Abstract
Background
Genetic alterations, including mutation of epidermal growth factor receptor or v-Ki-ras2 kirsten rat sarcoma viral oncogene homolog and fusion of anaplastic lymphoma kinase (ALK), RET proto-oncogene (RET), or v-ros UR2 sarcoma virus oncogene homolog 1 (ROS1), occur in non-small cell lung cancers, and these oncogenic drivers are important biomarkers for targeted therapies. A useful technique to screen for these fusions is the detection of native carboxy-terminal (C-terminal) protein by immunohistochemistry; however, the effects of other genetic alterations on C-terminal expression is not fully understood. In this study, we evaluated whether C-terminal expression is specifically elevated by fusion with or without typical genetic alterations of lung cancer.
Methods
In 37 human lung cancer cell lines and four tissue specimens, protein and mRNA levels were measured by capillary western blotting and reverse transcription–PCR, respectively.
Results
Compared with the median of all 37 cell lines, mRNA levels at the C-terminus of all five of the fusion-positive cell lines tested (three ALK, one RET, and one ROS1) were elevated at least 2000-, 300-, or 2000-fold, respectively, and high C-terminal protein expression was detected. In an ALK fusion–positive tissue specimen, the mRNA and protein levels of C-terminal ALK were also markedly elevated. Meanwhile, in one of 36 RET fusion–negative cell lines, RET mRNA levels at the C-terminus were elevated at least 500-fold compared with the median of all 37 cell lines, and high C-terminal protein expression was detected despite the absence of RET fusion.
Conclusions
This study of 37 cell lines and four tissue specimens shows the detection of C-terminal ALK or ROS1 proteins could be a comprehensive method to determine ALK or ROS1 fusion, whereas not only the detection of C-terminal RET protein but also other methods would be needed to determine RET fusion.
Electronic supplementary material
The online version of this article (10.1186/s12885-019-5527-2) contains supplementary material, which is available to authorized users.
Keywords: EML4, ALK, RET, ROS1, Lung cancer, Alectinib, Immunohistochemistry, Rearrangement, Fusion
Background
Molecular subsets of non-small cell lung cancer (NSCLC) have been defined by various types of driver gene mutations involving epidermal growth factor receptor (EGFR), v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS), and anaplastic lymphoma kinase (ALK) gene fusion. Specific tyrosine kinase inhibitors (TKIs), such as EGFR-TKIs (erlotinib and afatinib) and ALK-TKIs (crizotinib and alectinib), that inhibit the oncogenic activity of these genes have been developed and approved [1, 2]. A key issue in identifying patients that would be suitable for the targeted agents is precisely identifying the presence or absence of the driver gene mutations in a molecular diagnosis of the lung cancer. In approximately 5% of NSCLC, the rearrangement of the amino-terminal (N-terminal) region of echinoderm microtubule associated protein like 4 (EML4) with the carboxy-terminal (C-terminal) region of ALK occurs by inversion within the short arm of chromosome 2 [3]. In cancer cells with EML4-ALK, the transcription of the C-terminal region of ALK depends on the promoter activity of the fusion partner, EML4, which is a housekeeping gene for the stabilization of microtubules during mitosis, by which the C-terminal ALK protein level also becomes elevated [4, 5].
To detect ALK fusion, there are three different techniques: fluorescence in situ hybridization (FISH), immunohistochemistry (IHC) and reverse transcription–PCR (RT-PCR) are available for the detection of ALK fusion [3]. Although each test has some advantages and disadvantages, IHC is more useful as a routine screening method in clinical settings because of cost effectiveness and technical ease [3]. The ALK IHC method determines whether tumor cells are harboring an ALK fusion using an antibody directed to the C-terminal ALK protein, but unlike FISH tests, it has been reported to show not only positive results in patients with ALK fusion–positive cancer but also false-negative errors in some patients who actually have ALK fusion–positive cancer [3, 6–8]. In addition to ALK fusion, RET proto-oncogene (RET) or v-ros UR2 sarcoma virus oncogene homolog 1 (ROS1) are rearranged in approximately 1% of NSCLC. In consequence, RET-TKIs (such as alectinib or vandetanib) and ROS1-TKIs (such as crizotinib or lorlatinib) are under development for fusion-positive NSCLC patients, and precise diagnostic methods for these fusions are needed [2, 9–11].
In this study, we verified the reliability of IHC methods that target ALK, RET, and ROS1 C-terminal protein as diagnostic tools for lung cancer by investigating whether the expression at the C-terminal region is elevated in each of the fusion-positive lung cancer cells compared with that in fusion-negative cells.
Methods
Lung cancer cell lines
The histology, driver gene mutation, culture medium, and supplier of the 37 human lung cancer cell lines are listed in Table 1 [12–14]. Cells were purchased from the American Type Culture Collection (ATCC; Manassas, VA), Korean Cell Line Bank (KCLB; Seoul, South Korea), Riken Bioresource Center (Ibaraki, Japan), Health Science Research Resources Bank (HSRRB; Osaka, Japan), National Cancer Institute (NCI; Bethesda, MD), Immuno-Biological Laboratories (IBL; Gunma, Japan), and Leibniz Institute DSMZ-German Collection (DSMZ; Braunschweig, Germany) and were maintained using RPMI1640 (Sigma-Aldrich (SIGMA); St. Louis, MO), minimum essential medium (MEM; SIGMA), McCoy’s 5A (Thermo Fisher Scientific; Waltham, MA), or Ham’s nutrient mixture F12 (HAMF12; SIGMA) supplemented with fetal bovine serum (FBS; Japan Bio Serum, Hiroshima, Japan) at 37 °C in 5% CO2. Authentication of all cell lines was conducted by DNA fingerprinting with short tandem repeat profiling using the Powerplex 16 HS system (Promega; Madison, WI).
Table 1.
37 lung cancer cell lines
| N | Cell line | Histology | Driver gene mutation | Supplier | Catalogue number | Culture medium |
|---|---|---|---|---|---|---|
| 1 | NCI-H2228 | AD | EML4-ALK1 | ATCC | CRL-5935 | 10%FBS-RPMI1640 |
| 2 | SNU-2535 | NS | EML4-ALK, G1269A2 | KCLB | 02535 | 10%FBS-RPMI1640 |
| 3 | SNU-2292 | AD | EML4-ALK3 | KCLB | 02292 | 10%FBS-RPMI1640 |
| 4 | HCC827 | AD | EGFR E746_A750 DL1 | ATCC | CRL-2868 | 10%FBS-RPMI1640 |
| 5 | PC-9 | AD | EGFR E746_A750 DL1 | IBL | 37,012 | 10%FBS-RPMI1640 |
| 6 | B901L | AD | EGFR E746_A750 DL1 | RIKEN | RCB3530 | 10%FBS-RPMI1640 |
| 7 | HCC4006 | AD | EGFR L747_E749 DL, A750P4 | ATCC | CRL-2871 | 10%FBS-RPMI1640 |
| 8 | HCC2935 | AD | EGFR E746_T751 DL, S752I4 | ATCC | CRL-2869 | 10%FBS-RPMI1640 |
| 9 | PC-3 | AD | EGFR L747_A750 DL1 | HSRRB | JCRB0077 | 10%FBS-MEM |
| 10 | NCI-H1650 | AD | EGFR E746_A750 DL1 | ATCC | CRL-5883 | 10%FBS-RPMI1640 |
| 11 | II-18 | AD | EGFR L858R1 | RIKEN | RCB2093 | 10%FBS-RPMI1640 |
| 12 | NCI-H1975 | AD | EGFR L858R, T790M1 | ATCC | CRL-5908 | 10%FBS-RPMI1640 |
| 13 | NCI-H820 | AD | EGFR E746_T751 DL, T790M5 | ATCC | HTB-181 | 10%FBS-RPMI1640 |
| 14 | Calu-1 | SQ | KRAS G12C1 | ATCC | HTB-54 | 10%FBS-McCoy’s 5A |
| 15 | NCI-H358 | AD | KRAS G12C1 | ATCC | CRL-5807 | 10%FBS-RPMI1640 |
| 16 | HOP-62 | AD | KRAS G12C1 | NCI | 502,467 | 10%FBS-RPMI1640 |
| 17 | NCI-H2122 | AD | KRAS G12C1 | ATCC | CRL-5985 | 10%FBS-RPMI1640 |
| 18 | Calu-6 | AC | KRAS Q61K1 | ATCC | HTB-56 | 10%FBS-MEM |
| 19 | NCI-H460 | LC | KRAS Q61H, PIK3CA E545K1 | ATCC | HTB-177 | 10%FBS-RPMI1640 |
| 20 | NCI-H596 | AS | PIK3CA E545K1 | ATCC | HTB-178 | 10%FBS-RPMI1640 |
| 21 | NCI-H1781 | AD | ERBB2 G776VC1 | ATCC | CRL-5894 | 10%FBS-RPMI1640 |
| 22 | NCI-H1755 | AD | BRAF G469A1 | ATCC | CRL-5892 | 10%FBS-RPMI1640 |
| 23 | LC-2/ad | AD | CCDC6-RET6 | RIKEN | RCB0440 | 15%FBS-HAMF12 |
| 24 | HCC78 | AD | SLC34A2-ROS11 | DSMZ | ACC563 | 10%FBS-RPMI1640 |
| 25 | NCI-H2347 | AD | NRAS Q61R1 | ATCC | CRL-5942 | 10%FBS-RPMI1640 |
| 26 | NCI-H1993 | AD | MET amplification1 | ATCC | CRL-5909 | 10%FBS-RPMI1640 |
| 27 | NCI-H1568 | AD | ND1 | ATCC | CRL-5876 | 10%FBS-RPMI1640 |
| 28 | NCI-H522 | AD | ND1 | ATCC | CRL-5810 | 10%FBS-RPMI1640 |
| 29 | NCI-H838 | AD | ND1 | ATCC | CRL-5844 | 10%FBS-RPMI1640 |
| 30 | A529L | AS | ND1 | RIKEN | RCB2817 | 10%FBS-RPMI1640 |
| 31 | NCI-H1703 | AD | ND1 | ATCC | CRL-5889 | 10%FBS-RPMI1640 |
| 32 | NCI-H520 | SQ | ND1 | ATCC | HTB-182 | 10%FBS-RPMI1640 |
| 33 | NCI-H2170 | SQ | ND1 | ATCC | CRL-5928 | 10%FBS-RPMI1640 |
| 34 | NCI-H226 | SQ | ND1 | ATCC | CRL-5826 | 10%FBS-RPMI1640 |
| 35 | SK-MES-1 | SQ | ND1 | ATCC | HTB-58 | 10%FBS-MEM |
| 36 | NCI-H1915 | LC | ND1 | ATCC | CRL-5904 | 10%FBS-RPMI1640 |
| 37 | NCI-H292 | MC | ND1 | ATCC | CRL-1848 | 10%FBS-RPMI1640 |
AD Adenocarcinoma, NS Non-small cell carcinoma, SQ Squamous carcinoma, AC Anaplastic carcinoma, LC Large cell carcinoma, AS Adenosquamous carcinoma, MC Mucoepidermoid carcinoma, DL Deletion, ND Not detected. Mutations were referred from 1 COSMIC cell database, 2 Yoshimura Y. et al., 3 Additional file 5: Figure S1a,4 ATCC’s datasheet, 5 Shimamura T. et al., 6 Matsubara D. et al.
Lung cancer tissue specimens
Four frozen lung cancer tissue specimens with paired non-tumor normal adjacent tissue (NAT) were commercially obtained from BioreclamationIVT (Hicksville, NY), and kept at − 80 °C until analysis. The specimens and their clinical and pathological features are listed in Table 2. All studies were ethically reviewed and approved by the ethical review committee at Chugai Pharmaceutical Co., Ltd. The committee is independent from the commercial aspect of the company by involving third-party members.
Table 2.
Four tissue specimens with lung cancer
| N | Specimen ID | Driver gene mutation | Histology | Distance of NAT from tumor |
|---|---|---|---|---|
| 1 | ILS31007 | EML4-ALK | AD | 3 cm |
| 2 | ILS33976 | EGFR exon 19 deletion | AD | 3 cm |
| 3 | ILS31624 | KRAS G12C | AD | 6 cm |
| 4 | ILS31026 | ND | SQ | 5 cm |
Each driver gene mutation was determined by Cancer Personalized Profiling by Deep Sequencing or the cobas 4800 mutation test at BioreclamationIVT. ND Not detected, AD Adenocarcinoma, SQ Squamous carcinoma, NAT Normal adjacent tissue
RT-PCR assay
RNA was obtained from the cells using a SV total RNA isolation system (Promega), and the cDNA was synthesized using a PrimeScript RT reagent kit (Takara Bio; Shiga, Japan). RT-PCR was performed using the LightCycler 480 system (Roche Diagnostics; Basel, Switzerland) and Taqman probes (Thermo Fisher Scientific) (Additional file 1: Tables S1 and Additional file 2: Table S2). The value of target mRNA expression normalized by GAPDH mRNA was calculated from the crossing point PCR-cycle of each mRNA using the LightCycler 480 software.
Western blotting assay
Western blotting was performed by the capillary electrophoresis–based protein analysis system, Sally Sue (ProteinSimple; Santa Clara, CA), as described previously [15]. The same amount of protein lysate was loaded in each analysis. The antibodies were shown in Additional file 3: Table S3, and we determined which terminus of protein is recognized by the antibody with reference to the supplier’s datasheet and NCBI reference sequence database (RefSeq) (Additional file 3: Table S3 and Additional file 4: Table S4).
Results
Expression of EML4 or ALK in cancer cell lines with or without ALK fusion
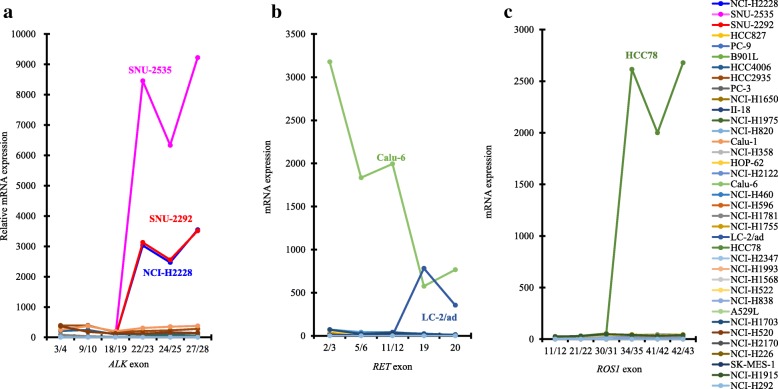
To examine ALK expression at the C-terminus in lung cancer cells with or without ALK fusion, we used 37 lung cancer cell lines (Table 1) that harbor already-known driver mutations, including ALK fusion and wild-type, to mimic the populations of patients with lung cancer as shown in Korpanty G.J. et al. [2]. Both the NCI-H2228 and SNU-2292 cell lines had EML4-ALK variant 3a, and the SNU-2535 cell line had EML4-ALK variant 1 (Additional file 5: Figure S1a and Additional file 6: Figure S2). All ALK-TKIs (alectinib, crizotinib, and ceritinib) inhibited the cell growth of all ALK fusion–positive cell lines (IC50: < 500 nM) (Additional file 7: Table S5). However, the EGFR-TKI, erlotinib, did not inhibit the cell growth of ALK fusion–positive cell lines (IC50: > 1000 nM) (Additional file 7: Table S5), indicating that growth of these ALK fusion–positive cell lines strongly depends on the signal from ALK kinase. Moreover, in all ALK fusion–positive cell lines, mRNA levels of ALK at the C-terminal region, which is backward from the breakpoint at exon 20 in ALK rearrangement [4], were elevated by at least 2000-fold more than the median of all 37 cell lines, and high protein expression at the C-terminus was detected (Figs. 1a and 2). None of the ALK fusion–negative cell lines expressed any mRNA or C-terminal ALK protein. Meanwhile, the protein expression of N-terminal EML4 was detected in each of the 37 cell lines, independent of ALK fusion status (Fig. 2 and Additional file 8: Figure S4a).
Fig. 1.
RT-PCR analysis of ALK, RET, or ROS1 mRNA in 37 cancer cell lines. The mRNA expression of ALK (a), RET (b), or ROS1 (c) was determined by RT-PCR using the aqman probes (Additional file 2: Tables S2). Exons of each probe are shown on the X-axis. The relative mRNA expression was calculated as the ratio of the normalized value with GAPDH mRNA to the median of that in 37 cell lines
Fig. 2.
Western blotting analysis in 37 lung cancer cell lines. The protein expression of ALK, RET, ROS1, KIF5B, or CCDC6 and phosphorylation of ALK, RET, or ROS1 in 37 cell lines was determined by western blotting using the antibodies (Additional file 3: Tables S3). Both actin and COX4 were used as loading controls
Expression of EML4 or ALK in tissues with or without ALK fusion
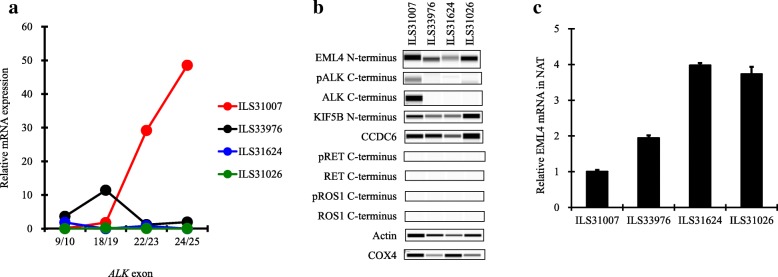
Next, we tested four lung cancer tissue specimens harboring ALK fusion, EGFR mutation, KRAS mutation, or none (Table 2). ILS31007 had EML4-ALK variant 3a, and markedly expressed mRNA, protein, and phosphorylation of C-terminal ALK compared with the other three ALK fusion–negative tissue specimens (Fig. 3a, b and Additional file 9: Figures S5a, b, c and Additional file 10: Figure S6). Expression of C-terminal ALK protein was not detected in the other three ALK fusion–negative tissues (Fig. 3b). Meanwhile, the N-terminal EML4 protein was expressed in all tumor tissues independent of ALK fusion status (Fig. 3b and Additional file 8: Figure S4b).
Fig. 3.
RT-PCR and western blotting analysis in four tissue specimens. The mRNA expression of ALK in the four tumor tissue specimens shown in Table 2 was determined by RT-PCR using the Taqman probes (Additional file 2: Tables S2). Exons of each probe are shown on the X-axis. The relative mRNA expression was calculated as the ratio of the normalized value with GAPDH mRNA to the median of that in the four tissue specimens (a). The protein expression of ALK, RET, ROS1, KIF5B, or CCDC6 and phosphorylation of ALK, RET, or ROS1 in the tumor tissue specimens were determined by western blotting using the antibodies (Additional file 3: Tables S3) (b). The relative EML4 mRNA expression at exon 1/2 in NAT specimens was calculated as the ratio of the normalized values with GAPDH mRNA to that in tumor tissue specimens (c). Each bar represents the mean + SD (n = 3)
Then, we used the non-tumor NAT specimens to investigate whether transcription of EML4 is constitutively activated even in normal lung cells. Firstly, in ALK fusion–positive ILS31007, we confirmed that the NAT specimens did not include any tumor cells (Additional file 11: Figure S7) and that the EML4 mRNA levels at exon1/2, 2/3, 5/6, or 6/7 in NAT were almost the same as those in the tumor tissues (Fig. 3c and Additional file 12: Figure S8). In the other ALK fusion–negative NAT specimens, more EML4 mRNA was expressed at each exon than in the tumor tissues. These data suggest that EML4 is constitutively transcribed by its own strong promoter activity in not only ALK fusion–positive and –negative lung cancer cells but also in non-tumor lung cells.
Expression of RET in cancer cell lines with or without RET fusion
RET is fused with genes such as KIF5B or CCDC6 in approximately 1% of lung cancers [2]. We therefore examined RET expression at the C-terminus in 37 lung cancer cell lines with or without RET fusion (Table 1). The LC-2/ad cell line contained CCDC6-RET fusion (Additional file 5: Figure S1b and Additional file 6: Figure S2). One of the ALK-TKIs, alectinib, which has the potential to inhibit RET kinase activity [16], suppressed the cell growth of the LC-2/ad cell (Additional file 7: Table S5). In the LC-2/ad cell, the mRNA level of the C-terminal RET region, which is backward from the breakpoint at exon 12 in RET rearrangement [14], was elevated by at least 300-fold more than the median of all 37 cell lines, and protein expression at the C-terminus was also high (Figs. 1b and 2). Interestingly, despite being a RET fusion–negative cell line, the Calu-6 cell markedly expressed RET mRNA from the N- to the C-terminal region and C-terminal RET protein, and the cell was insensitive to alectinib (Figs. 1b and 2, Additional file 7: Table S5 and Additional file 13: Figure S9b). None of the 35 cell lines other than LC-2/ad and Calu-6 expressed any mRNA or C-terminal RET protein. Regardless of RET status, the protein expression of CCDC6 and KIF5B was detected in all 37 cell lines (Figs. 1b, 2 and Additional file 8: Figure S4c).
Expression of ROS1 in cancer cell lines with or without ROS1 fusion
ROS1 is rearranged with genes such as SCL34A2 or CD73 in approximately 1% of lung cancers [2]. We therefore examined ROS1 expression at the C-terminus in 37 lung cancer cell lines with or without ROS1 fusion (Table 1). The HCC78 cell line contained SLC34A2-ROS1 fusion (Additional file 5: Figure S1c and Additional file 6: Figure S2). The ROS1-TKIs, crizotinib and ceritinib, suppressed cell growth (Additional file 7: Table S5). In HCC78 cells, the mRNA level of the C-terminal ROS1 region, which is backward from the breakpoint at exon 32 or 34 in ROS1 rearrangement [17], was elevated by at least 2000-fold more than the median of all 37 cell lines, and high protein expression at the C-terminus was detected (Figs. 1c and 2).
Discussion
Chromosomal rearrangements involving ALK, RET, and ROS1 are attractive anticancer targets that provide opportunities for therapies for patients with NSCLC [9]. As described in previous preclinical studies [18, 19], only the three ALK fusion–positive cell lines in the 12 cell lines tested were sensitive to all ALK-TKIs (alectinib, crizotinib and ceritinib) through the suppression of phosphorylation of ALK signaling pathways involving STAT3/AKT/ERK (Additional file 7: Table S5, Additional file 14: Figure S3, Additional file 13: Figure S9, Additional file 15: Figure S10, and Additional file 16: Figure S11), indicating that the growth of these ALK fusion–positive cell lines strongly depends on the signal from ALK. In the US, both a FISH test using the Vysis ALK break apart FISH probe kit (Abbott Laboratories) and an IHC test using the Ventana ALK (D5F3) CDx assay (Ventana Medical Systems; Tucson, AZ) have been approved as a companion diagnostic (CDx) test for crizotinib [20]. In Japan, the Vysis FISH test has been approved as a CDx test for crizotinib, and a diagnostic tool combining the Vysis FISH test with an IHC test using the N-Histofine ALK Detection kit (Nichirei Biosciences) has been approved as a CDx test for alectinib [6]. These two IHC kits, which respectively include clone D5F3 or 5A4 as a primary antibody directed to C-terminal ALK protein, were highly concordant with the ALK FISH tests. However, it was reported that IHC-positive and FISH-negative patients were occasionally present, with these discordant patients showing a clinical response to crizotinib [6, 21, 22]. To identify patients who would be suitable for ALK-TKIs, the accurate diagnosis of ALK fusion is a critical issue. In this study, we focused on the reliability of the detection of C-terminus ALK protein for the diagnosis of ALK fusion using various types of lung cancer cell lines and tissues. We found that the promoter of EML4 was constitutively activated in lung cancer as well as normal cells independent of ALK fusion, and C-terminal ALK protein level and phosphorylation were specifically elevated in ALK fusion–positive cancer cells (Figs. 2 and 3b). As previously demonstrated [4, 7], these findings suggest that wild-type ALK is silenced in normal lung cells because of lack of production, but when C-terminal ALK is fused to N-terminal EML4 in normal cells, the transcription of the kinase domain of ALK is activated by the constant promoter activity of EML4, and the resultant abundantly produced EML4-fused ALK leads to cancer through aberrant ALK signal transduction. Therefore, IHC tests for ALK, such as those using the Ventana and N-Histofine kits, could be sufficiently reliable diagnostic methods in the treatment of patients with lung cancer using ALK-TKIs.
As previously described in preclinical studies [16, 23, 24], 2 cell lines tested only LC-2/ad or HCC78 cell line with RET or ROS1 fusion were sensitive to RET-TKI (alectinib) or ROS1-TKIs (crizotinib and ceritinib) by suppressing the level of phosphorylation of STAT3/AKT/ERK, which are located downstream of RET or ROS1 kinase, respectively (Additional file 7: Table S5, Additional file 13: Figure S9a, Additional file 15: Figure S10 and Additional file 16: Figure S11). No IHC or FISH CDx tests that detect RET or ROS1 fusions have been approved, but an IHC test using an antibody clone, EPR2871 (Abcam), is under investigation [10]. Although RET expression was low in normal lung tissue [25], discordant results between the IHC test and the FISH test for RET have been reported [26, 27]. In this study, the promoter activity of KIF5B or CCDC6, which are two major genes involved in fusion with C-terminal RET, was constitutively activated in every cell line, and the expression of C-terminal RET was considerably high not only in the RET fusion–positive cell line LC-2/ad, but also in one of the five RET fusion–negative and KRAS-mutated cell lines Calu-6. Wild-type RET expression in Calu-6 cells has been also reported by Zhou et al. [28]. The weight of RET protein in LC-2/ad or Calu-6 cells was, respectively, 50 to 100 kDa or 100 to 200 kDa (Figs. 1b and 2), which is approximately the same weight as that reported previously for, respectively, CCDC6-fused RET or wild-type RET [29, 30]. Just as surprisingly, in 37 cell lines, phosphorylation of the C-terminal RET domain was only detected in the RET fusion–negative and MET-amplified NCI-H1993 cell line (Fig. 2), but that elevation of the level of MET phosphorylation by trans-phosphorylation of MET within RET and MET heterodimers was reported in NCI-H1993 cells [31]. Both Calu-6 and NCI-H1993 cells were completely insensitive to alectinib (Additional file 7: Table S5 and Additional file 13: Figure S9b and c), which means that cell growth with wild-type RET is independent of RET kinase even if the cells have high expression or high phosphorylation of RET, and RET kinase would be an oncogenic growth driver after fusion with, for example, CCDC6 or KIF5B. Taking all these findings together, the stand-alone RET IHC test for the detection of C-terminal RET protein may cause misleading judgments of RET fusion, and CDx tests with both IHC and FISH or RT-PCR would be needed in the treatment of patients with RET fusion–positive lung cancer using RET-TKIs.
Regarding the ROS1 IHC test using an antibody for the C-terminus, some patients were reported to show discordant results between IHC and FISH tests despite low expression of ROS1 protein in normal lung tissue [11, 32]. However, in this study, the ROS1 fusion–positive cell line, HCC78, harboring an SLC34A2-ROS1 fusion only showed protein expression at the C-terminal domain and a sensitivity to the ROS1-TKIs by inhibiting ROS1 signaling pathways involving STAT3/AKT/ERK (Figs. 1c and 2 and Additional file 7: Table S5 and Additional file 15: Figure S10). SLC34A2 and CD74 are two genes fused to C-terminal ROS1, and SLC34A2 mRNA expression was shown in tissues and cell lines of NSCLC as well as normal lung tissues [33–35]. CD74 protein was also strongly expressed in many lung cancer tissues [36]. Therefore, the C-terminal ROS1 protein level could only be elevated by the strong promoter activity of genes such as SLC34A2 or CD74 in ROS1 fusion–positive lung cancer cells, which suggests that the ROS1 IHC test is a reliable diagnostic test for the detection of patients with lung cancer who have ROS1 fusion.
On the other hand, ALK or ROS1 IHC tests occasionally showed positive results even in patient samples diagnosed by FISH or RT-PCR tests to be fusion-negative [6, 8, 11]. As one of the causes of this IHC+/FISH- discordance, it was reported that wild-type FISH signals in fusion–positive cases were caused by rare atypical chromosomal rearrangements with EML4 and ALK [6]. In addition, Takeuchi K et al. showed that ALK expression is detected in some ALK fusion–negative cases with small-cell carcinoma, large-cell neuroendocrine carcinoma, and poorly differentiated carcinoma [7]. Hyper-methylation of promoter and copy number gain of ROS1 were reported as one of the mechanisms that activate ROS1 expression in fusion-negative carcinomas [37]. However, the possible factors of discordances mentioned above have not been fully clarified at present. This study with 37 lung cancer cell lines and four tissues did not reproduce the phenomenon of discordance. Therefore, further studies using a larger panel with various types of lung cancer cell lines and tissues would be useful to elucidate the causes of discordance in clinical ALK or ROS1 IHC tests.
Next-generation sequencing (NGS) technology enables high-throughput and multiplex analysis of various driver oncogenes. For NSCLC, NGS-based tumor-profiling multiplex gene panels, such as Oncomine Dx target test or FoundationOne CDx, have recently been approved as companion diagnostics to detect mutations of EGFR and BRAF, or fusions of ALK and ROS1 in the US [38]. These NGS panels are also designed to detect RET fusions [39, 40]. At present, clinical diagnosis to select patients with ALK fusion–positive NSCLC is predominantly performed by IHC test, while NGS screening might have the potential to test for multiple gene alterations in a quick single analysis. In this study, we could not compare the analysis of fusions by IHC with that by NGS since we have no data on NGS. However, evaluations of the usability of diagnosis by NGS compared to IHC or FISH in NSCLC specimens showed that NGS screenings could provide an alternative method of detecting fusion genes to IHC or FISH tests [39–41]. Therefore, further studies of NGS in addition to C-terminal protein expression analysis using NSCLC cell lines would be a strong support to precise selection of NSCLC patients with fusion genes by NGS with or without IHC.
Conclusion
We demonstrated that the transcription levels of ALK- or RET-fusion partner genes, such as EML4, CCDC6 and KIF5B, were constitutively activated in lung cancer cells, and the expression at the C-terminal region of ALK, RET, or ROS1 was also markedly elevated in each fusion-positive lung cancer cell. Moreover, although the expression of ALK and ROS1 at the C-terminus was very limited in all fusion-negative cancer cells, the expression or phosphorylation of C-terminal RET was markedly elevated in 2 of the 36 RET fusion–negative cancer cells. Our findings suggest that the measurement of C-terminal ALK or ROS1 protein could be a reliable diagnostic method for each fusion, whereas measuring C-terminal RET protein might be a diagnostic method with a potential to give false-positive results when detecting RET fusion in patients with lung cancer.
Additional files
Table S1. Taqman probes for fusion (DOCX 26 kb)
Table S2. Taqman probes for exons of mRNA (DOCX 28 kb)
Table S3. Target terminus of antibody (DOCX 28 kb)
Table S4. Exon count of mRNA (DOCX 26 kb)
Figure S1. RT-PCR analysis of ALK, RET, or ROS1 fusion in 37 cancer cell lines. The mRNA expression of variant 1, 2, 3a, or 3b of EML4-ALK (a), KIF5B-RET, CCDC6-RET (b) or SLC34A2-ROS1 (c) was determined by RT-PCR using the Taqman probes shown in Additional file 1: Table S1. Red lines show fusion gene–positive cell lines and green lines show fusion gene–negative cell lines (PPTX 129 kb)
Figure S2. Summary of ALK, RET, or ROS1 fusion in 37 cancer cell lines. Plot of normalized values calculated from the data in Additional file 5: Figure S1 (PPTX 50 kb)
Table S5. IC50s of ALK-TKIs and erlotinib (DOCX 28 kb)
Figure S4. Comparison of molecular weights between wild-type and fusion protein. After high exposure of N-terminal EML4 in the results shown in Fig. 2, two sizes of EML4 protein were detected in NCI-H2228 and SNU-2292 cell lines (left-hand figure), and we assumed that the larger protein was wild-type EML4 (arrow) and the smaller protein was ALK-fused EML4 (arrowhead), after referring to the protein weight of EML4-ALK detected by an ALK C-terminus antibody (right-hand figure) (a). In the same way, we assumed that the larger and smaller proteins were respectively wild-type EML4 (arrow) and ALK-fused EML4 (arrowhead) in the results for the ILS31007 tumor tissue specimen shown in Fig. 3b (b), and the larger and smaller proteins were respectively wild-type CCDC6 (arrow) and RET-fused CCDC6 (arrowhead) in the results for CCDC6 in the LC-2/ad cell line shown in Fig. 2 (c) (PPTX 600 kb)
Figure S5. RT-PCR analysis of ALK, RET, or ROS1 fusion in 4 tumor tissues. The mRNA expression of variant 1, 2, 3a, or 3b of EML4-ALK (a), KIF5B-RET, CCDC6-RET (b) or SLC34A2-ROS1 (c) was determined by RT-PCR using the Taqman probes shown in Additional file 1: Table S1. Red lines show fusion gene–positive tumor tissue, and green lines show fusion gene–negative tumor tissue (PPTX 122 kb)
Figure S6. Summary of ALK, RET, or ROS1 fusion in 4 tumor tissue specimens. Plot of normalized values calculated from the data in Additional file 9: Figure S5 (PPTX 45 kb)
Figure S7. Analysis by IHC and FISH of ALK in ILS31007 tissue specimen. The FFPE specimens with tumor tissue (upper panel) and the paired NAT specimens (lower panel) were obtained from BioreclamationIVT. IHC (left panel) and FISH (right panel) analyses for ALK rearrangement were performed with, respectively, the N-Histofine ALK detection kit (Nichirei Biosciences; Tokyo, Japan) and the Vysis ALK break apart FISH probe kit (Abbott laboratories; Abbott Park, IL) at a commercial clinical laboratory, LSI Medience (Tokyo, Japan). Images were captured using standard settings by the BZ900 (Keyence; Osaka, Japan) (PPTX 754 kb)
Figure S8. RT-PCR analysis of EML4 in four tissue specimens. The relative EML4 mRNA expression at each exon in NAT specimens was calculated as the ratio of the normalized values with GAPDH mRNA to those in tumor tissues. Each bar represents the mean + SD (n = 3) (PPTX 45 kb)
Figure S9. Western blotting analysis in three cancer cell lines with or without RET fusion. Cell lysates were harvested after 2 h of treatment with each drug at the concentrations shown (nM). The levels of RET phosphorylation in LC-2/ad were undetectable by this western blotting system (PPTX 2307 kb)
Figure S3. Western blotting analysis in 3 cancer cell lines with ALK fusion. Cell lysates were harvested after 2 h of treatment with each drug at the concentration shown (nM). The antibodies were obtained from Cell signaling technology. (PPTX 2483 kb)
Figure S10. Western blotting analysis in a cancer cell line with ROS1 fusion. Cell lysates were harvested after 2 h of treatment with each drug at the concentrations shown (nM) (PPTX 797 kb)
Figure S11. Western blotting analysis in five cancer cell lines without any fusion gene. Cell lysates were harvested after 2 h of treatment with each drug at the concentrations shown (nM) (PPTX 2123 kb)
Acknowledgements
The authors thank KY and MK for their technical support in the experiments.
Funding
The study was funded by Chugai Pharmaceutical Co., Ltd., and no other specific funding was received for this study.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- ALK
Anaplastic lymphoma kinase
- BRAF
v-Raf murine sarcoma viral oncogene homolog B1
- CCDC6
Coiled-coil domain containing 6
- CDx
Companion diagnostics
- COSMIC
Catalogue of somatic mutations in cancer
- C-terminal
Carboxy-terminal
- EGFR
Epidermal growth factor receptor
- EML4
Echinoderm microtubule associated protein like 4
- FISH
Fluorescence in situ hybridization
- HER-2
Human epidermal growth factor receptor 2
- IHC
Immunohistochemistry
- KIF5B
Kinesin family member 5
- KRAS
v-Ki-ras2 kirsten rat sarcoma viral oncogene homolog
- MET
MET proto-oncogene
- NAT
Non-tumor normal adjacent tissue
- NRAS
Neuroblastoma RAS viral (v-ras) oncogene homolog
- NSCLC
Non-small cell lung cancer
- N-terminal
Amino-terminal
- PIK3CA
Phosphoinositide-3-kinase, catalytic, α polypeptide
- RET
RET proto-oncogene
- ROS1
v-Ros UR2 sarcoma virus oncogene homolog 1
- RT-PCR
Reverse transcription polymerase chain reaction
- SLC34A2
Solute carrier family 34 member 2
- TKI
Tyrosine kinase inhibitor
Authors’ contributions
KF performed all experiments, analyzed, interpreted the data, and was a major contributor in writing the manuscript. MM, MK and SS supported the experiments and analyses. NH and YY supported the interpretation and writing manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable because all materials were commercially available products. All studies were ethically reviewed and approved by the ethical review committee at Chugai Pharmaceutical Co., Ltd..
Consent for publication
Not applicable.
Competing interests
The authors are employees of Chugai Pharmaceutical Co., Ltd., and declare no conflict of interest.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Koh Furugaki, Email: furugakiku@chugai-pharm.co.jp.
Marie Mochizuki, Email: mochie115@hotmail.co.jp.
Mirei Kohno, Email: kouno.mirei98@chugai-pharm.co.jp.
Sei Shu, Email: shu.sei63@chugai-pharm.co.jp.
Naoki Harada, Email: haradanok@chugai-pharm.co.jp.
Yasushi Yoshimura, Phone: +81-467-47-6355, Email: yoshimurayss@chugai-pharm.co.jp.
References
- 1.Chan BA, Hughes BGM. Targeted therapy for non-small cell lung cancer: current standards and the promise of the future. Transl Lung Cancer Res. 2014;4(1):36–54. doi: 10.3978/j.issn.2218-6751.2014.05.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Korpanty GJ, Graham DM, Vincent MD, Leighl NB. Biomarkers That Currently Affect Clinical Practice in Lung Cancer: EGFR, ALK, MET, ROS-1, and KRAS. Front Oncol. 2014;4:204. doi: 10.3389/fonc.2014.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morán T, Quiroga V, Gil Mde L, Vilà L, Pardo N, Carcereny E, Capdevila L, Muñoz-Mármol AM, Rosell R. Targeting EML4-ALK driven non-small cell lung cancer (NSCLC) Transl Lung Cancer Res. 2013;2(2):128–141. doi: 10.3978/j.issn.2218-6751.2013.03.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448(7153):561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 5.Houtman SH, Rutteman M, De Zeeuw CI, French PJ. Echinoderm microtubule-associated protein like protein 4, a member of the echinoderm microtubule-associated protein family, stabilizes microtubules. Neuroscience. 2007;144(4):1373–1382. doi: 10.1016/j.neuroscience.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 6.Takeuchi K, Togashi Y, Kamihara Y, Fukuyama T, Yoshioka H, Inoue A, Katsuki H, Kiura K, Nakagawa K, Seto T, et al. Prospective and clinical validation of ALK immunohistochemistry: results from the phase I/II study of alectinib for ALK-positive lung cancer (AF-001JP study) Ann Oncol. 2016;27(1):185–192. doi: 10.1093/annonc/mdv501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takeuchi K. Interpretation of anti-ALK immunohistochemistry results. J Thorac Oncol. 2013;8(7):e67–e68. doi: 10.1097/JTO.0b013e318293e1ff. [DOI] [PubMed] [Google Scholar]
- 8.Cabillic F, Gros A, Dugay F, Begueret H, Mesturoux L, Chiforeanu DC, Dufrenot L, Jauffret V, Dachary D, Corre R, et al. Parallel FISH and Immunohistochemical studies of ALK status in 3244 non–small-cell lung cancers reveal major discordances. J Thorac Oncol. 2014;9(3):295–306. doi: 10.1097/JTO.0000000000000072. [DOI] [PubMed] [Google Scholar]
- 9.Tsao AS, Scagliotti GV, Bunn PA, Jr, Carbone DP, Warren GW, Bai C, de Koning HJ, Yousaf-Khan AU, McWilliams A, Tsao MS, et al. Scientific advances in lung Cancer 2015. J Thorac Oncol. 2016;11(5):613–638. doi: 10.1016/j.jtho.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 10.Lee SE, Lee B, Hong M, Song J-Y, Jung K, Lira ME, Mao M, Han J, Kim J, Choi Y-L. Comprehensive analysis of RET and ROS1 rearrangement in lung adenocarcinoma. Mod Pathol. 2015;28(4):468–479. doi: 10.1038/modpathol.2014.107. [DOI] [PubMed] [Google Scholar]
- 11.Cao B, Wei P, Liu Z, Bi R, Lu Y, Zhang L, Zhang J, Yang Y, Shen C, Du X, et al. Detection of lung adenocarcinoma with ROS1 rearrangement by IHC, FISH, and RT-PCR and analysis of its clinicopathologic features. Onco Targets Ther. 2016;9:131–138. doi: 10.2147/OTT.S94997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshimura Y, Kurasawa M, Yorozu K, Puig O, Bordogna W, Harada N. Antitumor activity of alectinib, a selective ALK inhibitor, in an ALK-positive NSCLC cell line harboring G1269A mutation. Cancer Chemother Pharmacol. 2016;77(3):623–628. doi: 10.1007/s00280-016-2977-y. [DOI] [PubMed] [Google Scholar]
- 13.Shimamura T, Li D, Ji H, Haringsma HJ, Liniker E, Borgman CL, Lowell AM, Minami Y, McNamara K, Perera SA, et al. Hsp90 inhibition suppresses mutant EGFR-T790M signaling and overcomes kinase inhibitor resistance. Cancer Res. 2008;68(14):5827–5838. doi: 10.1158/0008-5472.CAN-07-5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsubara D, Kanai Y, Ishikawa S, Ohara S, Yoshimoto T, Sakatani T, Oguni S, Tamura T, Kataoka H, Endo S, et al. Identification of CCDC6-RET Fusion in the Human Lung Adenocarcinoma Cell Line, LC-2/ad. J Thorac Oncol. 2012;7(12):1872–1876. doi: 10.1097/JTO.0b013e3182721ed1. [DOI] [PubMed] [Google Scholar]
- 15.Furugaki K, Fukumura J, Iwai T, Yorozu K, Kurasawa M, Yanagisawa M, Moriya Y, Yamamoto K, Suda K, Mizuuchi H, et al. Impact of bevacizumab in combination with erlotinib on EGFR-mutated non–small cell lung cancer xenograft models with T790M mutation or MET amplification. Int J Cancer. 2016;138(4):1024–1032. doi: 10.1002/ijc.29848. [DOI] [PubMed] [Google Scholar]
- 16.Kodama T, Tsukaguchi T, Satoh Y, Yoshida M, Watanabe Y, Kondoh O, Sakamoto H. Alectinib shows potent antitumor activity against <em>RET</em>−rearranged non–small cell lung Cancer. Mol Cancer Ther. 2014;13(12):2910–2918. doi: 10.1158/1535-7163.MCT-14-0274. [DOI] [PubMed] [Google Scholar]
- 17.Davies KD, Le AT, Theodoro MF, Skokan MC, Aisner DL, Berge EM, Terracciano LM, Incarbone M, Roncalli M, Cappuzzo F, et al. Identifying and targeting ROS1 gene fusions in non-small cell lung Cancer. Clin Cancer Res. 2012;18(17):4570–4579. doi: 10.1158/1078-0432.CCR-12-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marsilje TH, Pei W, Chen B, Lu W, Uno T, Jin Y, Jiang T, Kim S, Li N, Warmuth M, et al. Synthesis, structure–activity relationships, and in vivo efficacy of the novel potent and selective anaplastic lymphoma kinase (ALK) inhibitor 5-Chloro-N2-(2-isopropoxy-5-methyl-4-(piperidin-4-yl)phenyl)-N4-(2-(isopropylsulfonyl)phenyl)pyrimidine-2,4-diamine (LDK378) currently in phase 1 and phase 2 clinical trials. J Med Chem. 2013;56(14):5675–5690. doi: 10.1021/jm400402q. [DOI] [PubMed] [Google Scholar]
- 19.Sakamoto H, Tsukaguchi T, Hiroshima S, Kodama T, Kobayashi T, Fukami Takaaki A, Oikawa N, Tsukuda T, Ishii N, Aoki Y. CH5424802, a selective ALK inhibitor capable of blocking the resistant gatekeeper mutant. Cancer Cell. 2011;19(5):679–690. doi: 10.1016/j.ccr.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 20.Facchinetti F, Tiseo M, Di Maio M, Graziano P, Bria E, Rossi G, Novello S. Tackling ALK in non-small cell lung cancer: the role of novel inhibitors. Transl Lung Cancer Res. 2016;5(3):301–321. doi: 10.21037/tlcr.2016.06.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wynes MW, Sholl LM, Dietel M, Schuuring E, Tsao MS, Yatabe Y, Tubbs RR, Hirsch FR. An international interpretation study using the ALK IHC antibody D5F3 and a sensitive detection kit demonstrates high concordance between ALK IHC and ALK FISH and between evaluators. J Thorac Oncol. 2014;9(5):631–638. doi: 10.1097/JTO.0000000000000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun J-M, Choi Y-L, Won J-K, Hirsch FR, Ahn JS, Ahn M-J, Park K. A dramatic response to Crizotinib in a non–small-cell lung Cancer patient with IHC-positive and FISH-negative ALK. J Thorac Oncol. 2012;7(12):e36–e38. doi: 10.1097/JTO.0b013e318274694e. [DOI] [PubMed] [Google Scholar]
- 23.Zou Helen Y, Friboulet L, Kodack David P, Engstrom Lars D, Li Q, West M, Tang Ruth W, Wang H, Tsaparikos K, Wang J, et al. PF-06463922, an ALK/ROS1 inhibitor, overcomes resistance to first and second generation ALK inhibitors in preclinical models. Cancer Cell. 2015;28(1):70–81. doi: 10.1016/j.ccell.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katayama R, Kobayashi Y, Friboulet L, Lockerman EL, Koike S, Shaw AT, Engelman JA, Fujita N. Cabozantinib overcomes Crizotinib resistance in ROS1 fusion–positive Cancer. Clin Cancer Res. 2015;21(1):166–174. doi: 10.1158/1078-0432.CCR-14-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, et al. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci. 2002;99(7):4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang R, Hu H, Pan Y, Li Y, Ye T, Li C, Luo X, Wang L, Li H, Zhang Y, et al. RET fusions define a unique molecular and Clinicopathologic subtype of non–small-cell lung Cancer. J Clin Oncol. 2012;30(35):4352–4359. doi: 10.1200/JCO.2012.44.1477. [DOI] [PubMed] [Google Scholar]
- 27.Lipson D, Capelletti M, Yelensky R, Otto G, Parker A, Jarosz M, Curran JA, Balasubramanian S, Bloom T, Brennan KW, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med. 2012;18(3):382–384. doi: 10.1038/nm.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Y, Zhang Y, Zou H, Cai N, Chen X, Xu L, Kong X, Liu P. The multi-targeted tyrosine kinase inhibitor vandetanib plays a bifunctional role in non-small cell lung cancer cells. Sci Rep. 2015;5:8629. doi: 10.1038/srep08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiesner T, He J, Yelensky R, Esteve-Puig R, Botton T, Yeh I, Lipson D, Otto G, Brennan K, Murali R, et al. Kinase fusions are frequent in Spitz tumours and spitzoid melanomas. Nat Commun. 2014;5:3116. doi: 10.1038/ncomms4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki M, Makinoshima H, Matsumoto S, Suzuki A, Mimaki S, Matsushima K, Yoh K, Goto K, Suzuki Y, Ishii G, et al. Identification of a lung adenocarcinoma cell line with CCDC6-RET fusion gene and the effect of RET inhibitors in vitro and in vivo. Cancer Sci. 2013;104(7):896–903. doi: 10.1111/cas.12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanizaki J, Okamoto I, Sakai K, Nakagawa K. Differential roles of trans-phosphorylated EGFR, HER2, HER3, and RET as heterodimerisation partners of MET in lung cancer with MET amplification. Br J Cancer. 2011;105(6):807–813. doi: 10.1038/bjc.2011.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rimkunas VM, Crosby KE, Li D, Hu Y, Kelly ME, Gu T-L, Mack JS, Silver MR, Zhou X, Haack H. Analysis of receptor tyrosine kinase ROS1-positive tumors in non–small cell lung Cancer: identification of a FIG-ROS1 fusion. Clin Cancer Res. 2012;18(16):4449–4457. doi: 10.1158/1078-0432.CCR-11-3351. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, Yang W, Pu Q, Yang Y, Ye S, Ma Q, Ren J, Cao Z, Zhong G, Zhang X, et al. The effects and mechanisms of SLC34A2 in tumorigenesis and progression of human non-small cell lung cancer. J Biomed Sci. 2015;22(1):52. doi: 10.1186/s12929-015-0158-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gainor JF, Shaw AT. Novel targets in non-small cell lung Cancer: ROS1 and RET fusions. Oncologist. 2013;18(7):865–875. doi: 10.1634/theoncologist.2013-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kopantzev EP, Monastyrskaya GS, Vinogradova TV, Zinovyeva MV, Kostina MB, Filyukova OB, Tonevitsky AG, Sukhikh GT, Sverdlov ED. Differences in gene expression levels between early and later stages of human lung development are opposite to those between normal lung tissue and non-small lung cell carcinoma. Lung Cancer. 2008;62(1):23–34. doi: 10.1016/j.lungcan.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 36.McClelland M, Zhao L, Carskadon S, Arenberg D. Expression of CD74, the receptor for macrophage migration inhibitory factor, in non-small cell lung Cancer. Am J Pathol. 2009;174(2):638–646. doi: 10.2353/ajpath.2009.080463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee HJ, Seol HS, Kim JY, Chun SM, Suh YA, Park YS, Kim SW, Choi CM, Park SI, Kim DK, et al. ROS1 receptor tyrosine kinase, a Druggable target, is frequently overexpressed in non-small cell lung carcinomas via genetic and epigenetic mechanisms. Ann Surg Oncol. 2013;20(1):200–208. doi: 10.1245/s10434-012-2553-6. [DOI] [PubMed] [Google Scholar]
- 38.Kohno T. Implementation of “clinical sequencing” in cancer genome medicine in Japan. Cancer Sci. 2018;109(3):507–512. doi: 10.1111/cas.13486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Velizheva NP, Rechsteiner MP, Valtcheva N, Freiberger SN, Wong CE, Vrugt B, Zhong Q, Wagner U, Moch H, Hillinger S, et al. Targeted next-generation-sequencing for reliable detection of targetable rearrangements in lung adenocarcinoma—a single center retrospective study. Pathol - Res Pract. 2018;214(4):572–578. doi: 10.1016/j.prp.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ross JS, Wang K, Gay L, et al. Comprehensive genomic profiling of carcinoma of unknown primary site: new routes to targeted therapies. JAMA Oncol. 2015;1(1):40–49. doi: 10.1001/jamaoncol.2014.216. [DOI] [PubMed] [Google Scholar]
- 41.Vendrell JA, Taviaux S, Béganton B, Godreuil S, Audran P, Grand D, Clermont E, Serre I, Szablewski V, Coopman P, et al. Detection of known and novel ALK fusion transcripts in lung cancer patients using next-generation sequencing approaches. Sci Rep. 2017;7(1):12510. doi: 10.1038/s41598-017-12679-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Taqman probes for fusion (DOCX 26 kb)
Table S2. Taqman probes for exons of mRNA (DOCX 28 kb)
Table S3. Target terminus of antibody (DOCX 28 kb)
Table S4. Exon count of mRNA (DOCX 26 kb)
Figure S1. RT-PCR analysis of ALK, RET, or ROS1 fusion in 37 cancer cell lines. The mRNA expression of variant 1, 2, 3a, or 3b of EML4-ALK (a), KIF5B-RET, CCDC6-RET (b) or SLC34A2-ROS1 (c) was determined by RT-PCR using the Taqman probes shown in Additional file 1: Table S1. Red lines show fusion gene–positive cell lines and green lines show fusion gene–negative cell lines (PPTX 129 kb)
Figure S2. Summary of ALK, RET, or ROS1 fusion in 37 cancer cell lines. Plot of normalized values calculated from the data in Additional file 5: Figure S1 (PPTX 50 kb)
Table S5. IC50s of ALK-TKIs and erlotinib (DOCX 28 kb)
Figure S4. Comparison of molecular weights between wild-type and fusion protein. After high exposure of N-terminal EML4 in the results shown in Fig. 2, two sizes of EML4 protein were detected in NCI-H2228 and SNU-2292 cell lines (left-hand figure), and we assumed that the larger protein was wild-type EML4 (arrow) and the smaller protein was ALK-fused EML4 (arrowhead), after referring to the protein weight of EML4-ALK detected by an ALK C-terminus antibody (right-hand figure) (a). In the same way, we assumed that the larger and smaller proteins were respectively wild-type EML4 (arrow) and ALK-fused EML4 (arrowhead) in the results for the ILS31007 tumor tissue specimen shown in Fig. 3b (b), and the larger and smaller proteins were respectively wild-type CCDC6 (arrow) and RET-fused CCDC6 (arrowhead) in the results for CCDC6 in the LC-2/ad cell line shown in Fig. 2 (c) (PPTX 600 kb)
Figure S5. RT-PCR analysis of ALK, RET, or ROS1 fusion in 4 tumor tissues. The mRNA expression of variant 1, 2, 3a, or 3b of EML4-ALK (a), KIF5B-RET, CCDC6-RET (b) or SLC34A2-ROS1 (c) was determined by RT-PCR using the Taqman probes shown in Additional file 1: Table S1. Red lines show fusion gene–positive tumor tissue, and green lines show fusion gene–negative tumor tissue (PPTX 122 kb)
Figure S6. Summary of ALK, RET, or ROS1 fusion in 4 tumor tissue specimens. Plot of normalized values calculated from the data in Additional file 9: Figure S5 (PPTX 45 kb)
Figure S7. Analysis by IHC and FISH of ALK in ILS31007 tissue specimen. The FFPE specimens with tumor tissue (upper panel) and the paired NAT specimens (lower panel) were obtained from BioreclamationIVT. IHC (left panel) and FISH (right panel) analyses for ALK rearrangement were performed with, respectively, the N-Histofine ALK detection kit (Nichirei Biosciences; Tokyo, Japan) and the Vysis ALK break apart FISH probe kit (Abbott laboratories; Abbott Park, IL) at a commercial clinical laboratory, LSI Medience (Tokyo, Japan). Images were captured using standard settings by the BZ900 (Keyence; Osaka, Japan) (PPTX 754 kb)
Figure S8. RT-PCR analysis of EML4 in four tissue specimens. The relative EML4 mRNA expression at each exon in NAT specimens was calculated as the ratio of the normalized values with GAPDH mRNA to those in tumor tissues. Each bar represents the mean + SD (n = 3) (PPTX 45 kb)
Figure S9. Western blotting analysis in three cancer cell lines with or without RET fusion. Cell lysates were harvested after 2 h of treatment with each drug at the concentrations shown (nM). The levels of RET phosphorylation in LC-2/ad were undetectable by this western blotting system (PPTX 2307 kb)
Figure S3. Western blotting analysis in 3 cancer cell lines with ALK fusion. Cell lysates were harvested after 2 h of treatment with each drug at the concentration shown (nM). The antibodies were obtained from Cell signaling technology. (PPTX 2483 kb)
Figure S10. Western blotting analysis in a cancer cell line with ROS1 fusion. Cell lysates were harvested after 2 h of treatment with each drug at the concentrations shown (nM) (PPTX 797 kb)
Figure S11. Western blotting analysis in five cancer cell lines without any fusion gene. Cell lysates were harvested after 2 h of treatment with each drug at the concentrations shown (nM) (PPTX 2123 kb)
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.