Summary
Background:
Activated coagulation factor IX (FIXa) consists of a γ-carboxyglutamic acid domain, two epidermal growth factor-like (EGF) domains and C-terminal protease domain. Consensus sequence and biochemical data support the existence of Na+-site in FIXa protease domain. However, soaking experiments or crystals grown in high concentration of ammonium sulfate did not reveal Na+-site in wild-type or mutant FIXa EGF2/protease domain structure.
Objective:
Determine structure of FIXa EGF2/protease domain in presence of Na+; perform molecular dynamics (MD) simulations to explore the role of Na+ in stabilizing FIXa structure.
Methods:
Crystallography, MD simulations and modeling heparin binding to FIXa.
Results:
Crystal structure at 1.37 Å resolution revealed that Na+ is coordinated to carbonyl groups of residues 184A, 185, 221A and 224 in FIXa protease domain. Na+-site in FIXa is similar to that of FXa and is linked to Asp189 S1-site. In MD simulations, Na+ reduced fluctuations in residues 217-225 (Na+-loop) and 70-80 (Ca2+-loop), whereas Ca2+ reduced fluctuations only in residues of the Ca2+-loop. Ca2+ and Na+ together reduced fluctuations in residues of the Ca2+- and Na+-loops (residues 70-80, 183-194 and 217-225). Moreover, we observed four sulfate ions that make salt bridges with FIXa protease domain Arg/Lys residues, which have been implicated in heparin binding. Based upon locations of the sulfate ions, we modeled heparin binding to FIXa, which is similar to the heparin binding in thrombin.
Conclusions:
FIXa Na+-site in association with Ca2+ contributes to stabilization of the FIXa protease domain. The heparin binding mode in FIXa is similar to that in thrombin.
Keywords: Blood coagulation, factor IXa, Crystallography, Molecular dynamics simulation, Heparin
Introduction
Human factor (F) IX is a vitamin K-dependent single chain glycoprotein of 415 residues [1]. During normal hemostasis, it is activated by FXIa requiring Ca2+ [2] and by FVIIa requiring Ca2+ and tissue factor [3]. At physiologic Ca2+, these reactions are accelerated by Mg2+ [4–8]. Both enzymes cleave two peptide bonds (Arg145-Ala146 and Arg180-Val181) in FIX to yield a disulfide-linked serine protease FIXa, consisting of a light chain (residues 1-145) and a heavy chain (181-415) [1]. FIXa generated proteolytically converts FX to FXa in the coagulation cascade requiring Ca2+, phospholipid and FVIIIa leading to thrombin generation and fibrin formation [2]. The light chain of FIXa contains the γ-carboxyglutamic acid (Gla) domain followed by two epidermal growth factor-like (EGF1 and EGF2) domains, whereas the heavy chain contains the serine protease domain [1,9]. The Gla domain of FIX/FIXa possesses several Ca2+/Mg2+ sites [5,6,10], whereas the EGF1 and protease domains each have one Ca2+-binding site [6,11–15]. The Gla domain of FIX/FIXa is required for its binding to phospholipid or platelets, and the EGF1 domain of FIX is essential for activation by FVIIa/tissue factor [16–19]. Further, each domain of FIXa appears to play an important role in binding to FVIIIa [20–25].
In addition to the Ca2+-site, the serine protease domain of FIXa is predicted to contain a Na+-site [26,27] similar to that in FXa. The Na+ potentiation of FXa and thrombin amidolytic activity was first described by Orthner and Kosow [28,29]. Later, crystal structures by Di Cera and Tulinsky of thrombin and FXa provides the framework for defining the Na+-site in serine proteases [30–33]. Di Cera proposed and experimentally verified that residue 225 determines the presence of Na+-site in serine proteases [30,31]. Na+-binding is observed when Tyr or Phe is present at position 225, whereas Pro at this position obliterates the Na+-site [26,30,31]. Tulinksy expanded this definition and proposed that in addition to the 220-loop, Na+-binding to proteases such as FXa also involves the 183-loop [32,33]. These observations predict the existence of a Na+-site in the FIXa protease domain [26,27] similar to that outlined for FXa. However, crystal structures of wild-type or mutated FIXa protease domain or FIXa bound to antithrombin did not reveal the predicted Na+-site [13–15,34]. Nonetheless, Na+ increased the second-order rate constant for association of antithrombin to FIXa by one order of magnitude in the absence of Ca2+ [35]. However, in the presence of Ca2+, the catalytic activity of FIXa towards a chromogenic substrate is minimally affected by Na+ [35,36]. Here, we provide evidence for the existence of Na+-site in FIXa protease domain from the crystal structure solved at 1.37 Å. In addition, we performed molecular dynamics (MD) simulations studies to investigate the role of Na+ in the structural stability of FIXa protease domain in the absence/presence of Ca2+.
Materials and methods
Expression, Crystallization, Data Collection and Refinement
The FIX EGF2/Protease domain (residues 103-431, FIX-WTEGF2/P) was cloned, expressed in BL21 (DE3) cells, folded and purified using FPLC gel filtration and ion exchange chromatography as outlined by Hopfner et al. [37]. The isolated protein was activated with Russel viper venom (RVV-X) and the FIXa-WTEGF2/P was purified using FPLC HiTrap heparin and benzamidine column chromatography. The purified FIXa-WTEGF2/P was fully activated as analyzed by reduced SDS-PAGE [38]. The protein in 10 mM CaCl2, 1 M NaCl/50 mM Tris pH 8.0 containing 10 mM p-aminobenzamidine (pAB) was crystallized using PEG 4000 and 0.2 M ammonium sulfate as precipitants. Crystals were flash frozen without additional cryoprotectant and the data were collected at ESRF, Grenoble using the 0.93 Å wavelength at 100 K. The data were processed with XDS [39] and scaled with Aimless [40] and STARANISO [41] as implemented in autoProc [42]. The diffraction was anisotropic and for this reason we used anisotropic scaling. The crystal diffracted to 1.62 Å resolution in the a* and b* directions, and to 1.37 Å resolution in the c* direction. The structure was solved by molecular replacement using 2wpj as the search model [14]. Refinement was carried out using Buster and model building was done using COOT [43–45]. Data processing and refinement statistics are given in Table 1. The FIXa-WTEGF2/P coordinates and structure factors are deposited in the protein data bank.
Table 1.
Data collection and refinement statistics
| Crystal | FIXa EGF2/Protease domain |
|---|---|
| PDB code | 6MV4 |
| Space Group | I4122 |
| Unit cell parameters | |
| a (Å) | 109.75 |
| b (Å) | 109.75 |
| c (Å) | 112.45 |
| α, β, γ (°) | 90.0, 90.0, 90.0 |
| Data Collection | |
| Beam line | ESRF, ID14-3 |
| Wavelength (Å) | 0.93 |
| Resolution (Å) | 78.54 -1.37 |
| Molecules per asymmetric unit | 1 |
| Measured reflections | 337283 |
| Unique Reflections | 49105 |
| %Completeness (spherical) | 68.3 (12.1) |
| %Completeness (ellipsoidal) | 99.45(99.55) |
| Multiplicity* | 6.9 (5.2) |
| CC(1/2) | 1.0 (0.85) |
| Rmergea | 3.5(46.2) |
| Average I/σ(I) | 26.7(2.7) |
| Wilson B-factor | 18.6 |
| Refined statistics | |
| Resolution (Å) | 1.37 |
| No. of non-H atoms | |
| Total | 2854 |
| Protein | 2360 |
| Ligand/ion | 56 |
| Water | 438 |
| R-factor (%)b | 17.8 |
| Rfree (%)b | 21.0 |
| r.m.s.d. from ideal values | |
| Bond Lengths (Å) | 0.012 |
| Bond Angles (°) | 1.23 |
| Ramachandran plot (%) | |
| Most favored regions (%) | 83.9 |
| Additional allowed regions (%) | 13.7 |
| Generously allowed regions (%) | 0.4 |
| Disallowed regions (%) | 0.0 |
Values in the parenthesis are for the high resolution shell. ESRF, European Synchrotron Radiation Facility.
Rmerge=ΣhklΣi|Ii(hkl)−〈I(hkl)〉|/ΣhklΣiIi(hkl), where 〈I(hkl)〉 is the mean intensity of reflection hkl.
R-factor=Σhkl||Fobs|−|Fcalc||/Σhkl|Fobs|, where Fobs and Fcalc are the observed and calculated structure factors, respectively; Rfree is the same as the R-factor but is calculated for 10% of randomly selected reflections excluded from the refinement.
Molecular dynamics simulations
Molecular dynamics (MD) simulations studies were performed to investigate the effect of Na+- and Ca2+-binding to the protease domain of FIXa. MD simulations were carried out in the absence and presence of Na+, Ca2+ or Na+/Ca2+ using the AMBER 16 program [46]. Protease domain containing residues Val16 {181} to 248 {415} of FIXa was used for these studies. Since we are studying the effect of Na+ and Ca2+ binding to the protease domain, we simply wanted to use the protease domain in our simulation studies without the influence of other domains. After adding hydrogens, the protein structures were solvated in a truncated octahedral TIP3P box of 12 Å, and the system was neutralized with sodium ions. Periodic boundary conditions, Particle Mesh Ewald summation and SHAKE-enabled 2-femto seconds time steps were used in all MD simulations. Langevin dynamics temperature control was employed with a collision rate equal to 1.0 ps−1. A cutoff of 10 Å was used for nonbonding interactions. The metal ion parameters used were from the Amber force field [47] and metal interactions were treated using 12–6 Lennard-Jones nonbonded model. Initial configurations were subjected to a 1000-step minimization with the harmonic constraints of 10 kcal.mol−1.Å−2 on the protein heavy atoms. The systems were gradually heated from 0 to 300 K over a period of 50 ps with harmonic constraints. The simulations at 300 K were then continued for 50 ps during which the harmonic constraints were gradually lifted. The systems were then equilibrated for a period of 500 ps before the 50 ns production runs. All simulations were carried out in the NPT ensemble. Equilibration and production run simulations were carried out using the Sander and PMEMD modules (optimized for CUDA) of AMBER 16.0 (ff14SB), respectively [46,48]. Initial structures of the production runs were used as the reference structures for calculations of the root mean square deviations (RMSD) and root mean square fluctuations (RMSF). All analyses were performed using the cpptraj module of AmberTools 16.
Modeling heparin binding to protease domain of FIXa
To investigate whether the locations of the observed sulfate ions in the protease domain of FIXa mimic sulfates in heparin, we modeled heparin binding mode in FIXa using the heparin bound structure of thrombin [49]. First, the protease domain of FIXa and the structure of thrombin bound to heparin (PDB id 1xmn) were superimposed. Next, the heparin fragment as a whole was subjected to minor translational and rotational motions to overlay onto the sulfates observed in the FIXa structure. Finally, to relieve steric clashes, the modeled FIXa-heparin fragment complex was energy minimized with the backbone constraints using AMBER [46].
Results and Discussion
Na+-site in the protease domain of FIXa
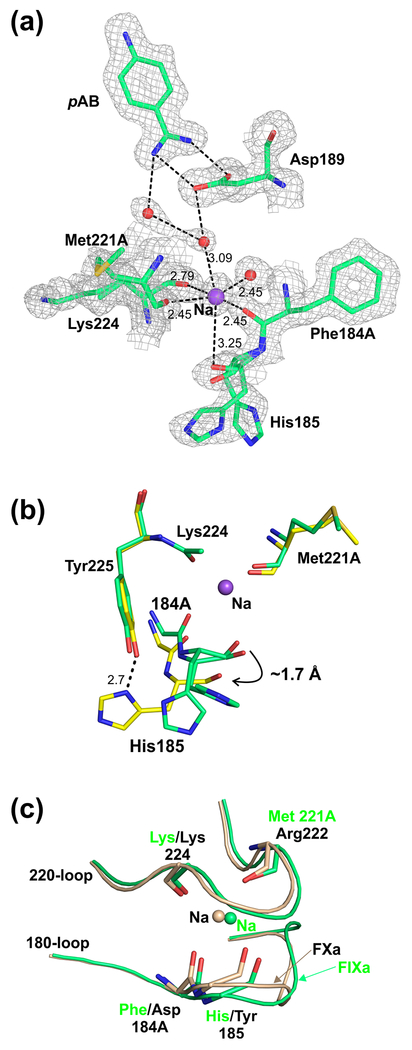
The structure of EGF2/protease domain of human wild-type FIXa (FIXa-WTEGF2/P) was determined at 1.37 Å resolution in the presence of pAB, Ca2+, Na+ and sulfate ions. Data processing and the structure refinement statistics are provided in Table 1. The overall structure of FIXa-WTEGF2/P is similar to the reported structures of wild-type FIXaEGF2/P as well as to the mutant FIXaEGF2/P (Y94F/K98T/Y177T), and the FIXa-WTEGF2/P bound to antithrombin with noticeable variations in the Na+- and Ca2+-binding loops. Unlike the previous structures, the present structure of FIXa-WTEGF2/P reveals the presence of Na+, which is coordinated to the main chain carbonyl oxygen atoms of residues Phe184A (chymotrypsin numbering) {353 amino acid in FIXa} and His185 {354}, Met221A {391} and Lys224 {394} as well as to two water molecules (Fig. 1a). Since it is difficult to distinguish between Na+ and a water molecule due to their identical number of electrons and comparable electron densities, we used the following criteria to assign Na+ at this position. First, coordination geometry—the assigned atom has six coordination ligands, four of which are main chain carbonyl groups; further, four of the six ligand atoms are within the target distances (2.5 Å-2.8 Å) while the remaining two are within the acceptable distance range for Na+ in protein structures [50,51]. Second, a water molecule can only coordinate to two main chain carbonyl oxygen atoms and interact with a maximum of four ligands. Importantly, most of the coordination distances observed in the present structure are shorter than the sum of the allowed van der Waals contact distances between the two oxygen atoms. These considerations favor placement of Na+ at this site instead of a water molecule. In addition, as in FXa and thrombin [30–33,52–55], the Na+-site in FIXa is linked to the S1-site Asp189 {359} through water molecules (Fig. 1a).
Fig. 1. Environment of the Na+-site in the protease domain of wild-type FIXa.
(a) Na+-site coordination. The FIXa residues 184A {353}, 185 {354}, 221A {391} and 224 {394} carbonyl groups and the two water molecules that serve as ligands for Na+ are shown. The two alternate conformations of His185 as well as the water mediated linkages between the Na+-site and the S1-site are depicted. The electron density (2Fobs–Fcalc) map is contoured at 1σ. The sodium (Na) and water molecules are shown as purple and red spheres, respectively. The salt-bridge between Asp189 {359} and pAB is also shown. The metal ion coordination to its ligands and H-bonds between the water molecules are shown with black dashed lines. (b) Overlay of the residues in the FIXa triple mutant (PDB id 2wpj) and in the wild-type FIXa involved in Na+ binding. As compared to wild-type FIXa, the carbonyl groups of 184A {353} and 185 {354} in the FIXa triple mutant [14] are farther away (indicated with a curved arrow) from the Na+-site (purple sphere). Dashed line indicates the H-bond between the side chains of His185 {354} and Tyr225 {395} in the FIXa triple mutant; this interaction is absent in wild-type FIXa. Also shown are Met221A {391} and Lys224 {394} residues of these structures in which the carbonyl groups occupy similar positions. The carbon atoms of the wild-type FIXa are shown in green and of the triple mutant in yellow. (c) Superposition of the Na+-sites in wild-type FIXa and FXa. Similar positions of the carbonyl groups of the 183-loop (residues 184A and 185) and of the 220-loop (residues 221A/222 and 224) in the two structures are shown. The residues in FIXa are labeled in green and in FIXa in black. Sodium (Na) in FIXa is shown in green and in FXa in wheat color.
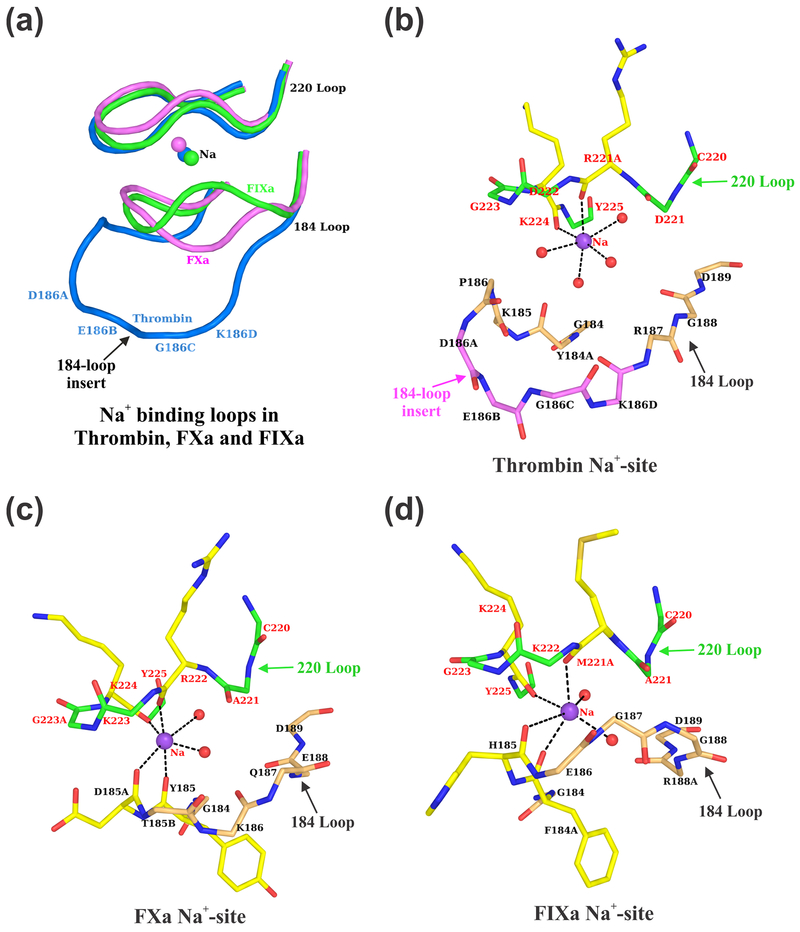
The Na+-site observed in FIXa is similar to that in FXa but not to thrombin (Fig. 2). In FXa and FIXa, residues of the 180-loop as well as of the 220-loop provide coordinating ligand atoms for the Na+-site whereas in thrombin, only residues of the 220-loop are involved in coordination with Na+ (Fig. 2a). In thrombin, four water molecules serve as ligands (Fig. 2b) as opposed to two water molecules in FXa (Fig. 2c) or FIXa (Fig. 2d). In the present FIXa structure, the Na+-site ligand atom distances vary between 2.5 Å to 3.25 Å. Similarly, in FXa structures, the Na+ coordination distances vary between 2.0 Å to 3.1 Å [32,33,52–54], while in thrombin, one of the coordination distance is always longer than 2.7 Å [30,31,33,55]. Although, the ideal coordination distance for Na+ is 2.38 ± 0.10, it is not always the case in protein structures; in many instances the distances are longer and there are less than six-coordinate ligand atoms [50]. Since the interactions between the Na+ and the ligand atoms from the protein or water molecule are more electrostatic and to some extent less covalent in nature, it is difficult to precisely define the interaction distances for Na+ [51]. Further in the present FIXa structure, the temperature factors of Na+ and its neighboring atoms are similar and do not show any large variations among them. The temperature factor of Na+ (38.18 Å2) and the average temperature factors of the non-bonded atoms within 6 Å of Na+ are ~37.0 Å2. This is consistent with the temperature factors observed earlier for the Na+-site in FXa [53].
Fig. 2. Na+ coordination ligands in Thrombin, FXa and FIXa.
(a) The Na+ binding loops of thrombin, FXa and FIXa. In thrombin, residues only of the 220-loop are involved in Na+ binding, whereas in FXa and FIXa, residues both from the 184- and the 220-loops are involved in Na+ binding. The four-residue insertion in thrombin (186A-186D) precludes the 184-loop to participate in providing ligands for Na+ binding [55]. The loops are blue in thrombin, magenta in FXa and green in FIXa. (b) Na+ coordination ligands in thrombin. The main chain carbonyl oxygen atoms of Arg221A and Lys224 and four water molecules provide ligand atoms for Na+ in thrombin [55]. Main chain carbon atoms are shown in magenta for the 184-loop insertion residues. (c) Na+ coordination ligands in FXa. The main chain carbonyl oxygen of Tyr185, Asp185A, Arg222 and Lys224 and two water molecules provide ligand atoms for Na+ in FXa. (d) Na+ coordination ligands in FIXa. The main chain carbonyl oxygen of Phe184A, His185, Met221A and Lys224 and two water molecules provide ligand atoms for Na+ in FIXa. In panels (b), (c) and (d), only the backbone atoms are shown for the 184- and 220-loops except for the residues providing ligands for Na+. The carbon atoms are in wheat for the 184-loop, in green for the 220-loop and in yellow for the residues that provide carbonyl oxygen ligands for Na+. The oxygen atoms are in red and nitrogen atoms are in blue.
In contrast to the present structure, Na+ was not observed previously in the FIXa crystal structures [13,34]. These include structures solved at medium (~2.8 Å) to high resolutions (1.3 Å) but none of them revealed the presence of Na+. These FIXa crystals were grown in the presence of high concentrations of ammonium sulfate (1.4 M) in the crystallization buffer [34] or in the absence of Na+ [13], which could explain the missing Na+ in these structures. Similarly, Na+-site was not observed in the FIXa-S195AEGF2/P bound to antithrombin [15] or in the FIXaEGF2/P triple mutant (Y94F/K98T/Y177T) [14]. Residue His185 {354} is unique to FIXa and its side chain makes a strong hydrogen bond with hydroxyl group of Tyr225 {395} in the FIXaEGF2/P triple mutant (Fig. 1b, ref 14) as well as in the FIXa-S195AEGF2/P bound to antithrombin [15]. Consequently, Na+ was not found in these structures due to the unfavorable positions of Phe184A {353} and His185 {354} carbonyl groups leading to an incomplete coordination sphere for Na+-binding. In the present structure, His185 {354} adopts two alternate conformations; however, in both conformations, the side chain of His185 does not make a hydrogen bond with Tyr225 {395} (Fig. 1b). Accordingly, the carbonyl groups of 184A {353} and 185 {354} in the present FIXa structure are favorably situated to serve as ligands for Na+. It should be noted that in the previous structures, the crystals were obtained in the absence of Na+, where the conformation of the 184-loop disfavors Na+-binding. Subsequent soaking of these crystals in Na+ containing buffer might not alter the conformation of the 184-loop and disrupt the existing hydrogen bond between His185 {354} and Tyr225 {395}. This could provide a reasonable explanation for the absence of Na+-site in these two previous FIXa structures [14,15].
Earlier, it has also been proposed that the difference in the orientation of the carbonyl groups at Gly187 {356} in FIXa versus Lys in FXa impairs the Na+-binding in FIXa [14]. Therefore, we compared the molecular environment of the Na+-binding sites in FIXa (present structure) and FXa structures [32,52–54]. The superimposed 184- and 220-loops involved in Na+-binding in these two proteins are shown in Fig. 1c. Except for the tripeptide region (186EGG188 in FIXa and the corresponding TKQ residues in FXa), no major differences were observed in the positions of the Na+-binding loops in these two proteins. Further, the main chain carbonyl atoms, which serve as ligands for Na+ occupy similar positions in both proteins. Thus, it would appear that the change in orientation of the carbonyl group at Gly187 {356} does not impair Na+-binding in FIXa.
Ca2+-site in the protease domain of FIXa
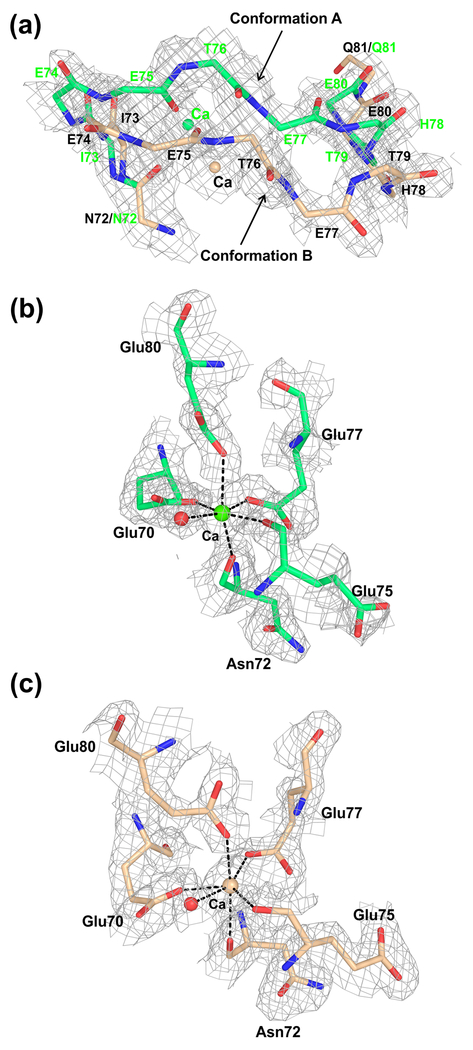
In the present structure, we observed two alternate conformations of the Ca2+-loop in FIXa (Fig. 3a). Conformation A is similar to the previous conformation of the Ca2+-loop [14,15], whereas the alternate conformation B begins at residue Asn72 {237} and ends at residue Gln81 {246} with a maximum deviation of ~3.6 Å at residue Thr76 {241} (Fig. 3a). However, the coordinating ligands for Ca2+ in each conformation are the same (Fig. 3b and 3c) and involve side chains of Glu70 {235}, Glu77 {242} and Glu80 {245} and carbonyl groups of Asn72 {237} and Glu75 {240} as well as a water molecule; these coordination ligands are the same as observed earlier [14,15]. Notably, the Ca2+-site in FIXa is similar to that in porcine pancreatic elastase [56], which involves the sidechain of Asn77 instead of a water molecule as in trypsin [57].
Fig. 3. Conformational flexibility of the Ca2+-loop in the protease domain of FIXa.
(a) The two alternate conformations of the Ca2+-loop observed in the FIXa structure. The backbone atoms of residues 72-81 are shown; the alternate conformations of the Ca2+-loop begin at residue 72 and end at residues 81. The calcium (Ca) and carbon atoms are in green in conformation A and wheat in conformation B. The electron density (2Fobs–Fcalc) map contoured at 1σ is shown for each alternate conformation of the 70-loop backbone atoms and of calcium. (b) Ca2+-binding site in conformation A. (c) Ca2+-binding site in conformation B. The alternate conformations of the protein ligands and the water molecule (red) involved in coordination to Ca2+ are depicted in panel (b) for conformation A and in panel (c) for conformation B. In both panels (b) and (c), the electron density (2Fobs–Fcalc) map is contoured at 1σ. Densities are carved to 1.7 Å around the selected atoms. The calcium (Ca) is shown in green in panel (b) and in wheat in panel (c), and the water molecule is shown in red in both panels.
Molecular dynamics studies
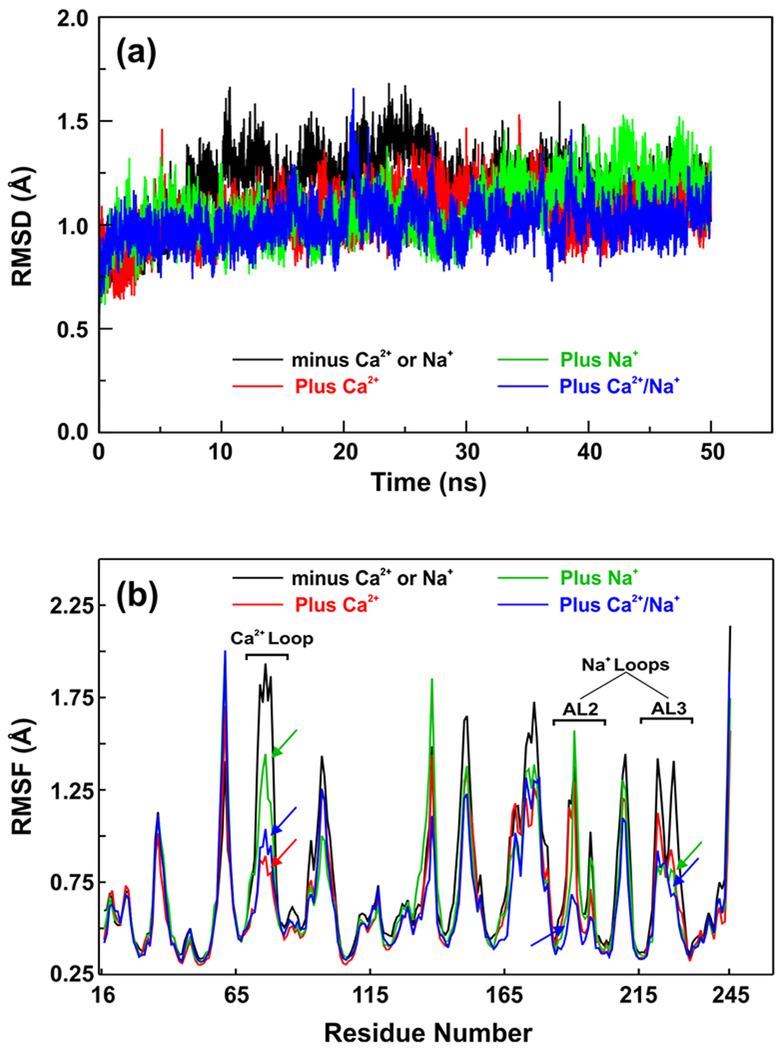
To understand the role of Na+ in FIXa, we performed MD simulations on the FIXa protease domain for 50 ns each in the presence of Na+, Ca2+, and Na+/Ca2+ as well as in the absence of these metals using the AMBER16 package [46]. The root mean square deviations (RMSD) and the average residue root mean square fluctuations (RMSF) for the backbone atoms of FIXa protease domain for the 50 ns MD data are presented in Fig. 4. The MD data indicate that the presence of Na+, Ca2+ or Na+/Ca2+ stabilizes the FIXa protease domain as compared to the metal-free form (Fig. 4a). The average RMSF for the backbone atoms of each residue presented in Fig. 4b indicate that Na+ reduces fluctuations (~0.6 Å) in the 216-225 residues (one of the Na+-loops) as well as in the 70-80 residues (Ca2+-loop), while Ca2+ reduces fluctuations (~1.0 Å) only in the Ca2+-loop residues. Presence of both Ca2+/Na+ reduces fluctuations in residues of the Ca2+-loop (~1.0 Å) and the two Na+-loops (~0.6 Å, residues 183-194 and residues 216-225). Cumulatively, the MD data suggest that Na+ plays an important role in stabilizing the FIXa protease domain including the Ca2+-binding loop as well as the two Na+-binding loops (residues 183-194 and residues 216-225), which are also involved in the conversion of zymogen to a protease [58,59].
Fig. 4. The root mean square deviations (RMSD) and average root mean square fluctuations (RMSF) of the backbone atoms of FIXa protease domain in the presence as well as absence of Na+ and Ca2+.
(a) RMSD of backbone atoms of FIXa over 50 ns molecular dynamics (MD) trajectories. (b) Comparative RMSF plots of backbone atoms of FIXa during 50 ns MD simulations. In both panels (a) and (b) – black, absence of Na+ and Ca2+; green, presence of Na+; red, presence of Ca2+; and blue, presence of both Na+ and Ca2+. Arrows in panel (b) indicate the reduced fluctuations in the residues of the Ca2+ and Na+ loops in the presence of Na+, Ca2+ as well as both Na+ and Ca2+.
Earlier studies support the functional role of Na+ for FIXa in the absence of Ca2+ [26,35,36]. Na+ increased the relative specificity (kcat/kM) of hydrolysis of two chromogenic substrates by FIXa ~4-fold [26,36], which was primarily reflected in the reduction of kM [36]. In the absence of Ca2+, Na+ also affected the reactivity of FIXa with antithrombin [35]. Inclusion of Na+ in the presence of Ca2+ reduced the kM of a chromogenic substrate by ~2-fold without affecting the kcat [36]; in another study, data were presented where the velocity (v) versus [S] plot (up to 8 mM) was linear [35]. These data represent first-order kinetics at ~8 mM substrate concentration of <0.1 kM [60] and predict a kM of >50 mM; this kM value differs from the reported values of ~2-5 mM [35,61,62]. Thus, the marginal enhancement (~2-fold) in relative specificity of FIXa by Na+ in the presence of Ca2+ [36] might not be detected under these conditions [35]. In additional studies, in reaction mixtures containing Ca2+ and 33% ethylene glycol, Na+ also had no effect on the chromogenic substrate hydrolysis by plasma FIXa [35] as well as by His185A, Glu186A and Arg188A mutants [63]; notably, ethylene glycol enhances amidolytic activity of FIXa several-fold [64] and could mask the effect of Na+ especially in the presence of Ca2+. Further, binding of cofactor FVIIIa to FIXa could rearrange the 184- and 220-loops and diminish the effect of Na+ during FX activation [35,63] as has been previously observed for FVa binding to FXa during prothrombin activation [65]; this concept was further elaborated by Page and Di Cera for both FIXa and FXa enzymes [66]. Thus, it would appear that Na+ contributes primarily towards structural stabilization of the FIXa protease domain.
Heparin binding mode in FIXa
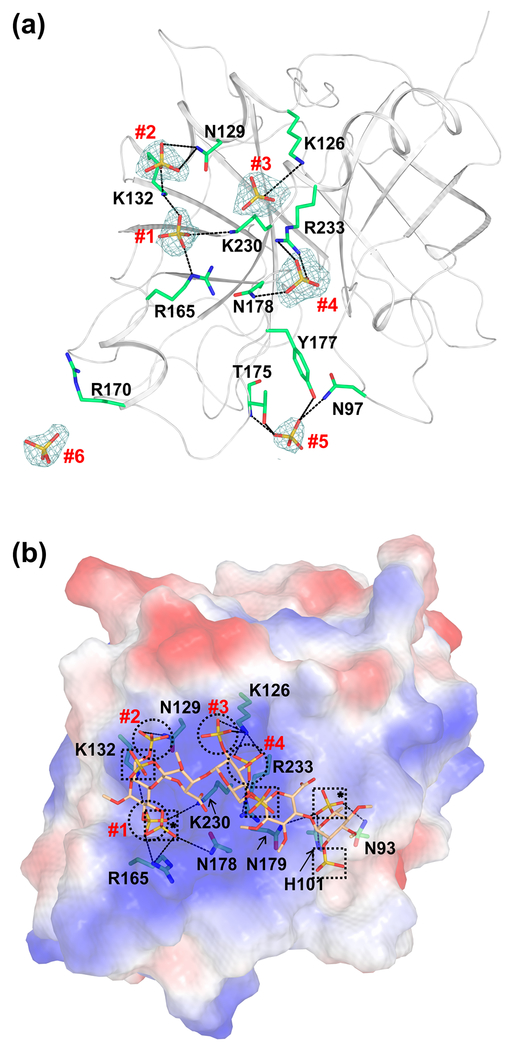
In the present FIXa-WTEGF2/P structure, we found six sulfate ions in the protease domain (Fig. 5a). These sulfates are located in the putative heparin binding exosite region of FIXa. Four of the six sulfates, numbered #1 to #4 in Fig. 5a, interact with Arg/Lys residues of FIXa that have been shown by mutagenesis to be involved in heparin binding to FIXa [67,68]. Sulfate #5 interacts with Asn97 {264}, Thr175 {343} and Tyr177 {345}; these residues have been implicated in interaction with heparin from the modeling studies [15]. Further, sulfates #4 and #6 are located at the interface between the symmetry related molecules. Sulfate #4 is involved in interactions with FIXa residues in the asymmetric unit and its symmetry mate, whereas sulfate #6 is involved in interactions with residues only from the symmetry related molecule. Details of the interactions between the sulfate ions and FIXa residues are provided in Table 2.
Fig. 5. Cartoon and electrostatic surface representations of the FIXa protease domain containing the sulfate ions and the modeled heparin binding mode.
(a) The sulfate ions found in the FIXa protease domain crystal structure. The sulfate ions and the side chain of residues involved in the salt-bridge and hydrogen bond interactions are shown in stick representation. The electron density (2Fobs−Fcalc) only of the sulfate ions (numbered #1 to #6) contoured at 1σ is shown. Densities are carved to 1.7 Å around the sulfate ions. (b) Mode of heparin binding to the FIXa protease domain. Electrostatic surface potential of the FIXa protease domain is shown with heparin fragment in stick representation. The FIXa heparin binding mode was modeled using the structure of thrombin bound to the heparin fragment [49]. Sulfates from the heparin fragment that occupy the same positions (#1 to #4) as the sulfates found in the crystal structure of FIXa are represented by circles. The other sulfates from the heparin fragment are marked with squares and those involved in interactions with FIXa are marked with asterisks. The salt-bridge interactions between FIXa residues and the sulfates in heparin are shown in dashed lines. Sulfates #5 and #6 of panel (a) do not match with any of the sulfates from the heparin fragment and thus are not depicted here.
Table 2.
The salt-bridge and hydrogen bond interactions between the observed sulfate ions and the protease domain residues of FIXa.
| Sulfate ion | Atom | FIXa Residue | Atom | Distance (Å) |
|---|---|---|---|---|
| #1 SO42− | O2 | Lys132 | NZ | 2.8 |
| O1 | Arg165 | NE | 2.8 | |
| O3 | Arg165 | NH2 | 2.9 | |
| O3 | Lys230 | NZ | 3.3 | |
| #2 SO42− | O2 | Asn129 | ND2 | 3.1 |
| O1 | Asn129 | ND2 | 3.2 | |
| O3 | Lys132 | NZ | 2.7 | |
| #3 SO42− | O1 | Lys126 | NZ | 3.6 |
| O1 | Arg233 | NH2 | 3.3 | |
| O3 | Arg233 | NH2 | 3.1 | |
| O4 | Arg233 | NE | 2.8 | |
| #4 SO42− | O4 | Asn178 | ND2 | 2.8 |
| O2 | Arg233 | NH2 | 3.0 | |
| O3 | Arg233 | NH1 | 2.9 | |
| O2 | Asn178* | ND2 | 2.8 | |
| O1 | Arg233* | NH2 | 3.2 | |
| O4 | Arg233* | NH1 | 2.8 | |
| #5 SO42− | O2 | Asn97 | ND2 | 3.2 |
| O3 | Thr175 | N | 3.0 | |
| O4 | Thr175 | OG1 | 2.7 | |
| O2 | Tyr177 | OH | 3.0 | |
| #6 SO42− | O4 | Asn92* | ND2 | 2.9 |
| O2 | Tyr128* | OH | 3.1 | |
| O4 | Tyr128* | OH | 3.1 |
Chymotrypsin numbering is used for the FIXa residues.
Residues from the symmetry related molecule
The heparin sulfate binding to FIXa plays a crucial role in physiologic thrombin generation [67–70]. In the present FIXa-WTEGF2/P structure, the observed locations of the four sulfate ions (#1 to #4) and their interatomic separation distances appear to mimic the sulfates in the thrombin-heparin structure. Thus, we modeled the heparin binding to FIXa using the structure of heparin bound to thrombin as a template [49]. The modeled heparin binding to FIXa is depicted in Fig. 5b. Importantly, the four sulfates (#1to #4) that interact with Arg/Lys residues overlay with sulfates from the heparin fragment bound to thrombin. Interaction of these four sulfates with Arg/Lys residues (Table 2) involved in heparin binding are depicted by circles. Four additional sulfates depicted by squares are part of the heparin fragment, two of which (marked with an asterisk) interact with FIXa residues Asn93 {260}, His101 {268} and Asn179 {347} that have also been implicated in heparin binding [15]. In the current model, sulfates #5 and #6 could not be aligned with the sulfates of the heparin fragment. Further work is needed to ascertain whether the residues these sulfates (#5 and #6) interact with constitute part of the heparin binding region in FIXa.
Previously, Johnson and coworkers modeled heparin binding to FIXa using the structure of FIXa bound to antithrombin-heparin complex [15]; this model was built based upon the crystal contacts observed between the heparin fragment and the FIXa protease domain. In this model, a majority of the interactions between heparin sulfate and FIXa residues are mediated through water molecules [15]. In the model presented here, sulfate ions interact directly with the FIXa residues implicated in heparin binding [67–70]. Further, the effects of mutagenesis on the solution binding of FIXa to hypersulfated low molecular weight heparin [70] are in agreement with the sulfate binding sites #1–4. Moreover, the present model resembles the mode of heparin binding to thrombin [49]. As pointed out by Johnson and coworkers [15], the FIXa-heparin binding mode in their model is orthogonal to thrombin and might not represent a favorable mode of binding in solution. Furthermore, our present model would predict that interaction of heparin sulfate with FIXa and thrombin are similar.
Conclusions
As predicted by Di Cera and Tulinsky [30–33], our crystal structure data provide evidence for the existence of Na+-site in FIXa. The linkage between Na+ and the S1-site in FIXa is consistent with the reduction in the kinetic constant kM, in the presence or absence of Ca2+. MD simulations data support that Na+ in conjunction with Ca2+ stabilizes the structure of protease domain in FIXa. As is the case with FXa, binding of cofactor FVIIIa to FIXa protease domain could provide conformational stability in the absence of Na+. This could be due to the evolutionary transitions where the original effect of Na+ is replaced by specialized protein-protein interactions as proposed by Page and Di Cera [66]. The heparin binding mode in FIXa presented here resembles the heparin binding mode in thombin. Similar to thrombin, the heparin binding residues in FIXa directly interact with the heparin sulfates providing support for the proposed model.
Essentials.
Consensus sequence and biochemical data suggest Na+-site in factor (F) IXa protease domain
X-ray structure of FIXa EGF2/protease domain at 1.37 Å reveals Na+-site not observed earlier
Molecular dynamics simulations data support that Na+±Ca2+ promote FIXa protease domain stability
Sulfate ions found in the protease domain mimic heparin sulfate binding mode in FIXa
Acknowledgements
This work was supported by National Heart, Lung and Blood Institute grant R01HL141850. We thank Dr. Michael Sawaya of UCLA for helpful discussions.
Dedication
We dedicate this paper to the memory of Alexander Tulinsky Ph.D., who made seminal contributions to the field of structural biology in hemostasis and fibrinolysis.
Footnotes
Present Address
Department of Pathology and Laboratory Medicine, University of Rochester Medical Center, Rochester, NY 14642, USA
Disclosure of conflict of interests
Herman Schreuder and Alexander Liesum are employed by the pharmaceutical company Sanofi. The other authors state that they have no conflict of interest.
Data access
Coordinates and structure factors have been deposited in the Protein Data Bank with accession number 6MV4.
References
- 1.Yoshitake S, Schach BG, Foster DC, Davie EW, Kurachi K. Nucleotide sequence of the gene for human factor IX (antihemophilic factor B). Biochemistry. 1985; 24: 3736–50. [DOI] [PubMed] [Google Scholar]
- 2.Davie EW, Fujikawa K, Kisiel W. The coagulation cascade: initiation, maintenance, and regulation. Biochemistry. 1991; 30: 10363–70. [DOI] [PubMed] [Google Scholar]
- 3.Osterud B, Rapaport SI. Activation of factor IX by the reaction product of tissue factor and factor VII: additional pathway for initiating blood coagulation. Proc Natl Acad Sci U S A. 1977; 74: 5260–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sekiya F, Yamashita T, Atoda H, Komiyama Y, Morita T. Regulation of the tertiary structure and function of coagulation factor IX by magnesium (II) ions. J Biol Chem 1995; 270: 14325–31. [DOI] [PubMed] [Google Scholar]
- 5.Shikamoto Y, Morita T, Fujimoto Z, Mizuno H. Crystal structure of Mg2+- and Ca2+-bound Gla domain of factor IX complexed with binding protein. J Biol Chem 2003; 278: 24090–4. [DOI] [PubMed] [Google Scholar]
- 6.Agah S, Bajaj SP. Role of magnesium in factor XIa catalyzed activation of factor IX: calcium binding to factor IX under physiologic magnesium. J Thromb Haemost 2009; 7: 1426–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sekiya F, Yoshida M, Yamashita T, Morita T. Magnesium(II) is a crucial constituent of the blood coagulation cascade. Potentiation of coagulant activities of factor IX by Mg2+ ions. J Biol Chem 1996; 271: 8541–4. [DOI] [PubMed] [Google Scholar]
- 8.Vadivel K, Agah S, Messer AS, Cascio D, Bajaj MS, Krishnaswamy S, Esmon CT, Padmanabhan K, Bajaj SP. Structural and functional studies of gamma-carboxyglutamic acid domains of factor VIIa and activated Protein C: role of magnesium at physiological calcium. J Mol Biol 2013; 425: 1961–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Di Scipio RG, Kurachi K, Davie EW. Activation of human factor IX (Christmas factor). J Clin Invest 1978; 61: 1528–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang M, Furie BC, Furie B. Crystal structure of the calcium-stabilized human factor IX Gla domain bound to a conformation-specific anti-factor IX antibody. J Biol Chem 2004; 279: 14338–46. [DOI] [PubMed] [Google Scholar]
- 11.Rao Z, Handford P, Mayhew M, Knott V, Brownlee GG, Stuart D. The structure of a Ca(2+)-binding epidermal growth factor-like domain: its role in protein-protein interactions. Cell. 1995; 82: 131–41. [DOI] [PubMed] [Google Scholar]
- 12.Bajaj SP, Sabharwal AK, Gorka J, Birktoft JJ. Antibody-probed conformational transitions in the protease domain of human factor IX upon calcium binding and zymogen activation: putative high-affinity Ca(2+)-binding site in the protease domain. Proc Natl Acad Sci U S A. 1992; 89: 152–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hopfner KP, Lang A, Karcher A, Sichler K, Kopetzki E, Brandstetter H, Huber R, Bode W, Engh RA. Coagulation factor IXa: the relaxed conformation of Tyr99 blocks substrate binding. Structure. 1999; 7: 989–96. [DOI] [PubMed] [Google Scholar]
- 14.Zogg T, Brandstetter H. Structural basis of the cofactor- and substrate-assisted activation of human coagulation factor IXa. Structure. 2009; 17: 1669–78. [DOI] [PubMed] [Google Scholar]
- 15.Johnson DJ, Langdown J, Huntington JA. Molecular basis of factor IXa recognition by heparin-activated antithrombin revealed by a 1.7-A structure of the ternary complex. Proc Natl Acad Sci U S A. 2010; 107: 645–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahmad SS, Rawala-Sheikh R, Cheung WF, Jameson BA, Stafford DW, Walsh PN. High-affinity, specific factor IXa binding to platelets is mediated in part by residues 3-11. Biochemistry. 1994; 33: 12048–55. [DOI] [PubMed] [Google Scholar]
- 17.Freedman SJ, Blostein MD, Baleja JD, Jacobs M, Furie BC, Furie B. Identification of the phospholipid binding site in the vitamin K-dependent blood coagulation protein factor IX. J Biol Chem 1996; 271: 16227–36. [DOI] [PubMed] [Google Scholar]
- 18.Grant MA, Baikeev RF, Gilbert GE, Rigby AC. Lysine 5 and phenylalanine 9 of the factor IX omega-loop interact with phosphatidylserine in a membrane-mimetic environment. Biochemistry. 2004; 43: 15367–78. [DOI] [PubMed] [Google Scholar]
- 19.Zhong D, Smith KJ, Birktoft JJ, Bajaj SP. First epidermal growth factor-like domain of human blood coagulation factor IX is required for its activation by factor VIIa/tissue factor but not by factor XIa. Proc Natl Acad Sci U S A. 1994; 91: 3574–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishimura H, Takeya H, Miyata T, Suehiro K, Okamura T, Niho Y, Iwanaga S. Factor IX Fukuoka. Substitution of ASN92 by His in the second epidermal growth factor-like domain results in defective interaction with factors VIIa/X. J Biol Chem 1993; 268: 24041–6. [PubMed] [Google Scholar]
- 21.Mertens K, Celie PH, Kolkman JA, Lenting PJ. Factor VIII-factor IX interactions: molecular sites involved in enzyme-cofactor complex assembly. Thromb Haemost 1999; 82: 209–17. [PubMed] [Google Scholar]
- 22.Mathur A, Bajaj SP. Protease and EGF1 domains of factor IXa play distinct roles in binding to factor VIIIa. Importance of helix 330 (helix 162 in chymotrypsin) of protease domain of factor IXa in its interaction with factor VIIIa. J Biol Chem 1999; 274: 18477–86. [DOI] [PubMed] [Google Scholar]
- 23.Chang YJ, Wu HL, Hamaguchi N, Hsu YC, Lin SW. Identification of functionally important residues of the epidermal growth factor-2 domain of factor IX by alanine-scanning mutagenesis. Residues Asn(89)-Gly(93) are critical for binding factor VIIIa. J Biol Chem 2002; 277: 25393–9. [DOI] [PubMed] [Google Scholar]
- 24.Ahmad SS, London FS, Walsh PN. Binding studies of the enzyme (factor IXa) with the cofactor (factor VIIIa) in the assembly of factor-X activating complex on the activated platelet surface. J Thromb Haemost 2003; 1: 2348–55. [DOI] [PubMed] [Google Scholar]
- 25.Ngo JC, Huang M, Roth DA, Furie BC, Furie B. Crystal structure of human factor VIII: implications for the formation of the factor IXa-factor VIIIa complex. Structure. 2008; 16: 597–606. [DOI] [PubMed] [Google Scholar]
- 26.Dang QD, Di Cera E. Residue 225 determines the Na(+)-induced allosteric regulation of catalytic activity in serine proteases. Proc Natl Acad Sci U S A. 1996; 93: 10653–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silva FP Jr., Antunes OA, de Alencastro RB, De Simone SG The Na+ binding channel of human coagulation proteases: novel insights on the structure and allosteric modulation revealed by molecular surface analysis. Biophys Chem 2006; 119: 282–94. [DOI] [PubMed] [Google Scholar]
- 28.Orthner CL, Kosow DP. The effect of metal ions on the amidolytic acitivity of human factor Xa (activated Stuart-Prower factor). Arch Biochem Biophys 1978; 185: 400–6. [DOI] [PubMed] [Google Scholar]
- 29.Orthner CL, Kosow DP. Evidence that human alpha-thrombin is a monovalent cation-activated enzyme. Arch Biochem Biophys 1980; 202: 63–75. [DOI] [PubMed] [Google Scholar]
- 30.Di Cera E, Guinto ER, Vindigni A, Dang QD, Ayala YM, Wuyi M, Tulinsky A. The Na+ binding site of thrombin. J Biol Chem 1995; 270: 22089–92. [DOI] [PubMed] [Google Scholar]
- 31.Guinto ER, Caccia S, Rose T, Futterer K, Waksman G, Di Cera E. Unexpected crucial role of residue 225 in serine proteases. Proc Natl Acad Sci U S A. 1999; 96: 1852–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Padmanabhan K, Padmanabhan KP, Tulinsky A, Park CH, Bode W, Huber R, Blankenship DT, Cardin AD, Kisiel W. Structure of human des(1-45) factor Xa at 2.2 A resolution. J Mol Biol 1993; 232: 947–66. [DOI] [PubMed] [Google Scholar]
- 33.Zhang E, Tulinsky A. The molecular environment of the Na+ binding site of thrombin. Biophys Chem 1997; 63: 185–200. [DOI] [PubMed] [Google Scholar]
- 34.Sakurada I, Endo T, Hikita K, Hirabayashi T, Hosaka Y, Kato Y, Maeda Y, Matsumoto S, Mizuno T, Nagasue H, Nishimura T, Shimada S, Shinozaki M, Taguchi K, Takeuchi K, Yokoyama T, Hruza A, Reichert P, Zhang T, Wood HB, Nakao K, Furusako S. Discovery of novel aminobenzisoxazole derivatives as orally available factor IXa inhibitors. Bioorg Med Chem Lett 2017; 27: 2622–8. [DOI] [PubMed] [Google Scholar]
- 35.Gopalakrishna K, Rezaie AR. The influence of sodium ion binding on factor IXa activity. Thromb Haemost 2006; 95: 936–41. [DOI] [PubMed] [Google Scholar]
- 36.Schmidt AE, Stewart JE, Mathur A, Krishnaswamy S, Bajaj SP. Na+ site in blood coagulation factor IXa: effect on catalysis and factor VIIIa binding. J Mol Biol 2005; 350: 78–91. [DOI] [PubMed] [Google Scholar]
- 37.Hopfner KP, Brandstetter H, Karcher A, Kopetzki E, Huber R, Engh RA, Bode W. Converting blood coagulation factor IXa into factor Xa: dramatic increase in amidolytic activity identifies important active site determinants. EMBO J 1997; 16: 6626–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970; 227: 680–5. [DOI] [PubMed] [Google Scholar]
- 39.Xds Kabsch W.. Acta Crystallogr D Biol Crystallogr 2010; 66: 125–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Evans P Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr 2006; 62: 72–82. [DOI] [PubMed] [Google Scholar]
- 41.Tickle IJ, Flensburg C, Keller P, Paciorek W, Sharff A, Vonrhein C, Bricogne. G STARANISO. 2017, Global Phasing Ltd: Cambridge, United Kingdom. [Google Scholar]
- 42.Vonrhein C, Flensburg C, Keller P, Sharff A, Smart O, Paciorek W, Womack T, Bricogne G. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr D Biol Crystallogr 2011; 67: 293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bricogne G, Blanc E, Brandl M, Flensburg C, Keller P, Paciorek W, Roversi P, Sharff A, Smart OS, Vonrhein C, Womack TO. (2017). BUSTER version 2.11.17. Cambridge, United Kingdom: Global Phasing Ltd. [Google Scholar]
- 44.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 2010; 66: 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 2011; 67: 235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Case DA, Ben-Shalom IY, Brozell SR, Cerutti DS, Cheatham TE III, Cruzeiro VWD, Darden TA, Duke RE, Ghoreishi D, Gilson MK, Gohlke H, Goetz AW, Greene D, Harris R, Homeyer N, Izadi S, Kovalenko A, Kurtzman T, Lee TS, LeGrand S et al. , AMBER. San Francisco: University of California 2016. [Google Scholar]
- 47.Li P, Roberts BP, Chakravorty DK, Merz KM Jr. Rational Design of Particle Mesh Ewald Compatible Lennard-Jones Parameters for +2 Metal Cations in Explicit Solvent. J Chem Theory Comput 2013; 9: 2733–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Le Grand S, Götz AW, Walker RC. SPFP: Speed without compromise—A mixed precision model for GPU accelerated molecular dynamics simulations. Comp Phy Comm 184 (2013) 374–380. [Google Scholar]
- 49.Carter WJ, Cama E, Huntington JA. Crystal structure of thrombin bound to heparin. J Biol Chem 2005; 280: 2745–9. [DOI] [PubMed] [Google Scholar]
- 50.Harding MM. Metal-ligand geometry relevant to proteins and in proteins: sodium and potassium. Acta Crystallogr D Biol Crystallogr 2002; 58: 872–4. [DOI] [PubMed] [Google Scholar]
- 51.Harding MM. Small revisions to predicted distances around metal sites in proteins. Acta Crystallogr D Biol Crystallogr 2006; 62: 678–82. [DOI] [PubMed] [Google Scholar]
- 52.Scharer K, Morgenthaler M, Paulini R, Obst-Sander U, Banner DW, Schlatter D, Benz J, Stihle M, Diederich F. Quantification of cation-pi interactions in protein-ligand complexes: crystal-structure analysis of Factor Xa bound to a quaternary ammonium ion ligand. Angew Chem Int Ed Engl 2005; 44: 4400–4. [DOI] [PubMed] [Google Scholar]
- 53.Murakami MT, Rios-Steiner J, Weaver SE, Tulinsky A, Geiger JH, Arni RK. Intermolecular interactions and characterization of the novel factor Xa exosite involved in macromolecular recognition and inhibition: crystal structure of human Gla-domainless factor Xa complexed with the anticoagulant protein NAPc2 from the hematophagous nematode Ancylostoma caninum. J Mol Biol 2007; 366: 602–10. [DOI] [PubMed] [Google Scholar]
- 54.Salonen LM, Bucher C, Banner DW, Haap W, Mary JL, Benz J, Kuster O, Seiler P, Schweizer WB, Diederich F. Cation-pi interactions at the active site of factor Xa: dramatic enhancement upon stepwise N-alkylation of ammonium ions. Angew Chem Int Ed Engl 2009; 48: 811–4. [DOI] [PubMed] [Google Scholar]
- 55.Pineda AO, Carrell CJ, Bush LA, Prasad S, Caccia S, Chen ZW, Mathews FS, Di Cera E. Molecular dissection of Na+ binding to thrombin. J Biol Chem 2004; 279: 31842–53. [DOI] [PubMed] [Google Scholar]
- 56.Meyer E, Cole G, Radhakrishnan R, Epp O. Structure of native porcine pancreatic elastase at 1.65 A resolutions. Acta Crystallogr B. 1988; 44 (Pt 1): 26–38. [DOI] [PubMed] [Google Scholar]
- 57.Bode W, Schwager P. The single calcium-binding site of crystallin bovin beta-trypsin. FEBS Lett 1975; 56: 139–43. [DOI] [PubMed] [Google Scholar]
- 58.Huber R, Bode W. Structural basis of the activation and action of trypsin. Acc Chem Res 1978; 11:114–122. [Google Scholar]
- 59.Perera L, Darden TA, Pedersen LG. Modeling human zymogen factor IX. Thromb Haemost 2001; 85: 596–603. [PubMed] [Google Scholar]
- 60.Segel IH. Enzyme Kinetics. John Wiley & Sons, New York: (1975) Chapter 2, pp 18–89. [Google Scholar]
- 61.Lenting PJ, ter Maat H, Clijsters PP, Donath MJ, van Mourik JA, Mertens K. Cleavage at arginine 145 in human blood coagulation factor IX converts the zymogen into a factor VIII binding enzyme. J Biol Chem 1995; 270: 14884–90. [DOI] [PubMed] [Google Scholar]
- 62.Christophe OD, Lenting PJ, Kolkman JA, Brownlee GG, Mertens K. Blood coagulation factor IX residues Glu78 and Arg94 provide a link between both epidermal growth factor-like domains that is crucial in the interaction with factor VIII light chain. J Biol Chem 1998; 273: 222–7. [DOI] [PubMed] [Google Scholar]
- 63.Yang L, Gopalakrishna K, Manithody C, Rezaie AR. Expression, purification and characterization of factor IX derivatives using a novel vector system. Protein Expr Purif 2006;50: 196–202. [DOI] [PubMed] [Google Scholar]
- 64.Sturzebecher J, Kopetzki E, Bode W, Hopfner KP. Dramatic enhancement of the catalytic activity of coagulation factor IXa by alcohols. FEBS Lett 1997; 412: 295–300. [DOI] [PubMed] [Google Scholar]
- 65.Camire RM. Prothrombinase assembly and S1 site occupation restore the catalytic activity of FXa impaired by mutation at the sodium-binding site. J Biol Chem 2002; 277: 37863–70. [DOI] [PubMed] [Google Scholar]
- 66.Page MJ, Di Cera E. Is Na+ a coagulation factor? Thromb Haemost 2006; 95: 920–1. [DOI] [PubMed] [Google Scholar]
- 67.Yang L, Manithody C, Rezaie AR. Localization of the heparin binding exosite of factor IXa. J Biol Chem 2002; 277: 50756–60. [DOI] [PubMed] [Google Scholar]
- 68.Buyue Y, Whinna HC, Sheehan JP. The heparin-binding exosite of factor IXa is a critical regulator of plasma thrombin generation and venous thrombosis. Blood. 2008; 112: 3234–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Misenheimer TM, Buyue Y, Sheehan JP. The heparin-binding exosite is critical to allosteric activation of factor IXa in the intrinsic tenase complex: the role of arginine 165 and factor X. Biochemistry. 2007; 46: 7886–95. [DOI] [PubMed] [Google Scholar]
- 70.Misenheimer TM, Sheehan JP. The regulation of factor IXa by supersulfated low molecular weight heparin. Biochemistry. 2010;49: 9997–10005. [DOI] [PMC free article] [PubMed] [Google Scholar]