Abstract
The Ebola virus is a zoonotic pathogen that can cause severe hemorrhagic fever in humans, with up to 90% lethality. The deadly 2014 Ebola outbreak quickly made an unprecedented impact on human lives. While several vaccines and therapeutics are under development, current approaches contain several limitations, such as virus mutational escape, need for formulation or refrigeration, poor scalability, long lead-time, and high cost. To address these challenges, we developed locked nucleic acid (LNA)-modified antisense oligonucleotides (ASOs) to target critical Ebola viral proteins and the human intracellular host protein Niemann-Pick C1 (NPC1), required for viral entry into infected cells. We generated noninfectious viral luciferase reporter assays to identify LNA ASOs that inhibit translation of Ebola viral proteins in vitro and in human cells. We demonstrated specific inhibition of key Ebola genes VP24 and nucleoprotein, which inhibit a proper immune response and promote Ebola virus replication, respectively. We also identified LNA ASOs targeting human host factor NPC1 and demonstrated reduced infection by chimeric vesicular stomatitis virus harboring the Ebola glycoprotein, which directly binds to NPC1 for viral infection. These results support further in vivo testing of LNA ASOs in infectious Ebola virus disease animal models as potential therapeutic modalities for treatment of Ebola.
Keywords: : antisense oligonucleotides, locked nucleic acids, Niemann-Pick C1 (NPC1), Ebola virus (EBOV), Ebola
Introduction
The Ebola virus is one of the deadliest human pathogens, with a case fatality rate of 25%–90% depending on the strain and outbreak [1]. The virus is transmitted through direct contact with blood or other body fluids such as saliva, mucus, feces, sweat, tears, urine, and vomit [1]. The outbreak that began in West Africa in 2014 infected more than 28,000 people and led to over 11,000 deaths. This is the largest outbreak since the Ebola virus was first identified in 1976; more humans have died in this single outbreak than all other outbreaks combined (World Health Organization, Ebola Situation Reports 2016).a Ebola virus disease (EVD) survivors frequently suffer from a number of complications due to the persistence of viral infection, including impaired vision, hearing loss, troubled sleep, memory loss, confusion, and joint pain (World Health Organization, Ebola Situation Reports 2016)b [2,3]. In addition, an emerging concern with more than 10,000 survivors is viral resurgence. When patients are in the last stages of the disease, viral loads can increase to billions of copies per/mL of blood, facilitating detection. However, during recovery the Ebola virus remains in survivors in what are known as immune-privileged sites such as the eye (ocular aqueous humor), breast milk, and semen, months after Ebola RNA is no longer detectable in blood [4–8].
In one documented case, a male EVD survivor sexually transmitted the virus to a partner 155 days after testing negative for circulating Ebola virus [4]. The partner later developed and died from EVD [4]. In another survivor case, the Ebola virus relapsed leading to hospitalization [9], and in yet another case, Ebola virus RNA was found in the fetal blood, amniotic fluid, placenta, and stool of a stillborn child from a pregnant woman who survived EVD and whose blood tested negative for Ebola RNA 8 days after symptoms started [10]. These cases and others suggest that Ebola virus can persist for months in tissue reservoirs and new outbreaks from transmitted infectious virus are still a dangerous possibility, eliciting a need for effective therapeutics targeting Ebola. Currently, there are no approved treatments or licensed vaccines.
Recent responses to the Ebola epidemic have largely relied on public health strategies for surveillance of affected areas, tracing exposure, containing confirmed cases of Ebola at treatment centers, testing suspected cases at mobile labs, and preventing confirmed cases or contacts of cases from spreading to densely populated areas/cities. There is an unlicensed vaccine being used on an exploratory basis for ring vaccination of those who have been in contact with confirmed cases of Ebola, and a few “investigational therapeutics being used on a compassionate use basis” (World Health Organization, Ebola Reports 2018).c However, the recent and unexpected resurgence of the Ebola virus in Congo (May 2018), with a rising death toll, reminds us how quickly and easily this virus can spread and kill (World Health Organization, Ebola Reports 2018),d and highlights the urgent need for novel therapeutic strategies.
There are four identified subtypes of Ebola virus according to the four outbreaks: Zaire, Sudan, Ivory Coast, and Reston strains [1]. Ebola virus is a member of the filovirus family, which are single strand negative RNA viruses [1]. Ebola virus exhibits a 19 kb long genome that encodes seven proteins: glycoprotein (GP), which produces small soluble GP and soluble GP through transcriptional editing [11,12], nucleoprotein (NP), VP35, VP40, VP30, VP24, and the viral polymerase L. NP coats the viral genome and forms a complex with VP30, VP35, and L to modulate transcription of the viral RNA. Phosphorylation of VP30 displaces VP30 from this complex allowing it to participate in viral replication, which is facilitated by NP, VP24, and VP35 [13]. VP40, a matrix protein, then associates with L for viral particle assembly, budding, and delivery. VP40 along with VP24 also play a role in inhibiting antiviral type I and II interferon response/signaling [14]. Additional details of the interaction between virus infection and innate or adaptive immune responses are further reviewed elsewhere [14–16].
Ebola virus infection has been shown to require the host factor Niemann-Pick C1 (NPC1) [17–19]. NPC1 is a 1,278 amino acid protein essential for cholesterol transport to the trans-Golgi and plasma membrane caveolae and is localized primarily in the late endosomal/lysosomal membrane compartment [20]. The structure of NPC1 includes 13 transmembrane domains, a cytoplasmic loop at the C-terminal end, 6 small loops in the cytoplasm, and 3 large and 4 small luminal loops. NPC1 has been demonstrated to facilitate virus entry into the host cell by direct binding of the EboV GP to one of the NPC1 luminal domains [21–23]. In fact, cells from species normally not susceptible to EVD were rendered permissive for infectivity by the expression of human NPC1 [24]. Indeed, mice heterozygous for NPC1 are completely protected from Ebola virus infection. Small molecules targeting NPC1 also offer protection from infection [25]. Studies have found that upon cell endocytosis, the Ebola virus GP is proteolyzed by the endosomal Cathepsin B protease, which then allows the N-terminal portion of GP to associate specifically with NPC1, permitting virus entry into the cytosolic compartment and viral replication [23,26–28]. Together, these findings suggest that NPC1 might represent an interesting therapeutic target to prevent Ebola virus infection.
While there are currently no vaccines or therapeutics approved for the prevention or treatment of EVD, a number of experimental strategies are at various stages of development [29,30]. These include several vaccines and therapeutic strategies aimed at directly targeting the Ebola virus. A monoclonal antibody cocktail (ZMapp) directed against the virus appears to exhibit efficacy in nonhuman primate (NHP) EVD models [31] and has been used to treat an EVD patient. In addition, a siRNA-based approach that targets the viral RNA-dependent RNA polymerase (L protein), VP24, and VP35 appears to exhibit some efficacy in animal models [32]. Peptide-conjugated and chemically modified morpholino antisense oligonucleotides (ASOs) directed to block translation of VP24 and VP35 have also been shown to be quite efficacious in rodent and NHP models [33]. Several companies have also tested nucleoside analogs originally designed to target other RNA viruses such as influenza virus, which may have activity against the Ebola virus RNA-dependent RNA polymerase (eg, favipiravir/Avigan) [34]. Although these strategies have shown promise to counter Ebola virus infection or transmission, they exhibit a number of limitations, such as the need for refrigeration, requirement for drug formulation, inability to scale production, lack of versatility beyond the current Ebola virus strain, and unknown off-target effects.
Locked nucleic acid (LNA)-modified phosphorothioate (PS) ASOs offer distinct advantages due to their properties that have been optimized over the years [35–38]. Large-scale manufacture of therapeutic LNA ASOs is well established, and LNA ASOs are very stable and therefore do not require refrigeration like biologic drugs, such as antibodies and vaccines. This is particularly useful given that most Ebola outbreaks originate in sub-Saharan Africa where regular temperatures are high. LNA ASOs can be stored lyophilized and are easily suspended in solution before injection. In addition, PS backbone modifications increase the biostability of LNA ASOs in the blood stream and tissues, and enhance their pharmacokinetic properties, thereby facilitating delivery to many peripheral tissues in vivo [35,37,39]. LNA-modified ASOs exhibit increased binding affinity and specificity toward their cognate target sequences in comparison with other ASO chemistries such as morpholinos, leading to improved potency and fewer off-target effects. Furthermore, LNA ASOs tend to accumulate in the liver, which is a primary target organ for EVD once it infected monocytes, dendrites, and macrophages [40,41]. LNA ASOs also accumulate in the kidney—another key organ targeted by Ebola virus—in addition to other tissues such as bone marrow, adipocytes, and lymph nodes [39,40].
We have devised a two-pronged approach for targeting Ebola virus infections using LNA ASOs targeting Ebola virus protein translation, and blocking production of the NPC1 host gene product required for virus cell entry. Translational blocking has previously been achieved using morpholino ASOs [33], however, this ASO chemistry is difficult to scale up for large-scale production. Using the sequence of the most virulent strain of Ebola virus from the recent outbreak [42], we designed LNA ASO mixmers harboring a PS backbone that target the mRNA sequence of key Ebola virus proteins. We also employed LNA gapmer ASOs to target the NPC1 mRNA. Mixmers have LNA modifications interspersed with DNA in the ASO sequence, while gapmers exhibit a central stretch of DNA sequence flanked by LNA wings, where the RNA in the DNA–RNA duplex formed after target engagement is subjected to RNase H cleavage [37].
Mixmer LNA ASOs were designed to target sequences overlapping with or downstream of the start codons of NP, VP24, VP35, and L to interfere with translation of these Ebola virus genes. Previous work suggested that these regions around the start codons, including parts of the 5′UTR, modulate the translation of Ebola virus proteins [43,44]. These Ebola virus genes were chosen for a number of reasons. NP has been noted as a high quality target for EVD therapeutics due to its crucial role in Ebola viral replication and transcription [45]. VP35 and VP30 decoy the host's immune response to EVD by blocking the normal RNAi response against viral gene expression [15]. VP24 along with VP40 play a role in inhibiting the type I and II interferon responses/signaling [14,15]. In addition, recent findings indicate that VP24 may be key to the ability of the virus to mutate [46]. NPC1 was targeted due to its vital role in facilitating Ebola virus infection through direct interaction with the Ebola virus GP protein as previously mentioned. Our studies revealed effective LNA ASO targeting of two Ebola viral genes, NP and VP24, and also demonstrated prevention of Ebola virus GP-dependent infectivity via NPC1 inhibition in human and mouse cells. Our approach thus attacks core components of all phases of virus replication and infection.
Materials and Methods
ASO design
ASOs capable of downregulating NPC1 were designed as LNA-modified DNA gapmer ASOs with a PS backbone. Three ASOs were designed with a perfect match to NPC1 in primates (human, chimpanzee, rhesus macaque, and African green monkey), and additional three ASOs with a perfect match to NPC1 in primates and mouse (see Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/nat). None of the ASOs had any predicted perfect match off-targets in any of the tested species when checked against the most recent version of the Ensembl database as of October 2017 (release 90), except LNA23, which had a single perfect match off-target in rhesus macaque. Furthermore, a random sequence LNA/DNA gapmer with no perfect match or 1-mismatched hits in any of the five species served as a negative control (LNA24, Supplementary Table S1).
ASOs capable of translational inhibition of Ebola virus transcripts were designed as LNA/DNA mixmer oligonucleotides with a PS backbone. Alignment of 101 Ebola virus genomes published by Gire et al. [42] (Supplementary material, File S1 in Gire et al. [42]) were used to extract sequences corresponding to 20 bases upstream and 35 bases downstream of the start codons of virus genes NP, VP35, VP24, and L. LNA ASOs were designed to target sites in these regions that are conserved across all Ebola variants from the 2014 outbreak and some also conserved across strains isolated earlier (1976, 1977, 1994, 1995, 1996, 2002, and 2008). ASOs were designed with a low propensity for secondary structure and self-hybridization, and a high LNA load to achieve high affinity. ASOs containing cg or cG (lowercase = DNA, uppercase = LNA) motifs were synthesized with methylated cytosine to mask the immunostimulatory motif and all LNA ASOs were obtained from Exiqon (Vedbaek, Denmark).
Ebola viral gene-luciferase fusion reporters
Luciferase reporters were designed as follows. The open reading frame (ORF) for each of the four Ebola virus genes used in this study was fused to the luciferase gene in the pSP-lucNF fusion vector (Promega). Reporters included 98 bp covering a portion of the 5′UTR plus mRNA sequence of the four Ebola virus genes fused to luciferase. Ninety-eight base pairs of ORF in frame with the proximal ATG (pATG) for each Ebola virus gene was used. Constructs were designed to ensure that the pATG of the 98 bp ORF was in frame with the luciferase gene. The ATG of the Luciferase gene was mutated to alanine to ensure that translation initiated only at the pATG of Ebola virus gene ORFs. While the 98 bp ORF used for L protein did include an upstream ATG (uATG), this uATG was not in frame and produced a truncated protein due to a premature stop codon. An uATG is also included in the VP35 sequence, however, this is immediately followed by a stop codon ensuring that no functional protein is made. Reporter sequences are provided in Supplementary Table S1.
VP24: 57 bp of protein (19 amino acids) +41 bp upstream of pORF region
NP: 39 bp of protein (13 amino acids) +59 bp upstream of uATG
VP35: 60 bp of protein (20 amino acids) +38 bp upstream of uATG
L: 60 bp of protein (20 amino acids) +38 bp upstream of uATG
Complementary single-stranded oligonucleotides were mixed at equal volume and annealed. Annealed oligonucleotides were cooled gradually: first at room temperature (23°C) for 1 h then at 4°C for up to 2 h and finally 1 h at −20°C. Double-stranded oligonucleotides were digested with Mung Bean nuclease to remove single-stranded overhangs as follows: double-stranded DNA was suspended in 1 × Mung Bean nuclease buffer and 1 μL Mung Bean Nuclease (NEB) and incubated at 30°C for 30 min. The enzyme was inactivated by addition of 0.01% sodium dodecyl sulfate and the reaction cleaned with PCR Clean-up Kit (Qiagen). Primers (see Supplementary Table S1 for sequences) were designed to add NheI and HindIII restriction enzyme sites to the double-stranded sequences by PCR and confirmed with agarose gel diagnostics. Primers were synthesized by Integrated DNA Technologies.
The Ebola virus sequences with NheI and HindIII restriction sites were inserted into a pcDNA3.1 vector that had the full luciferase gene encoded for expression under a CMV promoter. The start codon AUG (methionine) in the luciferase gene was mutated to GCT (alanine) using the Quick Change Site-Directed Mutagenesis Kit (Agilent Technologies). Mutagenic primers were designed to perform this codon substitution and to ensure that Ebola virus gene-luciferase fusion did not generate a frameshift. Primers for mutagenesis were synthesized by Invitrogen Life Technologies and purified by high performance liquid chromatography. Ebola virus gene ORF insertions and luciferase gene ATG to GCT substitution were confirmed by sequencing analysis (Genewiz).
In vitro transcription/translation assay
The in vitro transcription and translation assay was performed with the TNT T7 Quick Coupled Transcription/Translation System Catalog (Promega) according to manufacturer's guidelines. Concentrations of Ebola virus gene-luciferase fusion reporters and LNA ASOs to add to the TNT reaction were optimized by titration and LNA ASO:DNA molar ratio calculations. Ten microliter reactions of TNT Quick mix + DNA + 1 mM methionine ± LNA ASO in nuclease-free water was prepared for each reporter. Reactions were incubated at 30°C for 90 min, and the samples were placed at room temperature (23°C) for 30 min before addition of luciferase assay reagent. The luciferase assay reagent was thawed on ice ∼45 min and then warmed to 23°C for 30 min. About 2.5 μL of in vitro lysate was mixed with 50 μL of luciferase assay reagent, and luminescence immediately read (<30 s) in the luminometer, and then recorded and analyzed with Excel (Microsoft).
Cell culture
HeLa, HepG2, and 293T cells (ATCC) were maintained in Minimum Essential Media (MEM) with 5% penicillin streptomycin, 5% sodium pyruvate, and 10% fetal bovine serum (FBS). J774A.1 cells (ATCC T1B-67) were cultured per ATCC guidelines in Dulbecco's modified Eagle medium (DMEM) media supplemented with 10% FBS. Cell culture products were purchased from Thermo Fisher Scientific.
Transfection of HEK293T cells with Ebola-luciferase vectors and antiviral LNA ASOs
Transfections were performed with Opti-MEM media (ThermoFisher Scientific). On day 1, 293T cells were plated with Opti-MEM at 50,000 cells/mL in 24-well plates and incubated at 37°C for 24 h. On day 2, LNA ASOs were transfected with Lipofectamine RNAiMAX (Invitrogen). Cells transfected with LNA ASOs were incubated at 37°C for 4–6 h. Ebola virus-luciferase and β-galactosidase vectors were then transfected with Lipofectamine2000 (Invitrogen) using manufacturer's protocol and cells were incubated at 37°C. Cells were harvested 24 h later.
Cells were lysed with Glo-Lysis reagent (Promega) at room temperature. Luminescence was measured using Dual-Glo Reagent (Promega) and read with Centro LB 960 plate reader (Berthold Technologies). β-Galactosidase expression was measured using Assay 2 × Buffer (Promega) and read with SpectraMax M5 (Molecular Devices).
Transfection of HeLa cells with Ebola virus-luciferase vectors and antiviral LNA ASOs
Cells were plated at 60,000 cells/mL with MEM supplemented as described above in 24-well plates and incubated at 37°C for 24 h. Ebola virus-luciferase and β-galactosidase vectors and LNA ASOs were transfected with Opti-MEM using Lipofectamine2000 with EXIQON Co-Transfection Protocol. Cells were incubated at 37°C for 24 h. Cells were harvested and luminescence and β-galactosidase expression measured as previously described.
Transfection of HeLa and HepG2 cells with NPC1-targeting LNA ASOs for RNA extraction
HepG2 cells were transfected with NPC1 LNA ASOs at 50 and 100 nM in six-well plates with Lipofectamine RNAiMax (Invitrogen) as per manufacturer's instructions. RNA was harvested 48 h later with TRIzol Reagent (Invitrogen). HeLa cells were transfected with LNA ASOs at 50 and 100 nM by forward transfection with Lipofectamine RNAiMax. RNA was harvested 48 h later with TRIzol Reagent.
Transfection of mouse J774A.1 macrophages with NPC1-targeting LNA ASOs
J774A.1 cells (ATCC T1B-67) were cultured per ATCC guidelines in DMEM media supplemented with 10% FBS. NPC1 LNA ASOs and siRNAs were transfected with TransIT-TKO Transfection Reagent (Mirus Bio). NPC1, Accell Mouse Npc1 (18145) siRNA SMARTpool and nontargeting siRNA, ON-TARGETplus Nontargeting pool, were obtained commercially from GE Healthcare Dharmacon.
Cells were counted and plated at 100,000 cells/mL with complete media in six-well plates and incubated at 37°C on day 1. On day 2, transIT-TKO Reagent was warmed to room temperature for 30 min. and gently vortexed. Opti-MEM media were warmed at 37°C. Ten microliter of TransIT-TKO Reagent was added to 250 μL Opti-MEM per sample in separate tubes. Then siRNAs and LNA ASOs were added to final concentration of 100 nM and allowed to incubate at room temperature 15–30 min for complex formation. Cells plated on day 1 were removed from 37°C incubator and half the volume of media removed in each well. Trans-IT TKO Reagent and LNA ASO/siRNA complex mixture in media were added drop-wise to different areas of the wells and incubated at 37°C. RNA and protein were extracted 72 h later.
Western blot analysis
Protein was extracted with RIPA Buffer as follows. Briefly, cells were washed with 1 × phosphate-buffered saline (PBS), scraped, and spun down in 1 × PBS 610 g for 10 min. PBS was removed and cells resuspended in RIPA Buffer on ice ∼30 min. Lysates were sonicated for 30 s and then spun down 14,000 g for 15 min at 4°C, and protein quantified with Pierce BCA Protein Assay Kit No. 23227 and stored.
NPC1 antibody Anti-NPC1 (Catalog No. MABS739) was procured from Millipore. Secondary antibody (Catalog No. NA934V) was purchased from GE Healthcare. For human cell samples, monoclonal Anti-β-tubulin antibody (Catalog No. T7 B16) from Sigma Aldrich was used as loading control for western blot analysis. Actin antibody, β-actin (13E5) Rabbit mAb (Catalog No. 4970P) from Cell Signaling was used as a loading control for western blot analysis of mouse samples.
Quantitative reverse transcription-polymerase chair reaction analysis
RNA extraction was performed following TRIzol Reagent (Catalog No. 15596026) guidelines and quantified by Nanodrop. cDNA was synthesized with Roche Transcriptor First Strand cDNA synthesis Kit (Catalog No. 04379012001). qPCRs were performed using Roche LightCycler 480 SYBR Green I Master (Catalog No. 04707516001). The primers used for qPCR are listed in Supplementary Table S1.
Infection with vesicular stomatitis virus harboring EBOV-GP
NPC1 LNA ASOs along with siRNA and LNA ASO controls were added to HeLa cells at 100 nM by reverse transfection using Lipofectamine RNAiMax (Invitrogen). After 48 h, RNA and protein were purified from one group of transfected cells using TRIzol Reagent and RIPA Buffer and analyzed for expression of NPC1 mRNA by RT-PCR and NPC1 protein by western blotting. A second group of treated cells was incubated with vesicular stomatitis virus (VSV) encoding luciferase and pseudotyped with Ebola Zaire GP (VSVluc-EboV GP) [18]. Twenty-four hours later, expression of virus-encoded luciferase was measured (Promega).
Results
LNA ASOs significantly reduce translation of Ebola virus genes in vitro and in human cells
Using the Ebola virus sequences from the 2014 outbreak, LNA ASOs were designed to target conserved regions close to the translation start sites for the following Ebola viral proteins: NP, VP24, VP35, and L. One or two LNAs were designed per Ebola virus gene (Supplementary Table S1). The lengths of the LNA ASOs ranged from 16 to 20 nucleotides and targeted different regions near the translation start site of each gene (Fig. 1 and Supplementary Fig. S1; Supplementary Table S1). A control LNA ASO designed not to target any particular sequence in the Ebola virus genome served as a negative control. Six LNA ASOs targeting different sites in the human NPC1 mRNA sequence were also designed (Supplementary Fig. S2).
FIG. 1.
pATG and ORFs of Ebola proteins used to design Ebola luciferase reporters for cellular studies. Ninety-eight base pairs of ORF sequence that was in frame with the pATG was fused with the luciferase gene in all reporters. In other words, 98 bp encompassing the start codons in the ORF of NP, VP24, VP35, and L were fused to luciferase. The pATG (the start site of the main ORF of the Ebola protein) is used as the main start site to express the in frame Luciferase-Ebola ORF fusion reporters. The uATGs and Luciferase ATG are either nonfunctional or mutated in the reporters. The pATG used in the reporters, their locations in the reporters, and LNA ASO target sites are highlighted in this figure. Regions encompassing the start codons in the ORF of (a) L, (b) VP24, (c) VP35, and (d) NP were fused to luciferase (98 bp of 5′UTR—luciferase). The start codon (ATG) of luciferase was mutated to CTG (alanine). Reporter expression was under the control of the CMV promoter. (e) LNA ASOs targeting Ebola proteins reduce expression of Ebola reporters in vitro and in HeLa cells. Schematic of LNA ASOs designed to target translation of Ebola fusion proteins: NP, VP24, VP35, and L. (f) Two micromolar of each LNA ASOs tested inhibited the translation of Ebola fusion proteins in vitro. (g) Two LNA ASOs targeting Ebola VP24 protein reduce expression of the Ebola VP24-luciferase fusion in HeLa cells, as determined by measurement of luminescence 24 h post-transfection. LNA30 targeting Ebola VP35 protein and LNA38 targeting Ebola L protein were used as negative controls. Neither demonstrated inhibition of Ebola VP24 protein, highlighting the specificity of LNA ASOs. IC50 values were calculated for LNAs tested in HeLa cells and are provided in Supplementary Table S2. (h) Two LNA ASOs targeting Ebola NP protein reduce expression of Ebola NP-luciferase fusion in HeLa cells, as determined by measurement of luminescence 24 h post-transfection. LNA30 targeting Ebola VP25 protein and LNA38 targeting Ebola L protein were used as negative controls. Neither demonstrated inhibition of Ebola NP protein, indicating specificity of LNA ASOs. *P < 0.05, **P < 0.01, ***P < 0.001. All experiments were performed in biological triplicates; each biological replicate was performed in technical triplicates. pATG, proximal ATG; ORF, open reading frame; NP, nucleoprotein; uATG, upstream ATG; LNA, locked nucleic acid; ASO, antisense oligonucleotide.
Due to the requirement for a BL-4 facility to work with Ebola virus, we developed a noninfectious viral reporter system to study the effectiveness of the LNA ASOs in vitro and in mammalian cells in a BL-2+ setting. Previous work has established viral-like reporter systems as a valid method to investigate viral transcription, replication, and infection [14,15,34,39,43,47]. Reporters with 98 nucleotides covering a portion of the 5′UTR and mRNA sequence of each Ebola gene fused to luciferase were constructed (Fig. 1). The start codon of the luciferase gene was mutated to ensure that any luciferase read-out was due to expression of Ebola virus genes via usage of the Ebola virus gene start codons.
The reporters incubated with LNA ASOs were tested with an in vitro-coupled transcription/translation system using rabbit reticulocyte lysate as a proof-of-concept study for the efficacy of LNA ASO treatment. Expression of the Ebola virus gene reporters for NP, VP24, VP35, and L were measured by analyzing levels of luciferase activity. The optimum DNA template concentration was determined by dose titration to avoid variability in expression of the fusion reporters (Supplementary Fig. S3). Dose titration studies were also carried out to determine the optimal concentration of LNA ASOs for specific inhibition (Fig. 2 and Supplementary Figs. S4–S6).
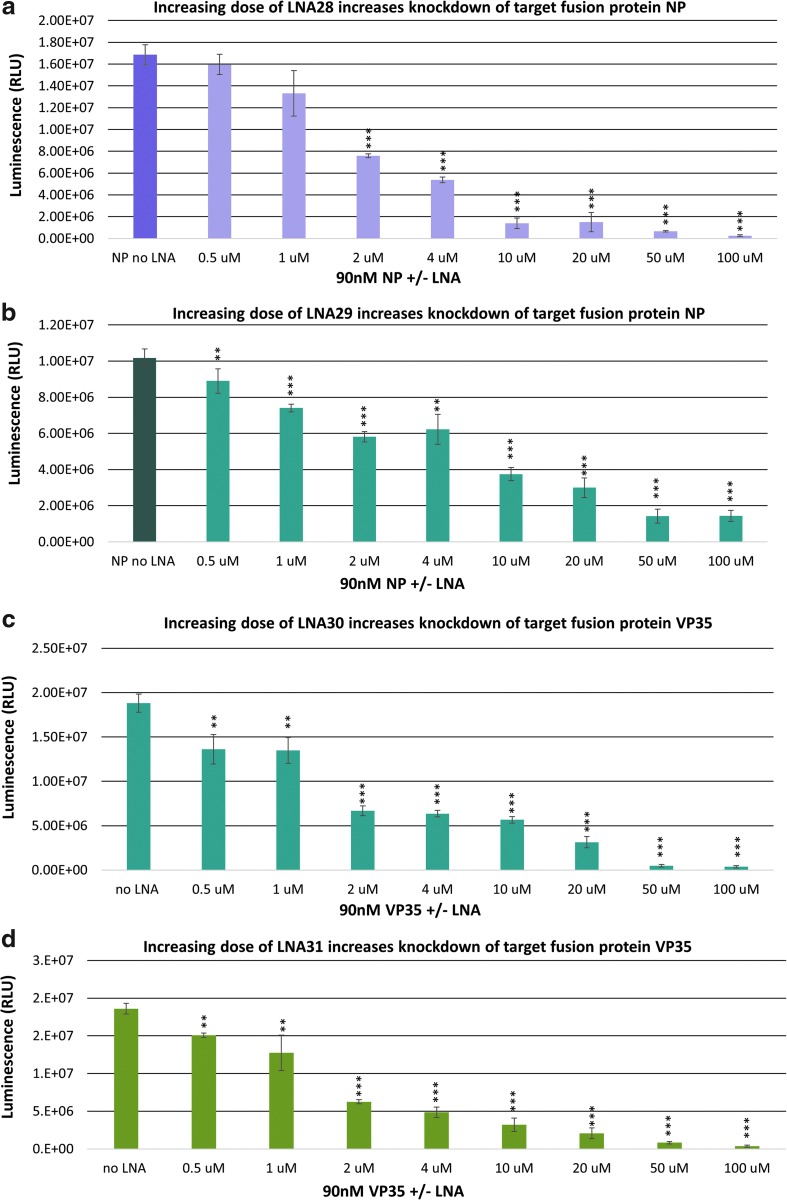
FIG. 2.
Titration of optimal concentration of LNA ASOs. A dose-dependent titration of LNA ASOs determined optimal concentration for specific and significant reduction of target Ebola genes. Individual coupled in vitro transcription/translation assays with rabbit reticulocyte lysate were performed at varying LNA ASO concentrations and fixed Ebola luciferase reporter concentrations (90 nM). Ebola luciferase reporter expression was measured and analyzed for each LNA ASO tested. (a) LNA28 targeting Ebola NP fusion protein dose dependently reduces expression of Ebola NP-luciferase fusion protein in vitro. (b) LNA29 targeting Ebola NP fusion protein dose dependently reduces expression of Ebola NP-luciferase fusion protein in vitro. (c) LNA30 targeting Ebola VP35 fusion protein dose dependently reduces expression of Ebola VP35-luciferase fusion protein in vitro. (d) LNA31 targeting Ebola VP35 fusion protein dose dependently reduces expression of Ebola VP35-luciferase fusion protein in vitro. Data were measured for LNA37, LNA36, and LNA38 (not reported); >50% knockdown of target reporters/fusion proteins was not observed until >2 μM of LNA ASO. IC50 values were calculated and are reported with graphs in Supplementary Fig. S4. *P < 0.05, **P < 0.01, ***P < 0.001.
All LNA ASOs tested showed significant reduction of the target Ebola virus protein translation reporters in vitro (Fig. 1). The negative control LNA ASO (LNA24) did not reduce expression of the luciferase gene and/or the Ebola virus reporters, until concentrations were beyond the range for specific knockdown (Supplementary Fig. S5). In addition, LNA ASOs were tested on the empty luciferase reporter cassette as a control at the specific concentrations where significant and specific reduction of Ebola virus NP, VP24, VP35, and L expression was observed (Supplementary Fig. S6). None of the LNA ASOs nonspecifically reduced expression of luciferase gene alone in the determined range (0.5–2 μM), indicating the specificity of the LNA ASOs for the targeted sequences within the translation start site of the selected Ebola virus genes (data not shown).
To test functionality of LNA ASOs for Ebola virus reporter inhibition in cultured human cells, LNA ASOs were co-transfected with Ebola virus gene-luciferase fusion reporters in HEK293T cells. LNA ASOs were transfected over a concentration range in HEK293T cells to determine the optimal concentration(s) for specific inhibition of Ebola virus gene expression. LNA ASOs effectively reduced in a dose-dependent manner (concentration range of 1–25 nM) target fusion luciferase reporter expression for Ebola virus genes NP, VP24, and VP35 (Supplementary Fig. S7). The LNA ASO designed to target Ebola viral polymerase L was used as a negative control in these assays. LNA ASOs targeting NP and VP24 were also shown to be effective in HeLa cells (Fig. 1).
LNA ASOs significantly reduce NPC1 mRNA and protein expression in human and mouse cells
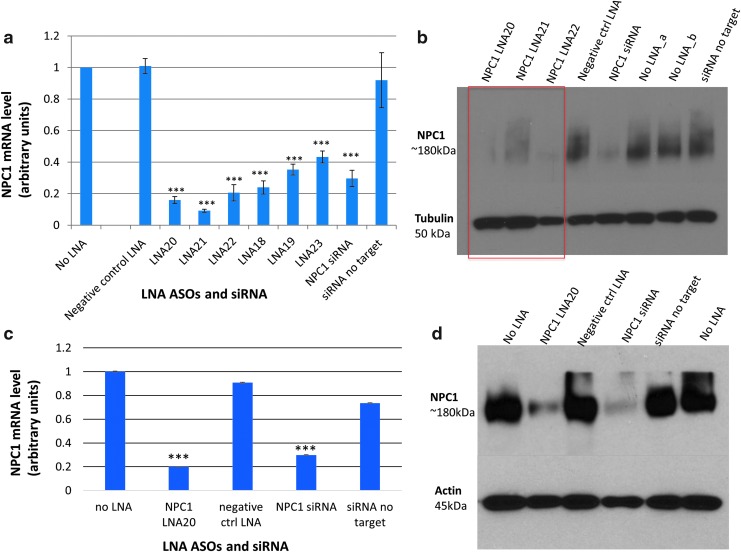
Six LNA ASOs targeting different sites in the NPC1 mRNA sequence were transfected into HepG2 and HeLa cells to screen for NPC1 knockdown (Supplementary Figs. S8 and S9). NPC1 siRNA was used as a positive control. Scrambled siRNA (labeled nontargeting siRNA) and LNA24 were included as negative controls. The potency of LNA ASOs in reducing NPC1 mRNA expression was evaluated by quantitative reverse transcription-polymerase chain reaction (qRT-PCR) at 50 and 100 nM LNA ASO concentration in both HeLa and HepG2 cells (Supplementary Figs. S8 and S9). Of the six LNA ASOs, three (LNA20, LNA21, and LNA22) showed the greatest (P < 0.001) reduction of NPC1 mRNA by qRT-PCR analysis (Fig. 3). Therefore, these three LNA ASOs were tested at 100 nM in HeLa cells to assess NPC1 protein knockdown. LNA20 treatment produced the greatest reduction in NPC1 protein levels 48 h after transfection (Fig. 3, Supplementary Fig. S10).
FIG. 3.
LNA ASOs targeting NPC1 reduce expression of NPC1 mRNA in HeLa cells. (a) Six LNA ASOs targeting NPC1 were transfected into HeLa cells and RNA harvested 48 h later to measure mRNA levels. The negative control LNA ASO, siRNAs targeting NPC1, and nontargeting siRNAs were included as controls. LNA20, LNA21, and LNA22 demonstrated greatest reduction in NPC1 mRNA levels. (b) LNA ASOs targeting NPC1 reduce NPC1 protein levels in HeLa cells. One hundred nanomolar of LNA ASOs targeting NPC1 (LNA20, 21, and 22) were transfected into HeLa cells, and protein harvested 48 h later and NPC1 levels measured by immunoblotting. Tubulin was used as loading control. (c) LNA20 targeting NPC1 reduces expression of NPC1 mRNA in mouse macrophages. NPC1 siRNA was used as positive control. The negative control LNA24 and nontargeting siRNA were used as negative controls. (d) LNA20 targeting Npc1 reduces Npc1 protein levels in mouse macrophages as assessed by immunoblotting. β-Tubulin was used as loading control. Npc1 siRNA was used as positive control for Npc1 knockdown. Negative control LNA ASO and nontargeting siRNA controls did not reduce Npc1 levels. ***P < 0.001. Experiments were performed in biological triplicates; each biological replicate was performed in technical triplicates. NPC1, Niemann-Pick C1.
LNA20 was further tested in mouse J774A.1 macrophages. This is pertinent to the overall goal of using LNA ASOs in an EVD setting given that one of the first cell types infected by the Ebola virus are macrophages. LNA20 was transfected at 100 nM in J774A.1 cells, and lysate harvested after 72 h to assess Npc1 reduction. Npc1 siRNA and nontargeting siRNA were used as controls, in addition to the negative control LNA24. LNA20 significantly (P < 0.001) knocked down Npc1 mRNA and protein levels in this mouse cell line as shown by qRT-PCR and western blot analysis (Fig. 3, Supplementary Fig. S10).
LNA ASO targeting NPC1 reduces VSVluc-Ebov GP in human cells
Since the NPC1 ASO LNA20 caused the greatest decrease in NPC1 mRNA and protein expression in both human and mouse cells, this LNA ASO was tested for its effects on virus infection. The NPC1 ASO LNA20 and the negative control LNA24 were transfected at a range of concentrations into HeLa cells and 48 h later challenged with VSVluc-EboV GP, a defective VSV encoding luciferase and pseudotyped with Ebola virus GP [22]. Transfection of NPC1 LNA20 was specifically correlated with markedly decreased VSVluc-EboV GP infection as measured by expression of virus-encoded luciferase activity (Fig. 4). In contrast, NPC1 LNA20 had no effect on infection by VSVluc pseudotyped with GP from Lassa fever virus (data not shown). The decrease in infection conferred by NPC1 LNA20 was comparable to the effect of NPC1-directed siRNA. To our knowledge, this represents the first successful use of ASOs to target the NPC1 host factor as a means to reduce or prevent Ebola virus GP-dependent viral infection of mammalian cells. Thus, in addition to viral targets, the host gene that encodes the essential Ebola virus host factor NPC1 is also a candidate target for therapeutic LNA ASOs.
FIG. 4.
VSVluc-EboV GP infection is reduced in the presence of LNA ASOs that inhibit NPC1 expression. NPC1 LNA20 and the negative control LNA24 were transfected into HeLa cells, which were then infected with a VSV vector encoding Ebola-GP (VSVluc-EboV GP) after 48 h. VSVluc-EboV GP luciferase levels were measured after 48 h. VSVluc-EboV GP luciferase levels decreased significantly in a dose-dependent manner after treatment with 25, 50, and 100 nM of NPC1-targeting LNA20. LNA20 data are presented relative to LNA24 (control LNA). NPC1 siRNA used as positive control for NPC1 knockdown also showed significant decrease in VSVluc-EboV GP luciferase levels. Negative control LNA ASO and nontargeting siRNA controls did not reduce VSVluc-EboV GP luciferase levels. IC50 for LNA20 was calculated and provided in Supplementary Table S2. **P < 0.01, ***P < 0.001. Experiments were performed in three biological replicates (triplicate). GP, glycoprotein; VSV, vesicular stomatitis virus.
Discussion
Although several therapeutic strategies have been attempted for EVD treatment [29,30], LNA ASOs offer distinct advantages due to properties that have been optimized over the years [35–39].
We are proposing a combinatorial therapeutic approach of using LNA ASOs to target key Ebola virus proteins NP and VP24, and host protein NPC1. We have shown that LNA ASOs can be used to decrease expression of the host factor NPC1 that is required to interact with Ebola virus GP, and demonstrated that LNA ASOs targeting NPC1 are effective for disrupting virus infection via GP. Inhibition of NPC1 as a means of preventing Ebola virus infection is a promising therapeutic approach. First, NPC1 has been documented to be required for filoviruses infection [19,48]. Moreover, small reductions in NPC1 can have a profound effect on the efficacy of Ebola virus infection, which is dependent on multivalent GP-NPC1 contacts. Thus, our approach of targeting NPC1 provides a powerful tool to bypass mutagenic adaptation of the Ebola virus, which represents a challenge for antibody-based therapeutic approaches that target Ebola virus GP [49–56].
We propose to also target multiple Ebola virus proteins with LNA ASOs as to increase therapeutic efficacy and decrease the emergence of viral escape mutations. Here, we have focused on Ebola virus NP as one of the key targets for therapeutics. We have demonstrated that we can use LNA ASOs to successfully inhibit NP expression as a means to prevent viral protein translation. Previous work has indicated that NP may not only be a suitable target for EVD treatment strategies, but also in other viruses within the same filovirus family. For example, recently a morpholino ASO targeting NP was shown to be protective against Marburg virus (MARV) in non-human primates [57]. This successful targeting of the MARV NP with morpholino ASOs provides strong support for the use of LNA ASOs to target not only Ebola virus NP, but also NPs of other viruses. However, morpholino ASOs lack some of the advantages of LNA ASOs, such as high affinity and specificity for target sequences. Another study reported the use of chemical ligands to disrupt the interaction of Ebola virus and MARV NPs with viral single-stranded RNA [45]. However, this approach is limited by the use of small molecules that lack the high affinity, specificity, and stability of LNA ASOs.
We have also targeted Ebola virus proteins such as VP24, which is essential for subverting the host immune response. The Ebola virus has evolved a decoy mechanism to counter the response mounted by the host immune system. Ebola virus VP24 along with VP40 inhibit interferon responses, facilitating virus replication that is disruptive to macrophages and dendritic cells [41]. Macrophages normally produce antiviral responses via innate immune system activation, whereas dendritic cells mount antiviral defense primarily via the adaptive immune system [41]. After Ebola virus infection the decoy activities of VP24 and VP40 render macrophages and dendritic cells unable to inhibit virus transmission to other cells despite their ability to trigger inflammation. This facilitates viral transmission to other tissues and organs, resulting in massive tissue injury and hemorrhage from Ebola virus replication, and frequent death of the host [14,15,41]. Given that many viruses act to disrupt the host immune response, a similar therapeutic strategy to the one reported here could possibly be approached with other viruses, using LNA ASOs designed to target the viral proteins responsible for interfering with the immune response. This would assist the host in mounting a proper immune response that can effectively counter the virus.
A combinatorial approach employing LNA ASOs may help to bypass the high rate of mutagenesis of the Ebola virus, and may overcome limitations inherent to other therapeutic strategies. For example, unlike vaccines and antibody-based approaches, LNA ASOs can be quickly designed based on available sequencing data of current viral strains and synthesized for rapid testing in model organisms and for clinical studies. LNA ASOs are also extremely stable and can be stored lyophilized at ambient temperatures and resuspended in saline solution for subcutaneous injection when needed. Moreover, LNA ASOs have high affinity for their targets and are highly stable in the circulation, with adequate pharmacokinetics/pharmacodynamics (PK/PD) and safety properties [58]. LNA ASOs have been used successfully in vivo from rodents to humans to inhibit microRNAs that function in diseases such as hepatitis C virus infection, cancers (eg, multiple myeloma), and cholesterol deregulation [58–65].
While our efforts have centered on targeting the Ebola virus, there are a number of additional RNA viruses that could be targeted using LNA ASOs, including other Ebola family viruses such as the Reston virus (RESTV), Tai Forest virus (TAFV), Sudan virus (SUDV), and Bundibogyo virus (BDBV/BEVD) [66]. Moreover, strains of the related filovirus MARV have caused considerable human death [66]. As further evidence of the broad applicability of targeting a host factor, Lassa virus, a negative strand RNA virus that also produces severe hemorrhagic fever like Ebola virus, encodes a GP that mediates virus entry via a host factor, the lysosome-associated membrane protein 1, allowing transport of its genome into the late endosomes [67–70]. GP binds to receptors on the cell surface facilitating transport of the viral particles to endosomes and fusion of the viral and host cell membranes. We propose that the successful inhibition of NPC1 in our Ebola virus model could be expanded to inhibit host receptors of other viruses as a means to prevent GP-mediated virus entry. Additionally, a new filovirus (Lloviu) was recently discovered in Spain, a country with no documented history of filovirus infections [71]. Lloviu is genetically different from Ebola virus and MARV, highlighting once again the need for new therapeutic strategies that are versatile enough for the sporadic occurrence of new viruses. LNA ASO-based antiviral strategies may thus have broad applicability.
Supplementary Material
Acknowledgments
This work was supported by MGH Institutional support to A.M.N. (MGH Research Scholar Award). S.K. and A.P. were supported by the Lundbeck Foundation and the Novo Nordisk Foundation.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Moghadam SRJ, Omidi N, Bayrami S, Moghadam SJ. and Seyed Alinaghi S. (2015). Ebola viral disease: a review literature. Asian Pac J Trop Biomed 5:260–267 [Google Scholar]
- 2.Billioux BJ, Smith B. and Nath A. (2016). Neurological complications of Ebola virus infection. Neurotherapeutics 13:461–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clark DV, Kibuuka H, Millard M, Wakabi S, Lukwago L, Taylor A, Eller MA, Eller LA, Michael NL, et al. (2015). Long-term sequelae after Ebola virus disease in Bundibugyo, Uganda: a retrospective cohort study. Lancet Infect Dis 15:905–912 [DOI] [PubMed] [Google Scholar]
- 4.Mate SE, Kugelman JR, Nyenswah TG, Ladner JT, Wiley MR, Cordier-Lassalle T, Christie A, Schroth GP, Gross SM, et al. (2015). Molecular evidence of sexual transmission of Ebola virus. N Engl J Med 373:2448–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deen GF, Knust B, Broutet N, Sesay FR, Formenty P, Ross C, Thorson AE, Massaquoi TA, Marrinan JE. (2015). Ebola RNA persistence in semen of Ebola virus disease survivors — preliminary report. N Engl J Med [Epub ahead of print]; DOI: 10.1056/ME/Moa1511410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Green E, Hunt L, Ross JCG, Nissen NM, Curran T, Badhan A, Sutherland KA, Richards J, Lee JS, et al. (2016). Viraemia and Ebola virus secretion in survivors of Ebola virus disease in Sierra Leone: a cross-sectional cohort study. Lancet Infect Dis 3099:3–7 [DOI] [PubMed] [Google Scholar]
- 7.Fischer RJ, Judson S, Miazgowicz K, Bushmaker T. and Munster VJ. (2016). Ebola virus persistence in semen ex vivo. Emerg Infect Dis 22:289–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sow MS, Etard J-F, Baize S, Magassouba N, Faye O, Msellati P, Touré AI, Savane I, Barry M, Delaporte and E. Postebogui Study Group. (2016). New evidence of long-lasting persistence of Ebola virus genetic material in semen of survivors. J Infect Dis 214:1475–1476 [DOI] [PubMed] [Google Scholar]
- 9.Jacobs M, Rodger A, Bell DJ, Bhagani S, Cropley I, Filipe A, Gifford RJ, Hopkins S, Hughes J, et al. (2016). Late Ebola virus relapse causing meningoencephalitis: a case report. Lancet 6736:1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.WHO. (2016). Interim Advice on the Sexual Transmission of the Ebola Virus Disease. WHO, Geneva, Switzerland [Google Scholar]
- 11.de La Vega M-A, Wong G, Kobinger GP, and Qiu X. (2015). The multiple roles of sGP in Ebola pathogenesis. Viral Immunol 28:3–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mehedi M, Falzarano D, Seebach J, Hu X, Carpenter MS, Schnittler H-J. and Feldmann H. (2011). A new Ebola virus nonstructural glycoprotein expressed through RNA editing. J Virol 85:5406–5414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biedenkopf N, Lier C. and Becker S. (2016). Dynamic phosphorylation of VP30 is essential for Ebola virus life cycle. J Virol 90:4914–4925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prescott JB, Marzi A, Safronetz D, Robertson SJ, Feldmann H. and Best SM. (2017). Immunobiology of Ebola and Lassa virus infections. Nat Rev Immunol 17:195–207 [DOI] [PubMed] [Google Scholar]
- 15.Misasi J. and Sullivan NJ. (2014). Camouflage and misdirection: the full-on assault of Ebola virus disease. Cell 159:477–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falasca L, Agrati C, Petrosillo N, Di Caro A, Capobianchi MR, Ippolito G. and Piacentini M. (2015). Molecular mechanisms of Ebola virus pathogenesis: focus on cell death. Cell Death Differ 22:1250–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee K, Ren T, Coîté M, Gholamreza B, Misasi J, Bruchez A. and Cunningham J. (2013). Inhibition of Ebola virus infection: identification of Niemann-Pick C1 as the target by optimization of a chemical probe. ACS Med Chem Lett 4:239–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Côté M, Misasi J, Ren T, Bruchez A, Lee K, Filone CM, Hensley L, Li Q, Ory D, Chandran K. and Cunningham J. (2011). Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature 477:344–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White JM. and Schornberg KL. (2012). A new player in the puzzle of filovirus entry. Nat Rev Microbiol 10:317–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garver WS, Krishnan K, Gallagos JR, Michikawa M, Francis GA. and Heidenreich RA. (2002). Niemann-Pick C1 protein regulates cholesterol transport to the trans-Golgi network and plasma membrane caveolae. J Lipid Res 43:579–589 [PubMed] [Google Scholar]
- 21.Carette JE, Raaben M, Wong AC, Herbert AS, Obernosterer G, Mulherkar N, Kuehne AI, Kranzusch PJ, Griffin AM, et al. (2011). Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 477:340–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller EH, Obernosterer G, Raaben M, Herbert AS, Deffieu MS, Krishnan A, Ndungo E, Sandesara RG, Carette JE, et al. (2012). Ebola virus entry requires the host-programmed recognition of an intracellular receptor. EMBO J 31:1947–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang H, Shi Y, Song J, Qi J, Lu G, Yan J. and Gao GF. (2016). Ebola viral glycoprotein bound to its endosomal receptor Niemann-Pick C1. Cell 164:258–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ng M, Ndungo E, Kaczmarek ME, Herbert AS, Binger T, Kuehne AI, Jangra RK, Hawkins JA, Gifford RJ, et al. (2015). Filovirus receptor NPC1 contributes to species-specific patterns of ebolavirus susceptibility in bats. ELife 4:pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johansen LM, Dewald LE, Shoemaker CJ, Hoffstrom BG, Lear-rooney CM, Stossel A, Nelson E, Delos SE, Simmons JA, et al. (2015). A screen of approved drugs and molecular probes identifies therapeutics with anti-Ebola virus activity. Sci Transl Med 7:1–14 [DOI] [PubMed] [Google Scholar]
- 26.Aleksandrowicz P, Marzi A, Biedenkopf N, Beimforde N, Becker S, Hoenen T, Feldmann H. and Schnittler HJ. (2011). Ebola virus enters host cells by macropinocytosis and clathrin-mediated endocytosis. J Infect Dis 204:S957–S967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mulherkar N, Raaben M, de la Torre JC, Whelan SP. and Chandran K. (2011). The Ebola virus glycoprotein mediates entry via a non-classical dynamin-dependent macropinocytic pathway. Virology 419:72–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nanbo A, Imai M, Watanabe S, Noda T, Takahashi K, Neumann G, Halfmann P. and Kawaoka Y. (2010). Ebolavirus is internalized into host cells via macropinocytosis in a viral glycoprotein-dependent manner. PLoS Pathog 6:e1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cardile AP, Warren TK, Martins KA, Reisler RB. and Bavari S. (2016). Will there be a cure for Ebola? Annu Rev Pharmacol Toxicol 57:329–348 [DOI] [PubMed] [Google Scholar]
- 30.Mendoza EJ, Qiu X. and Kobinger GP. (2016). Progression of Ebola therapeutics during the 2014–2015 outbreak. Trends Mol Med 22:164–173 [DOI] [PubMed] [Google Scholar]
- 31.Qiu X, Wong G, Audet J, Bello A, Fernando L, Alimonti JB, Fausther-Bovendo H, Wei H, Aviles J, et al. (2014). Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature 514:47–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geisbert TW, Lee AC, Robbins M, Geisbert JB, Honko AN, Sood V, Johnson JC, de Jong S, Tavakoli I, et al. (2010). Postexposure protection of non-human primates against a lethal Ebola virus challenge with RNA interference: a proof-of-concept study. Lancet 375:1896–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iversen PL, Warren TK, Wells JB, Garza NL, Mourich DV, Welch LS, Panchal RG. and Bavari S. (2012). Discovery and early development of AVI-7537 and AVI-7288 for the treatment of Ebola virus and Marburg virus infections. Viruses 4:2806–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oestereich L, Lüdtke A, Wurr S, Rieger T, Muñoz-Fontela C. and Günther S. (2014). Successful treatment of advanced Ebola virus infection with T-705 (favipiravir) in a small animal model. Antiviral Res 105:17–21 [DOI] [PubMed] [Google Scholar]
- 35.Sharma VK, Sharma RK. and Singh SK. (2014). Antisense oligonucleotides: modifications and clinical trials. Med Chem Commun 5:1454–1471 [Google Scholar]
- 36.McClorey G. and Wood MJ. (2015). An overview of the clinical application of antisense oligonucleotides for RNA-targeting therapies. Curr Opin Pharmacol 24:52–58 [DOI] [PubMed] [Google Scholar]
- 37.Lundin KE, Højland T, Hansen BR, Persson R, Bramsen JB, Kjems J, Koch T, Wengel J. and Smith CIE. (2013). Biological activity and biotechnological aspects of locked nucleic acids. Adv Genet 82:47–107 [DOI] [PubMed] [Google Scholar]
- 38.Yu RZ, Grundy JS. and Geary RS. (2013). Clinical pharmacokinetics of second generation antisense oligonucleotides. Expert Opin Drug Metab Toxicol 9:169–182 [DOI] [PubMed] [Google Scholar]
- 39.Geary RS, Norris D, Yu R. and Bennett CF. (2015). Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv Drug Deliv Rev 87:46–51 [DOI] [PubMed] [Google Scholar]
- 40.Feldmann H. and Geisbert TW. (2011). Ebola haemorrhagic fever. Lancet 377:849–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bray M. and Geisbert TW. (2005). Ebola virus: the role of macrophages and dendritic cells in the pathogenesis of Ebola hemorrhagic fever. Int J Biochem Cell Biol 37:1560–1566 [DOI] [PubMed] [Google Scholar]
- 42.Gire SK, Goba A, Andersen KG, Sealfon RSG, Park DJ, Kanneh L, Jalloh S, Momoh M, Fullah M, et al. (2014). Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science 345:1369–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neumann G, Watanabe S. and Kawaoka Y. (2009). Characterization of Ebola virus regulatory genomic regions. Virus Res 144:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shabman RS, Hoenen T, Groseth A, Jabado O, Binning JM, Amarasinghe GK, Feldmann H. and Basler CF. (2013). An upstream open reading frame modulates ebola virus polymerase translation and virus replication. PLoS Pathog 9:e1003147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu X, Wang Z, Li L, Dong S, Li Z, Jiang Z, Wang Y, Shui W. and Heymann DL. (2016). Novel chemical ligands to Ebola virus and Marburg virus nucleoproteins identified by combining affinity mass spectrometry and metabolomics approaches. Sci Rep 6:29680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pappalardo M, Juliá M, Howard MJ, Rossman JS, Michaelis M. and Wass MN. (2016). Conserved differences in protein sequence determine the human pathogenicity of Ebola viruses. Sci Rep 6:23743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoenen T, Watt A, Mora A. and Feldmann H. (2014). Modeling the lifecycle of Ebola virus under biosafety level 2 conditions with virus-like particles containing tetracistronic minigenomes. J Vis Exp 52381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ng M, Ndungo E, Jangra RK, Cai Y, Postnikova E, Radoshitzky SR, Dye JM, Ramírez de Arellano E, Negredo A, et al. (2014). Cell entry by a novel European filovirus requires host endosomal cysteine proteases and Niemann-Pick C1. Virology 468:637–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Q, Gui M, Niu X, He S, Wang R, Feng Y, Kroeker A, Zuo Y, Wang H, et al. (2016). Potent neutralizing monoclonal antibodies against Ebola virus infection. Sci Rep 6:25856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takada A, Ebihara H, Jones S, Feldmann H. and Kawaoka Y. (2007). Protective efficacy of neutralizing antibodies against Ebola virus infection. Vaccine 25:993–999 [DOI] [PubMed] [Google Scholar]
- 51.Corti D, Misasi J, Mulangu S, Stanley DA, Kanekiyo M, Wollen S, Ploquin A, Doria-Rose NA, Staupe RP, et al. (2016). Protective monotherapy against lethal Ebola virus infection by a potently neutralizing antibody. Science 351:1339–1342 [DOI] [PubMed] [Google Scholar]
- 52.Bornholdt ZA, Turner HL, Murin CD, Li W, Sok D, Souders CA, Piper AE, Goff A, Shamblin JD, et al. (2016). Isolation of potent neutralizing antibodies from a survivor of the 2014 Ebola virus outbreak. Science 351:1078–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qiu X, Audet J, Wong G, Pillet S, Bello A, Cabral T, Strong JE, Plummer F, Corbett CR, Alimonti JB. and Kobinger GP. (2012). Successful treatment of Ebola virus-infected cynomolgus macaques with monoclonal antibodies. Sci Transl Med 4:138ra81. [DOI] [PubMed] [Google Scholar]
- 54.Oswald WB, Geisbert TW, Davis KJ, Geisbert JB, Sullivan NJ, Jahrling PB, Parren PWHI. and Burton DR. (2007). Neutralizing antibody fails to impact the course of Ebola virus infection in monkeys. PLoS Pathog 3:62–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reynard O. and Volchkov VE. (2015). Characterization of a novel neutralizing monoclonal antibody against ebola virus GP. J Infect Dis 212(Suppl):S372–S378 [DOI] [PubMed] [Google Scholar]
- 56.Flyak AI, Shen X, Murin CD, Turner HL, David JA, Fusco ML, Lampley R, Kose N, Ilinykh PA, et al. (2016). Cross-reactive and potent neutralizing antibody responses in human survivors of natural Ebola virus infection. Cell 164:392–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Warren TK, Whitehouse CA, Wells J, Welch L, Charleston JS, Heald A, Nichols DK, Mattix ME, Palacios G, et al. (2016). Delayed time-to-treatment of an antisense morpholino oligomer is effective against lethal Marburg Virus Infection in cynomolgus macaques. PLoS Negl Trop Dis 10:e0004456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, et al. (2013). Treatment of HCV infection by targeting microRNA. N Engl J Med 368:1685–1694 [DOI] [PubMed] [Google Scholar]
- 59.Di Martino MT, Gull A, Cantafio MEG, Altomare E, Amodio N, Leone E, Morelli E, Lio SG, Caracciolo D, et al. (2014). In vitro and in vivo activity of a novel locked nucleic acid (LNA)-inhibitor-miR-221 against multiple myeloma cells. PLoS One 9:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Elmén J, Lindow M, Schütz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjärn M, Hansen HF, et al. (2008). LNA-mediated microRNA silencing in non-human primates. Nature 452:896–899 [DOI] [PubMed] [Google Scholar]
- 61.Straarup EM, Fisker N, Hedtj M, Lindholm MW, Rosenbohm C, Aarup V, Hansen HF, Ørum H, Hansen JB. and Koch T.rn. (2010). Short locked nucleic acid antisense oligonucleotides potently reduce apolipoprotein B mRNA and serum cholesterol in mice and non-human primates. Nucleic Acids Res 38:7100–7111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S. and Orum H. (2010). Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 327:198–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Obad S, dos Santos CO, Petri A, Heidenblad M, Broom O, Ruse C, Fu C, Lindow M, Stenvang J, et al. (2011). Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet 43:371–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bianchini D, Omlin A, Pezaro C, Lorente D, Ferraldeschi R, Mukherji D, Crespo M, Figueiredo I, Miranda S, et al. (2013). First-in-human phase I study of EZN-4176, a locked nucleic acid antisense oligonucleotide to exon 4 of the androgen receptor mRNA in patients with castration-resistant prostate cancer. Br J Cancer 109:2579–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rottiers V, Obad S, Petri A, McGarrah R, Lindholm MW, Black JC, Sinha S, Goody RJ, Lawrence MS, et al. (2013). Pharmacological inhibition of a microRNA family in nonhuman primates by a seed-targeting 8-mer antimiR. Sci Transl Med 5:212ra162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Anthony SM. and Bradfute SB. (2015). Filoviruses: one of these things is (not) like the other. Viruses 7:5172–5190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thomas CJ, Shankar S, Casquilho-Gray HE, York J, Sprang SR. and Nunberg JH. (2012). Biochemical reconstitution of hemorrhagic-fever arenavirus envelope glycoprotein-mediated membrane fusion. PLoS One 7:e51114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hastie KM, Zandonatti MA, Kleinfelter LM, Heinrich ML, Rowland MM, Chandran K, Branco LM, Robinson JE, Garry RF. and Saphire EO. (2017). Structural basis for antibody-mediated neutralization of Lassa virus. Science 356:923–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cohen-Dvashi H, Cohen N, Israeli H. and Diskin R. (2015). Molecular mechanism for LAMP1 recognition by lassa virus. J Virol 89:7584–7592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Di Simone C, Zandonatti MA. and Buchmeier MJ. (1994). Acidic pH triggers LCMV membrane fusion activity and conformational change in the glycoprotein spike. Virology 198:455–465 [DOI] [PubMed] [Google Scholar]
- 71.Negredo A, Palacios G, Vázquez-Morón S, González F, Dopazo H, Molero F, Juste J, Quetglas J, Savji N, et al. (2011). Discovery of an ebolavirus-like filovirus in Europe. PLoS Pathog 7:e1002304. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.