Abstract
The cotton‐melon aphid, Aphis gossypii Glover, is a worldwide‐spreading species, and pesticide‐resistant populations are increasing rapidly. In this study, investigations were performed based on Illumina HiSeq sequencing of the 16S rDNA V4 region for the bacterial communities embodied as intracellular symbionts under natural and in pesticide‐treated populations of A. gossypii. The results revealed that more than 82% of bacterial communities belonged to the phylum Proteobacteria in which the maximum proportion (53.24%) was of the genus Arsenophonus; Hamiltonella composed 22.31; and 1.37% was of the genus Acinetobacter. The relative abundance of Hamiltonella was obvious, vertically transmitted, divided into two groups, and its infection influenced the bacterial communities in A. gossypii. Symbiont density and composition were changed in samples tested on different days. Azadirachtin and phoxim influenced on the composition of bacterial communities. Different biomarkers were used for pesticide‐treated samples with LEfSe results. These findings will increase awareness regarding bacterial communities in naturally occurring populations of A. gossypii and pave the way to study the relationship between symbionts and pesticide resistance.
Keywords: 16S rDNA, agroecological environment, associated bacteria, azadirachtin, cotton‐melon aphid, Hamiltonella
1. INTRODUCTION
The cotton‐melon aphid, Aphis gossypii Glover, is a species that is increasing worldwide, colonizing more than 600 plant species (Blackman & Eastop, 2000). It causes serious economic losses in agriculture by feeding on plant foliage and spreading viral diseases in plants, such as cotton, cucurbits, and citrus (Thomas, Vanlerberghe‐Masutti, Mistral, Loiseau, & Boissot, 2016). It has been described as holocyclic in North China: eggs hatch in March and after two to three generations, late adults appear and move to cotton fields during late April to mid‐May. Then, its population increases rapidly and becomes the source of crop yield loss from the seedling stage (Wu & Guo, 2003; Xia, 1997).
Cotton‐melon aphid management is primarily dependent on the application of pesticides, such as the use of organophosphates, pyrethroids, and carbamates, but their indiscriminate use caused the development of resistance in its population, which is increasing rapidly (Cao, Zhang, Gao, Liang, & Guo, 2008). To date, the mechanism of pesticide resistance is mostly studied in aphids on the gene level. A. gossypii resistance to organophosphates is associated with esterase activity. It has been reported that overexpression of esterases shows higher esterase activity, and expression of mutant carboxylesterases shows lower esterase activity (Field, Blackman, Tyler‐Smith, & Devonshire, 1999; Sun, Zhou, Zhang, & Gao, 2005). Studies have reported that increased metabolic detoxification by increased esterase and oxidase enzymes or modification of voltage‐gated sodium channels decreases the target site sensitivity and induces resistance to pyrethroids (Carletto, Martin, Vanlerberghe‐Masutti, & Brevault, 2010). Similarly, it is stated that resistance mechanisms to neonicotinoid pesticides in aphids are based on enhanced detoxification of cytochrome P450 monooxygenases or reduced affinity of the nicotinic acetylcholine receptor for imidacloprid (Bass et al., 2011).
Including aphids, many insect species harbor endosymbionts known as primary endosymbionts and associated bacteria. The special primary endosymbiont Buchnera aphidicola is found in almost all aphid species. B. aphidicola is transmitted from mother to offspring with high fidelity and synthesizes essential amino acids and other nutrients that complement the host plant diets. Most aphid‐associated bacteria are thought to be vertically transmitted (Li et al., 2016) and have conditionally beneficial fitness consequences for the host, such as host adaptation (Tsuchida, Koga, & Fukatsu, 2004), increased resistance susceptibility to natural enemies and pathogenic fungi (Oliver, Russell, Moran, & Hunter, 2003; Scarborough, Ferrari, & Godfray, 2005; Vorburger & Rouchet, 2016), and tolerance to high heat shocks (Russell & Moran, 2006). A recent study stated that relative quantities of symbionts have been significantly affected by pesticide resistance (Pan et al., 2013), and gut symbionts enhance pesticide resistance in Bactrocera dorsalis (Cheng et al., 2017).
Some studies have been carried out on bacterial communities in A. gossypii populations, but bacterial communities and time trends in the natural populations are not clear. In this study, we used the Illumina HiSeq2500 platform, targeting the V4 region of the 16S rDNA to identify the bacterial communities of A. gossypii collected from the field at different times and compared them with bacterial communities of A. gossypii treated with various pesticides to verify whether the pesticides influence the composition of bacterial communities in A. gossypii.
2. MATERIALS AND METHODS
2.1. Experimental design
The non‐Bt cotton variety CCRI49 was sown in the field at a density of 45,000 hm2 in Anyang (36°5′34.8″N, 114°31′47.19″) during April 2016. The plot size for each treatment was kept to 8 × 5 m with three replications.
Five pesticides, including the target and nontarget, were applied against A. gossypii on 25 May 2016 when its peak reached to convergence and cotton has one true leaf, while the control treatment consisted of water only (Table 1).
Table 1.
Pesticide information in this study
| Pesticide (concentration) | Formulation | Dosage | Company |
|---|---|---|---|
| Deltamethrin (25 g/L) | EC | 450 ml/hm2 | Bayer Crop Science (China) Co., Ltd |
| Phoxim (40%) | EC | 450 ml/hm2 | Lianyungang Liben crop science and Technology Co., Ltd |
| Azadirachtin (100%) | / | 90 g/hm2 | Xi'an Realin Biotechnology Co., Ltd |
| Imidacloprid (10%) | WP | 225 g/hm2 | Jiangsu KWIN Group |
| Carbosulfan (20%) | EC | 225 ml/hm2 | Shandong Sino‐Agri United Biotechnology Co., Ltd |
EC, emulsifiable concentrate; WP, wettable powder.
Cotton seedlings along with A. gossypii were pulled out before pesticide application. Likewise, cotton seedlings along with A. gossypii were pulled out 3 days after and 7 days after pesticide application; then, apterous adult aphids were picked up in nuclease‐free Eppendorf tubes for 16S rDNA analysis. Fifty aphids from each were collected, mixed and considered as one sample; one individual per plant was collected to avoid the sampling of offspring from a single mother. To keep the endosymbiotic bacteria community stable, the aphids were picked up immediately after the cotton seedlings were removed, then the tubes with aphid samples were put into liquid nitrogen as soon as possible where they were stored until DNA extraction. The samples were processed one after another. In total, 54 samples were collected and immediately immersed in liquid nitrogen and stored at −80°C for further work.
2.2. DNA extraction
Prior to DNA extractions, each aphid sample was washed for 5 min in 70% ethanol and rinsed three times with sterile water in 2.0 ml Eppendorf tubes to remove surface contaminants. TIANamp Genomic DNA Kits (TIANGEN Biotech (Beijing) LTD., China) were used for DNA extractions. The protocol that was followed has been previously reported (Zhao et al., 2016). The quantity and quality of the DNA were measured with a NanoDrop 2000C spectrophotometer (Thermo Scientific, USA).
2.3. V4 region of 16S rDNA amplification and sequencing
The V4 region of the 16S rDNA were amplified using the 515F/806R primer; amplicon generation PCR products, quantification and qualification, PCR products mixing and purification, library preparation, and sequencing were done as previously reported (Zhao et al., 2016). Finally, the library was sequenced on an Illumina HiSeq2500 platform and 250 bp paired‐end reads were generated at Novogene Bioinformatics Technology (Beijing, China). Data from 54 samples were obtained independently.
2.4. Bioinformatics and statistical analysis
Paired‐end reads were assigned to samples based on their unique barcode and truncated by cutting off the barcode and primer sequence. Paired‐end reads were merged using FLASH (V1.2.7, http://ccb.jhu.edu/software/FLASH/) (Magoc & Salzberg, 2011). Quality filtering on the raw tags and sequence analyses were the same as previously reported (Zhao et al., 2016). Sequences with ≥97% similarity were assigned to the same OTUs. Each OTU was annotated as the taxonomic information used, based on the RDP classifier (Version 2.2, http://sourceforge.net/projects/rdp-classifier/) algorithm according to the GreenGene Database (Desantis et al., 2006; Wang, Garrity, Tiedje, & Cole, 2007).
To account for inequalities in the sequence read depths among the samples, the OTU abundance information was normalized by random selected sequences per sample according to the sequence number of the sample with the least sequences. Subsequent analysis of alpha diversity and beta diversity were performed based on this normalized output data. Alpha diversity analysis included observed species, ACE and Chao1 estimators, Simpson and Shannon diversity indices, and Good's coverage estimates. Linear discriminatory analysis effective size (LEfSe) analysis were performed (Segata et al., 2011). ANOSIM analysis was performed using R software (Version 2.15.3), with a statistical significance threshold of p < 0.05. SPSS 20.0 was used for data entry correlation analysis and one‐way ANOVA analysis.
3. RESULTS
3.1. Sequencing data
The Illumina HiSeq sequencing of the 16S rDNA V4 region amplicons from the natural population of A. gossypii yielded 137,579–194,290 raw reads per treatment (three repetitions) (Table 1). The quality filtering sequences were assigned to 264–1,981 OTUs at 97% sequence identity. The rarefaction curves tended to approach the saturation plateau for every sample (Table 2). Good's coverage estimations of sequencing data were above 99% obtained in all samples at a 0.03 dissimilarity cut‐off (Table 2).
Table 2.
Sequencing analysis of 16S rDNA of Aphis gossypii with diversity indices in each treatment
| Treatment name | Treatment time (d) | Total reads | Clean reads | OTU numbera | Observed species | Shannon | Simpson | Chao1 | Goods coverage |
|---|---|---|---|---|---|---|---|---|---|
| Control | 0 | 151,802 | 150,816 | 310 | 310 | 1.22 | 0.36 | 481.59 | 1.00 |
| 3 | 162,209 | 161,147 | 270 | 206 | 0.80 | 0.19 | 235.00 | 1.00 | |
| 7 | 165,958 | 164,615 | 1,631 | 1,379 | 3.48 | 0.69 | 1722.73 | 1.00 | |
| Azadirachtin | 0 | 175,807 | 174,467 | 1,178 | 973 | 1.87 | 0.40 | 1214.37 | 1.00 |
| 3 | 163,254 | 162,211 | 276 | 206 | 1.33 | 0.40 | 249.33 | 1.00 | |
| 7 | 151,671 | 150,444 | 1,266 | 1,135 | 3.49 | 0.73 | 1247.22 | 1.00 | |
| Carbosulfan | 0 | 137,579 | 136,384 | 1,506 | 1,349 | 3.07 | 0.55 | 1514.82 | 1.00 |
| 3 | 194,290 | 193,040 | 478 | 364 | 0.98 | 0.24 | 506.92 | 1.00 | |
| 7 | 145,628 | 144,637 | 517 | 460 | 3.06 | 0.77 | 495.13 | 1.00 | |
| Deltamethrin | 0 | 194,026 | 192,850 | 400 | 309 | 1.19 | 0.36 | 444.34 | 1.00 |
| 3 | 145,601 | 144,686 | 283 | 224 | 1.25 | 0.37 | 263.76 | 1.00 | |
| 7 | 155,283 | 153,899 | 1,981 | 1,751 | 4.03 | 0.68 | 1918.29 | 1.00 | |
| Imidacloprid | 0 | 162,818 | 161,677 | 1,264 | 1,060 | 1.74 | 0.35 | 1217.80 | 1.00 |
| 3 | 191,471 | 190,161 | 434 | 333 | 1.84 | 0.54 | 400.80 | 1.00 | |
| 7 | 151,407 | 150,106 | 1,040 | 954 | 3.95 | 0.79 | 1024.14 | 1.00 | |
| Phoxim | 0 | 180,764 | 179,574 | 375 | 296 | 1.92 | 0.63 | 344.12 | 1.00 |
| 3 | 172,909 | 171,790 | 264 | 192 | 0.84 | 0.21 | 277.41 | 1.00 | |
| 7 | 155,166 | 153,750 | 1,014 | 863 | 2.02 | 0.42 | 987.54 | 1.00 |
OTUs (operational taxonomic units) were defined with pairwise 97% sequence identity.
3.2. Overview of bacterial communities in Aphis gossypii
The average abundance values across 18 samples of aphid‐associated bacteria were analyzed before applying pesticides. Most bacterial communities were affiliated with the phylum Proteobacteria with a relative abundance of 82.44%. Three aphid‐associated dominant communities from Proteobacteria were Alphaproteobacteria, which remained at 1.79%, Betaproteobacteria at 0.87%, and Gammaproteobacteria at 79.78% (Figure 1). The most abundant associated bacterial families were Enterobacteriaceae, which accounted for 77.29%. In the genus type, there were 22 genera, which represented more than 0.1%. The most abundant genera were three from which the maximum proportion (53.24%) was of the genus Arsenophonus, while Hamiltonella remained at 22.31 and 1.37% for the genus Acinetobacter. The other genera were less than 1.00%. Unlike the associated bacteria, Buchnera was in the mycetocyte of aphids, usually known as primary symbionts, and its abundance in A. gossypii was 2.92‐fold of the total associated bacterial reads.
Figure 1.
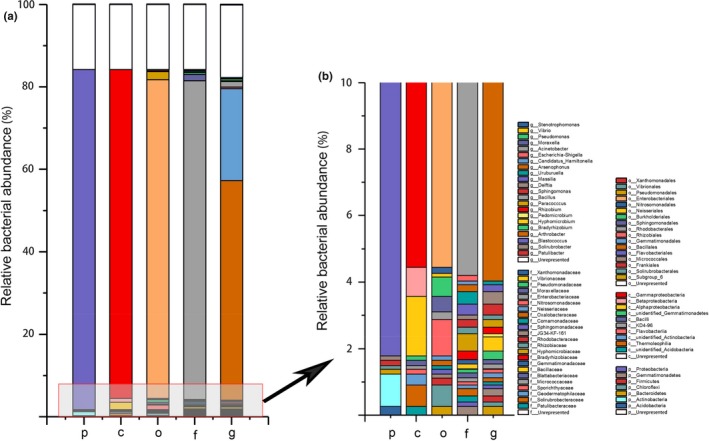
Associated bacteria in Aphis gossypii before pesticide application. Average relative abundance of bacterial operational taxonomic units (OTUs) identified across the 18 species before pesticide application on aphid samples at different levels (a) and enlarged part of the low abundance associated bacteria in A. gossypii (b). P: phylum‐level; C: class‐level; O: order‐level; F: family‐level; G: genus‐level
The abundance values of the top 10 most abundant OTUs were analyzed at the genus‐level across the pesticides and control. Generally, Arsenophonus, Hamiltonella, and Acinetobacter were the most abundant genera of aphid‐associated bacteria. Primary symbionts of Buchnera in A. gossypii were more abundant than total associated bacterial reads in most treatments. The abundance of Buchnera was decreased after insecticide applications except in phoxim‐treated samples, which have more abundant Hamiltonella before insecticide applications. Both in the insecticide treatment samples and control sample, the abundance of Acinetobacter was increased, but the abundance of Acinetobacter was higher in the insecticide treatment samples compared to the control sample, and there were 4.60–6.60% and 2.83%, respectively (Figure 2).
Figure 2.
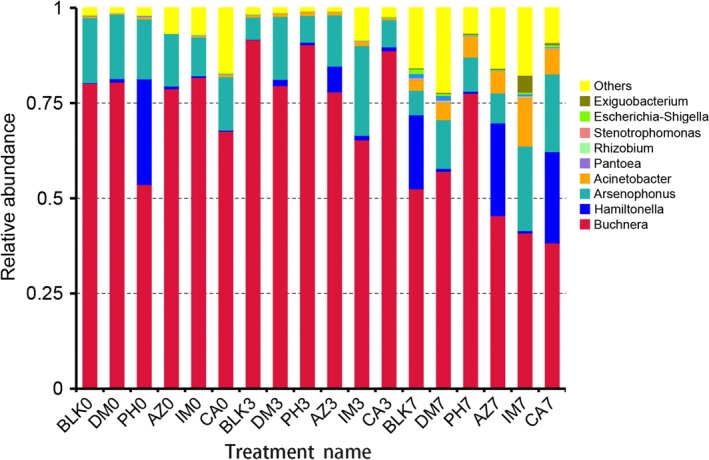
Top 10 most abundant associated bacteria in A. gossypii with different pesticides treatment. The abundance values of the OTUs were analyzed at the genus‐level across the pesticides and the control at the genus‐level. BLK0, BLK3, BLK7: used water as a control treatment plot before treatment and after treatment 3 and 7 days; AZ0, AZ3, AZ7: before application of azadirachtin and after application of azadirachtin 3 and 7 days; CA0, CA3, CA7: before application of carbosulfan and after application of carbosulfan 3 and 7 days; DM0, DM3, DM7: before application of deltamethrin and after application of deltamethrin 3 and 7 days; IM0, IM3, IM7: before application of imidacloprid and after application of imidacloprid 3 and 7 days; PH0, PH3, PH7: before application of phoxim and after application of phoxim 3 and 7 days
3.3. Infection of Hamiltonella
The relative abundance of Hamiltonella was divided into two groups of 54 samples, with 0.28–86.38% of the associated bacteria. The aphid population was divided into two groups by the relative abundance of Hamiltonella (p < 0.01). One group composed of eight samples had a high‐relative abundance (43.49–86.38%), and another group has a low‐relative abundance (0.28–17.04%) as shown in Figure 3. The eight high Hamiltonella‐infected samples included one sample collected before application of the pesticides, two samples that were collected at 3 days, and five samples that were collected at 7 days. The relative abundance was increased in the samples composed of a relatively high abundance of Hamiltonella (r = 0.51, p = 0.034), but not the samples that contained a relatively low abundance of Hamiltonella (r = 0.083 p = 0.316).
Figure 3.
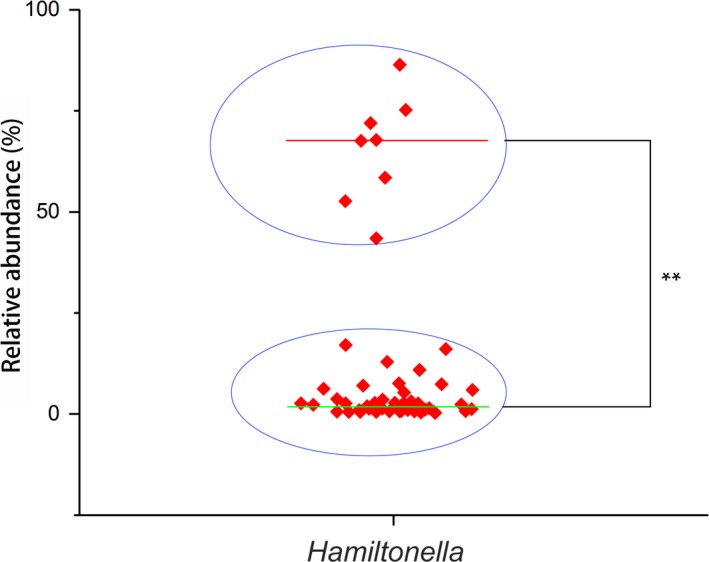
The relative abundance of Hamiltonella in A. gossypii samples. The 54 aphid populations were divided into two groups (as blue cycle show) by relative abundance of Hamiltonella. One group was composed of eight highly abundant populations, and another group has low relative abundance. “**”: one‐way ANOVA analysis use arcsin square root transformed data with SPSS 20.0, p < 0.01
The infection of Hamiltonella influenced the primary symbionts in aphids, the relative abundance of Buchnera, which were negative according to the relative abundance of Hamiltonella (r = −0.684, p < 0.01). In contrast, the infection of Hamiltonella did not influence the main aphid‐associated bacteria, such as Arsenophonus (r = −0.158, p = 0.253), Acinetobacter (r = −0.044, p = 0.749), Delftia (r = −0.033, p = 0.726), Escherichia‐Shigella (r = −0.034, p = 0.807), Pseudomonas (r = −0.011, p = 0.938), Rhizobium (r = 0.054, p = 0.700), Sphingomonas (r = 0.064, p = 0.643), and Vibrio (r = 0.113, p = 0.416).
3.4. The time trends of natural bacterial communities
In the plots that were used as controls with no pesticide application, the OTU numbers Shannon, Simpson, and Chao 1 decreased on day 3 and increased on day 7. This means that the most OTU numbers and most symbiont diversity were at day 7. The relative abundance of primary symbiont Buchnera remained stable among three sampling times (r = −0.489, p = 0.219).
At different sampling times, the relative abundance of Acinetobacter (r = 0.862, p = 0.006), Pseudomonas (r = 0.750, p = 0.032), and Rhizobium (r = 0.785, p = 0.021) were increased, whereas the relative abundance of Arsenophonus (r = 0.795, p = 0.018) was decreased, and the relative abundance of Sphingomonas (r = 0.575, p = 0.136) and Delftia (r = 0.518, p = 0.188) remained stable.
3.5. The influence of pesticide treatments on bacterial communities in Aphis gossypii
Bacterial communities between the naturally occurring aphid population and the pesticide‐treated population were analyzed. Most pesticides indicated no influence on the composition of bacterial communities (ANOSIM, p > 0.05). Bacterial communities in aphids were significantly affected by plant‐based pesticides, azadirachtin at 3 days (azadirachtin vs. carbosulfan p = 0.015, azadirachtin vs. phoxim p = 0.018). Bacterial community composition also varied between carbosulfan and imidacloprid (p = 0.041) at 3 days.
LEfSe results showed different biomarkers (LDA score > 2.0) in the pesticide‐treated samples and the control samples using the total bacterial communities or only the associated abundant bacterial features. In consideration of the total bacterial communities, biomarker Halomonas and Thermomonas were from plant‐based pesticides azadirachtin and phoxim‐treated samples, respectively. Thirty biomarkers were identified. When the associated bacteria were considered, there were 22 identified biomarkers that were identified from phoxim‐treated samples 5 days following azadirachtin treatment. Only one was from deltamethrin or imidacloprid or carbosulfan‐treated samples (Figure 4).
Figure 4.
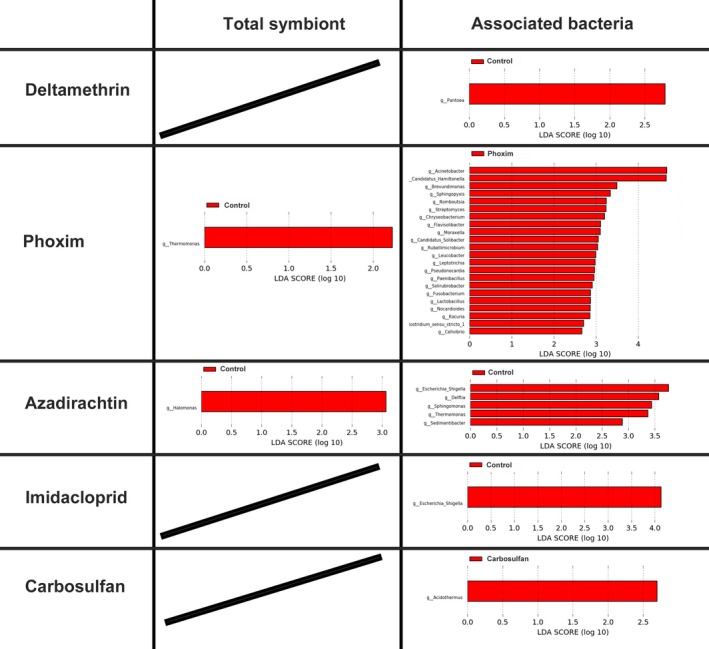
Pesticide treatment effects on bacterial diversity at the genus‐level. Total symbiont: both Buchnera primary symbionts and associated bacteria were included in the data analysis. Associated bacteria: only associated bacteria were included in the data analysis. Linear discriminant analysis (LDA) effect size (LEfSe) was used to identify specific phylotypes that were significantly associated with treatments at the genus‐level. An LDA more than two reflects significant differences between the groups. LEfSe analysis provided the list of phylotypes that are differential among dietary supplementations with statistical and biological significance
4. DISCUSSION
Taxonomic classification of insect symbionts revealed that bacterial communities were dominated by only a few phyla, including Proteobacteria, Bacteroidetes, Actinobacteria, and Firmicutes (Jones, Sanchez, & Fierer, 2013). Proteobacteria were the dominant bacterial phylum, including A. gossypii, in this study, with the dominant sub‐phylum Gammaproteobacteria. Similar to other insects, the Enterobacteriaceae family constituted the dominant populations in A. gossypii (Baumann, 2005; Yuval, Ben‐Ami, Behar, Ben‐Yosef, & Jurkevitch, 2013).
In addition to the Buchnera primary symbionts, aphids possess several associated bacteria, which show remarkable differences in morphology, localization, and quantity between lineages and were thought to be polyphyletic (Fukatsu, Tsuchida, Nikoh, & Koga, 2001). The model pea aphid species Acyrthosiphon pisum harbors several associated bacteria, including Serratia symbiotica, Hamiltonella defensa, Regiella insecticola, a Rickettsia sp., and Spiroplasma sp. (Fukatsu et al., 2001; Guay, Boudreault, Michaud, & Cloutier, 2009; Sakurai, Koga, Tsuchida, Meng, & Fukatsu, 2005; Tsuchida, Koga, Fujiwara, & Fukatsu, 2014). In DGGE, 5 bands closely related to Arsenophonus, Pseudomonas, Acinetobacter, Pelomonas, and Burkholderia were separated from malvaceous and cucurbitaceous populations within Japan and Australia (Najar‐Rodriguez, McGraw, Mensah, Pittman, & Walter, 2009). Eight bacterial groups, including Arsenophonus, Serratia, Rickettsia, and Wolbachia were cocolonized on Colocasia esculenta and Alpinia purpurata across the four Hawaiian Islands (Jones, Bressan, Greenwell, & Fierer, 2011). In this study, the relative abundances of 22 identified genera were more than 0.1%, including the three most abundant bacterial genera Arsenophonus, Hamiltonella and Acinetobacter and the three genera with low abundance (Pseudomonas, Burkholderia, Stenotrophomonas) that have been reported in A. gossypii (Jones et al., 2011; Najar‐Rodriguez et al., 2009; Zhao et al., 2016). None of the aphids surveyed were infected with the main endosymbiotic types (Serratia symbiotica, Regiella insecticola, Rickettsia, Spiroplasma) that were previously reported for other aphid species (Russell, Latorre, Sabater‐Munoz, Moya, & Moran, 2003).
Hamiltonella can be infected with bacteriophage APSEs that reduce the rate of successful parasitism by killing developing wasp larvae (Degnan & Moran, 2008; Laughton, Garcia, Altincicek, Strand, & Gerardo, 2011; Oliver et al., 2003; van der Wilk, Dullemans, Verbeek, & van den Heuvel, 1999). Hamiltonella appeared roughly in 14% aphid species (Oliver, Degnan, Burke, & Moran, 2010), it was present in all of our samples and the population was clearly divided into two groups by relative abundance. It indicated that Hamiltonella was vertically transmissible in A. gossypii, with lower abundance in the A. gossypii population; however, it is increasing rapidly in some populations for unknown reasons.
Hamiltonella provides strong protection against parasitoid wasps, but appears to have a negative effect on Aphis fabae longevity in the absence of parasitoids (Vorburger & Gouskov, 2011). In this study, we found that Hamiltonella has a negative effect on the abundance of the primary symbiont Buchnera in aphids, but not on the associated bacteria. As Buchnera synthesizes essential amino acids and other nutrients for their host aphids, we inferred that Hamiltonella has a negative effect on aphids, and reduces the abundance of Buchnera.
Environmental factors influenced the symbiont density and composition in normal plots, while both were changed depending on the sampled day. Symbiont density is found to differ significantly between the populations when reared under controlled environmental conditions in mealy bugs (Parkinson, Gobin, & Hughes, 2017). The density of the primary symbiont, Buchnera, in pea aphids tends to decrease with host age and increase with rearing temperature (Lu, Chiu, & Kuo, 2014). In this study, bacterial density variation was prospected in the natural adult A. gossypii population; the results showed the flexible dynamics of bacterial density over time, although the higher levels of symbionts have no clear benefit to the hosts and therefore appeared to be superfluous (Parkinson et al., 2017).
It was reported that the relative amounts of symbionts were affected significantly by pesticide resistance, and higher Wolbachia densities correlated with higher numbers of resistant genes in Culex pipiens (Duron et al., 2006). Similarly, high densities of Rickettsia were correlated with higher susceptibility to pesticides in Bemisia tabaci (Ghanim & Kontsedalov, 2009). Most studies have focused on the correlation of pesticide resistance to single symbiont densities, and the role of bacterial communities in insect pesticide resistance is still not clear. Because pesticide resistance of A. gossypii populations is rapidly increasing, we used low doses of pesticide sprayed on field aphids to study the bacterial communities' dynamics in A. gossypii. The results showed that the composition of bacterial communities was stable under four pesticide‐treated samples, but plant‐based pesticide azadirachtin depressed the bacterial communities in A. gossypii due to its bacteriostatic activity.
Insecticide applications enriched the insecticide‐degrading bacteria in the agroecosystem (Kikuchi et al., 2012), even though the composition of bacterial communities remained stable under phoxim treatment. There were 22 biomarkers identified at the genus‐level, all of them raised the relative abundance in the phoxim‐treated population. There were some types that are probably involved in insect resistance to pesticides, such as Acinetobacter, which was isolated from the midgut of a pesticide‐resistant H. armigera (Malhotra et al., 2012), and Chryseobacterium isolated from the pyridaben‐resistant population of Tetranychus urticae (Yoon et al., 2010). Many pesticides can be degraded by Kocuria, Nocardioides or Pseudonocardia (Bostanian & Akalach, 2006; Chakraborty & Das, 2016; Kumar, Kumar, & Sharma, 2016).
In this study, a broad characterization of the bacterial communities in naturally occurring populations and under pesticide‐treated populations of A. gossypii was achieved. Our sequencing data revealed that the relative abundances of 22 genera included more than 0.1% in A. gossypii from which Arsenophonus, Hamiltonella and Acinetobacter were the most common bacteria. The symbiont density and composition were changed on different sample days. Interestingly, Hamiltonella was obviously divided into two groups according to the relative abundance in A. gossypii. It indicated that Hamiltonella was vertically transmitted and infection of Hamiltonella could influence the bacterial communities in A. gossypii. Five pesticides were chosen, including azadirachtin and phoxim, which exhibited influence on the composition of bacterial communities. Different biomarkers were identified in the pesticide‐treated samples with LEfSe results. These findings contribute a new viewpoint to the bacterial communities in aphids, and pesticides can influence bacterial community composition in A. gossypii and pave the way to study the relationship between symbionts and pesticide resistance.
ACKNOWLEDGMENTS
This research was supported by Natural Science Foundation of China (No. 31572015) and National Special Transgenic Project of China (No. 2016ZX08012‐004).
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
AUTHOR CONTRIBUTIONS
JC and SZ conducted experiments. SZ and LW performed the experiments. SZ, JL, LZ, and XZ analyzed the data. SZ and WJ wrote the manuscript. All the authors reviewed the manuscript.
Zhang S, Luo J, Wang L, et al. Bacterial communities in natural versus pesticide‐treated Aphis gossypii populations in North China. MicrobiologyOpen. 2019;8:e652 10.1002/mbo3.652
REFERENCES
- Bass, C. , Puinean, A. M. , Andrews, M. , Cutler, P. , Daniels, M. , Elias, J. , … Field, L. M. (2011). Mutation of a nicotinic acetylcholine receptor β subunit is associated with resistance to neonicotinoid insecticides in the aphid Myzus persicae . Bmc Neuroscience, 12, 51. 10.1186/1471-2202-12-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann, P. (2005). Biology of bacteriocyte‐associated endosymbionts of plant sap‐sucking insects. Annual Review of Microbiology, 59, 155–189. 10.1146/annurev.micro.59.030804.121041 [DOI] [PubMed] [Google Scholar]
- Blackman, R. L. , & Eastop, V. F. (2000). Aphids on the World's crops. An identification guide. Chichester: Wiley‐Interscience. [Google Scholar]
- Bostanian, N. J. , & Akalach, M. (2006). The effect of indoxacarb and five other insecticides on Phytoseiulus persimilis (Acari: Phytoseiidae), Amblyseius fallacis (Acari: Phytoseiidae) and nymphs of Orius insidiosus (Hemiptera: Anthocoridae). Pest Management Science, 62, 334–339. 10.1002/(ISSN)1526-4998 [DOI] [PubMed] [Google Scholar]
- Cao, C. W. , Zhang, J. , Gao, X. W. , Liang, P. , & Guo, H. L. (2008). Overexpression of carboxylesterase gene associated with organophosphorous insecticide resistance in cotton aphids, Aphis gossypii (Glover). Pesticide Biochemistry and Physiology, 90, 175–180. 10.1016/j.pestbp.2007.11.004 [DOI] [Google Scholar]
- Carletto, J. , Martin, T. , Vanlerberghe‐Masutti, F. , & Brevault, T. (2010). Insecticide resistance traits differ among and within host races in Aphis gossypii . Pest Management Science, 66, 301–307. 10.1002/ps.1874 [DOI] [PubMed] [Google Scholar]
- Chakraborty, J. , & Das, S. (2016). Molecular perspectives and recent advances in microbial remediation of persistent organic pollutants. Environmental Science and Pollution Research, 23, 16883–16903. 10.1007/s11356-016-6887-7 [DOI] [PubMed] [Google Scholar]
- Cheng, D. F. , Guo, Z. J. , Riegler, M. , Xi, Z. Y. , Liang, G. W. , & Xu, Y. J. (2017). Gut symbiont enhances insecticide resistance in a significant pest, the oriental fruit fly Bactrocera dorsalis (Hendel). Microbiome, 5(1), 13 10.1186/s40168-017-0236-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan, P. H. , & Moran, N. A. (2008). Diverse phage‐encoded toxins in a protective insect endosymbiont. Applied and Environmental Microbiology, 74, 6782–6791. 10.1128/AEM.01285-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desantis, T. Z. , Hugenholtz, P. , Larsen, N. , Rojas, M. , Brodie, E. L. , Keller, K. , … Andersen, G. L. (2006). Greengenes, a chimera‐checked 16S rRNA gene database and workbench compatible with ARB. Applied and Environmental Microbiology, 72, 5069–5072. 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duron, O. , Labbe, P. , Berticat, C. , Rousset, F. , Guillot, S. , Raymond, M. , & Weill, M. (2006). High Wolbachia density correlates with cost of infection for insecticide resistant Culex pipiens mosquitoes. Evolution, 60, 303–314. 10.1111/j.0014-3820.2006.tb01108.x [DOI] [PubMed] [Google Scholar]
- Field, L. M. , Blackman, R. L. , Tyler‐Smith, C. , & Devonshire, A. L. (1999). Relationship between amount of esterase and gene copy number in insecticide‐resistant Myzus persicae (Sulzer). Biochemical Journal, 339, 737–742. 10.1042/bj3390737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukatsu, T. , Tsuchida, T. , Nikoh, N. , & Koga, R. (2001). Spiroplasma symbiont of the pea aphid, Acyrthosiphon pisum (Insecta: Homoptera). Applied and Environment Microbiology, 67, 1284–1291. 10.1128/AEM.67.3.1284-1291.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanim, M. , & Kontsedalov, S. (2009). Susceptibility to insecticides in the Q biotype of Bemisia tabaci is correlated with bacterial symbiont densities. Pest Management Science, 65, 939–942. 10.1002/ps.1795 [DOI] [PubMed] [Google Scholar]
- Guay, J. F. , Boudreault, S. , Michaud, D. , & Cloutier, C. (2009). Impact of environmental stress on aphid clonal resistance to parasitoids: Role of Hamiltonella defensa bacterial symbiosis in association with a new facultative symbiont of the pea aphid. Journal of Insect Physiology, 55, 919–926. 10.1016/j.jinsphys.2009.06.006 [DOI] [PubMed] [Google Scholar]
- Jones, R. T. , Bressan, A. , Greenwell, A. M. , & Fierer, N. (2011). Bacterial communities of two parthenogenetic aphid species cocolonizing two host plants across the Hawaiian islands. Applied and Environmental Microbiology, 77, 8345–8349. 10.1128/AEM.05974-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, R. T. , Sanchez, L. G. , & Fierer, N. (2013). A cross‐taxon analysis of insect‐associated bacterial diversity. PLoS ONE, 8, e61218 10.1371/journal.pone.0061218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi, Y. , Hayatsu, M. , Hosokawa, T. , Nagayama, A. , Tago, K. , & Fukatsu, T. (2012). Symbiont‐mediated insecticide resistance. Proceedings of the National Academy of Sciences of the United States of America, 109, 8618–8622. 10.1073/pnas.1200231109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, D. , Kumar, A. , & Sharma, J. (2016). Degradation study of lindane by novel strains Kocuria sp. DAB‐1Y and Staphylococcus sp. DAB‐1W. Bioresources and Bioprocessing, 3(1), 53 10.1186/s40643-016-0130-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughton, A. M. , Garcia, J. R. , Altincicek, B. , Strand, M. R. , & Gerardo, N. M. (2011). Characterisation of immune responses in the pea aphid, Acyrthosiphon pisum . Journal of Insect Physiology, 57, 830–839. 10.1016/j.jinsphys.2011.03.015 [DOI] [PubMed] [Google Scholar]
- Li, T. , Wu, X. J. , Jiang, Y. L. , Zhang, L. , Duan, Y. , Miao, J. , … Wu, Y. Q. (2016). The genetic diversity of SMLS (Sitobion miscanthi L type symbiont) and its effect on the fitness, mitochondrial DNA diversity and Buchnera aphidicola dynamic of wheat aphid, Sitobion miscanthi (Hemiptera: Aphididae). Molecular Ecology, 25, 3142–3151. 10.1111/mec.13669 [DOI] [PubMed] [Google Scholar]
- Lu, W. N. , Chiu, M. C. , & Kuo, M. H. (2014). Host life stage‐ and temperature‐dependent density of the symbiont Buchnera aphidicola in a subtropical pea aphid (Acyrthosiphon pisum) population. Journal of Asia‐Pacific Entomology, 17, 537–541. 10.1016/j.aspen.2014.03.012 [DOI] [Google Scholar]
- Magoc, T. , & Salzberg, S. L. (2011). FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics, 27, 2957–2963. 10.1093/bioinformatics/btr507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra, J. , Dua, A. , Saxena, A. , Sangwan, N. , Mukherjee, U. , Pandey, N. , … Lal, R. (2012). Genome sequence of Acinetobacter sp strain HA, isolated from the gut of the polyphagous insect pest Helicoverpa armigera . Journal of Bacteriology, 194, 5156 10.1128/JB.01194-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najar‐Rodriguez, A. L. , McGraw, E. A. , Mensah, R. K. , Pittman, G. W. , & Walter, G. H. (2009). The microbial flora of Aphis gossypii: Patterns across host plants and geographical space. Journal of Invertebrate Pathology, 100, 123–126. 10.1016/j.jip.2008.10.005 [DOI] [PubMed] [Google Scholar]
- Oliver, K. M. , Degnan, P. H. , Burke, G. R. , & Moran, N. A. (2010). Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annual Review of Entomology, 55, 247–266. 10.1146/annurev-ento-112408-085305 [DOI] [PubMed] [Google Scholar]
- Oliver, K. M. , Russell, J. A. , Moran, N. A. , & Hunter, M. S. (2003). Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proceedings of the National Academy of Sciences of the United States of America, 100, 1803–1807. 10.1073/pnas.0335320100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, H. P. , Chu, D. , Liu, B. M. , Xie, W. , Wang, S. L. , Wu, Q. J. , … Zhang, Y. J. (2013). Relative amount of symbionts in insect hosts changes with host‐plant adaptation and insecticide resistance. Environmental Entomology, 42, 74–78. 10.1603/EN12114 [DOI] [PubMed] [Google Scholar]
- Parkinson, J. F. , Gobin, B. , & Hughes, W. O. H. (2017). The more, the merrier? Obligate symbiont density changes over time under controlled environmental conditions, yet holds no clear fitness consequences. Physiological Entomology, 42, 163–172. 10.1111/phen.12186 [DOI] [Google Scholar]
- Russell, J. A. , Latorre, A. , Sabater‐Munoz, B. , Moya, A. , & Moran, N. A. (2003). Side‐stepping secondary symbionts: Widespread horizontal transfer across and beyond the Aphidoidea. Molecular Ecology, 12, 1061–1075. 10.1046/j.1365-294X.2003.01780.x [DOI] [PubMed] [Google Scholar]
- Russell, J. A. , & Moran, N. A. (2006). Costs and benefits of symbiont infection in aphids: Variation among symbionts and across temperatures. Proceedings of the Royal Society B‐Biological Sciences, 273, 603–610. 10.1098/rspb.2005.3348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai, M. , Koga, R. , Tsuchida, T. , Meng, X. Y. , & Fukatsu, T. (2005). Rickettsia symbiont in the pea aphid Acyrthosiphon pisum: Novel cellular tropism, effect on host fitness, and interaction with the essential symbiont Buchnera . Applied and Environmental Microbiology, 71, 4069–4075. 10.1128/AEM.71.7.4069-4075.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarborough, C. L. , Ferrari, J. , & Godfray, H. C. J. (2005). Aphid protected from pathogen by endosymbiont. Science, 310, 1781 10.1126/science.1120180 [DOI] [PubMed] [Google Scholar]
- Segata, N. , Izard, J. , Waldron, L. , Gevers, D. , Miropolsky, L. , Garrett, W. S. , & Huttenhower, C. (2011). Metagenomic biomarker discovery and explanation. Genome Biology, 12, R60 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, L. J. , Zhou, X. G. , Zhang, J. , & Gao, X. W. (2005). Polymorphisms in a carboxylesterase gene between organophosphate‐resistant and ‐susceptible Aphis gossypii (Homoptera: Aphididae). Journal of Economic Entomology, 98, 1325–1332. 10.1603/0022-0493-98.4.1325 [DOI] [PubMed] [Google Scholar]
- Thomas, S. , Vanlerberghe‐Masutti, F. , Mistral, P. , Loiseau, A. , & Boissot, N. (2016). Insight into the durability of plant resistance to aphids from a demo‐genetic study of Aphis gossypii in melon crops. Evolutionary Applications, 9, 756–768. 10.1111/eva.12382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida, T. , Koga, R. , Fujiwara, A. , & Fukatsu, T. (2014). Phenotypic effect of “Candidatus Rickettsiella viridis”, a facultative symbiont of the pea aphid (Acyrthosiphon pisum), and its interaction with a coexisting symbiont. Applied and Environmental Microbiology, 80, 525–533. 10.1128/AEM.03049-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida, T. , Koga, R. , & Fukatsu, T. (2004). Host plant specialization governed by facultative symbiont. Science, 303, 1989 10.1126/science.1094611 [DOI] [PubMed] [Google Scholar]
- Vorburger, C. , & Gouskov, A. (2011). Only helpful when required: A longevity cost of harbouring defensive symbionts. Journal of Evolutionary Biology, 24, 1611–1617. 10.1111/j.1420-9101.2011.02292.x [DOI] [PubMed] [Google Scholar]
- Vorburger, C. , & Rouchet, R. (2016). Are aphid parasitoids locally adapted to the prevalence of defensive symbionts in their hosts? Bmc Evolutionary Biology, 16, 271 10.1186/s12862-016-0811-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q. , Garrity, G. M. , Tiedje, J. M. , & Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and Environmental Microbiology, 73, 5261–5267. 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Wilk, F. , Dullemans, A. M. , Verbeek, M. , & van den Heuvel, J. F. (1999). Isolation and characterization of APSE‐1, a bacteriophage infecting the secondary endosymbiont of Acyrthosiphon pisum . Virology, 262, 104–113. 10.1006/viro.1999.9902 [DOI] [PubMed] [Google Scholar]
- Wu, K. M. , & Guo, Y. Y. (2003). Influences of Bacillus thuringiensis Berliner cotton planting on population dynamics of the cotton aphid, Aphis gossypii Glover, in northern China. Environmental Entomology, 32, 312–318. 10.1603/0046-225X-32.2.312 [DOI] [Google Scholar]
- Xia, J . 1997. Biological control of cotton aphid (Aphis gossypii Glover) in cotton (inter)cropping systems in China: a simulation study. Ph.D, Wageningen University.
- Yoon, C. , Indiragandhi, P. , Anandham, R. , Cho, S. , Sa, T. M. , & Kim, G. H. (2010). Bacterial diversity and distribution from the whole mite extracts in acaricide resistant and susceptible populations of twospotted spider mite‐Tetranychus urticae (Acari: Tetranychidae). Journal of the Korean Society for Applied Biological Chemistry, 53, 446–457. 10.3839/jksabc [DOI] [Google Scholar]
- Yuval, B. , Ben‐Ami, E. , Behar, A. , Ben‐Yosef, M. , & Jurkevitch, E. (2013). The Mediterranean fruit fly and its bacteria – potential for improving sterile insect technique operations. Journal of Applied Entomology, 137, 39–42. 10.1111/j.1439-0418.2010.01555.x [DOI] [Google Scholar]
- Zhao, Y. , Zhang, S. , Luo, J. Y. , Wang, C. Y. , Lv, L. M. , & Cui, J. J. (2016). Bacterial communities of the cotton aphid Aphis gossypii associated with Bt cotton in northern China. Scientific Reports, 6, 22958 10.1038/srep22958 [DOI] [PMC free article] [PubMed] [Google Scholar]


