Abstract
The relapsing fever spirochete, Borrelia hermsii, escapes immune selection by alternating expression of surface lipoprotein alleles. The switch results from a duplicative transposition of one of several surface lipoprotein-encoding nucleotide sequences into the singular expression site. These nucleotide sequences constitute a large gene family whose diversity originated, in some cases, before the major divergences of Borrelia species. We have examined the B. hermsii vsp subfamily of alleles, which are carried on linear plasmids within each cell and maintained in several diverse copies as an antigenic archive. Each encodes a distinct serotype-specific protein. We sequenced more than 90% of the alleles within a single strain—B. hermsii strain HS1. A preponderance of allelic mosaicism suggests that intragenic recombination, coupled with selection imposed by host immune response, has driven diversification of the archived ensemble of vsp alleles. The recombinational diversification of vsp alleles generates change in the associated serotypes of the magnitude (30–40% amino acid differentiation) necessary for overcoming cross-reactivity of neutralizing antibodies. We conclude that evolution of vsp has occurred by punctuated occurrence of allelic differentiation, rather than by gradual selection of incremental point mutations that do not meet the threshold for antigenic diversity.
Keywords: homologous recombination‖antigenic variation‖evolution
Borrelia hermsii is a tick-borne spirochetal agent of relapsing fever (1), which is characterized by recurrences of febrile illness during the infection's course. These recurrences are the result of sequential alteration of the spirochete's outer-membrane proteins in an animal host with an adaptive immune system (2). Under selection by antibodies, the B. hermsii population changes from one dominant serotype to another serotype that is unaffected by the first wave of antibodies. This population-level change is driven by selective forces that may be similar to those acting on the lipoprotein encoded by the ospC locus in the Lyme disease spirochete, Borrelia burgdorferi (3, 4). However, contrary to the situation of ospC in B. burgdorferi, B. hermsii maintains the entire repertoire of its antigenic variants as an archive within a single cell, whereas the homologous genes in B. burgdorferi are single-copy loci distributed among different strains. As a consequence, frequency-dependent selection in B. hermsii can occur even in a clonal lineage, with no requirement for novel alleles from outside the clone.
The phenomenon by which B. hermsii thus alters its expressed surface proteins is one example of a common immune-evasion strategy known as antigenic variation. The term antigenic variation encompasses a variety of mechanisms, which are used by such pathogens as Neisseria gonorrhoeae, African trypanosomes, and Plasmodium falciparum to overcome immune response of their respective hosts (2). However, B. hermsii antigenic variation is unique when compared with antigenic variation of most closely studied bacteria (5), because the set of B. hermsii's sequentially expressed alleles constitute an archive located among the linear plasmids of the genome (6–9). Serotype switching results when one of these serotype-specific variants replaces another by duplicative transposition into the single expression site (10, 11). This situation contrasts with antigenic variation of N. gonorrhoeae, wherein only portions of the pilin genes are exchanged between alleles to generate novel proteins (12). These pilin constructs are ephemeral and because N. gonorrhoeae has a nearly panmictic population structure, many donor sequences come from outside of the clone (13). B. hermsii is therefore exceptional in having a set of enduring archived alleles maintained even within a single clone.
From experimental infections of immunocompetent mice starting with a single B. hermsii (strain HS1) cell, 28 antigenically distinct serotypes were isolated, and the nucleotide sequences of variable antigen genes of 26 serotypes were determined (9, 14). At least 25 of the sequences are alternately expressed from a single expression site and constitute the repertoire for antigenic variation (5). These sequences constitute two allele families that are readily distinguished by their lengths, and have been designated as variable short protein (vsp; n = 11) and variable long protein (vlp; n = 14). A lone gene, vsp33, is expressed from a different promoter on a different plasmid and does not have an archived version (15). Unlike the other vsp as well as vlp genes, vsp33 is expressed in the tick vector rather than in the mammal host (16).
In the absence of mixed infection with the opportunity for genetic exchange, a B. hermsii clone overcomes immune selection by maintaining its diverse antigenic archive. We sought to determine the mechanisms by which such a clone, specifically strain HS1, generates and maintains this diversity. We hypothesized that recombination among the archived alleles has driven their diversification and that balancing selection has maintained the archive. For the present study we restricted our focus to the 11 vsp alleles that are present in the B. hermsii HS1 clonal strain and that are sequentially expressed during infections of mammals.
Because of the peculiarities of B. hermsii genomic structure, the designation of gene and allele is unprecedented and warrants discussion. In B. hermsii there is a single functional unit consisting of the telomeric expression site in combination with one of several inserted vsp nucleotide sequences. This functional unit is the gene. The set of homologous archived vsp nucleotide sequences are alleles. As with alleles distributed among a population of genomes, B. hermsii vsp alleles may be modified through time or may be lost from the population altogether. The plasmid-borne vsp alleles are normally inherited as a unit during reproduction, and accordingly, some alleles may be modified or lost through successive generations. Although the biology of vsp expression is unusual in that it involves expression of multiple alleles within a single cell, expectations of polymorphism among these alleles are subsumed by classical Fisher–Wright-like sampling models.
Materials and Methods
DNA Sequences.
We examined nucleotide sequences of 11 expressed vsp alleles, which were cloned from B. hermsii strain HS1. The alleles and their GenBank accession numbers are as follows: vsp1, L33870; vsp2, L33897; vsp3, L04789; vsp6, L33898; vsp8, L33899; vsp11, L33900; vsp13, L33901; vsp22, L33902; vsp24, L04786; vsp26, L26497; and vsp27, L33903. The original clone began as a single cell of serotype 1, which expresses vsp1. The isolation of the different serotypes is described in ref. 14. In brief, this cell line was passed in a series of immunocompetent mice. As the mice underwent recurrences of bacteremia, bacteria of each relapse serotype were isolated, cloned by limiting dilution, and then used to start another infection. Serotype identity was originally determined with antisera and then by sequence analysis. Each of the 11 vsp sequences was derived from within the telomeric expression site (9, 14). For comparison with B. hermsii, we examined these vsp orthologues of a clonal isolate (strain Oz1) of the closely related relapsing fever agent Borrelia turicatae: vspA, U85413; vspB, AF049852; vspD, AF129737; vspE, AF130138; and vspF, AF130139.
Sequence Analysis.
The 11 B. hermsii and 5 B. turicatae vsp nucleotide sequences were initially aligned with the clustalx program (17) and corrections were made manually where the alignment algorithm failed to produce suitable alignments. We used the geneconv (version 1.81) program developed by one of us (S.A.S.) to test for evidence of intragenic recombination (18, 19). Analysis by the Poisson Random Field (PRF) model was performed with the prfmle program. Complete documentation of the geneconv and prfmle procedures and the programs themselves are available on the World Wide Web (http://www.math.wustl.edu/∼sawyer/mbprogs/). Descriptive statistics of the aligned nucleotide sequences and Tajima tests for departure from neutrality were performed with the DnaSP computer program, version 3.14 (20). For detecting the occurrence of nucleotide repeats within individual vsp sequences, we used the repeat program of the GCG package.
Results
Nucleotide Polymorphism.
The 11 B. hermsii vsp alleles ranged in size from 618 to 657 bp (mean = 647 bp), whereas the lengths of the 5 B. turicatae alleles were between 645 and 666 bp (mean = 654 bp). The nucleotide alignment of these alleles required the incorporation of 52 codon gaps corresponding to putative insertion-deletions (indels). The complete alignment of vsp alleles is available on the World Wide Web (http://130.64.245.82/PAGES/STEVE/downloads.html). The alignment of all 16 sequences measures 693 bp in length. Descriptive statistics describing the degree of polymorphism within each species are summarized in Table 1. The numbers of parsimony-informative (where each variant is present in at least two alleles) and singleton (variant occurs in only one sequence among the set) polymorphic sites as well as the number of different nucleotides at that site are indicated. Also shown are the values of Ka (average number of nonsynonymous polymorphisms per nonsynonymous site) and Ks (average number of synonymous polymorphisms per synonymous site).
Table 1.
Summary of nucleotide polymorphism among sets of vsp alleles from two Borrelia species
| Species | No. sites | No. variants per site
|
Ks* | Ka† | |||||
|---|---|---|---|---|---|---|---|---|---|
| Informative
|
Singleton
|
||||||||
| 2 | 3 | 4 | 2 | 3 | 4 | ||||
| B. hermsii (n = 11) | 591 | 114 | 91 | 26 | 85 | 12 | 3 | 0.4217 | 0.2570 |
| B. turicatae (n = 5) | 610 | 91 | 23 | 0 | 107 | 33 | 7 | 0.4081 | 0.2531 |
Informative, variant(s) occurs in two or more sequences; Singleton, variant(s) occurs in only one sequence.
Average number of synonymous substitutions per synonymous site.
Average number of nonsynonymous substitutions per nonsynonymous site.
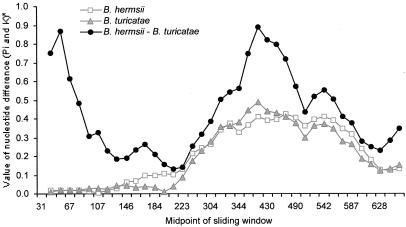
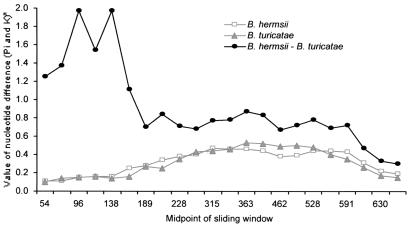
Sliding window analysis was applied to graphically depict the lack of uniformity of nucleotide differences across the length of the aligned vsp sequences. This method was performed separately for nonsynonymous (Fig. 1) and synonymous (Fig. 2) sites. Because there are an average of 3.5 times as many nonsynonymous sites as synonymous sites, we used different window sizes and sliding increments for each to give roughly the same degree of resolution along the length of the alignment. For nonsynonymous sites, the window measured 50 bp in length and was moved incrementally along the alignment in 10-bp steps. For synonymous sites, the window measured 20 bp in length and was moved incrementally along the alignment in 5-bp steps. The first 57 bp of these genes encode a signal peptide that is not present in the mature protein (21). As expected for a region under functional constraint, the first 200 bp are highly conserved within each species. The majority of intraspecific nucleotide polymorphism—both synonymous and nonsynonymous—is found in the region between nucleotides 200 and 620. In this region, the divergence between species exceeds the polymorphism within either species.
Figure 1.
Nonsynonymous nucleotide intraspecific polymorphism (Pi) and interspecific divergence (K) among vsp alleles based on a sliding window of 50 bp (moved in 10-bp increments). *The Jukes–Cantor correction.
Figure 2.
Synonymous nucleotide intraspecific polymorphism and interspecific divergence among vsp alleles based on a sliding window of 20 bp (moved in 5-bp increments). *The Jukes–Cantor correction.
Intragenic Recombination.
Recombination manifests itself by (i) breaking linkage associations among sites inside and outside of the recombinant fragment and (ii) making sequences identical at sites within the recombined region. Therefore, it is possible to detect recombinant events in the evolutionary history of a set of homologous gene sequences by analyzing the pattern of nucleotide similarity along their lengths.
We first tested whether nucleotide polymorphisms across the length of the 11 B. hermsii alleles are randomly associated with each other—i.e., in linkage equilibrium, as would be consistent with frequent recombination. We estimated a standard measure of linkage disequilibrium, D′ (22), for all 6,441 possible pairwise comparisons of the 114 phylogenetically informative sites with exactly two nucleotide variants. Among these comparisons, 84 were significantly different from random expectation by the two-tailed Fisher exact test (P < 0.05). The ratio 84/6441 = 0.0130 is less that the 0.05 proportion that one would expect under pure randomness. The smallest P value of the 6,441 comparisons was P = 0.0022, which, after the appropriate Bonferroni correction (23), is not remarkable for 6,441 tests or even for 114 polymorphic sites. The same procedure was applied to 10 random alignments that were generated by (i) constructing a random coalescent pedigree with 11 tips by using the Kingman–Hudson algorithm (24) and (ii) propagating a random 400-bp sequence from the root of the pedigree up to the tips, taking into account 275 possible mutations. The mutations were randomly placed on the pedigree and applied at randomly chosen sites. The randomization procedure generated a set of 10 randomly generated coalescent alignments with 11 sequences and roughly the same number of phylogenetically informative two-variant sites. These alignments yielded proportions of statistically significant 2 × 2 tables that varied between 0.2030 and 0.7303 as compared with the 0.0130 observed (data not shown). We therefore conclude that there is no evidence for linkage disequilibrium among the 11 B. hermsii vsp sequences as measured by this procedure. The same conclusion can be made for the alignment of 5 B. turicatae sequences, wherein 0 of the 4,095 pairwise D′ values were significant.
One of us (S.A.S.) has developed a means for discerning evidence of recombination among a set of aligned DNA or amino acid sequences (18, 25). The principle of the Sawyer test is that unidirectional intragenic exchange will result in regions of identity between the two sequences involved. The method is powerful because it is not contingent on assumptions about population dynamics and/or phylogeny. The only considerations are of the amount of DNA polymorphism and the pattern of similarity between paired sets of alleles. In brief, the method entails determining the lengths of identical fragments among paired alleles. The polymorphic sites of these alleles are then randomized 10,000 times to generate a random sample of fragments of identity given the observed number of polymorphic sites. The observed fragments are then compared with the permuted lengths to determine whether they are longer than expected by chance and to compute P values. The estimated P values are automatically multiple-comparison corrected for both the number of sequences and the sequence length (18, 24).
In our application of the Sawyer test to the vsp data, we excluded the first 57 bp, which correspond to the signal peptide. The sequence encoding the first several amino acids of the signal peptide is conserved among vsp and vlp alleles and is probably part of the upstream recombination regions (26). We identified 34 significant fragments (P < 0.05) in the remaining 636-bp alignment of B. hermsii and B. turicatae vsp alleles (Table 3, which is published as supporting information on the PNAS web site, www.pnas.org). Among the 34 significant fragments, 29 indicated gene conversion tracts between pairs of B. hermsii vsp alleles, whereas the other 5 gene conversions were among B. turicatae alleles. No gene conversion was detected between the two species' alleles. Each of the 11 B. hermsii vsp alleles shares at least one significant fragment with another allele from the same species, indicating that intragenic recombination is commonplace among the archive of a clonal lineage. Indeed, among all 55 pairwise allelic comparisons within B. hermsii, 27 (49%) involve at least one shared globally significant inner (GSI) fragment. This result affirms the hypothesis that intracellular recombination has occurred frequently in evolution of these alleles.
Repetitive Nucleotide Motifs.
Where sequence exchange between alleles is promiscuous, it may be difficult to discern the pattern of gene conversion by the Sawyer test or other statistical procedures. Among the vsp alleles only a single gene conversion—that of a 74-bp fragment between vsp22 and vsp24—was detected in the most variable region of the gene, i.e., between bases 237 and 572. If this region contains important epitopes, immune pressure may select against homogenizing effects of gene conversion. Alternatively, we hypothesized that the lack of statistically significant gene conversion tracts may be due to repeated occurrence of overlapping conversions and point mutations that mask their history.
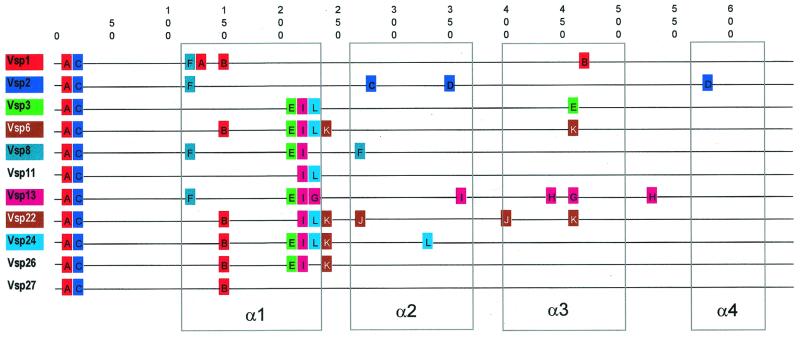
To detect recombination within the highly variable region, we incorporated a qualitative method that starts by searching for nonrandom occurrence of DNA repeats. We found several such repeats in the vsp alleles of both B. hermsii and B. turicatae (Table 2). We focused on the B. hermsii repeat sequences, which ranged in length from 10 to 12 bp. These sequences were arbitrarily assigned letter designations (A–L). Among the set of all B. hermsii vsp alleles, 8 of 11 contained one or more direct repeats. The probability of a chance occurrence of a 10-bp sequence appearing twice within a single sequence is about 10−6, and so the likelihood that the set of all repeats found among the 11 vsp alleles derives independently is vanishingly small. Each of these direct repeat DNA sequences also appeared as a single copy in at least one sequence other than that from which it occurs as a repeat. The distribution of these sequence motifs among the vsp alleles and their alignment is shown in Fig. 3. The positions of the sequence motifs are approximately to scale and their order is according to occurrence in respective alleles. Also shown are the relative positions of predicted α-helices in the mature proteins (27). Two of the repeats, A and C, occur first in the sequence encoding the signal peptide. The remainder, repeats B and D–L, occur only in sequence encoding the processed protein. The peptides corresponding to the repeats tend to cluster at the beginnings and ends of the α-helices as well as in the loops between helices.
Table 2.
Nucleotide repeats in B. hermsii vsp alleles
| Allele | Sequence | Length, bp | First occurrence | Second occurrence | Code |
|---|---|---|---|---|---|
| vsp1 | AATAAGTGCAA | 11 | 12–22 | 129–139 | A |
| AAGAGGCTAGTG | 12 | 146–157 | 452–463 | B | |
| vsp2 | AATAATAATG | 10 | 21–30 | 274–283 | C |
| GTTATTAAAAG | 11 | 342–352 | 558–568 | D | |
| vsp3 | CTTGCTAAAG | 10 | 211–220 | 445–454 | E |
| vsp8 | GCAAAAATAA | 10 | 124–133 | 272–281 | F |
| vsp13 | TTGGGAAAAA | 10 | 224–233 | 446–455 | G |
| TAAAGATAAA | 10 | 426–435 | 507–516 | H | |
| AAAGCTATTGG | 11 | 217–227 | 346–356 | I | |
| vsp22 | CTGATGCTGA | 10 | 263–272 | 392–401 | J |
| TAAAAATGATG | 11 | 237–247 | 456–466 | K | |
| vsp24 | GAAAATTAAA | 10 | 231–240 | 318–327 | L |
Figure 3.
Location of nucleotide motifs (as in Table 2) relative to predicted α-helical structures (α1–α4) of the 11 B. hermsii vsp alleles. Motifs are color-coded to match the vsp allele in which they occur more than once. Numbers indicate nucleotide position.
We concluded that the occurrence of these repeats is further evidence of past nonreciprocal exchange between sequences. Our reasoning is based on the assumption that those vsp sequences that contain repeats received these DNA fragments from some donor sequence among the archive. The most likely donor candidate is one of the vsp alleles that contains the sequence motif as a single copy. Alternatively, the recombination may derive from duplication of the motif in a given allele.
Test of Neutrality.
This genetic system—vsp sequences plus expression site—acts as a single, multiploid locus undergoing primarily vegetative reproduction. Only one allele at this locus is expressed at any one time, which is analogous to sex-chromosome silencing in diploid systems such as that observed for X chromosomes of Barr bodies in human females. While the biology of the system is unique, these conditions are subsumed by population genetic models such as the Fisher–Wright model by (i) correcting for effective population size where multiploidy is in evidence, and (ii) scaling the selection coefficients accordingly to account for gene silencing.
The terms population size and effective population size are commonly used in population genetics, but their definitions can be confusing because of the peculiar but standard way in which they are used. For example, a monoecious diploid animal population of size N with random selfing has effective populations size 2N. There are only N individuals, but the effective population size is 2N, because 2N is a count of the alleles at a given autosomal locus. However, the effective population size is not the same for all genes. Case in point, the effective population size at Y-chromosome loci is half the effective population size at autosomal loci, while loci located on the X chromosome have an effective population size that is somewhere in between. In the present case, if the Borrelia population consists of Nb organisms, each with an average “satchel” size of m vsp alleles, then the effective population size is approximately mNb. At other nonmultiploid genetic loci (i.e., where m is close to 1), the effective population size will be close to Nb. Because all of the arguments in our paper are concerned only with the vsp locus, all population scalings are in terms of the same effective population size.
We used the method of Tajima (28) to determine whether the pattern of nucleotide polymorphisms among the aligned set of vsp alleles is consistent with neutral expectations. The test is based on two different estimators of the scaled mutation rate θ. One estimator is the average heterozygosity (k). The second is S/a1, where S is the number of segregating sites and a1 is a constant. If the alleles are evolving neutrally without recombination, then the difference d = k − S/a1 will have mean zero. Tajima's statistic DT, which is a scaled version of d, can be used to determine whether nonneutral evolution is in evidence. If DT < 0 significantly, the difference is attributable to purifying selection. If DT > 0, balancing selection or selection for diversification of alleles is indicated. Although the distribution of the DT statistic is derived under the assumptions of panmixia and haploidy, it is often applied under related assumptions such as diploidy with an appropriate definition of the effective population size. Indeed, in his original description of the method, Tajima gave examples of the test applied to haploid and diploid systems (28). We surveyed the Institute for Scientific Information (ISI, Web of Science) citation database and found more than 480 publications citing this method. Among these applications, the Tajima test has been used to detect nonneutral evolution in genes of organisms that are haploid (e.g., ref. 29), diploid (e.g., ref. 30), of tetraploid (e.g., ref. 31). It has even been applied to mitochondrial genes (e.g., ref. 32), a situation that is probably most similar to ours, with one organism, indeed one cell, harboring many allelic copies of the gene in each mitochondrial genome. Ours is a unique application to a multiploid bacterial gene system, but it follows the precedent of other studies.
We applied the Tajima test to the entire aligned set of 11 aligned B. hermsii vsp alleles. No significant departure from neutral expectations was observed (DT = 1.21270, P > 0.10). However, if recombination is frequent among the vsp alleles, different regions may be evolving independently. If true, then selection may not be detected for the alignment as a whole, but certain portions of this alignment may be significant. Accordingly, we applied a sliding window approach to detect whether shorter regions may be significant. The sliding window size of 50 bp was chosen so that each window would be smaller than the mean length (77 bp) of the significant recombinant fragments detected by the Sawyer test. Sites with alignment gaps were excluded so that all windows have the same number of nucleotides. Among the complete set of 53 adjacent windows, 8 had values significant at the P < 0.10 level, and 4 were significant at the P < 0.05 level (28). One of the significant negative values of DT was that for the first window (base pairs 1–63), corresponding to the signal peptide. The remaining significant values of D were all positive and constituted 11 of the 12 windows between base pairs 241 and 543 in the alignment, the most polymorphic region.
Where there is apparent independence among the nucleotide polymorphisms, as seen in the vsp alleles, one can use the Poisson Random Field model to test whether these polymorphisms are evolving under neutral expectations (33, 34). Under the null model, new mutant codons arise at an expected rate μ > 0 and have a relative selective advantage of γ > 0 with respect to previously fixed codons at the same codon position. One can then estimate μ and γ to test the null hypothesis that γ = 0. For vsp alleles, the value of γ is essentially the selective advantage of the B. hermsii spirochete divided by the proportion of time that a particular allele is expressed. Scaling of selection coefficients amounts to multiplying γ by a constant value that corrects for the rates of gene silencing. Deriving this coefficient is nontrivial because several alleles may be expressed for various lengths of time in each B. hermsii, however for our application, it is only necessary to know the sign (+ or −) of the unscaled values to determine the nature the selective pressure. Analysis of the 11 B. hermsii vsp alleles leads to an estimate of γ = 2.70 ± 2.54 (95% confidence interval) for γ scaled by the effective population size, but rejects the null hypothesis γ = 0 with P = 4.0 × 10−4 by a likelihood ratio test. This result means that rare codons have a highly significantly tendency to be selectively advantageous. In contrast, most Escherichia coli gene alignments either show nonsignificant results for γ or else have a significant γ < 0, indicating purifying selection (35). Thus, as with the results from Tajima tests, the Poisson Random Field model indicates that diversifying selection is acting on the set of B. hermsii vsp alleles.
Discussion
Borrelia and other bacterial pathogens are confronted by the problem of maintaining a sufficient diversity of antigens to survive immune recognition long enough to be transmitted to a new host. One solution to this problem is the B. burgdorferi ospC system, in which allelic variants are maintained among populations of different strains. Because the vectors of B. burgdorferi feed on more than one host during their life cycle, these ticks and their different mammalian and avian hosts frequently harbor multiple strains of B. burgdorferi (36). This situation presents the opportunity for gene transfer of ospC between strains (3). When genetic exchange is coupled with frequency-dependent selection among strains, it acts to maintain a high degree of ospC variation in local populations of B. burgdorferi (3).
Transmission of B. hermsii and the other relapsing fever agents occurs within a different ecological context (1, 37). The soft tick vectors of relapsing fever spirochetes are locally restricted in their home ranges. Transmission occurs primarily within the nest of the mammalian host. The opportunities for horizontal exchange between B. hermsii strains are rare, because of the intimate association of the vector and host (38). A survey of polymorphism of four genetic loci sampled from 26 B. hermsii strains from California, Idaho, Washington, Utah, Colorado, and British Columbia showed that these populations are clonal with very little evidence of genetic exchange (S. Porcella and T. Schwan, personal communication). This sample covered nearly the entire range of the B. hermsii tick vector, Ornithodoros hermsi, and was therefore likely to be inclusive of the entire range of B. hermsii. In another study of isolates from Washington, California, and Idaho, Hinnebusch et al. (38) found that B. hermsii comprise three major groups based on their repertoire of vsp and vlp alleles. These three groups appear to be distinct, and although they may co-occur locally, there was little evidence that either vsp or vlp alleles are exchanged between them.
Lacking the opportunity for genetic exchange that other pathogens such as N. gonorrhoeae experience (13), B. hermsii has evolved genomic innovations that permit each and every cell to maintain an antigenic archive of its vsp alleles. Accordingly, B. hermsii lineages that maintain several, antigenically distinct vsp alleles within their genome will most successfully escape immune clearance. We found that B. hermsii generates and maintains its vsp allelic repertoire by mechanisms that are analogous to those by which B. burgdorferi ospC variation arises and persists. In both instances, genetic exchange generates novel gene sequences that are maintained by balancing selection. However, the exchange and selection among vsp occurs within a single cell of a single strain, rather than between cells of different strains.
Three lines of evidence indicate that genetic exchange, which is most consistent with gene conversion, has occurred among the B. hermsii vsp alleles: (i) The nucleotide polymorphisms along the length of these alleles are in linkage equilibrium. (ii) The Sawyer test reveals numerous tracts of identity between pairs of sequences that are not attributable to chance. (iii) Numerous short nucleotide repeats suggest a pattern of frequent intragenic recombination. While the pattern of nucleotide polymorphism among these alleles may indicate their past exchanges, it does not permit us to discern the molecular mechanisms of this exchange. The occurrence of the short repetitive motifs may prove interesting in the latter regard. These short motifs may be merely an artifact of past gene conversion, or they might play a more direct role in the molecular mechanisms of intragenic recombination. Interestingly, several of the repeat sequences we have identified in the B. hermsii vsp alleles also occur as single copies in the vsps of B. turicatae as well as in the B. hermsii vsp33. However, among these interspecific examples, the motifs occur in very different regions of the molecule so that while the sequences of these motifs are conserved their locations are not. The conservative nature of these sequences suggests that they may have some functional relevance, perhaps as targets of enzyme-mediated recombination. Other direct repeats have been demonstrated to be the substrates for a DNA rearrangement in B. hermsii (9). Furthermore, the single-strand annealing mechanisms of recombination in bacteriophage, such as the recE pathway of E. coli and analogous pathways in some other bacteria, typically act upon direct repeats (39).
Our conclusion that intragenic recombination of archived alleles within a cell is the primary source of variation does not exclude the possibility that some variation could have come from a remote lateral gene transfer. However, we have identified 34 nucleotide sequences that were most likely the result of a recombination, for which both the donor and recipient of gene conversion were present in the HS1 clone. We maintain that occurrence of these fragments by intraorganismal exchange is the more parsimonious interpretation than the alternative explanation whereby each fragment was introduced (at least) twice into the clone from some outside donor. Moreover, analyses of population structure among multiple strains of B. hermsii indicate that lateral transfer of variable surface proteins is extremely rare (38).
Although gene conversion does not increase overall nucleotide polymorphism per se, it may redistribute existing polymorphic sites into configurations that alter the encoded protein in an immunologically relevant manner. For example, novel arrangements of peptides in epitopes may abrogate antibody recognition. In the case of conformational epitopes, the polymorphisms need not be adjacent in primary structure to change the epitope. A single peptide occurring at different positions in different VSP proteins may create distinct epitopes.
Kitten et al. (40) showed that chimeric molecules form between expressed and archived copies of B. hermsii vlp alleles, but that the resulting mosaic alleles are not retained in the antigenic archive because the exchange occurs at the expression site. Here we have shown that the vsp alleles are also mosaic products of numerous recombinant events. Since many of the expressed vsp alleles have a nearly identical archived counterpart, the diversity-generating recombination occurs, at least in part, between archived vsp alleles (8).
During the recombination leading to mosaicism in archived vsp alleles, a certain proportion of the resulting allelic variants may acquire deleterious mutations (e.g., nonsense mutations resulting in a truncated reading frame) that are selected against as populations expand (8). On the other hand, rearrangements that result in a functional full-length protein that is not bound by circulating antibodies of the host will be favored. Presumably, cell lineages with more diverse and larger archives of antigens will fare better in confronting immune response than lineage with more limited ones.
For antigen switching to be effective, each allele of the antigenic ensemble must differ sufficiently from the others so as to present a distinct antigen and thus avoid the current immune response. Studies with polyclonal antisera indicate that nondenatured globular proteins differing by 30–40% in their amino acid sequences will not cross-react (41, 42). The pairwise percent difference in the amino acids of 11 VSP sequences range from 21.0% to 53.3%, with a mean value of 38.2%. In fact, only one pairwise comparison, that of VSP2–VSP22, is less than 27%. Therefore, it would appear that a switch involving any two vsp alleles would change the presented antigen sufficiently to avoid cross-reactivity and evade immune response. Empirical evidence shows a lack of antigenic cross-reactivity among the 11 B. hermsii serotypes associated with these vsp alleles (14).
In the context of selection by polyclonal antibodies, most individual nucleotide polymorphisms will be of little consequence because of the requirement for a minimum of 30–40% difference. Alleles would need to be retained for exceedingly long periods of time before they would reach the 30% amino acid differences necessary to confer advantage to the cell. If B. hermsii has evolved with the constant pressure of immune selection, such incremental single-nucleotide changes in vsp alleles seem an implausible model for explaining their diversity. This is not to say that point mutations are not relevant to the allelic diversification, but rather that they must occur in concert with the larger-magnitude mutations of intragenic recombination.
Immune evasion is not the only possible driving force of selection for a diverse array of vsp alleles. B. turicatae cells expressing either vspA or vspB alleles differ in their degree of virulence and tissue association (43). Those spirochetes expressing vspA appear to be more neurotropic, whereas those expressing vspB achieve high densities in the blood. Selection for a niche diversity in the host, such as the particular tissue tropism of cells expressing vspA, may also serve to maintain the diversity of the allelic archive.
In summary, we have found that B. hermsii seems to be fine-tuned to a clonal existence, wherein a complete, although perhaps minimal, ensemble of archived vsp alleles is maintained. In this genetic system, new vsp alleles arise by intracellular recombination among the archived set. To overcome barriers to genetic exchange between populations of cells, these clonal spirochetes harbor a “population” of archived antigenic determinants within the cell, and by a process analogous to that of sexual recombination, they generate novel allelic arrangements to confront pressure imposed by host immune systems. While such sex-like processes, including transformation and conjugation, are well known to occur in prokaryotes, B. hermsii is unusual in that the entire process takes place within a cell. These adaptations are most probably important contributors to the overall success of the species, and we speculate that similar recombinational processes may have led to gene diversification not only within B. hermsii but also in the genus as a whole.
Supplementary Material
Acknowledgments
We are grateful to Ana Rita Ponce and three anonymous reviewers for comments on early drafts of this manuscript. This work was supported by National Institutes of Health Grant AI60759 to S.M.R., National Science Foundation Grant DMS9707045 to S.A.S., and National Institutes of Health Grant AI24424 to A.G.B.
References
- 1.Barbour A G, Hayes S F. Microbiol Rev. 1986;50:381–400. doi: 10.1128/mr.50.4.381-400.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbour A G, Restrepo B I. Emerg Infect Dis. 2000;6:449–457. doi: 10.3201/eid0605.000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang I N, Dykhuizen D E, Qiu W, Dunn J J, Bosler E M, Luft B J. Genetics. 1999;151:15–30. doi: 10.1093/genetics/151.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qiu W G, Bosler E M, Campbell J R, Ugine G D, Wang I N, Luft B J, Dykhuizen D E. Hereditas. 1997;127:203–216. doi: 10.1111/j.1601-5223.1997.00203.x. [DOI] [PubMed] [Google Scholar]
- 5.Barbour A G. In: Mobile DNA II. Craig N L, Craigie R, Gellert M, Lambowitz A, editors. Washington, DC: Am. Soc. Microbiol.; 2001. , in press. [Google Scholar]
- 6.Barbour A G, Tessier S L, Stoenner H G. J Exp Med. 1982;156:1312–1324. doi: 10.1084/jem.156.5.1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plasterk R H, Simon M I, Barbour A G. Nature (London) 1985;318:257–263. doi: 10.1038/318257a0. [DOI] [PubMed] [Google Scholar]
- 8.Restrepo B I, Barbour A G. Cell. 1994;78:867–876. doi: 10.1016/s0092-8674(94)90642-4. [DOI] [PubMed] [Google Scholar]
- 9.Restrepo B I, Carter C J, Barbour A G. Mol Microbiol. 1994;13:287–299. doi: 10.1111/j.1365-2958.1994.tb00423.x. [DOI] [PubMed] [Google Scholar]
- 10.Kitten T, Barbour A G. Proc Natl Acad Sci USA. 1990;87:6077–6081. doi: 10.1073/pnas.87.16.6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kitten T, Barbour A G. Genetics. 1992;132:311–324. doi: 10.1093/genetics/132.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swanson J, Belland R J, Hill S A. Curr Opin Genet Dev. 1992;2:805–811. doi: 10.1016/s0959-437x(05)80143-1. [DOI] [PubMed] [Google Scholar]
- 13.Smith J M, Smith N H, O'Rourke M, Spratt B G. Proc Natl Acad Sci USA. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stoenner H G, Dodd T, Larsen C. J Exp Med. 1982;156:1297–1311. doi: 10.1084/jem.156.5.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barbour A G, Carter C J, Sohaskey C D. Infect Immun. 2000;68:7114–7121. doi: 10.1128/iai.68.12.7114-7121.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwan T G, Hinnebusch B J. Science. 1998;280:1938–1940. doi: 10.1126/science.280.5371.1938. [DOI] [PubMed] [Google Scholar]
- 17.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sawyer S. Mol Biol Evol. 1989;6:526–538. doi: 10.1093/oxfordjournals.molbev.a040567. [DOI] [PubMed] [Google Scholar]
- 19.Padidam M, Sawyer S, Fauquet C M. Virology. 1999;265:218–225. doi: 10.1006/viro.1999.0056. [DOI] [PubMed] [Google Scholar]
- 20.Rozas J, Rozas R. Comput Appl Biosci. 1997;13:307–311. [PubMed] [Google Scholar]
- 21.Carter C J, Bergström S, Norris S J, Barbour A G. Infect Immun. 1994;62:2792–2799. doi: 10.1128/iai.62.7.2792-2799.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewontin R C. Genetics. 1964;49:46–67. doi: 10.1093/genetics/49.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weir B S. Genetic Data Analysis II. Sunderland, MA: Sinauer; 1996. [Google Scholar]
- 24.Hudson R R. Oxford Surv Evol Biol. 1990;7:1–44. [Google Scholar]
- 25.Hartl D L, Sawyer S A. J Evol Biol. 1991;4:519–532. [Google Scholar]
- 26.Barbour A G, Burman N, Carter C J, Kitten T, Bergström S. Mol Microbiol. 1991;5:489–493. doi: 10.1111/j.1365-2958.1991.tb02132.x. [DOI] [PubMed] [Google Scholar]
- 27.Zückert W R, Kerentseva T A, Lawson C L, Barbour A G. J Biol Chem. 2001;276:457–463. doi: 10.1074/jbc.M008449200. [DOI] [PubMed] [Google Scholar]
- 28.Tajima F. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Escalante A A, Lal A A, Ayala F J. Genetics. 1998;149:189–202. doi: 10.1093/genetics/149.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsaur S C, Ting C T, Wu C I. Mol Biol Evol. 1998;15:1040–1046. doi: 10.1093/oxfordjournals.molbev.a026002. [DOI] [PubMed] [Google Scholar]
- 31.Kawabe A, Yamane K, Miyashita N T. Genetics. 2000;156:1339–1347. doi: 10.1093/genetics/156.3.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rand D M, Kann L M. Genetica. 1998;102–103:393–407. [PubMed] [Google Scholar]
- 33.Sawyer S A, Hartl D L. Genetics. 1992;132:1161–1176. doi: 10.1093/genetics/132.4.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sawyer S A. In: Progress in Population Genetics and Human Evolution. Donnelly P, Tavare S, editors. New York: Springer; 1997. pp. 193–205. [Google Scholar]
- 35.Hartl D L, Moriyama E N, Sawyer S A. Genetics. 1994;138:227–234. doi: 10.1093/genetics/138.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guttman D S, Wang P W, Wang I N, Bosler E M, Luft B J, Dykhuizen D E. J Clin Microbiol. 1996;34:652–656. doi: 10.1128/jcm.34.3.652-656.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Felsenfeld O. Borrelia: Strains, Vectors, Human and Animal Borreliosis. St. Louis: Greene; 1971. [Google Scholar]
- 38.Hinnebusch B J, Barbour A G, Restrepo B I, Schwan T G. Infect Immun. 1998;66:432–440. doi: 10.1128/iai.66.2.432-440.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuzminov A. Microbiol Mol Biol Rev. 1999;63:751–813. doi: 10.1128/mmbr.63.4.751-813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kitten T, Barrera A V, Barbour A G. J Bacteriol. 1993;175:2516–2522. doi: 10.1128/jb.175.9.2516-2522.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson A C, Carlson S S, White T J. Annu Rev Biochem. 1977;46:573–639. doi: 10.1146/annurev.bi.46.070177.003041. [DOI] [PubMed] [Google Scholar]
- 42.Reichlin M. Adv Immunol. 1975;20:71–123. doi: 10.1016/s0065-2776(08)60207-2. [DOI] [PubMed] [Google Scholar]
- 43.Pennington P M, Allred C D, West C S, Alvarez R, Barbour A G. Infect Immun. 1997;65:285–292. doi: 10.1128/iai.65.1.285-292.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.