Bacterial extracellular polysaccharides serve important cell protective, structural, and signaling roles. They have particularly attracted attention as adhesives and matrix components promoting biofilm formation, which significantly contributes to resistance against antibiotics. In the root nodule symbiosis between rhizobia and leguminous plants, extracellular polysaccharides have a signaling function. UDP-sugar 4-epimerases are important enzymes in the synthesis of the activated sugar substrates, which are frequently shared between multiple polysaccharide biosynthesis pathways. Thus, these enzymes are potential targets to interfere with these pathways. Our finding of a bifunctional UDP-sugar 4-epimerase in Sinorhizobium meliloti generally advances the knowledge of substrate promiscuity of such enzymes and specifically of the biosynthesis of extracellular polysaccharides involved in biofilm formation and symbiosis in this alphaproteobacterium.
KEYWORDS: ExoB, Sinorhizobium meliloti, bifunctional UDP-glucose/UDP-xylose 4-epimerase, biofilm formation, c-di-GMP, surface polysaccharide, symbiosis, transcriptional regulation
ABSTRACT
Sinorhizobium meliloti produces multiple extracellular glycans, including among others, lipopolysaccharides (LPS), and the exopolysaccharides (EPS) succinoglycan (SG) and galactoglucan (GG). These polysaccharides serve cell protective roles. Furthermore, SG and GG promote the interaction of S. meliloti with its host Medicago sativa in root nodule symbiosis. ExoB has been suggested to be the sole enzyme catalyzing synthesis of UDP-galactose in S. meliloti (A. M. Buendia, B. Enenkel, R. Köplin, K. Niehaus, et al. Mol Microbiol 5:1519–1530, 1991, https://doi.org/10.1111/j.1365-2958.1991.tb00799.x). Accordingly, exoB mutants were previously found to be affected in the synthesis of the galactose-containing glycans LPS, SG, and GG and consequently, in symbiosis. Here, we report that the S. meliloti Rm2011 uxs1-uxe-apsS-apsH1-apsE-apsH2 (SMb20458-63) gene cluster directs biosynthesis of an arabinose-containing polysaccharide (APS), which contributes to biofilm formation, and is solely or mainly composed of arabinose. Uxe has previously been identified as UDP-xylose 4-epimerase. Collectively, our data from mutational and overexpression analyses of the APS biosynthesis genes and in vitro enzymatic assays indicate that Uxe functions as UDP-xylose 4- and UDP-glucose 4-epimerase catalyzing UDP-xylose/UDP-arabinose and UDP-glucose/UDP-galactose interconversions, respectively. Overexpression of uxe suppressed the phenotypes of an exoB mutant, evidencing that Uxe can functionally replace ExoB. We suggest that under conditions stimulating expression of the APS biosynthesis operon, Uxe contributes to the synthesis of multiple glycans and thereby to cell protection, biofilm formation, and symbiosis. Furthermore, we show that the C2H2 zinc finger transcriptional regulator MucR counteracts the previously reported CuxR–c-di-GMP-mediated activation of the APS biosynthesis operon. This integrates the c-di-GMP-dependent control of APS production into the opposing regulation of EPS biosynthesis and swimming motility in S. meliloti.
IMPORTANCE Bacterial extracellular polysaccharides serve important cell protective, structural, and signaling roles. They have particularly attracted attention as adhesives and matrix components promoting biofilm formation, which significantly contributes to resistance against antibiotics. In the root nodule symbiosis between rhizobia and leguminous plants, extracellular polysaccharides have a signaling function. UDP-sugar 4-epimerases are important enzymes in the synthesis of the activated sugar substrates, which are frequently shared between multiple polysaccharide biosynthesis pathways. Thus, these enzymes are potential targets to interfere with these pathways. Our finding of a bifunctional UDP-sugar 4-epimerase in Sinorhizobium meliloti generally advances the knowledge of substrate promiscuity of such enzymes and specifically of the biosynthesis of extracellular polysaccharides involved in biofilm formation and symbiosis in this alphaproteobacterium.
INTRODUCTION
Nucleotide sugars are the activated sugar substrates of glycosyltransferases in glycosylation reactions, which add a monosaccharide to a glycosyl acceptor through the formation of a glycosidic bond. Glycosidically linked monosaccharides (glycans) serve a broad variety of structural, metabolic, and signaling roles in all domains of life (1).
The diastereomers xylose and arabinose are important components of various glycans. UDP-xylose is a sugar donor for the synthesis of a broad range of xylose-containing glycans in animals (2), fungi (3), and plants (4), while UDP-arabinose is a donor for the synthesis of pectic and hemicellulosic polysaccharides, proteoglycans, and glycoproteins in plants (5, 6). Xylose- and arabinose-containing glycans have also been reported in bacteria. For instance, arabinose is a component of the mycobacterial cell wall (7), and in several alphaproteobacterial rhizobial species, lipopolysaccharides (LPS) and exopolysaccharides (EPS) containing one or both of these sugars were identified (8–10).
Previously, biochemical evidence for bacterial UDP-xylose synthase and UDP-xylose 4-epimerase activities was provided in Sinorhizobium meliloti (11), an alphaproteobacterial nitrogen-fixing symbiont of plants from the genera Medicago, Melilotus, and Trigonella (12). This study identified the UDP-xylose synthase Uxs1 (SMb20458) and the UDP-xylose 4-epimerase Uxe (SMb20459) of S. meliloti for the first time. Recently, we reported that uxs1 and uxe are part of a six-gene operon (referred to as aps operon) governing biosynthesis of a Congo red (CR)-binding extracellular polysaccharide (13). Expression of this operon is induced by the c-di-GMP-responsive transcriptional activator CuxR, encoded by a gene located upstream and in reverse orientation to the biosynthesis operon (13).
S. meliloti produces various surface polysaccharides, such as LPS, and the EPS succinoglycan (SG) and galactoglucan (GG), which are important for the symbiotic interaction with the host plant and serve cell protective roles in the free-living state (14). SG and GG promote infection of Medicago sativa nodules by S. meliloti. Thus, mutants lacking both these EPS induce noninfected nodules on these host plants (15). Biosynthesis of S. meliloti LPS, SG, and GG requires UDP-galactose. UDP-glucose 4-epimerase ExoB has been suggested as the only enzyme catalyzing the biosynthesis of UDP-galactose in this bacterium, since UDP-glucose 4-epimerase activity was not detected in cell extracts of exoB mutants (16). Correspondingly, these mutants were found to be affected in the biosynthesis of multiple galactose-containing glycans, including SG, GG, and LPS, and consequently to be deficient in infecting the host plants (16–19).
MucR, a transcriptional regulator with a C2H2 zinc finger DNA binding domain, stimulates SG and represses GG production as well as swimming motility (20–24). Strains harboring a functional mucR gene have been reported to produce GG under low-phosphate or quorum sensing conditions, the latter requiring a functional expR gene encoding a LuxR-type regulator (15, 25, 26).
In this study, we explored transcriptional regulation, the nature of the polysaccharide product, and biological role of the aps operon in S. meliloti Rm2011. We show that this operon directs biosynthesis of an arabinose-containing polysaccharide (APS) and is oppositely regulated by MucR and CuxR, integrating c-di-GMP-dependent control of APS production into the opposing regulation of EPS biosynthesis and swimming motility. Furthermore, we report evidence that Uxe is a bifunctional enzyme with UDP-xylose 4- and UDP-glucose 4-epimerase activities, which can functionally replace ExoB.
RESULTS
The uxs1-apsH2 gene cluster directs biosynthesis of an arabinose-containing polysaccharide.
The CR-binding extracellular polysaccharide synthesized by the gene products of the uxs1-uxe-apsS (SMb20460)-apsH1 (SMb20461)-apsE (SMb20462)-apsH2 (SMb20463) gene cluster was isolated for biochemical characterization. The polysaccharide was purified by precipitation with ethanol from the supernatant of cultures of S. meliloti Rm2011 deficient in SG and GG biosynthesis and producing enhanced levels of the CR-binding polysaccharide due to induced overexpression of the aps operon (ΔexoP-Z wgeB/pWBT-pleD-cuxR) (see Fig. S1 in the supplemental material). This was achieved by ectopically induced overexpression of cuxR, encoding a c-di-GMP-responsive transcriptional activator of the aps operon, and pleD, encoding a diguanylate cyclase, whose overproduction results in high intracellular c-di-GMP levels (13, 27). Nevertheless, the amount of sugar of approximately 5 mg · liter−1 · OD600−1 in the culture supernatant, probably attributed to the CR-binding polysaccharide, was rather low (Fig. S1).
As determined by carbohydrate fingerprinting using the high-throughput–1-phenyl-3-methyl-5-pyrazolone (HT-PMP) method (28), polysaccharide precipitated from the supernatant of the S. meliloti ΔexoP-Z wgeB/pWBT-pleD-cuxR strain contained xylose and/or arabinose in addition to glucose and galactose (Table 1). No monomeric sugars were detected in nonhydrolyzed samples of the supernatant. Mainly galactose, but neither xylose/arabinose nor glucose, was lost after dialysis with a 7,000-Da molecular weight cutoff membrane, indicating that galactose probably is not a component of the CR-binding polysaccharide (Table 1). Polysaccharide obtained from the supernatants of the ΔexoP-Z wgeB/pWBT and ΔexoP-Z wgeB Δuxs1-apsH2/pWBT-pleD-cuxR control strains did not contain xylose/arabinose, and lack of the uxs1-apsH2 gene cluster did not result in a decrease in the glucose content (Table 1). This suggests that the CR-binding polysaccharide is mainly or exclusively composed of xylose/arabinose and that glucose was most likely derived from another surface polysaccharide.
TABLE 1.
Carbohydrate fingerprints of S. melilotia
| Genetic background | pleD++ cuxR++b | Mean (range) content (mg/liter) forc: |
||||
|---|---|---|---|---|---|---|
| Glucose | Galactose | Xylose/arabinose | Cellobiose | Gentiobiose | ||
| ΔexoP-Z wgeB | − | 124 (7) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| ΔexoP-Z wgeB | + | 143 (18) | 9 (5) | 54 (1) | 0 (0) | 0 (0) |
| ΔexoP-Z wgeB | + | 317 (195)d | 0 (0)d | 145 (14)d | 0 (0)d | 0 (0)d |
| ΔexoP-Z wgeB Δuxs1-apsH2 | + | 317 (17) | 11 (0) | 0 (0) | 0 (0) | 0 (0) |
| ΔexoB | + | 402 (13) | 54 (10) | 34 (2) | 15 (2) | 8 (2) |
S. meliloti Rm2011 mutants lacking the succinoglycan biosynthesis gene cluster (ΔexoP-Z), carrying a plasmid insertion inside the wgeB gene of the galactoglucan biosynthesis gene cluster (wgeB), containing a deletion of the exoB gene, and/or lacking the APS biosynthesis gene cluster (Δuxs1-apsH2) were used to determine carbohydrate fingerprints of culture supernatants.
Strains harbored either empty vector pWBT (−) or pWBT-pleD-cuxR (+) and were grown in liquid MM supplemented with 0.5 mM IPTG.
Values are from two independent biological replicates.
Sample dialysis with a 7,000-Da molecular weight cutoff membrane after ethanol precipitation.
Since the employed carbohydrate fingerprinting method is not able to distinguish xylose and arabinose, an additional analytical method was applied to identify C5 sugars of the dialyzed and hydrolyzed polysaccharide from the supernatant of the ΔexoP-Z wgeB/pWBT-pleD-cuxR strain. For this, high-pressure liquid chromatography (HPLC) analysis was performed using a Rezex ROA-H+ column and a photodiode array (PDA) detector after total hydrolysis by use of H2SO4. This analysis detected only arabinose and glucose but no xylose (see Fig. S2). Hence, we conclude that the polysaccharide synthesized by the uxs1-apsH2 gene products is mainly composed of arabinose and unlikely to contain xylose. We therefore named this polysaccharide APS (arabinose-containing polysaccharide) and the uxs1-apsH2 genes the aps operon.
Stimulated APS production promotes S. meliloti biofilm formation.
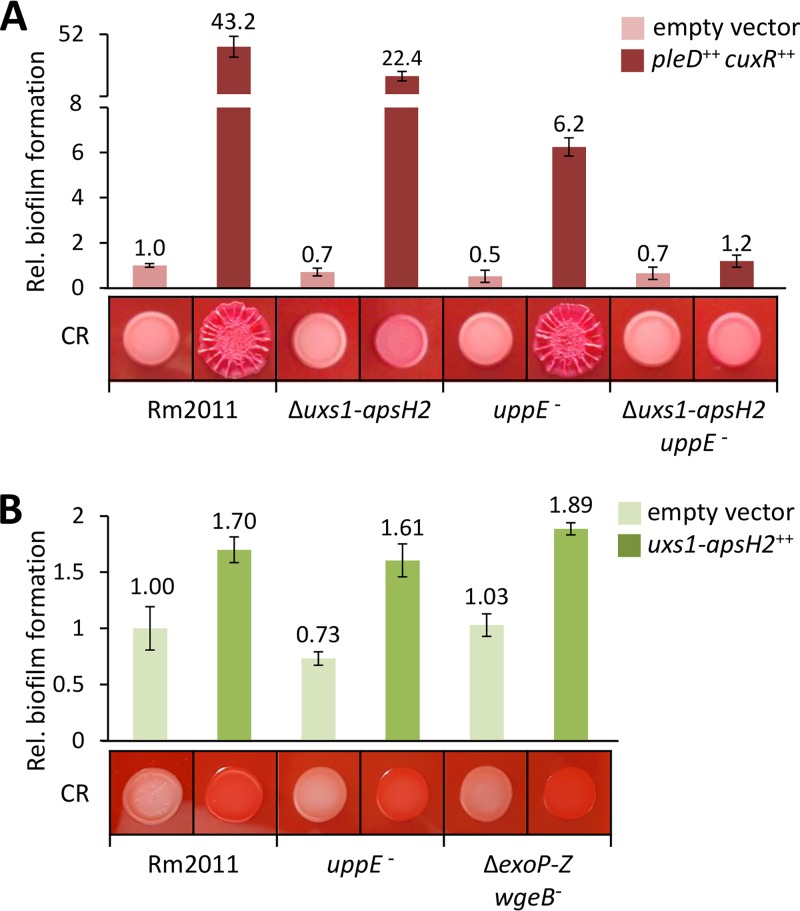
We tested the effect of APS production on biofilm formation of S. meliloti Rm2011 derivative strains on a polystyrene surface. Ectopically induced overexpression of pleD-cuxR resulted in a strong increase in biofilm formation and enlarged, wrinkled red-colored macrocolonies on CR-containing minimal medium (MM) agar (Fig. 1A). Previously, we showed that uppE (SMc01792) contributes to c-di-GMP-stimulated biofilm formation (27). This gene is part of the SMc01790-SMc01796 gene cluster, potentially directing biosynthesis of a unipolar polysaccharide (UPP) (27). An analysis of strains inactivated in the biosynthesis of APS and/or UPP indicated that when APS is produced at enhanced levels, this polysaccharide and UPP cumulatively contribute to c-di-GMP-stimulated biofilm formation (Fig. 1A).
FIG 1.
APS biosynthesis governed by the uxs1-apsH2 gene cluster facilitates biofilm formation of S. meliloti. Biofilm formation of S. meliloti strains, harboring empty vector pWBT and pWBT-pleD-cuxR (A) or empty vector pSRKGm and pSRKGm-uxs1-SMb20463 (B), assessed by crystal violet staining of cell material attached to polystyrene surface. Gene overexpression was induced by adding 0.5 mM IPTG to the medium. Biofilm formation is shown relative to that of the wild type carrying the respective empty vector. Error bars represent the standard deviations from four biological replicates. Congo red (CR)-stained macrocolonies of indicated strains are also shown.
This finding is further supported by the biofilm formation of strains expressing a plasmid-borne copy of the uxs1-apsH2 gene cluster from a heterologous inducible promoter. Induced APS production resulted in red-colored macrocolonies on CR-containing MM agar and an increase in biofilm formation in the Rm2011 wild type but also in strains either disabled for UPP (uppE) or SG and GG (ΔexoP-Z wgeB) biosynthesis (Fig. 1B). This suggests that APS contributes to biofilm formation independent of high c-di-GMP levels, UPP, SG, and GG.
uxs1, uxe, and apsS are important for production of APS in S. meliloti.
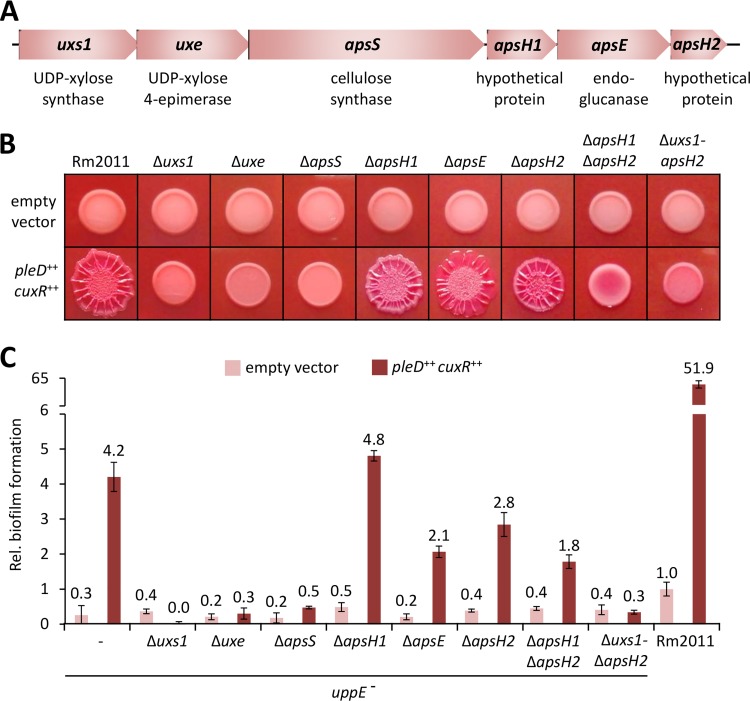
The biosynthesis gene cluster directing APS biosynthesis comprises uxs1 and uxe, involved in the synthesis of nucleotide-sugar precursors (11), apsS, coding for a putative family 2 glycosyltransferase with eight predicted transmembrane α-helices, and apsH1, apsE, and apsH2, encoding proteins with N-terminal signal peptides predicted to localize to the periplasmic space without a membrane-anchoring α-helix (Fig. 2A). ApsE was annotated as a member of glycosylhydrolase family 26 involved in the endohydrolysis of glucosidic linkages. ApsH1 and ApsH2 both contain a domain of unknown function (DUF995) and share 38% amino acid sequence identity with each other.
FIG 2.
uxs1, uxe, and apsS are important for APS-facilitated biofilm formation of S. meliloti. (A) Schematic representation of the APS biosynthesis gene cluster uxs1-apsH2 with annotated gene products. (B) Macrocolonies of Rm2011 and deletion strains, harboring either empty vector pWBT or pWBT-pleD-cuxR, grown on MM agar supplemented with CR and 0.5 mM IPTG. (C) Biofilm formation on polystyrene surface of Rm2011 and uppE mutant with indicated APS biosynthesis gene deletions, each harboring either empty vector pWBT or pWBT-pleD-cuxR. Gene overexpression was induced by adding 0.5 mM IPTG to the medium. Biofilm formation is shown relative to that of the wild type carrying the empty vector. Error bars represent the standard deviations from four biological replicates.
A phylogenetic analysis suggests that other species of the Rhizobiaceae are also able to produce APS. The complete uxs1-apsH2 gene cluster is conserved in Sinorhizobium medicae and Phyllobacterium sp. strain Tri-48, while in Sinorhizobium fredii, Sinorhizobium americanum, and Rhizobium leguminosarum, the corresponding gene clusters lack homologs of apsH1, apsH2, or both genes (see Fig. S3). Notably, two paralogous gene clusters were found in R. leguminosarum (11). However, one of these clusters does not include an endoglucanase-encoding gene and lacks a cuxR homolog upstream of the operon.
We determined the individual contributions of the uxs1-apsH2 cluster genes to APS biosynthesis in S. meliloti Rm2011 wild-type and uppE mutant backgrounds. To monitor APS-mediated phenotypes, the promoter of the aps operon was activated by induced overexpression of plasmid-borne copies of cuxR and pleD. This resulted in enlarged, wrinkled red-colored macrocolonies of the wild type grown on CR-containing MM agar and enhanced biofilm formation on a polystyrene surface by the uppE mutant (Fig. 2B and C). Individual deletions of uxs1, uxe, and apsS caused phenotypes similar to those of strains harboring the empty vector control or lacking the complete uxs1-apsH2 gene cluster (Fig. 2B and C). Macrocolony and CR-staining phenotypes of the uxs1 and uxe mutants were at least partially restored by individual ectopic expression of uxs1 and uxe, respectively (see Fig. S4). Partial complementation of mutant strains could be explained by low gene expression levels or undesired polar effects of gene deletions. The apsE mutant was not notably affected, and the apsH1 and apsH2 mutants only moderately affected, in macrocolony morphology and CR-staining. However, the apsH1 apsH2 double deletion mutant formed macrocolonies that were not expanded in size, lacked wrinkles, and were only moderately stained by CR (Fig. 2B). This mutant was partially complemented by ectopic expression of apsH1 (Fig. S4). While biofilm formation of the uppE apsH1 mutant was not affected, the uppE apsE, uppE apsH2, and uppE apsH1 apsH2 mutants showed reduced biofilm formation compared to that of the uppE mutant (Fig. 2C). Based on these results, we conclude that Uxs1, Uxe, and ApsS are very important for APS production, whereas ApsH1, ApsE, and ApsH2 likely have accessory functions. Furthermore, ApsH1 and ApsH2 are probably able to functionally replace each other.
Furthermore, we tested pleD-cuxR overexpression-induced APS-mediated phenotypes in strains deficient in the biosynthesis of other c-di-GMP-regulated polysaccharides. Macrocolony morphology and CR staining, indicating APS production, were not notably affected in strains unable to produce UPP (uppE) (27), mixed-linkage β-glucan (bgsA) (29), and SG and GG (ΔexoP-Z wgeB or ΔexoB) (see Fig. S5).
uxe overexpression suppresses the pleiotropic phenotype of a S. meliloti exoB mutant.
An S. meliloti strain lacking the UDP-glucose 4-epimerase-encoding exoB gene is affected in the biosynthesis of multiple galactose-containing glycans, including SG, GG, and LPS, and unable to infect host plants (16). Intriguingly, in the exoB mutant but not in an exoB uxe double mutant, overexpression of pleD-cuxR restored SG production as indicated by calcofluor (CF) staining of macrocolonies (Fig. S5). This finding is further supported by the carbohydrate fingerprint of the culture supernatant of the ΔexoB/pWBT-pleD-cuxR strain grown under phosphate-sufficient conditions not promoting GG production. SG is a polymer of octasaccharide repeating units composed of seven glucose molecules and one galactose molecule joined by β-1,4, β-1,3, and β-1,6 glycosidic linkages (30). This is consistent with the detected glucose-to-galactose ratio as well as with the presence of cellobiose and gentiobiose in the supernatant of this strain (Table 1). We therefore hypothesized that expression of the uxs1-apsH2 gene cluster suppressed SG deficiency in an exoB mutant.
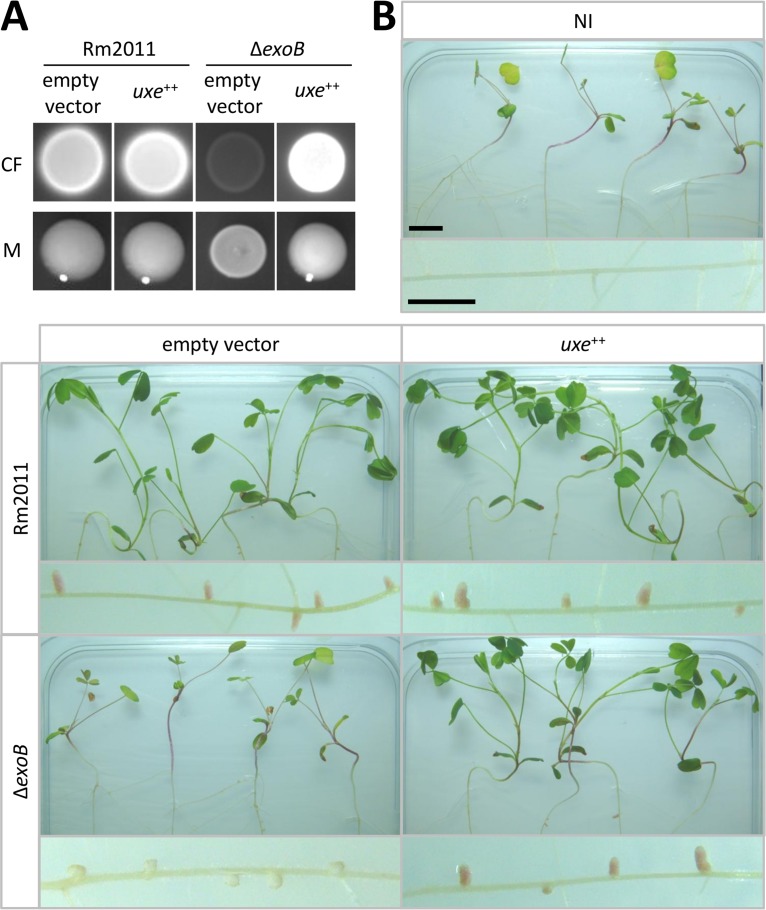
The best candidate for functional replacement of ExoB is the UDP-xylose 4-epimerase Uxe. Constitutive expression of a plasmid-borne copy of uxe restored CF brightness and mucoid phenotype of macrocolonies (Fig. 3A), a wild type-like LPS profile as determined by SDS-PAGE (see Fig. S6A), and root nodule symbiosis of the exoB mutant (Fig. 3B; Fig. S6B). On M. sativa roots, the ΔexoB strain induced white noninfected nodules, whereas this strain constitutively overexpressing uxe established red nitrogen-fixing nodules (Fig. 3B). Thus, ectopic expression of uxe suppressed all tested phenotypes of an exoB mutant. APS overproduction, caused by constitutive expression of the heterologous diguanylate cyclase dgcA from Caulobacter crescentus and cuxR, did not rescue the SG-deficient ΔexoP-Z strain for an efficient symbiosis with M. sativa (see Fig. S7). This indicates that APS itself cannot functionally replace the symbiotically active EPS.
FIG 3.
uxe complements pleiotropic S. meliloti exoB mutant phenotype. (A) Rm2011 wild-type and exoB deletion strains, harboring either empty vector pSRK or pSRK-uxe, grown on LB agar supplemented with calcofluor (CF) or on phosphate-limiting MM agar to evaluate mucoid phenotypes (M). (B) Plant shoots and roots of M. sativa inoculated with indicated S. meliloti strains. NI, not inoculated. Bars, 1 cm.
We also tested if ExoB can functionally replace Uxe in the APS biosynthesis pathway. Since the Δuxe strain did not show detectable pleD-cuxR overexpression-induced APS production, native levels of exoB expression were not sufficient for the production of detectable amounts of APS (Fig. 2B). However, induced overexpression of a plasmid-borne exoB copy partially restored pleD-cuxR overexpression-induced APS production of a Δuxe strain (Fig. S4), suggesting that ExoB has UDP-xylose 4-epimerase side activity.
Uxe has UDP-glucose 4-epimerase activity in vitro.
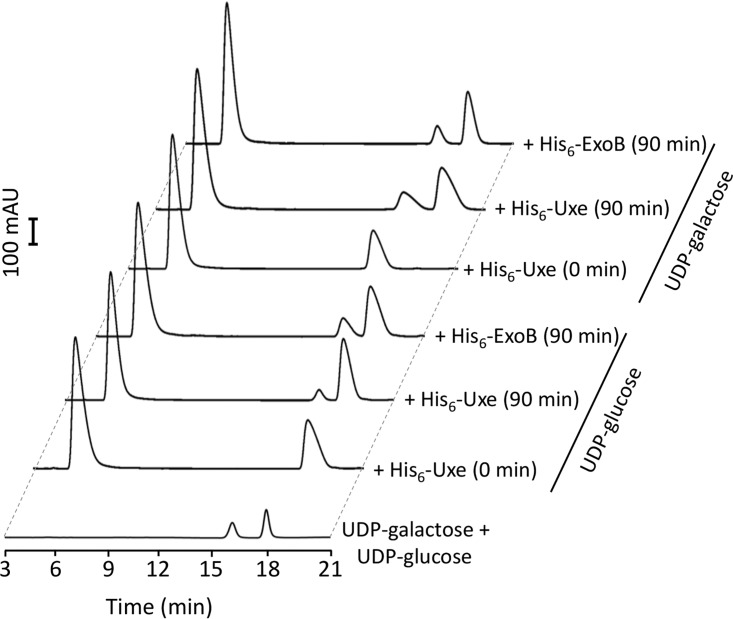
We hypothesized that Uxe is a bifunctional enzyme with UDP-xylose 4- (11) and UDP-glucose 4-epimerase activities. To test this hypothesis, His6-Uxe and His6-ExoB proteins were produced in Escherichia coli and purified (see Fig. S8). UDP-glucose 4-epimerase assays were performed in vitro (reaction conditions: 10 μM protein, 90 min, 30°C). These assays demonstrated that His6-Uxe and His6-ExoB both were able to interconvert UDP-glucose and UDP-galactose in vitro (Fig. 4). In line with the study of Gu et al. (11), the addition of NAD+, an enzyme-bound cofactor of short-chain dehydrogenase/reductase (SDR) proteins, to the reaction mixtures had no major effect on the enzyme activities of His6-Uxe (see Fig. S9A). In contrast, Gu et al. (11) did not detect UDP-glucose 4-epimerase activity for Uxe (reaction conditions: 0.5 μM protein, 15 min, 37°C). Consistently, using the same reaction conditions, we detected conversion of UDP-glucose to UDP-galactose by His6-ExoB but not by His6-Uxe nor by preboiled His6-ExoB (Fig. S9B). We did not follow up on the differences in reaction conditions responsible for the detection of His6-Uxe UDP-glucose 4-epimerase activity in our study but not in that by Gu et al. (11), but we speculate that the extension of reaction time and increase in enzyme concentration were probably instrumental.
FIG 4.
Uxe interconverts UDP-glucose and UDP-galactose in vitro. HPLC profiles of UDP-sugars after incubation of 10 μM indicated purified proteins with 0.5 mM either UDP-glucose or UDP-galactose at 30°C for indicated time periods. UDP-glucose and UDP-galactose standards were used at 0.1 mM. His6-ExoB served as the positive control.
MucR counteracts CuxR–c-di-GMP-mediated transcriptional activation of the uxs1 promoter.
Our findings imply a role of uxs1-apsH2-directed APS biosynthesis in biofilm formation and a contribution of uxe to the biosynthesis of multiple polysaccharides. To obtain hints to the conditions allowing for maximal expression of this gene cluster, we dissected its transcriptional regulation. In our previous study, we showed that pleD overexpression results in a stronger CR phenotype of a S. meliloti mucR mutant than the wild-type strain (13).
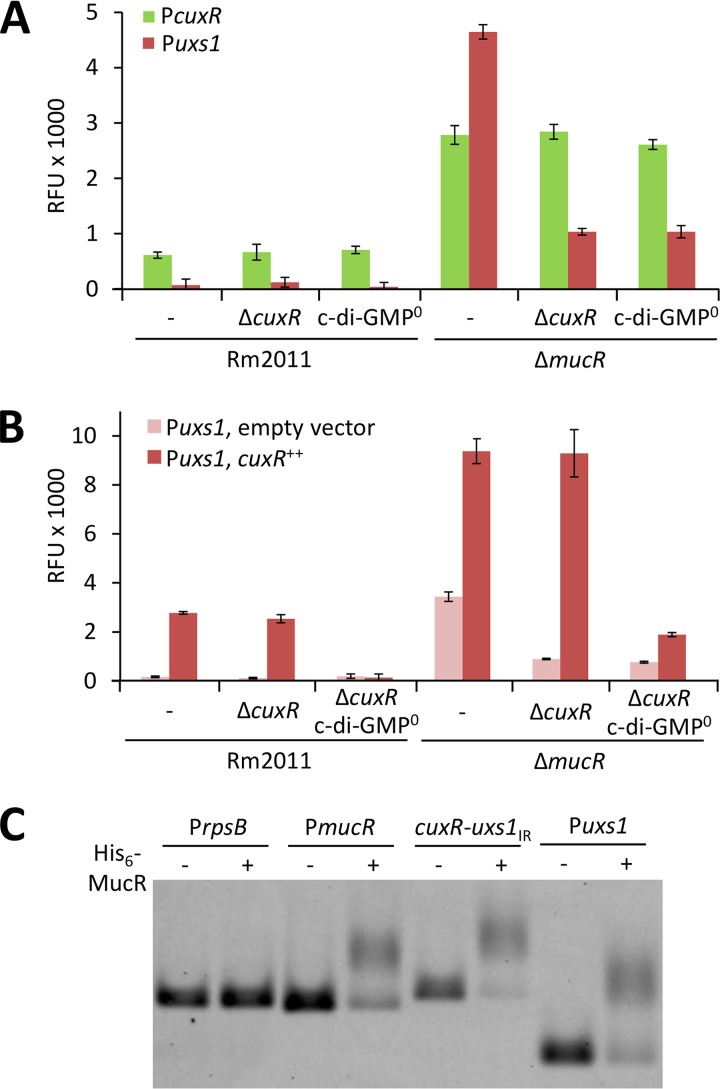
Here, we found that uxs1 and cuxR promoter activities were negatively affected by the presence of the mucR gene, as determined by egfp fusions to Puxs1 and PcuxR (Fig. 5A). Increased Puxs1 activity resulting from deletion of mucR was largely dependent on functional cuxR and the ability to synthesize c-di-GMP (Fig. 5A). In contrast, PcuxR activity was independent of cuxR and c-di-GMP in the mucR mutant (Fig. 5A). We uncoupled cuxR expression from MucR-dependent regulation of PcuxR by constitutive expression of a plasmid-borne copy of cuxR. Overexpression of cuxR induced activity of a Puxs1-egfp promoter fusion in a c-di-GMP-dependent manner, while under these conditions, the mucR mutant showed approximately 3.5-fold higher Puxs1 activity than the wild type (Fig. 5B). These levels of activation were unaffected by deletion of the genomic cuxR locus (Fig. 5B).
FIG 5.
MucR counteracts c-di-GMP- and CuxR-mediated activation of the uxs1 promoter. (A) cuxR and uxs1 promoter activities in different S. meliloti genetic backgrounds measured using promoter-egfp fusions on medium-copy-number plasmid pSRKGm-egfp. (B) uxs1 promoter activity in different S. meliloti genetic backgrounds with (pABC-Psyn-cuxR) or without (empty vector pABC-Psyn) constitutively expressed cuxR. (A and B) c-di-GMP0, Rm2011 ΔXVI unable to produce detectable levels of c-di-GMP (27), was used as a background strain. Error bars represent the standard deviations from four biological replicates. (C) Interaction of His6-MucR with DNA containing the cuxR-uxs1 intergenic region (cuxR-uxs1IR) or a 196-bp region upstream of uxs1 (Puxs1) assayed with EMSA. DNA containing either the upstream region of rpsB or that of mucR served as negative or positive control, respectively.
His6-MucR was produced in E. coli and purified (Fig. S8). Direct interaction between His6-MucR and the cuxR-uxs1 intergenic region was confirmed by electrophoretic mobility shift assay (EMSA) (Fig. 5C). A putative MucR binding motif was identified between the transcriptional start sites of the divergently oriented aps operon and cuxR by in silico comparisons (see Fig. S10).
Collectively, these data indicate that MucR represses both Puxs1 and PcuxR, while CuxR–c-di-GMP activates Puxs1 but not PcuxR.
DISCUSSION
In bacteria, the ubiquitous second messenger c-di-GMP is a key player in controlling the switch between planktonic motile and surface-associated sedentary lifestyles (31). Recently, we reported the activation of the otherwise silent putative extracellular polysaccharide biosynthesis SMb20458-63 (aps) operon by a c-di-GMP-responsive AraC-like transcription factor in S. meliloti (13). Here, we unraveled the monosaccharide composition and biological function of the corresponding polysaccharide product and gained further insights into its biosynthesis pathway and regulation.
APS is a novel component of the sinorhizobial extracellular matrix contributing to biofilm formation.
Our carbohydrate compositional analysis of polysaccharide purified from the supernatant of aps operon-overexpressing S. meliloti cells indicates a polysaccharide mainly composed of arabinose. Bacterial polysaccharides are frequently found in both soluble and cell-associated forms (32). Strong CR binding and wrinkled morphology of macrocolonies formed by these cells imply the production of considerable amounts of APS. However, only small amounts of APS were present in the culture supernatant, suggesting that most APS is not released from the cell but rather is bound to the cell surface.
Only few examples of arabinose-containing polysaccharides have been reported in the bacterial kingdom, including the mycobacterial cell wall, Bradyrhizobium japonicum and Burkholderia cenocepacia LPS, and Azorhizobium caulinodans Nod factors (7, 9, 33, 34). APS is a novel component of the sinorhizobial extracellular matrix, broadening the spectrum of arabinose-containing glycans in bacteria.
Bacterial extracellular polysaccharides contribute to cell protection against various environmental stresses, such as extreme pH, antibiotics, or desiccation, and serve as adhesives promoting surface attachment. Furthermore, these polysaccharides play essential roles in host-pathogen interactions and as components of the biofilm matrix (35, 36). Consistent with these functional roles of extracellular polysaccharides, APS-producing S. meliloti cells showed enhanced biofilm formation.
APS biosynthesis employs a synthase-dependent pathway.
The biosynthesis of most microbial homopolymeric polysaccharides is directed by the so-called synthase-dependent pathway (37). Synthase-dependent systems of Gram-negative bacteria employ a transenvelope multiprotein complex typically consisting of a glycosyltransferase and a translocation pore in the cytoplasmic membrane (A), periplasmic polysaccharide-modifying enzymes (B), β-barrel porins in the outer membrane (C), tetratricopeptide repeat (TPR)-containing periplasmic scaffold proteins (D), and a c-di-GMP regulatory component (E) (32). The proteins required for polymerization, modification, and export are often encoded by a single operon, whereas the genes required for the synthesis of the nucleotide-sugar precursor molecules (F) more typically are encoded elsewhere on the chromosome (32). Polysaccharides produced by the synthase-dependent pathway include alginate (e.g., Pseudomonas aeruginosa) and cellulose (e.g., E. coli) (32).
The architecture of the APS biosynthesis operon and the upstream neighboring regulatory gene cuxR is mostly conserved in several rhizobial species. The aps operon encodes the putative components A, B, C, and F, consistent with a synthase-dependent APS biosynthesis pathway (Fig. 6). The cytoplasmic nucleotide-sugar precursor synthesis proteins Uxs1 and Uxe (F) catalyze synthesis of UDP-xylose and epimerization of this nucleotide sugar to UDP-arabinose, respectively (11). The membrane-bound family 2 glycosyltransferase ApsS (A) probably directs polysaccharide synthesis and its translocation across the inner membrane, similarly to cellulose synthase BcsA in E. coli and Rhodobacter sphaeroides (38, 39). ApsS is most likely specific for the substrate UDP-arabinose, as suggested by the monosaccharide composition of APS and the requirement of the UDP-xylose 4-epimerase Uxe for APS production. The aps operon further encodes the putative periplasmic polysaccharide hydrolase ApsE (B), which may regulate polymer length and export, similarly to AlgL in P. aeruginosa (40) and BcsZ in E. coli (41). At least one of the secreted proteins ApsH1 and ApsH2 (C) is required for APS production. ApsH1 and ApsH2 both contain eight predicted short sequential β-strands. These proteins may facilitate polysaccharide transport across the outer membrane, facilitate protein-protein interactions, or introduce postsynthetic modifications to the glycan, as reported for AlgE (42), AlgF (32), or AlgX (43) in P. aeruginosa, respectively. The aps operon does not encode a TPR domain protein (D), suggesting that this component is encoded elsewhere in the S. meliloti genome.
FIG 6.
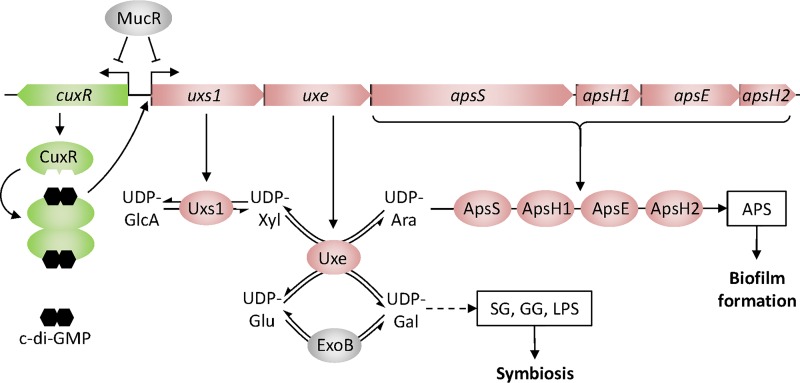
Model of Uxe function in biosynthesis of cell surface polysaccharides in S. meliloti. The combined activities of UDP-xylose synthase Uxs1 and UDP-xylose 4-epimerase Uxe result in the formation of UDP-arabinose, a nucleotide-sugar precursor in the biosynthesis of an arabinose-containing polysaccharide (APS) that promotes biofilm formation. Similar to ExoB and in addition to its UDP-xylose 4-epimerase activity, Uxe catalyzes epimerization between UDP-glucose and UDP-galactose, thereby supporting biosynthesis of the symbiotically relevant polysaccharides succinoglycan (SG), galactoglucan (GG), and lipopolysaccharide (LPS). Opposite transcriptional regulation of the aps operon by CuxR–c-di-GMP and MucR integrates the APS biosynthesis pathway into the global regulatory network of exopolysaccharide biosynthesis and swimming motility. UDP-GlcA, UDP-glucuronate; UDP-Xyl, UDP-xylose; UDP-Ara, UDP-arabinose; UDP-Glu, UDP-glucose; UDP-Gal, UDP-galactose.
Uxe is a bacterial UDP-sugar 4-epimerase with activity on both UDP-xylose and UDP-glucose.
In S. meliloti, the UDP-glucose 4-epimerase ExoB, providing the major activity for UDP-glucose/UDP-galactose interconversion, is important for SG and GG biosynthesis, LPS composition, and root nodule infection (16–19). We showed that the S. meliloti Uxe protein has UDP-xylose 4- and UDP-glucose 4-epimerase activities, thus catalyzing UDP-xylose/UDP-arabinose and UDP-glucose/UDP-galactose interconversions, respectively. Accordingly, uxe overexpression suppressed the pleiotropic phenotypes of an S. meliloti exoB mutant. Reversely, exoB partially complemented the uxe mutant, suggesting that ExoB is also able to catalyze UDP-xylose/UDP-arabinose epimerization. Thus, ExoB and Uxe can contribute to the production of multiple surface polysaccharides and thereby to biofilm formation and symbiosis in S. meliloti (Fig. 6).
A variety of UDP-sugar 4-epimerases with different substrate specificities and promiscuities have been reported. Among these are epimerases active on UDP-glucose/UDP-galactose (44), their acetylated and uronic acid forms (45, 46), the corresponding pentoses UDP-arabinose/UDP-xylose (11), or multiples of these substrates (47, 48). Among the biochemically characterized bifunctional UDP-glucose/UDP-xylose 4-epimerases are PsUGE1 from Pisum sativum L. (47), AtUGE1/3 from Arabidopsis thaliana (47), and OcUGE1/2 from Ornithogalum caudatum (49).
Studies on the substrate promiscuity of UDP-sugar 4-epimerases have mainly focused on the determinants that cause the epimerases to discriminate between N-acetylated and nonacetylated UDP-glucose and UDP-galactose. A “gatekeeper” residue determines substrate specificity for N-acetylated and nonacetylated substrates (50). However, current models lack an explanation for substrate specificity on UDP-pentoses and uronic acid forms of UDP-sugars (51). Members of the SDR superfamily share some characteristic features, including the two signature sequences YXXXK and GXXGXXG (51). The former plays a key role in the activation of the cofactor, while the repetitive glycine motif participates in NAD+ binding. Both motifs are conserved in Uxe and ExoB from S. meliloti.
The active site of UDP-sugar 4-epimerases is located between an N-terminal nucleotide binding domain and a smaller C-terminal domain responsible for the correct positioning of the UDP-sugar substrate (44). The number of N-terminal enzymatic residues that form hydrogen bonding contacts to the adenosyl group determines the strength of NAD+ binding (52, 53). In agreement with the study by Gu et al. (11), the addition of NAD+ had no effect on the activity of purified Uxe from S. meliloti, suggesting that NAD+ is tightly bound to the protein and therefore was not lost during protein purification.
Control of APS production is integrated into the global regulatory network of EPS biosynthesis and swimming motility.
In contrast to synthase-dependent pathways, posttranscriptionally regulated through a c-di-GMP-responsive component, such as the biosynthesis of a mixed linkage β-glucan in S. meliloti (29), c-di-GMP-dependent regulation of APS synthesis is controlled by the c-di-GMP-responsive transcriptional regulator CuxR (13). However, additional regulation of APS biosynthesis by c-di-GMP at the posttranslational level cannot be ruled out, e.g., by binding of c-di-GMP to the cytoplasmic portion of ApsS. Unlike ApsS, the alginate and cellulose synthase complexes contain PilZ domains dedicated to allosteric control of glycosyltransferase activity through c-di-GMP (32).
Full activation of aps operon expression requires the presence of CuxR–c-di-GMP and absence of the zinc-finger transcriptional regulator MucR. While in S. meliloti, MucR regulates EPS biosynthesis and swimming motility (23), transcriptional regulation by MucR orthologs in C. crescentus and S. fredii furthermore involves cell cycle-related genes (54). It was suggested that MucR primarily supports the S→G1 phase transcriptional switch and integrates regulation of symbiosis- or virulence-related genes with the cell cycle in several alphaproteobacteria (54). The experimentally verified binding site of CuxR–c-di-GMP (13) is just 27 bp from the in silico-deduced binding site of MucR within the cuxR-uxs1 intergenic region. The positions of these binding sites and our results from promoter activity studies are consistent with a model of PcuxR and Puxs1 regulation in which MucR bound to the cuxR-uxs1 intergenic region represses PcuxR and prevents CuxR–c-di-GMP-mediated activation of aps operon transcription. In agreement with this model, transcriptome profiling of S. meliloti lacking MucR revealed a 7.13- and 2.08-fold accumulation of uxs1 and cuxR transcripts, respectively (24).
In S. meliloti, flagellum-based motility and biosynthesis of SG, GG, and APS are regulated by MucR and c-di-GMP-dependent signaling pathways (13, 23, 27). Moreover, we showed that transcriptional activation of the aps operon not only results in APS production but also affects the production of SG, GG, and LPS, thereby promoting biofilm formation and symbiosis with leguminous host plants. The high complexity and connectivity in the regulation of extracellular matrix components probably allows for the integration of multiple environmental stimuli and adaptability of S. meliloti under changing environmental conditions.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are shown in Table S1 in the supplemental material. S. meliloti was grown at 30°C in tryptone-yeast extract (TY) medium (55), LB medium (56), modified morpholinepropanesulfonic acid (MOPS)-buffered minimal medium (MM) (25), and nutrient-depleted 30% MM (nitrogen, carbon, and phosphate sources reduced to 30%). The medium compositions are provided in the supplemental material. E. coli was grown in LB medium at 37°C. For S. meliloti, antibiotics were used at the following concentrations (mg/liter; respectively liquid and solid media): kanamycin, 100 and 200; gentamicin, 20 and 40; tetracycline, 5 and 10; spectinomycin 200 and 200; and streptomycin, 600 and 600. For E. coli, the following concentrations were used: kanamycin, 50 and 50; gentamicin, 5 and 8; tetracycline, 5 and 10; spectinomycin, 100 and 100; and ampicillin, 100 and 100. Isopropyl-β-d-1-thiogalactopyranoside (IPTG) was added at 0.5 mM.
Construction of strains and plasmids.
The constructs used in this work were generated using standard cloning techniques. The primers used are listed in Table S2. All constructs were verified by sequencing. Plasmids were transferred to S. meliloti by E. coli S17-1-mediated conjugation (57) as previously described (58). Correct plasmid integrations were verified by PCR.
For the construction of gene deletion mutants, gene-flanking regions of circa 700 to 900 bp were cloned into suicide plasmid pK18mobsacB. The resulting constructs were integrated into the S. meliloti genome, and transconjugants were subjected to sucrose selection as previously described (59). Gene deletions were verified by PCR and following agarose gel DNA fragment size analysis. The DNA sequence of the aps gene cluster was confirmed in all strains containing single or double deletions of aps genes.
The promoter-egfp fusions were generated by inserting a 368-bp fragment including the cuxR-uxs1 intergenic region (or a 509-bp fragment including the mucR upstream region) and the three first codons of the gene of interest into the replicative medium-copy-number plasmid pSRKGm-egfp (or the low-copy-number plasmid pPHU231-egfp). This generated an in-frame fusion of these first codons of the gene of interest to egfp.
Gene overexpression constructs were generated by inserting the full-length coding sequence into the replicative medium-copy-number vectors pWBT and pSRK, the low-copy-number vector pR-egfp, the single-copy vector pABC-Psyn, and the integrative vector pSM10. Constructs for production of N-terminally His6-tagged Uxe and ExoB from S. meliloti were generated by inserting the respective coding sequence excluding the start codon into expression vector pWH844.
Isolation of APS and total sugar determination.
For the analysis of APS polysaccharide, TY precultures of the S. meliloti ΔexoP-Z wgeB, ΔexoP-Z wgeB Δuxs1-apsH2, and ΔexoB strains, harboring either empty vector pWBT or pWBT-pleD-cuxR, were diluted 1:60 in 300 ml MM supplemented with IPTG in duplicates and incubated for 3 days at 30°C under shaking conditions. Cells were harvested by centrifugation (30 min, 4,000 × g, 4°C), and the supernatants were collected, mixed with four volumes of ethanol, and incubated at 4°C overnight. Precipitate was collected and either dried at 30°C or dialyzed with distilled water using SnakeSkin dialysis tubing with a molecular weight cutoff (MWCO) of 7,000 Da (Thermo Fisher). Dialyzed samples were precipitated a second time with four volumes of ethanol prior to drying. Dried samples were stored at 4°C.
Total sugar determination was carried out for S. meliloti culture supernatants from 5-ml cultures, in triplicates, prepared as described above. One milliliter of culture supernatant was mixed with 200 μl reagent solution (0.7 mg/ml l-cysteine hydrochloride [Sigma-Aldrich] dissolved in 86% [vol/vol] H2SO4) and incubated for 4 min at 100°C. After cooling the samples, the optical density (OD) at 415 nm was measured using an Infinite 200 Pro multimode reader (Tecan). Absolute sugar contents were calculated based on a standard curve generated with d-glucose (Roth).
Carbohydrate fingerprint.
Simultaneous high-resolution detection of carbohydrates which can be derivatized with 1-phenyl-3-methyl-5-pyrazolone (PMP) was performed via HT-PMP as previously described (28). Briefly, 1-g/liter solutions containing APS were hydrolyzed in 2 M trifluoroacetic acid (TFA) for 90 min at 121°C and subsequently neutralized with 3.2% (vol/vol) ammonium hydroxide. Thereafter, 25 μl neutralized sample was mixed with 75 μl derivatization reagent (0.1 M methanolic-PMP and 0.4% [vol/vol] ammonium hydroxide solutions mixed at a 2:1 ratio) and incubated for 100 min at 70°C. Finally, 130 μl of 19.23 mM acetic acid was added to 20 μl cooled sample, and HPLC separation was performed on a reverse-phase column (Gravity C18, 100-mm length, 2-mm inside diamter [i.d.], 1.8-μm particle size; Macherey-Nagel) tempered to 50°C. For gradient elution, mobile phase A (5 mM ammonium acetate buffer [pH 5.6] with 15% acetonitrile) and mobile phase B (pure acetonitrile) were pumped at a flow rate of 0.6 ml/min. The HPLC system (Ultimate 3000RS; Dionex) was composed of a degasser (SRD 3400), a pump module (HPG 3400RS), auto sampler (WPS 3000TRS), a column compartment (TCC 3000RS), a diode array detector (DAD 3000RS), and an electrospray ionization (ESI)-ion-trap unit (HCT; Bruker). Standards for each sugar (2, 3, 4, 5, 10, 20, 30, 40, 50, and 200 mg/liter) were processed as the samples, starting with the derivatization step. Data were collected and analyzed with BrukerHystar, QuantAnalysis, and Dionex Chromelion software. Since this method is not able to distinguish xylose and arabinose, an additional measurement by the use of a Rezex ROA-H+ column (Phenomenex, Torrance, CA), a refractive index detector (RI 101; Shodex, Tokyo, Japan) and a PDA detector (210/278 nm; Dionex, Sunnyvale, CA) was performed after total hydrolysis at 120°C for 1 h with 72% (vol/vol) H2SO4 and following neutralization with BaCO3.
Phenotypic assays.
Congo red (CR) staining was assayed on TY or MM agar containing 120 mg/liter CR. Calcofluor (CF) brightness was analyzed on LB agar containing 200 mg/liter CF. Fresh precultures grown on TY agar were resuspended in 0.9% (wt/vol) NaCl to an optical density at 600 nm (OD600) of 0.1, and 50 μl was dropped onto the agar medium. Plates were incubated at 30°C and documented after 2 to 3 days. Biofilm formation was determined in flat-bottom polystyrene 96-well plates (Greiner) in 30% MM, in triplicates, as previously described (27, 60). Briefly, a preculture grown in liquid TY medium was diluted circa 10-fold in 30% MM (final volume of 100 μl) and grown without shaking for 48 h at 30°C. Growth was recorded by determining the OD600. The culture medium and nonattached cells were removed. Plate wells were washed with water, stained with 200 μl of 0.1% (wt/vol) crystal violet (Sigma-Aldrich) for 30 min with gentle agitation, and washed twice with water. Stained attached cell material was dissolved in 200 μl of detaching solution (80% ethanol, 20% acetone) for 20 min upon shaking. Biofilm formation was calculated as the ratio of crystal violet-bound attached cell material at A570 to the OD600 of the culture. The ability of strains to form symbiosis with the host Medicago sativa cv. Eugenia was assessed as previously described (27). The nodulation frequency of a single bacterial strain was determined by counting the nodules on 48 plant roots in a 2-week time period after inoculation.
Rapid preparation of LPS samples.
LPS was extracted from S. meliloti cells according to a published protocol (61) omitting the polymyxin B purification step. Briefly, S. meliloti Rm2011 wild-type and exoB deletion strains, harboring pSRK or pSRK-uxe, were grown in liquid TY medium overnight. One milliliter of each culture was centrifuged (2 min, 10,000 × g) at room temperature (RT), and cell pellets were resuspended in 50 μl of 100 mM EDTA titrated with triethylamine (TEA) to pH 7.0. Cell suspensions were mixed with gentle agitation and incubated at 20°C for 15 min. Cells were centrifuged (2 min, 10,000 × g, RT), and 50 μl of the supernatants was incubated with 10 μl of 2.5 μg/μl proteinase K in EDTA-TEA solution for 1 h at 60°C. LPS extracts were boiled in Laemmli sample buffer and fractionated by SDS-PAGE with 16% acrylamide gels, followed by visualization with silver nitrate staining.
Fluorescence measurements.
For promoter-egfp assays, TY precultures were diluted 1:500 in 100 μl 30% MM and grown to exponential growth phase in 96-well plates at 30°C with shaking. Enhanced green fluorescent protein (EGFP) fluorescence (excitation wavelength [λex] of 488 ± 9 nm; emission wavelength [λem] of 522 ± 20 nm; gain, 82) and growth (OD600) were recorded using an Infinite 200 Pro multimode reader (Tecan). Strains carrying the empty vector pSRKGm-egfp were used for measuring background fluorescence. Relative fluorescence units (RFU), calculated as EGFP signals, were normalized to the OD600 (n = 4).
Protein purification.
Purified His6-Uxe and His6-ExoB proteins were obtained according to a described protocol (27) while using a modified lysis buffer (20 mM HEPES [pH 7.5], 250 mM NaCl, 40 mM imidazole, 20 mM MgCl2, 20 mM KCl). For His6-MucR purification, the pellet obtained from 2 liters of culture was resuspended in 30 ml lysis buffer (50 mM MOPS [pH 7.5], 500 mM NaCl, 40 mM imidazole), and the lysate was filtered (0.2-μm pore size) and applied to a 1-ml HisTrap HP column (GE Healthcare) at a flow rate of 0.025 ml/s. The column was washed and His6-MucR was eluted with lysis buffer containing 100 mM and 500 mM imidazole, while samples were collected in 1-ml fractions. After SDS-PAGE analysis, fractions containing purified His6-MucR were pooled, concentrated using centrifugal filters (Amicon) with a MWCO of 10,000 Da, frozen in liquid nitrogen, and stored at −80°C. Absolute protein concentrations were determined using Bradford reagent (Bio-Rad) and bovine serum albumin (BSA; Sigma-Aldrich) as the standard.
Electrophoretic mobility shift assay.
An EMSA reaction mixture contained 50 mM KCl, 1.7 μg sonicated salmon sperm DNA (Invitrogen), 100 μg BSA (Sigma-Aldrich), and 15 ng Cy3-labeled DNA in a final volume of 10 μl. Cy3-labeled DNA fragments were obtained by PCR with primer Cy3-egfp-28-rev and a promoter-specific primer (Table S2) using pSRKGm-egfp- and pPHU231-egfp-based constructs as the templates. Two microliters of 0.6-mg/ml His6-MucR was added to the reaction mixture. The reaction mixtures were incubated at RT for 30 min; 1.5 μl of 90% glycerol was added and the reaction was loaded onto a 1.5% agarose gel in Tris-acetate-EDTA (TAE) buffer. After electrophoresis at 90 V for 1 h, gel images were scanned using a Typhoon 8600 variable-mode imager (Amersham Bioscience).
UDP-glucose 4-epimerase activity assay.
Standard reaction mixtures (50 μl final volume) contained reaction buffer (20 mM HEPES [pH 7.5], 250 mM NaCl, 20 mM MgCl2, 20 mM KCl), 0.5 mM UDP-glucose (Sigma-Aldrich) or UDP-galactose (Sigma-Aldrich), and 10 μM His6-Uxe or His6-ExoB. Unless otherwise specified, reaction mixtures were incubated for 90 min at 30°C. To remove proteins from the reaction mixtures prior to HPLC analysis, samples were mixed with 50 μl chloroform and centrifuged (5 min, 21,000 × g, RT), and the aqueous phase was collected and stored at −20°C.
Separation of UDP-glucose and UDP-galactose was achieved according to a method described by Behmüller et al. (62) using an Agilent 1100 quaternary HPLC system equipped with a HYPERCARB 150-mm by 1-mm column (Thermo Scientific) at a temperature of 60°C, a flow rate of 0.1 ml/min with a gradient of A (water), B (water-0.3% formic acid [pH adjusted to 9 using NH3; aqueous]), C (acetonitrile), and D (79.95% acetonitrile-19.95% water-0.1% TFA). The gradient is shown in Table S3. Analytes were detected at a UV wavelength of 254 nm.
Supplementary Material
ACKNOWLEDGMENTS
We thank Uwe Linne for support with HPLC-based analysis of UDP-sugars, Broder Rühmann and Manuel Döring for assistance in carbohydrate monomer composition analysis, and Elizaveta Krol, Matthew McIntosh, Pornsri Charoenpanich, and Helena Meier for sharing plasmids.
Financial support for this work was from the German Research Foundation (Collaborative Research Center 987) and the LOEWE program of the State of Hesse (Germany) (A.B.).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00801-18.
REFERENCES
- 1.Varki A. 2017. Biological roles of glycans. Glycobiology 27:3–49. doi: 10.1093/glycob/cww086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Götting C, Kuhn J, Zahn R, Brinkmann T, Kleesiek K. 2000. Molecular cloning and expression of human UDP-d-xylose:proteoglycan core protein beta-d-xylosyltransferase and its first isoform XT-II. J Mol Biol 304:517–528. doi: 10.1006/jmbi.2000.4261. [DOI] [PubMed] [Google Scholar]
- 3.Klutts JS, Doering TL. 2008. Cryptococcal xylosyltransferase 1 (Cxt1p) from Cryptococcus neoformans plays a direct role in the synthesis of capsule polysaccharides. J Biol Chem 283:14327–14334. doi: 10.1074/jbc.M708927200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Egelund J, Petersen BL, Motawia MS, Damager I, Faik A, Olsen CE, Ishii T, Clausen H, Ulvskov P, Geshi N. 2006. Arabidopsis thaliana RGXT1 and RGXT2 encode Golgi-localized (1,3)-alpha-d-xylosyltransferases involved in the synthesis of pectic rhamnogalacturonan-II. Plant Cell 18:2593–2607. doi: 10.1105/tpc.105.036566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Porchia AC, Sørensen SO, Scheller HV. 2002. Arabinoxylan biosynthesis in wheat. Characterization of arabinosyltransferase activity in Golgi membranes. Plant Physiol 130:432–441. doi: 10.1104/pp.003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Konishi T, Takeda T, Miyazaki Y, Ohnishi-Kameyama M, Hayashi T, O'Neill MA, Ishii T. 2007. A plant mutase that interconverts UDP-arabinofuranose and UDP-arabinopyranose. Glycobiology 17:345–354. doi: 10.1093/glycob/cwl081. [DOI] [PubMed] [Google Scholar]
- 7.Jankute M, Cox JA, Harrison J, Besra GS. 2015. Assembly of the mycobacterial cell wall. Annu Rev Microbiol 69:405–423. doi: 10.1146/annurev-micro-091014-104121. [DOI] [PubMed] [Google Scholar]
- 8.De Leizola M, Dedonder R. 1955. Various polysaccharides produced by Rhizobium strains. C R Hebd Seances Acad Sci 240:1825–1827. (In French). [PubMed] [Google Scholar]
- 9.Puvanesarajah V, Schell FM, Gerhold D, Stacey G. 1987. Cell surface polysaccharides from Bradyrhizobium japonicum and a nonnodulating mutant. J Bacteriol 169:137–141. doi: 10.1128/jb.169.1.137-141.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell GR, Reuhs BL, Walker GC. 2002. Chronic intracellular infection of alfalfa nodules by Sinorhizobium meliloti requires correct lipopolysaccharide core. Proc Natl Acad Sci U S A 99:3938–3943. doi: 10.1073/pnas.062425699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu X, Lee SG, Bar-Peled M. 2011. Biosynthesis of UDP-xylose and UDP-arabinose in Sinorhizobium meliloti 1021: first characterization of a bacterial UDP-xylose synthase, and UDP-xylose 4-epimerase. Microbiology 157:260–269. doi: 10.1099/mic.0.040758-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones KM, Kobayashi H, Davies BW, Taga ME, Walker GC. 2007. How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat Rev Microbiol 5:619–633. doi: 10.1038/nrmicro1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schäper S, Steinchen W, Krol E, Altegoer F, Skotnicka D, Søgaard-Andersen L, Bange G, Becker A. 2017. AraC-like transcriptional activator CuxR binds c-di-GMP by a PilZ-like mechanism to regulate extracellular polysaccharide production. Proc Natl Acad Sci U S A 114:E4822–E4831. doi: 10.1073/pnas.1702435114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niehaus K, Becker A. 1998. The role of microbial surface polysaccharides in the Rhizobium-legume interaction. Subcell Biochem 29:73–116. doi: 10.1007/978-1-4899-1707-2_3. [DOI] [PubMed] [Google Scholar]
- 15.Glazebrook J, Walker GC. 1989. A novel exopolysaccharide can function in place of the calcofluor-binding exopolysaccharide in nodulation of alfalfa by Rhizobium meliloti. Cell 56:661–672. doi: 10.1016/0092-8674(89)90588-6. [DOI] [PubMed] [Google Scholar]
- 16.Buendia AM, Enenkel B, Köplin R, Niehaus K, Arnold W, Pühler A. 1991. The Rhizobium meliloti exoZl exoB fragment of megaplasmid 2: ExoB functions as a UDP-glucose 4-epimerase and ExoZ shows homology to NodX of Rhizobium leguminosarum biovar viciae strain TOM. Mol Microbiol 5:1519–1530. doi: 10.1111/j.1365-2958.1991.tb00799.x. [DOI] [PubMed] [Google Scholar]
- 17.Putnoky P, Petrovics G, Kereszt A, Grosskopf E, Ha DT, Bánfalvi Z, Kondorosi A. 1990. Rhizobium meliloti lipopolysaccharide and exopolysaccharide can have the same function in the plant-bacterium interaction. J Bacteriol 172:5450–5458. doi: 10.1128/jb.172.9.5450-5458.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams MN, Hollingsworth RI, Brzoska PM, Signer ER. 1990. Rhizobium meliloti chromosomal loci required for suppression of exopolysaccharide mutations by lipopolysaccharide. J Bacteriol 172:6596–6598. doi: 10.1128/jb.172.11.6596-6598.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reuhs BL, Williams MN, Kim JS, Carlson RW, Côté F. 1995. Suppression of the Fix- phenotype of Rhizobium meliloti exoB mutants by lpsZ is correlated to a modified expression of the K polysaccharide. J Bacteriol 177:4289–4296. doi: 10.1128/jb.177.15.4289-4296.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keller M, Roxlau A, Weng WM, Schmidt M, Quandt J, Niehaus K, Jording D, Arnold W, Pühler A. 1995. Molecular analysis of the Rhizobium meliloti mucR gene regulating the biosynthesis of the exopolysaccharides succinoglycan and galactoglucan. Mol Plant Microbe Interact 8:267–277. doi: 10.1094/MPMI-8-0267. [DOI] [PubMed] [Google Scholar]
- 21.Bertram-Drogatz PA, Quester I, Becker A, Pühler A. 1998. The Sinorhizobium meliloti MucR protein, which is essential for the production of high-molecular-weight succinoglycan exopolysaccharide, binds to short DNA regions upstream of exoH and exoY. Mol Gen Genet 257:433–441. doi: 10.1007/s004380050667. [DOI] [PubMed] [Google Scholar]
- 22.Bahlawane C, Baumgarth B, Serrania J, Rüberg S, Becker A. 2008. Fine-tuning of galactoglucan biosynthesis in Sinorhizobium meliloti by differential WggR (ExpG)-, PhoB-, and MucR-dependent regulation of two promoters. J Bacteriol 190:3456–3466. doi: 10.1128/JB.00062-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bahlawane C, McIntosh M, Krol E, Becker A. 2008. Sinorhizobium meliloti regulator MucR couples exopolysaccharide synthesis and motility. Mol Plant Microbe Interact 21:1498–1509. doi: 10.1094/MPMI-21-11-1498. [DOI] [PubMed] [Google Scholar]
- 24.Mueller K, González JE. 2011. Complex regulation of symbiotic functions is coordinated by MucR and quorum sensing in Sinorhizobium meliloti. J Bacteriol 193:485–496. doi: 10.1128/JB.01129-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhan HJ, Lee CC, Leigh JA. 1991. Induction of the second exopolysaccharide (EPSb) in Rhizobium meliloti SU47 by low phosphate concentrations. J Bacteriol 173:7391–7394. doi: 10.1128/jb.173.22.7391-7394.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Charoenpanich P, Meyer S, Becker A, McIntosh M. 2013. Temporal expression program of quorum sensing-based transcription regulation in Sinorhizobium meliloti. J Bacteriol 195:3224–3236. doi: 10.1128/JB.00234-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schäper S, Krol E, Skotnicka D, Kaever V, Hilker R, Søgaard-Andersen L, Becker A. 2016. Cyclic di-GMP regulates multiple cellular functions in the symbiotic Alphaproteobacterium Sinorhizobium meliloti. J Bacteriol 198:521–535. doi: 10.1128/JB.00795-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rühmann B, Schmid J, Sieber V. 2014. Fast carbohydrate analysis via liquid chromatography coupled with ultra violet and electrospray ionization ion trap detection in 96-well format. J Chromatogr A 1350:44–50. doi: 10.1016/j.chroma.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 29.Pérez-Mendoza D, Rodríguez-Carvajal M, Romero-Jiménez L, Farias GA, Lloret J, Gallegos MT, Sanjuán J. 2015. Novel mixed-linkage β-glucan activated by c-di-GMP in Sinorhizobium meliloti. Proc Natl Acad Sci U S A 112:E757–E765. doi: 10.1073/pnas.1421748112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reinhold BB, Chan SY, Reuber TL, Marra A, Walker GC, Reinhold VN. 1994. Detailed structural characterization of succinoglycan, the major exopolysaccharide of Rhizobium meliloti Rm1021. J Bacteriol 176:1997–2002. doi: 10.1128/jb.176.7.1997-2002.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jenal U, Reinders A, Lori C. 2017. Cyclic di-GMP: second messenger extraordinaire. Nat Rev Microbiol 15:271–284. doi: 10.1038/nrmicro.2016.190. [DOI] [PubMed] [Google Scholar]
- 32.Low KE, Howell PL. 2018. Gram-negative synthase-dependent exopolysaccharide biosynthetic machines. Curr Opin Struct Biol 53:32–44. doi: 10.1016/j.sbi.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 33.Hamad MA, Di Lorenzo F, Molinaro A, Valvano MA. 2012. Aminoarabinose is essential for lipopolysaccharide export and intrinsic antimicrobial peptide resistance in Burkholderia cenocepacia. Mol Microbiol 85:962–974. doi: 10.1111/j.1365-2958.2012.08154.x. [DOI] [PubMed] [Google Scholar]
- 34.Mergaert P, Van Montagu M, Promé JC, Holsters M. 1993. Three unusual modifications, a d-arabinosyl, an N-methyl, and a carbamoyl group, are present on the Nod factors of Azorhizobium caulinodans strain ORS571. Proc Natl Acad Sci U S A 90:1551–1555. doi: 10.1073/pnas.90.4.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corbett D, Roberts IS. 2009. The role of microbial polysaccharides in host-pathogen interaction. F1000 Biol Rep 1:30. doi: 10.3410/B1-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Limoli DH, Jones CJ, Wozniak DJ. 2015. Bacterial extracellular polysaccharides in biofilm formation and function. Microbiol Spectr 3:MB-0011-2014. doi: 10.1128/microbiolspec.MB-0011-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmid J. 2018. Recent insights in microbial exopolysaccharide biosynthesis and engineering strategies. Curr Opin Biotechnol 53:130–136. doi: 10.1016/j.copbio.2018.01.005. [DOI] [PubMed] [Google Scholar]
- 38.Morgan JL, McNamara JT, Fischer M, Rich J, Chen HM, Withers SG, Zimmer J. 2016. Observing cellulose biosynthesis and membrane translocation in crystallo. Nature 531:329–334. doi: 10.1038/nature16966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krasteva PV, Bernal-Bayard J, Travier L, Martin FA, Kaminski PA, Karimova G, Fronzes R, Ghigo JM. 2017. Insights into the structure and assembly of a bacterial cellulose secretion system. Nat Commun 8:2065. doi: 10.1038/s41467-017-01523-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y, Moradali MF, Goudarztalejerdi A, Sims IM, Rehm BH. 2016. Biological function of a polysaccharide degrading enzyme in the periplasm. Sci Rep 6:31249. doi: 10.1038/srep31249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mazur O, Zimmer J. 2011. Apo- and cellopentaose-bound structures of the bacterial cellulose synthase subunit BcsZ. J Biol Chem 286:17601–17606. doi: 10.1074/jbc.M111.227660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whitney JC, Hay ID, Li C, Eckford PD, Robinson H, Amaya MF, Wood LF, Ohman DE, Bear CE, Rehm BH, Howell PL. 2011. Structural basis for alginate secretion across the bacterial outer membrane. Proc Natl Acad Sci U S A 108:13083–13088. doi: 10.1073/pnas.1104984108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Riley LM, Weadge JT, Baker P, Robinson H, Codée JD, Tipton PA, Ohman DE, Howell PL. 2013. Structural and functional characterization of Pseudomonas aeruginosa AlgX: role of AlgX in alginate acetylation. J Biol Chem 288:22299–22314. doi: 10.1074/jbc.M113.484931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thoden JB, Holden HM. 1998. Dramatic differences in the binding of UDP-galactose and UDP-glucose to UDP-galactose 4-epimerase from Escherichia coli. Biochemistry 37:11469–11477. doi: 10.1021/bi9808969. [DOI] [PubMed] [Google Scholar]
- 45.Frirdich E, Whitfield C. 2005. Characterization of Gla(KP), a UDP-galacturonic acid C4-epimerase from Klebsiella pneumoniae with extended substrate specificity. J Bacteriol 187:4104–4115. doi: 10.1128/JB.187.12.4104-4115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bhatt VS, Guo CY, Guan W, Zhao G, Yi W, Liu ZJ, Wang PG. 2011. Altered architecture of substrate binding region defines the unique specificity of UDP-GalNAc 4-epimerases. Protein Sci 20:856–866. doi: 10.1002/pro.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kotake T, Takata R, Verma R, Takaba M, Yamaguchi D, Orita T, Kaneko S, Matsuoka K, Koyama T, Reiter WD, Tsumuraya Y. 2009. Bifunctional cytosolic UDP-glucose 4-epimerases catalyse the interconversion between UDP-d-xylose and UDP-l-arabinose in plants. Biochem J 424:169–177. doi: 10.1042/BJ20091025. [DOI] [PubMed] [Google Scholar]
- 48.Bernatchez S, Szymanski CM, Ishiyama N, Li J, Jarrell HC, Lau PC, Berghuis AM, Young NM, Wakarchuk WW. 2005. A single bifunctional UDP-GlcNAc/Glc 4-epimerase supports the synthesis of three cell surface glycoconjugates in Campylobacter jejuni. J Biol Chem 280:4792–4802. doi: 10.1074/jbc.M407767200. [DOI] [PubMed] [Google Scholar]
- 49.Yin S, Kong JQ. 2016. Transcriptome-guided discovery and functional characterization of two UDP-sugar 4-epimerase families involved in the biosynthesis of anti-tumor polysaccharides in Ornithogalum caudatum. RSC Adv 6:37370–37384. doi: 10.1039/C6RA03817D. [DOI] [Google Scholar]
- 50.Schulz JM, Watson AL, Sanders R, Ross KL, Thoden JB, Holden HM, Fridovich-Keil JL. 2004. Determinants of function and substrate specificity in human UDP-galactose 4'-epimerase. J Biol Chem 279:32796–32803. doi: 10.1074/jbc.M405005200. [DOI] [PubMed] [Google Scholar]
- 51.Beerens K, Soetaert W, Desmet T. 2015. UDP-hexose 4-epimerases: a view on structure, mechanism and substrate specificity. Carbohydr Res 414:8–14. doi: 10.1016/j.carres.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 52.Thoden JB, Frey PA, Holden HM. 1996. Crystal structures of the oxidized and reduced forms of UDP-galactose 4-epimerase isolated from Escherichia coli. Biochemistry 35:2557–2566. doi: 10.1021/bi952715y. [DOI] [PubMed] [Google Scholar]
- 53.Frey PA, Hegeman AD. 2013. Chemical and stereochemical actions of UDP-galactose 4-epimerase. Acc Chem Res 46:1417–1426. doi: 10.1021/ar300246k. [DOI] [PubMed] [Google Scholar]
- 54.Fumeaux C, Radhakrishnan SK, Ardissone S, Théraulaz L, Frandi A, Martins D, Nesper J, Abel S, Jenal U, Viollier PH. 2014. Cell cycle transition from S-phase to G1 in Caulobacter is mediated by ancestral virulence regulators. Nat Commun 5:4081. doi: 10.1038/ncomms5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beringer JE. 1974. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol 84:188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- 56.Green MR, Sambrook J. 2012. Molecular cloning: a laboratory manual, 4th ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 57.Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Nat Biotechnol 1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 58.Krol E, Becker A. 2014. Rhizobial homologs of the fatty acid transporter FadL facilitate perception of long-chain acyl-homoserine lactone signals. Proc Natl Acad Sci U S A 111:10702–10707. doi: 10.1073/pnas.1404929111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 60.Robledo M, Rivera L, Jiménez-Zurdo JI, Rivas R, Dazzo F, Velázquez E, Martínez-Molina E, Hirsch AM, Mateos PF. 2012. Role of Rhizobium endoglucanase CelC2 in cellulose biosynthesis and biofilm formation on plant roots and abiotic surfaces. Microb Cell Fact 11:125. doi: 10.1186/1475-2859-11-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Valverde C, Hozbor DF, Lagares A. 1997. Rapid preparation of affinity-purified lipopolysaccharide samples for electrophoretic analysis. Biotechniques 22:230–236. doi: 10.2144/97222bm07. [DOI] [PubMed] [Google Scholar]
- 62.Behmüller R, Forstenlehner IC, Tenhaken R, Huber CG. 2014. Quantitative HPLC-MS analysis of nucleotide sugars in plant cells following off-line SPE sample preparation. Anal Bioanal Chem 406:3229–3237. doi: 10.1007/s00216-014-7746-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.