Abstract
Protein biosynthesis is energetically costly, is tightly regulated and is coupled to stress conditions including glucose deprivation. RNA polymerase III (RNAP III)-driven transcription of tDNA genes for production of tRNAs is a key element in efficient protein biosynthesis. Here we present an analysis of the effects of altered RNAP III activity on the Saccharomyces cerevisiae proteome and metabolism under glucose-rich conditions. We show for the first time that RNAP III is tightly coupled to the glycolytic system at the molecular systems level. Decreased RNAP III activity or the absence of the RNAP III negative regulator, Maf1 elicit broad changes in the abundance profiles of enzymes engaged in fundamental metabolism in S. cerevisiae. In a mutant compromised in RNAP III activity, there is a repartitioning towards amino acids synthesis de novo at the expense of glycolytic throughput. Conversely, cells lacking Maf1 protein have greater potential for glycolytic flux.
Keywords: amino acid metabolism, comparative proteomics, glycolysis, maf1, RNA polymerase III
Introduction
Regulation of glycolytic flux is a long-standing, but still highly relevant, issue in biology and pathobiology. Glycolytic performance is connected to enzymes abundance, cell fermentative activity and proliferation, all hallmarks of the ‘Warburg effect’. Both Saccharomyces cerevisiae and mammalian cells can sense glycolytic state/flux intracellularly, a dominant signal over that of external nutritional status [1–4]. In S. cerevisiae under favorable growth conditions, high glycolytic activity elicits rapid cell growth, due to the robust synthesis of proteins and biomass expansion [5–7]. Nutrient-limited growth, on the other hand, is associated with a down-regulation of transcription and protein synthesis to reduce demands on the ribosomal machinery and an appropriate supply of amino acids and tRNAs.
As key players in protein synthesis, transfer RNAs are synthesized by RNA polymerase III (RNAP III), which is also responsible for the transcription of other specific products such as ribosomal 5S rRNA and spliceosomal U6 snRNA. RNAP III activity is regulated by extracellular glucose levels [8,9]. The only known direct regulatory factor of RNAP III in S. cerevisiae is the protein Maf1, a mediator of a range of stress signals [10–13] conserved from yeast to human [14]. Yeast Maf1 inhibits RNAP III activity reversibly under carbon source starvation and oxidative stress, reducing tRNA transcript levels [15]. Although the MAF1 gene is not essential for yeast viability, maf1Δ cells are unable to repress RNAP III [15–18].
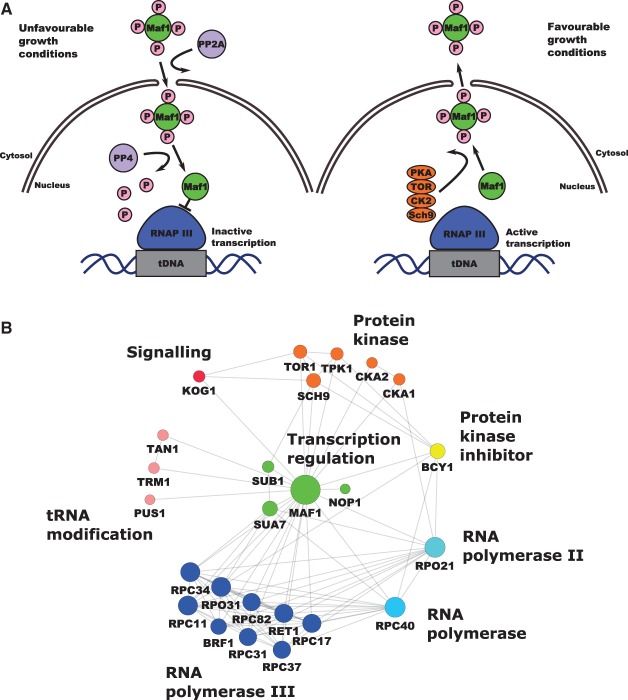
Under favorable growth conditions, Maf1 is an interaction partner of several cytoplasmic proteins playing different biological functions (Figure 1A,B), but its function in the cytoplasm is unknown. Maf1 is a target of several kinases and phosphorylation patterns may dictate cellular localization [15,41–47] (Figure 1B).
Figure 1. RNAP III regulation by Maf1 and Maf1 interaction network.
(A) RNAP III transcription repression is regulated by Maf1. Phosphorylation and dephosphorylation events are involved in the mobility and transportation of Maf1 through the nuclear membrane in which a group of protein kinases are involved in the control of Maf1 nuclear localization responding to stress events. Maf1 produces transcriptional repression on RNAP III by inducing conformational changes. (B) Maf1 protein–protein interaction network. Experimental interactions from STRING database are shown. Nodes have been colored by protein activity in which different protein complexes related to tRNA modification and transportation can be observed. green: transcription regulation; MAF1: negative regulator of RNAP III, SUB1: Sub1 transcriptional regulator facilitating elongation through factors that modify RNAP II, role in hyper-osmotic stress response through RANP II and RNAP III, negatively regulates sporulation [19–21], NOP1: Nop1, histone glutamine methyltransferase, modifies H2A at Q105 in nucleolus that regulates transcription from the RNAP I promoter involved in C/D snoRNA 3′-end processing. Essential gene leads to reduced levels of pre-rRNA species and defects in small ribosomal subunits biogenesis [22–24], SUA7: transcriptional factor TFIIB, a general transcription factor required for transcription initiation and start site selection by RNAP II [25,26] — Sub1 interaction with TFIIB [27]. Marine blue: RNAP III holoenzyme subunits, red: protein kinases, KOG1: Kog1 the component of the TPR complex, Kog1 depletion display the starvation-like phenotypes — cell growth arrest, reduction in protein synthesis, glycogen accumulation, up-regulation in the transcription of nitrogen catabolite repressed and retrograde responses genes conserved in from yeast to man is the homolog of the mammalian TORC1 regulatory protein RAPTOR/mKOG1 [28,29], TOR1 mediates cell growth in response to nutrient availability and cellular stress by regulating protein synthesis, ribosome biogenesis, autophagy, transcription activation cell cycle [30,31] . Yellow: PKA kinase inhibitor protein BCY1, pink: tRNA modification TAN1: tRNA modifying proteins Tan1 (responsible for tRNASER turnover [32]), TRM1: Trm1 tRNA methyltranspherase produces modified base N2, n2 dimethylguanosine in tRNA in nucleus and mitochondrion [33], PUS1: PUS1 associated with human disease [34], introduces pseudouridines in tRNA, also as on U2 snRNA and pseudouridylation of some mRNA [35,36]. Blue: RPC40 (AC40) is a common subunit to RNAP I and III conserve in all eukaryotes [37,38]. Light blue: RPO21: largest subunit of RNAP II, which produces all nuclear mRNAs, most snoRNAs and snRNA and the telomerase RNA encoded by TLC1 [39,40] (according to Saccharomyces Genome Database).
Growth-limiting conditions result in dephosphorylation of Maf1 by the PP2A and PP4 phosphatases [15,48]. Under unfavorable growth conditions, dephosphorylated Maf1 protein is imported to the nucleus in a process driven by nuclear localization sequences (NLS) [15,42]. In the nucleus, Maf1 binds to the RNAP III complex after transcription initiation (for review, see Boguta [49]). Such interaction results in RNAP III dissociation from tRNA promoter sequences [14,15,43]. On the other hand, favorable growth conditions induce Maf1 inactivation as a repressor by multiple phosphorylation due to casein kinase II (CK2), protein kinase A (PKA), Sch9 and TOR following specific stress signals, which affects adversely the binding of Maf1 to RNAP III [47,50]. In favorable growth conditions, Maf1 is localized predominantly in the cytoplasm, although is never fully excluded from the nucleus [51].
Although MAF1 deletion is not lethal under optimal growth conditions, deletion mutants display high tRNA transcription with diminished growth on non-fermenting carbon sources at 30°C; it becomes lethal, however, at elevated temperatures. The low growth rate results from a decrease in steady-state mRNA levels of FBP1 and PCK1 genes encoding the key gluconeogenesis enzymes fructose 1,6-bisphosphatase (Fbp1) and phosphoenolpyruvate carboxykinase (Pck1) [10,52]. Intriguingly, this maf1Δ growth defect on non-fermentable carbon sources is suppressed by point mutation (rpc128-1007) in the second largest RNAP III subunit RET1/C128 [13]. tRNA transcription levels in this rpc128-1007 mutant are very low, which suggests that the temperature-sensitive lethality of maf1Δ can be rescued by attendant reduction in RNAP III activity, or a critical process affected by this transcription. The maf1Δ and rpc128-1007 strains have different phenotypes, not only in growth on non-fermentable carbon sources but also in preference towards glucose utilization, in excess glucose [13,53]. Transcription of the high-affinity glucose transporter genes HXT6, HXT7 is decreased in maf1Δ, but increased over WT in the maf1Δ second-site suppressor rpc128-1007 [53], suggesting differences in glucose utilization.
We wished to explore the potential for a feedback loop between control of glycolytic flux and RNAP III in yeast cells by label-free proteomics, which revealed changes in abundance of a large group of proteins in maf1Δ and rpc128-1007 strains, supported with targeted analysis of specific metabolites. We provide novel molecular data which is able to explain the severe reduction in growth rate caused by RNAP III mutation rpc128-1007 through cellular processes that facilitate efficient glucose metabolism in the MAF1 deletion strain on glucose. Changes in protein profiles impact several metabolic pathways, suggesting differences in cellular metabolic homeostasis in the mutant strains and providing an alternative explanation for maf1Δ lethality on non-fermentable carbon sources. Finally, using yeast as a model organism, which is often used for studies of the ‘Warburg effect’, we established direct metabolic relationship between the capacity of the glycolytic pathway and transcription of non-coding genes, which can explain why several cancerous cell lines exhibit higher RNAP III activity, creating a new perspective on glucose flux modification via manipulation of the RNAP III holoenzyme as a novel therapeutic strategy.
Materials and methods
Yeast strains and media
The following strains were used: wild-type MB159-4D [54] with unchanged RNAP III activity, the MA159-4D maf1::URA3 [53] MAF1-deficient mutant with elevated RNAP III activity and MJ15-9C mutant [13] with a single point mutation in the RET1/C128 RNAP III subunit with reduced polymerase activity. Yeast strains were cultured in rich medium (YP; 1% yeast extract, 1% peptone) supplemented with either 2% glucose (YPD) or 2% glycerol (YPGly) as a carbon source. Overnight cell cultures were grown in YPD medium. Cells were harvested by centrifugation (2000 rpm, RT) and washed twice with fresh, sterile YPD or YPGly medium. Yeast cells were diluted to D600 ≈ 0.1 and grown in YPD or YPGly until exponential phase (D600 ≈ 1.0). All yeast cultures were incubated at 30°C with agitation 250 rpm. GCN4-3HA DNA construct for chromosomal C-terminus fusion was prepared as described previously [53,55]. Hexokinase isoforms (HXK1, HXK2, GLK1) single and double gene deletions were created by transforming haploid yeast strains with appropriate PCR fragments. For HXK1 deletion amplification of His3MX6 cassette on pFA6-VC155-His3MX6 plasmid DNA was done. DNA constructs for obtaining HXK2- and GLK1-deficient strains were amplified on gDNA of BY4741 glk1Δ and BY4741 hxk2Δ (Euroscarf). High-efficiency yeast transformation using LiAc/SS carrier DNA/PEG method was used according to Gietz and Schiestl [56]. All yeast strains are listed in Table 1.
Table 1. Yeast strains used in the study.
| Strain | Genotype | Reference/source |
|---|---|---|
| MB159-4D | MATa SUP11 ura3 leu2-3, 112 ade2-1 lys2-1 trp | [54] |
| MA159-4D maf1Δ | MATa SUP11 ura3 leu2-3, 112 ade2-1 lys2-1 trp maf1::URA3 | [53] |
| MB159-4D maf1Δ | MATa SUP11 ura3 leu2-3, 112 ade2-1 lys2-1 trp maf1::kanMX6 | [54] |
| MJ15-9C | MATa rpc128-1007 SUP11 ura3 leu2-3, 112 ade2-1 lys2-1 trp | [13] |
| RS159-4D Gcn4-3HA | MATa SUP11 ura3 leu2-3, 112 ade2-1 lys2-1 trp Gcn4-3HA-kanMX6 | This study |
| RS15-9C Gcn4-3HA | MATa rpc128-1007 SUP11 ura3 leu2-3, 112 ade2-1 lys2-1 trp Gcn4-3HA-kanMX6 | This study |
| RS159-4D maf1Δ Gcn4-3HA | MATa SUP11 ura3 leu2-3, 112 ade2-1 lys2-1 trp maf1::URA3 Gcn4-3HA-kanMX6 | This study |
| KR159-4D hxk1Δ | MATa SUP11 ura3 leu2-3, 112 ade2-1 lys2-1 trp hxk1::HIS3MX6 | This study |
| KR159-4D maf1Δ hxk1Δ | MATa SUP11 ura3 leu2-3, 112 ade2-1 lys2-1 trp maf1::URA3 hxk1::HIS3MX6 | This study |
| KR15-9C hxk1Δ | MATa rpc128-1007 SUP11 ura3 leu2-3, 112 ade2-1 lys2-1 trp hxk1::HIS3MX6 | This study |
| KR159-4D hxk1Δ hxk2Δ | MATa SUP11 ura3 leu2-3, 112 ade2-1 lys2-1 trp hxk1::HIS3MX6 hxk2::kanMX4 | This study |
| KR159-4D maf1Δ hxk1Δ hxk2Δ | MATa SUP11 ura3 leu2-3, 112 ade2-1 lys2-1 trp maf1::URA3 hxk1::HIS3MX6 hxk2::kanMX4 | This study |
| KR15-9C hxk1Δ hxk2Δ | MATa rpc128-1007 SUP11 ura3 leu2-3, 112 ade2-1 lys2-1 trp hxk1::HIS3MX6 hxk2::kanMX4 | This study |
| KR159-4D hxk2Δ | MATa SUP11 ura3 leu2-3, 112 ade2-1 lys2-1 trp hxk2::kanMX4 | This study |
| KR159-4D maf1Δ hxk2Δ | MATa SUP11 ura3 leu2-3, 112 ade2-1 lys2-1 trp maf1::URA3 hxk2::kanMX4 | This study |
| KR15-9C hxk2Δ | MATa rpc128-1007 SUP11 ura3 leu2-3, 112 ade2-1 lys2-1 trp hxk2::kanMX4 | This study |
| KR159-4D hxk1Δ glk1Δ | MATa SUP11 ura3 leu2-3, 112 ade2-1 lys2-1 trp hxk1::HIS3MX6 glk1::kanMX4 | This study |
| KR159-4D maf1Δ hxk1Δ glk1Δ | MATa SUP11 ura3 leu2-3, 112 ade2-1 lys2-1 trp maf1::URA3 hxk1::HIS3MX6 glk1::kanMX4 | This study |
| KR15-9C hxk1Δ glk1Δ | MATa rpc128-1007 SUP11 ura3 leu2-3, 112 ade2-1 lys2-1 trp hxk1::HIS3MX6 glk1::kanMX4 | This study |
| RS159-4D glk1Δ | MATa SUP11 ura3 leu2-3, 112 ade2-1 lys2-1 trp glk1::kanMX4 | This study |
| RS159-4D maf1Δ glk1Δ | MATa SUP11 ura3 leu2-3, 112 ade2-1 lys2-1 trp maf1::URA3 glk1::kanMX4 | This study |
| RS15-9C glk1Δ | MATa rpc128-1007 SUP11 ura3 leu2-3, 112 ade2-1 lys2-1 trp glk1::kanMX4 | This study |
| BY4741 hxk2Δ | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hxk2::kanMX4 | Euroscarf |
| BY4741 glk1Δ | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 glk1::kanMX4 | Euroscarf |
| BY4741 reg1Δ | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 reg1::kanMX4 | Euroscarf |
| MB159-4D [pBM2636] | MATa SUP11 ura3 leu2-3, 112 ade2-1 lys2-1 trp [HXT1::lacZ] | This study |
| MB159-4D maf1Δ [pBM2636] | MATa SUP11 ura3 leu2-3, 112 ade2-1 lys2-1 trp maf1::KanMX6 [HXT1::lacZ] | This study |
| MB15-9C [pBM2636] | MATa rpc128-1007 SUP11 ura3 leu2-3, 112 ade2-1 lys2-1 trp [HXT1::lacZ] | This study |
Proteomic analysis
Samples (∼15 ml culture medium) — corresponding to 25 × 106 cells as determined in a cell count on hemocytometer were analyzed by global label-free proteomics (four biological replicates per strain). Cells were spun down (10 min, 4°C) and the pellets flash frozen in liquid N2 for storage. Cells were resuspended in 50 mM NH4HCO3, protease inhibitor ROCHE mini complete protease inhibitor. The samples were homogenized with Mini-Beadbeater 24 (Biospec products) using the 200 µl of glass beads (425–600 µm; Sigma–Aldrich) 15 times (3000 hits per min) with a duration of 30 s each with a 1 min cool down period in between each cycle. The cells were further centrifuged (10 min, 13 000 rpm, 4°C). 250 µl fresh breaking buffer was added to pellets and cells were washed by vigorous vortexing. The wash and cell debris were collected as flow through. Each flow through and supernatants from previous steps were combined. Protein concentration was determined with a Bradford assay [57].
A volume equivalent to 25 × 106 cells of each homogenate was removed, diluted with 25 mM AMBIC containing 0.05% Rapigest (Waters, Manchester) and shaken (550 rpm, 10 min, 80°C). The samples were then reduced (addition of 10 µl of 60 mM DTT and incubation at 60°C for 10 min) and alkylated (addition of 10 µl of 180 mM iodoacetamide and incubation at room temperature for 30 min in the dark). Trypsin (Sigma, Poole, U.K., proteomics grade) was reconstituted in 50 mM acetic acid to a concentration of 0.2 µg/µl and 10 µl was added to the sample followed by overnight incubation at 37°C. The digestion was terminated and RapiGest™ removed by acidification (1 µl of TFA and incubation at 37°C for 45 min) and centrifugation (15 000×g, 15 min). To check for complete digestion each sample was analyzed pre- and post-acidification by SDS–PAGE.
For LC–MS/MS analysis, a 2 µl injection of each digest, corresponding to approximately 25 × 104 cells, was analyzed using an Ultimate 3000 RSLC™nano system (Thermo Scientific, Hemel Hempstead) coupled to a QExactive™ mass spectrometer (Thermo Scientific). The sample was loaded onto the trapping column (Thermo Scientific, PepMap100, C18, 300 µm × 5 mm), using partial loop injection, for 7 min at a flow rate of 4 µl/min with 0.1% (v/v) FA. The sample was resolved on the analytical column (Easy-Spray C18 75 µm × 500 mm 2 µm column) using a gradient of 97% A (99.9% water: 0.1% formic acid) 3% B (99.9% ACN: 0.1% formic acid) to 60% A: 40% B over 90 min at a flow rate of 300 nL min−1. The data-dependent program used for data acquisition consisted of a 70 000 resolution full-scan MS scan (AGC set to 1e6 ions with a maximum fill time of 250 ms) the 10 most abundant peaks were selected for MS/MS using a 17 000 resolution scan (AGC set to 5e4 ions with a maximum fill time of 250 ms) with an ion selection window of 3 m/z and a normalized collision energy of 30. To avoid repeated selection of peptides for MS/MS the program used a 30 s dynamic exclusion window.
Label-free quantification
The raw data from the mass spectrometer was then processed using MaxQuant (MQ) software version 1.5.3.30 [58]. Protein identification was performed with the built-in Andromeda search engine, searching MS/MS spectra vs. the S. cerevisiae strain ATCC 204508/S288c downloaded from UniProt (https://www.uniprot.org/proteomes/UP000002311). The following parameters were used; digest reagent: trypsin, maximum missed cleavages: 2, modifications: protein N-terminal acetylation and methionine oxidation, with a maximum of five modifications per peptide. The false discovery rate (FDR) for accepted peptide-spectrum matches and protein matches was set to 1%. For protein quantification, the ‘match between runs’ options was selected. Label-free quantification was performed with the MaxLFQ algorithm within MaxQuant (MQ), based on razor and unique peptides. All other MQ parameters were left at default values.
Protein significance testing
To determine statistically significantly changing proteins with respect to the wild-type strain we used the MSstats package [59] in the R environment. Protein identities, conditions, biological replicates and intensities were directly uploaded from the MaxQuant output. Protein ID information was obtained from the ‘proteinGroups.txt’ file, conditions and biological replicates from the ‘annotation.csv’ file, and intensities from the ‘evidence.txt’ file. Data normalization was performed using the ‘equalizeMedians’ option and summarization using the Tukey's median polish option. Following this, a condition comparison was performed using the ‘groupComparison’ option from where the log2 fold changes and adjusted P-values were obtained. Adjusted P-values are used for statistically significance. The adjusted P-values are the P-values adjusted among all the proteins present in the specific comparisons using the Benjamini and Hochberg approach.
Functional analysis
Gene ontology enrichment analysis was performed with the online application Panther [60], directly on the Gene Ontology Consortium webpage (http://pantherdb.org/). The background set consisted of all proteins identified in a given MS experiment. Protein changes were mapped to central carbon and amino acid metabolic pathways following KEGG database [61] guidelines. Maf1 protein–protein interactions were obtained from the STRING [62] database and clustered with the Cytoscape [63] tool.
Transcription factor target enrichment analysis
For transcription factor (TF) target enrichment analysis, all proteins with an adjusted P-value below 0.05 from both comparisons (WT — rpc128-1007 and WT — maf1Δ) were uploaded to the GeneCodis tool [64]. Proteins were classified according to their positive or negative fold change and the background set consisted of all proteins identified in the given MS experiment. All statistical parameters were left as default. Adjusted P-values were obtained indicating those statistically significantly TFs being active according to their known target proteins.
Western blotting
The total cellular proteins from Gcn4-3HA expressing yeast cells were extracted as described previously [53]. Protein extracts were separated by 12% SDS–PAGE and transferred to nitrocellulose membrane by electrotransfer (1 h, 400 mM, 4°C). For detection of HA-tagged proteins, monoclonal mouse anti-HA (1:3330, Sigma, H3663) and polyclonal goat anti-mouse antibodies (1:2000, Dako P0447) conjugated with horseradish peroxidase (HRP) were used. For Vma2 protein detection, monoclonal mouse anti-Vma2 antibodies (1:4000, Life Technologies, A6427) were used.
RNA isolation and real-time PCR quantification
RNA isolation and real-time PCR amplification were performed as described previously [53]. Isolated RNAs were examined by SYBR-Green-based real-time PCR. Oligonucleotide sequences of the primers used in real-time PCR experiment for GCN4 were taken from Cankorur-Cetinkaya et al. [65]. Samples were normalized to two reference genes — U2 spliceosomal RNA (U2) and small cytosolic RNA (SCR1). Expression levels in WT strain (MB159-4D) were taken as 1.0. The relative expression (mean ± SD) was calculated for at least three independent biological replicates. Statistical significance of P-values was determined by Student's t-test.
Enzymatic assays
All yeast strains including transformants carrying pBM2636 plasmid [66] for measurement of β-galactosidase activity were cultivated in rich medium supplemented with 2% glucose (YPD) or 2% glycerol (YPGly) at 30°C with agitation of 250 rpm until reached D600 ≈ 1.0. Yeast cultures were harvested at 5000 rpm at 4°C and washed twice with 10 mM potassium phosphate buffer (pH 7.5). Cells for Hxk, Tdh1-3, Cdc19 and Zwf1 activity assays were suspended in 100 mM potassium phosphate buffer (pH 7.5), for β-galactosidase in 50 mM potassium phosphate buffer (pH 7.0), rapidly frozen in liquid nitrogen and stored at −20°C. Samples were washed twice with sonication buffer (100 mM potassium phosphate buffer (pH 7.5), 2 mM MgCl2) or 50 mM potassium phosphate buffer (pH 7.0) in the second case and disintegrated with Mini-Beadbeater 24 (Biospec products) using glass beads (425–600 µm; Sigma–Aldrich). Hexokinase (EC 2.7.1.1) activity was measured according to Adamczyk et al. [67], glyceraldehyde-3-phosphate dehydrogenase (Tdh1-3, EC 1.2.1.12) according to van Hoek et al. [68], pyruvate kinase (Cdc19; EC 2.7.1.40) according to Grüning et al. [69], glucose-6-phosphate dehydrogenase (Zwf1, EC 1.1.1.49) according to Postma et al. [70], β-galactosidase according to Smale [71], and catalase (EC 1.11.1.6) according to Beers and Sizer [72]. All assays were performed for a minimum of three independent biological replicates.
Glycogen, trehalose, fructose 1,6-bisphosphate and glutathione measurement
The glycogen and trehalose content was measured in yeast cells, grown in YPD until D600 ≈ 1.0. Cell preparation and extraction was as described in Rossouw et al. [73]. Glycogen determination was as described by Parrou and François [74]. Glucose concentration from glycogen enzymatic breakdown was determined by the glucose (HK) Assay Kit according to the manufacturer's protocol (Sigma–Aldrich, GAHK-20). Trehalose content was measured using Trehalose Assay Kit (Megazyme International Ireland, Wicklow, Ireland) according to the manufacturer's protocol. Fructose 1,6-bisphosphate was measured according to Peeters et al. [4] with minor modifications. Oxidized (GSSG), reduced (GSH) and total glutathione (oxidized and reduced) levels were measured according to Quantification kit for oxidized and reduced glutathione (Sigma–Aldrich, 38185) as stated in manufacturer's protocol.
Determination of yeast fermentative capacity
Fermentative capacity assays were performed as described by van Hoek et al. [75] with minor changes. The fermentative capacity can be defined as the specific maximal production rate of ethanol per gram of biomass (mmol/g/h) under anaerobic conditions at excess of glucose. Samples corresponding to 60–70 mg dry weight were harvested by centrifugation at 5000 rpm at 4°C. Cells were washed twice with synthetic medium CBS-without carbon source (CBS-C) and resuspended in CBS (-C) to make 2% wet weight suspensions. Analysis was performed in a thermostatted (30°C) vessel. Cells were flushed with N2 gas at a flow rate of ∼0.6 L/h and glucose was added to a final concentration of 10 g/liter. Samples for measurement of ethanol were collected every 5 min, incubated with 35% (w/v) perchloric acid on ice for 10 min and neutralized with KOH before centrifugation at 13 000 rpm and stored in −20°C freezer. The ethanol production of each strain was normalized to the dry weight of the culture. Ethanol and glycerol in supernatants were determined with enzymatic assays according to the manufacturer (Megazyme International Ireland, Wicklow, Ireland).
Biomass determination
Sample suspensions of 1 ml volume in duplicates were filtered over pre-weighted nitrocellulose filters (pore size, 0.45 µm; HAWP04700). After removal of medium, the filters were washed with demineralized water, dried in an oven overnight and weighed [75].
Results
Overall proteome profiling and changes
We hypothesized that perturbations in RNAP III activity would impact on global expression of the proteome. The lack of the negative regulator of RNAP III, Maf1, as well as the loss of function due to a point mutation in the RNAP III RET1/C128 subunit, would be expected to elicit broad changes in the S. cerevisiae proteome. We aimed to identify proteins, the changed abundance of which, could explain the diminished growth of rpc128-1007 on glucose and de-repression of HXT2 and HXT6/7 genes that encode high-affinity glucose transporters, when compared with maf1Δ under glucose-rich conditions [53]. We, therefore, performed a systematic comparative analysis of maf1Δ and rpc128-1007 mutants using label-free proteomics.
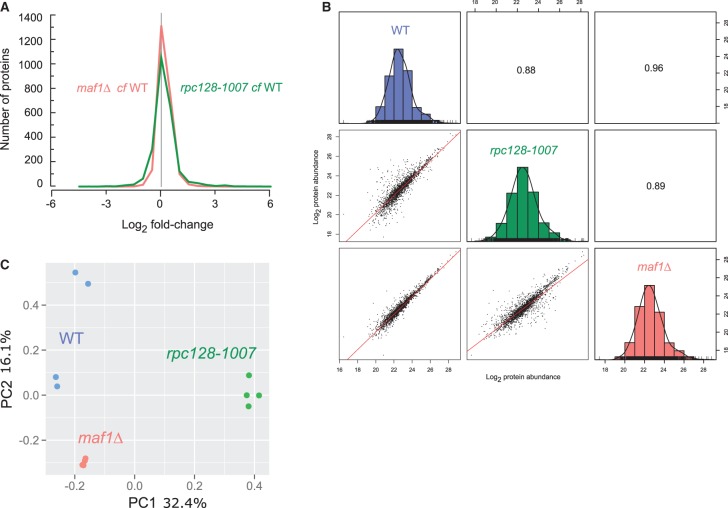
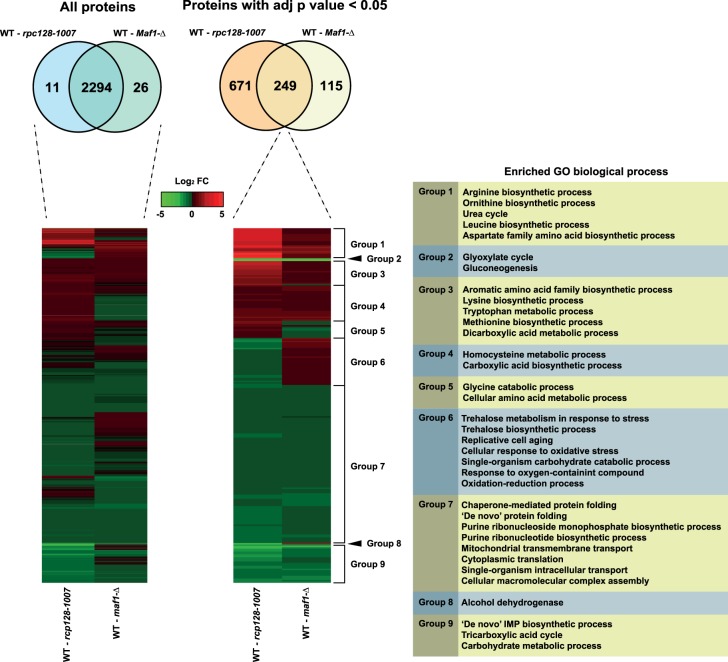
Reproducible, deep proteome coverage was obtained (Figure 2A) for maf1Δ and rpc128-1007 mutants grown under glucose-rich conditions. The proteomics data were of high quality, with replicates clustered together and there was no systematic difference reflecting sample preparation bias (Figure 2). In total, over 2300 protein groups were identified and quantified. As anticipated, there was considerable overlap between the maf1Δ and rpc128-1007 proteomes (Figure 3). Differential proteome analysis was carried out pairwise in two sets as follows: first: WT vs rpc128-1007 mutant and secondly: WT vs. maf1Δ mutant, resulting in 2294 quantified proteins common to all strains. A subset of statistically significant changes revealed 249 proteins that were common to both comparisons (with an adjusted P-value < 0.05). This subset of 249 proteins were clustered into coherent groups (see Materials and methods) that display common Gene Ontology (GO) annotation, consistent with co-ordinated regulation of relevant biological processes (Figure 3). Some of the groups exhibited parallel changes in the two strains (Groups 1–4) whereas others highlighted divergent, essentially reciprocal, functions in the two strain (i.e. Groups 5–6). These unbiased clusters show enrichments for concerted biological functions, embodied by the limited subset of GO term enrichments listed in Figure 3, including elements of amino acid and monosaccharide/carbohydrate metabolism. In both strains, enzymes of gluconeogenesis and the glyoxylate cycle were decreased when compared with the reference strain grown under the same glucose repression conditions (Group 2). Groups 7 and 9 also show decreased protein abundance with respect to wild type and are similarly enriched in enzymes from the TCA cycle, purine ribonucleotide biosynthetic pathways, inosine monophosphate biosynthesis de novo, and also mitochondrial transmembrane transport. In contrast, many of the enzymes involved in amino acid synthesis de novo were increased in abundance in both maf1Δ and rpc128-1007 (Groups 1, 3 and 4). The negatively correlated, reciprocally altered groups (Groups 5, 6 and 8) were consistent with shifts in trehalose biosynthesis, pentose phosphate pathway (PPP) activity, oxidative stress, oxidation–reduction processes, glycine catabolic processes, replicative cell aging and alcohol production.
Figure 2. Proteome signature of maf1Δ and rpc128-1007 mutants compared with the wild-type strain.
(A) Histogram of proteins present on both mutants organized according to their corresponding Log2 fold change expression. (B) Comparative scatter plots and histograms of the different strains. The Log2 transformed protein abundances of proteins present in the WT, rpc128-1007 and maf1Δ strains are plotted against one another along with their distribution. The number shown is the Pearson correlation coefficient between the two relevant strains. (C) Principal component analysis (PCA) based on proteins present on all four biological replicates.
Figure 3. Increased and decreased protein abundance is presented relative to the wild-type strain for both maf1Δ and rpc128-1007 mutants.
All those statically significant proteins with an adjusted P-value < 0.05 overlapped between both comparisons were then subjected to hierarchical clustering. This clustering analysis created different groups showing the similarities and differences between both mutants with clusters enriching to biological processes related to amino acid and carbohydrate metabolism, response to stress, and respiratory processes.
Key enzymes of the glyoxylate cycle are reduced in abundance in both maf1Δ and rpc128-1007 mutants.
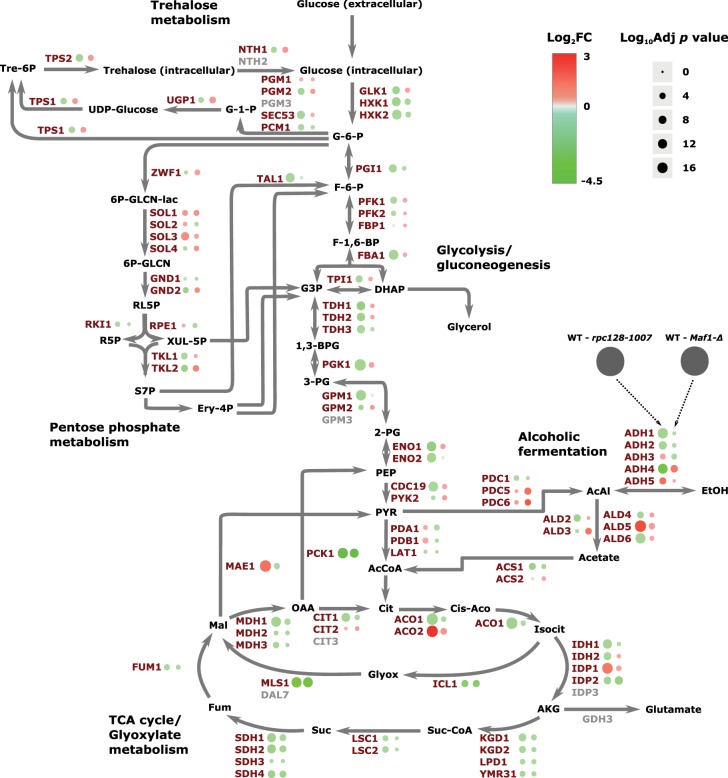
In both strains, phosphoenolpyruvate carboxykinase (Pck1) the gluconeogenetic enzyme and malate synthase 1 (Mls1) were reduced (Figure 4). These enzymes direct acetyl-CoA to malate and oxaloacetate that in turn can be metabolized to phosphoenolpyruvate for gluconeogenesis. Mls1 is a component of the glyoxylate cycle that allows yeast cells to metabolize non-fermentable carbon sources, including fatty acids. The mechanism governing glucose-repressed genes is particularly important in the RNAP III compromised mutant as well as in Maf1-deprived cells due to the previously reported growth perturbations of maf1Δ on non-fermentable carbon source. Two-fold decrease in maf1Δ PCK1 mRNA was reported [52], though under inducing conditions on glycerol. Notably, the relative decrease in Pck1 abundance in Maf1-deficient cells is the largest in our proteomic dataset. The very much decreased Pck1 abundance proves that the enzyme is subject to degradation in a glucose-dependent manner [76] and the mechanism is not perturbed in both the mutants. Other enzymes of the glyoxylate cycle are concomitantly reduced in maf1Δ cells, including malate dehydrogenases Mdh2, Mdh3, isocitrate lyase (Icl1) and glyoxylate aminotransferase (Agx1), the last implicated in glycine synthesis from glyoxylate. In rpc128-1007, most of the glyoxylate enzymes as well as those of the TCA cycle were also decreased. To our knowledge the decrease in abundance of TCA cycle enzymes in these mutants does not activate retrograde signaling, otherwise, we would have observed auxotrophy for glutamate in the mutant strains (Supplementary Figure S1).
Figure 4. Comparative proteomic profiling of maf1Δ and rpc128-1007 mutants when compared with the wild-type strain.
The differences in protein abundances are presented on a schematic representation of the central carbon metabolism. Those proteins with an increased abundance are presented in red and those with a decreased abundance in green.
Enzymes of the TCA and glyoxylate cycles undergo co-ordinated transcriptional down-regulation [76,77] induced by glucose through the master kinase Snf1/AMPK [78,79] therefore strongly suggesting unperturbed functioning of Snf1 signaling on glucose. In contrast, the other key enzyme of gluconeogenesis, Fbp1 (fructose 1,6-bisphosphatase) that bypasses the physiologically irreversible step in the glycolytic pathway, was increased under glucose deprivation in maf1Δ, but was unchanged in the rpc128-1007 mutant.
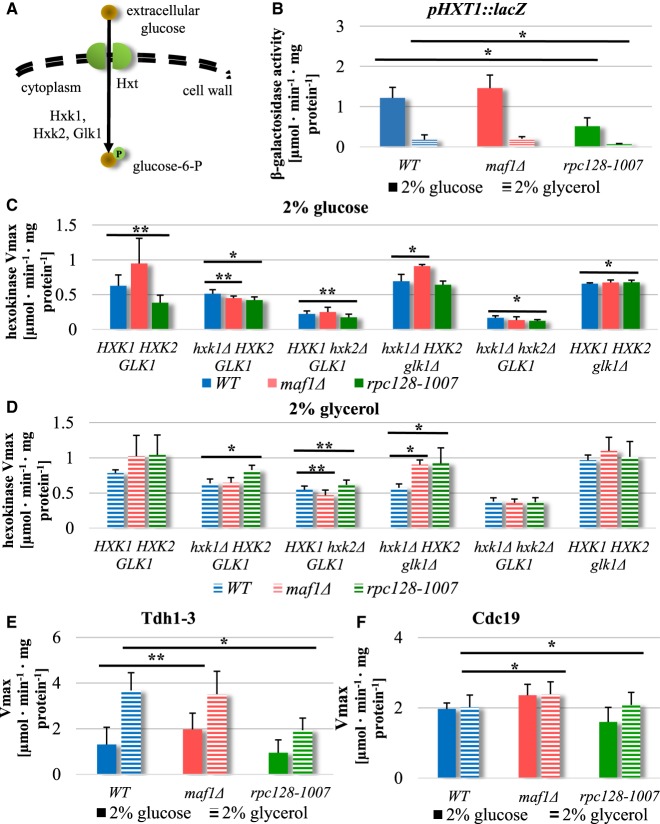
Reduction in glycolytic enzymes in a RNAP III compromised strain correlates with lower activity of the glucose transporter Hxt1
Proteome analysis captured changes in relative cellular abundances of all glycolytic enzymes and implied a reduced capacity for glycolysis in rpc128-1007 cells, but an unchanged glycolytic capacity in maf1Δ cells (Figure 4). In rpc128-1007, enzymes that were significantly decreased (between 2- and 2.6-fold) included glyceraldehyde-3-phosphate dehydrogenase isozyme 1 (Tdh1), enolase (Eno1), glyceraldehyde-3-phosphate dehydrogenase isozyme 2 (Tdh2), 3-phosphoglycerate kinase (Pgk1) and glucokinase (Glk1). The lower abundance of the entire complement of glycolytic enzymes is consistent with reduced glycolytic performance in rpc128-1007.
Glycolytic flux in S. cerevisiae can regulate glucose uptake, at least in part through the activity of glucose uptake mechanisms [80] and in particular, induction and increases of membrane internalization of low-affinity glucose transporters [66,81–84]. The principal example is Hxt1, only activated when yeast grow in glucose-rich media [66]. We explored the potential for changes in glucose transport by measurement of transcriptional de-repression of the key gene encoding the major low-affinity, high-capacity glucose transporter HXT1, in maf1Δ and rpc128-1007 using a HXT1-lacZ reporter plasmid (Figure 5B). The HXT1 gene was strongly activated in maf1Δ under glucose-rich conditions, whereas a 3-fold lower activity of the HXT1 promoter was observed in rpc128-1007 cells in the same conditions. When assessed in cells grown on glycerol, expression of HXT1-lacZ reporter was decreased in all strains (Figure 5B) as expected [66]. Consistent with the reports on mutants in genes of the glycolytic pathway, which are blocked in glycolysis [85], the HXT1 expression was reduced in RNAP III compromised yeast suggesting that in this mutant, the supply of glucose for a functional glycolytic pathway cannot be maintained.
Figure 5. Opposite effects have been observed in HXT1 promoter activity in strains with altered RNAP III.
(A) Schematic representation of glucose uptake and phosphorylation in yeast cells. Hxt, hexose transporter; P, phosphorylation; Hxk1, hexokinase 1; Hxk2, hexokinase 2; Glk1, glucokinase 1. WT, maf1Δ, rpc128-1007 yeast cells and single or double HXK1, HXK2, GLK1 knockouts strains in WT, maf1Δ and rpc128-1007 genetic background were cultured in YPD (C) or YPGly (D) rich medium under either inducing (2% glucose) or repressing (2% glycerol) conditions. Maf1 deficiency increases HXT1 expression (B) on glucose and Hxk2 activity regardless of carbon source (C, D). Metabolic effects observed in rpc128-1007 correlate with decreased HXT1 expression (B) and decreased hexokinase activity in glucose-rich medium (C), but increased hexokinase activity in glycerol rich medium (D). Compromised RNAP III and maf1Δ have an effect on enzymes in lower glycolysis: Tdh1-3 and Cdc19 activities (E, F). The WT strain (MB159-4D), maf1Δ and rpc128-1007 mutant strains were grown under 2% glucose and 2% glycerol conditions. The experiment was performed in cell-free extracts isolated from the aforementioned strains. Data are expressed as the mean obtained from at least three independent experiments measured in triplicate. The standard deviations are shown. Enzymatic assays were performed in cell-free extracts. The reaction rates were monitored by measuring NADH concentration change over time at 340 nm. Vmax mean value is expressed as µmol·min−1·mg−1 protein (C, D, E, F). (B) HXT1 expression was measured in WT [pBM2636], maf1Δ [pBM2636] and rpc128-1007 [pBM2636] strains by using the lacZ reporter gene system [66]. β-galactosidase activity was assayed in cell-free extracts. The error bars indicate the standard deviation from three independent transformants assayed in triplicate. Asterisk (*) indicate P-value <0.05 and double asterisk (**) illustrate P-values <0.1 according to Student's t-test averaged from all technical repeats.
Positive relationship between Hxk2, Tdh1-3 and Cdc19 enzyme activities and the potency of RNAP III-dependent transcription
Since HXT1 gene expression was elevated in maf1Δ, but decreased in rpc128-1007, we measured the activity of selected glycolytic enzymes in vitro [67] in both mutant strains and evaluated the relationship between activity changes and changes in protein abundance assessed by proteomics. S. cerevisiae encodes three isoenzymes with hexokinase activity (Figure 5A). Proteomic analysis showed that Glk1 was the only isoenzyme phosphorylating glucose in maf1Δ (Figure 4), with increased abundance whereas Hxk1 and Hxk2 were observed decreased in this strain. For rpc128-1007, the protein abundance of all enzymes conferring hexokinase activity was decreased (Figure 4). We, therefore, grew the three strains in rich media supplemented with 2% (w/v) glucose and measured the hexokinase reaction (Vmax) in cell-free extracts. Total hexokinase activity was increased in maf1Δ and reduced in rpc128-1007 (Figure 5C, solid bars).
To quantify the activities of the individual hexokinase enzymes we designed and constructed deletion mutants of hexokinases in the three strains (Figure 5C,D). Quantification of glucose phosphorylation activity in single and double null mutants of genes encoding hexokinases clarifies that hexokinase 2 (Hxk2) is the predominant isoenzyme engaged in glucose phosphorylation in maf1Δ. The triple deletion maf1Δ hxk1Δ glk1Δ, in which the only isoform left intact is Hxk2, results in comparable hexokinase activity to the observed in maf1Δ deletion strain with all the isoforms present (Figure 5C). In contrast, the mutants in whom we observe the reverse trend in the enzymatic activity are the maf1Δ hxk2Δ double mutant and maf1Δ hxk1Δ hxk2Δ triple mutant. An increase in glycolytic flux is possible to achieve in cells lacking Maf1 despite a decrease in Hxk2 abundance and only a slight increase in Glk1 cellular concentration. Under growth on glycerol, there was an increased contribution of Hxk1 to total hexokinase activity in wild-type maf1Δ and rpc128-1007 (Figure 5D), suggesting that the compensation regulatory mechanisms are not perturbed in the two mutant strains. HXK1 induction by non-fermentable carbon source has previously been reported [86]. Interestingly, on glycerol growth, the total hexokinase activity in rpc128-1007 was higher than in maf1Δ.
Since glyceraldehyde-3-phosphate dehydrogenase (Tdh1-3) and pyruvate kinase (Cdc19) are important providers of NADH and ATP respectively, these enzymes were also assayed. Measuring the activity of controlling and rate-limiting glycolytic enzymes is one of the techniques to estimate carbon flux through the entire pathway. In yeast, glyceraldehyde-3-phosphate dehydrogenase, placed between upper and lower segments of glycolysis, is considered a rate controlling step of glycolysis [87,88], whereas Cdc19 kinase levels affect the rate of carbon flux and its direction towards pyruvate (PYR) or phosphoenolpyruvate (PEP) under fermentative conditions. The activity of Cdc19 is sufficient to cause a shift from fermentative to oxidative metabolism in S. cerevisiae [69,89] and controls glycolytic rate during growth on glucose [90].
In maf1Δ, in which there was a small increase in Tdh1, 2 abundance (Tdh1; 0.21 log2FC and 0.3 adjusted P-value, Tdh2: 0.16 log2FC and 0.37 adjusted P-value), in vitro activity was 2-fold higher (Figure 5E). From proteomics, Tdh1-3 activity was slightly lower in rpc128-1007 cells under the same growth conditions, but Tdh1-3 activity measured in rpc128-1007 grown on glycerol was significantly lower when compared with the reference strain, which suggests that the catalytic activity of the enzyme decreases in vivo while the enzyme converts 1,3-bisphosphoglycerate (1,3-BPG) into glyceraldehydes-3-phosphate (G3P) in the reverse direction to glycolysis, when the enzymes are involved in gluconeogenesis in the presence of non-fermentable carbon sources in the medium. This shuttle between the cytosol and the nucleus linking metabolic redox status to gene transcription [91] and contributes to tRNA transport [92].
We further measured the activity of the final enzyme in the glycolytic pathway, pyruvate kinase (Cdc19) activity. Cdc19 protein abundance is comparable in maf1Δ and its parental strain. We found (Figure 5F), that Cdc19 shows significantly lower enzymatic activity in rpc128-1007 compared with the reference strain grown on glucose, but a slightly elevated activity in Maf1-deficient cells both on glucose and glycerol. Overall, in maf1Δ the glycolytic enzymes show higher activity than originally thought judging by proteomics data, whereas Hxk2, Tdh1-3 and Cdc19 protein decreased abundance in rpc128-1007 is fully in agreement with their reduced enzymatic activity. This leads us to the conclusion that glycolytic flux is diminished in rpc128-1007, whereas in maf1Δ it is not only higher than in rpc128-1007 but also than in WT. Due to the fact that F16BP mediated allosteric control [93–95], but not Cdc19 abundance or phosphorylation, was reported as having a predominant role in regulating the metabolic flux through the pyruvate kinase Cdc19 [96], we decided to measure F16BP intracellular concentration.
Fructose 1,6-bisphosphate intracellular concentration does not reflect differences in RNAP III activity and glycolytic flux in the mutant strains
The glycolytic metabolite F16BP is a molecule triggering of the metabolic switch from respiration to fermentation in unicellular and higher organisms [3,4,90]. We reasoned that lower glycolytic flux in rpc128-1007 would result in lower fructose 1,6-bisphosphate (F16BP) concentration in this mutant. We measured F16BP in rpc128-1007 grown under anaerobic conditions. After the addition of 100 mM glucose, F16BP concentration sharply increased during the first two minutes to the physiological level observed in the wild-type cells but then declines to a new steady state. In maf1Δ cells, the concentration of F16BP is similar to rpc128-1007 but there is no initial overshoot (Figure 6). The intracellular level of F16BP is therefore unlikely to be the trigger for the perturbed metabolic switching in cells with different RNAP III activity. Another product of glycolysis (or its side branches) may control the transcriptional reprogramming in yeast, particularly when there are multiple-metabolite-responsive elements present at promoters to sense diverse metabolic signals. Additionally, F16BP concentration may affect Cdc19 activity in vivo, and thus, the flux direction.
Figure 6. Changes in intracellular concentration of fructose 1,6-bisphosphate (F16BP).
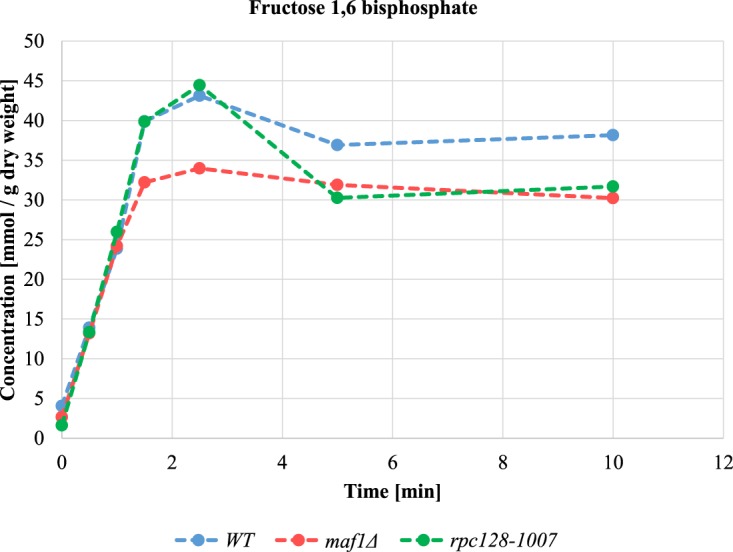
Intracellular fructose 1,6-bisphosphate concentration is lowered in cells with changed RNAP III activity under glucose pulse experiment. Cells were grown in YPD until reaching D600 ≈ 1.0, collected washed in minimal medium lacking carbon source (CBS-C) and resuspended in CBS (-C). Analysis was performed in a thermostatted vessel at 30°C. Cells were flushed with Ar2 gas and glucose was added to a final concentration of 2%. Cell samples suspension were collected in time. Fructose 1,6-bisphosphate content was measured by enzymatic breakdown of NADH monitored by changed absorbance at 340 nm in time according to [4]. Fructose 1,6-biphosphate concentration was calculated from a standard curve and standardized to cells dry weight expressed in g. Results are shown as mean value for four biological replicates.
Higher glucose flux in Maf1-deficient cells results in activation of glycogen and trehalose shunts
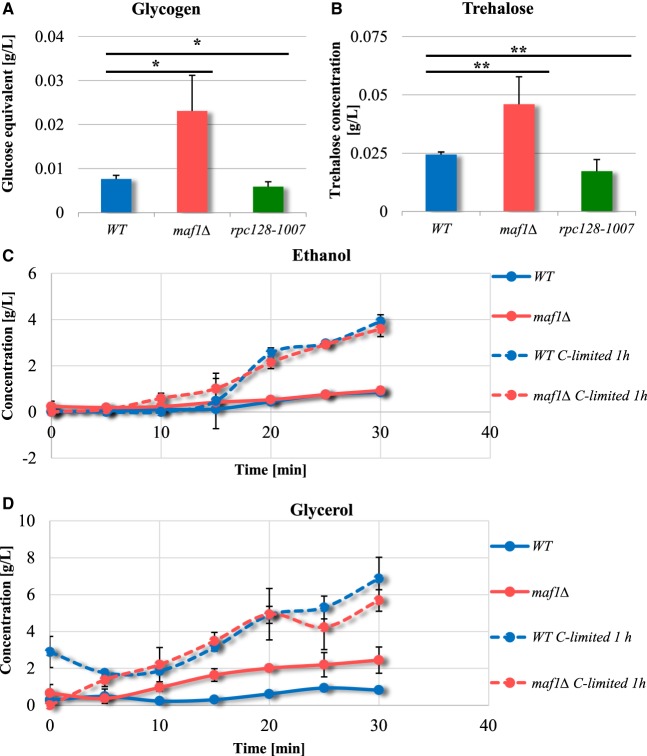
We explored the direction of carbon flux in maf1Δ in the absence of any increase in F16BP concentration. Yeast cells are equipped to counteract excessive influx of glucose by the diversion of glucose into glycogen and trehalose [97]. During exponential growth, glycogen and trehalose biosynthesis play additional roles as part of an adaptive response facilitating survival when the cell is challenged with increased glycolytic flux as a consequence of glucose overflux into a cell. The glycogen shunt prevents accumulation of glycolytic intermediates, particularly F16BP and ATP that otherwise would ultimately lead to perturbation in cell metabolic homeostasis [98–101].
The proteomics data confirmed a strong, negative correlation between RNAP III activity and enzyme abundance in the trehalose and glycogen synthesis pathways, which share common enzymes. UDP-glucose pyrophosphorylase (Ugp1), trehalose-6-P synthases (Tps1 and Tps2) and glycogen synthase (Gsy2) increase in maf1Δ, whilst the same proteins were markedly reduced in rpc128-1007 cells (Figure 3, group 6; Figure 4). The product of Tps1 activity, trehalose-6-phosphate, controls glycolysis by restricting the flow of glucose into the pathway and is an allosteric inhibitor of hexokinase 2 activity [98,102]. We assessed the metabolic allocation of glucose through quantification of trehalose and glycogen content during exponential growth. Both metabolites were 2.5-fold higher in maf1Δ in the presence of high glucose. In contrast, rpc128-1007 cells could accumulate neither glycogen nor trehalose (Figure 7A,B).
Figure 7. Maf1-deficient yeast strain accumulates glycogen (A) and trehalose (B) during the exponential phase.
Yeasts were cultivated in rich medium supplemented with 2% glucose and harvested by centrifugation at D600 ≈ 1.0. Glucose concentration from enzymatic breakdown of glycogen (A) by amyloglucosidase from A. niger, was determined by the Glucose (HK) Assay Kit (GAHK-20, Sigma). Trehalose (B) content determination assay was performed using Trehalose Assay Kit (Megazyme International Ireland, Wicklow, Ireland) according to the manufacturer's protocol. Trehalose and glycogen content is presented as a mean value of at least three independent biological replicates with standard deviations. There were no significant changes in the ethanol production rate between wild-type (MB159-4D) and maf1Δ strain (C). maf1Δ accumulated glycerol (D). Ethanol and glycerol concentration was determined under Fermentative capacity assay (FCA) conditions in maf1Δ strain (C and D). Fermentative capacity assay was performed as described by van Hoek et al. [75] with modifications (for details, see Materials and methods section). All assays were performed in triplicates. Results are shown as mean concentration (g/L) value with the standard deviation in time (min). ‘C-limited’ stands for ‘carbon-limited conditions’. Asterisk (*) indicate P-value <0.05 and double asterisk (**) illustrate P-values <0.1 according to Student's t-test calculated from biological replicates.
In summary, the results are consistent with increased glycolytic flux in maf1Δ and conversely, a diminished flux in rpc128-1007. Metabolic overflow in maf1Δ leads to flux redistribution into the trehalose pathway, to protect the cells from either an increase in intracellular glucose concentration or accumulation of glycolytic intermediates downstream from glucose-6-phosphate as observed in wild-type budding yeast [103].
Ethanol overproduction is not observed in cells lacking Maf1 during logarithmic growth
In yeast, glucose is fermented to ethanol for energy production, as it is often used as a measure of increased glycolytic flux. We examined, whether maf1Δ produces ethanol more efficiently than the wild type, as might be predicted from the Group 8 GO terms (Figure 2). Pyruvate decarboxylases isoenzymes Pdc5 and Pdc6 (the key enzymes in alcohol fermentation) are increased in abundance in maf1Δ (1.7 and 2.3 Log2FC, respectively), suggested that Maf1 deficiency should lead to increased ethanol synthesis. We performed a fermentative capacity assay under anaerobic conditions; without cells pretreatment or with the pretreatment, when cells were glucose starved for 10 min. Under both condition, there was no evidence of enhanced ethanol production in maf1Δ (Figure 7C, Supplementary Table S1). Instead, accumulation of the fermentation by-product, glycerol was observed (Figure 7D). This is consistent with increased glycolytic flux being rerouted upstream of pyruvate or downstream from acetaldehyde by the enzymes of the pyruvate dehydrogenase bypass [75].
Glycerol rather than ethanol production was also evident under aerobic conditions suggesting that access to oxygen does not affect the glycolytic flux redirection towards glycerol biosynthetic pathway in maf1Δ. The maf1Δ mutant is possibly under oxidative stress since glycerol production has a role in response to the stress. Evidence for oxidative stress in maf1Δ also derives from increased protein abundance for the pentose phosphate pathway (PPP) enzymes in this mutant, which balances the systemic manifestation of reactive oxygen species and the ability to detoxify reactive intermediates [104].
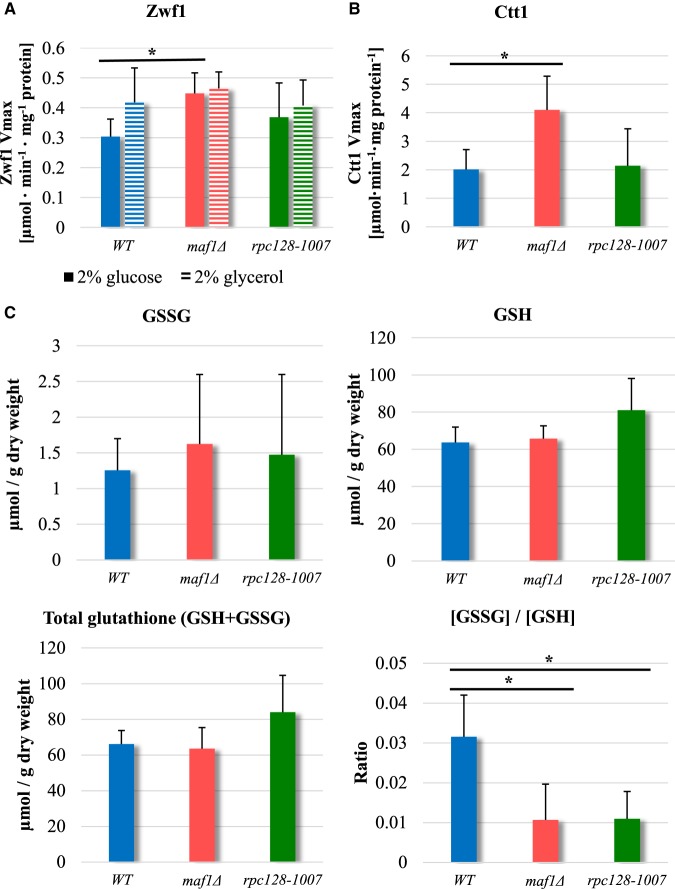
Activation of pentose phosphate pathway in cells deprived of Maf1 regulator
The comparative proteomic analysis suggests reciprocal modulation of the pentose phosphate pathway (PPP), in rpc128-1007 and maf1Δ (Figures 4 and 9). PPP is a source of NADPH during oxidative stress conditions. The data are consistent with an increase in flux through the PPP in maf1Δ, which can be achieved by increased glucose-6-phosphate dehydrogenase Zwf1 abundance, the enzyme catalyzing the rate-limiting, irreversible step of the pathway. Downstream enzymes including 6-phosphogluconolactonase (Sol3, Sol4) and 6-phosphogluconate dehydrogenase Gnd2 that balance the redox potential via the cytosolic NADPH/NADP+ ratio in native yeast cells and both isoforms of transketolase (Tkl1, Tkl2) are increased in maf1Δ. Conversely, depletion of Zwf1, Sol4, Tkl1 and Tkl2 in rpc128-1007 is consistent with a reduced potential of this mutant to redirect carbon flux from glucose-6-phosphate (G6P) towards 6-phosphogluconolactone (6PG) and downstream metabolic intermediates (Figures 4 and 9).
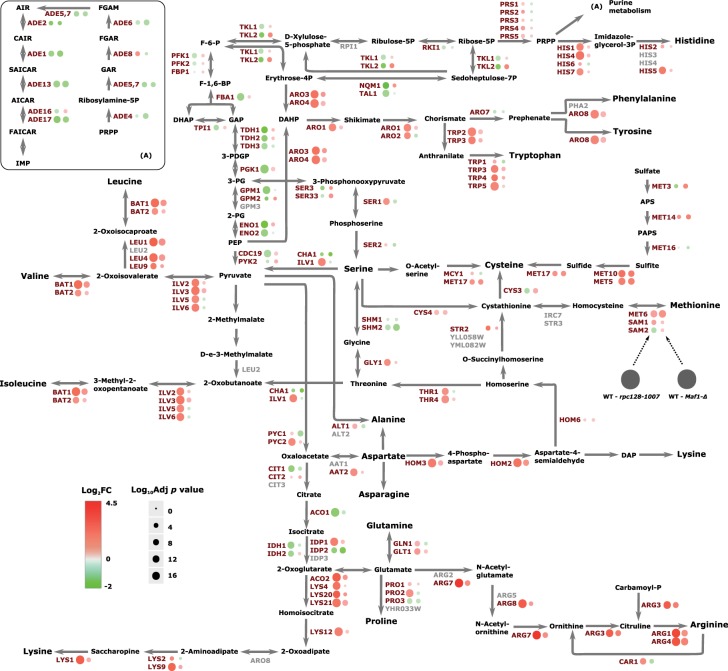
Figure 9. Amino acid biosynthesis and associated proteome signature.
Abundance protein patterns for amino acid metabolism are presented showing those proteins with an increased abundance in red and those with a decreased abundance in green.
We grew yeast in rich medium supplemented with 2% glucose as previously and measured the Zwf1 glucose-6-phosphate dehydrogenase reaction rates (Vmax) in cell-free extracts to check the potential to produce NADPH. This enzyme is highly regulated and is critical in determining the overall flow of glucose into the pentose phosphate pathway [105,106]. Zwf1 activity in maf1Δ, was elevated not only on glucose, as a carbon source, but also on glycerol whereas the enzyme activity in rpc128-1007 remains essentially unchanged (Figure 8A). To further corroborate the relationship between MAF1 deletion and the oxidative stress response other proteins, such as Ctt1 stress inducible cytosolic catalase T were elevated (Supplementary Table S2). Furthermore, total catalase activity was higher in maf1Δ, when compared with WT or rpc128-1007 (Figure 8B).
Figure 8. Zwf1 and the Ctt1 catalase activity is increased in Maf1-deficient mutant.
Yeast cells logarithmically growing in YPD (A, B, C) or YPGly (A) medium were harvested at D600 ≈ 1.0. (A) For Zwf1 activity assay, NADH breakdown was measured at 340 nm in time at 30°C. For catalase activity (B), hydrogen peroxide decomposition in reaction mixtures containing yeast cell-free extracts was monitored as a change in absorbance at 240 nm in time at 30°C. Results are presented as total mean enzymatic activity from five independent biological replicates with standard deviation expressed as µmol·min−1·mg−1 protein. Asterisk (*) indicate P-value <0.05 according to Student's t-test for biological replicates. (C) Glutathione GSSG/GSH ratio in maf1Δ and rpc128-1007 does not indicate oxidative stress. GSSG, GSH and total glutathione levels were measured according to Quantification kit for oxidized and reduced glutathione (Sigma–Aldrich, 38185) from the kinetic method in agreement with the manufacturer's protocol. Yeast strains were grown in rich medium supplemented with 2% glucose until D600 ≈ 1.0. The absorbance was obtained for four biological samples assayed in duplicate at 405 nm. [GSSG]/[GSH] ratio was measured separately for each sample and averaged. Results are shown as a mean value with standard deviation from four biological replicates. Asterisk (*) indicate P-value <0.05 according to Student's t-test for biological replicates.
The magnitude of the abundance changes in enzymes of the oxidative stress response is usually observed during carbon source downshift. Here, however, it occurs in the presence of glucose during the exponential phase, indicates that MAF1 gene deletion elicits broader metabolic reprogramming than originally thought and may present a particular case of a cell's steady-state with high capacity of antioxidant defense.
Additionally, we measured the ratio of oxidized/reduced glutathione in both mutant strains (Figure 8C) and observed no increase in GSSG/GSH ratio, but a higher overall concentration of the total glutathione in rpc128-1007. This suggests that maf1Δ is protected efficiently from oxidative stress by the reactive oxygen species (ROS) scavenging enzymatic cytoprotective system (Supplementary Table S2) whereas rpc128-1007 may predominantly use the back-up chemical system (GSSG/GSH) to control redox state and to reprogram metabolism in order to efficiently maintain the redox homeostasis. The increased levels of total glutathione (γ-glutamyl-l-cysteinyl-glycine) in rpc128-1007 may indicate changes in turnover profile of this compound, that consist of glutamic acid, cysteine and glycine; from which the first and the latter are precursors in purine nucleotides biosynthesis. The increased level of glutathione has been previously observed under dietary restriction in rodents [107,108]. It is not entirely clear how nutrients limitation such as glucose contribute to this effect.
RNAP III subunit RET1/C128 point mutation is associated with metabolic reprogramming dependent on transcriptional and translational induction by Gcn4
Glycolytic intermediates are precursors of the carbon skeletons of several amino acids. Thus, lowered glucose flux could result in the amino acid starvation response in yeast. We identified a large group of proteins that were substantially increased in abundance, that are involved in amino acid biosynthesis. The relative abundance of those was increased in rpc128-1007 relative to WT. However, the same set of enzymes in maf1Δ were increased in some cases and unchanged in others. The magnitude of the increases was generally much higher in the rpc128-1007 compared with the maf1Δ mutant. In rpc128-1007, over 30 proteins in the pathways for arginine, lysine, leucine, isoleucine, and valine biosynthesis de novo, along with aromatic amino acids such as histidine, tryptophan, tyrosine and threonine or their precursors were elevated (Figure 9).
In rpc128-1007, the decreased cellular concentration was observed in methionine biosynthesis subpathway, that is for ATP sulfurylase the product of MET3 gene essential to catalyze the first step for assimilatory reduction in sulfate to sulfide, involved in methionine metabolism. The other proteins diminished in rpc128-1007 were Ser3, Ser33, Cys3 and Shm2 contributing to serine and cysteine biosynthesis.
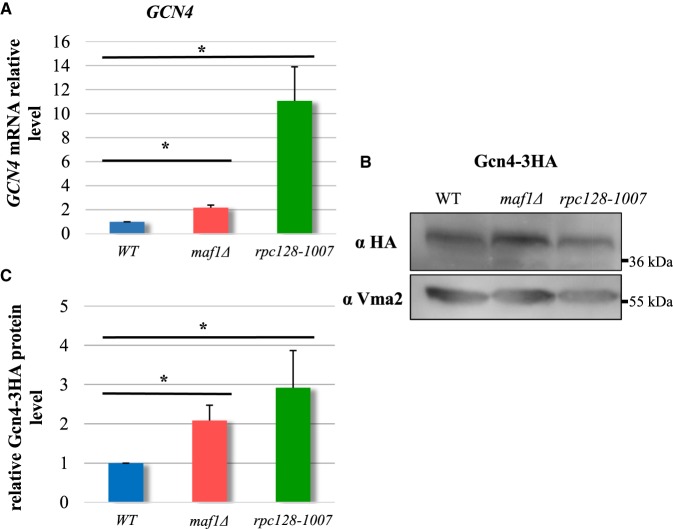
In maf1Δ, the proteins Arg4, Arg3, Cpa2, Bat1, Bat2, Leu1, Leu4, Leu9 and Ilv3 were elevated; all are components of the metabolic branch that is part of l-arginine and l-leucine biosynthesis (Figure 9). In contrast rpc128-1007, maf1Δ cells exhibited enrichment in the branch of serine/cysteine, methionine biosynthesis pathway and sulfate metabolism (Met3, Met5, Met10, Met14), and these abundance changes were amongst the largest in the proteomic dataset. The expression of most genes involved in amino acid biosynthesis is under the control of the major Gcn4 transcriptional activator, part of the general amino acids control (GAAC) regulon [28,109]. GCN4 transcription is stimulated by starvation for amino acids, purine, glucose limitation and specifically by initiator tRNAMet depletion [110,111]. Therefore, we evaluated the mRNA levels of GCN4 by RT-PCR (Figure 10A).
Figure 10. GCN4 transcripts and Gcn4 protein relative levels are significantly increased rpc128-1007 yeast cells and moderately in maf1Δ cells.
Yeast cells were grown in rich medium supplemented with 2% glucose until reached exponential growth phase (D600 ≈ 1.0). SYBR-Green-based real-time PCR (A) showed that GCN4 transcript increased 2-fold in maf1Δ and by 11-fold in rpc128-1007. Wild-type strain expression level was taken as 1.0. Samples were normalized to two reference genes — U2 spliceosomal RNA (U2) and small cytosolic RNA (SCR1). Asterisk (*) indicates P-values lowered than 0.05 according to Student's t-test using all technical replicates of biological samples. Western blotting assay (B) showed increased stability of Gcn4-3HA protein in maf1Δ and rpc128-1007 mutant strains expressing chromosomally encoded Gcn4-3HA. Total cell protein extracts were subjected to SDS–PAGE and examined by Western blotting with anti-HA antibodies (B). Quantitative relative level of Gcn4-3HA protein in comparison with yeast Vma2 protein level was calculated for at least three independent biological replicates (C).
The GCN4 mRNA steady-state levels were 11-fold higher in the rpc128-1007 and, 2-fold elevated in maf1Δ compared with WT cells. The difference between these two mutants is reflected in the extent of the response in the proteomics analysis rpc128-1007 had much stronger phenotypic change than maf1Δ. We constructed mutant strains with chromosomally encoded GCN4-3HA protein fusions to assess Gcn4 protein abundance by immunoblotting. Gcn4 abundance, normalized to Vma2 level in the total protein extracts, was elevated 3-fold in rpc128-1007 and 2-fold in maf1Δ, when compared with the reference strain (Figure 10B,C). The marked decoupling between transcript and protein changes in the strains, rpc128-1007 (11-fold mRNA, 3-fold protein) and maf1Δ (2-fold mRNA, 2-fold protein), also suggests that other regulatory factors are in operation.
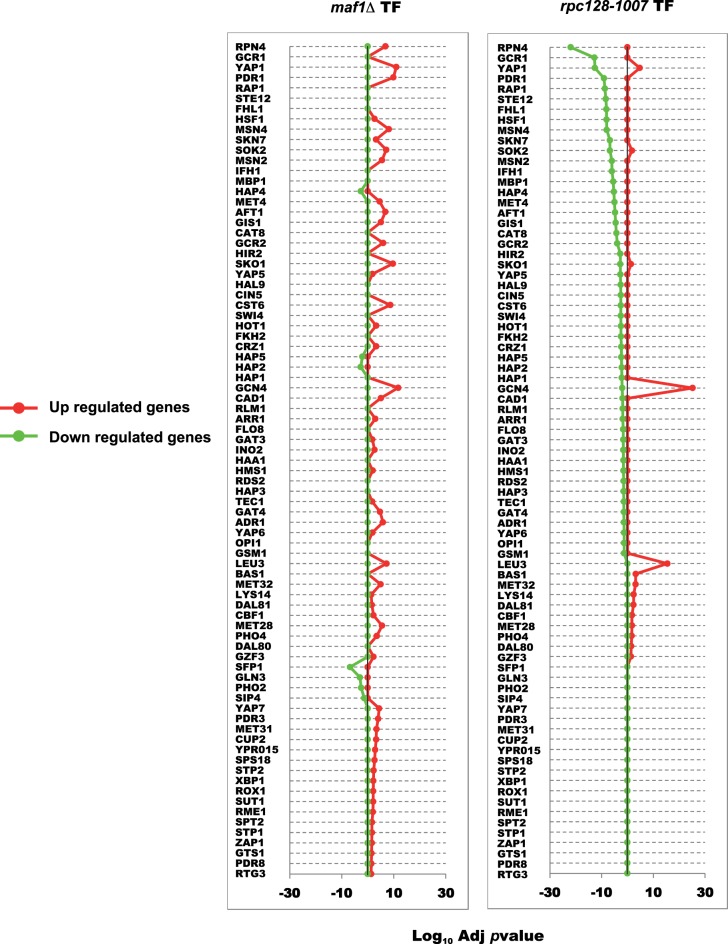
For the entire proteome data set, we were able to perform transcription factor enrichment analysis, using the web-based GeneCodis tool to identify over-representation of the targets of given transcription factors in the differentially abundant proteomes (Figure 11). For rpc128-1007, Gcn4 has the most significant enrichment of its targets genes in the proteins which increase in abundance shown by the red line. The second most significant transcription factor in gene activation relationship was observed for Leu3 (Supplementary Figure S2) followed by Yap1 and Bas1. We also identified a group of genes with motifs enriched for GATA transcriptional factors such as Dal81, Dal80 and Gzf3 regulating genes by nitrogen catabolite repression (NCR). The increase in Gcn4 levels we observed may contribute to activation of some of the NCR sensitive genes alone or in concert with Gln3 (FT gene regulator in response to nitrogen limitation or starvation). Some genes possess both Gcn4 and Gln3 binding sites in their promoter [112]. As these transcriptional factors were represented by a much smaller group of up-regulated genes and the yeast cells were grown in the rich medium it seems that these genes up-regulation is not the result of increased NCR activity. We also note that RPN4, the FT of genes encoding protein degradation machinery, including the 26S proteasome genes [113], is most significant in the down-regulated targets in rpc128-1007 (Supplementary Table S3).
Figure 11. Transcription factor enrichment analysis.
Enrichment in the proteome sets for individual transcription factors was calculated using the GeneCodis website taking the sets of proteins with an adjusted P-value of <0.05 from both strains maf1Δ and rpc128-1007 compared against the wild-type. Proteins were classified according to their positive or negative fold change and the background set consisted of all proteins identified in the given MS experiment.
The outcome of experimental and in silico analyses is that the dramatic increase in specific protein changes observed in rpc128-1007 can be largely attributed to the GCN4 stress response to low glucose flux and GCN4 de-repression on glucose as a compensatory effect. By contrast with the highly focused changes in rpc128-1007, the gene regulatory network in maf1Δ (which liberates RNAP III from regulatory circuits and nutrient signaling) (Figure 1), exhibits a broad spectrum of modest changes (including more balanced GCN4 up-regulation) across several cellular processes, to provide the mutant with better adaptation/selective advantage to growth in glucose-rich environment.
Discussion
Regulation of the central carbon metabolism of S. cerevisiae has always been a topic of considerable interest. Here, we present for the first time, the evidence that glycolytic flux in yeast can be modulated accordingly to the RNAP III activity and that central carbon metabolism adjusts to the activity of RNAP III in yeast. Our results clearly indicate the new connections between RNAP III and cellular metabolism. On the basis of the previous and the present findings, we propose that there is internal signaling in the yeast cells that competes with the extracellular nutrients-sensing, when a cell faces non-optimal RNAP III activity.
A RNAP III point mutation in the RET1/C128 subunit is correlated with lower efficiency of glycolysis and up-regulation of GCN4-dependent genes of amino acid metabolism
In a previous report, we have shown that the rpc128-1007 strain is insensitive to external glucose concentration cues, which manifests via constitutive overexpression of the HXT2 gene, whether in glucose or glycerol growth conditions [53]. Our new data suggest that the rpc128-1007 mutant operates under substrate limitation, such as glucose, even though the sugar is present in excess in the growth medium. RNAP III activity inhibition results in diminished abundance of enzymes in central carbon metabolism and elicits preferential accumulation of proteins involved in amino acid biosynthesis (Figure 9) attributable to the GCN4 response. The molecular events that induce Gcn4 translation also reduce the rate of general protein synthesis [109]. Gcn4 is translationally up-regulated in response to numerous tRNA perturbations [111,114] and yeast cells with the RET1/C128 point mutation produce as much as 1.6-fold less tRNA molecules compared with WT [13]. This suppresses the defect of Maf1 inactivation, caused by increased or unbalanced levels of various tRNAs including increased tRNAMet levels [13]. Initiator tRNAMet depletion triggers a GCN4-dependent reprogramming of global genome expression in response to decreased RNAP III transcription in rpc160-112 mutant (in the largest C160 subunit) [111]. However, HXT2 transcription is not dependent on Gcn4 transcriptional activity [111] in the rpc160-122 mutant. HXT2 overexpression in rpc128-1007 is most likely related to lower glycolytic efficiency in the mutant strain [85], but the direct regulator of the phenomenon still remains to be discovered.
The biological significance of the increase in enzymes abundance in amino acids biosynthesis pathways, let us speculate that in rpc128-1007 de novo synthesized amino acids provide an alternative source of metabolic intermediates to other processes. It is possible that these mutant cells reprogram their metabolism in order to guarantee that there are sufficient biosynthetic precursors for replication. This is indicated in Figure 3, group 6 in which some genes involved in replicative aging are not abundant. De novo synthesis of nucleotides occurs from readily available components such as glutamine, aspartate (indicated in Figure 3 group 1 with genes involved in aspartate biosynthesis are strongly represented) and glycine (indicated in Figure 3 group 5 genes in glycine catabolic processes are strongly represented). Further in de novo IMP synthesis, the first nucleotide formed that is further converted to AMP or GMP (indicated in Figure 3 group 9, in which gene involved in synthesis are drastically decreased by log2FC-5), relies not only on availability of amino acids but also components of the folate-one-carbon pool (10-formyl THF) and ribose 5-P derived from glucose in PPP pathway.
In the wild-type yeast cells, most glycolytic enzymes exist with significant overcapacity, regardless of the carbon source. The enzymes are present in the cells at generally fixed concentration, even if reverse glycolytic processes take place [115]. However, in the rpc128-1007 mutant, the abundance of all the glycolytic isoenzymes is reduced and concomitantly with a decrease in abundance of several proteins engaged in the side branches of the glycolytic pathway (trehalose/glycogen shunt and PP pathway). The two enzymes, fructose 1,6-bisphosphatase Fbp1 and pyruvate kinase 2 Pyk2, that catalyze the reactions in gluconeogenesis are the least affected in rpc128-1007 in agreement with the preference of this mutant to grow on respiratory carbon sources such as glycerol [13].
In a Drosophila melanogaster gut model [116], reduction in RNAP III activity through controlled degradation of the C160 subunit (C160 encoded by RPC160) leads to diminished protein synthesis. Inhibition of RNAP III affects RNAP I, but not RNAP II-generated transcripts, suggesting that translation is the major factor regulating protein abundance in RNAP III compromised cells [116]. This further suggests that the rpc128-1007 mutant, might be limited at translation and indeed the rpc128-1007 mutant has reduced tRNA levels [13]. In rpc128-1007, translation seems to be selective towards enzymes in de novo amino acids synthesis at the expense of the full complement of glycolytic enzymes. As a consequence of decreased abundance of the glycolytic enzymes, the glycolytic flux is very likely to decrease.
The reduction in abundance of selected glycolytic enzymes, such as Hxk2, Tdh1-3 and Cdc19 in rpc129-1007 is followed by the decrease in their enzymatic activity at high concentrations of glucose (Figures 5 and 6). Lowering the intracellular Cdc19 concentration is sufficient to shift from fermentative to oxidative metabolism in yeast, which reduced flux towards pyruvate [69]. The lower abundance and activity of the first (Hxk2) and the last (Cdc19) enzymes in the glycolytic pathway can be expected to reduce glycolytic flux in the mutant.
The RNAP III RET1/C128 mutation elicits a diminution of the low-affinity glucose transporter 1 (activated on high glucose), the major glucose transporter facilitating glucose uptake under glucose-rich conditions [82] and which is under control of Rgt2 (low affinity) glucose sensor (Figure 5B). External glucose signaling, may not be a dominant factor in reprogrammed HXT genes expression in rpc128-1007, glucose metabolism may dominate in this case [53]. Our data are consistent with the postulate that ‘glucose sensing’ could occur intracellularly [1–4], but not yet (so far) linked to metabolic reprogramming upon change in RNAP III activity. There is some debate as to whether the signaling molecule for metabolism switching is fructose 1,6-bisphosphate [4,82]. F16BP triggers a switch in metabolism from respiration to fermentation in unicellular and higher organisms. It is the key metabolic factor determining AMPK/Snf1 kinase activity and is a potent activator of Ras pathway [3,4,90]. If F16BP plays such key roles, we reasoned that the intracellular concentration should be lower rpc128-1007 and higher in maf1Δ.
Although significantly lower than in the wild-type cells, the levels of F16BP in both mutant strains are comparable (Figure 6). This suggested that in maf1Δ, the mechanisms protecting cells from damaging increased concentrations of glycolytic intermediates are unperturbed thus increasing cells survival; cells capable of increased glucose consumption.
Despite the initial rise in F16BP after a glucose pulse, the metabolite attains a steady-state concentration at levels that are not likely to be toxic to the rpc128-1007 mutant. In rpc128-1007 cells, we presume that there is no direct relationship between intracellular F16BP and the growth defect on glucose medium exhibited by these cells, however alternative scenario is possible if taking into account the overall abundance of the glycolytic enzymes and an excess of F16BP which may affect Ras proteins. These cells also exhibit a large decrease in ribosomal proteins (RP) (Supplementary Table S4). RNAP III transcription is co-ordinately regulated with transcription of rDNA and ribosomal protein-coding genes [12,117]. Normally ribosomal proteins and their mRNAs are stabilized when yeast is subject to increased glucose [118]. Arguably, the lower abundance of RP proteins in rpc128-1007 could reduce the energy expense of cells that are unable to metabolize available extracellular glucose. For the rpc128-1007, all of the changes at the proteome level are reminiscent of the global changes in yeast cells in response to environmentally stressful, glucose-deprived conditions. An exception is the cohort of proteins of the TCA cycle. RNAP III compromised cells have reduced glycolysis but this reduction does not lead to enhanced oxidative metabolism.
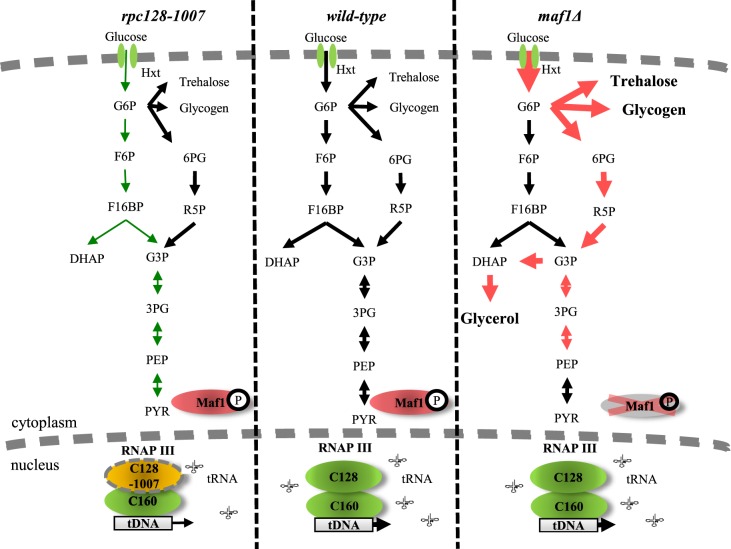
maf1Δ cells preferentially metabolizes glucose, which results in carbon overflux fueling the side pathways dependent on glycolytic intermediates as precursors
Lack of Maf1 causes cells to reprogram their metabolism towards higher glycolytic activity when grown under high glucose conditions. This response is not reflected in an increase in abundance of the glycolytic enzymes but rather in enzymatic activity. Of course, the profile of activity modulating posttranslational modifications of these enzymes could well be different in maf1Δ but this was beyond the scope of this study. For example, hexokinase 2 exhibited higher activity in maf1Δ even though the protein abundance was reduced and the activity of this enzyme is regulated by phosphorylation [119,120]. Higher hexokinase activity should lead to increased flux into glycolysis. However, due to robustness of flux regulation, carbon is redistributed in maf1Δ into the side branches of the glycolytic pathway at glucose-6P (Figure 12). The maf1Δ shows the increased capability to direct carbon into all the side branch pathways as suggested by the proteomic data and confirmed by direct metabolite assay (Figure 7A). Glucokinase (Glk1), increased in abundance in maf1Δ, may redirect glucose toward glycogen storage as previously postulated [121]. The enzymes of glycogen trehalose and central carbon metabolism may be altered in maf1Δ as these are dependent on control by the major nutrient-sensing protein kinases TOR, PKA, Snf1, Pho85 and the energy sensor Pas kinase [119,122,123].
Figure 12. Proposed model of carbon flow in rpc128-1007 and maf1Δ yeast cells.
Altered RNAP III activity affects carbon flux. Low activity of RNAP III in rpc128-1007 strain is correlated with decreased carbon flow through glycolysis in comparison with reference strain. In contrast, maf1Δ cells demonstrate increased carbon flow through hexokinase step and lower glycolysis compared with the control strain. In maf1Δ, excess glucose-6-P (G6P) is redirected into PPP and trehalose and glycogen biosynthesis. As a result, fructose 1,6-bisphosphate (F16BP) concentration decreases in maf1Δ. From the increased glycerol concentration, carbon flux is partially redirected towards upper glycolysis at PEP. Green: decrease in carbon flux; Red: increase in carbon flux. Legend: glucose-6-phosphate (G6P); fructose-6-phosphate (F6P); fructose 1,6-bisphosphate (F16BP); dihydroxyacetone phosphate (DHAP); glyceraldehyde-3-phosphate (G3P); 6-phosphogluconate (6PG); ribose-5-phosphate (R5P); 3-phosphoglyceric acid (3PG); phosphoenolpyruvate (PEP).
In rat hepatoma cells, glucose import, and the activity of hexokinase, hexose phosphate isomerase and the glucose-6P branches that generate F16BP exert most of the flux control [124]. We believe that in S. cerevisiae, hexokinase 2 and increased activity of the low-affinity glucose transporter Hxt1 have the greatest potential to contribute to flux rerouting in maf1Δ. At this stage, we do not know the intracellular signal. The activity of Hxk2 is elevated even though the trehalose shunt, which acts as a safety valve against excessive supply of glucose, may correct glucose influx through allosteric inhibition of Hxk2 by trehalose-6P [98,102]. Furthermore, the glycolytic flux towards pyruvate could be enhanced by Tdh over-activation. However, carbon flux in maf1Δ fuel the glycerol synthesis pathway rather than causing ethanol accumulation. Glycerol accumulation in maf1Δ must serve as a drain for excess reducing power, to ameliorate redox imbalance in the cells. MAF1 deletion in yeast cells is associated with redox imbalance as has previously reported by Bonhoure et al. [125] in MAF1 knockout mice. In S. cerevisiae, this is efficiently counteracted by NADH-consuming glycerol formation [126].
Activation of the branch pathways of the central carbon metabolism seen in maf1Δ is a hallmark of cancer cells that reprogram glycolytic activity towards the synthesis of metabolites required in excess when cells rapidly divide. For instance, the PP pathway provides precursors for nucleotide and amino acid biosynthesis. This pathway, also referred to as a metabolic redox sensor, is important to maintain carbon homeostasis, is highly correlated to oncogenic, nutrient response signaling pathways [104,127–129] and is required for NADPH regeneration. It supports metabolic reconfiguration in rapidly proliferating cells since NADPH is a ubiquitous cofactor for most anabolic reductive reactions and got scavenging of reactive oxygen species (ROS) that cause oxidative damage to DNA and proteins and which reduce protein synthesis [130,131]. ROS scavenging enzymes also increased in abundance in maf1Δ, suggesting a role of Maf1 in the regulation of intracellular redox potential.
How is the glucose flux distributed in Maf1-deficient yeast cells? We propose a scenario, built on our observation of the decreased levels of the allosteric activator of pyruvate kinase Cdc19. The reduced F16BP concentration in maf1Δ cells could adversely affect Cdc19 activity. Cdc19 activity dependency on the F16BP availability for binding [93–95], but not Cdc19 abundance per se or phosphorylation, plays a predominant role in regulating the metabolic flux through the pyruvate kinase Cdc19 [96]. Previously published data, on the effect of MAF1 deletion on Cdc19 activity did not agree, possible because that study lacked estimation of the intracellular concentration of F16BP (involved in Cdc19 regulation by ultrasensitive allostery) but also lacked data on fluxes into the exometabolome, specifically trehalose, glycogen and glycerol fluxes [132]. The previous result [132], is controversial since there is much lower activity of Cdc19 in the reference strain on glycerol in in vitro assay done under conditions of saturating concentration of F16BP. This impacts on the overall activity of Cdc19 in maf1Δ cells, which is compared with Cdc19 activity in the reference strain.
Since F16BP levels are low in maf1Δ (Figure 6), this may further lead to decrease in Cdc19 activity in vivo and pushing the glycolytic intermediated of lower glycolysis back to upper side branches of the pathway since lower PEP/pyruvate conversion, catalyzed by pyruvate kinase, favors accumulation of glycolytic intermediates, refueling diverging anabolic pathways, such as the pentose phosphate pathway (PPP) and serine biosynthesis [90,133].
In our model, the glycolytic flux bypasses the steps in upper glycolysis between G6P and G3P, achieved via improved flux through PPP. This pathway operates in three modes, depending on a cell demand for metabolic intermediates and cofactors. To avoid extensive F16BP synthesis, which would improve cells survival, the flux should be directed towards glyceraldehydes (G3P) [4]. This scenario is supported by our observation of glycerol accumulation and no change in ethanol production in maf1Δ cells. This glucose flux redistribution towards the PPP shunt, amino acids and nucleotide biosynthesis, due to Cdc19 action has been reported for cancer cells [134]. Furthermore, human fibroblasts exposed to hydrogen peroxide elicit enhanced carbon flow through upper glycolysis and the oxidative branch of PPP, causing reduction in lower glycolysis activity [135].
The characteristics of maf1Δ in central carbon metabolism is reminiscent of cancer proliferating mammalian cells which are stimulated in the early part of glycolysis via PI3K/AKT activation, and making glycolytic intermediates available for macromolecular synthesis due to the low-activity isoform of PK-M2 pyruvate kinase and producing NADPH due to mutated p53 tumor suppressor [133,134].
There are further similarities between maf1Δ and mammalian cells after oncogenic transformation. We noted increased potential for amino acid biosynthesis, including the arginine and leucine metabolic pathways. Arg and Leu are crucial for TORC1 signaling and activation of protein translation in yeast and higher eukaryotes. Leucine is the most frequently encoded amino acid in eukaryotic genomes and its levels are sensed by leucyl-tRNA synthetase to activate TORC1 kinase [136,137]. How yeast TORC1 integrates arginine signals is presently unknown. In mammals, arginine levels are communicated by two mechanisms, involving Rag GTPases mediating amino acids signals to control mTORC1 and by a cytoplasmic mechanism that involves arginine signaling by a sensor called CASTOR [138,139].
maf1Δ cells have also a strong enrichment in the branch of serine/cysteine, methionine biosynthesis pathway and in sulfate metabolism by contrast with rpc128-1007 cells. Methionine biosynthesis is connected to tRNA quality control [140]. A crucial contribution of serine/glycine to cellular metabolism is through the glycine cleavage system, which resupplies once carbon units for one-carbon metabolism. The importance of serine/glycine metabolism is emphasized by genetic and functional evidence indicating that hyperactivation of the serine/glycine biosynthetic pathway drives oncogenesis. During growth on a fermentable carbon source, most serine is derived from the phosphoglycerate-3P by the gene products Ser3 and Ser33. Ser3 is a cytosolic enzyme with the dual function of phosphoglycerate dehydrogenase and α-ketoglutarate reductase [141]. Known for oxidizing 3-phosphoglycerate in the main serine biosynthesis pathway Ser3 also reduces α-ketoglutarate to D-2-hydroxyglutarate (D-2HG) using NADH, the major intracellular source of D-2HG in yeast. High levels of intracellular D-2HG are found in several types of cancer including gliomas and acute myelogenous leukemia [142].
RNAP III genes are not equally regulated by Maf1. Comparison of expression of selected tDNA genes in maf1Δ on glucose has revealed elevated tRNAMet levels [13]. Methionine is a proteinogenic amino acid. Overexpression of initiator methionine tRNA (tRNAMet) leads to reprogramming of tRNA expression and increased cell metabolic activity in Drosophila and proliferation in human epithelial cells [143,144]. Moreover, methionine metabolism influences genomic architecture via H3K4me3 histone methylation to alter chromatin dynamics and cancer-associated gene expression [145]. Furthermore, high methionine metabolism and sulfur utilization is intertwined with high Tkl1 transketolase activity and is dependent on the non-oxidative phase of the PPP, Tkl1 abundance increased under our study in maf1Δ [146]. Methionine biosynthesis, via the assimilation of inorganic sulfate, requires three molecules of NADPH per molecule of methionine [106]. The data presented here suggest that efficient supply of NADPH derived from the PPP in maf1 may also support methionine biosynthesis in the mutant.
Finally, the proteomics data obtained here with glucose-grown maf1Δ, suggest an alternative explanation to the reduced fitness of this strain on non-fermentable carbon sources. Down-regulation of FBP1 transcription [52] is unlikely to be the major cause of maf1Δ lethality when grown on glycerol. The FBP1 gene is expressed only in the absence of glucose, and even if it is not expressed under glucose-rich condition, that is, under this study the Fbp1 protein is still present in maf1Δ at wild-type levels. It is possible that the growth defect in maf1Δ on glycerol is due to decrease in abundance of the enzymes involved in the glyoxylate cycle.
In conclusion, global label-free profiling of enzymatic proteins in yeast has provided new insight into metabolic physiology. Protein abundance patterns characterized in the mutant strains that show different phenotypes on fermentable and non-fermentable carbon source highlighted metabolic pathways that could now be the target for further genetic or metabolic analysis. This work emphasizes S. cerevisiae as a very good model organism for systems-level studies on the dynamics of cellular networks. There is growing evidence for contradictory observations in cultured human cancer cells and in multicellular organisms including mouse models [125,145]. We anticipate that yeast cells will continue to be appreciated as a source of basic biological information in building an integrated picture of metabolism and gene regulation.
Conclusions
The capacity of the glycolytic pathway can be altered by manipulation of RNAP III activity.
Lack of Maf1, the negative regulator of RNAP III driven non-coding RNA transcription, enhances glycolytic flux and results in accumulation of end products upstream of pyruvate.
Severe reduction in growth rate caused by RNAP III mutation rpc128-1007 on glucose is correlated with a decrease in the abundance of glycolytic enzymes.
The translation machinery in the rpc128-1007 mutant seems to be selective towards mRNA coding for enzymes for amino acids synthesis de novo at the expense of full complement of glycolytic enzymes.
The critical decrease in abundance of glyoxylate cycle enzymes reduced the ability to convert non-fermentable substrates in MAF1 knockout yeast.
Acknowledgements
We want to thank Mark Johnston (Dept. of Biochemistry and Molecular Genetics, University of Colorado Denver, US) for providing pBM2636 plasmid, Jennifer A. Tate (Department of Microbiology, Immunology and Biochemistry, University of Tennessee Health Science Center, US) and Wojciech Bal (Institute of Biochemistry and Biophysics, PAS) for discussion on this manuscript. We are grateful to Dr. Philip Brownridge for excellent instrument support. The corresponding author, MA, dedicates this work to her baby son, Piotr.
Abbreviations
- 1,3-BPG
1,3-bisphosphoglycerate
- 3PG
3-phosphoglyceric acid (3PG)
- 6PG
6-phosphogluconolactone
- Agx1
glyoxylate aminotransferase
- Arg4
argininosuccinate lyase
- ATP
adenosine triphosphate
- Bas1
myb-related transcription factor 1
- Bat1
mitochondrial branched-chain amino acid (BCAA) aminotransferase
- Bat2
cytosolic branched-chain amino acid (BCAA) aminotransferase
- Bcy1
regulatory subunit of the cyclic AMP-dependent protein kinase
- Cdc19
pyruvate kinase
- Cpa2
large subunit of carbamoyl phosphate synthetase
- Ctt1
cytosolic catalase T
- Cys3
cystathionine gamma-lyase 3
- D-2HG
D-2-hydroxyglutarate
- DHAP
dihydroxyacetone phosphate
- Eno1
enolase
- F16BP
fructose 1,6-bisphosphate
- F6P
fructose-6-phosphate
- Fbp1
fructose 1,6-bisphosphatase 1
- G3P
glyceraldehydes-3-phosphate
- G6P
glucose-6-phosphate
- Glk1
glucokinase
- Gnd2
6-phosphogluconate dehydrogenase
- GO
gene ontology
- Gsy2
glycogen synthase 2
- Gzf3
GATA zinc finger protein
- Hxk2
hexokinase
- Hxt
hexose transporter
- Icl1
isocitrate lyase
- Ilv3
dihydroxy-acid dehydratase 3
- Kog1
subunit of TORC1
- Leu
isopropylmalate isomerase
- Mdh
malate dehydrogenase
- Met3
ATP sulfurylase
- Mls1
malate synthase 1
- mRNA
messenger RNA
- NADH
nicotinamide adenine dinucleotide - hydrogen
- NADPH
nicotinamide adenine dinucleotide phosphate - hydrogen
- NCR
nitrogen catabolite repression
- Nop1
histone glutamine methyltransferase
- PCA
principal component analysis
- Pck1
phosphoenolpyruvate carboxykinase 1
- Pdc1
pyruvate decarboxylase isoenzyme
- PEP
phosphoenolpyruvate
- Pgk1
3-phosphoglycerate kinase
- PKA
cAMP-dependent protein kinase
- PPP
pentose phosphate pathway
- Pus1
tRNA:pseudouridine synthase
- PYR
pyruvate
- R5P
ribose 5-phosphate (R5P)
- RET1/RPC128
second largest subunit of RNAP III
- RNAP II
RNA Polymerase II
- RNAP III
RNA polymerase III
- ROS
reactive oxygen species
- RP
ribosomal proteins
- RPC160
the largest subunit of RNAP III
- RPC40 (AC40)
a common subunit to RNAP I and III
- RPO21
largest subunit of RNAP II
- rRNA
ribosomal RNA
- Ser
3-phosphoglycerate dehydrogenase and α-ketoglutarate reductase
- Shm2
cytosolic serine hydroxymethyltransferase 2
- Snf1/AMPK
AMP-activated S/T protein kinase
- snoRNA
small nucleolar RNA
- snRNA
small nuclear RNA
- Sol3
6-phosphogluconolactonase
- Sua7
transcriptional factor TFIIB
- Sub1
transcriptional regulator facilitating elongation through factors that modify RNAP II
- Tan1
putative tRNA acetyltransferase
- TCA
tricarboxylic acid cycle
- Tdh
glyceraldehyde-3-phosphate dehydrogenase isozyme
- Tdh1
glyceraldehyde-3-phosphate dehydrogenase isozyme 1
- TF
transcription factor
- Tkl1
transketolase
- TOR1
target of rapamycin
- TORC1
complex of TOR1 and Raptor
- Tps1
trehalose-6-P synthase
- Trm1
tRNA methyltransferase 1
- tRNA
transfer RNA
- Ugp1
UDP-glucose pyrophosphorylase
- Yap1
basic leucine zipper (bZIP) transcription factor 1
- YPD
yeast extract peptone dextrose medium supplemented with 2% glucose
- YPGly
yeast extract peptone medium supplemented with 2% glycerol
- Zwf1
glucose-6-phosphate dehydrogenase
Author contribution
Conceptualization: M.A., R.J.B., S.J.H.; Data curation: M.G.A.; Formal analysis: R.Sz, K.R., S.W.H., E.F.; Funding acquisition: M.A.; Investigation: M.A., R.J.B., S.W.H; Methodology: M.A., R.Sz, R.J.B., S.W.H.; Project administration: M.A., R.J.B., S.J.H.; Resources: M.A., R.J.B.; Software: M.G.A.; Supervision: M.A., R.J.B., S.J.H.; Visualization: R.Sz, M.A., M.G.A., S.J.H., E.F.; Writing — original draft: M.A., R.J.B., S.J.H.; Writing — review and editing M.A., R.J.B., S.J.H., R.Sz, M.G.A.
Funding
This work was supported by National Science Centre, Poland grant 2012/05/E/NZ2/00583 to M.A. and by funding from Faculty of Chemistry, Warsaw University of Technology, Poland. We also acknowledge funding from the BBSRC, in the form of an ERA-IB grant, supporting RJB and SWH (BB/M025756/1) and to MGA and SJH (BB/M025748/1).
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
Supplementary Material
References
- 1.Otterstedt K., Larsson C., Bill R.M., Ståhlberg A., Boles E., Hohmann S. et al. (2004) Switching the mode of metabolism in the yeast Saccharomyces cerevisiae. EMBO Rep. 5, 532–537 10.1038/sj.embor.7400132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dechant R., Binda M., Lee S.S., Pelet S., Winderickx J. and Peter M. (2010) Cytosolic pH is a second messenger for glucose and regulates the PKA pathway through V-ATPase. EMBO J. 29, 2515–2526 10.1038/emboj.2010.138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang C.-S., Hawley S.A., Zong Y., Li M., Wang Z., Gray A. et al. (2017) Fructose-1,6-bisphosphate and aldolase mediate glucose sensing by AMPK. Nature 548, 112–116 10.1038/nature23275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peeters K., Van Leemputte F.L., Fischer B., Bonini B.M., Quezada H., Tsytlonok M. et al. (2017) Fructose-1,6-bisphosphate couples glycolytic flux to activation of Ras. Nat. Commun. 8, 922–922 10.1038/s41467-017-01019-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitchison J.M. The Biology of the Cell Cycle. CUP Archive; 1971. 324 p.
- 6.Elliott S.G. and McLaughlin C.S. (1978) Rate of macromolecular synthesis through the cell cycle of the yeast Saccharomyces cerevisiae. Proc. Natl Acad. Sci. U.S.A. 75, 4384–4388 10.1073/pnas.75.9.4384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Polymenis M. and Aramayo R. (2015) Translate to divide: сontrol of the cell cycle by protein synthesis. Microb. Cell. 2, 94–104 10.15698/mic2015.04.198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waldron C. (1977) Synthesis of ribosomal and transfer ribonucleic acids in yeast during a nutritional shift-up. J. Gen. Microbiol. 98, 215–221 10.1099/00221287-98-1-215 [DOI] [PubMed] [Google Scholar]
- 9.Kief D.R. and Warner J.R. (1981) Coordinate control of syntheses of ribosomal ribonucleic acid and ribosomal proteins during nutritional shift-up in Saccharomyces cerevisiae. Mol. Cell. Biol. 1, 1007–1015 10.1128/MCB.1.11.1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boguta M., Czerska K. and Żołądek T. (1997) Mutation in a new gene MAF1 affects tRNA suppressor efficiency in Saccharomyces cerevisiae. Gene 185, 291–296 10.1016/S0378-1119(96)00669-5 [DOI] [PubMed] [Google Scholar]
- 11.Pluta K., Lefebvre O., Martin N.C., Smagowicz W.J., Stanford D.R., Ellis S.R. et al. (2001) Maf1p, a negative effector of RNA polymerase III in Saccharomyces cerevisiae. Mol. Cell. Biol. 21, 5031–5040 10.1128/MCB.21.15.5031-5040.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Upadhya R., Lee J. and Willis I.M. (2002) Maf1 is an essential mediator of diverse signals that repress RNA polymerase III transcription. Mol. Cell. 10, 1489–1494 10.1016/S1097-2765(02)00787-6 [DOI] [PubMed] [Google Scholar]
- 13.Cieśla M., Towpik J., Graczyk D., Oficjalska-Pham D., Harismendy O., Suleau A. et al. (2007. Nov) Maf1 is involved in coupling carbon metabolism to RNA polymerase III transcription. Mol. Cell. Biol. 27, 7693–7702 10.1128/MCB.01051-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vannini A., Ringel R., Kusser A.G., Berninghausen O., Kassavetis G.A. and Cramer P. (2010) Molecular basis of RNA polymerase III transcription repression by Maf1. Cell 143, 59–70 10.1016/j.cell.2010.09.002 [DOI] [PubMed] [Google Scholar]
- 15.Oficjalska-Pham D., Harismendy O., Smagowicz W.J., Gonzalez de Peredo A., Boguta M., Sentenac A. et al. (2006) General repression of RNA polymerase III transcription is triggered by protein phosphatase type 2A-mediated dephosphorylation of Maf1. Mol. Cell. 22, 623–632 10.1016/j.molcel.2006.04.008 [DOI] [PubMed] [Google Scholar]
- 16.Zaragoza O. and Gancedo J.M. (2001) Elements from the cAMP signaling pathway are involved in the control of expression of the yeast gluconeogenic gene FBP1. FEBS Lett. 506, 262–266 10.1016/S0014-5793(01)02922-2 [DOI] [PubMed] [Google Scholar]
- 17.Harismendy O., Gendrel C.-G., Soularue P., Gidrol X., Sentenac A., Werner M. et al. (2003) Genome-wide location of yeast RNA polymerase III transcription machinery. EMBO J. 22, 4738–4747 10.1093/emboj/cdg466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reina J.H., Azzouz T.N. and Hernandez N. (2006) Maf1, a new player in the regulation of human RNA polymerase III transcription. PLoS ONE. 1, e134 10.1371/journal.pone.0000134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knaus R., Pollock R. and Guarente L. (1996) Yeast SUB1 is a suppressor of TFIIB mutations and has homology to the human co-activator PC4. EMBO J. 15, 1933–1940 10.1002/j.1460-2075.1996.tb00544.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calvo O. and Manley J.L. (2005) The transcriptional coactivator PC4/Sub1 has multiple functions in RNA polymerase II transcription. EMBO J. 24, 1009–1020 10.1038/sj.emboj.7600575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosonina E., Willis I.M. and Manley J.L. (2009) Sub1 functions in osmoregulation and in transcription by both RNA polymerases II and III. Mol. Cell. Biol. 29, 2308–2321 10.1128/MCB.01841-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henríquez R., Blobel G. and Aris J.P. (1990) Isolation and sequencing of NOP1. A yeast gene encoding a nucleolar protein homologous to a human autoimmune antigen. J. Biol. Chem. 265, 2209–2215 PMID: [PubMed] [Google Scholar]
- 23.Schimmang T., Tollervey D., Kern H., Frank R. and Hurt E.C. (1989) A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. EMBO J. 8, 4015–4024 10.1002/j.1460-2075.1989.tb08584.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tessarz P., Santos-Rosa H., Robson S.C., Sylvestersen K.B., Nelson C.J., Nielsen M.L. et al. (2014) Glutamine methylation in histone H2A is an RNA-polymerase-I-dedicated modification. Nature 505, 564–568 10.1038/nature12819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pinto I., Ware D.E. and Hampsey M. (1992) The yeast SUA7 gene encodes a homolog of human transcription factor TFIIB and is required for normal start site selection in vivo. Cell 68, 977–988 10.1016/0092-8674(92)90040-J [DOI] [PubMed] [Google Scholar]
- 26.Wu W.H., Pinto I., Chen B.S. and Hampsey M. (1999) Mutational analysis of yeast TFIIB. A functional relationship between Ssu72 and Sub1/Tsp1 defined by allele-specific interactions with TFIIB. Genetics 153, 643–652 PMC:1460761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cho E.J. and Buratowski S. (1999) Evidence that transcription factor IIB is required for a post-assembly step in transcription initiation. J. Biol. Chem. 274, 25807–25813 10.1074/jbc.274.36.25807 [DOI] [PubMed] [Google Scholar]
- 28.Loewith R. and Hall M.N. (2011. Dec) Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 189, 1177–1201 10.1534/genetics.111.133363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim D.-H., Sarbassov D.D., Ali S.M., King J.E., Latek R.R., Erdjument-Bromage H. et al. (2002) mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110, 163–175 10.1016/S0092-8674(02)00808-5 [DOI] [PubMed] [Google Scholar]
- 30.Inoki K., Ouyang H., Li Y. and Guan K.-L. (2005) Signaling by target of rapamycin proteins in cell growth control. Microbiol. Mol. Biol. Rev. 69, 79–100 10.1128/MMBR.69.1.79-100.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin D.E. and Hall M.N. (2005) The expanding TOR signaling network. Curr. Opin. Cell Biol. 17, 158–166 10.1016/j.ceb.2005.02.008 [DOI] [PubMed] [Google Scholar]
- 32.Turowski T.W., Karkusiewicz I., Kowal J. and Boguta M. (2012) Maf1-mediated repression of RNA polymerase III transcription inhibits tRNA degradation via RTD pathway. RNA 18, 1823–1832 10.1261/rna.033597.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ellis S.R., Morales M.J., Li J.M., Hopper A.K. and Martin N.C. (1986) Isolation and characterization of the TRM1 locus, a gene essential for the N2,N2-dimethylguanosine modification of both mitochondrial and cytoplasmic tRNA in Saccharomyces cerevisiae. J. Biol. Chem. 261, 9703–9709 PMID: [PubMed] [Google Scholar]
- 34.Powell C.A., Nicholls T.J. and Minczuk M. (2015) Nuclear-encoded factors involved in post-transcriptional processing and modification of mitochondrial tRNAs in human disease. Front. Genet. 6, 79 10.3389/fgene.2015.00079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Massenet S., Motorin Y., Lafontaine D.L., Hurt E.C., Grosjean H. and Branlant C. (1999) Pseudouridine mapping in the Saccharomyces cerevisiae spliceosomal U small nuclear RNAs (snRNAs) reveals that pseudouridine synthase pus1p exhibits a dual substrate specificity for U2 snRNA and tRNA. Mol. Cell. Biol. 19, 2142–2154 10.1128/MCB.19.3.2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carlile T.M., Rojas-Duran M.F., Zinshteyn B., Shin H., Bartoli K.M. and Gilbert W.V. (2014) Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515, 143–146 10.1038/nature13802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lalo D., Carles C., Sentenac A. and Thuriaux P. (1993) Interactions between three common subunits of yeast RNA polymerases I and III. Proc. Natl Acad. Sci. U.S.A. 90, 5524–5528 10.1073/pnas.90.12.5524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang Y. and Maraia R.J. (2001) Comparison of the RNA polymerase III transcription machinery in Schizosaccharomyces pombe, Saccharomyces cerevisiae and human. Nucleic Acids Res. 29, 2675–2690 10.1093/nar/29.13.2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cramer P., Bushnell D.A., Fu J., Gnatt A.L., Maier-Davis B., Thompson N.E. et al. (2000) Architecture of RNA polymerase II and implications for the transcription mechanism. Science 288, 640–649 10.1126/science.288.5466.640 [DOI] [PubMed] [Google Scholar]
- 40.Chapon C., Cech T.R. and Zaug A.J. (1997) Polyadenylation of telomerase RNA in budding yeast. RNA 3, 1337–1351 PMID: [PMC free article] [PubMed] [Google Scholar]
- 41.Desai N., Lee J., Upadhya R., Chu Y., Moir R.D. and Willis I.M. (2005) Two steps in Maf1-dependent repression of transcription by RNA polymerase III. J. Biol. Chem. 280, 6455–6462 10.1074/jbc.M412375200 [DOI] [PubMed] [Google Scholar]
- 42.Moir R.D., Lee J., Haeusler R.A., Desai N., Engelke D.R. and Willis I.M. (2006) Protein kinase A regulates RNA polymerase III transcription through the nuclear localization of Maf1. Proc. Natl Acad. Sci. U.S.A. 103, 15044–15049 10.1073/pnas.0607129103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roberts D.N., Wilson B., Huff J.T., Stewart A.J. and Cairns B.R. (2006) Dephosphorylation and genome-wide association of Maf1 with Pol III-transcribed genes during repression. Mol. Cell 22, 633–644 10.1016/j.molcel.2006.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boisnard S., Lagniel G., Garmendia-Torres C., Molin M., Boy-Marcotte E., Jacquet M. et al. (2009) H2O2 activates the nuclear localization of Msn2 and Maf1 through thioredoxins in Saccharomyces cerevisiae. Eukaryot. Cell 8, 1429–1438 10.1128/EC.00106-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee J., Moir R.D. and Willis I.M. (2009) Regulation of RNA polymerase III transcription involves SCH9-dependent and SCH9-independent branches of the target of rapamycin (TOR) pathway. J. Biol. Chem. 284, 12604–12608 10.1074/jbc.C900020200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei Y., Tsang C.K. and Zheng X.F.S. (2009) Mechanisms of regulation of RNA polymerase III-dependent transcription by TORC1. EMBO J. 28, 2220–2230 10.1038/emboj.2009.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Graczyk D., Debski J., Muszyńska G., Bretner M., Lefebvre O. and Boguta M. (2011) Casein kinase II-mediated phosphorylation of general repressor Maf1 triggers RNA polymerase III activation. Proc. Natl Acad. Sci. U.S.A. 108, 4926–4931 10.1073/pnas.1010010108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oler A.J. and Cairns B.R. (2012) PP4 dephosphorylates Maf1 to couple multiple stress conditions to RNA polymerase III repression. EMBO J. 31, 1440–1452 10.1038/emboj.2011.501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boguta M. (2013) Maf1, a general negative regulator of RNA polymerase III in yeast. Biochim. Biophys. Acta 1829, 376–384 10.1016/j.bbagrm.2012.11.004 [DOI] [PubMed] [Google Scholar]
- 50.Huber A., Bodenmiller B., Uotila A., Stahl M., Wanka S., Gerrits B. et al. (2009) Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev. 23, 1929–1943 10.1101/gad.532109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Towpik J., Graczyk D., Gajda A., Lefebvre O. and Boguta M. (2008) Derepression of RNA polymerase III transcription by phosphorylation and nuclear export of its negative regulator, Maf1. J. Biol. Chem. 283, 17168–17174 10.1074/jbc.M709157200 [DOI] [PubMed] [Google Scholar]
- 52.Morawiec E., Wichtowska D., Graczyk D., Conesa C., Lefebvre O. and Boguta M. (2013) Maf1, repressor of tRNA transcription, is involved in the control of gluconeogenetic genes in Saccharomyces cerevisiae. Gene 526, 16–22 10.1016/j.gene.2013.04.055 [DOI] [PubMed] [Google Scholar]
- 53.Adamczyk M. and Szatkowska R. (2017) Low RNA polymerase III activity results in up regulation of HXT2 glucose transporter independently of glucose signaling and despite changing environment. PLoS ONE 12, e0185516 10.1371/journal.pone.0185516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kwapisz M., Smagowicz W.J., Oficjalska D., Hatin I., Rousset J.-P., Żołądek T. et al. (2002) Up-regulation of tRNA biosynthesis affects translational readthrough in maf1-Δ mutant of Saccharomyces cerevisiae. Curr. Genet. 42, 147–152 10.1007/s00294-002-0342-7 [DOI] [PubMed] [Google Scholar]
- 55.Longtine M.S., McKenzie A. III, Demarini D.J., Shah N.G., Wach A., Brachat A. et al. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 56.Gietz R.D. and Schiestl R.H. (2007) Large-scale high-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 38–41 10.1038/nprot.2007.15 [DOI] [PubMed] [Google Scholar]
- 57.Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 58.Cox J. and Mann M. (2008) Maxquant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- 59.Choi M., Chang C.-Y., Clough T., Broudy D., Killeen T., MacLean B. et al. (2014) MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics 30, 2524–2526 10.1093/bioinformatics/btu305 [DOI] [PubMed] [Google Scholar]
- 60.Mi H., Muruganujan A. and Thomas P.D. (2013) PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 41, D377–D386 10.1093/nar/gks1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kanehisa M., Furumichi M., Tanabe M., Sato Y. and Morishima K. (2017) KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45, D353–D361 10.1093/nar/gkw1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jensen L.J., Kuhn M., Stark M., Chaffron S., Creevey C., Muller J. et al. (2009) STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 37, D412–D416 10.1093/nar/gkn760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D. et al. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carmona-Saez P., Chagoyen M., Tirado F., Carazo J.M. and Pascual-Montano A. (2007) GENECODIS: a web-based tool for finding significant concurrent annotations in gene lists. Genome Biol. 8, R3 10.1186/gb-2007-8-1-r3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cankorur-Cetinkaya A., Dereli E., Eraslan S., Karabekmez E., Dikicioglu D. and Kirdar B. (2012) A novel strategy for selection and validation of reference genes in dynamic multidimensional experimental design in yeast. PLoS ONE 7, e38351 10.1371/journal.pone.0038351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ozcan S. and Johnston M. (1995) Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Mol. Cell. Biol. 15, 1564–1572 10.1128/MCB.15.3.1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Adamczyk M., van Eunen K., Bakker B.M. and Westerhoff H.V. (2011) Enzyme kinetics for systems biology. Methods Enzymol. 500, 233–257 10.1016/B978-0-12-385118-5.00013-X [DOI] [PubMed] [Google Scholar]
- 68.van Hoek P., van Dijken J.P. and Pronk J.T. (2000) Regulation of fermentative capacity and levels of glycolytic enzymes in chemostat cultures of Saccharomyces cerevisiae. Enzyme Microb. Technol. 26, 724–736 10.1016/S0141-0229(00)00164-2 [DOI] [PubMed] [Google Scholar]
- 69.Grüning N.-M., Rinnerthaler M., Bluemlein K., Mülleder M., Wamelink M.M.C., Lehrach H. et al. (2011) Pyruvate kinase triggers a metabolic feedback loop that controls redox metabolism in respiring cells. Cell Metab. 14, 415–427 10.1016/j.cmet.2011.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Postma E., Verduyn C., Scheffers W.A. and Dijken J.P.V. (1989) Enzymic analysis of the crabtree effect in glucose-limited chemostat cultures of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 55, 468 PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Smale S.T. (2010) Beta-galactosidase assay. Cold Spring Harb. Protoc. 2010, pdb.prot5423 10.1101/pdb.prot5423 [DOI] [PubMed] [Google Scholar]
- 72.Beers R.F. and Sizer I.W. (1952) A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J. Biol. Chem. 195, 133–140 PMID: [PubMed] [Google Scholar]
- 73.Rossouw D., Heyns E.H., Setati M.E., Bosch S. and Bauer F.F. (2013) Adjustment of trehalose metabolism in wine Saccharomyces cerevisiae strains to modify ethanol yields. Appl. Environ. Microbiol. 79, 5197–5207 10.1128/AEM.00964-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Parrou J.L. and François J. (1997) A simplified procedure for a rapid and reliable assay of both glycogen and trehalose in whole yeast cells. Anal. Biochem. 248, 186–188 10.1006/abio.1997.2138 [DOI] [PubMed] [Google Scholar]
- 75.Van Hoek P., Van Dijken J.P. and Pronk J.T. (1998) Effect of specific growth rate on fermentative capacity of baker's yeast. Appl. Environ. Microbiol. 64, 4226–4233 PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Picotti P., Bodenmiller B., Mueller L.N., Domon B. and Aebersold R. (2009) Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 138, 795–806 10.1016/j.cell.2009.05.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Feng Y., De Franceschi G., Kahraman A., Soste M., Melnik A., Boersema P.J. et al. (2014) Global analysis of protein structural changes in complex proteomes. Nat. Biotechnol. 32, 1036–1044 10.1038/nbt.2999 [DOI] [PubMed] [Google Scholar]
- 78.Carlson M. (1999) Glucose repression in yeast. Curr. Opin. Microbiol. 2, 202–207 10.1016/S1369-5274(99)80035-6 [DOI] [PubMed] [Google Scholar]
- 79.Young E.T., Dombek K.M., Tachibana C. and Ideker T. (2003) Multiple pathways are co-regulated by the protein kinase Snf1 and the transcription factors Adr1 and Cat8. J. Biol. Chem. 278, 26146–26158 10.1074/jbc.M301981200 [DOI] [PubMed] [Google Scholar]
- 80.Wendell D.L. and Bisson L.F. (1994) Expression of high-affinity glucose transport protein Hxt2p of Saccharomyces cerevisiae is both repressed and induced by glucose and appears to be regulated posttranslationally. J. Bacteriol. 176, 3730–3737 10.1128/jb.176.12.3730-3737.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Galazzo J.L. and Bailey J.E.. Growing Saccharomyces cerevisiae in calcium-alginate beads induces cell alterations which accelerate glucose conversion to ethanol. Biotechnol. Bioeng. 36, 417–426 10.1002/bit.260360413 [DOI] [PubMed] [Google Scholar]
- 82.Rolland F., Winderickx J. and Thevelein J.M. (2002) Glucose-sensing and -signalling mechanisms in yeast. FEMS Yeast Res. 2, 183–201 10.1111/j.1567-1364.2002.tb00084.x [DOI] [PubMed] [Google Scholar]
- 83.Elbing K., Larsson C., Bill R.M., Albers E., Snoep J.L., Boles E. et al. (2004) Role of hexose transport in control of glycolytic flux in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 70, 5323–5330 10.1128/AEM.70.9.5323-5330.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Guillaume C., Delobel P., Sablayrolles J.-M. and Blondin B. (2007) Molecular basis of fructose utilization by the wine yeast Saccharomyces cerevisiae: a mutated HXT3 allele enhances fructose fermentation. Appl. Environ. Microbiol. 73, 2432–2439 10.1128/AEM.02269-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lane S., Xu H., Oh E.J., Kim H., Lesmana A., Jeong D. et al. (2018) Glucose repression can be alleviated by reducing glucose phosphorylation rate in Saccharomyces cerevisiae. Sci. Rep. 8, 2613 10.1038/s41598-018-20804-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Herrero P., Galíndez J., Ruiz N., Martínez-Campa C. and Moreno F. (1995) Transcriptional regulation of the Saccharomyces cerevisiae HXK1, HXK2 and GLK1 genes. Yeast 11, 137–144 10.1002/yea.320110205 [DOI] [PubMed] [Google Scholar]
- 87.Bakker B.M., Walsh M.C., ter Kuile B.H., Mensonides F.I.C., Michels P.A.M., Opperdoes F.R. et al. (1999) Contribution of glucose transport to the control of the glycolytic flux in Trypanosoma brucei. Proc. Natl Acad. Sci. U.S.A. 96, 10098–10103 10.1073/pnas.96.18.10098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shestov A.A., Liu X., Ser Z., Cluntun A.A., Hung Y.P., Huang L. et al. (2014) Quantitative determinants of aerobic glycolysis identify flux through the enzyme GAPDH as a limiting step. eLife 3, e03342 10.7554/eLife.03342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pearce A.K., Crimmins K., Groussac E., Hewlins M.J.E., Dickinson J.R., Francois J. et al. (2001) Pyruvate kinase (Pyk1) levels influence both the rate and direction of carbon flux in yeast under fermentative conditions. Microbiology 147, 391–401 10.1099/00221287-147-2-391 [DOI] [PubMed] [Google Scholar]
- 90.Zampar G.G., Kümmel A., Ewald J., Jol S., Niebel B., Picotti P. et al. (2013) Temporal system-level organization of the switch from glycolytic to gluconeogenic operation in yeast. Mol. Syst. Biol. 9, 651 10.1038/msb.2013.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zheng L., Roeder R.G. and Luo Y. (2003) S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell 114, 255–266 10.1016/S0092-8674(03)00552-X [DOI] [PubMed] [Google Scholar]
- 92.White M.R. and Garcin E.D. (2016) The sweet side of RNA regulation: glyceraldehyde-3-phosphate dehydrogenase as a noncanonical RNA-binding protein. Wiley Interdiscip. Rev. RNA 7, 53–70 10.1002/wrna.1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Blair J.B. and Walker R.G. (1984) Rat liver pyruvate kinase: influence of ligands on activity and fructose 1,6-bisphosphate binding. Arch. Biochem. Biophys. 232, 202–213 10.1016/0003-9861(84)90536-8 [DOI] [PubMed] [Google Scholar]
- 94.Jurica M.S., Mesecar A., Heath P.J., Shi W., Nowak T. and Stoddard B.L. (1998) The allosteric regulation of pyruvate kinase by fructose-1,6-bisphosphate. Structure 6, 195–210 10.1016/S0969-2126(98)00021-5 [DOI] [PubMed] [Google Scholar]
- 95.Dombrauckas J.D., Santarsiero B.D. and Mesecar A.D. (2005) Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis. Biochemistry 44, 9417–9429 10.1021/bi0474923 [DOI] [PubMed] [Google Scholar]
- 96.Xu Y.-F., Zhao X., Glass D.S., Absalan F., Perlman D.H., Broach J.R. et al. (2012) Regulation of yeast pyruvate kinase by ultrasensitive allostery independent of phosphorylation. Mol. Cell. 48, 52–62 10.1016/j.molcel.2012.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Apweiler E., Sameith K., Margaritis T., Brabers N., van de Pasch L., Bakker L.V. et al. (2012) Yeast glucose pathways converge on the transcriptional regulation of trehalose biosynthesis. BMC Genomics 13, 239 10.1186/1471-2164-13-239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hohmann S., Bell W., NevesA M.J., Valckx D. and Thevelein J.M. (1996) Evidence for trehalose-6-phosphate-dependent and -independent mechanisms in the control of sugar influx into yeast glycolysis. Mol. Microbiol. 20, 981–991 10.1111/j.1365-2958.1996.tb02539.x [DOI] [PubMed] [Google Scholar]
- 99.Teusink B., Walsh M.C., van Dam K. and Westerhoff H.V. (1998) The danger of metabolic pathways with turbo design. Trends Biochem. Sci. 23, 162–169 10.1016/S0968-0004(98)01205-5 [DOI] [PubMed] [Google Scholar]
- 100.van Heerden J.H., Wortel M.T., Bruggeman F.J., Heijnen J.J., Bollen Y.J.M., Planqué R. et al. (2014) Lost in transition: start-up of glycolysis yields subpopulations of nongrowing cells. Science 343, 1245114 10.1126/science.1245114 [DOI] [PubMed] [Google Scholar]
- 101.Shulman R.G. and Rothman D.L. (2015) Homeostasis and the glycogen shunt explains aerobic ethanol production in yeast. Proc. Natl Acad. Sci. U.S.A. 112, 10902–10907 10.1073/pnas.1510730112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Blázquez M.A., Lagunas R., Gancedo C. and Gancedo J.M. (1993) Trehalose-6-phosphate, a new regulator of yeast glycolysis that inhibits hexokinases. FEBS Lett. 329, 51–54 10.1016/0014-5793(93)80191-V [DOI] [PubMed] [Google Scholar]
- 103.Gancedo C. and Flores C.-L. (2004) The importance of a functional trehalose biosynthetic pathway for the life of yeasts and fungi. FEMS Yeast Res. 4, 351–359 10.1016/S1567-1356(03)00222-8 [DOI] [PubMed] [Google Scholar]
- 104.Krüger A., Grüning N.-M., Wamelink M.M.C., Kerick M., Kirpy A., Parkhomchuk D. et al. (2011) The pentose phosphate pathway is a metabolic redox sensor and regulates transcription during the antioxidant response. Antioxid. Redox Signal. 15, 311–324 10.1089/ars.2010.3797 [DOI] [PubMed] [Google Scholar]
- 105.Stanton R.C. (2012) Glucose-6-phosphate dehydrogenase, NADPH, and cell survival. IUBMB Life 64, 362–369 10.1002/iub.1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stincone A., Prigione A., Cramer T., Wamelink M.M.C., Campbell K., Cheung E. et al. (2015) The return of metabolism: biochemistry and physiology of the pentose phosphate pathway. Biol. Rev. Camb. Philos. Soc. 90, 927–963 10.1111/brv.12140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rebrin I., Kamzalov S. and Sohal R.S. (2003) Effects of age and caloric restriction on glutathione redox state in mice. Free Radic. Biol. Med. 35, 626–635 10.1016/S0891-5849(03)00388-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Walsh M.E., Shi Y. and Van Remmen H. (2014) The effects of dietary restriction on oxidative stress in rodents. Free Radic. Biol. Med. 66, 88–99 10.1016/j.freeradbiomed.2013.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hinnebusch A.G. (2005) Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59, 407–450 10.1146/annurev.micro.59.031805.133833 [DOI] [PubMed] [Google Scholar]
- 110.Natarajan K., Meyer M.R., Jackson B.M., Slade D., Roberts C., Hinnebusch A.G. et al. (2001) Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 21, 4347–4368 10.1128/MCB.21.13.4347-4368.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Conesa C., Ruotolo R., Soularue P., Simms T.A., Donze D., Sentenac A. et al. (2005) Modulation of yeast genome expression in response to defective RNA polymerase III-dependent transcription. Mol. Cell. Biol. 25, 8631–8642 10.1128/MCB.25.19.8631-8642.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tate J.J., Rai R. and Cooper T.G. (2018) More than one way in: three Gln3 sequences required to relieve negative Ure2 regulation and support nuclear Gln3 import in Saccharomyces cerevisiae. Genetics 208, 207–227 10.1534/genetics.117.300457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Xie Y. and Varshavsky A. (2001) RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: a negative feedback circuit. Proc. Natl Acad. Sci. U.S.A. 98, 3056–3061 10.1073/pnas.071022298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chou H.-J., Donnard E., Gustafsson H.T., Garber M. and Rando O.J. (2017) Transcriptome-wide analysis of roles for tRNA modifications in translational regulation. Mol. Cell 68, 978–992.e4 10.1016/j.molcel.2017.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vilela C., Linz B., Rodrigues-Pousada C. and McCarthy J.E. (1998) The yeast transcription factor genes YAP1 and YAP2 are subject to differential control at the levels of both translation and mRNA stability. Nucleic Acids Res. 26, 1150–1159 10.1093/nar/26.5.1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Filer D., Thompson M.A., Takhaveev V., Dobson A.J., Kotronaki I., Green J.W.M. et al. (2017) RNA polymerase III limits longevity downstream of TORC1. Nature 552, 263–267 10.1038/nature25007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li Y., Moir R.D., Sethy-Coraci I.K., Warner J.R. and Willis I.M. (2000) Repression of ribosome and tRNA synthesis in secretion-defective cells is signaled by a novel branch of the cell integrity pathway. Mol. Cell Biol. 20, 3843–3851 10.1128/MCB.20.11.3843-3851.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yin Z., Wilson S., Hauser N.C., Tournu H., Hoheisel J.D. and Brown A.J.P. (2003) Glucose triggers different global responses in yeast, depending on the strength of the signal, and transiently stabilizes ribosomal protein mRNAs. Mol. Microbiol. 48, 713–724 10.1046/j.1365-2958.2003.03478.x [DOI] [PubMed] [Google Scholar]
- 119.Oliveira A.P., Ludwig C., Picotti P., Kogadeeva M., Aebersold R. and Sauer U. (2012) Regulation of yeast central metabolism by enzyme phosphorylation. Mol. Syst. Biol. 8, 623 10.1038/msb.2012.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Barbosa A.D., Pereira C., Osório H., Moradas-Ferreira P. and Costa V. (2016) The ceramide-activated protein phosphatase Sit4p controls lifespan, mitochondrial function and cell cycle progression by regulating hexokinase 2 phosphorylation. Cell Cycle 15, 1620–1630 10.1080/15384101.2016.1183846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ihmels J.H. and Bergmann S. (2004) Challenges and prospects in the analysis of large-scale gene expression data. Brief. Bioinformatics 5, 313–327 10.1093/bib/5.4.313 [DOI] [PubMed] [Google Scholar]
- 122.François J. and Parrou J.L.) Reserve carbohydrates metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 25, 125–145 10.1111/j.1574-6976.2001.tb00574.x [DOI] [PubMed] [Google Scholar]
- 123.Ptacek J., Devgan G., Michaud G., Zhu H., Zhu X., Fasolo J. et al. (2005) Global analysis of protein phosphorylation in yeast. Nature 438, 679–684 10.1038/nature04187 [DOI] [PubMed] [Google Scholar]
- 124.Marín-Hernández A., Rodríguez-Enríquez S., Vital-González P.A., Flores-Rodríguez F.L., Macías-Silva M., Sosa-Garrocho M. et al. (2006) Determining and understanding the control of glycolysis in fast-growth tumor cells. Flux control by an over-expressed but strongly product-inhibited hexokinase. FEBS J. 273, 1975–1988 10.1111/j.1742-4658.2006.05214.x [DOI] [PubMed] [Google Scholar]
- 125.Bonhoure N., Byrnes A., Moir R.D., Hodroj W., Preitner F., Praz V. et al. (2015) Loss of the RNA polymerase III repressor MAF1 confers obesity resistance. Genes Dev. 29, 934–947 10.1101/gad.258350.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.van Dijken J.P., van den Bosch E., Hermans J.J., de Miranda L.R. and Scheffers W.A. (1986) Alcoholic fermentation by ‘non-fermentative’ yeasts. Yeast 2, 123–127 10.1002/yea.320020208 [DOI] [PubMed] [Google Scholar]
- 127.Godon C., Lagniel G., Lee J., Buhler J.M., Kieffer S., Perrot M. et al. (1998) The H2O2 stimulon in Saccharomyces cerevisiae. J. Biol. Chem. 273, 22480–22489 10.1074/jbc.273.35.22480 [DOI] [PubMed] [Google Scholar]
- 128.Ralser M., Wamelink M.M., Kowald A., Gerisch B., Heeren G., Struys E.A. et al. (2007) Dynamic rerouting of the carbohydrate flux is key to counteracting oxidative stress. J. Biol. 6, 10 10.1186/jbiol61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Patra K.C. and Hay N. (2014) The pentose phosphate pathway and cancer. Trends Biochem. Sci. 39, 347–354 10.1016/j.tibs.2014.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Delaunay A., Pflieger D., Barrault M.B., Vinh J. and Toledano M.B. (2002) A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell 111, 471–481 10.1016/S0092-8674(02)01048-6 [DOI] [PubMed] [Google Scholar]
- 131.Shenton D. and Grant C.M. (2003) Protein S-thiolation targets glycolysis and protein synthesis in response to oxidative stress in the yeast Saccharomyces cerevisiae. Biochem. J. 374, 513–519 10.1042/bj20030414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mierzejewska J. and Chreptowicz K. (2016) Lack of Maf1 enhances pyruvate kinase activity and fermentative metabolism while influencing lipid homeostasis in Saccharomyces cerevisiae. FEBS Lett. 590, 93–100 10.1002/1873-3468.12033 [DOI] [PubMed] [Google Scholar]
- 133.Amelio I., Cutruzzolá F., Antonov A., Agostini M. and Melino G. (2014) Serine and glycine metabolism in cancer. Trends Biochem. Sci. 39, 191–198 10.1016/j.tibs.2014.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Vander Heiden M.G., Cantley L.C. and Thompson C.B. (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 10.1126/science.1160809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kuehne A., Emmert H., Soehle J., Winnefeld M., Fischer F., Wenck H. et al. (2015) Acute activation of oxidative pentose phosphate pathway as first-line response to oxidative stress in human skin cells. Mol. Cell 59, 359–371 10.1016/j.molcel.2015.06.017 [DOI] [PubMed] [Google Scholar]
- 136.Echols N., Harrison P., Balasubramanian S., Luscombe N.M., Bertone P., Zhang Z. et al. (2002) Comprehensive analysis of amino acid and nucleotide composition in eukaryotic genomes, comparing genes and pseudogenes. Nucleic Acids Res. 30, 2515–2523 10.1093/nar/30.11.2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Avruch J., Long X., Ortiz-Vega S., Rapley J., Papageorgiou A. and Dai N. (2009) Amino acid regulation of TOR complex 1. Am. J. Physiol. Endocrinol. Metab. 296, E592–E602 10.1152/ajpendo.90645.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sancak Y., Peterson T.R., Shaul Y.D., Lindquist R.A., Thoreen C.C., Bar-Peled L. et al. (2008) The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501 10.1126/science.1157535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Saxton R.A., Chantranupong L., Knockenhauer K.E., Schwartz T.U. and Sabatini D.M. (2016) Mechanism of arginine sensing by CASTOR1 upstream of mTORC1. Nature 536, 229–233 10.1038/nature19079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hopper A.K. (2013) Transfer RNA post-transcriptional processing, turnover, and subcellular dynamics in the yeast Saccharomyces cerevisiae. Genetics 194, 43–67 10.1534/genetics.112.147470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Becker-Kettern J., Paczia N., Conrotte J.-F., Kay D.P., Guignard C., Jung P.P. et al. (2016) Saccharomyces cerevisiae forms d-2-hydroxyglutarate and couples its degradation to d-lactate formation via a cytosolic transhydrogenase. J. Biol. Chem. 291, 6036–6058 10.1074/jbc.M115.704494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Losman J.-A. and Kaelin W.G. (2013) What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev. 27, 836–852 10.1101/gad.217406.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Rideout E.J., Marshall L. and Grewal S.S. (2012) Drosophila RNA polymerase III repressor Maf1 controls body size and developmental timing by modulating tRNAiMet synthesis and systemic insulin signaling. Proc. Natl Acad. Sci. U.S.A. 109, 1139–1144 10.1073/pnas.1113311109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pavon-Eternod M., Gomes S., Rosner M.R. and Pan T. (2013) Overexpression of initiator methionine tRNA leads to global reprogramming of tRNA expression and increased proliferation in human epithelial cells. RNA 19, 461–466 10.1261/rna.037507.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Dai Z., Mentch S.J., Gao X., Nichenametla S.N. and Locasale J.W. (2018) Methionine metabolism influences genomic architecture and gene expression through H3K4me3 peak width. Nat. Commun. 9, 1955 10.1038/s41467-018-04426-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Campbell K., Vowinckel J., Keller M.A. and Ralser M. (2016) Methionine metabolism alters oxidative stress resistance via the pentose phosphate pathway. Antioxid. Redox Signal. 24, 543–547 10.1089/ars.2015.6516 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.