Abstract
Methionine in proteins is often thought to be a generic hydrophobic residue, functionally replaceable with another hydrophobic residue such as valine or leucine. This is not the case, and the reason is that methionine contains sulfur that confers special properties on methionine. The sulfur can be oxidized, converting methionine to methionine sulfoxide, and ubiquitous methionine sulfoxide reductases can reduce the sulfoxide back to methionine. This redox cycle enables methionine residues to provide a catalytically efficient antioxidant defense by reacting with oxidizing species. The cycle also constitutes a reversible post-translational covalent modification analogous to phosphorylation. As with phosphorylation, enzymatically-mediated oxidation and reduction of specific methionine residues functions as a regulatory process in the cell. Methionine residues also form bonds with aromatic residues that contribute significantly to protein stability. Given these important functions, alteration of the methionine-methionine sulfoxide balance in proteins has been correlated with disease processes, including cardiovascular and neurodegenerative diseases. Methionine isn’t just for protein initiation.
Keywords: Methionine, Methionine Sulfoxide, Methionine Sulfoxide Reductase, Oxidative Defenses, Protein Structure, Cellular Regulation
Introduction
We are pleased to join the colleagues, students, and admirers of Elias Michaelis in this special issue honoring him. At first glance, a mini-review on the roles of methionine in proteins may seem tenuously connected to his many contributions to neuroscience. Actually, it connects directly through his interest in oxidative stress, one of the important areas Dr. Michaelis has advanced our knowledge through his research.
When many of us took our first course in biochemistry, we learned that methionine was one of several hydrophobic residues in proteins and, except for its role in protein initiation, we were taught that these hydrophobic residues were pretty much interchangeable. Such was not the case for the other sulfur-containing residue, cysteine. This important amino acid was recognized as having roles as an antioxidant -- especially in the tripeptide glutathione, in protein structure through disulfide bond formation, in catalysis at the active site of several classes of enzymes such as proteases, oxidoreductases, phosphatases, and peroxiredoxins, and in cellular regulation through its reversible oxidation and reduction. Research from many investigators in the last years has revealed that methionine in proteins shares many of these functions [1–8]. The results of these studies make clear that each of the two sulfur-containing amino acids function in antioxidant defense, protein structure, and redox sensing and regulation. Experimental investigation of methionine’s roles in those functions thus has a high probability of identifying important cellular mechanisms as well as diseases caused by defects in their function. With the recognition that oxidative defense, protein structure, and cellular regulation are mediated by methionine residues in proteins, it is not surprising that impairment of those functions has also been associated with several disease processes, including neurodegeneration, cancer, and cardiovascular disease. In this article, we provide examples of the expanded role of methionine and summarize the mechanisms by which methionine is thought to perform these functions.
Oxidation of Methionine to Methionine Sulfoxide and Reduction Back to Methionine
One of the important properties of both cysteine and methionine residues in proteins is that they are subject to reversible oxidation and reduction, mediated either enzymatically or non-enzymatically. While cysteine is well-recognized for the ease of its oxidation, it is often not appreciated that methionine can be readily oxidized to methionine sulfoxide (MetO) [9, 10]. Indeed, the standard redox potential for the two electron reduction of dimethyl sulfoxide is +160 mV [11] while that for cystine is +220 mV [12]. Cysteine is easily oxidized when ionized to its thiolate, but is difficult to oxidize when in the thiol form [13]. However, most cysteine residues, including those in glutathione, have a pKa around 8.3–8.7 and are not easily oxidized at physiological pH, unless the oxidation is catalyzed by an enzyme. In contrast, oxidation of methionine residues is essentially independent of pH [14]. In vitro, hypochlorous acid (HOCl), a major halogenated oxidant generated by leukocytes, reacts rapidly with methionine at physiological pH [14, 15], but hydrogen peroxide does not, although the rate can be accelerated by the bicarbonate/carbon dioxide present in vivo [16].
MetO is reduced back to methionine by the methionine sulfoxide reductases, thioredoxin-dependent enzymes that are virtually universal among aerobic organisms [17, 18]. Oxidation of methionine to MetO introduces a chiral center at the sulfur atom so there are two epimers of MetO, R-MetO and S-MetO [19]. While an epimerase that interconverts the forms could theoretically exist, none has been found so far. Instead, organisms have two types of methionine sulfoxide reductases (msr). MsrA specifically reduces S-MetO, but not R-MetO. Conversely, MsrB reduces R-MetO, but not S-MetO. Recycling by the reductases allows the methionine residue to react again with oxidizing species, creating a system with catalytic efficiency in scavenging reactive species. The reducing power is ultimately provided by NADPH (Fig. 1).
Fig. 1.
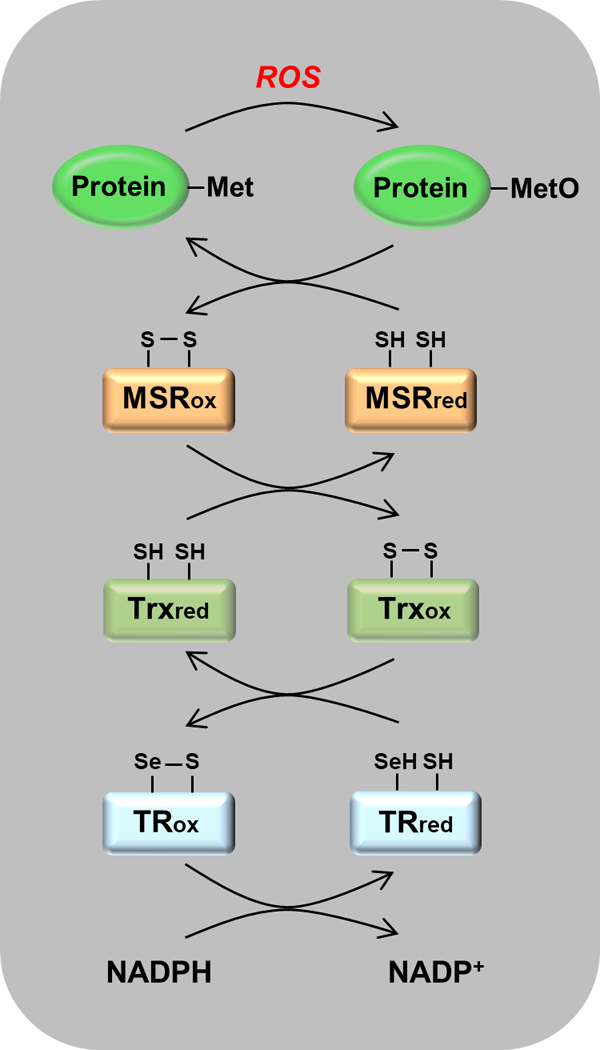
Scavenging of reactive oxygen species (ROS) by the msr-dependent catalytic cascade. Reduced forms of the proteins carry the subscript “red” and oxidized forms carry “ox”. Reading from top to bottom, an ROS is intercepted by a Met residue that is oxidized to MetO. MetO is reduced back to Met by msr, with the formation of a disulfide bond. The oxidized msr is reduced by thioredoxin (Trx), which now carries the disulfide bond. It is reduced by thioredoxin reductase (TR), which in mammals contains a selenocysteine residue that is oxidized, forming a selenocysteine-cysteine bond. This disulfide analogue is then reduced by NADPH. The net result is that ROS is reduced at the expense of NADPH.
The A class of reductases was described some years ago and has been characterized in considerable detail, especially by Weissbach, Brot, and colleagues [20]. The B class of reductases, some of which are selenoproteins in higher animals, was discovered more recently but has also been studied intensively [21, 22]. Mammals have 3 isoforms of the B class, and one of the A class. While there is only one gene for msrA, the enzyme has been reported to be in both the cytoplasm and in mitochondria, although we are re-evaluating the subcellular localization in our laboratory.
The in vivo importance of the reductases is well established, particularly for msrA. Knocking out the enzyme caused increased susceptibility to oxidative stress in mice [23], yeast [24], and bacteria [25–28]. Conversely, overexpressing msrA conferred increased resistance to oxidative stress in Drosophila [29], Saccharomyces [30], Arabidopsis [31], PC-12 cells [32], human T cells [30], and microglial-mediated neuroinflammation [33]. Helicobacter pylori, a causative agent of gastric ulcers and carcinoma, requires msrA for protection against oxidative stress and appears to act through reduction of methionine sulfoxide in the bacterial catalase [28, 34].
Overexpression of bovine msrA in Drosophila almost doubled the lifespan of the flies [29], and this impressive result was replicated using Drosophila msrA in an independent laboratory [35]. However, overexpression of msrA in mice does not increase lifespan [36]. Although an initial report with a small number of mice suggested that knocking out msrA caused neurological abnormalities and drastically reduced the lifespan of mice [23], studies with appropriate numbers of animals found no change in lifespan nor neurological abnormalities [36].
Solvent Exposed Methionines as Antioxidants
α−2-macroglobulin is a physiologically important proteinase inhibitor, often acting at sites of inflammation where reactive oxygen and nitrogen species are at relatively high concentration. It had been thought that α−2-macroglobulin was resistant to oxidative modification, but studies by Weiss and colleagues demonstrated that the protein was consuming oxidizing species, initially without loss of anti-proteinase activity [37]. More detailed studies established that while activity was indeed retained, consumption of oxidant was stoichiometrically accounted for by oxidation of methionine residues to the sulfoxide [38]. With continued exposure to an oxidizing environment, a single tryptophan residue was eventually oxidized with concomitant loss of anti-proteinase activity. These observations led to the proposal that certain methionine residues of α−2-macroglobulin served as antioxidants, protecting the critical tryptophan from damage.
The protecting methionines were presumed to be surface exposed, but no structure of α−2-macroglobulin was then available to determine whether that was the case. We therefore examined the effect of hydrogen peroxide on glutamine synthetase, since the crystal structure for this 12-subunit enzyme was solved by Eisenberg and colleagues [39] and because the active site was known to be susceptible to oxidative inactivation [40]. Exposure of the enzyme to varying concentrations of hydrogen peroxide generated a series of preparations with increasing content of methionine sulfoxide; no other covalent modifications were detected [1]. Eight of the 16 methionine residues could be oxidized without loss of catalytic activity. Mapping of the oxidizable methionine residues revealed that all were surface exposed; conversely, the residues that remained unoxidized were buried. More detailed examination of the topographic distribution of the oxidizable methionine residues was intriguing as these residues were found to line the bay leading from the surface of the enzyme to its active site (Fig. 4 in [1]). In other words, these methionine residues are mustered in a phalanx guarding the active site where they function as macromolecular bodyguards.
The effective concentration of exposed methionines is extremely high near the protein surface, greater than 1 M with certain assumptions (1). Recognizing the ease of oxidation of methionines already mentioned, surface-exposed methionines constitute a formidable antioxidant defense mechanism, capable of protecting critical residues within the protein as well as other cellular components. Since the methionine sulfoxide reductases can reduce MetO back to methionine, this antioxidant defense gains catalytic efficiency. One of a number of examples of the system’s function comes from Stocker and colleagues who established that high density lipoproteins reduce toxic cholesteryl ester hydroperoxides to alcohols, with the concomitant oxidation of two methionine residues to the sulfoxides [41]. This system can function catalytically as shown by Sigalov and Stern, who demonstrated that the oxidized apolipoprotein could be reduced by methionine sulfoxide reductase [42].
As summarized in the introduction, there exists considerable experimental evidence compatible with the proposal that methionines in proteins have an important antioxidant function, analogous to that of cysteine in glutathione. However, all of the studies describe correlations and do not assess causality. Direct experimental testing of the methionine antioxidant hypothesis is difficult in eukaryotic cells, but it has been done with E. coli by altering the global content of methionine in the bacterial cellular proteins. It is well established that the selenium analogue of methionine, selenomethionine, can replace methionine in proteins with little or no effect on protein structure and function. It is not so well known that the carbon analogue, norleucine, can also replace methionine, at least in bacteria. This was shown by Georges Cohen and his colleagues in 1959 [43]. Barker and Bruton then demonstrated that norleucine was charged onto both methionyl-tRNA and formylmethionyl-tRNA [44]. After charging with norleucine, the latter undergoes formylation, allowing N-formylnorleucine to initiate protein synthesis. When grown in a medium with a high ratio of norleucine to methionine, norleucine substitutes for methionine residues globally. As with selenomethionine substitution, the incorporation of norleucine does not alter the activity of enzymes that have been assayed to date.
The methionine as antioxidant hypothesis was tested by comparing the survival of control and norleucine-substituted cells, with and without oxidative stress. If there were little difference in survival, the hypothesis would be rejected. If there were a difference, the hypothesis would live to face other tests. We were able to replace 40% of the methionine residues in E. coli with norleucine [45]. Control and norleucine-grown cells had almost identical growth rates, and neither free methionine nor S-adenosylmethionine levels were altered by growth on norleucine. When left unstressed, both control and norleucine-substituted cells survived equally well in stationary phase for at least 32 hours. However, when challenged by exposure to hypochlorite, hydrogen peroxide, or ionizing radiation, the norleucine-substituted cells died more rapidly than the control cells. For example, 10 μM hypochlorite did not kill any control cells while it killed 100% of the norleucine-substituted cells.
Further support for the methionine as antioxidant hypothesis comes from more recent investigations in several laboratories that establish the surprising ability of eukaryotes, including mammals, to increase the content of methionine in their proteins in response to oxidative stress [7, 46]. The mechanism was elucidated by Kim and colleagues [8] who showed that ERK1/2 phosphorylates methionyl-tRNA synthetase in cells experiencing oxidative stress. The phosphorylation renders the synthetase promiscuous, so that it acylates non-methionine tRNAs with methionine, thus increasing the methionine content of proteins during oxidative stress (Fig. 2).
Fig. 2.
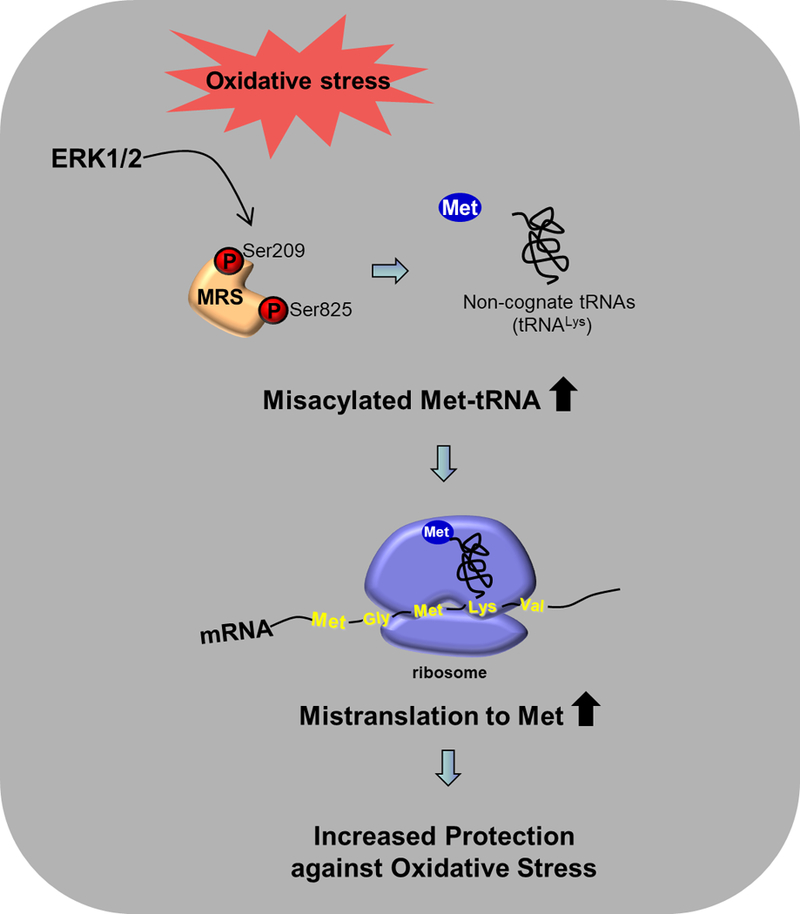
The mechanism by which oxidative stress increases the methionine content of proteins [8]. In response to oxidative stress, ERK1/2 phosphorylates methionyl tRNA synthetase (MRS). This renders the synthetase promiscuous so that it charges non-cognate tRNAs with Met, as shown here for tRNALys. In this example, the Lys codon leads to insertion of Met, thus increasing the total methionine content of the protein to provide additional protection against oxidative stress.
Studies on the evolution of mitochondria and their use of an alternate genetic code also support the proposition that methionine in proteins acts as an antioxidant [2, 47, 48]. Bender and colleagues noted that AUA codes for isoleucine in the nucleus, but it specifies methionine in the mitochondria of animals using the modified code. Looking at a large number of species not using the modified mitochondrial code, they established that the average methionine content in mitochondrially encoded proteins is 2%, which is the same as that for nuclear proteins encoded in those organisms. However, in organisms whose mitochondrial code evolved to specify methionine by AUA, the average mitochondrial methionine content jumped 3 fold to 6%. Moreover, the additional methionine residues were topographically arranged on the surface of the proteins, nicely positioned to intercept reactive oxygen species generated by mitochondrial respiration [2].
Cellular Regulation through Redox Cycling of Methionine Residues
Progress in understanding the physiological and pathological effects of methionine oxidation is severely hampered by the lack of analytical tools for detecting and quantitating MetO content of individual proteins in complex mixtures such as those found in tissue homogenates. The absence of immunochemical or chemical methods for detecting and quantitating MetO is particularly vexing [49, 50].
Just as is the case for phosphoserine and phosphothreonine, no general anti-MetO antibody exists, but one can raise sequence specific anti-MetO or anti-methionine antibodies by immunization with a peptide or protein containing MetO or methionine [50, 51], [Lim JC, Kim G, and Levine RL, unpublished]. An antiserum sold by several vendors is claimed to specifically recognize MetO [52]. A number of publications, particularly in the area of neurodegeneration, have reported using it for that purpose [52–64]. Without confirmation by an independent method of analysis, one must be extremely cautious about results from that antiserum [49].
Like phosphorylation, methionine oxidation is a reversible covalent modification. Thus, cyclic oxidation and reduction of methionine residues could function as a regulatory or signaling mechanism [65, 66]. Although oxidation could occur non-enzymatically, the products would then be a mixture of the R and S epimers. Reversal would require coordinated action of msrA and msrB which is not an attractive regulatory mechanism. Enzymatic oxidation would likely be stereospecific and thus require coupling to only one reductase to complete the regulatory cycle.
Despite these limitations, many examples of regulation by methionine oxidation have been published, although in most cases it is unknown whether the oxidation is reversed in vivo by msr. Ciorba and colleagues reported that the inactivation of a potassium channel by nitric oxide was likely due to oxidation of an essential methionine residue in the channel [67]. Similarly, Sroussi et al presented evidence that the ability of the calcium-binding proteins to direct leukocyte migration was abolished by oxidation of specific methionine residues [68]. Interestingly, consistent with the notion that methionine oxidation does not invariably link to enzyme inactivation, Erickson and collaborators convincingly identified a calcium-independent pathway for activation of Ca2+/calmodulin-dependent protein kinase that was mediated by oxidation of specific methionine residue in vitro and in vivo [69]. In yet another example of a MetO “activation” process, oxidation of Met80, an iron ligand in cytochrome c, increases cytoplasmic translocation of the cytochrome as part of a potential defense system against nitrative stress in non-apoptotic cells [70]. Likewise, in plants, hydrogen peroxide-triggered protein phosphorylation can be regulated by oxidation of a specific methionine residue in the substrate recognition site of kinases [71]. Alternatively, methionine oxidation may lead to enhanced function via indirect routes. For example, the blood clotting protein, von Willebrand factor, undergoes HOCl-dependent methionine oxidation that renders the protein resistant to proteolysis by the metalloprotease, ADAMTS123, thereby endowing it with increased activity [72].
The Terman laboratory identified an NADPH oxidoreductase, MICAL, that specifically oxidizes a methionine residue in actin that induces filament severing and decreases actin polymerization [73]. Subsequently that group and Gladyshev’s laboratory demonstrated that the oxidation was stereospecific and generated only the R-MetO [4, 5]. Importantly, the modification was fully reversible by msrB1, a cytosolic msrB. Thus, oxidation of Met44 in actin by MICAL induces de-polymerization of actin and reduction of MetO44 by msrB1 restores the ability to polymerize (Fig. 3).
Fig. 3.
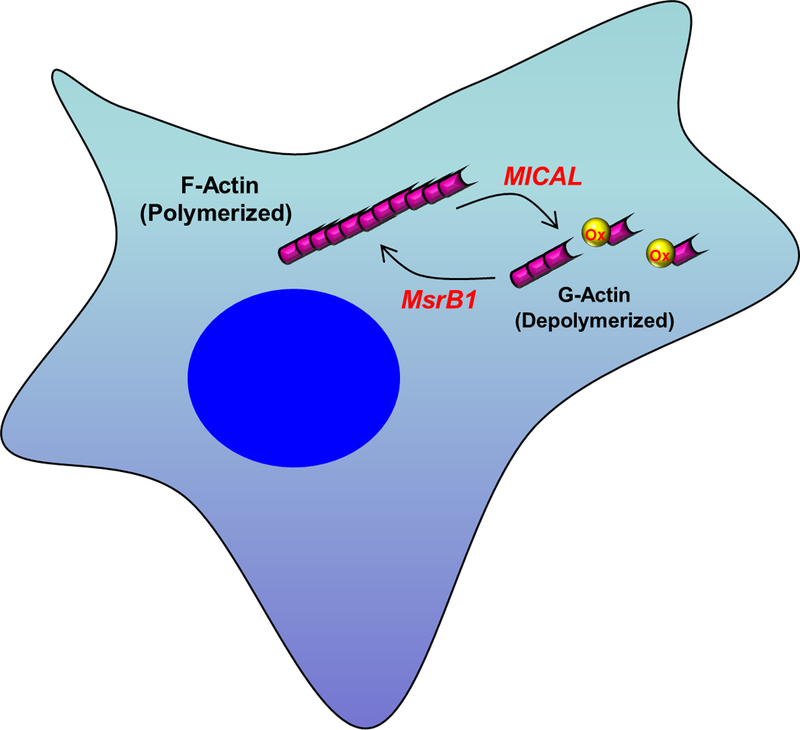
Redox regulation of actin polymerization by oxidation and reduction of methionine. Met44 of actin is oxidized by the monooxygenase MICAL causing depolymerization. Reduction of MetO44 by MsrB1 restores the ability to polymerize.
msrA operating in oxidase mode stereospecifically oxidizes just one of the 9 methionine residues in calmodulin [74]. Met77 is oxidized to S-MetO77. When msrA operates in reductase mode, MetO77 is fully reduced back to methionine. However, to date, no in vivo targets of calmodulin affected by this oxidation have been identified.
Methionine and Protein Structure
It has long been appreciated that the sulfur atom of methionine is a ligand to the heme in cytochrome c [75]. In myeloperoxidase, the heme is linked to methionine via a sulfonium ionic bond [76, 77]. More recently, a sulfilimine bond (-S=N-), the nitrogen analogue of a sulfoxide (-S=O-), was discovered in type IV collagen [78]. In a mechanism that is conserved from flies to humans, the carboxyl-terminal methionine of one type IV collagen subunit is covalently linked to a lysine of another subunit. Formation of the sulfilimine is catalyzed by a specific peroxidase, termed peroxidasin, that appears to generate the sulfilimine by formation of hypohalous acids as a reactive intermediate [79]. Drosophila with mutant peroxidasin fail to generate sulfilimine cross-links and display disorganized collagen IV networks with associated defects in basement membrane structure [79].
An interaction of methionine residues with nearby aromatic residues was pointed out 30 years ago [80], and crystallographic data suggested that the interaction contributed to protein stability [81]. More recent work of Valley and colleagues established that methionine, like cysteine, has a substantive role in stabilizing protein structure and in protein-protein interactions [3]. While the methionine-aromatic interaction occurs at a greater distance (~–6 Å) than that of a salt bridge (<4 Å), the energies associated with either interaction are comparable. These bonds are very common in proteins and thus contribute significantly to the stabilization of the native protein structure [3].
Oxidation of methionine to its sulfoxide provides a simple mechanism for an on-off switch for cellular regulation. For example, the binding of lymphotoxin-α to the tumor necrosis factor receptor 1 requires a methionine-aromatic bond between Met120 of lymphotoxin-α and Trp107 of tumor necrosis factor receptor 1 [3]. Oxidation of Met120 prevents binding [82]. Other hydrophobic amino acids are unable to form the bond with aromatic rings, potentially explaining some of the many examples in the literature in which leucine, isoleucine, or valine cannot functionally replace a methionine.
While oxidation may strengthen rather than weaken the methionine-aromatic bond [82], oxidation of a sufficient number of methionine residues is expected to perturb the native structure and expose otherwise normally buried residues, explaining both the association of methionine oxidation with increased surface hydrophobicity of proteins and their increased susceptibility to proteolytic degradation [1, 83]. This effect may be substantial during aging in which progressive increases in the surface hydrophobicity of proteins correlate with an age-related increase in MetO content [83]. However, in general, the changes in MetO content in aging tissues are rather modest. Thus far, no studies have validated total MetO content as a marker of biological aging.
Methionine Sulfoxide Reductases and Disease
Cardiopulmonary disease
In humans, a G to A polymorphism in msrA is associated with an increased risk of cardiovascular disease [84, 85]. In another genome-wide association study, an A to G intro variant was associated with hypertension, a known risk factor for cardiovascular disease [86]. In apolipoprotein E deficient mice, feeding a high fat “Western diet” causes atherosclerosis and hepatic steatosis. Hepatic overexpression of msrA in those mice reduced the plasma VLDL/LDL levels, hepatic steatosis, and aortic atherosclerosis [87]. As mentioned above, msrA in mammals has been localized to mitochondria and the cytosol. Because mitochondria are a major source of reactive oxygen species, it was hypothesized that overexpression of msrA targeted to the mitochondria would protect against cardiac ischemia-reperfusion while cytosolic msrA would not [88]. Notably, the opposite was found: mitochondrial overexpression of msrA provided no protection while cytosolic overexpression gave substantial protection. Moreover, the cytosolic form required myristoylation to be protective, an observation that is not yet explained mechanistically.
α1-antitrypsin is a member of the serpin family that inhibits serine proteases. The main physiological target of α1-antitrypsin is neutrophil elastase. A genetic deficiency in the synthesis of α1-antitrypsin is associated with the development of emphysema by the third or fourth decade of life, and 15 years earlier if the affected person is a smoker [89]. Deficiency of α1-antitrypsin causes a protease:antiprotease imbalance in which lung elastase damages tissue. Proteolytic injury to the lung is also a feature of diseases such as cystic fibrosis and cigarette-smoking in which serum levels of α1-antitrypsin are normal and therefore should theoretically provide a protective antiprotease [90, 91]. However, the specific activity of the α1-antitrypsin in the lung is decreased in these disorders [91, 92] because inflammation that occurs produces reactive oxygen species that oxidize a surface exposed methionine residue that is required for α1-antitrypsin activity [91, 93, 94]. This mechanism also accounts for the much earlier onset of emphysema in α1-antitrypsin deficient individuals who smoke. Inactivated α1-antitrypsin can be reactivated in vitro by msrA [95], suggesting that increasing the activity of msrA could be therapeutic [96].
Neurodegenerative disease
Oxidative damage in the brain is a well-established feature of Alzheimer’s disease [97], and the activity of msrA is decreased in the brain of Alzheimer’s patients [98]. Aβ peptides, particularly Aβ(1–42), are a major component of the senile plaques found in Alzheimer’s brains, and oxidative damage is distinctly increased in regions rich in Aβ [99]. Aβ peptides are toxic in a variety of in vivo and in vitro systems [100]. There are no reports of the activity of the msrB isozymes in Alzheimer’s disease, although it has been shown that msrB1 interacts with an Aβ peptide [101]. The Aβ(1–42) peptide contains one methionine, at position 35. Butterfield and his colleagues have reported strong evidence supporting their proposal that oxidation of Met35 is required for the toxicity of Aβ(1–42) [102, 103]. For example, substituting norleucine for methionine at position 35 abolishes toxicity. Thus, the decrease in msrA associated with Alzheimer’s disease could lead to an increased concentration of MetO-containing Aβ(1–42) that mediates toxicity.
Oxidative stress is also a feature of Parkinson’s disease [104]. Lewy bodies, a pathognomonic feature of Parkinson’s pathology, are rich in aggregated α-synuclein, and familial mutations in α-synuclein or elevated levels of wild-type α-synuclein cause Parkinson’s disease [105]. Dopaminergic cells in rat midbrain cultures were protected from oxidative-stress induced damage by transfection with msrA [106]. α-synuclein contains 4 methionine residues, two of which are particularly susceptible to oxidation that is reversible by msrA [107]. Overexpression of human α-synuclein in Drosophila creates a model of Parkinson’s disease with loss of dopaminergic neurons and the appearance of locomotor defects [108]. When bovine msrA was overexpressed along with the α-synuclein, the locomotor defects were almost completely suppressed [109].
Cancer
Following up on their studies that associated a deletion on chromosome 8 with metastatic spread of hepatocellular carcinoma, Lei and colleagues identified msrA as a candidate metastasis suppressor gene [110]. They then measured the mRNA levels for msrA in 40 human hepatocarcinoma tissue samples, half with metastasis and half without. The mean messenger RNA level was lower in tissue from patients with metastasis. They also transfected a human hepatocarcinoma cell line with msrA and found suppression of colony formation as well as decreased invasion in a 3-dimensional Matrigel assay. There was no effect on cell proliferation itself. These results are consistent with an effect on metastasis but not on cell division itself.
MsrA has also been found to be down regulated in human breast cancer, with the decrease being greater in tumors of advanced grade [111]. This observation led the investigators to investigate the effect of reducing msrA levels in a human breast cancer cell line, MDA-MB231. They found that knocking down msrA with shRNA caused an increase in cellular reactive oxygen species and oxidative damage to cellular proteins. Further, since msrA levels were decreased, the effects on tissue invasion and 3-dimensional growth were the opposite of those in the hepatocarcinoma study mentioned above – the breast cancer cells exhibited increased invasiveness and increased 3-dimensional growth.
Deafness
Loss of msrB3 causes congenital deafness, as first established by Riazuddin and her colleagues through genetic studies of Pakistani families [112]. These investigators also showed that msrB3 is present in the auditory and vestibular sensory epithelia of the inner ear. Knocking down msrB3 in zebrafish caused apototic death of hair cells in the fish neuromasts, providing a mechanistic basis for deafness [113]. Apototic death of hair cells also occurs during gestation in the msrB3 knockout mouse, causing deafness [114]. Lack of msrB3 may cause a failure of reduction of R-MetO in a specific protein, thus causing the apototic death. Neither the hypothesized protein nor the stereospecific methionine oxidase that generates R-MetO has been identified.
Other disorders
A genome-wide association study in a Chinese population was carried out to identify single nucleotide polymorphisms (SNPs) associated with schizophrenia [115]. The study demonstrated an association of SNPs in msrA with schizophrenia. The same group reported an association of SNPs in msrA with bipolar disorder, although the number of subjects in the study was relatively small [116].
A genome-wide association study of French and German populations reported an association of extreme obesity in children and adolescents with a locus at or near the msrA gene. The association with central adiposity was confirmed by a meta-analysis of genome-wide associations [117]. Another genome-wide association study found an association of the msrB3 locus with delayed development of teeth in human infants [118].
Conclusion
Methionine isn’t just for protein initiation anymore.
Acknowledgment
This research was supported by the Intramural Research Division of the National Heart, Lung, and Blood Institute (ZIA HL000225).
Abbreviations
- msr
Methionine sulfoxide reductase
- MetO
Methionine sulfoxide
- ROS
Reactive oxygen species
References
- 1.Levine RL, Mosoni L, Berlett BS, Stadtman ER (1996) Methionine residues as endogenous antioxidants in proteins. Proc Natl Acad Sci USA 93:15036–15040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bender A, Hajieva P, Moosmann B (2008) Adaptive antioxidant methionine accumulation in respiratory chain complexes explains the use of a deviant genetic code in mitochondria. Proc Natl Acad Sci USA 105:16496–16501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valley CC, Cembran A, Perlmutter JD, Lewis AK, Labello NP, Gao J, Sachs JN (2012) The Methionine-aromatic Motif Plays a Unique Role in Stabilizing Protein Structure. J Biol Chem 287:34979–34991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee BC, Peterfi Z, Hoffmann FW, Moore RE, Kaya A, Avanesov A, Tarrago L, Zhou Y, Weerapana E, Fomenko DE, Hoffmann PR, Gladyshev VN (2013) MsrB1 and MICALs regulate actin assembly and macrophage function via reversible stereoselective methionine oxidation. Mol Cell 51:397–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hung RJ, Spaeth CS, Yesilyurt HG, Terman JR (2013) SelR reverses Mical-mediated oxidation of actin to regulate F-actin dynamics. Nat Cell Biol 15:1445–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hung R-J, Pak CW, Terman JR (2011) Direct Redox Regulation of F-Actin Assembly and Disassembly by Mical. Science 334:1710–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Netzer N, Goodenbour JM, David A, Dittmar KA, Jones RB, Schneider JR, Boone D, Eves EM, Rosner MR, Gibbs JS, Embry A, Dolan B, Das S, Hickman HD, Berglund P, Bennink JR, Yewdell JW, Pan T (2009) Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature 462:522–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee JY, Kim DG, Kim BG, Yang WS, Hong J, Kang T, Oh YS, Kim KR, Han BW, Hwang BJ, Kang BS, Kang MS, Kim MH, Kwon NH, Kim S (2014) Promiscuous methionyl-tRNA synthetase mediates adaptive mistranslation to protect cells against oxidative stress. J Cell Sci 127:4234–4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lavine TF (1947) The formation, resolution, and optical properties of the diasteriomeric sulfoxides derived from L-methionine. J Biol Chem 169:477–491 [PubMed] [Google Scholar]
- 10.Vogt W (1995) Oxidation of methionine residues in proteins: Tools, targets, and reversal. Free Radic Biol Med 18:93–105 [DOI] [PubMed] [Google Scholar]
- 11.Wood PM (1981) The redox potential for dimethyl sulphoxide reduction to dimethyl sulphide: evaluation and biochemical implications. FEBS Lett 124:11–14 [DOI] [PubMed] [Google Scholar]
- 12.Jocelyn PC (1967) The Standard Redox Potential of Cysteine-Cystine from the Thiol-Disulphide Exchange Reaction with Glutathione and Lipoic Acid. EurJBiochem 2:327–331 [DOI] [PubMed] [Google Scholar]
- 13.Barton JP, Packer JE, Sims RJ (1973) Kinetics of reaction of hydrogen-peroxide with cysteine and cysteamine. J Chem Soc Perkin Trans 2:1547–1549 [Google Scholar]
- 14.Peskin AV, Winterbourn CC (2001) Kinetics of the reactions of hypochlorous acid and amino acid chloramines with thiols, methionine, and ascorbate. Free Radic Biol Med 30:572–579 [DOI] [PubMed] [Google Scholar]
- 15.Winterbourn CC Comparative reactivities of various biological compounds with myeloperoxidase-hydrogen peroxide-chloride, and similarity of the oxidant to hypochlorite. Biochim Biophys Acta 840:204–210 [DOI] [PubMed] [Google Scholar]
- 16.Richardson DE, Regino CA, Yao H, Johnson JV (2003) Methionine oxidation by peroxymonocarbonate, a reactive oxygen species formed from CO2/bicarbonate and hydrogen peroxide. Free Radic Biol Med 35:1538–1550 [DOI] [PubMed] [Google Scholar]
- 17.Brot N, Weissbach H (1983) Biochemistry and physiological role of methionine sulfoxide reductase in proteins. Arch Biochem Biophys 223:271–281 [DOI] [PubMed] [Google Scholar]
- 18.Zhang XH, Weissbach H (2008) Origin and evolution of the protein-repairing enzymes methionine sulphoxide reductases. Biol Rev Camb Philos Soc 83:249–257 [DOI] [PubMed] [Google Scholar]
- 19.Toennies G, Kolb JJ (1939) Methionine studies. J Biol Chem 128:399–405 [Google Scholar]
- 20.Weissbach H, Resnick L, Brot N (2005) Methionine sulfoxide reductases: history and cellular role in protecting against oxidative damage. Biochim Biophys Acta 1703:203–212 [DOI] [PubMed] [Google Scholar]
- 21.Lee BC, Dikiy A, Kim HY, Gladyshev VN (2009) Functions and evolution of selenoprotein methionine sulfoxide reductases. Biochim Biophys Acta 1790:1471–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boschi-Muller S, Gand A, Branlant G (2008) The methionine sulfoxide reductases: Catalysis and substrate specificities. Arch Biochem Biophys 474:266–273 [DOI] [PubMed] [Google Scholar]
- 23.Moskovitz J, Bar-Noy S, Williams WM, Requena J, Berlett BS, Stadtman ER (2001) Methionine sulfoxide reductase (MsrA) is a regulator of antioxidant defense and lifespan in mammals. Proc Natl Acad Sci USA 98:12920–12925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moskovitz J, Berlett BS, Poston JM, Stadtman ER (1997) The yeast peptide-methionine sulfoxide reductase functions as an antioxidant in vivo. Proc Natl Acad Sci USA 94:9585–9589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moskovitz J, Rahman MA, Strassman J, Yancey SO, Kushner SR, Brot N, Weissbach H (1995) Escherichia coli peptide methionine sulfoxide reductase gene: regulation of expression and role in protecting against oxidative damage. J Bacteriol 177:502–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Douglas T, Daniel DS, Parida BK, Jagannath C, Dhandayuthapani S (2004) Methionine sulfoxide reductase A (MsrA) deficiency affects the survival of Mycobacterium smegmatis within macrophages. J Bacteriol 186:3590–3598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.St John G, Brot N, Ruan J, Erdjument-Bromage H, Tempst P, Weissbach H, Nathan C (2001) Peptide methionine sulfoxide reductase from Escherichia coli and Mycobacterium tuberculosis protects bacteria against oxidative damage from reactive nitrogen intermediates. Proc Natl Acad Sci USA 98:9901–9906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahawar M, Tran V, Sharp JS, Maier RJ (2011) Synergistic roles of Helicobacter pylori methionine sulfoxide reductase and GroEL in repairing oxidant-damaged catalase. J Biol Chem 286:19159–19169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruan H, Tang XD, Chen ML, Joiner ML, Sun G, Brot N, Weissbach H, Heinemann SH, Iverson L, Wu CF, Hoshi T, Chen ML, Joiner MA, Heinemann SH (2002) High-quality life extension by the enzyme peptide methionine sulfoxide reductase. Proc Natl Acad Sci USA 99:2748–2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moskovitz J, Flescher E, Berlett BS, Azare J, Poston JM, Stadtman ER (1998) Overexpression of peptide-methionine sulfoxide reductase in Saccharomyces cerevisiae and human T cells provides them with high resistance to oxidative stress. Proc Natl Acad Sci USA 95:14071–14075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romero HM, Berlett BS, Jensen PJ, Pell EJ, Tien M (2004) Investigations into the role of the plastidial peptide methionine sulfoxide reductase in response to oxidative stress in Arabidopsis. Plant Physiol 136:3784–3794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yermolaieva O, Xu R, Schinstock C, Brot N, Weissbach H, Heinemann SH, Hoshi T (2004) Methionine sulfoxide reductase A protects neuronal cells against brief hypoxia/reoxygenation. Proc Natl Acad Sci USA 101:1159–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fan H, Wu PF, Zhang L, Hu ZL, Wang W, Guan XL, Luo H, Ni M, Yang JW, Li MX, Chen JG, Wang F (2015) Methionine sulfoxide reductase A negatively controls microglia-mediated neuroinflammation via inhibiting ROS/MAPKs/NF-kappaB signaling pathways through a catalytic antioxidant function. Antioxid Redox Signal 22:832–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benoit SL, Maier RJ (2016) Helicobacter catalase devoid of catalytic activity protects the bacterium against oxidative stress. J Biol Chem [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chung H, Kim AK, Jung SA, Kim SW, Yu K, Lee JH (2010) The Drosophila homolog of methionine sulfoxide reductase A extends lifespan and increases nuclear localization of FOXO. FEBS Lett 584:3609–3614 [DOI] [PubMed] [Google Scholar]
- 36.Salmon AB, Kim G, Liu C, Wren JD, Georgescu C, Richardson A, Levine RL (2016) Effects of transgenic methionine sulfoxide reductase A (MsrA) expression on lifespan and age-dependent changes in metabolic function in mice. Redox Biol 10:251–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reddy VY, Pizzo SV, Weiss SJ (1989) Functional inactivation and structural disruption of human alpha 2-macroglobulin by neutrophils and eosinophils. J Biol Chem 264:13801–13809 [PubMed] [Google Scholar]
- 38.Reddy VY, Desrochers PE, Pizzo SV, Gonias SL, Sahakian JA, Levine RL, Weiss SJ (1994) Oxidative dissociation of human a2macroglobulin tetramers into dysfunctional dimers. J Biol Chem 269:4683–4691 [PubMed] [Google Scholar]
- 39.Almassy RJ, Janson CA, Hamlin R, Xuong NH, Eisenberg D (1986) Novel subunit-subunit interactions in the structure of glutamine synthetase. Nature 323:304–309 [DOI] [PubMed] [Google Scholar]
- 40.Levine RL (1983) Oxidative modification of glutamine synthetase. I. Inactivation is due to loss of one histidine residue. J Biol Chem 258:11823–11827 [PubMed] [Google Scholar]
- 41.Garner B, Waldeck AR, Witting PK, Rye KA, Stocker R (1998) Oxidation of high density lipoproteins. II. Evidence for direct reduction of lipid hydroperoxides by methionine residues of apolipoproteins AI and AII. J Biol Chem 273:6088–6095 [DOI] [PubMed] [Google Scholar]
- 42.Sigalov AB, Stern LJ (1998) Enzymatic repair of oxidative damage to human apolipoprotein A-I. FEBS Lett 433:196–200 [DOI] [PubMed] [Google Scholar]
- 43.Cowie DB, Cohen GN, Bolton ET, De Robichon-Szulmajster H (1959) Amino acid analog incorporation into bacterial proteins. Biochim Biophys Acta 34:39–46 [DOI] [PubMed] [Google Scholar]
- 44.Barker DG, Bruton CJ (1979) The fate of norleucine as a replacement for methionine in protein synthesis. J Mol Biol 133:217–231 [DOI] [PubMed] [Google Scholar]
- 45.Luo S, Levine RL (2008) Testing The Hypothesis That “Methionine Residues In Proteins Are Antioxidants”. FASEB J 22 [Google Scholar]
- 46.Wang X, Pan T (2016) Stress Response and Adaptation Mediated by Amino Acid Misincorporation during Protein Synthesis. Advances in nutrition (Bethesda, Md) 7:773s–779s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aledo JC, Li Y, de Magalhaes JP, Ruiz-Camacho M, Perez-Claros JA (2011) Mitochondrially encoded methionine is inversely related to longevity in mammals. Aging Cell 10:198–207 [DOI] [PubMed] [Google Scholar]
- 48.Schindeldecker M, Moosmann B (2015) Protein-borne methionine residues as structural antioxidants in mitochondria. Amino Acids 47:1421–1432 [DOI] [PubMed] [Google Scholar]
- 49.Wehr NB, Levine RL (2012) Wanted and wanting: Antibody against rnethionine sulfoxide. Free Radic Biol Med 53:1222–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Le DT, Liang X, Fomenko DE, Raza AS, Chong C-K, Carlson BA, Hatfield DL, Gladyshev VN (2008) Analysis of Methionine/Selenomethionine Oxidation and Methionine Sulfoxide Reductase Function Using Methionine-Rich Proteins and Antibodies against Their Oxidized Forms. Biochemistry 47:6685–6694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liang X, Zhang Y, Le DT, Gladyshev VN (2009) Characterization of peptide Methionine oxidation and Methionine sulfoxide reduction using Methionine-rich proteins. The FASEB Journal 23:855–858.18987303 [Google Scholar]
- 52.Oien DB, Canello T, Gabizon R, Gasset M, Lundquist BL, Burns JM, Moskovitz J (2009) Detection of oxidized methionine in selected proteins, cellular extracts and blood serums by novel anti-methionine sulfoxide antibodies. Arch Biochem Biophys 485:35–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haigh CL, Drew SC (2015) Cavitation during the protein misfolding cyclic amplification (PMCA) method – The trigger for de novo prion generation? Biochemical and Biophysical Research Communications 461:494–500 [DOI] [PubMed] [Google Scholar]
- 54.Fan H, Wu P-F, Zhang L, Hu Z-L, Wang W, Guan X-L, Luo H, Ni M, Yang J-W, Li M-X, Chen J-G, Wang F (2015) Methionine Sulfoxide Reductase A Negatively Controls Microglia-Mediated Neuroinflammation via Inhibiting ROS/MAPKs/NF-κB Signaling Pathways Through a Catalytic Antioxidant Function. Antioxid Redox Signal 22:832–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mochin MT, Underwood KF, Cooper B, McLenithan JC, Pierce AD, Nalvarte C, Arbiser J, Karlsson AI, Moise AR, Moskovitz J, Passaniti A (2015) Hyperglycemia and redox status regulate RUNX2 DNA-binding and an angiogenic phenotype in endothelial cells. Microvascular Research 97:55–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim JS, Park HM, Chae S, Lee TH, Hwang DJ, Oh SD, Park JS, Song DG, Pan CH, Choi D, Kim YH, Nahm BH, Kim YK (2014) A Pepper MSRB2 Gene Confers Drought Tolerance in Rice through the Protection of Chloroplast-Targeted Genes. PLoS One 9:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moskovitz J, Du F, Bowman CF, Yan SS (2016) Methionine sulfoxide reductase A affects beta-amyloid solubility and mitochondrial function in a mouse model of Alzheimer’s disease. American Journal of Physiology-Endocrinology and Metabolism 310:E388–E393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moskovitz J (2014) Detection and Localization of Methionine Sulfoxide Residues of Specific Proteins in Brain Tissue. Protein and Peptide Letters 21:52–55 [DOI] [PubMed] [Google Scholar]
- 59.Salama SA, Snapkai RM (2012) Amino Acid Chloramine Damage to Proliferating Cell Nuclear Antigen in Mammalian Cells. In Vivo 26:501–517 [PubMed] [Google Scholar]
- 60.Moskovitz J, Malik A, Hernandez A, Band M, Avivi A (2012) Methionine sulfoxide reductases and methionine sulfoxide in the subterranean mole rat (Spalax): Characterization of expression under various oxygen conditions. Comparative Biochemistry and Physiology a-Molecular & Integrative Physiology 161:406–414 [DOI] [PubMed] [Google Scholar]
- 61.Ringman JM, Fithian AT, Gylys K, Cummings JL, Coppola G, Elashoff D, Pratico D, Moskovitz J, Bitan G (2012) Plasma Methionine Sulfoxide in Persons with Familial Alzheimer’s Disease Mutations. Dementia and Geriatric Cognitive Disorders 33:219–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oien DB, Osterhaus GL, Lundquist BL, Fowler SC, Moskovitz J (2010) Caloric restriction alleviates abnormal locomotor activity and dopamine levels in the brain of the methionine sulfoxide reductase A knockout mouse. Neurosci Lett 468:38–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ringman JM, Fithian AT, Gylys K, Cummings JL, Coppola G, Elashoff D, Pratico D, Moskovitz J, Bitan G (2012) Plasma Methionine Sulfoxide in Persons with Familial Alzheimer’s Disease Mutations. Dement Geriatr Cogn Disord 33:219–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Day AM, Brown JD, Taylor SR, Rand JD, Morgan BA, Veal EA (2012) Inactivation of a peroxiredoxin by hydrogen peroxide is critical for thioredoxin-mediated repair of oxidized proteins and cell survival. Mol Cell 45:398–408 [DOI] [PubMed] [Google Scholar]
- 65.Hoshi T, Heinemann S (2001) Regulation of cell function by methionine oxidation and reduction. JPhysiol 531:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bigelow DJ, Squier TC (2005) Redox modulation of cellular signaling and metabolism through reversible oxidation of methionine sensors in calcium regulatory proteins. Biochim Biophys Acta 1703:121–134 [DOI] [PubMed] [Google Scholar]
- 67.Ciorba MA, Heinemann SH, Weissbach H, Brot N, Hoshi T (1999) Regulation of voltage-dependent K+ channels by methionine oxidation: effect of nitric oxide and vitamin C. FEBS Lett 442:48–52 [DOI] [PubMed] [Google Scholar]
- 68.Sroussi HY, Berline J, Palefsky JM (2007) Oxidation of methionine 63 and 83 regulates the effect of S100A9 on the migration of neutrophils in vitro. JLeukocBiol 81:818–824 [DOI] [PubMed] [Google Scholar]
- 69.Erickson JR, Joiner ML, Guan X, Kutschke W, Yang J, Oddis CV, Bartlett RK, Lowe JS, O’Donnell SE, Aykin-Burns N, Zimmerman MC, Zimmerman K, Ham AJ, Weiss RM, Spitz DR, Shea MA, Colbran RJ, Mohler PJ, Anderson ME (2008) A dynamic pathway for calcium-independent activation of CaMKII by methionine oxidation. Cell 133:462–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Godoy LC, Munoz-Pinedo C, Castro L, Cardaci S, Schonhoff CM, King M, Tortora V, Marin M, Miao Q, Jiang JF, Kapralov A, Jemmerson R, Silkstone GG, Patel JN, Evans JE, Wilson MT, Green DR, Kagan VE, Radi R, Mannick JB (2009) Disruption of the M80-Fe ligation stimulates the translocation of cytochrome c to the cytoplasm and nucleus in nonapoptotic cells. Proc Natl Acad Sci USA 106:2653–2658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hardin SC, Larue CT, Oh MH, Jain V, Huber SC (2009) Coupling oxidative signals to protein phosphorylation via methionine oxidation in Arabidopsis. BiochemJ 422:305–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fu X, Chen J, Gallagher R, Zheng Y, Chung DW, Lopez JA (2011) Shear stress-induced unfolding of VWF accelerates oxidation of key methionine residues in the A1A2A3 region. Blood 118:5283–5291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hung RJ, Pak CW, Terman JR (2011) Direct redox regulation of F-actin assembly and disassembly by Mical. Science 334:1710–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lim JC, Kim G, Levine RL (2013) Stereospecific oxidation of calmodulin by methionine sulfoxide reductase A. Free Radic Biol Med 61:257–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Senn H, Wüthrich K (1985) Amino acid sequence, haem-iron co-ordination geometry and functional properties of mitochondrial and bacterial c-type cytochromes. Quarterly Reviews of Biophysics 18:111–134 [DOI] [PubMed] [Google Scholar]
- 76.Taylor KL, Pohl J, Kinkade JM (1992) Unique autolytic cleavage of human myeloperoxidase. Implications for the involvement of active site MET409. J Biol Chem 267:25282–25288 [PubMed] [Google Scholar]
- 77.Kooter IM, Moguilevsky N, Bollen A, van der Veen LA, Otto C, Dekker HL, Wever R (1999) The Sulfonium Ion Linkage in Myeloperoxidase: Direct Spectroscopic Detection by Isotopic Labeling and Effect of Mutation. J Biol Chem 274:26794–26802 [DOI] [PubMed] [Google Scholar]
- 78.Vanacore R, Ham AJ, Voehler M, Sanders CR, Conrads TP, Veenstra TD, Sharpless KB, Dawson PE, Hudson BG (2009) A sulfilimine bond identified in collagen IV. Science 325:1230–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bhave G, Cummings CF, Vanacore RM, Kumagai-Cresse C, Ero-Tolliver IA, Rafi M, Kang JS, Pedchenko V, Fessler LI, Fessler JH, Hudson BG (2012) Peroxidasin forms sulfilimine chemical bonds using hypohalous acids in tissue genesis. NatChemBiol 8:784–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Reid KSC, Lindley PF, Thornton JM (1985) Sulfur-aromatic interactions in proteins. FEBS Lett 190:209–213 [Google Scholar]
- 81.Zauhar RJ, Colbert CL, Morgan RS, Welsh WJ (2000) Evidence for a strong sulfur-aromatic interaction derived from crystallographic data. Biopolymers 53:233–248 [DOI] [PubMed] [Google Scholar]
- 82.Lewis AK, Dunleavy KM, Senkow TL, Her C, Horn BT, Jersett MA, Mahling R, McCarthy MR, Perell GT, Valley CC, Karim CB, Gao J, Pomerantz WCK, Thomas DD, Cembran A, Hinderliter A, Sachs JN (2016) Oxidation increases the strength of the methionine-aromatic interaction. Nat Chem Biol 12:860–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chao CC, Ma YS, Stadtman ER (1997) Modification of protein surface hydrophobicity and methionine oxidation by oxidative systems. Proc Natl Acad Sci USA 94:2969–2974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.García-Bermúdez M, López-Mejías R, González-Juanatey C, Castañeda S, Miranda-Filloy JA, Blanco R, Fernández-Gutiérrez B, Balsa A, González-Álvaro I, Gómez-Vaquero C, Llorca J, Martín J, González-Gay MA (2012) Association of the methionine sulfoxide reductase A rs10903323 gene polymorphism with cardiovascular disease in patients with rheumatoid arthritis. Scand J Rheumatol 41:350–353 [DOI] [PubMed] [Google Scholar]
- 85.Gu H, Chen W, Yin J, Chen S, Zhang J, Gong J (2013) Methionine sulfoxide reductase A rs10903323 G/A polymorphism is associated with increased risk of coronary artery disease in a Chinese population. Clin Biochem 46:1668–1672 [DOI] [PubMed] [Google Scholar]
- 86.Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, Glazer NL, Morrison AC, Johnson AD, Aspelund T, Aulchenko Y, Lumley T, Kottgen A, Vasan RS, Rivadeneira F, Eiriksdottir G, Guo X, Arking DE, Mitchell GF, Mattace-Raso FUS, Smith AV, Taylor K, Scharpf RB, Hwang S-J, Sijbrands EJG, Bis J, Harris TB, Ganesh SK, O’Donnell CJ, Hofman A, Rotter JI, Coresh J, Benjamin EJ, Uitterlinden AG, Heiss G, Fox CS, Witteman JCM, Boerwinkle E, Wang TJ, Gudnason V, Larson MG, Chakravarti A, Psaty BM, van Duijn CM (2009) Genome-wide association study of blood pressure and hypertension. Nat Genet 41:677–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xu YY, Du F, Meng B, Xie GH, Cao J, Fan D, Yu H (2015) Hepatic overexpression of methionine sulfoxide reductase A reduces atherosclerosis in apolipoprotein E-deficient mice. Journal of lipid research 56:1891–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhao H, Sun J, Deschamps AM, Kim G, Liu C, Murphy E, Levine RL (2011) Myristoylated Methionine Sulfoxide Reductase A Protects the Heart from Ischemia-Reperfusion Injury. Am J Physiol Heart Circ Physiol 301:H1513–H1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Larsson C (1978) Natural history and life expectancy in severe alpha1-antitrypsin deficiency, Pi Z. Acta medica Scandinavica 204:345–351 [DOI] [PubMed] [Google Scholar]
- 90.Gadek JE, Fells GA, Crystal RG (1979) Cigarette smoking induces functional antiprotease deficiency in the lower respiratory tract of humans. Science 206:1315–1316 [DOI] [PubMed] [Google Scholar]
- 91.Carp H, Miller F, Hoidal JR, Janoff A (1982) Potential mechanism of emphysema: alpha 1-proteinase inhibitor recovered from lungs of cigarette smokers contains oxidized methionine and has decreased elastase inhibitory capacity. Proc Natl Acad Sci USA 79:2041–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McGuire WW, Spragg RG, Cohen AB, Cochrane CG (1982) Studies on the pathogenesis of the adult respiratory distress syndrome. J Clin Invest 69:543–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Johnson D, Travis J (1979) The oxidative inactivation of human alpha-1-proteinase inhibitor. Further evidence for methionine at the reactive center. J Biol Chem 254:4022–4026 [PubMed] [Google Scholar]
- 94.Taggart C, Cervantes-Laurean D, Kim G, McElvaney NG, Wehr N, Moss J, Levine RL (2000) Oxidation of either methionine 351 or methionine 358 in alpha 1-antitrypsin causes loss of anti-neutrophil elastase activity. J Biol Chem 275:27258–27265 [DOI] [PubMed] [Google Scholar]
- 95.Carp H, Janoff A, Abrams W, Weinbaum G, Drew RT, Weissbach H, Brot N (1983) Human methionine sulfoxide-peptide reductase, an enzyme capable of reactivating oxidized alpha-1-proteinase inhibitor in vitro. Am Rev Respir Dis 127:301–305 [DOI] [PubMed] [Google Scholar]
- 96.Cudic P, Joshi N, Sagher D, Williams BT, Stawikowski MJ, Weissbach H (2016) Identification of activators of methionine sulfoxide reductases A and B. Biochem Biophys Res Commun 469:863–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Markesbery WR, Carney JM (1999) Oxidative alterations in Alzheimer’s disease. Brain pathology (Zurich, Switzerland) 9:133–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gabbita SP, Aksenov MY, Lovell MA, Markesbery WR (1999) Decrease in peptide methionine sulfoxide reductase in Alzheimer’s disease brain. J Neurochem 73:1660–1666 [DOI] [PubMed] [Google Scholar]
- 99.Hensley K, Hall N, Subramaniam R, Cole P, Harris M, Aksenov M, Aksenova M, Gabbita SP, Wu JF, Carney JM, et al. (1995) Brain regional correspondence between Alzheimer’s disease histopathology and biomarkers of protein oxidation. J Neurochem 65:2146–2156 [DOI] [PubMed] [Google Scholar]
- 100.Price DL, Borchelt DR, Walker LC, Sisodia SS (1992) Toxicity of synthetic aβ peptides and modeling of alzheimer’s disease. Neurobiology of Aging 13:623–625 [DOI] [PubMed] [Google Scholar]
- 101.Wang C, Chen P, He X, Peng Z, Chen S, Zhang R, Cheng J, Liu Q (2017) Direct interaction between selenoprotein R and Aβ42. Biochemical and Biophysical Research Communications 489:509–514 [DOI] [PubMed] [Google Scholar]
- 102.Varadarajan S, Yatin S, Aksenova M, Butterfield DA (2000) Review: Alzheimer’s amyloid beta-peptide-associated free radical oxidative stress and neurotoxicity. Journal of structural biology 130:184–208 [DOI] [PubMed] [Google Scholar]
- 103.Butterfield DA (2014) The 2013 SFRBM discovery award: Selected discoveries from the butterfield laboratory of oxidative stress and its sequela in brain in cognitive disorders exemplified by Alzheimer disease and chemotherapy induced cognitive impairment. Free Radic Biol Med 74:157–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Litvan I, Chesselet MF, Gasser T, Di Monte DA, Parker D Jr., Hagg T, Hardy J, Jenner P, Myers RH, Price D, Hallett M, Langston WJ, Lang AE, Halliday G, Rocca W, Duyckaerts C, Dickson DW, Ben-Shlomo Y, Goetz CG, Melamed E (2007) The etiopathogenesis of Parkinson disease and suggestions for future research. Part II. J Neuropathol Exp Neurol 66:329–336 [DOI] [PubMed] [Google Scholar]
- 105.Eriksen JL, Dawson TM, Dickson DW, Petrucelli L (2003) Caught in the act: alpha-synuclein is the culprit in Parkinson’s disease. Neuron 40:453–456 [DOI] [PubMed] [Google Scholar]
- 106.Liu F, Hindupur J, Nguyen JL, Ruf KJ, Zhu J, Schieler JL, Bonham CC, Wood KV, Davisson VJ, Rochet J-C (2008) Methionine sulfoxide reductase A protects dopaminergic cells from Parkinson’s disease-related insults. Free Radic Biol Med 45:242–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Maltsev AS, Chen J, Levine RL, Bax A (2013) Site-specific interaction between alpha-synuclein and membranes probed by NMR-observed methionine oxidation rates. J Am Chem Soc 135:2943–2946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Feany MB, Bender WW (2000) A Drosophila model of Parkinson’s disease. Nature 404:394–398 [DOI] [PubMed] [Google Scholar]
- 109.Wassef R, Haenold R, Hansel A, Brot N, Heinemann SH, Hoshi T (2007) Methionine sulfoxide reductase A and a dietary supplement S-methyl-L-cysteine prevent Parkinson’s-like symptoms. J Neurosci 27:12808–12816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lei KF, Wang YF, Zhu XQ, Lu PC, Sun BS, Jia HL, Ren N, Ye QH, Sun HC, Wang L, Tang ZY, Qin LX (2007) Identification of MSRA gene on chromosome 8p as a candidate metastasis suppressor for human hepatitis B virus-positive hepatocellular carcinoma. BMC cancer 7:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.De Luca A, Sanna F, Sallese M, Ruggiero C, Grossi M, Sacchetta P, Rossi C, De Laurenzi V, Di Ilio C, Favaloro B (2010) Methionine sulfoxide reductase A down-regulation in human breast cancer cells results in a more aggressive phenotype. Proc Natl Acad Sci USA 107:18628–18633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ahmed ZM, Yousaf R, Lee BC, Khan SN, Lee S, Lee K, Husnain T, Rehman AU, Bonneux S, Ansar M, Ahmad W, Leal SM, Gladyshev VN, Belyantseva IA, Van Camp G, Riazuddin S, Friedman TB, Riazuddin S (2011) Functional Null Mutations of MSRB3 Encoding Methionine Sulfoxide Reductase Are Associated with Human Deafness DFNB74. The American Journal of Human Genetics 88:19–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shen X, Liu F, Wang Y, Wang H, Ma J, Xia W, Zhang J, Jiang N, Sun S, Wang X, Ma D (2015) Down-regulation of msrb3 and destruction of normal auditory system development through hair cell apoptosis in zebrafish. The International journal of developmental biology 59:195–203 [DOI] [PubMed] [Google Scholar]
- 114.Kwon TJ, Cho HJ, Kim UK, Lee E, Oh SK, Bok J, Bae YC, Yi JK, Lee JW, Ryoo ZY, Lee SH, Lee KY, Kim HY (2014) Methionine sulfoxide reductase B3 deficiency causes hearing loss due to stereocilia degeneration and apoptotic cell death in cochlear hair cells. Hum Mol Genet 23:1591–1601 [DOI] [PubMed] [Google Scholar]
- 115.Ma X, Deng W, Liu X, Li M, Chen Z, He Z, Wang Y, Wang Q, Hu X, Collier DA, Li T (2011) A genome-wide association study for quantitative traits in schizophrenia in China. Genes, brain, and behavior 10:734–739 [DOI] [PubMed] [Google Scholar]
- 116.Ni P, Ma X, Lin Y, Lao G, Hao X, Guan L, Li X, Jiang Z, Liu Y, Ye B, Liu X, Wang Y, Zhao L, Cao L, Li T (2015) Methionine sulfoxide reductase A (MsrA) associated with bipolar I disorder and executive functions in A Han Chinese population. Journal of affective disorders 184:235–238 [DOI] [PubMed] [Google Scholar]
- 117.Lindgren CM, Heid IM, Randall JC, Lamina C, Steinthorsdottir V, Qi L, Speliotes EK, Thorleifsson G, Willer CJ, Herrera BM, Jackson AU, Lim N, Scheet P, Soranzo N, Amin N, Aulchenko YS, Chambers JC, Drong A, Luan J, Lyon HN, Rivadeneira F, Sanna S, Timpson NJ, Zillikens MC, Zhao JH, Almgren P, Bandinelli S, Bennett AJ, Bergman RN, Bonnycastle LL, Bumpstead SJ, Chanock SJ, Cherkas L, Chines P, Coin L, Cooper C, Crawford G, Doering A, Dominiczak A, Doney AS, Ebrahim S, Elliott P, Erdos MR, Estrada K, Ferrucci L, Fischer G, Forouhi NG, Gieger C, Grallert H, Groves CJ, Grundy S, Guiducci C, Hadley D, Hamsten A, Havulinna AS, Hofman A, Holle R, Holloway JW, Illig T, Isomaa B, Jacobs LC, Jameson K, Jousilahti P, Karpe F, Kuusisto J, Laitinen J, Lathrop GM, Lawlor DA, Mangino M, McArdle WL, Meitinger T, Morken MA, Morris AP, Munroe P, Narisu N, Nordstrom A, Nordstrom P, Oostra BA, Palmer CN, Payne F, Peden JF, Prokopenko I, Renstrom F, Ruokonen A, Salomaa V, Sandhu MS, Scott LJ, Scuteri A, Silander K, Song K, Yuan X, Stringham HM, Swift AJ, Tuomi T, Uda M, Vollenweider P, Waeber G, Wallace C, Walters GB, Weedon MN, Witteman JC, Zhang C, Zhang W, Caulfield MJ, Collins FS, Davey Smith G, Day IN, Franks PW, Hattersley AT, Hu FB, Jarvelin MR, Kong A, Kooner JS, Laakso M, Lakatta E, Mooser V, Morris AD, Peltonen L, Samani NJ, Spector TD, Strachan DP, Tanaka T, Tuomilehto J, Uitterlinden AG, van Duijn CM, Wareham NJ, Hugh W, Waterworth DM, Boehnke M, Deloukas P, Groop L, Hunter DJ, Thorsteinsdottir U, Schlessinger D, Wichmann HE, Frayling TM, Abecasis GR, Hirschhorn JN, Loos RJ, Stefansson K, Mohlke KL, Barroso I, McCarthy MI (2009) Genome-wide association scan meta-analysis identifies three Loci influencing adiposity and fat distribution. PLoS genetics 5:e1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pillas D, Hoggart CJ, Evans DM, O’Reilly PF, Sipila K, Lahdesmaki R, Millwood IY, Kaakinen M, Netuveli G, Blane D, Charoen P, Sovio U, Pouta A, Freimer N, Hartikainen AL, Laitinen J, Vaara S, Glaser B, Crawford P, Timpson NJ, Ring SM, Deng G, Zhang W, McCarthy MI, Deloukas P, Peltonen L, Elliott P, Coin LJ, Smith GD, Jarvelin MR (2010) Genome-wide association study reveals multiple loci associated with primary tooth development during infancy. PLoS genetics 6:e1000856. [DOI] [PMC free article] [PubMed] [Google Scholar]


