Enteropathogenic Bacillus cereus causes foodborne infections due to the production of pore-forming enterotoxins in the intestine. Before that, spores have to be ingested, survive the stomach passage, and germinate.
KEYWORDS: Bacillus cereus, enterotoxins, food infection, mucus layer, porcine gastric mucin, transcriptome
ABSTRACT
Enteropathogenic Bacillus cereus causes foodborne infections due to the production of pore-forming enterotoxins in the intestine. Before that, spores have to be ingested, survive the stomach passage, and germinate. Thus, before reaching epithelial cells, B. cereus comes in contact with the intestinal mucus layer. In the present study, different aspects of this interaction were analyzed. Total RNA sequencing revealed major transcriptional changes of B. cereus strain F837/76 upon incubation with porcine gastric mucin (PGM), comprising genes encoding enterotoxins and further putative virulence factors, as well as proteins involved in adhesion to and degradation of mucin. Indeed, PGM was partially degraded by B. cereus via secreted, EDTA-sensitive proteases. The amount of enterotoxins detectable in culture media supplemented with PGM was also clearly increased. Tests of further strains revealed that enhancement of enterotoxin production upon contact with PGM is broadly distributed among B. cereus strains. Interestingly, evidence was found that PGM can also strain-specifically trigger germination of B. cereus spores and that vegetative cells actively move toward mucin. Overall, our data suggest that B. cereus is well adapted to the host environment due to massive transcriptome changes upon contact with PGM, attributing mucin an important and, thus far, neglected role in pathogenesis.
INTRODUCTION
Mucus is a highly complex viscous substance that covers gastrointestinal cells. Besides salts, lipids, and several proteins involved in defense mechanisms, it consists mainly of water (approximately 95%) and the glycoprotein mucin (1, 2). A large number of different mucins has been discovered. These can be divided into membrane-bound and secretory mucins, which form the mucus layers (3, 4). Mucins have a molecular weight of 0.5 to 20 MDa and consist of ∼20% proteins and ∼80% carbohydrates (1). The protein core is organized in tandem repeats of serine-, threonine-, and proline-rich regions, which are mostly O-glycosylated. At the C and N termini, large amounts of cysteine are found. The carbohydrates include N-acetylgalactosamine, N-acetylglucosamine, N-acetylneuraminic acid, fucose, galactose, and mannose. Mucin monomers dimerize and multimerize via disulfide bonds (1).
Mucus functions as a lubricant in the gastrointestinal tract, facilitating the passage of food. It is also important for growth, adhesion, and protection of the intestinal microbiota (2). Furthermore, it represents a biophysical barrier between epithelial cells and the environment, including chemical and mechanical insults as well as the commensal microbiota and pathogens (2, 5, 6). The mucus layer is a reservoir for many antimicrobial molecules, and various mucin oligosaccharides themselves show antimicrobial activity (5). By adhesion to mucin oligosaccharides, pathogens are trapped and removed due to the constant renewal of the mucus layer (5, 6). Cell surface-bound mucins can even initiate intracellular signaling in response to bacteria (5).
On the other hand, pathogens, especially invasive bacteria, have developed various strategies to antagonize the mucosal defenses. Helicobacter pylori, Campylobacter jejuni, Salmonella enterica serovar Typhimurium, Yersinia enterocolitica, enterohemorrhagic Escherichia coli (EHEC), enteropathogenic E. coli (EPEC), and others use the mucus glycoproteins for colonization by adhesion to the oligosaccharides (5, 7, 8). H. pylori, Salmonella, and C. jejuni are able to penetrate the mucus barrier (9, 10). Y. enterocolitica, Shigella, streptococci, and others produce glycosidases and proteases to degrade mucins (7, 9). H. pylori can also induce downregulation of mucin gene expression or alteration of the glycosylation profile (7, 11). Another strategy is to avoid the mucin barrier by invading intestinal M cells or by disrupting the tight junctions between mucosal epithelial cells (5, 9).
Little is known about the interaction of Bacillus cereus with the mucus layer or mucins. In vitro assays demonstrated adhesion of nonpathogenic and pathogenic B. cereus strains to porcine gastric mucin (PGM) (12–14). Furthermore, induction of fucosidase production suggests that B. cereus degrades and assimilates PGM (15). Our own studies concentrate on enteropathogenic B. cereus, which is responsible for a diarrheal type of food poisoning. Before causing diarrhea, ingested spores survive the stomach passage, germinate in the intestine, and produce locally large amounts of enterotoxins, which, as pore-forming toxins, harm epithelial cells (16–19). Most important are the two three-component enterotoxin complexes Nhe (nonhemolytic enterotoxin [20, 21]) and Hbl (hemolysin BL [22]). So far, adhesion (23) as well as toxicity studies (24, 25) have been performed only with cell lines, neglecting the possible role of mucus in the gut. Recently, we showed that incubation of enteropathogenic B. cereus under simulated intestinal conditions results in increased enterotoxin gene expression and enhanced toxin production (26). The mucus layer was not considered.
In the present study, it was discovered for the first time that enterotoxin gene expression and toxin production of B. cereus are enhanced in the presence of PGM. Furthermore, total RNA sequencing revealed that various genes involved in adhesion to and degradation of mucin, as well as genes encoding further putative virulence factors, are upregulated upon contact with mucin. We also showed that B. cereus is able to degrade PGM and further use it as a growth substrate. This was supported by the active movement of vegetative cells toward mucin. Moreover, PGM strain specifically triggered germination of spores. Interestingly, mucin also protected the enterotoxins from enzymatic degradation.
RESULTS
Differential gene expression upon contact with PGM.
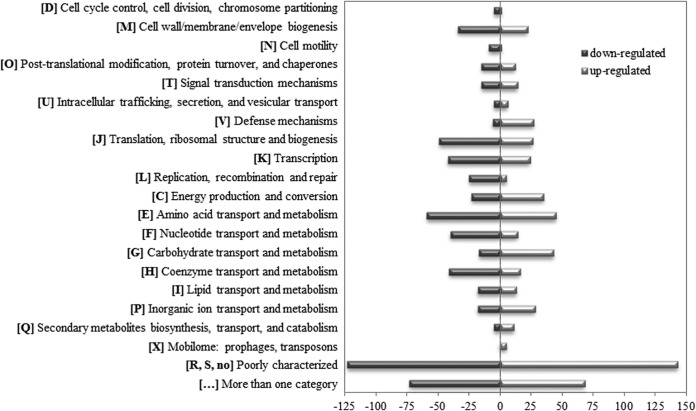
It is not clear which signals trigger enterotoxin production of B. cereus in the host environment. Before reaching its target, the epithelial cells, B. cereus comes in contact with the intestinal mucus layer. To investigate the overall response to that contact, total transcriptome analyses were performed using PGM, which is a commonly applied model for mucus interactions. Reference strain F837/76 was grown in minimal medium with or without 0.25% PGM, and after 3 h samples for RNA sequencing were taken. Differentially expressed genes were allocated to Clusters of Orthologous Groups (COG) categories (Fig. 1) (27, 28). Five hundred fifty-nine genes were upregulated in the presence of PGM. Except genes with no clear COG definition, the groups amino acid transport and metabolism (45 genes), carbohydrate transport and metabolism (43 genes), and energy production and conversion (35 genes) were most strongly represented. On the other hand, 622 genes were downregulated in the presence of PGM. Most strongly represented were the groups amino acid transport and metabolism (59 genes), translation, ribosomal structure, and biogenesis (49 genes), transcription (42 genes), coenzyme transport and metabolism (41 genes), and nucleotide transport and metabolism (40 genes).
FIG 1.
Differential gene regulation in B. cereus strain F837/76 after 3 h of growth in mMOD minimal medium with 0.25% PGM. Genes with detected P values of ≤0.05 and a fold change of >2 were considered. Hypothetical proteins were excluded. Differentially regulated genes were allocated to Clusters of Orthologous Groups (COG) categories (27, 28) using NCBI Conserved Domains Search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Five hundred fifty-nine genes were upregulated in the presence of PGM, of which 68 belong to more than one COG category. These genes are shown as light gray bars. Six hundred twenty-two genes were downregulated in the presence of PGM, of which 73 are assigned to more than one COG category. They are shown as dark gray bars.
We were especially interested in genes putatively involved in virulence (Table 1). The binding components of the enterotoxin complexes Nhe and Hbl were upregulated, as were putative additional virulence factors, such as proteases, collagenase, flagellin, internalins, and S-layer proteins. Interestingly, genes encoding phospholipase C were downregulated, as was bcf_15280, the fourth component of the hbl operon, encoding a hemolysin BL binding component precursor. Differential regulation was confirmed by quantitative PCR (qPCR) (Table 1; see also Tables S1 and S2 in the supplemental material for details).
TABLE 1.
Genes of interest differentially expressed in B. cereus strain F837/76a
| Gene | Fold change | P value | Gene product | COG | qPCR |
|---|---|---|---|---|---|
| Upregulated | |||||
| bcf_04565 | 534.8 | 5.00E−05 | S-layer protein | No | ✓ |
| bcf_04560 | 469.2 | 5.00E−05 | S-layer protein | No | ✓ |
| bcf_27370 | 462.9 | 5.00E−05 | Hemolysin III-like protein | U | ✓ |
| bcf_09850 | 252.4 | 5.00E−05 | Serine protease | O | ✓ |
| bcf_16240 | 221.2 | 5.00E−05 | Internalin | No | ✓ |
| bcf_08380 | 214.5 | 5.00E−05 | Flagellin protein FlaA | N | ✓ |
| bcf_13835 | 207.8 | 5.00E−05 | Chitin binding protein | R | ✓ |
| bcf_05660 | 200.3 | 5.00E−05 | S-layer protein | No | ✓ |
| bcf_17385 | 195.0 | 5.00E−05 | Collagenase | S | ✓ |
| bcf_26895 | 193.5 | 5.00E−05 | Collagen adhesion protein | S | ND |
| bcf_17575 | 192.2 | 5.00E−05 | Internalin | No | ✘ |
| bcf_01910 | 170.5 | 5.00E−05 | Zinc metalloprotease | R | ✓ |
| bcf_24405 | 161.7 | 0.01445 | Cell envelope-bound metalloprotease | No | ✓ |
| bcf_24230 | 154.5 | 5.00E−05 | Bacitracin export permease protein BceB like protein | V | ✓ |
| bcf_17530 | 141.8 | 5.00E−05 | Cell wall endopeptidase, family M23/M37 | M | ✓ |
| bcf_19505 | 138.7 | 5.00E−05 | Lon-like protease with PDZ domain | T | ND |
| bcf_18185 | 138.4 | 5.00E−05 | Internalin | S | ✘ |
| bcf_17340 | 117.9 | 5.00E−05 | Bile acid sodium symporter | R | ✓ |
| bcf_15285 | 109.6 | 0.00035 | HblB protein | No | ✘ |
| bcf_00530 | 109.3 | 0.00115 | ATP-dependent Clp protease, ATP-binding subunit ClpC | O | ✓ |
| bcf_09265 | 100.4 | 0.00335 | Putative nonhemolytic enterotoxin lytic component L1 (NheB) | No | ✓ |
| bcf_13655 | 15.0 | 5.00E−05 | ATP-dependent Clp protease proteolytic subunit | O | ND |
| bcf_08015 | 14.4 | 5.00E−05 | Internalin | No | ✘ |
| Downregulated | |||||
| bcf_03465 | 182.5 | 5.00E−05 | Broad-substrate-range phospholipase C | No | ✓ |
| bcf_04545 | 159.9 | 5.00E−05 | Protein export cytoplasm protein SecA ATPase RNA helicase | U | ✓ |
| bcf_16685 | 154.2 | 5.00E−05 | Hemolysin-like protein containing CBS domains | R | ✓ |
| bcf_15280 | 126.1 | 0.0001 | Hemolysin BL binding component precursor | No | ✓ |
| bcf_13360 | 107.9 | 0.00045 | Bacillolysin | O | ✓ |
| bcf_18650 | 106.3 | 5.00E−05 | Phosphatidylinositol-specific phospholipase C | No | ✓ |
Measurements were taken after 3 h of incubation in mMOD minimal medium supplemented with 0.25% PGM. Genes are sorted according to their fold change in expression. Allocation to the COG categories is shown (27, 28), as is the confirmation of differential expression by qRT-PCR. ✓, differential expression confirmed by qRT-PCR. ✘, differential expression not confirmed by qRT-PCR. ND, not determined.
Increased enterotoxin production under addition of PGM.
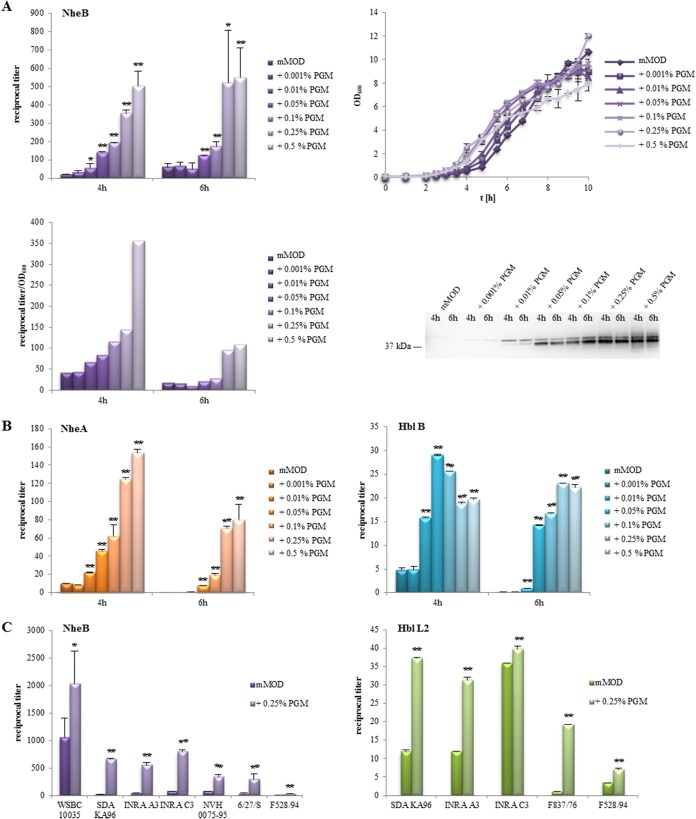
Transcriptome studies showed that genes encoding enterotoxin components are upregulated in the presence of PGM (Table 1). Thus, we investigated the presence of enterotoxins in B. cereus culture supernatants after growth in minimal medium supplied with PGM. First, the B component of the nonhemolytic enterotoxin (21) served as an indicator for toxin production and was determined in culture supernatants of strain F837/76 after 4 and 6 h of growth. Rising PGM concentrations moderately enhanced growth of strain F837/76. On the other hand, PGM concentrations of 0.01% and higher resulted in a significant increase of NheB titers after 4 h. Furthermore, the productivity for NheB was determined as titer per optical density at 600 nm (OD600) to exclude growth effects, and enzyme immunoassay (EIA) data were additionally confirmed in a Western blot (Fig. 2A). All experiments described in this study were conducted with PGM type III, which is a processed and partially purified variant of PGM type II, a crude mucin preparation. Figure S1 shows that PGM type II also enhances growth and especially enterotoxin production of B. cereus. In contrast to these findings, only large amounts of glucose resulted in enhanced growth rates and enhanced NheB productivity (Fig. S2). Further experiments showed that these observations are not limited to NheB or strain F837/76. Increasing amounts of NheA or Hbl B, the binding component of hemolysin BL (22), were also detected depending on PGM concentrations (Fig. 2B). An increase of NheB and Hbl L2 in culture supernatants of further enteropathogenic or nonpathogenic B. cereus strains (25, 26) after 6 h of incubation with 0.25% PGM was also observed (Fig. 2C). Thus, enhanced enterotoxin production upon contact with PGM is broadly distributed among B. cereus strains.
FIG 2.
Growth and toxin production of B. cereus in mMOD minimal medium. Toxin production was determined from culture supernatants after 4 and 6 h using specific EIAs. Reciprocal titers are shown. *, significant difference from growth in mMOD alone with a P value of ≤0.05. **, P ≤ 0.01. (A) Growth and NheB production of strain F837/76 at different PGM concentrations added to the minimal medium. Reciprocal NheB titers per OD600 unit were also determined. NheB was additionally detected in a Western blot. (B) NheA and Hbl B production of strain F837/76 at different PGM concentrations. (C) Increased enterotoxin production of various B. cereus strains upon incubation with PGM. Enterotoxin components NheB and Hbl L2 were detected in culture supernatants after 6 h of incubation in minimal medium with 0.25% PGM. Toxin production of these strains under standard and simulated intestinal conditions has already been well investigated (25, 26).
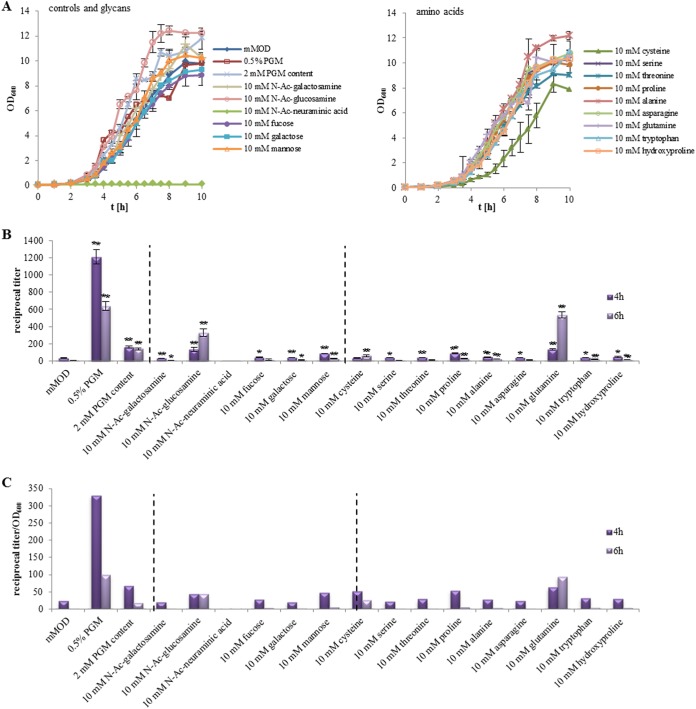
Consequently, we raised the question of which mucin components might be responsible for the enhanced toxin production. Strain F837/76 was again cultivated in minimal medium supplemented with 10 mM of the single components. The different amino acids and sugars known to be found in mucins (1) were tested. Compared to growth in minimal medium, N-acetylglucosamine, alanine, and a mixture of all components (2 mM each) enhanced growth, while cysteine reduced and N-acetylneuraminic acid inhibited it (Fig. 3A). Significant changes in NheB production compared to that in modified MOD (mMOD) were detected for nearly all tested components (Fig. 3B), with the 2 mM PGM mixture, N-acetylglucosamine, mannose, proline, and glutamine showing the biggest increase (Fig. 3B and C). Nevertheless, none of the tested substances increased NheB titers in the magnitude of PGM, which was applied as a positive control. Thus, the origin of PGM-enhanced enterotoxin production by B. cereus is not fully clear, but first hints were collected regarding which substances might be involved. These tests were repeated using emetic B. cereus strain F4810/72, which bears not only the nhe operon but also a variety of glycosyl hydrolases as well as fucose transporters and degrading enzymes (www.cazy.org). Interestingly, growth and toxin production were not enhanced with 10 mM fucose compared to that of F837/76, but with 0.5% PGM, N-acetylglucosamine, and the 2 mM PGM content, accelerated growth rates were detected (Fig. S3A). These data match the results of Warda and coworkers, who stated that genotypes and fucose utilization do not necessarily correlate (29). With 10 mM cysteine, growth was not as reduced as it was in F837/76, and after 4 h, toxin production was enhanced, which was the only significant difference from growth in F837/76 (Fig. S3B and C).
FIG 3.
Growth and toxin production of B. cereus strain F837/76, grown in mMOD minimal medium supplied with different amino acids and sugars. (A) Growth. (B) NheB production as determined in sandwich EIAs. PGM (0.5%) was used as a positive control. Titers vary from those of earlier experiments, as they were obtained in an independent approach. *, significant difference from growth in mMOD alone with a P value of ≤0.05; **, P ≤ 0.01. (C) Relative productivity of NheB calculated as reciprocal titer/OD600.
Mucin degradation by B. cereus.
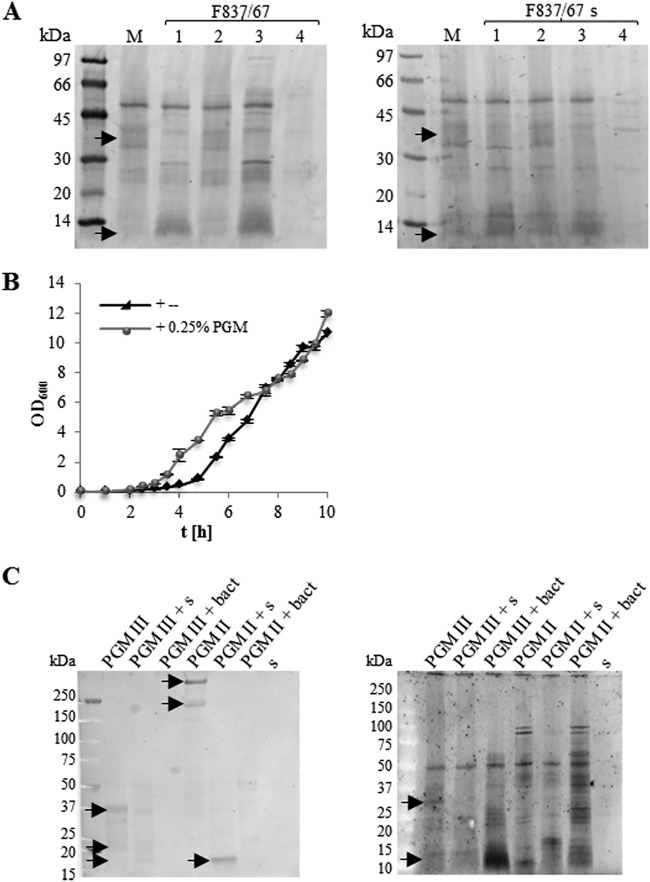
Mucin degradation assays showed that F837/76 (Fig. 4A) as well as other B. cereus strains (Fig. S4) are able to partially degrade PGM. Addition of B. cereus cultures or supernatants led to a decrease of protein bands of approximately 30 to 45 kDa and to an accumulation of protein bands at <14 kDa. This effect was abolished by addition of a protease inhibitor and could be restored when EDTA-free protease inhibitor was used (Fig. 4A and Fig. S4). B. cereus might further use PGM components for growth, which is suggested by an accelerated growth rate upon addition of PGM (Fig. 2A and 4B). These experiments allow us to conclude that B. cereus partially degrades PGM via secreted, EDTA-sensitive proteases. To make conclusions from gastric to intestinal mucus, which consist mainly of MUC2 (30–32), a Western blot with a MUC2-specific antibody was added to the degradation experiments. It was shown that MUC2 is indeed present in PGM types II and III and that it is degraded by B. cereus (Fig. 4C).
FIG 4.
Mucin degradation. (A) Mucin degradation by F837/76 cells (left) and culture supernatant (right). Lanes: M, PGM in Tris buffer; 1, PGM plus F837/76; 2, PGM plus F837/76 plus protease inhibitor; 3, PGM plus F837/76 plus protease inhibitor, EDTA free; 4, Tris buffer plus F837/76. Arrows highlight differing protein bands. (B) Growth of strain F837/76 in mMOD minimal medium after addition of 0.25% PGM. (C) Mucin degradation by F837/76 cells and supernatant (s). PGM type II and type III were used. (Left) Western blot with MUC2 specific antibody. (Right) Sypro Ruby staining of the same samples as a control. Arrows indicate MUC2 as well as differing protein bands.
Furthermore, in addition to a variety of proteases, several genes encoding glycosyl hydrolases were differentially regulated in the presence of PGM (Table 1 and Table S3). Some of them might play a role in mucin degradation. In the Carbohydrate-Active enZYmes Database (www.cazy.org), their prevalence in 48 B. cereus strains is indicated. Of 153 glycosyl hydrolase families and various subfamilies, 30 occur in B. cereus, as well as several hydrolases not yet assigned to a family. Most abundant are hydrolases of family no. 13, comprising glucosidases, amylases, pullulanases, and others (7 to 11 per strain), and no. 18, comprising chitinases, N-acetyl-glucosaminidases, and others (3 to 4 per strain). Other families known to be involved in O-glycan mucin degradation appear rather rarely. For instance, families no. 33 (sialidases), no. 29 (fucosidases), no. 2 (galactosidases), no. 84, no. 85, no. 89 (N-acetyl-glucosaminidases), and no. 129 (N-acetyl-galactosaminidases) are not present at all. Family no. 42 (galactosidases) appears only twice in a single strain, family no. 101 (N-acetyl-galactosaminidases) once in 4 out of 48 strains, and family no. 95 (fucosidases) once in 6 out of 48 strains. The B. cereus strain tested in this study, F837/76, harbors a total of 25 glycosyl hydrolases, which are allocated to 9 families. The genes encoding 10 of these were upregulated in the presence of PGM (Table S3).
Germination is strain specific and partly PGM dependent.
Next to genes encoding toxins and further putative virulence factors, a series of genes involved in sporulation and germination were differentially expressed upon contact with PGM (Table S3). Moreover, germination is a prerequisite for the establishment of B. cereus infections. Thus, we subsequently tested if PGM triggers germination of B. cereus spores by monitoring the reduction of the OD620 in 3-min intervals for 30 to 60 min. In casein-glucose-yeast (CGY) medium, the OD620 of all tested strains showed the strongest reduction, indicating that these spores were able to germinate in full medium (Fig. 5). Reduction was enhanced for strains F837/76 and MHI 226 when the spores were previously heat treated for 10 min at 80°C (Fig. 5A and B). In mMOD medium, spores of all three strains showed minimal or no germination. Heat treatment reduced the OD620 of F837/76 and SDA KA 96 by 10 and 15%, respectively (Fig. 5A and C). Addition of PGM to the minimal medium resulted in approximately 10% (F837/76; Fig. 5A) or no (MHI 226 and SDA KA 96; Fig. 5B and C) reduction. Here, heat treatment enhanced germination of F837/76 to 15%, germination of MHI 226 to 10%, and germination of SDA KA 96 to approximately 15%.
FIG 5.
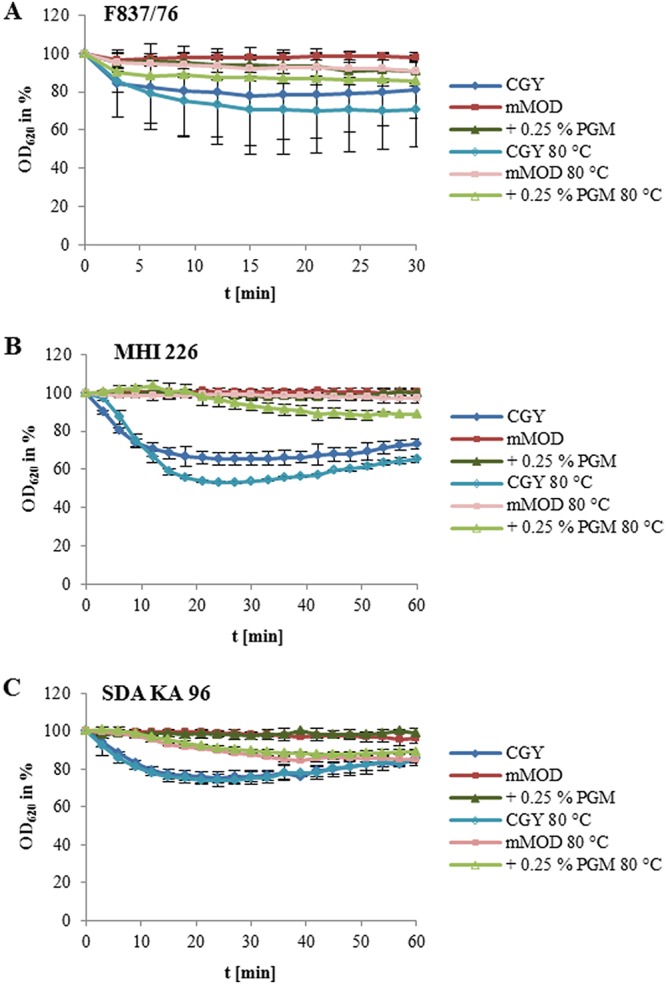
Germination of spores in response to CGY full medium, mMOD minimal medium, addition of 0.25% PGM, and heat treatment (10 min at 80°C). Germination is depicted as the decrease of OD620 per unit of time. OD620 at time point 0 was set to 100%. (A) Strain F837/76. (B) Strain MHI 226. (C) Strain SDA KA 96.
Motility under addition of PGM.
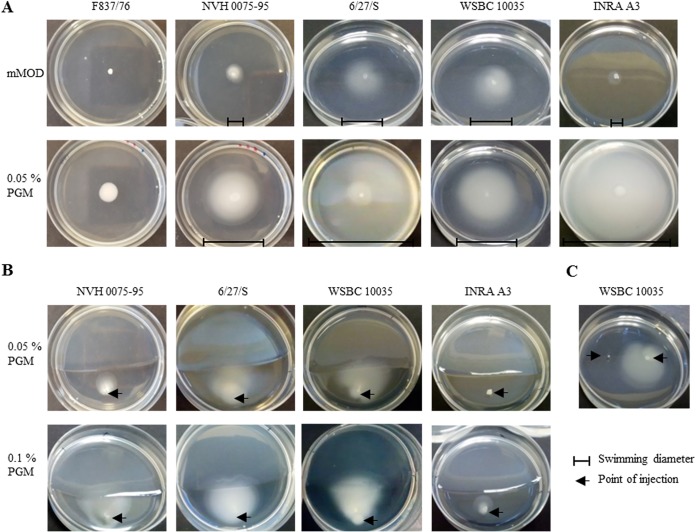
Total transcriptome analyses further revealed that genes involved in motility and chemotaxis were differentially regulated upon contact with PGM, including flagellar proteins, methyl-accepting chemotaxis proteins, and others (Table S3). Thus, motility of B. cereus in minimal medium containing 0.25% agar as well as 0.05% PGM was investigated. Surprisingly, strain F837/76 showed swimming neither under the conditions chosen in this study (Fig. 6A) nor in CGY agar (data not shown), while other tested strains were able to move. Swimming diameters were enhanced in the presence of PGM (Fig. 6A).
FIG 6.
Motility of B. cereus under addition of PGM. (A) One μl of overnight culture was injected at the center of the plates, which contained mMOD medium with either 0.25% agar (upper row) or 0.05% PGM and 0.25% agar (lower row). (B) One μl of overnight culture was injected approximately 1 cm from a line dividing sole mMOD agar (lower part) from mMOD agar supplemented with PGM (upper part). (C) One μl of 0.25% PGM (left side) and 1 μl of culture (right side) were injected into mMOD agar with approximately 2 cm distance. Pictures were taken after 24 h of incubation at 37°C.
However, the more interesting finding was that, when applied to plates with two separate agars, strains NVH 0075-95, 6/27/S, WSBC 10035, and INRA A3 moved toward PGM, with swimming radius partially depending on the mucin concentration (Fig. 6B). This was also seen when 1 μl 0.25% PGM and 1 μl culture were injected into mMOD agar, as shown for WSBC 10035 (Fig. 6C).
Enterotoxin protection by PGM.
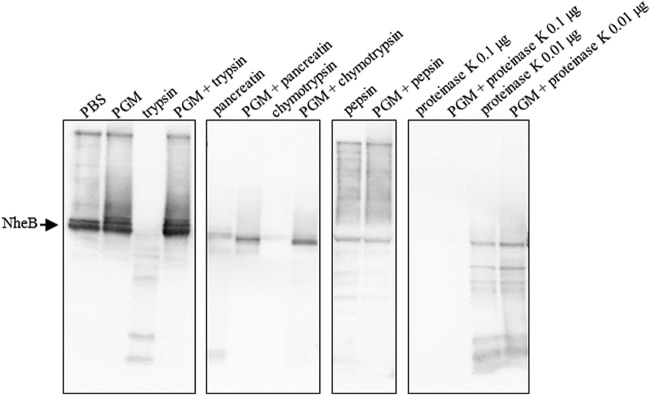
Subsequently, we were interested if PGM for its part might be able to protect the enterotoxins produced by B. cereus from enzymatic degradation. For that purpose, supernatant of strain F837/76 was incubated at 37°C with different enzymes before Western blot analyses specific for NheB were performed. NheB is the component of the tripartite nonhemolytic enterotoxin of B. cereus known to correlate with cytotoxicity (24, 33). Preliminary tests revealed that trypsin, chymotrypsin, pepsin, pancreatin, and proteinase K were able to degrade NheB depending on enzyme concentrations and incubation time (data not shown). In the following step, 0.25% PGM in phosphate-buffered saline (PBS) was added to the samples. Under addition of PGM, enzymatic digestion of NheB by trypsin, chymotrypsin, and pancreatin was clearly diminished. On the other hand, PGM had no detectable effect on pepsin or proteinase K (Fig. 7).
FIG 7.
Decreased enterotoxin digestion under addition of PGM. Supernatant of strain F837/76 was incubated with PBS, 0.25% PGM in PBS, 1 μg/μl trypsin, pancreatin, chymotrypsin or pepsin, or a mixture of the enzymes and 0.25% PGM in PBS for 2 h at 37°C. Proteinase K was used at concentrations of 0.1 and 0.01 μg/μl, and the incubation time was 15 min. Subsequently, Western blot analyses specific for NheB were performed.
DISCUSSION
Before B. cereus reaches the epithelial cells in the intestine, it comes in contact with the mucus layer. In the present study, we detected lively interaction between B. cereus and PGM and analyzed several aspects of this interaction, such as toxin production and mucin degradation. In this context, the first total transcriptome study of a pathogenic bacterium after contact with mucin was performed. As the production of cytotoxins is commonly seen as central for enteropathogenicity of B. cereus, differential regulation of genes encoding Nhe and Hbl components was analyzed. The nonhemolytic enterotoxin (Nhe) consists of the components A, B, and C, which are encoded by the nheABC operon (34). The hbl genes are organized in the operon hblCDA, encoding the components Hbl L2, L1, and B (35). Thus, it was rather unexpected that only the binding components NheB (nheB) and Hbl B (hblA) were significantly overexpressed under the chosen conditions. The hbl operon in strain F837/76 also includes hblB, which encodes a protein described as a hemolysin BL binding component precursor. Only in 2010 was it shown that the gene is indeed transcribed and the corresponding protein secreted by B. cereus (36). Here, we found that, in contrast to hblA, hblB was downregulated upon contact with PGM (Table 1). This finding demonstrates once more that although it is part of the hbl operon, it underlies a different regulatory mechanism. Earlier studies identified a stem-loop upstream and downstream of hblB that might act as a transcriptional terminator (37). A separate promoter structure for hblB also has been suggested (36).
We further showed that PGM also massively enhances the amount of toxins in the supernatant of B. cereus. Depending on PGM concentrations, NheB titers were increased more than 10-fold (Fig. 2A). This disagrees with prior studies in which NheB could not be detected in the supernatant of four B. cereus strains after they had been incubated in the presence of PGM (13, 14). The authors concluded that Nhe was not detected due to insufficient bacterial concentrations or toxin degradation (13). In contrast to that, our studies proved that toxins accumulate in the supernatant and that this effect is not limited to a single toxin component. Besides NheB, increased levels of NheA, Hbl B, and Hbl L2 were also found (Fig. 2B and C). Furthermore, this enhancement was observed for various strains, including NVH 0075-95, which has also been used by Tsilia and coworkers (Fig. 2B and C) (14). This suggests that B. cereus is able to rapidly sense the host environment and subsequently activate its toxin production machinery, assigning mucin an important role in pathogenesis.
Besides the enterotoxins, other B. cereus virulence factors also were overexpressed after contact with mucin, such as S-layer proteins, hemolysin III-like protein, proteases, chitin binding protein, and flagellin (Table 1). Flagella play an important role in virulence of various pathogens in processes such as motility and chemotaxis, colonization, adhesion, invasion, biofilm formation, secretion of virulence factors, and activation of proinflammatory responses or phagocytosis (38–40). Upregulation of flagellin in response to human mucin has been previously described for H. pylori and C. jejuni (41, 42). Surface-associated proteases as well as flagellin and an S-layer protein have also been found to be involved in adhesion of probiotic B. cereus to PGM (12). For Pseudomonas aeruginosa it has been shown that flagellin can bind mucin (43). Similar findings were reported for EPEC, which adhere to the intestinal mucosa with the help of flagella (44). In parallel to the high expression of flagellin, we even observed a directed movement of motile B. cereus toward mucin (Fig. 6). The active swimming toward PGM seems to depend on the ability of the single strains to move as well as to sense mucin, but together with the differential regulation of genes involved in motility and chemotaxis, it emphasizes once more the importance of mucin for B. cereus, either as a nutrient source or as a target location for adhesion, toxin production, etc. Overall, mucin-induced upregulation of genes important for pathogenicity has been reported for several bacteria, such as H. pylori, Vibrio cholerae, and C. jejuni (41, 42, 45). For the latter, upregulation of genes encoding toxins, Campylobacter invasion antigen, a multidrug efflux system, flagellin, and mucin-degrading enzymes was found (42).
Various commensal and pathogenic microorganisms degrade mucins via specific saccharolytic and proteolytic enzymes (proteases, glycosidases, sialidases, and sulfatases) (2, 6). A study by Fang and coworkers on Bacillus thuringiensis indicated that members of the B. cereus group are generally able to degrade mucins (46). Our own experiments confirmed this assumption. The B. cereus strains used in our study were able to partially degrade PGM via secreted, EDTA-sensitive proteases and to use it as a growth substrate (Fig. 4). We used, as have many others, porcine gastric mucin as a model for host-pathogen interactions. Gastric mucin consists mainly of MUC5AC, MUC5B, and MUC6. Intestinal mucus, on the contrary, primarily consists of MUC2 (30–32). By proving that PGM also contains MUC2 and that MUC2 is degraded by B. cereus (see Fig. 4C), it is quite evident that B. cereus senses and responds not only to gastric but also to intestinal mucus. The specific mucin components triggering enhanced enterotoxin production were not definitively identified, but our studies pointed to an involvement of N-acetylglucosamine, mannose, proline, and glutamine (Fig. 3; see also Fig. S3 in the supplemental material).
Exoproteases contribute to virulence of B. cereus (47–49) and other pathogens, such as Bacillus anthracis (50) and Staphylococcus aureus (51). Of the proteases upregulated in this study, bcf_17385, encoding collagenase, and bcf_24405, encoding a cell envelope-bound metalloprotease, code for secretion signal peptides (52). Chitin binding proteins and chitinases are used in the first place for chitin degradation but can also be important virulence factors for bacterial pathogens, as described for Legionella pneumophila, Salmonella spp., and Listeria monocytogenes (53). In addition, N-acetylgalactosamine of mucin is a target for V. cholerae chitin binding protein (53). Thus, one can assume that bcf_13835 (chitin binding protein), bcf_02170, and bcf_18465 (chitinases) (Table 1 and Table S3) were upregulated as additional virulence factors, and that they also contribute to mucin degradation.
A hemolysin III-like protein and, surprisingly, four genes encoding internalins were also upregulated (Table 1). Preliminary studies showed that hemolysin III forms transmembrane pores (54). Its gene expression is suggested to be PlcR independent (55, 56), and the corresponding protein is suggested to be membrane bound rather than secreted (57). Its particular function with regard to mucin contact remains unclear so far. Internalins are proteins found on the cell surface of invasive pathogens such as L. monocytogenes, which are involved in internalization into mammalian cells (58). Although B. cereus is generally considered a noninvasive pathogen, there are publications suggesting invasive strategies (59, 60).
Conclusions.
Our data suggest that once B. cereus reaches the gastrointestinal tract and senses the host environment (mucin in this case), the gene expression pattern of the pathogen is significantly altered, leading to the best possible adaptation to the host environment. After sensing the gastrointestinal environment, enterotoxin gene expression as well as toxin production is massively increased, attributing mucin an important and, thus far, neglected role in pathogenesis. Overall, the rapid, mucin-induced activation of the toxin production machinery might also be an explanation for the relatively short incubation time of B. cereus-associated food infections compared to that of other pathogens with a similar pathogenicity mechanism.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
In this study, the B. cereus strains F837/76 (DSM 4222), F528/95, SDA KA 96, INRA A3, INRA C3, WSBC 10035, 6/27/S, NVH 0075-95, MHI 226, and emetic F4810/72 (AH187) were used. Enterotoxin production of most strains under standard laboratory and simulated intestinal conditions has been characterized in detail in earlier studies (25, 26). Overnight cultures were grown in CGY (casein-glucose-yeast) medium with 1% glucose at 32°C under continuous agitation (24). B. cereus strains were further cultivated in modified MOD (mMOD) minimal medium (61, 62) at 32°C under continuous agitation. Porcine gastric mucin type II and type III were purchased from Sigma-Aldrich. Single mucin components were added as stated in Results, either individually or as a mixture of all components (2 mM each). For collection of toxin-rich supernatants, cells were treated as previously described (24). For RNA preparation, strain F837/76 was grown in 50 ml mMOD medium with or without 0.25% PGM, each in triplicates. The initial OD600 was 0.05. After 3 h, 10-ml samples were centrifuged for 10 min at 4,000 × g and 4°C. Cell pellets were stored at −80°C.
Sample preparation for RNA sequencing.
Total RNA preparation and on-column DNase digestion were performed using an RNeasy minikit (Qiagen) according to the instructions of the manufacturer. RNA quality was tested via spectrophotometer and agarose gel electrophoresis. RNA purity was confirmed via PCR for 16S rRNA (see Table S2 in the supplemental material for primers).
RNA sequencing.
RNA sequencing was performed at GATC Biotech (Konstanz, Germany). After initial quality control on an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Waldbronn, Germany) and rRNA depletion, random primed cDNA libraries were constructed according to the Illumina Genome Analyzer II protocol. Samples were sequenced via an Illumina HiSeq 2500 genome sequencer in 50-bp paired-end mode. On average, 20 million read pairs were achieved for each of the six samples. Initial data analysis included mapping of the samples against a reference genome (B. cereus F837/76, GenBank accession no. CP003187) and determination of gene expression. Differentially expressed genes between mMOD and PGM samples were further allocated to functional categories (Clusters of Orthologous Groups [27, 28]) using NCBI Conserved Domains Search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi).
qRT-PCR.
Double-stranded cDNA was synthesized using the ProtoScript first-strand cDNA synthesis kit (New England Biolabs, Inc.) with random primer mix according to the instructions of the manufacturer. One hundred ng RNA was used as the template. Quantitative real-time PCR (qRT-PCR) was carried out on a LightCycler 480 instrument (Roche). Samples were composed of 10 μl KAPA SYBR mix (Sigma-Aldrich), 0.4 μl forward primer, 0.4 μl reverse primer, 8.2 μl H2O, and 1 μl cDNA. The PCR program setup included the following steps: activation, 3 min at 95°C; amplification (40 cycles), 5 s at 95°C, 20 s at Ta (52 to 60°C [see Table S2]), and 8 s at 72 C° (2 s at 77/78°C). For some analyses, a fourth step was added to the amplification procedure to exclude primer dimers (Table S2). The melting curve was performed for 5 s at 95°C, 1 min at 60°C, and then continuously at 99°C. To determine primer efficiencies, dilution series of template cDNA were applied at least in duplicates (50, 10, 1, 0.1, 0.01, and 0 ng). Primer efficiencies were calculated from amplification curves using LightCycler 480 software (Roche) and are listed in Tables S1 and S2. To verify differentially expressed genes, generally 5 ng cDNA was applied. Other cDNA amounts are listed in Table S2. adk (adenylate kinase) and gatB (GatB/YqeY domain-containing protein) gene expression was used as a reference for normalization (63). Primer details are listed in Table S2. Differential gene expression was calculated using REST software (Qiagen) (64).
EIAs.
Indirect and sandwich enzyme immunoassays (EIAs) for the detection of enterotoxin components were performed as described before (65–67). The following monoclonal antibodies (MAbs) were used for detection: 5 μg/ml MAb 2B11/1E11-HRP, 1:4,000 (NheB), 1 μg/ml MAb 1A8/rabbit-anti-mouse-horseradish peroxidase (HRP) conjugate (Sigma), 1:2,000 (NheA), 10 μg/ml MAb 1A12/1H9-HRP, 1:2,000 (Hbl L2), and 5 μg/ml MAb 1D12/1B8-HRP, 1:2,000 (Hbl B). Results are shown as titers, which are defined as the reciprocal of the highest dilutions resulting in an absorbance value of ≥1.0.
Western blotting.
SDS-PAGE was performed on 12% Bis-Tris gels in a Criterion Cell (Bio-Rad) for 1 h at 200 V. Proteins were blotted to a polyvinylidene fluoride P membrane (Millipore). After 1 h of blocking in 3% casein-PBS, the membrane was incubated with 2 μg/ml NheB-specific MAb 1E11 (66) or with 1 μg/ml MUC2-specific antibody (abx177613; Abbexa Ltd.) for 1 h at room temperature. Three washing steps in PBS with 0.1% Tween 20 followed. After that, rabbit anti-mouse- or swine anti-rabbit-horseradish peroxidase conjugate (Dako) was applied diluted 1:2,000 in 1% casein-PBS. Three further washing steps in PBS with 0.1% Tween 20 and two in PBS followed. After that, Super Signal Western Femto maximum sensitivity substrate (Pierce) was applied, and chemiluminescence signals were detected on a Kodak imager (Eastman Kodak Company).
Mucin degradation assays.
Mucin degradation assays were performed according to reference 68. Briefly, 5 mg/ml (0.5%) PGM was dissolved in 0.5 ml Tris buffer (0.05 M Tris, pH 7.5). If appropriate, 50 μl protease inhibitor (cOmplete, with and without EDTA; Roche) were added. B. cereus overnight cultures were diluted in the samples to OD600 of 0.2. Alternatively, 50 μl B. cereus culture supernatants was added. Samples were incubated overnight at 37°C under gentle shaking and afterwards applied to SDS-PAGE on a PhastGel gradient (10% to 15%) minigel system (GE Healthcare). Proteins were fixed on the gel for two cycles of 30 min each in 50% methanol (MeOH) and 7% acetic acid and then incubated with 2 ml Sypro Ruby protein stain (Thermo Fisher) overnight at room temperature. The gel was washed in 10% MeOH and 7% acetic acid for 30 min and additionally in H2O for 10 min before fluorescence signals were detected on a Kodak imager (Eastman Kodak Company).
Germination.
B. cereus spores were prepared as previously described (69). Media to be tested were inoculated with the spores to an optical density of 1. Two hundred-μl samples (3 technical replicates each) were incubated at 37°C for 30 min to 1 h in 96-well plates. Every 3 min the optical density at 620 nm was measured in a Tecan photometer using Ridawin software. At least 2 replicates per plate were measured. Germination is depicted as the decrease of OD620 per unit of time. The OD620 at time point 0 was set to 100%.
Motility.
Motility of the B. cereus strains was assessed by investigating swimming behavior on mMOD medium supplemented with 0.25% agar. If appropriate, 0.05 or 0.1% PGM was added. To test the general swimming ability, 1 μl of an overnight culture (grown in CGY medium, OD600 adjusted to 20) was injected at the center of the plate. To test directed movement toward mucin, plates were divided into two halves, one containing mMOD with 0.25% agar and the other containing mMOD with 0.25% agar plus PGM. The overnight culture was injected into the sole mMOD side approximately 1 cm from the dividing line. Alternatively, 1 μl 0.25% PGM and 1 μl culture were injected into mMOD agar with approximately 2-cm distance. After 24 h of incubation at 37°C, growth and/or swimming were monitored. Motility assays were performed twice with at least three replicates for each condition at a time.
Enzymatic digestion of B. cereus culture supernatant.
Ten μl PBS or 0.25% PGM in PBS was added to 20 μl supernatant of B. cereus strain F837/76. The mixture was incubated for 2 h at 37°C with 1 μg/μl trypsin, chymotrypsin, pepsin, or pancreatin or for 15 min at 37°C with 0.1 and 0.01 μg/μl proteinase K. Subsequently, samples were mixed with loading buffer, incubated for 10 min at 95°C, and applied to SDS-PAGE and Western blotting with NheB-specific MAb 1E11 (described above).
In silico analysis of glycosyl hydrolases.
In silico analysis of the occurrence of glycosyl hydrolases involved in O-glycan mucin degradation in 48 B. cereus strains was performed using the Carbohydrate-Active enZYmes Database (www.cazy.org).
Statistical analyses.
Reciprocal titers obtained in all EIAs were statistically validated. For this purpose, data were analyzed using the column statistics program of GraphPad Prism, version 5.00, for Windows (GraphPad Software, San Diego, CA; www.graphpad.com). Unpaired t test was applied with two-tailed P values and 95% confidence intervals. All data were compared to the respective negative control (mMOD minimal medium). Significant differences were identified at a P value of <0.05 (*) or <0.01 (**).
Data availability.
RNA sequencing data have been deposited in the NCBI GEO database under accession number GSE127189.
Supplementary Material
ACKNOWLEDGMENTS
N.J. was responsible for bacterial growth and sample preparation, mucin and toxin degradation studies, germination and motility, RNA preparation, analysis of the transcriptome studies, and writing of the manuscript. A.-K.M. performed EIAs and Western blotting. C.D.R. contributed to bacterial growth and sample preparation and analysis of the transcriptome studies. R.D. and E.M. were involved in experimental setup and writing of the manuscript.
The IGF Project 18677 N of the FEI was supported via AiF within the program for promoting Industrial Collective Research (IGF) of the German Ministry of Economic Affairs and Energy (BMWi), based on a resolution of the German Parliament.
We thank Nele Maxseiner for excellent technical assistance.
We declare that the research was conducted in the absence of any conflict of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00765-18.
REFERENCES
- 1.Bansil R, Turner BS. 2006. Mucin structure, aggregation, physiological functions and biomedical applications. Curr Opin Colloid Interface Sci 11:164–170. doi: 10.1016/j.cocis.2005.11.001. [DOI] [Google Scholar]
- 2.Derrien M, van Passel MW, van de Bovenkamp JH, Schipper RG, de Vos WM, Dekker J. 2010. Mucin-bacterial interactions in the human oral cavity and digestive tract. Gut Microbes 1:254–268. doi: 10.4161/gmic.1.4.12778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dekker J, Rossen JW, Buller HA, Einerhand AW. 2002. The MUC family: an obituary. Trends Biochem Sci 27:126–131. doi: 10.1016/S0968-0004(01)02052-7. [DOI] [PubMed] [Google Scholar]
- 4.Desseyn JL, Tetaert D, Gouyer V. 2008. Architecture of the large membrane-bound mucins. Gene 410:215–222. doi: 10.1016/j.gene.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 5.Linden SK, Sutton P, Karlsson NG, Korolik V, McGuckin MA. 2008. Mucins in the mucosal barrier to infection. Mucosal Immunol 1:183–197. doi: 10.1038/mi.2008.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McGuckin MA, Linden SK, Sutton P, Florin TH. 2011. Mucin dynamics and enteric pathogens. Nat Rev Microbiol 9:265–278. doi: 10.1038/nrmicro2538. [DOI] [PubMed] [Google Scholar]
- 7.Naughton J, Duggan G, Bourke B, Clyne M. 2014. Interaction of microbes with mucus and mucins: recent developments. Gut Microbes 5:48–52. doi: 10.4161/gmic.26680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naughton JA, Marino K, Dolan B, Reid C, Gough R, Gallagher ME, Kilcoyne M, Gerlach JQ, Joshi L, Rudd P, Carrington S, Bourke B, Clyne M. 2013. Divergent mechanisms of interaction of Helicobacter pylori and Campylobacter jejuni with mucus and mucins. Infect Immun 81:2838–2850. doi: 10.1128/IAI.00415-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sperandio B, Fischer N, Sansonetti PJ. 2015. Mucosal physical and chemical innate barriers: Lessons from microbial evasion strategies. Semin Immunol 27:111–118. doi: 10.1016/j.smim.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 10.Alemka A, Corcionivoschi N, Bourke B. 2012. Defense and adaptation: the complex inter-relationship between Campylobacter jejuni and mucus. Front Cell Infect Microbiol 2:15. doi: 10.3389/fcimb.2012.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooke CL, An HJ, Kim J, Canfield DR, Torres J, Lebrilla CB, Solnick JV. 2009. Modification of gastric mucin oligosaccharide expression in rhesus macaques after infection with Helicobacter pylori. Gastroenterology 137:1061–1071. 1071.e1-8. doi: 10.1053/j.gastro.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 12.Sanchez B, Arias S, Chaignepain S, Denayrolles M, Schmitter JM, Bressollier P, Urdaci MC. 2009. Identification of surface proteins involved in the adhesion of a probiotic Bacillus cereus strain to mucin and fibronectin. Microbiology 155:1708–1716. doi: 10.1099/mic.0.025288-0. [DOI] [PubMed] [Google Scholar]
- 13.Tsilia V, Kerckhof FM, Rajkovic A, Heyndrickx M, Van de Wiele T. 2016. Bacillus cereus NVH 0500/00 can adhere to mucin but cannot produce enterotoxins during gastrointestinal simulation. Appl Environ Microbiol 82:289–296. doi: 10.1128/AEM.02940-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsilia V, Uyttendaele M, Kerckhof FM, Rajkovic A, Heyndrickx M, Van de Wiele T. 2015. Bacillus cereus adhesion to simulated intestinal mucus is determined by its growth on mucin, rather than intestinal environmental parameters. Foodborne Pathog Dis 12:904–913. doi: 10.1089/fpd.2014.1926. [DOI] [PubMed] [Google Scholar]
- 15.Miura T, Okamoto K, Yanase H. 2005. Purification and characterization of extracellular 1,2-alpha-L-fucosidase from Bacillus cereus. J Biosci Bioeng 99:629–635. doi: 10.1263/jbb.99.629. [DOI] [PubMed] [Google Scholar]
- 16.Clavel T, Carlin F, Lairon D, Nguyen-The C, Schmitt P. 2004. Survival of Bacillus cereus spores and vegetative cells in acid media simulating human stomach. J Appl Microbiol 97:214–219. doi: 10.1111/j.1365-2672.2004.02292.x. [DOI] [PubMed] [Google Scholar]
- 17.Senesi S, Ghelardi E. 2010. Production, secretion and biological activity of Bacillus cereus enterotoxins. Toxins (Basel) 2:1690–1703. doi: 10.3390/toxins2071690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stenfors Arnesen LP, Fagerlund A, Granum PE. 2008. From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS Microbiol Rev 32:579–606. doi: 10.1111/j.1574-6976.2008.00112.x. [DOI] [PubMed] [Google Scholar]
- 19.Wijnands LM, Dufrenne JB, van Leusden FM, Abee T. 2007. Germination of Bacillus cereus spores is induced by germinants from differentiated Caco-2 Cells, a human cell line mimicking the epithelial cells of the small intestine. Appl Environ Microbiol 73:5052–5054. doi: 10.1128/AEM.02390-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Didier A, Dietrich R, Märtlbauer E. 2016. Antibody binding studies reveal conformational flexibility of the Bacillus cereus non-hemolytic enterotoxin (Nhe) A-component. PLoS One 11:e0165135. doi: 10.1371/journal.pone.0165135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lund T, Granum PE. 1996. Characterisation of a non-haemolytic enterotoxin complex from Bacillus cereus isolated after a foodborne outbreak. FEMS Microbiol Lett 141:151–156. doi: 10.1111/j.1574-6968.1996.tb08377.x. [DOI] [PubMed] [Google Scholar]
- 22.Beecher DJ, Schoeni JL, Wong AC. 1995. Enterotoxic activity of hemolysin BL from Bacillus cereus. Infect Immun 63:4423–4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramarao N, Lereclus D. 2006. Adhesion and cytotoxicity of Bacillus cereus and Bacillus thuringiensis to epithelial cells are FlhA and PlcR dependent, respectively. Microbes Infect 8:1483–1491. doi: 10.1016/j.micinf.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 24.Jeßberger N, Dietrich R, Bock S, Didier A, Märtlbauer E. 2014. Bacillus cereus enterotoxins act as major virulence factors and exhibit distinct cytotoxicity to different human cell lines. Toxicon 77:49–57. doi: 10.1016/j.toxicon.2013.10.028. [DOI] [PubMed] [Google Scholar]
- 25.Jeßberger N, Krey VM, Rademacher C, Böhm M-E, Mohr A-K, Ehling-Schulz M, Scherer S, Märtlbauer E. 2015. From genome to toxicity: a combinatory approach highlights the complexity of enterotoxin production in Bacillus cereus. Front Microbiol 6:560. doi: 10.3389/fmicb.2015.00560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeßberger N, Rademacher C, Krey VM, Dietrich R, Mohr A-K, Böhm M-E, Scherer S, Ehling-Schulz M, Märtlbauer E. 2017. Simulating intestinal growth conditions enhances toxin production of enteropathogenic Bacillus cereus. Front Microbiol 8:627. doi: 10.3389/fmicb.2017.00627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tatusov RL, Galperin MY, Natale DA, Koonin EV. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tatusov RL, Koonin EV, Lipman DJ. 1997. A genomic perspective on protein families. Science 278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 29.Warda AK, Siezen RJ, Boekhorst J, Wells-Bennik MH, de Jong A, Kuipers OP, Nierop Groot MN, Abee T. 2016. Linking Bacillus cereus genotypes and carbohydrate utilization capacity. PLoS One 11:e0156796. doi: 10.1371/journal.pone.0156796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ho SB, Roberton AM, Shekels LL, Lyftogt CT, Niehans GA, Toribara NW. 1995. Expression cloning of gastric mucin complementary DNA and localization of mucin gene expression. Gastroenterology 109:735–747. doi: 10.1016/0016-5085(95)90380-1. [DOI] [PubMed] [Google Scholar]
- 31.Ho SB, Shekels LL, Toribara NW, Kim YS, Lyftogt C, Cherwitz DL, Niehans GA. 1995. Mucin gene expression in normal, preneoplastic, and neoplastic human gastric epithelium. Cancer Res 55:2681–2690. [PubMed] [Google Scholar]
- 32.Hollingsworth MA, Swanson BJ. 2004. Mucins in cancer: protection and control of the cell surface. Nat Rev Cancer 4:45–60. doi: 10.1038/nrc1251. [DOI] [PubMed] [Google Scholar]
- 33.Moravek M, Dietrich R, Buerk C, Broussolle V, Guinebretiére MH, Granum PE, Nguyen-The C, Märtlbauer E. 2006. Determination of the toxic potential of Bacillus cereus isolates by quantitative enterotoxin analyses. FEMS Microbiol Lett 257:293–298. doi: 10.1111/j.1574-6968.2006.00185.x. [DOI] [PubMed] [Google Scholar]
- 34.Granum PE, O'Sullivan K, Lund T. 1999. The sequence of the non-haemolytic enterotoxin operon from Bacillus cereus. FEMS Microbiol Lett 177:225–229. doi: 10.1111/j.1574-6968.1999.tb13736.x. [DOI] [PubMed] [Google Scholar]
- 35.Ryan PA, Macmillan JD, Zilinskas BA. 1997. Molecular cloning and characterization of the genes encoding the L1 and L2 components of hemolysin BL from Bacillus cereus. J Bacteriol 179:2551–2556. doi: 10.1128/jb.179.8.2551-2556.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clair G, Roussi S, Armengaud J, Duport C. 2010. Expanding the known repertoire of virulence factors produced by Bacillus cereus through early secretome profiling in three redox conditions. Mol Cell Proteomics 9:1486–1498. doi: 10.1074/mcp.M000027-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sastalla I, Fattah R, Coppage N, Nandy P, Crown D, Pomerantsev AP, Leppla SH. 2013. The Bacillus cereus Hbl and Nhe tripartite enterotoxin components assemble sequentially on the surface of target cells and are not interchangeable. PLoS One 8:e76955. doi: 10.1371/journal.pone.0076955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chaban B, Hughes HV, Beeby M. 2015. The flagellum in bacterial pathogens: for motility and a whole lot more. Semin Cell Dev Biol 46:91–103. doi: 10.1016/j.semcdb.2015.10.032. [DOI] [PubMed] [Google Scholar]
- 39.Duan Q, Zhou M, Zhu L, Zhu G. 2013. Flagella and bacterial pathogenicity. J Basic Microbiol 53:1–8. doi: 10.1002/jobm.201100335. [DOI] [PubMed] [Google Scholar]
- 40.Haiko J, Westerlund-Wikström B. 2013. The role of the bacterial flagellum in adhesion and virulence. Biology (Basel) 2:1242–1267. doi: 10.3390/biology2041242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skoog EC, Sjoling A, Navabi N, Holgersson J, Lundin SB, Linden SK. 2012. Human gastric mucins differently regulate Helicobacter pylori proliferation, gene expression and interactions with host cells. PLoS One 7:e36378. doi: 10.1371/journal.pone.0036378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tu QV, McGuckin MA, Mendz GL. 2008. Campylobacter jejuni response to human mucin MUC2: modulation of colonization and pathogenicity determinants. J Med Microbiol 57:795–802. doi: 10.1099/jmm.0.47752-0. [DOI] [PubMed] [Google Scholar]
- 43.Lillehoj EP, Kim BT, Kim KC. 2002. Identification of Pseudomonas aeruginosa flagellin as an adhesin for Muc1 mucin. Am J Physiol Lung Cell Mol Physiol 282:L751–L756. doi: 10.1152/ajplung.00383.2001. [DOI] [PubMed] [Google Scholar]
- 44.Giron JA, Torres AG, Freer E, Kaper JB. 2002. The flagella of enteropathogenic Escherichia coli mediate adherence to epithelial cells. Mol Microbiol 44:361–379. doi: 10.1046/j.1365-2958.2002.02899.x. [DOI] [PubMed] [Google Scholar]
- 45.Silva AJ, Pham K, Benitez JA. 2003. Haemagglutinin/protease expression and mucin gel penetration in El Tor biotype Vibrio cholerae. Microbiology 149:1883–1891. doi: 10.1099/mic.0.26086-0. [DOI] [PubMed] [Google Scholar]
- 46.Fang S, Wang L, Guo W, Zhang X, Peng D, Luo C, Yu Z, Sun M. 2009. Bacillus thuringiensis bel protein enhances the toxicity of Cry1Ac protein to Helicoverpa armigera larvae by degrading insect intestinal mucin. Appl Environ Microbiol 75:5237–5243. doi: 10.1128/AEM.00532-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cadot C, Tran S-L, Vignaud M-L, De Buyser M-L, Kolstø A-B, Brisabois A, Nguyen-Thé C, Lereclus D, Guinebretière M-H, Ramarao N. 2010. InhA1, NprA, and HlyII as candidates for markers to differentiate pathogenic from nonpathogenic Bacillus cereus strains. J Clin Microbiol 48:1358–1365. doi: 10.1128/JCM.02123-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guillemet E, Cadot C, Tran SL, Guinebretiére MH, Lereclus D, Ramarao N. 2010. The InhA metalloproteases of Bacillus cereus contribute concomitantly to virulence. J Bacteriol 192:286–294. doi: 10.1128/JB.00264-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tran SL, Guillemet E, Gohar M, Lereclus D, Ramarao N. 2010. CwpFM (EntFM) is a Bacillus cereus potential cell wall peptidase implicated in adhesion, biofilm formation, and virulence. J Bacteriol 192:2638–2642. doi: 10.1128/JB.01315-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chung MC, Popova TG, Millis BA, Mukherjee DV, Zhou W, Liotta LA, Petricoin EF, Chandhoke V, Bailey C, Popov SG. 2006. Secreted neutral metalloproteases of Bacillus anthracis as candidate pathogenic factors. J Biol Chem 281:31408–31418. doi: 10.1074/jbc.M605526200. [DOI] [PubMed] [Google Scholar]
- 51.Kolar SL, Ibarra JA, Rivera FE, Mootz JM, Davenport JE, Stevens SM, Horswill AR, Shaw LN. 2013. Extracellular proteases are key mediators of Staphylococcus aureus virulence via the global modulation of virulence-determinant stability. Microbiologyopen 2:18–34. doi: 10.1002/mbo3.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 53.Frederiksen RF, Paspaliari DK, Larsen T, Storgaard BG, Larsen MH, Ingmer H, Palcic MM, Leisner JJ. 2013. Bacterial chitinases and chitin-binding proteins as virulence factors. Microbiology 159:833–847. doi: 10.1099/mic.0.051839-0. [DOI] [PubMed] [Google Scholar]
- 54.Baida GE, Kuzmin NP. 1996. Mechanism of action of hemolysin III from Bacillus cereus. Biochim Biophys Acta 1284:122–124. doi: 10.1016/S0005-2736(96)00168-X. [DOI] [PubMed] [Google Scholar]
- 55.Gohar M, Faegri K, Perchat S, Ravnum S, Økstad OA, Gominet M, Kolsto AB, Lereclus D. 2008. The PlcR virulence regulon of Bacillus cereus. PLoS One 3:e2793. doi: 10.1371/journal.pone.0002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ramarao N, Sanchis V. 2013. The pore-forming haemolysins of bacillus cereus: a review. Toxins (Basel) 5:1119–1139. doi: 10.3390/toxins5061119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baida GE, Kuzmin NP. 1995. Cloning and primary structure of a new hemolysin gene from Bacillus cereus. Biochim Biophys Acta 1264:151–154. doi: 10.1016/0167-4781(95)00150-F. [DOI] [PubMed] [Google Scholar]
- 58.Bierne H, Sabet C, Personnic N, Cossart P. 2007. Internalins: a complex family of leucine-rich repeat-containing proteins in Listeria monocytogenes. Microbes Infect 9:1156–1166. doi: 10.1016/j.micinf.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 59.Minnaard J, Lievin-Le Moal V, Coconnier M-H, Servin AL, Pérez PF. 2004. Disassembly of F-actin cytoskeleton after interaction of Bacillus cereus with fully differentiated human intestinal Caco-2 cells. Infect Immun 72:3106–3112. doi: 10.1128/IAI.72.6.3106-3112.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Minnaard J, Rolny IS, Perez PF. 2013. Interaction between Bacillus cereus and cultured human enterocytes: effect of calcium, cell differentiation, and bacterial extracellular factors. J Food Prot 76:820–826. doi: 10.4315/0362-028X.JFP-12-294. [DOI] [PubMed] [Google Scholar]
- 61.Glatz BA, Goepfert JM. 1977. Production of Bacillus cereus enterotoxin in defined media in fermenter-grown cultures. J Food Prot 40:472–474. doi: 10.4315/0362-028X-40.7.472. [DOI] [PubMed] [Google Scholar]
- 62.Rosenfeld E, Duport C, Zigha A, Schmitt P. 2005. Characterization of aerobic and anaerobic vegetative growth of the food-borne pathogen Bacillus cereus F4430/73 strain. Can J Microbiol 51:149–158. doi: 10.1139/w04-132. [DOI] [PubMed] [Google Scholar]
- 63.Reiter L, Kolsto AB, Piehler AP. 2011. Reference genes for quantitative, reverse-transcription PCR in Bacillus cereus group strains throughout the bacterial life cycle. J Microbiol Methods 86:210–217. doi: 10.1016/j.mimet.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 64.Pfaffl MW, Horgan GW, Dempfle L. 2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dietrich R, Fella C, Strich S, Märtlbauer E. 1999. Production and characterization of monoclonal antibodies against the hemolysin BL enterotoxin complex produced by Bacillus cereus. Appl Environ Microbiol 65:4470–4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dietrich R, Moravek M, Burk C, Granum PE, Martlbauer E. 2005. Production and characterization of antibodies against each of the three subunits of the Bacillus cereus nonhemolytic enterotoxin complex. Appl Environ Microbiol 71:8214–8220. doi: 10.1128/AEM.71.12.8214-8220.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tausch F, Dietrich R, Schauer K, Janowski R, Niessing D, Märtlbauer E, Jessberger N. 2017. Evidence for complex formation of the Bacillus cereus haemolysin BL components in solution. Toxins (Basel) 9:E288. doi: 10.3390/toxins9090288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang P, Granados RR. 1997. An intestinal mucin is the target substrate for a baculovirus enhancin. Proc Natl Acad Sci U S A 94:6977–6982. doi: 10.1073/pnas.94.13.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Riol CD, Dietrich R, Märtlbauer E, Jessberger N. 2018. Consumed foodstuffs have a crucial impact on the toxic activity of enteropathogenic Bacillus cereus. Front Microbiol 9:1946. doi: 10.3389/fmicb.2018.01946. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data have been deposited in the NCBI GEO database under accession number GSE127189.