ATRA, a biologically active metabolite of vitamin A, exerts pleiotropic biological effects, including the activation of both innate and adaptive immunity, thereby serving as a potent antimicrobial compound against numerous viral pathogens. Despite the enrichment of hepatocytes with vitamin A, HCV still establishes an efficient viral life cycle. Here, we discovered that the hepatocellular response to ATRA creates either a proviral or an antiviral environment depending on its engagement with CRABP1 or -2, respectively. CRABP1 supports the robust replication of HCV, while CRABP2 potently inhibits the efficiency of viral replication. Our biochemical, genetic, and microscopic analyses reveal that the pro- and antiviral effects of CRABPs are mediated by modulation of LD abundance, where HCV establishes the platform for viral replication and assembly on the LD-associated ER membrane. This study uncovered a cell-intrinsic mechanism by which HCV exploits the proviral function of CRABP1 to establish an efficient viral life cycle.
KEYWORDS: cellular retinoic acid binding protein, all-trans retinoic acid, hepatitis C virus, lipid droplet, vitamin A
ABSTRACT
Retinoid (vitamin A) is an essential diet constituent that governs a broad range of biological processes. Its biologically active metabolite, all-trans retinoic acid (ATRA), exhibits a potent antiviral property by enhancing both innate and adaptive antiviral immunity against a variety of viral pathogens, such as, but not limited to, HIV, respiratory syncytial virus (RSV), herpes simplex virus (HSV), and measles. Even though the hepatocyte is highly enriched with retinoid and its metabolite ATRA, it supports the establishment of efficient hepatitis C virus (HCV) replication. Here, we demonstrate the hepatocyte-specific cell-intrinsic mechanism by which ATRA exerts either a proviral or antiviral effect, depending on how it engages cellular retinoic acid binding proteins (CRABPs). We found that the engagement of CRABP1 by ATRA potently supported viral infection by promoting the accumulation of lipid droplets (LDs), which robustly enhanced the formation of a replication complex on the LD-associated endoplasmic reticulum (ER) membrane. In contrast, ATRA binding to CRABP2 potently inhibited HCV via suppression of LD accumulation. However, this antiviral effect of CRABP2 was abrogated due to the functional and quantitative predominance of CRABP1 in the hepatocytes. In summary, our study demonstrates that CRABPs serve as an on-off switch that modulates the efficiency of the HCV life cycle and elucidates how HCV evades the antiviral properties of ATRA via the exploitation of CRABP1 functionality.
IMPORTANCE ATRA, a biologically active metabolite of vitamin A, exerts pleiotropic biological effects, including the activation of both innate and adaptive immunity, thereby serving as a potent antimicrobial compound against numerous viral pathogens. Despite the enrichment of hepatocytes with vitamin A, HCV still establishes an efficient viral life cycle. Here, we discovered that the hepatocellular response to ATRA creates either a proviral or an antiviral environment depending on its engagement with CRABP1 or -2, respectively. CRABP1 supports the robust replication of HCV, while CRABP2 potently inhibits the efficiency of viral replication. Our biochemical, genetic, and microscopic analyses reveal that the pro- and antiviral effects of CRABPs are mediated by modulation of LD abundance, where HCV establishes the platform for viral replication and assembly on the LD-associated ER membrane. This study uncovered a cell-intrinsic mechanism by which HCV exploits the proviral function of CRABP1 to establish an efficient viral life cycle.
INTRODUCTION
Retinoid, also known as vitamin A, is an essential diet constituent that governs a broad range of biological processes, including the antimicrobial defense program. Accordingly, hypovitaminosis A is associated with enhanced virulence, while supplementation improves the clinical outcomes of a variety of viral infectious diseases (1). The biologically active metabolite of vitamin A, all-trans retinoic acid (ATRA), contributes to the antiviral defense program by regulating the differentiation, growth, and migration of both innate and adaptive immune cells. It has also been shown that the antiviral effect of ATRA is partially mediated by the enhanced expression of antiviral genes, referred to as interferon (IFN)-stimulated genes (ISGs), which cooperatively govern intracellular antiviral innate immunity (2–6).
The liver plays a central role in the regulation of systemic retinoid homeostasis, as it stores up to 80% of total body retinoid (7). The diet-derived retinoid is first taken up by hepatocytes and stored in the cytoplasm in the form of retinol (ROL) or retinyl ester (RE). Hepatocytes then secrete ROL or RE into the systemic circulation for the maintenance of the serum retinoid concentration. Alternatively, an excessive amount of retinoid in hepatocytes is transferred to hepatic stellate cells (HSC), a nonparenchymal cell of the liver, for additional storage (8). These notions indicate that hepatocytes and HSC contain a substantially high quantity and concentration of retinoid compared with other cell types. In hepatocytes, ROL receives two steps of oxidative metabolism by medium-chain dehydrogenases, such as alcohol dehydrogenases (ADHs) and aldehyde dehydrogenases (ALDHs), for the biogenesis of ATRA (9, 10).
ATRA regulates the transcription of more than 500 genes through the activation of the nuclear receptor complex retinoic acid receptor (RAR)-retinoid X receptor (RXR) heterodimer (11). Given the hydrophobic nature of ATRA, it requires a chaperone molecule that captures and shuttles it from the cytoplasm to the nucleus in order to exert gene regulatory activity. Cellular retinoic acid binding proteins (CRABPs), CRABP1 and -2, are the chaperone molecules that capture ATRA in the cytoplasm (12). Although these two proteins share extremely high homology, a few previous studies suggested that cellular responses to ATRA exhibit partially differential biological effects depending on its engagement with CRABP1 or CRABP2. For example, CRABP1 expression appears to upregulate cellular proliferation, and thus, its abundant expression in cancerous tissue is associated with poor prognosis (13). In contrast, the abundant expression of CRABP2 is coupled with growth inhibition and apoptosis of tumor cells (14). Based on these observations, CRABP1 and -2 appear to have inverse functionality, at least in the field of cancer biology. To date, however, it remains unexplored how CRABPs modulate the hepatocellular response to ATRA or whether CRABPs influence the antiviral properties of ATRA.
Consequently, this study investigated the underlying mechanism by which HCV establishes an efficient viral life cycle despite hepatocytes consisting of a substantial amount of antiviral vitamin by focusing on the differential roles of CRABP1 and -2 in hepatocytes. Of great interest, we found that CRABP1 is a potent proviral host factor, while CRABP2 greatly limits the hepatitis C virus (HCV) life cycle. Our nutrigenomics studies of CRABP1- and -2-expressing cells illustrated a grossly redundant transcriptome profile with exceptions seen in a few subsets of genes. Our bioinformatics analysis followed by in vitro biochemical and virological studies suggested that the proviral property of CRABP1 is mediated by its capacity to moderate the cellular response to ATRA. This event results in the accumulation of lipid droplets (LDs), which enhanced the efficiency of the viral life cyle. In contrast, the antiviral activity of CRABP2 resulted from the inhibition of LD accumulation. Finally, we found that the effect of CRABP1 surpasses that of CRABP2 on the regulation of LD abundance. In summary, our results revealed a novel aspect of retinoid biology regulated by CRABPs, which modulate the efficiency of the HCV life cycle through the regulation of hepatic lipid homeostasis.
RESULTS
CRABP1 and -2 differentially regulate HCV infection in hepatocytes.
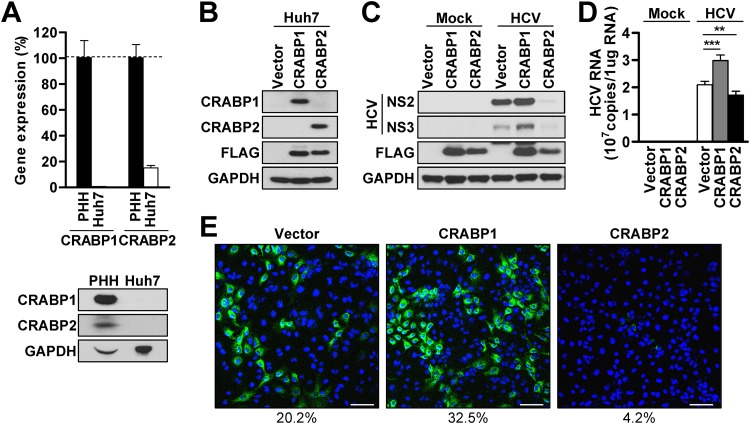
Both CRABP1 and -2 are known to serve as chaperone molecules of ATRA in the cytoplasm and modulate the cellular responses to vitamin A (12). As in vitro study of the HCV life cycle is restricted to the use of Huh7-derived cells, we first evaluated the relative expression of CRABP1 and -2 in Huh7 cells in comparison with that in freshly isolated primary human hepatocytes (PHH). The result demonstrated that Huh7 cells barely express CRABP1 and -2 at both the transcript and protein levels (Fig. 1A). In contrast, PHH abundantly expressed CRABP1 and, to a much lesser extent, CRABP2 (Fig. 1A; see also Fig. S1A in the supplemental material). To investigate the significance of CRABPs in the HCV life cycle, we established a set of Huh7 cells that were individually reconstituted for the expression of CRABP1 or -2 (Fig. 1B and Fig. S1B and S1C). First, we assessed whether the expression of CRABPs in Huh7 cells influences cell proliferation or susceptibility to cell death, which may nonspecifically impact the efficiency of the viral life cycle given its heavy reliance on the general cellular machinery. The result demonstrated that the reconstitution of CRABP1 and -2 in Huh7 cells had a negligible influence on cell viability and the rate of growth/proliferation (Fig. S1D and S1E). Next, we challenged Huh7-CRAPB1 or -2 cells with an HCV infectious clone, and infectivity was compared with that in control cells by assessing the abundances of viral RNA and protein. The results demonstrated that CRABP1 expression in Huh7 cells substantially enhances the efficiency of HCV infection, while CRABP2 expression is coupled with a potent antiviral effect (Fig. 1C to E). These results indicate that CRABP1 and -2 have an inverse biological effect, at least for the regulation of the HCV life cycle in hepatocytes.
FIG 1.
CRABP1 and -2 differently regulate HCV infection. (A) Freshly isolated primary human hepatocytes (PHH) and Huh7 cells were used for the assessment of CRABP1 and -2 expression levels via RT-qPCR (top panel) and immunoblot analysis (bottom panel). The relative CRABP transcript abundances in Huh7 cells compared to those in PHH are shown as percentages after normalization with GAPDH of each cell type. (B) Immunoblot analysis for the detection of stably expressed Flag-tagged CRABPs in Huh7 cells. (C and D) Immunoblot analysis of HCV proteins (NS2 and NS3) (C) or relative HCV genome abundances (D) in CRABP-expressing Huh7 cells infected with HCV (MOI of 0.1) for 48 h. **, P < 0.01; ***, P < 0.001. (E) Immunofluorescence microscopic analysis of CRABP-expressing Huh7 cells infected with HCV (MOI of 0.1) for 48 h. Blue, DAPI; green, anti-HCV core. The percentages shown below the images indicate the number of HCV-positive cells/total number of cells. Bars, 100 μm. See also Fig. S1 in the supplemental material.
CRABPs regulate HCV infection at the stage of viral replication.
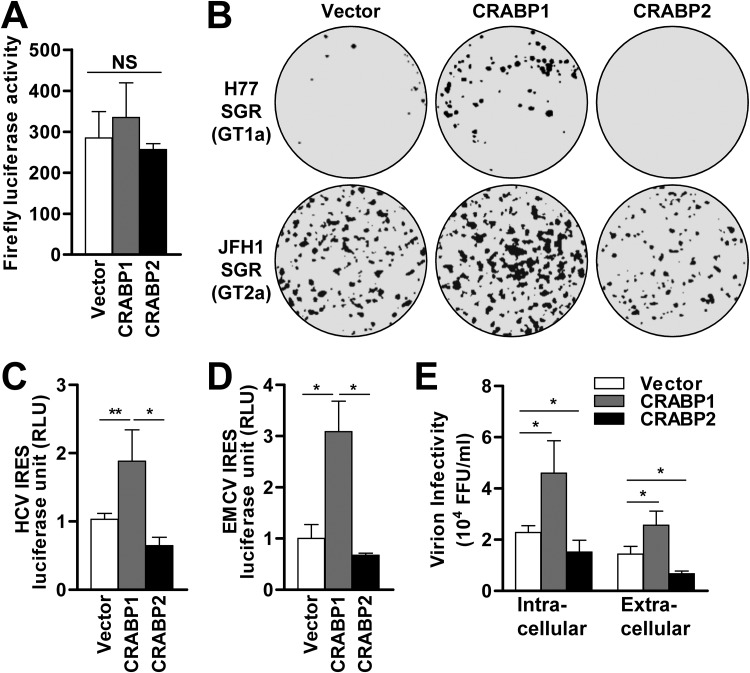
To further understand how the expression of CRABPs in Huh7 cells differentially regulates the efficiency of HCV infection, we tested their effect at each stage of the viral life cycle. First, an HCV pseudoparticle (HCVpp) system that encodes a firefly luciferase reporter was utilized to test the effect of CRABPs on viral entry. The result indicated that the expression of CRABPs has a negligible effect at the stage of viral entry (Fig. 2A). Next, an HCV subgenomic replicon (SGR) transduction assay was employed to assess the influence of CRABPs on HCV replication, which involves both viral protein translation and genome amplification. Similar to the result from the HCV infectious clone experiment (Fig. 1C to E), the results with the HCV SGR transduction assay with two distinct viral genotypes (GTs) (H77, GT1a; JFH1, GT2a) demonstrated the proviral effect of CRABP1 as well as the antiviral effect of CRABP2 (Fig. 2B and Fig. S2A and S2B).
FIG 2.
CRABPs regulate HCV infection at the stage of viral replication. (A) Huh7 cells stably expressing CRABPs were challenged with HCVpp (p24; 20 ng/ml), followed by a firefly luciferase reporter assay 96 h after transduction. NS, not significant. (B) Huh7 cells stably expressing CRABPs were electroporated with in vitro-transcribed HCV subgenomic replicon (SGR) genomes (top, GT1a H77; bottom, GT2a JFH1) followed by G418 selection for 3 weeks prior to crystal violet staining. (C and D) Huh7 cells stably expressing CRABPs were transfected with a bicistronic vector containing cap-dependent Renilla and HCV IRES-dependent firefly luciferases (C) or EMCV IRES-dependent firefly luciferase (D). *, P < 0.05; **, P < 0.01. Relative luciferase activity is presented as the ratio of IRES-dependent luciferase activity to cap-dependent luciferase activity. RLU, relative luciferase units. (E) Cell lysates and supernatants of CRABP-expressing cells infected with HCV for 72 h at an MOI of 0.1 were subjected to an HCV infection titer assay using Huh7.5 cells. The titers of intracellular and extracellular infectious particles were determined as focus-forming units (FFU) per milliliter. *, P < 0.05. See also Fig. S2 in the supplemental material.
Consequently, we investigated whether CRABPs modulate the HCV life cycle at the stage of viral protein translation using a luciferase reporter system that is regulated by the HCV internal ribosome entry site (IRES) (Fig. 2C and D and Fig. S2C). The result indicated that CRABP1 expression greatly upregulated the efficiency of HCV IRES-dependent protein translation, while it showed a modest suppression of cap-dependent protein translation (Fig. 2C and Fig. S2C). Of note, we also found that the enhancement of IRES-dependent translation by CRABP1 is not unique to the HCV IRES, as an experiment using an encephalomyocarditis virus (EMCV) IRES luciferase reporter system also demonstrated a similar outcome (Fig. 2D and Fig. S2D). In addition, we observed that the expression of CRABP2 showed an inhibitory effect on both IRES- and cap-dependent protein translation (Fig. S2C and S2D). Finally, we evaluated the influence of CRABP expression on the abundance of intracellular and extracellular infectious particles (Fig. 2E). We found that CRABP1-expressing cells had increased abundances of both intracellular and extracellular virions, while CRABP2-expressing cells contained significantly smaller amounts of infectious particles. Taken together, these observations suggest that CRABP1 and -2 inversely modulate the efficiency of the HCV life cycle at the stage of viral replication, likely at IRES-dependent protein translation.
HCV-specific regulation of the viral life cycle by CRABPs.
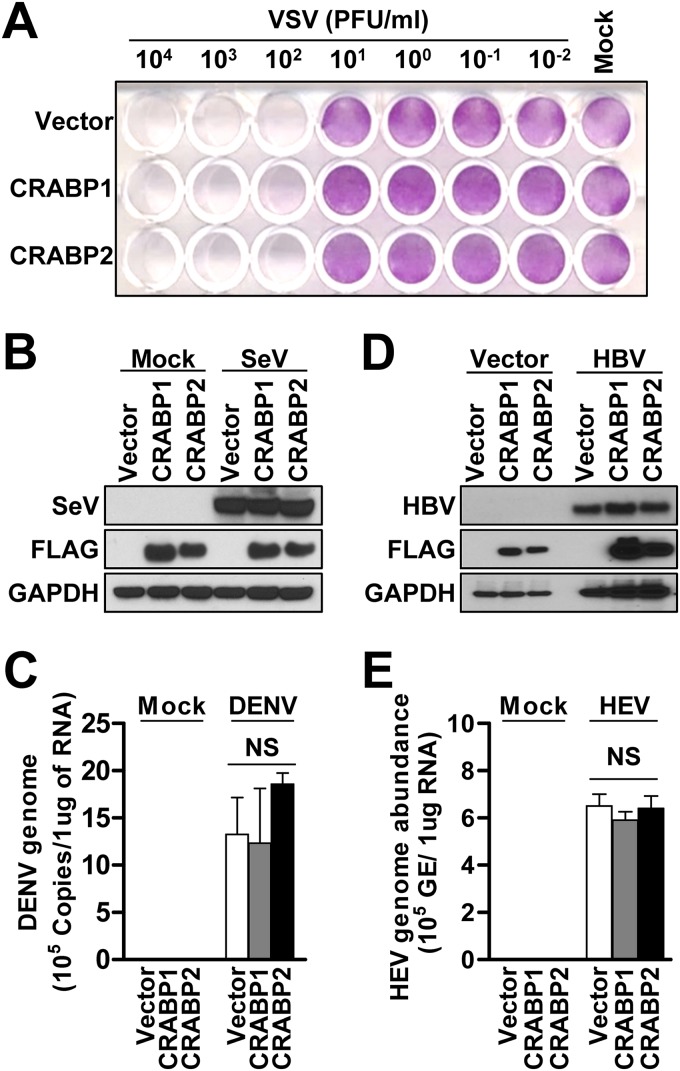
We next tested whether the pro- and antiviral effects of CRABPs could be generalized to other RNA viruses that belong to different viral families, such as vesicular stomatitis virus (VSV) and Sendai virus (SeV). The results showed that neither the cytopathic effects nor the replication efficiencies of these pathogens were influenced by the expression of CRABPs (Fig. 3A and B and Fig. S3A). Next, we tested the pro- and antiviral effects of CRABPs on another member of the Flaviviridae, dengue virus (DENV). The result showed that the expression of CRABPs had no significant effect on the efficiency of DENV infection (Fig. 3C and Fig. S3B and S3C). Subsequently, we extended our investigations to test whether the regulatory effects of CRABPs are specific to the life cycle of hepatotropic viral pathogens. To test this possibility, we overexpressed the hepatitis B virus (HBV) genome or performed a challenge with hepatitis E virus (HEV) in Huh7-CRABP1 and -2 cells. Similarly to VSV and SeV, the expression of CRABP1 and -2 had a negligible effect on HBV replication and HEV infection (Fig. 3D and E and Fig. S3D and S3E). Taken together, our data suggest that the regulatory effect of CRABP expression is unique to the HCV life cycle but not other DNA or RNA viruses.
FIG 3.
HCV-specific regulation of the viral life cycle by CRABPs. (A) Huh7 cells stably expressing CRABPs were infected with vesicular stomatitis virus (VSV) at the indicated PFU per milliliter for 24 h, followed by crystal violet staining. Images are representative of results from three independent experiments. (B) Cell lysates from Huh7 cells stably expressing CRABPs and infected with Sendai virus (SeV) at 100 hemagglutinating units (HAU)/ml for 24 h were subjected to immunoblot analysis for the detection of the indicated proteins. (C) Cell lysates from Huh7 cells stably expressing CRABPs and infected with dengue virus (DENV) at an MOI of 0.2 for 72 h were subjected to assessment of viral genome abundance by RT-qPCR. NS, not significant. (D) Huh7 cells stably expressing CRABPs were transfected with the p1.3x HBV DNA-encoding plasmid (HBV) or pUC19 (vector control) for 48 h, followed by immunoblot analysis for the detection of HBV core proteins. (E) Total RNAs extracted from Huh7 cells stably expressing CRABPs and infected with hepatitis E virus (HEV) at 6 × 108 genome equivalents (GE)/ml for 5 days were subjected to a probe-based RT-qPCR assay for the quantification of HEV genome copies. Each bar represents the mean ± SD from triplicate samples. NS, not significant. See also Fig. S3 in the supplemental material.
Transcriptome analysis reveals the cell biology governed by CRABP1 and -2.
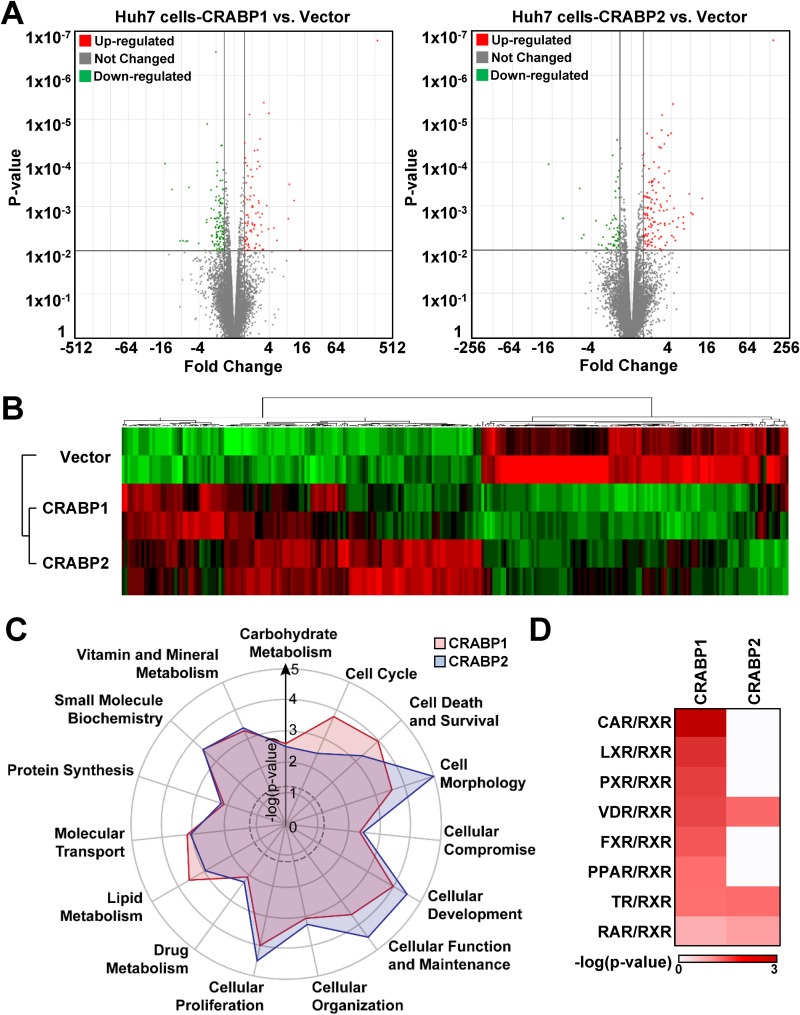
In order to better understand the mechanism by which the expression of CRABP1 and -2 inversely regulates the viral life cycle in an HCV-specific manner, we conducted an RNA sequencing (RNA-seq)-based transcriptome analysis of Huh7-CRABP1 and -2 cells. The volcano analysis revealed that Huh7-CRABP1 and -2 cells differentially express 168 and 148 genes, respectively (Fig. 4A). The subsequent heat map analysis indicated that these genes differentially expressed in CRABP1 and -2 cells grossly overlap (Fig. 4B and Fig. S4A), suggesting that the functions of CRABP1 and -2 may largely be redundant. Of note, however, the heat map analysis also demonstrated that there are subsets of genes that are uniquely up- or downregulated in cells stably expressing CRABP1 or -2 (Fig. 4B and Tables S1 to S3).
FIG 4.
Transcriptome analysis reveals the cell biology governed by CRABP1 and -2. (A and B) Total RNAs extracted from Huh7 cells stably expressing the control vector or CRABPs were subjected to mRNA sequencing. (A) A volcano plot illustrating the gene-specific analysis results, which revealed the genes differentially expressed in CRABP1- or -2-expressing cells. Cutoffs used were a P value of 0.01 and a fold change of >1.5 or −1.5. (B) Hierarchical clustering demonstrates the differentially regulated genes in CRABP1- or -2-expressing cells. (C) Functional enrichment analyses of the differentially expressed genes in either CRABP1- or -2-expressing cells were performed using IPA. Overrepresented functional processes are shown in a radar plot, with their enrichment P values. The dotted line indicates the threshold for statistical significance (enrichment P value of <0.05). (D) IPA canonical pathway analysis shows the enrichment of selective retinoid-relative pathways in CRABP-expressing cells. The color intensity reflects the −log of the enrichment P values. See also Fig. S4 and Tables S1 to S3 in the supplemental material.
It is plausible that these differences seen between Huh7-CRABP1 and -2 cells are likely the factors, at least in part, that inversely regulate the efficiency of the HCV life cycle. We therefore conducted an Ingenuity Pathway Analysis (IPA) for the cellular and molecular functions of CRABP1- and -2-expressing cells using the genes that are differentially expressed in these cells (Fig. 4C and Tables S1 to S3). The result recapitulated the redundancy seen in the heat map analysis, with a few exceptions, such as lipid metabolism, cell death, cell morphology, and the cell cycle (Fig. 4B and C). To further characterize the functional difference between CRABP1 and -2, the uniquely expressed genes were subjected to canonical signaling pathway analysis using IPA software (Fig. 4D and Tables S1 and S2). The result indicated that the expression of CRABP1 has broader influences on the nuclear receptor pathways involving heterodimer complex formation with RXR (Fig. 4D). In contrast, CRABP2 expression had a selective influence on a few RXR heterodimer complex pathways, such as the conventional ATRA response pathway, which utilizes the RAR-RXR heterodimer complex (Fig. 4D). Finally, a disease association analysis of the genes uniquely expressed in CRABP1- and -2-expressing cells showed that the expression of CRABP1 is significantly associated with liver steatosis and cholestasis, which are both known to be associated with lipid metabolism (Fig. S4B).
Given that the association noted in the pathway analysis could be either positive or negative regulation, further functional assessment is required to better understand the differential functionality between CRABP1 and -2. For assessment of the conventional ATRA response pathway, which was influenced by both CRABP1 and -2 expressions (Fig. 4D), we employed a RARE-DR5 luciferase reporter construct that is regulated by the RAR-RXR heterodimer complex. The result showed that CRABP2 expression enhanced the induction of the luciferase reporter both under homeostatic conditions and in response to exogenous ATRA treatment (Fig. S4C). This result suggests that CRABP2 indeed serves as a chaperone that supports the translocation of ATRA into the nucleus, thereby potentiating the ATRA response pathway. In contrast, the expression of CRABP1 modestly but significantly suppressed the activation of luciferase reporter activity under basal conditions or upon exogenous ATRA treatment, indicating that CRABP1 negatively regulates the ATRA response (Fig. S4C). Next, we assessed the pathways that are uniquely influenced by CRABP1, such as the farnesoid X receptor (FXR)-RXR, peroxisome proliferator-activated receptor (PPAR)-RXR, and liver X receptor (LXR)-RXR pathways, using a luciferase reporter system (Fig. S4D to S4F). We found that CRABP1 expression has an inhibitory effect on FXR-RXR and PPAR-RXR. In contrast, the expression of CRABP1 enhanced the activation of the LXR pathway. It is known that the activation of the FXR-RXR and PPAR-RXR pathways suppresses the accumulation of LDs, while the activation of the LXR-RXR pathway results in an increase in the abundance of LDs (15–17). Thus, the results from our bioinformatics analyses and the reporter assays collectively suggest that the expression of CRABP1 may result in the accumulation of LDs in hepatocytes.
CRABP1 and -2 differentially regulate the abundance of LDs.
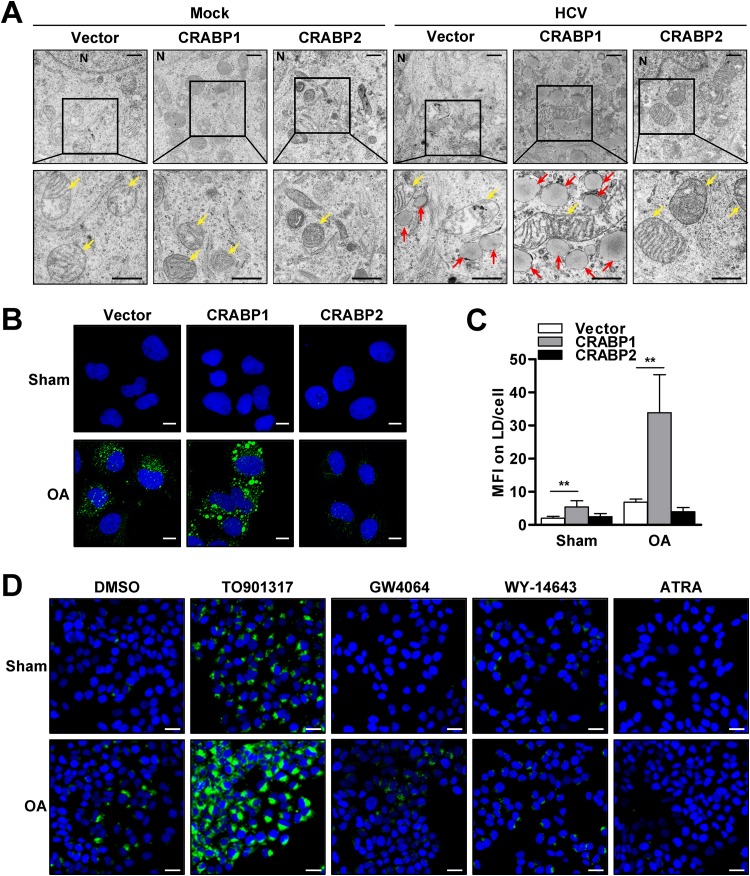
It is well known that HCV infection induces the accumulation of LDs, a triglyceride-rich cellular organelle, and exploits the accumulated LDs as a platform for the formation of the viral replicase complex and for virion assembly (18–23). Thus, we hypothesized that the proviral effect of CRABP1 might result from an alteration in the abundance of LDs. Consequently, we tested this idea by the visualization of CRABP-expressing Huh7 cells by a transmission electron microscopy (TEM) approach. The result demonstrated that the expression of CRABP itself did not show notable alterations in the internal structures of the cells and organelles under homeostatic conditions. However, of great interest, the expression of CRABP1 markedly increased the number and size of LDs upon HCV infection (Fig. 5A). In contrast, CRABP2-expressing cells did not demonstrate an accumulation of LDs both under resting conditions and upon HCV infection. This result suggests that the expression of CRABP1 lowers the threshold for the accumulation of LDs upon HCV infection, while the expression of CRABP2 results in the suppression of LD accumulation.
FIG 5.
CRABPs inversely regulate the abundance of LDs. (A) TEM analysis of CRABP-expressing Huh7 cells infected with HCV for 48 h. N, nucleus; yellow arrows, mitochondria; red arrows, lipid droplets. Bars, 1 μm (top panels) and 600 nm (bottom panels). (B) Huh7 cells stably expressing CRABPs were treated with or without oleic acid (OA) (50 μM) for 24 h, followed by fluorescence microscopic analysis for the assessment of LD abundance via BODIPY (green) and DAPI (blue) staining. Bars, 10 µm. (C) The abundances of LDs in CRABP-expressing cells were quantified as median fluorescence intensity (MFI) values for 50 representative cells. **, P < 0.01. (D) Huh7 cells were cultured with the indicated nuclear receptor agonists (LXR, TO901317 [10 μM]; FXR, GW4064 [10 μM]; PPAR, WY-14643 [100 μM]; RAR, ATRA [1 μM]) for 72 h in the presence of OA (50 μM), followed by BODIPY (green) and DAPI staining for fluorescence microscopic analysis. Bars, 20 μm. DMSO, dimethyl sulfoxide. See also Fig. S5 in the supplemental material.
Next, we investigated whether the enhanced LD accumulation in CRABP1-expressing cells is unique to HCV infection, which is known to induce the accumulation of LDs through the upregulation of the de novo fatty acid synthesis pathway in an SREBP1-dependent manner (20). Our fluorescence microscopic analysis demonstrated that the cells expressing CRABP1 substantially increased the abundance of LDs, while the expression of CRABP2 suppressed LD accumulation in response to monounsaturated free fatty acid (FFA) treatment, such as that with oleic acid (OA) (Fig. 5B and C).
Our reporter analysis indicated that CRABP1 expression enhances the prosteatotic (LXR-RXR) pathway while suppressing the antisteatosis (FXR-RXR and PPAR-RXR) pathways (Fig. S4D to S4F). As noted above, we also found that CRABP1 attenuates, but CRABP2 augments, the conventional ATRA response (RAR-RXR) pathway (Fig. S4C). Accordingly, we assessed the significance of these pathways in the regulation of LD abundance. Naive Huh7 cells were cultured with the ligands for LXR, FXR, PPAR, and RAR in the absence or presence of OA. The result showed that the LXR agonist significantly augmented the accumulation of LDs, while FXR, PPAR, and RAR agonists all reduced the abundance of LDs (Fig. 5D and Fig. S5B). Similar to the experiments described above, we assessed the potential involvement of the other RXR-related pathways, which are influenced by the expression of CRABPs (Fig. 4D), in the regulation of LD abundance. Our results indicate that the activation of the constitutive androstane receptor (CAR)-RXR, pregnane X receptor (PXR)-RXR, thyroid receptor (TR)-RXR, and vitamin D receptor (VDR)-RXR pathways has a negligible influence on the modulation of LD abundance (Fig. S5A and S5B). Taken together, our results suggest that CRABP1 expression promotes the accumulation of LDs, likely via the activation of the LXR-RXR pathway along with the suppression of the PPAR-RXR, FXR-RXR, and RAR-RXR pathways. In addition, our results also suggest that CRABP2 expression likely inhibits LD accumulation via stimulation of the RAR-RXR pathway.
HCV exploits the function of CRABP1 for the establishment of the viral replicase complex.
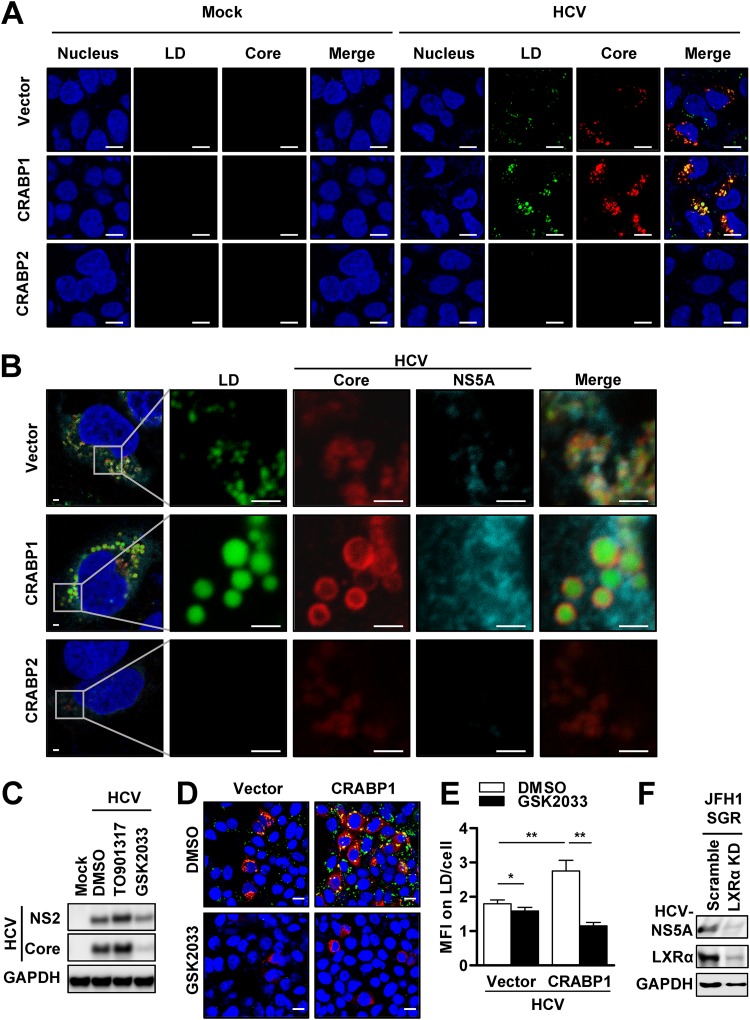
Our results together led us to hypothesize that HCV takes advantage of the prosteatotic function of CRABP1 for the efficient establishment of the viral life cycle. To test this idea, we investigated whether the formation of LDs upregulated by the expression of CRABP1 indeed supports the establishment of an HCV replicase complex. Consistent with data from previous studies by others (20), HCV infection of Huh7 cells increased the abundance of LDs (Fig. 6A and Fig. S6A). As seen in the experiment with TEM (Fig. 5A), the expression of CRABP1 robustly accentuated the accumulation of LDs upon HCV infection. We also noted that the size of LDs formed in HCV-infected CRABP1-expressing cells was substantially larger than that of LDs in control cells (Fig. 6A and Fig. S6B). Of note, it was impossible to observe the change in the degree of LD accumulation in CRABP2-expressing cells upon HCV challenge, as it did not support the establishment of efficient and stable infection (Fig. 6A).
FIG 6.
CRABPs modulate lipid droplet formation, which is a platform of HCV replicase. (A and B) Huh7 cells stably expressing CRABPs were either mock infected or infected with HCV JFH1 for 48 h at an MOI of 0.1. (A) The cells were stained with HCV core (red), and LDs were stained with BODIPY (green) and DAPI (blue), to evaluate LD formation in uninfected and infected cells. Bars, 10 μm. (B) Cells were stained with HCV core (red) and HCV NS5A (cyan), and LDs were stained with BODIPY (green) and DAPI (blue), to evaluate the colocalization of HCV proteins and LDs. Bars, 2 μm. (C) Huh7 cells were treated with either an LXR agonist (TO901317; 10 μM) or inhibitor (GSK2033; 10 μM) for 3 days, followed by HCV infection at an MOI of 0.1 for 48 h. The infected cells were subjected to immunoblot analysis for the detection of the indicated proteins. (D) Huh7 cells stably expressing CRABP1 were pretreated with an LXR inhibitor (GSK2033; 10 μM) for 3 days and infected with HCV at an MOI of 0.1 for 48 h, followed by immunofluorescence staining with HCV core (red) and staining of LDs with BODIPY (green) and DAPI (blue). Bars, 20 μm. (E) The abundances of LDs in CRABP1-expressing cells were quantified as median fluorescence intensity (MFI) values for 100 representative cells. *, P < 0.05; **, P < 0.01. (F) Huh7 JFH1 SGR cells stably expressing scrambled shRNA or shRNA against LXR-α via lentiviral particle transduction were subjected to immunoblot analysis for the detection of the indicated proteins. See also Fig. S6 in the supplemental material.
The HCV replicase complex, comprised of nonstructural (NS) proteins, is anchored on the endoplasmic reticulum (ER) membrane adjacent to HCV core protein-coated LDs (19). Of note, it has been believed that the proximity between the HCV core-coated LDs and the replicase complex is essential for efficient viral replication and virion assembly (19, 22, 23). Accordingly, we investigated whether the pronounced LDs in CRABP1-expressing cells are indeed being utilized for the HCV life cycle. Our confocal microscopic analysis demonstrated that the LDs were surrounded by HCV core protein in CRABP1-expressing cells and, to a much lesser extent, in control cells (Fig. 6B). We also found a marked increase in the abundance of HCV NS5A protein on the edge of the LDs coated with HCV core protein in CRABP1-expressing cells. These results indicate that the enhanced accumulation of LDs serves, at least in part, as the explanation for the proviral properties of CRABP1.
Based on our study results (Fig. 4 and 5), we extended our hypothesis that HCV exploits the enhanced accumulation of LDs resulting from the activation of the LXR-RXR pathway in CRABP1-expressing cells. To further assess the significance of the enhanced LXR-RXR pathway in the regulation of HCV infection, we first challenged naive Huh7 cells with HCV in the presence of an LXR agonist or antagonist. We found that Huh7 cells treated with an LXR agonist or antagonist changed the efficiency of HCV infection (Fig. 6C). Subsequently, we examined whether the LXR signaling pathway is indeed involved in the regulation of HCV infection in CRABP1-expressing cells. As expected, we found that LXR antagonist treatment or LXR knockdown (KD) via short hairpin RNA (shRNA) substantially suppressed HCV infection, which appeared to be in parallel with the change in LD abundance (Fig. 6D and E). In addition, the LXR-α KD substantially suppressed the replication of the HCV SGR (Fig. 6F and Fig. S6C and S6D). These observations suggest that CRABP1 exhibits its proviral property by the enhancement of LD formation, likely via the activation of the LXR-RXR pathway. Moreover, we also observed that the accumulation of LDs via supplementation with exogenous OA overpowers the proviral effect of CRABP1 (Fig. S6E to S6G), further indicating that the regulation of HCV by CRABPs primarily results from its effect on the modulation of LD abundance. Finally, the induction of LDs by OA treatment also potentiated HCV IRES-dependent protein translation (Fig. S6H). Taken together, our results suggest that the activation of the LXR-RXR pathway by CRABP1 supports HCV replication by providing the platform for the formation of the HCV replicase complex as well as the enhancement of IRES-dependent viral protein translation.
The CRABP2-RAR-RXR pathway inhibits HCV via reduction of LD abundance.
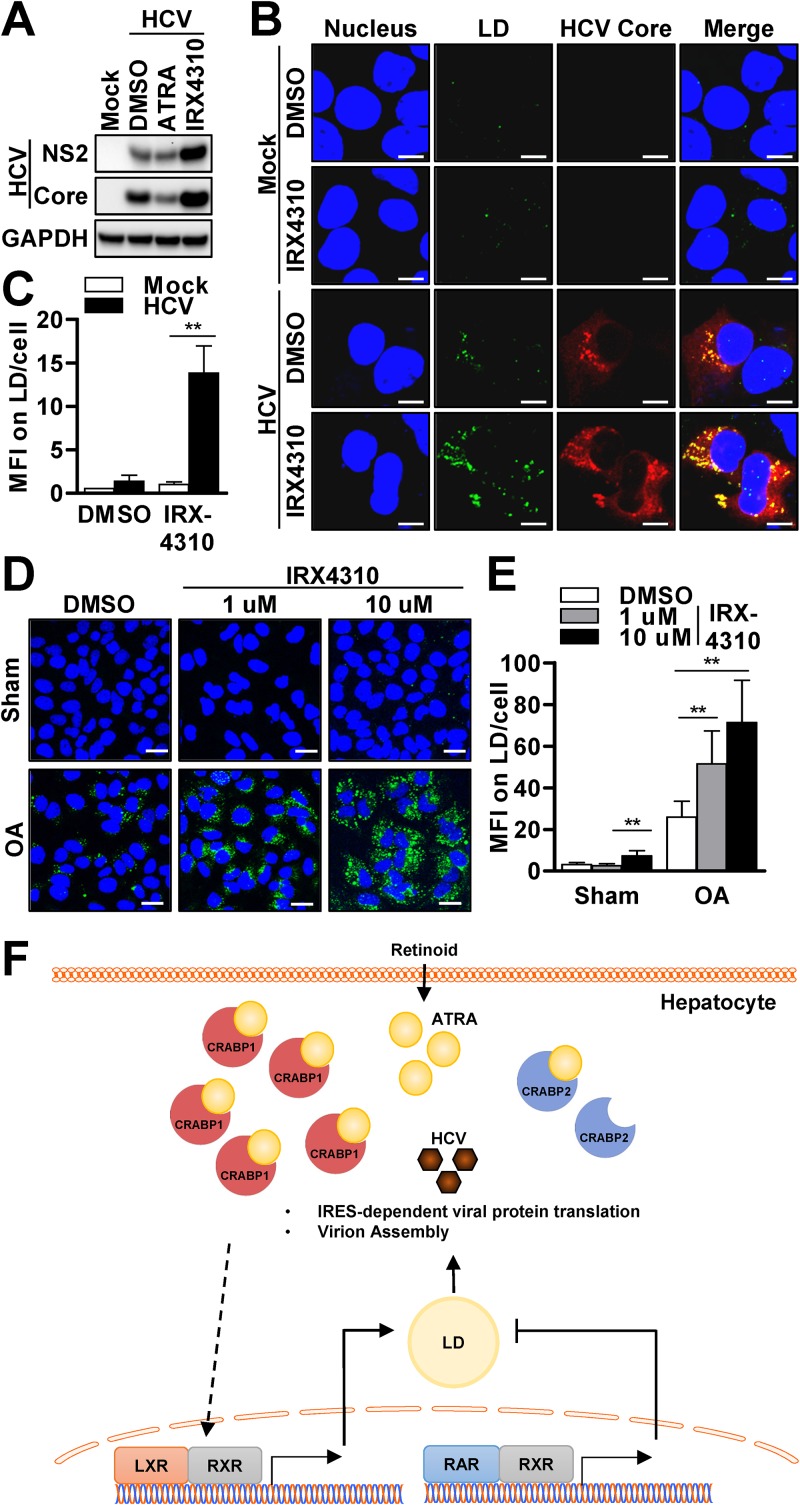
Our results indicated that the expression of CRABP2 is coupled with resistance to the accumulation of LDs upon HCV infection as well as FFA treatment (Fig. 5 and 6). We also found that the antisteatotic effect of CRABP2 is primarily governed by the activation of the RAR-RXR pathway (Fig. 5). To better understand whether the inhibition of LD accumulation by the RAR-RXR pathway serves as an explanation for the suppression of HCV infection, we conducted a series of investigations. First, we assessed the effect of a pan-RAR inhibitor, IRX4310, on the efficiency of HCV infection. The inhibition of the RAR-RXR pathway resulted in a marked enhancement of the viral replication efficiency, while the activation of RAR-RXR with ATRA resulted in the potent suppression of HCV (Fig. 7A). Moreover, the activation of the RAR-RXR pathway with a synthetic ATRA (EC23), which is known to have a much longer half-life, demonstrated further enhancement of the antiviral property (Fig. S7A). Next, we assessed the change in LD abundance upon IRX4310 treatment. Although we did not observe a significant change in LD abundance under resting conditions, the inhibition of the RAR-RXR pathway resulted in a marked increase in the quantity of LDs upon HCV infection (Fig. 7B and C).
FIG 7.
The CRABP2-RAR-RXR pathway inhibits HCV via reduction of LD abundance. (A to C) Huh7 cells were pretreated with an RAR agonist (ATRA; 1 μM) or an RAR inhibitor (IRX4310; 1 μM) for 72 h, followed by HCV infection at an MOI of 0.1 for 48 h. (A and B) Cell lysates were subjected to immunoblot analysis for the detection of the indicated proteins (A), or the cells were costained with LDs with BODIPY (green), anti-HCV core (red), and DAPI (blue) (B). Bars, 10 μm. (C) Fluorescence microscopic images of 50 representative cells were used for analysis of the MFI of LDs per cell. **, P < 0.01. (D and E) Huh7 cells were treated with an RAR inhibitor (IRX4310; 1 μM) at the indicated concentrations for 48 h, followed by oleic acid (OA) (50 μM) treatment for 24 h. (D) The cells were then stained with BODIPY (green) and DAPI (blue). Bars, 20 μm. (E) Fluorescence microscopic images of 50 representative cells were used for analysis of the MFI values of LDs per cell. **, P < 0.01. (F) Proposed model of CRABP regulation of the HCV life cycle via modulation of the nuclear receptor (NR) complex pathways, which governs the abundance of LDs in hepatocytes. See also Fig. S7 in the supplemental material.
Finally, we examined whether the inhibition of the RAR-RXR pathway results in the accumulation of LDs in response to exogenous OA treatment. Similar to the results seen with HCV infection, Huh7 cells treated with IRX4310 increased the abundance of LDs (Fig. 7D and E). Taken together, our results suggest that CRABP2 expression achieves the suppression of HCV infection via the suppression of LD accumulation through the activation of the RAR-RXR pathway.
DISCUSSION
The cellular response to nutrients significantly influences various aspects of cell biology beyond its fundamental role for homeostatic energy production. One of the essential micronutrients, vitamin A, also referred to as retinoid, has been known as an antimicrobial compound for many decades, since 1928 (24). Accordingly, numerous epidemiological studies have demonstrated that supplementation with vitamin A or its final metabolite, ATRA, improves the clinical outcomes of numerous viral infectious diseases, such as, but not limited to, measles, herpes simplex virus (HSV), HIV, and respiratory syncytial virus (RSV) infections (1, 25–28). The antiviral properties of ATRA have been explained, at least in part, by its effect on the activation of innate and adaptive immune cells (29). Also, several studies have shown that ATRA potentiates the expression of ISGs, which play a central role in intracellular antiviral innate immunity (2, 3, 5, 6, 30).
The antiviral effect of retinoid has also been implicated in the suppression of HCV infection (2, 6, 31, 32). On the other hand, it is also true that HCV establishes a highly efficient viral life cycle in hepatocytes, which store a substantial amount of retinoid. These somewhat conflicting notions might be attributed to the experimental conditions employed in in vitro studies wherein the physiological intracellular trafficking of ATRA had not been taken into consideration. Upon uptake by hepatocytes, the pro- or preformed vitamin A undergoes oxidative metabolism, resulting in the biogenesis of ATRA in the cytoplasm. Given the hydrophobic characteristics of ATRA, it requires a chaperone molecule that facilitates its translocation into the nucleus for the activation of nuclear receptor complexes (12). CRABPs have been considered the carrier proteins that shuttle ATRA from the cytoplasm to the nuclear receptors. As CRABP1 and -2 share high amino acid sequence identity (76%) (33), it has been considered that CRABP1 and -2 play redundant roles. In fact, our transcriptome analysis of CRABP-expressing cells demonstrated a grossly identical pattern of gene expression except for a few subsets of genes (Fig. 4). Of note, however, previous studies by others also suggested that CRABPs may have differential or perhaps opposed functions. First, a few studies demonstrated that CRABP1 has a much higher affinity for ATRA than does CRABP2 (34, 35). In addition, apo-CRABP2 predominantly localizes in the cytoplasm and rapidly translocates into the nucleus upon binding of ATRA, whereas apo- and holo-CRABP1s tend to remain in the cytoplasm, even in the presence of ATRA at a high concentration (36). Furthermore, CRABP2 is capable of directly transferring ATRA to RAR through protein-protein interaction, whereas CRABP1 delivers ATRA to the nuclear receptor in a diffusion-dependent manner (36, 37). Finally, it has been suggested that CRABP1 not only halts ATRA but also allocates it to cytochrome P450 enzymes, thereby promoting the conversion of ATRA to 9-cis RA or to polar metabolites such as 4-OH RA (12, 38). These observations together suggest that CRABP1 moderates the cellular response to ATRA via sequestrations; in contrast, CRABP2 shuttles ATRA from the cytoplasm to the RAR-RXR complex, thereby augmenting the cellular response to vitamin A. In fact, our reporter analysis well supported this notion (see Fig. S4C in the supplemental material).
Consequently, our study discovered that CRABP1 and -2 exhibit potent pro- and antiviral properties, respectively. We found that the proviral effect of CRABP1 is coupled with an increased abundance of a triglyceride-rich organelle, the LD, which has been known to serve as a platform for the HCV life cycle, especially for the stages of viral replication and virion assembly (19, 22, 23). Accordingly, the antiviral effect of CRABP2 is accompanied by a significant reduction of LDs (Fig. 5B and Fig. 6A and B). Thus, the pro- and antiviral effects of CRABPs appeared to correlate well with their influence on the abundance of LDs. This notion is also well supported by our study results demonstrating that the expression of CRABPs had a negligible effect on the viral pathogens that are not dependent on LDs (Fig. 3). Based on these observations, we conclude that the regulation of viral life cycle efficiency by CRABPs is unique and specific to HCV given its life cycle’s heavy reliance on LD abundance in hepatocytes.
Regarding the potential mechanism by which CRABP expression modulates the abundance of LDs, it has been suggested that ATRA enhances lipolysis via activation of the PPARβ/δ-RXR and RAR-RXR pathways, thereby depleting lipid stores (39). Moreover, the inhibition of C/EBP by ATRA serves as an alternative explanation for the inhibition of liver steatosis (40–42). In fact, the significance of intrahepatic ATRA in the suppression of LDs is well supported by an in vivo study that showed that vitamin A deficiency in mice promoted a marked accumulation of macrocytic LDs in the liver (43). Conversely, ATRA-treated mice showed a reduction of adiposity with enhanced gene expression of UCP1, which serves as the signature gene of PPARα activation (44, 45). Thus, it is plausible to conclude that CRABP2 expression inhibits HCV by decreasing the abundance of LDs through the enhancement of the antisteatotic effect of ATRA. Accordingly, we propose that the expression of CRABP1 supports the HCV life cycle by increasing the abundance of LDs. Our data suggest that there may be at least two mechanisms that govern the prosteatotic effect of CRABP1: (i) the sequestration of ATRA and (ii) the upregulation of the LXR pathway. In fact, our results (Fig. 6C to F) and observations by others support our conclusion well, as these studies demonstrated that the LXR-RXR pathway modulates the efficiency of HCV infection via LD abundance or the formation of virus-associated membrane compartments (46, 47). We believe that these observations justify our proposed mechanisms well.
Based on a seminal study by others, it has been thought that LD abundance plays a critical role in the regulation of HCV virion assembly (19). Of great interest, however, our data indicated that the accumulation of LDs by CRABP1 expression greatly enhances the efficiency of IRES-dependent, but not cap-dependent, protein translation (Fig. 2C and D). This result suggests that LD abundance modulates the efficiency of the HCV life cycle, at least in part, at the stage of viral protein translation in addition to its influence on HCV virion assembly. In fact, our result with the HCV SGR system (Fig. 2B), which does not involve virion assembly, showed marked changes in HCV replication efficiency in the CRABP-expressing cells. While it is beyond the scope of this study to delineate the underlying mechanism of this interesting phenomenon, previous studies implied that FFA treatment, perhaps the accumulation of LDs, enhances IRES-dependent translation via induction of an IRES trans-acting factor (ITAF) (48–50). While we believe that this mechanism serves as an underlying mechanism by which CRABPs regulate HCV replication, previous studies suggested that components of the LD, such as Rab18 and TIP47, but not the LD itself, support HCV replication by enhancing the amplification of the HCV genome (22, 23). Thus, further investigation is required to precisely understand the complexity of how LDs regulate the HCV life cycle.
Previous studies, including ours, also indicated that the antiviral effect of ATRA is partially explained by the enhanced expression of innate immune genes, ISGs (2, 31, 32), which are comprised of over 300 antiviral effectors (51). Therefore, we initially hypothesized that CRABP2 expression, which augments the cellular response to ATRA, may exhibit an antiviral effect by enhancing the expression of ISGs. However, our transcriptome analysis of CRAPB2-expressing cells did not demonstrate the upregulation of any ISGs under resting conditions (Table S2). Similarly, CRABP1-expressing cells also did not show a reduction of ISG expression under basal conditions (Table S1). On the other hand, we found that CRABP2-expressing cells have greater capacity for the induction of some but not all ISGs in response to IFN treatment and, to a lesser extent, treatment with RIG-I ligands (52) (Fig. S7B and S7C). Based on these results, we propose that the augmentation of ISG expression by ATRA, or CRABP2 expression, does not serve as the primary means to restrict HCV infection; however, this mechanism may play a role in the event that infected cells are exposed to IFN secreted from surrounding innate immune cells (52–54).
While our observations led us to conclude that the regulation of HCV by CRABPs is primarily mediated by their effect on the modulation of LD abundance, further studies are required to precisely understand the mechanisms by which CRABPs regulate the abundance of LDs via regulation of the nuclear receptor pathways. In addition, in vitro and in vivo studies with loss-of-function approaches are necessary to validate the importance of our study results.
In summary, our observations collectively indicated that the cellular environment favors HCV in the event that the abundance of CRABP1 dominates that of CRABP2. With regard to the potential explanations of how HCV establishes an efficient life cycle in hepatocytes, our data suggest the predominant abundance of CRABP1 over CRABP2 (Fig. 1A and Fig. S1A) as well as CRABP1’s high capacity to attenuate the ATRA response pathway (Fig. S4C and S6I). This notion also leads to a new concept: CRABPs serve as an on-off switch for the regulation of the HCV life cycle and thus can be exploitable for the development of a novel antiviral strategy. Moreover, our discovery also has a great implication in the broader context, such as for the management of metabolic liver diseases, beyond its implication in the management of HCV infection.
MATERIALS AND METHODS
Cells.
Primary human hepatocytes were obatined from Lonza. The human hepatoma Huh7 cell line (a gift from T. Wakita, Japan), the Huh7-derived Huh7.5 cell line (a gift from T. Wakita, Japan), and human embryonic kidney 293T cells (ATCC) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with l-glutamine (2 mM), sodium pyruvate (1 mM), HEPES (10 mM), penicillin (10,000 IU/ml), streptomycin (10,000 µg/ml), amphotericin (25 µg/ml), and 10% fetal bovine serum (FBS) at 37°C with 5% CO2. For the establishment and maintenance of cells stably expressing CRABPs, puromycin (2 µg/ml) was added to the culture medium.
Nucleic acid.
(i) Plasmids. The transient-expression or lentiviral expression constructs were generated in pEF Myc/His Version C (Life Technologies) with or without the Flag gene or in pCDH-CMV-MCS-EF1-Puro (System Bioscience), respectively. Human CRABP1 and CRABP2 were cloned using total RNA of primary human hepatocytes. All plasmids were propagated in Escherichia coli DH5α cells under ampicillin (100 µg/ml) selection and prepped with a ZR plasmid miniprep or a ZymoPure plasmid midiprep kit (Zymo Research).
(ii) RT-qPCR. RNA extraction was carried out using the Quick-RNA miniprep kit followed by cDNA synthesis using qScript cDNA SuperMix (Quantabio). Quantitative PCR (qPCR) was conducted using SYBR green supermix (Quantabio) along with the primers for human CRABP1 (forward primer 5′-TCAACTTCAAGGTCGGAGAAG-3′ and reverse primer 5′-CTCCCAAGTGGCTAAACTCCT-3′), human CRABP2 (forward primer 5′-ATCGGAAAACTTCGAGGAATTGC-3′ and reverse primer 5′-AGGCTCTTACAGGGCCTCC-3′), human LXR-α (forward primer 5′-GTTATAACCGGGAAGACTTTGCCA-3′ and reverse primer 5′-GCCTCTCTACCTGGAGCTGGT-3′), human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (forward primer 5′-CTGGGCTACACTGAGCACCAG-3′ and reverse primer 5′-CCAGCGTCAAAGGTGGAG-3′), and SeV (forward primer 5′-GACGCGAGTTATGTGTTTGC-3′ and reverse primer 5′-TTCCACGCTCTCTTGGATCT-3′). One-step reverse transcription-qPCR (RT-qPCR) was performed using qScript XLT 1-Step RT-qPCR ToughMix ROX (Quantabio) along with primers and probes for HCV (forward primer 5′-CGGGAGAGCCATAGTGG-3′, reverse primer 5′-AGTACCACAAGGCCTTTCG-3′, and probe 5′-FAM [6-carboxyfluorescein]-CTGCGGAAC-ZEN-CGGTGAGTACAC-3IABkFQ [Iowa Black FQ]-3′), HEV (forward primer 5′-GGTGGTTTCTGGGGTGAC-3′, reverse primer 5′-AGGGGTTGGTTGGATGAA-3′, and probe 5′-FAM-TGATTCTCAGCCCTTCGC-TAMRA [6-carboxytetramethylrhodamine]-3′), and DENV (forward primer 5′-CCGTCACACTGGGTTCCAA-3′ and reverse primer 5′-CCGTCGTCATCCATTCATGCT-3′). The serially diluted in vitro-transcribed 5′ untranslated region (UTR) of the HCV genome, the full-length HEV genome encoding the Kernow C1 P6 plasmid, and a DENV2-GFP (green fluorescent protein) plasmid were used for the determination of the standard curve.
Chemicals.
Agonists and inhibitors of nuclear receptors, including a CAR agonist (Citco), an FXR agonist (GW4064), an LXR agonist (TO901317), a PPAR agonist (WY-14643), a PXR agonist (rifampin), an RAR agonist (ATRA), a TR agonist (T3), a VDR agonist (vitamin D3 [Vit D3]), and an LXR inhibitor (GSK2033), were purchased from MilliporeSigma. The RAR agonist (EC23) was purchased from Tocris, and the RAR inhibitor (IRX4310) was a gift from Michael Underhill (University of British Columbia, Canada). Free fatty acid treatment was conducted with oleic acid (MilliporeSigma) following serum starvation for 8 h at the concentrations and durations indicated in the corresponding figure legends.
Virus infection.
(i) HCV-JFH1 infectious clone propagation and infection. Infectious JFH1 particles, which were a gift from Takaji Wakita (Tokyo, Japan), were propagated and titers were determined as described previously (55). Briefly, Huh7.5 cell transfected with the in vitro-transcribed HCV-JFH1 genome were incubated in DMEM containing 2% FBS for 48 h, and the HCV-containing supernatant was then filtered through a 0.22-μm filter and concentrated 100-fold using Centricon 100,000-molecular-weight (MW)-cutoff filters (Millipore). Titers of virus stocks were determined with naive Huh 7.5 cells using a focus-forming unit assay and stored at −80°C.
(ii) HCV pseudoparticle propagation and infection. HCV pseudoparticles (HCVpp) were propagated as previously described (56). Briefly, HCVpp were generated by cotransfection of plasmids (pJFH1pp-E1E2, pMD2-VSVG, and pNL4.3_Luc-R-E-) into 293T cells for 48 h, followed by concentration of supernatants (100-fold) by ultracentrifugation (47,000 × g for 2 h at 16°C). The titer of the concentrated HCVpp was determined by using an HIV p24 enzyme-linked immunosorbent assay (ELISA) kit (Lenti-X p24 rapid titer kit; TaKaRa). HCVpp transduction was carried out with 20 ng/ml p24 for 5 days in the presence of Polybrene (10 μg/ml).
(iii) HCV subgenomic replicon transduction assay. H77 H/SG-Neo(L+1) and pSGR JFH1 plasmids, which were gifts from Charles M. Rice (Rockefeller University, NY), were linearized using Xbal followed by in vitro transcription of the subgenomic replicon (SGR) genome using a MEGAscript T7 transcription kit (Life Technologies). The purification of SGR RNA was carried out with the phenol-chloroform-ethanol precipitation method. The SGR genome (1 µg) was transduced into Huh7 cells (4 × 106 cells) via RNA transfection with a TransIT-mRNA transfection kit (Mirus) or electroporation and cultured in the presence of G418 (400 μg/ml).
(iv) Lentiviral particles. The propagation and titer determination of lentiviral particles were carried out as described previously (55). In brief, the pCDH-CMV-MCS-EF1-Puro (System Bioscience) vector harboring open reading frames (ORFs) of CRABPs and pPACK packaging mix were cotransfected into the 293T cell line for 72 h. For the production of shRNA-expressing stable cell lines, pLKO.1 vectors encoding scrambled shRNA and LXR-α (clone 1, 5′-CCGGAGTTCTCCAGGGCCATGAATGCTCGAGCATTCATGGCCCTGGAGAACTTTTTTTG-3′; clone 2, 5′-CCGGCCGACTGATGTTCCCACGGATCTCGAGATCCGTGGGAACATCAGTCGGTTTTT-3′; clone 3, 5′-CCGGCCTTCCTCAAGGATTTCAGTTCTCGAGAACTGAAATCCTTGAGGAAGGTTTTT-3′) (MilliporeSigma) were transfected into 293T cells according to the manufacturer’s protocol. The culture supernatant was collected and filtered with a 0.45-μm filter to remove cell debris, followed by infection of Huh7 cells at a multiplicity of infection (MOI) of 5 × 107 transduction units (TU)/ml, determined by using an HIV p24 ELISA kit (Lenti-X p24 rapid titer kit; TaKaRa), followed by selection in the presence of puromycin (2 μg/ml) without clonal selection. Upon completion of selection, the bulk puromycin-resistant cell population was expanded and used for in vitro studies.
(v) SeV, VSV, HEV, and DENV infection and HBV transfection. Vesicular stomatitis virus (VSV) and EMCV were gifts from Michael Gale, Jr. (University of Washington, Seattle, WA). HEV was a gift from Zongdi Feng (Nationwide Children’s Hospital, OH). The infection titers of Sendai virus (SeV; Charles River Laboratories), VSV, hepatitis E virus (HEV), and dengue virus (DENV2 strain 16681) are described in the corresponding figure legends. An HBV replication assay was conducted by the transfection of plasmid p1.3x HBV, encoding the 1.3-mer overlength HBV genome, with Bio-T (Bioland).
Luciferase reporter assay.
pRL-HL and pIRES-Luc plasmids were gifts from Stanley M. Lemon (University of North Carolina) and Michael Gale, Jr. (University of Washington), respectively. Plasmids (LXRE-Luc [LXR-response element-luciferase], FXRE-Luc [FXR-response element-luciferase], and PPRE-Luc [PPAR-response element-luciferase]) for luciferase reporter assays were kindly provided by Steven L. Sabol (NIH) (57), Jongsook Kim Kemper (University of Illinois at Urbana-Champaign), and Christopher K. Glass (UCSD), respectively. Cells were either transfected with a bicistronic IRES reporter (pRL-HL or pIRES-Luc) or cotransfected with nuclear receptor response element-regulated firefly luciferase plasmids (LXRE-Luc, FXRE-Luc, and PPRE-Luc) and a cytomegalovirus (CMV) promoter-regulated Renilla luciferase plasmid. The firefly luciferase activity was normalized with Renilla luciferase activity to determine relative luciferase units using a dual-luciferase reporter assay system (Promega).
Transcriptome analysis.
RNA extraction was carried out with a Quick-RNA miniprep kit (Zymo Research), followed by RNA quality assessment using the Agilent Bioanalyzer 2100 system (Agilent Technologies). cDNA libraries were prepared with a Kapa mRNA HyperPrep kit (Kapa Biosystems) and then quality controlled via a Qubit 2.0 fluorometer (Thermo Fisher Scientific Inc.) and the Bioanalyzer. The libraries were then pooled equimolarly, and the final pool was quantified via qPCR using the Kapa Biosystems library quantification kit, according to the manufacturer’s instructions. The pool was sequenced across two lanes of high-output paired-end (PE) 50 cycles on an Illumina HiSeq 2500 platform. Library preparation, pooling, quality control, and sequencing were all performed at the USC Genome Core (University of Southern California, Los Angeles, CA, USA).
Bioinformatics.
RNA-Seq data were analyzed with Partek Flow version 6 (Partek Inc.). Raw sequencing reads were first trimmed from both ends with a quality score method (bases with a quality score of <20 were trimmed from both ends, and trimmed reads shorter than 25 nucleotides [nt] were excluded from downstream analyses). Trimmed reads were then mapped to human genome hg38 using Star version 2.4.1d with default parameter settings and using Gencode v25 annotation as guidance (58). Gencode v25 annotation was used to quantify the aligned reads to genes using Partek's optimization of the expectation-maximization algorithm (Partek E/M) method. Genes with fewer than 10 raw reads in all samples were excluded from downstream analysis. Finally, read counts per gene in all samples were normalized using upper quartile normalization (59) and analyzed for differential expression using the Partek gene-specific analysis method. Significantly differentially expressed genes were selected using a P value of <0.01 and a fold change (FC) of more than 1.5 (or less than −1.5).
Protein analysis.
(i) Immunoblotting. Cell lysates were prepared in modified radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris [pH 7.5], 150 mM NaCl, 0.5% sodium deoxycholate, and 1% Triton X-100) supplemented with a protease inhibitor mixture (Roche) and phosphatase inhibitor mixture II (Calbiochem), followed by SDS-PAGE-based immunoblot analysis with the indicated primary antibodies along with horseradish peroxidase (HRP) conjugated with a secondary antibody. The specific protein signal was detected using SuperSignal West Pico chemiluminescent substrate (Thermo Fisher Scientific Inc.).
(ii) Immunofluorescence microscopic analysis. Cells plated in 8-chamber slides (Falcon) were fixed with 4% paraformaldehyde (PFA), followed by permeabilization with 0.2% Triton X-100 in phosphate-buffered saline (PBS). The slides were then treated with blocking buffer prior to incubation with primary and secondary antibodies. The immunostained slides were mounted with 4′,6-diamidino-2-phenylindole (DAPI)-containing mounting medium (Vector Laboratories). The images from the immunofluorescence confocal microscopy analysis were acquired using a Leica confocal microscope system. The mean fluorescence intensity (MFI) of lipid droplets was measured using ImageJ (NIH), and the diameter of LDs was measured using LAS X software (Leica).
(iii) Antibodies. The following antibodies were used for this study: mouse monoclonal anti-GAPDH (clone GT239; GeneTex), mouse monoclonal anti-CRABP1 (clone C-1; GeneTex), mouse monoclonal anti-CRABP2 (Millipore), mouse monoclonal anti-Flag (clone M2; Sigma), mouse monoclonal anti-HCV NS3 (clone 8 G-2; Abcam), Mouse monoclonal anti-HCV core (clone C7-50), rabbit polyclonal anti-HCV core, rabbit polyclonal anti-HBV core, rabbit whole-serum anti-SeV (MBL), mouse anti-DENV (MilliporeSigma), Rabbit polyclonal anti-IFITM1 (ProSci), rabbit anti-ISG15 (Cell Signaling Technology), and mouse monoclonal anti-OAS1 (clone 1.3.3; Kineta). Mouse monoclonal anti-HCV NS2 (clone 6H6) and mouse monoclonal anti-HCV NS5A (clone 9E10) were gifts from Charles M. Rice (Rockefeller University). Rabbit polyclonal anti-ISG56 was a gift from Ganes C. Sen (Lerner Research Institute).
Electron microscopy.
Cells were fixed with 2.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4) for 30 min at room temperature and scraped off from the culture dish. The samples were then kept in fixative solution for another 90 min at 4°C followed by incubation in 1% OsO4 in phosphate buffer (pH 7.4) for 2 h, embedded in 2% agar, dehydrated in ethanol, and embedded in Epon 812. Ultrathin sections were double stained with uranyl acetate and lead citrate and examined using a transmission electron microscope (H-7100; Hitachi Ltd., Tokyo, Japan).
Cell growth rate and viability.
To assess the cell growth rate, cells plated in a 96-well plate were cultured for 24, 48, and 72 h, followed by assessment of cell viability, and the average cell growth rate was then calculated with the cell viability at each time point. To assess the sensitivity of cell death to tumor necrosis factor (TNF), cells plated in a 96-well plate were treated with human recombinant TNF in the absence or presence of actinomycin D (ActD) or cycloheximide (CHX) for 24 h, followed by assessment of cell viability. Cell viability was measured by using a CellTiter 96 nonradioactive cell proliferation assay [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT)] (Promega).
Statistical analysis.
All data are presented as means ± standard deviations (SD) and were analyzed by two-tailed Student’s t test using Prism version 5 (GraphPad). P values of <0.05 were considered significant.
Data availability.
The RNA-seq data have been deposited in the NCBI Gene Expression Omnibus (GEO) under accession number GSE101934.
Supplementary Material
ACKNOWLEDGMENTS
Microscopy services were provided by the Cell and Tissue Imaging Core at the USC Research Center for Liver Diseases (NIH P30DK048522). This work was supported by funds from the NIH NIAAA (R21AA022751 [T.S.]), the NIH NIAID (R21AI139954 [T.S.]), the NIH NIDDK (RO1DK101773 [T.S.]), the Civilian Research and Development Foundation (DAA3-17-62993-1), and research funding from PhoenixBio.
We thank Zongdi Feng (Nationwide Children’s Hospital) for technical support. Finally, we thank Jeffrey Sun for critical reading of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.02302-18.
REFERENCES
- 1.Ross AC, Stephensen CB. 1996. Vitamin A and retinoids in antiviral responses. FASEB J 10:979–985. doi: 10.1096/fasebj.10.9.8801180. [DOI] [PubMed] [Google Scholar]
- 2.Cho NE, Bang BR, Gurung P, Li M, Clemens DL, Underhill TM, James LP, Chase JR, Saito T. 2016. Retinoid regulation of antiviral innate immunity in hepatocytes. Hepatology 63:1783–1795. doi: 10.1002/hep.28380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matikainen S, Ronni T, Hurme M, Pine R, Julkunen I. 1996. Retinoic acid activates interferon regulatory factor-1 gene expression in myeloid cells. Blood 88:114–123. [PubMed] [Google Scholar]
- 4.Trottier C, Colombo M, Mann KK, Miller WH Jr, Ward BJ. 2009. Retinoids inhibit measles virus through a type I IFN-dependent bystander effect. FASEB J 23:3203–3212. doi: 10.1096/fj.09-129288. [DOI] [PubMed] [Google Scholar]
- 5.Xiao S, Li D, Zhu HQ, Song MG, Pan XR, Jia PM, Peng LL, Dou AX, Chen GQ, Chen SJ, Chen Z, Tong JH. 2006. RIG-G as a key mediator of the antiproliferative activity of interferon-related pathways through enhancing p21 and p27 proteins. Proc Natl Acad Sci U S A 103:16448–16453. doi: 10.1073/pnas.0607830103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamamoto S, Fukuda R, Ishimura N, Rumi MA, Kazumori H, Uchida Y, Kadowaki Y, Ishihara S, Kinoshita Y. 2003. 9-cis retinoic acid enhances the antiviral effect of interferon on hepatitis C virus replication through increased expression of type I interferon receptor. J Lab Clin Med 141:58–66. doi: 10.1067/mlc.2003.8. [DOI] [PubMed] [Google Scholar]
- 7.Clugston RD, Blaner WS. 2012. The adverse effects of alcohol on vitamin A metabolism. Nutrients 4:356–371. doi: 10.3390/nu4050356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedman SL. 2008. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol Rev 88:125–172. doi: 10.1152/physrev.00013.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Napoli JL. 1999. Retinoic acid: its biosynthesis and metabolism, p 139–188. In Kivie M. (ed), Progress in nucleic acid research and molecular biology, vol 63 Academic Press, San Diego, CA. [DOI] [PubMed] [Google Scholar]
- 10.Chase JR, Poolman MG, Fell DA. 2009. Contribution of NADH increases to ethanol’s inhibition of retinol oxidation by human ADH isoforms. Alcohol Clin Exp Res 33:571–580. doi: 10.1111/j.1530-0277.2008.00871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allenby G, Bocquel MT, Saunders M, Kazmer S, Speck J, Rosenberger M, Lovey A, Kastner P, Grippo JF, Chambon P, Levin AA. 1993. Retinoic acid receptors and retinoid X receptors: interactions with endogenous retinoic acids. Proc Natl Acad Sci U S A 90:30–34. doi: 10.1073/pnas.90.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Napoli JL. 2017. Cellular retinoid binding-proteins, CRBP, CRABP, FABP5: effects on retinoid metabolism, function and related diseases. Pharmacol Ther 173:19–33. doi: 10.1016/j.pharmthera.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu RZ, Garcia E, Glubrecht DD, Poon HY, Mackey JR, Godbout R. 2015. CRABP1 is associated with a poor prognosis in breast cancer: adding to the complexity of breast cancer cell response to retinoic acid. Mol Cancer 14:129. doi: 10.1186/s12943-015-0380-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Q, Wang R, Xiao W, Sun F, Yuan H, Pan Q. 2016. Cellular retinoic acid binding protein 2 is strikingly downregulated in human esophageal squamous cell carcinoma and functions as a tumor suppressor. PLoS One 11:e0148381. doi: 10.1371/journal.pone.0148381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watanabe M, Houten SM, Wang L, Moschetta A, Mangelsdorf DJ, Heyman RA, Moore DD, Auwerx J. 2004. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J Clin Invest 113:1408–1418. doi: 10.1172/JCI21025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karimian Azari E, Leitner C, Jaggi T, Langhans W, Mansouri A. 2013. Possible role of intestinal fatty acid oxidation in the eating-inhibitory effect of the PPAR-alpha agonist Wy-14643 in high-fat diet fed rats. PLoS One 8:e74869. doi: 10.1371/journal.pone.0074869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li M, Meng X, Xu J, Huang X, Li H, Li G, Wang S, Man Y, Tang W, Li J. 2016. GPR40 agonist ameliorates liver X receptor-induced lipid accumulation in liver by activating AMPK pathway. Sci Rep 6:25237. doi: 10.1038/srep25237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adinolfi LE, Gambardella M, Andreana A, Tripodi MF, Utili R, Ruggiero G. 2001. Steatosis accelerates the progression of liver damage of chronic hepatitis C patients and correlates with specific HCV genotype and visceral obesity. Hepatology 33:1358–1364. doi: 10.1053/jhep.2001.24432. [DOI] [PubMed] [Google Scholar]
- 19.Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, Wakita T, Hijikata M, Shimotohno K. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol 9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 20.Li Q, Pene V, Krishnamurthy S, Cha H, Liang TJ. 2013. Hepatitis C virus infection activates an innate pathway involving IKK-alpha in lipogenesis and viral assembly. Nat Med 19:722–729. doi: 10.1038/nm.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kapadia SB, Chisari FV. 2005. Hepatitis C virus RNA replication is regulated by host geranylgeranylation and fatty acids. Proc Natl Acad Sci U S A 102:2561–2566. doi: 10.1073/pnas.0409834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vogt DA, Camus G, Herker E, Webster BR, Tsou CL, Greene WC, Yen TS, Ott M. 2013. Lipid droplet-binding protein TIP47 regulates hepatitis C virus RNA replication through interaction with the viral NS5A protein. PLoS Pathog 9:e1003302. doi: 10.1371/journal.ppat.1003302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salloum S, Wang H, Ferguson C, Parton RG, Tai AW. 2013. Rab18 binds to hepatitis C virus NS5A and promotes interaction between sites of viral replication and lipid droplets. PLoS Pathog 9:e1003513. doi: 10.1371/journal.ppat.1003513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Green HN, Mellanby E. 1928. Vitamin a as an anti-infective agent. Br Med J ii:691–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Semba RD. 1994. Vitamin A, immunity, and infection. Clin Infect Dis 19:489–499. doi: 10.1093/clinids/19.3.489. [DOI] [PubMed] [Google Scholar]
- 26.Mostad SB, Kreiss JK, Ryncarz AJ, Mandaliya K, Chohan B, Ndinya-Achola J, Bwayo JJ, Corey L. 2000. Cervical shedding of herpes simplex virus in human immunodeficiency virus-infected women: effects of hormonal contraception, pregnancy, and vitamin A deficiency. J Infect Dis 181:58–63. doi: 10.1086/315188. [DOI] [PubMed] [Google Scholar]
- 27.Villamor E, Koulinska IN, Aboud S, Murrin C, Bosch RJ, Manji KP, Fawzi WW. 2010. Effect of vitamin supplements on HIV shedding in breast milk. Am J Clin Nutr 92:881–886. doi: 10.3945/ajcn.2010.29339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neuzil KM, Gruber WC, Chytil F, Stahlman MT, Graham BS. 1995. Safety and pharmacokinetics of vitamin A therapy for infants with respiratory syncytial virus infections. Antimicrob Agents Chemother 39:1191–1193. doi: 10.1128/AAC.39.5.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mora JR, Iwata M, von Andrian UH. 2008. Vitamin effects on the immune system: vitamins A and D take centre stage. Nat Rev Immunol 8:685–698. doi: 10.1038/nri2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matikainen S, Lehtonen A, Sareneva T, Julkunen I. 1998. Regulation of IRF and STAT gene expression by retinoic acid. Leuk Lymphoma 30:63–71. doi: 10.3109/10428199809050930. [DOI] [PubMed] [Google Scholar]
- 31.Yano M, Ikeda M, Abe K, Dansako H, Ohkoshi S, Aoyagi Y, Kato N. 2007. Comprehensive analysis of the effects of ordinary nutrients on hepatitis C virus RNA replication in cell culture. Antimicrob Agents Chemother 51:2016–2027. doi: 10.1128/AAC.01426-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bocher WO, Wallasch C, Hohler T, Galle PR. 2008. All-trans retinoic acid for treatment of chronic hepatitis C. Liver Int 28:347–354. doi: 10.1111/j.1478-3231.2007.01666.x. [DOI] [PubMed] [Google Scholar]
- 33.Kleywegt GJ, Bergfors T, Senn H, Le Motte P, Gsell B, Shudo K, Jones TA. 1994. Crystal structures of cellular retinoic acid binding proteins I and II in complex with all-trans-retinoic acid and a synthetic retinoid. Structure 2:1241–1258. doi: 10.1016/S0969-2126(94)00125-1. [DOI] [PubMed] [Google Scholar]
- 34.Fogh K, Voorhees JJ, Astrom A. 1993. Expression, purification, and binding properties of human cellular retinoic acid-binding protein type I and type II. Arch Biochem Biophys 300:751–755. doi: 10.1006/abbi.1993.1104. [DOI] [PubMed] [Google Scholar]
- 35.Norris AW, Cheng L, Giguere V, Rosenberger M, Li E. 1994. Measurement of subnanomolar retinoic acid binding affinities for cellular retinoic acid binding proteins by fluorometric titration. Biochim Biophys Acta 1209:10–18. doi: 10.1016/0167-4838(94)90130-9. [DOI] [PubMed] [Google Scholar]
- 36.Budhu AS, Noy N. 2002. Direct channeling of retinoic acid between cellular retinoic acid-binding protein II and retinoic acid receptor sensitizes mammary carcinoma cells to retinoic acid-induced growth arrest. Mol Cell Biol 22:2632–2641. doi: 10.1128/MCB.22.8.2632-2641.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kane MA, Folias AE, Pingitore A, Perri M, Krois CR, Ryu JY, Cione E, Napoli JL. 2011. CrbpI modulates glucose homeostasis and pancreas 9-cis-retinoic acid concentrations. Mol Cell Biol 31:3277–3285. doi: 10.1128/MCB.05516-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boylan JF, Gudas LJ. 1992. The level of CRABP-I expression influences the amounts and types of all-trans-retinoic acid metabolites in F9 teratocarcinoma stem cells. J Biol Chem 267:21486–21491. [PubMed] [Google Scholar]
- 39.Berry DC, Noy N. 2009. All-trans-retinoic acid represses obesity and insulin resistance by activating both peroxisome proliferation-activated receptor beta/delta and retinoic acid receptor. Mol Cell Biol 29:3286–3296. doi: 10.1128/MCB.01742-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bonet ML, Ribot J, Palou A. 2012. Lipid metabolism in mammalian tissues and its control by retinoic acid. Biochim Biophys Acta 1821:177–189. doi: 10.1016/j.bbalip.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 41.Inoue Y, Inoue J, Lambert G, Yim SH, Gonzalez FJ. 2004. Disruption of hepatic C/EBPalpha results in impaired glucose tolerance and age-dependent hepatosteatosis. J Biol Chem 279:44740–44748. doi: 10.1074/jbc.M405177200. [DOI] [PubMed] [Google Scholar]
- 42.Siersbæk M, Varticovski L, Yang S, Baek S, Nielsen R, Mandrup S, Hager GL, Chung JH, Grøntved L. 2017. High fat diet-induced changes of mouse hepatic transcription and enhancer activity can be reversed by subsequent weight loss. Sci Rep 7:40220. doi: 10.1038/srep40220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kang HW, Bhimidi GR, Odom DP, Brun PJ, Fernandez ML, McGrane MM. 2007. Altered lipid catabolism in the vitamin A deficient liver. Mol Cell Endocrinol 271:18–27. doi: 10.1016/j.mce.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 44.Mercader J, Ribot J, Murano I, Felipe F, Cinti S, Bonet ML, Palou A. 2006. Remodeling of white adipose tissue after retinoic acid administration in mice. Endocrinology 147:5325–5332. doi: 10.1210/en.2006-0760. [DOI] [PubMed] [Google Scholar]
- 45.Amengual J, Ribot J, Bonet ML, Palou A. 2010. Retinoic acid treatment enhances lipid oxidation and inhibits lipid biosynthesis capacities in the liver of mice. Cell Physiol Biochem 25:657–666. doi: 10.1159/000315085. [DOI] [PubMed] [Google Scholar]
- 46.García-Mediavilla MV, Pisonero-Vaquero S, Lima-Cabello E, Benedicto I, Majano PL, Jorquera F, González-Gallego J, Sánchez-Campos S. 2012. Liver X receptor alpha-mediated regulation of lipogenesis by core and NS5A proteins contributes to HCV-induced liver steatosis and HCV replication. Lab Invest 92:1191–1202. doi: 10.1038/labinvest.2012.88. [DOI] [PubMed] [Google Scholar]
- 47.Nakajima S, Watashi K, Ohashi H, Kamisuki S, Izaguirre-Carbonell J, Kwon AT, Suzuki H, Kataoka M, Tsukuda S, Okada M, Moi ML, Takeuchi T, Arita M, Suzuki R, Aizaki H, Kato T, Suzuki T, Hasegawa H, Takasaki T, Sugawara F, Wakita T. 2016. Fungus-derived neoechinulin B as a novel antagonist of liver X receptor, identified by chemical genetics using a hepatitis C virus cell culture system. J Virol 90:9058–9074. doi: 10.1128/JVI.00856-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Damiano F, Alemanno S, Gnoni GV, Siculella L. 2010. Translational control of the sterol-regulatory transcription factor SREBP-1 mRNA in response to serum starvation or ER stress is mediated by an internal ribosome entry site. Biochem J 429:603–612. doi: 10.1042/BJ20091827. [DOI] [PubMed] [Google Scholar]
- 49.Damiano F, Rochira A, Tocci R, Alemanno S, Gnoni A, Siculella L. 2013. hnRNP A1 mediates the activation of the IRES-dependent SREBP-1a mRNA translation in response to endoplasmic reticulum stress. Biochem J 449:543–553. doi: 10.1042/BJ20120906. [DOI] [PubMed] [Google Scholar]
- 50.Siculella L, Tocci R, Rochira A, Testini M, Gnoni A, Damiano F. 2016. Lipid accumulation stimulates the cap-independent translation of SREBP-1a mRNA by promoting hnRNP A1 binding to its 5′-UTR in a cellular model of hepatic steatosis. Biochim Biophys Acta 1861:471–481. doi: 10.1016/j.bbalip.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 51.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. 2011. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M Jr.. 2008. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature 454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takahashi K, Asabe S, Wieland S, Garaigorta U, Gastaminza P, Isogawa M, Chisari FV. 2010. Plasmacytoid dendritic cells sense hepatitis C virus-infected cells, produce interferon, and inhibit infection. Proc Natl Acad Sci U S A 107:7431–7436. doi: 10.1073/pnas.1002301107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoshio S, Kanto T, Kuroda S, Matsubara T, Higashitani K, Kakita N, Ishida H, Hiramatsu N, Nagano H, Sugiyama M, Murata K, Fukuhara T, Matsuura Y, Hayashi N, Mizokami M, Takehara T. 2013. Human blood dendritic cell antigen 3 (BDCA3)(+) dendritic cells are a potent producer of interferon-lambda in response to hepatitis C virus. Hepatology 57:1705–1715. doi: 10.1002/hep.26182. [DOI] [PubMed] [Google Scholar]
- 55.Ooi EL, Chan ST, Cho NE, Wilkins C, Woodward J, Li M, Kikkawa U, Tellinghuisen T, Gale M Jr, Saito T. 2014. Novel antiviral host factor, TNK1, regulates IFN signaling through serine phosphorylation of STAT1. Proc Natl Acad Sci U S A 111:1909–1914. doi: 10.1073/pnas.1314268111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bartosch B, Dubuisson J, Cosset FL. 2003. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J Exp Med 197:633–642. doi: 10.1084/jem.20021756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sabol SL, Brewer HB Jr, Santamarina-Fojo S. 2005. The human ABCG1 gene: identification of LXR response elements that modulate expression in macrophages and liver. J Lipid Res 46:2151–2167. doi: 10.1194/jlr.M500080-JLR200. [DOI] [PubMed] [Google Scholar]
- 58.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bullard JH, Purdom E, Hansen KD, Dudoit S. 2010. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics 11:94. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data have been deposited in the NCBI Gene Expression Omnibus (GEO) under accession number GSE101934.