Abstract
Selective pressure exerted by the widespread use of antibacterial drugs is accelerating the development of resistant bacterial populations. The purpose of this scoping review was to summarise the range of studies that use dynamic models to analyse the problem of bacterial resistance in relation to antibacterial use in human and animal populations. A comprehensive search of the peer-reviewed literature was performed and non-duplicate articles (n = 1486) were screened in several stages. Charting questions were used to extract information from the articles included in the final subset (n = 81). Most studies (86%) represent the system of interest with an aggregate model; individual-based models are constructed in only seven articles. There are few examples of inter-host models outside of human healthcare (41%) and community settings (38%). Resistance is modelled for a non-specific bacterial organism and/or antibiotic in 40% and 74% of the included articles, respectively. Interventions with implications for antibacterial use were investigated in 67 articles and included changes to total antibiotic consumption, strategies for drug management and shifts in category/class use. The quality of documentation related to model assumptions and uncertainty varies considerably across this subset of articles. There is substantial room to improve the transparency of reporting in the antibacterial resistance modelling literature as is recommended by best practice guidelines.
Key words: Antibiotic resistance, modelling, transmission
Introduction
The World Health Organisation (WHO) describes antimicrobial resistance (AMR) as ‘a problem so serious that it threatens the achievements of modern medicine’ [1]. The effective prevention and treatment of an increasing range of bacterial, viral, fungal and parasitic infections is a challenge in many parts of the world where high rates of resistance are observed. Increases in mortality, length of hospitalisation and costs of care are associated with the development of resistance in several important bacterial pathogens [2]. A recent report on AMR [3] estimates that 10 million more people are expected to die every year by 2050 if resistance is left unchecked; while the size of this figure has been met with skepticism [4], there is clearly a large clinical and public health burden related to AMR. Indeed, the World Bank estimates that the annual global gross domestic product (GDP) would fall by 1.1% by 2050 under even a modest ‘low impact’ AMR scenario [5].
Selective pressure exerted by the widespread use of antimicrobial drugs is accelerating the development of resistant populations of bacteria and other pathogens [1, 3]. The development of resistance is a normal evolutionary process for microorganisms [1] and even the ‘appropriate’ use of antimicrobials selects for preexisting resistant populations in nature [3, 6]. The AMR issue is exacerbated by the ongoing misuse and overuse of antibiotic drugs in humans and food-producing animals [1, 3]; further, the ‘collapse of the antibiotic research-and-development pipeline’ [6] is a related problem underscoring the need for urgent solutions. The epidemiology of AMR is exceedingly complex and effective solutions must necessarily involve a coordinated effort by many sectors. Resistant bacteria and resistant genes do not recognise ecologic and geographic borders [7] and the transmission between human, animal [8, 9] and environmental [10] domains is of shared concern. The WHO, World Organisation for Animal Health (OIE) and Food and Agriculture Organisation are thus mobilising a ‘One Health’ response in a tripartite collaboration on AMR [7].
Systems science and AMR
AMR has been framed by several experts as a ‘super-wicked’ problem [11], a specific type of policy challenge defined by the problem's inherent complexity, the interrelatedness of multiple fields and the potential for conflict arising from competing goals. There is a critical role for the ‘systems science approach’ to optimise intervention strategies for problems of this nature [11, 12]. While traditional scientific approaches might examine how mechanisms work in isolation, a systems science approach makes it possible to examine the relationships between many interdependent components by integrating existing epidemiological data [12–14]. Dynamic modelling methods are used to develop mathematical representations of systems characterised by nonlinearities, feedback loops and multiple variables that evolve over time [15]. Representations of this type are necessarily a simplification of the system of interest [14, 16], but can help identify the critical functional aspects of a system via experimentation with scenarios under various conditions [15, 16]. By defining the conditions under which an intervention is likely to work [17], dynamic or mathematical models offer insights on which to build effective policy responses [12, 18]. Furthermore, models of this sort challenge researchers to both share and scrutinise their assumptions and/or hypotheses about how a system works. Risk assessment (RA) is a related and complementary approach for data integration and was the focus of a parallel review (unpublished).
Temime et al. [19] previously demonstrated the growing interest in mathematical modelling approaches to evaluate antibiotic resistance. In particular, these models have been used extensively to explore the development and spread of multi-drug resistant bacteria in hospital settings [14, 16, 20, 21]. Interventions of interest include those with implications for physical infection control strategies (e.g. isolation, hand hygiene) [16] and to a lesser extent, antibiotic restriction [21] and other prescription strategies [14]. Mathematical modelling is a valuable tool with which to generate hypotheses about the relationship between antimicrobial use (AMU) and AMR at the population level [17]. However, the scope of work undertaken for this purpose in both healthcare and other relevant domains has yet to be fully characterised [22, 23]. Of special interest to this research team is the use of dynamic models as they pertain to AMR in agricultural and food system settings; this is notably relevant in light of the recent suggestion that veterinary epidemiologists should progress to the multi-scale modelling of food animal systems [12].
Scope of inquiry
The purpose of this scoping review was to identify and describe how researchers have used dynamic modelling techniques to explore and advance policies that limit AMR-related risk arising from AMU. To establish an effective search strategy, a broad research question should be combined with a clearly articulated scope of inquiry, including the concept, target population and health outcomes of interest [24]. This review was thus conceived as a means to summarise the range of studies that use dynamic models to analyse the problem of bacterial resistance in relation to antibacterial use in humans and animals. The intended focus was studies which generate and draw conclusions from simulated data using aggregate or individual-based dynamic models of resistance development and spread. Population-level outcomes of interest included the frequency of resistant hosts, the likelihood of morbidity and mortality arising from resistant infections and/or the costs associated with resistant infections.
Arksey and O'Malley [25] describe several reasons for conducting a scoping review, several of which may be relevant for the overall purpose [24]. This review combines multiple objectives, namely to describe the specific contexts to which these models are applied, to summarise the approches to model building and to identify where gaps exist in the peer-reviewed literature. Furthermore, the effort was motivated in part by this group's interest in the documentation and reporting of models in relation to recommended guidelines. In 2012, the Good Research Practices in Modeling Task Force published a series of articles describing best practices for designing and building dynamic models [26], analysing uncertainty [27] and achieving transparency [28]. Several of the questions in the ‘Data charting’ section was developed with reference to this framework [26–28] and to related materials concerning the responsible conduct of research with simulation models [29].
Methods
The scoping study team consisted of members with the appropriate content and methodological knowledge to perform this review [24] and included expertise in the areas of AMR, infectious and veterinary epidemiology, food system safety, dynamic modelling and RA techniques and scoping review methodology. As is typical of scoping reviews, the process of selecting relevant studies and charting the data was non-linear [24, 25]; frequent team discussions were necessary to refine the inclusion/exclusion criteria and extraction variables over the course of this review. The criteria and questions described below reflect the multiple iterations and improvements gleaned through the team's increasing familiarity with the literature.
Search terms and strategy
In consultation with a research librarian, the team developed a strategy to search the peer-reviewed literature for studies modelling AMR in humans, animals and the environments with which they interact. The search was limited to English language publications but was otherwise deliberately broad to ensure a comprehensive review of the literature. There were no limitations on the date nor geographical origin of publication. Four databases were included in the search (PubMed, Scopus, Agricola and CAB Abstracts) and identical search terms were modified only to reflect the differing indexing schemes. The PubMed search in particular made use of Medical Subject Headings (MeSH) terms, the US National Library of Medicine's controlled vocabulary thesaurus.
The search was comprised of concatenated descriptors from three general areas: the first concerned study subject and included variations on the terms humans, patients, hospitals, domestic and companion animals, farms, water and soils; the second concerned methodological approach and included variations on the terms simulation and dynamic models; the third concerned general content area and included variations on the terms antibacterial use and resistance. A full list of terms included in the search is available in the online Supplementary Material. The search was initially conducted on 28 February 2016 and updated with additional terms on 5 November 2016; in an effort to establish reproducibility, the search was performed independently by two reviewers (JI and DR). Reference information for the identified articles was exported to and de-duplicated using Zotero, a web-based reference management program. De-duplicated references were exported to an Excel spreadsheet for sorting and screening.
In order to identify relevant studies missed by the database search, the team conducted a supplementary review of reference lists from pertinent articles. Included in this subset of articles were previous reviews of studies modelling bacterial resistance [30–32] and studies published between 2013 and 2016 and retained for inclusion after full-text review (see the ‘Article screening’ section). The reference lists of retained studies concerning food-producing animals [e.g. 33–34], cost outcomes [e.g. 35] and network modelling [e.g. 36, 37] were likewise reviewed, given that these were comparably unique and suggestive of missed search terms. Retained studies published prior to 2013 were not reviewed further, as this time period was comprehensively covered by [30–32] and new articles were no longer being identified. As the focus of this review was peer-reviewed literature, the team chose not to include grey literature in the search.
Article screening
Two reviewers (JI and DR) independently performed the title and abstract screens. A third reviewer (CW) was consulted when agreement could not be reached on a particular reference at either the title or abstract screening stages. Articles were excluded if they were not in English, if they were not published in a peer-reviewed journal and if they did not present novel findings (e.g. literature reviews, commentaries, editorials or methodological descriptions). Because the focus of this review was bacterial resistance, studies which were specific to non-bacterial pathogens (e.g. viruses, fungi, parasites or mycobacteria) were excluded, as were those addressing non-infectious disease outcomes. Studies which were strictly observational in nature, including case series and prevalence surveys, were likewise excluded at this stage.
Where the article title was not sufficiently informative to make an inclusion/exclusion decision, the reference was retained for abstract screening. In addition to the aforementioned criteria, studies were excluded at this stage if they concerned pharmacodynamic-pharmacokinetic models, or if they focused exclusively on susceptible bacterial variants. Studies which were strictly experimental in nature, including microbiological investigations, were likewise excluded at this stage.
An additional level of screening was performed at the stage of full-text review and data extraction/charting. A series of questions was applied to each remaining reference to ensure the final subset was reflective of the review's scope of inquiry. The list of questions appears below and evolved over several iterations of review, analysis and discussion.
-
(1)
Does this study possess a dynamic component (i.e. does the outcome vary over time, in contrast to studies where the outcome is independent of time, as in static assessments of risk)?
-
(2)
Does this study generate data using simulation(s) and/or draw conclusions from simulated data?
-
(3)
Does this study concern the development or spread of bacterial strains with inherited antibiotic resistance (i.e. is not exclusive to susceptible bacteria and/or bacterial strains with non-inherited antibiotic resistance)?
-
(4)
Does this study investigate the dynamics of resistant bacteria in the presence of antibacterial treatment/use, either in the baseline or counterfactual scenarios?
-
(5)
Does this study endeavour to examine the dynamics of resistant bacteria at the population or inter-host level (i.e. is not strictly an intra-host model)?
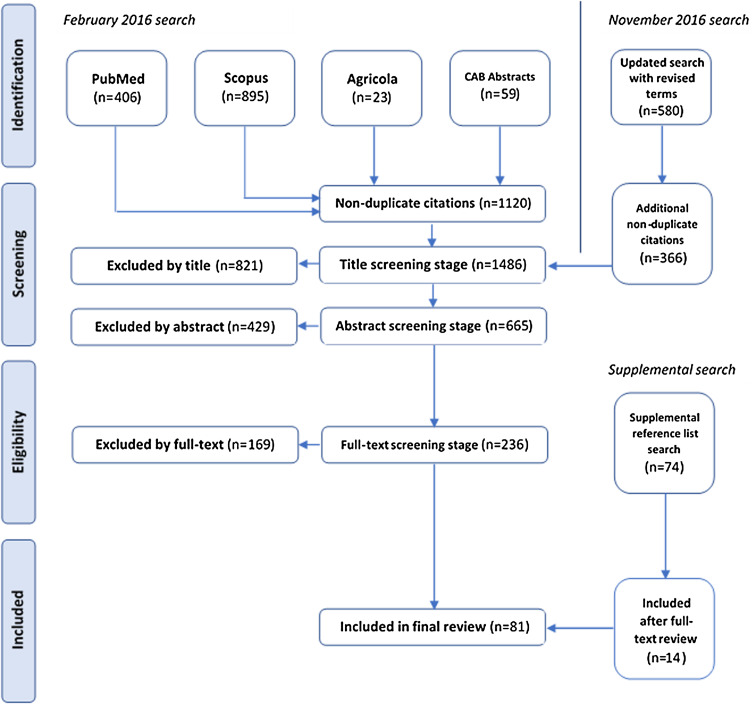
The reference was excluded if a ‘no’ answer was observed for any of the above questions. A sample of 30 full-text articles over several iterations was screened independently by two reviewers (CW and DR) to evaluate reviewer agreement. As there were no significant disagreements or challenges related to study selection using this framework, a single reviewer (DR) screened the full texts of remaining articles. When the decision to include/exclude was uncertain, a second reviewer (CW) determined the final inclusion. Articles were not excluded on the basis of quality. A PRISMA diagram depicts the flow of information through the identification, screening and eligibility stages in Figure 1.
Fig. 1.
PRISMA flow diagram depicting the number of articles at each of the identification, screening and eligibility stages.
Data charting
The review team developed a data charting form in Microsoft Excel in which to capture variables relevant to the study's scope. As is recommended by Levac et al. [24], the data charting form was frequently updated in concert with the team's increasing familiarity with the studies retained for inclusion (n = 81). The charting questions appear below and are organised by category to highlight distinct contextual and methodological model features.
Model type and context
-
(1)
What type of model was built for this study?
-
(2)
What is the setting or context for the system represented by this model?
-
(3)
Which bacterial host(s) is/are represented in this model, if any?
-
(4)
Which bacterial organism(s) is/are of interest in this model, if any?
-
(5)
Which resistance pattern(s) (e.g. cephalosporin resistance) is/are of interest in this model, if any?
-
(6)
Which particular antibiotic treatment/application(s) is/are simulated in this model, if any?
Model construction and parameterisation
-
(1)
Does the study describe and/or justify the assumptions made in constructing this model?
-
(2)
Does the paper provide a visual depiction of the system represented by this model (e.g. compartmental diagram)?
-
(3)
Does the paper detail and/or solve the differential equations for the system represented by this model (where applicable, as in compartmental models)?
-
(4)
What source(s) of data are used to generate input values for this model?
-
(5)
Do the authors use a range or distribution of values for one or several parameters in the baseline scenario?
-
(6)
Do the authors calibrate (i.e. ‘fit’) the model to empirical data in order to generate input values?
Model outputs and validation
-
(1)
Which emergent property or endogenous variable(s) (i.e. outputs) of interest is/are generated by this model?
-
(2)
For a particular set of assumptions, is the output of this model represented by a single possible outcome (arising from deterministic models) or a distribution of possible outcomes (arising from stochastic processes)?
-
(3)
Does the paper report the time units and/or length of time over which the output is observed (i.e. the simulation length)?
-
(4)
Do the authors perform sensitivity analyses to identify parameters which strongly affect the outputs of this model (i.e. explicitly characterise the uncertainty associated with model inputs)?
-
(5)
Do the authors attempt to validate this model by reproducing observed or empirical data?
Model applications
-
(1)
Does this study attempt to explain an observed epidemiological phenomenon or investigate a future and/or counterfactual state, given a set of initial conditions?
-
(2)
If the model's purpose is explicative, do the authors compare competing hypotheses for observed phenomena?
-
(3)
If the model's purpose is investigative, do the authors examine the impact of changes to the system in alternative or counterfactual scenarios?
-
(4)
What type of system change(s) and/or intervention(s) are of interest in this study, if any?
-
(5)
Do the results or conclusions reported in the paper have the potential to inform policy development or decision-making?
The required responses were either binary (i.e. yes/no) or restricted to a defined list of descriptors. Data from a sample of 10 included studies were extracted independently by two reviewers (CW and DR) to evaluate reviewer agreement and consistency. As there were no significant disagreements or challenges related to data extraction using the charting form, a single reviewer (DR) charted the data for the remaining articles. The responses were summarised across the included studies and reported in a tabular format where appropriate (see the ‘Results’ section).
Results
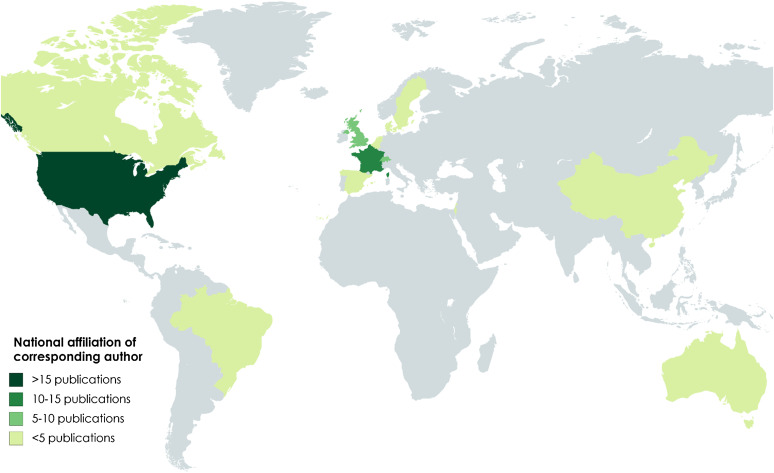
The review identified 81 articles for inclusion at the stage of full-text screening and data charting [33–113]. Nine articles (11%) were published before 2000, 25 articles (31%) were published between 2000 and 2009 (inclusive) and 47 (58%) articles were published between 2010 and the most recent database search in November 2016. Twenty-two of the included articles (27%) were published after 2012, the most recent year covered by a review in our supplemental search [32]. The corresponding author in nearly half of the articles (43%) was affiliated with an American institution; 12 articles (15%) and eight articles (10%) were affiliated with corresponding authors based in France and the UK, respectively. The map in Figure 2 highlights the national affiliation of the corresponding author for articles included in the review.
Fig. 2.
National affiliation of corresponding author for articles included in the scoping review.
Model type and context
The studies are summarised by model setting and bacterial host(s) in Table 1 and are further subdivided by model type and behaviour. Thirty-three articles (41%) feature models relevant to healthcare settings (i.e. hospitals or clinics), while 31 articles (38%) feature models based in human communities; six additional articles (7%) model the interaction between healthcare and community settings [56, 57, 59, 66, 71, 73]. Only two articles examine antibacterial resistance in an exclusively agricultural setting at the inter-host level [33, 34]. Humans are the relevant bacterial hosts in most of the included studies, with some notable exceptions where pigs [33, 34], dogs [72] and food-producing animals [85] are the hosts of interest. In eight articles with non-specific settings, the model simulates antibacterial resistance development/spread in a generic ‘host’ population.
Table 1.
Summary of studies included in the final review (n = 81), organised by model context and type
| System context | Bacterial host(s) | Model type | Dynamic behaviour | Reference(s) |
|---|---|---|---|---|
| Hospital or clinic | Inpatient humans | Compartmental | Deterministic | [39–48, 50, 51, 53, 54, 59–68, 70, 71, 73–75] |
| Stochastic | [38, 49, 57, 58] | |||
| Both formulations | [52, 76] | |||
| Individual-based | Stochastic | [47, 55, 56, 69] | ||
| Healthcare workers | Compartmental | Deterministic | [44, 46, 47, 50, 51, 64, 70] | |
| Stochastic | [38] | |||
| Individual-based | Stochastic | [47, 55, 56, 69, 72] | ||
| Inpatient dogs | Individual-based | Stochastic | [72] | |
| Community or household | Humans | Compartmental | Deterministic | [35, 56, 59, 66, 71, 73, 77–82, 84, 85, 88–90, 92–97, 99, 101–105] |
| Stochastic | [57, 91] | |||
| Both formulations | [86, 98, 100] | |||
| Individual-based | Stochastic | [87, 96] | ||
| Social network | Stochastic | [36] | ||
| Neural network | Deterministic | [37] | ||
| Agricultural setting | Pigs | Compartmental | Deterministic | [33] |
| Stochastic components | [34] | |||
| Food-producing animals | Compartmental | Deterministic | [85] | |
| Non-specific setting | Humans | Compartmental | Deterministic | [108, 111] |
| Monte Carlo | Stochastic | [110] | ||
| Generic hosts | Compartmental | Deterministic | [106, 107, 109, 112, 113] |
Most of the included studies (86%) represent the system of interest with a compartmental or aggregate model. Of the 70 compartmental-type models, 84% exhibit exclusively deterministic behaviour (i.e. a single possible outcome is determined by the system's initial conditions). Six models treat the transitions between compartments as random phenomena and thus exhibit stochastic behaviour [34, 38, 49, 57, 58, 91]. In five studies, the authors perform both deterministic and stochastic simulations of the same compartmental model [52, 76, 86, 98, 100]. Individual or agent-based models (ABM) are described in four studies [55, 69, 72, 87]; a single study combines compartmental and individual-based components in a hybrid model [56]. In [47] and [96], the authors compare simulations arising from compartmental and individual-based models representing the same system.
The studies are summarised by the bacterial organism(s) and resistance pattern(s) of interest in Table 2; an organism was considered to be ‘of interest’ if it was the motivation for model building [e.g. 84, 97, 104], or if the model was parameterised and/or validated with data specific to that bacteria. In 40% of the articles (n = 32), resistance is modelled for a generic or non-specific bacterial organism. An additional six articles model resistance in otherwise unspecified Enterobacteriaceae species [49, 72] or commensals [61, 77, 110, 112]. Vancomycin-resistant enterococci (n = 5) and methicillin-resistant Staphylococcus aureus (MRSA) (n = 4) are of frequent interest in hospital settings. In the community setting, the bacteria of interest are most often Streptococcus pneumoniae (n = 18) and Neisseria gonorrhoeae (n = 6).
Table 2.
Bacterial organism(s) and resistance pattern(s) of interest for studies included in the final review (n = 81)
| System context | Organism(s) | Resistance pattern(s) | Reference(s) | |
|---|---|---|---|---|
| Antibacterial class | Antibacterial drug | |||
| Hospital or clinic | Not specified | – | – | [39–43, 45, 47, 48, 50, 52, 54, 57, 58, 65, 66, 68–70, 74, 76] |
| Commensals, unspecified | – | – | [61] | |
| Enterobacteriaceae, unspecified | Beta-lactamsa | – | [49] | |
| Ampicillin | [72] | |||
| Quinolones | Nalidixic acid | [72] | ||
| Enterococcus spp. | Glycopeptides | Vancomycinb | [38, 46, 51, 64, 75] | |
| Streptogramins | Virginiamycin | [71] | ||
| Escherichia coli | Aminoglycosides | Amikacin | [67] | |
| Gentamicin | [67] | |||
| Beta-lactamsa | Cefuroxime | [73] | ||
| Fluoroquinolones | Ciprofloxacin | [67] | ||
| Klebsiella pneumoniae | Aminoglycosides | Amikacin | [62] | |
| Pseudomonas aeruginosa | Aminoglycosides | Gentamicin | [60] | |
| Tobramycin | [60] | |||
| Beta-lactams | Ceftazidime | [53] | ||
| Carbapenems | Meropenem | [53] | ||
| Fluoroquinolones | Ciprofloxacin | [53] | ||
| Staphylococcus aureus | Penicillins | Methicillinc | [44, 55, 56, 59] | |
| Streptococcus pneumoniae | Penicillins | Penicillin | [63] | |
| Community or household | Not specified | – | – | [57, 66, 81, 85, 89, 95] |
| Commensals, unspecified | – | – | [77] | |
| Enterococcus spp. | Glycopeptides | Vancomycinb | [97] | |
| Streptogramins | Virginiamycin | [71] | ||
| Escherichia coli | Beta-lactamsa | – | [73] | |
| Moraxella catarrhalis | Beta-lactamsa | – | [78] | |
| Neisseria gonorrhoeae | – | – | [79, 83, 86, 87, 102, 105] | |
| Neisseria meningitidis | Beta-lactams | – | [98] | |
| Staphylococcus aureus | Penicillins | Methicillinc | [56, 59, 92] | |
| Streptococcus pneumoniae | – | – | [80, 91] | |
| Beta-lactams | – | [82, 93, 96] | ||
| Penicillin | [36, 37, 78, 90, 94, 98–101] | |||
| Amoxicillin | [103] | |||
| Fluoroquinolones | – | [104] | ||
| Ketolides | – | [93] | ||
| Macrolides | – | [82, 84, 93] | ||
| Erythromycin | [90, 103] | |||
| Mixed infections | Carbapenems | Ertapenem | [35, 88] | |
| Penicillins | Piperacillin | [35, 88] | ||
| Agricultural setting | Not specified | – | – | [33, 85] |
| Escherichia coli | Tetracyclines | Tetracycline | [34] | |
| Non-specific setting | Not specified | – | – | [106–109, 111, 113] |
| Commensals, unspecified | – | – | [110, 112] | |
‘–’ indicates the antimicrobial class and/or antimicrobial drug was not specified in the study.
Beta-lactamase or extended-spectrum beta-lactamase-producing bacteria.
Vancomycin-resistant enterococci (VRE).
Methicillin-resistant staphylococcus aureus (MRSA).
Of the 43 articles which specify the bacteria of interest, eight (19%) do not indicate the resistance pattern of interest. Resistance to penicillin in Streptococcus pneumoniae is the most frequently modeled phenotype (n = 10); beta-lactamase-producing bacteria are considered in only three studies [49, 73, 78]. In 74% of the articles (n = 60), the antibiotic applied to the bacterial host(s) in the model is generic or non-specific. Twelve studies simulate treatment with antibiotics from multiple classes [35, 53, 55, 56, 67, 73, 82, 88, 90, 93, 96, 103]. Treatment with beta-lactams (including carbapenems, cephalosporins and penicillins) is most frequently simulated (n = 17), while fluoroquinolone [53, 67, 73] and macrolide [82, 90, 93, 103] treatments are simulated less often. The single example of treatment with tetracycline concerns the spread of tetracycline-resistant Escherichia coli in pigs [34].
Model construction and parameterisation
Many of the included articles (77%) provide a visual depiction of the system represented by the model (e.g. a compartmental diagram or other schematic representation, as in [55, 69, 72]). In 84% of the 70 strictly compartmental-style models, the paper details the differential equations representing the system of interest; the relevant equations are also highlighted for the deterministic component or formulation in [47, 56, 96]. While most articles describe to some extent the source(s) of data used to parameterise the model (86%), 11 articles do not comment on the source of the input values [41, 68, 71, 74, 77, 84, 85, 89, 107, 108, 113]. An additional eight articles rely solely on expert opinion or ‘guesstimates’ to parameterise the model and/or purposively choose values in order to highlight specific dynamic behaviours [33, 40, 52, 81, 86, 95, 106, 110].
Of the 70 articles for which the source(s) of data are described, 50% make use of empirical data either collected by the study's authors (n = 11) or published in a report of particular relevance (n = 25). In 22 studies, the authors estimate previously unknown parameters by calibrating or ‘fitting’ model outputs to empirical data. The published literature and/or expert opinion was used to generate at least one parameter in 74% and 64% of studies for which the input value source could be identified with reasonable confidence, respectively. These percentages should be interpreted as relative trends and not conclusive findings, given that the data source(s) for unique inputs were frequently unreported or otherwise not easily gleaned from the article.
Model output and validation
In most of the included articles (84%), the proportion of resistant patients is an emergent property of primary or secondary interest. Variations of the term proportion in this subset include ‘fraction’, ‘frequency’, or ‘prevalence’ of resistance; likewise, variants of the term resistant patients including ‘colonisation with resistant bacteria’, ‘resistant infections’ or ‘resistant strains’ were assigned to this outcome category. Additional outcomes of interest include the likelihood of and/or time to resistance emergence or extinction [65, 66, 78, 83, 86, 87, 91, 93, 97, 98, 110], the likelihood of coexistence (i.e. of sensitive and resistant strains) [59, 62, 80, 106], the fraction of resistant infections attributable to antibiotic use [71, 96, 97, 101] and the number of treatment failures or inappropriately-treated patients [58, 66, 104]. The outcome of interest in four articles [94, 98–100] is the distribution of resistance levels determined by the minimum inhibitory concentration.
The studies are summarised by the characteristics of their reported outputs in Table 3. ‘Uniquely determined’ outputs (i.e. outcomes with a single possible value) are observed for models with deterministic behaviour and inputs represented by point estimates (n = 52). Conversely, distributions of output values are observed for models with stochastic behaviour (see Table 1), or for models with deterministic behaviour that draw multiple parameter sets from input distributions [e.g. 65, 81, 104]. In order to highlight different scales of temporal abstraction, the articles are further subdivided by the length of time over which the output is observed (i.e. the simulation time). Models which were simulated to equilibrium or ‘steady state’ values were classified based on the time reported in the associated graphical output [e.g. 44, 54, 77]. There was no specific time associated with the equilibrium state in eight articles; in six additional articles, time is measured in otherwise undefined ‘steps’.
Table 3.
Characteristics of the reported output arising from dynamic modelling effort in included studies (n = 81)
| Article focus | Output value | Time scale for output | Reference(s) |
|---|---|---|---|
| Analytical study | Equilibrium behaviour | – | [43, 106, 108] |
| Numerical simulations | Uniquely determined value | 1 week ⩽ x <1 month | [75] |
| 1 month ⩽ x <1 year | [42] | ||
| 1 year ⩽ x <10 years | [35, 37, 41, 42, 64, 67, 70, 71, 73, 88, 90, 97] | ||
| 10 years ⩽ x <50 years | [59, 66, 89, 92, 93, 99, 101, 103] | ||
| 50 years ⩽ x | [59, 66, 79, 93, 94, 102] | ||
| Undefined time steps | [109] | ||
| Undefined steady state | [46, 51, 53, 63, 110] | ||
| Distribution of values reported as mean or range | 1 week ⩽ x <1 month | [55] | |
| 1 month ⩽ x <1 year | [82] | ||
| 1 year ⩽ x <10 years | [49, 56, 69, 72, 83] | ||
| 10 years ⩽ x <50 years | [36, 57, 58, 81, 83, 91, 105] | ||
| 50 years ⩽ x | [80, 87, 104] | ||
| Both formulations | 1 month ⩽ x <1 year | [34, 96] | |
| 1 year ⩽ x <10 years | [52, 98] | ||
| 10 years ⩽ x <50 years | [76, 98] | ||
| Both analytical study and numerical simulations | Uniquely determined value | 1 month ⩽ x <1 year | [33, 44, 50, 54, 61, 74] |
| 1 year ⩽ x <10 years | [44, 45, 48, 60, 62, 112] | ||
| 10 years ⩽ x <50 years | [77, 78] | ||
| 50 years ⩽ x | [111] | ||
| Undefined time steps | [39, 40, 85, 107, 113] | ||
| Undefined steady state | [68, 84, 95] | ||
| Distribution of values reported as mean or range | 1 month ⩽ x <1 year | [38] | |
| 1 year ⩽ x <10 years | [65] | ||
| Both formulations | 1 year ⩽ x <10 years | [47, 86] | |
| 10 years ⩽ x <50 years | [86, 100] |
In 53% of included articles, the author(s) perform sensitivity analyses to identify parameters and/or structural features which exert a strong effect on the model's outputs. In addition to the term sensitivity, related concepts including ‘uncertainty’ or ‘robust(ness)’ were suggestive of an attempt to formally characterise the impact on the outcome of model assumptions. As with the sources for input data, relevant analyses are often incompletely described and this estimate should be interpreted in that context. An effort is made to formally validate the relevant model by reproducing empirical data in 17 of the included articles [37, 38, 49, 53, 56, 62, 63, 78, 82, 83, 87, 90, 93, 94, 98, 103, 105]. Less formal comparisons between model outputs and historical trends are explored in seven additional articles [34, 41, 72, 73, 76, 92, 99].
Model applications
The reported study purpose is to investigate one or several counterfactual and/or future states in 75% of the included articles; in five articles, the purpose of the study is to explain or interpret an epidemiological phenomenon [59, 62, 80, 82, 83]. In 15 additional articles, the model is used both to explain phenomena and to examine the system's dynamics in competing or alternative scenarios. ‘Intervention(s)’ representing theoretically modifiable system change(s) were explored in 90% of the included articles (n = 73). The intervention type and potential relevance of study results and/or conclusions for policy development are summarised in Table 4. Note that in 25 articles, the author(s) examine the effect of multiple intervention types on the system of interest; the total number of interventions reported in the table is thus larger than 73.
Table 4.
Modifiable intervention types investigated in hypothetical and/or counterfactual scenarios (n = 73)
| Intervention type | Number of articles | Potential policy implications | Reference(s) | |
|---|---|---|---|---|
| Antibiotic use | Other policy | |||
| Change to overall consumption of antibiotics | 37 | X | [34, 36, 38, 46–48, 50, 51, 53, 56, 63, 71–75, 77, 78, 81, 85, 86, 91, 92, 94, 95, 97, 98, 101, 105, 106, 108, 110–112] | |
| X | X | [44, 61, 89] | ||
| Change to antibiotic management strategy | 21 | X | [39–43, 45, 54, 61, 65, 66, 68, 70, 76, 79, 84, 102, 105, 107, 109, 113] | |
| X | X | [58] | ||
| Change to antibiotic category, class or type | 15 | X | [35, 53, 55, 56, 60, 67, 79, 88, 93, 96, 103–105] | |
| X | X | [33, 52] | ||
| Change to hygiene or infection control protocols | 17 | X | [38, 44–46, 48, 50, 54, 61, 64, 69, 70, 72–75, 105] | |
| X | X | [49] | ||
| Change to patient management or other operational functions | 6 | X | [46, 48, 64, 72, 75] | |
| X | X | [57] | ||
| Change to human behaviour impacting antibiotic use | 4 | X | [45, 84, 95, 108] | |
| Change to pattern of vaccination, including coverage or efficacy | 4 | X | [91, 99, 101, 103] | |
| Change to diagnostic capacity impacting antibiotic use decisions | 3 | X | [79, 87, 111] | |
Interventions involving a change to the overall consumption of antibiotics were the most frequently investigated (n = 37); pertinent examples explore changes in antibiotic use frequency, duration and dose. Other interventions with particular relevance for antibiotic use policy include changes to antibiotic management strategies (n = 21) and changes to antibiotic category or class (n = 15). In the former category, deployment strategies of interest include cycling, mixing or combining antibiotics at the institutional or population level; in the latter, shifts in antibiotic use within and between classes are highlighted. Eight (10%) of the included studies do not impose a theoretically modifiable change on the system of interest. A subset of these articles nevertheless has the potential to inform antibiotic use [37, 80, 83, 90] and/or hygiene and infection control [80, 83] policies. The results from six articles have relevant implications for antibiotic use in companion animals [72] and food-producing animals [33, 34, 71, 85, 97].
Discussion and implications
Given the increasing interest in [19] and projected value of [17] mathematical modelling to generate hypotheses about the AMU/AMR relationship, the authors endeavoured to better characterise the peer-reviewed literature in this domain. This review identified 81 studies which used simulated data from dynamic models to analyse the problem of bacterial resistance in relation to antibiotic use in humans or animals [33–113]. Note that the focus of this work was resistance dynamics at the population or inter-host level and thus strictly intra-host models are excluded from this count. As was expected, the number of included articles grew in each 5-year increment since 2000, in parallel with the AMR modelling literature more generally. The articles’ corresponding authors are affiliated with 14 countries on five continents, although most articles (70%) emerged out of three countries (USA, UK and France). This observation is likely explained in part by the English language limitations of our search, but might also indicate that well-resourced institutions are better able to engage in the ‘systems science approach’ and with its associated methodologies. In particular, the National Institutes of Health (NIH) has played a leading role in the promotion and funding of systems science projects at American institutions.
Model type and context
Aggregate models comprise the vast majority of studies included in this subset of articles (86%). In most instances and unless they are simulated stochastically as in [38, 39, 57, 58, 91], compartmental models exhibit deterministic behaviour. Models of this type describe the average behaviour of a system and are most appropriate when the population is sufficiently large to minimise the impact of differences between individuals. While compartmental models have the advantage of being comparably easy to parameterise and calibrate, they cannot account for individual heterogeneity nor the substantial impact of chance events in smaller populations (e.g. closed hospital wards). In contrast, individual or ABM represent the system as a population of heterogeneous entities engaged in local interactions; models of this type better capture the stochastic behaviour of a system but impose opportunity costs related to model construction, parameterisation and computing time [114]. Opatowski and colleagues [30] argue that increasingly realistic predictions arising from ABM models are likely to advance the utility of computer simulations for decision support. Only seven articles in this series develop individual-based representations of the system of interest [47, 55, 56, 69, 72, 87, 96], suggesting that ABMs have been infrequently used to examine the AMU/AMR relationship at the population level.
While experts decidedly agree that AMR is a ‘One Health’ problem requiring multi-sectoral action [7], there are few examples of inter-host models outside of strictly human contexts. Further, there are only two models in this subset which incorporate multiple host species in shared environments [72, 85]. Antibiotic use in agricultural settings accounts for a substantial proportion of global antimicrobial consumption [8] and global AMU in livestock is expected to rise significantly through 2030 [115]. These observations highlight the potential role for dynamic models to inform strategies that limit the inter-ecosystem spread of AMR arising from AMU in food-producing animals. Recent and impressive examples exist at the within-host level; for instance, Volkova and colleagues examine the effect of antibiotic therapy on the dynamics of ceftiofur-resistant commensal Escherichia coli in the large intestine of cattle in [116, 117]. At the inter-host level, dynamic models have been used to investigate the persistence of resistant bacteria following a clinical outbreak of Salmonella typhimurium in dairy cows [118] and in the context of non-AMU interventions targeting Escherichia coli commensals in beef cattle [119].
Model applications
Models included in the final subset range from general or hypothetical [e.g. 39, 41] to those with a single, specific application [e.g. 46, 53]. The purpose of the model is primarily theoretical in 40% and 74% of articles where resistance is modelled for a generic bacterial organism and/or a generic antibiotic, respectively. Articles in which the organism of interest is explicitly identified are more useful for decision support [30] and frequently concern ‘priority’ [120] or ‘urgent’ [121] AMR disease threats. Indeed, drug-resistant Streptococcus pneumoniae (n = 18), Neisseria gonnorhoeae (n = 6) and MRSA (n = 5) are well represented in this dataset and appear on WHO's list of bacteria of international concern [1]. Extended-spectrum beta-lactamase-producing Enterobacteriaceae are routinely identified as high priority threats [1, 120, 121], but are the focus of only two studies reported here [49, 73]; no studies examine the urgent threat of carbapenem-resistant Enterobacteriaceae. Further, there are no studies which model resistance in zoonotic bacterial pathogens that are important sources of infection for humans (e.g. Salmonella spp. or Campylobacter jejuni). Normally commensal bacteria that may harbour resistant genes, including Enterococcus spp. (n = 7), are better represented in this dataset.
This review was limited to models in which antibacterial treatment or use is directly simulated; the inclusion criteria are thus reflected in the scope and types of interventions applied in this subset of articles. Sixty-seven articles (83%) impose at least one antibiotic use-related intervention, ranging from institutional drug management strategies (e.g. cycling antibiotics) to drug category changes (e.g. switching from a first to a second-line drug). Antibiotic stewardship is one component of a necessarily multifaceted approach to the AMR problem [122]; there are only 22 articles (27%) which examine an antibacterial use intervention in combination with another approach. Future consideration should be given to the impact on system behaviour of the interactions between prudent antibiotic use and other biological or behavioural interventions. While they do not simulate treatment directly, several related models estimate the relative importance of resistant bacterial acquisition by the endogenous route (i.e. arising from the selective pressure of antibiotics) [e.g. 123, 124]. There are likewise numerous articles which model the acquisition of resistant bacteria via the exogenous route (i.e. via direct or indirect transmission) in the absence of selective treatment pressure [e.g. 125, 126].
Model transparency and uncertainty
Responses to the charting questions were limited to yes/no selections or an otherwise restricted set of descriptors; this review thus summarises but does not offer a critical appraisal of the included studies. Nevertheless, some broad generalisations can be made with respect to the achievement of transparency and management of uncertainty in this set of articles. Modelers can enhance credibility by clearly describing the model structure, assumptions and parameter values [27, 28]. While several studies offer robust, defensible explanations for the assumed model properties [e.g. 83, 87], others provide only limited support in this regard. Rather than assume a particular model structure, some notable examples [e.g. 37, 80] compare several possible configurations to arrive at the best representation. The articles likewise vary considerably by both the source of model inputs (see the ‘Model construction and parameterisation’ section) and the clarity with which the sources are reported. Readers should be provided with sufficient information to understand the model's limitations, including those arising from uncertain or estimated inputs [28]; parameter tables are used in many studies [e.g. 48, 72] and are a particularly transparent means of reporting input values.
The Good Research Practices in Modeling Task Force describes the importance of reporting uncertainty analyses in modeling studies [26, 27]. As a complement to uncertainty or sensitivity analyses, ‘scenario analyses’ report alternative outcomes under discrete assumptions (i.e. values) for a parameter of primary interest [27]. Only half of the included articles (n = 43) performed and clearly reported a deterministic or probabilistic sensitivity analyses [e.g. 99, 104]. In many articles, it is difficult to distinguish sensitivity analyses from attempts to examine the impact of interventions in unique ‘scenarios’. Sensitivity analyses are one component of the formal process for external validation [28], wherein a model's results are compared to actual event data. Simulated outcomes are compared to observed results in only 21% of the articles, suggesting that models in this domain are not routinely assessed for accuracy in making relevant predictions. Most examples in this dataset use the same source to estimate model parameters and to validate the model (i.e. dependent validation) [e.g. 63, 94], which is less rigorous than independent or blinded validation [28]. In the absence of a full sensitivity analysis and validation effort, it is a challenge for decision-makers to ascertain the value of the model for informing a course of action [27, 28].
Limitations and conclusions
Several limitations of this work deserve mentioning. In addition to the English language restriction, the team did not search the grey literature for non-peer-reviewed studies. The inadvertent exclusion of relevant search terms could have similarly impacted the number of database ‘hits’. For both reasons, it is possible that models fitting the inclusion criteria were overlooked in this review. Models specific to bacteria with non-inherited resistance (i.e. phenotypically resistant or ‘persister’ cells) rather than inherited resistance were likewise excluded from consideration [e.g. 127, 128]. The data charting questions were refined and updated over several iterations, but nevertheless, reflect in part the particular interests of this review team. The extent to which these models are constructed in line with best practices [26, 28] may require further scrutiny. Dynamic models capture the non-linear relationships and feedback loops characteristic of communicable disease spread. ABM are only infrequently used to model the AMU/AMR relationship at the population level, limiting their usefulness for decision support; the popularity of ABM is likely to increase as ‘big data’ becomes more readily accessible. Models investigating AMR in agricultural and/or environmental settings are critically underrepresented in this dataset, in spite of recent efforts to promote farm animal populations as models for disease transmission [129]. Finally, there is substantial room to improve the clarity and transparency of reporting in the AMR modelling literature.
Acknowledgement
This work was supported by Alberta Agriculture and Forestry (grant number 2015E017R).
Conflict of interest
None.
Supplementary material
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0950268818002091.
click here to view supplementary material
References
- 1.World Health Organization (2014) Antimicrobial Resistance: Global Report on Surveillance. Geneva, Switzerland: World Health Organization. [Google Scholar]
- 2.Cosgrove SE (2006) The relationship between antimicrobial resistance and patient outcomes: mortality, length of hospital stay, and health care costs. Clinical Infectious Diseases 42(Suppl. 2), S82-S89. [DOI] [PubMed] [Google Scholar]
- 3.O'Neill J (2014) Review on Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. London, UK: The Review on Antimicrobial Resistance. [Google Scholar]
- 4.de Kraker MEA, Stewardson AJ and Harbarth S (2016) Will 10 million people die a year due to antimicrobial resistance by 2050? PLoS Medicine 13, e1002184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Bank (2017) Drug-resistant Infections: a Threat to Our Economic Future (Report no. 114679). Washington, DC, USA: World Bank,.
- 6.Spellberg B, Bartlett JG and Glibert DN (2013) The future of antibiotics and resistance. New England Journal of Medicine 368, 299–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.World Health Organization. One health collaboration to combat antimicrobial resistance. Available at http://www.oie.int/eng/BIOTHREAT2015/Presentations/6_Awa_Aidara-Kane_presentation_10.pdf (Accessed 5 September 2017).
- 8.Silbergeld EK, Graham J and Price LB (2008) Industrial food animal production, antimicrobial resistance and human health. Annual Review of Public Health 29, 151–169. [DOI] [PubMed] [Google Scholar]
- 9.Landers TF et al. (2012) A review of antibiotic use in food animals: perspective, policy and potential. Public Health Reports 127, 4–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allen HK et al. (2010) Call of the wild: antibiotic resistance genes in natural environments. Nature Reviews Microbiology 8, 251–259. [DOI] [PubMed] [Google Scholar]
- 11.Littman JR (2014) Antimicrobial Resistance and Distributive Justice (dissertation). University College London, London, UK. [Google Scholar]
- 12.Gröhn YT (2015) Progression to multi-scale models and the application to food system intervention strategies. Preventive Veterinary Medicine 118, 238–246. [DOI] [PubMed] [Google Scholar]
- 13.Armeanu E, Bonten MJM and Weinstein RA (2005) Control of vancomycin-resistant enterococci: one size fits all? Clinical Infectious Diseases 41, 210–216. [DOI] [PubMed] [Google Scholar]
- 14.Bonten MJM, Austin DJ and Lipsitch M (2001) Understanding the spread of antibiotic resistant pathogens in hospitals: mathematical models as tools for control. Clinical Infectious Diseases 33, 1739–1746. [DOI] [PubMed] [Google Scholar]
- 15.Marshall DA et al. (2015) Applying dynamic simulation modeling methods in healthcare delivery research – the SIMULATE checklist: report of the ISPOR simulation modeling emerging good practices task force. Value in Health 18, 5–16. [DOI] [PubMed] [Google Scholar]
- 16.Grundmann H and Hellriegel B (2006) Mathematical modelling: a tool for hospital infection control. Lancet Infectious Diseases 6, 39–45. [DOI] [PubMed] [Google Scholar]
- 17.Lipsitch M and Samore MH (2002) Antimicrobial use and resistance: a population perspective. Emerging Infectious Diseases 8, 347–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith DL, Dushoff J and Morris JG Jr (2005) Agricultural antibiotics and human health. PLoS Medicine 2, e232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Temime L et al. (2008) The rising impact of mathematical modelling in epidemiology: antibiotic resistance research as a case study. Epidemiology and Infection 136, 289–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Opatowski L et al. (2013) Contributions of mathematical models to the understanding of the dynamics of nosocomial pathogens in hospitals. Journal des Anti-infectieux 15, 193–203. [Google Scholar]
- 21.Doan TN et al. (2014) Optimizing hospital infection control: the role of mathematical modeling. Infection Control and Hospital Epidemiology 35, 1521–1530. [DOI] [PubMed] [Google Scholar]
- 22.Austin DJ and Anderson RM (1999) Studies of antibiotic resistance within the patient, hospitals and the community using simple mathematical models. Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences 354, 721–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levin BR (2001) Minimizing potential resistance: a population dynamics view. Clinical Infectious Diseases 33(Suppl. 3), S161–S169. [DOI] [PubMed] [Google Scholar]
- 24.Levac D, Colquhoun H and O'Brien KK (2010) Scoping studies: advancing the methodology. Implementation Science 5, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arksey H and O'Malley L (2005) Scoping studies: towards a methodological framework. International Journal of Social Research Methodology 8, 19–32. [Google Scholar]
- 26.Pitman R et al. (2012) Dynamic transmission modeling: a report of the ISPOR-SMDM modeling good research practices task force-5. Value in Health 15, 828–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Briggs AH et al. (2012) Model parameter estimation and uncertainty: a report of the ISPOR-SMDM modeling good research practices task force working group-6. Value in Health 15, 835–842. [DOI] [PubMed] [Google Scholar]
- 28.Eddy DM et al. (2012) Model transparency and validation: a report of the ISPOR-SMDM modeling good research practices task force-7. Value in Health 15, 843–850. [DOI] [PubMed] [Google Scholar]
- 29.Kijowski DJ (2014) Responsible Conduct of Research with Computational Models and Simulations (thesis). University of Illinois at Urbana-Champaign, Champaign, IL, USA. [Google Scholar]
- 30.Opatowski L et al. (2011) Contribution of mathematical modeling to the fight against bacterial antibiotic resistance. Current Opinion in Infectious Diseases 24, 279–287. [DOI] [PubMed] [Google Scholar]
- 31.Spicknall IH et al. (2013) A modeling framework for the evolution and spread of antibiotic resistance: literature review and model categorization. American Journal of Epidemiology 178, 508–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arepeva M et al. (2015) What should be considered if you decide to build your own mathematical model for predicting the development of bacterial resistance? Recommendations based on a systematic review of the literature. Frontiers in Microbiology 6, 352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abatih EN et al. (2009) Impact of antimicrobial usage on the transmission dynamics of antimicrobial resistant bacteria among pigs. Journal of Theoretical Biology 256, 561–573. [DOI] [PubMed] [Google Scholar]
- 34.Graesbøll K et al. (2014) How fitness reduced, antimicrobial resistant bacteria survive and spread: a multiple pig – multiple bacterial strain model. PLoS ONE 9, e100458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jansen JP, Kumar R and Carmeli Y (2009) Cost-effectiveness evaluation of ertapenem versus piperacillin/tazobactam in the treatment of complicated intraabdominal infections accounting for antibiotic resistance. Value in Health 12, 234–244. [DOI] [PubMed] [Google Scholar]
- 36.Karlsson D (2011) Probabilistic network modelling of the impact of penicillin consumption on spread of pneumococci. Epidemiology and Infection 139, 1351–1360. [DOI] [PubMed] [Google Scholar]
- 37.Geli P et al. (2006) Modeling pneumococcal resistance to penicillin in southern Sweden using artificial neural networks. Microbial Drug Resistance 12, 149–157. [DOI] [PubMed] [Google Scholar]
- 38.Austin DJ et al. (1999) Vancomycin-resistant enterococci in intensive-care hospital settings: transmission dynamics, persistence, and the impact of infection control programs. Proceedings of the National Academy of Sciences of the United States of America 96, 6908–6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beardmore R and Pena-Miller R (2010) Rotating antibiotics selects optimally against antibiotic resistance. Mathematical Biosciences and Engineering 7, 527–552. [DOI] [PubMed] [Google Scholar]
- 40.Beardmore R and Pena-Miller R (2010) Antibiotic cycling versus mixing: the difficulty of using mathematical models to definitively quantify their relative merits. Mathematical Biosciences and Engineering 7, 923–933. [DOI] [PubMed] [Google Scholar]
- 41.Bergstrom CT, Lo M and Lipsitch M (2004) Ecological theory suggests that antimicrobial cycling will not reduce antimicrobial resistance in hospitals. Proceedings of the National Academy of Sciences of the United States of America 101, 13285–13290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bonhoeffer S, Zur Wiesch PA and Kouyos RD (2010) Rotating antibiotics does not minimize selection for resistance. Mathematical Biosciences and Engineering 7, 919–922. [DOI] [PubMed] [Google Scholar]
- 43.Campbell EM and Chao L (2014) A population model evaluating the consequences of the evolution of double-resistance and tradeoffs on the benefits of two-drug antibiotic treatments. PLoS ONE 9, e86971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chamchod F and Ruan S (2012) Modeling methicillin-resistant Staphylococcus aureus in hospitals: transmission dynamics, antibiotic usage and its history. Theoretical Biology and Medical Modelling 9, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chow K et al. (2011) Evaluating the efficacy of antimicrobial cycling programmes and patient isolation on dual resistance in hospitals. Journal of Biological Dynamics 5, 27–43. [DOI] [PubMed] [Google Scholar]
- 46.D'Agata EMC, Webb G and Horn M (2005) A mathematical model quantifying the impact of antibiotic exposure and other interventions on the endemic prevalence of vancomycin-resistant enterococci. Journal of Infectious Diseases 192, 2004–2011. [DOI] [PubMed] [Google Scholar]
- 47.D'Agata EMC et al. (2007) Modeling antibiotic resistance in hospitals: the impact of minimizing treatment duration. Journal of Theoretical Biology 249, 487–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.D'Agata EMC et al. (2012) Efficacy of infection control interventions in reducing the spread of multi-drug resistant organisms in the hospital setting. PLoS ONE 7, e30170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Domenech de Cellès M et al. (2013) Limits of patient isolation measures to control extended-spectrum beta-lactamase-producing Enterobacteriaceae: model-based analysis of clinical data in a pediatric ward. BMC Infectious Diseases 13, 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Friedman A, Ziyadi N and Boushaba K (2010) A model of drug resistance with infection by health care workers. Mathematical Biosciences and Engineering 7, 779–792. [DOI] [PubMed] [Google Scholar]
- 51.Grima DT, Webb GF and D'Agata EMC (2012) Mathematical model of the impact of a nonantibiotic treatment for Clostridium difficile on the endemic prevalence of vancomycin-resistant enterococci in a hospital setting. Computational and Mathematical Models in Medicine 2012, 605861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haber M, Levin BR and Kramarz P (2010) Antibiotic control of antibiotic resistance in hospitals: a simulation study. BMC Infectious Diseases 10, 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hurford A et al. (2012) Linking antimicrobial prescribing to antimicrobial resistance in the ICU: before and after an antimicrobial stewardship program. Epidemics 4, 203–210. [DOI] [PubMed] [Google Scholar]
- 54.Joyner ML, Manning CC and Canter BN (2012) Modeling the effects of introducing a new antibiotic in a hospital setting: a case study. Mathematical Biosciences and Engineering 9, 601–625. [DOI] [PubMed] [Google Scholar]
- 55.Kardas-Sloma L et al. (2011) Impact of antibiotic exposure patterns on selection of community-associated methicillin-resistant Staphylococcus aureus in hospital settings. Antimicrobial Agents and Chemotherapy 55, 4888–4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kardas-Sloma L et al. (2013) Antibiotic reduction campaigns do not necessarily decrease bacterial resistance: the example of methicillin-resistant Staphylococcus aureus. Antimicrobial Agents and Chemotherapy 57, 4410–4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kouyos RD, zur Wiesch PA and Bonhoeffer S (2011) On being the right size: the impact of population size and stochastic effects on the evolution of drug resistance in hospitals and the community. PLoS Pathogens 7, e1001334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kouyos RD, zur Wiesch PA and Bonhoeffer S (2011) Informed switching strongly decreases the prevalence of antibiotic resistance in hospital wards. PLoS Computational Biology 7, e1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kouyos R, Klein E and Grenfell B (2013) Hospital-community interactions foster coexistence between methicillin-resistant strains of Staphylococcus aureus. PLoS Pathogens 9, e1003134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Laxminarayan R and Brown GM (2001) Economics of antibiotic resistance: a theory of optimal use. Journal of Environmental Economics and Management 42, 183–206. [Google Scholar]
- 61.Lipsitch M, Bergstrom CT and Levin BR (2000) The epidemiology of antibiotic resistance in hospitals: paradoxes and prescriptions. Proceedings of the National Academy of Sciences of the United States of America 97, 1938–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Massad E, Lundberg S and Yang HM (1993) Modeling and simulating the evolution of resistance against antibiotics. International Journal of Bio-Medical Computing 33, 65–81. [DOI] [PubMed] [Google Scholar]
- 63.Massad E, Burattini MN and Coutinho FAB (2008) An optimization model for antibiotic use. Applied Mathematics and Computation 201, 161–167. [Google Scholar]
- 64.McBryde ES and McElwain DLS (2006) A mathematical model investigating the impact of an environmental reservoir on the prevalence and control of vancomycin-resistant enterococci. Journal of Infectious Diseases 193, 1473–1474. [DOI] [PubMed] [Google Scholar]
- 65.Obolski U and Hadany L (2012) Implications of stress-induced genetic variation for minimizing multidrug resistance in bacteria. BMC Medicine 10, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Obolski U, Stein GY and Hadany L (2015) Antibiotic restriction might facilitate the emergence of multi-drug resistance. PLoS Computational Biology 11, e1004340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Obolski U et al. (2016) Antibiotic cross-resistance in the lab and resistance co-occurrence in the clinic: discrepancies and implications in E. coli. Infection, Genetics and Evolution 40, 155–161. [DOI] [PubMed] [Google Scholar]
- 68.Reluga TC (2005) Simple models of antibiotic cycling. Mathematical Medicine and Biology 22, 187–208. [DOI] [PubMed] [Google Scholar]
- 69.Sébille V and Valleron AJ (1997) A computer simulation model for the spread of nosocomial infections caused by multidrug-resistant pathogens. Computers and Biomedical Research 30, 307–322. [DOI] [PubMed] [Google Scholar]
- 70.Sébille V, Chevret S and Valleron A (1997) Modeling the spread of resistant nosocomial pathogens in an intensive-care unit. Infection Control and Hospital Epidemiology 18, 84–92. [DOI] [PubMed] [Google Scholar]
- 71.Smith DL et al. (2003) Assessing risks for a pre-emergent pathogen: virginiamycin use and the emergence of streptogramin resistance in Enterococcus faecium. Lancet Infectious Diseases 3, 241–249. [DOI] [PubMed] [Google Scholar]
- 72.Suthar N et al. (2014) An individual-based model of transmission of resistant bacteria in a veterinary teaching hospital. PLoS ONE 9, e98589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Talaminos A et al. (2016) Modelling the epidemiology of Escherichia coli ST131 and the impact of interventions on the community and healthcare centres. Epidemiology and Infection 144, 1974–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Webb GF et al. (2005) A model of antibiotic-resistant bacterial epidemics in hospitals. Proceedings of the National Academy of Sciences of the United States of America 102, 13343–13348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yahdi M et al. (2012) Vancomycin-resistant enterococci colonization-infection model: parameter impacts and outbreak risks. Journal of Biological Dynamics 6, 645–662. [DOI] [PubMed] [Google Scholar]
- 76.zur Wiesch PA et al. (2014) Cycling empirical antibiotic therapy in hospitals: meta-analysis and models. PLoS Pathogens 10, e1004225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Austin DJ, Kakehashi M and Anderson RM (1997) The transmission dynamics of antibiotic-resistant bacteria: the relationship between resistance in commensal organisms and antibiotic consumption. Proceedings of the Royal Society B: Biological Sciences 264, 1629–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Austin DJ, Kristinsson KG and Anderson RM (1999) The relationship between the volume of antimicrobial consumption in human communities and the frequency of resistance. Proceedings of the National Academy of Sciences of the United States of America 96, 1152–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chan CH, McCabe CJ and Fisman DN (2012) Core groups, antimicrobial resistance and rebound in gonorrhoea in North America. Sexually Transmitted Infections 88, 200–204. [DOI] [PubMed] [Google Scholar]
- 80.Colijn C et al. (2010) What is the mechanism for persistent coexistence of drug-susceptible and drug-resistant strains of Streptococcus pneumoniae? Journal of the Royal Society Interface 7, 905–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Colijn C and Cohen T (2015) How competition governs whether moderate or aggressive treatment minimizes antibiotic resistance. eLife 4, e10559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Domenech de Celles M et al. (2011) Intrinsic epidemicity of Streptococcus pneumoniae depends on strain serotype and antibiotic susceptibility pattern. Antimicrobial Agents and Chemotherapy 55, 5255–5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fingerhuth SM et al. (2016) Antibiotic-resistant Neisseria gonorrhoeae spread faster with more treatment, not more sexual partners. PLoS Pathogens 12, e1005611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gao D, Lietman TM and Porco TC (2015) Antibiotic resistance as collateral damage: the tragedy of the commons in a two-disease setting. Mathematical Biosciences 263, 121–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gao X, Pan Q and He M (2015) Transmission dynamics of resistant bacteria in a predator-prey system. Computational and Mathematical Models in Medicine 2015, 638074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Handel A, Regoes RR and Antia R (2006) The role of compensatory mutations in the emergence of drug resistance. PLoS Computational Biology 2, e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hui BB et al. (2015) Exploring the benefits of molecular testing for gonorrhoea antibiotic resistance surveillance in remote settings. PLoS ONE 10, e0133202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jansen JP, Kumar R and Carmeli Y (2009) Accounting for the development of antibacterial resistance in the cost effectiveness of ertapenem versus piperacillin/tazobactam in the treatment of diabetic foot infections in the UK. Pharmacoeconomics 27, 1045–1056. [DOI] [PubMed] [Google Scholar]
- 89.Levin BR, Baquero F and Johnsen PJ (2014) A model-guided analysis and perspective on the evolution and epidemiology of antibiotic resistance and its future. Current Opinion in Microbiology 19, 83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McCormick AW et al. (2003) Geographic diversity and temporal trends of antimicrobial resistance in Streptococcus pneumoniae in the United States. Nature Medicine 9, 424–430. [DOI] [PubMed] [Google Scholar]
- 91.Mitchell PK, Lipsitch M and Hanage WP (2015) Carriage burden, multiple colonization and antibiotic pressure promote emergence of resistant vaccine escape pneumococci. Philosophical Transactions of the Royal Society of London Series B, Biological Sciences 370, 20140342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nielsen KL et al. (2012) Fitness cost: a bacteriological explanation for the demise of the first international methicillin-resistant Staphylococcus aureus epidemic. Journal of Antimicrobial Chemotherapy 67, 1325–1332. [DOI] [PubMed] [Google Scholar]
- 93.Opatowski L et al. (2008) Antibiotic innovation may contribute to slowing the dissemination of multiresistant Streptococcus pneumoniae: the example of ketolides. PLoS ONE 3, e2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Opatowski L et al. (2010) Antibiotic dose impact on resistance selection in the community: a mathematical model of beta-lactams and Streptococcus pneumoniae dynamics. Antimicrobial Agents and Chemotherapy 54, 2330–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Porco TC et al. (2012) When does overuse of antibiotics become a tragedy of the commons? PLoS ONE 7, e46505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Samore MH et al. (2006) Mechanisms by which antibiotics promote dissemination of resistant pneumococci in human populations. American Journal of Epidemiology 163, 160–170. [DOI] [PubMed] [Google Scholar]
- 97.Smith DL et al. (2002) Animal antibiotic use has an early but important impact on the emergence of antibiotic resistance in human commensal bacteria. Proceedings of the National Academy of Sciences of the United States of America 99, 6434–6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Temime L et al. (2003) Bacterial resistance to penicillin G by decreased affinity of penicillin-binding proteins: a mathematical model. Emerging Infectious Diseases 9, 411–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Temime L, Guillemot D and Boëlle PY (2004) Short- and long-term effects of pneumococcal conjugate vaccination of children on penicillin resistance. Antimicrobial Agents and Chemotherapy 48, 2206–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Temime L, Boëlle P and Thomas G (2005) Deterministic and stochastic modeling of pneumococcal resistance to penicillin. Mathematical Population Studies 12, 1–16. [Google Scholar]
- 101.Temime L et al. (2005) Penicillin-resistant pneumococcal meningitis: high antibiotic exposure impedes new vaccine protection. Epidemiology and Infection 133, 493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Trecker MA et al. (2015) Revised simulation model does not predict rebound in gonorrhoea prevalence where core groups are treated in the presence of antimicrobial resistance. Sexually Transmitted Infections 91, 300–302. [DOI] [PubMed] [Google Scholar]
- 103.Van Effelterre T et al. (2010) A dynamic model of pneumococcal infection in the United States: implications for prevention through vaccination. Vaccine 28, 3650–3660. [DOI] [PubMed] [Google Scholar]
- 104.Wang YC and Lipsitch M (2006) Upgrading antibiotic use within a class: tradeoff between resistance and treatment success. Proceedings of the National Academy of Sciences of the United States of America 103, 9655–9660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xiridou M et al. (2015) Public health measures to control the spread of antimicrobial resistance in Neisseria gonorrhoeae in men who have sex with men. Epidemiology and Infection 143, 1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Beams AB et al. (2016) Harnessing intra-host strain competition to limit antibiotic resistance: mathematical model results. Bulletin of Mathematical Biology 78, 1828–1846. [DOI] [PubMed] [Google Scholar]
- 107.Bonhoeffer S, Lipsitch M and Levin BR (1997) Evaluating treatment protocols to prevent antibiotic resistance. Proceedings of the National Academy of Sciences of the United States of America 94, 12106–12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Boni MF and Feldman MW (2005) Evolution of antibiotic resistance by human and bacterial niche construction. Evolution 59, 477–491. [PubMed] [Google Scholar]
- 109.Gomes ALC, Galagan JE and Segrè D (2013) Resource competition may lead to effective treatment of antibiotic resistant infections. PLoS ONE 8, e80775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Levin BR et al. (1997) The population genetics of antibiotic resistance. Clinical Infectious Diseases 24(Suppl. 1), S9–S16. [DOI] [PubMed] [Google Scholar]
- 111.Saddler CA et al. (2013) Epidemiological control of drug resistance and compensatory mutation under resistance testing and second-line therapy. Epidemics 5, 164–173. [DOI] [PubMed] [Google Scholar]
- 112.Stewart FM et al. (1998) The population genetics of antibiotic resistance II: analytic theory for sustained populations of bacteria in a community of hosts. Theoretical Population Biology 53, 152–165. [DOI] [PubMed] [Google Scholar]
- 113.Sun H, Lu X and Ruan S (2010) Qualitative analysis of models with different treatment protocols to prevent antibiotic resistance. Mathematical Biosciences 227, 56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Osgood N (2008) Using traditional and agent based toolsets for system dynamics: present tradeoffs and future evolution In Sterman J (ed.) Proceedings of the International Conference of the System Dynamics Society, pp. 21. Boston: Curran Associates, Inc. [Google Scholar]
- 115.Van Boeckel TP et al. (2015) Global trends in antimicrobial use in food animals. Proceedings of the National Academy of Sciences of the United States of America 112, 5649–5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Volkova VV et al. (2012) Mathematical model of plasmid-mediated resistance to ceftiofur in commensal enteric Escherichia coli of cattle. PLoS ONE 7, e36738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Volkova VV et al. (2013) Modelling dynamics of plasmid-gene mediated antimicrobial resistance in enteric bacteria using stochastic differential equations. Scientific Reports 3, 2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lanzas C et al. (2010) Transmission dynamics of a multidrug-resistant Salmonella typhimurium outbreak in a dairy farm. Foodborne Pathogens and Disease 7, 467–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Volkova VV et al. (2013) Evaluating targets for control of plasmid-mediated antimicrobial resistance in enteric commensals of beef cattle: a modelling approach. Epidemiology and Infection 141, 2294–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Garner MJ et al. (2015) An assessment of antimicrobial resistant disease threats in Canada. PLoS ONE 10, e0125155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Centers for Disease Control and Prevention (2013) Antibiotic Disease Threats in the United States, 2013. Atlanta, GA, USA: Centers for Disease Control. [Google Scholar]
- 122.Fishman N (2006) Antimicrobial stewardship. American Journal of Infection Control 34(Suppl.), S55-S63. [DOI] [PubMed] [Google Scholar]
- 123.Pelupessy I, Bonten MJM and Diekmann O (2002) How to assess the relative importance of different colonization routes of pathogens within hospital settings. Proceedings of the National Academy of Sciences of the United States of America 99, 5601–5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bootsma MCJ et al. (2007) An algorithm to estimate the importance of bacterial acquisition routes in hospital settings. American Journal of Epidemiology 166, 841–851. [DOI] [PubMed] [Google Scholar]
- 125.Raboud J et al. (2005) Modeling the transmission of methicillin-resistant Staphylococcus aureus among patients admitted to a hospital. Infection Control and Hospital Epidemiology 26, 607–615. [DOI] [PubMed] [Google Scholar]
- 126.Barnes SL et al. (2014) Preventing the transmission of multidrug-resistant organisms: modelling the relative importance of hand hygiene and environmental cleaning interventions. Infection Control and Hospital Epidemiology 35, 1156–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Levin BR and Rozen DE (2006) Non-inherited antibiotic resistance. Nature Reviews Microbiology 4, 556–562. [DOI] [PubMed] [Google Scholar]
- 128.Bootsma MCJ et al. (2012) Modeling non-inherited antibiotic resistance. Bulletin of Mathematical Biology 74, 1691–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lanzas C et al. (2010) Model or meal? Farm animal populations as models for infectious diseases of humans. Nature Reviews Microbiology 8, 139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0950268818002091.
click here to view supplementary material