Abstract
Background
Triple-negative breast cancer (TNBC) is a poor prognostic breast cancer with the highest mutations and limited therapeutic choices. Cytokine networking between cancer cells and the tumor microenvironment (TME) maintains the self-renewing subpopulation of breast cancer stem cells (BCSCs) that mediate tumor heterogeneity, resistance and recurrence. Immunotherapy of those factors combined with targeted therapy or chemoagents may advantage TNBC treatment.
Results
We found that the oncogene Multiple Copies in T-cell Malignancy 1 (MCT-1/MCTS1) expression is a new poor-prognosis marker in patients with aggressive breast cancers. Overexpressing MCT-1 perturbed the oncogenic breast epithelial acini morphogenesis and stimulated epithelial-mesenchymal transition and matrix metalloproteinase activation in invasive TNBC cells, which were repressed after MCT-1 gene silencing. As mammary tumor progression was promoted by oncogenic MCT-1 activation, tumor-promoting M2 macrophages were enriched in TME, whereas M2 macrophages were decreased and tumor-suppressive M1 macrophages were increased as the tumor was repressed via MCT-1 knockdown. MCT-1 stimulated interleukin-6 (IL-6) secretion that promoted monocytic THP-1 polarization into M2-like macrophages to increase TNBC cell invasiveness. In addition, MCT-1 elevated the soluble IL-6 receptor levels, and thus, IL-6R antibodies antagonized the effect of MCT-1 on promoting M2-like polarization and cancer cell invasion. Notably, MCT-1 increased the features of BCSCs, which were further advanced by IL-6 but prevented by tocilizumab, a humanized IL-6R antibody, thus MCT-1 knockdown and tocilizumab synergistically inhibited TNBC stemness. Tumor suppressor miR-34a was induced upon MCT-1 knockdown that inhibited IL-6R expression and activated M1 polarization.
Conclusions
The MCT-1 pathway is a novel and promising therapeutic target for TNBC.
Electronic supplementary material
The online version of this article (10.1186/s12943-019-0988-0) contains supplementary material, which is available to authorized users.
Keywords: MCT-1/miR-34a/IL-6/IL-6R pathway, EMT, M2 macrophages, Cancer stemness
Background
Cancer cell-intrinsic mechanisms and cell-extrinsic factors determine tumor development and aggressiveness [1]. Epithelial-mesenchymal transition (EMT) induces epithelial cells transforming into mesenchymal cells [2], cancer cell movement, cancer progression, metastasis and stemness. Tumor-associated macrophages (TAMs) derived from peripheral blood monocytes are recruited to microenvironment and polarized into M1 or M2 macrophages in response to secreted factors from cancer cells or microenvironmental cells [3]. M1 macrophages highly express inducible nitric oxide synthase (iNOS) and Tumor Necrosis Factor (TNF)-α and promote pro-inflammatory and immune responses that prevent oncogenic effects [4], while M2 macrophages express Arginase 1 (ARG1) and highly produce cytokines, growth factors and protease that are crucial for pro-tumorigenic processes. Furthermore, M2 macrophages stimulate tumor angiogenesis [5, 6], cancer cell migration/invasion, immunosuppression and marix remodeling.
IL-6 is involved in immune regulation [7], inflammation and oncogenesis. IL-6 and IL-6R interaction induces the dimerization of glycoprotein 130 (gp130) to activate signal transducer and activator of transcription 3 (Stat3) [8]. However, signaling activation of IL-6/Stat3 repressed the p53-mediated miR-34a expression [8], and miR-34a-deficient mice exhibited the stimulation of Stat3, IL-6R and Snail as well as the increase in colorectal cancer invasion/metastasis. IL-6 induction is associated with a poorer prognosis in patients with breast cancer and serum IL-6 levels are increased with pathological grades [9]. Clinical trials of IL-6 and IL-6R immunotherapy have been widely studied in multicentric Castleman’s disease [7], multiple myeloma and solid tumors including renal prostate, lung, colorectal and ovarian cancers. However, the studies of IL-6/IL-6R/gp130 immunotherapy for treatment of breast cancers are limited. IL-6/IL-6R/gp130 pathway communicates between breast tumor and immune cells [10], resulting in tumor promotion and enriched effect on BCSCs. Therefore, targeting IL-6/Sta3 signaling axis potentially improve the efficacy of cancer immunotherapy [11]. Supporting this notion, the nanoparticles of anti- CD44 Ab encapsulating anti-IL-6R Ab target the TME and CD44+ BCSCs that inhibit the metastatic niche of triple-negative and luminal breast cancer in two mouse models, namely syngeneic BALB/c mice bearing 4 T1 cells and transgenic MMTV-PyMT mice [12]. Also, the use of nanoparticle-based system for CD44 and IL-6R immunotherapy suppresses Stat3, Sox2, VEGF-A, MMP-9 and CD206 expression in breast tissues as well as Sox2+/CD206+ stem cells in lung metastatic foci.
MCT-1 is a ribosome binding protein encoded by MCTS1 gene [13, 14], which orchestrates ribosomal recycling, translation reinitiation and tissue growth. Density regulated protein and the MCT-1 heterodimer bind to the 40S ribosomal subunit and, with the recruitment of tRNA, cooperatively regulate noncanonical translation initiation [15, 16]. Moreover, MCT-1 affects mitotic progression via interacting with γ-tubulin molecule and Src/p190B complex [17, 18]. MCT-1 destabilizes p53 and PTEN in a ubiquitin-dependent proteasome pathway [18, 19]. Consequently, MCT-1 expression advances the p53-null or PTEN-null cancer cell progression and chromosomal/nuclear aberrations [17, 18, 20, 21]. Importantly, targeting MCT-1 suppresses genomic instability and tumorigenicity [18, 22]. MCT-1 overexpression also induces ROS generation [23], leading to YY-1/EGFR/MnSOD signaling amplification and cancer cell invasion. Here, we first demonstrate that MCT-1 induces IL-6/IL-6R/Stat3 pathway, M2 macrophage polarization, TNBC progression and stemness.
Results
MCT-1 is a poor-prognosis marker of aggressive breast cancer
Oncogenic MCT-1 (also known as MCTS1) activation in breast cancer was investigated using the Kaplan-Meier Plotter database [24], and we observed that high MCT-1 expression in patients was associated with lower overall survival (OS) in overall breast cancer (p = 0.0053) as well as in TP53 wild type (p = 0.024) (Additional file 1: Figure S1A), lymph node metastasis-free (p = 0.001), HER2-negative (p = 0.0067), luminal-A (p = 0.026) and luminal-B (p = 0.043) breast cancers than that of patients with low-level MCT-1. Patients with MCT-1 gene overexpression also exhibited lower recurrence-free survival (RFS) in overall breast cancer (p = 1E-16) as well as in TP53 wild type (p = 0.029) (Additional file 1: Figure S1B), lymph node metastasis (p = 0.017) and metastasis-free (p = 0.00096), ER-negative (p = 0.015), HER2-negative (p = 1E-06), TNBC (p = 0.046), luminal-A (p = 5E-08) and luminal-B (p = 2.3E-06) breast cancer than did patients with low MCT-1 levels.
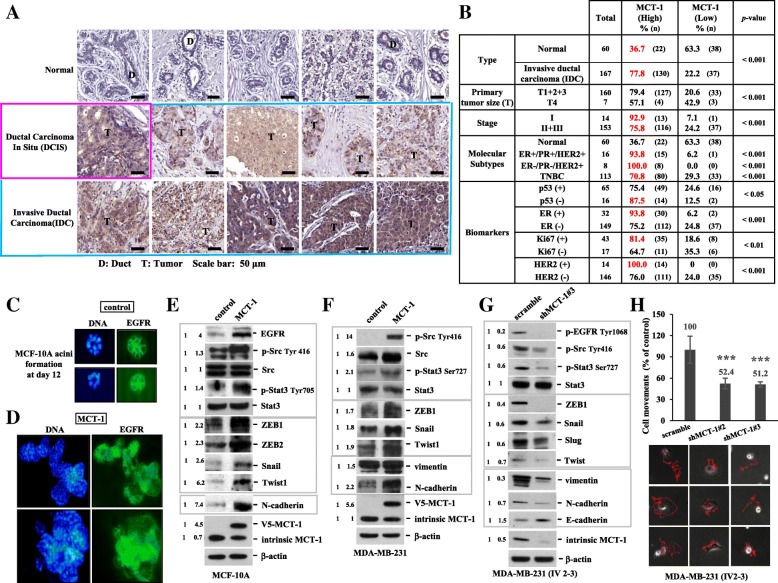
Immunohistostaining revealed MCT-1 protein enrichment in ductal carcinoma in situ (DCIS) and in invasive ductal carcinoma (IDC) of the breast comparative to that in normal breast tissue and the adjacent stroma (Fig. 1a). Moreover, MCT-1 protein was often enriched in IDC (77.8%, n = 167) as well as in the majority of patients at stage I (92.9%, n = 14) and stage II/III (75.8%, n = 153) but was less identified in normal breast tissues (36.7%, n = 60) (Fig. 1b). Characterized by the molecular subtypes, we found MCT-1 induction in 93.8% of ER+/PR+/HER2+ cancers (n = 16), 100% of ER−/PR−/HER2+ (n = 8) cancers and 70.8% of TNBC (ER−/PR−/HER2−) (n = 113). MCT-1 protein is also enriched in p53-negative (87.5%, n = 16), ER-positive (93.8%, n = 32), Ki67-positive (81.4%, n = 43) and HER2-positive (100%, n = 14) breast carcinomas. Conceivably, MCT-1 gene activation and protein increase implicate in breast carcinogenesis.
Fig. 1.
MCT-1 is a prognostic marker of human breast carcinoma. (a) Immunohistology was used to analyze MCT-1 protein levels in normal tissues, DCIS and IDC of the breast. T: tumor. D: Duct. Scale bars, 50 μm. (b) MCT-1 protein levels in breast cancers were classified based on pathologic stages, molecular subtypes and biomarkers. The results were evaluated from 6 to 8 randomly chosen imaging fields per sample. The Fisher exact probability test of independence was used to calculate the clinical significance. (c and d) MCF-10A acini morphogenesis was studied on a Matrigel matrix culture for 12 days followed by EGFR Ab (green) and DNA (blue) staining. (e and f) EGFR expression, Src/Stat3 activation and EMT molecules were analyzed in MCF-10A and MDA-MB-231 cells without (control) or with MCT-1 induction (MCT-1). (g) EMT molecules and EGFR/Src/Stat3 activation were assessed in MDA-MB-231 (IV2–3) without (scramble) or with MCT-1 knockdown (shMCT-1#3). (h) Time-lapse imaging recorded movements of MDA-MB-231 (IV2–3) (scramble vs. shMCT-1#2 and #3). Different blots were normalized with β-actin and then the relative protein amounts were compared with the corresponding controls. One-way ANOVA with a post hoc two-tailed t-test was used to calculate the statistical significance of the cell migration assays (n = 12). (*** p < 0.001)
MCT-1 promotes mammary acini oncogenesis, EMT progression and MMP activation
Normal breast epithelial MCF-10A cells grown on a basement membrane matrix form polarized, growth-arrested acini-like spheroids that recapitulate the glandular architecture in vivo [25]. Examining MCF-10A acini morphogenesis on a growth factor-reduced Matrigel for 12 days, the control cells assembled in a regular acini-like, spheroid with well-organized nuclear arrangement and cell-cell interaction (Fig. 1c). MCF-10A cells were virally transfected with pLXSN or pLXSN/V5-MCT-1 (V5-tagged MCT-1) using the retroviral supernatants. Upon MCT-1 overexpression (V5-MCT-1), MCF-10A acinarization was perturbed with aggressive cellular growth and proliferation that produced a multiacinar structure with a disorganized nuclear arrangement and poor cell-cell interaction (Fig. 1d), implying that MCT-1 overexpression disturbed the early stage of mammary acinar morphogenesis. Furthermore, the development of MCF-10A acini on Matrigel matrix at day 3 and day 5 were examined (Additional file 2: Figure S2A and B). The diameters of MCF-10A acini at day 5 were quantified (Additional file 2: Figure S2C), and it showed that MCT-1 overexpression more promoted MCF-10A acini development than control cells.
EMT disrupts MCF-10A polarity and growth-arrested spheroid formation [26]. Consistently, overexpressing MCT-1 in MCF-10A cells induced EGFR (Fig. 1e), phospho-activation of Src (p-Src Tyr416) and Stat3 (p-Stat3 Tyr705), EMT transcription factors (ZEB1, ZEB2, Snail, Twist1) and N-cadherin. Moreover, MCT-1 overexpression in human TNBC (MDA-MB-231) cells increased Src/Stat3 activation as well as the levels of ZEB1 (Fig. 1f), Snail, Twist1, vimentin and N-cadherin. Conversely, MCT-1 gene was targeted by a small hairpin RNA (shMCT-1) in a highly invasive MDA-MB-231 (IV2–3) subline that has been isolated from two rounds of in vivo selection from lung metastases of the parental MDA-MB-231 cells [27]. Silencing MCT-1 (shMCT-1#3) repressed EGFR/Src activation as well as the levels of ZEB1 (Fig. 1g), Snail, Slug, Twist, vimentin and N-cadherin but increased E-cadherin. Similar results were also identified in the shMCT-1#2 clone of IV2–3 (Additional file 2: Figure S2D). Corresponding to mesenchymal-epithelial transition, the migratory abilities were significantly reduced in the shMCT-1 clones (#2 and #3) compared with those scramble control cells (Fig. 1h).
An examination of precursor (pro) and active matrix metalloproteinases (MMPs) and their inhibitors (TIMPs) revealed that MCT-1 increased the amounts of vascular endothelial growth factor (VEGF) alongside pro- and active- MMP2 and MMP9 elevation but reduced TIMP-1 and TIMP-2 levels (Additional file 3: Figure S3A). However, shMCT-1#3 increased TIMP-1 and TIMP-2 but decreased pro- and active- MMP2 and MMP9 (Additional file 3: Figure S3B). Consistently, MCT-1 promoted MDA-MB-231 invasiveness through a Boyden chamber-coating Matrigel matrix (Additional file 3: Figure S3C), whereas shMCT-1#3 repressed the high invasive ability of MDA-MB-231 (IV2–3) (Additional file 3: Figure S3D). Using gelatin zymography to evaluate gelatinases in the conditioned medium (CM) (Additional file 3: Figure S3E), induced MMP2 and MMP9 activities were detected while MCT-1 overexpressing but these activities were decreased in shMCT-1#3 condition. Likewise, shMCT-1 (#3–28) inhibited the wound closure ability (Additional file 3: Figure S3G) and the invasiveness of murine TNBC (4 T1) cells (Additional file 3: Figure S3H) alongside reducing MMP2 and MMP9 but increasing TIMP-1 and TIMP-3 (Additional file 3: Figure S3F). Confirmatively, MCT-1 promotes EMT processes and MMP activities which enhance cancer cell migration/invasion.
MCT-1 induces the IL-6/IL-6R signaling
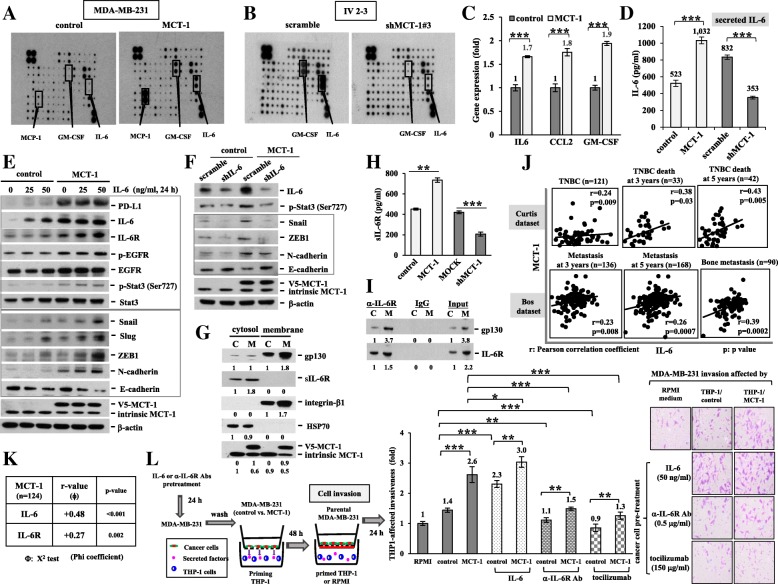
The cytokine arrays were incubated with the CM of the MDA-MB-231 cells to identify the factors that communicate between cancer cells and immune cells. The results exhibited that there were more IL-6, CCL2 and GM-CSF secretion from MCT-1-overexpressing cells relative to control cells (Fig. 2a). However, only IL-6 and GM-CSF secretion were dramatically reduced after loss-function of MCT-1 (shMCT-1#3) compared with that of scramble control of the high-invasive IV2–3 cells (Fig. 2b). The IL-6, CCL2 and GM-CSF mRNA levels induced by MCT-1 were also detected by quantitative RT-PCR (qRT-PCR) (Fig. 2c). An enzyme-linked immunosorbent assay (ELISA) with the specific anti-IL-6 Ab confirmed that IL-6 secretion was promoted by MCT-1 overexpression (Fig. 2d), but it was significantly repressed in shMCT#3 cells.
Fig. 2.
The MCT-1/IL-6/IL-6R pathway activates THP-1 differentiation that stimulates cancer cell invasion. (a and b) The cytokine arrays were incubated with the CM of the MDA-MB-231 cells (control vs. MCT-1) and IV2–3 subline (scramble vs. shMCT-1#3) to identify the secreted factors specific to the indicated cellular conditions. (c) Quantitative RT-PCR evaluated IL-6, CCL2 and GM-CSF mRNA levels in the MDA-MB-231 cells. (d) The IL-6 secreted from the MDA-MB-231 cells (control vs. MCT-1) and IV2–3 subline (scramble vs. shMCT-1#3) were quantified by ELISA using the anti-IL-6 mAb. The results were normalized with the plated cell numbers (2 × 105). (e) PD-L1, IL-6/IL-6R, EGFR, Stat3 and EMT molecules were analyzed upon different doses of IL-6 simulation for 24 h. Different blots with normalized internal control were used to present the data of the experiment. (f) The effect of IL-6 knockdown (shIL-6) on EMT signaling molecules were characterized. (g) The proteins in the cytosol and membrane fractions were isolated from 5 × 106 cells and then compared between control (c) and MCT-1 (m) groups. (h) The conditioned media of the MDA-MB-231 cells with different MCT-1 expression conditions were harvested after 48 h culture. The ELISA measured soluble IL-6R (sIL-6R) levels, and the results were normalized with the plated cell numbers. (i) The interaction of gp130 and IL-6R in vivo were identified by IL-6R immunoprecipitation assay. (j) The clinical relevance of MCT-1 and IL-6 was examined in TNBC (Curtis database) and in metastatic breast cancer (Bos dataset) using the Oncomine database. Pearson’s correlation coefficient indicates the statistical significance. (k) MCT-1 associated with IL-6/IL-6R was studied using the breast cancer cDNA arrays (n = 124). The Chi-square test was used to calculate the statistical significance. (l) The MDA-MB-231 cells were pretreated by IL-6, anti-IL-6R mAb or tocilizumab for 24 h followed by THP-1 coculture in the transwell for 48 h. Parental MDA-MB-231 invasiveness was examined as cocultured with the preconditioned THP-1 cells in a Boyden chamber-coating Matrigel matrix for 24 h. The results are expressed as the mean ± SD of three independent assays (n = 3). One-way ANOVA with a post hoc two-tailed t-test was used to calculate the statistical significance of pairwise comparisons. (*p < 0.05; **p < 0.01; *** p < 0.001)
The IL-6-STAT3-Twist circuit stimulates the EMT process and M2 macrophage polarization [28]. We found that MCT-1 increased the programmed death-ligand 1 (PD-L1), IL-6 and IL-6R amounts (Fig. 2e), and IL-6 further advanced MCT-1-induced EGFR and Stat3 phospho-activation as well as Snail, Slug, ZEB1 and N-cadherin but further suppressed E-cadherin in a dose-dependent manner. MCT-1 and IL-6 additively stimulated EMT molecules (EGFR, p-Stat3, Snail, Slug, ZEB1 and N-caherin), and PD-L1 was induced by MCT-1 but not further stimulated by IL-6. Silencing IL-6 (shIL-6) expression also reduced the Stat3 activation and EMT signaling molecules (Snail, ZEB1 and N-cadherin) but induced E-cadherin in the MCT-1-overexpressing cells (Fig. 2f). Thus, MCT-1 enhances EMT process via IL-6 pathway. Furthermore, subcellular fractionation identified the levels of membrane-bound gp130 and soluble IL-6R (sIL-6R) were most elevated (Fig. 2g) and sIL-6R amounts present in the CM were also induced (Fig. 2h) upon MCT-1 overexpression. We speculate that MCT-1 regulates the proteolytic cleavage of IL-6R or the alternative splicing of IL-6R mRNA [29], by which amplifies IL-6 trans-signaling through sIL-6R rather than classic IL-6 signaling by membrane-bound IL-6R [30]. In the IL-6R pull-down assay, gp130 was increased by 3.7 fold while IL-6R was increased by 1.5 fold in the MCT-1-overexpressing cells (M) over the control cells (C) (Fig. 2i), indicating that MCT-1 overexpression induced more gp130 molecules interaction with IL-6R in vivo.
An assessment of the clinical connection between MCT-1 and IL-6, the positive correlations between MCT-1 and IL-6 gene activation in overall TNBC patients (r = 0.24, p = 0.009) as well as in TNBC patient deaths at 3 years (r = 0.38, p = 0.03) and at 5 years (r = 0.43, p = 0.005) were recognized in the Curtis dataset of the Oncomine database (Fig. 2j). Moreover, MCT-1 and IL-6 gene promotion was correlated in breast cancer patients with 3-year metastasis (r = 0.23, p = 0.008), 5-year metastasis (r = 0.26, p = 0.007) and bone metastasis (r = 0.39, p = 0.0002, n = 90) in the Bos dataset. Further analysis of the breast cancer cDNA arrays (n = 124) confirmed positive correlations of MCT-1 expression with that of IL-6 (r = 0.48, p < 0.001) and IL-6R (r = 0.27, p = 0.002) (Fig. 2k).
To inspect whether cancer cell invasion is affected by the macrophages, the MDA-MB-231 cells were pretreated with IL-6 (Fig. 2l), the anti-IL-6R mAb or tocilizumab before priming THP-1 monocytes. The preconditioned THP-1 cells were placed in the lower chamber to test the invasion ability of MDA-MB-231 cells from the upper chamber. The cell invasiveness was more enhanced by the THP-1 which were primed by MCT-1-overexpressing cells (THP-1/MCT-1) than which were primed by the control cells (THP-1/control). Furthermore, the invasiveness was advanced with THP-1 priming by IL-6-stimulated MDA-MB-231, whereas the pretreatment with the anti-IL-6R mAb or with tocilizumab failed to activate THP-1, thus suppressing the invasion.
MCT-1 promotes mammary tumor progression and TAM polarization
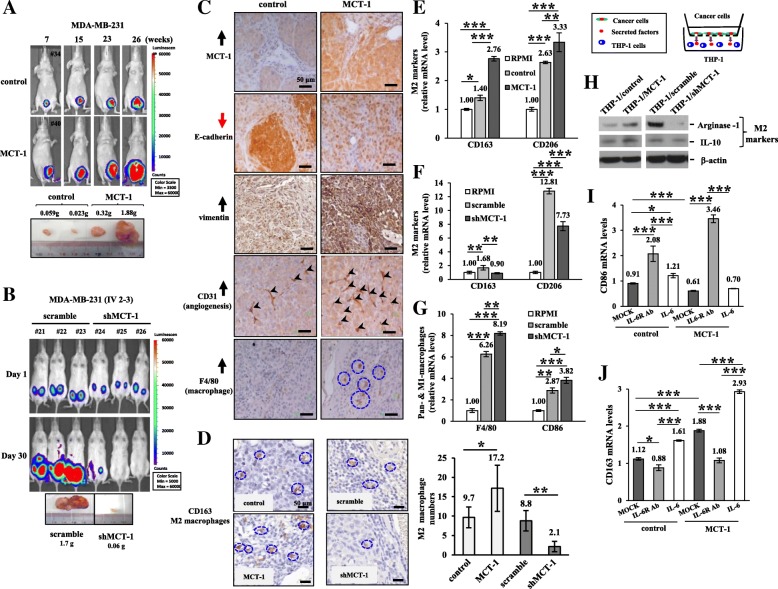
The bioluminescent MDA-MB-231 cells (1 × 105) bearing a pGL3-luciferase reporter were injected into the mammary fat pad of immunodeficient BALB/c nude mice and monitored by an in vivo imaging system (Fig. 3a). At week 26, the MCT-1-overexpressing tumor burdens were dramatically increased up to 26-fold in the maximal tumor masses compared with those of the controls. However, MCT-1 knockdown (shMCT-1#3) greatly reduced the tumor development of aggressive IV2–3 cells (1 × 106) after inoculation for 30 days (Fig. 3b), and the greatest difference in shMCT-1 tumor burdens showed a 28.3-fold reduction relative to scramble groups. MCT-1 and vimentin were increased but E-cadherin was reduced (Fig. 3c), together with enriched angiogenesis (CD31) and macrophages (F4/80) in the TME, as detected by immunohistostaining.
Fig. 3.
MCT-1 promotes breast tumor progression and M2 macrophage polarization. (a and b) MDA-MB-231 (control vs. MCT-1) and IV2–3 (scramble vs. shMCT-1#3) cells were injected into the mammary fat pad of nude mice. Tumor growth and burdens were analyzed at the indicated time. (c) Tumor immunohistology revealed the levels of MCT-1, E-cadherin, vimentin, CD31 (indicated by arrowheads) and F4/80 (enclosed in circles). Scale bars, 50 μm. (d) Immunohistochemistry characterized the tumor-associated CD163-positive M2 macrophages (enclosed in circles). The images were captured with 40X objective lens and macrophage numbers were counted (n = 6). (E and F) Levels of the M2-specific markers CD163 and CD206 were compared after THP-1 coculture with RPMI medium, MDA-MB-231 (control vs. MCT-1) or IV2–3 (scramble vs. shMCT-1#3) cells for 48 h. (g) The pan-macrophage (F4/80) and M1-like macrophage (CD86) markers were analyzed after THP-1 coculture with RPMI or with MDA-MB-231 cells (scramble vs. shMCT-1#3) for 48 h in a Boyden chamber. (h) THP-1 polarization into M2-like macrophages was signified by an increase in Arginase-1 and IL-10 after coculture with the indicated cells (control vs. MCT-1; scramble vs. shMCT-1#3). Different blots with normalized internal control were used to present the data of the experiment. (I and J) Quantitative RT-PCR analysis evaluated the CD86 and CD163 expression after THP-1 priming by RPMI or by the MDA-MB-231 cells (control vs. MCT-1) pretreated with an IL-6R mAb (tocilizumab) or stimulated with IL-6 for 48 h. The results are expressed as the mean ± SD (n = 3). One-way ANOVA with a post hoc two-tailed t-test was used to calculate the statistical significance of pairwise comparisons. (*p < 0.05; **p < 0.01; *** p < 0.001)
Tumor progression is prompted by M2 macrophages but repressed by M1 macrophages [31]. When the M1 marker (CD80) was assessed by immunohistochemistry, the CD80-positive M1 macrophages were more accumulated in the stroma of shMCT-1#3 tumors than in the scramble shRNA treated tumors (Additional file 4: Figure S4A). However, the CD80-positive M1 macrophages present in the stroma of MCT-1-overexpressing tumors were less than the control group (Additional file 4: Figure S4B). The CD163-positive M2 macrophages were more abundant in MCT-1-promoted tumors but were less in shMCT-1 tumors compared with the scramble control tumors (Fig. 3d), demonstrating MCT-1 impacts on the recruitment and polarization of TAMs. Further surveillance of THP-1 polarity in coculture with the MCT-1-overexpressed cells showed that the M2 markers (CD163 and CD206) were upregulated than co-cultured with control MDA-MB-231 cells or RPMI medium (Fig. 3e). However, the M2-like polarization was reduced by coculture with shMCT-1#3 cells relative to scramble shRNA-treated cells or RPMI medium (Fig. 3f), as reflected in CD163 and CD206 reduction. Upon coculture with shMCT-1#3 cells, THP-1 cells were preferentially polarized into pan-macrophages and M1-like macrophages (Fig. 3g), in which F4/80 and CD86 were significantly activated. Consistently, the M2 markers Arginase-1 and IL-10 in the polarized THP-1 cells were more induced after priming by the MCT-1-overexpressing cells (THP-1/MCT-1) than after priming by control cells (THP-1/control) (Fig. 3h), but Arginase-1 and IL-10 were greatly reduced in the THP-1 cells while cocultured with shMCT-1#3 cells (THP-1/shMCT-1) than with scramble control cells (THP-1/scramble).
Furthermore, when MDA-MB-231 (control vs, MCT-1) cells were pretreated with an IL-6R mAb (tocilizumab) and then co-cultured with THP-1 cells (Fig. 3I), the CD86 expression was induced in the polarized M1-like macrophages. However, the IL-6 pretreatment did not significantly increase CD86-positived M1 macrophage differentiation. In addition, the M2-like polarization revealed by CD163 expression was induced when THP-1 cells were cocultured with the IL-6-treated cancer cells but was inhibited when THP-1 cells were cocultured with tocilizumab-treated cancer cells (Fig. 3j). Accordingly, MCT-1-overexpressing breast cancer cells promotes M2-like macrophage polarization in vitro. Tocilizumab antagonizes the MCT-1 function to enhance M1 polarity, whereas IL-6 stimulates MCT-1 effect on M2 promotion.
MCT-1/IL-6/IL-6R signaling mediates breast cancer stemness
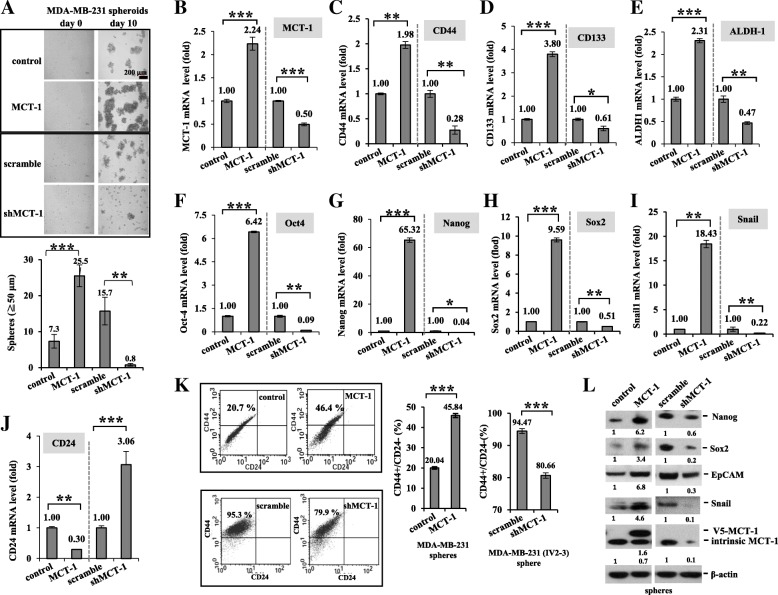
IL-6/Stat3 signaling promotes breast cancer stemness [32]. Investigating BCSCs derived from MDA-MB-231 cells, we first identified that MCT-1 overexpression promoted MDA-MB-231 mammospheriods (3.49-fold) but that were significantly reduced after MCT-1 depletion in IV2–3 (shMCT-1#3) (0.05-fold) compared with that of the control and the scramble groups (Fig. 4a). Cancer stemness marker CD44 level was also elevated through MCT-1 induction (1.98-fold) (Fig. 4b) but decreased upon MCT-1 knockdown (0.28-fold) than the controls in mammospheroids as detected by qRT-PCR (Fig. 4c). Similarly, MCT-1 promoted CD133 (Fig. 4d), ALDH-1 (Fig. 4e), Oct4 (Fig. 4f), Nanog (Fig. 4g), Sox2 (Fig. 4h) and Snail (Fig. 4i) mRNA levels in the mammospheres, but those were all greatly reduced in shMCT-1#3. Conversely, CD24 expression was increased in shMCT-1#3 mammospheres (3.06-fold) but suppressed upon MCT-1 induction (0.3-fold) (Fig. 4j). Analyzing the mammospheres by flow cytometry, the CD24(−)/CD44(+) subpopulations were more abundant with overexpressed MCT-1 (46.4%) than the controls (20.7%) (Fig. 4k). Contrarily, the abundant CD24(−)/CD44(+) subpopulations in the high metastatic IV2–3 mammospheres (scramble, 95.3%) were decreased by shMCT-1#3 (79.9%). Because the MDA-MB-231 (IV2–3) subline has been selected from two rounds of lung metastasis; thus, it enriched higher cancer stem cells (95.3%) than the MCT-1-overexpressing cells (46.4%). Consistently, Nanog (Fig. 4l), Sox2, EpCAM and Snail proteins were induced by MCT-1 but suppressed in shMCT-1#3.
Fig. 4.
MCT-1 promotes MDA-MB-231 breast cancer stemness. (a) MDA-MB-231 sphere formation at day 10 was studied in different MCT-1 circumstances (control vs. MCT-1; scramble vs. shMCT-1#3). Mammospheroids (≧50 μm in diameter) were measured. The mRNA levels of the indicated gene were quantified by qRT-PCR in day 10–14 mammospheres. (b) MCT-1. (c) CD44. (d) CD133. (e) ALDH-1. (f) Oct4. (G) Nanog. (h) Sox2. (i) Snail. (j) CD24. (k) Flow cytometry was used to evaluate CD44(+)/CD24(−) subpopulations in the mammospheres. The results are expressed as the mean ± SD (n = 3). One-way ANOVA with a post hoc two-tailed t-test was used to calculate the statistical significance of pairwise comparisons. (*p < 0.05; **p < 0.01; *** p < 0.001) (l) Nanog, Sox2, EpCAM and Snail proteins were evaluated in the mammospheres. Relative protein amounts were compared with the controls. Different blots with normalized internal control were used to present the data of the experiment
Also, we found that IL-6 treatment indeed further stimulated MDA-MB-231 mammosphere formation (Additional file 5: Figure S5A) along with increase of MCT-1 (Additional file 5: Figure S5B) CD44 (Additional file 5: Figure S5C), CD133 (Additional file 5: Figure S5D), ALDH-1 (Additional file 5: Figure S5E), Oct-4 (Additional file 5: Figure S5F), Sox2 (Additional file 5: Figure S5G) and Nanog (Additional file 5: Fig. 5h) mRNAs, particularly in an oncogenic MCT-1 background. Hence, IL-6 and MCT-1 collaboratively advance cancer stemness.
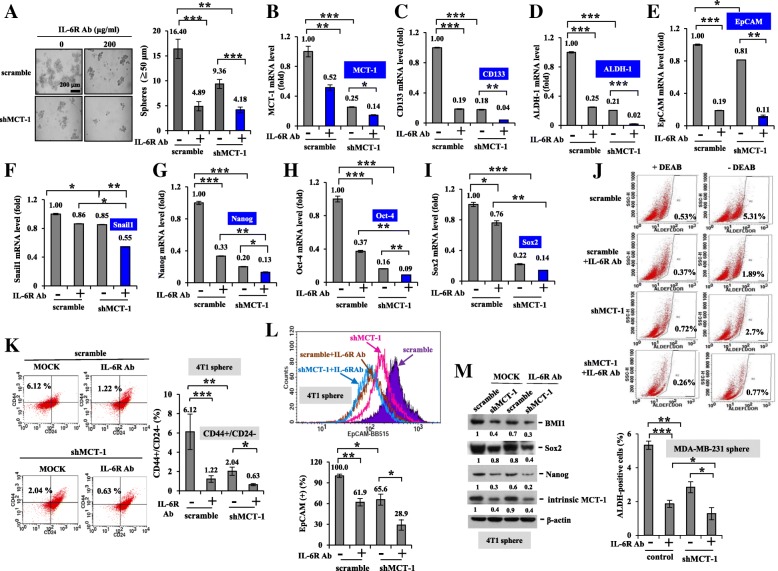
Fig. 5.
Tocilizumab and shMCT-1 synergistically inhibit breast cancer stemness. (a) The mammospheres derived from IV2–3 subline (scramble vs. shMCT-1#3) were evaluated upon tocilizumab (200 μg/ml) treatment for 20 days. The mRNA levels of the indicated gene were examined in MDA-MB-231 mammospheres and after tocilizumab challenge. (b) MCT-1. (c) CD133. (D) ALDH-1. (e) EpCAMP. (f) Snail. (g) Nanog. (H) Oct-4. (i) Sox2. (j) ALDH(+) cancer stem cells were detected by an ALDEFLUOR assay as DEAB inhibited ALDH activity, to accurately analyze ALDH activity in day 14 mammospheres and after tocilizumab treatment. (k) CD44-FITC and CD24-Alexa staining identified CD24(−)/CD44(+) subpopulations in the 4 T1 mammospheres (scramble and shMCT-1#3–28) and upon tocilizumab treatment for 6 days. (l) EpCAM(+) cancer stem cells were evaluated in the 4 T1 mammospheres and upon tocilizumab treatment for 6 days using EpCAM-BB515 staining. The results are expressed as the mean ± SD (n = 3). One-way ANOVA with a post hoc two-tailed t-test was used to calculate the statistical significance of pairwise comparisons. (*p < 0.05; **p < 0.01; *** p < 0.001) (m) The cancer stemness molecules in scramble and shMCT-1#3–28 cells were examined before and after tocilizumab treatment
Consistent with an IL-6R increase (Fig. 2e), IL-6R mRNA levels were elevated by MCT-1 but inhibited by shMCT-1#3 in mammospheres (Additional file 5: Figure S5I). Tocilizumab (IL-6R Ab) treatment suppressed MCT-1-induced mammosphere formation (Additional file 5: Figure S5J) combined with decrease of IL-6R (Additional file 5: Figure S5K), IL-6 (Additional file 5: Figure S5L), CD44 (Additional file 5: Figure S5M), ALDH-1 (Additional file 5: Figure S5N), EpCAM (Additional file 5: Figure S5O) and Oct-4 (Additional file 5: Figure S5P) mRNAs. Moreover, tocilizumab repressed the MCT-1-induced CD44(+)/CD24(−) subpopulations (55.7%) to the control degree (32.5%) (Additional file 5: Figure S5Q), confirming that IL-6R immunotherapy inhibited MCT-1-promoted cancer stemness.
Importantly, tocilizumab (IL-6R Ab) treatment further inhibited the MDA-MB-231 mammospheroids (Fig. 5a) as well as levels of MCT-1 (Fig. 5b), CD133 (Fig. 5c), ALDH-1 (Fig. 5d), EpCAM (Fig. 5e), Snail (Fig. 5f), Nanog (Fig. 5g), Oct-4 (Fig. 5h) and Sox2 (Fig. 5i) mRNAs in the shMCT-1#3 cellular background. These results indicate that MCT-1 induces cancer stemness via multiple ways in addition to IL-6R. MCT-1 and IL-6/IL-6R pathway can together advance cancer stemness effects. As aldehyde dehydrogenase (ALDH) activity reduced by diemethylamino-benzaldehyde (DEAB) (Fig. 5j), it specifically inhibited ALDH to accurately identify the ALDH(+) cancer stem cells in each group. The ALDH(+) cells were found to be fewer in the shMCT-1#3 group (2.7%) than in the scramble set (5.31%), and the ALDH(+) cells in the shMCT-1 condition were further suppressed by tocilizumab (0.77%).
Similarly, tocilizumab treatment of 4 T1 cells further repressed MCT-1 (Additional file 6: Figure S6A), CD133 (Additional file 6: Figure S6B), Snail (Additional file 6: Figure S6C), Nanog (Additional file 6: Figure S6D) and Oct-4 (Additional file 6: Figure S6E) mRNA levels, particularly in shMCT-1 mammospheres. Furthermore, shMCT-1 (#3–28) reduced the CD24(−)/CD44(+) subpopulations (2.04%) than that identified in the scramble set (6.12%) of the 4 T1 mammospheres (Fig. 5k), and tocilizumab further reduce the CD24(−)/CD44(+) subpopulations in the shMCT-1 (#3–28) background (0.63%). Using EpCAM to define 4 T1 cancer stemness (Fig. 5l), EpCAM(+) cells were reduced in shMCT-1 (#3–28) mammospheres (65.6%) than in the scramble control (100%), and tocilizumab further decreased the EpCAM(+) populations (28.9%) in the shMCT-1 background. In consistence, tocilizumab further decreased BMI-1 (Fig. 5m), Sox2 and Nanog amounts in the shMCT-1 (#3–28) mammospheres. Consequently, IL-6R immunotherapy inhibits the oncogenicity of MCT-1 and cooperates with MCT-1 knockdown to profoundly suppress cancer stemness.
miR-34a inhibits MCT-1-promoted IL-6R expression and M2 polarization
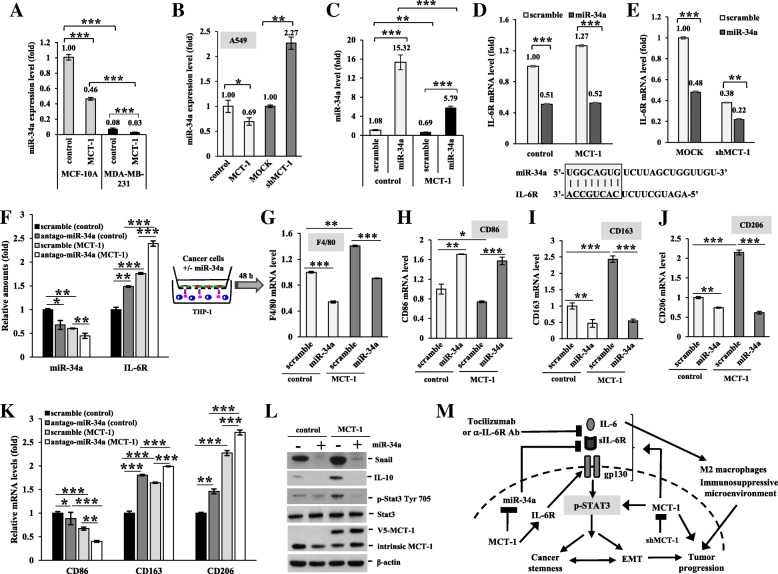
MicroRNAs (miRNAs) communicate between tumor cells and their microenvironment; thus, the miRNAs are used for anti-cancer therapy/regimens [33]. To identify global microRNA profiling in the MCT-1 pathway, RNA samples isolated from the tumors of TNBC cells (MDA-MB-468) [18] were subjected to the direct, digital counting of miRNA levels by an nCounter miRNA Expression Assay (NanoString Technologies), as described before [34]. Screening of the 800-miRNA panel revealed 21 downregulated miRNAs and 43 upregulated miRNAs in the shMCT-1 tumor compared with the scramble group (Additional file 7: Figure S7A). The levels of miRNAs altered in the shMCT-1 tumor are listed (Additional file 7: Figure S7B). Among the upregulated 43 miRNAs, the tumor-suppressive miRNAs (miR-34a, miR-99a, miR-125b) were increased in shMCT-1 (#3) of IV2–3 subline as detected by qRT-PCR analysis (Additional file 7: Figure S7C and D), showing that shMCT-1 induced the tumor suppressor miRNAs.
We found that miR-34a levels were more abundant in MCF-10A cells than in MDA-MB-231 cells but significantly repressed upon MCT-1 overexpression (Fig. 6a). Intriguingly, MCT-1 depletion in IV2–3 subline (p53-mutant) restored the miR-34a level independent of p53 function (Additional file 7: Figure S7D) similar to that of invasive lung cancer A549 cells (p53-wildtype) upon MCT-1 knockdown (Fig. 6b). Even after introducing pre-miR-34a oligonucleotides into MDA-MB-231 cells (Fig. 6c), the mature miR-34a levels were still much suppressed in the MCT-1-overexpressing cells (5.79-fold) than in control cells (15.32-fold). The IL-6R/STAT3/miR-34a loop mediates cancer invasion and metastasis [8]. Upon miR-34a re-expression, the MCT-1-induced IL-6R expression was decreased (Fig. 6d) and IL-6R was further abrogated in shMCT-1#3 cells (Fig. 6e). These indicate that miR-34a and shMCT-1 can synergistically inhibit IL-6R function. To further investigate whether inhibition of miR-34a induces IL-6R expression in MCT-1 pathway, we found that antago-miR-34a transfection into the cells effectively suppressed miR-34a level that further promoted IL-6R in MCT-1 overexpression background (Fig. 6f), comparing with scramble-miR transfectant. Because overexpressing MCT-1 in a loss-of-function miR34 condition still highly induces IL-6R, MCT-1 promotes IL-6R at least in part independently of miR-34.
Fig. 6.
miR-34a suppresses IL-6R expression and M2-like macrophage polarization. (a and b) MiR-34a levels were examined by qRT-PCR analysis in MCF-10A, MDA-MB-231 and A549 cells with different MCT-1 expression conditions. (c) The mature miR-34a levels were compared in the MDA-MB-231 cells (control vs. MCT-1) after transfection of pre-miR-34a and scramble-miR oligonucleotides. (d and e) IL-6R mRNA levels were compared in different MCT-1 backgrounds (control vs. MCT-1; scramble vs. shMCT-1#3) after transfection of pre-miR-34a and scramble-miR oligonucleotides. (f) IL-6R mRNA levels were compared in different MCT-1 backgrounds (control vs. MCT-1; scramble vs. shMCT-1#3) after transfection of pLemiR-antago-miR-34a or pLemiR-scramble. The MDA-MB-231 cells were virally transfected with pLemiR-scramble or pLemiR-pre-miR-34a, and then were co-incubated with THP-1 for 48 h to analyze the macrophage differentiation markers. (g) Pan-macrophages (indicated by F4/80). (h) M1-like macrophages (shown by CD86). (i and j) M2-like macrophages (revealed by CD163 and CD206). The results are expressed as the mean ± SD (n = 3). One-way ANOVA with a post hoc two-tailed t-test was used to calculate the statistical significance of pairwise comparisons. (*p < 0.05; **p < 0.01; *** p < 0.001). (k) THP-1 differentiation into M1 (CD86) or M2 (CD163 and CD206) macrophages were examined by qRT-PCR after culture with the MDA-MB-231 cells (control vs. MCT-1) introduced with pLemiR-antago-miR-34a or pLemiR-scramble. (l) The amounts of Snail, IL-10 and Stat3 were inspected in the THP-1 cells. Different blots with normalized internal control were used to present the data of the experiment. (m) Implications of MCT-1-targeted therapies. The oncogenic MCT-1 pathway in TNBC can be retracted by MCT-1 inhibitor(s), IL-6R immunotherapy or miR-34a expression, together they can further prevent EMT progression, cancer stemness, M2 macrophage polarization and tumor progression
To explore the miR-34a role in macrophage differentiation, MDA-MB-231 cells were virally transduced with pLemiR-scramble-miR (scramble) or with pLemiR-pre-miR-34a (miR-34a) followed by priming THP-1 cells. The miR-34a-expressed cancer cells more induced THP-1 polarization into M1-like CD86-positive macrophages than that of the scramble cells (Fig. 6h). Inversely, miR-34a induction suppressed the polarization of pan-macrophages (F4/80) (Fig. 6g) and M2-like macrophages promoted by MCT-1, as characterized by decrease in CD163 (Fig. 6i) and CD206 (Fig. 6j). However, M2 macrophage markers (CD163 and CD206) were advanced and M1 macrophage marker (CD86) was suppressed in THP-1 cells after co-culture with the cancer cells transfected with antago-miR-34a compared with scramble-miR transfectant (Fig. 6k). Since CD163 and CD206 expression are constitutively activated while overexpressing MCT-1 in a loss-of-function miR34 condition, MCT-1 also promotes the M2 markers independent of miR-34 pathway.
Similarly, the inducers of M2 differentiation (Snail, IL-10 and phospho-active Stat3) which were promoted by MCT-1 were significantly reduced upon miR-34a expression (Fig. 6l). Hence, miR-34a expression in TNBC cells mediates M1 polarization but antago-miR-34a promotes M2 plasticity.
Collectively, oncogenic MCT-1 activation stimulates the IL-6/IL-6R/Stat3 axis that enhances EMT progression (Fig. 6m), cancer stemness and M2 polarization in the TNBC system. IL-6 enhanced the effect of MCT-1 on EMT plasticity, M2 polarity and cancer stemness, which were suppressed by tocilizumab. The combination of MCT-1 and IL-6/IL-6R pathway cooperatively enhanced cancer stemness and the oncogenic effects. Furthermore, targeting MCT-1 induced miR-34a that may reprogram EMT and macrophage plasticity and inhibit TNBC stemness and tumor progression. Therefore, MCT-1 inhibition combined with IL-6R antagonist or miR-34a expression may further renovate non-BCSC effect and tumor-suppressive M1 macrophages in TNBC.
Discussion
Inflammatory microenvironment plays an important role for cancer progression [35]. Immunotherapy is emerging as a novel promising strategy for TNBC treatment [36]. The TNBC cells with MCT-1 overexpression secrete more inflammatory cytokines IL-6, MCP-1 and GM-CSF. GM-CSF and IL-6 influences TAM polarity, and MCP-1 functionally promotes osteoclast differentiation from monocytes [28, 37, 38]. Moreover, IL-6 and MCP-1/CCL2 regulate the fate of CSCs and the TME [39]. Therefore, immunotherapeutic interventions of the cytokine pathways may modify the TME and eradicate CSCs.
Serum IL-6 levels are increased with advanced stages and related to poor survival in various cancers [40], and IL-6 drives breast cancer metastasis and stemness [41]. IL-6/IL-6R antagonists as anti-breast cancer agents have not been broadly investigated and are even less studied in TNBC. Tocilizumab, a recent FDA-approved humanized mAb used to treat autoimmune and inflammatory diseases, has been proposed to inhibit the trastuzumab-resistant HER2(+) breast cancer [42].
Abundant IL-6 released from aggressive cancer cells stimulates angiogenesis and tumor evasion from immune surveillance [43]. Nevertheless, IL-6 promotes antitumor effect by boosting T-cell immunity and by trafficking antitumor T cells to lymph nodes and tumor sites, executing the cytotoxic effects. It is unclear whether MCT-1 also influences Th1-Th2 polarization. Reflecting the Th1-Th2 polarization of T cells [44], the activation of M1 (pro-inflammatory) and M2 (anti-inflammatory) macrophages are functionally modified by Th1 and Th2 cytokines. Th1 cytokines such as IFN-γ and GM-CSF induce M1 polarization, which produces pro-inflammatory cytokines (IL-1β, IL-6, IL-12, IL-23 and TNF-α). Th2 cytokines such as IL-4 and IL-13 promote M2 polarization, which produces anti-inflammatory cytokines (IL-10 and TGF-β).
YY1 transcriptionally activates IL-6 gene expression [45], and the EGFR signaling triggers IL-6 production via NF-kB activation [46]. Oncogenic MCT-1 activation promotes the expression of YY1 and EGFR [23], suggesting that MCT-1 may increase IL-6 expression via the YY1-EGFR signaling amplification. NOTCH activation through NO facilitates constitutive IL-6-dependent STAT3 activation [32], promoting breast cancer stemness. MCT-1 stimulated IL-6/Stat3 signaling (Fig. 2e), suggesting that MCT-1 may also stimulate the NO/NOTCH pathway to mediate breast cancer metastasis and recurrence. In addition, systematic administration of IL-6/IL-6R antagonist(s) with MCT-1 inhibitor(s) may promote immune cell infiltration to advance therapeutics against tumor heterogeneity and aggressiveness, with fewer adverse effect(s).
MCT-1 induces PD-L1 but reduces miR-34a. Targeting PD-L1 by miR-34a in the cancer cells prevent the PD-1/PD-L1 interaction that increases anti-tumor activity [47, 48]. miR-34a inhibits cancer stemness via targeting CD44 [49]; miR-34a expression inhibits TGF-β-induced EMT and downregulates Snail [50], Slug and ZEB1 as well as the stemness factors (BMI1, CD44, CD133, OLFM4 and c-MYC). Reciprocally, Snail and ZEB1 repress the miR-34a function to promote EMT [50, 51]. To sustain the immune escape mechanism, the TME recruits and changes myeloid cells to TAMs [52], dendritic cells, myeloid-derived suppressor cells and neutrophils. Macrophage colony-stimulating factor (M-CSF) induces M2 polarization [53], and miR-34a targets receptor of M-CSF, which regulates dendritic cell maturation to maintain a proper immune balance in anti-Th2 response, immune stimulation and tumor resistance. We now identify that miR-34a expression in p53-mutant TNBC cells promotes M1 polarization, emphasizing that miR-34a potentially modifies the tumor immunity and heterogenicity. MCT-1 antagonist combined with miR-34a expression may alter the polarity and activation of the immune cells, thus improving the efficacy of TNBC treatment.
Conclusions
MCT-1/miR-34a/IL-6/IL-6R is a novel signaling axis identified in TNBC. MCT-1 inhibition combined with IL-6/IL-6R immunotherapy or with miR-34a expression would be a new stratagem for administration of TNBC. Better understanding the circuits between cytokines and microRNAs orchestrated by the oncogenic activity will facilitate breast cancer diagnosis, prevention and therapeutics.
Methods
THP-1 polarization and cancer cell invasion
Cancer cells (1 × 105) were seeded into the upper chamber of Falcon® Cell Culture Inserts (Corning, Corning, NY) and cocultured with THP-1 monocytes (1 × 106) in the bottom chamber for 48 h. A control experiment was conducted as THP-1 cells co-incubated with RPMI medium alone. The markers of pan-macrophages (F4/80), M1 macrophage (CD86) and M2 macrophages (CD163 and CD206) were analyzed in the primed THP-1 cells by qRT-PCR using the synthesized primers (MDBio) listed in Additional file 8: Table S1.
Moreover, MDA-MB-231 cells (1 × 105) were pretreated with IL-6 (50 ng/ml), IL-6R mAb (0.5 μg/ml) (PeproTech, Rocky Hill, NJ) or tocilizumab (150 μg/ml) (CHUGAI, Tochigi, Japan) for 24 h; next, the pretreated MDA-MB-231 cells (2 × 104) were used to prime THP-1 cells (1 × 106) in Cell Culture Inserts for 48 h. The preconditioned THP-1 cells with 10% FBS/RPMI or the medium alone were placed into the lower chamber of Cell Invasion Inserts (Corning), and the invasiveness of serum-free parental MDA-MB-231 cells (2 × 104) in the upper chamber were analyzed for 24 h. Supplementary methods can be found in Additional file 9.
Cytokine array analysis
Cells were seeded in 10 cm plates (1 × 106 cells/plate) with serum-free RPMI for 24 h. The condition medium (CM) was centrifuged for 20 min at 1000 x g at 4 °C and then collected the supernatant to perform the assay. Human cytokine array C6 membranes (RayBiotech, Norcross, GA) were incubated overnight with 1 ml of CM at 4 °C. After washing with the buffer, the membranes were incubated overnight with biotin-conjugated Abs (human cytokine antibody cocktail), washed thrice, reacted with horseradish peroxidase-conjugated streptavidin in blocking buffer for 2 h and performed photography on X-ray film.
Analysis of the secretory IL-6 and IL-6R
Human IL-6 and IL-6R ELISA MAX™ Deluxe (BioLegend, San Diego, CA) analyzed the secreted IL-6 amounts. Briefly, the indicated cells were seeded in 6-well plates (2 × 105 cells/well) with serum-free RPMI medium for 24 h. The condition medium was centrifuged for 20 min at 1000×g at 4 °C and then collected the supernatant to carry out the assay. The ELISA plates were coated with the diluted IL-6 or IL-6R A, incubated overnight at 4 °C, washed 4 times with the buffer (0.05% Tween-20 in PBS) and incubated with the diluted buffer (included in the kit) for 1 h. After washing for 4 times, the plates were incubated with the diluted standards (included in the kit) and the 100 μl conditioned medium for 2 h, rinsed 4 times, incubated with the diluted detection Ab (included in the kit) for 1 h, washed 5 times and incubated with 3, 3′, 5, 5′-Tetramethylbenzidine substrate solution (included in the kit) in the dark for 15 min. Reactions were stopped and detected the absorbance at 450 nm within 15 min. The results were normalized with the numbers of the seeded cancer cells.
Cancer stemnness analysis
A single-cell suspension was cultured on 6-well ultralow-attachment plate (Corning) at a density of 4 × 104 cells/well in serum-free DMEM/F12 medium with 1% L-glutamine, 1% penicillin/streptomycin, 2% B27 (Invitrogen), 20 ng/ml EGF (Sigma-Aldrich) and 20 ng/ml FGFb (PeproTech). IL-6 (50 μg/ml) (PeproTech) or tocilizumab (200 μg/ml) was added to examine the mammosphere formation. Mammospheroids were photographed at a magnification of 200× using a Nikon DIAPHOT300 microscope at the indicated time. Mammosphere cells (1 × 105) were further stained with anti-human CD24-PE (BD Pharmingen), anti-human CD44-FITC (BD Pharmingen), CD24-Alexa 647 (BD Pharmingen) or EpCAM-BB515 (BD Pharmingen) for 1 h at 4 °C, PBS rinsed and resuspended in 500 μl PBS. CD44-FITC and EpCAM-BB515 were excited at 490 nm, and the emissions were determined by FL1 PMT (515–545 nm bandpass filter). CD24-Alexa 647 was excited at 633 nm, and the emissions were determined by FL-4 PMT (653–669 nm bandpass filter). BD FACSCalibur flow cytometry (BD Bioscience) and Cell Quest software (BD Biosciences) were used to identify CD44(+)/CD24(−) and EpCAM(+) subpopulations.
Tumor progression
Six to eight week-old female BALB/c nude mice (BALB/cAnN.Cg-Foxn1nu/CrlNarl) were purchased from the National Laboratory Animal Center (Taipei, Taiwan), according to the Animal Use Protocol approved by the National Health Research Institutes (NHRI-IACUC-106012-A). MDA-MB-231 cells (1 × 105) or MDA-MB-231 (IV2–3) cells (1 × 106) bearing a pcDNA3.1-luciferase reporter were injected into the fourth mammary fat pads of the mice for a tumor progression study. Luminescent tumor images were checked weekly after the intraperitoneal injection of luciferin (150 mg/kg) (PerkinElmer, Waltham, MA) for 10 min and detected by a Xenogen IVIS 200 bioluminescence imaging system (Caliper LifeSciences, Hopkinton, MA).
Clinical study
Human breast tissue microarrays (BR1503c, BR953 and BRN801a) were obtained from US Biomax (Rockville, MD), the TNBC tissue microarray was obtained from Pantomics (BRC964) (Richmond, CA) and the adjacent normal breast tissue microarray was obtained from SOBC (Hbre-Duc052Bch-01) (Pudong, Shanghai, China). Samples were stained with MCT-1 Ab (1:200, GeneTex, GTX117793) using a Discovery XT Automated IHC/ISH Slide Staining System (Ventana Medical System, Tucson, AZ) and an UltraView Universal DAB Detection Kit (Ventana Medical System). The results were classified according to the clinical and pathology information provided by the companies.
The MCT-1, IL-6 and IL-6R mRNA expression levels in breast carcinomas versus normal breast tissues were analyzed in the Oncomine database (http://www.oncomine.org). The Kaplan-Meier Plotter (http://www.kmplot.com) analyzed the probability of OS and RFS in breast cancer patients. The survival curve was exported using GraphPad Prism software. MCT-1, IL-6 and IL-6R mRNA levels were quantified by using TissueScan Breast Cancer Tissue qPCR Panels (I, III and IV) (OriGene Technologies, Inc., Rockville, MD). Relative mRNA levels were calculated: ΔΔCT = ΔCt cancer-ΔCt normal tissue. The fold change in the gene was calculated using the formula 2−ΔΔCT.
MicroRNA profiling
The nCounter® Human v2 miRNA Panel (nanoString, Seattle, WA) containing 800 unique miRNA barcodes was used. RNA samples were extracted from the MDA-MB-468 tumors (MOCK vs. shMCT-1) by TRIzol™ reagent (Invitrogen). Total RNAs (100 ng) were used as input for the nanoString platform. Mature miRNAs were multiplied by annealing to a human-specific tag sequence (miRtag) via melting-temperature-controlled splinted ligation onto the 3′-end. Excess unligated miRtags were then removed by enzymatic purification, and the resulting material was hybridized at 65 °C for 16 h with a panel of miR: tag-specific nCounter capture and barcoded reporter probes. The raw data were normalized by 6 positive-control and 8 negative-control probe pairs. All the samples were analyzed in triplicate. The miRNA amounts were quantified by a nanoString nCounter Digital Analyzer and gene-expression system.
Expression of miR-34a and antago-miR-34a
Scramble-miR and pre-miR-34a oligonucleotides (Biotools, New Taipei City, Taiwan) were transiently transfected into cells using Lipofectamine 3000 (Thermo Fisher Scientific) for 48 h. The pre-miR-34a was cloned from pcDNA3-miR-34a using the primers (forward: 5′-ggctcgagTAGTTGCCTGGG CTGGTCTT; reverse: 5′-gggcggccgcCCTGTGCCTTTTTCCTTCC). The thermal cycling conditions were conducted at 94 °C for 2 min, followed by 35 cycles at 94 °C for 15 s, 55 °C for 1 min and 68 °C for 1 min. The amplicon was constructed into the XhoI and NotI sites of a pLemiR-NS (nonspecific hairpin) vector (Open Biosystem, Huntsville, AL, USA). The scramble-miR was also constructed into a pLemiR-NS vector. The 293 T cells were transfected with pLemiR-pre-miR-34a and pLemiR-scramble-miR using the TransIT-LT1 Transfection Reagent (Mirus Bio LLC, Madison, WI). The lentiviral supernatants were infected the indicated cells.
The antago-miR-34a was constructed by the primers (forward: 5′-gatccTGGCCAGTGTCTTAGCTGGTTGTttcaagagaACAACCAGCTAAGACACTGGCCAttttt; reverse:5′-agcttaaaaaTGGCCAGTGTCTTAGCTGGTTGTtctcttgaaACAACCAGCTAAGACACTGGCCA). The primers were annealed in the buffer (0.1 M potassium acetate, 30 mM HEPES KOH, and 2 mM magnesium acetate) at 95 °C for 2 min and then cool down to 25 °C within 50 min. The products were cloned into the BamHI and HindIII sites of a pRNA-U6.1 vector (GeneScript, Piscataway, NJ) and confirmed by DNA sequencing. Cells were transfected with Lipofectamine 3000 reagent (Thermo Fisher Scientific) according to the manufacturer’s protocol.
Additional files
Figure S1. MCT-1 is a prognostic marker of human breast carcinoma. (PDF 215 kb)
Figure S2. MCT-1 overexpression increases size of the MCF-10A acini. (PDF 223 kb)
Figure S3. MCT-1 expression regulates MMP activity and breast cancer cell movements. (PDF 260 kb)
Figure S4. CD80-positive M1 macrophages enriched in the TME after MCT-1 knockdown. (PDF 289 kb)
Figure S5. MCT-1/IL-6/IL-6R signaling promotes breast cancer stemness. (PDF 259 kb)
Figure S6. Targeting MCT-1 and tocilizumab treatment together suppress 4 T1 cancer stemness. (PDF 144 kb)
Figure S7. MicroRNA profiling in the MCT-1 pathway. (PDF 48 kb)
Table S1. Primer sequences. (PDF 104 kb)
Supplementary Methods. Cell culture. Plasmid construction and transfection. Targeting MCT-1 gene. Targeting IL-6 gene. Antibodies (Abs) and protein analysis. MCF-10A acinar morphogenesis. Cell invasion and migration assays. Gelatin zymography assay. Cell fractionation. Quantitative RT-PCR analysis of cancer stemness markers. ALDEFLUOR assay. Immunohistochemistry study. Quantification of miR-34a levels. Statistical analysis. (PDF 227 kb)
Acknowledgements
We thank Dr. C.C. Hong for providing the pcDNA3-miR-34a vector (National Health Research Institutes) and thank Dr. L.H. Wang (China Medical University) for giving valuable comments and providing MDA-MB-231 subline (IV2-3) as a gift.
Funding
This work was funded by Ministry of Science and Technology of Taiwan (MOST) 106–2320-B-400-002 HLH) and supported by the National Health Research Institutes MG-107-PP-05 (HLH) and EX107-10523EI (YCT).
Availability of data and materials
The datasets used for the current study are available from the corresponding author on reasonable request.
Abbreviations
- BCSCs
breast cancer stem cells
- CM
conditioned medium
- DCIS
ductal carcinoma in situ
- DEAB
diemethylamino-benzaldehyde
- ELISA
enzyme-linked immunosorbent assay
- EMT
epithelial-mesenchymal transition
- gp130
glycoprotein 130
- IDC
invasive ductal carcinoma
- IL-6
interleukin-6
- IL-6R
IL-6 receptor
- M-CSF
macrophage colony-stimulating factor
- MCT-1
Multiple Copies in T-cell Malignancy 1
- miR-34a
microRNA-34a
- MMPs
matrix metalloproteinases
- OS
overall survival
- PD-L1
programmed death-ligand 1
- qRT-PCR
quantitative RT-PCR
- RFS
recurrence-free survival
- sIL-6R
soluble IL-R
- Stat3
activate signal transducer and activator of transcription 3
- TAMs
tumor-associated macrophages
- TIMs
tissue inhibitors of metalloproteinase inhibitors
- TME
tumor microenvironment
- TNBC
triple-negative breast cancer
Authors’ contributions
Conception, study supervision, writing and revision of the manuscript: H.L. Hsu. Methodology development: Y.S. Weng, H. Y Tseng, Y.A. Chen, P.C. Shen. Data acquisition, analysis and interpretation: Y.S. Weng, H.Y. Tseng, Y.A. Chen, P.C. Shen, A.T.A. Haq, L.M. Chen, Y.C. Tung.
Ethics approval and consent to participate
All authors approved and directly participated in the planning, execution and/or analysis of the data presented herein. The animal studies were conducted in accordance with the Animal Use Protocol approved by the National Health Research Institutes. Clinical studies were approved by the Institutional Review Board (IRB) of the National Health Research Institutes.
Consent for publication
The content of this manuscript has not been previously published and is not under consideration for publication elsewhere.
Competing interests
The authors declare no conflicts of interest.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yueh-Shan Weng, Email: joyceweng@nhri.org.tw.
Hong-Yu Tseng, Email: hytseng@nhri.org.tw.
Yen-An Chen, Email: neuropt438@nhri.org.tw.
Pei-Chun Shen, Email: green9250@nhri.org.tw.
Aushia Tanzih Al Haq, Email: 054094@nhri.org.tw.
Li-Mei Chen, Email: limei@nhri.org.tw.
Yi-Chung Tung, Email: tungy@gate.sinica.edu.tw.
Hsin-Ling Hsu, Phone: 886-37-246-166, Email: hsinling88@nhri.org.tw.
References
- 1.Pradella D, Naro C, Sette C, Ghigna C. EMT and stemness: flexible processes tuned by alternative splicing in development and cancer progression. Mol Cancer. 2017;16(8). [DOI] [PMC free article] [PubMed]
- 2.Lopez-Soto A, Gonzalez S, Smyth MJ, Galluzzi L. Control of metastasis by NK cells. Cancer Cell. 2017;32:135–154. doi: 10.1016/j.ccell.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 3.Solinas G, Schiarea S, Liguori M, Fabbri M, Pesce S, Zammataro L, Pasqualini F, Nebuloni M, Chiabrando C, Mantovani A, Allavena P. Tumor-conditioned macrophages secrete migration-stimulating factor: a new marker for M2-polarization, influencing tumor cell motility. J Immunol. 2010;185:642–652. doi: 10.4049/jimmunol.1000413. [DOI] [PubMed] [Google Scholar]
- 4.Solinas G, Germano G, Mantovani A, Allavena P. Tumor-associated macrophages (TAM) as major players of the cancer-related inflammation. J Leukoc Biol. 2009;86:1065–1073. doi: 10.1189/jlb.0609385. [DOI] [PubMed] [Google Scholar]
- 5.Santoni M, Romagnoli E, Saladino T, Foghini L, Guarino S, Capponi M, Giannini M, Cognigni PD, Ferrara G, Battelli N. Triple negative breast cancer: key role of tumor-associated macrophages in regulating the activity of anti-PD-1/PD-L1 agents. Biochim Biophys Acta. 2018;1869:78–84. doi: 10.1016/j.bbcan.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 6.Goswami KK, Ghosh T, Ghosh S, Sarkar M, Bose A, Baral R. Tumor promoting role of anti-tumor macrophages in tumor microenvironment. Cell Immunol. 2017;316:1–10. doi: 10.1016/j.cellimm.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 7.Kampan NC, Xiang SD, McNally OM, Stephens AN, Quinn MA, Plebanski M. Immunotherapeutic Interleukin-6 or Interleukin-6 receptor blockade in cancer: challenges and opportunities. Curr Med Chem. 2017. [DOI] [PubMed]
- 8.Rokavec M, Oner MG, Li H, Jackstadt R, Jiang L, Lodygin D, Kaller M, Horst D, Ziegler PK, Schwitalla S, et al. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J Clin Invest. 2014;124:1853–1867. doi: 10.1172/JCI73531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noman AS, Uddin M, Chowdhury AA, Nayeem MJ, Raihan Z, Rashid MI, Azad AK, Rahman ML, Barua D, Sultana A, et al. Serum sonic hedgehog (SHH) and interleukin-(IL-6) as dual prognostic biomarkers in progressive metastatic breast cancer. Sci Rep. 2017;7:1796. doi: 10.1038/s41598-017-01268-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heo TH, Wahler J, Suh N. Potential therapeutic implications of IL-6/IL-6R/gp130-targeting agents in breast cancer. Oncotarget. 2016;7:15460–15473. doi: 10.18632/oncotarget.7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kitamura H, Ohno Y, Toyoshima Y, Ohtake J, Homma S, Kawamura H, Takahashi N, Taketomi A. Interleukin-6/STAT3 signaling as a promising target to improve the efficacy of cancer immunotherapy. Cancer Sci. 2017;108:1947–1952. doi: 10.1111/cas.13332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo C, Chen Y, Gao W, Chang A, Ye Y, Shen W, Luo Y, Yang S, Sun P, Xiang R, Li N. Liposomal nanoparticles carrying anti-IL6R antibody to the tumour microenvironment inhibit metastasis in two molecular subtypes of breast Cancer mouse models. Theranostics. 2017;7:775–788. doi: 10.7150/thno.17237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schleich S, Strassburger K, Janiesch PC, Koledachkina T, Miller KK, Haneke K, Cheng YS, Kuechler K, Stoecklin G, Duncan KE, Teleman AA. DENR-MCT-1 promotes translation re-initiation downstream of uORFs to control tissue growth. Nature. 2014;512:208–212. doi: 10.1038/nature13401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skabkin MA, Skabkina OV, Dhote V, Komar AA, Hellen CU, Pestova TV. Activities of Ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev. 2010;24:1787–1801. doi: 10.1101/gad.1957510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lomakin IB, Stolboushkina EA, Vaidya AT, Zhao C, Garber MB, Dmitriev SE, Steitz TA. Crystal structure of the human ribosome in complex with DENR-MCT-1. Cell Rep. 2017;20:521–528. doi: 10.1016/j.celrep.2017.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weisser M, Schafer T, Leibundgut M, Bohringer D, Aylett CHS, Ban N. Structural and functional insights into human re-initiation complexes. Mol Cell. 2017;67:447–456 e447. doi: 10.1016/j.molcel.2017.06.032. [DOI] [PubMed] [Google Scholar]
- 17.Shih HJ, Chu KL, Wu MH, Wu PH, Chang WW, Chu JS, Wang LH, Takeuchi H, Ouchi T, Hsu HL. The involvement of MCT-1 oncoprotein in inducing mitotic catastrophe and nuclear abnormalities. Cell Cycle. 2012;11:934–952. doi: 10.4161/cc.11.5.19452. [DOI] [PubMed] [Google Scholar]
- 18.Wu MH, Chen YA, Chen HH, Chang KW, Chang IS, Wang LH, Hsu HL. MCT-1 expression and PTEN deficiency synergistically promote neoplastic multinucleation through the Src/p190B signaling activation. Oncogene. 2014;33:5109–5120. doi: 10.1038/onc.2014.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu HL, Choy CO, Kasiappan R, Shih HJ, Sawyer JR, Shu CL, Chu KL, Chen YR, Hsu HF, Gartenhaus RB. MCT-1 oncogene downregulates p53 and destabilizes genome structure in the response to DNA double-strand damage. DNA Repair (Amst) 2007;6:1319–1332. doi: 10.1016/j.dnarep.2007.02.028. [DOI] [PubMed] [Google Scholar]
- 20.Kasiappan R, Shih HJ, Chu KL, Chen WT, Liu HP, Huang SF, Choy CO, Shu CL, Din R, Chu JS, Hsu HL. Loss of p53 and MCT-1 overexpression synergistically promote chromosome instability and tumorigenicity. Mol Cancer Res. 2009;7:536–548. doi: 10.1158/1541-7786.MCR-08-0422. [DOI] [PubMed] [Google Scholar]
- 21.Kasiappan R, Shih HJ, Wu MH, Choy C, Lin TD, Chen L, Hsu HL. The antagonism between MCT-1 and p53 affects the tumorigenic outcomes. Mol Cancer. 2010;9:311. doi: 10.1186/1476-4598-9-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shih HJ, Chen HH, Chen YA, Wu MH, Liou GG, Chang WW, Chen L, Wang LH, Hsu HL. Targeting MCT-1 oncogene inhibits Shc pathway and xenograft tumorigenicity. Oncotarget. 2012;3:1401–1415. doi: 10.18632/oncotarget.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tseng HY, Chen YA, Jen J, Shen PC, Chen LM, Lin TD, Wang YC, Hsu HL. Oncogenic MCT-1 activation promotes YY1-EGFR-MnSOD signaling and tumor progression. Oncogenesis. 2017;6:e313. doi: 10.1038/oncsis.2017.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 25.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/S1046-2023(03)00032-X. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Yan W, Chen X. Mutant p53 disrupts MCF-10A cell polarity in three-dimensional culture via epithelial-to-mesenchymal transitions. J Biol Chem. 2011;286:16218–16228. doi: 10.1074/jbc.M110.214585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan SH, Huang WC, Chang JW, Chang KJ, Kuo WH, Wang MY, Lin KY, Uen YH, Hou MF, Lin CM, et al. MicroRNA-149 targets GIT1 to suppress integrin signaling and breast cancer metastasis. Oncogene. 2014;33:4496–4507. doi: 10.1038/onc.2014.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu XL, Duan W, Su CY, Mao FY, Lv YP, Teng YS, Yu PW, Zhuang Y, Zhao YL. Interleukin 6 induces M2 macrophage differentiation by STAT3 activation that correlates with gastric cancer progression. Cancer Immunol Immunother. 2017;66:1597–1608. doi: 10.1007/s00262-017-2052-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones SA, Rose-John S. The role of soluble receptors in cytokine biology: the agonistic properties of the sIL-6R/IL-6 complex. Biochim Biophys Acta. 2002;1592:251–263. doi: 10.1016/S0167-4889(02)00319-1. [DOI] [PubMed] [Google Scholar]
- 30.Lazzaro L, Tonkin BA, Poulton IJ, McGregor NE, Ferlin W, Sims NA. IL-6 trans-signalling mediates trabecular, but not cortical, bone loss after ovariectomy. Bone. 2018;112:120–127. doi: 10.1016/j.bone.2018.04.015. [DOI] [PubMed] [Google Scholar]
- 31.Genard G, Lucas S, Michiels C. Reprogramming of tumor-associated macrophages with anticancer therapies: radiotherapy versus chemo- and immunotherapies. Front Immunol. 2017;8:828. doi: 10.3389/fimmu.2017.00828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng D, Tanikawa T, Li W, Zhao L, Vatan L, Szeliga W, Wan S, Wei S, Wang Y, Liu Y, et al. Myeloid-derived suppressor cells endow stem-like qualities to breast Cancer cells through IL6/STAT3 and NO/NOTCH cross-talk signaling. Cancer Res. 2016;76:3156–3165. doi: 10.1158/0008-5472.CAN-15-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 34.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 35.Kantono M, Guo B. Inflammasomes and Cancer: the dynamic role of the Inflammasome in tumor development. Front Immunol. 2017;8:1132. doi: 10.3389/fimmu.2017.01132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Z, Qiu Y, Lu W, Jiang Y, Wang J. Immunotherapeutic interventions of triple negative breast Cancer. J Transl Med. 2018;16:147. doi: 10.1186/s12967-018-1514-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Solary E. When monocyte life hangs by a thread. Blood. 2012;119:2699–2700. doi: 10.1182/blood-2012-01-403709. [DOI] [PubMed] [Google Scholar]
- 38.Duluc D, Delneste Y, Tan F, Moles MP, Grimaud L, Lenoir J, Preisser L, Anegon I, Catala L, Ifrah N, et al. Tumor-associated leukemia inhibitory factor and IL-6 skew monocyte differentiation into tumor-associated macrophage-like cells. Blood. 2007;110:4319–4330. doi: 10.1182/blood-2007-02-072587. [DOI] [PubMed] [Google Scholar]
- 39.Chen K, Huang YH, Chen JL. Understanding and targeting cancer stem cells: therapeutic implications and challenges. Acta Pharmacol Sin. 2013;34:732–740. doi: 10.1038/aps.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo Y, Xu F, Lu T, Duan Z, Zhang Z. Interleukin-6 signaling pathway in targeted therapy for cancer. Cancer Treat Rev. 2012;38:904–910. doi: 10.1016/j.ctrv.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 41.Chin AR, Wang SE. Cytokines driving breast cancer stemness. Mol Cell Endocrinol. 2014;382:598–602. doi: 10.1016/j.mce.2013.03.024. [DOI] [PubMed] [Google Scholar]
- 42.Korkaya H, Kim GI, Davis A, Malik F, Henry NL, Ithimakin S, Quraishi AA, Tawakkol N, D'Angelo R, Paulson AK, et al. Activation of an IL6 inflammatory loop mediates trastuzumab resistance in HER2+ breast cancer by expanding the cancer stem cell population. Mol Cell. 2012;47:570–584. doi: 10.1016/j.molcel.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fisher DT, Appenheimer MM, Evans SS. The two faces of IL-6 in the tumor microenvironment. Semin Immunol. 2014;26:38–47. doi: 10.1016/j.smim.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang N, Liang H, Zen K. Molecular mechanisms that influence the macrophage m1-m2 polarization balance. Front Immunol. 2014;5:614. doi: 10.3389/fimmu.2014.00614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin J, He Y, Chen J, Zeng Z, Yang B, Ou Q. A critical role of transcription factor YY1 in rheumatoid arthritis by regulation of interleukin-6. J Autoimmun. 2017;77:67–75. doi: 10.1016/j.jaut.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 46.Alberti C, Pinciroli P, Valeri B, Ferri R, Ditto A, Umezawa K, Sensi M, Canevari S, Tomassetti A. Ligand-dependent EGFR activation induces the co-expression of IL-6 and PAI-1 via the NFkB pathway in advanced-stage epithelial ovarian cancer. Oncogene. 2012;31:4139–4149. doi: 10.1038/onc.2011.572. [DOI] [PubMed] [Google Scholar]
- 47.Cortez MA, Ivan C, Valdecanas D, Wang X, Peltier HJ, Ye Y, Araujo L, Carbone DP, Shilo K, Giri DK, et al. PDL1 regulation by p53 via miR-34. J Natl Cancer Inst. 2016;108. [DOI] [PMC free article] [PubMed]
- 48.Wang X, Li J, Dong K, Lin F, Long M, Ouyang Y, Wei J, Chen X, Weng Y, He T, Zhang H. Tumor suppressor miR-34a targets PD-L1 and functions as a potential immunotherapeutic target in acute myeloid leukemia. Cell Signal. 2015;27:443–452. doi: 10.1016/j.cellsig.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 49.Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H, Patrawala L, Yan H, Jeter C, Honorio S, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2011;17:211–215. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Siemens H, Jackstadt R, Hunten S, Kaller M, Menssen A, Gotz U, Hermeking H. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle. 2011;10:4256–4271. doi: 10.4161/cc.10.24.18552. [DOI] [PubMed] [Google Scholar]
- 51.Hahn S, Jackstadt R, Siemens H, Hunten S, Hermeking H. SNAIL and miR-34a feed-forward regulation of ZNF281/ZBP99 promotes epithelial-mesenchymal transition. EMBO J. 2013;32:3079–3095. doi: 10.1038/emboj.2013.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schupp J, Krebs FK, Zimmer N, Trzeciak E, Schuppan D, Tuettenberg A. Targeting myeloid cells in the tumor sustaining microenvironment. Cell Immunol. 2017. [DOI] [PubMed]
- 53.Self-Fordham JB, Naqvi AR, Uttamani JR, Kulkarni V, Nares S. MicroRNA: dynamic regulators of macrophage polarization and plasticity. Front Immunol. 2017;8:1062. doi: 10.3389/fimmu.2017.01062. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. MCT-1 is a prognostic marker of human breast carcinoma. (PDF 215 kb)
Figure S2. MCT-1 overexpression increases size of the MCF-10A acini. (PDF 223 kb)
Figure S3. MCT-1 expression regulates MMP activity and breast cancer cell movements. (PDF 260 kb)
Figure S4. CD80-positive M1 macrophages enriched in the TME after MCT-1 knockdown. (PDF 289 kb)
Figure S5. MCT-1/IL-6/IL-6R signaling promotes breast cancer stemness. (PDF 259 kb)
Figure S6. Targeting MCT-1 and tocilizumab treatment together suppress 4 T1 cancer stemness. (PDF 144 kb)
Figure S7. MicroRNA profiling in the MCT-1 pathway. (PDF 48 kb)
Table S1. Primer sequences. (PDF 104 kb)
Supplementary Methods. Cell culture. Plasmid construction and transfection. Targeting MCT-1 gene. Targeting IL-6 gene. Antibodies (Abs) and protein analysis. MCF-10A acinar morphogenesis. Cell invasion and migration assays. Gelatin zymography assay. Cell fractionation. Quantitative RT-PCR analysis of cancer stemness markers. ALDEFLUOR assay. Immunohistochemistry study. Quantification of miR-34a levels. Statistical analysis. (PDF 227 kb)
Data Availability Statement
The datasets used for the current study are available from the corresponding author on reasonable request.