Abstract
Isolates of the gastric pathogen Helicobacter pylori harvested from different individuals are highly polymorphic. Strain variation also has been observed within a single host. To more fully ascertain the extent of H. pylori genetic diversity within the ecological niche of its natural host, we harvested additional isolates of the sequenced H. pylori strain J99 from its human source patient after a 6-year interval. Randomly amplified polymorphic DNA PCR and DNA sequencing of four unlinked loci indicated that these isolates were closely related to the original strain. In contrast, microarray analysis revealed differences in genetic content among all of the isolates that were not detected by randomly amplified polymorphic DNA PCR or sequence analysis. Several ORFs from loci scattered throughout the chromosome in the archival strain did not hybridize with DNA from the recent strains, including multiple ORFs within the J99 plasticity zone. In addition, DNA from the recent isolates hybridized with probes for ORFs specific for the other fully sequenced H. pylori strain 26695, including a putative traG homolog. Among the additional J99 isolates, patterns of genetic diversity were distinct both when compared with each other and to the original prototype isolate. These results indicate that within an apparently homogeneous population, as determined by macroscale comparison and nucleotide sequence analysis, remarkable genetic differences exist among single-colony isolates of H. pylori. Direct evidence that H. pylori has the capacity to lose and possibly acquire exogenous DNA is consistent with a model of continuous microevolution within its cognate host.
The highly diverse bacteria Helicobacter pylori persist in the human stomach and induce chronic gastritis for the lifetime of their hosts, a process that increases risk for developing peptic ulceration, noncardia gastric adenocarcinoma, and gastric lymphoma (1–4). However, clinical sequelae develop in only a fraction of colonized individuals (1, 2, 5) and likely depend on differentially represented bacterial determinants, host characteristics that are governed by genetic polymorphisms (6), and/or the specific interactions between a particular strain and its host that occur during decades of coexistence.
Although the H. pylori population structure appears to be clonal over short periods of time, isolates obtained from different individuals exhibit substantial genetic diversity, consistent with extensive recombination and a panmictic population structure (7–12). Differences among strains include point mutations in highly conserved genes (11, 13), the presence of nonconserved (14, 15) and/or mosaic forms of genes (16, 17), and chromosomal organization (18–20). Putative mechanisms for the generation of diversity within H. pylori include frequent horizontal genetic exchange among strains and a high level of spontaneous mutation occurring over a long evolutionary time period within a highly restricted niche (9, 21, 22). To examine the extent and types of genetic diversity that are present during chronic colonization of the human stomach, we harvested additional H. pylori isolates from the source patient from whom the completely sequenced strain J99 was obtained, 6 years after the original isolate was cultured. Differences in genetic content among recent and archival J99 single-colony isolates were examined by randomly amplified polymorphic DNA (RAPD) PCR analysis, DNA sequencing, and hybridization to an H. pylori microarray in an effort to take advantage of this unique opportunity to study the composite genetic profile of an H. pylori strain within the gastric niche of a single individual.
Methods
Bacterial Strains and Culture.
The human source patient of the fully sequenced H. pylori strain J99 (20) was a 48-year-old Caucasian male resident of Tennessee with no family history of peptic ulceration. He underwent upper gastrointestinal endoscopy initially in 1994 and was found to have a duodenal ulcer, and strain J99 was isolated from a gastric antral biopsy as described (23). For the current study, 12 additional single-colony isolates were purified from the original culture sweep recovered from this biopsy specimen. The J99 source patient refused antimicrobial treatment and, because of persistent dyspepsia, he underwent repeat upper endoscopy in 2000, 6 years after the original J99 strain was isolated. Thirty single-colony isolates were isolated from gastric cardia (n = 1), corpus (n = 12), antral (n = 12), and duodenal biopsies (n = 5). All procedures were approved by the Vanderbilt University and Nashville Department of Veterans Affairs institutional review boards. Bacteria were routinely grown on trypticase soy agar plates with 5% sheep blood (BBL) at 37°C with 5% CO2. Antibiotic resistance was determined on Brucella agar plates with 5% FCS containing either metronidazole (16 μg/ml), clarithromycin (1.0 μg/ml), or ampicillin (1.0 μg/ml).
Molecular Biology.
Chromosomal DNA was prepared from 48-h plate-grown bacteria with the cetyltrimethylammonium bromide method (24). Randomly amplified polymorphic DNA (RAPD) PCR analysis was performed as described by using primers D11344 and 14307 (7). Additional PCRs were performed by using primers based on the sequence of strain J99 (Table 2, which is published as supporting information on the PNAS web site, www.pnas.org) under typical conditions, and products were analyzed by agarose gel electrophoresis.
DNA Sequence Analysis.
Purified PCR (QiaQuick PCR purification, Qiagen, Chatsworth, CA) was sequenced on both strands by using an automated sequencer (ABI 377, Applied Biosystems). Regions sequenced correspond to nucleotides 156616–157275 (recA), 147484–146787 (mutY), 44524–45009 (JHP0039–001), and 1344381–1345021 (JHPJHP1211–1212) of the sequence J99 chromosome (20) (GenBank accession no. AE001439). Sequence analysis was performed with vector nti (InforMax, Bethesda, MD), and homology searches were performed by blast (25).
Microarray Analysis.
Microarray design and hybridization conditions have been described (26, 27). Briefly, the array consists of probes representing ORFs of the H. pylori 26695 genome supplemented with probes for J99-specific ORFs printed in duplicate. Cy3- or Cy5-labeled probes were generated by priming of 250 ng genomic DNA with random octamers and extension with Klenow. Labeled DNA from each isolate was hybridized on multiple H. pylori microarray as described (26, 27). Data points were excluded because of low signal or slide abnormalities. For this analysis three measurements were required for each gene of each strain, and only those genes with three measurements in 80% of the strains were included. Data were normalized by using the default-computed normalization of the Stanford Microarray Database (http://genome-www4.stanford.edu/MicroArray/help/results_normalization.html) and the mean of the log2(red/green ratio) was computed. The cutoff for absence of a gene was defined as a log2(red/green) of <−1.0 based on test hybridizations (26). Data were simplified into a binary score, analyzed with xcluster software (http://genome-www.stanford.edu/∼sherlock/cluster.html), and displayed with treeview (28). The false positive and false negative rates were determined to be 3.5% and 0.34%, respectively. The complete data set is available as supporting information on the PNAS web site.
Results
RAPD PCR Analysis of H. pylori Strain J99 Isolates.
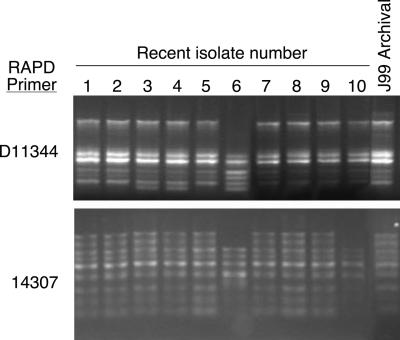
To determine whether the same strain persisted in the J99 source patient 6 years after primary isolation, we obtained 30 additional single colony isolates from distinct biopsy sites (antrum, corpus, duodenum, cardia). Because RAPD PCR analysis can reliably distinguish H. pylori strains isolated from different individuals (7), we explored whether this method also could distinguish isolates from a single individual, by performing RAPD PCR analysis on the new isolates as well as a representative isolate of the archival strain. Results using two different primers demonstrated that all of the recent isolates have similar amplification profiles. This finding indicates that they represent a closely related population and the similarity of these profiles to that of archival J99 also indicates that this patient was colonized by the same strain at both time points (Fig. 1). However, patterns of amplification were not identical and subtle differences in RAPD profiles of recent isolates were present when compared with the archival J99 isolate (Fig. 1). These data suggest that, although this patient was persistently colonized by a single strain of H. pylori, genetic variants may exist among these isolates.
Figure 1.
RAPD PCR analysis of recent and archival H. pylori J99 isolates from the original human source patient. RAPD PCR patterns of 10 representative recent isolates and the archival J99 strain are shown. PCR was performed with primers D11344 and 14307, and products were analyzed by agarose gel electrophoresis. Results for corpus samples C1–C10 are shown in lanes 1–10, respectively, and results for the archival strain are indicated.
Sequence Analysis of Four Unlinked Chromosomal Loci.
Another indicator of H. pylori genetic diversity is nucleotide sequence variation. Therefore, DNA sequences from four different intragenic and intergenic chromosomal loci from a subset of recent isolates (C3, C6, and C10, Fig. 1) and the archival J99 isolate were compared. Selected intragenic regions were within the highly conserved gene recA (660 bp) and the more variable gene mutY (698 bp) (11). Because intergenic regions that do not include promoter elements should be subject to less selective pressure and may have greater potential to accumulate spontaneous point mutations, the regions between JHP0039 and JHP0040 (486 bp) and between JHP1211 and JHP1212 (641 bp), which are flanked by convergent operons, were chosen for analysis (20). In all cases, the nucleotide sequences of recent isolates were 100% identical to archival J99, supporting the RAPD PCR results indicating that the recent isolates are very closely related to the original J99 isolate.
Antibiotic Susceptibility of Recent Isolates.
The above analyses suggested genotypic relatedness between original and recent H. pylori J99 isolates. We next sought to determine whether phenotype was similarly conserved by testing 30 recent isolates and 12 single-colony isolates from the archival sweep for susceptibility to metronidazole, ampicillin, and clarithromycin. All isolates were susceptible to metronidazole and ampicillin; however, four recent isolates (three antral, one duodenal) were resistant to clarithromycin. Because specific point mutations in the H. pylori 23S rRNA sequence confer resistance to clarithromycin (29–31), this region was sequenced for each of the clarithromycin-resistant isolates along with three recent clarithromycin-sensitive isolates and the archival J99. The sequences were identical except at position 2142 where resistant isolates were found to have an A2142G transition to which clarithromycin resistance has previously been attributed, confirming the significance of this locus for clarithromycin susceptibility (29–31).
Analysis of Recent and Archival J99 Isolates by Whole-Genome Microarray.
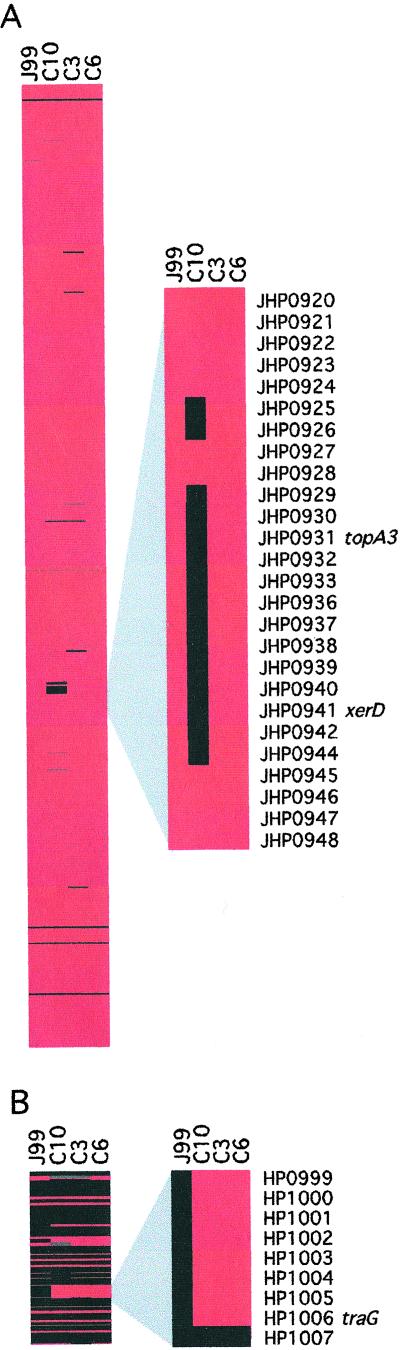
RAPD PCR and sequence analyses indicated that recent and archival J99 isolates belong to the same strain. However, subtypic RAPD amplification patterns also suggested that subtle genetic differences may exist among isolates. Therefore, we used whole-genome microarray hybridization to more comprehensively assess differences in genetic content among three recent isolates (C3, C6, and C10, Fig. 1) compared with the original J99. DNA hybridization revealed several differences in genetic content between the archival and new isolates as well as among the new isolates themselves (Fig. 2, Table 1), and some of these differences included regions of linked genes. One such region in recent isolate C10 was of particular interest, as a group of contiguous ORFs within the J99 plasticity zone was absent (Fig. 2A). PCR analysis of selected ORFs within this region confirmed that isolate C10 lacks ORFs JHP0925, JHP0926, JHP0929–0931, JHP0941, JHP0942, and JHP0944. All of the missing ORFs in this region are predicted to encode hypothetical proteins with the exception of JHP0931 and JHP0941, which are predicted to encode a topoisomerase (topA3) and recombinase (xerD), respectively. Gene-specific PCR analysis also indicated that JHP0929–0931 and JHP0944 were present in 100% of 12 archival single colony isolates tested, but were absent in 59% and 30% of recent isolates, respectively. Notably, despite the absence of numerous ORFs, all of the genes within the cag island were present in all of the recent isolates tested.
Figure 2.
Absence and presence of ORFs in recent J99 isolates as determined by microarray analysis. Representative isolates C10, C3, and C6 were compared with archival J99 by whole-genome microarray analysis. The presence (red) or absence (black) of genes is displayed according to their position on the chromosome for each isolate (missing data are gray). (A) The ORFs of archival J99 are represented vertically in chromosomal order. (Left) The entire chromosome starting at JHP0001. (Right) An enlarged image showing a region of contiguous ORFs within the J99 plasticity zone missing in isolate C10. (B) Variably present ORFs specific to H. pylori strain 26695. (Left) All of the 26695 ORFs not present in sequenced strain J99. The red ORFs shown in the archival J99 lane are indicative of false positive hybridization. (Right) An enlarged image showing a region of linked ORFs present in all three recent J99 isolates.
Table 1.
ORFs that have putative identities that were present or absent in recent isolates of J99 as compared with archival J99 based on microarray hybridization
| J99 ORF | 26695 ORF | Putative gene | C10 | C3 | C6 | J99 archival |
|---|---|---|---|---|---|---|
| ORFs lost | ||||||
| JHP0673 | HP0736 | serC | − | − | + | + |
| JHP0976 | HP0941 | alr | + | − | + | + |
| JHP0931 | N/A | topA3 | − | + | + | + |
| JHP0941 | HP0995 | xerD | − | + | + | + |
| ORFs gained | ||||||
| N/A | HP0459 | virB4 | + | + | + | − |
| N/A | HP1006 | traG | + | + | + | − |
| N/A | HP1000 | parA | + | + | + | − |
N/A, not applicable.
The published genomic sequence of H. pylori strain 26695 includes many ORFs not present in the sequenced J99 isolate, and the inclusion of these ORFs on the microarray provided the ability to examine whether sequences absent in the sequenced J99 genome might be present in the recent isolates (20, 26, 32). DNA from the recent J99 isolates hybridized with several 26695-specific ORFs, whereas archival J99 DNA did not (Fig. 2B, Table 1, and Table 3, which is published as supporting information on the PNAS web site). The presence of one such locus (HP1000–HP1006) was confirmed by PCR in three recent J99 isolates (C3, C6, and C10) and found to be organized in the same order found in sequenced H. pylori strain 26695. These ORFs are predicted to encode hypothetical proteins with the exceptions of HP1000 and HP1006, which are predicted to encode a putative partitioning protein, ParA, and a putative conjugal transfer protein, TraG (32), respectively. PCR analysis of the 12 archival single-colony isolates with primer pairs for HP1000 and HP1006 yielded no product, whereas products for HP1000 and HP1006 were obtained for 100% of recent isolates (n = 30), supporting findings from the microarray analysis.
A Subset of Recent J99 Isolates Has Novel Sequences.
In the process of confirming microarray data by PCR, an amplified product spanning JHP0929 to JHP0931 was noted to be larger in isolates C3 and C6 relative to the archival J99 isolate (Fig. 3). As predicted by microarray data, isolate C10 lacked this region (Fig. 2A). Sequence analysis of this region from isolates C3 and C6 revealed a 674-bp insertion between ORFs JHP0929 and JHP0930 (Fig. 3; GenBank accession numbers AF433040 and AF433041). This insertion does not appear to contain an ORF, nor does it exhibit homology to any sequence in the nonredundant or microbial genomes National Center for Biotechnology Information databases. PCR analysis of this region indicated that 59% of the recent isolates lack this locus, whereas the remaining 41% yielded products of similar size as for isolates C3 and C6 (data not shown). The archival J99 single-colony isolates tested did not yield the larger product indicative of this insertion. Thus, microarray-based genetic comparisons led to the identification of H. pylori sequences that are not present in the genome sequences of J99 or 26695 (20, 32).
Figure 3.
Identification of an insertion sequence between JHP0929 and JHP0930 in recent isolates. (A) PCR products generated by amplification with primers within JHP0929 and JHP0931 were analyzed by agarose gel electrophoresis. MW, 1-kb ladder (Promega); lane 1, J99-archival; lane 2, C3; lane 3, C6; lane 4, C10; lane 5, negative control. (B) Location of insertion as determined by sequence analysis is indicated between ORFs JHP0929 and JHP0930.
All Isolates Examined by Microarray Analysis Are Unique.
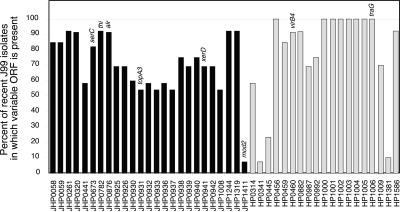
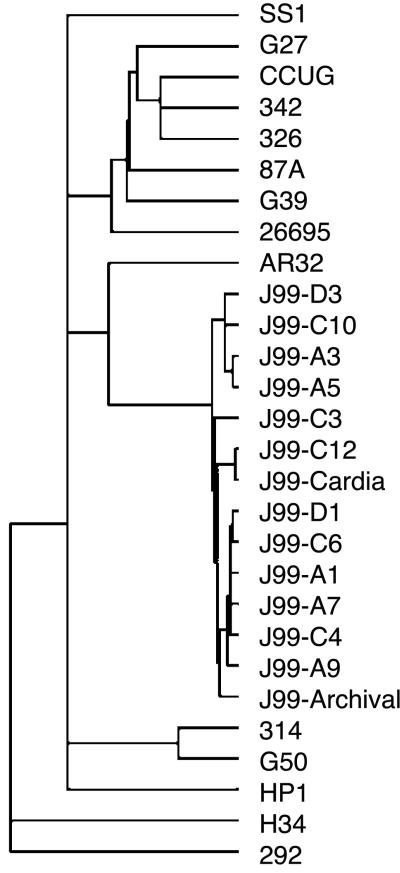
Having demonstrated both the absence and the presence of sequences in recent isolates versus the archival strain, we extended this analysis by performing microarray hybridizations on 10 additional recent J99 isolates. This group included isolates from the gastric corpus as well as the cardia, antrum, and duodenum. A total of 44 variable loci were identified that were either absent or present among this population as compared with the archival J99 (Fig. 4). There was no single locus universally absent from the recent isolates; conversely, homologues of eight ORFs from strain 26695 not present in the original isolate were present in all 13 recent isolates (Fig. 4). Cluster analysis to group strains based on their gene content combined with previously reported data from 14 unrelated strains (26) indicated that, despite the differences between J99 variants, these isolates are much more closely related to each other than to the independent strains (Fig. 5). Further, anatomic distribution of J99 isolates within the gastric niche is not reflected in the patterns of genetic diversity as isolates harvested from the cardia, corpus, antrum, and duodenum were distributed heterogeneously throughout the J99 branch of the cluster (Fig. 5).
Figure 4.
Presence of variable loci among a population of recent J99 isolates (n = 13). The presence of loci that varied between the archival and recent J99 isolates as determined by microarray analysis are shown. Black bars represent the percentage of recent J99 isolates that possess ORFs present in the archival J99 and gray bars indicate the percentage of recent isolates that have DNA with homology to 26695-specific genes.
Figure 5.
Relatedness of archival and recent J99 isolates as compared with each other and to independent H. pylori strains. Cluster analysis of microarray data for J99 isolates and 14 unrelated strains (26) shows that all J99 isolates cluster together on a branch distinct from other isolates.
Discussion
Remarkable genetic variation exists between H. pylori strains isolated from unique individuals, yet the origins and determinants of this diversity are not completely understood. Previous data indicate that although point mutation may be one source of generating diversity, recombination is the predominant mechanism within this species (9, 11). Further, although isolates from a single individual or family members are frequently clonal, the overall population structure of H. pylori is panmictic (9).
There have been few reports of H. pylori genetic changes that may occur during long-term colonization within the gastric niche of a single individual. Kuipers et al. (33) studied pairs of isolates obtained 7–10 years apart from individual patients and, using RAPD PCR and amplified fragment length polymorphism analyses, identified differences in amplification profiles, suggesting that genetic changes develop during colonization of a single host. However, neither the identity nor the genomic location of the variable genes was characterized. Another study that examined strains isolated from members of a single family found evidence that DNA exchange occurred in vivo, resulting in loss of the cag pathogenicity island (34). The data presented here extend these studies by implementing both traditional molecular and whole-genome microarray techniques to examine the extent of genetic diversity present in the fully sequenced strain H. pylori strain J99 during long-term colonization of its natural host.
As expected for most individuals persistently colonized by a single H. pylori strain, RAPD PCR amplification patterns were similar for archival and recent isolates. However, subtle differences in amplification profiles suggested that although the population consisted of very closely related bacteria, it was not completely homogeneous (Fig. 1). We further confirmed relatedness of new and archival J99 isolates by demonstrating an absolute concordance in DNA sequence of four unlinked intergenic and intragenic chromosomal regions (Table 1). Bereswill et al. (35) recently reported that polymorphisms within regions upstream of ribA and vacA and downstream of cagA could be used to differentiate H. pylori isolates from the corpus and antrum of individual patients. In contrast, PCR analysis of these loci in the current study revealed no differences among recent and archival J99 isolates, regardless of the anatomical site of isolation (data not shown). These results, along with the finding that three recent isolates had absolute sequence identity to the archival J99 over 2,485 bp, confirmed that they are indeed derivatives of the same strain.
Although sequence analysis from these unlinked loci revealed no point mutations, antibiotic susceptibility testing reiterated the population heterogeneity inferred from RAPD PCR analysis. Four (13%) of 30 recent isolates were resistant to clarithromycin, data that are concordant with previous reports indicating that antibiotic sensitive and resistant isolates of a single H. pylori strain may exist within a single bacterial population (36–46). Whether the J99 source patient was exposed to clarithromycin or another macrolide antibiotic (e.g., azithromycin) over the 6-year interval is unknown, but this represents a plausible hypothesis for the development of clarithromycin-resistant isolates.
Analysis by whole-genome microarray revealed both the differences that exist among the J99 isolates (recent and archival) as well as the relatedness of these isolates to each other when compared with H. pylori isolates from other individuals (Fig. 5). Despite a difference in genomic composition in 3% of J99 loci, the extent of variation is much less than between a similar number of strains from different individuals for which 22% of loci showed variation (26). Although it has been proposed that the cag island in an unstable locus, subject to either deletion or loss by homologous recombination (15, 34, 47), all of the recent J99 isolates tested possess an intact cag island as did the archival isolate. Cluster analysis also indicated that H. pylori isolates harvested from the same region of the stomach are no more similar to each other than to isolates from an anatomically distinct site, suggesting that, at least within this patient, genetic variation is not related to bacterial adaptation exerted by conditions in a particular gastric microniche.
Microarray analysis revealed further evidence of diversity by demonstrating both the absence of sequences and presence of additional DNA relative to the archival J99. The total number of ORFs lost by at least one of the recent isolates represents 2.3% of the J99 ORFs assayed and each individual isolate had lost between 0.28 and 1.52% (mean ± SEM = 0.83 ± 0.45%). Isolate C10, which appeared most similar to the archival strain by RAPD PCR analysis (Fig. 1), lacks a large group of contiguous genes within the J99 plasticity zone (Fig. 2A). ORFs with homology to those in this region of J99 have been found in numerous H. pylori strains tested (26, 48); however, the number of these ORFs varies dramatically among unrelated strains (48), confirming the pliant nature of this region. In addition to the absence of ORFs, all of the recent isolates appear to possess DNA with homology to ORFs HP1000–HP1006 of sequenced strain 26695 (Figs. 2B and 4). In contrast, none of the 12 archival J99 single-colony isolates possessed either HP1000 or HP1006, suggesting that these ORFs were not present within the archival population colonizing the specific site of original biopsy harvest. A potential explanation for the presence of these previously undetected sequences is that a distinct J99 subpopulation possessing these ORFs was present in a different gastric location at the time of the original endoscopy and during chronic colonization, these sequences were disseminated throughout the population. Alternatively, it is possible that these loci were acquired in the 6-year interval by means of horizontal gene transfer from another strain of H. pylori or closely related organism that was transiently present in the stomach. Based on the data presented in this study and sample availability, neither possibility can be eliminated. Most of the newly identified ORFs are hypothetical genes, with the notable exception of HP1006, predicted to encode a protein belonging to the TraG family (32). In Escherichia coli and Agrobacterium tumefaciens, TraG family members are required for the delivery of DNA to the transport machinery for conjugation (49–51). Although it has been demonstrated that HP1006 is not required for transformation in H. pylori, its potential role in H. pylori conjugation has not been determined (52, 53). If this protein does participate in DNA transfer, acquisition of this gene might confer an evolutionary advantage by providing an additional mechanism through which DNA exchange may occur. Based on %GC content, codon usage, and amino acid usage, Garcia-Vallve et al. (54) have suggested that strain 26695 originally acquired ORFs H1002-HP1006 by horizontal gene transfer (http://www.fut.es/∼debb/HGT). This hypothesis, together with the newly identified presence of these ORFs as a contiguous locus in recent J99 isolates, suggests that this may represent a highly mobile genetic element.
The microarray used for this study was designed to assay ORFs of sequenced H. pylori strain 26695 and J99 (26) and other H. pylori sequences that may exist, but that are not present within either of these genomes, cannot be detected by this method. One such sequence was serendipitously identified during the course of confirming the microarray data (Fig. 3). It is likely that additional sequences without homology to 26695 or J99 ORFs are present within the genomes of the recent J99 isolates, but remain to be identified.
In conclusion, these results indicate that H. pylori exists within its ecological niche as a bacterial population in a continuous state of genetic flux, which may allow the bacteria to rapidly adapt to changing conditions in its current host as well as being poised to colonize a new host (55–57). Further, these data also suggest that the proposed concept of core and flexible gene pools may not be applicable only to the genomes of bacterial species, but also to bacterial strain populations (58–61). Genes common to all isolates of a strain may comprise a core strain genome, whereas the remainder of the genes retained in the population comprise the flexible, or auxiliary, gene pool, with the sum of both of these groups of genes constituting the composite strain genotype. In the case of H. pylori, it is likely that the natural competence and potential for conjugation of this species facilitates the formation of recombinants within an individual strain population, which subsequently results in the varied distribution of the genes belonging to the flexible genome.
Supplementary Material
Acknowledgments
We thank Cathy Kelton and Inna Billis for excellent technical assistance. This work was supported in part by the National Institutes of Health (R29 CA77955, R01 DK58587, and R01 AI38459), F. D. H. N. Fiterman Foundation Award, Vanderbilt University Clinical Research Center, the Medical Research Service of the Department of Veterans Affairs, and the Jane Coffin Childs Memorial Fund for Medical Research.
Abbreviation
- RAPD
randomly amplified polymorphic DNA
Footnotes
References
- 1.Blaser M J, Perez-Perez G I, Kleanthous H, Cover T L, Peek R M, Chyou P H, Stemmermann G N, Nomura A. Cancer Res. 1995;55:2111–2115. [PubMed] [Google Scholar]
- 2.Nomura A, Stemmermann G N, Chyou P H, Kato I, Pérez-Pérez G I, Blaser M J. N Engl J Med. 1991;325:1132–1136. doi: 10.1056/NEJM199110173251604. [DOI] [PubMed] [Google Scholar]
- 3.Parsonnet J, Friedman G D, Vandersteen D P, Chang Y, Vogelman J H, Orentreich N, Sibley R K. N Engl J Med. 1991;325:1127–1131. doi: 10.1056/NEJM199110173251603. [DOI] [PubMed] [Google Scholar]
- 4.Peterson W L. N Engl J Med. 1991;324:1043–1048. doi: 10.1056/NEJM199104113241507. [DOI] [PubMed] [Google Scholar]
- 5.Taylor D N, Blaser M J. Epidemiol Rev. 1991;13:42–59. doi: 10.1093/oxfordjournals.epirev.a036078. [DOI] [PubMed] [Google Scholar]
- 6.El-Omar E M. Gut. 2001;48:743–747. doi: 10.1136/gut.48.6.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akopyanz N, Bukanov N O, Westblom T U, Kresovich S, Berg D E. Nucleic Acids Res. 1992;20:5137–5142. doi: 10.1093/nar/20.19.5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Doorn N E, Namavar F, Kusters J G, van Rees E P, Kuipers E J, de Graaff J. FEMS Microbiol Lett. 1998;160:145–150. doi: 10.1111/j.1574-6968.1998.tb12904.x. [DOI] [PubMed] [Google Scholar]
- 9.Suerbaum S, Smith J M, Bapumia K, Morelli G, Smith N H, Kunstmann E, Dyrek I, Achtman M. Proc Natl Acad Sci USA. 1998;95:12619–12624. doi: 10.1073/pnas.95.21.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salaun L, Audibert C, Le Lay G, Burucoa C, Fauchere J L, Picard B. FEMS Microbiol Lett. 1998;161:231–239. doi: 10.1111/j.1574-6968.1998.tb12953.x. [DOI] [PubMed] [Google Scholar]
- 11.Achtman M, Azuma T, Berg D E, Ito Y, Morelli G, Pan Z J, Suerbaum S, Thompson S A, van der Ende A, van Doorn L J. Mol Microbiol. 1999;32:459–470. doi: 10.1046/j.1365-2958.1999.01382.x. [DOI] [PubMed] [Google Scholar]
- 12.Go M F, Kapur V, Graham D Y, Musser J M. J Bacteriol. 1996;178:3934–3938. doi: 10.1128/jb.178.13.3934-3938.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kansau I, Raymond J, Bingen E, Courcoux P, Kalach N, Bergeret M, Braimi N, Dupont C, Labigne A. Res Microbiol. 1996;147:661–669. doi: 10.1016/0923-2508(96)84023-x. [DOI] [PubMed] [Google Scholar]
- 14.Akopyants N S, Clifton S W, Kersulyte D, Crabtree J E, Youree B E, Reece C A, Bukanov N O, Drazek E S, Roe B A, Berg D E. Mol Microbiol. 1998;28:37–53. doi: 10.1046/j.1365-2958.1998.00770.x. [DOI] [PubMed] [Google Scholar]
- 15.Censini S, Lange C, Xiang Z, Crabtree J E, Ghiara P, Borodovsky M, Rappuoli R, Covacci A. Proc Natl Acad Sci USA. 1996;93:14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peek R M, Jr, Thompson S A, Donahue J P, Tham K T, Atherton J C, Blaser M J, Miller G G. Proc Assoc Am Phys. 1998;110:531–544. [PubMed] [Google Scholar]
- 17.Pride D T, Meinersmann R J, Blaser M J. Infect Immun. 2001;69:1160–1171. doi: 10.1128/IAI.69.2.1160-1171.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor D E, Eaton M, Chang N, Salama S M. J Bacteriol. 1992;174:6800–6806. doi: 10.1128/jb.174.21.6800-6806.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang Q, Hiratsuka K, Taylor D E. Mol Microbiol. 1996;20:833–842. doi: 10.1111/j.1365-2958.1996.tb02521.x. [DOI] [PubMed] [Google Scholar]
- 20.Alm R A, Ling L S, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noonan B, Guild B C, deJonge B L, et al. Nature (London) 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 21.Blaser M J. In: Helicobacter pylori: Basic Mechanisms to Clinical Cure. Hunt R H, Tytgat G N H, editors. Dordrecht, The Netherlands: Kluwer; 1996. pp. 33–39. [Google Scholar]
- 22.Wang G, Humayun M Z, Taylor D E. Trends Microbiol. 1999;7:488–493. doi: 10.1016/s0966-842x(99)01632-7. [DOI] [PubMed] [Google Scholar]
- 23.Peek R M, Jr, Miller G G, Tham K T, Perez-Perez G I, Cover T L, Atherton J C, Dunn G D, Blaser M J. J Clin Microbiol. 1995;33:28–32. doi: 10.1128/jcm.33.1.28-32.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson K. In: Current Protocols in Molecular Biology. Ausubel F M, Brent R, Kingston R R, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Vol. 1. New York: Wiley; 1995. pp. 2.4.1–2.4.5. [Google Scholar]
- 25.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salama N, Guillemin K, McDaniel T K, Sherlock G, Tompkins L, Falkow S. Proc Natl Acad Sci USA. 2000;97:14668–14673. doi: 10.1073/pnas.97.26.14668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Israel D A, Salama N, Arnold C N, Moss S F, Ando T, Wirth H P, Tham K T, Camorlinga M, Blaser M J, Falkow S, Peek R M., Jr J Clin Invest. 2001;107:611–620. doi: 10.1172/JCI11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eisen M B, Spellman P T, Brown P O, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Versalovic J, Shortridge D, Kibler K, Griffy M V, Beyer J, Flamm R K, Tanaka S K, Graham D Y, Go M F. Antimicrob Agents Chemother. 1996;40:477–480. doi: 10.1128/aac.40.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Debets-Ossenkopp Y J, Sparrius M, Kusters J G, Kolkman J J, Vandenbroucke-Grauls C M. FEMS Microbiol Lett. 1996;142:37–42. doi: 10.1111/j.1574-6968.1996.tb08404.x. [DOI] [PubMed] [Google Scholar]
- 31.Taylor D E, Ge Z, Purych D, Lo T, Hiratsuka K. Antimicrob Agents Chemother. 1997;41:2621–2628. doi: 10.1128/aac.41.12.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, et al. Nature (London) 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 33.Kuipers E J, Israel D A, Kusters J G, Gerrits M M, Weel J, van Der Ende A, van Der Hulst R W, Wirth H P, Hook-Nikanne J, Thompson S A, Blaser M J. J Infect Dis. 2000;181:273–282. doi: 10.1086/315173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kersulyte D, Chalkauskas H, Berg D E. Mol Microbiol. 1999;31:31–43. doi: 10.1046/j.1365-2958.1999.01140.x. [DOI] [PubMed] [Google Scholar]
- 35.Bereswill S, Schonenberger R, Thies C, Stahler F, Strobel S, Pfefferle P, Wille L, Kist M. Med Microbiol Immunol. 2000;189:105–113. doi: 10.1007/s004300000049. [DOI] [PubMed] [Google Scholar]
- 36.Arents N L, Smeets L C, van Zwet A A, Thijs J C, van der Wouden E J, de Jong A, Degener J E, Kusters J G. Eur J Clin Microbiol Infect Dis. 2001;20:418–420. doi: 10.1007/pl00011283. [DOI] [PubMed] [Google Scholar]
- 37.Taylor N S, Fox J G, Akopyants N S, Berg D E, Thompson N, Shames B, Yan L, Fontham E, Janney F, Hunter F M, et al. J Clin Microbiol. 1995;33:918–923. doi: 10.1128/jcm.33.4.918-923.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berg D E, Gilman R H, Lelwala-Guruge J, Srivastava K, Valdez Y, Watanabe J, Miyagi J, Akopyants N S, Ramirez-Ramos A, Yoshiwara T H, et al. Clin Infect Dis. 1997;25:996–1002. doi: 10.1086/516081. [DOI] [PubMed] [Google Scholar]
- 39.Wong B C, Wang W H, Berg D E, Fung F M, Wong K W, Wong W M, Lai K C, Cho C H, Hui W M, Lam S K. Aliment Pharmacol Ther. 2001;15:493–503. doi: 10.1046/j.1365-2036.2001.00949.x. [DOI] [PubMed] [Google Scholar]
- 40.Dore M P, Osato M S, Kwon D H, Graham D Y, el-Zaatari F A. Clin Infect Dis. 1998;27:84–89. doi: 10.1086/514640. [DOI] [PubMed] [Google Scholar]
- 41.Weel J F, van der Hulst R W, Gerrits Y, Tytgat G N, van der Ende A, Dankert J. J Clin Microbiol. 1996;34:2158–2162. doi: 10.1128/jcm.34.9.2158-2162.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jorgensen M, Daskalopoulos G, Warburton V, Mitchell H M, Hazell S L. J Infect Dis. 1996;174:631–635. doi: 10.1093/infdis/174.3.631. [DOI] [PubMed] [Google Scholar]
- 43.van der Ende A, van Doorn L-J, Rooijakkers S, Feller M, Tytgat G N J, Dankert J. J Clin Microbiol. 2001;39:2648–2651. doi: 10.1128/JCM.39.7.2648-2651.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maeda S, Yoshida H, Matsunaga H, Ogura K, Kawamata O, Shiratori Y, Omata M. J Clin Microbiol. 2000;38:210–214. doi: 10.1128/jcm.38.1.210-214.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marais A, Monteiro L, Occhialini A, Pina M, Lamouliatte H, Megraud F. Gut. 1999;44:463–467. doi: 10.1136/gut.44.4.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Doorn L J, Debets-Ossenkopp Y J, Marais A, Sanna R, Megraud F, Kusters J G, Quint W G. Antimicrob Agents Chemother. 1999;43:1779–1782. doi: 10.1128/aac.43.7.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hacker J, Kaper J B. Annu Rev Microbiol. 2000;54:641–679. doi: 10.1146/annurev.micro.54.1.641. [DOI] [PubMed] [Google Scholar]
- 48.Occhialini A, Marais A, Alm R, Garcia F, Sierra R, Megraud F. Infect Immun. 2000;68:6240–6249. doi: 10.1128/iai.68.11.6240-6249.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cabezon E, Sastre J I, de la Cruz F. Mol Gen Genet. 1997;254:400–406. doi: 10.1007/s004380050432. [DOI] [PubMed] [Google Scholar]
- 50.Szpirer C Y, Faelen M, Couturier M. Mol Microbiol. 2000;37:1283–1292. doi: 10.1046/j.1365-2958.2000.02077.x. [DOI] [PubMed] [Google Scholar]
- 51.Christie P J. J Bacteriol. 1997;179:3085–3094. doi: 10.1128/jb.179.10.3085-3094.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smeets L C, Bijlsma J J, Kuipers E J, Vandenbroucke-Grauls C M, Kusters J G. FEMS Immunol Med Microbiol. 2000;27:99–102. doi: 10.1111/j.1574-695X.2000.tb01418.x. [DOI] [PubMed] [Google Scholar]
- 53.Kuipers E J, Israel D A, Kusters J G, Blaser M J. J Bacteriol. 1998;180:2901–2905. doi: 10.1128/jb.180.11.2901-2905.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garcia-Vallve S, Romeu A, Palau J. Genome Res. 2000;10:1719–1725. doi: 10.1101/gr.130000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Covacci A, Rappuoli R. Curr Opin Microbiol. 1998;1:96–102. doi: 10.1016/s1369-5274(98)80148-3. [DOI] [PubMed] [Google Scholar]
- 56.Blaser M J. J Clin Invest. 1997;100:759–762. doi: 10.1172/JCI119588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blaser M J, Berg D E. J Clin Invest. 2001;107:767–773. doi: 10.1172/JCI12672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hacker J, Carniel E. EMBO Rep. 2001;2:376–381. doi: 10.1093/embo-reports/kve097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morschhauser J, Kohler G, Ziebuhr W, Blum-Oehler G, Dobrindt U, Hacker J. Philos Trans R Soc London B. 2000;355:695–704. doi: 10.1098/rstb.2000.0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boucher Y, Nesbo C L, Doolittle W F. Curr Opin Microbiol. 2001;4:285–289. doi: 10.1016/s1369-5274(00)00204-6. [DOI] [PubMed] [Google Scholar]
- 61.Lan R, Reeves P R. Trends Microbiol. 2000;8:396–401. doi: 10.1016/s0966-842x(00)01791-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.