closely with neighboring populations.
In addition to using FST to infer relationships between populations, we examined the distribution of the number and length of IBD tracts between individuals across populations to identify recent shared ancestry (40, 41). We explored the possibility that the signal of EHG common ancestry represents shared gene flow with Cushitic- and Bantu-speaking populations who expanded into East Africa within the past 5 kya (42). We used a distance measure, based on the ratio of IBD tracts (≥2 cM) found within and between populations, to construct a population tree (Fig. 2B). We compared FST- and IBD-based distances between populations (SI Appendix), as the latter measure is more sensitive to recent gene flow originating 25–50 generations ago (43). Unlike the FST-based tree, the EHG populations do not form a clade in the IBD-based tree, instead clustering with geographically proximate populations (Fig. 2B), indicating an increase in interactions between the EHG and neighboring agriculturalist and pastoralist populations in the recent past (18). Notably, the Boni and Dahalo, who neighbor each other, cluster together on the IBD tree, which explains the frequency with which they appear together in bootstrap replicates of the FST-based tree. Furthermore, in the IBD tree, the Dinka form a clade with the Sabue, consistent with the genotypic PCA (Fig. 1C), suggesting recent gene flow and/or shared ancestry. These observations are consistent with a model suggested by some linguists in which the Shabo language is classified with the NS language family (13), although other linguists (14) find inadequate evidence for a connection between the Shabo language and proto-NS. This uncertainty could be due to the age of the linguistic relationship between Shabo and proto-NS, which may predate the upper bound for linguistic reconstruction (∼10 kya) (44).
The lack of clustering of the EHG in the IBD-based distance tree indicates that the pattern of shared ancestry among the EHG populations observed in the reconstructed FST -based population tree (Fig. 2A) is not due to recent events. We used an approach introduced by Fearnhead and Prangle (45) to construct summary statistics for approximate Bayesian computation (ABC) inference based not only on allele frequency differences between populations, but also on patterns of LD and admixture LD (i.e., LD weighted by differences in allele frequencies between populations) to infer divergence times between pairs of EHG populations (31, 46–51) (SI Appendix). The demographic model we employed (SI Appendix, Fig. S8) included changes in Ne, gene flow from populations speaking NC, NS, or AA languages, and ascertainment bias due to SNPs on the Illumina 1M array that were identified primarily in non-African populations.
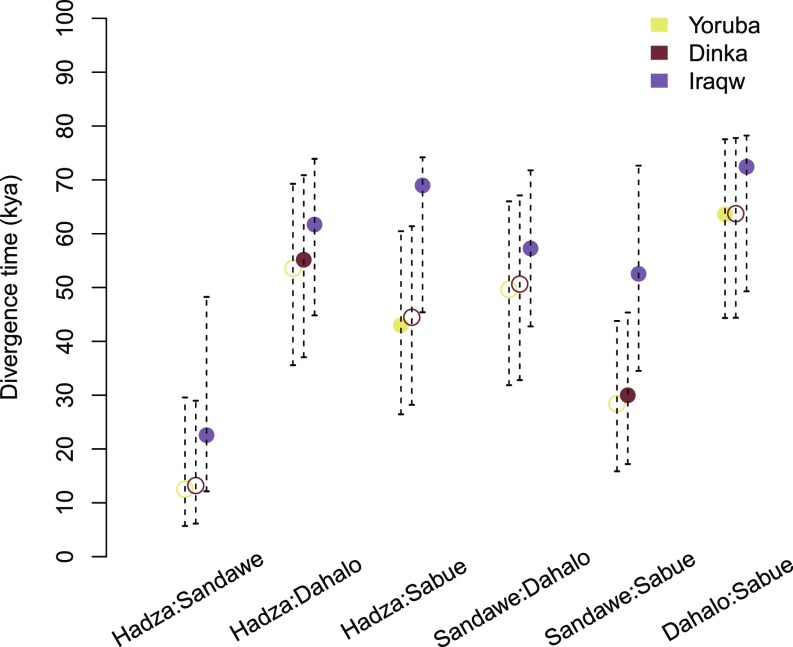
The maximum a posteriori estimate and 95% credible interval for pairwise divergence time estimates are shown in Fig. 3. The maximum a posteriori divergence time estimates for the Hadza and Sandawe were 13 or 22 kya when accounting for different primary sources of admixture based on STRUCTURE analysis (Fig. 1D) (NC or AA, respectively); these estimates overlap with previous studies (17). The Hadza split times with other populations were older; the divergence time estimates with the Sabue (NC or AA gene flow) were 44 or 61 kya, respectively, and with the Dahalo (NC or AA gene flow) were 55 or 61 kya, respectively. Sandawe population divergence time estimates with the Sabue (NS or AA admixture) were 30 or 52 kya, respectively, and with the Dahalo (NS or AA gene flow) were 50 or 57 kya, respectively. The estimated times of divergence of the Sabue and Dahalo (NC or AA gene flow) were 63 or 72 kya, respectively. These results are consistent with a model in which population divergence between the Dahalo and Sabue and the ancestors of the Sandawe and Hadza occurred >30 kya, whereas the Hadza and Sandawe divergence was more recent. Whole-genome sequence analyses will be informative for more accurately resolving the time of population divergence among EHGs.
Fig. 3.
Divergence time estimates. The maximum a posteriori estimates and 95% credible intervals for pairwise divergence time estimates are displayed for each set of population samples. Estimates incorporated shared gene flow with the Yoruba, Iraqw, and Dinka, representing NC, AA, and NS source populations, and are color-coded as yellow, purple, and red, respectively. The closed circles represent population combinations for which we believe the included source population likely contributed migrants to either HG population in the past.
Genome-Wide Patterns of Adaptation.
Over the past two decades, several genome-wide scans for selection have been developed and applied to worldwide human genetic data (52–54). Fewer studies, however, have focused on variation within Africa (24, 35, 36, 38, 55–57). These studies have tended to focus on specific regions (e.g., Southern Africa or Ethiopia) in Sub-Saharan Africa (24, 36, 55) or specific populations of interest living in Africa (35, 38, 57). Thus, the pattern and distribution of adaptive candidate loci among geographically and culturally diverse African populations is not well understood. We used three complementary statistical tests of neutrality to characterize African genome-wide signatures of adaptation. We combined individuals into larger population groupings based on shared ethno-linguistic affiliation and on shared ancestry, as inferred from PCA clustering (SI Appendix, Fig. S9) for the subsequent analyses.
Shared Adaptive Signals.
Given the wide range of diverse populations sampled in the study, we were interested in studying the distribution of adaptive candidate genes within and among population groupings. We first employed the D statistic, an extension of the locus-specific branch length statistic that includes more than three population samples (58, 59), to identify signatures of regionally restricted adaptation within population groupings. We identified genes near (within 100 kb) SNPs in the top 0.1% of the empirical distribution of results for the D statistic test (expected to be enriched for targets of natural selection) for each population grouping, and we performed pathway-enrichment analyses of these adaptive candidate genes (SI Appendix, Table S3). Because the D statistic identifies SNPs with allele frequencies that are unusual in one sample relative to all others in the analysis (58), it was not surprising that the majority of top (0.1%) candidate genes (93%) occur in only a single population grouping (SI Appendix, Fig. S10 and Dataset S1).
We next employed the integrated haplotype score [iHS; a within-population statistic (54)] to identify relatively recent signatures of selective sweeps within population groupings based on extended haplotype homozygosity, and a cross-population composite likelihood ratio test [XP-CLR; a between-population statistic using the NC-west grouping as the reference population (60)] to identify older signatures of adaptation and signatures of selection from standing variation (genes near SNPs in the top 0.1% of the empirical distributions are shown in Datasets S2 and S3 for iHS and XP-CLR, respectively). When we looked at the degree to which top iHS and XP-CLR candidate genes were shared across population groupings, we found that the majority occur in only a single grouping (57% and 66%, respectively) (SI Appendix, Figs. S11 and S12), and this prevalence of population-specific signatures is significantly more than would expected to occur by chance (bootstrap P < 1e-06).
As expected, we identified the MCM6 locus upstream of lactase (LCT), which contains SNPs associated with regulating lactase gene expression (61, 62) in the top 0.1% of candidate loci identified by all three tests in several pastoral population groupings: Eastern-Cushitic, Beja, Datog, Southern-Nilotic, and Fulani (as well as in populations that have experienced recent gene flow with pastoralists). This result supports previous work demonstrating the MCM6 region to have one of the strongest signals of adaptation in East African pastoralists (62, 63), and validates the sensitivity of our chosen methods for detecting adaptation.
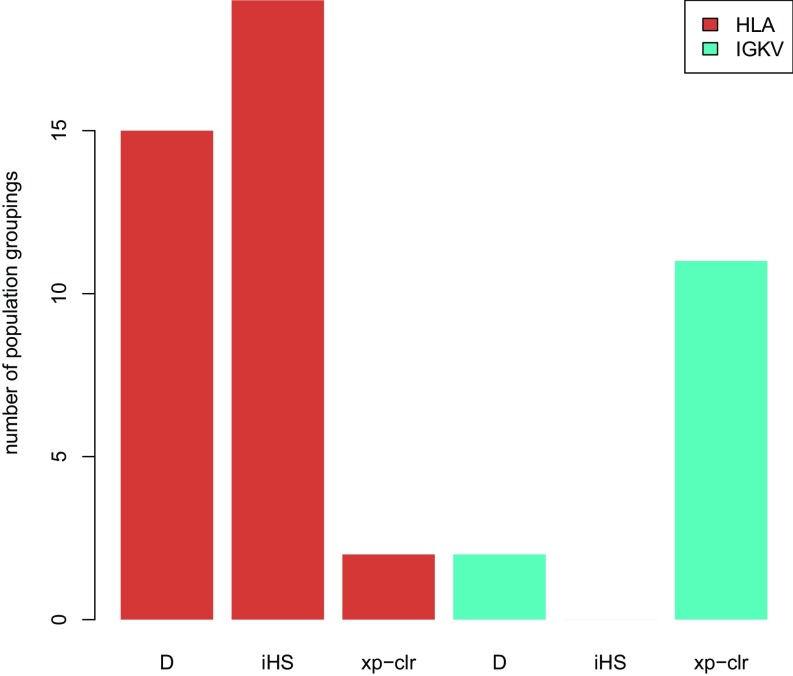
In addition, we identified a number of immune-related candidate loci that show shared signatures of selection in several population groupings. Seven of the 52 candidate loci identified using iHS candidate genes that are present in many population groupings (≥10) belong to the histocompatibility complex (HLA) gene family, which is known to be critical to immune function (64), and 14 of the 32 XP-CLR candidate genes that are present in at least 10 population groupings belong to the Igκ chain variable (IGKV) gene cluster, which is known to have been subjected to positive selection in humans (65) (Fig. 4). Because many of the adaptive candidate genes across the population groupings in the study are involved in immune function, we more formally tested for enrichment of gene ontology (GO) immune system process terms (GO:0002376) (66). We found significant enrichment in all three sets of results (Methods): XP-CLR (P < 10e-05), iHS (P < 10e-05), and D (P < 10e-05).
Fig. 4.
Signatures of selection shared among population groupings. These shared signals among population groupings include candidate loci in the top 0.01% of the empirical distribution of each neutrality test statistic (D, iHS, XP-CLR, respectively). The HLA and IGKV gene families are displayed along the x axis for each neutrality test. The y axis displays the number of population groupings that share signatures of selection at these loci.
Given the significant enrichment of GO immune system process genes in each set (XP-CLR, iHS, D) of adaptive candidate loci pooled across population groupings, we were interested in testing whether particular environmental variables have impacted the degree to which immune function genes are overrepresented in adaptive candidate genes among population groupings. Because our study includes populations living in diverse environments with a range of malaria endemicities and practicing a wide range of subsistence strategies, we tested whether these two variables were associated with immune function enrichment (Methods). We found that the degree to which adaptive candidate genes identified with iHS (which is sensitive to the most recent signals of adaptation relative to the D statistic and XP-CLR) are enriched for immune function genes is significantly correlated with both subsistence and malaria endemicity (R2 = 0.59, P = 0.021) (Methods). This result is also significant for adaptive candidate genes identified with the D statistic (R2 = 0.52, P = 0.038), but is not significant for adaptive candidate genes identified with XP-CLR (R2 = 0.29, P = 0.42). One possible explanation for the lack of XP-CLR significance is that this test is more sensitive to older adaptive signatures (60) that may predate the emergence of malaria as a strong selective pressure in Africa.
Adaptive Signals Present Within Population Groupings.
Given the extent of population-specific signals of adaptation in the data, we explored the genes near (within 100 kb) SNPs in the extreme tails (top 100 loci) of the population-specific results in more detail (Dataset S1). As noted above, many of the strongest signals of population-specific adaptation are involved in immune function (Table 1). These include genes involved in innate and adaptive immune function, which have been shown to be important in resistance to malaria and other infectious diseases (67–75). More specifically, we have identified genes involved in the production and regulation of B and T cells (76–80), genes involved in resistance to malaria and viral infections (including HIV-1) (81–85), genes involved in resistance to bacterial infection (86, 87), and genes involved in inflammatory response (88, 89). We additionally observed significant pathway enrichment of inflammation mediated by chemokine and cytokine signaling in the El Molo population grouping (SI Appendix, Table S3).
Table 1.
Signatures of adaptation within population groupings
| Biological role and locus | Population grouping |
| Innate and adaptive immune function | |
| MYLK | Fulani |
| TRAF3 | Amhara |
| IL6 | Bulala |
| TRAF3IP2 | Hadza |
| RAG2 | Niger-Congo–east |
| NFX1 | Eastern-Cushitic |
| IL2RA | Sandawe |
| LGALS3 | Elmolo |
| NCAM1 | Bulala |
| MAVS | Eastern-Cushitic |
| GAB2 | Niger-Congo–east |
| ISCU | Dinka |
| ICAM1 | Bulala |
| CD46 | Sabue |
| FCGR3A | Southern-Nilotic |
| FCGR2B | Southern-Nilotic |
| IFNGR1 | Eastern-Cushitic |
| COLEC11 | Ogiek |
| ORM1 | Sabue |
| TFCP2 | Ogiek |
| Digestion and metabolism | |
| SLC2A10 | Boni |
| PPARGC1A | Iraqw |
| IDE | Luo |
| PSMB9 | Mada |
| ALMS1 | Niger-Congo–west |
| FBP1 | Amhara |
| LDHB | Iraqw |
| PNPLA2 | Yaaku |
| LPIN2 | Hadza |
| PLTP | Southern Nilotic |
These include genes within 100 kb of candidate loci in the most extreme 100 D test statistic results.
In addition, we observed candidate loci that may play a role in adaptation to diverse diets and climates (Datasets S1–S3). For example, the D and the XP-CLR statistics identified loci near CISH and DOCK3 on chromosome 3, which are highly differentiated in WRHG and were previously identified as targets of selection and associated with stature in the same population, thought to be an adaptation to a tropical environment (38). The D and XP-CLR statistics identified a cluster of taste receptor loci on chromosome 12, and XP-CLR identified the amylase gene cluster, which plays a role in starch digestion (90), as targets of selection in the WRHG. Additionally, several of the strongest candidates for selection we identified encode proteins involved in insulin resistance (91–95), hypoglycemia (96), lactate dehydrogenase B deficiency (97), as well as lipid metabolism, transfer, and storage (98–100) (Table 1). Additionally, we observed significant enrichment of the cholesterol biosynthesis pathway in the southern-Nilotic population grouping living in Kenya, who are predominantly pastoralists (SI Appendix, Table S3).
Discussion
In this study we have characterized genomic variation in sub-Saharan populations representing a breadth of cultural and geographic diversity. The results of the study support the influence of geographic proximity, as well as cultural affiliation (e.g., language and subsistence patterns), in defining the complex relationships among populations. In particular, the Hadza, Sandawe, Dahalo, and Sabue live relatively far apart from each other in Tanzania, Kenya, and Ethiopia; however, we show that there is a closer genetic relationship among these populations than would be expected based on their geographic residences alone. They all either currently, or until very recently, have employed hunting and gathering as a primary subsistence strategy, and three of the languages spoken by these populations contain click consonants. Our results indicate that these HG populations, like the San and rain forest HG, are not impoverished agriculturalists or pastoralists who have lost their land or livestock; instead, they likely have remained relatively isolated for an extended period of time and have only come into contact with other populations in the more recent past. On the other hand, other East African populations who practice an HG lifestyle and speak AA or NS languages, appear to be genetically similar to neighboring non-HG populations. This could either be due to the loss of domestication or may reflect older ancestral subsistence patterns (20).
These relationships are consistent with a demographic history in which structure among EHG populations emerged before the Last Glacial Maximum (∼21 kya). This period has been identified as one of increased aridity and reduced temperatures in East Africa; these climatic conditions were accompanied by shifts in vegetation, particularly reduced forest coverage (101, 102), and these environmental changes are thought to have triggered human dispersals into environmental refugia (103). Thus, we have uncovered a connection among geographically disparate HG populations in East Africa, consistent with a broad geographic distribution of their ancestors in the late Pleistocene before 30 kya.
Our analysis of signals of positive selection in geographically and ethnically diverse African population samples highlights the degree to which recent, regionally restricted positive selection has shaped patterns of variation in contemporary Africans. We have identified candidate loci that may be targets of natural selection; future in vitro and in vivo studies will be necessary to determine functional impact. We found that the majority of genes near SNPs showing the strongest signals of positive selection occurred in only one of the population groupings included in the analysis. This result is consistent with previous work that found a minority of overlapping signals of adaptation across continental groups (44–12%) (104). Because of the ascertainment strategy used for the Illumina 1M-Duo SNP array—common variants were prioritized, and these variants tend to be older (arose before the out of Africa migration) and may exclude population-specific SNPs—future studies based on high coverage whole-genome sequencing are likely to uncover additional loci that play a role in adaptation to diverse diets, climates, and infectious diseases across sub-Saharan Africa. Given our results, we argue that the common practice of using only one or a handful of population samples to represent an entire continent is inadequate, and this is especially true for sub-Saharan Africa, which harbors the largest proportion of human genetic variation relative to other regions across the world.
Our study includes populations living in highly diverse environments, with variable pathogen exposure, and practicing a wide range of subsistence strategies. Therefore, we were able to explore whether this diversity has had an impact on the ways in which adaptation has shaped variation in Africa. The loci that were identified as putative targets of selection are significantly enriched for genes that play a role in immune function. It is not especially surprising that loci that play a role in response to infectious disease have had such a large impact on variation among African genomes, given that infectious disease mortality is one of the strongest selective pressures identified in contemporary populations (105). Additionally, we identified candidate adaptive loci that play a role in cholesterol and glucose metabolism, taste perception, and starch digestion, many of which are specific to population groupings. Loci that may be adaptive in indigenous environments could be associated with disease in urban environments (106); therefore, it is critical to include diverse populations in studies of human adaptation, especially when the results have implications for human health and disease.
Conclusion
Human demographic history in Africa involves a complex tapestry of population movements, admixture, and adaptations to diverse environments that have shaped the genomic landscape of Africa. We have used patterns of genomic variation to investigate the demographic history of HG populations living in East Africa, demonstrating ancient common ancestry. Changes in environment and subsistence within Africa have resulted in novel and distinct selective pressures. While these biological pressures appear consistent across African populations, the specific genetic regions affected by selective pressures often vary across populations. These combined results demonstrate the importance of including ethnically diverse sub-Saharan African populations in human genetic studies to improve our understanding of complex population histories. Finally, these data demonstrate the critical importance of including African populations in biomedical studies to best encompass the full range of human diversity.
Methods
Sample Acquisition and Genotyping.
Institutional Review Board approval for this project was obtained from the University of Maryland at College Park and the University of Pennsylvania. Written informed consent was obtained from all participants and research/ethics approval and permits were obtained from the following institutions before sample collection: COSTECH (the Tanzania Commission for Science and Technology) and the National Institute of Medical Research in Dar es Salaam, Tanzania; the Kenya Medical Research Institute in Nairobi, Kenya; the University of Khartoum in Sudan; the Nigerian Institute for Research and Pharmacological Development, Abuja, Nigeria; the Ministry of Health and National Committee of Ethics, Cameroon; the University of Addis Ababa and the Federal Democratic Republic of Ethiopia Ministry of Science and Technology National Health Research Ethics Review Committee. A total of 816 samples were genotyped on the Illumina 1M-Duo Bead Array SNP chip. We removed individuals with <95% successfully genotyped SNPs. We also removed related individuals as inferred by PLINK ( > 0.25). A total of 697 individuals passed these filters; this sample was then merged with data from Li et al. (23) and Henn et al. (107), resulting in 840 individuals with genotypes available at ∼621,000 SNPs used for further analyses.
Principal Components Analysis.
The smartpca program provided in EIGENSOFT 4.2 was used to calculate principal components of the sample genotype matrix of all 840 individuals at all SNPS; to account for LD, the regress option of smartpca was utilized.
Bayesian Clustering.
For analysis with STRUCTURE, the full complement of SNPs was pruned to a smaller set of 20,000 SNPs using PLINK with the goal of minimizing LD. The model was run at K values from two through nine; each chain was run 10 times. Results from different runs were aligned using the CLUMPP software; the modal configuration for ancestry was identified visually and presented using the DISTRUCT software. Haplotypes were phased using the algorithm implemented in the BEAGLE 3.3.2 software suite (29). We inferred phase and haplotype clusters using all SNPs and then reran the k-allele STRUCTURE analysis with haplotype clusters at the same sites as with the biallelic STRUCTURE analysis.
LD Decay and Ne.
LD decay was calculated by sampling pairs of SNPs within 20 kbp of each other and calculating the genotypic correlation, which approximates r2. SNPs were placed into bins based on their distance: SNPs 0–1 kbp apart were placed into one bin; SNPs 1–2 kbp were placed into another bin, and so forth. The r2 values of pairs of SNPs within bins were then averaged to obtain E[r2]. The relationship between E[r2] and Ne derived by Tenesa et al. (46) was used to estimate Ne via nonlinear least squares.
Population Tree.
The FST statistic as defined by Weir (108) was implemented in R and calculated for population samples with ≥10 individuals using the same set of 20,000 SNPs used for STRUCTURE analysis. The NJ algorithm was used to estimate a population tree from pairwise distances between populations. The pairwise distance employed between populations i and j was defined as follows: ; here, is the harmonic mean of the Ne estimates for populations i and j, which helps mitigate the potential for long-branch attraction due to bottlenecks or population expansions and concomitant changes in allele frequencies. The bootstrap support of the tree was estimate by resampling SNPs as well as individuals.
Identity-By-Descent.
Haplotypes were phased using the algorithm implemented in the BEAGLE 3.3.2 software suite. To infer IBD tracts between pairs of individuals, we used the GERMLINE v2.2 software. The lengths and number of IBD tracts between pairs of individuals were used to calculate a distance based on the model of Huff et al. (109). A statistic FIBD, analogous to FST, was calculated between populations by averaging the IBD-based distances between pairs of individuals within populations and between populations. The NJ algorithm was used to reconstruct a population tree from the IBD-based distance matrix.
Inference of Divergence Time.
We employed the ABC approach to infer the time of divergence between EHG populations; specifically, we used rejection sampling with local linear regression adjustment (110). For simulations, we utilized a realistic demographic model representative of four contemporary populations: two EHG populations, an agriculturalist or pastoralist (A/P) population, and a non-African population. The parameters of the demographic history of the A/P population and the non-African population were based on previous results (111, 112). The unknown parameters in this model included not only time of divergence, but also gene flow rates from the A/P population to the two simulated EHG populations as well as EHG Ne, the population size of the population ancestral to the EHG, and finally the population size of the simulated ancestral African population. Each of these parameters was sampled from our prior distributions. Gene flow from the population representing the A/P was introduced into the EHG populations 100–200 generations in the past, approximately the time populations in Neolithic populations in African began expanding (8, 10). For the EHG population, likely sources of gene flow from A/P population (i.e., representative NC, AA, or NS populations) were identified from STRUCTURE results. We also fixed parameters regarding the evolution of Ne in the A/P and non-African population. In addition, the A/P population diverged from other populations 4,500 generations in the past and the non-African population diverged 3,500 generations in the past. To simulate the effect of SNP ascertainment bias, only SNPs with a frequency >5% in the non-African population were retained from the simulated African populations (113–115). In addition, we accounted for the ascertainment bias introduced by choosing tag SNPs: we removed SNPs from analysis if they were in high LD (r2 > 0.70) with other SNPs in the simulated non-African population. The demographic model was simulated using the coalescent framework (116); for each simulation (10,000 replicates), a total of 200 regions, 50 kbp in length, were generated. The mutation rate was fixed for each region (1.1 × 10−8 mutations per base pair per generation). The recombination rate was allowed to vary; we matched the average local recombination rates in 200 randomly selected 50-kbp regions in the deCode recombination map (117).
We constructed summary statistics using the approach of Fearnhead and Prangle (45). We proposed an initial set of summary statistics S(Xsim) based on the f2 distances between the EHG and between each and A/P population, LD decay, and admixture LD decay (SI Appendix) (46, 50, 51, 118–120). We simulated these summary statistics in a pilot stage of 10,000 simulations. We estimated the functional relationship between each of the seven parameters and corresponding summary statistics: that is, θp ∼ ĝp(S(Xsim)) using gradient boosting machines, an ensemble method that constructs a functional approximation by iteratively combining regression trees while minimizing the squared error with respect to the target function (121–125). We then ran a second stage of simulations (10,000 replicates); summary statistics were transformed using the functional approximations obtained by gradient boosting machines in the pilot stage, ĝp(S(Xobs)). We used ABC with local linear regression adjustment to draw samples from the posterior distribution f(θ | ĝp(S(Xobs))).
Selection Scan Population Groupings.
When we grouped two or more population samples, we used shared ethno-linguistic affiliations among the included population samples to refer to these population groupings in the text (SI Appendix, Table S2). In particular, we grouped the Gabra, Gurreh, and Rendille into an eastern-Cushitic population grouping; the Baniamer and Hadandawa into a Beja population grouping; the Cameroon Fulani, Nigeria Fulani, and Mbororo Fulani into a Fulani population grouping; the Lemande, Ngumba, southern Tikar, and Yoruba into an NC-west population grouping; the Pare, Taita, and Taveta into an NC-east population grouping; the Pokot and Sengwer into a southern-Nilotic population grouping; and the Aari and Hamer into an Omotic population grouping. Because the Baka, Bakola, and Bedzan are thought to have adopted languages that belong to the NC language family, we refer to them as the WRHG grouping in place of linguistic affiliation. All other ethno-linguistic populations are referred to individually.
Genome-Wide Tests of Neutrality.
We utilized three complementary statistics for identifying regions of the genome that deviate from neutral expectations: the D statistic (58), XP-CLR (60), and the iHS (54). The D statistic leverages information across all of the included population groupings so that the SNPs with the most extreme values will have allele frequencies that are distinct only in our reference population sample; this method therefore, is designed to identify regions of the genome that are highly differentiated in one population sample. XP-CLR leverages pairwise populations sample comparisons to identify regions of the genome that contain highly differentiated regions of LD. XP-CLR has also been shown to be sensitive to selection from standing variation. The iHS complements the other strategies by identifying regions that contain extended haplotype homozygosity within a given population, a classic signature of selective sweeps.
Following Akey and colleagues (58, 59), we calculated pairwise FST among all of the 23 population groupings using the method described in Weir (108). We then calculated the D statistic, and identified the top 0.1% of SNPs and genes100 kb up and downstream as our candidate regions (to account for regulatory SNPs that are typically within 100 kb of genes which they regulate; we refer to these as “candidate genes”) (Dataset S1).
We employed the (iHS) test of neutrality within each of the 23 groupings as described previously (38). Briefly, we used the software package BEAGLE v3.3.2 to infer phase (29), and we generated a fine-scale recombination map relevant to the African populations with LDhat v2.1 (126). Individuals used to generate the recombination map were 100 unrelated samples, 25 males and 25 females, each from two populations in HapMap3 Release 2: the Yoruba from Ibadan, Nigeria (YRI) and the Luhya from Webuye, Kenya (LWK) (127). We estimated a genetic map in Morgan units of r from ρ = 4Ner units using an Ne of 15,700, consistent with the estimation in Myers et al. (128). We used genome-wide sequence data from several nonhuman primates (chimpanzee, orangutan, and rhesus macaque) downloaded from the University of California, Santa Cruz Genome Browser website (129) to establish the ancestral allele for each of the SNPs included in our iHS analysis. Approximately 5% of the SNPs in our data could not be assigned an unambiguous ancestral state and were removed before our iHS analysis. In addition, SNPs with minor allele frequencies less than 5% in population samples were removed from the phased dataset used in the iHS analysis, in agreement with other publications (e.g., ref. 54). Finally, we removed SNPs containing missing data. The unstandardized scores returned by the iHS binary executable were adjusted such that all scores had zero means and unit variances with respect to SNPs with similar derived allele frequencies (for iHS, as described in ref. 54). We considered all of the results (for iHS we took the absolute values) in the top 0.1% of the distribution to be the top candidates (Dataset S2).
We additionally performed XP-CLR because it has been shown to be robust to ascertainment bias and because it has been shown to be sensitive to detecting selection from standing variation (60). Using the recombination map described above, we ran the XP-CLR software package (60) with 0.005-cM sliding windows and a between-window distance of 5 kb. Previous work has shown that many if not all of the included population groupings, have experienced recent gene flow resulting from the Neolithic expansion of peoples, technologies, and Bantu languages, often referred to as the Bantu expansion (8, 18, 130). Therefore, we wanted to minimize the effects of this gene flow on the XP-CLR results. Thus, in this analysis we used the NC-west grouping as our comparison population for each of the other population groupings to highlight regions of the genome that are unusually structured between the NC-west agriculturalists and the other diverse population groupings included in the study. We considered all of the results in the top 0.1% of the distribution to be the top candidates (Dataset S3).
Pathway Enrichment.
We tested for significantly overrepresented Panther biological pathways (131) in our top candidate regions for each of the three genome-wide tests of neutrality. For each genome-wide scan of selection we generated a list of genes [annotated with Biomart (132)] within 100 kb of a top 0.1% SNP and tested whether this list contained more Panther pathway genes that would be expected by chance using a χ2 test. We corrected the pathway results for multiple testing with a Bonferroni correction. We used a range of 100 kb because we were interested in retaining potential cis-regulatory variants in our analysis.
Bootstrap Analysis.
To test the null hypothesis that the number of candidate genes that are present among population groupings could be explained just by chance, we randomly sampled 1,000 SNPs from our empirical data for 24 population groupings, including all genes 100 kb up and downstream of the SNPs, and then quantified the overlap among population groupings. We assessed the significance of this result with 1,000,000 bootstraps and found that all bootstrap runs resulted in a minority (<34%) of candidate genes occurring in a single population grouping (P < 1e-06).
Enrichment of GO Immune Function Genes.
Given the prevalence of immune-related genes in our top adaptive candidate genes, we more formally tested whether this enrichment was statistically significant. We used the set of immune system process terms (GO:0002376) defined by the GO website (66), and tested for overrepresentation in each set of unique candidate genes identified with a given statistic (XP-CLR, iHS, D) across population groupings. We assessed statistical significance with 1,000,000 bootstrap runs, all of which resulted in lower levels of enrichment than our empirical results for candidate genes identified with each of the three statistics (XP-CLR, iHS, D) (P < 1e-06).
We were also interested in any variability in the degree to which sets of candidate genes were enriched for particular biological processes among population groupings. For this analysis we tested whether environmental variables (malaria endemicity and subsistence strategy) had any impact on the degree to which a given set of adaptive candidate genes identified with a given statistic (XP-CLR, iHS, D) in a given population was enriched for GO immune system genes. We used linear modeling with the χ2 measure of enrichment as our dependent variable and malaria endemicity (estimated from information available through the Malaria Atlas Project, https://map.ox.ac.uk/) (133) and subsistence strategy as our explanatory variables (χ2 ∼ malaria_endemicity + subsistence_strategy + malaria_endemicity × subsistence_strategy). Because the residuals of the linear model were not normally distributed, we bootstrapped both malaria endemicity and subsistence strategy 1,000 times to generate statistical significance.
Supplementary Material
Acknowledgments
We thank Joseph Lachance for helpful comments and discussion; and the African volunteers for samples. Genotyping services were provided by Hakon Hakonarsson of the Center for Applied Genomics at the Children’s Hospital of Philadelphia. This research was funded by National Science Foundation Grants BCS-0196183 and BCS-0827436 and National Institutes of Health Grants 8DP1ES022577, 5-R01-GM076637, 1R01DK104339, and 1R01GM113657 (to S.A.T.).
Footnotes
The authors declare no conflict of interest.
Data deposition: Genotype data from this study have been deposited in the NIH dbGAP repository, https://www.ncbi.nlm.nih.gov/gap (accession no. phs001780.v1.p1).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1817678116/-/DCSupplemental.
References
- 1.McDougall I, Brown FH, Fleagle JG. Stratigraphic placement and age of modern humans from Kibish, Ethiopia. Nature. 2005;433:733–736. doi: 10.1038/nature03258. [DOI] [PubMed] [Google Scholar]
- 2.McDermott F, et al. New Late-Pleistocene uranium–thorium and ESR dates for the Singa hominid (Sudan) J Hum Evol. 1996;31:507–516. [Google Scholar]
- 3.Hublin JJ, et al. New fossils from Jebel Irhoud, Morocco and the pan-African origin of Homo sapiens. Nature. 2017;546:289–292. doi: 10.1038/nature22336. [DOI] [PubMed] [Google Scholar]
- 4.Scheinfeldt LB, Soi S, Tishkoff SA. Colloquium paper: Working toward a synthesis of archaeological, linguistic, and genetic data for inferring African population history. Proc Natl Acad Sci USA. 2010;107:8931–8938. doi: 10.1073/pnas.1002563107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mcbrearty S, Brooks AS. The revolution that wasn’t: A new interpretation of the origin of modern human behavior. J Hum Evol. 2000;39:453–563. doi: 10.1006/jhev.2000.0435. [DOI] [PubMed] [Google Scholar]
- 6.Nurse D. The contributions of linguistics to the study of history in Africa. J Afr Hist. 1997;38:355–391. [Google Scholar]
- 7.Philipson D. The chronology of the Iron Age in Bantu Africa. J Afr Hist. 1975;16:321–342. [Google Scholar]
- 8.de Filippo C, et al. Y-chromosomal variation in sub-Saharan Africa: Insights into the history of Niger-Congo groups. Mol Biol Evol. 2011;28:1255–1269. doi: 10.1093/molbev/msq312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patin E, et al. Dispersals and genetic adaptation of Bantu-speaking populations in Africa and North America. Science. 2017;356:543–546. doi: 10.1126/science.aal1988. [DOI] [PubMed] [Google Scholar]
- 10.Bower J. The pastoral neolithic of East Africa. J World Prehist. 1991;5:49–82. [Google Scholar]
- 11.Ehret C. Language and history. In: Heine B, Nurse D, editors. African Languages: An Introduction. Cambridge Univ Press; Cambridge, UK: 2000. pp. 272–297. [Google Scholar]
- 12.Guldemann T, Stoneking M. A historical appraisal of clicks: A linguistic and genetic population perspective. Annu Rev Anthropol. 2008;37:93–109. [Google Scholar]
- 13.Blench R. Archaeology, Language, and the African Past. Altamira Press; Lanham, MD: 2006. [Google Scholar]
- 14.Ehret C. Proceedings of the Fifth Nilo-Saharan Linguistics Colloquium, Nice. Rudiger Koppe Verlag; Cologne, Germany: 1992. Do Krongo and Shabo belong in Nilo-Saharan; pp. 169–193. [Google Scholar]
- 15.Nurse D. Sugia: Sprache und Geschichte in Afrika. 1986. Reconstruction of Dahalo history through evidence from loanwords. (Rudiger Koppe Verlag, Cologne, Germany), Vol 7, pp 267–305. [Google Scholar]
- 16.Morris AG. The myth of the East African ‘Bushmen’. S Afr Archaeol Bull. 2003;58:85–90. [Google Scholar]
- 17.Tishkoff SA, et al. History of click-speaking populations of Africa inferred from mtDNA and Y chromosome genetic variation. Mol Biol Evol. 2007;24:2180–2195. doi: 10.1093/molbev/msm155. [DOI] [PubMed] [Google Scholar]
- 18.Tishkoff SA, et al. The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edwards A, Cavalli-Sforza L. Genetics Today. Proceedings, 11th International Congress of Genetics, The Hague. Pergamon Press; Oxford: 1963. Analysis of Human Evolution; pp. 923–933. [Google Scholar]
- 20.Stiles D. The hunter-gatherer ‘revisionist’debate. Anthropol Today. 1992;8:13–17. [Google Scholar]
- 21.Ambrose SH. Archaeology and linguistic reconstructions of history in East Africa. In: Ehret C, Posnansky M, editors. The Archaeological and Linguistic Reconstruction of African History. Univ of California Press; Berkeley, CA: 1982. pp. 104–157. [Google Scholar]
- 22.Semino O, Santachiara-Benerecetti AS, Falaschi F, Cavalli-Sforza LL, Underhill PA. Ethiopians and Khoisan share the deepest clades of the human Y-chromosome phylogeny. Am J Hum Genet. 2002;70:265–268. doi: 10.1086/338306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li JZ, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 24.Henn BM, et al. Hunter-gatherer genomic diversity suggests a southern African origin for modern humans. Proc Natl Acad Sci USA. 2011;108:5154–5162. doi: 10.1073/pnas.1017511108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 26.McVean G. A genealogical interpretation of principal components analysis. PLoS Genet. 2009;5:e1000686. doi: 10.1371/journal.pgen.1000686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pickrell JK, et al. The genetic prehistory of southern Africa. Nat Commun. 2012;3:1143. doi: 10.1038/ncomms2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosenberg NA, et al. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- 31.Sved JA. Linkage disequilibrium and homozygosity of chromosome segments in finite populations. Theor Popul Biol. 1971;2:125–141. doi: 10.1016/0040-5809(71)90011-6. [DOI] [PubMed] [Google Scholar]
- 32.Marlowe FW. Mate preferences among Hadza hunter-gatherers. Hum Nat. 2004;15:365–376. doi: 10.1007/s12110-004-1014-8. [DOI] [PubMed] [Google Scholar]
- 33.Bahuchet S. Languages of African rainforest “Pygmy” hunter-gatherers: Lanugage shifts without cultural admixture. In: Güldemann T, McConvell P, Rhodes R, editors. Hunter-Gatherers and Linguistic History: A Global Perspective. Cambridge Univ Press; Cambridge, UK: 2006. [Google Scholar]
- 34.Newman JL. The Ecological Basis for Subsistence Change Among the Sandawe of Tanzania. National Academies; Washington, DC: 1970. [Google Scholar]
- 35.Lachance J, et al. Evolutionary history and adaptation from high-coverage whole-genome sequences of diverse African hunter-gatherers. Cell. 2012;150:457–469. doi: 10.1016/j.cell.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schlebusch CM, et al. Genomic variation in seven Khoe-San groups reveals adaptation and complex African history. Science. 2012;338:374–379. doi: 10.1126/science.1227721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saha N, et al. A study of some genetic characteristics of the population of the Sudan. Ann Hum Biol. 1978;5:569–575. doi: 10.1080/03014467800003251. [DOI] [PubMed] [Google Scholar]
- 38.Jarvis JP, et al. Patterns of ancestry, signatures of natural selection, and genetic association with stature in Western African pygmies. PLoS Genet. 2012;8:e1002641. doi: 10.1371/journal.pgen.1002641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lopez M, et al. The demographic history and mutational load of African hunter-gatherers and farmers. Nat Ecol Evol. 2018;2:721–730. doi: 10.1038/s41559-018-0496-4. [DOI] [PubMed] [Google Scholar]
- 40.Ralph P, Coop G. The geography of recent genetic ancestry across Europe. PLoS Biol. 2013;11:e1001555. doi: 10.1371/journal.pbio.1001555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gusev A, et al. The architecture of long-range haplotypes shared within and across populations. Mol Biol Evol. 2012;29:473–486. doi: 10.1093/molbev/msr133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ehret C, Keita SO, Newman P. The origins of Afroasiatic. Science. 2004;306:1680; author reply 1680. doi: 10.1126/science.306.5702.1680c. [DOI] [PubMed] [Google Scholar]
- 43.Browning SR, Browning BL. Identity by descent between distant relatives: Detection and applications. Annu Rev Genet. 2012;46:617–633. doi: 10.1146/annurev-genet-110711-155534. [DOI] [PubMed] [Google Scholar]
- 44.Dunn M, Terrill A, Reesink G, Foley RA, Levinson SC. Structural phylogenetics and the reconstruction of ancient language history. Science. 2005;309:2072–2075. doi: 10.1126/science.1114615. [DOI] [PubMed] [Google Scholar]
- 45.Fearnhead P, Prangle D. Constructing summary statistics for approximate Bayesian computation: Semi‐automatic approximate Bayesian computation. J R Stat Soc Series B Stat Methodol. 2012;74:419–474. [Google Scholar]
- 46.Tenesa A, et al. Recent human effective population size estimated from linkage disequilibrium. Genome Res. 2007;17:520–526. doi: 10.1101/gr.6023607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McEvoy BP, Powell JE, Goddard ME, Visscher PM. Human population dispersal “Out of Africa” estimated from linkage disequilibrium and allele frequencies of SNPs. Genome Res. 2011;21:821–829. doi: 10.1101/gr.119636.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chakraborty R, Smouse PE. Recombination of haplotypes leads to biased estimates of admixture proportions in human populations. Proc Natl Acad Sci USA. 1988;85:3071–3074. doi: 10.1073/pnas.85.9.3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pfaff CL, et al. Population structure in admixed populations: Effect of admixture dynamics on the pattern of linkage disequilibrium. Am J Hum Genet. 2001;68:198–207. doi: 10.1086/316935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moorjani P, et al. The history of African gene flow into Southern Europeans, Levantines, and Jews. PLoS Genet. 2011;7:e1001373. doi: 10.1371/journal.pgen.1001373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loh P-R, et al. Inferring admixture histories of human populations using linkage disequilibrium. Genetics. 2013;193:1233–1254. doi: 10.1534/genetics.112.147330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kelley JL, Madeoy J, Calhoun JC, Swanson W, Akey JM. Genomic signatures of positive selection in humans and the limits of outlier approaches. Genome Res. 2006;16:980–989. doi: 10.1101/gr.5157306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nielsen R, et al. Genomic scans for selective sweeps using SNP data. Genome Res. 2005;15:1566–1575. doi: 10.1101/gr.4252305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pagani L, et al. Ethiopian genetic diversity reveals linguistic stratification and complex influences on the Ethiopian gene pool. Am J Hum Genet. 2012;91:83–96. doi: 10.1016/j.ajhg.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Granka JM, et al. Limited evidence for classic selective sweeps in African populations. Genetics. 2012;192:1049–1064. doi: 10.1534/genetics.112.144071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scheinfeldt LB, et al. Genetic adaptation to high altitude in the Ethiopian highlands. Genome Biol. 2012;13:R1. doi: 10.1186/gb-2012-13-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Akey JM, et al. Tracking footprints of artificial selection in the dog genome. Proc Natl Acad Sci USA. 2010;107:1160–1165. doi: 10.1073/pnas.0909918107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shriver MD, et al. The genomic distribution of population substructure in four populations using 8,525 autosomal SNPs. Hum Genomics. 2004;1:274–286. doi: 10.1186/1479-7364-1-4-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen H, Patterson N, Reich D. Population differentiation as a test for selective sweeps. Genome Res. 2010;20:393–402. doi: 10.1101/gr.100545.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Enattah NS, et al. Identification of a variant associated with adult-type hypolactasia. Nat Genet. 2002;30:233–237. doi: 10.1038/ng826. [DOI] [PubMed] [Google Scholar]
- 62.Tishkoff SA, et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet. 2007;39:31–40. doi: 10.1038/ng1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ranciaro A, et al. Genetic origins of lactase persistence and the spread of pastoralism in Africa. Am J Hum Genet. 2014;94:496–510. doi: 10.1016/j.ajhg.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gras S, et al. A structural voyage toward an understanding of the MHC-I-restricted immune response: Lessons learned and much to be learned. Immunol Rev. 2012;250:61–81. doi: 10.1111/j.1600-065X.2012.01159.x. [DOI] [PubMed] [Google Scholar]
- 65.Sitnikova T, Nei M. Evolution of immunoglobulin kappa chain variable region genes in vertebrates. Mol Biol Evol. 1998;15:50–60. doi: 10.1093/oxfordjournals.molbev.a025846. [DOI] [PubMed] [Google Scholar]
- 66.Ashburner M, et al. The Gene Ontology Consortium Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Finkelman FD, Vercelli D. Advances in asthma, allergy mechanisms, and genetics in 2006. J Allergy Clin Immunol. 2007;120:544–550. doi: 10.1016/j.jaci.2007.05.025. [DOI] [PubMed] [Google Scholar]
- 68.Dhiman N, et al. Associations between cytokine/cytokine receptor single nucleotide polymorphisms and humoral immunity to measles, mumps and rubella in a Somali population. Tissue Antigens. 2008;72:211–220. doi: 10.1111/j.1399-0039.2008.01097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jones SA. Directing transition from innate to acquired immunity: Defining a role for IL-6. J Immunol. 2005;175:3463–3468. doi: 10.4049/jimmunol.175.6.3463. [DOI] [PubMed] [Google Scholar]
- 70.Klareskog L, Padyukov L, Rönnelid J, Alfredsson L. Genes, environment and immunity in the development of rheumatoid arthritis. Curr Opin Immunol. 2006;18:650–655. doi: 10.1016/j.coi.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 71.Liang HE, et al. The “dispensable” portion of RAG2 is necessary for efficient V-to-DJ rearrangement during B and T cell development. Immunity. 2002;17:639–651. doi: 10.1016/s1074-7613(02)00448-x. [DOI] [PubMed] [Google Scholar]
- 72.Simmons WA, et al. Novel HY peptide antigens presented by HLA-B27. J Immunol. 1997;159:2750–2759. [PubMed] [Google Scholar]
- 73.Tsoi LC, et al. Collaborative Association Study of Psoriasis (CASP); Genetic Analysis of Psoriasis Consortium; Psoriasis Association Genetics Extension; Wellcome Trust Case Control Consortium 2 Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat Genet. 2012;44:1341–1348. doi: 10.1038/ng.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu Y, Cheng G, Baltimore D. Targeted disruption of TRAF3 leads to postnatal lethality and defective T-dependent immune responses. Immunity. 1996;5:407–415. doi: 10.1016/s1074-7613(00)80497-5. [DOI] [PubMed] [Google Scholar]
- 75.Stevenson MM, Riley EM. Innate immunity to malaria. Nat Rev Immunol. 2004;4:169–180. doi: 10.1038/nri1311. [DOI] [PubMed] [Google Scholar]
- 76.Ocklenburg F, et al. UBD, a downstream element of FOXP3, allows the identification of LGALS3, a new marker of human regulatory T cells. Lab Invest. 2006;86:724–737. doi: 10.1038/labinvest.3700432. [DOI] [PubMed] [Google Scholar]
- 77.Bochud PY, Bochud M, Telenti A, Calandra T. Innate immunogenetics: A tool for exploring new frontiers of host defence. Lancet Infect Dis. 2007;7:531–542. doi: 10.1016/S1473-3099(07)70185-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chtanova T, et al. T follicular helper cells express a distinctive transcriptional profile, reflecting their role as non-Th1/Th2 effector cells that provide help for B cells. J Immunol. 2004;173:68–78. doi: 10.4049/jimmunol.173.1.68. [DOI] [PubMed] [Google Scholar]
- 79.Hidalgo LG, Einecke G, Allanach K, Halloran PF. The transcriptome of human cytotoxic T cells: Similarities and disparities among allostimulated CD4(+) CTL, CD8(+) CTL and NK cells. Am J Transplant. 2008;8:627–636. doi: 10.1111/j.1600-6143.2007.02128.x. [DOI] [PubMed] [Google Scholar]
- 80.Nishida K, et al. Gab-family adapter proteins act downstream of cytokine and growth factor receptors and T- and B-cell antigen receptors. Blood. 1999;93:1809–1816. [PubMed] [Google Scholar]
- 81.Milet J, et al. Genome wide linkage study, using a 250K SNP map, of Plasmodium falciparum infection and mild malaria attack in a Senegalese population. PLoS One. 2010;5:e11616. doi: 10.1371/journal.pone.0011616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Favre N, et al. Role of ICAM-1 (CD54) in the development of murine cerebral malaria. Microbes Infect. 1999;1:961–968. doi: 10.1016/s1286-4579(99)80513-9. [DOI] [PubMed] [Google Scholar]
- 83.Nam DH, Ge X. Development of a periplasmic FRET screening method for protease inhibitory antibodies. Biotechnol Bioeng. 2013;110:2856–2864. doi: 10.1002/bit.24964. [DOI] [PubMed] [Google Scholar]
- 84.Niederer HA, et al. Copy number, linkage disequilibrium and disease association in the FCGR locus. Hum Mol Genet. 2010;19:3282–3294. doi: 10.1093/hmg/ddq216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Saifuddin M, et al. Human immunodeficiency virus type 1 incorporates both glycosyl phosphatidylinositol-anchored CD55 and CD59 and integral membrane CD46 at levels that protect from complement-mediated destruction. J Gen Virol. 1997;78:1907–1911. doi: 10.1099/0022-1317-78-8-1907. [DOI] [PubMed] [Google Scholar]
- 86.Decker T, Müller M, Stockinger S. The yin and yang of type I interferon activity in bacterial infection. Nat Rev Immunol. 2005;5:675–687. doi: 10.1038/nri1684. [DOI] [PubMed] [Google Scholar]
- 87.Devuyst O, Dahan K, Pirson Y. Tamm-Horsfall protein or uromodulin: New ideas about an old molecule. Nephrol Dial Transplant. 2005;20:1290–1294. doi: 10.1093/ndt/gfh851. [DOI] [PubMed] [Google Scholar]
- 88.van Dijk W, et al. Inflammation-induced changes in expression and glycosylation of genetic variants of alpha 1-acid glycoprotein. Studies with human sera, primary cultures of human hepatocytes and transgenic mice. Biochem J. 1991;276:343–347. doi: 10.1042/bj2760343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Randall CN, et al. Cluster analysis of risk factor genetic polymorphisms in Alzheimer’s disease. Neurochem Res. 2009;34:23–28. doi: 10.1007/s11064-008-9626-8. [DOI] [PubMed] [Google Scholar]
- 90.Perry GH, et al. Diet and the evolution of human amylase gene copy number variation. Nat Genet. 2007;39:1256–1260. doi: 10.1038/ng2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Deng GY, Muir A, Maclaren NK, She JX. Association of LMP2 and LMP7 genes within the major histocompatibility complex with insulin-dependent diabetes mellitus: Population and family studies. Am J Hum Genet. 1995;56:528–534. [PMC free article] [PubMed] [Google Scholar]
- 92.Farris W, et al. Insulin-degrading enzyme regulates the levels of insulin, amyloid beta-protein, and the beta-amyloid precursor protein intracellular domain in vivo. Proc Natl Acad Sci USA. 2003;100:4162–4167. doi: 10.1073/pnas.0230450100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Scheepers A, Joost HG, Schürmann A. The glucose transporter families SGLT and GLUT: Molecular basis of normal and aberrant function. JPEN J Parenter Enteral Nutr. 2004;28:364–371. doi: 10.1177/0148607104028005364. [DOI] [PubMed] [Google Scholar]
- 94.Yoneda M, et al. Association between PPARGC1A polymorphisms and the occurrence of nonalcoholic fatty liver disease (NAFLD) BMC Gastroenterol. 2008;8:27. doi: 10.1186/1471-230X-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Scheinfeldt LB, et al. Population genomic analysis of ALMS1 in humans reveals a surprisingly complex evolutionary history. Mol Biol Evol. 2009;26:1357–1367. doi: 10.1093/molbev/msp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Moon S, et al. Novel compound heterozygous mutations in the fructose-1,6-bisphosphatase gene cause hypoglycemia and lactic acidosis. Metabolism. 2011;60:107–113. doi: 10.1016/j.metabol.2009.12.021. [DOI] [PubMed] [Google Scholar]
- 97.Maekawa M, et al. Detection and characterization of new genetic mutations in individuals heterozygous for lactate dehydrogenase-B(H) deficiency using DNA conformation polymorphism analysis and silver staining. Hum Genet. 1993;91:163–168. doi: 10.1007/BF00222718. [DOI] [PubMed] [Google Scholar]
- 98.Fischer J, et al. The gene encoding adipose triglyceride lipase (PNPLA2) is mutated in neutral lipid storage disease with myopathy. Nat Genet. 2007;39:28–30. doi: 10.1038/ng1951. [DOI] [PubMed] [Google Scholar]
- 99.Kirschning CJ, et al. Similar organization of the lipopolysaccharide-binding protein (LBP) and phospholipid transfer protein (PLTP) genes suggests a common gene family of lipid-binding proteins. Genomics. 1997;46:416–425. doi: 10.1006/geno.1997.5030. [DOI] [PubMed] [Google Scholar]
- 100.Reue K, Zhang P. The lipin protein family: Dual roles in lipid biosynthesis and gene expression. FEBS Lett. 2008;582:90–96. doi: 10.1016/j.febslet.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Felton AA, et al. Paleolimnological evidence for the onset and termination of glacial aridity from Lake Tanganyika, Tropical East Africa. Palaeogeogr Palaeoclimatol Palaeoecol. 2007;252:405–423. [Google Scholar]
- 102.Hetherington R, et al. Climate, African and Beringian subaerial continental shelves, and migration of early peoples. Quat Int. 2008;183:83–101. [Google Scholar]
- 103.Carto SL, Weaver AJ, Hetherington R, Lam Y, Wiebe EC. Out of Africa and into an ice age: On the role of global climate change in the late Pleistocene migration of early modern humans out of Africa. J Hum Evol. 2009;56:139–151. doi: 10.1016/j.jhevol.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 104.Pickrell JK, et al. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 2009;19:826–837. doi: 10.1101/gr.087577.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fumagalli M, et al. Signatures of environmental genetic adaptation pinpoint pathogens as the main selective pressure through human evolution. PLoS Genet. 2011;7:e1002355, and erratum (2011) 7. doi: 10.1371/journal.pgen.1002355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Neel JV. The “thrifty genotype” in 1998. Nutr Rev. 1999;57:S2–S9. doi: 10.1111/j.1753-4887.1999.tb01782.x. [DOI] [PubMed] [Google Scholar]
- 107.Henn BM, et al. Genomic ancestry of North Africans supports back-to-Africa migrations. PLoS Genet. 2012;8:e1002397. doi: 10.1371/journal.pgen.1002397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Weir BS. Genetic Data Analysis II: Methods for Discrete Population Genetic Data. Sinauer Associates; Sunderland, MA: 1996. [Google Scholar]
- 109.Huff CD, et al. Maximum-likelihood estimation of recent shared ancestry (ERSA) Genome Res. 2011;21:768–774. doi: 10.1101/gr.115972.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Beaumont MA, Zhang W, Balding DJ. Approximate Bayesian computation in population genetics. Genetics. 2002;162:2025–2035. doi: 10.1093/genetics/162.4.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schaffner SF, et al. Calibrating a coalescent simulation of human genome sequence variation. Genome Res. 2005;15:1576–1583. doi: 10.1101/gr.3709305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Emery LS, Felsenstein J, Akey JM. Estimators of the human effective sex ratio detect sex biases on different timescales. Am J Hum Genet. 2010;87:848–856. doi: 10.1016/j.ajhg.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Clark AG, Hubisz MJ, Bustamante CD, Williamson SH, Nielsen R. Ascertainment bias in studies of human genome-wide polymorphism. Genome Res. 2005;15:1496–1502. doi: 10.1101/gr.4107905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lohmueller KE, Bustamante CD, Clark AG. Methods for human demographic inference using haplotype patterns from genomewide single-nucleotide polymorphism data. Genetics. 2009;182:217–231. doi: 10.1534/genetics.108.099275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li S, Jakobsson M. Estimating demographic parameters from large-scale population genomic data using Approximate Bayesian Computation. BMC Genet. 2012;13:22. doi: 10.1186/1471-2156-13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hudson RR. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- 117.Kong A, et al. Fine-scale recombination rate differences between sexes, populations and individuals. Nature. 2010;467:1099–1103. doi: 10.1038/nature09525. [DOI] [PubMed] [Google Scholar]
- 118.Pritchard JK, Przeworski M. Linkage disequilibrium in humans: Models and data. Am J Hum Genet. 2001;69:1–14. doi: 10.1086/321275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wright S. Isolation by distance. Genetics. 1943;28:114–138. doi: 10.1093/genetics/28.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Holsinger KE, Weir BS. Genetics in geographically structured populations: Defining, estimating and interpreting F(ST) Nat Rev Genet. 2009;10:639–650. doi: 10.1038/nrg2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Friedman JH. Greedy function approximation: A gradient boosting machine. Ann Stat. 2001;29:1189–1232. [Google Scholar]
- 122.Natekin A, Knoll A. Gradient boosting machines, a tutorial. Front Neurorobot. 2013;7:21. doi: 10.3389/fnbot.2013.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lin K, Li H, Schlötterer C, Futschik A. Distinguishing positive selection from neutral evolution: Boosting the performance of summary statistics. Genetics. 2011;187:229–244. doi: 10.1534/genetics.110.122614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Aeschbacher S, Beaumont MA, Futschik A. A novel approach for choosing summary statistics in approximate Bayesian computation. Genetics. 2012;192:1027–1047. doi: 10.1534/genetics.112.143164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lin K, Futschik A, Li H. A fast estimate for the population recombination rate based on regression. Genetics. 2013;194:473–484. doi: 10.1534/genetics.113.150201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.McVean GA, et al. The fine-scale structure of recombination rate variation in the human genome. Science. 2004;304:581–584. doi: 10.1126/science.1092500. [DOI] [PubMed] [Google Scholar]
- 127.Altshuler DM, et al. International HapMap 3 Consortium Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- 129.Fujita PA, et al. The UCSC Genome Browser database: Update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Salas A, et al. The making of the African mtDNA landscape. Am J Hum Genet. 2002;71:1082–1111. doi: 10.1086/344348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Thomas PD, et al. PANTHER: A library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kasprzyk A. BioMart: Driving a paradigm change in biological data management. Database (Oxford) 2011;2011:bar049. doi: 10.1093/database/bar049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gething PW, et al. A new world malaria map: Plasmodium falciparum endemicity in 2010. Malar J. 2011;10:378. doi: 10.1186/1475-2875-10-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.