Abstract
Sexual selection can accelerate speciation by driving the evolution of reproductive isolation, but forces driving speciation could also reciprocally feedback on sexual selection. This might be particularly important in the context of ‘reinforcement’, where selection acts directly to increase prezygotic barriers to reduce the cost of heterospecific matings. Using assays of sperm competition within and between two sister species, we show a signature of reinforcement where these species interact: populations of Drosophila pseudoobscura that co-occur with sister species D. persimilis have an elevated ability to outcompete heterospecific sperm, consistent with selection for increased postcopulatory isolation. We also find these D. pseudoobscura populations have decreased sperm competitive ability against conspecifics, reducing the opportunity for sexual selection within these populations. Our findings demonstrate that direct selection to increase reproductive isolation against other species can compromise the efficacy of sexual selection within species, a collateral effect of reproductive traits responding to heterospecific interactions.
Keywords: speciation, sexual selection, sperm competition
1. Introduction
The presence of heterospecifics can radically alter the selective environment of closely related species, including altering sexual interactions and changing patterns of selection on reproductive traits. In cases where these species have the potential to interbreed, selection can favour divergence in sexual traits to avoid costs of heterospecific mating, a type of reproductive character displacement commonly called reinforcement [1–3]. The frequency at which reinforcement contributes to speciation is still under debate [3,4], although several recent examples provide strong evidence for reinforcement acting on mating traits [5–8]. Regardless, trait evolution in response to reinforcement can have collateral effects on intraspecific sexual dynamics [5]. This can in turn alter the magnitude and efficacy of sexual selection specifically within populations exposed to heterospecifics. These potential reciprocal interactions between sexual selection and reproductive isolation remain relatively untested [5], but can have important consequences for how we interpret evolution of sexual traits. For example, in rapid radiations where sexual selection is thought to be the primary driver, patterns of reproductive trait evolution may be misinterpreted if they do not take into account species interactions.
For reinforcement and sexual selection to reciprocally affect the evolution of sexual traits, these traits must be involved in both processes and share a genetic basis. Currently, the best example of a shared genetic basis for sexual selection and reproductive isolation comes from Drosophila sperm competition genes, some of which have been shown to mediate both sexual selection through intraspecies sperm competition (ISC) and reproductive isolation via conspecific sperm precedence (CSP) [9,10]. CSP occurs when a female mates with both heterospecific and conspecific males, yet most of the progeny are sired by the conspecific male. CSP has proven to be a strong reproductive isolation barrier among species in Drosophila [11,12] and in many other plant and animal species ([4], and references therein). Moreover, although reinforcement studies have overwhelmingly focused on premating traits, postcopulatory prezygotic traits including CSP can also be the target of reinforcement [13,14]. Nonetheless, because CSP involves inconspicuous phenotypes that are not readily observed in the field [4,15], and because no single study has simultaneously compared levels of CSP in allopatric and sympatric populations, empirical evidence for increased CSP specifically in sympatry remains equivocal (references within [4,16–18]).
While reinforcing selection (acting on CSP) and sexual selection (acting on ISC) could interact to influence evolutionary change in postcopulatory traits, the outcomes of this interaction clearly will depend upon whether these forces act in concert or in opposition. When they act in concert, trait evolution can proceed faster than otherwise expected, but the direction of evolution remains unchanged. By contrast, when sexual selection and reproductive isolation act at cross-purposes, the potential feedback between these forces can generate complex evolutionary outcomes. For example, sperm competition is expected to maintain high variance in the affected traits because it is shaped by sexual conflict between males and females [19–22]; indeed, sperm competition genes are often highly variable both in terms of molecular and phenotypic variation [20,23]. By contrast, traits that act as barriers to reproduction are generally expected to have reduced genetic variance, and an overall shift in trait means between lineages, especially in cases where isolation is generated by strong disruptive selection between populations [24,25]. Accordingly, the net effect of selection imposed by intrapopulation sexual interactions and by reinforcement could together produce phenotypic and genetic variation in sperm competition traits that is different from the optimal variation when sexual selection acts alone.
One way these potentially antagonistic optima could play out is when reinforcement-mediated changes alter the opportunity for sexual selection among conspecifics [5]. Sexual selection is the strongest when the variance in among-male reproductive success is large. Because sperm competition amplifies among-male variance in reproductive success, elevated sperm competition leads to greater opportunity for sexual selection compared to scenarios where males have equal probability of siring offspring [26,27]. Predictions of the possible effects of reinforcement on sexual selection depend on the specific genetic correlation between CSP and ISC. First, if CSP and ISC are positively genetically correlated, strong directional selection from reinforcement could lead to enhanced sperm competition when specific male genotypes excel at both CSP and ISC. An initial increase in the frequency of a strong CSP/ISC allele increases variance in reproductive success because when one male ‘wins’ at sperm competition, its competitor ‘loses’. This increased variance in reproductive success could persist depending on the relative strength of frequency-dependent selection and reinforcement (similar to [28]). Any increase in the variance of reproductive success increases the opportunity for sexual selection. Alternatively, a trade-off or negative genetic correlation between CSP and ISC could result in reduced intraspecific sperm competition as a collateral effect of strong directional selection for increased CSP. Lower mean ISC would equalize fitness among males, thus reducing the opportunity for sexual selection.
In this study, we evaluated the interaction between selection for increased reproductive isolation (i.e. reinforcement) and sexual selection on postcopulatory sperm competition genes by estimating CSP and ISC phenotypes in parallel across a common set of genotypes. Both CSP and ISC are estimated by allowing females to mate sequentially with two different male genotypes and scoring the paternity of the resulting progeny. Comparing the relative competitive success of replicate male lines against a common set of heterospecific and conspecific male tester genotypes, in parallel, allowed us to estimate which genotype effects (male genotype, female genotype, or the interaction) might shape both CSP and ISC. Previous studies indicate that both male and female genetic effects, and their interaction, significantly influence ISC [21,29]; however, equivalent phenotypic and genetic variance for CSP has not been empirically investigated. Nonetheless, because females experience the greater cost of heterospecific matings [30,31] and could control CSP via cryptic female choice [32], we might expect strong female genotype effects on CSP.
To evaluate these expectations, here, we examine evidence for reinforcement of CSP among populations of Drosophila pseudoobscura that are allopatric or sympatric with their closely related sister species D. persimilis, and assess the potential consequences of these heterospecific interactions for ISC and sexual selection within D. pseudoobscura populations. This species pair is estimated to have diverged approximately 589 000 years ago [33] and provided one of the first clear empirical demonstrations of reinforcement on premating isolation [34], a finding that suggests heterospecific interactions and matings are frequent and sustained over evolutionary time in this system, and can act as a substantial selective agent on reproductive traits. Here, we determine whether there is evidence that heterospecific interactions have selected for increased CSP, by evaluating whether CSP is stronger specifically in sympatry—consistent with reinforcement. Our design also allows us to estimate premating reproductive isolation in the same experiment, and compare its strength in sympatry and allopatry. Second, we evaluate whether selection for strong CSP in sympatry has affected ISC, and thereby postcopulatory sexual selection, as might occur when CSP and ISC have shared genetic architecture. Throughout, we test for differences in trait variation across a set of distinct genotypes which allows us to specifically evaluate which sex is playing a more critical role in determining variation in heterospecific and conspecific postcopulatory interactions.
2. Material and methods
(a). Wild-type fly stocks
All stocks were reared on standard media prepared by the Bloomington Drosophila Stock Center, and were kept at room temperature (approx. 22°C). At this temperature, D. pseudoobscura and D. persimilis have a 21-day generation time. We used a set of isofemale lines collected from four natural populations in the summers of 2013 and 2014. Sperm competition and mating experiments (below) were completed once stable lines were established, starting in August 2015 and ending January 2016. Allopatric D. pseudoobscura were collected at Zion National Park, Utah (latitude = 37.217, longitude = −112.976 provided by N. Phadnis) and Lamoille Canyon, Nevada (latitude = 40.649, longitude = −115.405). Sympatric D. pseudoobscura and D. persimilis were collected at two sites: Mt St Helena, California (Latitude = 38.652, longitude = −122.608, D. pseudoobscura collected by A. Hish/M. Noor and D. Castillo) and, near Meadow Vista and Forest Hill, California (latitude = 39.012, longitude = −121.004; called here ‘Sierra’). For both sympatric populations, both species were present in field collections and can be considered truly co-occurring. Our replication of sympatric populations is consistent with previous studies in this system [34] and other Drosophila pairs [5], and generally exceeds these and other reinforcement studies in terms of the number of genotypes assessed per population [5–8]. Nonetheless, it is important to consider the potential effects of sampling error and whether our sample of genotypes represents the full range of genetic variation within and between natural populations of these species. Several lines of evidence—including consistent sympatric–allopatric differences across multiple reproductive phenotypes—indicate that the influence of sampling error on observed population phenotypic differences is small compared to the biological differences predicted between allopatry and sympatry. To address these, we provide a detailed discussion of how population sampling, genotype replication per population, and post-collection laboratory conditions could affect our observations and inferences, in the electronic supplementary material.
(b). Conspecific sperm competition assay
Sperm competition assays involve mating an individual female sequentially with two distinct male genotypes. In all experimental crosses between species, females were paired first with a D. persimilis male and second with a D. pseudoobscura male in order to evaluate the ‘offensive’ sperm competitive ability of conspecific males to displace heterospecific sperm (equivalent to ‘P2’, or second male siring ability; [35,36]). We focused on ‘offensive’ sperm competition because D. pseudoobscura females do not remate with D. persimilis males if they have first mated with a conspecific male. In this experiment, we partitioned the variance in CSP due to male genotype, female genotype, and the male × female genotype interaction using a ‘diallel-like’ crossing design where all parental genotypes were crossed in all combinations, as in other similar studies [22] (electronic supplementary material, figure S1). We completed separate CSP experiments for each of our four D. pseudoobscura collection locations (Sympatric = Sierra and Mt St Helena, Allopatric = Zion and Lamoille). For each population, we used a 4 × 4×4 design: four D. pseudoobscura female genotypes from that population, four D. persimilis genotypes as first males (tester males), and four D. pseudoobscura male genotypes as second males from the same population as females. Each 4 × 4×4 combination was replicated once (n = 64 unique cross-combinations for each population). If CSP is important for reproductive isolation in sympatry, it should be consistently strong across multiple heterospecific genotypes. Accordingly, we used multiple wild-collected D. persimilis tester male lines for our experiments, two lines at the Sierra location and two lines at Mt St Helena. Specific details of the sperm competition design are reported in the electronic supplementary material, Methods.
(c). Intrapopulation sperm competition assay
The design for ISC assay mirrored the experimental design for CSP except that, rather than a D. persimilis tester male, the first male was a D. pseudoobscura tester male derived from the same population as the D. pseudoobscura female and second male genotypes in the trial. For each population, we used a 4 × 2×4 design: four D. pseudoobscura female genotypes, two D. pseudoobscura green fluorescent protein (GFP) genotypes as first males, and four D. pseudoobscura male genotypes as second males. The same female×second male genotypes were used in ISC and CSP experiments. Each combination was replicated twice (n = 64 for each population, with 32 unique cross-combinations). This resulted in a total sample size per population that matched the CSP experiment (64 replicates per population, 256 replicates across all populations).
The details of the mating scheme are identical to the CSP experiment. We did not observe matings directly, but an average refractory period of 4 days for D. pseudoobscura [37] means that on average only a single mating occurred in the 24 h co-housing timeframe. Each individual female was randomly assigned one of the two D. pseudoobscura first male (tester) genotypes, against which we determined the strength of P2 (second male siring ability) of our four focal second male genotypes.
(d). Generating visibly marked tester males for quantifying conspecific sperm precedence and intraspecies sperm competition
Paternity was scored with the aid of visible markers in both CSP and ISC experiments. This required us to generate marked male tester lines with wild-caught D. persimilis (for CSP tester males) and D. pseudoobscura (for ISC tester males) lines from each study population. For CSP, we introgressed an X-linked marker (‘short’ or sh) from a D. persimilis line (UCSD stock centre 14011-0111.57) into four of our collected D. persimilis genotypes (electronic supplementary material, Methods and figure S2). For ISC experiments, the marked tester males were created by introgressing a GFP marker into two new wild-type D. pseudoobscura strains per location (electronic supplementary material, Methods). The original GFP strain was obtained from the UCSD stock centre (14011-0121.166), the creation of which is described in Holtzman et al. [38].
(e). Scoring conspecific sperm precedence
CSP was scored by differentiating the progeny of first (heterospecific) from second (conspecific) mated males, for each replicate female. Hybrid male progeny from D. pseudoobscura×D. persimilis crosses are sterile (there are no motile sperm, observable by dissecting the testes), so we used this sterility phenotype to determine the sire of each F1 male, in 10 dissected male progeny produced after the second mating (electronic supplementary material, Methods). F1 males with no motile sperm were scored as hybrid. Because female hybrids are fertile in these crosses, the sh allele was used to differentiate the female progeny. Any F1 female that produced sh progeny was considered hybrid. We required each F1 female to produce at least 10 progeny to be used in scoring CSP (electronic supplementary material, Methods). Further details are reported in the electronic supplementary material, Methods.
(f). Scoring intrapopulation sperm competition
We scored all progeny that eclosed in the 5 days after the second mating for the presence/absence of the GFP phenotype. Our measure of sperm competition (P2) for ISC was then the number of wild-type (non-GFP) progeny out of the total number of progeny scored for a particular cross. Any replicate in which all progeny were GFP was excluded in our analyses because we could not ensure that a second mating had taken place. (As with CSP, the estimated proportion of females that did not remate was not significantly different between populations [39].) Individuals were scored as they eclosed, using a Leica M205FA Stereo Microscope that has an Hg fluorescent lamp attached and GFP filter, as described in Castillo & Moyle [9].
(g). Statistical analyses
All analyses were completed in R v. 3.01.
(h). Differences in the probability of first mating with heterospecifics
We evaluated evidence for reinforcement acting on first mating (premating isolation) in two ways. First, we used a χ2 test of independence to test the null hypothesis that the mating rate with heterospecifics was the same for allopatry versus sympatry, after combining both allopatric and both sympatric populations for this single comparison (pairwise tests among individual populations gave the same result; electronic supplementary material, table S1). Second, coding mating events as a binary variable (0, no mating; 1, successful mating), we used a logistic regression model with all four populations represented by a categorical variable, using the glmer function. We then tested for differences in heterospecific mating between populations by conducting a Wald's test (using the wald.test function from the aod package; [40]).
To evaluate whether there was significant variation within each population (i.e. among isofemale line genotypes) in the probability of mating with a heterospecific, we used logistic regression, and tested significance of isofemale line and D. persimilis tester line effects using a Wald's test.
(i). Differences in mean and variance of conspecific sperm precedence and intraspecies sperm competition between populations
To evaluate evidence for reinforcement acting on CSP, we assessed whether the allopatric and sympatric populations had a mean difference in CSP or whether they differed in variance. To evaluate differences in the mean CSP, we used a Welch's t-test that accounts for unequal variances between samples, and confirmed our results with a Wilcoxon ranked-sum test, because our data are not normally distributed. To evaluate differences in variance, we compared the total phenotypic variation between geographical classes of population with a Levene-type test implemented in the lawstat package in R ([41]; details provided in electronic supplementary material, Methods). Using the same statistical approach, we tested for differences in the mean and variance of ISC between sympatric and allopatric populations. Results were not different whether populations were analysed pairwise or pooled (electronic supplementary material, Methods).
(j). Genetic variation and genotype effects on conspecific sperm precedence and intraspecies sperm competition
Within each population, we assessed whether female, male, or female×male genotype predicted variation in the strength of CSP and ISC. Because binomial data typically violate the assumptions of ANOVA [42,43], we used binomial regression to fit a model of the form
where α and β are each categorical variables with four levels that represent male and female genotype, respectively, and the variable (αβ) represents the male × female genotype interactions. Since we were interested in partitioning the variance and estimating the variance components (, , ), we assumed that each variable was a random variable. To test the significance of each variance component, we used a binomial regression in a mixed modelling framework with parametric bootstrap ([44]; electronic supplementary material, Methods).
(k). Genetic association between conspecific sperm precedence and intraspecies sperm competition
We conducted an exploratory analysis of the potential molecular genetic association between CSP and ISC by estimating the correlations between each phenotype and single-nucleotide polymorphisms (SNPs) within a set of 14 candidate genes and 13 control genes. The candidate loci are orthologues to known sperm competition genes in D. melanogaster (electronic supplementary material, table S2; [45]). The control genes have been used previously as controls for studies of molecular evolution (electronic supplementary material, table S3; [46–48]). After removing SNPs that were strongly associated with population structure, we estimated the association between individual SNPs and either the CSP or ISC phenotype while accounting for population of origin and mating partner (see Results; electronic supplementary material, Methods).
(l). Quantifying the opportunity for sexual selection and variance in male reproductive success
To evaluate whether the intensity/opportunity for sexual selection differs among populations, we determined whether their variance in male reproductive success differed [49]. We estimated male fitness as the proportion of progeny sired, taking into consideration that we had two distinct classes of males—tester first (defensive) males and second (offensive) males—that may differ in their frequency and variance in fitness in the experiment. Following Shuster et al. [50], we define total variance in male reproductive success as the sum of within (first two terms) and between (last term in the equation) male class variance
We were interested in reproductive variance at the level of male genotype, so we averaged biological replicates to generate mean fitness values for each individual genotype. We used empirical bootstrap confidence intervals to estimate error that may have been a product of averaging over replicates [51,52] (as described in the electronic supplementary material, Methods); significantly different variances have non-overlapping bootstrap confidence intervals.
3. Results
(a). No difference between allopatric and sympatric populations in premating isolation
We found that the average probability of heterospecific matings ranged from 46 to 52% between populations, and did not differ between allopatric and sympatric populations (χ2 test of independence: χ2 = 1.185, d.f. = 1, p = 0.2763; Wald's test: χ2 = 1.9, d.f. = 4, p = 0.75; electronic supplementary material, table S4). Pairwise tests between each allopatric and sympatric population also failed to reject the null hypothesis. Although we did not detect a signal of reinforcement, there was ample genetic variance in heterospecific mating rate between female genotypes available for selection within each population (figure 1; electronic supplementary material, table S5). Only in one of the populations (Lamoille, which is allopatric) did the identity of the D. persimilis tester line affect variation in premating isolation (electronic supplementary material, table S5).
Figure 1.
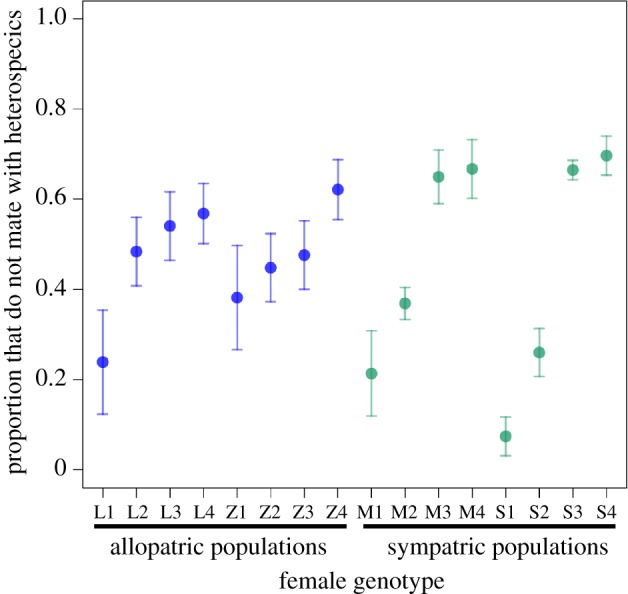
Prezygotic reproductive isolation via female mating preference does not show a pattern consistent with reinforcement. Reproductive isolation is measured by the proportion of females that did not mate with heterospecifics in individual no-choice trials. Significant variation among D. pseudoobscura female genotypes in female preference occurs in each population (electronic supplementary material, table S1). Each point is the mean reproductive isolation for each isofemale line tested against each of four D. persimilis tester males. Error bars represent ± one standard error.
(b). Reinforcement acts on conspecific sperm precedence
Unlike premating isolation, we observed a pattern consistent with reinforcement for CSP. Specifically, in sympatry, we find both greater average CSP (t = −6.59, d.f. = 210.92, p < 0.001; Wilcox W = 4427.5, p < 0.001) and less phenotypic variation in this trait (Levene-type test χ2 = 22.82, p < 0.0001) when data were pooled by geographical region (allopatry versus sympatry) (electronic supplementary material, table S4; figure 2a). These differences in the mean and variance of CSP were also observed in pairwise tests between individual allopatric and sympatric populations (electronic supplementary material, tables S6 and S7).
Figure 2.
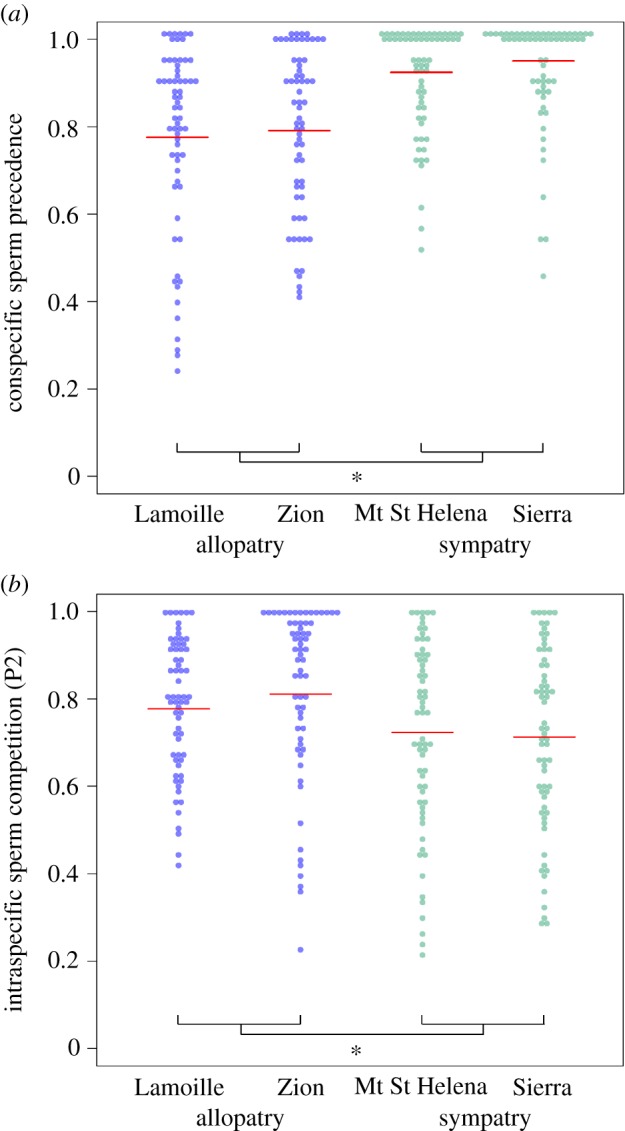
The phenotypic distributions of CSP (a) are consistent with a pattern of reinforcement. The distribution of ISC (b) shows a shift in ISC in the opposite direction compared to CSP for sympatric populations. CSP is defined as the proportion of progeny sired by the D. pseudoobscura male competing against a previously mated D. persimilis male. ISC is defined as the progeny sired by a D. pseudoobscura male competing against a previously mated D. pseudoobscura male. The red line in each distribution represents the mean value. Significant differences determined by Welch's t-test and Wilcox tests between the allopatric and sympatric populations are denoted by *.
(c). Reinforcement has collateral effects on intrapopulation sperm competition
ISC also differed between allopatric and sympatric populations, in both mean and variance (electronic supplementary material, table S4; figure 2b). First, the mean offensive ability for ISC was significantly lower in sympatric populations (t = 3.73, d.f. = 246.55, p = 0.0002; Wilcox's W = 10280, p = 0.0004). This contrasts with the observed increase in offensive CSP in sympatric populations. Second, there was more variation in ISC in the sympatric populations compared to the allopatric populations (Leven-type test χ2 = 5.74, p = 0.0172). Given the differences in ISC and CSP across populations, we used the mean CSP and ISC phenotype for each male×female genotype combination within a population (i.e. each cell within the diallel crossing design) to examine the relationship between the two phenotypes across the four populations. We observed a significant negative relationship between CSP and ISC (Pearson's r = −0.31, p = 0.01; electronic supplementary material, figure S3). Because each male or female genotype is represented in multiple combinations, we further controlled for non-independence using a linear mixed effect model and confirmed that the negative slope of the relationship was significantly different from zero (profiled CI = −0.451, −0.028).
(d). Female genotype effects contribute to conspecific sperm precedence and male×female genotype effects explain both conspecific sperm precedence and intraspecies sperm competition
Of male, female, and male×female genotype effects that could contribute to explaining the variance in CSP, we found that three of the four populations had a significant female genotype effect (electronic supplementary material, table S8 and figure S4), and all populations had a significant male×female genotype interaction effect. The D. persimilis tester male line was also significant in three of four populations. There was no consistent pattern among populations in which effect explained the largest proportion of variance; in some populations, this was largest for the female genotype effect, while in others, the male×female genotype interaction effect was larger (electronic supplementary material, table S8). By contrast, for ISC in all four populations, we only observed significant male×female genotype interaction effects and a significant effect of the first-tester male genotype (electronic supplementary material, table S9 and figure S5). In every case, the male×female genotype effect explained more variance than the identity of the tester male genotype.
(e). Conspecific sperm precedence and intraspecies sperm competition are genetically associated at a specific locus
After accounting for population structure, no SNPs in the control genes had a significant correlation for CSP or ISC. For our candidate genes, only a single SNP—in the gene Esp—retained a significant correlation. Interestingly, this SNP had pleiotropic effects, with a positive effect on CSP and a negative effect on ISC.
(f). The opportunity for sexual selection is decreased in sympatry
We found that the sympatric populations had significantly lower variance for reproductive success compared to the allopatric populations (figure 3; electronic supplementary material, table S10). The variance in reproductive success across all male genotypes in the allopatric Lamoille population was significantly greater than both sympatric populations (Mt St Helena F = 1.96, bootstrap p = 0.003; Sierra F = 2.08, bootstrap p = 0.008), as was the variance in reproductive success in the allopatric Zion population compared to the sympatric populations (Mt St Helena F = 2.65, bootstrap p = 0.003; Sierra F = 2.83, bootstrap p = 0.004). The reduced variance in reproductive success in sympatry is completely a product of lower offensive sperm competition values in sympatry (figure 2b), that result in equalized differences in the siring success between offensive and defensive males.
Figure 3.
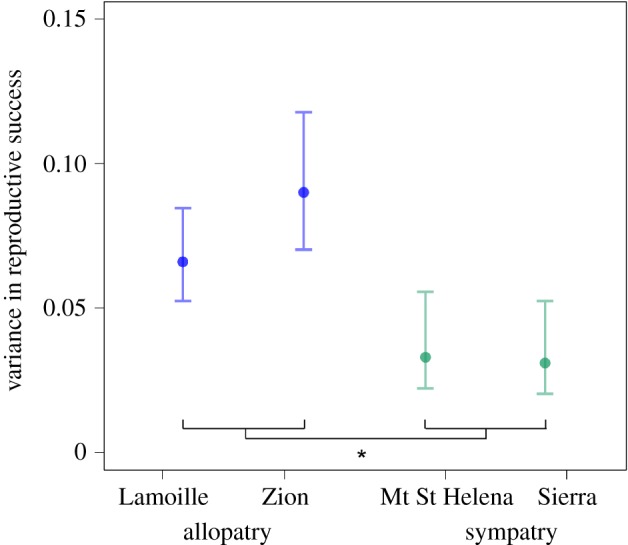
The variance in reproductive success across populations calculated in the framework that combines offensive and defensive males. Each point represents the estimate for the variance in fitness for each population. The error bars are confidence intervals generated from the empirical bootstrap distribution. Significance, denoted by *, was assessed in pairwise comparisons between allopatric and sympatric populations using empirical bootstrap hypothesis testing.
4. Discussion
Interactions with heterospecifics have the potential to drive divergent sexual selection and the evolution of reproductive isolation, via reproductive character displacement and reinforcement [5,53]. Using D. pseudoobscura and D. persimilis, we assessed evidence for reinforcement of species barriers in sympatry via elevated female mate preference or CSP. Premating isolation is historically considered a strong barrier to isolation between these species, and one that reinforcing selection has acted on [34], but we saw no evidence for reproductive character displacement for this trait. By contrast, we found that the average CSP was higher and the overall level of phenotypic variation was lower, specifically in sympatric populations, a pattern consistent with recent or recurrent directional selection acting on CSP in these populations. We further asked whether reinforcement could have had collateral effects on intraspecific sperm competition, given that this trait is mechanistically associated with CSP [32]. We found that our two sympatric populations also had lower offensive sperm competitive ability against conspecifics, in comparison to our two allopatric populations, consistent with weakened sexual selection in sympatry.
Our results are consistent with the evolution of significantly strengthened reproductive isolation in the form of CSP in response to reinforcing selection. While CSP is known to be a barrier to gene flow in Drosophila [11,12] and other taxa [2], its importance in nature has been difficult to ascertain [13,15]. Moreover, even though models of speciation by sexual selection predict that strong divergent selection will erode phenotypic variation in selected traits [54,55], trait variance is not typically quantified [7,14]. Our observations of both increased mean CSP and reduced variation specifically in sympatry provide strong support for the inference that CSP has responded to selection imposed by heterospecific interactions, and underscore the important role that CSP could play in maintaining species boundaries.
While other factors have been proposed to account for reproductive character displacement, including differential fusion [56] or ecological differences that have collateral effects on mating traits [53,57], additional data indicate that the pattern we observe for CSP is most consistent with reinforcement. For example, differential fusion predicts that reproductive isolation evolves between species in allopatry and merely acts to prevent species collapse upon subsequent secondary contact, creating the incidental appearance of stronger isolation in sympatry [58,59]. However, if differential fusion operates specifically in sympatric populations, we would expect sympatric CSP values to be a subset of allopatric CSP values [14], and this is not the case; sympatric values of CSP are systematically higher than in allopatry (figure 2).
Similarly, several lines of evidence argue that systematic ecological differences between allopatry and sympatry are unlikely to explain our observed postcopulatory differences. First, there is little evidence that differential natural selection acts consistently on sympatric versus allopatric populations; ecological differences across the range of D. pseudoobscura are largely continuous, rather than uniquely differentiating regions of allopatry and sympatry with D. persimilis. Moreover, although both our sympatric populations are located in California, they are collected from environmentally distinct mountain ranges [60], whose habitat and ecological variation maintains stable differences in inversion frequencies between populations of D. pseudoobscura in these locations [61]. Second, it is unclear how external ecological factors would produce our patterns of differential sperm competition. Indirect effects of diet and nutrition can affect sperm competition outcomes [62,63], but should not persist in a laboratory environment. Moreover, if ecological mechanisms existed, there is no a priori reason to expect they would act in the specific direction we observed. Given this, while the ecological alternative to reinforcement might be plausible for some premating phenotypes, it is unlikely to explain the postcopulatory phenotypes that we examine here.
Our second major inference is that the response to reinforcing selection observed in CSP has had a collateral effect on the magnitude of offensive ISC because of a negative genetic correlation between CSP and ISC. This has, in turn, reduced the opportunity for sexual selection in these sympatric populations. Sperm competition strongly contributes to sexual selection in D. pseudoobscura where multiple mating is frequent in wild-caught females [64], and male mating success, including sperm competition, is a major component of selection in natural populations [65]. We observed that, on average, ISC was higher for our allopatric populations compared to sympatric populations—where we found little difference in postcopulatory fitness among male genotypes (ISC close to 0.5). An alternative explanation is that directional selection for weaker ISC indirectly increased CSP but only in sympatry; however, as well as being generally implausible, in that case, we should have observed reduced phenotypic variation in ISC, but did not.
Instead, we infer that selection for stronger CSP in sympatry has reduced mean ISC in sympatric populations via a negative genetic correlation between these two sperm competitive phenotypes. Consistent with this, one female-specific gene (Esp) showed a significant association with postcopulatory phenotypes, via an SNP that had opposite effects for CSP and ISC. This analysis applied a very conservative correction for population structure and likely underestimates the number of SNPs contributing to CSP and ISC. Regardless, any shared genetic basis between CSP and ISC allows selection on one trait to have collateral effects on the other, when the strength of selection acting on each trait has different magnitudes and directions.
For reinforcing selection to interfere with sexual selection as we propose, selection favouring increased CSP must outweigh selection acting to maximize ISC. This requires sufficient heterospecific contact for interspecific matings to impose a significant selective burden on D. pseudoobscura. The rate of heterospecific contact can vary across the sympatric range as a product of the relative frequency of each species [66,67]. In our no-choice experiments, D. pseudoobscura females accepted D. persimilis males at relatively high frequencies, suggesting the potential for heterospecific matings is substantial. Interestingly, these data also indicated that female acceptance of D. persimilis first males did not differ between sympatry and allopatry, although elevated premating isolation has previously been observed in D. pseudoobscura that are sympatric with D. persimilis ([34]; although see also [68–70]). While there are no prior data on CSP from these species, our observations here—including that the average allopatric premating isolation in our experiment is similar to previous reports [34]—suggest that the relative contribution of different barriers in sympatry might have changed over time.
Comparative analysis across Drosophila species suggests that, on average, CSP evolves more slowly than premating isolation [71]. However, specific conditions could increase the rate of CSP evolution. For example, a cost of remaining unmated (‘the wallflower effect’; [72]) can lead to reductions in female pre-copulatory choice; under these circumstances, CSP could evolve faster than premating isolation. In addition, ‘swamping effects’ of gene flow from allopatric populations [73] might explain a temporal shift to reduce premating isolation in sympatry, but it is difficult to disentangle this from costs associated with preference. Evidence for both gene flow between sympatric and allopatric populations of D. pseudoobscura [74,75] as well as a cost of female preference in experimental populations of D. pseudoobscura [76] suggest that both processes could contribute to the patterns we observe. Specifically, sympatric populations had a comparatively large variance in female mating rates, as evidenced by a bimodal distribution, compared to the more narrow female preferences observed in allopatry (figure 1).
Additional evidence for interspecific hybridization also suggests that heterospecific mating rates are sufficiently common between these two species to impose selection on CSP. F1 progeny, while rare, have been identified from wild collections [77], and genomic data suggest a history of post-speciation heterospecific mating and introgression between D. pseudoobscura and D. persimilis [74,78]. Notably, these estimates of hybridization only capture events that successfully produced F1 progeny and/or later generation hybrids, so they systematically underestimate the rate of heterospecific matings; for example, given the presence of strong CSP, many heterospecific matings may never produce hybrid progeny.
Given this sustained heterospecific contact, one clear way fitness effects of CSP could outweigh ISC is via a higher selective premium specifically for females. Weaker CSP imposes substantial fitness costs on females because it results in reproductive investment in low fitness hybrids, whereas weaker ISC likely has a comparatively marginal effect on female fitness outcomes. We were able to evaluate the expectation that females face more costs of hybridization [30,31] and that choice manifests as female control of sperm use patterns [79–81] by contrasting the genotype effects (male, female, and male×female genotype effects) between CSP and ISC. We detected male×female interactions for both CSP and ISC but, interestingly, only saw significant female genotype effects for CSP. This latter finding suggests that cryptic female choice might operate similarly to premating isolation mechanisms where females are often the more ‘choosy’ sex and female effects control the level of reproductive isolation more so than male effects [82].
Overall, our data suggest that strong reinforcing selection for reproductive isolation can have consequences for sexual selection and sexual interactions, in these important postmating sperm competition traits. The direction of this interaction inverts standard expectations about the connection between sexual selection and speciation. Sexual selection is often thought of as a driver of sexual characteristics whose evolutionary divergence then contributes to reproductive isolation. But a direct genetic connection between these processes implies reproductive isolation also has the reciprocal potential to shape sexual selection [83]. Based on our observations of higher mean but lower variance in CSP in sympatry, a negative correlation between CSP and ISC, and reduced variance in reproductive success via ISC among sympatric conspecific males, we infer that strong selection for reproductive isolation within populations exposed to heterospecific species has reduced the efficacy of sexual selection in these populations, a collateral effect of reinforcing selection that has not previously been demonstrated.
Supplementary Material
Acknowledgements
We thank E. Walburn and J. Roesener for assistance with crosses and scoring progeny, J. Powers and the IU Light Microscopy Imaging Center for assistance with the Leica microscope, M. Noor, A. Hish, and N. Phadnis for providing several strains, and Donn Castillo for help with collecting strains.
Data accessibility
All data and R code used in this analysis are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.n88kb1m [84].
Competing interests
We declare we have no competing interests.
Funding
Collections were supported by IU Biology Department travel awards to D.M.C. Research was supported by Indiana University Department of Biology funding to L.C.M. and an American Society of Naturalists student research award to D.M.C. D.M.C. was supported by a President's Diversity Initiative Dissertation Fellowship from the Indiana University Graduate School.
References
- 1.Dobzhansky T. 1951. Genetics and the origin of species. New York, NY: Columbia University Press. [Google Scholar]
- 2.Howard DJ. 1993. Reinforcement: origins, dynamics, and the fate of an evolutionary hypothesis. In Hybrid zones and evolutionary process (ed. Harrison RG.), pp. 46–69. Oxford, UK: Oxford University Press. [Google Scholar]
- 3.Servedio MR, Noor MAF. 2003. The role of reinforcement in speciation: theory and data. Annu. Rev. Ecol. Evol. 34, 339–364. ( 10.1146/annurev.ecolsys.34.011802.132412) [DOI] [Google Scholar]
- 4.Howard DJ. 1999. Conspecific sperm and pollen precedence and speciation. Annu. Rev. Ecol. Evol. 30, 109–132. ( 10.1146/annurev.ecolsys.30.1.109) [DOI] [Google Scholar]
- 5.Higgie M, Blows MW. 2008. The evolution of reproductive character displacement conflicts with how sexual selection operates within a species. Evolution 62, 1192–1203. ( 10.1111/j.1558-5646.2008.00357.x) [DOI] [PubMed] [Google Scholar]
- 6.Porretta D, Urbanelli S. 2012. Evolution of premating reproductive isolation among conspecific populations of the sea rock-pool beetle Ochthebius urbanelliae driven by reinforcing natural selection. Evolution 66, 1284–1295. ( 10.1111/j.1558-5646.2011.01535.x) [DOI] [PubMed] [Google Scholar]
- 7.Dyer KA, White BE, Sztepanacz JL, Bewick ER, Rundle HD. 2014. Reproductive character displacement of epicuticular compounds and their contribution to mate choice in Drosophila subquinaria and Drosophila recens. Evolution 68, 1163–1175. ( 10.1111/evo.12335) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kozak G, Rolan MG, Rankhorn C, Falater A, Berdan EL, Fuller RC. 2015. Behavioral isolation due to cascade reinforcement in Lucania killifish. Am. Nat. 185, 491–506. ( 10.1086/680023) [DOI] [PubMed] [Google Scholar]
- 9.Castillo DM, Moyle LC. 2014. Intraspecific sperm competition genes enforce post-mating species barriers in Drosophila. Proc. R. Soc. B 281, 20142050 ( 10.1098/rspb.2014.2050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Civetta A, Finn S. 2014. Do candidate genes mediating conspecific sperm precedence affect sperm competitive ability within species? A test case in Drosophila. G3 4, 1701–1707. ( 10.1534/g3.114.012476) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Price CSC. 1997. Conspecific sperm precedence in Drosophila. Nature 388, 663–666. ( 10.1038/41753) [DOI] [PubMed] [Google Scholar]
- 12.Chang AS. 2004. Conspecific sperm precedence in sister species of Drosophila with overlapping ranges. Evolution 58, 781–789. ( 10.1111/j.0014-3820.2004.tb00411.x) [DOI] [PubMed] [Google Scholar]
- 13.Servedio MR. 2001. Beyond reinforcement: the evolution of premating isolation by direct selection on preferences and postmating, prezygotic incompatibilities. Evolution 55, 1909–1920. ( 10.1111/j.0014-3820.2001.tb01309.x) [DOI] [PubMed] [Google Scholar]
- 14.Matute DR. 2010. Reinforcement of gametic isolation in Drosophila. PLoS Biol. 8, 3 ( 10.1371/journal.pbio.1000341) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marshall JL, Arnold ML, Howard DJ. 2002. Reinforcement: the road not taken. Trends Ecol. Evol. 17, 558–563. ( 10.1016/S0169-5347(02)02636-8) [DOI] [Google Scholar]
- 16.Williams JH Jr, Friedman WE, Arnold ML. 1999. Developmental selection within the angiosperm style: using gamete DNA to visualize interspecific pollen competition. Proc. Natl Acad. Sci. USA 96, 9201–9206. ( 10.1073/pnas.96.16.9201) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geyer JB, Palumbi SR. 2005. Conspecific sperm precedence in two species of tropical sea urchins. Evolution 59, 97–105. ( 10.1111/j.0014-3820.2005.tb00897.x) [DOI] [PubMed] [Google Scholar]
- 18.Peterson MA, et al. 2011. Cryptic gametic interactions confer both conspecific and heterospecific advantages in the Chrysochus (Coloptera: Chrysomleidae) hybrid zone. Genetica 139, 663–676. ( 10.1007/s10709-011-9567-z) [DOI] [PubMed] [Google Scholar]
- 19.Fiumera AC, Dumont BL, Clark AG. 2006. Natural variation in male-induced ‘cost-of-mating’ and allele-specific association with male reproductive genes in Drosophila melanogaster. Phil. Trans. R. Soc. B 361, 355–361. ( 10.1098/rstb.2005.1791) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang R, Clark AG, Fiumera AC. 2012. Natural genetic variation in male reproductive genes contributes to non-transitivity of sperm competitive ability in Drosophila melanogaster. Mol. Ecol. 22, 1400–1415. ( 10.1111/mec.12113) [DOI] [PubMed] [Google Scholar]
- 21.Bjork A, Starmer WT, Higginson DM, Rhodes CJ, Pitnick S. 2007. Complex interactions with females and rival males limit the evolution of sperm offence and defence. Proc. R. Soc. B 274, 1779–1788. ( 10.1098/rspb.2007.0293) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chow CY, Wolfner MF, Clark AG. 2010. The genetic basis for male×female interactions underlying variation in reproductive phenotypes of Drosophila. Genetics 186, 1355–1365. ( 10.1534/genetics.110.123174) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong A, Turchin MC, Wolfner MF, Aquadro CF. 2008. Evidence for positive selection on Drosophila melanogaster seminal fluid protease homologs. Mol. Biol. Evol. 25, 497–506. ( 10.1093/molbev/msm270) [DOI] [PubMed] [Google Scholar]
- 24.Panhuis TM, Butlin R, Zuk M, Tregenza T. 2001. Sexual selection and speciation. Trends Ecol. Evol. 16, 364–371. ( 10.1016/S0169-5347(01)02160-7) [DOI] [PubMed] [Google Scholar]
- 25.Kirkpatrick M, Ravigne V. 2002. Speciation by natural and sexual selection: models and experiments. Am. Nat. 159, S22–S35. ( 10.1086/338370) [DOI] [PubMed] [Google Scholar]
- 26.Levitan DR. 2008. Gamete traits influence the variance in reproductive success, the intensity of sexual selection, and the outcome of sexual conflict among congeneric sea urchins. Evolution 62, 1305–1316. ( 10.1111/j.1558-5646.2008.00378.x) [DOI] [PubMed] [Google Scholar]
- 27.Pischedda A, Rice WR. 2012. Partitioning sexual selection into its mating success and fertilization success components. Proc. Natl Acad. Sci. USA 109, 2049–2053. ( 10.1073/pnas.1110841109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otto SP, Servedio MR, Nuismer SL. 2008. Frequency-dependent selection and the evolution of assortative mating. Genetics 179, 2091–2112. ( 10.1534/genetics.107.084418) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clark AG, Begun DJ, Prout T. 1999. Female×male interactions in Drosophila sperm competition. Science 283, 217–220. ( 10.1126/science.283.5399.217) [DOI] [PubMed] [Google Scholar]
- 30.Trivers RL. 1972. Parental investment and sexual selection. In Sexual selection and the descent of mane (ed. Campbell B.), pp. 137–179. Chicago, IL: Aldine. [Google Scholar]
- 31.Bonduriansky R. 2011. Sexual selection and conflict as engines of ecological diversification. Am. Nat. 178, 729–745. ( 10.1086/662665) [DOI] [PubMed] [Google Scholar]
- 32.Manier MK, et al. 2013. Postcopulatory sexual selection generates speciation phenotypes in Drosophila. Curr. Biol. 23, 1853–1862. ( 10.1016/j.cub.2013.07.086) [DOI] [PubMed] [Google Scholar]
- 33.Hey J, Nielsen R. 2004. Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics 167, 747–760. ( 10.1534/genetics.103.024182) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noor MAF. 1995. Speciation driven by natural selection in Drosophila. Nature 375, 674–675. ( 10.1038/375674a0) [DOI] [PubMed] [Google Scholar]
- 35.Boorman E, Parker GA. 1976. Sperm (ejaculate) competition in Drosophila melanogaster, and reproductive value of females to males in relation to female age and mating status. Ecol. Entomol. 1, 145–155. ( 10.1111/j.1365-2311.1976.tb01217.x) [DOI] [Google Scholar]
- 36.Dixon SM, Coyne JA, Noor MAF. 2003. The evolution of conspecific sperm precedence in Drosophila. Mol. Ecol. 12, 1179–1184. ( 10.1046/j.1365-294X.2003.01742.x) [DOI] [PubMed] [Google Scholar]
- 37.Markow TA, O'Grady PM. 2005. Drosophila: a guide to species identification and use. Waltham, MA: Academic Press. [Google Scholar]
- 38.Holtzman S, Miller D, Eisman R, Kuwayama H, Niimi T, Kaufman T. 2010. Transgenic tools for members of the genus Drosophila with sequenced genomes. Fly 4, 349–362. ( 10.4161/fly.4.4.13304) [DOI] [PubMed] [Google Scholar]
- 39.Davis JS, Castillo DM, Moyle LC. 2017. Remating responses are shaped by male post-copulatory manipulation but not reinforcement in D. pseudoobscura. Ecol. Evol. 7, 507–515. ( 10.1002/ece3.2628) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lesnoff M, Lancelot R. 2012. aod: analysis of overdispersed data. R package version 1.3.
- 41.Hui W, Gel YR, Gastwirth JL. 2008. lawstat: an R package for law, public policy and biostatistics. J. Stat. Softw. 28, 1–26. ( 10.18637/jss.v028.i03)27774042 [DOI] [Google Scholar]
- 42.Fox J. 2008. Applied regression analysis and generalized linear models, 2nd edn Thousand Oaks, CA: Sage Publications, Inc. [Google Scholar]
- 43.Menard SW. 2010. Logistic regression: from introductory to advanced concepts and applications. Thousand Oaks, CA: Sage Publications, Inc. [Google Scholar]
- 44.Halekoh U, Højsgaar S. 2014. A Kenward-Roger approximation and parametric bootstrap methods for tests in linear mixed models—the R Package pbkrtest. J. Stat. Softw. 59, 1–30. ( 10.18637/jss.v059.i09)26917999 [DOI] [Google Scholar]
- 45.Findlay GD, Sitnik JL, Wang W, Aquadro CF, Clark NL, Wolfner MF. 2014. Evolutionary rate covariation identifies new members of a protein network required for Drosophila melanogaster female post-mating responses. PLoS Genet. 10, e1004108 ( 10.1371/journal.pgen.1004108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Begun DJ, Whitley P. 2000. Reduced X-linked nucleotide polymorphism in Drosophila simulans. Proc. Natl Acad. Sci. USA 97, 5960–5965. ( 10.1073/pnas.97.11.5960) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Begun DJ, Whitley P, Todd BL, Waldrip-Dail HM, Clark AG. 2000. Molecular population genetics of male accessory gland proteins in Drosophila. Genetics 156, 1879–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swanson WJ, Clark AG, Waldrip-Dail HM, Wolfner MF, Aquadro CF. 2001. Evolutionary EST analysis identifies rapidly evolving male reproductive proteins in Drosophila. Proc. Natl Acad. Sci. USA 98, 7375–7379. ( 10.1073/pnas.131568198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wade MJ. 1979. Sexual selection and variance in reproductive success. Am. Nat. 114, 742–747. ( 10.1086/283520) [DOI] [PubMed] [Google Scholar]
- 50.Shuster SM, Briggs WR, Dennis PA. 2013. How multiple mating by females affects sexual selection. Phil. Trans. R. Soc. B 368, 20120046 ( 10.1098/rstb.2012.0046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Efron B, Tibshirani RJ. 1993. An introduction to the bootstrap. London, UK: Chapman and Hall, Inc. [Google Scholar]
- 52.Davison AC, Hinkley CV. 1997. Bootstrap methods and their application. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 53.Pfennig KS, Pfennig DW. 2009. Character displacement: ecological and reproductive responses to a common evolutionary problem. Q. Rev. Biol. 84, 253–276. ( 10.1086/605079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lande R. 1981. Models of speciation by sexual selection on polygenic traits. Proc. Natl Acad. Sci. USA 78, 3721–3725. ( 10.1073/pnas.78.6.3721) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kirkpatrick M. 1982. Sexual selection and the evolution of female choice. Evolution 36, 1–12. ( 10.1111/j.1558-5646.1982.tb05003.x) [DOI] [PubMed] [Google Scholar]
- 56.Templeton AR. 1981. Mechanisms of speciation—a population genetic approach. Annu. Rev. Ecol. Evol. 12, 23–48. ( 10.1146/annurev.es.12.110181.000323) [DOI] [Google Scholar]
- 57.Schluter D. 2000. Ecological character displacement in adaptive radiation. Am. Nat. 156, S4–S16. ( 10.1086/303412) [DOI] [Google Scholar]
- 58.Noor MAF. 1999. Reinforcement and other consequences of sympatry. Heredity 83, 503–508. ( 10.1038/sj.hdy.6886320) [DOI] [PubMed] [Google Scholar]
- 59.Yukilevich R. 2012. Asymmetrical patterns of speciation uniquely support reinforcement in Drosophila. Evolution 66, 1430–1446. ( 10.1111/j.1558-5646.2011.01534.x) [DOI] [PubMed] [Google Scholar]
- 60.Baldwin BG, Goldman DH, Keil DJ, Patterson R, Rosatti TJ, Wilken DH. 2012. The Jepson manual: vascular plants of California, 2nd edn Berkeley, CA: University of California Press. [Google Scholar]
- 61.Schaeffer SW. 2008. Selection in heterogenous environments maintains the gene arrangement polymorphism of Drosophila pseudoobscura. Evolution 62, 3082–3099. ( 10.1111/j.1558-5646.2008.00504.x) [DOI] [PubMed] [Google Scholar]
- 62.Clark SCA, Sharp NP, Rowe L, Agrawal AF. 2012. Relative effectiveness of mating success and sperm competition at eliminating deleterious mutations in Drosophila melanogaster. PLoS One 7, e37351 ( 10.1371/journal.pone.0037351) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zajitschek F, Zajitschek S, Manier M. 2017. High-protein paternal diet confers an advantage to sons in sperm competition. Biol. Lett. 13, 20160914 ( 10.1098/rsbl.2016.0914) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anderson WA. 1974. Frequent multiple insemination in a natural population of Drosphila pseudobscura. Am. Nat. 108, 709–711. ( 10.1086/282949) [DOI] [Google Scholar]
- 65.Anderson WA, Levine L, Olvera O, Powell JR, de la Rosa ME, Salceda VM, Gaso MI, Guzman J. 1979. Evidence for selection by male mating success in natural populations of Drosophila pseudoobscura. Proc. Natl Acad. Sci. USA 76, 1519–1523. ( 10.1073/pnas.76.3.1519) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peterson MA, et al. 2005. Relative abundance and the species-specific reinforcement of male mating preference in the Chrysochus (Coleoptera: Chrysomelidae) hybrid zone. Evolution 59, 2639–2655. ( 10.1111/j.0014-3820.2005.tb00976.x) [DOI] [PubMed] [Google Scholar]
- 67.Matute DR. 2014. The magnitude of behavioral isolation is affected by characteristics of the mating community. Ecol. Evol. 4, 2945–2956. ( 10.1002/ece3.1142) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Anderson WW, Kim YK. 2005. Sexual isolation between sympatric and allopatric populations of Drosophila pseudoobscura and D. persimilis. Behav. Genet. 35, 305–312. ( 10.1007/s10519-005-3222-3) [DOI] [PubMed] [Google Scholar]
- 69.Anderson WW, Kim YK. 2006. A further analysis of sexual isolation between sympatric and allopatric populations of Drosophila pseudoobscura and D. persimilis—rejoinder to Noor and Ortiz-Barrientos. Behav. Genet. 36, 328–330. ( 10.1007/s10519-005-9030-y) [DOI] [PubMed] [Google Scholar]
- 70.Noor MAF, Ortiz-Barrientos D. 2006. Simulating natural conditions in the laboratory: a re-examination of sexual isolation between sympatric and allopatric populations of Drosophila pseudoobscura and D. persimilis. Behav. Genet. 36, 322–327. ( 10.1007/s10519-005-9033-8) [DOI] [PubMed] [Google Scholar]
- 71.Turissini DA, McGirr JA, Patel SS, David JR, Matute DR. 2018. The rate of evolution of postmating-prezygotic reproductive isolation in Drosophila. Mol. Biol. Evol. 35, 312–334. ( 10.1093/molbev/msx271) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kokko H, Mappes J. 2005. Sexual selection when fertilization is not guaranteed. Evolution 59, 1876–1885. ( 10.1111/j.0014-3820.2005.tb01058.x) [DOI] [PubMed] [Google Scholar]
- 73.Sanderson N. 1989. Can gene flow prevent reinforcement? Evolution 43, 1223–1235. ( 10.1111/j.1558-5646.1989.tb02570.x) [DOI] [PubMed] [Google Scholar]
- 74.Kulathinal RJ, Stevison LS, Noor MAF. 2009. The genomics of speciation in Drosophila: diversity, divergence and introgression on a genome-wide scale. PLoS Genet. 5, e1000550 ( 10.1371/journal.pgen.1000550) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McGaugh SE, Heil CSS, Manzano-Winkler B, Loewe L, Goldstein S, Himmel TL, Noor MAF. 2012. Recombination modulates how selection affects linked sites in Drosophila. PLoS Biol. 10, e1001423 ( 10.1371/journal.pbio.1001422) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Myers EM, Frankino WA. 2012. Time in a bottle: the evolutionary fate of species discrimination in sibling Drosophila species. PLoS One 2, e31759 ( 10.1371/journal.pone.0031759) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dobzhansky T. 1973. Is there gene exchange between Drosophila pseudoobscura and D. persimilis in their natural habitats? Am. Nat. 107, 312–314. ( 10.1086/282833) [DOI] [Google Scholar]
- 78.Powell JR. 1983. Interspecific cytoplasmic gene flow in the absence of nuclear gene flow: evidence from Drosophila. Proc. Natl Acad. Sci. USA 80, 492–495. ( 10.1073/pnas.80.2.492) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Eberhard WG. 1996. Female control: sexual selection by crypitc female choice. Princeton, NJ: Princeton University Press. [Google Scholar]
- 80.Manier MK, Belote JM, Lüpold S, Berben KS, Ala-Honkola O, Collins WF, Pitnick S. 2013. Rapid diversification of sperm precedence traits and processes among three sibling Drosophila species. Evolution 67, 2348–2362. ( 10.1111/evo.12117) [DOI] [PubMed] [Google Scholar]
- 81.Tyler F, Harrison XA, Bretman A, Veen T, Rodriguez-Munoz R, Trezenga T. 2013. Multiple post-mating barriers to hybridization in field crickets. Mol. Ecol. 22, 1640–1649. ( 10.1111/mec.12187) [DOI] [PubMed] [Google Scholar]
- 82.Andersson MM, Simmons LW. 2006. Sexual selection and mate choice. Trends Ecol. Evol. 21, 296–302. ( 10.1016/j.tree.2006.03.015) [DOI] [PubMed] [Google Scholar]
- 83.Servedio MR, Burger R. 2014. The counterintuitive role of sexual selection in species maintenance and speciation. Proc. Natl Acad. Sci. USA 111, 8113–8118. ( 10.1073/pnas.1316484111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Castillo DM, Moyle LC. 2019. Data from: Conspecific sperm precedence is reinforced, but postcopulatory sexual selection weakened, in sympatric populations of Drosophila Dryad Digital Repository. ( 10.5061/dryad.n88kb1m) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Castillo DM, Moyle LC. 2019. Data from: Conspecific sperm precedence is reinforced, but postcopulatory sexual selection weakened, in sympatric populations of Drosophila Dryad Digital Repository. ( 10.5061/dryad.n88kb1m) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
All data and R code used in this analysis are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.n88kb1m [84].


