Abstract
In Drosophila, enhancer trap strategies allow rapid access to expression patterns, molecular data, and mutations in trapped genes. However, they do not give any information at the protein level, e.g., about the protein subcellular localization. Using the green fluorescent protein (GFP) as a mobile artificial exon carried by a transposable P-element, we have developed a protein trap system. We screened for individual flies, in which GFP tags full-length endogenous proteins expressed from their endogenous locus, allowing us to observe their cellular and subcellular distribution. GFP fusions are targeted to virtually any compartment of the cell. In the case of insertions in previously known genes, we observe that the subcellular localization of the fusion protein corresponds to the described distribution of the endogenous protein. The artificial GFP exon does not disturb upstream and downstream splicing events. Many insertions correspond to genes not predicted by the Drosophila Genome Project. Our results show the feasibility of a protein trap in Drosophila. GFP reveals in real time the dynamics of protein's distribution in the whole, live organism and provides useful markers for a number of cellular structures and compartments.
A key to understanding the mechanisms of development of an organism is to detect the dynamic changes of gene expression in its different territories. The clarification of the function of a gene also requires the knowledge of the subcellular localization of its protein product. Although antibodies that specifically recognize a protein provide a great amount of information, their generation requires molecular information about the gene and they can be used only in fixed tissues. Ectopic expression of tagged versions of the protein, in particular fusions to autofluorescent tags such as the green fluorescent protein (GFP; ref. 1) and its rainbow of derivatives, allows a dynamic study of the fusion product's behavior in unfixed, living cells and tissues, but still relies on molecular information.
Several groups have reported the generation of cDNA–GFP fusion libraries and their ectopic expression in cultured mammalian cells and plants (2, 3), allowing the generation of information about protein localization on a large scale. These systems use ubiquitous promoters and do not provide any information about endogenous transcriptional regulations during cell cycle or developmental stages. In yeast, a large-scale protein trap screen was performed by using genomic fragments fused to a GFP reporter, providing information on both the protein subcellular localization and its developmental regulation, albeit in a unicellular organism (4).
Insertional mutagenesis, using the random insertion in a genome of a promoter-less reporter to detect a gene or a protein's expression pattern, has been used in a wide range of organisms, including plants (5, 6), mice (7, 8), frogs (9), and fish (10–12). The gene trap reporter is expressed as a fusion with the endogenous messenger transcribed from its own promoter. In some “protein trap” schemes, the reporter lacks an initiation codon and is fused with the N-terminus portion of the endogenous protein. The fusion retains localization sequences contained in the amino-terminal region of the trapped protein. This approach has been used in the mouse by using β-galactosidase (13, 14) and in cultured cells by using GFP (15).
In Drosophila, enhancer trap has been the preferred insertional mutagenesis method for over a decade (16–20). A reporter flanked by a weak promoter, usually carried by a P-element transposon, is transposed randomly to a large number of chromosomal locations. When it integrates near a gene enhancer sequence, the reporter is expressed in the same pattern as the endogenous gene controlled by the enhancer. Recently, a gene trap has been developed, in which the reporter gene does not contain a minimal promoter and is expressed only when it integrates within the trapped gene's expressed sequences (21). In this case, the reporter is expected to reproduce the complete transcription pattern of the trapped gene. No bona fide protein trap, which has the potential of reporting the subcellular localization of the endogenous proteins, has been described so far in flies.
In this article, we show that a protein trap approach, in which full-length endogenous proteins are expressed as GFP fusion proteins from their endogenous promoters, is feasible in Drosophila. We describe the generation of a transposable artificial exon encoding a GFP reporter. Devoid of initiation and stop codons and flanked by splice acceptor and donor sites, its insertion into an intron separating coding exons results in the production of a chimeric protein in which GFP is fused with both the amino and carboxyl termini of the trapped protein. We generated several hundred independent lines and show, in the case of known molecules, that the chimera's subcellular distribution reflects that of the wild-type endogenous protein. The use of GFP allows a dynamic study of this distribution in live tissues. Interestingly, we find that many insertions lie in loci that were not predicted by the algorithms used in the Drosophila Genome Project. We report on a system that allows detection of the distribution of “full-length” fusion proteins expressed from their own promoter in a living multicellular organism.
Methods
DNA Constructs.
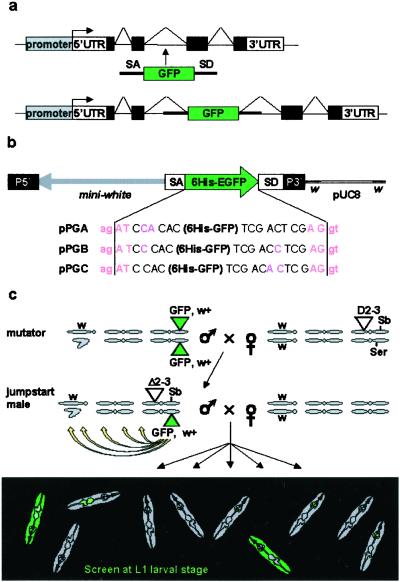
The three vectors are described in Fig. 1b. The GFP used is enhanced GFP from CLONTECH. Details of the construction scheme are available on request.
Figure 1.
The protein trap screen strategy. (a) Principle of the artificial exon: see text for details. (b) The PTTs. In addition to the 6His-GFP reporter flanked by splicing sequences, the P-element contains a miniwhite selection gene in the opposite orientation. In each of the three constructs GA, GB, and GC, the splice acceptor (ag | AT) and splice donor (AG | gt) consensus sequences are in a different reading frame relative to the 6His-GFP sequence. Although slightly different from the AG/GT acceptor splice consensus, AG/AT is the second most commonly found in Drosophila (31). (c) Crossing scheme used to generate GFP-positive flies. Flies are selected on the occurrence of a GFP signal. We used mutator lines with a “nonfluorescent” insertion on the third chromosome and no counter selection against the transposase or the starting chromosome. As a result, insertions on all three chromosomes can be recovered, including unstable insertions on the Delta2–3Sb chromosome or new insertions on the starting chromosome.
Screening Procedure.
Embryos were collected for 24 h on 2.5% agarose/grape juice plates, aged for 24 h into L1, and screened directly under a Wild MZ12 FlIII dissecting microscope (Leica, Deerfield, IL) at high magnification. Larvae were starved between hatching and screening to avoid autofluorescence caused by food ingestion. Daily egg collections were obtained over 7–10 days from cages of 15 mutator males mated with 30–40 yw females. Five thousand larvae could be routinely screened in 1 h. To minimize redundancy in our collection, we tried to select from individual cages only larvae with different patterns. GFP-positive larvae were recovered, and surviving adults were mated to yw flies. After a secondary screening, GFP+ progeny with the clearest eye color were selected to reduce the occurrence of multiple insertions and balanced.
Confocal Imaging of Living Embryos and Tissues.
Embryos were dechorionated manually and mounted in halocarbon oil between slide and coverslips separated by a coverslip spacer. Muscle fibers were dissected from adult thoracic indirect flight muscles and observed in 80% glycerol. Images were acquired with Bio-Rad MRC 600, Bio-Rad MRC 1024, or Olympus SV500 laser confocal systems.
Identification of the Trapped Genes.
Genomic sequences flanking the P-element insertion site were recovered by inverse PCR as described by the Berkeley Drosophila Genome Project, with the set of oligonucleotides used for EP constructs (http://www.fruitfly.org/about/methods/inverse.pcr.html). These sequences were used in blast searches against the Drosophila Genome Database.
Reverse Transcriptase–PCR.
Poly(A)+-RNA was isolated from late-stage embryos or larvae, by using a QuickPrep Micro mRNA purification kit (Amersham Pharmacia). cDNAs were prepared by using Superscript II Reverse Transcriptase (GIBCO/BRL). Oligonucleotide sequences and PCR conditions are available on request.
Results
Construction of the Protein Trap Transposon (PTT) and Generation of GFP-Positive Lines.
The PTT is a P-element designed to randomly tag proteins with an enhanced GFP, without disrupting their subcellular localization. It carries an artificial exon encoding GFP, deprived of initiation and stop codons, and flanked by splice acceptor and donor sequences (Fig. 1 a and b). Upon insertion into an intron, the splice donor and acceptor sequences regenerate an intron on each side of the GFP. GFP sequences are conserved in the mature mRNA. Translation results in a fusion of the GFP to both the amino- and carboxyl-terminal parts of the trapped protein. The chimera retains localization properties of the wild-type protein, except when the GFP disrupts a domain necessary for subcellular targeting. Because exon-intron boundaries can occur in each of the three reading frames, we constructed three vectors (Fig. 1b) with GFP in each reading frame relative to both splice sites. We used “strong” splice sites known to trigger preferential splicing of exon 17 to exon 19 over exon 18 in the fly myosin heavy chain II gene (22).
The three constructs were introduced into the fly germ line. Introns represent approximately one-sixth of the genome (20 of 120 Mb of euchromatin; ref. 23), but because P-element transposons tend to integrate preferentially into 5′ regions of genes (24), we anticipated a relatively low frequency of GFP-positive integrations. Besides, some introns are located outside of the protein coding sequences, and only one of six insertions in the remaining set of introns is expected to produce an in-frame GFP fusion. To counterbalance these limiting factors, we selected “mutator” lines with the highest frequency of transposition to new chromosomal positions (Table 1). These mutator lines do not express any detectable levels of GFP. The PTT was then mobilized to create GFP-positive insertions (see crossing scheme in Fig. 1c and Methods). GFP-positive larvae were recovered at first-instar larval stage at a frequency of 1/1,540–1,800 (Table 1). More than 600 lines obtained from independent parents were conserved.
Table 1.
Transposition rate and frequency of GFP+ insertions
| Construct | Mutator line | Sb-w+/Sb tot | Tranposition efficiency (%) | Green frequency |
|---|---|---|---|---|
| P-GA | GAIII-1b | 41/252 | 16.3 | 1/1,540 |
| P-GB | GBIII-3a | 5/144 | 3.5 | nd |
| GBIII-3b | 24/246 | 9.6 | 1/1,785 | |
| GBIII-5 | 5/183 | 2.7 | nd | |
| P-GC | GCIII-1 | 2/228 | 0.9 | nd |
| GCIII-3 | 4/294 | 1.4 | nd | |
| GCIII-4a | 2/104 | 1.9 | nd | |
| GCIII-4b | 41/227 | 18.1 | 1/1,600 | |
| GCIII-5 | 2/124 | 1.6 | nd |
To select mutator lines with high transposition frequency, jumpstart males with the PTT, w+/Delta2-3Sb genotype were mated with yw virgin females (see Fig. 1c). The transposition frequency was scored among the Sb progeny as the percentage of individuals showing a variegated eye phenotype. Sb flies have inherited the Delta2-3Sb chromosome III, and not the jumpstart chromosome III, from their father. The presence of the w+ marker in Sb individuals can therefore only result from a transposition of the PTT-w+ from its original localization on the jumpstart chromosome to a new position. The green frequency is the number of GFP-positive insertions divided by the total number of larvae screened. For each mutator line, the figures were calculated in the beginning of the screen out of a total number of approximately 40,000 larvae. nd, not determined.
Trapped Proteins Are Targeted to Specific Subcellular Compartments.
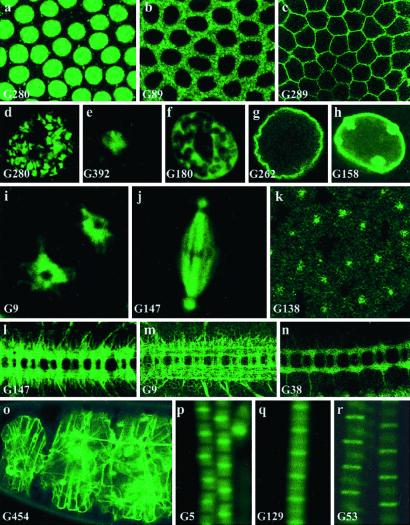
Using confocal microscopy, we investigated the subcellular distribution of the GFP reporter during embryonic stages of development in 380 of the fluorescent lines generated. As expected, a GFP signal could be detected in different cellular compartments; a few examples are shown in Fig. 2. Fig. 2 a–c shows signals specifically located in the nucleus (Fig. 2a), cytoplasm (Fig. 2b), and plasma membrane (Fig. 2c). Within the nucleus, targeting to the chromatin, nucleolus, nuclear matrix, and nuclear membrane were observed (Fig. 2 d–h). We found molecules associated with different organelles and cellular compartments, such as endoplasmic reticulum (Fig. 2i), microtubules (Fig. 2j), and centrosomes (Fig. 2k). Many lines show GFP fusions targeted to axons (Fig. 2 l–n); some lines harbor signals in the extracellular matrix (Fig. 2o). We also observed a number of fusion proteins distributed to different bands of the complex sarcomeric units found in muscle fibers (Fig. 2 p–r).
Figure 2.
Subcellular distribution of trapped proteins. (a–c) Examples of targeting of the trapped protein to the nucleus (a, line G280, His2Av), cytoplasm (b, line G89), and membrane (c, line G289). a and b are just before cellularization, and c is just after cellularization. (d–h) GFP distribution in the giant nuclei of third-instar larval salivary glands of different “nuclear” lines. These cells contain polytene chromosome arms that retain the arrangement that they adopt in diploid interphase nuclei. Their nuclear architecture is easily visualized and consists of a chromosomal domain (d, line G280, His2Av:GFP), a large central domain occupied by the nucleolus (e, line G392), a meshwork-like extra-chromosomal nuclear domain (32) (f, line G180), delimited by the nuclear envelope (g, line G262, lamin:GFP and h, line G158, lamin C:GFP). Note the large nuclear dots in h. (i) In line G9, GFP is detected in the endoplasmic reticulum, surrounding a prophase nucleus in the syncitial blastoderm. “Holes” corresponding to the position of the two centrosomes within the endoplasmic reticulum can be seen. (j–k) G147 produces a microtubule-associated fusion, seen here in a metaphase nucleus before cellularization (j) whereas the product of G138 is found in centrosomes only at a similar stage (k; the magnification is different between j and k). (l–n) G9, G147, and G38 show a predominant GFP signal in axons in stage 16 embryos. (o) In G454, an insertion in Viking, a collagen IV type molecule, GFP labels the extracellular matrix. (p–r) Insertions G5 (p, tropomyosin2), G129 (q), and G53 (r, kettin) reveal different subunits of the sarcomeric complex in adult thoracic indirect flight muscle fibers. (Magnifications: a–c and k, ×500; d–h, ×300; i–j, ×1,000; l–n, ×160; o, ×100; p–r, ×1,000.)
Splicing of the Fusion Transcripts Occurs Correctly and GFP Fusions Recapitulate the Expression of the Endogenous Trapped Protein.
Sequences flanking the insertion point of 102 independent lines were recovered by using inverse PCR. Using blast searches in the Drosophila genome databases, we identified insertions in several known or predicted genes (Table 2). Using reverse transcription followed by PCR, we assessed whether the insertion of a long exogenous sequence (>5 kb) in the transcript would interfere with the splicing characteristics of ductin (line G8), CG17238 (line G147), and the nonmuscle and muscle-specific isoforms of tropomyosin II (line G5). We did not detect any aberrations in the splicing of the exons located downstream of the insertion points (data not shown).
Table 2.
Summary of the known and predicted genes identified
| Line | Cytology | Gene | Intron size | Insertion point* | Dup |
|---|---|---|---|---|---|
| Known genes | |||||
| G5 | 3R, 88E11-12 | tropomyosinII | 3.6 kb | AE003708, s, 94200 | 5 |
| G7 | 3R, 42B2 | Vha16, ductin, vacuolar H+ ATPase | 4 kb | AE003789, a, 140890 | |
| G29 | — | Eif-4a | — | — | |
| G33 | X, 3B2-3 | shaggy | 1.6 kb | AE003425, s, 13241 | |
| G44 | 2R, 49A6-9 | lachesin | 10.25 kb | AE003822, a, 71330 | |
| G53 | 3L, 62C2-3 | kettin | 6.3 kb | AE003473, a, 266941 | 1 |
| G74 | 2L, 27B1 | nervana2 Na-K ATPase | 1.4 kb | AE003615, a, 35458 | |
| G109 | 3R, 93A7-B1 | ATPalpha | 13.4 kb | AE003732, s, 208589 | 1 |
| G126 | 3L, 65D5 | sugarless | 2.4 kb | AE003560, s, 245000 | |
| G129 | 2L; 25C6-7 | Possibly Msp300 | AE003608, s, 167972 | 3 | |
| G138 | X, 3A10-B1 | shaggy (different from 33) | >3.2 kb | AE003424, s, 286224 | 2 |
| G158 | 2R; 51B1 | laminC | 2.4 kb | AE003814, s, 61032 | |
| G169 | 3R, 82D2 | karybeta3 | 800 bp | AE003605, a, 193456 | |
| G259 | 2L; 36A7 | VhaSFD vacuolar H+ ATPase | 144 bp | AE003652, a, app 111600 | |
| G262 | 2L; 25E6-F1 | lamin | 660 bp | AE003610, s, 104227 | |
| G280 | 3R, 97D2 | His2Av | 183 bp | AE003758, s, 79583 | |
| G305 | X; 7F1 | Neuroglian | 1.5 kb | AE003444, s, 133625 | |
| G409 | 2L; 33E1-6 | bunched | 75 kb | AE003636, a, 96208 | |
| G430 | 2R, 47A7-8 | Go-alpha 47A (5′isoform) | 8.6 kb | AE003829, a, 184947 | |
| G454 | 2L; 25C1 | Viking collagen type IV | 8 kb | AE003608, a, 84156 | |
| Predicted genes | |||||
| G9 | 2L; 25B10-C1 | CG8895 | 9.1 kb | AE003608, a, 59877 | 9 |
| G38 | 3R, 89B17-19 | CG6963, casein kinase | 14.5 kb | AE003712, s, 164508 | 1 |
| G88 | 3R; 86E13-14 | CG6783, fatty acid binding protein | 2.2 kb | AE003692, a, 43275 | 3 |
| G89 | 3L, 69C2-4 | CG10686, hom to yeast SCD6 and pleur Rap55 | 1 kb | AE003541, a, 60796 | 1 |
| G93 | X, 12B8 | CG10990, homology to mouse apoptosis protein MA3 | 3.4 kb | AE003493, a, 192168 | |
| G112 | 3L, 68C9-10 | CG6084, aldose reductase | <1.4 kb | AE003544, s, 112017 | |
| G119 | 2R; 53D13-14 | CG5935, homology to DEK oncogene | <600 bp | AE003805, a, 138771 | 3 |
| G147 | 3R; 86E15-17 | CG17238 | 15-26 kb | AE003692, a, 81655 | 2 |
| G180 | 2L; 23B1 | CG9894 | <2.4 kb | AE003582, a, 73988 | 1 |
| G189 | 2R, 52C7-8 | CG12969, LIM and PDZ domains | 20 kb | AE003809, s, 147222 | 1 |
| G196 | 2L, 39E3 | CG2207, l(2)k05815 | 1.5 kb | AE003781, a, 73505 | 1 |
| G198 | 3L, 71B2 | CG6988, Pdi, prot disulfide isomerase | 2.7 kb | AE003532, s, 76056 | |
| G245 | 3R; 92F13 | CG17273, BcDNA:LD32788 | <2.2 kb | AE003732, a, 80766 | |
| G264 | X; 12B9 | CG10997, Cl- channel homology | 7 kb | AE003493, a, 266426 | |
| G271 | 2R, 52F7 | CG8443 | 1.4 kb | AE003808, a, app 8580 | |
| G282 | X; 11E9-10 | CG1640, alanine aminotransferase | 3.4 kb | AE003492, s, 117333 | |
| G365 | X, 11B7-9 | CG2556 | 17 kb | AE003489, s, 19=186911 |
Dup, number of sequenced duplicates.
AE00xxxx is the GenBank accession number of the genomic scaffold the insertion matches to. s and a mean that GFP is in the sense or antisense orientation on the scaffold, respectively, and the number corresponds to the insertion point. app, approximately.
When genes were previously known, the distribution of the chimeric protein corresponds to the distribution described, as shown for GFP-tropomyosin II (line G5) and GFP-kettin (line G53) fusions in adult thoracic indirect flight muscles (Fig. 2 p and r). Fig. 2d shows the distribution of the trapped His2Av (G280) in salivary gland giant nuclei: like the wild-type protein and previous GFP-His2Av fusions (25), the fusion is associated with chromosomes. A similar distribution was found for a fusion expressed from a locus predicted to encode a protein homologous to the human DEK protooncogene (G119, not shown). DEK is a nuclear protein known to interact specifically with histones H2A and H2B (26). We identified an insertion in the Drosophila lamin gene (G262). As expected, lamin-GFP is detected at the nuclear envelope in the lamin insertion (Fig. 2g).
It is likely that in some cases, random insertion of the GFP exon will disrupt a localization signal or interfere with the proper delivery of a protein to its destination compartment. One possible example in our limited set of data is the case of an insertion in lamin C: lamin C-GFP is mostly visible as bright nuclear granules in addition to the previously described signal at the nuclear envelope (Fig. 2h). However, it is reminiscent of what has been described for its vertebrate homolog lamin A: buried in dense chromatin, internal lamin A is normally inaccessible to antibodies and can be detected only by removing chromatin (27). A fusion with GFP may circumvent this technical limitation in the lamin C line and reveal new aspects of the protein's distribution.
The Protein Trap Method Reveals Genes Not Predicted by the Genome Project.
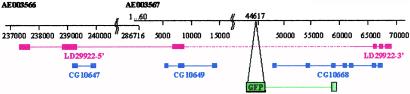
Despite our secondary screening against multiple insertions (see Methods), we found that 20 of the 102 insertions for which we have obtained sequence data have double or triple insertions, based on the occurrence of multiple bands in the inverse PCR. However, only three lines carry two independent new integrations, whereas in all of the other cases, one insertion corresponds to the “silent” jumpstart insertion. In these three cases, only one of the two insertions falls into a known or predicted locus. We therefore can reliably link each pattern with a cytological position. The 102 sequenced insertions correspond to 67 independent loci. Twenty correspond to known genes and 17 to genes predicted by the Drosophila Genome Project (Table 2), whereas 30 (44%) do not correspond to any known or predicted gene (Table 3). We isolated the 3′ region of the GFP–cDNA fusion from several of these lines (not shown). In all cases, the cDNA sequence flanking GFP corresponds to genomic sequences located downstream of the P-element insertion point; some of them do not match any expressed sequence tag (EST) or predictions, and some correspond to parts of EST sequences that have been associated with a prediction entirely located downstream of the insertion. Although these GFP signals could be caused by splicing artefacts generated by the protein trap method, they also could reveal genes with unusual structure, poorly represented in cDNA libraries, or resulting from the use of unpredicted alternative promoters. Indeed, closer inspection of the sequences surrounding several of these insertions reveals that segments of ESTs matching the 5′ side of the insertion have not been included in the genome annotation. For example, line G108 carries such an insertion. Fig. 3 shows that parts of the three predicted genes (CG10647, CG10649, and CG10668) belong to a single gene, whose sequence is contained in EST LD29922 and whose expression pattern is revealed by our insertion G108.
Table 3.
Summary of the unpredicted genes
| Line | Cytology | Insertion point* | Dup |
|---|---|---|---|
| G50 | 2R, 48F5 | AE003822, a, 266019 | |
| G108 | 3L; 64C13 | AE003567, s, 44617 | 3 |
| G116 | 3R; 88A10 | AE003703, s, 63951 | |
| G123 | 3L; 70C11 | AE003536, s, app 53200 | |
| G154 | X; 14A6-8 | AE003501, s, 71697 | |
| G157 | X, 11B7 | AE003489, s, 151058 | |
| G231 | 2R, 48F5 | AE003822, s, app 265962 | |
| G258 | 2L; 36F4-6 | AE003658, s, 186632 | |
| G270 | 2L, 28E5-6 | AE003620, s, 4037 | 2 |
| G318 | 2R, 52D10-12 | AE003805, s, 167069 | |
| G357bis | 2L, 26A8 | AE003611, a, app 247620 | |
| G145 | 2R, 54C3-5 | AE003803, s, 76977 | |
| G215 | 3L; 77D1-4 | AE003591, s, 290886 | |
| G214 | X, 3D1 | AE003427, s, 46536 | |
| G260 | 3R, 89B13 | AE003712, a, app 73692 | |
| G276 | 3L, 61A | AE003467, s, 204864 | |
| G281 | ///// | Multiple hits: subtelomeric heterochromatin repeat | |
| G284 | 2L; 26A8 | AE003611, a, 248506 | |
| G287 | X, 14F2 | AE003502, a, 251633 | |
| G304 | X, 9E7-9 | AE003484, a, 36343 | |
| G341 | 3L, 66F2 | AE003553, s, 131273 | |
| G357 | X; 1C1 | AE003418, a, 222735 | |
| G360 | 3R, 82A4 | AE003606, a, app 287800 | |
| G361 | X, 12B8 | AE003493, s, 200162 | |
| G370 | 2R; 50C23 | AE003816, s, 110448 | |
| G377 | 3R, 85E2 | AE003693, s, 168116 | |
| G392 | 3R; 83D1 | AE003601, s, 33991 | |
| G413 | 2L, 28E3 | AE003619, s, 273106 | |
| G419 | 3L, 75D8 | AE003519, s, 78791 | |
| G428 | 2R, 48F5 | AE003822, a, 265512 |
Legend as in Table 2. The cDNA sequences fused to the GFP 3′ end were identified by 3′ rapid amplification of cDNA ends-PCR for the 11 first lines (G50-G357bis). All matched several hundred base pairs or kilobases downstream of the insertion point.
Figure 3.
Protein trap lines reveal genes not predicted in the genome annotation database. In line G108, the PTT is inserted at position 44617 of the genomic scaffold AE003567, downstream of predicted gene CG10649 and upstream of CG10668. blast searches of EST databases with CG10649 and CG10668 identify regions on the 5′ and 3′ ends of EST LD29922, respectively. Besides, the 5′-most part of LD29922 matches a third prediction, CG10647, further upstream, on the adjacent scaffold AE003566. Therefore, segments of all three predictions (CG10647, CG10649, and CG10668) are part of a single gene, which spans ≈120 kb. The insertion in line G108 reveals the expression of this gene: 3′ cDNA sequences fused to GFP match sequences of CG10668. Predicted genes are in blue, sequenced parts of the EST are in red, and the region found to be fused with GFP in the 3′ rapid amplification of cDNA ends experiment is in green.
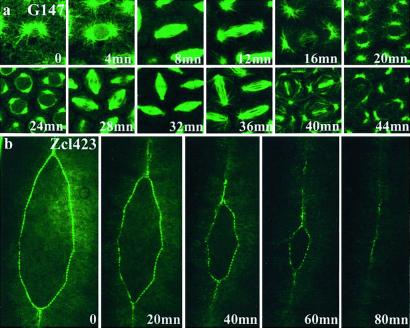
Dynamics of GFP Trapped Proteins Can Be Studied in Vivo in Real Time.
One of the most useful aspects of the GFP protein trap is the ability to follow in live animals the behavior of subcellular structures or cell populations during developmental events. Fig. 4a shows time-lapse imaging of a microtubule-associated protein during the last precellular divisions of a syncitial embryo. Fig. 4b shows the movement of the epithelial cells during dorsal closure, revealed by a GFP fusion with an unidentified molecule, which is targeted to the leading edge.
Figure 4.
Dynamics of GFP fusion distribution. (a) The distribution of the protein fusions produced in line G147 (microtubule-associated protein) was observed at different times during cell division in the syncitial embryo. (b) In line Zcl423, the GFP fusion is specifically expressed at the leading edge of epithelial cells during the zipper-like cell movements of dorsal closure. Anterior is up. (Magnifications: a, ×500; b, ×150.) Video versions of these and other time-lapse experiments can be viewed as Movies 1–4, which are published as supporting information on the PNAS web site, www.pnas.org.
Discussion
In this article, we describe a protein trap system in Drosophila, based on the insertion of a GFP reporter into proteins expressed from their endogenous locus. This method was designed to identify new genes and study in live animals the subcellular distribution of the proteins encoded at the trapped loci.
Sensitivity of the System and Frequency of Protein Trap Events.
P-elements integrate preferentially into 5′ regions of genes and often upstream of the transcription start (24), and our screen relies on the direct visual selection of comparatively rare insertions of the transposon into introns. By screening “en masse” the progeny from medium-sized crosses with a binocular equipped for GFP detection, we have identified up to 20 positive events per day. Although a significant fraction of our protein trap lines display restricted expression patterns, the main limitation is our ability to detect weak GFP signals. Preliminary results obtained with an automated sorter for fluorescent embryos suggest that it could be up to three times more sensitive than the human eye (M. Buszczak and L. Cooley, personal communication). Combined with new generations of brighter GFP, these machines could allow the detection of weaker and more restricted patterns. We also found a significant amount of redundancies. Together, these data suggest that the use of new transposable elements with different insertion specificity could improve the system. More than 50% of the protein trap events are found in genes with introns larger than 2.5 kb, whereas very few insertions are found in introns shorter than 200 bp. This finding does not reflect the distribution of intron size in Drosophila, where a majority of genes have only very short introns and their average size is less than 100 bp, but it is statistically not surprising that one would find more frequently insertions in long rather than short introns. Generating more lines, although it will produce more copies of redundant events, also will increase the number of rare events in the collection.
Fidelity and Accuracy.
The aim of the protein trap is to detect accurately the dynamics of the spatial distribution of the trapped protein during cell cycle and developmental events. Contrary to existing systems, our reporter is expressed from the endogenous promoter as part of the wild-type transcript, so that important transcriptional and translational regulatory mechanisms are reflected in the pattern of the trapped protein. One potential limitation is that the folding time of GFP may introduce some delays in the detection of fast changes in protein expression levels. It is also important that the half-life of the fusion should be similar to the half-life of the wild-type protein. GFP has a relatively long half-life of its own (4 h), but can be efficiently destabilized by the adjunction of protein degradation sequences (28). We therefore anticipate that very unstable trapped proteins confer their intrinsic short life to the GFP fusion.
The adjunction of a GFP module at either the N- or C-terminal end of a protein usually does not significantly affect its structure and function, and GFP fusion proteins have been shown to rescue mutant phenotypes (25). The protein trap events are insertions into the protein and are more likely to disrupt important domains and interfere with the normal function. In the cases of insertions into known genes, we find that the distribution of the fusion protein corresponds to previous descriptions, and we think that the great majority of subcellular distribution that we observe is also correct for new and unknown molecules. However, given that less than a third of the genes are essential for viability, we find a surprisingly high rate of lethality (17%) in our collection. This figure is only an estimate, based on our collection of 215 insertions on the second chromosome, not cleared from potential duplicates. We have not assessed whether lethality is caused by the insertion itself or secondary mutations on the chromosome, which are common in screens based on P-element mobilization. This approximate rate may appear high, but it should be noted that our collection is a selected subset of insertions of the PTT. All our lines affect the coding region of a gene, as opposed to previous P-elements for which lethality has been assessed in random collections with a bias toward 5′ untranslated region insertions and a high incidence of insertions between genes (29). Even though the distribution of the trapped proteins may not be altered, protein trap insertions could interfere with their correct function and be more mutagenic than nonselected random insertions obtained with this or other types of P-elements. In some cases, deleterious effects of a GFP insertion on the function of the trapped protein may be masked because some residual wild-type protein is produced by alternative splicing at levels sufficient to maintain a minimal wild-type activity in a mutated background.
In conclusion, it seems likely that in the majority of cases the distribution of GFP fusion proteins is correct, although their function might often be partially or totally impaired.
Identification of New Genes.
The analysis of our sequencing data were greatly enhanced by the availability of the Drosophila genome sequence. The annotation helped us to assign a gene identity to many of our insertions. However, we found that a surprising proportion of the sequenced insertions does not correspond to any predicted genes. Although we have not formally excluded that GFP expression might, in some cases, be an artifact, closer inspection of the data provided in Flybase (http://flybase.bio.indiana.edu:82/) reveals some prediction errors. Our observations are consistent with the results of the Genome Annotation Assessment Project, which evaluated different annotation tools on the well-characterized Adh region (30). Moreover, they are reminiscent of data found in the Drosophila gene trap description, whose authors also have identified a significant number of fusions with transcripts absent from the databases (21). These results suggest that the algorithms used to predict genes from genomic databases have missed a significant number of genes. The protein trap method may be useful in identifying unsuspected novel genes and functions. As noted previously, the screen is biased toward genes with long introns, which may be more difficult to predict, and these figures may not reflect the actual proportion of unpredicted genes in the whole genome.
Real-Time Imaging.
Protein trap events provide essential information on the protein's distribution and its dynamics, as exemplified by the time-lapse experiments presented here. As the study of developmental processes relies more and more on the observation of events occurring inside and between living cells, our collection of several hundred fly lines represents a unique and valuable source of in vivo markers (microtubule dynamics, nuclear architecture, sarcomere architecture, etc.) for the future of developmental and cell biology.
Supplementary Material
Acknowledgments
We thank Gerald Udolph for the injection of the GA construct, Zalina Osman and Lee Chai Lin for excellent technical assistance, and members of the lab for their persistent enthusiasm and daily inquiries about this project. This work was funded by the Institute of Molecular and Cell Biology, a Marie Curie Category 30 Postdoctoral Fellowship (to X.M.) and a Wellcome Trust Principal Research Fellowship and Program Grant (to W.C.).
Abbreviations
- GFP
green fluorescent protein
- PTT
protein trap transposon
- EST
expressed sequence tag
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 2.Cutler S R, Ehrhardt D W, Griffitts J S, Somerville C R. Proc Natl Acad Sci USA. 2000;97:3718–3723. doi: 10.1073/pnas.97.7.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Misawa K, Nosaka T, Morita S, Kaneko A, Nakahata T, Asano S, Kitamura T. Proc Natl Acad Sci USA. 2000;97:3062–3066. doi: 10.1073/pnas.060489597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding D Q, Tomita Y, Yamamoto A, Chikashige Y, Haraguchi T, Hiraoka Y. Genes Cells. 2000;5:169–190. doi: 10.1046/j.1365-2443.2000.00317.x. [DOI] [PubMed] [Google Scholar]
- 5.Lindsey K, Wei W, Clarke M C, McArdle H F, Rooke L M, Topping J F. Transgenic Res. 1993;2:33–47. doi: 10.1007/BF01977679. [DOI] [PubMed] [Google Scholar]
- 6.Sundaresan V, Springer P, Volpe T, Haward S, Jones J D, Dean C, Ma H, Martienssen R. Genes Dev. 1995;9:1797–1810. doi: 10.1101/gad.9.14.1797. [DOI] [PubMed] [Google Scholar]
- 7.Zambrowicz B P, Friedrich G A, Buxton E C, Lilleberg S L, Person C, Sands A T. Nature (London) 1998;392:608–611. doi: 10.1038/33423. [DOI] [PubMed] [Google Scholar]
- 8.Skarnes W C, Auerbach B A, Joyner A L. Genes Dev. 1992;6:903–918. doi: 10.1101/gad.6.6.903. [DOI] [PubMed] [Google Scholar]
- 9.Bronchain O J, Hartley K O, Amaya E. Curr Biol. 1999;9:1195–1198. doi: 10.1016/S0960-9822(00)80025-1. [DOI] [PubMed] [Google Scholar]
- 10.Bayer T A, Campos-Ortega J A. Development (Cambridge, UK) 1992;115:421–426. doi: 10.1242/dev.115.2.421. [DOI] [PubMed] [Google Scholar]
- 11.Amsterdam A, Burgess S, Golling G, Chen W, Sun Z, Townsend K, Farrington S, Haldi M, Hopkins N. Genes Dev. 1999;13:2713–2724. doi: 10.1101/gad.13.20.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaiano N, Amsterdam A, Kawakami K, Allende M, Becker T, Hopkins N. Nature (London) 1996;383:829–832. doi: 10.1038/383829a0. [DOI] [PubMed] [Google Scholar]
- 13.Skarnes W C, Moss J E, Hurtley S M, Beddington R S. Proc Natl Acad Sci USA. 1995;92:6592–6596. doi: 10.1073/pnas.92.14.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tate P, Lee M, Tweedie S, Skarnes W C, Bickmore W A. J Cell Sci. 1998;111:2575–2585. doi: 10.1242/jcs.111.17.2575. [DOI] [PubMed] [Google Scholar]
- 15.Zheng X H, Hughes S H. J Virol. 1999;73:6946–6952. doi: 10.1128/jvi.73.8.6946-6952.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Kane C J, Gehring W J. Proc Natl Acad Sci USA. 1987;84:9123–9127. doi: 10.1073/pnas.84.24.9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brand A H, Perrimon N. Development (Cambridge, UK) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 18.Bellen H J, O'Kane C J, Wilson C, Grossniklaus U, Pearson R K, Gehring W J. Genes Dev. 1989;3:1288–1300. doi: 10.1101/gad.3.9.1288. [DOI] [PubMed] [Google Scholar]
- 19.Wilson C, Pearson R K, Bellen H J, O'Kane C J, Grossniklaus U, Gehring W J. Genes Dev. 1989;3:1301–1313. doi: 10.1101/gad.3.9.1301. [DOI] [PubMed] [Google Scholar]
- 20.Bier E, Vaessin H, Shepherd S, Lee K, McCall K, Barbel S, Ackerman L, Carretto R, Uemura T, Grell E, et al. Genes Dev. 1989;3:1273–1287. doi: 10.1101/gad.3.9.1273. [DOI] [PubMed] [Google Scholar]
- 21.Lukacsovich T, Asztalos Z, Awano W, Baba K, Kondo S, Niwa S, Yamamoto D. Genetics. 2001;157:727–742. doi: 10.1093/genetics/157.2.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hodges D, Bernstein S I. Mech Dev. 1992;37:127–140. doi: 10.1016/0925-4773(92)90075-u. [DOI] [PubMed] [Google Scholar]
- 23.Adams M D, Celniker S E, Holt R A, Evans C A, Gocayne J D, Amanatides P G, Scherer S E, Li P W, Hoskins R A, Galle R F, et al. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 24.Spradling A C, Stern D M, Kiss I, Roote J, Laverty T, Rubin G M. Proc Natl Acad Sci USA. 1995;92:10824–10830. doi: 10.1073/pnas.92.24.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clarkson M, Saint R. DNA Cell Biol. 1999;18:457–462. doi: 10.1089/104454999315178. [DOI] [PubMed] [Google Scholar]
- 26.Alexiadis V, Waldmann T, Andersen J, Mann M, Knippers R, Gruss C. Genes Dev. 2000;14:1308–1312. [PMC free article] [PubMed] [Google Scholar]
- 27.Hozak P, Sasseville A M, Raymond Y, Cook P R. J Cell Sci. 1995;108:635–644. doi: 10.1242/jcs.108.2.635. [DOI] [PubMed] [Google Scholar]
- 28.Li X, Zhao X, Fang Y, Jiang X, Duong T, Fan C, Huang C C, Kain S R. J Biol Chem. 1998;273:34970–34975. doi: 10.1074/jbc.273.52.34970. [DOI] [PubMed] [Google Scholar]
- 29.Roseman R R, Johnson E A, Rodesch C K, Bjerke M, Nagoshi R N, Geyer P K. Genetics. 1995;141:1061–1074. doi: 10.1093/genetics/141.3.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reese M G, Hartzell G, Harris N L, Ohler U, Abril J F, Lewis S E. Genome Res. 2000;10:483–501. doi: 10.1101/gr.10.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mount S M, Burks C, Hertz G, Stormo G D, White O, Fields C. Nucleic Acids Res. 1992;20:4255–4262. doi: 10.1093/nar/20.16.4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wasser M, Chia W. Nat Cell Biol. 2000;2:268–275. doi: 10.1038/35010535. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.