Batista et al. show that increased auxin biosynthesis in the endosperm prevents its cellularization, leading to seed arrest.
Keywords: auxin, cellularization, endosperm, hybridization barrier, seed development, triploid block
Abstract
The endosperm is an ephemeral tissue that nourishes the developing embryo, similar to the placenta in mammals. In most angiosperms, endosperm development starts as a syncytium, in which nuclear divisions are not followed by cytokinesis. The timing of endosperm cellularization largely varies between species, and the event triggering this transition remains unknown. Here we show that increased auxin biosynthesis in the endosperm prevents its cellularization, leading to seed arrest. Auxin-overproducing seeds phenocopy paternal-excess triploid seeds derived from hybridizations of diploid maternal plants with tetraploid fathers. Concurrently, auxin-related genes are strongly overexpressed in triploid seeds, correlating with increased auxin activity. Reducing auxin biosynthesis and signaling reestablishes endosperm cellularization in triploid seeds and restores their viability, highlighting a causal role of increased auxin in preventing endosperm cellularization. We propose that auxin determines the time of endosperm cellularization, and thereby uncovered a central role of auxin in establishing hybridization barriers in plants.
In flowering plants, seed development is initiated by the fertilization of two maternal gametes, egg cell and central cell, by two paternal sperm cells (Dresselhaus et al. 2016). This double-fertilization event originates two fertilization products: the embryo, which will form a new plant, and the endosperm, a nourishing tissue that ensures adequate nutrient transfer from the mother plant to the developing embryo (Bleckmann et al. 2014). The endosperm of most angiosperms is a triploid tissue, derived after fertilization of the diploid central cell. It thus contains two maternal and one paternal (2M:1P) genome copies. In Arabidopsis, like in most angiosperms, the endosperm initially develops as a syncytium, in which nuclear divisions are not followed by cytokinesis (Li and Berger 2012). After a defined number of nuclear divisions, the endosperm cellularizes (Boisnard-Lorig et al. 2001); however, the pathways regulating this transition remain unknown. The balance of 2M:1P genome copies in the endosperm is crucial for reproductive success. Deviation from this ratio in response to hybridizations of plants that differ in ploidy frequently leads to unviable seeds, a phenomenon referred to as triploid block (von Wangenheim 1962; Scott et al. 1998; Leblanc et al. 2002; Stoute et al. 2012; Sekine et al. 2013). Importantly, interploidy hybridizations affect endosperm cellularization: Although maternal excess crosses (4x × 2x; by convention the maternal parent is always mentioned first) shift the cellularization to earlier timepoints, paternal-excess hybridization (2x × 4x) causes a delay or complete failure of endosperm cellularization (Scott et al. 1998; Lafon-Placette and Köhler 2016). In Arabidopsis, the triploid (3x) embryos resulting from 2x × 4x crosses are viable and produce healthy plants when transferred to nutritive medium, revealing that failure of endosperm cellularization impairs embryo viability (Hehenberger et al. 2012; Lafon-Placette and Köhler 2016). Mutations in the paternally expressed imprinted genes (PEGs) ADMETOS (ADM), SUVH7, PEG2, and PEG9 restore endosperm cellularization and viability of paternal-excess 3x seeds (Kradolfer et al. 2013b; Wolff et al. 2015; Jiang et al. 2017).
In this study we show that auxin activity is strongly increased in paternal-excess 3x seeds and that the 3x seed phenotype can be phenocopied by overproduction of auxin in the endosperm of diploid seeds. Furthermore, we show that down-regulating auxin biosynthesis or signaling can partly restore 3x seed viability. Overall, our data link auxin activity with endosperm cellularization and show that increased auxin activity in the endosperm establishes a postzygotic hybridization barrier in Arabidopsis.
Results
Paternal-excess crosses lead to increased auxin activity after fertilization
Triploid seed abortion in paternal-excess (2x × 4x) crosses is characterized by the overproliferation of the endosperm, which fails to cellularize (Scott et al. 1998), but the molecular mechanisms that lead to this phenotype are yet to be elucidated. To search for pathways potentially involved in 3x seed abortion, we compared gene expression data of wild-type seeds at 6 d after pollination (DAP) with that of wild-type maternal plants pollinated with pollen of omission of second division 1 (osd1) (d'Erfurth et al. 2009). Mutants for osd1 form unreduced diploid gametes (2n) and therefore can be used to mimic paternal-excess crosses when used as a pollen donor to a wild-type mother (d'Erfurth et al. 2009; Kradolfer et al. 2013a). We found genes involved in auxin homeostasis to be significantly enriched among those genes that were up-regulated in 3x seeds (Table 1). In particular, genes involved in auxin biosynthesis (TAA/TAR and YUCCA) (Cheng et al. 2007; Stepanova et al. 2008), auxin transport (PIN and PGP-type) (Lin and Wang 2005; Křeček et al. 2009), and auxin response factors (ARFs) (Boer et al. 2014; Weijers and Wagner 2016) were highly up-regulated in 3x seeds compared with 2x seeds (Fig. 1A; Supplemental Fig. S1). Consistent with the transcriptome data, we found a marked increase in the activity of the auxin sensor DR5v2::VENUS (Liao et al. 2015) in 3x seeds that was most prominent in the seed coat, suggesting that increased auxin generated in the fertilization products in response to osd1 pollination is rapidly transported to the seed coat (Fig. 1B,C), in line with previous reports (Figueiredo et al. 2016). Indeed, genes coding for auxin biosynthesis, as well as auxin signaling, are strongly up-regulated in the endosperm of 3x seeds compared with that of 2x seeds (Supplemental Fig. S2). These observations indicate that paternal-excess crosses induce increased auxin production and signaling in the endosperm of 3x seeds.
Table 1.
Significantly enriched biological processes for genes up-regulated at 6 DAP in 3x seeds (Ler × osd1 cross) compared with 2x seeds (Ler × Col cross) (log2 fold change > 1, P-value <0.05)
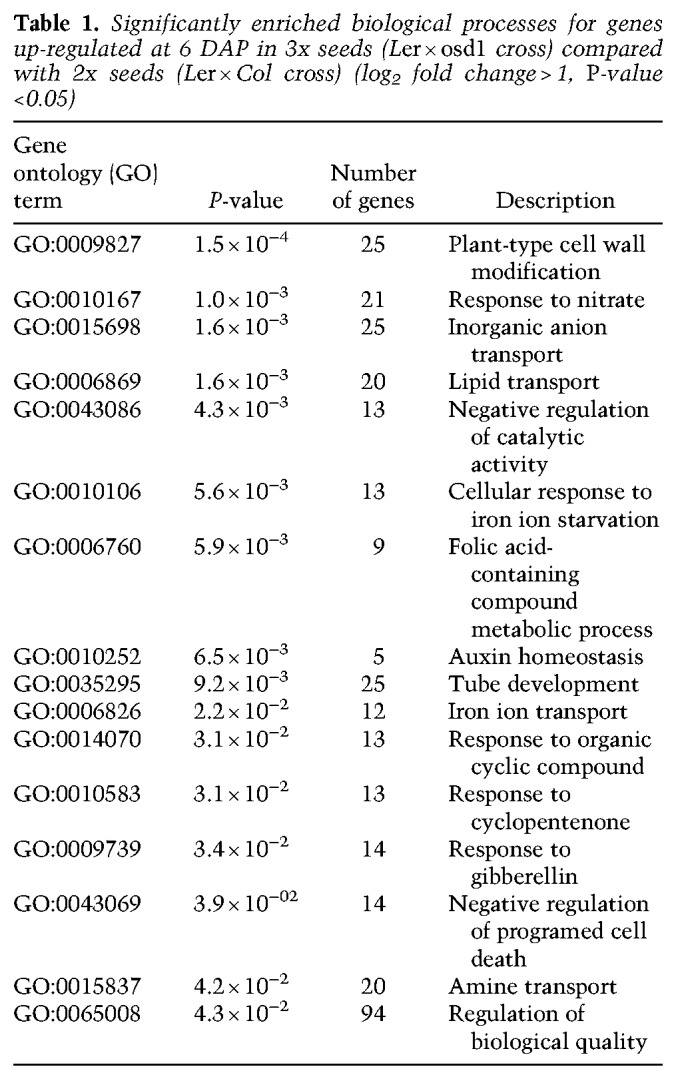
Figure 1.
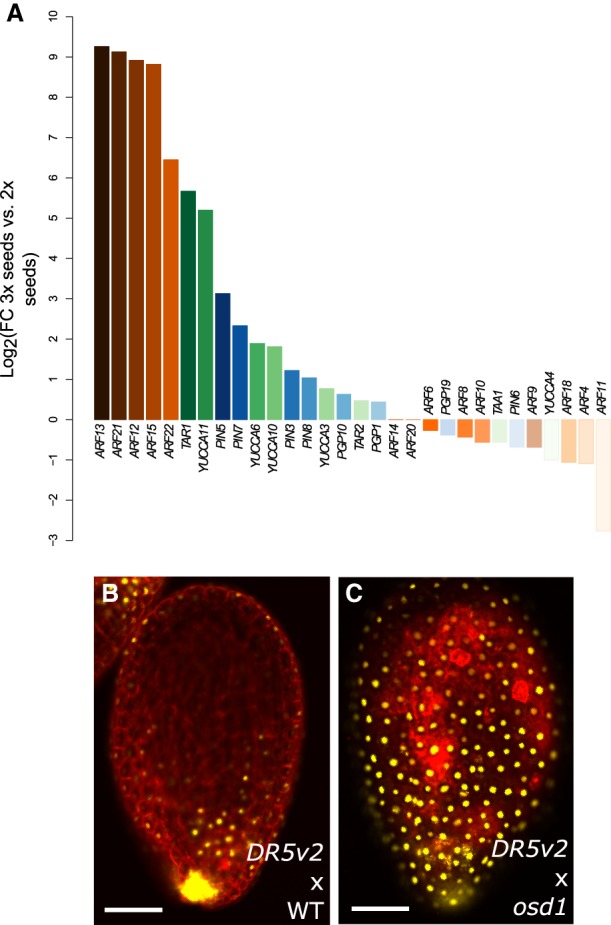
Auxin is overproduced in 3x seeds. (A) Log2-fold expression change between 3x and 2x seeds of genes coding for auxin biosynthesis (green bars), signaling (orange bars), and transport proteins (blue bars). (B,C) Auxin activity as measured by expression of DR5v2::VENUS in 2x (B) and 3x (C) seeds at 5 DAP. Pictures show representative seeds of three independent siliques per cross. Red staining is propidium iodide. Bars, 100 µm.
Overproduction of auxin in the endosperm phenocopies paternal-excess triploid seeds
Based on the finding that auxin activity is increased in 3x seeds, we addressed the question whether overproduction of auxin is responsible for the endosperm developmental defects leading to 3x seed abortion. To test this hypothesis, we raised transgenic plants overexpressing the bacterial auxin biosynthesis gene Indole Acetimide Hydrolase (IaaH) under the control of the early-endosperm specific promoter DD25 (Tinland et al. 1991; Steffen et al. 2007). The production of the active auxin indole 3-acetic acid (IAA) by IaaH relies on the availability of indole 3-acetamide (IAM), which was previously shown to be present in Arabidopsis (Pollmann et al. 2002; Sugawara et al. 2009). Furthermore, genes coding for IAM-synthetizing enzymes are strongly expressed in the endosperm (Supplemental Fig. S3). Strikingly, out of 31 transgenic lines expressing DD25::IaaH, all showed aborting seeds that closely resembled paternal-excess 3x seeds by their dark and shriveled appearance (Fig. 2A–C). In seven lines that were analyzed in detail, we found that the frequency of either partially or fully collapsed seeds ranged between 10% and 40%, which largely corresponded with the rate of nongerminating seeds (Supplemental Figs. S4, S5). Embryos of DD25::IaaH-expressing lines were retarded in growth, similar to 3x embryos (Fig. 2D–L). In both 3x seeds and those expressing DD25::IaaH, embryo development progressed up to the early heart stage without noticeable differences compared with 2x wild-type seeds (5-DAP time point) (Fig. 2). However, from 6 DAP onward, the embryos of 3x and DD25::IaaH transgenic seeds were delayed in development and did not progress beyond the torpedo stage (Fig. 2D–L).
Figure 2.
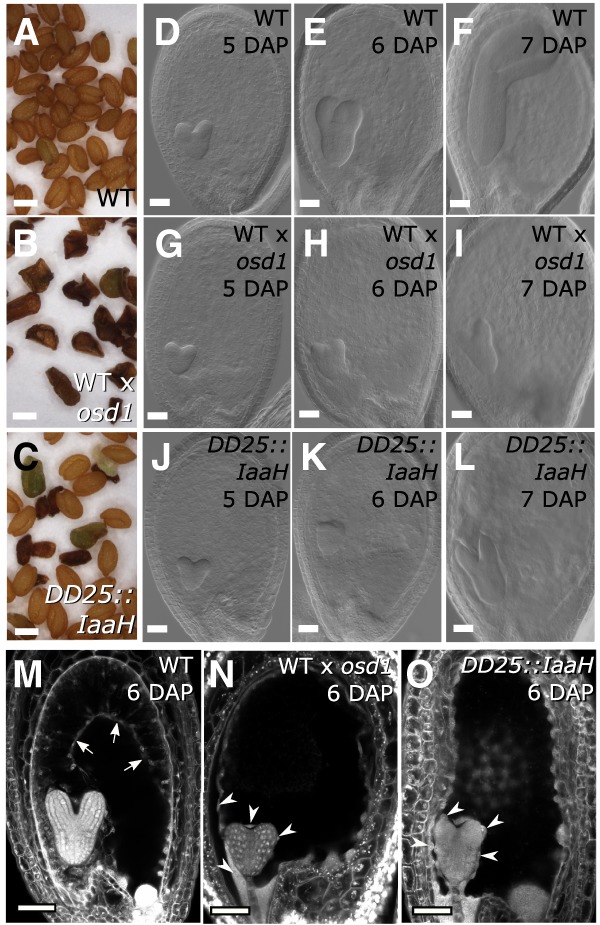
Increased auxin in the endosperm prevents cellularization. (A–C) Dry seed morphology of wild-type 2x (A), wild-type 3x (B), and DD25::IaaH 2x seeds (C). Bars, 0.5 mm. (D–L) Clearings of wild-type 2x (D–F), wild-type 3x (G–I), and DD25::IaaH 2x seeds (J–L) from 5 to 7 DAP. Pictures show representative seeds of three independent siliques per cross. Bars, 50 µm. (M–O) Endosperm cellularization as determined by Feulgen staining at 6 DAP for 2x seeds (M), 3x seeds (N), and 2x seeds expressing DD25::IaaH (O). Pictures show representative seeds of 10 independent siliques per cross. Arrows indicate cellularized peripheral endosperm, and arrowheads indicate free endosperm nuclei surrounding the embryo. Bars, 50 µm. (WT) Wild type.
The endosperm of seeds derived from paternal-excess crosses fails to cellularize (Scott et al. 1998); therefore, we tested whether seeds expressing DD25::IaaH showed a similar developmental defect (Fig. 2M–O; Supplemental Fig. S6). Endosperm cellularization of 2x wild-type seeds initiated around 5 DAP and was almost complete at 7 DAP (Supplemental Fig. S6). Consistent with previous reports (Scott et al. 1998), in 3x seeds derived from paternal-excess crosses, the endosperm failed to cellularize, and free endosperm nuclei could be seen surrounding the embryo. Importantly, DD25::IaaH expression induced a similar phenotype, and many seeds showed no signs of endosperm cellularization even at 7 DAP (Supplemental Fig. S6). These observations indicate that overproduction of auxin in the endosperm is sufficient to impair its cellularization.
To test whether the phenotypes observed in DD25::IaaH lines are indeed caused by overproduction of auxin in the endosperm, we crossed wild plants with pollen from DD25::IaaH plants. Indeed, we observed the same seed phenotypes when the transgene was inherited through pollen, confirming that endosperm-produced auxin is causal to this phenotype and ruling out that the effect originates in maternal sporophyte tissues (Supplemental Fig. S4). Furthermore, when crossing maternal plants expressing the auxin reporter DR5v2::VENUS with pollen carrying the DD25::IaaH transgene, we observed a significant increase in VENUS fluorescence, similar to what is observed in paternal-excess crosses (Fig. 1B,C; Supplemental Fig. S4). Together, we conclude that increased auxin production in the endosperm prevents endosperm cellularization, leading to a phenocopy of paternal-excess 3x seeds.
PEGs and AGAMOUS-LIKE genes are partly deregulated by auxin overproduction
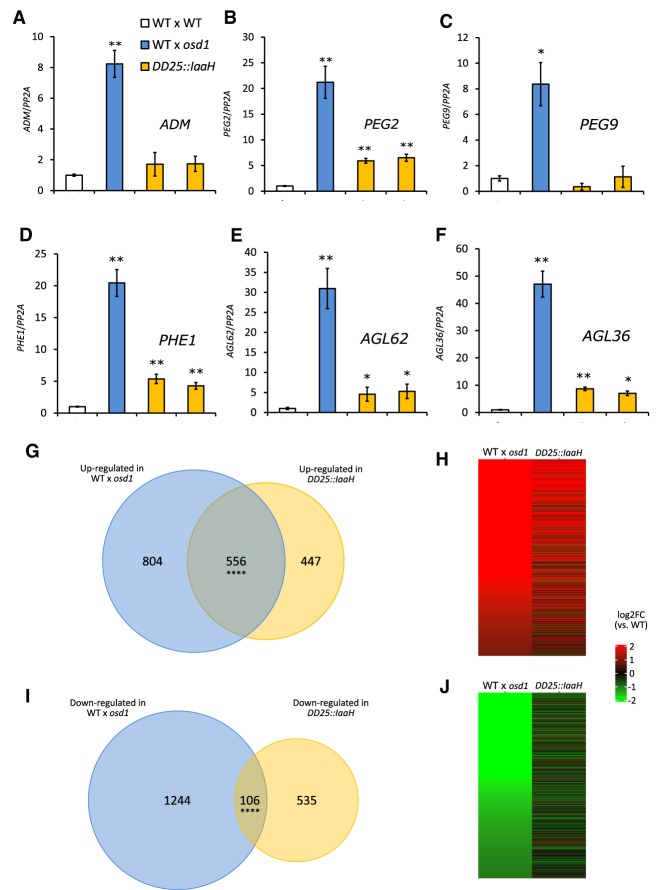
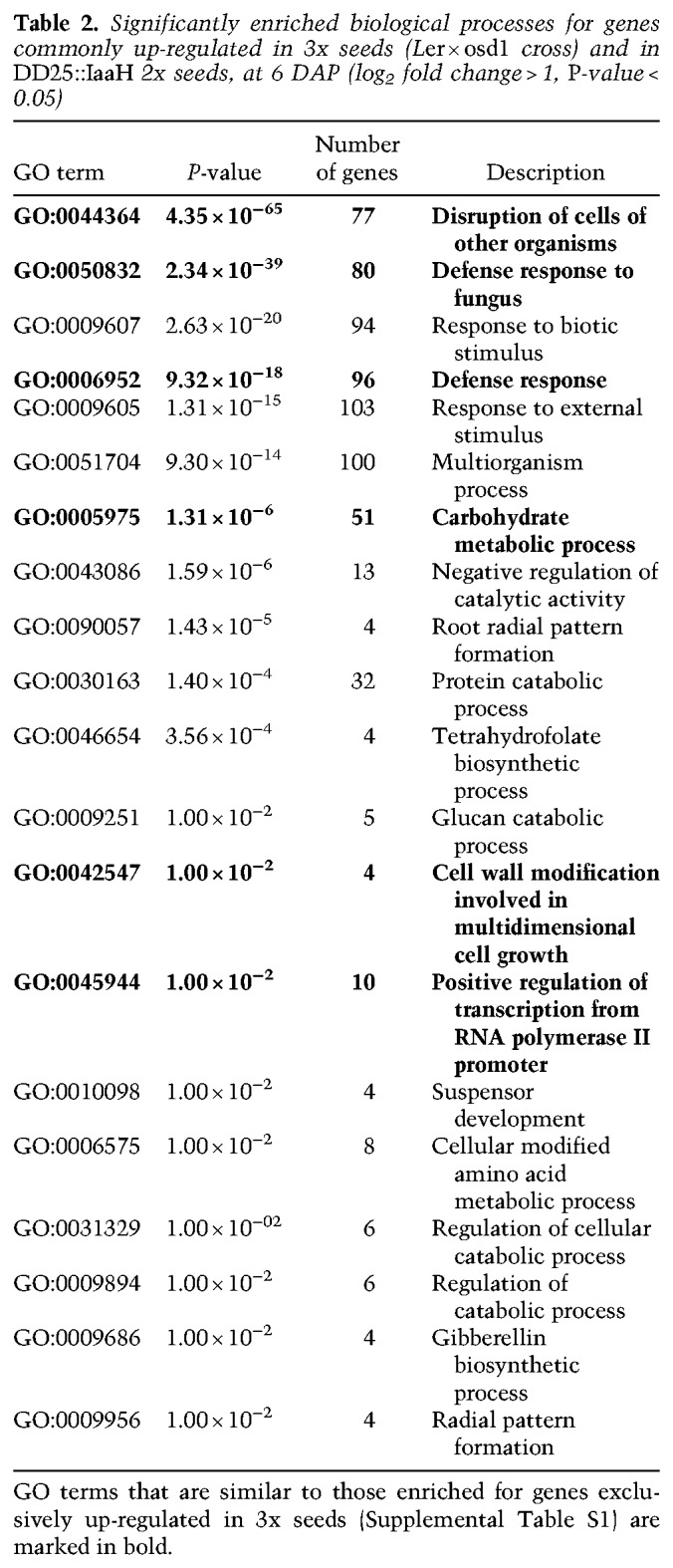
Triploid paternal-excess seeds are characterized by a strong deregulation of PEGs and genes coding for AGAMOUS-LIKE (AGL) MADS-box transcription factors (Erilova et al. 2009; Kradolfer et al. 2013a). We tested whether overproduction of auxin causes a transcriptional phenocopy of 3x paternal-excess seeds by analyzing expression of PEGs and AGLs that were previously shown to be strongly deregulated in 3x seeds (Kradolfer et al. 2013b; Wolff et al. 2015). Although PEGs ADM and PEG9 were not significantly deregulated in seeds of DD25::IaaH-expressing plants compared with 2x wild-type seeds, PEG2 and the AGL genes PHE1, AGL62, and AGL36 were expressed at significantly higher levels in auxin-overproducing seeds (Fig. 3A–F). However, their level of deregulation remained substantially lower compared with 3x seeds. To further explore the molecular bases of seed abortion in auxin-overexpressing lines, we performed RNAseq on 6-DAP seeds from lines expressing DD25::IaaH and asked which genes were deregulated compared with wild-type seeds. We then compared this list to that of genes deregulated in 3x seeds (Schatlowski et al. 2014), and observed that out of 1003 genes up-regulated in seeds expressing DD25::IaaH, compared with wild-type, 556 genes (55%) were also up-regulated in 3x seeds (Fig. 3G,H). Among these genes we observed an enrichment of gene ontology (GO) terms related to cell wall modification, transcription, and hormone responses (Table 2). Genes belonging to these GO categories were mainly expressed in the endosperm (Supplemental Fig. S7), indicating that the transcriptional deregulation observed in 3x seeds and auxin-overproducing seeds is predominantly endosperm based. Analysis of genes specifically up-regulated in 3x osd1 seeds revealed an enrichment in GO terms related to cell wall modification and transcription (Supplemental Table S1), similar to what is observed in genes commonly up-regulated in 3x seeds and auxin-overproducing seeds (Table 2). Interestingly, <20% of genes down-regulated in DD25::IaaH seeds were commonly deregulated in 3x seeds (Fig. 3I,J). These results indicate that overproduction of auxin in the endosperm leads to a partial molecular phenocopy of the transcriptome of 3x seeds but that several pathways are exclusively deregulated in a paternal-excess situation. This is consistent with our qPCR analysis, which showed that overproduction of auxin caused only a partial deregulation of PEGs, compared with paternal-excess crosses (Fig. 3A–F). Together, these data suggest either that auxin acts independently of the pathways previously shown to affect 3x seed abortion (Kradolfer et al. 2013b; Wolff et al. 2015; Jiang et al. 2017) or, alternatively, that auxin signaling is downstream from PEG and AGL functions in the endosperm.
Figure 3.
PEGs and AGL genes are not substantially deregulated in DD25::IaaH transgenic seeds. (A–F) Relative gene expression in seeds at 6 DAP, as determined by RT-qPCR, in 2x wild-type, 3x wild-type, and 2x DD25::IaaH transgenic seeds of two independent lines for ADM (A), PEG2 (B), PEG9 (C), PHE1 (D), AGL62 (E), and AGL36 (F). Results from a representative biological replicate are shown. Three technical replicates were performed; error bars, SD. Differences are significant for Student's t-test for P < 0.05 (*) or P < 0.001 (**). (WT) Wild-type. (G–J) Comparison of genes deregulated at 6 DAP, as determined by RNAseq, in 3x (WT × osd1) and 2x DD25::IaaH seeds. (G,I) The Venn diagrams show the overlap of genes that are commonly up-regulated (G) or down-regulated (I) in 3x and in 2x DD25::IaaH seeds. Overlaps between deregulated genes are significantly higher than expected; hypergeometric distribution, P < 0.0001 (****). (H,J) Each line in each plot indicates one gene that is deregulated in 3x seeds (left) and its corresponding expression in 2x seeds expressing DD25::IaaH (right).
Table 2.
Significantly enriched biological processes for genes commonly up-regulated in 3x seeds (Ler × osd1 cross) and in DD25::IaaH 2x seeds, at 6 DAP (log2 fold change > 1, P-value < 0.05)
Decreased auxin biosynthesis and signaling suppress triploid seed abortion
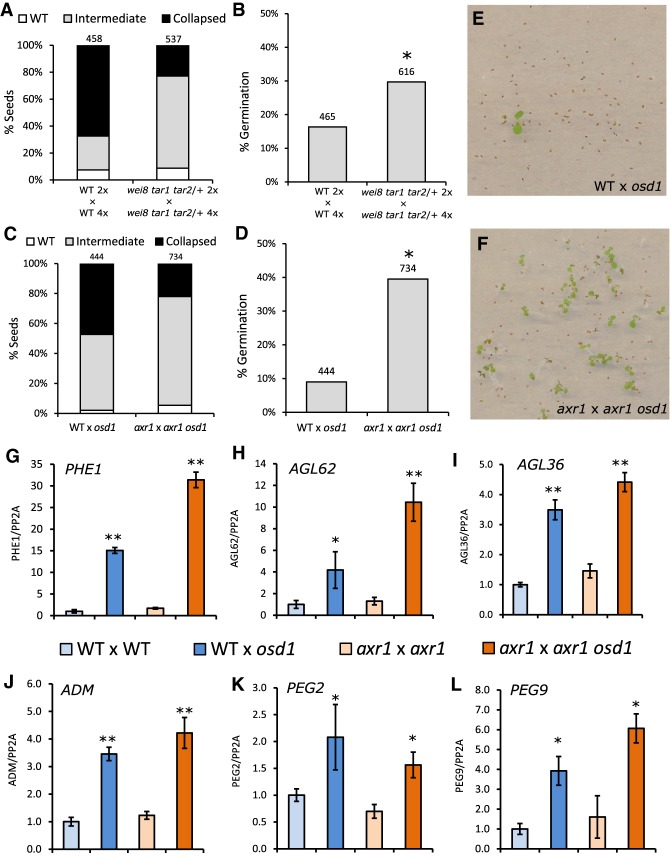
To address the question whether endosperm failure in 3x paternal-excess seeds is because of overproduction of auxin, we analyzed whether mutants for either auxin biosynthesis (wei8 tar1 tar2-1/+) (Stepanova et al. 2008) or auxin signaling (axr1) (Dharmasiri et al. 2007) could suppress 3x seed abortion. We generated 4x wild-type and wei8 tar1 tar2/+ plants by colchicine treatment and used these plants as a pollen donor in crosses with 2x wild-type or wei8 tar1 tar2/+ mutant maternal plants. In the 2x × 4x wild-type cross, ∼70% of the seeds were fully collapsed (Fig. 4A). In contrast, only 20% of 3x wei8 tar1 tar2/+ seeds were fully collapsed, and the germination rate of mutant 3x seeds was nearly doubled compared with that of the wild type (Fig. 4B), revealing that decreased auxin biosynthesis can suppress 3x seed abortion. To substantiate these findings, we tested the effect of the auxin-signaling-mutant axr1 in suppressing 3x seed abortion. Using the osd1 mutant as a pollen donor resulted in ∼50% of fully collapsed 3x seeds, whereas only 20% of 3x seeds were fully collapsed when using the axr1 osd1 double mutant as a pollen donor (Fig. 4C). The axr1 3x seeds were phenotypically distinct from 2x wild-type seeds by having a box-shaped phenotype (Supplemental Fig. S8); however, many of these seeds were viable and germinated at a rate of 40% compared with 9% of 3x wild-type seeds (Fig. 4D–F). These results show that decreased auxin signaling can suppress paternal-excess seed abortion. The fact that 2x axr1 seeds are largely viable, compared with only 80% viability of 2x wei8 tar1 tar2/+ seeds (Supplemental Fig. S8), likely accounts for the decreased viability of 3x wei8 tar1 tar2/+ seeds compared with 3x axr1 (Fig. 4B,D). Given that 3x seed abortion is characterized by a failure of the endosperm to cellularize, we thus tested whether this process was restored in the axr1 mutant background (Supplemental Fig. S9). Endosperm cellularization dynamics in 2x axr1 seeds was similar to that of 2x wild-type seeds (Fig. 2; Supplemental Fig. S9), and the endosperm was almost fully cellularized at 7 DAP. Although cellularization in 3x axr1 seeds was delayed compared with 2x seeds, signs of endosperm cellularization in this mutant were clearly visible at 7 DAP, as opposed to 3x wild-type seeds. We thus conclude that rescue of the 3x seed abortion by reduced auxin signaling occurs by restoration of endosperm cellularization.
Figure 4.
Mutants in auxin biosynthesis and signaling suppress triploid seed abortion. (A,B) Phenotypic classification of 3x seeds in wild-type background and in the wei8 tar1 tar2 auxin biosynthesis mutant (A) and their corresponding germination rate (B). (C,D) Same as for A and B, but for the auxin-signaling-mutant axr1. Seed classification was performed according to that in Supplemental Figure S4. Numbers at the top indicate the number of seeds assayed. Differences between wild-type and mutant seed germination in B and D are significant for χ2 test for P < 0.0001 (*). (E,F) Representative image of germinating triploid seedlings in wild-type (E) and axr1 (F). (G–L) Relative gene expression in 6-DAP seeds, as determined by RT-qPCR, in 2x and 3x seeds in wild-type and axr1 mutant backgrounds, for PHE1 (G), AGL62 (H), AGL36 (I), ADM (J), PEG2 (K), and PEG9 (L). Results of a representative biological replicate are shown. Three technical replicates were performed; error bars, SD. Differences between 3x seeds and each respective 2x control are significant for Student's t-test for P < 0.05 (*) or P < 0.001(**).
As discussed above, AGL genes and PEGs are strongly up-regulated in 3x seeds (Fig. 3A–F). We addressed the question whether the rescue of 3x seeds by reduced auxin signaling restored gene expression to wild-type levels. Thus, we tested the expression of AGL genes and PEGs in 2x and 3x wild-type and axr1 seeds. For all genes tested, their expression remained significantly increased in 3x axr1 seeds and, with the exception of PEG2, was even higher in 3x axr1 seeds than 3x wild-type seeds (Fig. 4G–L). These data reveal that 3x seed rescue by decreased auxin signaling occurs independently of PEGs and AGLs, supporting the notion that auxin acts either downstream from or independently of those pathways during endosperm development.
Triploid seeds show increased auxin activity in the sporophyte tissues (Fig. 1B,C), which is consistent with previous observations that auxin produced in the endosperm is transported to the developing seed coat (Figueiredo et al. 2016). One possible scenario is that increased seed coat expansion, because of enhanced auxin activity, prevents endosperm cellularization in 3x seeds. To test this hypothesis, we crossed maternal plants mutant for axr1 with pollen mutant for osd1. In this cross, the endosperm is genotypically wild type, whereas the seed coat is impaired in auxin signaling. Nevertheless, we did not observe an increase in seed viability compared with a wild-type × osd1 cross (Supplemental Fig. S8). Accordingly, we did not observe a rescue of 3x seed abortion in crosses of maternal kluh (klu) mutants to an osd1 pollen donor (Supplemental Fig. S8). KLU codes for a cytochrome P450 monooxygenase, and the klu mutation causes reduced seed coat growth (Adamski et al. 2009). Thus, these observations indicate that decreasing seed coat expansion is not sufficient to rescue 3x seed abortion and restore endosperm cellularization. However, we did observe a small but significant rescue of 3x-like seed abortion when crossing maternal plants mutant for either AXR1 or KLU to a DD25::IaaH pollen donor (Supplemental Fig. S8). We thus tested whether overproducing auxin specifically in the seed coat would cause a phenocopy of the 3x seed phenotype. For this, we developed transgenic lines overexpressing the auxin biosynthesis gene YUC6 under the seed coat-specific promoter KLU (Adamski et al. 2009). Indeed, auxin overproduction in the seed coat was sufficient to recapitulate a 3x-like seed abortion (Supplemental Fig. S10). This suggests that the increased auxin activity in the seed coat of 3x seeds is one of the components of the triploid seed phenotype but that additional factors acting in the endosperm likely contribute to abortion of 3x seeds.
Auxin activity is necessary for endosperm proliferation (Figueiredo et al. 2015). Thus, we hypothesized that 3x seed abortion could be caused by auxin-induced overproliferation of the endosperm and, consequently, its failure to cellularize. Therefore, we tested whether the miniseed3 (mini3) mutation, which reduces endosperm proliferation (Luo et al. 2005), was sufficient to rescue the 3x seed abortion phenotype. However, we did not observe increased survival of mini3 3x seeds compared with 3x wild-type seeds (Supplemental Fig. S8). This indicates that reducing the endosperm rate of proliferation by itself is not sufficient to rescue the 3x seed abortion phenotype.
Altogether, our data reveal that the effect of auxin in the seed coat is sufficient to phenocopy a 3x seed but that the phenotype of 3x seeds is likely a result of the joint deregulation of molecular pathways both at the sporophytic and at the zygotic level.
Down-regulation of auxin biosynthesis and signaling genes coincides with endosperm cellularization
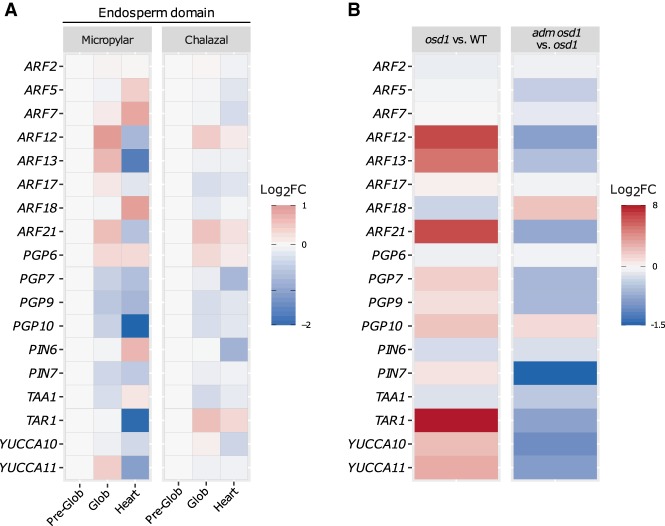
The observation that overproduction of auxin prevents endosperm cellularization suggests that auxin levels have to decrease below a certain threshold in order for the endosperm to cellularize. To test this hypothesis, we analyzed the expression of auxin biosynthesis, transport, and signaling genes in the micropylar and chalazal domains of the endosperm. Endosperm cellularization is initiated in the micropylar domain of the endosperm at around the heart stage of embryo development, whereas cellularization in the chalazal domain occurs later when the embryo has reached the torpedo stage (Mansfield and Briarty 1990). Indeed, the expression level of the PEGs YUC10 and TAR1 was significantly lower in the micropylar endosperm of heart-stage 2x embryos, compared with earlier timepoints (Fig. 5A; Supplemental Fig. S11), consistent with the initiation of cellularization in 2x seeds. The same expression pattern was observed for genes coding for YUC11, for PGP and PIN-type transporters, and for several ARFs (Fig. 5A; Supplemental Fig. S11). This was further confirmed by reduced activity of the fluorescent markers YUC10::YUC10:GFP (Robert et al. 2013) and PGP10::GFP (Figueiredo et al. 2016) at the timing of cellularization (Supplemental Fig. S11). Down-regulation of auxin-related genes was substantially less pronounced in the chalazal endosperm domain, correlating with its delayed cellularization (Fig. 5A). Furthermore, in 3x seeds, in which the endosperm fails to cellularize, there was a strong increase in expression of several auxin-related genes (Fig. 5B). Coinciding with rescued endosperm cellularization in 3x adm seeds (Kradolfer et al. 2013b), expression of auxin-related genes became down-regulated (Fig. 5B). Together, we conclude that increased auxin activity prevents endosperm cellularization, revealing a central regulatory role of auxin in the transition from the syncytial to the cellularized endosperm state.
Figure 5.
Endosperm cellularization is associated with down-regulation of auxin-related gene expression. (A) Expression of auxin-related genes in the micropylar and chalazal endosperm domains throughout different stages of seed development. Expression in each domain is normalized to the preglobular stage and expressed as a log2-fold change relative to that stage. (B) Log2-fold change of expression of auxin-related genes in 3x (osd1) versus 2x seeds and in 3x adm seeds (osd1 adm) versus 3x seeds (osd1).
Discussion
In many plant species, crosses between individuals of different ploidies have long been known to result in abortion of the progeny because of failure of endosperm cellularization, a critical process in seed development (Scott et al. 1998; Sekine et al. 2013; Lafon-Placette and Köhler 2016). Nevertheless, the molecular mechanisms underlying this developmental transition have remained elusive. Here we show that increased production of the plant hormone auxin prevents endosperm cellularization in 3x seeds of Arabidopsis thaliana. Thus, we propose, in addition to its known role to initiate endosperm development and seed coat formation (Figueiredo et al. 2015, 2016), that auxin levels need to be tightly controlled at later stages of seed development to allow the endosperm to cellularize. This hypothesis is strongly supported by our findings that overproduction of auxin prevents endosperm cellularization in 2x seeds and that down-regulation of auxin activity in 3x seeds restores cellularization and, consequently, seed viability. Importantly, the auxin-induced endosperm phenotype is characteristic of paternal-excess crosses, leading to uncellularized inviable seeds (Scott et al. 1998; Sekine et al. 2013). Auxin biosynthesis genes YUC10 and TAR1 are imprinted and paternally expressed in the endosperm (Hsieh et al. 2011; Wolff et al. 2011; Figueiredo et al. 2015). Like many other PEGs, YUC10 and TAR1 are up-regulated in the endosperm of 3x seeds (Wolff et al. 2015), likely causing increased auxin biosynthesis. The observed strong increase of ARF expression may be a consequence of a positive feedback loop, similar to the self-sustained activation of the ARF MONOPTEROS during early embryogenesis (Lau et al. 2011). ARFs are transcription factors that regulate the expression of auxin-responsive genes (Li et al. 2016; Weijers and Wagner 2016) and thus are able to amplify the response to increased auxin levels in the endosperm. The overexpression of ARFs in the endosperm, together with the increased auxin activity in the seed coat of 3x seeds, suggests that the triploid seed phenotype is because of a combined action of zygotic and sporophytic factors. This is in line with our observations that reducing endosperm proliferation or seed coat expansion, by themselves, is not sufficient to rescue the 3x seed abortion. This was only achieved by blocking auxin activity concomitantly in the sporophyte and in the fertilization products.
Our data suggest that increased auxin activity in the endosperm is likely downstream from or independent of AGLs and the known suppressors of 3x seed abortion ADM, PEG2, and PEG9 (Kradolfer et al. 2013b; Wolff et al. 2015). This conclusion is based on the fact that increased auxin could induce a 3x seed-like phenotype without causing increased suppressor gene expression. Furthermore, reduced auxin signaling in axr1 could suppress 3x seed abortion despite high expression levels of ADM, PEG2, and PEG9. Consistent with auxin acting downstream from ADM, most auxin-related genes being up-regulated in 3x seeds became repressed in 3x adm seeds. We propose that endosperm cellularization can only take place when auxin levels are below a certain threshold. If this threshold is not reached, like in 3x seeds, the endosperm will fail to cellularize and the seed aborts. Interestingly, endosperm cellularization in maize occurs at around 3 DAP, clearly before the increase of auxin levels at around 9 DAP (Lur and Setter 1993). The increase in auxin levels coincides with the onset of endoreduplication and cellular differentiation in the endosperm, whereas proliferation rates decrease. It therefore seems unlikely that endosperm cellularization failure in 3x seeds is a consequence of auxin-induced nuclear overproliferation but that nuclear proliferation and endosperm cellularization are mechanistically unlinked. This is consistent with data showing that both processes can be uncoupled in response to interspecies hybridization in rice (Ishikawa et al. 2011) and also with our data showing that the mini3 mutation that, despite impairing endosperm proliferation, does not rescue 3x seed abortion. Auxin is well known to induce changes in cell wall mechanical properties and cell wall synthesis. Auxin-induced organ outgrowth requires demethylesterification of pectin, which causes cell wall loosening (Braybrook and Peaucelle 2013). Auxin could have a similar role in the endosperm and, by inducing demethylesterification and pectin degradation, inhibits endosperm cellularization. We recently found increased demethylesterification activity in the endosperm of 3x seeds, adding support to this idea (Wolff et al. 2015).
In conclusion, we have shown that auxin regulates endosperm cellularization in Arabidopsis. Increased auxin levels in 3x seeds negatively interfere with endosperm cellularization, uncovering a central role of auxin in establishing hybridization barriers by changing the timing of endosperm cellularization.
Materials and methods
Plant material, growth conditions, and treatments
The A. thaliana mutant and reporter lines used were described previously: wei8-1/− tar1/− tar2-1/+ and wei8-1/− tar1/- tar2-2/+ (Stepanova et al. 2008), axr1-12/+ (Dharmasiri et al. 2007), osd1-1 (d'Erfurth et al. 2009), osd1-3 (Heyman et al. 2011), and klu-4 (Adamski et al. 2009).
Seeds were sterilized in 5% commercial bleach and 0.01% Tween-20 for 10 min and washed three times in sterile ddH2O. Sterile seeds were plated on one-half MS-medium (0.43% MS-salts, 0.8% Bacto agar, 0.19% MES hydrate, and 1% sucrose; when necessary, the medium was supplemented with the appropriate antibiotics) and stratified in the dark for 48 h at 4°C. Plates were then transferred to a growth chamber (16 h light/8 h dark; 110 µmol/s/m2; 21°C; 70% humidity). After 10-d seedlings were transferred to soil and grown in a growth chamber (16 h light/8 h dark; 110 µmol/s/m2; 21°C; 70% humidity).
Tetraploid plants were generated by treating 2-wk-old seedlings with 7 µL of 0.25% colchicine. Treated plants were grown to maturity and scored for alterations in pollen size. Seeds of plants showing enlarged pollen grains were collected, and the ploidy of the subsequent generation was determined in a Cyflow ploidy analyzer, using the Cystain UV precise P kit (Sysmex).
Transcriptome analysis
To determine changes in gene expression for lines expressing DD25::IaaH, we extracted total RNA from 6-DAP wild-type and DD25::IaaH seeds, using the MagJET plant RNA purification kit (Thermo Fisher Scientific). We then purified messenger RNA using the NEBNext poly(A) mRNA magnetic isolation module and used this RNA to prepare sequencing libraries with the NEBNext ultra II library preparation kit for Illumina. Three biological replicates for each sample were submitted for Illumina HiSeq 125-bp paired-end sequencing. Mapping was performed according to the method of Moreno-Romero et al. (2016), and differentially regulated genes across the three replicates were detected using DESeq (v1.12.0) (Anders and Huber 2010) as implemented in R (version 3.3.2) (R Development Core Team 2016). The transcriptome sequencing data set of DD25::IaaH seeds has been submitted to the National Center for Biotechnology Information's Gene Expression Omnibus repository (http://www.ncbi.nlm.nih.gov/geo) and is available under accession number GSE124108. Analysis of deregulated genes in 3x seeds was performed using previously published RNAseq data (Schatlowski et al. 2014). We generated lists of overexpressed genes in 3x seeds and DD25::IaaH seeds by filtering all genes with log2 fold change > 1 and P-value < 0.05. These lists were then used to determine enriched GO terms. Significantly enriched biological processes were identified with AtCOECIS (Vandepoele et al. 2009) and further summarized using REVIGO (Supek et al. 2011).
To assess the individual behavior of auxin-related genes in 3x seeds, genes involved in biosynthesis, signaling, and transport of auxin were selected among the gene expression data produced by Schatlowski et al. (2014), and their log2 fold change (3x seeds vs. 2x seeds) values were plotted. Endosperm-specific expression of these genes was assessed using the transcriptome data of isolated endosperm from 3x and 2x seeds (Martinez et al. 2018).
To evaluate expression changes of auxin-related genes throughout endosperm development, we used published transcriptome data (Belmonte et al. 2013). Only auxin-related genes that were expressed at the preglobular stage, in a given endosperm domain, were considered for further analysis. Gene expression values in each endosperm domain and for each time point were then normalized to the preglobular stage and subsequently log transformed. To determine how the expression of these genes is affected in 3x seeds, in which endosperm cellularization is restored, we used previously published transcriptome data of seeds corresponding to the cross Ler × osd1 adm-2 (Wolff et al. 2015).
Expression profiles for genes belonging to GO categories enriched among commonly up-regulated genes in 3x seeds and DD25::IaaH seeds were derived from published data (Belmonte et al. 2013). A gene was considered to be expressed in the endosperm if expression could be detected in at least one endosperm domain within a given time point.
Cloning and generation of transgenic plants
To clone the promoter of DD25 (Steffen et al. 2007), wild-type Col-0 genomic DNA was used as a template. The amplified fragment was purified from the gel, recombined into the donor vector pDONR221, and sequenced. The insert was excised using the restriction sites SacI and SpeI introduced in the primer adaptors and used to replace the CaMV35 promoter in the vector pB7WG2 (Karimi et al. 2002). The IaaH coding sequence was then recombined from an entry vector into pB7WG2, downstream from the DD25 promoter, forming the DD25::IaaH construct. To clone the KLU::YUC6 construct, both the promoter of KLU and the coding sequence of YUC6 were amplified from Col-0 genomic DNA. The amplified fragments were purified, recombined into the donor vector pDONR221, and sequenced. The KLU insert was excised from pDONR221 using the restriction sites SacI and NheI introduced in the primer adaptors and used to replace the CaMV35 promoter in the vector pB7WG2 (Karimi et al. 2002). The YUC6 coding region was then recombined from an entry vector into pB7WG2, downstream from the KLU promoter, forming the KLU::YUC6 construct. Gateway cloning was performed according to the manufacturer's instructions (Life Technologies). All primer sequences can be found in Supplemental Table S2.
The constructs were transformed into Agrobacterium tumefaciens strain GV3101, and Arabidopsis plants were transformed using the floral dip method (Clough and Bent 1998). Transformants were selected with the appropriate antibiotics.
Histological and fluorescence analyses
For clearing of ovules and seeds, the whole pistils/siliques were fixed with EtOH:acetic acid (9:1), washed for 10 min in 90% EtOH and 10 min in 70% EtOH and cleared overnight in chloralhydrate solution (66.7% [w/w] chloralhydrate, 8.3% [w/w] glycerol). The ovules/seeds were observed under differential interference contrast (DIC) optics using a Zeiss Axioplan or Axioscope A1 microscopes. Images were recorded using a Leica DFC295 camera with a 0.63× optical adapter.
For fluorescence analysis, seeds were mounted in 7% glucose. Where indicated, 0.1 mg/mL propidium iodide (PI) was used. Samples were analyzed under confocal microscopy on a Zeiss 780 inverted Axio observer with a supersensitive GaASp detector with the following settings (in nanometers; excitation [ex] and emission [em]): GFP, ex 488 and em 499–525; PI, ex 488/514 and em 635–719; YFP (VENUS), ex 514 and em 499–552 for DR5v2. Images were acquired, analyzed, and exported using Zeiss ZEN software.
For Feulgen staining of seeds, whole siliques were fixed in ethanol:acetic acid (3:1) overnight. The samples were washed three times for 15 min in water, followed by 1-h incubation in freshly prepared 5 N HCl, and were washed again three times for 15 min in water. Staining was performed for 4 h in Schiff reagent, followed by three 15-min washes in cold water and a series of 10-min washes in a series of ethanol dilutions (10%, 30%, and 50%). The samples were then incubated in 70% ethanol overnight, which was followed by a 10-min wash in 95% ethanol and 1 h in 99.5% ethanol. Embedding of the seeds was performed in a dilution series of ethanol:LR White resin (1:3, 1:2, 1:1, and 2:1) for 1 h each. The samples were then incubated overnight in LR White resin, mounted in LR White plus accelerator, and baked overnight at 60°C for polymerization. The seeds were imaged in a Zeiss multiphoton LSM 710 NLO with excitation at 800 nm and emission between 565–610 nm. The images were treated using the ZEN software.
RT-qPCR analyses
For the determination of gene expression of PEGs and AGLs, 10 whole siliques were collected for each cross and frozen in liquid nitrogen. All samples were collected in duplicate. Total RNA was extracted using the MagJET plant RNA purification kit (Thermo Fisher Scientific), and 200 ng of total RNA were used to synthesize cDNA using the RevertAid first strand cDNA synthesis kit (Thermo Fisher Scientific) using an oligo dT primer. Maxima SYBR green qPCR master mix (Thermo Fisher Scientific) was used to perform the qPCR in a CFX Connect system (Bio-Rad). The primers used for the RT-qPCR are described in Supplemental Table S2. PP2A was used as the reference gene. Relative quantification of gene expression was performed as described (Pfaffl 2001).
Supplementary Material
Acknowledgments
We thank Dolf Weijers for providing the DR5v2 reporter before publication, Adrien Sicard for the klu-4 mutant line, and Eva Sundberg and Izabela Cierlik for providing the IaaH pENTRY vector.
Author contributions: D.D.F. and R.A.B. performed the experiments. D.D.F., R.A.B., J.S.-G., and C.K. analyzed the data. D.D.F., R.A.B., and C.K. wrote the paper.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.316554.118.
References
- Adamski NM, Anastasiou E, Eriksson S, O'Neill CM, Lenhard M. 2009. Local maternal control of seed size by KLUH/CYP78A5-dependent growth signaling. Proc Natl Acad Sci 106: 20115–20120. 10.1073/pnas.0907024106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11: R106 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmonte MF, Kirkbride RC, Stone SL, Pelletier JM, Bui AQ, Yeung EC, Hashimoto M, Fei J, Harada CM, Munoz MD, et al. 2013. Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc Natl Acad Sci 110: E435–E444. 10.1073/pnas.1222061110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleckmann A, Alter S, Dresselhaus T. 2014. The beginning of a seed: regulatory mechanisms of double fertilization. Front Plant Sci 5: 452 10.3389/fpls.2014.00452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer DR, Freire-Rios A, van den Berg WAM, Saaki T, Manfield IW, Kepinski S, López-Vidrieo I, Franco-Zorrilla JM, de Vries SC, Solano R, et al. 2014. Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell 156: 577–589. 10.1016/j.cell.2013.12.027 [DOI] [PubMed] [Google Scholar]
- Boisnard-Lorig C, Colon-Carmona A, Bauch M, Hodge S, Doerner P, Bancharel E, Dumas C, Haseloff J, Berger F. 2001. Dynamic analyses of the expression of the HISTONE::YFP fusion protein in Arabidopsis show that syncytial endosperm is divided in mitotic domains. Plant Cell 13: 495–509. 10.1105/tpc.13.3.495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braybrook SA, Peaucelle A. 2013. Mechano-chemical aspects of organ formation in Arabidopsis thaliana: the relationship between auxin and pectin. PLoS One 8: e57813 10.1371/journal.pone.0057813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y. 2007. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19: 2430–2439. 10.1105/tpc.107.053009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- d'Erfurth I, Jolivet S, Froger N, Catrice O, Novatchkova M, Mercier R. 2009. Turning meiosis into mitosis. PLoS Biol 7: e1000124 10.1371/journal.pbio.1000124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Karunarathna N, Jurgens G, Estelle M. 2007. AXL and AXR1 have redundant functions in RUB conjugation and growth and development in Arabidopsis. Plant J 52: 114–123. 10.1111/j.1365-313X.2007.03211.x [DOI] [PubMed] [Google Scholar]
- Dresselhaus T, Sprunck S, Wessel GM. 2016. Fertilization mechanisms in flowering plants. Curr Biol 26: R125–R139. 10.1016/j.cub.2015.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erilova A, Brownfield L, Exner V, Rosa M, Twell D, Mittelsten Scheid O, Hennig L, Köhler C. 2009. Imprinting of the polycomb group gene MEDEA serves as a ploidy sensor in Arabidopsis. PLoS Genet 5: e1000663 10.1371/journal.pgen.1000663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo DD, Batista RA, Roszak PJ, Köhler C. 2015. Auxin production couples endosperm development to fertilization. Nat Plants 1: 15184 10.1038/nplants.2015.184 [DOI] [PubMed] [Google Scholar]
- Figueiredo DD, Batista RA, Roszak PJ, Hennig L, Köhler C. 2016. Auxin production in the endosperm drives seed coat development in Arabidopsis. Elife 5: e20542 10.7554/eLife.20542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehenberger E, Kradolfer D, Köhler C. 2012. Endosperm cellularization defines an important developmental transition for embryo development. Development 139: 2031–2039. 10.1242/dev.077057 [DOI] [PubMed] [Google Scholar]
- Heyman J, Van den Daele H, De Wit K, Boudolf V, Berckmans B, Verkest A, Kamei CLA, De Jaeger G, Koncz C, De Veylder L. 2011. Arabidopsis ULTRAVIOLET-B-INSENSITIVE4 maintains cell division activity by temporal inhibition of the anaphase-promoting complex/cyclosome. Plant Cell 23: 4394–4410. 10.1105/tpc.111.091793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh TF, Shin J, Uzawa R, Silva P, Cohen S, Bauer MJ, Hashimoto M, Kirkbride RC, Harada JJ, Zilberman D, et al. 2011. Regulation of imprinted gene expression in Arabidopsis endosperm. Proc Natl Acad Sci 108: 1755–1762. 10.1073/pnas.1019273108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa R, Ohnishi T, Kinoshita Y, Eiguchi M, Kurata N, Kinoshita T. 2011. Rice interspecies hybrids show precocious or delayed developmental transitions in the endosperm without change to the rate of syncytial nuclear division. Plant J 65: 798–806. 10.1111/j.1365-313X.2010.04466.x [DOI] [PubMed] [Google Scholar]
- Jiang H, Moreno-Romero J, Santos-González J, De Jaeger G, Gevaert K, Van De Slijke E, Köhler C. 2017. Ectopic application of the repressive histone modification H3K9me2 establishes post-zygotic reproductive isolation in Arabidopsis thaliana. Genes Dev 31: 1272–1287. 10.1101/gad.299347.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A. 2002. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195. 10.1016/S1360-1385(02)02251-3 [DOI] [PubMed] [Google Scholar]
- Kradolfer D, Hennig L, Köhler C. 2013a. Increased maternal genome dosage bypasses the requirement of the FIS polycomb repressive complex 2 in Arabidopsis seed development. PLoS Genet 9: e1003163 10.1371/journal.pgen.1003163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kradolfer D, Wolff P, Jiang H, Siretskiy A, Köhler C. 2013b. An imprinted gene underlies postzygotic reproductive isolation in Arabidopsis thaliana. Dev Cell 26: 525–535. 10.1016/j.devcel.2013.08.006 [DOI] [PubMed] [Google Scholar]
- Křeček P, Skůpa P, Libus J, Naramoto S, Tejos R, Friml J, Zažímalová E. 2009. The PIN-FORMED (PIN) protein family of auxin transporters. Genome Biol 10: 249 10.1186/gb-2009-10-12-249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafon-Placette C, Köhler C. 2016. Endosperm-based postzygotic hybridization barriers: developmental mechanisms and evolutionary drivers. Mol Ecol 25: 2620–2629. 10.1111/mec.13552 [DOI] [PubMed] [Google Scholar]
- Lau S, De Smet I, Kolb M, Meinhardt H, Jurgens G. 2011. Auxin triggers a genetic switch. Nat Cell Biol 13: 611–615. 10.1038/ncb2212 [DOI] [PubMed] [Google Scholar]
- Leblanc O, Pointe C, Hernandez M. 2002. Cell cycle progression during endosperm development in Zea mays depends on parental dosage effects. Plant J 32: 1057–1066. 10.1046/j.1365-313X.2002.01491.x [DOI] [PubMed] [Google Scholar]
- Li J, Berger F. 2012. Endosperm: food for humankind and fodder for scientific discoveries. New Phytol 195: 290–305. 10.1111/j.1469-8137.2012.04182.x [DOI] [PubMed] [Google Scholar]
- Li SB, Xie ZZ, Hu CG, Zhang JZ. 2016. A review of auxin response factors (ARFs) in plants. Front Plant Sci 7: 47 10.3389/fpls.2016.00047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao C-Y, Smet W, Brunoud G, Yoshida S, Vernoux T, Weijers D. 2015. Reporters for sensitive and quantitative measurement of auxin response. Nat Methods 12: 207–210. 10.1038/nmeth.3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Wang H. 2005. Two homologous ATP-binding cassette transporter proteins, AtMDR1 and AtPGP1, regulate Arabidopsis photomorphogenesis and root development by mediating polar auxin transport. Plant Physiol 138: 949–964. 10.1104/pp.105.061572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Dennis ES, Berger F, Peacock WJ, Chaudhury A. 2005. MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc Natl Acad Sci 102: 17531–17536. 10.1073/pnas.0508418102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lur HS, Setter TL. 1993. Role of auxin in maize endosperm development (timing of nuclear DNA endoreduplication, zein expression, and cytokinin). Plant Physiol 103: 273–280. 10.1104/pp.103.1.273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield S, Briarty L. 1990. Endosperm cellularization in Arabidopsis thaliana (L.). Arab Inf Serv 27: 65–72. [Google Scholar]
- Martinez G, Wolff P, Wang Z, Moreno-Romero J, Santos-González J, Conze LL, DeFraia C, Slotkin RK, Köhler C. 2018. Paternal easiRNAs regulate parental genome dosage in Arabidopsis. Nat Genet 50: 193–198. 10.1038/s41588-017-0033-4 [DOI] [PubMed] [Google Scholar]
- Moreno-Romero J, Jiang H, Santos-González J, Köhler C. 2016. Parental epigenetic asymmetry of PRC2-mediated histone modifications in the Arabidopsis endosperm. EMBO J 35: 1298–1311. 10.15252/embj.201593534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollmann S, Müller A, Piotrowski M, Weiler EW. 2002. Occurrence and formation of indole-3-acetamide in Arabidopsis thaliana. Planta 216: 155–161. 10.1007/s00425-002-0868-4 [DOI] [PubMed] [Google Scholar]
- R Development Core Team. 2016. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. [Google Scholar]
- Robert HS, Grones P, Stepanova AN, Robles LM, Lokerse AS, Alonso JM, Weijers D, Friml J. 2013. Local auxin sources orient the apical-basal axis in Arabidopsis embryos. Curr Biol 23: 2506–2512. 10.1016/j.cub.2013.09.039 [DOI] [PubMed] [Google Scholar]
- Schatlowski N, Wolff P, Santos-González J, Schoft V, Siretskiy A, Scott R, Tamaru H, Köhler C. 2014. Hypomethylated pollen bypasses the interploidy hybridization barrier in Arabidopsis. Plant Cell 26: 3556–3568. 10.1105/tpc.114.130120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RJ, Spielman M, Bailey J, Dickinson HG. 1998. Parent-of-origin effects on seed development in Arabidopsis thaliana. Development 125: 3329–3341. [DOI] [PubMed] [Google Scholar]
- Sekine D, Ohnishi T, Furuumi H, Ono A, Yamada T, Kurata N, Kinoshita T. 2013. Dissection of two major components of the post-zygotic hybridization barrier in rice endosperm. Plant J 76: 792–799. 10.1111/tpj.12333 [DOI] [PubMed] [Google Scholar]
- Steffen JG, Kang IH, Macfarlane J, Drews GN. 2007. Identification of genes expressed in the Arabidopsis female gametophyte. Plant J 51: 281–292. 10.1111/j.1365-313X.2007.03137.x [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Doležal K, Schlereth A, Jürgens G, Alonso JM. 2008. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133: 177–191. 10.1016/j.cell.2008.01.047 [DOI] [PubMed] [Google Scholar]
- Stoute AI, Varenko V, King GJ, Scott RJ, Kurup S. 2012. Parental genome imbalance in Brassica oleracea causes asymmetric triploid block. Plant J 71: 503–516. 10.1111/j.1365-313X.2012.05015.x [DOI] [PubMed] [Google Scholar]
- Sugawara S, Hishiyama S, Jikumaru Y, Hanada A, Nishimura T, Koshiba T, Zhao Y, Kamiya Y, Kasahara H. 2009. Biochemical analyses of indole-3-acetaldoxime-dependent auxin biosynthesis in Arabidopsis. Proc Natl Acad Sci 106: 5430–5435. 10.1073/pnas.0811226106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Škunca N, Šmuc T. 2011. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6: e21800 10.1371/journal.pone.0021800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinland B, Kares C, Herrmann A, Otten L. 1991. 35S-β-glucuronidase gene blocks biological effects of cotransferred iaa genes. Plant Mol Biol 16: 853–864. 10.1007/BF00015077 [DOI] [PubMed] [Google Scholar]
- Vandepoele K, Quimbaya M, Casneuf T, De Veylder L, Van de Peer Y. 2009. Unraveling transcriptional control in Arabidopsis using cis-regulatory elements and coexpression networks. Plant Physiol 150: 535–546. 10.1104/pp.109.136028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wangenheim KH. 1962. Zur ursache der abortion von samenanlagen in diploid-polyploid kreuzungen. Zeitschrfft lfi r Vererbungslehre 93: 319–334. [Google Scholar]
- Weijers D, Wagner D. 2016. Transcriptional responses to the auxin hormone. Annu Rev Plant Biol 67: 539–574. 10.1146/annurev-arplant-043015-112122 [DOI] [PubMed] [Google Scholar]
- Wolff P, Weinhofer I, Seguin J, Roszak P, Beisel C, Donoghue MT, Spillane C, Nordborg M, Rehmsmeier M, Köhler C. 2011. High-resolution analysis of parent-of-origin allelic expression in the Arabidopsis endosperm. PLoS Genet 7: e1002126 10.1371/journal.pgen.1002126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff P, Jiang H, Wang G, Santos-González J, Köhler C. 2015. Paternally expressed imprinted genes establish postzygotic hybridization barriers in Arabidopsis thaliana. Elife 4: e10074 10.7554/eLife.10074 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.