Abstract
Cantharidin (CTD) is a potent anticancer small molecule produced by several species of blister beetle. It has been a traditional medicine for the management of warts and tumors for many decades. CTD suppresses tumor growth by inducing apoptosis, cell cycle arrest, and DNA damage and inhibits protein phosphatase 2 phosphatase activator (PP2A) and protein phosphatase 1 (PP1). CTD also alters lipid homeostasis, cell wall integrity, endocytosis, adhesion, and invasion in yeast cells. In this study, we identified additional molecular targets of CTD using a Saccharomyces cerevisiae strain that expresses a cantharidin resistance gene (CRG1), encoding a SAM-dependent methyltransferase that methylates and inactivates CTD. We found that CTD specifically affects phosphatidylethanolamine (PE)-associated functions that can be rescued by supplementing the growth media with ethanolamine (ETA). CTD also perturbed endoplasmic reticulum (ER) homeostasis and cell wall integrity by altering the sorting of glycosylphosphatidylinositol (GPI)-anchored proteins. A CTD-dependent genetic interaction profile of CRG1 revealed that the activity of the lipid phosphatase cell division control protein 1 (Cdc1) in GPI-anchor remodeling is the key target of CTD, independently of PP2A and PP1 activities. Moreover, experiments with human cells further suggested that CTD functions through a conserved mechanism in higher eukaryotes. Altogether, we conclude that CTD induces cytotoxicity by targeting Cdc1 activity in GPI-anchor remodeling in the ER.
Keywords: glycosylphosphatidylinositol (GPI-anchor), yeast genetics, anticancer drug, phospholipid, unfolded protein response (UPR), cell signaling, lipid metabolism, cantharidin, Cdc1, ER homeostasis, ER-Golgi transport, GPI-anchor remodeling, lipid phosphatase
Introduction
Glycosylphosphatidylinositol (GPI)3 anchor biosynthesis is an essential and conserved pathway in eukaryotes. GPI-anchoring is a type of post-translational modification of proteins destined to the plasma membrane or cell wall. This modification takes place in the endoplasmic reticulum (ER), which acts as a signal for sorting of the proteins to the cell surface. The GPI-anchored protein sorting occurs via the ER–Golgi traffic system (1). The GPI-anchor biosynthesis takes place in the inner membrane of ER through a series of enzymatic reactions, which is subsequently incorporated onto the C terminus of protein (1). Post-synthesis, the GPI-anchor undergoes several steps of modifications in the ER (yeast) or Golgi (mammals). This sequential process of modifications is called GPI-anchor remodeling. Bst1, Cdc1, Ted1, Per1, Gup1, and Cwh43 are the key factors that mediate the process of GPI-anchor remodeling in yeast (1, 2). Cdc1 acts as a Mn2+-dependent (EtNP) phosphodiesterase that removes EtNP from the first mannose of the GPI (3, 4). CDC1 is a homolog of human PGAP5 and is essential for cell survival (4, 5). Therefore, different point mutants have been created to characterize the CDC1 function (3, 4, 6). Previous studies have reported that cdc1-314 mutant exhibits a defect in GPI-anchored protein sorting, temperature sensitivity, cell wall damage, actin depolarization, increased Ca2+ ion signaling, and unfolded protein response (UPR) (3, 4). GPI-anchored proteins have diverse biological functions in different organisms. In yeast, they regulate cell wall biosynthesis, flocculation, adhesion, and invasion (7). In protozoa (Trypanosoma brucei), GPI-anchored proteins form a protective layer on the cell surface, which helps in the virulence of the parasite (8, 9). In plants, it is required for the cell wall biosynthesis, developmental morphogenesis, pollen tube germination, etc. (2). In mammals, it regulates embryogenesis, fertilization, immune response, neurogenesis, etc. (2, 9). GPI-anchored proteins are also associated with the progression, invasion, and metastasis of malignant cells (10–12). A few GPI-anchored proteins have been found to serve as markers for the specific stages of tumors (13, 14).
Cantharidin (CTD) is a terpenoid produced by blister beetles as a defense molecule. The people of a few Asian countries have been using it as a traditional medicine for the treatment of warts and molluscum contagiosum for more than 2,000 years (15). In the last few decades, many studies have demonstrated the anticancer property of CTD. It has been shown to inhibit the growth of hepatocellular carcinoma (16), leukemia (17), pancreatic (18), colorectal (19), gallbladder (20), oral (21), and breast cancer (22). The serine-threonine protein phosphatases, PP1 and PP2A, are the only reported molecular targets of CTD (23, 24). The inhibition of PP2A causes cell cycle arrest (25, 26) and apoptosis (27, 28). CTD also impairs different cellular processes such as heat shock response (29), autophagy (22), DNA damage response, and mitogen-activated protein kinase signaling (18, 21). One of these studies also demonstrated a PP2A- or PP1-independent alteration in heat shock response (29), suggesting the existence of additional molecular targets of CTD (29, 30). Most of the studies performed with CTD were based on mammalian cell lines, making it difficult to decipher a conserved mechanism of action of the drug due to their tissue-specific origin and differential gene regulation. Hence, yeast (Saccharomyces cerevisiae) serves as an appropriate model system to identify the conserved molecular targets of the drug (31–33). Previous studies showed that yeast YHR209W gene was essentially required for CTD resistance (34), which was subsequently named as cantharidin-resistant gene (CRG1) (35). Later, Crg1 was characterized as a SAM-dependent methyltransferase that detoxifies CTD by methylation (30). Deletion of CRG1 enables the identification of the molecular targets of CTD more easily, so we utilized budding yeast as a model organism to dissect the molecular mechanism of CTD toxicity.
Our study was focused on the identification of the conserved cellular pathways targeted by CTD. Interestingly, we found that CTD impaired the GPI-anchored protein sorting by targeting the remodeling process in ER. More specifically, it affected the Cdc1 activity, leading to multiple cellular changes, such as missorting and aggregation of GPI-anchored proteins, temperature sensitivity, cell wall damage, and decreased UPR. Most of the CTD-induced phenotypes observed in yeast cells were also reproducible in human cells. Our comprehensive genetic and cell biology–based experiments revealed that the Cdc1 activity is a molecular target of CTD in eukaryotic cells. Overall, we identified the GPI-anchor remodeling as a direct target of CTD.
Results
Supplementation of ethanolamine (ETA) suppresses the cytotoxic effect of CTD
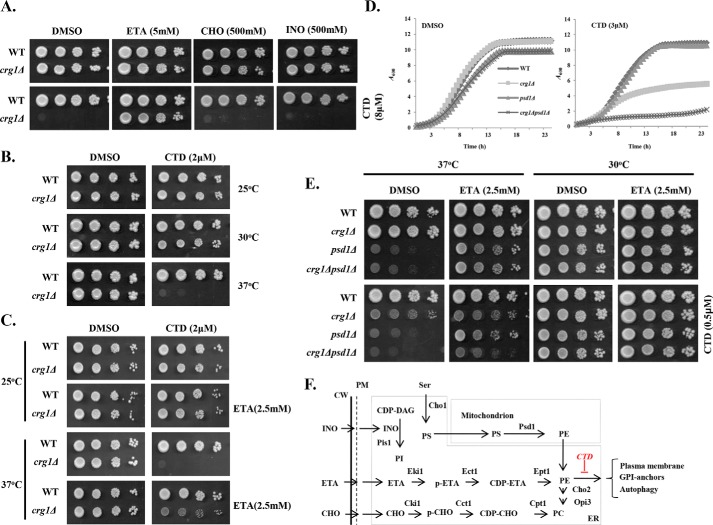
Previous studies have shown that CTD treatment affects the lipid homeostasis in budding yeast by inhibition of the elongation of short-chain phospholipids to long-chain phospholipids (30). The phospholipid imbalance can be restored with exogenous supplementation of the precursor molecules. For example, supplementation of ETA and choline (CHO) activates the synthesis of phosphatidylethanolamine (PE) and phosphatidylcholine (PC), respectively, via an alternative pathway, i.e. the Kennedy Pathway (Fig. 1F) (36). Inositol (INO) and Ser enter into the canonical pathways of phosphatidylinositol (PI) and phosphatidylserine (PS) biosynthesis, respectively (Fig. 1F) (37, 38). Based on these phenomena, we sought to identify the specific phospholipid affected by CTD. We supplemented the medium with specific precursor molecules, ETA, CHO, and INO, with or without CTD and measured the growth of WT and crg1Δ strains (Fig. 1A and Fig. S10). CTD exposure produced a lethal effect on crg1Δ mutant compared with WT (30). However, ETA supplementation completely rescued the growth of the crg1Δ strain from CTD cytotoxicity (Fig. 1A and Fig. S1, A–D). On the other hand, CHO and INO supplementation failed to rescue the growth of crg1Δ strain in CTD-containing medium (Fig. 1A). This observation suggests that CTD specifically targets PE. The exclusive rescue in the growth of the CTD-treated cells by ETA supplementation was a surprising phenomenon, because PE and PC are both synthesized in the same pathway (37). Thus, we believe that CTD may not affect the PE biosynthesis pathway, but it might be altering the PE-associated structures or functions. PE plays an essential role in maintaining membrane and cell wall integrity under heat stress (38, 39), so we examined the fitness profile of WT and crg1Δ strains in heat stress (37 °C) with a permissible dose of CTD (2 μm). Interestingly, we found complete inhibition of growth of crg1Δ mutant at 37 °C in the presence of CTD, whereas the growth was unaffected at optimum (30 °C) or below the optimum (25 °C) temperature (Fig. 1B). CTD cytotoxicity was suppressed again at 37 °C by supplementation of ETA (Fig. 1C). PE biosynthesis takes place in mitochondria and Golgi/vacuole with the help of Psd1 and Psd2, respectively (40). A major fraction (>90%) of the net PE in a cell is synthesized by Psd1 in mitochondria (40), so we created a double-deletion mutant, crg1Δpsd1Δ, to check synthetic lethality between PSD1 and CRG1 in the presence of CTD. For this purpose, WT, crg1Δ, psd1Δ, and crg1Δpsd1Δ strains were grown in CTD-containing medium. We found that the crg1Δpsd1Δ mutant was hypersensitive to CTD than crg1Δ, suggesting that PE is essentially required to tolerate CTD toxicity (Fig. 1D and Fig. S1, E–H). crg1Δpsd1Δ mutant followed the same trend at higher temperature as well (37 °C) (Fig. 1E). The synthetic lethality between CRG1 and PSD1 in the presence of CTD suggests an essential role of PE to tolerate CTD toxicity. These observations suggest that CTD affects the PE-associated functions (Fig. 1F); therefore, enhanced synthesis of PE helps to overcome the CTD toxicity.
Figure 1.
CTD specifically targets PE in crg1Δ cells. A, B, C, and E, growth sensitivity assays. Equal numbers of cells were serially diluted and spotted on SC agar medium. Images were captured after 72 h of incubation. A, supplementation of ETA rescues crg1Δ mutant from CTD toxicity. The phospholipid precursors ETA, INO, and CHO were added into SC agar medium with or without CTD. WT and crg1Δ cells were spotted and incubated at 30 °C. B, CTD toxicity increases with rising temperature. WT and crg1Δ cells were spotted on SC agar medium containing CTD and incubated at different temperatures (25, 30, and 37 °C). C, ETA supplementation rescues the crg1Δ mutant from CTD toxicity at higher temperature. WT and crg1Δ cells were spotted on SC agar medium containing CTD with and without ETA supplementation and incubated at different temperatures (25, 30, and 37 °C). D and E, CRG1 shows synthetic lethality with PSD1 under CTD stress. D, growth curve assay. Equal numbers of cells of WT, crg1Δ, psd1Δ, and crg1Δpsd1Δ were grown at 30 °C with or without CTD in liquid medium. A600 was measured at the time interval of 30 min using an automated plate reader for 23 h. E, WT, crg1Δ, psd1Δ, and crg1Δpsd1Δ cells were spotted on SC agar medium containing CTD with or without ETA and incubated at two different temperatures (30 and 37 °C). F, phospholipid biosynthesis pathways in yeast (37, 66, 77, 78). INO and Ser in medium are directly utilized to synthesize PI and PS with the help of Pis1 and Cho1, respectively. PE and PC biosynthesis has two pathways. The first pathway is canonical biosynthesis of PE/PC, which takes place in mitochondria and the ER. The first reaction starts in the ER, where Cho2 synthesizes PS from Ser. PS is transported to mitochondria, where Psd1 catalyzes its decarboxylation to synthesize PE. (A similar mechanism also takes place in Golgi and vacuole by Psd2, which contributes a very minor fraction of the net PE content). Next, PE is transported again to the ER, where Cho2 and Opi3 convert it into PC via a sequence of methylation reactions. The second pathway is noncanonical PE or PC synthesis, also known as the Kennedy pathway. In this pathway, externally supplemented precursors (ETA/CHO) are utilized and converted into PE or PC, respectively, via a series of enzymatic reactions.
CTD alters ER homeostasis by inhibition of UPR
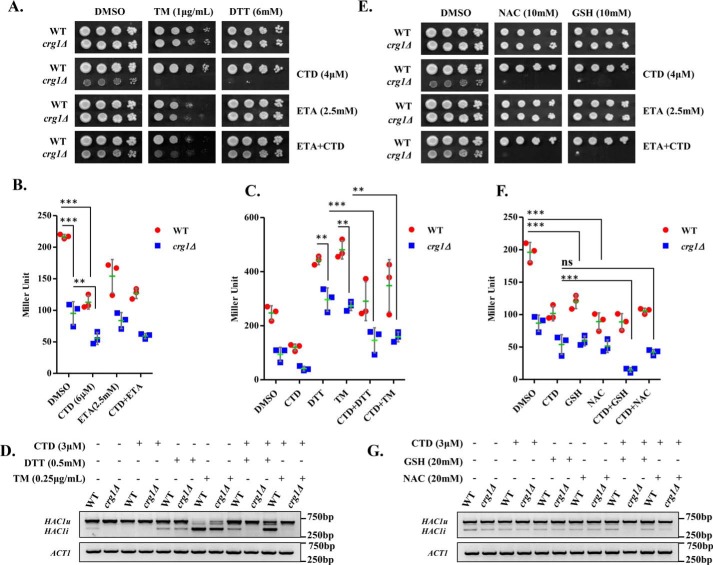
ER is the organelle for the synthesis of the major phospholipids. Imbalance in the phospholipid composition of lipid bilayer membrane is reported to induce ER stress (41–44). Existing evidence suggests that CTD also perturbs ER-synthesized phospholipids (30); thus, we proposed that CTD might be altering the ER homeostasis. We examined ER stress in crg1Δ cells in the presence of CTD. First, WT and crg1Δ cells were co-treated with CTD and ER stress (or UPR) inducers, dithiothreitol (DTT) or tunicamycin (TM), to check whether there was any synergistic effect between the two molecules. For this, we chose a permissible dose of CTD (4 μm) for the crg1Δ mutant, at which it survived, but survival was lower than that of the WT. Both the strains were spotted on CTD-containing medium, with and without TM or DTT. Interestingly, the co-treatments (CTD + TM and CTD + DTT) inhibited the growth of crg1Δ cells more severely compared with only CTD treatment (Fig. 2A and Fig. S1, I–L and Fig. S11, A). The synergistic lethal effect on the growth of crg1Δ cells upon co-treatments suggests that CTD perturbs ER homeostasis. Next, we measured UPR by β-gal assay with the help of UPRE-LacZ reporter plasmid (45, 46). We found that the basal level of UPR was lower in crg1Δ cells compared with WT, and CTD treatment further inhibited UPR in both the strains (WT and crg1Δ) (Fig. 2B). Because ETA supplementation rescues the yeast cells from CTD toxicity, we decided to measure UPR upon ETA supplementation. Surprisingly, ETA supplementation could not rescue UPR inhibited by CTD (Fig. 2B), suggesting that CTD inhibits UPR via a distinct mechanism independent of PE in ER. To gain more insight into this mechanism, UPR was measured upon co-treatments of cells with CTD + DTT and CTD + TM. We found decreased UPR levels in crg1Δ cells upon DTT and TM treatment (Fig. 2C). Moreover, CTD treatment strongly inhibited UPR induced by DTT or TM in WT as well as crg1Δ mutant (Fig. 2C). Consistent with these observations, we also found inhibition of HAC1 mRNA splicing in crg1Δ cells compared with WT. The splicing of HAC1 mRNA was further inhibited in both of the strains, WT and crg1Δ, upon CTD treatment (Fig. 2D). DTT and TM treatment strongly induced HAC1 mRNA splicing; however, the presence of CTD with DTT or TM suppressed HAC1 mRNA splicing (Fig. 2D). These results suggest that CTD inhibits UPR by making an obstruction in HAC1 mRNA splicing, although the mechanism remains unclear.
Figure 2.
CTD treatment inhibits UPR by alteration of the ER-redox homeostasis. A, UPR inducers (DTT/TM) synergistically enhance CTD toxicity. Equal numbers of serially diluted WT and crg1Δ cells were spotted on CTD-containing SC agar medium with or without DTT/TM in the presence or absence of ETA and incubated at 30 °C for 72 h. B, CTD inhibits UPR. WT and crg1Δ strains transformed with pPW344 (UPRE-LacZ) plasmid were grown in SC-URA medium at 30 °C. Cells were treated with CTD (6 μm) with or without ETA (2.5 mm) at the mid-exponential phase (A600 = 0.8) and incubated for 2 h. A β-gal assay was performed to measure the UPR. The graph shows a scatter plot of each data point of three independent experiments with mean (horizontal green line) ± S.D. (error bars). Statistical analysis was done with GraphPad Prism version 5, applying two-way ANOVA and Bonferroni post hoc test, where p ≤ 0.05 (*), p ≤ 0.01 (**), and p ≤ 0.001 (***). C, CTD inhibits UPR in presence of DTT and TM. WT and crg1Δ strains carrying pPW344 vector were grown until the mid-exponential phase and treated with CTD (3 μm) in combination with DTT (0.5 mm) or TM (0.25 μg/ml) for 2 h. A β-gal assay was performed to measure the UPR. The graph shows a scatter plot of each data point of three independent experiments with mean ± S.D. (error bars). Statistical analysis was done using GraphPad Prism version 5, applying two-way ANOVA and Bonferroni post hoc test, where p ≤ 0.05 (*), p ≤ 0.01 (**), and p ≤ 0.001 (***). D, CTD inhibits HAC1 mRNA splicing. WT and crg1Δ strains were grown in the conditions mentioned above (C), and HAC1 mRNA splicing was measured by RT-PCR. HAC1(u), unspliced HAC1; HAC1(i), spliced HAC1. The figure represents one of the three independently performed experiments. E, GSH or NAC supplementation enhances the CTD cytotoxicity. Equal numbers of WT and crg1Δ cells were serially diluted and spotted on CTD-containing SC agar medium with or without reducing agents (GSH and NAC) in the presence or absence of ETA, incubated at 30 °C for 72 h. F, GSH and NAC supplementation reduces UPR. WT and crg1Δ strains transformed with pPW344 (UPRE-LacZ) were grown in SC-URA medium at 30 °C until mid-exponential phase. The cells were treated with CTD (3 μm) in the presence or absence of GSH (20 mm) or NAC (20 mm) for 2 h and processed for the β-gal assay. The graph shows a scatter plot of each data point of three independent experiments with mean ± S.D. (error bars). Statistical analysis was done using GraphPad Prism version 5, applying two-way ANOVA and Bonferroni post hoc test, where p ≤ 0.05 (*), p ≤ 0.01 (**), and p ≤ 0.001 (***). G, GSH and NAC supplementation enhances the CTD-mediated inhibition of HAC1 splicing. WT and crg1Δ cells were grown in SC medium at 30 °C until mid-exponential phase under the same conditions mentioned above (F), and the HAC1 mRNA splicing was measured by RT-PCR. The figure represents one of the three independently performed experiments.
Our data suggest that CTD exposure leads to ER stress that cannot be rescued by ETA supplementation. The ER-lumen maintains higher oxidation potential with the help of a low GSH/GSSG ratio (1:1 to 3:1) compared with the high GSH/GSSG ratio (30:1 to 100:1) of the cytosol (47). GSH provides a redox buffer for the catalytic activity of the protein-folding enzymes in the ER (48, 49). The imbalance in GSH/GSSG ratio in ER impairs oxidative protein folding that causes ER stress (50, 51). Based on these previous findings, we predicted that CTD-induced ER stress might be due to imbalance in the GSH/GSSG ratio in ER. To test this hypothesis, we checked the effect of GSH on CTD toxicity. We used the permissible dose of CTD (4 μm) for crg1Δ mutant and supplemented the medium with a high dose of GSH and NAC. We found that the growth of crg1Δ mutant was suppressed in the presence of either of the two reducing molecules, GSH or NAC, along with CTD. However, GSH or NAC alone did not show any effect on the growth of crg1Δ mutant (Fig. 2E and Fig. S11, B). Furthermore, the supplementation of ETA did not rescue the growth of crg1Δ mutant upon CTD + GSH or CTD + NAC treatments. This result supports the previous observation where, upon ETA supplementation, UPR suppressed by CTD treatment could not be rescued (Fig. 2B). Similar observations were also made in liquid growth culture (Fig. S2, A–F). Next, we measured UPR using a β-gal assay. Interestingly, we observed that GSH or NAC supplementation results in the reduction in UPR in WT and crg1Δ cells (Fig. 2F). As CTD treatment also inhibits UPR, we observed severe reduction in UPR upon co-treatments with CTD + GSH or CTD + NAC (Fig. 2F). We also found an inhibition in HAC1 mRNA splicing upon the addition of GSH and NAC (Fig. 2G), which was more when the cells were co-treated with CTD + GSH or CTD + NAC (Fig. 2G). These observations suggest that CTD-mediated inhibition of UPR is probably due to imbalance in ER-redox homeostasis, which is enhanced with the addition of GSH. It also explains the reason why ETA supplementation failed to rescue the UPR.
CTD exposure perturbs the cell wall integrity via ER stress
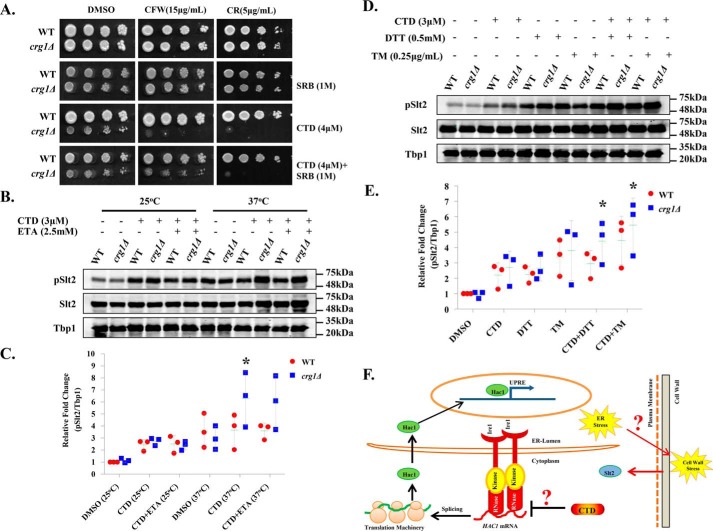
Yeast cell wall biosynthesis and maintenance largely depend on functional ER (7, 52, 53). Dysfunctional ER affects the synthesis, modifications, folding, and transport of the proteins destined to the plasma membrane or cell wall. Based on our results, we proposed that CTD-induced ER stress could also perturb cell wall integrity. To examine the effects of CTD on cell wall integrity, we measured chitin content in the cell wall of WT and crg1Δ cells by calcofluor white (CFW) staining (54). We found substantial increase in chitin content in crg1Δ cells upon CTD treatment, suggesting that CTD treatment induced cell wall damage (Fig. S3A). To gain more insight on the effect of CTD on cell wall integrity, we co-treated the cells with CTD and cell wall–perturbing agents, Congo red (CR) and CFW. We used a permissible dose of CTD (4 μm) in combination with cell wall–perturbing agents to measure the growth of WT and crg1Δ cells. We found that crg1Δ mutant did not grow in either of the co-treatments (CTD + CR or CTD + CFW), whereas the growth was unaffected in individual treatments (Fig. 3A and Fig. S11, C and D). We also supplemented sorbitol (SRB) into the medium to maintain the osmotic balance across the cell membrane. SRB rescued the growth of crg1Δ mutant upon CTD + CFW treatment, but not upon CTD + CR treatment. That suggests the CTD + CR–induced cell wall damage is irreversible, although the mechanism remains to be identified (Fig. 3A). We obtained similar results in liquid growth culture under similar conditions (Fig. S3, B–E). Yeast cell wall damage is sensed by many sensor proteins residing in the cell wall, which in turn activate downstream signaling via Slt2 (55). Activation of Slt2 triggers the transcription of cell wall maintenance genes via Rlm1 and Swi4–Swi6 transcription factors (52, 56). Hence, we did Western blot analysis of Slt2 phosphorylation in WT and crg1Δ cells upon CTD treatment. We observed increased phosphorylation of Slt2 in crg1Δ cells upon CTD treatment at 25 °C. Slt2 phosphorylation increased further when the cells were grown at 37 °C, and CTD treatment induced Slt2 phosphorylation strongly in crg1Δ cells at this temperature (Fig. 3, B and C). As we knew that CTD cytotoxicity could be neutralized by ETA supplementation, we decided to measure Slt2 phosphorylation in CTD-treated cells supplemented with ETA. We did not find significant decrease in Slt2 phosphorylation upon ETA supplementation in CTD-treated cells (Fig. 3, B and C). Next, we challenged the WT and crg1Δ cells with the combined treatment of CTD and UPR inducers (DTT and TM) to measure the synergistic effect on Slt2 phosphorylation. We found strong induction in Slt2 phosphorylation upon co-treatments with CTD + DTT or CTD + TM compared with individual treatments (CTD/DTT/TM) (Fig. 3, D and E). This observation suggests that CTD-induced cell wall damage might be due to ER stress. Furthermore, we checked Slt2 phosphorylation upon co-treatments with CTD + GSH and CTD + NAC. We found that both the co-treatments did not cause any significant change in Slt2 phosphorylation compared with CTD alone. Moreover, only GSH or NAC did not affect Slt2 phosphorylation (Fig. S12, A and B), suggesting a distinct mechanism of GSH-induced ER stress unlike DTT, TM, and CTD. We conclude that CTD perturbs cell wall integrity via ER stress. Our data indicate the linked phenotypes of ER stress and cell wall damage, which is illustrated briefly in the schematic diagram (Fig. 3F).
Figure 3.
CTD-induced ER stress perturbs the cell wall integrity. A, CTD and cell wall–perturbing agents (CR or CFW) are synergistically lethal to crg1Δ mutant. Equal numbers of WT and crg1Δ cells were serially diluted and spotted on SC agar medium containing CTD with and without CR or CFW. The cells were incubated at 30 °C for 72 h. B–E, Western blot analysis of the Slt2 phosphorylation. Whole-cell lysates were prepared from WT and crg1Δ cells grown in different conditions. Tbp1 was taken as a loading control. B, CTD-induced cell wall damage increases with heat stress. WT and crg1Δ strains were grown at two different temperatures, 24 and 37 °C, until mid-exponential phase (0.8 A600) and then treated with CTD in the presence or absence of ETA for 2 h. The data represent one of the three independently performed experiments. C, densitometry quantification of the three biological repeats of the Western blots shown in B, with the help of ImageJ software. The graph shows a scatter plot of each data point of three independent experiments with mean ± S.D. (error bars). Statistical analysis was done, applying Student's t test, where p ≤ 0.05 (*), p ≤ 0.01 (**), and p ≤ 0.001 (***). D, CTD-induced cell wall damage increases with UPR induction. WT and crg1Δ strains were grown at 24 °C until mid-exponential phase and treated with CTD with or without DTT or TM for 2 h. The figure represents one of the three independently performed experiments. E, densitometry quantification of the three biological repeats of the western blots shown in D, with the help of ImageJ software. The graph shows a scatter plot of each data point of three independent experiments with mean ± S.D. (error bars). Statistical analysis was done, applying Student's t test, where p ≤ 0.05 (*), p ≤ 0.01 (**), and p ≤ 0.001 (***). F, a hypothetical model connecting two majorly affected pathways, UPR and CWI, by CTD. The CTD inhibits HAC1 mRNA splicing and subsequent UPRE activation, which promotes ER stress. Yeast cell wall biosynthesis is an ER-dependent process; the CTD-induced ER stress may alter the cell wall integrity, evident in this study by Slt2 activation. The probable link between ER stress and cell wall damage could be the GPI-anchored protein sorting, and it might be the direct target of CTD.
CTD alters GPI-anchored protein sorting
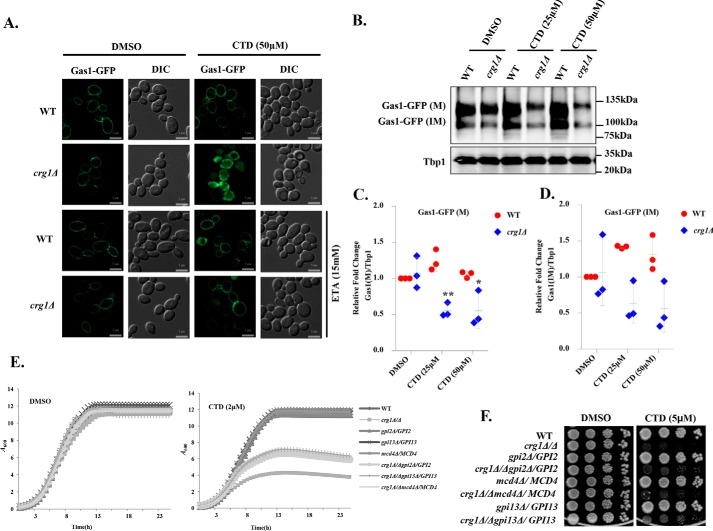
To identify the major pathway affected by CTD treatment, we did functional clustering (57) of the genetic interactors of CRG1 that show synthetic lethality in the presence of CTD (30). We found that the majorly affected pathways were associated with the ER–Golgi traffic system (Table S4). Yeast cell wall biosynthesis and maintenance mainly depend on the GPI-anchored proteins, sorted by the ER–Golgi traffic system (1, 7, 58). PE also plays a crucial role in the regulation of this traffic system (4, 7, 44, 58). Thus, we hypothesized that the CTD-induced cell wall damage might be due to the defect in GPI-anchored protein sorting. We decided to study the GPI-anchored protein sorting upon CTD treatment. We used Gas1-GFP as a model GPI-anchored protein and tracked its localization in WT and crg1Δ cells upon CTD treatment (3, 4, 59). We observed that CTD induced missorting and aggregation of Gas1-GFP in crg1Δ cells (Fig. 4A). Additionally, Gas1-GFP protein, mature (M) and immature (IM), decreased considerably after CTD treatment in crg1Δ mutant, probably due to the degradation of the aggregated proteins (Fig. 4, B–D) (60–62). We also observed slower migration and shift of the Gas1-GFP(M) band in SDS-PAGE (Fig. 4B) upon CTD treatment in crg1Δ mutant, indicating the direct effect on the maturity of GPI-anchored proteins (Figs. 4B and 6E). Supplementation of ETA completely rescued the sorting of Gas1-GFP in CTD-treated crg1Δ cells (Fig. 4A). This might be the reason for ETA-mediated rescue against CTD treatment (Fig. 1, A and C). Furthermore, we measured DTT-extractable cell surface proteins (CSPs) integrated into the cell wall and plasma membrane through GPI-anchors. These proteins were extracted as described previously (63). We found a high yield of CSPs from CTD-treated crg1Δ cells compared with WT cells. However, the supplementation of ETA restored the cell surface proteins to normal level, equal to that of WT (Fig. S4, A and B). High yield of CSPs from CTD-treated crg1Δ cells are maybe due to inappropriate anchorage to cell wall or cell membrane; hence, they become easily extractable from the surface. In contrast, ETA supplementation restabilizes the binding of the GPI-anchors, reversing the phenotype to normal and equivalent to WT. To further ascertain the role of CTD on GPI-anchor biosynthesis, we checked the genetic interaction of CRG1 with a few GPI-anchor biosynthesis genes (GPI2, GPI13, and MCD4) (1). Because these genes are essential for the cell survival, we used their heterozygous deletion mutants (gpi2Δ/GPI2, gpi13Δ/GPI13, and mcd4Δ/MCD4). We deleted CRG1 to create double-deletion mutants, crg1Δ/Δgpi2Δ/GPI2, crg1Δ/Δgpi13Δ/GPI13, and crg1Δ/Δmcd4Δ/MCD4, and performed a growth assay upon CTD treatment. Surprisingly, the double-deletion mutants (crg1Δ/Δgpi2Δ/GPI2, crg1Δ/Δgpi13Δ/GPI13, and crg1Δ/Δmcd4Δ/MCD4) showed better growth compared with single-deletion mutant, crg1Δ/Δ, in CTD-treated medium (Fig. 4 (E and F) and Fig. S3, F–I). That suggests that the molecular target of CTD may be downstream of the GPI biosynthesis cascade (4). We conclude that CTD alters the GPI-anchored protein sorting, which can be rescued by ETA supplementation.
Figure 4.
CTD alters GPI-anchored protein sorting. A, CTD treatment induces missorting of Gas1-GFP. WT and crg1Δ cells were transformed with YEp24-GAS1-GFP plasmid. Cells were grown in YPD at 30 °C until mid-exponential phase, treated with CTD with or without ETA, and incubated for 6 h before imaging. Subcellular localization of Gas1-GFP was observed by using a ZEISS-Apotome fluorescence microscope. B, CTD treatment decreases Gas1-GFP expression. WT and crg1Δ strains expressing Gas1-GFP were grown in YPD at 30 °C until mid-exponential phase and then treated with CTD. Cells were harvested after 3 h of incubation to analyze the expression of Gas1-GFP. In this data, the mature form of Gas1-GFP is represented as Gas1-GFP(M), whereas the immature Gas1-GFP is shown is Gas1-GFP(IM). Tbp1 was used as a loading control. The figure represents one of the three independently performed experiments. C and D, densitometry quantification of the three biological repeats of the Western blotting shown in B, with the help of ImageJ software. C, level of Gas1-GFP(M). D, level of Gas1-GFP(IM). The graph shows a scatter plot of each data point of three independent experiments with mean ± S.D. (error bars). Statistical analysis was done, applying Student's t test, where p ≤ 0.05 (*), p ≤ 0.01 (**), and p ≤ 0.001 (***). E and F, GPI biosynthesis genes show synthetic rescue with CRG1 under CTD stress. E, growth curve assay to compare the sensitivity of crg1Δ/Δ, gpi2Δ/GPI2, gpi13Δ/GPI13, mcd4Δ/MCD4, crg1Δ/Δgpi2Δ/GPI2, crg1Δ/Δgpi13Δ/GPI13, and crg1Δ/Δmcd4Δ/MCD4 mutants to CTD. F, growth sensitivity spot assay of WT, crg1Δ/Δ, gpi2Δ/GPI2, gpi13Δ/GPI13, mcd4Δ/MCD4, crg1Δ/Δgpi2Δ/GPI2, crg1Δ/Δgpi13Δ/GPI13, and crg1Δ/Δmcd4Δ/MCD4 mutants. Equal numbers of WT and mutant cells were serially diluted and spotted on the CTD-containing SC agar medium. The spotted cells were incubated at 30 °C for 72 h.
Figure 6.
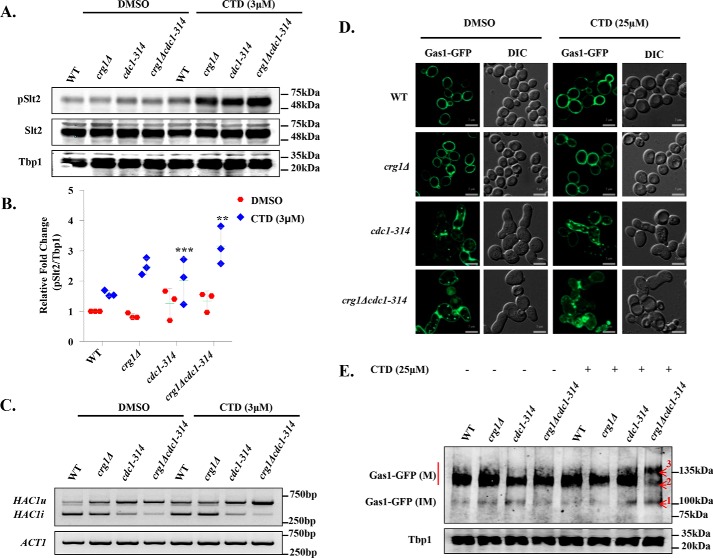
CTD treatment mimics CDC1 mutation (cdc1-314). A, CTD treatment induces Slt2 phosphorylation in crg1Δ and cdc1-314 mutant. Western blot analysis of Slt2 phosphorylation in WT, crg1Δ, cdc1-314, and crg1Δcdc1-314 strains. Cells were grown at 25 °C until mid-exponential phase and then treated with CTD for 2 h. The figure represents one of the three independently performed experiments. B, densitometry quantification of the three biological repeats of the Western blots shown in A, with the help of ImageJ software. The graph shows a scatter plot of each data point of three independent experiments with mean ± S.D. (error bars). Statistical analysis was done, applying Student's t test, where p ≤ 0.05 (*), p ≤ 0.01 (**), and p ≤ 0.001 (***). C, synergistic inhibition of HAC1 mRNA splicing in crg1Δcdc1-314 mutant upon CTD treatment. RT-PCR analysis of HAC1 mRNA in WT, crg1Δ, cdc1-314, and crg1Δcdc1-314 mutants. The cells were grown at 25 °C until mid-exponential phase and then treated with CTD for 2 h. The figure represents one of the three independently performed experiments. D, CTD induces Gas1-GFP missorting. Shown is subcellular localization of Gas1-GFP in WT, crg1Δ, cdc1-314, and crg1Δcdc1-314. Cells were transformed with YEp24-GAS1-GFP and grown in YPD medium with and without CTD treatment for 6 h at 25 °C. E, CTD alters GPI-anchor maturation of Gas1-GFP. Shown is Western blot analysis of Gas1-GFP in crg1Δcdc1-314 mutant. Numbers 1, 2, and 3 in the Gas-GFP bands represent the protein retardation and altered maturation of the GPI-anchor due to CTD treatment. The data represent one of the three independently performed experiments.
Cdc1-mediated GPI-anchor remodeling is the major target of CTD
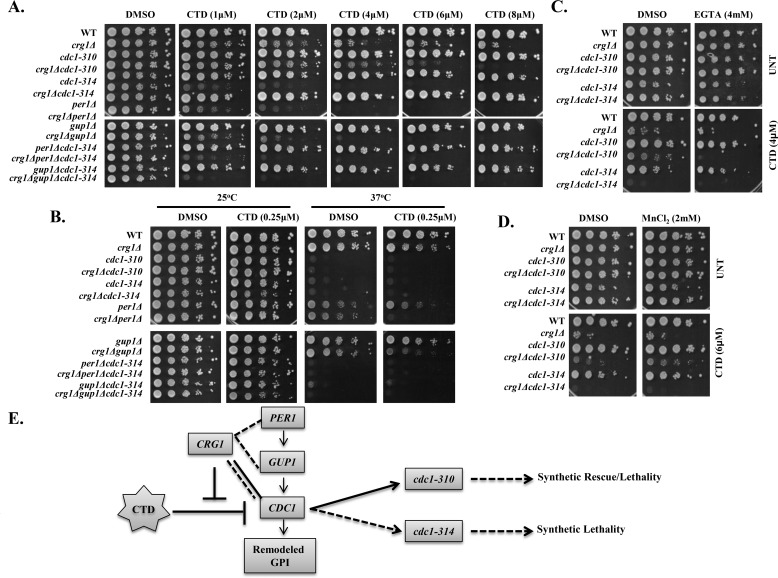
GPI-anchor remodeling is the step successive to biosynthesis. Based on the results discussed above, we hypothesized that GPI-anchor remodeling may be the target of CTD (30). We performed experiments to find synthetic lethality between CRG1 and GPI-anchor–remodeling genes (CDC1, PER1, and GUP1). First, we deleted CRG1 in the mutants of remodeling factors (per1Δ, gup1Δ, cdc1-310, cdc1-314, per1Δcdc1-314, and gup1Δcdc1-314) and measured their fitness profile upon CTD treatment. We found that the double mutants (crg1Δcdc1-314, crg1Δcdc1-310, crg1Δgup1Δ, and crg1Δper1Δ) were hypersensitive to CTD compared with the single mutants (crg1Δ, per1Δ, gup1Δ, cdc1-310, and cdc1-314) (Fig. 5A). Additionally, the triple mutants (crg1Δper1Δcdc1-314 and crg1Δgup1Δcdc1-314) showed even more sensitivity to CTD than single or double mutants (Fig. 5A and Fig. S5). Additionally, two different alleles of CDC1 (cdc1-310 and cdc1-314) showed contrasting phenotypes. cdc1-314 showed synthetic lethality, whereas cdc1-310 showed a dose-dependent behavior. It showed synthetic rescue at lower dose and synthetic lethality at higher dose (Fig. 5A and Fig. S5). We also checked their fitness profile upon CTD treatment at a higher temperature (37 °C), and we found that cdc1 mutants showed temperature sensitivity, whereas the growth of per1Δ and gup1Δ was unaffected (Fig. 5B) (3, 4). However, the double mutants crg1Δgup1Δ and crg1Δper1Δ were found to be hypersensitive compared with the single-deletion mutant crg1Δ at a very low dose of CTD (0.25 μm) (Fig. 5B). The results suggest that CRG1 shows synthetic lethality with GPI-anchor–remodeling genes, stronger with CDC1 than PER1 or GUP1 in the presence of CTD. Based on our data and the genetic interaction study performed between CDC1 and MCD4 (4), we propose that down-regulation of GPI-anchor biosynthesis genes or decreased GPI biosynthesis can rescue the growth defect of the mutants lacking GPI-anchor remodeling, perhaps by decreasing the GPI traffic on the remodeling factors. Because of the dynamic behavior of cdc1 alleles (cdc1-314 and cdc1-310) against CTD, we hypothesized that the Cdc1 activity could be a specific target of CTD in the remodeling process (Fig. 5A and Figs. S5 and S6 (C–F)). As the activity of Cdc1 is Mn2+-dependent (3, 5, 6), we decided to examine the effect of CTD by controlling Mn2+ concentration in the medium. We added the di-ionic chelator EGTA into the medium along with CTD and checked the fitness profile of the mutants. We observed that the growth of single and double mutants (crg1Δ, crg1Δcdc1-310, and crg1Δcdc1-314) was suppressed gradually with increasing concentration of EGTA (Fig. 5C and Fig. S6A). Moreover, exogenous supplementation of MnCl2 recovered the growth of crg1Δcdc1-310 mutant in CTD-treated medium (Fig. 5D and Fig. S6 (B and G–J)). In this study, the two cdc1 alleles exhibit reproducible phenotype in a condition different from that reported previously (3, 6). In summary, we conclude that CRG1 and CDC1 work in two different axes; CRG1 works as a guard to resist CTD, whereas CDC1 participates in the remodeling process. Loss of CRG1 results in the increased availability of active CTD that impairs the remodeling process by targeting the Cdc1 activity (Fig. 5E). Although both of the genes work in two different axes, they are required in parallel to tolerate CTD toxicity. We conclude that Cdc1 activity is essential to tolerate CTD cytotoxicity, and it may serve as a mechanistic target of the drug.
Figure 5.
CTD targets Cdc1 activity involved in GPI-anchor remodeling. A–D, growth sensitivity assay. Equal numbers of serially diluted cells of WT and the indicated mutants were spotted on SC agar medium with various treatments. Images were captured after 72 h of incubation. A, CRG1 shows synthetic lethality with GPI-anchor–remodeling genes under CTD stress. A spot assay on medium with increasing doses of CTD (1–8 μm) was followed by incubation at 25 °C. B, CRG1 shows synthetic lethality with GPI-anchor–remodeling genes under CTD and heat stress. A spot assay was done on medium containing CTD and incubated at 25 and 37 °C. C, Mn2+ chelation increases CTD toxicity. The yeast strains indicated above were spotted on medium containing CTD with and without EGTA and incubated at 25 °C. D, Mn2+ supplementation decreases CTD toxicity. Yeast strains indicated above were spotted on medium containing CTD and MnCl2 and incubated at 25 °C. E, schematic representation of CTD-dependent genetic interaction of CRG1 with GPI-anchor–remodeling genes; PER1, GUP1, and CDC1.CRG1 show synthetic lethality with PER1, GUP1, and CDC1. cdc1-310 shows dose-dependent interaction with crg1Δ: synthetic rescue at lower dose (2–4 μm) and synthetic lethality at higher dose (6–8 μm).
CTD-induced phenotypes strongly correlate with the loss of Cdc1 activity
Analysis of the phenotypes observed in this study and the investigations conducted previously suggest that CTD treatment induces phenotypes similar to cdc1 mutants (cdc1-314, cdc1-310, etc.) (3, 4, 30). To obtain further evidence to support this hypothesis, we employed genetic and cell biology–based experimental approaches. First, we measured the Slt2 phosphorylation upon CTD treatment. We observed that the cdc1-314 mutant showed increased phosphorylation of Slt2 compared with WT in untreated condition. CTD treatment induced the Slt2 phosphorylation in crg1Δ, cdc1-314, and crg1Δcdc1-314 (Fig. 6, A and B). Although CTD induced Slt2 phosphorylation in all of the three mutants, the maximum level was measured in the double mutant crg1Δcdc1-314 (Fig. 6, A and B). We also observed decreased UPR in cdc1-314 due to inhibition of HAC1 mRNA splicing in untreated condition (Fig. 6C) (4). The splicing of HAC1 mRNA further decreased in the double mutant crg1Δcdc1-314 with and without CTD treatment (Fig. 6C). We also observed a defect in GPI-anchored protein sorting (Gas1-GFP) in cdc1-314 (Fig. 6D) (3, 4), which became worse if treated with CTD (Fig. 6D). Additionally, we found decreased expression of Gas1-GFP in cdc1-314 (4). CTD treatment decreased Gas1-GFP expression in crg1Δcdc1-314 more effectively than single-mutant crg1Δ (Fig. 6E). Interestingly, crg1Δcdc1-314 mutant showed increased retardation of Gas1-GFP(M) in SDS-PAGE upon CTD treatment. The retardation was so prominent that three bands appeared for Gas1-GFP, band-1 corresponding to immature Gas1-GFP(IM) and band-2 and band-3 corresponding to mature Gas1-GFP(M) (Fig. 6E). We observed synthetic phenotypes for Slt2 phosphorylation, UPR, Gas1-GFP expression, maturation, and localization, suggesting that Cdc1 can be the specific target of CTD.
Next, we measured the growth of GPI-anchor–remodeling mutants in presence of CTD or antioxidants with increasing temperature. We found that the mutants of GPI-remodeling genes (per1Δ, gup1Δ, cdc1-310, cdc1-314, per1Δcdc1-314, and gup1Δcdc1-314) were sensitive to the higher doses of CTD, and the sensitivity increased with increasing temperature (Fig. S7). We also found these mutants to be hypersensitive to a reducing environment developed by supplementation of GSH or NAC into the medium (Fig. S7). The sensitivity to GSH as well as NAC increased again with elevated temperature. The result showed the essential role of redox balance in the remodeling process of the GPI-anchors and described the synergistic lethal phenotype generated by the co-treatments with CTD + GSH or CTD + NAC (Fig. 2E). Additionally, ETA supplementation did not rescue the growth defect of cdc1-314 and cdc1-310 at higher temperature (Fig. S7), suggesting that ETA-mediated rescue in Gas1-GFP sorting in CTD-treated cells did not occur via the GPI-anchor–remodeling mechanism. The hypersensitivity of the single mutant cdc1-314 to the higher doses of CTD indicates the involvement of a CRG1-independent pathway targeted by the drug (Fig. S7). We also observed that a higher dose of CTD (300 μm) altered Gas1-GFP sorting even in WT strain (Fig. S8A). Furthermore, to investigate whether CTD-induced alteration in GPI-anchored protein sorting was PP2A/PP1-dependent or not (24), we analyzed Gas1-GFP localization in sit4Δ (PP2A) and GLC7/glc7Δ (PP1) strains (Fig. S8B). We did not find any defect in Gas1-GFP localization in both of the mutants, implying the CTD-induced alteration in GPI-anchored protein sorting was independent of PP2A and PP1. Overall, the CTD-induced phenotypes strongly correlate to that of the cdc1-314 allele, so perhaps CTD targets Cdc1 activity. The entire molecular mechanism of CTD cytotoxicity can be illustrated in a graphical model that summarizes the complete sequence of events induced by CTD (see Fig. 8, A and B).
Figure 8.
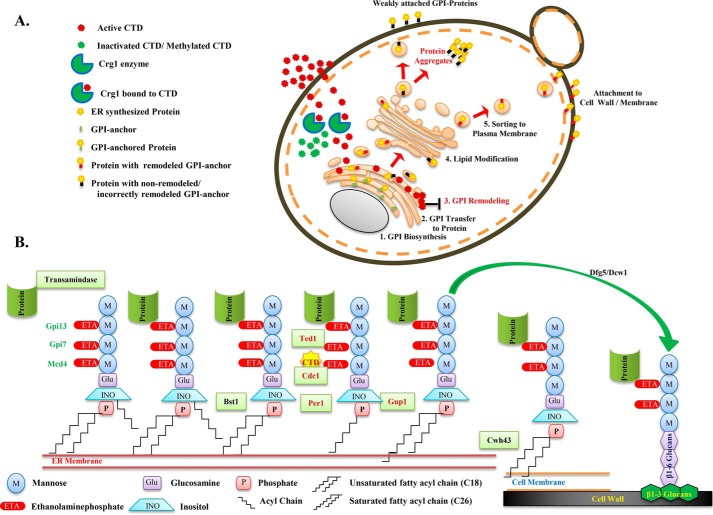
Schematic model illustrating the molecular targets and mechanism of CTD toxicity in yeast and higher eukaryotes. A, the model describes yeast Crg1 as a key defense molecule, localized in the cytoplasm, which protects the cell from CTD-induced cytotoxicity by methyltransferase activity. Loss of Crg1 enhances the binding of CTD to its molecular targets and perturbs the related biological functions. In the absence of Crg1, CTD enters into the ER and disturbs the ER homeostasis by altering the GSH/GSSG ratio and GPI-anchor remodeling, leading to missorting and aggregation of the proteins in the cytoplasm. B, illustration of the GPI-anchor–remodeling process in budding yeast. The C-terminal end of the protein is transferred to the ethanolamine phosphate of the third mannose of the GPI-anchor, catalyzed by a complex of enzymes, GPI–transamidase. In the subsequent process, Bst1 removes the acyl group from the inositol of GPI, Cdc1 removes ethanolamine phosphate from the first mannose, Per1 removes the unsaturated fatty acid (C18:1) from the sn-2 position of the GPI–lipid, Gup1 adds C26:0 saturated fatty acid at the sn-2 position of the GPI–lipid, and at last Cwh43 replaces the diacyglycerol type lipid with ceramide in GPI. Finally, the GPI-anchor is transferred to the plasma membrane or cell wall by Dfg5 or Dcw1. In this sequence of events, CTD targets Cdc1 activity, resulting in mislocalization and aggregation of GPI-anchored proteins.
CTD alters GPI-anchored protein sorting in human cancer cells
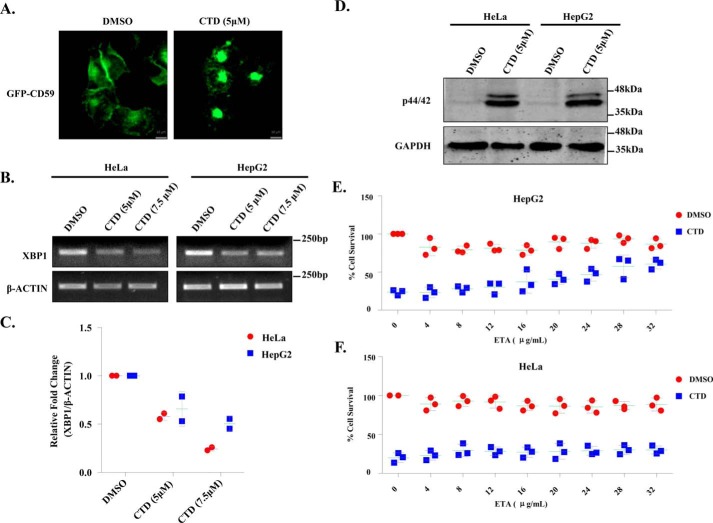
The pathway for the biosynthesis and sorting of GPI-anchored proteins is conserved from yeast to higher eukaryotes (1, 64). Therefore, we reasoned that the fundamental mechanism of action of CTD would be similar in yeast and human cells. To study the GPI-anchored protein sorting in human cells, we used GFP-CD59 as a model GPI-anchored protein (5). We observed that CTD induced aggregation of GFP-CD59 in HeLa cells, whereas the untreated cells showed normal distribution of the protein (Fig. 7A and Fig. S8C). This observation suggests that the molecular mechanism of action of CTD is conserved from yeast to human cells. Furthermore, we also checked the total expression of GFP-CD59 in HeLa cells upon treatment with CTD. Unlike yeast, we did not find any change in GFP-CD59 expression (Fig. S13C), suggesting a distinct mechanism for the clearance of aggregated proteins in human cells. Next, we measured the phosphorylation of p44/42 (a human homologue of yeast Slt2). We found a significant induction in p44/42 phosphorylation in HeLa and HepG2 cells upon CTD treatment (Fig. 7D). CTD treatment also decreased the XBP1 mRNA expression (Fig. 7, B and C), suggesting a down-regulation of UPR similar to the yeast cells (Fig. 2, B and C) (65). To rescue the cells from CTD toxicity, we supplemented ETA, CHO, and INO into the Dulbecco's modified Eagle's medium. Interestingly, ETA supplementation rescued the HepG2 cells from CTD-induced cell death, but the rescue of HeLa cells was not considerable (Fig. 7, E and F), suggesting a cell type–specific utilization of ETA perhaps due to diverse origin. On the other hand, CHO and INO supplementation could not rescue the human cells (Fig. S9, A–D) from CTD toxicity as observed in yeast cells (Fig. 1A). Because the phenotypes induced by CTD treatment in human and yeast cells are quite similar, we propose a conserved mechanism of action of CTD in eukaryotes (Fig. 8).
Figure 7.
Conserved mechanism of CTD cytotoxicity in human cancer cells (HeLa and HepG2). A, CTD alters GPI-anchored protein sorting. Microscopic visualization of GFP-CD59, stably expressing in HeLa cells with or without CTD treatment for 12 h. B, CTD treatment down-regulates XBP1 expression. Semiquantitative RT-PCR analysis of XBP1 expression in HeLa and HepG2 cell lines treated with CTD for 48 h is shown. The data represent one of the two independent experiments. C, densitometry quantification of the two biological repeats of the semiquantitative PCR shown in B, with the help of ImageJ software. The graph shows a scatter plot of each data point of three independent experiments with mean ± S.D. (error bars). D, CTD treatment induces p44/42 (Slt2) phosphorylation. Shown is Western blot analysis of p44/42 phosphorylation in HeLa and HepG2 cell lines after 48 h of CTD treatment. The figure represents one of the three independent experiments. E, ETA supplementation rescues HepG2 cells from CTD cytotoxicity. MTT cell survival assay of HepG2 cells treated with CTD, supplemented with increasing concentrations of ETA. F, ETA supplementation does not rescue HeLa cells from CTD cytotoxicity. Shown is an MTT cell survival assay of HeLa cells treated with CTD, supplemented with increasing concentrations of ETA.
Discussion
GPI-anchored proteins control essential biological functions in animal cells by regulating the cell-to-cell communication, adhesion, and signal transduction (2, 9). GPI-anchored proteins are also shown to be involved in tumorigenesis and metastasis (10–12). Targeting an essential cellular pathway is one of the key aspects of anticancer chemotherapeutics. In this study, employing extensive genetic and cell biological approaches, we identified Cdc1 (yeast homologue of human PGAP5)-mediated GPI-anchor remodeling as a mechanistic target of CTD in addition to PP2A and PP1. However, biochemical validations will further support our observations.
CTD has been shown to disturb phospholipid homeostasis in crg1Δ mutant (30). To understand the underlying mechanism of its action, we screened the crg1Δ mutant for the auxotrophy of different phospholipids upon CTD treatment. This approach helped us to conclude that CTD specifically affects PE, which can be rescued by exogenous supplementation of ETA. CTD treatment induced phenotypes similar to psd1Δ (39, 44), and we found that PSD1 was synthetically lethal in combination with CRG1 under CTD stress. The reason for PE auxotrophy upon CTD treatment may be either inhibition of PE biosynthesis or alteration in PE-associated structures (e.g. GPI-anchors). The biosynthesis of PC mainly depends on the availability of PE in ER, which suggests that PE deficiency can lead to the deficiency of PC as well (66). However, the supplementation with PC did not rescue the growth defect induced upon CTD treatment. Thus, we conclude that CTD probably alters the PE-associated structures or functions rather than PE biosynthesis. PE deficiency is also known to induce ER stress and UPR in yeast (44). On the contrary, we found decreased UPR upon CTD treatment in the absence as well as the presence of the UPR inducers (TM and DTT). Our further investigations revealed that the drop in UPR upon CTD treatment was due to alteration in ER-redox homeostasis and Cdc1 activity (4), where we found that increased GSH level or lack of Cdc1 activity diminished UPR. The oxidative environment in ER is maintained by low GSH/GSSG (1:1 to 3:1) ratio for correct folding and modifications of the proteins (47, 49, 50). Our study demonstrates that the oxidative environment is also essential for the process of GPI-anchor remodeling (Fig. S7). ER is the site of synthesis and fate determination of the secretory proteins in the cell. Biosynthesis and maintenance of the yeast cell wall majorly depends on these secretory proteins (1). We believe that the CTD-induced cell wall damage (30, 34) is due to alteration in ER homeostasis. The synergistic lethal effect of CTD with ER stress inducers (heat, DTT, and TM) and cell wall–perturbing agents (CR and CFW) supports this hypothesis. CTD-induced Slt2 phosphorylation also increases synergistically with ER stress inducers. Thus, we conclude that CTD-induced ER stress triggers cell wall damage. We also found rescue from CTD-induced cytotoxicity upon ETA supplementation, the reason for which may be the restoration of the GPI-anchored protein sorting (38, 44). However, we do not know the exact mechanism by which the increased PE level restores the GPI-anchored protein sorting against CTD.
Next, we investigated the molecular mechanism for the ER stress and cell wall damage upon CTD treatment. The genetic interaction profile of CRG1 suggests that the ER–Golgi traffic system is a major pathway affected by CTD (30). In yeast cells, the proteins that travel from the ER to the cell wall are mostly the GPI-anchored proteins. GPI-anchored proteins constitute a major part of the total cell wall proteins and are required for the biosynthesis and maintenance of the yeast cell wall (1, 2, 7, 58, 64). Alteration in biosynthesis or sorting of GPI-anchored proteins induces ER stress and cell wall damage (4). Interestingly, we observed missorting and aggregation of the GPI-anchored protein (Gas1-GFP) upon CTD treatment. CRG1 showed synthetic rescue with GPI-anchor biosynthesis genes (GPI2, GPI13, and MCD4) and synthetic lethality with GPI-anchor–remodeling genes (GUP1, PER1, and CDC1) upon CTD stress, indicating that the CTD alters GPI-anchored protein sorting by targeting the remodeling process (4). These results also support the genetic interaction profile of CRG1 reported previously (30). In addition, we identified CDC1 as an additional new gene that showed synthetic lethality with CRG1 in the presence of CTD. CDC1 encodes for Mn2+-dependent mannose-EtNP phosphodiesterase required for the removal of EtNP from the first mannose of the GPI-anchor (4). The crg1Δcdc1-314 double mutant shows strong sensitivity to CTD compared with crg1Δ and cdc1-314 single mutants. The triple mutant strains crg1Δ per1Δcdc1-314 and crg1Δgup1Δcdc1-314 were found to be even more sensitive to CTD compared with single (crg1Δ, per1Δ, gup1Δ, cdc1-310, and cdc1-314) and double mutants (crg1Δper1Δ and crg1Δgup1Δ), suggesting that GPI-anchor remodeling is the major target of CTD. On the contrary, another allele of CDC1, cdc1-310, shows a dynamic phenotype upon CTD treatment. It shows synthetic rescue at lower dose and synthetic lethality at higher dose of CTD. Such dynamic and contrasting phenotypes of the two different alleles of CDC1 suggest a possibility of direct interaction of the enzyme with the small molecule CTD. To obtain more evidence in support of this hypothesis, we manipulated the Mn2+ concentrations in the medium. We found that CTD toxicity enhanced with decreasing concentrations of Mn2+ in the medium and vice versa, indicating an essential requirement of the Cdc1 activity to tolerate the CTD toxicity. Based on these results, we believe that CTD inhibits Cdc1 activity. CTD shows stronger affinity to Cdc1-314 than Cdc1-310, probably due to the specific protein confirmation. Previous studies suggest that CTD acts as a potent inhibitor of protein phosphatases PP2A and PP1 (23, 24, 67). However, our observations suggest that it can also inhibit lipid phosphatases such as Cdc1. CTD-dependent synthetic lethality of SAC1 (phosphatidylinositol phosphate phosphatase) with CRG1 supports this hypothesis (30, 68). Furthermore, the sit4Δ (PP2A) and GLC7/glc7Δ (PP1) mutants do not show any defect in GPI-anchored protein sorting, suggesting that CTD-induced alteration in GPI-anchored protein sorting is independent of its known protein targets PP2A and PP1 (23, 24, 30). Moreover, we also found that the higher dose of CTD induces the same phenotypes in WT and cdc1-314 as it does in crg1Δ mutant at a sublethal dose, suggesting that CTD-targeted pathways are independent of CRG1.
The enzymes involved in GPI-anchor biosynthesis and remodeling in yeast are mostly conserved in higher eukaryotes, suggesting that CTD can act through a similar mechanism in higher eukaryotes. To validate the existence of a conserved mechanism of CTD toxicity, we extended our studies to human cell lines HeLa and HepG2. We observed similar phenotypes induced by CTD in human cells. We found missorting and aggregation of GPI-anchored GFP-CD59 in the cytoplasm of HeLa cells upon CTD treatment, which was very similar to that of Gas1-GFP in yeast. Similarly, CTD also induced phosphorylation of p44/42 (yeast Slt2), supporting the previous observations of CTD-mediated activation of different mitogen-activated protein kinases (18, 21). We also found decreased expression of XBP1 (yeast HAC1) upon CTD treatment, which might be via ATF6 signaling that regulates the target gene XBP1 (65). The similar phenotypes produced by CTD in yeast and human cell lines suggest that the drug functions through a conserved mechanism.
Our study provides explanations for various observations reported upon CTD treatment in different organisms. CTD-induced alteration in GPI-anchored protein sorting can be a reason for the acantholysis (69–71) and inhibition of cancer metastasis (12). CTD-induced perturbation in adhesion, morphogenesis, and membrane trafficking in Candida albicans may be due to alteration in GPI-anchored protein sorting (72). The G2/M cell cycle arrest by CTD (18, 19, 67) is probably due to inhibition of Cdc1 activity, as the loss of Cdc1 functions also induces G2/M cell cycle arrest (73). CTD has been a traditional medicine to cure warts and molluscum contagiosum caused by viral infections. Our study suggests that CTD can be further explored as an antifungal, antiviral, or antiprotozoan drug, utilizing its property of altering the ER–Golgi traffic system (8, 15, 74).
Because CTD targets a conserved and essential pathway, its exposure can also lead to lethal side effects. Therefore, the drug delivery is required to be very specific. A cancer- or tumor-specific delivery of CTD is the only way to make it a successful chemotherapeutic anticancer drug. Similarly, the poisoning of cattle foods by the contamination with the blister beetle is another challenge, as there is no antidote available against the beetle toxin. Our study suggests that ETA can serve as a potent antidote against CTD poisoning. In summary, we identified a novel target of CTD in addition to PP2A/PP1 and a potent antidote that neutralizes its lethal cytotoxicity.
Materials and methods
Yeast strains, plasmids, and growth conditions
Unless otherwise stated, S. cerevisiae strains used in this study were isogenic with S288c (BY4741 or BY4743). All of the strains, plasmids, and primers used in this work are listed in Tables S1–S3, respectively. Yeast strains were grown in synthetic complete (SC) or yeast peptone dextrose (YPD) medium at 30 °C, maintaining the optimum growth excluding some temporal stress conditions. Various reagents used in different experiments were purchased from Sigma, Merck, Himedia, Invitrogen, Bio-Rad, and Applied Biosystems.
Growth sensitivity assays
Serial dilution assay
Equal number (A600 = 1.0) of overnight grown yeast cells were serially diluted, 10-fold, five times and then spotted on SC agar medium. The spotted cells were incubated at different temperatures according to the various experimental conditions.
Growth curve assay
An equal number (A600 = 0.2) of exponentially growing yeast cells were inoculated in 96-well plates with and without different treatments and grown for 23–28 h in the automated plate reader (Biotek) acquiring a reading at A600 in an interval of every 30 min.
Preparation of yeast whole-cell protein extract for Western blot analysis
Protein extraction from yeast cells was done by following the TCA protein extraction protocol (75). The equal number of cells were harvested and washed twice with 20% of TCA. Cell pellets were resuspended in 20% TCA with an equal volume of glass beads and vortexed rigorously to lyse the cells. TCA-precipitated protein extract was washed with ethanol and resuspended in 0.5 m Tris-Cl (pH 7.5) with 2× loading buffer. The sample was boiled at 100 °C for 10 min and centrifuged at maximum for 10 min to remove the debris. The supernatant was taken ahead for SDS-PAGE and Western blot analysis. The primary antibodies used in this study for immunoblotting experiments were as follows: anti-phospho-p44/42 (Cell Signaling, catalogue no. 4370S) to detect human phospho-44/42 and yeast phospho-Slt2 (pSlt2), anti-Mpk1 (Santa Cruz Biotechnology, Inc., catalogue no. SC-6803) to detect total Slt2 (Mpk1) in the cells, anti-GFP (Sigma, catalogue no. G1544) to detect Gas1-GFP and GFP-CD59, and anti-GAPDH (Cell Signaling, catalogue no. 5174S) as loading control for human cells. Primary antibody used for Tbp1 was the yeast loading control; it is a polyclonal antisera raised in rabbit. Densitometry quantification was performed with the help of ImageJ software in which the protein of interest was normalized with Tbp1 loading control.
Cell surface protein extraction
Yeast cells were washed twice with sodium phosphate buffer (0.1 m, pH 8.0). Wash-out solution was kept at 4 °C. Collected cells were resuspended again in sodium phosphate buffer with 2 mm DTT and incubated at 4 °C for 2 h, maintaining gentle agitation. Now the cells were pelleted down by centrifugation, and the supernatant along with the washout fraction was precipitated using 20% TCA in the final volume. TCA-precipitated cell surface proteins were separated via 8% SDS-PAGE and stained with 0.1% Coomassie Brilliant Blue R-250 (63).
RNA extraction, cDNA synthesis, and quantitative PCR for HAC1 mRNA splicing
RNA isolation was performed by using the heat/freeze RNA isolation protocol (76). Briefly, cells were grown until mid-exponential phase, harvested by centrifugation, and washed twice with 1× PBS. Harvested cells were lysed with 1% of SDS in AE buffer (50 mm sodium acetate, 10 mm EDTA, pH 5.3). An equal volume of acidic phenol of pH 4.2 was added and incubated at 65 °C for 4 min, followed by freezing at −85 °C for 4 min, centrifuged for 2 min at maximum speed to separate the aqueous layer. The aqueous phase was mixed with an equal volume of phenol/chloroform/isoamyl alcohol (25:24:1) and separated again from the phenol phase. The total RNA present in the aqueous phase was precipitated by adding sodium acetate (0.3 m) and 2.5 volumes of absolute ethanol. The cDNA synthesis was done by following the standard protocol provided by the iScriptTM cDNA synthesis kit (Bio-Rad, catalogue no. 1708891). HAC1 mRNA splicing was measured using primers specified in Table S3, following the PCR conditions as described previously (53).
β-gal assay
Exponentially growing yeast cells were harvested and washed twice with LacZ buffer (10 mm KCl, 1 mm MgSO4, 50 mm β-mercaptoethanol, and 100 mm NaPO4, pH 7.0). Cells were lysed using 0.01% SDS, and 22.7% chloroform in LacZ buffer in a final volume of 250 μl. Subsequently, 500 μl of ortho-nitrophenyl-β-d-galactoside (2 mg/ml) was added and incubated at 30 °C until the appearance of pale yellow color. The reaction was quenched by adding 500 μl of sodium bicarbonate (1 m). The reaction mixture was centrifuged at maximum speed for 15 min, and the supernatant was collected to measure the absorbance at a wavelength of 420 nm. Miller unit for the β-gal activity was determined by applying the following formula: Miller unit = (A420/A600 × time (min)) × 1,000 (45).
Fluorescence microscopy
Fluorescence microscopy was done to study the sorting of GPI-anchored protein (Gas1-GFP) with or without CTD. Yeast cells were grown in YPD medium for different time points and harvested by centrifugation (6,000 rpm, 2 min, 4 °C). Cell pellets were resuspended in PBS and kept on ice for at least 30 min (59). Gas1-GFP localization was observed using a ZEISS-Apotome.2 fluorescence microscope under a ×60 oil emulsion objective lens. For the microscopic localization study of the GFP-CD59, the overnight-grown HeLa cells with 50% confluence were treated with CTD (5 μm) for 12 h and visualized under a ×20 emulsion oil objective lens (5).
Cell culture and maintenance of human cell lines
Human cell lines (HeLa and HepG2) were maintained in Dulbecco's modified Eagle's medium (Lonza) having 10% fetal bovine serum (Gibco) and antibiotics (i.e. penicillin (100 units/ml) and streptomycin (100 μg/ml). Both of the cell lines were grown at 37 °C with 5% CO2.
Cell survival assay
Percentage survivability of the cells against CTD exposure was measured by an MTT assay. HeLa and HepG2 cells were seeded in a 96-well plate equal in number (5,000 cells) in each well. Cells were incubated for 24 h. Medium was removed, and fresh medium was added to the cells; simultaneously, cells were also challenged with CTD with or without supplementation of ETA for 48 h. 10 μl of MTT solution (5 mg/ml in 1× PBS) was added and incubated at growth conditions for 4 h. 100 μl of DMSO was added and mixed well. Absorbance was recorded at 570 nm using a microplate reader (Biotek) (16).
Statistical data analysis
Statistical analysis for the β-gal assay was performed by using GraphPad Prism version 5 software. Each graph shows the individual data points with mean value as a horizontal green line. The error bars represent S.D. of a minimum of three individual repeats. We applied two-way ANOVA and Bonferroni post hoc test, where p ≤ 0.05 (*), p ≤ 0.01 (**), and p ≤ 0.001 (***). Statistical significance of the relative -fold change in Slt2 phosphorylation or Gas1 expression with and without CTD treatment was calculated by applying Student's t test, where p ≤ 0.05 (*), p ≤ 0.01 (**), and p ≤ 0.001 (***).
Author contributions
The hypothesis and experiments were designed by R. S. T. and P. K. S. Experiments were performed by P. K. S., and the results were analyzed by P. K. S. and R. S. T. The manuscript was written by P. K. S. and R. S. T.
Supplementary Material
Acknowledgments
We thank Benjamin S. Glick for providing the mutants of the GPI-anchor–remodeling pathway (cdc1-310, cdc1-314, per1Δ, gup1Δ, per1Δcdc1-314, gup1Δcdc1-314, and WT-JK9-3d). We also thank Vishal M. Gohila for the gift of psd1Δ and WT (BY4741). We are also thankful to Peter Walter, Laura Popolo, and Morihisa Fujita for the gifts of pPW344 (UPRE-LacZ), YEp24-GAS1-GFP, and pME-NeodH-mEGFP-F-CD59_hGHss, respectively. We acknowledge all of the members of the laboratory for critical suggestions and helpful discussions throughout the work.
This work was supported by funds from the Indian Institute of Science Education and Research Bhopal and partly by SERB Government of India Grant EMR/2015/001797 (to R. S. T.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1–S4 and Figs. S1–S13.
- GPI
- glycophosphatidylinositol
- ER
- endoplasmic reticulum
- EtNP
- ethanolaminephosphate
- UPR
- unfolded protein response
- CTD
- C-terminal domain
- PP1
- protein phosphatase 1
- PP2A
- protein phosphatase 2 phosphatase activator
- ETA
- ethanolamine
- CHO
- choline
- PE
- phosphatidylethanolamine
- PC
- phosphatidylcholine
- INO
- inositol
- PI
- phosphatidylinositol
- PS
- phosphatidylserine
- TM
- tunicamycin
- NAC
- N-acetylcysteine
- CFW
- calcofluor white
- CR
- Congo red
- SRB
- sorbitol
- CSP
- cell surface protein
- SC
- synthetic complete
- YPD
- yeast peptone dextrose
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide.
References
- 1. Orlean P., and Menon A. K. (2007) Thematic review series: lipid posttranslational modifications. GPI anchoring of protein in yeast and mammalian cells, or: how we learned to stop worrying and love glycophospholipids. J. Lipid Res. 48, 993–1011 10.1194/jlr.R700002-JLR200 [DOI] [PubMed] [Google Scholar]
- 2. Fujita M., and Kinoshita T. (2012) GPI-anchor remodeling: potential functions of GPI-anchors in intracellular trafficking and membrane dynamics. Biochim. Biophys. Acta 1821, 1050–1058 10.1016/j.bbalip.2012.01.004 [DOI] [PubMed] [Google Scholar]
- 3. Losev E., Papanikou E., Rossanese O. W., and Glick B. S. (2008) Cdc1p is an endoplasmic reticulum-localized putative lipid phosphatase that affects Golgi inheritance and actin polarization by activating Ca2+ signaling. Mol. Cell. Biol. 28, 3336–3343 10.1128/MCB.00567-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vazquez H. M., Vionnet C., Roubaty C., and Conzelmann A. (2014) Cdc1 removes the ethanolamine phosphate of the first mannose of GPI anchors and thereby facilitates the integration of GPI proteins into the yeast cell wall. Mol. Biol. Cell 25, 3375–3388 10.1091/mbc.e14-06-1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fujita M., Maeda Y., Ra M., Yamaguchi Y., Taguchi R., and Kinoshita T. (2009) GPI glycan remodeling by PGAP5 regulates transport of GPI-anchored proteins from the ER to the Golgi. Cell 139, 352–365 10.1016/j.cell.2009.08.040 [DOI] [PubMed] [Google Scholar]
- 6. Paidhungat M., and Garrett S. (1998) Cdc1 is required for growth and Mn2+ regulation in Saccharomyces cerevisiae. Genetics 148, 1777–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pittet M., and Conzelmann A. (2007) Biosynthesis and function of GPI proteins in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1771, 405–420 10.1016/j.bbalip.2006.05.015 [DOI] [PubMed] [Google Scholar]
- 8. Nagamune K., Nozaki T., Maeda Y., Ohishi K., Fukuma T., Hara T., Schwarz R. T., Sutterlin C., Brun R., Riezman H., and Kinoshita T. (2000) Critical roles of glycosylphosphatidylinositol for Trypanosoma brucei. Proc. Natl. Acad. Sci. U.S.A. 97, 10336–10341 10.1073/pnas.180230697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Paulick M. G., and Bertozzi C. R. (2008) The glycosylphosphatidylinositol anchor: a complex membrane-anchoring structure for proteins. Biochemistry 47, 6991–7000 10.1021/bi8006324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mierke C. T., Bretz N., and Altevogt P. (2011) Contractile forces contribute to increased glycosylphosphatidylinositol-anchored receptor CD24-facilitated cancer cell invasion. J. Biol. Chem. 286, 34858–34871 10.1074/jbc.M111.245183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lubeseder-Martellato C., Hidalgo-Sastre A., Hartmann C., Alexandrow K., Kamyabi-Moghaddam Z., Sipos B., Wirth M., Neff F., Reichert M., Heid I., Schneider G., Braren R., Schmid R. M., and Siveke J. T. (2016) Membranous CD24 drives the epithelial phenotype of pancreatic cancer. Oncotarget 7, 49156–49168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang L., Gao Z., Hu L., Wu G., Yang X., Zhang L., Zhu Y., Wong B. S., Xin W., Sy M. S., and Li C. (2016) Glycosylphosphatidylinositol anchor modification machinery deficiency is responsible for the formation of pro-prion protein (PrP) in BxPC-3 protein and increases cancer cell motility. J. Biol. Chem. 291, 3905–3917 10.1074/jbc.M115.705830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhao P., Nairn A. V., Hester S., Moremen K. W., O'Regan R. M., Oprea G., Wells L., Pierce M., and Abbott K. L. (2012) Proteomic identification of glycosylphosphatidylinositol anchor-dependent membrane proteins elevated in breast carcinoma. J. Biol. Chem. 287, 25230–25240 10.1074/jbc.M112.339465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wu G., Guo Z., Chatterjee A., Huang X., Rubin E., Wu F., Mambo E., Chang X., Osada M., Sook Kim M., Moon C., Califano J. A., Ratovitski E. A., Gollin S. M., Sukumar S., et al. (2006) Overexpression of glycosylphosphatidylinositol (GPI) transamidase subunits phosphatidylinositol glycan class T and/or GPI anchor attachment 1 induces tumorigenesis and contributes to invasion in human breast cancer. Cancer Res. 66, 9829–9836 10.1158/0008-5472.CAN-06-0506 [DOI] [PubMed] [Google Scholar]
- 15. Wang G. S. (1989) Medical uses of mylabris in ancient China and recent studies. J. Ethnopharmacol. 26, 147–162 10.1016/0378-8741(89)90062-7 [DOI] [PubMed] [Google Scholar]
- 16. Zheng L. H., Bao Y. L., Wu Y., Yu C. L., Meng X., and Li Y. X. (2008) Cantharidin reverses multidrug resistance of human hepatoma HepG2/ADM cells via down-regulation of P-glycoprotein expression. Cancer Lett. 272, 102–109 10.1016/j.canlet.2008.06.029 [DOI] [PubMed] [Google Scholar]
- 17. Sun X., Cai X., Yang J., Chen J., Guo C., and Cao P. (2016) Cantharidin overcomes imatinib resistance by depleting BCR-ABL in chronic myeloid leukemia. Mol. Cells 39, 869–876 10.14348/molcells.2016.0023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li W., Xie L., Chen Z., Zhu Y., Sun Y., Miao Y., Xu Z., and Han X. (2010) Cantharidin, a potent and selective PP2A inhibitor, induces an oxidative stress-independent growth inhibition of pancreatic cancer cells through G2/M cell-cycle arrest and apoptosis. Cancer Sci. 101, 1226–1233 10.1111/j.1349-7006.2010.01523.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang W. W., Ko S. W., Tsai H. Y., Chung J. G., Chiang J. H., Chen K. T., Chen Y. C., Chen H. Y., Chen Y. F., and Yang J. S. (2011) Cantharidin induces G2/M phase arrest and apoptosis in human colorectal cancer colo 205 cells through inhibition of CDK1 activity and caspase-dependent signaling pathways. Int. J. Oncol. 38, 1067–1073 [DOI] [PubMed] [Google Scholar]
- 20. Zhang J. T., Sun W., Zhang W. Z., Ge C. Y., Liu Z. Y., Zhao Z. M., Lu X. S., and Fan Y. Z. (2014) Norcantharidin inhibits tumor growth and vasculogenic mimicry of human gallbladder carcinomas by suppression of the PI3-K/MMPs/Ln-5γ2 signaling pathway. BMC cancer 14, 193 10.1186/1471-2407-14-193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Su C. C., Lee K. I., Chen M. K., Kuo C. Y., Tang C. H., and Liu S. H. (2016) Cantharidin induced oral squamous cell carcinoma cell apoptosis via the JNK-regulated mitochondria and endoplasmic reticulum stress-related signaling pathways. PLoS One 11, e0168095 10.1371/journal.pone.0168095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li H. C., Xia Z. H., Chen Y. F., Yang F., Feng W., Cai H., Mei Y., Jiang Y. M., Xu K., and Feng D. X. (2017) Cantharidin inhibits the growth of triple-negative breast cancer cells by suppressing autophagy and inducing apoptosis in vitro and in vivo. Cell. Physiol. Biochem. 43, 1829–1840 10.1159/000484069 [DOI] [PubMed] [Google Scholar]
- 23. Li Y. M., and Casida J. E. (1992) Cantharidin-binding protein: identification as protein phosphatase 2A. Proc. Natl. Acad. Sci. U.S.A. 89, 11867–11870 10.1073/pnas.89.24.11867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Honkanen R. E. (1993) Cantharidin, another natural toxin that inhibits the activity of serine/threonine protein phosphatases types 1 and 2A. FEBS Lett. 330, 283–286 10.1016/0014-5793(93)80889-3 [DOI] [PubMed] [Google Scholar]
- 25. Jiang Y. (2006) Regulation of the cell cycle by protein phosphatase 2A in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70, 440–449 10.1128/MMBR.00049-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wlodarchak N., and Xing Y. (2016) PP2A as a master regulator of the cell cycle. Crit. Rev. Biochem. Mol. Biol. 51, 162–184 10.3109/10409238.2016.1143913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ray R. M., Bhattacharya S., and Johnson L. R. (2005) Protein phosphatase 2A regulates apoptosis in intestinal epithelial cells. J. Biol. Chem. 280, 31091–31100 10.1074/jbc.M503041200 [DOI] [PubMed] [Google Scholar]
- 28. Zimmerman R., Peng D. J., Lanz H., Zhang Y. H., Danen-Van Oorschot A., Qu S., Backendorf C., and Noteborn M. (2012) PP2A inactivation is a crucial step in triggering apoptin-induced tumor-selective cell killing. Cell Death Dis. 3, e291 10.1038/cddis.2012.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim J. A., Kim Y., Kwon B. M., and Han D. C. (2013) The natural compound cantharidin induces cancer cell death through inhibition of heat shock protein 70 (HSP70) and Bcl-2-associated athanogene domain 3 (BAG3) expression by blocking heat shock factor 1 (HSF1) binding to promoters. J. Biol. Chem. 288, 28713–28726 10.1074/jbc.M113.488346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lissina E., Young B., Urbanus M. L., Guan X. L., Lowenson J., Hoon S., Baryshnikova A., Riezman I., Michaut M., Riezman H., Cowen L. E., Wenk M. R., Clarke S. G., Giaever G., and Nislow C. (2011) A systems biology approach reveals the role of a novel methyltransferase in response to chemical stress and lipid homeostasis. PLoS Genet. 7, e1002332 10.1371/journal.pgen.1002332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Duffy S., Fam H. K., Wang Y. K., Styles E. B., Kim J. H., Ang J. S., Singh T., Larionov V., Shah S. P., Andrews B., Boerkoel C. F., and Hieter P. (2016) Overexpression screens identify conserved dosage chromosome instability genes in yeast and human cancer. Proc. Natl. Acad. Sci. U.S.A. 113, 9967–9976 10.1073/pnas.1611839113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Srivas R., Shen J. P., Yang C. C., Sun S. M., Li J., Gross A. M., Jensen J., Licon K., Bojorquez-Gomez A., Klepper K., Huang J., Pekin D., Xu J. L., Yeerna H., Sivaganesh V., et al. (2016) A network of conserved synthetic lethal interactions for exploration of precision cancer therapy. Mol. Cell 63, 514–525 10.1016/j.molcel.2016.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. van Pel D. M., Barrett I. J., Shimizu Y., Sajesh B. V., Guppy B. J., Pfeifer T., McManus K. J., and Hieter P. (2013) An evolutionarily conserved synthetic lethal interaction network identifies FEN1 as a broad-spectrum target for anticancer therapeutic development. PLoS Genet. 9, e1003254 10.1371/journal.pgen.1003254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hoon S., Smith A. M., Wallace I. M., Suresh S., Miranda M., Fung E., Proctor M., Shokat K. M., Zhang C., Davis R. W., Giaever G., St. Onge R. P., and Nislow C. (2008) An integrated platform of genomic assays reveals small-molecule bioactivities. Nat. Chem. Biol. 4, 498–506 10.1038/nchembio.100 [DOI] [PubMed] [Google Scholar]
- 35. Niewmierzycka A., and Clarke S. (1999) S-Adenosylmethionine-dependent methylation in Saccharomyces cerevisiae. Identification of a novel protein arginine methyltransferase. J. Biol. Chem. 274, 814–824 10.1074/jbc.274.2.814 [DOI] [PubMed] [Google Scholar]
- 36. Kennedy E. P., and Weiss S. B. (1956) The function of cytidine coenzymes in the biosynthesis of phospholipides. J. Biol. Chem. 222, 193–214 [PubMed] [Google Scholar]
- 37. Henry S. A., Kohlwein S. D., and Carman G. M. (2012) Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics 190, 317–349 10.1534/genetics.111.130286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen Y. L., Montedonico A. E., Kauffman S., Dunlap J. R., Menn F. M., and Reynolds T. B. (2010) Phosphatidylserine synthase and phosphatidylserine decarboxylase are essential for cell wall integrity and virulence in Candida albicans. Mol. Microbiol. 75, 1112–1132 10.1111/j.1365-2958.2009.07018.x [DOI] [PubMed] [Google Scholar]
- 39. Storey M. K., Clay K. L., Kutateladze T., Murphy R. C., Overduin M., and Voelker D. R. (2001) Phosphatidylethanolamine has an essential role in Saccharomyces cerevisiae that is independent of its ability to form hexagonal phase structures. J. Biol. Chem. 276, 48539–48548 10.1074/jbc.M109043200 [DOI] [PubMed] [Google Scholar]
- 40. Trotter P. J., and Voelker D. R. (1995) Identification of a non-mitochondrial phosphatidylserine decarboxylase activity (PSD2) in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 270, 6062–6070 10.1074/jbc.270.11.6062 [DOI] [PubMed] [Google Scholar]
- 41. Halbleib K., Pesek K., Covino R., Hofbauer H. F., Wunnicke D., Hänelt I., Hummer G., and Ernst R. (2017) Activation of the unfolded protein response by lipid bilayer stress. Mol. Cell 67, 673–684.e8 10.1016/j.molcel.2017.06.012 [DOI] [PubMed] [Google Scholar]
- 42. Volmer R., van der Ploeg K., and Ron D. (2013) Membrane lipid saturation activates endoplasmic reticulum unfolded protein response transducers through their transmembrane domains. Proc. Natl. Acad. Sci. U.S.A. 110, 4628–4633 10.1073/pnas.1217611110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ariyama H., Kono N., Matsuda S., Inoue T., and Arai H. (2010) Decrease in membrane phospholipid unsaturation induces unfolded protein response. J. Biol. Chem. 285, 22027–22035 10.1074/jbc.M110.126870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang S., Zhang S., Liou L. C., Ren Q., Zhang Z., Caldwell G. A., Caldwell K. A., and Witt S. N. (2014) Phosphatidylethanolamine deficiency disrupts α-synuclein homeostasis in yeast and worm models of Parkinson disease. Proc. Natl. Acad. Sci. U.S.A. 111, E3976–E3985 10.1073/pnas.1411694111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Garabedian M. J., and Yamamoto K. R. (1992) Genetic dissection of the signaling domain of a mammalian steroid receptor in yeast. Mol. Biol. Cell 3, 1245–1257 10.1091/mbc.3.11.1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cox J. S., and Walter P. (1996) A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87, 391–404 10.1016/S0092-8674(00)81360-4 [DOI] [PubMed] [Google Scholar]
- 47. Hwang C., Sinskey A. J., and Lodish H. F. (1992) Oxidized redox state of glutathione in the endoplasmic reticulum. Science 257, 1496–1502 10.1126/science.1523409 [DOI] [PubMed] [Google Scholar]
- 48. Malhotra J. D., and Kaufman R. J. (2007) The endoplasmic reticulum and the unfolded protein response. Semin. Cell Dev. Biol. 18, 716–731 10.1016/j.semcdb.2007.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zeeshan H. M., Lee G. H., Kim H. R., and Chae H. J. (2016) Endoplasmic reticulum stress and associated ROS. Int. J. Mol. Sci. 17, 327 10.3390/ijms17030327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Delic M., Rebnegger C., Wanka F., Puxbaum V., Haberhauer-Troyer C., Hann S., Köllensperger G., Mattanovich D., and Gasser B. (2012) Oxidative protein folding and unfolded protein response elicit differing redox regulation in endoplasmic reticulum and cytosol of yeast. Free Rad. Biol. Med. 52, 2000–2012 10.1016/j.freeradbiomed.2012.02.048 [DOI] [PubMed] [Google Scholar]
- 51. Malhotra J. D., Miao H., Zhang K., Wolfson A., Pennathur S., Pipe S. W., and Kaufman R. J. (2008) Antioxidants reduce endoplasmic reticulum stress and improve protein secretion. Proc. Natl. Acad. Sci. U.S.A. 105, 18525–18530 10.1073/pnas.0809677105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Levin D. E. (2011) Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway. Genetics 189, 1145–1175 10.1534/genetics.111.128264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Scrimale T., Didone L., de Mesy Bentley K. L., and Krysan D. J. (2009) The unfolded protein response is induced by the cell wall integrity mitogen-activated protein kinase signaling cascade and is required for cell wall integrity in Saccharomyces cerevisiae. Mol. Biol. Cell 20, 164–175 10.1091/mbc.e08-08-0809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Costa-de-Oliveira S., Silva A. P., Miranda I. M., Salvador A., Azevedo M. M., Munro C. A., Rodrigues A. G., and Pina-Vaz C. (2013) Determination of chitin content in fungal cell wall: an alternative flow cytometric method. Cytometry 83, 324–328 10.1002/cyto.a.22250 [DOI] [PubMed] [Google Scholar]
- 55. Rodicio R., and Heinisch J. J. (2010) Together we are strong–cell wall integrity sensors in yeasts. Yeast 27, 531–540 10.1002/yea.1785 [DOI] [PubMed] [Google Scholar]
- 56. Verna J., Lodder A., Lee K., Vagts A., and Ballester R. (1997) A family of genes required for maintenance of cell wall integrity and for the stress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 94, 13804–13809 10.1073/pnas.94.25.13804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mewes H. W., Frishman D., Güldener U., Mannhaupt G., Mayer K., Mokrejs M., Morgenstern B., Münsterkötter M., Rudd S., and Weil B. (2002) MIPS: a database for genomes and protein sequences. Nucleic Acids Res. 30, 31–34 10.1093/nar/30.1.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fujita M., and Jigami Y. (2008) Lipid remodeling of GPI-anchored proteins and its function. Biochim. Biophys. Acta 1780, 410–420 10.1016/j.bbagen.2007.08.009 [DOI] [PubMed] [Google Scholar]
- 59. Rolli E., Ragni E., Calderon J., Porello S., Fascio U., and Popolo L. (2009) Immobilization of the glycosylphosphatidylinositol-anchored Gas1 protein into the chitin ring and septum is required for proper morphogenesis in yeast. Mol. Biol. Cell 20, 4856–4870 10.1091/mbc.e08-11-1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ruan L., Zhou C., Jin E., Kucharavy A., Zhang Y., Wen Z., Florens L., and Li R. (2017) Cytosolic proteostasis through importing of misfolded proteins into mitochondria. Nature 543, 443–446 10.1038/nature21695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Amm I., Sommer T., and Wolf D. H. (2014) Protein quality control and elimination of protein waste: the role of the ubiquitin-proteasome system. Biochim. Biophys. Acta 1843, 182–196 10.1016/j.bbamcr.2013.06.031 [DOI] [PubMed] [Google Scholar]
- 62. Tyedmers J., Mogk A., and Bukau B. (2010) Cellular strategies for controlling protein aggregation. Nat. Rev. Mol. Cell Biol. 11, 777–788 10.1038/nrm2993 [DOI] [PubMed] [Google Scholar]
- 63. Klis F. M., de Jong M., Brul S., and de Groot P. W. (2007) Extraction of cell surface-associated proteins from living yeast cells. Yeast 24, 253–258 10.1002/yea.1476 [DOI] [PubMed] [Google Scholar]
- 64. Muñiz M., and Zurzolo C. (2014) Sorting of GPI-anchored proteins from yeast to mammals–common pathways at different sites? J. Cell Sci. 127, 2793–2801 10.1242/jcs.148056 [DOI] [PubMed] [Google Scholar]
- 65. Yoshida H., Matsui T., Yamamoto A., Okada T., and Mori K. (2001) XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107, 881–891 10.1016/S0092-8674(01)00611-0 [DOI] [PubMed] [Google Scholar]
- 66. Gibellini F., and Smith T. K. (2010) The Kennedy pathway–de novo synthesis of phosphatidylethanolamine and phosphatidylcholine. IUBMB Life 62, 414–428 [DOI] [PubMed] [Google Scholar]
- 67. Sakoff J. A., Ackland S. P., Baldwin M. L., Keane M. A., and McCluskey A. (2002) Anticancer activity and protein phosphatase 1 and 2A inhibition of a new generation of cantharidin analogues. Invest. New Drugs 20, 1–11 10.1023/A:1014460818734 [DOI] [PubMed] [Google Scholar]
- 68. Hughes W. E., Woscholski R., Cooke F. T., Patrick R. S., Dove S. K., McDonald N. Q., and Parker P. J. (2000) SAC1 encodes a regulated lipid phosphoinositide phosphatase, defects in which can be suppressed by the homologous Inp52p and Inp53p phosphatases. J. Biol. Chem. 275, 801–808 10.1074/jbc.275.2.801 [DOI] [PubMed] [Google Scholar]
- 69. Yell J. A., Burge S. M., and Dean D. (1994) Cantharidin-induced acantholysis: adhesion molecules, proteases, and related proteins. Br. J. Dermatol. 130, 148–157 10.1111/j.1365-2133.1994.tb02893.x [DOI] [PubMed] [Google Scholar]
- 70. Stoughton R. B., and Bagatell F. (1959) The nature of cantharidin acantholysis. J. Invest. Dermatol. 33, 287–292 10.1038/jid.1959.152 [DOI] [PubMed] [Google Scholar]
- 71. Weakley D. R., and Einbinder J. M. (1962) The mechanism of cantharidin acantholysis. J. Invest. Dermatol. 39, 39–45 10.1038/jid.1962.78 [DOI] [PubMed] [Google Scholar]
- 72. Lissina E., Weiss D., Young B., Rella A., Cheung-Ong K., Del Poeta M., Clarke S. G., Giaever G., and Nislow C. (2013) A novel small molecule methyltransferase is important for virulence in Candida albicans. ACS Chem. Biol. 8, 2785–2793 10.1021/cb400607h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Loukin S., and Kung C. (1995) Manganese effectively supports yeast cell-cycle progression in place of calcium. J. Cell Biol. 131, 1025–1037 10.1083/jcb.131.4.1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Leung A. K. C., Barankin B., and Hon K. L. E. (2017) Molluscum contagiosum: an update. Recent Pat. Inflamm. Allergy Drug Discov. 11, 22–31 [DOI] [PubMed] [Google Scholar]
- 75. Keogh M. C., Kim J. A., Downey M., Fillingham J., Chowdhury D., Harrison J. C., Onishi M., Datta N., Galicia S., Emili A., Lieberman J., Shen X., Buratowski S., Haber J. E., Durocher D., et al. (2006) A phosphatase complex that dephosphorylates γH2AX regulates DNA damage checkpoint recovery. Nature 439, 497–501 10.1038/nature04384 [DOI] [PubMed] [Google Scholar]
- 76. Schmitt M. E., Brown T. A., and Trumpower B. L. (1990) A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18, 3091–3092 10.1093/nar/18.10.3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Carman G. M., and Han G. S. (2009) Regulation of phospholipid synthesis in yeast. J. Lipid Res. 50, S69–S73 10.1194/jlr.R800043-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Carman G. M., and Han G. S. (2011) Regulation of phospholipid synthesis in the yeast Saccharomyces cerevisiae. Annu. Rev. Biochem. 80, 859–883 10.1146/annurev-biochem-060409-092229 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.