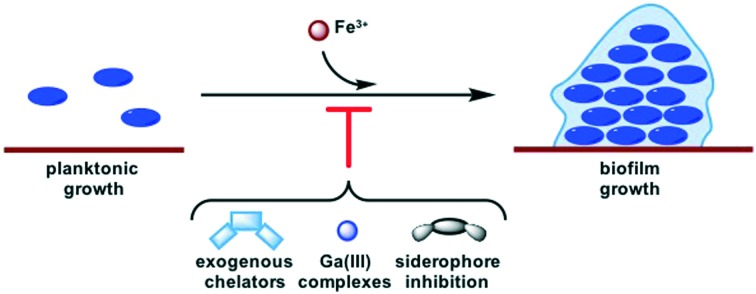
 Biofilms are linked to resistance development in the ESKAPE pathogens. This perspective summarizes several strategies for affecting iron homeostasis that have been implicated in biofilm inhibition.
Biofilms are linked to resistance development in the ESKAPE pathogens. This perspective summarizes several strategies for affecting iron homeostasis that have been implicated in biofilm inhibition.
Abstract
The rise of antibiotic resistant bacteria has become a problem of global concern. Of particular interest are the ESKAPE pathogens, species with high rates of multi-drug resistant infections. Novel antibiotic mechanisms of action are necessary to compliment traditional therapeutics. Recent research has focused on targeting virulence factors as a method of combatting infection without creating selective pressure for resistance or damaging the host commensal microbiome. Some investigations into one such virulence behavior, iron acquisition, have displayed additional effects on another virulence behavior, biofilm formation. The use of exogenous iron-chelators, gallium as an iron mimic, and inhibition of siderophore-mediated iron acquisition are all strategies for disturbing iron-homeostasis that have implicated effects on biofilms. However, the exact nature of this connection remains ambiguous. Herein we summarize these findings and identify opportunities for further investigation.
Introduction
Since the initial discovery of penicillin in 1928,1 antibiotics have made a significant impact in the healthcare2 and agriculture3 industries. However, widespread use has accelerated the spread of antibiotic resistance4–6 through the rapid evolutionary selection for antibiotic-resistance genes.7 The ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) have become especially threatening. Together, these species represent the most common cause of nosocomial infections and are especially prevalent in immunocompromised patients.8,9 These organisms have a high incidence of multidrug resistant (MDR) strains9–14 and utilize virulence behaviors such as the formation of biofilms15–17 and the expression of nutrient acquisition systems,18–20 and have thus been the focus of a growing body of research.21
Despite the surge of antibiotic resistance, more and more pharmaceutical companies are shutting down their antibiotic development divisions due to the high cost and low success rate of bringing a drug to market.22 This has resulted in fewer resources devoted to research on novel therapeutics, while bacterial resistance continues to rise at an alarming rate.23 This has caused speculation of a post-antibiotic era,24 in which the ability to treat bacterial infections would mirror that of the pre-antibiotic era when a bacterial infection was often a death sentence.2 This would negate decades of progress in modern medicine, which has inspired the search for new strategies to treat infection that are less prone to resistance development.
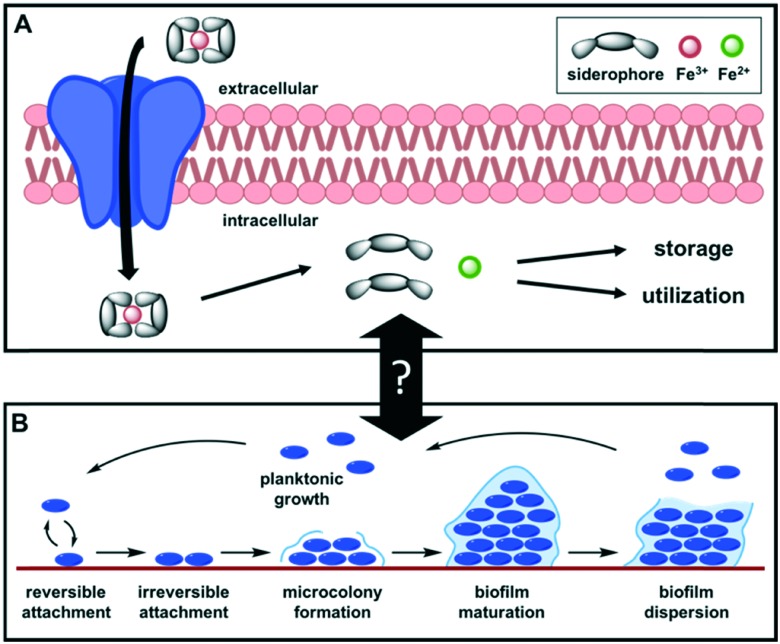
One potential strategy to circumvent resistance is to target bacterial virulence behaviors, which refer to an organism's ability to establish infection. This strategy does not target vital life processes of the bacteria, which results in less selective pressure for resistant mutations.25 Additionally, this would cause less damage to the host commensal microbiome, which is implicated in every realm of human health.26–28 One such virulence behavior is iron acquisition. Iron is a necessity for nearly all living systems, and iron limitation has been shown to reduce acute infection in numerous bacterial species.29,30 In aerobic conditions, iron primarily exists as Fe3+, which is poorly soluble in aqueous environments. Additionally, animals have evolved systems to sequester iron away from bacteria, providing a form of innate immunity to infection. To gain a competitive advantage, pathogens have evolved methods for obtaining this micronutrient in iron-deficient conditions. One of the most crucial strategies is the production of siderophores, a diverse array of iron-binding small molecules (270 structurally characterized)31 produced by bacteria,32 plants,33 and fungi.34 Siderophores are biosynthesized in the bacterial cell, excreted into the extracellular space, and form high affinity iron(iii) chelate complexes. These complexes are recognized by receptors on the bacterial surface and actively transported into the cell where the iron is extracted (either by reduction from Fe3+ to Fe2+ or chemical degradation of the siderophore) to be used or stored (Fig. 1A). Beyond their iron-affinity, alternative roles for siderophore-like molecules include cell signaling, detoxification, oxidative-stress response, and antibacterial activity.32 Due to the imminent and growing threat of MDR pathogens, the antibiotic potential of this class of molecules is of immediate and broad impact.
Fig. 1. (A) Overview of iron acquisition by siderophores. (B) Steps in biofilm formation.
Through investigations into siderophores and other iron-chelating molecules, additional effects on biofilms have been observed. The formation of biofilms is a virulence behavior in which an extracellular polymeric matrix is excreted by the bacteria to form a three-dimensional microbial community (Fig. 1B). Biofilm cells enter a lower metabolic state35 and adhere to surfaces to form protective barriers. Together, these mechanisms make antibiotics and host immune defenses less effective,16 and are thus a major contributor to resistance. As such, investigations of anti-biofilm compounds have garnered wide interest. The evidence supporting a link between iron acquisition and biofilm formation has been rudimentary, and the exact details of this connection are unclear. In some pathogens it is well-documented, but in many cases the literature is sparse and contradictory. Herein we highlight some of the major findings that explore the relationship between iron depletion and biofilm formation in the ESKAPE pathogens and define the areas that require further investigation.
Exogenous Iron chelators
Exogenous iron chelators have been studied as therapeutics for many years,36–39 but experiments looking at the effect of iron depravation on biofilm formation did not begin until the early 2000s. In their initial report, Singh and coworkers discovered that lactoferrin, a human iron transport protein, hindered biofilm formation in Gram-negative P. aeruginosa at concentrations below those required to inhibit planktonic growth.40 Chelation of iron by lactoferrin induced the cellular twitching response, which prevented the planktonic cells from attaching to surfaces (the first step of biofilm formation) (Fig. 2A). By contrast, an earlier report found that P. aeruginosa was able to utilize iron from transferrin and lactoferrin for planktonic growth in an iron-poor environment.41 These findings emphasize that processes for iron utilization have key differences in planktonic growth and biofilm formation.
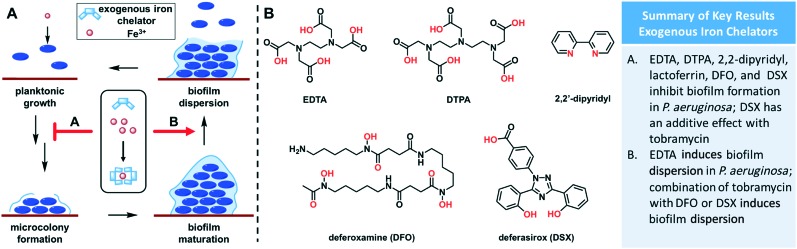
Fig. 2. (A) Effect of exogenous iron chelators on biofilm formation. Some compounds inhibit formation of biofilms, while others induce biofilm dispersion. (B) Structures of relevant exogenous iron chelators. Chelating atoms are highlighted in red.
In another investigation, Banin and coworkers determined that known iron-chelator EDTA (Fig. 2B) was capable of disrupting and killing mature biofilm cells in P. aeruginosa.42 EDTA was 1000-fold more effective at killing P. aeruginosa biofilm cells than gentamycin, and when administered together complete biofilm eradication was achieved. Another study observed strong inhibition of biofilm formation in P. aeruginosa by EDTA at 0.1 mM, but this only occurred during the early stages of biofilm formation.43 If EDTA was administered after 72 hours of growth, no inhibition was observed. Additionally, they found that saturation of EDTA with cations did not attenuate biofilm inhibition and that EDTA was able to inhibit cell-to-surface and cell-to-cell interactions. Together, these data suggest that EDTA inhibits formation of biofilms by blocking initial surface adhesion, rather than by simply sequestering iron. Other organoferric complexes have also been shown to inhibit biofilm formation,44,45 including the human iron-transport protein transferrin.46 Following these investigations, work from several groups determined that a number of synthetic iron chelators are able to inhibit biofilm formation in P. aeruginosa to various extents. Two FDA-approved drugs, deferoxamine (DFO) and deferasirox (DSX) (Fig. 2B), were able to reduce biofilm formation in cystic fibrosis (CF) cells by 49% and 99%, respectively.47 Additionally, DSX had an additive effect with tobramycin in the inhibition of biofilm formation. Furthermore, combination of tobramycin with DFO or DSX reduced established biofilms by 90%. A follow up investigation found that 2,2′-dipyridyl and DTPA (Fig. 2B) were effective at preventing biofilm formation in CF isolates, but it was highly strain dependent, and 2,2′-dipyridyl could disrupt mature biofilms.48 These results provide a basis for further investigation of exogenous iron-chelators as a novel therapeutic strategy.
Studies have shown that several known iron chelators inhibit planktonic growth of different E. faecium, Enterococcus faecalis,49 and A. baumannii strains to various extents50 in both iron-rich and iron-poor conditions. Additionally, several compounds being studied in pharmaceutical companies for other ailments were found to inhibit planktonic growth of A. baumannii in iron-poor media.51 However, a more recent report contradicted these claims, stating that planktonic and biofilm growth of A. baumannii were minimally affected by exogenous iron chelators.52 They reported that the sensitivity to iron levels is highly dependent on strain, as was the innate ability to grow biofilms. They also observed high variability in growth based on media composition. They conclude that rigorous standards should be set when conducting biofilm growth assays, to allow for reproducibility. These data, while inconsistent, provide a starting point for future investigations into the effect of exogenous iron chelators on biofilm formation in the other ESKAPE pathogens.
An innate limitation of using exogenous iron chelators to disrupt bacterial functions is that different species are able to sequester iron from different sources. Some studies have shown secondary infections after treatment with iron chelators, resulting from a new species that is able to utilize iron from this complex gaining a competitive advantage over the initial pathogen.53 As such, choice of a chelator with activity against multiple pathogens is critical. Alternatively, multiple iron chelators that are structurally diverse could be co-administered, in an effort to mitigate the effects of a broad range of species. Overall, initial results show some promise as a strategy for combatting MDR infections, and the repurposing of previously approved drugs provides a streamlined approach to obtaining a viable therapeutic, although further investigation is required.
Gallium as an iron mimic
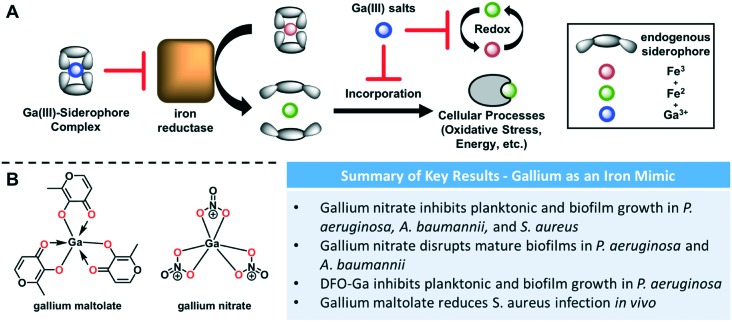
Another strategy for disrupting iron homeostasis has been the use of gallium as a redox-stable iron mimic. Gallium(iii) can compete with iron(iii) in many binding interactions due to its similar atomic radius and valence.54 However, because the gallium(ii) oxidation state has never been observed, it does not have the redox properties required for the biological activity of iron and can thus inhibit iron-dependent enzymes. For example, an iron(iii) reductase responsible for extraction of the metal from a siderophore-chelate would be ineffective on a gallium-bound complex (Fig. 3A). Furthermore, iron-dependent enzymes in critical life-sustaining processes such as respiration require the redox-cycling between iron(ii) and iron(iii) and are thus potentially susceptible to gallium inhibition (Fig. 3A). Kaneko and coworkers found that gallium nitrate (Fig. 3B), which is already FDA-approved for the treatment of hypercalcemia in cancer patients,55 was effective against P. aeruginosa and could be tuned for either planktonic or biofilm inhibition depending on concentration.56 Low concentrations prevented formation of biofilms, while high concentrations inhibited planktonic growth and eradicated existing biofilms. They also found that gallium was effective in treating MDR isolates from CF lungs, as well as lung infection mouse models. These data provide support for the versatility of this approach. Not long after, Banin et al. described the use of a desferrioxamine–gallium complex to inhibit planktonic and biofilm growth.57P. aeruginosa is believed to have two uptake systems for desferrioxamine–iron complexes,58 and thus this gallium complex serves as a mimic to deliver gallium into the cell through a “Trojan horse” mechanism.
Fig. 3. (A) Effect of gallium treatments on iron utilization. Gallium-siderophore complexes inhibit iron-reductase. Gallium salts inhibit other iron-dependent redox processes. (B) Structures of relevant gallium and iron complexes. Chelating atoms are highlighted in red.
A growing body of work using gallium as a treatment for A. baumannii has focused on planktonic growth. Gallium nitrate was able to inhibit 90% of A. baumannii growth at 3.1 μM in serum,50 but was minimally effective in chemically defined iron-rich or iron-poor media. They hypothesize that this is because extracellular transferrin, a human iron-transport protein, can act synergistically with gallium. In a murine model there was a 100-fold reduction in CFU in mice treated with gallium nitrate 48 hours post-infection with A. baumannii compared to placebo. Antunes and coworkers studied the effects of gallium nitrate on 58 A. baumannii strains (mostly MDR clinical isolates)59 and found that growth was inhibited in a dose- and time-dependent manner in iron-poor media and human serum. Finally, they performed a checkerboard assay which displayed synergy between gallium nitrate and colistin across a variety of strains. Colistin is an antibiotic often used as a last line of defense against MDR bacteria due to its nephrotoxicity. For this reason, a combination therapy to reduce doses (and thus side effects) is an appealing strategy. Recently these experiments have been expanded to include effects on biofilm growth, with a report of planktonic and biofilm inhibition with 16 μM gallium nitrate in human serum, and disruption of existing biofilms at 64 μM.60 These preliminary results, along with the success of gallium treatments for biofilm inhibition in other bacteria, provide a basis for further investigations in A. baumannii.
Gallium-based therapies have also been studied in S. aureus. One report showed that gallium nitrate was able to effectively inhibit both planktonic and biofilm growth.61 Importantly, it was effective against stationary phase cells, which are often at the center of biofilms and can be the most difficult to eradicate.56 Gallium maltolate (Fig. 3B), which was shown to have higher efficacy than gallium nitrate against P. aeruginosa, was able to reduce colonization of S. aureus in the wounds of infected mice.62 There has been little investigation into this type of treatment in the other ESKAPE pathogens, but these results show the potential of gallium as a therapeutic approach. Similar to exogenous iron chelators, repurposing FDA-approved drugs for this strategy would also serve to expedite the process of bringing a new antimicrobial to market.
While the use of gallium to interfere with iron-dependent processes in bacteria has been widespread, there are some challenges to consider. Iron is crucial for many human processes, so some cytotoxicity has been observed with gallium treatments in the past.55 Despite this, gallium therapies have already been FDA-approved because these side effects are generally minimal, and the benefits outweigh the costs. Another obstacle is that gallium salts are not very soluble in the gut, reducing their bioavailability.63 However, the use of siderophore-gallium “Trojan-horse” strategies serve to overcome this limitation and deliver the gallium into cells. The use of gallium is especially attractive in light of results showing synergism between gallium and known antibiotics.59 This provides the added benefit of lowering required doses for the antibiotics, which is especially important for many of the antibiotics with poor therapeutic indexes used as a last line of defense against MDR infections. Additionally, the use of gallium is less likely to develop resistance, because any change resulting in decreased binding or uptake of gallium would also reduce uptake of iron that is crucial to the survival of the bacteria.64,65 Even with these limitations, gallium therapeutics have been effective at inhibiting biofilms in many species, and remain a viable option for further investigation of novel antimicrobials.
Siderophore-mediated iron acquisition
Rather than targeting iron directly, an alternative approach is to inhibit bacterial methods of iron acquisition or utilization. Potential sites for inhibition that have been studied in various bacteria include siderophore biosynthesis,66 outer membrane receptors,67 iron release enzymes,68 and signaling receptors69 (Fig. 4A). Compared to the use of exogenous iron chelators and gallium complexes, targeting siderophore-mediated processes directly has been less studied. The data presented herein are more preliminary and very few compounds are known to target these processes directly. However, these results demonstrate the potential for future investigations using small molecule inhibitors.
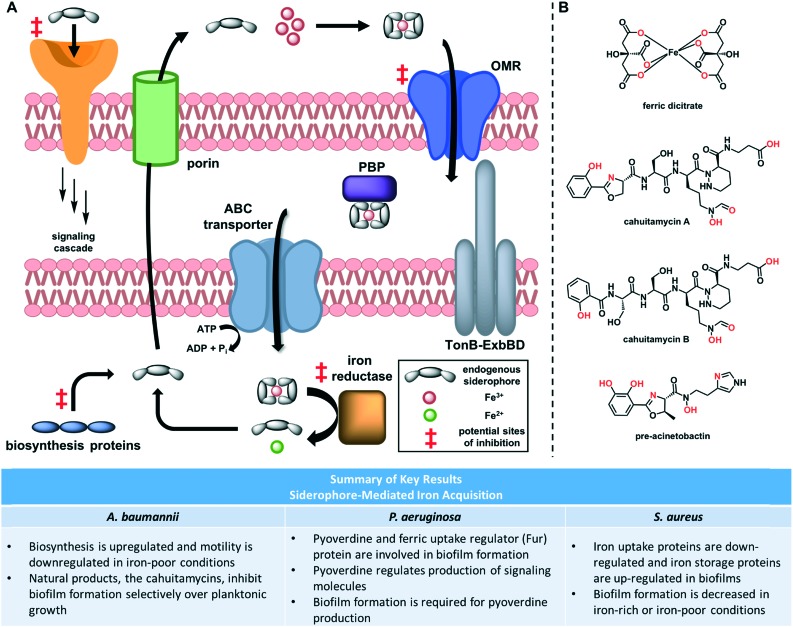
Fig. 4. (A) Overview of siderophore-mediated iron acquisition. Following biosynthesis, siderophores are exported through porins. They bind iron in the extracellular space and these complexes are taken into the periplasm through outer membrane receptors (OMRs). Periplasmic binding proteins (PBPs) transfer the complex to ATP-binding cassette (ABC) transporters, which bring them into the cytoplasm. Iron reductase catalyses the reduction of iron(iii) to iron(ii), which induces release by the siderophore. (B) Structures of siderophores and siderophore mimics. Chelating atoms are highlighted in red.
Siderophore biosynthesis is perhaps the most studied step in this process. Brown and coworkers performed transcriptomic analysis and unsurprisingly found that three A. baumannii siderophore biosynthesis gene clusters were significantly upregulated under iron-poor conditions.70 However, many additional genes involved in motility were down-regulated, and this decrease in motility was confirmed through phenotypic assays. Of particular interest among the down-regulated genes were those that encode for type I and type IV pili. Homologous structures have been associated with motility and biofilm formation in other species, including E. coli and P. aeruginosa.71–73 These results hint that both siderophore production and motility-related biofilm formation in A. baumannii are mediated by iron levels, but the details of this connection and how it translates to a phenotypic relationship between iron and biofilm formation necessitates further study.
A more recent study conducted proteomic analysis of both S. aureus planktonic and biofilm cells.74 The results indicate that biofilms contain a decreased amount of iron-uptake proteins and an increased amount of the iron-storage protein FtnA, which suggests that biofilms may require lower levels of iron. This is not surprising, as iron is required for many cellular processes involved in replication and growth,45 and biofilm cells often down-regulate metabolism. This gives further support to the transcriptomic results in A. baumannii, suggesting that low iron levels may trigger the cells to form biofilms and enter this lower metabolic state. In a highly virulent strain of Gram-positive S. aureus, iron-poor conditions induce biofilm formation.75 Subsequently, two virulence determinants for this strain were identified, which allow the formation of biofilms in iron-poor conditions.76 These findings suggests that iron-regulated biofilm formation may be highly strain-dependent. Additional studies looking at the formation of S. aureus biofilms found that they form more readily in plasma with normal iron levels, compared to plasma that is iron-deficient or rich in iron.77 This may suggest a “goldilocks” relationship between iron and biofilms, in which too much iron does not signal for biofilm formation, too little iron does not allow cell growth, but at just the right iron concentration biofilm formation is stimulated.
In P. aeruginosa, the link between iron utilization systems and biofilm formation has been studied more directly, with a minireview on the topic published in 2018.78 Meyer and coworkers confirmed that pyoverdine, the endogenous siderophore in P. aeruginosa, is a virulence factor.79 These results were observed in vitro and in vivo and have been validated by in vivo follow-up studies.80,81 All of these data matched previous reports that pyoverdine, but not pyochelin (another P. aeruginosa siderophore), is required for growth in human serum.82 Building on this foundation, Banin and coworkers found that a genetic mutant unable to produce pyoverdine, while exhibiting planktonic growth similar to that of the parent, formed only thin, uniform biofilms under iron-poor conditions, in contrast to the mushroom-like shape that is typical of P. aeruginosa biofilms grown under flow conditions.58 Furthermore, supplementation of DFO or ferric dicitrate (Fig. 4B) restores the mushroom-like shape of biofilms, indicating the bacteria was able to utilize iron from these complexes. Subsequently, they identified three possible gene clusters that may be responsible for the cellular uptake of these exogenous iron-chelators. Finally, they provided preliminary data showing that the ferric uptake regulator (Fur) protein plays a role in biofilm formation, but additional work is required to elucidate the mechanism and validate the results in vivo.
Another key process involved in virulence and biofilm formation in which siderophores have been implicated is intercellular signaling. Lamont et al. showed that pyoverdine regulates its own production, as well as the production of exotoxin A and PrpL protease, all of which are virulence factors in P. aeruginosa.83 Subsequent identification of pyoverdine outer-membrane receptors in P. aeruginosa,84 followed by the discovery of a bacteriocin that kills P. aeruginosa by selectively targeting these receptors,85,86 provide a basis for future investigation of novel small molecules that target this uptake. In a recent high-throughput screen for pyoverdine biosynthesis genes, Kang and Kirienko report that early biofilm formation is required for complete pyoverdine biosynthesis.87 This suggests that siderophore production may signal biofilm formation. Further investigation is necessary to determine if one of these virulence traits has a direct, causative effect on the other.
Only preliminary results have been reported on the relationship between siderophores and virulence in the other ESKAPE pathogens. Yersiniabactin, an endogenous siderophore of K. pneumoniae, was confirmed as a virulence factor in vivo.88 Additionally, a hypervirulent strain of K. pneumoniae produced more siderophores than the wild type.89 Another group screened 55 E. faecium and E. faecalis strains for antimicrobial resistance and siderophore production.90 They found a direct correlation between siderophore production and fluoroquinolone resistance, and found that siderophores were present in large quantities in ciprofloxacin- and norfloxacin-resistant strains. Thus, their results provided initial evidence that siderophore production may contribute to virulence in Enterococcus species and aid in their resistance to various antibiotics. Additional studies should be executed to investigate whether this virulence is directly related to biofilm formation. Overall, targeting the synthesis siderophores will have a number of effects on the pathogenicity of the bacteria, and represents a viable strategy to disarm them.
A recent report disclosed a group of newly discovered natural products called the cahuitamycins (Fig. 4B), which show promise as biofilm inhibitors.91 Sherman and coworkers show quantitative and qualitative inhibition of A. baumannii biofilm formation when grown in the presence of these molecules, without any inhibition of planktonic growth. Furthermore, these molecules show moderate to strong iron chelation and possess canonical hydroxamate and phenolate–oxazoline moieties typically found in siderophores. The exception is cahuitamycin B (Fig. 4B), which contains no oxazoline and does not inhibit biofilm growth. However, when biofilms were grown in the presence of DFO no inhibition was observed. Together, these results suggest that the mechanism of these molecules is more complex than merely sequestering iron. Pre-acinetobactin (Fig. 4B), an endogenous siderophore of A. baumannii, also possesses a phenolate–oxazoline moiety. It is thus plausible that the cahuitamycins are recognized by acinetobactin uptake proteins. However, it remains unclear why the bacteria would be unable to utilize the iron once the complex is brought inside the cell. Nevertheless, these molecules are a launching point to probe the relationship between iron chelation and biofilm formation in A. baumannii and provide rationale for using small molecules to disrupt these virulence pathways.
Targeting siderophore pathways has enormous potential as a therapeutic due to the substantial evidence that they play a role in biofilm formation. However, to date there are very few small molecules that are known to target a specific protein in this pathway. One possible explanation is that some species may have redundancy in this area. For example, P. aeruginosa has over 30 genes that encode for iron receptors, although there is homology between many of them.92 Thus, if this strategy is to be utilized, it is vital that the approach be broad enough to target multiple receptors to avoid simple upregulation of alternative iron import pathways. The use of small molecules that mimic endogenous siderophores has shown potential in this area, as they utilize the innate machinery of the bacteria to gain entrance into the cell. Overall, these results provide evidence that inhibition of siderophore pathways may be a viable strategy to inhibit biofilm formation as an alternative to traditional therapeutic strategies.
Conclusions
One problem in the field of biofilm inhibition is consistency and reproducibility. Future investigations must carefully track the effects of iron depletion on planktonic growth, biofilm formation, or eradication of mature biofilms. Standardized experimental conditions could help ease this burden, but implementation across many different research areas/groups can be challenging. It is crucial to understand what part of the process is being affected in order to accurately target it as a viable therapeutic. Additionally, due to the strain-dependent nature of this phenomenon, it is increasingly important to use clinical isolates in laboratory experiments, especially those that exhibit MDR phenotypes.
While the outlook may seem grim due to the continued emergence of new MDR strains, there are many viable strategies discussed herein that have been underutilized and deserve increased attention. The success of such strategies has been mostly limited to S. aureus, A. baumannii, and P. aeruginosa, with very little investigation into the other ESKAPE pathogens. Thus, the investigation of these strategies in E. faecium, K. pneumoniae, and Enterobacter species offers a plethora of information yet to be discovered. Overall, the connection between iron acquisition and biofilm formation should be investigated further as it holds great therapeutic potential. In response to the growing threat posed by the ESKAPE pathogens, novel therapeutic strategies are sorely needed, and a fundamental understanding of these biochemical processes is critical to the future development of therapeutics.
Conflicts of interest
The authors declare no conflicts.
Acknowledgments
We gratefully acknowledge funding from the National Institute of General Medical Sciences (GM119426) and the National Science Foundation (CHE1755698). We would also like to thank Dr. William Shafer, Dr. Taylor Hari, Amy Solinski, and Dr. Colleen Keohane for their helpful feedback.
References
- Sengupta S., Chattopadhyay M. K., Grossart H. Front. Microbiol. 2013;4:1–13. doi: 10.3389/fmicb.2013.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aminov R. I. Front. Microbiol. 2010;1:1–7. doi: 10.3389/fmicb.2010.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael C. A., Dominey-howes D., Labbate M., Maria C., Elisabeth J. Front. Public Health. 2014;2:1–8. doi: 10.3389/fpubh.2014.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright G. D. Curr. Opin. Microbiol. 2010;13:589–594. doi: 10.1016/j.mib.2010.08.005. [DOI] [PubMed] [Google Scholar]
- Nesme J., Cecillon S., Delmont T. O., Monier J., Vogel T. M., Simonet P. Curr. Biol. 2014;24:1096–1100. doi: 10.1016/j.cub.2014.03.036. [DOI] [PubMed] [Google Scholar]
- Lipsitch M., Samore M. H. Emerging Infect. Dis. 2002;8:347–354. doi: 10.3201/eid0804.010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin B. R., Bergstrom C. T. Proc. Natl. Acad. Sci. U. S. A. 2000;97:6981–6985. doi: 10.1073/pnas.97.13.6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fick R. B., Pseudomonas aeruginosa, the opportunist: pathogenesis and disease, 1993. [Google Scholar]
- Rice L. B. Infect. Control Hosp. Epidemiol. 2010;31:7–10. doi: 10.1086/655995. [DOI] [PubMed] [Google Scholar]
- Moskowitz S. M., Emerson J. C., McNamara S., Shell R. D., Orenstein D. M., Rosenbluth D., Katz M. F., Ahrens R., Hornick D., Joseph P. M., Gibson R. L., Aitken M. L., Benton W. W., Burns J. L. Pediatr. Pulmonol. 2011;46:184–192. doi: 10.1002/ppul.21350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskowitz S. M., Foster J. M., Emerson J. C., Gibson R. L., Burns J. L. J. Antimicrob. Chemother. 2005;56:879–886. doi: 10.1093/jac/dki338. [DOI] [PubMed] [Google Scholar]
- Lewis K. Curr. Top. Microbiol. Immunol. 2008;322:107–131. doi: 10.1007/978-3-540-75418-3_6. [DOI] [PubMed] [Google Scholar]
- Gould I. M. Int. J. Antimicrob. Agents. 2013;42:17–21. doi: 10.1016/j.ijantimicag.2013.04.006. [DOI] [PubMed] [Google Scholar]
- Gardete S., Tomasz A. J. Clin. Invest. 2014;124:2836–2840. doi: 10.1172/JCI68834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkshoorn L., Nemec A., Seifert H. Nat. Rev. Microbiol. 2007;5:939–951. doi: 10.1038/nrmicro1789. [DOI] [PubMed] [Google Scholar]
- Parsek M. R., Singh P. K. Annu. Rev. Microbiol. 2003;57:677–701. doi: 10.1146/annurev.micro.57.030502.090720. [DOI] [PubMed] [Google Scholar]
- Götz F. Mol. Microbiol. 2002;43:1367–1378. doi: 10.1046/j.1365-2958.2002.02827.x. [DOI] [PubMed] [Google Scholar]
- Minandri F., Imperi F., Frangipani E., Bonchi C., Visaggio D., Facchini M., Pasquali P., Bragonzi A., Visca P. Infect. Immun. 2016;84:2324–2335. doi: 10.1128/IAI.00098-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caza M., Kronstad J. W. Front. Cell. Infect. Microbiol. 2013;3:1–23. doi: 10.3389/fcimb.2013.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer N. D., Skaar E. P. Annu. Rev. Microbiol. 2011;65:129–147. doi: 10.1146/annurev-micro-090110-102851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendleton J., Gorman S., Gilmore B. Expert Rev. Anti-infect. Ther. 2013;11:297–308. doi: 10.1586/eri.13.12. [DOI] [PubMed] [Google Scholar]
- DiMasi J. A., Hansen R. W., Grabowski H. G. J. Health Econ. 2002;22:151–185. doi: 10.1016/S0167-6296(02)00126-1. [DOI] [PubMed] [Google Scholar]
- Spellberg B., Guidos R., Gilbert D., Bradley J., Boucher H. W., Scheld W. M., Bartlett J. G., Edwards J., Diseases I. Clin. Infect. Dis. 2008;46:155–164. doi: 10.1086/524891. [DOI] [PubMed] [Google Scholar]
- Bragg R. R., Meyburgh C. M., Lee J. Y., Coetzee M. Adv. Exp. Med. Biol. 2018;1052:51–61. doi: 10.1007/978-981-10-7572-8_5. [DOI] [PubMed] [Google Scholar]
- Clatworthy A. E., Pierson E., Hung D. T. Nat. Chem. Biol. 2007;3:541–548. doi: 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- Yatsunenko T., Rey F. E., Manary M. J., Trehan I., Dominguez-Bello M. G., Contreras M., Magris M., Hidalgo G., Baldassano R. N., Anokhin A. P., Heath A. C., Warner B., Reeder J., Kuczynski J., Caporaso J. G., Lozupone C. A., Lauber C., Clemente J. C., Knights D., Knight R., Gordon J. I. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R. E., Turnbaugh P. J., Klein S., Gordon J. I. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- Kau A. L., Ahern P. P., Griffin N. W., Goodman A. L., Gordon J. I. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullen J. J., Rogers H. J., Spalding P. B., Ward C. G. FEMS Immunol. Med. Microbiol. 2005;43:325–330. doi: 10.1016/j.femsim.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Griffiths E., in Iron and infection: molecular, physiological and clinical aspects, ed. D. J. Bullen and E. Griffiths, John Wiley & Sons Ltd, Chichester, 2nd edn, 1999, pp. 1–26. [Google Scholar]
- Hider R. C., Kong X. Nat. Prod. Rep. 2010;27:637–657. doi: 10.1039/b906679a. [DOI] [PubMed] [Google Scholar]
- Johnstone T. C., Nolan E. M. Dalton Trans. 2015;44:6320–6339. doi: 10.1039/c4dt03559c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloepper J. W., Leong J., Teintze M., Schroth M. N. Nature. 1980;286:885–886. [Google Scholar]
- Haas H., Eisendle M., Turgeon B. G. Annu. Rev. Phytopathol. 2008;46:149–187. doi: 10.1146/annurev.phyto.45.062806.094338. [DOI] [PubMed] [Google Scholar]
- Singh S., Singh S. K., Chowdhury I., Singh R. Open Microbiol. J. 2017;11:53–62. doi: 10.2174/1874285801711010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler P. Diabetes. 1989;38:1207–1210. doi: 10.2337/diab.38.10.1207. [DOI] [PubMed] [Google Scholar]
- Bergeron R., Wiegand J., Dionis J., Al E. J. Med. Chem. 1991;34:2072–2078. doi: 10.1021/jm00111a023. [DOI] [PubMed] [Google Scholar]
- Buss J., Greene B., Turner J., Torti F., Torti S. Curr. Top. Med. Chem. 2004;4:1623–1635. doi: 10.2174/1568026043387269. [DOI] [PubMed] [Google Scholar]
- Borgna-Pignatti C., Cappellini M., De Stefano P., Del Vecchio G., Forni G., Gamberini M., Ghilardi R., Piga A., Romeo M., Zhao H., Cnaan A. Blood. 2006;107:3733–3737. doi: 10.1182/blood-2005-07-2933. [DOI] [PubMed] [Google Scholar]
- Singh P. K., Parsek M. R., Greenberg E. P., Welsh M. J. Nature. 2002;417:552. doi: 10.1038/417552a. [DOI] [PubMed] [Google Scholar]
- Xiao R., Kisaalita W. S. Microbiology. 1997;143:2509–2515. [Google Scholar]
- Banin E., Brady K. M., Greenberg E. P. Appl. Environ. Microbiol. 2006;72:2064–2069. doi: 10.1128/AEM.72.3.2064-2069.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L., Cai W., Qu H., Liu J., Lan J., Lu J., Lan T., Li J. Food Sci. Technol. Res. 2013;19:323–330. [Google Scholar]
- Badea M., Crasanda A. M., Chifiriuc M. C., Marutescu L., Lazar V., Marinescu D., Olar R. J. Therm. Anal. Calorim. 2013;111:1743–1751. [Google Scholar]
- Lin M. H., Shu J. C., Huang H. Y., Cheng Y. C. PLoS One. 2012;7:1–7. [Google Scholar]
- Ardehali R., Shi L., Janatova J., Mohammad F., Burns G. L. Artif. Organs. 2002;26:512–520. doi: 10.1046/j.1525-1594.2002.06923.x. [DOI] [PubMed] [Google Scholar]
- Moreau-Marquis S., O'Toole G. A., Stanton B. A. Am. J. Respir. Cell Mol. Biol. 2009;41:305–313. doi: 10.1165/rcmb.2008-0299OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'May C. Y., Sanderson K., Roddam L. F., Kirov S. M., Reid D. W. J. Med. Microbiol. 2009;58:765–773. doi: 10.1099/jmm.0.004416-0. [DOI] [PubMed] [Google Scholar]
- Lisiecki P. Med. Dosw. Mikrobiol. 2010;62:271–280. [PubMed] [Google Scholar]
- De Léséleuc L., Harris G., KuoLee R., Chen W. Antimicrob. Agents Chemother. 2012;56:5397–5400. doi: 10.1128/AAC.00778-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M. G., Corey B. W., Si Y., Craft D. W., Zurawski D. V. Antimicrob. Agents Chemother. 2012;56:5419–5421. doi: 10.1128/AAC.01197-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentile V., Frangipani E., Bonchi C., Minandri F., Runci F., Visca P. Pathogens. 2014;3:704–719. doi: 10.3390/pathogens3030704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windus D. W., Stokes T. J., Julian B. A., Fenves A. Z. Ann. Intern. Med. 1987;107:678–680. doi: 10.7326/0003-4819-107-5-678. [DOI] [PubMed] [Google Scholar]
- García-Quintanilla M., Pulido M. R., López-Rojas R., Pachón J., McConnell M. J. Trends Microbiol. 2013;21:157–163. doi: 10.1016/j.tim.2012.12.002. [DOI] [PubMed] [Google Scholar]
- Chitambar C. R. Int. J. Environ. Res. Public Health. 2010;7:2337–2361. doi: 10.3390/ijerph7052337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko Y., Thoendel M., Olakanmi O., Britigan B. E., Singh P. K. J. Clin. Invest. 2007;117:877–888. doi: 10.1172/JCI30783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banin E., Lozinski A., Brady K. M., Berenshtein E., Butterfield P. W., Moshe M., Chevion M., Greenberg E. P., Banin E. Proc. Natl. Acad. Sci. U. S. A. 2008;105:16761–16766. doi: 10.1073/pnas.0808608105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banin E., Vasil M. L., Greenberg E. P. Proc. Natl. Acad. Sci. U. S. A. 2005;102:11076–11081. doi: 10.1073/pnas.0504266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes L. C. S., Imperi F., Minandri F., Visca P. Antimicrob. Agents Chemother. 2012;56:5961–5970. doi: 10.1128/AAC.01519-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runci F., Bonchi C., Frangipani E., Visaggio D., Visca P. Antimicrob. Agents Chemother. 2017;61:1–8. doi: 10.1128/AAC.01563-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia R. A., Tennent D. J., Chang D., Wenke J. C., Sanchez C. J. Biomed Res. Int. 2016:1–11. doi: 10.1155/2016/7078989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLeon K., Balldin F., Watters C., Hamood A., Griswold J., Sreedharan S., Rumbaugh K. P. Antimicrob. Agents Chemother. 2009;3:1331–1337. doi: 10.1128/AAC.01330-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein L. R. Pharmacol. Rev. 1998;50:665–682. [PubMed] [Google Scholar]
- Alvarez-Ortega C., Wiegand I., Olivares J., Hancock R. E. W., Martínez J. L. Virulence. 2011;2:144–146. doi: 10.4161/viru.2.2.15014. [DOI] [PubMed] [Google Scholar]
- Costa S. S., Viveiros M., Amaral L., Couto I. Open Microbiol. J. 2013;7:59–71. doi: 10.2174/1874285801307010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb A. L. Biochim. Biophys. Acta. 2015;1854:1054–1070. doi: 10.1016/j.bbapap.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleuel C., Grosse C., Taudte N., Scherer J., Wesenberg D., Krauss G. J., Nies D. H., Grass G. J. Bacteriol. 2005;187:6701–6707. doi: 10.1128/JB.187.19.6701-6707.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinella D., Albrecht C., Cashel M., D'Ari R. Mol. Microbiol. 2005;56:958–970. doi: 10.1111/j.1365-2958.2005.04601.x. [DOI] [PubMed] [Google Scholar]
- Theriault J., Wurst J., Jewett I., Verplank L., Perez J., Gulick A., Drake E., Palmer M., Moskowitz S., Dasgupta N., Brannon M., Dandapani S., Munoz B. and Schreiber S., Probe Reports from NIH Mol. Libr. Progr..
- Eijkelkamp B. A., Hassan K. A., Paulsen I. T., Brown M. H. BMC Genomics. 2011;12:1–14. doi: 10.1186/1471-2164-12-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häußler S. Environ. Microbiol. 2004;6:546–551. doi: 10.1111/j.1462-2920.2004.00618.x. [DOI] [PubMed] [Google Scholar]
- Waksman G., Hultgren S. J. Nat. Rev. Microbiol. 2009;7:765–774. doi: 10.1038/nrmicro2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudleman E., Kaiser D. J. Mol. Microbiol. Biotechnol. 2004;7:52–62. doi: 10.1159/000077869. [DOI] [PubMed] [Google Scholar]
- Moche M., Schlüter R., Bernhardt J., Plate K., Riedel K., Hecker M., Becher D. J. Proteome Res. 2015;14:3804–3822. doi: 10.1021/acs.jproteome.5b00148. [DOI] [PubMed] [Google Scholar]
- Johnson M., Cockayne A., Williams P. H., Morrissey J. A. J. Bacteriol. 2005;187:8211–8215. doi: 10.1128/JB.187.23.8211-8215.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M., Cockayne A., Morrissey J. A. Infect. Immun. 2008;76:1756–1765. doi: 10.1128/IAI.01635-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonov V. V., Mironov A. Y. Klin. Lab. Diagn. 2016;61:52–54. [PubMed] [Google Scholar]
- Kang D., Kirienko N. V. J. Microbiol. 2018;56:449–457. doi: 10.1007/s12275-018-8114-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer J. M., Neely A., Stintzi A., Georges C., Holder I. A. Infect. Immun. 1996;64:518–523. doi: 10.1128/iai.64.2.518-523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takase H., Nitanai H., Hoshino K., Otani T. Infect. Immun. 2000;68:1834–1839. doi: 10.1128/iai.68.4.1834-1839.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minandri F., Imperi F., Frangipani E., Bonchi C., Visaggio D., Facchini M., Pasquali P., Bragonzi A., Visca P. Infect. Immun. 2016;84:2324–2335. doi: 10.1128/IAI.00098-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ankenbauer R., Skiyosachati S., Cox C. D. Infect. Immun. 1985;45:132–140. doi: 10.1128/iai.49.1.132-140.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont I. L., Beare P. A., Ochsner U., Vasil A. I., Vasil M. L. Proc. Natl. Acad. Sci. U. S. A. 2002;99:7072–7077. doi: 10.1073/pnas.092016999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Chial M., Ghysels B., Beatson S. A., Geoffroy V., Meyer J. M., Pattery T., Baysse C., Chablain P., Parsons Y. N., Winstanley C., Cordwell S. J., Cornelis P. Microbiology. 2003;149:821–831. doi: 10.1099/mic.0.26136-0. [DOI] [PubMed] [Google Scholar]
- Denayer S., Matthijs S., Cornelis P. J. Bacteriol. 2007;189:7663–7668. doi: 10.1128/JB.00992-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfarash A., Wei Q., Cornelis P. Microbiology. 2012;1:268–275. doi: 10.1002/mbo3.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D., Kirienko N. V. Front. Microbiol. 2017;8:1707. doi: 10.3389/fmicb.2017.01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor M. S., O'Connor C., Miller V. L. Infect. Immun. 2007;75:1463–1472. doi: 10.1128/IAI.00372-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo T. A., Shon A. S., Beanan J. M., Olson R., MacDonald U., Pomakov A. O., Visitacion M. P. PLoS One. 2011;6:1–13. doi: 10.1371/journal.pone.0026734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisiecki P. Med. Dosw. Mikrobiol. 2014;66:1–10. [PubMed] [Google Scholar]
- Park S. R., Tripathi A., Wu J., Schultz P. J., Yim I., McQuade T. J., Yu F., Arevang C. J., Mensah A. Y., Tamayo-Castillo G., Xi C., Sherman D. H. Nat. Commun. 2016;7:1–11. doi: 10.1038/ncomms10710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis P., Matthijs S. Environ. Microbiol. 2002;4:787–798. doi: 10.1046/j.1462-2920.2002.00369.x. [DOI] [PubMed] [Google Scholar]