Abstract
Sleep disordered breathing (SDB)-related overnight hypoxemia is associated with cardiometabolic disease and other comorbidities. Understanding the genetic bases for variations in nocturnal hypoxemia may help understand mechanisms influencing oxygenation and SDB-related mortality. We conducted genome-wide association tests across 10 cohorts and 4 populations to identify genetic variants associated with three correlated measures of overnight oxyhemoglobin saturation: average and minimum oxyhemoglobin saturation during sleep and the percent of sleep with oxyhemoglobin saturation under 90%. The discovery sample consisted of 8,326 individuals. Variants with p < 1 × 10−6 were analyzed in a replication group of 14,410 individuals. We identified 3 significantly associated regions, including 2 regions in multi-ethnic analyses (2q12, 10q22). SNPs in the 2q12 region associated with minimum SpO2 (rs78136548 p = 2.70 × 10−10). SNPs at 10q22 were associated with all three traits including average SpO2 (rs72805692 p = 4.58 × 10−8). SNPs in both regions were associated in over 20,000 individuals and are supported by prior associations or functional evidence. Four additional significant regions were detected in secondary sex-stratified and combined discovery and replication analyses, including a region overlapping Reelin, a known marker of respiratory complex neurons.These are the first genome-wide significant findings reported for oxyhemoglobin saturation during sleep, a phenotype of high clinical interest. Our replicated associations with HK1 and IL18R1 suggest that variants in inflammatory pathways, such as the biologically-plausible NLRP3 inflammasome, may contribute to nocturnal hypoxemia.
Author summary
Variation in oxyhemoglobin saturation, the proportion of oxygen-saturated to total hemoglobin in the blood, is associated with numerous disorders and is a predictor of health outcomes including mortality, incident heart failure, and dementia. Despite the fundamental role of oxygen saturation in normal and abnormal physiology, there are few large-scale genetic studies of oxygen saturation performed across populations. Overnight measurements provide more variability than daytime levels due to the “stresses” associated with normal and disordered breathing, and also provide an important measure of sleep apnea severity, a common disorder in the population that is associated with considerable morbidity. In this study, for the first time, we identified multiple replicated genome-significant associations based on up to 22,736 individuals from 10 cohort studies. Our findings suggest a contribution of inflammatory genes such as the Interleukin 18 receptor subunit genes to the genetic architecture of sleep-disordered breathing. These results extend our understanding of the genetics of oxyhemoglobin saturation and sleep-disordered breathing and may provide further insight into the biology of associated diseases.
Introduction
Arterial oxyhemoglobin saturation is a fundamental physiological trait that is tightly regulated at cellular and systemic levels to optimize tissue oxygen delivery. Reduced values, or hypoxemia, occurs secondary to acute and chronic respiratory or cardiovascular diseases, and rarely due to hemoglobin protein mutations. Chronically, low oxygen saturation predicts cognitive deficits in patients with chronic obstructive pulmonary disease (COPD) and in sleep apnea (SA) [1,2].
Oxyhemoglobin saturation (SpO2), the proportion of oxygen-saturated to total hemoglobin in the blood, is most commonly measured using non-invasive equipment (oximeters). Oximetry is used to screen and monitor a wide range of health conditions. Normal SpO2 values range from 95% to 100% during wakefulness and normally fall by 2 to 4% during sleep. Oxygen saturation is reduced in individuals living at high altitude and in patients with cardiopulmonary diseases. However, even within specific disease groups, there is variation in SpO2 that is not explained by factors such as age, obesity, lung function or tobacco exposure [3]. Twin studies indicate that as much as 26% of the variation in waking SpO2 can be explained by genetic factors [4]. Population studies also indicate that genetic effects contribute to variation in waking SpO2 among Tibetan highlanders and in COPD [3,5].
Sleep disordered breathing (SDB) is a common disorder characterized by recurrent falls of SpO2 during sleep due to repetitive episodes of apneas (no airflow) or partial airflow (hypopneas), most often due to recurrent collapse of the upper airway. Our prior family-based studies have indicated that average SpO2 during sleep is significantly heritable [6]. Sleep-related hypoxemia is a key component of the pathophysiology of the disorder and variations in SpO2 during sleep in patients with SDB are predictive of incident atrial fibrillation [7], certain types of cancer [8–10], and death [7]. Minimum nocturnal SpO2 predicted future carotid plaque burden in the Wisconsin sleep cohort, even after adjusting for traditional risk factors [11]. In cohorts of patients with both heart failure and SDB, overnight hypoxemia is a stronger risk factor for incident cardiovascular events and death than is the apnea hypopnea index (AHI, a count of the number of breathing pauses per sleep hour) [12–14]. Mean oxygen saturation and acute hypoxemia during sleep have more significant associations with liver steatosis than the AHI [15]. Sleep-related hypoxemia also may significantly influence prognosis of patients with COPD, asthma, and interstitial lung disease [16–19]. Adverse effects of sleep-related hypoxemia include those directly related to tissue ischemia as well as to the effects related to activation of hypoxia-inducible factor-1 (HIF1A) mediated and NF-kB pathways, that then activate the sympathetic nervous system, stimulate release of angiogenic and inflammatory factors, cause oxidative stress, reduce insulin sensitivity, and cause endothelial dysfunction. Therefore, understanding variation in nocturnal SpO2 is important for improving our understanding of variation in risk of a wide range of chronic health outcomes.
In this study, we conducted the first multi-ethnic genome-wide association study (GWAS) of 3 nocturnal oxygen hemoglobin saturation (SpO2) traits: average and minimum SpO2, and the percent of sleep time under 90% SpO2 (Per90). (Lower values of Per90 are better while higher values are better for the other two measures.) These measures provide complementary information on hypoxemic burden across the sleep episode and can be derived from oximetry, which is potentially scalable for large-scale studies.
We analyzed data from 10 cohort studies and four ethnic groups and focused on identifying obesity-independent loci by adjusting for body mass index (BMI). We also considered sex differences in associations given the growing interest in sexual dimorphism in genetic analyses [20]. Furthermore, SDB prevalence varies by sex [21,22]), as do SDB risk factors such as the ventilatory response to arousal and regional fat distributions [23,24], and sex differences have also been reported for the relationship between chronic hypoxemia and cardiovascular events [12]. These analyses extend our prior report of results for SDB in Hispanic/Latino-Americans [25].
Results
Study sample and cross-phenotype correlations
Descriptive characteristics of the discovery and replication study samples are provided in Tables 1 and 2. Collectively, we studied 22,736 individuals. The discovery sample consisted of 8,326 individuals across 6 studies and 4 populations (1,209 African-Americans [AA]; 228 Asian-Americans [AsA]; 5,649 European-Americans [EA]; 1,240 Hispanic/Latino-Americans [HA]). Replication cohorts included 14,410 individuals (681 AAs, 2,378 EAs and European-Australians, and 11,351 HA) from 4 cohorts. Across cohorts, mean age ranged from 37.8 (CFS African-Americans) to 77.7 years (CHS European-Americans). Female participation ranged from 0% (MrOS) to 72% (Starr). The mean BMI ranged from 24.1 kg/m2 (MESA Asian-Americans) to 32.3 (JHS). Waking SpO2 values, which were typically measured prior to the sleep episode on the same equipment, ranged from 94.96% (in the older MrOS cohort) to 97.78% (among relatively young CFS African-Americans). Average SpO2 during sleep ranged from 93.74% (CFS European-Americans) to 96.45% (HCHS/SOL). The median Apnea Hypopnea Index ranged from 1.97 (HCHS/SOL, a relatively young cohort) to 24.60 (WASHS, a sleep clinic-derived cohort). Average forced vital capacity (FVC; percent predicted value) exceeded 90% in cohorts in which these data were available. The prevalence of chronic lung diseases (asthma, COPD) and diabetes varied across cohorts, likely reflecting differences in age, ascertainment and possible disease definitions.
Table 1. Discovery sample description.
| Ethnic Group | Cohort | N | Age | Percent Female | BMI | Sleep Episode SpO2 | Minimum Sleep Episode SpO2 | Percent Sleep Under 90% SpO2 | Apnea Hypopnea Index | AHI (% <5, 5–15, > = 15) | Waking SpO2 | FVC (% Predicted) | % Asthma | COPD | % Diabetes |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| African-American | CFS* | 719 | 37.8 (19.4) | 55.6 | 31.6 (9.6) | 94.57 (3.77) | 85.82 (9.88) | 4.58 (13.14) | 5.61 (19.68) | 47.1, 21.2, 31.7 | 97.78 (1.91) | 95.10 (19.67) | 21.5 | 15.9 | 15.8 |
| MESA | 490 | 69.1 (9.1) | 54.3 | 30.4 (5.7) | 94.44 (1.96) | 82.93 (8.17) | 4.01 (9.46) | 13.34 (21.16) | 21.2, 32.2, 46.5 | 96.11 (1.35) | 96.02 (17.62) | 5.3 | 14.1 | 28.0 | |
| Asian-American | MESA | 228 | 68.1 (9.2) | 50.4 | 24.1 (3.2) | 94.96 (1.22) | 83.42 (7.38) | 2.17 (4.30) | 13.97 (23.90) | 22.4, 30.3, 47.4 | 96.18 (1.01) | 98.45 (15.49) | 4.4 | 16.6 | 16.0 |
| European-American | ARIC | 1,432 | 62.4 (5.7) | 51.5 | 28.8 (5.1) | 94.47 (1.99) | 85.62 (6.12) | 3.36 (10.33) | 8.67 (15.55) | 3.0, 34.5, 32.5 | 96.08 (1.70) | 102.39 (13.48) | 7.3 | 0.9 | 6.1 |
| CFS* | 692 | 41.6 (19.5) | 52.8 | 30.2 (8.7) | 93.74 (3.67) | 86.46 (9.07) | 4.29 (12.30) | 5.52 (18.17) | 48.6, 20.7, 30.8 | 96.96 (2.01) | 95.68 (18.05) | 15.4 | 18.6 | 9.2 | |
| FHS* | 640 | 59.4 (9.0) | 50.0 | 28.5 (5.0) | 94.68 (1.96) | 85.71 (6.07) | 2.79 (8.16) | 8.20 (14.42) | 34.5, 35.5, 30.0 | 96.15 (1.86) | 101.45 (13.41) | 7.7 | 0.4 | 5.3 | |
| MESA | 707 | 68.5 (9.1) | 53.6 | 28.0 (5.2) | 93.93 (1.75) | 83.49 (7.41) | 4.36 (10.84) | 12.62 (20.67) | 21.6, 34.4, 44.0 | 95.70 (1.40) | 94.43 (14.08) | 2.0 | 13.6 | 11.1 | |
| MrOS | 2,178 | 76.7 (5.7) | 0.0 | 27.2 (3.7) | 93.85 (1.73) | 84.30 (6.08) | 4.41 (9.89) | 12.74 (18.13) | 21.2, 35.0, 43.8 | 94.96 (1.63) | 98.99 (18.33) | 7.5 | 5.3 | 13.0 | |
| Hispanic/ Latino-American | MESA | 458 | 68.3 (9.2) | 52.8 | 30.1 (5.5) | 94.33 (1.56) | 81.50 (9.38) | 3.84 (7.35) | 16.94 (23.05) | 17.2, 27.9, 54.8 | 96.12 (1.37) | 94.42 (14.91) | 5.5 | 9.2 | 27.6 |
| Starr | 782 | 52.3 (11.3) | 71.9 | 32.2 (6.8) | 94.65 (2.09) | 85.78 (7.50) | 2.83 (8.79) | 10.35 (17.18) | 31.5, 31.5, 37.1 | 95.93 (2.43) | N/A | N/A | N/A | 47.90 |
Six studies included 8,326 individuals with genotypes and phenotypes (1,209 African-Americans; 228 Asian-Americans; 5,649 European-Americans; 1,240 Hispanic/Latino-Americans). Values are displayed as mean (SD), except for the skewed Apnea Hypopnea Index, which is displayed as median (IQR). Waking O2 saturation values were based on point measurements collected prior to the sleep episode.
*: Family cohort.
Table 2. Replication sample description.
| Ethnic Group | Cohort | N | Age | Percent Female | BMI | Sleep Episode SpO2 | Minimum Sleep Episode SpO2 | Percent Sleep Under 90% SpO2 | Apnea Hypopnea Index | AHI (% <5, 5–15, > = 15) | Waking SpO2 | FVC (% Predicted) | % Asthma | % COPD | % Diabetes |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| African-American | CHS | 185 | 75.7 (4.8) | 59.5 | 28.7 (4.8) | 95.01 (2.07) | 85.69 (5.34) | 3.16 (8.84) | 11.42 (16.67) | 25.4, 36.8, 37.8 | 96.16 (1.99) | 96.30 (23.21) | 11.29 | 3.33 | 22.58 |
| JHS | 496 | 62.7 (10.8) | 63.1 | 32.3 (7.0) | 94.72 (2.06) | 84.07 (6.52) | 3.20 (9.19) | 10.87 (14.65) | 24.7, 39.6, 35.7 | N/A | N/A | 8.23 | 4.32 | 22.43 | |
| European-American | CHS | 731 | 77.7 (4.2) | 60.6 | 27.3 (4.4) | 94.13 (1.91) | 84.77 (6.39) | 4.24 (11.32) | 10.82 (15.47) | 48.6, 20.7, 30.8 | 95.45 (1.87) | 90.69 (18.80) | 6.72 | 1.81 | 10.25 |
| European-Australian | WASHS | 1,647 | 52.1 (13.7) | 40.2 | 32.0 (7.8) | N/A | 83.95 (9.39) | 6.11 (15.10) | 24.60 (30.80) | 4.7, 24.5, 70.9 | 95.13 (2.48) | 91.68 (14.59) | 25.8 | 17.0 | 13.96 |
| Hispanic/ Latino-American | HCHS/SOL | 11,351 |
46.2 (13.8) | 59.1 | 29.8 (6.0) | 96.45 (0.95) | 87.07 (6.05) | 0.85 (3.14) | 1.97 (6.20) | 68.9, 19.4, 11.7 | 96.94 (3.13) | 94.34 (15.71) | 7.70 | 2.78 | 19.55 |
Four studies included 14,410 individuals with genotypes and phenotypes (681 African-Americans; 2,378 European-Americans and European-Australians; 11,351 Hispanic/Latino-Americans). Values are displayed as mean (SD), except for the skewed Apnea Hypopnea Index, which is displayed as median (IQR). Waking O2 saturation values were based on point measurements collected prior to the sleep episode.
Pairwise correlations among phenotypes and selective demographic variables are shown in S1 Table. As expected, the three overnight oxyhemoglobin saturation traits are strongly correlated (average SpO2 –minimal SpO2 ρ = 0.61; average SpO2 –Per90 ρ = -0.73; minimal SpO2 –Per90 ρ = -0.86). Waking oxyhemoglobin saturation correlates with average nocturnal SpO2 (ρ = 0.59), minimal nocturnal SpO2 (ρ = 0.35) and Per90 (ρ = -0.40). The AHI was also correlated with minimal SpO2 (ρ = -0.71), Per90 (ρ = 0.70), and average SpO2 (ρ = -0.55). Lung function (percent predicted FEV1 and FVC [26]) correlated modestly with each of the overnight oxygen saturation measures (ρ = -0.20 –+0.23).
Meta-analysis results overview
Manhattan and QQ plots for the overall sample and population-specific primary discovery analyses are provided in S1–S3 Figs. The maximum lambda value was 1.02, in the multi-ethnic average SpO2 analysis, suggesting that our analysis results were largely free of technical artifacts and corrected appropriately for population structure within each ethnic group. We analyzed SNPs in loci with discovery p-values < 1 × 10−6 in our replication cohorts and identified 6 significant (p < 5.0 × 10−8) and 1 suggestive (p < 1.0 × 10−6) associations in joint discovery and replication analyses spanning 5 regions; 2q12 (IL18R1; Fig 1), 10q22 (HK1; Fig 2), 3p24 (intergenic region; S8 Fig), 4q35 (RP11-242J7.1, S9 Fig); S2 Table), with several associations specific to given population backgrounds. Effect estimates and directions of allelic effect were consistent in the discovery and replication stages for all SNPs (METAL heterogeneity p > 0.1).
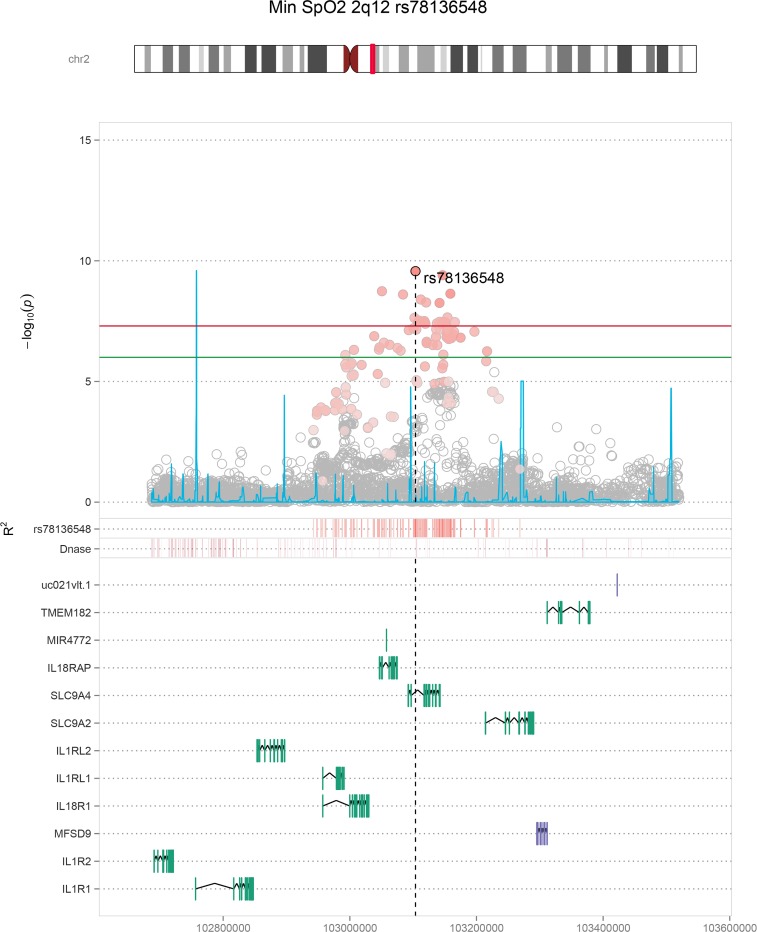
Fig 1. Minimum oxygen saturation 2q12 regional association plot.
Physical positions (Build 37 coordinates) are shown on the X-axis. The main graphic shows log-transformed p-values for individual SNPs on the Y-axis. SNP colors indicate the degree of linkage disequilibrium (LD) with the lead SNP rs78136548 (based on combined 1000 Genomes AFR, AMR, and EUR populations). The significance cut-off of p = 5 × 10−8 is shown with a horizontal red line. The blue line denotes recombination rates. Lower tracks indicate positions of SNPs with strong LD with rs78136548, regions of Dnase hypersensitivity sites, and exon positions for local genes.
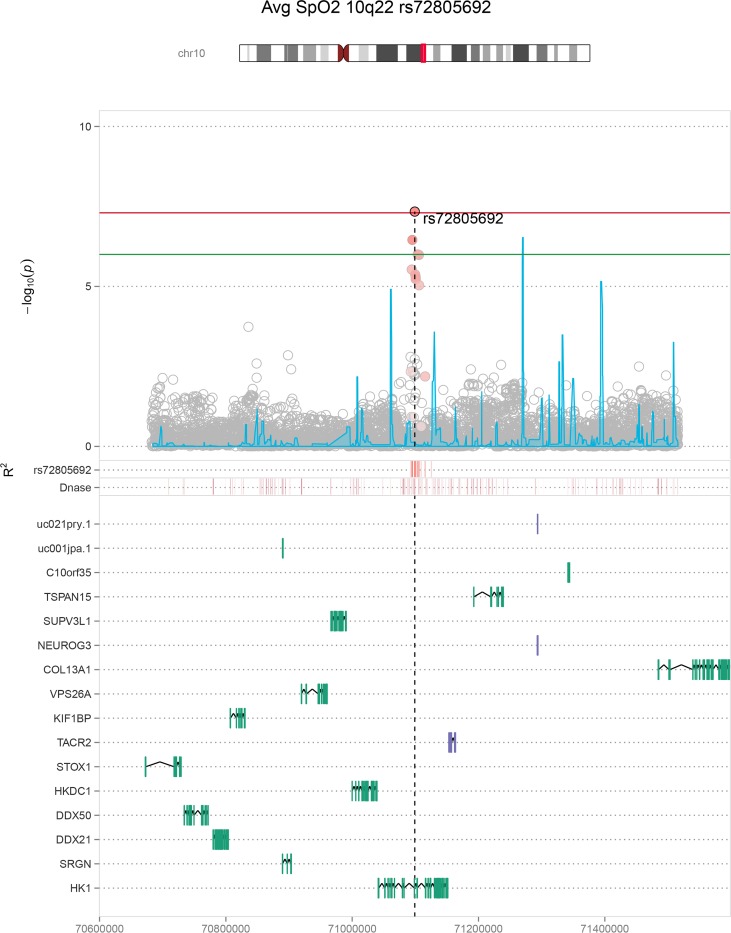
Fig 2. Average oxygen saturation 10q22 regional association plot.
This figure depicts the multi-ethnic average oxygen saturation results in the HK1 (hexokinase 1) region, which had the highest sample size among all significant HK1 region associations (n = 20,676). Other significant and suggestive HK1 regional associations are shown in S7, S11 and S17 Figs.
Interleukin 18 Receptor 1 and Hexokinase 1 region meta-analysis results
In the multi-ethnic combined discovery and replication meta-analysis (n>20,000), genome-wide significant associations were identified with: a) minimum SpO2 in the IL18R1 region (rs78136548 discovery, p = 2.66 × 10−7, combined p = 2.70 × 10−10); and b) average SpO2 in the HK1 region (rs72805692 discovery p = 7.20 × 10−8, combined p = 4.58 × 10−8) (Figs 1 and 2, Table 3). Lead population- and sex-specific SNPs from each locus meeting our replication criteria definitions are also presented (S2 Table). Consistent negative effect directionality was observed for the IL18R1 region SNP rs78136548 T across all 13 available association tests in African, European, and Hispanic/Latino ancestral populations (S3 Table). The association was largely driven by males (females beta(se) -0.065 (0.023), p = 0.005, males beta(se) -0.131 (0.022), p = 2.69 × 10−9; S4 Table). Average SpO2 was significantly associated with the HK1 region in a European-American analysis (rs16926246 n = 5,649; p = 2.46 × 10−8; 1000G EUR rs72805692 r2 = 0.625). The HK1 region was also notable for a second European-American significant association using the complementary phenotype Per90 (percentage of sleep with oxyhemoglobin saturation below 90%; rs148471505 p = 3.08 × 10−8; 1000G EUR rs72805692 r2 = 0.679; S2 Table). Minimum SpO2 was also suggestively associated with the HK1 region in European-American males (rs17476364 p = 6.79 × 10−8; 1000G EUR rs72805692 r2 = 0.937; S3 Table).
Table 3. Significant IL18R1 and HK1 region meta-analysis results.
| Region | Phenotype | Model | SNP | Suggestive Regional SNPs | Combined N | CAF | Discovery Beta (SE) | Discovery P | Replication Beta (SE) | Replication P | Combined Beta (SE) | Combined P |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IL18R1 | Min SpO2 | All | rs78136548 T | 66 | 22,333 | 0.866–0.953 | -0.124 (0.024) | 2.66 × 10−7 | -0.082 (0.021) | 1.01 × 10−4 | -0.101 (0.016) | 2.70 × 10−10 |
| HK1 | Avg SpO2 | All | rs72805692 G | 2 | 20,676 | 0.017–0.112 | 0.141 (0.026) | 7.20 × 10−8 | 0.060 (0.025) | 1.46 × 10−2 | 0.098 (0.018) | 4.58 × 10−8 |
Lead significant (p < 5.0 × 10−8) IL18R1 (2q12) and HK1 (10q22) region SNP multi-ethnic (n > 20,000) meta-analysis associations are shown. Suggestive regional SNPs denotes the count of SNPs with p < 1.0 × 10−6. CAF indicates coded allele frequency range. SNP columns include the coded allelle. Individual regional SNP results are provided in S3 Table, with sex-stratified analyses of each locus SNP provided in S4 Table. Stage-specific analyses (lead loci SNPs only) are provided in S5 Table.
Given that sleep disordered breathing and respiratory control vary by sleep state [27,28], we also explored whether associations for oxygen saturation differed when using measurements specific to non rapid eye movement (NREM) versus rapid eye movement (REM) sleep in cohorts with sleep state information (S5 Table). Several SNPs showed associations with lower p-values and/or higher point estimates for stage-specific results. The minimum SpO2 within NREM rs72805692 association result showed the lowest p-value for any HK1 locus SNP across all analyses (p = 1.60 × 10−9) and further indicated that the HK1 region SNPs were significantly associated with all three traits.
Additional analyses
The secondary sex-stratified discovery analyses (Miami plots, S4–S6 Figs) identified 6 additional independent loci associated in males with consistent effects in replication and suggestive p-values < 1.0 × 10−6 in joint analyses (S3 Table; S11–S17 Figs). A combined secondary meta-analysis of the discovery and replication cohorts for SNPs with initial discovery phase p-values ≥ 1.0 × 10−6 identified four additional significantly associated loci (22q11, 6q25, 17p13 and 7q22 including new candidate genes CHRNE and RELN; S6 Table and S18–S22 Figs). Joint analysis results of the lead loci are provided for each SNP in S3 Table, while a comparison of individual SNP results using combined-sex and sex-stratified models can be found in S4 Table.
GWAS results overlap across oxygen saturation traits, apnea hypopnea index, and pulmonary traits
Although overnight oxygen saturation values most strongly correlate with measurements of SDB, they also may be influenced by pulmonary function, waking oxygen saturation, and hemoglobin levels. We therefore tested associations of the lead loci for sleep SpO2 traits with waking SpO2, the AHI (the clinical metric for SDB), forced vital capacity (FVC, percent predicted, a measure of pulmonary function), and hemoglobin concentration (S5 Table). Sample sizes for the comparison traits varied based on the availability of these exploratory phenotypes, with hemoglobin and FVC collected at different exams from the sleep recordings in a subset of cohorts. Consistent with the strong correlation between sleep period SpO2 and AHI phenotypes (S1 Table), several lead oxygen saturation SNPs also displayed modest to strong associations with the AHI (12 of 17 available p-values < 0.05; minimum AHI p = 5.3 × 10−5). Weaker associations were observed with the other traits: waking SpO2 (p generally > 0.01 and < 0.05; minimum p = 0.005); FVC (p generally > 0.05; minimum p = 0.025); and hemoglobin concentration (p generally > 0.05; minimum p = 0.003). We next evaluated whether associations between sleep SpO2 values and lead SNPs persisted after adjusting for AHI, FVC percent predicted, waking SpO2, asthma history, COPD history, current smoking status, and hemoglobin concentration (S8 Table). Analyses restricted to individuals with these available covariates showed that neither FVC, asthma, COPD, current smoker status or hemoglobin concentration changed the estimated SNP effects for sleep SpO2 by more than 10% for any SNP, suggesting that lung function and lung disease did not mediate the associations between sleep oxygen saturation and each SNP. In contrast, adjustment for AHI reduced the SNP association effect estimates by more than 10% in most of the models tested. Adjustment for waking SpO2, which correlated with nocturnal oxygen saturation, reduced the effect estimates by 0.2 to 35% (S8 Table), although the statistical significance of direct associations of these SNPs with waking SpO2 was modest (p = 0.003–0.93; S5 Table).
Correlated functional and eQTL SNPs and cell line enhancer enrichment: Bioinformatics data
We searched for SNPs in our top loci that overlap regulatory regions as determined by the ENCODE and Roadmap Epigenomics Consortia and collated by HaploReg. S9 Table lists the 144 of 227 unique SNPs with p-values < 1 × 10−6 that overlap regulatory regions (promoter or enhancer marks; DNase I hypersensitivity sites; or protein binding regions) in at least 1 cell line. Our lead HK1 region SNP in multi-ethnic meta-analysis, rs72805692 (average SpO2 multi-ethnic p = 4.58 × 10−8) overlapped enhancer marks in 107 cell lines across 21 organs. Other notable genome-level significant SNPs in the HK1 locus include rs16926246 and rs148471505 (overlapping enhancer marks in 91 and 71 cell lines, respectively). We further queried for Blueprint Consortium promoters and enhancers (largely blood cell lines) and Vermunt et al. brain region enhancers (S10 Table). 104 of the 227 unique replication and combined meta-analysis SNPs with p-values < 1 × 10−6 overlapped at least one regulatory region. rs72805692 additionally overlapped 30 Blueprint and 98 Vermunt enhancer regions, and rs16926246 overlapped 106 combined enhancer regions.
We also queried overlap with published expression quantitative trait loci (eQTL) associations. 182 of the 227 unique SNPs with p-values < 1 × 10−6 were eQTL SNPs for at least one of 42 genes (S11 Table). 13 SNPs that were genome-level significant SNPs in the IL18R1 region were also eQTL SNPs for both interleukin 18 receptor subunits in whole blood (IL18R1 eQTL p < 4.9 × 10−11; IL18RAP eQTL p < 1.6 × 10−46), indicating a possible role for interleukin 18 signaling in the as yet unknown causal tissue(s). No significant colocalization was observed when testing this region using Blueprint Consortium eQTL signals. The lead significant HK1 region SNP rs16926246 was also associated with HK1 expression in whole blood (EA average SpO2 p = 2.46 × 10−8; HK1 eQTL p = 9.64 × 10−13).
Gene and pathway analyses
We performed a multi-variate GWAS of the three SpO2 traits in the European-ancestry samples using MTAG [29]. Lead results (p < 1 × 10−6) are shown in S12 Table. No novel genome-level significant loci were detected.
We used our European-American GWAS meta-analysis results to impute gene-level expression differences in a subset of 6 GTEx-assayed tissues and Depression and Genes and Networks (DGN) whole blood using MetaXcan. Tissue-specific results are presented in S13–S19 Tables. Three genes were associated at either a Bonferroni-adjusted significance level (p < 4.01 × 10−7) or at a suggestive level within an order of magnitude, all in the minimum SpO2 analysis: CHRNE (minimum p = 7.61 × 10−8 in subcutaneous adipose tissue), C17orf107 (overlapping and antisense to CHRNE; minimum p = 2.68 × 10−7 in visceral omentum adipose tissue), and IL18R1 (minimum p = 6.28 × 10−7 in subcutaneous adipose tissue).
We carried the whole blood MetaXcan results forward for pathway analyses (DGN sample size = 922). GIGSEA analyses of KEGG pathways and Molecular Signatures Database curated microRNAs and transcription factors are presented in S20–S22 Tables respectively. The most enriched KEGG pathway was steroid hormone biosynthesis (Average SpO2 empirical p-value = “0” following 10,000 permutations). This pathway was observed twice with empirical p-values < 0.05, as were the KEGG asthma and ribosome pathways. The most significantly observed miRNA binding site was for MIR-380-3P (average SpO2 empirical p-value = 0.006). MIR-140 and MIR-190 displayed empirical p-values < 0.05 in two analyses. PPARG transcription factor binding sites were enriched in all three analyses, while PPAR signaling was the most enriched Per90 KEGG pathway. NHLH1 (formerly HEN1) transcription factor binding sites were enriched in all three analyses, and six transcription factor binding sites were enriched in two analyses.
Discussion
Novel associations were identified for several genetic loci with traits measuring oxyhemoglobin saturation during sleep in a large, multi-ethnic population. The traits examined quantify overnight hypoxemia, a key component of sleep disordered breathing that predicts risk of developing cognitive impairment, cardiovascular disease, atrial fibrillation, and mortality in community and clinical cohorts [2,7,12–14]. Although nocturnal hemoglobin oxygen saturation level inversely correlates with the number of breathing pauses at night (apneas, hypopneas), there is much individual variation in sleep-associated hypoxemia that is not well understood. For the first time, this study identified genetic variants associated with oxyhemoglobin saturation traits measured during sleep. Specifically, we identified and replicated variants in two gene regions- hexokinase 1 (HK1) and interleukin 18 receptor (IL18R1)- that individually and together are of potential high relevance to lung and ventilatory-control pathobiology. In addition, we identified several other loci of potential biological significance.
HK1 and IL18R1 regional associations
We identified significant associations between HK1 SNPs and average oxygen saturation (SpO2) during sleep and percentage of sleep with SpO2 < 90% (Table 3). State-specific results also indicate a genome-level association with minimum SpO2 during NREM sleep (S5 Table). Hexokinase is the first enzyme and the rate-limiting step in the glycolysis pathway [30,31] and its activity is regulated by hypoxia inducible factor 1a (HIF1A) [5,32,33]. Obstructive sleep apnea following CPAP withdrawal increases glucose during sleep [34]. Rs16926246 (significantly associated with average SpO2) and rs10159477 (among our suggestive average SpO2 SNPs, S3 Table) are associated with HK1 expression in whole blood ([35], S11 Table) and also have been associated with hemoglobin concentration [36]. Rs16926246 was the study-wide lead SNP and previously found to be highly significantly associated with hemoglobin A1c (HbA1c) levels, a marker of glucose homeostasis, through an erythrocytic pathway [37]. Rs72805692, our lead multi-ethnic average SpO2 SNP, has also been associated with HbA1c levels [38]. Both SNPs overlap enhancer marks in ≥ 197 ENCODE, Roadmap Epigenomics, Blueprint, and Vermunt et al. cell lines and/or brain regions (including erythroblasts; S9 and S10 Tables). The Rapoport–Luebering glycolytic shunt affects erythrocytic oxygen capacity through allosteric binding of 2,3-bisphosphoglycerate (2,3-BPG, also known as 2,3-diphosphoglycerate or 2,3-DPG) to hemoglobin. The concentration of glycolytic pathway intermediates can impact 2,3-BPG concentration, partially mediated by hexokinase [39–41]. Rs72805692 was marginally associated with hemoglobin concentration in our sample for individuals with available measurements (p = 0.0034). The A allele was associated with reduced hemoglobin concentration, average sleeping and waking SpO2 and increased sleep time with oxygen saturation under 90%. However, the association with average sleeping SpO2 was not appreciably changed when adjusting for hemoglobin concentration (S5 Table).
Alternatively, HK1, in concert with cytokines including IL18, may influence overnight oxygen saturation through effects on pulmonary inflammation and ventilation-perfusion mismatch. Hexokinase-1 mediated glycolysis regulates the NLR Family Pyrin Domain Containing 3 (NLRP3) inflammasome [42], a multiprotein complex implicated in obesity-related inflammation [43] as well as several pulmonary diseases [44–50]. The NLRP3 inflammasome activates caspase-1, resulting in cleavage of pro-IL1B and pro-IL18 into their mature forms, amplifying inflammation [50,51]. The NLRP3 inflammasome is proposed to play a critical role in lung injury occurring in response to exposures to inflammatory mediators, oxidative stress and mechanical ventilation, including cyclic pulmonary stretching, which induces NLRP3 inflammasome activation in alveolar macrophages [52]. Patients with SDB, particularly obstructive sleep apnea, experience oxidative stress and pulmonary inflammation [53], as well as swings in intrathoracic pressure, potentially causing pulmonary strain. Our data suggest the possibility that variations in HK1 (and possibly IL18) pathways may contribute to individual differences in pulmonary gas exchange occurring during sleep, possibly through pulmonary inflammation and/or subclinical pulmonary injury. Circulating markers of alveolar epithelial injury, including KL-6, surfactant protein-A, and matrix metalloproteinase-7, correlate with degree of overnight hypoxemia and AHI in patients with SDB [54,55]. Chronic intermittent hypoxia induces physiological deficits in rats with allergen-induced airway inflammation, due to collagen deposition and other effects [56]. Although the mechanisms underlying these associations are unclear, lung imaging studies show an increase in subclinical interstitial lung abnormalities in individuals with sleep apnea [55]. Our data suggest the possibility that variations in HK1 pathways may contribute to differences in pulmonary gas exchange that occurs during sleep, possibly through effects on ventilation-perfusion mismatch due to subclinical pulmonary inflammation. Recent population-based studies found that sleep apnea associates with both elevated pulmonary inflammatory markers and imaging evidence of interstitial lung abnormalities [55]. Finally, the NLRP3 inflammasome has been shown to influence brain tissue, and changes in sleep delta power have been observed in knock-out mice [57].
Another and correlated mechanism could be through HIF1A. In addition to oxygen sensing effects in the carotid body [58], HIF1A regulates HK1 in human alveolar cells [59], is regulated by PFKM (a downstream glycolysis enzyme) in macrophages [60], and is involved with metabolic reprogramming of macrophages [61]. Activation of glycolytic enzymes in pulmonary epithelial cells exposed to cyclic mechanical stretching is abrogated with HIF1A repression [62]. Ventilatory differences in responses to intermittent hypoxemia secondary to SDB could influence several measures of overnight SpO2, as observed in our analyses.
The second set of significant SNPs implicated in overnight SpO2 levels were in the IL18R1 region, with external evidence indicating eQTL associations with both IL18 receptor subunit genes (IL18R, IL18RAP) in over a dozen genome-level significant SNPs (S8 Table). Minor alleles were associated with an increase in minimum oxygen saturation, an increase in IL18RAP expression, and a decrease in IL18R1 expression in whole blood [35]. IL18R1 expression was also suggestively associated with minimum oxygen saturation in a gene-level analysis using MetaXcan. These genes are essential for IL18 signaling [63,64], suggesting that the IL18 pathway may partially mediate the association at this locus. IL18 is a pro-inflammatory cytokine produced by macrophages and is involved in multiple inflammatory disorders [65]. This region has been associated with Blautia genus microbiota abundance in the gut (lead locus SNP rs79387448 Min SpO2 p = 8.61 x 10–8 [S3 Table]) [66]. Il18 over-expression in mice leads to chronic pulmonary inflammation, including the increased levels of CD4+, CD8+ CD19+, eosinophils, macrophages, NK1.1+, and neutrophils; along with alveolar destruction, fibrosis, and other effects [67,68]. In humans, IL18 plasma concentration levels are elevated in acute respiratory distress syndrome [69]. As described above, IL18 as well as IL1B are the inflammatory proteins activated by caspase 1 in HK1-regulated NLRP3 inflammasome activation [42]. MTOR, a member of the complex that induces this HK1-mediated activation, is the 6th most associated gene in our analyses of Per90 using MetaXcan whole blood analysis (p = 4.5 × 10−4, S18 Table). Mechanical stretching-induced NLRP3 inflammasome activation induces activated IL18 release from alveolar macrophages [52]. Casp1- (required for Il18 activation) and Nlrp3-knockout mice are protected from hypoxemia accompanying mechanical ventilation [70]. Models of cyclic stretching induce the release of Il18 in mouse alveolar macrophages, mediated by Nlrp3 [52]. Serum IL18 concentrations are also significantly higher in patients with SDB compared to obese controls and correlated with serum concentrations of C reactive protein and interleukin 6 [71]. Il18r1 is among the genes with the most robust circadian rhythmic profiles in healthy mouse lung [72]; it is possible that timing-specific gene expression may influence the nocturnal hypoxemia phenotype we studied. An association between asthma and a SNP in the IL18R1 region is reported [73]. Our lead SNPs had reduced linkage disequilibrium with this SNP (rs3771166 minimum p in any model = 5.6 x 10−5 for minimum SpO2 in EA males). Analyses adjusted for asthma did not significantly attenuate our associations (rs78136548 minimum SpO2 p adjusted for asthma = 1.21 x 10−7, unadjusted p = 1.01 x 10−7 in equivalent samples). Therefore, different variants in the IL18R1 region may influence pulmonary and sleep related hypoxemia traits. The circadian timing of sleep may impact IL18 pathway-specific effects on pulmonary-related traits. The associations with HK1 further also implicate the possibility that variants in both genes may contribute to SpO2 levels. IL18 has also been shown to regulate HIF1A [74].
Other regional associations
The protein coding gene most proximal to the Per90 association with the 4q35 region in African-Americans (S2 Table, S9 Fig) was CASP3, a second caspase gene involved with alveolar wall destruction [75]. Despite the modest sample size, the p-value almost met genome-wide significance (p = 9.39 x 10−7), suggesting the utility of future studies of the role of this gene in influencing nocturnal saturation.
We also detected multiple genome-level significant associations following a combined discovery plus replication cohort meta-analysis (S7 Table). Although these associations require independent evidence for replication, two of these associations are of particular interest. In European-Americans, minimum oxygen saturation was associated with 2 genome-level significant and 65 suggestive SNPs that are associated with CHRNE (acetylcholine receptor, nicotinic epsilon [muscle]) expression in 19 GTEx tissues and monocytes (p < 5 x 10−8; S11 Table). CHRNE and the proximal gene C17orf107 had the lowest p-values in the expression-based MetaXcan gene tests. Phospholipidase D2 (PLD2), another gene associated with the locus through expression (eQTL) SNPs, is required for hypoxia-induced expression of HIF1A. Hypoxia-induced gene expression in mouse lung endothelial cells is reduced in Pld2 knockout mice [76]. A multi-ethnic Per90 association physically overlaps RELN (reelin). While no expression or epigenetic evidence was available, this association is of interest given the suggested respiratory role of reelin within the Pre-Bötzinger complex, a major center for respiratory control [77].
No novel genome-level significant loci were detected in our multi-variate MTAG analyses (S12 Table). As expected from the univariate results, HK1 was significantly associated and the lead SNP rs72805692 had reduced p-values for average and minimum SpO2 (p = 4.0 × 10−9 and 1.1 × 10−9 respectively). Among the genes in novel suggestive regions was WLS (formerly GPR177; rs17481104 minimum SpO2 p = 5.8 × 10−7), which is involved in pulmonary vascular development and has been suggestively associated with airflow obstruction in COPD [78,79]. Enrichment of the KEGG asthma pathway for both minimum SpO2 and Per90 (S20 Table) lends support to the ‘overlap syndrome’ of these two pulmonary diseases [80]. PPAR-gamma transcription factor binding site enrichment in all three analyses (S22 Table) suggests the potential importance of future mechanistic studies of this inflammation-related transcription factor. PPAR signaling was the most enriched KEGG pathway in the Per90 analysis (S20 Table). PPAR signaling and PPARG expression in visceral adipose tissue have previously been associated with obstructive sleep apnea [81].
Sex- and state-specific effects
Sex-stratified analyses identified stronger associations among males compared to females for the IL18R1 signal (rs78136548 p females = 0.005, males = 2.69 × 10−9; S4 Table) and for several SNPs within the HK1 locus (e.g. rs17476364 EA min SpO2 p females = 0.19, males = 6.79 × 10−8). In addition, sex-stratified analyses identified two other loci of interest (S6 Table). Among EA males only, a suggestive association with Per90 in the IL1RAPL1 region of the X chromosome was identified. The region has recently been suggestively associated with asthma in Hispanic/Latino children, with one replication cohort indicating potential male-specific effects [82]. An association in the PPP4R1 region (protein phosphatase 4 regulatory subunit 1) was nearly genome-level significant in EA males (p = 5.4 × 10−8). Ten suggestive SNPs in this locus were also PPP4R1 eQTL SNPs in whole blood (p < 1 × 10−40; S11 Table). PPP4R1 regulates HDAC3 (histone deacetylase 3), an epigenetic modulator of circadian lipid metabolism [83,84].
Respiratory control and neuromuscular activation vary between NREM and REM sleep. Analyses restricting to these states may reduce heterogeneity due to differences in state that influence airway patency or respiratory chemosensitivity. The PPP4R1 locus p-value lowered to genome-level significance when analyzed using NREM sleep data (rs78805840 p = 1.81 × 10−8; S5 Table). The lowest overall p-value in the HK1 locus was a minimum SpO2 within NREM association with rs27805692 (all population combined-sex p = 1.60 × 10−9).
Pulmonary trait effects
The three traits that we analyzed (average and minimum SpO2 during sleep, percent of sleep with SpO2 < 90%) each are commonly measured and reported in evaluation of patients with sleep apnea. Although correlated, they each measure somewhat different aspects of oxygen saturation. Notably, associations for the Hexokinase 1 (HK1) region showed associations with several measures of oxygen saturation. Consistency of these findings across phenotypes suggests the importance of the HK1 pathway in influencing several aspects of oxygenation during sleep, including severity of response to an airway occlusion (i.e., as measured by minimal saturation), overall severity (Per90), and overall level (average). Although overnight hypoxemia can occur with underlying pulmonary disease, our analyses also showed that each SpO2 association typically associated with the AHI, a primary index of sleep apnea, and associations were not appreciably influenced after considering effects of lung disease, tobacco use, lung function, and hemoglobin level (S8 Table). As expected, baseline oxygen saturation also correlated with average SpO2, consistent with an influence of waking SpO2 on overall nocturnal levels. Notably, none of our sleep-related trait associations overlapped published associations for resting oxygen saturation in COPD.
Across our cohorts, average level of lung function was within the normal range, and prevalence of lung diseases was low. These findings, as well as our analyses that adjusted for several factors and independently assessed genetic signals for the other pulmonary traits, indicate that the variations in SpO2 traits in our cohorts predominantly reflected differences in SDB-related levels of hypoxemia. Relevance of our results to SDB is supported by finding that 12 of the 17 available lead loci SNPs across all analyses had at least a nominal association with the AHI (S5 Table), including the associations for HK1 rs72805692 and IL18R1 region rs78136568 SNPs (p = 8.1 × 10−5 and 7.2 × 10−5 respectively). SDB is a common disorder affecting 17% of middle-aged men and 9% of middle-aged women and characterized by repetitive episodes of upper airway obstruction resulting in intermittent hypoxemia, sleep disruption, and profound physiological disturbances [85]. Past genetic analyses of sleep apnea have mainly focused on the AHI, which does not fully describe the broad range of physiologic stressors that occur in SA, including patterns of SpO2 desaturations [86]. Overnight SpO2 analysis provides clinically relevant information [2,7–9,12–14] and can be measured relatively simply, and thus can be scaled for future genetic studies and precision medicine.
Strengths and weaknesses
Our study has several strengths. The sample size of over 22,000 is among the largest available GWAS analyses of any trait associated with objectively recorded sleep disordered breathing. The associations were observed in cohorts with varying demographics and ascertainment strategies (Tables 1 and 2), and as such are likely generalizable to diverse populations. We used a stringent imputation quality threshold (0.88) to reduce random error, using a 1000 Genomes Project or denser template in all studies. Several of our associations are supported by published gene expression, bioinformatics evidence, and/or physiological studies.
Several weaknesses also need to be acknowledged. While we have not performed functional assays as part of the present analysis, the most biologically compelling candidates are supported by several lines of evidence from the literature and will require future experimental validation. Data on potential mediators (e.g., lung function, hemoglobin) were collected in different visits or were available only in a subset of our cohorts. Some promising findings did not meet genome-wide significance criteria or could not be replicated across cohorts. While this first multi-ethnic meta-analysis of the three traits included over 22,000 individuals, weak or population-specific associations were likely to be missed due to power limitations.
In conclusion, we have performed the first genome-wide association analysis of clinically relevant sleep disordered breathing traits, specifically measures of nocturnal oxygen saturation, and identified several novel associations that are of potential biological relevance. Of particular interest were variants in the HK1 and IL18R1 regions. Understanding the genetic underpinnings of these sleep-related traits may guide future studies investigating the contribution of sleep disordered breathing to the hypoxemic burden of pulmonary disorders, and identify common mechanisms such as activation of the NLRP3-inflammasome pathway.
Full meta-analysis results are freely available from http://www.sleepdisordergenetics.org/informational/data.
Material and methods
This research was approved by the Partners Healthcare IRB committee (protocol # 2010P001765). Participant consent was obtained through written documents.
Discovery group studies
The Atherosclerosis Risk in Communities Study (ARIC; n = 1,432) and Framingham Heart Study (FHS; n = 640) cohorts participating in the Sleep Heart Health Study (SHHS) were analyzed with available polysomnography (PSG) and genotype data [87–89]. This community-based study included a baseline examination (1995–1998) that included in-home polysomnography, and questionnaires [89]. Polysomnography from the baseline examination was collected using the Compumedics PS-2 system (Abbotsford, AU) [90]. Oxyhemoglobin saturation was measured with finger pulse oximetry over the sleep episode and cleaned of signal artifact. The 3 parent cohorts are described below, with CHS used as a replication cohort in the current study (genetic data from this cohort was obtained after the other two studies). In ARIC, genotyping was performed using the Affymetrix 6.0 array. In FHS, genotyping was performed using the Affymetrix 500k and Illumina Omni 5M arrays (obtained from dbGaP; pht000395.v7.p8).
The Cleveland Family Study (CFS) is examining the genetic and familial basis of sleep apnea with 2,534 African- and European-American individuals from 356 families. Four visits occurred from 1990–2006, with a final visit at a clinical research center (2000–2006). Index probands with confirmed sleep apnea were recruited from sleep centers in northern Ohio. Additional family members and neighborhood control families were also studied [91]. Measurements including sleep apnea monitoring, anthropometry, other related phenotypes, and questionnaires. Before 2000, an Edentrace Type 3 home sleep apnea device was used (Eden Prairie, MN). The final examination used 14-channel polysomnography (Compumedics E series, Abottsford, AU). Genotyping was based on the Affymetrix 6.0 and Illumina OmniExpress, Exome, and IBC chip arrays. Data were based on 1,411 individuals with both genotypes and sleep data from either the home sleep study (n = 784) or the lab-based study (n = 627).
The Multi-Ethnic Study of Atherosclerosis (MESA) is examining the risk factors of clinical cardiovascular disease [92]. The baseline examination in 2000 included 6,814 participants ages 45 to 84 from 6 communities: Baltimore MD, Chicago IL, Los Angeles CA, New York NY, Minneapolis/St. Paul MN, and Winston-Salem NC. Four ethnicities are being studied: African-, Asian-, European-, and Hispanic/Latino-Americans. An ancillary sleep study of 2,060 individuals who did not use nightly CPAP, overnight oxygen, or an oral device for sleep apnea occurred between 2010–2013. Sleep measurements included in-home PSG, actigraphy, and a questionnaire adapted from the SHHS questionnaire [93]. Unattended polysomnography used a 15-channel monitor (Compumedics Somte System, Abbotsford, AU). Final study inclusion for individuals with an Affymetrix 6.0 assay was 1,883.
The Osteoporotic Fractures in Men Study (MrOS) is a prospective cohort study examining the risk factors for fractures, osteoporosis, and prostate cancer [94,95] in males age 65 or older from six U.S. communities. An ancillary sleep study of 3,135 individuals was conducted between 2003 and 2005, including in-home PSG (Compumedics Safiro system; Abbotsford AU), anthropometry, and questionnaires. Genotyping was performed with the Illumina Human Omni 1 Quad v1-0 H array. A total of 2,178 European ancestry individuals had PSG and genotype data.
The Starr County Health Studies (Starr) have been examining the risk factors of diabetes in a predominantly Mexican-American border county in Texas since 1981 [96,97]. The sleep apnea assessment occurred between 2010 and 2014 and included a questionnaire and home sleep apnea testing using the WatchPAT-200 device (Itamar-Medical Ltd., Caesarea, Israel), with recording of finger pulse oximetry, actigraphy, body position, peripheral arterial tonometry, and snoring. It has previously been validated using polysomnography [98]. The current analysis included 782 individuals with valid oximetry and Affymetrix 6.0 data.
Replication group studies
Data from the Sleep Heart Health Study Cardiovascular Health Study (CHS) [99] was available after ARIC and FHS and used in replication analysis. Sleep phenotyping was performed as described earlier. 185 African-American and 731 European-Americans with available polysomnography and Illumina CNV370, and/or Omni1M plus IBC genotypes obtained through dbGaP (pht003699.v1.p1) were analyzed.
The Hispanic Community Health Study/Study of Latinos (HCHS/SOL) is studying risk and protective factors of multiple health conditions in Hispanics/Latinos [100,101]. 16,415 community members from randomly selected households aged 18–74 from 4 cities (Chicago, IL; Miami, FL; Bronx, NY; San Diego, CA) were examined in a baseline exam between 2008–2011. The sample design consisted of a stratified two-stage area probability sample of household addresses. Six cohort backgrounds were represented: Central American (n = 1,730), Cuban (n = 2,348), Dominican (n = 1,460), Mexican (n = 6,471), Puerto-Rican (n = 2,728), and South American (n = 1,068). The exam included anthropometry, questionnaires, and home sleep apnea testing using the ARES Unicorder 5.2 (B-Alert, Carlsbad, CA), which records measurements of airflow using a nasal pressure cannula and pressure transducer; oxyhemoglobin saturation and pulse rate using a forehead-based reflectance oximeter; head movements and position using an accelerometer; and snoring levels using a microphone. The device has undergone previous validation for in-home use [102]. Records were manually scored and cleaned of artifacts at a central sleep reading center [101]. The current study includes 11,351 non-Asian ancestry individuals with oxyhemoglobin saturation values during sleep and Illumina Omni 2.5 genotyping.
The Jackson Heart Study (JHS) is a population-based prospective investigation of cardiovascular disease [103,104]. The ancillary sleep study occurred from 2012–2016, and included home sleep apnea testing with the Embla Embletta Gold, a 6-channel device that includes an oximeter (Broomfield, CO). The device has been validated previously [105]. Additional collected measures include sleep questionnaires and anthropometry. 496 African-American individuals with phenotyping and Affymetrix 6.0 genotyping were included in this study, reflecting a dataset freeze at the time of analysis.
The Western Australian Sleep Health Study (WASHS) is examining the epidemiology and genetics of sleep apnea and related comorbidities [106]. This clinic-based study examines patients presenting to the sole public sleep clinic in Western Australia, located in Perth. 91% of patients were referred for SDB. Data collection for individuals in the current analysis occurred from 2006–2010. In-lab, attended polysomnography was performed using the Compumedics Series E device (Abbotsford, AU). After excluding principal component (PC) outliers (see below), valid oximetry data and genotype data (Illumina Omni 2.5) were available for 1,647 European ancestry patients.
Phenotype and covariate definitions
The quantitative phenotypic outcome was oxyhemoglobin saturation during sleep (SpO2), measured as an average, minimum, or as a percentage of the night with SpO2 < 90% (Per90) measured using finger pulse oximetry (all using NONIN oximetry boards) or transcutaneous oximetry (HCHS/SOL only) measured continuously as part of polysomnography or home sleep apnea testing. Other than the WASHS clinical cohort, all sleep data were scored by a central reading center with high levels of established reliability [107] by scorers blinded to all other data. Intermittent waking and SpO2 artifact were manually edited from all records. Covariates were obtained by questionnaires, direct measurement (BMI), and oximetry (waking oxygen saturation was measured prior to the sleep recording). Secondary measures such as hemoglobin concentration and spirometry were collected from the same visit whenever possible, however this was not possible for all cohorts (most notably hemoglobin was collected years prior to the sleep exam in some cohorts). Potential device differences were minimized by both performing analyses at a cohort level and using a rank-normal phenotype transformation to reduce the impact of phenotypic outliers. Our analysis focused on identifying potential loci operating in obesity-independent pathways. Hispanic/Latino-specific results have been reported previously for average SpO2 [25].
Genotyping and statistical analyses
Genotypes from all cohorts were imputed to at least a 1000 Genomes Phase 1 density. ARIC, JHS, and HCHS/SOL were imputed using a 1000 Genomes Phase 1 version 3 template. WASHS was imputed using a Haplotype Reference Consortium version 1.0 template [108]. All other cohorts were imputed using a 1000 Genomes Phase 3 version 5 template [109,110]. Single nucleotide polymorphisms (SNPs) and insertions/deletions with minor allele frequency < 0.01, minor allele counts < 20 within a cohort, or an IMPUTE2/PBWT info score < 0.88 were removed from the analysis. Sample sizes and variant counts for each cohort for the three primary phenotype analyses are provided in S23 Table.
We explored ancestry-specific associations given past SDB trait differences (e.g. [93,101]) and linkage disequilibrium differences [109]. Data were analyzed at a cohort- and population-specific level (e.g. 4 separate analyses for the MESA cohort). Population structure was controlled for using linear mixed models followed by genomic control. Population structure principal components were calculated for the minimally-admixed, self-reported Asian-American and European-American/Australian population groups within individual studies using TRACE [111]. WASHS initial self-reported European ancestry was based on classification of the patient's parents [106]. Individuals were defined as population outliers and removed from analysis if any coordinate from PC 1–4 was greater than 5 standard deviations from the population mean. Individuals self-reporting into groups with modest sample size within a cohort (e.g. MrOS Asian-Americans) were excluded from study.
Our analysis focused on identifying potential loci operating in obesity-independent pathways. We adjusted for age, age2, sex, age × sex, BMI, and BMI2 to address known demographic factors and potential non-linear effects of age and BMI. Phenotypes, adjusted for age and sex, were rank-normalized. Residuals were then calculated by further adjusting for BMI. Primary analyses were performed using GEMMA, which incorporates linear mixed models that control for the relatedness structure [112]. HCHS/SOL analyses were performed using the GENESIS Bioconductor package [113] (DOI:10.18129/B9.bioc.GENESIS). A fixed effects, inverse variance weighted meta-analysis was performed using METAL with genomic control applied in each analysis [114]. Variants with p-values < 1 × 10−6 in the discovery cohort meta-analysis were carried forward to replication and combined discovery/replication analysis. To reduce the influence of small studies possibly leading to spurious findings, we only present meta-analysis results where 1,000 or more individuals contributed. Individual SNPs in the multi-ethnic analyses were only analyzed if they remained unfiltered in two or more populations. The three traits differed in their final variant counts due to phenotype missingness and ascertainment (no average SpO2 was available for WASHS in this analysis). In aggregate for the discovery cohorts, there were 11,297,250–11,298,080 AA; 8,956,016–8,958,150 EA; and 9,481,040–9,481,751 multi-ethnic variants. The replication cohorts included 12,243,361–12,346,361 AA; 7,448,148–8,707,439 EA; and 10,173,881–10,480,925 multi-ethnic variants. Visualizations were constructed using LocusExplorer and EasyStrata [115,116].
Sleep and sleep disordered breathing may be regulated by multiple tissues [86,117]. Epigenetic database queries were performed using HaploReg version 4 using an imputed model and exact SNPs. HaploReg data included ENCODE and Roadmap Epigenomics consortia data [118–120]. Additional queries examined non-cancerous Blueprint Consortium data (largely related to blood cell lines) and Vermunt et al. brain region enhancer data[121–123]. Gene expression data used in expression quantitative trait loci (eQTL) lookups were obtained from multiple studies, including a seven-cohort consortium investigating whole blood (Westra et al.) [35,124–130]. The Westra data were pruned to include only eQTL SNPs with FDR < 0.05. Moloc was used to test colocalization [131]. Gene-level analyses used MetaXcan to impute expression levels based on GTEx tissues and Depression and Genes and Networks (DGN) whole blood [132]. GIGSEA, which is designed to work with MetaXcan output, was used for pathway analyses using the whole blood results (queried due to improved power from increased sample size) [133]. We used the weighted linear regression model with 10,000 permutations.
Supporting information
Top: African-Americans; Middle: European-Americans; Bottom: Multi-ethnic. Variants in the multi-ethnic results had to have results from cohorts in two or more populations.
(PDF)
Top: African-Americans; Middle: European-Americans; Bottom: Multi-ethnic (2 or more populations for each variant).
(PDF)
Top: African-Americans; Middle: European-Americans; Bottom: Multi-ethnic (2 or more populations for each variant).
(PDF)
Top: Females; Bottom: Males.
(PDF)
Top: Females; Bottom: Males.
(PDF)
Top: Females; Bottom: Males.
(PDF)
The supplemental regional plot order corresponds to the order found in S2, S3 and S4 Tables, minus the associations shown in Figs 1 and 2.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Spearman correlations between phenotypes and covariates across all discovery cohorts and ethnic groups are shown. Correlations with the spirometry measures FEV1 and FVC are also shown. Correlations were pooled using Fisher Z-transformations weighted by sample size. 95% confidence intervals are listed in parentheses.
(XLS)
Lead SNPs are shown for regions with significant (p < 5.0 × 10–8) and suggestive (p < 1.0 × 10–6) p-values. Discovery regions were carried forward if the combined discovery and replication analysis results indicated a lower p-value, retained genome-level significance, or replication was unavailable (i.e. due to frequency or imputation filters). SNPs denotes the count of regional SNPs with p < 1.0 × 10–6. Genes indicates overlapping Ensembl genes within 5 kb of the p < 1 × 10–6 SNPs. CAF indicates coded allele frequency range. Individual regional SNP results are provided in S3 Table. Individual regions were minimally 500 kb apart.
(XLS)
“Locus Class” indicates “Discovery/Replication” for main analyses or “Combined meta-analysis” for regions not clearing initial discovery p-value thresholds (i.e. S7 Table loci).
(XLS)
Sex-stratified samples with n < 1000 are included here for comparison but were not included in the main analyses. An individual variant present in the combined-sex analysis for a given cohort may be missing from the equivalent sex-stratified analysis due to analysis-specific minor allele count thresholds of 20. “Locus Class” indicates “Discovery/Replication” for main analyses or “Combined meta-analysis” for regions not clearing initial discovery p-value thresholds (i.e. S7 Table loci).
(XLS)
Each lead SNP from the “Original Phenotype” and “Original Model” columns was analyzed using equivalent individuals and models for different phenotypes. Available sample size will vary depending on the availability of the phenotype. AHI values are taken from Chen, Cade, et al. (submitted) “Locus Class” indicates “Discovery/Replication” for main analyses or “Combined meta-analysis” for regions not clearing initial discovery p-value thresholds (i.e. S7 Table loci).
(XLS)
Lead SNPs are shown for regions with suggestive (p < 1.0 × 10–6) p-values. Discovery regions were carried forward if the combined discovery and replication analysis results indicated a lower p-value, retained genome-level significance, or replication was unavailable (i.e. due to frequency or imputation filters). SNPs denotes the count of regional SNPs with p < 1.0 × 10–6. Genes indicates overlapping Ensembl genes within 5 kb of the p < 1 × 10–6 SNPs. CAF indicates coded allele frequency range. Individual regional SNP results are provided in S3 Table. Individual regions were minimally 500 kb apart.
(XLS)
Lead SNPs are shown for regions with significant (p < 5.0 x 10–8) combined-analysis p-values with p > 1 x 10–6 in the discovery phase. SNPs denotes the count of regional SNPs with p < 1.0 x 10–6. Genes indicates overlapping Ensembl genes within 5 kb of the p < 1 x 10–6 SNPs. CAF indicates coded allele frequency range. Individual regional SNP results are provided in S3 Table. Individual regions were minimally 500 kb apart.
(XLS)
Lead variants were re-analyzed with standard models and additional covariates as listed. As these covariates were not collected in all cohorts and individuals, the analysis were also performed with the same individuals for direct comparisons (matched N control columns).
(XLS)
Data were obtained from HaploReg 4.1 (https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php) using exact SNPs (LD threshold = NA) and a ChromHMM 15-state model, followed by extraction from HTML. “Locus Class” indicates “Discovery/Replication” for main analyses or “Combined meta-analysis” for regions not clearing initial discovery p-value thresholds (i.e. S7 Table loci).
(XLS)
Blueprint cell lines are a non-cancerous sub-set of largely blood cells. Vermunt brain region samples were not assayed for promoter regions. “Locus Class” indicates “Discovery/Replication” for main analyses or “Combined meta-analysis” for regions not clearing initial discovery p-value thresholds (i.e. S7 Table loci).
(XLS)
Data were obtained from Fairfax et al. (monocytes), Geuvadis (LCLs), GTEx, Hao et al. (lung), Muther (adipose, LCLs, skin), Raj et al. (CD4, monocytes), Westra et al. (whole blood), and Zeller et al. (monocytes). “Locus Class” indicates “Discovery/Replication” for main analyses or “Combined meta-analysis” for regions not clearing initial discovery p-value thresholds (i.e. S7 Table loci).
(XLS)
MTAG analysis was performed on European ancestry samples using our summary statistics for the three traits simultaneously. Lead results (p < 1 x 10–6) are shown.
(XLS)
Lead results (p < 0.05) are shown. Note that Ensembl IDs obtained from MetaXcan are occasionally unavailable.
(XLS)
Lead results (p < 0.05) are shown. Note that Ensembl IDs obtained from MetaXcan are occasionally unavailable.
(XLS)
Lead results (p < 0.05) are shown. Note that Ensembl IDs obtained from MetaXcan are occasionally unavailable.
(XLS)
Lead results (p < 0.05) are shown. Note that Ensembl IDs obtained from MetaXcan are occasionally unavailable.
(XLS)
Lead results (p < 0.05) are shown. Note that Ensembl IDs obtained from MetaXcan are occasionally unavailable.
(XLS)
Lead results (p < 0.05) are shown. Note that Ensembl IDs obtained from MetaXcan are occasionally unavailable.
(XLS)
Lead results (p < 0.05) are shown. Note that Ensembl IDs obtained from MetaXcan are occasionally unavailable.
(XLS)
MetaXcan gene-level results for DGN whole blood (S19 Table, all p-values) were used as input.
(XLS)
MetaXcan gene-level results for DGN whole blood (S19 Table, all p-values) were used as input. MicroRNAs are curated by the Molecular Signatures Database (http://software.broadinstitute.org/gsea/msigdb/index.jsp).
(XLS)
MetaXcan gene-level results for DGN whole blood (S19 Table, all p-values) were used as input. Transcription factor binding sites are curated by the Molecular Signatures Database (http://software.broadinstitute.org/gsea/msigdb/index.jsp).
(XLS)
Cohort-level information is shown for the three primary analyses, including discovery/replication category, the number of individuals with available phenotyping and genotyping, genotyping platform, imputation panel, and the number of imputed variants tested in each analysis following info score, MAC, and MAF filtering.
(XLS)
Acknowledgments
The authors wish to thank the participants and study staff of all of our cohorts for their important contributions. The authors thank the staff and participants of the ARIC study for their important contribution. This manuscript was not prepared in collaboration with investigators of the ARIC Study, and does not necessarily reflect the opinions or conclusions of the ARIC Study or the NHLBI. The authors thank the staff and participants of HCHS/SOL for their important contributions. Investigators website– http://www.cscc.unc.edu/hchs/. The authors thank the participants and data collection staff of the Jackson Heart Study. The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Heart, Lung, and Blood Institute; the National Institutes of Health; or the U.S. Department of Health and Human Services. The authors thank the investigators, the staff, and the participants of the MESA study for their valuable contributions. A full list of participating MESA investigators and institutions can be found at http://www.mesa-nhlbi.org. We thank the field staff in Starr County for their careful collection of these data and are especially grateful to the participants who so graciously cooperated and gave of their time.
Data Availability
Full meta-analysis results are freely available from http://www.sleepdisordergenetics.org/informational/data
Funding Statement
Brian Cade is supported by grants from the National Institutes of Health [K01-HL135405-01, R01-HL113338-04, R35-HL135818-01] and the American Thoracic Society Foundation (http://foundation.thoracic.org). The Sleep Reading Center of Brigham and Women's Hospital has been supported by National Institutes of Health grants [5-R01-HL046380-15 and 5-KL2-RR024990-05]. The Atherosclerosis Risk in Communities (ARIC) Study is conducted and supported by the National Heart, Lung, and Blood Institute in collaboration with the University of North Carolina (N01-HC-55015, N01-HC-55018), Baylor College of Medicine (N01-HC-55016), University of Minnesota (N01-HC-55019), Johns Hopkins University (N01-HC-55020), and University of Mississippi Medical Center (N01-HC-55021). This Cardiovascular Health Study (CHS) research was supported by NHLBI contracts HHSN268201200036C, HHSN268200800007C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086; and NHLBI grants U01HL080295, R01HL087652, R01HL105756, R01HL103612, R01HL120393, and R01HL130114 with additional contribution from the National Institute of Neurological Disorders and Stroke (NINDS). Genotyping among the African-American cohort was supported in part by HL085251Additional support was provided through R01AG023629 from the National Institute on Aging (NIA). A full list of principal CHS investigators and institutions can be found at CHS-NHLBI.org. The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, CTSI grant UL1TR000124, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center. The Framingham Heart Study is conducted and supported by NHLBI in collaboration with Boston University (Contract No. N01-HC-25195). Funding for SHARe Affymetrix genotyping was provided by NHLBI Contract N02-HL- 64278. SHARe Illumina genotyping was provided under an agreement between Illumina and Boston University. Funding support for the Framingham Sleep Heart Health Study was provided by NIH/NHLBI grant U01 HL 53941. The Hispanic Community Health Study/Study of Latinos is a collaborative study supported by contracts from the National Heart, Lung, and Blood Institute (NHLBI) to the University of North Carolina (HHSN268201300001I / N01-HC-65233), University of Miami (HHSN268201300004I / N01-HC-65234), Albert Einstein College of Medicine (HHSN268201300002I / N01-HC-65235), University of Illinois at Chicago – HHSN268201300003I / N01-HC-65236 Northwestern Univ), and San Diego State University (HHSN268201300005I / N01-HC-65237). The following Institutes/Centers/Offices have contributed to the HCHS/SOL through a transfer of funds to the NHLBI: National Institute on Minority Health and Health Disparities, National Institute on Deafness and Other Communication Disorders, National Institute of Dental and Craniofacial Research, National Institute of Diabetes and Digestive and Kidney Diseases, National Institute of Neurological Disorders and Stroke, NIH Institution-Office of Dietary Supplements. The Genetic Analysis Center at Washington University was supported by NHLBI and NIDCR contracts (HHSN268201300005C AM03 and MOD03). The Jackson Heart Study is supported and conducted in collaboration with Jackson State University (HHSN268201300049C and HHSN268201300050C), Tougaloo College (HHSN268201300048C), and the University of Mississippi Medical Center (HHSN268201300046C and HHSN268201300047C) contracts from the National Heart, Lung, and Blood Institute (NHLBI) and the National Institute for Minority Health and Health Disparities (NIMHD). Dr. Wilson is supported by U54GM115428 from the National Institute of General Medical Sciences. The authors thank the participants and data collection staff of the Jackson Heart Study. The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Heart, Lung, and Blood Institute; the National Institutes of Health; or the U.S. Department of Health and Human Services. The Multi-Ethnic Study of Atherosclerosis (MESA) is conducted and supported by the NHLBI in collaboration with MESA investigators. MESA and the MESA SHARe project are conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with MESA investigators. Support for MESA is provided by contracts HHSN268201500003I, N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-000040, UL1-TR-001079, UL1-TR-001420, UL1-TR-001881, and DK063491. Funding for SHARe genotyping was provided by NHLBI Contract N02-HL-64278. Genotyping was performed at Affymetrix (Santa Clara, California, USA) and the Broad Institute of Harvard and MIT (Boston, Massachusetts, USA) using the Affymetrix Genome-Wide Human SNP Array 6.0. Funding support for the Sleep Polysomnography dataset was provided by grant HL56984. Provision of genotyping services supported in part by NCATS CTSI grant UL1TR000124 and NIDDK DRC grant DK063491. The Osteoporotic Fractures in Men (MrOS) Study is supported by NIH funding. The following institutes provide support: the National Institute on Aging (NIA), the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), NCATS, and NIH Roadmap for Medical Research under the following grant numbers: U01 AG027810, U01 AG042124, U01 AG042139, U01 AG042140, U01 AG042143, U01 AG042145, U01 AG042168, U01 AR066160, and UL1 TR000128. The NHLBI provides funding for the MrOS Sleep ancillary study "Outcomes of Sleep Disorders in Older Men" under the following grant numbers: R01 HL071194, R01 HL070848, R01 HL070847, R01 HL070842, R01 HL070841, R01 HL070837, R01 HL070838, and R01 HL070839. The NIAMS provides funding for the MrOS ancillary study ‘Replication of candidate gene associations and bone strength phenotype in MrOS’ under the grant number R01 AR051124. The NIAMS provides funding for the MrOS ancillary study ‘GWAS in MrOS and SOF’ under the grant number RC2 AR058973. The Starr County Health Studies is supported in part by grants R01 DK073541, U01 DK085501, R01 AI085014, and R01 HL102830 from the National Institutes of Health, and funds from the University of Texas Health Science Center at Houston. Funding for the Western Australian Sleep Health Study was obtained from the Sir Charles Gairdner and Hollywood Private Hospital Research Foundations, the Western Australian Sleep Disorders Research Institute, and the Centre for Genetic Epidemiology and Biostatistics at the University of Western Australia. Funding for the GWAS genotyping obtained from the Ontario Institute for Cancer Research and a McLaughlin Centre Accelerator Grant from the University of Toronto. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Antonelli Incalzi R., Marra C., Giordano A., Calcagni M.L., Cappa A., Basso S., Pagliari G., and Fuso L. (2003). Cognitive impairment in chronic obstructive pulmonary disease–a neuropsychological and spect study. J Neurol 250, 325–332. 10.1007/s00415-003-1005-4 [DOI] [PubMed] [Google Scholar]
- 2.Yaffe K., Laffan A.M., Harrison S.L., Redline S., Spira A.P., Ensrud K.E., Ancoli-Israel S., and Stone K.L. (2011). Sleep-disordered breathing, hypoxia, and risk of mild cognitive impairment and dementia in older women. JAMA 306, 613–619. 10.1001/jama.2011.1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McDonald M.-L.N., Cho M.H., Sørheim I.-C., Lutz S.M., Castaldi P.J., Lomas D.A., Coxson H.O., Edwards L.D., MacNee W., Vestbo J., et al. (2014). Common genetic variants associated with resting oxygenation in chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol 51, 678–687. 10.1165/rcmb.2014-0135OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tarnoki D.L., Medda E., Tarnoki A.D., Lazar Z., Fagnani C., Stazi M.A., Karlinger K., Torzsa P., Kalabay L., Garami Z., et al. (2014). Genetic influence on capillary oxygen saturation: a twin study. Lung 192, 429–434. 10.1007/s00408-014-9563-z [DOI] [PubMed] [Google Scholar]
- 5.Beall C.M. (2006). Andean, Tibetan, and Ethiopian patterns of adaptation to high-altitude hypoxia. Integr Comp Biol 46, 18–24. 10.1093/icb/icj004 [DOI] [PubMed] [Google Scholar]
- 6.Liang J., Cade B.E., Wang H., Chen H., Gleason K.J., Larkin E.K., Saxena R., Lin X., Redline S., and Zhu X. (2016). Comparison of Heritability Estimation and Linkage Analysis for Multiple Traits Using Principal Component Analyses. Genet Epidemiol 40, 222–232. 10.1002/gepi.21957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gami A.S., Hodge D.O., Herges R.M., Olson E.J., Nykodym J., Kara T., and Somers V.K. (2007). Obstructive sleep apnea, obesity, and the risk of incident atrial fibrillation. J Am Coll Cardiol 49, 565–571. 10.1016/j.jacc.2006.08.060 [DOI] [PubMed] [Google Scholar]
- 8.Nieto F.J., Peppard P.E., Young T., Finn L., Hla K.M., and Farré R. (2012). Sleep-disordered breathing and cancer mortality: results from the Wisconsin Sleep Cohort Study. Am J Respir Crit Care Med 186, 190–194. 10.1164/rccm.201201-0130OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campos-Rodriguez F., Martinez-Garcia M.A., Martinez M., Duran-Cantolla J., Peña M. de la, Masdeu M.J., Gonzalez M., F. del Campo, Gallego I., Marin J.M., et al. (2013). Association between obstructive sleep apnea and cancer incidence in a large multicenter Spanish cohort. Am J Respir Crit Care Med 187, 99–105. 10.1164/rccm.201209-1671OC [DOI] [PubMed] [Google Scholar]
- 10.Gozal D., Ham S.A., and Mokhlesi B. (2016). Sleep Apnea and Cancer: Analysis of a Nationwide Population Sample. Sleep 39, 1493–1500. 10.5665/sleep.6004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gunnarsson S.I., Peppard P.E., Korcarz C.E., Barnet J.H., Hagen E.W., Hla K.M., Palta M., Young T., and Stein J.H. (2015). Minimal nocturnal oxygen saturation predicts future subclinical carotid atherosclerosis: the Wisconsin sleep cohort. J. Sleep Res. 24, 680–686. 10.1111/jsr.12321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kendzerska T., Gershon A.S., Hawker G., Leung R.S., and Tomlinson G. (2014). Obstructive sleep apnea and risk of cardiovascular events and all-cause mortality: a decade-long historical cohort study. PLoS Med 11, e1001599 10.1371/journal.pmed.1001599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oldenburg O., Wellmann B., Buchholz A., Bitter T., Fox H., Thiem U., Horstkotte D., and Wegscheider K. (2016). Nocturnal hypoxaemia is associated with increased mortality in stable heart failure patients. Eur Heart J 37, 1695–1703. 10.1093/eurheartj/ehv624 [DOI] [PubMed] [Google Scholar]
- 14.Gellen B., Canouï-Poitrine F., Boyer L., Drouot X., Le Thuaut A., Bodez D., Covali-Noroc A., D’ortho M.P., Guendouz S., Rappeneau S., et al. (2016). Apnea-hypopnea and desaturations in heart failure with reduced ejection fraction: Are we aiming at the right target? Int J Cardiol 203, 1022–1028. 10.1016/j.ijcard.2015.11.108 [DOI] [PubMed] [Google Scholar]
- 15.Minville C., Hilleret M.-N., Tamisier R., Aron-Wisnewsky J., Clement K., Trocme C., Borel J.-C., Lévy P., Zarski J.-P., and Pépin J.-L. (2014). Nonalcoholic fatty liver disease, nocturnal hypoxia, and endothelial function in patients with sleep apnea. Chest 145, 525–533. 10.1378/chest.13-0938 [DOI] [PubMed] [Google Scholar]
- 16.Lacasse Y., Sériès F., Vujovic-Zotovic N., Goldstein R., Bourbeau J., Lecours R., Aaron S.D., and Maltais F. (2011). Evaluating nocturnal oxygen desaturation in COPD–revised. Respir Med 105, 1331–1337. 10.1016/j.rmed.2011.04.003 [DOI] [PubMed] [Google Scholar]
- 17.Ross K.R., Storfer-Isser A., Hart M.A., Kibler A.M.V., Rueschman M., Rosen C.L., Kercsmar C.M., and Redline S. (2012). Sleep-disordered breathing is associated with asthma severity in children. J Pediatr 160, 736–742. 10.1016/j.jpeds.2011.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corte T.J., Wort S.J., Talbot S., Macdonald P.M., Hansel D.M., Polkey M., Renzoni E., Maher T.M., Nicholson A.G., and Wells A.U. (2012). Elevated nocturnal desaturation index predicts mortality in interstitial lung disease. Sarcoidosis Vasc Diffuse Lung Dis 29, 41–50. [PubMed] [Google Scholar]
- 19.Kolilekas L., Manali E., Vlami K.A., Lyberopoulos P., Triantafillidou C., Kagouridis K., Baou K., Gyftopoulos S., Vougas K.N., Karakatsani A., et al. (2013). Sleep oxygen desaturation predicts survival in idiopathic pulmonary fibrosis. J Clin Sleep Med 9, 593–601. 10.5664/jcsm.2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shungin D., Winkler T.W., Croteau-Chonka D.C., Ferreira T., Locke A.E., Mägi R., Strawbridge R.J., Pers T.H., Fischer K., Justice A.E., et al. (2015). New genetic loci link adipose and insulin biology to body fat distribution. Nature 518, 187–196. 10.1038/nature14132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bixler E.O., Vgontzas A.N., Lin H.M., Ten Have T., Rein J., Vela-Bueno A., and Kales A. (2001). Prevalence of sleep-disordered breathing in women: effects of gender. Am J Respir Crit Care Med 163, 608–613. 10.1164/ajrccm.163.3.9911064 [DOI] [PubMed] [Google Scholar]
- 22.Cairns A., Poulos G., and Bogan R. (2016). Sex differences in sleep apnea predictors and outcomes from home sleep apnea testing. Nat Sci Sleep 8, 197–205. 10.2147/NSS.S101186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jordan A.S., McEvoy R.D., Edwards J.K., Schory K., Yang C.-K., Catcheside P.G., Fogel R.B., Malhotra A., and White D.P. (2004). The influence of gender and upper airway resistance on the ventilatory response to arousal in obstructive sleep apnoea in humans. J Physiol 558, 993–1004. 10.1113/jphysiol.2004.064238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simpson L., Mukherjee S., Cooper M.N., Ward K.L., Lee J.D., Fedson A.C., Potter J., Hillman D.R., Eastwood P., Palmer L.J., et al. (2010). Sex differences in the association of regional fat distribution with the severity of obstructive sleep apnea. Sleep 33, 467–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cade B.E., Chen H., Stilp A.M., Gleason K.J., Sofer T., Ancoli-Israel S., Arens R., Bell G.I., Below J.E., Bjonnes A.C., et al. (2016). Genetic Associations with Obstructive Sleep Apnea Traits in Hispanic/Latino Americans. Am J Respir Crit Care Med 194, 886–897. 10.1164/rccm.201512-2431OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hankinson J.L., Odencrantz J.R., and Fedan K.B. (1999). Spirometric reference values from a sample of the general U.S. population. Am J Respir Crit Care Med 159, 179–187. 10.1164/ajrccm.159.1.9712108 [DOI] [PubMed] [Google Scholar]
- 27.Gugger M., Bögershausen S., and Schäffler L. (1993). Arousal responses to added inspiratory resistance during REM and non-REM sleep in normal subjects. Thorax 48, 125–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carberry J.C., Jordan A.S., White D.P., Wellman A., and Eckert D.J. (2016). Upper Airway Collapsibility (Pcrit) and Pharyngeal Dilator Muscle Activity are Sleep-Stage Dependent. Sleep 39, 511–521. 10.5665/sleep.5516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turley P., Walters R.K., Maghzian O., Okbay A., Lee J.J., Fontana M.A., Nguyen-Viet T.A., Wedow R., Zacher M., Furlotte N.A., et al. (2018). Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat. Genet. 50, 229–237. 10.1038/s41588-017-0009-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rapoport S. (1968). The regulation of glycolysis in mammalian erythrocytes. Essays Biochem 4, 69–103. [PubMed] [Google Scholar]
- 31.Lowry O.H., and Passonneau J.V. (1964). The Relationships between Substrates and Enzymes of Glycolysis in Brain. J Biol Chem 239, 31–42. [PubMed] [Google Scholar]
- 32.Hoffstein V., Herridge M., Mateika S., Redline S., and Strohl K.P. (1994). Hematocrit levels in sleep apnea. Chest 106, 787–791. [DOI] [PubMed] [Google Scholar]
- 33.Wagner P.D., Simonson T.S., Wei G., Wagner H.E., Wuren T., Qin G., Yan M., and Ge R.L. (2015). Sea-level haemoglobin concentration is associated with greater exercise capacity in Tibetan males at 4200 m. Exp Physiol 100, 1256–1262. 10.1113/EP085036 [DOI] [PubMed] [Google Scholar]
- 34.Chopra S., Rathore A., Younas H., Pham L.V., Gu C., Beselman A., Kim I.-Y., Wolfe R.R., Perin J., Polotsky V.Y., et al. (2017). Obstructive Sleep Apnea Dynamically Increases Nocturnal Plasma Free Fatty Acids, Glucose, and Cortisol During Sleep. J. Clin. Endocrinol. Metab. 102, 3172–3181. 10.1210/jc.2017-00619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Westra H.-J., Peters M.J., Esko T., Yaghootkar H., Schurmann C., Kettunen J., Christiansen M.W., Fairfax B.P., Schramm K., Powell J.E., et al. (2013). Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet 45, 1238–1243. 10.1038/ng.2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Harst P., Zhang W., Mateo Leach I., Rendon A., Verweij N., Sehmi J., Paul D.S., Elling U., Allayee H., Li X., et al. (2012). Seventy-five genetic loci influencing the human red blood cell. Nature 492, 369–375. 10.1038/nature11677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soranzo N., Sanna S., Wheeler E., Gieger C., Radke D., Dupuis J., Bouatia-Naji N., Langenberg C., Prokopenko I., Stolerman E., et al. (2010). Common variants at 10 genomic loci influence hemoglobin A₁(C) levels via glycemic and nonglycemic pathways. Diabetes 59, 3229–3239. 10.2337/db10-0502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.An P., Miljkovic I., Thyagarajan B., Kraja A.T., Daw E.W., Pankow J.S., Selvin E., Kao W.H.L., Maruthur N.M., Nalls M.A., et al. (2014). Genome-wide association study identifies common loci influencing circulating glycated hemoglobin (HbA1c) levels in non-diabetic subjects: the Long Life Family Study (LLFS). Metabolism 63, 461–468. 10.1016/j.metabol.2013.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benesch R., Benesch R.E., and Yu C.I. (1968). Reciprocal binding of oxygen and diphosphoglycerate by human hemoglobin. Proc Natl Acad Sci U A 59, 526–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Wijk R., and van Solinge W.W. (2005). The energy-less red blood cell is lost: erythrocyte enzyme abnormalities of glycolysis. Blood 106, 4034–4042. 10.1182/blood-2005-04-1622 [DOI] [PubMed] [Google Scholar]
- 41.Mulquiney P.J., and Kuchel P.W. (1999). Model of 2,3-bisphosphoglycerate metabolism in the human erythrocyte based on detailed enzyme kinetic equations: computer simulation and metabolic control analysis. Biochem J 342 Pt 3, 597–604. [PMC free article] [PubMed] [Google Scholar]
- 42.Moon J.-S., Hisata S., Park M.-A., DeNicola G.M., Ryter S.W., Nakahira K., and Choi A.M.K. (2015). mTORC1-Induced HK1-Dependent Glycolysis Regulates NLRP3 Inflammasome Activation. Cell Rep 12, 102–115. 10.1016/j.celrep.2015.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Traba J., Kwarteng-Siaw M., Okoli T.C., Li J., Huffstutler R.D., Bray A., Waclawiw M.A., Han K., Pelletier M., Sauve A.A., et al. (2015). Fasting and refeeding differentially regulate NLRP3 inflammasome activation in human subjects. J Clin Invest 125, 4592–4600. 10.1172/JCI83260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lasithiotaki I., Giannarakis I., Tsitoura E., Samara K.D., Margaritopoulos G.A., Choulaki C., Vasarmidi E., Tzanakis N., Voloudaki A., Sidiropoulos P., et al. (2016). NLRP3 inflammasome expression in idiopathic pulmonary fibrosis and rheumatoid lung. Eur Respir J 47, 910–918. 10.1183/13993003.00564-2015 [DOI] [PubMed] [Google Scholar]
- 45.Grailer J.J., Canning B.A., Kalbitz M., Haggadone M.D., Dhond R.M., Andjelkovic A.V., Zetoune F.S., and Ward P.A. (2014). Critical role for the NLRP3 inflammasome during acute lung injury. J Immunol 192, 5974–5983. 10.4049/jimmunol.1400368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sayan M., and Mossman B.T. (2016). The NLRP3 inflammasome in pathogenic particle and fibre-associated lung inflammation and diseases. Part Fibre Toxicol 13, 51 10.1186/s12989-016-0162-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stout-Delgado H.W., Cho S.J., Chu S.G., Mitzel D.N., Villalba J., El-Chemaly S., Ryter S.W., Choi A.M.K., and Rosas I.O. (2016). Age-Dependent Susceptibility to Pulmonary Fibrosis Is Associated with NLRP3 Inflammasome Activation. Am J Respir Cell Mol Biol 55, 252–263. 10.1165/rcmb.2015-0222OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hosseinian N., Cho Y., Lockey R.F., and Kolliputi N. (2015). The role of the NLRP3 inflammasome in pulmonary diseases. Ther Adv Respir Dis 9, 188–197. 10.1177/1753465815586335 [DOI] [PubMed] [Google Scholar]
- 49.Besnard A.-G., Guillou N., Tschopp J., Erard F., Couillin I., Iwakura Y., Quesniaux V., Ryffel B., and Togbe D. (2011). NLRP3 inflammasome is required in murine asthma in the absence of aluminum adjuvant. Allergy 66, 1047–1057. 10.1111/j.1398-9995.2011.02586.x [DOI] [PubMed] [Google Scholar]
- 50.Lee T.-H., Song H.J., and Park C.-S. (2014). Role of inflammasome activation in development and exacerbation of asthma. Asia Pac Allergy 4, 187–196. 10.5415/apallergy.2014.4.4.187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Im H., and Ammit A.J. (2014). The NLRP3 inflammasome: role in airway inflammation. Clin Exp Allergy 44, 160–172. 10.1111/cea.12206 [DOI] [PubMed] [Google Scholar]
- 52.Wu J., Yan Z., Schwartz D.E., Yu J., Malik A.B., and Hu G. (2013). Activation of NLRP3 inflammasome in alveolar macrophages contributes to mechanical stretch-induced lung inflammation and injury. J Immunol 190, 3590–3599. 10.4049/jimmunol.1200860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lavie L. (2015). Oxidative stress in obstructive sleep apnea and intermittent hypoxia–revisited–the bad ugly and good: implications to the heart and brain. Sleep Med Rev 20, 27–45. 10.1016/j.smrv.2014.07.003 [DOI] [PubMed] [Google Scholar]
- 54.Lederer D.J., Jelic S., Basner R.C., Ishizaka A., and Bhattacharya J. (2009). Circulating KL-6, a biomarker of lung injury, in obstructive sleep apnoea. Eur Respir J 33, 793–796. 10.1183/09031936.00150708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim J.S., Podolanczuk A.J., Borker P., Kawut S.M., Raghu G., Kaufman J.D., Hinckley Stukovsky K.D., Hoffman E.A., Barr R.G., Gottlieb D.J., et al. (2017). Obstructive Sleep Apnea and Subclinical Interstitial Lung Disease in MESA. Ann. Am. Thorac. Soc. 14, 1786–1795. 10.1513/AnnalsATS.201701-091OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Broytman O., Braun R.K., Morgan B.J., Pegelow D.F., Hsu P.-N., Mei L.S., Koya A.K., Eldridge M., and Teodorescu M. (2015). Effects of chronic intermittent hypoxia on allergen-induced airway inflammation in rats. Am. J. Respir. Cell Mol. Biol. 52, 162–170. 10.1165/rcmb.2014-0213OC [DOI] [PubMed] [Google Scholar]
- 57.Zielinski M.R., Gerashchenko D., Karpova S.A., Konanki V., McCarley R.W., Sutterwala F.S., Strecker R.E., and Basheer R. (2017). The NLRP3 inflammasome modulates sleep and NREM sleep delta power induced by spontaneous wakefulness, sleep deprivation and lipopolysaccharide. Brain Behav Immun. 62, 137–150. 10.1016/j.bbi.2017.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Prabhakar N.R., and Semenza G.L. (2016). Regulation of carotid body oxygen sensing by hypoxia-inducible factors. Pflugers Arch. 468, 71–75. 10.1007/s00424-015-1719-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luo F., Liu X., Yan N., Li S., Cao G., Cheng Q., Xia Q., and Wang H. (2006). Hypoxia-inducible transcription factor-1alpha promotes hypoxia-induced A549 apoptosis via a mechanism that involves the glycolysis pathway. BMC Cancer 6, 26 10.1186/1471-2407-6-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Palsson-McDermott E.M., Curtis A.M., Goel G., Lauterbach M.A.R., Sheedy F.J., Gleeson L.E., van den Bosch M.W.M., Quinn S.R., Domingo-Fernandez R., Johnston D.G.W., et al. (2015). Pyruvate kinase M2 regulates Hif-1α activity and IL-1β induction and is a critical determinant of the warburg effect in LPS-activated macrophages. Cell Metab 21, 65–80. 10.1016/j.cmet.2014.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Corcoran S.E., and O’Neill L.A.J. (2016). HIF1α and metabolic reprogramming in inflammation. J Clin Invest 126, 3699–3707. 10.1172/JCI84431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eckle T., Brodsky K., Bonney M., Packard T., Han J., Borchers C.H., Mariani T.J., Kominsky D.J., Mittelbronn M., and Eltzschig H.K. (2013). HIF1A reduces acute lung injury by optimizing carbohydrate metabolism in the alveolar epithelium. PLoS Biol 11, e1001665 10.1371/journal.pbio.1001665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hoshino K., Tsutsui H., Kawai T., Takeda K., Nakanishi K., Takeda Y., and Akira S. (1999). Cutting edge: generation of IL-18 receptor-deficient mice: evidence for IL-1 receptor-related protein as an essential IL-18 binding receptor. J Immunol 162, 5041–5044. [PubMed] [Google Scholar]
- 64.Cheung H., Chen N.-J., Cao Z., Ono N., Ohashi P.S., and Yeh W.-C. (2005). Accessory protein-like is essential for IL-18-mediated signaling. J Immunol 174, 5351–5357. [DOI] [PubMed] [Google Scholar]
- 65.Dinarello C.A., Novick D., Kim S., and Kaplanski G. (2013). Interleukin-18 and IL-18 binding protein. Front Immunol 4, 289 10.3389/fimmu.2013.00289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang J., Thingholm L.B., Skiecevičienė J., Rausch P., Kummen M., Hov J.R., Degenhardt F., Heinsen F.-A., Rühlemann M.C., Szymczak S., et al. (2016). Genome-wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat. Genet. 48, 1396–1406. 10.1038/ng.3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hoshino T., Kato S., Oka N., Imaoka H., Kinoshita T., Takei S., Kitasato Y., Kawayama T., Imaizumi T., Yamada K., et al. (2007). Pulmonary inflammation and emphysema: role of the cytokines IL-18 and IL-13. Am J Respir Crit Care Med 176, 49–62. 10.1164/rccm.200603-316OC [DOI] [PubMed] [Google Scholar]
- 68.Kang M.-J., Choi J.-M., Kim B.H., Lee C.-M., Cho W.-K., Choe G., Kim D.-H., Lee C.G., and Elias J.A. (2012). IL-18 induces emphysema and airway and vascular remodeling via IFN-γ, IL-17A, and IL-13. Am J Respir Crit Care Med 185, 1205–1217. 10.1164/rccm.201108-1545OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dolinay T., Kim Y.S., Howrylak J., Hunninghake G.M., An C.H., Fredenburgh L., Massaro A.F., Rogers A., Gazourian L., Nakahira K., et al. (2012). Inflammasome-regulated cytokines are critical mediators of acute lung injury. Am J Respir Crit Care Med 185, 1225–1234. 10.1164/rccm.201201-0003OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jones H.D., Crother T.R., Gonzalez-Villalobos R.A., Jupelli M., Chen S., Dagvadorj J., Arditi M., and Shimada K. (2014). The NLRP3 inflammasome is required for the development of hypoxemia in LPS/mechanical ventilation acute lung injury. Am J Respir Cell Mol Biol 50, 270–280. 10.1165/rcmb.2013-0087OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Minoguchi K., Yokoe T., Tazaki T., Minoguchi H., Tanaka A., Oda N., Okada S., Ohta S., Naito H., and Adachi M. (2005). Increased carotid intima-media thickness and serum inflammatory markers in obstructive sleep apnea. Am J Respir Crit Care Med 172, 625–630. 10.1164/rccm.200412-1652OC [DOI] [PubMed] [Google Scholar]
- 72.Haspel J.A., Chettimada S., Shaik R.S., Chu J.-H., Raby B.A., Cernadas M., Carey V., Process V., Hunninghake G.M., Ifedigbo E., et al. (2014). Circadian rhythm reprogramming during lung inflammation. Nat Commun 5, 4753 10.1038/ncomms5753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moffatt M.F., Gut I.G., Demenais F., Strachan D.P., Bouzigon E., Heath S., von Mutius E., Farrall M., Lathrop M., Cookson W.O., et al. (2010). A large-scale, consortium-based genomewide association study of asthma. N Engl J Med 363, 1211–1221. 10.1056/NEJMoa0906312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim J., Shao Y., Kim S.Y., Kim S., Song H.K., Jeon J.H., Suh H.W., Chung J.W., Yoon S.R., Kim Y.S., et al. (2008). Hypoxia-induced IL-18 increases hypoxia-inducible factor-1alpha expression through a Rac1-dependent NF-kappaB pathway. Mol Biol Cell 19, 433–444. 10.1091/mbc.E07-02-0182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Petrache I., Fijalkowska I., Medler T.R., Skirball J., Cruz P., Zhen L., Petrache H.I., Flotte T.R., and Tuder R.M. (2006). alpha-1 antitrypsin inhibits caspase-3 activity, preventing lung endothelial cell apoptosis. Am J Pathol 169, 1155–1166. 10.2353/ajpath.2006.060058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ghim J., Moon J.-S., Lee C.S., Lee J., Song P., Lee A., Jang J.-H., Kim D., Yoon J.H., Koh Y.J., et al. (2014). Endothelial deletion of phospholipase D2 reduces hypoxic response and pathological angiogenesis. Arter. Thromb Vasc Biol 34, 1697–1703. 10.1161/ATVBAHA.114.303416 [DOI] [PubMed] [Google Scholar]
- 77.Tan W., Sherman D., Turesson J., Shao X.M., Janczewski W.A., and Feldman J.L. (2012). Reelin demarcates a subset of pre-Bötzinger complex neurons in adult rat. J Comp Neurol 520, 606–619. 10.1002/cne.22753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jiang M., Ku W., Fu J., Offermanns S., Hsu W., and Que J. (2013). Gpr177 regulates pulmonary vasculature development. Dev. Camb. Engl. 140, 3589–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wilk J.B., Shrine N.R.G., Loehr L.R., Zhao J.H., Manichaikul A., Lopez L.M., Smith A.V., Heckbert S.R., Smolonska J., Tang W., et al. (2012). Genome-wide association studies identify CHRNA5/3 and HTR4 in the development of airflow obstruction. Am. J. Respir. Crit. Care Med. 186, 622–632. 10.1164/rccm.201202-0366OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Owens R.L., Macrea M.M., and Teodorescu M. (2017). The overlaps of asthma or COPD with OSA: A focused review. Respirol. Carlton Vic 22, 1073–1083. 10.1111/resp.13107 [DOI] [PubMed] [Google Scholar]
- 81.Gharib S.A., Hayes A.L., Rosen M.J., and Patel S.R. (2013). A pathway-based analysis on the effects of obstructive sleep apnea in modulating visceral fat transcriptome. Sleep 36, 23–30. 10.5665/sleep.2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Marques C.R., Costa G.N., da Silva T.M., Oliveira P., Cruz A.A., Alcantara-Neves N.M., Fiaccone R.L., Horta B.L., Hartwig F.P., Burchard E.G., et al. (2017). Suggestive association between variants in IL1RAPL and asthma symptoms in Latin American children. Eur J Hum Genet 25, 439–445. 10.1038/ejhg.2016.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang X., Ozawa Y., Lee H., Wen Y.-D., Tan T.-H., Wadzinski B.E., and Seto E. (2005). Histone deacetylase 3 (HDAC3) activity is regulated by interaction with protein serine/threonine phosphatase 4. Genes Dev 19, 827–839. 10.1101/gad.1286005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Feng D., Liu T., Sun Z., Bugge A., Mullican S.E., Alenghat T., Liu X.S., and Lazar M.A. (2011). A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 331, 1315–1319. 10.1126/science.1198125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Peppard P.E., Young T., Barnet J.H., Palta M., Hagen E.W., and Hla K.M. (2013). Increased prevalence of sleep-disordered breathing in adults. Am J Epidemiol 177, 1006–1014. 10.1093/aje/kws342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Redline S. (2011). Genetics of Obstructive Sleep Apnea In Principles and Practice of Sleep Medicine, Kryger Meir H W.C.;Roth, Thomas Dement, ed. (St. Louis, MO: Saunders; ), pp. 1183–1193. [Google Scholar]
- 87.The ARIC Investigators (1989). The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. The ARIC investigators. Am. J. Epidemiol. 129, 687–702. [PubMed] [Google Scholar]
- 88.Feinleib M. (1985). The Framingham Study: sample selection, follow-up, and methods of analyses. Natl Cancer Inst Monogr 67, 59–64. [PubMed] [Google Scholar]
- 89.Quan S.F., Howard B.V., Iber C., Kiley J.P., Nieto F.J., O’Connor G.T., Rapoport D.M., Redline S., Robbins J., Samet J.M., et al. (1997). The Sleep Heart Health Study: design, rationale, and methods. Sleep 20, 1077–1085. [PubMed] [Google Scholar]
- 90.Redline S., Sanders M.H., Lind B.K., Quan S.F., Iber C., Gottlieb D.J., Bonekat W.H., Rapoport D.M., Smith P.L., and Kiley J.P. (1998). Methods for obtaining and analyzing unattended polysomnography data for a multicenter study. Sleep Heart Health Research Group. Sleep 21, 759–767. [PubMed] [Google Scholar]
- 91.Redline S., Tishler P.V., Tosteson T.D., Williamson J., Kump K., Browner I., Ferrette V., and Krejci P. (1995). The familial aggregation of obstructive sleep apnea. Am J Respir Crit Care Med 151, 682–687. 10.1164/ajrccm.151.3.7881656 [DOI] [PubMed] [Google Scholar]
- 92.Bild D.E., Bluemke D.A., Burke G.L., Detrano R., Diez Roux A.V., Folsom A.R., Greenland P., Jacob D.R.J., Kronmal R., Liu K., et al. (2002). Multi-Ethnic Study of Atherosclerosis: objectives and design. Am J Epidemiol 156, 871–881. [DOI] [PubMed] [Google Scholar]
- 93.Chen X., Wang R., Zee P., Lutsey P.L., Javaheri S., Alcántara C., Jackson C.L., Williams M.A., and Redline S. (2015). Racial/Ethnic Differences in Sleep Disturbances: The Multi-Ethnic Study of Atherosclerosis (MESA). Sleep 38, 877–888. 10.5665/sleep.4732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Orwoll E., Blank J.B., Barrett-Connor E., Cauley J., Cummings S., Ensrud K., Lewis C., Cawthon P.M., Marcus R., Marshall L.M., et al. (2005). Design and baseline characteristics of the osteoporotic fractures in men (MrOS) study–a large observational study of the determinants of fracture in older men. Contemp Clin Trials 26, 569–585. 10.1016/j.cct.2005.05.006 [DOI] [PubMed] [Google Scholar]
- 95.Mehra R., Stone K.L., Blackwell T., Ancoli Israel S., Dam T.-T.L., Stefanick M.L., and Redline S. (2007). Prevalence and correlates of sleep-disordered breathing in older men: osteoporotic fractures in men sleep study. J Am Geriatr Soc 55, 1356–1364. 10.1111/j.1532-5415.2007.01290.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hanis C.L., Ferrell R.E., Barton S.A., Aguilar L., Garza-Ibarra A., Tulloch B.R., Garcia C.A., and Schull W.J. (1983). Diabetes among Mexican Americans in Starr County, Texas. Am J Epidemiol 118, 659–672. [DOI] [PubMed] [Google Scholar]
- 97.Hanis C.L., Redline S., Cade B.E., Bell G.I., Cox N.J., Below J.E., Brown E.L., and Aguilar D. (2016). Beyond type 2 diabetes, obesity and hypertension: an axis including sleep apnea, left ventricular hypertrophy, endothelial dysfunction, and aortic stiffness among Mexican Americans in Starr County, Texas. Cardiovasc Diabetol 15, 86 10.1186/s12933-016-0405-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Choi J.H., Kim E.J., Kim Y.S., Choi J., Kim T.H., Kwon S.Y., Lee H.M., Lee S.H., Shin C., and Lee S.H. (2010). Validation study of portable device for the diagnosis of obstructive sleep apnea according to the new AASM scoring criteria: Watch-PAT 100. Acta Otolaryngol. (Stockh.) 130, 838–843. 10.3109/00016480903431139 [DOI] [PubMed] [Google Scholar]
- 99.Fried L.P., Borhani N.O., Enright P., Furberg C.D., Gardin J.M., Kronmal R.A., Kuller L.H., Manolio T.A., Mittelmark M.B., Newman A., et al. (1991). The Cardiovascular Health Study: design and rationale. Ann Epidemiol 1, 263–276. [DOI] [PubMed] [Google Scholar]
- 100.Sorlie P.D., Avilés-Santa L.M., Wassertheil-Smoller S., Kaplan R.C., Daviglus M.L., Giachello A.L., Schneiderman N., Raij L., Talavera G., Allison M., et al. (2010). Design and implementation of the Hispanic Community Health Study/Study of Latinos. Ann Epidemiol 20, 629–641. 10.1016/j.annepidem.2010.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Redline S., Sotres-Alvarez D., Loredo J., Hall M., Patel S.R., Ramos A., Shah N., Ries A., Arens R., Barnhart J., et al. (2014). Sleep-disordered breathing in Hispanic/Latino individuals of diverse backgrounds. The Hispanic Community Health Study/Study of Latinos. Am J Respir Crit Care Med 189, 335–344. 10.1164/rccm.201309-1735OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Westbrook P.R., Levendowski D.J., Cvetinovic M., Zavora T., Velimirovic V., Henninger D., and Nicholson D. (2005). Description and validation of the apnea risk evaluation system: a novel method to diagnose sleep apnea-hypopnea in the home. Chest 128, 2166–2175. 10.1378/chest.128.4.2166 [DOI] [PubMed] [Google Scholar]
- 103.Taylor H.A.J., Wilson J.G., Jones D.W., Sarpong D.F., Srinivasan A., Garrison R.J., Nelson C., and Wyatt S.B. (2005). Toward resolution of cardiovascular health disparities in African Americans: design and methods of the Jackson Heart Study. Ethn Dis 15, S6-4–17 [PubMed] [Google Scholar]
- 104.Wilson J.G., Rotimi C.N., Ekunwe L., Royal C.D.M., Crump M.E., Wyatt S.B., Steffes M.W., Adeyemo A., Zhou J., Taylor H.A.J., et al. (2005). Study design for genetic analysis in the Jackson Heart Study. Ethn Dis 15, S6-30–37. [PubMed] [Google Scholar]
- 105.Ng S.S.S., Chan T.-O., To K.-W., Ngai J., Tung A., Ko F.W.S., and Hui D.S.C. (2010). Validation of Embletta portable diagnostic system for identifying patients with suspected obstructive sleep apnoea syndrome (OSAS). Respirol. Carlton Vic 15, 336–342. 10.1111/j.1440-1843.2009.01697.x [DOI] [PubMed] [Google Scholar]
- 106.Mukherjee S., Hillman D., Lee J., Fedson A., Simpson L., Ward K., Love G., Edwards C., Szegner B., and Palmer L.J. (2012). Cohort profile: the Western Australian Sleep Health Study. Sleep Breath 16, 205–215. 10.1007/s11325-011-0491-3 [DOI] [PubMed] [Google Scholar]
- 107.Whitney C.W., Gottlieb D.J., Redline S., Norman R.G., Dodge R.R., Shahar E., Surovec S., and Nieto F.J. (1998). Reliability of scoring respiratory disturbance indices and sleep staging. Sleep 21, 749–757. [DOI] [PubMed] [Google Scholar]
- 108.McCarthy S., Das S., Kretzschmar W., Durbin R., Abecasis G., and Marchini J. (2015). A reference panel of 64,976 haplotypes for genotype imputation. BioRxiv 035170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.1000 Genomes Project Consortium (2015). A global reference for human genetic variation. Nature 526, 68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Howie B.N., Donnelly P., and Marchini J. (2009). A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 5, e1000529 10.1371/journal.pgen.1000529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang C., Zhan X., Liang L., Abecasis G.R., and Lin X. (2015). Improved ancestry estimation for both genotyping and sequencing data using projection procrustes analysis and genotype imputation. Am J Hum Genet 96, 926–937. 10.1016/j.ajhg.2015.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhou X., and Stephens M. (2012). Genome-wide efficient mixed-model analysis for association studies. Nat Genet 44, 821–824. 10.1038/ng.2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Conomos, M.P., Thornton, T.A., Gogarten, S.M., and Brown, L. (2017). GENESIS: GENetic EStimation and Inference in Structured samples (GENESIS): Statistical methods for analyzing genetic data from samples with population structure and/or relatedness. R package version 2.8.0. 10.18129/B9.bioc.GENESIS [DOI]
- 114.Willer C.J., Li Y., and Abecasis G.R. (2010). METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191. 10.1093/bioinformatics/btq340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dadaev T., Leongamornlert D.A., Saunders E.J., Eeles R., and Kote-Jarai Z. (2016). LocusExplorer: a user-friendly tool for integrated visualization of human genetic association data and biological annotations. Bioinformatics 32, 949–951. 10.1093/bioinformatics/btv690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Winkler T.W., Kutalik Z., Gorski M., Lottaz C., Kronenberg F., and Heid I.M. (2015). EasyStrata: evaluation and visualization of stratified genome-wide association meta-analysis data. Bioinformatics 31, 259–261. 10.1093/bioinformatics/btu621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ehlen J.C., Brager A.J., Baggs J., Pinckney L., Gray C.L., DeBruyne J.P., Esser K.A., Takahashi J.S., and Paul K.N. (2017). Bmal1 function in skeletal muscle regulates sleep. ELife 6, e26557 10.7554/eLife.26557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ward L.D., and Kellis M. (2012). HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 40, D930–4. 10.1093/nar/gkr917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.ENCODE Project Consortium (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Roadmap Epigenomics Consortium, Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., et al. (2015). Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330. 10.1038/nature14248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Javierre B.M., Burren O.S., Wilder S.P., Kreuzhuber R., Hill S.M., Sewitz S., Cairns J., Wingett S.W., Várnai C., Thiecke M.J., et al. (2016). Lineage-Specific Genome Architecture Links Enhancers and Non-coding Disease Variants to Target Gene Promoters. Cell 167, 1369–1384.e19. 10.1016/j.cell.2016.09.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chen L., Ge B., Casale F.P., Vasquez L., Kwan T., Garrido-Martín D., Watt S., Yan Y., Kundu K., Ecker S., et al. (2016). Genetic Drivers of Epigenetic and Transcriptional Variation in Human Immune Cells. Cell 167, 1398–1414.e24. 10.1016/j.cell.2016.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Vermunt M.W., Reinink P., Korving J., de Bruijn E., Creyghton P.M., Basak O., Geeven G., Toonen P.W., Lansu N., Meunier C., et al. (2014). Large-scale identification of coregulated enhancer networks in the adult human brain. Cell Rep 9, 767–779. 10.1016/j.celrep.2014.09.023 [DOI] [PubMed] [Google Scholar]
- 124.Lappalainen T., Sammeth M., Friedländer M.R., ‘t Hoen P.A.C., Monlong J., Rivas M.A., Gonzàlez-Porta M., Kurbatova N., Griebel T., Ferreira P.G., et al. (2013). Transcriptome and genome sequencing uncovers functional variation in humans. Nature 501, 506–511. 10.1038/nature12531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.GTEx Consortium (2015). Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660. 10.1126/science.1262110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Fairfax B.P., Humburg P., Makino S., Naranbhai V., Wong D., Lau E., Jostins L., Plant K., Andrews R., McGee C., et al. (2014). Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science 343, 1246949 10.1126/science.1246949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Raj T., Rothamel K., Mostafavi S., Ye C., Lee M.N., Replogle J.M., Feng T., Lee M., Asinovski N., Frohlich I., et al. (2014). Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science 344, 519–523. 10.1126/science.1249547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zeller T., Wild P., Szymczak S., Rotival M., Schillert A., Castagne R., Maouche S., Germain M., Lackner K., Rossmann H., et al. (2010). Genetics and beyond–the transcriptome of human monocytes and disease susceptibility. PLoS One 5, e10693 10.1371/journal.pone.0010693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hao K., Bossé Y., Nickle D.C., Paré P.D., Postma D.S., Laviolette M., Sandford A., Hackett T.L., Daley D., Hogg J.C., et al. (2012). Lung eQTLs to help reveal the molecular underpinnings of asthma. PLoS Genet. 8, e1003029 10.1371/journal.pgen.1003029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Grundberg E., Small K.S., Hedman Å.K., Nica A.C., Buil A., Keildson S., Bell J.T., Yang T.-P., Meduri E., Barrett A., et al. (2012). Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat. Genet. 44, 1084–1089. 10.1038/ng.2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Giambartolomei C., Zhenli Liu J., Zhang W., Hauberg M., Shi H., Boocock J., Pickrell J., Jaffe A.E., CommonMind Consortium, Pasaniuc B., et al. (2018). A Bayesian framework for multiple trait colocalization from summary association statistics. Bioinformatics 34, 2538–2545. 10.1093/bioinformatics/bty147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Barbeira A.N., Dickinson S.P., Bonazzola R., Zheng J., Wheeler H.E., Torres J.M., Torstenson E.S., Shah K.P., Garcia T., Edwards T.L., et al. (2018). Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat Commun. 9, 1825 10.1038/s41467-018-03621-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhu S., Qian T., Hoshida Y., Shen Y., Yu J., and Hao K. (2018). GIGSEA: Genotype Imputed Gene Set Enrichment Analysis using GWAS Summary Level Data. Bioinformatics 35, 160–163. 10.1093/bioinformatics/bty529 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Top: African-Americans; Middle: European-Americans; Bottom: Multi-ethnic. Variants in the multi-ethnic results had to have results from cohorts in two or more populations.
(PDF)
Top: African-Americans; Middle: European-Americans; Bottom: Multi-ethnic (2 or more populations for each variant).
(PDF)
Top: African-Americans; Middle: European-Americans; Bottom: Multi-ethnic (2 or more populations for each variant).
(PDF)
Top: Females; Bottom: Males.
(PDF)
Top: Females; Bottom: Males.
(PDF)
Top: Females; Bottom: Males.
(PDF)
The supplemental regional plot order corresponds to the order found in S2, S3 and S4 Tables, minus the associations shown in Figs 1 and 2.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Spearman correlations between phenotypes and covariates across all discovery cohorts and ethnic groups are shown. Correlations with the spirometry measures FEV1 and FVC are also shown. Correlations were pooled using Fisher Z-transformations weighted by sample size. 95% confidence intervals are listed in parentheses.
(XLS)
Lead SNPs are shown for regions with significant (p < 5.0 × 10–8) and suggestive (p < 1.0 × 10–6) p-values. Discovery regions were carried forward if the combined discovery and replication analysis results indicated a lower p-value, retained genome-level significance, or replication was unavailable (i.e. due to frequency or imputation filters). SNPs denotes the count of regional SNPs with p < 1.0 × 10–6. Genes indicates overlapping Ensembl genes within 5 kb of the p < 1 × 10–6 SNPs. CAF indicates coded allele frequency range. Individual regional SNP results are provided in S3 Table. Individual regions were minimally 500 kb apart.
(XLS)
“Locus Class” indicates “Discovery/Replication” for main analyses or “Combined meta-analysis” for regions not clearing initial discovery p-value thresholds (i.e. S7 Table loci).
(XLS)
Sex-stratified samples with n < 1000 are included here for comparison but were not included in the main analyses. An individual variant present in the combined-sex analysis for a given cohort may be missing from the equivalent sex-stratified analysis due to analysis-specific minor allele count thresholds of 20. “Locus Class” indicates “Discovery/Replication” for main analyses or “Combined meta-analysis” for regions not clearing initial discovery p-value thresholds (i.e. S7 Table loci).
(XLS)
Each lead SNP from the “Original Phenotype” and “Original Model” columns was analyzed using equivalent individuals and models for different phenotypes. Available sample size will vary depending on the availability of the phenotype. AHI values are taken from Chen, Cade, et al. (submitted) “Locus Class” indicates “Discovery/Replication” for main analyses or “Combined meta-analysis” for regions not clearing initial discovery p-value thresholds (i.e. S7 Table loci).
(XLS)
Lead SNPs are shown for regions with suggestive (p < 1.0 × 10–6) p-values. Discovery regions were carried forward if the combined discovery and replication analysis results indicated a lower p-value, retained genome-level significance, or replication was unavailable (i.e. due to frequency or imputation filters). SNPs denotes the count of regional SNPs with p < 1.0 × 10–6. Genes indicates overlapping Ensembl genes within 5 kb of the p < 1 × 10–6 SNPs. CAF indicates coded allele frequency range. Individual regional SNP results are provided in S3 Table. Individual regions were minimally 500 kb apart.
(XLS)
Lead SNPs are shown for regions with significant (p < 5.0 x 10–8) combined-analysis p-values with p > 1 x 10–6 in the discovery phase. SNPs denotes the count of regional SNPs with p < 1.0 x 10–6. Genes indicates overlapping Ensembl genes within 5 kb of the p < 1 x 10–6 SNPs. CAF indicates coded allele frequency range. Individual regional SNP results are provided in S3 Table. Individual regions were minimally 500 kb apart.
(XLS)
Lead variants were re-analyzed with standard models and additional covariates as listed. As these covariates were not collected in all cohorts and individuals, the analysis were also performed with the same individuals for direct comparisons (matched N control columns).
(XLS)
Data were obtained from HaploReg 4.1 (https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php) using exact SNPs (LD threshold = NA) and a ChromHMM 15-state model, followed by extraction from HTML. “Locus Class” indicates “Discovery/Replication” for main analyses or “Combined meta-analysis” for regions not clearing initial discovery p-value thresholds (i.e. S7 Table loci).
(XLS)
Blueprint cell lines are a non-cancerous sub-set of largely blood cells. Vermunt brain region samples were not assayed for promoter regions. “Locus Class” indicates “Discovery/Replication” for main analyses or “Combined meta-analysis” for regions not clearing initial discovery p-value thresholds (i.e. S7 Table loci).
(XLS)
Data were obtained from Fairfax et al. (monocytes), Geuvadis (LCLs), GTEx, Hao et al. (lung), Muther (adipose, LCLs, skin), Raj et al. (CD4, monocytes), Westra et al. (whole blood), and Zeller et al. (monocytes). “Locus Class” indicates “Discovery/Replication” for main analyses or “Combined meta-analysis” for regions not clearing initial discovery p-value thresholds (i.e. S7 Table loci).
(XLS)
MTAG analysis was performed on European ancestry samples using our summary statistics for the three traits simultaneously. Lead results (p < 1 x 10–6) are shown.
(XLS)
Lead results (p < 0.05) are shown. Note that Ensembl IDs obtained from MetaXcan are occasionally unavailable.
(XLS)
Lead results (p < 0.05) are shown. Note that Ensembl IDs obtained from MetaXcan are occasionally unavailable.
(XLS)
Lead results (p < 0.05) are shown. Note that Ensembl IDs obtained from MetaXcan are occasionally unavailable.
(XLS)
Lead results (p < 0.05) are shown. Note that Ensembl IDs obtained from MetaXcan are occasionally unavailable.
(XLS)
Lead results (p < 0.05) are shown. Note that Ensembl IDs obtained from MetaXcan are occasionally unavailable.
(XLS)
Lead results (p < 0.05) are shown. Note that Ensembl IDs obtained from MetaXcan are occasionally unavailable.
(XLS)
Lead results (p < 0.05) are shown. Note that Ensembl IDs obtained from MetaXcan are occasionally unavailable.
(XLS)
MetaXcan gene-level results for DGN whole blood (S19 Table, all p-values) were used as input.
(XLS)
MetaXcan gene-level results for DGN whole blood (S19 Table, all p-values) were used as input. MicroRNAs are curated by the Molecular Signatures Database (http://software.broadinstitute.org/gsea/msigdb/index.jsp).
(XLS)
MetaXcan gene-level results for DGN whole blood (S19 Table, all p-values) were used as input. Transcription factor binding sites are curated by the Molecular Signatures Database (http://software.broadinstitute.org/gsea/msigdb/index.jsp).
(XLS)
Cohort-level information is shown for the three primary analyses, including discovery/replication category, the number of individuals with available phenotyping and genotyping, genotyping platform, imputation panel, and the number of imputed variants tested in each analysis following info score, MAC, and MAF filtering.
(XLS)
Data Availability Statement
Full meta-analysis results are freely available from http://www.sleepdisordergenetics.org/informational/data