Abstract
Mammalian hosts constantly interact with diverse exogenous microbes, but only a subset of the microbes manage to colonize due to selective colonization resistance exerted by host genetic factors as well as the native microbiota of the host. An important question in microbial ecology and medical science is if such colonization resistance can discriminate closely related microbial species, or even closely related strains of the same species. Using human-mouse fecal microbiota transplantation and metagenomic shotgun sequencing, we reconstructed colonization patterns of human fecal microbes in mice with different genotypes (C57BL6/J vs. NSG) and with or without an intact gut microbiota. We found that mouse genotypes and the native mouse gut microbiota both exerted different selective pressures on exogenous colonizers: human fecal Bacteroides successfully established in the mice gut, however, different species of Bacteroides selectively enriched under different gut conditions, potentially due to a multitude of functional differences, ranging from versatility in nutrient acquisition to stress responses. Additionally, different clades of Bacteroides cellulosilyticus strains were selectively enriched in different gut conditions, suggesting that the fitness of conspecific microbial strains in a novel host environment could differ.
Subject terms: Metagenomics, Microbiome
Introduction
A fundamental question in microbial ecology is how exogenous microbes interact with an established host-associated microbial community. Ecological competition and cooperation for resources [1–5], indirect intervention by modulated host factors (e.g., gut commensals stimulate dendritic cells, impairing the colonization of vancomycin-resistant Enterococcus [6]), or incompatibility to host niche characteristics [7–9] can all impact the successful colonization of exogenous microbes. This question is also crucial to the biomedical field because of its direct relevance to understanding host defense and colonization resistance against pathogens, estimating efficacy of therapeutic probiotics, and improving human microbiota-associated mouse models to study human diseases.
Colonization ability of exogenous microbes in a host environment has been extensively studied in humans (e.g., refs.10, 11) and mice (e.g., refs. 12, 13). Multiple factors have been identified that collectively determine the fate of exogenous colonizers into an established microbial community. First, the composition of the host’s existing community can influence colonization, often referred to as colonization resistance [14, 15]. For example, the host native microbiota could resist the colonization of the majority of bacterial phylotypes [13]; host communities with higher diversity are more resistant to colonization by multiple pathogens, including Campylobacter jejuni [16] and Clostridium difficile [17, 18]. Interspecies antagonistic relationships are also common, such as the release of antimicrobials [3, 19–21] and the injection of toxic effectors [22]. Second, the composition of the colonizers can also influence colonization ability. For example, Streptococcus pneumoniae and Staphylococcus aureus can form dual-species biofilms leading to stable co-colonization of the upper respiratory tract, even in a vaccinated host [23]. Finally, intrinsic host factors other than the microbiome can also influence colonization ability of exogenous microbes, for not all microbes can colonize the gut of a germ-free mouse [13]. The genetic background of the host is known to shape and influence the establishment of colonizing microbes [24–26], with the most common example being the selectivity of the host immune system against many microbial pathogens [27]. Nonetheless, previous studies have not studied these factors in an integrative manner: first, it is unclear how differently the host microbiome and intrinsic host factors influence exogenous colonizers. Second, the composition of the exogenous colonizers is often restricted to a single microbe (typically a pathogen of interest), while host niches in nature likely interact with complex communities of exogenous microbes.
In addition, previous studies have had limited ability to compare colonization abilities of closely related microbial species or strains, due to the resolution afforded by conventional approaches to assaying microbial community composition, such as 16S rRNA sequencing. Understanding species and importantly, strain-level dynamics is critical because different microbial species of the same genus, and even different strains of the same species, can have fundamentally divergent phenotypes. For example, different Escherichia coli strains exhibit highly versatile metabolism, pathogenicity, and ability in colonizing and adapting to new niches [28, 29]. We hypothesize that closely related microbial species and strains can exhibit different abilities in colonizing a new host environment.
In contrast to common tools like marker gene-based amplicon sequencing, metagenomic whole-genome shotgun sequencing (mWGS) directly sequences the full complement of genetic material in a microbial community to provide a comprehensive and in-depth portrait of the community. Facilitated by state-of-the-art computational analyses, mWGS can be used to analyze the interaction between host environments and microbial colonizers at high taxonomic resolution, even to the strain level [30–32], allowing relatively unbiased profiling of strain composition compared with culture-based methods. Recent studies have tracked the fate of colonizing microbial strains in the host [10, 11], but it is unclear whether and how host environments select for colonization of specific strain types. Here, we used mWGS to study how host phenotype and endogenous microbiota affect exogenous colonizers at species-resolution and strain-resolution. To model the colonization of exogenous microbes, we transplanted human fecal microbiota (FMT) into two types of mice: the severely immunodeficient NSG mice (NOD/ShiLTJ background) and immunocompetent C57BL/6 J mice. This allowed us to investigate the effect of host genetic differences that influence immunity on engraftment of exogenous human microbiota into mice. Then, to partition the host genotype effect from the host microbiome effect (i.e., potential interspecies competition or incompatibility), we depleted by antibiotic treatment (ABT) the native gut microbes in half of the mice before FMT. The system is analogous to a competitive growth assay, in which human fecal microbes that have higher fitness in the mouse gut environment are enriched and the ones with lower fitness diminish or go extinct. We found that mouse genotypes and the composition of the endogenous mouse gut microbiome both influence the colonization of human fecal species, while such influences are significantly different among closely related microbial species. In addition, we present evidence that closely related strains of the same microbial species can have diverse colonizing abilities in a new host environment.
Materials and methods
Human FMT in C57BL6/J and NSG mice
In total, 12 C57BL6/J and 12 NSG mice (female, 5 weeks of age) were purchased from the Jackson Laboratory (Bar Harbor, ME, USA) from two Bar Harbor production rooms (C57BL6/J mice were from room AX-27 and NSG mice were from room MP-14), and raised as shown in Fig. 2a. For ABT, antibiotic cocktail (1 mg/mL ampicillin; 5 mg/mL streptomycin; 1 mg/mL colistin; 0.25 mg/mL vancomycin) were added directly into the drinking water, and the drinking water was changed with fresh antibiotics once per week for 2 weeks. Healthy human stools samples were purchased from OpenBiome (Somerville, MA, USA) stool collection. For FMT, the mice were oral gavaged with 0.2 mL/10 g of stool sample (mixed from six healthy human donors) resuspended in glycerol. Note that, we used a mixture of human fecal samples to explore competitions between the microbial strains in different human donors. Therefore, the mixed samples were more diverse and did not mimic any single fecal sample.
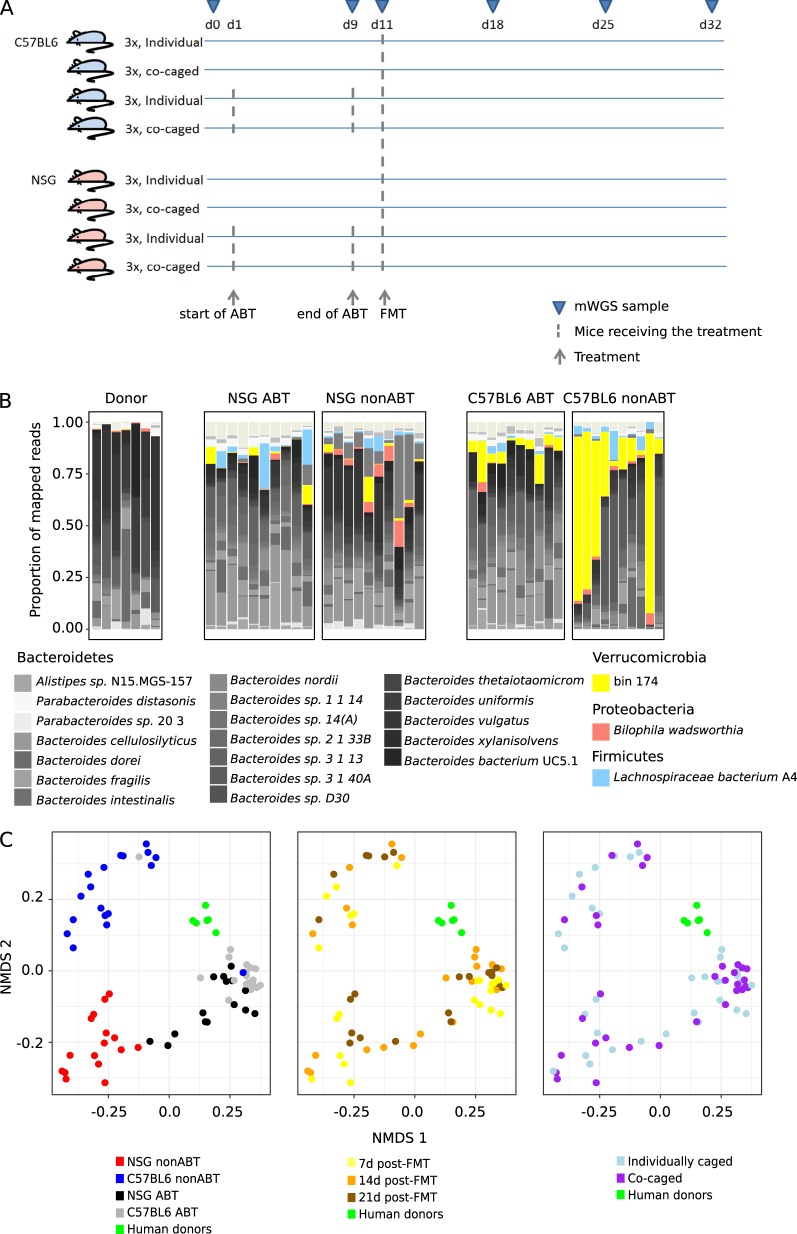
Fig. 2.
Compositional profiling of the FMT-established microbial communities. a The timeline of treatments and data collection. b The proportion of reads from mice that had received human FMT mapped to the most abundant microbial species. Here, “species” refers to both microbial species in panDB and the high-quality genome bins. 10 randomly chosen mWGS samples (i.e., random engraftment time and caging) were shown for each type of mouse gut environment. c Multidimensional scaling of pairwise Bray–curtis dissimilarities of the FMT-established microbial communities. Samples are grouped and color coded based on host, gut conditions, time post-FMT, and caging. The two axes explained 42.5% of the total variance
Total DNA extraction from mouse stool samples and quantification of microbial DNA
Mouse stool was collected into Cell & Tissue Lysis buffer (Ambion, Austin, TX, USA), and it was homogenized with a pestle before being frozen at −80 °C. DNA was extracted using the Qiagen (Germantown, MD, USA) QIAamp 96 DNA QIAcube HT Kit with the following modifications: enzymatic digestion with 50 μg of lysozyme (Sigma, St. Louis, MO, USA) and 5U each of lysostaphin and mutanolysin (Sigma) for 30 min at 37 °C followed by beadbeating with 50 μg 0.1 mm of zirconium beads for 6 min on the Tissuelyzer II (Qiagen) prior to loading onto the Qiacube HT. DNA concentration was measured using the Qubit high sensitivity dsDNA kit (Invitrogen, Carlsbad, CA, USA).
Quantitative PCR was performed in triplicate on 1 ng extracted DNA using PowerUp™ SYBR® Green Master Mix (Thermofisher, Waltham, MA, USA) on a ViiA 7 Real-Time PCR System (Thermofisher) using primers 357 F and 519 R [33] to amplify the 16S rRNA gene.
16S rRNA sequencing
The V1–V3 region of the 16S rRNA gene was amplified using primers 8 F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 534 R (5′-ATTACCGCGGCTGCTGG-3′) that were tailed with Illumina adapter sequences and index tags to facilitate sample pooling. A single-step amplification was used with the following PCR conditions: 95 C for 2 min followed by 30 cycles of 95 C for 20 s, 56 C for 30 s, and 72 C for 60 s. Amplicons were separated from unincorporated primers and nucleotides using 1.8X AMPure (Beckman Coulter, Brea, CA, USA) bead purification. Concentrations were determined using Qubit (Invitrogen), and equimolar amounts of each amplicon were pooled for sequencing on the Illumina MiSeq with 2 × 300 base reads. The resulting sequence reads were filtered to remove low-quality sequences and adapters were removed. Each forward/reverse pair was assembled into the full amplicon sequence using FLASH [34].
Metagenomic shotgun sequencing
Illumina libraries were created using Nextera XT DNA Library Prep Kit (Illumina, San Diego, CA, USA) with reduced reaction volumes: 200 pg of DNA were used (160 pg/μL × 1.25 μL), and tagmentation and PCR reagent volumes were reduced to 1/4 of the standard volumes. Tagmentation and PCR reactions were carried out according to the manufacturer’s instructions. The reaction mixtures were then adjusted to 50 μL by adding dH2O, and the AMPure (Beckman Coulter) Cleanup was carried out as per the manufacturer’s instructions. Libraries were then sequenced with 2 × 150 bp paired end reads on an Illumina HiSeq2500. For quality control, expected versus observed frequencies of species in sequencing of a bacterial mock community were closely matched (data not shown).
Sequencing adapters and low-quality bases were removed from the sequencing reads using scythe (v0.994) [35] and sickle (v1.33) [36], respectively, with default parameters. Host reads were then filtered by mapping all sequencing reads to the hg19 human reference genome or mm10 mouse reference genome using bowtie2 (v2.2.8) [37], under “very-sensitive” mode. Unmapped reads were used for downstream analyses. Characteristics of the shotgun metagenomic sequencing data were summarized in Supplementary File 1.
De novo assembly and binning
Sequencing reads from mouse samples before receiving ABT or FMT were pooled for de novo assembly using MEGAHIT (v1.0.6) [38, 39] with default parameters. The resulting contigs were filtered by size ( > 1000 bp), and sequencing reads were mapped back to the contigs using bowtie2 (v2.2.8) [37] under “very-sensitive” mode. Genome bins were constructed from the contigs using MetaBat [40] with the runMetaBat.sh wrapper using default parameters, accepting genome bins with mean depth of coverage > = 1 in each library. The resulting genome bins were estimated for genome completeness and contamination based on the presence or absence of universal single-copy orthologs using BUSCO (v1.22) [41] with default parameters. Genome bins with at least 70% completeness and at most 10% contamination (i.e., high-quality bins) were included for downstream analyses. High-quality bins were assigned taxonomic labels using the checkM pipeline (v1.0.9) [42]. To validate the taxonomic assignments, we additionally used Kraken (v0.10.6) [43] to classify the contigs with default parameters, and each genome bin was assigned the lowest taxonomic label that was assigned to at least 70% of the contigs in the genome bin.
Taxonomic composition profiling of 16S rRNA sequencing samples
Taxonomic compositions of 16S rRNA sequencing samples were profiled using VSEARCH [44]. First, reads shorter than 250, having more than one expected errors, or having over eight Nʼs were discarded. Reads were then dereplicated by collapsing identical sequences and discarding collapsed sequences that have fewer than five reads. Chimeric sequences were detected de novo and filtered using UCHIME. Next, OTUs were generated by clustering non-chimeric sequences at 0.97 sequence identity using cluster_fast, and the abundances of the the OTUs were estimated by aligning reads back to the centroids of the OTUs using usearch_global. Finally, the OTUs were taxonomically annotated using USEARCH sintax based on the Silva database [45–47].
Taxonomic composition profiling of mWGS samples
In this study, taxonomic compositions of mWGS samples were profiled in two ways, using either MetaPhlAn2 (v2.5.0) [48] or Pathoscope 2.0 [49] with a custom reference catalog. MetaPhlAn2 has been shown to be most accurate when profiling human-associated microbes [48], but is limited by the comprehensiveness of the reference database; on the other hand, Pathoscope 2.0 can take a more comprehensive reference database and account for microbes uncommon in the human microbiome. Our rationale is to use the more accurate profiling method (i.e., MetaPhlAn2) whenever a community is composed of predominantly human-associated microbes, including human donor communities and FMT-established communities in ABT mice. For all other communities where the native mouse gut microbes are abundant and the comprehensiveness of profiling is a major concern (Fig. 1a), Pathoscope 2.0 with a custom database was used to improve the comprehensiveness of the composition profiles. In this study, to maximize comprehensiveness, Pathoscope 2.0 used a custom database composed of PanDB [50]—a comprehensive and compact representation of all microbial assemblies in GenBank, and the high-quality de novo genome bins generated as described in the previous section. MetaPhlAn2 was used with default parameters to generate composition profiles leading to Fig. 3 and S2. Pathoscope 2.0 with default parameters was used to generate composition profiles leading to Figs. 1, 2, and 4.
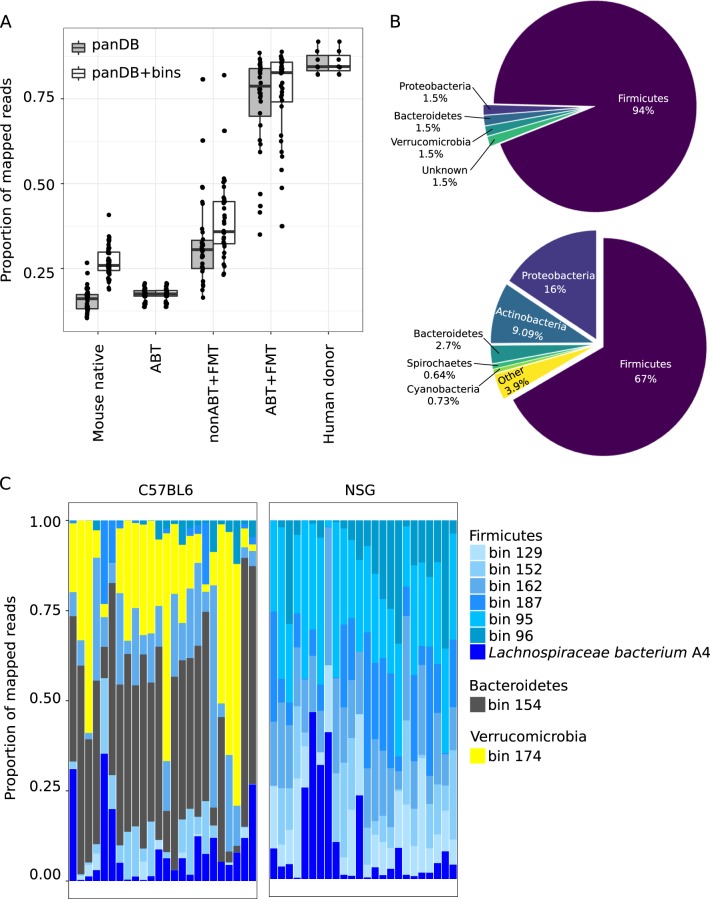
Fig. 1.
Reconstruction of the mouse gut microbiome using a combination of reference-based and de novo profiling. a the proportion of reads mapped to the reference pan-genomes in the database panDB and high-quality genome bins reconstructed de novo from native mouse gut metagenomic reads. b Phylum-level taxonomic distribution of the high-quality genome bins (upper) and assembled contigs that were not assigned into the genome bins (lower). c Mouse gut metagenomic reads mapped to the most abundant species. Here, “species” refers to both microbial species in panDB and the high-quality genome bins. Each column corresponds to one mWGS sample, the proportion of reads mapped to the most abundant taxa shown in the graph were rescaled so that they added up to 1
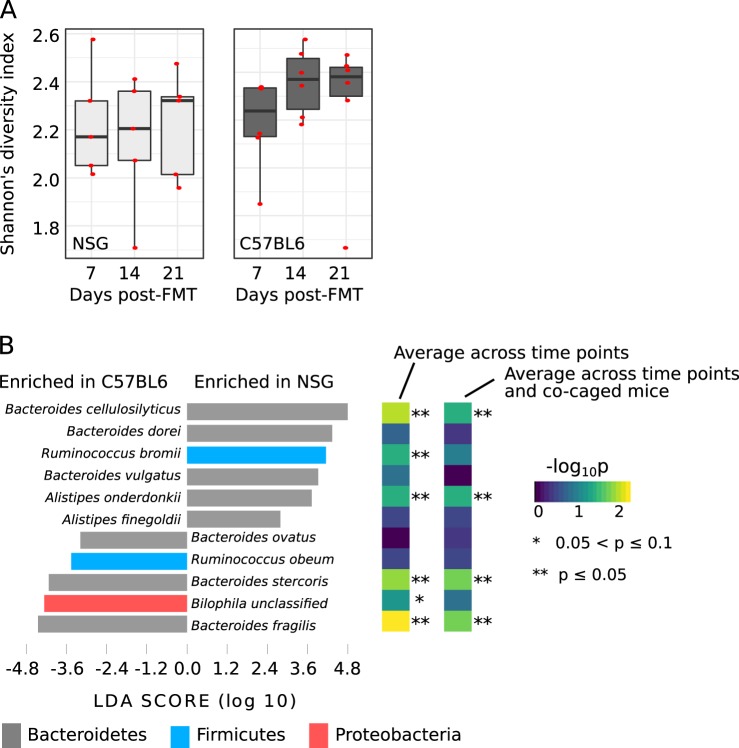
Fig. 3.
Enrichment of human fecal microbial species in different mouse genotypes. a Species diversity (Shannon’s diversity index) of human fecal microbes established in NSG ABT and C57BL6/J ABT mice. b Human fecal microbial species enriched in NSG ABT versus C57BL6/J ABT mice. Linear Discriminant Analysis score were shown for taxa showing significant enrichment (p < 0.05 and |LDA| > 2) when all samples were treated as independent. Additionally, the significance of enrichment were also shown with different sample pooling strategies: “average across time points”—samples representing time points of a same mouse were combined, “average across time points and co-caged mice”—samples representing time points of all mice in a same cage were combined. LDA linear discriminant analysis
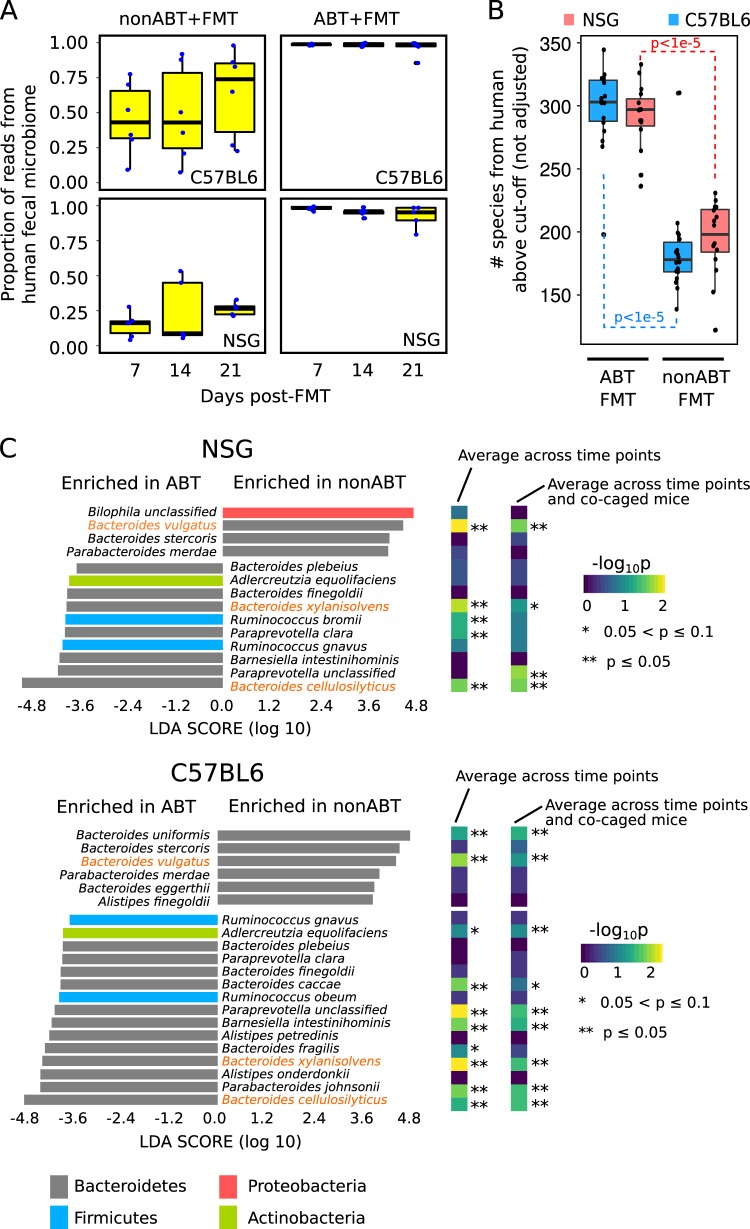
Fig. 4.
Influence of mouse gut microbiome on human fecal species. a Relative abundances of the total human fecal microbes that colonized different mouse gut environments. b Species richness of human fecal microbes in the FMT-established microbial communities. p-values of Wilcoxon rank sum test were shown. c Human fecal species enriched in nonABT versus ABT mice. Linear Discriminant Analysis score were shown for taxa showing significant enrichment (p < 0.05 and |LDA| > 2) when all samples were treated as independent. Additionally, the significance of enrichment were also shown with different sample pooling strategies: “average across time points” – samples representing time points of a same mouse were combined, “average across time points and co-caged mice”—samples representing time points of all mice in a same cage were combined. Names of species that were consistently enriched (p < 0.1) in nonABT or ABT mice for both mouse genotypes were highlighted in orange. LDA linear discriminant analysis
Enrichment and colonization resistance of microbes in different host environments
The proportion of human versus mouse microbes in each sample was estimated using the R package SourceTracker [51] (using alpha1 = alpha2 = 0.001). From compositional profiles generated as described above, extremely rare species with an average relative abundance lower than 0.001% were excluded from the analysis. When identifying differentially abundant human fecal microbes, we filtered the compositional profiles so that only human fecal species were included in the profile. This was done by removing species that had (1) an average relative abundance higher than 0.001% in the mouse samples without ABT and FMT, OR (2) an average relative abundance lower than 0.1% in the human donor samples from the profile. The relative abundance estimates of the remaining species in each sample were then scaled such that they summed up to one. Differentially enriched human fecal species in different host conditions were identified using LEfSe [52] (with the argument –o 1000000, as a normalization factor) based on the filtered and rescaled compositional profiles.
Compilation of gene catalogs
Four types of gene catalogs were used in this study. First, a metagenomic gene catalog of human fecal microbes were generated by pooling human donor mWGS reads and assembling the pooled reads de novo using MEGAHIT (v1.0.6) [38, 39] with default parameters. Genes were then predicted from the resulting contigs using prodigal (v2.6.3) [53] with default parameters and aligned to the prokaryotic KEGG gene database (ublast with at least 50% sequence identity), which contains genes annotated in each KEGG organism representing different microbial strains [54, 55], in order to be assigned KEGG ortholog numbers.
Next, a previously reported mouse metagenomic gene catalog [56] was combined with the above mentioned human metagenomic gene catalog to compare abundances of genes in the human and mouse native gut microbiota.
Additionally, a gene catalog was compiled for six Bacteroides species (B. stercoris, B. vulgatus, B. plebeius, B. finelgoldii, B. xylanisolvens, and B. cellulosilyticus) by directly combining annotated gene sequences from all 39 assemblies of the six species available in GenBank. A fourth gene catalog was compiled in the same way but only for the five Bacteroides cellulosilyticus strains.
Differential abundances of genes and pathways
For a given mWGS sample, abundances of genes of interest were computed by mapping the mWGS reads to a gene catalog (bowtie2 [v2.2.8] [37], very-sensitive mode) and counting the reads mapped to each gene using SAMtools (v1.5) [57, 58]. Abundances of KEGG orthologs were estimated by summing the reads mapped to all genes that were assigned the corresponding KEGG ortholog number. Differentially abundant KEGG orthologs were then inferred using the DESeq2 package (v1.18.1) [59], which has superior performance among other statistical tools for comparative metagenomics [60]. Based on the DESeq2 results, the differentially abundant KEGG pathways were consequently inferred using the GAGE package (v2.28.2) [61] and visualized using the Pathview package (v1.18.2) [62], according to the suggested workflow [63].
Combination of samples for differential enrichment analyses
MWGS samples representing different time points collected from a single mouse or representing co-caged mice could be inter-correlated and might inflate the sample size. Therefore, differential enrichment analyses of microbes, genes, and pathways were performed using three different strategies to account for potential inter-correlation: (1) all samples were treated as independent samples, (2) samples representing different time points collected from the same mouse were combined (i.e., each mouse has an effective sample size of 1), and (3) all samples collected from co-caged mice were combined (i.e., each cage has an effective sample size of 1). To combine mWGS samples, each sample were first rarefied to 100,000 reads before pooled into a new sample representing an averaged metagenome.
Host influence on conspecific strains
For each sample, the haplotype sequence of the dominant strain of a given species was reconstructed using StrainPhlAn [32] by concatenating the highest-coverage base type at each SNP position. Each consensus sequence represents the haplotype of the dominant strain of the species in the sample; the more similar two consensus sequences are, the more likely the dominant strains have descended from a recent common ancestor. The evolutionary similarities of the consensus sequences were visualized in form of a maximum-likelihood phylogeny as part of the StrainPhlAn pipeline.
Statistical analyses
Wilcoxon rank sum test and PERMANOVA were performed in R with the standard wilcox.test function and the adonis function in the R package vegan [64], respectively. Multidimensional scaling was conducted using the standard cmdscale function in R. Bray–curtis distance was computed using the function vegdist in the R package vegan [64]. Shannon’s diversity index (H) was computed by:
where s is the total number of taxa and pi is the relative abundance of the ith taxon.
Results
Native microbiota of C57BL/6J and NSG mice profiled using a combination of reference-based and de novo assembly-based approaches
Shotgun metagenomic sequencing was conducted on the fecal samples of 24 C57BL/6 J and 24 NSG mice before any treatments were applied to the mice. However, only 13.7 ± 3.3% of the mWGS reads could be taxonomically classified based on panDB, a comprehensive pangenome microbial reference database, we developed that was previously demonstrated to be capable of profiling over 70% of human fecal mWGS reads [50] (Fig. 1a). This suggests a broad lack of mouse microbial genomes in current reference databases. To reduce the uncharacterized space in the mouse fecal metagenome, we reconstructed genome sequences from the mWGS samples using pooled de novo assembly and binning. This process generated 194 binned genome sequences, out of which 65 binned genome sequences were deemed high quality (i.e., genome completeness over 70% and contamination, that is, duplicated single-copy marker genes, lower than 10%). Each of the 65 high-quality genome bins presumably corresponded to a microbial species draft genome; however, most could not be assigned a taxonomic label at species resolution (Supplementary File 2) although the majority (61 out of 65 genome bins) represented bacteria from the phylum Firmicutes (Fig. 1b, Supplementary File 2). On the other hand, contigs that were not included in the 65 high-quality bins (427 Mbp DNA from 160,012 contigs) represented a greater taxonomic diversity, including bacteria from the phyla Firmicutes, Proteobacteria, Actinobacteria, Cyanobacteria, Spirochaetes, and Bacteroidetes (Fig. 1b). We then used the 65 high-quality genome bins as a reference catalog to classify reads from the mouse native metagenome. These genome bins explained an additional 11.2 ± 5.0% of the mWGS reads when combined with panDB (Fig. 1a).
With the additional discriminatory resolution provided by including the 65 high-quality bins, C57BL/6 J and NSG mice showed different community composition (Fig. 1c), potentially due to differential host immune selection, although additional factors, such as mouse room origin, may also impact initial composition. The most abundant taxa were mostly represented by the high-quality genome bins, but not panDB (Fig. 1c), underscoring the need for classification approaches that incorporate not only reference-based as well as reference-independent methods. The native gut communities of C57BL6/J showed an overrepresentation of non-Firmicutes bacteria (bin 154 that represents Bacteroides thetaiotaomicron and bin 174 that represents Akkermansia muciniphila) compared with NSG mice (Fig. 1c). Community composition profiles of the mouse gut communities generated using mWGS and 16S rRNA sequencing were generally consistent (Spearman’s correlation coefficient = 0.83 ± 0.06) (Figure S1A), while the mWGS profiles showed an overrepresentation of Firmicutes and under representation of Bacteroidetes and Tenericutes compared with profiles generated using 16S rRNA sequencing, suggesting that the uncharacterized sequence information in the mWGS samples likely represent non-Firmicutes bacteria, such as Bacteroidetes and Tenericutes. In terms of biological functions, no statistically significant differences were identified between the native mouse gut microbiota and human gut microbiota on pathway level, but the abundances of many KEGG orthologs showed statistically significant difference between human and mouse gut microbiota (see Supplementary File 3 for a list of the top 10 most significantly different KEGG orthologs between human donors and the C57BL6/J or NSG mice).
Transplantation of diverse healthy human fecal microbiota (FMT) into the mice
Half of the C57BL6/J mice and NSG mice were then fed a broad spectrum antibiotic cocktail (ABT) for 8 days with 2 days of clear out of the residual antibiotics. The remaining half were not treated by antibiotics (nonABT), creating a total of four host conditions (ABT NSG, nonABT NSG, ABT C57BL6/J, and nonABT C57BL6/J). The experimental timeline was summarized in Fig. 2a. ABT effectively depleted the gut communities in both C57BL6/J and NSG mice, demonstrated by quantitative PCR against the 16S rRNA gene (Figure S1B). This effective depletion of endogenous microbiota allowed us to largely remove effects of room origin and existing microbiota and assess the effects of genotype/immunity on colonization.
Then, to establish to what degree the presence of native mouse microbiota affected the engraftment of exogenous microbiota, all mice, both ABT and nonABT, received the same high-diversity FMT from a bulk fecal sample aggregated from six healthy human donors. The mixture of human fecal samples were used to model competitions between the microbial strains in different human donors, but were not used to simulate a real biological fecal sample. The engrafted microbiota showed increased diversity when at least 1 day was available to clear out residual antibiotics, while antibiotic clear out time ranging from 1–4 days did not affect the diversity of the engrafted microbiota (Figure S1C). Community composition profiles, generated using mWGS of the engrafted gut communities, as well as the donor human gut communities, were consistent with 16S rRNA sequencing (Spearman’s correlation coefficient = 0.88 ± 0.07 and 0.89 ± 0.09, respectively) (Figure S1A). Mouse genotypes (C57BL6/J and NSG) and antibiotic treatments (ABT and nonABT) both influenced the composition of the FMT-established mouse gut community (Figs. 2b, c). Remarkably, many donor Bacteroides species were successfully colonized in all mice (Fig. 2b), while human fecal species that did not colonize the mouse gut ( < 0.001% relative abundance in the FMT samples), including multiple genera predominantly from the Firmicutes phylum (Figure S2A). The major sources of variation among the FMT-established communities were mouse genotype (p = 0.001, PERMANOVA based on Bray–Curtis distance) and ABT (p = 0.001, PERMANOVA based on Bray–Curtis distance) (Fig. 2c), as well as their interaction effect (p = 0.001, PERMANOVA based on Bray–Curtis distance). Sampling time and caging effects were also significant after adjusted for genotype and ABT effects (p = 0.009 and p = 0.014, respectively, PERMANOVA based on Bray–Curtis distance) but were not significant without adjusting for genotype and ABT effects (p = 0.26 and p = 0.24, respectively, PERMANOVA based on Bray–Curtis distance) (Fig. 2c). However, co-caging did not always increase microbiome similarity (Figure S2B, no comparisons between the co-caged mice and individually caged mice were statistically significant due to the small sample size). To adjust for these sources of variations, downstream differential abundance analyses were performed using three different strategies: (1) all samples were treated as independent samples, (2) samples representing different time points collected from the same mouse were combined (i.e., each mouse has an effective sample size of 1), and (3) all samples collected from co-caged mice were combined (i.e., each cage has an effective sample size of 1).
Effect of mouse genotype on the colonization of human fecal microbes
Next, we investigated how mouse genotypes (NSG and C57BL6/J) influence colonization of human fecal microbes by comparing the human FMT-established communities in ABT C57BL6/J mice with ABT NSG mice. We did not detect a significant difference between the overall diversity of the colonized human species in the two mouse genotypes (Wilcoxon rank sum test p = 0.26 with all samples treated as independent, p = 0.79 with combined time points, and p = 0.2 with combined co-caging samples) (Fig. 3a). Composition-wise, multiple human-associated Bacteroidetes species were differentially enriched in C57BL6/J and NSG mice: B. cellulosilyticus and Alistipes onderdonkii were enriched in the NSG mice, and B. stercoris and B. fragilis were enriched in the C57BL6/J mice (Fig. 3b). These conclusions were consistently observed when all samples were treated independently, when samples representing different time points were combined, and when samples representing co-caged mice were combined, suggesting that even closely related species can have different fitness in hosts with distinct immune phenotypes.
Effect of mouse native gut microbiome on the colonization of human fecal microbes
We then asked how the mouse native gut microbiome influences colonization of human fecal microbes. We first estimated the proportion of human fecal species in the FMT-established communities using sourceTracker [51], which uses the Bayesian method to partition components of a mixed community to different source communities that may contain overlapping species. The FMT-established communities in the ABT mice were dominated by human fecal microbes (Fig. 4a), while for the nonABT mice, the FMT-established communities contained significantly fewer human fecal microbes species (Wilcoxon rank sum test p < 10−5, Fig. 4a). Additionally, the richness of human species engrafted was also significantly larger in the ABT mice than the nonABT mice (Wilcoxon rank sum test p < 10−5, Fig. 4b), confirming that depleting the mouse gut microbiome promotes colonization of human microbes. Next, we investigated if there were differentially enriched human fecal species in ABT and nonABT mice. As expected, a larger number of human fecal species were enriched in the ABT mice compared with the nonABT mice (Fig. 4c), consistent with our previous conclusions that the native mouse gut microbiome resists colonization of human fecal microbes. We also observed enrichments that were specific to mouse gut conditions: different species from the phylum Bacteroidetes were selectively enriched in ABT or nonABT mice, and the association is highly consistent between NSG and C57BL6/J mice. For closely related species under the genus Bacteroides, B. vulgatus was associated with nonABT mice, while B. cellulosilyticus and B. xylanisolvens were associated with ABT mice for both mice genotypes and all sample combination strategies (Fig. 4c). Collectively, these findings suggest that ABT mice accept a wider range of exogenous colonizers in general, but some Bacteroides species colonize consistently well in the nonABT mice, exhibiting a species-level diversity and versatility in the ability to colonize different gut environments.
Bacterial genes and pathways associated with colonization ability in different gut environments
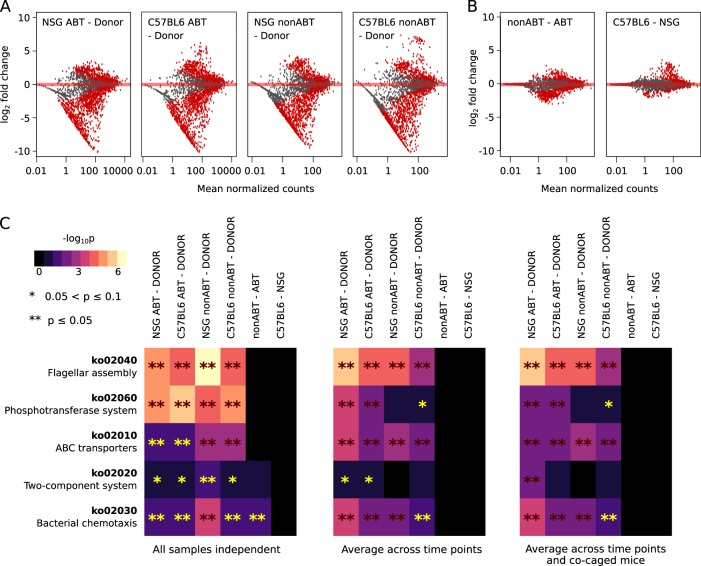
To infer microbial functional pathways that influenced colonizing abilities of human fecal species in different mouse gut environments, we generated a gene catalog from the human fecal metagenome and annotated the catalog using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database [54, 55], as a reference (223,237 non-redundant genes with 81 795 successfully assigned a KEGG ortholog number), and identified differentially abundant gene sets in different mouse gut conditions. A great diversity of KEGG orthologs were lower in abundance in the successfully colonized human fecal species compared with the original human fecal community (Benjamin–Hochberg adjusted p < 0.05, Fig. 5a), including genes involved in flagellar assembly (ko02040), bacterial chemotaxis (ko02030), and phosphotransferase (ko02060) systems (Fig. 5c and Figure S3A, B). These pathways were common bacterial pathways but were lacking in the dominant colonizer—Bacteroides bacteria, explaining, at least partially, the decreased abundances of the pathways in the colonized community.
Fig. 5.
Functional genes and pathways enriched in the successful colonizers of different mouse gut environments. a MA plot (log fold change versus mean normalized counts) showing the difference in KEGG ortholog abundances in the successful colonizers compared to the human donor samples. KEGG orthologs with significantly different abundances were shown in red (Benjamin–Hochberg adjusted p < 0.05). As an example, all samples were treated as independent. b MA plot showing KEGG orthologs enriched in the successful colonizers of the nonABT mice compared to ABT mice and of the C57BL6/J mice compared to NSG mice. KEGG orthologs with significantly different abundances were shown in red (Benjamin–Hochberg adjusted p < 0.05). As an example, all samples were treated as independent. c Differentially abundant KEGG pathways in the successful colonizers. Benjamin–Hochberg adjusted p-values were shown for different sample pooling strategies: panel 1—all samples were treated as independent (no pooling), panel 2—samples representing time points of a same mouse were combined, and panel 3—samples representing time points of all mice in a same cage were combined. Differentially abundant KEGG pathways were compared between successful colonizers and the original human samples (first four columns of each panel), between successful colonizers of the nonABT and ABT mice (column five of each panel) and between successful colonizers of the C57BL6/J and NSG mice (column six of each panel)
Different classes of ABC transporters (ko02010) exhibited varying abundances in the successful colonizers compared with the original human fecal metagenome (Fig. 5c and Figure S3C). For example, two component systems responsible for nitrogen assimilation, including genes involved in nitrogen fixation (NtrY, NtrX, and NifA) and nitrate reduction (NarL, NarG, NarH, and NarI) were enriched in human microbes that colonize NSG ABT and C57BL6/J ABT mice (Figure S3C), potentially due to the need to actively synthesize amino acids and nitrogenous bases or conducting nitrate respiration. The dlt operon (dltABCD), involved in a two component system that incorporates D-ala into Gram-positive cell walls, was significantly reduced in the successful colonizers in all mouse gut environments (Figure S3C), likely because of the enrichment of Gram-negative Bacteroidetes. Collectively, these differences in nutrient acquisition abilities potentially reflect the difference in nutrient usage requirements of the human fecal microbes as well as the availability of the nutrients in the mouse and human gut environments.
Next, we examined pathways that were differentially abundant between successful colonizers of ABT and nonABT mice, which indicate functions important for the colonization ability of exogenous microbes when interacting with the native gut microbiota. We also investigated pathways that were differentially abundant between microbial communities that successfully colonize NSG and C57BL6/J mice to infer functions that increase fitness of microbes in each mouse genotype. Despite the presence of various differentially abundant genes (KEGG orthologs) between colonizers of NSG and C57BL6/J mice (Benjamin–Hochberg adjusted p < 0.05, Fig. 5a), these genes did not cluster into pathways, and therefore no significant differences were identified at the pathway level.
Taxonomic profiling had revealed a selective enrichment of B. vulgatus in nonABT mice of both genotypes, while B. cellulosilyticus and B. xylanisolvens showed reduced abundances under the same conditions (Fig. 4c). It is intriguing what biological functions could be related with the difference in colonization ability between closely related species. Thus, we pooled genes from 29 sequenced draft genomes of the three Bacteroides species, generating a catalog of 138,055 genes, in which 44,378 genes were successfully assigned a KEGG ortholog. The catalog allowed us to compare KEGG functions constrained within these three closely related species, which consistently showed differential abundances in nonABT and ABT mice. We examined which of the KEGG orthologs were significantly enriched in nonABT mice to infer functions important for colonizing a gut environment with a native microbiome. To further restrict our targets to functions that are associated with B. vulgatus, we extracted only those KEGG orthologs that were present in at least one B. vulgatus strain, while absent in at least one strain of B. xylanisolvens and B. cellulosilyticus. This filtering step ensured that there is at least one combination of strains from the three Bacteroides species, in which the KEGG ortholog is present in the B. vulgatus strains but absent in B. xylanisolvens and B. cellulosilyticus strains. A total of 20 KEGG orthologs were enriched in nonABT mice and associated with B. vulgatus, regardless of the sample pooling strategy used (Fig. 6b). Most of these orthologs represented enzymes with biosynthetic and genetic information editing functions, while we also identified several transporters with substrates including amino acids, polyamine, metal ions, and carbohydrates, highlighting the potential importance of nutrient acquisition abilities in colonizing a native gut environment. These findings collectively suggest that colonization ability in a native gut environment could be attributed to a multitude of biological functions, including nutrient acquisition, biosynthesis and genetic information editing.
Fig. 6.
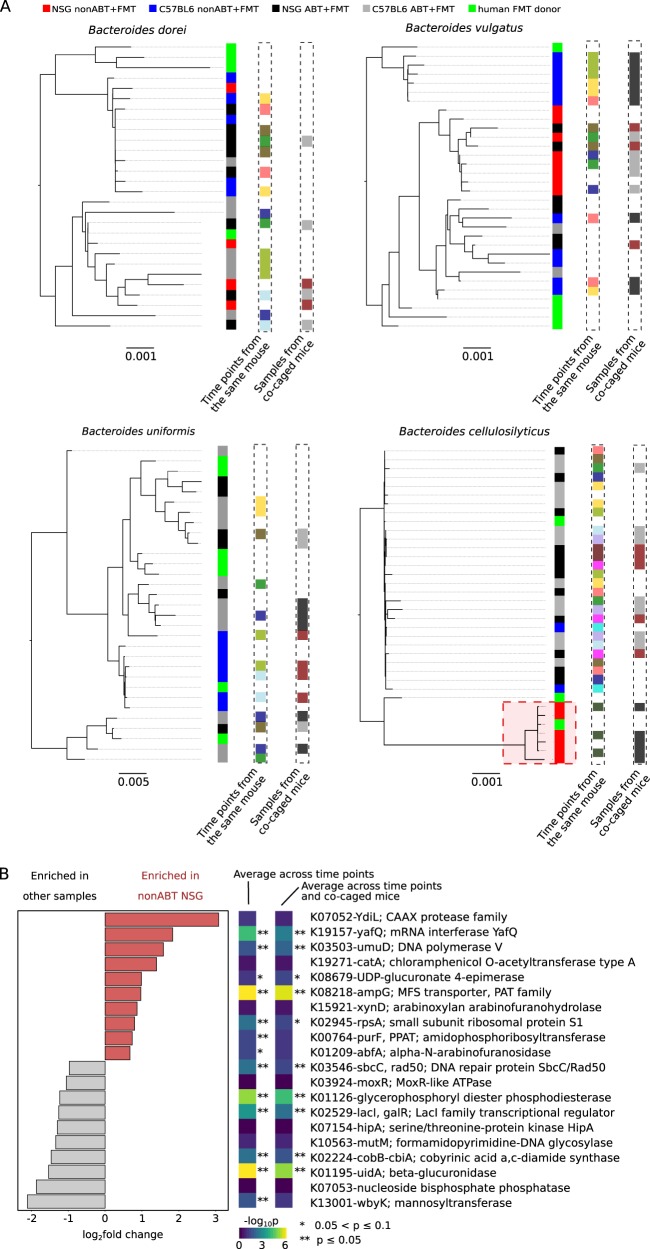
Genes associated with the ability of B. vulgatus to colonize a native gut environment with an intact gut microbiome. KEGG orthologs associated with human fecal B. vulgatus selectively enriched in nonABT mice. Presence/absence of the KEGG orthologs in different Bacteroides strains, as well as the fold enrichment of the KEGG orthologs in nonABT mice was shown. All KEGG orthologs were significantly enriched in nonABT mice (Benjamin–Hochberg adjusted p < 0.05) regardless of sample pooling strategies. GenBank assembly accession numbers were shown for each strain below each column. B. xylanisolvens and B. cellulosilyticus were Bacteroides species consistently enriched in ABT mice while B. vulgatus (highlighted in orange) was consistently enriched in nonABT mice
Selective colonization ability of conspecific bacterial strains in different gut conditions
Closely related, conspecific strains—microbial lineages belonging to a single species—could have diverse phenotypes and preference for environmental conditions, which could result in differential colonizing abilities in novel environments. Indeed, different strains were previously shown to be associated with different environmental types [65, 66], but the colonization abilities of strains under different environments have not been explicitly compared. Leveraging recent developments in mWGS analyses that enabled examination of microbial interactions at strain-resolution, we asked how well-conspecific human fecal strains colonize in different mouse gut environments. We restricted our study targets to human fecal strains, but not native mouse gut strains by reconstructing strain haplotypes from four species (and the only four species) that were abundant in both the donor fecal samples and the FMT-established mouse communities, but were absent in the mouse native gut microbiome. Importantly, the system is analogous to a competitive growth assay among multiple human fecal strains under different mouse gut conditions, but the system is not used to model the fate of strains within a real fecal microbial community of a single individual, for which many species may have only one dominant strain.
We used StrainPhlAn [32] to study the strain diversity of the four species in FMT mice. StrainPhlAn reconstructs strain haplotypes based on single-nucleotide variants observed within species–specific marker gene regions to avoid interference between closely related species. Importantly, StrainPhlAn reconstructs one haplotype for each metagenomic sample, corresponding to the dominant strain type in the sample, which allows robust phylogenetic comparison and population genetics among multiple samples [30, 32, 65], leading to inferences of how well different strain types fit various environmental conditions. Samples representing different time points or co-caged mice did not always share the same dominant strain type (Fig. 7a). We found that strain types could be restricted to specific mouse gut environments. For example, phylogenetic comparison of the reconstructed strain haplotypes showed that nonABT NSG samples were dominated by a distinct cluster of B. cellulosilyticus strains with a very close evolutionary relationship (the red cluster in Fig. 7a). Interestingly, this cluster of B. cellulosilyticus strains is specifically and consistently associated with all nonABT NSG mice, while all other experimental groups are dominated by other B. cellulosilyticus strain types (Fig. 7a). It is unlikely that this observation resulted purely from chance: the probability that the clustering of strains under a specific gut condition, completely resulted from chance, conditional on the phylogeny (i.e., uncorrected p-value) is 9.0 × 10−7 (6C6/33C6, the probability that the distinct cluster in the phylogeny contained all, and only, strains reconstructed from a specific gut environment, among all other possible combinations of strains) when treating all samples as independent, 4.2 × 10−4 (4C4/17C4) when each mouse has an effective sample size of 1, and 0.015 (2C2/12C2) when each cage has an effective sample size of 1. The finding suggests that the cluster of B. cellulosilyticus has high-colonization ability when, and only when the native microbiota of NSG mouse is present. Also, the cluster of B. cellulosilyticus can be traced to a closely related strain haplotype reconstructed from one of the six human donors (Fig. 7a). The findings suggest that although the human fecal sample contained multiple conspecific strains of B. cellulosilyticus as it was an aggregate from multiple individuals, it is possible that the mouse native gut microbiome specifically selected for only a small subset of them. We do note that, however, when corrected for familywise error rate (Šidák correction, at least one of the four species exhibited the pattern by chance), the case where each cage has an effective sample size of 1 is not statistically significant (corrected p = 0.059). Although this is the minimal p-value achievable given the phylogeny and the sample size, the low statistical power suggests the need for an increased sample size to better capture the strain clustering.
Fig. 7.
Selection on conspecific strains by the mouse gut environments. a Maximum-likelihood phylogeny of the reconstructed strain haplotypes. Each strain is color coded by its host condition (the first column to the right of each phylogeny). Additionally, strains sharing the same color code (white not included) in columns labeled with “time points from the same mouse” or “samples from co-caged mouse” were reconstructed from samples representing time points from a same mouse or co-caged mice, respectively. A distinct cluster of B. cellulosilyticus strains that were exclusively dominant in the nonABT NSG mice were highlighted in a red square. b B. cellulosilyticus accessory genes that had significantly higher or lower abundances in the nonABT NSG mice. Top 10 enriched or depleted genes with the largest fold changes were shown; all genes satisfied Benjamin–Hochberg adjusted p < 10−5 when all samples were treated as independent. Additionally, the Benjamin–Hochberg adjusted p-values were also shown with different sample pooling strategies: “average across time points”—samples representing time points of a same mouse were combined, “average across time points and co-caged mice” —samples representing time points of all mice in a same cage were combined
The fact that the B. cellulosilyticus strains dominating the nonABT NSG mice were phylogenetically similar suggested that they likely share genes that facilitated engraftment even when the native mouse gut microbiome is present. To explore genes that are potentially related to the engraftment ability, we compiled an accessory gene catalog for all five sequenced B. cellulosilyticus assemblies (as listed in Fig. 6b), in order to explore B. cellulosilyticus-specific functions that are enriched or depleted in nonABT NSG samples. We found a variety of B. cellulosilyticus accessory genes that were differentially abundant in nonABT NSG mice and other mouse conditions (Fig. 7b). Although inconclusive, several differentially abundant genes were previously reported to influence bacterial colonization abilities. For example, genes in the porphyrin and chlorophyll metabolism pathway (KEGG: ko00860), such as the uidA and cobB-cbiA genes (Fig. 7b) among others (Figure S4), had significantly lower abundances in nonABT NSG samples (pathway differential abundance Benjamin–Hochberg adjusted p = 0.036 when all samples were treated as independent, p = 0.094 when each mouse had an effective sample size of 1, and p = 0.061 when each cage had an effective sample size of 1), although these genes were present in all five sequenced B. cellulocilyticus strains deposited in GenBank. The pathway synthesizes, among other products, Vitamin B12 (Vb12) coenzyme, a compound known to regulate mouse gut microbiota compositions [67, 68]. The observation raised the possibility that the nonABT NSG mice were colonized by novel B. cellulocilyticus strains, which had lost porphyrin and chlorophyll biosynthetic genes. Additionally, yafQ, a gene highly enriched in nonABT NSG samples, has been shown to facilitate the general stress responses of bacteria [69]. These genes are potential determinants of the observed strain-level selectivity, underscoring the value of strain-level metagenomic analyses for generating hypotheses for future experimental validation.
Discussion
In this study, we analyzed the interaction between host genotypes, the native host microbiome, and exogenous colonizers at high taxonomic resolution. We found that both the host genotypes and the native host microbiome can each exert different selective pressures on colonizer species. The mouse gut microbiota resists the colonization of human fecal species, resulting in the decreased abundances and taxonomic diversity of the engrafted human fecal species in the mice not pre-treated with antibiotics. Nonetheless, multiple human fecal species from the Bacteroides genera were able to successfully invade the existing microbial communities of both the ABT and nonABT mice. Moreover, different Bacteroides species are selectively enriched under different gut conditions: for both mouse genotypes, B. xylanisolvens and B. cellulosilyticus were consistently enriched in the ABT mice while B. vulgatus was consistently enriched in the nonABT mice, demonstrating strong colonization ability despite resistance exerted by the mouse gut microbiome. The differential colonization abilities of these closely related species were likely due to a multitude of functional differences, including versatility in nutrient acquisition, biosynthetic abilities, and genetic information editing. Such colonization abilities are unlikely due to an evolutionary adaptation to the new environment, because adaptation commonly occurs over a much longer evolutionary time scales [70]. This observation can be better described by ecological fitting [71], in which organisms successfully colonize new environments using traits already in hand from their native environment. Using a mixture of human fecal samples, we tracked the differential colonization ability of different human fecal microbial strains in the mouse gut environments. Our data suggested that even strains from the same bacterial species could have distinct fitness in a novel host environment. For example, the native gut microbiome of NSG mice accept a specific subset of B. cellulosilyticus strains while resisting colonization of other strains of the same species—a pattern consistently observed across all native NSG mice. The strain-level selectivity is potentially due to the bioavailability of important nutrients such as Vb12 and the differential ability of the strains to synthesize them. Putting together, this study demonstrated interactions between host conditions and exogenous colonizers that could not be discovered without profiling microbial communities at species and strain resolution.
Previous studies have demonstrated the differential colonization ability of diverse microbial taxa in the mouse gut environments [12, 13], and more recent studies have revealed that the gut space can host different strains of the same exogenous species [10, 11]. Nonetheless, it was unclear whether and how exogenous colonizers are influenced by host genotypes and the native host microbiome, and how different strains of the same species colonize in different gut environments. Therefore, our findings are novel in several ways. First, we showed that both host genotype and host microbiota influence the colonization ability of exogenous microbes. Second, we showed that different conspecific strain types dominate different gut environments. Additionally, we showed that the different colonization ability of closely related species and strains could be related with functional genes and pathways that synthesize important nutrients or are essential to stress responses.
Previous studies have shown that immunodeficiency reshapes the native microbiome [72, 73], and the microbiota plays a significant role in immunodeficiency diseases such as common variable immunodeficiency [74], which increases the host’s susceptibility to infections. Additionally, ABT is able to modulate immune responses in mice (e.g., ref. [75]), potentially magnifying the difference between immunodeficient and immunocompetent mice. However, it was unclear how an immunodeficiency phenotype would shape an exogenous colonizing community. For example, would the lack of major mouse immune compartments allow a greater number of microbes to engraft due to reduced immune selectivity? We indeed identified differential engraftment by FMT in the severely immunodeficient NSG mice versus immunocompetent C57BL6/J mice, although in unexpected ways. Interestingly, no significant difference in overall species diversity was observed in the FMT-established communities in NSG versus C57BL6/J mice whose native microbiota were depleted by ABT, suggesting that a fully functional immune system does not simply act as a resistance mechanism against exogenous colonizers. Despite no differences in the species diversity in NSG and C57BL6/J mice, we observed differential enrichment of closely related human Bacteroides species from the same genus in the two mouse genotypes, underscoring the value of species and strain-level analyses. These findings suggest that the selection exerted by the host immune phenotypes is likely highly specific - at least specific enough to result in differential enrichment of closely related microbial species in NSG and C57BL6/J mice.
In addition to species-level selectivity, our results suggested the presence of strain-level selection on exogenous species exerted by the mouse gut microbiome, potentially due to functional differences among the strains. It has been well-established that conspecific strains can diverge significantly in their functional capacity, leading to varying fitness in new host environments. For example, different Escherichia coli strains have diverse colonization abilities in the mouse gut [76] and enteric pathogens such as C. jejuni also show strain-level divergence in colonization ability in animal models [77]. In other ecosystems, such as human skin, functionally diverse strains of Propionibacterium acnes and Staphylococcus epidermidis can stably co-exist in the same niche, suggesting a saturation of pangenome functions that results in a homeostatic community that likely has colonization resistance against other strains [78, 79]. Strain-level fitness variations identified in this study were restricted to Bacteroides species due to insufficient sequencing coverage of other colonizers. The human gut hosts a wide variety of strains in Bacteroides species [80], which have diverse metabolic and immune-modulatory properties (e.g., ref. [81]), which could manifest as unusually flexible colonizing ability. Moreover, the transmission of gut Bacteroides strains was also found between mother and infants, highlighting their natural colonization ability [30, 82]. Among the Bacteroides species, B. cellulocilyticus strains showed an especially strong environmental selectivity: a distinct cluster of B. cellulocilyticus strains dominated and only dominated the nonABT NSG mice, suggesting for the presence of important functional factors that result in this evolutionary pattern. B. cellulocilyticus accessory genes that were significantly enriched or depleted in the nonABT NSG mice samples covered a large spectrum of biological functions from metabolic potential to stress responses. One factor that potentially contributed to the environmental selectivity is the biosynthetic pathway of porphyrin and chlorophyll, which is responsible for the biosynthesis of Vb12, a known modulator of gut microbiota [67]. Although most bacterial species may require some variant of Vb12 [68], the biosynthetic pathway was present in only half of the bacteria species [83], suggesting the ability of other bacteria to uptake Vb12 from the environment. Likewise, for B. cellulosilyticus, the Vb12 biosynthetic pathway is not essential for in vivo fitness [84], due to its ability to uptake environmental Vb12 through transporters BtuB1234. Therefore, when environmental Vb12 was not depleted, B. cellulosilyticus strains without the biosynthetic pathway could gain fitness by avoiding the energy cost of synthesizing the gene products in the pathway.
In this study, we identified human fecal species and strains that exhibited differential fitness under different gut conditions, and inferred functional factors that could influence their interaction with the mouse gut environment. Nonetheless, it is important to note that the statistical power of the study is restricted by the relatively small sample size (24 mice in 16 cages), allowing us to focus only on patterns consistently observed across sample pooling strategies. Similarly, our data suggested strain-level selectivity of the mouse gut environments on certain exogenous bacterial species, but the pattern was not consistently significant across sample pooling strategies (corrected p = 0.059 when pooling all samples in each cage) due to the limitation of sample sizes, indicating the need of an increased effective sample size for future strain tracking analyses. Moreover, functional inferences based on metagenomic sequencing are not conclusive by nature—their strength lies in hypothesis generation that require experimental validation. Additionally, a better understanding of these functional interactions requires comprehensive profiling of the native mouse gut microbiome, which is yet poorly understood given the under representation of mouse-associated microbes in the open-access databases. Indeed, even with a combination of a comprehensive microbial reference database and de novo reconstruction, we found that the majority (over 70%) of the mouse gut microbiome still remained uncharacterized. Based on our results, the characterized fraction of the mouse gut microbiome was dominated by bacteria from the phylum Firmicutes, consistent with findings from recent efforts to profile the mouse gut microbiome using culturomics [85] and combining previous 16S sequencing data [86]. However, our 16S rRNA sequencing results (Figure S1A), as well as several other studies including mWGS-based profiling [56], and analyses using full-length 16S rRNA [87], identified a larger proportion of non-Firmicutes bacteria, suggesting the under representation of mouse-specific non-Firmicutes in present whole-genome reference databases. To control for these complications, future studies seeking to elucidate the interaction between host microbiome and exogenous colonizers could use models with defined microbiota, such as gnotobiotic mouse models.
Finally, the differential colonization ability of closely related bacterial species and strains is especially relevant for medical treatments such as FMT and probiotic applications, which involve the application of live bacteria. Given the diversity in functionality and colonization ability between closely related bacteria—particularly at the strain level, where the greatest interindividual variability likely exists—an important consideration for probiotic and FMT-based medication is how to successfully introduce a species/strain with the desired function but also robust colonization ability into a wide range of recipient communities. Based on our findings, a strong colonizer, and therefore a strong therapeutic candidate, requires the ability to synthesize nutrients that might be lacking in the target host environment as well as the ability to respond to stress conditions caused by the host microbiome. These data provide initial insights to guide which microbiota can be selectively isolated or engineered to optimize these characteristics.
Electronic supplementary material
Acknowledgements
We would like to thank Bonnie Lyons and Dina Baker, and Kin-hoe Chow’s teams for their valuable support in mouse experiments. This work was supported by Mackenzie Foundation #270271-1217 and Global Probiotics Council #270250-0617.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0312-9) contains supplementary material, which is available to authorized users.
References
- 1.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol. 2010;8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sung J, Kim S, Cabatbat JJT, Jang S, Jin YS, Jung GY, et al. Global metabolic interaction network of the human gut microbiota for context-specific community-scale analysis. Nat Commun. 2017;8:15393. doi: 10.1038/ncomms15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Little AE, Robinson CJ, Peterson SB, Raffa KF, Handelsman J. Rules of engagement: interspecies interactions that regulate microbial communities. Annu Rev Microbiol. 2008;62:375–401. doi: 10.1146/annurev.micro.030608.101423. [DOI] [PubMed] [Google Scholar]
- 4.Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol. 2012;10:323–35. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dostal A, Chassard C, Hilty FM, Zimmermann MB, Jaeggi T, Rossi S, et al. Iron depletion and repletion with ferrous sulfate or electrolytic iron modifies the composition and metabolic activity of the gut microbiota in rats. J Nutr. 2012;142:271–7. doi: 10.3945/jn.111.148643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. 2013;13:790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisher CK, Mora T, Walczak AM. Variable habitat conditions drive species covariation in the human microbiota. PLoS Comput Biol. 2017;13:e1005435. doi: 10.1371/journal.pcbi.1005435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stecher B, Berry D, Loy A. Colonization resistance and microbial ecophysiology: using gnotobiotic mouse models and single-cell technology to explore the intestinal jungle. FEMS Microbiol Rev. 2013;37:793–829. doi: 10.1111/1574-6976.12024. [DOI] [PubMed] [Google Scholar]
- 9.Sharon I, Morowitz MJ, Thomas BC, Costello EK, Relman DA, Banfield JF. Time series community genomics analysis reveals rapid shifts in bacterial species, strains, and phage during infant gut colonization. Genome Res. 2013;23:111–20. doi: 10.1101/gr.142315.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li SS, Zhu A, Benes V, Costea PI, Hercog R, Hildebrand F, et al. Durable coexistence of donor and recipient strains after fecal microbiota transplantation. Science. 2016;352:586–9. doi: 10.1126/science.aad8852. [DOI] [PubMed] [Google Scholar]
- 11.Smillie CS, Sauk J, Gevers D, Friedman J, Sung J, Youngster I, et al. Strain Tracking Reveals the Determinants of Bacterial Engraftment in the Human Gut Following Fecal Microbiota Transplantation. Cell Host Microbe. 2018;23:229–40. doi: 10.1016/j.chom.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Staley C, Kaiser T, Beura LK, Hamilton MJ, Weingarden AR, Bobr A, et al. Stable engraftment of human microbiota into mice with a single oral gavage following antibiotic conditioning. Microbiome. 2017;5:87. doi: 10.1186/s40168-017-0306-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seedorf H, Griffin NW, Ridaura VK, Reyes A, Cheng J, Rey FE, et al. Bacteria from diverse habitats colonize and compete in the mouse gut. Cell. 2014;159:253–66. doi: 10.1016/j.cell.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mushin R, Dubos R. Colonization of the mouse intestine with Escherichia coli. J Exp Med. 1965;122:745–57. doi: 10.1084/jem.122.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freter R, Abrams GD. Function of various intestinal bacteria in converting germfree mice to the normal state. Infect Immun. 1972;6:119–26. doi: 10.1128/iai.6.2.119-126.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang C, Miller JF. Campylobacter jejuni colonization of mice with limited enteric flora. Infect Immun. 2006;74:5261–71. doi: 10.1128/IAI.01094-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ross CL, Spinler JK, Savidge TC. Structural and functional changes within the gut microbiota and susceptibility to Clostridium difficile infection. Anaerobe. 2016;41:37–43. doi: 10.1016/j.anaerobe.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seekatz AM, Young VB. Clostridium difficile and the microbiota. J Clin Invest. 2014;124:4182–9. doi: 10.1172/JCI72336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zipperer A, Konnerth MC, Laux C, Berscheid A, Janek D, Weidenmaier C, et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature. 2016;535:511–6. doi: 10.1038/nature18634. [DOI] [PubMed] [Google Scholar]
- 20.Mota-Meira M, LaPointe G, Lacroix C, Lavoie MC. MICs of mutacin B-Ny266, nisin A, vancomycin, and oxacillin against bacterial pathogens. Antimicrob Agents Chemother. 2000;44:24–29. doi: 10.1128/aac.44.1.24-29.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rea MC, Sit CS, Clayton E, O’Connor PM, Whittal RM, Zheng J, et al. Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile. Proc Natl Acad Sci USA. 2010;107:9352–7. doi: 10.1073/pnas.0913554107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong M, Liang X, Smart M, Tang L, Moore R, Ingalls B, et al. Microbial herd protection mediated by antagonistic interaction in polymicrobial communities. Appl Environ Microbiol. 2016;82:6881-6888. [DOI] [PMC free article] [PubMed]
- 23.Reddinger RM, Luke-Marshall NR, Sauberan SL, Hakansson AP, Campagnari AA. Streptococcus pneumoniae modulates Staphylococcus aureus biofilm dispersion and the transition from colonization to invasive disease. MBio. 2018;9:e02089-17. [DOI] [PMC free article] [PubMed]
- 24.Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci USA. 2010;107:18933–8. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Esworthy RS, Smith DD, Chu FF. A strong impact of genetic background on gut microflora in mice. Int J Inflam. 2010;2010:986046. doi: 10.4061/2010/986046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vaahtovuo J, Toivanen P, Eerola E. Bacterial composition of murine fecal microflora is indigenous and genetically guided. FEMS Microbiol Ecol. 2003;44:131–6. doi: 10.1016/S0168-6496(02)00460-9. [DOI] [PubMed] [Google Scholar]
- 27.Lawley TD, Walker AW. Intestinal colonization resistance. Immunology. 2013;138:1–11. doi: 10.1111/j.1365-2567.2012.03616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vieira G, Sabarly V, Bourguignon PY, Durot M, Le Fevre F, Mornico D, et al. Core and panmetabolism in. Escherichia coli J Bacteriol. 2011;193:1461–72. doi: 10.1128/JB.01192-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moriel DG, Rosini R, Seib KL, Serino L, Pizza M, Rappuoli R. Escherichia coli: great diversity around a common core. MBio. 2012;3:e00118–12. doi: 10.1128/mBio.00118-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asnicar F, Manara S, Zolfo M, Truong DT, Scholz M, Armanini F, et al. Studying vertical microbiome transmission from mothers to infants by strain-level metagenomic profiling. mSystems. 2017;2:e00164–16. doi: 10.1128/mSystems.00164-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scholz M, Ward DV, Pasolli E, Tolio T, Zolfo M, Asnicar F, et al. Strain-level microbial epidemiology and population genomics from shotgun metagenomics. Nat Methods. 2016;13:435–8. doi: 10.1038/nmeth.3802. [DOI] [PubMed] [Google Scholar]
- 32.Truong DT, Tett A, Pasolli E, Huttenhower C, Segata N. Microbial strain-level population structure and genetic diversity from metagenomes. Genome Res. 2017;27:626–38. doi: 10.1101/gr.216242.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turner S, Pryer KM, Miao VP, Palmer JD. Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J Eukaryot Microbiol. 1999;46:327–38. doi: 10.1111/j.1550-7408.1999.tb04612.x. [DOI] [PubMed] [Google Scholar]
- 34.Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–63. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buffalo V. Scythe - A Bayesian adapter trimmer [software]. https://github.com/vsbuffalo/scythe; 2011.
- 36.Joshi NAF, J.N. Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files [Software]. https://github.com/najoshi/sickle; 2011.
- 37.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li D, Liu CM, Luo R, Sadakane K, Lam TW. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–6. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 39.Li D, Luo R, Liu CM, Leung CM, Ting HF, Sadakane K, et al. MEGAHITv1.0: a fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods. 2016;102:3–11. doi: 10.1016/j.ymeth.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 40.Kang DD, Froula J, Egan R, Wang Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ. 2015;3:e1165. doi: 10.7717/peerj.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–2. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 42.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rognes T, Flouri T, Nichols B, Quince C, Mahe F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–6. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yilmaz P, Parfrey LW, Yarza P, Gerken J, Pruesse E, Quast C, et al. The SILVA and “all-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014;42:D643–8. doi: 10.1093/nar/gkt1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Glockner FO, Yilmaz P, Quast C, Gerken J, Beccati A, Ciuprina A, et al. 25 years of serving the community with ribosomal RNA gene reference databases and tools. J Biotechnol. 2017;261:169–76. doi: 10.1016/j.jbiotec.2017.06.1198. [DOI] [PubMed] [Google Scholar]
- 48.Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, Huttenhower C. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods. 2012;9:811–4. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hong C, Manimaran S, Shen Y, Perez-Rogers JF, Byrd AL, Castro-Nallar E, et al. PathoScope 2.0: a complete computational framework for strain identification in environmental or clinical sequencing samples. Microbiome. 2014;2:33. doi: 10.1186/2049-2618-2-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou W, Gay N, Oh J. ReprDB and panDB: minimalist databases with maximal microbial representation. Microbiome. 2018;6:15. doi: 10.1186/s40168-018-0399-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Knights D, Kuczynski J, Charlson ES, Zaneveld J, Mozer MC, Collman RG, et al. Bayesian community-wide culture-independent microbial source tracking. Nat Methods. 2011;8:761–3. doi: 10.1038/nmeth.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinforma. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xiao L, Feng Q, Liang S, Sonne SB, Xia Z, Qiu X, et al. A catalog of the mouse gut metagenome. Nat Biotechnol. 2015;33:1103–8. doi: 10.1038/nbt.3353. [DOI] [PubMed] [Google Scholar]
- 57.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–93. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jonsson V, Osterlund T, Nerman O, Kristiansson E. Statistical evaluation of methods for identification of differentially abundant genes in comparative metagenomics. BMC Genom. 2016;17:78. doi: 10.1186/s12864-016-2386-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luo W, Friedman MS, Shedden K, Hankenson KD, Woolf PJ. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinforma. 2009;10:161. doi: 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Luo W, Brouwer C. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics. 2013;29:1830–1. doi: 10.1093/bioinformatics/btt285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Luo W. RNA-Seq data pathway and gene-set analysis workflows [Web tutorial]. [https://bioconductor.org/packages/3.7/bioc/vignettes/gage/inst/doc/RNA-seqWorkflow.pdf].
- 64.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, et al. vegan: Community Ecology Package (R package version 2.4-6). https://CRAN.R-project.org/package=vegan; 2018.
- 65.Lloyd-Price J, Mahurkar A, Rahnavard G, Crabtree J, Orvis J, Hall AB, et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature. 2017;550:61–66. doi: 10.1038/nature23889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.O’Donnell MM, Harris HM, Lynch DB, Ross RP, O’Toole PW. Lactobacillus ruminis strains cluster according to their mammalian gut source. BMC Microbiol. 2015;15:80. doi: 10.1186/s12866-015-0403-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Degnan PH, Taga ME, Goodman AL. Vitamin B12 as a modulator of gut microbial ecology. Cell Metab. 2014;20:769–78. doi: 10.1016/j.cmet.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Frank SA. Receptor uptake arrays for vitamin B12, siderophores, and glycans shape bacterial communities. Ecol Evol. 2017;7:10175–95. doi: 10.1002/ece3.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang X, Wood TK. Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl Environ Microbiol. 2011;77:5577–83. doi: 10.1128/AEM.05068-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Slobodkin LB. Growth and regulation of animal populations. Science. 1961;134:1. [Google Scholar]
- 71.Janzen DH. On ecological fitting. Oikos. 1985;45:3. [Google Scholar]
- 72.Lane JP, Stewart CJ, Cummings SP, Gennery AR. Gut microbiome variations during hematopoietic stem cell transplant in severe combined immunodeficiency. J Allergy Clin Immunol. 2015;135:1654–6. doi: 10.1016/j.jaci.2015.01.024. [DOI] [PubMed] [Google Scholar]
- 73.Selvanantham T, Lin Q, Guo CX, Surendra A, Fieve S, Escalante NK, et al. NKT cell-deficient mice harbor an altered microbiota that fuels intestinal inflammation during chemically induced colitis. J Immunol. 2016;197:4464–72. doi: 10.4049/jimmunol.1601410. [DOI] [PubMed] [Google Scholar]
- 74.Berbers RM, Nierkens S, van Laar JM, Bogaert D, Leavis HL. Microbial dysbiosis in common variable immune deficiencies: evidence, causes, and consequences. Trends Immunol. 2017;38:206–16. doi: 10.1016/j.it.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 75.Knoop KA, McDonald KG, Kulkarni DH, Newberry RD. Antibiotics promote inflammation through the translocation of native commensal colonic bacteria. Gut. 2016;65:1100–9. doi: 10.1136/gutjnl-2014-309059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Myhal ML, Laux DC, Cohen PS. Relative colonizing abilities of human fecal and K 12 strains of Escherichia coli in the large intestines of streptomycin-treated mice. Eur J Clin Microbiol. 1982;1:186–92. doi: 10.1007/BF02019621. [DOI] [PubMed] [Google Scholar]
- 77.Hanel I, Muller J, Muller W, Schulze F. Correlation between invasion of Caco-2 eukaryotic cells and colonization ability in the chick gut in Campylobacter jejuni. Vet Microbiol. 2004;101:75–82. doi: 10.1016/j.vetmic.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 78.Oh J, Byrd AL, Deming C, Conlan S, Program NCS, Kong HH, et al. Biogeography and individuality shape function in the human skin metagenome. Nature. 2014;514:59–64. doi: 10.1038/nature13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Oh J, Byrd AL, Park M, Program NCS, Kong HH, Segre JA. Temporal Stability of the Human Skin Microbiome. Cell. 2016;165:854–66. doi: 10.1016/j.cell.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kraal L, Abubucker S, Kota K, Fischbach MA, Mitreva M. The prevalence of species and strains in the human microbiome: a resource for experimental efforts. PLoS ONE. 2014;9:e97279. doi: 10.1371/journal.pone.0097279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Neff CP, Rhodes ME, Arnolds KL, Collins CB, Donnelly J, Nusbacher N, et al. Diverse intestinal bacteria contain putative zwitterionic capsular polysaccharides with anti-inflammatory properties. Cell Host Microbe. 2016;20:535–47. doi: 10.1016/j.chom.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ferretti P, Pasolli E, Tett A, Asnicar F, Gorfer V, Fedi S, et al. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe. 2018;24:133–45. doi: 10.1016/j.chom.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Magnusdottir S, Ravcheev D, de Crecy-Lagard V, Thiele I. Systematic genome assessment of B-vitamin biosynthesis suggests co-operation among gut microbes. Front Genet. 2015;6:148. doi: 10.3389/fgene.2015.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu M, McNulty NP, Rodionov DA, Khoroshkin MS, Griffin NW, Cheng J, et al. Genetic determinants of in vivo fitness and diet responsiveness in multiple human gut Bacteroides. Science. 2015;350:aac5992. doi: 10.1126/science.aac5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lagkouvardos I, Pukall R, Abt B, Foesel BU, Meier-Kolthoff JP, Kumar N, et al. The Mouse Intestinal Bacterial Collection (miBC) provides host-specific insight into cultured diversity and functional potential of the gut microbiota. Nat Microbiol. 2016;1:16131. doi: 10.1038/nmicrobiol.2016.131. [DOI] [PubMed] [Google Scholar]
- 86.Clavel T, Lagkouvardos I, Blaut M, Stecher B. The mouse gut microbiome revisited: From complex diversity to model ecosystems. Int J Med Microbiol. 2016;306:316–27. doi: 10.1016/j.ijmm.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 87.Shin J, Lee S, Go MJ, Lee SY, Kim SC, Lee CH, et al. Analysis of the mouse gut microbiome using full-length 16S rRNA amplicon sequencing. Sci Rep. 2016;6:29681. doi: 10.1038/srep29681. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.