Abstract
The clinical translation of promising products, technologies and interventions from the disciplines of nanomedicine and cell therapy has been slow and inefficient. In part, translation has been hampered by suboptimal research practices that propagate biases and hinder reproducibility. These include the publication of small and underpowered preclinical studies, suboptimal study design (in particular, biased allocation of experimental groups, experimenter bias and lack of necessary controls), the use of uncharacterized or poorly characterized materials, poor understanding of the relevant biology and mechanisms, poor use of statistics, large between-model heterogeneity, absence of replication, lack of interdisciplinarity, poor scientific training in study design and methods, a culture that does not incentivize transparency and sharing, poor or selective reporting, misaligned incentives and rewards, high costs of materials and protocols, and complexity of the developed products, technologies and interventions. In this Perspective, we discuss special manifestations of these problems in nanomedicine and in cell therapy, and describe mitigating strategies. Progress on reducing bias and enhancing reproducibility early on ought to enhance the translational potential of biomedical findings and technologies.
For a product, technology or intervention to warrant clinical trials, there must be sufficient preclinical evidence of safety and efficacy. However, the clinical translation of promising fundamental discoveries and preclinical approaches in nanomedicine and cell therapy, which hold great promise for the design of future medical interventions and for the improvement of current medical technologies, has been challenging and inefficient1,2. Much of the difficulty to achieve the desirable clinical translation may stem from lack of reproducibility and from biases in the early stages of the translational pipeline3,4. Lack of reproducibility does not necessarily mean that research done in these fields is flawed. It may herald genuine heterogeneity in biological and experimental systems5,6 that is poorly controlled or not well understood. It may also point to the presence of biases that are identifiable and correctable preemptively. Biases may pertain to how single studies are designed, reported and disseminated, or used for building future work. In this Perspective, we define criteria for designing preclinical studies that minimize bias and maximize reproducibility, with a focus on studies in the active and promising disciplines of nanomedicine and cell therapy. We also discuss the potential sources of genuine heterogeneity and bias that arise in typical experimental studies in these two disciplines, and how to handle these to improve the prospects of clinical translation. Because we examine the issues side-by-side, we hope that the lessons learnt can be extrapolated to other fields in biomedicine and biomedical engineering.
Biases and lack of reproducibility
Several empirical studies have evaluated problems of reproducibility and the presence of major biases in diverse types of preclinical research4–15. One approach is the conduct of reproducibility checks, where investigators try to repeat previously published experimental studies, following as closely as possible the methods, materials, procedures and analyses used in the original study. This typically involves communication with the original investigators to clarify how exactly to design and execute the experiments, and feedback ensures that the reproducibility check is a close replica of the original. Nevertheless, the level of involvement and prior endorsement of the original investigators can vary. This leaves room for debate when results are not reproduced7,8. For some early reproducibility checks, full data have not been made available9,10; yet those that are ongoing, especially in cancer biology11, are more transparent, providing thorough protocols and statements of data availability, and even making use of pre-registration7,11. Even then, results that can’t be reproduced can create debate and emotional reactions. Allowing for these caveats, reproducibility checks in preclinical biomedical research have yielded very low rates of successful replication. For example, only 20–25% of the 67 preclinical studies in general biology that were being considered for translational efforts in oncology (47 of them), or in applications in women’s health (12 studies) and cardiovascular disease (8 studies), could be reproduced9. Typically, inconsistencies between published data and in-house data resulted in termination of the projects because of halted investment (in this case, from industry). Similarly, only 11% (6 of 53) of oncology drug-target studies published by academic investigators could be reproduced10. Moreover, the first released results of the Reproducibility: Cancer Biology project8,11 have shown that among the first five highly cited studies assessed only two could be reproduced as originally planned. To date, reproducibility checks are available in relatively small numbers, and they cover some selected fields of preclinical research, with oncology having the lions share, and research in cardiovascular and neurological diseases having smaller numbers of attempts. There is different sensitization across preclinical disciplines about the need to probe the status quo of the reproducibility of highly influential research.
There is far more evidence that indirectly suggests that reproducibility in preclinical research may be low because of the high prevalence of biases and of suboptimal research practices12. Several evaluations have shown that most preclinical studies are too small, which increases the chances of false-negative and false-positive results and of exaggerated conclusions. For example, in neuroscience, even with lenient assumptions, the average power of an experiment is about 20%13,14. Although most animal experiments in the context of neurological diseases find significant results, very few of these materialize in human applications15. Some pivotal aspects of study design, such as randomization and the blinding of investigators who analyse the results of animal experiments, are used in less than 20% of studies16–18, even though they are easy to adopt and are indispensable in order to reduce experimenter bias. The use of statistical methods has been demonstrated to be suboptimal in some fields. Major threats that have high prevalence include the misuse and misinterpretation of null-hypothesis testing methods19,20, the over-reliance on P values with lenient thresholds (such as P < 0.05)21, P-hacking22,23 (where investigators perform multiple analyses sequentially, for example by adding more samples or excluding some potential outliers until the desired thresholds of statistical significance are achieved), and insufficient consideration of the multiplicity involved in statistical analyses (many fundamental and preclinical experiments involve concurrent testing of multiple hypotheses). These deficiencies are not the result of fraud, which is rare in science at large, but reflect questionable and detrimental research practices, which most researchers unfortunately adopt24. The use of questionable and detrimental research practices is probably caused by the lack of sufficient methodological and statistical expertise, and can be affected by the expectation and pressure to deliver significant results. Substandard practices that are overspread create a vicious circle, because currently published work refers to previously published studies to justify the perpetuation of such practices: for example, a small sample size or the absence of adjustment for multiplicity is justified on the basis of small sample sizes and the lack of adjustment for multiplicity in previous studies in the field.
Empirical studies of biases have been performed across disciplines. It is however difficult to compare notes between different empirical studies and conclude that one field of preclinical research employs better research practices than another. Nevertheless, the challenges seem to be widespread and pervasive enough to warrant immediate and careful attention. Various calculations estimate that tens of billions of US dollars are wasted annually owing to lack of reproducibility in preclinical research25,26.
Strategies to minimize bias and to facilitate reproducibility
In preclinical research, it is essential that the design of a study includes sample-size calculations to ensure the adequate power to detect what might be considered to be effect sizes for the intervention that would warrant optimism in further clinical testing, proper randomization (for experimental work), the blinding of the investigators who read outcomes of experiments so as to obviate experimenter bias, the use of appropriate positive and negative controls, pre-specified criteria for any data exclusions to avoid post-hoc exclusion of samples and data points, the use of suitable statistical tests, with full consideration of the multiplicity of the analyses that are expected to be performed, and the avoidance of P-hacking. Ideally, all of these considerations can be pre-specified and even registered ahead of performing the experiments, thus enhancing trust in the ensuing results. Nevertheless, sometimes not all study-design features may be possible to pre-specify. This may be far more common in exploratory or discovery-driven research than in late preclinical research, when the goals of the experiments can be predetermined more clearly. It is thus essential to be transparent about which aspects of the study and analyses are exploratory and which are pre-specified27. Exploratory studies and analyses should be seen as hypothesis-generating, and the findings would require explicit confirmation in rigorous, hypothesis-testing evaluations.
Depending on the type of experimental system, standardization and verification of the key materials used (such as cell types or specimens) is indispensable to minimize noise and errors28. Careful thinking should go into capturing and removing known sources of biological heterogeneity by standardizing experiments for these factors. Often, there may be many different pathways of experimentation in different models. Priority should be given to models that come as close as possible to human applications in terms of relevance, and to capturing more closely the disease mechanisms. When several models are available for testing, their results can be compared, as they offer some sort of conceptual replication. Consistent results in different, independent models strengthen the evidence. However, for pivotal results that are considered influential in the decision to move a technology to clinical experimentation in humans, it is essential to also replicate the results of each pivotal model independently in new samples or animals, and ideally by independent investigators.
Collaboration between investigators can increase both the credibility and trust of the research and its translational potential. Joining efforts from specialists in different disciplines may promote out-of-the-box interdisciplinary approaches. Furthermore, for investigators who do similar work (for example, on similar animal models), collaborative multi-site studies29 may allow for the optimization of power and for the evaluation of the consistency of results across multiple laboratories. Results that show consistent promise across several laboratories may have better chances of success in further translational efforts. Moreover, comparing notes when results from different laboratories diverge can reveal potential sources of heterogeneity and lead to the design of future experiments that account for such heterogeneity.
Design options and statistical options are becoming increasingly complex for some types of experiment. Therefore, it is essential that either the experimenters have sufficient training in the methods that they use, or that an expert who is experienced in the employed methodologies is included in the research team as early as possible (when the study is being designed, and certainly before actual experiments are run). This may spare embarrassments and wasted effort and resources. Given that methods evolve and need to be adapted to new measurement tools and increasingly complex technologies, investigators require continuing methodological education. Statistical choices need to consider whether the question of interest is suitably answered with null-hypothesis significance testing or by other approaches, such as Bayesian30 or false-discovery-rate31 methods. Also, they need to account properly for any multiplicity involved in the testing and for the correlation structure (lack of independence) between multiple hypotheses. Given how frequently P-values are misinterpreted32,33, it would be best to shift attention away from them and towards more meaningful alternatives that present the magnitude of the effects, their uncertainty, and how big the risk of false positives is34,35. Sharing of data, protocols, materials, software and code enhances the trust in the research and its results, and allows other scientists to use these resources in further experiments, to verify, extend and expand previous work36,37. Therefore, a culture of open sharing is likely to accelerate the translation of successful lines of investigation, may allow picking errors and sources of heterogeneity and bias earlier in the process, and may help abandon non-working options faster.
Besides improved and standardized design and analysis, the reporting of research in scientific publications can also be improved and standardized. The EQUATOR website38 includes multiple reporting guidance and recommendations for different types of research. For preclinical animal experiments, the ARRIVE39 guidelines are most relevant, and most journals would benefit from adopting them.
Study registration ahead of study execution may help to improve transparency and to differentiate between unspecified post-hoc explorations and pre-specified design and analysis features. Registration can be performed with various levels of pre-specified detail. One thorough option is the registration of full reports, where practically the paper is peer-reviewed and accepted by a journal before the experiments are done on the basis of explicit, detailed descriptions of the proposed design, experiments and analyses. After running the experiments, the proposed tables and figures are then populated according to the pre-specified plan, and the paper is peer-reviewed again and published40.
One cannot overstress the importance of having proper incentives and rewards that place emphasis on efficient clinical translation rather than just on publishing nice-looking, statistically significant results. There is always the risk that academic investigators are rewarded mostly for publishing their work in high-impact journals and for attracting funding, rather than for high-quality work, for sharing their resources and obtaining reproducible results, and eventually for delivering discoveries that can be translated into effective human treatments41,42.
Because preclinical studies can sometimes be expensive to carry out, it is important to have a clear sense ahead of time, in the early phases of designing an experiment or a series of experiments, of what the cost is expected to be. It is useful also to try to anticipate whether this cost may be higher, and by how much, if different aspects of the experimentation do not proceed as originally planned. This will allow assessing the feasibility of the proposed experiments and ensuring that the experimentation course is likely to be completed. Furthermore, one should consider the complexity of the developed product and try to think from the early steps what approaches might make the desired product simpler yet still effective, and what might the realistic requirements for its synthesis and fabrication be? Making choices between different avenues of experimentation should be guided by the anticipated clinical use of the developed product, technology or intervention. Except for early exploratory science, which should be unconstrained by pre-conceptions, once specific hypotheses and experiments are conceived one should always keep in mind the real clinical need of the new product, technology or intervention.
Each of the approaches discussed in this section, and summarized in Table 1, may help contain and minimize the threats of bias and lack of reproducibility, and improve clinical translatability. In what follows, we discuss from a biomedical engineering perspective (rather than from fundamental or exploratory standpoints) reproducibility and translatability challenges and solutions that are specific to nanomedicine and cell therapy.
Table 1 |.
Reproducibility threats and solutions in preclinical work
| Threat | Solutions |
|---|---|
| Small underpowered studies | Proper sample size (power calculations) Collaborative multi-site studies |
| Biased allocation | Proper randomization (unbiased generation of the randomization sequence) |
| Experimenter bias | Blinding of the investigators reading the outcomes |
| Lack of sufficient controls | Proper use of positive and negative controls |
| Uncharacterized or poorly characterized materials | Standardization and verification of materials |
| Poor understanding of the biology or its mechanisms | Understanding biological heterogeneity |
| Irrelevance to humans | Use of best approximate to the human application |
| Large between-model heterogeneity | Replication in different models |
| Absence of replication | Independent replication of the same model |
| Lack of interdisciplinarity | Collaborative studies with diversity in expertise |
| Poor training of scientists in study design and methods | Design literacy, continuing methodological education and involvement of methodologists |
| Poor use of statistics | Statistical literacy and numeracy |
| Lack of transparency and data sharing | Sharing of data, protocols and materials |
| Poorly reported research | Reporting standards (such as the ARRIVE guidelines) |
| Publication bias and other selective reporting | Study registration, registered reports |
| Misaligned incentives and rewards | Focus on clinical translation rather than just publication |
| High costs of materials and protocols | Early design considerations in terms of the overall costs |
| Complexity of the product | Early design considerations in terms of the overall simplicity of the product and of the synthesis and fabrication protocols to make it |
Tackling reproducibility and bias in nanomedicine
Nanomedicine aims to develop therapeutic or diagnostic systems by using synthetic or naturally derived nanomaterials to address biomedical problems related to human diseases. Significant effort has been dedicated to developing nanomedicines for autoimmune and infectious diseases43–45, and the past decade has witnessed an explosion in the development of nanomedicines for cancer therapy and diagnostics (and for theranostics)46–48. Nanomedicine offers unique advantages over conventional approaches, including the versatility of nanoparticle design and the ease of modifying the physical and chemical properties of nanomaterials to achieve a specific function. However, the diversity in the physical, chemical and biological properties of material substrates used for nanomedicines also highlights the potential for significant heterogeneity and for bias, making the generalization of observed phenomena difficult, and limiting the translational potential of discoveries47,48.
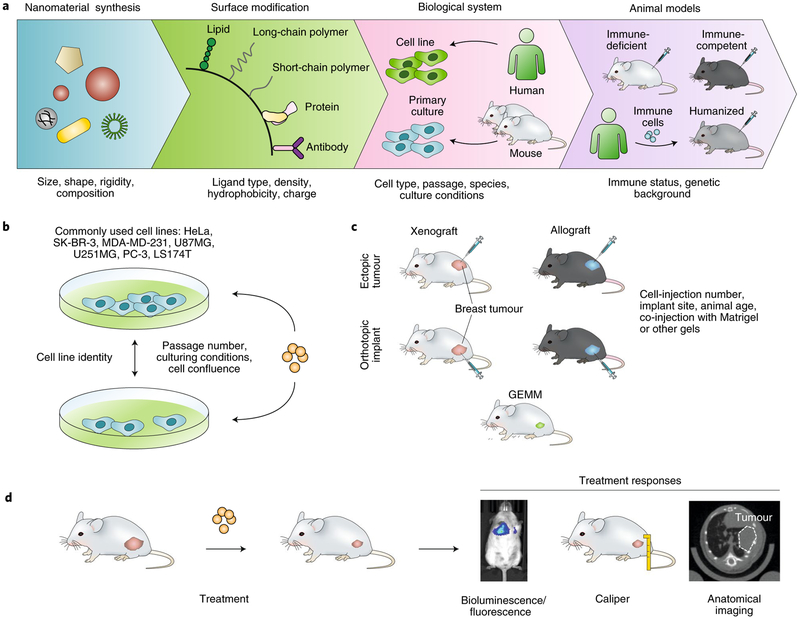
Figure 1a shows a typical workflow for the development of a complex multifunctional nanomedicine. From the initial step of material synthesis to the preclinical testing of candidate systems in vivo, experimental variations along each developmental stage can contribute to potential biases and reduce reproducibility. Biases may be further amplified during successive steps. Although the versatility of nanomaterial synthesis enables researchers to readily develop nanomedicines with unique physicochemical properties tailored to specific biological applications, it also makes the generalizability of nanomedicine studies difficult. For example, nanomaterial properties such as size, geometry, material composition and surface chemistry can greatly influence the interactions of the nanomaterials with biological systems49,50. Indeed, these properties strongly dictate nanoparticle interactions with proteins and cells, their distribution within tissues, and their clearance by the renal and hepatic systems in vivo. Therefore, many studies that examine nanomedicine behaviour in biological systems apply only to the specific nanomaterial used, with limited generalizability. Hence, if any unknown or uncontrolled variations in the properties of nanomaterials are present, the experimental outcome may differ.
Fig. 1 |. Examples of heterogeneity and potential bias arising at each step of the preclinical research process in nanomedicine.
a, Nanomedicine studies face challenges in controlling for differences in (from left to right): the physical and chemical properties of the nanomaterial substrate, surface chemistries, interactions with specific biological systems and the animal models used to characterize in vivo end points. Humanized mice are generated by grafting human haematopoietic stem or progenitor cells into immune-deficient animals to reconstitute a functional human immune system. b, For cancer-nanomedicine studies, standardization of culturing conditions and validation of the identity of commonly used cancer cell lines are critical for minimizing variations for in vitro experiments. c, For in vivo experiments, different preclinical models can produce unique biological responses for the same nanomedicine because of heterogeneities introduced in establishing the tumour models. GEMM, genetically engineered mouse model. d, Measurements of treatment-response parameters also need to be standardized across different studies. This is especially important for the correlation of physical tumour growth with tumour growth measured with imaging-based techniques, for example via expression levels of luciferase or other fluorescent proteins in transfected cell lines and via the normalization of attenuation coefficients in bioluminescence imaging (left) or computed tomography (right). Tumours are in pink and nanomaterials are in orange.
Although some heterogeneity across different nanomedicine systems is unavoidable, nanomedicine interventions are often intentionally designed to achieve specific experimental outcomes. Batch-to-batch variability on the other hand, is a direct result of the lack of precision in the synthesis of the nanomaterial. For example, geometric uniformity is a key feature for nanomaterial synthesis. For colloidal nanoparticles, synthetic techniques aim to produce a monodisperse nanoparticle population with a narrow size distribution. However, current methods for nanomaterial synthesis depend predominantly on bulk solution chemistry, which limits the production of highly monodisperse and homogenous nanomaterials on a large scale. Recent advances in synthetic techniques using high-precision fabrication, such as particle replication in non-wetting templates and microfluidic-based reaction chemistry, show promise for the production of more precisely defined microparticles and nanoparticles51,52. These technological advancements now offer protocols for nanomaterial synthesis that are compliant with good manufacturing practice (GMP), thus greatly reducing inter-batch variability and potentially improving the reproducibility of nanomedicine studies.
Beyond the intrinsic heterogeneities of synthetic nanomaterials, a significant degree of diversity also exists in the surface chemistry of nanomaterials designed for biomedical applications. The wide variety of surface-modification techniques enables nanomaterials to be designed and tailored for specific applications, such as the targeting of specific cells or tissues, or the evasion of systemic clearance or of physiological barriers47,53–56. However, the diverse selection of nanoparticle-surface modifications also magnifies the issues associated with sample heterogeneity and experimental reproducibility. For example, small variations in the density and distribution patterns of ligands on nanoparticle surfaces can significantly influence the way that nanoparticles interact with biological systems57,58. Variations introduced in the surface chemistry of nanomaterials can be further augmented by differences in the intrinsic properties of the nanomaterial itself to produce a synergistic effect in which changes in both nanomaterial size and ligand density can have a more profound effect on the binding kinetics to specific cell-membrane receptors when compared with changes to only one of these variables57,59. International standards in measuring the material and biological properties of nanoparticles have been established by organizations such as the International Organization for Standardization (ISO)60. Similarly, validation studies should be carried out by comparing independent experimental duplicates against reference materials and by measuring the most relevant performance characteristics, including nanomaterial size, material composition and surface charge, and their distribution profiles. This will require the establishment of a large library of reference materials for nanoparticles, such as the libraries available from the National Institute of Standards and Technology (NIST)61.
Experimental reproducibility can be further improved by standardizing the biological model systems used to study the translational potential of nanomedicines. Earlier studies in cancer nanomedicine, for example, largely rely on in vitro characterization of nanomedicine effects against long-term cultured immortalized human cancer cell lines (Fig. 1b). Although these cell lines are publicly available from the American Type Culture Collection and from other repositories, there are growing concerns regarding the true identity of many publicly available human-tumour-derived cell lines62. Careful validation of cell lines used for nanomedicine studies is thus essential to ensure experimental reproducibility. Further complicating in vitro studies is the lack of standardized culturing conditions. For example, studies using the classic U87MG and U251MG glioma cell lines have shown that when grown in serum-containing medium these glioma cells exhibit transcrip-tomic deviation from the original tumour from which they were derived63. Only cells maintained in serum-free media retain their original tumour-specific phenotype64. Therefore, small differences in cell-culturing conditions, such as serum supplementation, passage numbers and cell confluency, may result in different biological effects for the nanomedicine under study. These parameters need to be explicitly reported and properly controlled during experimentation. The variability in in vitro model systems also affects the reproducibility of nanomedicine studies beyond cancer. For example, multiple studies on nanoparticle clearance by the mononuclear phagocytosis system were conducted using immortalized human THP-1 cells (THP-1 is a human monocytic cell line derived from the peripheral blood of an acute monocytic leukaemia patient65). The phagocytic capacity of these human cells may differ significantly from that of murine bone marrow-derived macrophages or of tissue-specific phagocytes such as hepatic Kupffer cells or alveolar macrophages, thus resulting in potential conflicting results under different experimental conditions.
Beyond cell-culture studies, preclinical in vivo experiments using nanomedicines should be carefully designed and the results reported in sufficient detail. A significant degree of experimental heterogeneity exists in the nanomedicine literature, in particular for cancer-nanomedicine studies, owing to the wide range of tumour models used. A large amount of reported cancer-nanomedicine studies rely on the use of human-xenograft tumours, derived from long-term cultured cell lines that are implanted in immune-compromised mice. Variations in the site of tumour inoculation (orthotopic versus ectopic sites), number of passages of the cell line and whether extracellular-matrix materials were used to assist tumour cell implantation, can all affect the biological properties of the tumour and result in different experimental outcomes. In addition, many of the conclusions drawn from xenograft studies in immune-compromised animals are unlikely to be reproducible in syngeneic tumours or in genetically engineered mouse models (Fig. 1c). The importance of a functional immune system cannot be overstated48. Since nanomaterials possess a much higher affinity for interacting with immune cells than do conventional systemic therapies, the use of immune-competent hosts is essential for eventual clinical translatability. As more sophisticated animal models are being employed in nanomedicine studies, the use of patient-derived xenograft (PDX) tumours is also gaining increased interest, as these models closely resemble the disease in patients. Establishing a PDX model is time consuming and expensive, and requires rigorous molecular characterizations. However, the information generated from PDX-based studies can provide highly relevant and valuable evidence to support the clinical translation of the nanomedicine.
Importantly, both the choice of primary outcomes and their measurement require standardization. For nanomedicine studies focusing on drug delivery and tissue-specific targeting, the demonstration of such capabilities is essential. This requires detailed characterization of nanoparticle distributions within the targeted tissues as well as in organs at risk for non-specific accumulation. Although fluorescence labelling or radioisotope labelling have been commonly used to quantify the tissue-specific distribution of nanoparticles in vivo, newer technologies, such as imaging mass spectrometry, enable more powerful and label-free methods for the characterization of the distribution of nanomedicines in tissues66. Beyond functional endpoints, therapeutic outcomes for nanomedicine studies also require standardized quantification. Tumour growth, for example, is calculated from caliper measurements of the dimensions of superficial palpable tumours. For deep-seated tumours, size measurements rely on tissue contrast generated by imaging modalities, including computed tomography, magnetic resonance imaging, fluorescence imaging and bioluminescence imaging (Fig. 1d). Because these techniques rely on tissue contrast to delineate the boundaries between tumour tissue and normal tissue, it is critical to establish proper thresholds to distinguish the signal from background noise. Similarly, calibrations between fluorescence intensity or bioluminescence intensity and tumour-volume measurements should be conducted to adjust for different expression levels of reporter proteins across different tumour models. This is especially the case for tumour cells with low luciferase-transfection efficiency, which results in discordance between the actual tumour size and the bioluminescence-intensity profile in vivo. Moreover, studies involving immune nanomedicines need to account for the potential immunogenicity of the firefly luciferase protein, which may result in murine T-cell-mediated responses, leading to variations in tumour growth67,68. Therefore, the careful selection of the proper study endpoints, the reporter systems and the measurement techniques is crucial to ensure the reproducibility and validity of nanomedicine studies.
Tackling reproducibility and bias in cell therapy
Cell therapies involve the use of large populations of cells selected or manufactured for reconstructive purposes (regenerative medicine), for containment (the control of inflammatory processes or the arrest of degenerative processes) or for targeting pathological infection, or the growth of abnormal cells (such as cancerous cells). There remain numerous challenges for the reliable and reproducible development of cell products69. Table 2 summarizes some primary issues, which revolve around the themes of cell-composition heterogeneity, selection drift of cells cultured in vitro, teratoma formation, cell type and cell-maturity stage, adjunct cell types included in the cell product, manufacturing variance, underpowered efficacy, cell cryopreservation, reference standards and quality-management systems.
Table 2 |.
Sources of variability that can impact reproducibility in cell therapy
| Parameter | Action |
|---|---|
| Heterogeneity of cell composition | Selection of cell markers and gene expression to identify the functional cohort. |
| Selection drift of cells cultured in vitro | Screening for mutations in oncogenes and in other functional genes associated with the targeted repair of diseased or damaged tissue. |
| Teratoma formation | Selection of cells against pluripotent cell markers. |
| Cell type and stage of maturity | Ensuring that the progenitor cell type is within the lineage appropriate for the end-differentiated cell type needed for targeted tissue repair. |
| Adjunct cell types included in the product | Determination of the cells required for adjunct function to optimize the benefit of therapy (note that immune cells for cell therapy typically contain a mixture of cell types). |
| Manufacturing variance | Ensuring that the manufacturing process is repeatable and that the bioactivity of the product is within the predetermined range for therapeutic benefit. |
| Underpowered efficacy | Demonstration of robust preclinical efficacy in vitro and in animal models (note that efficacy generally decays in human studies because of genetic heterogeneity and patient-response variability). |
| Cell cryopreservation | Minimization of cell death and loss of function (the freezing and thawing of cells can reduce their viability and distort cell type). |
| Reference standards | Expression of data on cell bioactivity and longevity as a fraction or percentage of the agreed reference standards (which are yet to be developed for various cell types used in therapies). Generation of bioequivalence data for different seed banks of cells used in therapies. |
| Quality-management systems | Agreement on a set of standards for quality management and documentation. |
Populations of cells are rarely synchronized in their mitotic cell cycle and often include a variety of cell types or subtypes that may be in various stages of maturity or differentiation. Cellular heterogeneity may not be an issue, or a certain level of heterogeneity may be tolerable for therapeutic purposes, provided that safety and some measure of efficacy can be demonstrated in vitro or in animal models of the targeted disease. Microfluidic separation and gene-expression analyses of single cells have shown significant variability, which enables automated partitional clustering of component cells into populations that have significant clinical relevance. For example, this has been demonstrated with bone-marrow-derived mesenchymal stem cells (MSCs) and neural stem cells70. Because of the repeated observation of cell-product variation (for instance, individual stem cell lines vary significantly in their ability to differentiate into different lineages71, and MSC cell lines exhibit variety in functional properties such as immunomodulatory cytokine expression72), it may be important to analyse cell lines for their expression phenotypes and to select only those in the clustered group that have the desired clinical properties and the specific capacity to influence the targeted disease or injury. Making these choices requires careful experimental design and sound statistical evaluation70.
For autologous cell therapies, probably more heterogeneity is tolerated because the patient receives their own cells and therefore their body is less likely to mount an immune response against the transplanted cells. There is no regulatory guidance about the degree of acceptable heterogeneity of cells used in cell therapies. An example is the use of MSCs derived from the bone marrow, fat tissue or umbilical cord tissue, or differentiated from pluripotent stem cells. They are all phenotypically different but share common marker antigens73. MSCs are not clonally derived and there is usually no attempt to control for heterogeneity in gene expression or in functionality, except when carrying out cell-surface-marker screens or when measuring the cells’ adherence to plastic surfaces or other general properties. It is unknown how much cellular heterogeneity contributes to the lack of functional reproducibility of different cell types.
Cultured (or expanded) cells can develop mutations and other genetic errors such as genomic deletions or rearrangements. For pluripotent stem cells—embryonic stem cells (ESCs) or induced pluripotent stem cells (iPSCs)—there appears to be a selection advantage for mutations of some oncogenes, in particular for TP53 and BCOR, which can become fixed in culture74,75. Oncogene mutations need to be screened for, and the mutated cell lines excluded from clinical use. Malignant transformations also happen to other cell types cultured in vitro76, so screening for potential oncogene mutations during culture is desirable for all cell-therapy products. Mutations may also affect genes that are critical for the specific functions of the transplanted cells. Pluripotent stem cells may form teratomas in vivo, and need to be differentiated into cell types that are unable to form these precancerous structures. This can be achieved after cell differentiation by selecting against residual pluripotent cell markers77.
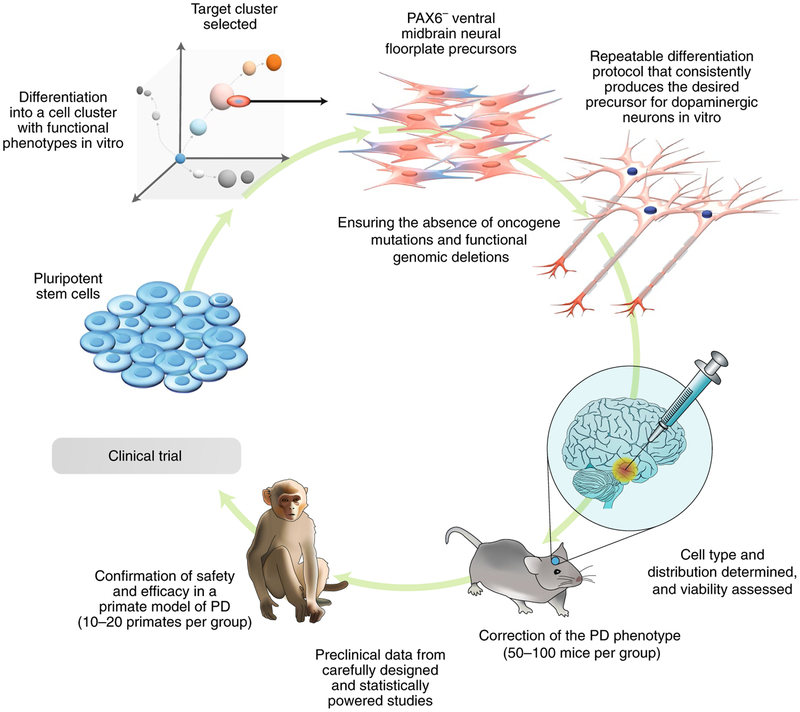
The stage of maturity of cells produced via directed differentiation of stem cells may also add to cellular heterogeneity, and can be critical to the outcome of therapy. In some cases, it is argued that specific functional endpoints (for example, in animal models of Parkinson’s disease, the capacity to produce dopamine and to correct a specific function; Fig. 2) is desired for therapeutic benefit, whereas others argue that employing a specific stem-cell type would be better. Arguments concerning this have arisen recently in a clinical trial for Parkinson’s disease, where parthenogenetic ESCs differentiated into a primitive neural-stem-cell type are considered to involve the wrong cell type78. Although there are concerns about the normality of the parthenogenetic origin of these cells, perhaps more importantly, these transplanted neural stem cells are of a fate (PAX6+ dorsal neural) that is unlikely to produce dopaminergic neurons (instead, the PAX6− ventral midbrain neural floor plate precursor cells usually produce the dopaminergic neurons that are necessary for correcting Parkinson’s disease79). Studies using these cells in non-human primates80 haven’t resolved the concerns because the evidence for any behavioural change in the monkeys was ambivalent (some positive change at low doses but not at high doses) and because post-mortem evidence of the co-localization of transplanted-cell markers with dopamine didn’t exclude cell fusion, which is not uncommon81.
Fig. 2 |. Cell therapy for Parkinson’s disease (PD).
Pluripotent stem cells, either embryonic stem cells or induced pluripotent stem cells, can be directed towards the ventral neural floor plate lineage (PAX6−) and multiplied as dopaminergic precursors capable of the efficient production of dopamine-secreting neurons. The differentiation protocol needs to be robust and capable of being adapted for the large-scale manufacturing of a cell product that can be defined and whose biological activity is determined by dose. Product-release criteria need to be developed in order to provide consistency of function for the dose of cells to be transplanted. The selection of the cells expressing markers consistent with the ventral neural floor plate is critical for obtaining highly functioning dopamine neurons. The ability of the cells to populate the putamen and striatum and integrate into the host neural networks to produce dopamine as required is also essential. Tests for dopamine response in transplanted cells are necessary, as are for the correction of PD behavioural abnormalities (such as rotational motions). The safety and efficacy of neural transplants in the brain usually requires confirmation in a nonhuman primate PD model because rodents may insufficiently represent the human PD condition. Shown are the potential numbers of animals involved in the required preclinical studies. The target product profile and release criteria need to be developed from robust and repeatable data derived from the biological, molecular, immune and functional properties of the cells (including genomic integrity and freedom from oncogenic mutation) if the cell population is to be used for clinical trials.
A comprehensive study of human iPSC-derived dopamine neuron progenitor cells expressing specific markers of the neural floor plate (CORIN+; FOXA2+; TUJ1+; PAX6−) and transplanted into a monkey model of Parkinson’s disease showed stronger evidence of behavioural benefits, of cellular integration of mature dopamine neurons within the putamen, and of extension of their dense neurites into the host striatum, consistent with expectations82. Most of the transplanted cells expressed FOXA2 and a third expressed tyrosine hydroxylase consistent with mature functional dopamine neurons. These data underline the need to transplant cells that are capable of efficiently forming the specific lineage needed in situ.
Taking cells developed from ESCs for type I diabetes as an example, it has been argued that, for establishing long-term function, progenitors of the insulin-secreting β-islet cells are preferable than β-islet cells themselves83; yet others prefer the mature insulinsecreting β-islet cell type84. Cell maturity should thus contribute to the design of manufacturing for cell therapies. It is unclear how well the entire cell population should be confined to a particular stage of maturity for clinical function because it is often difficult to hold cells specifically in particular differentiation states.
In the case of cell products used for targeting cancer cells or infected cells, these may be autologous or allogeneic and can also be genetically engineered to include chimeric antigen receptors (CARs) targeting tumour antigens85. This includes immune T cells or natural killer (NK) cells, but they may also be produced together with a variety of other immune cells that have different phenotypes and function86. In patients, CAR-T cells function for about 3–8 weeks. It is unknown if immune-cell heterogeneity for CAR-T cells contributes to lack of reproducibility in patients, but it is probably desirable to transplant genetically transduced effector or memory T cells only.
Because of the lack of a complete human immune system in animal models, extrapolating efficacy parameters from animal models is problematic. Indeed, cell therapies may succeed in an animal model but the same cells may provide little benefit or no benefit in clinical studies (and vice versa). These disparities raise concerns about cell manufacture and the final release criteria for the identity, purity and potency of the cell product. An example concerns the lack of reproducibility of transplants of fetal neural stem cells for spinal-cord repair in humans87. The cells may not have been equivalent to those in earlier preclinical studies, or the animal model used wasn’t ideal for predicting human benefit. Clinical benefit in patients occurred up to 9 months after cell transplantation, but was lost after 12 months perhaps owing to a delayed immune response that rejected the allogeneic transplant. Further complications for outcomes can arise when combining cells with biomaterials, which can assist cells to remain and develop in specific sites, usually for regenerative purposes88. The biomaterials may impart different characteristics and properties to the cell product and may thus further affect clinical outcomes.
Well-defined cell-quality attributes are needed to reach agreement on how to improve confidence in the clinical benefits of cell therapies. Cell products should include standards for the cells, potential impurities and the characteristics of the suspension medium: composition of the cell population, potential adventitious agents and biological/non-biological impurities, and biological and non-biological media components. A third tier of standards for user-defined cell populations may include viability, morphology and cell-surface markers. The Foundation for the Accreditation of Cellular Therapies (FACT; http://www.factwebsite.org) defined common standards for cellular therapy that include quality management for clinical processes and documentation; still, it doesn’t yet address many of the quality issues affecting reproducibility.
Replication studies and translational successes
To our knowledge there is no systematic empirical study that has assessed what proportion of preclinical studies in nanomedicine or cell therapy have had attempts at replication, and how many of those have failed. Such systematic empirical evaluations on a sample of preclinical investigations may be useful to set an empirical level about the reproducibility status of these fields, as has been done also in other disciplines. Similarly, there is no systematic assessment of what proportion of the initial discoveries in these fields have reached the stage of human trials and what proportion has been successful (that is, licensed). However, given the vast amount of work done in these fields and the relative dearth of high-profile clinical applications, it is highly probable that the translation rate is low; certainly, there is plenty of room for improvement. In fact, a comprehensive account of cancer-nanomedicine products approved by the United States Food and Drug Administration (FDA) lists 14 products and another 35 that are in different phases of clinical trials for cancer therapy50. And Table 3 provides a list of the currently market-approved cell therapies (excluding umbilical cord blood products). Only 8 such therapies have received approval to date by the FDA, and another 4 have received approval from the European Medicines Agency (EMA), despite extensive preclinical work and many early clinical trials on cell therapies89. Box 1 provides a guiding checklist for translational studies in both nanomedicine and in cell therapy. There are clear parallels in both fields as well as some aspects that are topic-specific. Common threads are the optimization of technical facets, the relevance of the models used, data transparency and sharing, and the optimization of the study design and statistical methods.
Table 3 |.
Market-approved cell therapies (excluding umbilical cord blood products)
| Product | Drug name and application | Company |
|---|---|---|
| FDA approved | ||
| LAVIV | Azficel-T (autologous skin fibroblasts for aesthetic applications) | Fibrocell Technologies |
| MACI | Autologous cultured chondrocytes on a porcine collagen membrane | Vericel |
| GINTUIT | Allogeneic cultured keratinocytes and fibroblasts in bovine collagen | Organogenesis |
| IMLYGIC | Talimogene laherparepyec (oncolytic viral therapy) | BioVex (Amgen) |
| KYMRIAH | Tisagenlecleucel (CAR-T cancer therapy for B-cell tumours) | Novartis |
| LUXTURNA | Voretigene neparvovec-rzyl (adeno-associated virus vector gene therapy) | Spark Therapeutics |
| PROVENGE | Sipuleucel-T (autologous dendritic cell therapy for prostate cancer) | Dendreon |
| YESCARTA | Axicabtagepe ciloleucel (CAR-T cancer therapy for B-cell tumours) | Kite |
| EMA approved | ||
| HOLOCLAR | Ex vivo expanded autologous corneal epithelial cells for corneal burns | Holostem TA |
| STRIMVELIS | CD34+ cells transduced with ADA cDNA for severe combined immunodeficiency gene therapy | GlaxoSmithKline |
| ZALMOXIS | Allogeneic T cells transduced with nerve growth factor and HSV-TK Mut2 for graft-versus-host disease | MolMed SpA |
| CX601 | Allogeneic adipose-derived ‘stem’ cells for perinatal fistulas and Crohn’s disease | Tigenix |
ADA, adenosine deaminase; CAR, chimeric antigen receptor.
Box 1 |. Checklist for improving the translational potential of preclinical studies in nanomedicine and cell therapy
Are the critical technical aspects optimized? Technical considerations are, for example, reproducible synthesis and manufacturing (in nanomedicine), and cell preparation, cell screening and cell-product development (in cell therapy).
Is there sufficient understanding of the relevant biology, with strong evidence for safety and efficacy in relevant settings? Such understanding is typically based on relevant animal or in vitro models of human disease; for example, humanized mice models with an intact immune system, or tissue-on-a-chip systems.
Is there sufficient data transparency and sharing of resources?
Are the study design and statistical analyses clearly defined and optimal?
Moving forward in nanomedicine
In nanomedicine, maximizing the translational potential of preclinical studies should involve improved reproducibility of the synthesis of nanomaterials, the assessment of their biological effects, proper study design, and enhanced datatransparency and data sharing.
Reproducible synthesis and manufacturing.
At the most fundamental level, more controllable and reproducible methods for the synthesis of nanomaterials for biomedical applications are still needed. Compared with bulk-synthesis techniques that largely rely on solution-based chemistry, high-precision microfluidic and micropatterning techniques for the production of highly mono-disperse nanoparticles with uniform physicochemical properties and greatly reduced batch-to-batch variability are rapidly gaining attention90,91. In addition, the continued development and scaling-up of the manufacturing of nanomaterial products that meet prespecified quality standards is needed92. Multiple companies have already produced nanomedicine products that have resulted in clinical trials, providing further evidence that tackling the issues of GMP is feasible, at least for simple nanomedicine formulations such as PEGylated-proteins or liposomes (that is, functionalized with poly(ethylene) glycol; PEG) incorporating specific active pharmaceutical ingredients that are aimed to achieve desired pharmacokinetic and pharmacodynamics properties. However, as nanomedicine design becomes increasingly complex to accomplish a wider range of functional specifications, new manufacturing processes will be required to ensure successful transition for clinical testing.
Evaluation of biological effects.
The field of nanomedicine is beginning to adopt changes in how the biological effects of nanomaterials are evaluated and subsequently validated. More in-depth analyses of the biological mechanisms involved in the biological activities of nanomedicines, rather than just proof-of-principle demonstrations, are however needed. This transition is not easy, as it typically requires in-depth understanding of chemistry, nanoscience, biology, drug delivery, immunology and imaging. Critically, an improved understanding of complex biological mechanisms will help the reproducibility of the results.
Better model systems for evaluating the biological activities of nanomaterials are also emerging. Unlike simple two-dimensional cell cultures, the development of biomimetic organ-on-a-chip and tissue-on-a-chip systems may enable researchers to more accurately predict the behaviour of nanomedicines in a controlled setting by considering cell-cell interactions, interstitial flow and nanoparticle transport in three dimensions93,94. The availability of these systems allows for increased accuracy in the selection of nanomedicine candidates for in vivo testing. Nanomedicine will also benefit from more relevant animal models. For cancer-nanomedicine studies, xenograft human tumour models in immune-deficient mice are beginning to be replaced by PDX and genetically engineered mouse models to study nanomedicine actions in more clinically relevant settings. Similarly, mice reconstituted with a functional human immune system (humanized mice) can be used for studying the therapeutic effects of nanomedicines against autoimmune diseases such as diabetes, or to harbour patient-derived tumours for preclinical testing of cancer immune nanomedicines95.
Data transparency and sharing of resources.
An on-going effort to promote data transparency and to improve experimental reproducibility is the establishment of freely accessible databases with detailed experimental protocols and data on the biological responses to nanomaterials according to their specific physicochemical properties. A cancer-nanomedicine repository (http://inbs.med.utoronto.ca/cgi-bin/Repository.cgi) spearheaded by the University of Toronto enables researchers to share their own experimental data, and has allowed for the determination of the delivery efficiency of specific nanomaterials into tumours46. Multiple other initiatives have also been launched to make specific experimental protocols and raw experimental data publicly accessible, and to drive the direct deposition of primary data at the point of collection91. One main criterion for any open-access database, however, is that the shared data must be screened and validated. Such curation requires a major investment, typically attainable only through cooperative consortia.
Study design and statistical analyses.
An often-overlooked aspect in nanomedicine studies is the robustness of the statistical design of the experiments and of the analyses of the data. It is important that proficiency in statistics is available in the research team, and that proper statistical design for each experiment is carefully thought out before experimentation begins. Questions that may need to be asked are: what is the most appropriate primary objective and which are the endpoints of the study? What statistical power is acceptable? What is the sample size needed to achieve that power? Are the statistical methods used for data analyses appropriate, and are proper actions taken to adjust for potential confounders or to correct for multiple comparisons?
Moving forward in cell therapy
In cell therapy, maximizing the translational potential of preclinical studies should involve proper study design, improved standards for cell preparation and screening, and improved animal models of human disease. Preclinical developments that have led to registered cell products might serve as a guide.
Study design and statistical analyses.
Given the cell variability inherent in cell therapies, studies must be adequately powered and include the necessary controls. ‘Clustergrams’ (graphs for visualizing hierarchical and non-hierarchical cluster analyses) enable the visualization of biomarker and transcriptional data patterns70. Cell selection based on function potential is essential for the repeatability of outcomes in vitro and in vivo. With large datasets, it is difficult to design preclinical studies without statistical input to address the influence of multiple factors and their interactions on signalling-pathway analyses. Many models have continuous-response variables associated with achieving the desired therapeutic correction; these require ordinal regression models for predicting cumulative probability outcomes96. In addition, some statistical models allow for the prediction of optimal therapy parameters by using time-course experimental data and statistical analyses for identifying robust and optimal treatment protocols (see ref.97 for a proof of concept). These methods enhance the repeatability of cell-therapy experiments and should be considered in experimental study design.
Improving standards for cell preparation and screening.
Standards have been proposed but need to be agreed by all for cell source and quality, including age of donor, process of cell production and of screening for therapeutic purposes, cell viability, cell pluripotency, DNA mutations, chromatin deletions, chromosomal rearrangements (particularly in oncogenes), cell-surface-marker antigen expression, functional capacity in vitro98, stage of maturity and homology, absence of viruses and other potential immunogens, and bioequivalence in repeated production lines. Equivalences against established industry standards (such as those for immune effector cells)99 for similar preparations in academic laboratories, in companies and in clinics are needed. Product profiles need to provide details against these standards. At present, there is little consensus on processing and manufacturing standards100, on cryopreservation101 and on the degree of heterogeneity of the cell population that is acceptable102. Unless these standards evolve, the reproducibility levels of cell therapies will likely remain suboptimal.
Drawing from experience and knowledge in the development of the few registered cell products.
Studies that have developed limbal cells to a registered product for patients with limbal-stem-cell deficiency can provide a useful roadmap for other cell therapies. This is an autologous cell therapy involving the selection and surgical collection of viable limbal cells that exist in the eyes of a patient, expansion of the cells in vitro and the return of the cells as a monolayer on a scaffold. Data were repeatable and consistently high efficacy in vitro and in animal models was obtained. One year after autologous cell grafting, the cells were 80% successful or partially successful; three years later 68% were judged to be successful103. These are robust data that encourage widespread clinical adoption. For allogeneic therapy, cells are taken surgically from compatible donors and expanded in vitro and transplanted with some local immune suppression.
The recent approval by the FDA of CAR-T cell therapy for patients 25 years or younger is a product that is effective for B-cell (CD19) acute lymphoblastic leukaemia (~80% clearance rate of cancers)104, but is expensive and has dangerous side effects (cytokine release syndrome and neurological symptoms). The high efficacy has been critical for the rapid adoption and registration of these CD19 CAR-T cells, but there has been some considerable variation in patient response and some serious neurological side-effects of the treatments that could not have been predicted from animal studies (rodents)105. Predicting the reproducibility of cancer treatments even by experts has not always been accurate106. Matching the patient to the cell capability is central. CAR-T cells with the same antibody have been less successful and less reproducible for other B-cell tumours (such as chronic lymphoblastic leukaemia) and for other antibodies and solid tumours107. The lack of reproducibility may indicate a mismatch of the CAR-T antibody design and tumour recognition or access, or may result from yet unknown factors.
Cell products tend to evolve from laboratories and be adopted by companies, with little reference to other similar or different cell products. Mistakes are often repeated. To try and avoid the repetition or errors and the associated costs, some institutions have merged their common experiences (notable examples are California’s Alpha Stem Cell Clinics, supported by the California Institute for Regenerative Medicine108,109, the UK Catapult Program in Cell Therapies and the Ontario Institute for Regenerative Medicine). These organizations claim to have improved time-to-clinical-trial and the reproducibility of cell products, but empirical data to this effect have not been published.
Data sharing between major pharmaceutical companies can accelerate clinical drug development110 but there isn’t any evidence of this happening in cell therapies (although there are numerous publications about progress in data sharing for immune cell therapies)111,112. The commercialization process does not favour data sharing because of intellectual-property issues; this suggests that standards that have failed to evolve can underpin clinical reproducibility.
Improving animal and in vitro models of human disease.
Many animal models do not represent the human condition sufficiently to be used for reliable predictions of efficacy in human patients. The absence of a competent human immune system can make animal studies inadequate for genuine safety issues. Treatments for autoimmune diseases, such as type 1 diabetes or multiple sclerosis, are difficult to model in a predictable way. In many cases, there is no reliable animal model because the mechanism of the disease is unknown or the condition cannot be replicated in animals. There is a real need to improve animal models and to further develop in vitro models, in particular by using organoids113 and microphysiological systems114.
Case studies
In trying to improve the translational efficiency of both nanomedicine and cell therapies, one can also learn by examining past case studies where interventions moved, with variable success, from the bench to licensed clinical use. Box 2 describes two case studies of the translational trajectories for trastuzumab emtansine (an anti-body-drug conjugate) and sipuleucel-T (a cell-therapy product). These cases illustrate that the preclinical phase can be instrumental in determining the subsequent clinical success of the intervention.
Box 2 |. Two examples of translational trajectories
An antibody-drug conjugate for cancer treatment: trastuzum-ab emtansine.
Early cancer nanomedicines approved by the FDA for treating human cancers (such as Abraxane and Doxil) were relatively simple formulations designed to provide more favourable pharmacokinetic properties to the attached drug. Recently, the development of cancer nanomedicines has expanded on this concept, with the goal of improving therapeutic potency of an existing drug, or to achieve new therapeutic efficacy by combining components that are individually ineffective. Trastuzumab emtansine (T-DM1, marketed as Kadcyla) is an example of this design concept. It is a drug conjugate that links, via a thioether linker, an antihuman epidermal growth factor receptor-2 (HER2) monoclonal antibody with mertansine (also called DM1), a cytotoxin that inhibits the assembly of microtubules. In a phase-III randomized clinical trial, T-DM1 improved both progression-free survival and overall survival, with respect to second-line chemotherapy or other HER2-directed therapies, in patients with unresectable, locally advanced or metastatic HER2-positive breast cancer who were previously treated with trastuzumab and taxane chemotherapy115. The development of T-DM1 was based on the urgent and critical unmet clinical need for treating HER2-positive breast cancer patients whose cancer progressed on trastuzumab (Herceptin) or lapatinib therapy. With this clearly defined objective, early preclinical studies on the antibody-drug conjugate focused on the identification of the ideal linker for enabling optimal release of the drug in targeted cells, with minimal cross-contamination into surrounding tissues116,117. Similarly, efficacy studies examining the antitumour activities of T-DM1 were conducted in experimental models refractory to trastuzumab and lapatinib treatments, with elucidation of the molecular processes involved in the cytotoxic effect of the drug117. The preclinical efficacy studies were conducted in a way that mirrored their clinical experimental design. With additional preclinical and pharmacological assessments completed, the initial clinical evaluation was conducted in a dose-escalation phase-I trial in patients with HER2-positive metastatic breast cancer who were previously treated with trastuzumab, which ultimately identified the optimal dose regimen of T-DM1, for further clinical development118. The successful development of T-DM1 was initiated with a clearly defined objective that focused on a specific clinical scenario, with preclinical experiments designed to achieve the endpoints that would provide a strong rationale for initiating an early-phase clinical trial.
A therapeutic cellular vaccine: sipuleucel-T.
The prostate-cancer cellular vaccine sipuleucel-T (marketed by Dendreon as Provenge) was developed by using the specific tumour target prostatic acid phosphatase (PAP), which is expressed in 95% of prostate tumours, fused to granulocyte macrophage colony stimulating factor (GM-CSF), which stimulates dendritic cell (DC) activation. Studies in rodent models of prostate cancer had shown statistically significant and repeatable tumour inhibition in the Dunning rat model and in a transgenic mouse model of prostate cancer using an irradiated tumour-cell vaccine expressing PAP-GM-CSF119,120. These preclinical data, which were in agreement, set the opportunity for clinical trials of antigen-presenting DCs harvested from patients with prostate cancer. The DCs are isolated by leukapheresis and sent to a central laboratory facility for their transduction of the immunogen, and then returned to the patient to activate and stimulate T-cell-mediated tumour killing. The early clinical trials showed some transient side effects, evidence of immunological response, and in some patients, lowered the tumour marker prostate-specific antigen; however, few patients experienced tumour regression. Randomized phase-III trials showed no delay in time to disease regression, but a three-year follow up reported a 4.5-month survival benefit compared with the placebo group121,122. Yet there were criticisms regarding data pooled from separate independent studies and the possible lack of equivalence of baseline disease states in these studies. There were doubts also about the magnitude of clinical benefit, and discordance about the results for different endpoints. Even though sipuleucel-T was licensed, Dendreon filed for bankruptcy five years after obtaining FDA approval for the drug. This case shows how a translational effort that reached fruition for an important clinical indication failed in part123–126. Sipuleucel-T was marketed at high prices, and met delays and negative decisions for reimbursement. The criticism of the cost-benefit of Provenge therapy was further anchored in the relatively limited magnitude of benefit and in concerns about the management of the therapy (leukapheresis procedure, therapeutic window and manufacturing variances, in particular) and about its high cost (~US$100,000 per treatment). In retrospect, the earlier concerns about the major response difference between the rodent models used and the autologous human therapy (lack of exactness of model and between-species variations) should have been seriously considered, in particular because they were reinforced by the lack of sufficient and consistent efficacy in the clinical trials.
Outlook
To our knowledge, a systematic assessment of whether successful translational efforts in nanomedicine or cell therapy (or in other bench-discovery fields) were linked to particularly pristine and meticulously executed preclinical studies has not been carried out. With more attention to high-quality study design and optimal research practices, it is possible that the rate of clinical translation would have been higher. Conformity to better research-practice criteria should not be a doctrinaire impediment to scientific work, but a facilitator of efficient clinical translation.
Nevertheless, expectations need to be tempered by realism. Not all of the items in the checklist for improving the translational potential of preclinical studies in nanomedicine and cell therapy (Box 1) will be equally easy to tackle in all settings and for all types of translational effort. Improving some fundamental processes in the way research is done in these fields may have a greater impact than circumscribed technical advances. Coordination of efforts for teamwork and standardization and the use of minimal acceptable standards may further accelerate the pace of translatable discoveries. The use of optimal study designs and statistical analyses should be a relatively straightforward fix in theory, since the methods are already available and they are simply underused or ignored. In practice, it is unlikely that all research efforts in these fields will use optimal designs and analyses, yet even if only an increasing fraction adopts such practices, there will be a clear gain. Transparency and sharing may require appropriate resources and, ideally, some centralized efforts that are supported by funders, research institutions and other research stakeholders. Once design, statistical analyses and sharing practices have been optimized, research communities can focus on reaping the benefits from improving technical facets and the relevance of the experimental models used, towards obtaining the best possible biological evidence.
Acknowledgements
We thank N. Boyd for helping create Fig. 2. We also acknowledge funding from the Mayo Clinic Center for Regenerative Medicine (B.Y.S.K.), the National Institute of Neurological Disorders and Stroke Grant R01 NS104315 (B.Y.S.K.) and the Laura and John Arnold Foundation for providing funding for the Meta-Research Innovation Center at Stanford (METRICS) (J.P.A.I.).
Footnotes
Competing interests
The authors declare no competing interests.
Publisher's Disclaimer: Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bowen A & Casadevall A Increasing disparities between resource inputs and outcomes, as measured by certain health deliverables, in biomedical research. Proc. Natl Acad. Sci. USA 112, 11335–11340 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Contopoulos-Ioannidis DG, Ntzani E & Ioannidis JP Translation of highly promising basic science research into clinical applications. Am. J. Med 114, 477–484 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Baker M 1,500 scientists lift the lid on reproducibility. Nature 533, 452–454 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Allison DB, Brown AW, George BJ & Kaiser KA Reproducibility: a tragedy of errors. Nature 530, 27–29 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lithgow GJ, Driscoll M & Phillips P A long journey to reproducible results. Nature 548, 387–388 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bissell M Reproducibility: the risks of the replication drive. Nature 503, 333–334 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Ioannidis JP The reproducibility wars: successful, unsuccessful, uninterpretable, exact, conceptual, triangulated, contested replication. Clin. Chem 63, 943–945 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Ioannidis JP Acknowledging and overcoming nonreproducibility in basic and preclinical research. JAMA 317, 1019–1020 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Prinz F, Schlange T & Asadullah K Believe it or not: how much can we rely on published data on potential drug targets? Nat. Rev. Drug Discov 10, 712 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Begley CG & Ellis LM Drug development: raise standards for preclinical cancer research. Nature 483, 531–533 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Nosek BA & Errington TM Making sense of replications. eLife 6, e23383 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Begley CG & Ioannidis JP Reproducibility in science: improving the standard for basic and preclinical research. Circ. Res 116, 116–126 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Button KS et al. Power failure: why small sample size undermines the reliability of neuroscience. Nat. Rev. Neurosci 14, 365–376 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Szucs D & Ioannidis JP Empirical assessment of published effect sizes and power in the recent cognitive neuroscience and psychology literature. PLoS Biol. 15, e2000797 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsilidis KK et al. Evaluation of excess significance bias in animal studies of neurological diseases. PLoS Biol. 11, e1001609 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hess KR Statistical design considerations in animal studies published recently in Cancer Research. Cancer Res. 71, 625 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Kilkenny C et al. Survey of the quality of experimental design, statistical analysis and reporting of research using animals. PLoS ONE 4, e7824 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steward O, Popovich PG, Dietrich WD & Kleitman N Replication and reproducibility in spinal cord injury research. Exp. Neurol 233, 597–605 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Szucs D & Ioannidis JPA When null hypothesis significance testing is unsuitable for research: a reassessment. Front. Hum. Neurosci 11, 390 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greenland S et al. Statistical tests, P values, confidence intervals, and power: a guide to misinterpretations. Eur. J. Epidemiol 31, 337–350 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benjamin DJ et al. Redefine statistical significance. Nat. Hum. Behav 2, 6–10 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Bruns SB & Ioannidis JP p-curve and p-hacking in observational research. PLoS ONE 11, e0149144 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Veresoglou SD P hacking in biology: an open secret. Proc. Natl Acad. Sci. USA 112, E5112–E5113 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fanelli D How many scientists fabricate and falsify research? A systematic review and meta-analysis of survey data. PLoS ONE 4, e5738 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freedman LP, Cockburn IM & Simcoe TS The economics of reproducibility in preclinical research. PLoS Biol. 13, e1002165 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ioannidis JPA et al. Increasing value and reducing waste in research design, conduct, and analysis. Lancet 383, 166–175 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimmelman J, Mogil JS & Dirnagl U Distinguishing between exploratory and confirmatory preclinical research will improve translation. PLoS Biol. 12, e1001863 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simeon-Dubach D, Burt AD & Hall PA Quality really matters: the need to improve specimen quality in biomedical research. J. Pathol 228, 431–433 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Dirnagl U et al. A concerted appeal for international cooperation in preclinical stroke research. Stroke 44, 1754–1760 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goodman SN Introduction to Bayesian methods I: measuring the strength of evidence. Clin. Trials 2, 282–290 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Benjamini Y & Hochberg Y Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995). [Google Scholar]
- 32.Wasserstein RL & Lazar NA The ASAs statement on p-values: context, process, and purpose. Am. Statistician 70, 129–133 (2016). [Google Scholar]
- 33.Colquhoun D The reproducibility of research and the misinterpretation of p-values. R. Soc. Open Sci 4, 171085 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Colquhoun D The false positive risk: a proposal concerning what to do about p values. Preprint at https://arXiv.org/abs/1802.04888 (2018).
- 35.Ioannidis JP The proposal to lower P value thresholds to .005. JAMA 319, 1429–1430 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Nosek BA et al. Promoting an open research culture. Science 348, 1422–1425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stodden V et al. Enhancing reproducibility for computational methods. Science 354, 1240–1241 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Simera I, Moher D, Hoey J, Schulz KF & Altman DG A catalogue of reporting guidelines for health research. Eur. J. Clin. Invest 40, 35–53 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Henderson VC, Kimmelman J, Fergusson D, Grimshaw JM & Hackam DG Threats to validity in the design and conduct of preclinical efficacy studies: a systematic review of guidelines for in vivo animal experiments. PLoS Med. 10, e1001489 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chambers CD, Dienes Z, McIntosh RD, Rotshtein P & Willmes K Registered reports: realigning incentives in scientific publishing. Cortex 66, A1–A2 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Moher D, Goodman SN & Ioannidis JP Academic criteria for appointment, promotion and rewards in medical research: where’s the evidence? Eur. J. Clin. Invest 46, 383–385 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Ioannidis JP & Khoury MJ Assessing value in biomedical research: the PQRST of appraisal and reward. JAMA 312, 483–484 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jackman JA, Lee J & Cho NJ Nanomedicine for infectious disease applications: innovation towards broad-spectrum treatment of viral infections. Small 12, 1133–1139 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Barranco C Nanomedicine, meet autoimmune disease. Nat. Rev. Rheum 12, 193 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Clemente-Casares X et al. Expanding antigen-specific regulatory networks to treat autoimmunity. Nature 530, 434–440 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Wilhelm S et al. Analysis of nanoparticle delivery to tumours. Nat. Rev. Mater 1, 16014 (2016). [Google Scholar]
- 47.Shi J, Kantoff PW, Wooster R & Farokhzad OC Cancer nanomedicine: progress, challenges and opportunities. Nat. Rev. Cancer 17, 20–37 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang W et al. Designing nanomedicine for immuno-oncology. Nat. Biomed. Eng 1, 0029 (2017). [Google Scholar]
- 49.Blanco E, Shen H & Ferrari M Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat. Biotechnol 33, 941–951 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.von Roemeling CA, Jiang W, Chan CK, Weissman IL & Kim BYS Breaking down the barriers to precision cancer nanomedicine. Trends Biotechnol. 35, 159–171 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Rolland JP, Hagberg EC, Denison GM, Carter KR & De Simone JM High-resolution soft lithography: enabling materials for nanotechnologies. Angew. Chem. Int. Ed 43, 5796–5799 (2004). [DOI] [PubMed] [Google Scholar]
- 52.Xu J et al. Future of the particle replication in nonwetting templates (PRINT) technology. Angew. Chem. Int. Ed 52, 6580–6589 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peer D et al. Nanocarriers as an emerging platform for cancer therapy. Nat. Nanotech 2, 751–760 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Rodriguez PL et al. Minimal “self” peptides that inhibit phagocytic clearance and enhance delivery of nanoparticles. Science 339, 971–975 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Monopoli MP, Åberg C, Salvati A & Dawson KA Biomolecular coronas provide the biological identity of nanosized materials. Nat. Nanotech 7, 779–786 (2012). [DOI] [PubMed] [Google Scholar]
- 56.Albanese A, Tang PS & Chan WCW The effect of nanoparticle size, shape, and surface chemistry on biological systems. Annu. Rev. Biomed. Eng 14, 1–16 (2012). [DOI] [PubMed] [Google Scholar]
- 57.Walkey CD, Olsen JB, Guo H, Emili A & Chan W, C. W. Nanoparticle size and surface chemistry determine serum protein adsorption and macrophage uptake. J. Am. Chem. Soc 134, 2139–2147 (2012). [DOI] [PubMed] [Google Scholar]
- 58.Salvati A et al. Transferrin-functionalized nanoparticles lose their targeting capabilities when a biomolecule corona adsorbs on the surface. Nat. Nanotech 8, 137–143 (2013). [DOI] [PubMed] [Google Scholar]
- 59.Jiang W, Kim BYS, Rutka JTR & Chan WCW Nanoparticle-mediated cellular response is size-dependent. Nat. Nanotech 3, 145–150 (2008). [DOI] [PubMed] [Google Scholar]
- 60.Rice SB et al. Particle size distributions by transmission electron microscopy: an interlaboratory comparison case study. Metrologia 50, 663–678 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krystek P, Ulrich A, Garcia CC, Manohar S & Ritsema R Application of plasma spectrometry for the analysis of engineered nanoparticles in suspensions and products. J. Anal. Atom. Spectrom 26, 1701–1721 (2011). [Google Scholar]
- 62.Masters JR Cell-line authentication: end the scandal of false cell lines. Nature 492, 186 (2012). [DOI] [PubMed] [Google Scholar]
- 63.Allen M, Bjerke M, Edlund H, Nelander S & Westermark B Origin of the U87MG glioma cell line: good news and bad news. Sci. Transl. Med 8, 354re3 (2016). [DOI] [PubMed] [Google Scholar]
- 64.Pollard SM et al. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell 4, 568–580 (2009). [DOI] [PubMed] [Google Scholar]
- 65.Tsuchiya S et al. Establishment and characterization of a human acute monocytic leukemia cell line (THP-1). Int. J. Cancer 26, 171–176 (1980). [DOI] [PubMed] [Google Scholar]
- 66.Prideaux B & Stoeckli M Mass spectrometry imaging for drug distribution studies. J. Proteomics 75, 4999–5013 (2012). [DOI] [PubMed] [Google Scholar]
- 67.Limberis MP, Bell CL & Wilson JM Identification of the murine firefly luciferase-specific CD8 T-cell epitopes. Gene Therapy 16, 441–447 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baklaushev VP et al. Luciferase expression allows bioluminescence imaging but imposes limitations on the orthotopic mouse (4T1) model of breast cancer. Sci. Rep 7, 7715 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dimmeler S, Ding S, Rando TA & Trounson A Translational strategies and challenges in regenerative medicine. Nat. Med 20, 814–821 (2014). [DOI] [PubMed] [Google Scholar]
- 70.Rennert RC et al. High-resolution microfluidic single-cell transcriptional profiling reveals clinically relevant subtypes among human stem cell populations commonly utilized in cell-based therapies. Front. Neurology 7, 41 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ortmann D & Vallier L Variability of human pluripotent stem cell lines. Curr. Opin. Genet. Dev 46, 179–185 (2017). [DOI] [PubMed] [Google Scholar]
- 72.Paladino FV, Sardinha LR, Piccinato CA & Goldberg AC Intrinsic variability present in Wharton’s jelly mesenchymal stem cells and T cell responses may impact cell therapy. Stem Cells Int. 2017, 8492797 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bianco P “Mesenchymal” stem cells. Annu. Rev. Cell Dev. Biol 30, 677–704 (2014). [DOI] [PubMed] [Google Scholar]
- 74.Merkle FT et al. Human pluripotent stem cells recurrently acquire and expand dominant P53 mutations. Nature 545, 229–233 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Trounson A Potential pitfall of pluripotent stem cells. N. Engl. J. Med 377, 490–491 (2017). [DOI] [PubMed] [Google Scholar]
- 76.Wang X et al. Tumor suppressor gene alterations of spontaneously transformed cells from human embryonic muscle in vitro. Oncol. Rep 24, 555–561 (2010). [PubMed] [Google Scholar]
- 77.Tang C, Weissman IL & Drukker M The safety of embryonic stem cell therapy relies on teratoma removal. Oncotarget 3, 7–8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Barker RA et al. Are stem cell-based therapies for Parkinson’s disease ready for the clinic in 2016? J. Parkinsons Dis. 6, 57–63 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kriks S et al. Floor plate-derived dopamine neurons from hESCs efficiently engraft in animal models of PD. Nature 480, 547–551 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gonzalez R et al. Neural stem cells from human parthenogenetic stem cells engraft and promote recovery in a nonhuman primate model of Parkinson’s disease. Cell Transplant. 25, 1945–1966 (2016). [DOI] [PubMed] [Google Scholar]
- 81.Wagers AJ & Weissman IL Plasticity of adult stem cells. Cell 116, 639–648 (2004). [DOI] [PubMed] [Google Scholar]
- 82.Kikuchi T et al. Human iPS cell-derived dopaminergic neurons function in a primate Parkinson’s disease model. Nature 548, 592–596 (2017). [DOI] [PubMed] [Google Scholar]
- 83.Schulz TC Concise review: manufacture of pancreatic endoderm cells for clinical trials in type 1 diabetes. Stem Cells Transl. Med 4, 927–931 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vegas AJ et al. Long term glycemic control using polymer encapsulated human stem-cell derived β-cells in immune competent mice. Nat. Med 22, 306–311 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brudno JN & Kochenderfer JN Chimeric antigen receptor T-cell therapies for lymphoma. Nat. Rev. Clin. Oncol 15, 31–46 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hombach AA & Abken H Most do, but some do not: CD4+CD25− T cells, but not CD4+CD25+ Treg cells, are cytolytic when redirected by chimeric antigen receptor (CAR). Cancers (Basel) 9, 112 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Temple S & Studder L Lessons learned from pioneering neural stem cell studies. Stem Cell Rep. 8, 191–193 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Prestwich GD et al. What is the greatest regulatory challenge in the translation of biomaterials to the clinic? Sci. Transl. Med 4, 160cm14 (2012). [DOI] [PubMed] [Google Scholar]
- 89.Trounson A & McDonald C Stem cell therapies in clinical trials: progress and challenges. Cell Stem Cell 17, 11–22 (2015). [DOI] [PubMed] [Google Scholar]
- 90.Karnik R et al. Microfluidic platform for controlled synthesis of polymeric nanoparticles. Nano Lett. 8, 2906–2912 (2008). [DOI] [PubMed] [Google Scholar]
- 91.Tropsha A, Mills KC & Hickey AJ Reproducibility, sharing and progress in nanomaterial databases. Nat. Nanotech 12, 1111–1114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rolland JP et al. Direct fabrication and harvesting of monodisperse, shape-specific nanobiomaterials. J. Am. Chem. Soc 127, 10096–10100 (2005). [DOI] [PubMed] [Google Scholar]
- 93.Huh D et al. Reconstituting organ-level lung functions on a chip. Science 328, 1662–1668 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Albanese A, Lam AK, Sykes EA, Rocheleau JV & Chan WC Tumour-on-a-chip provides an optical window into nanoparticle tissue transport. Nat. Commun 4, 2718 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rongvaux A et al. Development and function of human innate immune cells in a humanized mouse model. Nat. Biotechnol 32, 364–372 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu Q, Shepherd BE, Li C & Harrell FE Jr Modeling continuous response using ordinal regression. Stat. Med 10.1002/sim.7433 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Barish S, Ochs MF, Sontag ED & Gevertz JL Evaluating optimal therapy robustness by virtual expansion of a sample population, with a case study in cancer immunotherapy. Proc. Natl Acad. Sci. USA 114, E6277–E6286 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lin-Gibson S, Sarkar S & Ito Y Defining quality attributes to enable measurement assurance for cell therapy products. Cytotherapy 18, 1241–1244 (2016). [DOI] [PubMed] [Google Scholar]
- 99.Maus MV & Kikiforow S The why, what, and how of the new FACT standards for immune effector cells. J. Immunother. Cancer 5, 36 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dropulic B Reference standards for gene and cell therapy products. Mol. Therapy 25, 1259–1260 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Stacey GN et al. Preservation and stability of cell therapy products: recommendations from an expert workshop. Regen. Med 12, 553–564 (2017). [DOI] [PubMed] [Google Scholar]
- 102.Williams DJ et al. Comparability: manufacturing, characterization and controls, report of a UK regenerative medicine platform pluripotent stem cell platform workshop, Trinity Hall, Cambridge, 14–15 September 2015. Regen. Med 11, 483–492 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rama P, Ferrari G & Pellegrini G Cultivated limbal epithelial transplantation. Curr. Opin. Ophthalmol 28, 387–389 (2017). [DOI] [PubMed] [Google Scholar]
- 104.Wei G, Wang J, Huang H & Zhao Y Novel immunotherapies for adult patients with B-lineage acute lymphoblastic leukemia. J. Hemat. Oncol 10, 150 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Brudno JN & Kochenderfer JN Toxicities of chimeric antigen receptor T cells: recognition and management. Blood 127, 3321–3330 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Benjamin D, Mandel DR & Kimmelman J Can cancer researchers accurately judge whether preclinical reports will reproduce? PLoS Biol. 15, e2002212 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fesnak A, June CH & Levine BL Engineered T cells: the promise and challenges of cancer immunotherapy. Nat. Rev. Cancer 16, 566–681 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Trounson A, DeWitt ND & Feigal EG The alpha stem cell clinic: a model for evaluating and delivering stem cell-based therapies. Stem Cells Transl. Med 1, 9–14 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lomax GP et al. Accelerating stem cell treatments for patients: the value of networks and collaborations. Stem Cells Portal http://stemcellsportal.com/content/2015-0090 (2015). [Google Scholar]
- 110.Mayo-Wilson E, Doshi P & Dickersin K Are manufacturers sharing data as promised?. Br. Med. J 351, h4169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Guinney J et al. Prediction of overall survival for patients with metastatic castration-resistant prostate cancer: development of a prognostic model through a crowdsourced challenge with open clinical trial data. Lancet Oncol. 18, 132–142 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Corrie BD et al. iReceptor: a platform for querying and analysing antibody/B-cell and T-cell receptor repertoire data across federated repositories. Immunol. Rev 284, 24–41 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Clevers H Modeling development and disease with organoids. Cell 165, 1586–1597 (2016). [DOI] [PubMed] [Google Scholar]
- 114.Esch EW, Bahinski A & Huh D Organs-on-chips at the frontiers of drug discovery. Nat. Rev. Drug Discov. 14, 248–260 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Verma S et al. Trastuzumab emtansine for HER2-positive advanced breast cancer. N. Engl. J. Med 367, 1783–1791 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lewis Phillips GD et al. Targeting HER2-positive breast cancer with trastuzumab-DM1, an antibody-cytotoxic drug conjugate. Cancer Res. 68, 9280–9290 (2008). [DOI] [PubMed] [Google Scholar]
- 117.Erickson HK et al. Antibody-maytansinoid conjugates are activated in targeted cancer cells by lysosomal degradation and linker-dependent intracellular processing. Cancer Res. 66, 4426–4433 (2006). [DOI] [PubMed] [Google Scholar]
- 118.Holden SN et al. A phase I study of weekly dosing of trastuzumab-DM1 (T-DM1) in patients (pts) with advanced HER2+ breast cancer. J. Clin. Oncol 26, 1029 (2008). [Google Scholar]
- 119.Vieweg J et al. Immunotherapy of prostate cancer in the Dunning rat model: use of cytokine gene modified tumor vaccines. Cancer Res. 54, 1760–1765 (1994). [PubMed] [Google Scholar]
- 120.Hurwitz AA et al. Combination immunotherapy of primary prostate cancer in a transgenic mouse model using CTLA-4 blockade. Cancer Res. 60, 2444–2448 (2000). [PubMed] [Google Scholar]
- 121.Kantoff PW et al. Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N. Engl. J. Med 363, 411–422 (2010). [DOI] [PubMed] [Google Scholar]
- 122.Higano CS et al. Integrated data from 2 randomized, double-blind, placebo-controlled, phase 3 trials of active cellular immunotherapy with sipuleucel-T in advanced prostate cancer. Cancer 115, 3670–3679 (2009). [DOI] [PubMed] [Google Scholar]
- 123.Gong CL & Hay JW Cost-effectiveness analysis of abiraterone and sipuleucel-T in asymptomatic metastatic castration-resistant prostate cancer. J. Natl Compr. Canc. Netw 12, 1417–1425 (2014). [DOI] [PubMed] [Google Scholar]
- 124.Geynisman DM, Chien CR, Smieliauskas F, Shen C & Shih YC Economic evaluation of therapeutic cancer vaccines and immunotherapy: a systematic review. Hum. Vaccin. Immunother 10, 3415–3424 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jaroslawski S & Toumi M Sipuleucel-T (Provenge)-autopsy of an innovative paradigm change in cancer treatment: why a single-product biotech company failed to capitalize on its breakthrough invention. BioDrugs 29, 301–307 (2015). [DOI] [PubMed] [Google Scholar]
- 126.Simpson EL, Davis S, Thokala P, Breeze PR, Bryden P & Wong R Sipuleucel-T for the treatment of metastatic hormone-relapsed prostate cancer: a NICE single technology appraisal; an evidence review group perspective. Pharmacoeconomics 33, 1187–1194 (2015). [DOI] [PubMed] [Google Scholar]