SUMMARY
Despite the known causality of copy number variations (CNVs) to human neurodevelopmental disorders, the mechanisms behind each genes’ contribution to the constellation of neural phenotypes remains elusive. Here, we investigated the 7q11.23 CNV, whose hemideletion causes Williams syndrome (WS), and uncovered mitochondrial dysfunction participates in WS pathogenesis. Dysfunction is facilitated in part by the 7q11.23 protein DNAJC30, which interacts with mitochondrial ATP synthase machinery. Removal of Dnajc30 in mice resulted in hypofunctional mitochondria, diminished morphological features of neocortical pyramidal neurons, and altered behaviors reminiscent of WS. The mitochondrial features are consistent with the decreased integrity of oxidative phosphorylation supercomplexes and ATP synthase dimers we observed in WS. Thus, we reveal DNAJC30 as a novel auxiliary component of ATP synthase machinery, and link mitochondrial maladies as underlying certain defects in brain development and function associated with WS.
Keywords: Neurodevelopment, neurodevelopmental disorders, Williams syndrome, intellectual disability, autism spectrum disorder, genomics, mitochondria, CNV, ATP synthesis, neocortex
INTRODUCTION
Copy number variations (CNVs) are major contributors to disorders of the nervous system including intellectual disability, autism spectrum disorder (ASD), and schizophrenia (SCZ) (Geschwind and Flint, 2015; Girirajan et al., 2011; Hu et al., 2014; Pescosolido et al., 2013; Sebat et al., 2007). A fundamental challenge to understanding these disorders is identifying critical genes within a CNV locus whose deletions or duplications drive specific neural phenotypes. This challenge is further complicated by the highly dynamic and precisely coordinated spatiotemporal transcriptional landscape of the human brain (Silbereis et al., 2016). Previous efforts to identify genotype-phenotype correlations have largely relied on clinical assessments of patients harboring partial CNV locus deletions or duplications of otherwise well characterized syndromes (Fusco et al., 2014; Hoeft et al., 2014). One of the best characterized CNVs and resulting syndromes is 7q11.23, where a hemizygous deletion of the 26–28 genes results in Williams-Beuren syndrome (WS) (Dutly and Schinzel, 1996; Ewart et al., 1993; Frangiskakis et al., 1996; Gilbert-Dussardier et al., 1995; Osborne et al., 1996; Perez Jurado et al., 1996) and their duplication is associated with ASD or SCZ (7q11dup) (Depienne et al., 2007; Merla et al., 2010; Mulle et al., 2014; Sanders et al., 2011; Somerville et al., 2005; Van der Aa et al., 2009).
The most prominent features of WS include cardiovascular, metabolic, and brain abnormalities (Beuren et al., 1962; Meyer-Lindenberg et al., 2006; Morris, 2010; Pober, 2010; Stinton et al., 2010; Williams et al., 1961). Specific metabolic deficits include diabetes, impaired glucose tolerance, and higher resting energy expenditure (Cherniske et al., 2004; Kaplan et al., 1998; Palacios-Verdu et al., 2015; Pober et al., 2010). Brain abnormalities include decreased cerebral volume, smaller neurons, and abnormal gyrification, (Galaburda et al., 2002; Jackowski et al., 2009; Meyer-Lindenberg et al., 2006; Reiss et al., 2000; Schmitt et al., 2001; Van Essen et al., 2006). While imaging studies have shown that several white matter axonal pathways, including long-range projection systems such as the anterior commissure and corticospinal tract, have an altered microstructure (Arlinghaus et al., 2011; Campbell et al., 2009; Faria et al., 2012; Fung et al., 2012; Haas et al.; Hoeft et al., 2007), it is also appreciated that the corpus callosum (CC) is smaller and abnormally shaped in WS (Faria et al., 2012; Luders et al., 2007; Marenco et al., 2007; Sampaio et al., 2013; Schmitt et al., 2001; Tomaiuolo et al., 2002). These anatomical abnormalities likely play a major role in WS manifestations of intellectual disability, increased anxiety, attention deficit hyperactivity disorder, and hypersociability (Meyer-Lindenberg et al., 2006).
The relationship between individual genes on 7q11.23 and the neuroanatomical and functional alterations as well as neuropsychiatric symptoms associated with WS, ASD, or SCZ are under investigation (Abbas et al., 2016; Barak and Feng, 2016). Systems biology approaches have made some progress in understanding the role of individual CNV-related genes in a range of neurodevelopmental disorders (Golzio et al., 2012; Malhotra and Sebat, 2012; Tebbenkamp et al., 2014). For example, one can identify misregulated transcripts from induced pluripotent stem (iPS) cells generated from 7q11.23 deletion and duplication patients (Adamo et al., 2015), and query the spatiotemporal profile of those transcripts within the normally developing human brain to potentially reveal effects of gene dosage on disease and neurodevelopmentally relevant processes. Combining such datasets has the potential to glean novel insights into several CNVs or disorders with multi-genic contributions (Gulsuner et al., 2013; Lin et al., 2015; Parikshak et al., 2013; Willsey et al., 2013). These strategies can provide some spatial and temporal resolution of disease processes, but still cannot reveal the role of individual genes. Pursuing genotype-phenotype correlations is a promising strategy to not only understand mechanisms of these disorders, but to make inroads towards identifying novel therapeutic interventions.
The goal of this study was to identify and functionally validate underexplored neurobiological processes uniquely affected in 7q11.23 syndromes. We employed systems biology analyses utilizing brain gene expression and protein-protein interaction datasets, a strategy which revealed molecular coordination and spatio-temporal specificity of gene expression within, and ontologies derived from, the 7q11.23 locus. By looking at both coordination within the locus and downstream ontologies we were able to identify mitochondrial processes as dysregulated in WS, as well as highlight the functionally uncharacterized gene DNAJC30 (previously known as WBSCR18). We describe the spatiotemporal expression profile of DNAJC30, and show in comprehensive detail the function of DNAJC30 - from whole brain to neural circuits and behavior to its molecular interactions. Specifically, DNAJC30 is enriched in developing and mature neurons where it interacts with the mitochondrial ATP synthase machinery and facilitates ATP synthesis. We also found that decreased DNAJC30 expression in mice correlates with altered mitochondrial and neuronal function and morphology. Finally, we observed that several aberrations seen in Dnajc30 knockout mice are also present in post-mortem brain tissue and primary cells from WS patients, thus validating the participation of DNAJC30 in WS phenotypes.
RESULTS
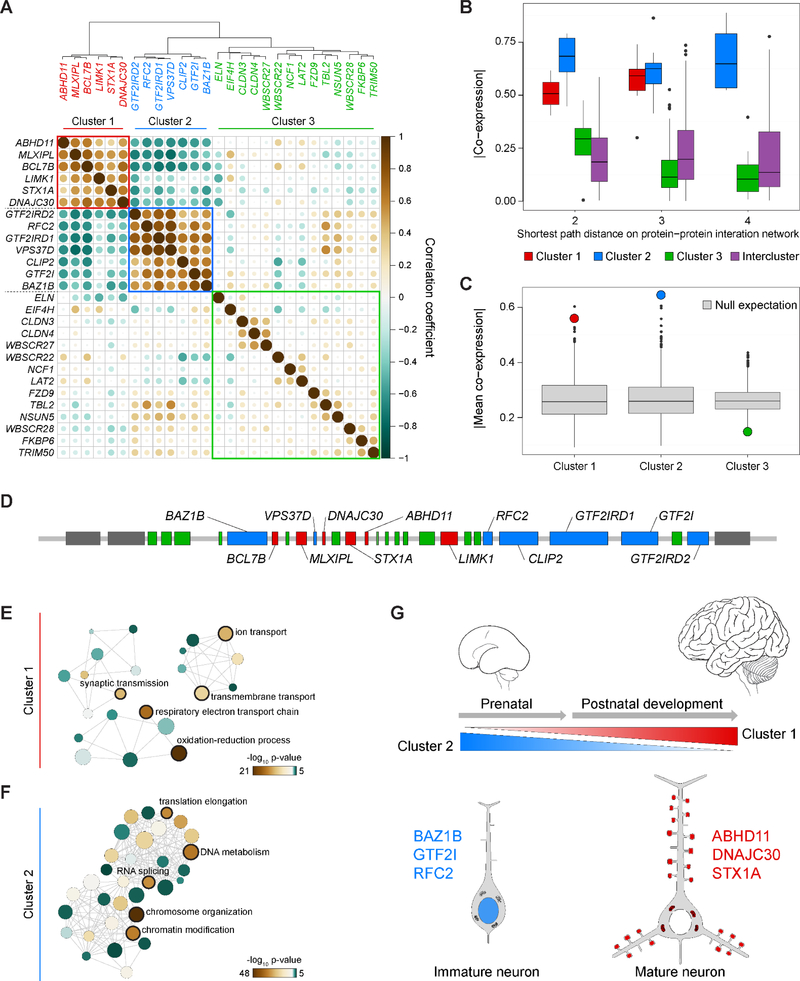
Integrated Co-Expression and PPI Analyses Identify Three 7q11.23 Gene Clusters with Distinct Functions
Our previous studies of gene expression across different regions of the developing human brain have revealed that the transcriptome can be cataloged into modules of highly co-expressed genes associated with distinct biological pathways and processes (Kang et al., 2011; Pletikos et al., 2014). Moreover, these and other studies have also shown that many genes associated with neurodevelopmental disorders are expressed in specific cell types and developmental periods, and are enriched in modules associated with specific pathways (Gulsuner et al., 2013; Lin et al., 2015; Parikshak et al., 2013; State and Sestan, 2012; Willsey et al., 2013). The human spatiotemporal expression patterns of the 26–28 protein coding genes within the 7q11.23 CNV have not been analyzed in great detail. Therefore, we analyzed RNA-seq data from the developing human brain, using the publicly available BrainSpan resource (www.brainspan.org). This transcriptomic resource was generated from 607 high-quality, histologically verified tissue samples representing sixteen regions (11 areas of the neocortex [NCX], hippocampus, amygdala, striatum, mediodorsal nucleus of thalamus, and cerebellar cortex) systematically dissected from 41 brains (18 females and 23 males) of clinically and histopathologically unremarkable donors of different ancestry, ranging in age from 5 postconception weeks (pcw) to 40 years (see BrainSpan resource for further information). The sampled brain regions are critically involved in perception, cognition, memory, emotional regulation, and complex behavior, and are affected in major neurological and psychiatric disorders.
We first calculated pairwise co-expression among all 7q11.23 genes followed by hierarchical clustering, from which we identified three clusters (Figure 1A). A cutoff of log2 (RPKM+1) ≥1 showed brain expression of five of six genes from cluster 1 and six of seven genes from cluster 2, while only six of 14 genes from cluster 3 were expressed (Figure S1A-S1B). To determine if each cluster was merely co-expressed or represented a deeper molecular coordination with interacting proteins, we looked at co-expression of each cluster with nearest neighbors on the BioGRID protein-protein interaction (PPI) network (Stark et al., 2006). No proteins encoded in 7q11.23 are known to directly interact with each other, but by extending the PPI network to second and third order interacting proteins, we still observed high co-expression for clusters 1 and 2, while cluster 3 and intercluster interacting proteins had much lower co-expression (Figure 1B). This molecular coordination of clusters 1 and 2 could not be fortuitous given that their co-expression was higher than that of randomly sampled genes across the genome with matched number of interacting proteins (Figure 1C). The coordination is also unlikely explained by genomic adjacency given the genes of any one cluster are scattered across the 7q11.23 locus (Figure 1D).
Figure 1. Top Functions of 7q11.23 Brain-Expressed Genes are Nuclear, Synaptic, and Mitochondrial.
(A) A dendrogram and heatmap of brain transcriptional similarities for all 7q11.23 genes highlights three clusters. (B) The co-expression of all genes in each cluster and their PPI network. Data are plotted as ±SEM and outliers as points. All of the cluster 1 proteins are connected by a distance of 3 on the PPI network. (C) The mean co-expression for clusters 1 and 2 are higher than predicted based on randomly selected genes having identical numbers of protein-protein interactors (null expectation). (D) Genes of the 7q11.23 locus are colored by cluster from part A, with gene names from clusters 1 and 2. (E-F) GO analysis of the top 1000 co-expressed genes for each gene in clusters 1 and 2, respectively. Larger nodes indicate higher term frequency. (G) Summary of Figure 1 and Supplementary Figure 1 showing the spatiotemporal contributions of cluster 1 and cluster 2 genes to neurodevelopmental processes. See also Tables S1–S4 and Figure S1.
We next sought to determine more specifically where and when the expression of genes within these clusters might have more pronounced effects. Using the GTEx dataset (GTEx Consortium, 2013), we found variations in cluster expression across non-central nervous system (CNS) tissues, but clusters 1 and 2 are enriched over cluster 3 in many adult brain regions (Figure S1C and other tissues not shown). Next, using BrainSpan data, we created four spatial groups ([1] neocortex (NCX), [2] hippocampus and amygdala, [3] striatum and thalamus, and [4] cerebellum) across nine time windows to look for cluster expression specificity, and found that whereas cluster 2 is enriched across all brain regions prenatally, cluster 1 is enriched in the NCX postnatally (Figure S1D). While the magnitude of this result is less after including PPI partners, the trend remains (Figure S1E). This result is consistent with the spatio-temporal enrichment observed previously (Lin et al., 2015), with slight differences in our results likely explained by our use of gene clusters, modified spatial groups, and specific interacting proteins.
We next focused on clusters 1 and 2 given their enriched expression in the human brain. To investigate the molecular functions within these clusters, we identified the top 1000 co-expressed genes for each gene in a cluster, and performed Gene Ontology (GO) enrichment analysis for each set of genes. The list of GO terms (FDR <0.01) was then combined and summarized using REViGO (Supek et al., 2011) to identify clustered and enriched GO terms. For cluster 2, we found enrichment for gene regulation and mitosis processes driven by BAZ1B, RFC2, and GTF2I (Figure 1F and Supplemental Tables 1–4). The transcription factor GTF2I has been reported to regulate some of the cognitive defects and separation anxiety in WS and ASD (Malenfant et al., 2012; Mervis et al., 2012; Morris et al., 2003). By contrast, enriched processes for cluster 1 were synaptic transmission and mitochondrial respiration, the latter being a novel potential participant of 7q11.23 phenotypes. (Figure 1E and Supplemental Tables 1–4). The genes driving the enrichment for these processes were STX1A, ABHD11, and DNAJC30, and this was consistent using either BrainSpan’s RNA-seq dataset or an independent exon array dataset from developmental human brains (Kang et al., 2011). This confluence of synaptic transmission and mitochondrial respiration ontologies is consistent with the high metabolic demand and necessarily coordinated calcium flux needed for proper neuronal excitation (Kwon et al., 2016).
Taking these analyses together, we provide evidence for prenatal gene regulation, postnatal synaptic transmission and mitochondrial respiration as potential contributors to 7q11.23 phenotypes (Figure 1G). Considering these processes prospectively as therapeutic targets, prenatal gene regulation is not ideal given the several layers of upstream gene regulation and downstream transcriptional targets. Furthermore, given that most 7q11.23 cases happen de novo and are not detected until after birth, that therapeutic option is impractical. Alternatively, these analyses also identified mitochondrial processes as potentially novel participants in 7q11.23 syndromes, and these processes exert their biggest effects after birth when therapeutic options are more tractable following a diagnosis. Among the genes associated with mitochondria GO terms, DNAJC30 (DnaJ (Hsp40) Homolog, Subfamily C, Member 30) was an intriguing candidate to pursue. DNAJC30 is of the J-domain containing family of proteins that have a vast array of functions (Kampinga and Craig, 2010), yet itself has an unknown function. Due to our motivation to identify unexplored mechanisms of 7q11.23 syndromes, and that DNAJC30 may play important roles in brain development, we chose to focus on unraveling the neurobiology of DNAJC30.
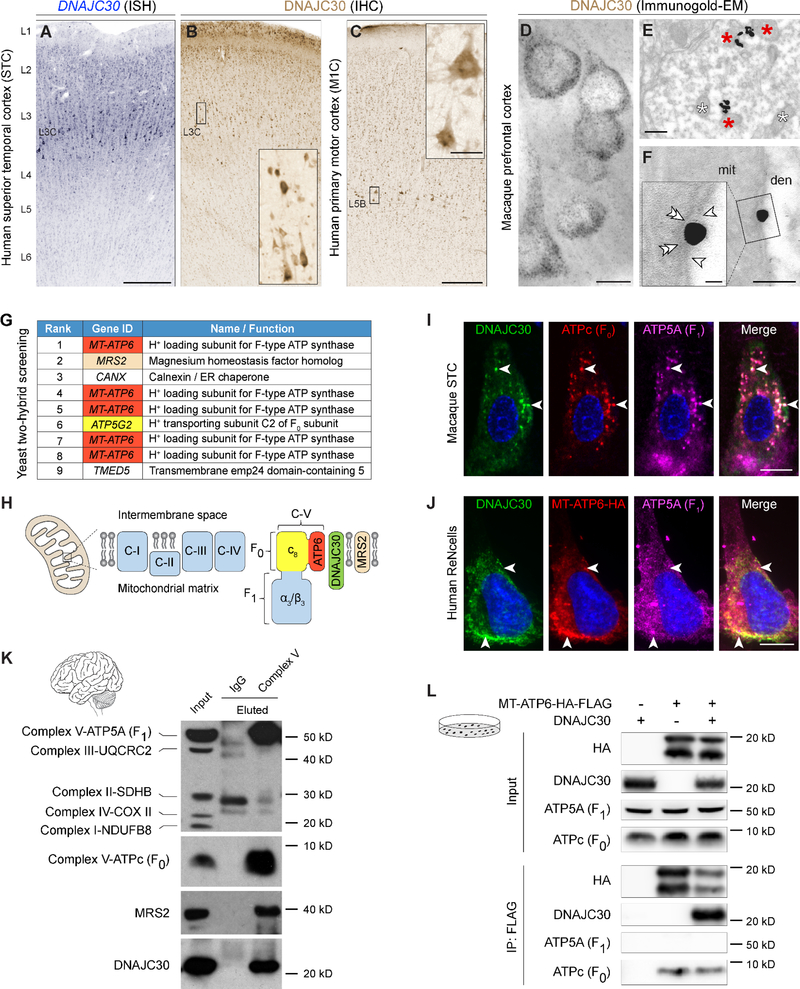
DNAJC30 Localizes to Mitochondrial ATP Synthase Machinery in Neocortical Neurons
Expression of DNAJC30 mRNA steadily increases from early human fetal development throughout the brain (Figure S2A). We confirmed the developmental increase of DNAJC30 protein by immunoblotting lysates from human superior temporal cortex (STC; the posterior part of Brodmann area 22) (Figure S2B), using that region because it was previously shown to have reduced asymmetric folding in WS patients (Van Essen et al., 2006). We found the onset of appreciable levels of DNAJC30 protein during the perinatal period (i.e., late fetal development to early infancy), which coincided with the peak period of human neocortical dendritogenesis and synaptogenesis (Silbereis et al., 2016) (Figure S2C). To identify which cells in the neocortex express DNAJC30, we performed in situ hybridization and detected mRNA in virtually all glutamatergic excitatory projection (i.e. pyramidal) neurons (PNs) and many interneurons with the strongest signal noticeably in large PNs of layer (L) 3C (Figure 2A). Using immunohistochemistry, we confirmed increased DNAJC30 immunoreactivity in large L3C PNs of the STC. Additionally, we observed the strongest immunoreactivity in large PNs (Betz cells) of L5B primary motor cortex (M1C) (Figure 2B-2C), but not L3 PNs in M1C, which are smaller in size compared to L3 PNs in STC and other association areas (Arion et al., 2015; Elston, 2003; Rivara et al., 2003). Interestingly, Betz cells are among the largest neurons in the central nervous system, and have the longest axons, which extend to the spinal cord and form the corticospinal tract, and is altered in WS (Faria et al., 2012; Marenco et al., 2007). Additionally, PNs in L3C of the association areas, such as STC, have the second largest somata in the NCX and form either ipsilateral or contralateral cortico-cortical projections, the latter of which comprise the anterior commissure or the CC, both of which are smaller in size in WS (Faria et al., 2012; Luders et al., 2007; Marenco et al., 2007; Sampaio et al., 2013; Schmitt et al., 2001; Tomaiuolo et al., 2002). Double immunofluorescence staining of STC showed that DNAJC30 is present in virtually all neurogranin-(NRGN) positive PNs (Figure S2D), as well as glutamic acid decarboxylase 1- (GAD1) positive inhibitory neurons (Figure S2E). In contrast, there was a paucity of DNAJC30 signal in GFAP-positive neocortical astrocytes or CNP-positive oligodendrocytes, suggesting DNAJC30 is not expressed in these cells or is below our level of detection (Figure S2F-S2G). Next, we performed immunogold electron microscopy (EM) to precisely identify the subcellular localization of neocortical DNAJC30. In both dendrites and somata of PNs, gold particles labeled the inner mitochondrial membrane (Figure 2D-2F), building upon recent high throughput studies that detected DNAJC30 in the mitochondrial proteome (Calvo et al., 2016; Huttlin et al., 2015) and supporting our hypothesis that mitochondria malfunction may contribute to 7q11.23 disorders.
Figure 2. DNAJC30 Interacts with Mitochondrial ATP Synthase of Neocortical Neurons.
(A) In situ hybridization for DNAJC30 in human superior temporal cortex (STC). (B-C) STC and human primary motor cortex (M1C), respectively, were immunolabeled against DNAJC30 and visualized with DAB. (D-E) Macaque dorsolateral prefrontal cortex labeled using anti-DNAJC30 and silver-enhanced 1.4nm gold. Red and white asterisks denote DNAJC30-reactive and unlabeled mitochondria, respectively. (F) High power electron micrograph captures DNAJC30 in the inner mitochondrial membrane (double arrowheads) where the outer mitochondrial membrane is fractured (arrowheads). (G) Results from a yeast-two-hybrid screen, ranked qualitatively by strength of interaction. (H) Illustration of a mitochondrion and proteins investigated at the inner mitochondrial membrane. (I-J) Double immunofluorescence staining of macaque STC and transfected the ReNcell human neocortical neural cell line, respectively, with white arrows showing co-localization of mitochondrial proteins. Nuclei (blue) are visualized with DAPI. (K) Immunoprecipitations and western blots of mitochondria isolated from human STC. (L) Immunoprecipitations and western blots of transfected HEK293T cells. Scale bars: A, 250μm; B-C, 500μm; inset, 50μm; D, 10μm; E, 200nm; F, 100nm; inset, 20nm; I-J, 10μm. mit, mitochondrion; den, dendrite. See also Figures S2-S3.
To investigate DNAJC30 interacting proteins, we performed a yeast-two-hybrid (Y2H) screen against an adult human brain library. The protein that interacted with DNAJC30 most frequently and with the highest affinity was the mitochondrially encoded ATP synthase 6 (MT-ATP6; also known as ATP6) (Figure 2G). MT-ATP6 is the H+_loading component of the F-type ATP synthase (Complex V), which is also located at the inner mitochondrial membrane, and is composed of the transmembrane H+-translocating F0 subunit and the ATP-generating F1 subunit in the matrix (Figure 2H) (Walker, 2013). Mutations in MT-ATP6 are associated with Leigh syndrome (de Vries et al., 1993), a condition characterized by progressive loss of mental and movement abilities (psychomotor regression) during early postnatal development, the developmental syndrome of neuropathy, ataxia, and retinitis pigmentosa (NARP) (Holt et al., 1990), and Charcot-Marie-Tooth progressive axonal neuropathy (Pitceathly et al., 2012). Another interacting protein from the Y2H screen was ATP5G2, which encodes a c-ring protein that is also part of the F0 subunit of ATP synthase, further suggesting DNAJC30 is a part of the mitochondrial ATP synthase complex. We then used double immunofluorescence staining in STC (Figure 2I), and in transfected human cells (Figure 2J and Figure S3A-S3C) to confirm DNAJC30 co-localization and co-fractionation with mitochondrial ATP synthase subunits. Of note, DNAJC30 is predicted to only exist in tetrapods, and while protein homology is high across mouse, cow, monkey, and human, some variations exist that may alter interactions or functions across species (Figure S3D-S3E). To further confirm the interaction between DNAJC30 and ATP synthase, we isolated mitochondria from adult human STC, and performed immunoprecipitation with an antibody against the F1 subunit of the intact ATP synthase. Whereas the control IgG did not pull-down any proteins of the ATP synthase complex, the ATP synthase antibody specifically captured proteins of both F0 and F1 subunits as well as DNAJC30 (Figure 2K), confirming its proximity to the mitochondrial ATP synthase complex. We were also able to confirm that another interacting protein from the Y2H screen, the magnesium transporter MRS2, also immunoprecipitated with ATP synthase and DNAJC30. This result was expected, in part, given that when ATP is synthesized, it is complexed with Mg2+ to make it biologically active. We also overexpressed MT-ATP6 either by itself, or in combination with DNAJC30, and found that MT-ATP6 was properly localized to the F0 subunit ATPc, and that MT-ATP6 co-immunoprecipated DNAJC30 (Figure 2L). These data use several complementary approaches to localize the interaction of DNAJC30 and mitochondrial ATP synthase in human neocortical neurons. Comparing the expression of DNAJC30 to ATP synthase and other genes in the oxidative phosphorylation (OXPHOS) pathway, we found DNAJC30 does not follow the trend of the majority of OXHPOS genes, which is to decrease their expression in late adulthood. Many other OXPHOS genes exhibit spatio-temporal specificity (Figure S2H-S2I).
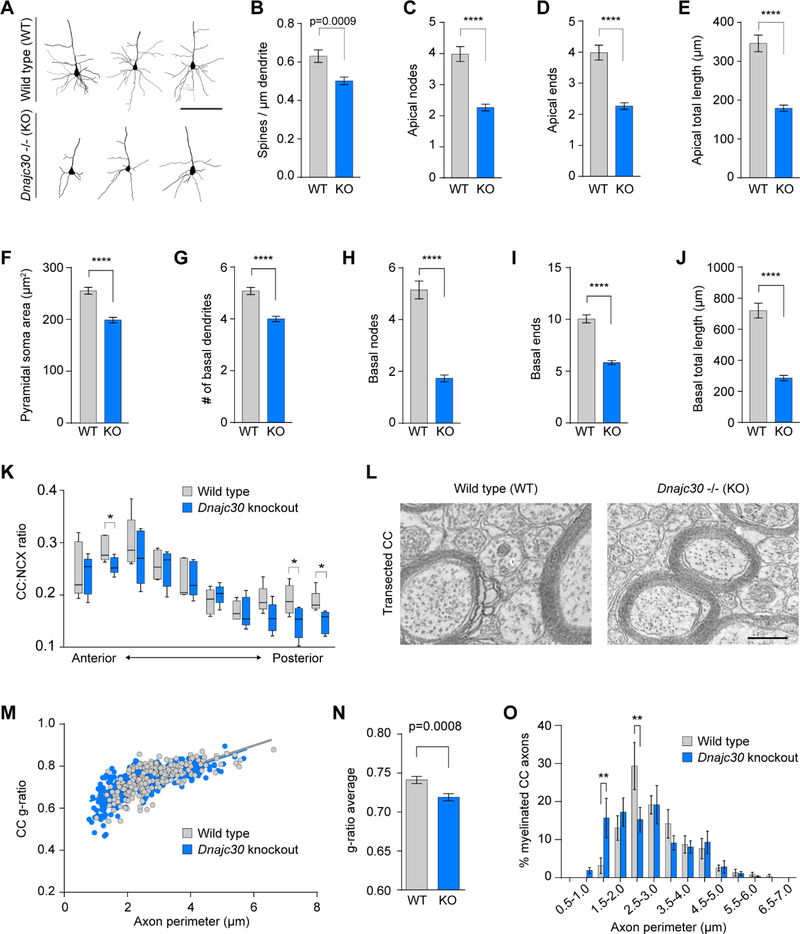
Morphological Defects of Neocortical Projection Neurons in the Dnajc30-Deficient Mouse
Having established that DNAJC30 is localized to ATP synthase, we next examined the effects of decreased Dnajc30 expression in knockout (KO) mice. We generated two independent lines of KO mice and confirmed their decreased gene expression by droplet digital PCR (ddPCR) (Figure S4A). These mice contained a β-galactosidase cassette in place of the single Dnajc30 exon and, using an X-gal stain to visualize Dnajc30 expression, we saw no reactivity in wildtype (WT) mice and increasing reactivity from heterozygotes to KO mice. X-gal staining of the forebrain revealed expression in gray matter structures, with the highest amount in the NCX. Within the NCX, X-gal reactivity was higher in L2–3 and L5 than in L4 and L6, consistent with the human expression pattern of more DNAJC30 expression in larger PNs. Also consistent with our previous human cell-type expression data, we observed X-gal in both NRGN-positive PNs and GAD1-immunopositive interneurons, but not in ALDH1L1-immunopositive astrocytes (Figure S4B-S4D). Macroscopic measurements revealed that KO mice display ~20% decrease in body weight beginning at postnatal days (P) 9–10 and persisting until at least 2 months old (Figure S4E and S4G). Similarly, extracted brains from both lines of these mice were also ~20% smaller in KO compared to WT littermates (Figure S4F). We performed all subsequent experiments on line 2, convinced that both lines were similar in expression and gross phenotypes. We also focused on comparisons between WT and KO mice given that the differences observed in heterozygous mice were not frequently significant (see also Figure S4H, explained below). Despite WS being a hemizygous genetic condition, we required a homozygous KO mouse to provide the necessary resolution to understand DNAJC30 function.
To investigate the effects of Dnajc30 on neuronal morphology, we performed Golgi staining and then used Neurolucida to reconstruct and quantify dendrites of PNs (Figure 3A). Morphometric analyses showed that KO mice have fewer spines on the principal apical dendrite (Figure 3B), while the total number of nodes, ends, and total length were also significantly reduced by roughly 50% (Figures 3C-3E). Additionally, the number of basal dendrites (Figure 3G), as well as the number of nodes, ends, and total dendritic length (Figures 3H-3J) were significantly reduced in KO mice, which also have smaller soma area (Figure 3F). These results are consistent with, and build upon, those found in mice hemizygous for all 26 WS genes (Li et al., 2009). Those mice exhibited craniofacial and behavioral defects, and like Dnajc30 KO mice, have smaller brains and increased neuronal packing which reflects smaller neuron size.
Figure 3. Dnajc30 Knockout Mice have Reduced Pyramidal Soma Area, Dendritic Morphology, and Callosal Axon Thickness.
(A) Reconstructed neurons stained using the Golgi-Cox technique (scale = 100μm). (B-J) Quantification of Golgi-Cox stained neuronal architecture. WT, n=64 neurons/4 mice; KO, n=93 neurons/6 mice. (K) Serial coronal brain sections (age P9–10) of corpus callosum:NCX thickness. n=5 for WT and KO. (L) Representative electron micrographs of transected CC (scale = 500nm). (M) CC g-ratio and axon perimeter distribution (age 3–4 weeks). (N) Average g-ratio between WT and KO mice (WT, n = 209 axons/5 mice; KO, n = 265 axons/5 mice). (O) Percentage distribution of axon perimeter populations. *, p<0.05; **, p<0.01; ****, p<0.0001. Statistical analyses were performed using student’s t-tests and multiple t-tests and corrections using the Holm-Sidak method. See also Figure S4.
We also bred Dnajc30 mice to Thy1-YFP-H reporter mice, to specifically label the soma and all processes of neocortical L5 PNs. L5 neocortical PNs have apical dendrites that can extend to L1 of the NCX, and axons that can project to subcerebral brain regions or intracortically, including via the corpus callosum (CC). To determine the effect of Dnajc30 levels on these specific neurons, we made serial coronal sections and measured soma size and thicknesses of the NCX, CC, and L5. While the overall thicknesses of the NCX, CC, and L5 trended towards a reduction, soma size was significantly reduced in KO mice, as was seen with Golgi measurements (data not shown). However, we found that KO mice had a disproportionately lower ratio of CC:NCX (Figure 3K). Given that this difference was at P9–10, an age before any appreciable myelination (Sturrock, 1980), it is most likely explained by fewer axons and/or thinner axon calibers. To determine whether axon calibers are altered after myelination, we performed electron microscopy (EM) on transected callosal axons of 3–4 week old mice (Figure 3L). At high magnification (9300x), we measured the g-ratio (axon perimeter/axon+myelin perimeter) and found it was significantly reduced in KO mice (Figure 3M-3N), confirming they have smaller axon calibers. This result was further supported by a significant and population-wide shift toward smaller axon perimeter in KO mice (Figure 3O). These data are noteworthy in that they reveal that Dnajc30 expression contributes to axonal caliber, and that its decreased expression causes a smaller corpus callosum, recapitulating one of the major neuroanatomical features of WS.
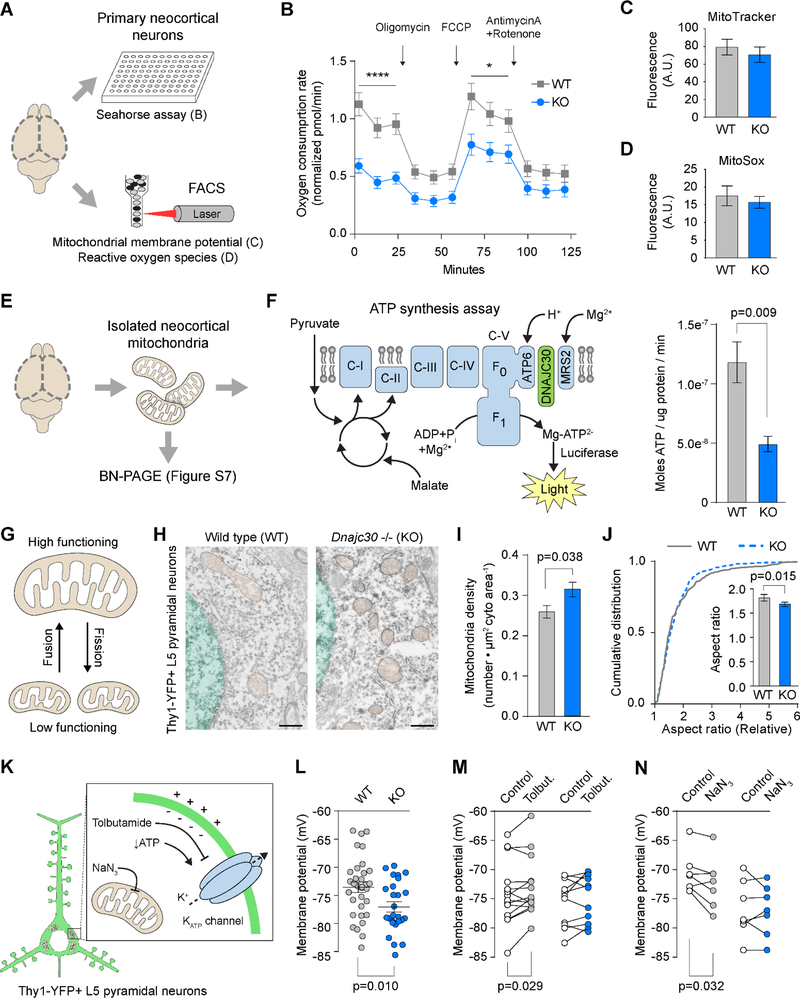
Dnajc30 is a Regulator of Mitochondrial Respiration
We have shown that Dnajc30 KO mice exhibit neuro-morphological defects. To explain these defects, we wanted to identify an upstream molecular contributor, and confirm that other cellular processes were affected accordingly. We analyzed cultured primary neurons for their oxygen consumption based on mitochondrial function, their mitochondrial membrane potential, and their reactive oxygen species (ROS) generation (Figure 4A). Using the Seahorse Mito Stress assay, we found basal oxygen consumption rate was significantly reduced in Dnajc30 KO neurons. We then sequentially added inhibitors to stress mitochondria, and while oligomycin and AntimycinA/Rotenone treatment (OXPHOS inhibiting drugs), revealed no differences between genotypes, FCCP treatment (maximizes mitochondrial respiration) revealed KO neurons cannot respire at as high of a rate as WT neurons (Figure 4B). We then measured the mitochondrial membrane potential and ROS production, byproducts of ATP synthesis that can function in a variety of pathways throughout the organism (Shadel and Horvath, 2015); finding however, no difference between genotypes in either test (Figure 4C-4D). Given cellular respiration is affected in KO neurons, we next sought to determine if ATP production was affected. Fresh mouse cortices were dissected from the brain followed by mitochondria extraction and analysis (Figure 4E), where ATP production was stimulated then quantified by a kinetic luciferase activity assay. In this minimalized system, we found ATP production is decreased in mitochondria without Dnajc30 (Figure 4F).
Figure 4. Hypofunctional and Dysmorphic Mitochondria in Neocortex of Dnajc30 Knockout Mice.
(A) Illustration of experiments utilizing P0 primary neocortical neurons. (B) Seahorse assay-generated oxygen consumption rates of neurons following pharmacological modifications (arrows). (C-D) Mitochondrial membrane potential and reactive oxygen species, respectively. (E) Illustration of mitochondria utilized for subsequent experiments. (F) Design and results of luciferase-based ATP synthesis assay. (G) Schematic showing mitochondrial dynamics. (H) Representative EM images from Thy-1-YFP-DAB-labeled L5 neurons. Mitochondria are shaded brown and the nucleus is shaded green (scale = 500nm). (I-J) Mitochondrial density and aspect ratio, respectively, were significantly different in KO mice. WT, n=394 mitochondria/25 cells/5 mice; KO, n=501 mitochondria/30 cells/6 mice. (K) Illustration of the ATP-sensitive K+ channel (blue) and effects of compounds used in this study. (L) Resting membrane potential (WT, n=33 cells/15 mice; KO, n=29 cells/17 mice). (M) Effect of tolbutamide (0.1mM) on membrane potential (WT, n=14 cells/8 mice; KO, n=11 cells/7 mice). (N) Effect of NaN3 (1mM) on membrane potential (WT, n=7 cells/4 mice; KO, n=7 cells/6 mice). *, p<0.05; ****, p<0.0001. Statistical analyses were performed using student’s t-tests and multiple t-tests and corrections using the Holm-Sidak method.
Having observed altered mitochondrial function in Dnajc30 KO mice, we next investigated corroborating features of altered function. We first examined mitochondrial morphology given the location of DNAJC30 and because its morphology reflects function (Figure 4G). We performed EM on L5 Thy1-YFP-H-expressing PNs specifically immunolabeled for YFP, and quantified several features. Compared to WT, the L5 Thy1-YFP-expressing PNs in KO mice exhibited an increase in mitochondria density and a more rounded shape (decreased aspect ratio) (Figure 4H-4J). These morphological characteristics are consistent with increased mitochondrial fission, a response to lower functioning mitochondria. Although cumulative mitochondrial coverage, area, and perimeter, remained statistically unchanged (data not shown), the collective EM data suggest mitochondria are more abundant and rounded in shape, which is indicative of, and consistent with, their decreased function.
A major ATP-demanding process in PNs is maintaining a negative resting membrane potential, thus providing another reliable readout for a consequence of altered mitochondrial function. To determine if these KO mice have altered membrane potentials, we used perforated patch recordings to examine the electrophysiology of Thy1-YFP-positive L5 PN before and after manipulations of the ATP-sensitive potassium (K+ATP) channel, a key player in setting the resting membrane potential (Figure 4K). After a long stabilization period, resting membrane potential in KO mice was significantly more negative, consistent with lower intracellular ATP levels (Figure 4L). When we applied the K+ATP channel inhibitor tolbutamide, membrane potential became more positive in WT PNs, while there was no effect in KO PNs (Figure 4M). This result may reflect that the decreased ATP in KO L5 PNs has a stronger influence on K+ATP channel permeability than the concentration of tolbutamide. Furthermore, applying sodium azide (NaN3), a Complex IV inhibitor which indirectly decreases ATP production, significantly lowered membrane potential in WT PNs, but not in KO PNs (Figure 4N), effectively rendering WT PNs similar to untreated KO PNs, and suggesting that KO PNs have severely decreased levels of ATP.
Systemic Metabolic Defects are Minimal in Dnajc30 Knockout Mice
We observed that deleting Dnajc30 in neocortical PNs results in diminished mitochondrial and neuronal function. These results have also been observed in specific cell populations within the hypothalamus, and correlate with obesity, diabetes, and glucose regulation (Nasrallah and Horvath, 2014). Given that 1) WS patients also present with overlapping systemic metabolic abnormalities; 2) that DNAJC30 and 7q11.23 cluster 1 genes have enriched expression in the hypothalamus (Figure S1C), and to a lesser degree in the pituitary, adrenal glands, and thyroid glands; and 3) that Dnajc30 KO mice have decreased body weight, we examined whether KO mice display peripheral metabolic deficits. We first challenged mice with a glucose tolerance test (GTT), and found a trend for female KO mice to have lower blood glucose levels, and for male KO mice to have higher blood glucose levels over the two-hour experimental window (Figure S5A-S5D). We also challenged mice with an insulin tolerance test (ITT), and found no difference in the response in females, but a stronger trend for male KO mice to have higher blood glucose levels (Figure S5E-S5H). Though no differences were observed, probing with specific diets might uncover potential differences currently on the threshold of glucose metabolism in male Dnajc30 KO mice.
To complement the GTT and ITT studies, we also analyzed mice using metabolic chambers. Here, male mice were individually housed in a controlled and measured environment to record several physiological features. While we found no differences across all features, including respirator exchange ratio (VCO2/VO2) (Figure S6A-S6E), food intake (Figure S6F), and locomotor activity (Figure S6I), two interesting trends emerged. First, Dnajc30 KO mice consumed ~25% less water over a 24-hour period (Figure S6G), and second, KO mice had higher energy expenditure (Figure S6H). This latter feature is observed in the WS population, and perhaps with deeper investigations, true differences between genotypes in mice may come to light. Additionally, diet and activity measures are inherently difficult to compare across mice and humans, and therefore not every human feature should be expected to be fully reproduced in model organisms. Taken together, while we observed no differences in peripheral metabolism, some of the more deviating features may work cumulatively to produce mice with lower brain and body weights.
Social Alterations and Anxiety in Mice Lacking Dnajc30
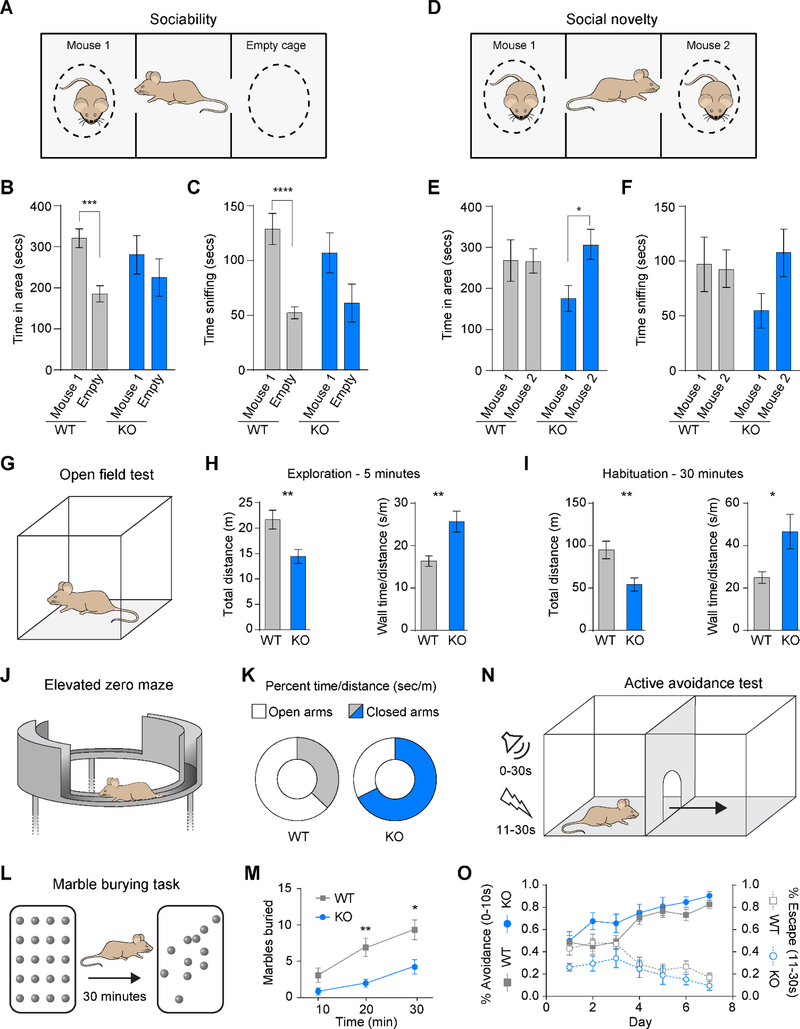
One of the more prominent behavioral features of WS patients is hypersociability. To determine if Dnajc30 KO mice also displayed this behavior, we employed sociability and social novelty tests (Moy et al., 2007). Here, mice are tested in three phases: 1) acclimation to an empty 3-chambered arena, 2) a choice between an isolated stranger mouse and an empty cage, 3) a choice between the same stranger mouse and a new stranger mouse (Figure 5A and 5D). As has been reported in several studies, we found WT mice spent significantly more time in the area and sniffing the stranger mouse than the empty cage (Figure 5B-5C). The KO mice, though showing a strong trend, had no statistical preference between the mouse and the cage. Conversely, when the second stranger mouse was introduced, KO mice had increased preference for time spent with that mouse, and a trend for increased sniffing of that mouse (Figure 5E-5F), which might reflect an interest in a novel social interaction.
Figure 5. Dnajc30 Knockout Mice Exhibit Hypersociability and Increased Anxiety.
(A and D) Illustrations of sociability and social novelty tests, respectively. (B-C) Time spent in area and time spent sniffing, respectively, in the sociability test. (E-F) Time spent in area and time spent sniffing, respectively, in social novelty test (WT, n=11; KO, n=9). (G) Illustration of an open field test. (H) Distance traveled and time spent along the wall during the first 5 minutes of OFT. (I) Distance traveled and time spent along the wall during the total 30 minutes of OFT. (J-K) Illustration and results of elevated zero maze (WT, n=10; KO, n=7; popen=0.29; pclosed=0.25). (L-M) Illustration and results of the Marble burying task (WT, n=12; KO, n=8). (N-O) Illustration and results of the Active avoidance test (WT, n=12; KO, n=7). m, meters; min, minutes; s/sec, seconds. Statistical analyses were performed using student’s t-test with corrections using the Holm-Sidak method, and using repeated measures ANOVA. *, p<0.05; **, p<0.005; ***, p<0.0005; ****, p<0.0001. See also Figure S5 and S6.
Another behavioral feature of WS patients is increased anxiety. We tested whether Dnajc30 KO mice model this behavior using the open field test, in which mice freely explore a novel empty area (Figure 5G). Consistent with an anxiety-like phenotype, KO mice exhibited less exploratory behavior (traveled less distance) and spent more time close to the walls during the 5-minute test period (Figure 5H). Decreased distance traveled and more wall time in KO mice persisted after 30 minutes of habituation (Figure 5I). We also employed another test for anxiety-like behavior, the elevated zero maze, and found a strong trend for KO mice to spend more time in the closed arms (Figure 5J-5K). We probed this phenotype further using a marble burying task (Figure 5L). Although this task has been used to measure anxiety-like behavior, it more accurately reflects repetitive, perseverative behaviors reflective of an obsessive-compulsive phenotype (Thomas et al., 2009). Therefore, if KO mice display more anxiety-like behavior, their thigmotaxis would override their innate digging behavior. Indeed, KO mice buried fewer marbles than WT counterparts (Figure 5M). Of note, the increased anxiety-like phenotype may also confound social interactions, contributing to more variable responses in the social tests.
These anxiety tests can be confounded by hypomobility. To determine if KO mice are hypomobile, we trained them in an active avoidance test, which requires mice to learn and remember that a tone will precede a foot shock. Movement to another chamber within ten seconds avoids the foot shock, but the mouse can also escape to another chamber while receiving the foot shock (Figure 5N). For each of the seven days, we found no difference between WT and KO mice in successful avoidance or percentage of escapes (Figure 5O), suggesting KO mice exhibit normal mobility and can learn, consistent with normal home cage locomotor activity observed in in KO mice (Figure S6I). Finally, to determine if the above behaviors might also be influenced by motor coordination, we tested mice on the rotarod and found no significant difference between WT and KO mice in latency to fall (Figure S6J-S6K). Together, these results suggest that the loss of Dnajc30 alters the neural circuitry related to social and anxiety-like behaviors, while sparing circuitry related to learning and memory, and movement and motor coordination.
Neuronal and Molecular Deficits in Williams Syndrome
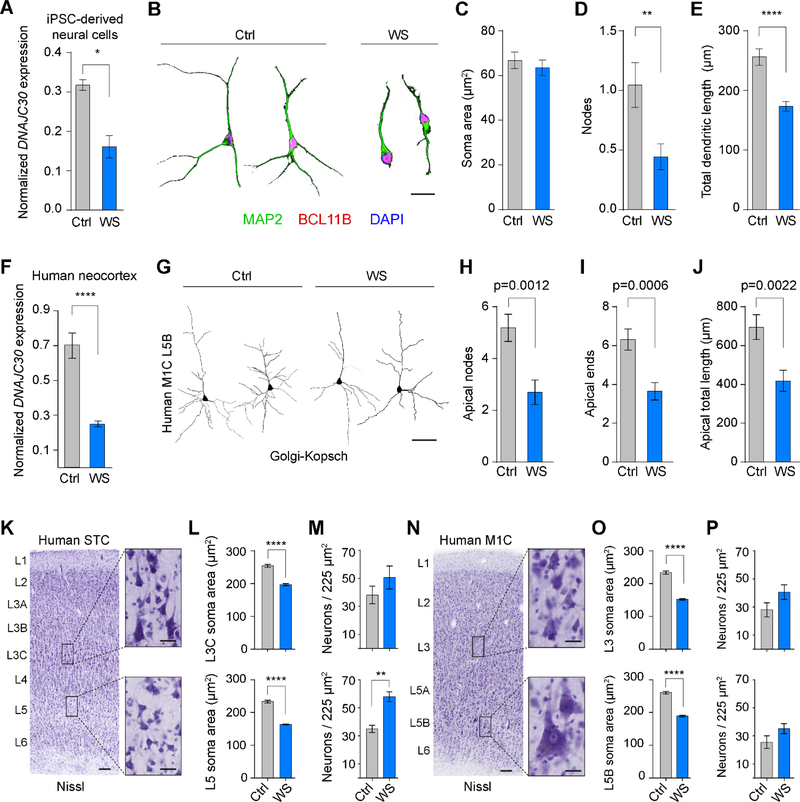
We have made substantial inroads to understanding the relationship between ATP synthesis, mitochondrial function, and cortical PNs physiomorphology, and how Dnajc30 is integral to this biology. Given that Dnajc30 KO mice phenocopy the CC defect and some of the behavioral deviations in WS patients, we wanted to determine whether these newly discovered processes are at work in WS tissue. First, we acquired skin fibroblasts from unaffected donors and patients having a typical WS deletion, and used episomal-based iPS cell reprogramming and directed differentiation to generate cortical neural cells (Figure S7A-S7B). We then confirmed using ddPCR that DNAJC30 expression is decreased in iPSC-derived neural cells and fibroblasts by about 60% (Figure 6A and Figure 7A). We then performed morphometric analysis on immunolabeled iPSC-derived neurons (Figure 6B) and found that while not every metric was decreased in WS cells, the number of nodes and total dendritic length were reduced, as seen in Dnajc30 KO neurons (Figure 6C-6E and data not shown).
Figure 6. Neuronal Structural Deficits in Williams Syndrome Cells and Brains.
(A) ddPCR showing expression ratio of DNAJC30:TBP in human iPSC-derived neural cells (Ctrl, n=3; WS, n=4). (B) Representative iPSC-derived neurons immunofluorescently labeled for morphological analysis. Scale bar = 40um. (C-E) Quantification of soma area, nodes, and total dendritic length, respectively, of MAP2- and BCL11B-positive neurons. (F) Expression ratio (DNAJC30:TBP) in human NCX samples (n=3 for each genotype). (G) Representative images from Golgi-stained M1C deep layer neurons in Ctrl (n=3) and WS (n=3) NCX. Scale bar = 100um. (H-J) Quantification of neuronal apical architecture from Golgi-stained NCX (Ctrl, n=19 neurons/3 specimens; WS, n=22 neurons/3 specimens). (K) Nissl stain and regions of interest from human STC. Low magnification scale bar = 100um, high magnification insets = 20um. (L) Soma area from L3C and L5 Nissl-stained neurons (Ctrl L3, n=286 neurons/3 specimens; WS L3, n=298 neurons/3 specimens; Ctrl L5, n=301 neurons/3 specimens; WS L5, n=293 neurons/3 specimens). (M) Neuron density in L3C and L5 STC (Ctrl, n=2 areas each for 3 specimens; WS, n=2 areas each for 3 specimens). (N) Same as K, but for human M1C. (O) Soma area from L3 and L5B Nissl-stained neurons (Ctrl L3, n=185 neurons/3 specimens; WS L3, n=190 neurons/3 specimens; Ctrl L5, n=199 neurons/3 specimens; WS L5, n=200 neurons/3 specimens). (P) Neuron density in L3 and L5B M1C (Ctrl, n=2 areas each for 3 specimens; WS, n=2 areas each for 3 specimens). *, p<0.05; **, p<0.01; ****, p<0.0001 using a student’s t-test.
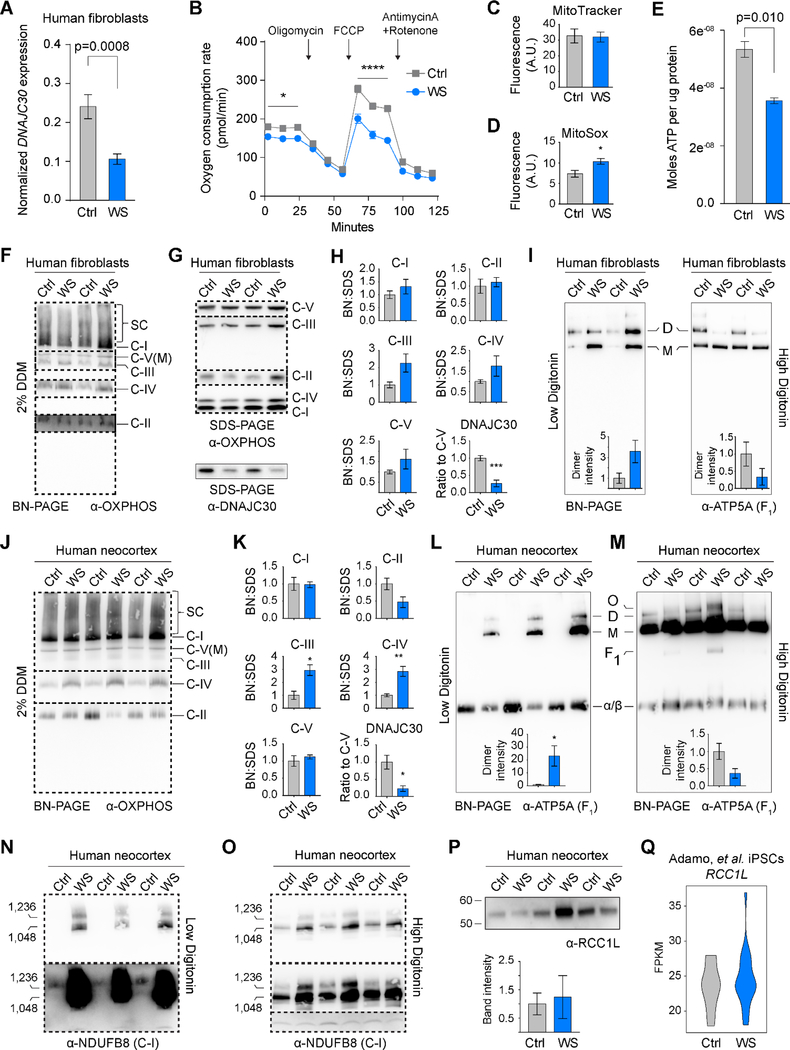
Figure 7. Oxidative Phosphorylation Supercomplexes are Dissociated in Williams Syndrome.
(A) Expression ratio (DNAJC30:TBP) in human fibroblasts (Ctrl, n=3; WS, n=9). (B) Seahorse Mito Stress Kit assay on Ctrl (n=4) and WS (n=4) fibroblasts. Arrows denote timing of pharmacological modification. (C) Relative mitochondrial membrane potential as measured by MitoTracker labeling. (D) Relative ROS levels as measured by MitoSOX labeling. (E) Luciferase assay shows decreased ATP synthesis in fibroblasts from patients affected with WS (Ctrl, n=3; WS, n=9). (F) OXPHOS cocktail western blot following BN-PAGE of human fibroblast mitochondria treated with 2% Dodecyl β-D-maltoside (DDM). Different dashed boxes indicate different exposures of the same membrane. (G) SDS-PAGE and western blots of human fibroblast samples. (H) Ratios of OXPHOS complexes and DNAJC30 as measured in BN- and SDS-PAGE. DNAJC30 was normalized to ATP5A of the SDS-PAGE western blot. (I) BN-PAGE and western blots for ATP5A on human fibroblast mitochondria treated with low or high digitonin concentrations Complex V dimers (D), monomers (M) are seen (p=0.097 and p=0.194, for left and right histograms, respectively). (J-K) As in F and H, but with human neocortical samples. (L) Western blot for ATP5A after low detergent treatment shows Complex V dimers, monomers, and the alpha and beta hetero-hexamer component of F1 subunit (α/β). The inset shows relative band intensity of dimers. (M) As in panel L, but with a higher detergent treatment (p=0.077) (O, oligomer; F1, intact subunit: α/β hexamer and central stalk). (N-O) Western blots for NDUFB8 after low and high digitonin treatments, respectively. (P) Western blot of RCC1L in WS NCX, and quantified after normalizing to ATP5A (p=0.790). (Q) Violin plots for RCC1L RNA-seq expression obtained from previously published Adamo, et al. (meanCtrl=23.56536, meanWS=24.52450, Welch two-sample t-test p=0.4405). *, p<0.05; **, p<0.01; ****, p<0.0001 using a student’s t-test. See also Figure S7.
After confirming DNAJC30 expression is also decreased in post-mortem WS NCX tissue (Figure 6F) we used a modified Golgi-Kopsch stain to compare PN morphologies in situ (Figure 6G). While silver impregnation was thorough only in deep layers of primary motor cortex (M1C), there was statistically significant reduction in apical dendrite nodes, ends, and total length (Figure 6H-6J), and trending reduction between 5%−15% in soma area and basal dendritic architecture (data not shown). Because the Golgi-Kopsch stain only sparsely labeled cells whose laminar position is not obvious, we performed Nissl stains on STC and M1C from control and WS brains. Cell area measurements revealed that L3C and L5 PNs from STC, and L3 and L5B cortical PNs from M1C, were significantly smaller in WS brains (Figure 6K-6L and 6N-6O). We also saw a trend for increased neuron density in all measured layers of WS NCX, with a significant difference in L5 STC neurons (Figure 6M and 6P). Taking these data together, we found cellular deficits in WS tissue similar to Dnajc30 KO mice, and these parallels demonstrate the role mitochondria may play in WS.
To draw better parallels between Dnajc30 KO mice and WS cells, we a performed suite of mitochondrial analyses on primary fibroblasts. Similar to Dnajc30 KO neurons, we found decreased basal respiration and maximal respiratory capacity in WS cells, while observing no difference in membrane potential, increased ROS generation, and decreased ATP synthesis (Figures 7B-E). The increased ROS, though not observed in KO mice, may reveal more broad OXPHOS aberrations in WS. These similarities further demonstrate mitochondrial malfunction as participating in WS pathogenesis.
Having observed functional differences in the OXPHOS pathway, we then searched for potential differences in the integrity or abundance within OXPHOS complexes. OXPHOS complexes are known to work most efficiently as supercomplexes (multimers), where ATP synthase forms a dimer at the center-most folds of cristae (Guo et al., 2017; Hahn et al., 2016; He et al., 2018), and Complexes I, III, and IV form supercomplexes in a range of ratios such as 1:2:0 or 1:2:4, respectively (Chaban et al., 2014; Schagger, 2002). Here, fibroblast mitochondrial proteins were differentially solubilized in Dodecyl β-D-maltoside (DDM), separated through BlueNative-PAGE, and analyzed by western blot. Though our evidence suggests DNAJC30 interacts with ATP synthase, we did not observe any difference in its monomeric state abundance (Figure 7F). However, there was a trend for increased monomeric Complexes III and IV in WS, and this difference was not due to expression differences in the specific proteins to which the antibodies were targeted (Figure 7G-7H). It has been shown that, because ATP synthase dimers help anchor cristae folding, and therefore stabilize supercomplexes, destabilizing the dimer can indirectly disrupt supercomplexes (Cogliati et al., 2016). To determine if ATP synthase dimers are altered in WS, and thus affecting the supercomplex abundances containing Complexes III and IV, we used low and high digitonin treatments to liberate ATP synthases from the inner mitochondrial membrane. In low digitonin, we see a trend for more dimers in WS, and this trend is reversed in high digitonin (Figure 7I), suggesting that dimers are less stable in WS, and that the decreased stabilization can affect supercomplex formation. We subsequently found that the changes in Complex III and IV abundance were statistically significant in WS neocortex (Figures 7J-7K and Figures S7C-S7E), perhaps highlighting the sensitivity of brain tissue to mitochondrial aberrations. Furthermore, the shift in ATP synthase dimer stabilization was also significant in WS neocortex (Figures 7L-7M). In support of DNAJC30 facilitating dimerization, DNAJC30 was observed, in differing detergent treatments, to migrate to a similar level as Complex V monomers (Figures S7F-S7G), and was significantly less abundant in WS samples. If DNAJC30 is a node explaining, at least partially, the disruption of OXPHOS supercomplex formation, then we would expect similar results in Dnajc30 KO mice. However, no differences were observed between WT and KO mitochondria in Coomassie-stained gels, or in western blots of these native gels for specific proteins expected to be in close, or direct, interaction with Dnajc30 (Figures S7H-S7I). These results suggest that while mitochondrial function is compromised in mice without Dnajc30, it may not be solely due to its interaction with ATP synthase. To that end, we also observed in human NCX BN-PAGE western blots of DNAJC30 that it also co-migrated with supercomplexes and some lower molecular weight species (Figure S7G). Given that most supercomplexes contain Complex I, we performed immunoblots for the Complex I protein, NDUFB8, on low and high digitonin treated samples, and found a trend for changes in the largest Complex I-containing supercomplexes (Figures S7J-S7K). Performing the same experiment on human NCX preparations revealed decidedly apparent differences in Complex I-containing supercomplexes (Figures 7N-7O).
To reconcile the difference in mouse and human ATP synthase dimerization, we searched the literature and came across a report that identified RCC1L as a guanine nucleotide exchange factor for OPA1, the known mitochondrial fusion protein (Cipolat et al., 2004; Huang et al., 2017). This report showed that decreased RCC1L can phenocopy mitochondrial defects observed in Opa1 knockout cells, which themselves can exhibit decreased ATP synthase dimers and dissociation of OXPHOS supercomplexes (Cogliati et al., 2013). RCC1L lies ~225kb away from the telomeric boundary of a typical WS deletion (Figure S7L), and thus was not included in our initial analysis. We conceived that RCC1L is regulated by elements within the WS locus, or is regulated by a GTF2 gene product within the typical WS locus. We measured RCC1L expression by western blot, and did not find a difference between control and WS NCX (Figure 7P). We also did not find RCC1L to be differentially expressed between control and WS samples in published gene expression datasets (Adamo et al., 2015; Chailangkarn et al., 2016) (Figure 7Q). While these results do not completely dispel the possibility that RCC1L and DNAJC30 function in concert to preserve OXPHOS supercomplexes (Figure S7L), they do further highlight the importance of DNAJC30 as having a role in maintaining OXPHOS supercomplex stability.
DISCUSSION
In this study, we combined human brain transcriptomics and PPI datasets, with functional characterizations to reveal a novel role for mitochondria in normal human brain development and WS. Mitochondrial participation is driven in part by the heretofore functionally uncharacterized gene DNAJC30. We revealed that DNAJC30 is highly enriched in neurons and is a novel auxiliary protein of the mitochondrial ATP synthase complex. Decreased DNAJC30, whether in KO mice or tissue from WS patients, resulted in decreased ATP as well as deviations in neuronal morphology, electrophysiology, and mitochondrial function. Specifically, decreased Dnajc30 in mice causes thinner callosal axons, social aberrations, and increased anxiety, all major hallmarks of WS. To the best of our knowledge, these data are the first to demonstrate mitochondrial dysfunction as part of the pathogenesis of WS, and they also have implications for the role of mitochondria in 7q11.23 duplication syndromes and similar neurodevelopmental disorders.
Mitochondrial contribution to aging and neurological and psychiatric disorders, such as Huntington’s disease (HD), Alzheimer’s disease (AD), Parkinson’s disease (PD), and SCZ, has been previously established (Manji et al., 2012; Riera and Dillin, 2015; Wallace, 2005). For example in HD and AD, mitochondria propagate secondary deleterious effects initiated by, respectively, the mutant HTT gene and Aβ peptides, which are cleaved from the AD-linked amyloid precursor protein (APP). Interestingly, Aβ peptides are also reported to be direct interacting partners with 7q11.23 proteins ABHD11, LIMK1, and DNAJC30. These protein interactions cluster within our ‘mitochondrial’ GO categories and, in light of mitochondria playing a role in 7q11.23 syndromes and AD, there may be some genetic interactions between the 7q11.23 locus and AD-linked loci. This hypothesis is substantiated in a report describing pathological hallmarks of Aβ plaques in a patient with WS (Golden et al., 1995). Alternatively, it is reported that WS patients also exhibit several features of mild-accelerated aging (Cherniske et al., 2004; Pober, 2010), which may be caused by dysfunctional mitochondria, and this dysfunction may subsequently facilitate neurodegenerative hallmarks.
In SCZ, several pieces of evidence suggest mitochondria can have an instrumental role (Clay et al., 2011). Studies have shown that metabolic genes, including those of the transcarboxylic acid cycle and from the mitochondrial genome, have significantly decreased expression in the prefrontal cortex in postmortem SCZ tissue (Arion et al., 2015; Middleton et al., 2002). Furthermore, the transcriptome alterations in mitochondrial energy production genes found in the dorso-lateral prefrontal cortex layer 3 neurons from SCZ are distinct compared to bipolar disorder and major depressive disorder (Arion et al., 2017). It is also reported that complex I genes of the electron transport chain are decreased in the prefrontal cortex (Karry et al., 2004), bolstering a hypothesis of hypofunctionality of prefrontal cognitive processes in SCZ. It is also known that variants in mitochondrial DNA (mtDNA) that cause bona fide mitochondrial disorders such as Leigh syndrome or mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes (MELAS) often display psychiatric comorbidities including SCZ (Verge et al., 2011). Other mtDNA variants, including in the aforementioned MT-ATP6 gene, have also been associated with SCZ outright (Ueno et al., 2009). Interestingly, alteration in the levels of the mitochondrial protein uncoupling protein 2 (UCP2), which controls substrate oxidation of the mitochondria in neurons (Andrews et al., 2008), as well as mitochondrial fidelity overall, have significant impact on cortical structure and function (Hermes et al., 2016; Kann and Kovacs, 2007; Li et al., 2004; Varela et al., 2016). These findings illustrate that mitochondrial function can facilitate neuropsychiatric disorders, including major depressive disorder (Cai et al., 2015; Converge Consortium, 2015) and intellectual and developmental disability (Ouyang et al., 2016), and substantiate the need for further investigations into similar neurodevelopmental disorders such as ASD.
Investigations into mitochondria and ASD are relatively unchartered (Naviaux, 2013; Smith et al., 2012). Early epidemiology of co-occurrences of mitochondrial disease and ASD have been followed by identifying disease-causing variants in mtDNA, thus laying the foundation for subsequent gene expression studies that revealed misexpression of several nuclear and mtDNA genes in postmortem ASD brains (Legido et al., 2013). Others have used the human brain transcriptome to investigate unifying mechanisms of ASD and found that ASD candidate genes were enriched in co-expression modules of mitochondrial function (Mahfouz et al., 2015). But fewer studies have revealed molecular mechanisms for how mitochondria can impact ASD (Ebrahimi-Fakhari et al., 2016; Lin-Hendel et al., 2016; Novarino et al., 2012), which is a daunting task given that mitochondrial disorders and autism both exist on spectrums (Amaral et al., 2008).
Our results provide the basis for exploring novel therapeutic options for WS, several of which are already being tested for mitochondrial disorders (DiMauro et al., 2013). One example is Leigh syndrome, which, as stated, can be caused by mutations in MT-ATP6. One study showed enhanced survival and attenuated disease in a mouse model of Leigh syndrome using the mTOR inhibitor rapamycin (Johnson et al., 2013). Other frequently used pharmacological options for mitochondrial disorders include creatine, N-Acetyl Cysteine, and Coenzyme Q10 (CoQ10). While benefits varied in clinical trials, two synthetic analogs of CoQ10, idebenone and parabenzoquinone, have reversed vision loss in four of five patients with Leber hereditary optic neuropathy, and halted or reversed progression in children with Leigh syndrome (Enns et al., 2012; Martinelli et al., 2012; Sadun et al., 2012). Because DNAJC30 is an auxiliary component of ATP synthase, compared to the essential protein MT-ATP6, it might be expected that the metabolic phenotypes in WS would be milder than in Leigh syndrome. Nevertheless, the molecular process is similar and thus it is anticipated that therapies for Leigh syndrome might also benefit WS. The potential of therapeutic intervention in WS, pharmacological or otherwise, is buoyed by our finding that cluster 1 genes, of which DNAJC30 is a member, are enriched postnatally, providing a larger time window to maximize treatments.
One specific open question remains about how removing Dnajc30 in mice resulted in mitochondrial malfunction, if not through obvious changes in OXPHOS supercomplexes. We are unfortunately limited in visualizing with which proteins the mouse ortholog of DNAJC30 interacts. Neither antibody we used, which reliably detects the human and monkey protein in native or denatured conditions, provides any signal for the mouse protein. Although these proteins share high homology, there are enough amino acid differences that may lose the epitope in mice, and similarly, may alter its function and interacting partners. For example, there is a seven residue deletion in mouse of the human sequence T37YSQGDC43, which contains a number of polar residues just N-terminal to the J-domain. There are also variations S82P and T105I (residues 75 and 98 in mouse, respectively), in the middle of the J-domain, and several variations between the J-domain and transmembrane region. One can hypothesize several effects of variations, but their elucidation may require structural biology techniques.
STAR * METHODS
CONTACT FOR REAGENTS AND RESOURCE SHARING
Further information and requests for reagents may be directed to, and will be fulfilled by the Lead Contact, Nenad Sestan (nenad.sestan@yale.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human tissue
De-identified post-mortem brain tissue was from the NIH Neurobiobank (neurobiobank.nih.gov), or from the Sestan lab collection, details of which have been published (Kang et al., 2011). Tissue was collected after obtaining parental or next of kin consent and with approval by the institutional review boards. The postmortem interval (PMI) was defined as hours between time of death and time when tissue samples were frozen. Tissue was handled in accordance with ethical guidelines and regulations for the research use of human brain tissue set forth by the NIH (http://bioethics.od.nih.gov/humantissue.html) and the WMA Declaration of Helsinki (http://www.wma.net/en/30publications/10policies/b3/index.html). Appropriate informed consent was obtained and all available non-identifying information was recorded for each specimen. Meta-data on the samples, all of which are de-identified, can be found in Table S6.
Rhesus macaque tissue
Post-mortem brain tissue was from the Sestan lab or Arnsten lab collections. Tissue was collected at pre-determined ages and in accordance with a protocol approved by Yale University’s Committee on Animal Research and NIH guidelines. The sample used for immunofluorescence was from an experimentally naïve 1 year old male where the brain was extracted and immersion fixed in 4% paraformaldehyde/PBS followed by 30% sucrose/PBS. The samples used for immuno-gold EM were from two experimentally naïve female young-adult (9 and 11-year-old) rhesus macaques housed in the Yale Department of Neuroscience non-human primate colony. Macaques were anesthetized with sodium pentobarbital (30mg/kg, i.v.), and perfused transcardially with oxygenated artificial cerebrospinal fluid, followed by 4% paraformaldehyde/0.05% glutaraldehyde, plus 0.2% picric acid in 0.1 M phosphate buffer (PB). All perfusates were administered ice-cold. The brains were blocked coronally, vibrosliced at 60μm, cryoprotected in sucrose, and stored at −80°C.
Cell lines
ReNcell CX cells (human neural progenitor cell line) were purchased from Millipore and cultured in ReNcell NSC medium with fibroblast growth factor-basic (1ng/mL) and epidermal growth factor (2ng/mL). HEK293T cells were purchased from ATCC and cultured in DMEM supplemented with 10% FBS, 1% penicillin/streptomycin, and 1% L-glutamine. Human primary fibroblasts were obtained from NIH Neurobiobank and from the Genomic and Genetic Disorders Biobank (GGDB, Network of Telethon Genetic Biobanks, project no. GTB07001G). All specimens were collected under guidelines approved by institutional review boards and anonymized prior to our receipt. Fibroblasts were cultured in aforementioned DMEM. DPBS (1x) was used to wash cells of media, and 0.25% trypsin was used to passage cells. All cellular reprogramming was initiated on low passage cells (≤8) (except lines GDB361, passage ≤12; and UMB5437, passage unknown). Cells were maintained in 5% CO2 and humidified 37°C incubators. Fibroblast reprogramming and culture conditions are described below, but was performed as previously published (Shi et al., 2012a; Shi et al., 2012b).
Mice
All experiments using animals were carried out in accordance with a protocol approved by Yale University’s Committee on Animal Research and NIH guidelines. The Dnajc30 knockout mouse strains used for this research project were created from ES cell clones 16795A-A9 and 16795A-F1, obtained from the NCRR-NIH supported KOMP Repository (www.komp.org) and generated by Regeneron Pharmaceuticals, Inc. for the NIH funded Knockout Mouse Project (KOMP). Methods used to create the Velocigene targeted alleles have been published (Valenzuela et al., 2003). The ES cells were used to create chimera at Yale Genome Editing Center, which were then backcrossed to make congenic C57BL/6N mice. Some heterozygous males were returned to KOMP as a part of their resubmission and sharing program. The Thy-1-YFP-H transgenic mice were obtained from the Jackson Laboratory.
Mice were reared in group housing in a 12h light:12h dark cycle and provided food and water ad libitum with veterinary care provided by Yale Animal Resource Center. Multiple breeding pairs were maintained and siblings were never mated to increase genetic diversity, and prevent unintended selection for features that could affect results. Both sexes were used and randomly assigned for all experiments. Different postnatal ages were also used, depending on the assay (details below). No drugs were used in this study, and mice were naïve for the open field test, but the same cohort was used for social interaction and rotarod behavior tests.
METHOD DETAILS
Expression and protein network analysis
Gene expression analysis was performed on the exon array (Kang et al., 2011) and publicly available BrainSpan RNA-seq datasets (www.BrainSpan.org) using the RNA-seq data for data presentation. We calculated the pairwise co-expression coefficient (Pearson correlation coefficient) among all 7q11.23 CNV genes and performed hierarchical clustering (Ward’s method) with the distance being defined as 1 – co-expression coefficient. We used log2 transformed RPKM values for the co-expression calculation. The three clusters were identified by visual inspection of the dendrogram and heatmap.
Integration of the brain transcriptome and protein-protein interaction network was done using the non-redundant physical BioGRID protein-protein interaction network (3.4.132, Jan. 2016) as a backbone interactome. We confined the interactome space based on the availability and the abundance of the BrainSpan RNA-seq dataset. We first removed all the nodes (proteins and corresponding genes) that are not profiled in the BrainSpan RNA-seq dataset from the interactome. We further refine it by removing genes with noise expression levels by grouping as expressed vs. non-expressed using a mixture model. This resulted in an interactome network of 12,367 nodes and 159,353 edges. We found that within the resulting interactome network, the pairwise co-expression of nodes significantly decayed over the shortest path distance between the nodes, consistent with several reports showing that the proximity of genes in the interactome network is indicative of the functional similarity of genes. We found similar results when using expression level cutoff of RPKM>1 (data not shown). Data presented are without removing non-expressed genes to demonstrate the contribution of all genes in the 7q11.23 locus.
We tested the significance of the observed mean pairwise co-expression of gene set (i.e., the group of genes belong to a cluster) against an empirical null expectation. To draw a null expected mean co-expression for a set of genes in a cluster, we randomly sampled a set of matched number of genes having the same number of protein interacting partners from the interactome network and averaged the absolute value of the pairwise co-expression coefficients. The significance p-value of the co-expression of the cluster is then determined by 1 - the number of times the mean absolute co-expression of null sets larger than that of a cluster gene set divided by the number of randomly sampled null sets. Here we used 1000 null sets.
We examined the relative tissue-specific gene expression enrichment between groups of genes using Genotype-Tissue Expression (GTEx) project RNA-Seq dataset (GTEx Consortium, 2013). We downloaded GTEx V6 Analysis RNA-Seq dataset available from http://www.gtexportal.org/home/datasets/GTEx_Analysis_v6_RNA-seq_RNA-SeQCv1.1.8_gene_rpkm.gct.gz. We summarized the expression values of genes per tissue group and performed the Wilcoxon rank sum test between two groups (i.e., C1 and C2 combined vs. C3 cluster) upon the quantile-normalization of the per-tissue mean expression values.
Identifying spatio-temporal context of 7q11.23 genes was done by grouping all the BrainSpan RNA-seq dataset into 36 spatio-temporal intervals: 4 spatial groups [(i)neocortex, (ii)hippocampus and amygdala, (iii)striatum and thalamus, and (iv)cerebellum)] and 9 temporal windows (8 pcw – 40 years; see www.BrainSpan.org). We then evaluated the specificity of gene expression of all genes in the interactome network using geneEnrichment profiler (Benita et al., 2010), which provides a quantitative measure of up-down regulation in a given spatio-temporal interval with respect to all the rest intervals. We standardized the specificity scores with mean=0 and standard deviation=1 in each spatio-temporal interval. Therefore, the resulting gene-expression specificity score of each gene provides a deviation (Z-score) from the normalized specificity score distribution in a given spatio-temporal interval.
Analysis of co-expressed genes was performed by retrieving normalized log2 signal intensity and RPKM values from BrainSpan datasets (exon array and RNA-seq, respectively), and then using a Pearson correlation, identifying the top 1000 correlation coefficients for each 7q11.23 gene. These gene lists were analyzed for GO terms using the Functional Annotation Tool within DAVID v6.7 (Huang da et al., 2009). We reached similar observations from both exon array and RNA-seq platforms. All results were then filtered for FDR <0.01, and all subsequent GO terms associated with genes from one cluster were submitted simultaneously to REVIGO (Supek et al., 2011), using medium similarity (0.7), and SimRel semantic similarity measures. The provided .xgmml files were imported into Cytoscape v3.3 for plotting (Shannon et al., 2003).
DNAJC30 protein sequences of human (Uniprot ID: Q96LL9), rhesus macaque (F6WFD4), cow (Q3SZ23), and mouse (P59041) were aligned using ClustalW (Li et al., 2015), and analyzed with Jalview (Waterhouse et al.).
Western blotting
Unless otherwise described, general western blotting procedures were followed. Cells or tissue were lysed by sonication in PBS or TBS with 0.01% Tween-20 and protease and phosphatase inhibitors. Following centrifugation at 20,000×g, supernatant protein concentrations were quantified and mixed with Laemmli loading buffer (Laemmli, 1970), boiled, and electrophoresed on NuPage Bis-Tris gels, followed by protein transfer to PVDF membranes. Membranes were incubated in 5% milk or 5% BSA, incubated with primary and secondary antibodies, and developed using chemiluminescence. Bands were imaged and quantified using a G:Box Chemi XRQ and GeneTools software, respectively.
To immunoprecipitate the ATP synthase complex, intact mitochondria were isolated from human superior temporal cortex in mitochondrial extraction (ME) buffer + protease inhibitor cocktail (225mM mannitol, 75mM sucrose, 50mM KCl, 10mM KH2PO4, 10mM Tris pH 7.4, and 0.5mM EGTA) by Dounce homogenization (Pestle Ax10, Pestle Bx30). Homogenate was centrifuged at 1000×g for 10 minutes, and the resulting supernatant centrifuged at 12,000×g for 10 minutes. Pelleted mitochondria were washed and resuspended in ME buffer + 0.05% n-Dodecyl β-D-maltoside and incubated at 4°C for 1 hour to liberate protein complexes from membranes. Proteins were then added to antibody-conjugated Protein G beads following the manufacturer’s protocol for overnight incubation at 4°C. Following bead washing (3×100 volumes of TBS+0.1% Tween-20), proteins were eluted in 2X Laemmli buffer without β-mercaptoethanol. For these and all subsequent single-image experiments, the data presented are representative of at least three independent experiments.
For Blue-Native gel electrophoresis, mitochondria were isolated in ME buffer as before with modification. Following the final high speed mitochondrial pelleting centrifugation, protein concentrations were determined and equal mass of protein was centrifuged in a new tube. The resulting pellet was resuspended in 50mM Tris, 50mM NaCl, and 1mM EDTA and incubated in specified detergents (DDM or Digitonin) for 30 minutes at 4°C. Detergent concentrations were determined empirically, and we identified that 2% DDM, 4g/g Digitonin (low), or 13g/g Digitonin (high) produced the required solubilization of protein complexes. Treated samples were then centrifuged at 20,000×g for 30 minutes, and the supernatant was diluted in 4x NativePage sample buffer and Coomassie G-250. NativePage gels were run according to manufacturer’s protocol. For western blots, proteins were transferred as before, membranes were rinsed for approximately 5 minutes in 100% methanol before washing in TBST and then milk, and continuing with antibody incubation.
Cloning and cell culture
Human DNAJC30 cDNA was purchased from OriGene (SC319163) and subcloned into a pCAGEN expression plasmid. Mouse Dnajc30 cDNA was cloned from a mouse brain-derived cDNA library. Both human and mouse sequences were amplified using PCR where the 3’ reverse primer had significant overlapping sequence with the 5’ forward primer for GFP sequence (see Table S5). Separately generated human or mouse DNAJC30 and GFP amplicons were then used as templates in a second PCR that amplified across both sequences. MT-ATP6-HA-FLAG cDNA was synthesized by GenScript into pcDNA3.1(+). It was designed with an N-terminal mitochondrial targeting sequence from COX VIII and C-terminal epitope tags HA and FLAG using optimized nuclear codon usage.
Human fetal neocortical neural cells were cultured in ReNcell media supplemented with EGF and FGF on mouse laminin coated dishes. We followed the recommended protocol for Amaxa Nucleofection, using the mouse neural stem cell kit. Cells were plated onto chamber slides, and then 16–24 hours post-transfection cells were fixed with 4% paraformaldehyde (PFA) and incubated in blocking buffer (5% BSA, 1% normal donkey serum in PBS) with 0.01% n-Dodecyl β-D-maltoside before adding primary antibodies. HEK293T cells were cultured in DMEM with 10% FBS on lysine-coated dishes until ~80% confluency when plasmids were transfected using Lipofectamine 2000. Cells were analyzed 16–24 hours post-transfection. For immunoprecipitation of MT-ATP6 and DNAJC30, whole cell lysates were prepared in ME buffer with Dounce homogenization followed immediately by adding n-Dodecyl β-D-maltoside to a final concentration of 0.05%. Lysate was then incubated with antibody-conjugated Protein G beads and continued as described above. For GFP and mitochondria co-fluorescence visualization, cells were incubated with MitoTracker (100nM) for 30 minutes, following the manufacturer’s protocol. For subcellular fractionation, a 10cm dish of transfected cells were collected and resuspended in 400μL of TBS + protease inhibitor for Dounce homogenization (10× Pestle A, 30× Pestle B). Homogenate was centrifuged at 4°C for 5 min. at 1000×g creating a pellet 1 (P1) and supernatant 1 (S1). S1 was put in a new tube for centrifugation at 14000×g for 5 minutes creating P2 and S2. P2 was washed twice with TBS and S2 was centrifuged again to enhance purity of each fraction before mixing with Laemmli buffer and loading on polyacrylamide gels.
Immunohistochemistry
Post-mortem brain tissue from adult humans, rhesus macaque, or mouse was used for immunohistochemistry (IHC), immuno-electron microscopy (immuno-EM), and immunofluorescence (IF). Standard procedures were followed for each application. For IHC, human sections were pre-treated with 0.3% H2O2 followed by incubation in blocking buffer (5% BSA, 1% normal donkey serum in PBS + 0.05% Tween-20) at room temperature and then incubation for 24–48 hours at 4°C in primary antibodies. Tissue was then incubated with biotin-labeled secondary antibodies, conjugated with Avidin-Biotin-Peroxidase Complex, and visualized with DAB-nickel following the manufacturer’s recommended protocol. Developed sections were mounted on Superfrost Plus charged slides, dehydrated through an ethanol series, cleared with xylenes, and preserved with Permount and glass coverslips. Images were taken using Aperio ScanScope digital scanner.
For immunofluorescence, a block of adult rhesus macaque STC was fixed and incubated in 30% sucrose. The block was then embedded in agarose, sectioned at 25μm, and perforated by sucrose-protected freeze/thaw treatment. Sections were blocked in blocking buffer using 0.05% n-Dodecyl β-D-maltoside, which was used for subsequent antibody dilutions. DAPI was used at 1ng/μL. Finished sections were mounted on slides and covered with Aqua-Poly/Mount. Fluorescent images were acquired on a Zeiss LSM 510 multi-photon microscope and modified using Zen software.
Electron microscopy
Sections of the macaque dorsolateral prefrontal cortex (DFC) underwent 3 freeze–thaw cycles in liquid nitrogen to facilitate penetration of immunoreagents, and were subsequently processed free-floating for DNAJC30-immunocytochemistry. Non-specific reactivity was suppressed with 10% non-immune goat serum (NGS) and 2% IgG-free bovine serum albumin in 50 mM Tris-buffered saline (TBS). The sections were additionally treated with 0.5% sodium borohydride in TBS to quench non-reactive aldehydes prior to protein blocking.
For labeling with clustered immunogold, sections were incubated for 36 h at 4°C in anti-DNAJC30 mouse polyclonal antibody, 1:500 in TBS, plus 2% NGS (NTBS), and transferred for 2 h at room temperature (RT) to goat anti-mouse biotinylated F(ab′)2 fragments, 1:500 in NTBS, and finally to goat anti-biotin IgG conjugated to 1.4 nm gold, 1:200 in NTBS supplemented with 0.07% Tween 20 and 0.1% BSA-c (gold-buffer). To eliminate traces of endogenous biotin, sections were additionally treated with an avidin/biotin blocker prior to the immunoprocedure. After fixation in 1% buffered glutaraldehyde, gold was enhanced under a mercury-vapor safelight for 8–10 min on ice with the HQ Silver autometallographic developer. For labeling with single gold immunoparticles, the sections were washed after the primary antibody in gold-buffer (see above), and incubated for 2 h at RT with goat anti-mouse Fab′ conjugated to 1.4 nm gold. Gold was fixed and enhanced as described above.
Labeled sections were mounted on glass slides for imaging under an Axiophot photomicroscope equipped with Axiocam HRc digital camera or processed for electron microscopy and embedded in Durkupan epoxy resin. Layers II/III of the DFC, were sampled for re-sectioning and analysis under a JEM1010 transmission electron microscope operating at 80 kV, with or without lead counterstaining. Images were captured at ×60,000 to 200,000 original magnification with a digital camera (Gatan, Pleasanton, CA), and micrographs were edited using Photoshop CS4 (Adobe Systems, San Jose, CA) for brightness and contrast (applied to the entire panel). No other manipulation was made to raw images.
Yeast-two-hybrid
Yeast-two-hybrid experiments were performed by ProteinLinks, Inc. (Pasadena, CA). We supplied human DNAJC30 cDNA, the entirety of which was used for the bait in a screen against an adult human brain library. Briefly, this Y2H system utilized a tetracycline inducible DNA-binding domain and a tetracycline-induced operon-reporter system. Semi-quantitative measurements and raw pictures were provided to the investigators.
Droplet digital PCR
RNA was isolated from tissues/cells essentially as previously described (Kang et al., 2011). For frozen Dnajc30 mouse cortices and WS neocortex, tissue was pulverized in a chilled mortar and pestle, and homogenized using chilled steel beads and a bead mill homogenizer. All lysates were then subjected to RNA purification using the RNeasy Plus Mini Kit. Concentration and RIN values were obtained using R6K ScreenTape and a TapeStation 2200. One μg of total RNA was used to create cDNA using SuperScript III reverse transcriptase. One ng of cDNA was used for template in droplet digital PCR. Primer/probe sets are available in Supplemental Table S5.
LacZ staining
One month old mice were anesthetized and transcardially perfused with PBS followed by 4% PFA in PBS. Extracted brains were then incubated at 4°C in 4% PFA for four hours and then washed and stored in PBS with 0.01% sodium azide. Brains were sectioned at 50μm, washed in LacZ buffer (100mM phosphate buffer, 2mM MgCl2, 0.02% NP-40), then incubated in the dark at 37°C in LacZ buffer with 5mM potassium ferricyanide, 5mM potassium ferrocyanide, and 1mg/mL X-gal. After color development, sections were washed in LacZ buffer, postfixed in 4% PFA for 1 hour, and stored in PBS at 4°C. Sections were then used for immunohistochemistry and detection using DAB without nickel. For this staining, neuronal and mitochondrial mophometrics, and electrophysiology, we studied dorsal frontal/motor and anterior somatosensory mouse brain regions.
Neuronal morphometrics and corpus callosum analysis
A FD Rapid GolgiStain kit was used to compare neuronal morphologies (~15 neurons per mouse) throughout the neocortex of WT (n=4) and KO (n=6) mice aged 9–10 days. The manufacturer’s protocol was closely followed, incubating brains in solution A+B for 10 days and solution C for 3 days before sectioning and mounting on gelatin-coated slides. After placing on coverslips, sections were viewed on a Zeiss Axio Imager.A1 and neuronal morphology analyzed with Neurolucida. For this and all other person-based quantification, researchers were blinded to genotype before their analyses.
Mice were crossed with Thy-1-YFP-H to label L5 cortical PNs and their processes. To measure thicknesses of the neocortex, corpus callosum, and L5, serial coronal sections from P9-P10 mice, 5 of each genotype, were mounted and covered on microscope slides, and tiled images taken with a 5X objective on a Zeiss Imager M2 fluorescent microscope. Thickness measurements were made using Zeiss AxioVision SE64 software. To measure soma sizes, mice at 2 months of age were used and images were captured at 40X. ImageJ was used to trace YFP-positive cells and measure their somata. We used five WT mice and eight KO mice, and measured 40 cells per mouse (data not shown but described in text).
To measure axon perimeter of the corpus callosum, 3-week old mice were transcardially perfused with 4% paraformaldehyde and 2.5% glutaraldehyde, and extracted brains were incubated in fixative overnight or longer at 4°C. Sagittal sections (100μm) were generated on a vibratome (Leica), starting most medial, and placed on microscopes before processing for electron microscopy. Standard osmication and an ethanol dehydration series was performed with 1% uranyl acetate added to 70% ethanol to enhance membrane contrast. Five mice of each genotype were used. Sections were imaged at high magnification (9300X), and measured using the g-ratio plugin for ImageJ (http://gratio.efil.de).
Mitochondrial functional analyses
Primary neuron cultures were made from P0 cortices as described previously (Sestan et al., 1999). The Seahorse XF Cell Mito Stress Test Kit was utilized per the manufacturer’s recommendation. For Seahorse assays, 7.5×103 P0 neurons were plated on lysine- and laminin-coated Seahorse assay plates, and analyzed between 4–7 days in culture. To calculate maximum respiration, averaged Antimycin A + Rotenone values were subtracted from averaged FCCP values; the differences were compared across genotypes to determine statistical significane. For flow cytometry analysis of MitoSOX and MitoTracker Orange CMTMRos, cells were loaded with 5×10−4ug/uL or 3×10−6ug/uL of dye, respectively. After washings to remove dyes, cells were suspended in PBS + 0.1% BSA and analyzed on a BD Biosciences LSR II Cytometer. For these functional analyses performed on fibroblasts, cells were grown in low glucose DMEM, with supplements as describe above. Fibroblasts were seeded on gelatin-coated Seahorse assay plates at 3.5×104/well four hours prior experimentation. For flow cytometry analyses, cells were grown to ~80% confluency in a 24-well plate.
ATP measurements
To measure ATP production in age P9–10 mice, mitochondria were extracted as above and maintained on ice. After adjusting volumes to make equal protein concentrations, equal volumes of mitochondria were plated in duplicate in a 96-well plate and loaded into a GloMax®-Multi+ Microplate Multimode Reader (Promega). A timed injection of respiration buffer (ME buffer with 1mM malate, 1mM pyruvate, 0.1mM ADP) was followed by a 1 second delay, and then a luciferase injection (Roche, ATP CLS II kit), which allowed for determining ATP production over time. Luminescence was integrated for 1 second, every 10 seconds for 20 total readings (~3.5 minutes). This protocol was adapted from (Vives-Bauza et al., 2007).
To measure ATP content in fibroblasts, 1.0×104 cells were plated in triplicate wells of 96well white plates. After allowing 12 hours to re-establish homeostasis, cells were provided fresh media, incubated for 4 hours, and then analyzed by CellTiter-Glo 2.0 following the manufacturer’s protocol. Duplicate plates were also made in clear 96-well plates to confirm cell adherence and subsequently measure protein concentration for standardization. A standard curve of ATP molar concentrations 10−12–10−3 was used for all ATP experiments.
Mitochondrial morphometrics
Electron microscopic analysis of mitochondria was performed as above and previously described (Dietrich et al., 2012). Briefly, YFP-positive cortical PNs were immunolabeled with primary and secondary antibodies, and then visualized with glucose oxidase-DAB. Stained sections were further processed before ultrathin sectioning, placing in Formvar-coated single-slot grids, and analyzed with a Tecnai 12 Biotwin electron microscope (FEI). We analyzed five WT mice and six KO mice, aged 2 months, and five cells from each mouse. Measurements of mitochondria were performed using ImageJ.
Electrophysiology
We used 3–4 week old mice to perform electrophysiology similar to as previously described (Dietrich et al., 2013; Toda et al., 2016). Briefly, after mice were anaesthetized with isoflurane and decapitated, the brains were rapidly removed and immersed in cold (4°C) oxygenated highsucrose solution containing (in mM): sucrose 220, KCl 2.5, NaH2PO4 1.23, NaHCO3 26, CaCl2 1, MgCl2 6, and glucose 10, pH (7.3) with NaOH. Coronal slices (300 μm thick) were cut on a vibratome and maintained in a holding chamber with artificial cerebrospinal fluid (ACSF, bubbled with 5% CO2 and 95% O2) containing (in mM): NaCl 124, KCl 3, CaCl2 2, MgCl2 2, NaH2PO4 1.23, NaHCO3 25, and glucose 2.5, (pH) 7.4 with NaOH. After a 1-hr recovery period, slices were transferred to a recording chamber and were constantly perfused with ACSF (33°C) at a rate of 2 mL/min. Both input resistance and series resistance were monitored throughout the experiments, and keeping only those that were stable. Initial resting membrane potential measurements were done after a 10 minute stabilization period from time of patching onto YFP-positive neurons. For these cells, the compounds tolbutamide (0.1mM) or NaN3 (1mM) were applied in the bath perfusion in the appropriate experiments. At the end of the perforated patch recordings, the membrane of every cell was ruptured and whole-cell patch recording was measured to check current-voltage relationship. All data were analyzed with an Apple Macintosh computer using AxoGraph X.
Systemic physiology and metabolism
Glucose tolerance and insulin tolerance tests were performed as described previously (Jeong et al., 2012). Briefly, baseline glucose levels were measured followed by an intraperitoneal injection of 20% glucose (10mL/kg) or insulin (0.75U/kg), respectively. Glucose tolerance tested mice were fasted overnight. Blood glucose levels were measured at 15, 30, 60, and 120 minutes after injection. Five male mice of each genotype were used.
Metabolic cages were used as described previously (Jeong et al., 2012). Briefly, seven WT mice and five KO male mice were individually housed and acclimated in metabolic chambers for seven days before 48 consecutive hours of measurements. Weights were recorded prior to acclimation. All gas exchange, diet, and locomotion measures were recorded for analysis by the metabolic chamber system.
Behavioral analysis
Mice between 2–4 months old were used for social interaction, open-field, active avoidance, and rotarod tests, essentially as described previously (Dietrich et al., 2015; Moy et al., 2007). For the sociability tests, mice had one training day of 10 minutes with no stimuli in the 3-chambered apparatus, which had retractable doorways to separate the chambers. The next day, mice were given one, ten minute exploratory trial in the apparatus, followed by 10 minutes with stranger mouse 1, and then 10 minutes with stranger mice 1 and 2. For the open-field test, naïve mice were placed in a black-walled, 37 × 37 × 37 cm box and monitored for 30 minutes. Times, distances, and areas were videoed and analyzed using ANY-maze software.
The Active avoidance test was based on previous studies {Martinez, de Graaf, 1985; moscarello, ledoux; 2013}. On day 0, mice were habituated to the chamber with an open door and no stimuli for 10 minutes. On day 1, mice were placed on the “shock” side for 10 seconds, after which the door was opened and tone was initiated (t=0). At t=10 seconds, a mild foot shock was initiated (0.15 mA) and sustained until the mouse crossed to the safe chamber, or until t=30 seconds, at which point the shock was terminated and door closed. If the mouse crossed to the safe chamber before t=30, it was given 10–30 seconds of rest and then placed on the shock side before starting the next trial after another 20–30 seconds. Day 1 consisted of six trials and Days 2–7 consisted of fifteen trials each day.
For rotarod tests, mice were given three consecutive days of training on the apparatus, three trials each day, and were tested on the fourth day. The rotarod parameters were acceleration from 0 r.p.m. to a maximum 40 r.p.m. over 90 seconds.
iPS cell reprogramming and differentiation
Human iPSCs were generated by reprogramming of human skin fibroblasts with episomal vectors pCXLE-hOCT3.4-shp53-F, pCXLE-hSK, pCXLE-hUL, and pCXLE-EGFP obtained from Addgene. Human fibroblasts (2×106) collected from unaffected donors (n=3) and patients affected with WS (n=4) were nucleofected with 1.5 ug of each episome using Amaxa Nucleofector II and Amaxa NHDF nucleofector kit, and then seeded onto 100-mm dishes coated with 1:50 diluted growth factor reduced Matrigel. Cultures were grown under hypoxic conditions in Essential 6 medium supplemented with 100 ng/mL bFGF, hydrocortisone 100 nM, and sodium butyrate 0.5 mM. Cells were re-plated 10 days after transduction onto 100-mm dishes coated with Matrigel at 5000 cells/cm2 density and cultured under hypoxic conditions in Essential 6 medium supplemented with 100 ng/mL bFGF. Colonies were selected for further expansion and evaluation 24–34 days after plating.
iPSC lines were cultured under hypoxic conditions in Essential 8 Flex medium on plates coated with Matrigel diluted 1:100, and were passaged using a 0.5 mM EDTA solution for longterm expansion. To validate iPSC lines, we used PCR to confirm electroporated plasmids were not integrated, karyotyping by Yale Cytogenetics Lab to confirm no chromosomal rearrangements had occurred, and teratoma assays by Yale Mouse Research Pathology Service and immunofluorescence of marker genes to confirm pluripotency. All measures were passed before a cell line was used for subsequent differentiation.
Human iPSCs directed differentiation into neocortical neurons was achieved by applying a monolayer, feeder-free, dual SMAD inhibition based differentiation protocol (Okita et al., 2011; Onorati et al., 2016; Shi et al., 2012a). Briefly, iPSC were dissociated to single cells through incubation with Accutase at 37 °C, seeded at 0.7–2 × 105 cells/cm2 density on plates coated with Matrigel diluted 1:50 and cultured in Essential 8 Flex medium supplemented with 10 μM ROCK inhibitor Y-27632. ROCK inhibitor was withdrawn the next day, and the iPSCs were allowed to expand in Essential 8 FLEX medium until the cells were nearly confluent. Neural differentiation was started by changing the culture medium to neural induction medium composed of a 1:1 mixture of DMEM/F-12 and Neurobasal supplemented with N-2, B-27, 20 μg/ml Insulin, 1% L-glutamine 200 mM, 1% MEM Non-Essential Amino Acids, 0.1 mM 2-mercaptoethanol, 100 nM LDN-193189, 10 μM SB-431542, and 2 μM XAV 939. Cells were cultured in neural induction medium for 11 days changing the medium daily. The efficiency of neural induction was monitored visually by assessing the appearance of cells with characteristic neuroepithelial morphology and the formation of neural rosettes. On day 12, neuroepithelial cells were harvested through incubation with Accutase at 37 °C for approximately 15–20 minutes and replated onto on 13 mm glass coverslips or 6-well plates previously coated with 10 μg/mL Poly-D-lysine and 3 μg/mL Laminin at 0.7 cells/cm2 density in neural induction medium supplemented with 100 nM LDN-193189 and 10 uM ROCK inhibitor. After replating, the medium was progressively switched through partial media changes to neural differentiation medium which contained Neurobasal supplemented with N-2, B-27, 1% Penicillin/Streptomycin, 1% GlutaMAX, and 30 ng/mL BDNF human recombinant protein. Cultures were maintained for up to 80 days of differentiation changing the medium partially every 2–3 days.
Histology
For the Golgi-Kopsch stain, we followed a protocol previously described (Riley, 1979). Briefly, blocks ~0.5–1.0cm3 of cortex were incubated in 3.5% potassium dichromate, 15% sucrose, and 0.6% formaldehyde for 7 days, followed by incubation in 0.75% silver nitrate for 3 days, and finally dehydrated overnight in 95% ethanol. Blocks were then sectioned on a vibratome at 200μm, cleared in Histo-Clear, and mounted on microscope slides with Permount and coverslips. Silver impregnation was generally poor in STC areas of all brains. The best cells that appeared fully impregnated (several branched apical and basal dendrites from easily identifiable soma) were in the deep layers of M1C, and for this reason only these cells were analyzed. The impregnation was not sufficient to reliably quantify spines/dendritic protrusions.
For Nissl staining, 50μm brain sections were mounted on microscope slides, and once dried, slides were rinsed in water. Slides were then immersed in 0.1% cresyl violet acetate at room temperature until most cells were fully penetrated (~30 minutes). Slides were rinsed in 70% ethanol, then 90% ethanol, 2× 100% ethanol, and finally xylenes before covering with a coverslip and Permount mounting media. Soma area was counted by first identifying the layer within the region of interest and tracing pyramidal-shaped cells with Neurolucida software. Motor cortex layer 5B was identified by the presence of Betz cells, however these cells were not measured because a disproportionate number of cells of such large size could easily skew data. Also, soma area greater or less than two standard deviations from average were removed before final statistical analysis. Cell density was performed using ImageJ to place standardized grids over low magnification images of Nissl-stained sections. Two non-adjacent grids were used to count pyramidal-shaped cells and the average of the two grids were used to represent the density for that layer and cortical area.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistics were performed using Microsoft Excel and GraphPad Prism software. Where only two groups were compared, a two-tailed Student’s t-test was used. The exception was in the electrophysiology studies where a one-tailed t-test was used because values were expected to move in only one direction of the mean following drug treatment. Where there were three or more groups compared, one-way ANOVA followed by Dunnett’s multiple comparisons test was used, and repeated measure ANOVA was used for the active avoidance test. For the corpus callosum and axon distribution measurements, and for behavior tests, multiple t-tests were performed and significance determined using the Holm-Sidak method. P<0.05 was considered significant for all studies. Figures were organized with Adobe Illustrator and Adobe Photoshop.
DATA AND SOFTWARE AVAILABILITY
We generated custom scripts to identify or provide information listed below. All scripts will be made available to investigators who request them. The scripts are: (1) Pearson correlation coefficient for exon-array-generated brain transcriptome, (2) Pearson correlation coefficient for RNA-seq-generated brain transcriptome, (3) co-expression cluster analysis, (4) co-expression significance analysis using protein-protein interaction network, (5) tissue specificity analysis across multiple tissues using GTEx RNAseq data, (6) co-expression cluster significance against null expectation, (7) tissue specificity across spatio-time interval (BrainSpan RNAseq data) with 7q11.23 CNV genes, (8) tissue specificity across spatio-time interval (BrainSpan RNAseq data) with 7q11.23 CNV genes + network neighbors.
Supplementary Material
Table S1 (related to Figure 1). RNA-seq Generated Top 1000 Co-Expressed 7q11.23 Genes.
Table S2 (related to Figure 1). Exon-Array Generated Top 1000 Co-Expressed 7q11.23 Genes.
Table S3 (related to Figure 1). RNA-seq Generated GO Analysis on BrainSpan Expressed 7q11.23 Genes.
Table S4 (related to Figure 1). Exon-Array Generated GO Analysis on BrainSpan Expressed 7q11.23 Genes.
Table S5 (related to STAR methods). Primer Names and Sequences.
Table S6 (related to STAR methods). Human Tissue Meta-Data.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-DNAJC30 | Novus | Cat#H00084277-B01P; RRID:AB_2095222 |
| Rabbit anti-DNAJC30 | Novus | Cat#NBP1–80621; RRID:AB_11040278 |
| Sheep anti-NRGN | R&D Systems | Cat#AF7947 |
| Goat anti-GAD1 | R&D Systems | Cat#AF2086; RRID:AB_2107724 |
| Rabbit anti-GFAP | Sigma | Cat#G9269; RRID:AB_477035 |
| Mouse anti-ALDH1L1 | Encor | Cat#MCA-4A12; RRID:AB_2572221 |
| Rabbit anti-GFAP | Encor | Cat#RPCA-GFAP; RRID:AB_2572310 |
| Chicken anti-CNP | Encor | Cat#CPCA-CNP; RRID:AB_2572249 |
| Mouse anti-GAPDH | EnCor | Cat#MCA-1D4; RRID:AB_2107599 |
| Rabbit anti-ATP synthase C | Abcam | Cat#ab181243 |
| Rabbit anti-ATP5A1 | Abcam | Cat#ab176569 |
| Goat anti-ATP5A1 | Abcam | Cat#ab188107 |
| Mouse anti-ATP synthase immunocapture | Abcam | Cat#ab109867; RRID:AB_10866627 |
| Mouse total OXPHOS human cocktail | Abcam | Cat#ab110411 |
| Rabbit anti-MRS2 | Novus | Cat#NBP2–34200 |
| Mouse anti-NDUFB8 | Abcam | Cat#ab110242; RRID:AB_1085912 |
| Rabbit anti-WBSCR16 | ThermoFisher | Cat#PA5–48652, RRID:AB_2634109 |
| Mouse anti-FLAG M2 | Sigma | Cat#F1804; RRID:AB_262044 |
| Rabbit anti-HA tag | Sigma | Cat#SAB430603 |
| Mouse anti-GFP | Sigma | Cat#11814460001; RRID:AB_390913 |
| Rabbit anti-Nanog | Stemgent | Cat#09–0020; RRID:AB_2298294 |
| Mouse anti-TRA-1–60 | Stemgent | Cat#09–0010; RRID:AB_1512170 |
| Rat anti-BCL11B (CTIP2) | Abcam | Cat#ab18465; RRID:AB_2064130 |
| Rabbit anti-CUX1 (CDP) | SantaCruz | Cat#sc-13024; RRID:AB_2261231 |
| Rabbit anti-TBR1 | SantaCruz | Cat#sc-48816; RRID:AB_2287060 |
| Mouse anti-SATB2 | GenWay | Cat#20–372-60065; RRID:AB_1029576 |
| Mouse anti-beta III-tubulin (TuJ1) | Covance | Cat#MMS-43SP |
| Rabbit anti-MAP2 | Millipore | Cat#ab5622; RRID:AB_11213363 |
| Mouse anti-NF1 | Millipore | Cat#MAB1615; RRID:AB_94285 |
| ChromPure Mouse IgG | Jackson ImmunoResearch | Cat#015–000-003 |
| Biotin-SP donkey anti-mouse IgG | Jackson ImmunoResearch | Cat#715–065-150 |
| Biotin-SP donkey anti-sheep IgG | Jackson ImmunoResearch | Cat#713–065-003 |
| Biotin-SP donkey anti-goat IgG | Jackson ImmunoResearch | Cat#705–065-003 |
| Goat anti-mouse biotinylated F(ab’)2 fragments | Jackson ImmunoResearch | Cat#115–066-146 |
| Goat anti-biotin IgG conjugated to 1.4 nm gold cluster | Nanoprobes | Cat#2015 |
| Goat anti-mouse Fab’ conjugated to 1.4 nm gold cluster | Nanoprobes | Cat#2002 |
| Alexa 488 donkey anti-mouse IgG | Jackson ImmunoResearch | Cat#715–545-150 |
| Alexa 594 donkey anti-rabbit IgG | Jackson ImmunoResearch | Cat#711–585-152 |
| Alexa 647 donkey anti-goat IgG | Jackson ImmunoResearch | Cat#705–605-003 |
| Alexa 594 donkey anti-sheep IgG | Jackson ImmunoResearch | Cat#713–585-003 |
| Alexa 594 donkey anti-goat IgG | Jackson ImmunoResearch | Cat#705–585-003 |
| HRP goat anti-mouse IgG | Jackson ImmunoResearch | Cat#115–035-003 |
| HRP goat anti-rabbit IgG | Jackson ImmunoResearch | Cat#111–035-003 |
| Biological Samples | ||
| Healthy human neocortex, fixed | Sestan lab | N/A |
| Healthy human neocortex, frozen | Sestan lab | N/A |
| Healthy monkey neocortex, fixed | Sestan lab | N/A |
| Healthy monkey neocortex, fixed | Arnsten lab | N/A |
| Williams syndrome neocortex, fixed | University of Maryland Brain & Tissue Bank; http://medschool.umaryland.edu/btbank/ | Cat.# UMB5638; UMB5291; UMB5530 |
| Williams syndrome neocortex, frozen | University of Maryland Brain & Tissue Bank; http://medschool.umaryland.edu/btbank/ | Cat.# UMB1574; UMB5437; UMB5857 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Phosphate-buffered saline | ThermoFisher | Cat#14190–144 |
| 1M Tris pH 7.4 | American Bioanalytical | Cat#AB14044 |
| 5M Sodium chloride | American Bioanalytical | Cat#AB13198 |
| Protease inhibitor cocktail | Sigma Aldrich | Cat#P8340 |
| PhosSTOP phosphatase inhibitor | Roche | Cat#04906845001 |
| 4X Laemmli buffer (with and without beta-mercaptoethanol) | Sestan lab | N/A |
| NuPage Bis-Tris gels | ThermoFisher | Cat#NP0322 |
| NativePage gels | ThermoFisher | Cat#BN1002 |
| PVDF membranes | Bio-Rad | Cat#162–0218 |
| SuperSignal™ West Pico Chemiluminescent Substrate | ThermoFisher | Cat#34080 |
| Mitochondrial extraction buffer | This paper | N/A |
| Malic acid | Sigma Aldrich | Cat#M1000 |
| Sodium pyruvate | ThermoFisher | Cat#11360–070 |
| Adenosine diphosphate | Sigma Aldrich | Cat#A2754 |
| n-Dodecyl β-D-maltoside | MP Biomedicals | Cat#0215101391 |
| Digitonin | Millipore | Cat#300410 |
| Protein G Dynabeads | ThermoFisher | Cat#10004D |
| Dulbecco’s Modified Eagle Medium | ThermoFisher | Cat#11965–092 |
| Low glucose DMEM | ThermoFisher | Cat#11885–084 |
| L-glutamine | ThermoFisher | Cat#25030–081 |
| Heat inactivated fetal bovine serum | ThermoFisher | Cat#16140–071 |
| Penicillin and streptomycin | ThermoFisher | Cat#15140–122 |
| ReNcell NSC Medium | Millipore | Cat#SCM005 |
| Epidermal growth factor | Millipore | Cat#01–107 |
| Basic fibroblast growth factor | Millipore | Cat#GF003 |
| Lipofectamine 2000 | ThermoFisher | Cat#11668–019 |
| Mouse Neural Stem Cell Nucleofector® Kit | Lonza | Cat#VAPG-1004 |
| NHDF Nucleofector® Kit | Lonza | Cat#VPD-1001 |
| Oligomycin | Sigma Aldrich | Cat#75351 |
| FCCP | Sigma Aldrich | Cat#C2920 |
| HRP | Sigma Aldrich | Cat#P-6782 |
| DCF | ThermoFisher | Cat#D399 |
| Pyruvate | Sigma Aldrich | Cat#P-2256 |
| Malate | Sigma Aldrich | Cat#M-7397 |
| Paraformaldehyde | J.T. Baker | Cat#S898–07 |
| Bovine serum albumin | Sigma | Cat#A9647 |
| Normal donkey serum | Jackson ImmunoResearch | Cat#017–000-121 |
| Hydrogen peroxide | J.T. Baker | Cat#2186 |
| Avidin-Biotin-Peroxidase Complex | Vector Laboratories | Cat#PK-6100 |
| Avidin-Biotin blocker | Vector Laboratories | Cat#SP-2001 |
| DAB-nickel | Vector Laboratories | Cat#SK-4100 |
| MitoTracker Deep Red FM | ThermoFisher | Cat#M22426 |
| MitoTracker Orange CMTMRos | ThermoFisher | Cat#M7510 |
| MitoSOX | ThermoFisher | Cat#M36008 |
| DAPI | ThermoFisher | Cat#D3571 |
| Aqua-Poly/Mount | Polysciences | Cat#18606 |
| X-gal | American Bioanalytical | Cat#AB02400 |
| Matrigel | Corning | Cat#354230 |
| Essential 6 medium | ThermoFisher | Cat#A1516401 |
| bFGF | R&D Systems | Cat#4114-TC |
| Hydrocortisone | Sigma Aldrich | Cat#H0888 |
| Sodium butyrate | Stemgent | Cat#04–0005 |
| Essential 8 Flex medium | ThermoFisher | Cat#A2858501 |
| Accutase | ThermoFisher | Cat#A11105–01 |
| ROCK inhibitor Y-27632 | StemCell | Cat#72308 |
| DMEM/F-12 medium | ThermoFisher | Cat#11330 |
| Neurobasal medium | ThermoFisher | Cat#21103–049 |
| N-2 supplement | ThermoFisher | Cat#17502–048 |
| B-27 supplement | ThermoFisher | Cat#17504–044 |
| Insulin solution, human | Sigma Aldrich | Cat#I9278 |
| MEM non-essential amino acids | ThermoFisher | Cat#11140–050 |
| 2-mercaptoethanol | ThermoFisher | Cat#21985 |
| LDN-193189 | Stemgent | Cat#04–0074 |
| SB-431542 | TOCRIS | Cat#1614 |
| XAV 939 | Stemgent | Cat#04–0046 |
| Poly-D-lysine | Sigma Aldrich | Cat#P6407 |
| Mouse Natural Laminin | ThermoFisher | Cat#23017–015 |
| BDNF, human recombinant | ThermoFisher | Cat#PHC7074 |
| Gelatin subbed slides | SouthernBiotech | Cat#SLD01 |
| Tolbutamide | Sigma Aldrich | Cat#T0891 |
| Sodium azide | Alfa Aesar | Cat#14314 |
| Cresyl violet acetate | MP Biomedicals | Cat#150727 |
| Potassium dichromate | Acros | Cat#42411–0500 |
| Silver nitrate | Sigma-Aldrich | Cat#209139 |
| Critical Commercial Assays | ||
| Yeast-2-hybrid, custom | ProteinLinks, Inc. (Pasadena, CA) | Bait: DNAJC30; Prey: Adult human brain library |
| FD Rapid GolgiStain Kit | FD Neurotechnologies | Cat#PK401 |
| Seahorse XF Cell Mito Stress Test Kit | Agilent | Cat#103015–100 |
| ATP CLS II kit | Roche | Cat#11699695001 |
| CellTiter-Glo® 2.0 Assay | Promega | Cat#G9242 |
| WestPico Chemiluminescent substrate | ThermoFisher | Cat#34080 |
| BCA Protein Assay | ThermoFisher | Cat#23227 |
| RNeasy Plus Mini Kit | Qiagen | Cat#74134 |
| SuperScript III | ThermoFisher | Cat#18080 |
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| HEK293T | ATCC | Cat#CRL-11268; RRID:CVCL_0063 |
| ReNcell CX | Millipore | Cat#SCC007; RRID:CVCL_E922 |
| Primary fibroblasts | EuroBioBank, Fondazione Telethon (GGDB) (http://www.eurobiobank.org/) | Sample ID# GDB101, GDB306, GDB316, GDB361, GDB361–1, GDB682, GDB683, GDB728, GDB728–1, GDB242, GDB314, GDB809 |
| Primary fibroblasts | Coriell Institute for Medical Research | Sample ID# GM08680, GM05399, GM08399 |
| Primary fibroblasts | University of Maryland Brain & Tissue Bank; http://medschool.umaryland.edu/btbank/ | Cat# UMB1574, UMB5437 |
| Primary fibroblasts | Onorati et al., 2016 | N/A |
| Experimental Models: Organisms/Strains | ||
| Dnajc30 knockout mice | KOMP (https://www.komp.org/index.php) | Cat# 16795A-F1 (line 1); 16795A-A9 (line 2) |
| Thy-1-YFP-H mice | The Jackson Laboratory | B6.Cg-Tg(Thy1-YFP)HJrs/J |
| Recombinant DNA | ||
| human DNAJC30 | OriGene | Cat#SC319163 |
| mouse Dnajc30 | This paper | N/A |
| ATP6-HA-FLAG | GenScript | custom design |
| human DNAJC30-GFP | This paper | N/A |
| mouse Dnajc30-GFP | This paper | N/A |
| pCXLE-hOCT3.4-shp53-F | Okita et al., 2011 | Addgene #27077 |
| pCXLE-hSK | Okita et al., 2011 | Addgene #27078 |
| pCXLE-hUL | Okita et al., 2011 | Addgene #27080 |
| pCXLE-EGFP | Okita et al., 2011 | Addgene #27082 |
| Sequence-Based Reagents | ||
| Primers for cloning and ddPCR | See Table S5 | N/A |
| Software and Algorithms | ||
| BioGRID v3.4.132 (Jan. 2016) | Stark et al., 2006 | https://thebiogrid.org/ |
| GTEx | GTEx consortium, 2013 | http://www.gtexportal.org/home/ |
| DAVID Bioinformatics Resources v6.7 | Huang da et al., 2009 | https://david.ncifcrf.gov/ |
| REVIGO | Supek et al., 2011 | http://revigo.irb.hr/ |
| Cytoscape v3.3 | Shannon et al., 2003 | http://www.cytoscape.org/ |
| GeneTools | Syngene | N/A |
| ZEN | Zeiss Microscopy | N/A |
| Neurolucida | MBF Bioscience | N/A |
| Aperio ImageScope v12 | Leica | N/A |
| ImageJ | NIH | http://imagej.nih.gov/ij |
| AxoGraph X | AxoGraph Scientific | N/A |
| ANY-maze behavior tracking software | ANY-maze | http://www.anymaze.co.uk/ |
| Jalview | Waterhouse et al., 2009 | http://www.jalview.org/ |
| PhenoMaster Software | TSE System | https://www.tse-systems.com/ |
| Clustal Omega | Li et al., 2015 | http://www.ebi.ac.uk/Tools/msa/clustalo/ |
| GraphPad Prism 7 | GraphPad | N/A |
| Other | ||
| BrainSpan exon array data | Kang et al., 2011 | N/A |
| BrainSpan RNA-seq data | http://www.BrainSpan.org/ | N/A |
| G:Box Chemi XRQ | Syngene | N/A |
| droplet digital PCR QX100 | Bio-Rad | N/A |
| Zeiss LSM 510 confocal | Zeiss Microscopy | N/A |
| Zeiss Imager M2 fluorescence/brightfield | Zeiss Microscopy | N/A |
| Tecnai 12 transmission electron microscope | FEI | N/A |
| Seahorse XFe96 Analyzer | Agilent | N/A |
| Axiophotomicroscope with Axio HRc camera | Zeiss Microscopy | N/A |
| JEM1010 transmission electron microscope | Jeol | N/A |
| Aperio ScanScope | Leica | N/A |
| 96/384 microplate reader | Ascent | N/A |
| TSE LabMaster System | TSE System | https://www.tse-systems.com/ |
| Behavior apparati | Yale Electronics and Machine Shop | N/A |
| AccuRotor rotarod | Accuscan Instruments Inc. | N/A |
| GloMax Mulit-detection system | Promega | Cat#E7081 |
HIGHLIGHTS.
Systems biology analyses reveal mitochondrial contribution to 7q11.23 CNV syndromes
The 7q11.23 gene product DNAJC30 facilitates ATP synthesis in neocortical neurons
Mitochondrial and neuronal physiomorphology and social and anxiety-related behaviors are altered in Dnajc30−/− mice
Altered mitochondrial function and OXPHOS architecture in WS cells and post-mortem brain
ACKNOWLEDGMENTS
Human tissue was obtained from the University of Maryland Brain and Tissue Bank, which is a Brain and Tissue Repository of the NIH NeuroBioBank. We thank The Genomic and Genetic Disorders Biobank, a member of the Telethon Network of Genetic Biobanks funded by Telethon Italy (project no. GTB12001G) for the banking of biospecimens. We thank Angus Nairn and Arthur Horwich for helpful discussions. This work was supported by the Brown-Coxe and 5T32NS007224–27 fellowships (A.T.N.T.), Telethon Foundation – Italy (Grant no. GGP14265), Italian Ministry of Health (Ricerca Corrente) (G.M.), Pioneer Award DP1AG047744–01 (A.F.T.A.), AG052005, AG052986, AG051459, DK111178 from NIH and NKFI-126998 from the Hungarian National Research, Development and Innovation Office (T.L.H), and MH106934, MH106874, MH116488, MH110926 and MH109904 (N.S.).
REFERENCES
- Abbas E, Cox DM, Smith T, and Butler MG (2016). The 7q11.23 Microduplication Syndrome: A Clinical Report with Review of Literature. J Pediatr Genet 5, 129–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamo A, Atashpaz S, Germain PL, Zanella M, D’Agostino G, Albertin V, Chenoweth J, Micale L, Fusco C, Unger C, et al. (2015). 7q11.23 dosage-dependent dysregulation in human pluripotent stem cells affects transcriptional programs in disease-relevant lineages. Nature genetics 47, 132–141. [DOI] [PubMed] [Google Scholar]
- Amaral DG, Schumann CM, and Nordahl CW (2008). Neuroanatomy of autism. Trends Neurosci 31, 137–145. [DOI] [PubMed] [Google Scholar]
- Andrews ZB, Liu ZW, Walllingford N, Erion DM, Borok E, Friedman JM, Tschop MH, Shanabrough M, Cline G, Shulman GI, et al. (2008). UCP2 mediates ghrelin’s action on NPY/AgRP neurons by lowering free radicals. Nature 454, 846–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arion D, Corradi JP, Tang S, Datta D, Boothe F, He A, Cacace AM, Zaczek R, Albright CF, Tseng G, et al. (2015). Distinctive transcriptome alterations of prefrontal pyramidal neurons in schizophrenia and schizoaffective disorder. Molecular psychiatry 20, 1397–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arion D, Huo Z, Enwright JF, Corradi JP, Tseng G, and Lewis DA (2017). Transcriptome Alterations in Prefrontal Pyramidal Cells Distinguish Schizophrenia From Bipolar and Major Depressive Disorders. Biol Psychiatry 82, 594–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlinghaus LR, Thornton-Wells TA, Dykens EM, and Anderson AW (2011). Alterations in diffusion properties of white matter in Williams syndrome. Magnetic resonance imaging 29, 1165–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak B, and Feng G (2016). Neurobiology of social behavior abnormalities in autism and Williams syndrome. Nat Neurosci 19, 647–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benita Y, Cao Z, Giallourakis C, Li C, Gardet A, and Xavier RJ (2010). Gene enrichment profiles reveal T-cell development, differentiation, and lineage-specific transcription factors including ZBTB25 as a novel NF-AT repressor. Blood 115, 5376–5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuren AJ, Apitz J, and Harmjanz D (1962). Supravalvular aortic stenosis in association with mental retardation and a certain facial appearance. Circulation 26, 1235–1240. [DOI] [PubMed] [Google Scholar]
- Cai N, Li Y, Chang S, Liang J, Lin C, Zhang X, Liang L, Hu J, Chan W, Kendler KS, et al. (2015). Genetic Control over mtDNA and Its Relationship to Major Depressive Disorder. Curr Biol 25, 3170–3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo SE, Clauser KR, and Mootha VK (2016). MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic acids research 44, D1251–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell LE, Daly E, Toal F, Stevens A, Azuma R, Karmiloff-Smith A, Murphy DG, and Murphy KC (2009). Brain structural differences associated with the behavioural phenotype in children with Williams syndrome. Brain research 1258, 96–107. [DOI] [PubMed] [Google Scholar]
- Chaban Y, Boekema EJ, and Dudkina NV (2014). Structures of mitochondrial oxidative phosphorylation supercomplexes and mechanisms for their stabilisation. Biochimica et biophysica acta 1837, 418–426. [DOI] [PubMed] [Google Scholar]
- Chailangkarn T, Trujillo CA, Freitas BC, Hrvoj-Mihic B, Herai RH, Yu DX, Brown TT, Marchetto MC, Bardy C, McHenry L, et al. (2016). A human neurodevelopmental model for Williams syndrome. Nature 536, 338–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherniske EM, Carpenter TO, Klaiman C, Young E, Bregman J, Insogna K, Schultz RT, and Pober BR (2004). Multisystem study of 20 older adults with Williams syndrome. American journal of medical genetics Part A 131, 255–264. [DOI] [PubMed] [Google Scholar]
- Cipolat S, Martins de Brito O, Dal Zilio B, and Scorrano L (2004). OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc Natl Acad Sci U S A 101, 15927–15932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay HB, Sillivan S, and Konradi C (2011). Mitochondrial dysfunction and pathology in bipolar disorder and schizophrenia. Int J Dev Neurosci 29, 311–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogliati S, Enriquez JA, and Scorrano L (2016). Mitochondrial Cristae: Where Beauty Meets Functionality. Trends Biochem Sci 41, 261–273. [DOI] [PubMed] [Google Scholar]
- Cogliati S, Frezza C, Soriano ME, Varanita T, Quintana-Cabrera R, Corrado M, Cipolat S, Costa V, Casarin A, Gomes LC, et al. (2013). Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency. Cell 155, 160–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Converge Consortium. (2015). Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature 523, 588–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium. (2013). The Genotype-Tissue Expression (GTEx) project. Nature genetics 45, 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries DD, van Engelen BG, Gabreels FJ, Ruitenbeek W, and van Oost BA (1993). A second missense mutation in the mitochondrial ATPase 6 gene in Leigh’s syndrome. Ann Neurol 34, 410–412. [DOI] [PubMed] [Google Scholar]
- Depienne C, Heron D, Betancur C, Benyahia B, Trouillard O, Bouteiller D, Verloes A, LeGuern E, Leboyer M, and Brice A (2007). Autism, language delay and mental retardation in a patient with 7q11 duplication. Journal of medical genetics 44, 452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich MO, Bober J, Ferreira JG, Tellez LA, Mineur YS, Souza DO, Gao XB, Picciotto MR, Araujo I, Liu ZW, et al. (2012). AgRP neurons regulate development of dopamine neuronal plasticity and nonfood-associated behaviors. Nat Neurosci 15, 1108–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich MO, Liu ZW, and Horvath TL (2013). Mitochondrial dynamics controlled by mitofusins regulate Agrp neuronal activity and diet-induced obesity. Cell 155, 188–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich MO, Zimmer MR, Bober J, and Horvath TL (2015). Hypothalamic Agrp neurons drive stereotypic behaviors beyond feeding. Cell 160, 1222–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMauro S, Schon EA, Carelli V, and Hirano M (2013). The clinical maze of mitochondrial neurology. Nat Rev Neurol 9, 429–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutly F, and Schinzel A (1996). Unequal interchromosomal rearrangements may result in elastin gene deletions causing the Williams-Beuren syndrome. Human molecular genetics 5, 1893–1898. [DOI] [PubMed] [Google Scholar]
- Ebrahimi-Fakhari D, Saffari A, Wahlster L, Di Nardo A, Turner D, Lewis TL Jr., Conrad C, Rothberg JM, Lipton JO, Kolker S, et al. (2016). Impaired Mitochondrial Dynamics and Mitophagy in Neuronal Models of Tuberous Sclerosis Complex. Cell reports 17, 1053–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elston GN (2003). Cortex, cognition and the cell: new insights into the pyramidal neuron and prefrontal function. Cereb Cortex 13, 1124–1138. [DOI] [PubMed] [Google Scholar]
- Enns GM, Kinsman SL, Perlman SL, Spicer KM, Abdenur JE, Cohen BH, Amagata A, Barnes A, Kheifets V, Shrader WD, et al. (2012). Initial experience in the treatment of inherited mitochondrial disease with EPI-743. Mol Genet Metab 105, 91–102. [DOI] [PubMed] [Google Scholar]
- Ewart AK, Morris CA, Atkinson D, Jin W, Sternes K, Spallone P, Stock AD, Leppert M, and Keating MT (1993). Hemizygosity at the elastin locus in a developmental disorder, Williams syndrome. Nature genetics 5, 11–16. [DOI] [PubMed] [Google Scholar]
- Faria AV, Landau B, O’Hearn KM, Li X, Jiang H, Oishi K, Zhang J, and Mori S (2012). Quantitative analysis of gray and white matter in Williams syndrome. Neuroreport 23, 283–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangiskakis JM, Ewart AK, Morris CA, Mervis CB, Bertrand J, Robinson BF, Klein BP, Ensing GJ, Everett LA, Green ED, et al. (1996). LIM-kinase1 hemizygosity implicated in impaired visuospatial constructive cognition. Cell 86, 59–69. [DOI] [PubMed] [Google Scholar]
- Fung LK, Quintin EM, Haas BW, and Reiss AL (2012). Conceptualizing neurodevelopmental disorders through a mechanistic understanding of fragile X syndrome and Williams syndrome. Current opinion in neurology 25, 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusco C, Micale L, Augello B, Teresa Pellico M, Menghini D, Alfieri P, Cristina Digilio M, Mandriani B, Carella M, Palumbo O, et al. (2014). Smaller and larger deletions of the Williams Beuren syndrome region implicate genes involved in mild facial phenotype, epilepsy and autistic traits. Eur J Hum Genet 22, 64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galaburda AM, Holinger DP, Bellugi U, and Sherman GF (2002). Williams syndrome: neuronal size and neuronal-packing density in primary visual cortex. Arch Neurol 59, 1461–1467. [DOI] [PubMed] [Google Scholar]
- Geschwind DH, and Flint J (2015). Genetics and genomics of psychiatric disease. Science 349, 1489–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert-Dussardier B, Bonneau D, Gigarel N, Le Merrer M, Bonnet D, Philip N, Serville F, Verloes A, Rossi A, Ayme S, et al. (1995). A novel microsatellite DNA marker at locus D7S1870 detects hemizygosity in 75% of patients with Williams syndrome. American journal of human genetics 56, 542–544. [PMC free article] [PubMed] [Google Scholar]
- Girirajan S, Campbell CD, and Eichler EE (2011). Human copy number variation and complex genetic disease. Annu Rev Genet 45, 203–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden JA, Nielsen GP, Pober BR, and Hyman BT (1995). The neuropathology of Williams syndrome. Report of a 35-year-old man with presenile beta/A4 amyloid plaques and neurofibrillary tangles. Arch Neurol 52, 209–212. [DOI] [PubMed] [Google Scholar]
- Golzio C, Willer J, Talkowski ME, Oh EC, Taniguchi Y, Jacquemont S, Reymond A, Sun M, Sawa A, Gusella JF, et al. (2012). KCTD13 is a major driver of mirrored neuroanatomical phenotypes of the 16p11.2 copy number variant. Nature 485, 363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulsuner S, Walsh T, Watts AC, Lee MK, Thornton AM, Casadei S, Rippey C, Shahin H, Consortium on the Genetics of, S., Group, P.S., et al. (2013). Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell 154, 518–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Bueler SA, and Rubinstein JL (2017). Atomic model for the dimeric FO region of mitochondrial ATP synthase. Science 358, 936–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BW, Barnea-Goraly N, Sheau KE, Yamagata B, Ullas S, and Reiss AL (2014). Altered microstructure within social-cognitive brain networks during childhood in Williams syndrome. Cereb Cortex 24, 2796–2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn A, Parey K, Bublitz M, Mills DJ, Zickermann V, Vonck J, Kuhlbrandt W, and Meier T (2016). Structure of a Complete ATP Synthase Dimer Reveals the Molecular Basis of Inner Mitochondrial Membrane Morphology. Mol Cell 63, 445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Ford HC, Carroll J, Douglas C, Gonzales E, Ding S, Fearnley IM, and Walker JE (2018). Assembly of the membrane domain of ATP synthase in human mitochondria. Proc Natl Acad Sci U S A 115, 2988–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermes G, Nagy D, Waterson M, Zsarnovszky A, Varela L, Hajos M, and Horvath TL (2016). Role of mitochondrial uncoupling protein-2 (UCP2) in higher brain functions, neuronal plasticity and network oscillation. Mol Metab 5, 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeft F, Barnea-Goraly N, Haas BW, Golarai G, Ng D, Mills D, Korenberg J, Bellugi U, Galaburda A, and Reiss AL (2007). More is not always better: increased fractional anisotropy of superior longitudinal fasciculus associated with poor visuospatial abilities in Williams syndrome. J Neurosci 27, 11960–11965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeft F, Dai L, Haas BW, Sheau K, Mimura M, Mills D, Galaburda A, Bellugi U, Korenberg JR, and Reiss AL (2014). Mapping genetically controlled neural circuits of social behavior and visuo-motor integration by a preliminary examination of atypical deletions with Williams syndrome. PloS one 9, e104088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt IJ, Harding AE, Petty RK, and Morgan-Hughes JA (1990). A new mitochondrial disease associated with mitochondrial DNA heteroplasmy. American journal of human genetics 46, 428–433. [PMC free article] [PubMed] [Google Scholar]
- Hu WF, Chahrour MH, and Walsh CA (2014). The diverse genetic landscape of neurodevelopmental disorders. Annu Rev Genomics Hum Genet 15, 195–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, and Lempicki RA (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Huang G, Massoudi D, Muir AM, Joshi DC, Zhang CL, Chiu SY, and Greenspan DS (2017). WBSCR16 Is a Guanine Nucleotide Exchange Factor Important for Mitochondrial Fusion. Cell reports 20, 923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, Tam S, Zarraga G, Colby G, Baltier K, et al. (2015). The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell 162, 425–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackowski AP, Rando K, Maria de Araujo C, Del Cole CG, Silva I, and Tavares de Lacerda AL (2009). Brain abnormalities in Williams syndrome: a review of structural and functional magnetic resonance imaging findings. European journal of paediatric neurology: 13, 305–316. [DOI] [PubMed] [Google Scholar]
- Jeong JK, Szabo G, Raso GM, Meli R, and Diano S (2012). Deletion of prolyl carboxypeptidase attenuates the metabolic effects of diet-induced obesity. Am J Physiol Endocrinol Metab 302, E1502–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SC, Yanos ME, Kayser EB, Quintana A, Sangesland M, Castanza A, Uhde L, Hui J, Wall VZ, Gagnidze A, et al. (2013). mTOR inhibition alleviates mitochondrial disease in a mouse model of Leigh syndrome. Science 342, 1524–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga HH, and Craig EA (2010). The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nature reviews Molecular cell biology 11, 579–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AM, Pletikos M, Meyer KA, Sedmak G, et al. (2011). Spatio-temporal transcriptome of the human brain. Nature 478, 483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kann O, and Kovacs R (2007). Mitochondria and neuronal activity. Am J Physiol Cell Physiol 292, C641–657. [DOI] [PubMed] [Google Scholar]
- Kaplan AS, Stallings VA, Zemel BS, Green KA, and Kaplan P (1998). Body composition, energy expenditure, and energy intake in patients with Williams syndrome. The Journal of pediatrics 132, 223–227. [DOI] [PubMed] [Google Scholar]
- Karry R, Klein E, and Ben Shachar D (2004). Mitochondrial complex I subunits expression is altered in schizophrenia: a postmortem study. Biol Psychiatry 55, 676–684. [DOI] [PubMed] [Google Scholar]
- Kwon SK, Sando R 3rd, Lewis TL, Hirabayashi Y, Maximov A, and Polleux F (2016). LKB1 Regulates Mitochondria-Dependent Presynaptic Calcium Clearance and Neurotransmitter Release Properties at Excitatory Synapses along Cortical Axons. PLoS Biol 14, e1002516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Legido A, Jethva R, and Goldenthal MJ (2013). Mitochondrial dysfunction in autism. Semin Pediatr Neurol 20, 163–175. [DOI] [PubMed] [Google Scholar]
- Li HH, Roy M, Kuscuoglu U, Spencer CM, Halm B, Harrison KC, Bayle JH, Splendore A, Ding F, Meltzer LA, et al. (2009). Induced chromosome deletions cause hypersociability and other features of Williams-Beuren syndrome in mice. EMBO molecular medicine 1, 50–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Cowley A, Uludag M, Gur T, McWilliam H, Squizzato S, Park YM, Buso N, and Lopez R (2015). The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic acids research 43, W580–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Okamoto K, Hayashi Y, and Sheng M (2004). The importance of dendritic mitochondria in the morphogenesis and plasticity of spines and synapses. Cell 119, 873–887. [DOI] [PubMed] [Google Scholar]
- Lin-Hendel EG, McManus MJ, Wallace DC, Anderson SA, and Golden JA (2016). Differential Mitochondrial Requirements for Radially and Non-radially Migrating Cortical Neurons: Implications for Mitochondrial Disorders. Cell reports 15, 229–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin GN, Corominas R, Lemmens I, Yang X, Tavernier J, Hill DE, Vidal M, Sebat J, and Iakoucheva LM (2015). Spatiotemporal 16p11.2 protein network implicates cortical late midfetal brain development and KCTD13-Cul3-RhoA pathway in psychiatric diseases. Neuron 85, 742–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luders E, Di Paola M, Tomaiuolo F, Thompson PM, Toga AW, Vicari S, Petrides M, and Caltagirone C (2007). Callosal morphology in Williams syndrome: a new evaluation of shape and thickness. Neuroreport 18, 203–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahfouz A, Ziats MN, Rennert OM, Lelieveldt BP, and Reinders MJ (2015). Shared Pathways Among Autism Candidate Genes Determined by Co-expression Network Analysis of the Developing Human Brain Transcriptome. J Mol Neurosci 57, 580–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malenfant P, Liu X, Hudson ML, Qiao Y, Hrynchak M, Riendeau N, Hildebrand MJ, Cohen IL, Chudley AE, Forster-Gibson C, et al. (2012). Association of GTF2i in the Williams-Beuren syndrome critical region with autism spectrum disorders. J Autism Dev Disord 42, 1459–1469. [DOI] [PubMed] [Google Scholar]
- Malhotra D, and Sebat J (2012). CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell 148, 1223–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manji H, Kato T, Di Prospero NA, Ness S, Beal MF, Krams M, and Chen G (2012). Impaired mitochondrial function in psychiatric disorders. Nat Rev Neurosci 13, 293–307. [DOI] [PubMed] [Google Scholar]
- Marenco S, Siuta MA, Kippenhan JS, Grodofsky S, Chang WL, Kohn P, Mervis CB, Morris CA, Weinberger DR, Meyer-Lindenberg A, et al. (2007). Genetic contributions to white matter architecture revealed by diffusion tensor imaging in Williams syndrome. Proc Natl Acad Sci U S A 104, 15117–15122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli D, Catteruccia M, Piemonte F, Pastore A, Tozzi G, Dionisi-Vici C, Pontrelli G, Corsetti T, Livadiotti S, Kheifets V, et al. (2012). EPI-743 reverses the progression of the pediatric mitochondrial disease--genetically defined Leigh Syndrome. Mol Genet Metab 107, 383–388. [DOI] [PubMed] [Google Scholar]
- Merla G, Brunetti-Pierri N, Micale L, and Fusco C (2010). Copy number variants at Williams-Beuren syndrome 7q11.23 region. Human genetics 128, 3–26. [DOI] [PubMed] [Google Scholar]
- Mervis CB, Dida J, Lam E, Crawford-Zelli NA, Young EJ, Henderson DR, Onay T, Morris CA, Woodruff-Borden J, Yeomans J, et al. (2012). Duplication of GTF2I results in separation anxiety in mice and humans. American journal of human genetics 90, 1064–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Lindenberg A, Mervis CB, and Berman KF (2006). Neural mechanisms in Williams syndrome: a unique window to genetic influences on cognition and behaviour. Nat Rev Neurosci 7, 380–393. [DOI] [PubMed] [Google Scholar]
- Middleton FA, Mirnics K, Pierri JN, Lewis DA, and Levitt P (2002). Gene expression profiling reveals alterations of specific metabolic pathways in schizophrenia. J Neurosci 22, 2718–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris CA (2010). The behavioral phenotype of Williams syndrome: A recognizable pattern of neurodevelopment. American journal of medical genetics Part C, Seminars in medical genetics 154C, 427–431. [DOI] [PubMed] [Google Scholar]
- Morris CA, Mervis CB, Hobart HH, Gregg RG, Bertrand J, Ensing GJ, Sommer A, Moore CA, Hopkin RJ, Spallone PA, et al. (2003). GTF2I hemizygosity implicated in mental retardation in Williams syndrome: genotype-phenotype analysis of five families with deletions in the Williams syndrome region. American journal of medical genetics Part A 123A, 45–59. [DOI] [PubMed] [Google Scholar]
- Moy SS, Nadler JJ, Young NB, Perez A, Holloway LP, Barbaro RP, Barbaro JR, Wilson LM, Threadgill DW, Lauder JM, et al. (2007). Mouse behavioral tasks relevant to autism: phenotypes of 10 inbred strains. Behav Brain Res 176, 4–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulle JG, Pulver AE, McGrath JA, Wolyniec PS, Dodd AF, Cutler DJ, Sebat J, Malhotra D, Nestadt G, Conrad DF, et al. (2014). Reciprocal duplication of the Williams-Beuren syndrome deletion on chromosome 7q11.23 is associated with schizophrenia. Biol Psychiatry 75, 371–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrallah CM, and Horvath TL (2014). Mitochondrial dynamics in the central regulation of metabolism. Nat Rev Endocrinol 10, 650–658. [DOI] [PubMed] [Google Scholar]
- Naviaux RK (2013). Mitochondria and Autism Spectrum Disorders In The Neuroscience of Autism Spectrum Disorders, Buxbaum JD, and Hof PR, eds. (Elsevier; ), pp. 179–193. [Google Scholar]
- Novarino G, El-Fishawy P, Kayserili H, Meguid NA, Scott EM, Schroth J, Silhavy JL, Kara M, Khalil RO, Ben-Omran T, et al. (2012). Mutations in BCKD-kinase lead to a potentially treatable form of autism with epilepsy. Science 338, 394–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K, Matsumura Y, Sato Y, Okada A, Morizane A, Okamoto S, Hong H, Nakagawa M, Tanabe K, Tezuka K, et al. (2011). A more efficient method to generate integration-free human iPS cells. Nat Methods 8, 409–412. [DOI] [PubMed] [Google Scholar]
- Onorati M, Li Z, Liu F, Sousa AM, Nakagawa N, Li M, Dell’Anno MT, Gulden FO, Pochareddy S, Tebbenkamp AT, et al. (2016). Zika Virus Disrupts Phospho-TBK1 Localization and Mitosis in Human Neuroepithelial Stem Cells and Radial Glia. Cell reports 16, 2576–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne LR, Martindale D, Scherer SW, Shi XM, Huizenga J, Heng HH, Costa T, Pober B, Lew L, Brinkman J, et al. (1996). Identification of genes from a 500-kb region at 7q11.23 that is commonly deleted in Williams syndrome patients. Genomics 36, 328–336. [DOI] [PubMed] [Google Scholar]
- Palacios-Verdu MG, Segura-Puimedon M, Borralleras C, Flores R, Del Campo M, Campuzano V, and Perez-Jurado LA (2015). Metabolic abnormalities in Williams-Beuren syndrome. Journal of medical genetics 52, 248–255. [DOI] [PubMed] [Google Scholar]
- Parikshak NN, Luo R, Zhang A, Won H, Lowe JK, Chandran V, Horvath S, and Geschwind DH (2013). Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell 155, 1008–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez Jurado LA, Peoples R, Kaplan P, Hamel BC, and Francke U (1996). Molecular definition of the chromosome 7 deletion in Williams syndrome and parent-of-origin effects on growth. American journal of human genetics 59, 781–792. [PMC free article] [PubMed] [Google Scholar]
- Pescosolido MF, Gamsiz ED, Nagpal S, and Morrow EM (2013). Distribution of disease-associated copy number variants across distinct disorders of cognitive development. J Am Acad Child Adolesc Psychiatry 52, 414–430 e414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitceathly RD, Murphy SM, Cottenie E, Chalasani A, Sweeney MG, Woodward C, Mudanohwo EE, Hargreaves I, Heales S, Land J, et al. (2012). Genetic dysfunction of MT-ATP6 causes axonal Charcot-Marie-Tooth disease. Neurology 79, 1145–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletikos M, Sousa AM, Sedmak G, Meyer KA, Zhu Y, Cheng F, Li M, Kawasawa YI, and Sestan N (2014). Temporal specification and bilaterality of human neocortical topographic gene expression. Neuron 81, 321–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pober BR (2010). Williams-Beuren syndrome. N Engl J Med 362, 239–252. [DOI] [PubMed] [Google Scholar]
- Pober BR, Wang E, Caprio S, Petersen KF, Brandt C, Stanley T, Osborne LR, Dzuria J, and Gulanski B (2010). High prevalence of diabetes and pre-diabetes in adults with Williams syndrome. American journal of medical genetics Part C, Seminars in medical genetics 154C, 291–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss AL, Eliez S, Schmitt JE, Straus E, Lai Z, Jones W, and Bellugi U (2000). IV. Neuroanatomy of Williams syndrome: a high-resolution MRI study. Journal of cognitive neuroscience 12 Suppl 1, 65–73. [DOI] [PubMed] [Google Scholar]
- Riera CE, and Dillin A (2015). Tipping the metabolic scales towards increased longevity in mammals. Nat Cell Biol 17, 196–203. [DOI] [PubMed] [Google Scholar]
- Riley JN (1979). A reliable Golgi-Kopsch modification. Brain Res Bull 4, 127–129. [DOI] [PubMed] [Google Scholar]
- Rivara CB, Sherwood CC, Bouras C, and Hof PR (2003). Stereologic characterization and spatial distribution patterns of Betz cells in the human primary motor cortex. Anat Rec A Discov Mol Cell Evol Biol 270, 137–151. [DOI] [PubMed] [Google Scholar]
- Sadun AA, Chicani CF, Ross-Cisneros FN, Barboni P, Thoolen M, Shrader WD, Kubis K, Carelli V, and Miller G (2012). Effect of EPI-743 on the clinical course of the mitochondrial disease Leber hereditary optic neuropathy. Arch Neurol 69, 331–338. [DOI] [PubMed] [Google Scholar]
- Sampaio A, Bouix S, Sousa N, Vasconcelos C, Fernandez M, Shenton ME, and Goncalves OF (2013). Morphometry of corpus callosum in Williams syndrome: shape as an index of neural development. Brain structure & function 218, 711–720. [DOI] [PubMed] [Google Scholar]
- Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, Chu SH, Moreau MP, Gupta AR, Thomson SA, et al. (2011). Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron 70, 863–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schagger H (2002). Respiratory chain supercomplexes of mitochondria and bacteria. Biochimica et biophysica acta 1555, 154–159. [DOI] [PubMed] [Google Scholar]
- Schmitt JE, Eliez S, Warsofsky IS, Bellugi U, and Reiss AL (2001). Corpus callosum morphology of Williams syndrome: relation to genetics and behavior. Developmental medicine and child neurology 43, 155–159. [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, et al. (2007). Strong association of de novo copy number mutations with autism. Science 316, 445–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sestan N, Artavanis-Tsakonas S, and Rakic P (1999). Contact-dependent inhibition of cortical neurite growth mediated by notch signaling. Science 286, 741–746. [DOI] [PubMed] [Google Scholar]
- Shadel GS, and Horvath TL (2015). Mitochondrial ROS signaling in organismal homeostasis. Cell 163, 560–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, and Ideker T (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Kirwan P, and Livesey FJ (2012a). Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nature protocols 7, 1836–1846. [DOI] [PubMed] [Google Scholar]
- Shi Y, Kirwan P, Smith J, Robinson HP, and Livesey FJ (2012b). Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nat Neurosci 15, 477–486, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silbereis JC, Pochareddy S, Zhu Y, Li M, and Sestan N (2016). The Cellular and Molecular Landscapes of the Developing Human Central Nervous System. Neuron 89, 248–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M, Flodman PL, Gargus JJ, Simon MT, Verrell K, Haas R, Reiner GE, Naviaux R, Osann K, Spence MA, et al. (2012). Mitochondrial and ion channel gene alterations in autism. Biochimica et biophysica acta 1817, 1796–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville MJ, Mervis CB, Young EJ, Seo EJ, del Campo M, Bamforth S, Peregrine E, Loo W, Lilley M, Perez-Jurado LA, et al. (2005). Severe expressive-language delay related to duplication of the Williams-Beuren locus. N Engl J Med 353, 1694–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, and Tyers M (2006). BioGRID: a general repository for interaction datasets. Nucleic acids research 34, D535–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- State MW, and Sestan N (2012). Neuroscience. The emerging biology of autism spectrum disorders. Science 337, 1301–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinton C, Elison S, and Howlin P (2010). Mental health problems in adults with Williams syndrome. Am J Intellect Dev Disabil 115, 3–18. [DOI] [PubMed] [Google Scholar]
- Sturrock RR (1980). Myelination of the mouse corpus callosum. Neuropathol Appl Neurobiol 6, 415–420. [DOI] [PubMed] [Google Scholar]
- Supek F, Bosnjak M, Skunca N, and Smuc T (2011). REVIGO summarizes and visualizes long lists of gene ontology terms. PloS one 6, e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebbenkamp AT, Willsey AJ, State MW, and Sestan N (2014). The developmental transcriptome of the human brain: implications for neurodevelopmental disorders. Current opinion in neurology 27, 149–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas A, Burant A, Bui N, Graham D, Yuva-Paylor LA, and Paylor R (2009). Marble burying reflects a repetitive and perseverative behavior more than novelty-induced anxiety. Psychopharmacology (Berl) 204, 361–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda C, Kim JD, Impellizzeri D, Cuzzocrea S, Liu ZW, and Diano S (2016). UCP2 Regulates Mitochondrial Fission and Ventromedial Nucleus Control of Glucose Responsiveness. Cell 164, 872–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaiuolo F, Di Paola M, Caravale B, Vicari S, Petrides M, and Caltagirone C (2002). Morphology and morphometry of the corpus callosum in Williams syndrome: a T1-weighted MRI study. Neuroreport 13, 2281–2284. [DOI] [PubMed] [Google Scholar]
- Ueno H, Nishigaki Y, Kong QP, Fuku N, Kojima S, Iwata N, Ozaki N, and Tanaka M (2009). Analysis of mitochondrial DNA variants in Japanese patients with schizophrenia. Mitochondrion 9, 385–393. [DOI] [PubMed] [Google Scholar]
- Valenzuela DM, Murphy AJ, Frendewey D, Gale NW, Economides AN, Auerbach W, Poueymirou WT, Adams NC, Rojas J, Yasenchak J, et al. (2003). High-throughput engineering of the mouse genome coupled with high-resolution expression analysis. Nat Biotechnol 21, 652–659. [DOI] [PubMed] [Google Scholar]
- Van der Aa N, Rooms L, Vandeweyer G, van den Ende J, Reyniers E, Fichera M, Romano C, Delle Chiaie B, Mortier G, Menten B, et al. (2009). Fourteen new cases contribute to the characterization of the 7q11.23 microduplication syndrome. Eur J Med Genet 52, 94–100. [DOI] [PubMed] [Google Scholar]
- Van Essen DC, Dierker D, Snyder AZ, Raichle ME, Reiss AL, and Korenberg J (2006). Symmetry of cortical folding abnormalities in Williams syndrome revealed by surface-based analyses. J Neurosci 26, 5470–5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela L, Schwartz ML, and Horvath TL (2016). Mitochondria controlled by UCP2 determine hypoxia-induced synaptic remodeling in the cortex and hippocampus. Neurobiol Dis 90, 68–74. [DOI] [PubMed] [Google Scholar]
- Verge B, Alonso Y, Valero J, Miralles C, Vilella E, and Martorell L (2011). Mitochondrial DNA (mtDNA) and schizophrenia. Eur Psychiatry 26, 45–56. [DOI] [PubMed] [Google Scholar]
- Vives-Bauza C, Yang L, and Manfredi G (2007). Assay of mitochondrial ATP synthesis in animal cells and tissues. Methods in cell biology 80, 155–171. [DOI] [PubMed] [Google Scholar]
- Walker JE (2013). The ATP synthase: the understood, the uncertain and the unknown. Biochem Soc Trans 41, 1–16. [DOI] [PubMed] [Google Scholar]
- Wallace DC (2005). A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet 39, 359–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, and Barton GJ (2009). Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JC, Barratt-Boyes BG, and Lowe JB (1961). Supravalvular aortic stenosis. Circulation 24, 1311–1318. [DOI] [PubMed] [Google Scholar]
- Willsey AJ, Sanders SJ, Li M, Dong S, Tebbenkamp AT, Muhle RA, Reilly SK, Lin L, Fertuzinhos S, Miller JA, et al. (2013). Co-expression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell 155, 997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 (related to Figure 1). RNA-seq Generated Top 1000 Co-Expressed 7q11.23 Genes.
Table S2 (related to Figure 1). Exon-Array Generated Top 1000 Co-Expressed 7q11.23 Genes.
Table S3 (related to Figure 1). RNA-seq Generated GO Analysis on BrainSpan Expressed 7q11.23 Genes.
Table S4 (related to Figure 1). Exon-Array Generated GO Analysis on BrainSpan Expressed 7q11.23 Genes.
Table S5 (related to STAR methods). Primer Names and Sequences.
Table S6 (related to STAR methods). Human Tissue Meta-Data.
Data Availability Statement
We generated custom scripts to identify or provide information listed below. All scripts will be made available to investigators who request them. The scripts are: (1) Pearson correlation coefficient for exon-array-generated brain transcriptome, (2) Pearson correlation coefficient for RNA-seq-generated brain transcriptome, (3) co-expression cluster analysis, (4) co-expression significance analysis using protein-protein interaction network, (5) tissue specificity analysis across multiple tissues using GTEx RNAseq data, (6) co-expression cluster significance against null expectation, (7) tissue specificity across spatio-time interval (BrainSpan RNAseq data) with 7q11.23 CNV genes, (8) tissue specificity across spatio-time interval (BrainSpan RNAseq data) with 7q11.23 CNV genes + network neighbors.