Abstract
Acquired uniparental disomy (aUPD, also known as copy-neutral loss of heterozygosity) is a common feature of cancer cells and characterized by extended tracts of somatically-acquired homozygosity without any concurrent loss or gain of genetic material. The presumed genetic targets of many regions of aUPD remain unknown. Here we describe the association of chromosome 22 aUPD with mutations that delete the C-terminus of PRR14L in patients with chronic myelomonocytic leukemia (CMML), related myeloid neoplasms and age-related clonal hematopoiesis (ARCH). Myeloid panel analysis identified a median of 3 additional mutated genes (range 1-6) in cases with a myeloid neoplasm (n=8), but no additional mutations in cases with ARCH (n=2) suggesting that mutated PRR14L alone may be sufficient to drive clonality. PRR14L has very limited homology to other proteins and its function is unknown. ShRNA knockdown of PRR14L in human CD34+ cells followed by in vitro growth and differentiation assays showed an increase in monocytes and decrease in neutrophils consistent, with a CMML-like phenotype. RNA-Seq and cellular localization studies suggest a role for PRR14L in cell division. PRR14L is thus a novel, biallelically mutated gene and potential founding abnormality in myeloid neoplasms.
Keywords: PRR14L, CMML, myeloid neoplasms
Introduction
Uniparental disomy refers to the situation in which both copies of a chromosomal region or an entire chromosome originate from one parent. Acquired uniparental disomy (aUPD; also known as copy-neutral loss of heterozygosity) is a common feature of cancer cells and characterized by extended tracts of somatically-acquired homozygosity without any concurrent loss or gain of genetic material.1 Typically, aUPD is initiated by mitotic crossing over or aneuploidy rescue and acts to convert a somatically-acquired heterozygous driver mutation to homozygosity, an event that confers a further growth advantage to the mutant clone. In myeloid neoplasms, aUPD is seen in up to a third of cases and is strongly associated with a range of somatically mutated genes such as MPL at chromosome 1p34 (aUPD1p), TET2 (aUPD4q), EZH2 (aUPD7q), JAK2 (aUPD9p), CBL (aUPD11q), FLT3 (aUPD13q) and CALR (aUPD19p).2–9 Occasionally, aUPD targets inherited variants or imprinted regions rather than somatic mutations, e.g. aUPD7q in myeloid malignancies may lead to loss of inherited mutations in SAMD9L,10 and aUPD14q leads to homozygosity for the paternal copy of the imprinted MEG3-DLK1 locus.11 In addition, aUPD and associated mutations can be found in elderly populations unselected for hematological disorders, a finding known as age-related clonal hematopoiesis (ARCH), clonal hematopoiesis of indeterminate potential (CHIP) or, more broadly, aberrant clonal expansions (ACE).12–16
Although it is possible that aUPD might occasionally be a passenger event in cancer, it seems more likely that these regions are only apparent because of the selective advantage they confer in conjunction with specific mutations or imprinted loci.15 Nevertheless, several regions of aUPD have not yet been associated with specific genetic targets, one of the most prominent being aUPD22q. Although uncommon, this abnormality is clearly recurrent in myeloid neoplasms as well as ARCH. We previously identified 3/148 (2%) cases of myelodysplastic/myeloproliferative neoplasms (MDS/MPN) with chromosome 22 aUPD by genome-wide single nucleotide polymorphism (SNP) analysis,5 and other studies identified cases with MDS/MPN or myelodysplastic syndrome (MDS) with complete or partial aUPD22 of the q arm.6, 17 Two independent studies of >50,000 subjects recruited for genome-wide association studies revealed a total of 424 instances of aUPD, of which 16 (4%) involved chromosome 22q.12, 13 Finally, we reported an analysis of 1141 cancer-free elderly men from the Uppsala Longitudinal Study of Adult Men (ULSAM) cohort and identified aUPD >2Mb in 16 (1.4%) individuals, of which 3 (19% of those with aUPD) involved chromosome 22.14 Here we identify the target of aUPD22 as PRR14L, a gene not previously recognized as being mutated in cancer.
Methods
Study cohorts
Our study focused on two major groups: (i) patients diagnosed with a myeloid neoplasm according to standard morphological, hematologic and laboratory criteria; (ii) the Uppsala Longitudinal Study of Adult Men, an ongoing longitudinal epidemiologic study based on all available men born between 1920 and 1924 and living in Uppsala County, Sweden. Genome wide SNP analysis of most of these subjects has been published previously and used to identify cases with aUPD22.4, 5, 9, 11, 14 The studies were approved by the UK National Research Ethics Service (NRES) Committee South West and NRES East Midlands, the Uppsala Regional Ethical Review Board, the Ethics Committee of the Biomedical Research Foundation of the Academy of Athens.
Whole exome sequencing
For cases E4051 and E6526 we sequenced both tumor (peripheral blood leucocyte) and constitutional (cultured T-cell) DNA. T-cells were isolated from anticoagulated peripheral blood using CD3 MicroBeads and cultured using a T-cell Activation/Expansion kit (Miltenyi Biotec, Bisley, UK). Samples were prepared for exome sequencing using the Agilent SureSelect kit (Agilent Technologies, Palo Alto, CA, USA) (Human All Exon 50 Mb) and then sequenced on an Illumina HiSeq 2000 (Illumina, Great Abington, UK) at the Wellcome Trust Centre for Human Genetics, Oxford, UK. Exome sequencing of ULSAM1182, ULSAM1242 and ULSAM1356 (peripheral blood leukocyte DNA only) was performed by SciLifeLab (Stockholm, Sweden). Sequence data from all five samples were analyzed using a custom bioinformatic pipeline as previously described.18
Mutation screening
We screened all coding exons of PRR14L in 115 patient samples (CMML, n=52; atypical chronic myeloid leukemia, n=46, myelodysplastic/myeloproliferative neoplasm-unclassified, n=7, myeloproliferative neoplasm, n=10) and 20 myeloid cell lines (Supplementary Table 1) using a combination of Sanger sequencing and a custom designed Illumina TruSeq panel. Samples were processed according to the manufacturer’s protocol and run on an Illumina Miseq. The Illumina Trusight Myeloid Sequencing Panel (Illumina) was used to screen PRR14L mutated samples for additional pathogenic mutations. Samples were processed according to the manufacturer’s protocol and run on an Illumina Miseq.
Cell lines and expression constructs
The HEK293F cell line (Thermo Fisher Scientific, Waltham, MA, USA) was grown in DMEM plus 10% fetal calf serum and transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). Full length PRR14L (Genbank accession: NM_173566) was synthesized (Genescript, New Jersey, USA) and transferred to pCMV6-Entry (C-terminal Myc-DDK tags; Origene Technologies Inc., Maryland, USA). Wildtype and mutant PRR14L were then transferred to pCMV6-AN-Myc-DDK, pCMV6-AC-GFP and pCMV6-AN-GFP (Origene Technologies Inc, Maryland, USA).
Lentiviral knockdown of PRR14L expression
Lentivirus was produced by transfection of pLKO.1 shRNA plasmids with the Mission Lentiviral packaging mix (Sigma-Aldrich Company Ltd., Poole, UK) into HEK293F cells and harvested on day 3. Lentiviruses were harvested after 48h and 72h post-transfection and concentrated by ultracentrifugation (Beckman Coulter, Brea, CA, USA; Ultracentrifuge Rotor SW28) at 28000 rpm for 3h at 4°C. To test knockdown efficiency, five pLK0.1 shRNAs were transduced into HEK293F cells with 8 μg/ml polybrene and selected in with 1 μg/ml puromycin (Thermo Fisher Scientific) for 5 days. PRR14L expression was measured by Taqman assay (Assay Hs00543135_m1, Thermo Fisher Scientific) with GUSB (assay Hs00939627_m1) and B2M (assay Hs99999907_m1) as internal normalisation controls. Two clones showing the greatest degree of knockdown (TRCN0000422471 and TRCN0000135981; Sigma Mission, Sigma-Aldrich Company Ltd.) were used for all further experiments. Healthy CD34+ cells were infected with virus in presence of polybrene (8µg/ml). Cells were spinoculated at 800Xg for 2 h at 32°C. Cells were replenished with fresh medium after 24 h of viral infection. Selection of transduced cells was performed with puromycin (0.65µg/ml) and the cells were kept in selection medium throughout the experiment. Details of all antibodies and flow cytometric analysis are given in the Supplementary Material.
Immunofluorescence
Cells were cytospun onto slides, dried briefly then fixed in methanol at -20 °C for 10 mins. Blocking and antibody dilutions were in 1% BSA in PBS. Primary antibodies were incubated at 4°C overnight and secondary antibodies for 1 hour at room temperature.
Immunoprecipitation
Lysates were prepared for immunoprecipitation either with a Nuclear Complex Co-IP kit (Active Motif, Carlsbad, CA, USA) using the low stringency buffer without additional detergent or salt, or by lysing in Tris/Triton buffer (20 mM Tris-Cl pH 7.6, 150 nM NaCl, 1% Triton-X, 1 mM EDTA plus protease inhibitors). Potential interactions between PRR14L and ASXL1 or BAP1 were tested in (i) unmanipulated HL60, K562 and MARIMO cells and (ii) HEK293F cells transiently transfected with PRR14L-N-FLAG/MYC or PRR14L-C-FLAG/MYC expression constructs.
Cell culture
CD34+ cells from healthy donors were obtained from Lonza (Basel, Switzerland). The cells were cultured in erythroid or granulomonocytic differentiation media for 14 day as described previously.19, 20 For cell growth assays, on day 7 of culture transduced cells were seeded in 96 well plates (10000 cells/200µl). Viable cells were counted by using trypan blue exclusion assay. For colony assays, on day 7 of culture 3000 transduced cells were plated on methylcellulose (MethoCult H4434 Classic, Stemcell Technologies) containing 0.65µg/ml puromycin according to the manufacturer’s protocol. Colonies were counted after 14 days. May-Grünwald and Giemsa stains were used to stain cytospin slides of granulomonocytic and erythroid cells according to the manufacturer’s protocol (Sigma Aldrich).
RNA-seq
Individual CFU-GM colonies expressing PRR14L shRNA (n=3) or a scramble shRNA control (n=2) were resuspended in TrIzol reagent and total RNA was extracted according to the manufacturer’s protocol (Thermo Fisher Scientific). RNA was treated with DNase and purified using Agencourt RNAClean XP beads (Beckman Coulter). Libraries were produced using SMART-Seq2 library preparation protocol21 and sequencing was performed on an Illumina HiSeq4000 with 75bp paired-end reads. Reads were trimmed for Nextera, Smart-seq2, Illumina adapter sequences using skewer-v0.1.125. Trimmed read pairs were mapped to human genome hg38.ERCC using HISAT2 version 2.0.422. Uniquely mapped read pairs were counted by using featureCounts, subread-1.5.0, using exons annotated in ENSEMBL annotations, release 75.23, 24
Data analysis
Differential gene expression analysis was performed using edgeR.25 Analysis of gene set up- or downregulation was performed using Gene Set Association Analysis for RNA-Seq with Sample Permutation (GSAASeqSP).26 Ingenuity Pathway Analysis (IPA) software (Qiagen, Hilden, Germany) was used for the identification of significantly dysregulated pathways, and of significant upstream regulators and downstream biological functions. The significance of the comparisons between cells expressing PRR14L shRNAs or the scramble control was determined by repeated-measures 1-way ANOVA with Tukey’s post-hoc tests. The measurements of cell growth over time for cells expressing PRR14L shRNAs or the scramble control were compared using 2-way ANOVA and Bonferroni’s post tests. P values of less than 0.05 were considered statistically significant.
Results
Recurrent mutations of PRRL14 associated with aUPD22q
Whole exome sequence (WES) analysis of 5 cases with aUPD22q (3 from the ULSAM cohort and 2 with MDS/MPN) revealed that 4 had inactivating mutations in PRR14L (nonsense, n=2; frameshift, n=2). No other gene was found to have inactivating mutations in >2 cases indicating that PRR14L is the likely target of aUPD22q (Supplementary Table 1). Analysis of a further 6 cases with MDS/MPN with aUPD22q for PRR14L mutations by a combination of Sanger sequencing and targeted next generation sequencing revealed an additional four mutated individuals. We then screened PRR14L in unselected cases with myeloid neoplasia (n=115) and identified two further mutated cases. These cases were not tested for aUPD22q but one of these cases (E6353) had a PRR14L variant allele frequency (vaf) of 0.93, indicating homozygosity or hemizygosity in the great majority of cells.
All 10 PRR14L mutations were frameshift or nonsense mutations resulting in loss of the C-terminus of the protein (Figure 1a; Table 1). In 2/2 cases tested we found that the PRR14L mutation was absent in cultured T-cells, indicating a somatic origin (Supplementary Figure 1). We found no PRR14L mutations in 20 myeloid cell lines (Supplementary Table 2) but inspection of the Cosmic database (https://cancer.sanger.ac.uk/cosmic; accessed 3rd May 2018) revealed 28 entries with truncating PRR14L mutations associated with a wide range of cancers but mainly solid tumors (Supplementary Table 3), thus demonstrating that pathogenic abnormalities of this gene are not limited to hematological malignancies.
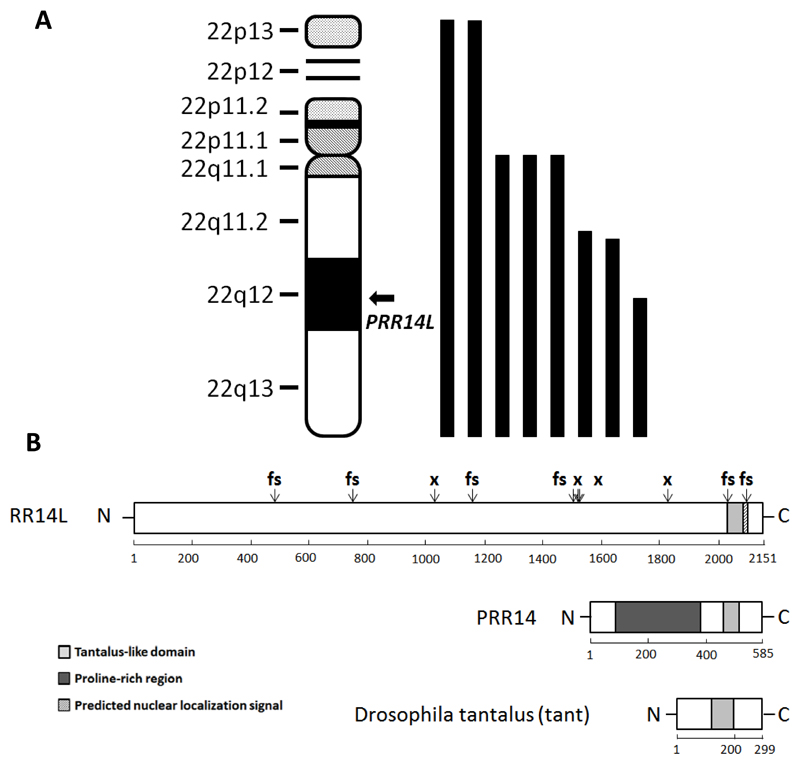
Figure 1. Acquired UPD at chromosome 22q and PRR14L mutations.
(A) Location of PRR14L on chromosome 22 and positions of the 8 detected regions of aUPD22 overlapping this gene (B). The positions of 6 frameshift (fs) and 4 nonsense (x) PRR14L mutations are indicated. PRR14L has no recognised motifs apart from a predicted nuclear localization signal and a tantalus-like domain also seen in PRR14 and the Drosophilia protein tant.
Table 1. Cases with aUPD22 and/or mutations of PRR14L.
| UPN | aUPD22 | Disorder | PRR14L mutation (vaf) | Additional mutations (vaf) |
|---|---|---|---|---|
| E2633 | chr22:15685581-qter | MPN-U | exon4: c.5497C>T p.R1833* (0.90) | CBL p.I383M (0.79) ETV6 p.S47* (0.21) GATA2 p.K390del (0.41) PHF6 p.G10Rfs*12 (0.80) U2AF1 p.Q157R (0.50) |
| E4051 | chr22:16604328-qter | CMML | exon4: c.3081T>A p.C1027* (0.94) | NRAS p.G12S (0.50) ASXL1 p.Q748* (0.46) GATA2 p.L386_E391del (0.24) |
| E5317 | chr22:pter-qter | CMML | exon 4: c.2223_2236del p.E741Dfs*3 (0.95) | NRAS p.G12S (0.49) TET2 p.Q1603* (0.50) RUNX1 p.L82Rfs*41 (0.47) |
| E5319 | chr22:pter-qter | CMML | exon 4: c.1446delC p.H482Qfs*6 (0.95) | EZH2 p.Y731D (0.46) RUNX1 p.G122Qfs12 (0.46) PTPN11 p.G503R (0.46) |
| E6526 | chr22:31496485-qter | MDS/MPN | exon4: c.3489delA p.E1163Dfs*10 (0.75) | EZH2 p.I109* (0.45) TET2 p.K1208* (0.53) TET2 p.N1610Kfs*4 (0.50) |
| E12759 | chr22:23855603-qter | MDS RAEB1 | exon 9: c.6287delC p.P2096Rfs*33 (N/A) | U2AF1 p.Q157P (0.33) |
| ULSAM1242 | chr22:17000000-qter | ARCH | exon4: c.4561C>T p.Q1521* (0.58) | None |
| ULSAM1182 | chr22:24700000-qter | ARCH | exon4: c.4518delA p.H1507Mfs*12 (0.37) | None |
| E3765 | N/A | CMML | exon 4: c.4570C>T p.R1524* (0.47) | NRAS p.G12D (0.38) SETBP1 p.S869N (0.48) |
| E6353 | N/A | CMML | exon 7: c.6084delC p.M2029Wfs*9 (0.93) | ASXL1 p.T794Nfs*6 (0.44) CSF3R p.E815* (0.44) CSF3R p.T618I (0.40) SRSF2 p.P95H (0.27) TET2 p.K318Nfs*29 (0.45) TET2 p.K318* (0.45) |
vaf, variant allele frequency; N/A, not available
Association of PRR14L mutations with ARCH and CMML
Of the 10 PRR14L mutated cases, 8 had a diagnosis of a myeloid neoplasm and 5 of these had chronic myelomonocytic leukemia (CMML). The other 2 mutated cases were recruited from the ULSAM cohort: ULSAM1182 died at 91 years of age, 11 years after detection of aUPD22q/PRR14L, without any evidence of malignancy. ULSAM1242 died at the age of 77 of prostate cancer but also had a ‘secondary malignant neoplasm of bone and bone marrow’. Both malignancies were diagnosed 4 years after the date of the sample in which we detected aUPD22q/PRR14L. Of note, the third individual from the ULSAM cohort (ULSAM1356) with aUPD22q but without a detectable PRR14L mutation developed an unspecified myeloid leukemia after two years. WES revealed a frameshift mutation in CHEK2 at 22q12 (NM_007194: c.550_551insAT; p.N184fs), suggesting that aUPD22q might occasionally target this gene.
Myeloid neoplasms are often characterised by somatic mutations in multiple genes.27, 28 We searched for additional mutations in PRRL14 mutated cases using the Illumina TruSight Myeloid Sequencing Panel and/or WES data. All 8 cases tested with a diagnosed myeloid neoplasm showed additional mutations (median=3, range 1-6) but there was no gene that was recurrently co-mutated with PRR14L (Table 1). Strikingly, no additional mutations were detected in ULSAM1182 or ULSAM1242, suggesting that mutated PRR14L alone may be sufficient to drive ARCH. Comparison of the variant allele frequencies (VAF) between additional mutations and PRR14L in cases with a myeloid neoplasm did not allow us to infer the order in which some of the mutations were acquired (Supplementary Figure 2), but there was no case in which a PRR14L mutation was clearly a late event. Our data thus suggest that PRR14L mutations may be founding events in the multistep pathogenesis of myeloid neoplasms.
PRR14L is located at the midbody in dividing cells
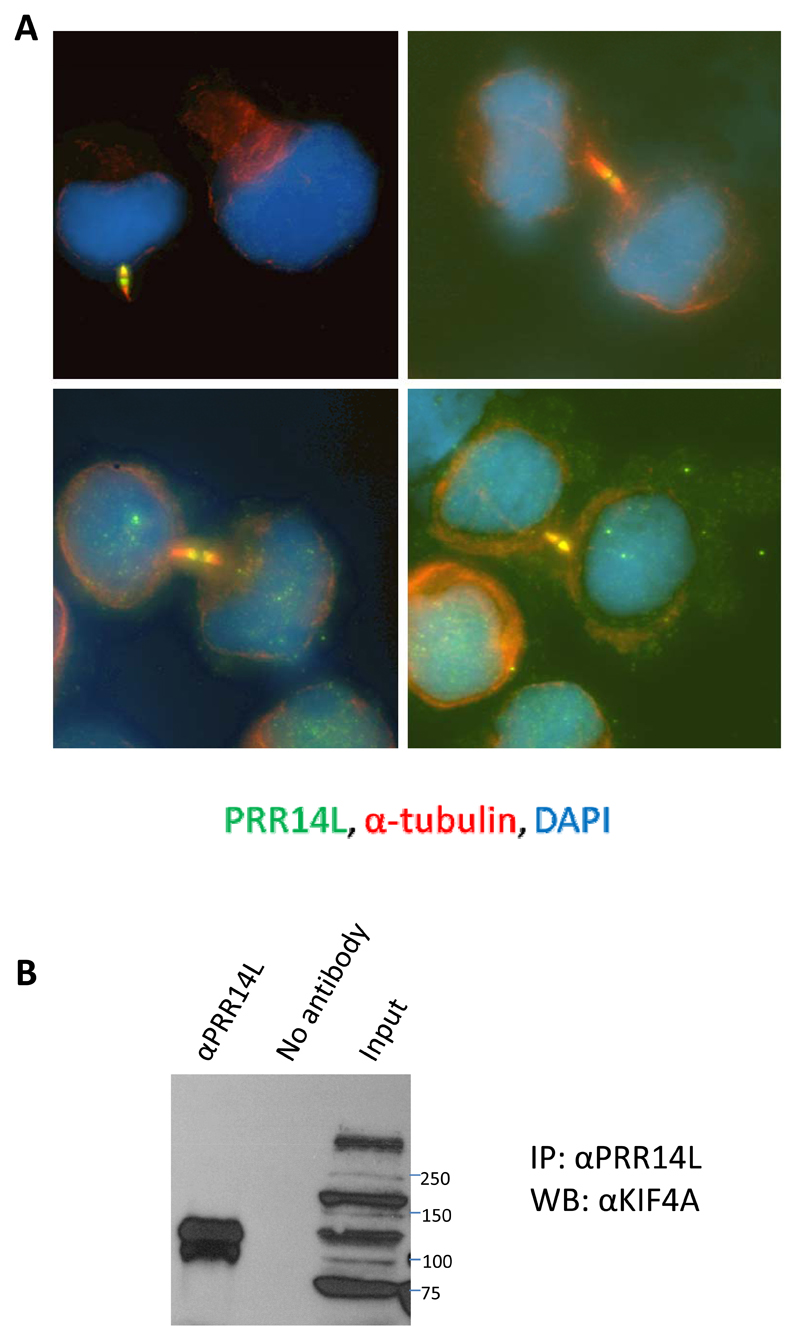
PRR14L (proline rich repeat 14-like) is predicted to encode a widely expressed 2151 amino acid (237KDa) protein of unknown function and no recognised domains except for a 49 amino acid region of homology with the Drosophila protein tantalus (tant;29) and PRR14, an independent gene on chromosome 16 (Figure 1 and Supplementary Figure 3). The function of this tantalus-like domain is unknown. Tant has either a cytoplasmic or nuclear localisation depending on cellular context29 whereas the Protein Atlas (https://www.proteinatlas.org/ENSG00000183530-PRR14L/cell) indicates PRR14L is nuclear in SiHa and U-2 OS cells. Consistent with this, we identified a classical bipartite nuclear localisation signal30 at amino acids 2079-2104 that is predicted to be lost or disrupted in all 10 mutants (Figure 1). Strikingly, using the same antibody (HPA062645) we found that PRR14L was located at the midbody in dividing HEK293F cells (Figure 2A). In support of this finding, mass spectrometry31 identified two midbody proteins, KIF4A and KIF23 as PRR14L interacting partners, and we confirmed an interaction between PRR14L and KIF4A by co-immunoprecipitation (Figure 2B).
Figure 2. PRR14L is located at the midbody and interacts with KIF4A.
(A) HEK293 cells were immunostained with PRR14L (HPA062645 antibody; green) and α-tubulin (red) and counterstained with DAPI. Confirmatory evidence for midbody localization of PRR14L in dividing HEK-293F is that the midbody signal can be blocked by the PrEST antigen for the HPA062645 antibody (not shown).
(B) HEK293F lysates were immunoprecipitated (IP) with the PRR14L antibody HPA062645 using the Nuclear Complex Co-IP kit and blotted (WB) with anti-KIF4A (GTX115759). The identity of KIF4A was confirmed by repeating with a second antibody (ab3815) (not shown). The PRR14L midbody staining was reproducible and seen in the majority of midbodies identified by tubulin staining and was also seen in HL60 and K562 cells (not shown).
Downregulation of PRR14L induces a CMML-like phenotype in vitro
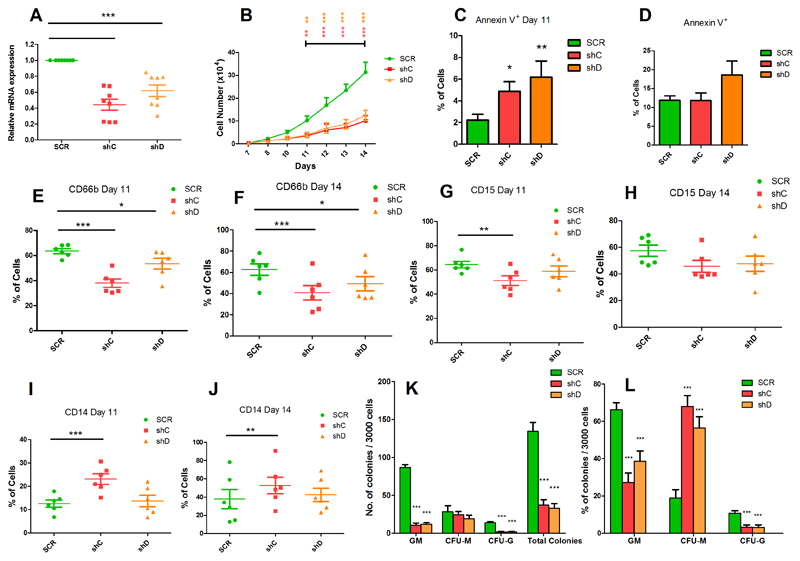
The PRRL14 mutations we identified suggest a loss of function. To study the impact of PRR14L inactivation on granulomonocytic and erythroid differentiation, we knocked down PRR14L in primary human bone marrow CD34+ cells using two different shRNAs. Transduced cells were cultured for 14 days under conditions for differentiation into erythroid and granulomonocytic cells and PRR14L knockdown was confirmed in granulomonocytic and erythroid cells by real-time quantitative PCR (Figures 3A & 4A). Granulomonocytic cells with PRR14L knockdown showed impaired cell growth and an increase in apoptosis at day 11 (Figure 3B-D) compared to the scramble control. The effects of PRR14L knockdown on granulomonocytic differentiation were studied by flow cytometric analysis of granulomonocytic cell surface markers (granulocytes, CD66b and CD15; monocytes, CD14) using cells harvested on day 11 and day 14 of culture under granulomonocytic differentiating conditions. We observed a significant decrease in the cell population expressing CD66b (Figure 3E-F) and a reduction of cells expressing CD15 (Figure 3G-H), indicating a decrease in the percentage of granulocytes, in cells with PRR14L knockdown compared to the scramble control. We also observed a decrease in the CD15+CD66b+ population (Supplementary Figure 4A-B). A significant increase in the population expressing CD14 was observed, indicating an increase in the percentage of monocytes in the granulomonocytic cultures following PRR14L knockdown (Figure 3I-J). Morphological examination of granulomonocytic cells cytospins showed that PRR14L knockdown resulted in a decrease in the number of granulocytes (mainly neutrophils) and an increase in the number of macrophages (Supplementary figure 4C). In colony forming assays PRR14L knockdown resulted in a significant reduction in the total number of colonies compared to the scramble control with a higher relative proportion of CFU-M compared to CFU-GM and CFU-G (Figure 3 K-L). Thus, knockdown of PRR14L results in a relative increase in the monocyte/macrophage population, consistent with a CMML phenotype.
Figure 3. Effects of PRR14L knockdown on granulomonocytic differentiation.
(A) Real-time quantitative PCR showing knockdown of PRR14L in granulomonocytic cells. (B) Cell growth curves obtained by trypan blue exclusion assay from day 7 to day 14 of the granulomonocytic cultures. (C-D) Apoptosis measured by Annexin V staining using flow cytometry on (C) day 11 and (D) day 14 of culture. (E-J) Flow cytometry analysis of CD66b, CD15 and CD14 for evaluation of granulomonocytic differentiation: CD66b+ cells on (E) day 11 and (F) day 14 of culture, CD15+ cells on (G) day 11 and (H) day 14 of culture, and CD14+ cells on (I) day 11 and (J) day 14 of culture. (K-L) Colony forming cell assays for evaluation of graulomonocytic differentiation: (K) total number of colonies, CFU-GM, CFU-M and CFU-G, and (L) relative proportion of the different types of colonies. Results shown in panel A, B, C-J, K-L are from 8, 7, 6, 7 independent experiments respectively. Data are represented as Mean ± SEM. P values were obtained by repeated-measures 1-way ANOVA with Tukey’s post-hoc tests. P values in panel (B) were calculated by 2-way ANOVA with Bonferroni’s post test.*P<0.05, **P< 0.01, ***P< 0.001.
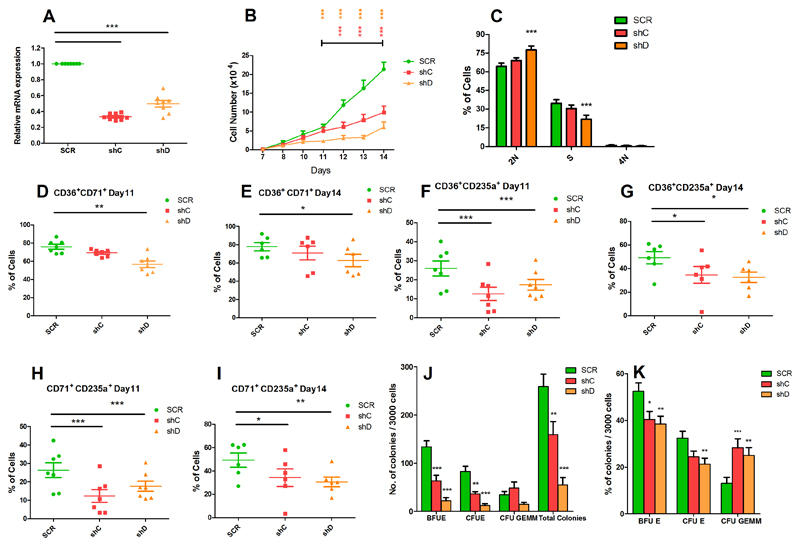
Figure 4. Effects of PRR14L knockdown on erythroid differentiation.
(A) Real-time quantitative PCR showing knockdown of PRR14L in erythroid cells. (B) Cell growth curves obtained by trypan blue exclusion assay from day 7 to day 14 of the erythroid cultures. (C) Cell cycle analysis of erythroid cells. (D-I) Flow cytometry analysis of CD36, CD71 and CD235a for evaluation of erythroid differentiation: CD36+CD71+cells on (D) day 11 and (E) day 14 of culture, CD36+CD235a+ cells on (F) day 11 and (G) day 14 of culture, CD71+CD235a+ cells on (H) day 11 and (I) day 14 of culture. (J-K) Colony forming cell assay for evaluation of erythroid differentiation: (J) total number of colonies, BFU-E, CFU-E and CFU-GEMM, and (K) relative proportion of the different types of colonies. Results shown in panel A, B, C, D, E, F, G, H, I, J-K are from 8, 7, 6, 7, 6, 7, 6, 7, 6, 7 independent experiments respectively. Data are represented as Mean ± SEM. P values for panel (A, C-K) were obtained by repeated-measures 1-way ANOVA with Tukey’s post-hoc tests. P values in panel (B) were calculated by 2-way ANOVA with Bonferroni’s post test.*P<0.05, **P< 0.01, ***P<0.001
Erythroid cells with PRR14L knockdown showed impaired cell growth compared to the scramble control (Figure 4B). No change in apoptosis was found (data not shown), however PRR14L knockdown resulted in cell cycle arrest at the G1 phase (Figure 4C). The effects of PRR14L knockdown on erythroid differentiation were studied by flow cytometry analysis of erythroid cell surface markers (CD36, CD71 and CD235a) using cells harvested on day 11 and day 14 of culture under erythroid differentiating conditions. We observed a decrease in the cell population expressing early erythroid markers (CD36+CD71+) (Figure 4D-E) and a concomitant significant decrease in the intermediate erythroid cell population (CD71+CD235a+ and CD36+CD235a+) (Figure 4F-I). We also observed a decrease in the CD36+, CD71+ and CD235a+ cell populations in erythroid cells with PRR14L knockdown (Supplementary figure 5A-F). PRR14L knockdown resulted in a significant reduction in the total number of BFU-E and of CFU-E compared to the scramble control (Figure 4J). In colony forming assays, the relative proportion of BFU-E and CFU-E obtained from cells with PRR14L knockdown was lower compared to the scramble control, while the relative proportion of CFU-GEMM was higher (Figure 4K).
Gene expression analysis implicates PRR14L in cell division
To determine the effects of PRR14L knockdown on the transcriptome of hematopoietic cells differentiated towards the granulomonocytic lineage, RNAseq was performed on individual CFU-GM with shRNA-mediated PRR14L knockdown and CFU-GM colonies expressing the scramble shRNA control. A total of 104 significantly differentially expressed genes (22 upregulated and 82 downregulated) were identified in cells with PRR14L knockdown (Supplementary Table 4). We analyzed specific panels of genes described in the literature as involved in granulocyte32 and monocyte33 function. Differentially expressed genes in cells with PRR14L knockdown include DEFA3, DEFA4 and MPO from the list of granulocyte-relevant genes, and SERPINB2 and CSF1R from the list of monocyte-relevant genes (Supplementary Table 5). GSAASeqSP showed significant association of genes upregulated in CFU-GM with PRR14L knockdown and many enriched gene sets, including several related to cell cycle and mitosis (approximately 30% of the significant gene sets) (Supplementary Table 6).
Differentially expressed genes were analyzed by Ingenuity Pathway Analysis (IPA) to identify enriched biological functions. Hematological System Development and Function was the top ranking function (p-value range 9.6x10-3-1.4x10-13), with significantly enriched subcategories including several neutrophil-related functions among those with a negative activation score (Supplementary Table 7). These data are consistent with our observations that PRR14L knockdown results in a decrease of granulocyte populations in granulomonocytic cultures.
Using IPA we performed an analysis of upstream transcriptional regulators to determine if the differentially expressed genes were connected by a common regulatory process. Several potential transcriptional regulators were identified, the most significant being MKL1, MKL2 and SRF (Supplementary Table 8).
Further analysis of the significantly differentially expressed genes showed dysregulation of several pathways (Supplementary Table 9), with retinol biosynthesis and Gαi signalling amongst the top 5. Gαi signalling was also dysregulated in CFU-GM with PRR14L knockdown. Gαi proteins localize in the centrosomes and at the midbody and altered expression or function of these proteins leads to defective cell division.34 Moreover, Gαi signalling is required for the asymmetric positioning of the spindle in the process of asymmetric cell division.35 Some genes encoding proteins localized to the midbody and/or spindle structures (obtained from MiCroKiTS)36 were significantly differentially expressed (MYO6, FAM83D and TUBA4A) in CFU-GM with PRR14L knockdown. The observed inhibition of Gαi signalling in CFU-GM with PRR14L knockdown points to a role of PRR14L in cell division and links to our finding that PRR14L localises to the midbody in dividing cells.
Discussion
Characterization of recurrent chromosome abnormalities has formed the foundation of our understanding of the molecular genetics of hematological malignancies. Large regions of cytogenetically cryptic somatically acquired UPD, however, escaped attention until the large scale application of genome wide single nucleotide polymorphism arrays. Surprisingly, perhaps, there are still a number of targets of recurrent aUPD that remain to be identified. We report here the finding of truncating mutations of PRR14L, a gene of unknown function, associated with a chromosome 22 aUPD. Of particular interest, PRR14L mutations were identified as sole abnormalities in ARCH suggesting that mutated PRR14L alone may be sufficient to drive clonality. ARCH is associated with several age related conditions, including inflammation, vascular diseases and a high risk of developing hematologic malignancies. It has been suggested that identification and treatment of ARCH may have wide benefits for human health 16. Our findings expand the mutational repertoire associated with clonal hematopoiesis and will help to identify individuals who may benefit from early intervention.
PRR14 is the only human protein with recognisable homology to PRRL14L, with the two both having a tantulus domain, a motif of unknown function first identified in the Drosophilia gene tant. Despite its name, PRR14L has no homology with the proline-rich region of PRR14 and is not itself proline rich. PRR14 has been reported to tether heterochromatin to the nuclear lamina during interphase and mitotic exit37 and promote tumorigenesis by activating the PI3K pathway.38 Of particular interest, Drosophilia tant was initially identified as encoding a protein that interacts specifically with the polycomb/trithorax group protein Additional Sex Combs (Asx).29 ASXL1, a human orthologue of Asx, is frequently mutated in myeloid neoplasms, including CMML,27, 39 resulting in global loss of H3K27 methylation.40, 41 Furthermore, a broad interactome screen using mass spectrometry identified a potential interaction between BAP1 (an ASXL1 binding protein) and PRR14L.31 We were unable, however, to confirm an interaction between human PRR14L and either BAP1 or ASXL1 in several cell lines either by pull down of native or tagged PRR14L with BAP1 or ASXL1 antibodies, or by the reverse pull down of BAP1 or ASXL1 by PRR14L or tag antibodies (not shown). Gene expression profiling after knockdown of PRR14L, however, revealed two potential links between PRR14L and ASXL1.
First, we identified a number of candidate transcriptional regulators linked to the observed pattern of differentially expressed genes (Supplementary Table 8), one of which was PPARG (p=0.00077). ASXL1 is known to be involved in transcriptional regulation mediated by ligand-bound nuclear hormone receptors, such as PPARG and retinoic acid receptors.42, 43 Specifically, ASXL1 acts as corepressor of PPARG and coactivator of RARs and PPARG modulates gene networks involved in controlling growth, cellular differentiation, and apoptosis.44 We observed up-regulation of several PPARG target genes in CFU-GM, suggesting that PRR14L knockdown might affect ASXL1 function resulting in loss of repression of PPARG.
Second, analysis of the significantly differentially expressed genes showed dysregulation of retinol biosynthesis (Supplementary Table 9). Retinoic acid receptor alpha (RARα) plays an important role in regulating myeloid development, especially along the granulocytic lineage. Chromosomal translocations involving RARα result in dysregulation of this process in acute promyelocytic leukemia, a disease characterized by a block in granulocytic differentiation.45 We have previously reported dysregulation of the RAR activation pathway in CD34+ progenitor cells of patients with myeloid malignancies with ASXL1 mutations46 and of the RXR activation pathway in ASXL1-deficient cells.19 The RXR activation pathway was significantly dysregulated in CFU-GM with PRR14L knockdown (Supplementary Table 9), suggesting a potential link with ASXL1 function.
Gene expression analysis identified MKL1, MKL2 and SRF as the most significant candidate upstream transcriptional regulators (Supplementary Table 8). MKL1 and MKL2 are coactivators of the nuclear transcription factor serum response factor (SRF) and are found in complex with globular actin (G-actin) in the cytoplasm, preventing their nuclear localization. Incorporation of G-actin into filamentous actin (F-actin) in response to stimuli liberates MKL1/2, enabling their nuclear translocation and interaction with SRF. This stimulates expression of cytoskeletal genes, including actin and actin regulatory genes. Thereby, the actin-MKL-SRF circuit modulates gene expression along with cytoskeletal dynamics for the regulation of cell motility as well as cell survival, proliferation, and differentiation.47 MKL1 loss of function results in severe defects in actin rearrangement causing clinical presentation of neutrophil defects.48 Our data thus indicate that PRR14L knockdown results in downregulation of several genes belonging to the MKL-SRF circuit, potentially impacting neutrophil function.
We located PRR14L to the midbody in dividing cells. The midbody is a transient structure that forms during cell division and appears to direct abscission, the final stage in which the two daughter cells separate.49, 50 One of the daughter cells retains the midbody and there is evidence that midbody retention or release may influence cell fate. Altered midbody dynamics has been linked to oncogenesis since midbody retention or accumulation has been associated with a stem cell-like phenotype51, a point that is clearly of potential relevance to the finding of PRR14L mutations as sole abnormalities in ARCH.
In summary, we report for the first time the finding of loss of function PRR14L mutations in myeloid neoplasia and ARCH. Knockdown of PRR14L results in altered myeloid differentiation and cell growth in vitro and our data suggest that PRR14L may play a role in cell division. These findings increase our knowledge of the mechanisms by which myeloid malignancies arise and may have important implications for the treatment of CMML and related conditions, as well as the identification and potential management of ARCH.
Supplementary Material
Acknowledgements
This work was funded by Bloodwise Specialist Programme Grants 13002 to NCPC, AC and WT, and 13042 to JB and AP. We are grateful to the Central England Haemato-Oncology Research Biobank for providing DNA from case D14.31916.
Footnotes
Conflict-of-interest disclosure
The authors declare no competing financial interests.
References
- 1.Score J, Cross NC. Acquired uniparental disomy in myeloproliferative neoplasms. Hematol Oncol Clin North Am. 2012;26:981–991. doi: 10.1016/j.hoc.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 2.Kralovics R, Passamonti F, Buser AS, Teo SS, Tiedt R, Passweg JR, et al. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352:1779–1790. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- 3.Raghavan M, Smith LL, Lillington DM, Chaplin T, Kakkas I, Molloy G, et al. Segmental uniparental disomy is a commonly acquired genetic event in relapsed acute myeloid leukemia. Blood. 2008;112:814–821. doi: 10.1182/blood-2008-01-132431. [DOI] [PubMed] [Google Scholar]
- 4.Grand FH, Hidalgo-Curtis CE, Ernst T, Zoi K, Zoi C, McGuire C, et al. Frequent CBL mutations associated with 11q acquired uniparental disomy in myeloproliferative neoplasms. Blood. 2009;113:6182–6192. doi: 10.1182/blood-2008-12-194548. [DOI] [PubMed] [Google Scholar]
- 5.Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42:722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 6.Sanada M, Suzuki T, Shih LY, Otsu M, Kato M, Yamazaki S, et al. Gain-of-function of mutated C-CBL tumour suppressor in myeloid neoplasms. Nature. 2009;460:904–908. doi: 10.1038/nature08240. [DOI] [PubMed] [Google Scholar]
- 7.Langemeijer SM, Kuiper RP, Berends M, Knops R, Aslanyan MG, Massop M, et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet. 2009;41:838–842. doi: 10.1038/ng.391. [DOI] [PubMed] [Google Scholar]
- 8.Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360:2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 9.Tapper W, Jones AV, Kralovics R, Harutyunyan AS, Zoi K, Leung W, et al. Genetic variation at MECOM, TERT, JAK2 and HBS1L-MYB predisposes to myeloproliferative neoplasms. Nat Commun. 2015;6 doi: 10.1038/ncomms7691. 6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tesi B, Davidsson J, Voss M, Rahikkala E, Holmes TD, Chiang SCC, et al. Gain-of-function SAMD9L mutations cause a syndrome of cytopenia, immunodeficiency, MDS, and neurological symptoms. Blood. 2017;129:2266–2279. doi: 10.1182/blood-2016-10-743302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chase A, Leung W, Tapper W, Jones AV, Knoops L, Rasi C, et al. Profound parental bias associated with chromosome 14 acquired uniparental disomy indicates targeting of an imprinted locus. Leukemia. 2015;29:2069–2074. doi: 10.1038/leu.2015.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laurie CC, Laurie CA, Rice K, Doheny KF, Zelnick LR, McHugh CP, et al. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat Genet. 2012;44:642–650. doi: 10.1038/ng.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacobs KB, Yeager M, Zhou W, Wacholder S, Wang Z, Rodriguez-Santiago B, et al. Detectable clonal mosaicism and its relationship to aging and cancer. Nat Genet. 2012;44:651–658. doi: 10.1038/ng.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forsberg LA, Rasi C, Malmqvist N, Davies H, Pasupulati S, Pakalapati G, et al. Mosaic loss of chromosome Y in peripheral blood is associated with shorter survival and higher risk of cancer. Nat Genet. 2014;46:624–628. doi: 10.1038/ng.2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forsberg LA, Gisselsson D, Dumanski JP. Mosaicism in health and disease - clones picking up speed. Nat Rev Genet. 2017;18:128–142. doi: 10.1038/nrg.2016.145. [DOI] [PubMed] [Google Scholar]
- 16.Shlush LI. Age-related clonal hematopoiesis. Blood. 2018;131:496–504. doi: 10.1182/blood-2017-07-746453. [DOI] [PubMed] [Google Scholar]
- 17.Gondek LP, Dunbar AJ, Szpurka H, McDevitt MA, Maciejewski JP. SNP array karyotyping allows for the detection of uniparental disomy and cryptic chromosomal abnormalities in MDS/MPD-U and MPD. PLoS One. 2007;2:e1225. doi: 10.1371/journal.pone.0001225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tapper WJ, Foulds N, Cross NC, Aranaz P, Score J, Hidalgo-Curtis C, et al. Megalencephaly syndromes: exome pipeline strategies for detecting low-level mosaic mutations. PLoS One. 2014;9:e86940. doi: 10.1371/journal.pone.0086940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davies C, Yip BH, Fernandez-Mercado M, Woll PS, Agirre X, Prosper F, et al. Silencing of ASXL1 impairs the granulomonocytic lineage potential of human CD34(+) progenitor cells. Br J Haematol. 2013;160:842–850. doi: 10.1111/bjh.12217. [DOI] [PubMed] [Google Scholar]
- 20.Yip BH, Steeples V, Repapi E, Armstrong RN, Llorian M, Roy S, et al. The U2AF1S34F mutation induces lineage-specific splicing alterations in myelodysplastic syndromes. J Clin Invest. 2017;127:2206–2221. doi: 10.1172/JCI91363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Picelli S, Faridani OR, Bjorklund AK, Winberg G, Sagasser S, Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 22.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 24.Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41:e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiong Q, Mukherjee S, Furey TS. GSAASeqSP: a toolset for gene set association analysis of RNA-Seq data. Sci Rep. 2014;4 doi: 10.1038/srep06347. 6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Itzykson R, Kosmider O, Renneville A, Gelsi-Boyer V, Meggendorfer M, Morabito M, et al. Prognostic score including gene mutations in chronic myelomonocytic leukemia. J Clin Oncol. 2013;31:2428–2436. doi: 10.1200/JCO.2012.47.3314. [DOI] [PubMed] [Google Scholar]
- 28.Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122:3616–3627. doi: 10.1182/blood-2013-08-518886. quiz 3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dietrich BH, Moore J, Kyba M, dosSantos G, McCloskey F, Milne TA, et al. Tantalus, a novel ASX-interacting protein with tissue-specific functions. Dev Biol. 2001;234:441–453. doi: 10.1006/dbio.2001.0255. [DOI] [PubMed] [Google Scholar]
- 30.Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J Biol Chem. 2007;282:5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hein MY, Hubner NC, Poser I, Cox J, Nagaraj N, Toyoda Y, et al. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell. 2015;163:712–723. doi: 10.1016/j.cell.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 32.Naranbhai V, Fairfax BP, Makino S, Humburg P, Wong D, Ng E, et al. Genomic modulators of gene expression in human neutrophils. Nat Commun. 2015;6 doi: 10.1038/ncomms8545. 7545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gren ST, Rasmussen TB, Janciauskiene S, Hakansson K, Gerwien JG, Grip O. A Single-Cell Gene-Expression Profile Reveals Inter-Cellular Heterogeneity within Human Monocyte Subsets. PLoS One. 2015;10:e0144351. doi: 10.1371/journal.pone.0144351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho H, Kehrl JH. Localization of Gi alpha proteins in the centrosomes and at the midbody: implication for their role in cell division. J Cell Biol. 2007;178:245–255. doi: 10.1083/jcb.200604114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knust E. G protein signaling and asymmetric cell division. Cell. 2001;107:125–128. doi: 10.1016/s0092-8674(01)00534-7. [DOI] [PubMed] [Google Scholar]
- 36.Huang Z, Ma L, Wang Y, Pan Z, Ren J, Liu Z, et al. MiCroKiTS 4.0: a database of midbody, centrosome, kinetochore, telomere and spindle. Nucleic Acids Res. 2015;43:D328–334. doi: 10.1093/nar/gku1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poleshko A, Mansfield KM, Burlingame CC, Andrake MD, Shah NR, Katz RA. The human protein PRR14 tethers heterochromatin to the nuclear lamina during interphase and mitotic exit. Cell Rep. 2013;5:292–301. doi: 10.1016/j.celrep.2013.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang M, Lewinska M, Fan X, Zhu J, Yuan ZM. PRR14 is a novel activator of the PI3K pathway promoting lung carcinogenesis. Oncogene. 2016;35:5527–5538. doi: 10.1038/onc.2016.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boultwood J, Perry J, Pellagatti A, Fernandez-Mercado M, Fernandez-Santamaria C, Calasanz MJ, et al. Frequent mutation of the polycomb-associated gene ASXL1 in the myelodysplastic syndromes and in acute myeloid leukemia. Leukemia. 2010;24:1062–1065. doi: 10.1038/leu.2010.20. [DOI] [PubMed] [Google Scholar]
- 40.Abdel-Wahab O, Gao J, Adli M, Dey A, Trimarchi T, Chung YR, et al. Deletion of Asxl1 results in myelodysplasia and severe developmental defects in vivo. J Exp Med. 2013;210:2641–2659. doi: 10.1084/jem.20131141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.LaFave LM, Beguelin W, Koche R, Teater M, Spitzer B, Chramiec A, et al. Loss of BAP1 function leads to EZH2-dependent transformation. Nat Med. 2015;21:1344–1349. doi: 10.1038/nm.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cho YS, Kim EJ, Park UH, Sin HS, Um SJ. Additional sex comb-like 1 (ASXL1), in cooperation with SRC-1, acts as a ligand-dependent coactivator for retinoic acid receptor. J Biol Chem. 2006;281:17588–17598. doi: 10.1074/jbc.M512616200. [DOI] [PubMed] [Google Scholar]
- 43.Park UH, Seong MR, Kim EJ, Hur W, Kim SW, Yoon SK, et al. Reciprocal regulation of LXRalpha activity by ASXL1 and ASXL2 in lipogenesis. Biochem Biophys Res Commun. 2014;443:489–494. doi: 10.1016/j.bbrc.2013.11.124. [DOI] [PubMed] [Google Scholar]
- 44.Rosen ED, Spiegelman BM. PPARgamma : a nuclear regulator of metabolism, differentiation, and cell growth. J Biol Chem. 2001;276:37731–37734. doi: 10.1074/jbc.R100034200. [DOI] [PubMed] [Google Scholar]
- 45.de The H, Pandolfi PP, Chen Z. Acute Promyelocytic Leukemia: A Paradigm for Oncoprotein-Targeted Cure. Cancer Cell. 2017;32:552–560. doi: 10.1016/j.ccell.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 46.Boultwood J, Perry J, Zaman R, Fernandez-Santamaria C, Littlewood T, Kusec R, et al. High-density single nucleotide polymorphism array analysis and ASXL1 gene mutation screening in chronic myeloid leukemia during disease progression. Leukemia. 2010;24:1139–1145. doi: 10.1038/leu.2010.65. [DOI] [PubMed] [Google Scholar]
- 47.Olson EN, Nordheim A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat Rev Mol Cell Biol. 2010;11:353–365. doi: 10.1038/nrm2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Record J, Malinova D, Zenner HL, Plagnol V, Nowak K, Syed F, et al. Immunodeficiency and severe susceptibility to bacterial infection associated with a loss-of-function homozygous mutation of MKL1. Blood. 2015;126:1527–1535. doi: 10.1182/blood-2014-12-611012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steigemann P, Gerlich DW. Cytokinetic abscission: cellular dynamics at the midbody. Trends Cell Biol. 2009;19:606–616. doi: 10.1016/j.tcb.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 50.Zheng Y, Guo J, Li X, Xie Y, Hou M, Fu X, et al. An integrated overview of spatiotemporal organization and regulation in mitosis in terms of the proteins in the functional supercomplexes. Front Microbiol. 2014;5:573. doi: 10.3389/fmicb.2014.00573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dionne LK, Wang XJ, Prekeris R. Midbody: from cellular junk to regulator of cell polarity and cell fate. Curr Opin Cell Biol. 2015;35:51–58. doi: 10.1016/j.ceb.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.