Abstract
The deubiquitylation of target proteins is mediated by deubiquitylating enzymes (DUB) such as OTUB1, which plays an important role in immune response, cell cycle progression, and DNA repair. Within these processes, OTUB1 reduces the ubiquitylation of target proteins in two distinct ways, either by using its catalytic DUB activity or in a noncatalytic manner by inhibiting the E2-conjugating enzyme. Here, we show that the ubiquitin-like modifier FAT10 regulates OTUB1 stability and functionality in different ways. Covalent FAT10ylation of OTUB1 resulted in its proteasomal degradation, whereas a noncovalent interaction stabilized OTUB1. We provide evidence that OTUB1 interacts directly with FAT10 and the E2-conjugating enzyme USE1. This interaction strongly stimulated OTUB1 DUB activity toward Lys-48–linked diubiquitin. Furthermore, the noncovalent interaction between FAT10 and OTUB1 not only enhanced its isopeptidase activity toward Lys-48–linked ubiquitin moieties but also strengthened its noncatalytic activity in reducing Lys-63 polyubiquitylation of its target protein TRAF3 (TNF receptor–associated factor 3). Additionally, the cellular clearance of overall polyubiquitylation by OTUB1 was strongly stimulated through the presence of FAT10. The addition of FAT10 also led to an increased interaction between OTUB1 and its cognate E2 UbcH5B, implying a function of FAT10 in the inhibition of polyubiquitylation. Overall, these data indicate that FAT10 not only plays a role in covalent modification, leading its substrates to proteasomal degradation, but also regulates the stability and functionality of target proteins by interacting in a noncovalent manner. FAT10 is thereby able to exert a major influence on ubiquitylation processes.
Keywords: ubiquitin, ubiquitin thioesterase (OTUB1), ubiquitin-conjugating enzyme (E2 enzyme), ubiquitylation (ubiquitination), proteasome, enzyme activity, FAT10, inflammation, ubiquitin-like protein, USE1
Introduction
The homeostasis of protein synthesis and degradation under normal and under stress conditions is one of the most tightly regulated processes in mammalian cells. A central regulator of proteostasis is the ubiquitin–proteasome system. Substrate proteins become post-translationally modified by ubiquitin and, depending on chain type and length, are targeted for degradation by the 26S proteasome (1). The family of ubiquitin and ubiquitin-like modifiers contains several other members such as SUMO, Nedd8, and ISG15 (2). Although all members share the ubiquitin fold as a structural similarity, the functional consequences of modifications by ubiquitin-like modifiers are extremely diverse (2). Protein stability or activity, subcellular localization, or protein-protein interactions are altered as well as regulatory processes such as DNA repair, cell cycle progression, and apoptosis (2, 3).
FAT10 (human leukocyte antigen-F (HLA-F) adjacent transcript 10) also belongs to the family of ubiquitin-like proteins, as it possesses two of the conserved β-grasp ubiquitin folds composed in a head-to-tail formation combined by a short linker (4, 5). The N- and C-terminal domains of FAT10 share sequence homologies of 29% and 36% with ubiquitin, respectively. The C-terminal domain contains a diglycine motif, required for conjugation to substrate proteins (6–8). In contrast to ubiquitin, FAT10 expression is restricted largely to the organs of the immune system such as thymus, spleen, and lymph nodes (9–11). Also numerous different cancer types highly express FAT10 (12). In tissues that do not express FAT10 without stimulation, endogenous expression can be induced synergistically by the proinflammatory cytokines interferon-γ (IFNγ)4 and tumor necrosis factor-α (TNFα) (13, 14).
The conjugation of FAT10 to an internal lysine residue of target proteins, similar to the ubiquitin cascade, is mediated by the E1-activating enzyme UBA6 (15–17), the E2-conjugating enzyme USE1 (15, 18, 19), and probably so far unknown E3 ligases. FAT10ylated proteins, such as USE1 itself (19), the tumor suppressor p53 (20), the ubiquitin-activating enzyme UBE1 (21), and the autophagy marker p62 (22), become degraded by the 26S proteasome, whereas FAT10 is degraded along with its substrates (23). Besides covalent interactions, FAT10 interacts prominently with proteins in a noncovalent manner. The presence of FAT10 influences the localization and functionality of noncovalent interaction partners like spindle assembly checkpoint protein MAD2 or stress granule protein HDAC6 (13, 24, 25).
To learn more about the functional consequences of FAT10 interacting covalently and noncovalently with target proteins, we recently performed a yeast two-hybrid screen with USE1 as a bait and a human prey thymus library. While searching for new substrates and interaction partners, we identified members of the ubiquitin conjugation family. One of the candidates was the deubiquitylating enzyme otubain 1 (OTUB1) belonging to the ovarian tumor family (OTU) of cysteine proteases (26). OTUB1 possesses a specificity to clear Lys-48 polyubiquitin chains from substrate proteins such as the tumor suppressor p53 (26). The removal of polyubiquitin chains from p53 by OTUB1 prevents p53 from proteasomal degradation and leads to stabilization of the protein (27).
Furthermore, OTUB1 inhibits polyubiquitylation in a noncatalytic manner. Interaction with its cognate E2-conjugating enzymes UbcH5b and Ubc13 leads to suppression of the conjugating activity of the E2 enzymes and thereby to a reduced transfer of ubiquitin onto E3 ligases and substrate proteins (28, 29). Additionally, OTUB1 plays a role in multiple biological processes such as immune response and DNA damage response as well as pathogen biology; however, its catalytic activity is not required in every one of these processes (30–34).
Because only a few interaction partners of the E2-conjugating enzyme USE1 are known so far and no FAT10 deconjugating enzyme has been published as yet, we investigated several different possibilities of how USE1, FAT10, and OTUB1 could influence each other. We examined whether OTUB1 possesses a deFAT10ylating activity or whether it inhibits FAT10 conjugation by interacting with the E2-conjugating enzyme USE1. We further analyzed whether FAT10 has an impact on the functionality of OTUB1 regarding its isopeptidase activity or its inhibitory noncatalytic activity.
In this study, we elucidated USE1 and FAT10 as new interaction partners of OTUB1. We show that the direct interaction of FAT10 with OTUB1 stimulates the OTUB1 isopeptidase activity toward Lys-48–linked diubiquitin. Furthermore, we demonstrate that a covalent FAT10ylation of OTUB1 in cellulo and in vitro targets the conjugate for proteasomal degradation, whereas a direct, noncovalent interaction between FAT10 and OTUB1 stabilizes the protein. Thereby, FAT10 positively affects both the catalytic and noncatalytic activities of OTUB1 in reducing the polyubiquitylation of its substrate protein TRAF3 (TNF receptor–associated factor 3) and of the general formation of ubiquitin conjugates in cellulo. Here, we have proposed a new bilateral regulatory effect of FAT10 on its substrate protein OTUB1 whereby it influences its target in both a covalent and noncovalent manner.
Results
OTUB1 does not act as deconjugating enzyme for FAT10 but is a new interaction partner of USE1
To search for interaction partners of the FAT10 E2-conjugating enzyme USE1, we performed a yeast two-hybrid screen using USE1 as bait and a thymus cDNA library as prey (Clontech, Takara; data not shown). Besides candidates for a putative FAT10-specific E3 ligase, we started to investigate the deubiquitylating enzyme (DUB) otubain 1 (OTUB1) as a new interaction partner of USE1. We asked whether OTUB1 might function as a deFAT10ylating enzyme or whether FAT10 and USE1 might regulate the stability and functionality of OTUB1.
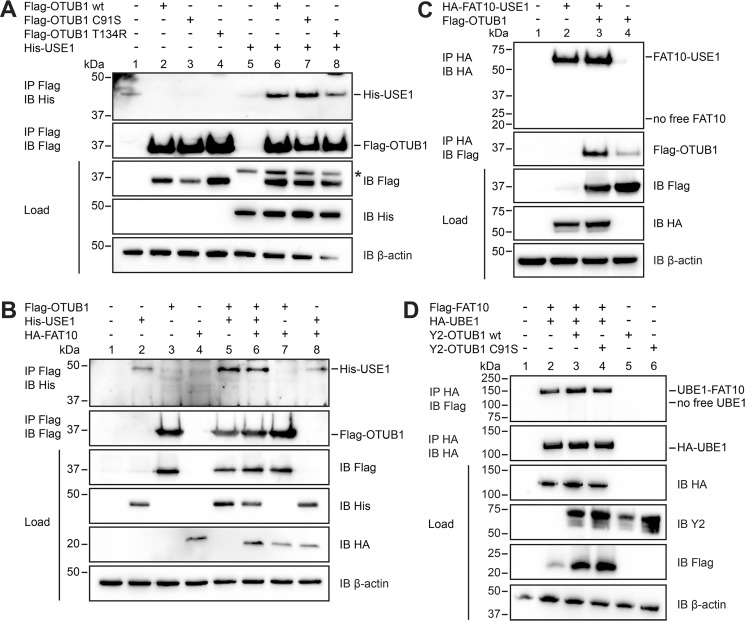
To confirm OTUB1 as a new interaction partner of USE1, HEK293 cells were transiently transfected with expression plasmids for His-USE1 and the Flag-tagged OTUB1 versions of the WT, the active-site cysteine mutant (C91S), and an E2 interaction deletion mutant (T134R). The point mutation T134R in the catalytic OTU domain of OTUB1 has been shown to disrupt the interaction with the E2-conjugating enzyme UbcH5B and thereby diminish the inhibitory effect of OTUB1 on ubiquitin conjugation (28). Our subsequent co-immunoprecipitation revealed a USE1 interaction with OTUB1 WT as well as with the C91S variant (Fig. 1A). The interaction of OTUB1 T134R with USE1 was reduced (Figs. 1A and S1), supporting a structural similarity between USE1 and UbcH5B as published recently (35).
Figure 1.
OTUB1 does not act as a deconjugating enzyme for FAT10 but is a novel interaction partner of USE1. A, HEK293 cells were transiently transfected with expression plasmids for Flag-OTUB1 WT, active-site cysteine mutant (Flag-OTUB1 C91S), OTU domain mutant T134R (Flag-OTUB1 T134R), and His-USE1. Cells were lysed in lysis buffer containing 1% NP-40, and cleared lysates were subjected to immunoprecipitation (IP) with anti-Flag M2 antibody–coupled agarose. Proteins were separated on 4–12% NuPAGE bis-Tris gels, and Western blot (IB) analysis was performed with antibodies reactive against His or Flag. β-Actin was used as loading control. The two upper panels show the immunoprecipitated proteins, and the three lower panels show the total protein expression in the cell lysates (Load). The asterisk marks the remaining His-USE1 signals after stripping. B, transiently transfected HEK293 cells were harvested, lysed, and subjected to immunoprecipitation, and Western blot analysis with antibodies against His, Flag, or HA was performed as described in A. C, HEK293 cells were transiently transfected with expression plasmids for Flag-OTUB1 WT and a linear fusion protein consisting of FAT10 C terminally fused to USE1 (HA-FAT10-USE1). Immunoprecipitation using anti-HA–agarose with subsequent Western blot analysis against Flag and HA was performed as described in A. D, Flag-FAT10, HA-UBE1, and Y2-tagged OTUB1 variants (WT or C91S) were transiently overexpressed in HEK293 cells. Lysis, immunoprecipitation using HA–agarose, and Western blot analysis were performed as described in A. One representative experiment of three experiments with similar outcomes is shown.
Former publications discuss an increased interaction of OTUB1 and its cognate E2-conjugating enzyme UbcH5B in the presence of ubiquitin (28, 36). To investigate whether ubiquitin or FAT10 could increase the interaction intensity of OTUB1 and USE1, both proteins were co-expressed in HEK293 cells in the presence or absence of one of the two modifiers (Fig. 1B; data not shown for ubiquitin). Overexpression of FAT10 did not change the interaction between USE1 and OTUB1 compared with the interaction in the absence of FAT10 in cellulo (Fig. 1B, lanes 5 and 6). Likewise, the presence of ubiquitin had also no influence on the interplay between E2 and DUB in this experimental setup (data not shown). As OTUB1 is known to function as a deconjugating enzyme for ubiquitin (26), we asked whether OTUB1 could also remove FAT10 from substrate proteins. To answer this question a linear fusion construct (HA-FAT10-USE1) was used to mimic a peptide-linked FAT10ylated USE1. In HEK293 cells this fusion construct was co-expressed in the presence or absence of Flag-tagged OTUB1, and a co-immunoprecipitation with subsequent Western blot analysis was performed (Fig. 1C). Although a high amount of linear HA-FAT10-USE1 was observed (Fig. 1C, top panel), no free HA-FAT10 and thereby no removal of FAT10 from USE1 was detectable, indicating that OTUB1 does not function as a deconjugating enzyme for linearly conjugated FAT10. As it is published that OTUB1 interacts with certain E2-conjugating enzymes when loaded with ubiquitin to regulate ubiquitin conjugation to substrate proteins (28, 36, 37), we tested whether OTUB1 interacts with the FAT10-USE1 fusion. Western blot analysis against Flag-OTUB1 in the same experimental setup revealed an interaction between OTUB1 and the linear fusion protein (Fig. 1C, middle panel), indicating that OTUB1 still can interact with FAT10 and USE1 when both proteins are bound in a complex.
Next, we examined whether OTUB1 was able to clear FAT10 from a known isopeptide-linked substrate. Therefore, HA-UBE1 and FAT10 were transiently expressed in HEK293 cells in the absence or presence of Y2-tagged OTUB1 WT or C91S (C-terminal part of YFP at the N terminus of OTUB1), and a co-immunoprecipitation was performed (Fig. 1D). Western blot analysis detecting FAT10ylated UBE1 did not show any differences in the conjugate amount when either OTUB1 WT or C91S was present (Fig. 1D, lanes 2, 3, and 4). These data supported our finding that OTUB1 does not possess isopeptidase activity toward FAT10ylated proteins. To further confirm this hypothesis, we tested whether the DUB activity of OTUB1 has an impact on the overall FAT10 conjugation to substrate proteins. Flag-tagged OTUB1 variants were overexpressed together with HA-tagged FAT10, and a co-immunoprecipitation with subsequent Western blot analysis was performed. A reduction in the FAT10 conjugation pattern in the presence of OTUB1 WT was not observable (Fig. 2A, middle panel, lanes 2 and 3). FAT10ylation of substrate proteins also was not affected by co-expression of the OTUB1 active-site cysteine mutant C91S or the T134R E2 interaction mutant (Fig. 2A, middle panel, lanes 4 and 5). These data indicate that the new USE1 interaction partner OTUB1 does not function as a deFAT10ylating enzyme in cellulo and that this interaction has no inhibitory effect on the FAT10 conjugation pattern as shown for UbcH5B and ubiquitin conjugation (28, 36).
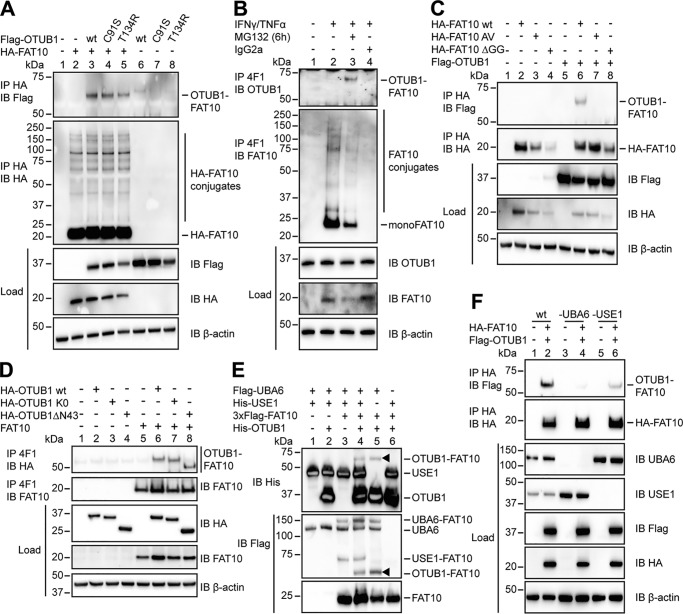
Figure 2.
OTUB1 becomes FAT10ylated in cellulo and in vitro. A, 24 h after transient transfection of HEK293 cells with the indicated expression plasmids, lysis in 1% NP-40 lysis buffer was performed. Cleared lysates were subjected to anti-HA immunoprecipitation (IP), and proteins were separated on 4–12% NuPAGE bis-Tris gels with subsequent Western blot (IB) analysis using anti-Flag M2 or anti-HA directly labeled antibodies. B, endogenous expression of FAT10 was induced by treating HEK293 cells with IFNγ and TNFα for 24 h. Where indicated, cells were treated additionally with proteasome inhibitor MG132 (10 μm) for 6 h. After lysis in 1% NP-40 lysis buffer, the cleared lysates were incubated with protein A–Sepharose and FAT10 mAb 4F1 or an unspecific IgG2a isotype control antibody. Western blot analysis was performed using a rabbit mAb against endogenous OTUB1 or a polyclonal rabbit antibody against endogenous FAT10. C and D, whole cell extracts from HEK293 cells transiently transfected with the indicated expression plasmids were treated and subsequently used in Western blot analysis as described in A and B. E, Western blot analysis of in vitro conjugation experiments where recombinant proteins (Flag-UBA6, 1 μg (0.1 mg/ml); 6His-USE1, 6 μg (0.6 mg/ml); 3xFlag-FAT10, 4 μg (0.4 mg/ml); and His-OTUB1 2.5 μg (1.25 mg/ml)) were incubated at 30 °C for 60 min, and the reaction was stopped by the addition of 5× gel sample buffer containing 4% 2-mercaptoethanol. Arrowheads indicate the OTUB1–FAT10 conjugate. F, HEK293 wt, HEK293 UBA6 KO, and HEK293 USE1 KO cells were transiently transfected with expression plasmids for HA-FAT10 and Flag-OTUB1. Subsequent lysis, immunoprecipitation, and Western blot analysis were performed as described in A. In all cellular experiments detection of endogenous β-actin was used as a loading control. One representative experiment of three experiments with similar outcomes is shown.
OTUB1 becomes FAT10ylated in cellulo and in vitro
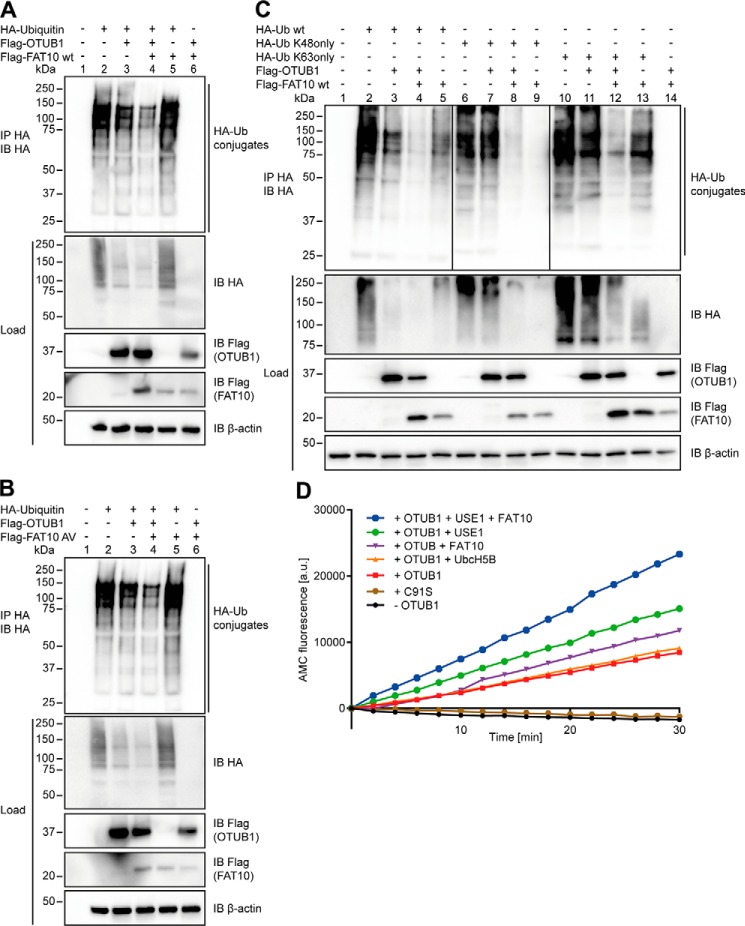
Having demonstrated that OTUB1 has no impact on FAT10 conjugation, we wondered whether FAT10 has an influence on OTUB1 stability and functionality. Indeed, in overexpression experiments with Flag-OTUB1 and HA-FAT10 and subsequent co-immunoprecipitation and Western blot analysis, OTUB1 became modified by a single FAT10 moiety (Fig. 2A, top panel). This FAT10ylation still occurred when OTUB1 possessed a point mutation in either the active site (C91S) or the catalytic OTU domain (T134R), indicating that these residues do not have a relation to the FAT10 binding to OTUB1 (Fig. 2A, top panel).
To examine the interaction between FAT10 and OTUB1 under endogenous conditions, FAT10 expression was induced by treating HEK293 cells with the proinflammatory cytokines IFNγ and TNFα. An immunoprecipitation of FAT10 with the FAT10-specific mAb 4F1 and subsequent Western blot analysis using a rabbit mAb against OTUB1 were performed. Upon additional inhibition of the 26S proteasome by treatment with MG132 for 6 h, OTUB1 became FAT10ylated under endogenous conditions (Fig. 2B) suggesting a rapid degradation of the conjugate. Next, we tested whether this covalent interaction relies on the C-terminal diglycine motif of FAT10 as shown previously for other proteins targeted by FAT10 (19, 21, 22). Therefore, two different mutant variants of FAT10 were used: HA-FAT10 AV, which carries AV instead of GG at its C terminus, and HA-FAT10 ΔGG, which is lacking the diglycine motif completely (Fig. 2C). In HEK293 cells these variants were co-expressed together with Flag-OTUB1 WT, and Western blot analysis showed that only FAT10 WT was conjugated to OTUB1 (Fig. 2C, lane 6). In contrast, both FAT10 variants carrying the mutations at the C terminus could no longer be conjugated to OTUB1 (Fig. 2C, lanes 7 and 8), showing that FAT10 conjugation is strictly dependent on the diglycine motif at the very C terminus of FAT10.
Additionally, former publications show that a lysine-less variant of a target protein such as USE1 no longer becomes FAT10ylated to the same extent as the WT protein (19). To test whether this holds true as well for OTUB1 FAT10ylation, an HA-tagged lysine-less (K0) mutant of OTUB1 was overexpressed and compared with HA-OTUB1 WT in the absence or presence of tagless FAT10 (Fig. 2D). In contrast to those former publications, the FAT10ylation of OTUB1 was not reduced when all of the lysine residues were mutated to arginines (Fig. 2D, lane 7), leaving the question open as to which type of residue FAT10 modifies in OTUB1.
Furthermore, by removing the first 43 amino acids from the N terminus of OTUB1 (OTUB1 ΔN43), the covalent FAT10ylation of this mutant OTUB1 could not be reduced (Fig. 2D, lane 8). The N terminus of OTUB1 is required for the binding of ubiquitin as shown by Wolberger and colleagues (37), indicating that binding of FAT10 may happen in another region.
To investigate whether OTUB1 becomes FAT10ylated not only in cellulo but also in vitro, Flag-UBA6, His-USE1, and His3xFlag-FAT10 were incubated together with His6-tagged OTUB1 (Enzo LifeScience) for 60 min at 30 °C. In a subsequent Western blot analysis, an OTUB1–FAT10 conjugate was detected in the presence of UBA6 and USE1 under these in vitro conditions (Fig. 2E, lane 4). Furthermore, the conjugate was also present in the absence of the E2-conjugating enzyme USE1 (Fig. 2E, lane 5, arrowhead), indicating that the FAT10ylation of OTUB1 was mediated solely by UBA6, independent of USE1. To confirm these findings in cellulo, UBA6- and USE1-CRISPR/Cas9 knockout cell lines of HEK293 parental cells were produced, and the co-immunoprecipitation experiments with overexpressed FAT10 and OTUB1 were repeated. In HEK293 wt cells the FAT10ylated OTUB1 was present as shown previously (Fig. 2F, top panel, lane 2), whereas it was almost completely absent in UBA6 KO cells (Fig. 2F, top panel, lane 4 and Fig. S2). In contrast to the in vitro experiments, the conjugate was clearly reduced in USE1 KO cells (Fig. 2F, top panel, lane 6 and Fig. S2).
Taken together, our data confirm a mono-FAT10ylation of OTUB1, which depends on the C-terminal diglycine motif of FAT10. Furthermore we show that this modification is mediated by the E1-activating enzyme UBA6 and the E2-conjugating enzyme USE1.
FAT10 targets OTUB1 for proteasomal degradation, but free OTUB1 becomes stabilized in the presence of FAT10
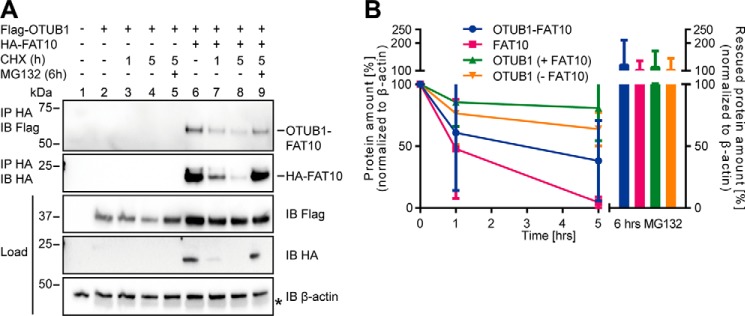
Because substrate proteins of FAT10 are described mainly as targets for degradation by the 26S proteasome (19, 21, 23, 38), we examined whether the same holds true for the OTUB1–FAT10 conjugate. FAT10 and OTUB1 overexpressing HEK293 cells were treated with cycloheximide (CHX) for the indicated time periods to inhibit protein de novo synthesis. As a control, MG132 was added to block the catalytic activity of the 26S proteasome (Fig. 3). FAT10 was almost completely degraded within 5 h, and upon inhibition of the proteasome a recovery of FAT10 protein amount was detected (Fig. 3A, middle panel). The OTUB1–FAT10 conjugate was degraded more slowly; however, the MG132-mediated rescue was almost as strong as shown for monomeric FAT10 (Fig. 3A, top panel).
Figure 3.
FAT10ylated OTUB1 is degraded by the 26S proteasome. A, transiently HA-FAT10 and Flag-OTUB1 expressing HEK293 cells were treated for the indicated time periods with cycloheximide and with proteasome inhibitor MG132 (10 μm) for another 6 h where indicated. Cell lysates were subjected to immunoprecipitation (IP) and Western blot (IB) analysis by using directly labeled antibodies detecting HA or Flag. The asterisk marks the remaining ECL signals of Flag-OTUB1 detection. One representative experiment of three experiments with similar outcomes is shown. B, Western blotting signals of OTUB1–FAT10 conjugate, FAT10, and OTUB1 in the absence or presence of FAT10 were quantified by using densitometric determinations of the signal intensity. Values (n = 3) were normalized to Western blotting signals of β-actin.
The Western blotting signals were analyzed by densitometry with normalization to protein amounts of β-actin (Fig. 3B). The calculated half-life of FAT10 is published to be ∼1.5 h (23), which we confirmed with these data. The OTUB1–FAT10 conjugate degraded more slowly, and the half-life was about 3.5 h (Fig. 3B). In previous projects the half-life of substrate proteins in the presence and absence of FAT10 was evaluated, and no difference could be detected.5 Here, we show that presence of FAT10 may have an impact on overall substrate protein stability. Contrary to expectations, the half-life of unconjugated OTUB1 increased in the presence of FAT10, and OTUB1 was stabilized as compared with OTUB1 in the absence of FAT10 (Fig. 3B).
Taken together our data imply that FAT10 influences its substrate protein, OTUB1, in two different ways. On the one hand it modifies OTUB1 in a covalent manner, to a minor extent, and targets it for proteasomal degradation. On the other hand the presence of FAT10, and thereby a putative noncovalent interaction with OTUB1, is sufficient to stabilize the substrate protein.
FAT10 interacts with USE1, UbcH5B, and OTUB1 in a direct, noncovalent manner
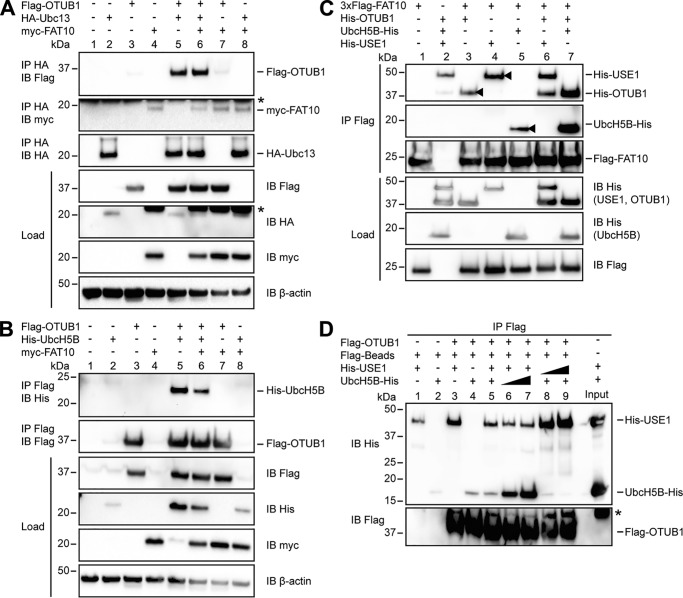
OTUB1 has been found to interact with E2-conjugating enzymes such as UbcH5B and Ubc13 (28, 36, 37). This interaction leads to an inhibition of ubiquitin transfer and additionally stimulates the OTUB1-mediated deubiquitylation of target proteins (28, 36, 37). To investigate whether FAT10 has an impact on this regulatory mechanism, we examined its possible influence on the interaction between OTUB1 and the E2-conjugating enzymes UbcH5B and Ubc13. In HEK293 cells, Flag-tagged OTUB1 and HA-tagged Ubc13 were overexpressed in the presence or absence of myc-FAT10, and a co-immunoprecipitation with subsequent Western blot analysis was performed (Fig. 4A). A noncovalent interaction between OTUB1 and Ubc13 was observed; however, this interaction did not change in the presence of FAT10 (Fig. 4A, top panel). Additionally, in this experimental setup no interaction between FAT10 and Ubc13 could be detected (Fig. 4A, middle panel).
Figure 4.
FAT10 interacts with USE1, UbcH5B and OTUB1 in a direct, noncovalent manner. A, HEK293 cells were transiently transfected with Flag-OTUB1, HA-Ubc13, and myc-FAT10. After lysis in buffer containing 1% NP-40, the cleared lysates were subjected to immunoprecipitation (IP) using HA-coupled agarose. Western blot (IB) analysis was performed by using directly labeled antibodies reactive against HA or Flag or a mouse primary antibody against myc (clone 9E10). Endogenous β-actin was used as a loading control. B, extracts from HEK293 cells transiently transfected with the indicated expression plasmids were subjected to immunoprecipitation using Flag M2–coupled agarose. Western blot analysis was performed as described in A. Asterisks in A and B mark the remaining ECL signals. C, Western blot analysis of in vitro interaction experiments where equal molar ratios (8 μg) of recombinant proteins (3xFlag-FAT10 (0.4 mg/ml), His-OTUB1 (1.25 mg/ml), His-USE1 (0.6 mg/ml), and UbcH5B-His (0.6 mg/ml)) were mixed with Flag M2–coupled agarose and incubated for 60 min at 8 °C. Western blot analysis was performed by using directly labeled antibodies detecting His or Flag. The arrowheads indicate the direct interaction of FAT10 with OTUB1, USE1, or UbcH5B. D, transiently overexpressed Flag-OTUB1 was purified from HEK293 cells by immunoprecipitation using Flag M2–coupled agarose. For in vitro competition experiments increasing amounts of UbcH5B-His (8, 42, and 84 μg; 0.6 mg/ml) and His-USE1 (4, 22, and 44 μg; 0.6 mg/ml) were added to agarose-bound Flag-OTUB1 for 60 min at 37 °C. Western blot analysis was performed as described in C. One representative experiment of three experiments with similar outcomes is shown.
We also examined the interaction between Flag-tagged OTUB1 and His-tagged UbcH5B in co-immunoprecipitation experiments (Fig. 4B). Here, an interaction between OTUB1 and UbcH5B could be detected although it was not changed with FAT10 co-expression (Fig. 4B, top panel). To investigate whether the interaction between FAT10 and OTUB1 occurs in a direct, noncovalent manner, an in vitro interaction assay was performed. Recombinant Flag-FAT10 bound to Flag M2–agarose was incubated with His-OTUB1, and a noncovalent interaction between both proteins was observed (Fig. 4C, top panel, lane 3, arrowhead). Because there was no activating E1 enzyme in the reaction mix, the interaction of OTUB1 with the modifier must have occurred in a direct and noncovalent manner.
To investigate the influence of E2-conjugating enzymes on the interaction strength of OTUB1 and FAT10, the assay was also performed with recombinant USE1 or UbcH5B. Western blot analysis showed that the direct interaction between OTUB1 and FAT10 was slightly increased by the presence of USE1 (Fig. 4C, top panel, lane 6). In former experiments the binding between FAT10 and USE1 was described as a covalent isopeptide-linked auto-modification (19). Here, for the first time, a direct, noncovalent and UBA6-independent interaction of USE1 and FAT10 could be detected (Fig. 4C, top panel, lane 4, arrowhead). However, this interaction was not increased in the presence of OTUB1 (Fig. 4C, top panel, lane 6). Furthermore, UbcH5B could be shown to interact directly with FAT10 (Fig. 4C, second panel, lane 5, arrowhead) and this interaction was strongly increased in the presence of OTUB1 (Fig. 4C, second panel, lane 7). And vice versa, the interaction of OTUB1 and FAT10 was strongly increased when UbcH5B was present (Fig. 4C, top panel, lane 7). Taken together, our data demonstrate that the presence of USE1 or UbcH5B strengthens the direct, noncovalent, UBA6-independent interaction between OTUB1 and FAT10, indicating that FAT10 and OTUB1 form a native complex, which could be a binding platform for an E2-conjugating enzyme, and thereby providing a first hint as to how FAT10 influences OTUB1 functionality.
To further investigate the interaction between OTUB1 and USE1 or UbcH5B, an in vitro competition assay was performed (Fig. 4D). Flag-OTUB1 was overexpressed in HEK293 cells and purified by using Flag M2 antibody–coupled agarose. Next, the recombinant E2-conjugating enzymes USE1 and UbcH5B were added in equimolar ratios of 1:1 (USE1, 4 μg:UbcH5B, 8 μg), 1:5 (4:42 μg), and 5:1 (22:8 μg), or 1:10 (4:84 μg) and 10:1 (44:8 μg), and incubated for 60 min at 37 °C. Western blot analysis showed a slightly reduced interaction between OTUB1 and USE1 when UbcH5B was increased (Fig. 4D, lanes 6 and 7). However, increasing amounts of USE1 competed away UbcH5B from OTUB1 (Fig. 4D, lanes 8 and 9). These data indicate that the E2-conjugating enzymes USE1 and UbcH5B competed for the same binding site within OTUB1, confirming our in cellulo data with the OTUB1 T134R mutant (Fig. 1) and implying the regulatory role of USE1 in OTUB1 functionality.
FAT10 enhances the OTUB1-mediated deubiquitylation of TRAF3
The E3 ligase TRAF3 is described as a substrate of OTUB1-mediated deubiquitylation (39, 40). Polyubiquitylation of TRAF3 occurs in two distinct manners. Degradative Lys-48 polyubiquitylation of TRAF3 during Toll-like receptor (TLR) signaling is described as essential for the induction of proinflammatory cytokine production (41). In contrast, Lys-63 autoubiquitylation of TRAF3 stimulated by other TLR triggers the noncanonical NF-κB pathway and thereby the expression of inflammatory type I interferons (41). In addition, FAT10 plays a role in the antiviral immune response mediated by TLR signaling via RIG-I (retinoic acid–inducible gene I) occurring upstream of TRAF3 (42, 43).
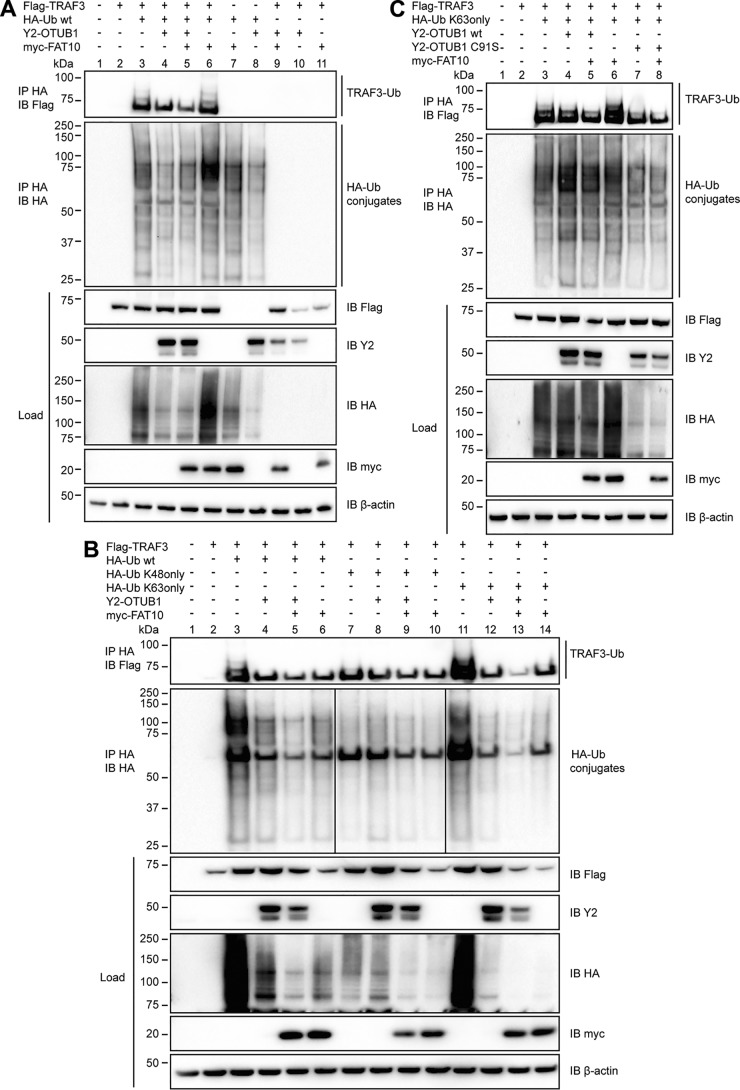
Based on this knowledge, we investigated whether FAT10 might affect the OTUB1-mediated deubiquitylation of TRAF3. In HEK293 cells Flag-TRAF3, HA-tagged ubiquitin, myc-tagged FAT10, and Y2-OTUB1 were overexpressed and an immunoprecipitation of HA tagged proteins was performed (Fig. 5A). Upon co-expression with HA-ubiquitin, Flag-TRAF3 became polyubiquitylated and this ubiquitylation pattern was reduced in the presence of OTUB1 (Fig. 5A, top panel, lanes 3 and 4). This reduction was even more pronounced when FAT10 was co-expressed (Fig. 5A, top panel, lane 5) indicating that FAT10 supports the deubiquitylating function of OTUB1 on TRAF3.
Figure 5.
FAT10 stimulates both the inhibitory and deubiquitylating activity of OTUB1 toward TRAF3. A, lysates of HEK293 cells transiently expressing Flag-TRAF3, HA-ubiquitin, Y2-OTUB1, and myc-FAT10 were subjected to immunoprecipitation (IP) using anti-HA–agarose followed by Western blot (IB) analysis using directly labeled anti-Flag or anti-HA antibodies or primary antibodies against myc or the C-terminal part of GFP. β-Actin was used as a loading control. B, HEK293 cells were transiently transfected with the indicated expression plasmids, and lysates were subjected to anti-HA immunoprecipitation and Western blot analysis as described in A. C, after transient transfection with the indicated expression plasmids, HEK293 cells were subjected to procedures and treatments as described in A. One representative experiment of three experiments with similar outcomes is shown.
To further investigate our hypothesis that OTUB1 and FAT10 have an impact on the ubiquitylation of TRAF3, the ubiquitin moieties K48only and K63only were overexpressed in the presence of TRAF3, OTUB1 and FAT10 in HEK293 cells (Fig. 5B). In both mutants all lysine residues are exchanged to arginines except for the indicated one (44). Under these conditions, TRAF3 mainly became ubiquitylated by WT or K63only ubiquitin, and this ubiquitylation was reduced by OTUB1 (Fig. 5B, top panel). Furthermore, the addition of FAT10 enhanced the OTUB1-dependent reduction of TRAF3 ubiquitylation independently of the overexpressed ubiquitin variant (Fig. 5B, top panel, lanes 8 and 9 versus 4 and 5 and 12 and 13).
Because OTUB1 mediates Lys-48–linked polyubiquitin removal (26) and inhibits Lys-63–linked polyubiquitylation (37) our data imply a more relevant role for OTUB1 and FAT10 in the latter regulatory process. To demonstrate this, the catalytically inactive mutant C91S and WT OTUB1 were expressed with TRAF3 and K63only ubiquitin (Fig. 5C). Although WT OTUB1 had only a minor impact on the ubiquitylation of TRAF3 in this particular experiment, OTUB1 C91S diminished the ubiquitylation of TRAF3 (Fig. 5C, top panel, lanes 4 and 7) indicating that the noncatalytic activity of OTUB1 toward TRAF3 was pivotal in clearing TRAF3 ubiquitylation. The additional expression of FAT10 increased the OTUB1-mediated effect in the presence of WT or C91S OTUB1 (Fig. 5C, top panel, lanes 5 and 8).
Taken together our data indicate that OTUB1 does not only partially deubiquitylate TRAF3 but also inhibits the ubiquitylation of TRAF3 in a noncatalytic manner. Moreover, the presence of FAT10 stimulated both activities of OTUB1 toward TRAF3.
FAT10 and USE1 enhance the cleavage activity of OTUB1 in vitro
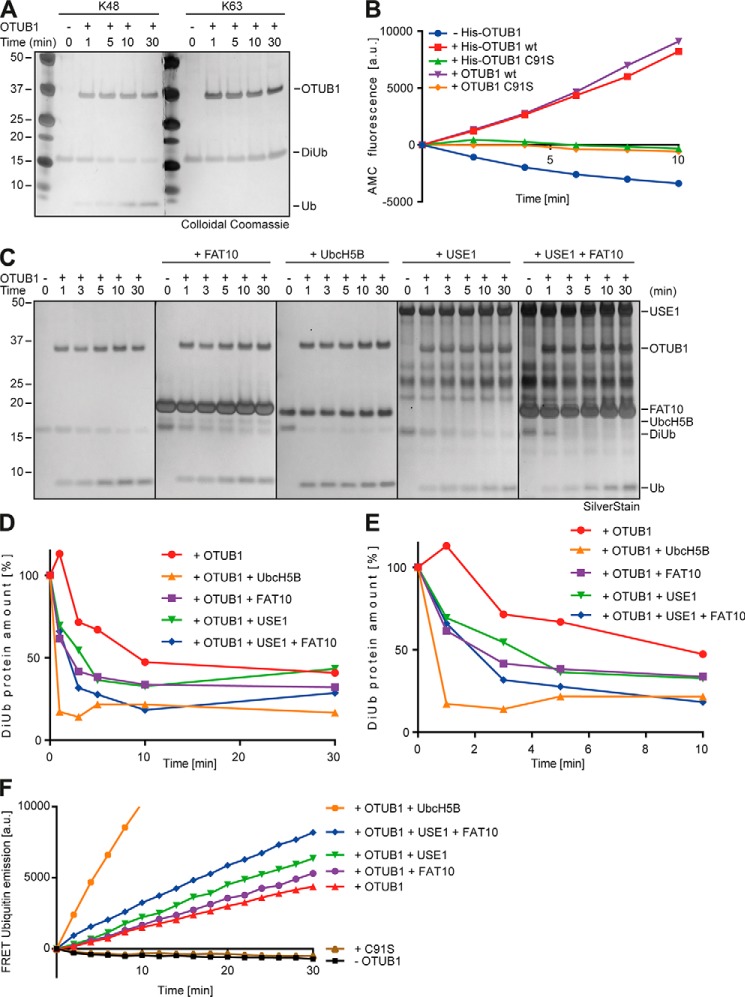
OTUB1 is described to specifically remove Lys-48–linked polyubiquitin chains from target proteins (26). To confirm this, recombinant Lys-48–linked diubiquitin was incubated with recombinant OTUB1 for the indicated time periods and SDS-PAGE with subsequent colloidal Coomassie staining was performed. Lys-63–linked diubiquitin was applied as control. Lys-48 diubiquitin amount was rapidly diminished and in parallel monoubiquitin increased, indicating a cleavage by OTUB1 over time (Fig. 6A, left panel). In contrast, Lys-63–linked diubiquitin did not serve as a substrate for OTUB1 (Fig. 6A, right panel).
Figure 6.
FAT10 and USE1 enhance the cleavage activity of OTUB1 in vitro. A, recombinant Lys-48 or Lys-63 DiUb (5 μm) was incubated with tagless OTUB1 (5 μm) for the indicated time periods at 37 °C, and reactions were stopped by the addition of gel sample buffer supplemented with 4% 2-mercaptoethanol. Proteins were separated on 4–12% NuPAGE bis-Tris gels with subsequent colloidal Coomassie staining. B, quantitative analysis of AMC fluorescence (ex. 360 nm, em. 465 nm) of ubiquitin-AMC (5 μm) incubated with His-tagged or tagless OTUB1 WT or C91S (all 10 μm) for the indicated time periods at 30 °C. C, silver-stained gel of recombinant Lys-48–linked DiUb (5 μm) incubated for the indicated time periods with tagless OTUB1 (5 μm) and tagless FAT10, UbcH5B-His, and His-USE1 (all 10 μm) at 37 °C. The reactions were stopped by the addition of gel sample buffer supplemented with 4% 2-mercaptoethanol. D, densitometric analysis of cleaved Lys-48–linked DiUb by OTUB1 in the presence of FAT10, USE1, and UbcH5B. The DiUb protein amount at time point 0 was set to 100%. E, enlarged focus on the first 10 min of the densitometric analysis in D. F, FRET-based analysis for the cleavage of internally quenched Lys-48 DiUb (400 nm) by OTUB1 WT or C91S (both 30 nm) in the absence or presence of FAT10, USE1, and UbcH5B (all 5 μm). One representative experiment of three experiments with similar outcomes is shown.
Furthermore OTUB1 WT (tagless and His6-tagged) was compared with its active-site cysteine mutant C91S (tagless and His6-tagged) in their activity to cleave ubiquitin-AMC. The removal of AMC from ubiquitin by isopeptidases leads to an alteration of the charged electron cloud around free AMC which is measurable in an increased fluorescence. The negative controls His-or tagless OTUB1 C91S did not induce AMC fluorescence (Fig. 6B) proving them as catalytically inactive. However, both WT OTUB1 variants (His6-tagged and tagless) cleaved AMC off from ubiquitin (Fig. 6B), again confirming the catalytic activity of OTUB1 toward ubiquitin.
With these active recombinant OTUB1 versions in hand we investigated in vitro whether FAT10 had an impact on the isopeptidase activity of OTUB1 toward Lys-48–linked diubiquitin. Recombinant OTUB1 and its interaction partners UbcH5B, USE1 and FAT10 were added for the indicated time periods to recombinant Lys-48–linked diubiquitin and SDS-PAGE with subsequent silver staining was performed (Fig. 6C). OTUB1-mediated cleavage of diubiquitin could be detected and was strongly stimulated by the presence of UbcH5B as published by the Wolberger group (Fig. 6C, first and third panels (36)). FAT10 or USE1 stimulated the isopeptidase activity of OTUB1 almost as strongly as UbcH5B (Fig. 6C, second and fourth panels). Moreover, combined addition of FAT10 and USE1 enhanced the stimulatory effect on OTUB1-mediated cleavage (Fig. 6C, rightmost panel). Densitometric analyses confirmed the gel-based results (Fig. 6D) and the enlarged focus into the first 10 mins supported the detected effects (Fig. 6E). These data already mirrored the in cellulo data and confirmed that FAT10 stimulated the OTUB1 isopeptidase activity. Additionally, free USE1 was almost as effective as free UbcH5B in stimulating the DUB activity of OTUB1.
To further confirm that USE1 and FAT10 stimulate the DUB activity of OTUB1, FRET Lys-48–linked diubiquitin internally quenched fluorescent (IQF DiUb) substrate 5 (LifeSensors, Malvern, MA) was used where only upon cleavage one ubiquitin moiety becomes fluorescent (Figs. 6F and S3). Without OTUB1 WT or with OTUB1 C91S, diubiquitin was not cleaved and did not show any signal. In contrast, fluorescence measured in the presence of OTUB1 was strongly stimulated when UbcH5B was added (Figs. 6F and S3). Moreover, FAT10 or USE1 enhanced the fluorescence intensity, and the addition of both USE1 and FAT10 increased the fluorescence signal even further (Figs. 6F and S3). Taken together, our data show that the presence of FAT10 and USE1 stimulated the isopeptidase activity of OTUB1 toward Lys-48–linked diubiquitin, indicating that not only does free UbcH5B or free ubiquitin influence OTUB1 but also free FAT10 and USE1 (an enzyme involved in FAT10 conjugation).
FAT10 stimulates the OTUB1-mediated reduction of overall ubiquitylation
Because we could show an impact of FAT10 on the OTUB1-mediated removal of polyubiquitin from substrates as well as on the cleavage of diubiquitin, we investigated its role in the deconjugation of total ubiquitin chains in cells. As found by Wiener et al., OTUB1 shows a high potential to remove ubiquitin chains without necessarily having a specific substrate protein. Therefore, we overexpressed HA-ubiquitin and examined the impact of OTUB1 in the presence or absence of FAT10 on the overall ubiquitin conjugates. OTUB1 clearly reduced the overall ubiquitylation, an effect that was even more pronounced upon expression of FAT10 (Fig. 7A). Likewise, FAT10 AV increased the OTUB1-mediated deconjugation of ubiquitin (Fig. 7B), again supporting the hypothesis that the noncovalent interaction of OTUB1 and FAT10 is necessary and sufficient to enhance the functionality of OTUB1.
Figure 7.
The noncovalent interaction of FAT10 with OTUB1 stimulates its DUB activity in cellulo and in vitro. A and B, HEK293 cells were transiently transfected with HA-ubiquitin, Flag-OTUB1 and Flag-FAT10 WT, or Flag-FAT10 AV. Cell lysates were subjected to anti-HA immunoprecipitation (IP) and Western blot (IB) analysis using anti-HA and anti-Flag peroxidase–conjugated antibodies. C, lysates of HEK293 cells transiently expressing HA-ubiquitin WT, HA-ubiquitin K48only, HA-ubiquitin K63only (remaining Lys residues were mutated to Arg), Flag-OTUB1, and Flag-FAT10 were subjected to immunoprecipitation using anti-HA–coupled agarose and Western blot analysis. β-Actin was used as the loading control in A–C. D, quantitative analysis of AMC fluorescence (ex. 360 nm, em. 465 nm) of ubiquitin-AMC (5 μm) incubated with OTUB1 (10 μm) in the absence or presence of FAT10, UbcH5B, and USE1 (5 μm) for the indicated time periods at 30 °C. One representative experiment of three experiments with similar outcomes is shown.
Next, we investigated the influence of FAT10 on the activity of OTUB1 toward different polyubiquitin chains. In line with our previous results, OTUB1 reduced the polyubiquitin chains of ubiquitin WT as well as of ubiquitin K48only and K63only ubiquitin chains (Fig. 7C). This OTUB1 activity toward all three chain types was again strongly enhanced in the presence of FAT10 (Fig. 7C). However, we cannot exclude the possibility that endogenous ubiquitin participated in the chain formation resulting in mixed chains with K48only or K63only, respectively. Because we detected no difference in the amount of chains (Lys-48, Lys-63, or WT), we suggest that the possible contribution of endogenous ubiquitin to the formation of mixed chains can be considered equal.
Finally, the impact of FAT10 on OTUB1-mediated cleavage of ubiquitin-AMC was investigated in the presence or absence of FAT10, USE1, and UbcH5B (Fig. 7D). Ubiquitin-AMC is a substrate that binds to the distal ubiquitin-binding site of OTUB1 but is lacking a proximal ubiquitin (36). Wolberger and colleagues (36) propose that UbcH5B increases OTUB1 cleavage activity toward Lys-48 diubiquitin via the proximal ubiquitin binding site in OTUB1. This led to the assumption that UbcH5B would not stimulate OTUB1 affinity to ubiquitin-AMC but to diubiquitin IQF (36). In the absence of OTUB1 WT or in the presence of C91S OTUB1 mutant, no AMC fluorescence was measured (Fig. 7D). However, upon the addition of OTUB1, AMC was cleaved off from ubiquitin and the fluorescence intensity increased (Fig. 7D). This isopeptidase activity of OTUB1 was not enhanced by the presence of UbcH5B (as shown previously (36)) but was stimulated when FAT10 or USE1 was added. The AMC fluorescence was even more elevated when USE1 plus FAT10 was present (Fig. 7D), indicating that free FAT10 and free USE1 are able to stimulate the isopeptidase activity of OTUB1 by increasing its affinity for Lys-48 diubiquitin via the proximal and distal ubiquitin binding site in OTUB1.
Taken together, we have shown that OTUB1 is regulated by FAT10 via two different mechanisms. On the one hand, OTUB1 becomes covalently FAT10ylated and degraded by the 26S proteasome. This modification depends on the C-terminal diglycine motif of FAT10. On the other hand, free OTUB1 becomes stabilized by the presence of noncovalently interacting FAT10. This interaction induces an enhanced deconjugation of ubiquitin from either specific substrates like TRAF3 or from overall ubiquitin conjugates. The presence of FAT10 or its conjugating enzyme USE1 stimulates the DUB activity of OTUB1 toward Lys-48–linked diubiquitin or polyubiquitin chains. Moreover, by interacting in a noncovalent manner with OTUB1, FAT10 is able to influence not only the DUB activity of OTUB1 but also the noncatalytic inhibitory function of OTUB1. With this understanding we present a new mechanism of how FAT10 regulates the stability and functionality of its target protein, OTUB1.
Discussion
We have recently used a yeast two-hybrid approach with the E2-conjugating enzyme USE1 (22), and here OTUB1 was identified as a new interaction partner of USE1 (Fig. 1). Because USE1 transfers not only ubiquitin but also FAT10 (15, 18, 19), we focused on the interplay among OTUB1, USE1, and FAT10. We investigated different scenarios of how OTUB1 and FAT10 might influence each other. Either OTUB1 could have an impact on FAT10 and its conjugation or FAT10 could change the stability and functionality of OTUB1. We examined whether OTUB1 might function as a FAT10-deconjugating enzyme or whether it could bind the FAT10-loaded USE1, thereby inhibiting the further conjugation of FAT10. On the other hand, FAT10 could covalently modify OTUB1 or interact with it in a noncovalent manner to influence the DUB activity or the noncatalytic inhibitory activity of OTUB1.
In the present study, we have shown that OTUB1 was not able to cleave off FAT10 from substrates and did not display an impact on the overall FAT10ylation pattern (Figs. 1 and 2). Because neither peptide- nor isopeptide-conjugated FAT10 was removed from its substrates USE1 and UBE1, respectively, we suggest that OTUB1 does not serve as a deconjugating enzyme for FAT10. As FAT10 is published as being degraded by the 26S proteasome along with its substrates (38), a deconjugation from substrates through OTUB1 might not necessarily be required. Furthermore, we have no evidence that OTUB1 preferentially inhibits FAT10 conjugation by interacting with FAT10-USE1. An inhibition of conjugation in the presence of OTUB1 was not observable (Fig. 2). These findings argue for the hypothesis that FAT10 influences OTUB1 rather than for OTUB1 influencing FAT10.
Indeed, we have shown here that OTUB1 becomes covalently FAT10ylated and that this modification depends on the classical conjugation characteristics (Fig. 2) as published previously for other FAT10 substrates (19, 21, 22). As roughly only 5% of the substrate protein becomes FAT10ylated (as estimated), we speculated whether the noncovalent interaction between FAT10 and OTUB1 could play the more pivotal role. In line with this thinking, we confirmed that the presence of FAT10 but not its conjugation led to a stabilization of total cellular OTUB1 (Fig. 3), indicating that FAT10 influences OTUB1 in two different manners. The covalent modification with FAT10 leads OTUB1 to proteasomal degradation, whereas the noncovalent interaction stabilizes OTUB1 and thereby influences the functionality of OTUB1.
Because OTUB1 and FAT10 interact in a direct manner under in vitro conditions (Fig. 4), we investigated the interaction among OTUB1, FAT10, and the two E2-conjugating enzymes UbcH5B and USE1. We asked whether the presence of E2-conjugating enzymes could strengthen the interaction between OTUB1 and FAT10, or vice versa, and whether FAT10 is able to improve the binding of OTUB1 to the E2 enzymes as published for OTUB1, UbcH5B, and ubiquitin (28, 36). Although the interaction between OTUB1 and USE1 or UbcH5B was not changed in cells (Figs. 1 and 4), in vitro data indicated a stronger binding of OTUB1 to FAT10 in the presence of USE1 or UbcH5B (Fig. 4). Additionally, the binding of those E2 enzymes to FAT10 was increased when OTUB1 was added (Fig. 4). From these data we suggest that OTUB1, FAT10, and an E2-conjugating enzyme may form a trimeric complex.
In line with this proposal, we further show that FAT10 and USE1 improved the OTUB1 isopeptidase activity toward Lys-48–linked DiUb (Figs. 6 and 7). Not only was the degradation speed increased but also the amount of cleaved FRET-based, Lys-48—linked, internally quenched DiUb was enhanced (Figs. 6 and 7), further supporting our hypothesis that free uncharged USE1 and free FAT10 stimulate the DUB activity of OTUB1.
Because USE1 and UbcH5B are structurally very similar (35), we can explain why the interaction of USE1 with the OTUB1 T134R mutant was decreased (Fig. 1) as published previously for UbcH5B (28, 36). In contrast to UbcH5B, the USE1 interaction with OTUB1 T134R was not completely abolished, implying that there must be more residues or surface patches on OTUB1 involved in USE1 binding. This finding is supported by the data showing that free USE1 stimulates OTUB1 isopeptidase activity toward ubiquitin-AMC (Fig. 7), which is not the case for UbcH5B (36). Nevertheless, similar binding modes may lead to similar action modes, further strengthening the hypothesis that USE1 stimulates the Lys-48 DUB activity of OTUB1 as shown for UbcH5B (36) and by our in vitro experiments (Fig. 6).
Competition of USE1 with UbcH5B for the same binding site in OTUB1 could also diminish the transfer of ubiquitin via UbcH5B, ending up in less Lys-48 polyubiquitin chains on certain substrate proteins. As USE1 is also proposed to support the Lys-48–linked polyubiquitylation of target proteins, a competition of both E2 enzymes for ubiquitylating target proteins could occur in a substrate-specific manner. Under inflammatory conditions, where UBA6 preferentially activates FAT10 rather than ubiquitin (45), the competition between USE1 and UbcH5B could be a regulatory mechanism to control the ubiquitylation and FAT10ylation of target proteins specifically important under these stress conditions.
A possible regulatory impact could further explain why the interaction between FAT10 and OTUB1 is improved when UbcH5B is present (Fig. 4). In the case of FAT10 and OTUB1 both binding to UbcH5B, the E2 might be kept in a more inactive state, and ubiquitin cannot be transferred onto target proteins. The stabilization of this trimeric complex could be a basic requirement for FAT10 to support the noncatalytic function of OTUB1 in inhibiting the ubiquitylation of target proteins as shown for UbcH5B or Ubc13 (28, 36, 37).
One target protein of OTUB1 is TRAF3, which becomes polyubiquitylated in two distinct ways related to certain cellular stress conditions. Dependent on the type of TLR, TRAF3 becomes Lys-48–polyubiquitylated and degraded by the proteasome, a mechanism described as essential for the expression of proinflammatory cytokines (41). Alternatively, TRAF3 autoubiquitylates itself with Lys-63-linked ubiquitin chains inducing the noncanonical NF-κB pathway and thereby leading to the production of inflammatory type I interferons (41). Because FAT10 expression is up-regulated by proinflammatory conditions (11) and has been shown to play a role in the antiviral immune response mediated by RIG-I upstream of TRAF3 (42, 43), we investigated the impact of FAT10 on the OTUB1-mediated change in TRAF3 ubiquitylation.
Our data point to a dual role for FAT10 during the deubiquitylation of TRAF3. On the one hand, OTUB1 removes Lys-48 polyubiquitin chains from TRAF3, which is further improved by the presence of FAT10 (Fig. 5). On the other hand, Lys-63 polyubiquitylation of TRAF3 is reduced in the presence of OTUB1 WT or C91S, which is again stimulated in the presence of FAT10 (Fig. 5). This dual effect might be a hint to a regulatory mechanism of how FAT10 could influence the ubiquitin pathway under inflammatory conditions. Thereby, FAT10 could shape the antiviral immune response by stimulating the OTUB1-mediated noncatalytic deubiquitylation of TRAF3. As even the presence of OTUB1 reduces the production of type I interferon upon viral infection to prevent a prolonged harmful antiviral immune response (39, 40), FAT10 overexpression might further reduce type I interferon production. As FAT10 stimulates the isopeptidase activity of OTUB1 toward TRAF3 (Fig. 5), it might therefore also influence the immune response regarding the expression of proinflammatory cytokines.
After examining a specific OTUB1 substrate, we investigated the impact of FAT10 and OTUB1 on overall ubiquitylation in HEK293 cells. There, the addition of FAT10 WT or nonconjugatable FAT10 AV increased the OTUB1-mediated reduction of ubiquitin conjugates (Fig. 7). Taking into account that there are ∼100 DUB existing in humans (46), we postulated that the strong effect we observed on overall ubiquitylation was because of FAT10 stimulating not only the DUB activity of OTUB1 toward Lys-48–linked polyubiquitylation but also the noncatalytic activity toward Lys-48– or Lys-63–linked polyubiquitylation.
In conclusion, FAT10 seems to affect its target proteins in a more differentiated manner than only via covalent modification. Although the FAT10ylation and subsequent degradation of substrate proteins may play a role during certain processes, our data imply that the noncovalent interaction with target proteins influences their stability and functionality. Therefore FAT10 may shape many more regulatory processes within the cell than anticipated previously.
Experimental procedures
Overexpression constructs
HEK293 cells were transiently transfected with TransIT-LTI transfection reagent (Mirus Bio LLC, Madison, WI) and expression plasmids pcDNA3.1-HA-FAT10-USE1,6 pcDNA3.1-HA-FAT10 (23), pCMV-myc-FAT10,7 pcDNA6.1-FAT10 tagless (5), pcDNA3.1-His3xFlag-FAT10 (15), pcDNA3.1-His3xFlag-FAT10 AV (AV instead of C-terminal GG (22)), pcDNA3.1-His-UbcH5B (C. Pelzer,8 University of Konstanz), pcDNA3.1-His-USE1 (19), and pcDNA3-HA-UBE1 (21). For the expression of Flag-tagged OTUB1 WT, the active-site cysteine mutant C91S, and the OTU domain mutant T134R, expression plasmids pCMV6-Flag-OTUB1 wt, C91S, and T134R were kindly provided by Cynthia Wolberger (Johns Hopkins University, Baltimore, MD). For expression of Flag-tagged OTUB1 lysine-less mutant pcDNA3–2Flag-OTUB1 K0 was prepared by Daniel Li and kindly provided by Mu-Shui Dai (Oregon Health & Science University, Portland, OR). For expression of HA-tagged Ubc13, the expression plasmid pcDNA3.0-HA-Ubc13 was a kind gift from Dong Er-Zhang (Addgene plasmid 12461) (47). The plasmid pRK-Flag-TRAF3 for expression of Flag-tagged TRAF3 was a kind gift from Xiaofeng Zheng (40). For the expression of HA-tagged ubiquitin WT, the K48only mutant, and the K63only mutant, the expression plasmids pRK5-HA-ubiquitin wt, pRK5-HA-Ub K48only, and pRK5-HA-Ub K63only were a kind gift from Ted Dawson (Addgene plasmids 17608, 17605, and 17606) (44).
Generation of CRISPR knockout cell lines
CRISPR Cas9 knockout HEK293 cells lines of FAT10, USE1, and UBA6 were generated by A. Aichem et al. (5).
Cloning and mutagenesis
For the generation of Flag-FAT10 without the C-terminal diglycine motif (Flag-FAT10 ΔGG) a site-directed mutagenesis with pcDNA3.0-Y1-FAT105 as template and the following primers was performed: forward, 5′-GCA TCT TAT TGT ATT TGA TCT AGA GGG CCC TAT TC-3′, and reverse, 5′-GAA TAG GGC CCT CTA GAT CAA ATA CAA TAA GAT GC-3′. The generated FAT10 ΔGG was amplified by PCR using the following primers: forward, 5′-CGG GGT ACC TAT GGC TCC CAA TGC TTC C-3′, and reverse, 5′-CTA TAG ACT CAA ATA CAA TAA CAT GCC AGG AGG AG-3′. Via restriction digest with KpnI (5′) and XbaI (3′), FAT10 ΔGG was inserted into pcDNA3.1-His3xFlag expression vector (15).
OTUB1 N-terminally fused to the C-terminal part of YFP (Y2-OTUB1) was generated by introducing the PCR product of OTUB1 into the pcDNA3.0-Y2 expression vector (48) by restriction digest using the following primers and restriction enzymes, respectively: forward, 5′-ATA AGA ATG CGG CCG CGA TGG CGG CGG AGG AAC-3′ (NotI), and reverse, 5′-TTG GGC CCC TAT TTG TAG AGG ATA TCG TAG TG-3′ (ApaI). To generate active-site cysteine-mutated Y2-OTUB1 and recombinantly expressed His-tagged and tagless OTUB1, a site-directed mutagenesis with pcDNA3.0-Y2-OTUB1 and pPRO-Ex-His-TEV-OTUB1 (a kind gift from Cynthia Wolberger, Addgene 26959) (49), respectively, was performed. The following primers were used: forward, 5′-AGC CCG ATA GAA ACT GTT GCC GTC AGG CC-3′, and reverse, 5′-GGC CTG ACG GCA ACA GTT TCT ATC GGG CT-3′.
To generate pCMV6-Flag-OTUB1 ΔN43 lacking the first 43 amino acids at the N terminus, PCR and restriction digest with the following primers and enzymes was performed, respectively: forward, 5′-GAC ATC GAT TAT AAA GAT GAT GAT GAT AAA AAT TCA CAG AAC CCT CTG GTG TCA GAG C-3′ (ClaI), and reverse, 5′-ATC CTC TAG AGT CGA CTA TTT GTA GAG GAT ATC GTA GTG TCC AG-3′ (XbaI).
The generation of pcDNA3.1-HA OTUB1 wt, K0, and ΔN43 was performed by PCR and restriction digest using the following primers and enzymes, respectively: forward, 5′-CGG GGT ACC AAT GGC GGC GGA GGA ACC TCA G-3′ (wt and K0; KpnI) and 5′-CGG GGT ACC ACA GAA CCC TCT GGT GTC AGA G-3′ (dN43) (KpnI); and reverse, 5′-CCG CTC GAG CTA TTT GTA GAG GAT ATC GTA G-3′ (wt and dN43; XhoI) and 5′-CCG CTC GAG CTA TCT GTA GAG GAT ATC GTA G-3′ (K0; XhoI). All plasmids were verified by sequencing (Microsynth AG, Balgach, Switzerland).
Induction of endogenous FAT10 expression, CHX chase experiments, and immunoprecipitation
Endogenous FAT10 expression was induced by treating cells with the proinflammatory cytokines IFNγ (200 units/ml) and TNFα (400 units/ml) (both from Peprotech GmbH, Hamburg, Germany) as described recently (50). Before harvesting, 50 μg/ml CHX (Sigma) was added for the dedicated time periods as indicated. In parallel, 10 μm proteasome inhibitor MG132 (Enzo Lifesciences) was added as indicated, and the cells were incubated for a maximum of 6 h. The cells were harvested and lysed for 30 min on ice in lysis buffer containing 20 mm Tris-HCl (pH 7.6), 50 mm NaCl, 10 mm MgCl2, and 1% Nonidet P-40 supplemented with 1× protease inhibitor mix (Complete mini EDTA-free protease inhibitor mixture, Roche). The cleared lysates were subjected to pulldown using His60 Ni Superflow Resin (Takara, Saint-Germain-en-Leye, France) or to immunoprecipitation using the anti-HA–agarose conjugate HA-7 (Sigma-Aldrich), the Red Anti-Flag M2 affinity gel clone (Sigma-Aldrich), or protein A–Sepharose (Sigma-Aldrich) in combination with monoclonal mouse FAT10 antibody 4F1 (38). Proteins in 5× gel sample buffer containing 4% β-mercaptoethanol (2-ME), were separated on 4–12% NuPAGE bis-Tris SDS gradient gels (Invitrogen) and subjected to Western blot analysis with directly labeled peroxidase-conjugated antibodies (mAb against HA-7, mAb against Flag M2, and mAb against polyhistidine HIS-1 (all mouse, from Sigma-Aldrich)). Unlabeled antibodies such as mouse mAb against c-myc clone 9E10 (Sigma-Aldrich), rabbit mAb against OTUB1 (RabMAb EPR13028(B), Abcam, Cambridge, UK), FAT10 rabbit polyclonal antibody (23), and anti-GFP mouse monoclonal (clones 7.1 and 13.1, Roche) were used where indicated. An antibody against β-actin (AC-15, Abcam) was used for loading control detection. For the determination of protein half-life, Western blotting signals were analyzed using densitometric calculations (ImageLab Software, Bio-Rad), and values were normalized to β-actin signals. Statistical analysis was performed by using GraphPad Prism software (GraphPad Software Inc., San Diego, CA).
Protein expression and purification
All proteins were expressed in the bacterial expression strain Escherichia coli BL21(DE3) grown in modified LB medium (13.5 g/liter peptone, 7 g/liter yeast extract, 14.9 g/liter glycerol, 2.5 g/liter NaCl, 2.3 g/liter K2HPO4, 1.5 g/liter KH2PO4, and 0.14 g/liter MgSO4 × 7H2O (pH 7.0)). Cultures were inoculated to an A600 of 0.2 using saturated cultures and grown at 37 °C to an A600 of 0.7. Protein expression was induced at 16 °C overnight by the addition of 0.4 mm isopropyl β-d-thiogalactopyranoside. Cells were harvested by centrifugation (5000 × g, 30 min, 8 °C) and lysed immediately. FAT10 variants were purified as described previously (18). OTUB1 enzymes were purified basically as published by Wang et al. (49). His60 Ni Superflow Resin (Takara) was used to isolate His-TEV–tagged OTUB1 from bacterial lysates. The His tag was either cleaved off by incubation with His-tagged TEV protease overnight at 8 °C with parallel dialysis followed by a second run at the His60 Ni Superflow Resin (Takara) or His-OTUB1 and tagless OTUB1 were directly subjected to a Hiload 26/60 Superdex 75 gel filtration column (GE Healthcare) and eluted with 50 mm Tris-HCl (pH 7.5), 0.2 m NaCl, 1 mm tris(2-carboxyethyl)phosphine, and 1 mm EDTA. Peak fractions were combined, and stored at −80 °C. Tagless Ubc13 was purified basically by following the protocol published by Wiener et al. (37) except for using His60 Ni Superflow Resin (Takara) in the initial purification step. The recombinant UbcH5B-His expression plasmid, a kind gift by Martin Scheffner (University of Konstanz, Germany), was purified by using His60 Ni Superflow Resin. The purified proteins were stored in 20 mm Tris (pH 7.5), 100 mm NaCl, and 10% glycerol at −80 °C.
In vitro conjugation experiments
The buffer for in vitro reactions contained 20 mm Tris (pH 7.6), 50 mm NaCl, 10 mm MgCl2, 4 mm ATP, and 0.1 mm DTT (all from Sigma-Aldrich) supplemented with 1× protease inhibitor mix (Complete mini EDTA-free protease inhibitor mixture, Roche). In a final volume of 40 μl the 1× reaction buffer recombinant proteins were added in the following amounts: Flag-UBA6, 1 μg (0.1 mg/ml, BML-UW0350, Enzo LifeSciences), 6His-USE1, 6 μg (0.6 mg/ml) (19), 3xFlag-FAT10, 4 μg (0.4 mg/ml) (21), tagless FAT10, 4 μg (1.8 mg/ml) (18), and His-OTUB1, 2.5 μg (1.25 mg/ml). The reaction mixture was incubated at 30 °C for 60 min with shaking, and the reaction was stopped by the addition of 5× gel sample buffer containing 4% 2-ME. Proteins were separated on 4–12% NuPAGE bis-Tris SDS gradient gels (Invitrogen) with subsequent Western blot analysis using the directly labeled peroxidase-conjugated antibodies mAb against Flag M2 or mAb against polyhistidine HIS-1 (Sigma-Aldrich). For analysis of ubiquitin conjugates, a polyclonal rabbit antibody against ubiquitin was applied (Z0458, DakoCytomation, Hamburg, Germany).
In vitro interaction experiments
In vitro interaction assays were performed in buffer containing 20 mm Tris (pH 7.6), 50 mm NaCl, 10 mm MgCl2, 4 mm ATP, and 0.1 mm DTT, (all from Sigma-Aldrich) supplemented with 1× protease inhibitor mix (Roche). Equal amounts (8 μg) of His-OTUB1 (1.25 mg/ml), Flag-tagged FAT10 (0.4 mg/ml) (21), UbcH5B-His (0.6 mg/ml), tagless Ubc13 (0.6 mg/ml) (37), and His-USE1 (0.6 mg/ml) (19) with additional Red Anti Flag affinity gel clone M2 (Sigma-Aldrich) were incubated for 60 min at 8 °C. After the addition of 5× gel sample buffer containing 4% 2-ME, proteins were separated on 4–12% NuPAGE bis-Tris gradient gels (Invitrogen). Subsequent Western blot analysis was performed by using the following antibodies: directly labeled peroxidase-conjugated antibodies mAb against Flag M2 and mAb against polyhistidine HIS-1 (both mouse, both Sigma-Aldrich) and RabMAb against Ubc13 (Abcam).
In vitro competition experiments
Transiently overexpressed Flag-tagged OTUB1 WT was purified from HEK293 cells by immunoprecipitation using Red Anti Flag affinity gel clone M2 (Sigma-Aldrich) as described above. Agarose-bound Flag-OTUB1 was incubated for in vitro competition assays in buffer containing 20 mm Tris (pH 7.6), 50 mm NaCl, 10 mm MgCl2, 4 mm ATP, and 0.1 mm DTT (all from Sigma-Aldrich) supplemented with 1× protease inhibitor mix (Roche). Increasing amounts of UbcH5B-His (0.6 mg/ml) (8, 42, and 84 μg) and His-USE1 (0.6 mg/ml) (4, 22, and 44 μg) (19) were added for 60 min at 7 °C. Following the addition of 5× gel sample buffer containing 4% 2-ME, proteins were separated on 4–12% NuPAGE bis-Tris gradient gels (Invitrogen). Subsequent Western blot analysis was performed by using directly labeled peroxidase-conjugated mAb against Flag M2 and mAb against polyhistidine HIS-1 (both mouse, both from Sigma-Aldrich).
Assays of OTUB1 cleavage activity
Assays of OTUB1 cleavage of ubiquitin variants were performed as described previously (36). Briefly, ubiquitin-AMC (5 μm, Boston Biochem, Cambridge, MA) was cleaved by OTUB1 (10 μm) at 30 °C in buffer containing 20 mm HEPES (pH 7.5), 100 mm NaCl, 5 mm DTT, and 0.01% BSA. After starting the reactions by adding OTUB1, the experiments were carried out in the presence or absence of 5 μm FAT10, UbcH5B, and USE1. AMC fluorescence (ex. 360 nm, em. 465 nm) was monitored using a Tecan SPARK 10M plate reader (Tecan Group Ltd., Maennedorf, Switzerland). Briefly, FRET-based assays monitoring OTUB1 (30 nm) cleavage of Lys-48 or Lys-63 diubiquitin (400 nm, IQF substrate 5) were performed at 30 °C in buffer containing 20 mm HEPES (pH 7.5), 100 mm NaCl, 5 mm DTT, and 0.01% BSA in the absence or presence of 5 μm FAT10, UbcH5B, and USE1. The reaction was initiated by the addition of OTUB1. Fluorescence (ex. 535 nm, em. 595 nm) was monitored using a Tecan SPARK 10M plate reader (Tecan Group Ltd.). The rate of Lys-48 diubiquitin cleavage was calculated using the slope of a standard curve prepared with the indicated percentages of diubiquitin.
Gel-based assays for FAT10 as stimulator of OTUB1 isopeptidase activity were performed at 37 °C in reaction buffer containing 20 mm HEPES (pH 7.5), 100 mm NaCl, 5 mm DTT, and 0.01% BSA. 5 μm OTUB1 was mixed with 5 μm unlabeled Lys-48 or Lys-63 diubiquitin (AQUApure, Boston Biochem) in the presence and absence of 10 μm FAT10, USE1, and UbcH5B. Reactions were initiated by the addition of OTUB1. Samples were removed at the specific time points, and the reactions were stopped by the addition of 5× gel sample buffer containing 4% 2-ME. Samples were analyzed by gel electrophoresis on 4–12% NuPAGE bis-Tris gradient gels (Invitrogen). Gels were stained with colloidal Coomassie (Instant Blue, Gentaur, Aachen, Germany) or silver stain (Thermo Scientific).
Author contributions
J. B. and A. A. conceptualization; J. B., A. N. B., and N. C. investigation; J. B. methodology; J. B. writing-original draft; J. B., A. A., A. N. B., and M. G. writing-review and editing; A. N. B. and N. C. validation; A. A. and M. G. resources; A. A. and M. G. supervision; A. A. and M. G. funding acquisition.
Supplementary Material
Acknowledgments
We thank C. Wolberger (Johns Hopkins University, Baltimore, MD), M. Dai (Oregon Health & Science University, Portland, OR), X. Zheng (Peking University, Beijing, China), M. Scheffner, and E. Deuerling (both from University of Konstanz, Germany) for their kind contributions of plasmids.
This work was supported by the German Research Foundation (DFG) Collaborative Research Center SFB969, project C01 (to M. G.) and by Grant 1029 from the Velux Foundation (to A. A. and M. G.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S3.
J. Bialas, unpublished data.
A. Aichem, unpublished data.
N. Catone, unpublished data.
C. Pelzer, unpublished data.
- IFNγ
- interferon-γ
- TNF
- tumor necrosis factor
- DUB
- deubiquitylating enzyme(s)
- DiUb
- diubiquitin
- OTU
- ovarian tumor family
- OTUB1
- otubain 1
- UBA6
- ubiquitin-like modifier–activating enzyme 6
- USE1
- Uba6-specific E2-conjugating enzyme 1
- TRAF3
- TNF receptor–associated factor 3
- YFP
- yellow fluorescent protein
- CHX
- cycloheximide
- TLR
- Toll-like receptor
- AMC
- 7-amido-4-methylcoumarin
- IQF
- internally quenched fluorescent
- 2-ME
- β-mercaptoethanol
- bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- BSA
- bovine serum albumin.
References
- 1. Collins G. A., and Goldberg A. L. (2017) The logic of the 26S proteasome. Cell 169, 792–806 10.1016/j.cell.2017.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. van der Veen A. G., and Ploegh H. L. (2012) Ubiquitin-like proteins. Annu. Rev. Biochem. 81, 323–357 10.1146/annurev-biochem-093010-153308 [DOI] [PubMed] [Google Scholar]
- 3. Kerscher O., Felberbaum R., and Hochstrasser M. (2006) Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22, 159–180 10.1146/annurev.cellbio.22.010605.093503 [DOI] [PubMed] [Google Scholar]
- 4. Fan W., Cai W., Parimoo S., Schwarz D. C., Lennon G. G., and Weissman S. M. (1996) Identification of seven new human MHC class I region genes around the HLA-F locus. Immunogenetics 44, 97–103 10.1007/BF02660056 [DOI] [PubMed] [Google Scholar]
- 5. Aichem A., Anders S., Catone N., Rossler P., Stotz S., Berg A., Schwab R., Scheuermann S., Bialas J., Schutz-Stoffregen M. C., Schmidtke G., Peter C., Groettrup M., and Wiesner S. (2018) The structure of the ubiquitin-like modifier FAT10 reveals an alternative targeting mechanism for proteasomal degradation. Nat. Commun. 9, 3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bates E. E., Ravel O., Dieu M. C., Ho S., Guret C., Bridon J. M., Ait-Yahia S., Brière F., Caux C., Banchereau J., and Lebecque S. (1997) Identification and analysis of a novel member of the ubiquitin family expressed in dendritic cells and mature B cells. Eur. J. Immunol. 27, 2471–2477 10.1002/eji.1830271002 [DOI] [PubMed] [Google Scholar]
- 7. Groettrup M., Pelzer C., Schmidtke G., and Hofmann K. (2008) Activating the ubiquitin family: UBA6 challenges the field. Trends Biochem. Sci. 33, 230–237 10.1016/j.tibs.2008.01.005 [DOI] [PubMed] [Google Scholar]
- 8. Raasi S., Schmidtke G., and Groettrup M. (2001) The ubiquitin-like protein FAT10 forms covalent conjugates and induces apoptosis. J. Biol. Chem. 276, 35334–35343 10.1074/jbc.M105139200 [DOI] [PubMed] [Google Scholar]
- 9. Canaan A., Yu X., Booth C. J., Lian J., Lazar I., Gamfi S. L., Castille K., Kohya N., Nakayama Y., Liu Y. C., Eynon E., Flavell R., and Weissman S. M. (2006) FAT10/diubiquitin-like protein-deficient mice exhibit minimal phenotypic differences. Mol. Cell. Biol. 26, 5180–5189 10.1128/MCB.00966-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee C. G., Ren J., Cheong I. S., Ban K. H., Ooi L. L., Yong Tan S., Kan A., Nuchprayoon I., Jin R., Lee K. H., Choti M., and Lee L. A. (2003) Expression of the FAT10 gene is highly upregulated in hepatocellular carcinoma and other gastrointestinal and gynecological cancers. Oncogene 22, 2592–2603 10.1038/sj.onc.1206337 [DOI] [PubMed] [Google Scholar]
- 11. Lukasiak S., Schiller C., Oehlschlaeger P., Schmidtke G., Krause P., Legler D. F., Autschbach F., Schirmacher P., Breuhahn K., and Groettrup M. (2008) Proinflammatory cytokines cause FAT10 upregulation in cancers of liver and colon. Oncogene 27, 6068–6074 10.1038/onc.2008.201 [DOI] [PubMed] [Google Scholar]
- 12. Aichem A., and Groettrup M. (2016) The ubiquitin-like modifier FAT10 in cancer development. Int. J. Biochem. Cell Biol. 79, 451–461 10.1016/j.biocel.2016.07.001 [DOI] [PubMed] [Google Scholar]
- 13. Liu Y. C., Pan J., Zhang C., Fan W., Collinge M., Bender J. R., and Weissman S. M. (1999) A MHC-encoded ubiquitin-like protein (FAT10) binds noncovalently to the spindle assembly checkpoint protein MAD2. Proc. Natl. Acad. Sci. U.S.A. 96, 4313–4318 10.1073/pnas.96.8.4313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Raasi S., Schmidtke G., de Giuli R., and Groettrup M. (1999) A ubiquitin-like protein which is synergistically inducible by interferon-γ and tumor necrosis factor-α. Eur. J. Immunol. 29, 4030–4036 [DOI] [PubMed] [Google Scholar]
- 15. Chiu Y. H., Sun Q., and Chen Z. J. (2007) E1-L2 activates both ubiquitin and FAT10. Mol. Cell 27, 1014–1023 10.1016/j.molcel.2007.08.020 [DOI] [PubMed] [Google Scholar]
- 16. Jin J., Li X., Gygi S. P., and Harper J. W. (2007) Dual E1 activation systems for ubiquitin differentially regulate E2 enzyme charging. Nature 447, 1135–1138 10.1038/nature05902 [DOI] [PubMed] [Google Scholar]
- 17. Pelzer C., Kassner I., Matentzoglu K., Singh R. K., Wollscheid H. P., Scheffner M., Schmidtke G., and Groettrup M. (2007) UBE1L2, a novel E1 enzyme specific for ubiquitin. J. Biol. Chem. 282, 23010–23014 10.1074/jbc.C700111200 [DOI] [PubMed] [Google Scholar]
- 18. Aichem A., Catone N., and Groettrup M. (2014) Investigations into the auto-FAT10ylation of the bispecific E2 conjugating enzyme UBA6-specific E2 enzyme 1. FEBS J. 281, 1848–1859 10.1111/febs.12745 [DOI] [PubMed] [Google Scholar]
- 19. Aichem A., Pelzer C., Lukasiak S., Kalveram B., Sheppard P. W., Rani N., Schmidtke G., and Groettrup M. (2010) USE1 is a bispecific conjugating enzyme for ubiquitin and FAT10, which FAT10ylates itself in cis. Nat. Commun. 1, 13 [DOI] [PubMed] [Google Scholar]
- 20. Li T., Santockyte R., Yu S., Shen R. F., Tekle E., Lee C. G., Yang D. C., and Chock P. B. (2011) FAT10 modifies p53 and upregulates its transcriptional activity. Arch. Biochem. Biophys. 509, 164–169 10.1016/j.abb.2011.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bialas J., Groettrup M., and Aichem A. (2015) Conjugation of the ubiquitin activating enzyme UBE1 with the ubiquitin-like modifier FAT10 targets it for proteasomal degradation. PLoS ONE 10, e0120329 10.1371/journal.pone.0120329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Aichem A., Kalveram B., Spinnenhirn V., Kluge K., Catone N., Johansen T., and Groettrup M. (2012) The proteomic analysis of endogenous FAT10 substrates identifies p62/SQSTM1 as a substrate of FAT10ylation. J. Cell Sci. 125, 4576–4585 10.1242/jcs.107789 [DOI] [PubMed] [Google Scholar]
- 23. Hipp M. S., Kalveram B., Raasi S., Groettrup M., and Schmidtke G. (2005) FAT10, a ubiquitin-independent signal for proteasomal degradation. Mol. Cell. Biol. 25, 3483–3491 10.1128/MCB.25.9.3483-3491.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kalveram B., Schmidtke G., and Groettrup M. (2008) The ubiquitin-like modifier FAT10 interacts with HDAC6 and localizes to aggresomes under proteasome inhibition. J. Cell Sci. 121, 4079–4088 10.1242/jcs.035006 [DOI] [PubMed] [Google Scholar]
- 25. Theng S. S., Wang W., Mah W. C., Chan C., Zhuo J., Gao Y., Qin H., Lim L., Chong S. S., Song J., and Lee C. G. (2014) Disruption of FAT10-MAD2 binding inhibits tumor progression. Proc. Natl. Acad. Sci. U.S.A. 111, E5282–5291 10.1073/pnas.1403383111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Komander D., Clague M. J., and Urbé S. (2009) Breaking the chains: Structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10, 550–563 10.1038/nrm2731 [DOI] [PubMed] [Google Scholar]
- 27. Sun X. X., Challagundla K. B., and Dai M. S. (2012) Positive regulation of p53 stability and activity by the deubiquitinating enzyme Otubain 1. EMBO J. 31, 576–592 10.1038/emboj.2011.434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Juang Y. C., Landry M. C., Sanches M., Vittal V., Leung C. C., Ceccarelli D. F., Mateo A. R., Pruneda J. N., Mao D. Y., Szilard R. K., Orlicky S., Munro M., Brzovic P. S., Klevit R. E., Sicheri F., and Durocher D. (2012) OTUB1 co-opts Lys48-linked ubiquitin recognition to suppress E2 enzyme function. Mol. Cell 45, 384–397 10.1016/j.molcel.2012.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sato Y., Yoshikawa A., Yamagata A., Mimura H., Yamashita M., Ookata K., Nureki O., Iwai K., Komada M., and Fukai S. (2008) Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature 455, 358–362 10.1038/nature07254 [DOI] [PubMed] [Google Scholar]
- 30. Edelmann M. J., Kramer H. B., Altun M., and Kessler B. M. (2010) Post-translational modification of the deubiquitinating enzyme otubain 1 modulates active RhoA levels and susceptibility to Yersinia invasion. FEBS J. 277, 2515–2530 10.1111/j.1742-4658.2010.07665.x [DOI] [PubMed] [Google Scholar]
- 31. Herrador A., Léon S., Haguenauer-Tsapis R., and Vincent O. (2013) A mechanism for protein monoubiquitination dependent on a trans-acting ubiquitin-binding domain. J. Biol. Chem. 288, 16206–16211 10.1074/jbc.C113.452250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Juris S. J., Shah K., Shokat K., Dixon J. E., and Vacratsis P. O. (2006) Identification of otubain 1 as a novel substrate for the Yersinia protein kinase using chemical genetics and mass spectrometry. FEBS Lett. 580, 179–183 10.1016/j.febslet.2005.11.071 [DOI] [PubMed] [Google Scholar]
- 33. Nakada S., Tai I., Panier S., Al-Hakim A., Iemura S., Juang Y. C., O'Donnell L., Kumakubo A., Munro M., Sicheri F., Gingras A. C., Natsume T., Suda T., and Durocher D. (2010) Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature 466, 941–946 10.1038/nature09297 [DOI] [PubMed] [Google Scholar]
- 34. Soares L., Seroogy C., Skrenta H., Anandasabapathy N., Lovelace P., Chung C. D., Engleman E., and Fathman C. G. (2004) Two isoforms of otubain 1 regulate T cell anergy via GRAIL. Nat. Immunol. 5, 45–54 10.1038/nrm1276,10.1038/ni1017 [DOI] [PubMed] [Google Scholar]
- 35. Schelpe J., Monté D., Dewitte F., Sixma T. K., and Rucktooa P. (2016) Structure of UBE2Z enzyme provides functional insight into specificity in the FAT10 protein conjugation machinery. J. Biol. Chem. 291, 630–639 10.1074/jbc.M115.671545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wiener R., DiBello A. T., Lombardi P. M., Guzzo C. M., Zhang X., Matunis M. J., and Wolberger C. (2013) E2 ubiquitin-conjugating enzymes regulate the deubiquitinating activity of OTUB1. Nat. Struct. Mol. Biol. 20, 1033–1039 10.1038/nsmb.2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wiener R., Zhang X., Wang T., and Wolberger C. (2012) The mechanism of OTUB1-mediated inhibition of ubiquitination. Nature 483, 618–622 10.1038/nature10911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hipp M. S., Raasi S., Groettrup M., and Schmidtke G. (2004) NEDD8 ultimate buster-1L interacts with the ubiquitin-like protein FAT10 and accelerates its degradation. J. Biol. Chem. 279, 16503–16510 10.1074/jbc.M310114200 [DOI] [PubMed] [Google Scholar]
- 39. Li S., Zheng H., Mao A. P., Zhong B., Li Y., Liu Y., Gao Y., Ran Y., Tien P., and Shu H. B. (2010) Regulation of virus-triggered signaling by OTUB1- and OTUB2-mediated deubiquitination of TRAF3 and TRAF6. J. Biol. Chem. 285, 4291–4297 10.1074/jbc.M109.074971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Peng Y., Xu R., and Zheng X. (2014) HSCARG negatively regulates the cellular antiviral RIG-I like receptor signaling pathway by inhibiting TRAF3 ubiquitination via recruiting OTUB1. PLoS Pathog. 10, e1004041 10.1371/journal.ppat.1004041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tseng P. H., Matsuzawa A., Zhang W., Mino T., Vignali D. A., and Karin M. (2010) Different modes of ubiquitination of the adaptor TRAF3 selectively activate the expression of type I interferons and proinflammatory cytokines. Nat. Immunol. 11, 70–75 10.1038/ni.1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nguyen N. T., Now H., Kim W. J., Kim N., and Yoo J. Y. (2016) Ubiquitin-like modifier FAT10 attenuates RIG-I mediated antiviral signaling by segregating activated RIG-I from its signaling platform. Sci. Rep. 6, 23377 10.1038/srep23377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang Y., Tang J., Yang N., Liu Q., Zhang Q., Zhang Y., Li N., Zhao Y., Li S., Liu S., Zhou H., Li X., Tian M., Deng J., Xie P., et al. (2016) FAT10 is critical in influenza A virus replication by inhibiting type I IFN. J. Immunol. 197, 824–833 10.4049/jimmunol.1501563 [DOI] [PubMed] [Google Scholar]
- 44. Lim K. L., Chew K. C., Tan J. M., Wang C., Chung K. K., Zhang Y., Tanaka Y., Smith W., Engelender S., Ross C. A., Dawson V. L., and Dawson T. M. (2005) Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1: Implications for Lewy body formation. J. Neurosci. 25, 2002–2009 10.1523/JNEUROSCI.4474-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gavin J. M., Chen J. J., Liao H., Rollins N., Yang X., Xu Q., Ma J., Loke H. K., Lingaraj T., Brownell J. E., Mallender W. D., Gould A. E., Amidon B. S., and Dick L. R. (2012) Mechanistic studies on activation of ubiquitin and di-ubiquitin-like protein, FAT10, by ubiquitin-like modifier activating enzyme 6, Uba6. J. Biol. Chem. 287, 15512–15522 10.1074/jbc.M111.336198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mevissen T. E. T., and Komander D. (2017) Mechanisms of deubiquitinase specificity and regulation. Annu. Rev. Biochem. 86, 159–192 10.1146/annurev-biochem-061516-044916 [DOI] [PubMed] [Google Scholar]
- 47. Zou W., Papov V., Malakhova O., Kim K. I., Dao C., Li J., and Zhang D. E. (2005) ISG15 modification of ubiquitin E2 Ubc13 disrupts its ability to form thioester bond with ubiquitin. Biochem. Biophys. Res. Commun. 336, 61–68 10.1016/j.bbrc.2005.08.038 [DOI] [PubMed] [Google Scholar]
- 48. Nyfeler B., Michnick S. W., and Hauri H. P. (2005) Capturing protein interactions in the secretory pathway of living cells. Proc. Natl. Acad. Sci. U.S.A. 102, 6350–6355 10.1073/pnas.0501976102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang T., Yin L., Cooper E. M., Lai M. Y., Dickey S., Pickart C. M., Fushman D., Wilkinson K. D., Cohen R. E., and Wolberger C. (2009) Evidence for bidentate substrate binding as the basis for the K48 linkage specificity of otubain 1. J. Mol. Biol. 386, 1011–1023 10.1016/j.jmb.2008.12.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Aichem A., and Groettrup M. (2012) Detection and analysis of FAT10 modification. Methods Mol. Biol. 832, 125–132 10.1007/978-1-61779-474-2_7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.