High resolution structures reveal the mechanisms of sodium, potassium, and proton pumping by a light-driven microbial rhodopsin.
Abstract
Rhodopsins are the most universal biological light-energy transducers and abundant phototrophic mechanisms that evolved on Earth and have a remarkable diversity and potential for biotechnological applications. Recently, the first sodium-pumping rhodopsin KR2 from Krokinobacter eikastus was discovered and characterized. However, the existing structures of KR2 are contradictory, and the mechanism of Na+ pumping is not yet understood. Here, we present a structure of the cationic (non H+) light-driven pump at physiological pH in its pentameric form. We also present 13 atomic structures and functional data on the KR2 and its mutants, including potassium pumps, which show that oligomerization of the microbial rhodopsin is obligatory for its biological function. The studies reveal the structure of KR2 at nonphysiological low pH where it acts as a proton pump. The structure provides new insights into the mechanisms of microbial rhodopsins and opens the way to a rational design of novel cation pumps for optogenetics.
INTRODUCTION
The first microbial rhodopsin (bacteriorhodopsin) was discovered in the early 1970s in halophilic archaea (salt-loving microbes that live in saturated brines) (1). It was shown that bacteriorhodopsin is a light-driven proton pump used to generate a light-driven proton gradient as a source of energy for the cell (2). Soon after bacteriorhodopsin, two sensory rhodopsins (also known as slow rhodopsins) were found in the same microbe (Halobacterium salinarum) (3, 4). Then, again in the same organism, a fourth rhodopsin (5) was found that functions as an inward-directed chloride pump (halorhodopsin) that maintains the proper electrochemical balance (6). Because of their light activation via the chromophore retinal, all rhodopsins undergo a photocycle, which is connected to the function of these proteins. Therefore, rhodopsins played a key role in membrane protein research, extending our understanding of the molecular mechanisms of bioenergetics and transmembrane signaling and in the development of new biophysical, biochemical, and structural biology (x-ray crystallography and electron microscopy) methods, approaches, and potential applications (7).
In 2013, a new microbial rhodopsin was found in marine bacteria Krokinobacter eikastus, which can pump either lithium or sodium cations, but it pumps protons in the absence of these ions (8). This K. eikastus rhodopsin 2 (KR2) pumps sodium under physiological conditions (~500 mM NaCl and pH around 8 in the ocean) and belongs to a family of rhodopsins containing a unique NDQ motif, which is formed by the Asn112, Asp116, and Gln123 triad. The unique residue in Na+-pumping rhodopsins, Asp116, was suggested to serve as a proton acceptor from the retinal Schiff base (RSB) (8). The photocycle of KR2 indicated that Na+ uptake occurs with the rise of the O intermediate. It was shown by Fourier transform infrared spectroscopy, however, that the ion already binds to the protein in the resting state at pH 8.0 and that the binding site is located distant from the RSB region. In 2015, crystal structures of KR2 were obtained simultaneously by Kato et al. (9) and our group (10). All structures were determined using crystals initially grown at a pH not higher than 5.6. The structures of KR2 solved by both groups at acidic pH to 1.45 (pH 4.3) and 2.3 Å (pH 4.0) coincide, except for the presence of additional water molecules and the proposed earlier sodium ion bound to the protein surface in the model with higher resolution. The proteins in the crystals were in monomeric form. Optical properties of the protein are altered at acidic pH, and later in 2015, it was shown that KR2 pumps sodium ions and protons competitively and that the rate constant for Na+ uptake is much smaller than that of H+ (11). Under physiological conditions, when [Na+] is about 107 times higher than [H+], the protein almost exclusively pumps sodium ions, so we will call this KR2 form as a Na+-pumping form. Determining the structure of Na+-pumping form is of high importance for understanding the mechanism of sodium translocation. Previously, this was approached in two different ways. In (9), the structure was obtained at pH 7.5 to 8.5, with crystals grown at pH 4.0 and soaked in buffer solution with pH 8.0 to 9.0 (9). The only difference between the pH 4 and pH 7.5 to 8.5 structures is in their Asp116 side chain orientation. While at acidic pH, Asp116 does not interact with the RSB, at high pH, its side chain is flipped to the cytoplasmic side, creating a strong hydrogen bond with protonated RSB. Thus, this structure was claimed to be the model of the ground state of the Na+-pumping form of KR2. In (10), the crystals were grown at pH 4.9 and 5.6 (10). KR2 forms pentamers in the crystals with a sodium ion bound at the oligomerization interface. Asp116 is directly hydrogen bonded to RSB; however, it remains in the same orientation as at pH 4.3. In opposite, positions of the Asn112-Leu74 pair in KR2 protomers vary within a pentameric assembly between two extreme conformations. The first one is similar to that in the monomeric form, with one water molecule in the Schiff base region. In the second, the Asn112 side chain is flipped out of the RSB region, creating space for three water molecules between Asp116 and Asp251 residues. We named these conformations compact and expanded, respectively (12). Two different groups proposed two putative Na+-pumping mechanisms. In (9), it was suggested that after photon absorption, the chromophore isomerizes and the proton is moved from RSB to Asp116, followed by Asp116 side chain flip toward Asn112 and Ser70 in the M state, creating space for Na+ translocation near the deprotonated RSB to the Asp251 and Arg109 region in the O state. After that, the retinal reisomerizes and the proton is returned to RSB from Asp116. The importance of pentameric organization for Na+-pumping was stressed in (12), and the following mechanism was suggested. Photon absorption causes retinal isomerization, followed by H+ transport to the Asp116 side chain, neutralizing RSB in the M state. Na+ passes the deprotonated RSB with the rise of the O state, and the ion uptake occurs, when Asn112 is flipped out of the Schiff base region, so that the place for Na+ binding site is created near Asp251 and Asp116. After that, the proton is returned to RSB from Asp116, Na+ is released to the extracellular side, and Asn112 flips back toward the Asp116 and Ser70.
Recently, it was shown by high-speed atomic force microscopy (HS-AFM) and circular dichroism spectroscopy that KR2 forms pentamers in a biological membrane under both acidic and neutral conditions. It was suggested that the monomeric form of KR2 is the artifact of crystallization, corresponding to neither the H+ nor the Na+-pumping form of the protein (13).
Rational design of new cation pumps based on the KR2 structure has been actively performed, and it was shown that modification of the ion uptake cavity (particularly residues Asn61 and Gly263) leads to a change of protein selectivity (9, 10, 14). Recently, it was shown that a single mutation of the π-bulge of helix G of Dokdonia sp. PRO95 Na+-rhodopsin [comprising residues 253 to 257 in the case of KR2 (10) and also found in bacteriorhodopsin (15)] affects the selectivity of the protein (16). It was also proposed in (17) that Ser254 of that π-bulge of KR2 matches the Asn310 of the Na+ binding pocket of the heterotrimeric guanine nucleotide–binding protein–coupled receptor δ-opioid receptor [Protein Data Bank (PDB) 4N6H]. However, no structural information on any sodium pump mutants is available at the moment.
Thus, although a lot of functional and structural information of KR2 and other Na+- and K+-pumping rhodopsins has been obtained, it is not yet clear what the structure of the ion-pumping form of KR2 is and what mechanism lies behind Na+ translocation and the switch from the Na+-pumping form at physiological pH to the H+-pumping form at acidic pH. Resolving the existing discrepancy of structural data and deciphering the KR2 structure using crystals grown at pH 8.0 are crucial for revealing the sodium ion pumping mechanism and protein organization in its functional form. The need for a structure and a deep understanding of the mechanisms of cation pumping by microbial rhodopsins is also highly motivated by a recent demonstration that KR2 can be used as an optogenetic tool (9, 18). Thus, the structure will help to engineer new optogenetic tools.
RESULTS
Structure of Na+-pumping form of KR2
We present here the crystal structure of the KR2 ground state at 2.2-Å resolution obtained with the crystals grown at pH 8.0. The protein was crystallized using the in meso approach, similar to our previous work (10). The plate-like crystals of type I were red (fig. S5E) in color (maximum absorption at 528 nm), which corresponds to the absorption of light by the protein Na+-pumping form. These crystals contain one KR2 pentamer per asymmetric unit. Opposite to the previous results, all the five protomers in the asymmetric unit have the same conformation, which is similar to that of protomers A and D of the 4XTN model. Since the structure is pentameric, obtained at pH 8.0 and at a sufficient sodium concentration, we conclude that it represents the correct sodium-pumping conformation.
Each KR2 protomer has seven transmembrane helices, named A to G, which are connected by three intracellular loops, three cytoplasmic loops, and an N-terminal α helix, capping the protein. Retinal is in the all-trans configuration and covalently bound to Lys255 of the helix G via the Schiff base. The structure corresponds to the expanded conformation of KR2 (12). As in the previously determined structure of the expanded conformation of KR2 (10), there are three cavities present in the protomer (Fig. 1A).
Fig. 1. Architecture and cavities of KR2 protomers.
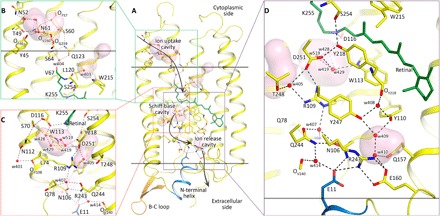
(A) Overall view. Helices F and G are not shown. (B) Detailed view of the cytoplasmic part of KR2 protomer. Helix G is shown with 90% transparency. (C) Detailed view of the Schiff base region. Helix B is shown with 90% transparency, and helix A is not shown. (D) Detailed view of the extracellular part of KR2 protomer. The hydrophobic membrane core boundaries were calculated using the PPM server (48) and are shown as solid horizontal lines. The cavities were calculated using HOLLOW (49) and are pink colored.
One of the cavities is placed at the cytoplasmic side (Fig. 1B). This water-filled cavity protrudes from the protein surface looking from the hydrophilic part of the lipid bilayer to Gln123. Mutagenesis shows that this cavity is the ion uptake vestibule (9, 10, 14). There is, however, no notable difference between structures of the cavity and that of the protein in the monomeric form. Gln123 is located at the level of hydrophobic/hydrophilic border of the protein surface and is separated from the Schiff base by the hydrophobic residues Val67 and Leu120 (Fig. 1B).
Second, there is a large cavity between the Schiff base and Arg109, which is surrounded by Trp113, Ser70, Asp116, Asp251, Asn112, Arg109, and Leu74. We named it the Schiff base cavity (12). It is filled with four water molecules (Fig. 1C). Three of the water molecules (Wat419, Wat428, and Wat429) are well ordered and were observed previously (10), unlike the fourth molecule Wat519. In the new structure, the presence of Wat519 is indicated by strong 4.0σ difference electron densities (fig. S10, B and C). However, the molecule is relatively poorly ordered, and its position slightly varies within the pentameric assembly. The RSB is hydrogen bonded to the Asp116 side chain, which is the primary acceptor of the proton. The Ser254 side chain is connected to the hydrogen bond network through Tyr218 and Asp251 side chains and therefore may play an important role in protein function (Fig. 1C).
Third, the ion-release cavity is placed in the extracellular part of the protein (Fig. 1D). Hydrogen bonds protrude from the Schiff base through Asp116, Asp251, Arg109, Gln244, Asn106, and water molecules to the ion-release cavity, separated from the bulk with the Arg243, Glu11, and Glu160 triad.
Pentamer oligomerization interface
Each pair of the protomers interacts through transmembrane helices B, C, D, A′, and B′ with an extended hydrogen bond network (Fig. 2). The connection between protomers at the cytoplasmic side is completely mediated by water molecules (Wat506′, Wat515′, Wat488′, and Wat434) (Fig. 2C). The hydrogen bond network at the extracellular side consists of two regions (Fig. 2, D and E). The view from outside of the pentamer shows tight connection of oxygen of His30′ with Asn112 side chain and Tyr108 oxygen through Wat401 and His30′ side chain with Tyr154 from helix D and Gln26′ from the same helix A′, which is further bonded to Glu22′. Both side chain and backbone of Glu22′ interact with Asn105, Asn7, and the backbone of the N terminus of the nearby protomer through water molecules Wat411, Wat412, Wat448′, and Wat491. Inside of the pentamer, Asn81 side chain interacts with Gln80′, Ser29′, Thr83′, Thr33′, and Tyr36′ side chains through several water molecules. Backbone atoms of N terminus and BC′-loop are also hydrogen bonded via Wat514′, stabilizing the structure. In addition, Tyr108 is directly hydrogen bonded to Ser29′. An important part of interprotomeric connection is the sodium ion binding site, comprising Asp102 and Tyr25′ side chains, Phe86′ and Thr83′ main chain oxygens, and Wat420′ and placed at the extracellular side of the protein interface.
Fig. 2. Overall architecture of KR2 pentamer and structure of the oligomerization interface.
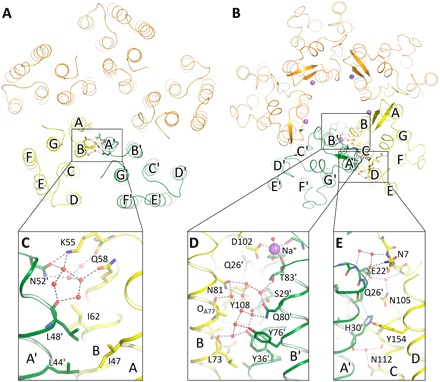
(A and B) View from the cytoplasmic and extracellular sides, respectively. Only contacts between chains A (yellow) and E (green) are shown. (C) Cytoplasmic side of the oligomerization interface. (D and E) Extracellular side of the oligomerization interface.
Comparison with known KR2 structures
The structure of the Na+-pumping form differs considerably from those obtained with the crystals grown at low pH. The structure is also very different from that obtained with crystals grown at pH 4.0 and soaked at pH 8.0 to 9.0 (Fig. 3) (9). It means that soaking of the crystals, which were grown at low pH, even at pH 8.0 to 9.0 cannot transfer the protein to Na+-pumping form in the case of KR2. A notable difference of monomeric and pentameric forms is the different orientation of the protein relative to membrane plane. In the monomeric form, the protein is tilted about 70° to the membrane plane. In opposite, the protein in the Na+-pumping pentameric form is oriented nearly perpendicular to the membrane plane. The considerable difference results in the position of ion uptake cavity relative to hydrophobic/hydrophilic membrane boundary. In Na+-pumping pentameric KR2, the cavity is placed in the hydrophilic part. In opposite, in monomeric KR2, the cavity is expected to be buried into the hydrophobic part (figs. S1 and S2).
Fig. 3. Comparison of Schiff base region of different KR2 structures.
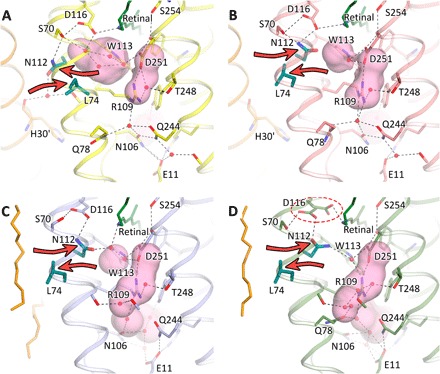
(A) Chain A of pentameric Na+-pumping form (expanded conformation, pH 8.0) is shown in yellow. (B) Chain E of 4XTN model (compact conformation, pH 4.9) is shown in salmon. (C) The 4XTL model (one of two closely related to compact conformations pH 4.3) is shown in light blue. (D) The 3X3C model (closely related to compact conformation, soaked at pH 8.0 to 9.0) is shown in green. The red dashed ellipse shows double conformation of the Asp116 side chain. Red contoured arrows show the important displacement of the Asn112-Leu74 pair (colored teal). Helix A′ of nearby protomer and fragments of lipid molecules are shown in orange for pentameric and monomeric models, respectively. The cavities are colored pink. The cartoon representation of helix A is hidden for clarity. A prosthetic group retinal is colored dark green.
The present structure allows the reinterpretation of solid state nuclear magnetic resonance (NMR), time-resolved absorption spectroscopy, and other data that recently have become available (19–22). The structure shows that the Asp116 orientation is uniform within the pentameric assembly under physiological conditions, which is in agreement with the observed homogeneity of the retinal-binding pocket at pH 8.0 (19, 20). Table S4 shows that in pentameric KR2 at pH 8.0, the Asp116 oxygen is 2.8 Å away from the Schiff base nitrogen and that in monomeric KR2 at neutral pH, two conformations were observed with the distance of 2.5 and 3.3 Å, respectively (9). In pentameric models at pH 4.9 and 5.6 (PDB: 4XTN and 4XTO, respectively), this distance varies within protomers from 2.9 to 3.9 Å (10).
In addition, opposite to the previously obtained structures, orientation of the two key residues, Asn112 and Leu74, is identical in all the five protomers and is the same for the expanded conformation of KR2 (12). The distance between the Cα atoms of Asn112 and Asp251 is 12.2 Å. The Asn112 side chain is stabilized by an additional hydrogen bond with the water molecule Wat401, which is present only in pentameric models and also makes hydrogen bonds with the main chain oxygens of Tyr108 and His30′ of the oligomerization interface (Fig. 3). Consequently, the Schiff base cavity, suggested to be the Na+ binding site in the O state (12), is considerably larger in the case of the expanded conformation of the KR2 protomer, while it is completely absent in the KR2 structure, obtained from crystals grown at pH 4.0 and soaked at pH 8.0 to 9.0 (Fig. 3).
The extracellular parts of the expanded and compact conformations in pentameric models of KR2 are nearly the same (fig. S3). On the contrary, there is a notable difference between pentameric and monomeric forms of KR2 in this part of the protein, mainly caused by the displacement of the helix C and the N-terminal helix. In particular, the size of ion-release cavity is larger in the case of monomeric form, and positions of the Glu11, Gln157, and Glu160 side chains differ considerably.
Structural basis of the switch from Na+- to H+-pumping forms
It was shown previously that KR2 pumps sodium ions when their concentration is much higher than that of protons, that is, at least at pH 6.4 to 8.0 (11). This means that under physiological conditions, KR2 is in the Na+-pumping form. However, no information on Na+ transport at acidic pH is available in literature. We studied the pumping activity of KR2 that was reconstituted into lipid vesicles at pH 8.0, 6.0, and 4.3 in the same manner, as described elsewhere (23, 24). The data indicate that KR2 is able to pump sodium ions at pH 6.0 and 8.0 and pumps protons at pH 4.3 (fig. S9). As the protein acts as the H+ pump at acidic pH even at physiological concentration of sodium, we define the low pH KR2 form as the H+-pumping form. In addition, optical properties of the protein are altered with the decrease of pH (8), and structural rearrangements in the retinal-binding pocket were recently reported (19). Structural insights into the nature of transitions in KR2 with the pH change may help to better understand the mechanism of Na+ pumping. To discover the dependence of KR2 conformations on the pH, we have determined crystal structures of KR2 using crystals initially grown at pH 8.0 and soaked for 48 hours in buffer solutions with pH 4.3, 5.0, 6.0, and 8.0 and in the buffer solution described in (9) (table S1).
After soaking at pH 6.0, crystals remain red and diffract up to 2.7 Å, revealing the same structure as at pH 8.0 (fig. S7F). This result is in accordance with solid-state NMR structural studies of the retinal-binding pocket, where no substantial changes were observed between pH 6.0 and 8.0 (19).
After soaking at pH 4.3, the structure of the KR2 pentameric form could not be determined. The crystals changed their color from red to purple (absorption maximum at 550 nm) (fig. S7H). However, they lost their quality during the soaking procedure. To decrease the stress on crystals, we soaked them at pH 4.3 with a very gradual exchange of the buffer during 48 hours. However, even such gentle manipulations led to a loss of diffraction quality; the resolution dropped to 7 Å, and the mosaicity was extremely high. Thus, we decided to soak crystals at a higher pH.
The structure of KR2 that was obtained with the crystals soaked at pH 5.0 was solved to 2.6 Å. Crystals also changed their color from red to purple after soaking (fig. S7G). The location of most of the residues remains the same as in the model corresponding to pH 8.0. However, the region comprising residues 109 to 115 of helix C is slightly displaced. The distance between the Asp116 oxygen and the Schiff base nitrogen is increased to 3.2 Å, and at pH 8.0, it is only 2.8 Å. The weakening of Asp116 interactions with RSB around pH 5.0 was also observed with the solid-state NMR study of KR2 retinal-binding pocket and was assigned to the protonation of Asp116 (19). The change in protonation of Asp116 can also be indicated by the shift of absorption maximum of KR2 in crystals from 528 to 550 nm. At the same time, Leu74 and Asn112 side chains occupy two alternative conformations: The first is the same as that in the expanded conformation of KR2, and the second is similar to that of the compact conformation of KR2 (Fig. 4B and fig. S10, D and E). Consequently, the Schiff base cavity is reduced in volume in the KR2 fraction at pH 5.0 (Fig. 4B).
Fig. 4. Schiff base region structures in pentameric KR2 at different pH.
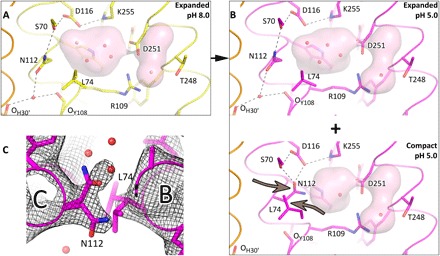
(A) Detailed view of the RSB region of KR2 at pH 8.0 (yellow). Hydrogen bonds, stabilizing the expanded conformation, are shown as black dashed lines. (B) Detailed view of the RSB region of KR2 at pH 5.0 (magenta). The two parts show expanded and compact conformations, which coexist in KR2 at pH 5.0. Arrows show the important displacement of the Asn112-Leu74 pair. Helix A′ of nearby protomer is shown in orange. Helices A and B are not shown. The cavities are colored pink. Hydrogen bonds, stabilizing the compact conformation, are shown as black dashed lines. (C) 2Fo-Fc electron density maps of the double conformation of the Asn112-Leu74 pair in the KR2 model at pH 5.0. The maps are contoured at the level of 1.0σ.
In summary, according to our functional and structural analysis at different pH, KR2 acts as a H+ pump at acidic pH and under the same conditions, a compact conformation of KR2 appears. Thus, we suggest that the compact conformation corresponds to the H+-pumping form. Therefore, we conclude that the switch from the expanded to the compact conformation results in the corresponding switch from the Na+- to H+-pumping forms in KR2.
Effects of dehydration on KR2 crystals
A previously published structure of the pentameric form of KR2 at pH 4.9 (PDB: 4XTN) contains protomers in both compact and expanded conformations, when the structure at pH 5.6 (PDB: 4XTO) contains only protomers in the compact conformation. We assume that this inconsistency is mainly due to the crystals drying, as the crystallization drop was not covered with additional buffer solution while fishing the crystals in our previous work. To check this hypothesis and resolve the controversy, we fished wizened crystals initially grown at pH 8.0, which also turned from red color to purple with time, without addition of the buffer solution. After that, we added buffer solution to the same drop and let it equilibrate for 30 min. Crystal color turned back to the red within 1 min. Last, we fished the soaked crystals and solved the structure of both pentameric “dry” and “wet” forms at 3.0 and 2.6 Å, respectively. The structure leaves no doubt that in the dry form, all the protomers inside the KR2 pentamer are present in the compact conformation, while after soaking, they switch back to the expanded one (fig. S8). The structure of the dry form is almost identical to the one deposited earlier (PDB: 4XTO). These observations stress the importance of humidity control during crystal growth and during their fishing.
The structure of KR2 monomeric form at different pH
To resolve the controversy of structural results obtained by two groups, we also studied structures of monomeric form at different pH. To find out the dependence of KR2 monomeric form conformations on pH, we determined crystal structures of KR2 using crystals that were initially grown at acidic pH 4.3 and blue-colored under the initial crystallization conditions (fig. S7A). These crystals were grown as described previously (10). Before fishing, crystals were soaked for 48 hours in buffer solutions with pH 4.3, 6.0, and 8.0 and in the buffer solution described in (9) (table S1).
All the structures are nearly the same as the existing model of the monomeric form of protein (PDB: 4XTL). There are no differences in the conformations of the residues, important for protein functioning. We did not observe that KR2 may occupy the expanded conformation when in the monomeric form. There is no flip of Asp116 at neutral pH even at 1.8 Å, as it was described in (9). It can be explained by the presence of radiation damage like in the case of bacteriorhodopsin (25–27) or protein disturbance due to soaking of the crystals initially grown at low pH. In that study, strong negative electron densities around the Asp116 residue were observed; however, no complementary positive densities were found (9).
Crystal structure of K+-pumping mutants
To better understand the mechanisms and to additionally verify the proposed key determinants of Na+ pumping, to and check whether they are also universal for pumping of other cations, we solved the structures of KR2 K+-pumping mutants.
Extensive mutational analysis of KR2 indicates that the S254A mutant has the capacity to pump not only protons and sodium but also potassium ions (fig. S4). Spectroscopic analysis also indicates the shift of maximum absorption wavelength from 525 to 545 nm in the case of this mutant. G263F was already demonstrated to pump K+ ions in our previous work (10). Here, we present structures of both monomeric and pentameric forms of the ground state of G263F and S254A mutants.
Monomeric blue-form structures of G263F and S254A were both determined at 2.0 and 2.1 Å, respectively, with the crystals grown at pH 4.3 and are nearly the same as that of monomeric wild-type KR2. The situation is the same with the pentameric form of the mutants. Structures were determined using crystals grown at pH 8.0, and the resolution in this case was 2.4 Å for both G263F and S254A mutants. Structures are very similar to that of the Na+-pumping form of KR2. The only differences are in the mutation regions (figs. S5 and S6). In the case of the G263F mutant, ion uptake cavity is dramatically decreased and separated from the solvent by the bulky side chain of Phe263 in both the monomeric and pentameric forms.
We observed the presence of a large Schiff base cavity filled with four water molecules in structures of the pentameric ion-pumping form. The cavity diminishes markedly in the monomeric form of both mutants because of the rearrangement of the Asn112-Leu74 pair in the same manner, as described for the wild-type protein (Fig. 5).
Fig. 5. Schiff base cavity in K+-pumping mutants of KR2.
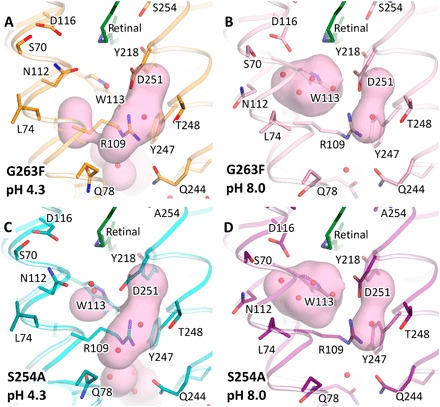
(A) Monomeric blue form of G263F. (B) Pentameric red form of G263F. (C) Monomeric blue form of S254A. (D) Pentameric red form of S254A. The cavities are colored pink. A prosthetic group retinal is colored dark green.
Role of the pentamerization on function of KR2
As structure of KR2 depends dramatically on the oligomeric state, we decided to mutate the oligomerization interface to destroy the pentameric assembly and simultaneously check the functional activity of the protein variants. Recently, it was shown that the alterations at the oligomerization interface around His30 affect the oligomeric assembly and change protein selectivity and Schiff base–counterion interaction (8, 20, 28). Consequently, we generated H30L, H30K, and Y154F mutants of KR2, which were aimed at breaking the His30-Tyr154 hydrogen bond, located in the middle of the oligomerization interface and therefore stabilizing the pentamer.
Size exclusion chromatography (SEC) profiles of detergent-purified protein in Fig. 6A demonstrate dependence of the wild-type KR2 oligomerization on pH. It qualitatively corresponds to that in the crystals. At pH 8.0, SEC profiles of the mutants in comparison with the wild type indicate that for H30K and Y154F mutants, pentameric assembly is negligible when H30L was notably unstable and had a large peak corresponding to the protein aggregates (Fig. 6B). Absence of pentameric organization was also recently demonstrated for Y154A (in detergent micelles) but not for the H30A mutant (20). Pumping activity studies in Escherichia coli cell suspension were done as described elsewhere (23) and showed that Na+ translocation is notably decreased in the case of H30K and Y154F, when it is completely absent for H30L (Fig. 6C). Y154F and H30K were successfully crystallized at pH 8.0, and the structures were determined at 1.8 and 2.2 Å, respectively. The proteins form monomers in the asymmetric unit, and the space group is the same as that of the monomeric form of wild-type KR2. Both structures do not differ from the structure of the monomeric form of KR2, except for the mutation region (Fig. 6, D to F).
Fig. 6. Effects of mutations of KR2 at oligomerization interface.
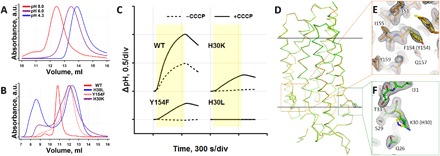
(A) SEC profiles of wild-type (WT) KR2 at pH 4.3, 6.0, and 8.0. a.u, arbitrary units. (B) SEC profiles of wild-type KR2 and its mutants at pH 8.0. Protein with an initial concentration of 70 mg/ml was dissolved in buffer solution with 0.1% n-dodecyl β-d-maltoside (DDM) to a final concentration of 1 to 2 mg/ml and incubated for 72 hours. (C) Na+-pumping activity of KR2 and its mutants measured in E. coli cell suspension. The solutions contain 100 mM NaCl (black and dashed) and 100 mM NaCl and 30 μM carbonyl cyanide m-chlorophenyl hydrazone (CCCP) (black and solid). The cells were illuminated for 300 s (yellow area on the plots). (D) Ribbon representation of the structure alignment of H30K (green) and Y154F (orange) mutants and wild type (yellow) of KR2. Side chains at positions 30 and 154 are shown as sticks. The hydrophobic membrane core boundaries are shown as solid horizontal lines. (E and F) Y154F and H30K mutation region aligned with the wild-type protein, respectively. 2Fo-Fc electron density maps are shown for the mutant structures and are contoured at the level of 1.5σ (Y154F) and 1.0σ (H30K).
These results evidence strong correlation between the oligomeric state and ion-pumping activity of KR2. The obstruction of the pentameric assembly, observed both in detergent and in crystals, leads to the decrease or complete absence of Na+ transport activity. Thus, we suggest that pentamerization is a key determinant of Na+-pumping activity of KR2.
DISCUSSION
Here, we presented a high-resolution structure of light-driven Na+-pump KR2 in its functional pentameric form. The structure of the KR2 Na+-pumping form suggests a putative ion translocation pathway. The first pore in the cytoplasmic part extending from the polar bulk to Gln123 serves as a vestibule for sodium translocation. Photon absorption leads to retinal isomerization from the all-trans to 13-cis configuration. In M intermediate, the proton from RSB is moved to Asp116, which is deprotonated and hydrogen bonded to RSB in the ground state. We suggest that the gate around Gln123 opens with the M-to-O transition and Na+ is translocated to the Schiff base cavity where it is bound transiently in the O state. Then, with the decay of the O intermediate, the Arg109 side chain is flipped to the extracellular side and opens the way for sodium translocation to the ion-release cavity. After that, the retinal reisomerizes and RSB reprotonates from Asp116. Na+ is translocated to the ion-release cavity, and Arg109 flips back, forming hydrogen bonds with the Asp251 side chain as in the ground state. Last, the sodium ion is released to the bulk through the Arg243, Glu11, and Glu160 cluster.
The structure of KR2 pentamer at pH 8.0 also shows that the five protomers in the pentamer are in the expanded conformation. The expanded conformations similar to that are found in the structures of all ion-pumping rhodopsins with the ion bound in the active center (12). We suggest that the expanded conformation is a key determinant of Na+ pumping and that, correspondingly, the large Schiff base cavity serves as a putative Na+ binding site in KR2 (29). The same enlarged cavity was found in the pentameric K+-pumping variants of KR2, stressing the universality of this determinant.
On the basis of our data, we propose a putative mechanism of the switch from Na+ pumping (at physiological pH) to H+ pumping (at acidic pH): The decrease in pH leads to a switch from the expanded to the compact conformations in pentameric KR2. This switch is possibly triggered by the protonation of Asp116 at low pH, which leads to the rearrangements of residues 109 to 116 of helix C, including Asn112. It results in the appearance of the compact conformation, in which the Asp116 side chain is hydrogen bonded with the Asn112 and Ser70 but not with the RSB, as it was also shown by the NMR studies of the retinal-binding pocket of KR2 (19). We believe that the decrease of the Schiff base cavity in compact conformation obstructs Na+ passage through the protein but does not interrupt H+ transport as the proton is smaller than the sodium ion. Consequently, KR2 acts as a H+ pump at acidic pH. We estimate the “switch pH” to be around 5.0, which is in agreement with the conducted studies of KR2 (19, 22).
As our data indicate low functional activity of the protein at acidic pH, we can speculate that the compact conformation does not correspond to the Na+- and/or H+-pumping form and may then be assigned as a low-pH inactive form of KR2. In this approximation, reduced pumping activity is provided by the existing fraction of the expanded conformation. The compact conformation may correspond to the structure of KR2 with protonated Asp116. It was shown that D116N mutant depicts the wild-type KR2 with the protonated Asp and does not pump either H+ or Na+ (8). Additional accurate functional and structural studies are needed to better understand the KR2 behavior at low pH.
The similarity of KR2 monomeric form structures at all pH levels has three possible explanations: first, the tight packing of proteins in crystals, which blocks any conformational rearrangements of the protein; second, the absence of any structural changes in the protein ground state at different pH levels; and third, monomeric form is present only in the crystals grown under acidic conditions and does not at all represent the H+ or Na+ transporting conformation. The latter assumption was recently corroborated by HS-AFM studies of KR2 reconstituted into lipids, which showed that the pentamers, not the monomers, are formed in a lipid bilayer at both pH 8.0 and 4.3 (13). Our additional studies of KR2 mutants with an eliminated pentamer formation showed that, unlike in bacteriorhodopsin (30) and other rhodopsins, the oligomeric organization is vital for protein function. Therefore, we suggest that the pentameric architecture of KR2 is essential for the formation and stabilization of the expanded conformation and the positioning of the entrance of the cytoplasmic pore to the hydrophilic part of the membrane and therefore is functionally important for ion translocation by the protein.
An interesting question that remains is the oligomeric state of the sodium pumps in native membranes. Closely related bacterial proton pumps, proteorhodopsins, were shown to be pentameric or hexameric in crystals (31) and in native-like membranes (32), whereas the best-known archaeal proton pump, bacteriorhodopsin, is trimeric (33). The current understanding of the role of oligomeric organization of microbial rhodopsins is limited; many pumps are also functional as monomers (34).
For KR2, unfortunately, neither the amount of proteins per single K. eikastus cell nor their dissociation constant is known. Thus, the oligomeric state of KR2 in native membranes cannot be predicted, although we expect it to be pentameric because close correspondence between oligomer formation in crystallographic structures and in vivo was shown for other pump rhodopsins (13). Yet another interesting question is whether the KR2 protomers get excited simultaneously or only a fraction of protomers is excited under natural light conditions. For bacteriorhodopsin, it was shown that, depending on light intensity, one, two, or all three protomers may be excited at the same time within the trimer (35). However, there are no data for KR2 at the moment.
After solving the structure of KR2 in its functional form, we believe that this pump can be useful as an optogenetic tool. The construction of structure-based new variants is now possible. Here, the K+-transporting variant might be of special interest because it could mimic, in a certain sense, the function of an outwardly directed K+ channel for the silencing of neurons by hyperpolarization of the cell in a minimally invasive manner.
MATERIALS AND METHODS
Expression plasmid
K. eikastus sodium-pumping rhodopsin gene (UniProt ID N0DKS8)–coding DNA was synthesized de novo. The nucleotide sequence was optimized for E. coli expression using the GeneOptimizer software (Life Technologies, USA). The gene was introduced into the pSCodon1.2 expression vector (Staby Codon T7, Eurogentec, Belgium) via Nde I and Xho I restriction sites. Consequently, the expressed construct harbored an additional C-terminal tag with a sequence LEHHHHHH.
Protein expression and purification
E. coli cells of strain SE1 (Staby Codon T7, Eurogentec, Belgium) were transformed with the KR2 expression plasmid. Transformed cells were grown at 37°C in shaking baffled flasks in an autoinducing medium ZYP-5052 (36) containing ampicillin (100 mg/liter). When glucose level in the growing bacterial culture dropped below 10 mg/liter, 10 μM all-trans retinal (Sigma-Aldrich, Germany) was added, the incubation temperature was reduced to 20°C, and incubation continued overnight. Collected cells were disrupted in an M-110P Lab Homogenizer (Microfluidics, USA) at 172.3 MPa in a buffer containing 20 mM tris-HCl (pH 8.0), 5% glycerol, 0.5% Triton X-100 (Sigma-Aldrich, USA), and deoxyribonuclease I (50 mg/liter) (Sigma-Aldrich, USA). The membrane fraction of cell lysate was isolated by ultracentrifugation at 90,000g for 1 hour at 4°C. The pellet was resuspended in a buffer containing 50 mM NaH2PO4/Na2HPO4 (pH 8.0), 0.1 M NaCl, and 1% DDM (Anatrace, Affymetrix, USA) and stirred overnight for solubilization. Insoluble fraction was removed by ultracentrifugation at 90,000g for 1 hour at 4°C. The supernatant was loaded on a Ni–nitrilotriacetic acid column (Qiagen, Germany), and KR2 was eluted in a buffer containing 50 mM NaH2PO4/Na2HPO4 (pH 7.5), 0.1 M NaCl, 0.5 M imidazole, and 0.1% DDM. The eluate was subjected to SEC on a 125-ml Superdex 200 prep grade column (GE Healthcare Life Sciences, USA) in a buffer containing 50 mM NaH2PO4/Na2HPO4 (pH 7.5), 0.1 M NaCl, and 0.05% DDM. Protein-containing fractions with the minimal A280/A525 absorbance ratio were pooled and concentrated to 60 mg/ml for crystallization.
Liposome preparation
Phospholipids (asolectin from soybean, Sigma-Aldrich) were dissolved in CHCl3 (chloroform ultrapure, PanReac AppliChem) and dried under a stream of N2 in a glass vial. Residual solvent was removed using a vacuum pump overnight. The dried lipids were resuspended at a final concentration of 1% (w/v) in 0.15 M NaCl supplemented with 2% (w/v) sodium cholate. The mixture was clarified by sonication at 4°C, and KR2 was added at a protein/lipid ratio of 7:100 (w/w). The detergent was removed by overnight stirring with detergent-absorbing beads (Amberlite XAD-2, Supelco). The mixture was dialyzed against 0.15 M NaCl (adjusted to a desired pH) at 4°C for 1 day (four 200-ml changes) to obtain a certain pH.
Spectroscopic characterization
Absorption spectra of KR2 in solution were collected using the UV-2401PC Spectrophotometer (Shimadzu, Japan). Absorption spectra of KR2 in crystals were collected at the ID-29S Cryobench laboratory at the European Synchrotron Radiation Facility (ESRF), Grenoble, France (37). Absorption maxima were determined by fitting the absorption spectra with skewed Gaussian functions.
Crystallization details and crystal preparation
The crystals were grown using the in meso approach (38, 39), similar to our previous work (23, 40–42). The solubilized protein in the crystallization buffer was added to the monooleoyl-formed lipidic phase (Nu-Chek Prep, USA). The best crystals were obtained using the protein concentration of 25 mg/ml. The blue and red crystals were grown using the precipitate 1.0 M sodium malonate (pH 4.3) and 1.2 M sodium malonate (pH 8.0), respectively (Hampton Research, USA). Crystallization probes were set up using the NT8 robotic system (Formulatrix, USA). The crystals were grown at 22°C and appeared in 2 to 4 weeks.
To obtain the structures of pentameric form of KR2 at different pH, the crystals that were initially grown at pH 8.0 were transferred from the crystallization drop to the bath filled with 3.4 M sodium malonate (pH 8.0). The pH was changed by iterative substitution of the buffer solution by 3.4 M sodium malonate at pH 4.3, 5.0, and 6.0. The crystals were equilibrated between iterations for at least 1 hour in the hermetic bath to avoid drying. The total time of soaking was not less than 48 hours.
To obtain the structures of monomeric form of KR2 at different pH, the crystals of KR2 that were initially grown at pH 4.3 were transferred from crystallization drop to the bath filled with 3.4 M sodium malonate (pH 4.3) (Hampton Research, USA). The crystals were the same as the type A crystals from our previous work (10). The soaking procedure was the same for red crystals. For the soaking, 3.4 M sodium malonate (pH 6.0), 8.0 and 42% polyethylene glycol (molecular weight 200), 0.1 M tris-HCl (pH 8.9), and 50 mM MgCl2 solutions were used. All crystals were harvested using MicroMounts (MiTeGen, USA) and were flash-cooled and stored in liquid nitrogen for further crystallographic analysis.
Acquisition and treatment of diffraction data
X-ray diffraction data (wavelengths 0.969 and 0.972 Å) were collected at the beamlines ID23-1 and ID29 of the ESRF using a PILATUS 6M detector. Diffraction images were processed using XDS (43). The reflection intensities were scaled using the SCALA software from the CCP4 program suite. There is no possibility of twinning for the crystals. Diffraction data from 13 crystals were used for the structure determination of the wild-type KR2 at pH 8.0. In all other cases, diffraction data from one crystal were used. The data statistics are presented in table S2.
Structure determination and refinement
Initial phases were successfully obtained in the C2221 space group by molecular replacement (MR) using MOLREP (44) using the chain A from 4XTN structure as a search model. Initial phases for pentameric KR2 at different pH were successfully obtained in the C2221 space group by MR using the crystal structure of KR2 at pH 8.0. Initial phases for monomeric KR2 at different pH were successfully obtained in the I222 space group by MR using the 4XTL structure as a search model. All models, which were used for MR, lacked side chains of residues Leu74, Arg109, Asn112, Asp116, and Asp251 to avoid appearance of bias electron densities. The initial MR models were iteratively refined using REFMAC5 (45), PHENIX (46), and Coot (47).
Measurements of the pump activity in E. coli
E. coli cells of strain C41(DE3) (Lucigen, USA) were transformed with the KR2 expression plasmid. Transformed cells were grown at 37°C in shaking baffled flasks in an autoinducing medium ZYP-5052 (36) containing ampicillin (100 mg/liter) and induced at the optical density OD600 of 0.6 to 0.7 with 1 mM isopropyl β-d-thiogalactopyranoside and 10 μM all-trans retinal. Three hours after the induction, the cells were collected by centrifugation at 3000g for 10 min and washed three times with unbuffered salt solution (100 mM NaCl or KCl and10 mM MgCl2) with 30-min intervals between the washes to allow exchange of the ions inside the cells with the bulk. After that, the cells were resuspended in 100 mM NaCl or KCl solution, respectively, and adjusted to an OD600 of 8.5. The measurements were performed on 3 ml of stirred cell suspension kept at 1°C. The cells were illuminated for 5 min using the halogen lamp Intralux 5000-1 (Volpi, Switzerland), and the light-induced pH changes were monitored with a pH meter (Lab 850, SCHOTT Instruments, Germany). Measurements were repeated under the same conditions after addition of 30 μM CCCP.
Measurements of the pump activity in liposomes
The measurements were performed on 2 ml of stirred proteoliposome suspension at 0°C. Proteoliposomes were illuminated for 18 min using a halogen lamp (Intralux 5000-1, Volpi) and then were kept in the dark for another 18 min. Changes in pH were monitored using a pH meter (Lab 850, SCHOTT Instruments). Measurements were repeated for different starting pH and in the presence of 30 μM CCCP under the same conditions.
Supplementary Material
Acknowledgments
We acknowledge the Structural Biology Group of the European Synchrotron Radiation Facility for granting access to the synchrotron beamlines. We are grateful to G. Gotthard, D. von Stetten, and A. Royant (ID29S Cryobench laboratory, ESRF) for help with collection of the KR2 crystal optical absorption spectra. Funding: This work was supported by the common program of Agence Nationale de la Recherche (ANR), France and Deutsche Forschungsgemeinschaft, Germany (ANR-15-CE11-0029-02), as well as Ministry of Education and Science of the Russian Federation (grant no. 6.3157.2017/PP) and by funding from Frankfurt: Cluster of Excellence Frankfurt Macromolecular Complexes (to E.B.) by the Max Planck Society (to E.B.) and by the Commissariat à l’Energie Atomique et aux Energies Alternatives (Institut de Biologie Structurale)–Helmholtz-Gemeinschaft Deutscher Forschungszentren (Forschungszentrum Jülich) Special Terms and Conditions 5.1 specific agreement. This work used the platforms of the Grenoble Instruct Centre (ISBG; UMS 3518 CNRS-CEA-UJF-EMBL) with support from the French Infrastructure for Integrated Structural Biology (ANR-10-INSB-05-02) and GRAL (ANR-10-LABX-49-01) within the Grenoble Partnership for Structural Biology. Data collection, data treatment, structure solution, and refinement as well as manuscript preparation were supported by RSF 16-15-00242. Author contributions: V.S., K.K., A.A., D.B., and S.V. expressed and purified the proteins. T.B. supervised the expression and purification; I.G. and V.P. proposed the S254A mutation. A.A. and K.K. performed functional studies. V.P. crystallized the protein and collected absorption spectra. K.K. and R.A. helped with crystallization. K.K. and V.P. collected the diffraction data and solved the structures. I.G. supervised structure refinement and analysis. D.B. and A.P. helped with data collection. V.B., V.C., E.B., and G.B. helped with data analysis. V.G. supervised the project. K.K., V.P., I.G., and V.G. analyzed the results and prepared the manuscript with input from all the other authors. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/4/eaav2671/DC1
Fig. S1. KR2 ion uptake pore.
Fig. S2. Comparison of orientations and cavities inside different KR2 structures.
Fig. S3. Comparison of extracellular regions of different KR2 structures.
Fig. S4. Activity tests of KR2 potassium-pumping mutants.
Fig. S5. Structural alignment of potassium-pumping KR2 mutants with the model of wild-type protein.
Fig. S6. Structures of potassium-pumping KR2 mutants.
Fig. S7. KR2 crystal soaking.
Fig. S8. KR2 dry and wet forms.
Fig. S9. Activity tests of KR2 at different pH.
Fig. S10. Electron density maps of KR2 structures.
Table S1. Summary information of crystal structures obtained.
Table S2. Data collection and refinement statistics of the wild-type KR2.
Table S3. Data collection and refinement statistics of KR2 mutants.
Table S4. Distance between Asp116 oxygen and RSB nitrogen atoms in KR2 models.
REFERENCES AND NOTES
- 1.Oesterhelt D., Stoeckenius W., Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nat. New Biol. 233, 149–152 (1971). [DOI] [PubMed] [Google Scholar]
- 2.Racker E., Stoeckenius W., Reconstitution of purple membrane vesicles catalyzing light-driven proton uptake and adenosine triphosphate formation. J. Biol. Chem. 249, 662–663 (1974). [PubMed] [Google Scholar]
- 3.Hildebrand E., Dencher N., Two photosystems controlling behavioural responses of Halobacterium halobium. Nature 257, 46–48 (1975). [DOI] [PubMed] [Google Scholar]
- 4.Takahashi T., Tomioka H., Kamo N., Kobatake Y., A photosystem other than PS370 also mediates the negative phototaxis of Halobacterium halobium. FEMS Microbiol. Lett. 28, 161–164 (1985). [Google Scholar]
- 5.Matsuno-Yagi A., Mukohata Y., Two possible roles of bacteriorhodopsin; a comparative study of strains of Halobacterium halobium differing in pigmentation. Biochem. Biophys. Res. Commun. 78, 237–243 (1977). [DOI] [PubMed] [Google Scholar]
- 6.Schobert B., Lanyi J. K., Halorhodopsin is a light-driven chloride pump. J. Biol. Chem. 257, 10306–10313 (1982). [PubMed] [Google Scholar]
- 7.I. Gushchin, V. Gordeliy, Microbial rhodopsins, in Membrane Protein Complexes: Structure and Function, J. Robin Harris, Egbert J. Boekema, Eds. (Springer, 2018), pp. 19–56. [Google Scholar]
- 8.Inoue K., Ono H., Abe-Yoshizumi R., Yoshizawa S., Ito H., Kogure K., Kandori H., A light-driven sodium ion pump in marine bacteria. Nat. Commun. 4, 1678 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Kato H. E., Inoue K., Abe-Yoshizumi R., Kato Y., Ono H., Konno M., Hososhima S., Ishizuka T., Hoque M. R., Kunitomo H., Ito J., Yoshizawa S., Yamashita K., Takemoto M., Nishizawa T., Taniguchi R., Kogure K., Maturana A. D., Iino Y., Yawo H., Ishitani R., Kandori H., Nureki O., Structural basis for Na+ transport mechanism by a light-driven Na+ pump. Nature 521, 48–53 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Gushchin I., Shevchenko V., Polovinkin V., Kovalev K., Alekseev A., Round E., Borshchevskiy V., Balandin T., Popov A., Gensch T., Fahlke C., Bamann C., Willbold D., Büldt G., Bamberg E., Gordeliy V., Crystal structure of a light-driven sodium pump. Nat. Struct. Mol. Biol. 22, 390–395 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Kato Y., Inoue K., Kandori H., Kinetic analysis of H+–Na+ selectivity in a light-driven Na+-pumping rhodopsin. J. Phys. Chem. Lett. 6, 5111–5115 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Gushchin I., Shevchenko V., Polovinkin V., Borshchevskiy V., Buslaev P., Bamberg E., Gordeliy V., Structure of the light-driven sodium pump KR2 and its implications for optogenetics. FEBS J. 283, 1232–1238 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Shibata M., Inoue K., Ikeda K., Konno M., Singh M., Kataoka C., Abe-Yoshizumi R., Kandori H., Uchihashi T., Oligomeric states of microbial rhodopsins determined by high-speed atomic force microscopy and circular dichroic spectroscopy. Sci. Rep. 8, 8262 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Konno M., Kato Y., Kato H. E., Inoue K., Nureki O., Kandori H., Mutant of a light-driven sodium ion pump can transport cesium ions. J. Phys. Chem. Lett. 7, 51–55 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Luecke H., Schobert B., Richter H.-T., Cartailler J.-P., Lanyi J. K., Structure of bacteriorhodopsin at 1.55 Å resolution. J. Mol. Biol. 291, 899–911 (1999). [DOI] [PubMed] [Google Scholar]
- 16.Mamedov A. M., Bertsova Y. V., Anashkin V. A., Mamedov M. D., Baykov A. A., Bogachev A. V., Identification of the key determinant of the transport promiscuity in Na+-translocating rhodopsins. Biochem. Biophys. Res. Commun. 499, 600–604 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Shalaeva D. N., Galperin M. Y., Mulkidjanian A. Y., Eukaryotic G protein-coupled receptors as descendants of prokaryotic sodium-translocating rhodopsins. Biol. Direct 10, 63 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grimm C., Silapetere A., Vogt A., Bernal Sierra Y. A., Hegemann P., Electrical properties, substrate specificity and optogenetic potential of the engineered light-driven sodium pump eKR2. Sci. Rep. 8, 9316 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shigeta A., Ito S., Inoue K., Okitsu T., Wada A., Kandori H., Kawamura I., Solid-state nuclear magnetic resonance structural study of the retinal-binding pocket in sodium ion pump rhodopsin. Biochemistry 56, 543–550 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Kaur J., Kriebel C. N., Eberhardt P., Jakdetchai O., Leeder A. J., Weber I., Brown L. J., Brown R. C. D., Becker-Baldus J., Bamann C., Wachtveitl J., Glaubitz C., Solid-state NMR analysis of the sodium pump Krokinobacter rhodopsin 2 and its H30A mutant. J. Struct. Biol. (2018). [DOI] [PubMed] [Google Scholar]
- 21.Tahara S., Takeuchi S., Abe-Yoshizumi R., Inoue K., Ohtani H., Kandori H., Tahara T., Ultrafast photoreaction dynamics of a light-driven sodium-ion-pumping retinal protein from Krokinobacter eikastus revealed by femtosecond time-resolved absorption spectroscopy. J. Phys. Chem. Lett. 6, 4481–4486 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Tahara S., Takeuchi S., Abe-Yoshizumi R., Inoue K., Ohtani H., Kandori H., Tahara T., Origin of the reactive and nonreactive excited states in the primary reaction of rhodopsins: pH dependence of femtosecond absorption of light-driven sodium ion pump rhodopsin KR2. J. Phys. Chem. B 122, 4784–4792 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Shevchenko V., Mager T., Kovalev K., Polovinkin V., Alekseev A., Juettner J., Chizhov I., Bamann C., Vavourakis C., Ghai R., Gushchin I., Borshchevskiy V., Rogachev A., Melnikov I., Popov A., Balandin T., Rodriguez-Valera F., Manstein D. J., Bueldt G., Bamberg E., Gordeliy V., Inward H+pump xenorhodopsin: Mechanism and alternative optogenetic approach. Sci. Adv. 3, e1603187 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H., Sineshchekov O. A., Da Silva G. F. Z., Spudich J. L., In vitro demonstration of dual light-driven Na+/H+ pumping by a microbial rhodopsin. Biophys. J. 109, 1446–1453 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borshchevskiy V., Round E., Erofeev I., Weik M., Ishchenko A., Gushchin I., Mishin A., Willbold D., Büldt G., Gordeliy V., Low-dose X-ray radiation induces structural alterations in proteins. Acta Crystallogr. D Biol. Crystallogr. 70, 2675–2685 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Borshchevskiy V. I., Round E. S., Popov A. N., Büldt G., Gordeliy V. I., X-ray-radiation-induced changes in bacteriorhodopsin structure. J. Mol. Biol. 409, 813–825 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Matsui Y., Sakai K., Murakami M., Shiro Y., Adachi S.-i., Okumura H., Kouyama T., Specific damage induced by X-ray radiation and structural changes in the primary photoreaction of bacteriorhodopsin. J. Mol. Biol. 324, 469–481 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Shigeta A., Ito S., Kaneko R., Tomida S., Inoue K., Kandori H., Kawamura I., Long-distance perturbation on Schiff base–counterion interactions by His30 and the extracellular Na+-binding site in Krokinobacter rhodopsin 2. Phys. Chem. Chem. Phys.. 20, 8450–8455 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Abe-Yoshizumi R., Inoue K., Kato H. E., Nureki O., Kandori H., Role of Asn112 in a light-driven sodium ion-pumping rhodopsin. Biochemistry 55, 5790–5797 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Dencher N. A., Heyn M. P., Bacteriorhodopsin monomers pump protons. FEBS Lett. 108, 307–310 (1979). [DOI] [PubMed] [Google Scholar]
- 31.Ran T., Ozorowski G., Gao Y., Sineshchekov O. A., Wang W., Spudich J. L., Luecke H., Cross-protomer interaction with the photoactive site in oligomeric proteorhodopsin complexes. Acta Crystallogr. D Biol. Crystallogr. 69, 1965–1980 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Klyszejko A. L., Shastri S., Mari S. A., Grubmüller H., Muller D. J., Glaubitz C., Folding and assembly of proteorhodopsin. J. Mol. Biol. 376, 35–41 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Henderson R., Unwin P. N. T., Three-dimensional model of purple membrane obtained by electron microscopy. Nature 257, 28–32 (1975). [DOI] [PubMed] [Google Scholar]
- 34.Brown L. S., Ernst O. P., Recent advances in biophysical studies of rhodopsins – Oligomerization, folding, and structure. Biochim. Biophys. Acta Proteins Proteom. 1865, 1512–1521 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Komrakov A. Y., Kaulen A. D., M-decay in the bacteriorhodopsin photocycle: Effect of cooperativity and pH. Biophys. Chem. 56, 113–119 (1995). [DOI] [PubMed] [Google Scholar]
- 36.Studier F. W., Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 41, 207–234 (2005). [DOI] [PubMed] [Google Scholar]
- 37.von Stetten D., Giraud T., Carpentier P., Sever F., Terrien M., Dobias F., Juers D. H., Flot D., Mueller-Dieckmann C., Leonard G. A., de Sanctis D., Royant A., In crystallo optical spectroscopy (icOS) as a complementary tool on the macromolecular crystallography beamlines of the ESRF. Acta Crystallogr. D Biol. Crystallogr. 71, 15–26 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Landau E. M., Rosenbusch J. P., Lipidic cubic phases: A novel concept for the crystallization of membrane proteins. Proc. Natl. Acad. Sci. U.S.A. 93, 14532–14535 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caffrey M., Cherezov V., Crystallizing membrane proteins using lipidic mesophases. Nat. Protoc. 4, 706–731 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bratanov D., Balandin T., Round E., Shevchenko V., Gushchin I., Polovinkin V., Borshchevskiy V., Gordeliy V., An approach to heterologous expression of membrane proteins. The case of bacteriorhodopsin. PLOS ONE 10, e0128390 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gushchin I., Chervakov P., Kuzmichev P., Popov A. N., Round E., Borshchevskiy V., Ishchenko A., Petrovskaya L., Chupin V., Dolgikh D. A., Arseniev A. S., Kirpichnikov M., Gordeliy V., Structural insights into the proton pumping by unusual proteorhodopsin from nonmarine bacteria. Proc. Natl. Acad. Sci. U.S.A. 110, 12631–12636 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gordeliy V. I., Labahn J., Moukhametzianov R., Efremov R., Granzin J., Schlesinger R., Büldt G., Savopol T., Scheidig A. J., Klare J. P., Engelhard M., Molecular basis of transmembrane signalling by sensory rhodopsin II-transducer complex. Nature 419, 484–487 (2002). [DOI] [PubMed] [Google Scholar]
- 43.Kabsch W., XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vagin A., Teplyakov A., Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66, 22–25 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A., REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67 (Pt 4), 355–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Lomize M. A., Pogozheva I. D., Joo H., Mosberg H. I., Lomize A. L., OPM database and PPM web server: Resources for positioning of proteins in membranes. Nucleic Acids Res. 40, D370–D376 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ho B. K., Gruswitz F., HOLLOW: Generating accurate representations of channel and interior surfaces in molecular structures. BMC Struct. Biol. 8, 49 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/4/eaav2671/DC1
Fig. S1. KR2 ion uptake pore.
Fig. S2. Comparison of orientations and cavities inside different KR2 structures.
Fig. S3. Comparison of extracellular regions of different KR2 structures.
Fig. S4. Activity tests of KR2 potassium-pumping mutants.
Fig. S5. Structural alignment of potassium-pumping KR2 mutants with the model of wild-type protein.
Fig. S6. Structures of potassium-pumping KR2 mutants.
Fig. S7. KR2 crystal soaking.
Fig. S8. KR2 dry and wet forms.
Fig. S9. Activity tests of KR2 at different pH.
Fig. S10. Electron density maps of KR2 structures.
Table S1. Summary information of crystal structures obtained.
Table S2. Data collection and refinement statistics of the wild-type KR2.
Table S3. Data collection and refinement statistics of KR2 mutants.
Table S4. Distance between Asp116 oxygen and RSB nitrogen atoms in KR2 models.


