Abstract
While substantial advances in the identification of cytogenomic subtypes of childhood acute lymphoblastic leukemia (ALL) have been made in recent decades, epidemiologic research characterizing the etiologic heterogeneity of ALL by subtype has not kept pace. The purpose of this review is to summarize the current literature concerning subtype-specific epidemiologic risk factor associations with ALL subtype defined by immunophenotype (e.g. B-cell vs T-cell) and cytogenomics (including gross chromosomal events characterized by recurring numerical and structural abnormalities, along with cryptic balanced rearrangements, and focal gene deletions). In case-control analyses investigating non-genetic risk factors, home paint exposure is associated with hyperdiploid, MLL-rearranged, and ETV6-RUNX1 subtypes, yet there are few differences in risk factor associations between T- and B-ALL. While the association between maternal smoking and ALL overall has been null, maternal smoking is associated with an increasing number of gene deletions among cases. GWAS-identified variants in ARID5B have been the most extensively studied and are strongly associated with hyperdiploid B-ALL. GATA3 single nucleotide variant rs3824662 shows a strong association with Ph-like ALL (OR=3.14). However, there have been relatively few population-based studies of adequate sample size to uncover risk factors that may define etiologic heterogeneity between and within the currently defined cytogenomic ALL subtypes.
Keywords: childhood acute lymphoblastic leukemia, epidemiology, subtype
Introduction
Cancer, regardless of age at diagnosis, is universally heterogeneous with variation in incidence by both immunophenotype and molecular subtype within tumor types. Studies of adult neoplasms including breast (1–5), colon (6–8), and prostate (9,10), have shown that subtyping, whether molecular or histologic, leads to identification of etiologic heterogeneity between tumor subtypes that may inform targeted public health prevention strategies. For example, in breast cancer, there are robust findings across multiple study populations showing evidence of etiologic heterogeneity by reproductive risk factors such as age at menarche, first birth, and menopause, as well as combined menopausal hormone therapy use for lobular, compared to ductal carcinoma, and by molecular breast cancer subtype.(1–5) Similarly, there is etiologic variation by histologic and molecular subtype for colorectal adenomas concerning combined hormone therapy use (6) and cigarette smoking.(6–8) Finally, there is reported variability in risk of prostate tumors by TMPRSS2-ERG fusion status concerning age at diagnosis (9), height, and obesity (10).
These examples highlight the presence of etiologic heterogeneity in adult breast, colon, and prostate tumors where once histology or molecular subtype are considered, differences in risk factor associations become clearer. Therefore, it is reasonable to hypothesize that studying variation in susceptibility to childhood acute lymphoblastic leukemia (ALL) by immunophenotype, and cytogenomic subtype, may similarly identify etiologic variation of this most common malignancy diagnosed in children aged 0–14 years.(11) The purpose of this review is to summarize the current state of the literature regarding etiologic variation in ALL subtypes. We will discuss the literature on non-genetic and genetic risk factors for ALL overall and by subtype as defined by immunophenotype and cytogenomic subtype.
Descriptive epidemiology of childhood acute lymphoblastic leukemia
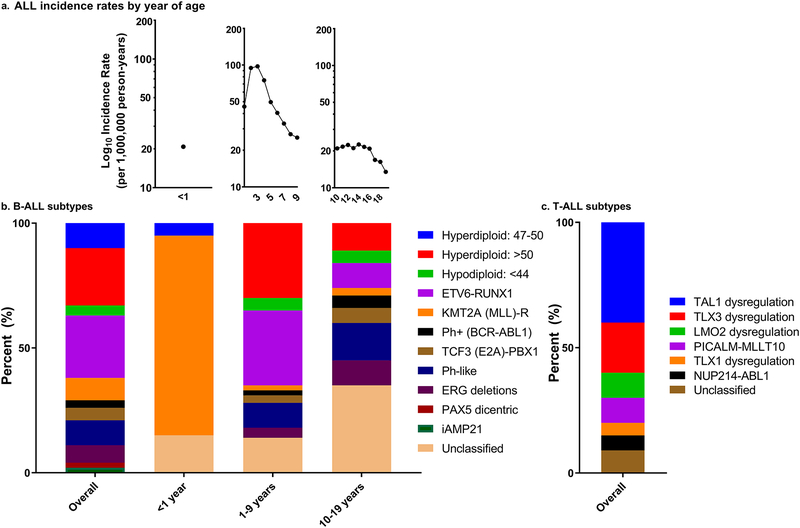
Acute lymphoblastic leukemia (ALL) is the most common childhood malignancy in the United States (US), with approximately 3,000 children and adolescents aged 0–19 years diagnosed annually.(11) As shown in Figure 1a, among children (aged 0–19 years) from the SEER 18 registries (2000–2014), there is notable variation in the age-specific incidence rates over the childhood and adolescent periods with the highest incidence rate for ALL between ages 2–4 years. Incidence rates of ALL continue to slowly increase necessitating the identification of modifiable risk factors.(11,12)
Figure 1.
contains childhood acute lymphoblastic leukemia (ALL) incidence rates from the Surveillance, Epidemiology, and End Results program (SEER) 18 registries (2000–2014) for all ages (0–19 years) and stratified by age group (<1 year, 1–9 years, 10–19 years) (a). The cytogenomic subtypes of childhood B-ALL immunophenotype overall and by age at diagnosis categories (<1 year, 1–9 years, 10–19 years) are presented (b). The cytogenomic subtypes of childhood T-ALL immunophenotype overall are presented (c). Categories assumed to be mutually exclusive.
Even though 5-year overall survival rates of ALL have drastically improved since the 1970’s and are currently at approximately 90% (13), there are documented differences in survival by age at diagnosis, race, and subtype.(13–15) Generally, once differences in survival and clinical outcome by disease subtype are reported, epidemiologic analyses are conducted on individual subtypes to identify etiologic differences.(16) However, despite documented differences in prognosis and biology between pediatric ALL subtypes, there is a paucity of studies aimed at identifying unique risk factor profiles for these subtypes. This is likely due to older and registry-based studies lacking cytogenomic and molecular genetic data and/or inadequate sample sizes.
Biology of acute lymphoblastic leukemia
Immunophenotypes
ALL is composed of two main immunophenotypes that are identified by distinctive hematopoietic lineage markers: B- and T-ALL.(17) Pediatric B-ALL is diagnosed in up to 85% of cases while T-ALL comprises the remaining 15% (18,19) and is more common among males.(20) There is racial/ethnic variation in the incidence of B- and T-ALL. Approximately 85% of white, 87% of Hispanic, 81% of Asian, and 75% of black children are diagnosed with B-ALL and the remaining proportion of children in each race/ethnicity are diagnosed with T-ALL (21–24), which has lower survival than B-ALL.(13,15)
Of the 15% of pediatric cases diagnosed with T-ALL, approximately 15% of these patients are diagnosed early T-precursor ALL (EPT-ALL).(25–28) EPT-ALL cells have retained myeloid and stem cell characteristics making it a distinct entity from T-ALL.(27,29–31) The recent identification of effective therapies for ETP-ALL has shown it to be similar to T-ALL in outcome and prognosis.(25,26,32) As observed for T-ALL, ETP-ALL is more common among males (ETP-ALL: 63% and T-ALL: 74%).(25) The difference in etiology of ETP-ALL and T-ALL is unclear.
Cytogenomic subtypes
Cytogenomic subtypes refer to specific numerical and/or structural chromosomal abnormalities that are well documented to recur in different subgroups of ALL and are used in the clinic to risk-stratify patients. Cytogenomic subtyping usually occurs through visual chromosome assessment by use of fluorescence in situ hybridization (FISH), karyotyping via G-banding, and/or chromosomal microarray.(14)
Figure 1b displays the overall and age specific distributions of cytogenomic subtypes of B-ALL and the overall cytogenomic distribution for T-ALL (Figure 1c) (cytogenomics subtypes assumed to be mutually exclusive).(20,21,23,31,33–36) Describing the differences in clinical characteristics of cytogenetic subtypes is beyond the scope of this review and can be found in greater detail elsewhere.(31,36) The most marked variation in B-ALL incidence patterns is observed by age at diagnosis, which may also serve as a proxy for some cytogenomic subtypes. A prime example of this is observed among infant ALL, diagnosed in children less than 1 year of age, where 80% of cases have an MLL-rearrangement (Figure 2). Extending beyond infants, approximately 35% of cases with B-ALL aged 1–9 years are diagnosed with the hyperdiploid subtype and another 30% are diagnosed with ETV6-RUNX1 fusions.(28)
Figure 2.
displays the odds ratios and 95% confidence intervals for associations by childhood acute lymphoblastic leukemia (ALL) immunophenotype and cytogenomic subtype for maternal and paternal exposures during the preconception period (a). Next, maternal and paternal exposures during pregnancy in association with childhood ALL by subtype are presented (b). Then, maternal, paternal, and birth characteristics of the child in association with childhood ALL by subtype are presented (c). Finally, childhood exposures in association with childhood ALL by subtype are displayed (d). Reference numbers are presented in brackets.
Ploidy changes, the most common chromosomal abnormality in childhood B-ALL, are observed in approximately 35% cases.(21,28,33–35,37) Hyperdiploidy, the most common subtype, involves acquiring extra chromosomes including 4, 6, 10, 14, 17, 18, 21, and X chromosomes.(23,31,38,39) Chromosomal translocations, which often place transcription factors (e.g. ETV6, RUNX1) next to one another leading to production of novel functional proteins, dysregulation of gene expression, or activation of an oncogene (19,31,40–43), are the second most frequent gross chromosomal changes in ALL. In B-ALL, the ETV6-RUNX1 subtype, which typically arises from a cryptic t(12;21) fusion, that can be visualized by FISH, is diagnosed in approximately 30% of cases.(28,34,44,45) The second most common translocation, involves MLL-rearrangement (MLLr) (also referred to as KMT2A-rearrangement) [t(4;11)(q21;q23)], is diagnosed in approximately 10% of cases overall but 80% of infant B-ALL cases.(20,28,34,44–48) MLL is known to have more than 100 partner genes with which it can pair, and the specific pairing impacts prognosis.(31) There are a number of less frequently diagnosed cytogenomic subtypes involving gene fusions and the presence of dicentric chromosomes (28,49), that vary by age at diagnosis as illustrated in Figure 1b.
While much work identifying cytogenomic subtypes has focused on the more common B-ALL immunophenotype of childhood ALL, cytogenomic subtypes have also been identified for T-ALL. T-ALL cases are rarely hyperdiploid and do not displayed the pattern of chromosomal gains seen in B-ALL, but can be triploid or tetraploid.(31,33) More often they have only 46 chromosomes, but harbor structural abnormalities and can be grouped into 4 main categories involving translocations of 1) TAL or LMO; 2) TLX1; 3) TLX3; and 4) HOXA genes.(31) T-ALL chromosomal translocations are characterized by transcription factor dysregulation through gene fusions.(34) TAL1 fusions [t(1;7)(p32;q35) and t(1:14)(p32;q11)] are diagnosed in an estimated 40% of cases.(26,28,34) In approximately 20% of cases, TLX3 [HOX11L2; t(5;14)(q35;32)] dysregulation is reported.(26,28,31,34,50) The remaining fusions in Figure 2 are diagnosed in 10% or less of pediatric cases each.(26,28,34,50) There is insufficient information on variation of these T-ALL subtypes by sex and race/ethnicity for the most part.
Molecular genomic abnormalities
Molecular genomic abnormalities, or genetic events that occur at sub-chromosomal levels, are detected by gene sequencing or gene expression patterns and have also been characterized for childhood ALL.(28,51) Most recently, the Philadelphia-like subtype (Ph+-like, BCR-ABL1-like), B-ALL has been identified by gene expression patterns similar to that found in BCR-ABL1 positive B-ALL and has been further shown to have mutations affecting genes important in leukemogenesis such as IKZF1 (52) and CDKN2A/B.(31) BCR-ABL1-like ALL has been identified in 10–15% of B-ALL cases >1 year of age, but is more frequent in adolescents and young adults.(28,31,45,53,54) Similarly, focal deletions of the oncogene ERG, now recognized as DUX4-rearranged, have been identified in 4–7% of childhood B-ALL patients and 10% of adolescent B-ALL patients.(20,28,55,56)
Chromosomal microarrays, and other cytogenomic and molecular genetic techniques have highlighted the occurrence of gene deletions in the tumor suppressor gene, CDKN2A, the B-cell lineage regulator PAX5, and Ikaros family members IKZF1 and IKZF2 in B-ALL. All have been studied with respect to childhood ALL immunophenotype and cytogenomic subtype. Biallelic CDKN2A deletions have been reported in approximately 20% of B-ALL cases at the time of diagnosis with marked variation between cytogenomic subtypes.(57) Hyperdiploid, MLL-rearranged, and ETV6-RUNX1 cases experience lower deletion rates (<15% each) while over 40% of cases with TCF3 (E2A)-PBX1 or Ph+ (BCR-ABL1) harbor CDKN2A deletions.(57) In T-ALL cases, biallelic CDKN2A deletions are more common than observed for B-ALL with approximately 50% of cases harboring a deletion and there is variation by subtype: 43% of TAL1-rearranged cases had a CDKN2A deletion as did 76% of cases with TLX3 rearrangements.(57)
Deletions involving PAX5 have been observed in nearly 1/3 of B-ALL cases with variation in frequency by ALL cytogenomic subtype.(58,59) Approximately 12% of ALL cases have deletions of IKZF1.(58,60) Deletions are more common among B-ALL (13%) than T-ALL (5%) cases and variation by cytogenomic subtype has been reported with over 80% BCR-ABL1 positive (58–60) and 70–80% of BCR-ABL1-like ALL (53) cases harboring an IKZF1 deletions. Mutations in Ras family members are also found in 20–30% of childhood ALL cases.(61) Mutations are found more frequently in Latino children and among children diagnosed with high-hyperdiploid ALL.(61) While characterizing deletion and mutation frequencies of the aforementioned genes in conjunction with cytogenomic abnormalities has begun, data on variation in deletion status by age, race, and sex are lacking.
Analytic epidemiology of acute lymphoblastic leukemia
Due to the hypothesized in utero origin of childhood leukemia, as well as prima facie evidence based on the detection of fusion genes including MLL-AF4, ETV6-RUNX1, and E2A-PBX1 in the neonatal blood spots of children diagnosed with ALL (40–42,62,63), many epidemiologic studies examine exposures from the preconception period through early life. Generally, gestational risk factors are thought to influence development of a “first hit” while early life risk factors are thought to impart the “second hit” necessary for overt leukemic transformation. This may be especially relevant for the ETV6-RUNX1 subtype where it is believed that this fusion is necessary, but insufficient by itself, to lead to leukemia (64); whereas, MLL-rearrangement alone appears to be sufficient to cause leukemia based on the identification of MLL-rearrangements in blood spots of children diagnosed with ALL and the short latency between birth and diagnosis for a majority of cases with this subtype.(42) The risk factor associations reported for ALL combined vary by window of exposure and by parent exposed. Figure 2 and Supplemental Table 1 display the measures of association for risk factors in association with ALL overall by timing of the exposure during the preconception, pregnancy, birth, and postnatal periods. To identify articles for non-genetic risk factors we gathered the meta- or pooled-analyses measures of association for each exposure from the reports with the largest number of studies on each topic from PubMed (last accessed August 31, 2018). We used the search terms “acute lymphoblastic leukemia”, “meta-analysis”, and/or “Childhood International Leukemia Consortium”. Only the most recent meta-analyses and pooled-analyses with 3 or more studies are displayed in Figure 2 and Supplemental Table 1.(65,66,75–84,67,85–92,68–74)
The risk factors imparting an increased risk of childhood ALL occur for parental exposures to chemicals including pesticides (maternal residential pesticide exposure during pregnancy OR: 1.43; 95% CI: 1.32–1.54) (66,73,93) and paint (any parental residential paint exposure preconception OR: 1.54; 95% CI: 1.28–1.85) (72) may impart the necessary ‘second hits’ for some leukemic clones or lead to the accumulation of somatic mutations leading to disease initiation. Non-chemical exposures associated with an increased risk of ALL include maternal coffee intake during pregnancy of greater than or equal to 2 cups per day (OR: 1.27; 95% CI: 1.09–1.48) (74), increasing maternal age (5-year increased OR: 1.05; 95% CI: 1.01–1.10) (81), high birth weight (1kg increase OR: 1.18; 95% CI: 1.12–1.32) (82), large for gestational age at birth (OR: 1.21; 95% CI: 1.11–1.32) (83), and pre-labor cesarean delivery (OR: 1.23; 95% CI: 1.04–1.47) (84). Conversely, postnatal exposures such as daycare attendance (any vs none OR: 0.76; 95% CI: 0.67–0.87) (91), being breastfed (any vs none OR: 0.91; 95% CI: 0.84–0.98) (88,94), contact with dogs (any vs none OR: 0.92; 95% CI: 0.86–0.99) and cats (any vs none OR: 0.87; 95% CI: 0.80–0.94) (90) in the first year of life, and the presence of allergies (any vs none OR: 0.67; 95% CI: 0.54–0.82) (86) have been found to reduce the risk of childhood ALL. The findings for daycare attendance and being breast fed are robust across multiple epidemiologic studies and are thought to provide a protective effect by priming the immune system early in life thereby allowing the individual to effectively control the development of leukemia through enhanced immune function as reviewed by Greaves (2018).(95) The associations with maternal alcohol use (65) and smoking during pregnancy and ALL overall are null. (80)
Analytic epidemiology of acute lymphoblastic leukemia by subtype
This section summarizes the literature on risk factors for ALL first by age at diagnosis and then by immunophenotype and cytogenomic subtype where available (Supplemental Table 1, Figure 2). There may be differences in subtyping methods where CGH or FISH have evolved over time; therefore, between-study variation in subtyping may exist. Conversely, karyotyping methods, which have not changed dramatically over time, may be more consistent between studies. While studying differences in risk factor associations by subtype can identify etiologic variation among tumors, amassing enough childhood ALL cases, uniformly subtyping cases using contemporary definitions based on molecular profiling techniques, and obtaining detailed risk factor data have posed a tremendous challenge for the field as evidenced by the small number of studies with risk factor information for each clinically-relevant subtype presented in Supplemental Table 1 and Figure 2 a–d. To identify articles for non-genetic risk factors stratified by immunophenotype and cytogenetic subtype, we first examined the meta- or pooled-analyses included, as described above, for each risk factor to determine if risk estimates were given by ALL subtype. If no subtype-specific risk estimates were listed in the main text or available supplemental materials, we then examined the component studies included in each meta- or pooled-analysis to see if they contained risk estimates stratified by subtype. If none of the component studies presented subtype-stratified risk estimates, we searched in PubMed (last accessed August 31, 2018) using the search terms “childhood acute lymphoblastic leukemia”, “risk factors”, and “subtype” to identify other studies where subtype-stratified risk estimates were presented either in the main text or supplemental materials. If more than one study reported subtype-stratified estimates, the risk estimates from the study with the most subtypes reported in their paper or the study with the largest population size was presented.
Risk of acute lymphoblastic leukemia by age at diagnosis
Studies stratified by age at diagnosis were the first to shed light on the etiologic heterogeneity between subtypes of ALL since, as previously mentioned, age may serve as a proxy for some cytogenomic subtypes. For example, high birthweight was associated with an increased risk of infant ALL, which is dominated by the MLL-rearranged subtype, (OR=2.8 for highest birth weight category in children <2 years of age).(96,97) In another study examining the trend in increasing birth weight by age at diagnosis, there were significant associations between each 1-kg increase and the risk of childhood ALL for children ages 1–5 years of age (OR: 1.24, 95% CI: 1.08–1.42) and 6–9 years of age (OR: 1.35, 95% CI: 1.05–1.74). (98) This association was elevated, though not significant for infants aged <1 years (OR: 1.62, 95% CI: 0.89–2.96), which may be due to a low sample size of low birth weight children <1 year of age, and was not associated with ALL among children ≥10 years of age at diagnosis (OR: 1.21, 95% CI: 0.90–1.63).(98)
Paternal smoking prior to conception has shown variation in risk by age at ALL diagnosis in the offspring with strong effects observed for children diagnosed between 0–4 years of age (smokers vs nonsmokers OR: 1.8, 95% CI: 1.2–2.6), null effects in children aged 5–9 years at diagnosis (OR: 0.9, 95% CI: 0.5–1.5), and 10–14 years at diagnosis (OR: 1.9, 95% CI: 0.5–1.8).(99) It has also been observed that older maternal age and higher maternal education are associated with ALL in children aged 2–4 years (maternal age ≥35 vs 20–34 years OR: 1.31, 95% CI: 1.06–1.63; maternal education ≥16 vs 12–15 years OR: 1.48, 95% CI: 1.14–1.93) but not children aged 0-<2 years at diagnosis (maternal age ≥35 vs 20–34 years OR: 1.07, 95% CI: 0.71–1.62; maternal education ≥16 vs 12–15 years OR: 1.01, 95% CI: 0.65–1.56).(100) As hinted at in analyses examining differences in risk factors by age at diagnosis, there appears to be considerable etiologic heterogeneity in childhood ALL. This becomes even more apparent as we examine risk factor associations by the established immunophenotype and cytogenomic subtypes of ALL.
Risk of acute lymphoblastic leukemia by immunophenotype
Concerning B- and T-ALL, some studies report risk factor associations stratified by immunophenotype (Figure 2 a–d); however, while some risk factors are significantly associated with B-ALL, few risk factors show significant associations with T-ALL. The lack of significant risk factor associations by T-ALL may be due to the small sample size available for this minor subtype, which can be inferred from the increased width of the 95% confidence intervals for T-ALL associations (sample sizes listed in Supplemental Table 1, where available). Additionally, a number of risk factors are associated with ALL overall but are not associated with the ALL immunophenotypes, which makes it hard to discern whether there may be etiologic heterogeneity by ALL immunophenotype.(73,77,81,101–106) A number of risk factors display significant associations with B-ALL but not T-ALL, as shown in Figure 2 a–d and Supplemental Table 1, including home paint exposure prior to conception, during pregnancy, and after birth (72), preconception paternal smoking (107), older paternal age (105), pre-labor caesarean delivery (84), increasing birth weight (98,108), increasing birth order (98), being breast fed (109), infections during childhood (89,110) and daycare attendance.(91,111–113)
A number of risk factors show similar associations with B-ALL and T-ALL (Figure 2 a–d, Supplemental Table 1). Exposures associated with significantly increased risks of both B- and T-ALL include large for gestational age (83), paternal occupational pesticide exposure (66), residential pesticide exposure during the preconception period and during childhood.(73) Exposures associated with significantly reduced risks of B- and T-ALL include any contact with cats (90) and prenatal vitamin use.(114)
Risk of acute lymphoblastic leukemia by cytogenomic subtype
When cytogenomic subtype of B-ALL is considered, there is some variation in risk (Supplemental Table 1 and Figure 2 a–d), but only a small number of studies report analyses stratified by these characteristics and consistency of the subtypes included is lacking across studies. Home paint exposure during pregnancy is associated with ETV6-RUNX1 (OR: 1.51; 95% CI: 1.08–2.11) and any MLL-rearranged ALL (OR: 3.30; 95% CI: 1.71–6.35).(72) Home paint exposure after birth is also associated with low hyperdiploid (OR: 1.94; 95% CI: 1.32–2.89) and ETV6-RUNX1 B-ALL (OR: 1.60; 95% CI: 1.16–2.21).(72) Daycare attendance is inversely associated with hyperdiploid B-ALL (OR: 0.38; 95% CI: 0.24–0.59) (113) and ETV6-RUNX1 B-ALL (OR: 0.47; 95% CI: 0.24–0.94).(113) Being breastfed is inversely associated with hyperdiploidy (OR: 0.37; 95% CI: 0.18–0.77).(115) Increasing birth weight is significantly associated with hyperdiploid (OR: 1.12; 95% CI: 1.05–1.20) and TCF3 (E2A)-PBX1; t(1;19) ALL (OR: 1.41; 95% CI: 1.09–1.84).(116) While maternal tea intake during pregnancy is not associated with an increased risk of ALL overall or by either immunophenotype, it was found to be associated with any MLL-rearranged B-ALL (≥2 cups/day vs none OR: 2.69, 95% CI:1.17–6.21); however, this association is based on a small number of cases (n=9) born to mothers that consumed ≥2 cups/day and should be interpreted cautiously.(74)
Risk of acute lymphoblastic leukemia by molecularly-defined mutation subtype
Maternal smoking is one of the most commonly studied risk factors for childhood ALL to date and has also been investigated in a molecular ALL subtype analysis, apparently the first study of its kind, in relation to any exposure. In a case-only study, that assessed the association between tobacco exposure during the early-life period and deletions in 8 genes commonly lost somatically in childhood ALL (CDKN2A, ETV6, IKXF1, PAX5, RB1, BTG1, PAR1 region and EBF1), a positive association was observed between maternal smoking and the total number of deletions observed in these genes in tumor samples from cases of childhood ALL.(117) Interestingly, the authors observed that B-ALL and hyperdiploid ALL experienced fewer deletions of these genes while T-ALL and ETV6-RUNX1 B-ALL cases experienced one or more deletions in the genes under study. These findings suggest that maternal smoking may exert subtype-specific effects on ALL development but require validation in a case-control study. In a different analysis on parental exposures to solvents, plastic materials and hydrocarbons, maternal exposure to these chemicals during pregnancy was associated with K-ras mutations among childhood ALL cases.(118) These studies highlight the possibility of the simultaneous evaluation of cytogenomic and molecular subtypes in association with epidemiologic risk factors.
Germline variants display subtype-specific variation in association with acute lymphoblastic leukemia
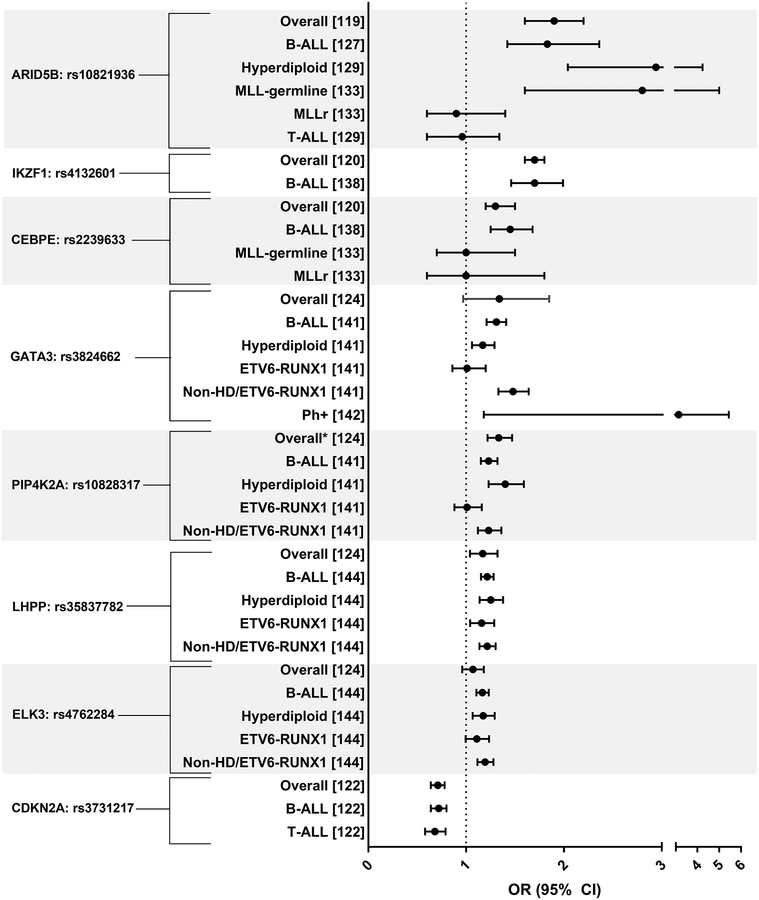
Investigations of germline risk for ALL have generally involved genome-wide association studies (GWAS), which examine common variants, and more recently, whole exome or genome sequencing studies, which examine rare variants. Currently, GWAS comprise the bulk of germline genetic research on ALL.(37,119–124) The GWAS studies have been conducted among French (37), European (119,120,122), Australian (121), American (white, black, and Hispanic (123); and Latino and non-Latino (124)) children. Additionally, numerous replication studies have been done whereby stratification by immunophenotype and cytogenomic subtype has been performed, including some analyses in non-white populations. We present single nucleotide variants (SNVs) identified in discovery GWAS analyses, reported in replication analyses where stratification by subtype was performed among Caucasian children only, as there was little racial variation observed in risk for most of the reported SNVs (Supplemental Table 2), and where cell counts were available to allow for the calculation of the per variant allele odds ratios (OR) and 95% confidence intervals (95% CI) (Figure 3). Supplemental Table 2 contains per allele ORs for SNVs in association with ALL overall and by subtype. GWAS studies were identified in PubMed (last accessed August 31, 2018) using the search terms “acute lymphoblastic leukemia” and “genome wide association study” and from references lists of current publications. Validation studies of individual SNVs were identified by searching for the specific gene along with the terms “acute lymphoblastic leukemia” and “subtype” in PubMed.
Figure 3.
displays the allelic odds ratios and 95% confidence intervals for single nucleotide variants (SNVs) associated with childhood acute lymphoblastic leukemia (ALL) overall and by immunophenotype and cytogenomic subtype from discovery genome-wide association studies (GWAS) and replication analyses. Reference numbers are presented in brackets. Estimates reported for Caucasians only. Non-HD refers to non-hyperdiploid ALL.
ARID5B, AT-Rich Interaction Domain 5B, SNVs were identified in multiple GWAS studies with effect estimates (ORs) ranging from 1.5–1.9 per risk allele.(37,119–121,124) ARID5B, which is involved in differentiation of B- ALL progenitors (119), SNV rs10821936 is associated with a significantly increased risk of ALL overall (OR: 1.9; 95% CI: 1.6–2.2) (119) and this finding is consistent across race groups and sexes.(24,123,133,125–132) This association persists among B-ALL only cases (OR: 1.83; 95% CI: 1.42–2.36; (127)).(128,129,132,134,135) The association is null between ARID5B SNV rs10821936 and T-ALL (OR: 0.96; 95%CI: 0.60–1.34) among Caucasian children (129,132), but shows an increased association among black children (OR: 2.80; 95% CI: 1.16–6.77).(24) Strong associations between rs10821936 and hyperdiploid B-ALL (OR: 2.94; 95%C: 2.04–4.24; (129)) (24,127,132) and MLL-germline (OR: 2.80; 95% CI: 1.60–5.00), but not MLL-rearranged (OR: 0.90; 95% CI: 0.60–1.40) cases have been reported.(133)
The IKAROS family zinc finger 1, IKZF1, gene, which is a transcription factor involved in lymphoid cell differentiation, particularly of T cells (136,137), confers risk of ALL in pediatric cases (SNV rs4132601 OR: 1.7; 95% CI: 1.6–1.8; (120)).(37) When stratified by subtype, rs4132601 is associated with an increased risk of B-ALL regardless of race/ethnicity (Caucasian OR: 1.70; 95% CI: 1.46–1.99; (138)).(132,134) Another gene harboring germline variants found to increase the risk of childhood ALL is CEBPE, which has a role in the determination of myeloid cells.(139) GWAS-identified CEBPE SNV rs2239633 is associated with an increased risk of ALL overall (OR:1.3; 95% CI: 1.2–1.5; (120)) (124) and among Caucasian B-ALL cases only (OR: 1.45; 95% CI: 1.25–1.68).(138) In analyses where subtype of ALL was considered, associations with rs2239633 and disease status are largely null among infants and young children with and without MLL rearrangements.(133,140)
GATA3 SNV rs3824662 has been studied in association with ALL overall (124) and by numerous cytogenomic subtypes.(141,142) Although GATA3 plays a role in T-cell differentiation (143), inherited germline variation in rs3824662 was significantly associated with B-ALL (OR: 1.31; 95% CI: 1.21–1.41) and hyperdiploid ALL (OR: 1.17; 95% CI: 1.06, 1.29) (141), though the strongest association was observed for Ph-like ALL (OR: 3.14; 95% CI:1.18–5.44).(142) Lastly, SNVs within the following genes: PIP4K2A (rs10828317), LHPP (rs35837782), ELK3 (rs4762284), and CDKN2A (rs3731217) have been identified in GWAS studies as increasing the risk of childhood ALL (122,124); however, none of these exhibit notable variation in association when considered by ALL overall (122,124) relative to associations by immunophenotype or cytogenomic subtype.(122,144)
Studies examining rare germline variants have assessed mutations in TP53 in association with childhood ALL (145) and PAX5 in association with familial ALL.(146) In a study of pathogenic TP53 variants among childhood ALL cases, 2% of cases had pathogenic TP53 mutations and 65% of pediatric patients harboring TP53 pathogenic mutations also had hypodiploid ALL compared to 15% cases with pathogenic mutations having hyperdiploid ALL.(145) This study demonstrates the possibility of studying the interplay between germline and somatic genetic aberrations, which may place some children at a higher risk for developing ALL.
Challenges in identifying etiologic heterogeneity in acute lymphoblastic leukemia
Tremendous progress has been made in the treatment of and subsequently the survival from childhood ALL over the past few decades; however, less progress has been made in identifying modifiable risk factors, which may shed light on the initiation of leukemogenesis or aid in prevention. Meta- and pooled-analyses, which allow for increased sample sizes, have been vital in identifying risk factors for childhood ALL overall and have identified pesticide exposure, paternal smoking, paint exposure, solvent exposure, and maternal coffee intake during pregnancy as potentially modifiable risk factors. On the other hand, immune-stimulating exposures such as being breastfed, attending daycare, having contact with cats and dogs, and the presence of allergies display inverse associations childhood ALL. In studies where stratification by subtype of ALL has been presented, there is suggested variation in association by subtype; however, studies with larger sample sizes to accommodate the less frequent subtypes, such as T-ALL, are necessary to uncover any true etiologic heterogeneity by ALL subtype. Further, the paucity of epidemiologic risk factor studies examining variation in risk by subtype highlights the need for future studies to examine etiologic heterogeneity using modern subtype definitions to keep pace with the clinically-recognized subgroups. Despite the cytogenomic and molecular subtype definitions being clinically useful and prognostically different, studying risk factor differences between cytogenomic subgroups of ALL remains a challenge due to sample size constraints.
In the modern era of epidemiology, where molecular epidemiology aids in the discovery of etiologic heterogeneity, the study of childhood ALL presents opportunities to characterize risk factor differences by clinically-recognized subtypes, that may help to identify risk-reduction strategies. The challenge of applying molecular subtyping methods to childhood ALL, as have been done in adult tumors, such as breast cancer, lies in the rarity of the disease and the rarity of each subtype. Therefore, a comprehensive childhood ALL study that has an adequate sample size to allow for stratification by the clinically-relevant subtypes discussed herein and that also has performed uniform cytogenetic subtyping will be necessary to study etiologic heterogeneity by ALL subtype. To overcome the issues of small sample size and to achieve uniform subtyping, we recommend a nationwide recruitment strategy through clinical trial organizations such as the Children’s Oncology Group in the United States and the International BFM study group in Europe.(147) Additionally, it is important to develop a means to investigate immune contribution to ALL by way of monitoring the immune response post-infection, which may shed light on the biologic underpinnings of ALL initiation. As we have presented, the summation of the previous work in the field provides the impetus for future studies to be initiated that will focus on thoroughly studying the etiologic heterogeneity of childhood ALL by immunophenotype and cytogenetic subtype, which has been slower than the progress observed in adult tumor types.
Supplementary Material
Funding:
This work was supported by the National Institutes of Health (grant numbers T32 CA099936 to LA Williams)
Abbreviation list:
- ALL
acute lymphoblastic leukemia
- B-ALL
B-cell ALL
- T-ALL
T-cell ALL
- FAB
French-American-British
- EPT-ALL
early T-precursor ALL
- FISH
fluorescence in situ hybridization
- GWA
genome-wide association studies
- SNV
single nucleotide variant
- OR
odds ratios
- 95% CI
95% confidence interval
Footnotes
Conflict of Interest: The authors have no financial conflicts of interest to disclose.
REFERENCES
- 1.Millikan RC, Newman B, Tse C-K, Moorman PG, Conway K, Dressler LG, et al. Epidemiology of basal-like breast cancer. Breast Cancer Res Treat [Internet]. 2008. [cited 2013 Aug 21];109:123–39. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2443103&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sisti JS, Collins LC, Beck AH, Tamimi RM, Rosner BA, Eliassen AH. Reproductive risk factors in relation to molecular subtypes of breast cancer: Results from the nurses’ health studies. Int J Cancer. 2016;138:2346–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Phipps AI, Li CI, Kerlikowske K, Barlow WE, Buist DSM. Risk factors for ductal, lobular, and mixed ductal-lobular breast cancer in a screening population. Cancer Epidemiol Biomarkers Prev [Internet]. 2010;19:1643–54. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2882996&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li CI, Daling JR, Haugen KL, Tang MTC, Porter PL, Malone KE. Use of menopausal hormone therapy and risk of ductal and lobular breast cancer among women 55–74 years of age. 2013;18:1199–216. [Google Scholar]
- 5.Williams LA, Nichols HB, Hoadley KA, Tse CK, Geradts J, Bell ME, et al. Reproductive risk factor associations with lobular and ductal carcinoma in the Carolina Breast Cancer Study. Cancer Causes Control [Internet]. Springer International Publishing; 2018;29:25–32. Available from: 10.1007/s10552-017-0977-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burnett-Hartman AN, Passarelli MN, Adams SV, Upton MP, Zhu LC, Potter JD, et al. Differences in epidemiologic risk factors for colorectal adenomas and serrated polyps by lesion severity and anatomical site. Am J Epidemiol. 2013;177:625–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Limsui D, Vierkant RA, Tillmans LS, Wang AH, Weisenberger DJ, Laird PW, et al. Cigarette smoking and colorectal cancer risk by molecularly defined subtypes. J Natl Cancer Inst. 2010;102:1012–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishihara R, Morikawa T, Kuchiba A, Lochhead P, Yamauchi M, Liao X, et al. A prospective study of duration of smoking cessation and colorectal cancer risk by epigenetics-related tumor classification. Am J Epidemiol. 2013;178:84–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steurer S, Mayer PS, Adam M, Krohn A, Koop C, Ospina-Klinck D, et al. TMPRSS2-ERG fusions are strongly linked to young patient age in low-grade prostate cancer. Eur Urol [Internet]. European Association of Urology; 2014;66:978–81. Available from: 10.1016/j.eururo.2014.06.027 [DOI] [PubMed] [Google Scholar]
- 10.Graff RE, Ahearn TU, Pettersson A, Ebot EM, Gerke T, Penney KL, et al. Height, obesity, and the risk of TMPRSS2: ERG-Defined prostate cancer. Cancer Epidemiol Biomarkers Prev. 2018;27:193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.American Cancer Society. Cancer in Children & Adolescents. Spec Sect Cancer Child Adolesc [Internet]. 2014;1:25–42. Available from: http://www.cancer.org/research/cancerfactsstatistics/cancerfactsfigures2014/ [Google Scholar]
- 12.Linabery AM, Ross JA. Trends in childhood cancer incidence in the U.S. (1992–2004). Cancer. 2008;112:416–32. [DOI] [PubMed] [Google Scholar]
- 13.Hunger SP, Lu X, Devidas M, Camitta BM, Gaynon PS, Winick NJ, et al. Improved survival for children and adolescents with acute lymphoblastic leukemia between 1990 and 2005: A report from the Children’s Oncology Group. J Clin Oncol. 2012;30:1663–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mrozek K, Harper DP, Aplan PD. Cytogenetics and molecular genetics of acute lymphoblastic leukemia. Hematol Oncol Clin North Am [Internet]. 2009;23:991–1010, v. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19825449%5Cnhttp://www.ncbi.nlm.nih.gov/pmc/articles/PMC3607311/pdf/nihms132175.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nguyen K, Devidas M, Cheng S-C, La M, Raetz EA, Carroll WL, et al. Factors influencing survival after relapse from acute lymphoblastic leukemia: a Children’s Oncology Group study. Leukemia [Internet]. 2008;22:2142–50. Available from: http://www.nature.com/doifinder/10.1038/leu.2008.251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khoury MJ, Moore CA, James LM, Cordero JF. The interaction between dysmorphology and epidemiology: Methodologic issues of lumping and splitting. Teratology. 1992;45:133–8. [DOI] [PubMed] [Google Scholar]
- 17.Huntly BJP, Gilliland DG. Leukaemia stem cells and the evolution of cancer-stem-cell research. Nat Rev Cancer [Internet]. 2005;5:311–21. Available from: http://www.nature.com/doifinder/10.1038/nrc1592 [DOI] [PubMed] [Google Scholar]
- 18.Pui C-H, Yang JJ, Hunger SP, Pieters R, Schrappe M, Biondi A, et al. Childhood Acute Lymphoblastic Leukemia: Progress Through Collaboration. J Clin Oncol [Internet]. 2015;33:JCO.2014.59.1636. Available from: http://jco.ascopubs.org/content/early/2015/08/21/JCO.2014.59.1636%5Cnhttp://www.ncbi.nlm.nih.gov/pubmed/26304874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Margolin JF, Rabin KR, Stueber P, Poplack DG. Principles and Practice of Pediatric Oncology Sixth. Pizzo P, Poplack D, editors. Philadelphia, PA: Lippincott Williams & Wilkins; 2011. [Google Scholar]
- 20.Hunger SP, Mullighan CG. Acute Lymphoblastic Leukemia in Children. N Engl J Med [Internet]. 2015;373:1541–52. Available from: http://www.nejm.org/doi/abs/10.1056/NEJMra1400972 [DOI] [PubMed] [Google Scholar]
- 21.Bhatia S, Sather HN, Heerema NA, Trigg ME, Gaynon PS, Robison LL. Racial and ethnic differences in survival of children with acute lymphoblastic leukemia. Blood. 2002;100:1957–64. [DOI] [PubMed] [Google Scholar]
- 22.Kadan-Lottick NS, Ness KK, Bhatia S, Gurney JG. Survival variability by race and ethnicity in childhood acute lymphoblastic leukemia. JAMA [Internet]. 2003;290:2008–14. Available from: http://jama.ama-assn.org/content/290/15/2008.abstract%5Cnpapers://bc21232e-1126-4152-95cc-c2784186e2af/Paper/p2856 [DOI] [PubMed] [Google Scholar]
- 23.Pui C-H, Sandlund JT, Rivera GK, Howard SC, Ribeiro RC, Rubnitz JE, et al. Results of Therapy for Acute Lymphoblastic in Black and White Children. JAMA. 2003;290:2001–7. [DOI] [PubMed] [Google Scholar]
- 24.Yang W, Treviño LR, Yang JJ, Scheet P, Pui C-H, Evans WE, et al. ARID5B SNP rs10821936 is associated with risk of childhood acute lymphoblastic leukemia in blacks and contributes to racial differences in leukemia incidence. Leukemia [Internet]. 2010;24:894–6. Available from: http://www.nature.com/leu/journal/v24/n4/full/leu2009277a.html%5Cnhttp://www.ncbi.nlm.nih.gov/pubmed/20054350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patrick K, Wade R, Goulden N, Mitchell C, Moorman AV., Rowntree C, et al. Outcome for children and young people with Early T-cell precursor acute lymphoblastic leukaemia treated on a contemporary protocol, UKALL 2003. Br J Haematol. 2014;166:421–4. [DOI] [PubMed] [Google Scholar]
- 26.Karrman K, Johansson B. Pediatric T-cell acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2017;56:89–116. [DOI] [PubMed] [Google Scholar]
- 27.Coustan-Smith E, Mullighan CG, Onciu M, Behm FG, Raimondi SC, Pei D, et al. Early T-Cell Precursor Leukemia: A Subtype of Very High-Risk Acute Lymphoblastic Leukemia Identified in Two Independent Cohorts. Lancet Oncol [Internet]. 2009;10:147–56. Available from: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19147408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hunger SP, Mullighan CG. Redefining ALL classification: toward detecting high-risk ALL and implementing precision medicine. Blood. 2015;125:3977–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–405. [DOI] [PubMed] [Google Scholar]
- 30.Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Easton J, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borowitz M, Chan J, Downing J, Le Beau M, Arber D. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Revised 4t. Swerdlow S, Campo E, Harris N, Jaffe E, Pileri S, Stein H, et al. , editors. Lyon: International Agency for Research on Cancer; 2017. [Google Scholar]
- 32.Wood BL, Winter SS, Dunsmore KP, Devidas M, Chen S, Asselin B, et al. T-Lymphoblastic Leukemia (T-ALL) Shows Excellent Outcome, Lack of Significance of the Early Thymic Precursor (ETP) Immunophenotype, and Validation of the Prognostic Value of End-Induction Minimal Residual Disease (MRD) in Children’s Oncology Group (COG) S. Blood. 2014;124:[Abstract].25359993 [Google Scholar]
- 33.Williams DL, Tsiatis A, Brodeur GM, Look AT, Melvin SL, Bowman WP, et al. Prognostic importance of chromosome number of 136 untreated children with acute lymphoblastic leukemia. Blood. 1982;60:864–71. [PubMed] [Google Scholar]
- 34.Armstrong SA, Look AT. Molecular genetics of acute lymphoblastic leukemia. J Clin Oncol. 2005;23:6306–15. [DOI] [PubMed] [Google Scholar]
- 35.Femmdo AA, Look AT. Clinical implications of recurring chromosomal and associated molecular abnormalities in acute lymphoblastaic leukemia. Semin Hematol. 2000;37:381–95. [DOI] [PubMed] [Google Scholar]
- 36.Moorman AV The clinical relevance of chromosomal and genomic abnormalities in B-cell precursor acute lymphoblastic leukaemia. Blood Rev [Internet]. Elsevier Ltd; 2012;26:123–35. Available from: 10.1016/j.blre.2012.01.001 [DOI] [PubMed] [Google Scholar]
- 37.Orsi L, Rudant J, Bonaventure A, Goujon-Bellec S, Corda E, Evans T-J, et al. Genetic polymorphisms and childhood acute lymphoblastic leukemia: GWAS of the ESCALE study (SFCE). Leukemia [Internet]. 2012;26:2561–4. Available from: http://www.nature.com/leu/journal/v26/n12/full/leu2012148a.html%5Cnhttp://www.ncbi.nlm.nih.gov/pubmed/22660188 [DOI] [PubMed] [Google Scholar]
- 38.Heerema NA, Raimondi SC, Anderson JR, Biegel J, Camitta BM, Cooley LD, et al. Specific extra chromosomes occur in a model number dependent pattern in pediatric acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2007;46:684–93. [DOI] [PubMed] [Google Scholar]
- 39.Raimondi SC, Roberson PK, Pui C-H, Behm FG, Rivera GK. Hyperdiploid (47–50) Acute Lymphoblastic Leukemia in Children. Blood. 1992;79:3245–52. [PubMed] [Google Scholar]
- 40.Greaves MF, Wiemels J. Origins of chromosome translocations in childhood leukaemia. Nat Rev Cancer [Internet]. 2003;3:639–49. Available from: http://www.nature.com/doifinder/10.1038/nrc1164 [DOI] [PubMed] [Google Scholar]
- 41.Greaves M In utero origins of childhood leukaemia. Early Hum Dev. 2005;81:123–9. [DOI] [PubMed] [Google Scholar]
- 42.Gale KB, Ford AM, Repp R, Borkhardt A, Keller C, Eden OB, et al. Backtracking leukemia to birth: Identification of clonotypic gene fusion sequences in neonatal blood spots. Proc Natl Acad Sci U S A [Internet]. 1997;94:13950–4. Available from: http://www.ncbi.nlm.nih.gov/pubmed/9391133%5Cnhttp://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC28413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Greaves MF, Maia AT, Wiemels JL, Ford AM. Leukemia in twins: Lessons in natural history. Blood. 2003;102:2321–33. [DOI] [PubMed] [Google Scholar]
- 44.Wartenberg D, Groves FD, Adelman AS. Hematologic Malignancies: Acute Leukemias. Estey E, Faderl S, Kantarjian H, editors. Springer Berlin Heidelberg New York; 2008. [Google Scholar]
- 45.Loh M, Raetz E, Devidas M, Dai Y, Borowitz M, Carroll A, et al. Outcomes of children, adolescents, and young adults with acute lymphoblastic leukemia based on blast genotype at diagnosis: A report from the Children’s Oncology Group. [Abstract]. 2017;
- 46.Hunger SP, Loh ML, Whitlock JA, Winick NJ, Carroll WL, Devidas M, et al. Children’s Oncology Group’s 2013 Blueprint for Research: Acute Lymphoblastic Leukemia. Pediatr Blood Cancer. 2013;60:957–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carroll WL. Race and outcome in childhood acute lymphoblastic leukemia. JAMA [Internet]. 2003;290:2061–3. Available from: http://www.ncbi.nlm.nih.gov/pubmed/14559962 [DOI] [PubMed] [Google Scholar]
- 48.Heerema N, Sather H, Ge J, Arthur D, Hilden J, Trigg ME, et al. Cytogenetic studies of infant acute lymphoblastic leukemia: poor prognosis of infants with t(4;11) - a report of the Children’s Cancer Group. Leukemia [Internet]. 1999;13:679–86. Available from: 10.1038/sj.leu.2401413 [DOI] [PubMed] [Google Scholar]
- 49.Strehl S, König M, Dworzak MN, Kalwak K, Haas O a. PAX5/ETV6 fusion defines cytogenetic entity dic(9;12)(p13;p13). Leuk Off J Leuk Soc Am Leuk Res Fund, UK [Internet]. 2003;17:1121–3. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12764378 [DOI] [PubMed] [Google Scholar]
- 50.Chiaretti S, Foà R. T-cell acute lymphoblastic leukemia. Haematologica. 2009;94:160–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paulsson K High hyperdiploid childhood acute lymphoblastic leukemia: Chromosomal gains as the main driver event. Mol Cell Oncol [Internet]. Taylor & Francis; 2016;3:e1064555 Available from: https://www.tandfonline.com/doi/full/10.1080/23723556.2015.1064555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Churchman ML, Qian M, te Kronnie G, Zhang R, Yang W, Zhang H, et al. Germline Genetic IKZF1 Variation and Predisposition to Childhood Acute Lymphoblastic Leukemia. Cancer Cell [Internet]. 2018;33:1–12. Available from: 10.1016/j.ccell.2018.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heatley SL, Sadras T, Kok CH, Nievergall E, Quek K, Dang P, et al. High prevalence of relapse in children with Philadelphia-like acute lymphoblastic leukemia despite risk-adapted treatment. Haematologica. 2017;102:e490–e493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roberts KG, Reshmi SC, Harvey RC, Chen I-M, Patel K, Stonerock E, et al. Genomic and outcome analyses of Ph-like ALL in NCI standard-risk patients: a report from the Children’s Oncology Group. Blood [Internet]. 2018;132: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clappier E, Auclerc MF, Rapion J, Bakkus M, Caye A, Khemiri A, et al. An intragenic ERG deletion is a marker of an oncogenic subtype of B-cell precursor acute lymphoblastic leukemia with a favorable outcome despite frequent IKZF1 deletions. Leukemia. 2014;28:70–7. [DOI] [PubMed] [Google Scholar]
- 56.Zhang J, McCastlain K, Yoshihara H, Xu B, Chang Y, Churchman ML, et al. Deregulation of DUX4 and ERG in acute lymphoblastic leukemia. Nat Genet. 2016;48:1481–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sulong S, Moorman AV, Irving J a E, Strefford JC, Konn ZJ, Case MC, et al. A comprehensive analysis of the CDKN2A gene in childhood acute lymphoblastic leukemia reveals genomic deletion, copy number neutral loss of heterozygosity, and association with specific cytogenetic subgroups. Blood. 2009;113:100–7. [DOI] [PubMed] [Google Scholar]
- 58.Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–64. [DOI] [PubMed] [Google Scholar]
- 59.Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453:110–4. [DOI] [PubMed] [Google Scholar]
- 60.Dörge P, Meissner B, Zimmermann M, Möricke A, Schrauder A, Bouquin JP, et al. IKZF1 deletion is an independent predictor of outcome in pediatric acute lymphoblastic leukemia treated according to the ALL-BFM 2000 protocol. Haematologica. 2013;98:428–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kaur M, de Smith AJ, Selvin S, Zhang L, Cunningham M, Kang MW, et al. Tobacco Smoke and Ras Mutations Among Latino and Non-Latino Children with Acute Lymphoblastic Leukemia. Arch Med Res [Internet]. Elsevier Inc; 2016;47:677–83. Available from: 10.1016/j.arcmed.2016.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Teuffel O, Betts DR, Dettling M, Schaub R, Schäfer BW, Niggli FK. Prenatal origin of separate evolution of leukemia in identical twins. Leuk Off J Leuk Soc Am Leuk Res Fund, UK [Internet]. 2004;18:1624–9. pm:15356660%5Cn Available from: http://www.ncbi.nlm.nih.gov/pubmed/15356660 [DOI] [PubMed] [Google Scholar]
- 63.Fasching K, Panzer S, Haas O a, Marschalek R, Gadner H, Panzer-Grümayer ER. Presence of clone-specific antigen receptor gene rearrangements at birth indicates an in utero origin of diverse types of early childhood acute lymphoblastic leukemia. Blood. 2000;95:2722–4. [PubMed] [Google Scholar]
- 64.Wiemels JL, Cazzaniga G, Daniotti M, Eden OB, Addison GM, Masera G, et al. Prenatal origin of acute lymphoblastic leukaemia in children. Lancet. 1999;354:1499–503. [DOI] [PubMed] [Google Scholar]
- 65.Karalexi MA, Dessypris N, Thomopoulos TP, Ntouvelis E, Kantzanou M, Diamantaras A-A, et al. Parental alcohol consumption and risk of leukemia in the offspring: A systematic review and meta-analysis. Eur J Cancer Prev. 2017;April 4. [DOI] [PubMed] [Google Scholar]
- 66.Bailey HD, Fritschi L, Metayer C, Infante-Rivard C, Magnani C, Petridou E, et al. Parental occupational paint exposure and risk of childhood leukemia in the offspring: findings from the Childhood Leukemia International Consortium. Cancer Causes Control. 2014;25:1351–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bailey HD, Fritschi L, Infante-Rivard C, Glass DC, Miligi L, Dockerty JD, et al. Parental occupational pesticide exposure and the risk of childhood leukemia in the offspring: Findings from the childhood leukemia international consortium. Int J Cancer. 2014;135:2157–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu R, Zhang L, McHale CM, Hammond SK. Paternal smoking and risk of childhood acute lymphoblastic leukemia: Systematic review and meta-analysis. J Oncol. 2011;2011:854584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hargreave M, Sc M, Jensen A, Sc M, Ph D, Toender A, et al. Fertility treatment and childhood cancer risk: a systematic meta-analysis. Fertil Steril. 2013;100:150–61. [DOI] [PubMed] [Google Scholar]
- 70.Dessypris N, Karalexi MA, Ntouvelis E, Diamantaras A-A, Papadakis V, Baka M, et al. Association of maternal and index child’s diet with subsequent leukemia risk: A systematic review and meta analysis. Cancer Epidemiol [Internet]. Elsevier Ltd; 2017;47:64–75. Available from: 10.1016/j.canep.2017.01.003 [DOI] [PubMed] [Google Scholar]
- 71.Karalexi MA, Dessypris N, Skalkidou A, Biniaris-Georgallis S-I, Kalogirou EI, Thomopoulos TP, et al. Maternal fetal loss history and increased acute leukemia subtype risk in subsequent offspring: A systematic review and meta-analysis. Cancer Causes Control. Springer International Publishing; 2017;28:599–624. [DOI] [PubMed] [Google Scholar]
- 72.Bailey HD, Metayer C, Milne E, Petridou E, Infante-Rivard C, Spector LG, et al. Home paint exposures and risk of childhood acute lymphoblastic leukemia: Findings from the Childhood Leukemia Internal Consortium. Cancer Causes Control. 2015;26:1257–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bailey HD, Infante-Rivard C, Metayer C, Clavel J, Lightfoot T, Kaatsch P, et al. Home pesticide exposures and risk of childhood leukemia: Findings from the Childhood Leukemia International Consortium. Int J Cancer. 2016;137:2644–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Milne E, Greenop KR, Petridou E, Bailey HD, Orsi L, Kang AY, et al. Maternal consumption of coffee and tea during pregnancy and risk of childhood ALL: a pooled analysis from the childhood Leukemia International Consortium. Cancer Causes Control [Internet]. Springer International Publishing; 2018;29:539–50. Available from: http://link.springer.com/10.1007/s10552-018-1024-1 [DOI] [PubMed] [Google Scholar]
- 75.Thomopoulos TP, Ntouvelis E, Diamantaras A-A, Tzanoudaki M, Baka M, Hatzipantelis E, et al. Maternal and childhood consumption of coffee, tea and cola beverages in association with childhood leukemia: a meta-analysis. Cancer Epidemiol [Internet]. Elsevier Ltd; 2015;39:1047–59. Available from: 10.1016/j.canep.2015.08.009 [DOI] [PubMed] [Google Scholar]
- 76.Zhao L, Liu X, Wang C, Yan K, Lin X, Li S, et al. Magnetic fields exposure and childhood leukemia risk: a meta-analysis based on 11,699 cases and 13,194 controls. Leuk Res [Internet]. Elsevier Ltd; 2014;38:269–74. Available from: 10.1016/j.leukres.2013.12.008 [DOI] [PubMed] [Google Scholar]
- 77.Zhou Y, Zhang S, Li Z, Zhu J, Bi Y, Bai Y, et al. Maternal benzene exposure during pregnancy and risk of childhood acute lymphoblastic leukemia: a meta-analysis of epidemiologic studies. PLoS One. 2014;15:e110466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Goh YI, Bollano E, Einarson TR, Koren G. Prenatal multivitamin supplementation and rates of congenital anomalies: A meta-analysis. J Obstet Gynaecol Canada [Internet]. Elsevier Masson SAS; 2006;28:680–9. Available from: 10.1016/S1701-2163(16)32227-7 [DOI] [PubMed] [Google Scholar]
- 79.Boothe VL, Boehmer TK, Wendel AM, Yip FY. Residential traffic exposure and childhood leukemia: a systematic review and meta-analysis. Am J Prev Med. 2014;46:413–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Klimentopoulou A, Antonopoulos C, Papadopoulou C, Kanavidis P, Tourvas A-D, Polychronopoulou S, et al. Maternal smoking during pregnancy and risk for childhood leukemia: A nation-wide case-control study in Greece and meta-analysis. Pediatr Blood Cancer. 2012;58:334–51. [DOI] [PubMed] [Google Scholar]
- 81.Sergentanis TN, Thomopoulos TP, Gialamas SP, Karalexi MA, Biniaris-Georgallis SI, Kontogeorgi E, et al. Risk for childhood leukemia associated with maternal and paternal age Eur. J. Epidemiol. Springer Netherlands; 2015. [DOI] [PubMed] [Google Scholar]
- 82.Caughey RW, Michels KB. Birth weight and childhood leukemia: a meta-analysis and review of the current evidence. Int J Cancer. 2009;124:2658–70. [DOI] [PubMed] [Google Scholar]
- 83.Milne E, Greenop KR, Metayer C, Schuz J, Pteridou E, Pombo-de-Oliveira MS, et al. Fetal growth and childhood acute lymphoblastic leukemia: findings from the childhood leukemia international consortium. Int J Breast Cancer. 2013;133:2968–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Marcotte EL, Thomopoulos TP, Infante-Rivard C, Clavel J, Petridou ET, Schuz J, et al. Caesarean delivery and risk of childhood leukaemia: A pooled analysis from the Childhood Leukemia International Consortium (CLIC). Lancet Haematol. 2016;3:e176–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huang Q, Gao Y, Zhong M, Yu Y. Preterm Birth and Subsequent Risk of Acute Childhood Leukemia: a Meta- Analysis of Observational Studies. Cell Physiol Biochem. 2016;39:1229–38. [DOI] [PubMed] [Google Scholar]
- 86.Dahl S, Schmidt LS, Vestergaard T, Schüz J, Schmiegelow K. Allergy and the risk of childhood leukemia: a meta-analysis. Leukemia [Internet]. 2009;23:2300–4. Available from: http://www.nature.com/doifinder/10.1038/leu.2009.162 [DOI] [PubMed] [Google Scholar]
- 87.Von Behren J, Spector LG, Mueller BA, Carozza SE, Chow EJ, Fox EE, et al. Birth order and risk of childhood cancer: a pooled analysis from five US States. Int J Cancer. 2011;128:2709–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Martin RM, Gunnell D, Owen CG, Smith GD. Breast-feeding and childhood cancer: a systematic review with metaanalysis. Int J Cancer. 2005;117:1020–31. [DOI] [PubMed] [Google Scholar]
- 89.Hwee J, Tait C, Sung L, Kwong JC, Sutradhar R, Pole JD. A systematic review and meta-analysis of the association between childhood infections and the risk of childhood acute lymphoblastic leukaemia. Br J Cancer [Internet]. Nature Publishing Group; 2017;1–11. Available from: http://www.nature.com/doifinder/10.1038/bjc.2017.360%0Ahttp://www.ncbi.nlm.nih.gov/pubmed/29065105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Orsi L, Magnani C, Petridou ET, Dockerty JD, Metayer C, Milne E, et al. Living on a farm, contact with farm animals and pets, and childhood acute lymphoblastic leukemia: pooled and meta-analyses from the Childhood Leukemia International Consortium. Cancer Med. 2018;7:2665–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Urayama KY, Buffler PA, Gallagher ER, Ayoob JM, Ma X. A meta-analysis of the association between day-care attendance and childhood acute lymphoblastic leukaemia. Int J Epidemiol. 2010;39:718–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Carlos-Wallace FM, Zhang L, Smith MT, Rader G, Steinmaus C. Parental, In Utero, and Early-Life Exposure to Benzene and the Risk of Childhood Leukemia: A Meta-Analysis. Am J Epidemiol. 2015;183:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Turner MC, Wigle DT, Krewski D. Residential pesticides and childhood leukemia: a systematic review and meta-analysis. Environ Health Perspect. 2010;118:33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kwan ML, Buffler PA, Abrams B, Kiley VA. Breastfeeding and the risk of childhood leukemia: A meta-analysis. Public Health Rep. 2004;119:521–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Greaves M A causal mechanism for childhood acute lymphoblastic leukaemia. Nat Rev Cancer [Internet]. Springer US; 2018;1 Available from: http://www.nature.com/articles/s41568-018-0015-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cnattingius S, Zack MM, Ekbom A, Gunnarskog J, Kreuger A, Linet M, et al. Prenatal and neonatal risk factors for childhood lymphatic leukemia. J Natl Cancer Inst. 1995;87:908–14. [DOI] [PubMed] [Google Scholar]
- 97.Yeazel MW, Ross JA, Buckley JD, Woods WG, Ruccione K, Robison LL. High birth weight and risk of specific childhood cancers: a report from the Children’s Cancer Group. J Pediatr [Internet]. 1997;131:671–7. Available from: http://www.ncbi.nlm.nih.gov/pubmed/9403644 [DOI] [PubMed] [Google Scholar]
- 98.Hjalgrim LL, Rostgaard K, Hjalgrim H, Westergaard T, Thomassen H, Forestier E, et al. Birth weight and risk for childhood leukemia in Denmark, Sweden, Norway, and Iceland. J Natl Cancer Inst. 2004;96:1549–56. [DOI] [PubMed] [Google Scholar]
- 99.Ji BT, Shu XO, Linet MS, Zheng W, Wacholder S, Gao YT, et al. Paternal cigarette smoking and the risk of childhood cancer among offspring of nonsmoking mothers. J Natl Cancer Inst [Internet]. 1997;89:238–44. Available from: http://www.ncbi.nlm.nih.gov/pubmed/9017004 [DOI] [PubMed] [Google Scholar]
- 100.Reynolds P, Von Behren J, Elkin EP. Birth Characteristics and Leukemima in Young Children. Am J Epidemiol. 2002;155:603–13. [DOI] [PubMed] [Google Scholar]
- 101.Karalexi MA, Skalkidou A, Thomopoulos TP, Belechri M, Biniaris-Georgallis S-I, Bouka E, et al. History of maternal fetal loss and childhood leukaemia risk in subsequent offspring: differentials by miscarriage or stillbirth history and disease subtype. Paediatr Perinat Epidemiol. 2015;29:453–61. [DOI] [PubMed] [Google Scholar]
- 102.Metayer C, Zhang L, Wiemels JL, Bartley K, Schiffman J, Ma X, et al. Tobacco smoke exposure and risk of childhood acute lymphoblastic and myeloid leukemias by cytogenetic subtype. Cancer Epidemiol Biomarkers Prev. 2013;22:1600–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Milne E, Greenop KR, Scott RJ, Bailey HD, Attia J, Dalla-Pozza L, et al. Parental prenatal smoking and risk of childhood acute lymphoblastic leukemia. Am J Epidemiol. 2012;175:43–53. [DOI] [PubMed] [Google Scholar]
- 104.Scelo G, Metayer C, Zhang L, Wiemels JL, Aldrich MC, Selvin S, et al. Household Exposure to paint and petroleum solvents, chromosomal translocations, and the risk of childhood Leukemia. Environ Health Perspect. 2009;117:133–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shu XO, Han D, Severson RK, Chen Z, Neglia JP, Reaman GH, et al. Birth characteristics, maternal reproductive history, hormone use during pregnancy, and risk of childhood acute lymphocytic leukemia by immunophenotype (United States). Cancer Causes Control [Internet]. 2002;13:15–25. Available from: http://www.issues4life.org/pdfs/cancercausesandcontrol.pdf%5Cnhttp://www.ncbi.nlm.nih.gov/pubmed/11895036%5Cnhttp://www.ncbi.nlm.nih.gov/pubmed/11899114 [DOI] [PubMed] [Google Scholar]
- 106.Slater ME, Linabery AM, Blair CK, Spector LG, Heerema NA, Robison LL, et al. Maternal prenatal cigarette, alcohol and illicit drug use and risk of infant leukaemia: a report from the Children’s Oncology Group. Paediatr Perinat Epidemiol. 2011;25:559–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rudant J, Menegaux F, Leverger G, Baruchel A, Lambilliotte A, Bertrand Y, et al. Childhood hematopoietic malignancies and parental use of tobacco and alcohol: The ESCALE study (SFCE). Cancer Causes Control. 2008;19:1277–90. [DOI] [PubMed] [Google Scholar]
- 108.O’Neill KA, Murphy MFG, Bunch KJ, Puumala SE, Carozza SE, Chow EJ, et al. Infant birthweight and risk of childhood cancer: international population-based case control studies of 40 000 cases. Int J Epidemiol. 2015;44:153–68. [DOI] [PubMed] [Google Scholar]
- 109.Shu XO, Linet MS, Steinbuch M, Wen WQ, Buckley JD, Neglia JP, et al. Breast-Feeding and Risk of Childhood Acute Leukemia. JNCI J Natl Cancer Inst [Internet]. 1999;91:1765–72. Available from: http://jnci.oxfordjournals.org/content/91/20/1765.long [DOI] [PubMed] [Google Scholar]
- 110.Rudant J, Lightfoot T, Urayama KY, Petridou E, Dockerty JD, Magnani C, et al. Childhood acute lymphoblastic leukemia and indicators of early immune stimulation: A childhood leukemia international consortium study. Am J Epidemiol. 2015;181:549–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kamper-Jorgensen M, Woodward A, Wohlfahrt J, Benn CS, Simonsen J, Hjalgrim H, et al. Childcare in the first 2 years of life reduces the risk of childhood acute lymphoblastic leukemia. Leukemia [Internet]. 2008;22:189–93. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17690702%5Cnhttp://www.nature.com/leu/journal/v22/n1/pdf/2404884a.pdf [DOI] [PubMed] [Google Scholar]
- 112.Ma X, Buffler PA, Wiemels JL, Selvin S, Metayer C, Loh M, et al. Ethnic difference in daycare attendance, early infections, and risk of childhood acute lymphoblastic leukemia. Cancer Epidemiol Biomarkers Prev. 2005;14:1928–34. [DOI] [PubMed] [Google Scholar]
- 113.Gilham C, Peto J, Simpson J, Roman E, Eden TOB, Greaves MF, et al. Day care in infancy and risk of childhood acute lymphoblastic leukaemia: findings from UK case-control study. BMJ [Internet]. 2005;330:1294 Available from: http://www.ncbi.nlm.nih.gov/pubmed/15849205%5Cnhttp://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC558199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Metayer C, Milne E, Dockerty JD, Clavel J, Pombo-de-Oliveira MS, Wesseling C, et al. Maternal Supplementation with Folic Acid and Other Vitamins and Risk of Leukemia in Offspring. Epidemiology [Internet]. 2014;25:811–22. Available from: http://content.wkhealth.com/linkback/openurl?sid=WKPTLP:landingpage&an=00001648-201411000-00005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Greenop KR, Bailey HD, Miller M, Scott RJ, Attia J, Ashton LJ, et al. Breastfeeding and nutrition to 2 years of age and risk of childhood acute lymphoblastic leukemia and brain tumors. Nutr Cancer. 2015;67:431–41. [DOI] [PubMed] [Google Scholar]
- 116.O’Neill KA, Bunch KJ, Vincent TJ, Spector LG, Moorman AV., Murphy MFG. Immunophenotype and cytogenetic characteristics in relationship between birth weight and childhood leukemia. Pediatr Blood Cancer. 2012;58:7–11. [DOI] [PubMed] [Google Scholar]
- 117.de Smith AJ, Kaur M, Gonseth S, Endicott A, Selvin S, Zhang L, et al. Correlates of prenatal and early-life tobacco smoke exposure and frequency of common gene deletions in childhood acute lymphoblastic leukemia. Cancer Res. 2017;77:1674–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shu XO, Perentesis JP, Wen W, Buckley JD, Boyle E, Ross JA, et al. Parental exposure to medications and hydrocarbons and ras mutations in children with acute lymphoblastic leukemia: a report from the Children’s Oncology Group. Cancer Epidemiol Biomarkers Prev [Internet]. 2004;13:1230–5. Available from: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=15247135%5Cnhttp://cebp.aacrjournals.org/content/13/7/1230.full.pdf [PubMed] [Google Scholar]
- 119.Treviño LR, Yang W, French D, Hunger SP, Carroll WL, Devidas M, et al. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat Genet [Internet]. 2009;41:1001–5. Available from: http://www.nature.com/doifinder/10.1038/ng.432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Papaemmanuil E, Hosking FJ, Vijayakrishnan J, Price A, Olver B, Sheridan E, et al. Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nat Genet [Internet]. 2009;41:1006–10. Available from: http://www.nature.com/doifinder/10.1038/ng.430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Evans T-J, Milne E, Anderson D, De Klerk NH, Jamieson SE, Talseth-Palmer BA, et al. Confirmation of Childhood Acute Lymphoblastic Leukemia Variants, ARID5B and IKZF1, and Interaction with Parental Environmental Exposures. PLoS One. 2014;9:e110255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sherborne AL, Hosking FJ, Prasad RB, Kumar R, Koehler R, Vijayakrishnan J, et al. Variation in CDKN2A at 9p21.3 influences childhood acute lymphoblastic leukemia risk. Nat Genet [Internet]. 2010;42:492–4. Available from: http://www.nature.com/doifinder/10.1038/ng.585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Xu H, Yang W, Perez-Andreu V, Devidas M, Fan Y, Cheng C, et al. Novel Susceptibility Variants at 10p12.31–12.2 for Childhood Acute Lymphoblastic Leukemia in Ethnically Diverse Populations. J Natl Cancer Inst. 2013;105:733–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wiemels JL, Walsh KM, de Smith AJ, Metayer C, Gonseth S, Hansen HM, et al. GWAS in childhood acute lymphoblastic leukemia reveals novel genetic associations at chromosomes 17q12 and 8q24.21. Nat Commun [Internet]. 2018;9:286 Available from: http://www.nature.com/articles/s41467-017-02596-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Al-absi B, Noor SM, Saif-Ali R, Salem SD, Ahmed RH, Razif MF, et al. Association of ARID5B gene variants with acute lymphoblastic leukemia in Yemeni children. Tumor Biol [Internet]. 2017;39:101042831769757 Available from: http://www.ncbi.nlm.nih.gov/pubmed/28381164%5Cnhttp://journals.sagepub.com/doi/10.1177/1010428317697573 [DOI] [PubMed] [Google Scholar]
- 126.Bekker-Méndez VC, Núñez-Enríquez JC, Torres Escalante JL, Alvarez-Olmos E, González-Montalvoc PM, Jiménez-Hernández E, et al. ARID5B, CEBPE and PIP4K2A Germline Genetic Polymorphisms and Risk of Childhood Acute Lymphoblastic Leukemia in Mexican Patients: AMIGICCL Study. Arch Med Res. 2016;47:623–8. [DOI] [PubMed] [Google Scholar]
- 127.Chokkalingam AP, Hsu L-I, Metayer C, Hansen HM, Month SR, Barcellos LF, et al. Genetic variants in ARID5B and CEBPE are childhood ALL susceptibility loci in Hispanics. Cancer Causes Control. 2013;24:1789–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Xu H, Cheng C, Devidas M, Pei D, Fan Y, Yang W, et al. ARID5B genetic polymorphisms contribute to racial disparities in the incidence and treatment outcome of childhood acute lymphoblastic leukemia. J Clin Oncol. 2012;30:751–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Linabery AM, Blommer CN, Spector LG, Davies SM, Robison LL, Ross JA. ARID5B and IKZF1 variants, selected demographic factors, and childhood acute lymphoblastic leukemia: A report from the Children’s Oncology Group. Leuk Res. 2013;37:936–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kennedy AE, Kamdar KY, Lupo PJ, Okcu MF, Scheurer ME, Dorak MT. Genetic markers in a multi-ethnic sample for childhood acute lymphoblastic leukemia risk. Leuk Lymphoma [Internet]. 2015;56:169–74. Available from: http://informahealthcare.com/doi/abs/10.3109/10428194.2014.910662%5Cnhttp://informahealthcare.com/doi/abs/10.3109/10428194.2014.910662?journalCode=lal%5Cnhttp://www.ncbi.nlm.nih.gov/pubmed/24707947 [DOI] [PubMed] [Google Scholar]
- 131.Kreile M, Piekuse L, Rots D, Dobele Z, Kovalova Z, Lace B. Analysis of possible genetic risk factors contributing to development of childhood acute lymphoblastic leukaemia in the Latvian population. Arch Med Sci. 2016;12:479–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lautner-Csorba O, Gézsi A, Semsei AF, Antal P, Erdélyi DJ, Schermann G, et al. Candidate gene association study in pediatric acute lymphoblastic leukemia evaluated by Bayesian network based Bayesian multilevel analysis of relevance. BMC Med Genomics [Internet]. 2012;5:42 Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3542204&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ross JA, Linabery AM, Blommer CN, Langer EK, Spector LG, Hilden JM, et al. Genetic Variants Modify Susceptibility to Leukemia in Infants: A Children’s Oncology Group Report. Pediatr Blood Cancer. 2013;60:31–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bhandari P, Ahmad F, Mandava S, Das BR. Association of genetic variants in ARID5B, IKZF1 and CEBPE with risk of childhood de novo B-lineage acute lymphoblastic leukemia in India. Asian Pacific J Cancer Prev. 2016;17:3989–95. [PubMed] [Google Scholar]
- 135.Healy J, Richer C, Bourgey M, Kritikou EA, Sinnett D. Replication analysis confirms the association of ARID5B with childhood B-cell acute lymphoblastic leukemia. Haematologica. 2010;95:1608–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Georgopoulos K, Bigby M, Wang J, Molnar A, Wu P, Winandy S, et al. The lkaros Gene Is Required for the Development of All Lymphoid Lineages. Cell. 1994;79:143–56. [DOI] [PubMed] [Google Scholar]
- 137.Harker N, Naito T, Cortes M, Hostert A, Hirschberg S, Tolaini M, et al. The CD8alpha Gene Locus Is Regulated by the Ikaros Family of Proteins. Mol Cell. 2002;10:1403–15. [DOI] [PubMed] [Google Scholar]
- 138.Vijayakrishnan J, Sherborne AL, Sawangpanich R, Hongeng S, Houlston RS, Pakakasama S. Variation at 7p12.2 and 10q21.2 influences childhood acute lymphoblastic leukemia risk in the Thai population and may contribute to racial differences in leukemia incidence. Leuk Lymphoma [Internet]. 2010;51:1870–4. Available from: http://www.tandfonline.com/doi/full/10.3109/10428194.2010.511356 [DOI] [PubMed] [Google Scholar]
- 139.Akasaka T, Balasas T, Russell LJ, Sugimoto K, Majid A, Walewska R, et al. Five members of the CEBP transcription factor family are targeted by recurrent IGH translocations in B-cell precursor acute lymphoblastic leukemia (BCP-ALL). Blood. 2007;109:3451–62. [DOI] [PubMed] [Google Scholar]
- 140.Emerenciano M, Barbosa TC, Lopes BA, Blunck CB, Faro A, Andrade C, et al. ARID5B polymorphism confers an increased risk to acquire specific MLL rearrangements in early childhood leukemia. BMC Cancer [Internet]. 2014;14:127 Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3948138&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Migliorini G, Fiege B, Hosking FJ, Ma Y, Kumar R, Sherborne AL, et al. Variation at 10p12.2 and 10p14 influences risk of childhood B-cell acute lymphoblastic leukemia and phenotype. Blood. 2013;122:3298–307. [DOI] [PubMed] [Google Scholar]
- 142.Perez-Andreu V, Roberts KG, Harvey RC, Yang W, Cheng C, Pei D, et al. Inherited GATA3 variants are associated with Ph-like childhood acute lymphoblastic leukemia and risk of relapse. Nat Genet [Internet]. Nature Publishing Group; 2013;45:1494–8. Available from: http://www.nature.com/doifinder/10.1038/ng.2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wei G, Abraham BJ, Yagi R, Jothi R, Cui K, Sharma S, et al. Genome-wide Analyses of Transcription Factor GATA3-Mediated Gene Regulation in Distinct T Cell Types. Immunity [Internet]. Elsevier Inc; 2011;35:299–311. Available from: 10.1016/j.immuni.2011.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Vijayakrishnan J, Kumar R, Henrion MYR, Moorman AV, Rachakonda PS, Hosen I, et al. A genome-wide association study identifies risk loci for childhood acute lymphoblastic leukemia at 10q26.13 and 12q23.1. Leukemia [Internet]. 2017;31:573–9. Available from: http://www.nature.com/doifinder/10.1038/leu.2016.271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Qian M, Cao X, Devidas M, Yang W, Cheng C, Dai Y, et al. TP53 Germline Variations Influence the Predisposition and Prognosis of B-Cell Acute Lymphoblastic Leukemia in Children. J Clin Oncol [Internet]. 2018;36:JCO.2017.75.521. Available from: http://ascopubs.org/doi/10.1200/JCO.2017.75.5215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Shah S, Schrader KA, Waanders E, Timms AE, Vijai J, Miething C, et al. A recurrent germline PAX5 mutation confers susceptibility to pre-B cell acute lymphoblastic leukemia. Nat Genet [Internet]. Nature Publishing Group; 2013;45:1226–31. Available from: 10.1038/ng.2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Spector LG, Ross JA, Olshan AF. Children’s Oncology Group’s 2013 Blueprint for Research: Epidemiology. Pediatr Blood Cancer. 2013;60:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.