Abstract
Aims: Activation of the nuclear factor (erythroid-derived 2)-like 2 (Nrf2) pathway in normal cells inhibits carcinogenesis, whereas constitutive activation of Nrf2 in cancer cells promotes tumor growth and chemoresistance. However, the effects of Nrf2 activation in immune cells during lung carcinogenesis are poorly defined and could either promote or inhibit cancer growth. Our studies were designed to evaluate tumor burden and identify immune cell populations in the lungs of Nrf2 knockout (KO) versus wild-type (WT) mice challenged with vinyl carbamate.
Results: Nrf2 KO mice developed lung tumors earlier than the WT mice and exhibited more and larger tumors over time, even at late stages. T cell populations were lower in the lungs of Nrf2 KO mice, whereas tumor-promoting macrophages and myeloid-derived suppressor cells were elevated in the lungs and spleen, respectively, of Nrf2 KO mice relative to WT mice. Moreover, 34 immune response genes were significantly upregulated in tumors from Nrf2 KO mice, especially a series of cytokines (Cxcl1, Csf1, Ccl9, Cxcl12, etc.) and major histocompatibility complex antigens that promote tumor growth.
Innovation: Our studies discovered a novel immune signature, characterized by the infiltration of tumor-promoting immune cells, elevated cytokines, and increased expression of immune response genes in the lungs and tumors of Nrf2 KO mice. A complementary profile was also found in lung cancer patients, supporting the clinical significance of our findings.
Conclusion: Overall, our results confirmed a protective role for Nrf2 in late-stage carcinogenesis and, unexpectedly, suggest that activation of Nrf2 in immune cells may be advantageous for preventing or treating lung cancer.
Keywords: : Nrf2, vinyl carbamate, lung carcinogenesis, immune signature, inflammatory cytokines
Introduction
The Nrf2-Keap1-antioxidant response element pathway is a master defense mechanism protecting against oxidative and electrophilic stress. It helps regulate numerous cellular processes as diverse as metabolism, detoxification, redox balancing, and autophagy (50). Under basal conditions, Nrf2 [nuclear factor (erythroid-derived 2)-like 2] is bound to its internal negative regulator Keap1 (Kelch-like ECH-associated protein 1) and targeted to the proteasome for degradation (34). Under stress conditions such as increased reactive oxygen species (ROS), Nrf2 is released from Keap1 and translocates to the nucleus to initiate transcription of a variety of downstream genes (50). These target genes encode for proteins including (i) phase I/II/III metabolism enzymes that detoxify xenobiotics and enhance their elimination (33, 64, 80), (ii) proteins to maintain cellular redox homeostasis (17), (iii) enzymes involved in heme, lipid, and glucose metabolism (2, 4, 85), (iv) enzymes that regulate nicotinamide adenine dinucleotide phosphate generation and pentose synthesis (40), and (v) proteins involved in apoptosis and autophagy (55, 88). In addition to interaction with Keap1, additional mechanisms have been reported to regulate Nrf2 activity, including phosphorylation by kinases [protein kinase C, mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinases/c-Jun N-terminal kinases, JUN/MYC], protein–protein interactions (e.g., retinoid X receptor α [RXRα]), and epigenetic modifications (microRNAs and acetylation) (25). The regulation of the Nrf2 pathway, therefore, is complex but profoundly important for numerous cellular processes (50).
Innovation.
Little is known regarding the effect of Nrf2 [nuclear factor (erythroid-derived 2)-like 2] activation in immune cells during carcinogenesis. Our studies discovered a pattern of infiltrating tumor-promoting immune cells, elevated cytokines, and increased expression of immune response genes in the lungs and tumors of Nrf2 knockout mice. A similar profile was also found in lung cancer patients, suggesting that Nrf2 regulates the immune cells and cytokines that can regulate tumor progression. Thus, our data suggest that activation of Nrf2 in immune cells likely has different effects than activation of Nrf2 in tumor cells and could be advantageous for preventing or treating lung cancer.
As a “multiorgan protector” (43), activation of Nrf2 protects against many diseases driven by unresolved inflammation including cancer, cardiovascular diseases, and neurodegenerative diseases. Recently, the role of Nrf2 in cancer has been the topic of numerous interesting studies. Activation of Nrf2 was traditionally considered beneficial for the prevention of cancer. As Nrf2 is the main cellular defense mechanism against both endogenous and exogenous insults, Nrf2 deficiency enhances the susceptibility to carcinogens (71). This effect is not limited to certain types of cancer or restricted to certain types of insults. Carcinogenesis is consistently exacerbated in Nrf2 knockout (KO) versus wild-type (WT) mice, whether induced by ultraviolet light or chemicals in skin, aflatoxin in the liver, polycyclic hydrocarbons in the forestomach, nitrosamines in the bladder, or inflammation in the colon (26, 29, 31, 37, 79). Moreover, knockdown (KD) of Keap1, which elevates Nrf2 levels, increases resistance to cancer metastasis (65, 66). In addition to affecting cancer susceptibility, Nrf2 also plays a crucial role in chemoprevention. Many Nrf2 activators, including a variety of natural products, can be used to prevent or delay tumor development, and these chemopreventive effects are greatly dampened in Nrf2 KO mice (31, 62).
However, increasing number of studies suggest a tumor-promoting role for Nrf2 (50). The detoxifying and cytoprotective environment created by activation of the Nrf2 pathway helps tumor cells eliminate hypoxia and elevated ROS levels, thus promoting survival of tumor cells (21). Gain-of-function mutations in the Nfe2l2 gene that encodes for Nrf2 and loss-of-function mutations in the Keap1 gene are found in a subset of advanced cancers of the lung, liver, esophagus, bladder and other organs, but lung cancer has the highest frequency of Nfe2l2 or Keap1 alterations (50). Epigenetic modifications of the Keap1 or Nfe2l2 promoters have also been found. All of these genetic and epigenetic alterations result in constitutively high levels of Nrf2 expression and activity, which is associated with chemoresistance and poor prognosis (50, 71, 76). More recently, Ngo et al. (54) reported that Nrf2 drives hepatocarcinogenesis, and Bauer et al. (6) reported that deletion of Nrf2 reduces lung tumor development induced by urethane. Although these studies suggest that the role of Nrf2 may be model and context dependent, the authors conclude that inhibiting Nrf2 activity should be beneficial in treating advanced cancers.
The complex tumor-promoting and tumor-suppressing dual roles of Nrf2 in cancer have generated a great deal of interest, especially regarding the safety of long-term use of Nrf2 activators and the need to develop Nrf2 inhibitors for treating cancer. We recently reported that two drugs that can activate the Nrf2 pathway have opposite effects in a lung carcinogenesis model (73). Dimethyl fumarate or Tecfidera®, approved by the Food and Drug Administration for the treatment of multiple sclerosis, increased the number and pathological grade of lung tumors. In contrast, synthetic oleanane triterpenoids (47) currently being tested in clinical trials for the treatment of chronic kidney disease, pulmonary hypertension, and cancer, reduced the number, size, and severity of lung tumors (73). Additional experiments are needed to determine whether these contradictory results are dependent on Nrf2, as both drugs are also potent anti-inflammatory agents that target the immune system and other specific protein targets.
Considering the conflicting reports regarding the role of Nrf2 in lung cancer and the importance of immunotherapy for treating lung cancer, understanding the regulation of immune cells by Nrf2 during carcinogenesis is necessary. The microenvironment regulates tumorigenesis, and a broad-spectrum integrative approach using natural products or pharmacological activators could be used to modulate this microenvironment for both cancer prevention and treatment (13). As many Nrf2 modulators have anti-inflammatory effects, Nrf2 may be a potential target to modulate the microenvironment. In our current studies, we used a relevant preclinical model to investigate the Nrf2 pathway in lung carcinogenesis. The potent carcinogen vinyl carbamate induces Kras mutations (28) and lung adenocarcinomas (45). Kras mutations are the most common mutation found in lung cancer, especially in smokers, and adenocarcinomas are the most frequent type of lung cancer (23). Notably, tumors driven by Kras mutations have been considered “undruggable” and are resistant to standard and targeted chemotherapies (23). Although carcinogens found in cigarettes can also induce lung tumors, the traditional NNK [4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone] model induces adenomas instead of adenocarcinomas (49). Kras transgenic mice also develop lung cancer (35) but the rapid development of tumors and high-tumor burden (35, 68) limit their utility for studying carcinogenesis and changes in the microenvironment over time. Here, we describe changes in tumor burden, immune cell populations, inflammatory cytokines, and immune signatures in the lungs of Nrf2 KO versus WT mice challenged with vinyl carbamate.
Results
Lung carcinogenesis exacerbated in Nrf2−/− mice challenged with vinyl carbamate
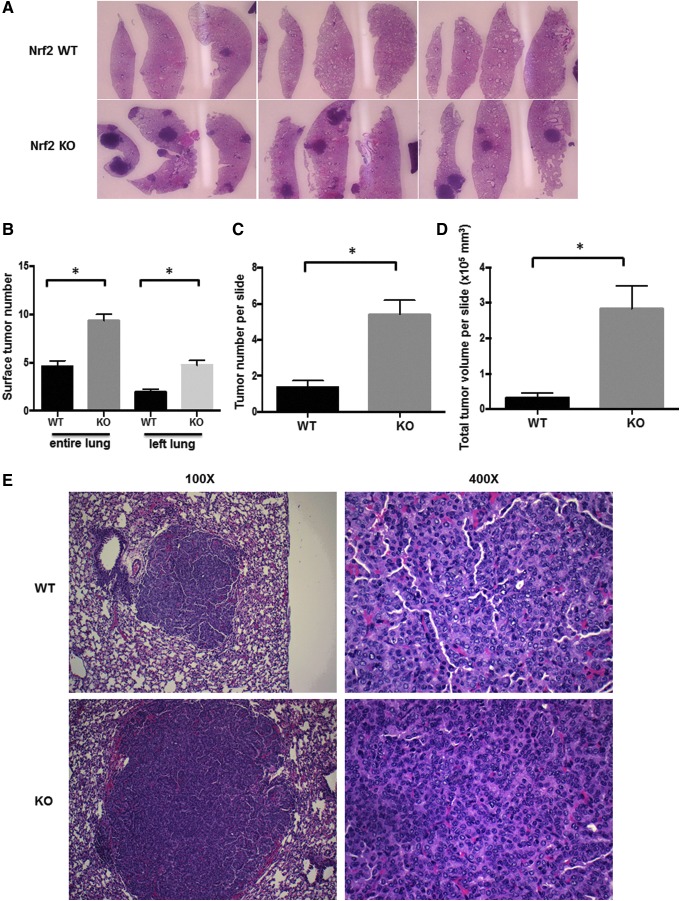
To determine the effects of vinyl carbamate on Nrf2 KO mice on a BALB/c background, female WT and KO mice were injected i.p. with two to four doses of vinyl carbamate, using protocols previously used with Nrf2 KO mice (65) or with vinyl carbamate (46). Age-matched cohorts of Nrf2 WT and KO mice (four mice per group per time point) were harvested beginning at 20 weeks after initiation and then every 4 (24, 28, and 32) weeks. More lung tumors were consistently visible over time (20–36 weeks) both (i) in Nrf2 KO mice versus WT mice and (ii) in mice injected with vinyl carbamate four times versus two times in both WT and KO mice (average number = 2.7 ± 0.7 tumors with two doses vinyl carbamate vs. 6.6 ± 0.6 tumors with four doses in WT mice, p < 0.05; 7.95 ± 0.7 tumors with two doses vs. 10.9 ± 1.0 tumors with four doses in Nrf2 KO mice, p < 0.05). Although the number and size of the tumors increased over time in both groups, these parameters were consistently higher in the Nrf2 KO mice (Fig. 1A). At 20 weeks, 4–12 tumors were visible on the surface of lungs in Nrf2 KO mice, whereas almost no visible tumors (0–1) were observed at this time in WT mice. By 32 weeks, an average of 6.5 ± 0.95 tumors per lung were found in the WT mice and 12.25 ± 2.6 tumors were found in the Nrf2 KO group. For all time points, the number of grossly visible surface tumors was significantly (p < 0.05) higher in Nrf2 KO mice than in WT mice (Fig. 1B), in the entire lung (unfixed) or in the intact left lung (after formalin fixation but before sectioning).
FIG. 1.
Nrf2 deficiency promotes lung carcinogenesis. Nrf2 KO and WT mice were injected with vinyl carbamate between 6 and 8 weeks of age. Lungs from age-matched Nrf2 KO and WT mice were harvested for a period of 20–40 weeks after initiation with the carcinogen to examine tumor burden. (A) Representative images of lung sections stained with hematoxylin and eosin; tumors are darkly stained purple areas (10 × magnification). (B) The average number of tumors on the surface of the entire lungs or on the left lung after fixation in formalin were counted, n = 31–42 mice per group (C). Average number of tumors (C) and total tumor volume (D) per slide were calculated in both Nrf2 WT and KO groups. n = 20 mice per group. *p < 0.05. (E) Representative images of lung histopathology in WT and KO groups. KO, knockout; Nrf2, nuclear factor (erythroid-derived 2)-like 2; WT, wild-type.
After gross evaluation, each left lung was then sectioned and stained with hematoxylin and eosin. The average number of tumors (Fig. 1C) and total tumor volume (Fig. 1D) per slide were also significantly (p < 0.05) higher in the lungs of Nrf2 KO mice versus WT mice. There were no significant differences in tumor histopathology between groups as almost all of the tumors were high grade (tumefactive architecture, fused trabeculae, and distinct nucleoli and conspicuous mitoses within nuclei) by 20 weeks after initiation. Representative images demonstrating this histopathology are shown in Figure 1E. As expected based on the larger tumor size, the tumors in the Nrf2 KO mice proliferate more rapidly than the WT tumors, as the percentage of proliferating cell nuclear antigen (PCNA) positive cells is significantly (p < 0.05) higher in the Nrf2 KO tumors (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/ars).
Nrf2 deficiency decreases T cell populations during lung carcinogenesis
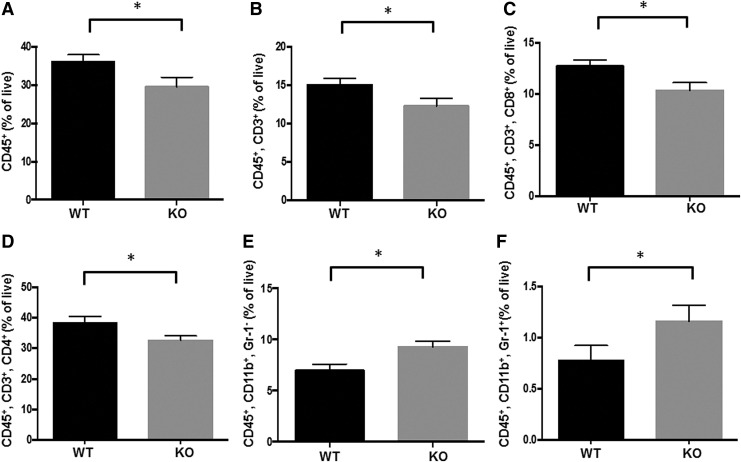
Next, we investigated immune cell populations in the lungs and spleens of Nrf2 WT and KO mice challenged with vinyl carbamate. The same two lobes of the right lung were always collected, and the fresh tissues were processed for flow cytometry using optimized antibody panels (42). To obtain an overview of the immune cell populations in the lung, the percentages of CD45+ immune cells as well as total T cells (CD45+ and CD3+), CD4 helper T cells (CD45+, CD3+, and CD4+), and CD8 cytotoxic T cells (CD45+, CD3+, and CD8+), B cells (CD45+, CD19+, and B220+), macrophages (CD45+, CD11b+, and Gr-1−), and myeloid-derived suppressor cells (MDSCs) (CD45+, CD11b+, and Gr-1+) were analyzed. Although cohorts of paired, age-matched lungs (four mice per group) were collected 20–36 weeks after initiation with vinyl carbamate, unless otherwise noted, the changes in the Nrf2 KO versus WT lungs were consistent over time, and Nrf2 KO and WT groups were pooled when appropriate.
As shown in Figure 2A, the percentage of CD45+ immune cells was significantly (p < 0.05) lower in the lungs of Nrf2 KO mice than that in WT mice, primarily because of changes in T cell populations. Nrf2 KO mice had a significantly (p < 0.05) lower number of total T cells (CD45+, CD3+, 20–32 weeks; Fig. 2B), CD8 cytotoxic T cells (28–32 weeks; Fig. 2C), and CD4 helper T cells (24–32 weeks; Fig. 2D). CD4 T cells play a central role in immunity, but their contribution to antitumor immunity is complex as different types of CD4 T cells have diverse functions (86). CD8 cytotoxic T cells recognize tumor cells and are essential for the response to immunotherapies such as PD-1/PD-L1 (36).
FIG. 2.
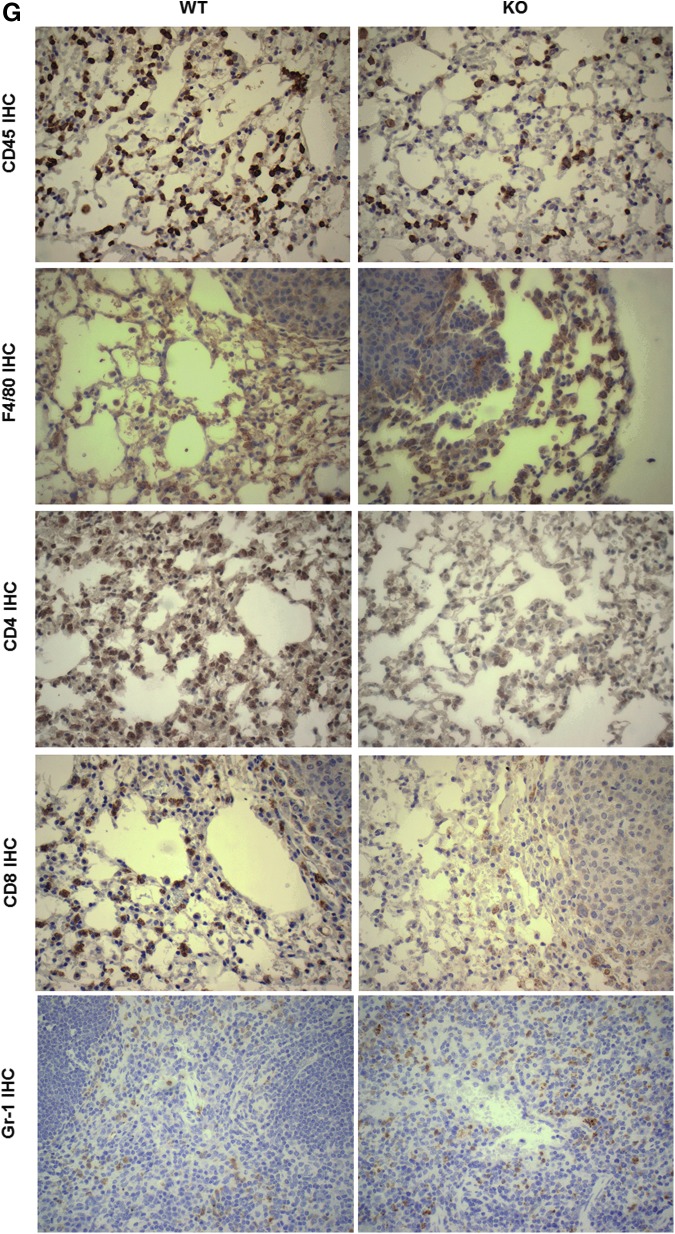
Nrf2 deficiency alters the immune cell populations in the lung and spleen of mice challenged with vinyl carbamate. The immune cell populations in the same two lobes of right lung (A–E) or spleen (F) from Nrf2 KO and Nrf2 WT were analyzed by flow cytometry. Percentages of total immune cells (CD45+), total T cells (CD45+, CD3+), cytotoxic T cells (CD45+, CD3+, CD8+), T helper cells (CD45+, CD3+, CD4+), and macrophages (CD45+, CD11b+, Gr-1−) in the lung (A–E) and myeloid-derived suppressor cells (CD45+, CD11b+, Gr-1+) in the spleen (F) are shown. *p < 0.05. The changes in immune cell populations were confirmed by IHC (G). F4/80 is a macrophage marker.
The percentage of macrophages was significantly (p < 0.05) higher in the lungs of Nrf2 KO mice before 24 weeks (Fig. 2E), but this difference disappeared at later time points. Macrophages play a critical role in promotion of urethane-induced lung carcinogenesis (87). Fewer and smaller lung tumors will develop if macrophages are depleted during tumor initiation and early promotion in this model. There was no significant difference in B cells between the two groups in the lung (data not shown). In the spleen, the only change was a small but significant (p < 0.05; 20–40 weeks) increase in MDSCs in Nrf2 KO mice (Fig. 2F). MDSCs play important roles in immunosuppression. High infiltration of these cells is associated with poor prognosis in cancer patients (32, 56, 75), and the survival and function of MDSCs are regulated by Nrf2 (7). All of the changes in immune populations were confirmed by immunohistochemistry and representative pictures are shown in Figure 2G. As indicated in the pictures, the macrophages and CD8 cytotoxic T cells are located around the periphery of the tumors.
In addition, the decrease of the CD45+ population, CD3+ T cells, and CD4+ T cells was similar at early time points (4 and 8 weeks after initiation; data not shown), whereas the percentage of CD8+ T cells was significantly (p < 0.05) higher in the lungs of Nrf2 KO mice than in those of WT mice at early time points (Supplementary Fig. S2). There was no significant difference in immune populations between saline-injected mice and vinyl carbamate-injected mice at these time points (data not shown).
Altered gene signatures in lung tumors from Nrf2 KO versus WT mice
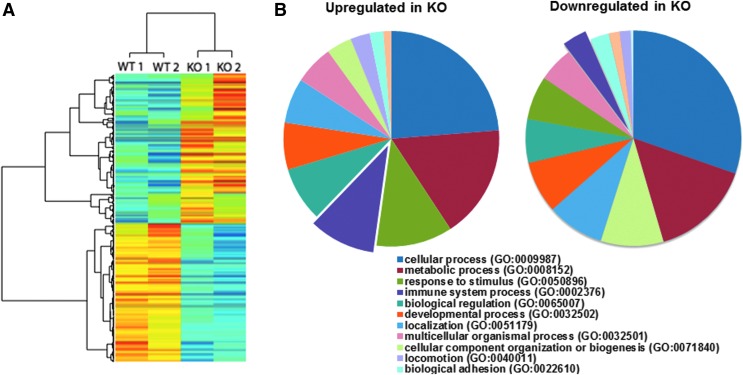
To further investigate the differences between Nrf2 WT and KO mice at a molecular level, lung tumors were dissected out from the lung tissue and pooled (three to four tumors). Total RNA from two independent samples per group was analyzed by RNA sequencing (RNAseq). Significantly (p < 0.05) different expression patterns were detected for 376 genes in tumors from the lungs of Nrf2 WT versus KO mice. Slightly more than half (197 out of 376) of the altered genes in the Nrf2 KO tumors were upregulated compared with WT tumors, and the rest were significantly downregulated (Fig. 3A).
FIG. 3.
Nrf2 deficiency alters global transcription in tumors in the lungs of mice challenged with vinyl carbamate. (A) An unsupervised-hierarchical clustering of Nrf2 WT and KO tumors by differentially regulated genes shows unique transcriptional profile in the two tumor types. The Nrf2 KO tumors have a distinct profile of upregulated (red) and downregulated (blue) genes, which is reversed in the WT tumors. (B) PANTHER analysis was performed on the upregulated genes (left) and downregulated genes (right) with Nrf2 deletion. This analysis identified a number of significantly overrepresented gene ontologies between the WT and KO tumors, especially in the immune system-related genes (GO:0002376) (purple).
Among the differentially regulated genes, the NFE2L2 (Nrf2 gene) was the most significantly downregulated gene in Nrf2 KO tumors, which confirms the deletion of Nrf2. Additional confirmation that Nrf2 was deleted was obtained by genotyping the mice and analyzing lung extracts by Western blotting (data not shown). Differentially regulated genes were then processed to identify over-represented genes using PANTHER analysis (Fig. 3B). As the Nrf2 cytoprotective mechanism was knocked out, the “response to stimuli gene set” (GO:0050896) including extracellular superoxide dismutase (Sod3) and glutathione peroxidase 3 (Gpx3) was downregulated in Nrf2 KO tumors. Notably, the “immune system process” (GO:0002376) was the gene set with the most significant change between groups. As listed in Table 1, a series of cytokines, chemokines, and peptide antigens in this gene set were significantly upregulated in tumors from Nrf2 KO mice. Many of these factors, including Cxcl (1, 3, 12) and Ccl (9, 11, 17) are important for regulation and chemotaxis of immune cells, especially MDSCs, and the activation of M2 macrophages (52, 89). Cytokines can be released in response to inflammation to inhibit tumor development and progression, but alternatively, cancer cells can respond to cytokines that promote growth, inhibit apoptosis, and facilitate metastasis (20). In addition to the striking effects on the immune system, a metastatic signature also emerged. Genes involved in localization (GO:0051179; Fig. 3B) and extracellular matrix (GO:0031012 and GO:0044421; Table 1), which regulate cell migration and metastasis, were differently expressed in these two groups. Overexpression of matrix metalloproteinase (MMP)12, which is upregulated in KO tumors, plays a key role in modulating myelopoiesis, immune suppression, and lung tumorigenesis (61). MMP25, which is elevated in many cancer types and promotes tumor invasion and metastasis through activation of MMP2, was also upregulated in Nrf2 KO tumors (69). Notch signaling (Table 1) is known to crosstalk with the Nrf2 pathway, and aberrant crosstalk between Nrf2 and notch plays an important role in lung carcinogenesis (70). The differential expression of Notch1 in the lung of Nrf2 WT versus KO mice was confirmed by Western blotting (Supplementary Fig. S3). Consistent with the observation of more and larger tumors in Nrf2 KO versus WT mice, cell growth related genes [insulin-like growth factor-binding proteins 5 and 6 (Igfbp5, Igfbp6) and bone morphogenetic protein 6 (Bmp6)] were upregulated and cell death related genes (Srcap and Thsd4) were downregulated in the Nrf2 KO lung tumors (Table 1). P27, an indicator of cell cycle arrest, was significantly downregulated in Nrf2 KO mice versus Nrf2 WT mice (Supplementary Fig. S3). Numerous genes involved in metabolic pathways were also significantly over represented, including heme transport (GO:0015886), negative regulation of cellular amide metabolic process (GO:0034249), and aldonic acid metabolic process (GO:0019520). The whole list of significantly altered genes can be found in Supplementary Table S1. The raw data and processed data are available on Gene Expression Omnibus (GEO) (GSE99338).
Table 1.
Over-Represented, Differentially Regulated Genes in Lung Tumors From Nrf2 Wild-Type Versus Knockout Mice
| Description | Genes upregulated in KO mice | Genes downregulated in KO mice | GO accession | p-Value (over-represented) |
|---|---|---|---|---|
| Chemokine activity | Cxcl1, Cxcl12, Ccl17, Ccl9, Cxcl3, Ccl11 | GO:0008009 | 1.07E-06 | |
| Chemokine receptor binding | Cxcl3, Ccl11, Cxcl12, Ccl9, Ccl17, Cxcl1 | GO:0042379 | 1.07E-06 | |
| Cytokine activity | Csf1, Ccl11, Cxcl3, Cxcl1, Ccl9, Ccl17, Cxcl12 | GO:0005125 | 3.35E-06 | |
| Immune response | Ltb, Cxcl3, Ccl9, H2-Q6, H2-K1, H2-M5, Ccl11, Cxcl1, H2-M2, Ccl17, Cxcl12, Ms4a1 | Dsp | GO:0006955 | 9.43E-06 |
| Immune system process | Ltb, Cxcl3, Ccl9, H2-K1, H2-Q6, H2-M5, Ccl11, Cxcl1, H2-M2, Ccl17, Cxcl12, Ms4a1 | Dsp | GO:0002376 | 3.47E-05 |
| Cytokine receptor binding | Cxcl3, Ccl11, Ltb, Cxcl12, Ccl17, Ccl9, Cxcl1 | GO:0005126 | 0.0008711 | |
| Antigen processing and presentation | H2-Q6, H2-K1, H2-M2, H2-M5 | GO:0019882 | 0.0087215 | |
| Extracellular matrix | S100g, Des, S100a9, Mfap5, Mmp12, Mmp25, Fcer2a, Fgl2 | Gpc3, Thsd4, Ppp3r2, Pls1, Gpc6 | GO:0031012 | 0.00026772 |
| Extracellular region part | Tmcc2, S100a9, S100g, Des, Fcer2a, Fgl2, Gm26788, Mfap5 | Myof, Gpc3, Afp, Pls1, Gpc6, Ppp3r2 | GO:0044421 | 0.00036759 |
| Proteinaceous extracellular matrix | S100a9, S100g, Des, Fcer2a, Fgl2, Mfap5 | Gpc3, Gpc6, Pls1, Ppp3r2 | GO:0005578 | 0.00053867 |
| Notch signaling pathway | Notch1, Notch2, Jag1 | GO:0007219 | 0.01156 | |
| Regulation of cell growth | Igfbp5, Igfbp6 | GO:0001558 | 0.013648 | |
| Insulin-like growth factor binding | Igfbp5, Igfbp6 | GO:0005520 | 0.013648 | |
| Cell growth | Igfbp5, Igfbp6 | GO:0016049 | 0.013648 | |
| Regulation of growth | Igfbp5, Igfbp6 | GO:0040008 | 0.017012 | |
| Growth | Igfbp5, Bmp6, Igfbp6 | GO:0040007 | 0.030478 | |
| Growth factor binding | Igfbp6, Igfbp5 | GO:0019838 | 0.028955 | |
| Positive regulation of cell death | Srcap, Thsd4 | GO:0010942 | 0.021795 | |
| Positive regulation of apoptotic process | Srcap, Thsd4 | GO:0043065 | 0.021795 | |
| Positive regulation of programmed cell death | Srcap, Thsd4 | GO:0043068 | 0.021795 |
Total RNA from two independent samples per group was analyzed by RNA sequencing. Routine identification of differentially expressed genes was performed using DESeq2. A total of 376 genes in tumors were detected with significantly (p < 0.05) different expression patterns from the lungs of Nrf2 WT versus KO mice. Cancer-relevant gene sets are emphasized in this table.
Bmp6, bone morphogenetic protein 6; Igfbp5, insulin-like growth factor-binding protein 5; Igfbp6, insulin-like growth factor-binding protein 6; KO, knockout; MMP, matrix metalloproteinase; Nrf2, nuclear factor (erythroid-derived 2)-like 2; WT, wild-type.
Cytokines are upregulated in tumors and lungs from Nrf2 KO mice
Both the flow cytometry data and RNAseq data revealed the important role of Nrf2 on the immune system in this lung cancer model. To confirm these results, four cytokines listed in Table 1 were selected to validate ex vivo: Cxcl1, Cxcl12, Ccl9, and Csf1. These cytokines and chemokines are all highly relevant in cancer. Chemokine (C-X-C motif) ligand 1 (Cxcl1) attracts MDSCs (CD11b+ and Gr1+) into the tumor, which then produce chemokines that enhance tumor survival and inhibit CD8 cytotoxic T cells. It also recruits tumor-associated macrophages (TAMs) and cancer-associated fibroblasts. High Cxcl1 levels are associated with higher metastatic potential and poor prognosis (52, 74, 84). Chemokine (C-X-C motif) ligand 12, Cxcl12, which is the sole ligand of CXCR4 (C-X-C chemokine receptor type 4), provides an attractive niche for tumor cell migration and colonization. It is highly expressed not only in primary lung cancer cells but also in the brain, liver, bone marrow, and adrenal glands, which are all common sites for lung cancer metastasis (24, 77, 78, 82). Chemokine (C-C motif) ligand 9 (Ccl9) is highly induced in myeloid cells by transforming growth factor-β signaling and is also secreted by premetastatic tumor cells to recruit more myeloid cells, which enhances tumor cell survival and metastasis (81). Colony stimulating factor 1 (Csf1) is involved in cancer growth, survival, and metastasis. Blockade of CSF1/CSF1R reprograms TAMs to enhance antigen presentation, thus improving the therapeutic effects of immunotherapies (1, 19, 30, 89).
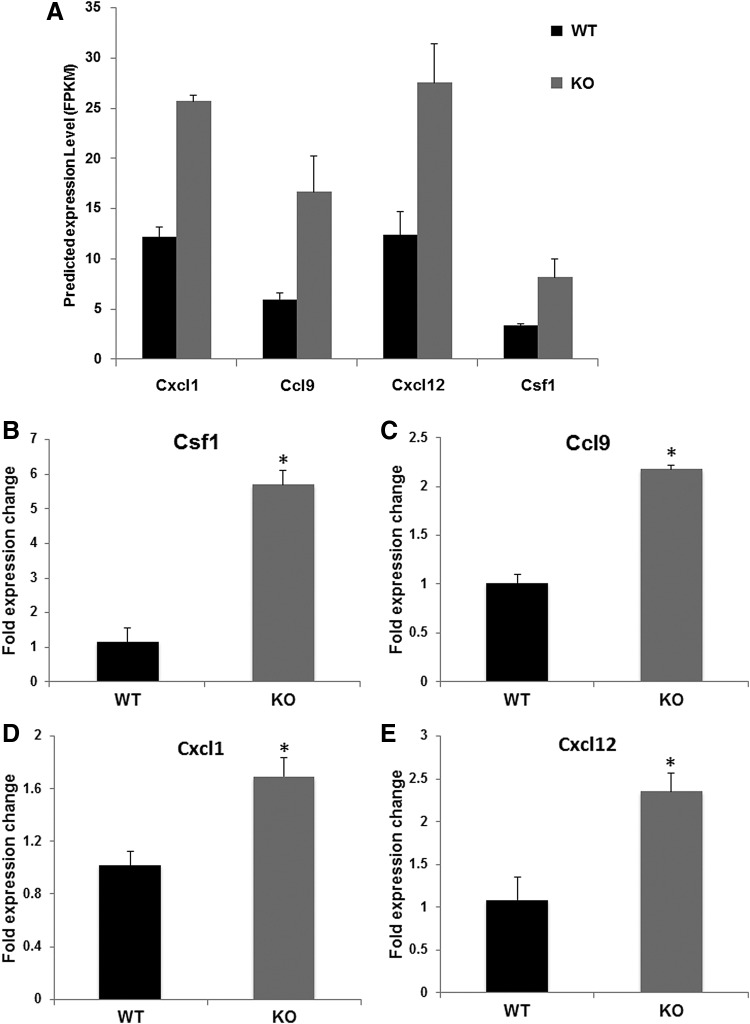
To validate the bioinformatics predictions (Fig. 4A) of the upregulation of these cytokines and chemokines, aliquots of the same RNA samples analyzed by RNAseq were used for real-time PCR. All four cytokines were significantly upregulated in Nrf2 KO versus WT tumors (Fig. 4B–E, p < 0.05). To confirm these changes, total RNA was isolated from lung tissue, and real-time PCR was used to evaluate relative gene expression of the cytokines. Notably, significant increases in expression of these cytokines in Nrf2 KO lungs were still detected (Fig. 4F). Cxcl1, Csf1, Cxcl12 (24 weeks), and Ccl9 (32 weeks) were significantly upregulated in the lungs of Nrf2 KO mice (Fig. 4F). The expression of Cxcl1 and Cxcl12 protein was detected by enzyme-linked immunosorbent assay (ELISA). Cxcl1 (24–32 weeks; Fig. 4G) and Cxcl12 (32 weeks; Fig. 4H) are significantly upregulated in the Nrf2 KO lungs compared with those in the WT lungs.
FIG. 4.
Differential gene expression of cytokines in the tumors and lungs of Nrf2 WT versus KO mice. (A) Predicted gene expression levels (FPKM) based on RNAseq data of lung tumors from Nrf2 WT and KO mice for Cxcl1, Ccl9, CXcl12, and CSF1. The same tumor RNA analyzed by RNAseq was used to confirm the cytokine gene expression using real-time PCR (B–E). *p < 0.05 WT versus KO. In (F), lung tissues from mice challenged with vinyl carbamate were harvested at three time points, 24–32 weeks after initiation. Total RNA was extracted from the lung and relative gene expression of Csf1, Cxcl1, Cxcl12, and Ccl9 was analyzed by real-time PCR. n = 4 lungs per group at each time point. *p < 0.05 WT versus KO at 24 weeks (Csf1, Cxcl1, Cxcl12) or 32 weeks (Ccl9). The production of Cxcl1 (G) and Cxcl12 (H) in the lung was detected by enzyme-linked immunosorbent assay. *p < 0.05 WT versus KO at 24–32 weeks with Cxcl1 and 32 weeks with Cxcl12. Data are presented as mean ± standard error of the mean. Ccl9, chemokine (C-C motif) ligand 9; Csf1, colony stimulating factor 1; Cxcl1, chemokine (C-X-C motif) ligand 1; Cxcl12, chemokine (C-X-C motif) ligand 12; FPKM, fragments per kilo bases per million reads; RNAseq, RNA sequencing.
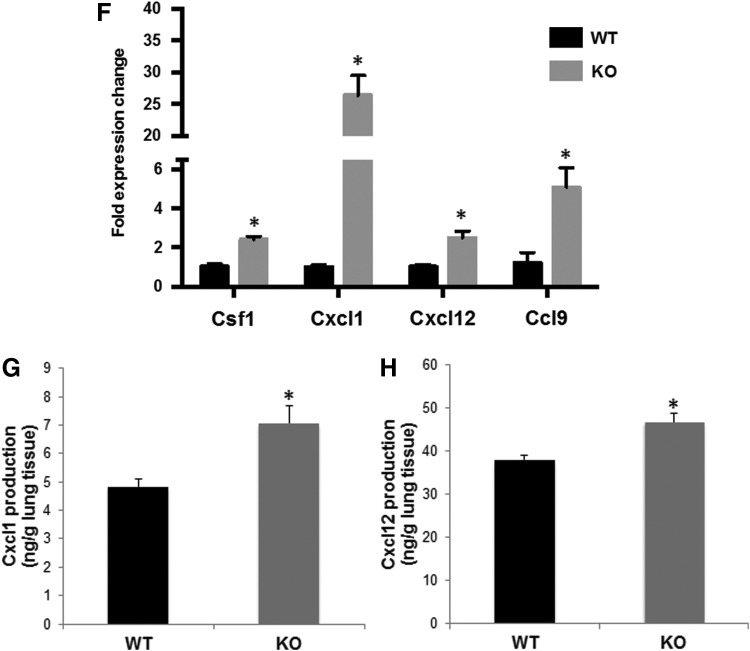
To further validate the regulation of Nrf2 on cytokine expression and identify which cell type is expressing the cytokines, in vitro studies with both tumor cells and immune cells were performed. VC1 cells, a primary lung tumor cell line extracted from the vinyl carbamate-induced lung cancer model (46), were treated with CDDO-Im (a synthetic triterpenoid and potent Nrf2 activator), and the expression of the cytokines was detected by real-time PCR. Treatment with the Nrf2 activation significantly (p < 0.05) decreased messenger RNA (mRNA) expression of these four cytokines in the lung cancer cells (Fig. 5A). Similarly, in RAW264.7 macrophage-like cells stimulated with lipopolysaccharide (LPS), activation of Nrf2 by CDDO-Im significantly (p < 0.05) decreased Csf1 mRNA expression (Fig. 5B). Cxcl1 and Cxcl12 were expressed at very low levels in RAW264.7 cells, even after treatment with LPS or conditioned media from VC1 cells (data not shown), and there was no significant change in Ccl9 expression after treatment with CDDO-Im (Fig. 5B). These experiments suggest that Nrf2 regulates cytokines in both tumor cells and immune cells and that the expression of different cytokines is cell type dependent.
FIG. 5.
Nrf2 activation decreased cytokine production in both tumor cells and immune cells. VC1 (primary mouse lung cancer cells), (A) and RAW264.7 (macrophage-like cells), (B) were treated with 300 nM CDDO-Im (a potent Nrf2 activator) for 24 h, and RAW264.7 cells were stimulated with 1 ng/mL LPS 20 mins after CDDO-Im treatment. Total RNA was extracted from the cells, and gene expression of Cxcl1, Csf1, Ccl9, and Cxcl12 was detected by real-time PCR. *p < 0.05 CDDO-Im treated group versus control for each cytokine. LPS, lipopolysaccharide.
The regulation of immune response in the mouse model is consistent with data from patients with lung adenocarcinomas
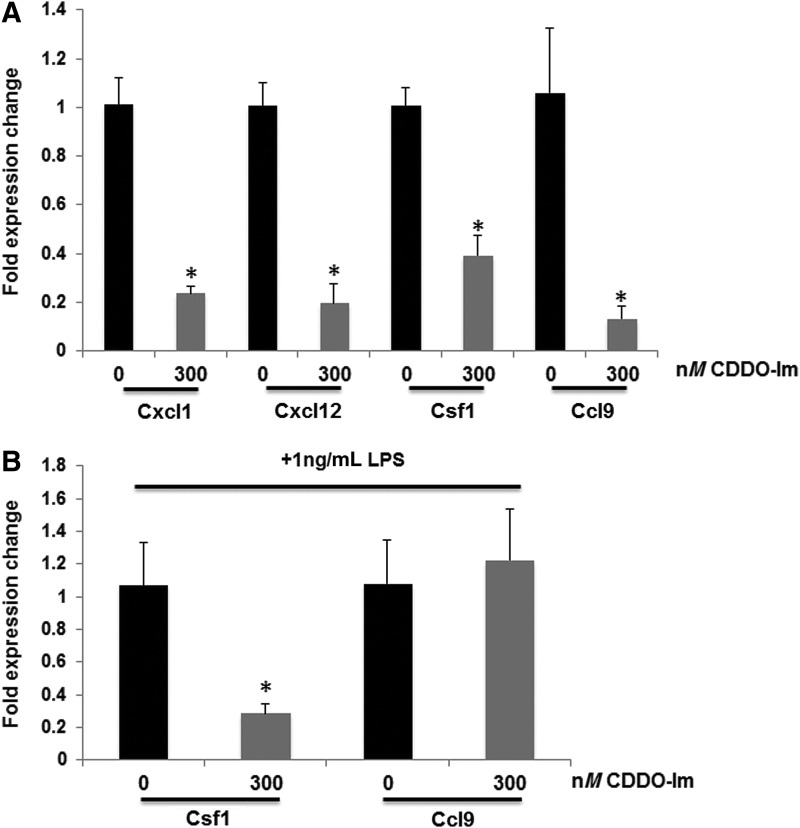
As a striking upregulation of immune responses was found in lung tumors from Nrf2 KO mice, we wanted to verify whether these results were consistent in human patients. We accessed genomic data from lung cancer patients through the TCGA human cancer database, and cBioPortal was used to analyze the genetic alterations in Nfe2l2 and Keap1 within lung cancer patients. As has been reported (71), Keap1 alterations (mutations and deletions) are most frequent in lung adenocarcinoma patients, whereas Nfe2l2 alterations (mutations and amplifications) are more frequent in lung squamous carcinoma. Both loss-of-function alterations of Keap1 and gain-of-function alterations of Nfe2l2 induce high constitutive Nrf2 activity. Oncoprints of Keap1 genetic alterations in lung adenocarcinoma patients (12) (230 samples) and Nfe2l2 genetic alterations in lung squamous carcinoma patients (11) (178 samples) are shown in Figure 6A and C, respectively. Enrichment of gene ontologies was identified through Bayes factor. As shown in Table 2, the expression of immune response-related gene ontologies (GO:0006955, GO:0019882, and GO:0006959) was consistently preserved in human tumors. Then, gene expression of individual cytokines was analyzed, as cytokines were differentially regulated in our mouse model. Cxcl1, 2, 10, 11, 12, 14, Ccl2, 3, 4L1, 13, 22, and 28 and Csf1, 1R, 2RB, and 3R were all significantly downregulated in Keap1-altered adenocarcinoma patients. Cxcl1, 2, 3, 6, 12, 17, Ccl15, 17, 22, 28, and Csf2, 3R were significantly downregulated in Nrf2-altered squamous carcinoma patients. Cxcl1, Cxcl12, and Csf1, the three cytokines upregulated in Nrf2 KO mice with adenocarcinomas, were consistently downregulated in Keap1 loss-of-function patients (Fig. 6B). These results are consistent with the function of Keap1 as a negative regulator of Nrf2, as Nrf2 would be constitutively active in these samples instead of deleted as in the mouse model. Cxcl1 was also downregulated in patients with gain-of-function Nrf2 amplifications and alterations (Fig. 6D). In summary, our findings suggest that Nrf2 regulation of cytokines in lung cancer can be found in both mice and humans.
FIG. 6.
The regulation of immune response by Nrf2 in the mouse model is consistent with data in lung patients. (A) Oncoprint of Keap1 genetic alterations in patients with lung adenocarcinoma (12) (230 samples). Unaltered patients are not shown. Alterations of Keap1 are mainly deletions and mutations. (B) Individual genes (Cxcl12, Cxcl1, Csf1) were downregulated in lung adenocarcinoma patients with Keap1 alterations. p < 0.05. Results are presented as box plot with maximum, third quartile, median, first quartile, and minimum (top to bottom). (C) Oncoprint of Nfe2l2 genetic alterations in lung squamous carcinoma patients (11) (178 samples). Unaltered patients are not shown. Alterations of Nfe2l2 are mainly amplifications and mutations. (D) Cxcl1 was downregulated in lung squamous carcinoma patients with Nfe2l2 alterations. p < 0.05. Results are presented as box plot with maximum, third quartile, median, first quartile, and minimum (top to bottom). Keap1, Kelch-like ECH-associated protein 1; mRNA, messenger RNA; NFE2L2, Nrf2 gene.
Table 2.
Differentially Regulated Gene Ontologies in Lung Adenocarcinoma Patients
| Annotation | Total genes with annotation | ln(Bayes factor) |
|---|---|---|
| GO:0007186: G-protein coupled receptor protein signaling pathway | 116 | 52.22 |
| GO:0050877: neurophysiological process | 90 | 45.06 |
| GO:0007600: sensory perception | 59 | 33.18 |
| GO:0009581: detection of external stimulus | 70 | 27.69 |
| GO:0006955: immune response | 271 | 22.42 |
| GO:0050875: cellular physiological process | 2391 | 22.28 |
| GO:0006952: defense response | 289 | 17.6 |
| GO:0009607: response to biotic stimulus | 323 | 17.37 |
| GO:0044249: cellular biosynthesis | 317 | 16.41 |
| GO:0009058: biosynthesis | 326 | 14.67 |
| GO:0050794: regulation of cellular process | 268 | 13.74 |
| GO:0006412: protein biosynthesis | 187 | 11.01 |
| GO:0009059: macromolecule biosynthesis | 204 | 10.46 |
| GO:0007166: cell surface receptor-linked signal transduction | 272 | 9.62 |
| GO:0051244: regulation of cellular physiological process | 191 | 7.79 |
| GO:0007155: cell adhesion | 195 | 7.66 |
| GO:0019882: antigen presentation | 27 | 7.05 |
| GO:0019226: transmission of nerve impulse | 32 | 6.83 |
| GO:0007268: synaptic transmission | 31 | 6.58 |
| GO:0007242: intracellular signaling cascade | 306 | 6.56 |
| GO:0006959: humoral immune response | 67 | 5.78 |
| GO:0006915: apoptosis | 150 | 5.67 |
| GO:0012501: programmed cell death | 150 | 5.4 |
| GO:0044260: cellular macromolecule metabolism | 824 | 5.28 |
GATHER analysis was performed on an mRNA expression data set from patients with lung adenocarcinomas (12) (230 samples). Enrichment of gene ontologies was identified through the Bayes factor. Differential expression of immune response-related gene ontologies was consistently and significantly preserved in the human tumors. Immune response related gene ontologies are shown in bold.
mRNA, messenger RNA.
Discussion
This study confirmed the importance of Nrf2 as a cytoprotective mechanism for reducing tumor development in a relevant model of lung carcinogenesis. Nrf2 KO mice were more sensitive to vinyl carbamate-induced lung adenocarcinomas and developed more and larger tumors than Nrf2 WT mice. More importantly, this study provided evidence that Nrf2 regulated the immune cell infiltration that contributes to tumorigenesis. Deletion of Nrf2 significantly elevated the production of many cytokines and genes involved in antigen presentation and processing. Fewer T cells, including both CD8 cytotoxic T cells and CD4 helper T cells, were found in the lung, but higher populations of tumor-promoting macrophages and MDSCs were found in the lung and spleen, respectively, in Nrf2 KO mice than in WT mice with advanced tumors. These striking effects of Nrf2 on immune processes are consistent with changes found in lung cancer patients, making these findings clinically relevant.
There are many inconsistent results regarding the complex role of Nrf2 in tumor initiation and development, especially in lung cancer. Our study confirmed that Nrf2 deficiency makes mice more susceptible to the carcinogen vinyl carbamate, as more and larger tumors were found in the Nrf2 KO mice (Fig. 1). These results are consistent with the observation that Nrf2-deficient mice are more sensitive to carcinogens (62), as the enzymes that would normally detoxify the carcinogen are missing. Satoh et al. (67) reported that Nrf2 prevented initiation of lung carcinogenesis, as Nrf2-deficient mice exhibited more tumor nodules than the wild-type mice 4–8 weeks after urethane injection. However, at later time points (16 weeks after initiation), the number and size of the lung tumors in the Nrf2 KO mice were reduced, but the tumors were more aggressive in the Nrf2 KO mice than in WT mice (67). KD of Keap1 in mice, which results in constitutive Nrf2 expression, led to resistance to urethane-induced carcinogenesis and fewer and smaller surface tumors at an early stage (up to 16 weeks). In contrast to the results cited previously, Bauer et al. (6) reported that deletion of Nrf2 reduced the number of urethane-induced lung tumors, as Nrf2 had prosurvival effects in tumor cells and the lack of Nrf2 enhanced apoptosis in the lung.
There are a number of possible reasons for these discrepancies. The studies described previously used mice on a variety of genetic backgrounds, with different carcinogens and protocols for initiating lung carcinogenesis, and with diverse time points chosen after initiation for final analysis. Although urethane is commonly used to induce experimental lung cancer, initiation with urethane yields adenomas. In contrast, vinyl carbamate, the carcinogen used in our studies, bypasses the first step oxidation of urethane, and induces invasive adenocarcinomas (46). The most common mutations in non-small cell lung cancer are in the Kras gene, and Kras mutations are prognostic for a poor outcome (63). Both urethane and vinyl carbamate induce mutations in Kras (28, 83), but lung carcinogenesis is more aggressive when induced by vinyl carbamate, as the number, size, and pathological grade of the tumors (44) increase in a reproducible, time-dependent manner. Moreover, vinyl carbamate is much more potent, as only 16 mg/kg is needed to induce lung carcinogenesis versus 1 g/kg for urethane (65).
In addition to the choice of carcinogen, a variety of mouse strains were used in these published studies, including BALB/cCR (6), ICR/CD-1 (67), and BALB/c strains. Mouse strains vary in their susceptibility to lung carcinogenesis, likely as a result of their response to inflammation (72). Lungs were also harvested at different time points, and Nrf2 has different roles during early versus late tumor progression (67). In our studies, tumors were not harvested until 20 or more weeks after initiation, during late stage carcinogenesis, and thus the majority of tumors were high-grade tumors (Fig. 1E).
Our most important finding is that Nrf2 regulates the immune cells and cytokines that can contribute to cancer development (Figs. 2–5). Nrf2 has been shown to directly inhibit the transcription of proinflammatory cytokines (39). To date, however, only a limited number of studies have investigated the effects of Nrf2 on the host immune system during carcinogenesis. Satoh et al. have shown that more inflammatory cells infiltrated into lungs of Keap1-KD mice than in WT Keap1 mice (65, 66), and these changes provided a more permissive environment for lung metastasis in the Lewis lung carcinoma model (66). Moreover, MDSCs, which suppress the activity of cytotoxic CD8 T cells, are regulated by Nrf2 (7), and the number and activity of splenic CD8 T cells were markedly diminished in tumor-bearing Nrf2 WT mice but not in Nrf2 KO mice (67). However, in our study, we observed a reduced percentage of CD8 T cells in Nrf2 KO lungs compared with WT lungs (Fig. 2C), which is consistent with a higher tumor burden in the Nrf2 KO group. CD8 T cells are a major effector of antitumor immunity and are necessary for T cell-based immunotherapies against lung cancer. Dysfunctions in CD8+ T cells in lung tumors are associated with a poor clinical response (60). MDSCs also contribute to tumor growth by inhibiting CD8+ cytotoxic T cells (21).
TAMs are another important immune cell that can drive tumor progression (59), as long-term depletion of macrophages markedly reduced lung tumorigenesis induced by urethane (87). The contribution of Nrf2 and other redox signaling for macrophage activation and polarization is an emerging area of investigation, in both health and disease (8). Both the abundance and activation state of different cell types in the tumor microenvironment influence the balance between tumor-promoting and tumor-suppressing phenotypes. Although we have not yet explored the role of Nrf2 in macrophage polarization in our lung cancer model, additional studies will address this important question. However, the phenotype of increased percentages of macrophages and MDSCs and decreased CD8 T cell populations found in our model (Fig. 2) are associated with a poor prognosis in humans and are consistent with an important beneficial regulatory role for Nrf2 in cancer immunity. Moreover, the enhanced cytokine production (Figs. 3–5) in Nrf2 KO mice further confirmed the unfavorable immune signature in the Nrf2 KO group. Elevated production of cytokines in Nrf2 KO tumors likely recruits macrophages and MDSCs, thus promoting survival and growth of the tumor. These cytokines could be secreted by either tumors or other cells, including macrophages or other immune cells and even fibroblasts (27).
The striking immune signatures found in our RNAseq analysis (Fig. 3) should draw more attention to the relatively unexplored function of Nrf2 on the immune system in lung carcinogenesis. A series of cytokines and major histocompatibility complex antigen genes are differentially expressed between Nrf2 WT and KO tumors (Table 1), and higher expression levels in Nrf2 KO mice are associated with more and larger tumors (Fig. 1). Admittedly, our approach did not differentiate the source of the immune signatures, but future studies will identify the relative contributions of cancer cells versus immune cells. However, the regulation of immune responses by Nrf2 is also found in patients with lung cancer (Fig. 6), indicating the potential value of the Nrf2 status and immune signatures for predicting disease prognosis or treatment. With greater availability of sequencing or RNAseq analysis, tumor mutations and transcriptional profiles are being studied and are providing useful information. Gene signatures can potentially be used as diagnostic or prognostic markers and have been used successfully in breast cancer to guide clinical decisions (5). In lung cancer, research into genetic signatures has been focused on early diagnosis of curable tumors, the need for new treatment regimens for patients with inoperable tumors, and the selection of effective therapies (41). As diagnostic techniques advance and understanding of biology accumulates, targeting Nrf2 under more circumstances in cancer will become more practical.
Because of the complexity of Nrf2 in cancer, targeting Nrf2 requires a deeper understanding of the role of Nrf2 at different stages of cancer (especially between tumor initiation and tumor progression) and on different components of the tumor microenvironment (including tumor cells and immune cells). This information is especially relevant as there are already several compounds that activate the Nrf2 pathway that are being evaluated in the clinic. Dimethyl fumarate has been approved for relapsing multiple sclerosis. Synthetic triterpenoids are in clinical trials for pulmonary diseases, chronic kidney disease, melanoma, etc. Natural compounds including sulforaphane and curcumin are being tested in clinical trials for cancer prevention. In contrast, there is still not a specific, potent Nrf2 inhibitor available, even though accumulating evidence suggests a tumor-promoting role for Nrf2 in advanced cancers. Treating patients appropriately with Nrf2 activators or inhibitors for prevention or treatment of cancer will require careful consideration of the genetic status of Nrf2/Keap1 and knowledge of the tumor stage.
It is also necessary to better understand the mechanisms of compounds that activate Nrf2 because of their potential off-target effects and the complex crosstalk between Nrf2 and other important pathways in cancer. It is well known that Nrf2 reciprocally interacts with HIF-1α, NF-κB, MAPK, and phosphatases, and many of these important signaling pathways are enriched in redox-active cysteines that can be targeted by natural products and other redox-sensitive drugs (9, 58). Some of these proteins, such as HIF-1 and NF-κB, also regulate inflammation and immune responses (18, 57), and additional studies are needed to elucidate the crosstalk between Nrf2 and these pathways, especially regarding the regulation of the immune system in cancer.
In summary, our study demonstrates a protective role for Nrf2 in lung cancer and identifies an immune phenotype in tumors generated using a different model system than has been previously reported. Future studies will investigate the effects Nrf2 has on different types of immune cells and determine whether Nrf2 status is correlated with the response to immunotherapy or other therapies.
Materials and Methods
In vivo lung carcinogenesis studies
All animal studies were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee at Michigan State University (MSU). Nrf2−/− mice on a mixed C57Bl/6 and AKR background were generated as previously described and received as a generous gift from Dr. Jefferson Chan at the University of California San Francisco (15). These mice were backcrossed onto a BALB/c background for eight generations and were found to be 99% congenic (analysis performed by Jackson Laboratory, Bar Harbor, ME). Age-matched female Nrf2 KO female mice and Nrf2 WT BALB/c mice (JAX) were injected i.p. once a week with two to four doses of vinyl carbamate (0.32 mg per mouse per dose, ∼16 mg per kg per dose), beginning when the mice were 6–8 weeks old. The mice were fed AIN-93G diet (BioServ, Flemington, NJ) throughout the study and were weighed weekly. Age-matched cohorts of Nrf2 WT and KO mice (four mice per group) were harvested from 4 to 8 weeks after initiation with carcinogen in the short-term study or 20–40 weeks after initiation with the carcinogen in the long-term study. Lungs were harvested en bloc, inflated with phosphate buffered saline, and tied off in two perpendicular directions. Left lungs were fixed in neutral buffered formalin for histopathology. Right lungs were not fixed but instead immediately homogenized and processed for flow cytometry (two lobes) or were flash frozen and saved at −80°C (the other two lobes). Tumor parameters were assessed as previously described (44, 46) and include the number, size, and classification of the tumors.
RNA extraction and real-time PCR analysis
Lung tumors were dissected and total RNA was isolated using the RNeasy Mini Kit (Qiagen, Valencia, CA). RNA concentrations are determined by NanoDrop, and complementary DNAs (cDNAs) were synthesized by TaqMan Reverse Transcription reagents (Life Technologies, Carlsbad, CA). Total RNA from lung tissue was isolated with TRIzol (Invitrogen, Carlsbad, CA). Two micrograms RNA was used to synthesize cDNA using SuperScript III reverse transcriptase (Invitrogen). Validated Cxcl1, Ccl9, Csf1, and Cxcl12 primers were purchased from Qiagen. iQ SYBR Green Supermix (Bio-Rad, Berkeley, CA) and the ABI 7500 FAST Real-Time PCR system were used to detect gene expression. The delta–delta Ct method was used to calculate relative gene expression (48). Values were normalized to the reference gene actin and expressed as fold induction compared with WT samples.
ELISA
Lung extracts were homogenized in EBC lysis buffer. Cytokine levels were detected in lung homogenates using ELISA kits and the manufacturer's instructions (R&D Systems).
Flow cytometry
The same two lobes of the unfixed right lung were harvested from each mouse for flow cytometry. Freshly harvested lung tissue and half of the spleen were homogenized and incubated in digestion media containing collagenase (300 U/mL; Sigma), dispase (1 U/mL; Worthington), and DNAse (2 U/mL; Calbiochem) for 30 min at 37°C. Cells were then passed through a 40 μm cell strainer (BD Falcon). Lysis solution (eBioscience) was used to eliminate red blood cells. Single cell suspensions were stained with 5 μg/mL antimouse Fc block antibody (eBioscience) and two optimized panels of antibodies (42) for 30 min at 4°C. Panel 1: CD45-VioGreen (Miltenyi, clone:30F11, 3 μg/mL), Gr-1-PE (Miltenyi, RB6-8C5, 3 μg/mL), CD11b-FITC (Miltenyi, clone:M1/70, 3 μg/mL), CD19-APC (BioLegend, clone:1D3/CD19, 2 μg/mL), B220-PerCP/Cy5.5 (BioLegend, clone:RA3-6B2, 2 μg/mL). Panel 2: CD4-FITC (Miltenyi, clone:GK1.5, 3 μg/mL), CD3-PE (BioLegend, clone:145-2C11, 2 μg/mL), CD8-PerCP/Cy5.5 (BioLegend, clone:53–6.7, 2 μg/mL). Live cells were determined by propidium iodide staining (BioLegend, 5 μg/mL), and only live cells were included in the analysis. Flow cytometry was performed using an LSR II flow cytometer with DIVA 6.2 software (BD Falcon) and three laser sources (488 nm, 633 nm, and 407 nm); data were analyzed by FlowJo x.10.0.7r2 software (Tree Star).
RNA sequencing
Lung tumors were dissected out from the lung tissue and were pooled (three to four tumors) before isolating total RNA using the RNeasy Mini Kit (Qiagen). The RNA integrity number was measured using the Aligent Bioanalyzer at the MSU Research Technology Support Facility Genomics Core facility. RNAseq and the bioinformatics analysis from two independent pools were performed by Novogene (Sacramento, CA). In brief, reads were processed and mapped. Raw data were stored in FASTQ format. After the removal of adaptors and low-quality reads, reads were mapped to the mm 10 mouse reference genome using TopHat2 (38). Reads that mapped to genes were counted and normalized to gene length, then reported in FPKM (fragments per kilo bases per million reads) as previously described (53). Routine identification of differentially expressed genes was performed using DESeq2 (3). Raw and processed data were deposited on GEO and can be retrieved though record number GSE99338.
Over-representation analysis of differentially expressed genes
Up- and downregulated genes in tumors from NFR2 KO versus WT mice were processed to identify over-represented genes using PANTHER analysis as previously described (51). To identify those over-represented genes upregulated with the alteration of Keap1 in human adenocarcinomas as identified by cBioPortal, significantly enriched genes were queried for over-represented gene ontologies using GATHER (16).
Human data sets
The impact of gene loss in human lung cancer patients was identified using the TCGA pan (10), adenocarcinoma (12), and squamous cell (11) lung cancer data sets. Figures were created using cBioPortal.org (14, 22).
Statistical analyses
The ex vivo experiments were performed in triplicate and were repeated independently at least three times. Unless noted otherwise, data are presented as mean ± standard error of the mean. Results were analyzed using the t-test or one-way analysis of variance (ANOVA) (SigmaStat 3.5). In vivo data were analyzed by one-way ANOVA followed by Tukey test, or one-way ANOVA on ranks and the Dunn test if the data did not fit a normal distribution (SigmaStat 3.5). A p-value <0.05 was considered statistically significant. For RNAseq data, differential expression analysis of two conditions/groups was performed using the DESeq2 R package (3). It provides accepted and routinely used statistical analysis for determining differential expression in digital gene expression data using a model based on the negative binomial distribution. If the readcount of the ith gene in the jth sample is Kij, there is Kij NB(μij, σij2). The resulting p-values were adjusted using the Benjamini and Hochberg's approach for controlling the false discovery rate.
Supplementary Material
Acknowledgments
The authors thank Sarah Carapellucci, Kayla Zydeck, and Nicole Chaaban for their assistance with the lung carcinogenesis studies and Ana S. Leal for her assistance with the immune population studies. The flow cytometry analysis was done in the Flow Cytometry Core at Michigan State University (MSU), and they also thank Dr. Louis King, Director of the MSU core, for his assistance. Startup funds provided by MSU and a research grant from Geisel School of Medicine at Dartmouth (K.T.L.) were used to complete these studies. D.Z. was supported by the Integrative Pharmacology Sciences Training Program (IPSTP) fellowship 5T32GM092715-07.
Abbreviations Used
- ANOVA
analysis of variance
- Bmp6
bone morphogenetic protein 6
- Ccl9
chemokine (C-C motif) ligand 9
- cDNAs
complementary DNAs
- Csf1
colony stimulating factor 1
- Cxcl1
chemokine (C-X-C motif) ligand 1
- Cxcl12
chemokine (C-X-C motif) ligand 12
- ELISA
enzyme-linked immunosorbent assay
- FPKM
fragments per kilo bases per million reads
- Igfbp5
insulin-like growth factor-binding protein 5
- Igfbp6
insulin-like growth factor-binding protein 6
- KD
knockdown
- Keap1
Kelch-like ECH-associated protein 1
- KO
knockout
- LPS
lipopolysaccharide
- MAPK
mitogen-activated protein kinase
- MDSCs
myeloid-derived suppressor cells
- MMP
matrix metalloproteinase
- mRNA
messenger RNA
- MSU
Michigan State University
- NEF2L2
Nrf2 gene
- Nrf2
nuclear factor (erythroid-derived 2)-like 2
- PCNA
proliferating cell nuclear antigen
- ROS
reactive oxygen species
- RNAseq
RNA sequencing
- TAMs
tumor-associated macrophages
- WT
wild-type
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Aharinejad S, Salama M, Paulus P, Zins K, Berger A, and Singer CF. Elevated CSF1 serum concentration predicts poor overall survival in women with early breast cancer. Endocr Relat Cancer 20: 777–783, 2013 [DOI] [PubMed] [Google Scholar]
- 2.Alam J, Stewart D, Touchard C, Boinapally S, Choi AM, and Cook JL. Nrf2, a Cap'n’Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J Biol Chem 274: 26071–26078, 1999 [DOI] [PubMed] [Google Scholar]
- 3.Anders S, Reyes A, and Huber W. Detecting differential usage of exons from RNA-Seq data. Genome Res 22: 2008–2017, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anwar AA, Li FY, Leake DS, Ishii T, Mann GE, and Siow RC. Induction of heme oxygenase 1 by moderately oxidized low-density lipoproteins in human vascular smooth muscle cells: role of mitogen-activated protein kinases and Nrf2. Free Radic Biol Med 39: 227–236, 2005 [DOI] [PubMed] [Google Scholar]
- 5.Arranz EE, Vara JA, Gamez-Pozo A, and Zamora P. Gene signatures in breast cancer: current and future uses. Transl Oncol 5: 398–403, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bauer AK, Cho HY, Miller-Degraff L, Walker C, Helms K, Fostel J, Yamamoto M, and Kleeberger SR. Targeted deletion of Nrf2 reduces urethane-induced lung tumor development in mice. PLoS One 6: e26590, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beury DW, Carter KA, Nelson C, Sinha P, Hanson E, Nyandjo M, Fitzgerald PJ, Majeed A, Wali N, and Ostrand-Rosenberg S. Myeloid-derived suppressor cell survival and function are regulated by the transcription factor Nrf2. J Immunol 196: 3470–3478, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brune B, Dehne N, Grossmann N, Jung M, Namgaladze D, Schmid T, von Knethen A, and Weigert A. Redox control of inflammation in macrophages. Antioxid Redox Signal 19: 595–637, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bryan HK, Olayanju A, Goldring CE, and Park BK. The Nrf2 cell defence pathway: Keap1-dependent and -independent mechanisms of regulation. Biochem Pharmacol 85: 705–717, 2013 [DOI] [PubMed] [Google Scholar]
- 10.Campbell JD, Alexandrov A, Kim J, Wala J, Berger AH, Pedamallu CS, Shukla SA, Guo G, Brooks AN, Murray BA, Imielinski M, Hu X, Ling S, Akbani R, Rosenberg M, Cibulskis C, Ramachandran A, Collisson EA, Kwiatkowski DJ, Lawrence MS, Weinstein JN, Verhaak RG, Wu CJ, Hammerman PS, Cherniack AD, Getz G, Cancer Genome Atlas Research Network, Artyomov MN, Schreiber R, Govindan R, and Meyerson M. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet 48: 607–616, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 489: 519–525, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 511: 543–550, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Casey SC, Amedei A, Aquilano K, Azmi AS, Benencia F, Bhakta D, Bilsland AE, Boosani CS, Chen S, Ciriolo MR, Crawford S, Fujii H, Georgakilas AG, Guha G, Halicka D, Helferich WG, Heneberg P, Honoki K, Keith WN, Kerkar SP, Mohammed SI, Niccolai E, Nowsheen S, Vasantha Rupasinghe HP, Samadi A, Singh N, Talib WH, Venkateswaran V, Whelan RL, Yang X, and Felsher DW. Cancer prevention and therapy through the modulation of the tumor microenvironment. Semin Cancer Biol 35(Suppl): S199–S223, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, and Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2: 401–404, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan K, Lu R, Chang JC, and Kan YW. NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proc Natl Acad Sci U S A 93: 13943–13948, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang JT. and Nevins JR. GATHER: a systems approach to interpreting genomic signatures. Bioinformatics 22: 2926–2933, 2006 [DOI] [PubMed] [Google Scholar]
- 17.Cullinan SB. and Diehl JA. PERK-dependent activation of Nrf2 contributes to redox homeostasis and cell survival following endoplasmic reticulum stress. J Biol Chem 279: 20108–20117, 2004 [DOI] [PubMed] [Google Scholar]
- 18.Del Prete A, Allavena P, Santoro G, Fumarulo R, Corsi MM, and Mantovani A. Molecular pathways in cancer-related inflammation. Biochem Med (Zagreb) 21: 264–275, 2011 [DOI] [PubMed] [Google Scholar]
- 19.Ding J, Guo C, Hu P, Chen J, Liu Q, Wu X, Cao Y, and Wu J. CSF1 is involved in breast cancer progression through inducing monocyte differentiation and homing. Int J Oncol 49: 2064–2074, 2016 [DOI] [PubMed] [Google Scholar]
- 20.Dranoff G. Cytokines in cancer pathogenesis and cancer therapy. Nat Rev Cancer 4: 11–22, 2004 [DOI] [PubMed] [Google Scholar]
- 21.Ganan-Gomez I, Wei Y, Yang H, Boyano-Adanez MC, and Garcia-Manero G. Oncogenic functions of the transcription factor Nrf2. Free Radic Biol Med 65: 750–764, 2013 [DOI] [PubMed] [Google Scholar]
- 22.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, and Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6: pl1, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guibert N, Ilie M, Long E, Hofman V, Bouhlel L, Brest P, Mograbi B, Marquette CH, Didier A, Mazieres J, and Hofman P. KRAS mutations in lung adenocarcinoma: molecular and epidemiological characteristics, methods for detection, and therapeutic strategy perspectives. Curr Mol Med 15: 418–432, 2015 [DOI] [PubMed] [Google Scholar]
- 24.Guo Q, Gao BL, Zhang XJ, Liu GC, Xu F, Fan QY, Zhang SJ, Yang B, and Wu XH. CXCL12-CXCR4 axis promotes proliferation, migration, invasion, and metastasis of ovarian cancer. Oncol Res 22: 247–258, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo Y, Yu S, Zhang C, and Kong AN. Epigenetic regulation of Keap1-Nrf2 signaling. Free Radic Biol Med 88: 337–349, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayes JD, McMahon M, Chowdhry S, and Dinkova-Kostova AT. Cancer chemoprevention mechanisms mediated through the Keap1-Nrf2 pathway. Antioxid Redox Signal 13: 1713–1748, 2010 [DOI] [PubMed] [Google Scholar]
- 27.Heneberg P. Paracrine tumor signaling induces transdifferentiation of surrounding fibroblasts. Crit Rev Oncol Hematol 97: 303–311, 2016 [DOI] [PubMed] [Google Scholar]
- 28.Hernandez LG. and Forkert PG. Inhibition of vinyl carbamate-induced lung tumors and Kras2 mutations by the garlic derivative diallyl sulfone. Mutat Res 662: 16–21, 2009 [DOI] [PubMed] [Google Scholar]
- 29.Hu R, Saw CL, Yu R, and Kong AN. Regulation of NF-E2-related factor 2 signaling for cancer chemoprevention: antioxidant coupled with antiinflammatory. Antioxid Redox Signal 13: 1679–1698, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hung JY, Horn D, Woodruff K, Prihoda T, LeSaux C, Peters J, Tio F, and Abboud-Werner SL. Colony-stimulating factor 1 potentiates lung cancer bone metastasis. Lab Invest 94: 371–381, 2014 [DOI] [PubMed] [Google Scholar]
- 31.Iida K, Itoh K, Kumagai Y, Oyasu R, Hattori K, Kawai K, Shimazui T, Akaza H, and Yamamoto M. Nrf2 is essential for the chemopreventive efficacy of oltipraz against urinary bladder carcinogenesis. Cancer Res 64: 6424–6431, 2004 [DOI] [PubMed] [Google Scholar]
- 32.Iriki T, Ohnishi K, Fujiwara Y, Horlad H, Saito Y, Pan C, Ikeda K, Mori T, Suzuki M, Ichiyasu H, Kohrogi H, Takeya M, and Komohara Y. The cell–cell interaction between tumor-associated macrophages and small cell lung cancer cells is involved in tumor progression via STAT3 activation. Lung Cancer 106: 22–32, 2017 [DOI] [PubMed] [Google Scholar]
- 33.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, and Nabeshima Y. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun 236: 313–322, 1997 [DOI] [PubMed] [Google Scholar]
- 34.Itoh K, Ishii T, Wakabayashi N, and Yamamoto M. Regulatory mechanisms of cellular response to oxidative stress. Free Radic Res 31: 319–324, 1999 [DOI] [PubMed] [Google Scholar]
- 35.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, and Tuveson DA. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev 15: 3243–3248, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamphorst AO, Pillai RN, Yang S, Nasti TH, Akondy RS, Wieland A, Sica GL, Yu K, Koenig L, Patel NT, Behera M, Wu H, McCausland M, Chen Z, Zhang C, Khuri FR, Owonikoko TK, Ahmed R, and Ramalingam SS. Proliferation of PD-1+ CD8 T cells in peripheral blood after PD-1-targeted therapy in lung cancer patients. Proc Natl Acad Sci U S A 114: 4993–4998, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khor TO, Huang MT, Prawan A, Liu Y, Hao X, Yu S, Cheung WK, Chan JY, Reddy BS, Yang CS, and Kong AN. Increased susceptibility of Nrf2 knockout mice to colitis-associated colorectal cancer. Cancer Prev Res (Phila) 1: 187–191, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, and Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kobayashi EH, Suzuki T, Funayama R, Nagashima T, Hayashi M, Sekine H, Tanaka N, Moriguchi T, Motohashi H, Nakayama K, and Yamamoto M. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat Commun 7: 11624, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kovac S, Angelova PR, Holmstrom KM, Zhang Y, Dinkova-Kostova AT, and Abramov AY. Nrf2 regulates ROS production by mitochondria and NADPH oxidase. Biochim Biophys Acta 1850: 794–801, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuner R. Lung cancer gene signatures and clinical perspectives. Microarrays (Basel) 2: 318–339, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leal AS, Williams CR, Royce DB, Pioli PA, Sporn MB, and Liby KT. Bromodomain inhibitors, JQ1 and I-BET 762, as potential therapies for pancreatic cancer. Cancer Lett 394: 76–87, 2017 [DOI] [PubMed] [Google Scholar]
- 43.Lee JM, Li J, Johnson DA, Stein TD, Kraft AD, Calkins MJ, Jakel RJ, and Johnson JA. Nrf2, a multi-organ protector? FASEB J 19: 1061–1066, 2005 [DOI] [PubMed] [Google Scholar]
- 44.Liby K, Black CC, Royce DB, Williams CR, Risingsong R, Yore MM, Liu X, Honda T, Gribble GW, Lamph WW, Sporn TA, Dmitrovsky E, and Sporn MB. The rexinoid LG100268 and the synthetic triterpenoid CDDO-methyl amide are more potent than erlotinib for prevention of mouse lung carcinogenesis. Mol Cancer Ther 7: 1251–1257, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liby K, Royce DB, Risingsong R, Williams CR, Wood MD, Chandraratna RA, and Sporn MB. A new rexinoid, NRX194204, prevents carcinogenesis in both the lung and mammary gland. Clin Cancer Res 13: 6237–6243, 2007 [DOI] [PubMed] [Google Scholar]
- 46.Liby K, Royce DB, Williams CR, Risingsong R, Yore MM, Honda T, Gribble GW, Dmitrovsky E, Sporn TA, and Sporn MB. The synthetic triterpenoids CDDO-methyl ester and CDDO-ethyl amide prevent lung cancer induced by vinyl carbamate in A/J mice. Cancer Res 67: 2414–2419, 2007 [DOI] [PubMed] [Google Scholar]
- 47.Liby KT. and Sporn MB. Synthetic oleanane triterpenoids: multifunctional drugs with a broad range of applications for prevention and treatment of chronic disease. Pharmacol Rev 64: 972–1003, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Livak KJ. and Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408, 2001 [DOI] [PubMed] [Google Scholar]
- 49.Melkamu T, Qian X, Upadhyaya P, O'Sullivan MG, and Kassie F. Lipopolysaccharide enhances mouse lung tumorigenesis: a model for inflammation-driven lung cancer. Vet Pathol 50: 895–902, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Menegon S, Columbano A, and Giordano S. The dual roles of NRF2 in cancer. Trends Mol Med 22: 578–593, 2016 [DOI] [PubMed] [Google Scholar]
- 51.Mi H, Muruganujan A, Casagrande JT, and Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc 8: 1551–1566, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miyake M, Hori S, Morizawa Y, Tatsumi Y, Nakai Y, Anai S, Torimoto K, Aoki K, Tanaka N, Shimada K, Konishi N, Toritsuka M, Kishimoto T, Rosser CJ, and Fujimoto K. CXCL1-mediated interaction of cancer cells with tumor-associated macrophages and cancer-associated fibroblasts promotes tumor progression in human bladder cancer. Neoplasia 18: 636–646, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mortazavi A, Williams BA, McCue K, Schaeffer L, and Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628, 2008 [DOI] [PubMed] [Google Scholar]
- 54.Ngo HKC, Kim DH, Cha YN. HK, and Surh YJ. Nrf2 mutagenic activation drives hepatocarcinogenesis. Cancer Res 77: 4797–4808, 2017 [DOI] [PubMed] [Google Scholar]
- 55.Ohtsubo T, Kamada S, Mikami T, Murakami H, and Tsujimoto Y. Identification of NRF2, a member of the NF-E2 family of transcription factors, as a substrate for caspase-3(-like) proteases. Cell Death Differ 6: 865–872, 1999 [DOI] [PubMed] [Google Scholar]
- 56.Ortiz ML, Lu L, Ramachandran I, and Gabrilovich DI. Myeloid-derived suppressor cells in the development of lung cancer. Cancer Immunol Res 2: 50–58, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Palazon A, Goldrath AW, Nizet V, and Johnson RS. HIF transcription factors, inflammation, and immunity. Immunity 41: 518–528, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pantano C, Reynaert NL, van der Vliet A, and Janssen-Heininger YM. Redox-sensitive kinases of the nuclear factor-kappaB signaling pathway. Antioxid Redox Signal 8: 1791–1806, 2006 [DOI] [PubMed] [Google Scholar]
- 59.Poczobutt JM, De S, Yadav VK, Nguyen TT, Li H, Sippel TR, Weiser-Evans MC, and Nemenoff RA. Expression profiling of macrophages reveals multiple populations with distinct biological roles in an immunocompetent orthotopic model of lung cancer. J Immunol 196: 2847–2859, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Prado-Garcia H, Romero-Garcia S, Aguilar-Cazares D, Meneses-Flores M, and Lopez-Gonzalez JS. Tumor-induced CD8+ T-cell dysfunction in lung cancer patients. Clin Dev Immunol 2012: 741741, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qu P, Yan C, and Du H. Matrix metalloproteinase 12 overexpression in myeloid lineage cells plays a key role in modulating myelopoiesis, immune suppression, and lung tumorigenesis. Blood 117: 4476–4489, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ramos-Gomez M, Kwak MK, Dolan PM, Itoh K, Yamamoto M, Talalay P, and Kensler TW. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc Natl Acad Sci U S A 98: 3410–3415, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roberts PJ. and Stinchcombe TE. KRAS mutation: should we test for it, and does it matter? J Clin Oncol 31: 1112–1121, 2013 [DOI] [PubMed] [Google Scholar]
- 64.Rushmore TH. and Kong AN. Pharmacogenomics, regulation and signaling pathways of phase I and II drug metabolizing enzymes. Curr Drug Metab 3: 481–490, 2002 [DOI] [PubMed] [Google Scholar]
- 65.Satoh H, Moriguchi T, Saigusa D, Baird L, Yu L, Rokutan H, Igarashi K, Ebina M, Shibata T, and Yamamoto M. NRF2 intensifies host defense systems to prevent lung carcinogenesis, but after tumor initiation accelerates malignant cell growth. Cancer Res 76: 3088–3096, 2016 [DOI] [PubMed] [Google Scholar]
- 66.Satoh H, Moriguchi T, Taguchi K, Takai J, Maher JM, Suzuki T, Winnard PT, Jr., Raman V, Ebina M, Nukiwa T, and Yamamoto M. Nrf2-deficiency creates a responsive microenvironment for metastasis to the lung. Carcinogenesis 31: 1833–1843, 2010 [DOI] [PubMed] [Google Scholar]
- 67.Satoh H, Moriguchi T, Takai J, Ebina M, and Yamamoto M. Nrf2 prevents initiation but accelerates progression through the Kras signaling pathway during lung carcinogenesis. Cancer Res 73: 4158–4168, 2013 [DOI] [PubMed] [Google Scholar]
- 68.Sheridan C. and Downward J. Overview of KRAS-driven genetically engineered mouse models of non-small cell lung cancer. Curr Protoc Pharmacol 70: 14.35.1–14.35.16, 2015 [DOI] [PubMed] [Google Scholar]
- 69.Sohail A, Sun Q, Zhao H, Bernardo MM, Cho JA, and Fridman R. MT4-(MMP17) and MT6-MMP (MMP25), a unique set of membrane-anchored matrix metalloproteinases: properties and expression in cancer. Cancer Metastasis Rev 27: 289–302, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sparaneo A, Fabrizio FP, and Muscarella LA. Nrf2 and Notch signaling in lung cancer: near the crossroad. Oxid Med Cell Longev 2016: 1–17, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sporn MB. and Liby KT. NRF2 and cancer: the good, the bad and the importance of context. Nat Rev Cancer 12: 564–571, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stathopoulos GT, Sherrill TP, Cheng DS, Scoggins RM, Han W, Polosukhin VV, Connelly L, Yull FE, Fingleton B, and Blackwell TS. Epithelial NF-kappaB activation promotes urethane-induced lung carcinogenesis. Proc Natl Acad Sci U S A 104: 18514–18519, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.To C, Ringelberg CS, Royce DB, Williams CR, Risingsong R, Sporn MB, and Liby KT. Dimethyl fumarate and the oleanane triterpenoids, CDDO-imidazolide and CDDO-methyl ester, both activate the Nrf2 pathway but have opposite effects in the A/J model of lung carcinogenesis. Carcinogenesis 36: 769–781, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang D, Sun H, Wei J, Cen B, and DuBois RN. CXCL1 is critical for premetastatic niche formation and metastasis in colorectal cancer. Cancer Res 77: 3655–3665, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang R, Zhang J, Chen S, Lu M, Luo X, Yao S, Liu S, Qin Y, and Chen H. Tumor-associated macrophages provide a suitable microenvironment for non-small lung cancer invasion and progression. Lung Cancer 74: 188–196, 2011 [DOI] [PubMed] [Google Scholar]
- 76.Wang XJ, Sun Z, Villeneuve NF, Zhang S, Zhao F, Li Y, Chen W, Yi X, Zheng W, Wondrak GT, Wong PK, and Zhang DD. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis 29: 1235–1243, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang Z, Sun J, Feng Y, Tian X, Wang B, and Zhou Y. Oncogenic roles and drug target of CXCR4/CXCL12 axis in lung cancer and cancer stem cell. Tumour Biol 37: 8515–8528, 2016 [DOI] [PubMed] [Google Scholar]
- 78.Xie S, Zeng W, Fan G, Huang J, Kang G, Geng Q, Cheng B, Wang W, and Dong P. Effect of CXCL12/CXCR4 on increasing the metastatic potential of non-small cell lung cancer in vitro is inhibited through the downregulation of CXCR4 chemokine receptor expression. Oncol Lett 7: 941–947, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xu C, Huang MT, Shen G, Yuan X, Lin W, Khor TO, Conney AH, and Kong AN. Inhibition of 7,12-dimethylbenz(a)anthracene-induced skin tumorigenesis in C57BL/6 mice by sulforaphane is mediated by nuclear factor E2-related factor 2. Cancer Res 66: 8293–8296, 2006 [DOI] [PubMed] [Google Scholar]
- 80.Xu C, Li CY, and Kong AN. Induction of phase I, II and III drug metabolism/transport by xenobiotics. Arch Pharm Res 28: 249–268, 2005 [DOI] [PubMed] [Google Scholar]
- 81.Yan HH, Jiang J, Pang Y, Achyut BR, Lizardo M, Liang X, Hunter K, Khanna C, Hollander C, and Yang L. CCL9 induced by TGFbeta signaling in myeloid cells enhances tumor cell survival in the premetastatic organ. Cancer Res 75: 5283–5298, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang DL, Xin MM, Wang JS, Xu HY, Huo Q, Tang ZR, and Wang HF. Chemokine receptor CXCR4 and its ligand CXCL12 expressions and clinical significance in bladder cancer. Genet Mol Res 14: 17699–17707, 2015 [DOI] [PubMed] [Google Scholar]
- 83.You M, Candrian U, Maronpot RR, Stoner GD, and Anderson MW. Activation of the Ki-ras protooncogene in spontaneously occurring and chemically induced lung tumors of the strain A mouse. Proc Natl Acad Sci U S A 86: 3070–3074, 1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yuan M, Zhu H, Xu J, Zheng Y, Cao X, and Liu Q. Tumor-derived CXCL1 promotes lung cancer growth via recruitment of tumor-associated neutrophils. J Immunol Res 2016: 1–11, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yuan X, Huang H, Huang Y, Wang J, Yan J, Ding L, Zhang C, and Zhang L. Nuclear factor E2-related factor 2 knockdown enhances glucose uptake and alters glucose metabolism in AML12 hepatocytes. Exp Biol Med (Maywood) 242: 930–938, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zanetti M. and Tapping CD. T cells for cancer immunotherapy: the choice of personalized genomics. J Immunol 194: 2049–2056, 2015 [DOI] [PubMed] [Google Scholar]
- 87.Zaynagetdinov R, Sherrill TP, Polosukhin VV, Han W, Ausborn JA, McLoed AG, McMahon FB, Gleaves LA, Degryse AL, Stathopoulos GT, Yull FE, and Blackwell TS. A critical role for macrophages in promotion of urethane-induced lung carcinogenesis. J Immunol 187: 5703–5711, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang L, Wang H, Fan Y, Gao Y, Li X, Hu Z, Ding K, Wang Y, and Wang X. Fucoxanthin provides neuroprotection in models of traumatic brain injury via the Nrf2-ARE and Nrf2-autophagy pathways. Sci Rep 7: 46763, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhu Y, Knolhoff BL, Meyer MA, Nywening TM, West BL, Luo J, Wang-Gillam A, Goedegebuure SP, Linehan DC, and DeNardo DG. CSF1/CSF1R blockade reprograms tumor-infiltrating macrophages and improves response to T-cell checkpoint immunotherapy in pancreatic cancer models. Cancer Res 74: 5057–5069, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.