Abstract
Purpose of review
Within-host diversity complicates transmission models because it recognizes that between-host virus phylogenies are not identical to the transmission history among the infected hosts. This review presents the biological and theoretical foundations for recent development in this field, and shows that modern phylodynamic methods are capable of inferring realistic transmission histories from HIV sequence data.
Recent findings
Transmission of single or multiple genetic variants from a donor’s HIV population results in donor-recipient phylogenies with combinations of mono-, para-, and poly-phyletic patterns. Large-scale simulations and analyses of many real HIV datasets have established that transmission direction, directness, or common source often can be inferred based on HIV sequence data. Phylodynamic reconstruction of HIV transmissions that include within-host HIV diversity have recently been established and made available in several software packages.
Summary
Phylodynamic methods that include realistic features of HIV genetic diversification have come of age, significantly improving inference of key epidemiological parameters. This opens the door to more accurate surveillance and better-informed prevention campaigns.
Keywords: transmission, phylogeny, phylodynamics, within-host evolution, transmission direction
Introduction
Phylogenetics is an attractive method to reconstruct HIV epidemics because 1) HIV evolves faster than it spreads, thus accumulating mutations as it spreads, 2) phylogenetics reconstructs evolutionary history, and 3) at the heart of any epidemic is transmission, which phylogenetics thusly can reconstruct. Here, we will review developments in the recent 18 months in phylogenetic inference of HIV transmissions, explicitly taking within-host diversity into account when reconstructing transmission events.
It is well known that HIV both diversifies and diverges in an infected person (1–4). Diversification is the process of generating genetic variability that exist at any given time point. It is from this diversity that immune escape and antiviral resistance is selected. The diversity can be thought of as a cloud of genetic variants. Divergence is the movement of this cloud away from the infecting variant(s). The rate of the divergence is described by a molecular clock. The evolutionary process that dictates HIV evolution is a complex interaction between virus and host factors, as well as outside factors such as antiviral treatment. The outcome of these interactions is patient specific (5–7), disease stage dependent (8), and virus related (9, 10). From a modeling point of view, HIV evolution is described by a combination of neutral and selective processes (11).
Because HIV within-host diversity can be very large, joint HIV phylogenies from patients that are linked through transmissions are not identical to their transmission history (12–14). Note that while within-host diversity can be summarized by a single statistic such as mean pair-wise genetic distance, it can also be described in more detail by a within-host phylogeny of the sampled virus variants, i.e., a dendogram that shows how the sampled HIV sequences are related to each other by descent (tree topology) and extent of nucleotide substitutions or time (branch lengths). Consequently, a joint phylogeny shows how sampled virus variants from more than one patient are evolutionarily linked to each other.
The time point when a transmission occurred is not directly displayed in a joint HIV phylogeny; instead, the location in the joint phylogeny where the recipient’s HIV population joins the donor’s HIV population simply relates to a random time point that indicates when a transmitted lineage coalesces with some lineage that can be reconstructed from the available sample in the donor. The difference in transmission time point and the corresponding coalescence time point is known as the pre-transmission interval (13, 14), which quantifies the bias backwards in time. The magnitude of this bias is determined by the level of diversity in the donor at time of transmission (12). Another effect of the within-host diversity is known as incomplete lineage sorting (15, 16), resulting in disordered transmission events, e.g., where it may seem as if a newborn child infected her mother (14, 17). The probability of such disordering also depends on the level of diversity and additionally how much time that has passed between the two transmission events (12). While these effects of within-host diversity may seem discouraging for phylogenetic transmission reconstructions, counterfactually, the within-host diversity also provides an opportunity to better reconstruct the transmission history.
Taking within-host diversity into account enhances transmission reconstruction
At transmission, a relatively small number of virus particles is typically transmitted. Thus, with any diversity present in the donor, only a subset of the available genetic variants is transferred from one host (donor) to a new host (recipient), known as a genetic bottleneck. Consequently, the new HIV population in the recipient will typically have a smaller level of diversity, and importantly for our purpose of reconstructing the transmission event, the recipient’s HIV population will sit inside the diversity of the donor’s HIV phylogeny. Phylogenetically, the donor’s HIV population is paraphyletic to the recipient’s HIV population. While this relationship has been described and observed in the past (14, 18), recent research has systematically investigated what type of phylogenetic relationship to expect in different types of transmission.
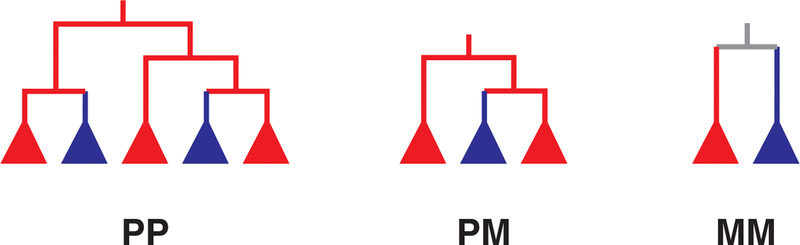
Based on a coalescent model of within-host HIV evolution, systematic simulation experiments have shown that direct transmission (from host A to B) typically results in either a paraphyletic-monophyletic (PM) or a paraphyletic-polyphyletic (PP) donor-recipient joint phylogeny (19). Figure 1 shows the prototypic joint phylogenies that can result from reconstructing the evolutionary history of the HIV populations in two epidemiologically linked hosts. Recipient monophyly results from transmission of a single genetic variant from the donor, and polyphyly when more than one variant is transmitted. When an intermediary link exists (A infects X, who is not sampled, and X later infects B), typically a PM donor-recipient joint phylogeny appears. Finally, when both A and B are infected by a common source, the joint A+B phylogeny is typically monophyletic-monophyletic (MM). Theoretically, it was found that these expected relationships were robust under many parameter settings, when at least 20 HIV variants were sequenced from each host (A and B), and sampling occurred at about the same time in both hosts. It was also shown that with inadequate sampling of genetic variants, the phylogenetic relationships would be harder to correctly recover (19). Similarly, with time, lineage death in the ongoing evolutionary process will eventually result in MM phylogenies regardless of transmission history. Lineage death results from the fact that not all viruses produce viable offspring.
Figure 1. Prototypic joint HIV phylogenies from two epidemiologically linked hosts.
These are the 3 possible topological classes that can be observed in a phylogenetic reconstruction of HIV transmission between two hosts (subtrees are colored according to which host HIV was sampled from, red or blue). The filled triangles mean that there can be any number of taxa (sequences) from only one host therein. In the PP phylogeny, the red population is paraphyletic and the blue population is polyphyletic, meaning that blue has more than one origin in the red population. Using parsimony, the root state in the PP phylogeny is red, suggesting that the red host infected the blue host in this example (but see main text for a discussion about transmission direction reconstruction). PP trees can also be ambiguous about root state, meaning both red and blue populations are polyphyletic. In the PM phylogeny, red is again paraphyletic while blue is monophyletic. Monophyletic means that all taxa from the same population have a single ancestor. The MM phylogeny thus has two monophyletic populations, one from each host, and is always ambiguous about root state (grey).
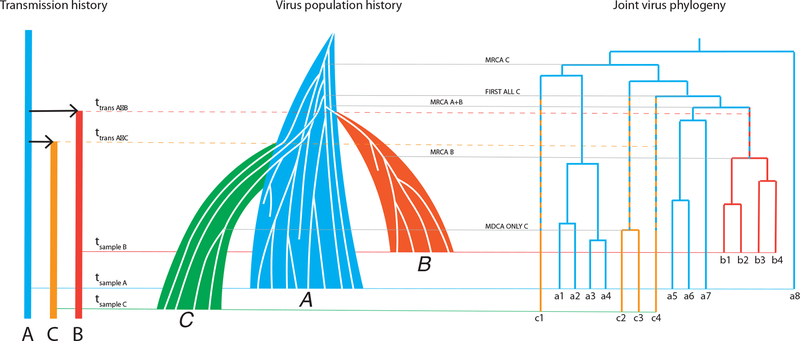
Figure 2 shows how a joint virus phylogeny from three hosts and the underlying transmission history are combined in the corresponding virus population history. In this example, A first infects B, and later C. Later yet, samples are collected from first B, then A and last C. From each such sample we retrieve 4, 8, and 4 sequences, respectively, from each host. Note that the number of sequences is too small for a real study (it should be aiming for at least 20); we show a small number here merely to keep the illustration simple. The sequences result in a joint phylogeny that is a sample from the HIV populations in A, B, and C. The actual HIV populations in A, B, and C have many more genetic variants in them than we have sampled. Using a coalescent framework, we can model the size and diversity of these populations based on the sample using a simple linear growth model, Nei(t) = αi + βit, where Nei(t) is the effective population size of host i = (A,B,C) at t, αi is the size of the infecting population, and βi is the slope of the population growth (12, 19). Note that the effective population size is related to the level of diversity in a host, and not to the census size that can be estimated from the viral load.
Figure 2. The connection between transmission history, virus population history, and the sampled phylogeny.
In this cartoon, person A (blue) infects first person B (red) at time ttrans A->B and later person C (green) at time ttrans A->C. Time flows from top to bottom. The resulting transmission history thus describes who-infected-whom and when. Each person can be described with a (colored) line that starts at the time of infection. If we draw a sample from each infected person, we can indicate the sampling times in the transmission history (at times tsample B, tsample A, and tsample C, respectively, in that order). From each sample, we extract a relatively small number of virus sequences (labelled as a1–a8, b1–b4, and c1–c4) from typically very large virus populations. Each person hosts a continuously evolving virus population (A, B, C), where new genetic virus variants are born and die. Based on the sample of virus sequences from each infected person, we can infer a joint virus phylogeny that reconstructs how the virus populations share a common descent. Based on the phylogeny, we can infer when certain events occurred in the tree, such as most recent common ancestors (MRCAs), discussed in the main text.
Due to the pre-transmission interval, none of the transmission times are represented by any node in the joint phylogeny. The phylogeny does provide limits of when transmission could have occurred, however. When only one variant is transmitted, as in the A to B transmission, the most recent time is limited by the most recent common ancestor of all phylogenetic lineages that were sampled in B (MRCA B). Clearly, a larger sample may push this limit further back in time. The most distant time point when transmission could have occurred is estimated by MRCA A+B, and similarly a larger sample from A may push this limit towards the present. The possible time interval of when A infected B is shown as an alternating blue-red branch segment. When two, or more, unique variants are transmitted, as in the A to C transmission, the most recent time of transmission is limited by the most distant common ancestor that occurs among lineages found only in C (MDCA ONLY C). The most distant time point of the possible transmission interval is estimated by the first occurrence of all unique lineages that end up in C (FIRST ALL C). The possible time interval when A infected C is shown as alternating blue-green branch segments leading to all green segments when the lineages must be in host C. Note also that the MRCA C has nothing to do with the time of transmission as this happens between random lineages in host A. Again, sampling more variants in A and C may reduce the possible time interval during which transmission could have occurred. Notice that a naïve interpretation, looking for when the transmitted lineages originate in A, would mislead about the order and timing of transmissions to B and C because C gets infected with lineages that happen to go further back in A even though C was infected after B.
The overall HIV population history shows how we are able to model and reconstruct the transmission history from a joint virus phylogeny that is based on a limited sample of HIV variants from each infected host (Fig 2). A and B are related to each other by a PM phylogeny, A and C to each other by a PP phylogeny, and B and C by a MM phylogeny. These types of pairwise relationships were confirmed by massive simulations under different parameter values of α, β, and t (19). The figure also shows that lineages that existed before sampling may have died out. The coalescent model can recapture the diversity that such lost variants may have contributed to at some point in the past, e.g., estimating the diversity at time of transmission (20).
A recent study evaluating 955 transmission pair datasets with known epidemiological linkage confirmed the theoretical expectations of mainly observing PM and PP phylogenies in direct transmissions (from A to B directly), and MM phylogenies when A and B had a common source (21). The study involved 272 previously published transmission chains, often sequenced in more than one genomic region and containing more than two hosts, decomposed into 955 genomic regions with a transmission pair. Overall, 52% of direct transmissions resulted in a detected PP phylogeny, 37% in a PM phylogeny and 11% in a MM phylogeny, while 76% of common source transmissions resulted in a MM phylogeny. Interestingly, PP phylogenies dominated (66%) among mother-to-child transmissions, and PP phylogenies were also more common in MSM (52%) than in HET transmission (19%). Even though PP phylogenies were observed quite frequently, transmission of more than one genetic variant is likely more common than suggested by the fraction of PP phylogenies, and previously expected, because of insufficient variant sampling and lineage death before samples were taken. The study also found that rooting the joint phylogeny is crucial as PM phylogenies could be mis-rooted as MM phylogenies or transmission direction could be reversed. The best rooting was achieved by using several sequences from the matching HIV-1 subtype or circulating recombinant form (CRF) as outgroup.
Lineage death over time, as well as the time of sampling relative to when transmissions occurred, may play unexpected tricks on the resulting joint phylogeny. In a recent study of a transmission that involved multiple variants, sampling of the donor much later than that of the recipient, and very different Ne growth rates in the hosts, lead to that the resulting PP phylogeny suggested that the recipient had infected the donor (20). Thus, this study presents a cautionary tale that even though PP phylogenies may indicate direct transmission, interpretation of the direction of transmission (A to B or B to A) must be done carefully. In this case, extensive simulations of the exact epidemiological scenario, explicitly taking sampling times into account, could reveal the true donor. When many phylogenetic lineages are transmitted the chance of transmitting old lineages increases, hence increasing the probability that the recipient carries an older lineage than the donor at time of sampling; indeed, in the study of the 955 real transmission pairs, it was found that PP phylogenies proposed the wrong direction of transmission in 24% of the cases (21). Similarly, a recent study of 33 index-partner pairs in the HPTN052 cohort also showed that simple root state reconstruction may mislead or be insufficient to determine the donor in linked transmissions (22). This points out that caution must be exercised when evaluating individual cases, as case details may be very different from previous studies.
Taking within-host diversity into account reveals underlying epidemic processes
While it may be interesting to analyze individual transmission events between a donor and a recipient for a wide range of reasons, the application of using HIV sequence data to identify how HIV spreads in a human population is particularly important as it can access otherwise difficult to estimate epidemiological parameters such as transmission risks, underlying contact networks, and numbers of infections over time. Phylodynamic methods have shown much promise, but until recently they typically ignored within-host diversity and evolution. Recently, however, several efforts have been made to allow for within-host diversity of a transmitted pathogen, either with HIV in mind, generically, or other pathogens in mind (23–25).
De Maio et al developed SCOTTI (26), a generic method to take within-host diversity into account when reconstructing transmission events in outbreak investigations using sequence data. SCOTTI is available as a BEAST 2 module (27), taking advantage of the rich BEAST environment. More recently, with deep next-generation sequence (NGS) data in mind, Skums et al developed QUENTIN (28), a software that reconstructs transmission histories among multiple hosts while taking within-host evolution into account. Instead of a conventional phylogenetic approach, QUENTIN uses a graph-based approach that can estimate transmission direction from sequence data. Favorizing scale-free transmission networks (29, 30), it maximizes the probability of observing a transmission tree given the genetic network of observed virus sequences and an arc weight function. The transmission tree identifies transmission directions, the genetic network describes how the sequences are related to each other by single mutations, and the arc weights are equal to genetic distances between the virus populations studied.
Anticipating very large data sets from epidemics, as well as virus full genome NGS data, Wymant et al recently expanded the above described donor-recipient phylogenetic patterns (Fig. 1) to the epidemic level in the software PHYLOSCANNER (31). This software performs phylogenetic reconstructions in multiple windows across aligned HIV genomes from multiple patients. Based on the most parsimonious host label at nodes joining sequences from different patients, transmission directions among the patients are reconstructed. Aggregating the results from all windows, PHYLOSCANNER constructs possible transmission histories displayed as relationship graphs. This approach mitigates random reconstruction errors, where greater credibility is given to those relationships that are observed more frequently. Because NGS data can have high error rates and may be sensitive to contamination, PHYLOSCANNER also identifies suspicious signals that typically are different from true phylogenetic events stemming from the within-host evolutionary process and transmission(s).
Within-host diversity was also recently taken into account when analyzing phylogenetic patterns that may result from different transmission network types. Giardina et al showed that HIV sequences sampled from epidemics that spread in archetypical network structures, characterized by different degree distributions and amount of clustering, result in different phylogenetic patterns, thus making it possible to infer general epidemic transmission histories (32). This study showed that a HIV time-scaled phylogeny from many patients may be substantially different than the between-host transmission history, and by not taking within-host diversity into account, the phylogeny may get misinterpreted leading to erroneous inference about the underlying epidemic contact network. While within-host evolution may display a disordered phylogeny vis-a-vis the transmission events, importantly, the diversification process also adds discriminatory power to differentiate between different types of contact networks.
Because within-host diversity causes significant backwards bias on infection times and may disorder transmission events compared to the actual transmission history, phylodynamic estimates of numbers of infected hosts may become severely overestimated if within-host HIV diversity is ignored. Volz et al showed that a multi-scale coalescent, which takes within-host diversity into account, can accurately estimate the number of infected hosts in growing HIV epidemics (33). In an analysis of a large outbreak among intravenous drug users in Latvia (30, 34), the multi-scale coalescent gave estimates of cumulative numbers of infected hosts close to the cumulative number of diagnoses in the epidemic. Surprisingly, working with only a single sequence per host (which currently is the standard in public health databases), the multi-scale coalescent is capable of estimating the within-host diversity in infected patients in an epidemic.
Conclusion
Given adequate data, phylogenetic analysis of HIV sequences can often reconstruct transmission direction. Typically, HIV sequence data can infer that direct transmission has occurred when a PP tree is observed, that transmission occurred either directly or indirectly from A to B when a PM tree is observed, and that an unsampled person infected both A and B when a MM tree is observed. With either too few sequences per host, or too long time since transmission, these patterns become more uncertain. Additional caution is also called for when sampling times vary between hosts, and when diversification rates are very different between hosts. In-depth analyses, which at this time are computationally expensive, may reveal transmission direction when the epidemiological and phylogenetic patterns are complicated. Recent softwares have exploited the theoretical expectations resulting from HIV transmission and added significant realism to modern phylodynamic inference of HIV epidemics. Future development of epidemiological models that include within-host evolution in even greater detail may further improve epidemiological reconstruction and prediction.
Key bullet points.
HIV phylogenies based on multiple clones from each host typically reveal the type of epidemiological linkage between the sampled hosts
Paraphyly typically reveals the donor in direct and directional transmissions
Polyphyly typically indicates direct transmission, but direction may be ambiguous depending on evolutionary and epidemiological parameters
A common source among sampled hosts typically results in monophyletic clades of each host’s HIV population
Several softwares have been developed to take within-host evolution into account to probabilistically infer the transmission history among sampled hosts
Acknowledgements
Financial support
NIH NIAID grant R01 AI087520
Footnotes
Conflicts of interest
The author has no conflicts of interest.
References and recommended reading
Papers of particular interest, published within the 18-month period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Shankarappa R, Margolick JB, Gange SJ, Rodrigo AG, Upchurch D, Farzadegan H, Gupta P, Rinaldo CR, Learn GH, He X, Huang X-L, Mullins JI. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol 73:10489–10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leitner T, Halapi E, Scarlatti G, Rossi P, Albert J, Fenyö EM, Uhlén M. 1993. Analysis of heterogeneous viral populations by direct DNA sequencing. BioTechniques 15:120–126. [PubMed] [Google Scholar]
- 3.Wolfs TFW, Zwart G, Bakker M, Goudsmit J. 1992. HIV-1 genomic RNA diversification following sexual parenteral virus transmission. Virology 189:103–110. [DOI] [PubMed] [Google Scholar]
- 4.McNearney T, Hornickova Z, Markham R, Birdwell A, Arens M, Saah A, Ratner L. 1992. Relationship of human immunodeficiency virus type 1 sequence heterogeneity to stage of disease. Proceedings of the National Academy of Sciences of the United States of America 89:10247–10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halapi E, Leitner T, Jansson M, Scarlatti G, Orlandi P, Plebani A, Romiti L, Albert J, Wigzell H, Rossi P. 1997. Correlation between HIV sequence evolution, specific immune response and clinical outcome in vertically infected infants. AIDS 11:1709–1717. [DOI] [PubMed] [Google Scholar]
- 6.Bagnarelli P, Mazzola F, Menzo S, Montroni M, Butini L, Clementi M. 1999. Host-specific modulation of the selective constraints driving human immunodeficiency virus type 1 env gene evolution. J Virol 73:3764–3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salemi M 2013. The intra-host evolutionary and population dynamics of human immunodeficiency virus type 1: a phylogenetic perspective. Infect Dis Rep 5:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee HY, Perelson AS, Park SC, Leitner T. 2008. Dynamic correlation between intrahost HIV-1 quasispecies evolution and disease progression. PLoS Comput Biol 4:e1000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hollingsworth TD, Anderson RM, Fraser C. 2008. HIV-1 transmission, by stage of infection. J Infect Dis 198:687–693. [DOI] [PubMed] [Google Scholar]
- 10.Lythgoe KA, Fraser C. 2012, p 3367–3375. Proc. R. Soc. B [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leitner T 2018. The Puzzle of HIV Neutral and Selective Evolution. Mol Biol Evol 35:1355–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Romero-Severson E, Skar H, Bulla I, Albert J, Leitner T. 2014. Timing and Order of Transmission Events Is Not Directly Reflected in a Pathogen Phylogeny. Mol Biol Evol 31:2472–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leitner T, Albert J. 1999. The molecular clock of HIV-1 unveiled through analysis of a known transmission history. Proc. Natl. Acad. Sci. USA 96:10752–10757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leitner T, Fitch WM. 1999. The phylogenetics of known transmission histories In Crandall KA (ed.), The evolution of HIV. Johns Hopkins Univ. Press, Baltimore, MD. [Google Scholar]
- 15.Pamilo P, Nei M. 1988. Relationships between gene trees and species trees. Mol. Biol. Evol 5:568–583. [DOI] [PubMed] [Google Scholar]
- 16.Degnan JH, Rosenberg NA. 2006. Discordance of species trees with their most likely gene trees. PLoS Genet 2:e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leitner T, Escanilla D, Franzén C, Uhlén M, Albert J. 1996. Accurate reconstruction of a known HIV-1 transmission history by phylogenetic tree analysis. Proc. Natl. Acad. Sci. USA 93:10864–10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scaduto DI, Brown JM, Haaland WC, Zwickl DJ, Hillis DM, Metzker ML. 2010. Source identification in two criminal cases using phylogenetic analysis of HIV-1 DNA sequences. Proc Natl Acad Sci U S A 107:21242–21247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romero-Severson EO, Bulla I, Leitner T. 2016. Phylogenetically resolving epidemiologic linkage. Proc Natl Acad Sci U S A 113:2690–2695.** Establishes expected phylogenetic patterns in direct, indirect, and common source transmissions by simulations under a wide range of transmission situations.
- 20.Romero-Severson EO, Bulla I, Hengartner N, Bartolo I, Abecasis A, Azevedo-Pereira JM, Taveira N, Leitner T. 2017. Donor-Recipient Identification in Para- and Poly-phyletic Trees Under Alternative HIV-1 Transmission Hypotheses Using Approximate Bayesian Computation. Genetics.* Highlights that simple phylogenetic interpretation of apparent phylogenetic patterns may mislead about transmission direction. In-depth analyses can nevertheless recover transmission direction.
- 21.Leitner T, Romero-Severson E. 2018. Phylogenetic patterns recover known HIV epidemiological relationships and reveal common transmission of multiple variants. Nat Microbiol 3:983–988.** Evaluates phylogenetic patterns in real HIV sequence data from 955 transmission pair genomic regions. Theoretical expectations of typical phylogenetic patterns are confirmed and transmission of multiple variants appears more common than previously thought.
- 22.Rose R, Hall M, Redd AD, Lamers S, Barbier AE, Porcella SF, Hudelson SE, Piwowar-Manning E, McCauley M, Gamble T, Wilson EA, Kumwenda J, Hosseinipour MC, Hakim JG, Kumarasamy N, Chariyalertsak S, Pilotto JH, Grinsztejn B, Mills LA, Makhema J, Santos BR, Chen YQ, Quinn TC, Fraser C, Cohen MS, Eshleman SH, Laeyendecker O. 2018. Phylogenetic methods inconsistently predict direction of HIV transmission among heterosexual pairs in the HPTN052 cohort. J Infect Dis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klinkenberg D, Backer JA, Didelot X, Colijn C, Wallinga J. 2017. Simultaneous inference of phylogenetic and transmission trees in infectious disease outbreaks. PLoS Comput Biol 13:e1005495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Didelot X, Fraser C, Gardy J, Colijn C. 2017. Genomic infectious disease epidemiology in partially sampled and ongoing outbreaks. Mol Biol Evol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ypma RJ, van Ballegooijen WM, Wallinga J. 2013. Relating phylogenetic trees to transmission trees of infectious disease outbreaks. Genetics 195:1055–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Maio N, Wu CH, Wilson DJ. 2016. SCOTTI: Efficient Reconstruction of Transmission within Outbreaks with the Structured Coalescent. PLoS Comput Biol 12:e1005130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D, Suchard MA, Rambaut A, Drummond AJ. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol 10:e1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skums P, Zelikovsky A, Singh R, Gussler W, Dimitrova Z, Knyazev S, Mandric I, Ramachandran S, Campo D, Jha D, Bunimovich L, Costenbader E, Sexton C, O’Connor S, Xia GL, Khudyakov Y. 2018. QUENTIN: reconstruction of disease transmissions from viral quasispecies genomic data. Bioinformatics 34:163–170.* Development of a software that takes within-host diversity into account from deep NGS data when reconstructing transmission histories.
- 29.Leigh Brown AJ, Lycett SJ, Weinert L, Hughes GJ, Fearnhill E, Dunn DT, Collaboration UHDR. 2011. Transmission network parameters estimated from HIV sequences for a nationwide epidemic. J Infect Dis 204:1463–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Graw F, Leitner T, Ribeiro RM. 2012. Agent-based and phylogenetic analyses reveal how HIV-1 moves between risk groups: injecting drug users sustain the heterosexual epidemic in Latvia. Epidemics 4:104–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wymant C, Hall M, Ratmann O, Bonsall D, Golubchik T, de Cesare M, Gall A, Cornelissen M, Fraser C, Stop-Hcv Consortium TMPC, The BC. 2017. PHYLOSCANNER: Inferring Transmission from Within- and Between-Host Pathogen Genetic Diversity. Mol Biol Evol.** Development of a software for large-scale analyses of epidemics with many hosts with full genome NGS data that takes within-host diversity into account.
- 32.Giardina F, Romero-Severson EO, Albert J, Britton T, Leitner T. 2017. Inference of Transmission Network Structure from HIV Phylogenetic Trees. PLoS Comput Biol 13:e1005316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Volz EM, Romero-Severson E, Leitner T. 2017. Phylodynamic inference across epidemic scales. Mol Biol Evol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balode D, Ferdats A, Dievberna I, Viksna L, Rozentale B, Kolupajeva T, Konicheva V, Leitner T. 2004. Rapid epidemic spread of HIV type 1 subtype A1 among intravenous drug users in Latvia and slower spread of subtype B among other risk groups. AIDS Res Hum Retroviruses 20:245–249. [DOI] [PubMed] [Google Scholar]