Abstract
Retinal gene therapy is leading the neurological gene therapy field, with 32 ongoing clinical trials of recombinant adeno-associated virus (rAAV)–based therapies. Importantly, over 50% of those trials are using restricted promoters from human genes. Promoters that restrict expression have demonstrated increased efficacy and can limit the therapeutic to the target cells thereby reducing unwanted off-target effects. Retinal ganglion cells are a critical target in ocular gene therapy; they are involved in common diseases such as glaucoma, rare diseases such as Leber's hereditary optic neuropathy, and in revolutionary optogenetic treatments. Here, we used computational biology and mined the human genome for the best genes from which to develop a novel minimal promoter element(s) designed for expression in restricted cell types (MiniPromoter) to improve the safety and efficacy of retinal ganglion cell gene therapy. Gene selection included the use of the first available droplet-based single-cell RNA sequencing (Drop-seq) dataset, and promoter design was bioinformatically driven and informed by a wide range of genomics datasets. We tested seven promoter designs from four genes in rAAV for specificity and quantified expression strength in retinal ganglion cells in mouse, and then the single best in nonhuman primate retina. Thus, we developed a new human-DNA MiniPromoter, Ple345 (NEFL), which in combination with intravitreal delivery in rAAV9 showed specific and robust expression in the retinal ganglion cells of the nonhuman-primate rhesus macaque retina. In mouse, we also developed MiniPromoters expressing in retinal ganglion cells, the hippocampus of the brain, a pan neuronal pattern in the brain, and peripheral nerves. As single-cell transcriptomics such as Drop-seq become available for other cell types, many new opportunities for additional novel restricted MiniPromoters will present.
Keywords: gene therapy, rAAV9, promoter, enhancer, intravitreal injection, brain
Introduction
Recently, there has been a promising revival of gene therapy for diseases with unmet treatment needs.1 Four gene therapies have been fully approved by either the U.S. Food and Drug Administration or the European Medicines Agency, including Luxturna, a one-time recombinant adeno-associated virus (rAAV)–augmentation gene therapy to improve and maintain vision in patients with inherited retinal diseases.2 Retinal gene therapy is leading the field, with 32 ongoing clinical trials of rAAV-based ocular therapies,3 of which just over 50% use restricted promoters generated from human genes.4–8 Promoters that restrict expression have already demonstrated increased efficacy9–11 and can limit the therapeutic to the target cells, thereby reducing unwanted off-target effects.9 Restricted promoters may come from either the therapeutic gene or, more often, from an unrelated gene with the appropriate expression for the therapy.4,6–8 This latter approach may be essential due to the complexity of the endogenous promoter, the unsuitability of the endogenous promoter due to the disease or developmental state of the target cell, or the therapeutic being unrelated to the disease gene, such as with neuroprotectants.9
Capturing the appropriate expression for a gene therapy in a small promoter is challenging. Human promoters are generally large and complex, and in contrast, rAAV (the gene-therapy vector of choice for many applications) is small, with a packaging capacity of only ∼4.9 kb.12 Minimization of a human promoter to make a minimal promoter element(s) designed for expression in restricted cell types (MiniPromoter) may inadvertently destroy specificity and weaken expression. To answer this challenge, we have established bioinformatic methodology using a wide variety of information resources devised to identify both genomic regulatory regions including the transcription start site (prom) and the enhancers that regulate it.13,14 For such endeavors, recently available single-cell transcriptomics data has created a powerful opportunity for selecting the very best possible genes and expression patterns for a specific cell type.15,16
The first droplet-based single-cell RNA sequencing (Drop-seq) dataset included information for retinal ganglion cells (RGCs), a critical target in ocular gene therapy. RGCs are involved in both common (glaucoma17) and rare (Leber's hereditary optic neuropathy18) diseases, and in treatment strategies such as optogenetics.11 Glaucoma is the second most common cause of blindness, with the death of RGCs being the main mechanism.19 Leber's hereditary optic neuropathy is a maternally inherited blinding disorder resulting in RGC degeneration, optic nerve atrophy, and consequent blindness;18 gene therapy trials are currently underway for Leber's hereditary optic neuropathy.20
In this work, we used computational biology and mined the human genome for the best candidate genes, using them to develop a novel MiniPromoter to improve the safety and efficacy of RGC gene therapy. Gene selection included use of the first Drop-seq data, and promoter design was bioinformatically driven. We empirically tested the designs in rAAV for specificity and strength with a three-tiered system including intravenous injection in neonatal mice, subretinal and intravitreal injection in adult mice eye, and finally intravitreal injection in nonhuman primate eye. Developing the appropriate delivery method and serotype combination for rAAV to target the primate RGCs has been challenging.21 Here we explored the rAAV9 capsid, which has not been used clinically for the eye and has no previous evidence for targeting primate RGCs22 but is in clinical trial in the brain.23 rAAV9 was chosen primarily due to its ability to cross the blood–brain barrier and minimal capsid-based cellular specificity,24 allowing any observed restriction of expression to reside primarily with the promoter.
Materials and Methods
MiniPromoter design
Bioinformatics design for each MiniPromoter was done as follows. Candidate genes were chosen from commonly used RGC markers in the literature or from Drop-Seq data. For the chosen genes, the identification of regulatory regions was limited to within neighboring-gene boundaries or regulatory confinements by topologically associating domains as identified by Hi-C data.25 It then relied on the integration of multiple sources of evidence: transcribed promoter and enhancer regions (identified through cap analysis of gene expression [CAGE]26,27 and genomic run-on sequence [GRO-seq] data28), predicted promoter and enhancer regions (ChromHMM and Segway29,30), chromatin accessibility (DNaseI data31), transcription factor–bound regions and histone modifications (identified by chromatin immunoprecipitation sequencing [ChIP-seq data31]), and multispecies conservation.32 The final MiniPromoter design sequence was obtained by placing one promoter region at the 3′ end and adding additional enhancer regions as allowed by the desired final size of the construct.
Cloning and virus production
Small chicken beta-actin promoter/cytomegalovirus enhancer (smCBA) promoter, used as a ubiquitous control, and MiniPromoter design sequences were ordered for direct DNA synthesis (GenScript, Inc., Piscataway, NJ) and cloned into the multiple cloning site (AvrII, FseI, MluI, and AscI) of our “plug and play” rAAV2 backbone plasmid (pEMS2131).13 This genome plasmid includes: an intron (optimized chimeric; 173 bp) (Promega, Madison, MI);33 NotI flanked emerald green fluorescent protein (EmGFP) (720 bp);34 AsiSI flanked the woodchuck hepatitis virus post-transcriptional regulatory element (WPRE) mut6 (587 bp);35 and SV40 polyA (222 bp) (Promega) sequences. Plasmids were propagated in Escherichia coli SURE cells (Agilent Technologies, Santa Clara, CA). DNA was prepared by QIAgen Spin MiniPrep Kit (QIAgen, Germantown, MD); plasmids were confirmed free of rearrangements by AhdI digest, inverted terminal repeats were verified by SmaI digest, and cloning sites verified by sequencing. The resulting smCBA containing plasmid was pEMS2143, and MiniPromoter plasmids are listed in Table 1. Confirmed plasmids were sent to the University of Pennsylvania Vector Core (Philadelphia, PA) for large-scale DNA amplification using the EndoFree Plasmid Mega Kit (QIAgen, Hilden, Germany). Quality control on the plasmid preparation was done by SmaI, PvuII, and SnaBI digests, and confirmed plasmids were packaged into rAAV9 capsid. Plasmids used for cloning, Pleiades MiniPromoters, and viruses are available to the research community through Addgene (Cambridge, MA).
Table 1.
Summary of seven MiniPromoters from four retinal ganglion cell genes
| Pleiades number | Gene | MiniPromoter size (bp) | PlasmidpEMS numbera | MiniPromoter design type | Figure showing expression |
|---|---|---|---|---|---|
| Ple320 | POU4F1 | 1,969 | 2241 | New | 2, 3, 4, S2 |
| Ple321 | TUBB3 | 1,829 | 2242 | New | 2, 4, 5, S2 |
| Ple342 | TUBB3 | 1,992 | 2272 | New | 2, S2 |
| Ple343 | TUBB3 | 2,669 | 2273 | New | 2, S2 |
| Ple344 | TUBB3 | 801 | 2274 | Cutdown of Ple321 | 2, 3, 4, 5, S2 |
| Ple345 | NEFL | 2,693 | 2280 | New | 2, 3, 4, 5, 6, 7, S2 |
| Ple346 | NEFM | 2,771 | 2281 | New | 2, 4, S2 |
Plasmid constructs available from Addgene.
NEFL; neurofilament, light polypeptide; NEFM; neurofilament, medium polypeptide; Ple, pleiades MiniPromoter drove emerald green fluorescent protein–woodchuck hepatitis virus post-transcriptional regulatory element; POU4F1, POU domain, class 4, transcription factor 1; TUBB3; tubulin, beta 3 class III; pEMS, plasmid.
Intravenous injection and histology in neonatal mice
All experimental mice were B6129F1 hybrids produced as the first generation of crossing C57BL/6J (JAX stock No. 000664) to 129S1/SvImJ (JAX stock No. 002448). Timed pregnancies were achieved using crowded females, experienced studs, and plug checking of females such that the day of birth could be accurately predicted. Postnatal day (P) 0 and P4 pup injections were into the superficial temporal vein; a 31-gauge needle on a 0.33 cc syringe (BD, Franklin Lakes, NJ) is inserted under the skin approximately 1–2 mm parallel to the vessel and then advanced into the vein, 50 μL of virus at a titre of 1 × 1013 genome copies/mL (GC/mL) (3.33 × 1012 GC/mL for smCBA) in phosphate buffered saline (PBS) and 0.001% pluronic acid (together, PBS+P) is then slowly injected. Following injection, the pups were tattooed for identification and returned to their home cage with the dam and a nanny (companion female). At 4 weeks, a minimum of two mice per timepoint were given a lethal dose of avertin (MilliporeSigma, Burlington, MA) injected intraperitoneally and perfused transcardially with 1 × PBS for 2 min and 4% paraformaldehyde for 10 min. The eyes, brain, spinal cord, liver, heart, and pancreas were dissected, and tissues were post fixed for 2 h at 4°C.
Tissues were cryoprotected with 25% sucrose overnight at 4°C and then embedded in optimal cutting temperature compound (Tissue-Tek, Sakura Finetek, Torrance, CA) on dry ice, and 20 μm cryosections were directly mounted onto slides. Sections were blocked in PBS +0.4% Triton X-100 (MilliporeSigma) +0.3% bovine serum albumin (MilliporeSigma) for 20 minutes and incubated in primary antibody at 4°C overnight. Primary antibodies used were anti-GFP (GFP-1020; Aves Labs Inc., Tigard, OR; 1:500), anti-neuronal nuclei (NeuN) (MAB377, MilliporeSigma; 1:500), and anti-glial fibrillary acid protein (GFAP) (G3893, MiliporeSigma; 1:500). Slides were washed 3 × with PBS +0.4% Triton X-100 and incubated with secondary antibody and Hoechst 33342 for 2 h at room temperature. Secondary antibodies used were Alexa Fluor 488 and Alexa Fluor 594 (Life Technologies, Carlsbad, CA, 1:1000). Sections were washed 3 × in 0.1 M phosphate buffer (PB), 1 × in 0.01 M PB, mounted in ProLong Gold (P36930; Thermo Fisher Scientific, Waltham, MA) and coverslipped. A minimum of two animals were studied for each injection timepoint for each MiniPromoter (i.e., a minimum of four animals per MiniPromoter), and unique expression patterns of the promoters were determined by microscope and image analysis. The images shown are typically from mice that had the most successful injections and thus the full dose of rAAV, and therefore the greatest likelihood to observe off-target expression if present. Fluorescent images were standardly taken using an Olympus BX61 motorized microscope (Olympus, Shinjuku, Japan). For the cornea and co-labeling, z-stack images were taken on a TCS SP8 Super-Resolution Confocal Microscope (Leica Microsystems, Wetzlar, Germany).
Mice were housed in the pathogen-free Centre for Molecular Medicine and Therapeutics facility on a 7 a.m.–8 p.m. light cycle, 20 ± 2°C with 50 ± 5% relative humidity, with food and water ad libitum. All procedures involving mice were in accordance with the guidelines of the Canadian Council on Animal Care for the Use of Experimental Animals and approved by the University of British Columbia Animal Care Committee (Protocols A14-0294 and A14-0295).
Subretinal and intravitreal injection, histology, and quantification in adult mice
For each virus, subretinal and intravitreal injections were into C57BL/6NCrl mice (19–22 g, Charles River International, Saint Constant, Quebec). The viruses were diluted in PBS+P. For the subretinal injection, 1 μL of virus at a titre of 1 × 1013 GC/mL were injected into the subretinal space using Wiretrol II capillary micropipettes (Drummond Scientific Co., Broomall, PA). The tip of the glass micropipette (diameter ∼100 μm) was inserted parallel to the eye wall into the subretinal space, between the retinal pigment epithelium and the retina. The use of the micropipette allows self sealing of the puncture site without reflux of the solution and with minimal injury. For intravitreal injections, 2 μL of virus at a titre of 1 × 1013 GC/mL were injected into the vitreous chamber. The injection was performed using a 10 μL Hamilton syringe (Hamilton Company, Reno, NV) adapted with a 32-gauge glass microneedle. The tip of the needle was inserted into the superior hemisphere of the eye at a ∼45° angle through the sclera into the vitreous body to avoid retinal detachment or injury to eye structures. All injections were performed in the right eye and treated with one drop of Vigamox (Alcon Laboratories, Fort Worth, TX) to minimize the risk of infection. The left eye was used as a negative control.
Four weeks after virus delivery, anesthetized animals were perfused transcardially with 4% paraformaldehyde, and both eyes were rapidly dissected and post fixed for 2–4 h at 4°C. The cornea and lens were removed and the eyecup was incubated overnight in 30% sucrose. The tissue was then embedded in optimal cutting temperature compound (Tissue-Tek, Miles Laboratories, Elkhart, IN) and retinal cross sections (16 μm) were generated and collected onto gelatin-coated slides (Fisherbrand, Pittsburgh, PA). Retinal sections were blocked for 1 h in blocking solution (PBS 1 × , 3% bovine serum albumin, 0.3% Triton X-100) at room temperature. The primary antibody anti-RNA-binding protein with multiple splicing (anti-RBPMS) (1 μg/μL; PhosphoSolutions, Aurora, CO) was added to the retinal sections and incubated overnight at 4°C. Sections were washed and incubated with secondary anti-rabbit IgG Alexa Fluor 594 (1 μg/mL; Life Technologies). Sections were washed and mounted in antifade reagent (Life Technologies) for visualization with an Axio Imager.M2 Microscope (Zeiss, Oberkochen, Germany).
For quantification of EmGFP epifluorescence intensity in mouse RGCs, four intravitreally injected mice were used per group, and individual EmGFP-positive cells co-labeled with the RGC marker anti-RBPMS were manually traced. The area and the integrity density (the summation of the pixels value in the traced area) were measured with ImageJ software.36 Retinal areas with no apparent fluorescence were also traced and used as background. As positive control and epifluorescence reference, we used retinas transduced with the ubiquitous control virus smCBA-EmGFP-WPRE. Importantly, images were taken at the same exposure time with no adjustments made and analyses were done at settings where there was a dynamic range of EmGFP expression in RGCs from weak to strong. Analysis and statistics were performed using the Graphpad Instat software, version 5.01 (GraphPad Software Inc., San Diego, CA) using a Student's t-test.
Mice were housed in the Centre de Recherche du Centre Hospitalier de l'Université de Montréal (CRCHUM) facility on a 12-hour light/dark cycle, with food and water ad libitum. All procedures involving mice were in accordance with the guidelines of the Canadian Council on Animal Care for the Use of Experimental Animals and approved by the CRCHUM Animal Care Committee (Protocol N15009ADPs).
Intravitreal injection, histology, and quantification in adult nonhuman primate
Prior to and repeatedly post injection, the eyes of the rhesus macaques (Macaca mulatta) were assessed by in vivo ophthalmologic examination, in vivo color fundus photography (FF450; Zeiss), in vivo spectral domain optical coherence tomography (sdOCT) and fundus autofluorescence (FAF) (Heidelberg Spectralis; Heidelberg Engineering, Inc., Heidelberg, Germany), and in vivo ultra-widefield retinal imaging (Optos, Marlborough, MA) to characterize the baseline status of the eye and allow for visualization of the injection site and any abnormalities, and OCT images further showed any damage due to surgery or virus induced toxicity. For each imaging session, monkeys were anesthetized either by an intramuscular injection of Telazol (1:1 mixture of tiletamine hydrochloride and zolazepam hydrochloride, 3.5–5 mg/kg) and maintained with ketamine (1–2 mg/kg) as required, or by inhalant isoflurane (1—2%) vaporized in oxygen. Supplemental oxygen was provided as needed via nasal cannula at 0.5—1.0 L/min, and heart rate and peripheral blood oxygen saturation were monitored by pulse oximetry. Rectal temperature was maintained between 37.0°C and 38.0°C by water-circulating heated pads placed under the animal. Animals were positioned prone with the head supported by a chinrest. Prior to image acquisition, the pupils were dilated to a minimum of 8 mm using phenylephrine (2.5%) and tropicamide (1%) eyedrops. Speculums were used to keep the eyelids open and clear plano contact lenses were inserted centered over the cornea. Following imaging, the contacts and speculums were removed and erythromycin ointment was applied to each eye. Animals were then recovered from sedation and returned to their home cages or enclosures.
Three female rhesus macaques (7, 11, and 15 years of age) that were negative for rAAV9 neutralizing antibodies were used for this study. AAV9 neutralizing antibody levels were assayed by testing the ability of serum to block the AAV9 luciferase-reporter virus transduction into susceptible CHO-Lec2 cells. Serum at various dilutions was pre-incubated with 1.00 × 109 GC of AAV9 luciferase reporter virus for 1 h at 37°C, and then added to 5.00 × 104 cells per 96 wells that had been pretransduced with adeno helper virus. After 48 h, Bright-Glo substrate (Promega, Madison, WI) was added to the cells and luciferase expression was quantitated using a Synergy Mx luminometer (BioTek, Winooski, VT).
For subretinal injections, after anesthesia was achieved, a pars plana without vitrectomy approach was used to create two 50 μL subretinal blebs per eye of virus at a titre of 1.88 × 1012 (left eye) and 1.88 × 1013 GC/mL (right eye) in sterile PBS + P using a 41-gauge cannula. The subretinal detachments in each eye were created superior and inferior to the fovea but encompassed area within the macula. For intravitreal injections, after anesthesia was achieved, approximately 150 μL of the aqueous was removed from the anterior chamber using a 29-gauge insulin syringe. An additional 29-gauge insulin syringe was used to inject into the vitreous directed towards the optic nerve to a depth of approximately 4 mm. For smCBA (ubiquitous), the left eye received 200 μL of virus at a titre of 2.04 × 1012 GC/mL in PBS + P and the right eye received 200 μL at 2.04 × 1013 GC/mL in PBS+P. For Ple345 (neurofilament, light polypeptide (NEFL)) the right eye only received 200 μL of virus at a titre of 1.013 × 1013 GC/mL in PBS+P. All eyes, regardless of surgical technique, received a single post-surgical subconjunctival injection of dexamethasone (0.5 mL, 10 mg/mL) and cefazolin (0.5 mL, 125 mg/ml). The animal that received the subretinal injections also received dexamethasone (1%) and ofloxacin (0.3%) eye drops applied twice daily for one week.
Retinal tissue was harvested at 4 weeks post injection, fixed in 4% paraformaldehyde and cut with a cryostat at 16 μM, stained with anti-RBPMS (ab194213, Abcam, Cambridge, UK) and AlexaFluor 568 anti-rabbit (Invitrogen), and imaged with a Leica confocal microscope (Leica, Wetzlar, Germany).
For quantification of EmGFP epifluorescence in monkey RGSs, the method paralleled that described for mouse, except a minimum of 10 cells from each of four histological slides were used.
Rhesus macaques were housed in the Oregon National Primate Research Center, with food and water ad libitum. All procedures involving monkeys were in accordance with the Association for Research in Vision and Ophthalmology Statement for the Use of Animals in Ophthalmic and Vision Research and approved by the Oregon Health and Science University Institutional Animal Care and Use Committee (Protocol IP00000768).
Image processing
Images were processed using ImageJ and Adobe Photoshop and Illustrator (Adobe, San Jose, CA). For the neonatal injected mouse tissues, brightness, contrast, and color balancing adjustments were made in ImageJ where necessary to improve visibility. For the adult injected mouse, histology, brightness, contrast, and color balancing adjustments were made in Photoshop where necessary to improve visibility. For the nonhuman primate, in vivo images were not adjusted (however, a physical neutral density filter was used when indicated. For the nonhuman primate, histology, brightness, contrast, and color balancing adjustments were made uniformly across all images in ImageJ except where necessary to improve visibility.
Results
Seven MiniPromoters designed from four retinal ganglion cells genes
Two rounds of RGC gene selection for MiniPromoter design were undertaken. In the first, candidate genes were those commonly used as RGC markers in the literature37–39 and with RGC expression in the Gene Expression Nervous System Atlas.40 This generated a list of 43 genes for further examination. Criteria for final selection included: strong and enriched expression in the retina with clear transcription start site prediction from FANTOM CAGE data,26,27 small gene size with low structural complexity from the University of California Santa Cruz (UCSC) genome browser,41 high sequence conservation from the UCSC genome browser, and expression conservation between mouse and primate from the literature. The two genes chosen were POU domain, class 4, transcription factor 1 (POU4F1), also known as BRN3A, and tubulin, beta 3 class III (TUBB3), also known as TUJ1, which are both commonly used as markers for RGCs in mouse and primate (Table 1).37,39,42,43 By the second round of gene selection, Drop-seq data had become available16 and was used as the primary criterion. The top 12 differentially expressed RGC genes relative to the rest of the Drop-seq dataset were further validated as candidates from the literature.44–46 The subsequent criteria for selection were as above, which led to two genes contained within the same topologically associating domain being chosen; NEFL and neurofilament, medium polypeptide (NEFM) (Table 1). NEFL and NEFM ranked 1 and 2 in the RGC Drop-seq dataset, respectively, and in retrospect TUBB3 and POU4F1 ranked 26 and 56 out of 174 differentially expressed RGC genes.16
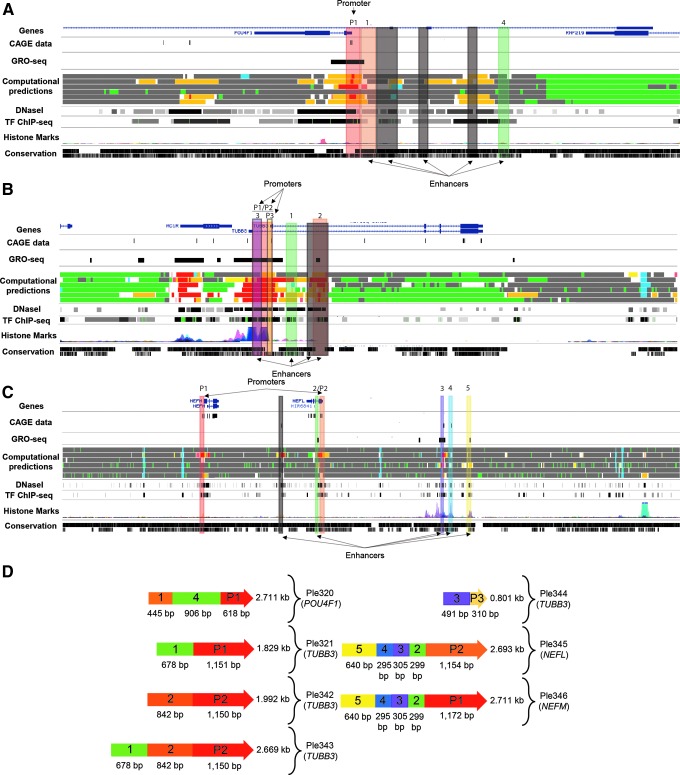
From the four chosen genes, seven MiniPromoters were designed: Ple320 (POU4F1); Ple321, 342, 343, and 344 (TUBB3); Ple345 (NEFL); and Ple346 (NEFM) (Table 1). Bioinformatic design of MiniPromoters was informed by a wide range of genomics datasets, including FANTOM5 CAGE data,26,27 ENCODE transcription factor and histone-mark ChIP-Seq and DNaseI data,31 computational predictions from ChromHMM and Segway,29,30 and multispecies conservation.32 Additionally, Ple342-346 (TUBB3, NEFL, and NEFM) included data from GRO-Seq.28 All these data were viewed on the UCSC genome browser and used to outline potential genomic regions indicative of regulatory activity. Regulatory regions were classified into promoters (regions overlapping a transcription start site, as indicated by CAGE data) or enhancers. For the genes POU4F1, TUBB3, NEFL, and NEFM, identified regulatory regions are shown in Fig. 1A–C, where regions were numbered, and the most promising regions were placed into MiniPromoter constructs, as seen in Fig. 1D. We used 2–2.5 kb as our target MiniPromoter size, with the aim to include enough regulatory sequence to have a successful promoter with restricted expression in the initial design, while being minimal enough for rAAV delivery of therapeutic RNAs and small protein-coding sequences.
Figure 1.
Bioinformatics design of seven MiniPromoters targeting retinal ganglion cells. (A–C) Promoter and enhancer elements potentially regulating the expression of selected genes are shown. Vertically highlighted colored regions correspond to their color-matched element included in (D) final MiniPromoter designs. Elements in black were not used in the final designs. (A) The identification of elements potentially regulating the expression of POU4F1. (B) The identification of elements potentially regulating the expression of TUBB3. (C) The identification of elements potentially regulating the expression of NEFL and NEFM. (D) Final MiniPromoter designs for Ple320 (POU4F1), Ple321 (TUBB3), Ple342 (TUBB3), Ple343 (TUBB3), Ple344 (TUBB3), Ple345 (NEFL), and Ple346 (NEFM). 1–5, enhancers; P1–3, proms. NEFL; neurofilament, light polypeptide; NEFM; neurofilament, medium polypeptide; Ple, pleiades MiniPromoter; POU4F1, POU domain, class 4, transcription factor 1; TUBB3; tubulin, beta 3 class III.
The final MiniPromoter design sequences were sent for direct DNA synthesis, cloned into our rAAV2 “plug and play” plasmid, and sent for packaging in rAAV9.
All seven MiniPromoters expressed in the ganglion cell layer, with variable retinal specificity, when delivered intravenously in mouse
Not every human MiniPromoter is expected to achieve endogenous-like expression in rAAV; therefore, it is ethically imperative to prescreen in mouse before testing in nonhuman primate, even though we may lose good promoters that only work in primates. Initial screening by intravenous delivery in mouse allowed us to examine not only expression in the eye, but peripheral tissues as well. Of two promoters equally suitable in the eye, the one with less peripheral expression would be the safer choice for gene therapy. Thus, we started by screening all seven MiniPromoters by intravenous delivery in neonatal mouse.
To evaluate the ability of the rAAV9 capsid to transduce different cell types and tissues by intravenous delivery in mouse, we used the ubiquitous smCBA promoter driving EmGFP with WPRE. Virus was injected at either P0 or P4 to evaluate the different layers of the retina, as it has previously been shown that the development of the blood–retina barrier changes dramatically at this time and results in different cell type expression.47 As previously published, P0 injection results primarily in ganglion, amacrine, horizontal, and corneal stroma cell expression, and P4 injection results in mainly ganglion, amacrine, bipolar, and Müller glia expression (Supplementary Fig. S1). Regardless of date of injection, positive staining was seen in all brain regions (including both neuronal and glial cells), and throughout the spinal cord, heart, liver, and pancreas (Supplementary Fig. S1).
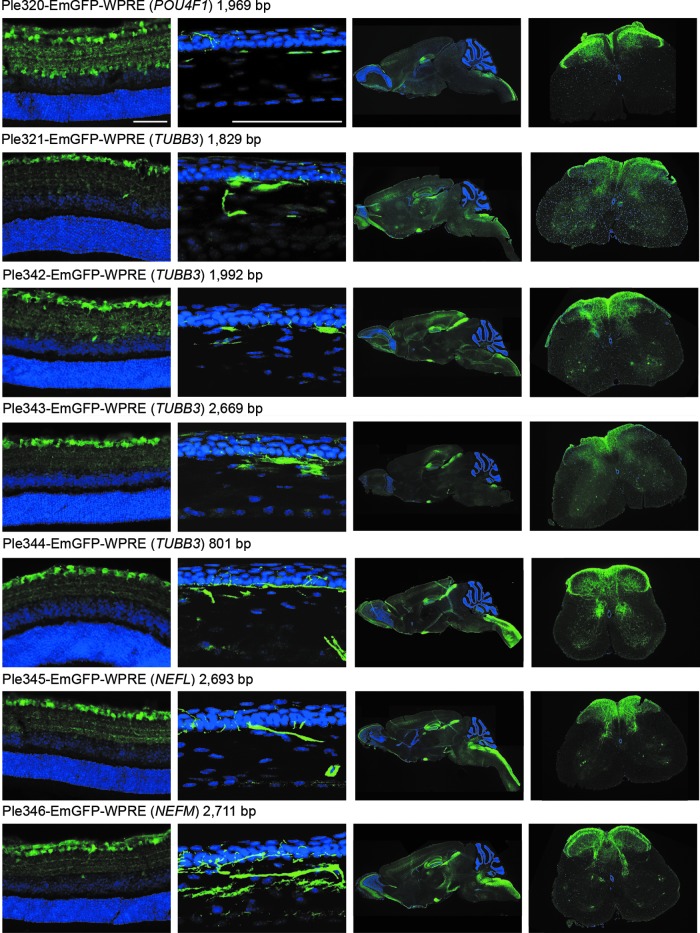
Figure 2 highlights the results of intravenous injection of the seven MiniPromoters designed for RGCs driving EmGFP-WPRE. Retinas, brains, and spinal cords shown are from P4 injections, and corneas are from P0 injections. Focusing on the eye, Ple320 (POU4F1) expresses in the ganglion cell layer (GCL) and inner nuclear layer (INL) of the retina and the corneal stroma and nerves. Ple321, 342, and 343 (TUBB3) all expressed in both the GCL and to a lesser extent the INL, and in the corneal stroma and nerves. Ple344 (TUBB3) was the most successful and smallest of the TUBB3 designs (801 bp), with expression mainly restricted to the GCL and corneal nerves. Ple345 (NEFL) and Ple346 (NEFM) shared very similar expression patterns, with both displaying robust and restricted expression in the GCL and in the corneal nerves.
Figure 2.
Seven MiniPromoters delivered by intravenous injection in neonatal mice showed expression in retinal ganglion cell layer and other tissues. MiniPromoters, each driving EmGFP-WPRE in rAAV9, were injected intravenously into neonatal mice, and harvested four weeks later. Tissues imaged are, from left to right: retina (ganglion cells up), cornea (epithelium up), whole sagittal brain, lumbar spinal cord (dorsal up) (See also Supplementary Figure S2.). EmGFP, emerald green fluorescent protein; rAAV9, recombinant adeno-associated virus packaged in capsid 9; WPRE, woodchuck hepatitis virus posttranscriptional regulatory element. Blue, DAPI; green, anti-GFP. Scale bars, 100 μm.
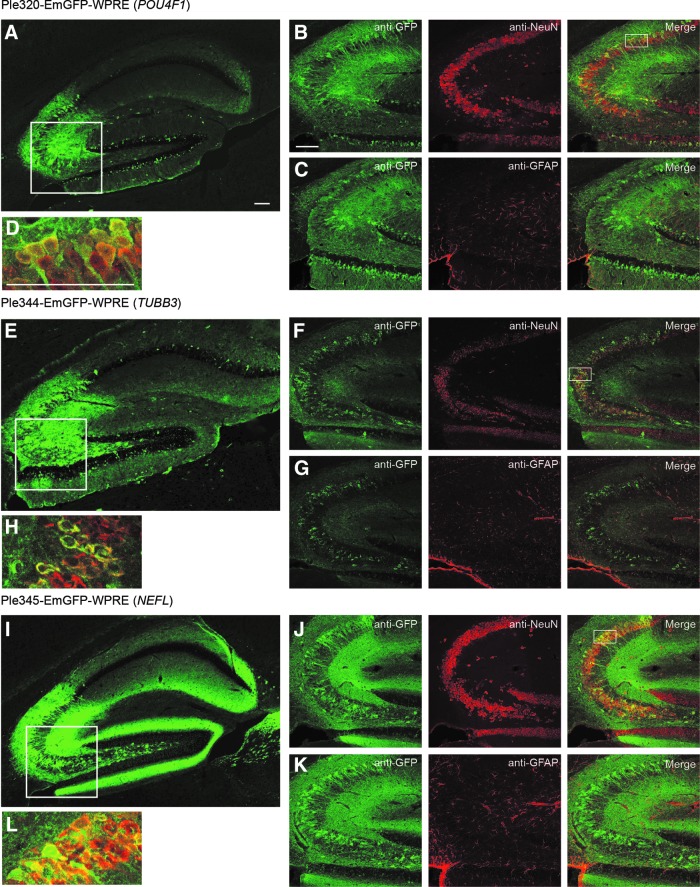
In the brain, Ple320 (POU4F1) expression is highly enriched in the hippocampus, an interesting but nonendogenous expression pattern, and in the spinal cord in the processes of the white matter (Fig. 2). Co-labeling with anti-NeuN a neuronal marker, and anti-GFAP an astrocyte marker, showed that the minor expression in the cortex and olfactory lobe was neuronal, with no co-labeling observed with GFAP and high levels of co-labeling observed with NeuN. In the hippocampus, there was neuronal-specific expression in the CA3 region, granule layer of the dentate gyrus, and sparse expression in the polymorph layer of the dentate gyrus (Fig. 3). Ple321, 342, 343, and 344 (TUBB3) all expressed in a broad pattern throughout the brain and spinal cord (Fig. 2). Co-labeling of Ple344 (TUBB3) with anti-NeuN and anti-GFAP showed the brain expression to be neuronal, with no co-labeling observed with GFAP and high levels of co-labeling observed with NeuN. In the hippocampus, there was neuronal-specific expression in the CA3 region, sparse expression in the granule layer of the dentate gyrus, and expression in the polymorph layer of the dentate gyrus (Fig. 3). Ple345 (NEFL) and Ple346 (NEFM) also displayed broad expression throughout the brain and spinal cord (Fig. 2). Co-labeling of Ple345 (NEFL) with anti-NeuN and anti-GFAP showed the brain expression to be neuronal, with no co-labeling observed with GFAP and high levels of co-labeling observed with NeuN. In the hippocampus, there was neuronal-specific expression in the CA3 region and polymorph layer of the dentate gyrus, and robust expression in the processes in the molecular layer of the dentate gyrus (Fig. 3).
Figure 3.
Three MiniPromoters delivered by intravenous injection in neonatal mice showed co-labeling with a neuronal cell marker in the hippocampus. MiniPromoters, each driving EmGFP-WPRE in rAAV9, were injected intravenously into neonatal mice and harvested 4 weeks later. (A) Ple320 (POU4F1) expression in the hippocampus; box indicates the region shown with co-labeling. (B) High level of co-staining with the neuronal marker NeuN indicates neuronal-specific expression; box indicates the region chosen for magnification. (C) Lack of co-staining with the astrocyte marker GFAP also indicates neuronal-specific expression. (D) Magnified image of NeuN co-labeling. (E) Ple344 (TUBB3) expression in the hippocampus; box indicates the region shown with co-labeling. (F) High level of co-staining with the neuronal marker NeuN indicates neuronal-specific expression; box indicates the region chosen for magnification. (G) Lack of co-staining with the astrocyte marker GFAP also indicates neuronal-specific expression. (H) Magnified image of NeuN co-labeling. (I) Ple345 (NEFL) expression in the hippocampus; box indicates the region shown with co-labeling. (J) High level of co-staining with the neuronal marker NeuN indicates neuronal-specific expression; box indicates the region chosen for magnification. (K) Lack of co-staining with the astrocyte marker GFAP also indicates neuronal-specific expression. (L) Magnified image of NeuN co-labeling. GFAP, anti-glial fibrillary acid protein; NeuN, anti-neuronal nuclei. Green, anti-GFP, red, anti-NeuN or anti-GFAP; yellow, merge. Scale bars, 100 μm.
In peripheral tissues, a very low expression level was observed for all MiniPromoters in the heart (Supplementary Fig. S2). A very low expression level was also observed for all MiniPromoters in the pancreas; however, this included the pancreatic nerves for all MiniPromoters except Ple320 (POU4F1). Finally, a moderate expression level was observed for all MiniPromoters in the liver.
All five MiniPromoters expressed in RGCs, three with retinal specificity, when delivered intravitreally in mouse
To evaluate the ability of the rAAV9 capsid to transduce different retinal cell types by subretinal and intravitreal injection into the adult mouse eye, we used the ubiquitous promoter smCBA driving EmGFP-WPRE (Supplementary Fig. S3). Our results demonstrated that this virus led to EmGFP expression in various different cell types in the mouse retina, regardless of injection route.
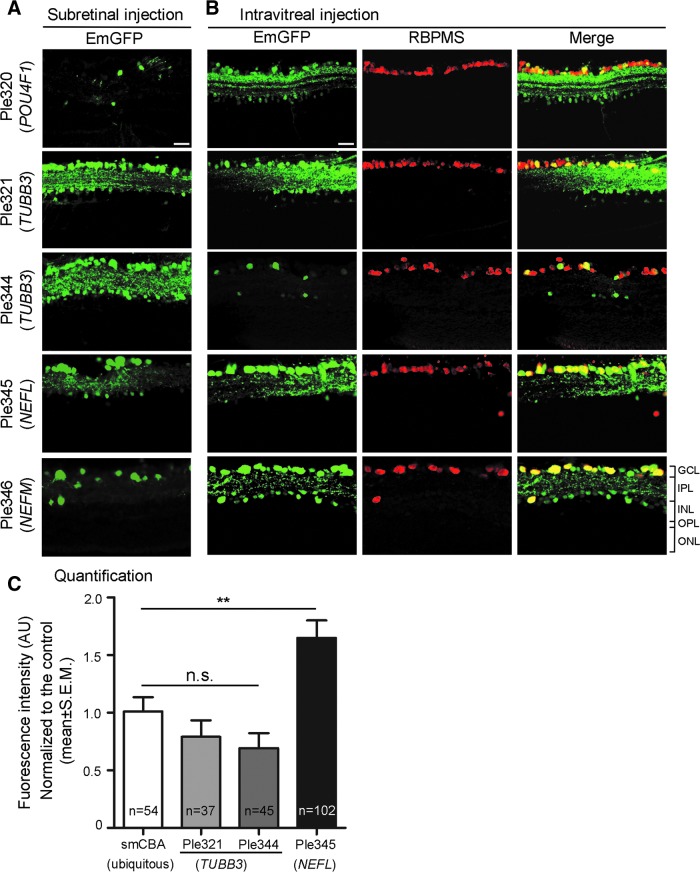
The five most GCL robust and enriched MiniPromoters, of the seven original designs tested intravenously, went on to be characterized in the adult mouse eye by subretinal and intravitreal injection: Ple320 (POU4F1); Ple321, and Ple344 (TUBB3); Ple345 (NEFL); and Ple346 (NEFM) (Fig. 4). Following subretinal delivery, all five MiniPromoters showed expression in the GCL. Some EmGFP-positive cells, presumably amacrines, were also observed in the INL (Fig. 4A). Ple320 (POU4F1) consistently resulted in low levels of expression, whereas the other four MiniPromoters resulted in robust and consistent EmGFP expression by subretinal injection. Following intravitreal delivery, all five MiniPromoters showed expression in RGCs, identified by their co-labeling with anti-RBPMS a ganglion cell marker. Although some EmGFP-positive RBPMS-negative cells were observed in the INL, these were particularly rare for MiniPromoters Ple321 (TUBB3), Ple344 (TUBB3), and Ple345 (NEFL). Ple344 (TUBB3) consistently resulted in low levels of expression, whereas the other four MiniPromoters resulted in robust and consistent EmGFP expression by intravitreal injection.
Figure 4.
Five MiniPromoters delivered by subretinal and intravitreal injection in adult mice showed co-labeling with a ganglion cell marker. MiniPromoters, each driving EmGFP-WPRE in rAAV9, were injected directly into adult mouse eyes, and harvested 4 weeks later. (A) Subretinal injection led to expression in the ganglion cell layer (GCL) and the inner nuclear layer (INL). (B) Intravitreal injection of MiniPromoters Ple320 (POU4F1), Ple321 (TUBB3), Ple344 (TUBB3), and Ple346 (NEFM) led to robust expression in retinal ganglion cells (RGCs), as indicated by co-staining with the ganglion cell marker RBPMS (RNA-binding protein with multiple splicing), and a few cells in the INL. MiniPromoter Ple345 (NEFL) led to robust expression only in RGCs. (C) Quantification of endogenous EmGFP fluorescence intensity in RBPMS-positive cells showed that Ple321 (TUBB3) and Ple344 (TUBB3) were not statistically different from the small chicken beta-actin promoter/CMV enhancer (smCBA) ubiquitous control (for histology see Supplementary Fig. S3). However, Ple345 (NEFL) drove significantly stronger endogenous EmGFP expression in RGCs compared to the smCBA ubiquitous control. Dose for all injected eyes was 2.00 × 1010 genome copies (GC)/eye. **p > 0.01; IPL, inner plexiform layer; n, number of cells counted; n.s., not significant; ONL, outer nuclear layer; OPL, outer plexiform layer. Green, anti-GFP; red, anti-RBPMS; yellow, merge. Scale bar, 20 μm.
Quantification was done for the three most specific MiniPromoters: Ple321 (TUBB3), Ple344 (TUBB3), and Ple345 (NEFL). Quantification of EmGFP epifluorescence intensity in individual RBPMS-positive RGCs transduced with the same dose of virus, showed that Ple321 (TUBB3) and Ple344 (TUBB3) were not significantly different from the ubiquitous control smCBA (p = 0.249 and 0.0786, respectively), whereas Ple345 (NEFL) drove significantly higher intensity levels at 1.7 × (p = 0.0058) (Fig. 4C). In addition, we observed approximately twice as many EmGFP-positive RGCs with Ple345 (NEFL) compared with smCBA, or the other two MiniPromoters.
MiniPromoter Ple345 (NEFL) expressed strongly in RGCs when delivered intravitreally in nonhuman primate
To evaluate the ability of the rAAV9 capsid to transduce different retinal cell types by subretinal and intravitreal injection into the adult rhesus macaque eye, we used the ubiquitous promoter smCBA driving EmGFP-WPRE. We first tested rAAV9 smCBA-EmGFP-WPRE by the subretinal delivery route (Supplementary Fig. S4). At 2 weeks post injection, in vivo FAF imaging revealed strong EmGFP expression mostly confined to the locations of the two subretinal blebs (one above and one below the fovea). By four weeks post injection, EmGFP expression was significantly higher than at two weeks and extended well beyond the borders of the surgically induced subretinal blebs with the strongest expression in the posterior retina. Histological assessment by cellular location and structure revealed tropism for all cell types studied including RGCs, bipolar cells, and photoreceptors. We subsequently tested rAAV9 smCBA-EmGFP-WPRE by the intravitreal delivery route (Supplementary Fig. S5). Robust EmGFP expression was observed in the iris, sclera, and choroid. In the retina, RGCs showed robust EmGFP expression in the perifoveal ring, and EmGFP expression was observed in patches throughout the peripheral retina, particularly around blood vessels. Histological assessment by cellular location and structure revealed broad tropism; EmGFP expression extended from RGCs to the external limiting membrane but in some cases also involved photoreceptor inner and outer segments.
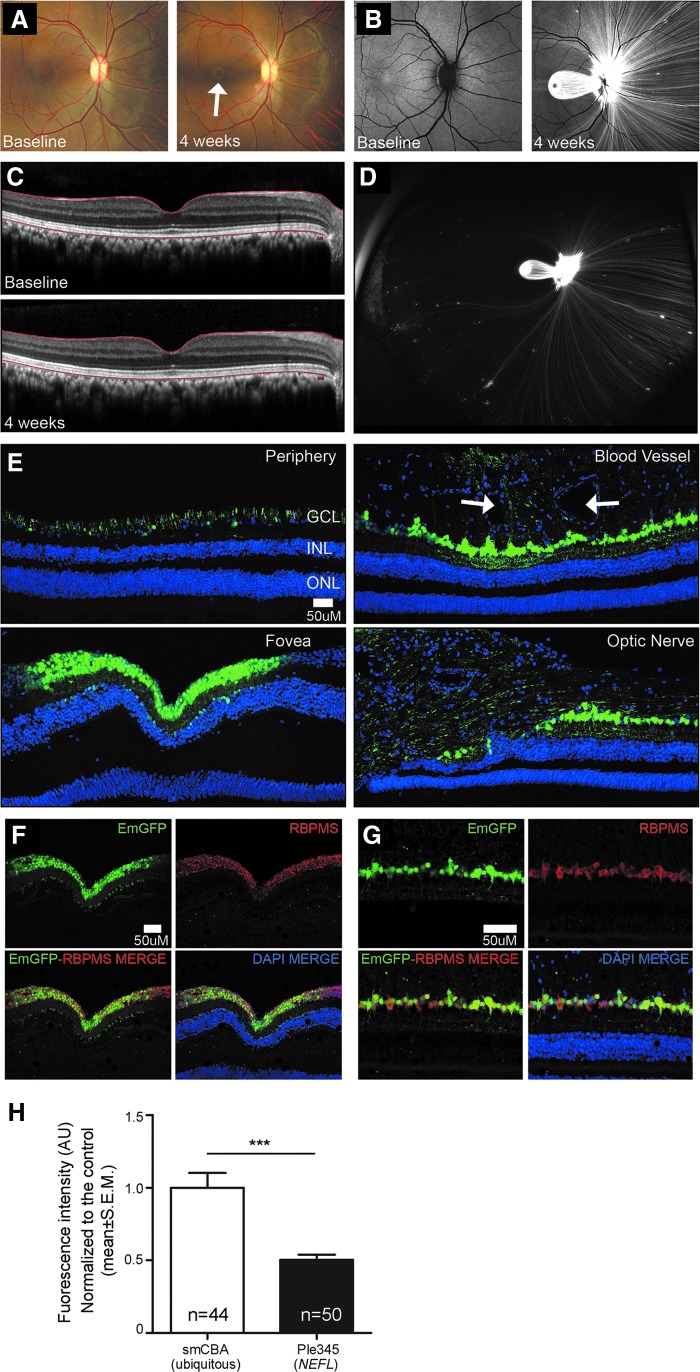
MiniPromoter Ple345 (NEFL), the most specific and strongest in the mouse retinal of the seven original designs, went on to be characterized in the nonhuman retina. Since both subretinal and intravitreal injection of rAAV9 smCBA-EmGFP-WPRE resulted in robust transduction of the GCL, and intravitreal injection is the preferred clinical route, it was chosen for delivery to the rhesus macaque eye. rAAV9 Ple345 (NEFL)-EmGFP-WPRE produced robust EmGFP transduction in RGC bodies and the nerve fiber layer (Fig. 5). EmGFP expression and effects on retinal structure were assessed weekly by in vivo color fundus photography (Fig. 5A), FAF (Fig. 5B), in vivo sdOCT (Fig. 5C), and in vivo ultra-widefield FAF (Fig. 5D). Robust EmGFP expression was seen in RGC bodies and nerve fibers extending both nasally and temporally from the optic nerve (Fig. 5E). Expression was especially robust in nerve fibers nasal to the optic nerve. In addition, RGCs showed very robust EmGFP expression in the perifoveal ring. EmGFP expression was also seen in patches throughout the periphery, particularly around blood vessels, where it generally extended from ganglion cells to the external limiting membrane. A small number of cells of the inner nuclear layer were also transduced at the foveal center. EmGFP expression was observed throughout the optic nerve and optic chiasm. A very small number EmGFP positive axons were observed in the lateral geniculate nucleus, but none in the visual cortex. Finally, we were able to confirm that within the retina, Ple345 (NEFL) expresses specifically in RGCs by showing EmGFP co-localization with anti-RBPMS (Fig. 5F–G).
Figure 5.
MiniPromoter Ple345 (NEFL) delivered by intravitreal injection in adult nonhuman primate showed restricted and strong expression in retinal ganglion cells. Ple345 (NEFL) driving EmGFP-WPRE in rAAV9 was injected directly into adult nonhuman primate eye and harvested 4 weeks later. (A) At harvest in vivo color fundus photograph showed intense expression in perifoveal ring compared to baseline (white arrow). (B) In vivo fundus autofluorescence (FAF) demonstrated intense expression (white) in the perifoveal ring, optic nerve, and nerve fibers. (C) In vivo spectral domain optical coherence tomography imaging showed no disruption of retinal structure or changes in retinal thickness. (D) In vivo ultrawidefield FAF showed expression (white) in patches of ganglion cells and their fibers in the far periphery, predominantly in the nasal retina. (E) Confocal images demonstrated expression in ganglion cells and nerve fibers of peripheral retina, around blood vessels (white arrows), in the fovea, and at the optic nerve. (F, G) Histological confocal images confirmed Ple345 (NEFL) specificity for ganglion cells in the retina by showing co-localization with the ganglion cell marker RBPMS in the (F) fovea and (G) between the fovea and optic nerve). (H) Quantification of EmGFP epifluorescence intensity in RBPMS-positive cells, showed that Ple345 (NEFL) drove significantly weaker expression in RGCs compared to the smCBA ubiquitous control (for histology see Supplementary Fig. S5). Dose for the Ple345 (NEFL) injected eye was 2.03 × 1012 GC/eye, and the dose for the smCBA injected eye was 4.08 × 1012 GC/eye. ***p > 0.001. Blue, DAPI; green, EmGFP epifluorescence; red, anti-RBPMS; yellow, merge.
Quantification of EmGFP epifluorescence intensity in individual RBPMS-positive RGCs showed that Ple345 (NEFL) drove significantly lower intensity levels at 0.5 × compared to the ubiquitous control smCBA (p = 2.39 × 10−5) (Fig. 5H). Since the viral dose given to the smCBA eye was 2 × higher than that given to the Ple345 (NEFL) eye, Ple345 (NEFL) expression may actually be stronger than measured, resulting in no difference in expression levels (i.e., 1 × ). Consistent with this, we did not observe a significant difference in EmGFP-positive RGCs with Ple345 (NEFL) compared to smCBA.
Discussion
We have developed a new human-DNA MiniPromoter, Ple345 (NEFL), which in combination with rAAV9 intravitreal delivery showed specific and robust expression in the RGCs of the nonhuman primate rhesus macaque retina. Expression was maximal at the perifoveal ring, but was also present to a lesser extent throughout the nasal and temporal retina, and even in patches in the periphery of the retina. This positive result with Ple345 (NEFL) may suggest that other human MiniPromoters we have developed would also be useful in primate.
Quantification of EmGFP epifluorescent intensity in RGC cells showed that Ple345 (NEFL) gave 1.7 × the intensity in mouse, and 0.5 × to perhaps 1 × the intensity in nonhuman primate, compared to the ubiquitous smCBA. These results support strong expression from Ple345 (NEFL) in both species. However, the expression levels were surprising given that the promoter is comprised of human DNA and a priori, we expected relative strength to be equal to or greater in primate. Thus, our results demonstrate that quantification in mouse alone is insufficient for predicting expression levels in primate. We further observed in mouse approximately twice as many EmGFP-positive RGCs with Ple345 (NEFL) compared to smCBA. This revealed that the transduction of cells with rAAV9 is actually much broader than we can detect with a commonly used ubiquitous promoter.
Interestingly, while our studies were ongoing, others were also tackling the same problem of restricted expression in RGCs. First, a mouse-DNA promoter was developed from the paralogue Nefh, based on microarray data and tested in mice only.48,49 Unlike NEFL and NEFM, which ranked 1 and 2 in the RGC Drop-seq dataset,16 this third paralogue NEFH ranked 137 of the 174 differentially expressed genes. Also, FANTOM526 data supports that, of the three paralogues, NEFH is the least strongly expressed in the retina. Second, a human-DNA promoter was developed from the gamma-synuclein gene (SNCG), then tested in mouse and subsequently proved restricted and useful in the primate cynomolgus macaques.11 SNCG ranks third, after NEFL and NEFM, of the 174 differentially expressed RGC Drop-seq dataset, and FANTOM5 supports strong retinal expression approaching that of NEFL.
We were unexpectedly pleased with the broad retinal transduction seen with rAAV9 in the nonhuman primate, indicated by the smCBA ubiquitous promoter driving EmGFP epifluorescence. With subretinal delivery, expression expanded well beyond the blebs, and extended to all layers of the retina. With intravitreal delivery, the less invasive and thus preferred route for clinical delivery, expression was even broader across the retina and extended from RGCs to external limiting membrane and in patches to photoreceptors. We attribute this difference from the more limited transduction reported in the literature22 to the 10 × to 100 × higher titres we injected. Studies conducted in nonhuman primate brains suggest that at very high titres, really only available for rAAV9 and its derivatives, toxic levels may be reached.50,51 However, we did not observe toxicity with intravitreal delivery.
This work has also provided additional new MiniPromoters with demonstrated use in mouse beyond the eye. By intravenous delivery, Ple320 (POU4F1) is highly enriched in the hippocampus of the brain. In contrast, using the same delivery method Ple344 (TUBB3) and Ple345 (NEFL) are broadly pan neuronal, although with distinct hippocampal patterns. Finally, all three of these MiniPromoters showed expression in the peripheral nerves.
Our bioinformatic design parameters resulted in MiniPromoters small enough for rAAV delivery of therapeutic RNAs and small protein-coding sequences; for example, Ple345 (NEFL) could drive a protein of 483 amino acids. Of course, much larger therapeutics could be accommodated in less restrictive vectors such as lentivirus, adenovirus, retrovirus, herpes simplex virus, plasmids, and nanoparticles. However, we do not know if we have reached the smallest possible size of any MiniPromoter, while maintaining specificity and strength in the retina. Previously, a 1.5 kb mouse Nefl promoter was used to generate a random insertion mouse, from which one of four integration sites showed neuronal enrichment.52 This may suggest that a further size restriction is possible for Ple345 (NEFL), but such cutdowns would be primarily empirical.
We do not expect, nor have we found, that every human gene has regulatory regions amenable to minimization to achieve endogenous-like expression in rAAV or other therapeutic delivery methods.13,14,53,54 Thus, identifying enough genes with both the desired cell-type restricted expression and strength has been a limiting challenge. The movement towards single-cell transcriptomics may substantially change the MiniPromoter landscape, as evidenced by this work, where the best candidate genes were identified using the first Drop-seq data available for the target cells.16 As single-cell transcriptomics becomes available for other cell types, and sub cell types as defined by expression,55 future opportunities for additional novel restricted MiniPromoters will present.
Supplementary Material
Acknowledgments
The authors thank Tess C. Lengyell for her assistance in establishing the neonatal intravenous injection pipeline; Dr. William W. Hauswirth for the generous gift of the smCBA promoter (University of Florida, 2011); David J. Arenillas for software development; Steven Bailey, Andreas Lauer, and David J. Wilson for their surgical expertise; the staff of the Department of Comparative Medicine at the Oregon National Primate Research Center for their valuable technical assistance and support; and Sara Vucetic for her technical assistance with perfusions, sectioning, and immunostaining.
This work was supported by Brain Canada through the Canada Brain Research Fund, with the financial support of Health Canada and the Consortium Québécois sur la Découverte du Médicament; the National Institutes of Health core grants (P30 EY010572 to the Casey Eye Institute, P51OD011092 to the Oregon National Primate Research Center); and Research to Prevent Blindness [unrestricted grant to the Casey Eye Institute]. The views expressed herein do not necessarily represent the views of the Minister of Health or the Government of Canada.
Author Disclosure
E.M.S., A.D.P., M.N., W.W.W., and their associated institutions are filing patents on these MiniPromoters. The remaining authors declare that no competing financial interests exist.
Supplementary Material
References
- 1. Keeler AM, ElMallah MK, Flotte TR. Gene therapy 2017: Progress and future directions. Clin Transl Sci 2017;10:242–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dias MF, Joo K, Kemp JA, et al. Molecular genetics and emerging therapies for retinitis pigmentosa: Basic research and clinical perspectives. Prog Retin Eye Res 2018;63:107–131 [DOI] [PubMed] [Google Scholar]
- 3. NIH. ClinicalTrials.gov U.S. National Library of Medicine. https://clinicaltrials.gov/ (last accessed March31, 2018)
- 4. Beltran WA, Cideciyan AV, Boye SE, et al. Optimization of retinal gene therapy for X-linked retinitis pigmentosa due to RPGR mutations. Mol Ther 2017;25:1866–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. MacLachlan TK, Milton MN, Turner O, et al. Nonclinical safety evaluation of scAAV8-RLBP1 for treatment of RLBP1 retinitis pigmentosa. Mol Ther Methods Clin Dev 2018;8:105–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pawlyk BS, Bulgakov OV, Sun X, et al. Photoreceptor rescue by an abbreviated human RPGR gene in a murine model of X-linked retinitis pigmentosa. Gene Ther 2016;23:196–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fischer MD, McClements ME, Martinez-Fernandez de la Camara C, et al. Codon-optimized RPGR improves stability and efficacy of AAV8 gene therapy in two mouse models of X-linked retinitis pigmentosa. Mol Ther 2017;25:1854–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bennett J. Taking stock of retinal gene therapy: Looking back and moving forward. Mol Ther 2017;25:1076–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fujita K, Nishiguchi KM, Shiga Y, et al. Spatially and temporally regulated NRF2 gene therapy using Mcp-1 promoter in retinal ganglion cell injury. Mol Ther Methods Clin Dev 2017;5:130–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Scalabrino ML, Boye SL, Fransen KM, et al. Intravitreal delivery of a novel AAV vector targets ON bipolar cells and restores visual function in a mouse model of complete congenital stationary night blindness. Hum Mol Genet 2015;24:6229–6239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chaffiol A, Caplette R, Jaillard C, et al. A new promoter allows optogenetic vision restoration with enhanced sensitivity in macaque retina. Mol Ther 2017;25:2546–2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dong JY, Fan PD, Frizzell RA. Quantitative analysis of the packaging capacity of recombinant adeno-associated virus. Hum Gene Ther 1996;7:2101–2112 [DOI] [PubMed] [Google Scholar]
- 13. de Leeuw CN, Korecki AJ, Berry GE, et al. rAAV-compatible MiniPromoters for restricted expression in the brain and eye. Molecular Brain 2016;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hickmott JW, Chen CY, Arenillas DJ, et al. PAX6 MiniPromoters drive restricted expression from rAAV in the adult mouse retina. Mol Ther Methods Clin Dev 2016;3:16051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Raj B, Wagner DE, McKenna A, et al. Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain. Nat Biotechnol 2018;36:442–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Macosko EZ, Basu A, Satija R, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 2015;161:1202–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Smith CA, Chauhan BC. In vivo imaging of adeno-associated viral vector labelled retinal ganglion cells. Sci Rep 2018;8:1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Carelli V, La Morgia C, Valentino ML, et al. Retinal ganglion cell neurodegeneration in mitochondrial inherited disorders. Biochim Biophys Acta 2009;1787:518–528 [DOI] [PubMed] [Google Scholar]
- 19. Daliri K, Ljubimov AV, Hekmatimoghaddam S. Glaucoma, stem cells, and gene therapy: Where are we now? Int J Stem Cells 2017;10:119–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Khatib TZ, Martin KR. Protecting retinal ganglion cells. Eye (Lond) 2017;31:218–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Boye S, Alexander JJ, Witherspoon CD, et al. Highly efficient delivery of AAV vectors to the primate retina. Hum Gene Ther 2016;27:580–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vandenberghe LH, Bell P, Maguire AM, et al. AAV9 targets cone photoreceptors in the nonhuman primate retina. PLoS One 2013;8:e53463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mendell JR, Al-Zaidy S, Shell R, et al. Single-dose gene-replacement therapy for spinal muscular atrophy. N Engl J Med 2017;377:1713–1722 [DOI] [PubMed] [Google Scholar]
- 24. Foust KD, Nurre E, Montgomery CL, et al. Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nat Biotechnol 2009;27:59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schmitt AD, Hu M, Jung I, et al. A compendium of chromatin contact maps reveals spatially active regions in the human genome. Cell Rep 2016;17:2042–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. The Fantom Consortium and the Riken PMI and CLST (DGT), Alistair RR, Forrest AR, et al. A promoter-level mammalian expression atlas. Nature 2014;507:462–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Andersson R, Gebhard C, Miguel-Escalada I, et al. An atlas of active enhancers across human cell types and tissues. Nature 2014;507:455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Danko CG, Hyland SL, Core LJ, et al. Identification of active transcriptional regulatory elements from GRO-seq data. Nat Methods 2015;12:433–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ernst J, Kellis M. ChromHMM: Automating chromatin-state discovery and characterization. Nat Methods 2012;9:215–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hoffman MM, Buske OJ, Wang J, et al. Unsupervised pattern discovery in human chromatin structure through genomic segmentation. Nat Methods 2012;9:473–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Blanchette M, Kent WJ, Riemer C, et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res 2004;14:708–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sakurai K, Shimoji M, Tahimic CG, et al. Efficient integration of transgenes into a defined locus in human embryonic stem cells. Nucleic Acids Res 2010;38:e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Teerawanichpan P, Hoffman T, Ashe P, et al. Investigations of combinations of mutations in the jellyfish green fluorescent protein (GFP) that afford brighter fluorescence, and use of a version (VisGreen) in plant, bacterial, and animal cells. Biochim Biophys Acta 2007;1770:1360–1368 [DOI] [PubMed] [Google Scholar]
- 35. Zanta-Boussif MA, Charrier S, Brice-Ouzet A, et al. Validation of a mutated PRE sequence allowing high and sustained transgene expression while abrogating WHV-X protein synthesis: application to the gene therapy of WAS. Gene Ther 2009;16:605–619 [DOI] [PubMed] [Google Scholar]
- 36. Rasband WS, Image J. U.S. National Institutes of Health, 1997–2016. https://imagej.nih.gov/ij/ (last accessed March2018)
- 37. Nadal-Nicolas FM, Jimenez-Lopez M, Sobrado-Calvo P, et al. Brn3a as a marker of retinal ganglion cells: qualitative and quantitative time course studies in naive and optic nerve-injured retinas. Invest Ophthalmol Vis Sci 2009;50:3860–3868 [DOI] [PubMed] [Google Scholar]
- 38. Sanes JR, Masland RH. The types of retinal ganglion cells: Current status and implications for neuronal classification. Annu Rev Neurosci 2015;38:221–246 [DOI] [PubMed] [Google Scholar]
- 39. Jiang SM, Zeng LP, Zeng JH, et al. beta-III-Tubulin: A reliable marker for retinal ganglion cell labeling in experimental models of glaucoma. Int J Ophthalmol 2015;8:643–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Siegert S, Scherf BG, Del Punta K, et al. Genetic address book for retinal cell types. Nat Neurosci 2009;12:1197–1204 [DOI] [PubMed] [Google Scholar]
- 41. Kent WJ, Sugnet CW, Furey TS, et al. The human genome browser at UCSC. Genome Res 2002;12:996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Koilkonda RD, Hauswirth WW, Guy J. Efficient expression of self-complementary AAV in ganglion cells of the ex vivo primate retina. Molecular vision 2009;15:2796–2802 [PMC free article] [PubMed] [Google Scholar]
- 43. Tshilenge KT, Ameline B, Weber M, et al. Vitrectomy before intravitreal injection of AAV2/2 vector promotes efficient transduction of retinal ganglion cells in dogs and nonhuman primates. Hum Gene Ther Methods 2016;27:122–134 [DOI] [PubMed] [Google Scholar]
- 44. Gill KP, Hung SS, Sharov A, et al. Enriched retinal ganglion cells derived from human embryonic stem cells. Sci Rep 2016;6:30552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rousso DL, Qiao M, Kagan RD, et al. Two pairs of ON and OFF retinal ganglion cells are defined by intersectional patterns of transcription factor expression. Cell Rep 2016;15:1930–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. de Souza CF, Nivison-Smith L, Christie DL, et al. Macromolecular markers in normal human retina and applications to human retinal disease. Exp Eye Res 2016;150:135–148 [DOI] [PubMed] [Google Scholar]
- 47. Byrne LC, Lin YJ, Lee T, et al. The expression pattern of systemically injected AAV9 in the developing mouse retina is determined by age. Mol Ther 2014;23:290–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hanlon KS, Chadderton N, Palfi A, et al. A novel retinal ganglion cell promoter for utility in AAV vectors. Front Neurosci 2017;11:521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kim CY, Kuehn MH, Clark AF, et al. Gene expression profile of the adult human retinal ganglion cell layer. Molecular vision 2006;12:1640–1648 [PubMed] [Google Scholar]
- 50. Samaranch L, San Sebastian W, Kells AP, et al. AAV9-mediated expression of a non-self protein in nonhuman primate central nervous system triggers widespread neuroinflammation driven by antigen-presenting cell transduction. Mol Ther 2014;22:329–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Flotte TR, Buning H. Severe Toxicity in non-human primates and piglets with systemic high-dose administration of AAV9-like vectors: Putting patients first. Hum Gene Ther 2018;29:283–284 [DOI] [PubMed] [Google Scholar]
- 52. Byrne GW, Ruddle FH. Multiplex gene regulation: A two-tiered approach to transgene regulation in transgenic mice. Proc Natl Acad Sci U S A 1989;86:5473–5477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Portales-Casamar E, Swanson DJ, Liu L, et al. A regulatory toolbox of MiniPromoters to drive selective expression in the brain. Proc Natl Acad Sci U S A 2010;107:16589–16594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. de Leeuw CN, Dyka FM, Boye SL, et al. Targeted CNS delivery using human MiniPromoters and demonstrated compatibility with adeno-associated viral vectors. Mol Ther Methods Clin Dev 2014;1:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mincarelli L, Lister A, Lipscombe J, et al. Defining cell identity with single cell omics. Proteomics 2018:e1700312. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.