Abstract
Introduction:
Targeted therapies for lung adenocarcinoma (LAC) have improved patient outcomes; however, drug resistance remains a major problem. One strategy to achieve durable response is to develop combination-based therapies that target both mutated oncogenes and key modifiers of oncogene-driven tumorigenesis. This is based on the premise that mutated oncogenes, while necessary, are not sufficient for malignant transformation. We aimed to uncover genetic alterations that cooperate with mutant EGFR during LAC development.
Methods and Results:
Through integrative genomic analyses of over 500 LAC tumors, we identified frequent amplifications/deletions of chromosomal regions affecting the activity of genes specifically in the context of EGFR mutation, including amplification of the mutant EGFR allele and deletion of the phosphatase DUSP4, which have both previously been reported. In addition, we identified the novel amplification of a segment of chromosome arm 16p in mutant EGFR tumors corresponding to increased expression of Golgi Associated, Gamma Adaptin Ear Containing, ARF Binding Protein 2 (GGA2), which functions in protein trafficking and sorting. Through co-immunoprecipitation and Western blot analysis, we found that GGA2 interacts with EGFR, increases EGFR protein levels and modifies EGFR degradation after ligand stimulation. Furthermore, we show that overexpression of GGA2 enhances EGFR mediated transformation while GGA2 knockdown reduces the colony and tumor forming ability of EGFR mutant LAC.
Conclusions:
These data suggest that overexpression of GGA2 in LAC tumors results in the accumulation of EGFR protein and increased EGFR signaling, which helps drive tumor progression. Thus, GGA2 plays a cooperative role with EGFR during LAC development and is a potential therapeutic target for combination-based strategies in LAC.
Keywords: GGA2, EGFR, targeted therapy, combination therapy, target validation, integrative genomics, oncogenes, signaling
Introduction
Lung cancer is the leading cause of cancer mortality worldwide, due to late stage of disease at the time of diagnosis and a lack of effective therapeutic strategies available to treat patients1. Lung adenocarcinoma (LAC) is the most common type of lung cancer, responsible for ~40% of all cases and, unlike other subtypes, is associated with both smokers and never smokers2. In order to identify biomarkers for diagnosis and targets for development of new therapies, substantial effort has been focused on identifying genes and pathways that are mutated or altered in human LAC. With the increasing understanding of LAC biology has come the advent of targeted therapies to combat this devastating disease. These therapies target mutated components of key cellular pathways on which tumor cells have become dependent on for survival, a phenomenon known as oncogene addiction3. For example, tyrosine kinase inhibitors (TKIs) targeting LACs driven by mutant EGFR (EGFRMUT) have been clinically successful, highlighting the potential of designing drugs to specifically target the molecular mechanisms driving cancer development, a concept often described as “personalized medicine” 3-5.
Despite these encouraging developments, significant problems remain. First, the majority of LAC patients are not candidates for these therapies as they have tumors without mutations in targetable genes, owing either to the lack of an identified driver or mutation in drivers such as mutant KRAS for which the development of inhibitors has proven elusive. Second, all patients eventually develop resistance to treatment with these targeted agents, either through secondary mutation of the target gene or activation of an alternative pathway that can sustain tumor growth6. Thus, while undoubtedly a major advancement in improving LAC patient outcomes and survival rates, targeted therapy has so far failed to achieve the major goal of curing lung cancer and a greater understanding of the determinants of response will be needed to improve the efficacy of these agents.
While our understanding of the biological basis and clinical treatment of LAC has rapidly advanced over the past decade, key questions have yet to be resolved. Perhaps the most outstanding issue is determining patterns of gene disruption that are selected for during tumorigenesis. This is an important consideration as experimental evidence suggests that multiple genetic alterations are required to transform normal lung cells and drive them to full malignancy7. For example, model systems have revealed that EGFRMUT, which is in ~10-20% of LACs, alone is not sufficient for tumorigenesis8, 9. This is exemplified by transgenic mouse models expressing EGFRMUT in the lung epithelium, where the variable latency period between transgene induction and the onset of lung tumors implies that secondary alterations are a requirement for full malignancy10. Furthermore, EGFRMUT has been detected in histologically normal lung epithelium in patients and immortalized lung epithelial cell lines transduced with EGFRmut fail to progress to a fully malignant phenotype8, 9. Therefore, although tumors expressing this oncogene are clearly dependent on its sustained expression for survival (as demonstrated by the clinical response of EGFRMUT tumor to TKIs such as gefitinib and erlotinib11), these findings suggest that additional genetic/epigenetic alterations cooperate with mutant oncogenes, activating/disrupting genes that “modify” tumorigenic capacity in LAC development. Identifying these modifiers of oncogene induced tumorigenesis is imperative in order to determine mechanisms of tumor progression and subsequently, for identifying new targets for anti-cancer agents that improve patient outcomes. For EGFRMUT, these modifiers may represent logical targets for the design combination-based therapies that counteract the inevitable drug resistance and tumor recurrence that occurs following treatment with singleagent TKIs.
In this study, we aimed to systematically identify genes that cooperate with EGFRMUT to drive lung tumorigenesis. Through the integrative genomic and gene expression analyses of >500 LAC tumors, we delineated regions of genetic alteration specific to tumors harbouring EGFR mutations and identified genes dysregulated by these changes. These included both known and novel EGFRMUT cooperators including amplification of EGFR itself, loss of the negative feedback regulator phosphatase DUSP4 and the novel activation of a gene involved in protein trafficking, Golgi Associated, Gamma Adaptin Ear Containing, ARF Binding Protein 2 (GGA2). Furthermore, we demonstrate that GGA2 interacts with EGFR, regulating its stability after activation and subsequently increasing its transformative capacity. Together, this work highlights that additional events are required for mutant EGFR to drive transformation, offering a potential candidate for the development of combination-based therapies to improve patient outcomes.
Results
Identification of recurrent genomic alterations specific to EGFR mutant lung adenocarcinomas
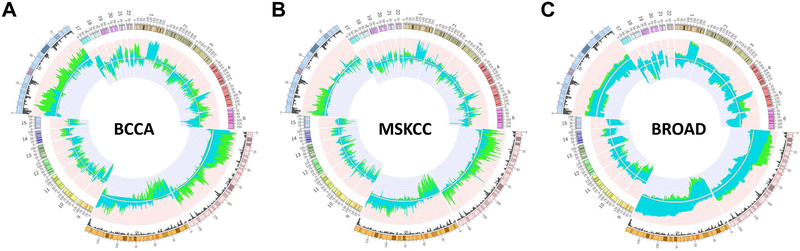
Previous studies have demonstrated that expression of mutant EGFR is not sufficient to drive lung epithelial cells to full malignancy8, 9. Thus, we hypothesized that secondary genomic alterations may activate/inactivate additional genes that cooperate with mutant EGFR during tumorigenesis. To identify such genes, we aimed to determine regions of copy number change significantly enriched in EGFR mutant (EGFRMUT) - in comparison to EGFR wild-type (EGFRWT) - LAC tumors. Using an approach we previously established12-15, we compared segmental copy number alterations between these groups in three independent cohorts: 83 tumors from the British Columbia Cancer Agency (BCCA, 20 EGFRMUT and 63 EGFRWT), 199 tumors from Memorial Sloan Kettering Cancer Center (MSKCC, 43 EGFRMUT and 156 EGFRWT) and 354 tumors from the Broad and collaborating institutes (29 EGFRMUT and 325 EGFRWT). Briefly, the frequency of alteration across the genome was determined for EGFRMUT and EGFRWT groups (Figure 1) and then compared to identify significantly differentially altered regions between the two groups that were common to all three datasets (see Methods). In total, 75 discrete regions comprising ~140 Mb of the genome were differentially altered according to EGFR status in all three groups, ranging in size from 0.5 Mb to 13.5 Mb (Supplemental Table 1). Significant differences were identified on 10 chromosomes in total, with major regions of disparity located on chromosome arms 7p (gained in an average of 64% of EGFRMUT tumors, range 48-74% in the BCCA dataset), 8p (lost in an average of 53% of EGFRMUT tumors, range 37-67% in the BCCA dataset) and 16p (gained in an average of 59% of EGFRMUT tumors, range 51-71% in the BCCA dataset), which are magnified in the Circos plots in Figure 1 (Supplemental Table 1 for specific regions and frequencies). While gain of 7p and loss of 8p have been previously reported to occur specifically in EGFR mutated adenocarcinomas16, 17, gain of 16p was a novel association.
Figure 1. Genomic alterations specific to EGFRMUT and EGFRWT lung adenocarcinoma tumors.
The frequency of copy number gain and loss were determined for the following datasets as described in the Methods Section: (A) 83 LAC tumors from the BCCA consisting of 20 EGFRMUT and 63 EGFRWT tumors; (B) 199 LAC tumors from MSKCC consisting of 43 EGFRMUT and 156 EGFRWT; and (C) 354 tumors from Broad Institute consisting of 29 EGFRMUT and 325 EGFRWT. Frequencies of copy number alteration for each dataset are plotted on circular representation of the human genome (hg18) with the outer circumference representing a ideogram displaying human autosomes. The copy-number status, expressed as the frequency of DNA gains (pointing outwards) and losses (inward) are plotted for EGFRMUT (green) and EGFRWT (blue) for each of the analyzed cohorts. Only regions exhibiting a frequency of 0.2 or higher for both copy number gains and losses are plotted. Regions containing major differences in copy number (chromosomes 7, 8 and 16) are magnified to highlight the changes specific to each group. The black histogram for these regions represents gene density.
Integration of genomic and gene expression data identifies candidate cooperating genes in EGFR mediated tumorigenesis
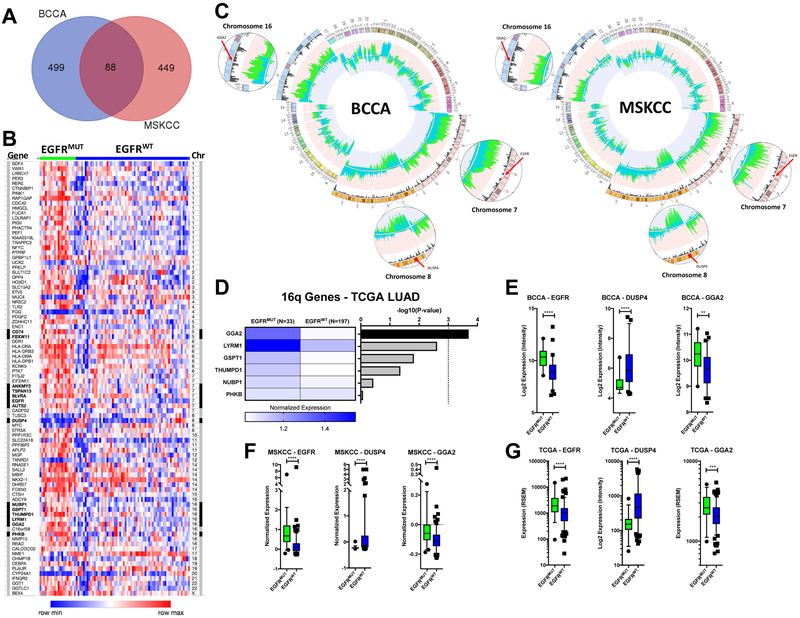
Alterations in gene dosage influence tumorigenesis through increasing or decreasing the transcription levels of the genes they contain. Therefore, in order to identify target genes within regions of copy number difference, we compared gene expression levels between EGFRMUT and EGFRWT tumors. The majority of BCCA (n=83, 20 EGFRMUT and 63 EGFRWT) and MSKCC (n=193, 39 EGFRMUT and 158 EGFRWT) samples from the above copy number cohorts had matching genome-wide expression data available and were used for analysis. Global gene expression profiles were compared between EGFRMUT and EGFRWT tumors for each dataset separately, then genes differentially expressed in both datasets were identified. In total, 587 and 537 genes were differentially expressed (see Methods) between the two groups in the BCCA and MSKCC cohorts, respectively, with 88 unique genes differentially regulated in both groups (Figure 2A and 2B, Supplemental Table 2). The 88 genes showed clear differential expression across EGFRMUT and EGFRWT tumors and contained known EGFRMUT associated genes such as ETV5 and NKX2-118, 19.
Figure 2. Integration of copy number and expression data reveals candidate EGFRMUT cooperating genes.
(A) Genes differentially expressed between EGFRMUT and EGFRWT tumors were determined using SAM in the BCCA and MSKCC datasets as described in the Methods section. 88 unique genes were significantly differentially expressed between the two datasets as indicated by the Venn diagram. (B) A heatmap indicating the relative expression of the 88 genes across the BCCA tumors. Tumors are separated left to right as EGFRMUT (green) and EGFRWT (blue) and genes are listed by relative chromosome position from chromosome 1 (top) to X (bottom). Genes mapping to regions of copy number disparity are indicated by the black highlighting to the left of the gene name and the right of the chromosome number. Expression levels were rank normalized for each gene across the samples using Morpheus software with blue indicating lower relative expression and red relative higher expression. There is a clear distinction in expression of the genes based on mutation status. (C) Copy number profiles for the BCCA datasets as in Figure 1 indicating the position of potential targets of EGFRMUT specific alterations on chromosomes 7 (EGFR), chromosome 8 (DUSP4) and chromosome 16 (GGA2). All genes show more frequent disruption in EGFRMUT tumors with expression matching the direction predicted by copy number. (D) To further identify the target of the chromosome 16 gain in EGFRMUT tumors, expression data for the significantly differentially expressed genes mapping to this region of copy number difference were compared between EGFRMUT and EGFRWT LACs in the TCGA dataset using a Mann-Whitney U-test. On the right, the resulting Bonferoni corrected p-values were –log 10 transformed and plotted in order of significance. On the left, the relative median normalized expression is plotted for each gene in each group, with blue indicating higher and white lower expression. GGA2 was the only gene to reach significant (corrected p<0.001) as indicated by the dashed line and was overexpressed in the EGFRMUT tumors, matching the direction predicted by copy number status (gained in EGFRMUT). (E-G) Box plots indicating the level of expression of EGFR, DUSP4 and GGA2 in EGFRMUT (green) and EGFRWT (blue) LAC tumors in the BCCA (E), MSKCC (F) and TCGA (G) datasets. Each gene is significantly differentially expressed (p<0.05, two-tailed Mann-Whitney U-Test) between EGFRMUT and EGFRWT tumors in each dataset in the direction predicted by copy number (higher in EGFRMUT for EGFR and GGA2, lower in EGFRMUT for DUSP4). Whiskers represent the 5-95 percentiles. *p<0.05, **p<0.01, ***p<0.001 and ****p<0.0001.
Mapping the 88 candidate genes to the regions of copy number difference revealed potential targets of the three main regions of EGFRMUT specific copy number alteration on chromosome arms 7p, 8p and 16p (Figure 2B and 2C). Of the eight differentially expressed genes located on chromosome 7, five mapped to regions of copy number difference (ANKMY2, TSPAN13, BLVRA, EGFR and AUTS2) and all demonstrated increased mRNA expression in EGFRMUT tumors, matching the direction of copy number change (gain/amplification). Of these, EGFR copy number increase has been previously shown to be associated with EGFRMUT tumors and our data further suggests that gain/amplification of the mutant EGFR allele is likely an important event in EGFRMUT tumorigenesis (Figure 2C). On chromosome 8, only a single differentially expressed gene mapped to a region of copy number change, DUSP4, which was located within a region of EGFRMUT specific chromosomal loss and showed concordant downregulation at the transcriptional level (Figure 2C). DUSP4 encodes a dual-specificity phosphatase that acts to downregulate EGFR signaling and has previously been identified as a potential cooperating tumor suppressor gene in EGFRMUT tumors16.
The remaining major regions of copy number difference were on chromosome arm 16p, were gained specifically in EGFRMUT tumors, and have not been previously described in this context. Ten genes on chromosome 16 showed a differential pattern of expression in EGFRMUT tumors, with six (GGA2, LYRM1, GSPT1, THUMPD1, NUBP1 and PHKB) mapping to regions of copy number difference and all demonstrating increased expression in EGFRMUT tumors, mirroring the copy number status (Figure 2B and 2D). In order to identify the potential target gene of this alteration, we next compared the expression of all six genes in an independent cohort of 230 LACs from The Cancer Genome Atlas (TCGA)20 consisting of 33 EGFRMUT and 197 EGFRWT tumors. While all genes showed higher expression in EGFRMUT tumors, matching the results from the original datasets, only one gene, GGA2, met our significance threshold (Bonferoni correct P<0.001, Figure 2D). As with EGFR (gained and overexpressed) and DUSP4 (lost and underexpressed), GGA2 was significantly differentially expressed in the direction predicted by copy number (gained and overexpressed) in the EGFRMUT tumors in all three datasets analyzed (Figures 2E, 2F, and 2G). These results were borne out by protein expression in EGFRMUT and EGFRWT cell lines (Supplemental Figure 1). Taken together, these data suggest that as with previously established EGFRMUT cooperating events including EGFR gain and DUSP4 loss, GGA2 gain and resulting overexpression could represent a candidate collaborating event in LACs driven by the mutant oncogene.
GGA2 is a candidate EGFR cooperating gene on chromosome arm 16p
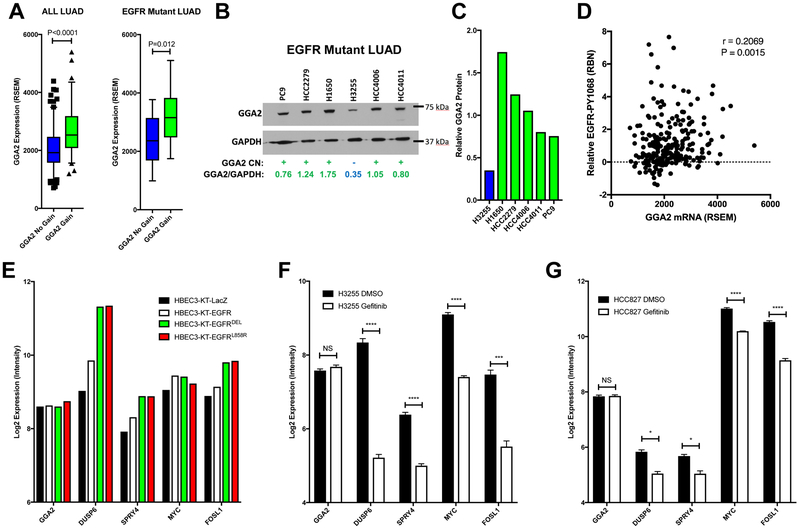
Although GGA2 is located on a region of EGFRMUT specific chromosomal alteration and is overexpressed in EGFRMUT tumors, we wanted to determine whether GGA2 activation is a true secondary event acquired during tumorigenesis or merely driven by mutant EGFR signaling. To confirm that GGA2 expression is driven by copy number alteration we assessed its expression in LAC tumors with different copy number status. As anticipated, TCGA tumors with increased GGA2 copy number showed higher levels of GGA2 mRNA suggesting the genetic alteration drives transcription (Figure 3A). This association was also true when only assessing the EGFRMUT tumors (Figure 3A). Furthermore, GGA2 protein levels were higher in EGFRMUT LAC cell lines with GGA2 gain compared to those without as determined by Western Blot, suggesting that these transcriptional differences are translated to the protein level (Figure 3B and 3C). Clinical LACs from TCGA with higher GGA2 mRNA also demonstrated higher levels of phosphorylated EGFR, confirming the association with active EGFR signaling as expected in EGFRMUT tumors (Figure 3D). Importantly, GGA2 transcription was not driven directly by EGFR signaling, as expression of oncogenic EGFR alleles in non-transformed Human Bronchial Epithelial Cells (HBECs), increased mRNA levels of known pathway targets such as DUSP67, SPRY421, MYC22 and FOSL123, but did not lead to increased GGA2 (Figure 2E). Likewise, treatment of two EGFRMUT LAC cell lines (HCC827 and H3255) with gefitinib did not impact GGA2 expression but did downregulate the same pathway targets described above (Figure 3F and 3G) – further suggesting that GGA2 expression is not dependent on active EGFR signaling in LAC. Similar results were also observed in EGFRMUT LAC cell lines that were treated with osimertinib (described below, Figure 7B). Taken together, we conclude that GGA2 transcriptional activation is not driven by mutant EGFR itself, but instead, is acquired independently through copy number alteration in EGFRMUT tumors.
Figure 3. GGA2 expression is driven by copy number gain in EGFRMUT lung adenocarcinomas.
(A) GGA2 expression is increased in tumors with copy number increased compared to those without. LAC tumors from the TCGA dataset were separated by their GGA2 copy number status and the expression of GGA2 compared between tumors with gain/amplification (green) and those without (blue) using a Mann-Whitney U-Test. Resulting box plots and p-values are provided for all LUAD (left) and only EGFRMUT LAC (right). As we predicted that GGA2 would be higher in cases with copy number increase, a one-tailed p-value was used. (B) GGA2 western blot for LAC cell lines. GGA2 copy number status for each line was defined based on our previous publication. Cell lines with GGA2 gain (+) are indicated in green and those without (−) are indicated in blue. Relative protein levels of GGA2 vs GAPDH were determined by dosimetry and are indicated for each cell line. (C) The relative GGA2 protein expression from (B) is plotted for the same cell lines. LACs with GGA2 gain have higher expression of GGA2 protein than those without. (D) GGA2 mRNA expression correlates with higher phospho-EGFR in LAC. GGA2 mRNA level (RSEM, x-axis) from RNA-Seq and EGFR-PY1068 level (RBN, y-axis) from RPPA are plotted and the resulting Spearman’s correlation coefficient and p-value are indicated revealing a significant positive association as predicted based on the association of GGA2 gain with EGFR mutation. (E) GGA2 expression is not driven by EGFR signaling. The expression of GGA2 and known EGFR regulated genes (DUSP6, SPRY4, MYC and FOSL1) are plotted for HBECs containing control (LacZ), EGFRWT (EGFR) and EGFRMUT (EGFRDEL and EGFRL858R) expression constructs. GGA2 levels do not change upon EGFR activation while known targets increase. (F-G) GGA2 expression is driven independent of EGFR signaling in LAC. Expression for the same genes in (E) are plotted in EGFRMUT LAC (H3255, F and HCC827, G) cell lines treated with the EGFR TKI gefitinib. GGA2 remains the same across all states in both cell lines whereas the known EGFR targets decrease upon receptor inhibition. *p<0.05, **p<0.01, ***p<0.001 and ****p<0.0001, two tailed unpaired t-test.
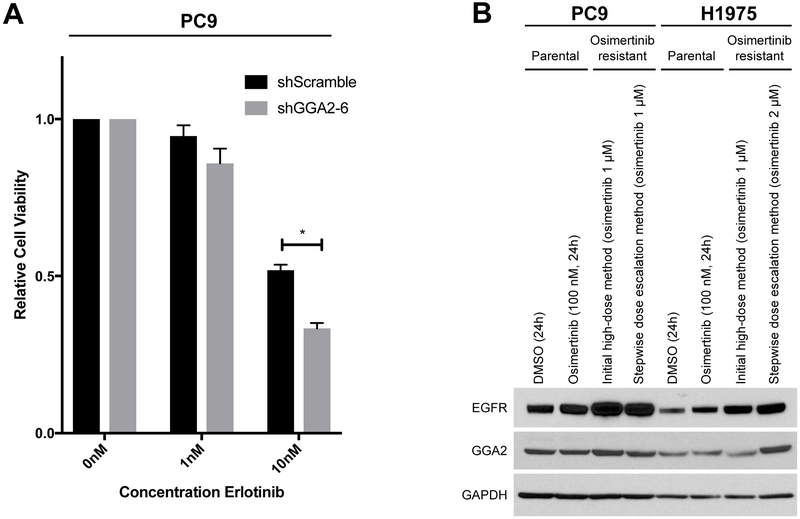
Figure 7. Activation of GGA2 increases EGFR mediated transformation.
(A) EGFRMUT PC9 cells expressing shGGA2-6 or shScramble were treated with the doses of erlotinib indicated for seven days and cell viability assessed by Alamar blue. GGA2 knockdown PC9 cells demonstrated significantly decreased viability at 10 nM erlotinib relative to control cells (*p<0.05, two tailed, unpaired t-test). Bars represent average of two biological replicates +/− SEM. (B) EGFRMUT PC9 and EGFRMUT H1975 cells were treated with osimertinib through different dosing strategies to derive resistant cell clones over time (see methods section). GGA2 levels were then assessed by Western blot and levels compared between resistant, parental and acutely treated sensitive cells for each cell line. GGA2 is overexpressed in the H1975 osimertinib resistant clone derived through dose escalation (far right) compared to the parental and acutely treated H1975 cells, suggesting it may play a role in TKI resistance.
GGA2 interacts with EGFR and stabilizes EGFR levels after activation
The genomic data suggests that activation of GGA2 may be a cooperating event in EGFRMUT tumorigenesis; however, its functional role in this process is unclear. GGA2 is a member of the Golgi-localized, gamma adaptin ear-containing, ARF-binding family (along with GGA1 and GGA3) that have been shown to regulate the trafficking of proteins, mainly between the trans-Golgi network and lysosome24. They are known to play a role in clathrin-coated vesicle assembly and in regulation of cargo proteins. For example, GGA3 interacts with the hepatocyte growth factor RTK (MET) after receptor activation and sorts it for recycling to the cell membrane, suggesting that GGA proteins may play an active role in stabilizing RTK levels by preventing lysosomal degradation25.
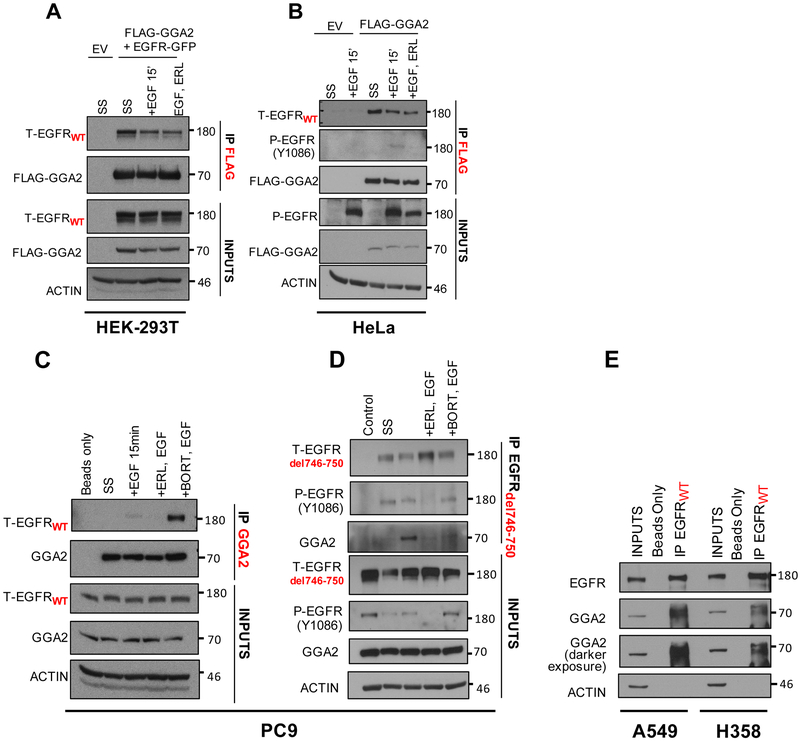
With this in mind, we hypothesized that GGA2 may interact with EGFR in a similar way as GGA3 does with MET. To test this, we first aimed to determine whether GGA2 and EGFR physically interact. Through co-transfection of HEK-293T cells with flag-tagged GGA2 (FLAG-GGA2) and wild-type EGFR (EGFR-GFP), cells were cultured in three different conditions - serum starved (SS), EGF stimulated (+EGF) and EGF stimulated with EGFR TKI treatment (+EGF, Erlotinib (ELR)). We found that overexpressed EGFR co-immunoprecipitated with exogenous GGA2 in all three conditions (Figure 4A). Furthermore, FLAG-GGA2 expressed in HeLa cells interacted with endogenous EGFR under all conditions tested, including the activated phosphorylated form after EGF stimulation, which was reversed by TKI treatment (Figure 4B). To confirm this association between endogenous proteins, we performed immunoprecipitations (IPs) with anti-GGA2 antibody and reverse IP’s with a mutant specific EGFR antibody in EGFR mutant LAC PC9 cells. GGA2 interacted with both endogenous EGFRWT (Figure 4C) and EGFRMUT (Figure 4D) especially after receptor stimulation with EGF ligand and upon inhibiting proteasomal degradation with Bortezomib. Lastly, endogenous EGFRWT also interacted with GGA2 in non-EGFRMUT LAC cells, as indicated by IPs in A549 and H358 cells (Figure 4E). Together, these data suggest that GGA2 interacts with EGFR-wild-type and mutant, which is enhanced upon EGFR stimulation in LAC cells.
Figure 4. EGFR and GGA2 physically interact in lung adenocarcinomas.
(A) HEK-293T cells were transfected with empty vector (EV) or FLAG-GGA2 and EGFR-GFP and (B) HeLa cells were transfected with empty vector (EV) or FLAG-GGA2. 48 hours later, cells were cultured 3 different conditions: SS: serum starvation for 2 hours, +EGF: serum starvation followed by stimulation with EGF ligand for 15 mins, +ERL,EGF: serum starvation followed by incubation with Erlotinib and subsequent stimulation with EGF ligand for 15 mins. FLAG-GGA2 was pulled down with anti-FLAG beads and probed for interaction with EGFR by Western Blot analysis. In HEK-293T cells (A), overexpressed GGA2 interacted with overexpressed EGFR in all 3 conditions. In Hela cells (B), overexpressed GGA2 interacted with endogenous EGFR in all 3 conditions. (C, D) PC9 cells were cultured 4 different conditions: SS: serum starvation for 2 hours, +EGF (50ng/ml): serum starvation followed by stimulation with EGF ligand for 15 mins, +ERL,EGF: serum starvation followed by incubation with Erlotinib (1uM) and subsequent stimulation with EGF ligand for 15 mins, +BORT,EGF: serum starvation followed by incubation with Bortezomib (2uM) and subsequent stimulation with EGF ligand for 15 mins. We performed IP’s with anti-GGA2 antibody (C) and reverse IP’s with anti-EGFRdel746-750 antibody (D). In PC9 cells, GGA2 interacted with both EGFRWT (C) and EGFRdel746-750 (D) especially after receptor stimulation with EGF ligand and upon inhibiting proteasomal degradation with Bortezomib. (E) When probed for interaction in A549 and H358 cell lines by IP GGA2/WB EGFRWT, endogenous GGA2 interacted with EGFRWT in both cell lines.
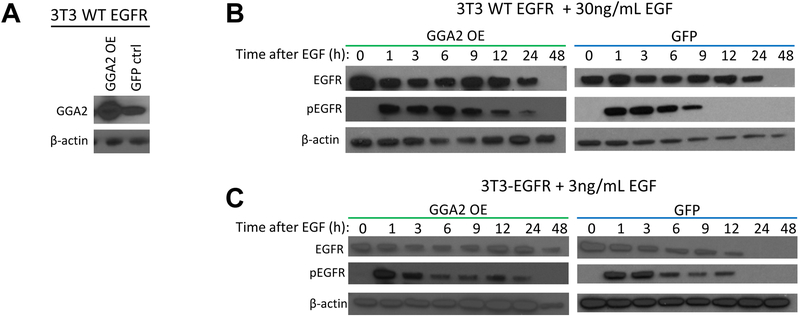
Next, we aimed to determine whether GGA2 stabilizes EGFR levels after activation. Human EGFRWT expressing NIH-3T3 cells were infected with lentiviral vectors expressing either GGA2 or GFP (Figure 5A). These lines were serum starved, treated with cyclohexamide (CHX) to block new protein production and stimulated with EGF for various amounts of time to assess EGFR levels (Figure 5B and 5C). Upon addition of EGF, EGFR becomes phosphorylated (pEGFR) and targeted for degradation at rate dependent on the level of ligand added. After treatment with 30ng/mL of EGF, GGA2 overexpressing (GGA2 OE) cells demonstrated marked enhancement of pEGFR at later time points (12 and 24 hours) compared to GFP expressing control cells (Figure 5B). Likewise, treatment with 3ng/mL of EGF lead to complete absence of both total and phosphorylated EGFR by 24 hours in GFP cells whereas GGA2 OE cells showed sustained EGFR expression at 24 and 48 hours with pEGFR detected at 24 hours as well (Figure 5C). Combined with the data from the IPs, this work suggests that endogenous GGA2 and EGFR interact with each other after receptor stimulation, with GGA2 promoting stability and thereby prolonging EGFR activity.
Figure 5. GGA2 stabilizes EGFR levels after ligand activation.
(A) NIH-3T3 cells expressing EGFRWT were infected with lentiviral constructs to stably overexpress GGA2 (GGA2 OE) or GFP leading to GGA2 overexpression in the former as indicated by Western blot. (B) GGA2 OE and GFP control cells were seeded in serum starved media before being subjected to 20uM cycloheximide for 30 minutes. Lysates were harvested for time point 0. Remaining cells were treated with 30ng/mL EGF and harvested at 1, 3, 6, 9, 12, 24 and 48 hours (h). Lysates were subjected to immunoblot analyses demonstrating that EGFR and pEGFR levels were more persistent in cells overexpressing GGA2. (C) The same experiment as described in (B), with 3ng/mL EGF also indicating increased EGFR stability and activity in the setting of GGA2 overexpresion.
Activation of GGA2 increases EGFR mediated transformation
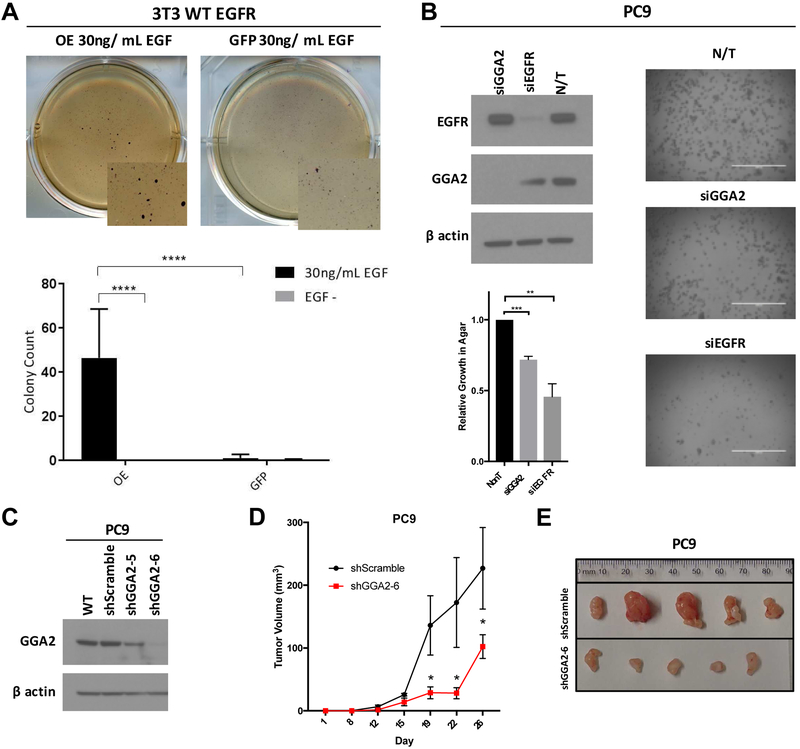
Based on the above findings, we next assessed whether GGA2 overexpression could enhance the transformative ability of EGFR. NIH-3T3 cells overexpressing human EGFR are not transformed and do not form colonies in soft agar until the addition of exogenous EGF (for 4 weeks), offering a useful system to assess the cooperativity of these two proteins. NIH-3T3s overexpressing EGFR and GGA2 formed numerous colonies in soft-agar after 2-weeks in the presence of EGF, whereas GFP control EGFR expressing cells demonstrated few colonies at this time point, even in the presence of EGF (Figure 6A). This suggests that GGA2 not only promotes EGFR stability and activity after EGF stimulation in this system, but it also enhances EGFR’s transformative potential, suggesting that GGA2 activation is a cooperative event in tumorigenesis. To confirm that this association also holds true in human LAC cells, we transfected EGFRMUT PC9 cells with siRNAs to GGA2, EGFR and a non-targeting control (NT) and assessed colony forming potential in soft-agar. As with inhibition of EGFR, knockdown of GGA2 also significantly impaired colony formation in PC9 cells (Figure 6B). Lastly, we attempted to achieve stable knockdown of GGA2 in PC9 cells using two different shRNAs, one of which (shGGA2-6) dramatically decreased GGA2 levels (Figure 6C) and led to a substantial decrease in xenograft tumor formation when implanted in the subcutaneous flank of immunocompromised mice (Figure 6D and 6F). Together, this functional work confirms that GGA2 gain and overexpression is a cooperative event in EGFR mediated tumorigenesis.
Figure 6. Activation of GGA2 increases EGFR mediated transformation.
(A) Overexpression of GGA2 cooperates with EGFR to transforms in NIH-3T3 cells. NIH-3T3 cells expressing WT EGFR were treated with EGF (30ng/mL) to activate pathway signaling with (GGA2 OE) or without (GFP) GGA2 activation and seeded in soft-agar to assess transformation. Above, representative images from soft agar plates 2-weeks after seeding are shown indicating colony formation in the GGA2 OE cells. Below, bar graph of average values +/− standard deviations from biological triplicate experiments. (****p<0.001, Bonferoni corrected ANOVA.) (B) Suppression of GGA2 inhibits growth of PC9 LAC cells. EGFRMUT PC9 cells were transfected with siRNAs targeting GGA2 (siGGA2), EGFR (siEGFR) or a non-targeting control (NT) and suppression of the respective targets confirmed by Western blot (top left). Cells were seeded in soft agar and representative images of resulting colony growth are shown on the right. The relative number of cells present in each condition was determined as described in the Methods section and the relative average compared to the NT control from biological triplicates is plotted in the bottom left, indicating decreased colony formation in the siGGA2 and siEGFR states (**p<0.01, ***p<0.001, one-sample t-test). (C) Western blots assessing knockdown of GGA2 in PC9 cells stably expressing shRNAs targeting GGA2 (shGGA2-5 and shGGA2-6) or a scramble control (shScramble). Dramatic GGA2 knockdown was observed in the shGGA2-6 expressing cell line which was used for subsequent analyses. (D) PC9 cells expressing shGGA2-6 or shScramble were subcutaneously transplanted in NRG mice on alternative flanks and tumor volume measured and plotted +/− SEM. Five independent experiments were performed with shGGA2-6 cells forming smaller tumors (*p<0.05, one-tailed Mann-Whitney U-Test). (E) Representative images of shGGA2-6 and shScramble PC9 cell tumors after resection at endpoint (day 26) demonstrates the smaller size of shGGA2-6 expressing tumors.
Suppression of GGA2 sensitizes EGFR mutant lung adenocarcinoma cells to tyrosine kinase inhibitor treatment
Based on the above findings, we next aimed to determine whether suppression of GGA2 could work in combination with EGFR TKIs to further inhibit EGFRMUT LAC cell growth. We treated PC9 cells with stable knockdown of GGA2 (Figure 6C) with different doses of erlotinib and assessed the impact on cell viability. After seven days of treatment, PC9 cells with GGA2 knockdown demonstrated an additional 30% decrease in cell viability in response to EGFR inhibition by erlotinib than GGA2-expressing control cells (Figure 7A), suggesting that combination-based strategies targeting both GGA2 and EGFR could provide greater TKI response rates in EGFRMUT LAC. Furthermore, we derived EGFRMUT cell lines (PC9 and H1975) resistant to osimertinib through two different dosing strategies (see Methods) and found that one resistant cell line (H1975 established through dose escalation) demonstrated increased GGA2 levels compared to the sensitive parental cells (Figure 7B). This provides preliminary evidence that GGA2 overexpression could be associated with TKI resistance, and further supports its potential as a target for combination-based strategies.
Discussion
The clinical importance and biological role of mutant EGFR in LAC is well established; however, many questions remain to be answered. As it has long been known that oncogenic EGFR -while essential in cancers in which it is mutated - is not sufficient for malignant transformation, one of the most pertinent questions is: what genomic alterations cooperate with mutant EGFR to drive LAC development? To identify such events, we performed a large scale integrative genomic and gene expression analyses of LACs and identified three main changes specific to EGFRMUT LAC tumors: 1) gain of chromosome arm 7p which increases the expression of EGFR, 2) loss of chromosome arm 8p leading to downregulation of DUSP4, and 3) gain of chromosome arm 16p causing the overexpression of GGA2. Gain/amplification of the mutant EGFR allele has previously been reported to frequently occur in LAC and is known to drive tumor progression17. Likewise, DUSP4 has been shown to suppress EGFRMUT LAC through negative feedback control of EGFR signaling and its loss relieves this suppression, increasing cell growth16. However, the finding that GGA2 is gained and overexpressed specifically in EGFRMUT LAC is novel, and prompted us to ask whether, like the EGFR and DUSP4 events, activation of GGA2 is involved in amplifying EGFR signaling to promote tumor progression. It should be noted that the gain of chromosome arm 16p has previously been described by our group and others in LAC and was found to be associated with tumors from never smokers15, 26-28. While the association of 16p amplification with EGFR mutation status was not assessed in these studies, tumors from never smokers are enriched in the frequency of EGFR mutation, providing a potential explanation for these previous observations.
Due to its essential role in regulating cellular growth and proliferation, EGFR signaling is normally controlled by a myriad of mechanisms - including negative feedback regulation through the induction of phosphatases and controlling EGFR protein levels – that become disrupted in cancer. DUSP4 loss would exploit the former category, while EGFR amplification would affect the later29. Another important process regulating EGFR protein levels is EGFR trafficking30. Typically, after stimulation of EGFR by ligand, the activated cell-surface receptor becomes internalized, localized in early endosomes and later in lysosomes, which mediate its degradation and ensure normal cellular homeostasis30. Mutant EGFR, despite being constitutively active, is also subjected to trafficking i.e. internalization and localization to the endosomes; but is often impaired in its interactions with Cbl, an E3 ubiquitin ligase, resulting in defective ubiquitination and degradation30. EGFR localized to the early endosomes can also be recycled back to the plasma membrane instead of proceeding to the lysosome, leading to sustained protein half-life and prolonged signaling30. However, the factors that promote receptor recycling versus degradation are still relatively unknown, especially in the context of LAC. Indeed, proteins promoting EGFR recycling would be expected to cooperate with mutant EGFR much like EGFR amplification and DUSP4 inactivation by increasing receptor signaling.
GGA2 and its related family members GGA1 and GGA3 are well known clathrin adapter proteins that function in protein sorting and trafficking, mainly between the trans-Golgi network and endosomes/lysosomes24. They do so by localizing to the trans-Golgi network from the cytosol through binding of their GGA and Tom1 (GAT) domain to ARF GTPases and PI4P, then binding acidic-cluster dileucine (ACLL) motifs in the cytoplasmic tails of cargo proteins through their VPS-27, Hrs and STAM domain (VHS), facilitating their incorporation into clathrin-coated vesicles for delivery to endosomes31. In this manner, GGAs have been shown to regulate the TGN-endosome trafficking of many cargo molecules with ACLL motifs including sortilin32, manose-6-phosphate receptors33, B-secretase34, stabilin-134, chloride channel 734, consortin35 and others. More recently, GGAs have also been implicated in regulating cell membrane trafficking. For example, GGA1-3 regulate the cell surface transport of α2B-adrenegic receptor (α2B-AR) - a G protein-coupled receptor – and their inhibition decreases the cell surface expression of α2B-AR and attenuates downstream signal transduction though ERK1/236. Likewise, GGA3 is known to promote the recycling of activated MET, promoting prolonged signalling and cell migration25.
Based on these observations, we hypothesized that GGA2 activation may promote the stability of activated EGFR – increasing receptor signalling and promoting EGFR driven tumorigenesis. To this end, we found that GGA2 physically interacts with both wild type and mutant EGFR and that this association is increased by ligand activation (Figure 4). Furthermore, overexpression of GGA2 stabilizes EGFR protein levels after activation (Figure 5), promoting EGFR driven transformation while inhibition of GGA2 supresses growth of EGFRMUT LAC cells (Figure 6). Together, these data suggest for the first time that genetic changes that increase GGA2 provide a selective advantage for EGFRMUT LAC cells, likely through impairing EGFR degradation and prolonging receptor signalling. In the process of assembling this manuscript, a hypothesis driven study by Uemura and colleagues was published that provide additional evidence that GGA2 interacts with the cytoplasmic domain of EGFR, stabilizing receptor expression and promoting growth of colorectal carcinoma cells37. Further, they showed that GGA2 is overexpressed in 23-30% of human hepatocellular carcinoma and colorectal cancers37. Combined with our comprehensive and unbiased genomic approach that uncovered GGA2 in EGFRMUT LAC, this provides compelling evidence that GGA2 functions to promote EGFR signalling after activation and enhances tumor development. We also found that GGA2 is expressed in several other cancer types to levels similar or higher than non-small cell lung cancer (Supplemental Figure 2), suggesting it may play a role in the development of other tumor types as well. However, the specific mechanism by which GGA2 regulates EGFR (or other oncogenic protein) trafficking in lung or other cancers remains to be elucidated and is subject to future investigation.
To conclude, our study demonstrates that EGFRMUT LACs contain frequent chromosomal alterations that affect EGFR, DUSP4 and GGA2 in a manner that promotes increased EGFR signaling. These genetic events likely occur after the acquisition of mutant EGFR and modify the transformative potential of oncogenes, facilitating tumor progression. Further, as GGA2 gain/amplification was not associated with patient outcome in EGFRMUT LACs in our datasets (data not shown), it is likely that each of these genetic events provides a similar advantage to EGFRMUT LACs during development. However, identification of these cooperating alterations, specifically GGA2, will likely have a significant impact on our understanding of EGFR signaling as well as translational opportunities for therapy. For example, developing inhibitors of GGA2 to use in combination with EGFR TKIs may provide an opportunity to combat both primary and secondary resistance and increase long term survival in LAC patients. Lastly, our work highlights the importance of alterations affecting EGFR trafficking in promoting tumor progression and suggest that a greater understanding of this process may offer new avenues for therapeutic intervention for LAC and other cancers dependent on TKI signaling.
Materials and Methods
Comparison of copy number alterations
Three datasets were used for the copy number comparison of EGFRMUT and EGFRWT LAC tumors. The first was from our institute, BCCA, and consisted of 83 LAC tumors, 20 EGFRMUT and 63 EGFRWT, which have been used in our previous studies15, 38. All tissues were collected from the Tumor Tissue Repository of the British Columbia Cancer Agency under informed, written patient consent and with approval from the University of British Columbia - BC Cancer Agency Research Ethics Board. Genomic DNA from these tumors and matched non-malignant lung tissues were hybridized to Affymetrix SNP 6.0 arrays according to the manufacturer's instructions and the resulting normalization and copy number segmentation was performed using Partek Genomics Suite Software (Partek Incorporated, Missouri) with the same settings and downstream processing we previously described15. The second dataset was from a study from MSKCC and consisted of 199 LAC tumors, 43 EGFRMUT and 156 EGFRWT collected at the time of resection under protocols approved by the MSKCC Institutional Review Board with selection criteria for study inclusion previously described16. Agilent 4×44 k array comparative genomic hybridization (aCGH) data and accompanying EGFR mutational status were obtained from http://cbio.mskcc.org/Public/lung_array_data/ and copy number profiles were generated using the segmentation algorithm, FACADE with default parameters and a baseline distribution of 10 kbp15, 38, 39. The last dataset was from the the Tumor Sequencing Project (TSP) and consisted of 354 LAC tumors, 29 EGFRMUT and 325 EGFRWT collected with consent of the human subjects Institutional Review Boards of participating institutions with selection criteria for study inclusion previously described40, 41. Affymetrix SNP 250 K array data for this cohort were downloaded from the Database of Genotypes and Phenotypes (Study Accession: Study Accession: phs000144.v1.p1) and processed in Partek as previously described15. EGFR mutation status was determined by linking to the data provided in the publication by Ding et.al 40
To identify regions of copy number disparity between EGFRMUT and EGFRWT tumors, we employed a strategy we previously used for the comparison of alterations between lung cancer subsets12-15. Briefly, segmental alterations identified in each tumor were parsed into typed copy numbers for each array element (1 - copy gain, 0 - copy neutral, or −1 - copy loss. Probes with similar copy number states within individual tumors were then collapsed into genomic regions across all tumor samples. The frequency of DNA gains, DNA losses, and neutral copy number were then compared in EGFRMUT and EGFRWT LAC genomes using a Fisher's Exact test performed in R with a 3×2 contingency table, with a p-value ≦0.05 considered significant. Significant regions within 1 Mbp of each other and with the same copy number status were merged into single regions. Differentially altered regions had to have a difference of at least 0.1 in frequency of alterations between EGFRMUT and EGFRWT, and a minimal observed frequency of 0.1 in at least one group to be considered. Using the above, the BCCA cohort resulted in 800 Mb of genomic DNA affected copy-number changes, TSP displayed 1,000 Mb of altered DNA and MSKCC 375 Mb. The intersection of genomic intervals amongst the three cohorts was determined using the “Intersect/Operate on Genomic Intervals/Intersect” function running on the public Galaxy server using Hg18 coordinates. Based on array resolutions, we only considered regions >500 kb in our final assessment. To plot the frequencies of copy number alteration for each dataset, circular representation of the human genome (hg18) was developed using the circos package implemented at the clicO platform.
Gene expression analysis and data integration
The BCCA and MSKCC datasets described above had matching gene expression profiles for all (BCCA) or a subset (n=193, 39 EGFRMUT and 158 EGFRWT) of tumors profiled for copy number, respectively. BCCA tumors were profiled on Illumina WG6-V3 expression arrays and normalized a previously described42. This data is available through GEO (accession number GSE75037). Data for MSKCC tumors profiled on Affymetrix (U133A and U133A 2.0) arrays were downloaded from http://cbio.mskcc.org/Public/lung_array_data/, normalized using GCRMA with the “affy” package from Bioconductor and z-transformed to allow comparisons across array platforms. Statistical Analysis of Microarrays (SAM)43 was used to compare global gene expression between EGFRMUT and EGFRWT groups for each dataset using parametric (BCCA, q<5) or non-parametric (MSKCC, q<1) settings as appropriate. In the instance of multiple significant probes for a given gene, the probe with the maximum intensity across the respective dataset was used44. Only probes mapping to annotated genes were considered. Significant genes in each dataset were then compared to identify those that overlapped. Morpheus from the Broad Institute was used to generate heatmaps (https://software.broadinstitute.org/morpheus/). Genes significantly differentially expressed in both datasets were then mapped to genomic region to identify those mapping to regions of copy number change (Supplemental Table 2).
Analysis of validation datasets
For genes of interest, mutation, copy number and gene expression data for 230 LAC tumors (TCGA Lung Adenocarcinoma Provisional dataset) profiled by TCGA20 were downloaded from the MSKCC cBIO Portal (http://www.cbioportal.org/)45. Expression (RNA Seq V2 RSEM) for each gene was compared between EGFRMUT and EGFRWT tumors using a Mann-Whitney U-Test in Graphpad Prism software. Similar analysis was performed for GGA2 by stratifying samples based on GGA2 copy number status (GISTIC values). Reverse Phase Protein Array (RPPA, replicate base normalized (RBN)) data for phospho-EGFR (Y1068) was available for a subset of tumors and downloaded from the UCSC Cancer Genomics Browser and correlated to GGA2 expression using a non-parametric Spearman’s correlation. Data from immortalized Human Bronchial Epithelial Cells (HBEC) expressing mutant EGFR alleles were profiled using Affymetrix U133A 2.0 array as previously described7. Data for gefitinib treated LAC cell lines profiled using Illumina Human HT-12 arrays were generated as previously described46.
Cell lines and culture conditions
All cell lines were obtained from American Type Tissue Culture or from Dr. Romel Somwar (MSKCC). Lung cancer cell lines were all maintained in RPMI-1640 media supplemented with 10% fetal bovine serum (FBS), 1% Glutamax and Pen/Strep. 3T3 WT EGFR cells were created by infecting NIH-3T3 cells with a retroviral construct expressing wild-type EGFR (Addgene #11011) and selecting resistant cells with puromycin. These cells were then engineered to express either human GGA2 (GGA2 OE) or GFP by infecting with lentiviral constructs driving these genes from a CMV promoter (pLenti-CMV-Blast, Addgene #17451) and selecting with blasticidin. All 3T3 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM), containing 10% FBS and Pen/Strep. PC9 cells were infected with lentiviral short hairpin RNA constructs targeting GGA2 - shGGA2-5 (TRCN0000065005) and shGGA2-6 (TRCN0000065006) - along with a scramble shRNA control (shScramble) and selected with puromycin to generate stable cell lines as previously described47. Knockdown was assessed by Western blot using the antibodies described below. shGGA2-6 resulted in the greatest degree of knockdown and was used for all subsequent experiments. All cell lines were grown in a humidified incubator at 37°C and with 5% CO 2.
Pulse chase assay
3T3 EGFRWT GGA2 overexpression (GGA2 OE) and GFP control cells were seeded in DMEM, 1% FBS at 2.5 – 3.5 × 105 cells/well in 6 well plates overnight. The next morning media was removed and replaced with 1mL fresh DMEM, 1% FBS with 20uM of cycloheximide (CHX). Cells were incubated at 37°C for 30 minutes, following addition of another 1mL media with 20uM CHX and 30ng/mL or 3ng/mL EGF (except for time-point 0, lysed immediately after 30-minute initial CHX stimulation, EGF-). Cells were lysed with RIPA buffer and protease phosphatase inhibitor (1:100) based on time-points (1hr, 3hr, 6hr, 9hr, 12hr, 24hr and 48hr). Lysates were cleared and prepped for immunoblotting analysis by SDS-PAGE.
Immunoprecipitation
PC9, A549 and H358 cells were grown as above. HEK-293T and HeLa cells were cultured in DMEM supplemented with 10% FBS. DNA transfections were performed using PEI (PEI 2.5:1 DNA). 48 hours post-transfection, cells were serum-starved for 2 hours. Next, EGF (50 ng/ml) was added to stimulate the cells. Erlotinib (1uM) or bortezomib (2 μM) were added 30 minutes prior to stimulating with EGF. At the end of stimulation, cells were harvested in CHAPS lysis buffer [1% CHAPS detergent, 150 mM NaCl, 50 mM Tris (pH 7) and 5 mM EDTA] supplemented with protease and phosphatase inhibitors. Total protein was quantified using BCA protein assay reagent kit from Pierce Biotechnology (Rockford, IL, U.S.A.) as per the manufacturer's protocol.
For immunoprecipitation, 400 μg of total cell lysate was incubated in 400 μl of total CHAPS buffer (1ug/ul) and incubated with indicated affinity matrix (anti-FLAG beads, 1 h at 4°C) or anti-GGA2 antibody (overnight, 4°C) and anti-EGFR antibody (overnight, 4°C) followed by Protein A/G agarose beads for one hour at 4°C. Following in cubation, the matrix was washed three times in CHAPS buffer, and then SDS loading buffer was added directly to washed matrix, boiled and loaded directly into the wells of a PAGE gel.
Western blot analysis
BCA assays were used to determine protein concentrations of lysates and consistent concentrations of protein lysate were ran on SDS-PAGE and immunoblotted with the following primary antibodies; wild-type EGFR (Cell Signaling Technology, CST # 2232), EGFRdel746-750 (CST # 2085), phospho-EGFR (Tyr1068, CST # 2234), GGA2 (Santa Cruz Biotechnology, SC # 133147 and Abcam, ab185557), FLAG (Sigma #F7425), and β-actin (13E5) (Cell Signaling Technology and Sigma # A5316).
Soft-agar anchorage-independent growth assays
Soft-agar anchorage-independent growth assays were performed using 3T3 wild-type EGFR cells expressing pLenti CMV BLAST GGA2 Over-expression (OE) and pLenti CMV BLAST GFP (GFP) were suspended in top agar layer containing DMEM, 10% FBS and 0.35% low melting point agar (Thermofisher/Invitrogen, Waltham, MA, USA). Bottom agar layer contained DMEM, 10% FBS and 0.7% Ultra pure agar (Thermofisher/Invitrogen, Waltham, MA, USA). 30ng/mL EGF was added to the bottom and top layers as well as top media for each cell line where indicated, and cells which were plated in triplicate. After a 2-week incubation period wells were stained with MTT (3-(4, 5 – Dimethyliazol-2-yl)-2,5-Diphenyltetrazolium Bromide) for 4 hours and the number of colonies determined from scans using ImageJ software, by altering the threshold to remove background, making a binary with mask overlay and manually counting colonies. Counts were then averaged between two independent experiments of triplicates and colony counts were graphed using GraphPad Prism version 7.0. Significance between EGF+ and EGF – conditions (p<0.05) determined by a Two-Way ANOVA and Bonferroni post test.
225,000 cells (PC9) per well were seeded in a 6-well plate. Cells were then transfected with ON-TARGETplus siRNA pools (Dharmacon) against the following targets as previously described48: EGFR (L-003114-00-0010), GGA2 (L-012908-01-0005) and Non-targeting control (D-001810-10-20). For consistent transfection efficiency across experiments, 10uL of 10uM siRNA pool was added to 190uL of OptiMEM (Life Technologies) and 5uL of Dharmafect 1 was added to 195uL of OptiMEM at room temperature. The siRNA and Dharmafect suspensions were mixed and incubated for 20 minutes prior to transfection. Media was changed 24 hours after transfection. The following day cells were trypsonized and counted. 80,000 cells from each transfection condition were seeded in a 6-well plate and harvested four days later to confirm siRNA knock down (western blots performed as described above). Soft agar assay was performed using CytoSelect 96-well cell transformation assay (CBA-130) from Cell Biolabs Inc. 2,000 cells (PC9) were seeded as described by manufacturer onto a 96-well plate (Falcon). The manufacturer’s protocol was followed except when culture medium was removed prior to agar solubilization. Culture medium was removed using a cotton swab to absorb medium rather than being removed by inverting the plate and blotting on paper towel as directed. Photos of the wells were taken before and after agar solubilization. Fluorescence measurement was performed using a Cytation 3 Multi Modal Reader with Gen5 software (BioTek). Values were normalized to the non-targeting control and compared between conditions using a one-sample t-test as previously described48.
In vivo tumor formation assays
Tumor forming ability of PC9 shGGA2-6 and shScramble cells was assessed in NRG (NOD-Rag1null IL2rgnull, NOD rag gamma) mice. Subcutaneous flank injections of 750,000 cells in 100ul of PBS were injected into four-month-old mice obtained from the BCCA Animal Resource Centre. Tumor size and volume was measured by palpation at 8 days following injection and every 3-4 days thereafter as previously described. Experiments were terminated 26 days after injection. Tumor burden between mice injected with shGGA2-6 cells and those expressing and shScramble control vector was assessed using a one-tailed Mann-Whitney T-test.
Tyrosine kinase inhibitor treatments and establishment of osimertinib-resistant cell lines
Ten thousand PC9 shGGA2-6 or shScramble cells were seeded in triplicate in 6-well plates. The next day, media was aspirated and replaced with media containing indicated final concentrations of erlotinib in 0.1% DMSO. Cells were grown for seven days with media/drug replenished every two days. At endpoint, media was aspirated and replaced with fresh media/drug containing Alamar Blue (Thermo Fisher), incubated and fluorescence read on a Cytation 3 Multi Modal Reader with Gen5 software (BioTek).
PC9 and H1975 cells were treated with osimertinib (Selleckchem, S7279) to generate resistant cells through two methods: first, both cell lines were treated with 1 μ M osimertinib (initial high-dose method), until cells resumed growth kinetics similar to that of drug-naïve cells: second, both cell lines were treated with gradually increasing concentrations of osimertinib (stepwise dose-escalation method), at the starting dose of 10 and 30 nM to the final dose of 1 and 2 μ M for PC9 and H1975, respectively. Osimertinib was refreshed every 72 to 96 hours. Resistant cells were maintained as polyclonal populations under the final drug concentrations. Lysates were collected from these cells under osimertinib treatment.
Supplementary Material
Supplemental Table 1: Significant regions of copy number difference between EGFRMUT and EGFRWT lung adenocarcinoma tumors.
Supplemental Table 2: Significant differentially expressed genes between EGFRMUT and EGFRWT lung adenocarcinoma tumors mapping to regions of copy number disparity.
Supplemental Figure 1: GGA2 expression in EGFRMUT and EGFRWT lung adenocarcinoma cell lines. (A) Western blot of GGA2 protein levels in EGFRMUT (left) and EGFRWT (represented by the KRASMUT and EGFR/KRASWT cell lines, right) lung adenocarcinoma cell lines. Samples were run on two separate gels, with H3255 cells run on each for comparison. (B) Dosimetry assessment of the blots from (A) showing the relative GGA2/GAPDH level for EGFRMUT and EGFRWT adenocarcinoma cell lines. H460 is a large cell carcinoma and was excluded. EGFRMUT cells demonstrate higher GGA2 protein on average, mirroring the transcript levels in human adenocarcinoma tumors.
Supplemental Figure 2: GGA2 expression across cancer cell lines. Microarray data was available for 967 cancer cell lines from the Broad Institute and Novartis Cancer Cell Line Encyclopedia and was downloaded through the cBioPortal (http://www.cbioportal.org/). mRNA levels (log2 fluorescence intensity) of GGA2 were median normalized across all cell lines and the resulting values plotted for each cancer type as box plots. Whiskers represent 10-90 percentiles with dots representing values beyond these limits.
Acknowledgements
The authors would like to thank Raj Chari for providing constructive feedback and technical assistance. This work was funded by the Canadian Institutes of Health Research (CIHR; PJT-148725), Terry Fox Research Institute and the BC Cancer Foundation. L. Beverly is supported by NIH NCI R01CA193220 and Kosair Pediatric Cancer Research Program. W. Lockwood is a Michael Smith Foundation for Health Research Scholar, CIHR New Investigator and International Association for the Study of Lung Cancer Young Investigator.
Financial Support: This work was funded by the Canadian Institutes of Health Research (CIHR; PJT-148725), Terry Fox Research Institute and the BC Cancer Foundation. L. Beverly is supported by NIH NCI R01CA193220 and Kosair Pediatric Cancer Research Program. W. Lockwood is a Michael Smith Foundation for Health Research Scholar, CIHR New Investigator and International Association for the Study of Lung Cancer Young Investigator.
Footnotes
RS has received research funding from Helsinn Healthcare. All other authors declare no potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sullivan JP, Minna JD, and Shay JW, Evidence for self-renewing lung cancer stem cells and their implications in tumor initiation, progression, and targeted therapy. Cancer Metastasis Rev, 2010. 29(1): p. 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pao W and Girard N, New driver mutations in non-small-cell lung cancer. Lancet Oncol, 2011. 12(2): p. 175–80. [DOI] [PubMed] [Google Scholar]
- 3.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, and Meyerson M, EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science, 2004. 304(5676): p. 1497–500. [DOI] [PubMed] [Google Scholar]
- 4.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, Mardis E, Kupfer D, Wilson R, Kris M, and Varmus H, EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A, 2004. 101(36): p. 13306–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mok T, Wu YL, and Zhang L, A small step towards personalized medicine for non-small cell lung cancer. Discov Med, 2009. 8(43): p. 227–31. [PubMed] [Google Scholar]
- 6.Pao W, and Chmielecki J, Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat Rev Cancer, 2010. 10(11): p. 760–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sato M, Vaughan MB, Girard L, Peyton M, Lee W, Shames DS, Ramirez RD, Sunaga N, Gazdar AF, Shay JW, and Minna JD, Multiple oncogenic changes (KRAS(V12), p53 knockdown, mutant EGFRs, p16 bypass, telomerase) are not sufficient to confer a full malignant phenotype on human bronchial epithelial cells. Cancer Res, 2006. 66(4): p. 2116–28. [DOI] [PubMed] [Google Scholar]
- 8.Gazdar AF, and Minna JD, Deregulated EGFR signaling during lung cancer progression: mutations, amplicons, and autocrine loops. Cancer Prev Res (Phila), 2008. 1(3): p. 156–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sato M, Larsen JE, Lee W, Sun H, Shames DS, Dalvi MP, Ramirez RD, Tang H, DiMaio JM, Gao B, Xie Y, Wistuba II, Gazdar AF, Shay JW, and Minna JD, Human lung epithelial cells progressed to malignancy through specific oncogenic manipulations. Mol Cancer Res, 2013. 11(6): p. 638–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Politi K, Zakowski MF, Fan PD, Schonfeld EA, Pao W, and Varmus HE, Lung adenocarcinomas induced in mice by mutant EGF receptors found in human lung cancers respond to a tyrosine kinase inhibitor or to down-regulation of the receptors. Genes Dev, 2006. 20(11): p. 1496–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pao W, Iafrate AJ, and Su Z, Genetically informed lung cancer medicine. J Pathol, 2011. 223(2): p. 230–40. [DOI] [PubMed] [Google Scholar]
- 12.Coe BP, Lockwood WW, Girard L, Chari R, Macaulay C, Lam S, Gazdar AF, Minna JD, and Lam WL, Differential disruption of cell cycle pathways in small cell and non-small cell lung cancer. Br J Cancer, 2006. 94(12): p. 1927–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lockwood WW, Chari R, Coe BP, Thu KL, Garnis C, Malloff CA, Campbell J, Williams AC, Hwang D, Zhu CQ, Buys TP, Yee J, English JC, Macaulay C, Tsao MS, Gazdar AF, Minna JD, Lam S, and Lam WL, Integrative genomic analyses identify BRF2 as a novel lineage-specific oncogene in lung squamous cell carcinoma. PLoS Med, 2010. 7(7): p. e1000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lockwood WW, Wilson IM, Coe BP, Chari R, Pikor LA, Thu KL, Solis LM, Nunez MI, Behrens C, Yee J, English J, Murray N, Tsao MS, Minna JD, Gazdar AF, II Wistuba, C.E MacAulay, S Lam, and W.L Lam, Divergent genomic and epigenomic landscapes of lung cancer subtypes underscore the selection of different oncogenic pathways during tumor development. PLoS One, 2012. 7(5): p. e37775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thu KL, Vucic EA, Chari R, Zhang W, Lockwood WW, English JC, Fu R, Wang P, Feng Z, MacAulay CE, Gazdar AF, Lam S, and Lam WL, Lung adenocarcinoma of never smokers and smokers harbor differential regions of genetic alteration and exhibit different levels of genomic instability. PLoS One, 2012. 7(3): p. e33003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chitale D, Gong Y, Taylor BS, Broderick S, Brennan C, Somwar R, Golas B, Wang L, Motoi N, Szoke J, Reinersman JM, Major J, Sander C, Seshan VE, Zakowski MF, Rusch V, Pao W, Gerald W, and Ladanyi M, An integrated genomic analysis of lung cancer reveals loss of DUSP4 in EGFR-mutant tumors. Oncogene, 2009. 28(31): p. 2773–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soh J, Okumura N, Lockwood WW, Yamamoto H, Shigematsu H, Zhang W, Chari R, Shames DS, Tang X, MacAulay C, Varella-Garcia M, Vooder T, Wistuba II, Lam S, Brekken R, Toyooka S, Minna JD, Lam WL, and Gazdar AF, Oncogene mutations, copy number gains and mutant allele specific imbalance (MASI) frequently occur together in tumor cells. PLoS One, 2009. 4(10): p. e7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liao S, Davoli T, Leng Y, Li MZ, Xu Q, and Elledge SJ, A genetic interaction analysis identifies cancer drivers that modify EGFR dependency. Genes Dev, 2017. 31(2): p. 184–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taguchi A, Politi K, Pitteri SJ, Lockwood WW, Faca VM, Kelly-Spratt K, Wong CH, Zhang Q, Chin A, Park KS, Goodman G, Gazdar AF, Sage J, Dinulescu DM, Kucherlapati R, Depinho RA, Kemp CJ, Varmus HE, and Hanash SM, Lung cancer signatures in plasma based on proteome profiling of mouse tumor models. Cancer Cell, 2011. 20(3): p. 289–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas Research, N., Comprehensive molecular profiling of lung adenocarcinoma. Nature, 2014. 511(7511): p. 543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masoumi-Moghaddam S, Amini A, and Morris DL, The developing story of Sprouty and cancer. Cancer Metastasis Rev, 2014. 33(2–3): p. 695–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chou YT, Lin HH, Lien YC, Wang YH, Hong CF, Kao YR, Lin SC, Chang YC, Lin SY, Chen SJ, Chen HC, Yeh SD, and Wu CW, EGFR promotes lung tumorigenesis by activating miR-7 through a Ras/ERK/Myc pathway that targets the Ets2 transcriptional repressor ERF. Cancer Res, 2010. 70(21): p. 8822–31. [DOI] [PubMed] [Google Scholar]
- 23.Ramnarain DB, Park S, Lee DY, Hatanpaa KJ, Scoggin SO, Otu H, Libermann TA, Raisanen JM, Ashfaq R, Wong ET, Wu J, Elliott R, and Habib AA, Differential gene expression analysis reveals generation of an autocrine loop by a mutant epidermal growth factor receptor in glioma cells. Cancer Res, 2006. 66(2): p. 867–74. [DOI] [PubMed] [Google Scholar]
- 24.Scott PM, Bilodeau PS, Zhdankina O, Winistorfer SC, Hauglund MJ, Allaman MM, Kearney WR, Robertson AD, Boman AL, and Piper RC, GGA proteins bind ubiquitin to facilitate sorting at the trans-Golgi network. Nat Cell Biol, 2004. 6(3): p. 252–9. [DOI] [PubMed] [Google Scholar]
- 25.Parachoniak CA, Luo Y, Abella JV, Keen JH, and Park M, GGA3 functions as a switch to promote Met receptor recycling, essential for sustained ERK and cell migration. Dev Cell, 2011. 20(6): p. 751–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong MP, Fung LF, Wang E, Chow WS, Chiu SW, Lam WK, Ho KK, Ma ES, Wan TS, and Chung LP, Chromosomal aberrations of primary lung adenocarcinomas in nonsmokers. Cancer, 2003. 97(5): p. 1263–70. [DOI] [PubMed] [Google Scholar]
- 27.Karlsson A, Ringner M, Lauss M, Botling J, Micke P, Planck M, and Staaf J, Genomic and transcriptional alterations in lung adenocarcinoma in relation to smoking history. Clin Cancer Res, 2014. 20(18): p. 4912–24. [DOI] [PubMed] [Google Scholar]
- 28.Job B, Bernheim A, Beau-Faller M, Camilleri-Broet S, Girard P, Hofman P, Mazieres J, Toujani S, Lacroix L, Laffaire J, Dessen P, Fouret P, and Investigators LG, Genomic aberrations in lung adenocarcinoma in never smokers. PLoS One, 2010. 5(12): p. e15145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Avraham R, and Yarden Y, Feedback regulation of EGFR signalling: decision making by early and delayed loops. Nat Rev Mol Cell Biol, 2011. 12(2): p. 104–17. [DOI] [PubMed] [Google Scholar]
- 30.Tomas A, Futter CE, and Eden ER, EGF receptor trafficking: consequences for signaling and cancer. Trends Cell Biol, 2014. 24(1): p. 26–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonifacino JS, The GGA proteins: adaptors on the move. Nat Rev Mol Cell Biol, 2004. 5(1): p. 23–32. [DOI] [PubMed] [Google Scholar]
- 32.Nielsen MS, Madsen P, Christensen EI, Nykjaer A, Gliemann J, Kasper D, Pohlmann R, and Petersen CM, The sortilin cytoplasmic tail conveys Golgi-endosome transport and binds the VHS domain of the GGA2 sorting protein. EMBO J, 2001. 20(9): p. 2180–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doray B, Ghosh P, Griffith J, Geuze HJ, and Kornfeld S, Cooperation of GGAs and AP-1 in packaging MPRs at the trans-Golgi network. Science, 2002. 297(5587): p. 1700–3. [DOI] [PubMed] [Google Scholar]
- 34.He X, Chang WP, Koelsch G, and Tang J, Memapsin 2 (beta-secretase) cytosolic domain binds to the VHS domains of GGA1 and GGA2: implications on the endocytosis mechanism of memapsin 2. FEBS Lett, 2002. 524(1-3): p. 183–7. [DOI] [PubMed] [Google Scholar]
- 35.del Castillo FJ, Cohen-Salmon M, Charollais A, Caille D, Lampe PD, Chavrier P, Meda P, and Petit C, Consortin, a trans-Golgi network cargo receptor for the plasma membrane targeting and recycling of connexins. Hum Mol Genet, 2010. 19(2): p. 262–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang M, Huang W, Gao J, Terry AV, and Wu G, Regulation of alpha2B-Adrenergic Receptor Cell Surface Transport by GGA1 and GGA2. Sci Rep, 2016. 6: p. 37921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uemura T, Kametaka S, and Waguri S, GGA2 interacts with EGFR cytoplasmic domain to stabilize the receptor expression and promote cell growth. Sci Rep, 2018. 8(1): p. 1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilson IM, et al. , EYA4 is inactivated biallelically at a high frequency in sporadic lung cancer and is associated with familial lung cancer risk. Oncogene, 2014. 33(36): p. 4464–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coe BP, Chari R, MacAulay C, and Lam WL, FACADE: a fast and sensitive algorithm for the segmentation and calling of high resolution array CGH data. Nucleic Acids Res, 2010. 38(15): p. e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding L, et al. , Somatic mutations affect key pathways in lung adenocarcinoma. Nature, 2008. 455(7216): p. 1069–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weir BA, et al. , Characterizing the cancer genome in lung adenocarcinoma. Nature, 2007. 450(7171): p. 893–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Girard L, Rodriguez-Canales J, Behrens C, Thompson DM, Botros IW, Tang H, Xie Y, Rekhtman N, Travis WD, Wistuba II, Minna JD, and Gazdar AF, An Expression Signature as an Aid to the Histologic Classification of Non-Small Cell Lung Cancer. Clin Cancer Res, 2016. 22(19): p. 4880–4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tusher VG, Tibshirani R, and Chu G, Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A, 2001. 98(9): p. 5116–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lockwood WW, Chari R, Coe BP, Girard L, Macaulay C, Lam S, Gazdar AF, Minna JD, and Lam WL, DNA amplification is a ubiquitous mechanism of oncogene activation in lung and other cancers. Oncogene, 2008. 27(33): p. 4615–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, and Schultz N, The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov, 2012. 2(5): p. 401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Somwar R, Erdjument-Bromage H, Larsson E, Shum D, Lockwood WW, Yang G, Sander C, Ouerfelli O, Tempst PJ, Djaballah H, and Varmus HE, Superoxide dismutase 1 (SOD1) is a target for a small molecule identified in a screen for inhibitors of the growth of lung adenocarcinoma cell lines. Proc Natl Acad Sci U S A, 2011. 108(39): p. 16375–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pikor LA, Lockwood WW, Thu KL, Vucic EA, Chari R, Gazdar AF, Lam S, and Lam WL, YEATS4 is a novel oncogene amplified in non-small cell lung cancer that regulates the p53 pathway. Cancer Res, 2013. 73(24): p. 7301–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lockwood WW, Zejnullahu K, Bradner JE, and Varmus H, Sensitivity of human lung adenocarcinoma cell lines to targeted inhibition of BET epigenetic signaling proteins. Proc Natl Acad Sci U S A, 2012. 109(47): p. 19408–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1: Significant regions of copy number difference between EGFRMUT and EGFRWT lung adenocarcinoma tumors.
Supplemental Table 2: Significant differentially expressed genes between EGFRMUT and EGFRWT lung adenocarcinoma tumors mapping to regions of copy number disparity.
Supplemental Figure 1: GGA2 expression in EGFRMUT and EGFRWT lung adenocarcinoma cell lines. (A) Western blot of GGA2 protein levels in EGFRMUT (left) and EGFRWT (represented by the KRASMUT and EGFR/KRASWT cell lines, right) lung adenocarcinoma cell lines. Samples were run on two separate gels, with H3255 cells run on each for comparison. (B) Dosimetry assessment of the blots from (A) showing the relative GGA2/GAPDH level for EGFRMUT and EGFRWT adenocarcinoma cell lines. H460 is a large cell carcinoma and was excluded. EGFRMUT cells demonstrate higher GGA2 protein on average, mirroring the transcript levels in human adenocarcinoma tumors.
Supplemental Figure 2: GGA2 expression across cancer cell lines. Microarray data was available for 967 cancer cell lines from the Broad Institute and Novartis Cancer Cell Line Encyclopedia and was downloaded through the cBioPortal (http://www.cbioportal.org/). mRNA levels (log2 fluorescence intensity) of GGA2 were median normalized across all cell lines and the resulting values plotted for each cancer type as box plots. Whiskers represent 10-90 percentiles with dots representing values beyond these limits.