Abstract
The metabolic serine hydrolases hydrolyze ester, amide, or thioester bonds found in broad small molecule substrates using a conserved activated serine nucleophile. The mammalian central nervous system (CNS) express a diverse repertoire of serine hydrolases that act as (phospho)lipases or lipid amidases to regulate lipid metabolism and signaling vital for normal neurocognitive function and CNS integrity. Advances in genomic DNA sequencing have provided evidence for the role of these lipid-metabolizing serine hydrolases in neurologic, psychiatric, and neurodegenerative disorders. This review briefly summarizes recent progress in understanding the biochemical and (patho)physiological roles of these lipid-metabolizing serine hydrolases in the mammalian CNS with a focus on serine hydrolases involved in the endocannabinoid system. The development and application of specific inhibitors for an individual serine hydrolase, if available, are also described.
1. Introduction
The mammalian central nervous system (CNS) exhibits a distinct lipid composition compared to other organs and tissues. This unique lipid composition is regulated and sustained by numerous lipidmetabolizing enzymes that are highly expressed in the CNS. One such superfamily are serine hydrolases, a class of enzymes that possesses the α/β hydrolase motif and a nucleophilic serine residue embedded in a Ser-His-Asp or Ser-Ser-Lys catalytic triad to enable cleavage of ester, amide, or thioester bonds of protein, peptide, and small molecule substrates [1]. Among more than 200 enzymes that belong to serine hydrolase, approximately half of the family are classified as metabolic serine hydrolases, whose substrates are generally small molecules including lipids. Several metabolic serine hydrolases are implicated in neurologic and psychiatric disorders [2]. Thus, these enzymes and their metabolic substrates or products play essential roles for the normal functions and development of the mammalian CNS.
Endocannabinoids [2-arachidonoylglycerol (2-AG) and arachidonoyl ethanolamine (AEA; also called anandamide)] are bioactive lipids that serve as endogenous ligands for the cannabinoid receptors CB1 and CB2, which are the molecular targets for the psychoactive agent THC (Δ9-tetrahydrocannabinol). Activation of these G protein-coupled receptors (GPCR) by endogenous (2-AG, AEA) and exogenous (e.g. THC) cannabinoids regulate a plethora of neuro-(patho)physiological processes. Recent advances in chemoproteomic technologies including activity-based protein profiling (ABPP [3]) led to identification and characterization of several serine hydrolases that play central roles in the biosynthesis and degradation of these lipid signaling molecules. These techniques also enabled development of a suite of mechanism-based covalent inhibitors that block serine hydrolase activity with high selectivity and potency in vitro and in vivo. Studies using serine hydrolase inhibitors have supported their potential therapeutic benefits on pathophysiological diseases and conditions of the CNS.
In this review, we summarize recent discoveries on metabolic serine hydrolases that regulate the biosynthesis and degradation of two major endocannabinoids, AEA and 2-AG. We also summarize other lipid-metabolizing serine hydrolases that are highly expressed in the CNS and implicated in CNS disorders.
2. Serine hydrolases involved in AEA synthesis and degradation
2.1. PLA2G4E
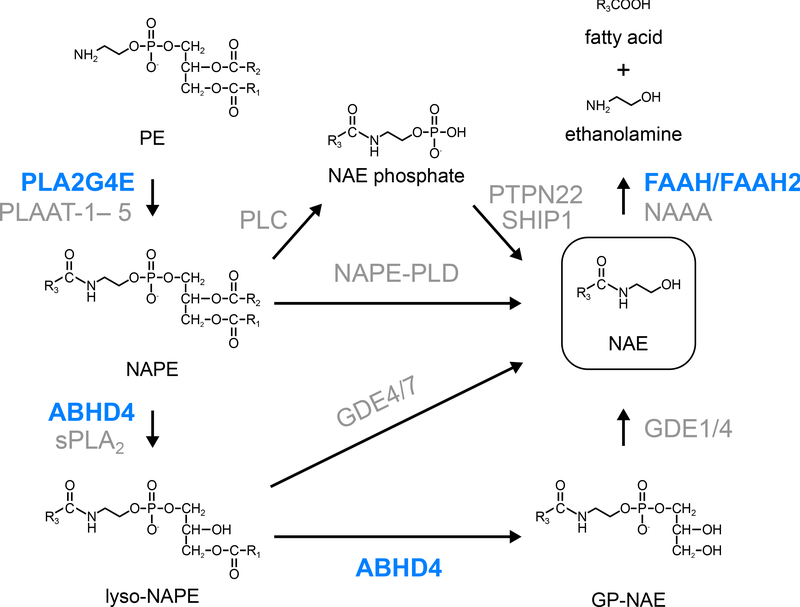
AEA is the first discovered endogenous ligand of the cannabinoid receptors, and is a member of the N-acyl ethanolamine (NAE) family of lipids. The first step of the NAE synthetic pathway is synthesis of N-acyl phosphatidylethanolamine (NAPE) by enzymatic reactions that transfer an acyl chain to the free amino nitrogen of phosphatidylethanolamine (PE) (Fig. 1). This enzymatic activity was first described in dog heart and brain tissues [4, 5], and was thereafter shown to be enriched in the rodent brain [6]. Biochemical analyses revealed that this enzyme requires calcium for its activity, and that it transfers the fatty acid attached to the sn-1 position of phosphatidylcholine (PC) to the amine of PE. Since this calcium-dependent N-acyltransferase (Ca-NAT) activity is sensitive to serine hydrolase inhibitors [7], it has been thought that this enzyme belongs to the serine hydrolase family. Many attempts to identify this enzyme have failed due potentially to its localization to the membrane, its low abundance, and its sensitivity to various kinds of detergent and separation methods using column chromatography.
Figure 1. Serine hydrolases involved in AEA synthesis and degradation.
PLA2G4E and PLAAT family enzymes (in human, PLAAT-1– 5; in mice, PLAAT-1, −3, and −5) transfer a fatty acyl chain at the sn-1 position of PCs to the free amino nitrogen of PEs to produce NAPEs. NAPEs are directly converted to NAEs by NAPE-PLD. The conversion of NAPEs to NAEs is also mediated by multiple metabolic enzymes including ABHD4. Glycerophosphodiesterase (GDE) family enzymes hydrolyze lyso-NAPEs or GP-NAEs to NAEs. NAEs are degraded by FAAH/FAAH2 and NAAA. Serine hydrolases and non-serine hydrolases are shown in blue and gray, respectively.
Recently, Ogura et al. found that this enzyme can be solubilized with a detergent IGEPAL CA-630, and that the enzyme activity can be separated and enriched by a sucrose gradient [8]. The subsequent ABPP and global correlation analysis between the quantity of serine hydrolases and the Ca-NAT activity in each fraction identified PLA2G4E (also known as cPLA2ϵ) as the long-soughtafter Ca-NAT [8]. PLA2G4E is a member of a cytosolic phospholipase A2 (cPLA2) family, and was originally characterized as an enzyme with considerably weaker PLA2 activity toward phospholipids compared to other members such as cPLA2α [9–11]. Consistent with previous research, the Ca-NAT activity of recombinant PLA2G4E was enhanced by addition of calcium ion and DTT, and was inhibited by serine hydrolase inhibitors and probes. Overexpression of PLA2G4E in HEK293T cells led to massive accumulation in NAPEs and the downstream N-acylated lipids such as glycerophospho (GP)-NAEs (GP-NAEs) and NAEs. Ionomycin treatment dramatically increased production of these N-acylated lipids in PLA2G4E-expressing cells. PLA2G4D (also known as Cpla2δ), the closest homolog of PLA2G4E with 43% sequence identity, did not show Ca-NAT activity. These results indicate that PLA2G4E is the bona fide Ca-NAT in the mouse brain. Recently, PLAAT (phospholipase A/acyltransferase) family enzymes have been shown to possess a calcium-independent NAPE-forming PE N-acyltransferase activity [12]. The contribution of PLA2G4E and the PLAAT family enzymes to endogenous levels of AEA and other NAEs in vivo should be determined by generating knockout animal models or pharmacological inhibition of these enzymes. Of note, a recent study suggested that single nucleotide variants in the human PLA2G4E gene may be linked to the risk of panic disorder [13]. Knockout animal models and inhibitors of PLA2G4E will be a new tool to uncover the physiological roles of NAPEs and their downstream lipids.
2.2. ABHD4
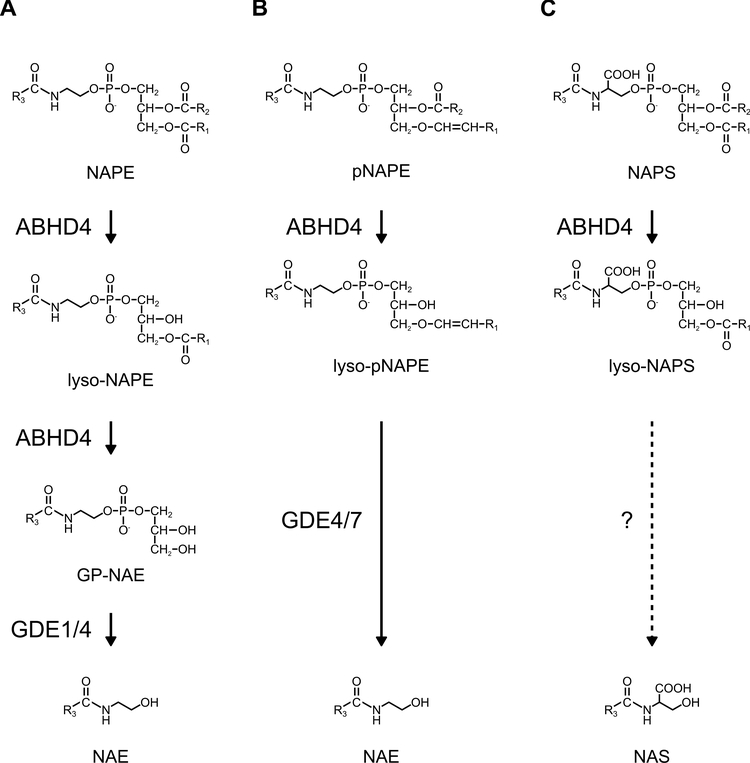
Multiple enzymatic pathways mediate the synthesis of NAEs from NAPEs (Fig. 1). NAPE-PLD, an enzyme that belongs to the zinc metallohydrolase family of the β-lactamase fold, exhibits a phospholipase D activity toward NAPEs and directly converts NAPEs into NAEs [14] (Fig. 1). Brains from Nape-pld−/− mice exhibit dramatic reductions in NAE species that contain saturated or monounsaturated fatty acids with more than 18 carbon chains such as C20:0, C22:0, C24:1, and C24:0; however, only modest reductions in major NAEs (~2-fold or less), including C16:0, C18:0, C18:1, and C20:4 NAEs, were observed, suggesting the existence of other metabolic pathways for NAE synthesis from NAPEs [15, 16]. A candidate pathway is the sequential hydrolysis of sn-1 and sn-2 ester bonds by PLA1/2 enzymes to yield lyso-NAPE and GP-NAE intermediates, followed by the removal of lysophosphatidic acid (LPA) and glycerol 3-phosphate [17] (Fig. 1). Protein purification combined with ABPP technique identified the serine hydrolase ABHD4 as the PLA1/2 (i.e., PLB) enzyme that cleaves both sn-1 and sn-2 O-acyl chains of NAPEs [18]. In mice, ABHD4 is highly expressed in the CNS and the testis, and moderately in other tissues such as the liver and the kidney [18, 19]. Recombinant ABHD4 protein exhibits both NAPE- and lyso-NAPE-lipase activity [18]. Abhd4−/− mouse brains show moderate reductions in lyso-NAPEs and GP-NAEs, and marked reductions in plasmalogen-type lyso-NAPEs (lyso-pNAPEs) [19]. However, NAE levels were not significantly altered compared to those in wild-type brains, probably due to the presence of redundant pathways in NAE synthesis. A non-targeted lipidomics analysis revealed that a novel class of lipids, N-acyl lysophosphatidylserine (lyso-NAPS) was dramatically reduced in Abhd4−/− mouse brains [19]. Biochemical analyses verified that NAPS lipids are also ABHD4 substrates (Fig. 2). Although ABHD4 catalyzes both PLA1 and PLA2 reactions in vitro, the results of the subsequent targeted lipidomics analysis indicated that it may mainly act as a PLA2 enzyme in vivo. While Abhd4−/− mouse brains showed dramatic reductions in lyso-NAPS species with O-saturated fatty acids, the levels of lyso-NAPSs with O-unsaturated fatty acids were not significantly affected. In general, saturated fatty acids are mainly found at sn-1 position of phospholipids, whereas unsaturated fatty acids are mainly attached to their sn-2 position. Thus, ABHD4 likely hydrolyzes the ester bond at sn-2 position more preferentially, where unsaturated fatty acids are attached, to produce sn-1-O-saturated lyso-NAPS species.
Figure 2. ABHD4 regulates multiple classes of N-acyl phospholipids.
(A) ABHD4 exhibits both NAPE- and lyso-NAPE-lipase activity to sequentially produce lyso-NAPEs and GP-NAEs. (B) ABHD4 also hydrolyzes plasmalogen-type NAPEs (pNAPEs) to produce lysopNAPEs. (C) ABHD4 predominantly hydrolyzes the fatty acyl ester bond at the sn-2 position of NAPS to produce lyso-NAPS. Whether N-acyl serine (NAS) is synthesized from NAPs or lyso-NAPS is not known.
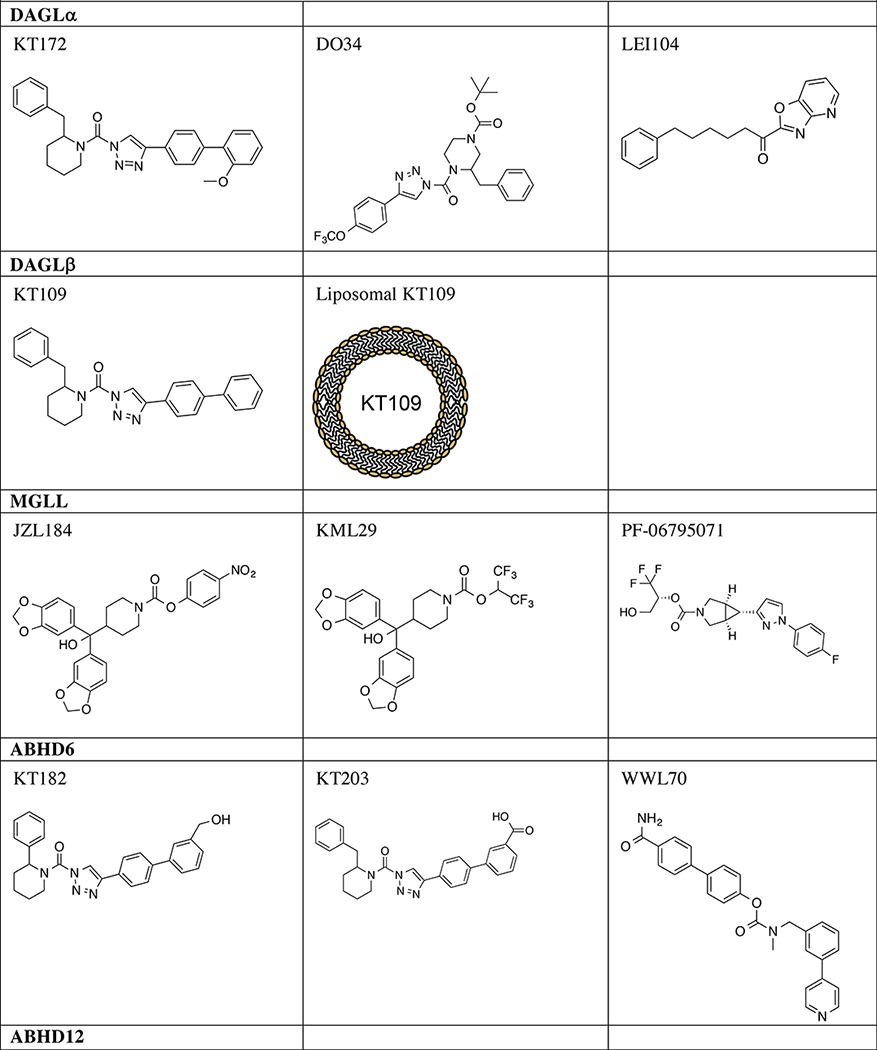
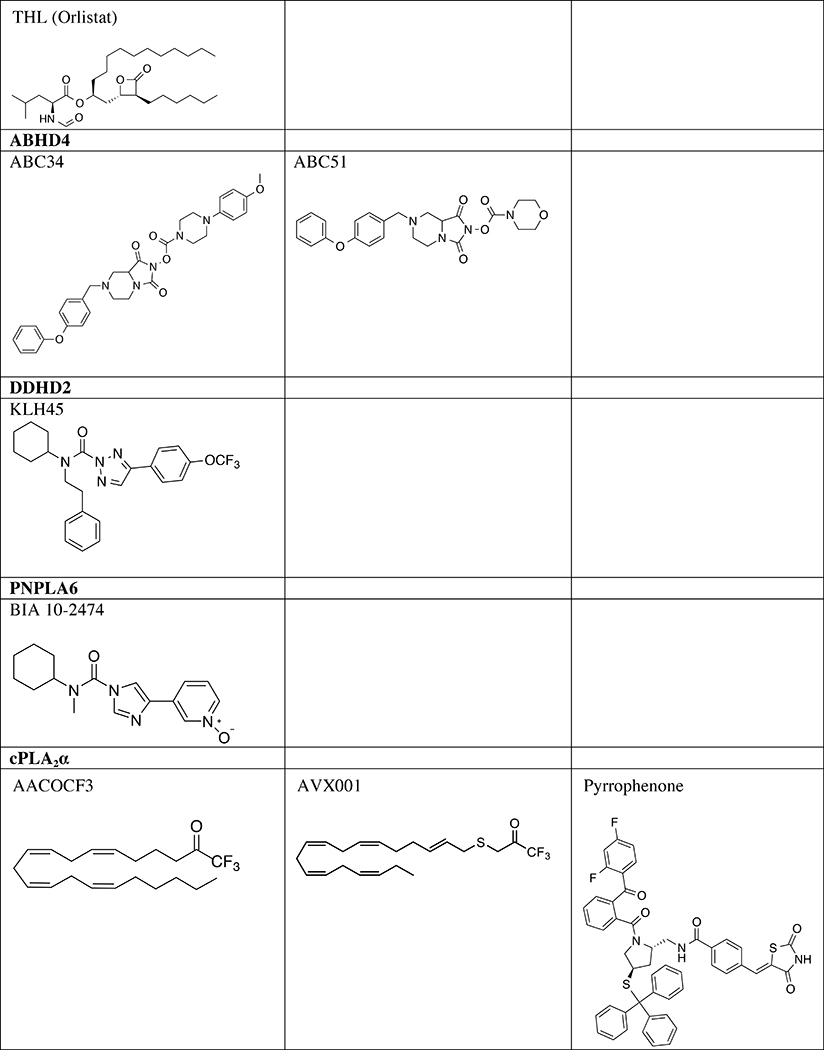
Taken together, ABHD4 regulates multiple classes of N-acyl phospholipids in the mammalian CNS (Fig. 2). Although ABHD4 is implicated in several pathophysiological events such as cancer and anoikis [20, 21], the contribution of its lipid metabolic activity in these events is not well understood. A study using competitive ABPP showed that several N-hydroxyhydantoin carbamates, a versatile class of irreversible serine hydrolase inhibitors, inhibit ABHD4 activity with good potency and selectivity [22] (Table 1). Further optimization of ABHD4-selective inhibitors will greatly enable functional studies of this serine hydrolase in the CNS.
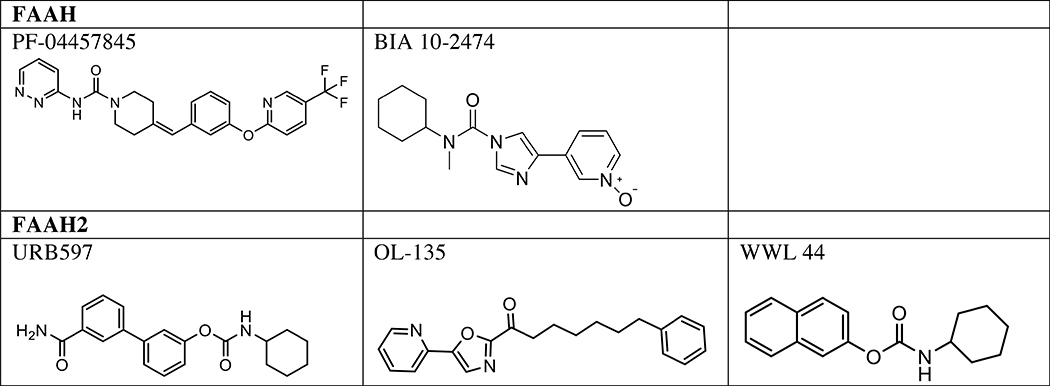
Table 1.
Serine hydrolase inhibitors for in vivo analysis of biology
 |
 |
 |
2.3. FAAH
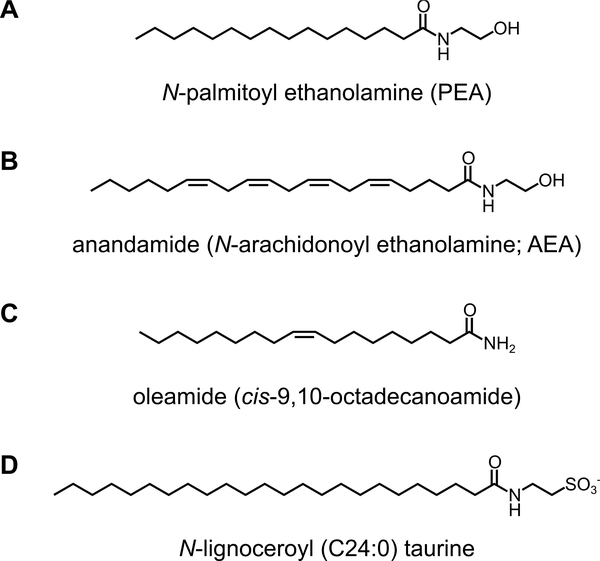
FAAH (Fatty acid amide hydrolase) is an integral membrane serine hydrolase that is highly expressed in the mammalian brain, liver, kidney, and testis. FAAH is localized to the ER and several transporters that are able to transport AEA to FAAH have been identified [23–25]. While serine hydrolases largely use a Ser-His-Asp triad for catalysis, FAAH forms an unusual Ser-Ser-Lys catalytic triad that is characteristic of amidase signature (AS) family enzymes. In vitro, FAAH most preferentially hydrolyzes C20:4 NAE (AEA), followed by C18:1 NAE (N-oleoyl ethanolamine; OEA) and C16:0 NAE (N-palmitoyl ethanolamine; PEA). The genetic ablation or pharmacological inhibition of FAAH lead to dramatic accumulation of AEA and related NAE congeners in the rodent brain, indicating that FAAH is the major NAE-degrading enzyme in the mammalian CNS (Fig. 1). FAAH also hydrolyzes oleamide (cis-9,10-octadecanoamide), a sleep-inducing lipid [26], at an equivalent rate to AEA (Fig. 3). A non-targeted lipidomics analysis on Faah−/− brains also identified a new class of lipids, N-acyl taurines (NATs) as another endogenous substrate of FAAH [27] (Fig. 3). Compared to wild-type brains, Faah−/− brains show 10–40 fold accumulation of very long chain NATs (more than 22 carbons). Since the hydrolytic activity of FAAH toward NATs is considerably lower than that toward NAEs in vitro, massive accumulation of NATs in Faah−/− is likely due to the absence of other NAT-degrading pathways [28]. NAAA (N-acyl ethanolamine-hydrolyzing acid amidase), a lysosomal enzyme that belongs to the choloylglycine hydrolase family, also has been shown to hydrolyze NAEs, with a preference for C16:0 NAE [29] (Fig. 1). However, its expression is mainly in peripheral tissues, and its contribution in the CNS is not well understood.
Figure 3. Structure of FAAH/FAAH2 substrates.
(A) N-palmitoyl ethanolamine (PEA), (B)anandamide (N-arachidonoyl ethanolamine; AEA), (C) oleamide (cis-9,10-octadecanoamide), and (D) N-lignoceroyl (C24:0) taurine.
In rodents, elevation of AEA levels by genetic or pharmacological disruption of FAAH leads to anxiolytic, anti-depressive, and analgesic phenotypes in a cannabinoid-receptor-dependent manner. FAAH blockade also leads to anti-inflammatory phenotypes in both a cannabinoid-receptordependent and -independent manners, the latter of which may include accumulation of PEA and OEA, endogenous agonists of peroxisome proliferator-activated receptor-α (PPARα) [30, 31]. A number of FAAH inhibitors have been developed and now under human clinical trials, and potential clinical efficacy of FAAH inhibition in pain and hyperalgesic has been reported [32] (Table 1). No severe side effects had been reported in these clinical trials, until the recent serious adverse events in a phase I clinical trial for BIA 10–2474, which include a single case of death and serious neurological damage [32, 33]. However, these adverse events may be due to this drug’s off targets, as other clinically tested FAAH inhibitors such as PF-04457845 have not yielded such neurotoxicity, and a recent ABPP analysis revealed that BIA 10–2474 and its major metabolite BIA 10–2639 also inhibit several lipidmetabolizing serine hydrolases including PNPLA6, disruption of which causes neurotoxicity and neurodegenerative disorders as discussed below [34].
2.4. FAAH2
FAAH2 is a serine hydrolase with ~20% sequence identity to FAAH, and is conserved in higher placental mammals, but not in rodents. In human, FAAH2 is located on the X chromosome, and is highly expressed in the liver, kidney, lung, and prostate, but only moderately expressed in the brain [28]. FAAH2 exhibits a distinct substrate preference compared to FAAH; FAAH2 preferentially hydrolyzes oleamide and N-oleoyl ethanolamine (C18:1 NAE), but shows considerably less activity toward other NAEs (C16:0 and C20:4) and no activity toward C18:1 N-acyl taurine (NAT) (Fig. 1 and 3). Our understanding of the physiological roles of FAAH2 has lagged behind that of FAAH because of the lack of animal knockout models and FAAH2-specific inhibitors. Although some FAAH inhibitors such as URB597 or OL-135 show enhanced potency for FAAH2 versus FAAH [28], more selective inhibitors, such as WWL44 [35], may be required to investigate FAAH2-specific functions (Table 1). Recent studies have suggested that genetic defects of FAAH2 are linked to neurologic and psychiatric disorders, such as ataxia, seizure, intellectual disability, and autism [36–38], Thus, in addition to FAAH, FAAH2 seem to play pivotal roles for the functions of the human CNS.
2.5. AEA signaling in other animal models
AEA signaling and related enzymes are conserved in other lower animals such as invertebrates and fish. Interestingly, these enzymes have been shown to also play essential roles in nervous system functions. A variety of NAE species, including AEA, have been identified in Caenorhabditis elegans, and inhibits dietary-restriction-induced lifespan extension [39]. A genome-wide RNA interference (RNAi) screen identified faah-1, a C. elegans ortholog of FAAH, as a regulator of axon regeneration [40]. The faah-1 deletion mutant shows a defect in axon regeneration after laser axotomy most likely due to the accumulation of endogenous AEA and eicosapentaenoyl ethanolamide (EPEA) [40], and this defect is suppressed by a deletion mutation of NAPE-PLD homologs [41]. The effects of these NAEs on axon regeneration are mediated by G protein-coupled receptors that couple to Goα and possess amino acid residues highly conserved or functionally important in human CB1/2 receptors [41] As mentioned earlier, FAAH2 is not conserved in rodents, which makes it difficult to investigate the physiological roles of FAAH2 in vivo. On the other hand, FAAH2 is conserved in non-rodent vertebrates such as zebrafish [42–44]. Krug et al. [45] generated a mutant line of faah2a, one of two zebrafísh FAAH2 paralogs, and found that these faah2a zebrafish mutants exhibited reduced stressassociated behavioral response, which is similar to the phenotypes observed in Faah−/− mice [46, 47]. Thus, these non-rodent animal models may serve as alternative tools to investigate the roles of AEA signaling and related enzymes in the CNS.
3. Serine hydrolases involved in 2-AG synthesis and degradation
3.1. DAGLα and DAGLβ
Diacylglycerol lipase-alpha and -beta (DAGLα and DAGLβ) are ~120 and ~70 kDa, respectively, serine hydrolases that hydrolyze AA (arachidonic acid)-esterified diacylglycerols (DAGs) to produce 2-AG (Fig. 4) [48, 49]. In vitro, DAGLs preferentially hydrolyze DAGs at the sn-1 position, can be stimulated with calcium, and have negligible activity against other lipids including monoacylglycerols and phospholipids [48]. DAGLs show high conservation between humans and mice (97% and 79% identity for alpha and beta, respectively [48]) but surprisingly low homology between isoforms within a given species (~20% sequence identity for human and mouse DAGLs, https://www.uniprot.org). Mammalian DAGLs are expressed as transmembrane proteins composed of a 4-transmembrane domain region at the N-terminus followed by a canonical α/β hydrolase domain (containing the nucleophilic serine residue). The C-terminal tail largely differentiates DAGLα and DAGLβ with the former isoform showing a more pronounced domain region. Activated calcium/calmodulin dependent protein kinase II (CaMKII) has been shown to phosphorylate the C-terminal domain of DAGLα (serine 782 and 808 of mouse DAGLα) to inhibit DAGLα activity in regulation of 2-AG metabolism and signaling in vivo [50]. The role of the C-terminal tail of DAGLβ is currently unknown. Several reports have identified cysteine palmitoylation of DAGLβ, which may serve as a novel posttranslational modification (PTM) to regulate function [51, 52]. Future studies aimed at studying posttranslation regulation of DAGL activity in vivo will be important to understand cross-talk between lipid and protein signaling pathways.
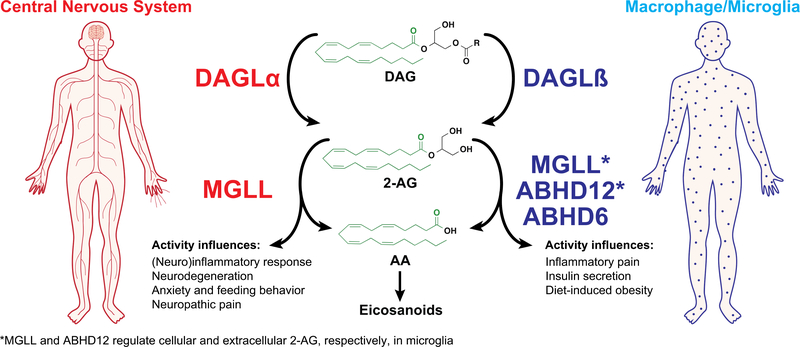
Figure 4. Tissue expression and activity of 2-AG biosynthetic and hydrolytic serine hydrolases that regulate endocannabinoid metabolism and signaling.
Schematic depicting 2-AG (2-arachidonoylglycerol) biosynthetic and degradation pathways and their respective tissue expression that is important for regulation of endocannabinoid signaling. DAGLs (DAGLα and DAGLβ) preferentially hydrolyze sn-1 fatty acids from diacylglycerols (DAG) that contain arachidonic acid (AA) esterified at the sn-2 position to biosynthesize 2-AG. DAGLα expression and activity is enriched in the central nervous system while DAGLβ is found largely in macrophages and microglia. MGLL is the principal 2-AG hydrolase in the central nervous system. In microglia and macrophages, DAGLβ produces 2-AG pools that are metabolized by downstream 2-AG hydrolases to release arachidonic acid (AA) utilized for production of proinflammatory lipids including eicosanoids. In microglia, MGLL, ABHD12, and ABHD6 have been show to function as 2-AG hydrolases.
3.1.1. DAGL lipid metabolism and signaling
In addition to differences in domains and PTMs, DAGL function is segregated by tissue- and cell type-specific expression (Fig. 4 and 5). DAGLα is expressed predominantly in central tissues (brain, spinal cord) and pancreas while DAGLβ expression is enriched in liver and immune cells including macrophages and microglia [53–56]. Consistent with gene expression profiles, lipid analyses of Dagla−/− mice showed 80–90% reductions in brain 2-AG as well as the downstream product arachidonic acid (AA) [53, 54, 57]. Daglb−/− mice showed 90% reductions in liver 2-AG and negligible effects on brain 2-AG and AA, although one report demonstrated modest changes in brain 2-AG [53].
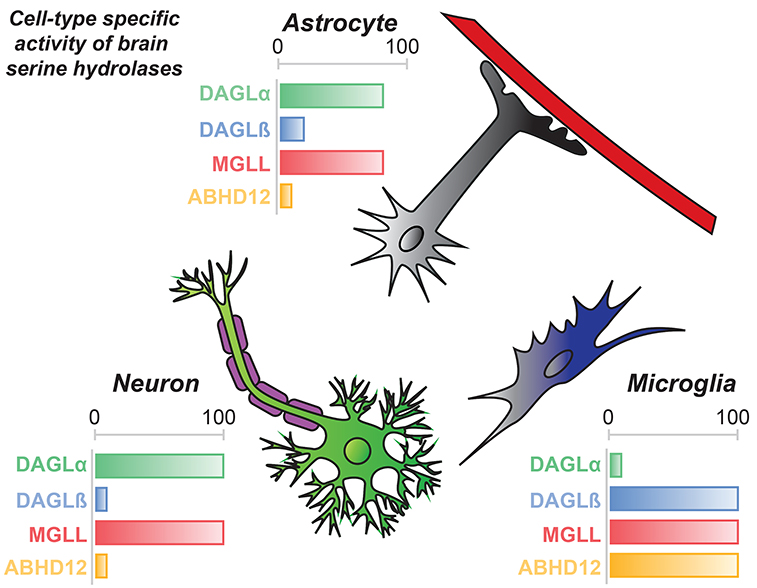
Figure 5. Distribution of 2-AG biosynthetic/hydrolytic serine hydrolases across cell types in brain.
2-AG signaling in the central nervous system is regulated by differential expression of metabolic enzymes across cell types in the mammalian brain. DAGLα and MGLL activity is enriched in neurons, while DAGLβ, MGLL, and ABHD12 regulate 2-AG metabolism and signaling in microglia. Astrocytes show similar activity profiles as neurons with the exception of increased contribution of DAGLβ activity in this cell type.
DAGL activity is further regulated by expression and activity within individual cell types of the brain (Fig. 5) [57]. Lipid profiles of neurons, astrocytes, and microglia revealed substantial reductions in 2-AG and AA (60–90%) in Dagla−/− neurons and astrocytes with negligible changes in these lipids measured in microglia. In contrast, lipid profiles of Daglb−/− neurons were not impacted; in contrast, Daglb−/− microglia showed ~50% reductions in 2-AG, AA, and prostaglandins (PGE2 and PGD2) compared with wild-type counterparts [57]. Astrocytes showed a mixed lipid profile with significant reductions in 2-AG, AA, and PGE2/D2 in Dagla−/− cells. Disruption of DAGLβ resulted in a modest (~20%) but significant reduction in astrocyte 2-AG. Taken together, these metabolomics findings illustrate the importance of DAGL expression and activity across tissues and individual celltypes of tissues to fine-tune 2-AG metabolism and signaling [57].
In the context of 2-AG signaling, disruption of DAGLα results in functional antagonism of CB1 through depletion of 2-AG available for CB1 signaling. Consistent with this hypothesis, Dagla−/− mice recapitulate many of the metabolic (lean, hypophagic, improved glycemic control in obesity) and behavior phenotypes (anxiety, depression) observed in CB1 knockout mice [58, 59]. In addition, reductions in adult neurogenesis were observed in Dagla−/− mice [53, 60]. Localization of DAGLα at the postsynaptic density [61], potentially through protein-protein interactions of its C-terminal tail with postsynaptic Homer adaptor proteins [62] and CaMKII [50], positions this lipase for regulation of synaptic plasticity (e.g. DSI and DSE) via retrograde signaling at presynaptic CB1 [53, 54]. In summary, the comparable phenotypes of knockout mice and complementary localization of DAGLα and CB1 supports a key role for DAGLα in regulating 2-AG signaling in the CNS [53, 60].
3.1.2. DAGLβ regulates macrophage and microglia inflammatory signaling
DAGLβ disruption does not result in the same metabolic and behavioral phenotypes observed in Dagla−/− mice [53, 54, 63]. Chemoproteomic studies have shown enrichment of DAGLβ activity in macrophages and microglia (brain macrophage subset) immune cells [55, 57]. Consistent with its activity profile, disruption of DAGLβ results in accumulation of AA-esterified DAGs (specifically the C18:0/C20:4 DAG species) and depletion of 2-AG, AA, and prostaglandins (PGE2 and PGD2) in macrophages [55] and microglia [57]. Regulation of both 2-AG and AA by DAGLβ revealed important cross-talk between endocannabinoid and eicosanoid signaling in macrophages. Disruption of DAGLβ in macrophages and microglia also attenuated proinflammatory cytokine (TNF-α) signaling in response to lipopolysaccharide stimulation [55, 57]. Thus, DAGLβ is a complementary pathway to classical cPLA2α pathways [64] for regulation of AA pools utilized by COX1/2 to generate lipid mediators of macrophage inflammatory responses (see section 4.1 for additional details).
3.1.3. DAGL inhibitors
Discovery of first-generation in vivo-active DAGL inhibitors (Table 1) was enabled by ABPP assays tailored for detection and quantitation of native DAGL activity [55, 65]. In addition to enhanced sensitivity, the DAGL-tailored activity-based probe HT-01 allowed rapid evaluation of potency and selectivity of DAGL inhibitors directly in complex proteomes in vitro and in vivo [55]. Using ABPPguided medicinal chemistry, the 1,2,3-triazole urea covalent inhibitor KT109 emerged as the first in vivo-active DAGLβ inhibitor suitable for cell (IC50 ~ 10 nM) and animal studies (EC50 ~10 mg kg−1 in animal pain models) [55, 57, 66]. A structurally analogous, DAGL-inactive negative control inhibitor KT195 was also developed to distinguish DAGLβ-specific from non-specific serine hydrolase inhibition [55, 67]. Liposomal encapsulation of KT109 (i.e. liposomal KT109) enabled targeted delivery of DAGLβ inhibitors to macrophages and dramatically reduced compound amounts required to elicit anti-inflammatory effects in animal pain models [68].
KT172, a structural analog of KT109, was developed as a dual DAGLα and DAGLβ inhibitor [55] (Table 1). Further medicinal chemistry around the KT172 scaffold produced the CNS-active variants, DH376 and DO34, which permitted in vivo analysis of DAGL biology in the brain [56]. DH376 and DO34 were used along with a negative-control probe (DO53) to demonstrate that acute blockade of DAGLs in mouse brain rapidly altered lipid profiles to impact synaptic plasticity, neuroinflammatory responses (50 mg kg−1) [56], and fasting-induced refeeding behavior (50 mg kg−1) [69]. Exploration of reversible DAGL inhibitors, as opposed to covalent irreversible 1,2,3-triazole ureas, identified the α-ketoheterocycle LEI104 (previously reported as a FAAH inhibitor) as a potential scaffold for developing DAGLα inhibitors [70] (Table 1). Future studies aimed at defining key structure-activity relationships that differentiate inhibitory activity against DAGLα versus DAGLβ, as well as common serine hydrolase off-targets (e.g. ABHD6), will help guide development of DAGLα-selective inhibitors suitable for in vivo use in the CNS.
3.2. MGLL
3.2.1. Principal 2-AG hydrolytic enzyme in the CNS
Monoacylglycerol lipase (MGLL or MAGL) is a ~33 kDa serine hydrolase that adopts an α/β hydrolase fold composed of an eight β-strand core (containing the conserved GXSXG motif) enclosed on two sides by α-helices; eight in the case of the human MGLL [71, 72]. MGLL was extracted and purified from rat adipose tissue where it displayed hydrolytic activity towards a diverse set of monoglyceride substrates [73]. MGLL was demonstrated to possess lipase activity with equal specificity for 2- and 1-monoglycerides and a preference for longer and more unsaturated MAG species [74, 75]. MGLL shows a strong preference for MAG versus other neutral lipids including DAGs and triacylglycerols (TAGs) in vitro [76]. In the context of brain lipid metabolism, MAGL accounts for roughly 85% of total 2-AG hydrolytic activity in mouse brain proteomes (Fig. 4 and 5), supporting this serine hydrolase as a critical regulator of endocannabinoid signaling in the CNS [77].
3.2.2. MGLL inhibitors
Competitive ABPP and medicinal chemistry enabled development of the O-aryl carbamate JZL184 as a potent and selective MGLL inhibitor (IC50 ~10 nM, Table 1) [78]. JZL184 treatments reduced 2-AG hydrolysis activity by >80%, resulting in an eight-fold enhancement in brain 2-AG without altering AEA in treated mice [78]. JZL184-treated mice showed a broad range of CB1-dependent behavioral effects including hypothermia, analgesia, and hypomotility. Long-term elevations in brain 2-AG from chronic MGLL blockade (Mgll−/− mice or JZL184 chronic treatments) desensitizes and downregulates brain CB1 receptors, further validating a key role of MGLL in regulating 2-AG-CB1 signaling in the CNS [79, 80]. In support of 2-AG as a primary source for AA in the CNS, disruption of brain MGLL with JZL184 treatments resulted in substantial decreases in AA and AA-derived prostaglandins (Fig. 4) [81]. The latter metabolic effect suppressed neuroinflammation and provided neuroprotection in a mouse model of Parkinson’s disease [81]. Thus, the positive effects of MGLL blockade, i.e. enhanced 2-AG signaling and suppression of inflammatory prostaglandins, has opened new therapeutic avenues for targeting this enzyme in neurodegeneration, inflammation, and pain [2].
While highly selective, JZL184 showed partial blockade of FAAH in brain and carboxylesterases in peripheral tissues of treated mice [78]. To improve selectivity in vivo, exploration of carbamate inhibitors bearing a hexafluoroisopropanol (HFIP)-leaving group produced KML29, which inactivated MGLL in vitro (IC50 ~15 nM) and in vivo with excellent potency and enhanced selectivity against FAAH [82] (Table 1). Recently, exploration of a trifluoromethyl glycol leaving group improved drug-like properties (solubility, chemical liability) of covalent MGLL inhibitors compared with HFIP counterparts (PF-06795071, Table 1) [83]. Following these discoveries, additional MGLL inhibitors have been pursued including novel compounds that block activity via a reversible mechanism [84].
3.2.3. Therapeutic potential of MGLL
3.2.3.1. Inflammation and pain
MGLL inactivation using JZL184 and KML29 attenuated inflammatory and neuropathic pain through enhanced brain 2-AG and activation of cannabinoid receptors CB1 and CB2 [85, 86]. Chronic treatments with high dose JZL184 or KML29 resulted in tolerance due to cannabinoid receptor downregulation and desensitization. However, repeated low dose administration of these compounds, to avoid functional antagonism of CB receptors [79], blocked pathogenic pain states without evidence of tolerance. Use of JZL184 and KML29 along with cannabinoid receptor knockout mice and antagonists supported differential involvement of cannabinoid receptor subtypes in the different pain models; the anti-allodynic effects of MGLL inhibitors require both CB1 and CB2 while only CB1 is necessary for neuropathic pain [85, 86]. Importantly, KML29 provided beneficial effects in mouse pain models without eliciting cannabimimetic effects (catalepsy, hypothermia and hypomotility) that are characteristic of full agonists of CB1 and CB2 like THC [87]. These results highlight the differential pharmacology of blocking a single endocannabinoid catabolic enzyme versus dual MGLL/FAAH inhibition, which elevates both 2-AG and AEA to elicit THC-like cannabimimetic effects [86].
3.2.3.2. Neurodegenerative diseases
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease that is marked by progressive deterioration of upper and lower motor neurons typically onset as an adult [88]. Mutations in superoxide dismutase 1 (SOD1) constitutes a substantial fraction of familial ALS and mutant SOD1 mice are capable of recapitulating clinical hallmarks and behaviors of ALS [89]. Using mutant (SOD1G93A) mice, researchers discovered accumulation of AEA and 2-AG in spinal cord at early symptomatic and late stage ALS, which may represent a neuroprotective mechanism to counteract disease progression [90, 91]. In support of this hypothesis, treatment of ALS SOD1G93A mice with KML29 (10 mg kg−1) resulted in accumulation of brain and spinal cord 2-AG, and delayed the onset of disease and extended life span up to 24 days [92]. KML29 treatments also reduced proinflammatory cytokines and enhanced brain-derived neurotrophic factor (BDNF) expression levels in the spinal cord, which is a major site of neurodegeneration in ALS [92].
3.3. ABHD12
3.3.1. Alternative 2-AG hydrolase in microglia
α/β-hydrolase domain-containing protein 12 (ABHD12) is expressed as a ~45-kDa membrane glycoprotein predicted to contain a single-pass transmembrane domain and face the extracellular/luminal cellular space [1]. From a functional proteomic study, ABHD12 was discovered to contribute ~9% of total 2-AG hydrolytic activity in mouse brain proteomes [77] (Fig. 4 and 5). However, mass spectrometry metabolomics studies of Abhd12−/− mice did not show changes in bulk levels of brain 2-AG [93]. Within individual brain cell types, ABHD12 contributed to substantial 2-AG metabolism in microglia but not neurons and astrocytes, suggesting a cell-type specific role for this lipase in brain 2-AG signaling (Fig. 5) [57]. Specifically, secreted 2-AG was much higher from Abhd12−/− microglia and unaltered in Mgll−/− microglia compared to wild type counterparts. These data support a primary role for ABHD12 in regulating extracellular 2-AG pools and is consistent with its luminal/extracellular orientation [77]. The enhanced 2-AG secretion in Abhd12−/− microglia increased cannabinoid receptor-dependent signaling as evidenced by ERK1/2 phosphorylation [57]. Abhd12−/− microglia did not show differences in AA or prostaglandins (either basal or LPS stimulated), supporting its minor role in regulation of 2-AG pools utilized for prostaglandin production [57].
3.3.2. Principal lysophosphatidylserine hydrolyzing enzyme in PHARC
The neurodegenerative disease polyneuropathy, hearing loss, ataxia, retinosis pigmentosa, and cataract (PHARC) is caused by mutations in the Abhd12 gene in humans. Five distinct ABHD12 mutations have been identified in PHARC patients; all mutations are expected to result in complete loss of ABHD12 expression [94, 95]. Recently, untargeted lipidomics was performed in mouse brain tissues fromAbhd12−/− mice to discover that ABHD12 is a principal lysophosphatidylserine (lysoPS) lipase in the mammalian brain [93]. Specifically, Abhd12−/− mice showed massive accumulation of a rare subset of long chain lysoPS lipids that are implicated in Toll-like receptor 2 signaling.
Biochemical assays confirmed that recombinant ABHD12 protein can robustly hydrolyze synthetic lysoPS, and this lysoPS hydrolytic activity is substantially reduced in Abhd12−/− brain proteomes. Brain lysoPS accumulation in Abhd12−/− mice occurs early in life followed by age-dependent increases in microglial activation and auditory and motor defects akin to behavioral phenotypes of human PHARC patients. Thus, a molecular model for PHARC emerges where ABDH12 disruption leads to aberrant lysoPS metabolism and accumulation of lysoPS, which has been shown to activate toll-like receptor (TLR)2 signaling of innate immune cells [96] and may promote pathogenic microglial and neurobehavioral alterations [93]. Further studies are needed to determine whether lysoPS accumulation results in microglial activation involved in neuroinflammation. Recently, ABHD16A was annotated as a phosphatidylserine (PS) lipase that generates lysoPS in mammalian systems and thus establishes a ABHD16A-ABHD12 metabolic axis for study and treatment of neuroimmunological disorders [97].
3.3.3. ABHD12 inhibitors
New pharmacological probes of ABHD12 are needed to complement the cell biology and animal physiology discovered using Abhd12−/− mice. To date, widely used ABHD12 inhibitors lack selectivity to enable functional studies in vivo. The lipase inhibitor tetrahydrolipstatin (THL, Table 1) is capable of inactivating ABHD12 but also shows cross-reactivity against other serine hydrolases including ABHD6 and pancreatic lipases [77]. In addition to THL, additional natural products including triterpenes have been shown to reversibly block ABHD12 activity with moderate selectivity against other serine hydrolases detected in mouse brain proteomes [98]. Future studies aimed at developing potent, selective, and in vivo-active inhibitors will continue to advance our understanding of ABHD12 function in physiology and PHARC pathogenesis.
3.4. ABHD6
3.4.1. Alternative 2-AG hydrolysis at the site of biosynthesis
ABHD6 is an ~30-kDa integral membrane protein that adopts the canonical α/β hydrolase fold containing the GXSXG motif that houses the serine nucleophile [1, 99]. Functional proteomics demonstrated that ABHD6 contributes to ~4% of total brain 2-AG hydrolysis activity in the mouse brain (Fig. 4). Short hairpin RNA (shRNA) knockdown of ABHD6 in BV-2 microglia cells reduced 2-AG hydrolysis activity in BV-2 lysates by ~50% [100]. ABHD6 knockdown also increased the efficacy of 2-AG-CB2 mediated cell migration. In Neuro2A cells, which lack MGLL, blockade of ABHD6 with a selective inhibitor resulted in accumulation of 2-AG in live cells [55]. In adult mouse cortex, ABHD6 is localized postsynaptically and has been shown to regulate 2-AG degradation and signaling in murine primary neurons and cortical slices [100]. Taken together, these findings support a role for ABHD6 in regulation of 2-AG metabolism and signaling.
3.4.2. Alternative lipid substrates of ABHD6
Antisense oligonucleotide (ASO) knockdown of ABHD6 in peripheral tissues of mice fed a high-fat diet resulted in protection against diet-induced body weight gain [101]. Liquid chromatography-mass spectrometry (LC-MS) metabolomics uncovered liver accumulation of lysophospholipids and phospholipids as a potential protective mechanism; lysophosphatidylglycerol (LPG) and phosphatidylglycerol (PG) lipid species showed the most prominent accumulation [101]. Candidate lipid substrates identified by metabolomics were validated by biochemical substrate assays using purified ABHD6. Recombinant ABHD6 displayed substantial lipase activity toward lysophospholipids (LPG, LPA, and LPE), but negligible activity against their major phospholipid counterparts (i.e. PG, phosphatidic acid (PA), and PE) [101]. The highest activity was observed using LPG as a substrate, matching the substrate preference for LPG observed in metabolomics experiments.
ABHD6 has also been implicated in hydrolysis of bis(monoacylglycero)phosphate (BMP) [102]. Treatment with ABHD6-specific inhibitor lowered BMP hydrolysis in both brain lysates and cultured cells revealing that ABHD6 is responsible for ~90% and ~40% of the BMP hydrolase activity detected in liver and brain, respectively [102]. Collectively, these data show that ABHD6 can hydrolyze 2-AG as well as broad range of lipids to regulate diverse mammalian biology.
3.4.3. ABHD6 inhibitors
Using ABPP, the carbamate inhibitor WWL70 was identified as a potent and selective ABHD6 inhibitor [103] (IC50 ~70 nM, Table 1). Competitive ABPP screening of a carbamate library resulted in discovery of an additional carbamate ABHD6 inhibitor WWL123 that showed lower potency (IC50 ~400 nM) but higher selectivity against other brain serine hydrolases [35]. Optimization of (2-substituted)-piperidyl-1,2,3-triazole urea inhibitors (KT182 and KT203, Table 1) resulted in the first in vivo-active ABHD6 inhibitors that were both potent (IC50 ~ 0.2 nM in live cells) and selective (negligible activity against >50 serine hydrolases) [104]. KT182 and KT203 serve as central and peripherally-restricted ABHD6 inhibitors, respectively, in treated mice (>90% inhibition at 1 mg kg−1). KT185 was discovered as an orally-bioavailable ABHD6 inhibitor (complete blockade of brain ABHD6 at 40 mg kg−1) that displayed excellent selectivity against other brain and liver serine hydrolases in vivo [104].
3.4.4. Therapeutic potential of ABHD6
3.4.4.1. Epilepsy
Pharmacological inhibition of ABHD6 (WWL123) decreased pentylenetetrazole (PTZ)-induced generalized tonic-clonic and myoclonic seizure incidence and severity. Effects were observed in cannabinoid receptor knockout mice (CB1 or CB2) but blocked by picrotoxin, a GABAa receptor antagonist, which suggests a GABAA-mediated mechanism [105]. Use of the tailored activity-based probe HT-01, which targets DAGLs and ABHD6 [55, 65, 104], enabled assessment of target engagement and validation of WWL123 selectivity in animal studies. One possible explanation for the observed pharmacology is that ABHD6 localizes to postsynaptic neurons, which places this serine hydrolase in an ideal location to regulate GABAA receptors through catalytic [105] and non-catalytic activity [106]. Chronic blockade of ABHD6 (daily WWL123 treatments for 5 weeks) did not produce tolerance as well as signs of motor or cognitive impairments. Collectively, these results suggest that ABHD6 inhibitors might show favorable safety profiles and be amenable to long-term use for the treatment of seizures.
3.4.4.2. Diabetes
ABHD6 is a key lipase in generating lipid signals that regulate glucose-stimulated insulin secretion (GSIS) of pancreatic β islet cells [107]. In β cells, glucose stimulation results in production of lipolysis-derived long-chain saturated 1-MAGs (C16:0 and C18:0 MAGs) and these lipid signals are enhanced with the ABHD6 inhibitor WWL70 [108]. Effects observed for 1-MAGs in β cells were not due to cannabinoid receptor signaling because inhibitors of CB1 and CB2 did not affect GSIS. Whole body and β cell-specific ABHD6 knockout mice show enhanced GSIS (islets showed 1-MAG accumulation and enhanced insulin secretion in responses to glucose) and ABHD6 inactivation of diabetic mice restored GSIS and improved glucose tolerance [108]. ABHD6 effects are partly explained by the role of MAGs in binding the C1 domain of Munc13–1 to enhance plasma membrane localization, which is an important step in insulin granule exocytosis [108]. Collectively, these studies support ABHD6 as a negative modulator of insulin secretion and promising target for development of antidiabetic agents through regulation of MAG lipid signals in pancreatic β islet cells.
3.4.4.3. Diet-induced obesity
ABHD6 disruption (ASO knockdown) protects against high-fat-diet-induced obesity, due in large part to a reduction in adipose tissue; effectively lowering body fat mass without altering lean body mass [101]. ABHD6 knockdown protects against metabolic disorders induced by high-fat feeding, such as hyperglycemia, hyperinsulinemia, hypercholesterolemia, and improved both glucose and insulin tolerance. ABHD6 knockdown increased and decreased expression of the serine hydrolases adipose triglyceride lipase (ATGL; TAG lipase; putative role in lipid metabolism at brain barriers [109]) and hormone-sensitive lipase (HSL; DAG lipase; putative role in hypothalamus lipid metabolism [110]), respectively, in the liver; the first two metabolic enzymes in the lipolysis pathway [101]. The authors also observed a downregulation of genes involved in de novo fatty acid synthesis in ABHD6 ASOtreated mouse liver with the in vivo rate of de novo fatty acid synthesis being reduced by ~60% [101]. Importantly these results demonstrate that following ABDH6 knockdown, mice were protected from high-fat-diet-induced hepatic steatosis and associated insulin resistance, canonical of type II diabetes [101].
4. Other serine hydrolases in the mammalian CNS
4.1. cPLA2α
Cytosolic phospholipase A2 (cPLA2α; gene name of PLA2G4A) is a ~85 kDa cytosolic enzyme that is a member of a larger phospholipase A2 superfamily that directly hydrolyze AA from the sn-2 position of membrane phospholipids [111, 112]. Cell activation results in localization of cPLA2α to the membrane and release of free AA, which are further converted to bioactive eicosanoid lipids that includes PGE2 [113]. cPLA2α preferentially hydrolyzes phosphatidylcholine (PC) with AA at the sn-2 position while still showing activity against PE and phosphatidylinositol (PI) [114–116]. The unique substrate specificity of cPLA2α can be partly explained by molecular dynamics (MD) simulations of cPLA2α-PAPC (1-palmitoyl-2-arachidonoyl-PC) interaction, which show a deep channel binding pocket that confers cPLA2α substrate specificity through π-π stacking unlike group VIA Ca2+-independent PLA2 (iPLA2β; gene name of PLA2G6), which exhibits little specificity for sn-2 specific hydrolysis [117].
4.1.1. Functional cross-talk of cPLA2α and 2-AG signaling pathways
Cross-talk between cPLA2α and DAGLβ pathways in macrophage lipid metabolism and signaling were recently revealed in model systems of inflammation. Blockade of either cPLA2α or DAGLβ pathways resulted in partial reductions in cellular AA while dual blockade of cPLA2α/DAGLβ resulted in near-complete depletion of AA in macrophages [55]. These metabolic effects were complemented by similar regulation of cytokine signaling where cPLA2α/DAGLβ-dual blockade provided a synergistic enhancement in TNF-alpha production. These metabolic and cell biological findings support distinct and complementary pathways for supplying AA utilized for COX-mediated production of proinflammatory lipid signals. cPLA2α responds to calcium signaling by nature of its Ca2+ binding C2 domain, which promotes membrane localization and direct release of AA from phospholipids. DAGLβ hydrolyzes AA-esterified DAGs that likely arise from activation of phospholipase C (PLC) which releases DAG and IP3 during signal transduction [118, 119]. Thus, release of AA can occur through cPLA2α hydrolysis of phospholipids or a PLC-DAGL-MGLL sequential metabolic pathway. Given the role of DAGLβ in microglia biology[57], cross-talk with cPLA2α pathways likely exists in the CNS where disruption of 2-AG metabolism (biosynthesis and degradation) can affect neuroinflammation[57, 81].
4.1.2. cPLA2α inhibitors
cPLA2α inhibitors are being pursued as novel anti-inflammatory agents in the clinic [120]. Kinetic studies of trifuoromethyl ketone (AACOCF3, Table 1) showed these compounds are tight- and slow-binding reversible cPLA2α inhibitors [121]. Substitution of AA with a saturated lipid counterpart resulted in lower potency, which supports the importance of substrate recognition of this ω6-polyunsaturated fatty acid (PUFA) [121]. To improve potency, trifluoromethyl ketones that substitute the methylene group β to the carbonyl group of the ketone with a sulphur atom to increase electrophilicity and potency. Further changes include substitution of AA for an ω3-PUFA (docosahexaenoic acid, DHA) to generate structurally related analogs AVX001 and AVX002 (differ by a single cis/trans configuration) that functioned as potent cPLA2α inhibitors (IC50 ~100 nM) [122] (Table 1). AVX001 has been evaluated in the clinic for treatment of atopic dermatitis [120]. Additional trifluoromethyl ketone and AA analogs are being pursued as novel cPLA2α inhibitors [123]. Additional cPLA2α inhibitors include methyl arachidonyl fluorophosphonate (MAFP), a phosphonate analog of AA, have been used as a covalent cPLA2α inhibitor [124]. Pyrrophenone is also a widely used cPLA2α inhibitor [125, 126] that has been applied to studies of reactive oxygen species (ROS) and nitric oxide (NO) signaling [127] as well as inflammasome activation [128] (Table 1). Further selectivity studies are needed to determine the full target profile of pyrrophenone, which was recently shown to exhibit cPLA2α-independent effects [129].
4.2. DDHD1 and DDHD2 (DDHD Domain Containing 1 and 2)
DDHD1, DDHD2, and SEC23IP belong to the iPLA1 (intracellular phospholipase A1) family. This family enzymes possess the serine hydrolase consensus sequence GXSXG and DDHD domain, the latter of which serves as a lipid binding domain that affects their intracellular localization via binding to lipids such as phosphatidylinositol 4-phosphate (PI(4)P). SAM domain, which is only found in DDHD2 and SEC23IP, are also necessary for their lipid binding and localization to Golgi/endoplasmic reticulum (ER)-Golgi intermediate compartment (ERGIC) and ER exit sites, respectively [130].
DDHD1, also called PA-selective phospholipase A1 (PA-PLA1) or iPLA1α, was originally purified and characterized as an enzyme that exhibits PLA1 activity toward PA [131–133], but has been also shown to hydrolyze other phospholipids such as PI and thus may contribute to the production of 2-arachidonoyl lysoPI and the activation of its receptor GPR55 [134]. DDHD1 is ubiquitously expressed throughout human tissues including the brain, with highest levels in the testis [133]. DDHD1 regulates mitochondrial membrane dynamics possibly by modulating the balance between PA and LPA, which are cone- and inverted cone-shaped lipids that favor negative and positive curvature in membranes, respectively [135]. Ddhd1−/− mice show mitochondrial disorganization during spermatogenesis, resulting in sperm malformation and male subinfertility [135]. DDHD1 was identified as the causative gene for a relatively non-complicated form of hereditary spastic paraplegia (HSP), type SPG28 [136–140]. However, a recent study reported that a patient harboring a novel homozygous mutation in DDHD1 presented a complex form of HSP with retinal dystrophy and neurodegeneration with brain iron accumulation (NBIA) [141]. DDHD1 is also implicated in juvenile amyotrophic lateral sclerosis (JALS) [142]. Thus, DDHD1 appear to be involved in a broader spectrum of (patho)physiological processes.
DDHD2, also called KIAA0725p or iPLA1γ, is ubiquitously expressed in human and rodent tissues with relatively higher levels in the brain [143, 144]. Initial biochemical analyses showed that DDHD2 hydrolyzes phospholipids such as PA, PE, PS, and PI [143, 145]. Similar to DDHD1, mutations in DDHD2 cause a complex form of HSP, designated SPG54 [144, 146–148]. This complex HSP subtype presents additional neurological symptoms such as intellectual disability, hypoplastic corpus callosum, and abnormal lipid accumulation [144]. Ddhd2−/− mice also showed cognitive and motor impairments that resemble complex HSP caused by DDHD2 mutations in human [149]. A nontargeted lipidomics analysis revealed that Ddhd2−/− mice exhibited drastic and selective elevations in TAG levels in the CNS, which led to ectopic lipid droplet (LD) accumulation in neuronal cell bodies [149]. Enzyme assays confirmed that DDHD2 is a principal TAG hydrolase in the mouse brain. In addition, DDHD2 may also hydrolyze DAGs [149–151]. Protein expression (TAG hydrolase activity) and the capacity to protect cells from LD accumulation was impaired in DDHD2 containing HSP-related mutations [152]. Taken together, these data suggest that DDHD2 is indispensable for the clearance of pathogenic LDs and the maintenance of lipid homeostasis in the mammalian CNS. ABPP techniques and DDHD2-directed serine hydrolase probe HT-01 [55, 65] led to the identification of a DDHD2-selective in vivo active inhibitor KLH45 [149, 152] that should be useful for further elucidation of DDHD2 biology (Table 1).
SEC23IP (Sec23p-Interacting Protein), also called p125 or iPLA1β, is another member of iPLA1 family that plays an important role in mouse spermiogenesis [153]. SEC23IP interacts with Sec23 and Sec31, components of the coat protein complex II (COPII), and is involved in the transport of vesicles from the ER membrane [154, 155]. Although SEC23IP belongs to iPLA1 family, its function as an enzyme, to the best of our knowledge, remains unknown. Recent exome sequence analyses suggested the potential involvement of SEC23IP in adult attention-deficit/hyperactivity disorder (ADHD) and neurodevelopmental disorders [156, 157]. Therefore, in addition to DDHD1 and DDHD2, SEC23IP may also play an important role in the mammalian CNS.
4.3. PNPLA6 (Neuropathy Target Esterase)
PNPLA6, also called PLA2G6C, iPLA2δ, or neuropathy target esterase (NTE), is a serine hydrolase highly expressed throughout the mammalian brain [158, 159]. PNPLA6 preferentially hydrolyzes lysophospholipids, but also hydrolyzes a variety of other endogenous and exogenous substrates, such as phospholipids, MAGs, acetylcholine, and phenyl valerate, indicating its broad substrate specificity [159–162]. In HeLa cells, knockdown of PNPLA6 impaired the turnover of phospholipids such as PC, PE, and PS [163]. Brain-specific Pnpla6 knockout mice show a variety of neuropathological symptoms including hippocampal and thalamic vacuolation, loss of Purkinje cells, and behavioral deficits [159]. PNPLA6 is the primary target of organophosphorus nerve agents [164, 165]. Exposure to organophosphorus compounds causes organophosphate-induced delayed neuropathy (OPIDN), characterized by axonal degeneration and lower limb paralysis. As mentioned earlier, PNPLA6 may be the culprit off-target of the side effects observed with BIA 10–2474 [34] (Table 1).
PNPLA6 is a causative gene for a broad spectrum of neurologic disorders including HSP (type SPG39), Gordon-Holmes syndrome, Boucher-Neuhäuser syndromes, Laurence-Moon syndrome, Oliver-McFarlane syndrome, and Leber’s congenital amarosis, pure cerebellar ataxia [166–172]. However, how the metabolic pathways regulated by PNPLA6 protect the mammalian CNS integrity remains to be investigated.
5. Conclusions
Genetic and pharmacological studies using knockout mice and selective inhibitors, respectively, combined with enabling ABPP and mass spectrometry lipidomics technologies, have greatly expanded our understanding of the biochemical and (patho)physiological functions of individual lipid-metabolizing serine hydrolases in the mammalian CNS. Advances in genome sequencing have implicated mutations and gene variants of these serine hydrolases in neurologic, psychiatric, and neurodegenerative disorders. However, the molecular mechanisms that link dysregulated lipid metabolism to pathogenesis and progression of CNS disorders due to perturbation of serine hydrolase functions remains poorly understood. In this review, we provide several key examples of using an integrated genetic and chemical approach to elucidate how serine hydrolases regulate lipid biology involved in pathogenesis of CNS disorders (e.g. ABHD12 - PHARC; DDHD2 - HSP). We also summarized recent findings in support of targeting key serine hydrolases in (neuro)inflammation (MGLL and DAGLβ), metabolic disease (DAGLα and ABHD6), and neurodegenerative conditions (MGLL and ABHD6). Given that many of these serine hydrolases remain poorly annotated, further studies are needed to understand the roles of metabolic serine hydrolases in the lipid signaling networks essential for maintaining the function and integrity of the mammalian CNS in normal physiology and disease.
Highlights.
Mammalian CNS express diverse serine hydrolases that act as (phospho)lipases
Serine lipases regulate metabolism of bioactive lipids including endocannabinoids
Genome sequencing have implicated mutations of serine lipases in CNS disorders
The role of serine lipases in CNS physiology and disease remains ill-defined
Chemoproteomics enable inhibitor discovery to probe serine lipase function in vivo
Acknowledgments
The authors thank Ben Cravatt for review of the manuscript. This work was supported by the University of Virginia (start-up funds to K.-L.H.), National Institutes of Health Grants DA035864 and DA043571 to K.-L.H., and GM801868 to T.B.W. This work was supported by JSPS KAKENHI Grant Number JP18K16246 and a grant from the Takeda Science Foundation (to H.-C.L).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Long JZ, Cravatt BF, The metabolic serine hydrolases and their functions in mammalian physiology and disease, Chem Rev, 111 (2011) 6022–6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Bachovchin DA, Cravatt BF, The pharmacological landscape and therapeutic potential of serine hydrolases, Nat Rev Drug Discov, 11 (2012) 52–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Niphakis MJ, Cravatt BF, Enzyme Inhibitor Discovery by Activity-Based Protein Profiling, Annual Review of Biochemistry, 83 (2014) 341–377. [DOI] [PubMed] [Google Scholar]
- [4].Reddy PV, Natarajan V, Schmid PC, Schmid HH, N-Acylation of dog heart ethanolamine phospholipids by transacylase activity, Biochim Biophys. Acta, 750 (1983) 472–480. [DOI] [PubMed] [Google Scholar]
- [5].Natarajan V, Schmid PC, Reddy PV, Zuzarte-Augustin ML, Schmid HH, Biosynthesis of Nacylethanolamine phospholipids by dog brain preparations, J. Neurochem, 41 (1983) 1303–1312. [DOI] [PubMed] [Google Scholar]
- [6].Cadas H, di Tomaso E, Piomelli D, Occurrence and biosynthesis of endogenous cannabinoid precursor, N-arachidonoyl phosphatidylethanolamine, in rat brain, J. Neurosci, 17 (1997) 1226–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Tully SE, Cravatt BF, Activity-based probes that target functional subclasses of phospholipases in proteomes, J. Am. Chem. Soc, 132 (2010) 3264–3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ogura Y, Parsons WH, Kamat SS, Cravatt BF, A calcium-dependent acyltransferase that produces N-acyl phosphatidylethanolamines, Nat. Chem. Biol, 12 (2016) 669–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ohto T, Uozumi N, Hirabayashi T, Shimizu T, Identification of novel cytosolic phospholipase A(2)s, murine cPLA(2){delta}, {epsilon}, and {zeta}, which form a gene cluster with cPLA(2){beta}, J.Biol. Chem, 280 (2005) 24576–24583. [DOI] [PubMed] [Google Scholar]
- [10].Ghomashchi F, Naika GS, Bollinger JG, Aloulou A, Lehr M, Leslie CC, Gelb MH, Interfacial kinetic and binding properties of mammalian group IVB phospholipase A2 (cPLA2beta) and comparison with the other cPLA2 isoforms, J. Biol. Chem, 285 (2010) 36100–36111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ghosh M, Tucker DE, Burchett SA, Leslie CC, Properties of the Group IV phospholipase A2 family, Prog Lipid Res, 45 (2006) 487–510. [DOI] [PubMed] [Google Scholar]
- [12].Hussain Z, Uyama T, Tsuboi K, Ueda N, Mammalian enzymes responsible for the biosynthesis of N-acylethanolamines, Biochim. Biophys. Acta, 1862 (2017) 1546–1561. [DOI] [PubMed] [Google Scholar]
- [13].Morimoto Y, Shimada-Sugimoto M, Otowa T, Yoshida S, Kinoshita A, Mishima H, Yamaguchi N, Mori T, Imamura A, Ozawa H, Kurotaki N, Ziegler C, Domschke K, Deckert J, Umekage T, Tochigi M, Kaiya H, Okazaki Y, Tokunaga K, Sasaki T, Yoshiura KI, Ono S, Wholeexome sequencing and gene-based rare variant association tests suggest that PLA2G4E might be a risk gene for panic disorder, Transl. Psychiatry, 8 (2018) 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Okamoto Y, Morishita J, Tsuboi K, Tonai T, Ueda N, Molecular characterization of a phospholipase D generating anandamide and its congeners, J. Biol. Chem, 279 (2004) 5298–5305. [DOI] [PubMed] [Google Scholar]
- [15].Leung D, Saghatelian A, Simon GM, Cravatt BF, Inactivation of N-acyl phosphatidylethanolamine phospholipase D reveals multiple mechanisms for the biosynthesis of endocannabinoids, Biochemistry, 45 (2006) 4720–4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Tsuboi K, Okamoto Y, Ikematsu N, Inoue M, Shimizu Y, Uyama T, Wang J, Deutsch DG, Burns MP, Ulloa NM, Tokumura A, Ueda N, Enzymatic formation of N-acylethanolamines from N-acylethanolamine plasmalogen through N-acylphosphatidylethanolamine-hydrolyzing phospholipase D-dependent and -independent pathways, Biochim. Biophys. Acta, 1811 (2011) 565–577. [DOI] [PubMed] [Google Scholar]
- [17].Tsuboi K, Okamoto Y, Rahman IA, Uyama T, Inoue T, Tokumura A, Ueda N, Glycerophosphodiesterase GDE4 as a novel lysophospholipase D: a possible involvement in bioactive N-acylethanolamine biosynthesis, Biochim. Biophys. Acta, 1851 (2015) 537–548. [DOI] [PubMed] [Google Scholar]
- [18].Simon GM, Cravatt BF, Endocannabinoid biosynthesis proceeding through glycerophospho-N-acyl ethanolamine and a role for alpha/beta-hydrolase 4 in this pathway, J. Biol. Chem, 281 (2006) 26465–26472. [DOI] [PubMed] [Google Scholar]
- [19].Lee HC, Simon GM, Cravatt BF, ABHD4 regulates multiple classes of N-acyl phospholipids in the mammalian central nervous system, Biochemistry, 54 (2015) 2539–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Brady CA, Jiang D, Mello SS, Johnson TM, Jarvis LA, Kozak MM, Kenzelmann Broz D, Basak S, Park EJ, McLaughlin ME, Karnezis AN, Attardi LD, Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression, Cell, 145 (2011) 571–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Simpson CD, Hurren R, Kasimer D, MacLean N, Eberhard Y, Ketela T, Moffat J, Schimmer AD, A genome wide shRNA screen identifies alpha/beta hydrolase domain containing 4 (ABHD4) as a novel regulator of anoikis resistance, Apoptosis, 17 (2012) 666–678. [DOI] [PubMed] [Google Scholar]
- [22].Cognetta AB 3rd, Niphakis MJ, Lee HC, Martini ML, Hulce JJ, Cravatt BF, Selective NHydroxyhydantoin Carbamate Inhibitors of Mammalian Serine Hydrolases, Chem Biol, 22 (2015) 928–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kaczocha M, Glaser ST, Deutsch DG, Identification of intracellular carriers for the endocannabinoid anandamide, Proc. Natl. Acad. Sci. U.S.A, 106 (2009) 6375–6380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Oddi S, Fezza F, Pasquariello N, D’Agostino A, Catanzaro G, De Simone C, Rapino C, Finazzi-Agro A, Maccarrone M, Molecular identification of albumin and Hsp70 as cytosolic anandamide-binding proteins, Chem. Biol, 16 (2009) 624–632. [DOI] [PubMed] [Google Scholar]
- [25].Niphakis Micah J., Lum Kenneth M., Cognetta Armand B. Iii, Correia Bruno E., Ichu T-A, Olucha J, Brown Steven J., Kundu S, Pisciteli F, Rosen H, Cravatt Benjamin F., A Global Map of Lipid-Binding Proteins and Their Ligandability in Cells, Cell, 161 (2015) 1668–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Cravatt BF, Prospero-Garcia O, Siuzdak G, Gilula NB, Henriksen SJ, Boger DL, Lerner RA, Chemical characterization of a family of brain lipids that induce sleep, Science, 268 (1995) 1506–1509. [DOI] [PubMed] [Google Scholar]
- [27].Saghatelian A, Trauger SA, Want EJ, Hawkins EG, Siuzdak G, Cravatt BF, Assignment of endogenous substrates to enzymes by global metabolite profiling, Biochemistry, 43 (2004) 14332–14339. [DOI] [PubMed] [Google Scholar]
- [28].Wei BQ, Mikkelsen TS, McKinney MK, Lander ES, Cravatt BF, A second fatty acid amide hydrolase with variable distribution among placental mammals, J. Biol. Chem, 281 (2006) 36569–36578. [DOI] [PubMed] [Google Scholar]
- [29].Tsuboi K, Sun YX, Okamoto Y, Araki N, Tonai T, Ueda N, Molecular characterization of N-acylethanolamine-hydrolyzing acid amidase, a novel member of the choloylglycine hydrolase family with structural and functional similarity to acid ceramidase, J. Biol. Chem, 280 (2005) 11082–11092. [DOI] [PubMed] [Google Scholar]
- [30].Petrosino S, Di Marzo V, The pharmacology of palmitoylethanolamide and first data on the therapeutic efficacy of some of its new formulations, Br J Pharmacol, 174 (2017) 1349–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Schlosburg JE, Kinsey SG, Lichtman AH, Targeting fatty acid amide hydrolase (FAAH) to treat pain and inflammation, AAPS J, 11 (2009) 39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Schaffer K, Yassen A, Reeh P, Passier P, A Randomized, Double-Blind, Placebo- and Active Comparator-Controlled Phase I Study of Analgesic/Antihyperalgesic Properties of ASP8477, a Fatty Acid Amide Hydrolase Inhibitor, in Healthy Female Subjects, Pain Med, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Mallet C, Dubray C, Duale C, FAAH inhibitors in the limelight, but regrettably, Int. J. Clin. Pharmacol. Ther, 54 (2016) 498–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].van Esbroeck ACM, Janssen APA, Cognetta AB 3rd, Ogasawara D, Shpak G, van der Kroeg M, Kantae V, Baggelaar MP, de Vrij FMS, Deng H, Allara M, Fezza F, Lin Z, van der Wel T, Soethoudt M, Mock ED, den Dulk H, Baak IL, Florea BI, Hendriks G, De Petrocellis L, Overkleeft HS, Hankemeier T, De Zeeuw CI, Di Marzo V, Maccarrone M, Cravatt BF, Kushner SA, van der Stelt M, Activity-based protein profiling reveals off-target proteins of the FAAH inhibitor BIA 10–2474, Science, 356 (2017) 1084–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Bachovchin DA, Ji T, Li W, Simon GM, Blankman JL, Adibekian A, Hoover H, Niessen S, Cravatt BF, Superfamily-wide portrait of serine hydrolase inhibition achieved by library-versus-library screening, Proc Natl Acad Sci U S A, 107 (2010) 20941–20946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Whibley AC, Plagnol V, Tarpey PS, Abidi F, Fullston T, Choma MK, Boucher CA, Shepherd L, Willatt L, Parkin G, Smith R, Futreal PA, Shaw M, Boyle J, Licata A, Skinner C, Stevenson RE, Turner G, Field M, Hackett A, Schwartz CE, Gecz J, Stratton MR, Raymond FL, Fine-scale survey of X chromosome copy number variants and indels underlying intellectual disability, Am. J. Hum. Genet, 87 (2010) 173–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lim ET, Raychaudhuri S, Sanders SJ, Stevens C, Sabo A, MacArthur DG, Neale BM, Kirby A, Ruderfer DM, Fromer M, Lek M, Liu L, Flannick J, Ripke S, Nagaswamy U, Muzny D, Reid JG, Hawes A, Newsham I, Wu Y, Lewis L, Dinh H, Gross S, Wang LS, Lin CF, Valladares O, Gabriel SB, dePristo M, Altshuler DM, Purcell SM, Project NES, State MW, Boerwinkle E, Buxbaum JD, Cook EH, Gibbs RA, Schellenberg GD, Sutcliffe JS, Devlin B, Roeder K, Daly MJ, Rare complete knockouts in humans: population distribution and significant role in autism spectrum disorders, Neuron, 77 (2013) 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Sirrs S, van Karnebeek CD, Peng X, Shyr C, Tarailo-Graovac M, Mandal R, Testa D, Dubin D, Carbonetti G, Glynn SE, Sayson B, Robinson WP, Han B, Wishart D, Ross CJ, Wasserman WW, Hurwitz TA, Sinclair G, Kaczocha M, Defects in fatty acid amide hydrolase 2 in a male with neurologic and psychiatric symptoms, Orphanet. J. Rare. Dis, 10 (2015) 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lucanic M, Held JM, Vantipalli MC, Klang IM, Graham JB, Gibson BW, Lithgow GJ, Gill MS, N-acylethanolamine signalling mediates the effect of diet on lifespan in Caenorhabditis elegans, Nature, 473 (2011) 226–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Pastuhov SI, Fujiki K, Nix P, Kanao S, Bastiani M, Matsumoto K, Hisamoto N, Endocannabinoid-Goalpha signalling inhibits axon regeneration in Caenorhabditis elegans by antagonizing Gqalpha-PKC-JNK signalling, Nat. Commun, 3 (2012) 1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Pastuhov SI, Matsumoto K, Hisamoto N, Endocannabinoid signaling regulates regenerative axon navigation in Caenorhabditis elegans via the GPCRs NPR-19 and NPR-32, Genes Cells, 21 (2016) 696–705. [DOI] [PubMed] [Google Scholar]
- [42].Klee EW, Schneider H, Clark KJ, Cousin MA, Ebbert JO, Hooten WM, Karpyak VM, Warner DO, Ekker SC, Zebrafish: a model for the study of addiction genetics, Hum. Genet, 131 (2012) 977–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Krug, RG 2nd, Clark KJ, Elucidating cannabinoid biology in zebrafish (Danio rerio), Gene, 570 (2015) 168–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].McPartland JM, Glass M, Matias I, Norris RW, Kilpatrick CW, A shifted repertoire of endocannabinoid genes in the zebrafish (Danio rerio), Mol. Genet. Genomics, 277 (2007) 555–570. [DOI] [PubMed] [Google Scholar]
- [45].Krug, RG 2nd, Lee HB, El Khoury LY, Sigafoos AN, Petersen MO, Clark KJ, The endocannabinoid gene faah2a modulates stress-associated behavior in zebrafish, PLoS One, 13 (2018) e0190897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Dincheva I, Drysdale AT, Hartley CA, Johnson DC, Jing D, King EC, Ra S, Gray JM, Yang R, DeGruccio AM, Huang C, Cravatt BF, Glatt CE, Hill MN, Casey BJ, Lee FS, FAAH genetic variation enhances fronto-amygdala function in mouse and human, Nat. Commun, 6 (2015) 6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Moreira FA, Kaiser N, Monory K, Lutz B, Reduced anxiety-like behaviour induced by genetic and pharmacological inhibition of the endocannabinoid-degrading enzyme fatty acid amide hydrolase (FAAH) is mediated by CB1 receptors, Neuropharmacology, 54 (2008) 141–150. [DOI] [PubMed] [Google Scholar]
- [48].Bisogno T, Howell F, Williams G, Minassi A, Cascio MG, Ligresti A, Matias I, Schiano-Moriello A, Paul P, Williams EJ, Gangadharan U, Hobbs C, Di Marzo V, Doherty P, Cloning of the first sn1-DAG lipases points to the spatial and temporal regulation of endocannabinoid signaling in the brain, J Cell Biol, 163 (2003) 463–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Reisenberg M, Singh PK, Williams G, Doherty P, The diacylglycerol lipases: structure, regulation and roles in and beyond endocannabinoid signalling, Philosophical Transactions of the Royal Society B: Biological Sciences, 367 (2012) 3264–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Shonesy BC, Wang X, Rose KL, Ramikie TS, Cavener VS, Rentz T, Baucum, AJ 2nd, Jalan-Sakrikar N, Mackie K, Winder DG, Patel S, Colbran RJ, CaMKII regulates diacylglycerol lipase-alpha and striatal endocannabinoid signaling, Nat Neurosci, 16 (2013) 456–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Martin BR, Cravatt BF, Large-scale profiling of protein palmitoylation in mammalian cells, Nat Methods, 6 (2009) 135–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Yang W, Di Vizio D, Kirchner M, Steen H, Freeman MR, Proteome scale characterization of human S-acylated proteins in lipid raft-enriched and non-raft membranes, Mol Cell Proteomics, 9 (2010) 54–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Gao Y, Vasilyev DV, Goncalves MB, Howell FV, Hobbs C, Reisenberg M, Shen R, Zhang MY, Strassle BW, Lu P, Mark L, Piesla MJ, Deng K, Kouranova EV, Ring RH, Whiteside GT, Bates B, Walsh FS, Williams G, Pangalos MN, Samad TA, Doherty P, Loss of Retrograde Endocannabinoid Signaling and Reduced Adult Neurogenesis in Diacylglycerol Lipase Knock-out Mice, J Neurosci, 30 (2010) 2017–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Tanimura A, Yamazaki M, Hashimotodani Y, Uchigashima M, Kawata S, Abe M, Kita Y, Hashimoto K, Shimizu T, Watanabe M, Sakimura K, Kano M, The endocannabinoid 2-arachidonoylglycerol produced by diacylglycerol lipase alpha mediates retrograde suppression of synaptic transmission, Neuron, 65 (2010) 320–327. [DOI] [PubMed] [Google Scholar]
- [55].Hsu KL, Tsuboi K, Adibekian A, Pugh H, Masuda K, Cravatt BF, DAGLbeta inhibition perturbs a lipid network involved in macrophage inflammatory responses, Nat Chem Biol, 8 (2012) 999–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Ogasawara D, Deng H, Viader A, Baggelaar MP, Breman A, den Dulk H, van den Nieuwendijk AM, Soethoudt M, van der Wel T, Zhou J, Overkleeft HS, Sanchez-Alavez M, Mori S, Nguyen W, Conti B, Liu X, Chen Y, Liu QS, Cravatt BF, van der Stelt M, Rapid and profound rewiring of brain lipid signaling networks by acute diacylglycerol lipase inhibition, Proc Natl Acad Sci U S A, 113 (2016) 26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Viader A, Ogasawara D, Joslyn CM, Sanchez-Alavez M, Mori S, Nguyen W, Conti B, Cravatt BF, A chemical proteomic atlas of brain serine hydrolases identifies cell type-specific pathways regulating neuroinflammation, eLife, 5 (2016) e12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Powell DR, Gay JP, Wilganowski N, Doree D, Savelieva KV, Lanthorn TH, Read R, Vogel P, Hansen GM, Brommage R, Ding ZM, Desai U, Zambrowicz B, Diacylglycerol Lipase alpha Knockout Mice Demonstrate Metabolic and Behavioral Phenotypes Similar to Those of Cannabinoid Receptor 1 Knockout Mice, Front Endocrinol (Lausanne), 6 (2015) 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Jenniches I, Ternes S, Albayram O, Otte DM, Bach K, Bindila L, Michel K, Lutz B, Bilkei-Gorzo A, Zimmer A, Anxiety, Stress, and Fear Response in Mice with Reduced Endocannabinoid Levels, Biol Psychiatry, (2015). [DOI] [PubMed] [Google Scholar]
- [60].Shonesy BC, Bluett RJ, Ramikie TS, Baldi R, Hermanson DJ, Kingsley PJ, Marnett LJ, Winder DG, Colbran RJ, Patel S, Genetic disruption of 2-arachidonoylglycerol synthesis reveals a key role for endocannabinoid signaling in anxiety modulation, Cell Rep, 9 (2014) 1644–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Zhou Y, Howell FV, Glebov OO, Albrecht D, Williams G, Doherty P, Regulated endosomal trafficking of Diacylglycerol lipase alpha (DAGLalpha) generates distinct cellular pools; implications for endocannabinoid signaling, Mol Cell Neurosci, 76 (2016) 76–86. [DOI] [PubMed] [Google Scholar]
- [62].Jung K-M, Astarita G, Zhu C, Wallace M, Mackie K, Piomelli D, A Key Role for Diacylglycerol Lipase-alpha in Metabotropic Glutamate Receptor-Dependent Endocannabinoid Mobilization, Mol Pharmacol, 72 (2007) 612–621. [DOI] [PubMed] [Google Scholar]
- [63].Powell DR, Gay JP, Wilganowski N, Doree D, Savelieva KV, Lanthorn TH, Read R, Vogel P, Hansen GM, Brommage R, Ding ZM, Desai U, Zambrowicz B, Diacylglycerol Lipase alpha Knockout Mice Demonstrate Metabolic and Behavioral Phenotypes Similar to Those of Cannabinoid Receptor 1 Knockout Mice, Front Endocrinol (Lausanne), 6 (2015) 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Leslie CC, Cytosolic phospholipase A(2): physiological function and role in disease, J Lipid Res, 56 (2015) 1386–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Hsu KL, Tsuboi K, Whitby LR, Speers AE, Pugh H, Inloes J, Cravatt BF, Development and optimization of piperidyl-1,2,3-triazole ureas as selective chemical probes of endocannabinoid biosynthesis, J Med Chem, 56 (2013) 8257–8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Wilkerson JL, Ghosh S, Bagdas D, Mason BL, Crowe MS, Hsu KL, Wise LE, Kinsey SG, Damaj MI, Cravatt BF, Lichtman AH, Diacylglycerol lipase beta inhibition reverses nociceptive behaviour in mouse models of inflammatory and neuropathic pain, Br J Pharmacol, 173 (2016) 1678–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Yun B, Lee H, Ghosh M, Cravatt BF, Hsu K-L, Bonventre JV, Ewing H, Gelb MH, Leslie CC, Serine Hydrolase Inhibitors Block Necrotic Cell Death by Preventing Calcium Overload of the Mitochondria and Permeability Transition Pore Formation, Journal of Biological Chemistry, 289 (2014) 1491–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Shin M, Snyder HW, Donvito G, Schurman LD, Fox TE, Lichtman AH, Kester M, Hsu KL, Liposomal Delivery of Diacylglycerol Lipase-Beta Inhibitors to Macrophages Dramatically Enhances Selectivity and Efficacy in Vivo, Mol Pharm, 15 (2018) 721–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Deng H, Kooijman S, van den Nieuwendijk AM, Ogasawara D, van der Wel T, van Dalen F, Baggelaar MP, Janssen FJ, van den Berg RJ, den Dulk H, Cravatt BF, Overkleeft HS, Rensen PC, van der Stelt M, Triazole Ureas Act as Diacylglycerol Lipase Inhibitors and Prevent Fasting-Induced Refeeding, J Med Chem, 60 (2017) 428–440. [DOI] [PubMed] [Google Scholar]
- [70].Baggelaar MP, Janssen FJ, van Esbroeck AC, den Dulk H, Allara M, Hoogendoorn S, McGuire R, Florea BI, Meeuwenoord N, van den Elst H, van der Marel GA, Brouwer J, Di Marzo V, Overkleeft HS, van der Stelt M, Development of an Activity-Based Probe and In Silico Design Reveal Highly Selective Inhibitors for Diacylglycerol Lipase-alpha in Brain, Angew Chem Int Ed Engl, (2013). [DOI] [PubMed] [Google Scholar]
- [71].Schalk-Hihi C, Schubert C, Alexander R, Bayoumy S, Clemente JC, Deckman I, DesJarlais RL, Dzordzorme KC, Flores CM, Grasberger B, Kranz JK, Lewandowski F, Liu L, Ma H, Maguire D, Macielag MJ, McDonnell ME, Mezzasalma Haarlander T, Miller R, Milligan C, Reynolds C, Kuo LC, Crystal structure of a soluble form of human monoglyceride lipase in complex with an inhibitor at 1.35 A resolution, Protein Sci, 20 (2011) 670–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Karlsson M, Contreras JA, Hellman U, Tornqvist H, Holm C, cDNA cloning, tissue distribution, and identification of the catalytic triad of monoglyceride lipase. Evolutionary relationship to esterases, lysophospholipases, and haloperoxidases, J Biol Chem, 272 (1997) 27218–27223. [DOI] [PubMed] [Google Scholar]
- [73].Kupiecki FP, Partial purification of monoglyceride lipase from adipose tissue, J Lipid Res, 7 (1966) 230–235. [PubMed] [Google Scholar]
- [74].Tornqvist H, Belfrage P, Purification and Some Properties of a Monoacylglycerol-Hydrolyzing Enzyme of Rat Adipose-Tissue, Journal of Biological Chemistry, 251 (1976) 813–819. [PubMed] [Google Scholar]
- [75].Vandevoorde S, Saha B, Mahadevan A, Razdan RK, Pertwee RG, Martin BR, Fowler CJ, Influence of the degree of unsaturation of the acyl side chain upon the interaction of analogues of 1-arachidonoylglycerol with monoacylglycerol lipase and fatty acid amide hydrolase, Biochem Biophys Res Commun, 337 (2005) 104–109. [DOI] [PubMed] [Google Scholar]
- [76].Vaughan M, Berger JE, Steinberg D, Hormone-Sensitive Lipase and Monoglyceride Lipase Activities in Adipose Tissue, J Biol Chem, 239 (1964) 401–409. [PubMed] [Google Scholar]
- [77].Blankman JL, Simon GM, Cravatt BF, A Comprehensive Profile of Brain Enzymes that Hydrolyze the Endocannabinoid 2-Arachidonoylglycerol, Chemistry & Biology, 14 (2007) 1347–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Long JZ, Li W, Booker L, Burston JJ, Kinsey SG, Schlosburg JE, Pavon FJ, Serrano AM, Selley DE, Parsons LH, Lichtman AH, Cravatt BF, Selective blockade of 2-arachidonoylglycerol hydrolysis produces cannabinoid behavioral effects, Nat Chem Biol, 5 (2009) 37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Schlosburg JE, Blankman JL, Long JZ, Nomura DK, Pan B, Kinsey SG, Nguyen PT, Ramesh D, Booker L, Burston JJ, Thomas EA, Selley DE, Sim-Selley LJ, Liu QS, Lichtman AH, Cravatt BF, Chronic monoacylglycerol lipase blockade causes functional antagonism of the endocannabinoid system, Nat Neurosci, 13 (2010) 1113–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Chanda PK, Gao Y, Mark L, Btesh J, Strassle BW, Lu P, Piesla MJ, Zhang M-Y, Bingham B, Uveges A, Kowal D, Garbe D, Kouranova EV, Ring RH, Bates B, Pangalos MN, Kennedy JD, Whiteside GT, Samad TA, Monoacylglycerol Lipase Activity Is a Critical Modulator of the Tone and Integrity of the Endocannabinoid System, Molecular Pharmacology, 78 (2010) 996–1003. [DOI] [PubMed] [Google Scholar]
- [81].Nomura DK, Morrison BE, Blankman JL, Long JZ, Kinsey SG, Marcondes MC, Ward AM, Hahn YK, Lichtman AH, Conti B, Cravatt BF, Endocannabinoid hydrolysis generates brain prostaglandins that promote neuroinflammation, Science, 334 (2011) 809–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Chang JW, Niphakis MJ, Lum KM, Cognetta AB 3rd, Wang C, Matthews ML, Niessen S, Buczynski MW, Parsons LH, Cravatt BF, Highly selective inhibitors of monoacylglycerol lipase bearing a reactive group that is bioisosteric with endocannabinoid substrates, Chem Biol, 19 (2012) 579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].McAllister LA, Butler CR, Mente S, O’Neil SV, Fonseca KR, Piro JR, Cianfrogna JA, Foley TL, Gilbert AM, Harris AR, Helal CJ, Johnson DS, Montgomery JI, Nason DM, Noell S, Pandit J, Rogers BN, Samad TA, Shaffer CL, da Silva RG, Uccello DP, Webb D, Brodney MA, Discovery of Trifluoromethyl Glycol Carbamates as Potent and Selective Covalent Monoacylglycerol Lipase (MAGL) Inhibitors for Treatment of Neuroinflammation, J Med Chem, 61 (2018) 3008–3026. [DOI] [PubMed] [Google Scholar]
- [84].Granchi C, Rizzolio F, Palazzolo S, Carmignani S, Macchia M, Saccomanni G, Manera C, Martinelli A, Minutolo F, Tuccinardi T, Structural Optimization of 4-Chlorobenzoylpiperidine Derivatives for the Development of Potent, Reversible, and Selective Monoacylglycerol Lipase (MAGL) Inhibitors, J Med Chem, 59 (2016) 10299–10314. [DOI] [PubMed] [Google Scholar]
- [85].Ghosh S, Wise LE, Chen Y, Gujjar R, Mahadevan A, Cravatt BF, Lichtman AH, The monoacylglycerol lipase inhibitor JZL184 suppresses inflammatory pain in the mouse carrageenan model, Life sciences, 92 (2013) 498–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Ignatowska-Jankowska BM, Ghosh S, Crowe MS, Kinsey SG, Niphakis MJ, Abdullah RA, Tao Q, ST ON, Walentiny DM, Wiley JL, Cravatt BF, Lichtman AH, In vivo characterization of the highly selective monoacylglycerol lipase inhibitor KML29: antinociceptive activity without cannabimimetic side effects, Br J Pharmacol, 171 (2014) 1392–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Mackie K, Cannabinoid receptors as therapeutic targets, Annu Rev Pharmacol Toxicol, 46 (2006) 101–122. [DOI] [PubMed] [Google Scholar]
- [88].Robberecht W, Philips T, The changing scene of amyotrophic lateral sclerosis, Nat Rev Neurosci, 14 (2013) 248–264. [DOI] [PubMed] [Google Scholar]
- [89].Turner BJ, Talbot K, Transgenics, toxicity and therapeutics in rodent models of mutant SOD1-mediated familial ALS, Prog Neurobiol, 85 (2008) 94–134. [DOI] [PubMed] [Google Scholar]
- [90].Bilsland LG, Dick JR, Pryce G, Petrosino S, Di Marzo V, Baker D, Greensmith L, Increasing cannabinoid levels by pharmacological and genetic manipulation delay disease progression in SOD1 mice, FASEB J, 20 (2006) 1003–1005. [DOI] [PubMed] [Google Scholar]
- [91].Witting A, Weydt P, Hong S, Kliot M, Moller T, Stella N, Endocannabinoids accumulate in spinal cord of SOD1 G93A transgenic mice, J Neurochem, 89 (2004) 1555–1557. [DOI] [PubMed] [Google Scholar]
- [92].Pasquarelli N, Engelskirchen M, Hanselmann J, Endres S, Porazik C, Bayer H, Buck E, Karsak M, Weydt P, Ferger B, Witting A, Evaluation of monoacylglycerol lipase as a therapeutic target in a transgenic mouse model of ALS, Neuropharmacology, 124 (2017) 157–169. [DOI] [PubMed] [Google Scholar]
- [93].Blankman JL, Long JZ, Trauger SA, Siuzdak G, Cravatt BF, ABHD12 controls brain lysophosphatidylserine pathways that are deregulated in a murine model of the neurodegenerative disease PHARC, Proc Natl Acad Sci U S A, 110 (2013) 1500–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Fiskerstrand T, H’Mida-Ben Brahim D, Johansson S, M’Zahem A, Haukanes BI, Drouot N, Zimmermann J, Cole AJ, Vedeler C, Bredrup C, Assoum M, Tazir M, Klockgether T, Hamri A, Steen VM, Boman H, Bindoff LA, Koenig M, Knappskog PM, Mutations in ABHD12 cause the neurodegenerative disease PHARC: An inborn error of endocannabinoid metabolism, Am J Hum Genet, 87 (2010) 410–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Eisenberger T, Slim R, Mansour A, Nauck M, Nurnberg G, Nurnberg P, Decker C, Dafinger C, Ebermann I, Bergmann C, Bolz HJ, Targeted next-generation sequencing identifies a homozygous nonsense mutation in ABHD12, the gene underlying PHARC, in a family clinically diagnosed with Usher syndrome type 3, Orphanet J Rare Dis, 7 (2012) 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].van der Kleij D, Latz E, Brouwers JF, Kruize YC, Schmitz M, Kurt-Jones EA, Espevik T, de Jong EC, Kapsenberg ML, Golenbock DT, Tielens AG, Yazdanbakhsh M, A novel host-parasite lipid cross-talk. Schistosomal lyso-phosphatidylserine activates toll-like receptor 2 and affects immune polarization, J Biol Chem, 277 (2002) 48122–48129. [DOI] [PubMed] [Google Scholar]
- [97].Kamat SS, Camara K, Parsons WH, Chen DH, Dix MM, Bird TD, Howell AR, Cravatt BF, Immunomodulatory lysophosphatidylserines are regulated by ABHD16A and ABHD12 interplay, Nat Chem Biol, 11 (2015) 164–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Parkkari T, Haavikko R, Laitinen T, Navia-Paldanius D, Rytilahti R, Vaara M, Lehtonen M, Alakurtti S, Yli-Kauhaluoma J, Nevalainen T, Savinainen JR, Laitinen JT, Discovery of triterpenoids as reversible inhibitors of alpha/beta-hydrolase domain containing 12 (ABHD12), PLoS One, 9 (2014) e98286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Navia-Paldanius D, Savinainen JR, Laitinen JT, Biochemical and pharmacological characterization of human alpha/beta-hydrolase domain containing 6 (ABHD6) and 12 (ABHD12), J Lipid Res, 53 (2012) 2413–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Marrs WR, Blankman JL, Horne EA, Thomazeau A, Lin YH, Coy J, Bodor AL, Muccioli GG, Hu SS, Woodruff G, Fung S, Lafourcade M, Alexander JP, Long JZ, Li W, Xu C, Moller T, Mackie K, Manzoni OJ, Cravatt BF, Stella N, The serine hydrolase ABHD6 controls the accumulation and efficacy of 2-AG at cannabinoid receptors, Nat Neurosci, 13 (2010) 951–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Thomas G, Betters JL, Lord CC, Brown AL, Marshall S, Ferguson D, Sawyer J, Davis MA, Melchior JT, Blume LC, Howlett AC, Ivanova PT, Milne SB, Myers DS, Mrak I, Leber V, Heier C, Taschler U, Blankman JL, Cravatt BF, Lee RG, Crooke RM, Graham MJ, Zimmermann R, Brown HA, Brown JM, The serine hydrolase ABHD6 Is a critical regulator of the metabolic syndrome, Cell Rep, 5 (2013) 508–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Pribasnig MA, Mrak I, Grabner GF, Taschler U, Knittelfelder O, Scherz B, Eichmann TO, Heier C, Grumet L, Kowaliuk J, Romauch M, Holler S, Anderl F, Wolinski H, Lass A, Breinbauer R, Marsche G, Brown JM, Zimmermann R, alpha/beta Hydrolase Domain-containing 6 (ABHD6) Degrades the Late Endosomal/Lysosomal Lipid Bis(monoacylglycero)phosphate, J Biol Chem, 290 (2015) 29869–29881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Li W, Blankman JL, Cravatt BF, A functional proteomic strategy to discover inhibitors for uncharacterized hydrolases, J Am Chem Soc, 129 (2007) 9594–9595. [DOI] [PubMed] [Google Scholar]
- [104].Hsu KL, Tsuboi K, Chang JW, Whitby LR, Speers AE, Pugh H, Cravatt BF, Discovery and optimization of piperidyl-1,2,3-triazole ureas as potent, selective, and in vivo-active inhibitors of alpha/beta-hydrolase domain containing 6 (ABHD6), J Med Chem, 56 (2013) 8270–8279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Naydenov AV, Horne EA, Cheah CS, Swinney K, Hsu KL, Cao JK, Marrs WR, Blankman JL, Tu S, Cherry AE, Fung S, Wen A, Li W, Saporito MS, Selley DE, Cravatt BF, Oakley JC, Stella N, ABHD6 blockade exerts antiepileptic activity in PTZ-induced seizures and in spontaneous seizures in R6/2 mice, Neuron, 83 (2014) 361–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Wei M, Zhang J, Jia M, Yang C, Pan Y, Li S, Luo Y, Zheng J, Ji J, Chen J, Hu X, Xiong J, Shi Y, Zhang C, alpha/beta-Hydrolase domain-containing 6 (ABHD6) negatively regulates the surface delivery and synaptic function of AMPA receptors, Proc Natl Acad Sci U S A, 113 (2016) E2695–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].MacDonald PE, Signal integration at the level of ion channel and exocytotic function in pancreatic beta-cells, Am J Physiol Endocrinol Metab, 301 (2011) E1065–1069. [DOI] [PubMed] [Google Scholar]
- [108].Zhao S, Mugabo Y, Iglesias J, Xie L, Delghingaro-Augusto V, Lussier R, Peyot ML, Joly E, Taib B, Davis MA, Brown JM, Abousalham A, Gaisano H, Madiraju SR, Prentki M, alpha/beta-Hydrolase domain-6-accessible monoacylglycerol controls glucose-stimulated insulin secretion, Cell metabolism, 19 (2014) 993–1007. [DOI] [PubMed] [Google Scholar]
- [109].Etschmaier K, Becker T, Eichmann TO, Schweinzer C, Scholler M, Tam-Amersdorfer C, Poeckl M, Schuligoi R, Kober A, Chirackal Manavalan AP, Rechberger GN, Streith IE, Zechner R, Zimmermann R, Panzenboeck U, Adipose triglyceride lipase affects triacylglycerol metabolism at brain barriers, J Neurochem, 119 (2011) 1016–1028. [DOI] [PubMed] [Google Scholar]
- [110].Sekiya M, Osuga J, Okazaki H, Yahagi N, Harada K, Shen WJ, Tamura Y, Tomita S, Iizuka Y, Ohashi K, Okazaki M, Sata M, Nagai R, Fujita T, Shimano H, Kraemer FB, Yamada N, Ishibashi S, Absence of hormone-sensitive lipase inhibits obesity and adipogenesis in Lep ob/ob mice, J Biol Chem, 279 (2004) 15084–15090. [DOI] [PubMed] [Google Scholar]
- [111].Balboa M.a.A., Varela-Nieto I, Killermann Lucas K, Dennis EA, Expression and function of phospholipase A2in brain, FEBS Letters, 531 (2002) 12–17. [DOI] [PubMed] [Google Scholar]
- [112].Balsinde J, Winstead MV, Dennis EA, Phospholipase A2regulation of arachidonic acid mobilization, FEBS Letters, 531 (2002) 2–6. [DOI] [PubMed] [Google Scholar]
- [113].Balsinde J, Balboa MA, Insel PA, Dennis EA, Regulation and inhibition of phospholipase A2, Annu Rev Pharmacol Toxicol, 39 (1999) 175–189. [DOI] [PubMed] [Google Scholar]
- [114].Diez E, Louis-Flamberg P, Hall RH, Mayer RJ, Substrate specificities and properties of human phospholipases A2 in a mixed vesicle model, J Biol Chem, 267 (1992) 18342–18348. [PubMed] [Google Scholar]
- [115].Li-Stiles B, Lo H-H, Fischer SM, Identification and characterization of several forms of phospholipase A2 in mouse epidermal keratinocytes, Journal of Lipid Research, 39 (1998) 569–582. [PubMed] [Google Scholar]
- [116].Burke JE, Hsu YH, Deems RA, Li S, Woods VL Jr., Dennis EA, A phospholipid substrate molecule residing in the membrane surface mediates opening of the lid region in group IVA cytosolic phospholipase A2, J Biol Chem, 283 (2008) 31227–31236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Mouchlis VD, Bucher D, McCammon JA, Dennis EA, Membranes serve as allosteric activators of phospholipase A2, enabling it to extract, bind, and hydrolyze phospholipid substrates, Proceedings of the National Academy of Sciences, 112 (2015) E516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Tang X, Edwards EM, Holmes BB, Falck JR, Campbell WB, Role of phospholipase C and diacylglyceride lipase pathway in arachidonic acid release and acetylcholine-induced vascular relaxation in rabbit aorta, Am J Physiol Heart Circ Physiol, 290 (2006) H37–45. [DOI] [PubMed] [Google Scholar]
- [119].Hokin LE, Effects of calcium omission on acetylcholine-stimulated amylase secretion and phospholipid synthesis in pigeon pancreas slices, Biochim Biophys Acta, 115 (1966) 219–221. [DOI] [PubMed] [Google Scholar]
- [120].Omland SH, Habicht A, Damsbo P, Wilms J, Johansen B, Gniadecki R, A randomized, double-blind, placebo-controlled, dose-escalation first-in-man study (phase 0) to assess the safety and efficacy of topical cytosolic phospholipase A2 inhibitor, AVX001, in patients with mild to moderate plaque psoriasis, J Eur Acad Dermatol Venereol, 31 (2017) 1161–1167. [DOI] [PubMed] [Google Scholar]
- [121].Street IP, Lin HK, Laliberte F, Ghomashchi F, Wang Z, Perrier H, Tremblay NM, Huang Z, Weech PK, Gelb MH, Slow- and tight-binding inhibitors of the 85-kDa human phospholipase A2, Biochemistry, 32 (1993) 5935–5940. [DOI] [PubMed] [Google Scholar]
- [122].Huwiler A, Feuerherm AJ, Sakem B, Pastukhov O, Filipenko I, Nguyen T, Johansen B, The omega3-polyunsaturated fatty acid derivatives AVX001 and AVX002 directly inhibit cytosolic phospholipase A(2) and suppress PGE(2) formation in mesangial cells, Br J Pharmacol, 167 (2012) 1691–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Ng CY, Kannan S, Chen YJ, Tan FCK, Ong WY, Go ML, Verma CS, Low CM, Lam Y, A New Generation of Arachidonic Acid Analogues as Potential Neurological Agent Targeting Cytosolic Phospholipase A2, Scientific reports, 7 (2017) 13683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Lio Y-C, Reynolds LJ, Balsinde J, Dennis EA, Irreversible inhibition of Ca2+-independent phospholipase A2 by methyl arachidonyl fuorophosphonate, Biochimica et Biophysica Acta (BBA)-Lipids and Lipid Metabolism, 1302 (1996) 55–60. [DOI] [PubMed] [Google Scholar]
- [125].Seno K, Okuno T, Nishi K, Murakami Y, Watanabe F, Matsuura T, Wada M, Fujii Y, Yamada M, Ogawa T, Okada T, Hashizume H, Kii M, Hara S.-i., Hagishita S, Nakamoto S, Yamada K, Chikazawa Y, Ueno M, Teshirogi I, Ono T, Ohtani M, Pyrrolidine Inhibitors of Human Cytosolic Phospholipase A2, Journal of Medicinal Chemistry, 43 (2000) 1041–1044. [DOI] [PubMed] [Google Scholar]
- [126].Seno K, Okuno T, Nishi K, Murakami Y, Yamada K, Nakamoto S, Ono T, Pyrrolidine inhibitors of human cytosolic phospholipase A2. Part 2, Bioorganic & Medicinal Chemistry Letters, 11 (2001) 587–590. [DOI] [PubMed] [Google Scholar]
- [127].Chuang DY, Simonyi A, Kotzbauer PT, Gu Z, Sun GY, Cytosolic phospholipase A2 plays a crucial role in ROS/NO signaling during microglial activation through the lipoxygenase pathway, J Neuroinflammation, 12 (2015) 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Norris PC, Gosselin D, Reichart D, Glass CK, Dennis EA, Phospholipase A2 regulates eicosanoid class switching during inflammasome activation, Proceedings of the National Academy of Sciences, 111 (2014) 12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Yun B, Lee H, Ewing H, Gelb MH, Leslie CC, Off-target effect of the cPLA2alpha inhibitor pyrrophenone: Inhibition of calcium release from the endoplasmic reticulum, Biochem Biophys Res Commun, 479 (2016) 61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Inoue H, Baba T, Sato S, Ohtsuki R, Takemori A, Watanabe T, Tagaya M, Tani K, Roles of SAM and DDHD domains in mammalian intracellular phospholipase A1 KIAA0725p, Biochim Biophys Acta, 1823 (2012) 930–939. [DOI] [PubMed] [Google Scholar]
- [131].Higgs HN, Glomset JA, Identification of a phosphatidic acid-preferring phospholipase A1 from bovine brain and testis, Proc Natl Acad Sci U S A, 91 (1994) 9574–9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Higgs HN, Glomset JA, Purification and properties of a phosphatidic acid-preferring phospholipase A1 from bovine testis. Examination of the molecular basis of its activation, J. Biol. Chem, 271 (1996) 10874–10883. [DOI] [PubMed] [Google Scholar]
- [133].Higgs HN, Han MH, Johnson GE, Glomset JA, Cloning of a phosphatidic acid-preferring phospholipase A1 from bovine testis, J. Biol. Chem, 273 (1998) 5468–5477. [DOI] [PubMed] [Google Scholar]
- [134].Yamashita A, Kumazawa T, Koga H, Suzuki N, Oka S, Sugiura T, Generation of lysophosphatidylinositol by DDHD domain containing 1 (DDHD1): Possible involvement of phospholipase D/phosphatidic acid in the activation of DDHD1, Biochim. Biophys. Acta, 1801 (2010) 711–720. [DOI] [PubMed] [Google Scholar]
- [135].Baba T, Kashiwagi Y, Arimitsu N, Kogure T, Edo A, Maruyama T, Nakao K, Nakanishi H, Kinoshita M, Frohman MA, Yamamoto A, Tani K, Phosphatidic acid (PA)-preferring phospholipase A1 regulates mitochondrial dynamics, J. Biol. Chem, 289 (2014) 11497–11511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Bouslam N, Benomar A, Azzedine H, Bouhouche A, Namekawa M, Klebe S, Charon C, Durr A, Ruberg M, Brice A, Yahyaoui M, Stevanin G, Mapping of a new form of pure autosomal recessive spastic paraplegia (SPG28), Ann. Neurol, 57 (2005) 567–571. [DOI] [PubMed] [Google Scholar]
- [137].Tesson C, Nawara M, Salih MA, Rossignol R, Zaki MS, Al Balwi M, Schule R, Mignot C, Obre E, Bouhouche A, Santorelli FM, Durand CM, Oteyza AC, El-Hachimi KH, Al Drees A, Bouslam N, Lamari F, Elmalik SA, Kabiraj MM, Seidahmed MZ, Esteves T, Gaussen M, Monin ML, Gyapay G, Lechner D, Gonzalez M, Depienne C, Mochel F, Lavie J, Schols L, Lacombe D, Yahyaoui M, Al Abdulkareem I, Zuchner S, Yamashita A, Benomar A, Goizet C, Durr A, Gleeson JG, Darios F, Brice A, Stevanin G, Alteration of fatty-acid-metabolizing enzymes affects mitochondrial form and function in hereditary spastic paraplegia, American journal of human genetics, 91 (2012) 1051–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Liguori R, Giannoccaro MP, Arnoldi A, Citterio A, Tonon C, Lodi R, Bresolin N, Bassi MT, Impairment of brain and muscle energy metabolism detected by magnetic resonance spectroscopy in hereditary spastic paraparesis type 28 patients with DDHD1 mutations, J. Neurol, 261 (2014) 1789–1793. [DOI] [PubMed] [Google Scholar]
- [139].Miura S, Morikawa T, Fujioka R, Kosaka K, Yamada K, Hattori G, Motomura M, Taniwaki T, Shibata H, A novel frameshift mutation of DDHD1 in a Japanese patient with autosomal recessive spastic paraplegia, Eur. J. Med. Genet, 59 (2016) 413–416. [DOI] [PubMed] [Google Scholar]
- [140].Mignarri A, Rubegni A, Tessa A, Stefanucci S, Malandrini A, Cardaioli E, Meschini MC, Stromillo ML, Doccini S, Federico A, Santorelli FM, Dotti MT, Mitochondrial dysfunction in hereditary spastic paraparesis with mutations in DDHD1/SPG28, J. Neurol. Sci, 362 (2016) 287–291. [DOI] [PubMed] [Google Scholar]
- [141].Dard R, Meyniel C, Touitou V, Stevanin G, Lamari F, Durr A, Ewenczyk C, Mochel F, Mutations in DDHD1, encoding a phospholipase A1, is a novel cause of retinopathy and neurodegeneration with brain iron accumulation, Eur. J. Med. Genet, 60 (2017) 639–642. [DOI] [PubMed] [Google Scholar]
- [142].Wu C, Fan D, A Novel Missense Mutation of the DDHD1 Gene Associated with Juvenile Amyotrophic Lateral Sclerosis, Front. Aging Neurosci, 8 (2016) 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Nakajima K, Sonoda H, Mizoguchi T, Aoki J, Arai H, Nagahama M, Tagaya M, Tani K, A novel phospholipase A1 with sequence homology to a mammalian Sec23p-interacting protein, p125, J. Biol. Chem, 277 (2002) 11329–11335. [DOI] [PubMed] [Google Scholar]
- [144].Schuurs-Hoeijmakers JH, Geraghty MT, Kamsteeg EJ, Ben-Salem S, de Bot ST, Nijhof B, van de V II, van der Graaf M, Nobau AC, Otte-Holler I, Vermeer S, Smith AC, Humphreys P, Schwartzentruber J, Ali BR, Al-Yahyaee SA, Tariq S, Pramathan T, Bayoumi R, Kremer HP, van de Warrenburg BP, van den Akker WM, Gilissen C, Veltman JA, Janssen IM, Vulto-van Silfhout AT, van der Velde-Visser S, Lefeber DJ, Diekstra A, Erasmus CE, Willemsen MA, Vissers LE, Lammens M, van Bokhoven H, Brunner HG, Wevers RA, Schenck A, Al-Gazali L, de Vries BB, de Brouwer AP, Mutations in DDHD2, encoding an intracellular phospholipase A(1), cause a recessive form of complex hereditary spastic paraplegia, American journal of human genetics, 91 (2012) 1073–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Morikawa R, Tsujimoto M, Arai H, Aoki J, Phospholipase A(1) assays using a radiolabeled substrate and mass spectrometry, Method Enzymol, 434 (2007) 1–13. [DOI] [PubMed] [Google Scholar]
- [146].Gonzalez M, Nampoothiri S, Kornblum C, Oteyza AC, Walter J, Konidari I, Hulme W, Speziani F, Schols L, Zuchner S, Schule R, Mutations in phospholipase DDHD2 cause autosomal recessive hereditary spastic paraplegia (SPG54), European journal of human genetics : EJHG, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Magariello A, Citrigno L, Zuchner S, Gonzalez M, Patitucci A, Sofia V, Conforti FL, Pappalardo I, Mazzei R, Ungaro C, Zappia M, Muglia M, Further evidence that DDHD2 gene mutations cause autosomal recessive hereditary spastic paraplegia with thin corpus callosum, Eur. J. Neurol, 21 (2014) e25–26. [DOI] [PubMed] [Google Scholar]
- [148].Doi H, Ushiyama M, Baba T, Tani K, Shiina M, Ogata K, Miyatake S, Fukuda-Yuzawa Y, Tsuji S, Nakashima M, Tsurusaki Y, Miyake N, Saitsu H, Ikeda S, Tanaka F, Matsumoto N, Yoshida K, Late-onset spastic ataxia phenotype in a patient with a homozygous DDHD2 mutation, Sci. Rep, 4 (2014) 7132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Inloes JM, Hsu KL, Dix MM, Viader A, Masuda K, Takei T, Wood MR, Cravatt BF, The hereditary spastic paraplegia-related enzyme DDHD2 is a principal brain triglyceride lipase, Proc Natl Acad Sci U S A, 111 (2014) 14924–14929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Araki M, Ohshima N, Aso C, Konishi A, Obinata H, Tatei K, Izumi T, Enzymatic characterization of recombinant rat DDHD2: a soluble diacylglycerol lipase, J. Biochem, 160 (2016) 269–279. [DOI] [PubMed] [Google Scholar]
- [151].Aso C, Araki M, Ohshima N, Tatei K, Hirano T, Obinata H, Kishi M, Kishimoto K, Konishi A, Goto F, Sugimoto H, Izumi T, Protein purification and cloning of diacylglycerol lipase from rat brain, J. Biochem, 159 (2016) 585–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Inloes JM, Kiosses WB, Wang H, Walther TC, Farese RV Jr., Cravatt BF, Functional Contribution of the Spastic Paraplegia-Related Triglyceride Hydrolase DDHD2 to the Formation and Content of Lipid Droplets, Biochemistry, 57 (2018) 827–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Arimitsu N, Kogure T, Baba T, Nakao K, Hamamoto H, Sekimizu K, Yamamoto A, Nakanishi H, Taguchi R, Tagaya M, Tani K, p125/Sec23-interacting protein (Sec23ip) is required for spermiogenesis, FEBS Lett, 585 (2011) 2171–2176. [DOI] [PubMed] [Google Scholar]
- [154].Tani K, Mizoguchi T, Iwamatsu A, Hatsuzawa K, Tagaya M, P125 is a novel mammalian Sec23p-interacting protein with structural similarity to phospholipid-modifying proteins, J. Biol. Chem, 274 (1999) 20505–20512. [DOI] [PubMed] [Google Scholar]
- [155].Ong YS, Tang BL, Loo LS, Hong W, p125A exists as part of the mammalian Sec13/Sec31 COPII subcomplex to facilitate ER-Golgi transport, J. Cell. Biol, 190 (2010) 331–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Zayats T, Jacobsen KK, Kleppe R, Jacob CP, Kittel-Schneider S, Ribases M, Ramos-Quiroga JA, Richarte V, Casas M, Mota NR, Grevet EH, Klein M, Corominas J, Bralten J, Galesloot T, Vasquez AA, Herms S, Forstner AJ, Larsson H, Breen G, Asherson P, Gross-Lesch S, Lesch KP, Cichon S, Gabrielsen MB, Holmen OL, Bau CH, Buitelaar J, Kiemeney L, Faraone SV, Cormand B, Franke B, Reif A, Haavik J, Johansson S, Exome chip analyses in adult attention deficit hyperactivity disorder, Transl. Psychiatry, 6 (2016) e923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Reuter MS, Tawamie H, Buchert R, Hosny Gebril O, Froukh T, Thiel C, Uebe S, Ekici AB, Krumbiegel M, Zweier C, Hoyer J, Eberlein K, Bauer J, Scheller U, Strom TM, Hoffjan S, Abdelraouf ER, Meguid NA, Abboud A, Al Khateeb MA, Fakher M, Hamdan S, Ismael A, Muhammad S, Abdallah E, Sticht H, Wieczorek D, Reis A, Abou Jamra R, Diagnostic Yield and Novel Candidate Genes by Exome Sequencing in 152 Consanguineous Families With Neurodevelopmental Disorders, JAmA Psychiatry, 74 (2017) 293–299. [DOI] [PubMed] [Google Scholar]
- [158].Moser M, Stempfl T, Li Y, Glynn P, Buttner R, Kretzschmar D, Cloning and expression of the murine sws/NTE gene, Mech. Dev, 90 (2000) 279–282. [DOI] [PubMed] [Google Scholar]
- [159].Akassoglou K, Malester B, Xu J, Tessarollo L, Rosenbluth J, Chao MV, Brain-specific deletion of neuropathy target esterase/swisscheese results in neurodegeneration, Proc. Natl. Acad. Sci. U.S.A, 101 (2004) 5075–5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Zaccheo O, Dinsdale D, Meacock PA, Glynn P, Neuropathy target esterase and its yeast homologue degrade phosphatidylcholine to glycerophosphocholine in living cells, J.Biol. Chem, 279 (2004) 24024–24033. [DOI] [PubMed] [Google Scholar]
- [161].van Tienhoven M, Atkins J, Li Y, Glynn P, Human neuropathy target esterase catalyzes hydrolysis of membrane lipids, J. Biol. Chem, 277 (2002) 20942–20948. [DOI] [PubMed] [Google Scholar]
- [162].Johnson MK, Improved assay of neurotoxic esterase for screening organophosphates for delayed neurotoxicity potential, Arch Toxicol, 37 (1977) 113–115. [DOI] [PubMed] [Google Scholar]
- [163].Hermansson M, Hanninen S, Hokynar K, Somerharju P, The PNPLA-family phospholipases involved in glycerophospholipid homeostasis of HeLa cells, Biochim. Biophys. Acta, 1861 (2016) 1058–1065. [DOI] [PubMed] [Google Scholar]
- [164].Glynn P, Neuropathy target esterase, Biochem. J, 344 Pt 3 (1999) 625–631. [PMC free article] [PubMed] [Google Scholar]
- [165].Johnson MK, The primary biochemical lesion leading to the delayed neurotoxic effects of some organophosphorus esters, J. Neurochem, 23 (1974) 785–789. [DOI] [PubMed] [Google Scholar]
- [166].Synofzik M, Gonzalez MA, Lourenco CM, Coutelier M, Haack TB, Rebelo A, Hannequin D, Strom TM, Prokisch H, Kernstock C, Durr A, Schols L, Lima-Martinez MM, Farooq A, Schule R, Stevanin G, Marques W Jr., Zuchner S, PNPLA6 mutations cause Boucher-Neuhauser and Gordon Holmes syndromes as part of a broad neurodegenerative spectrum, Brain, 137 (2014) 69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Rainier S, Bui M, Mark E, Thomas D, Tokarz D, Ming L, Delaney C, Richardson RJ, Albers JW, Matsunami N, Stevens J, Coon H, Leppert M, Fink JK, Neuropathy target esterase gene mutations cause motor neuron disease, American journal of human genetics, 82 (2008) 780–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Kmoch S, Majewski J, Ramamurthy V, Cao S, Fahiminiya S, Ren H, MacDonald IM, Lopez I, Sun V, Keser V, Khan A, Stranecky V, Hartmannova H, Pristoupilova A, Hodanova K, Piherova L, Kuchar L, Baxova A, Chen R, Barsottini OG, Pyle A, Griffin H, Splitt M, Sallum J, Tolmie JL, Sampson JR, Chinnery P, Care4Rare C, Banin E, Sharon D, Dutta S, Grebler R, Helfrich-Foerster C, Pedroso JL, Kretzschmar D, Cayouette M, Koenekoop RK, Mutations in PNPLA6 are linked to photoreceptor degeneration and various forms of childhood blindness, Nat. Commun, 6 (2015) 5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Hufnagel RB, Arno G, Hein ND, Hersheson J, Prasad M, Anderson Y, Krueger LA, Gregory LC, Stoetzel C, Jaworek TJ, Hull S, Li A, Plagnol V, Willen CM, Morgan TM, Prows CA, Hegde RS, Riazuddin S, Grabowski GA, Richardson RJ, Dieterich K, Huang T, Revesz T, Martinez-Barbera JP, Sisk RA, Jefferies C, Houlden H, Dattani MT, Fink JK, Dollfus H, Moore AT, Ahmed ZM, Neuropathy target esterase impairments cause Oliver-McFarlane and Laurence-Moon syndromes, J. Med. Genet, 52 (2015) 85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Wiethoff S, Bettencourt C, Paudel R, Madon P, Liu YT, Hersheson J, Wadia N, Desai J, Houlden H, Pure Cerebellar Ataxia with Homozygous Mutations in the PNPLA6 Gene, Cerebellum, 16 (2017) 262–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Langdahl JH, Frederiksen AL, Nguyen N, Brusgaard K, Juhl CB, Boucher Neuhauser Syndrome - A rare cause of inherited hypogonadotropic hypogonadism. A case of two adult siblings with two novel mutations in PNPLA6, Eur. J. Med. Genet, 60 (2017) 105–109. [DOI] [PubMed] [Google Scholar]
- [172].Teive HAG, Camargo CHF, Sato MT, Shiokawa N, Boguszewski CL, Raskin S, Buck C, Seminara SB, Munhoz RP, Different Cerebellar Ataxia Phenotypes Associated with Mutations of the PNPLA6 Gene in Brazilian Patients with Recessive Ataxias, Cerebellum, 17 (2018) 380–385. [DOI] [PMC free article] [PubMed] [Google Scholar]