Antibiotic resistance is arguably the biggest current threat to global health. An increasing number of infections are becoming harder or almost impossible to treat, carrying high morbidity, mortality, and financial cost.
KEYWORDS: antibiotic resistance, bacteriophage therapy, bacteriophages
SUMMARY
Antibiotic resistance is arguably the biggest current threat to global health. An increasing number of infections are becoming harder or almost impossible to treat, carrying high morbidity, mortality, and financial cost. The therapeutic use of bacteriophages, viruses that infect and kill bacteria, is well suited to be part of the multidimensional strategies to combat antibiotic resistance. Although phage therapy was first implemented almost a century ago, it was brought to a standstill after the successful introduction of antibiotics. Now, with the rise of antibiotic resistance, phage therapy is experiencing a well-deserved rebirth. Among the admittedly vast literature recently published on this topic, this review aims to provide a forward-looking perspective on phage therapy and its role in modern society. We cover the key points of the antibiotic resistance crisis and then explain the biological and evolutionary principles that support the use of phages, their interaction with the immune system, and a comparison with antibiotic therapy. By going through up-to-date reports and, whenever possible, human clinical trials, we examine the versatility of phage therapy. We discuss conventional approaches as well as novel strategies, including the use of phage-antibiotic combinations, phage-derived enzymes, exploitation of phage resistance mechanisms, and phage bioengineering. Finally, we discuss the benefits of phage therapy beyond the clinical perspective, including opportunities for scientific outreach and effective education, interdisciplinary collaboration, cultural and economic growth, and even innovative use of social media, making the case that phage therapy is more than just an alternative to antibiotics.
INTRODUCTION
Bacteriophages (phages) are viruses capable of infecting and replicating within bacterial cells. They are the most abundant and ubiquitous organisms on Earth, playing important roles in microbial physiology, population dynamics, evolution, and therapeutics (1). The first anecdotal observations that could be interpreted as phage activity have been traced back to ancient and biblical times (2), while the formal history of phages began over a century ago with the work of Hankin, Gamaleya, Twort, and d’Herelle (3, 4). Felix d’Herelle first coined the term “bacteriophage” (4), literally meaning “bacterium eater,” and began using phages to treat bacterial infections in human patients (Fig. 1). Phages replicate through two primary life cycles, the dynamics of which have important implications for their therapeutic application. Virulent or obligate lytic phages infect and quickly kill their bacterial host cell, whereas temperate or lysogenic phages may either stably integrate into their host’s genome or enter into the lytic life cycle. Temperate phages are capable of protecting their host from phage reinfection and may change the bacterial phenotype through the expression of viral genes, a process known as lysogenic conversion (1).
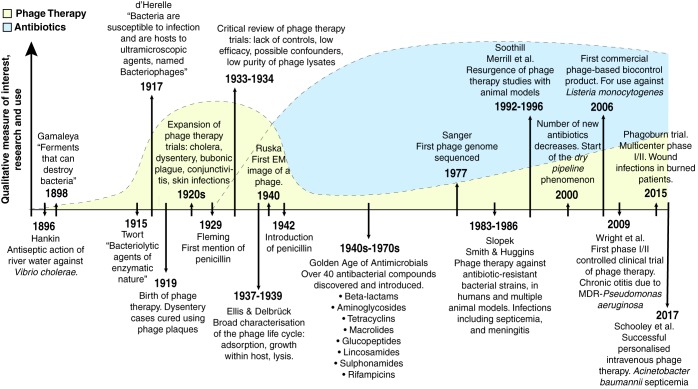
FIG 1.
Timeline of major events in the history of research on phages, phage therapy, and antibiotics. Background curves represent a qualitative measure of the overall interest, research, and use of phage therapy (yellow) and antibiotics (blue), showing how the introduction of antibiotics and the critical review of the early phage therapy studies coincided to bring phage therapy research and development to an almost complete standstill around the 1940s. (3, 4, 63–65, 67–69, 72, 73, 78, 169–174).
Phage therapy is defined as the administration of virulent phages directly to a patient with the purpose of lysing the bacterial pathogen that is causing a clinically relevant infection (5). The first reports on the effectiveness of phage therapy were met with great, albeit short-lived, enthusiasm, which was followed by a collapse that was driven primarily by the introduction of antibiotics (Fig. 1). However, phage therapy work was not completely abandoned. In places such as Georgia (part of the former Soviet Union) and Poland, phage therapy steadily flourished. Even though a substantial amount of the literature presented methodological flaws, it depicted extensive and mostly successful use of phage therapy across multiple medical specialties (3). Some of the longest-running institutions devoted to phage therapy are the Eliava Institute of Bacteriophage, Microbiology and Virology, founded by Georgian microbiologist George Eliava in 1923, and the Ludwik Hirszfeld Institute of Immunology and Experimental Therapy, founded in 1952 in Wroclaw, Poland.
Today, less than a century after the discovery of antibiotics, health care is facing a major threat from antimicrobial resistance. The crisis calls for urgent development, standardization, and implementation of new therapeutic strategies against infectious diseases, and the spotlight is shining once again on phage therapy. The aim of this review is to explore the role that phage therapy can play in the fight against the antimicrobial resistance crisis. We establish a dual focus: first, we discuss the antimicrobial resistance crisis and its causes and consequences, and second, we elaborate on the therapeutic use of phages. Finally, we provide our opinion-based perspective on the opportunities and challenges that phage therapy provides to modern society in relation to three issues: education, accessibility, and economic growth.
ANTIBIOTICS AND ANTIBIOTIC RESISTANCE: THE CRISIS
Antibiotic therapy is widely recognized as one of the most successful therapeutic interventions in the history of medicine. It has saved millions of lives and has been pivotal for the development of multiple medical breakthroughs, including organ transplantation and cancer chemotherapy (6). Antibiotics have truly revolutionized the world. Losing the effectiveness of antibiotic therapy in health care would be catastrophic, and we are quickly approaching such a crisis: a so-called “postantibiotic era” (7).
Resistance: Natural or Man-Made?
Antibiotic resistance is a naturally occurring phenomenon that predates the development and use of antibiotics by humans. Across diverse environments, both microorganisms and higher eukaryotes produce a plethora of biologically active molecules that have antibacterial properties, some of which have been repurposed as modern-day antibiotics (8). However, the concentrations of these chemicals within these natural environments are often below clinically relevant thresholds, suggesting that resistance does not arise exclusively to escape their toxic activity. It has been proposed that in nature, antibiotics and their interplay with antibiotic resistance mechanisms serve as a communication channel between the members of a microbial community rather than as strict antimicrobial agents (9). These compounds have been demonstrated to prompt adaptive phenotypic and genotypic responses and shape the composition of the community (9). When considering antibiotics from this biological perspective, it is unsurprising that genes conferring resistance to modern antibiotics have been identified in ancient microbial populations, such as unpolluted arctic permafrost, suggesting that antibiotic resistance exists even in the absence of anthropogenic influence (10–12).
Human activity, particularly clinical and industrial overuse of antibiotics, greatly aggravates the problem of antimicrobial resistance. Antibiotics are used to enhance the growth of livestock, treat crop and fish diseases in agriculture and aquaculture, respectively, and, of course, treat infectious diseases in humans (13). In fact, agricultural use of antibiotics has been estimated to be as high as 180 mg of active antibiotic agent per kg of meat produced in the United States and is reported to be even higher in other countries (14). An unintended side effect of their agricultural use is the release of millions of tons of antibiotics into water effluents and environmental reservoirs (15). This is further exacerbated by a lack of appropriate water treatment, the contribution of pharmaceutical waste, and the reduction in distance between farmlands and cities, all of which have resulted in increasing levels of antibiotic release and persistence in the environment. This continued exposure of environmental microbial communities to diverse antibiotics has led to the accelerated evolution and expanded repertoire of antibiotic resistance genes persisting in natural reservoirs (15).
In addition to spontaneous mutations in chromosomal genes selected by the pressure of the drug, resistance can arise from the process of horizontal gene transfer—the movement of genetic information between organisms—via transformation, conjugation, or transduction (16). Antibiotic-resistant bacteria selected for in the guts of humans and animals treated with antibiotics are a leading source of resistance determinants for horizontal transfer (17). Furthermore, bacterial cells can achieve transient, non-genetically encoded resistance through processes such as growth in biofilms, swarming adaptation, metabolic dormancy, and persistence (18). The myriad of resistance mechanisms can impede antibiotic action at every step of their passage through the bacterial cell: bacteria can alter the structure of their cell envelope to deny entrance of the drug or synthesize efflux pumps to expel it; they can modify (and even destroy) the compounds through the production of enzymes such as beta-lactamases or stop the production of enzymes required for antibiotic activation; they can modify, hide, or quantitatively adjust the intended target of the drug; or, finally, they can activate alternative metabolic pathways to circumvent the toxic action of the antibiotic (16, 19). For an illustrative graphical summary of antibiotic resistance mechanisms and the genotypic basis of antibiotic resistance, refer to the work of Yelin and Kishony (19).
From the Golden Age to the Dry Pipeline
The “golden age” of antibiotics began in the 1940s and continued for over four decades, with more than 40 antibiotics being discovered and introduced for clinical use (Fig. 1). During this period, emergence of resistance against a specific antibiotic was met with minimal concern, as newer compounds, more often than not exhibiting better pharmacokinetic and pharmacodynamic characteristics, were quickly developed, fueling a cycle of antibiotic discovery, use/overuse, and concomitant appearance of resistance (13, 20). From the decade of the 1990s, however, the consequences of this already-counterproductive cycle became more apparent, as the number of novel antibiotics introduced steadily decreased. The phenomenon has been described as a “dry pipeline” in antibiotic research and development, with the majority of recently introduced antibiotics being either modified or combined versions of previously known compounds.
Novel drugs must go through the challenging process of proving not only their efficacy but also their safety, favorable pharmacokinetic profile, and cost-effectiveness. It is estimated that only five out of 5,000 to 10,000 candidate molecules reach phase I studies and that only one out of those five receives regulatory approval for human use (21). The drug development process is as expensive as it is long, and pharmaceutical companies hesitate to invest in antibiotics when the odds are against them. Furthermore, antibiotics are consumed in short regimes, should be overseen by strict stewardship programs, and are vulnerable to the emergence of resistance, all of which can severely diminish the revenues a company receives (22). It is even believed that newly marketed antibiotics will be outpaced by the high rate at which antibiotic-resistant bacteria are emerging, highlighting the need for novel therapeutics (23). The dry pipeline aggravates the problem of antibiotic resistance, as it has stalled our arsenal of therapeutic options.
Extent and Consequences
The result of consecutive acquisition of antibiotic resistance traits is the generation of multidrug-resistant (MDR), extensively drug-resistant (XDR), and even pan-drug-resistant (PDR) pathogens (24). A group of bacterial species have caught the attention of researchers, clinicians, and public health officials, as they cause the most frequent and severe health care-associated MDR infections. This group includes the pathogens Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp. and is identified with the acronym ESKAPE, referencing the capacity of its members to escape the biocidal activity of antibiotics through multiple resistance mechanisms (25). Consequently, the last four of these pathogens, specifically the carbapenem- and cephalosporin-resistant strains, were recently listed by the World Health Organization (WHO) as being a critical priority for the research and development of new antibiotics (26). Additional species, listed under high priority, included the remaining ESKAPE members, the digestive tract pathogens Helicobacter pylori, Campylobacter spp., and Salmonella spp., and the sexually transmissible agent Neisseria gonorrhoeae (26). Patients with underlying medical conditions, those with any degree of immunocompromise, and those hospitalized (especially in intensive care units, surgical wards or burns units) are at a higher risk of contracting MDR infections. However, an ever-growing number of reports warn about “commonplace” community-acquired infections, in otherwise-healthy patients, becoming unresponsive to antibiotic treatment (27, 28). In the postantibiotic era, even common infections and minor injuries can kill (29).
The burden of antimicrobial resistance can be measured by morbidity and mortality rates and financial costs, but even a combination of these indicators fails to encompass its magnitude. From an oversimplified description, patients with MDR infections have poorer prognoses, higher mortality rates, lengthier hospital stays, higher risks of complications or permanent sequelae, and increased treatment failure compared with their counterparts with antibiotic-sensitive infections (7, 13, 29, 30). A recent review proposes that by the year 2050 ten million lives will be lost annually due to antimicrobial resistance, with a cost to the world economies equivalent to US$100 trillion (31). It is a major threat to global health that is capable of affecting all individuals, regardless of their age, socioeconomic status, or country of residence (32).
What Needs To Be Done?
Any strategy to combat the problem of antimicrobial resistance has to be multidimensional, multidisciplinary, and global. A key component to be included is the implementation of regulatory measures for the human and animal use of antibiotics (33). In many countries, particularly in the developing world, self-medication and easy purchase of antibiotics without a prescription are worryingly common and need to be addressed. Even more troubling is the everyday use of antibiotics in animal feeds and agriculture, which should be restricted. Furthermore, inappropriate prescription of antibiotics by health care professionals, such as prescribing antibiotics to treat viral or fungal infections, unnecessarily prolonging antibiotic courses, or using antimicrobials with a broader spectrum than reasonably needed for a specific infection, also occurs. The issue of the dry pipeline could be mitigated by providing administrative and financial stimuli to the field (22) or through the “revival” of old antibiotics. The latter is the case for polymyxins and chloramphenicol, which were abandoned due to toxicity concerns (nephrotoxicity/neurotoxicity and rare but potentially fatal hematological side effects, respectively) and are now making a comeback (34, 35). Most importantly, it is essential to encourage research on new therapeutic alternatives and to rekindle the interest in neglected ones, such as phage therapy.
PHAGES AS THERAPEUTIC AGENTS
Biological Considerations
Bacteriophage therapy, which was first used almost a century ago, is now going through a revival driven mostly by the antibiotic resistance crisis. This renewed interest in phage therapy has been facilitated by our improved understanding of phage biology, genetics, immunology, and pharmacology. Key aspects of phage therapy have now been standardized to improve treatment success. Minimum suggested regulatory requirements for the therapeutic use of phages call for strictly lytic phages, confirmed antimicrobial activity against the target pathogen, and removal of contaminating bacterial debris and endotoxins (36). In addition, the identity of the bacterial host receptor for any therapeutic phage should be established, which will provide important information on emergence of phage resistance, evolutionary trade-offs, and use of combination therapies that are less likely to generate phage-resistant hosts.
Lytic phage infection begins with adsorption to specific receptors on the bacterial host’s surface. These receptors may be located on either Gram-positive or -negative cell walls, as well as polysaccharide capsules or even appendages such as pili and flagella (37). The lock-and-key relationship between a phage and the bacterial receptors will typically determine the host range that the virus is able to infect, and the list of characterized phage receptors is constantly growing. After adsorption, the virus will eject its genetic material into the host. The majority of described lytic phages associated with human pathogens belong to the orders Caudovirales and Microviridae and have double- or single-stranded DNA genomes (38). Next, the virus takes over the bacterial replication machinery, creating the next generation of phage progeny in the process. Replication will continue until phage-encoded proteins (see “Phage-Derived Proteins” below) are activated in order to lyse the cell, effectively killing the host and allowing the newly synthesized viruses to escape and reinitiate the cycle. The lysis time, or latent period, is the amount of time taken by a phage to complete this intracellular life cycle.
As mentioned above, the use of temperate or lysogenic phages for phage therapy is normally inadvisable, not only because their killing capacity is hampered by the quick arise of homoimmunity but also because of the possible harmful consequences of lysogenic conversion. Through lysogenic conversion, bacteria can acquire new, often pathogenic, genetic traits, such as phage-encoded toxins capable of greatly enhancing their virulence (39) or potentially even antibiotic resistance determinants (40). However, for relevant pathogens such as Clostridium difficile no strictly lytic phages have been isolated, and the use of temperate phages may be necessary (41). Similarly, in cases of emergency, time constraints could justify the therapeutic use of temperate phages when lytic phages are unavailable. Unfortunately, this does not mean that the use of lytic phages is exempt from concerns. Lytic phage genomes can contain greater than 50% hypothetical genes with no known function (42) or encode auxiliary proteins that alter bacterial physiology in ways that are not fully understood. During abortive infection, where phage DNA is ejected into the cell and some genes expressed, without production of viral progeny, the bacterial host could potentially act as a reservoir for foreign DNA of unknown function. This reasoning should warrant continued research into phage genetics as a way to ensure the safety of phage therapy.
Comparison to Antibiotics
Although technically not living organisms, phages are certainly dynamic entities, and the lytic cycle is the cornerstone of phage-based therapeutics. In contrast, antibiotics are chemicals capable only of selective disruption of certain bacterial physiological processes, such as protein or cell wall synthesis. A quick comparison between phages and antibiotics demonstrates how strikingly different their mechanisms of action are. A summary of these differences, and some similarities, between the two can be found in Table 1 (43, 44). However, the following paragraphs elaborate on some of the most therapeutically relevant comparative points.
TABLE 1.
Advantages, disadvantages, and similarities of phage therapy compared to antibiotic therapy
| Advantages | Similarities | Disadvantages |
|---|---|---|
| Specificity: does not kill the microbiota Self-limitation: once the bacterial host is killed, it ceases to function Available for patients with antibiotic allergies Safety: no effects on mammalian cells Exponential reproduction allows for lower doses Evolution: if resistance arises, phages mutate alongside bacteria Antibiofilm activity Simple and inexpensive to produce Ubiquity |
Administration requires a neutralized-pH environment Therapeutic success depends on variables such as time of treatment initiation Activity is influenced by the immune system of the patient Versatility in routes of administration Occurrence of bacterial resistance to the therapeutic agent |
Specificity: causative bacterium must be identified beforehand, narrow spectrum of action Induction of phage-neutralizing antibody production (clinical relevance to be determined) Significantly smaller body of evidence and correctly designed clinical trials supporting its effectiveness Lack of a specific regulatory framework, and legal issues regarding intellectual property |
Phages have been classically described as highly specific for their hosts. However, it was recently demonstrated that phages are able to “jump” hosts, and in the gut that process is facilitated by the microbiota (45). Thus, phage-host specificity may evolve and adapt over time. This specificity is simultaneously the greatest advantage and greatest disadvantage of phage therapy. Phage therapy aims directly at the pathogenic bacteria, whereas antibiotic treatment carries collateral damage as it disrupts the microbiome. Due to its lack of off-target effects, phage therapy is exempt from side effects related to microbiome disturbances, such as mucosal candidiasis, antibiotic-associated diarrhea, pseudomembranous colitis caused by Clostridium difficile, and even long-term metabolic and immunological disorders (46). Conversely, that specificity also demands accurate diagnosis of the infection and identification of the etiological agent, sometimes to the strain level, a process that can be difficult and time- and resource-consuming (47). Moreover, early initiation of phage therapy has been shown to be critical to its success, and delays as short as 6 h can result in a significant decline in treatment effectiveness (48). Together, these facts justify the practice of establishing and expanding phage collections or automated pipelines to quickly isolate and identify candidate phages. The collections, or libraries, are made up of readily available, well-characterized phages, isolated from natural sources, that exhibit biological traits theoretically desirable for phage therapy, such as short latency time, large burst size, and broad host range (49).
Both antibiotics and phages interact with the immune system. First, the most severe, and potentially lethal, side effect of antibiotic therapy is hypersensitivity, a group of immune-mediated phenomena that include IgE-mediated reactions, anaphylaxis, Stevens-Johnson syndrome, and toxic epidermal necrolysis (50). A 24-year retrospective study in a tertiary care hospital found that antibiotics, particularly beta-lactams, were the most common triggers of anaphylaxis (51). Comparatively, there have been no documented adverse anaphylaxis cases associated with phage therapy in humans (52, 53), although the collective body of evidence is decidedly smaller. Second, the action of the immune system complements the antibacterial activity of both antibiotics and phages. Bactericidal agents are able to kill bacteria, whereas bacteriostatic agents only prevent their growth. These effects usually depend on the mechanism of action and dose of a given antibiotic, but at therapeutic doses, protein synthesis inhibitors such as macrolides and tetracyclines are mostly bacteriostatic (54). Bacteriostatic antibiotics heavily rely on the immune system of a patient to clear the infection (55). Although lytic phages are by definition “natural killers,” phage therapy on its own will not completely clear an infection either, at least theoretically. This is because eradication of the host would also result in termination of viral replication. Instead, phages engage in “kill-the-winner” dynamics with their bacterial hosts, rapidly reducing host abundance before reaching a dynamic equilibrium (56, 57). The immune system is still needed to eliminate the lingering bacterial population for phage therapy to be successful. This collaboration between phages and the immune system has been termed “immunophage synergy” (58). It could be argued that every treatment of an infectious disease in an immunocompetent patient (and to a lesser extent in an immunocompromised patient) is, by definition, combinational therapy due to the natural action of the immune system. Finally, it has been demonstrated that phages elicit both innate and acquired immune responses against them (59). More interestingly, the immune activation and inflammatory environment created by the bacterial infection could heighten the inhibition of phage therapy, although the clinical relevance of these phenomena remains to be determined (59).
As with antibiotics, phage therapy is affected by bacterial resistance. Lytic phages impart strong antimicrobial selective pressures on their hosts that rapidly select for phage-resistant bacterial mutants (2). Bacterial antiphage systems can be encountered along every step of the phage replication cycle. The best-known processes include modification of the receptors used during phage adsorption, superinfection exclusion (Sie) systems to prevent viral DNA entry, restriction-modification systems that protect host DNA while leaving foreign DNA vulnerable to the action of restriction enzymes, and clustered regularly interspaced short palindromic repeat (CRISPR)-Cas systems that recognize and degrade previously encountered foreign DNA (60). The sum of phage resistance mechanisms constitutes a true prokaryotic “immune system.” However, the characterization of these mechanisms has come from experiments using a small number of model phage-host pairs and may not necessarily be applicable to less-investigated pathogens and their phages. Regarding this issue, Doron et al. (61) recently searched more than 45,000 bacterial and archaeal genomes and discovered nine new families of antiphage defense systems, whose molecular mechanisms are yet to be fully understood. Closing the knowledge gap about phage-resistance mechanisms may allow for therapeutic and biotechnological exploitation (see “Exploitation of Phage Resistance” below).
A final benefit of phage therapy is its versatility. Due to their genetic diversity, abundance, and ubiquity, there is a virtually limitless source of phages. Furthermore, phage therapy can be delivered through different approaches, each one adaptable to the available resources, type of infections, and characteristics of the patients. Phages can be administered as a tailor-made, personalized therapy or, conversely, subjected to automated high-throughput production of phage cocktails. During the last decades of work with phages for therapeutic purposes, the in vitro and preclinical findings have begun to be translated into carefully designed clinical trials and case studies, highlighting important lessons to be considered moving forward.
Experience in Clinical Trials
In the early years of human phage therapy (Fig. 1), phages were used to treat conditions including typhoid fever, dysentery, skin and surgical wound infections, peritonitis, septicemia, urinary tract infections, and external otitis (62). During the 1930s, however, a series of analytical reviews posed important questions about the validity of the presented results (Fig. 1). Criticism was aimed at the lack of proper methodological design, controls, and standardized production and characterization of the phage preparations, as well as contradictory results (63–65). After the introduction of antibiotics, when the efforts in phage therapy studies were primarily relegated to a few countries in Eastern Europe, language also became a barrier for the widespread dissemination of the results. Sulakvelidze et al. (66) reviewed studies from the Georgian, Russian, and Polish literature, finding successful use of phages for the treatment of the previously mentioned conditions, as well as pneumonia, meningitis, osteomyelitis, and postsurgical infections in cancer patients. Although some early and encouraging work at the end of the 20th century presented the use of phage therapy in animal models (67–69), it was not until the start of the new millennium that the English literature rediscovered human phage therapy trials (62). Here, we discuss some of the results of these contemporary studies.
The results of the first phase I randomized, placebo-controlled phage therapy trial conducted in the United States were published by Rhoads et al. (70) in 2009. The study investigated the safety of a 12-week topical treatment, with a total follow-up period of 24 weeks, of a phage preparation targeting S. aureus, P. aeruginosa, and Escherichia coli in chronic venous leg ulcers. Their results showed no significant difference in the incidence of adverse events between groups. With a sample size of only 42 patients and no previous in vitro demonstration of susceptibility of the patients’ infectious agents to the phage preparation, the study was not designed to assess effectiveness of the intervention. Expectedly, the rate and frequency of healing were the same between treatment groups. Previously, in Georgia, Markoishvili et al. (71) had demonstrated successful healing of poorly vascularized ulcers with the application of a biodegradable polymer impregnated with antibiotic and lytic phages. However, the independent effect of the phages could not be discriminated from that of the antibiotic, and the authors recommended further studies.
The effectiveness of topical administration of phage therapy has been assessed by at least two phase I/II trials. In 2009, Wright et al. (72) established a randomized, double-blind, placebo-controlled study where they tested the action of a phage cocktail in the treatment of chronic otitis due to antibiotic-resistant P. aeruginosa in 24 patients. The measured outcomes included physician-assessed signs of inflammation, patient-reported symptoms, and quantification of the bacterial and viral loads. The phage preparation improved all of the measured outcomes compared to placebo after the 42-day follow-up period. In the second case, the randomized, multicenter, single-blind, and open study Phagoburn ran from 2015 to 2017 and evaluated the treatment of burn wound infections by P. aeruginosa in 25 patients, using a cocktail of 12 phages. The published report highlights this as being the first clinical trial of phage therapy ever performed according to both good manufacturing practices (GMP) and good clinical practices (GCP) (73). Additionally, no unwanted side effects attributable to the phage cocktail were reported. The authors also showed that the intervention significantly decreased the pathogen load in the wounds, but unfortunately, this happened at a lower rate than in the control arm, which consisted of standard care with 1% sulfadiazine silver emulsion cream. The authors explained these results with three claims. First, after delays associated with manufacturing and administrative challenges, the length of the recruitment period was nearly halved, leading to a small patient sample size. Second, the titer of the phage cocktail was found to have significantly decreased after manufacturing, leading to patients receiving a lower dose of phages than originally intended. Third, bacteria from patients in whom the phage treatment failed were shown to be resistant to low phage doses (73). According to the authors, further studies that address these issues are warranted. Finally, an ongoing phase II trial (ClinicalTrials registration no. NCT02664740) is looking at the topical treatment of diabetic foot ulcers infected by S. aureus with a phage cocktail.
Studies focusing on the oral administration of phages have also been carried out. A T4-like phage preparation, targeted against E. coli and designed for the treatment of diarrheal disease, has been assessed in phase I placebo-controlled trials in healthy adults from Switzerland (74) and Bangladesh (52), in 2005 and 2012, respectively, and in healthy and diseased children from Bangladesh in 2017 (75). No adverse effects from oral administration of phages were found by self-report, physical examination, and laboratory testing of hepatic, renal, or hematological function. The studies also provided insights into the bioavailability and activity of oral phage preparations. Phages did not amplify in the gut of healthy individuals, and only small, dose-dependent fractions of the initial phage dose were recovered from their feces. There was no evidence of phages or phage-specific antibodies in the bloodstream, and the phage preparation did not disturb the composition of the gut microbiota. However, the same lack of viral replication was seen in the children with diarrheal disease, and the treatment did not have significant favorable effects compared to standard rehydration therapy. While the studies had been designed primarily around safety, the latter observations prompted the authors to question the value of phage therapy to treat these low-abundance infections within the gut (75). In a different approach, Gindin et al. (76) recruited 32 adults with mild to moderate gastrointestinal complaints, to carry out a phase I randomized, double blind, placebo-controlled, crossover trial in 2018. The study was based on the premise that the gut microbiota can regulate human health, that dysbiosis can lead to disease states, and that modulation of its components, namely, eradication of specific detrimental organisms, can have beneficial effects in patients suffering from gastrointestinal distress. The intervention, a 28-day oral treatment with capsules containing 4 strains of bacteriophages targeting recognized gastrointestinal pathogens, was proven to be safe and tolerable. After controlling for sequence effects given the crossover design of the study, the treatment was more effective than placebo at reducing symptoms of colon pain and abnormal gastric function, comparable to placebo in reducing small-intestine pain, but ineffective in decreasing perceived gastrointestinal inflammation. Further studies could open the door for phages to be used as novel prebiotics.
Regarding intravenous phage therapy in humans, Speck and Smithyman (53) recently reviewed the evidence supporting its use. Their primary focus was on severe infections such as typhoid fever and S. aureus bacteremia, with data obtained from reports spanning the last 80 years. They reference studies that collectively account for almost 1,000 patients successfully treated with intravenous phage therapy, with a negligible number of side effects. In addition, all of the severe side effects (reactions resembling shock or serum sickness) could be attributed to contaminants from early phage preparations. Even though historical evidence tends to be dismissed because of its age or lack of compliance with modern clinical standards, many of the described studies came from groups in France, Canada, and the United States, countries with well-regulated medical systems. The authors concluded that there is a strong possibility that intravenous administration of phages will be safe and efficacious (53). Moreover, an ongoing phase II/III randomized, double-blind clinical trial (ClinicalTrials registration no. NCT03140085) is addressing an alternative route of parenteral phage administration. The trial will look at the efficacy of intravesical administration of the commercial phage cocktail Pyophage (PYO) versus oral antibiotics or intravesical placebo for the treatment of urinary tract infections in patients undergoing transurethral prostatectomy.
Parallel to full-scale clinical trials, the pathway toward widespread use of phage therapy and its translation from the lab bench to the patient’s bedside could be shortened by cases of compassionate use. In the absence of alternative treatments, or in terminally ill patients, phage therapy can obtain “off-license” approval for use. The advantages of the approach include immediate clinical usage, obtaining data that could be used to inform future work, and that it can be used for all forms of phage therapy (77). Nevertheless, the results from this approach are usually hard to replicate as they are limited to a single patient and do not directly lead to the approval of the therapeutic intervention.
Further use of intravenous and intracavitary phage therapy in humans has been assessed primarily through individual case studies of compassionate use. In 2017, Schooley et al. (78) reported the first case of successful intravenous and intracavitary phage therapy targeting a systemic MDR infection in the United States. The patient was a 68-year-old diabetic man with necrotizing pancreatitis complicated by an MDR A. baumannii infection. After the infection completely stopped responding to antibiotic therapy and the patient’s condition severely deteriorated, phage therapy was initiated. Several aspects of this case report are worth highlighting. First, treatment was personalized and quickly available, as the bacterial strain causing the infection was tested against preestablished A. baumannii-specific phage libraries. Second, treatment consisted of sequential administration of phage cocktails, which is necessary for countering the emergence of phage resistance (see “Conventional Phage Therapy” below). Third, phage therapy was well tolerated by the patient. In 2018, Chan et al. (79) reported a comparable case. A 76-year-old man underwent aortic arch replacement surgery with a Dacron graft and was later diagnosed with an MDR P. aeruginosa graft infection. As the patient was considered at too high risk for surgical replacement of the graft and treatment with intravenous ceftazidime and superficial chest wall debridement proved to be unsatisfactory, phage therapy was used. The intervention consisted of the simultaneous, single intracavitary application of phage OMKO1 and ceftazidime. The treatment was well tolerated, and even though the patient exhibited further complications expected in aortic graft carriers, the P. aeruginosa infection receded without any recurrence despite the discontinuation of antibiotics. Additional mechanistic aspects that contributed to the success of these two cases is discussed in “Exploitation of Phage Resistance” below. Lastly, Jennes et al. (80) reported the case of a 61-year-old patient who developed septicemia caused by a P. aeruginosa strain that was sensitive exclusively to colistin, which likely originated from local colonization of pressure sores. When the antibiotic treatment, along with the patient’s underlying conditions, led to acute kidney injury, therapy with a phage cocktail was initiated. The two-phage cocktail was administered intravenously every 6 h, and in topical irrigation of the wounds every 8 h, for a total of 10 days. Phage therapy immediately turned blood cultures negative and reduced the levels of C-reactive protein and the patient’s temperature, with kidney function being restored after a few days. Despite this favorable clinical course, the patient’s multiple comorbidities remained, leading to his death four months after phage therapy due to sepsis by a different pathogen.
In the coming years, we certainly expect to see an increase in phase I phage therapy clinical trials and their progress toward phases II and III. Notably, most of the presented studies have primarily assessed the so-called “conventional” approach to phage therapy. In the next section, we explore the notions behind this approach, but more importantly, we include innovative approaches that will progressively reach clinical use, including phage-antibiotic combinations, phage-derived enzymes, exploitation of phage resistance, and phage bioengineering.
APPROACHES TO PHAGE THERAPY DELIVERY
“Conventional” Phage Therapy
The use of phages as the only therapeutic agents administered to a patient during the course of a bacterial infection is referred to as conventional phage therapy (Fig. 2A). Additionally, the terms monophage and polyphage therapy clarify whether a single phage or a combination of phages is used, respectively (81).
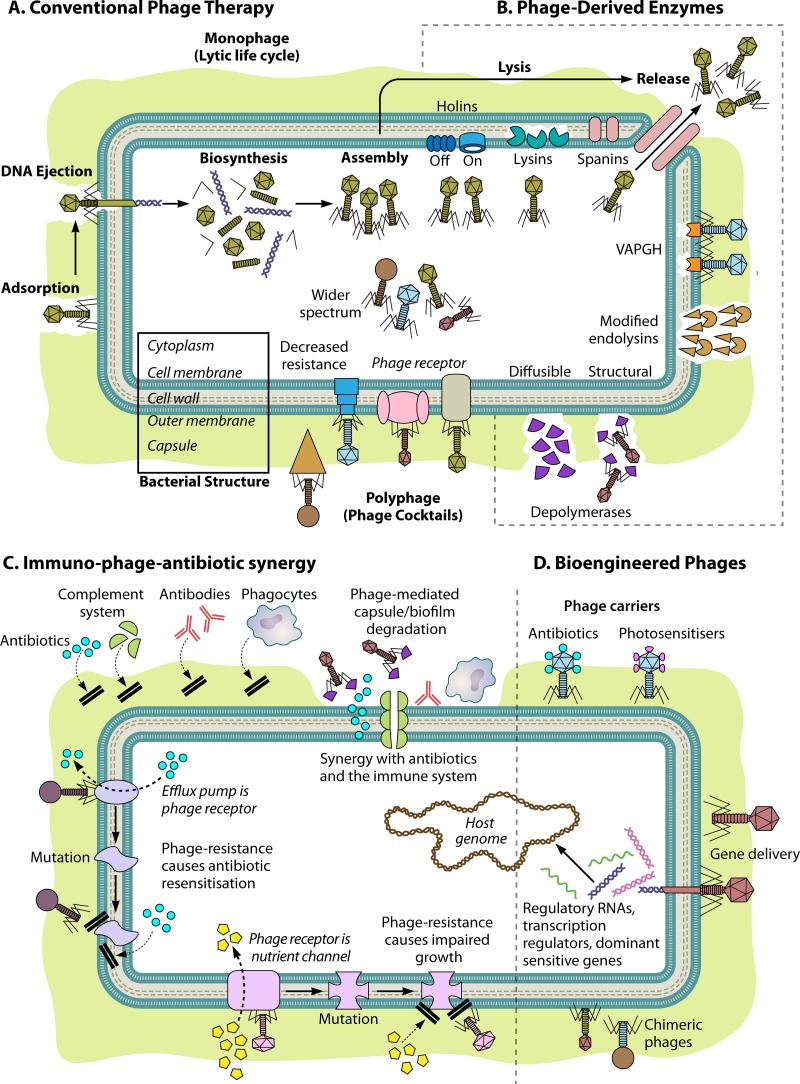
FIG 2.
Approaches to phage therapy delivery. A schematic representation of a bacterial cell consisting of capsule, outer membrane, cell wall, inner membrane, and cytoplasm is shown. (A) Phage lytic life cycle as the basis for conventional monophage therapy, from adsorption to lysis of the host cell, and polyphage therapy, or use of phage cocktails targeting different receptors on the same host cell, limiting the occurrence of resistance and expanding the therapeutic spectrum. (B) Use of phage-derived enzymes such as depolymerases (to target capsules and biofilm structures) and cell wall-degrading endolysins. Holins, spanins, and virion-associated peptidoglycan hydrolases (VAPGH) are represented as components of the lytic life cycle, without current therapeutic applications. (C) Mechanisms for synergy between phages, antibiotics, and the immune response include phage-mediated capsule or biofilm degradation, which enables the action of antibiotics, antibodies, the complement system, and phagocytes, and the exploitation of the evolutionary trade-offs of phage resistance, such as antibiotic resensitization and impaired bacterial growth. (D) Bioengineering of phages for therapeutic purposes includes the attachment of antibiotics or photosensitizing agents to phage capsids for targeted release into bacterial cells, delivery of genes to reverse/cancel antibiotic resistance and virulence determinants, and use of chimeric phages. Labels in italic represent bacterial structures.
Monophage therapy has been used primarily in laboratory settings with animal models, as proof of concept during the design and testing of phage preparations. As expected, focus is placed on troublesome infections by MDR pathogens, such as carbapenem-resistant A. baumannii pneumonia (82, 83) and vancomycin-resistant E. faecium septicemia (84). Nevertheless, the usefulness of monophage therapy in clinical practice may be hampered by the emergence of bacterial phage resistance, as it can appear relatively quickly, even in small bacterial populations (85, 86). Within a natural environment, phages and their bacterial host will continually evolve and adapt in order to preserve infective capacity or withstand it, respectively, entering an evolutionary arms race (87). Even though this evolutionary capacity of phages to overcome bacterial resistance is a further advantage over antibiotic therapy, the process may not be efficient enough to warrant the clinical use of monophage therapy. A further disadvantage of monophage therapy is that it requires precise matching between the etiological agent and the phage. This match is typically performed in a laboratory setting, yet the in vitro and in vivo lytic properties of a phage may not always coincide (88).
To address the issues of monophage therapy, polyphage therapy, also known as the use of phage cocktails, is a commonly used strategy (Fig. 2A). Phage cocktails can be designed to target a single bacterial strain, multiple strains of a single bacterial species, or multiple species, typically grouped by the clinical syndromes they cause. Unfortunately, these cocktails require longer and more complex preparation and purification processes, increasing the likelihood of eliciting immune responses, and have reduced predictability of phage pharmacokinetic and pharmacodynamic properties (81). To limit these shortcomings, the comparative efficacy of the individual phages within a cocktail should be assessed (89) and unnecessary, poorly active phages removed. A final approach combines the notions of monophage and polyphage therapies by using sequential, instead of simultaneous, administration of phage cocktails. With sequential administration, even if resistance arises, bacteria are continually exposed to new phages to which they were not initially resistant. This strategy would maintain the bacterial population density at a lower level for longer, facilitating the action of the immune system. An in vivo study using a wax moth larva model suggests that sequential administration of phage cocktails may provide better results over longer time scales regarding reduction of bacterial populations and emergence of phage resistance (90).
Phage cocktails are available as over-the-counter medication in Russia and Georgia for the treatment of bacterial infections of broad etiology. The Eliava Institute is associated with the production and biannual update of two of the most widely used generic phage cocktails: Pyophage (PYO) and Intestiphage (91). PYO contains phages targeting S. aureus, E. coli, P. aeruginosa, Proteus vulgaris, Proteus mirabilis, and Streptococcus pyogenes, whereas Intestiphage targets approximately 23 different enteric bacteria; they are marketed for the treatment of pyoinflammatory and enteric diseases, respectively (91). Western regulatory agencies demand, among other requirements, a thorough characterization of these products before approving them for use in humans. Recently, metagenomic analyses of PYO and an additional phage cocktail have been carried out (92, 93). The first of these studies, by Villarroel et al. (92), compared the compositions of three batches of PYO from a period between 1997 and 2014, finding sequences from up to 30 different phages and a surprisingly stable composition throughout the years, despite the aforementioned biannual “upgrade” of the product. However, most commercially available phage products in Eastern Europe, at least historically, have not been produced under the full manufacture controls required by these regulatory agencies.
In addition to the previously discussed clinical trials (see “Experience in Clinical Trials” above), results from preclinical studies of polyphage therapy have greatly increased in recent years. The targeted conditions or microorganisms include methicillin-resistant S. aureus (MRSA) osteomyelitis (94), P. aeruginosa colonization of the sputa of patients with cystic fibrosis (95), catheter-associated urinary tract infections by Proteus mirabilis (96), and C. difficile infection (97). The consensus demonstrates favorable safety profiles and encouraging evidence of efficacy.
Phages and Antibiotics Combined: Two Is Better than One
In clinical practice many conditions are treated, sometimes exclusively, using combination therapy. Highly active antiretroviral therapy (HAART) against HIV and multiple-drug therapy for tuberculosis or malaria are examples where multiple treatment approaches are used to combat a disease. Yet the concept transcends the field of infectious diseases, with similar tactics being used in chemotherapy protocols against specific types of cancer or in the control of refractory hypertension. Amid the looming threat of bacterial resistance, a commonly used strategy is the combination of two or more antibiotics (98) with the hope of achieving a synergistic effect. Pharmacological synergy occurs when the combined effect of two therapeutic agents is greater than the sum of their individual effects, a phenomenon that brings about significantly higher rates of treatment success (99). In some cases, even the addition of compounds that lack an inherent antimicrobial activity can be favorable; this is the case for adjuvants that can block certain resistance mechanisms or pharmacokinetically improve the action of the drug (100). All these premises can be summarized by the evolutionary principle that dictates that “two sufficiently different selective pressures are likely to be more effective than either alone” (99). This logic supports the practice of combining phages and antibiotics as a therapeutic strategy against MDR infections.
The term phage-antibiotic synergy (PAS) was first used by Comeau et al. (101) to describe the serendipitous discovery that sublethal concentrations of antibiotics could greatly increase the bacterial production of lytic phages. The phenomenon was attributed to the increased biomass and biosynthetic potential of bacteria in the presence of antibiotic concentrations high enough to inhibit cell division but not to cause cell death. For the virus, this resulted in a shorter latent period and larger burst size, allowing quicker spread and reduction of the bacterial population. This initial description, made with beta-lactams and quinolones on E. coli, has since been replicated with species including P. aeruginosa, Burkholderia cepacia, and methicillin-resistant S. aureus (MRSA) and with additional antibiotic groups, such as tetracyclines and oxazolidinones (102–104). We believe that this traditional concept of PAS can be greatly expanded, as additional mechanisms through which synergy between phages and antibiotics occurs have since been discovered (see “Phage-Derived Proteins” and “Exploitation of Phage Resistance” below). Finally, it has been shown that an “order” effect can arise when combining phages and antibiotics, whereby treatment with phages before antibiotics may achieve maximum killing. The effect has been described using protein synthesis inhibitors with bacteriostatic effects, reflecting the need of actively replicating hosts for phages to propagate and kill. The results show how optimizing the timing of administration of combinational therapy can potentiate its efficacy (105, 106).
The clinical exploitation of PAS would have two additional benefits. First, by limiting the amount of antibiotics used, it would assist with antibiotic stewardship and managing emergence of resistance. Second, it would provide antibiotics a second wind against MDR pathogens through combination treatments with phages (99). Previous studies have demonstrated the effectiveness of phage-antibiotic combinational therapy, both in vitro and in vivo, for ESKAPE bacteria. Chhibber et al. (107) demonstrated that the combination of a lytic phage with linezolid was more effective than either treatment alone in managing MRSA infections of diabetic foot ulcers in a murine model. The results are particularly remarkable as they were observed in a model of diabetes, a disease characterized by systemic immunosuppression, where reduced therapeutic effects would have been expected. Oechslin et al. (108) showed synergism between a single-dose phage cocktail and ciprofloxacin treatment against experimental endocarditis caused by P. aeruginosa in rats. One of the hallmarks of endocarditis is the formation of vegetations that protect bacteria from antibiotics and the host’s immune response. The combination treatment killed >6 log CFU per gram of vegetations, as opposed to 2.5 log CFU/g of vegetations for each of the single-agent treatments (108). Additionally, the study showed that emergence of phage-resistant mutants occurred exclusively in vitro, suggesting that those mutations had detrimental effects on the bacterial population in vivo. Finally, Valério et al. (109) demonstrated increased synergy between a monophage therapy and bactericidal agents against E. coli grown in human urine samples, with a significantly lower emergence of both singly and doubly resistant bacterial mutants when combination therapy was used. Together, the findings underpin the idea that phage therapy should not aim to replace antibiotics. The combination therapy with antibiotics and phages can work exceedingly well against complicated infections.
Phage-Derived Proteins
Phage genomes encode a number of proteins and enzymes required to breach the bacterial cell during infection (Fig. 2B). During phage adsorption to the host and ejection of its genome, two groups of proteins are typically required: virion-associated peptidoglycan hydrolases (VAPGH) and polysaccharide depolymerases (58). VAPGH are structural components of the virus that are typically located on the phage base plate and act to locally degrade the peptidoglycan layer, thereby allowing the phage tail tube structure to eject its genomic material into the host. The other major proteins are phage-encoded depolymerases, which target the polysaccharide components of the bacterial cell envelope, such as the bacterial capsule, the Gram-negative lipopolysaccharide (LPS), or the extracellular matrix of biofilms. Phage depolymerase activity facilitates access to secondary host receptors located at the cell wall by degrading the structural polysaccharide components and may have broader effects for the degradation of biofilms (110).
Bacterial capsule and biofilm structures are important virulence determinants that can block the action of antibiotics, disinfectants, and the immune response. As such, the use of phage-derived depolymerases to target and remove these structures is a viable treatment option. Bacterial capsules are essential virulence determinants for many important pathogenic bacterial species, including Streptococcus pneumoniae, Haemophilus influenzae, and E. coli. Capsules help the pathogen escape the action of phagocytes and the complement system and facilitate epithelial colonization, cell invasion, and intravascular survival (111). Similarly, biofilm structures protect bacterial communities from the immune response and, in addition, can increase their metabolic efficiency and genetic exchange, among multiple other benefits. Bedi et al. (112) demonstrated synergy between monophage therapy and amoxicillin for eradication of K. pneumoniae biofilm, which they attributed to the action of phage-encoded depolymerases breaking down the extracellular matrix of the biofilm, allowing better penetration of the antibiotic and whole phage particles. Relatedly, Mushtaq et al. (113) observed how stripping off the capsule of E. coli K1, a common causative agent of neonatal meningitis, using a phage-derived depolymerase increased the phagocytic action of macrophages and protected against bacteremia in rat models. Born et al. (114) demonstrated that a tail-associated phage depolymerase could degrade the capsule of the plant pathogen Erwinia amylovora and, by doing so, increase its susceptibility to phages it was previously resistant against. These studies suggest that phage-derived depolymerases are the protagonists of numerous mechanisms for synergy between phages, antibiotics, and the immune system (Fig. 2C).
Towards the end of the lytic cycle, phages use a different set of enzymes to facilitate lysis of the host and release of the viral progeny (Fig. 2B). Even though single-gene lysis systems have been characterized for phages with small, single-strand genomes (115), the systems used by tailed phages rely on the coordinated action of at least two types of proteins: holins and endolysins. Holins are hydrophobic proteins that passively accumulate in the inner bacterial cell membrane until reaching a specific concentration that triggers their arrangement into holes. The timing of holin triggering is allele specific, in that it can be advanced or retarded by missense mutations that can result in fine-tuning of phage lysis times (116). Holins thus are responsible for a generalized permeabilization of the inner cell membrane but are not independently capable of cell lysis (58). Next, endolysins use those holes to translocate from the cytoplasm to the periplasmic space, gaining access to their substrate: the bacterial cell wall polymer peptidoglycan. For Gram-positive bacteria, degradation of the bacterial cell wall is sufficient to cause cell lysis and release of the viral progeny. For Gram-negative bacteria, however, the outer membrane must be disrupted to allow for efficient cell lysis and release of viruses. This is achieved by the action of spanins, which catalyze the fusion of the inner and outer membranes, leading to cell lysis (117). A variation of this pathway is represented by the pinholin/signal anchor release (SAR) endolysin system. Here, inactive SAR endolysins are secreted by the host sec system (118). Along with pinholins, they accumulate on the inner cell membrane until the pinholins trigger its depolarization, which in turn prompts the activation of the SAR endolysins to begin the enzymatic destruction of the cell wall (117).
Endolysins are classified according to the peptidoglycan structure they target, with three major classes being glycosidases, amidases, and endopeptidases (58). Furthermore, their host range has been shown to be variable, with serovar-specific (119), multispecies (120), and even multigenus (121) endolysins having been isolated. The natural cell wall-lysing properties of endolysins have given them great potential as antimicrobial agents. Nevertheless, the majority of reports of successful endolysin use for therapeutic purposes have been for use of the enzymes against Gram-positive pathogens only. This is unsurprising, as the outer membranes of Gram-negative organisms act as a barrier to these enzymes. Few natural endolysins have the ability to bypass the outer membrane and thus possess relevant anti-Gram-negative activity (122). For this reason, additional tactics to bypass the outer membrane obstacle have been devised, including the coadministration of chemicals that permeabilize the outer membrane, such as polymyxins, aminoglycosides, or chelating agents such as EDTA (123), and bioengineered endolysins. One example of such bioengineering is seen in artilysins, where the addition of polycationic oligopeptides to the endolysin grants outer membrane-breaching ability and the capacity to kill Gram-negative MDR pathogens such as A. baumannii and P. aeruginosa with a 4- to 5-log reduction within 30 min in vitro (124).
One prospective target for endolysin therapy is the eradication of S. aureus, including MRSA, from the nose and skin (125, 126). Nasal and skin carriage of S. aureus in patients and health care workers is a risk factor for infections by this pathogen, and the topical antibiotic mupirocin has been recommended to eradicate it (127). A study by Pastagia et al. (128) demonstrated how an engineered lysin achieved an additional 1-log reduction in CFU of S. aureus compared to mupirocin treatment in a murine model. Similar results have been reported for the eradication of Streptococcus pneumoniae from the upper respiratory tract (129) and of Streptococcus agalactiae, which is associated with preterm labor and neonatal infection, from the urogenital tract of mice (130). Experiments have also assessed the protective effect and safety of intraperitoneal endolysin administration against S. aureus septicemia in mice (126, 131). Remarkably, one of the studies demonstrated synergy between an endolysin and the antibiotic oxacillin, as doses that were not protective individually efficiently protected from septic death when combined. One current antistaphylococcal intravenous endolysin therapy (SAL200) is currently in phase I clinical trials to assess its pharmacokinetic profile, establish its ideal dosing schedule, and evaluate its safety, with encouraging preliminary results (132, 133). Another example of current endolysin therapies is the topical use of the recombinant endolysin Staphefekt to treat skin flora dysbiosis caused by overgrowth of S. aureus. Staphefekt was able to decrease the load of S. aureus, restore the balance of the local flora, and clinically improve the condition in three patients (134) and has now progressed through to phase II clinical trials (135).
Exploitation of Phage Resistance
The use of phages combined with antibiotics and the natural immune response can be expanded beyond the classic concept of PAS. Here, we focus on the “trade-offs” of bacterial evolution toward phage resistance, following the evolutionary rationale proposed by Torres-Barceló and Hochberg (99). In this rationale, it is suggested that bacterial evolution has genetic constraints whereby acquisition of resistance against phages may come with a cost. The diverse range of evolutionary trade-offs has now been demonstrated in numerous studies. Using a fitness competition experiment with Pseudomonas fluorescens in soil, Gómez and Buckling demonstrated that acquisition of phage resistance reduced bacterial fitness, with a decrease in growth of 36% (136). Through coincubation of lytic phages with their corresponding bacterial hosts, Capparelli et al. were able to obtain phage-resistant S. aureus mutants with reduced growth rates and impaired production of capsular polysaccharide (137) and Salmonella enterica serovar Paratyphi B mutants that had completely lost virulence and had a shorter life span (138). Evans et al. (139) observed how resistance against a flagellatropic phage in the plant pathogen Erwinia carotovora subsp. atroseptica impaired bacterial motility, consequently reducing its virulence and resulting in easier-to-control infections. Finally, and perhaps more interestingly, Chan et al. (140), demonstrated that applying the selective pressure of phage OMKO1 to a population of MDR P. aeruginosa resulted in the selection of mutants that had a significant increase in antibiotic sensitivity. A review by León and Bastías (141) discusses how, in many cases, the trade-offs of phage resistance can be traced back to mutations in the phage receptors. For example, if a phage uses bacterial pili as its receptor, then a mutation in those pili could result in loss of adsorption, leading to phage resistance but also loss of function of the pilus structure, the consequences of which may include impaired bacterial adherence to epithelial surfaces or reduced virulence. For the case of antibiotic resensitization presented by Chan et al., it was shown that the receptor-binding site for phage OMKO1 was part of two multidrug efflux systems and that phage resistance caused loss of function of the efflux pumps, restoring antibiotic sensitivity. In summary, the natural evolution of phage and bacteria presents us with a number of opportunities to take advantage of bacterial phage resistance.
The clinical translation of the aforementioned in vitro studies has taken its first steps. The cases presented by Schooley et al. (78) and Chan et al. (79), described in “Experience in Clinical Trials” above, contain interesting insights related to this therapeutic approach. In the former, when the A. baumannii strain causing the infection acquired phage resistance, two fitness costs were observed: loss of the bacterial capsule and resensitization to minocycline. The trade-offs were effectively exploited in order to eradicate the infection, as minocycline was added to the antibiotic regime and the patient’s immune system could have theoretically improved its phagocytic capacity against the mutant pathogen. In the latter, the phage that was used, OMKO1, had been previously shown to direct the evolution of P. aeruginosa toward antibiotic resensitization in vitro. The phage was administered to the patient along with ceftazidime, an antibiotic that the pathogen was resistant against. Even though the phage-antibiotic combination was shown to reduce bacterial densities in the graft material in vitro, and ultimately a favorable outcome in the patient was achieved, the authors did not recapitulate the resensitization event in vivo.
This evolutionary trade-off approach could usher in the next generation of phage therapy: the use of selected phages that force an evolutionary cost on their bacterial host. In order to consistently and reliably take advantage of the evolutionary trade-offs associated with phage resistance in the clinical setting, we must use well-characterized phages with defined host receptors and molecular mechanisms of infection and resistance. Current research suggests that phage resistance is a nonphylogenetically conserved, highly species-specific, evolutionary trait (142). Indeed, a number of studies have highlighted that phage resistance can repeatably and predictably occur, as seen in Enterococcus faecalis mutations within the integral outer membrane protein granting phage resistance (143). However, further research is required to assess the broad applicability of these evolutionary mechanisms across critical MDR bacterial pathogens within a therapeutic setting.
Novel Approaches: Bioengineering and Vaccines
We have entered the age of “synthetic biology,” where the principles of biology and engineering are conjugated to design, create, or modify existing biological entities to perform tasks they would not naturally do (144). Bioengineering of phages could dramatically increase their therapeutic potential via a range of mechanisms, including expanded host range, switching host tropism, delivery of exogenous genes, or modification of phage capsids (Fig. 2D). Mahichi et al. (145) were able to expand the host range of E. coli phage T2 by incorporating the long tail fiber genes of phage IP008 through homologous recombination. The resulting chimeric phage exhibited the broader host range of phage IP008, while still maintaining the strong lytic activity of phage T2 (145). Furthermore, Lu and Collins (146) modified E. coli phage T7 to express the enzyme dispersin B, which is capable of degrading one of the key components of bacterial biofilms, within the bacterial host cell upon infection. The engineered virus was capable of reducing biofilm cell counts by >100-fold compared to the wild-type phage. Finally, Kim et al. (147) found that by conjugating polyethylene glycol (PEG) to phages, their blood circulation time was increased, possibly through evasion of the T-cell-mediated immune response. Similar modifications could be used to improve other pharmacokinetic parameters of phage therapy (148, 149).
Bioengineered phages have also been used to increase antibiotic specificity and to combat MDR bacterial pathogens. Yacoby et al. (150) devised a way to attach chloramphenicol molecules to lytic phages and used the viral specificity to deliver the antibiotic directly to bacterial cells, improving the in vitro potency of the drug by a factor of ∼20,000 and potentially eliminating the side effects caused by its interaction with human cells and the microbiota. Similarly, phages have been used to deliver photosensitizing agents to target bacteria, making them susceptible to photodynamic inactivation without disturbing the rest of the microbial community, an approach that has been demonstrated against MRSA (151) and the fungal pathogen Candida albicans (152). Finally, genetic engineering can utilize phage-mediated gene delivery for therapeutic purposes (Fig. 2D). The genetic material that can be delivered or inserted into the bacterial cell includes dominant sensitive genes to reverse antibiotic resistance (153), CRISPR/Cas9 sequences to inactivate virulence genes (154), modified lethal transcription regulators (155), small regulatory RNAs to silence antibiotic resistance determinants (156), and even genes that code for proteins capable of increasing the susceptibility to specific antibiotics (157).
Bioengineered phages have also found use in the field of vaccinology. Vaccines have arguably had a larger positive effect on global health than antibiotics, and vaccine development has a leading role in the fight against antibiotic resistance (158). Phage display is the most significant strategy for phage-based vaccine design and antigen expression. Here, the vaccine sequence, or antigen, is cloned into one of the phage capsid proteins, typically either the major capsid protein or an accessory protein. As a result, the antigen is expressed on the phage surface and readily presented to the immune system (159). Conversely, in phage DNA vaccines a eukaryotic gene expression cassette contains the antigen sequence and is cloned into the phage genome. After administration, the phages are recognized and taken up by antigen-presenting cells (APCs) such as macrophages and dendritic cells, which will in turn express the antigen and initiate an adaptive immune response (159). An unrelated mechanism is the use of bacteria that have lost their virulence, as a result of acquisition of phage resistance, as attenuated vaccines, since they retain their immunogenicity (137, 138). With innovative approaches being constantly tested, we foresee a remarkably vast potential in therapeutic phage bioengineering.
PHAGE THERAPY: OPPORTUNITIES AND CHALLENGES IN MODERN SOCIETY
Since their discovery over a century ago, phages have transformed biology (160). Phage research led to the discovery and understanding of major biological processes, including the fundamental principles of molecular biology, ecology, and evolution, and has led to the development of modern techniques such as genetic sequencing and genome editing (161). Phage research is moving quickly, and its results are being applied to an ever-expanding list of fields, including medical diagnostics, biocontrol and agriculture, nanotechnology, and drug discovery (162). It is time to embrace phage therapy with attention, determination, and enthusiasm, not only because it could potentially save innumerable patients from dying of resistant bacterial infections (163) but also because fully embracing phage therapy will carry a significant added value across a range of human endeavors. Conversely, attaining widespread use of phage therapy will pose a number of challenges that will have to be creatively addressed. Here, we discuss some of the opportunities and challenges that phage therapy presents to modern society in relation to education, accessibility, and economic growth.
Using Phage Therapy for Education and Outreach
We can use phage therapy as a proficient vehicle for education and scientific outreach.
Although efficacy is the keystone for adequate translation, education plays an important role in the acceptance, uptake, and dissemination of new therapeutic strategies by a population. Conversely, poor health literacy can lead to frank misuse of health care resources (as exemplified by antibiotics) or to dissemination of nonfactual information and active dissent (as seen with the antivaccine movement). Phage therapy is at a critical point where we are seeing increased therapeutic applications, media attention, and public awareness. There needs to be a concerted effort to promote education on phage therapy and avoid the pitfalls seen with other health care resources. With the rebirth of phage therapy, we propose four guiding educational principles to maximize its benefits. First, we must encourage, maintain, and capitalize on the extensive media attention phage therapy is already receiving. Second, we should direct outreach to all stakeholders: scientists, clinicians, pharmaceutical and biotechnology companies, public health officials, policy makers and lawmakers, politicians, and of course, the broader community. Education must be pervasive through all social layers. Third, we must send messages that are clear, approachable, and honest. Improving the public perception of phage therapy is certainly desirable, but transparency will ultimately gain the trust of the public. Fourth, we should incorporate all the fields of human knowledge in the initiative. Artistic and cultural approaches—museum exhibitions, narrative literature, filmmaking, music—have great potential for receiving inspiration from and disseminating information on phage therapy. The Pulitzer Award-winning novel Arrowsmith, for example, tells the story of a medical doctor using phages to control an outbreak of bubonic plague. However, we must be careful not to overhype the findings and translation possibilities. Complete information must be shared, contrasting success stories with the obstacles that are encountered.
Accessibility to Phage Therapy: Overcoming Equity Issues
The pathway of phage therapy from the lab bench to the bedside is still hindered. Current accessibility to phage therapy for a given patient is extremely low, with only three options presently available. (i) Medical tourism to established phage therapy clinics is possible, with destinations including Georgia, Poland, Russia, or the United States. This treatment approach incurs a high financial cost (typically at the patient’s expense), sometimes requires intricate paperwork regarding the transport of clinical samples and phages, and may require multiple visits to be effective. (ii) There are extended-access (compassionate-use) programs in some countries, including the United States, Canada, Australia, Japan, China, the United Kingdom, France, Belgium, and Italy (164). Unfortunately, the legal process can be long and convoluted, the programs are currently limited to life-threatening cases, they require a physician willing to consider and apply phage therapy, and there may be limited access to clinically relevant phages. (iii) Clinical trials are currently operating. There are databases of privately and publicly funded studies conducted around the world (such as clinicaltrials.gov). Nevertheless, the recruitment processes are typically difficult and highly selective, studies are limited to specific infections, and the patients face the possibility of being randomized to the placebo control groups.
Equity issues associated with phage therapy arise when considering, for example, the high personal expense associated with medical tourism. There is an existing public perception that phage therapy is available only to highly connected, wealthy individuals with the ability to mobilize large medical teams. Consequently, we recognize that significant efforts will be required to make phage therapy scalable to a larger population and not exclusively a personalized medicine intervention. In any case, increased accessibility must be accompanied by patient education, creating realistic expectations on the possibilities and limitations of therapeutic success.
Interestingly, social media, online platforms, and educational programs can facilitate the communication between patients and phage researchers, helping improve accessibility to phage therapy. Phage Directory (@phagedirectory), for example, is an online “catalogue” of researchers working on open phage therapy for streamlined collaborations between researchers, patients, doctors, and regulatory bodies. Likewise, Phages for Global Health (phagesforglobalhealth.org) develops programs aimed at teaching phage biology to scientists in developing countries and at creating international, multidisciplinary teams that codevelop phage products for specific applications, also in developing countries. Alternatively, the program SEA-PHAGES (Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science; seaphages.org) has engaged undergraduate students from over 73 institutions in phage research by allowing them to isolate, characterize, and name new phages (165). These resources can act as matchmaking applications between phage experts and people needing phages or interested in them, and they have the potential to empower the patients, generate new leaders in the field, and expand the availability of phages, in both the short and long terms.
Financial and Regulatory Considerations
Efficacy, education, accessibility, and public awareness will all effectively increase the pressure on regulatory agencies to accelerate the establishment of preliminary legal frameworks on the extensive production, testing, use, and vigilance of phage therapy. An approach that has been devised in Belgium is considering tailor-made phage medicines as magistral preparations, thus enabling a preexisting legal framework to be applied (166). Each country should take insights from its particular constitution and medical and industrial systems to develop the regulatory frameworks needed for the use of phage therapy. One of the expected challenges will be the management of intellectual property (IP) rights. Naturally occurring biological organisms, including phages, cannot be patented. As such, IP will likely be focused around methodologies, with first-mover advantage and clinical data on the efficacy of the phage product establishing the market value.
On a brighter side, we believe that the widespread use of phage therapy has the potential to catalyze economic growth. The established pharmaceutical industry has been called a dormant stakeholder for phage therapy whose involvement is limited by the lower degree of urgency it perceives (167). This is the reason why the primary investment into phage therapy translation will likely come from government bodies (168), private foundations, investment firms, commercial linkages, and universities. The pipeline of therapeutic phage manufacturing can be divided into smaller enterprises, including phage isolation, phage library expansion, purification and safety assessment, screening and personalized therapy, automation of methods, large-scale production, storage, and transport. This means that the bulk of this new economic activity could be undertaken by emerging start-ups and biotechnology companies, calling for experts in multiple fields and creating new job opportunities. We acknowledge that the field of phage therapy manufacture can be described as high-risk/high-reward, but successful trailblazers stand to gain considerable revenues and market share.
CONCLUDING REMARKS
The looming threat of antibiotic resistance calls for immediate action. Phage therapy is well suited to be part of the multidimensional strategies to fight against it. Simply put, phage therapy needs to be included in our repertoire of treatments against antibiotic-resistant pathogens, and the sooner the better. Additionally, there is no singular effective approach to clinical use of phage therapy, and in fact, its diversity and adaptability are among its greatest advantages. Although some gaps in knowledge must be filled before we can standardize the use of phage therapy, the field is rapidly advancing. Finally, we believe that although the widespread use of phage therapy seems to be a challenging process, undertaking it will bring along societal, commercial, and economic benefits that will far outreach those from the clinical standpoint alone.
ACKNOWLEDGMENTS
We gratefully acknowledge Christian Vásconez for his input regarding the clarity and structure of the manuscript.
Fernando L. Gordillo Altamirano acknowledges the support received from Monash University through the Monash Postgraduate Research Scholarship funding his doctoral studies. This work, including the efforts of Jeremy J. Barr, was funded by the Australian Research Council (ARC) Discovery Early Career Researcher Award (DECRA) (DE170100525), the National Health and Medical Research Council (NHMRC: 1156588), and the Perpetual Trustees Australia award (2018HIG00007).
Biographies

Fernando L. Gordillo Altamirano, M.D., M.Med., obtained his medical degree from the Pontificia Universidad Católica del Ecuador (PUCE) and his master of medicine (in infection and immunity) degree from the University of Sydney. He is currently a second-year Ph.D. student and a graduate teaching associate in medical microbiology and genetics at Monash University. With an exceptional academic record, he graduated valedictorian from medical school and was a finalist for the John C Harsanyi Medal for international student achievement at the Sydney Alumni Awards in 2018, and his doctoral studies are supported by a Monash postgraduate research scholarship. His research activities have focused on the social and psychological variables affecting health care for patients with chronic conditions and, since 2014, the antibiotic resistance crisis. His Ph.D. project looks into a novel phage therapy approach against Acinetobacter baumannii. He is also a TEDX speaker (“The Dark Side of Medicine,” TEDXQuito 2017) and a mental health activist.

Jeremy J. Barr, B.Biot., Hons., Ph.D., graduated with a bachelor’s degree in microbial biotechnology honors class I, a graduate certificate in research commercialization, and a Ph.D. in environmental microbiology from the University of Queensland (UQ), Australia. He then completed a five-year postdoctoral research fellowship at San Diego State University (SDSU), USA, where he still holds an adjunct professorship, in bacteriophage biology. In 2016, he joined the Monash University School of Biological Sciences as a lecturer and group leader, where he leads a research group investigating the tripartite interactions between bacteriophages and their bacterial and human hosts (https://thebarrlab.org). He is further involved in projects implementing phage therapy to combat multidrug-resistant bacterial pathogens. He has published numerous scientific articles and is listed as a coinventor on patent applications. Finally, he is involved with scientific outreach and education on bacteriophages, including coauthoring a graphic novel and online editorials on phages.
REFERENCES
- 1.Clokie MRJ, Millard AD, Letarov AV, Heaphy S. 2011. Phages in nature. Bacteriophage 1:31–45. doi: 10.4161/bact.1.1.14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Golkar Z, Bagasra O, Pace DG. 2014. Bacteriophage therapy: a potential solution for the antibiotic resistance crisis. J Infect Dev Ctries 8:129–136. doi: 10.3855/jidc.3573. [DOI] [PubMed] [Google Scholar]
- 3.Chanishvili N. 2012. Phage therapy—history from Twort and d'Herelle through Soviet experience to current approaches. Adv Virus Res 83:3–40. doi: 10.1016/B978-0-12-394438-2.00001-3. [DOI] [PubMed] [Google Scholar]
- 4.Duckworth DH. 1976. Who discovered bacteriophage? Bacteriol Rev 40:793–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Viertel TM, Ritter K, Horz H-P. 2014. Viruses versus bacteria—novel approaches to phage therapy as a tool against multidrug-resistant pathogens. J Antimicrob Chemother 69:2326–2336. doi: 10.1093/jac/dku173. [DOI] [PubMed] [Google Scholar]
- 6.Aminov RI. 2010. A brief history of the antibiotic era: lessons learned and challenges for the future. Front Microbiol 1:134. doi: 10.3389/fmicb.2010.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alanis AJ. 2005. Resistance to antibiotics: are we in the post-antibiotic era? Arch Med Res 36:697–705. doi: 10.1016/j.arcmed.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 8.Ganz T, Lehrer RI. 1999. Antibiotic peptides from higher eukaryotes: biology and applications. Mol Med Today 5:292–297. doi: 10.1016/S1357-4310(99)01490-2. [DOI] [PubMed] [Google Scholar]
- 9.Aminov RI. 2009. The role of antibiotics and antibiotic resistance in nature. Environ Microbiol 11:2970–2988. doi: 10.1111/j.1462-2920.2009.01972.x. [DOI] [PubMed] [Google Scholar]
- 10.Perron GG, Whyte L, Turnbaugh PJ, Goordial J, Hanage WP, Dantas G, Desai MM. 2015. Functional characterization of bacteria isolated from ancient arctic soil exposes diverse resistance mechanisms to modern antibiotics. PLoS One 10:e0069533. doi: 10.1371/journal.pone.0069533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allen HK, Moe LA, Rodbumrer J, Gaarder A, Handelsman J. 2009. Functional metagenomics reveals diverse beta-lactamases in a remote Alaskan soil. ISME J 3:243–251. doi: 10.1038/ismej.2008.86. [DOI] [PubMed] [Google Scholar]
- 12.D'Costa VM, King CE, Kalan L, Morar M, Sung WW, Schwarz C, Froese D, Zazula G, Calmels F, Debruyne R, Golding GB, Poinar HN, Wright GD. 2011. Antibiotic resistance is ancient. Nature 477:457–461. doi: 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- 13.Ventola CL. 2015. The antibiotic resistance crisis. 1. Causes and threats. P T 40:277–283. [PMC free article] [PubMed] [Google Scholar]
- 14.Price LB, Newland J, Bole A, Bortolaia V, Larsen J, Loneragan GH, Roach S, Smith TC, So AD, Tollefson L, Wagenaar J, Zaoutis T. 2017. Combating antibiotic resistance—a policy roadmap to reduce use of medically important antibiotics in livestock. George Washington University, Washington, DC. [Google Scholar]
- 15.Finley RL, Collignon P, Larsson DG, McEwen SA, Li XZ, Gaze WH, Reid-Smith R, Timinouni M, Graham DW, Topp E. 2013. The scourge of antibiotic resistance: the important role of the environment. Clin Infect Dis 57:704–710. doi: 10.1093/cid/cit355. [DOI] [PubMed] [Google Scholar]
- 16.Munita JM, Arias CA. 2016. Mechanisms of antibiotic resistance, p 481–511. In Kudva I, Cornick N, Plummer P, Zhang Q, Nicholson T, Bannarantine J, Bellaire G (ed), Virulence mechanisms of bacterial pathogens, 5th ed ASM Press, Washington, DC. [Google Scholar]
- 17.D'Costa VM, McGrann KM, Hughes DW, Wright GD. 2006. Sampling the antibiotic resistome. Science 311:374–377. doi: 10.1126/science.1120800. [DOI] [PubMed] [Google Scholar]
- 18.Olivares J, Bernardini A, Garcia-Leon G, Corona F, Sanchez M B, Martinez JL. 2013. The intrinsic resistome of bacterial pathogens. Front Microbiol 4:103. doi: 10.3389/fmicb.2013.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yelin I, Kishony R. 2018. Antibiotic resistance. Cell 172:1136–1136. doi: 10.1016/j.cell.2018.02.018. [DOI] [PubMed] [Google Scholar]
- 20.Davies J, Davies D. 2010. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev 74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Institute of Medicine Forum on Microbial Threats. 2010. Antibiotic resistance: implications for global health and novel intervention strategies: workshop summary. National Academies Press, Washington, DC. [PubMed] [Google Scholar]
- 22.Luepke KH, Mohr JF. 2017. The antibiotic pipeline: reviving research and development and speeding drugs to market. Expert Rev Anti Infect Ther 15:425–433. doi: 10.1080/14787210.2017.1308251. [DOI] [PubMed] [Google Scholar]
- 23.Simpkin VL, Renwick MJ, Kelly R, Mossialos E. 2017. Incentivising innovation in antibiotic drug discovery and development: progress, challenges and next steps. J Antibiot 70:1087–1096. doi: 10.1038/ja.2017.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 25.Pendleton JN, Gorman SP, Gilmore BF. 2013. Clinical relevance of the ESKAPE pathogens. Expert Rev Anti Infect Ther 11:297–308. doi: 10.1586/eri.13.12. [DOI] [PubMed] [Google Scholar]
- 26.World Health Organization. 2017. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. WHO Press, Geneva, Switzerland. [Google Scholar]
- 27.Fleming V, Buck B, Nix N, Kumar P, Southwood R. 2013. Community-acquired pneumonia with risk for drug-resistant pathogens. South Med J 106:209–216. doi: 10.1097/SMJ.0b013e318287fe71. [DOI] [PubMed] [Google Scholar]
- 28.Bours PH, Polak R, Hoepelman AI, Delgado E, Jarquin A, Matute AJ. 2010. Increasing resistance in community-acquired urinary tract infections in Latin America, five years after the implementation of national therapeutic guidelines. Int J Infect Dis 14:e770–e774. doi: 10.1016/j.ijid.2010.02.2264. [DOI] [PubMed] [Google Scholar]
- 29.World Health Organization. 2014. Antimicrobial resistance: global report on surveillance 2014. WHO Press, Geneva, Switzerland. [Google Scholar]
- 30.Cosgrove SE. 2006. The relationship between antimicrobial resistance and patient outcomes: mortality, length of hospital stay, and health care costs. Clin Infect Dis 42:S82–S89. doi: 10.1086/499406. [DOI] [PubMed] [Google Scholar]
- 31.The Review on Antimicrobial Resistance, chaired by Jim O’Neill. 2014. Antimicrobial resistance: tackling a crisis for the health and wealth of nations. http://www.jpiamr.eu/wp-content/uploads/2014/12/AMR-Review-Paper-Tackling-a-crisis-for-the-health-and-wealth-of-nations_1-2.pdf.
- 32.World Health Organization 2015. Global action plan on antimicrobial resistance. WHO Press, Geneva, Switzerland. [Google Scholar]
- 33.Doron S, Davidson LE. 2011. Antimicrobial stewardship. Mayo Clin Proc 86:1113–1123. doi: 10.4065/mcp.2011.0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cassir N, Rolain JM, Brouqui P. 2014. A new strategy to fight antimicrobial resistance: the revival of old antibiotics. Front Microbiol 5:551. doi: 10.3389/fmicb.2014.00551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Falagas ME, Kasiakou SK, Saravolatz LD. 2005. Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin Infect Dis 40:1333–1341. doi: 10.1086/429323. [DOI] [PubMed] [Google Scholar]
- 36.Young R, Gill JJ. 2015. Phage therapy redux—what is to be done? Science 350:1163–1164. doi: 10.1126/science.aad6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bertozzi Silva J, Storms Z, Sauvageau D. 2016. Host receptors for bacteriophage adsorption. FEMS Microbiol Lett 363:fnw002. doi: 10.1093/femsle/fnw002. [DOI] [PubMed] [Google Scholar]
- 38.Lin DM, Koskella B, Lin HC. 2017. Phage therapy: an alternative to antibiotics in the age of multi-drug resistance. World J Gastrointest Pharmacol Ther 8:162–173. doi: 10.4292/wjgpt.v8.i3.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fortier L-C, Sekulovic O. 2013. Importance of prophages to evolution and virulence of bacterial pathogens. Virulence 4:354–365. doi: 10.4161/viru.24498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haaber J, Leisner JJ, Cohn MT, Catalan-Moreno A, Nielsen JB, Westh H, Penadés JR, Ingmer H. 2016. Bacterial viruses enable their host to acquire antibiotic resistance genes from neighbouring cells. Nat Commun 7:13333. doi: 10.1038/ncomms13333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hargreaves KR, Clokie MRJ. 2014. Clostridium difficile phages: still difficult? Front Microbiol 5:184. doi: 10.3389/fmicb.2014.00184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Philipson CW, Voegtly LJ, Lueder MR, Long KA, Rice GK, Frey KG, Biswas B, Cer RZ, Hamilton T, Bishop-Lilly KA. 2018. Characterizing phage genomes for therapeutic applications. Viruses 10:188. doi: 10.3390/v10040188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Loc-Carrillo C, Abedon ST. 2011. Pros and cons of phage therapy. Bacteriophage 1:111–114. doi: 10.4161/bact.1.2.14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elbreki M, Ross RP, Hill C, O'Mahony J, McAuliffe O, Coffey A. 2014. Bacteriophages and their derivatives as biotherapeutic agents in disease prevention and treatment. J Viruses 2014:1. doi: 10.1155/2014/382539. [DOI] [Google Scholar]
- 45.De Sordi L, Khanna V, Debarbieux L. 2017. The gut microbiota facilitates drifts in the genetic diversity and infectivity of bacterial viruses. Cell Host Microbe 22:801–808. doi: 10.1016/j.chom.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 46.Langdon A, Crook N, Dantas G. 2016. The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome Med 8:39. doi: 10.1186/s13073-016-0294-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caliendo AM, Gilbert DN, Ginocchio CC, Hanson KE, May L, Quinn TC, Tenover FC, Alland D, Blaschke AJ, Bonomo RA, Carroll KC, Ferraro MJ, Hirschhorn LR, Joseph WP, Karchmer T, MacIntyre AT, Reller LB, Jackson AF. 2013. Better tests, better care: improved diagnostics for infectious diseases. Clin Infect Dis 57:S139–S170. doi: 10.1093/cid/cit578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chhibber S, Kaur S, Kumari S. 2008. Therapeutic potential of bacteriophage in treating Klebsiella pneumoniae B5055-mediated lobar pneumonia in mice. J Med Microbiol 57:1508–1513. doi: 10.1099/jmm.0.2008/002873-0. [DOI] [PubMed] [Google Scholar]
- 49.Nilsson AS. 2014. Phage therapy—constraints and possibilities. Ups J Med Sci 119:192–198. doi: 10.3109/03009734.2014.902878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Legendre DP, Muzny CA, Marshall GD, Swiatlo E. 2014. Antibiotic hypersensitivity reactions and approaches to desensitization. Clin Infect Dis 58:1140–1148. doi: 10.1093/cid/cit949. [DOI] [PubMed] [Google Scholar]
- 51.Khan NU, Shakeel N, Makda A, Mallick AS, Ali Memon M, Hashmi SH, Khan UR, Razzak JA. 2013. Anaphylaxis: incidence, presentation, causes and outcome in patients in a tertiary-care hospital in Karachi, Pakistan. QJM Int J Med 106:1095–1101. doi: 10.1093/qjmed/hct179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sarker SA, McCallin S, Barretto C, Berger B, Pittet AC, Sultana S, Krause L, Huq S, Bibiloni R, Bruttin A, Reuteler G, Brussow H. 2012. Oral T4-like phage cocktail application to healthy adult volunteers from Bangladesh. Virology 434:222–232. doi: 10.1016/j.virol.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 53.Speck P, Smithyman A. 2016. Safety and efficacy of phage therapy via the intravenous route. FEMS Microbiol Lett 363:fnv242. doi: 10.1093/femsle/fnv242. [DOI] [PubMed] [Google Scholar]
- 54.Kohanski MA, Dwyer DJ, Collins JJ. 2010. How antibiotics kill bacteria: from targets to networks. Nat Rev Microbiol 8:423–435. doi: 10.1038/nrmicro2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baca QJ, Coen DM, Golan DE. 2012. Principles of combination chemotherapy, p 716–727. In Golan DE, Tashjian AH, Jr, Armstrong EJ, Armstrong AW (ed), Principles of pharmacology: the pathophysiologic basis of drug therapy, 3rd ed Wolters Kluwer Health/Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 56.Campbell A. 1961. Conditions for the existence of bacteriophage. Evolution 15:153–165. doi: 10.1111/j.1558-5646.1961.tb03139.x. [DOI] [Google Scholar]
- 57.Maslov S, Sneppen K. 2017. Population cycles and species diversity in dynamic kill-the-winner model of microbial ecosystems. Sci Rep 7:39642. doi: 10.1038/srep39642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roach DR, Leung CY, Henry M, Morello E, Singh D, Di Santo JP, Weitz JS, Debarbieux L. 2017. Synergy between the host immune system and bacteriophage is essential for successful phage therapy against an acute respiratory pathogen. Cell Host Microbe 22:38–47. doi: 10.1016/j.chom.2017.06.018. [DOI] [PubMed] [Google Scholar]
- 59.Hodyra-Stefaniak K, Miernikiewicz P, Drapała J, Drab M, Jończyk-Matysiak E, Lecion D, Kaźmierczak Z, Beta W, Majewska J, Harhala M, Bubak B, Kłopot A, Górski A, Dąbrowska K. 2015. Mammalian host-versus-phage immune response determines phage fate in vivo. Sci Rep 5:14802. doi: 10.1038/srep14802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Labrie SJ, Samson JE, Moineau S. 2010. Bacteriophage resistance mechanisms. Nat Rev Microbiol 8:317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 61.Doron S, Melamed S, Ofir G, Leavitt A, Lopatina A, Keren M, Amitai G, Sorek R. 2018. Systematic discovery of antiphage defense systems in the microbial pangenome. Science 359:eaar4120. doi: 10.1126/science.aar4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wittebole X, De Roock S, Opal SM. 2014. A historical overview of bacteriophage therapy as an alternative to antibiotics for the treatment of bacterial pathogens. Virulence 5:226–235. doi: 10.4161/viru.25991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eaton MD, Bayne-Jones S. 1934. Bacteriophage therapy: review of the principles and results of the use of bacteriophage in the treatment of infections. JAMA 103:1769–1776. doi: 10.1001/jama.1934.72750490003007. [DOI] [Google Scholar]
- 64.Eaton MD, Bayne-Jones S. 1934. Bacteriophage therapy: Review of the principles and results of the use of bacteriophage in the treatment of infections. JAMA 103:1847–1853. doi: 10.1001/jama.1934.72750500003009. [DOI] [Google Scholar]
- 65.Eaton MD, Bayne-Jones S. 1934. Bacteriophage therapy: review of the principles and results of the use of bacteriophage in the treatment of infections. JAMA 103:1934–1939. doi: 10.1001/jama.1934.72750510005009. [DOI] [Google Scholar]
- 66.Sulakvelidze A, Alavidze Z, Morris JG Jr.. 2001. Bacteriophage therapy. Antimicrob Agents Chemother 45:649–659. doi: 10.1128/AAC.45.3.649-659.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Soothill JS. 1992. Treatment of experimental infections of mice with bacteriophages. J Med Microbiol 37:258–261. doi: 10.1099/00222615-37-4-258. [DOI] [PubMed] [Google Scholar]
- 68.Merril CR, Biswas B, Carlton R, Jensen NC, Creed GJ, Zullo S, Adhya S. 1996. Long-circulating bacteriophage as antibacterial agents. Proc Natl Acad Sci U S A 93:3188–3192. doi: 10.1073/pnas.93.8.3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smith HW, Huggins MB. 1983. Effectiveness of phages in treating experimental Escherichia coli diarrhoea in calves, piglets and lambs. J Gen Microbiol 129:2659–2675. doi: 10.1099/00221287-129-8-2659. [DOI] [PubMed] [Google Scholar]
- 70.Rhoads DD, Wolcott RD, Kuskowski MA, Wolcott BM, Ward LS, Sulakvelidze A. 2009. Bacteriophage therapy of venous leg ulcers in humans: results of a phase I safety trial. J Wound Care 18:237. doi: 10.12968/jowc.2009.18.6.42801. [DOI] [PubMed] [Google Scholar]
- 71.Markoishvili K, Tsitlanadze G, Katsarava R, Glenn J, Sulakvelidze A. 2002. A novel sustained-release matrix based on biodegradable poly(ester amide)s and impregnated with bacteriophages and an antibiotic shows promise in management of infected venous stasis ulcers and other poorly healing wounds. Int J Dermatol 41:453–458. doi: 10.1046/j.1365-4362.2002.01451.x. [DOI] [PubMed] [Google Scholar]
- 72.Wright A, Hawkins CH, Änggård EE, Harper DR. 2009. A controlled clinical trial of a therapeutic bacteriophage preparation in chronic otitis due to antibiotic‐resistant Pseudomonas aeruginosa; a preliminary report of efficacy. Clin Otolaryngol 34:349–357. doi: 10.1111/j.1749-4486.2009.01973.x. [DOI] [PubMed] [Google Scholar]
- 73.Jault P, Leclerc T, Jennes S, Pirnay JP, Que YA, Resch G, Rousseau AF, Ravat F, Carsin H, Le Floch R, Schaal JV, Soler C, Fevre C, Arnaud I, Bretaudeau L, Gabard J. 2018. Efficacy and tolerability of a cocktail of bacteriophages to treat burn wounds infected by Pseudomonas aeruginosa (PhagoBurn): a randomised, controlled, double-blind phase 1/2 trial. Lancet Infect Dis 19:35–45. doi: 10.1016/S1473-3099(18)30482-1. [DOI] [PubMed] [Google Scholar]
- 74.Bruttin A, Brussow H. 2005. Human volunteers receiving Escherichia coli phage T4 orally: a safety test of phage therapy. Antimicrob Agents Chemother 49:2874–2878. doi: 10.1128/AAC.49.7.2874-2878.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sarker SA, Berger B, Deng Y, Kieser S, Foata F, Moine D, Descombes P, Sultana S, Huq S, Bardhan PK, Vuillet V, Praplan F, Brussow H. 2017. Oral application of Escherichia coli bacteriophage: safety tests in healthy and diarrheal children from Bangladesh. Environ Microbiol 19:237–250. doi: 10.1111/1462-2920.13574. [DOI] [PubMed] [Google Scholar]
- 76.Gindin M, Febvre HP, Rao S, Wallace TC, Weir TL. 2018. Bacteriophage for gastrointestinal health (PHAGE) study: evaluating the safety and tolerability of supplemental bacteriophage consumption. J Am Coll Nutr 29:1–8. doi: 10.1080/07315724.2018.1483783. [DOI] [PubMed] [Google Scholar]
- 77.Cooper CJ, Khan Mirzaei M, Nilsson AS. 2016. Adapting drug approval pathways for bacteriophage-based therapeutics. Front Microbiol 7:1209. doi: 10.3389/fmicb.2016.01209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schooley RT, Biswas B, Gill JJ, Hernandez-Morales A, Lancaster J, Lessor L, Barr JJ, Reed SL, Rohwer F, Benler S, Segall AM, Taplitz R, Smith DM, Kerr K, Kumaraswamy M, Nizet V, Lin L, McCauley MD, Strathdee SA, Benson CA, Pope RK, Leroux BM, Picel AC, Mateczun AJ, Cilwa KE, Regeimbal JM, Estrella LA, Wolfe DM, Henry MS, Quinones J, Salka S, Bishop-Lilly KA, Young R, Hamilton T. 2017. Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob Agents Chemother 61:e0095417. doi: 10.1128/AAC.00954-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chan BK, Turner PE, Kim S, Mojibian HR, Elefteriades JA, Narayan D. 2018. Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol Med Public Health 2018:60–66. doi: 10.1093/emph/eoy005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jennes S, Merabishvili M, Soentjens P, Pang KW, Rose T, Keersebilck E, Soete O, François P-M, Teodorescu S, Verween G, Verbeken G, De Vos D, Pirnay J-P. 2017. Use of bacteriophages in the treatment of colistin-only-sensitive Pseudomonas aeruginosa septicaemia in a patient with acute kidney injury-a case report. Crit Care 21:129. doi: 10.1186/s13054-017-1709-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chan BK, Abedon ST, Loc-Carrillo C. 2013. Phage cocktails and the future of phage therapy. Future Microbiol 8:769–783. doi: 10.2217/fmb.13.47. [DOI] [PubMed] [Google Scholar]
- 82.Hua Y, Luo T, Yang Y, Dong D, Wang R, Wang Y, Xu M, Guo X, Hu F, He P. 2017. Phage therapy as a promising new treatment for lung infection caused by carbapenem-resistant Acinetobacter baumannii in mice. Front Microbiol 8:2659. doi: 10.3389/fmicb.2017.02659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jeon J, Ryu CM, Lee JY, Park JH, Yong D, Lee K. 2016. In vivo application of bacteriophage as a potential therapeutic agent to control OXA-66-like carbapenemase-producing Acinetobacter baumannii strains belonging to sequence type 357. Appl Environ Microbiol 82:4200–4208. doi: 10.1128/AEM.00526-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Biswas B, Adhya S, Washart P, Paul B, Trostel AN, Powell B, Carlton R, Merril CR. 2002. Bacteriophage therapy rescues mice bacteremic from a clinical isolate of vancomycin-resistant Enterococcus faecium. Infect Immun 70:204–210. doi: 10.1128/IAI.70.1.204-210.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Levin BR, Bull JJ. 2004. Population and evolutionary dynamics of phage therapy. Nat Rev Microbiol 2:166–173. doi: 10.1038/nrmicro822. [DOI] [PubMed] [Google Scholar]
- 86.Lenski RE. 1984. Two-step resistance by Escherichia coli B to bacteriophage T2. Genetics 107:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stern A, Sorek R. 2011. The phage-host arms-race: shaping the evolution of microbes. Bioessays 33:43–51. doi: 10.1002/bies.201000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chibani-Chennoufi S, Sidoti J, Bruttin A, Kutter E, Sarker S, Brüssow H. 2004. In vitro and in vivo bacteriolytic activities of Escherichia coli phages: implications for phage therapy. Antimicrob Agents Chemother 48:2558–2569. doi: 10.1128/AAC.48.7.2558-2569.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Abedon ST. 2017. Information phage therapy research should report. Pharmaceuticals (Basel) 10:43. doi: 10.3390/ph10020043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hall AR, De Vos D, Friman VP, Pirnay JP, Buckling A. 2012. Effects of sequential and simultaneous applications of bacteriophages on populations of Pseudomonas aeruginosa in vitro and in wax moth larvae. Appl Environ Microbiol 78:5646–5652. doi: 10.1128/AEM.00757-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kutter E, De Vos D, Gvasalia G, Alavidze Z, Gogokhia L, Kuhl S, Abedon ST. 2010. Phage therapy in clinical practice: treatment of human infections. Curr Pharm Biotechnol 11:69–86. doi: 10.2174/138920110790725401. [DOI] [PubMed] [Google Scholar]
- 92.Villarroel J, Larsen MV, Kilstrup M, Nielsen M. 2017. Metagenomic analysis of therapeutic PYO phage cocktails from 1997 to 2014. Viruses 9:328. doi: 10.3390/v9110328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.McCallin S, Alam Sarker S, Barretto C, Sultana S, Berger B, Huq S, Krause L, Bibiloni R, Schmitt B, Reuteler G, Brüssow H. 2013. Safety analysis of a Russian phage cocktail: from metagenomic analysis to oral application in healthy human subjects. Virology 443:187–196. doi: 10.1016/j.virol.2013.05.022. [DOI] [PubMed] [Google Scholar]
- 94.Kishor C, Mishra RR, Saraf SK, Kumar M, Srivastav AK, Nath G. 2016. Phage therapy of staphylococcal chronic osteomyelitis in experimental animal model. Indian J Med Res 143:87–94. doi: 10.4103/0971-5916.178615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Saussereau E, Vachier I, Chiron R, Godbert B, Sermet I, Dufour N, Pirnay JP, De Vos D, Carrie F, Molinari N, Debarbieux L. 2014. Effectiveness of bacteriophages in the sputum of cystic fibrosis patients. Clin Microbiol Infect 20:O983–O990. doi: 10.1111/1469-0691.12712. [DOI] [PubMed] [Google Scholar]
- 96.Melo LDR, Veiga P, Cerca N, Kropinski AM, Almeida C, Azeredo J, Sillankorva S. 2016. Development of a phage cocktail to control Proteus mirabilis catheter-associated urinary tract infections. Front Microbiol 7:1024. doi: 10.3389/fmicb.2016.01024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nale JY, Spencer J, Hargreaves KR, Buckley AM, Trzepiński P, Douce GR, Clokie MRJ. 2016. Bacteriophage combinations significantly reduce Clostridium difficile growth in vitro and proliferation in vivo. Antimicrob Agents Chemother 60:968–981. doi: 10.1128/AAC.01774-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Worthington RJ, Melander C. 2013. Combination approaches to combat multi-drug resistant bacteria. Trends Biotechnol 31:177–184. doi: 10.1016/j.tibtech.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Torres-Barceló C, Hochberg ME. 2016. Evolutionary rationale for phages as complements of antibiotics. Trends Microbiol 24:249–256. doi: 10.1016/j.tim.2015.12.011. [DOI] [PubMed] [Google Scholar]
- 100.Wright GD. 2016. Antibiotic adjuvants: rescuing antibiotics from resistance. Trends Microbiol 24:862–871. doi: 10.1016/j.tim.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 101.Comeau AM, Tetart F, Trojet SN, Prere MF, Krisch HM. 2007. Phage-antibiotic synergy (PAS): beta-lactam and quinolone antibiotics stimulate virulent phage growth. PLoS One 2:3799. doi: 10.1371/journal.pone.0000799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Knezevic P, Curcin S, Aleksic V, Petrusic M, Vlaski L. 2013. Phage-antibiotic synergism: a possible approach to combatting Pseudomonas aeruginosa. Res Microbiol 164:55–60. doi: 10.1016/j.resmic.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 103.Kamal F, Dennis J. 2015. Burkholderia cepacia complex phage-antibiotic synergy (PAS): antibiotics stimulate lytic phage activity. Appl Environ Microbiol 81:1132–1138. doi: 10.1128/AEM.02850-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kaur S, Harjai K, Chhibber S. 2012. Methicillin-resistant Staphylococcus aureus phage plaque size enhancement using sublethal concentrations of antibiotics. Appl Environ Microbiol 78:8227–8233. doi: 10.1128/AEM.02371-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chaudhry WN, Concepción-Acevedo J, Park T, Andleeb S, Bull JJ, Levin BR. 2017. Synergy and order effects of antibiotics and phages in killing Pseudomonas aeruginosa biofilms. PLoS One 12:e0168615. doi: 10.1371/journal.pone.0168615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Torres-Barceló C, Arias-Sánchez FI, Vasse M, Ramsayer J, Kaltz O, Hochberg ME. 2014. A window of opportunity to control the bacterial pathogen Pseudomonas aeruginosa combining antibiotics and phages. PLoS One 9:e106628. doi: 10.1371/journal.pone.0106628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chhibber S, Kaur T, Sandeep K. 2013. Co-therapy using lytic bacteriophage and linezolid: effective treatment in eliminating methicillin resistant Staphylococcus aureus (MRSA) from diabetic foot infections. PLoS One 8:e56022. doi: 10.1371/journal.pone.0056022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Oechslin F, Piccardi P, Mancini S, Gabard J, Moreillon P, Entenza JM, Resch G, Que Y-A. 2017. Synergistic interaction between phage therapy and antibiotics clears Pseudomonas aeruginosa infection in endocarditis and reduces virulence. J Infect Dis 215:703–712. doi: 10.1093/infdis/jiw632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Valerio N, Oliveira C, Jesus V, Branco T, Pereira C, Moreirinha C, Almeida A. 2017. Effects of single and combined use of bacteriophages and antibiotics to inactivate Escherichia coli. Virus Res 240:8–17. doi: 10.1016/j.virusres.2017.07.015. [DOI] [PubMed] [Google Scholar]
- 110.Maciejewska B, Olszak T, Drulis-Kawa Z. 2018. Applications of bacteriophages versus phage enzymes to combat and cure bacterial infections: an ambitious and also a realistic application? Appl Microbiol Biotechnol 102:2563–2581. doi: 10.1007/s00253-018-8811-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Moxon ER, Kroll JS. 1990. The role of bacterial polysaccharide capsules as virulence factors. Curr Top Microbiol Immunol 150:65–85. [DOI] [PubMed] [Google Scholar]
- 112.Bedi MS, Verma V, Chhibber S. 2009. Amoxicillin and specific bacteriophage can be used together for eradication of biofilm of Klebsiella pneumoniae B5055. World J Microbiol Biotechnol 25:1145. doi: 10.1007/s11274-009-9991-8. [DOI] [Google Scholar]
- 113.Mushtaq N, Redpath MB, Luzio JP, Taylor PW. 2005. Treatment of experimental Escherichia coli infection with recombinant bacteriophage-derived capsule depolymerase. J Antimicrob Chemother 56:160–165. doi: 10.1093/jac/dki177. [DOI] [PubMed] [Google Scholar]
- 114.Born Y, Fieseler L, Klumpp J, Eugster MR, Zurfluh K, Duffy B, Loessner MJ. 2014. The tail-associated depolymerase of Erwinia amylovora phage L1 mediates host cell adsorption and enzymatic capsule removal, which can enhance infection by other phage. Environ Microbiol 16:2168–2180. doi: 10.1111/1462-2920.12212. [DOI] [PubMed] [Google Scholar]
- 115.Bernhardt TG, Wang I-N, Struck DK, Young R. 2002. Breaking free: “protein antibiotics” and phage lysis. Res Microbiol 153:493–501. doi: 10.1016/S0923-2508(02)01330-X. [DOI] [PubMed] [Google Scholar]
- 116.Gründling A, Bläsi U, Young R. 2000. Genetic and biochemical analysis of dimer and oligomer interactions of the λ S holin. J Bacteriol 182:6082–6090. doi: 10.1128/JB.182.21.6082-6090.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Young R. 2013. Phage lysis: do we have the hole story yet? Curr Opin Microbiol 16:790–797. doi: 10.1016/j.mib.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Xu M, Struck DK, Deaton J, Wang I-N, Young R. 2004. A signal-arrest-release sequence mediates export and control of the phage P1 endolysin. Proc Natl Acad Sci U S A 101:6415–6420. doi: 10.1073/pnas.0400957101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Eugster MR, Loessner MJ. 2012. Wall teichoic acids restrict access of bacteriophage endolysin Ply118, Ply511, and PlyP40 cell wall binding domains to the Listeria monocytogenes peptidoglycan. J Bacteriol 194:6498–6506. doi: 10.1128/JB.00808-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nelson D, Schuch R, Chahales P, Zhu S, Fischetti VA. 2006. PlyC: a multimeric bacteriophage lysin. Proc Natl Acad Sci U S A 103:10765–10770. doi: 10.1073/pnas.0604521103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yoong P, Schuch R, Nelson D, Fischetti VA. 2004. Identification of a broadly active phage lytic enzyme with lethal activity against antibiotic-resistant Enterococcus faecalis and Enterococcus faecium. J Bacteriol 186:4808–4812. doi: 10.1128/JB.186.14.4808-4812.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yang H, Wang M, Yu J, Wei H. 2015. Antibacterial activity of a novel peptide-modified lysin against Acinetobacter baumannii and Pseudomonas aeruginosa. Front Microbiol 6:1471. doi: 10.3389/fmicb.2015.01471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Briers Y, Lavigne R. 2015. Breaking barriers: expansion of the use of endolysins as novel antibacterials against Gram-negative bacteria. Future Microbiol 10:377–390. doi: 10.2217/fmb.15.8. [DOI] [PubMed] [Google Scholar]
- 124.Briers Y, Walmagh M, Van Puyenbroeck V, Cornelissen A, Cenens W, Aertsen A, Oliveira H, Azeredo J, Verween G, Pirnay JP, Miller S, Volckaert G, Lavigne R. 2014. Engineered endolysin-based “artilysins” to combat multidrug-resistant gram-negative pathogens. mBio 5:e01379-14. doi: 10.1128/mBio.01379-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fenton M, Casey PG, Hill C, Gahan CGM, Ross RP, McAuliffe O, O'Mahony J, Maher F, Coffey A. 2010. The truncated phage lysin CHAP(k) eliminates Staphylococcus aureus in the nares of mice. Bioeng Bugs 1:404–407. doi: 10.4161/bbug.1.6.13422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rashel M, Uchiyama J, Ujihara T, Uehara Y, Kuramoto S, Sugihara S, Yagyu K, Muraoka A, Sugai M, Hiramatsu K, Honke K, Matsuzaki S. 2007. Efficient elimination of multidrug-resistant Staphylococcus aureus by cloned lysin derived from bacteriophage phi MR11. J Infect Dis 196:1237–1247. doi: 10.1086/521305. [DOI] [PubMed] [Google Scholar]
- 127.van Rijen MM, Bonten M, Wenzel RP, Kluytmans JA. 2008. Intranasal mupirocin for reduction of Staphylococcus aureus infections in surgical patients with nasal carriage: a systematic review. J Antimicrob Chemother 61:254–261. doi: 10.1093/jac/dkm480. [DOI] [PubMed] [Google Scholar]
- 128.Pastagia M, Euler C, Chahales P, Fuentes-Duculan J, Krueger JG, Fischetti VA. 2011. A novel chimeric lysin shows superiority to mupirocin for skin decolonization of methicillin-resistant and -sensitive Staphylococcus aureus strains. Antimicrob Agents Chemother 55:738–744. doi: 10.1128/AAC.00890-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Loeffler JM, Nelson D, Fischetti VA. 2001. Rapid killing of Streptococcus pneumoniae with a bacteriophage cell wall hydrolase. Science 294:2170–2172. doi: 10.1126/science.1066869. [DOI] [PubMed] [Google Scholar]
- 130.Cheng Q, Nelson D, Zhu S, Fischetti VA. 2005. Removal of group B streptococci colonizing the vagina and oropharynx of mice with a bacteriophage lytic enzyme. Antimicrob Agents Chemother 49:111–117. doi: 10.1128/AAC.49.1.111-117.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Daniel A, Euler C, Collin M, Chahales P, Gorelick KJ, Fischetti VA. 2010. Synergism between a novel chimeric lysin and oxacillin protects against infection by methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 54:1603–1612. doi: 10.1128/AAC.01625-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jun SY, Jang IJ, Yoon S, Jang K, Yu K-S, Cho JY, Seong M-W, Jung GM, Yoon SJ, Kang SH. 2017. Pharmacokinetics and tolerance of the phage endolysin-based candidate drug SAL200 after a single intravenous administration among healthy volunteers. Antimicrob Agents Chemother 61:e02629–e02616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jun SY, Jung GM, Yoon SJ, Youm SY, Han HY, Lee JH, Kang SH. 2016. Pharmacokinetics of the phage endolysin-based candidate drug SAL200 in monkeys and its appropriate intravenous dosing period. Clin Exp Pharmacol Physiol 43:1013–1016. doi: 10.1111/1440-1681.12613. [DOI] [PubMed] [Google Scholar]
- 134.Totté JEE, van Doorn MB, Pasmans SGMA. 2017. Successful treatment of chronic Staphylococcus aureus-related dermatoses with the topical endolysin Staphefekt SA.100: a report of 3 cases. Case Rep Dermatol 9:19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Totté J, de Wit J, Pardo L, Schuren F, van Doorn M, Pasmans S. 2017. Targeted anti-staphylococcal therapy with endolysins in atopic dermatitis and the effect on steroid use, disease severity and the microbiome: study protocol for a randomized controlled trial (MAAS trial). Trials 18:404. doi: 10.1186/s13063-017-2118-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gomez P, Buckling A. 2011. Bacteria-phage antagonistic coevolution in soil. Science 332:106–109. doi: 10.1126/science.1198767. [DOI] [PubMed] [Google Scholar]
- 137.Capparelli R, Nocerino N, Lanzetta R, Silipo A, Amoresano A, Giangrande C, Becker K, Blaiotta G, Evidente A, Cimmino A, Iannaccone M, Parlato M, Medaglia C, Roperto S, Roperto F, Ramunno L, Iannelli D. 2010. Bacteriophage-resistant Staphylococcus aureus mutant confers broad immunity against staphylococcal infection in mice. PLoS One 5:e11720. doi: 10.1371/journal.pone.0011720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Capparelli R, Nocerino N, Iannaccone M, Ercolini D, Parlato M, Chiara M, Iannelli D. 2010. Bacteriophage therapy of Salmonella enterica: a fresh appraisal of bacteriophage therapy. J Infect Dis 201:52–61. doi: 10.1086/648478. [DOI] [PubMed] [Google Scholar]
- 139.Evans TJ, Trauner A, Komitopoulou E, Salmond GP. 2010. Exploitation of a new flagellatropic phage of Erwinia for positive selection of bacterial mutants attenuated in plant virulence: towards phage therapy. J Appl Microbiol 108:676–685. doi: 10.1111/j.1365-2672.2009.04462.x. [DOI] [PubMed] [Google Scholar]
- 140.Chan BK, Sistrom M, Wertz JE, Kortright KE, Narayan D, Turner PE. 2016. Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci Rep 6:26717. doi: 10.1038/srep26717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.León M, Bastías R. 2015. Virulence reduction in bacteriophage resistant bacteria. Front Microbiol 6:343. doi: 10.3389/fmicb.2015.00343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Martiny JB, Jones SE, Lennon JT, Martiny AC. 2015. Microbiomes in light of traits: a phylogenetic perspective. Science 350:aac9323. doi: 10.1126/science.aac9323. [DOI] [PubMed] [Google Scholar]
- 143.Duerkop BA, Huo W, Bhardwaj P, Palmer KL, Hooper LV. 2016. Molecular basis for lytic bacteriophage resistance in enterococci. mBio 7:e01304-16. doi: 10.1128/mBio.01304-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Cameron DE, Bashor CJ, Collins JJ. 2014. A brief history of synthetic biology. Nat Rev Microbiol 12:381–390. doi: 10.1038/nrmicro3239. [DOI] [PubMed] [Google Scholar]
- 145.Mahichi F, Synnott AJ, Yamamichi K, Osada T, Tanji Y. 2009. Site-specific recombination of T2 phage using IP008 long tail fiber genes provides a targeted method for expanding host range while retaining lytic activity. FEMS Microbiol Lett 295:211–217. doi: 10.1111/j.1574-6968.2009.01588.x. [DOI] [PubMed] [Google Scholar]
- 146.Lu TK, Collins JJ. 2007. Dispersing biofilms with engineered enzymatic bacteriophage. Proc Natl Acad Sci U S A 104:11197–11202. doi: 10.1073/pnas.0704624104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kim KP, Cha JD, Jang EH, Klumpp J, Hagens S, Hardt WD, Lee KY, Loessner MJ. 2008. PEGylation of bacteriophages increases blood circulation time and reduces T-helper type 1 immune response. Microbial Biotech 1:247–257. doi: 10.1111/j.1751-7915.2008.00028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Nobrega FL, Costa AR, Santos JF, Siliakus MF, van Lent JWM, Kengen SWM, Azeredo J, Kluskens LD. 2016. Genetically manipulated phages with improved pH resistance for oral administration in veterinary medicine. Sci Rep 6:39235. doi: 10.1038/srep39235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Vitiello CL, Merril CR, Adhya S. 2005. An amino acid substitution in a capsid protein enhances phage survival in mouse circulatory system more than a 1000-fold. Virus Res 114:101–103. doi: 10.1016/j.virusres.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 150.Yacoby I, Bar H, Benhar I. 2007. Targeted drug-carrying bacteriophages as antibacterial nanomedicines. Antimicrob Agents Chemother 51:2156–2163. doi: 10.1128/AAC.00163-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hope CK, Packer S, Wilson M, Nair SP. 2009. The inability of a bacteriophage to infect Staphylococcus aureus does not prevent it from specifically delivering a photosensitizer to the bacterium enabling its lethal photosensitization. J Antimicrob Chemother 64:59–61. doi: 10.1093/jac/dkp157. [DOI] [PubMed] [Google Scholar]
- 152.Dong S, Shi H, Zhang X, Chen X, Cao D, Mao C, Gao X, Wang L. 2018. Difunctional bacteriophage conjugated with photosensitizers for Candida albicans-targeting photodynamic inactivation. Int J Nanomed (Lond) 13:2199–2216. doi: 10.2147/IJN.S156815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Edgar R, Friedman N, Molshanski-Mor S, Qimron U. 2012. Reversing bacterial resistance to antibiotics by phage-mediated delivery of dominant sensitive genes. Appl Environ Microbiol 78:744–751. doi: 10.1128/AEM.05741-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Park JY, Moon BY, Park JW, Thornton JA, Park YH, Seo KS. 2017. Genetic engineering of a temperate phage-based delivery system for CRISPR/Cas9 antimicrobials against Staphylococcus aureus. Sci Rep 7:44929. doi: 10.1038/srep44929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Moradpour Z, Sepehrizadeh Z, Rahbarizadeh F, Ghasemian A, Yazdi MT, Shahverdi AR. 2009. Genetically engineered phage harbouring the lethal catabolite gene activator protein gene with an inducer-independent promoter for biocontrol of Escherichia coli. FEMS Microbiol Lett 296:67–71. doi: 10.1111/j.1574-6968.2009.01620.x. [DOI] [PubMed] [Google Scholar]
- 156.Libis VK, Bernheim AG, Basier C, Jaramillo-Riveri S, Deyell M, Aghoghogbe I, Atanaskovic I, Bencherif AC, Benony M, Koutsoubelis N, Lochner AC, Marinkovic ZS, Zahra S, Zegman Y, Lindner AB, Wintermute EH. 2014. Silencing of antibiotic resistance in E. coli with engineered phage bearing small regulatory RNAs. ACS Synth Biol 3:1003–1006. doi: 10.1021/sb500033d. [DOI] [PubMed] [Google Scholar]
- 157.Ronayne EA, Wan YCS, Boudreau BA, Landick R, Cox MM. 2016. P1 Ref endonuclease: a molecular mechanism for phage-enhanced antibiotic lethality. PLoS Genet 12:44929. doi: 10.1371/journal.pgen.1005797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Jansen KU, Knirsch C, Anderson AS. 2018. The role of vaccines in preventing bacterial antimicrobial resistance. Nat Med 24:10–19. doi: 10.1038/nm.4465. [DOI] [PubMed] [Google Scholar]
- 159.Aghebati-Maleki L, Bakhshinejad B, Baradaran B, Motallebnezhad M, Aghebati-Maleki A, Nickho H, Yousefi M, Majidi J. 2016. Phage display as a promising approach for vaccine development. J Biomed Sci 23:66. doi: 10.1186/s12929-016-0285-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Rohwer F, Segall AM. 2015. In retrospect: a century of phage lessons. Nature 528:46–48. doi: 10.1038/528046a. [DOI] [PubMed] [Google Scholar]
- 161.Keen EC. 2015. A century of phage research: bacteriophages and the shaping of modern biology. Bioessays 37:6–9. doi: 10.1002/bies.201400152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.O'Sullivan L, Buttimer C, McAuliffe O, Bolton D, Coffey A. 2016. Bacteriophage-based tools: recent advances and novel applications. F1000Research 5:2782. doi: 10.12688/f1000research.9705.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Henein A. 2013. What are the limitations on the wider therapeutic use of phage? Bacteriophage 3:e24872. doi: 10.4161/bact.24872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Górski A, Międzybrodzki R, Weber-Dąbrowska B, Fortuna W, Letkiewicz S, Rogóż P, Jończyk-Matysiak E, Dąbrowska K, Majewska J, Borysowski J. 2016. Phage therapy: combating infections with potential for evolving from merely a treatment for complications to targeting diseases. Front Microbiol 7:1515. doi: 10.3389/fmicb.2016.01515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Jordan TC, Burnett SH, Carson S, Caruso SM, Clase K, DeJong RJ, Dennehy JJ, Denver DR, Dunbar D, Elgin SCR, Findley AM, Gissendanner CR, Golebiewska UP, Guild N, Hartzog GA, Grillo WH, Hollowell GP, Hughes LE, Johnson A, King RA, Lewis LO, Li W, Rosenzweig F, Rubin MR, Saha MS, Sandoz J, Shaffer CD, Taylor B, Temple L, Vazquez E, Ware VC, Barker LP, Bradley KW, Jacobs-Sera D, Pope WH, Russell DA, Cresawn SG, Lopatto D, Bailey CP, Hatfull GF. 2014. A broadly implementable research course in phage discovery and genomics for first-year undergraduate students. mBio 5:e01051-13. doi: 10.1128/mBio.01051-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Pirnay J-P, Verbeken G, Ceyssens P-J, Huys I, De Vos D, Ameloot C, Fauconnier A. 2018. The magistral phage. Viruses 10:64. doi: 10.3390/v10020064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Expert Round Table on Acceptance and Re-implementation of Bacteriophage Therapy. 2016. Silk route to the acceptance and re-implementation of bacteriophage therapy. Biotechnol J 11:595–600. doi: 10.1002/biot.201600023. [DOI] [PubMed] [Google Scholar]
- 168.Presidential Advisory Council on Combating Antibiotic-Resistant Bacteria 2017. Recommendations for incentivizing the development of vaccines, diagnostics and therapeutics to combat antibiotic resistance. US Department of Health and Human Services, Washington, DC. [Google Scholar]
- 169.Leverentz B, Conway WS, Camp MJ, Janisiewicz WJ, Abuladze T, Yang M, Saftner R, Sulakvelidze A. 2003. Biocontrol of Listeria monocytogenes on fresh-cut produce by treatment with lytic bacteriophages and a bacteriocin. Appl Environ Microbiol 69:4519–4526. doi: 10.1128/AEM.69.8.4519-4526.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Slopek S, Kucharewicz-Krukowska A, Weber-Dabrowska B, Dabrowski M. 1985. Results of bacteriophage treatment of suppurative bacterial infections. IV. Evaluation of the results obtained in 370 cases. Arch Immunol Ther Exp (Warsz) 33:219–240. [PubMed] [Google Scholar]
- 171.Sanger F, Air GM, Barrell BG, Brown NL, Coulson AR, Fiddes JC, Hutchison CA, Slocombe PM, Smith M. 1977. Nucleotide sequence of bacteriophage φX174 DNA. Nature 265:687–695. doi: 10.1038/265687a0. [DOI] [PubMed] [Google Scholar]
- 172.Ackermann H-W. 2011. The first phage electron micrographs. Bacteriophage 1:225–227. doi: 10.4161/bact.1.4.17280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ellis EL, Delbrück M. 1939. The growth of bacteriophage. J Gen Physiol 22:365–384. doi: 10.1085/jgp.22.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Abedon ST, Thomas-Abedon C, Thomas A, Mazure H. 2011. Bacteriophage prehistory: is or is not Hankin, 1896, a phage reference? Bacteriophage 1:174–178. doi: 10.4161/bact.1.3.16591. [DOI] [PMC free article] [PubMed] [Google Scholar]