Abstract
There are several plausible abiotic synthetic routes from prebiotic chemical materials to ribonucleotides and even short RNA oligomers. However, for refinement of the RNA World hypothesis to help explain the origins of life on the Earth, there needs to be a manner by which such oligomers can increase their length and expand their sequence diversity. Oligomers longer than at least 10–20 nucleotides would be needed for raw material for subsequent natural selection. Here, we explore spontaneous RNA–RNA recombination as a facile means by which such length and diversity enhancement could have been realized. Motivated by the discovery that RNA oligomers stored for long periods of time in the freezer expand their lengths, we systematically investigated RNA–RNA recombination processes. In addition to one known mechanism, we discovered at least three new mechanisms. In these, one RNA oligomer acts as a splint to catalyze the hybridization of two other oligomers and facilitates the attack of a 5′-OH, a 3′-OH, or a 2′-OH nucleophile of one oligomer onto a target atom of another. This leads to the displacement of one RNA fragment and the production of new recombinant oligomers. We show that this process can explain the spontaneous emergence of sequence complexity, both in vitro and in silico.
Keywords: RNA, oligomers, origins of life, recombination, ribozymes
INTRODUCTION
On the primordial Earth, during abiogenesis, informational polymers would have had to undergo some processes both to elongate and to enrich their sequence diversity. The complexity of living systems required a supply of polymers long enough to possess phenotypic function and diverse enough to provide raw material for natural selection. Prior to Darwinian evolution through mutation, amplification, and selection, there would have been a state of “prelife” in which chemical processes drove populational evolution without template-directed replication (Nowak and Ohtsuki 2008). Recombination, the swapping of large blocks of polymeric information through near energy-neutral reactions, offers a facile means to accomplish these goals (Lehman 2003, 2008; Lehman et al. 2011). In an RNA or RNA-like world, these reactions can occur via trans-esterification reactions, which can be cooperatively facilitated by other RNAs (Higgs and Lehman 2015).
Recombinogenic events involving RNA on the early Earth could have allowed for the disproportionation of short polymers (e.g., 2-mer + 2-mer → 3-mer + 1-mer, etc.) and consequent length increase and diversification, provided there was a source of polymers to begin with. Several theoretical and experimental results have given us plausible scenarios for the existence of oligomeric RNAs. For example, Orgel demonstrated the possibility of nonenzymatic template-directed production of ∼2–30-mers from activated nucleotides (Lohrmann et al. 1980; Wu and Orgel 1992). Later Ferris showed that montmorillonite clay could greatly enhance the lengths of such oligomers under environmental cycling conditions (Ferris et al. 1996). Abiotic polymerization could also be lipid assisted in wet-dry cycles (Rajamani et al. 2008; De Guzman et al. 2014). Recently Szostak's group has provided alternative mechanistic scenarios for oligomer production from activated monomers and trimers (Prywes et al. 2016). Furthermore, Krishnamurthy has showcased the use of diamidophosphate to polymerize RNA monomers into oligomers of at least 4 nucleotides (nt) long (Gibard et al. 2018), while Hud discovered that intercalating agents such as ethidium bromide can enhance 3-mer deoxynucleotide polymerization into much longer DNAs (Horowitz et al. 2010). Thus, the synthetic routes to short random RNAs seem abundant.
With oligomers as substrates, both ligation (requiring the use of an activated, or “high-energy”, leaving group) and recombination can take place to diversify the population (Briones et al. 2009). Intriguingly, a variety of RNA-directed recombination events have been detected under laboratory conditions (Chetverin et al. 1997; Chetverin 1999; Hayden et al. 2005; Lutay et al. 2007; Nechaev et al. 2009; Pino et al. 2013), but these have not been investigated in a systematic fashion. The propensity of randomly generated pools of RNA oligomers to possess an intrinsic ability to change their length and compositional distributions has been shown in a contemporaneous study, although the mechanisms require further investigation (Mutschler et al. 2018). The rationale for investigating this phenomenon experimentally has been heightened by a statistical-thermodynamic analysis that shows that, counterintuitively, entropic factors would be expected to drive an increase in polymer length through recombination (Blokhuis and Lacoste 2017).
Here, we investigate the response of both random and designed RNA populations to conditions under which RNA–RNA strand exchange can be promoted. Notably, we find that recombination is quite robust and facile in a range of environmental settings. We induce the production of novel sequences with potentially catalytic structures, and we show evidence that heretofore unknown mechanisms of spontaneous RNA recombination can operate efficiently. Computer simulations support these findings, and they depict a virile and dynamic prebiotic scenario in which catalytic function can bootstrap itself out of random chemistry. There may also exist complementary processes that include templated end-to-end ligation, but we will not focus on these, as the nature of the leaving group in such reactions would differ from those involved in recombination. Our work indicates that the spontaneous emergence of an information-rich RNA world may have been far more probable than previously believed.
RESULTS
Long-term incubations
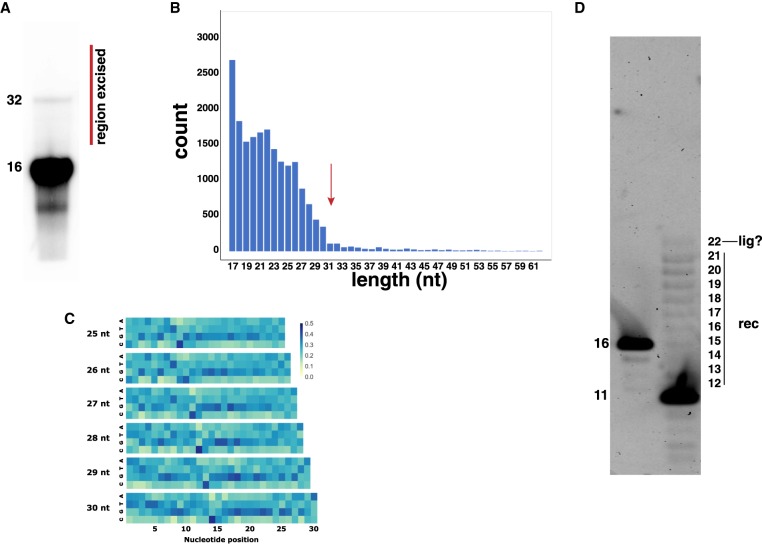
To begin, we examined the results of a long-term (2-yr) incubation of fully random RNA 16-mers at −15°C. When incubated under these conditions, with sporadic (approximately five times) thaw-and-refreeze cycles that transited the RNA in aqueous solution at −15°C to 28°C and back again at roughly 100 µM and in 0.1 mM Na2EDTA, we were curious as to whether any RNA at or near twice the input length (i.e., 32 nucleotides [nt]) could arise. This could occur via RNA–RNA recombination, and/or be consistent with an end-to-end ligation (Nechaev et al. 2009), even among oligomers that ostensibly contained no 2′,3′-cyclic phosphate (“<p”) activation on their 3′ ends (Pino et al. 2013). We thus ran the RNA on a 15% denaturing polyacrylamide gel, excised a region of this gel spanning the 32-mer product ± ca. 16 nt (Fig. 1A), and then subjected the resulting RNA to a high-throughput nucleotide sequence analysis. From 20,904 reads of RNAs of 17 nt in length or greater, we detected a distribution of oligomer lengths ranging from 17 to over 50 nt in length (Fig. 1B). As the bulk of these RNAs were 17–30 nt, with a sharp drop-off above the 31 nt value, and, because the initial pool contained RNAs with only 5′- and 3′-OH groups, these products must result from recombination, rather than ligation. A single RNA–RNA recombination among 16-mers could lead to RNAs 17–31 nt long (Lehman 2003). Among the products 25–30 nt long, we detected a significant enrichment of RNAs that would have contained a cytidine at a putative recombination junction. Likewise, there appears to be a paucity of the adenosine nucleotide at a putative recombination junction (Fig. 1C).
FIGURE 1.
Long-term incubation of RNA at −15°C leads to recombination. (A) Denaturing polyacrylamide gel electrophoresis of random 16-mers after 2 yr. A range of products spanning roughly 17–48 nt were excised from the gel and subjected to high-throughput sequence (HTS) analysis. A faint band at 32 nt can be seen, presumably from highly complementary duplex RNAs resistant to denaturation. (B) Distribution of sequence lengths determined by HTS analysis. Arrow denotes the dropoff in counts above one recombination event (16-mer + 16-mer → 31-mer + 1-mer). (C) Heat maps of nucleotide identities from HTS analyses of products in the 25–30 nt range. An enrichment in cytosine and a deficit of adenosine can be seen at the putative recombination junction, 16 nt from the 3′ terminus. (D) Comparison of fresh 16-mer LO1 (CUC UCC UUC CUG AAA A; pyrimidine-rich) RNA and 2-yr-old LO2 (GAG AGC AGG AA; purine-rich) RNA as visualized by SYBR Green staining after electrophoresis through a 15% denaturing polyacrylamide gel. Numbers refer to RNA lengths, as denoted in nucleotides. The extreme purine/pyrimidine bias of these oligomers explains their slight mobility difference.
By comparison, we were also able to examine the effects of long-term cold incubation on a specific oligomer sequence, the 11-mer LO2 (5′–GAG AGC AGG AA–3′). After 2 yr at −15°C with occasional (roughly 20 times) thaw and refreeze cycles akin to those described above, a series of distinctly sized products from 12 to 21 nt clearly appears, along with a fainter 22-mer product (Fig. 1D). Given that this product distribution is biased toward products up to 2N − 1 nt in length, these products are also the result of recombination rather than ligation; there is a product at approximately 22 nt, and it is unclear what ligation mechanism could be operative on RNAs with hydroxyl groups on their 5′ and 3′ termini.
Confirmation of known spontaneous RNA–RNA recombination
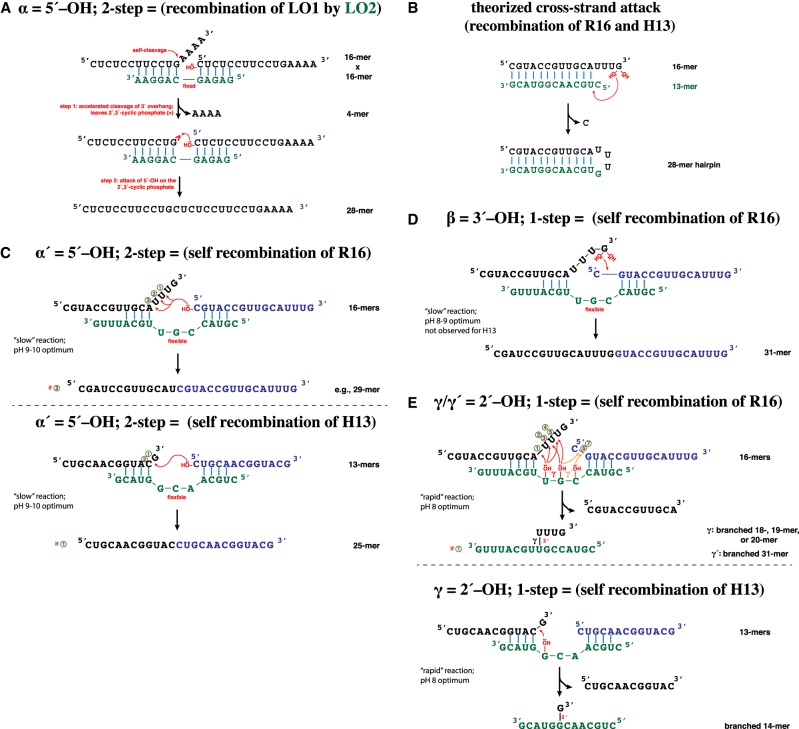
To investigate RNA recombination more directly, we then compared the spontaneous RNA–RNA recombination mechanisms proposed by Vlassov (Lutay et al. 2007; Nechaev et al. 2009) and Di Mauro (Pino et al. 2013). Using synthetic RNAs with specific sequences, we confirmed that the former works well in our hands, and we optimized the reaction conditions (Supplemental Fig. 1). This reaction requires a splint oligomer to pair simultaneously with two substrate oligomers, catalyzing a two-step recombination (Fig. 2A). In the first step, the rate of spontaneous self-cleavage of a 3′-overhang of one fragment is enhanced, followed by a second, slower step, in which the 5′-OH of the 5′-nucleotide on the other fragment attacks the resulting 2′,3′-cyclic phosphate. The cleavage/ligation events together produce a recombination of sequences in the substrates. Here, we will designate this as the α mechanism. Using a variety of rationally designed oligomers, including the sequences employed previously (Lutay et al. 2007), we were able to achieve a ∼2% product yield using pH 9.9 in a 3-d reaction at 22°C (Supplemental Fig. 1). Notably the inclusion of 5–100 mM MgCl2 in the reaction was greatly beneficial (Supplemental Fig. 2). We were unable to confirm the mechanism proposed by Di Mauro (Pino et al. 2013), which would be a one-step cross-strand attack of a 3′-OH (or 2′-OH) on a phosphor-ester center, with a nucleotide/polynucleotide leaving group to form a hairpin (Fig. 2B).
FIGURE 2.
Spontaneous RNA–RNA recombination schemes. (A) Two-step cleavage-ligation mechanism shown previously (Lutay et al. 2007) and confirmed herein and designated the α mechanism. In the first step, a splint (green) oligomer hybridizes to two substrate oligomers, and the overhanging portion of the left substrate (black) undergoes spontaneous cleavage to generate a 2′,3′-cyclic phosphate on the resulting 3′-nucleotide. In the second step, the 5′-OH of the right substrate (blue) attacks, resulting in a new phosphoester bond. (B) Theorized cross-strand attack proposed previously (Pino et al. 2013). The 16-mer R16 and 13-mer H13 were designed to test one arrangement of this reaction, but no reaction products have been detected in our hands (Supplemental Fig. 4). (C) A variant of the α mechanism in which a 3-nt bulge is created at the recombination junction when a ternary complex of either R16 (top) or H13 (bottom) forms. The recombination reaction shown is designated the α′ mechanism. (D) Another variant of recombination that could take place upon formation of an R16 ternary complex. Here, the 3′-OH (or possibly the 2′-OH) of one substrate strand displaces the terminal 5′-nucleotide of the other substrate strand. This reaction shown is designated the β mechanism. (E) Another detected recombination reaction that produces branched products. Upon ternary complex formation of either R16 or H13, attack of a 2′-OH on a hybridized strand at a variety of locations generates a suite of products that are branched and migrate slowly on a denaturing polyacrylamide gel. These reactions shown are designated the γ or γ′ mechanism, depending on the substrate.
New mechanisms of spontaneous RNA–RNA recombination
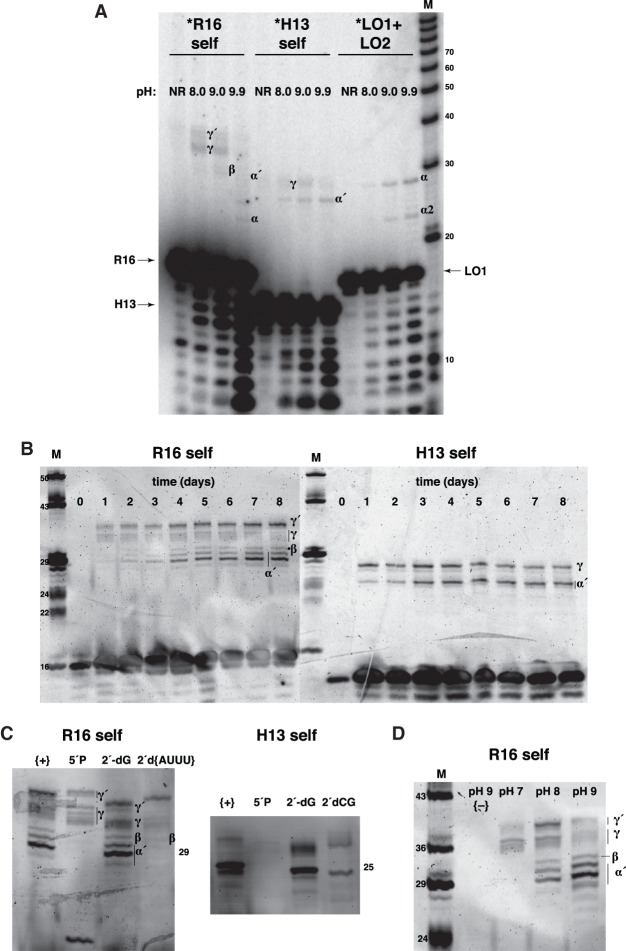
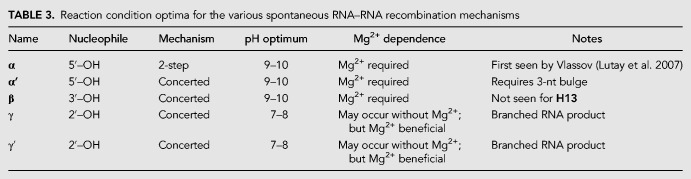
We did however detect three novel recombination mechanisms that are of great potential significance to the systems chemistry of a prebiotic soup (Fig. 2). The first is a variant of the previously reported α mechanism (Lutay et al. 2007). We designed two pairs of RNA oligomers that are partially complementary; the design was chosen to hinder the 2-step attack of the 5′-OH of one splinted oligomer on any <p of another splinted oligomer because of a lack of proximity, but allow for a putative cross-strand attack (Fig. 2B). The 3-d self-incubation of one oligomer (the 16-mer, R16) showed a trace of the α recombination to produce a 23-mer at pH 9.9 (Fig. 3A). The self-incubation of another oligomer (the 13-mer, H13) did not show evidence of this α recombination under the same conditions. However, both self-incubations gave other significant products (Fig. 3A). A time course showed the relative appearance of these products over 1–8 d at pH 8.0 (Fig. 3B). Both the ca. 29-mer products in the R16 self-incubation and the 25-mer product in the H13 self-recombination can be explained by a pairing topology that produces a 3-nt unpaired region on the splint oligomer (Fig. 2C). The resulting flexibility in the helical arrangement could lead to variability in the attack site of the 5′-OH, and we in fact see a range of product sizes from 28–30 nt with the R16 self-incubation, and two products at 24 and 25 nt with the H13 self-incubation (Fig. 3B). Given that the pH profile of this reaction is similar to that of the α reaction (Supplemental Fig. 3), we designated this reaction as α′ to reflect a similar mechanism as that of α but with a different geometry.
FIGURE 3.
Gel electrophoretograms of spontaneous RNA–RNA recombination. (A) Recombination of RNAs for 3 d at 22°C in 100 mM MgCl2 using 30 µM each with one 5′-32P-labeled oligomer (designated by *) on the gel. Product RNA was electrophoresed through a 15% denaturing polyacrylamide gel and visualized by phosphorimaging. Note that the 10-nt ladder (M, lengths in nt) has a 3′-phosphate, so it migrates ∼1 nt faster than the other RNA on the gel. (B) Several-day time course of 100 µM R16 (left) and H13 self-incubations at 0°/22°C in 100 mM MgCl2 at pH 8.0. Product RNA was electrophoresed through a 15% denaturing polyacrylamide gel and stained with SYBR Gold. The marker lane (M) was a mixture of 5′-OH-containing RNAs of known lengths. Proposed mechanisms that generate the product bands are indicated. (C) Effects of 5′ phosphorylation and inclusion of deoxy residues in R16 (left) and H13 (right). Phosphorylation inhibits the α′ (and β) mechanisms, but not the γ or γ′ mechanisms (at least in R16). Sequences of the deoxy-containing oligomers are as follows: R16 (2′-dG) = CGU ACC GUU GCA UUU dG; R16 (2′-d{AUUU}) = CGU ACC GUU GCdA dUdUdU G; H13 (2′-dG) = CUG CAA CGG UAC dG; H13 (2′-dG) = CUG CAA CGG UAdC G. {+} lanes are positive controls, with the all ribose oligomers with 5′-OH utilized. Product RNA was electrophoresed through a 15% denaturing polyacrylamide gel and stained with SYBR Gold. (D) Reaction products after a 5-d incubation of R16 at 0°C/22°C in 100 mM MgCl2 at various pH values. Negative control {−} was un-incubated R16 RNA. Product RNA was electrophoresed through a 15% denaturing polyacrylamide gel and stained with SYBR Gold.
We propose that the other two novel mechanisms are one-step (SN2-like) concerted mechanisms, which we will designate β and γ. A larger product can be seen in the R16 self-incubation at ∼31 nt. We posit that this new product is the result of the attack of the 3′–OH of the terminal nucleotide of R16 on the first phosphor-ester bond of the downstream splinted oligomer (Fig. 2D). As there is no cyclic phosphate involved in this reaction, this (β) mechanism, if confirmed, would be of a concerted (1-step) type, akin to the reaction catalyzed by group I (or group II) self-splicing introns. Use of an R16 oligomer with a 2′-deoxyguanosine residing on its 3′ terminus still produces the 31-mer β product, thus we conclude that the nucleophile is the 3′-OH rather than the 2′-OH (Fig 3C). Note that this mechanism is not the same as that suggested by Di Mauro (Pino et al. 2013). On the one hand, the β mechanism is strongly Mg2+-dependent (Supplemental Fig. 2), and on the other hand the geometry of the catalytic complex in the β mechanism requires a splinting of two oligomers with a 3-nt bulge (as in the α′ mechanism). Di Mauro proposed a mechanism that occurs in pure water and is a cross-strand attack to produce a hairpin (Pino et al. 2013). Our reaction between equimolar R16 and H13, which should allow for this type of reaction, did not show any such products (Supplemental Fig. 4); we conclude that this cross-strand attack mechanism is improbable.
The final product type we detected appears relatively rapidly (ca. 1 d) when R16 or H13 are self-incubated at high (ca. 10–100 µM) concentrations. We designate these products as the result of the γ mechanism. The apparent size (based on migration rate) of this product exceeds twice the input oligomer size (Fig. 3B), implying at first glance that it could not be either an end-to-end ligation product or a single-step recombination product. However, because there are no products of higher molecular mass that can be observed within the few apparent half-times of the reaction, a recursive mechanism seemed unlikely. Thus, we pursued the possibility that these are branched RNAs, resulting from an attack of an internal 2′-OH of one oligomer on a phosphate linkage of another, in a fashion similar to the intermediate that is formed during group II intron- or spliceosomal-directed splicing. In fact, we detected two general products in the 34–38-nt size range on the R16 self-incubation gel, and we designated the upper band(s) as γ′, to allow for the possibility of different target phosphates. Upon further analyses we conclude that there are approximately seven γ-like products (Fig. 3D), five (γ) of which are the result of the attack of the 2′-OH of G7 or U8 of the splint strand on the 3′-overhang of the left substrate, giving 19–21-nt branched products. The remaining two (γ′) could be the result of the attack of the 2′-OH of C6 or G7 of the splint strand on the 5′-overhang of the right substrate, giving 31-nt branched products (Fig. 2E). We were unable to obtain any evidence for the products being circular RNAs.
Nucleotide-sequence analyses
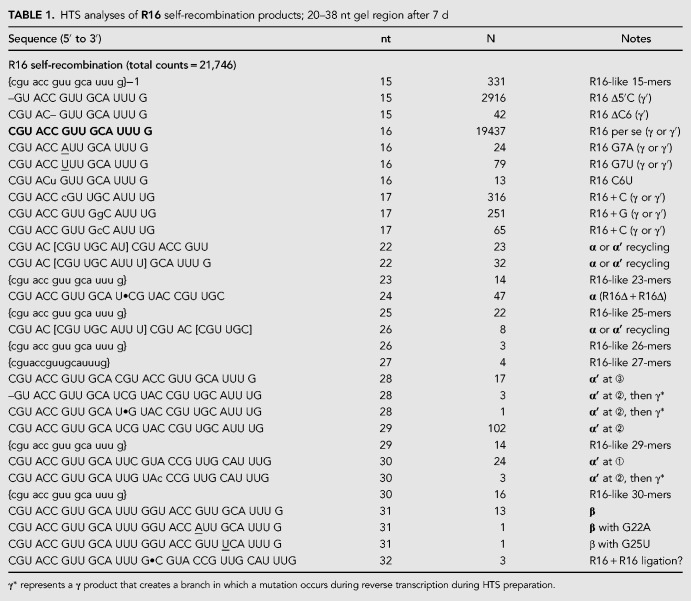
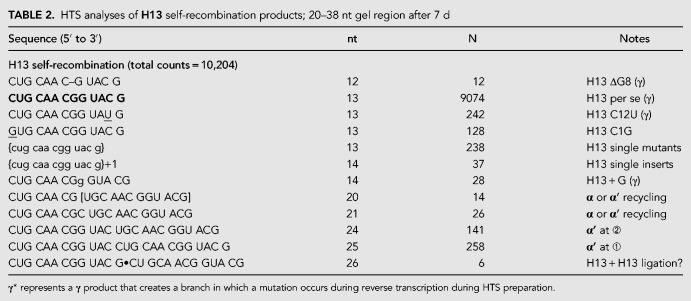
We excised the R16 and H13 self-incubation products from a 15% denaturing polyacrylamide gel, taking extreme care to only cut products in the size range from 20–38 nt, i.e., those that could be products of a single recombination but are not starting material. High-throughput nucleotide-sequence (HTS) analyses of these RNAs (Tables 1, 2) led to us to confirm the existence of the α′ products with high certainty for both the self-incubations. From 21,746 reads resulting from ≥15-mers from the R16 self-incubation after seven days, 17, 102, and 24 products had the expected sequences that would result from an α′ recombination at the ➂, ➁, and ➀ junctions, respectively (Fig. 2C). Though these are small numbers, they do mirror the observation that the 29-mer product (i.e., attack at R16 junction ➁) is the most intense band in this region of the gel (Fig. 4). Likewise, from 10,204 reads resulting from the H13 self-incubation, 141 and 258 products had the expected sequences that would result from an α′ recombination at the ➁ and ➀ junctions, respectively (Table 2; Fig. 2C). Again, these values parallel the relative band intensities, with the 25-mer (i.e., attack at H13 junction ➀) being favored. We note that the low yields of these reactions, even after 1 wk (∼1%–2% conversion at most of starting material into product), coupled with the need to gel purify the products and subject them to HTS preparation, including reverse transcription, led to far fewer HTS hits than one would expect for the sequencing of genomic DNA or cellular RNAs (tens of millions). Nevertheless, both the R16 and H13 self-incubations gave many products that can be explained if expected α′ products were then subsequently recombined with a γ mechanism (see Discussion). We designate these as “recycling” reactions that are evidence of more than one recombination event occurring in the time frame of the experiment.
TABLE 1.
HTS analyses of R16 self-recombination products; 20–38 nt gel region after 7 d
TABLE 2.
HTS analyses of H13 self-recombination products; 20–38 nt gel region after 7 d
FIGURE 4.
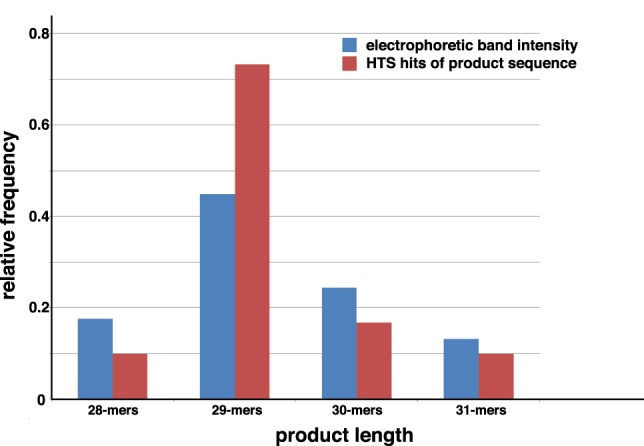
Comparison of relative electrophoretic band intensity and HTS frequency for R16 self-incubation products. (Blue bars) Relative band intensities for 28-, 29-, and 30-mers (α′ products), along with 31-mers (β products) from the image in Figure 3B at 7 d. (Red bars) Relative number of HTS hits from Table 1. Values scaled to sum equals 1.0.
The HTS analyses also gave support, albeit weaker, for the β mechanism, which was clearly fainter on the electrophoretic images (Figs. 3D, 4). Only 13 examples of the expected β RNA sequence were detected among the R16 gel extracts, plus two sequences that contained point mutations in the downstream oligomer (Table 1). A β-derived RNA sequence was not detected among the H13 gel extracts. Nonetheless this mechanism remains plausible for R16; it is a 31-mer product whose existence from any α′ mechanism was not detected (Table 1).
However, by far the most dominant sequences that we detected were ones that were either R16 (or H13) exactly, or were close variants thereof, despite being excised from a gel region that excluded these sizes (Tables 1, 2). Approximately 89% of products from each reaction fit this pattern. While many of these could be the result of fragmentation during the preparation for the HTS analysis, or from contamination of the starting material during gel purification (despite the many countermeasures employed), we propose that a more likely explanation for their abundance is the tendency of RNase-H deficient reverse transcriptases such as Superscript IV (which we used to obtain the data in Tables 1, 2) to generate both truncated products at a 2′–5′–3′ branched junction and full-length cDNA upon reading through the 3′,5′ side of such branches (Döring and Hurek 2017). This would result in the fragments we observed. Moreover, some RNase H-deficient M-MLV reverse transcriptases frequently cause mutations and/or deletions around the branch point (Döring and Hurek 2017), which is consistent with our detection of the slight variants of R16 and H13 (Tables 1, 2). The observation that the γ/γ′ products appear prior to any other product in both self-incubations (Fig. 3B) helps to explain the dominance of these fragments in the HTS analyses. In the same vein, several α′ sequences exhibit single-nucleotide insertions or deletions, which could be the result of reverse transcriptase stalling at a 2′–5′ linkage (Lorsch et al. 1995), which is the dominant product from the α mechanism (Lutay et al. 2007).
Mechanistic data
We sought additional confirmation of the proposed mechanisms. Prior phosphorylation of the 5′ nucleotide shuts down the α′ mechanism as expected; the 5′-OH nucleophile is removed (Fig. 3C). Interestingly, 5′ phosphorylation also suppresses the β mechanism, possibly through electrostatic interference of the 5′-nucleotide during attack of the 3′-OH (Fig. 3C). However, as expected, 5′ phosphorylation does not inhibit the production of the γ/γ′ products (Fig. 3C). Use of a 2′deoxy residue at the cytosine that is the target of the H13 α′ reaction at junction ➁ completely inhibits reaction at this junction, while permitting reaction at junction ➀ (cf. Fig. 2C); this would a be consequence of the inability to form the 2′,3′-cyclic phosphate intermediate during the spontaneous cleavage step of this mechanism. Note that when the 3′-terminal guanosine in H13 was converted to a 2′ deoxyG, recombination at neither junction was inhibited. We obtained similar data for the R16 self-incubation (Fig. 3C); in fact, with this deoxy construct, a band remains at 31 nt, providing support for the β mechanism. Lastly, should the canonical α mechanism operate weakly or the newly discovered α′ mechanism operate in the R16 or H13 self-incubations, as it does for the LO2-directed recombination of LO1 (Fig. 3A), recycling products (α2) should be seen if the hydrolyzed left substrate were to be released prior to ligation and instead move to become a right substrate. We see both electrophoretic and HTS evidence of this process occurring occasionally (Tables 1, 2; Fig. 3A). This portends the ability of multiple rounds of recombination to produce a wide variety of RNA sequences from a limited starting pool. After investigating the optimal reaction conditions for each product, we were able to summarize the differences among the various mechanisms (Table 3).
TABLE 3.
Reaction condition optima for the various spontaneous RNA–RNA recombination mechanisms
In silico simulations
Finally, to address the possibility of complex structure and function arising spontaneously out of random sequence pools via recombination, we constructed a sequence-explicit computer simulation of an RNA population undergoing iterative α recombination. The model (see Materials and Methods) starts with an abiotic polymerization of monomers to create short oligomers (i.e., ≤15-mers), which can then undergo iterative cycles of hybridization, recombination, degradation by hydrolysis, and dissociation. Herein, we examined only how the length distribution could change after many generations of the α recombination being the only chemical process (Fig. 5). Beginning with a random length distribution that is negatively exponentially populated as a function of length, after many generations the length distribution became skewed toward longer RNAs, as predicted from an entropic viewpoint (Blokhuis and Lacoste 2017). After 2 billion cycles, RNAs as long as 61 nt can be observed (Fig. 5). Future work will be to construct a series of computer simulations to examine the fate of RNA populations under the influence of all the various recombination mechanisms, and to add functional feedback mechanisms (e.g., catalysis). However, with even only the α recombination mechanism in operation, it is clear that RNAs in the size range of known ribozymes can be generated from random pools of short oligomers that are prebiotically plausible.
FIGURE 5.
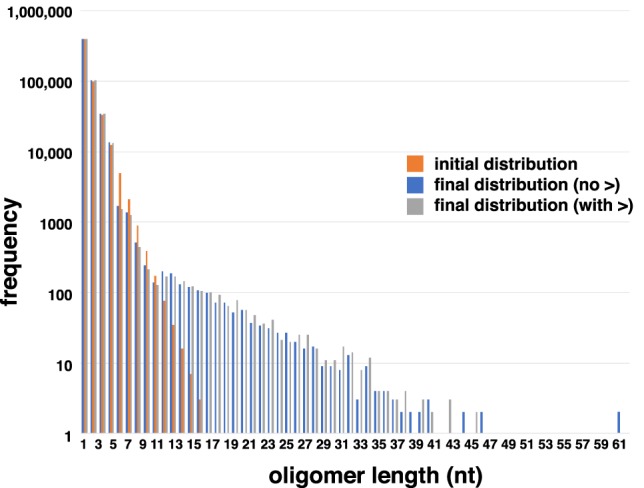
Nucleotide-explicit computer simulations of iterative cycles of recombination on a random pool of RNA oligomers. Frequency distributions of initial pool of random oligomers 1–15 nt (orange) was compared to final distributions after 2 billion cycles of iterated recombination emulating the α mechanism from initial pools that were not (blue) or were (gray) preactivated with 2′,3′ cyclic phosphates (>).
DISCUSSION
We have demonstrated that spontaneous RNA–RNA recombination should allow short RNA oligomers to rapidly increase their length and sequence diversity. The existence of the α′, β, and γ/γ′ mechanisms has the consequence that a far wider range of RNAs in a random pool should be able to spontaneously recombine than by the α mechanism alone. Note that in all cases, the splint oligomer would be behaving as a true enzyme, because it satisfies the criteria for such a substance: It speeds up the rate of the reaction (by lowering the activation energy barrier of the transition state by bringing the substrates in proximity), it is not consumed in the reaction, and it is specific to substrates and products (via nucleotide pairing). In the case of the new mechanisms described here (α′, β, and γ/γ′), an additional phenomenon exists in that the enzyme is identical to its substrates. This is distinct from self-cleavage, as in the hammerhead ribozyme, and from autocatalysis, where the enzyme is identical to one of its products. But it is analogous to trans autophosphorylation, where one protein (or DNazyme or ribozyme) kinase can phosphorylate another, identical kinase molecule.
Should random RNAs of these lengths (ca. 10–16 nt) be abiotically available, they could have been the first biological enzymes. The low yields of the α mechanism are in part a consequence of poor product release, because of the large numbers of consecutive nucleotide pairs involved (cf. Fig. 2A). Yet in the case of the α′ mechanism, the formation of a 3-nt bulge in the product promotes product release, as this bulge should not be thermodynamically favorable (Mathews et al. 1999). There are about 7 million possible 16-mer sequences that could form α′ geometries such as R16 shown in Figure 2C by satisfying the following criteria:
where N1 is anything, A is complementary to D, B is complementary to C, N1 is not complementary to N2, N2 is not complementary to N7, N3 is not complementary to N6, N4 is not complementary to N5, W is complementary to Z, X is complementary to Y, and N8 is anything.
These 7 million self-recombining sequences are out of about 416 = 4.3 billion possible 16-mers; this represents 0.16% of all sequences in 16-mer sequence space, or about a 1/600 chance that a single RNA could satisfy the criteria needed to form such a catalytic complex. Although further study is needed, it is likely that a 3-nt gap alone possesses the geometry needed for this reaction. We tested B16, a 16-mer (5′-CAG CUU AGU CCG GUU C-3′) that would give a 4-nt bulge, but it did not give an α′ product, though it did produce a weak γ-like product (Supplemental Fig. 5).
In the case of the α mechanism, the original discovery (Lutay et al. 2007) provided strong evidence that the nascent bond formed was primarily (ca. 95%) a 2′–5′ linkage, rather than a 3′–5′ linkage. To test this for the case of our newly discovered reaction variant, the α′ mechanism, we assayed the relative lability of the new bond to alkaline treatment (Supplemental Fig. 6). A 2′–5′ bond has been shown to be more susceptible to spontaneous cleavage (Lutay et al. 2006). While the experimental methods we used (1–2 min hot NaOH digestion of the isolated reaction products from LO1 × LO2 compared to those of R16 or H13 self-incubation) did support a predominance of a 2′–5′ linkage for the α mechanism, there was less such evidence for the α′ mechanism products, suggesting that the 3-nt bulge creates a geometry that favors a 3′–5′ linkage. One implication of this finding is that the products, being mostly 3′–5′ linked, would be a homogenous genetic population capable of open-ended evolution.
Cold cycling with occasional or sporadic thaws has been suggested as a means to promote several types of RNA chemistries (Vlassov et al. 2005; Yarus 2012; Mutschler et al. 2015). Presumably the cold temperatures and/or eutectic phases favor the formation of otherwise weak oligomer duplexes and facilitate in-line attack by limiting conformational flexibility. And the warming phases of such cycles would promote product release and lower thermally dependent activation energy barriers. Yet these specific effects on oligomer recombination have yet to be investigated systematically.
It is tempting to draw parallels between the simple RNA-oligomer-catalyzed mechanisms described herein and later, evolved catalytic activity. The most primitive of the recombination mechanisms is the α or α′ 2-step reaction. Vlassov and colleagues first recognized that the α reaction could be a form of simple RNA–RNA recombination (Lutay et al. 2005, 2007; Nechaev et al. 2009; Staroseletz et al. 2018), yet they have not mentioned that the splint oligomer behaves as a true enzyme. The net effect of this process is indeed a recombination, although the 2-step mechanism involves a facilitated site-specific cleavage, followed by a ligation promoted by the existence of a <p moiety. Given that hydrolysis of RNA would be occurring continuously and more-or-less independently of sequence in short prebiotic oligomers, the templated ligation of such fragments would be the most accessible form of sequence length enhancement available to the RNA world. The β mechanism, with the attack of a 3′-OH on a phosphor-ester center positioned nearby, would represent the next step in complexity. This mechanism awaits more formal confirmation, but its concerted nature would allow for more control over the specificity of the substrates involved, and it is the apparent precursor to group I intron self-splicing. The γ mechanism would perhaps be the least useful in propagating selectable oligomeric information, as a consequence of its production of branched products, but it does serve as the obvious precursor to group II and spliceosomal RNA splicing, and thus serves as a mechanism that biology could refine and exploit later for intronic shuffling. Moreover, even branched products can potentially serve as catalysts for further recombination of linear substrates.
Our results suggest that not only can recombination produce length increases in a single step, but also that iterative recombination could further enhance diversity and lead to more complex function. These results complement the recent findings of Mutschler et al. (2018) who show that ligation, along with nonenzymatic recombination can diversify N20 random pools of RNA. Iterative recombination is essentially a global reorganization of raw materials, or prebiotic recycling, which has been suggested previously as a means to provide a foundation for subsequent selection events (King 1982; Jain and Krishna 2001; Vaidya et al. 2013; Nghe et al. 2015; Arsène et al. 2018). Thinking beyond RNA, recombination of peptides has a similar potential to explore functional space effectively (Raches and Gazit 2003; Forsythe et al. 2015; Pappas et al. 2016).
The next stage in this line of investigation is to observe––or direct––the spontaneous appearance of RNAs with novel function (besides recombination) resulting from the shuffling of oligomeric fragments, which is apparently an inherent property of RNA. This function could first come in the form of a more complex recombination-promoting enzyme, such as a primordial group I intron catalytic core. Or it could first come in the form of another activity altogether, such as a replicase ribozyme (Mutschler et al. 2015) or in a peptide-bond-forming RNA core (Petrov et al. 2014). In any event, our work here depicts scenarios in which larger RNAs––some of which potentially could have useful functionalities––can be readily generated from shorter RNAs.
MATERIALS AND METHODS
RNAs and chemicals
Synthetic RNAs were purchased from TriLink (San Diego, CA) as HPLC purified and rehydrated in 10 mM Tris (pH 8.0) and 0.1 mM EDTA. Concentrations were calculated by UV spectroscopy at 260 nm using molar extinction coefficients calculated from the Williamson lab at the Scripps Research Institute: https://www.scripps.edu/researchservices/old/corefac/biopolymercalc2.html. RNAs were stored at 100 µM concentrations at −80°C until use. Buffers, salts, and polyacrylamide were purchased in the highest purity from Sigma Aldrich.
RNA–RNA recombination reactions
RNAs were diluted to the working concentration in RNase-free water (Invitrogen), mixed (if appropriate) heated to 65°C for 120 sec, cooled in incubation solution (1× = 100 mM MgCl2 and 30 mM pH buffer), and then incubated for the appropriate length of time. Typically, 3-h 22°C/0°C cycles were performed over the course of the incubation in a thermocyler, although isothermal trials at 4°C and at room temperature (22°C) were performed, with a loss of product yield. For pH-buffered reactions at 7.0 and 8.0, the buffer Tris(HCl), was used, while for those at pH 9.0, and 9.9, the buffers (Na)CHES, and (Na)CHES were used, respectively. Reactions were quenched with 100 mM Na2EDTA, ethanol precipitated, and stored at −80°C until gel electrophoresis.
Gel electrophoresis
RNA products were resuspended in 96% (v/v) formamide, 10 mM EDTA, and 0.1% bromphenol blue. They were then heated to 80°C for 1–2 min and loaded onto 15% polyacrylamide (19:1::acrylamide:bis-acrylamide)/8M urea gels and electrophoresed at 800V for ∼4 h. For radioimaging, the 5′-OH of one (or more) RNA fragments had been previously radiolabeled with γ32P·[ATP] and OptiKinase (USB), and bands were visualized by phosphorimaging on a Typhoon Trio+ imager (GE Healthcare). For non-radioactive imaging, gels were stained in 1X SYBR Gold or SYBR Green (Life Technologies) for 20–40 min prior to destaining with water and imaging on a Typhoon Trio+ imager.
High-throughput nucleotide sequence analysis
RNA products were excised from a polyacrylamide gel from a certain size range. These products were then prepared for sequence analysis using the Illumina TruSeq small RNA Library Preparation Kit by ligating on a 3′ adaptor, a 5′ adaptor, and performing RT-PCR using Superscript IV reverse transcriptase (Invitrogen) and Taq DNA polymerase (Fermentas). All PCR reactions were set up in a dedicated laminar flow hood with strict attention to PCR contamination concerns; for example, PCR buffers, primers, enzymes were all stored in a dedicated PCR set-up room, a one-way flow of materials from manufacturer to PCR set up was employed, and barrier tips were always used. These protocols have been in place for the application of in vitro selection of RNA in the Lehman lab for many years (cf. Lehman 2004). Nucleotide sequence analysis was carried out on MiSeq instruments with the v2 chip technology at either the University of Houston (long-term incubations) or the Oregon State University (R16 and H13 self-incubations) sequencing core facilities. In each case prior to sequencing, the library was quantified by qPCR using KAPA Biosystem's Library Quantification Kit on an Applied Biosystem's 7500 Fast Real-Time PCR instrument. Also the library was run on an Agilent BioAnalyzer 2100 using a high sensitivity DNA chip to verify fragment size. Standard operating procedures were followed to load the Illumina MiSeq run. The MiSeq run was a single end 50 bp run using 17.5pM as the loading concentration. All RNAs equal or greater in length than the experimental input (i.e., 16 or 13 nt) were included in the analyses (Tables 1, 2) by application of a manual filter to the raw sequence data files.
Simulations
A nucleotide-explicit computer simulation of the effects of iterative α recombination was performed using a program written in C++ and implemented on an AVA Direct Dual 2.4 GHz Processor desktop computer with 128 GB of RAM. The simulation emulated RNA growth and dynamics in an aqueous pond milieu with day-night cycling as envisioned previously (Damer and Deamer 2015). The logic behind the model is that an abiotic pool of short oligomers would experience cycles in an aqueous pond on the prebiotic Earth. The initial pool begins with 800,000 monomers and is halved with each increasing length and divided by size. Thus, for oligomers of length L, the number N of each length is N = (800,000)/(2L − 1 × L), working out to 200,000 dimers, 66,667 trimers, 25,000 tetramers, 10,000 pentamers, and so on. The maximum size in the simulation was 15 nt long. The monomers can be either nucleosides or nucleotides; in the latter case they were assumed to be terminated with a 2′,3′-cyclic phosphate.
RNA oligomers were simulated as RNA strand objects (RNAStrand) with properties of sequence (string), length (integer), cyclic phosphate (true or false), structure (true or false), ribozyme activity (true or false), type (None, GNRA, UNCG, CUUG, AANA, Tetraloop, Pentaloop, Hexaloop, Heptaloop), which represents the type of structure, and kcal (integer) which represents the total energy of all hydrogen bonds in the highest energy structure. A pool (vector) containing RNA oligomers (RNA Strand objects) of sizes 5–15 was created. Sizes of 1–4 were omitted due to computational considerations and the fact that these are too small to react via the α mechanism of Vlassov (Lutay et al. 2007). In each cycle, three strands were chosen at random, a template strand and two substrate strands. The substrate strands must be at least five nt long and the template strand must be at least 8 nt long to allow for sufficient Watson–Crick binding and at least one nucleotide in the leaving group. If the upper left substrate is terminated with a cyclic phosphate, a leaving group is not necessary.
An α arrangement occurs at random approximately 1/100,000 cycles; using our strict criteria, which resembles the known mechanism of Vlassov (Lutay et al. 2007; Nechaev et al. 2009), an α-permissive arrangement of oligomers can be found roughly once for every 100,000 attempts, implying that a 1 µM solution of random RNA should contain roughly 1 pmole of α arrangements. The RNA strands chosen at random are chosen to fixed positions (template, upper left, upper right) and all possible conformations are explored in order to find an alpha setup. Because two double strands of length X and Y with overlapping regions of Z can form no more than (X + Y − Z) conformations, the overall α arrangement requires relatively few computational resources to explore all possible conformations. If there is more than one different setup, the one with highest binding energy is chosen.
Once an α setup is found, it undergoes strand cleavage with an 80% probability. If it successfully undergoes strand cleavage, the leaving group becomes a new RNA strand within the pool. The remaining α setup can subsequently undergo ligation with a 30% chance. If it undergoes ligation, the two strands become one and one strand is removed from the pool. If it does not ligate, the upper left strand gains a cyclic phosphate (assumed due to cleavage) and the complex fully dissociates back into the pool as three distinct strands. For one-step recombination, the ligation and strand cleavage events are combined for a 10% overall chance. If a strand cannot undergo strand cleavage because it has no leaving group (e.g., the α setup has no tail) it can still undergo ligation with a 30% probability if the upper left strand has a cyclic phosphate. If not, the complex dissociates.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank P. Nghe, D. Lacoste, A. Blokhuis, S. Ameta, W. Hordijk, and P. Unrau for useful discussions, and A. Gustafson, C. Díaz Arenas, R. Gysbers, and M. Tirumalai for laboratory assistance. This project/publication was made possible through the support of a grant from the John Templeton Foundation (grant no. 54466). The opinions expressed in this publication are those of the author(s) and do not necessarily reflect the views of the John Templeton Foundation.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.068908.118.
REFERENCES
- Arsène S, Ameta S, Lehman N, Griffiths AD, Nghe P. 2018. Coupled catabolism and anabolism in autocatalytic RNA sets. Nucleic Acids Res 46: 9660–9666. 10.1093/nar/gky598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokhuis A, Lacoste D. 2017. Length and sequence relaxation of copolymers under recombination reactions. J Chem Phys 147: 094905 10.1063/1.5001021 [DOI] [PubMed] [Google Scholar]
- Briones C, Stich M, Manrubia SC. 2009. The dawn of the RNA World: toward functional complexity through ligation of random RNA oligomers. RNA 15: 743–749. 10.1261/rna.1488609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetverin AB. 1999. The puzzle of RNA recombination. FEBS Lett 460: 1–5. 10.1016/S0014-5793(99)01282-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetverin AB, Chetverina HV, Demidenko AA, Ugarov VI. 1997. Nonhomologous RNA recombination in a cell-free system: evidence for a transesterification mechanism guided by secondary structure. Cell 88: 503–513. 10.1016/S0092-8674(00)81890-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damer B, Deamer D. 2015. Coupled phases and combinatorial selection in fluctuating hydrothermal pools: a scenario to guide experimental approaches to the origin of cellular life. Life (Basel) 5: 872–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Guzman V, Shenasa H, Vercoutere W, Deamer D. 2014. Generation of oligonucleotides under hydrothermal conditions by non-enzymatic polymerization. J Mol Evol 78: 251–262. 10.1007/s00239-014-9623-2 [DOI] [PubMed] [Google Scholar]
- Döring J, Hurek T. 2017. Arm-specific cleavage and mutation during reverse transcription of 2′,5′-branched RNA by Moloney murine leukemia virus reverse transcriptase. Nucleic Acids Res 45: 3967–3984. 10.1093/nar/gkx073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris JP, Hill AR Jr, Liu R, Orgel LE. 1996. Synthesis of long prebiotic oligomers on mineral surfaces. Nature 381: 59–61. 10.1038/381059a0 [DOI] [PubMed] [Google Scholar]
- Forsythe JG, Yu S-S, Mamajanov I, Grover MA, Krishnamurthy R, Fernández FM, Hud NV. 2015. Ester-mediated amide bond formation driven by wet-dry cycles: a possible path to polypeptides on the prebiotic Earth. Angew Chem Int Ed 54: 9871–9875. 10.1002/anie.201503792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibard C, Bhowmik S, Karki M, Kim E-K, Krishnamurthy R. 2018. Phosphorylation, oligomerization and self-assembly in water under potential prebiotic conditions. Nat Chem 10: 212–217. 10.1038/nchem.2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden EJ, Riley CA, Burton AS, Lehman N. 2005. RNA-directed construction of structurally complex and active ligase ribozymes through recombination. RNA 11: 1678–1687. 10.1261/rna.2125305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs PG, Lehman N. 2015. The RNA World: molecular cooperation at the origins of life. Nat Rev Genet 16: 7–17. 10.1038/nrg3841 [DOI] [PubMed] [Google Scholar]
- Horowitz ED, Engelhart AE, Chen MC, Quarles KA, Smith MW, Lynn DG, Hud NV. 2010. Intercalation as a means to suppress cyclization and promote polymerization of base-pairing oligonucleotides in a prebiotic world. Proc Natl Acad Sci 107: 5288–5293. 10.1073/pnas.0914172107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Krishna S. 2001. A model for the emergence of cooperation, interdependence, and structure in evolving networks. Proc Natl Acad Sci 98: 543–547. 10.1073/pnas.98.2.543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King GA. 1982. Recycling, reproduction, and life's origins. Biosystems 15: 89–97. 10.1016/0303-2647(82)90022-3 [DOI] [PubMed] [Google Scholar]
- Lehman N. 2003. A case for the extreme antiquity of recombination. J Mol Evol 56: 770–777. 10.1007/s00239-003-2454-1 [DOI] [PubMed] [Google Scholar]
- Lehman N. 2004. Assessing the likelihood of recurrence during RNA evolution in vitro. Artif Life 10: 1–22. 10.1162/106454604322875887 [DOI] [PubMed] [Google Scholar]
- Lehman N. 2008. A recombination-based model for the origin and early evolution of genetic information. Chem Biodivers 5: 1707–1717. 10.1002/cbdv.200890159 [DOI] [PubMed] [Google Scholar]
- Lehman N, Arenas CD, White WA, Schmidt FJ. 2011. Complexity through recombination: from chemistry to biology. Entropy 13: 17–37. 10.3390/e13010017 [DOI] [Google Scholar]
- Lohrmann R, Bridson PK, Orgel LE. 1980. Efficient metal-ion catalyzed template-directed oligonucleotide synthesis. Science 208: 1464–1465. 10.1126/science.6247762 [DOI] [PubMed] [Google Scholar]
- Lorsch JR, Bartel DP, Szostak JW. 1995. Reverse transcriptase reads through a 2′–5′ linkage and a 2′-thiophosphate in a template. Nucleic Acids Res 23: 2811–2814. 10.1093/nar/23.15.2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutay AV, Chernolovskaya EL, Zenkova MA, Vlassov VV. 2005. Nonenzymatic template-dependent ligation of 2′,3′-cyclic phosphate-containing oligonucleotides catalyzed by metal ions. Dokl Akad Nauk SSSR 401: 705–708. [DOI] [PubMed] [Google Scholar]
- Lutay AV, Chernolovskaya EL, Zenkova MA, Vlassov VV. 2006. The nonenzymatic template-directed ligation of oligonucleotides. Biogeosciences 3: 243–249. 10.5194/bg-3-243-2006 [DOI] [Google Scholar]
- Lutay AV, Zenkova MA, Vlassov VV. 2007. Nonenzymatic recombination of RNA: possible mechanism for the formation of novel sequences. Chem Biodivers 4: 762–767. 10.1002/cbdv.200790062 [DOI] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol 288: 911–940. 10.1006/jmbi.1999.2700 [DOI] [PubMed] [Google Scholar]
- Mutschler H, Wochner A, Holliger P. 2015. Freeze-thaw cycles as drivers of complex ribozyme assembly. Nat Chem 7: 502–508. 10.1038/nchem.2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutschler H, Taylor AI, Porebski BT, Lightowers A, Houlihan G, Abramov M, Herdewijn P, Holliger P. 2018. Random-sequence genetic oligomer pools display an innate potential for ligation and recombination. eLife 7: e43022 10.7554/eLife.43022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev SY, Lutay AV, Vlassov VV, Zenkova MA. 2009. Non-enzymatic template-directed recombination of RNAs. Int J Mol Sci 10: 1788–1807. 10.3390/ijms10041788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nghe P, Hordijk W, Kauffman SA, Walker SI, Schmidt FJ, Kemble H, Yeates JAM, Lehman N. 2015. Prebiotic network evolution: six key parameters. Mol Biosyst 11: 3206–3217. 10.1039/C5MB00593K [DOI] [PubMed] [Google Scholar]
- Nowak MA, Ohtsuki H. 2008. Prevolutionary dynamics and the origin of evolution. Proc Natl Acad Sci 105: 14924–14927. 10.1073/pnas.0806714105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas CG, Shafi R, Sasselli IR, Siccardi H, Wang T, Narang V, Abzalimov R, Wijerathne N, Ulijn RV. 2016. Dynamic peptide libraries for the discovery of supramolecular nanomaterials. Nat Nanotechnol 11: 960–967. 10.1038/nnano.2016.169 [DOI] [PubMed] [Google Scholar]
- Petrov AS, Bernier CR, Hsiao C, Norris AM, Kovacs NA, Waterbury CC, Stepanov VG, Harvey SC, Fox GE, Wartell RM, et al. 2014. Evolution of the ribosome at atomic resolution. Proc Natl Acad Sci 111: 10251–10256. 10.1073/pnas.1407205111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pino S, Costanzo G, Giorgi A, Šponer J, Šponer J, Mauro E. 2013. Ribozyme activity of RNA nonenzymatically polymerized from 3′,5′-cyclic GMP. Entropy 15: 5362–5383. 10.3390/e15125362 [DOI] [Google Scholar]
- Prywes N, Blain JC, Del Frate F, Szostak JW. 2016. Nonenzymatic copying of RNA templates containing all four letters is catalyzed by activated oligonucleotides. Elife 5: e17756 10.7554/eLife.17756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raches M, Gazit E. 2003. Casting metal nanowires within discrete self-assembled peptidenanotubes. Science 300: 625–627. 10.1126/science.1082387 [DOI] [PubMed] [Google Scholar]
- Rajamani S, Vlassov A, Benner S, Coombs A, Olasagasti F, Deamer D. 2008. Lipid-assisted synthesis of RNA-like polymers from mononucleotides. Orig Life Evol Biosph 38: 57–74. 10.1007/s11084-007-9113-2 [DOI] [PubMed] [Google Scholar]
- Staroseletz Y, Nechaev S, Bichenkova E, Bryce R, Watson C, Vlassov V. 2018. Non-enzymatic recombination of RNA: ligation in loops. Biochim Biophys Acta 1862: 705–725. 10.1016/j.bbagen.2017.10.019 [DOI] [PubMed] [Google Scholar]
- Vaidya N, Walker SI, Lehman N. 2013. Recycling of informational units leads to selection of replicators in a prebiotic soup. Chem Biol 20: 241–252. 10.1016/j.chembiol.2013.01.007 [DOI] [PubMed] [Google Scholar]
- Vlassov AV, Kazakov S, Johnston BH, Landweber LF. 2005. The RNA world on ice: a new scenario for the emergence of RNA information. J Mol Evol 61: 264–273. 10.1007/s00239-004-0362-7 [DOI] [PubMed] [Google Scholar]
- Wu T, Orgel LE. 1992. Nonenzymatic template-directed synthesis on oligodeoxycytidylate sequences in hairpin oligonucleotides. J Am Chem Soc 114: 317–322. 10.1021/ja00027a040 [DOI] [PubMed] [Google Scholar]
- Yarus M. 2012. Darwinean behavior in a cold, sporadically fed pool of ribonucleotides. Astrobiology 12: 870–883. 10.1089/ast.2012.0860 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.