Abstract
Critical illness, a constellation of interrelated inflammatory and physiological derangements occurring subsequent to severe infection or injury, affects a large number of individuals in both developed and developing countries. The prototypical complex system embodied in critical illness has largely defied therapy beyond supportive care. We have focused on the utility of data-driven and mechanistic computational modelling to help address the complexity of critical illness and provide pathways towards discovering potential therapeutic options and combinations. Herein, we review recent progress in this field, with a focus on both animal and computational models of critical illness. We suggest that therapy for critical illness can be posed as a model-based dynamic control problem, and discuss novel theoretical and experimental approaches involving biohybrid devices aimed at reprogramming inflammation dynamically. Together, these advances offer the potential for Model-based Precision Medicine for critical illness.
Introduction
Critical illness refers to the constellation of acute inflammatory and pathophysiologic consequences that occur subsequent to sepsis, trauma/haemorrhage, and related events. Sepsis alone is responsible for more than 215,000 deaths in the United States per year and an annual healthcare cost of over $16 billion [1], whereas trauma/haemorrhage is the most common cause of death for young people in the United States, costing over $400 billion annually [2–4]. There is currently not a single drug approved by the U.S. Food and Drug Administration that specifically targets pathophysiological processes resulting in the dysregulated inflammatory response in trauma and sepsis [5, *6]. This is in stark contrast with the series of inflammation-directed biologics for chronic inflammatory conditions, such as rheumatoid arthritis and inflammatory bowel disease, many of which have their origins as failed anti-sepsis drugs [7]. We suggest that it is the acute, dynamic nature of dysregulated acute inflammation seen in sepsis and trauma that calls for a more precise characterization of both the system’s dynamics and any putative control/therapy.
The acute inflammatory response is a key generative factor which drives outcomes in critical illness. Though properly regulated inflammation allows for timely recognition and effective reaction to injury or infection, the immune dysregulation that ensues can impair physiological function, leading to a progressive Multiple Organ Dysfunction Syndrome (MODS)1. Moreover, the acute inflammatory response is not in and of itself detrimental: well-regulated, self-resolving acute inflammation is necessary for the appropriate resolution of injury, and for maintenance of proper physiology and homeostasis. An evolving view of the acute inflammatory response presents the paradox of a beneficial, robust, evolutionarily conserved network, and yet the structure of which may lead to disease [8–10]. Increasingly, this structure is being viewed as an interplay among the initiating insult itself, the ensuing inflammatory response, and the intertwined activation of dendritic cells and attendant activation of lymphoid cells, leading to a self-sustaining positive feedback loop in which inflammation-induced damage drives additional inflammation: inflammation → damage → inflammation [*11, *12].
Inflammation is driven and regulated by context-dependent chemokines and cytokines [13, 14], which are in turn negatively regulated via other cytokines [14] or via the production of cytokine antagonists [15]. The complex and paradoxical nature of the acute inflammatory response in critical illness has defied purely reductionist approaches to therapy. Though numerous promising candidate therapies have emerged from decades of pre-clinical studies, none have stood up under rigorous examination in the context of Phase III clinical trials as the therapy [5, 16]. We and others have hypothesized that the very features that make acute inflammation and critical illness a complex system are a key part of this failure, and that precision medicine for critical illness will necessitate the use of systems and computational biology approaches in order to arrive at effective treatment strategies [*6, 17–20]. Combining the tools of data-driven and mechanistic modelling with experimental studies has provided a deeper understanding of the dynamically changing nature of the acute inflammatory response and introduced methods for discovering effective control modalities [20–**26]. The synergy of these approaches reveals that one approach in isolation cannot provide what is needed to change the status quo in the treatment of acute illness. Here, we review the latest experimental and computational approaches in this endeavour, describe how they have yielded novel insights into the acute inflammatory response and its impact on critical illness, and suggest how these approaches can be combined to form a framework for Model-based Precision Medicine (Figure 1).
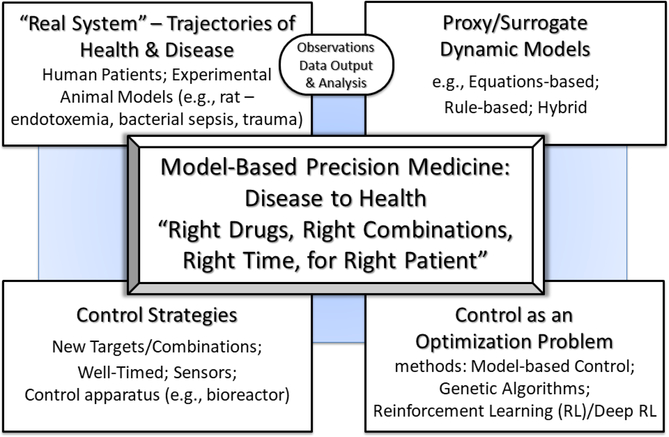
Figure 1:
This figure depicts the structure of what we term Model-based Precision Medicine: The right drugs in the right combinations at the right time for the right patient in order to control a disease trajectory back to a state of health. Model-based Precision Medicine starts with dynamic models as a proxy/surrogate of a real system (i.e. patient), informed by and updated with data streams coming from the patient as well as knowledge acquired from data-driven analyses. These dynamic models can be subjected to methods which allow the control discovery process to be formulated as an optimization problem not feasible/tractable for the real 97 elusive “golden bullet” world system. Subsequently, optimal therapeutic strategies/policies are identified that can incorporate multi-modal agents being adjusted/adapted based on patient trajectories/responses.
Important Developments in Experimental Work
The use of animal models in pre-clinical sepsis research is a controversial topic, with some groups suggesting a discordance between the response of experimental animals and humans at the transcriptomic level [27], and others contradicting this finding [28]. However, to date, the fact of the matter is that preclinical animal models represent a necessary phase of drug discovery and development. Rather than focus on whether or not such experimental models completely reflect clinical disease (they do not), we would turn our attention to how the information generated by these experiments can be best leveraged to enhance clinical success, e.g. by serving to generate, calibrate, and validate dynamic computational models that, in turn, could be used to predict the actions of drugs in simulated clinical trials [10, 29].
Animal models of sepsis:
Sepsis is the systemic manifestation of an out of control infection, typically a bacterial infection (though fungal and viral infections can lead to sepsis as well). There are a variety of pathophysiological criteria for sepsis, regarding which consensus is at times difficult to achieve [*30, 31]. However, sepsis is most typically caused by Gram-negative bacteria such as E. coli, of which the main immunostimulant is endotoxin. Originally, this, combined with an attempt to reduce experimental variability, led to various animal models of endotoxemia in mice, rats, dogs, swine, and non-human primates due to its convenience and reproducibility [32, 33]. Endotoxemia has also been carried out experimentally in humans, thereby allowing for “docking” of pre-clinical to human data [34]. Endotoxemia has yielded many important insights about acute inflammation, some of which are also applicable to true Gram-negative sepsis; as discussed below, endotoxemia proved to be an invaluable starting point for computational modelling of acute inflammation. However, experimental models involving administration of Gram-negative bacteria either directly or encapsulated in fibrin clots to mimic the physiological setting can offer greater realism [35]. The “gold standard” for experimental polymicrobial sepsis, however, is the experimental model of cecal ligation and puncture (CLP), in which the cecum is ligated surgically and punctured with a syringe [32, 33]. This animal model has worked well for predicting which sepsis therapies might work and which will not (mostly the latter) [36]. However, even these more realistic experimental models do not necessarily consider chronic disease burden in addition to the infection or a traumatic injury. Moreover, aging – and the associated phenomenon of “inflammaging” – further complicates diagnosis and therapy of critical illness in the elderly [37–39], though computational modelling of sepsis has been carried out using data from both small- and large-animal models of sepsis as well as studies involving aged animals [40].
Animal models of traumatic injury:
Polytrauma refers to severe injury to multiple body parts as a consequence of motor vehicle accidents, falls, etc. as well as due to intentional injuries caused by firearms, explosives, etc. These injuries lead to critical illness characterized by dysregulated inflammation and MODS [2–4]. Polytrauma usually presents with haemorrhagic shock, and thus various animal models of trauma/haemorrhage have been developed in order to study the pathophysiology of this aspect of critical illness [41, 42]. As is the case for sepsis, animal models have included mice, rats, dogs, swine, and (less commonly) non-human primates. Rodent models are the most common due to ease of manipulation and ability to obtain large numbers of experimental repeats. However, rodents typically do not exhibit MODS, whether following trauma/haemorrhage or models of sepsis described above; rather, they tend to exhibit an “all or none” phenomenon wherein the rodents survive increasingly severe trauma/haemorrhage until they die above a certain threshold. Thus, large animal models (especially non-human primate models) may better reflect the complex, individual-specific, and intertwined inflammatory and organ dysfunction phenotypes of humans [*6]. As mentioned above, age can also critically influence the response to traumatic injury [37, 38], though trauma patients are often fairly young and otherwise healthy, and yet still undergo a Systemic Inflammatory Response Syndrome and critical illness. Computational modelling of trauma/haemorrhage has also been carried out using data from both small and large animals, as well as humans.
Failure of therapies for acute inflammation in experimental critical illness:
The past several decades have seen a plethora of putative interventions for sepsis-induced (and to a lesser degree, trauma-induced) acute inflammation and MODS. While space constraints preclude a detailed discussion of these studies, these include numerous innate and adaptive immune pathways that culminate in the production of cytokines, chemokines, damage-associated molecular pattern molecules, oxygen and nitrogen free radicals, coagulation pathway intermediates, vasoactive peptides and lipids, and various organ-supportive strategies spanning the gamut from fluid resuscitation to organ/organism hibernation. Additionally, pro-inflammatory therapies (e.g. GMCSF) have also been tested. As mentioned above, all of these therapies have failed clinically.
We suggest that this failure stems from the fact that acute inflammation evolves too rapidly, with too much individual variation to be modulated appropriately given the current constraints on time and lack of strategies necessary for proper diagnosis and administration of therapy, and we offer Model-based Precision Medicine (Figure 1) as a possible solution. In the context of inflammatory diseases, we suggest that the current therapeutic approach – extinguishing inflammation to the greatest degree possible – is misguided. Rather, the therapeutic goal should not be to abolish inflammation, but rather to attenuate the vicious positive feedback cycle of inflammation→damage→inflammation, in essence, accomplishing control aimed at exploiting the system response to guide it back to a state of health (Figure 1).
To accomplish this goal, we hypothesize that sepsis is driven by existing and programmed pathways performing in a disordered fashion. Given the complexity and internal robustness of the system, control strategies should aim at redirecting these entangled control structures towards configurations in which the evolutionarily selected-for beneficial effects of these systems can come to the fore, thus allowing the body to re-equilibrate its inflammatory response incrementally and gradually. As one potential instantiation of the Control Strategies outlined in Figure 1, we have developed a self-regulating device for patient-specific, incremental regulation of inflammation, which is predicated on the hypothesis that the most efficient mechanism for doing so involves using gene-modified cells that are active in an extracorporeal support device and temporarily in exchange with the blood circulation via a bioreactor.
As a proof of concept, we targeted a key inflammatory cytokine involved in the aforementioned positive feedback loop, namely tumour necrosis factor-α (TNF-α). The bioreactor device was seeded with human HepG2 cells stably modified using lentiviral constructs to counter TNF-α through the constitutive or TNF-α-dependent production of soluble TNF-α receptor (sTNFR) [14]. Both constitutive and TNF-α-inducible sTNFR devices could reprogram dynamic networks of inflammation, and modulate key physiological outcomes in both endotoxemic [43] and septic [**44] rats.
Important Developments in Computational and Modelling Approaches in the Study of the Acute Inflammatory Response
Data-driven approaches in the context of the immunomodulatory bioreactor.
In earlier studies and in the bioreactor experimental work described above, several data-driven approaches were employed. At their core, data-driven modelling approaches are not mechanistic [45]. However, in this era of “big data”, incorporating a data-driven component is a rational first step towards defining putative mechanisms (via hypothesis generation). As such, data-driven approaches are an integral component of the Scientific Cycle of Observation, Analysis/Hypothesis generation and Hypothesis Testing [46]. In addition, the data-drive approaches can also provide prognosis/disease classification/outcome prediction metrics that would aid in building a case for clinical validity/utility, since prognosis/prediction need not account for mechanism.
Principal Component Analysis (PCA) was used in experimental trauma/haemorrhage in mice [47], endotoxemia in swine [48], and Gram-negative sepsis in rats [49] to define the subsets of mediators that are most strongly correlated with the inflammatory response under different experimental conditions, accounting for most of the variability seen in the dataset. This analysis was used to define the impact of experimental sepsis therapy using hemoadsorption [49], as well as the impact of inflammation-regulating bioreactors on endotoxemia and Gram-negative sepsis [**44]. PCA was also used as a data reduction tool to aid in the construction of a two-compartment, equation-based mechanistic model of endotoxin-induced inflammation in swine [48].
Data-driven methods often glean important information from network discovery methods. Dynamic Bayesian Network (DyBN) inference was used to identify the single most likely structure characterizing the dynamically changing inflammatory signalling network over all time points, including putative positive central nodes and negative feedback. This analysis suggested a potential “chemokine switch” architecture that might regulate the initial process of acute inflammation in humans [50]; proposed a novel role for the chemokine IP-10/CXCL-10 in spinal cord injury-induced systemic inflammation [51]; highlighted the structural similarity in the dynamic inflammatory responses of traumatic brain injury survivors and non-survivors which also offered a rational pathway from data, through data-driven modelling, to dynamic mechanistic models [*52]; and defined the dynamic inflammatory responses in subsets of patients with Paediatric Acute Liver Failure (PALF) [53, 54]. In the context of the inflammation-regulating bioreactor, DyBN suggested that sTNFR was a central node in experimental endotoxemia as well as Gram-negative sepsis in rats [**44].
Another network discovery method, Dynamic Network Analysis (DyNA) [47], defines central network nodes but, unlike DyBN, can give more granular information about network evolution over distinct time intervals for different experimental groups. In the context of the acute inflammatory response, DyNA can help suggest how these networks of inflammatory mediators change in complexity or connectively over certain time intervals. Examples of the utility of DyNA for critical illness include defining key structural differences in the inflammatory responses of mice undergoing trauma/haemorrhage vs. trauma alone [47]; the finding that human blunt trauma non-survivors [*11] as well as PALF non-survivors [54] exhibit dynamic networks that suggest ever-amplifying inflammation; the finding that the chemokine MCP-1/CCL2 is a central driver of stress induced hepatocyte inflammation in vitro [55]; and the demonstration of divergent inflammatory responses in various subsets of highly matched trauma patients [56, 57].
Mechanistic modelling and theoretical control approaches in the context of critical illness.
The aforementioned dynamic data-driven modelling tools extract from time-course data the crucial connections and relationships between the various mediators involved in the complex acute inflammatory response and suggest how these may change over time. These insights can be integrated directly into the development or calibration of mathematical models built from first-principles of the biological processes, such as those based on ordinary differential equations (ODE) or agent-based/rule-based simulations, [48, *52, 58]. These models are the foundation of Model-based Precision Medicine (Figure 1). Modelling approaches can help augment the development or testing of hypotheses regarding the mechanisms behind the observed outcomes among various experimental groups, including the potential role of gene knockouts [59] and experimental therapies [48, 60, 61]. The models also provide an important in silico testing environment for evaluating clinical trial-like scenarios [62–64]. Indeed, mathematical models consisting of nonlinear ODE as well as agent-based models (ABM) were constructed to capture the evolution of the response based on interactions among various key cell populations and inflammatory cytokines involved in the acute inflammatory response to a generic Gram-negative bacteria or bacterial endotoxin as well as traumatic insult [62, 65–70]. Together, these models provide preliminary qualitative or quantitative understanding of the dynamics of the inflammatory response and the importance of the timing of mediators at different stages of the response.
Given the paucity of effective therapies to treat sepsis as well as the potential capabilities of mechanism-based simulations of sepsis, we consider the role of modelling in exploring putative beneficial interventions. Specifically, we seek a means of identifying sets of interventions that can alter a detrimental individual system trajectory into a beneficial one; in this fashion we consider a therapeutic regimen as a “control” strategy for the system. Our suite of mechanism-based models crosses a range of methods, some of which (e.g. the ODE models) are suitable to classical control theoretic analysis, while other more complex stochastic models (e.g. the ABMs) can be subjected to optimization search methods that leverage advances in both high-performance computing platforms and machine-based learning algorithms.
The model-based control methodology of Model Predictive Control (MPC) was used to determine appropriate therapeutic inputs to correct dysfunctional immune responses in an in silico clinical trial of sepsis using heterogeneous virtual patients defined from an ODE model [22, 24], and in simulations with an ODE model of endotoxemia in rats [23]. In addition to control-theory based analysis, increased computational capabilities offered by advances in high performance computing (HPC) have now made feasible the application of simulation-based optimization and control discovery. Genetic algorithms, a computational method that employs synthetic genomes representing different potential therapeutic combinations and uses evolutionary principles to optimize control strategies, were applied to an ABM in the setting of an in silico clinical trial of sepsis [**26]. Also, by treating the management of sepsis as a “control game,” the advanced machine learning method known as deep reinforcement learning, which has previously been employed to train artificial intelligence (AI) systems to play games (most notably AlphaGo Zero [71]), has been used to train an AI to modulate the sepsis ABM to produce nearly 0% mortality across a simulated population [**25]. Results from these studies demonstrated that successful therapeutic strategies require targeting multiple mediators to varying degrees and at different times in order to modulate the complex, evolving inflammatory response to infection across heterogeneous simulations.
We note that these are proof-of-concept studies, as current experimental methodologies do not generate the data necessary to either comprehensively validate computational models and their associated uncertainties or sample combinatorically large spaces of treatment possibilities. These proof-of-concept studies highlight a role for mathematical models and control discovery methodologies in the pursuit of Model-based Precision Medicine (Figure 1), intended here as a framework for addressing key issues about data limitations and model uncertainty. By utilizing an iterative process of model development, testing, and refinement, the added component of examining control structures/strategies can both 1) provide investigatory insight by examining system response to perturbation/intervention, as is the case in nearly all experimental biology; and 2) provide potential high-level insight into boundary conditions for clinically-relevant investigation.
Important Developments in Synergistic Combination of Approaches in the Study of the Acute Inflammatory Response
In the setting of Gram-negative sepsis, a novel and synergistic strategy to study the acute inflammatory response is realized in combining the theoretical groundwork from the mathematical modelling and dynamic control studies with the experimental inflammation-regulating bioreactor protocol and represents an instantiation of Model-based Precision Medicine (Figure 1). The complex nonlinear dynamics of the immune response of the animal model (considered the ‘real system’) are captured by ABM and ODE model representations. These computational models are updated as often as possible with measurements from the real system in order to refine parameter selections or state trajectories for better prediction of the real system to baseline infection initiation and/or changes in the feedforward actions of the bioreactor. To assist in tuning the output of the bioreactor for optimal adaptive modulation of the response of the real system, the dynamics of the bioreactor environment are represented by another ODE model. Control discovery strategies are then implemented on the models of the real system and that of the bioreactor to periodically determine how to best tune the bioreactor such that its feedforward actions to the real system drive its response closer to a resolution trajectory. In this way, the framework seeks to realize an optimized reprograming of dynamic inflammation networks in vivo. The MPC methodology utilized in [22–24] provides the model-based control architecture that can more robustly tune and implement the immunomodulatory device on a patient-to-patient basis. Additionally, the framework allows for a closer integration of the modelling work with the data, promoting the necessary iterative process described above and providing an enhanced understanding of the acute inflammatory response in Gram-negative sepsis.
Conclusions
The experimental and computational approaches discussed here all provide essential components of an interdisciplinary strategy required to drive forward our understanding and control of the acute inflammatory response and can be envisioned to advance research of other inflammatory diseases. The development of suitable mechanistic models requires an interactive and sometimes lengthy cycle of development, calibration, evaluation, and back again to development. However, despite taking longer to develop and implement than some of the data-driven modelling approaches, mechanistic modelling provides direct exploration of the dynamics of an evolving and complex response. Additionally, the plethora of prior work in the area of mechanistic models of critical illness has laid a solid foundation from which to more easily move forward. Moreover, the ability to apply mathematical and engineering methodologies on such models provides an additional set of tools to tackle the complex problem of successfully modulating diverse responses in a customized and individualized fashion. A rational integration of the experimental bioreactor device with the data-driven computational modelling and mechanistic modelling and control work provides an exciting opportunity to tailor theoretical work in a practical setting with potential for large impact on personalized solutions for critical care support. This interdisciplinary framework provides the components necessary to pave a path toward Model-based Precision Medicine for acute inflammatory diseases.
Table 1.
Listing of acronyms used in the manuscript.
| Acronym | Meaning |
| ABM | Agent Based Model |
| AI | Artificial Intelligence |
| CLP | Cecal Ligation & Puncture |
| DRL | Deep Reinforcement Learning |
| DyBN | Dynamic Bayesian Network inference |
| DyNA | Dynamic Network Analysis |
| HepG2 | Human Hepatocytes |
| HPC | High Performance Computing |
| MCP-1/CCL2 | Monocyte Chemotactic Protein-1 |
| MODS | Multi Organ Dysfunction Syndrome |
| MPC | Model Predictive Control |
| ODE | Ordinary Differential Equations |
| PALF | Paediatric Acute Liver Failure |
| PCA | Principal Component Analysis |
| RL | Reinforcement Learning |
| sTNFR | soluble TNF-α receptor |
| TNF-α | Tumour Necrosis Factor - alpha |
Highlights.
Acute inflammation is a highly dynamic phenomenon that ensues as a response to infection or injury, and can be mimicked experimentally by the administration of agents such as Gram-negative bacterial endotoxin.
The complexity of acute inflammation is a key reason for the lack of therapies for sepsis, traumatic injury, and related phenomena known as critical illness.
Dynamic, data-driven approaches such as Principal Component Analysis, Dynamic Network Analysis, and Dynamic Bayesian Network inference, as well as dynamic mechanistic models using equations or agent-based rules, have been used to elucidate changes in complexity of inflammatory cytokine/chemokine signalling networks both in experimental models of critical illness (trauma/haemorrhage, endotoxemia, and Gram-negative sepsis), as well as in human trauma patients.
Mechanistic mathematical modelling of disease pathways – involving both equation- and agent-based frameworks – is based on first-principles biological knowledge. When combined with model-based control theoretic approaches and advanced computational methods such as genetic algorithms and deep reinforcement learning, such models may provide strategies for precision medicine in critical illness.
A novel class of engineered biohybrid devices may allow for personalized, fine-grained dynamic control of acute inflammation, and could be improved iteratively via model-based control strategies formulated from computational modelling.
Acknowledgements
JD was supported by U.S. National Science Foundation under Contract NSF-DMS1122462. RN, RZ, and YV were supported by U.S. National Institutes of Health grants RO1-GM107231–01A1, U01EB021960–01A1, UO1-DK072146, and P50-GM-53789, as well as U.S. Department of Defence grants W81 XWH-15–1-0336, W81XWH-15-PRORP-OCRCA, W81 XWH-13–2-0061, and W911 QY-14–1-0003. GA and CC were supported by high performance computing resources of the National Energy Research Scientific Computing Center, a DOE Office of Science User Facility supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02–05CH11231. Additionally, this research was supported in part by NIH through resources provided by the University of Chicago Computation Institute (Beagle2) and the Biological Sciences Division of the University of Chicago and Argonne National Laboratory, under grant 1S10OD018495–01. This work was also supported by funds from Lawrence Livermore National Laboratory under Award #B616283. This work was also motivated by the Lymphoid Cells in Acute Inflammation Investigative Workshop at the National Institute for Mathematical and Biological Synthesis (NIMBioS) Sponsored by the National Science Foundation through NSF Award # DBI-1300426.
Footnotes
Table 1 provides a listing of all acronyms used in the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Angus DC, et al. , Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit Care Med., 2001. 29(7): p. 1303–1310. [DOI] [PubMed] [Google Scholar]
- 2.Namas R, et al. , The acute inflammatory response in trauma / hemorrhage and traumatic brain injury: Current state and emerging prospects. Libyan J.Med., 2009. 4: p. 97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patton GC, et al. , Global patterns of mortality in young people: a systematic analysis of population health data. Lancet, 2009. 374(9693): p. 881–892. [DOI] [PubMed] [Google Scholar]
- 4.World Health Organization report: Young People: Health Risks and Solutions. 2011.
- 5.Angus DC, The search for effective therapy for sepsis: back to the drawing board? JAMA, 2011. 306(23): p. 2614–2615. [DOI] [PubMed] [Google Scholar]
- *6.Buchman TG, et al. , Precision medicine for critical illness and injury. Crit Care Med, 2016. 44(9): p. 1635–1638. This paper reviews the existing approaches for Precision Medicine as seen in oncology and cardiology, and poses a pathway for adopting those principles in critical illness and injury. This pathway involves increasing integration among the clinical collection of data, the analysis of that data using advanced computational methods and incorporation of knowledge generated from well-vetted and reproducible pre-clinical laboratory investigations. [DOI] [PubMed] [Google Scholar]
- 7.Brenner D, Blaser H, and Mak TW, Regulation of tumour necrosis factor signalling: live or let die. Nat Rev Immunol, 2015. 15(6): p. 362–74. [DOI] [PubMed] [Google Scholar]
- 8.Vodovotz Y, et al. , Mechanistic simulations of inflammation: Current state and future prospects. Math.Biosci., 2009. 217: p. 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vodovotz Y and An G, Systems Biology and Inflammation, in Systems Biology in Drug Discovery and Development: Methods and Protocols, Q. Yan, Editor. 2009, Springer Science & Business Media: Totowa, NJ: p. 181–201. [Google Scholar]
- 10.Vodovotz Y, et al. , Translational systems approaches to the biology of inflammation and healing Immunopharmacol.Immunotoxicol, 2010. 32: p. 181–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *11.Abboud AN, et al. , Computational analysis supports an early, type 17 cell-associated divergence of blunt trauma survival and mortality. Crit Care Med, 2016. 44: p. e1074–e1081. This paper describes the use of dynamic network analysis using data on circulating inflammatory mediators in trauma patients to derive a hypothesis regarding a heretofore-unrecognized role in acute inflammation for a particular inflammatory pathway (type 17 immunity) previously only associated with chronic inflammation; this hypothesis was validated partially in prospective experiments in mice subjected to experimental trauma/haemorrhage. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *12.Seshadri A, et al. , Phenotyping the immune response to trauma: A multiparametric systems immunology approach. Crit Care Med, 2017. This paper reports a systems-based analysis of circulating peripheral blood mononuclear cells from trauma patients, in which mass cytometry was used to define a broad array of inflammatory cells based on a large panel of cell markers. This study also verified the findings reported in Abboud et al., Crit. Care Med. 2016 [11] regarding an early role for type 17 immunity following trauma in humans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nathan C and Sporn M, Cytokines in context. J.Cell Biol, 1991. 113: p. 981–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nathan C, Points of control in inflammation. Nature, 2002. 420(6917): p. 846–852. [DOI] [PubMed] [Google Scholar]
- 15.Larrick JW and Wright SC, Native cytokine antagonists. Baillieres Clin.Haematol, 1992. 5(3): p. 681–702. [DOI] [PubMed] [Google Scholar]
- 16.Lord JM, et al. , The systemic immune response to trauma: an overview of pathophysiology and treatment. Lancet, 2014. 384(9952): p. 1455–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buchman TG, et al. , Complex systems analysis: a tool for shock research. Shock, 2001. 16(4): p. 248–251. [DOI] [PubMed] [Google Scholar]
- 18.Cobb JP and O’Keefe GE, Injury research in the genomic era. Lancet, 2004. 363(9426): p. 2076–2083. [DOI] [PubMed] [Google Scholar]
- 19.Vodovotz Y, et al. , Translational systems biology of inflammation. PLoS.Comput.Biol, 2008. 4: p. 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Complex Systems and Computational Biology Approaches to Acute Inflammation. 2013, New York, NY: Springer. [Google Scholar]
- 21.An G and Vodovotz Y, Translational Systems Biology: Concepts and Practice for the Future of Biomedical Research. 2014, New York, NY: Elsevier. [Google Scholar]
- 22.Day J, Rubin J, and Clermont G, Using nonlinear model predictive control to find optimal therapeutic strategies to modulate inflammation. Math.Biosci.Eng, 2010. 7(4): p. 739–763. [DOI] [PubMed] [Google Scholar]
- 23.Hogg JS, Clermont G, and Parker RS, Acute inflammation treatment via particle filter state estimation and MPC. IFAC Proceedings Volumes, 2010. 43: p. 272–277. [Google Scholar]
- 24.Zitelli G, Djouadi SM, and Day JD, Combining robust state estimation with nonlinear model predictive control to regulate the acute inflammatory response to pathogen. Math Biosci Eng, 2015. 12(5): p. 1127–39. [DOI] [PubMed] [Google Scholar]
- **25.Petersen BK, et al. , Precision medicine as a control problem: Using simulation and deep reinforcement learning to discover adaptive, personalized multi-cytokine therapy for sepsis. ArXiv 2018. This study is, to the best of our knowledge, the first application of deep reinforcement learning (DRL) to train an artificial intelligence on a biomedical simulation for purposes of control. This paper used DRL on a previously developed agent-based model that simulates the innate immune response to infection, the Innate Immune Response agent-based model (IIRABM), to discover an effective cytokine mediation treatment strategy for sepsis. The learned policy achieved 0.8% mortality over 500 randomly selected patient parameterizations with baseline mortalities ranging from 1–99% (with an average of 49%) spanning the entire clinically plausible parameter space of the IIRABM. The results suggest that adaptive, personalized multi-cytokine mediation therapy could be a promising approach for treating sepsis and that simulation-guided control policies could aid in the design of future clinical trials. [Google Scholar]
- **26.Cockrell RC and An G, Examining the controllability of sepsis using genetic algorithms on an agent-based model of systemic inflammation. PLoS Comput Biol, 2018. 14(2): p. e1005876 This work used evolutionary computation algorithms to explore drug-intervention space in a computational model, the Innate Immune Response agent-based model (IIRABM), serving as a surrogate system for human sepsis. The study identified the scale and scope of interventions needed to successfully control sepsis, as well as the types of data needed to derive these interventions. It was demonstrated that multi-point and time dependent varying controls are necessary and able to control the cytokine network dynamics of the immune system. This work provided insight into the scope of the clinical challenge, which can serve as a guide on the path towards true “precision control” of sepsis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seok J, et al. , Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takao K and Miyakawa T, Genomic responses in mouse models greatly mimic human inflammatory diseases. 2014. [DOI] [PMC free article] [PubMed]
- 29.An G, Bartels J, and Vodovotz Y, In silico augmentation of the drug development pipeline: Examples from the study of acute inflammation. Drug Dev.Res., 2011. 72: p. 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *30.Seymour CW, et al. , Assessment of Clinical Criteria for Sepsis: For the Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). Jama, 2016. 315(8): p. 762–74. This paper details the latest consensus definition of sepsis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Angus DC, et al. , A Framework for the Development and Interpretation of Different Sepsis Definitions and Clinical Criteria. Crit Care Med, 2016. 44(3): p. e113–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parker SJ and Watkins PE, Experimental models of gram-negative sepsis. Br.J.Surg, 2001. 88(1): p. 22–30. [DOI] [PubMed] [Google Scholar]
- 33.Marshall JC, et al. , Preclinical models of shock and sepsis: what can they tell us? Shock, 2005. 24 Suppl 1: p. 1–6. [DOI] [PubMed] [Google Scholar]
- 34.Martich GD, Boujoukos AJ, and Suffredini AF, Response of man to endotoxin. Immunobiology, 1993. 187(3–5): p. 403–416. [DOI] [PubMed] [Google Scholar]
- 35.Dunn DL, Rotstein OD, and Simmons RL, Fibrin in peritonitis. IV. Synergistic intraperitoneal infection caused by Escherichia coli and Bacteroides fragilis within fibrin clots. Arch.Surg., 1984. 119(2): p. 139–144. [DOI] [PubMed] [Google Scholar]
- 36.Remick DG and Ward PA, Evaluation of endotoxin models for the study of sepsis. Shock, 2005. 24 Suppl 1: p. 7–11. [DOI] [PubMed] [Google Scholar]
- 37.Horiguchi H, et al. , Innate immunity in the Persistent nflammation, mmunosuppression, and Catabolism Syndrome and its implications for therapy. Front Immunol, 2018. 9: p. 595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huber-Lang M, Lambris JD, and Ward PA, Innate immune responses to trauma. Nat Immunol, 2018. 19(4): p. 327–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Efron PA, et al. , Persistent inflammation, immunosuppression, and catabolism and the development of chronic critical illness after surgery. Surgery, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Namas RA, et al. , Combined in silico, in vivo, and in vitro studies shed insights into the acute inflammatory response in middle-aged mice. PLoS ONE, 2013. 8: p. e67419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lomas-Niera JL, et al. , Shock and hemorrhage: an overview of animal models. Shock, 2005. 24 Suppl 1: p. 33–39. [DOI] [PubMed] [Google Scholar]
- 42.Valparaiso AP, et al. , Modeling acute traumatic injury. J Surg Res, 2015. 194(1): p. 220–32. [DOI] [PubMed] [Google Scholar]
- 43.Namas R, et al. , A biohybrid device for the systemic control of acute inflammation. Disrupt.Science & Technol, 2012. 1: p. 20–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **44.Namas R, et al. , An adaptive, negative feedback circuit in a biohybrid device reprograms 491 dynamic inflammation networks in vivo. npj Systems Biology & Applications 2018. In Revision. This paper extends the work of Namas et al., Disrupt. Sci. Technol. 2012 [37] to define dynamic networks and principal characteristics of systemic acute inflammation in rat endotoxemia and Gram-negative sepsis, and the impact of a bioreactor producing soluble tumor necrosis factor-α receptor (TNF-α) – either constitutively or in a manner regulated by the animal’s circulating TNF-α – on these dynamic networks. [Google Scholar]
- 45.Janes KA and Yaffe MB, Data-driven modelling of signal-transduction networks. Nat.Rev.Mol.Cell Biol., 2006. 7(11): p. 820–828. [DOI] [PubMed] [Google Scholar]
- 46.An G, Closing the Scientific Loop: Bridging Correlation and Causality in the Petaflop Age. Science Translational Medicine, 2010. 2(41ps34): p. 1–6. [DOI] [PubMed] [Google Scholar]
- 47.Mi Q, et al. , A dynamic view of trauma/hemorrhage-induced inflammation in mice: Principal drivers and networks. PLoS ONE, 2011. 6: p. e19424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nieman K, et al. , A two-compartment mathematical model of endotoxin-induced inflammatory and physiologic alterations in swine. Crit.Care Med, 2012. 40: p. 1052–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Namas R, et al. , Hemoadsorption reprograms inflammation in experimental Gram negative septic fibrin peritonitis: Insights from in vivo and in silico studies. Mol.Med, 2012. 18: p. 1366–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Namas R, et al. , Insights into the role of chemokines, damage-associated molecular patterns, and lymphocyte-derived mediators from computational models of trauma-induced inflammation. Antiox. Redox Signaling, 2015. 10: p. 1370–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zaaqoq AM, et al. , Inducible protein-10, a potential driver of neurally-controlled IL-10 and morbidity in human blunt trauma. Crit.Care Med, 2014. 42: p. 1487–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *52.Abboud A, et al. , Inflammation following traumatic brain injury in humans: Insights from data-driven and mechanistic models into survival and death. Frontiers in Pharmacology, 2016. 7(342). This study demonstrated the potential utility of integrating data-driven and mechanistic modeling approaches based on 1) data on inflammatory mediators assayed in the cerebrospinal fluid of traumatic brain injury patients, 2) modeling the data using data driven modelling, and 3) abstraction of key insights from those data-driven models to generate mechanistic models based on ordinary differential equations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Azhar N, et al. , Analysis of serum inflammatory mediators identifies unique dynamic networks associated with death and spontaneous survival in pediatric acute liver failure. PLoS ONE, 2013. 8: p. e78202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zamora R, et al. , Data-driven modeling for precision medicine in pediatric ccute liver failure. Mol Med, 2016. 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ziraldo C, et al. , Central role for MCP-1/CCL2 in injury-induced inflammation revealed by in vitro, in silico, and clinical studies. PLoS ONE, 2013. 8(12): p. e79804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Namas RA, et al. , Temporal patterns of circulating inflammation biomarker networks differentiate susceptibility to nosocomial infection following blunt trauma in humans. Ann Surg, 2016. 263: p. 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abdul-Malak O, et al. , Elevated admission base deficit is associated with a complex dynamic network of systemic inflammation which drives clinical trajectories in blunt trauma patients. Mediators of Inflammation, 2016. 2016: p. 7950374 10.1155/2016/7950374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chow CC, et al. , The acute inflammatory response in diverse shock states. Shock, 2005. 24: p. 74–84. [DOI] [PubMed] [Google Scholar]
- 59.Prince JM, et al. , In silico and in vivo approach to elucidate the inflammatory complexity of CD14-deficient mice. Mol.Med., 2006. 12: p. 88–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Song SO, et al. , Ensemble models of neutrophil trafficking in severe sepsis. PLoS Comput Biol, 2012. 8(3): p. e1002422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Malkin AD, et al. , A neutrophil phenotype model for extracorporeal treatment of sepsis. PLoS Comput Biol, 2015. 11(10): p. e1004314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.An G, In-silico experiments of existing and hypothetical cytokine-directed clinical trials using agent based modeling. Crit Care Med., 2004. 32: p. 2050–2060. [DOI] [PubMed] [Google Scholar]
- 63.Clermont G, et al. , In silico design of clinical trials: a method coming of age. Crit Care Med., 2004. 32: p. 2061–2070. [DOI] [PubMed] [Google Scholar]
- 64.Brown D, et al. , Trauma in silico: individual-specific mathematical models and virtual clinical populations. Sci Transl Med, 2015. 7: p. 285ra61. [DOI] [PubMed] [Google Scholar]
- 65.Reynolds A, et al. , A reduced mathematical model of the acute inflammatory response: I. Derivation of model and analysis of anti-inflammation. J.Theor.Biol, 2006. 242: p. 220–236. [DOI] [PubMed] [Google Scholar]
- 66.Daun S, et al. , An ensemble of models of the acute inflammatory response to bacterial lipopolysaccharide in rats: Results from parameter space reduction. J.Theor.Biol, 2008. 253: p. 843–853. [DOI] [PubMed] [Google Scholar]
- 67.Torres A, et al. , Mathematical modeling of post-hemorrhage inflammation in mice: Studies using a novel, computer-controlled, closed-loop hemorrhage apparatus. Shock, 2009. 32: p. 172–178. [DOI] [PubMed] [Google Scholar]
- **68.Cockrell C and An G, Sepsis reconsidered: Identifying novel metrics for behavioral landscape characterization with a high-performance computing implementation of an agent-based model. J Theor Biol, 2017. 430: p. 157–168. This work provided a novel investigatory and analytical approach, derived from how High Performance Computing resources and simulation are used in the physical sciences, to identify the epistemic boundary conditions of the study of clinical sepsis via the use of a proxy agent-based model of systemic inflammation, the Innate Immune Response agent-based model. Two novel methods were developed for characterizing the behavior of a RDS: Probabilistic Basins of Attraction (PBoA) and Stochastic Trajectory Analysis (STA). Computationally generated behavioral landscapes demonstrated attractor structures involving stochastic regions of behavior, which highlighted the challenge for correlative attempts to characterize and classify clinical sepsis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.An G, Agent-based computer simulation and SIRS: building a bridge between basic science and clinical trials. Shock, 2001. 16(4): p. 266–273. [DOI] [PubMed] [Google Scholar]
- 70.Day J, et al. , A reduced mathematical model of the acute inflammatory response: II. Capturing scenarios of repeated endotoxin administration. J.Theor.Biol, 2006. 242: p. 237–256. [DOI] [PubMed] [Google Scholar]
- 71.Silver D, et al. , Mastering the game of Go without human knowledge. Nature, 2017. 550(7676): p. 354–359. [DOI] [PubMed] [Google Scholar]