Mucorales are a group of ancient saprophytic fungi that cause neglected infectious diseases collectively known as mucormycoses. The molecular processes underlying the establishment and progression of this disease are largely unknown. Our work presents a transcriptomic study to unveil the Mucor circinelloides genetic network triggered in fungal spores in response to phagocytosis by macrophages and the transcriptional response of the host cells. Functional characterization of differentially expressed fungal genes revealed three transcription factors and three extracellular proteins essential for the fungus to survive and germinate inside the phagosome and to cause disease in mice. Two of the transcription factors, highly similar to activating transcription factors (ATFs), coordinate a complex secondary gene response involved in pathogenesis. The significance of our research is in characterizing the initial stages that lead to evasion of the host innate immune response and, in consequence, the dissemination of the infection. This genetic study offers possible targets for novel antifungal drugs against these opportunistic human pathogens.
KEYWORDS: emerging pathogens, host-pathogen interaction, innate immunity, mucormycosis, transcriptomics
ABSTRACT
Mucormycosis is an emerging fungal infection that is often lethal due to the ineffectiveness of current therapies. Here, we have studied the first stage of this infection—the germination of Mucor circinelloides spores inside phagocytic cells—from an integrated transcriptomic and functional perspective. A relevant fungal gene network is remodeled in response to phagocytosis, being enriched in crucial functions to survive and germinate inside the phagosome, such as nutritional adaptation and response to oxidative stress. Correspondingly, the phagocytic cells induced a specific proinflammatory and apoptotic response to the pathogenic strain. Deletion of fungal genes encoding putative transcription factors (atf1, atf2, and gcn4), extracellular proteins (chi1 and pps1), and an aquaporin (aqp1) revealed that these genes perform important roles in survival following phagocytosis, germination inside the phagosome, and virulence in mice. atf1 and atf2 play a major role in these pathogenic processes, since their mutants showed the strongest phenotypes and both genes control a complex gene network of secondarily regulated genes, including chi1 and aqp1. These new insights into the initial phase of mucormycosis define genetic regulators and molecular processes that could serve as pharmacological targets.
INTRODUCTION
Mucormycosis is an angioinvasive fungal infection with mortality rates that can reach up to 90% in cases of disseminated infection (1–3). This infection is caused by diverse species belonging to the order Mucorales, a group of early-diverging fungi (4). The most common species causing mucormycosis belong to the Rhizopus, Mucor, and Lichtheimia genera. These ubiquitous saprophytes can act as opportunistic human pathogens due to the capacity for rapid vascular dissemination with subsequent tissue necrosis (5, 6). Mucormycosis is considered an emerging infection due to a steady increment in incidence among immunocompromised patients, along with the increasing diagnosis of mucormycosis in immunocompetent individuals, especially associated with trauma (7, 8).
A critical aspect of these neglected fungal pathogens is their innate resistance to current antifungal drugs, which partially explains the elevated mortality rates (9, 10). Antifungal therapies with voriconazole and fluconazole have poor outcomes against Mucorales, and other antifungals, such as posaconazole or amphotericin B, are used as salvage therapy and cause serious adverse effects (10, 11). This antifungal resistance in Mucorales may be due to an evolutionarily conserved amino acid substitution in lanosterol 14α-demethylase, the enzyme targeted by short-tailed azole drugs (10). In addition, Mucorales also possess an RNA interference (RNAi)-based resistance mechanism that specifically silences the expression of the gene targeted by the antifungal compound, preventing the formation of a target-antifungal complex that inhibits hyphal growth (9, 12).
Research has focused on finding new targets for antifungal drugs in Mucorales. However, molecular research is challenging because most Mucorales are not amenable to molecular genetic tools. Despite these difficulties, genetic studies in these pathogens have identified dimorphism, iron uptake, spore size, and spore coat proteins as virulence factors in mucormycosis (13–18).
Mucor circinelloides, herein referred to as Mucor, offers multiple advantages as a genetic model for pathogenesis among the typically intractable Mucorales. Along with the availability of an efficient genetic transformation procedure, its RNAi mechanism is well characterized and has become a useful tool for functional genetics (19, 20). Thus, an RNAi-based genome-wide screening identified two virulence determinants in mucormycosis: a myosin class V gene involved in intracellular transport and a phospholipase D gene involved in signaling (21). Another advantage of this model is the availability of two wild-type strains with clear, distinct pathogenic potentials in wax moth (16) and mouse (21) models. Indeed, a comparative genomic analysis revealed specific genomic variations between both pathotypes, which could be associated with virulence (22).
Mucoralean asexual spores can reach host tissues via inhalation or open wounds, which are the most frequent routes of colonization leading to mucormycosis. Once inside the host, the spores’ ability to germinate and invade the host defines the establishment of the infection (23). The innate immune response is the first barrier at the site of the infection, where phagocytic cells are recruited to internalize spores and form granulomas in rabbit, mouse, and zebrafish models (24–26). Phagocytosis prevents spore germination in these animal models, and failure of this early immune response leads to disseminated infections (23). Thus, deficiencies in innate immunity are risk factors associated with mucormycosis (2, 27), emphasizing the interaction between Mucorales and their hosts as a crucial stage in the invasive development of mucormycosis.
The molecular and cellular responses of Mucorales to host phagocytic cells are not well defined. Characterization of genes differentially regulated at this stage of the infection by transcriptomic analysis has been postulated as a straightforward strategy to elucidate key molecular aspects of the interaction. The first host-mucoral transcriptomic study was conducted in the fruit fly, in which several host genes related to pathogen recognition and stress response were selectively downregulated after infection (28). Other transcriptomic studies with airway epithelial cells and Rhizopus and Mucor species revealed that platelet-derived growth factor receptor B signaling participates in the core response to mucoralean infection that damages barrier host cells (29). However, these studies were mainly focused on the host response, and no follow-up analysis of fungal genes was performed.
Here, we conducted an integrated genetic study to detail the response of Mucor to host innate immunity. After macrophage phagocytosis, a transcriptomic analysis of spores from two different Mucor pathotypes revealed an intricate gene response that could account for the distinctive features of mucormycosis. In particular, a functional enrichment analysis of this response exposed biological and molecular functions required to survive phagocytosis. Deletion mutants of key genes participating in these functions were severely compromised after phagocytosis by macrophages and showed decreased virulence in a murine model. Transcriptomic analyses on mutants in transcription factors involved in virulence revealed a secondary gene network during the initial interaction between macrophages and germinating spores. Taken together, this work contributes to a better understanding of the mechanisms by which members of Mucorales overcome innate immune responses to cause mucormycosis, providing putative targets for new antifungal therapies against this often fatal infection.
RESULTS
Mucor pathotypes coordinate different transcriptional responses against macrophage phagocytosis.
Previous host-pathogen survival assays have shown significant differences in virulence between two Mucor strains. Immunosuppressed mice infected with the R7B strain show high mortality rates, whereas the NRRL3631 strain is completely avirulent (22). The virulence of these strains is related to their spores’ ability to germinate inside host phagocytic cells, since the virulent R7B strain is able to germinate and survive within mouse macrophages while NRRL3631 shows a strong delay in germination (21). To explore the transcriptional differences between the strains in response to host innate immunity, we performed an in vitro host-pathogen RNA-sequencing experiment on fungal spores interacting with mouse macrophages. R7B or NRRL3631 spores were cocultured with a cell line of mouse macrophages (J774A.1; ATCC T1B-67) for 5 h, ensuring that all the spores were phagocytized and able to start the germination process inside the macrophages. At this point, the differences in germination inside the phagosome between strains were most evident. Indeed, R7B spores had fully undergone the germination process and produced germ tubes, while NRRL3631 spores were still in a stage of isotropic growth. Noninteraction controls consisted of single cultures of either fungal spores or uninfected macrophages in the same cell medium used in the interaction. These controls were used to exclude differences in gene expression in response to the medium. Total RNA was isolated from both fungus-host interaction and control samples and then sequenced to perform a transcriptomic analysis.
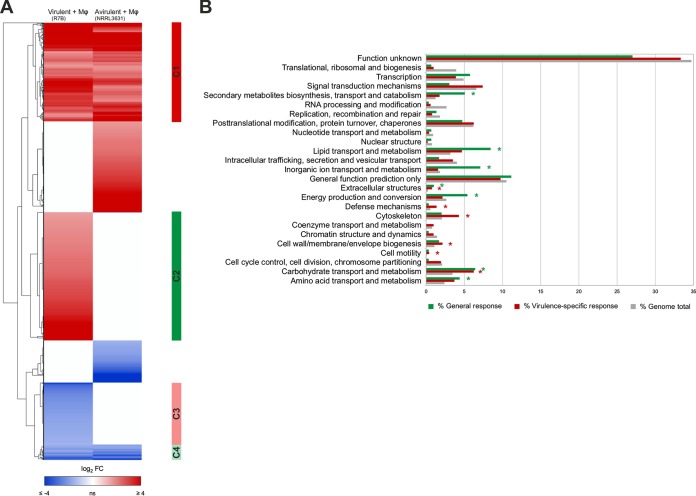
This analysis identified 1,068 genes (>9% of total genes) that were differentially expressed (Q ≤ 0.05) in the two Mucor strains after 5 h of macrophage interaction (Data Set S1 in the supplemental material). Expression values were clustered to discriminate between shared and strain-specific gene sets (Fig. 1A). This classification revealed a general response to the macrophages, comprised of genes that were either up- or downregulated in both the virulent (R7B) and the avirulent (NRRL3631) strain (Fig. 1A, gene clusters C1 and C4, respectively). A functional enrichment analysis of this general response determined that nutrient assimilation and metabolism were strongly induced as a result of the interaction, as well as the production of secondary metabolites and extracellular structures (Fig. 1B, general response), suggesting that nutrient competition in an impoverished environment, such as the phagosome, is critical for the spore to begin germination. These cellular processes, though essential, do not explain the exact mechanism for surviving the host innate immune response. We focused closely on the transcriptional profile of the R7B strain to characterize a virulence-specific response, which contained 464 genes that were differentially expressed in response to the macrophages (Fig. 1A, clusters C2 and C3). Gene clusters involved in the virulence-specific response were enhanced in functions associated with carbohydrate metabolism, cell wall and membrane biogenesis, cytoskeleton, cell motility, extracellular structures, and defense mechanisms (Fig. 1B, data for virulence-specific response). Moreover, we discovered an increase in biological processes involved in regulating cAMP-dependent intracellular signal transduction pathways and the response to oxidative stress (Data Set S1). These functions are in accord with the germination and hyphal development processes observed for the phagocytized spores and could explain their ability to survive macrophage attack.
FIG 1.
Mucor response to mouse macrophages. (A) Heat map showing Mucor genes that are differentially expressed (Q ≤ 0.05) in virulent R7B and avirulent NRRL3631 spores phagocytosed by mouse macrophages (Mφ) (cell line J774A.1) during coculture for 5 h in L15 cell culture medium. Differential expression values with respect to noninteraction cultures (without macrophages) were calculated for each strain and hierarchically clustered according to similarities. Red and blue represent up- and downregulated genes, respectively. Clusters 1 (C1) and 4 (C4) contain genes up- and downregulated, respectively, in both Mucor strains during phagocytosis, which was defined as the general response. Clusters 2 (C2) and 3 (C3) contain genes up- and downregulated, respectively, in the virulent Mucor strain R7B during phagocytosis, comprising the virulence-specific response. (B) Functional enrichment analysis showing the percentages of genes for each KOG class found in the Mucor genome (gray) and in the general (green) and virulence-specific (red) responses. Asterisks (*) indicate a significant enrichment (P ≤ 0.05, Fisher’s exact test) in the corresponding KOG class with respect to the expected number of genes found in the Mucor genome. FC, fold change.
Differentially expressed genes in Mucor strains after 5-h interaction with mouse macrophages. Expression values of virulent Mucor strain R7B spores (R7BMφ) interacting with mouse macrophages and noninteraction controls (R7Bc) and of avirulent strain NRRL3631 spores (NRRL3631Mφ and NRRL3631c, respectively, for interaction and noninteraction controls). Download Data Set S1, XLSX file, 3.1 MB (3.1MB, xlsx) .
Copyright © 2019 Pérez-Arques et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Virulent Mucor strain induces inflammation and apoptotic pathways in mouse macrophages.
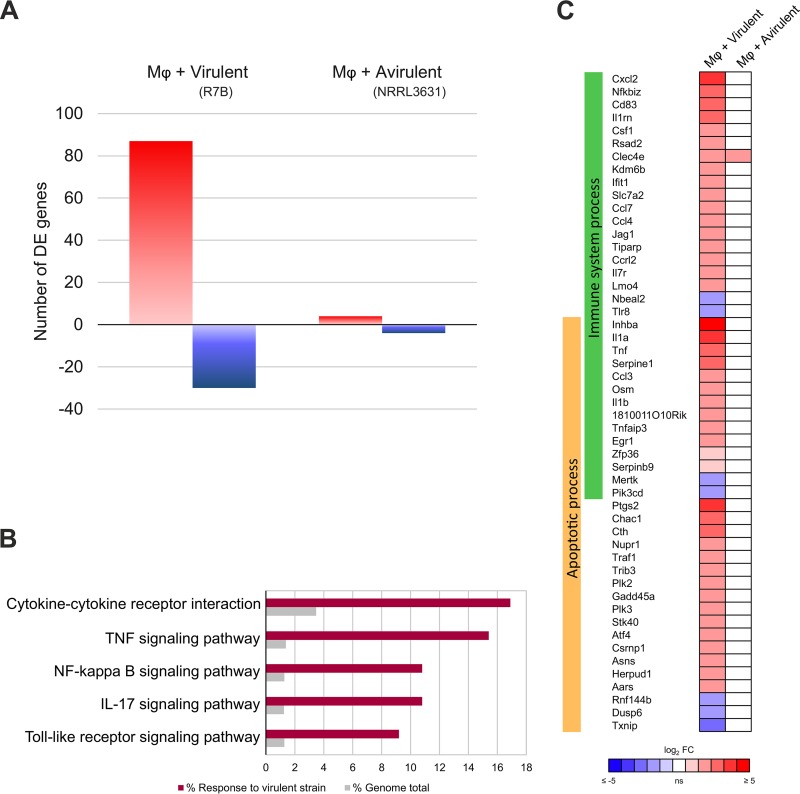
Gene expression in macrophages responding to each Mucor strain was compared to identify a specific reaction to either pathotype that could account for their differences in virulence. A differential expression analysis (Q ≤ 0.05) revealed 117 genes that responded to the virulent Mucor strain after 5 h of interaction compared to their expression in macrophage cultures without spores. In contrast, reaction to the avirulent Mucor strain was largely absent, consisting of 8 differentially expressed genes (Fig. 2A and Data Set S2).
FIG 2.
Macrophages respond exclusively to the virulent Mucor strain. (A) Differential expression (Q ≤ 0.05) of upregulated (red) and downregulated (blue) mouse macrophage (Mφ) (cell line J774A.1) genes induced after phagocytosing spores from each depicted Mucor strain in L15 cell culture medium for 5 h. Differential expression values were calculated by comparison with expression in noninteraction cultures (without spores). (B) Percentage of genes for each significantly enriched (adjusted P value, ≤0.01) KEGG pathway found in the macrophage response to the virulent Mucor R7B strain compared to the expected percentage of genes found in the mouse genome. (C) Differential expression values (Q ≤ 0.05) of selected mouse macrophage (Mφ) (cell line J774A.1) genes induced after phagocytosing spores from each depicted Mucor strain for 5 h. The selected genes are involved in two enriched GO Biological Processes (adjusted P value, ≤0.05), Immune System Process and Apoptotic Process. FC, fold change.
Differentially expressed genes in mouse macrophages after 5-h interaction with Mucor. Expression values of mouse macrophages interacting either with virulent Mucor strain R7B spores (MφR7B) or avirulent strain NRRL3631 spores (MφNRRL3631) or without spores (Mφ). Genes with a mean FPKM of <1.0 were filtered out. Download Data Set S2, XLSX file, 1.3 MB (1.3MB, xlsx) .
Copyright © 2019 Pérez-Arques et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A functional analysis of the macrophage response to the virulent strain revealed an enrichment in signaling pathways related to defense against pathogens, particularly inflammation signaling pathways (Fig. 2B). Most of these genes were involved in biological processes related to immunity and apoptosis (Fig. 2C and Data Set S2). This exclusive response to the virulent strain was confirmed by reverse transcription-quantitative PCR (RT-qPCR) of well-known proinflammatory cytokines. Thus, Il1a and Tnf expression was significantly upregulated in the macrophages (P < 0.005, unpaired t test) after 5 h of interaction with the virulent Mucor strain (Fig. S1), and their expression was significantly higher than in the cells interacting with the avirulent strain.
Relative levels of expression of proinflammatory cytokines in response to Mucor virulent strain. RT-qPCR-determined expression differences of Il1a and Tnf mouse macrophage genes during interaction with virulent and avirulent Mucor strains. Transcript levels were quantified in cDNA obtained from mouse macrophages (Mφ) (cell line J774A.1) cocultured with either virulent R7B spores or avirulent NRRL3631 spores in L15 cell culture medium for 5 h. Values were normalized using Actb as an internal control, and log2-fold change differential expression was calculated with respect to expression values in response to the avirulent Mucor strain. Error bars correspond to the SD from technical triplicates, and asterisks indicate a significant difference determined by unpaired t test (**, P ≤ 0.005; ***, P < 0.0001). Download FIG S1, PDF file, 0.03 MB (29.3KB, pdf) .
Copyright © 2019 Pérez-Arques et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genes induced in phagocytosed spores are expressed during in vivo infection.
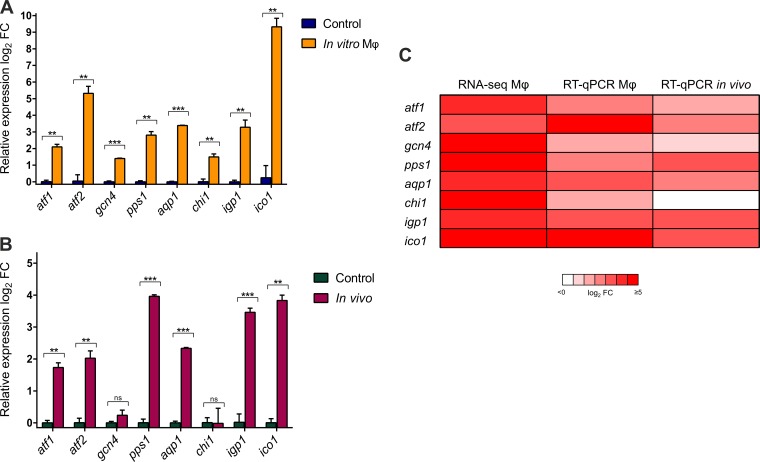
To validate the transcriptomic response and to further characterize the response of Mucor to the host innate immune system, we designed a thorough genetic analysis that combined quantification of both in vitro and in vivo gene expression with the generation of deletion mutants. The selection criteria focused on genes that were highly upregulated in response to the macrophages (i.e., log2-fold change of ≥3.5) and that could regulate the functional categories attributed to the virulence-specific response (Table 1). Thus, we selected the following genes: atf1 (activating transcription factor 1; Joint Genome Institute Mucor circinelloides v2.0 identification number [ID] 190180) and atf2 (activating transcription factor 2; ID 156289), which encode basic leucine zipper activating transcription factors (ATFs) that could regulate the response to oxidative stress; gcn4 (general control nonderepressible 4; ID 85517), which encodes a basic leucine zipper transcription factor that could be involved in inducing the robust metabolic response observed in the general response to macrophages; aqp1 (aquaporin 1; ID 167023), which encodes a putative aquaporin that could be implicated in cell wall and membrane processes; ico1 (isochorismatase 1; ID 155573), which encodes a putative isochorismatase that could participate in the biosynthesis of siderophores and other extracellular structures and defense mechanisms; and igp1 (immunoglobulin-like protein; ID 154866), chi1 (ID 189685), and pps1 (signal peptide-containing protein; ID 115037), genes encoding proteins of unknown function but with predicted secretion signals that could be Mucorales-specific surface antigens or secreted proteins in direct interaction with macrophages. The expression of these selected genes was analyzed by RT-qPCR, replicating the transcriptome sequencing (RNA-seq) conditions (gene-specific primers are in Table S1). The analysis confirmed the expression pattern of the selected genes, all of which were significantly upregulated (P ≤ 0.05, unpaired t test) after 5 h of interacting with macrophages, validating the RNA-seq data (Fig. 3A and C).
TABLE 1.
Candidates for gene deletion
| Gene | ID | Secretion prediction |
TMHMMa | Predicted domain (Pfam or InterPro accession no.) |
Putative protein homolog (reference) (% identity; alignment score; E value) |
|---|---|---|---|---|---|
| atf1 | 190180 | No | None | bZIP_1 (PF00170) | Atf1 (45) (30.4; 200; 1.4e−15) |
| atf2 | 156289 | No | None | bZIP_1 (PF00170) | Atf1 (45) (31.1; 180; 5.7e−14) |
| gcn4 | 85517 | No | None | bZIP_1 (PF00170) | GCN4 (62) (50; 100; 1.4e−3) |
| pps1 | 115037 | Yes | None | None predicted | MP88 (40) (29; 190; 7e−17) |
| aqp1 | 167023 | Yes | 6 | Aquaporin-like (IPR023271) | AQY1 (63) (32; 309; 3.7e−31) |
| chi1 | 189685 | NCSb | None | None predicted | None |
| igp1 | 154866 | Yes | None | None predicted | None |
| ico1 | 155573 | Yes | None | Isochorismatase (PF00857) | YcaC (64) (61.9; 655; 2.2e−84) |
TMHMM, Transmembrane Helix Prediction based on a hidden Markov Model.
Putative secretion by noncanonical secretion (NCS) pathway.
FIG 3.
In vitro and in vivo gene expression analysis of selected Mucor genes. (A) RT-qPCR-determined differences in the expression of selected Mucor genes during mouse macrophage phagocytosis. Transcript levels were quantified in cDNA obtained from virulent Mucor strain R7B spores cocultured with mouse macrophages (Mφ) (cell line J774A.1) in L15 cell culture medium for 5 h and from the same spores cultured in noninteraction controls (without macrophages). Values were normalized using 18S rRNA as an internal control, and the differential expression (log2-fold change) was calculated with respect to expression in noninteraction samples. (B) RT-qPCR-determined differences in the expression of the selected genes during interaction with peritoneal macrophages. Transcript levels were quantified in cDNA obtained from virulent Mucor strain R7B spores that were injected into the peritoneal cavity of OF-1 mice and recovered after 5 h, as well as from the same spores incubated in L15 cell culture medium as a control. Values were normalized using 18S rRNA as an internal control, and the differential expression (log2-fold change) was calculated with respect to the expression in L15 control samples. (C) Comparison of differential expression of selected genes during in vitro interaction with mouse macrophages, quantified by both RNA-seq and RT-qPCR, and during in vivo interaction with peritoneal macrophages, quantified by RT-qPCR. Error bars correspond to the standard deviations (SD) of the results from technical triplicates, and asterisks indicate a significant difference determined by unpaired t test (*, P ≤ 0.05; **, P ≤ 0.005; ***, P < 0.0001).
Primers used in the study. Download Table S1, DOCX file, 0.02 MB (17.2KB, docx) .
Copyright © 2019 Pérez-Arques et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The expression of the in vitro-validated genes was further analyzed in vivo. Spores of the virulent strain R7B were injected into the peritoneal cavity of mice and recovered after 5 h of interaction with recruited primary phagocytic cells. Total RNA samples were extracted to quantify gene expression by RT-qPCR using the same set of specific primer pairs (Table S1). Genes atf1, atf2, gcn4, pps1, aqp1, igp1, and ico1 were significantly upregulated (P ≤ 0.05, unpaired t test) (Fig. 3B). Conversely, chi1 expression was not affected by the peritoneal phagocytic cells (Fig. 3B). The accurate correlation between in vitro expression differences during macrophage phagocytosis and in vivo expression for most of the selected genes (Fig. 3C) indicated that the transcriptomic data can be useful in identifying genes that specifically respond to host innate immune cells.
Deletion of key genes involved in the response to macrophages decreases survival during both phagocytosis and virulence.
Because these genes were highly upregulated in Mucor after spores were phagocytosed, we hypothesized that their absence could impact the ability of spores to survive and escape macrophage phagocytosis. To test this hypothesis, single deletion mutants of each of the selected genes were generated by double homologous recombination with a disruption cassette, which replaced their native loci with the pyrG gene used as a selectable marker for uridine auxotrophy (Fig. S2). Mucor auxotrophic protoplasts were transformed by electroporation with each of the disruption cassettes. Subsequently, transformants that were able to grow on selective medium were selected as putative deletion mutants. Gene deletion was confirmed by PCR (specific primers are in Table S1). Because Mucor spores are multinucleated, 10 selective cycles of vegetative growth were conducted to yield homokaryotic deletion mutants, which were confirmed by Southern blot analysis (Fig. S2). To ensure that the phenotypes observed in the mutants were due to deletion of the targeted genes, all further phenotypic assays were conducted with two independently generated mutant strains for each deleted gene (Table S2).
Disruptions of atf1, atf2, gcn4, pps1, aqp1, chi1, igp1, and ico1 genes. Schematic representation of the wild-type (WT) locus and the mutant (MUT) locus after homologous recombination with the disruption fragment for each gene. Restriction sites for each restriction enzyme used in the Southern blot assays are indicated, showing the expected size of the fragment generated after digestion. The positions of the probes used are indicated for each locus. Southern blot analyses are shown below the schemes. For this, genomic DNA (gDNA) of a WT strain and the gDNA of mutants obtained for each gene were digested with the depicted restriction enzyme. Hybridizations were made with the respective probes, which allowed discrimination between disrupted and WT alleles. The positions and sizes of the GeneRuler DNA ladder mixture (M) (Fermentas) are indicated. Download FIG S2, PDF file, 2.2 MB (2.2MB, pdf) .
Copyright © 2019 Pérez-Arques et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
M. circinelloides f. lusitanicus strains used in this study. Download Table S2, DOCX file, 0.01 MB (14.9KB, docx) .
Copyright © 2019 Pérez-Arques et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
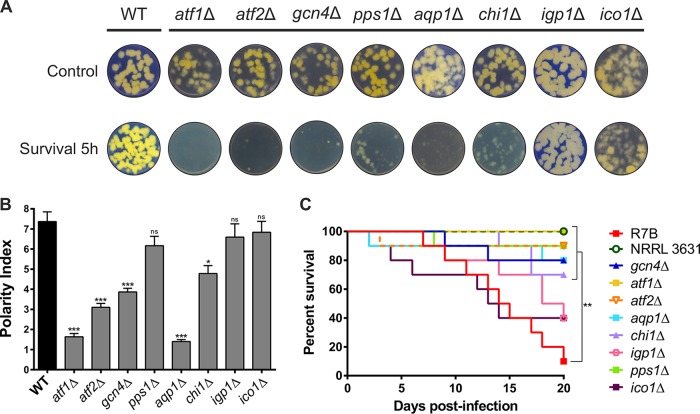
The role of the selected genes in evading the host innate immune response was analyzed using two different approaches. First, the fitness of the mutant strains was assessed after phagocytosis to examine their capacity to endure macrophage phagocytosis. Spores from each deletion mutant strain were cocultured with mouse macrophages for 5 h in mammalian cell culture medium and then plated in minimal medium with Casamino Acids (MMC), pH 3.2, for 48 h to test their growth rates and capacity to develop healthy colonies. Simultaneously, the spores were cultured under the same conditions but without macrophages to ensure that the observed growth defects were exclusively due to the fungus-host cell interaction. Deletion mutants in atf1 (atf1Δ), atf2 (atf2Δ), and gcn4 (gcn4Δ) showed clear defects in growth and development after 5 h of macrophage interaction, while deletion mutants in pps1 (pps1Δ), aqp1 (aqp1Δ), and chi1 (chi1Δ) had defects that were similar but relatively minor (Fig. 4A). On the other hand, deletion mutants in igp1 (igp1Δ) and ico1 (ico1Δ) were able to develop colonies as healthy as those of the wild-type virulent strain.
FIG 4.
Phenotypic analyses of Mucor deletion mutants in selected genes during phagocytosis and mouse infection. (A) Spore fitness of strains with the indicated mutations after 5 h of macrophage phagocytosis, assessed by their ability to develop healthy colonies on MMC medium. Control plates contain spores obtained after growing in cell culture medium for 5 h without macrophages. R7B was used as the wild-type (WT) strain. All MMC plates were incubated for 48 h. (B) Polarity index measures after phagocytosis for all deletion strains generated compared to those of the wild-type control (R7B). Error bars correspond to the standard errors of the means (SEM) from technical replicates (n = 50 spores), and statistical significance was analyzed by the unpaired t test. (C) Virulence assays with immunosuppressed mice. Each color shows the survival curve for a group of 10 mice infected with 1 × 106 spores from one of the deletion mutant strains. Survival rates were compared to the results for mice infected with a virulent control strain (R7B) and statistically analyzed by a Mantel-Cox test. NRRL3631 was used as an avirulent control strain. Asterisks (*) indicate a significant difference determined by the unpaired t test (*, P ≤ 0.05; **, P ≤ 0.005; ***, P < 0.0001).
Second, the germination capacity of the mutant spores was measured inside the phagosome. Polar growth of the deletion mutants was quantified as the ratio between the germ tube length and spore width, considering differences to be significant at a P value of ≤0.05 (unpaired t test). After 5 h of interaction with macrophages, spores from the virulent strain were fully germinated inside the phagosome, while spores from atf1Δ (P < 0.0001), atf2Δ (P < 0.0001), gcn4Δ (P < 0.0001), aqp1Δ (P < 0.0001), and chi1Δ (P = 0.0011) mutants exhibited strong and significant delays in germination (Fig. 4B and Fig. S3). In contrast, pps1Δ (P = 0.1058), igp1Δ (P = 0.3778), and ico1Δ (P = 0.5006) mutants did not show significant differences in polar growth compared to that of the wild-type virulent strain. Together, these germination and developmental defects suggest that the atf1, atf2, gcn4, aqp1, pps1, and chi1 genes, which were upregulated during phagocytosis, have a major role in Mucor surviving and escaping the phagosome.
Interaction of macrophages with deletion mutant spores. Micrographs of spores from each deletion mutant after 5 h inside the phagosome of mouse macrophages. Download FIG S3, PDF file, 1.8 MB (1.8MB, pdf) .
Copyright © 2019 Pérez-Arques et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In vitro defects of the mutant strains could have an impact on virulence because the ability to circumvent the host innate immunity is critical for dissemination. To determine the influence of these genes on infection, we performed virulence assays in a host-pathogen infection model. Immunosuppressed mice were challenged with spores from the mutants, the virulent strain R7B, and the avirulent strain NRRL3631, and survival was monitored for 20 days (Fig. 4C). Differences in survival rates, represented in a Kaplan-Meier curve for each group of mice, were considered statistically significant with a P value of ≤0.05 in a Mantel-Cox test. atf1Δ (P < 0.0001), atf2Δ (P = 0.0008), gcn4Δ (P = 0.0037), pps1Δ (P = 0.0007), aqp1Δ (P = 0.0021), and chi1Δ (P = 0.0040) mutants were significantly attenuated in virulence compared to the virulent strain R7B; conversely, mutants with igp1Δ (P = 0.1021) and ico1Δ (P = 0.5104) mutations did not show significant differences in virulence. These differences in virulence were replicated in a second virulence assay using independently generated mutants (Fig. S4), supporting the idea that the attenuation in virulence was the result of gene deletion. This correlation between in vitro and in vivo virulence phenotypes highlights the importance of the fungal response to host innate immunity.
Virulence assay in mice with independently generated homokaryons. Each color shows the survival curve for a group of 10 immunosuppressed mice infected with 1 × 106 spores from one of the deletion mutant strains. Survival rates were compared to the survival of mice infected with a virulent control strain (R7B) and statistically analyzed by a Mantel-Cox test. NRRL3631 was used as an avirulent control strain. Asterisks indicate a significant difference (*, P ≤ 0.05; **, P ≤ 0.005). Download FIG S4, PDF file, 0.06 MB (59KB, pdf) .
Copyright © 2019 Pérez-Arques et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
atf1 and atf2 genes induce pathways related to virulence processes in mucormycosis.
The Atf1 and Atf2 transcription factors appear to be important in the response to host innate immunity, considering that their deletion results in the highest reduction in virulence (Fig. 4C). Knowing which genes are under their control could provide new insights into specific cellular processes involved in virulence. Thus, to increase the scope of the analysis, we analyzed the transcriptomes of atf1Δ and atf2Δ mutants under two different conditions: cocultured with mouse macrophages for 5 h as previously described and in liquid MMC medium (pH 4.5) for 24 h. Total RNA was extracted, purified, and sequenced as in previous RNA-seq experiments with wild-type strains.
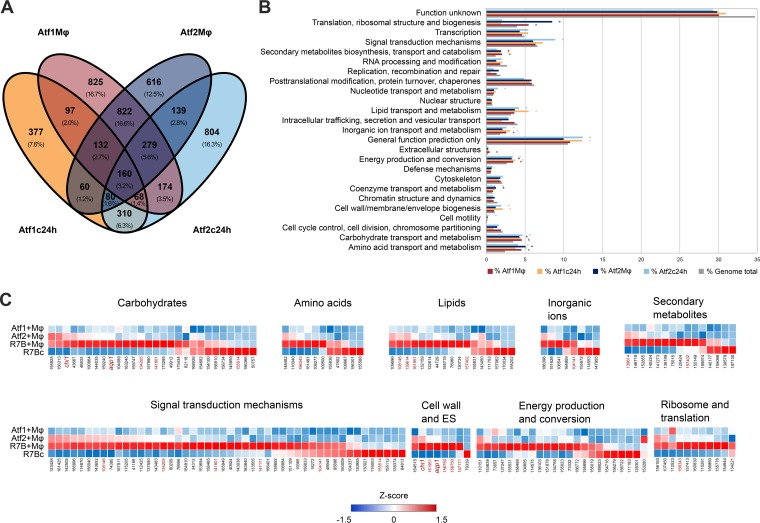
Under these conditions, 3,384 genes were differentially expressed in the atf1Δ mutant compared to their expression in the wild-type virulent strain (R7B). Similarly, 3,644 genes were differentially expressed in the atf2Δ mutant (Q ≤ 0.05) (Data Set S3). These results revealed that atf1 and atf2 encode transcription factors that control the expression of a considerable number of target genes. Interestingly, more than 40% of these genes are similarly regulated by both transcription factors (Fig. 5A), suggesting that they function in interacting pathways or are partially redundant.
FIG 5.
Atf1- and Atf2-regulated genes. (A) Venn diagram showing the relationships between differentially expressed genes (Q ≤ 0.05) in Mucor atf1Δ and atf2Δ mutants. Differential expression values were calculated as the ratio between the expression in virulent wild-type strain R7B and in each depicted mutant, either cocultured with mouse macrophages (cell line J774A.1) in L15 cell culture medium for 5 h (Mφ) or grown in liquid MMC, pH 4.5, for 24 h (c24h). Overlapping genes shared similar expression values (up- or downregulation) under the depicted conditions. (B) Percentages of genes regulated by Atf1 (red and orange) and Atf2 (dark and light blue) and of all genes found in the genome (gray) for each KOG class. Asterisks (*) indicate a significant enrichment (P ≤ 0.05, Fisher’s exact test) in the corresponding KOG class with respect to the expected number of genes found in the Mucor genome. (C) Analysis of genes upregulated (Q ≤ 0.05, fold change of ≥2.0) in the virulent wild-type strain R7B compared to their expression in either the Mucor atf1Δ or atf2Δ mutant cocultured with mouse macrophages (cell line J774A.1) for each enriched KOG class found in panel B. Z score was calculated as the deviation from the mean FPKM (fragments per kilobase of transcript per million mapped reads) value in standard deviation units for Mucor atf1Δ and atf2Δ mutants and strain R7B cocultured with mouse macrophages (Atf1+Mφ, Atf2+Mφ, and R7B+Mφ) and for R7B without mouse macrophages (R7Bc). Red gene IDs appear in more than one KOG class.
Differentially expressed genes in Mucor atf1 and atf2 mutant strains after 5-h interaction with mouse macrophages. Expression values of Mucor R7B wild-type strain and atf1 and atf2 mutants interacting with mouse macrophages for 5 h (R7BMφ, Atf1Mφ, and Atf2Mφ, respectively) and grown in solid MMC medium for 24 h (R7Bc24h, Atf1c24h, and Atf2c24h, respectively). Download Data Set S3, XLSX file, 4.0 MB (4MB, xlsx) .
Copyright © 2019 Pérez-Arques et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A functional enrichment analysis of differentially expressed genes was performed to determine the biological processes controlled by atf1 and atf2 (Fig. 5B and Data Set S3). As expected by the high proportion of genes regulated by both atf1 and atf2, they appear to share functions. After 5 h inside the phagosome, both transcription factors induced translation and ribosomal processes, carbon and nitrogen metabolism, oxidation-reduction processes, and carotenoid biosynthesis. atf1 and atf2 also promoted cell wall and membrane biogenesis and lipid metabolism when the spores were allowed to germinate and grow for 24 h in MMC medium. Regarding nonshared functions, atf2 controlled specific sets of genes involved in the high-affinity iron uptake mechanism and cytoskeletal changes that could be mediated by cAMP-dependent signaling during macrophage interaction (Data Set S3). High percentages of genes found in these enriched functional categories are upregulated by both transcription factors, being induced in the wild-type virulent strain when challenged by macrophages (Fig. 5C). This analysis revealed that the chi1 gene is upregulated by Atf1, whereas aqp1 is upregulated by both Atf1 and Atf2, when Mucor spores are internalized by the macrophages.
After phagocytosis, Mucor spores must confront a cytotoxic environment within the phagosome that includes acidification, nutrient starvation, oxidation, and antimicrobial proteins. To unveil the role of this ATF pathway in surviving phagocytosis, the germination of the mutants in atf1 and atf2 and in their regulated genes was tested at different pHs in yeast nitrogen base (YNB) minimal medium. After a 5-h incubation, atf1Δ and atf2Δ mutants, as well as chi1Δ and aqp1Δ mutants, showed significant decreases in their polar growth indices at acidic pH compared to the results for the virulent wild-type strain (Fig. S5A). These differences were not present either when the spores were cultured at neutral pH or in mutants of genes which are not controlled by either ATF (Fig. S5A and B). These results suggest that germination at acidic pH, equivalent to the intraphagosomal environment (30), relies on the ATF pathway.
Acidic pH affects germination in mutants involved in the ATF pathway. Polarity index measures after 5-h cultures of the depicted mutants in YNB at pH 4.0 (A) and pH 7.0 (B). Error bars correspond to the SEM of the results from technical replicates (n = 40 spores), and asterisks indicate a significant difference compared to the results for the virulent wild-type strain as determined by the unpaired t test (*, P ≤ 0.05; **, P ≤ 0.005; ***, P < 0.0001). Download FIG S5, PDF file, 0.2 MB (155KB, pdf) .
Copyright © 2019 Pérez-Arques et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
In this study, we have identified and functionally characterized the Mucor response to host innate immune cells, unveiling key elements involved in mucoralean virulence. The transcriptomic profiles of two Mucor strains—one virulent and one avirulent (22)— during mouse macrophage phagocytosis have identified a robust and mutual response between the virulent fungal strain and its host. Prompted by virulent Mucor spores, the macrophages induce proinflammatory cytokines and apoptotic processes.
Triggering programmed cell death to evade the immune response is a common mechanism in well-studied fungal pathogens (31–33). Recent studies have described in vivo formation of innate granulomas in zebrafish upon Mucor infection, formed by phagocytic cells recruited early to the site of infection (34) and apoptosis of infected macrophages (35). The link connecting macrophage apoptosis and the formation of innate granulomas in mucormycosis remains unexplored and represents a promising field to address in future research.
We have thoroughly studied Mucor transcriptomic responses to macrophages, revealing for the first time which molecular processes are necessary to survive phagocytosis. Macronutrient metabolic pathways are greatly upregulated in the phagocytosed spores, suggesting that Mucor is able to shift toward alternative nutrient sources as a consequence of nutrient scarcity within the phagosome (36). Such changes induced by phagocytosis were previously observed in Candida albicans, which shifts from a glycolytic program to gluconeogenesis, activation of the glyoxylate cycle, and β-oxidation of fatty acids (37). The gcn4 gene in engulfed Mucor spores was upregulated; gcn4 is a putative homolog of the Saccharomyces cerevisiae gene encoding the essential transcriptional activator of amino acid biosynthesis GCN4 (38). Given that amino acid starvation is a well-studied morphogenetic signal that induces hyphal development in Candida albicans (39), gcn4 could be orchestrating this metabolic shift that allows Mucor spores to fully germinate inside the phagosome and escape phagocytosis. Indeed, gcn4 deletion causes severe germination and growth defects in the spores after phagocytosis, having a significant impact on the development of mucormycosis in mice.
Surface or secreted proteins could be involved in defense mechanisms and mediate host-pathogen interaction (22). Among genes encoding proteins with a secretion signal, those with no predicted domains were selected under the assumption that they could be specific to Mucorales and therefore participate in the unique properties of mucormycosis. Deletion mutants lacking the chi1 gene were affected in survival after phagocytosis and germination inside the phagosome, showing a delay in polar growth that could explain their reduced virulence phenotype in vivo. Similarly, pps1Δ mutants had growth defects after being phagocytosed and a drastic reduction in virulence potential in vivo. However, germination in chi1Δ mutants was impaired inside the phagosome, whereas pps1Δ mutants were not affected in this process. Pps1 is highly conserved among mucormycosis agents (≥70% identity) and is a putative ortholog of the Cryptococcus neoformans immunoreactive mannoprotein MP88, which elicits an immune response in T cells (40). Resembling MP88, Pps1 contains a serine-/threonine-rich region that could serve as a site for extensive O-mannosylation (41), allowing its recognition by macrophage mannose receptors (42). Taken together, these results point to a possible role in mediating cell-to-cell processes involved in the interaction of Mucor with macrophages, rather than germination and hyphal growth.
Virulent Mucor spores can germinate inside the phagosome, first undergoing an isotropic stage during which spores increase their diameter, followed by rapid germ tube elongation that results in hyphal polar growth (16). During these growth stages inside the phagosome, functions related to cell wall and membrane biogenesis were upregulated in Mucor spores, indicating a crucial role in remodeling the cell surface for rapid polar growth inside the phagosome. We generated deletion mutants lacking aqp1, a putative aquaporin that could be related to membrane and cell wall modifications during pathogenesis. Several aquaporins are involved during osmotic and oxidative stresses to regulate competitive fitness in fungi (43, 44). Deletion of Mucor aqp1 resulted in severe defects in germination and polar growth inside the phagosome, suggesting that water or small molecule transport through the cell is essential for Mucor spores to start swelling at the isometric stage before hyphal growth.
Two basic leucine zipper transcription factors were highly induced during in vitro and in vivo macrophage phagocytosis; the genes encoding them were named atf1 and atf2 due to their high similarity with Schizosaccharomyces pombe Atf1 (45). ATF/CREB transcription factors are critical for the response to oxidative stress in many fungi, especially filamentous species (46, 47). Both atf1 and atf2 are needed to germinate and develop hyphae when the spores are engulfed by macrophages, and the lack of either of them results in a critical decrease in fitness after phagocytosis. The inability of these mutants to escape the host innate immune response leads to a drastic reduction in virulence, highlighting their importance in Mucor pathogenesis. As expected, the transcriptomic analyses of these two mutants suggest not only that atf1 and atf2 are involved in the response to oxidative stress but also that they participate in macronutrient metabolism. Both transcription factors appear to contribute to the control of these processes, because they regulate a broad number of shared genes in a manner similar to that of Aspergillus atfA and atfB (48) or S. pombe atf1 and pcr1 (49). More importantly, aqp1 and chi1 are induced by these transcription factors when Mucor spores are phagocytosed, indicating that they are involved in the same signaling pathway which is activated to respond to the macrophages. We hypothesized that this ATF-mediated pathway is induced by the acidic intraphagosomal environment and has a role in germination at low pH, since aqp1 is regulated by both ATFs and similar mucoralean aquaporins are expressed at acidic pH (50). Indeed, both atf1 and atf2 and chi1 and aqp1 mutants showed germination defects only at acidic pH, indicating that these genes are involved in the germination process at the phagosomal pH. Our findings support this hypothesis, since mutants in all four genes are also unable to germinate inside the phagosome. This inability to survive the phagosome correlates with a drastic reduction in virulence, suggesting that germinating inside the phagosome at early stages is crucial to evade the host innate defenses and disseminate the infection.
In summary, this study presents a thorough transcriptomic and functional analysis of the interaction of mucoralean spores with phagocytic cells. Mucoralean spores adapt to the host-cell environment by developing an intricate gene response during phagocytosis, which coordinates vital cellular functions involved in germinating within the phagosome and surviving its hostile environment. Indeed, a functional analysis of key genes involved in these cellular functions revealed their role in germinating inside host cells, surviving phagocytosis, and causing infection in mammalian models. Overall, our results provide a better understanding of the molecular mechanisms exploited by these pathogens to evade the innate immune response and invade the host, which could contribute to the development of efficient antifungal therapies.
MATERIALS AND METHODS
Fungal strains, cell cultures, and growth conditions.
M. circinelloides f. lusitanicus strains are listed in Table S2 in the supplemental material. Strain R7B (51), a leucine auxotroph derived from strain CBS277.49, was used as a wild-type virulent strain, whereas wild-type strain NRRL3631, an environmental isolate, was used as an avirulent strain (22). All mutant strains generated in this work derived from strain MU402 (52), a double auxotroph for uracil and leucine that shows virulence similar to that of its parental strain R7B (53).
Mucor cultures were grown at 26°C in yeast nitrogen base (YNB) medium (19), minimal medium with Casamino Acids (MMC) (52), or rich yeast extract-peptone-glucose (YPG) (54) medium, with pH 4.5 for optimal growth and sporulation or pH 3.2 for colonial growth. Media were supplemented with uridine (200 mg/liter) or leucine (20 mg/liter) when required for auxotrophy. For germination assays, YNB liquid medium was adjusted to pHs 4.0 and 7.0 using 1 M NaOH or HCl.
In vitro host-pathogen interaction assays were prepared by coculturing a mouse macrophage cell line (J774A.1; ATCC TIB-67) with Mucor spores in cell culture medium L15 (Capricorn Scientific GmbH) at 37°C, supplemented with 10% fetal bovine serum (FBS). Amounts of approximately 1.5 × 107 spores of the designated strains were added to the cell cultures, maintaining a proportion of 1.5 spores per macrophage cell. For noninteraction control samples, the same amounts of Mucor spores and macrophage cells were cultured separately under the same conditions as the cocultures.
RNA sequencing and analysis.
In vitro host-pathogen interaction assays were cultured for 5 h with macrophages and spores from designated Mucor strains, along with their appropriate noninteraction controls. When required as an additional control condition, Mucor cultures were grown in liquid MMC (pH 4.5) at 26°C for 24 h. Two replicates of each coculture and control samples were scraped from the plates, and total RNA was extracted using the RNeasy plant minikit (Qiagen, Hilden, Germany) following the supplier’s recommendations. mRNA enrichment by poly(A) purification capture and cDNA libraries were prepared at BaseClear (Leiden, The Netherlands) using TruSeq RNA library preparation kits. The cDNA library was sequenced using the Illumina HiSeq 2500 system.
Quality control of the sequence data sets was conducted with FastQC and Trim Galore! (http://www.bioinformatics.babraham.ac.uk/projects/). Since coculture data sets contained a mixture of fungal and mouse sequences, reads were aligned to each of the respective reference genomes, M. circinelloides version 2.0 and Mus musculus GRCm38, using STAR (55). Differential expression analyses were performed by CuffDiff (56) to calculate significant log2-fold changes in gene expression. The M. circinelloides version 2.0 consensus gene annotation or Mus musculus mm10 gene annotation was used accordingly to provide functional information on the gene sets from each species. Functional enrichment of Gene Ontology (GO) terms and KEGG pathways was determined in selected mouse gene sets using g:Profiler (57), while enrichment of Eukaryotic Orthologous Groups (KOG) classes, GO temrs, and KEGG pathways was analyzed on selected Mucor gene clusters at FungiDB (58).
RT-qPCR gene expression analysis.
The expression values of selected genes were measured by RT-qPCR, both in vitro and in vivo. Total RNA was isolated from in vitro host-pathogen interaction replicates in which Mucor wild-type strains R7B and NRRL3631 and atf1Δ and atf2Δ mutants were cultured with mouse macrophages, as described above.
For in vivo quantification of gene expression, amounts of 1 × 107 spores of each tested strain were injected intraperitoneally into OF-1 mice. Mice were euthanized 5 h postinjection, and 5 ml of L15 cell culture medium was injected into the peritoneal cavity to recover primary macrophage cells. Peritoneal fluid was collected after a brief abdominal massage and examined under the microscope for cell identification and fungal spore quantification. Total RNA was isolated from pooled samples from three infected mice for each fungal strain.
For RT-qPCR assays, cDNA was synthesized from 1 µg of total RNA using the iScript cDNA synthesis kit (Bio-Rad). Real-time PCR was performed in triplicate with a QuantStudio real-time PCR system (Applied Biosystems) using 2× SYBR green PCR master mix (Applied Biosystems) following the supplier’s recommendations. Primer sequences are shown in Table S1. In order to confirm the absence of nonspecific amplification, a nontemplate control and a melting curve were included. The efficiency of the target gene amplification and the efficiency of the 18S RNA gene (endogenous control) amplification were approximately equal, so the relative expression of the candidate gene could be obtained using the cycle threshold (ΔΔCT) method, normalizing for 18S RNA.
Mutant generation by genetic transformation.
Single deletion mutants were obtained by gene replacement through homologous recombination. Disruption fragments contained a 2-kb fragment of the selectable marker pyrG flanked by 1-kb upstream and downstream sequences of the targeted gene’s open reading frame. These fragments were PCR amplified separately and then fused by overlapping PCR using specific primers for each deleted gene (Table S1).
MU402 (pyrG− leuA−) protoplasts were transformed by electroporation with approximately 5 µg of a linear disruption fragment following a standardized procedure (59). Transformed protoplasts were grown in MMC selective medium (i.e., without uracil). Genomic DNA (gDNA) was extracted from selected uracil-prototrophic transformants as previously described (18). Deletions were confirmed by PCR amplification with discriminatory primers (Table S1). Mutants underwent 10 cycles of vegetative sporulation and single-colony isolation in selective medium to achieve homokaryosis, which was confirmed by Southern blotting. Specific DNA probes that discriminate between the wild-type and mutant alleles of each gene were obtained by PCR amplification and labeled with [α-32P]dCTP using Ready-To-Go labeling beads (GE Healthcare Life Science). After stringent hybridization (60), membranes were developed in a phosphor screen using a Personal Molecular Imager system (Bio-Rad).
In vitro host-pathogen survival assay.
The survival of all mutants was analyzed by conducting in vitro host-pathogen assays as described previously. Spores from two independently generated homokaryotic mutants for each gene were added to the macrophage cells (four interaction replicates per deleted gene). After 30 min of incubation, wells were washed with phosphate-buffered saline (PBS) to ensure that every spore remaining was phagocytosed. Then, the interaction was maintained for 5 h. As a noninteraction control, spores were incubated for 5 h in the same cell culture medium but without macrophages. Then, 10 micrographs were taken from each well in which the 5-h-interaction assay was conducted to measure 50 germinating spores using ImageJ software. The polarity index was calculated as the quotient between spore length and width; significant differences were estimated with an unpaired t test. Subsequently, macrophages were lysed with 0.1% NP-40 (Sigma-Aldrich) to release Mucor spores, which were collected and plated in MMC medium at pH 3.2 (three replicates of 500 spores per independent mutant and condition). The survival of the spores was assessed by their ability to germinate and develop healthy colonies after 48 h of incubation at 26°C.
Virulence assays.
OF-1 male mice weighing ≥30 g (Charles River, Barcelona, Spain) were used as the host model for virulence assays. Groups of 10 mice were immunosuppressed 2 days prior to infection by intraperitoneal administration of cyclophosphamide (200 mg/kg of body weight) and once every 5 days thereafter. Animals were housed under established conditions with free access to food and autoclaved water. Groups of mice were challenged intravenously with suspensions of 1 × 106 spores collected from each of the mutant or control strains. The survival rate of each group of mice was monitored twice a day, and animals meeting criteria for the endpoint were euthanized by CO2 inhalation.
Ethics statement.
The experiments performed complied with the Guidelines of the European Union Council (Directive 2010/63/EU) and the Spanish RD 53/2013, which is intended to ensure the welfare of animals and the ethics of any procedure related to animal experimentation. Experiments and procedures were supervised and approved by the University of Murcia Animal Welfare and Ethics Committee and the Consejería de Agua, Agricultura, Ganadería y Pesca de la CARM, Spain (authorization number REGA ES300305440012).
Data availability.
All data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (61) and are accessible through GEO Series accession number GSE117385.
ACKNOWLEDGMENTS
This work was funded by Ministerio de Economía y Competitividad, Spain (grant number BFU2015-65501-P, cofinanced by FEDER and RYC-2014-15844) and Ministerio de Educación, Cultura y Deporte, Spain (grants number FPU14/01832, FPU14/01983, and FPU17/05814).
Author contributions were as follows: conceptualization, F.E.N., V.G., J.H., C.P.-A., and M.I.N.-M.; methodology, F.E.N., M.I.N.-M., C.P.-A., and L.M.; investigation, C.P.-A., M.I.N.-M., L.M., F.E.N., C.L., and P.M.-G.; writing of the original draft, C.P.-A., M.I.N.-M., and F.E.N.; review and editing of the manuscript, C.P.-A., M.I.N.-M., L.M., F.E.N., V.G., and J.H.; visualization, C.P.-A. and M.I.N.-M.; and supervision and funding acquisition, V.G. and F.E.N.
The authors declare no competing interests.
Footnotes
Citation Pérez-Arques C, Navarro-Mendoza MI, Murcia L, Lax C, Martínez-García P, Heitman J, Nicolás FE, Garre V. 2019. Mucor circinelloides thrives inside the phagosome through an Atf-mediated germination pathway. mBio 10:e02765-18. https://doi.org/10.1128/mBio.02765-18.
Contributor Information
Anuradha Chowdhary, Vallabhbhai Patel Chest Institute.
Alexander Idnurm, University of Melbourne.
James Kronstad, University of British Columbia.
REFERENCES
- 1.Lackner M, Caramalho R, Lass-Flörl C. 2014. Laboratory diagnosis of mucormycosis: current status and future perspectives. Future Microbiol 9:683–695. doi: 10.2217/fmb.14.23. [DOI] [PubMed] [Google Scholar]
- 2.Petrikkos G, Skiada A, Lortholary O, Roilides E, Walsh TJ, Kontoyiannis DP. 2012. Epidemiology and clinical manifestations of mucormycosis. Clin Infect Dis 54:S23–S34. doi: 10.1093/cid/cir866. [DOI] [PubMed] [Google Scholar]
- 3.Roden MM, Zaoutis TE, Buchanan WL, Knudsen TA, Sarkisova TA, Schaufele RL, Sein M, Sein T, Chiou CC, Chu JH, Kontoyiannis DP, Walsh TJ. 2005. Epidemiology and outcome of zygomycosis: a review of 929 reported cases. Clin Infect Dis 41:634–653. doi: 10.1086/432579. [DOI] [PubMed] [Google Scholar]
- 4.Spatafora JW, Chang Y, Benny GL, Lazarus K, Smith ME, Berbee ML, Bonito G, Corradi N, Grigoriev I, Gryganskyi A, James TY, O’Donnell K, Roberson RW, Taylor TN, Uehling J, Vilgalys R, White MM, Stajich JE. 2016. A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 108:1028–1046. doi: 10.3852/16-042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann K, Pawłowska J, Walther G, Wrzosek M, de Hoog GS, Benny GL, Kirk PM, Voigt K. 2013. The family structure of the Mucorales: a synoptic revision based on comprehensive multigene-genealogies. Persoonia 30:57–76. doi: 10.3767/003158513X666259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ribes JA, Vanover-Sams CL, Baker DJ. 2000. Zygomycetes in human disease. Clin Microbiol Rev 13:236–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kontoyiannis DP, Yang H, Song J, Kelkar SS, Yang X, Azie N, Harrington R, Fan A, Lee E, Spalding JR. 2016. Prevalence, clinical and economic burden of mucormycosis-related hospitalizations in the United States: a retrospective study. BMC Infect Dis 16:730. doi: 10.1186/s12879-016-2023-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mignogna MD, Fortuna G, Leuci S, Adamo D, Ruoppo E, Siano M, Mariani U. 2011. Mucormycosis in immunocompetent patients: a case-series of patients with maxillary sinus involvement and a critical review of the literature. Int J Infect Dis 15:e533–e540. doi: 10.1016/j.ijid.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 9.Calo S, Shertz-Wall C, Lee SC, Bastidas RJ, Nicolás FE, Granek JA, Mieczkowski P, Torres-Martínez S, Ruiz-Vázquez RM, Cardenas ME, Heitman J. 2014. Antifungal drug resistance evoked via RNAi-dependent epimutations. Nature 513:555–558. doi: 10.1038/nature13575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caramalho R, Tyndall JDA, Monk BC, Larentis T, Lass-Flörl C, Lackner M. 2017. Intrinsic short-tailed azole resistance in mucormycetes is due to an evolutionary conserved aminoacid substitution of the lanosterol 14α-demethylase. Sci Rep 7:3–12. doi: 10.1038/s41598-017-16123-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cornely OA, Arikan-Akdagli S, Dannaoui E, Groll AH, Lagrou K, Chakrabarti A, Lanternier F, Pagano L, Skiada A, Akova M, Arendrup MC, Boekhout T, Chowdhary A, Cuenca-Estrella M, Freiberger T, Guinea J, Guarro J, de Hoog S, Hope W, Johnson E, Kathuria S, Lackner M, Lass-Flörl C, Lortholary O, Meis JF, Meletiadis J, Muñoz P, Richardson M, Roilides E, Tortorano AM, Ullmann AJ, van Diepeningen A, Verweij P, Petrikkos G. 2014. ESCMID and ECMM joint clinical guidelines for the diagnosis and management of mucormycosis 2013. Clin Microbiol Infect 20:5–26. doi: 10.1111/1469-0691.12371. [DOI] [PubMed] [Google Scholar]
- 12.Calo S, Nicolás FE, Lee SC, Vila A, Cervantes M, Torres-Martinez S, Ruiz-Vazquez RM, Cardenas ME, Heitman J. 2017. A non-canonical RNA degradation pathway suppresses RNAi-dependent epimutations in the human fungal pathogen Mucor circinelloides. PLoS Genet 13:e1006686. doi: 10.1371/journal.pgen.1006686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gebremariam T, Liu M, Luo G, Bruno V, Phan QT, Waring AJ, Edwards JE, Filler SG, Yeaman MR, Ibrahim AS. 2014. CotH3 mediates fungal invasion of host cells during mucormycosis. J Clin Invest 124:237–250. doi: 10.1172/JCI71349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ibrahim AS, Spellberg B, Edwards J. 2008. Iron acquisition: a novel perspective on mucormycosis pathogenesis and treatment. Curr Opin Infect Dis 21:620–625. doi: 10.1097/QCO.0b013e3283165fd1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee SC, Li A, Calo S, Heitman J. 2013. Calcineurin plays key roles in the dimorphic transition and virulence of the human pathogenic zygomycete Mucor circinelloides. PLoS Pathog 9:e1003625. doi: 10.1371/journal.ppat.1003625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li CH, Cervantes M, Springer DJ, Boekhout T, Ruiz-Vazquez RM, Torres-Martinez SR, Heitman J, Lee SC. 2011. Sporangiospore size dimorphism is linked to virulence of Mucor circinelloides. PLoS Pathog 7:e1002086. doi: 10.1371/journal.ppat.1002086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu M, Lin L, Gebremariam T, Luo G, Skory CD, French SW, Chou TF, Edwards JE, Ibrahim AS. 2015. Fob1 and Fob2 proteins are virulence determinants of Rhizopus oryzae via facilitating iron uptake from ferrioxamine. PLoS Pathog 11:e1004842. doi: 10.1371/journal.ppat.1004842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Navarro-Mendoza MI, Pérez-Arques C, Murcia L, Martínez-García P, Lax C, Sanchis M, Capilla J, Nicolás FE, Garre V. 2018. Components of a new gene family of ferroxidases involved in virulence are functionally specialized in fungal dimorphism. Sci Rep 8:7660. doi: 10.1038/s41598-018-26051-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicolás FE, Navarro-Mendoza MI, Pérez-Arques C, López-García S, Navarro E, Torres-Martínez S, Garre V. 2018. Molecular tools for carotenogenesis analysis in the mucoral Mucor circinelloides. Methods Mol Biol 1852:221–237. doi: 10.1007/978-1-4939-8742-9_13. [DOI] [PubMed] [Google Scholar]
- 20.Vellanki S, Navarro-Mendoza MI, Garcia A, Murcia L, Perez-Arques C, Garre V, Nicolas FE, Lee SC. 2018. Mucor circinelloides: growth, maintenance, and genetic manipulation. Curr Protoc Microbiol 49:e53. doi: 10.1002/cpmc.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trieu TA, Navarro-Mendoza MI, Pérez-Arques C, Sanchis M, Capilla J, Navarro-Rodriguez P, Lopez-Fernandez L, Torres-Martínez S, Garre V, Ruiz-Vázquez RM, Nicolás FE. 2017. RNAi-based functional genomics identifies new virulence determinants in mucormycosis. PLoS Pathog 13:e1006150. doi: 10.1371/journal.ppat.1006150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.López-Fernández L, Sanchis M, Navarro-Rodríguez P, Nicolás FE, Silva-Franco F, Guarro J, Garre V, Navarro-Mendoza MI, Pérez-Arques C, Capilla J. 2018. Understanding Mucor circinelloides pathogenesis by comparative genomics and phenotypical studies. Virulence 9:707–720. doi: 10.1080/21505594.2018.1435249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ibrahim AS, Voelz K. 2017. The mucormycete–host interface. Curr Opin Microbiol 40:40–45. doi: 10.1016/j.mib.2017.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheldon WH, Bauer H. 1959. The development of the acute inflammatory response to experimental cutaneous mucormycosis in normal and diabetic rabbits. J Exp Med 110:845–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waldorf AR, Levitz SM, Diamond RD. 1984. In vivo bronchoalveolar macrophage defense against Rhizopus oryzae and Aspergillus fumigatus. J Infect Dis 150:752–760. [DOI] [PubMed] [Google Scholar]
- 26.Voelz K, Gratacap RL, Wheeler RT. 2015. A zebrafish larval model reveals early tissue-specific innate immune responses to Mucor circinelloides. Dis Model Mech 8:1375–1388. doi: 10.1242/dmm.019992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katragkou A, Walsh TJ, Roilides E. 2014. Why is mucormycosis more difficult to cure than more common mycoses? Clin Microbiol Infect 20:74–81. doi: 10.1111/1469-0691.12466. [DOI] [PubMed] [Google Scholar]
- 28.Chamilos G, Lewis RE, Hu J, Xiao L, Zal T, Gilliet M, Halder G, Kontoyiannis DP. 2008. Drosophila melanogaster as a model host to dissect the immunopathogenesis of zygomycosis. Proc Natl Acad Sci U S A 105:9367–9372. doi: 10.1073/pnas.0709578105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chibucos MC, Soliman S, Gebremariam T, Lee H, Daugherty S, Orvis J, Shetty AC, Crabtree J, Hazen TH, Etienne KA, Kumari P, O’Connor TD, Rasko DA, Filler SG, Fraser CM, Lockhart SR, Skory CD, Ibrahim AS, Bruno VM. 2016. An integrated genomic and transcriptomic survey of mucormycosis-causing fungi. Nat Commun 7:12218. doi: 10.1038/ncomms12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geisow MJ, D’Arcy Hart P, Young MR. 1981. Temporal changes of lysosome and phagosome pH during phagolysosome formation in macrophages: studies by fluorescence spectroscopy. J Cell Biol 89:645–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shlezinger N, Irmer H, Dhingra S, Beattie SR, Cramer RA, Braus GH, Sharon A, Hohl TM. 2017. Sterilizing immunity in the lung relies on targeting fungal apoptosis-like programmed cell death. Science 357:1037–1041. doi: 10.1126/science.aan0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ibata-Ombetta S, Idziorek T, Trinel P-A, Poulain D, Jouault T. 2003. Candida albicans phospholipomannan promotes survival of phagocytosed yeasts through modulation of bad phosphorylation and macrophage apoptosis. J Biol Chem 278:13086–13093. doi: 10.1074/jbc.M210680200. [DOI] [PubMed] [Google Scholar]
- 33.Chiapello LS, Aoki MP, Rubinstein HR, Masih DT. 2003. Apoptosis induction by glucuronoxylomannan of Cryptococcus neoformans. Med Mycol 41:347–353. [DOI] [PubMed] [Google Scholar]
- 34.Inglesfield S, Jasiulewicz A, Hopwood M, Tyrrell J, Youlden G, Mazon-Moya M, Millington OR, Mostowy S, Jabbari S, Voelz K. 2018. Robust phagocyte recruitment controls the opportunistic fungal pathogen Mucor circinelloides in innate granulomas in vivo. mBio 9:e02010-17. doi: 10.1128/mBio.02010-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.López-Muñoz A, Nicolás FE, García-Moreno D, Pérez-Oliva AB, Navarro-Mendoza MI, Hernández-Oñate MA, Herrera-Estrella A, Torres-Martínez S, Ruiz-Vázquez RM, Garre V, Mulero V. 2018. An adult zebrafish model reveals that mucormycosis induces apoptosis of infected macrophages. Sci Rep 8:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Casadevall A. 2008. Evolution of intracellular pathogens. Annu Rev Microbiol 62:19–33. doi: 10.1146/annurev.micro.61.080706.093305. [DOI] [PubMed] [Google Scholar]
- 37.Fernández-Arenas E, Cabezón V, Bermejo C, Arroyo J, Nombela C, Diez-Orejas R, Gil C. 2007. Integrated proteomics and genomics strategies bring new insight into Candida albicans response upon macrophage interaction. Mol Cell Proteomics 6:460–478. doi: 10.1074/mcp.M600210-MCP200. [DOI] [PubMed] [Google Scholar]
- 38.Natarajan K, Meyer MR, Belinda M, Slade D, Roberts C, Alan G, Marton MJ, Jackson BM, Hinnebusch AG. 2001. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol Cell Biol 21:4347–4368. doi: 10.1128/MCB.21.13.4347-4368.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tripathi G, Wiltshire C, Macaskill S, Tournu H, Budge S, Brown AJP. 2002. Gcn4 co-ordinates morphogenetic and metabolic responses to amino acid starvation in Candida albicans. EMBO J 21:5448–5456. doi: 10.1093/emboj/cdf507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang C, Nong S-H, Mansour MK, Specht CA, Levitz SM. 2002. Purification and characterization of a second immunoreactive mannoprotein from Cryptococcus neoformans that stimulates T-cell responses. Infect Immun 70:5485–5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loibl M, Strahl S. 2013. Protein O-mannosylation: what we have learned from baker’s yeast. Biochim Biophys Acta 1833:2438–2446. doi: 10.1016/j.bbamcr.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 42.Levitz SM. 2010. Innate recognition of fungal cell walls. PLoS Pathog 6:e1000758. doi: 10.1371/journal.ppat.1000758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meyers GL, Jung K, Bang S, Kim J, Kim S, Hong J, Cheong E, Kim KH, Bahn Y. 2017. The water channel protein aquaporin 1 regulates cellular metabolism and competitive fitness in a global fungal pathogen Cryptococcus neoformans. Environ Microbiol Rep 9:268–278. doi: 10.1111/1758-2229.12527. [DOI] [PubMed] [Google Scholar]
- 44.Sabir F, Loureiro-Dias MC, Soveral G, Prista C. 2017. Functional relevance of water and glycerol channels in Saccharomyces cerevisiae. FEMS Microbiol Lett 364:fnx080. doi: 10.1093/femsle/fnx080. [DOI] [PubMed] [Google Scholar]
- 45.Shiozaki K, Russell P. 1996. Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes Dev 10:2276–2288. [DOI] [PubMed] [Google Scholar]
- 46.Brown AJP, Cowen LE, di Pietro A, Quinn J. 2017. Stress adaptation, p 463–485. In Heitman J, Howlett B, Crous P, Stukenbrock E, James T, Gow N (ed), The fungal kingdom. ASM Press, Washington, DC. [Google Scholar]
- 47.Linz JE, Hong SY, Roze LV. 2013. Oxidative stress-related transcription factors in the regulation of secondary metabolism. Toxins (Basel) 5:683–702. doi: 10.3390/toxins5040683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lara-Rojas F, Sánchez O, Kawasaki L, Aguirre J. 2011. Aspergillus nidulans transcription factor AtfA interacts with the MAPK SakA to regulate general stress responses, development and spore functions. Mol Microbiol 80:436–454. doi: 10.1111/j.1365-2958.2011.07581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanso M, Gogol M, Ayte J, Seidel C, Hidalgo E. 2008. Transcription factors Pcr1 and Atf1 have distinct roles in stress and Sty1-dependent gene regulation. Eukaryot Cell 7:826–835. doi: 10.1128/EC.00465-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Turgeman T, Shatil-Cohen A, Moshelion M, Teper-Bamnolker P, Skory CD, Lichter A, Eshel D. 2016. The role of aquaporins in pH-dependent germination of Rhizopus delemar spores. PLoS One 11:e0150543. doi: 10.1371/journal.pone.0150543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roncero MI. 1984. Enrichment method for the isolation of auxotrophic mutants of Mucor using the polyene antibiotic N-glycosyl-polifungin. Carlsberg Res Commun 49:685–690. doi: 10.1007/BF02907499. [DOI] [Google Scholar]
- 52.Nicolás FE, de Haro JP, Torres-Martínez S, Ruiz-Vázquez RM. 2007. Mutants defective in a Mucor circinelloides dicer-like gene are not compromised in siRNA silencing but display developmental defects. Fungal Genet Biol 44:504–516. doi: 10.1016/j.fgb.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 53.Patiño-Medina JA, Maldonado-Herrera G, Pérez-Arques C, Alejandre-Castañeda V, Reyes-Mares NY, Valle-Maldonado MI, Campos-García J, Ortiz-Alvarado R, Jácome-Galarza IE, Ramírez-Díaz MI, Garre V, Meza-Carmen V. 2018. Control of morphology and virulence by ADP-ribosylation factors (Arf) in Mucor circinelloides. Curr Genet 64:853–869. doi: 10.1007/s00294-017-0798-0. [DOI] [PubMed] [Google Scholar]
- 54.Bartnicki-Garcia S, Nickerson WJ. 1962. Nutrition, growth, and morphogenesis of Mucor rouxii. J Bacteriol 84:841–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dobin A, Gingeras TR. 2015. Mapping RNA-seq reads with STAR. Curr Protoc Bioinform 51:11.14.1–11.14.19. doi: 10.1002/0471250953.bi1114s51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reimand J, Arak T, Adler P, Kolberg L, Reisberg S, Peterson H, Vilo J. 2016. g:Profiler—a Web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res 44:W83–W89. doi: 10.1093/nar/gkw199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stajich JE, Harris T, Brunk BP, Brestelli J, Fischer S, Harb OS, Kissinger JC, Li W, Nayak V, Pinney DF, Stoeckert CJ, Roos DS. 2012. FungiDB: an integrated functional genomics database for fungi. Nucleic Acids Res 40:D675–D681. doi: 10.1093/nar/gkr918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gutiérrez A, López-García S, Garre V. 2011. High reliability transformation of the basal fungus Mucor circinelloides by electroporation. J Microbiol Methods 84:442–446. doi: 10.1016/j.mimet.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 60.Nicolás FE, Torres-Martínez S, Ruiz-Vázquez RM. 2003. Two classes of small antisense RNAs in fungal RNA silencing triggered by non-integrative transgenes. EMBO J 22:3983–3991. doi: 10.1093/emboj/cdg384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Edgar R, Domrachev M, Lash AE. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30:207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hinnebusch AG. 1984. Evidence for translational regulation of the activator of general amino acid control in yeast. Proc Natl Acad Sci U S A 81:6442–6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Laizé V, Gobin R, Rousselet G, Badier C, Hohmann S, Ripoche P, Tacnet F. 1999. Molecular and functional study of AQY1 from Saccharomyces cerevisiae: role of the C-terminal domain. Biochem Biophys Res Commun 257:139–144. doi: 10.1006/bbrc.1999.0425. [DOI] [PubMed] [Google Scholar]
- 64.Colovos C, Cascio D, Yeates TO. 1998. The 1.8 Å crystal structure of the ycaC gene product from Escherichia coli reveals an octameric hydrolase of unknown specificity. Structure 6:1329–1337. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Differentially expressed genes in Mucor strains after 5-h interaction with mouse macrophages. Expression values of virulent Mucor strain R7B spores (R7BMφ) interacting with mouse macrophages and noninteraction controls (R7Bc) and of avirulent strain NRRL3631 spores (NRRL3631Mφ and NRRL3631c, respectively, for interaction and noninteraction controls). Download Data Set S1, XLSX file, 3.1 MB (3.1MB, xlsx) .
Copyright © 2019 Pérez-Arques et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Differentially expressed genes in mouse macrophages after 5-h interaction with Mucor. Expression values of mouse macrophages interacting either with virulent Mucor strain R7B spores (MφR7B) or avirulent strain NRRL3631 spores (MφNRRL3631) or without spores (Mφ). Genes with a mean FPKM of <1.0 were filtered out. Download Data Set S2, XLSX file, 1.3 MB (1.3MB, xlsx) .
Copyright © 2019 Pérez-Arques et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Relative levels of expression of proinflammatory cytokines in response to Mucor virulent strain. RT-qPCR-determined expression differences of Il1a and Tnf mouse macrophage genes during interaction with virulent and avirulent Mucor strains. Transcript levels were quantified in cDNA obtained from mouse macrophages (Mφ) (cell line J774A.1) cocultured with either virulent R7B spores or avirulent NRRL3631 spores in L15 cell culture medium for 5 h. Values were normalized using Actb as an internal control, and log2-fold change differential expression was calculated with respect to expression values in response to the avirulent Mucor strain. Error bars correspond to the SD from technical triplicates, and asterisks indicate a significant difference determined by unpaired t test (**, P ≤ 0.005; ***, P < 0.0001). Download FIG S1, PDF file, 0.03 MB (29.3KB, pdf) .
Copyright © 2019 Pérez-Arques et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in the study. Download Table S1, DOCX file, 0.02 MB (17.2KB, docx) .
Copyright © 2019 Pérez-Arques et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Disruptions of atf1, atf2, gcn4, pps1, aqp1, chi1, igp1, and ico1 genes. Schematic representation of the wild-type (WT) locus and the mutant (MUT) locus after homologous recombination with the disruption fragment for each gene. Restriction sites for each restriction enzyme used in the Southern blot assays are indicated, showing the expected size of the fragment generated after digestion. The positions of the probes used are indicated for each locus. Southern blot analyses are shown below the schemes. For this, genomic DNA (gDNA) of a WT strain and the gDNA of mutants obtained for each gene were digested with the depicted restriction enzyme. Hybridizations were made with the respective probes, which allowed discrimination between disrupted and WT alleles. The positions and sizes of the GeneRuler DNA ladder mixture (M) (Fermentas) are indicated. Download FIG S2, PDF file, 2.2 MB (2.2MB, pdf) .
Copyright © 2019 Pérez-Arques et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
M. circinelloides f. lusitanicus strains used in this study. Download Table S2, DOCX file, 0.01 MB (14.9KB, docx) .
Copyright © 2019 Pérez-Arques et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Interaction of macrophages with deletion mutant spores. Micrographs of spores from each deletion mutant after 5 h inside the phagosome of mouse macrophages. Download FIG S3, PDF file, 1.8 MB (1.8MB, pdf) .
Copyright © 2019 Pérez-Arques et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Virulence assay in mice with independently generated homokaryons. Each color shows the survival curve for a group of 10 immunosuppressed mice infected with 1 × 106 spores from one of the deletion mutant strains. Survival rates were compared to the survival of mice infected with a virulent control strain (R7B) and statistically analyzed by a Mantel-Cox test. NRRL3631 was used as an avirulent control strain. Asterisks indicate a significant difference (*, P ≤ 0.05; **, P ≤ 0.005). Download FIG S4, PDF file, 0.06 MB (59KB, pdf) .
Copyright © 2019 Pérez-Arques et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Differentially expressed genes in Mucor atf1 and atf2 mutant strains after 5-h interaction with mouse macrophages. Expression values of Mucor R7B wild-type strain and atf1 and atf2 mutants interacting with mouse macrophages for 5 h (R7BMφ, Atf1Mφ, and Atf2Mφ, respectively) and grown in solid MMC medium for 24 h (R7Bc24h, Atf1c24h, and Atf2c24h, respectively). Download Data Set S3, XLSX file, 4.0 MB (4MB, xlsx) .
Copyright © 2019 Pérez-Arques et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Acidic pH affects germination in mutants involved in the ATF pathway. Polarity index measures after 5-h cultures of the depicted mutants in YNB at pH 4.0 (A) and pH 7.0 (B). Error bars correspond to the SEM of the results from technical replicates (n = 40 spores), and asterisks indicate a significant difference compared to the results for the virulent wild-type strain as determined by the unpaired t test (*, P ≤ 0.05; **, P ≤ 0.005; ***, P < 0.0001). Download FIG S5, PDF file, 0.2 MB (155KB, pdf) .
Copyright © 2019 Pérez-Arques et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
All data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (61) and are accessible through GEO Series accession number GSE117385.